 ,1,2,3
,1,2,3Advances of functional consequences and regulation mechanisms of alternative cleavage and polyadenylation
Haidong Xu1,2,3, Bolin Ning1,2,3, Fang Mu1,2,3, Hui Li1,2,3, Ning Wang ,1,2,3
,1,2,3通讯作者: 王宁,博士,教授,博士生导师,研究方向:动物遗传育种与繁殖。E-mail:wangning@neau.edu.cn
编委: 杨昭庆
收稿日期:2020-07-7修回日期:2020-10-19网络出版日期:2021-01-20
| 基金资助: |
Received:2020-07-7Revised:2020-10-19Online:2021-01-20
| Fund supported: |
作者简介 About authors
徐海冬,在读博士研究生,专业方向:动物遗传育种与繁殖。E-mail:

摘要
真核生物基因的前体mRNA (pre-mRNA)及一些lncRNA在成熟过程中其3'端会发生剪切和多聚腺苷酸化反应(cleavage and polyadenylation, C/P),C/P的发生需要多聚腺苷酸化信号(polyadenylation signal, PAS)的存在。选择性多聚腺苷酸化(alternative cleavage and polyadenylation, APA)是指具有多个PAS的基因,在其mRNA 3?端成熟过程中,由于选择不同的PAS,导致产生出多个3'UTR长度和序列组成不同的转录异构体。3?UTR长度和序列的不同会影响mRNA的稳定性、翻译效率、运输和细胞定位等,因此APA是真核生物的一个重要转录后调控方式。近年来,对大量动物、植物及酵母的基因组测序分析发现,APA在真核生物广泛存在,针对APA的生物学效应和调控机制开展了一系列研究。目前已鉴定出许多APA调控的顺式调控元件和反式作用因子。本文重点介绍了APA生物学效应和调控机制的最新研究进展,并探讨了未来APA调控的研究方向。
关键词:
Abstract
During the maturation of pre-mRNAs and some lncRNAs, their 3'ends are cleaved and polyadenylated. The cleavage and polyadenylation (C/P) require the presence of a polyadenylation signal (PAS) at the RNA 3?end. Most eukaryotic genes have multiple PASs, resulting in alternative cleavage and polyadenylation (APA). APA leads to transcript isoforms with different coding potentials and/or variable 3?UTRs. The 3'UTR affects mRNA stability, translation, transportation, and cellular localization. Therefore, APA is an important mechanism of posttranscriptional gene regulation in eukaryotes. In recent years, whole genome sequencing of animals, plants and yeast has revealed that APA is pervasive in eukaryotes, and the functional consequences and regulation of APA have been studied. To date, many cis-acting regulatory elements and trans-acting factors for APA regulation have been identified. In this review, we summarize the recent advances in the functional consequences and regulation of APA and discuss the future directions, aiming to provide clues and references for future APA study.
Keywords:
PDF (793KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
徐海冬, 宁博林, 牟芳, 李辉, 王宁. 选择性多聚腺苷酸化的生物学效应及其调控机制研究进展. 遗传[J], 2021, 43(1): 4-15 doi:10.16288/j.yczz.20-200
Haidong Xu.
真核生物的mRNA前体(pre-RNA)和一些长链非编码RNA前体(pre-lncRNA)在成熟过程中其3?端会被核酸内切酶剪切(cleavage),然后由模板非依赖的RNA聚合酶—poly(A)聚合酶(PAP)在3?端合成一个多聚腺苷酸尾巴(polyadenylation)。真核生物mRNA前体3?端的剪切和多聚腺苷酸化反应(cleavage and polyadenylation, C/P)需要多聚腺苷酸化信号(polyadenylation signal,PAS),PAS通常位于剪切位点上游10~30个核苷酸处。选择性多聚腺苷酸化(alternative cleavage and polyadenylation, APA)是指含有多个PAS的基因,在mRNA 3?端成熟过程中,不同PAS的选择利用将导致一个基因产生多个转录异构体,这些转录异构体具有长短不一的3'末端[1]。近年来,由于高通量测序技术的发展和研究的深入,人们发现APA在真核生物中普遍存在,例如人类超过70%的基因都存在APA现象[2]。APA在不同类型的基因中分布不同,其中,在蛋白编码基因中分布最为广泛,其次为lncRNA和假基因,而在小RNA (small RNA)和微小RNA (microRNA)中的分布最少[3]。大多数APA仅会导致mRNA含有不同长度的3?UTR,但有少数APA还会改变mRNA编码区序列,导致编码不同的蛋白。3?UTR存在大量基因表达的顺式调控元件,这些顺式调控元件广泛参与mRNA的稳定性、翻译、运输和细胞定位[1]。APA使得同一基因的不同转录本具有不同的顺式调控元件,因而导致不同转录本的表达不同。目前已知APA是真核生物基因表达调控的重要调控机制之一,它在许多生物学过程中发挥重要的调控作用;其调控异常会导致癌症等多种疾病发生[4,5,6,7]。近年来,由于APA功能的多样性及其调控的重要性,针对APA的研究引起了人们广泛关注。本文综述了APA调控研究的最新进展,旨在为深入解析APA的调控机制和生物学效应提供参考。
1 C/P反应的顺式调控元件和反式作用因子
C/P反应是mRNA前体成熟的一个重要步骤,关系到mRNA的核外运输和胞质翻译过程。C/P反应需要位于mRNA前体的顺式元件及许多反式作用因子的参与。目前已鉴定出许多C/P反应的顺式调控元件、反式作用因子以及多种核酸酶(图1)[1,8~10]。图1
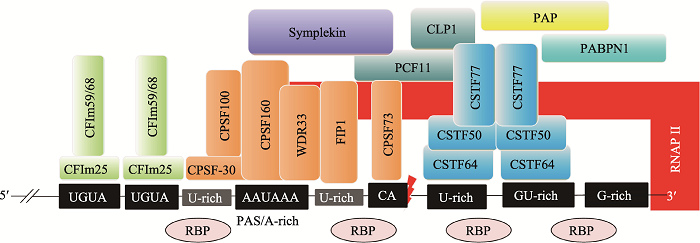 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1C/P反应的顺式调控元件和反式作用因子
CPSF:剪切多聚腺苷酸化特异因子(cleavage and polyadenylation specificity factor);CSTF:剪切激活因子(cleavage stimulation factor);CFIIm:剪切因子复合物I (cleavage factor I m);RBP:RNA结合蛋白(RNA binding protein);RNAP II:RNA聚合酶II(RNA polymerase II);PAP:poly(A)聚合酶(poly(A) polymerase);PABPN1:核poly(A)结合蛋白(nuclear poly(A)-binding protein 1);WDR33:WD重复域33 (WD repeat domain 33);FIP1:PAP互作因子(factor interacting with PAP);CLP1:C/P因子亚基1 (cleavage factor polyribonucleotide kinase subunit 1);PCF11:C/P因子亚基(cleavage and polyadenylation factor subunit);PAS:多聚腺苷酸化信号(polyadenylation signal)。根据参考文献[1, 8~10]总结绘制。
Fig. 1The cis-acting regulatory elements and trans-acting factors involved in cleavage and polyadenylation (C/P)
1.1 C/P反应的顺式调控元件
PAS是mRNA前体进行C/P反应的信号,它位于mRNA前体的3?端。根据PAS的序列特征和功能,可将其组合并归类为核心元件和辅助元件两部分。其中,核心元件包括距剪切位点上游约20~22个核苷酸处的A-rich六聚体、剪切位点下游约10~30个核苷酸处的U/GU-rich元件和临近剪切位点的CA (或UA)序列[9]。AAUAAA是典型的A-rich六聚体序列,其单碱基突变所形成的非经典序列仍具有AAUAAA的部分活性[5]。下游U/GU-rich元件的单个核苷酸突变体同样也具有一定的活性。CA (或UA)是临近剪切位点前常见的两个核苷酸,它们控制剪切效率。辅助元件包括剪切位点下游的G-rich元件和剪切位点上游的UGUA元件[9]。其中G-rich元件影响剪切效率;UGUA元件促进C/P反应[10]。此外,剪切位点上游的U-rich元件也参与剪切调控。U-rich元件有两类:一类是位于A-rich六聚体和剪切位点之间的U-rich元件;另一类是位于A-rich六聚体上游的U-rich元件,这两种U-rich元件都可以与CPSF复合体互作,促进C/P反应[10]。1.2 C/P反应的反式作用因子
目前已知有4种蛋白复合物和多个蛋白质单体参与C/P反应。参与C/P反应的4种蛋白质复合物分别是剪切多聚腺苷酸化特异因子(cleavage and polyadenylation specificity factor, CPSF)复合物、剪切激活因子(cleavage stimulation factor, CSTF)复合物、剪切因子复合物I(cleavage factor I m, CFIm)和剪切因子复合物II(cleavage factor II m, CFIIm)[1,9,11]。CPSF复合物主要由CPSF160 (CPSF1)、CPSF100 (CPSF2)、CPSF73 (CPSF3)、CPSF30 (CPSF4)、PAP互作因子(factor interacting with PAP, FIP1)和WD重复域33 (WD repeat domain 33, WDR33) 6种蛋白构成[9,11]。CPSF160通过β螺旋结构域特异性结合AAUAAA[12];CPSF30的N端含有5个保守的锌指结构域,其中第2个和第3个锌指结构域能与PAS上游的U-rich特异性结合;CPSF73是一种锌依赖性核酸内切酶,其金属-β-内酰胺酶结构域和β-CASP结构域可精准识别CA(或UA)位点并对其临近的位点进行精准剪切[9]。FIP1的C端精氨酸富集区可与PAS下游的U-rich序列结合,并同时招募PAP[9,12]。WDR33具有富含β螺旋的WD40保守结构域,能与AAUAAA直接结合,在蛋白-蛋白和蛋白-RNA互作中发挥重要功能[12,13]。
CSTF复合物是由CSTF77 (CSTF1)、CSTF64 (CSTF2)和CSTF50 (CSTF3) 3种蛋白质亚基组成的六聚体复合物[1,11]。CSTF64和CSTF64τ (CSTF64的旁系同源体)的N端具有U-/GU-rich的识别基序(RNA recognition motif, RRM)[9]。CSTF77蛋白C端的脯氨酸富集结构域(monkey-tail, MT)与RRM附近的铰链区(hinge结构域)相互作用,促进CSTF64与RNA的结合[9];CSTF77的N端具有一个螺旋HAT结构域,可分别结合CSTF64和CSTF50,可作为支架蛋白为其他反式作用因子提供结合平台。CSTF50的保守性较低,仅存在于少数高等动物中,可通过WD40结构域与CSTF77的MT结构域相互作用,也与RNAP II的羧基末端域(carboxy-terminal domain, CTD)互作,增强其他反式作用因子的活性[1]。
CFIm复合物主要包括CFIm25、CFIm59(或CFIm68)[9,11]。CFIm25具有RRM结构域,可特异性识别PAS上游的UGUA元件,进而招募CFIm59/ CFIm68,形成CFIm复合物[9]。CFIm59的RRM结构域与CFIm25二聚体结合,增强CFIm25的RNA结合活性,同时也可促进RNA环化[10]。CFIIm复合物主要包括PCF11 (PCF11 cleavage and polyadenylation factor subunit)和CLP1 (CLP1 cleavage factor polyribonucleotide kinase subunit 1)两种蛋白质,PCF11结合在PAS的下游,在RNAP II转录终止中发挥作用,CLP1可作为连接CPSF复合物和CFIm复合物的衔接蛋白[11]。
参与C/P反应的蛋白质单体包括偶对蛋白(symplekin)、RNA聚合酶II(RNA polymerase II,RNAP II)、poly(A)聚合酶(poly(A) polymerase, PAP)和核poly(A)结合蛋白(nuclear poly(A)-binding protein 1, PABPN1)等。RNAP II的C端结构域(CTD)能招募多个蛋白复合物,为C/P反应提供酶促反应的平台。
另外,有些反式作用因子蛋白本身不能直接结合RNA,但它们可以作为支架蛋白结合上述各复合物,从而发挥调控C/P的作用,例如,Symplekin作为连接CPSF和CSTF复合物的连接蛋白。
1.3 转录与C/P反应
真核生物基因的转录延伸、RNA前体的选择性拼接以及C/P反应相偶联。RNAP II在转录延伸、选择性拼接以及C/P反应中都发挥重要作用。RNAP II的N端控制RNA的合成,RNAP II的CTD结构域可募集C/P反应所需的多种反式作用因子,为mRNA前体的C/P反应提供平台。RNAP II的CTD结构域与C/P反应的反式作用因子的结合是一个动态过程,其中CPSF复合物中的WDR33和CPSF30在其他蛋白质的参与下特异性识别AAUAAA保守基序[9]。同时CFIm25和CSTF64分别与PAS上游UGUA元件和下游U/GU-rich元件等保守基序特异性结合,并在CSTF77、Symplekin、CFIm68/CFIm59和FIP1等蛋白的作用下,CSTF、CPSF和CFIm等形成一个复合体,随后CPSF73对PAS下游剪切位点进行精准剪切[9]。CPSF和FIP1互作可招募PAP,招募的PAP会在剪切后的前体mRNA的3'端缓慢合成10~15个腺苷酸残基。此后,PABPN1会结合到短的poly(A)序列上,同时锚定PAP,快速合成poly(A),形成长约200个腺苷酸的poly(A)尾巴[1]。转录延伸与C/P反应偶联保证了RNA的精细化加工,这是一种快速精确的调控方式,以最小的耗能方式为RNA转录加工提供保障。1.4 胞质多聚腺苷酸化
除了上述细胞核内多聚腺苷酸化,还存在胞质多聚腺苷酸化(cytoplasmic polyadenylation)。与核内多聚腺苷酸化不同,胞质多聚腺苷酸化发生于胞质内成熟的mRNA,它不与转录相偶联,也不存在剪切反应。这类mRNA没有翻译活性,它们的poly(A)尾巴通常都很短,大约有20多个核苷酸。胞质多聚腺苷酸化能延长这类mRNA的poly(A)尾巴,从而激活其翻译活性。胞质多聚腺苷酸化是调控mRNA翻译的一个重要机制,多见于卵子发生和早期发育胚胎。例如,在卵母细胞受精后,卵母细胞质存留的无翻译活性的mRNA因发生胞质多聚腺苷酸化而激活其活性,表达出合子基因组表达所需的酶和转录因子[14,15]。在胚胎期晶状体纤维分化过程中,由于晶状体蛋白相关基因的mRNA发生胞质多聚腺苷酸化,从而提高了这些mRNA的翻译效率[16]。与核内多聚腺苷酸化相同,胞质多聚腺苷酸化也同样受到许多顺式调控元件和反式作用因子的调控,其中,顺式调控元件包括胞质多聚腺苷酸化元件(cytoplasmic polyadenylation element, CPE)、CPE上游的Pumilio结合元件(pumilio binding element, PBE)以及CPE下游的PAS[17,18];已知的反式作用因子有CPE结合蛋白(CPE binding proteins, CPEBs)、Pumilio蛋白以及CPSF160/100/30[19]。反式作用因子CPEBs能结合CPE元件,Pumilio能结合PBE元件,而CPSF160/100/30能结合PAS元件。非磷酸化的CPEB1结合Pumilio,招募脱腺苷化酶和翻译起始抑制因子,进而抑制翻译;但磷酸化的CPEB1会与脱腺苷酸化酶和翻译起始抑制因子解离,转而与CPSF互作,从而招募胞质poly(A)聚合酶PAPD4(GLD-2),快速合成80~250个核苷酸的poly(A)尾巴[17]。胞质poly(A)结合蛋白(cytoplasmic poly(A)-binding protein 1, PABPC1)结合poly(A)尾巴,它同时与mRNA 5?端的帽结构复合物亚基eIF4G互作,形成闭锁环复合体(closed loop complex),从而促进翻译[19]。胞质多聚腺苷酸化与核内多聚腺苷酸化的调控有相似之处,也有不同之处。两者的相似之处,例如,两者都需要顺式调控元件PAS和反式作用因子CPSF和symplekin的参与。两者的不同之处,例如,与核内多聚腺苷酸化不同,具有多个PAS的mRNA在胞质多聚腺苷酸化中,其近端PAS可远程调控mRNA 3'末端的多聚腺苷酸化[18]。
2 APA的生物学效应
根据所在位置不同,PAS可以分为非编码区PAS (untranslated region PAS, UTR-PAS)和编码区PAS (coding region PAS, CR-PAS)两大类[1,20],其中以UTR-PAS最为多见。一个基因如果有多个PAS,不同PAS的选择利用会使同一个基因产生3?末端序列组成和长度均不同的转录本。基因如果选择不同的UTR-PAS,形成的转录异构体之间3?UTR的序列组成和长度均存在差异,但所编码的蛋白质序列不会改变[20]。如果选择位于外显子或内含子的CR-PAS,所形成的转录本会编码出截短型蛋白质(图2)[1,20]。真核生物mRNA的3?UTR存在许多的基因表达顺式调控元件,例如miRNA、lncRNA和RNA结合蛋白(RNA binding protein, RBP)的结合位点(图2)。这些调控元件在mRNA稳定性、运输、翻译以及细胞定位中发挥重要作用。APA导致同一个基因产生多个具有不同3?UTR的转录本,由于3?UTR的差异,使得这些转录本具有不同的mRNA稳定性、翻译效率、运输以及细胞定位,并最终导致它们具有不同的生物学功能。
2.1 APA产生差异性蛋白亚型
基因利用CR-PAS会编码出C端不同的蛋白异构体,有些蛋白异构体可能会缺失跨膜区或DNA结合区等重要功能结构域(图2),不同蛋白异构体的功能可能相近也可能相反[1]。有研究发现,鸡生长激素受体(growth hormone receptor, GHR)基因存在CR-PAS和UTR-PAS。GHR基因利用UTR-PAS会产生完整的GHR蛋白,完整的GHR定位于细胞膜,作为GH-GHR-IGF1信号通路的重要分子;而利用CR-PAS则会形成疏水跨膜域缺失的分泌蛋白—生长激素结合蛋白(growth hormone binding protein, GHBP)[21,22],主要参与维持体内GH水平的稳定[23]。在B细胞和T细胞激活过程中,多数基因选择利用CR-PAS,使得编码的蛋白质由膜锚定型转向分泌型[24]。视网膜母细胞瘤结合蛋白(retinoblastoma- binding protein 6, RBBP6)利用CR-PAS产生截短型蛋白ISO6,它与全长的RBBP6竞争性参与PAS剪切[25]。图2
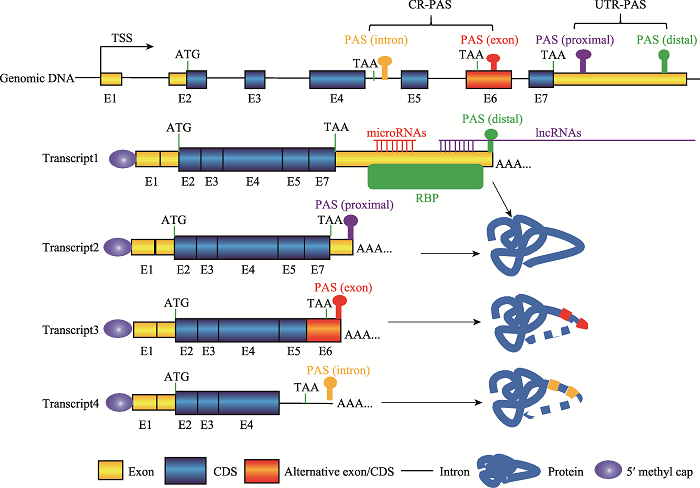 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2PAS类型及其APA示意图
PAS:多聚腺苷酸化信号(polyadenylation signal);CR-PAS:编码区PAS (coding region PAS);UTR-PAS:非编码区PAS (untranslated region PAS);TSS:转录起始位点(transcriptional start site);CDS:蛋白质编码区(coding sequence);RBP:RNA结合蛋白(RNA binding protein);lncRNA:长链非编码RNA (long non-coding RNA)。
Fig. 2Schematic of different types of PAS and the APA events
2.2 APA影响mRNA的稳定性
绝大多数APA可导致同一个基因转录出多个不同长度的3?UTR转录本。因为3?UTR存在调控稳定性的顺式调控元件,如miRNA的结合位点(图2),因此,同一个基因的不同转录本具有不同的稳定性[9]。一般来说,短3?UTR的转录本具有较高的稳定性和翻译效率,这类转录本多见于增殖期细胞;而长的3?UTR转录本则有利于转录后的精细化调控,这类转录本多见于细胞分化和组织发育等复杂生物学过程[26]。干扰素调节因子5 (interferon regulatory factor 5, IRF5)是系统性红斑狼疮的候选基因。双链RNA结合蛋白STAU1能结合IRF5长3?UTR亚型转录本的Alu元件,导致IRF5 mRNA快速降解;而IRF5短3?UTR亚型的转录本不存在Alu元件,因此稳定性较高[27]。Pax3是调节肌细胞和肝细胞增殖的关键基因,其长3?UTR亚型转录本含有miR-206结合位点,而其短3?UTR亚型转录本没有,因此Pax3的短3?UTR亚型转录本可逃避miR-206介导的基因沉默,稳定性较高[28,29]。
尽管短3?UTR亚型转录本可显著提升mRNA的翻译效率,但是,3?端测序和蛋白定量质谱分析发现小鼠和人的T细胞中短3'UTR对蛋白质表达丰度的影响有限[30,31,32],提示并非所有APA引起的短3?UTR亚型都可提高mRNA的翻译效率。
2.3 APA影响mRNA的运输和定位
mRNA的出核转运和胞质定位受到位于3?UTR的定位顺式调控元件(zip codes)和反式作用因子(zip codes结合蛋白)的调控[33]。同一基因3?UTR序列长度和组成不同的转录本可能具有不同的定位信号,这会导致同一个基因的不同转录本在细胞中的分布不同和功能差异。研究表明,出核蛋白(Aly/REF export factor, ALYREF)通过与PABPN1和CSTF64互作,并结合mRNA的3?UTR促进mRNA出核转运[34]。另外,研究发现,核RNA输出因子1 (nuclear RNA export factor1, NXF1)可与CFIm68互作,并结合于成熟的长3?UTR亚型转录本的UGUA元件协助其出核转运[35,36]。脑源性神经营养因子(brain-derived neurotrophic factor, BDNF)[37]和肌醇单磷酸酶(inositol monophosphatase 1)[38]的长3?UTR亚型转录本主要定位在神经细胞树突中,而其短3?UTR亚型转录本则主要分布在神经元胞体内[37]。长3?UTR亚型CD47转录本3?UTR的U-rich元件可结合Hu antigen R (HuR),HuR会招募SET蛋白,促进SET与新翻译出的CD47蛋白互作,并同时激活Rac家族GTP酶1 (Rac family small GTPase 1, RAC1),从而使CD47向质膜转移;短3'UTR亚型CD47转录本的3'UTR则缺少U-rich元件,不能与HuR结合,无法招募SET蛋白和激活RAC1,故造成CD47蛋白滞留在内质网中[39]。CD44、整合素α1 (integrin α1, ITGA1)和肿瘤坏死因子受体超家族成员13C (TNF receptor superfamily member 13C, TNFRSF13C)的不同蛋白质亚型也存在这种细胞定位机制[39]。虽然长3'UTR亚型转录本在细胞质中易降解,但却具有高效的出核方式,这保证了长3'UTR亚型转录本在细胞质中发挥作用,也保证了其转录后的精细调控。
3 APA调控机制
简单来说,一个具有多个PAS的基因,其mRNA前体在成熟过程中PAS的选择利用取决于各PAS发生C/P反应的时机。当mRNA前体具有两个PAS时,PAS的选择涉及Tp1、Tp2和Tt三个关键时间变量,Tp1和Tp2分别是在近端PAS和远端PAS进行C/P反应的时间,Tt是转录复合体通过两个PAS间的时间[40]。如果Tp1 < Tt + Tp2,近端PAS的利用率增加,会产生短3?UTR亚型转录本;如果Tp1 > Tt + Tp2,则远端PAS的利用率增加,产生长3?UTR亚型转录本[40]。这3个关键时间参数受到顺式调控元件信号强弱和反式作用因子作用强度等多个因素的调控。目前已知APA的调控受到PAS序列、PAS序列间的距离、染色质的修饰、转录酶的延伸速度、蛋白质-RNA互作强度和核心反式作用因子浓度的影响[1,9]。3.1 APA顺式调控
PAS是C/P反应的顺式调控元件,PAS序列信号的强弱影响其在APA中是否被选择和利用。远端PAS常具有经典的AAUAAA、U/GU-rich等元件[26,40],而近端PAS常含有这些元件的非经典序列,其剪切活性较远端PAS低,故在APA中通常选择和利用远端PAS,这种现象普遍存在于在果蝇、小鼠和人的大多数基因[2]。两个PAS间的序列也会影响PAS的选择。研究发现,在两个PAS间插入外源片段可提高近端PAS的利用[41]。两个PAS之间的DNA甲基化也会影响PAS的选择利用。研究发现,作为甲基化特异绝缘蛋白的CCCTC结合因子(CCCTC-binding factor, CTCF)会与DNA甲基化位点结合,并招募黏连蛋白复合物(cohesin complex)形成染色质环形结构,阻止RNAP II的转录延伸,从而促进近端PAS的选择利用[42]。
染色质空间结构也可影响PAS的选择,处在染色质较为松散区域的PAS,易被选择和利用。研究发现,酿酒酵母的组蛋白H3K4和H3K36甲基化酶SET1和SET2可调控APA,这是由于这两个甲基化酶修饰组蛋白H3后,H3K4me1和H3K36me3可增加核小体乙酰化,从而减少了核小体的位阻效应,使染色质处于松散的开放状态,有利于选择远端PAS[43]。还有研究发现APA的调控也与mRNA 3'UTR的腺嘌呤的甲基化(N6-methyladenosine, m6A)和RNA二级结构有关[44,45]。
3.2 APA反式调控
APA受到前述参与C/P反应的4个蛋白质复合物的核心蛋白因子、RNA拼接因子以及其他一些RNA结合蛋白等的调控。其中,RNAP II是APA的重要反式调控因子。黑腹果蝇中有一种突变型RNAP II,它的转录延伸效率较低,导致polo基因短3?UTR亚型转录本的表达量增加[46]。在紫外线诱导的酵母DNA损伤修复模型中,紫外线引起的RNAP II的C端高度磷酸化,在降低转录延伸效率的同时还会影响剪接体蛋白复合物的结合[47],结果导致RPB2基因利用近端PAS,产生短3?UTR亚型转录本[48]。
反式作用因子浓度的改变是调控APA的一个重要因素。有研究发现CSTF64/CSTF64τ或FIP1的缺失可增加多数基因远端高活性PAS的选择利用[49,50,51],敲低CFIm25和CFIm68会增加多数基因近端低活性PAS的选择利用[52]。在紫外线诱导DNA损伤修复的模型中,BRCA1/BARD1结合并隔绝CSTF50,使得CSTF50的浓度显著降低,造成较多基因选择利用远端PAS[53]。胚胎干细胞(ESCs)分化过程中,FIP1和CSTF各亚基的蛋白水平呈逐渐下降的趋势,同时较多的基因选择利用远端PAS。与此相一致,FIP1缺失会导致ESC细胞较多基因选择利用远端PAS[50]。降低RNA结合蛋白Nudt21会促进细胞重编程相关基因选择利用近端PAS,形成短3'UTR亚型转录本,提高其稳定性和编码能力,促进已分化细胞发生去分化,形成具有多能性的祖细胞[54]。
反式作用因子和顺式调控元件的互作强度减弱或丧失也是影响APA的重要因素。Robert等[27]发现IRF5基因近端PAS的经典序列AAUAAA突变为非经典AAGAAA后,CPSF160与其互作的能力下降,导致由选择近端PAS转而选择远端PAS,从而产生长3?UTR亚型的mRNA。异质性核糖核蛋白F(heterogeneous nuclear ribonucleoproteins F, hnRNP)和H/H[49]、多聚嘧啶序列结合蛋白(PTB)[55]和性别致死因子(sex lethal)[56]可结合CR-PAS下游的GU-rich元件,竞争性抑制CSTF64与GU-rich元件结合,导致UTR-PAS型选择利用增加。有报道发现PABPN1与近端PAS结合并抑制CPSF160与其互作,导致远端PAS的选择利用增加[57,58]。异质性核糖核蛋白K (heterogeneous nuclear ribonucleoptotein K, hnRNP K)可阻止CFIm68与CFIm25结合,抑制CFIm复合物与NEAT1的近端PAS上游的UGUA结合,导致远端PAS的选择利用增加,从而促进长3?UTR亚型lncRNA NEAT1_2的表达[59,60]。αCP胞嘧啶结合蛋白(αCP poly(C) binding protein, αCP PCBP)能结合C-rich元件,促进C-rich元件临近的PAS的选择利用[61]。
许多APA的调控因子自身也受到APA的调控。例如人和果蝇CSTF77基因利用CR-PAS产生没有编码能力的mRNA[62],导致CSTF77的表达降低,CSTF77表达下降使其对CR-PAS的选择利用减少,UTR-PAS选择利用增加,产生具有编码能力的长亚型转录本,提高细胞内CSTF77蛋白水平[63],而高水平的CSTF77会促进CR-PAS的选择利用,产生没有编码能力的mRNA。有报导表明CSTF64和CSTF64τ也存在这种表达互抑现象[64,65]。这种通过APA调节mRNA长度的机制可以维持C/P反应所需蛋白的生理浓度,并为其他基因的APA调控提供了保障。
4 APA异常与疾病
目前已知绝大多数的真核生物基因都存在APA。作为基因表达的一个重要调控机制,APA的异常与人类疾病密切相关。目前发现,血液、免疫、神经、癌症等多种疾病与APA的异常相关[4,5,6,7]。研究发现,在地中海贫血患者中,α2-globin和β-globin基因近端PAS的突变和缺失造成了远端PAS选择利用的增加[4];同样发现,在静脉血栓患者中,近端PAS突变引起的凝血酶原(prothrombin)基因远端PAS的利用增加[66]。在急性粒细胞白血病患者中,CPSF160的表达显著高于健康人群,造成白血病相关的融合基因产生高稳定性的短3'UTR亚型转录本,促进白血病细胞生长[67]。在动脉粥样硬化、II型糖尿病、结肠炎和关节炎等慢性炎症疾病患者中,炎症反应因子NLRP3的短3?UTR亚型转录本的表达水平较高,NLRP3的短3?UTR亚型转录本可逃避Tristetraprolin(TTP)和miR-223的负调控,导致NLRP3蛋白表达升高,诱导机体发生炎症反应[68]。胶质母细胞瘤的CFIm25的表达增加,导致致癌基因产生高稳定性的短3?UTR亚型转录本,增强了恶性胶质瘤细胞的增殖和侵袭能力[69]。在肌萎缩性脊髓侧索硬化症(amyotrophic lateral sclerosis, ALS)和额颞叶痴呆(frontotemporal dementia, FTD)患者中,抑微管装配蛋白(stathmin-2)基因利用CR-PAS所产生的转录本不能翻译出具有功能的蛋白,导致运动神经元轴突的再生被抑制[70]。在阿尔兹海默症(Alzheimer's disease,AD)患者中,微管相关蛋白的Tau基因选择利用近端PAS,避免了miR-34a的抑制作用,导致TAU蛋白的表达增加,加剧神经元内TAU蛋白凝聚,造成神经纤维缠结[71]。在帕金森疾病(Parkinson's disease, PD)患者中,细胞内多巴胺会促进α-突触蛋白(α-synuclein)基因产生长3'UTR亚型转录本,增强了α-突触蛋白转录本的稳定性和蛋白表达水平,造成α-突触蛋白由突触末端向线粒体和细胞体转移,蓄积的α-突触蛋白形成路易小体(lewy body)[72]。慢性淋巴细胞白血病和多发性骨髓瘤细胞的3?端转录组测序分析发现,肿瘤细胞的抑癌基因较多地选择利用CR-PAS,产生缺失DNA结合结构域或跨膜结构域的蛋白,这些蛋白往往丧失抑癌作用[73,74]。5 结语与展望
近来大量的研究表明,APA是真核生物中广泛存在的一个重要基因表达调控机制,但目前人们对于许多基因APA的生物学效应和调控机制了解还不够,特别是对特定组织和细胞在特定发育阶段的APA及其APA的生物学效应和调控尚不清楚。未来有必要开展以下4个方面的工作:(1)建立高通量、高准确率的APA测序分析平台。虽然目前已建立了多种3?端富集测序分析技术,但是这些方法仍存在不足之处,分析结果不够准确。未来,有必要采用第三代测序和单细胞测序技术开展特定组织或细胞在特定发育阶段的APA研究。(2)开展APA的调控机制研究。尽管目前人们已鉴定出了许多APA的顺式调控元件和反式作用因子,但还需进一步开展不同组织和细胞的特异性APA顺式调控元件和反式作用因子(包括非编码RNA)的鉴定和功能分析。(3)开展表观遗传修饰对APA的调控研究。目前已有研究发现,DNA、RNA和组蛋白的表观修饰影响基因的APA,但其作用机制尚不清楚,未来有必要开展DNA甲基化、组蛋白修饰以及RNA前体甲基化等表观修饰对APA的影响和作用机制的研究。(4)开展APA的生物学效应和调控机制的体内研究。目前,APA研究主要局限于体外试验,APA在体内真正的生物学效应和调控机制还有待阐明,未来需要利用基因敲除和基因编辑等技术,开展体内APA的生物学效应和调控机制研究。随着APA研究的不断深入,相信许多生物学过程的分子作用机制和一些人类疾病的发生机制终将会被阐明。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1038/nrm.2016.116URLPMID:27677860 [本文引用: 11]

Alternative polyadenylation (APA) is an RNA-processing mechanism that generates distinct 3' termini on mRNAs and other RNA polymerase II transcripts. It is widespread across all eukaryotic species and is recognized as a major mechanism of gene regulation. APA exhibits tissue specificity and is important for cell proliferation and differentiation. In this Review, we discuss the roles of APA in diverse cellular processes, including mRNA metabolism, protein diversification and protein localization, and more generally in gene regulation. We also discuss the molecular mechanisms underlying APA, such as variation in the concentration of core processing factors and RNA-binding proteins, as well as transcription-based regulation.
DOI:10.1101/gr.132563.111URL [本文引用: 2]

We developed PolyA-seq, a strand-specific and quantitative method for high-throughput sequencing of 3' ends of polyadenylated transcripts, and used it to globally map polyadenylation (polyA) sites in 24 matched tissues in human, rhesus, dog, mouse, and rat. We show that PolyA-seq is as accurate as existing RNA sequencing (RNA-seq) approaches for digital gene expression (DGE), enabling simultaneous mapping of polyA sites and quantitative measurement of their usage. In human, we confirmed 158,533 known sites and discovered 280,857 novel sites (FDR < 2.5%). On average 10% of novel human sites were also detected in matched tissues in other species. Most novel sites represent uncharacterized alternative polyA events and extensions of known transcripts in human and mouse, but primarily delineate novel transcripts in the other three species. A total of 69.1% of known human genes that we detected have multiple polyA sites in their 3'UTRs, with 49.3% having three or more. We also detected polyadenylation of noncoding and antisense transcripts, including constitutive and tissue-specific primary microRNAs. The canonical polyA signal was strongly enriched and positionally conserved in all species. In general, usage of polyA sites is more similar within the same tissues across different species than within a species. These quantitative maps of polyA usage in evolutionarily and functionally related samples constitute a resource for understanding the regulatory mechanisms underlying alternative polyadenylation.
DOI:10.1016/j.physbeh.2018.01.026URLPMID:29391168 [本文引用: 1]

RNA biogenesis has emerged as a powerful biological event that regulates energy homeostasis. In this context insertion of alternative polyadenylation sites (APSs) dictate the fate of newly synthesized RNA molecules and direct alternative splicing of nascent transcripts. Thus APSs serve a mechanistic function by regulating transcriptome expression and function. In this study we employed a novel RNA-Seq Next Generation Sequencing (NGS) approach that utilized the power of Whole Transcriptome Termini Site Sequencing (WTTS-Seq) to simultaneously measure APS events on multiple RNA biotypes. We used this technique to measure APS events in the hypothalamus of adult male Long Evans rats exposed to a palatable high fat diet (HFD) or chow. Rats maintained on HFD displayed typical hyperphagic feeding and ensuing body weight gain over the one-month manipulation period. Our WTTS-Seq analysis mapped approximately 89,000 unique hypothalamic APSs induced by HFD relative to chow fed controls. HFD exposure produced APSs on multiple RNA biotypes in the hypothalamus. The majority of detected APSs occur on mRNA transcripts that encode functional proteins. Notably we find APSs on micro (miRNA) and long non-coding RNAs (lncRNA), newly recognized transcription factors that regulate body weight in rodents. In addition we detect APSs on protein encoding mRNAs that control neuron projection development and synapse organization and glutamate signaling, key events hypothesized to maintain excess food intake. Importantly, quantitative real time PCR indicated that APS insertion led to increased hypothalamic expression of multiple RNA biotypes. Collectively these data highlight APS events as a novel genetic mechanism that directs hypothalamic RNA biogenesis stimulated by diet-induced obesity.
DOI:10.4161/nucl.36360URLPMID:25484187 [本文引用: 3]

Polyadenylation is the RNA processing step that completes the maturation of nearly all eukaryotic mRNAs. It is a two-step nuclear process that involves an endonucleolytic cleavage of the pre-mRNA at the 3'-end and the polymerization of a polyadenosine (polyA) tail, which is fundamental for mRNA stability, nuclear export and efficient translation during development. The core molecular machinery responsible for the definition of a polyA site includes several recognition, cleavage and polyadenylation factors that identify and act on a given polyA signal present in a pre-mRNA, usually an AAUAAA hexamer or similar sequence. This mechanism is tightly regulated by other cis-acting elements and trans-acting factors, and its misregulation can cause inefficient gene expression and may ultimately lead to disease. The majority of genes generate multiple mRNAs as a result of alternative polyadenylation in the 3'-untranslated region. The variable lengths of the 3' untranslated regions created by alternative polyadenylation are a recognizable target for differential regulation and clearly affect the fate of the transcript, ultimately modulating the expression of the gene. Over the past few years, several studies have highlighted the importance of polyadenylation and alternative polyadenylation in gene expression and their impact in a variety of physiological conditions, as well as in several illnesses. Abnormalities in the 3'-end processing mechanisms thus represent a common feature among many oncological, immunological, neurological and hematological disorders, but slight imbalances can lead to the natural establishment of a specific cellular state. This review addresses the key steps of polyadenylation and alternative polyadenylation in different cellular conditions and diseases focusing on the molecular effectors that ensure a faultless pre-mRNA 3' end formation.
DOI:10.1038/s41576-019-0145-zURLPMID:31267064 [本文引用: 3]

Most human genes have multiple sites at which RNA 3' end cleavage and polyadenylation can occur, enabling the expression of distinct transcript isoforms under different conditions. Novel methods to sequence RNA 3' ends have generated comprehensive catalogues of polyadenylation (poly(A)) sites; their analysis using innovative computational methods has revealed how poly(A) site choice is regulated by core RNA 3' end processing factors, such as cleavage factor I and cleavage and polyadenylation specificity factor, as well as by other RNA-binding proteins, particularly splicing factors. Here, we review the experimental and computational methods that have enabled the global mapping of mRNA and of long non-coding RNA 3' ends, quantification of the resulting isoforms and the discovery of regulators of alternative cleavage and polyadenylation (APA). We highlight the different types of APA-derived isoforms and their functional differences, and illustrate how APA contributes to human diseases, including cancer and haematological, immunological and neurological diseases.
DOI:10.1186/s12920-019-0509-4URLPMID:31072331 [本文引用: 2]

BACKGROUND: Transcriptome wide changes have been assessed extensively during the progression of neurodegenerative diseases. Alternative polyadenylation (APA) occurs in over 70% of human protein coding genes and it has recently been recognised as a critical regulator of gene expression during disease. However, the effect of APA in the context of neurodegenerative diseases, to date, has not been widely investigated. Dynamic Analysis of Alternative Polyadenylation from RNA-seq (DaPars) is a method by Xia and colleagues [Nat Commun. 5:5274, 2014] to investigate APA using standard RNA-seq data. Here, we employed this method to interrogate APA using publicly available RNA-seq data from Alzheimer's disease (AD), Parkinson's disease (PD) and Amyotrophic Lateral Sclerosis (ALS) patients and matched healthy individuals. RESULTS: For all three diseases, we found that APA profile changes were limited to a relative small number of genes suggesting that APA is not globally deregulated in neurodegenerative disease. However, for each disease phenotype we identified a subgroup of genes that showed disease-specific deregulation of APA. Whilst the affected genes differ between the RNA-seq datasets, in each cohort we identified an overrepresentation of genes that are associated with protein turnover pathways and mitochondrial function. CONCLUSIONS: Our findings, while drawn from a relatively small sample size, suggest that deregulation of APA may play a significant role in neurodegeneration by altering the expression of genes including UBR1 and OGDHL in AD, LONP1 in PD and UCHL1 in ALS. This report thus provides important novel insights into how APA can shape neurodegenerative disease characteristic transcriptomes.
DOI:10.3390/biom10060915URL [本文引用: 2]
DOI:10.1016/j.canlet.2019.114430URLPMID:31181319 [本文引用: 1]

Alternative polyadenylation (APA) is now widely recognized to regulate gene expression. APA is an RNA-processing mechanism that generates distinct 3' termini on mRNAs, producing mRNA isoforms. Different factors influence the initiation and development of this process. CFIm25 (among others) is a cleavage and polyadenylation factor that plays a key role in the regulation of APA. Shortening of the 3'UTRs on mRNAs leads to enhanced cellular proliferation and tumorigenicity. One reason may be the up-regulation of growth promoting factors, such as Cyclin D1. Different studies have reported a dual role of CFIm25 in cancer (both oncogenic and tumor suppressor). microRNAs (miRNAs) may be involved in CFIm25 function as well as competing endogenous RNAs (ceRNAs). The present review focuses on the role of CFIm25 in cancer, cancer treatment, and possible involvement in other human diseases. We highlight the involvement of miRNAs and ceRNAs in the function of CFIm25 to affect gene expression. The lack of understanding of the mechanisms and regulation of CFIm25 and APA has underscored the need for further research regarding their role in cancer and other diseases.
[本文引用: 14]
DOI:10.1007/978-1-4939-1221-6_3URL [本文引用: 4]

Almost all eukaryotic pre-mRNAs are processed at the 3' end by the cleavage and polyadenylation (C/P) reaction, which preludes termination of transcription and gives rise to the poly(A) tail of mature mRNA. Genomic studies in recent years have indicated that most eukaryotic mRNA genes have multiple cleavage and polyadenylation sites (pAs), leading to alternative cleavage and polyadenylation (APA) products. APA isoforms generally differ in their 3' untranslated regions (3' UTRs), but can also have different coding sequences (CDSs). APA expands the repertoire of transcripts expressed from the genome, and is highly regulated under various physiological and pathological conditions. Growing lines of evidence have shown that RNA-binding proteins (RBPs) play important roles in regulation of APA. Some RBPs are part of the machinery for C/P; others influence pA choice through binding to adjacent regions. In this chapter, we review cis elements and trans factors involved in C/P, the significance of APA, and increasingly elucidated roles of RBPs in APA regulation. We also discuss analysis of APA using transcriptome-wide techniques as well as molecular biology approaches.
[本文引用: 5]
DOI:10.1101/gad.250993.114URLPMID:25301780 [本文引用: 3]

AAUAAA is the most highly conserved motif in eukaryotic mRNA polyadenylation sites and, in mammals, is specifically recognized by the multisubunit CPSF (cleavage and polyadenylation specificity factor) complex. Despite its critical functions in mRNA 3' end formation, the molecular basis for CPSF-AAUAAA interaction remains poorly defined. The CPSF subunit CPSF160 has been implicated in AAUAAA recognition, but direct evidence has been lacking. Using in vitro and in vivo assays, we unexpectedly found that CPSF subunits CPSF30 and Wdr33 directly contact AAUAAA. Importantly, the CPSF30-RNA interaction is essential for mRNA 3' processing and is primarily mediated by its zinc fingers 2 and 3, which are specifically targeted by the influenza protein NS1A to suppress host mRNA 3' processing. Our data suggest that AAUAAA recognition in mammalian mRNA 3' processing is more complex than previously thought and involves multiple protein-RNA interactions.
DOI:10.1261/rna.070870.119URLPMID:31462423 [本文引用: 1]

Most eukaryotic messenger RNA precursors must undergo 3'-end cleavage and polyadenylation for maturation. We and others recently reported the structure of the AAUAAA polyadenylation signal (PAS) in complex with the protein factors CPSF-30, WDR33, and CPSF-160, revealing the molecular mechanism for this recognition. Here we have characterized in detail the interactions between the PAS RNA and the protein factors using fluorescence polarization experiments. Our studies show that AAUAAA is recognized with approximately 3 nM affinity by the CPSF-160-WDR33-CPSF-30 ternary complex. Variations in the RNA sequence can greatly reduce the affinity. Similarly, mutations of CPSF-30 residues that have van der Waals interactions with the bases of AAUAAA also lead to substantial reductions in affinity. Finally, our studies confirm that both CPSF-30 and WDR33 are required for high-affinity binding of the PAS RNA, while these two proteins alone and their binary complexes with CPSF-160 have much lower affinity for the RNA.
DOI:10.1093/biolre/ioz012URLPMID:30715134 [本文引用: 1]

In mammals, maternal-to-zygotic transition (MZT), or oocyte-to-embryo transition, begins with oocyte meiotic resumption due to the sequential translational activation and destabilization of dormant maternal transcripts stored in the ooplasm. It then continues with the elimination of maternal transcripts during oocyte maturation and fertilization and ends with the full transcriptional activation of the zygotic genome during embryonic development. A hallmark of MZT in mammals is its reliance on translation and the utilization of stored RNAs and proteins, rather than de novo transcription of genes, to sustain meiotic maturation and early development. Impaired maternal mRNA clearance at the onset of MZT prevents zygotic genome activation and causes early arrest of developing embryos. In this review, we discuss recent advances in our knowledge of the mechanisms whereby mRNA translation and degradation are controlled by cytoplasmic polyadenylation and deadenylation which set up the competence of maturing oocyte to accomplish MZT. The emphasis of this review is on the mouse as a model organism for mammals and BTG4 as a licensing factor of MZT under the translational control of the MAPK cascade.
DOI:10.1242/dev.160051URLPMID:29217751 [本文引用: 1]

The overall bauplan of the tetrapod brain is highly conserved, yet significant variations exist among species in terms of brain size, structural composition and cellular diversity. Understanding processes underlying neural and behavioral development in a wide range of species is important both from an evolutionary developmental perspective as well as for the identification of cell sources with post-developmental neurogenic potential. Here, we characterize germinal processes in the brain of Notophthalmus viridescens and Pleurodeles waltl during both development and adulthood. Using a combination of cell tracking tools, including clonal analyses in new transgenic salamander lines, we examine the origin of neural stem and progenitor cells found in the adult brain, determine regional variability in cell cycle length of progenitor cells, and show spatiotemporally orchestrated neurogenesis. We analyze how maturation of different brain regions and neuronal subpopulations are linked to the acquisition of complex behaviors, and how these behaviors are altered upon chemical ablation of dopamine neurons. Our data analyzed from an evolutionary perspective reveal both common and species-specific processes in tetrapod brain formation and function.
DOI:10.1073/pnas.1917922117URLPMID:32170011 [本文引用: 1]

Lens transparency is established by abundant accumulation of crystallin proteins and loss of organelles in the fiber cells. It requires an efficient translation of lens messenger RNAs (mRNAs) to overcome the progressively reduced transcriptional activity that results from denucleation. Inappropriate regulation of this process impairs lens differentiation and causes cataract formation. However, the regulatory mechanism promoting protein synthesis from lens-expressed mRNAs remains unclear. Here we show that in zebrafish, the RNA-binding protein Rbm24 is critically required for the accumulation of crystallin proteins and terminal differentiation of lens fiber cells. In the developing lens, Rbm24 binds to a wide spectrum of lens-specific mRNAs through the RNA recognition motif and interacts with cytoplasmic polyadenylation element-binding protein (Cpeb1b) and cytoplasmic poly(A)-binding protein (Pabpc1l) through the C-terminal region. Loss of Rbm24 reduces the stability of a subset of lens mRNAs encoding heat shock proteins and shortens the poly(A) tail length of crystallin mRNAs encoding lens structural components, thereby preventing their translation into functional proteins. This severely impairs lens transparency and results in blindness. Consistent with its highly conserved expression in differentiating lens fiber cells, the findings suggest that vertebrate Rbm24 represents a key regulator of cytoplasmic polyadenylation and plays an essential role in the posttranscriptional control of lens development.
DOI:10.1016/j.gde.2011.04.006URL [本文引用: 2]

Cytoplasmic polyadenylation is the process by which dormant, translationally inactive mRNAs become activated via the elongation of their poly(A) tails in the cytoplasm. This process is regulated by the conserved cytoplasmic polyadenylation element binding (CPEB) protein family. Recent studies have advanced our understanding of the molecular code that dictates the timing of CPEB-mediated poly(A) tail elongation and the extent of translational activation. In addition, evidence for CPEB-independent mechanisms has accumulated, and the breath of biological circumstances in which cytoplasmic polyadenylation plays a role has expanded. These observations underscore the versatility of CPEB as a translational regulator, and highlight the diversity of cytoplasmic polyadenylation mechanisms.
DOI:10.1093/nar/gky971URLPMID:30335155 [本文引用: 2]

Meiotic maturation of mammalian oocytes depends on the temporally and spatially regulated cytoplasmic polyadenylation and translational activation of maternal mRNAs. Cytoplasmic polyadenylation is controlled by cis-elements in the 3'-UTRs of mRNAs including the polyadenylation signal (PAS), which is bound by the cleavage and polyadenylation specificity factor (CPSF) and the cytoplasmic polyadenylation element (CPE), which recruits CPE binding proteins. Using the 3'-UTRs of mouse Cpeb1, Btg4 and Cnot6l mRNAs, we deciphered the combinatorial code that controls developmental stage-specific translation during meiotic maturation: (i) translation of a maternal transcript at the germinal vesicle (GV) stage requires one or more PASs that locate far away from CPEs; (ii) PASs distal and proximal to the 3'-end of the transcripts are equally effective in mediating translation at the GV stage, as long as they are not close to the CPEs; (iii) Both translational repression at the GV stage and activation after germinal vesicle breakdown require at least one CPE adjacent to the PAS; (iv) The numbers and positions of CPEs in relation to PASs within the 3'-UTR of a given transcript determines its repression efficiency in GV oocytes. This study reveals a previously unrecognized non-canonical mechanism by which the proximal PASs mediate 3'-terminal polyadenylation and translation of maternal transcripts.
DOI:10.1002/wrna.1171URLPMID:23776146 [本文引用: 2]

Poly(A) tail elongation after export of an messenger RNA (mRNA) to the cytoplasm is called cytoplasmic polyadenylation. It was first discovered in oocytes and embryos, where it has roles in meiosis and development. In recent years, however, has been implicated in many other processes, including synaptic plasticity and mitosis. This review aims to introduce cytoplasmic polyadenylation with an emphasis on the factors and elements mediating this process for different mRNAs and in different animal species. We will discuss the RNA sequence elements mediating cytoplasmic polyadenylation in the 3' untranslated regions of mRNAs, including the CPE, MBE, TCS, eCPE, and C-CPE. In addition to describing the role of general polyadenylation factors, we discuss the specific RNA binding protein families associated with cytoplasmic polyadenylation elements, including CPEB (CPEB1, CPEB2, CPEB3, and CPEB4), Pumilio (PUM2), Musashi (MSI1, MSI2), zygote arrest (ZAR2), ELAV like proteins (ELAVL1, HuR), poly(C) binding proteins (PCBP2, alphaCP2, hnRNP-E2), and Bicaudal C (BICC1). Some emerging themes in cytoplasmic polyadenylation will be highlighted. To facilitate understanding for those working in different organisms and fields, particularly those who are analyzing high throughput data, HUGO gene nomenclature for the human orthologs is used throughout. Where human orthologs have not been clearly identified, reference is made to protein families identified in man.
DOI:10.1016/j.gpb.2017.06.001URLPMID:29031844 [本文引用: 3]

Alternative polyadenylation (APA), a phenomenon that RNA molecules with different 3' ends originate from distinct polyadenylation sites of a single gene, is emerging as a mechanism widely used to regulate gene expression. In the present review, we first summarized various methods prevalently adopted in APA study, mainly focused on the next-generation sequencing (NGS)-based techniques specially designed for APA identification, the related bioinformatics methods, and the strategies for APA study in single cells. Then we summarized the main findings and advances so far based on these methods, including the preferences of alternative polyA (pA) site, the biological processes involved, and the corresponding consequences. We especially categorized the APA changes discovered so far and discussed their potential functions under given conditions, along with the possible underlying molecular mechanisms. With more in-depth studies on extensive samples, more signatures and functions of APA will be revealed, and its diverse roles will gradually heave in sight.
[本文引用: 1]
DOI:10.1016/j.domaniend.2006.04.012URLPMID:16814975 [本文引用: 1]

Growth hormone (GH) is indispensable for the growth of animals and its biological activity is mediated by binding to the growth hormone receptor (GHR) [Harvey S, Scanes CG, Daughaday WH. Growth hormone. Boca Raton: CRC Press; 1995]. GHR is a transmembrane protein responsible for signal transduction upon GH binding. GH also binds to the growth hormone binding protein (GHBP) which is the soluble form of GHR extracellular domain existing in circulation. Actions of GHBP include prolongation of GH bioavailability and prevention of GH signaling system from over-stimulation. To date, little is known about the mechanisms generating the chicken GHBP (cGHBP). Elucidating the genomic structure of cGHR will provide insights into such underlying mechanisms. Using polymerase chain reaction and library screening methods, we have characterized the genomic organization of chicken GHR (cGHR). The full-length coding region of the cGHR transcript is composed of eight exons (exons 2-10), lacking a human homolog exon 3 and spans at least 71 kb on the genome. A novel transcript of size 1.2kb was isolated from chicken liver total RNA using 5' and 3' rapid cDNA ends amplification (RACE). It was generated by utilizing a previously unknown polyadenylation signal located at the intron 6. Semi-quantitative reverse transcription polymerase chain reaction showed that this transcript is widely expressed in a variety of tissues. This transcript has an open reading frame comprising 203 amino acids. In vitro binding assay using ELISA demonstrated that Escherichia coli expressed recombinant protein encoded by this transcript was able to bind with chicken GH. Hence, this transcript is a potential candidate for cGHBP.
DOI:10.3389/fendo.2018.00035URL [本文引用: 1]
DOI:10.1038/s41467-020-16959-2URLPMID:32576858 [本文引用: 1]

Most eukaryotic genes produce alternative polyadenylation (APA) isoforms. Here we report that, unlike previously characterized cell lineages, differentiation of syncytiotrophoblast (SCT), a cell type critical for hormone production and secretion during pregnancy, elicits widespread transcript shortening through APA in 3'UTRs and in introns. This global APA change is observed in multiple in vitro trophoblast differentiation models, and in single cells from placentas at different stages of pregnancy. Strikingly, the transcript shortening is unrelated to cell proliferation, a feature previously associated with APA control, but instead accompanies increased secretory functions. We show that 3'UTR shortening leads to transcripts with higher mRNA stability, which augments transcriptional activation, especially for genes involved in secretion. Moreover, this mechanism, named secretion-coupled APA (SCAP), is also executed in B cell differentiation to plasma cells. Together, our data indicate that SCAP tailors the transcriptome during formation of secretory cells, boosting their protein production and secretion capacity.
DOI:10.1101/gad.245787.114URLPMID:25319826 [本文引用: 1]

Polyadenylation of mRNA precursors is mediated by a large multisubunit protein complex. Here we show that RBBP6 (retinoblastoma-binding protein 6), identified initially as an Rb- and p53-binding protein, is a component of this complex and functions in 3' processing in vitro and in vivo. RBBP6 associates with other core factors, and this interaction is mediated by an unusual ubiquitin-like domain, DWNN (
DOI:10.1016/j.semcdb.2017.08.056URLPMID:28867199 [本文引用: 2]

Transcriptional control shapes a cell's transcriptome composition, but it is RNA processing that refines its expression. The untranslated regions (UTRs) of mRNA are hotspots for regulatory control. Features in these can impact mRNA stability, localisation and translation. Here we describe how alternative cleavage and polyadenylation can change mRNA fate by changing the length of its 3'UTR.
DOI:10.1073/pnas.0701266104URLPMID:17412832 [本文引用: 2]

Systematic genome-wide studies to map genomic regions associated with human diseases are becoming more practical. Increasingly, efforts will be focused on the identification of the specific functional variants responsible for the disease. The challenges of identifying causal variants include the need for complete ascertainment of genetic variants and the need to consider the possibility of multiple causal alleles. We recently reported that risk of systemic lupus erythematosus (SLE) is strongly associated with a common SNP in IFN regulatory factor 5 (IRF5), and that this variant altered spicing in a way that might provide a functional explanation for the reproducible association to SLE risk. Here, by resequencing and genotyping in patients with SLE, we find evidence for three functional alleles of IRF5: the previously described exon 1B splice site variant, a 30-bp in-frame insertion/deletion variant of exon 6 that alters a proline-, glutamic acid-, serine- and threonine-rich domain region, and a variant in a conserved polyA+ signal sequence that alters the length of the 3' UTR and stability of IRF5 mRNAs. Haplotypes of these three variants define at least three distinct levels of risk to SLE. Understanding how combinations of variants influence IRF5 function may offer etiological and therapeutic insights in SLE; more generally, IRF5 and SLE illustrates how multiple common variants of the same gene can together influence risk of common disease.
DOI:10.1016/j.stem.2012.01.017URL [本文引用: 1]

Pax3, a key myogenic regulator, is transiently expressed during activation of adult muscle stem cells, or satellite cells (SCs), and is also expressed in a subset of quiescent SCs (QSCs), but only in specific muscles. The mechanisms regulating these variations in expression are not well understood. Here we show that Pax3 levels are regulated by miR-206, a miRNA with a previously demonstrated role in myogenic differentiation. In most QSCs and activated SCs, miR-206 expression suppresses Pax3 expression. Paradoxically, QSCs that express high levels of Pax3 also express high levels of miR-206. In these QSCs, Pax3 transcripts are subject to alternative polyadenylation, resulting in transcripts with shorter 3' untranslated regions (3'UTRs) that render them resistant to regulation by miR-206. Similar alternate polyadenylation of the Pax3 transcript also occurs in myogenic progenitors during development. Our findings may reflect a general role of alternative polyadenylation in circumventing miRNA-mediated regulation of stem cell function.
DOI:10.1126/science.aax1694URLPMID:31699935 [本文引用: 1]

Adult stem cells are essential for tissue homeostasis. In skeletal muscle, muscle stem cells (MuSCs) reside in a quiescent state, but little is known about the mechanisms that control homeostatic turnover. Here we show that, in mice, the variation in MuSC activation rate among different muscles (for example, limb versus diaphragm muscles) is determined by the levels of the transcription factor Pax3. We further show that Pax3 levels are controlled by alternative polyadenylation of its transcript, which is regulated by the small nucleolar RNA U1. Isoforms of the Pax3 messenger RNA that differ in their 3' untranslated regions are differentially susceptible to regulation by microRNA miR206, which results in varying levels of the Pax3 protein in vivo. These findings highlight a previously unrecognized mechanism of the homeostatic regulation of stem cell fate by multiple RNA species.
DOI:10.1038/ncomms6465URLPMID:25413384 [本文引用: 1]

Alternative polyadenylation is a cellular mechanism that generates mRNA isoforms differing in their 3' untranslated regions (3' UTRs). Changes in polyadenylation site usage have been described upon induction of proliferation in resting cells, but the underlying mechanism and functional significance of this phenomenon remain largely unknown. To understand the functional consequences of shortened 3' UTR isoforms in a physiological setting, we used 3' end sequencing and quantitative mass spectrometry to determine polyadenylation site usage, mRNA and protein levels in murine and human naive and activated T cells. Although 3' UTR shortening in proliferating cells is conserved between human and mouse, orthologous genes do not exhibit similar expression of alternative 3' UTR isoforms. We generally find that 3' UTR shortening is not accompanied by a corresponding change in mRNA and protein levels. This suggests that although 3' UTR shortening may lead to changes in the RNA-binding protein interactome, it has limited effects on protein output.
DOI:10.1101/gr.156919.113URL [本文引用: 1]

Variation in protein output across the genome is controlled at several levels, but the relative contributions of different regulatory mechanisms remain poorly understood. Here, we obtained global measurements of decay and translation rates for mRNAs with alternative 3' untranslated regions (3' UTRs) in murine 3T3 cells. Distal tandem isoforms had slightly but significantly lower mRNA stability and greater translational efficiency than proximal isoforms on average. The diversity of alternative 3' UTRs also enabled inference and evaluation of both positively and negatively acting cis-regulatory elements. The 3' UTR elements with the greatest implied influence were microRNA complementary sites, which were associated with repression of 32% and 4% at the stability and translational levels, respectively. Nonetheless, both the decay and translation rates were highly correlated for proximal and distal 3' UTR isoforms from the same genes, implying that in 3T3 cells, alternative 3' UTR sequences play a surprisingly small regulatory role compared to other mRNA regions.
DOI:10.1016/j.scr.2013.06.002URLPMID:23845413 [本文引用: 1]

Adipocyte stem cells (hASCs) can proliferate and self-renew and, due to their multipotent nature, they can differentiate into several tissue-specific lineages, making them ideal candidates for use in cell therapy. Most attempts to determine the mRNA profile of self-renewing or differentiating stem cells have made use of total RNA for gene expression analysis. Several lines of evidence suggest that self-renewal and differentiation are also dependent on the control of protein synthesis by posttranscriptional mechanisms. We used adipogenic differentiation as a model, to investigate the extent to which posttranscriptional regulation controlled gene expression in hASCs. We focused on the initial steps of differentiation and isolated both the total mRNA fraction and the subpopulation of mRNAs associated with translating ribosomes. We observed that adipogenesis is committed in the first days of induction and three days appears as the minimum time of induction necessary for efficient differentiation. RNA-seq analysis showed that a significant percentage of regulated mRNAs were posttranscriptionally controlled. Part of this regulation involves massive changes in transcript untranslated regions (UTR) length, with differential extension/reduction of the 3'UTR after induction. A slight correlation can be observed between the expression levels of differentially expressed genes and the 3'UTR length. When we considered association to polysomes, this correlation values increased. Changes in the half lives were related to the extension of the 3'UTR, with longer UTRs mainly stabilizing the transcripts. Thus, changes in the length of these extensions may be associated with changes in the ability to associate with polysomes or in half-life.
DOI:10.1261/rna.262607URLPMID:17449729 [本文引用: 1]

Asymmetric subcellular distribution of RNA is used by many organisms to establish cell polarity, differences in cell fate, or to sequester protein activity. Accurate localization of RNA requires specific sequence and/or structural elements in the localized RNA, as well as proteins that recognize these elements and link the RNA to the appropriate molecular motors. Recent advances in biochemistry, molecular biology, and cell imaging have enabled the identification of many RNA localization elements, or
DOI:10.1093/nar/gkx597URLPMID:28934468 [本文引用: 1]

The TREX complex (TREX) plays key roles in nuclear export of mRNAs. However, little is known about its transcriptome-wide binding targets. We used individual cross-linking and immunoprecipitation (iCLIP) to identify the binding sites of ALYREF, an mRNA export adaptor in TREX, in human cells. Consistent with previous in vitro studies, ALYREF binds to a region near the 5' end of the mRNA in a CBP80-dependent manner. Unexpectedly, we identified PABPN1-dependent ALYREF binding near the 3' end of the mRNA. Furthermore, the 3' processing factor CstF64 directly interacts with ALYREF and is required for the overall binding of ALYREF on the mRNA. In addition, we found that numerous middle exons harbor ALYREF binding sites and identified ALYREF-binding motifs that promote nuclear export of intronless mRNAs. Together, our study defines enrichment of ALYREF binding sites at the 5' and the 3' regions of the mRNA in vivo, identifies export-promoting ALYREF-binding motifs, and reveals CstF64- and PABPN1-mediated coupling of mRNA nuclear export to 3' processing.
DOI:10.1091/mbc.e09-05-0389URLPMID:19864460 [本文引用: 1]

Export of mRNA from the nucleus is linked to proper processing and packaging into ribonucleoprotein complexes. Although several observations indicate a coupling between mRNA 3' end formation and export, it is not known how these two processes are mechanistically connected. Here, we show that a subunit of the mammalian pre-mRNA 3' end processing complex, CF I(m)68, stimulates mRNA export. CF I(m)68 shuttles between the nucleus and the cytoplasm in a transcription-dependent manner and interacts with the mRNA export receptor NXF1/TAP. Consistent with the idea that CF I(m)68 may act as a novel adaptor for NXF1/TAP, we show that CF I(m)68 promotes the export of a reporter mRNA as well as of endogenous mRNAs, whereas silencing by RNAi results in the accumulation of mRNAs in the nucleus. Moreover, CF I(m)68 associates with 80S ribosomes but not polysomes, suggesting that it is part of the mRNP that is remodeled in the cytoplasm during the initial stages of translation. These results reveal a novel function for the pre-mRNA 3' end processing factor CF I(m)68 in mRNA export.
DOI:10.1016/j.molcel.2019.01.026URLPMID:30819645 [本文引用: 1]

Alternative polyadenylation (APA) produces mRNA isoforms with different 3' UTR lengths. Previous studies indicated that 3' end processing and mRNA export are intertwined in gene regulation. Here, we show that mRNA export factors generally facilitate usage of distal cleavage and polyadenylation sites (PASs), leading to long 3' UTR isoform expression. By focusing on the export receptor NXF1, which exhibits the most potent effect on APA in this study, we reveal several gene features that impact NXF1-dependent APA, including 3' UTR size, gene size, and AT content. Surprisingly, NXF1 downregulation results in RNA polymerase II (Pol II) accumulation at the 3' end of genes, correlating with its role in APA regulation. Moreover, NXF1 cooperates with CFI-68 to facilitate nuclear export of long 3' UTR isoform with UGUA motifs. Together, our work reveals important roles of NXF1 in coordinating transcriptional dynamics, 3' end processing, and nuclear export of long 3' UTR transcripts, implicating NXF1 as a nexus of gene regulation.
DOI:10.1016/j.cell.2008.05.045URLPMID:18614020 [本文引用: 2]

The brain produces two brain-derived neurotrophic factor (BDNF) transcripts, with either short or long 3' untranslated regions (3' UTRs). The physiological significance of the two forms of mRNAs encoding the same protein is unknown. Here, we show that the short and long 3' UTR BDNF mRNAs are involved in different cellular functions. The short 3' UTR mRNAs are restricted to somata, whereas the long 3' UTR mRNAs are also localized in dendrites. In a mouse mutant where the long 3' UTR is truncated, dendritic targeting of BDNF mRNAs is impaired. There is little BDNF in hippocampal dendrites despite normal levels of total BDNF protein. This mutant exhibits deficits in pruning and enlargement of dendritic spines, as well as selective impairment in long-term potentiation in dendrites, but not somata, of hippocampal neurons. These results provide insights into local and dendritic actions of BDNF and reveal a mechanism for differential regulation of subcellular functions of proteins.
DOI:10.1016/j.tcb.2009.06.001URL [本文引用: 1]

Translation of localized mRNA is a fast and efficient way of reacting to extracellular stimuli with the added benefit of providing spatial resolution to the cellular response. The efficacy of this adaptive response ultimately relies on the ability to express a particular protein at the right time and in the right place. Although mRNA localization is a mechanism shared by most organisms, it is especially relevant in highly polarized cells, such as differentiated neurons. 3′-Untranslated regions (3′UTRs) of mRNAs are critical both for the targeting of transcripts to specific subcellular compartments and for translational control. Here we review recent studies that indicate how, in response to extracellular cues, nuclear and cytoplasmic remodeling of the 3′UTR contributes to mRNA localization and local protein synthesis.
DOI:10.1038/nature14321URLPMID:25896326 [本文引用: 2]

About half of human genes use alternative cleavage and polyadenylation (ApA) to generate messenger RNA transcripts that differ in the length of their 3' untranslated regions (3' UTRs) while producing the same protein. Here we show in human cell lines that alternative 3' UTRs differentially regulate the localization of membrane proteins. The long 3' UTR of CD47 enables efficient cell surface expression of CD47 protein, whereas the short 3' UTR primarily localizes CD47 protein to the endoplasmic reticulum. CD47 protein localization occurs post-translationally and independently of RNA localization. In our model of 3' UTR-dependent protein localization, the long 3' UTR of CD47 acts as a scaffold to recruit a protein complex containing the RNA-binding protein HuR (also known as ELAVL1) and SET to the site of translation. This facilitates interaction of SET with the newly translated cytoplasmic domains of CD47 and results in subsequent translocation of CD47 to the plasma membrane via activated RAC1 (ref. 5). We also show that CD47 protein has different functions depending on whether it was generated by the short or long 3' UTR isoforms. Thus, ApA contributes to the functional diversity of the proteome without changing the amino acid sequence. 3' UTR-dependent protein localization has the potential to be a widespread trafficking mechanism for membrane proteins because HuR binds to thousands of mRNAs, and we show that the long 3' UTRs of CD44, ITGA1 and TNFRSF13C, which are bound by HuR, increase surface protein expression compared to their corresponding short 3' UTRs. We propose that during translation the scaffold function of 3' UTRs facilitates binding of proteins to nascent proteins to direct their transport or function--and this role of 3' UTRs can be regulated by ApA.
DOI:10.1631/jzus.B1400076URLPMID:24793760 [本文引用: 3]

The majority of eukaryotic genes produce multiple mRNA isoforms with distinct 3' ends through a process called mRNA alternative polyadenylation (APA). Recent studies have demonstrated that APA is dynamically regulated during development and in response to environmental stimuli. A number of mechanisms have been described for APA regulation. In this review, we attempt to integrate all the known mechanisms into a unified model. This model not only explains most of previous results, but also provides testable predictions that will improve our understanding of the mechanistic details of APA regulation. Finally, we briefly discuss the known and putative functions of APA regulation.
DOI:10.3390/ijms19051347URL [本文引用: 1]
DOI:10.1016/j.molcel.2020.03.024URLPMID:32333838 [本文引用: 1]

Dysregulation of DNA methylation and mRNA alternative cleavage and polyadenylation (APA) are both prevalent in cancer and have been studied as independent processes. We discovered a DNA methylation-regulated APA mechanism when we compared genome-wide DNA methylation and polyadenylation site usage between DNA methylation-competent and DNA methylation-deficient cells. Here, we show that removal of DNA methylation enables CTCF binding and recruitment of the cohesin complex, which, in turn, form chromatin loops that promote proximal polyadenylation site usage. In this DNA demethylated context, either deletion of the CTCF binding site or depletion of RAD21 cohesin complex protein can recover distal polyadenylation site usage. Using data from The Cancer Genome Atlas, we authenticated the relationship between DNA methylation and mRNA polyadenylation isoform expression in vivo. This DNA methylation-regulated APA mechanism demonstrates how aberrant DNA methylation impacts transcriptome diversity and highlights the potential sequelae of global DNA methylation inhibition as a cancer treatment.
DOI:10.1093/nar/gkaa292URLPMID:32356874 [本文引用: 1]

Adjusting DNA structure via epigenetic modifications, and altering polyadenylation (pA) sites at which precursor mRNA is cleaved and polyadenylated, allows cells to quickly respond to environmental stress. Since polyadenylation occurs co-transcriptionally, and specific patterns of nucleosome positioning and chromatin modifications correlate with pA site usage, epigenetic factors potentially affect alternative polyadenylation (APA). We report that the histone H3K4 methyltransferase Set1, and the histone H3K36 methyltransferase Set2, control choice of pA site in Saccharomyces cerevisiae, a powerful model for studying evolutionarily conserved eukaryotic processes. Deletion of SET1 or SET2 causes an increase in serine-2 phosphorylation within the C-terminal domain of RNA polymerase II (RNAP II) and in the recruitment of the cleavage/polyadenylation complex, both of which could cause the observed switch in pA site usage. Chemical inhibition of TOR signaling, which causes nutritional stress, results in Set1- and Set2-dependent APA. In addition, Set1 and Set2 decrease efficiency of using single pA sites, and control nucleosome occupancy around pA sites. Overall, our study suggests that the methyltransferases Set1 and Set2 regulate APA induced by nutritional stress, affect the RNAP II C-terminal domain phosphorylation at Ser2, and control recruitment of the 3' end processing machinery to the vicinity of pA sites.
DOI:10.1038/s41421-018-0019-0URLPMID:29507755 [本文引用: 1]

N(6)-methyladenosine (m(6)A) is enriched in 3'untranslated region (3'UTR) and near stop codon of mature polyadenylated mRNAs in mammalian systems and has regulatory roles in eukaryotic mRNA transcriptome switch. Significantly, the mechanism for this modification preference remains unknown, however. Herein we report a characterization of the full m(6)A methyltransferase complex in HeLa cells identifying METTL3/METTL14/WTAP/VIRMA/HAKAI/ZC3H13 as the key components, and we show that VIRMA mediates preferential mRNA methylation in 3'UTR and near stop codon. Biochemical studies reveal that VIRMA recruits the catalytic core components METTL3/METTL14/WTAP to guide region-selective methylations. Around 60% of VIRMA mRNA immunoprecipitation targets manifest strong m(6)A enrichment in 3'UTR. Depletions of VIRMA and METTL3 induce 3'UTR lengthening of several hundred mRNAs with over 50% targets in common. VIRMA associates with polyadenylation cleavage factors CPSF5 and CPSF6 in an RNA-dependent manner. Depletion of CPSF5 leads to significant shortening of 3'UTR of over 2800 mRNAs, 84% of which are modified with m(6)A and have increased m(6)A peak density in 3'UTR and near stop codon after CPSF5 knockdown. Together, our studies provide insights into m(6)A deposition specificity in 3'UTR and its correlation with alternative polyadenylation.
DOI:10.1093/molbev/msr073URL [本文引用: 1]

Hox genes encode a family of transcriptional regulators that operate differential developmental programs along the anteroposterior axis of bilateral animals. Regulatory changes affecting Hox gene expression are believed to have been crucial for the evolution of animal body plans. In Drosophila melanogaster, Hox expression is post-transcriptionally regulated by microRNAs (miRNAs) acting on target sites located in the 3' untranslated regions (3'UTRs) of Hox mRNAs. Notably, recent work has shown that during D. melanogaster development Hox genes produce mRNAs with variable 3'UTRs (short and long forms) in different sets of tissues as a result of alternative polyadenylation; importantly, Hox short and long 3'UTRs contain very different target sites for miRNAs. Here, we use a computational approach to explore the evolution of Hox 3'UTRs treated with especial regard to miRNA regulation. Our work is focused on the 12 Drosophila species for which genomic sequences are available and shows, first, that alternative polyadenylation of Hox transcripts is a feature shared by all drosophilids tested in the study. Second, that the regulatory impact of miRNAs is evolving very fast within the Drosophila group. Third, that in contrast to the low degree of primary sequence conservation, Hox 3'UTR regions within the group show very similar RNA topology indicating that RNA structure is under strong selective pressure. Finally, we also demonstrate that Hox alternative polyadenylation can remodel the control regions seen by miRNAs by at least two mechanisms: via adding new cis-regulatory sequences-in the form of miRNA target sites-to short 3'UTR forms as well as by modifying the regulatory impact of miRNA target sites in short 3'UTR forms through changes in RNA secondary structure caused by the use of distal polyadenylation signals.
DOI:10.1038/emboj.2011.156URL [本文引用: 1]

Regulated alternative polyadenylation is an important feature of gene expression, but how gene transcription rate affects this process remains to be investigated. polo is a cell-cycle gene that uses two poly(A) signals in the 30 untranslated region (UTR) to produce alternative messenger RNAs that differ in their 3'UTR length. Using a mutant Drosophila strain that has a lower transcriptional elongation rate, we show that transcription kinetics can determine alternative poly(A) site selection. The physiological consequences of incorrect polo poly(A) site choice are of vital importance; transgenic flies lacking the distal poly(A) signal cannot produce the longer transcript and die at the pupa stage due to a failure in the proliferation of the precursor cells of the abdomen, the histoblasts. This is due to the low translation efficiency of the shorter transcript produced by proximal poly(A) site usage. Our results show that correct polo poly(A) site selection functions to provide the correct levels of protein expression necessary for histoblast proliferation, and that the kinetics of RNA polymerase II have an important role in the mechanism of alternative polyadenylation. The EMBO Journal (2011) 30, 2431-2444. doi: 10.1038/emboj.2011.156; Published online 20 May 2011
DOI:10.1002/wrna.1574URLPMID:31680436 [本文引用: 1]

Phosphorylation of the RNA polymerase II C-terminal domain (Pol II CTD) has important roles in the kinetic coupling of splicing with transcription, which is essential for many genes to maintain correct splicing patterns. However, because of the extensively repeated low complexity sequences of Pol II CTD, it was unclear how phosphorylation-dependent molecular interactions were able to provide sufficient specificity to spatiotemporally partition various cotranscriptional events. Here we try to view the molecular mechanisms governing cotranscriptional splicing from the role of phase separation based on recent studies showing the ability of Pol II CTD to form droplets. This article is categorized under: RNA Processing > Splicing Regulation/Alternative Splicing RNA Processing > Splicing Mechanisms RNA Interactions with Proteins and Other Molecules > RNA-Protein Complexes.
DOI:10.1093/nar/gkt020URLPMID:23355614 [本文引用: 1]

Alternative polyadenylation (APA) is conserved in all eukaryotic cells. Selective use of polyadenylation sites appears to be a highly regulated process and contributes to human pathogenesis. In this article we report that the yeast RPB2 gene is alternatively polyadenylated, producing two mRNAs with different lengths of 3'UTR. In normally growing wild-type cells, polyadenylation preferentially uses the promoter-proximal poly(A) site. After UV damage transcription of RPB2 is initially inhibited. As transcription recovers, the promoter-distal poly(A) site is preferentially used instead, producing more of a longer form of RPB2 mRNA. We show that the relative increase in the long RPB2 mRNA is not caused by increased mRNA stability, supporting the preferential usage of the distal poly(A) site during transcription recovery. We demonstrate that the 3'UTR of RPB2 is sufficient for this UV-induced regulation of APA. We present evidence that while transcription initiation rates do not seem to influence selection of the poly(A) sites of RPB2, the rate of transcription elongation is an important determinant.
DOI:10.1093/nar/gkw823URLPMID:28180311 [本文引用: 2]

Acetylcholinesterase (AChE), encoded by the ACHE gene, hydrolyzes the neurotransmitter acetylcholine to terminate synaptic transmission. Alternative splicing close to the 3 end generates three distinct isoforms of AChET, AChEH and AChER. We found that hnRNP H binds to two specific G-runs in exon 5a of human ACHE and activates the distal alternative 3 splice site (ss) between exons 5a and 5b to generate AChET. Specific effect of hnRNP H was corroborated by siRNA-mediated knockdown and artificial tethering of hnRNP H. Furthermore, hnRNP H competes for binding of CstF64 to the overlapping binding sites in exon 5a, and suppresses the selection of a cryptic polyadenylation site (PAS), which additionally ensures transcription of the distal 3 ss required for the generation of AChET. Expression levels of hnRNP H were positively correlated with the proportions of the AChET isoform in three different cell lines. HnRNP H thus critically generates AChET by enhancing the distal 3 ss and by suppressing the cryptic PAS. Global analysis of CLIP-seq and RNA-seq also revealed that hnRNP H competitively regulates alternative 3 ss and alternative PAS in other genes. We propose that hnRNP H is an essential factor that competitively regulates alternative splicing and alternative polyadenylation.
DOI:10.1002/embj.201386537URL [本文引用: 2]

Synopsis;image;Alternative polyadenylation is an emerging key post-transcriptional regulatory mechanism. The essential role for mRNA 3 ' end processing factor Fip1 in stem cell self-renewal and pluripotency reveals the importance of poly(A) site choice.
mRNA alternative polyadenylation is critical for stem cell self-renewal and pluripotency;Fip1 is a key regulator of mRNA alternative polyadenylation;The direction of alternative polyadenylation switch is controlled by Fip1 binding and the distance between alternative poly(A) sites;Abstract;mRNA alternative polyadenylation (APA) plays a critical role in post-transcriptional gene control and is highly regulated during development and disease. However, the regulatory mechanisms and functional consequences of APA remain poorly understood. Here, we show that an mRNA 3 ' processing factor, Fip1, is essential for embryonic stem cell (ESC) self-renewal and somatic cell reprogramming. Fip1 promotes stem cell maintenance, in part, by activating the ESC-specific APA profiles to ensure the optimal expression of a specific set of genes, including critical self-renewal factors. Fip1 expression and the Fip1-dependent APA program change during ESC differentiation and are restored to an ESC-like state during somatic reprogramming. Mechanistically, we provide evidence that the specificity of Fip1-mediated APA regulation depends on multiple factors, including Fip1-RNA interactions and the distance between APA sites. Together, our data highlight the role for post-transcriptional control in stem cell self-renewal, provide mechanistic insight on APA regulation in development, and establish an important function for APA in cell fate specification.
DOI:10.1016/j.celrep.2016.03.023URLPMID:27050522 [本文引用: 1]

Accurate and precise annotation of 3' UTRs is critical for understanding how mRNAs are regulated by microRNAs (miRNAs) and RNA-binding proteins (RBPs). Here, we describe a method, poly(A) binding protein-mediated mRNA 3' end retrieval by crosslinking immunoprecipitation (PAPERCLIP), that shows high specificity for mRNA 3' ends and compares favorably with existing 3' end mapping methods. PAPERCLIP uncovers a previously unrecognized role of CstF64/64tau in promoting the usage of a selected group of non-canonical poly(A) sites, the majority of which contain a downstream GUKKU motif. Furthermore, in the mouse brain, PAPERCLIP discovers extended 3' UTR sequences harboring functional miRNA binding sites and reveals developmentally regulated APA shifts, including one in Atp2b2 that is evolutionarily conserved in humans and results in the gain of a functional binding site of miR-137. PAPERCLIP provides a powerful tool to decipher post-transcriptional regulation of mRNAs through APA in vivo.
DOI:10.1172/jci.insight.133972URL [本文引用: 1]
DOI:10.1093/nar/gkw780URLPMID:27591253 [本文引用: 1]

Alternative splicing of terminal exons increases transcript and protein diversity. How physiological and pathological stimuli regulate the choice between alternative terminal exons is, however, largely unknown. Here, we show that Brahma (BRM), the ATPase subunit of the hSWI/SNF chromatin-remodeling complex interacts with BRCA1/BARD1, which ubiquitinates the 50 kDa subunit of the 3' end processing factor CstF. This results in the inhibition of transcript cleavage at the proximal poly(A) site and a shift towards inclusion of the distal terminal exon. Upon oxidative stress, BRM is depleted, cleavage inhibition is released, and inclusion of the proximal last exon is favoored. Our findings elucidate a novel regulatory mechanism, distinct from the modulation of transcription elongation by BRM that controls alternative splicing of internal exons.
DOI:10.1016/j.cell.2017.11.023URLPMID:29249356 [本文引用: 1]

Cell fate transitions involve rapid gene expression changes and global chromatin remodeling, yet the underlying regulatory pathways remain incompletely understood. Here, we identified the RNA-processing factor Nudt21 as a novel regulator of cell fate change using transcription-factor-induced reprogramming as a screening assay. Suppression of Nudt21 enhanced the generation of induced pluripotent stem cells, facilitated transdifferentiation into trophoblast stem cells, and impaired differentiation of myeloid precursors and embryonic stem cells, suggesting a broader role for Nudt21 in cell fate change. We show that Nudt21 directs differential polyadenylation of over 1,500 transcripts in cells acquiring pluripotency, although only a fraction changed protein levels. Remarkably, these proteins were strongly enriched for chromatin regulators, and their suppression neutralized the effect of Nudt21 during reprogramming. Collectively, our data uncover Nudt21 as a novel post-transcriptional regulator of cell fate and establish a direct, previously unappreciated link between alternative polyadenylation and chromatin signaling.
DOI:10.1016/j.febslet.2013.10.005URL [本文引用: 1]

Retention of unspliced pre-messenger RNA (pre-mRNA) in the nucleus is essential for cell survival. Available nuclear factors must recognize and discern between diverse export signals present in pre-mRNA to establish an export inhibitory complex. We found that polypyrimidine domains present in both intron and exon were important for export inhibition of a minigene transcript based on hepatitis B virus pregenomic RNA. Overexpression of PTB drastically reduced export and presence of RRM4 domain seemed critical. This inhibitory network overrode stimulation from an exonic export-facilitating element. We posit that binding of PTB to multiple loci on pre-mRNA regulates nuclear retention. (C) 2013 Federation of European Biochemical Societies. Published by Elsevier B. V.
DOI:10.1038/sj.emboj.7601022URLPMID:16511567 [本文引用: 1]

The Drosophila master sex-switch protein Sex-lethal (SXL) regulates the splicing and/or translation of three known targets to mediate somatic sexual differentiation. Genetic studies suggest that additional target(s) of SXL exist, particularly in the female germline. Surprisingly, our detailed molecular characterization of a new potential target of SXL, enhancer of rudimentary (e(r)), reveals that SXL regulates e(r) by a novel mechanism--polyadenylation switching--specifically in the female germline. SXL binds to multiple SXL-binding sites, which include the GU-rich poly(A) enhancer, and competes for the binding of CstF64 in vitro. The SXL-binding sites are able to confer sex-specific poly(A) switching onto an otherwise nonresponsive polyadenylation signal in vivo. The sex-specific poly(A) switching of e(r) provides a means for translational regulation in germ cells. We present a model for the SXL-dependent poly(A) site choice in the female germline.
DOI:10.1016/j.cell.2012.03.022URLPMID:22502866 [本文引用: 1]

Alternative cleavage and polyadenylation (APA) is emerging as an important layer of gene regulation. Factors controlling APA are largely unknown. We developed a reporter-based RNAi screen for APA and identified PABPN1 as a regulator of this process. Genome-wide analysis of APA in human cells showed that loss of PABPN1 resulted in extensive 3' untranslated region shortening. Messenger RNA transcription, stability analyses, and in vitro cleavage assays indicated enhanced usage of proximal cleavage sites (CSs) as the underlying mechanism. Using Cyclin D1 as a test case, we demonstrated that enhanced usage of proximal CSs compromises microRNA-mediated repression. Triplet-repeat expansion in PABPN1 (trePABPN1) causes autosomal-dominant oculopharyngeal muscular dystrophy (OPMD). The expression of trePABPN1 in both a mouse model of OPMD and human cells elicited broad induction of proximal CS usage, linked to binding to endogenous PABPN1 and its sequestration in nuclear aggregates. Our results elucidate a novel function for PABPN1 as a suppressor of APA.
[本文引用: 1]
DOI:10.1038/emboj.2012.251URL [本文引用: 1]

Paraspeckles are unique subnuclear structures built around a specific long noncoding RNA, NEAT1, which is comprised of two isoforms produced by alternative 3'-end processing (NEAT1_1 and NEAT1_2). To address the precise molecular processes that lead to paraspeckle formation, we identified 35 paraspeckle proteins (PSPs), mainly by colocalization screening with a fluorescent protein-tagged full-length cDNA library. Most of the newly identified PSPs possessed various putative RNA-binding domains. Subsequent RNAi analyses identified seven essential PSPs for paraspeckle formation. One of the essential PSPs, HNRNPK, appeared to affect the production of the essential NEAT1_2 isoform by negatively regulating the 3'-end polyadenylation of the NEAT1_1 isoform. An in vitro 3'-end processing assay revealed that HNRNPK arrested binding of the CPSF6-NUDT21 (CFIm) complex in the vicinity of the alternative polyadenylation site of NEAT1_1. In vitro binding assays showed that HNRNPK competed with CPSF6 for binding to NUDT21, which was the underlying mechanism to arrest CFIm binding by HNRNPK. This HNRNPK function led to the preferential accumulation of NEAT1_2 and initiated paraspeckle construction with multiple PSPs. The EMBO Journal (2012) 31, 4020-4034. doi:10.1038/emboj.2012.251; Published online 7 September 2012
DOI:10.4161/rna.23547URL [本文引用: 1]

Paraspeckles are unique subnuclear structures that are built around a specific long non-coding RNA (lncRNA), NEAT1, which is comprised of two isoforms (NEAT1_1 and NEAT1_2) that are produced by alternative 3'-end processing. NEAT1 lncRNAs are unusual RNA polymerase II transcripts that lack introns. The non-polyadenylated 3'-end of NEAT1_2 is non-canonically processed by RNase P. NEAT1_2 is an essential component for paraspeckle formation. Paraspeckles form during the NEAT1_2 lncRNA biogenesis process, which encompasses transcription from its own chromosome locus through lncRNA processing and accumulation. Recent RNAi analyses of 40 paraspeckle proteins (PSPs) identified four PSPs that are required for paraspeckle formation by mediating NEAT1 processing and accumulation. In particular, HNRNPK was shown to arrest CFIm-dependent NEAT1_1 polyadenylation, leading to NEAT1_2 synthesis. The other three PSPs were required for paraspeckle formation, but did not affect NEAT1_2 expression. This observation suggests that NEAT1_2 accumulation is necessary but not sufficient for paraspeckle formation. An additional step, presumably the bundling of NEAT1 ribonucleoprotein sub-complexes, may be required for construction of the intact paraspeckle structure. NEAT1 expression is likely regulated at transcriptional and post-transcriptional steps under certain stress conditions, suggesting roles for paraspeckles in the lncRNA-mediated regulation of gene expression, such as the nucleocytoplasmic transport of mRNA in response to certain stimuli.
DOI:10.1128/MCB.01380-12URL [本文引用: 1]

We have previously demonstrated that the KH-domain protein alpha CP binds to a 3' untranslated region (3'UTR) C-rich motif of the nascent human alpha-globin (h alpha-globin) transcript and enhances the efficiency of 3' processing. Here we assess the genome-wide impact of alpha CP RNA-protein (RNP) complexes on 3' processing with a specific focus on its role in alternative polyadenylation (APA) site utilization. The major isoforms of alpha CP were acutely depleted from a human hematopoietic cell line, and the impact on mRNA representation and poly(A) site utilization was determined by direct RNA sequencing (DRS). Bioinformatic analysis revealed 357 significant alterations in poly(A) site utilization that could be specifically linked to the alpha CP depletion. These APA events correlated strongly with the presence of C-rich sequences in close proximity to the impacted poly(A) addition sites. The most significant linkage was the presence of a C-rich motif within a window 30 to 40 bases 5' to poly(A) signals (AAU AAA) that were repressed upon alpha CP depletion. This linkage is consistent with a general role for alpha CPs as enhancers of 3' processing. These findings predict a role for alpha CPs in posttranscriptional control pathways that can alter the coding potential and/or levels of expression of subsets of mRNAs in the mammalian transcriptome.
DOI:10.1017/s1355838200001266URLPMID:11105753 [本文引用: 1]

The Suppressor of forked protein is the Drosophila homolog of the 77K subunit of human cleavage stimulation factor, a complex required for the first step of the mRNA 3'-end-processing reaction. We have shown previously that wild-type su(f) function is required for the accumulation of a truncated su(f) transcript polyadenylated in intron 4 of the gene. This led us to propose a model in which the Su(f) protein would negatively regulate its own accumulation by stimulating 3'-end formation of this truncated su(f) RNA. In this article, we demonstrate this model and show that su(f) autoregulation is tissue specific. The Su(f) protein accumulates at a high level in dividing tissues, but not in nondividing tissues. We show that this distribution of the Su(f) protein results from stimulation by Su(f) of the tissue-specific utilization of the su(f) intronic poly(A) site, leading to the accumulation of the truncated su(f) transcript in nondividing tissues. Utilization of this intronic poly(A) site is affected in a su(f) mutant and restored in the mutant with a transgene encoding wild-type Su(f) protein. These data provide an in vivo example of cell-type-specific regulation of a protein level by poly(A) site choice, and confirm the role of Su(f) in regulation of poly(A) site utilization.
DOI:10.1371/journal.pgen.1003613URLPMID:23874216 [本文引用: 1]

The human gene encoding the cleavage/polyadenylation (C/P) factor CstF-77 contains 21 exons. However, intron 3 (In3) accounts for nearly half of the gene region, and contains a C/P site (pA) with medium strength, leading to short mRNA isoforms with no apparent protein products. This intron contains a weak 5' splice site (5'SS), opposite to the general trend for large introns in the human genome. Importantly, the intron size and strengths of 5'SS and pA are all highly conserved across vertebrates, and perturbation of these parameters drastically alters intronic C/P. We found that the usage of In3 pA is responsive to the expression level of CstF-77 as well as several other C/P factors, indicating it attenuates the expression of CstF-77 via a negative feedback mechanism. Significantly, intronic C/P of CstF-77 pre-mRNA correlates with global 3'UTR length across cells and tissues. In addition, inhibition of U1 snRNP also leads to regulation of the usage of In3 pA, suggesting that the C/P activity in the cell can be cross-regulated by splicing, leading to coordination between these two processes. Importantly, perturbation of CstF-77 expression leads to widespread alternative cleavage and polyadenylation (APA) and disturbance of cell proliferation and differentiation. Thus, the conserved intronic pA of the CstF-77 gene may function as a sensor for cellular C/P and splicing activities, controlling the homeostasis of CstF-77 and C/P activity and impacting cell proliferation and differentiation.
DOI:10.1073/pnas.1211101109URLPMID:23112178 [本文引用: 1]

Cleavage stimulation factor 64 kDa (CstF64) is an essential pre-mRNA 3' processing factor and an important regulator of alternative polyadenylation (APA). Here we characterized CstF64-RNA interactions in vivo at the transcriptome level and investigated the role of CstF64 in global APA regulation through individual nucleotide resolution UV crosslinking and immunoprecipitation sequencing and direct RNA sequencing analyses. We observed highly specific CstF64-RNA interactions at poly(A) sites (PASs), and we provide evidence that such interactions are widely variable in affinity and may be differentially required for PAS recognition. Depletion of CstF64 by RNAi has a relatively small effect on the global APA profile, but codepletion of the CstF64 paralog CstF64tau leads to greater APA changes, most of which are characterized by the increased relative use of distal PASs. Finally, we found that CstF64 binds to thousands of dormant intronic PASs that are suppressed, at least in part, by U1 small nuclear ribonucleoproteins. Taken together, our findings provide insight into the mechanisms of PAS recognition and identify CstF64 as an important global regulator of APA.
DOI:10.1261/rna.042317.113URL [本文引用: 1]

mRNA 3' processing is dynamically regulated spatially and temporally. However, the underlying mechanisms remain poorly understood. CstF64 tau is a paralog of the general mRNA 3' processing factor, CstF64, and has been implicated in mediating testis-specific mRNA alternative polyadenylation (APA). However, the functions of CstF64 tau in mRNA 3' processing have not been systematically investigated. We carried out a comprehensive characterization of CstF64 tau and compared its properties to those of CstF64. In contrast to previous reports, we found that both CstF64 and CstF64 tau are widely expressed in mammalian tissues, and their protein levels display tissue-specific variations. We further demonstrated that CstF64 and CstF64 tau have highly similar RNA-binding specificities both in vitro and in vivo. CstF64 and CstF64 tau modulate one another's expression and play overlapping as well as distinct roles in regulating global APA profiles. Interestingly, protein interactome analyses revealed key differences between CstF64 and CstF64 tau, including their interactions with another mRNA 3' processing factor, symplekin. Together, our study of CstF64 and CstF64 tau revealed both functional overlap and specificity of these two important mRNA 3' processing factors and provided new insights into the regulatory mechanisms of mRNA 3' processing.
DOI:10.1111/j.1538-7836.2003.00493.xURLPMID:14717975 [本文引用: 1]

BACKGROUND: The prothrombin G20210A mutation is associated with increased plasma prothrombin levels and risk of thrombosis. The mechanism by which this mutation leads to increased prothrombin expression is as yet unclear and still the subject of debate. OBJECTIVES: The aim of this study was to investigate the effect of the G20210A mutation on mRNA and protein expression. METHODS: We made a set of constructs containing the prothrombin 5'-regulatory region, the firefly luciferase reporter gene and the prothrombin 3'-UTR+ downstream region. The latter element contained either the 20210G or A allele and was inserted either as a single unit (constructs G1 and A1) or in tandem (A1A2, G1G2, A1G2, G1A2). Constructs were transiently expressed in HepG2 cells. Expression was evaluated by luciferase assays and reverse transcriptase-polymerase chain reaction (RT-PCR), followed by quantification of the products and determination of the ratio of poly(A)site usage. RT-PCR sequencing was used for determination of the actual site of polyadenylation in mRNAs from constructs G1 and A1 and from endogenous prothrombin mRNAs from HepG2 cells and human liver tissue. RESULTS: The A1 constructs expressed 1.2-fold more protein than the G1 constructs. The double constructs expressed 1.4-fold more protein (A1A2 vs. G1G2). Similar results were found in a set of constructs in which an SV40 promoter replaced the prothrombin 5'-regulatory region. Ratios of poly(A) site usage (expressed as ratio poly(A) site 1 and 2) for the tandem constructs were similar for constructs with two Gs or As at both poly(A)sites; 2.92 (95% confidence interval 2.39-3.45) and 2.75 (2.55-2.95). pA1/pA2 ratios were 1.46 (1.11-1.81) for G1A2 and 6.29 (5.48-7.10) for A1G2 constructs with different poly(A) sites, indicating that the poly(A)site with the 20210A variant is favored over the normal site. In 20210G mRNAs, the G at 20210 was the last non-A nucleotide in the majority of mRNAs, whereas in most 20210A mRNAs, the last non-A nucleotide was the C at 20209. Over 70% of the prothrombin 20210G mRNAs from HepG2 cells and human liver tissue is polyadenylated at position 20210. CONCLUSIONS: The 20210A variant has a more effective poly(A) site, leading to increased mRNA and protein expression, irrespective of the promoter and gene. It does not affect the position of poly(A) attachment.
[本文引用: 1]
DOI:10.1074/jbc.M116.772947URLPMID:28302726 [本文引用: 1]

The NLRP3 inflammasome is a central regulator of inflammation in many common diseases, including atherosclerosis and type 2 diabetes, driving the production of pro-inflammatory mediators such as IL-1beta and IL-18. Due to its function as an inflammatory gatekeeper, expression and activation of NLRP3 need to be tightly regulated. In this study, we highlight novel post-transcriptional mechanisms that can modulate NLRP3 expression. We have identified the RNA-binding protein Tristetraprolin (TTP) as a negative regulator of NLRP3 in human macrophages. TTP targets AU-rich elements in the NLRP3 3'-untranslated region (UTR) and represses NLRP3 expression. Knocking down TTP in primary macrophages leads to an increased induction of NLRP3 by LPS, which is also accompanied by increased Caspase-1 and IL-1beta cleavage upon NLRP3, but not AIM2 or NLRC4 inflammasome activation. Furthermore, we found that human NLRP3 can be alternatively polyadenylated, producing a short 3'-UTR isoform that excludes regulatory elements, including the TTP- and miRNA-223-binding sites. Because TTP also represses IL-1beta expression, it is a dual inhibitor of the IL-1beta system, regulating expression of the cytokine and the upstream controller NLRP3.
DOI:10.1038/nature13261URL [本文引用: 1]

The global shortening of messenger RNAs through alternative polyadenylation (APA) that occurs during enhanced cellular proliferation represents an important, yet poorly understood mechanism of regulated gene expression(1,2). The 3' untranslated region (UTR) truncation of growth-promoting mRNA transcripts that relieves intrinsic microRNA-and AU-rich-element-mediated repression has been observed to correlate with cellular transformation(3); however, the importance to tumorigenicity of RNA 3'-end-processing factors that potentially govern APA is unknown. Here we identify CFIm25 as a broad repressor of proximal poly(A) site usage that, when depleted, increases cell proliferation. Applying a regression model on standard RNA-sequencing data for novel APA events, we identified at least 1,450 genes with shortened 3'UTRs after CFIm25knockdown, representing 11% of significantly expressed mRNAs in human cells. Marked increases in the expression of several known oncogenes, including cyclin D1, are observed as a consequence of CFIm25 depletion. Importantly, we identified a subset of CFIm25-regulated APA genes with shortened 39 UTRs in glioblastoma tumours that have reduced CFIm25 expression. Downregulation of CFIm25 expression in glioblastoma cells enhances their tumorigenic properties and increases tumour size, where as CFIm25 overexpression reduces these properties and inhibits tumour growth. These findings identify a pivotal role of CFIm25 in governing APA and reveal a previously unknown connection between CFIm25 and glioblastoma tumorigenicity.
DOI:10.1038/s41593-018-0293-zURLPMID:30643298 [本文引用: 1]

Amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) are associated with loss of nuclear transactive response DNA-binding protein 43 (TDP-43). Here we identify that TDP-43 regulates expression of the neuronal growth-associated factor stathmin-2. Lowered TDP-43 levels, which reduce its binding to sites within the first intron of stathmin-2 pre-messenger RNA, uncover a cryptic polyadenylation site whose utilization produces a truncated, non-functional mRNA. Reduced stathmin-2 expression is found in neurons trans-differentiated from patient fibroblasts expressing an ALS-causing TDP-43 mutation, in motor cortex and spinal motor neurons from patients with sporadic ALS and familial ALS with GGGGCC repeat expansion in the C9orf72 gene, and in induced pluripotent stem cell (iPSC)-derived motor neurons depleted of TDP-43. Remarkably, while reduction in TDP-43 is shown to inhibit axonal regeneration of iPSC-derived motor neurons, rescue of stathmin-2 expression restores axonal regenerative capacity. Thus, premature polyadenylation-mediated reduction in stathmin-2 is a hallmark of ALS-FTD that functionally links reduced nuclear TDP-43 function to enhanced neuronal vulnerability.
DOI:10.1111/jnc.12437URLPMID:24032460 [本文引用: 1]

Tau pathologically aggregates in Alzheimer's disease, and evidence suggests that reducing tau expression may be safe and beneficial for the prevention or treatment of this disease. We sought to examine the role of the 3'-untranslated region (3'-UTR) of human tau mRNA in regulating tau expression. Tau expresses two 3'-UTR isoforms, long and short, as a result of alternative polyadenylation. Using luciferase reporter constructs, we found that expression from these isoforms is differentially controlled in human neuroblastoma cell lines M17D and SH-SY5Y. Several microRNAs were computationally identified as candidates that might bind the long, but not short, tau 3'-UTR isoform. A hit from a screen of candidates, miR-34a, was subsequently shown to repress the expression of endogenous tau protein in M17D cells. Conversely, inhibition of endogenously expressed miR-34 family members leads to increased endogenous tau expression. In addition, through an unbiased screen of fragments of the human tau 3'-UTR using a luciferase reporter assay, we identified several other regions in the long tau 3'-UTR isoform that contain regulatory cis-elements. Improved understanding of the regulation of tau expression by its 3'-UTR may ultimately lead to the development of novel therapeutic strategies for the treatment of Alzheimer's disease and other tauopathies. mRNA 3'-untranslated regions (3'-UTR) often regulate transcript stability or translation. Despite the centrality of the tau protein in Alzheimer's and other neurodegenerative diseases, the human tau 3'-UTR has been little studied. This report identifies regions of the tau 3'-UTR that influence expression and shows that microRNA (miR)-34a targets this 3'-UTR to lower expression, which is considered an important therapeutic goal.
DOI:10.1038/ncomms2032URLPMID:23011138 [本文引用: 1]

alpha-Synuclein is implicated both in physiological functions at neuronal synaptic terminals as well as in pathological processes in the context of Parkinson's disease. However, the molecular mechanisms for these apparently diverse roles are unclear. Here we show that specific RNA transcript isoforms of alpha-synuclein with an extended 3' untranslated region, termed aSynL, appear selectively linked to pathological processes, relative to shorter alpha-synuclein transcripts. Common variants in the aSynL 3' untranslated region associated with Parkinson's disease risk promote the accumulation and translation of aSynL transcripts. The presence of intracellular dopamine can further enhance the relative abundance of aSynL transcripts through alternative polyadenylation site selection. We demonstrate that the presence of the extended aSynL transcript 3' untranslated region impacts accumulation of alpha-synuclein protein, which appears redirected away from synaptic terminals and towards mitochondria, reminiscent of Parkinson's disease pathology. Taken together, these findings identify a novel mechanism for aSyn regulation in the context of Parkinson's disease-associated genetic and environmental variations.
DOI:10.1038/s41586-018-0465-8URLPMID:30150773 [本文引用: 1]

DNA mutations are known cancer drivers. Here we investigated whether mRNA events that are upregulated in cancer can functionally mimic the outcome of genetic alterations. RNA sequencing or 3'-end sequencing techniques were applied to normal and malignant B cells from 59 patients with chronic lymphocytic leukaemia (CLL)(1-3). We discovered widespread upregulation of truncated mRNAs and proteins in primary CLL cells that were not generated by genetic alterations but instead occurred by intronic polyadenylation. Truncated mRNAs caused by intronic polyadenylation were recurrent (n = 330) and predominantly affected genes with tumour-suppressive functions. The truncated proteins generated by intronic polyadenylation often lack the tumour-suppressive functions of the corresponding full-length proteins (such as DICER and FOXN3), and several even acted in an oncogenic manner (such as CARD11, MGA and CHST11). In CLL, the inactivation of tumour-suppressor genes by aberrant mRNA processing is substantially more prevalent than the functional loss of such genes through genetic events. We further identified new candidate tumour-suppressor genes that are inactivated by intronic polyadenylation in leukaemia and by truncating DNA mutations in solid tumours(4,5). These genes are understudied in cancer, as their overall mutation rates are lower than those of well-known tumour-suppressor genes. Our findings show the need to go beyond genomic analyses in cancer diagnostics, as mRNA events that are silent at the DNA level are widespread contributors to cancer pathogenesis through the inactivation of tumour-suppressor genes.
DOI:10.1038/s41467-018-04112-zURLPMID:29712909 [本文引用: 1]

Alternative cleavage and polyadenylation (ApA) is known to alter untranslated region (3'UTR) length but can also recognize intronic polyadenylation (IpA) signals to generate transcripts that lose part or all of the coding region. We analyzed 46 3'-seq and RNA-seq profiles from normal human tissues, primary immune cells, and multiple myeloma (MM) samples and created an atlas of 4927 high-confidence IpA events represented in these cell types. IpA isoforms are widely expressed in immune cells, differentially used during B-cell development or in different cellular environments, and can generate truncated proteins lacking C-terminal functional domains. This can mimic ectodomain shedding through loss of transmembrane domains or alter the binding specificity of proteins with DNA-binding or protein-protein interaction domains. MM cells display a striking loss of IpA isoforms expressed in plasma cells, associated with shorter progression-free survival and impacting key genes in MM biology and response to lenalidomide.
