 ,, 赵向前, 黄福灯, 李春寿
,, 赵向前, 黄福灯, 李春寿 ,浙江省农业科学院作物与核技术利用研究所,杭州 310021
,浙江省农业科学院作物与核技术利用研究所,杭州 310021The role of PPR proteins in posttranscriptional regulation of organelle components in plants
Yuanyuan Hao ,, Xiangqian Zhao, Fudeng Huang, Chunshou Li
,, Xiangqian Zhao, Fudeng Huang, Chunshou Li ,Institute of Crop and Nuclear Technology Utilization, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, China
,Institute of Crop and Nuclear Technology Utilization, Zhejiang Academy of Agricultural Sciences, Hangzhou 310021, China通讯作者: 李春寿,学士,研究员,研究方向:籼型杂交稻的选育。E-mail:lichunshou@126.com
编委: 宋任涛
收稿日期:2021-06-30修回日期:2021-08-20
| 基金资助: |
Received:2021-06-30Revised:2021-08-20
| Fund supported: |
作者简介 About authors
郝媛媛,博士,助理研究员,研究方向:稻米品质的改良和分子机理。E-mail:

摘要
PPR (pentatricopeptide repeats)蛋白是陆生植物中最大的蛋白家族之一,是一类RNA结合蛋白,参与细胞器基因的转录后加工过程,对细胞器的生物合成和功能具有深远影响。PPR突变后造成叶绿体光合电子传递链和线粒体呼吸链等进程受损,最终通过影响光合作用或者呼吸作用,造成植株生长发育异常,影响产量、育性、籽粒品质等。近年来,关于该蛋白家族成员参与植物生长发育的报道越来越多,但由于该家族庞大,大部分成员的功能还未被解析。本文系统总结了目前PPR蛋白调控细胞器基因转录后加工的分子机理及对细胞器和植株发育的影响,提出PPR家族还未解决的问题,为深入解析该基因家族的功能及育种应用提供理论基础。
关键词:
Abstract
Pentatricopeptide repeat (PPR) proteins constitute one of the largest protein families in land plants. They are sequence-specific RNA-binding proteins and play key roles in posttranscriptional processes within organelles. Their combined actions have profound effects on chloroplast photosynthetic electron transport chain and mitochondrial respiratory chain, affecting photosynthesis and respiration respectively, and ultimately on yield, fertility, and grain quality. Over the past decade, much has been learned about the molecular functions of these proteins on plant growth and development. However, due to the large size of this protein family, the functions of most members remain largely unknown. Here, we summarize the molecular mechanisms of PPR proteins functions on organelle genes, and effects on development of organelles and plants. Problems that need to be resolved are also identified. This article will provide a theoretical basis for understanding the functions of PPR protein family and genetic improvements of grain yield and quality.
Keywords:
PDF (838KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
郝媛媛, 赵向前, 黄福灯, 李春寿. PPR蛋白在植物细胞器组分转录后调控中的作用机制. 遗传[J], 2021, 43(11): 1050-1065 doi:10.16288/j.yczz.21-233
Yuanyuan Hao.
PPR (pentatricopeptide repeats)蛋白表达基因约在20多年前被发现[1,2],但是对PPR家族的系统研究,是从拟南芥(Arabidopsis thaliana)基因组测序之后才开始。对拟南芥基因组测序发现,将近31%的基因与已知功能的基因不同,而在未知功能的基因中,PPR基因占比6%,约有450个成员构成这个庞大的家族。由于与已知的TPR (tetratricopeptide repeat)蛋白结构域相似,****们将其命名为PPR蛋白,TPR蛋白是一类蛋白结合型蛋白家族,而PPR蛋白属于RNA结合蛋白,这两种蛋白可以通过保守的氨基酸残基进行区分。PPR蛋白广泛存在于真核细胞中,原核生物只有在少数病原体和共生菌中存在,因此PPR蛋白是一类真核细胞特有的蛋白家族[3]。
近年来,在研究各种类型突变体的过程中发现PPR蛋白在植物不同生长发育阶段发挥重要作用,因此有关植物PPR成员的功能研究越来越多。目前,PPR大部分成员被定位于线粒体和叶绿体,对细胞器基因的转录后调控起重要作用,主要包括RNA剪接、RNA稳定、RNA切割、RNA翻译和RNA编辑[3]。一些PPR基因突变后,下游的RNA底物加工异常,导致产生异常的转录本,进而影响了叶绿体和线粒体的正常发育,从而影响植物光合作用和呼吸作用等重要生物进程,导致植物生长发育受阻。如此,研究庞大的PPR家族成员影响植物生长的分子机理至关重要。
对PPR家族分子机理的研究关键是探寻其所作用的RNA底物及作用方式。植物线粒体及叶绿体的基因组较小,且各基因在基因组的位置已研究的较为清楚,利用成熟的分子生物学手段,可较快速地定位目标RNA,并确定影响植物生长发育的机理。因此,掌握PPR基因对底物的作用机理及作用方式有助于探究并总结PPR家族对植物生长发育的作用。本文以水稻(Oryza sativa)、拟南芥、玉米(Zea mays)为例,总结了目前多个PPR成员的分子作用机理及不同的PPR突变体对植物形态建成的影响,以期为未来研究庞大的PPR家族成员提供理论依据及研究方向。
1 PPR蛋白分类
PPR蛋白主要分为P类和PLS类,其中P类是由相对保守的35个氨基酸串联重复排列而成,PLS类成员则是由P基序(35个氨基酸)、L (longer)基序(35~36个氨基酸)和S (shorter) (31~32个氨基酸)基序为一个重复单位排列而成[3]。此外,PLS类成员按照其C端的不同氨基酸序列又分为E亚类、E+亚类和DYW亚类。E和E+结构域包含34个氨基酸,这种结构类似于TPR蛋白,DYW结构域包含胞苷脱氨酶序列,其命名来源于3个保守的氨基酸—天冬氨酸(D)、酪氨酸(Y)和色氨酸(W)[3]。2 PPR蛋白分子作用机理
PPR蛋白作为一种RNA结合蛋白,对mRNA转录后加工起着关键作用。P类成员主要参与RNA的剪接、稳定、切割和翻译过程,PLS类成员主要参与RNA的编辑过程。2.1 PPR蛋白参与顺式和反式剪接
RNA剪接是指把内含子从前体mRNA (pre- mRNA)上移除,并将外显子连接起来的过程。与细胞核基因不同,叶绿体和线粒体基因的外显子并非连续分布在环状基因组上,而是包含多个多顺反子结构,需要顺式剪切和反式剪切形成多个转录中间产物,并分别去除内含子,才能将外显子连接起来形成成熟mRNA。发生在一条前体mRNA之间的剪接,称为顺式剪接(cis-splicing),多见于真核生物细胞核基因的剪接;发生在不同前体mRNA之间的剪接,称为反式剪接(trans-splicing),反式剪接可产生某个成熟mRNA的中间转录物,反式剪接多见于对叶绿体和线粒体基因组的初始转录物进行加工的过程中[4]。需要剪切的叶绿体和线粒体基因内含子在不同物种中数量不同,被归类为I型内含子和II型内含子,其中大部分是II型内含子[4]。I型内含子由10个结构域构成(P1~P10),作用于RNA折叠,促进剪切和连接;II型内含子由6个双螺旋结构域构成(I~VI),形成复杂的空间结构,构成剪切的活性识别位点,每个结构域都有独特的作用,其剪切方式是经过两步转酯反应完成的,剪切机制类似于细胞核中的剪接体[5]。I型内含子和II型内含子的区别主要是二级结构和第一步剪切步骤的化学反应机制不同[6]。
II型内含子在植物中的分布较普遍,以水稻、玉米和拟南芥为例,拟南芥线粒体基因组9个基因共含有23个内含子,水稻和玉米线粒体基因组8个基因共含有22个内含子,且均为II型内含子。其中需要顺式剪切的内含子有17个(包含拟南芥中rpl2的一个内含子),需要反式剪切的内含子有6个[5]。目前,参与RNA剪接的PPR蛋白大多参与nad1、nad2、nad4的顺反式剪切,参与ccmFc、rps3、cox2 RNA剪切的PPR蛋白还未见报道(图1)[7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28]。拟南芥叶绿体基因组有20个II型内含子和1个I型内含子(trnL),分布在18个基因中;水稻和玉米叶绿体基因组有17个II型内含子和1个I型内含子(trnL),
图1
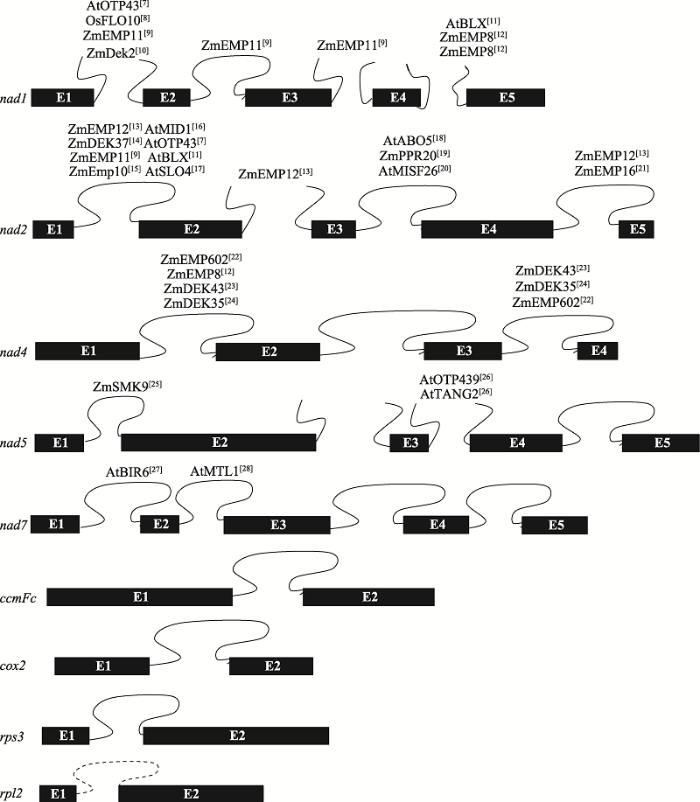 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1水稻、玉米和拟南芥中参与线粒体内含子剪接的PPR蛋白
水稻和玉米包含22个内含子,拟南芥包含23个内含子,蛋白名称右上角为参考文献。Zm:玉米;Os:水稻;At:拟南芥。图中展示了9个包含内含子的基因结构图,拟南芥比水稻和玉米多了1个内含子(rpl2的内含子,虚线标注),黑色方框代表外显子(E),闭合曲线表示顺式剪接的内含子,未闭合的曲线代表反式剪接的内含子。
Fig. 1PPR proteins involved in splicing of mitochondrial transcripts in rice, maize and Arabidopsis thaliana
分布在16个基因中[29]。这些内含子中只有trnL的一个内含子属于I型内含子,其余均为II型内含子。此外,根据结构特异性和功能差异性,II型内含子可分为IIA组、IIB组、IIC组和IIE/F组[30]。在植物中,依据内含子的二级结构不同,Michel等[31]将叶绿体的II型内含子分为IIA和IIB两组。叶绿体基因只有rps12的第一个内含子需反式剪切,且属于B组内含子,其余所有剪切方式均为顺式剪切。叶绿体中报道参与RNA剪接的PPR蛋白数量相对较少(图2)[32,33,34,35,36,37,38,39,40,41,42]。
图2
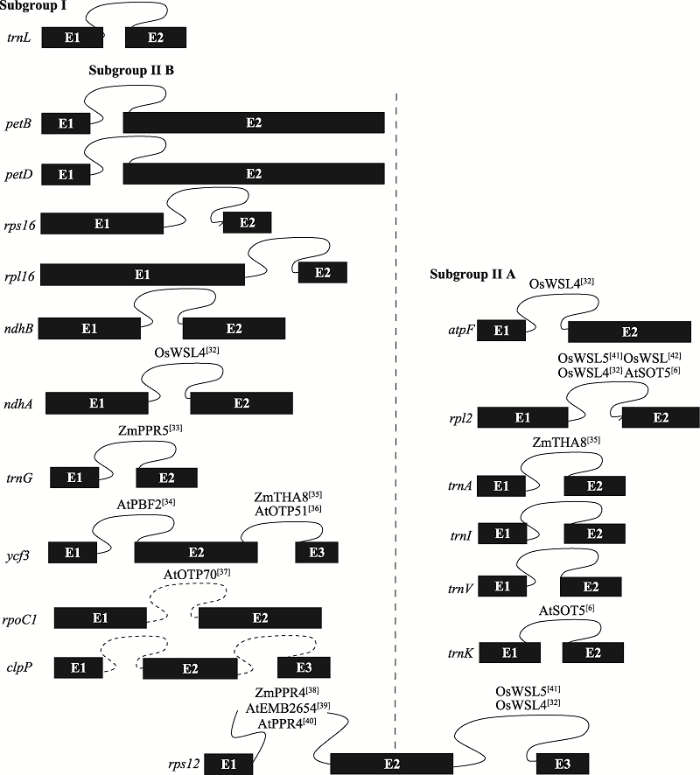 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2水稻、玉米和拟南芥中参与叶绿体内含子剪接的PPR蛋白
水稻和玉米包含18个内含子,拟南芥包含21个内含子,蛋白右上角为参考文献。Zm:玉米;Os:水稻;At:拟南芥。trnL是唯一一个I型内含子,其余均为II型内含子,且II型内含子分为两个亚组(Subgroup II A和Subgroup II B)。虚线左侧的内含子为B组内含子,虚线右侧的内含子为A组内含子。图中展示了包含内含子的基因结构图,拟南芥比水稻和玉米多了3个内含子(rpoC1的1个内含子,clpP的2个内含子,虚线标注)。黑色方框代表外显子(E),闭合曲线表示顺式剪接的内含子,未闭合的曲线代表反式剪接的内含子。
Fig. 2PPR proteins involved in the intron splicing of chloroplast transcripts in rice, maize and Arabidopsis thaliana
参与RNA剪接的PPR蛋白突变后,通常会造成成熟(spliced)的mRNA含量减少或缺失,而未剪接(unspliced)的前体mRNA含量增多。根据目前的报道,还没有一个PPR蛋白可参与某个基因所有内含子的剪接,可见一个基因,或者某个特定内含子,是由多个PPR蛋白和其他RNA加工类蛋白共同完成的,且这些RNA加工蛋白的氨基酸序列并不保守,这种现象反映了进化的多样性和剪接过程的复杂性。在进化过程中,不同物种线粒体和叶绿体基因组的内含子发生自然突变,进而招募相应的核编码蛋白(如PPR蛋白)进行剪接加工。若内含子在不同物种中的突变发生在单子叶和双子叶植物进化之后,则同一个内含子的剪接可能需要不同的PPR蛋白;若内含子的突变发生在单双子叶植物进化之前,则作用于内含子的PPR蛋白在不同物种中具有保守性。此外,同一个PPR蛋白也可参与多个基因内含子的剪切过程,这体现了PPR蛋白对下游底物作用机制的复杂性。
2.2 PPR蛋白参与mRNA稳定
PPR 蛋白稳定底物RNA主要是保护底物RNA免受核酸外切酶的非特异性切割。其作用机理是PPR蛋白结合于初始转录物的5ʹ或3ʹ末端,或者结合于多顺反子反式剪切中间转录物的5ʹ或3ʹ末端,作为一种“障碍”,阻挡5ʹ或3ʹ外切酶的非特异性切割,稳定mRNA不被降解。同时,由于所结合位置是固定的、特异性的,所以PPR也界定了转录物的5ʹ或3ʹ末端[43]。因此,PPR蛋白参与mRNA的稳定,既确定了转录物的末端,又保证了转录物免于降解,具有双重作用。由于线粒体和叶绿体基因组由多顺反子组成,因此界定正确的5ʹ或3ʹ末端这一功能对成熟mRNA的形成至关重要。参与RNA剪接的PPR蛋白功能缺失时,通常会造成成熟的底物RNA转录本减少或缺失,未剪切的中间转录本增加,表明剪接过程受阻。相反,参与mRNA稳定的PPR蛋白功能缺失时,虽然也会阻止mRNA成熟,但是未剪切的转录本并未增多,反而是减少的,由于未剪切的中间转录物被5ʹ或3ʹ外切酶非特异性切割,稳定性降低并缺失,导致不能形成功能正常的成熟mRNA。因此,该分子机理是鉴定PPR蛋白参与底物RNA的剪接或稳定进程的依据。
叶绿体中同时拥有5ʹ和3ʹ核酸外切酶,PPR蛋白可结合于转录物的5ʹ或3ʹ末端,参与mRNA的稳定(图3A)。玉米ZmPPR10可结合于atpI-atpH、psaJ-rpl33基因间区[44];此外,玉米ZmATP4、ZmCRP1、拟南芥AtHCF152可分别作用于psaJ-rpl33基因间区、petB-petD基因间区、psbH-petB基因间区[45,46,47],上述4个蛋白均能同时阻止5ʹ和3ʹ核酸外切酶的非特异性切割。拟南芥AtMRL1、AtPGR3、玉米ZmPPR103分别结合于rbcL、petL、rpl16的5ʹUTR,阻止5ʹ核酸外切酶的非特异性切割[48,49,50]。ZmPPR5较为特殊,它结合于trnG的3ʹ内含子,促进trnG的稳定,并促进剪切[33]。
线粒体基因组在进化过程中经过动态的进化和重排,只有3ʹ核酸外切酶,而无法检测到5ʹ核酸外切酶活性[51]。所以,在线粒体中,参与mRNA稳定的报道多集中于阻止3ʹ核酸外切酶的切割(图3B),而5ʹ端的加工为一种切割作用(见下文)。线粒体nad1的成熟过程经过3个反式剪切过程,所以其前体转录物包含4个中间产物,AtMTSF2和AtPPR19均可结合于第二个前体int1b-exon2-int2-exon3-int3a的内含子3a处,阻止3ʹ核酸外切酶的切割[52,53];AtMTSF1结合于nad4的3ʹUTR,免受3ʹ核酸外切酶的非特异性切割[54];ZmPPR78比较特殊,不参与稳定反式剪切的中间转录物,而是直接稳定成熟的nad5转录物,且突变后,成熟转录本中缺失5ʹATG和3ʹTGA的不完整转录本增多,猜测其可通过阻止3ʹ核酸外切酶的切割稳定3ʹ端,但是由于未能检测到5ʹ核酸外切酶活性,其缩短的5ʹ端转录本还未能解释。由此可见,在线粒体中仍然有未被发现的mRNA加工进程待挖掘[55]。
图3
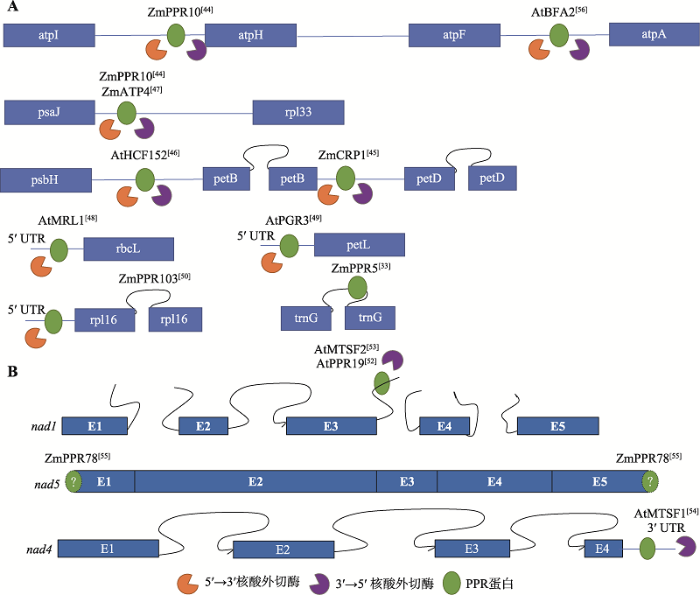 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3水稻、玉米和拟南芥中参与叶绿体(A)和线粒体(B)转录物稳定的PPR蛋白
蛋白右上角为参考文献。蓝色方框代表外显子,闭合曲线代表顺式剪接的内含子,未闭合的曲线代表反式剪接的内含子,蓝色直线代表基因间区。
Fig. 3PPR proteins involved in the RNA stabilization of chloroplast (A) and mitochondrial (B) transcripts in rice, maize and Arabidopsis thaliana
2.3 PPR蛋白参与mRNA切割
PPR蛋白参与RNA切割包含两种方式:核酸外切酶方式和核酸内切酶方式。较典型的是PPR编码的RF (restorers of fertility)蛋白可以切割育性相关的线粒体RNAs,在三系配套育种工作中发挥重要作用,也是系谱法选育品种的理论依据。细胞质雄性不育(cytoplasmic male sterility, CMS)是一种母系遗传的线粒体遗传缺陷,产生不育的花粉,目前已在200多种高等植物中观察到CMS。与CMS相关的线粒体基因均是以嵌合体的结构分布在线粒体基因组上[57],这种有缺陷的线粒体基因编码的蛋白,使植物特定发育阶段的组织生长受阻从而产生不育的花粉。用包含RF蛋白的恢复系与不育系杂交产生F1杂交种,在F1的植株中携带育性恢复基因,即可恢复植株的雄性育性,其分子机理即PPR蛋白可对线粒体目标RNA正确切割,进而不能产生有毒蛋白,从而恢复F1杂交种的育性。目前大部分的RF蛋白都是PPR蛋白,包括矮牵牛花(Petunia hybrida) RF[58]、油菜(Brassica napus) Rfk1[59]、萝卜(Raphanus sativus) Rfo[60]、高粱(Sorghum bicolor) PPR13[61]、水稻RF1[62]。以水稻BT-CMS育性恢复基因Rf-1为例,该基因包含两个位于第10号染色体的PPR基因Rf-1A和Rf-1B。BT-CMS线粒体基因组包含两个拷贝的atp6,分别为N-atp6和B-atp6。其中,B-atp6下游为orf79,ORF79蛋白是一个特异性表达于小孢子的毒性蛋白。B-atp6-orf79的初始转录物为2 kb,Rf-1A参与B-atp6 3ʹ末端和orf79 5ʹ端基因间区的切割,加工后分为1 kb B-atp6和0.4 kb orf79,而BT-CMS中初始转录物不能正确切割,产生毒性蛋白导致不育;Rf-1B降解B-atp6/orf79转录物,使植株不受毒性蛋白ORF79侵害,从而恢复育性,Rf-1A的作用上位于Rf-1B[63]。PPR蛋白参与5ʹ端切割的具体机制还未研究清楚,Barkan等[43]指出,可能的机制是切割位点的顺式作用元件可与PPR蛋白的结合位点形成一个RNA结构,当PPR蛋白结合于目标位点时,可暴露顺式作用元件,继而通过一种未知机制促进5ʹ端的加工成熟过程。拟南芥中与RF同源的蛋白称之为RFLs (restorers of fertility like),其中RPF1[64]作用于nad4 的5ʹUTR区、RPF2[65]作用于nad9和cox3的5'UTR区、RPF3[66]作用于ccmC的5ʹUTR区、RPF5[67]作用于nad6和26S RNAs的5ʹUTR区,促进邻近的5ʹ端形成。鉴于线粒体5ʹ→3ʹ核酸外切酶的缺失,这些蛋白功能丧失后,通常形成5ʹ端异常长的转录本。
除此之外,一些包含DYW结构域的PPR蛋白本身就具有核酸内切酶的活性。CRR2是一个DYW型的PPR蛋白,可作为一种核酸内切酶,切割rps7- ndhB基因间区,参与ndhB的成熟[68],CRR22和CRR28在体外也被证实具有核酸内切酶的活性[69]。此外,PPR蛋白家族的一小部分成员C端包含SMR (small MutS-related)结构域,SMR结构域具有核酸内切酶活性[70]。目前PPR成员中,只发现SOT1的C段SMR结构域具有内切酶的活性,AtSOT1可识别23S-4.5S rRNA前体5ʹ末端13 bp的序列,正确切割前体末端形成成熟的23S及4.5S rRNA[71]。ZmPPR53是AtSOT1的同源蛋白,PPR53直接结合于23S rRNA 5ʹ端上游70 nt的位点,稳定23S rRNA不被5ʹ→3ʹ外切酶切割,也可直接结合于ndhA 5ʹ端上游66 nt的位点,增加ndhA的翻译效率,但是其C端的SMR结构域功能还未能解释清楚[72]。
2.4 PPR蛋白参与mRNA翻译
PPR蛋白参与mRNA的翻译通常结合于底物的5ʹUTR区,促进底物RNA与核糖体的结合,从而激活或者抑制翻译。ZmPPR10[44]、AtPGR3[49]、ZmATP4[47]可分别激活atpH、petL、atpB的翻译过程,ZmCRP1[45]可同时激活psaC和petA的翻译。叶绿体中,PPR蛋白参与mRNA翻译的经典例子是PPR10。PPR10激活atpH翻译的研究为了解叶绿体PPR介导的翻译激活的潜在机制提供了基础。在PPR10未结合的情况下,PPR10的结合位点和atpH的核糖体结合位点(ribosome binding site, RBS)形成一个RNA发夹结构,PPR10结合后,可使RBS暴露,促进atpH和核糖体结合,激活翻译,同时结合到该位点的PPR10也阻断5ʹ→3ʹ外切核糖核酸酶活性,从而兼具稳定atpH的作用[43]。
线粒体中,PPR蛋白影响翻译的研究较少,且线粒体mRNA的核糖体结合位点不能被清楚认定[73],这就需要UTR特异性结合的PPR和其他蛋白质识别这些结合位点,对线粒体基因表达进行特异性的调节,例如ZmMPPR6结合于线粒体rps3 mRNA的5ʹUTR的3ʹ末端,rps3 5ʹUTR的RNA二级结构遮挡了起始密码子,从而抑制翻译起始,MPPR6可使这种二级结构松散,从而保证了与核糖体的正确结合,激活rps3翻译[74]。
除了激活翻译外,参与mRNA切割的RF蛋白可抑制底物RNA的翻译。萝卜的不育花器官中成熟毒蛋白ORF138的含量比根中高10倍,而含育性恢复基因Rfo的可育植株中,花器官和其他组织的毒蛋白含量均显著下降。但是,不管在可育或不育的植株中,ORF138转录物含量不受影响,这表明Rfo影响了ORF138的翻译或翻译后进程,使蛋白表达量在不育和可育植株中含量不同,但是调控翻译的机制还未可知[60]。
2.5 PPR蛋白参与mRNA编辑
RNA编辑是一种发生在转录后核苷酸特异位点的加工修饰现象,包括核苷酸的插入、删除和转换,其中核苷酸的转换包括两种方式,A→I和C→U。A→I的编辑仅由作用于RNA的腺苷脱氨酶(adenosine deaminase acting on RNA, ADAR)完成,主要发生在动物、真菌及细菌中[75];高等植物中RNA编辑主要发生在线粒体与叶绿体中,C→U核苷酸替换的RNA编辑是高等植物中的普遍方式,该方式是由RNA编辑体统筹,通过对初始转录物编码的C残基进行脱氨基来完成的[76]。C→U的改变,(1)一般发生在密码子的第一个或者第二个碱基位点,造成氨基酸的改变;(2)产生新的AUG起始密码子或者消除初始转录物的无义终止密码子,从而形成稳定成熟的mRNA,翻译为有功能的蛋白;(3)编辑后的位点可能产生RNA二级结构,影响mRNA的剪接和稳定[77]。C→U RNA 编辑可以影响细胞器蛋白质的序列、改变调节基序、RNA-蛋白质相互作用或RNA二级结构,在RNA 加工过程中发挥重要作用[77]。目前的研究已确定水稻、玉米、拟南芥中所有的编辑位点,以水稻为例,线粒体共485个编辑位点,分布在36个线粒体基因上(https://www.ncbi.nlm.nih.gov/nucleotide/BA000029);叶绿体共21个编辑位点,分布在11个叶绿体基因上[78],本文列出了目前在水稻、玉米、拟南芥中报道的部分参与RNA编辑的PPR蛋白及对应的编辑位点[79,80,81,82,83,84,85,86,87,88,89,90,91,92,93,94,95,96,97,98,99,100,101,102](表1)。Table 1
表1
表1水稻、玉米和拟南芥中部分参与叶绿体和线粒体RNA编辑的PPR蛋白
Table 1
| 玉米(Zea mays) | 拟南芥(Arabidopsis thaliana) | 水稻(Oryza sativa) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PPR编 辑蛋白 | 目的 基因 | 编辑 位点 | 蛋白 定位 | 参考文献 | PPR编 辑蛋白 | 目的 基因 | 编辑 位点 | 蛋白 定位 | 参考 文献 | PPR编 辑蛋白 | 目的 基因 | 编辑 位点 | 蛋白 定位 | 参考文献 |
| ZmSMK1 | nad7 | C836 | 线粒体 | [79] | PPR596 | rps3 | C1344 | 线粒体 | [89] | OGR1 | nad4 | C401/416/443 | 线粒体 | [100] |
| ZmSMK4 | cox1 | C1489 | 线粒体 | [80] | CRR4 | ndhD | C1 | 叶绿体 | [90] | nad2 | C1457 | |||
| ZmEMP5 | rpl16 | C458 | 线粒体 | [81] | CRR21 | ndhD | C2 | 叶绿体 | [91] | ccmC | C458 | |||
| nad9 | C190/C356 | SLO1 | nad4 | C449 | 线粒体 | [92] | cox2 | C167 | ||||||
| cox3 | C245/C257 | nad9 | C328 | cox3 | C572 | |||||||||
| ZmEMP7 | ccmFn | C1553 | 线粒体 | [82] | MEF3 | ATP4 | C89 | 线粒体 | [93] | MPR25 | nad5 | C1580 | 线粒体 | [101] |
| ZmEMP9 | ccMb | C43 | 线粒体 | [83] | CRR22 | ndhB | C7 | 叶绿体 | [94] | WSP1 | ndhD | C878 | 叶绿体 | [102] |
| rps4 | C335 | ndhD | C5 | rps14 | C80 | |||||||||
| ZmEMP18 | atp6 | C635 | 线粒体 | [84] | CRR28 | rpoB | C3 | 叶绿体 | [94] | rpoB | C467/545/560 | |||
| cox2 | C449 | ndhB | C2 | |||||||||||
| ZmDEK10 | cox2 | C550 | 线粒体 | [85] | ndhD | C3 | ||||||||
| ZmDEK39 | nad3 | C247/C275 | 线粒体 | [86] | OTP82 | ndhB | C9 | 叶绿体 | [95] | |||||
| ZmSMK6 | nad1 | C740 | 线粒体 | [87] | ndhG | C1 | ||||||||
| nad4L | C110 | OTP85 | ndhD | C116/494 | 叶绿体 | [96] | ||||||||
| nad7 | C739 | OTP86 | rps14 | C37161 | 叶绿体 | [96] | ||||||||
| mttb | C138/139 | MEF7 | cob | C206/325 | 线粒体 | [97] | ||||||||
| ZmEMP21 | nad7 | C77 | 线粒体 | [88] | ndh2 | C1433 | ||||||||
| atp1 | C1292 | nad4L | C41 | |||||||||||
| atp8 | C437 | MEF9 | nad7 | C200 | 线粒体 | [98] | ||||||||
| nad3 | C275 | MEF14 | matR | C1895 | 线粒体 | [99] | ||||||||
新窗口打开|下载CSV
RNA编辑体统筹RNA编辑过程,组成成员包括:PPR蛋白、MORFs (multiple organellar rna editing factors)蛋白、ORRM (organelle RNA recognition motif-containing)蛋白、PPO1 (protoporphyrinogen IX oxidase 1)蛋白、OZ1 (organelle zinc finger 1)蛋白[75]。其中PPR蛋白的PLS家族成员就是其中一种,主要作为反式作用因子,结合到编辑位点上游5~20个核苷酸的顺式作用元件上,使编辑位点被辨认出来,招募RNA编辑酶催化C→U的反应[91]。PLS家族成员C端的E1和E2结构域类似于PPR基序,可能通过蛋白互作或者结合RNA底物的方式参与编辑过程[103]。DYW结构域具有脱氨酶活性,可直接参与C→U编辑。
一个PPR蛋白可参与多个底物RNA的多个位点的编辑,同时某个特定RNA的编辑位点,可被不同的PPR蛋白识别。RNA编辑位点的编辑效率并不是100%,平均约有80%。不同组织或不同发育条件下,RNA编辑效率均会受影响[104]。
此外,PPR蛋白可与MORFs蛋白互作共同协作完成RNA的编辑过程。MORFs是叶绿体和线粒体编辑体成员,拟南芥中共10个成员,其中MORF1、3、4、6、7定位于线粒体,MORF2、9定位于叶绿体,MORF5、8双定位于线粒体和叶绿体。不同于PPR蛋白,MORF蛋白参与RNA编辑的过程是非特异性的,一个MORF蛋白即可参与相应细胞器中所有编辑位点的编辑过程。PPR蛋白的P基序或E1、E2结构域与MORF蛋白的N端或者中间区域互作,在不同的编辑位点可装配特异的蛋白复合体。MORF蛋白还可与PPR蛋白的L基序互作,减小PPR蛋白构象上P基序和S基序的距离,增加PLS结构域与底物RNA的亲和性[105]。
2.6 PPR蛋白通过特异的分子机制识别底物RNA
PPR蛋白的P、L、S基序分别具有保守的氨基酸位点,能区分不同的基序类别,并确定每个PPR重复的起始氨基酸[3]。Cheng等[106]发现,PPR基序识别下游RNA底物具有特异的分子机制。若PPR基序第五位的氨基酸是天冬酰胺(N),则对应的PPR基序结合嘧啶,若为丝氨酸(S)或苏氨酸(T),则结合嘌呤。此外,PPR基序的最后一位氨基酸是天冬氨酸(D),则结合尿嘧啶或鸟嘌呤,若为天冬酰胺(N),则结合胞嘧啶或腺苷酸。综上所述,每个PPR基序的第五位和最后一位氨基酸决定了所结合的RNA碱基。基于此规律,改变拟南芥PPR10对应位置的氨基酸就会改变对应的RNA识别位点[107]。这种识别方式也适用于PLS型PPR蛋白,该类型蛋白最后一个S基序与编辑位点前的第4位核苷酸结合,该位置允许胞苷脱氨酶活性特异性作用于待编辑的胞苷。位于编辑位点上游5~20个核苷酸的顺式作用元件是PLS型PPR蛋白的识别位点[77]。PPR56可作为PLS型PPR蛋白特异性识别底物的模式蛋白,PPR56第五位与最后一位氨基酸的组合与相对应碱基可归纳为以下规律:T/S+N: A, T/S+D: G, N+N: C/U, N+S: C>U, N+ D: U>C,这与其底物Nad3和Nad4对应的顺式作用元件相吻合[77]。
值得注意的是,这种结合机制并不适合所有的PPR蛋白。一些P类家族成员在每个重复之间可能存在多余的氨基酸残基,并不是保守的每35个氨基酸重复紧密相连。而且每个重复第五位的氨基酸和最后一个氨基酸并不是保守地按照上述规律排列,不同的PPR蛋白差异很大,所以还需进一步补充这种分子机制,完善PPR蛋白结合底物RNA的规律。
3 PPR蛋白对细胞器发育的影响
3.1 PPR蛋白影响线粒体和叶绿体发育
几乎所有PPR突变体表现出的表型均是由一种或几种线粒体或叶绿体基因产物的功能缺失。尽管PPR家族成员很多,但不同家族成员之间的功能几乎没有冗余,不同物种中某些同源的PPR蛋白也具有不同的作用底物。PPR突变体的表型主要包括:光合缺陷[35,45,48,68]、叶片发育异常[108]、色素积累[109,110]、生长受阻[8]、胚或胚乳发育异常[7,9,10]、脱落酸信号途径受损[111]和胞质雄性不育[58,59,60,61,62]。在真核细胞中,线粒体是一种半自主性细胞器,有自身的基因组。在进化过程中,线粒体大部分基因整合到宿主细胞核基因组中,经过转录翻译后,转运到线粒体,参与线粒体代谢和线粒体基因表达调控[112]。陆生植物线粒体基因组包含60~70个基因,这些基因编码的蛋白包括tRNAs、rRNAs、核糖体蛋白、复合体I(NADH脱氢酶)亚基、复合体III(细胞色素C还原酶)亚基、复合体IV(细胞色素C氧化酶)亚基、ATP合酶亚基和细胞色素C合成酶亚基等。
线粒体为植物发育提供了能量,其功能丧失对植物生长有害。线粒体PPR蛋白分子功能的确定有助于阐明RNA加工机制以及氧化磷酸化机制的组装。复合体I嵌入线粒体内膜并介导电子从NADH转移至泛醌[113],是电子进入电子传输链的主要入口点,多个研究表明PPR参与的剪接缺陷能影响复合体I的装配和稳定[13,114,115]。Nad1、Nad2和Nad4蛋白是复合物I的膜臂成分[116],Nad1形成醌结合位点,Nad2是复合物I中质子转移的位点,而Nad4、Nad5形成质子易位子之一,并且在结构上与K或Na+/H+逆向运输蛋白有关。由于需要复合物I中Nad1、Nad2、Nad4、Nad5参与质子转移和醌键结合,PPR蛋白表达受阻而导致的nad1、nad2、nad4、nad5异常转录都会严重降低复合物I的含量,使活性显著降低[10,12,22,25]。
PPR蛋白ZmEMP8、ZmDEK43、AtPPR19[12,23,52]功能异常时,复合体I亚基功能的缺失还可导致细胞色素途径受损,线粒体氧化磷酸化效率降低,导致植物出现代谢问题[117],诱发交替途径起始[24],导致交替氧化酶基因的转录水平提高。此外,OsNPPR1、OsFlo10[8,118]突变后,由于电子传递效率降低,使得呼吸链产生的ATP含量显著下降[117]。氧化磷酸化中产生的活性氧(reactive oxygen species, ROS)在触发植物细胞程序性死亡(programmed cell death, PCD)中起关键作用[119]。通常,PCD从胚乳的中部开始,然后扩散到外围[120]。PPR突变体由于氧化磷酸化进程的异常,导致产生过量的ROS,使突变体胚乳比野生型更早的发生PCD,这会干扰淀粉和贮藏蛋白的合成[121],所以定位于线粒体的PPR蛋白突变通常会造成胚乳粉质和皱缩的表型。可见,线粒体PPR功能缺陷型突变体,会导致线粒体发育受损,影响线粒体功能。
真核细胞中,叶绿体是光合作用的主要场所,其基因组包含约100个基因左右,这些基因主要参与光合作用、ATP合成和基质中蛋白的转录翻译和降解过程。叶绿体光合作用需要光合电子传递链,由3种复合物PS I、PS II和Cytb6f组成。PPR蛋白HCF152[46]、MRL1[48]、PGR3[49]等突变,通过影响光合电子传递链复合物的成员转录后加工,从而造成叶绿体发育异常;叶绿体ATP合酶是位于类囊体膜上的一个多亚基复合体,它利用光合作用电子传递产生的质子动力将ADP转化为ATP。叶绿体ATP合酶由CFo和CF1模块组成,其中由叶绿体基因参与的亚基产生于两个多顺反子叶绿体转录单位atpI- H-F-A和atpB-E[122]。PPR蛋白WSL4[32]、ATP4[47]通过影响合酶成员的剪接和翻译,影响叶绿体中ATP的合成过程;此外,在叶绿体基质中存在叶绿体蛋白酶体clp复合体、叶绿体RNA聚合酶复合体以及一些核糖体蛋白[56]。PPR蛋白WSL[42]、CRR2[68]、CLB19[109]的突变影响这些途径叶绿体mRNA的成熟过程。
3.2 PPR蛋白在细胞核中的功能
有一类特殊的PPR蛋白,亚细胞定位于细胞核,包括GRP23、PNM1、SORA1和OsNPPR1。其中GRP23只定位于细胞核,与RNA聚合酶II亚基III互作,突变后致死[123]。PNM1双定位于细胞核和线粒体,其突变体的致死表型只与线粒体定位有关,在细胞核中PNM1可与转录因子TCP互作,调控线粒体氧化磷酸化过程相关的核基因表达[124]。GRP23与细胞核RNA聚合酶II亚基III互作,PNM1与细胞核转录因子TCP8和NAP1互作,说明PPR定位于细胞核可以影响核mRNA的转录和转录后调控进程。此外,TCP8转录因子特异性的转录与线粒体氧化磷酸化途径相关的细胞核基因[125],说明定位于细胞核的PPR蛋白可能通过参与定位于细胞器的核基因的转录后调控过程,参与细胞器的代谢过程。在水稻中,OsNPPR1是定位于细胞核的PPR蛋白,但却影响了线粒体的功能,猜测可能是参与了细胞核中与线粒体发育相关基因的转录后调控而间接影响线粒体发育[118];SOAR1双定位于细胞核和胞质,参与ABA信号途径,作用于ABAR1下游和ABI5的上游[126]。4 PPR功能展望
本文系统综述了水稻、玉米、拟南芥中PPR蛋白参与转录后调控的分子机理和性状表现。PPR蛋白可参与整个植物生育期的发育过程,影响植株的生长,对植物的正常长成有重要作用。近年来,对PPR成员的功能研究也越来越多。但是,仍有一些重要的科学问题没有解决:(1)庞大的PPR蛋白家族成员是否仅调控100多个细胞器基因的表达;(2) E和E+型的PLS PPR蛋白结合底物后,招募编辑酶的分子机制还未知;(3)萝卜的不育花器官中成熟毒蛋白ORF138的含量极高,而含育性恢复基因Rfo的可育植株中,各组织毒蛋白含量均显著下降。但是,不管在可育或不育的植株中,ORF138转录物含量不受影响,虽然参与翻译的过程,但是调控翻译的机制还不清楚;(4)参与RNA切割的线粒体PPR蛋白的切割机制还未知;(5)虽然通过对PPR10的分子机理进行系统的研究,发现了可能存在的PPR基序与底物结合的“密码”[99],但是由于PPR家族各成员之间保守性相差较大,所以“密码”并不适用于所有的PPR成员,是否有更精细明确的结合方式存在还有待探索。细胞器基因的正常表达,需要PPR家族成员协同工作,研究PPR蛋白可定向控制细胞器基因的表达,为利用生物工程技术,改良作物光合作用和呼吸作用进程提供理论基础。
(责任编委: 宋任涛)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
PMID:7664742 [本文引用: 1]

Expression of the Saccharomyces cerevisiae mitochondrial COX1 locus, which contains several introns and is co-transcribed with the downstream genes AAP1, OLI2 and ENS2, is controlled by at least 18 nuclear-encoded proteins. The PET309 gene, encoding one of these proteins, was cloned, sequenced and shown to contain an open reading frame of 965 codons. Isonuclear PET309+ and delta pet309::URA3 strains carrying mitochondrial genomes that differ in the number of COX1 introns, were generated. Analysis of RNA species from these strains demonstrated an inverse relationship between the number of introns present in the precursor RNA and the amount of COX1 and AAP1/OLI2/ENS2 RNAs accumulated in a pet309 mutant. Hence, PET309 plays a role either in transcription of intron-containing primary transcripts from the COX1-AAP1-OLI2-ENS2 transcription unit or in stabilization of primary transcripts. PET309 is also required in translation of COX1 mRNA. A mitochondrial bypass suppressor of the pet309 deletion mutation was isolated, and shown to consist of a DNA rearrangement at the COX1 locus, such that the 5' untranslated leader region (UTR) of the COB gene was fused to COX1 at nucleotide -174 of its 5' UTR. This result suggests that Pet309p acts through the COX1 5' UTR to activate initiation of translation of the COX1 coding region.
PMID:9342407 [本文引用: 1]

The cya-5 nuclear mutant of Neurosopora crassa was previously shown to be deficient in cytochrome aa3, cytochrome c oxidase activity, and the immunologically detectable COXI protein. We have now demonstrated that the mitochondria of this mutant contain mRNA for the COXI protein and that COXI cannot be detected during pulse-chase labeling experiments of mitochondrial translation products. Cloning and analysis of the cya-5 gene reveal a long open reading frame capable of encoding a 1136 amino-acid protein. Sequence analysis suggests that the potential CYA-5 protein contains a mitochondrial targeting sequence at its amino-terminus. The long open reading frame also contains a 200 amino-acid region with homology to the PET309 protein, which is required for the production or stability of intron-containing coxI mRNAs, as well as the translation of mature coxI mRNAs, in the yeast Saccharomyces cerevisiae. These data suggest that the CYA-5 protein of N. crassa is required in a post-transcriptional step for COXI expression, most probably for the efficient translation of coxI mRNA.
DOI:10.1105/tpc.104.022236URL [本文引用: 5]
DOI:10.1016/j.mito.2007.09.005URL [本文引用: 2]
[本文引用: 2]
DOI:10.1104/pp.18.00406URL [本文引用: 1]
PMID:17965268 [本文引用: 2]

The mitochondrial NADH:ubiquinone oxidoreductase complex (Complex I) is a large protein complex formed from both nuclearly and mitochondrially encoded subunits. Subunit ND1 is encoded by a mitochondrial gene comprising five exons, and the mature transcript requires four RNA splicing events, two of which involve trans-splicing independently transcribed RNAs. We have identified a nuclear gene (OTP43) absolutely required for trans-splicing of intron 1 (and only intron 1) of Arabidopsis thaliana nad1 transcripts. This gene encodes a previously uncharacterized pentatricopeptide repeat protein. Mutant Arabidopsis plants with a disrupted OTP43 gene do not present detectable mitochondrial Complex I activity and show severe defects in seed development, germination, and to a lesser extent in plant growth. The alternative respiratory pathway involving alternative oxidase is significantly induced in the mutant.
DOI:10.1111/nph.2019.223.issue-2URL [本文引用: 3]
DOI:10.1093/jxb/erx212URL [本文引用: 2]
DOI:10.1534/genetics.116.196105URL [本文引用: 3]
DOI:10.1016/j.jgg.2018.01.006URL [本文引用: 1]
DOI:10.1111/tpj.2018.95.issue-5URL [本文引用: 3]
DOI:10.1093/jxb/ery432URL [本文引用: 2]
DOI:10.1534/genetics.117.300602URL [本文引用: 1]
DOI:10.1111/tpj.2017.91.issue-1URL [本文引用: 1]
DOI:10.1038/s41598-020-58495-5URL [本文引用: 1]
DOI:10.1007/s11103-016-0566-4URL [本文引用: 1]
DOI:10.1111/tpj.2010.63.issue-5URL [本文引用: 1]
DOI:10.1093/pcp/pcz204PMID:31670803 [本文引用: 1]

Pentatricopeptide repeat (PPR) proteins are helical repeat RNA-binding proteins that function in RNA processing by conferring sequence-specific RNA-binding activity. Owing to the lethality of PPR mutants, functions of many PPR proteins remain obscure. In this study, we report the function of PPR20 in intron splicing in mitochondria and its role in maize seed development. PPR20 is a P-type PPR protein targeted to mitochondria. The ppr20 mutants display slow embryo and endosperm development. Null mutation of PPR20 severely reduces the cis-splicing of mitochondrial nad2 intron 3, resulting in reduction in the assembly and activity of mitochondrial complex I. The ppr20-35 allele with a Mu insertion in the N-terminal region shows a much weaker phenotype. Molecular analyses revealed that the mutant produces a truncated transcript, coding for PPR20ΔN120 lacking the N-terminal 120 amino acids. Subcellular localization revealed that PPR20ΔN120:GFP is able to target to mitochondria as well, suggesting the sequence diversity of the mitochondrial targeting peptides. Another mutant zm_mterf15 was also found to be impaired in the splicing of mitochondrial nad2 intron 3. Further analyses are required to identify the exact function of PPR20 and Zm_mTERF15 in the splicing of nad2 intron 3.© The Author(s) 2019. Published by Oxford University Press on behalf of Japanese Society of Plant Physiologists. All rights reserved. For permissions, please email: journals.permissions@oup.com.
DOI:10.1093/jxb/ery275URL [本文引用: 1]
DOI:10.1111/tpj.13122URL [本文引用: 1]
DOI:10.1093/pcp/pcz083URL [本文引用: 2]
DOI:10.1111/jipb.v62.3URL [本文引用: 2]
DOI:10.1016/j.molp.2016.08.008URL [本文引用: 2]
DOI:10.1016/j.plantsci.2019.110205URL [本文引用: 2]
PMID:24958715 [本文引用: 1]

Pentatricopeptide repeat proteins constitute a large family of RNA-binding proteins in higher plants (around 450 genes in Arabidopsis [Arabidopsis thaliana]), mostly targeted to chloroplasts and mitochondria. Many of them are involved in organelle posttranscriptional processes, in a very specific manner. Splicing is necessary to remove the group II introns, which interrupt the coding sequences of several genes encoding components of the mitochondrial respiratory chain. The nad5 gene is fragmented in five exons, belonging to three distinct transcription units. Its maturation requires two cis- and two trans-splicing events. These steps need to be performed in a very precise order to generate a functional transcript. Here, we characterize two pentatricopeptide repeat proteins, ORGANELLE TRANSCRIPT PROCESSING439 and TANG2, and show that they are involved in the removal of nad5 introns 2 and 3, respectively. To our knowledge, they are the first two specific nad5 splicing factors found in plants so far.© 2014 American Society of Plant Biologists. All Rights Reserved.
DOI:10.1074/jbc.M110.147603PMID:20682769 [本文引用: 1]

Splicing of plant organellar transcripts is facilitated by members of a large protein family, the pentatricopeptide repeat proteins. We have identified a pentatricopeptide repeat protein in a genetic screen for mutants resistant to inhibition of root growth by buthionine sulfoximine (BSO), an inhibitor of glutathione synthesis and consequently named BIR6 (BSO-insensitive roots 6). BIR6 is involved in splicing of intron 1 of the mitochondrial nad7 transcript. Loss-of-function mutations in BIR6 result in a strongly reduced accumulation of fully processed nad7 transcript. This affects assembly of Complex I and results in moderate growth retardation. In agreement with disruption of Complex I function, the genes encoding alternative NADH oxidizing enzymes are induced in the mutant, and the mutant plants are less sensitive to mannitol and salt stress. Mutation in the BIR6 gene allowed normal root growth in presence of BSO and strongly attenuated depletion of glutathione content at these conditions. The same phenotype was observed with other mutants affected in function of Complex I, thus reinforcing the importance of Complex I function for cellular redox homeostasis.
DOI:10.1104/pp.15.01591URL [本文引用: 1]
DOI:10.1093/mp/ssq025PMID:20603383 [本文引用: 1]

Plant organelles arose from two independent endosymbiosis events. Throughout evolutionary history, tight control of chloroplasts and mitochondria has been gained by the nucleus, which regulates most steps of organelle genome expression and metabolism. In particular, RNA maturation, including RNA splicing, is highly dependent on nuclearly encoded splicing factors. Most introns in organelles are group II introns, whose catalytic mechanism closely resembles that of the nuclear spliceosome. Plant group II introns have lost the ability to self-splice in vivo and require nuclearly encoded proteins as cofactors. Since the first splicing factor was identified in chloroplasts more than 10 years ago, many other proteins have been shown to be involved in splicing of one or more introns in chloroplasts or mitochondria. These new proteins belong to a variety of different families of RNA binding proteins and provide new insights into ribonucleo-protein complexes and RNA splicing machineries in organelles. In this review, we describe how splicing factors, encoded by the nucleus and targeted to the organelles, take part in post-transcriptional steps in higher plant organelle gene expression. We go on to discuss the potential for these factors to regulate organelle gene expression.
DOI:10.1146/biophys.2016.45.issue-1URL [本文引用: 1]
DOI:10.1016/0378-1119(89)90026-7URL [本文引用: 1]
DOI:10.3389/fpls.2017.01116URL [本文引用: 2]
DOI:10.1128/MCB.00563-08URL [本文引用: 2]
DOI:10.1111/jipb.v62.11URL [本文引用: 1]
DOI:10.1261/rna.032623.112PMID:22495966 [本文引用: 2]

A maize gene designated thylakoid assembly 8 (tha8) emerged from a screen for nuclear mutations that cause defects in the biogenesis of chloroplast thylakoid membranes. The tha8 gene encodes an unusual member of the pentatricopeptide repeat (PPR) family, a family of helical repeat proteins that participate in various aspects of organellar RNA metabolism. THA8 localizes to chloroplasts, where it associates specifically with the ycf3-2 and trnA group II introns. The splicing of ycf3-2 is eliminated in tha8 mutants, and trnA splicing is strongly compromised. Reverse-genetic analysis of the tha8 ortholog in Arabidopsis thaliana showed that these molecular functions are conserved, although null alleles are embryo lethal in Arabidopsis and seedling lethal in maize. Whereas most PPR proteins have more than 10 PPR motifs, THA8 belongs to a subfamily of plant PPR proteins with only four PPR motifs and little else. THA8 is the first member of this subfamily with a defined molecular function, and illustrates that even small PPR proteins have the potential to mediate specific intermolecular interactions in vivo.
DOI:10.1111/tpj.2008.56.issue-1URL [本文引用: 1]
DOI:10.1111/tpj.2011.65.issue-4URL [本文引用: 1]
PMID:17041147 [本文引用: 1]

The pentatricopeptide repeat (PPR) is a degenerate 35-amino acid repeat motif that is widely distributed among eukaryotes. Genetic, biochemical, and bioinformatic data suggest that many PPR proteins influence specific posttranscriptional steps in mitochondrial or chloroplast gene expression and that they may typically bind RNA. However, biological functions have been determined for only a few PPR proteins, and with few exceptions, substrate RNAs are unknown. To gain insight into the functions and substrates of the PPR protein family, we characterized the maize (Zea mays) nuclear gene ppr4, which encodes a chloroplast-targeted protein harboring both a PPR tract and an RNA recognition motif. Microarray analysis of RNA that coimmunoprecipitates with PPR4 showed that PPR4 is associated in vivo with the first intron of the plastid rps12 pre-mRNA, a group II intron that is transcribed in segments and spliced in trans. ppr4 mutants were recovered through a reverse-genetic screen and shown to be defective for rps12 trans-splicing. The observations that PPR4 is associated in vivo with rps12-intron 1 and that it is also required for its splicing demonstrate that PPR4 is an rps12 trans-splicing factor. These findings add trans-splicing to the list of RNA-related functions associated with PPR proteins and suggest that plastid group II trans-splicing is performed by different machineries in vascular plants and algae.
DOI:10.1104/pp.16.01840PMID:28011633 [本文引用: 1]

We report the partial complementation and subsequent comparative molecular analysis of two nonviable mutants impaired in chloroplast translation, one (emb2394) lacking the RPL6 protein, and the other (emb2654) carrying a mutation in a gene encoding a P-class pentatricopeptide repeat protein. We show that EMB2654 is required for the trans-splicing of the plastid rps12 transcript and that therefore the emb2654 mutant lacks Rps12 protein and fails to assemble the small subunit of the plastid ribosome, explaining the loss of plastid translation and consequent embryo-lethal phenotype. Predictions of the EMB2654 binding site match a small RNA "footprint" located on the 5' half of the trans-spliced intron that is almost absent in the partially complemented mutant. EMB2654 binds sequence specifically to this target sequence in vitro. Altered patterns in nuclease-protected small RNA fragments in emb2654 show that EMB2654 binding must be an early step in, or prior to, the formation of a large protein-RNA complex covering the free ends of the two rps12 intron halves.© 2017 American Society of Plant Biologists. All Rights Reserved.
DOI:10.1111/tpj.v100.6URL [本文引用: 1]
DOI:10.1093/jxb/ery259PMID:30011040 [本文引用: 1]
DOI:10.1093/mp/ssu054URL [本文引用: 2]
DOI:10.1146/annurev-arplant-050213-040159PMID:24471833 [本文引用: 3]

Pentatricopeptide repeat (PPR) proteins constitute one of the largest protein families in land plants, with more than 400 members in most species. Over the past decade, much has been learned about the molecular functions of these proteins, where they act in the cell, and what physiological roles they play during plant growth and development. A typical PPR protein is targeted to mitochondria or chloroplasts, binds one or several organellar transcripts, and influences their expression by altering RNA sequence, turnover, processing, or translation. Their combined action has profound effects on organelle biogenesis and function and, consequently, on photosynthesis, respiration, plant development, and environmental responses. Recent breakthroughs in understanding how PPR proteins recognize RNA sequences through modular base-specific contacts will help match proteins to potential binding sites and provide a pathway toward designing synthetic RNA-binding proteins aimed at desired targets.
DOI:10.1038/emboj.2009.121URL [本文引用: 2]
PMID:8039510 [本文引用: 3]

A mutant designated crp1 (chloroplast RNA processing 1) was identified in a screen for transposon-induced maize mutants with defects in chloroplast gene expression. crp1 is a recessive, nuclear mutation that causes the loss of the cytochrome f/b6 complex and a reduction in photosystem I. The molecular basis for these protein losses is unique relative to previously described mutants with defects in organelle gene expression; it involves defects in the metabolism of two organellar mRNAs and in the translation of two organellar proteins. Mutants lack the monocistronic forms of the petB and petD mRNAs (encoding cytochrome f/b6 subunits), but contain normal levels of their polycistronic precursors. Pulse-labeling experiments revealed normal synthesis of the petB gene product, but a large decrease in synthesis of the petD gene product. These results suggest that petD sequences are more efficiently translated in a monocistronic than in a polycistronic context, thereby providing evidence that the elaborate RNA processing typical of chloroplast transcripts can play a role in controlling gene expression. Structural predictions suggest that the petD start codon lies in a stable hairpin in the polycistronic RNA, but remains unpaired in the monocistronic transcript. Thus, processing to a monocistronic form may increase translational efficiency by releasing the translation initiation region from inhibitory interactions with upstream RNA sequences. Synthesis of a third cytochrome f/b6 subunit, encoded by the petA gene, was undetectable in crp1, although its mRNA appeared unaltered. Two mechanisms are consistent with the simultaneous loss of both petA and petD protein synthesis: the translation of the petA and petD mRNAs might be coupled via a mechanism independent of crp1, or the crp1 gene may function to coordinate the expression of the two genes, which encode subunits of the same complex.
PMID:12782738 [本文引用: 2]

The psbB-psbT-psbH-petB-petD operon of higher plant chloroplasts is a heterogeneously composed transcriptional unit that undergoes complex RNA processing events until the mature oligocistronic RNAs are formed. To identify the nucleus-encoded factors required for the processing and expression of psbB-psbT-psbH-petB-petD transcripts, we performed mutational analysis using Arabidopsis. The allelic nuclear mutants hcf152-1 and hcf152-2 were identified that are affected specifically in the accumulation of the plastidial cytochrome b(6)f complex. In both mutants, reduced amounts of spliced petB RNAs (encoding the cytochrome b(6) subunit) were detected, thus explaining the observed protein deficiencies. Additionally, mutant hcf152-1 is affected in the accumulation of transcripts cleaved between the genes psbH and petB. As a result of a close T-DNA insertion, the HCF152 gene was cloned and its identity confirmed by complementation of homozygous mutant plants. HCF152 encodes a pentatricopeptide repeat (PPR) protein with 12 putative PPR motifs that is located inside the chloroplast. The protein shows a significant structural, but not primary, sequence similarity to the maize protein CRP1, which is involved in the processing and translation of the chloroplast petD and petA RNAs. In addition, we found that HCF152 is an RNA binding protein that binds certain areas of the petB transcript. The protein possibly exists in the chloroplast as a homodimer and is not associated with other proteins to form a high molecular mass complex.
DOI:10.1111/tpj.2012.72.issue-4URL [本文引用: 3]
DOI:10.1105/tpc.109.066266URL [本文引用: 3]
DOI:10.1111/tpj.2011.67.issue-2URL [本文引用: 3]
DOI:10.1093/nar/gkw270PMID:27095196 [本文引用: 1]

Pentatricopeptide repeat (PPR) proteins are a large family of helical-repeat proteins that bind RNA in mitochondria and chloroplasts. Precise RNA targets and functions have been assigned to only a small fraction of the >400 members of the PPR family in plants. We used the amino acid code governing the specificity of RNA binding by PPR repeats to infer candidate-binding sites for the maize protein PPR103 and its ortholog Arabidopsis EMB175. Genetic and biochemical data confirmed a predicted binding site in the chloroplast rpl16 5'UTR to be a site of PPR103 action. This site maps to the 5' end of transcripts that fail to accumulate in ppr103 mutants. A small RNA corresponding to the predicted PPR103 binding site accumulates in a PPR103-dependent fashion, as expected of PPR103's in vivo footprint. Recombinant PPR103 bound specifically to this sequence in vitro These observations imply that PPR103 stabilizes rpl16 mRNA by impeding 5'→3' RNA degradation. Previously described PPR proteins with this type of function consist of canonical PPR motifs. By contrast, PPR103 is a PLS-type protein, an architecture typically associated with proteins that specify sites of RNA editing. However, PPR103 is not required to specify editing sites in chloroplasts.© The Author(s) 2016. Published by Oxford University Press on behalf of Nucleic Acids Research.
[本文引用: 1]
DOI:10.1111/nph.2017.215.issue-1URL [本文引用: 2]
DOI:10.1093/nar/gkx162URL [本文引用: 1]
DOI:10.1093/nar/gkt337URL [本文引用: 1]
DOI:10.1016/j.molp.2017.09.009URL [本文引用: 1]
DOI:10.3389/fpls.2019.00446PMID:31031784 [本文引用: 1]

As a fascinating and complicated nanomotor, chloroplast ATP synthase comprises nine subunits encoded by both the nuclear and plastid genomes. Because of its uneven subunit stoichiometry, biogenesis of ATP synthase and expression of plastid-encoded ATP synthase genes requires assistance by nucleus-encoded factors involved in transcriptional, post-transcriptional, and translational steps. In this study, we report a P-class pentatricopeptide repeat (PPR) protein BFA2 (Biogenesis Factor required for ATP synthase 2) that is essential for accumulation of the dicistronic transcript in Arabidopsis chloroplasts. A loss-of-function mutation in results in a specific reduction of more than 3/4 of chloroplast ATP synthase, which is likely due to the absence of dicistronic transcript. BFA2 protein contains 22 putative PPR motifs and exclusively localizes in the chloroplast. Bioinformatics and Electrophoretic Mobility Shift Assays (EMSA) analysis showed that BFA2 binds to the consensus sequence of the intergenic region in a sequence-specific manner. However, translation initiation of the was not affected in the mutant. Thus, we propose that the chloroplast PPR protein BFA2 mainly acts as barrier to prevent the transcript degradation by exoribonucleases by binding to the consensus sequence of the intergenic region.
[本文引用: 1]
DOI:10.4161/rna.25568URL [本文引用: 2]
DOI:10.1073/pnas.102301599URL [本文引用: 2]
DOI:10.1046/j.1365-313X.2003.01799.xURL [本文引用: 3]
PMID:16078015 [本文引用: 2]

With an aim to clone the sorghum fertility restorer gene Rf1, a high-resolution genetic and physical map of the locus was constructed. The Rf1 locus was resolved to a 32-kb region spanning four open reading frames: a plasma membrane Ca(2+)-ATPase, a cyclin D-1, an unknown protein, and a pentatricopeptide repeat (PPR13) gene family member. An approximately 19-kb region spanning the cyclin D-1 and unknown protein genes was completely conserved between sterile and fertile plants as was the sequence spanning the coding region of the Ca(2+)-ATPase. In contrast, 19 sequence polymorphisms were located in an approximately 7-kb region spanning PPR13, and all markers cosegregated with the fertility restoration phenotype. PPR13 was predicted to encode a mitochondrial-targeted protein containing a single exon with 14 PPR repeats, and the protein is classified as an E-type PPR subfamily member. To permit sequence-based comparison of the sorghum and rice genomes in the Rf1 region, 0.53 Mb of sorghum chromosome 8 was sequenced and compared to the colinear region of rice chromosome 12. Genome comparison revealed a mosaic pattern of colinearity with an approximately 275-kb gene-poor region with little gene conservation and an adjacent, approximately 245-kb gene-rice region that is more highly conserved between rice and sorghum. Despite being located in a region of high gene conservation, sorghum PPR13 was not located in a colinear position on rice chromosome 12. The present results suggest that sorghum PPR13 represents a potential candidate for the sorghum Rf1 gene, and its presence in the sorghum genome indicates a single gene transposition event subsequent to the divergence of rice and sorghum ancestors.
DOI:10.1046/j.1365-313X.2003.01961.xURL [本文引用: 2]
DOI:10.1105/tpc.105.038240URL [本文引用: 1]
DOI:10.1111/tpj.2011.65.issue-5URL [本文引用: 1]
DOI:10.1105/tpc.109.066944URL [本文引用: 1]
DOI:10.1104/pp.111.181552URL [本文引用: 1]
DOI:10.1111/tpj.2013.74.issue-4URL [本文引用: 1]
PMID:14617084 [本文引用: 3]

The chloroplast NDH complex, NAD(P)H dehydrogenase, reduces the plastoquinone pool non-photochemically and is involved in cyclic electron flow around photosystem I (PSI). A transient increase in chlorophyll fluorescence after turning off actinic light is a result of NDH activity. We focused on this subtle change in chlorophyll fluorescence to isolate nuclear mutants affected in chloroplast NDH activity in Arabidopsis by using chlorophyll fluorescence imaging. crr2-1 and crr2-2 (chlororespiratory reduction) are recessive mutant alleles in which accumulation of the NDH complex is impaired. Except for the defect in NDH activity, photosynthetic electron transport was unaffected. CRR2 encodes a member of the plant combinatorial and modular protein (PCMP) family consisting of more than 200 genes in Arabidopsis. CRR2 functions in the intergenic processing of chloroplast RNA between rps7 and ndhB, which is possibly essential for ndhB translation. We have determined the function of a PCMP family member, indicating that the family is closely related to pentatrico-peptide PPR proteins involved in the maturation steps of organellar RNA.
DOI:10.1105/tpc.108.064667URL [本文引用: 1]
DOI:10.4161/rna.26172URL [本文引用: 1]
DOI:10.1073/pnas.1612460114URL [本文引用: 1]
DOI:10.1111/tpj.2016.85.issue-5URL [本文引用: 1]
DOI:10.1093/molbev/msm030URL [本文引用: 1]
DOI:10.1105/tpc.112.099051URL [本文引用: 1]
DOI:10.1007/s11427-017-9170-3URL [本文引用: 2]
PMID:1709762 [本文引用: 1]
DOI:10.1111/tpj.14578 [本文引用: 4]

The central dogma in biology defines the flow of genetic information from DNA to RNA to protein. Accordingly, RNA molecules generally accurately follow the sequences of the genes from which they are transcribed. This rule is transgressed by RNA editing, which creates RNA products that differ from their DNA templates. Analyses of the RNA landscapes of terrestrial plants have indicated that RNA editing (in the form of C-U base transitions) is highly prevalent within organelles (that is, mitochondria and chloroplasts). Numerous C -> U conversions (and in some plants also U -> C) alter the coding sequences of many of the organellar transcripts and can also produce translatable mRNAs by creating AUG start sites or eliminating premature stop codons, or affect the RNA structure, influence splicing and alter the stability of RNAs. RNA-binding proteins are at the heart of post-transcriptional RNA expression. The C-to-U RNA editing process in plant mitochondria involves numerous nuclear-encoded factors, many of which have been identified as pentatricopeptide repeat (PPR) proteins that target editing sites in a sequence-specific manner. In this review we report on major discoveries on RNA editing in plant organelles, since it was first documented 30 years ago.
DOI:10.1007/s004380000295URL [本文引用: 1]
DOI:10.1111/tpj.2014.79.issue-5URL [本文引用: 2]
DOI:10.1111/jipb.v62.6URL [本文引用: 2]
DOI:10.1105/tpc.112.106781URL [本文引用: 2]
DOI:10.1111/tpj.12993URL [本文引用: 2]
DOI:10.1111/nph.14424PMID:28121385 [本文引用: 2]

Pentatricopeptide repeat (PPR) proteins comprise a large family of sequence-specific RNA binding proteins in land plants. Because of its large family size and frequent embryo lethality in the mutants, molecular functions and physiological roles of many PPR proteins are unknown. Through characterization of an empty pericarp9 (emp9) mutant in maize (Zea mays), we defined the functions of EMP9 in mitochondrial RNA editing, respiratory complex formation and seed development. Mu insertions in different regions of Emp9 facilitated dissection of the domain functions of the EMP9. Through genetic and functional analyses of multiple alleles, we showed that deletions of two N-terminal PPR motifs and partial E+ domain do not eliminate the editing function of EMP9. Emp9 encodes an E+ subclass PPR protein that is localized in mitochondria. Loss of EMP9 function abolishes the C-to-U editing of ccmB-43 and rps4-335 sites in mitochondria. The loss of editing at ccmB-43 and rps4-335 affects the maturation of cytochrome c and impairs the biogenesis of mitochondrial respiratory complexes, particularly complex III. This work extends our understanding of PPR-E+ protein in editing function and seed development, and provides insights into the molecular function of mitochondrial CcmB protein in higher plants.© 2017 The Authors. New Phytologist © 2017 New Phytologist Trust.
DOI:10.1111/nph.2019.221.issue-2URL [本文引用: 2]
DOI:10.1534/genetics.116.199331URL [本文引用: 2]
DOI:10.1111/jipb.v60.1URL [本文引用: 2]
DOI:10.1016/j.jplph.2019.152992URL [本文引用: 2]
DOI:10.1371/journal.pgen.1008305URL [本文引用: 2]
DOI:10.1016/j.gene.2010.01.008PMID:20117193 [本文引用: 2]

C-to-U RNA editing (i.e., alteration of a C in the genomic sequence to U in the transcript) has been confirmed widely in angiosperm organellar genomes. During the C-to-U RNA editing event, incomplete edited transcripts have been observed at many sites in the steady-state mRNA population (partial editing). Here, by using coexpression analysis and the surveillance of whole editing status on the mitochondrial genome, we have revealed that a pentatricopeptide repeat (PPR) protein classified into the P subfamily (PPR596) has site-specific influence on the efficiency of C-to-U RNA editing events at partial editing sites on the Arabidopsis thaliana mitochondrial genome. Previous works have revealed that PPR proteins classified into the PLS subfamily containing the E or E and DYW motif are involved in RNA editing as trans-factors; they are believed to recruit deaminase at editing sites. In contrast with the mutant analyses of PLS-subfamily PPR proteins, the editing efficiency at rps3eU1344SS was revealed to be significantly increased in ppr596 mutants. Our study implies P-subfamily PPR protein is involved in the control of the degree of partial editing.
DOI:10.1105/tpc.112.099507URL [本文引用: 2]
DOI:10.1073/pnas.0700865104URL [本文引用: 3]
DOI:10.1111/tpj.2010.63.issue-3URL [本文引用: 2]
DOI:10.1093/pcp/pcr182PMID:22186180 [本文引用: 2]

In plants, RNA editing is observed in mitochondria and plastids, changing selected C nucleotides into Us in both organelles. We here identify the PPR (pentatricopeptide repeat) protein MEF3 (mitochondrial editing factor 3) of the E domain PPR subclass by genetic mapping of a variation between ecotypes Columbia (Col) and Landsberg erecta (Ler) in Arabidopsis thaliana to be required for a specific RNA editing event in mitochondria. The Ler variant of MEF3 differs from Col in two amino acids in repeats 9 and 10, which reduce RNA editing levels at site atp4-89 to about 50% in Ler. In a T-DNA insertion line, editing at this site is completely lost. In Vitis vinifera the gene most similar to MEF3 continues into a DYW extension beyond the common E domain. Complementation assays with various combinations of PPR and E domains from the vine and A. thaliana proteins show that the vine E region can substitute for the A. thaliana E region with or without the DYW domain. These findings suggest that the additional DYW domain does not disturb the MEF3 protein function in mitochondrial RNA editing in A. thaliana.
DOI:10.1105/tpc.108.064667URL [本文引用: 3]
DOI:10.1111/j.1365-313X.2009.04059.xURL [本文引用: 2]
DOI:10.1105/tpc.109.071472URL [本文引用: 3]
DOI:10.4161/rna.18644PMID:22258224 [本文引用: 2]

In plant mitochondria and plastids, RNA editing alters about 400 and about 35 C nucleotides into Us, respectively. Four of these RNA editing events in plant mitochondria specifically require the PPR protein MEF7, characterized by E and DYW extension domains. The gene for MEF7 was identified by genomic mapping of the locus mutated in plants from EMS treated seeds. The SNaPshot screen of the mutant plant population identified two independent EMS mutants with the same editing defects as a corresponding T-DNA insertion line of the MEF7 gene. Although the amino acid codons introduced by the editing events are conserved throughout flowering plants, even the combined failure of four editing events does not impair the growth efficiency of the mutant plants. Five nucleotides are conserved between the four affected editing sites, but are not sufficient for specific recognition by MEF7 since they are also present at three other sites which are unaffected in the mutants.
DOI:10.1104/pp.109.151175PMID:20018598 [本文引用: 2]

RNA editing in plants alters specific nucleotides from C to U in mRNAs in plastids and in mitochondria. I here characterize the nuclear gene MITOCHONDRIAL EDITING FACTOR9 (MEF9) that is required for RNA editing of the site nad7-200 in the nad7 mitochondrial mRNA in Arabidopsis (Arabidopsis thaliana). The MEF9 protein belongs to the E subfamily of pentatricopeptide repeat proteins and unlike the three previously identified mitochondrial editing factors MEF1 and MEF11 in Arabidopsis and OGR1 in rice (Oryza sativa) does not contain a DYW C-terminal domain. In addition, the E domain is incomplete, but seems to be functionally required, since one of the two independent EMS mutants encodes a MEF9 protein truncated by a stop codon at the beginning of the E domain. In both mutant plants premature stop codons in MEF9 inactivate RNA editing at site nad7-200. The homozygous mutant plants are viable and develop rather normally. The lack of RNA editing at site nad7-200 thus seems to be tolerated although this editing event is conserved in most plant species or the genomic sequence already codes for a T at this position, resulting in a generally conserved amino acid codon.
DOI:10.1016/j.febslet.2011.01.037PMID:21281638 [本文引用: 3]

We here identify the PPR protein MEF14 of the DYW subclass as a specific trans-factor required for C to U editing of site matR-1895 by genetic mapping of an EMS induced editing mutant in Arabidopsis thaliana. The wild type Col MEF14 gene complements mutant protoplasts. A T-DNA insertion in the MEF14 gene abolishes detectable editing at the matR-1895 site. Lack of RNA editing at the matR-1895 site does not alter the level of mature and precursor nad1 mRNA molecules. Such RNA editing mutants can be used to analyse the function of genes like this maturase related reading frame in plant mitochondria.Copyright © 2011 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
DOI:10.1111/tpj.2009.59.issue-5URL [本文引用: 1]
a.
DOI:10.1111/tpj.2012.72.issue-3URL [本文引用: 1]
DOI:10.1016/j.molp.2016.08.009URL [本文引用: 1]
DOI:10.1074/mcp.RA119.001396PMID:31023727 [本文引用: 1]

Mitochondrial transcripts are subject to a wealth of processing mechanisms including cis- and trans-splicing events, as well as base modifications (RNA editing). Hundreds of proteins are required for these processes in plant mitochondria, many of which belong to the pentatricopeptide repeat (PPR) protein superfamily. The structure, localization, and function of these proteins is only poorly understood. Here we present evidence that several PPR proteins are bound to mitoribosomes in plants. A novel complexome profiling strategy in combination with chemical crosslinking has been employed to systematically define the protein constituents of the large and the small ribosomal subunits in the mitochondria of plants. We identified more than 80 ribosomal proteins, which include several PPR proteins and other non-conventional ribosomal proteins. These findings reveal a potential coupling of transcriptional and translational events in the mitochondria of plants. Furthermore, the data indicate an extremely high molecular mass of the "small" subunit, even exceeding that of the "large" subunit.© 2019 Rugen et al.
PMID:12736308 [本文引用: 1]

In tobacco, 30 of 34 sites in chloroplast transcripts that undergo C-to-U RNA editing can be grouped into clusters of 2-5 sites based on sequence similarities immediately 5' to the edited C. According to a previous transgenic analysis, overexpression of transcripts representing one cluster member results in reduction in editing of all cluster members, suggesting that members of an individual cluster share a trans-factor that is present in limiting amounts. To compare leaves and roots, we quantified the editing extent at 34 sites in wild-type tobacco and at three sites in spinach and Arabidopsis. We observed that transcripts of most NADH dehydrogenase subunits are edited inefficiently in roots. With few exceptions, members of the same editing site cluster co-varied in editing extent in chloroplasts versus non-green root plastids, with members of most clusters uniformly exhibiting either a high or low editing extent in roots. The start codon of the ndhD transcript must be created by editing, but the C target is edited inefficiently in roots, and no NDH-D protein could be detected upon immunoblotting. Our data are consistent with the hypothesis that cluster-specific trans-factors exist and that some are less abundant in roots, limiting the editing extent of certain sites in root plastids.
DOI:10.1038/nplants.2017.37URL [本文引用: 1]
DOI:10.1111/tpj.2016.85.issue-4URL [本文引用: 1]
DOI:10.1371/journal.pgen.1002910URL [本文引用: 1]
DOI:10.1111/j.1365-313X.2008.03595.xURL [本文引用: 1]
DOI:10.1111/tpj.2008.56.issue-4URL [本文引用: 2]
DOI:10.1111/j.1365-313X.2008.03766.xURL [本文引用: 1]
DOI:10.1093/jxb/eru293URL [本文引用: 1]
DOI:10.1146/arplant.2011.62.issue-1URL [本文引用: 1]
[本文引用: 1]
DOI:10.1093/jxb/erz193URL [本文引用: 1]
DOI:10.1093/jxb/erz391URL [本文引用: 1]
DOI:10.1146/annurev-biochem-070511-103700PMID:23527692 [本文引用: 1]

Complex I (NADH:ubiquinone oxidoreductase) is crucial for respiration in many aerobic organisms. In mitochondria, it oxidizes NADH from the tricarboxylic acid cycle and β-oxidation, reduces ubiquinone, and transports protons across the inner membrane, contributing to the proton-motive force. It is also a major contributor to cellular production of reactive oxygen species. The redox reaction of complex I is catalyzed in the hydrophilic domain; it comprises NADH oxidation by a flavin mononucleotide, intramolecular electron transfer along a chain of iron-sulfur clusters, and ubiquinone reduction. Redox-coupled proton translocation in the membrane domain requires long-range energy transfer through the protein complex, and the molecular mechanisms that couple the redox and proton-transfer half-reactions are currently unknown. This review evaluates extant data on the mechanisms of energy transduction and superoxide production by complex I, discusses contemporary mechanistic models, and explores how mechanistic studies may contribute to understanding the roles of complex I dysfunctions in human diseases.
DOI:10.1016/j.tplants.2007.01.005URL [本文引用: 2]
DOI:10.1093/jxb/erz226URL [本文引用: 2]
DOI:10.1038/cr.2015.46URL [本文引用: 1]
PMID:10794539 [本文引用: 1]

Cereal endosperm undergoes programmed cell death (PCD) during its development, a process that is controlled, in part, by ethylene. Whether other hormones influence endosperm PCD has not been investigated. Abscisic acid (ABA) plays an essential role during late seed development that enables an embryo to survive desiccation. To examine whether ABA is also involved in regulating the onset of PCD during endosperm development, we have used genetic and biochemical means to disrupt ABA biosynthesis or perception during maize kernel development. The onset and progression of cell death, as determined by viability staining and the appearance of internucleosomal DNA fragmentation, was accelerated in developing endosperm of ABA-insensitive vp1 and ABA-deficient vp9 mutants. Ethylene was synthesized in vp1 and vp9 mutant kernels at levels that were 2-4-fold higher than in wild-type kernels. Moreover, the increase and timing of ethylene production correlated with the premature onset and accelerated progression of internucleosomal fragmentation in these mutants. Treatment of developing wild-type endosperm with fluridone, an inhibitor of ABA biosynthesis, recapitulated the increase in ethylene production and accelerated execution of the PCD program that was observed in the ABA mutant kernels. These data suggest that a balance between ABA and ethylene establishes the appropriate onset and progression of programmed cell death during maize endosperm development.
PMID:12223841 [本文引用: 1]

We characterized the progression of programmed cell death during maize (Zea mays L.) endosperm development of starchy (Su; wild-type) and shrunken2 (sh2) genotypes and tested the involve ment of ethylene in mediating this process. Histological and viability staining demonstrated that endosperm cell death was initiated earlier and progressed more rapidly in sh2 endosperm compared with Su endosperm. Internucleosomal DNA fragmentation accompanied endosperm cell death and occurred more extensively in sh2 endosperm. 1-Aminocyclopropane-1-carboxylic acid levels peaked approximately 16 d after pollination (dap) in Su endosperm and gradually decreased during subsequent development, whereas two large 1-aminocyclopropane-1-carboxylic acid peaks were observed in sh2 endosperm, the first between 16 and 20 dap and the second at 36 dap. Ethylene levels were elevated in sh2 kernels compared with Su kernels, with an initial peak 20 dap approximately 3-fold higher than in Su kernels and a second peak 36 dap approximately 5-fold higher than that in Su kernels. Ethylene treatment of Su kernels resulted in earlier and more extensive endosperm cell death and DNA fragmentation. Aminoethoxyvinylglycine treatment of sh2 kernels reduced the extent of DNA fragmentation. We conclude that ethylene is involved in triggering programmed cell death in developing maize endosperm and is responsible for the aberrant phenotype of sh2 kernels.
DOI:10.1126/science.aat4318URL [本文引用: 1]
DOI:10.1105/tpc.105.039495URL [本文引用: 1]
DOI:10.1105/tpc.110.081638URL [本文引用: 1]
DOI:10.1016/j.tplants.2009.11.003PMID:19963426 [本文引用: 1]

TCP genes encode plant-specific transcription factors with a bHLH motif that allows DNA binding and protein-protein interactions. The TCP gene family has five members in the lycophytes and >20 members in the eudicots. Gene duplication and diversification has generated two clades (class I and II) with slightly different TCP domains. Here, we summarize our current knowledge of the evolution of this family, their regulation, the biochemical activity of their proteins and the biological function of some members, in particular, in the control of cell proliferation in developing tissues. Increasing knowledge of the functions of TCP genes should enable their use as tools to modulate plant growth patterns and to generate novel morphologies in species of agronomical interest.
DOI:10.1093/jxb/eru293URL [本文引用: 1]
