 ,, 周强伟, 成盛, 李国亮
,, 周强伟, 成盛, 李国亮 ,华中农业大学信息学院,农业生物信息学湖北省重点实验室,农业大数据湖北省工程技术研究中心,三维基因组学研究中心,武汉 430070
,华中农业大学信息学院,农业生物信息学湖北省重点实验室,农业大数据湖北省工程技术研究中心,三维基因组学研究中心,武汉 430070Research progress of CTCF in mediating 3D genome formation and regulating gene expression
Cong Zhou ,, Qiangwei Zhou, Sheng Cheng, Guoliang Li
,, Qiangwei Zhou, Sheng Cheng, Guoliang Li ,Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China
,Agricultural Bioinformatics Key Laboratory of Hubei Province, Hubei Engineering Technology Research Center of Agricultural Big Data, 3D Genomics Research Center, College of Informatics, Huazhong Agricultural University, Wuhan 430070, China通讯作者: 李国亮,博士,教授,研究方向:生物信息学,三维基因组学。E-mail:guoliang.li@mail.hzau.edu.cn
编委: 吴强
收稿日期:2021-09-8修回日期:2021-09-13
| 基金资助: |
Received:2021-09-8Revised:2021-09-13
| Fund supported: |
作者简介 About authors
周聪,博士,在站博士后,研究方向:三维基因组学。E-mail:

摘要
真核细胞间期的染色质在细胞核中经过复杂的盘曲折叠,形成高级拓扑结构,这样的染色质结构空间组织对基因表达有重要影响。CTCF (CCCTC-binding factor)作为关键的染色质高级结构架构蛋白,对三维基因组结构的形成起到了重要作用。CTCF还可以与基因组内大量的绝缘子结合,影响染色质远程交互,实现对增强子和基因转录调控的绝缘效应。本文主要对近期美国圣裘德儿童研究医院Chunliang Li团队对于CTCF完全降解后发现染色质可及性发生变化的研究结果,上海交通大学系统生物医学研究院吴强团队、美国加州路德维希癌症研究所任兵团队对于CTCF结合位点充当绝缘子作用机制的最新结果进行部分点评及讨论。
关键词:
Abstract
In interphase eukaryotic nuclei, chromatin is folded to form a higher-order topological structure. The spatial organization of such chromatin domain has an important impact on the regulation of gene expression. As a key architectural structural protein, CTCF (CCCTC-binding factor) plays an important role in the formation of chromatin three-dimensional chromatin structure. CTCF can also bind to many insulator elements in the genome and insulate enhancers from activating target genes via modulating remote chromatin interactions. A recent study by Dr. Chunliang Li and his team at St. Jude Children’s Research Hospital in the United States showed that when CTCF was acutely degraded, significant changes were found in the three-dimensional structure of chromatin. The mechanism by which CTCF binding sites function as insulator elements was investigated by Prof. Qiang Wu’s team at Institute of Systems Biomedicine and Shanghai Jiao Tong University in China and Prof. Bing Ren’s team at Ludwig Institute for Cancer Research in the United States. Here we mainly review and discuss some of these latest progresses.
Keywords:
PDF (521KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
周聪, 周强伟, 成盛, 李国亮. CTCF在介导三维基因组形成及调控基因表达中的研究进展. 遗传[J], 2021, 43(9): 816-821 doi:10.16288/j.yczz.21-326
Cong Zhou.
真核细胞的基因组以线性DNA序列来存储遗传信息。然而,基因的精准表达,需要染色质折叠成复杂的基因组三维结构,进而形成DNA调控元件与基因之间的相互作用,达成精准的基因表达调控。在每条染色质内部,DNA包裹在组蛋白形成的八聚体周围,形成了10 nm的核小体,随后被高效、有序地包装成复杂的多层次结构。除了染色质的多层次结构外,基因组片段在三维细胞核空间中的空间定位也对基因组的功能产生了重要影响。这种由染色质折叠而形成的三维基因组结构,对于基因转录、DNA复制及修复、细胞分裂及分化等重要的生物学过程起到关键的调控作用[1,2,3]。特定远程交互的改变会导致发育和肿瘤基因表达的改变进而引起发育障碍及癌症的发生[4]。
CTCF (CCCTC-binding factor)作为一种关键的染色质架构蛋白,在染色质三维结构的形成中发挥着重要作用。CTCF可以在染色质黏连蛋白(cohesin)的协助下形成染色质环,cohesin顺着环状结构两端向外延伸移动挤压导致染色质环增大,直至遇到两个方向朝环内的CTCF结合位点 (CTCF binding site, CBS) 并和CTCF共同稳定该环状结构,进而介导基因组范围内的远距离染色质相互作用。这些相互作用会优先在正向-反向的CBS对之间形成,这就是环挤出模型[5,6,7,8]。此外,CTCF在脊椎动物中高度保守,在小鼠(Mus musculus)、鸡(Gallus gallus)和人(Homo sapiens)之间接近100%的同源性,具有11个与DNA结合的锌指结构,通过对这些锌指结构的组合使用,可以定向、动态地结合到基因组中的数万个CBS[9]。除了作为染色质架构蛋白,CTCF还能够与绝缘子结合调控增强子与启动子的交互从而进行转录绝缘,以及作为转录抑制因子调控基因表达,甚至根据基因组中序列环境,CTCF在基因激活、可变剪接和类别转换重组 (class switch recombination, CSR) 中发挥重要作用[10,11,12,13,14,15]。
虽然对CTCF已经有了大量的研究,关于CTCF如何以及在何处调节绝缘子,进而对染色质高级结构进行调节的具体机制及功能,仍然有很多不清楚的地方。近期有多篇关于CTCF功能和三维基因组学的科研论文发表,为更深入地理解CTCF介导的三维基因组机制和功能提供了新的视角。本文将进行部分点评和讨论,为大家提供参考。
1 CTCF作为架构蛋白在三维染色质折叠中行使关键作用
CTCF是脊椎动物中已知的最重要也是研究最多的绝缘体结合蛋白之一。CTCF最初是在鸡中被发现对c-myc基因具有抑制作用,后又被发现可以阻断增强子功能[11,16,17]。之后的研究表明,CTCF能够扮演多重角色,包括抑制子、增强子和绝缘子等[10]。近十几年的研究发现,CTCF可能主要参与形成三维基因组结构。在染色质三维结构中,CTCF能够通过阻碍cohesin蛋白在染色质上的滑动而形成染色质环结构,以此将线性距离较远的启动子与增强子拉到空间距离较近的交互区域内,对于基因组的三维架构以及关键基因的转录调控具有重要作用[8,18]。此外,CTCF可以介导一些转录相关因子(如RNAPII)调控的三维结构,进而影响基因的转录调控。在单倍型水平上研究发现,单个SNP的改变也能影响CTCF的结合进而导致CTCF维持的三维结构发生改变[19]。此前通过ChIA-PET、Hi-C等实验技术较多研究了CTCF在染色体三维结构中的重要作用,而有关研究表明CTCF的完全降解对全基因组转录的影响并没有那么显著[20,21]。2021年美国圣裘德儿童研究医院Chunliang Li博士团队进一步分析了在CTCF蛋白完全降解后染色质可及性的变化以及CTCF调节转录的共调节因子[22]。该研究发现CTCF蛋白降解后染色质结构发生了明显的重排,8876个区域的可及性显著降低,8042个区域的可及性显著增强。分析结果表明,可及性显著降低的区域中富集最高的特征序列 (motif) 就是CBS motif,并且在可及性降低区域中CBS是以双峰形式分布的,而在新产生的以及未发生变化的可及性区域中CBS并无双峰分布。此外,通过计算差异可及性区域(differential accessibility regions, DARs) 与附近CBS距离可知,可及性降低区域距离CBS最近 (0 bp),新生的可及性区域次之(间隔大约100 bp),而未发生变化的可及性区域附近未发现CTCF富集。该研究表明,这些新生的可及性区域在野生型细胞中可能起到绝缘附近其它转录因子的作用,而CTCF的去结合使得附近转录因子之间的交互成为可能,这与新生的可及性区域更多地覆盖在基因启动子附近相符合。
较多研究表明CTCF蛋白的结合受CBS motif区域DNA甲基化水平的影响,并且有研究报道CTCF单倍型剂量表达能够影响DNA甲基化的稳定性[23]。然而Chunliang Li团队通过全基因组DNA甲基化差异分析,发现CTCF在完全降解的情况下DNA甲基化并未发生明显变化。此外,与之前的报道相似[20,21],Chunliang Li团队发现在CTCF完全降解后,全基因组范围内基因的转录水平早期并未发生显著改变(仅有488个基因表达显著变化)。但是令人惊奇的是,在下游磷酸化以及蛋白水平上检测到了显著的差异。这些结果都表明CTCF对于DNA甲基化的维持和稳定,以及对于基因的转录和翻译的影响机制非常复杂,还需要后续更多针对性的分析和研究。
2 CTCF与绝缘子结合的功能及作用机制
绝缘子是一类能够保护基因不受周围染色质环境发出不当调控信号影响的DNA序列元件,对细胞类型特异性基因表达至关重要[24]。CTCF是目前研究最广泛的绝缘子结合蛋白之一,能够以其串联的锌指结构方向性地结合到CBS上[9]。上海交通大学系统生物医学研究院吴强团队近期利用CRISPR大片段编辑,结合染色质构象捕获技术对原钙粘蛋白 (protocadherin, Pcdh) 基因簇、HOXD基因簇等研究发现单个CTCF绝缘子能够确保准确的增强子绝缘和启动子激活,串联排列的CTCF拓扑绝缘体能够平衡染色质的空间接触和增强子与启动子的拓扑性选择[18,25,26]。位于增强子和启动子之间的绝缘子,可以通过与远端的CBS形成染色质环,抑制启动子与增强子的远程互作,进而抑制目标基因的表达。该研究还突破性地发现串联CTCF绝缘子具有拓扑性,能够促进远端基因与增强子的空间远程互作,进而可以平衡染色质环的形成和增强子-启动子的拓扑性选择(模式图如图1A所示)。在对Pcdh基因簇的研究中还发现,CBS的绝缘性与其方向无关,且与果蝇(Drosophila)中成对绝缘体会损坏彼此的绝缘活性所不同,哺乳动物中正向反向串联的CBS并不会损坏其绝缘活性。此外,串联的CBS具有叠加效应,即位点数目越多,调控范围越广,这与近期美国加州路德维希癌症研究所任兵团队通过利用小鼠胚胎干细胞开发的一种绝缘子报告系统所检测到的结果一致,多个串联的CBS具有更好的绝缘效应[10]。任兵团队对于单个CBS检测的结果显示,单个CBS具有较弱或者无绝缘活性。此外,吴强团队对于人类全基因组CBS进行计算模拟,推测人类全基因组范围内CTCF结合的CBS元件能够通过形成定向染色质环发挥绝缘子作用。任兵团队进一步研究了CTCF转录绝缘的序列环境依赖性,发现CBS核心序列上游10~20 bp的序列决定了绝缘子效应,该序列可能通过与CTCF第9~11个锌指结构结合从而行使绝缘子功能。针对CTCF在基因组内何处会行使绝缘功能,任兵团队还发现位于拓扑相关结构域(topological-associated domain, TAD)边界的CBS比非边界的CBS更有可能充当绝缘子且与结合强弱无关。这些结果推动了CTCF绝缘效应及作用机制方面的研究,也需要科研人员进一步深入探究。
吴强团队对HOXD基因簇调控区域内一系列串联反向排列的CTCF位点进行删除及反转的结果也再次证明,串联反向的 CTCF位点通过其绝缘子功能维持上下游增强子簇对HOXD基因簇表达调控的平衡。单个或者多个TAD边界的CBS删除,会改变HOXD基因簇启动子与增强子的远程交互,进而下调HOXD基因的表达。TAD边界处CBS的全部删除,会改变HOXD基因的转录调控模式,原本受到的下游增强子调控减弱,而上游增强子调控增强,调控平衡的破坏导致HOXD基因表达急剧下降。此外,TAD边界区域CBS反转,会改变原有的染色质环方向,导致TAD边界漂移至新形成的一对反向-正向CTCF位点处。为解释串联CTCF位点作为拓扑绝缘子的工作机制,吴强团队提出了葫芦模型,即由于CTCF与DNA结合的动态性,导致收敛方向的CTCF位点对于cohesin的阻碍并不完全,串联的CTCF位点就可能通过环挤出模型呈现出类似于“葫芦”结构(模式图如图1B所示)。
图1
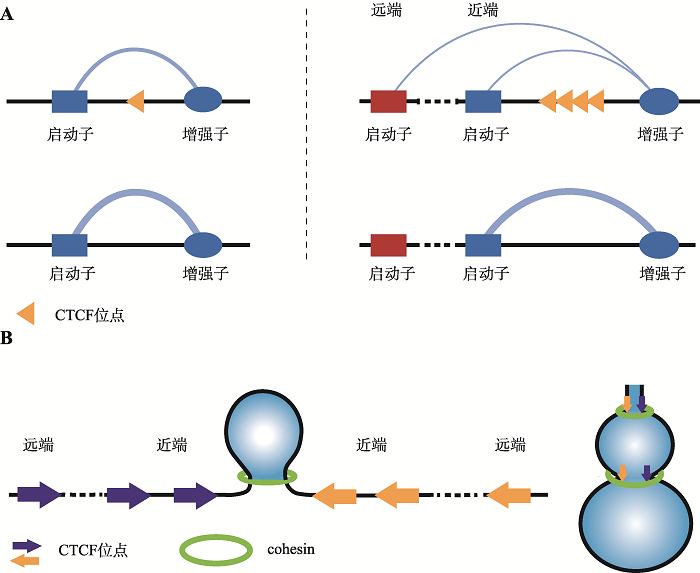 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1CTCF位点的拓扑绝缘性模式图
A:当CTCF位点位于启动子和增强子中间时能够抑制启动子和增强子的远程交互,单个CTCF位点主要行使该项功能(左图)。串联排列的CTCF位点不仅能够抑制两侧的启动子和增强子之间的染色质远程交互,还能够促进增强子与远端启动子的交互(右图),进而可以进行染色质空间接触的平衡和增强子-启动子的拓扑性选择,这就是该研究中的绝缘子拓扑性。B:串联CTCF位点通过环挤出模型呈现出的“葫芦”模型示意图。
Fig. 1The diagram of topological insulation at CTCF sites
3 结语与展望
早期研究发现,CTCF具有绝缘效应,随着三维基因组学的发展,证实CTCF作为染色质架构蛋白,在染色质三维结构折叠中发挥了关键作用。近几年很多研究报道了关于CTCF与绝缘子结合对染色质高级拓扑结构和基因表达的影响,这进一步推进了三维基因组折叠机制的研究进展。但是全基因组范围内CTCF对基因转录、翻译以及作为绝缘子、增强子的分布及作用机制仍有很多不清楚之处。例如,CTCF蛋白降解后对于基因的早期转录水平影响并不太显著,但是对于下游蛋白表达水平和磷酸化修饰具有较大影响,针对这些问题未来还需要更加深入的探究。(责任编委: 吴强)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1038/nrm.2016.104URL [本文引用: 1]
DOI:10.1038/s41576-019-0128-0PMID:31086298 [本文引用: 1]

Spatiotemporal gene expression programmes are orchestrated by transcriptional enhancers, which are key regulatory DNA elements that engage in physical contacts with their target-gene promoters, often bridging considerable genomic distances. Recent progress in genomics, genome editing and microscopy methodologies have enabled the genome-wide mapping of enhancer-promoter contacts and their functional dissection. In this Review, we discuss novel concepts on how enhancer-promoter interactions are established and maintained, how the 3D architecture of mammalian genomes both facilitates and constrains enhancer-promoter contacts, and the role they play in gene expression control during normal development and disease.
DOI:10.1038/s41580-019-0132-4URL [本文引用: 1]
DOI:10.1038/s41588-021-00878-zURL [本文引用: 1]
DOI:10.1073/pnas.1518552112URL [本文引用: 1]
DOI:10.1016/j.cell.2015.07.038PMID:26276636 [本文引用: 1]

CTCF and the associated cohesin complex play a central role in insulator function and higher-order chromatin organization of mammalian genomes. Recent studies identified a correlation between the orientation of CTCF-binding sites (CBSs) and chromatin loops. To test the functional significance of this observation, we combined CRISPR/Cas9-based genomic-DNA-fragment editing with chromosome-conformation-capture experiments to show that the location and relative orientations of CBSs determine the specificity of long-range chromatin looping in mammalian genomes, using protocadherin (Pcdh) and β-globin as model genes. Inversion of CBS elements within the Pcdh enhancer reconfigures the topology of chromatin loops between the distal enhancer and target promoters and alters gene-expression patterns. Thus, although enhancers can function in an orientation-independent manner in reporter assays, in the native chromosome context, the orientation of at least some enhancers carrying CBSs can determine both the architecture of topological chromatin domains and enhancer/promoter specificity. These findings reveal how 3D chromosome architecture can be encoded by linear genome sequences. Copyright © 2015 Elsevier Inc. All rights reserved.
DOI:10.1126/science.aaz4475URL [本文引用: 1]
DOI:10.1038/ng.857PMID:21685913 [本文引用: 2]

Mammalian genomes are viewed as functional organizations that orchestrate spatial and temporal gene regulation. CTCF, the most characterized insulator-binding protein, has been implicated as a key genome organizer. However, little is known about CTCF-associated higher-order chromatin structures at a global scale. Here we applied chromatin interaction analysis by paired-end tag (ChIA-PET) sequencing to elucidate the CTCF-chromatin interactome in pluripotent cells. From this analysis, we identified 1,480 cis- and 336 trans-interacting loci with high reproducibility and precision. Associating these chromatin interaction loci with their underlying epigenetic states, promoter activities, enhancer binding and nuclear lamina occupancy, we uncovered five distinct chromatin domains that suggest potential new models of CTCF function in chromatin organization and transcriptional control. Specifically, CTCF interactions demarcate chromatin-nuclear membrane attachments and influence proper gene expression through extensive cross-talk between promoters and regulatory elements. This highly complex nuclear organization offers insights toward the unifying principles that govern genome plasticity and function.
DOI:10.1016/j.cell.2009.06.001URL [本文引用: 2]
DOI:10.1038/s41588-021-00863-6URL [本文引用: 3]
DOI:10.1128/MCB.16.6.2802PMID:8649389 [本文引用: 2]

We have isolated and analyzed human CTCF cDNA clones and show here that the ubiquitously expressed 11-zinc-finger factor CTCF is an exceptionally highly conserved protein displaying 93% identity between avian and human amino acid sequences. It binds specifically to regulatory sequences in the promoter-proximal regions of chicken, mouse, and human c-myc oncogenes. CTCF contains two transcription repressor domains transferable to a heterologous DNA binding domain. One CTCF binding site, conserved in mouse and human c-myc genes, is found immediately downstream of the major P2 promoter at a sequence which maps precisely within the region of RNA polymerase II pausing and release. Gel shift assays of nuclear extracts from mouse and human cells show that CTCF is the predominant factor binding to this sequence. Mutational analysis of the P2-proximal CTCF binding site and transient-cotransfection experiments demonstrate that CTCF is a transcriptional repressor of the human c-myc gene. Although there is 100% sequence identity in the DNA binding domains of the avian and human CTCF proteins, the regulatory sequences recognized by CTCF in chicken and human c-myc promoters are clearly diverged. Mutating the contact nucleotides confirms that CTCF binding to the human c-myc P2 promoter requires a number of unique contact DNA bases that are absent in the chicken c-myc CTCF binding site. Moreover, proteolytic-protection assays indicate that several more CTCF Zn fingers are involved in contacting the human CTCF binding site than the chicken site. Gel shift assays utilizing successively deleted Zn finger domains indicate that CTCF Zn fingers 2 to 7 are involved in binding to the chicken c-myc promoter, while fingers 3 to 11 mediate CTCF binding to the human promoter. This flexibility in Zn finger usage reveals CTCF to be a unique "multivalent" transcriptional factor and provides the first feasible explanation of how certain homologous genes (i.e., c-myc) of different vertebrate species are regulated by the same factor and maintain similar expression patterns despite significant promoter sequence divergence.
DOI:S0092-8674(15)00377-3PMID:25959774 [本文引用: 1]

Mammalian genomes are organized into megabase-scale topologically associated domains (TADs). We demonstrate that disruption of TADs can rewire long-range regulatory architecture and result in pathogenic phenotypes. We show that distinct human limb malformations are caused by deletions, inversions, or duplications altering the structure of the TAD-spanning WNT6/IHH/EPHA4/PAX3 locus. Using CRISPR/Cas genome editing, we generated mice with corresponding rearrangements. Both in mouse limb tissue and patient-derived fibroblasts, disease-relevant structural changes cause ectopic interactions between promoters and non-coding DNA, and a cluster of limb enhancers normally associated with Epha4 is misplaced relative to TAD boundaries and drives ectopic limb expression of another gene in the locus. This rewiring occurred only if the variant disrupted a CTCF-associated boundary domain. Our results demonstrate the functional importance of TADs for orchestrating gene expression via genome architecture and indicate criteria for predicting the pathogenicity of human structural variants, particularly in non-coding regions of the human genome. Copyright © 2015 Elsevier Inc. All rights reserved.
DOI:10.1038/nature10442URL [本文引用: 1]
PMID:9407128 [本文引用: 1]

The promoter of the amyloid beta-protein precursor (APP) gene directs high levels of cell type-specific transcription with 94 base pairs 5' to the main transcriptional start site. An essential activator domain in this proximal APP promoter is a nuclear factor binding site designated as APBbeta. The recognition domain for the APBbeta binding factor is located between position -93 and -82 relative to the main transcriptional start site. The nuclear factor that binds to the APBbeta site was partially purified by multiple steps of ion exchange and hydroxyapatite chromatography. Based on UV cross-linking results, a protein with an apparent molecular mass of 140 kDa was selected as the putative APBbeta binding protein. After the final purification step consisting of preparative SDS-polyacrylamide gel electrophoresis, partial peptide sequences were obtained that completely matched the transcriptional factor CTCF. This protein is a known regulator of c-myc and lysozyme gene expression, and it binds to a variety of diverse DNA sequences. The binding of CTCF to the APBbeta domain was further established by competition with CTCF binding oligonucleotides in mobility shift electrophoresis. The identity was also confirmed by the observation that the APBbeta binding factor is recognized by antibodies against C- and N-terminal sequences of CTCF. In addition, oligonucleotide competition during in vitro transcription affirmed that CTCF acts as a transcriptional activator in the APP gene promoter.
DOI:10.1038/s41586-019-1723-0URL [本文引用: 1]
DOI:10.1038/35013100URL [本文引用: 1]
PMID:2284094 [本文引用: 1]

The chicken c-myc 5'-flanking sequence has previously been shown to bind multiple proteins present in undifferentiated and differentiated red blood cells. In this report the protein binding to one specific region within a hypersensitive site approximately 200 base pairs upstream of the start of transcription has been analysed in detail. Using a combination of a modified agarose gel retardation assay with O-phenanthroline-copper footprinting in situ, missing contact point and methylation interference techniques, two proteins were found to bind to overlapping sequences within 180-230 bp upstream of the start of transcription. One protein resembles the transcription factor Sp1, the other is a protein which binds to three regularly spaced repeats of the core sequence CCCTC. This CCCTC-binding factor was termed CTCF. It requires additional sequences outside the three recognition motifs for tight binding. CTCF was purified to near homogeneity by sequence-specific DNA chromatography. The approximate molecular weight of the CTCF was estimated to be 130,000. Removal of 110 bp sequence binding both CTCF and Sp1-like proteins leads to a 4 to 8-fold increase in transcription of stably transfected c-myc fusion constructs in chicken embryonic fibroblasts, suggesting that the CTCF is likely to be one of multiple nuclear factors involved in the transcriptional regulation of the chicken c-myc gene.
DOI:10.1186/s13059-020-01984-7URL [本文引用: 2]
DOI:10.1016/j.cell.2015.11.024URL [本文引用: 1]
DOI:10.1093/nar/gkz462URL [本文引用: 2]
DOI:10.1016/j.cell.2017.05.004 [本文引用: 2]

The molecular mechanisms underlying folding of mammalian chromosomes remain poorly understood. The transcription factor CTCF is a candidate regulator of chromosomal structure. Using the auxin-inducible degron system in mouse embryonic stem cells, we show that CTCF is absolutely and dose-dependently required for looping between CTCF target sites and insulation of topologically associating domains (TADs). Restoring CTCF reinstates proper architecture on altered chromosomes, indicating a powerful instructive function for CTCF in chromatin folding. CTCF remains essential for TAD organization in non-dividing cells. Surprisingly, active and inactive genome compartments remain properly segregated upon CTCF depletion, revealing that compartmentalization of mammalian chromosomes emerges independently of proper insulation of TADs. Furthermore, our data support that CTCF mediates transcriptional insulator function through enhancer blocking but not as a direct barrier to heterochromatin spreading. Beyond defining the functions of CTCF in chromosome folding, these results provide new fundamental insights into the rules governing mammalian genome organization.
DOI:10.1186/s13059-021-02466-0URL [本文引用: 1]
DOI:10.1016/j.celrep.2014.04.004URL [本文引用: 1]
DOI:10.1101/gad.954702URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
