 ,2, 陈金联
,2, 陈金联 ,3
,3Effect of Uhrf1 on intestinal development
Xinyue Wang1,3, Liang Li2,3, Qiuhui Duan2, Dali Li ,2, Jinlian Chen
,2, Jinlian Chen ,3
,3通讯作者: 李大力,博士,研究员,研究方向:基因编辑。E-mail:dlli@bio.ecnu.edu.cn
编委: 赵冰
收稿日期:2020-11-26修回日期:2020-12-22网络出版日期:2021-01-20
| 基金资助: |
Corresponding authors: 陈金联,博士,主任医师,研究方向:消化道肿瘤与肝病。E-mail:wqq_021002@163.com
Received:2020-11-26Revised:2020-12-22Online:2021-01-20
| Fund supported: |
作者简介 About authors
王芯悦,在读硕士研究生,专业方向:消化内科学。E-mail:

摘要
作为一种常见的表观遗传修饰类型,DNA甲基化对哺乳动物发育起着重要作用。Uhrf1作为重要的表观遗传调控因子,在DNA合成过程中可结合半甲基化的DNA同时招募DNA甲基转移酶1参与DNA甲基化的维持,保证遗传信息在细胞分裂前后的稳定传递。目前关于Uhrf1介导的DNA甲基化是否影响肠上皮发育过程尚不清楚。为探索Uhrf1在肠上皮发育中的作用,本研究成功构建了肠上皮特异性敲除Uhrf1的小鼠模型,利用HE染色对肠上皮组织形态学观察发现,与正常小鼠相比,敲除Uhrf1的小鼠肠上皮发育异常,主要表现为绒毛变短,数量减少,隐窝萎缩;通过表型分析发现,在小鼠肠上皮中特异性敲除Uhrf1后,细胞增殖明显受到抑制、凋亡细胞增加、细胞分化异常,同时肠干细胞相关基因表达降低。进一步对可能的分子机制进行初步探索发现Uhrf1缺失后 DNA甲基化水平大幅下降,诱发DNA损伤。本研究结果表明Uhrf1介导的DNA甲基化对肠上皮的正常发育成熟具有重要作用,有望丰富Uhrf1介导的DNA甲基化在体内的生物学功能,并为进一步明确Uhrf1介导的表观遗传调控机制提供实验依据。
关键词:
Abstract
As a best-characterized epigenetic modification, DNA methylation plays an important role in mammalian development. Uhrf1 is a critical epigenetic regulator that can bind to hemimethylated DNA and recruit DNA methyltransferase 1 to maintain DNA methylation. So far, the role of Uhrf1-mediated DNA methylation in intestinal development is still unknown. In order to investigate the impact of Uhrf1 deletion in intestinal development, we have successfully constructed the epithelial-specific Uhrf1 knockout mouse model. After Uhrf1 ablation, we found the mutant mice exhibited abnormal epithlial structure with less and shorter villi and shrinked crypts compared with wild type mice via hematoxylin-eosin staining. Further analysis showed that Uhrf1 deletion in the intestinal epithelium significantly decreased the cell proliferation and induced cell apoptosis. In addition, Uhrf1 deletion inhibited the normal epithelial differentiation and the expression of intestinal stem cell marker genes. Preliminary mechanism study revealed that loss of Uhrf1 caused global DNA hypomethylation which induced DNA damage in crypt cells. Taken together, our data suggested that DNA methylation mediated by Uhrf1 is vital for the normal intestinal development. Our results enriched the in vivo role of Uhrf1 and laid the foundation for further epigenetic regulatory mechanism exploration.
Keywords:
PDF (910KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王芯悦, 李亮, 段秋慧, 李大力, 陈金联. Uhrf1对肠上皮发育的影响. 遗传[J], 2021, 43(1): 84-93 doi:10.16288/j.yczz.20-337
Xinyue Wang.
DNA甲基化作为一种动态的和可逆的表观遗传修饰,是由甲基转移酶(DNA methyltransferases, DNMTs)介导的对胞嘧啶的第5位碳原子进行甲基修饰,形成5-甲基胞嘧啶(5-methylcytosine, 5mC),主要发生在CpG二核苷酸位点上[1]。DNA甲基化在个体发育、基因表达调控及基因组稳定性等多种生物学过程中发挥重要作用 [2,3,4,5]。
肠上皮作为人体更新较快的组织,上皮细胞的正常增殖与分化无论是在空间还是时间上都受到严格调控,增殖与分化间的失衡将会破坏上皮的完整性及其屏障功能,甚至引发肿瘤的形成。肠上皮细胞这种快速的更新速率与不同类型细胞自身的表观遗传状态密切相关,目前关于DNA甲基化如何调控肠上皮的早期发育和稳态建立仍不是很清楚。
UHRF1 (ubiquitin-like with PHD and RING finger domains 1)是泛素样含植物同源化结构域(plant homeodomain domain, PHD)和环指域蛋白家族的主要成员之一,对DNA甲基化的维持至关重要[6]。UHRF1作为一种多结构域蛋白,SRA结构域(set and ring associated domain)可以识别半甲基化的DNA,并招募DNMT1到复制叉处,保证在细胞复制时遗传信息由亲代向子代的稳定传递[7];PHD及Tudor结构域与甲基化组蛋白H3K9特异性地结合,参与异染色质的形成和维持,同时有利于DNMT1的正确定位,将组蛋白修饰与DNA甲基化紧密联系起来[8,9]。此外UHRF1的RING(really interesting new gene)结构域被证明是一种泛素连接酶,可以催化组蛋白H3K23等位点发生泛素化修饰,并被DNMT1所识别,促进DNMT1招募进而参与DNA甲基化的维持[10,11]。除了参与DNA甲基化外,UHRF1还与细胞周期调控及DNA损伤修复等诸多生物学过程相关[12,13]。随着UHRF1生物化学功能的不断阐明,关于由UHRF1介导的DNA甲基化在体内的生物学功能也得到研究****的广泛关注。研究发现Uhrf1决定卵母细胞质量,影响卵母细胞成熟,对胚胎着床前的发育至关重要[14];在斑马鱼(Danio rerio)中,Uhrf1的突变导致整体DNA甲基化水平降低,肝脏体积缩小,组织发育不全[15];在对胸腺细胞的研究中表明,敲除Uhrf1造成胸腺细胞数量明显降低,Uhrf1介导的表观遗传调控对胸腺细胞的正常发育必不可少[16];而在大脑皮层的研究中却发现Uhrf1的缺失对早期发育作用较小,不影响细胞的增殖[17]。为探讨Uhrf1及其介导的DNA甲基化在肠上皮发育中的作用,本研究构建了在肠上皮中特异性敲除Uhrf1的小鼠模型,并对表型及可能的分子机制进行分析,初步揭示了Uhrf1介导的DNA甲基化在肠组织系统中的功能。
1 材料与方法
1.1 实验动物
实验所用的小鼠遗传背景均C57BL/6J品系,饲养于华东师范大学实验动物中心SPF级清洁实验动物房。动物实验的设计与操作均符合华东师范大学动物伦理委员会相关规定并被授权,严格遵守实验动物3R原则。1.2 实验试剂
苏木精和伊红染液购自南京建成生物工程研究所;免疫组化二抗试剂盒和DAB显色试剂盒购自德国VECTOR公司;免疫组化及苏木精-伊红染色相关的无水酒精、二甲苯、甲醇等购自上海国药集团;EB染液、dNTP混合物、ExTaq DNA聚合酶和6×DNA loading buffer等购自北京天根生化科技有限公司;RNA反转录试剂盒和Trizol等购自日本TaKaRa 公司;SYBR green购自上海翊圣生物科技有限公司;DNA marker购自美国Thermo Fisher公司;Ki67抗体(RM9106,美国Thermo Fisher公司);Cleaved Caspase3抗体 (9664#,美国Cell Signaling Technology公司);Muc2 (Mucin 2)抗体(美国Santa Cruz Biotechnology公司);Dclk1(Doublecortin-like kinase 1)抗体(ab31704,英国Abcam),ChgA (Chromogranin A)抗体(ab15160,英国Abcam);Uhrf1抗体(61341,美国Active Motif公司);5mC抗体(39769,美国Active Motif公司);γH2AX (phosphorylation of histone H2AX)抗体(05636#,美国Millipore公司)。1.3 Uhrf1条件性敲除小鼠的构建
通过基因打靶技术,在Uhrf1基因的第3号外显子和第4号外显子之间的内含子区域插入可以表达β-半乳糖苷酶(LacZ)和neo抗性蛋白以及polyA,同时两端还有2个同向的FRT (short flippase recognition target)序列,另外在第4号外显子两侧插入了同向的loxp序列。通过移植该ES细胞到假孕小鼠的子宫,获得可以条件性敲除Uhrf1的小鼠。取Uhrf1-LacZ小鼠与Flp工具鼠交配,Flp(flippase)重组酶发挥作用,切除FRT之间序列,得到仅在第4号外显子两侧插入同向的loxp序列的小鼠,使用此小鼠与在肠上皮中特异性表达重组酶Cre的Villin- Cre工具鼠杂交,切割Uhrf1的第4号外显子,导致Uhrf1蛋白的翻译出现移码,从而敲除Uhrf1基因。1.4 聚合酶式反应(polymerase chainreaction, PCR)
剪取小鼠脚趾并标记放入1.5 mL无菌EP管中,按1:500比例加入蛋白酶K与消化液,置于55度水浴锅中消化过夜,次日100度煮沸5分钟后离心,进行PCR扩增。引物由上海铂尚生物技术有限公司合成,序列Uhrf1(F):5?- ACTCTTGATCTGTGCCCTGC-3?和Uhrf1(R):5?-ATCCCAGGCCTCCATACACT-3?。扩增体系(总体积为20 μL):2 μL 10×buffer,2 μL dNTP混合物,2 μL引物,3 μL模板,0.2 μL Easy Taq酶,11 μL ddH2O。反应条件:95℃ 5 min;95℃ 30 s,60℃ 30 s,72℃ 40 s;循环35次。扩增后进行凝胶电泳,凝胶自动成像仪记录电泳图像,比对条带大小,确定小鼠基因型。1.5 肠组织切片及苏木精-伊红染色(hematoxylin-eosin staining, HE)
采用颈椎脱臼法将小鼠处死后,分离小鼠肠组织,制成“瑞士卷”,用4%多聚甲醛固定过夜,包埋后将蜡块放入-20℃冰箱中保存,包埋好的组织制成5 μm切片,62℃烤片2 h,待干燥后进行脱蜡复水,具体过程为二甲苯I:7 min→二甲苯II:7 min→二甲苯:乙醇(1∶1):7 min→100%乙醇:4 min→95%乙醇:4 min→85%乙醇:4 min→75%乙醇:4 min→纯水:3 min→苏木精:5 min(具体视苏木精浓度而定)→1%盐酸酒精分色:10~30 s→蒸馏水中返蓝:15 min→伊红染色:30 s~1min→脱水及中性树脂封片→显微镜下拍照。1.6 免疫组织化学染色
组织切片脱蜡复水同HE染色→3%的过氧化氢避光处理13 min→抗原修复液置于100℃处理20 min→PBST润洗3次,每次5 min→封闭液室温封闭30 min以上→滴加一抗于4℃孵育过夜→回收一抗,PBST润洗3次→滴加HRP室温避光反应30 min→PBST润洗3次→显色3~5 min,深度适宜后蒸馏水中终止显色反应→苏木精复染→盐酸酒精分色→自来水中返蓝15 min→脱水封片同HE染色。1.7 实时荧光定量PCR (quantitative real-time PCR, qRT-PCR)分析
小鼠脱颈椎处死后取出肠组织液氮研磨后加入1 mLTrizol,按RNA抽提试剂盒说明书提取总 RNA,利用反转录试剂盒合成cDNA。qRT-PCR扩增体系为25 μL,包括:12.5 μL (2×) SYBR Premix ExTaq,2 μL引物,1 μL cDNA,9.5 μL RNase Free H2O。扩增条件:95℃ 5 min;95℃ 30 s,60℃ 30 s,72℃ 30 s,共40个循环,每个样进行3次重复。根据每个样品与内参基因β-actin所得的Ct值,利用2-ΔΔCt 公式分析目的基因相对表达量。基因的扩增引物序列见表1。Table 1
表1
表1qRT-PCR引物序列
Table 1
| 基因 | 引物序列(5′→3′) |
|---|---|
| Lgr5 | F: CAGTGTTGTGCATTTGGGGG |
| R: CAAGGTCCCGCTCATCTTGA | |
| Sox9 | F: CACAAGAAAGACCACCCCGA |
| R: GGACCCTGAGATTGCCCAGA | |
| Ascl2 | F: CGTGAAGCTGGTGAACTTGG |
| R: GGATGTACTCCACGGCTGAG | |
| Olfm4 | F: TCTTGGGCAGAAGGTGGGACT |
| R: GGACCGTCAGGTTCAGGAGC | |
| Uhrf1 | F: ACGGTGCCTACTCATTGGTC |
| R: GCTTCTGGTCAGAGGACTGG | |
| β-actin | F: CAGCCTTCCTTCTTGGGTAT |
| R: TGATCTTGATCTTCATGGTGC |
新窗口打开|下载CSV
1.8 统计方法
用Graphpad Prism 6软件进行统计学分析,各组之间的比较用平均数±标准差表示,P<0.05被认为有统计学意义。2 结果与分析
2.1 Uhrf1基因条件性敲除小鼠的构建与鉴定
Uhrf1fl/fl小鼠是以Uhrf1基因的第4号外显子为靶基因,在其两端插入loxp位点,与表达Cre酶的Villin-Cre工具鼠交配后获得VillinCre-Uhrf1fl/fl小鼠,从而切除相同方向的两个loxp位点间第4号外显子序列,导致UHRF1蛋白翻译时出现移码,从而实现Uhrf1靶基因的敲除(图1A)。设计引物针对loxp序列进行PCR,对子代进行基因型鉴定。如图1B所示,仅扩增出305 bp条带者为Uhrf1fl/fl,扩增出305 bp和106 bp两条带的为Uhrf1fl/+,仅扩增出106 bp条带者为Uhrf1+/+。通过对子代小鼠进行基因型的鉴定,观察发现VillinCre-Uhrf1fl/fl小鼠可以正常出生,但是出生率低于理论预期,部分小鼠在断奶前后出现显著死亡情况,很难发育到成年阶段,因此后续实验选取出生后3周小鼠作为实验研究对象。图1
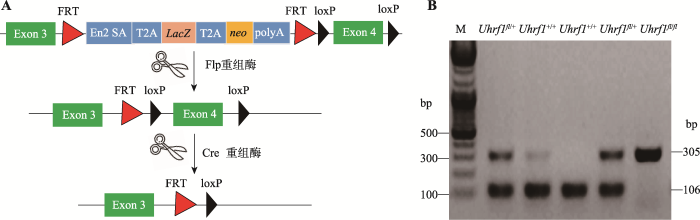 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1Uhrf1基因条件性敲除小鼠的构建与鉴定
A:Uhrf1基因条件性敲除小鼠构建示意图。B:PCR鉴定Uhrf1基因敲除结果。M:DNA marker。
Fig. 1The construction and identification of epithelial-specific Uhrf1 knockout mouse
2.2 Uhrf1基因条件性敲除小鼠的敲除效果
为了探索Uhrf1在肠发育及稳态建立中的作用,首先采用免疫组织化学染色法对Uhrf1表达量及位置进行检测,如图2A(左)染色结果显示,着色区域阳性细胞仅定位于肠隐窝部位,肠的其他部位未见有表达,说明Uhrf1主要存在于肠隐窝增殖性细胞中,即主要包含小肠干细胞及快速增殖的祖细胞。进一步采用Cre-loxp重组技术构建肠上皮中特异性敲除Uhrf1的突变小鼠,并采用免疫组织化学染色和qRT-PCR方法检测Uhrf1在肠组织中的表达情况,免疫组织化学技术结果显示在条件性敲除Uhrf1小鼠的肠隐窝底部区域并未检测到Uhrf1的表达(图2A,右);由图2B结果可见,Uhrf1基因敲除小鼠肠组织中Uhrf1 mRNA表达水平低于对照组小鼠。结果表明Cre重组酶可以发挥正常功能,导致Uhrf1基因敲除,证实Uhrf1基因条件性敲除小鼠构建成功。图2
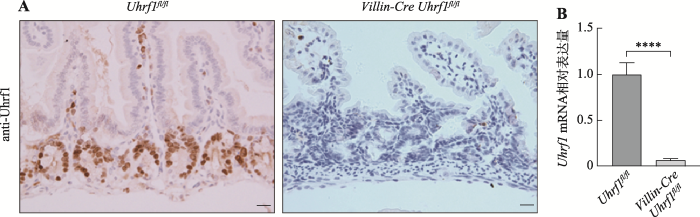 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2Uhrf1在肠组织中的表达情况
A:野生型和突变型小鼠肠组织中Uhrf1抗体免疫组织化学染色。标尺:20 μm。B:野生型与突变型小鼠肠组织中Uhrf1 mRNA表达量对比。****:P<0.0001。
Fig. 2The expression of Uhrf1 in intestinal tissue
2.3 敲除Uhrf1造成肠上皮组织形态异常
为探究Uhrf1对肠上皮组织形态学的影响,选取出生后3周的野生型与敲除Uhrf1小鼠,脱颈椎处死后取小肠组织制作石蜡切片,通过HE染色进行组织形态学分析发现,与对照组小鼠相比(图3A),Uhrf1的缺失导致肠绒毛长度明显变短,隐窝发生萎缩,同时两者数量减少,表明敲除Uhrf1后肠上皮发育过程受阻(图3B),肠上皮组织形态的建立依赖于Uhrf1。图3
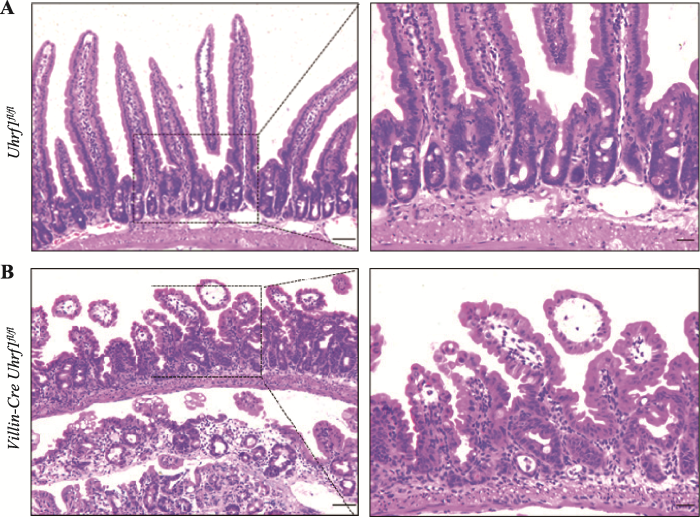 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3野生型及突变型小鼠肠组织HE染色
A:野生型小鼠肠组织切片观察。B:突变型小鼠肠组织切片观察。右图均是左图虚线区域的放大示意图。标尺左:50 μm,标尺右:20 μm。
Fig. 3HE staining of intesinal tissue from wild type and Uhrf1 mutant mice
2.4 Uhrf1的缺失影响肠上皮细胞的增殖与分化能力,诱导细胞凋亡
隐窝作为肠上皮的功能单位,负责维持细胞增殖与分化的平衡。Uhrf1特异性表达隐窝底部区域,暗示其可能影响上皮细胞的增殖和分化。因此选取3周龄VillinCre-Uhrf1fl/fl小鼠,同时选取同窝Uhrf1fl/fl的小鼠作为对照,采用免疫组织化学染色方法检测肠上皮细胞的增殖、凋亡和分化情况,并进一步对凋亡细胞及各类分化细胞数量进行检测,在20倍显微镜视野下拍照,随机选取3个形态相似视野,统计每个视野阳性细胞数。结果显示:与对照组小鼠相比,胚胎期敲除Uhrf1导致增殖标记蛋白Ki67阳性细胞数量显著减少(图4A),说明肠上皮细胞的正常增殖受到明显抑制。而凋亡相关蛋白Caspase3阳性细胞数量明显增加,并且主要集中在隐窝底部区域(图4,B和F),表明Uhrf1的缺失不仅影响了细胞增殖,而且促使肠上皮细胞发生凋亡。敲除Uhrf1后肠上皮细胞增殖受到抑制以及凋亡增加暗示分化过程可能也受到影响,因此为进一步了解Uhrf1对肠上皮分化细胞的影响,对杯状细胞-Muc2、内分泌细胞-ChgA、Tuft细胞-Dclk1标记阳性细胞数目进行检测,结果发现VillinCre-Uhrf1fl/fl小鼠肠组织Muc2 (图4,C和G)、ChgA (图4,D和H)和Dclk1 (图4,E和I)阳性细胞数减少,表达下降,说明敲除Uhrf1明显影响了肠上皮细胞的正常分化过程。因此,这些研究结果表明Uhrf1是调节肠上皮细胞增殖与分化的关键因子。图4
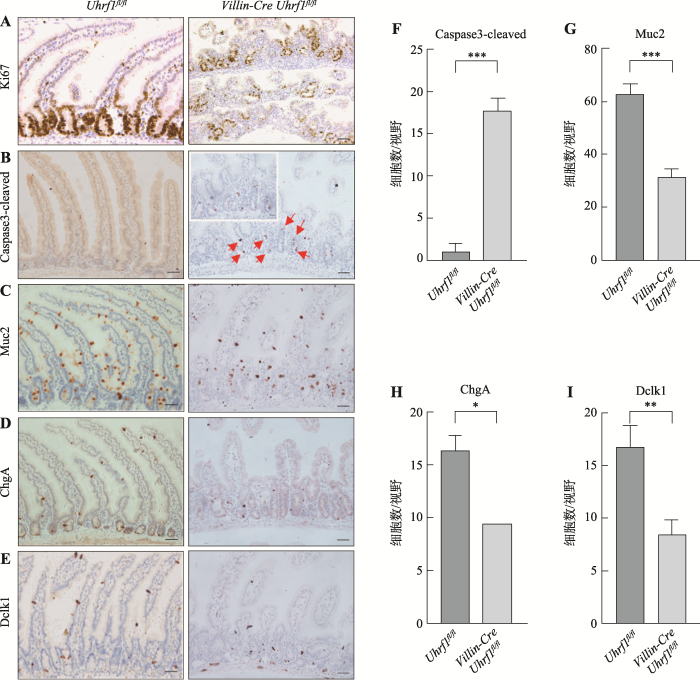 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4Uhrf1fl/fl和VillinCre-Uhrf1fl/fl小鼠肠组织中的表型分析
A:肠组织增殖标记蛋白Ki67免疫染色。B:肠组织凋亡标志蛋白Caspase3-cleaved免疫染色。红色箭头指示凋亡阳性细胞,左上角为红色箭头区域放大图。C:杯状细胞标记蛋白Muc2免疫染色。D:内分泌细胞标记蛋白ChgA免疫染色。E:Tuft细胞标记蛋白Dclk1免疫染色。标尺:50 μm F~I:野生型与突变型小鼠Caspase3-cleaved、Muc2、ChgA、Dclk1阳性细胞统计结果。*:P<0.05;**:P<0.01;***:P<0.001。
Fig. 4The phenotype analysis of wild type and Uhrf1 mutant intestine
2.5 敲除Uhrf1肠干细胞标志基因的表达降低
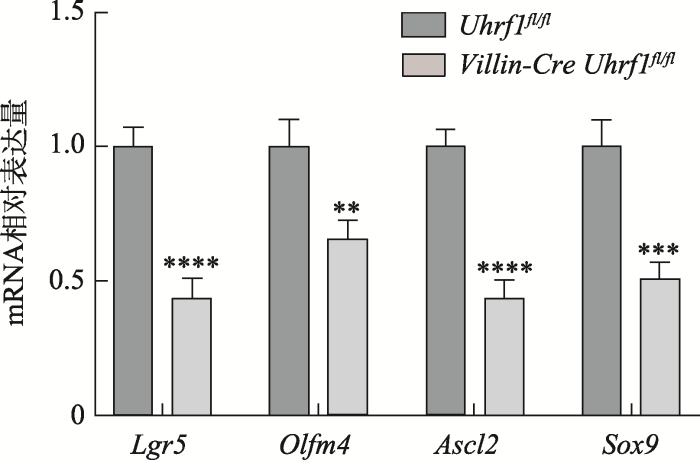
Lgr5+肠干细胞作为一种快速增殖的细胞,对肠组织生理功能及稳态的维持起着重要的作用,因此为了解Uhrf1基因敲除鼠中Lgr5+干细胞有没有受到影响,本研究进一步在分子水平上对Lgr5+干细胞相关的基因:Lgr5、Olfm4、Ascl2和Sox9表达情况进行检测,结果发现,与对照组小鼠相比,Uhrf1在肠组织中特异性敲除后,造成肠干细胞相关标志基因的表达降低(图5),表明Uhrf1可能对肠干细胞数量具有一定的影响。2.6 敲除Uhrf1导致DNA甲基化水平下降,诱导DNA损伤
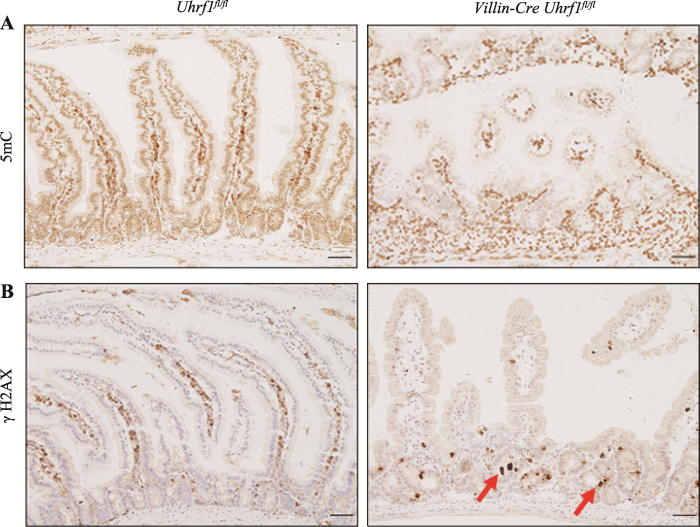
如前所述,UHRF1作为表观遗传调控因子,在DNA甲基化维持中起着关键作用。为确定Uhrf1的缺失是否会导致肠上皮基因组甲基化的丢失,采用免疫组织化学染色方法对5mC表达水平进行检测发现,与对照组小鼠相比,条件性敲除小鼠的肠组织中5mC水平明显降低,表明敲除Uhrf1会造成肠上皮组织甲基化水平显著下降(图5A)。研究发现DNA甲基化水平降低会引起DNA损伤应答反应,因此对DNA损伤标志物γH2AX进行免疫组织化学染色,发现敲除Uhrf1后在肠隐窝部位可以看到明显的DNA损伤(图5B),说明Uhrf1的缺失导致肠上皮组织DNA损伤增加。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5肠干细胞相关标志基因的表达
**:P<0.01;***:P<0.001;****:P<0.0001。
Fig. 5The expression of intestinal stem cell marker genes
3 讨论
DNA甲基化作为最稳定的表观遗传修饰,对体内多个自我更新组织中基因的表达调控至关重要,DNA甲基化的异常将造成基因表达的改变,从而导致疾病的发生[18,19]。多项研究证明Uhrf1可招募Dnmt1,在增殖性细胞中维持DNA甲基化信息在细胞分裂前后的稳定传递,缺失Uhrf1将会导致细胞内的DNA甲基化水平大幅下降[20]。本研究发现Uhrf1主要表达在肠隐窝底部区域主要是肠干细胞和快速增殖的祖细胞,而在绒毛等其他部位未见有表达,进一步证明Uhrf1在增殖性细胞中表达丰富,这与之前报道的Uhrf1在其他组织系统中的表达相一致[21]。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6Uhrf1介导的DNA甲基化对肠上皮细胞影响
A:野生型与条件性敲除Uhrf1小鼠肠上皮中5mC免疫组织化学染色。B:野生型与条件性敲除Uhrf1小鼠肠上皮中DNA损伤标志物γH2AX 免疫组织化学染色。红色箭头表示DNA损伤阳性细胞。标尺:50 μm。
Fig. 6The effect of Uhrf1-mediated DNA methylation in intestinal epithelial cells
研究表明Uhrf1可以影响不同组织细胞的增殖和分化并与凋亡过程相关。在对结肠调节性T细胞的研究中发现Uhrf1缺陷的小鼠表现为细胞增殖和正常发育成熟过程受阻,免疫功能降低,并自发形成结肠炎[22]。在斑马鱼肝脏中,敲减Uhrf1可显著抑制肝细胞增殖,造成凋亡增加 [23]。在四肢间充质细胞中敲除Uhrf1后,由于破坏了软骨细胞增殖及分化过程,造成小鼠肢体明显缩短[24]。同样,本研究结果发现Uhrf1的缺失抑制了肠上皮细胞的增殖与分化,诱导细胞凋亡,导致肠上皮发育异常, 并造成肠干细胞相关标记基因表达水平的降低。已有研究证明DNA甲基化水平的降低将会导致基因组不稳定,突变率增加,从而引起DNA损伤反应[25,26,27]。Amy等[28]在结肠癌细胞系HCT116中敲减Uhrf1发现,Uhrf1的丢失导致DNA损伤反应的激活,主要表现为组蛋白H2AX在第139位丝氨酸的磷酸化以及细胞周期检测点激酶2 (checkpoint kinase 2, CHK2)第68位苏氨酸的磷酸化等,并造成细胞通过caspases 8和3途径发生凋亡。为进一步探索Uhrf1基因是如何诱发细胞凋亡,通过免疫组织化学染色方法对肠上皮组织甲基化水平及DNA损伤标志物γH2AX进行检测,结果显示与对照组小鼠相比,Uhrf1的缺失导致肠上皮组织整体甲基化水平降低,DNA损伤增加,因此初步推测可能是Uhrf1的缺失造成DNA甲基化水平大幅下降,诱发DNA损伤,进而引起肠上皮细胞增殖分化减缓,凋亡增加。目前已有研究表明DNA甲基化可以通过调控发育过程中细胞增殖和分化间的平衡来调节肠上皮稳态的建立。Sheaffer等[29,30,31]研究发现Dnmt1介导的维持性DNA甲基化对肠上皮细胞分化过程中的基因表达调控起着关键作用,胚胎期敲除Dnmt1发现Dnmt1的缺失导致小鼠肠上皮细胞增殖下降、绒毛数量减少、基因组甲基化水平降低,并采用转录组学测序技术分析发现与DNA损伤及细胞周期相关的基因表达升高,造成细胞周期阻滞,从而引起细胞死亡,这与本研究中在小鼠胚胎期肠上皮细胞中特异性敲除Uhrf1的表型相类似;进一步在成体小鼠中的研究发现,尽管短期内敲除Dnmt1可导致小鼠体重下降,基因组稳定性及甲基化水平降低,但两个月左右由于Dnmt3b的激活,DNA甲基化水平及肠上皮完整性可以逐渐恢复到正常水平,Dnmt1和Dnmt3b共同负责成体小鼠肠上皮甲基化的维持,目前关于Uhrf1在成体小鼠肠组织中的功能还有待建立动物模型进行下一步的研究。据报道Uhrf1可以通过靶向多个信号通路发挥生物学功能。例如Chen等[32]证明Uhrf1可以通过影响细胞周期抑制蛋白CDKN1A (cyclin dependent kinase inhibitor 1A)以及调节B细胞增殖和衰老家族的Schlafen 1/2(Slfn1/2)表达水平来促进B细胞增殖,进一步应用亚硫酸盐测序分析发现Cdkn1a基因的CpG位点的DNA甲基化水平在Uhrf1敲除后明显降低。Xiang 等[33]发现Uhrf1的缺失导致基底细胞阻滞在G1期,损害了气道再生过程,并表明Uhrf1可能影响了与G1/S期转变相关的细胞周期蛋白依赖性激酶的活性。在自然杀伤性T细胞中Uhrf1被证明可以通过调节蛋白激酶B/哺乳动物雷帕霉素白蛋白(protein kinase B/mammalian target of rapamycin, Akt-mTOR)信号轴来控制细胞的分化,影响细胞的发育[34]。目前关于Uhrf1介导的DNA甲基化对肠上皮发育具体调控机制尚不清楚,考虑到Uhrf1和Dnmt1在甲基化维持中的作用,Uhrf1和Dnmt1是否具有类似的功能与机制,需要更进一步的探索。
综上所述,本研究通过构建在肠上皮中特异性敲除Uhrf1的小鼠模型,发现在胚胎期敲除Uhrf1后,肠上皮发育异常,上皮稳态失衡,Uhrf1的缺失导致DNA甲基化的维持过程被破坏,诱发DNA损伤反应,造成肠上皮细胞的增殖与分化受阻,细胞凋亡增加,首次揭示了Uhrf1介导的DNA甲基化在肠上皮发育中的重要作用,为进一步明确Uhrf1介导的表观遗传机制提供了实验依据。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1038/nrg3354URL [本文引用: 1]

DNA methylation is among the best studied epigenetic modifications and is essential to mammalian development. Although the methylation status of most CpG dinucteotides in the genome is stably propagated through mitosis, improvements to methods for measuring methylation have identified numerous regions in which it is dynamically regulated. In this Review, we discuss key concepts in the function of DNA methylation in mammals, stemming from more than two decades of research, including many recent studies that have elucidated when and where DNA methylation has a regulatory role in the genome. We include insights from early development, embryonic stem cells and adult Lineages, particularly haematopoiesis, to highlight the general features of this modification as it participates in both global and localized epigenetic regulation..
DOI:10.1038/s41580-019-0159-6URLPMID:31399642 [本文引用: 1]

DNA methylation is of paramount importance for mammalian embryonic development. DNA methylation has numerous functions: it is implicated in the repression of transposons and genes, but is also associated with actively transcribed gene bodies and, in some cases, with gene activation per se. In recent years, sensitive technologies have been developed that allow the interrogation of DNA methylation patterns from a small number of cells. The use of these technologies has greatly improved our knowledge of DNA methylation dynamics and heterogeneity in embryos and in specific tissues. Combined with genetic analyses, it is increasingly apparent that regulation of DNA methylation erasure and (re-)establishment varies considerably between different developmental stages. In this Review, we discuss the mechanisms and functions of DNA methylation and demethylation in both mice and humans at CpG-rich promoters, gene bodies and transposable elements. We highlight the dynamic erasure and re-establishment of DNA methylation in embryonic, germline and somatic cell development. Finally, we provide insights into DNA methylation gained from studying genetic diseases.
DOI:10.1038/nature14192URLPMID:25592537 [本文引用: 1]

Cytosine methylation is a DNA modification generally associated with transcriptional silencing. Factors that regulate methylation have been linked to human disease, yet how they contribute to malignances remains largely unknown. Genomic maps of DNA methylation have revealed unexpected dynamics at gene regulatory regions, including active demethylation by TET proteins at binding sites for transcription factors. These observations indicate that the underlying DNA sequence largely accounts for local patterns of methylation. As a result, this mark is highly informative when studying gene regulation in normal and diseased cells, and it can potentially function as a biomarker. Although these findings challenge the view that methylation is generally instructive for gene silencing, several open questions remain, including how methylation is targeted and recognized and in what context it affects genome readout.
DOI:10.1038/nrm.2016.6URLPMID:26883001 [本文引用: 1]

Differentiating somatic cells are progressively restricted to specialized functions during ontogeny, but they can be experimentally directed to form other cell types, including those with complete embryonic potential. Early nuclear reprogramming methods, such as somatic cell nuclear transfer (SCNT) and cell fusion, posed significant technical hurdles to precise dissection of the regulatory programmes governing cell identity. However, the discovery of reprogramming by ectopic expression of a defined set of transcription factors, known as direct reprogramming, provided a tractable platform to uncover molecular characteristics of cellular specification and differentiation, cell type stability and pluripotency. We discuss the control and maintenance of cellular identity during developmental transitions as they have been studied using direct reprogramming, with an emphasis on transcriptional and epigenetic regulation.
DOI:10.1093/biolre/ioaa026URLPMID:32101289 [本文引用: 1]
DOI:10.2147/OTT.S192234URLPMID:30666134 [本文引用: 1]

Ubiquitin-like with plant homeodomain and really interesting new gene finger domains 1 (UHRF1) functions as an epigenetic regulator recruiting PCNA, DNMT1, histone deacetylase 1, G9a, SuV39H, herpes virus-associated ubiquitin-specific protease, and Tat-interactive protein by multiple corresponding domains of DNA and H3 to maintain DNA methylation and histone modifications. Overexpression of UHRF1 has been found as a potential biomarker in various cancers resulting in either DNA hypermethylation or global DNA hypo-methylation, which participates in the occurrence, progression, and invasion of cancer. The role of UHRF1 in the reciprocal interaction between DNA methylation and histone modifications, the dynamic structural transformation of UHRF1 protein within epigenetic code replication machinery in epigenetic regulations, as well as modifications during cell cycle and chemotherapy targeting UHRF1 are evaluated in this study.
DOI:10.1038/nature06397URLPMID:17994007 [本文引用: 1]

DNA methyltransferase (cytosine-5) 1 (Dnmt1) is the principal enzyme responsible for maintenance of CpG methylation and is essential for the regulation of gene expression, silencing of parasitic DNA elements, genomic imprinting and embryogenesis. Dnmt1 is needed in S phase to methylate newly replicated CpGs occurring opposite methylated ones on the mother strand of the DNA, which is essential for the epigenetic inheritance of methylation patterns in the genome. Despite an intrinsic affinity of Dnmt1 for such hemi-methylated DNA, the molecular mechanisms that ensure the correct loading of Dnmt1 onto newly replicated DNA in vivo are not understood. The Np95 (also known as Uhrf1 and ICBP90) protein binds methylated CpG through its SET and RING finger-associated (SRA) domain. Here we show that localization of mouse Np95 to replicating heterochromatin is dependent on the presence of hemi-methylated DNA. Np95 forms complexes with Dnmt1 and mediates the loading of Dnmt1 to replicating heterochromatic regions. By using Np95-deficient embryonic stem cells and embryos, we show that Np95 is essential in vivo to maintain global and local DNA methylation and to repress transcription of retrotransposons and imprinted genes. The link between hemi-methylated DNA, Np95 and Dnmt1 thus establishes key steps of the mechanism for epigenetic inheritance of DNA methylation.
DOI:10.3390/genes9120600URL [本文引用: 1]
DOI:10.1074/jbc.M112.415398URLPMID:23161542 [本文引用: 1]

UHRF1 is an important epigenetic regulator connecting DNA methylation and histone methylations. UHRF1 is required for maintenance of DNA methylation through recruiting DNMT1 to DNA replication forks. Recent studies have shown that the plant homeodomain (PHD) of UHRF1 recognizes the N terminus of unmodified histone H3, and the interaction is inhibited by methylation of H3R2, whereas the tandem tudor domain (TTD) of UHRF1 recognizes trimethylated histone H3 lysine 9 (H3K9me3). However, how the two domains of UHRF1 coordinately recognize histone methylations remains elusive. In this report, we identified that PHD largely enhances the interaction between TTD and H3K9me3. We present the crystal structure of UHRF1 containing both TTD and PHD (TTD-PHD) in complex with H3K9m3 peptide at 3.0 A resolution. The structure shows that TTD-PHD binds to the H3K9me3 peptide with 1:1 stoichiometry with the two domains connected by the H3K9me3 peptide and a linker region. The TTD interacts with residues Arg-8 and trimethylated Lys-9, and the PHD interacts with residues Ala-1, Arg-2, and Lys-4 of the H3K9me3 peptide. The biochemical experiments indicate that PHD-mediated recognition of unmodified H3 is independent of the TTD, whereas TTD-mediated recognition of H3K9me3 PHD. Thus, both TTD and PHD are essential for specific recognition of H3K9me3 by UHRF1. Interestingly, the H3K9me3 peptide induces conformational changes of TTD-PHD, which do not affect the autoubiquitination activity or hemimethylated DNA binding affinity of UHRF1 in vitro. Taken together, our studies provide structural insight into the coordinated recognition of H3K9me3 by the TTD and PHD of UHRF1.
DOI:10.1038/nature12488URL [本文引用: 1]

Faithful propagation of DNA methylation patterns during DNA replication is critical for maintaining cellular phenotypes of individual differentiated cells(1-5). Although it is well established that Uhrf1 (ubiquitin-like with PHD and ring finger domains 1; also known as Np95 and ICBP90) specifically binds to hemi-methylated DNA through its SRA (SET and RING finger associated) domain and has an essential role in maintenance of DNA methylation by recruiting Dnmt1 to hemi-methylated DNA sites(6-10), the mechanism by which Uhrf1 coordinates the maintenance of DNA methylation and DNA replication is largely unknown. Here we show that Uhrf1-dependent histone H3 ubiquitylation has a prerequisite role in the maintenance DNA methylation. Using Xenopus egg extracts, we successfully reproduce maintenance DNA methylation in vitro. Dnmt1 depletion results in a marked accumulation of Uhrf1-dependent ubiquitylation of histone H3 at lysine 23. Dnmt1 preferentially associates with ubiquitylated H3 in vitro though a region previously identified as a replication foci targeting sequence(11). The RING finger mutant of Uhrf1 fails to recruit Dnmt1 to DNA replication sites and maintain DNA methylation in mammalian cultured cells. Our findings represent the first evidence, to our knowledge, of the mechanistic link between DNA methylation and DNA replication through histone H3 ubiquitylation.
DOI:10.7554/eLife.17101URLPMID:27595565 [本文引用: 1]

The epigenetic inheritance of DNA methylation requires UHRF1, a histone- and DNA-binding RING E3 ubiquitin ligase that recruits DNMT1 to sites of newly replicated DNA through ubiquitylation of histone H3. UHRF1 binds DNA with selectivity towards hemi-methylated CpGs (HeDNA); however, the contribution of HeDNA sensing to UHRF1 function remains elusive. Here, we reveal that the interaction of UHRF1 with HeDNA is required for DNA methylation but is dispensable for chromatin interaction, which is governed by reciprocal positive cooperativity between the UHRF1 histone- and DNA-binding domains. HeDNA recognition activates UHRF1 ubiquitylation towards multiple lysines on the H3 tail adjacent to the UHRF1 histone-binding site. Collectively, our studies are the first demonstrations of a DNA-protein interaction and an epigenetic modification directly regulating E3 ubiquitin ligase activity. They also define an orchestrated epigenetic control mechanism involving modifications both to histones and DNA that facilitate UHRF1 chromatin targeting, H3 ubiquitylation, and DNA methylation inheritance.
DOI:10.1038/sj.onc.1208878URLPMID:16007129 [本文引用: 1]

The retinoblastoma protein (pRB) is encoded by the RB1 gene whose promoter contains several putative binding sites for ICBP90 (Inverted CCAAT box Binding Protein of 90 kDa), a transcriptional regulator of the topoisomerase IIalpha gene. ICBP90 has two consensus binding sites for pRB in its primary sequence. Here, we show that pRB and ICBP90 co-immunoprecipitate in cell extracts of proliferating human lung fibroblasts and of proliferating or confluent Jurkat cells. GST pull-down assays and immunocytochemistry, after cell synchronization in late G1 phase, confirmed this interaction. Overexpression of ICBP90 induces downregulation of pRB expression in lung fibroblasts as a result of mRNA decrease. DNA chromatin immunoprecipitation experiment shows that ICBP90 binds to the RB1 gene promoter under its methylated status. Overexpression of ICBP90 increases the S and G2/M phase cell fractions of serum-starved lung fibroblasts as assessed by flow cytometry analysis and increases topoisomerase IIalpha expression. Together, these results show that ICBP90 regulates pRB at the protein and gene transcription levels, thus favoring the entry into the S phase of the cells. We propose that ICBP90 overexpression, found in cancer cells, is involved in the altered checkpoint controls occurring in cancerogenesis.
DOI:10.1016/j.celrep.2015.03.038URLPMID:25818288 [本文引用: 1]

We identified ubiquitin-like with PHD and RING finger domain 1 (UHRF1) as a binding factor for DNA interstrand crosslink (ICL) lesions through affinity purification of ICL-recognition activities. UHRF1 is recruited to DNA lesions in vivo and binds directly to ICL-containing DNA. UHRF1-deficient cells display increased sensitivity to a variety of DNA damages. We found that loss of UHRF1 led to retarded lesion processing and reduced recruitment of ICL repair nucleases to the site of DNA damage. UHRF1 interacts physically with both ERCC1 and MUS81, two nucleases involved in the repair of ICL lesions. Depletion of both UHRF1 and components of the Fanconi anemia (FA) pathway resulted in increased DNA damage sensitivity compared to defect of each mechanism alone. These results suggest that UHRF1 promotes recruitment of lesion-processing activities via its affinity to recognize DNA damage and functions as a nuclease recruitment scaffold in parallel to the FA pathway.
DOI:10.1371/journal.pgen.1007042URLPMID:28976982 [本文引用: 1]

The methylation of cytosine at CG sites in the mammalian genome is dynamically reprogrammed during gametogenesis and preimplantation development. It was previously shown that oocyte-derived DNMT1 (a maintenance methyltransferase) is essential for maintaining and propagating CG methylation at imprinting control regions in preimplantation embryos. In mammalian somatic cells, hemimethylated-CG-binding protein UHRF1 plays a critical role in maintaining CG methylation by recruiting DNMT1 to hemimethylated CG sites. However, the role of UHRF1 in oogenesis and preimplantation development is unknown. In the present study, we show that UHRF1 is mainly, but not exclusively, localized in the cytoplasm of oocytes and preimplantation embryos. However, smaller amounts of UHRF1 existed in the nucleus, consistent with the expected role in DNA methylation. We then generated oocyte-specific Uhrf1 knockout (KO) mice and found that, although oogenesis was itself unaffected, a large proportion of the embryos derived from the KO oocytes died before reaching the blastocyst stage (a maternal effect). Whole genome bisulfite sequencing revealed that blastocysts derived from KO oocytes have a greatly reduced level of CG methylation, suggesting that maternal UHRF1 is essential for maintaining CG methylation, particularly at the imprinting control regions, in preimplantation embryos. Surprisingly, UHRF1 was also found to contribute to de novo CG and non-CG methylation during oocyte growth: in Uhrf1 KO oocytes, transcriptionally-inactive regions gained less methylation, while actively transcribed regions, including the imprinting control regions, were unaffected or only slightly affected. We also found that de novo methylation was defective during the late stage of oocyte growth. To the best of our knowledge, this is the first study to demonstrate the role of UHRF1 in de novo DNA methylation in vivo. Our study reveals multiple functions of UHRF1 during the global epigenetic reprogramming of oocytes and early embryos.
DOI:10.1242/dev.115980URLPMID:25564650 [本文引用: 1]

UHRF1 (ubiquitin-like, containing PHD and RING finger domains, 1) recruits DNMT1 to hemimethylated DNA during replication and is essential for maintaining DNA methylation. uhrf1 mutant zebrafish have global DNA hypomethylation and display embryonic defects, including a small liver, and they die as larvae. We make the surprising finding that, despite their reduced organ size, uhrf1 mutants express high levels of genes controlling S-phase and have many more cells undergoing DNA replication, as measured by BrdU incorporation. In contrast to wild-type hepatocytes, which are continually dividing during hepatic outgrowth and thus dilute the BrdU label, uhrf1 mutant hepatocytes retain BrdU throughout outgrowth, reflecting cell cycle arrest. Pulse-chase-pulse experiments with BrdU and EdU, and DNA content analysis indicate that uhrf1 mutant cells undergo DNA re-replication and that apoptosis is the fate of many of the re-replicating and arrested hepatocytes. Importantly, the DNA re-replication phenotype and hepatic outgrowth failure are preceded by global loss of DNA methylation. Moreover, uhrf1 mutants are phenocopied by mutation of dnmt1, and Dnmt1 knockdown in uhrf1 mutants enhances their small liver phenotype. Together, these data indicate that unscheduled DNA replication and failed cell cycle progression leading to apoptosis are the mechanisms by which DNA hypomethylation prevents organ expansion in uhrf1 mutants. We propose that cell cycle arrest leading to apoptosis is a strategy that restricts propagation of epigenetically damaged cells during embryogenesis.
DOI:10.4049/jimmunol.1901471URLPMID:32358021 [本文引用: 1]

Thymocyte differentiation is a highly complex process that is accompanied by epigenetic changes. Ubiquitin-like containing PHD ring finger 1 (UHRF1) is a critical epigenetic modifier involved in various cellular processes. In this study, we demonstrated that it is highly expressed in T cell precursors of the thymus. Further, its deficiency results in significantly reduced thymocyte cellularity and thymus size in mice. Through systematic analysis based on single-cell RNA sequencing, we found that UHRF1 deficiency thwarts alphabeta T cell lineage development, whereas biasing gammadelta T lineage differentiation dampens the progression of immature single-positive cells. UHRF1 deficiency promotes the IL-17 secreting and RORgammat expression in gammadelta T cell, indicating a Tgammadelta17 phenotype. Further, the analysis of gene-regulatory networks demonstrated that UHRF1 controls the expression of early growth response 1 (EGR1). UHRF1 interacts with DNA methyltransferase 1 (DNMT1) at the CpG promoter region of Egr1 loci and affects the nearby chromatin modifications of H3K9me3 and H3K4me3. Taken together, our results demonstrate that UHRF1 is a key factor that mediates the epigenetic regulation of EGR1 and, consequently, thymocyte fate decisions.
DOI:10.1101/gad.284992.116URLPMID:27798843 [本文引用: 1]

In order to understand whether early epigenetic mechanisms instruct the long-term behavior of neural stem cells (NSCs) and their progeny, we examined Uhrf1 (ubiquitin-like PHD ring finger-1; also known as Np95), as it is highly expressed in NSCs of the developing brain and rapidly down-regulated upon differentiation. Conditional deletion of Uhrf1 in the developing cerebral cortex resulted in rather normal proliferation and neurogenesis but severe postnatal neurodegeneration. During development, deletion of Uhrf1 lead to global DNA hypomethylation with a strong activation of the intracisternal A particle (IAP) family of endogenous retroviral elements, accompanied by an increase in 5-hydroxymethylcytosine. Down-regulation of Tet enzymes rescued the IAP activation in Uhrf1 conditional knockout (cKO) cells, suggesting an antagonistic interplay between Uhrf1 and Tet on IAP regulation. As IAP up-regulation persists into postnatal stages in the Uhrf1 cKO mice, our data show the lack of means to repress IAPs in differentiating neurons that normally never express Uhrf1 The high load of viral proteins and other transcriptional deregulation ultimately led to postnatal neurodegeneration. Taken together, these data show that early developmental NSC factors can have long-term effects in neuronal differentiation and survival. Moreover, they highlight how specific the consequences of widespread changes in DNA methylation are for certain classes of retroviral elements.
DOI:10.1038/nature08683URLPMID:20081831 [本文引用: 1]

Progenitor cells maintain self-renewing tissues throughout life by sustaining their capacity for proliferation while suppressing cell cycle exit and terminal differentiation. DNA methylation provides a potential epigenetic mechanism for the cellular memory needed to preserve the somatic progenitor state through repeated cell divisions. DNA methyltransferase 1 (DNMT1) maintains DNA methylation patterns after cellular replication. Although dispensable for embryonic stem cell maintenance, the role for DNMT1 in maintaining the progenitor state in constantly replenished somatic tissues, such as mammalian epidermis, is unclear. Here we show that DNMT1 is essential for epidermal progenitor cell function. DNMT1 protein was found enriched in undifferentiated cells, where it was required to retain proliferative stamina and suppress differentiation. In tissue, DNMT1 depletion led to exit from the progenitor cell compartment, premature differentiation and eventual tissue loss. Genome-wide analysis showed that a significant portion of epidermal differentiation gene promoters were methylated in self-renewing conditions but were subsequently demethylated during differentiation. Furthermore, UHRF1 (refs 9, 10), a component of the DNA methylation machinery that targets DNMT1 to hemi-methylated DNA, is also necessary to suppress premature differentiation and sustain proliferation. In contrast, Gadd45A and B, which promote active DNA demethylation, are required for full epidermal differentiation gene induction. These data demonstrate that proteins involved in the dynamic regulation of DNA methylation patterns are required for progenitor maintenance and self-renewal in mammalian somatic tissue.
URLPMID:23017451 [本文引用: 1]

DNA methylation is an important epigenetic modification that regulates temporal and spatial expression of genes for controlling cell fate and differentiation. Recently, DNA methylation has been demonstrated to be required for vertebrate early embryogenesis. Loss of Dnmt genes in zebrafish and mice caused defects in organogenesis and tissue terminal differentiation. This paper summarizes the dynamic expression pattern of Dnmt genes and the roles of DNA methylation from early embryogenesis to organogenesis in both mice and zebrafish, specifically, how DNA methylation and histone modifications cooperatively regulate gene transcription during these processes. Better understanding of DNA methylation in vertebrate embryogenesis will provide insights for new treatment of DNA methylation-related human diseases.
URLPMID:23017451 [本文引用: 1]

DNA methylation is an important epigenetic modification that regulates temporal and spatial expression of genes for controlling cell fate and differentiation. Recently, DNA methylation has been demonstrated to be required for vertebrate early embryogenesis. Loss of Dnmt genes in zebrafish and mice caused defects in organogenesis and tissue terminal differentiation. This paper summarizes the dynamic expression pattern of Dnmt genes and the roles of DNA methylation from early embryogenesis to organogenesis in both mice and zebrafish, specifically, how DNA methylation and histone modifications cooperatively regulate gene transcription during these processes. Better understanding of DNA methylation in vertebrate embryogenesis will provide insights for new treatment of DNA methylation-related human diseases.
DOI:10.1126/science.1147939URLPMID:17673620 [本文引用: 1]

Epigenetic inheritance in mammals relies in part on robust propagation of DNA methylation patterns throughout development. We show that the protein UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1), also known as NP95 in mouse and ICBP90 in human, is required for maintaining DNA methylation. UHRF1 colocalizes with the maintenance DNA methyltransferase protein DNMT1 throughout S phase. UHRF1 appears to tether DNMT1 to chromatin through its direct interaction with DNMT1. Furthermore UHRF1 contains a methyl DNA binding domain, the SRA (SET and RING associated) domain, that shows strong preferential binding to hemimethylated CG sites, the physiological substrate for DNMT1. These data suggest that UHRF1 may help recruit DNMT1 to hemimethylated DNA to facilitate faithful maintenance of DNA methylation.
DOI:10.1002/stem.2889URLPMID:29999568 [本文引用: 1]

Adult neurogenesis in the brain continuously seeds new neurons throughout life, but how homeostasis of adult neural stem cells (NSCs) is maintained is incompletely understood. Here, we demonstrate that the DNA methylation adapter ubiquitin-like, containing PHD and RING finger domains-1 (UHRF1) is expressed in, and regulates proliferation of, the active but not quiescent pool of adult neural progenitor cells. Mice with a neural stem cell-specific deficiency in UHRF1 exhibit a massive depletion of neurogenesis resulting in a collapse of formation of new neurons. In the absence of UHRF1, NSCs unexpectedly remain in the cell cycle but with a 17-fold increased cell cycle length due to a failure of replication phase entry caused by promoter demethylation and derepression of Cdkn1a, which encodes the cyclin-dependent kinase inhibitor p21. UHRF1 does not affect the proportion progenitor cells active within the cell cycle but among these cells, UHRF1 is critical for licensing replication re-entry. Therefore, this study shows that a UHRF1-Cdkn1a axis is essential for the control of stem cell self-renewal and neurogenesis in the adult brain. Stem Cells 2018;36:1736-1751.
DOI:10.1038/ni.2886URL [本文引用: 1]

Intestinal regulatory T cells (T-reg cells) are necessary for the suppression of excessive immune responses to commensal bacteria. However, the molecular machinery that controls the homeostasis of intestinal ireg cells has remained largely unknown. Here we report that colonization of germ-free mice with gut microbiota upregulated expression of the DNA-methylation adaptor Uhrf1 in T-reg cells. Mice with T cell specific deficiency in Uhrf1 (Uhrf1(fl/fl)Cd4-Cre mice) showed defective proliferation and functional maturation of colonic T-reg cells. Uhrf1 deficiency resulted in derepression of the gene (Cdkn1a) that encodes the cyclin-dependent kinase inhibitor p21 due to hypomethylation of its promoter region, which resulted in cell-cycle arrest of T-reg cells. As a consequence, Uhrf1(fl/fl)Cd4-Cre mice spontaneously developed severe colitis. Thus, Uhrf1-dependent epigenetic silencing of Cdknla was required for the maintenance of gut immunological homeostasis. This mechanism enforces symbiotic host-microbe interactions without an inflammatory response.
DOI:10.1073/pnas.0610774104URLPMID:17242348 [本文引用: 1]

In contrast to the deregulated hepatocellular division that is a feature of many hepatic diseases and malignancies, physiologic liver growth during embryonic development and after partial hepatectomy (PH) in adults is characterized by tightly controlled cell proliferation. We used forward genetic screening in zebrafish to test the hypothesis that a similar genetic program governs physiologic liver growth during hepatogenesis and regeneration after PH. We identified the uhrf1 gene, a cell cycle regulator and transcriptional activator of top2a expression, as required for hepatic outgrowth and embryonic survival. By developing a methodology to perform PH on adult zebrafish, we found that liver regeneration inuhrf1+/- adult animals is impaired.uhrf1 transcript levels dramatically increase after PH in both mice, and zebrafish and top2a is not up-regulated in uhrf1+/- livers after PH. This indicates that uhrf1 is required for physiologic liver growth in both embryos and adults and illustrates that zebrafish livers regenerate.
DOI:10.1242/dev.160051URLPMID:29217751 [本文引用: 1]

The overall bauplan of the tetrapod brain is highly conserved, yet significant variations exist among species in terms of brain size, structural composition and cellular diversity. Understanding processes underlying neural and behavioral development in a wide range of species is important both from an evolutionary developmental perspective as well as for the identification of cell sources with post-developmental neurogenic potential. Here, we characterize germinal processes in the brain of Notophthalmus viridescens and Pleurodeles waltl during both development and adulthood. Using a combination of cell tracking tools, including clonal analyses in new transgenic salamander lines, we examine the origin of neural stem and progenitor cells found in the adult brain, determine regional variability in cell cycle length of progenitor cells, and show spatiotemporally orchestrated neurogenesis. We analyze how maturation of different brain regions and neuronal subpopulations are linked to the acquisition of complex behaviors, and how these behaviors are altered upon chemical ablation of dopamine neurons. Our data analyzed from an evolutionary perspective reveal both common and species-specific processes in tetrapod brain formation and function.
DOI:10.1091/mbc.e05-03-0194URLPMID:16195352 [本文引用: 1]

Early cellular events associated with tumorigenesis often include loss of cell cycle checkpoints or alteration in growth signaling pathways. Identification of novel genes involved in cellular proliferation may lead to new classes of cancer therapeutics. By screening a tetracycline-inducible cDNA library in A549 cells for genes that interfere with proliferation, we have identified a fragment of UHRF1 (ubiquitin-like protein containing PHD and RING domains 1), a nuclear RING finger protein, that acts as a dominant negative effector of cell growth. Reduction of UHRF1 levels using an UHRF1-specific shRNA decreased growth rates in several tumor cell lines. In addition, treatment of A549 cells with agents that activated different cell cycle checkpoints resulted in down-regulation of UHRF1. The primary sequence of UHRF1 contains a PHD and a RING motif, both of which are structural hallmarks of ubiquitin E3 ligases. We have confirmed using an in vitro autoubiquitination assay that UHRF1 displays RING-dependent E3 ligase activity. Overexpression of a GFP-fused UHRF1 RING mutant that lacks ligase activity sensitizes cells to treatment with various chemotherapeutics. Taken together, our results suggest a general requirement for UHRF1 in tumor cell proliferation and implicate the RING domain of UHRF1 as a functional determinant of growth regulation.
DOI:10.1073/pnas.1116349109URLPMID:22411829 [本文引用: 1]

UHRF1 (Ubiquitin-like, with PHD and RING finger domains 1) plays an important role in DNA CpG methylation, heterochromatin function and gene expression. Overexpression of UHRF1 has been suggested to contribute to tumorigenesis. However, regulation of UHRF1 is largely unknown. Here we show that the deubiquitylase USP7 interacts with UHRF1. Using interaction-defective and catalytic mutants of USP7 for complementation experiments, we demonstrate that both physical interaction and catalytic activity of USP7 are necessary for UHRF1 ubiquitylation and stability regulation. Mass spectrometry analysis identified phosphorylation of serine (S) 652 within the USP7-interacting domain of UHRF1, which was further confirmed by a UHRF1 S652 phosphor (S652ph)-specific antibody. Importantly, the S652ph antibody identifies phosphorylated UHRF1 in mitotic cells and consistently S652 can be phosphorylated by the M phase-specific kinase CDK1-cyclin B in vitro. UHRF1 S652 phosphorylation significantly reduces UHRF1 interaction with USP7 in vitro and in vivo, which is correlated with a decreased UHRF1 stability in the M phase of the cell cycle. In contrast, UHRF1 carrying the S652A mutation, which renders UHRF1 resistant to phosphorylation at S652, is more stable. Importantly, cells carrying the S652A mutant grow more slowly suggesting that maintaining an appropriate level of UHRF1 is important for cell proliferation regulation. Taken together, our findings uncovered a cell cycle-specific signaling event that relieves UHRF1 from its interaction with USP7, thus exposing UHRF1 to proteasome-mediated degradation. These findings identify a molecular mechanism by which cellular UHRF1 level is regulated, which may impact cell proliferation.
DOI:10.1093/hmg/ddr236URL [本文引用: 1]

DNA methyltransferase 1 (DNMT1) maintains methylation at CpG dinucleotides, important for transcriptional silencing at many loci. It is also implicated in stabilizing repeat sequences: DNMT1 deficiency causes microsatellite instability in mouse embryonic stem cells, but it is unclear how this occurs, how repeats lacking CpG become unstable and whether the effect is confined to stem cells. To address these questions, we transfected hTERT-immortalized normal human fibroblasts (hTERT-1604) with a short hairpin RNA construct targeting DNMT1 and isolated stable integrants with different levels of protein. DNMT1 expression levels agreed well with methylation levels at imprinted genes. Knockdown cells showed two key characteristics of mismatch repair (MMR) deficiency, namely resistance to the drug 6-thioguanine and up to 10-fold elevated mutation rates at a CA(17) microsatellite reporter, but had limited viability. The likely cause of MMR defects is a matching drop in steady-state protein levels for key repair components in DNMT1 knockdown cells, affecting both the MutL alpha and MutS alpha complexes. This indirect effect on MMR proteins was also seen using a different targeting method in HT29 colon cancer cells and did not involve transcriptional silencing of the respective genes. Decreased levels of MMR components follow activation of the DNA damage response and blocking this response, and in particular poly(ADP-ribose) polymerase (PARP) overactivation, rescues cell viability in DNMT1-depleted cells. These results offer an explanation for how and why unmethylated microsatellite repeats can be destabilized in cells with decreased DNMT1 levels and uncover a novel and important role for PARP in this process.
DOI:10.1042/BJ20100840URLPMID:21214517 [本文引用: 1]

UHRF1 [ubiquitin-like protein, containing PHD (plant homeodomain) and RING finger domains 1] is required for cell cycle progression and epigenetic regulation. In the present study, we show that depleting cancer cells of UHRF1 causes activation of the DNA damage response pathway, cell cycle arrest in G2/M-phase and apoptosis dependent on caspase 8. The DNA damage response in cells depleted of UHRF1 is illustrated by: phosphorylation of histone H2AX on Ser139, phosphorylation of CHK (checkpoint kinase) 2 on Thr68, phosphorylation of CDC25 (cell division control 25) on Ser216 and phosphorylation of CDK1 (cyclin-dependent kinase 1) on Tyr15. Moreover, we find that UHRF1 accumulates at sites of DNA damage suggesting that the cell cycle block in UHRF1-depleted cells is due to an important role in damage repair. The consequence of UHRF1 depletion is apoptosis; cells undergo activation of caspases 8 and 3, and depletion of caspase 8 prevents cell death induced by UHRF1 knockdown. Interestingly, the cell cycle block and apoptosis occurs in p53-containing and -deficient cells. From the present study we conclude that UHRF1 links epigenetic regulation with DNA replication.
DOI:10.1101/gad.230318.113URLPMID:24637118 [本文引用: 1]

The mammalian intestinal epithelium has a unique organization in which crypts harboring stem cells produce progenitors and finally clonal populations of differentiated cells. Remarkably, the epithelium is replaced every 3-5 d throughout adult life. Disrupted maintenance of the intricate balance of proliferation and differentiation leads to loss of epithelial integrity or barrier function or to cancer. There is a tight correlation between the epigenetic status of genes and expression changes during differentiation; however, the mechanism of how changes in DNA methylation direct gene expression and the progression from stem cells to their differentiated descendants is unclear. Using conditional gene ablation of the maintenance methyltransferase Dnmt1, we demonstrate that reducing DNA methylation causes intestinal crypt expansion in vivo. Determination of the base-resolution DNA methylome in intestinal stem cells and their differentiated descendants shows that DNA methylation is dynamic at enhancers, which are often associated with genes important for both stem cell maintenance and differentiation. We establish that the loss of DNA methylation at intestinal stem cell gene enhancers causes inappropriate gene expression and delayed differentiation.
DOI:10.1242/dev.117341URLPMID:26023099 [本文引用: 1]

The DNA methyltransferase Dnmt1 maintains DNA methylation patterns and genomic stability in several in vitro cell systems. Ablation of Dnmt1 in mouse embryos causes death at the post-gastrulation stage; however, the functions of Dnmt1 and DNA methylation in organogenesis remain unclear. Here, we report that Dnmt1 is crucial during perinatal intestinal development. Loss of Dnmt1 in intervillus progenitor cells causes global hypomethylation, DNA damage, premature differentiation, apoptosis and, consequently, loss of nascent villi. We further confirm the crucial role of Dnmt1 during crypt development using the in vitro organoid culture system, and illustrate a clear differential requirement for Dnmt1 in immature versus mature organoids. These results demonstrate an essential role for Dnmt1 in maintaining genomic stability during intestinal development and the establishment of intestinal crypts.
DOI:10.7554/eLife.12975URLPMID:26808831 [本文引用: 1]

Dnmt1 is critical for immediate postnatal intestinal development, but is not required for the survival of the adult intestinal epithelium, the only rapidly dividing somatic tissue for which this has been shown. Acute Dnmt1 deletion elicits dramatic hypomethylation and genomic instability. Recovery of DNA methylation state and intestinal health is dependent on the de novo methyltransferase Dnmt3b. Ablation of both Dnmt1 and Dnmt3b in the intestinal epithelium is lethal, while deletion of either Dnmt1 or Dnmt3b has no effect on survival. These results demonstrate that Dnmt1 and Dnmt3b cooperate to maintain DNA methylation and genomic integrity in the intestinal epithelium.
DOI:10.1084/jem.20171815URLPMID:29618490 [本文引用: 1]

The production of high-affinity antibody is essential for pathogen clearance. Antibody affinity is increased through germinal center (GC) affinity maturation, which relies on BCR somatic hypermutation (SHM) followed by antigen-based selection. GC B cell proliferation is essentially involved in these processes; it provides enough templates for SHM and also serves as a critical mechanism of positive selection. In this study, we show that expression of epigenetic regulator ubiquitin-like with PHD and RING finger domains 1 (Uhrf1) was markedly up-regulated by c-Myc-AP4 in GC B cells, and it was required for GC response. Uhrf1 regulates cell proliferation-associated genes including cdkn1a, slfn1, and slfn2 by DNA methylation, and its deficiency inhibited the GC B cell cycle at G1-S phase. Subsequently, GC B cell SHM and affinity maturation were impaired, and Uhrf1 GC B knockout mice were unable to control chronic virus infection. Collectively, our data suggest that Uhrf1 regulates GC B cell proliferation and affinity maturation, and its expression in GC B cells is required for virus clearance.
DOI:10.1038/celldisc.2017.19URLPMID:28626588 [本文引用: 1]

Cellular senescence is a cell fate characterized by an irreversible cell cycle arrest, but the molecular mechanism underlying this senescence hallmark remains poorly understood. Through an unbiased search for novel senescence regulators in airway basal cells, we discovered that the epigenetic regulator ubiquitin-like with PHD and ring finger domain-containing protein 1 (UHRF1) is critical for regulating cell cycle progression. Upon injury, basal cells in the mouse airway rapidly induce the expression of UHRF1 in order to stimulate stem cell proliferation and tissue repair. Targeted depletion of Uhrf1 specifically in airway basal cells causes a profound defect in cell cycle progression. Consistently, cultured primary human basal cells lacking UHRF1 do not exhibit cell death or differentiation phenotypes but undergo a spontaneous program of senescence. Mechanistically, UHRF1 loss induces G1 cell cycle arrest by abrogating DNA replication factory formation as evidenced by loss of proliferating cell nuclear antigen (PCNA) puncta and an inability to enter the first cell cycle. This proliferation defect is partially mediated by the p15 pathway. Overall, our study provides the first evidence of an indispensable role of UHRF1 in somatic stem cells proliferation during the process of airway regeneration.
DOI:10.1016/j.celrep.2016.03.016URLPMID:27050515 [本文引用: 1]

Uhrf1 (also known as Np95) is a regulator of DNA methylation and histone ubiquitination and plays an important role in embryogenesis and tumorigenesis. Here, we report that Uhrf1 is essential for invariant natural killer T (iNKT) cell development. We found that Uhrf1 was significantly upregulated in stage 1 iNKT cells. Targeted disruption of Uhrf1 resulted in stage 1-specific transition defects as observed by not only increased apoptosis, but also aberrant effector differentiation, which eventually led to the impaired generation of iNKT cells in Uhrf1-deficient mice. Notably, Uhrf1 deficiency resulted in attenuated activation of Akt-mTOR signaling in stage 1 iNKT cells and overexpression of active Akt rescued iNKT cell developmental defects. Collectively, our results suggest that Uhrf1 regulation of the Akt-mTOR signaling pathway is required for iNKT cell development.
