 ,1, 黄允智
,1, 黄允智 ,1, 刘学英2, 傅向东2,3
,1, 刘学英2, 傅向东2,3Genetic improvement of nitrogen use efficiency in crops
Shan Li ,1, Yunzhi Huang
,1, Yunzhi Huang ,1, Xueying Liu2, Xiangdong Fu2,3
,1, Xueying Liu2, Xiangdong Fu2,3通讯作者: 傅向东,研究员,博士生导师,研究方向:植物发育和环境适应的激素调控机理。E-mail:
第一联系人:
编委: 储成才
收稿日期:2021-02-18修回日期:2021-04-2网络出版日期:2021-07-20
| 基金资助: |
Received:2021-02-18Revised:2021-04-2Online:2021-07-20
| Fund supported: |
作者简介 About authors

李姗,博士,教授,研究方向:植物激素调控水稻养分高效利用。E-mail:
黄允智,在读博士研究生,研究方向:植物激素调控水稻养分高效利用。E-mail:

摘要
氮素是植物生长发育所需的大量元素之一,施用氮肥是农业生产中提高农作物产量的重要手段。自20世纪60年代以来,“绿色革命”半矮秆农作物品种的育成和大面积推广有效地解决了“高产与倒伏”之间的矛盾,提高了农作物的收获指数和产量。然而半矮秆水稻和小麦品种也表现出生长发育对氮肥响应减弱、根系对铵态氮和硝态氮的吸收能力下降以及氮肥利用效率(nitrogen use efficiency, NUE)低的弊病,其产量增加依赖于氮肥的大量投入,这不仅提高了种植成本还导致了严重的环境污染问题。因此,提高农作物氮肥利用效率对于保障国家粮食安全和农业可持续发展具有重要战略意义。本文概述了“绿色革命”与赤霉素的作用机理,系统总结了植物氮素吸收、同化和代谢调控方面的研究进展,并介绍了提高作物氮肥利用效率的最新研究发现,以期为作物氮肥高效利用的遗传改良提供参考。
关键词:
Abstract
Nitrogen (N) is an essential mineral nutrient for plant growth and development. N deficiency is the major factor limiting plant growth and crop production in most natural and agricultural soils. The green revolution of the 1960’s boosted crop yields through cultivation of semi-dwarf plant varieties. However, green revolution wheat and rice varieties have relatively poor nitrogen use efficiency (NUE), require a high N fertilizer supply to achieve maximum yield potential, and this leads to an increase in production costs and environmental problem. Therefore, a major challenge for sustainable agriculture is whether improvement of NUE through the reduction of N fertilizer supply can be achieved without yield penalty. In this review, we summarize the recent advances in understanding of molecular mechanisms underlying the regulation of N-responsive plant growth, utilization and possibility for improvements of NUE in crops, and new breeding strategies through modulation of N-responsive growth-metabolism coordination for future sustainable agriculture.
Keywords:
PDF (716KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李姗, 黄允智, 刘学英, 傅向东. 作物氮肥利用效率遗传改良研究进展. 遗传[J], 2021, 43(7): 629-641 doi:10.16288/j.yczz.21-064
Shan Li.
自20世纪60年代以来,以半矮秆小麦(Triticum aestivum L.)和水稻(Oryza sativaL.)等农作物品种的培育和推广为标志的“绿色革命”带来了全球粮食产量的飞跃,解决了由于人口快速增长而引发的粮食危机。水稻半矮秆基因sd1(semi-dwarf 1)和小麦半矮秆基因Rht-1(reduced height-1)的应用解决了因密植和大量施肥而导致的倒伏问题,提高了收获指数,使作物单产大幅度提升。然而,半矮秆品种也伴随着生长发育对氮肥响应减弱、根系对氮素吸收能力下降以及氮肥利用效率(nitrogen use efficiency, NUE)降低等问题[1]。联合国粮食及农业组织(Food and Agriculture Organization of the United Nations, FAO)的数据显示,在过去10年中,世界范围的氮肥消耗量显著增加,但粮食产量增速缓慢,氮肥的过量投入不但没有大幅度地提高产量,反而导致了经济效益和生态效益的下滑。目前农田氮肥利用效率仅为40.2%,大部分氮肥在土壤中累积,并随着水土而流失进江河湖海或经过微生物反硝化作用排入大气,这不仅浪费了资源和能源,还引起了土壤酸化、大气和水体污染等一系列生态环境问题。因此,在保证产量不减的基础上如何提高氮肥的利用效率,减少氮肥的使用量是我国农业可持续发展亟待解决的重要问题。
1 “绿色革命”与赤霉素的作用机理
“绿色革命”的分子本质归结于赤霉素(gibberellic acid, GA)的生物学效应。小麦“绿色革命”基因Rht-1编码赤霉素信号途径的负调控因子DELLA蛋白,育种中应用比较广泛的Rht-B1b和Rht-D1b均编码N端截断突变形式的DELLA蛋白[2],突变形式的DELLA蛋白本身不受GA诱导降解而积累,使小麦表现出半显性的、GA不敏感的半矮化表型[3]。水稻“绿色革命”基因sd1位于1号染色体的长臂上,编码GA合成途径的GA20氧化酶2 (GA20ox2)。在籼稻生产上广泛应用的是该基因缺失了383 bp的突变类型,突变导致蛋白翻译提前终止,阻断了GA53至GA20的合成过程,降低了水稻内源活性GA的含量,使水稻DELLA蛋白SLR1 (SLENDER RICE 1)高水平积累,进而导致水稻株高降低[4,5]。DELLA蛋白为植物特有的GRAS家族(由最初发现的3个家族成员GAI、RGA和SCR的特征字母命名)的一个亚家族,是GA信号途径的关键元件,在胞内起到阻遏植物生长发育的作用[6,7,8]。GA信号首先被赤霉素受体蛋白GID1 (GIBBERELLIN- INSENSITIVE DWARF1)感知并与之结合[9]。GID1中存在一个可以结合GA的口袋和一个可延展的N端区域[10],具有生物活性的GA与GID1结合后,GID1蛋白的N端区域就会盖住口袋,然后与DELLA蛋白的TVHYNP结构域互作形成GA-GID1-DELLA复合体[10,11],进一步使DELLA蛋白的GRAS结构域发生变化,增强DELLA蛋白与F-BOX蛋白(SCFSLY1/GID2)的互作强度[12],加速了DELLA蛋白的泛素化和26S蛋白酶体降解进程。植物生长阻遏蛋白DELLA被降解后,释放与其互作的转录因子,游离的转录因子启动GA信号途径相关基因的表达,进而促进植物的生长发育。DELLA蛋白N端的两个保守结构域DELLA和TVHYNP在拟南芥(Arabidopsis thaliana)、水稻、小麦、玉米(Zea maysLinn.)和大麦(Hordeum vulgareL.)等不同物种中高度保守。拟南芥基因组中的DELLA蛋白有5个成员,分别是GAI (GA-INSENSITIVE)、RGA (REPRESSOR OF ga1-3)、RGL1 (RGA-LIKE1)、RGL2 (RGA-LIKE2)和RGL3 (RGA-LIKE3)。小麦绿色革命基因Rht-1、玉米矮秆基因D8(Dwarf-8)、水稻SLR1和大麦SLN1(SLENDER 1)均为GAI的直系同源基因[6,7,13,14]。
水稻生长调节因子OsGRF4 (growth-regulating factor 4)能与GIF1 (GRF-interacting factor 1)蛋白形成转录激活复合体,该复合体可以直接结合多个氮代谢相关基因的启动子,并促进基因表达,从而促进氮的吸收同化和转运。而DELLA蛋白能与OsGRF4互作,并抑制OsGRF4-GIF1复合体的形成,进而抑制了氮代谢相关基因的表达[15]。NGR5 (nitrogen-mediated tiller growth response 5)是一个控制水稻分蘖响应土壤供氮水平变化的关键基因,其编码一个AP2 (APETALA2)结构域的转录因子。NGR5能够招募PRC2复合体(polycomb repressive complex)并通过介导组蛋白H3K27m3甲基化修饰来调控水稻分蘖相关基因的表达。此外,NGR5基因表达受氮素调控,并且NGR5蛋白还是GA-GID1促进的蛋白酶体降解的新靶标。DELLA能与NGR5蛋白互作并与之竞争性结合GID1蛋白来抑制NGR5降解,从而促进水稻分蘖增加[16]。
因此,“绿色革命”半矮秆的水稻和小麦因DELLA蛋白高水平积累而获得了3个方面的特性:第一,DELLA蛋白的积累因抑制植物的生长而降低了株高,提高了抗倒伏能力;第二,高水平的DELLA蛋白抑制了OsGRF4对下游氮响应因子的激活作用,降低了作物的氮肥利用效率;第三,高丰度的DELLA提高了NGR5蛋白的稳定性,促进了植物分蘖,提高了产量。因此,半矮秆的水稻、小麦品种在高水肥条件下能保持其半矮化、抗倒伏和多分蘖等高产特性,但同时也降低了作物的氮素响应和氮肥利用效率(图1)。
图1
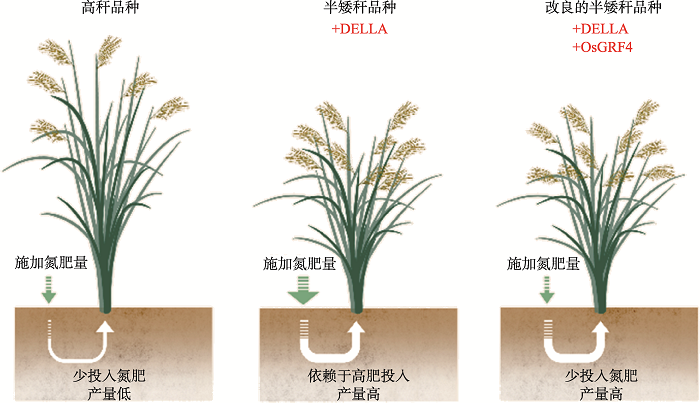 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1新“绿色革命”品种协同提升氮肥利用效率与产量
Fig. 1New green revolution cultivars synergistically improve N use efficiency and grain yield
2 植物的氮素吸收、转运与同化
氮素是植物生长发育所必需的大量元素之一,是体内氨基酸和蛋白质的重要构成组分,其平均含量约占植物干重的1.5%。为了获取充足的氮源以满足生长发育的需要,植物根系需要从土壤中吸收不同形式的氮素,包括硝态氮(NO3-)、铵态氮(NH4+)、可溶性多肽和复杂的不溶性含氮化合物等,其中以吸收NO3-和NH4+为主。2.1 NO3-的吸收和转运
在通气良好的旱田土壤中,植物主要以NO3-为氮源,植物根系吸收NO3-主要依靠硝酸盐转运蛋白(nitrate transporters, NRTs)来完成。从植物中分离鉴定出来的NO3-转运体大致分为4类:NRT1/PTR (nitrate transporter 1/peptide transport family)、NRT2 (nitrate transporter 2)、CLC (chloride channel)和SLAC1/SLAH (slow anion channel/slow anion channel homolog)。其中,大多数NRT1蛋白主要发挥低亲和力NO3-转运蛋白功能,但拟南芥AtNPF6.3/ AtNRT1.1、水稻OsNPF2.42和蒺藜苜蓿(Medicago truncatula) MtNRT1.3例外;NRT2主要负责低浓度NO3-的转运;CLC蛋白部分成员是液泡膜定位的NO3-/H+的逆向转运蛋白,在液泡的NO3-积累方面发挥作用[17];SLAC1/SLAH家族蛋白定位在保卫细胞和根的中柱细胞,可能参与气孔的关闭和NO3-从根到茎的运输[17,18]。在模式植物拟南芥中,有关NO3-吸收的分子机制研究比较深入。NRT1和PTR蛋白家族也被称为NPF (NRT1 PTR family)[19]。除AtNPF6.3/AtNRT1.1/ AtCHL1外,拟南芥NPF家族成员均属于低亲和力转运体。AtNPF6.3/AtNRT1.1/AtCHL1属于双亲和力转运体,在高浓度和低浓度的NO3-条件下均参与根系对NO3-的吸收,这主要取决于AtNRT1蛋白的第101位苏氨酸是否发生磷酸化。当T101位被磷酸化时,AtNPF6.3/AtNRT1.1作为高亲和力的NO3-转运体在低浓度NO3-的条件下发挥作用;当T101位没有被磷酸化时,AtNPF6.3/AtNRT1.1作为低亲和力的NO3-转运体在高浓度NO3-的条件下发挥作用[20],在此过程中蛋白激酶CIPK23 (calcineurin B-like interacting protein kinase 23)感应NO3-信号,能够对AtNRT1.1进行磷酸化修饰,进而调控AtNPF6.3/ AtNRT1.1对NO3-的亲和活性和吸收[21]。拟南芥NRT2家族至少有7个成员,其中6个成员发挥吸收NO3-的功能需要伴侣蛋白NAR2 (nitrate assimilation related protein 2)的存在,NRT2和NAR2家族成员均是NO3-高亲和力运输系统的组成部分[22,23]。
在厌氧条件下土壤中的氮素多以NH4+形式存在,但由于根系周围发生的硝化作用,仍有大量的氮以NO3-的形式被作物吸收,因此NO3-对于水稻等喜铵作物也十分重要[24]。水稻中NO3-转运蛋白大致可以分为低亲和力转运体OsNRT1和高亲和力转运体OsNRT2两个家族,而OsNPF6.5 (OsNRT1.1B)和OsNPF6.1例外。OsNRT1.1B编码双亲和的NO3-转运蛋白,受硝酸盐诱导表达,在高、低NO3-浓度下都会发挥吸收NO3-的功能,高表达后可以显著促进根系对NO3-的吸收,增加株高、产量和籽粒的氮积累[25]。此外,OsNRT1.1B能影响根系富集的、具有氮转化能力的微生物组,从而改变根际微环境,进而影响水稻的氮肥利用效率[26]。OsNPF6.1也是一个双亲和硝酸根转运蛋白,定位于质膜,在根部高表达,在硝酸盐的吸收和再分配中发挥作用,并且受氮响应的转录因子OsNAC42的调控[27]。OsNRT1.1A编码一个定位于液泡膜上的低亲和力NO3-转运蛋白,其表达不受硝酸盐的诱导,而受铵盐的诱导,主要参与细胞内硝酸盐、铵盐利用的调节[28]。在高浓度NO3-条件下低亲和力转运体OsNPF8.9 (OsNRT1.1) 和OsNPF2.4 (OsNRT1.6)发挥吸收NO3-的功能[29,30]。在低浓度NO3-条件下,NRT2家族成员的高亲和力转运蛋白OsNRT2.1、OsNRT2.2及其伴侣蛋白OsNAR2.1 起主导作用[31,32]。OsNRT2家族成员OsNRT2.3具有OsNRT2.3a和OsNRT2.3b两种不同的剪接形式。OsNRT2.3a编码一个由516个氨基酸残基组成的膜定位蛋白,负责NO3-在韧皮部的长距
离运输;OsNRT2.3b编码一个由486个氨基酸残基组成的膜定位蛋白,在茎韧皮部高表达,在根部低表达,是一个pH敏感的NO3-转运蛋白,其发挥功能不需要OsNAR2.1的协助[33,34]。
2.2 NH4+的吸收和转运
在淹水环境中生长的植物主要以NH4+为氮源。NH4+的摄入主要由NH4+转运体家族(ammonium transporter/methylammonium permease/rhesus, AMT/ MEP/Rh)来完成。在植物中,NH4+转运体主要有AMT1和AMT2/MEP两个蛋白亚家族。目前研究表明,AMT家族成员在拟南芥中有6个、衣藻(Chlamydomonas)中有8个、水稻中有10个、杨树(PopulusL.)中有14个[35,36]。拟南芥的根系吸收NH4+主要由AtAMT1.1、AtAMT1.2、AtAMT1.3和AtAMT1.5四个基因负责,它们均编码定位于细胞质膜的转运蛋白,并在根表皮细胞高表达,在氮源缺乏或供应蔗糖的情况下,这4个基因的表达均上调[37,38]。这4个成员的NH4+吸收能力排序为:AtAMT1.1 = AtAMT1.3 > AtAMT1.2 > AtAMT1.5,其中AtAMT1.1、AtAMT1.2 和AtAMT1.3三个成员承担了拟南芥根系对NH 4+吸收量的90%[39,40]。
水稻主要以吸收NH4+为主。目前发现水稻中至少有10个OsAMT成员,分为OsAMT1、OsAMT2、OsAMT3和OsAMT4四个家族[41]。其中OsAMT1、OsAMT2、OsAMT3各包含3个成员,OsAMT4只有1个成员。OsAMT1家族成员属于高亲和力NH4+转运体(high-affinity transport system, HATS)。在NH4+浓度较低时,高亲和力的铵转运体负责植物根系对NH4+的吸收,会表现饱和动力学特征;而当NH4+浓度较高(1~40 mmol/L)时,低亲和力NH4+转运体起主导作用,不会表现出饱和动力学特征[42]。OsAMT1.1在水稻根部和地上部组成型表达,其表达水平受底物积累的反馈调节;OsAMT1.2在根部特异性表达,其表达水平受NH4+的诱导;OsAMT1.3同样在根部特异性表达,其表达水平受高浓度铵的抑制。根部游离的谷氨酸盐抑制OsAMT1.3的表达,但可以促进OsAMT1.1和OsAMT1.2的表达[36]。
2.3 植物的氮素同化途径
植物根系吸收的不同形式的氮源需要经过一系列的同化作用才能被植物体利用。植物吸收的NO3-大部分被运送至地上部分进行同化与利用。首先在细胞质中,NO3-在硝酸还原酶(nitrate reductase, NR)的作用下被还原成NO2-,随后NO2-被运送至细胞质体中,在亚硝酸还原酶(nitrite reductase, NiR)的作用下转变成NH4+,最后NH4+在谷氨酰胺合成酶/谷氨酸合酶(glutamine synthetase/ferredoxin-glutamate synthase, GS/Fd-GOGAT)的作用下进入谷氨酸循环。首先GS催化NH4+与谷氨酸结合生成谷氨酰胺,随后谷氨酰胺与三羧酸循环的中间产物2-酮戊二酸(2-OG)在GOGAT催化作用下生成两个谷氨酸[43]。参与GS/GOGAT循环的酶主要是定位于叶绿体的GS2和Fd-GOGAT以及定位在细胞质中的GS1和NADH-GOGAT (nicotinamide adenine dinucleotide- glutamate synthase)。水稻GS1家族有3个成员。OsGS1.1对水稻的生长和灌浆非常重要,几乎在水稻的所有组织中都能检测到OsGS1.1的表达,在叶片中表达最高。OsGS1.2在根部特异性表达,不仅参与NH4+在根部的同化过程,还参与水稻对高温的响应;OsGS1.2的突变体其分蘖数、穗粒数和千粒重均显著减少,总氨基酸、谷氨酸和天冬酰胺含量显著下降,表明其在根部的NH4+同化过程中起着重要作用[44]。OsGS1.3主要在小穗中表达,并且OsGS1.2和OsGS1.3均不能补偿OsGS1.1的功能[45]。OsGS2主要在叶片的叶绿体中表达,在光呼吸作用释放出来的NH4+的再同化过程中起主导作用[46]。
由于NH4+对植物具有毒害作用,因此植物根系从土壤吸收的NH4+大部分在根部进行同化,然后主要以谷氨酸和谷氨酰胺的形式向地上部运输,在水稻中OsGS1.2和OsNADH-GOGAT1在这个过程中起主要作用。当外界NH4+浓度升高时,OsGS1.2和OsNADH-GOGAT2在根部表皮细胞和外皮层细胞中的表达量会显著上升,从而将NH4+快速同化,生成的谷氨酰胺和谷氨酸被运输到地上部[47]。
3 植物的氮素代谢调控
氮代谢是植物生命活动中最基本的物质代谢过程之一。植物在长期的进化过程中,形成了一套复杂的、精细的氮素代谢调控网络来感知、响应氮素水平的动态变化以快速获取生长所需的氮营养。目前研究发现,植物的氮素代谢也受到多种转录因子、miRNA、植物激素等的共同调控。3.1 转录因子调控植物氮代谢
转录因子是一类能识别基因上游启动子特异性基序,调控基因表达的蛋白质分子。研究表明,NAC (NAM, ATAF, CUC)、NLP (NIN-LIKE protein)、BTB (Bric-a-Brac/Tramtrack/Broad)、Dof (DNA-binding with one finger)、bZIP (basic leucine zipper)和MYB (v-Myb avian myeloblastosis viral oncogene homolog)等转录因子均参与植物氮代谢的调控过程。NAC是植物特有的一类转录因子。水稻OsNAC42基因表达受低氮诱导,OsNAC42蛋白可与OsNPF6.1启动子结合进而激活OsNPF6.1的表达[27]。NLP是一类调控氮诱导相关基因表达的转录因子[48],能够调控AtNPF6.3/AtNRT1.1、ANR1 (ANTHOCYANIDIN REDUCTASE1)、CIPK8和LBD37/38(lateral organ boundaries domain 37/38)等NO3-响应基因的表达。此外,Ca2+依赖蛋白激酶III亚族(subgroup III Ca2+- sensor protein kinases, CPKs) 中的成员CPK10/30/32能转导NO3-信号诱导产生的Ca2+浓度变化,通过磷酸化激活NLP蛋白的活性,组成NO3--CPK-NLP信号传导网络,进而促进根系和叶片发育[49]。拟南芥BT1/BT2编码BTB家族蛋白,其表达受转录因子NLP调控。在低氮条件下,BT1/BT2通过负调控NRT2基因的表达来影响氮吸收[50]。Dof是植物特有的一类转录因子。水稻中OsDOF18能调控下游多个AMT基因和氮同化基因的表达,从而调节NH4+的运输和氮分配[51]。bZIP转录因子在植物氮信号传导、氮代谢和碳-氮平衡过程中均发挥重要作用,如小麦TabZIP60负调控氮素利用[52]。水稻中的生长调控因子OsGRF4蛋白能够直接激活氮吸收和同化相关基因的表达,同时也能激活光合作用相关基因的表达,实现对植物碳-氮代谢平衡的调控[15]。MYB是一类包含Myb保守结构域、具有调控植物生长发育功能的转录因子。水稻MYB61是一种调节纤维素合成的转录因子,其基因表达受低氮诱导并受OsGRF4的调控,因此OsGRF4-MYB61信号通路将纤维素合成与氮响应代谢联系在一起[53]。Varala等[54]将机器学习引入到基于时间序列的氮反应转录组数据分析中,明确了氮信号相关转录因子的时序性调控关系,并发现146个能够响应氮信号的新转录因子。Gaudinier等[55]利用大规模酵母单杂交(yeast one- hybrid, Y1H)筛选调节氮代谢的转录因子,并结合大数据分析得到了一个氮代谢相关的酵母单杂交网络(yeast one-hybrid network for nitrogen-associated metabolism, YNM),并通过计算预测了在氮代谢中起关键作用的转录因子。这些研究为深入研究植物氮吸收、代谢调控网络提供了很大的帮助。3.2 MicroRNA调控植物氮代谢
MicroRNA (miRNA)是在真核生物中发现的一类内源性的具有调控功能的非编码RNA,长度约为20~25 bp。miRNA对靶基因mRNA稳定性及其翻译过程的作用,是植物在转录后水平调控基因功能的重要方式之一[56]。第一个将miRNA与氮素利用联系起来的基因是miRNA167靶标基因AtARF8(auxin response factor 8)。研究发现,在根的中柱鞘中,硝酸盐和谷氨酰胺/谷氨酸抑制miRNA167的表达,从而增强AtARF8的转录水平,最终导致侧生根数目增多[57]。AtABF3(auxin-signaling F-box protein 3)的表达受硝酸盐诱导,AtABF3高表达能抑制主根生长而促进侧根生长,而氮的代谢物促进miR393的表达,随后miR393会抑制AtABF3的表达[58]。miR167和miR393以及它们的下游基因形成了一个改变植物根系结构来响应氮信号的反馈机制。
在氮饥饿状态下,NFYA(nuclear factor Y, subunit A)家族成员的表达量上升,这是由于miR169的表达下降引起的。在低氮条件下,miR169在韧皮部的表达量下降,暗示着miR169能够在植物体内的氮含量信号从茎至根的长距离传输过程中发挥作用[59]。过表达OsmiR444a会抑制氮诱发的侧根生长,但能够通过增强磷转运基因的表达来提高作物的含磷量,这说明OsmiR444a能够联系氮、磷信号通路[60]。磷饥饿能诱导AtmiR827的表达,进而降低靶基因NLA(nitrogen limitation adaptation)的表达,而NLA参与植物低氮逆境胁迫响应。在低氮条件下,与野生型相比,nla突变体中能积累更多的磷[61]。因此,miRNA不仅通过改变植物根系构型和氮信号的长距离传输,还通过与磷信号通路互作来调节植物根系对氮的响应。
3.2 植物激素信号与氮信号的互作
环境中的氮源还是植物生长发育和响应环境胁迫的重要信号物质。许多氮代谢与激素信号转导相关的最新发现都将氮信号与植物激素信号联系在一起。生长素(Indole-3-acetic acid, IAA)的极性运输是由顶部运送至基部,促进侧根的起始和发育。因此,IAA一直被认为是介导氮信号从地上部传递至地下部的媒介[62]。NRT1.1可以通过调节硝酸盐依赖的生长素运输和硝酸盐信号途径来调控侧根的生长。模拟磷酸化的AtNPF6.3/AtNRT1.1T101D的突变表现出较强生长素运输调节能力,促进生长素快速横向流动,而非磷酸化的AtNPF6.3/AtNRT1.1T101A的突变表现出较弱的生长素运输调节能力,导致生长素在根尖积累,表明低NO3-诱导的T101磷酸化影响了生长素运输从而调节生长素介导的侧根生长[63]。NLP7能诱导生长素生物合成基因TAR2(tryptophan aminotransferase related 2)和生长素外排基因PIN7 (PIN-FORMED 3)的表达[49]。在低氮条件下,TAR2突变体在侧根原基中的生长素积累减少,侧根的形成受到抑制[64]。Gaudinier等[55]构建的酵母单杂交网络表明生长素响应因子ARF9能调控PNR (primary nitrate responses)基因(例如GS2和ASN2 (asparagine synthetase))的表达;ARF18能调控下游氮代谢基因NRT2.4和AMT1;2的表达。在水稻中,DNR1(dull nitrogen response 1)编码一个负调控生长素合成的酶,其表达水平受外界氮供应的调控从而改变植物体内的生长素含量,进而影响生长素响应因子OsARF6和OsARF17对下游氮代谢相关基因的调控作用,最终影响植物的氮代谢[65]。
脱落酸(abscisic acid, ABA)被认为是一种参与植物响应胁迫的应激激素。外界高浓度的氮素能够抑制拟南芥侧根发育,ABA信号途径可能参与其中[66]。ABA不敏感突变体(abi4-1、abi4-2、abi5-1)和ABA合成缺失突变体(aba1-1、aba2-3、aba2-4和aba3-2)的侧根生长对高浓度NO3-的抑制作用不敏感。在ABA信号转导关键调节因子ABI2的突变体abi2-2中,NO3-诱导AtNRT2.1表达和侧根生长的效应减弱,说明ABI2在NO3-信号传递途径中发挥作用[21]。赤霉素信号途径的DELLA蛋白调控植物
生长发育对氮肥的响应。一方面,DELLA蛋白可以与OsGRF4互作,抑制OsGRF4-GIF1转录激活复合体的形成,从而抑制OsGRF4对下游氮代谢基因的激活作用,降低作物的氮吸收能力[15];另一方面,DELLA蛋白结合GID1蛋白抑制氮响应因子NGR5降解,而NGR5能促进植物分蘖,调控植株在高氮水平下的分蘖响应。氮响应负调控因子LBD可以直接结合在OsTCP19启动子上调控OsTCP19的表达,而OsTCP19作为转录因子可以抑制油菜素内酯(brassinosteroids, BRs)信号途径中的关键组分DLT(DWARF AND LOW-TILLERING)的表达,从而调控水稻分蘖。LBD-OsTCP19-DLT通路证明了BR信号途径直接参与氮调控植物生长发育的过程[67]。茉莉酸甲酯(methyl jasmonate, MeJA)处理使水稻根系中GS1.2和NiR的转录水平和蛋白质水平均降低,并且根系中的NO3-和NH4+转运相关基因的表达水平显著下降,抑制了根中氮素的吸收和同化,并且MeJA 还会导致质体分解和谷氨酸脱氢酶上调,促进叶片中氮的再利用[68]。
4 提高作物氮肥利用效率
提高作物品种的氮肥利用效率,减少氮肥投入和环境污染已成为农业可持续发展中一个艰巨的挑战。影响作物氮利用效率的主要因素包括两个部分:氮吸收效率(nitrogen uptake efficiency, NUpE)和氮利用效率(nitrogen utilization efficiency, NUtE)。氮吸收效率是指成熟农作物地上部分的含氮量与土壤中含氮量的比值;氮利用效率是指单株粒重与成熟农作物地上部分的含氮量之比[69]。一般来说,在高氮条件下,氮吸收效率与氮效率的关联较大,而在低氮条件下,氮利用效率与氮效率的关联性较大[70]。提高氮的吸收、同化效率是提高作物氮效率最有效的途径。例如,高表达OsNRT1.1B能够增加作物产量并提高NUE,因此,将该基因表达水平较高的籼稻等位基因导入到表达水平较低的粳稻品种中,可以显著改良作物的NUE[25]。籼稻类型OsNR2基因编码具有更强活性的硝酸还原酶NR2,将籼型OsNR2等位基因导入到粳稻品种中能有效地提高分蘖数、产量和NUE[71]。野生稻来源的优异等位基因
OsNPF6.1HapB能提高氮吸收和NUE,并在低氮条件下提高水稻的产量[27]。过表达OsNRT1.1A和OsNRT2.3b均可以提高水稻的产量和NUE[28,33];使用OsNAR2.1的启动子驱动OsNRT2.1基因在根和茎中特异性高表达可以显著提高水稻的NUE和谷物产量[72]。OsPTR9过表达后能够促进侧根的形成,提高水稻NUE和产量[73]。OsNPF7.3和OsNPF7.7过表达后能增加水稻的分蘖、提高NUE和产量[74,75]。通过增加亚硝酸还原酶基因OsNiR启动子中的硝酸盐反应顺式元件(nitrate-responsive cis-element, NRE)可以显著地增强其氮同化的功能[76]。使用拟南芥NRT1.7的启动子驱动一个由CHL1和AtNRT1.2嵌合产生的硝酸盐转运蛋白基因NC4N的表达可以激活植物体内硝酸盐的再利用,从而提高NUE和产量[77]。在水稻中过量表达OsAMT1;1能够增强水稻根系对NH4+的吸收,在低浓度的NH4+环境下能促进水稻生长,提高NUE和产量;但在高浓度的NH4+环境下植株会表现出铵中毒[78]。过量表达OsGS1;2可以在高氮条件下协同提高水稻的NUE和产量[79],但也有研究表明过量表达GS后会破坏碳-氮代谢平衡,从而影响水稻的生长和产量[44]。
NUE是一个复杂性状,受植物生命周期中不同的代谢、发育和环境信号网络的协同控制。因此,仅仅通过提高植物的氮素吸收和同化来提高NUE存在着局限性,而通过优化植物氮素响应、代谢信号网络,系统性协调植物的生长发育和代谢过程来提高作物NUE是一个更加理想的策略。DEP1 (dense and erect panicle 1)基因编码的植物G蛋白的γ亚基,其通过与水稻Gα亚基(RGA1)和Gβ亚基(RGB1)互作共同调控植物的氮响应。显性dep1-1等位基因可使植物在营养生长期的地上部生长发育对氮不敏感,但能增强植物根系的氮吸收和同化能力,提高氮肥利用效率,进而提高收获指数和产量[80]。MYB61是联系碳-氮代谢的关键节点,受到OsGRF4的调控,其启动子区域存在一个helitron转座子插入的籼粳分化,将不含转座子插入的、籼稻来源的MYB61导入到粳稻品种中可以同时提高作物的纤维素合成水平和NUE,并在低氮条件下促进增产[53]。ARE1编码一个叶绿体蛋白,其功能缺失突变可延迟植物衰老,在低氮条件下提高NUE,携带启动子小片段插入的ARE1低表达的突变类型可在低氮条件下提高NUE和产量[81]。DNR1能够响应外界的氮信号,通过改变生长素的水平来调控氮代谢,DNR1启动子存在520 bp的片段插入/缺失的籼粳差异,将片段缺失型低表达的籼稻DNR1等位基因导入到粳稻中可以显著提高其NUE和产量[65]。OsTCP19启动子在不同品种中存在29 bp的插入/缺失,缺失型的启动子可以使氮响应负调控因子LBD蛋白高效地结合在该位点并抑制OsTCP19的表达,从而促进水稻的分蘖发育。而在我国现代水稻品种中这一氮高效的变异类型几乎全部丢失,将该等位基因导入现代水稻品种中可以在少施氮肥的条件下显著提高水稻的NUE[67]。DELLA蛋白是“绿色革命”的关键蛋白,其在作物中的高积累量赋予了“绿色革命”品种半矮化、抗倒伏、高产、低NUE等特性,其效应与关键作用因子OsGRF4和NGR5密切相关。OsGRF4的优异等位基因OsGRF4ngr2能显著增加自身的转录水平和蛋白水平,使得DELLA-OsGRF4平衡向OsGRF4丰度增加倾斜,可在不增加作物株高的情况下协同提高作物的光合作用和氮肥利用效率[15];NGR5能响应土壤的氮素水平变化,正向调控水稻的生长发育,提高NGR5的表达能够保持其半矮秆、耐倒伏、多分蘖等高产特性的同时,提升水稻的NUE;在“绿色革命”品种中聚合应用OsGRF4和NGR5优异等位基因可以进一步提高水稻产量和NUE[16]。
5 结语与展望
氮肥是促进作物生长发育和产量提高的重要因素。通过遗传改良培育氮肥高效利用的农作物新品种对农业可持续发展至关重要。利用各种种质资源材料和遗传材料,深入挖掘氮肥利用和吸收效率相关的关键基因、解析其遗传调控网络,并将优异等位基因用于育种,是培育氮高效品种的重要途径。近年来,随着功能基因组学的不断发展,植物的氮吸收、代谢及其信号转导和调控网络得到了不断的完善,越来越多具有改良作物NUE和提升产量潜力的基因被发现,但如何结合植物生长和代谢平衡的调控网络,将这些优良的基因应用到育种实践中,培育出“少投入、多产出、环境友好”的资源高效型作物品种还需要很大努力。总之,随着氮素吸收、代谢和响应的信号传导分子机制的不断解析,必将极大地促进农作物NUE和产量的协同改良、推动新一轮的“绿色革命”。
中国科学院遗传与发育生物学研究所傅向东课题组简介
中国科学院遗传与发育生物学研究所傅向东课题组成立于2005年,课题组长为傅向东研究员。课题组以水稻和拟南芥为研究对象,围绕植物激素与环境因子互作调控植物生长-代谢耦合的分子机制和农作物产量性状形成的遗传调控网络展开研究。在植物发育和环境适应的激素调控机理研究方面取得了一系列研究成果,在国际知名期刊Nature、Science、Nature Genetics、Nature Communications和Cell Research等发表论文50余篇,相关研究成果入选2018年度中国科学十大进展,2019年度中国农业科学十大进展和2020年度中国生命科学十大进展。与此同时,课题组承担了科技部、国家自然科学基金委及中国科学院等一系列重大课题。课题组网站:

(责任编委: 储成才)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1073/pnas.0912953109URL [本文引用: 1]
DOI:10.1016/j.molp.2021.01.002URL [本文引用: 1]
DOI:10.1038/22307URL [本文引用: 1]
DOI:10.1038/416701aURL [本文引用: 1]
DOI:10.1073/pnas.0708013104URL [本文引用: 1]
PMID:12011349 [本文引用: 2]

A dominant dwarf mutant of barley (Hordeum vulgare) that resembles dominant gibberellin (GA) "-insensitive" or "-nonresponsive" mutants in other species is described. alpha-Amylase production by endosperm half-grains of the mutant required GA3 at concentrations about 100 times that of the WT. The mutant showed only a slight growth response to GA3, even at very high concentrations. However, when additionally dwarfed, growth rate responded to GA3 over the normal concentration range, although only back to the original (dwarf) elongation rate. Genetic studies indicated that the dominant dwarf locus was either closely linked or identical to the Sln1 (Slender1) locus. A barley sequence related to Arabidopsis GAI/RGA was isolated, and shown to represent the Sln1 locus by the analysis of sln1 mutants. The dominant dwarf mutant was also altered in this sequence, indicating that it too is an allele at Sln1. Thus, mutations at Sln1 generate plants of radically different phenotypes; either dwarfs that are largely dominant and GA "-insensitive/-nonresponsive," or the recessive slender types in which GA responses appear to be constitutive. Immunoblotting studies showed that in growing leaves, SLN1 protein localized almost exclusively to the leaf elongation zone. In mutants at the Sln1 locus, there were differences in both the abundance and distribution of SLN1 protein, and large changes in the amounts of bioactive GAs, and of their metabolic precursors and catabolites. These results suggest that there are dynamic interactions between SLN1 protein and GA content in determining leaf elongation rate.
PMID:11340177 [本文引用: 2]

The rice slender mutant (slr1-1) is caused by a single recessive mutation and results in a constitutive gibberellin (GA) response phenotype. The mutant elongates as if saturated with GAs. In this mutant, (1) elongation was unaffected by an inhibitor of GA biosynthesis, (2) GA-inducible alpha-amylase was produced by the aleurone layers without gibberellic acid application, and (3) endogenous GA content was lower than in the wild-type plant. These results indicate that the product of the SLR1 gene is an intermediate of the GA signal transduction pathway. SLR1 maps to OsGAI in rice and has significant homology with height-regulating genes, such as RHT-1Da in wheat, D8 in maize, and GAI and RGA in Arabidopsis. The GAI gene family is likely to encode transcriptional factors belonging to the GRAS gene superfamily. DNA sequence analysis revealed that the slr1-1 mutation is a single basepair deletion of the nuclear localization signal domain, resulting in a frameshift mutation that abolishes protein production. Furthermore, introduction of a 6-kb genomic DNA fragment containing the wild-type SLR1 gene into the slr1-1 mutant restored GA sensitivity to normal. These results indicate that the slr1-1 mutant is caused by a loss-of-function mutation of the SLR1 gene, which is an ortholog of GAI, RGA, RHT, and D8. We also succeeded in producing GA-insensitive dwarf rice by transforming wild-type rice with a modified SLR1 gene construct that has a 17-amino acid deletion affecting the DELLA region. Thus, we demonstrate opposite GA response phenotypes depending on the type of mutations in SLR1.
PMID:10713441 [本文引用: 1]

This paper reports isolation and properties of a rice gene, OsGAI, a putative homolog of the GAI of Arabidopsis thaliana. OsGAI encodes a polypeptide of 625 amino acids, which shows 53-55% identity to GAI and RGA from A. thaliana, and 85% identity to wheat rht-D1a and maize d8. Genomic DNA blot analysis indicated the OsGAI to be a single-copy gene in the rice genome. RNA blot hybridization showed that OsGAI transcripts increased within 6h upon GA(3) but not ABA application. This GA-induced increment in OsGAI transcripts did not require de novo protein synthesis. High levels of OsGAI transcripts were detected in nodes, internodes, leaf sheaths and ears of adult plants and leaf sheaths of young seedlings, where GA enhances cell elongation and division. Transiently expressed OsGAI-GFP fusion protein located to the nucleus in onion epidermal cells. Transactivation assays clearly indicated that OsGAI protein is a transcriptional activator or a coactivator.
DOI:10.1038/nature04028URL [本文引用: 1]
DOI:10.1038/nature07546URL [本文引用: 2]
PMID:17644730 [本文引用: 1]

GIBBERELLIN INSENSITIVE DWARF1 (GID1) encodes a soluble gibberellin (GA) receptor that shares sequence similarity with a hormone-sensitive lipase (HSL). Previously, a yeast two-hybrid (Y2H) assay revealed that the GID1-GA complex directly interacts with SLENDER RICE1 (SLR1), a DELLA repressor protein in GA signaling. Here, we demonstrated, by pull-down and bimolecular fluorescence complementation (BiFC) experiments, that the GA-dependent GID1-SLR1 interaction also occurs in planta. GA(4) was found to have the highest affinity to GID1 in Y2H assays and is the most effective form of GA in planta. Domain analyses of SLR1 using Y2H, gel filtration, and BiFC methods revealed that the DELLA and TVHYNP domains of SLR1 are required for the GID1-SLR1 interaction. To identify the important regions of GID1 for GA and SLR1 interactions, we used many different mutant versions of GID1, such as the spontaneous mutant GID1s, N- and C-terminal truncated GID1s, and mutagenized GID1 proteins with conserved amino acids replaced with Ala. The amino acid residues important for SLR1 interaction completely overlapped the residues required for GA binding that were scattered throughout the GID1 molecule. When we plotted these residues on the GID1 structure predicted by analogy with HSL tertiary structure, many residues were located at regions corresponding to the substrate binding pocket and lid. Furthermore, the GA-GID1 interaction was stabilized by SLR1. Based on these observations, we proposed a molecular model for interaction between GA, GID1, and SLR1.
DOI:10.1105/tpc.110.075549URL [本文引用: 1]
DOI:10.1105/tpc.006197URL [本文引用: 1]
DOI:10.1101/gad.11.23.3194URL [本文引用: 1]
DOI:10.1038/s41586-018-0415-5URL [本文引用: 4]
DOI:10.1126/science.aaz2046URL [本文引用: 2]
DOI:10.1074/jbc.M901170200URL [本文引用: 2]
DOI:10.1007/s003440010038URL [本文引用: 1]
DOI:10.1016/j.tplants.2013.08.008URL [本文引用: 1]
DOI:10.1093/emboj/cdg118URL [本文引用: 1]
DOI:10.1126/scisignal.aaa4829URL [本文引用: 2]
DOI:10.1105/tpc.111.092221URL [本文引用: 1]
DOI:10.1111/nph.2012.194.issue-3URL [本文引用: 1]
[本文引用: 1]
DOI:10.1038/ng.3337 [本文引用: 2]

Hu, Bin; Wang, Wei; Ou, Shujun; Tang, Jiuyou; Li, Hua; Che, Ronghui; Zhang, Zhihua; Wang, Hongru; Wang, Yiqin; Liang, Chengzhen; Xu, Chi; Liang, Yan; Chu, Chengcai Chinese Acad Sci, Inst Genet & Dev Biol, Natl Ctr Plant Gene Res Beijing, State Key Lab Plant Gen, Beijing, Peoples R China. Wang, Wei; Li, Hua; Zhang, Zhihua; Wang, Hongru; Liang, Yan Univ Chinese Acad Sci, Coll Life Sci, Beijing, Peoples R China. Ou, Shujun Michigan State Univ, Dept Hort, E Lansing, MI 48824 USA. Chai, Xuyang Capital Normal Univ, Coll Life Sci, Beijing, Peoples R China. Liu, Linchuan Chinese Acad Sci, Shanghai Ctr Plant Stress Biol, Shanghai, Peoples R China. Piao, Zhongze Shanghai Acad Agr Sci, Crop Breeding & Cultivat Res Inst, Shanghai, Peoples R China. Deng, Qiyun China Natl Hybrid Rice Res, Changsha, Peoples R China. Deng, Qiyun Dev Ctr, Changsha, Peoples R China. Deng, Kun; Zhang, Lianhe Henan Univ Sci & Technol, Sch Agr, Luoyang, Peoples R China.
DOI:10.1038/s41587-019-0104-4URL [本文引用: 1]
DOI:10.1038/s41467-019-13187-1URL [本文引用: 3]
DOI:10.1105/tpc.17.00809URL [本文引用: 2]
DOI:10.1038/srep09635URL [本文引用: 1]
PMID:10677431 [本文引用: 1]

Elucidating how rice (Oryza sativa) takes up nitrate at the molecular level could help improve the low recovery rate (<50%) of nitrogen fertilizer in rice paddies. As a first step toward that goal, we have cloned a nitrate transporter gene from rice called OsNRT1. OsNRT1 is a new member of a growing transporter family called PTR, which consists not only of nitrate transporters from higher plants that are homologs of the Arabidopsis CHL1 (AtNRT1) protein, but also peptide transporters from a wide variety of genera including animals, plants, fungi, and bacteria. However, despite the fact that OsNRT1 shares a higher degree of sequence identity with the two peptide transporters from plants (approximately 50%) than with the nitrate transporters (approximately 40%) of the PTR family, no peptide transport activity was observed when OsNRT1 was expressed in either Xenopus oocytes or yeast. Furthermore, contrasting the dual-affinity nitrate transport activity of CHL1, OsNRT1 displayed only low-affinity nitrate transport activity in Xenopus oocytes, with a K(m) value of approximately 9 mM. Northern-blot and in situ hybridization analysis indicated that OsNRT1 is constitutively expressed in the most external layer of the root, epidermis and root hair. These data strongly indicate that OsNRT1 encodes a constitutive component of a low-affinity nitrate uptake system for rice.
DOI:10.1093/jxb/erq403URL [本文引用: 1]
DOI:10.1111/nph.2014.204.issue-1URL [本文引用: 1]
DOI:10.1073/pnas.1525184113URL [本文引用: 2]
DOI:10.1007/s00299-017-2153-9URL [本文引用: 1]
PMID:15821986 [本文引用: 1]

Ammonium transport is a key process in nitrogen metabolism. In the green alga Chlamydomonas, we have characterized molecularly the largest family of ammonium transporters (AMT1) so far described consisting of eight members. CrAmt1 genes have an interesting transcript structure with some very small exons. Differential expression patterns were found for each CrAmt1 gene in response to the nitrogen source by using Real Time PCR. These expression patterns were similar under high and low CO2 atmosphere. CrAmt1;1 expression was characterized in detail. It was repressed in both ammonium and nitrate medium, and strongly expressed in nitrogen-free media. Treatment with a Glutamine synthetase inhibitor released partially repression in ammonium and nitrate suggesting that ammonium and its derivatives participate in the observed repressing effects. By studying CrAmt1;1 expression in mutants deficient at different steps of the nitrate assimilation pathway, it has been shown that nitrate has a double negative effect on this gene expression; one related to its reduction to ammonium, and a second one by itself. This second effect of nitrate was dependent on the functionality of the regulatory gene Nit2, specific for nitrate assimilation. Thus, NIT2 would have a dual role on gene expression: the well-known positive one on nitrate assimilation and a novel negative one on Amt1;1 regulation.
DOI:10.1093/pcp/pcg083URL [本文引用: 2]
DOI:10.1105/tpc.013516URL [本文引用: 1]
PMID:10330477 [本文引用: 1]

Ammonium and nitrate are the prevalent nitrogen sources for growth and development of higher plants. 15N-uptake studies demonstrated that ammonium is preferred up to 20-fold over nitrate by Arabidopsis plants. To study the regulation and complex kinetics of ammonium uptake, we isolated two new ammonium transporter (AMT) genes and showed that they functionally complemented an ammonium uptake-deficient yeast mutant. Uptake studies with 14C-methylammonium and inhibition by ammonium yielded distinct substrate affinities between </=0.5 and 40 microM. Correlation of gene expression with 15NH4+ uptake into plant roots showed that nitrogen supply and time of day differentially regulated the individual carriers. Transcript levels of AtAMT1;1, which possesses an affinity in the nanomolar range, steeply increased with ammonium uptake in roots when nitrogen nutrition became limiting, whereas those of AtAMT1;3 increased slightly, with AtAMT1;2 being more constitutively expressed. All three ammonium transporters showed diurnal variation in expression, but AtAMT1;3 transcript levels peaked with ammonium uptake at the end of the light period, suggesting that AtAMT1;3 provides a link between nitrogen assimilation and carbon provision in roots. Our results show that high-affinity ammonium uptake in roots is regulated in relation to the physiological status of the plant at the transcriptional level and by substrate affinities of individual members of the AMT1 gene family.
PMID:10675553 [本文引用: 1]

We have cloned and characterized the first member of a novel family of ammonium transporters in plants: AtAMT2 from Arabidopsis thaliana. AtAMT2 is more closely related to bacterial ammonium transporters than to plant transporters of the AMT1 family. The protein was expressed and functionally characterized in yeast. AtAMT2 transported ammonium in an energy-dependent manner. In contrast to transporters of the AMT1 family, however, AtAMT2 did not transport the ammonium analogue, methylammonium. AtAMT2 was expressed more highly in shoots than roots and was subject to nitrogen regulation.
DOI:10.1105/tpc.107.052134URL [本文引用: 1]
DOI:10.1016/j.jgg.2016.11.001URL [本文引用: 1]
DOI:10.1007/s11033-011-0972-2URL [本文引用: 1]
DOI:10.1093/aob/mcq028URL [本文引用: 1]
DOI:10.1371/journal.pone.0095581URL [本文引用: 2]
PMID:15918879 [本文引用: 1]

Rice (Oryza sativa L.) plants possess three homologous but distinct genes for cytosolic glutamine synthetase (GS1): these are OsGS1;1, OsGS1;2, and OsGS1;3. OsGS1;1 was expressed in all organs tested with higher expression in leaf blades, while OsGS1;2, and OsGS1;3 were expressed mainly in roots and spikelets, respectively. We characterized knockout mutants caused by insertion of endogenous retrotransposon Tos17 into the exon-8 (lines ND8037 and ND9801) or the exon-10 (line NC2327) of OsGS1;1. Mendelian segregation occurred in each progeny. Homozygously inserted mutants showed severe retardation in growth rate and grain filling when grown at normal nitrogen concentrations. Abnormal mRNA for GS1;1 was transcribed, and the GS1 protein and its activity in the leaf blades were barely detectable in these mutants. The glutamine pool in the roots and leaf blades of the mutants was lower than that of the wild type. Re-introduction of OsGS1;1 cDNA under the control of its own promoter into the mutants successfully complemented these phenotypes. Progeny where Tos17 was heterozygously inserted or deleted during segregation showed normal phenotypes. The results indicate that GS1;1 is important for normal growth and grain filling in rice; GS1;2 and GS1;3 were not able to compensate for GS1;1 function.
PMID:16665193 [本文引用: 1]

Eight mutants of barley (Hordeum vulgare cv Maris Mink) lacking the chloroplast isozyme of glutamine synthetase (EC 6.3.1.2.) were isolated by their inability to grow under photorespiratory conditions. The cytoplasmic isozyme of glutamine synthetase was present in the leaves of all the mutants, with activities comparable to the wild-type (10-12 nanokatals per gram fresh weight). The mutant plants developed normally and were fully fertile under conditions that minimize photorespiration. In 1% O(2) the rate of CO(2) fixation in leaves of one of the mutants, RPr 83/32, was the same as the wild-type, but in air this rate declined to 60% of the wild-type after 30 minutes. During this time the ammonia concentration in leaves of the mutant rose from 1 to 50 micromoles per gram fresh weight. Such ammonia accumulation in air was found in all the mutant lines. In back-crosses with the parent line, F(1) plants were viable in air. In the F(2) generation, nonviability in air and the lack of chloroplast glutamine synthetase co-segregated, in both the lines tested. These two lines and four others proved to be allelic; we designate them gln 2a-f. The characteristics of these mutants conclusively demonstrate the major role of chloroplast glutamine synthetase in photorespiration and its associated nitrogen recycling.
[本文引用: 1]
DOI:10.1038/ncomms2621PMID:23511481 [本文引用: 1]

In plants, nitrate is not only a major nitrogen source but also a signalling molecule that modulates the expression of a wide range of genes and that regulates growth and development. The critical role of nitrate as a signalling molecule has been established for several decades. However, the molecular mechanisms underlying the nitrate response have remained elusive, as the transcription factor that primarily responds to nitrate signals has not yet been identified. Here we show that Arabidopsis NIN-LIKE PROTEIN (NLP) family proteins bind the nitrate-responsive cis-element and activate nitrate-responsive cis-element-dependent and nitrate-responsive transcription. Our results also suggest that the activity of NLPs is post-translationally modulated by nitrate signalling. Furthermore, the suppression of NLP function impairs the nitrate-inducible expression of a number of genes and causes severe growth inhibition. These results indicate that NLPs are the transcription factors mediating the nitrate signal and thereby function as master regulators of the nitrate response.
DOI:10.1038/nature22077URL [本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1111/pbi.v17.9URL [本文引用: 1]
DOI:10.1038/s41467-020-19019-xURL [本文引用: 2]
DOI:10.1073/pnas.1721487115URL [本文引用: 1]
DOI:10.1038/s41586-018-0656-3URL [本文引用: 2]
DOI:10.1038/nrm3838URL [本文引用: 1]
DOI:10.1073/pnas.0709559105URL [本文引用: 1]
DOI:10.1073/pnas.0909571107URL [本文引用: 1]
DOI:10.1104/pp.109.139139URL [本文引用: 1]
DOI:10.1111/tpj.2014.78.issue-1URL [本文引用: 1]
DOI:10.1111/nph.14938URL [本文引用: 1]
DOI:10.1007/s11103-008-9417-2URL [本文引用: 1]
DOI:10.1104/pp.19.00346PMID:31431511 [本文引用: 1]

The dual-affinity nitrate transceptor NITRATE TRANSPORTER1.1 (NRT1.1) has two modes of transport and signaling, governed by Thr-101 (T101) phosphorylation. NRT1.1 regulates lateral root (LR) development by modulating nitrate-dependent basipetal auxin export and nitrate-mediated signal transduction. Here, using the Arabidopsis () NRT1.1 phosphomimetic and NRT1.1 nonphosphorylatable mutants, we found that the phosphorylation state of NRT1.1 plays a key role in NRT1.1 function during LR development. Single-particle tracking revealed that phosphorylation affected NRT1.1 spatiotemporal dynamics. The phosphomimetic NRT1.1 form showed fast lateral mobility and membrane partitioning that facilitated auxin flux under low-nitrate conditions. By contrast, nonphosphorylatable NRT1.1 showed low lateral mobility and oligomerized at the plasma membrane (PM), where it induced endocytosis via the clathrin-mediated endocytosis and microdomain-mediated endocytosis pathways under high-nitrate conditions. These behaviors promoted LR development by suppressing NRT1.1-controlled auxin transport on the PM and stimulating Ca-ARABIDOPSIS NITRATE REGULATED1 signaling from the endosome. © 2019 American Society of Plant Biologists. All Rights Reserved.
DOI:10.1111/tpj.2014.78.issue-1URL [本文引用: 1]
DOI:10.1093/plcell/koaa037URL [本文引用: 2]
PMID:11851911 [本文引用: 1]

The formation of lateral roots (LR) is a major post-embryonic developmental event in plants. In Arabidopsis thaliana, LR development is inhibited by high concentrations of NO3(-). Here we present strong evidence that ABA plays an important role in mediating the effects of NO3(-) on LR formation. Firstly, the inhibitory effect of NO3(-) is significantly reduced in three ABA insensitive mutants, abi4-1, abi4-2 and abi5-1, but not in abi1-1, abi2-1 and abi3-1. Secondly, inhibition by NO3(-) is significantly reduced, but not completely abolished, in four ABA synthesis mutants, aba1-1, aba2-3, aba2-4 and aba3-2. These results indicate that there are two regulatory pathways mediating the inhibitory effects of NO3(-) in A. thaliana roots. One pathway is ABA-dependent and involves ABI4 and ABI5, whereas the second pathway is ABA-independent. In addition, ABA also plays a role in mediating the stimulation of LR elongation by local NO3(-) applications.
DOI:10.1038/s41586-020-03091-wURL [本文引用: 2]
DOI:10.1111/pce.v42.2URL [本文引用: 1]
DOI:10.2134/agronj1982.00021962007400030037xURL [本文引用: 1]
PMID:16660579 [本文引用: 1]

The uptake of nitrate and ammonium by rice (Oryza sativa) seedlings was compared under various conditions. Nitrate uptake showed a 1-hour lag phase and then a rapid absorption phase, whereas ammonium uptake showed passive absorption during the first hour, then a shoulder of absorption, followed by a rapid metabolism-dependent absorption phase. Light did not affect the uptake of nitrate or ammonium. The uptake of nitrate and ammonium was markedly suppressed by removal of the endosperm. After removal of the endosperm, the uptake was restored by exogenous supply of 30 mm sucrose. No appreciable nitrate uptake was detected at temperatures below 15 C, whereas appreciable ammonium uptake occurred at 15 C, although lowered passive absorption and complete inhibition of the rapid metabolism-dependent absorption phase were observed at 5 C.Nitrate uptake was decreased by aeration and also by bubbling the mixture with O(2). It was greatly depressed by bubbling the mixture with N(2). These results suggest that there is a low optimal O(2) concentration for nitrate uptake. In contrast, ammonium uptake was increased by aeration, not influenced by bubbling with O(2), and slightly decreased by bubbling the mixture with N(2).Nitrate uptake was almost entirely inhibited by addition of cycloheximide, KCN, or arsenate. These compounds had scarcely any effect on the passive absorption of ammonium, but entirely depressed its successive metabolism-dependent absorption. Ammonium uptake occurred before nitrate uptake in solution containing both nitrate and ammonium.
DOI:10.1038/s41467-019-13110-8URL [本文引用: 1]
DOI:10.1111/pbi.2016.14.issue-8URL [本文引用: 1]
DOI:10.1111/pbi.2013.11.issue-4URL [本文引用: 1]
DOI:10.3389/fpls.2017.01338URL [本文引用: 1]
DOI:10.3389/fpls.2018.00300URL [本文引用: 1]
DOI:10.1111/pbi.v19.1URL [本文引用: 1]
[本文引用: 1]
DOI:10.1093/jxb/ert458PMID:24420570 [本文引用: 1]

The major source of nitrogen for rice (Oryza sativa L.) is ammonium (NH4(+)). The NH4(+) uptake of roots is mainly governed by membrane transporters, with OsAMT1;1 being a prominent member of the OsAMT1 gene family that is known to be involved in NH4(+) transport in rice plants. However, little is known about its involvement in NH4(+) uptake in rice roots and subsequent effects on NH4(+) assimilation. This study shows that OsAMT1;1 is a constitutively expressed, nitrogen-responsive gene, and its protein product is localized in the plasma membrane. Its expression level is under the control of circadian rhythm. Transgenic rice lines (L-2 and L-3) overexpressing the OsAMT1;1 gene had the same root structure as the wild type (WT). However, they had 2-fold greater NH4(+) permeability than the WT, whereas OsAMT1;1 gene expression was 20-fold higher than in the WT. Analogous to the expression, transgenic lines had a higher NH4(+) content in the shoots and roots than the WT. Direct NH4(+) fluxes in the xylem showed that the transgenic lines had significantly greater uptake rates than the WT. Higher NH4(+) contents also promoted higher expression levels of genes in the nitrogen assimilation pathway, resulting in greater nitrogen assimilates, chlorophyll, starch, sugars, and grain yield in transgenic lines than in the WT under suboptimal and optimal nitrogen conditions. OsAMT1;1 also enhanced overall plant growth, especially under suboptimal NH4(+) levels. These results suggest that OsAMT1;1 has the potential for improving nitrogen use efficiency, plant growth, and grain yield under both suboptimal and optimal nitrogen fertilizer conditions.
DOI:10.1111/ppl.2011.141.issue-4URL [本文引用: 1]
DOI:10.1038/ng.2958URL [本文引用: 1]
DOI:10.1038/s41467-017-02781-wURL [本文引用: 1]
