 ,1,2, 孙昌春1,2, 薛思瑶1,2, 赵慧2, 江丽
,1,2, 孙昌春1,2, 薛思瑶1,2, 赵慧2, 江丽 ,2, 李彩霞
,2, 李彩霞 ,1,2
,1,249AISNP: a study on the ancestry inference of the three ethnic groups in the north of East Asia
Xiaoyuan Guo ,1,2, Changchun Sun1,2, Siyao Xue1,2, Hui Zhao2, Li Jiang
,1,2, Changchun Sun1,2, Siyao Xue1,2, Hui Zhao2, Li Jiang ,2, Caixia Li
,2, Caixia Li ,1,2
,1,2通讯作者: 江丽,博士,副主任法医师,研究方向:法医遗传学。E-mail:jl@mail.bnu.edu.cn;李彩霞,博士,主任法医师,研究方向:法医遗传学。E-mail:licaixia@tsinghua.org.cn
编委: 朱波峰
收稿日期:2021-02-25修回日期:2021-05-31
| 基金资助: |
Received:2021-02-25Revised:2021-05-31
| Fund supported: |
作者简介 About authors
郭晓媛,在读硕士研究生,专业方向:法医学。E-mail:

摘要
样本的族群来源推断在法医调查中可发挥重要作用,一个理想的推断体系是用一组较少的遗传标记实现较高的族群推断准确性。本研究调研搜集了区分东亚北方三个族群北方汉族、日本人和韩国人的428个祖先信息SNP (ancestry informative SNP, AISNP),获取了其在三个族群307份样本中的分型,通过位点Fst值及等位基因频率聚类等信息进一步精简位点,最终得到了一组49AISNP组合。基于307份样本利用留一法对49AISNP进行推断准确性验证,结果表明其在北方汉族、日本和韩国族群中的推断准确性均高于99%。49AISNP组合将有助于东亚地区亚族群的进一步区分。
关键词:
Abstract
The ancestry inference of unknown samples plays an important role in forensic investigations. An ideal panel is a set of few markers with high ancestry inference accuracy. We collected 428 AISNP (ancestry informative SNP) that can distinguish the three ethnic groups in north of East Asia, including northern Han, Japanese and Korean. The genotypes of 428 AISNP in 307 samples from these three ethnic groups were obtained. Based on the information of Fst value and clustering by allele frequency, the panel was further refined into 49AISNP smart panel. Inference accuracy of the 49AISNP was verified by the leave-one-out method with 307 samples, and the results showed that its accuracy was higher than 99% in the northern Han, Japanese and Korean ethnic groups. This panel can also be helpful to further distinguish the ethnic sub-groups in East Asia.
Keywords:
PDF (1129KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
郭晓媛, 孙昌春, 薛思瑶, 赵慧, 江丽, 李彩霞. 49AISNP:东亚北方三个族群遗传来源推断. 遗传[J], 2021, 43(9): 880-889 doi:10.16288/j.yczz.21-073
Xiaoyuan Guo.
单核苷酸多态性(single nucleotide polymorphisms, SNPs)是指基因组中单个碱基的变异形成的多态性,多数SNP表现为基因座上二等位基因的变化[1]。SNP在人类基因组中分布广泛,突变率低,具有较高的遗传稳定性。在不同族群之间频率分布差异大的SNP标记可用于预测族群来源,被称为祖先信息SNP (ancestry informative SNP, AISNP)[2,3,4]。利用一组AISNP即可判断个体的族源信息。目前已报道了大量洲际族群区分的AISNP体系[5,6,7,8],在洲际内部进行进一步细分的二级族群推断体系也有报道[9,10,11],例如可区分东亚、南亚、北亚的74-plex SNP等[9]。然而对东亚内部的区分推断仍有待进一步深入研究。
东亚人口众多[12,13],有研究表明东亚地区存在从南到北的遗传差异[14,15],且已发表不同的AISNP体系对其内部不同族群进行区分[11,16,17],这为研究东亚内部族群进化和遗传多样性奠定了基础。在东亚北方地区,北方汉族、日本人和韩国人有着相似的外貌特征,如黄皮肤、黑眼睛、鼻子短而扁平等;语言也有相似之处,如韩语和日语在结构、元音以及缺乏连词方面有着显著的共同特点[18]、汉字早在公元5世纪传入日本[19]。基于线粒体DNA (mitochondrial DNA, mtDNA)[20,21]、Y染色体[18]和短串联重复序列[22]的研究也表明三个族群间存在密切的遗传关系。最近有研究发现基于全基因组水平可区分汉族、日本和韩国族群[17,23],也有研究建立了包含142、98、59及36个AISNP的四组嵌套体系,可实现汉族、日本和韩国族群的区分,但是随着位点数目的减少,区分准确率会下降至90%[16]。对法医学应用而言,需筛选一组精简高效的AISNP,用于目标族群的推断区分,同时又可满足现场微量样本的检验需求。本研究从已发表文献中收集了东亚北方三个族群的428个AISNP及307份样本中的分型数据集,筛选了一组适用于法医学应用的AISNP,使其在这些族群中具有良好的区分度,以进一步提高族群推断技术在东亚北方的分辨率。
1 材料与方法
1.1 样本
参考数据库、测试样本群体信息见表1。参考数据库307份样本包括:来自千人基因组计划的103份北方汉族样本和104份日本样本[24],以及亚洲多样性计划 (分型基于affymetrix genome-wide human SNP array 6.0芯片)[17]的100份韩国样本。测试样本包括来自2个族群的38份样本:来自人类基因组多样性计划的10份北方汉族样本以及来自人类基因组多样性计划和西蒙斯基因组多样性计划的28份日本族群样本[25]。1.2 SNP位点来源
通过文献调研,从以下几个方面收集了428个AISNP位点:(1)中国科学院上海生命科学研究院徐书华课题组的203个AISNP[17],用于北方汉族、日本和韩国族群的两两区分;(2)中国科学院北京基因组研究所陈华课题组的142个AISNP[16],用于区分中国、日本和韩国族群的体系;(3)本课题组前期筛选的88个AISNP[26,27,28,29],针对北方汉族与日本族群以及东亚南北方族群区分的位点。具体信息见附表1。Table 1
表1
表1样本信息
Table 1
| 族群简称 | 族群信息 | 样本量 | 样本来源 |
|---|---|---|---|
| CHB | 中国北京的汉族人 | 103 | 千人基因组计划 |
| JPT | 东京的日本人 | 104 | 千人基因组计划 |
| KOR | 朝鲜人 | 100 | 亚洲多样性计划 |
| CHB-C | 中国北方汉族人 | 10 | 人类基因组多样性计划 |
| JPT-C | 日本人 | 28 | 人类基因组多样性计划、西蒙斯基因组多样性计划 |
新窗口打开|下载CSV
1.3 质量控制
去除分型缺失率超过10%的样本和AISNP位点。使用Haploview v4.1软件计算总族群中AISNP的连锁不平衡值[30],去除连锁不平衡(r2>0.2)的位点[31,32]。1.4 位点筛选及评估
在法医学应用中,具有相似信息量同时包含较少位点的组合最为实用。为了实现这个目标,需要剔除信息量较少或冗余的SNP以获得更为精简的体系。Fst值是群体遗传学中衡量群体间分化程度的一个重要指标,其范围是0~1,值越大表明群体间的遗传分化程度越大[33]。在族群推断应用方面,可以衡量一组AISNP位点的信息含量。使用Genepop v4.7.5软件计算每个位点在三个族群中总的Fst值以及等位基因频率[34],基于Fst值,以0.01为单位取不同范围的AISNP组合进行分析,确保优化后的AISNP组合提供与全部AISNP相似的族群推断信息量。使用R v3.3.2软件基于等位基因频率绘制热图并进行主成分分析(principal components analysis, PCA),使用R v4.0.2软件基于每个AISNP位点对PCA中样本聚类结果的贡献度值(Cos2值)进行可视化展示。基于热图聚类结果删除Fst值较小的位点来减少冗余信息[9]。使用Snipper网站(
1.5 推断方法准确性验证
使用DNA族群推断系统软件(DNA ancestry analyzer version1.0, DAA v1.0)[36]计算三个族群307份样本的群体匹配概率(population assignment match probability, AMP)和似然比(likelihood ratio, LR),并进行族群聚类分析,参数设置为K=3,运行20次,统计每个样本的AMP和LR值。通过样本数据库的留一法验证,对筛选的AISNP进行准确性评估,即依次从数据库取出一份样本作为未知族群来源的样本,其余作为参考数据库进行分析,以此类推,直到所有样本均被测试一次为止。基于留一法验证的族群推断结果有3种情况:(1)一致性结论,AMP排名第一位的人族群与样本信息来源一致且LR大于10;(2)不排除结论,LR小于或等于10表明不排除AMP排名前两位的族群;(3)错误结论,AMP排名第一位的族群与样本信息来源不一致且LR大于10。根据样本推断族群来源结果与其实际所属族群的一致与否,统计推断结果的准确性。测试样本准确性验证:以307份样本为参考数据库,38份测试集样本为未知族群来源的样本,计算每个测试集的AMP和LR值,并对推断结果的准确性进行统计。
1.6 位点的对比评估
为了验证本筛选方法的有效性,使用R v4.0.2软件中的sample函数进行随机抽样,从质控后的位点组合中随机抽取两组与最终组合数目相同的位点组合,并对其进行留一法准确性验证,与本研究所筛选的位点组合的准确性进行对比。2 结果与分析
2.1 位点筛选及评估
307份样本的分型缺失率均小于10%,在428AISNP中25个位点的分型缺失率大于10%,去除后得到403个AISNP。基于三个族群307份样本在403个AISNP上的分型,在三个族群中总Fst值见附表1。连锁不平衡分析结果如附图1所示,共有70个SNP位点分别位于24个单体型块中,每个单体型块代表着位点之间存在高度连锁。附表2列出了相互连锁位点间的r2值,统计各个单体型块中位点的Fst值,仅留下每个单体型块中Fst值最大的AISNP位点,共得到357个AISNP。Table 2
表2
表249AISNP位点信息
Table 2
| 序号 | rs号 | 染色体 | 第37版序列位置 | 等位基因 | Fst | 序号 | rs号 | 染色体 | 第37版序列位置 | 等位基因 | Fst |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs10088365 | 8 | 10097398 | G/A | 0.2709 | 26 | rs2622637 | 8 | 106505971 | G/A/T | 0.0951 |
| 2 | rs10489744 | 1 | 165380623 | G/A | 0.0690 | 27 | rs2642066 | 17 | 65639014 | G/T | 0.0910 |
| 3 | rs10521076 | 9 | 108878562 | C/T | 0.0775 | 28 | rs2838408 | 21 | 45246422 | A/G | 0.0750 |
| 4 | rs10894724 | 11 | 133547942 | T/A/G | 0.0812 | 29 | rs2976396 | 8 | 143764001 | G/A | 0.1521 |
| 5 | rs10973829 | 9 | 38446392 | C/T | 0.0781 | 30 | rs3111745 | 3 | 21827159 | G/T | 0.0751 |
| 6 | rs11034709 | 11 | 38428289 | A/G | 0.1083 | 31 | rs3217805 | 12 | 4388084 | C/A/G/T | 0.0844 |
| 7 | rs11841589 | 13 | 73814891 | G/T | 0.0862 | 32 | rs374722 | 2 | 147839830 | G/A | 0.0740 |
| 8 | rs11846710 | 14 | 58343352 | G/A | 0.0768 | 33 | rs3923736 | 7 | 155060730 | A/C/G/T | 0.0714 |
| 9 | rs11959012 | 5 | 167668843 | C/T | 0.0694 | 34 | rs4353835 | 3 | 32446775 | C/T | 0.0849 |
| 10 | rs12006467 | 9 | 35090720 | T/C | 0.0993 | 35 | rs4372441 | 11 | 7657259 | C/T | 0.0787 |
| 11 | rs12039715 | 1 | 242801261 | G/C | 0.1726 | 36 | rs4718412 | 7 | 66286867 | T/C | 0.0751 |
| 12 | rs12483769 | 22 | 47790072 | T/C | 0.0870 | 37 | rs5022079 | 18 | 76478188 | A/G | 0.0931 |
| 13 | rs1256519 | 14 | 65736324 | G/A | 0.1027 | 38 | rs6123723 | 20 | 37082145 | C/T | 0.0891 |
| 14 | rs1322944 | 13 | 53683163 | A/G | 0.0740 | 39 | rs6436971 | 2 | 231582797 | T/C | 0.0815 |
| 15 | rs1333099 | 13 | 73691236 | G/A | 0.0715 | 40 | rs6576127 | 14 | 106194763 | C/T | 0.1076 |
| 16 | rs1678537 | 12 | 57900341 | G/A | 0.0885 | 41 | rs7655849 | 4 | 161867415 | T/A | 0.0759 |
| 17 | rs17121800 | 10 | 108710873 | C/T | 0.1007 | 42 | rs928844 | 21 | 37999799 | C/T | 0.0846 |
| 18 | rs17207681 | 5 | 138168527 | A/G | 0.0823 | 43 | rs929115 | 1 | 171591189 | T/G | 0.0757 |
| 19 | rs17451739 | 5 | 144030993 | C/T | 0.0732 | 44 | rs9321180 | 6 | 130102959 | C/T | 0.0866 |
| 20 | rs174520 | 14 | 88006776 | T/G | 0.0899 | 45 | rs9549212 | 13 | 41022093 | C/T | 0.0708 |
| 21 | rs17599827 | 5 | 89518433 | A/C | 0.0810 | 46 | rs9896443 | 17 | 47125982 | T/C | 0.0738 |
| 22 | rs17631488 | 5 | 18777746 | A/G | 0.1030 | 47 | rs2770310 | 6 | 70165296 | C/A/T | 0.0924 |
| 23 | rs1981370 | 18 | 74070815 | A/G | 0.0722 | 48 | rs3027238 | 17 | 8135061 | T/C | 0.9803 |
| 24 | rs2269275 | 5 | 83261405 | A/G | 0.0688 | 49 | rs4578397 | 11 | 131832284 | G/T | 0.0737 |
| 25 | rs229562 | 22 | 37599065 | G/T | 0.0827 |
新窗口打开|下载CSV
基于357个AISNP在三个族群中等位基因频率绘制的热图见附图2。在热图中,颜色的深浅代表了AISNP位点的基因频率在不同群体中的相似性和差异性,颜色越红表示基因频率越高,越浅表示基因频率越低,且具有相似频率分布的AISNP通常处于聚类树的同一分枝中。去除信息含量较少及冗余的位点以获得具有相似信息量的较小的体系,例如图中红色标记位点处,rs11104947与rs229562处于同一聚类小枝中且颜色相似,而rs11104947的Fst值(0.0782)低于rs229562的Fst值(0.0827),所以可删除rs11104947位点。经过多轮测试,最终获得了一组49个AISNP(以下简称49AISNP),表2列出了49AISNP的详细信息,三个族群的总Fst值介于0.0688~0.9803,平均值为0.1083。图1显示基于49AISNP在三个族群中的等位基因频率热图。
图1
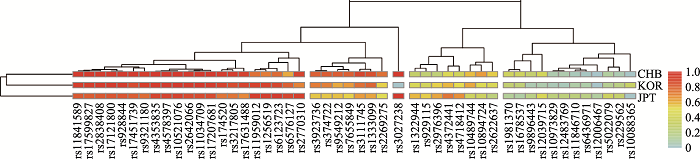 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1基于49AISNP的三个族群等位基因频率热图
Fig. 1Heatmap of 49AISNP based on the allele frequencies in three ethnic groups
表3显示了49个位点在各族群中的累计PSD值。可以看到三个族群之间的累计PSD值差异在1.4左右,说明该49AISNP在这三个族群之间的区分平衡度较好。
Table 3
表3
表349AISNP在每个族群中的累计PSD值
Table 3
| 49AISNP | 北方汉族 | 日本 | 韩国 |
|---|---|---|---|
| 累计PSD值 | 3.24566 | 3.96998 | 2.53635 |
新窗口打开|下载CSV
2.2 基于49AISNP的群体遗传结构
基于49AISNP的三个族群STRUCTURE聚类分析在K=3时的结果见图2,可观察到北方汉族、日本和韩国三个族群间明显的遗传差异。群体水平的祖先成分见饼状图,各颜色的比例代表该族群的祖先成分比例。如图所示,黄色、绿色和蓝色分别代表韩国、北方汉族和日本的祖先成分,其中,北方汉族族群中绿色成分占0.95;日本族群中蓝色成分占0.95;韩国族群中黄色成分占0.96。个体水平祖先成分见柱状图,其中每个柱形表示为一个样本,每种颜色代表一个祖先成分,每个条中各颜色的长度对应各样本的祖先成分比例。各颜色代表的祖先成分与饼状图相同。其中似然值最高的聚类结果显示:北方汉族族群样本的北方汉族成分中位数为0.98 (95%置信区间为0.94~0.97);日本族群样本的日本成分中位数为0.98 (95%置信区间为0.93~0.97);韩国族群样本的韩国成分中位数为0.98 (95%置信区间为0.95~0.98)。主成分分析(PCA)可反映个体水平上的族群结构,不同的颜色代表不同的族群,样本的聚类情况可反映族群间的遗传关系,同时根据不同的主成分可进行族群的区分。图3A展示了基于49AISNP的主成分分析图,前2个主成分占变异的23.36%。第一主成分(PC1)占变异的12.96%,可将北方汉族与日本和韩国族群区分开;第二主成分(PC2)占变异的10.40%,可将日本族群与其他两族群区分开。图3B展示了基于每个AISNP位点对PCA中样本聚类结果的Cos2值分析,图中颜色由绿色到红色以及箭头的长短均代表Cos2值的大小,图中只展示Cos2值排名前18个AISNP的名称,其余位点的Cos2值参照箭头长短及颜色变化。结合Fst值来看,每个位点对样本聚类的贡献值大小排序也对应该位点的Fst值大小排序。
图2
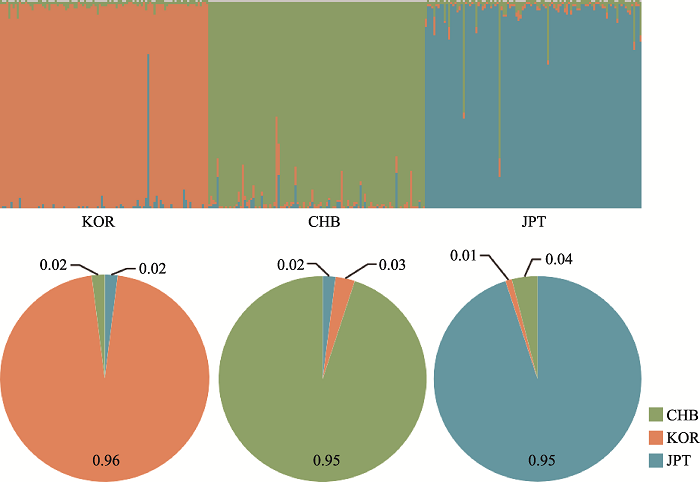 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图249AISNP在K=3时的三个族群STRUCTURE分析结果(Ln P(D)=-12,331.7)
Fig. 2The STRUCTURE analysis for three ethnic groups at K = 3 of 49AISNP(Ln P(D)= -12,331.7)
图3
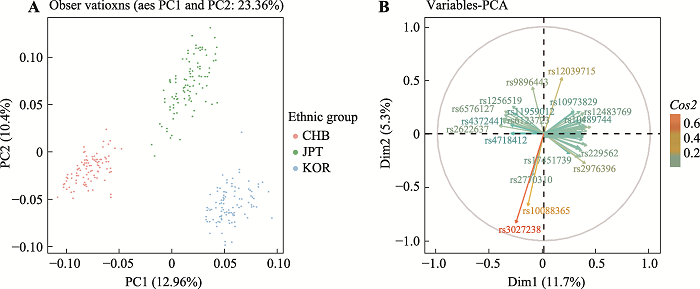 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3主成分分析及Cos2值分析
A:基于三个族群在49AISNP的等位基因频率进行主成分分析;B:基于每个AISNP位点对PCA中样本聚类结果的Cos2值分析。
Fig. 3Principal component analysis and Cos2 value analysis
2.3 留一法准确性验证
基于49AISNP的留一法准确性验证结果见表4,北方汉族、韩国样本族群推断结果一致性为100%;日本样本族群推断结果一致性为99.04%,错误率为0.96%,1例错判样本信息见表5。Table 4
表4
表4基于49AISNP的参考数据库族群推断结果统计
Table 4
| 族群 | 北方汉族 | 日本 | 韩国 | 总计 |
|---|---|---|---|---|
| 北方汉族 | 103(0) | 103 | ||
| 日本 | 1(0) | 103(0) | 104 | |
| 韩国 | 100(0) | 100 |
新窗口打开|下载CSV
Table 5
表5
表5基于参考数据库的错判样本信息
Table 5
| 样本来源 | 样本编号 | 族群 | 群体匹配概率 | 似然比 | 祖先成分 |
|---|---|---|---|---|---|
| 日本 | NA18976 | 北方汉族 | 1.96E-15 | 0.75 | |
| 日本 | 8.10E-17 | 2.43E+01 | 0.16 | ||
| 韩国 | 1.83E-19 | 1.07E+04 | 0.09 |
新窗口打开|下载CSV
2.4 测试样本验证
基于49AISNP的测试集准确性验证结果见表6,北方汉族族群推断结果一致性为80%,不排除结论为20%,错误率为0;日本样本族群推断结果一致性占82.15%,不排除结论占10.71%,错误率占7.14%。Table 6
表6
表6基于49AISNP的测试集族群推断结果统计
Table 6
| 族群 | 北方汉族 | 日本 | 韩国 | 总计 |
|---|---|---|---|---|
| 北方汉族 | 9(1) | 1(1) | 10 | |
| 日本 | 2(2) | 24(1) | 2(0) | 28 |
新窗口打开|下载CSV
2.5 49AISNP对比评估
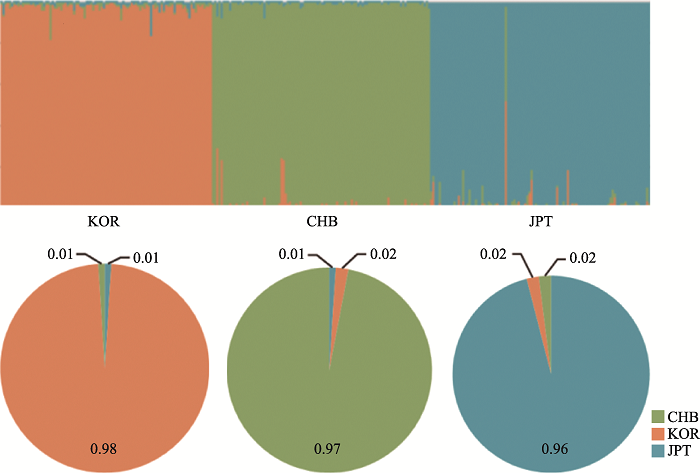
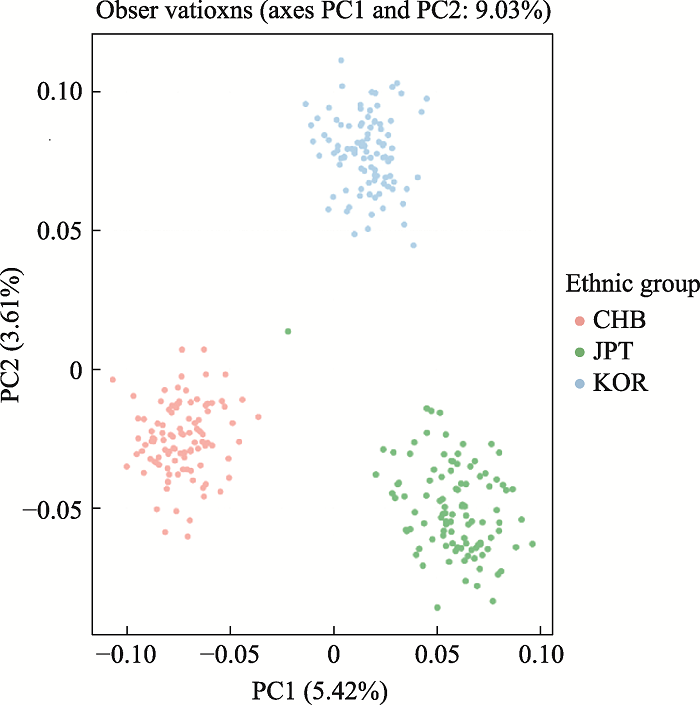
附图3显示了基于三个族群307份样本在357AISNP下的STRUCTURE聚类结果,在K=3时,可观察到三个族群明显的遗传差异,基于群体水平的祖先成分图显示北方汉族族群中绿色成分占0.97;日本族群中蓝色成分占0.96;韩国族群中黄色成分占0.98。基于个体水平祖先成分柱状图显示北方汉族族群样本的北方汉族成分中位数为0.99(95%置信区间为0.94~0.97);日本族群样本的日本成分中位数为0.99 (95%置信区间为0.93~0.97);韩国族群样本的韩国成分中位数为0.99 (95%置信区间为0.95~ 0.98)。同时利用主成分分析来评估357AISNP在个体水平上的族群结构,结果如附图4所示,前2个主成分占变异的9.03%。第一主成分(PC1)占变异的5.42%,可区分北方汉族和其他两族群;第二主成分(PC2)占变异的3.61%,可将韩国族群与其他两族群区分开。对比49AISNP与357AISNP的结果,两组AISNP均表现出良好的族群聚类能力,具有相似的族群推断准确性,且49AISNP的PC1和PC2总比例相较于357AISNP有所上升,证明通过一系列的筛选,我们成功地获得了一组具有相似信息量且位点数较少的组合。图4
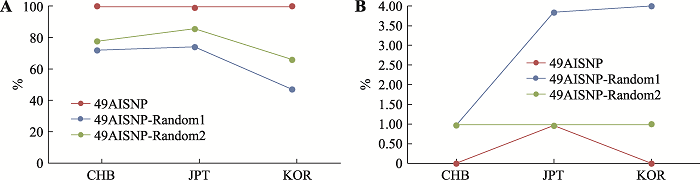 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图449AISNP与两组不同的、随机49AISNP在三个族群中的族群推断一致性和错误率比较
A:49AISNP与两组不同的、随机49AISNP在三个族群中的族群推断一致性比较;B:49AISNP与两组不同的、随机49AISNP在三个族群中的族群推断错误率比较。
Fig. 4Comparison of ancestry inference accuracy and error rate for the 49 AISNP with two different, random samplings of 49AISNP in the three ethnic groups
从357AISNP中用随机抽样的方法抽取的两组49AISNP,分别编为49AISNP-Random1和49AISNP- Random2。这两组的族群推断一致性和错误率与49AISNP的结果对比分别见图4A和图4B。结果显示两组随机49AISNP在三大族群中的族群推断一致性均比通过Fst值以及热图筛选出的49AISNP的低,且推断错误率较高,进一步说明我们的位点筛选方法是有效的。
3 讨论
汉族人口约占中国总人口的91.6%[37],是世界上最大的遗传群体,有研究表明汉族与韩国人或日本人间的遗传关系比与中国的少数民族间的关系更为密切[22]。现代日本人有双重血统,源于绳纹文化的狩猎采集者与弥生文化的移民之间的混合,他们从中国南部的长江口通过韩国带来了稻谷农业[38]。这种历史迁徙造成了如今汉族、日本与韩国之间的遗传差异,而近年来随着中、日、韩三国之间社会经济的频繁交流,大量人口离开其出生地,旅居他国,融入当地社会。使用遗传标记进行样本的族群来源推断在法医实践应用中起着关键作用。随着测序技术的发展,高密度遗传标记越来越多地被应用在族群推断上,因其能够获到高分辨率的区分度[39],然而,在实际应用中,高密度遗传标记并不适用于微量或降解检材的分型检测,位点数目少但信息量高的族群推断位点组合,对于法医学应用是最实用的。因此,我们需要在高密度遗传标记中剔除冗余的标记,本研究利用较高的Fst值筛选出了一组具有较高信息量且位点数较少的位点组合,为后续的体系构建及进一步验证奠定基础。在体系的测试评估方面,一般用以下几种方法评估族群推断的准确性:一是用随机抽样的方式,将现有数据库以一定比例划分为训练集和测试集,用训练集内样本数据进行位点筛选,用测试集内样本对筛选结果进行准确性验证[40];二是在应用现有数据库进行位点筛选后,新采集一部分样本,用新的测试集进行验证[41];三是用交叉验证的方法评估数据库的准确性[40],当样本量较大时,用十折交叉验证的方法,当样本量较少时,选择留一法评估数据库的准确性。但是将所有数据均用于训练集和测试集,容易造成筛选体系的过拟合,即最终模型在训练集上效果好;在测试集上效果差。为了避免过拟合的出现,一般可以重采样来评价模型效能,或者采用多次交叉验证的方法,求其均值估计算法准确性。由于本文中样本量较少,故未采用随机抽样的方法划分训练集和测试集,而是采用了留一法来逐一抽取测试样本,可以最大程度避免由于训练集样本量过少造成的偏差,同时另收集一部分样本做为测试集,以验证筛选体系的准确性。
在分析样本的族群来源时,应综合分析AMP、LR和族群成分[7,32,41],AMP值是在三个族群中AISNP组合的等位基因频率计算所得,AMP值最高的族群并不一定是真正的来源族群,当两个族群间AMP值的比值即似然比LR大于10时,可以认为AMP值第一位的族群是该样本来源族群,当似然比小于10时,要结合族群成分进行综合分析[41]。在49AISNP中,参考数据库族群来源推断错误的样本来自于日本族群中的NA18976,该样本的北方汉族和日本的似然比LR值计算结果为2.43E+01,其北方汉族、日本和韩国的族群成分分别为:0.75、0.16和0.09,该样本被推断为北方汉族。分析可能的原因如下:一是有可能推断错误,未来还需增加针对日本族群的特异性位点以达到更高的族群推断准确性;二是样本采样标签错误,或者由于历史或社会等原因,样本本身可能也不知道自己的族群来源[9]。
从测试集的验证结果来看,利用49AISNP基于307份参考数据库上的测试集族群推断一致性结论占81.58%,不排除结论占13.16%,错误率仅5.26%。证明我们筛选的49AISNP可以有效地区分东亚的这三个族群。综上所述,本研究获得了一组49AISNP可以进行东亚北方三个族群的区分,且具有高鉴别能力和区分平衡度。该组合将有助于东亚地区二级族群推断体系的发展,进一步提升东亚族群的内部区分力。但是本研究中涉及的样本量较少,未来可增加样本量,进一步验证该组合在北方汉族、日本和韩国族群中的推断能力。
附录:
附加材料详见文章电子版Supplementary Table 1
附表1
附表1428AISNP位点信息
Supplementary Table 1
| 序号 | rs号 | 染色体 | 第37版序列位置 | 等位基因 | Fst值 | 序号 | rs号 | 染色体 | 第37版序列位置 | 等位基因 | Fst值 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | rs10018432 | 4 | 189437086 | C/A | 0.0460 | 40 | rs10957985 | 8 | 81789921 | G/A | 0.0645 |
| 2 | rs10058739 | 5 | 144074365 | G/A | 0.0297 | 41 | rs10972006 | 9 | 3430684 | T/C | 0.0266 |
| 3 | rs10088365 | 8 | 10097398 | G/A | 0.2709 | 42 | rs10973829 | 9 | 38446392 | C/T | 0.0781 |
| 4 | rs1009544 | 22 | 42239348 | G/C | 0.0430 | 43 | rs10991718 | 9 | 93716118 | T/C | 0.0048 |
| 5 | rs10107492 | 8 | 80568935 | T/A/C | 0.0503 | 44 | rs11012716 | 10 | 21762267 | A/G | 0.0302 |
| 6 | rs10110194 | 8 | 119479124 | G/C/T | 0.0585 | 45 | rs11023463 | 11 | 15209506 | A/C | 0.0052 |
| 7 | rs10132336 | 14 | 74869017 | A/G | 0.0327 | 46 | rs11032331 | 11 | 33678161 | G/T | 0.0519 |
| 8 | rs10132483 | 14 | 102299420 | T/C | 0.0519 | 47 | rs11034709 | 11 | 38428289 | A/G | 0.1083 |
| 9 | rs10134903 | 14 | 95229777 | G/A | 0.0360 | 48 | rs11042911 | 11 | 10678785 | T/C | 0.0432 |
| 10 | rs10139575 | 14 | 57087118 | A/C/G | 0.0342 | 49 | rs11058961 | 12 | 127544998 | G/A | 0.0438 |
| 11 | rs10166397 | 2 | 44372909 | G/T | 0.0490 | 50 | rs11086012 | 19 | 16032643 | A/C | 0.0537 |
| 12 | rs10171891 | 2 | 195971878 | C/T | 0.0431 | 51 | rs11104947 | 12 | 88942980 | G/A | 0.0782 |
| 13 | rs10177273 | 2 | 122239287 | C/G | 0.0782 | 52 | rs1111212 | 2 | 159312733 | C/G/T | -0.0007 |
| 14 | rs1026975 | 4 | 113971374 | T/G | 0.0229 | 53 | rs11128125 | 3 | 69389614 | T/A/C/G | 0.0035 |
| 15 | rs10277413 | 7 | 55238464 | T/A/G | 0.0294 | 54 | rs11133144 | 4 | 177229719 | A/C/T | 0.0446 |
| 16 | rs10431079 | 11 | 116440678 | A/G | 0.0728 | 55 | rs11151527 | 18 | 67347133 | A/G | 0.0506 |
| 17 | rs10459664 | 15 | 35064934 | C/T | 0.0505 | 56 | rs11159882 | 14 | 89525994 | G/T | 0.0479 |
| 18 | rs10483991 | 14 | 88682616 | G/A | 0.0607 | 57 | rs11161614 | 1 | 85898160 | T/G | 0.0299 |
| 19 | rs10486260 | 7 | 8859616 | C/T | 0.0210 | 58 | rs11164354 | 1 | 102439329 | G/A | 0.0436 |
| 20 | rs10489744 | 1 | 165380623 | G/A | 0.0690 | 59 | rs11174261 | 12 | 62362843 | C/T | 0.0473 |
| 21 | rs10506294 | 12 | 51070554 | T/C | 0.0019 | 60 | rs11177698 | 12 | 69906287 | A/G | 0.0534 |
| 22 | rs10506725 | 12 | 77449514 | T/C | 0.0444 | 61 | rs11222851 | 11 | 131831335 | G/A | 0.0683 |
| 23 | rs10521076 | 9 | 108878562 | C/T | 0.0775 | 62 | rs11223550 | 11 | 133532372 | A/G | 0.0527 |
| 24 | rs10756234 | 9 | 11316481 | C/T | -0.0004 | 63 | rs11224765 | 11 | 101310590 | C/T | 0.0981 |
| 25 | rs10768017 | 11 | 33681025 | G/T | 0.0571 | 64 | rs11257349 | 10 | 6260717 | T/G | 0.0065 |
| 26 | rs10771923 | 12 | 32320912 | C/T | 0.0332 | 65 | rs1125832 | 16 | 52421140 | T/A | 0.0337 |
| 27 | rs10778125 | 12 | 101962634 | C/A | 0.0513 | 66 | rs11466640 | 4 | 38778903 | G/A/T | 0.0090 |
| 28 | rs10806975 | 6 | 23766367 | A/G/T | 0.0372 | 67 | rs11611033 | 12 | 63771136 | C/T | 0.0488 |
| 29 | rs10819066 | 9 | 101356091 | C/G | 0.0262 | 68 | rs11621121 | 14 | 65822493 | C/T | 0.0990 |
| 30 | rs10833071 | 11 | 19209050 | T/C | 0.0328 | 69 | rs11625485 | 14 | 80242132 | T/C | 0.0538 |
| 31 | rs10836092 | 11 | 33676946 | G/A/C | 0.0515 | 70 | rs11625876 | 14 | 35248467 | C/T | 0.0559 |
| 32 | rs10836093 | 11 | 33678526 | A/G | 0.0530 | 71 | rs11680024 | 2 | 49535856 | A/C | -0.0040 |
| 33 | rs10847616 | 12 | 128910954 | A/C/T | 0.0308 | 72 | rs1178148 | 7 | 18778113 | A/C | 0.0795 |
| 34 | rs10849181 | 12 | 638166 | C/T | 0.0525 | 73 | rs11841589 | 13 | 73814891 | G/T | 0.0862 |
| 35 | rs10858883 | 12 | 89893348 | T/C | -0.0004 | 74 | rs11846710 | 14 | 58343352 | G/A | 0.0768 |
| 36 | rs10883736 | 10 | 104320029 | G/T | 0.0365 | 75 | rs11847263 | 14 | 65775695 | T/A/G | 0.0635 |
| 37 | rs10890510 | 1 | 48334490 | A/C | 0.0495 | 76 | rs11874960 | 18 | 1948608 | A/C/G/T | 0.0049 |
| 38 | rs10894034 | 11 | 129255618 | C/T | 0.0521 | 77 | rs11959012 | 5 | 167668843 | C/T | 0.0694 |
| 39 | rs10894724 | 11 | 133547942 | T/A/G | 0.0812 | 78 | rs12006467 | 9 | 35090720 | T/C | 0.0993 |
| 79 | rs12039715 | 1 | 242801261 | G/C | 0.1726 | 119 | rs1391099 | 4 | 143373910 | A/G/T | 0.0025 |
| 80 | rs12045644 | 1 | 40084488 | G/T | 0.0393 | 120 | rs1420288 | 16 | 54477881 | T/C | 0.0519 |
| 81 | rs12095503 | 1 | 171653329 | C/T | 0.0346 | 121 | rs1420528 | 16 | 52417241 | C/G | 0.0374 |
| 82 | rs12134013 | 1 | 164459913 | C/T | 0.0586 | 122 | rs1420638 | 3 | 181497374 | G/A | 0.0407 |
| 83 | rs12141436 | 1 | 68463212 | G/A | 0.0418 | 123 | rs1422931 | 5 | 167397540 | C/T | 0.0462 |
| 84 | rs12151767 | 2 | 198274929 | G/A | 0.0548 | 124 | rs1428150 | 5 | 132490807 | A/G | 0.0666 |
| 85 | rs12231617 | 12 | 124930127 | C/G | 0.0458 | 125 | rs1442502 | 1 | 166548317 | G/A | 0.0277 |
| 86 | rs12351269 | 9 | 16806521 | T/C | 0.0558 | 126 | rs1453054 | 2 | 181359907 | T/C/G | 0.0119 |
| 87 | rs12459941 | 19 | 10666112 | G/A | 0.0631 | 127 | rs1465306 | 7 | 66699784 | T/C | 0.0672 |
| 88 | rs12483769 | 22 | 47790072 | T/C | 0.0870 | 128 | rs1465759 | 2 | 142576915 | T/C | 0.0365 |
| 89 | rs12488690 | 3 | 150490463 | G/A | 0.0502 | 129 | rs1488485 | 3 | 7951364 | T/C | 0.0125 |
| 90 | rs12522710 | 5 | 65051804 | G/A | 0.0125 | 130 | rs1522340 | 3 | 70369504 | G/A | 0.0320 |
| 91 | rs1256519 | 14 | 65736324 | G/A | 0.1027 | 131 | rs1524930 | 21 | 40968460 | A/G | 0.0649 |
| 92 | rs1256520 | 14 | 65737193 | C/A | 0.0958 | 132 | rs1537523 | 9 | 3649097 | A/C | 0.0512 |
| 93 | rs12567990 | 1 | 207681685 | C/T | -0.0038 | 133 | rs1538374 | 10 | 72718336 | G/A | 0.0663 |
| 94 | rs12571942 | 10 | 127954919 | G/A | 0.0056 | 134 | rs1551653 | 8 | 34324240 | A/G | 0.0084 |
| 95 | rs12586912 | 14 | 34023334 | G/T | 0.0422 | 135 | rs155872 | 3 | 1454063 | C/T | 0.0227 |
| 96 | rs12588061 | 14 | 50481926 | T/C | 0.0703 | 136 | rs1561201 | 18 | 38205375 | G/A | 0.0330 |
| 97 | rs12589835 | 14 | 30848932 | C/T | 0.0615 | 137 | rs1566857 | 4 | 46048880 | A/T | 0.0468 |
| 98 | rs12598852 | 16 | 13329469 | G/A | 0.0500 | 138 | rs160539 | 16 | 63353953 | C/G | 0.0641 |
| 99 | rs12630087 | 3 | 80935171 | T/C | 0.0429 | 139 | rs1609763 | 5 | 142018424 | T/G | 0.0232 |
| 100 | rs12632233 | 3 | 94033744 | G/T | 0.0334 | 140 | rs1613215 | 3 | 77738317 | T/C | 0.0509 |
| 101 | rs12676684 | 8 | 118857704 | G/C/T | 0.0584 | 141 | rs1652519 | 15 | 58612113 | G/A/C | 0.0219 |
| 102 | rs12691557 | 2 | 141469911 | G/A/T | 0.0461 | 142 | rs1678537 | 12 | 57900341 | G/A | 0.0885 |
| 103 | rs12768145 | 10 | 17610277 | G/A | 0.0532 | 143 | rs16834705 | 2 | 193385209 | A/G | 0.0168 |
| 104 | rs12776442 | 10 | 59297413 | A/G | 0.0075 | 144 | rs16850913 | 2 | 166421763 | A/G | 0.0450 |
| 105 | rs1284605 | 12 | 57921188 | C/T | 0.0863 | 145 | rs16862627 | 3 | 150114701 | G/A | -0.0022 |
| 106 | rs13076655 | 3 | 80920612 | C/T | 0.0412 | 146 | rs16893074 | 5 | 24074021 | T/A | 0.0042 |
| 107 | rs13094402 | 3 | 80710455 | A/G | 0.0467 | 147 | rs16895073 | 5 | 65596821 | T/C | 0.0302 |
| 108 | rs13160399 | 5 | 165711426 | G/A | 0.0550 | 148 | rs16944149 | 17 | 10938783 | T/G | 0.0223 |
| 109 | rs1317548 | 10 | 12245520 | G/A | 0.0575 | 149 | rs16951472 | 13 | 97073977 | C/T | 0.0503 |
| 110 | rs131864 | 22 | 47271217 | T/G | 0.0450 | 150 | rs16986850 | 20 | 41048928 | T/C | 0.0307 |
| 111 | rs1322944 | 13 | 53683163 | A/G | 0.0740 | 151 | rs16991180 | 20 | 10148468 | G/A | 0.0655 |
| 112 | rs13249584 | 8 | 9710943 | A/G | 0.0786 | 152 | rs16997770 | 4 | 124693346 | T/C | 0.0611 |
| 113 | rs1325192 | 1 | 199244248 | G/C/T | 0.0625 | 153 | rs17002737 | 22 | 42282012 | C/T | 0.0454 |
| 114 | rs1333099 | 13 | 73691236 | G/A | 0.0715 | 154 | rs17016175 | 4 | 143391299 | A/G | 0.0018 |
| 115 | rs13419695 | 2 | 7347211 | A/T | 0.0808 | 155 | rs17029405 | 3 | 32451697 | C/A | 0.0508 |
| 116 | rs1362553 | 16 | 52438954 | C/G | 0.0329 | 156 | rs17034099 | 3 | 11220006 | C/A | 0.0412 |
| 117 | rs1371566 | 2 | 119738187 | T/G | 0.0337 | 157 | rs17072984 | 18 | 62051788 | G/A | 0.0312 |
| 118 | rs1378365 | 4 | 61775645 | G/A | 0.0587 | 158 | rs17076328 | 5 | 173143508 | G/A | 0.0164 |
| 159 | rs17121800 | 10 | 108710873 | C/T | 0.1007 | 199 | rs2175743 | 1 | 70295901 | C/T | 0.0413 |
| 160 | rs17129041 | 1 | 66996037 | T/C | 0.0529 | 200 | rs2194757 | 2 | 21706029 | G/A | 0.0108 |
| 161 | rs17207681 | 5 | 138168527 | A/G | 0.0823 | 201 | rs2195856 | 1 | 187922876 | A/C | 0.0548 |
| 162 | rs17239258 | 4 | 183263055 | A/G | 0.0500 | 202 | rs2253974 | 21 | 23231832 | C/T | 0.0187 |
| 163 | rs17372695 | 4 | 155060317 | A/G | -0.0030 | 203 | rs2255957 | 22 | 42241372 | G/A/T | 0.0440 |
| 164 | rs17377643 | 22 | 42152988 | A/C | 0.0565 | 204 | rs2269275 | 5 | 83261405 | A/G | 0.0688 |
| 165 | rs17451739 | 5 | 144030993 | C/T | 0.0732 | 205 | rs2269658 | 22 | 42280618 | T/C | 0.0375 |
| 166 | rs174520 | 14 | 88006776 | T/G | 0.0899 | 206 | rs2278339 | 18 | 50866195 | T/G | 0.0298 |
| 167 | rs174534 | 11 | 61549458 | A/G | -0.0023 | 207 | rs229562 | 22 | 37599065 | G/T | 0.0827 |
| 168 | rs174583 | 11 | 61609750 | C/T | -0.0046 | 208 | rs2295756 | 11 | 35241229 | T/C | 0.0000 |
| 169 | rs17469810 | 3 | 80807568 | G/A | 0.0390 | 209 | rs2313427 | 16 | 8815000 | G/A | 0.0470 |
| 170 | rs17582830 | 4 | 38867427 | A/G | 0.0004 | 210 | rs2330015 | 22 | 22536956 | C/T | 0.0678 |
| 171 | rs17583068 | 4 | 38908616 | G/C/T | 0.0558 | 211 | rs2333656 | 17 | 58184540 | T/C | 0.0576 |
| 172 | rs17599827 | 5 | 89518433 | A/C | 0.0810 | 212 | rs2349093 | 12 | 128520953 | T/A | 0.0004 |
| 173 | rs17631488 | 5 | 18777746 | A/G | 0.1030 | 213 | rs2352181 | 10 | 87653766 | G/A | 0.0257 |
| 174 | rs17717717 | 3 | 71404405 | G/A | 0.0320 | 214 | rs2353686 | 16 | 86354605 | T/C | 0.0566 |
| 175 | rs17766621 | 14 | 71262932 | T/C | 0.0349 | 215 | rs2377962 | 18 | 9101722 | G/T | 0.0522 |
| 176 | rs17823795 | 14 | 63384260 | G/A | 0.0411 | 216 | rs2430184 | 1 | 187786661 | G/T | 0.0482 |
| 177 | rs1794072 | 11 | 61303803 | C/T | 0.0446 | 217 | rs2526371 | 17 | 56443545 | G/A | 0.0501 |
| 178 | rs1795704 | 12 | 58739693 | C/A | 0.0502 | 218 | rs2547353 | 19 | 58582797 | G/C/T | 0.0666 |
| 179 | rs1835298 | 11 | 112057620 | G/A | 0.0428 | 219 | rs255630 | 5 | 127677188 | T/C | 0.0133 |
| 180 | rs1841305 | 18 | 50163950 | A/T | 0.0556 | 220 | rs258728 | 7 | 81663275 | C/A | 0.0337 |
| 181 | rs1861693 | 12 | 107603157 | C/T | 0.0471 | 221 | rs2587694 | 2 | 120213497 | A/G | 0.0026 |
| 182 | rs1864307 | 18 | 57332158 | T/G | 0.0440 | 222 | rs2589787 | 5 | 38260303 | G/A | 0.0374 |
| 183 | rs1875174 | 10 | 2971488 | C/G/T | 0.0806 | 223 | rs2622637 | 8 | 106505971 | G/A/T | 0.0951 |
| 184 | rs1877696 | 8 | 9712822 | C/G/T | 0.0956 | 224 | rs2642066 | 17 | 65639014 | G/T | 0.0910 |
| 185 | rs189062 | 2 | 137349278 | G/A | 0.0429 | 225 | rs2645645 | 4 | 77859067 | A/G/T | 0.0469 |
| 186 | rs1898227 | 16 | 63282080 | A/G/T | 0.0546 | 226 | rs26461 | 5 | 11202649 | C/T | 0.0211 |
| 187 | rs1901786 | 3 | 130021945 | G/A | 0.0411 | 227 | rs27061 | 5 | 1362793 | T/C | 0.0761 |
| 188 | rs1944975 | 18 | 57276748 | T/C | 0.0369 | 228 | rs2770310 | 6 | 70165296 | C/A/T | 0.0924 |
| 189 | rs1981370 | 18 | 74070815 | A/G | 0.0722 | 229 | rs2793438 | 6 | 24461427 | G/C | 0.0382 |
| 190 | rs198464 | 11 | 61521621 | G/A | -0.0050 | 230 | rs2836749 | 21 | 40287238 | A/C/T | 0.0548 |
| 191 | rs2008819 | 7 | 39662268 | C/T | 0.0459 | 231 | rs2837352 | 21 | 41363778 | C/T | 0.0312 |
| 192 | rs2011071 | 5 | 88741077 | A/G | 0.0583 | 232 | rs2838408 | 21 | 45246422 | A/G | 0.0750 |
| 193 | rs2035023 | 4 | 137753683 | T/C | 0.0549 | 233 | rs2853668 | 5 | 1300025 | G/T | 0.0580 |
| 194 | rs2047297 | 4 | 121511851 | G/C/T | 0.0271 | 234 | rs2901142 | 2 | 124764392 | C/T | 0.0727 |
| 195 | rs2068746 | 17 | 11389480 | T/C | 0.0464 | 235 | rs2945733 | 8 | 134615750 | T/G | 0.0602 |
| 196 | rs2099563 | 3 | 80740366 | G/A | 0.0447 | 236 | rs2972336 | 5 | 76504150 | G/C | -0.0028 |
| 197 | rs2160913 | 18 | 8716090 | A/C/G | 0.0216 | 237 | rs2976396 | 8 | 143764001 | G/A | 0.1521 |
| 198 | rs2174739 | 6 | 79659170 | A/G | 0.0042 | 238 | rs3027238 | 17 | 8135061 | T/C | 0.9803 |
| 239 | rs3104517 | 15 | 27940339 | A/G | 0.0288 | 279 | rs4690508 | 4 | 178295766 | G/T | 0.0504 |
| 240 | rs3111745 | 3 | 21827159 | G/T | 0.0751 | 280 | rs4718412 | 7 | 66286867 | T/C | 0.0751 |
| 241 | rs315808 | 5 | 169750158 | G/T | 0.0626 | 281 | rs475235 | 18 | 10029988 | G/A/T | 0.0289 |
| 242 | rs3217805 | 12 | 4388084 | C/A/G/T | 0.0844 | 282 | rs4756305 | 11 | 36292065 | T/A/C | 0.0532 |
| 243 | rs321967 | 7 | 78314549 | G/A/C | 0.0454 | 283 | rs4790409 | 17 | 1243941 | C/T | 0.0600 |
| 244 | rs34052939 | 3 | 80765431 | C/G/T | 0.0447 | 284 | rs480631 | 6 | 138406328 | C/A/T | 0.0620 |
| 245 | rs34254286 | 4 | 164411734 | A/G | 0 | 285 | rs4820428 | 22 | 41537589 | A/G | 0.0762 |
| 246 | rs374722 | 2 | 147839830 | G/A | 0.0740 | 286 | rs4834226 | 4 | 128986874 | C/T | -0.0008 |
| 247 | rs3757419 | 7 | 66029429 | C/T | 0.0718 | 287 | rs4839523 | 1 | 116800016 | T/C | 0.0284 |
| 248 | rs3757425 | 7 | 150067640 | A/C | 0.0706 | 288 | rs4846992 | 1 | 230137518 | A/G | 0.0426 |
| 249 | rs3758229 | 9 | 95078852 | C/A/G | 0.0574 | 289 | rs4865142 | 4 | 57549583 | C/T | 0.0372 |
| 250 | rs3775539 | 4 | 99304600 | A/G | 0.0452 | 290 | rs4865290 | 4 | 53278689 | G/A | 0.0307 |
| 251 | rs3910142 | 20 | 12921966 | T/C | 0.0613 | 291 | rs4883926 | 13 | 73843655 | T/C | 0.0583 |
| 252 | rs3923084 | 2 | 227948726 | A/G/T | 0.0586 | 292 | rs4902391 | 14 | 65815979 | G/T | 0.0929 |
| 253 | rs3923736 | 7 | 155060730 | A/C/G/T | 0.0714 | 293 | rs4903754 | 14 | 40865770 | G/A | 0.0492 |
| 254 | rs3924308 | 9 | 130185385 | C/A | 0.0129 | 294 | rs4912933 | 5 | 143038150 | G/A/C/T | -0.0034 |
| 255 | rs4133446 | 5 | 86155113 | T/C | 0.0645 | 295 | rs4918000 | 10 | 94837743 | C/T | 0.0399 |
| 256 | rs4144800 | 4 | 38006916 | G/A | 0.0228 | 296 | rs4922234 | 8 | 16639271 | G/A/C/T | 0.0433 |
| 257 | rs4254643 | 3 | 792207 | G/A | 0.0307 | 297 | rs4938285 | 11 | 116440011 | T/C | 0.0697 |
| 258 | rs4263026 | 18 | 73309989 | G/C | 0.0466 | 298 | rs5022079 | 18 | 76478188 | A/G | 0.0931 |
| 259 | rs4280278 | 16 | 29439227 | C/G | 0.0841 | 299 | rs5030883 | 10 | 70480868 | A/C/T | 0.0759 |
| 260 | rs4325622 | 17 | 28526475 | T/C | 0.3617 | 300 | rs520605 | 1 | 184795779 | C/T | 0.0581 |
| 261 | rs4328700 | 20 | 23709545 | A/G | 0.0344 | 301 | rs536430 | 10 | 79001643 | C/T | 0.0107 |
| 262 | rs4353835 | 3 | 32446775 | C/T | 0.0849 | 302 | rs5770018 | 22 | 49595560 | C/T | 0.0411 |
| 263 | rs4355871 | 9 | 29653331 | G/A/C | 0.0665 | 303 | rs5996064 | 22 | 42172080 | C/T | 0.0476 |
| 264 | rs4372441 | 11 | 7657259 | C/T | 0.0787 | 304 | rs6000401 | 22 | 37149336 | C/T | 0.0426 |
| 265 | rs4393669 | 18 | 67296561 | T/C | 0.0330 | 305 | rs6007756 | 22 | 48282309 | G/A/C | 0.0571 |
| 266 | rs4414069 | 1 | 200391561 | T/C | 0.0478 | 306 | rs6016226 | 20 | 38661387 | C/T | 0.0181 |
| 267 | rs4466495 | 9 | 17496114 | C/A/T | 0.0489 | 307 | rs6030932 | 20 | 42146320 | T/G | 0.0659 |
| 268 | rs4486887 | 16 | 50677571 | C/T | 0.0579 | 308 | rs6031579 | 20 | 43015108 | C/G | 0.0654 |
| 269 | rs4491175 | 11 | 90956214 | T/C | 0.0485 | 309 | rs6049064 | 20 | 23727145 | C/T | 0.0360 |
| 270 | rs4533076 | 12 | 56069231 | C/A | 0.0410 | 310 | rs6074520 | 20 | 12905188 | T/A/C | 0.0559 |
| 271 | rs4543050 | 3 | 74954560 | A/C/T | 0.0194 | 311 | rs6117562 | 20 | 753310 | G/A | -0.0011 |
| 272 | rs4578397 | 11 | 131832284 | G/T | 0.0737 | 312 | rs6123723 | 20 | 37082145 | C/T | 0.0891 |
| 273 | rs4603782 | 2 | 102735809 | T/A | 0.0642 | 313 | rs6137010 | 20 | 2090118 | C/T | 0.0110 |
| 274 | rs4668680 | 2 | 10529961 | A/G | 0.0199 | 314 | rs6138046 | 20 | 23704388 | G/C/T | 0.0342 |
| 275 | rs4674759 | 2 | 224068666 | G/A | 0.0059 | 315 | rs616022 | 3 | 80711349 | G/C/T | 0.0253 |
| 276 | rs4677399 | 3 | 74519384 | T/C | 0.0307 | 316 | rs6436971 | 2 | 231582797 | T/C | 0.0815 |
| 277 | rs4678169 | 3 | 124543103 | C/A | 0.0557 | 317 | rs6450876 | 5 | 31925423 | A/G | -0.0024 |
| 278 | rs4681869 | 3 | 58573534 | C/T | 0.0583 | 318 | rs6465469 | 7 | 95179593 | G/A | 0.0611 |
| 319 | rs6478966 | 9 | 101776125 | G/A | 0.0192 | 359 | rs760873 | 20 | 45393072 | T/G | 0.0582 |
| 320 | rs6502840 | 17 | 4989857 | C/T | 0.0515 | 360 | rs7630111 | 3 | 114165901 | C/A | 0.0499 |
| 321 | rs6549596 | 3 | 74507710 | T/C | 0.0271 | 361 | rs7642488 | 3 | 84688723 | C/T | 0.0406 |
| 322 | rs6549601 | 3 | 74525187 | C/T | 0.0307 | 362 | rs7655849 | 4 | 161867415 | T/A | 0.0759 |
| 323 | rs6559935 | 9 | 89140339 | G/A | 0.0425 | 363 | rs7666030 | 4 | 24728480 | G/A | 0.0492 |
| 324 | rs6561294 | 13 | 46695350 | T/C | 0.0215 | 364 | rs7674135 | 4 | 84577025 | T/C | 0.0399 |
| 325 | rs6576127 | 14 | 106194763 | C/T | 0.1076 | 365 | rs769411 | 2 | 171693639 | C/G/T | 0.0537 |
| 326 | rs6589072 | 11 | 109322914 | C/G | 0.0501 | 366 | rs7698051 | 4 | 60278484 | A/G | 0.0445 |
| 327 | rs6599390 | 4 | 956047 | A/G | 0.0584 | 367 | rs770576 | 11 | 99884272 | A/G | 0.0565 |
| 328 | rs6707773 | 2 | 191344132 | C/T | 0.0279 | 368 | rs7802058 | 7 | 81697666 | A/C | 0.0512 |
| 329 | rs6717406 | 2 | 187743228 | T/G | 0.0326 | 369 | rs7845383 | 8 | 88289450 | G/T | 0.0361 |
| 330 | rs6738590 | 2 | 65059603 | A/G | 0.0429 | 370 | rs789378 | 3 | 154079645 | C/T | 0.0205 |
| 331 | rs6743998 | 2 | 196676489 | T/A/C/G | 0.0612 | 371 | rs8014475 | 14 | 65811537 | T/C | 0.0937 |
| 332 | rs6760967 | 2 | 4669324 | C/T | 0.0400 | 372 | rs8015594 | 14 | 40942142 | C/G | 0.0688 |
| 333 | rs6767410 | 3 | 80699208 | C/T | 0.0505 | 373 | rs8049660 | 16 | 89857700 | A/G | 0.0398 |
| 334 | rs6768622 | 3 | 74508075 | C/T | 0.0271 | 374 | rs8050932 | 16 | 50687673 | C/T | 0.0545 |
| 335 | rs6833150 | 4 | 61659236 | A/G | 0.0235 | 375 | rs8060207 | 16 | 82273372 | G/A | 0.0287 |
| 336 | rs6898653 | 5 | 115975656 | A/G | 0.0654 | 376 | rs807959 | 19 | 22115835 | A/G | 0.0561 |
| 337 | rs6924957 | 6 | 96853579 | G/T | 0.0527 | 377 | rs8084884 | 18 | 75513922 | A/T | 0.0597 |
| 338 | rs6955018 | 7 | 125115418 | A/G | 0.0474 | 378 | rs8107011 | 19 | 30841000 | A/C | 0.0341 |
| 339 | rs7006443 | 8 | 9290753 | T/C | 0.0383 | 379 | rs827287 | 10 | 72708276 | A/T | 0.0435 |
| 340 | rs7022178 | 9 | 29780195 | G/A | 0.0720 | 380 | rs883433 | 19 | 39206288 | C/T | 0.0580 |
| 341 | rs7032231 | 9 | 117025771 | C/A | 0.0706 | 381 | rs928844 | 21 | 37999799 | C/T | 0.0846 |
| 342 | rs7038964 | 9 | 104423907 | C/T | 0.0419 | 382 | rs929115 | 1 | 171591189 | T/G | 0.0757 |
| 343 | rs7097617 | 10 | 77504287 | A/G | 0.0447 | 383 | rs9302550 | 16 | 52405521 | C/T | 0.0374 |
| 344 | rs7117447 | 11 | 101351383 | G/A | 0.1118 | 384 | rs9303660 | 17 | 31420730 | C/T | 0.0614 |
| 345 | rs712645 | 5 | 110585627 | C/T | 0.0604 | 385 | rs9321180 | 6 | 130102959 | C/T | 0.0866 |
| 346 | rs713278 | 11 | 133563491 | G/A | 0.0619 | 386 | rs933199 | 6 | 26112893 | T/C | 0.0107 |
| 347 | rs7173716 | 15 | 70123826 | T/G | 0.0723 | 387 | rs9384981 | 6 | 116803328 | T/C | 0.0259 |
| 348 | rs7173982 | 15 | 36685718 | C/A/T | -0.0013 | 388 | rs9398434 | 6 | 116787017 | T/G | 0.0229 |
| 349 | rs7193505 | 16 | 52401477 | C/G | 0.0394 | 389 | rs943327 | 9 | 113599422 | T/C | 0.0695 |
| 350 | rs7195477 | 16 | 29288634 | T/C | 0.0020 | 390 | rs9532080 | 13 | 38126537 | G/A | 0.0395 |
| 351 | rs7211426 | 17 | 53654548 | G/A/C | 0.0037 | 391 | rs9549212 | 13 | 41022093 | C/T | 0.0708 |
| 352 | rs7268940 | 20 | 9961532 | C/T | 0.0461 | 392 | rs9590216 | 13 | 95905305 | T/C | -0.0035 |
| 353 | rs7271913 | 20 | 39263421 | C/A | 0.0155 | 393 | rs9663367 | 10 | 19776910 | A/C/T | 0.0370 |
| 354 | rs7317643 | 13 | 108536028 | C/T | 0.1665 | 394 | rs978605 | 10 | 94903693 | A/G | 0.0208 |
| 355 | rs738745 | 22 | 48246398 | A/G | 0.0255 | 395 | rs9825713 | 3 | 100936248 | C/A | 0.0450 |
| 356 | rs741245 | 12 | 4227248 | G/A | 0.0535 | 396 | rs9826148 | 3 | 114464858 | C/A/T | 0.0677 |
| 357 | rs7555405 | 1 | 31183486 | C/T | 0.0420 | 397 | rs9852677 | 3 | 50291617 | G/A/T | 0.0412 |
| 358 | rs758219 | 19 | 40294953 | T/C | 0.0461 | 398 | rs9857773 | 3 | 149344615 | C/A | 0.0547 |
| 399 | rs9860483 | 3 | 138730737 | C/T | 0.0985 | 414 | rs1991955 | 6 | 29884066 | A/C | 0.0938 |
| 400 | rs9896443 | 17 | 47125982 | T/C | 0.0738 | 415 | rs2182268 | 13 | 113170683 | G/A | 0.0573 |
| 401 | rs9941426 | 18 | 487281 | T/G | 0.0689 | 416 | rs2261033 | 6 | 31603591 | A/G | 0.1538 |
| 402 | rs9947737 | 18 | 455128 | A/G | 0.0477 | 417 | rs2524095 | 6 | 31266117 | A/C | 0.0456 |
| 403 | rs9979724 | 21 | 22447717 | A/G | 0.0383 | 418 | rs284784 | 4 | 100335874 | C/A | 0.0255 |
| 404 | rs10901244 | 9 | 136079463 | C/T | 0.0077 | 419 | rs3015224 | 9 | 72728388 | A/G | 0.1419 |
| 405 | rs11078951 | 17 | 38831415 | T/A/G | 0.1249 | 420 | rs3130688 | 6 | 31210216 | C/A/T | 0.0991 |
| 406 | rs11673276 | 19 | 55347963 | C/T | 0.0264 | 421 | rs3135402 | 6 | 33024654 | A/C | 0.1028 |
| 407 | rs12896399 | 14 | 92773663 | G/T | 0.0615 | 422 | rs376877 | 6 | 33024606 | C/G | 0.0902 |
| 408 | rs1414241 | 9 | 21744392 | G/A/T | 0.0756 | 423 | rs4800105 | 18 | 19651982 | C/G/T | 0.0906 |
| 409 | rs1431403 | 6 | 33047031 | T/C | -0.0048 | 424 | rs5758649 | 22 | 42605548 | C/T | 0.0059 |
| 410 | rs1496279 | 5 | 18680420 | A/T | - | 425 | rs6062275 | 20 | 61146909 | C/G/T | 0.1420 |
| 411 | rs1500127 | 5 | 165739982 | C/T | 0.0416 | 426 | rs7207346 | 17 | 75668619 | C/A/G/T | 0.0105 |
| 412 | rs174592 | 11 | 61618608 | A/G | -0.0049 | 427 | rs8074124 | 17 | 36531565 | C/T | 0.0457 |
| 413 | rs1927568 | 13 | 99740117 | T/C | 0.0137 | 428 | rs901134 | 3 | 84688173 | G/A | 0.0574 |
新窗口打开|下载CSV
Supplementary Table 2
附表2
附表2成对连锁不平衡位点间的r2值
Supplementary Table 2
| Block | rs号-1 | rs号-2 | r2值 | Block | rs号-1 | rs号-2 | r2值 |
|---|---|---|---|---|---|---|---|
| Block1 | rs2853668 | rs27061 | 0.243 | rs5996064 | rs2255957 | 0.869 | |
| Block2 | rs13249584 | rs1877696 | 0.885 | rs5996064 | rs2269658 | 0.695 | |
| Block3 | rs6074520 | rs3910142 | 0.771 | rs5996064 | rs17002737 | 0.574 | |
| Block4 | rs6138046 | rs4328700 | 1 | rs1009544 | rs2255957 | 0.991 | |
| rs6138046 | rs6049064 | 0.993 | rs1009544 | rs2269658 | 0.795 | ||
| rs4328700 | rs6049064 | 0.993 | rs1009544 | rs17002737 | 0.679 | ||
| Block5 | rs4355871 | rs7022178 | 0.221 | rs2255957 | rs2269658 | 0.802 | |
| Block6 | rs10836092 | rs11032331 | 0.98 | rs2255957 | rs17002737 | 0.686 | |
| rs10836092 | rs10836093 | 0.987 | rs2269658 | rs17002737 | 0.816 | ||
| rs10836092 | rs10768017 | 0.906 | Block10 | rs4486887 | rs8050932 | 0.371 | |
| rs11032331 | rs10836093 | 0.981 | Block11 | rs7193505 | rs9302550 | 0.959 | |
| rs11032331 | rs10768017 | 0.899 | rs7193505 | rs1420528 | 0.959 | ||
| rs10836093 | rs10768017 | 0.919 | rs7193505 | rs1125832 | 0.93 | ||
| Block7 | rs11466640 | rs17582830 | 0.449 | rs7193505 | rs1362553 | 0.911 | |
| Block8 | rs4903754 | rs8015594 | 0.285 | rs9302550 | rs1420528 | 1 | |
| Block9 | rs17377643 | rs5996064 | 0.849 | rs9302550 | rs1125832 | 0.95 | |
| rs17377643 | rs1009544 | 0.735 | rs9302550 | rs1362553 | 0.912 | ||
| rs17377643 | rs2255957 | 0.742 | rs1420528 | rs1125832 | 0.95 | ||
| rs17377643 | rs2269658 | 0.62 | rs1420528 | rs1362553 | 0.912 | ||
| rs17377643 | rs17002737 | 0.66 | rs1125832 | rs1362553 | 0.961 | ||
| rs5996064 | rs1009544 | 0.861 | Block12 | rs1678537 | rs1284605 | 0.965 | |
| Block13 | rs198464 | rs174534 | 0.388 | rs6767410 | rs34052939 | 0.96 | |
| Block14 | rs6833150 | rs1378365 | 0.278 | rs6767410 | rs17469810 | 0.944 | |
| Block15 | rs1898227 | rs160539 | 0.326 | rs6767410 | rs13076655 | 0.898 | |
| Block16 | rs1256519 | rs1256520 | 0.898 | rs6767410 | rs12630087 | 0.905 | |
| rs1256519 | rs11847263 | 0.565 | rs13094402 | rs616022 | 0.468 | ||
| rs1256519 | rs8014475 | 0.61 | rs13094402 | rs2099563 | 0.992 | ||
| rs1256519 | rs4902391 | 0.61 | rs13094402 | rs34052939 | 0.992 | ||
| rs1256519 | rs11621121 | 0.606 | rs13094402 | rs17469810 | 0.976 | ||
| rs1256520 | rs11847263 | 0.528 | rs13094402 | rs13076655 | 0.929 | ||
| rs1256520 | rs8014475 | 0.596 | rs13094402 | rs12630087 | 0.937 | ||
| rs1256520 | rs4902391 | 0.575 | rs616022 | rs2099563 | 0.472 | ||
| rs1256520 | rs11621121 | 0.572 | rs616022 | rs34052939 | 0.472 | ||
| rs11847263 | rs8014475 | 0.814 | rs616022 | rs17469810 | 0.466 | ||
| rs11847263 | rs4902391 | 0.791 | rs616022 | rs13076655 | 0.468 | ||
| rs11847263 | rs11621121 | 0.791 | rs616022 | rs12630087 | 0.476 | ||
| rs8014475 | rs4902391 | 0.95 | rs2099563 | rs34052939 | 1 | ||
| rs8014475 | rs11621121 | 0.925 | rs2099563 | rs17469810 | 0.984 | ||
| rs4902391 | rs11621121 | 0.975 | rs2099563 | rs13076655 | 0.937 | ||
| Block17 | rs3757419 | rs4718412 | 0.312 | rs2099563 | rs12630087 | 0.944 | |
| Block18 | rs827287 | rs1538374 | 0.258 | rs34052939 | rs17469810 | 0.984 | |
| Block19 | rs6549596 | rs6768622 | 1 | rs34052939 | rs13076655 | 0.937 | |
| rs6549596 | rs4677399 | 0.923 | rs34052939 | rs12630087 | 0.944 | ||
| rs6549596 | rs6549601 | 0.975 | rs17469810 | rs13076655 | 0.921 | ||
| rs6768622 | rs4677399 | 0.923 | rs17469810 | rs12630087 | 0.929 | ||
| rs6768622 | rs6549601 | 0.975 | rs13076655 | rs12630087 | 0.992 | ||
| rs4677399 | rs6549601 | 0.916 | Block21 | rs11224765 | rs7117447 | 0.442 | |
| Block20 | rs6767410 | rs13094402 | 0.952 | Block22 | rs4938285 | rs10431079 | 0.876 |
| rs6767410 | rs616022 | 0.468 | Block23 | rs9398434 | rs9384981 | 0.949 | |
| rs6767410 | rs2099563 | 0.96 | Block24 | rs1391099 | rs17016175 | 0.593 |
新窗口打开|下载CSV
附图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图1403AISNP连锁不平衡图
Supplementary Fig. 1Linkage disequilibrium diagram of 403AISNP
附图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图2基于357AISNP的3个族群等位基因频率热图
Supplementary Fig. 2Heatmap of 357AISNP based on the allele frequencies in 3 ethnic groups
附图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图3357AISNP在K=3时的3个族群STRUCTURE分析结果(Ln P(D)=-107,854.6)
Supplementary Fig. 3The STRUCTURE analysis for 3 ethnic groups at K=3 of 357AISNP(Ln P(D)= -107,854.6)
附图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图4基于3个族群在357AISNP的等位基因频率进行主成分分析
Supplementary Fig. 4Principal component analysis of 3 ethnic groups based on allele frequencies of 357AISNP
致谢:
感谢中国科学院上海生命科学研究院计算生物学研究所的徐书华老师在文章数据方面给予的帮助。(责任编委: 朱波峰)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.2144/000112806PMID:18474034 [本文引用: 1]

Forensic samples that contain too little template DNA or are too degraded require alternate genetic marker analyses or approaches to what is currently used for routine casework. Single nucleotide polymorphisms (SNPs) offer promise to support forensic DNA analyses because of an abundance of potential markers, amenability to automation, and potential reduction in required fragment length to only 60-80 bp. The SNP markers will serve an important role in analyzing challenging forensic samples, such as those that are very degraded, for augmenting the power of kinship analyses and family reconstructions for missing persons and unidentified human remains, as well as for providing investigative lead value in some cases without a suspect (and no genetic profile match in CODIS). The SNPs for forensic analyses can be divided into four categories: identity-testing SNPs; lineage informative SNPs; ancestry informative SNPs; and phenotype informative SNPs. In addition to discussing the applications of these different types of SNPs, this article provides some discussion on privacy issues so that society and policymakers can be more informed.
DOI:10.1016/j.fsigen.2015.05.012URL [本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.fsigen.2014.01.002URL [本文引用: 1]
DOI:10.1016/j.fsigen.2007.06.008URL [本文引用: 2]
DOI:10.1186/2041-2223-2-1URL [本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.fsigen.2016.04.002URL [本文引用: 4]
DOI:10.1016/j.fsigen.2019.05.001URL [本文引用: 1]
DOI:10.1007/s12024-018-0071-yURL [本文引用: 2]
DOI:10.1098/rstb.2007.2028URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/ejhg.2011.139PMID:21792231 [本文引用: 1]

Recent reports have identified a north-south cline in genetic variation in East and South-East Asia, but these studies have not formally explored the basis of these clinical differences. Understanding the origins of these variations may provide valuable insights in tracking down the functional variants in genomic regions identified by genetic association studies. Here we investigate the genetic basis of these differences with genome-wide data from the HapMap, the Human Genome Diversity Project and the Singapore Genome Variation Project. We implemented four bioinformatic measures to discover genomic regions that are considerably differentiated either between two Han Chinese populations in the north and south of China, or across 22 populations in East and South-East Asia. These measures prioritized genomic stretches with: (i) regional differences in the allelic spectrum for SNPs common to the two Han Chinese populations; (ii) differential evidence of positive selection between the two populations as quantified by integrated haplotype score (iHS) and cross-population extended haplotype homozygosity (XP-EHH); (iii) significant correlation between allele frequencies and geographical latitudes of the 22 populations. We also explored the extent of linkage disequilibrium variations in these regions, which is important in combining genetic association studies from North and South Chinese. Two of the regions that emerged are found in HLA class I and II, suggesting that the HLA imputation panel from the HapMap may not be directly applicable to every Chinese sample. This has important implications to autoimmune studies that plan to impute the classical HLA alleles to fine map the SNP association signals.
DOI:10.1126/science.1177074URL [本文引用: 1]
DOI:10.1111/ahg.v83.5URL [本文引用: 3]
DOI:10.1186/s41065-018-0057-5URL [本文引用: 4]
PMID:10721667 [本文引用: 2]

We have examined variations of five polymorphic loci (DYS287, DXYS5Y, SRY465, DYS19, and DXYS156Y) on the Y chromosome in samples from a total of 1260 males in eight ethnic groups of East Asia. We found four unique haplotypes constructed from three biallelic markers in these samples of East Asians. The Japanese population was characterized by a relatively high frequency of either the haplotype I-2b (-/Y2/T) or II-1 (+/Y1/C). These dual patterns of the distribution of Y chromosomes (I-2b/II-1) were also found in Korea, although they were present at relatively low frequencies. The haplotype II-1 was present in Northeast Asian populations (Chinese, Japanese, Koreans, and Mongolians) only, except for one male from the Thai population among the Southeast Asian populations (Indonesians, Philippines, Thais, and Vietnamese). The Japanese were revealed to have the highest frequency of this haplotype (27.5%), followed by Koreans (2.9%), Mongolians (2.6%), and mainland Chinese (2.2%). In contrast, the frequency of the haplotype I-2b was found to be 17.1% in the Japanese, 9.5% in Indonesian, 6.3% in Korean, 3.8% in Vietnamese, and 2.7% in Thai samples. These findings suggested that the chromosomes of haplotype I-2b were likely derived from certain areas of Northeast Asia, the region closest to Southeast Asia. Phylogenetic analysis using the neighbor-joining tree also reflected a general distinction between Southeast and Northeast Asian populations. The phylogeny revealed a closer genetic relationship between Japanese and Koreans than to the other surveyed Asian populations. Based on the result of the dual patterns of the haplotype distribution, it is more likely that the population structure of Koreans may not have evolved from a single ancient population derived from Northeast Asians, but through dual infusions of Y chromosomes entering Korea from two different waves of East Asians.
DOI:10.4236/aa.2011.12004URL [本文引用: 1]
PMID:2840820 [本文引用: 1]

Mitochondrial DNA (mtDNA) polymorphisms were detected using 13 restriction enzymes on the total DNA obtained from blood samples of five Asian populations: Japanese and Ainu of northern Japan, Korean, Negrito (Aeta) of the Philippines, and Vedda of Sri Lanka. Of a total of 28 restriction-enzyme morphs detected, eight had not been reported previously. By combining the morphs, we were able to classify mtDNAs of 243 individuals into 20 mtDNA types. Phylogenetic analyses using maximum parsimony and genetic distance methods both showed that the Japanese, Ainu, and Korean populations were closely related to each other. Aeta was found to show a relatively close relationship to these three populations, confirming the conclusion from previous studies of blood markers. In contrast, Vedda was quite different from the other four populations.
PMID:8751859 [本文引用: 1]

Nucleotide sequences of the major noncoding (D-loop) region of human mtDNA from five East Asian populations including mainland Japanese, Ainu, Ryukyuans, Koreans, and Chinese were analyzed. On the basis of a comparison of 482-bp sequences in 293 East Asians, 207 different sequence types were observed. Of these, 189 were unique to their respective populations, whereas 18 were shared between two or three populations. Among the shared types, eight were found in common between the mainland Japanese and Koreans, which is the largest number in the comparison. The intergenic COII/tRNA(Lys) 9-bp deletion was observed in every East Asian population with varying frequencies. The D-loop sequence variation suggests that the deletion event occurred only once in the ancestry of East Asians. Phylogenetic analysis revealed that East Asian lineages were classified into at least 18 monophyletic clusters, though lineages from the five populations were completely intermingled in the phylogenetic tree. However, we assigned 14 of the 18 clusters for their specificity on the basis of the population from which the maximum number of individuals in each cluster was derived. Of note is the finding that 50% of the mainland Japanese had continental specificity in which Chinese or Koreans were dominant, while < 20% of either Ryukyuans or Ainu possessed continental specificity. Phylogenetic analysis of the entire human population revealed the closest genetic affinity between the mainland Japanese and Koreans. Thus, the results of this study are compatible with the hybridization model on the origin of modern Japanese. It is suggested that approximately 65% of the gene pool in mainland Japanese was derived from the continental gene flow after the Yayoi Age.
PMID:9703426 [本文引用: 2]

Allelic frequencies for up to five short tandem repeat systems (HumTH01. HumVWA, HumF13B, HumCD4, HumD2111) were analyzed in seven population samples from Asia using the polymerase chain reaction and gel electrophoresis. No deviations from Hardy-Weinberg equilibrium were observed. Two new alleles of the CD4 and TH01 loci were detected, and sequenced and their molecular structure is presented. A phylogenetic tree based on Thai, Han Chinese (from the northeast of China), Japanese, German and Ovambo allelic frequencies was constructed and demonstrates the close relationship of the Asian populations. Additionally, allelic frequency data for the VWA and TH01 systems were determined for the south Chinese minorities Bai, Dai and Qiang and for Koreans and compared with the above data. The Bai and Dai populations were clear outliers of the cluster of all other Asians, indicating an unexpected pattern of genetic heterogeneity of the Chinese nation. Two clusters of Asian populations could be established: the Koreans and Japanese together with the Han and Qiang Chinese, and, forming a separate cluster, the Bai and Dai populations.
DOI:10.1126/sciadv.aaz7835URL [本文引用: 1]
DOI:10.1038/nature15393URL [本文引用: 1]
DOI:10.1093/nar/gkw829URL [本文引用: 1]
DOI:10.1016/j.ajhg.2009.10.015URL [本文引用: 1]
DOI:10.1038/jhg.2015.79URL [本文引用: 1]
DOI:10.1007/s00439-005-1334-8URL [本文引用: 1]
DOI:10.1038/ejhg.2013.111URL [本文引用: 1]
PMID:15297300 [本文引用: 1]

Research over the last few years has revealed significant haplotype structure in the human genome. The characterization of these patterns, particularly in the context of medical genetic association studies, is becoming a routine research activity. Haploview is a software package that provides computation of linkage disequilibrium statistics and population haplotype patterns from primary genotype data in a visually appealing and interactive interface.http://www.broad.mit.edu/mpg/haploview/jcbarret@broad.mit.edu
DOI:10.1086/381000URL [本文引用: 1]
DOI:10.1007/s00414-015-1183-5URL [本文引用: 2]
DOI:10.1073/pnas.70.12.3321URL [本文引用: 1]
DOI:10.1111/j.1471-8286.2007.01931.xURL [本文引用: 1]
DOI:10.1016/j.fsigen.2016.01.015URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.5260/chara.19.1.58URL [本文引用: 1]
DOI:10.1126/sciadv.1601877URL [本文引用: 1]
DOI:10.1186/s41065-020-00162-wURL [本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 3]
[本文引用: 3]
