 ,上海交通大学系统生物医学研究院比较生物医学研究中心,系统生物医学教育部重点实验室,上海 200240
,上海交通大学系统生物医学研究院比较生物医学研究中心,系统生物医学教育部重点实验室,上海 200240RFX5 regulates gene expression of the Pcdhα cluster
Na Wang, Zhilian Jia, Qiang Wu ,Center for Comparative Biomedicine, Key laboratory of Systems Biomedicine (Ministry of Education), Institute of Systems Biomedicine, Shanghai Jiao Tong University, Shanghai 200240, China
,Center for Comparative Biomedicine, Key laboratory of Systems Biomedicine (Ministry of Education), Institute of Systems Biomedicine, Shanghai Jiao Tong University, Shanghai 200240, China通讯作者: 吴强,博士,教授,研究方向:三维基因组架构与功能。E-mail:qwu123@gmail.com
编委: 方向东
收稿日期:2020-06-19修回日期:2020-07-8网络出版日期:2020-08-20
| 基金资助: |
Received:2020-06-19Revised:2020-07-8Online:2020-08-20
| Fund supported: |
作者简介 About authors
王娜,在读硕士研究生,专业方向:生物学。E-mail:

摘要
基因的表达调控与基因组在细胞核内的三维空间架构相辅相成,原钙粘蛋白(protocadherin, Pcdh)基因簇在大脑发育中起到关键作用,可以作为研究基因表达调控机制的模式基因。转录因子RFX5 (regulatory factor x 5)是翼螺旋家族(winged HLH family)的成员,其蛋白由寡聚化结构域、DNA结合域、螺旋结构域和激活域组成,在调控免疫系统的主要组织相容性复合物II类(major histocompatibility complex class II, MHC II)的表达中起着至关重要的作用。本研究发现RFX5与CTCF在全基因组上结合的位点有部分重叠,利用CRISPR/Cas9 DNA大片段编辑技术,构建了RFX5基因缺失的HEC-1-B细胞系。通过RNA-seq实验,发现RFX5敲除能够显著升高Pcdhα6、Pcdhα12、Pcdhαc2的表达水平。通过ChIP-nexus实验,发现敲除RFX5导致染色质架构蛋白CTCF和cohesin在原钙粘蛋白α基因簇处的结合增加。最后,染色质构象捕获QHR-4C实验发现Pcdhα6、Pcdhα12启动子与远端增强子HS5-1的染色质远距离相互作用增强。上述研究表明RFX5蛋白可能通过调控染色质高级结构影响原钙粘蛋白α基因簇的表达,为未来进一步探索RFX5的功能提供了参考。
关键词:
Abstract
Gene expression and three-dimensional (3D) genome organization are thought to be closely related. The protocadherin (Pcdh) clusters are central for neuronal self-avoidance in brain development and have been used as model genes to explore the role of 3D genome structures in gene regulation. Transcription factor RFX5 (regulatory factor x 5) is a member of the winged-helix subfamily of the helix-turn-helix superfamily proteins. The RFX5 protein contains four domains: oligomerization, DNA-binding, helical, and transactivation domains. RFX5 plays an important role in regulating expression of the major histocompatibility complex class II (MHC II) gene complex. Here we report a role of RFX5 in the regulation of clustered Pcdh genes. We first knocked out the RFX5 gene by using CRISPR/Cas9 DNA-fragment editing in HEC-1-B cell line. By RNA-seq experiments, we found that RFX5 deletion results in a significant increase of expression levels of the Pcdhα6, Pcdhα12 and Pcdhαc2 genes. By ChIP-nexus experiments, we found that RFX5 deletion leads to the increased binding of CTCF and RAD21 in the Pcdhα cluster. Finally, through quantitative high-resolution chromosome conformation capture copy (QHR-4C) experiments, we found that RFX5 deletion results in a significant increase of long-distance chromatin interactions between the HS5-1 enhancer and its target promoters in the Pcdhα cluster. In conclusion, RFX5 regulates gene expression of the Pcdhα cluster through higher-order chromatin structure.
Keywords:
PDF (1566KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王娜, 甲芝莲, 吴强. RFX5调控原钙粘蛋白α基因簇的表达. 遗传[J], 2020, 42(8): 760-774 doi:10.16288/j.yczz.20-184
Na Wang.
人类基因组由超过30亿的核苷酸所组成,长度超过2 m,而细胞核的大小仅有数微米,因此基因组经过有规律地折叠形成复杂的三维空间结构,最终被包含在细胞核内[1]。然而,关于基因组如何折叠形成三维空间结构及其对基因组功能有何影响,仍有待进一步研究。原钙粘蛋白(protocadherin, Pcdh)基因簇是研究三维基因组空间结构与转录调控的模式基因。人类原钙粘蛋白基因簇位于5号染色体上,线性长度横跨将近1 Mb,由原钙粘蛋白α、原钙粘蛋白β和原钙粘蛋白γ3个基因簇组成,共包含53个基因[2,3,4,5,6,7,8,9]。原钙粘蛋白α基因簇与原钙粘蛋白γ基因簇的基因排列类似,均包含可变区外显子和恒定区外显子,并且每一个可变区外显子都有各自独立的启动子,转录产物可以顺式剪接到下游的3个恒定区外显子[10]。原钙粘蛋白β基因簇则仅包含可变区外显子,并无恒定区外显子。原钙粘蛋白基因簇在神经系统中发挥多种功能,包括中间神经元的存活、皮层神经元的迁移、神经元的自我识别、突触的形成、树突的自我回避以及轴突的发育等[11],因此研究原钙粘蛋白的表达调控机制对理解大脑的正常发育以及认识脑疾病的发生发展十分重要。
人类原钙粘蛋白α基因簇由13个随机表达的可变区外显子(α1~α13),2个C型可变区外显子(αc1和αc2),以及3个恒定区外显子构成(图1A)[2,6]。原钙粘蛋白α基因簇的表达受下游的增强子HS5-1调控[12]。在一维线性状态下,HS5-1与所调控的原钙粘蛋白α基因的线性距离在115~250 kb之间(与最远的Pcdhα1相距250 kb,与最近的Pcdhαc1相距115 kb)。因此,增强子HS5-1必须通过远距离染色质环形成在空间上靠近启动子的三维高级结构。CTCF (CCCTC-binding factor)蛋白是一种转录因子,与DNA结合具有方向性。原钙粘蛋白α1到α13的每一个启动子上都有两个正向的CTCF结合位点(CTCF binding site, CBS):CSE (conserved sequence element)和eCBS (exonic CBS),原钙粘蛋白αc1的启动子上有一个正向的CBS,但是原钙粘蛋白αc2的启动子上无CBS。有趣的是,下游的增强子HS5-1上有两个反向的CBS HS5-1a和HS5-1b (图1A)[13,14,15]。CTCF蛋白通过锌指结构域结合在这些CBS上,滑动的cohesin蛋白复合体会停留在启动子的正向CBS和增强子的反向CBS上,从而形成染色质环,使得增强子靠近并激活Pcdhα基因的表达[13,14,16]。本课题组前期发现在增强子HS5-1处有RFX5 (regulatory factor x 5)蛋白结合,并且与CTCF的结合区域相互重叠。由此推测RFX5可能通过影响三维基因组的空间构象调控基因的表达。
RFX5是一种DNA结合蛋白[17],属于螺旋-转角-螺旋家族中翼螺旋亚家族[18]。RFX5的首次报道见于对II型裸淋巴细胞综合征(the bare lymphocyte syndrome, BLS)的研究[19]。BLS是一种罕见的常染色体隐性遗传病,该病的特征在于缺乏主要组织相容性复合物II类(major histocompatibility complex class Ⅱ, MHC II)分子的表达,从而导致了严重的免疫缺陷,机体反复遭受感染,患者常在婴幼儿期死亡[20,21,22,23,24,25]。研究表明,MHC II类分子缺乏的根本原因是调节基因表达的相关因子RFX5蛋白存在缺陷,致使MHC II类分子无法在免疫细胞中表达或表达降低[26,27],由此开启了转录因子RFX5在基因表达调控方面的研究[28,29,30,31,32]。RFX5基因包含11个外显子,编码一个含有616个氨基酸的蛋白质。该蛋白包含4个结构域:寡聚化结构域、DNA结合域、螺旋结构域和激活域[33,34]。RFX5蛋白的寡聚化结构域由一个较长 的中央螺旋和两个较短的螺旋组成。当RFX5单独存在时,两个RFX5分子的中心螺旋形成一个反平行的盘绕线圈,侧翼螺旋垂直排列在长螺旋的末端,使得RFX5二聚体的形状如钉书钉[35]。此时高度保守的DNA结合域被遮蔽,无法与染色体结合。只有在寡聚化结构域及螺旋结构域通过与配体RFXANK、RFXAP的相互作用形成RFX复合物,暴露出包裹在内部的DNA结合域时,RFX5才能与启动子、增强子等结合,从而发挥调控功能[33]。关于RFX5的研究主要集中在MHC II类分子的表达调控方面,以及RFX复合体如何相互作用[26,36~39],鲜有涉及RFX5在三维基因组空间结构形成方面的作用。
本研究利用II型成簇规律间隔短回文重复序列CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated nuclease 9)系统发展起来的DNA大片段编辑技术[40,41,42,43,44,45,46,47],通过引入两个sgRNA (single-guide RNA),将Cas9蛋白定位到靶点位置并进行切割,造成DNA双链断裂,经过细胞自我修复后,可以获得基因片段的删除、反转、重复和插入[42,46]。本研究通过向细胞中导入两个sgRNA,得到RFX5基因敲除的单细胞克隆株。然后利用RNA-seq实验检测敲除细胞中基因表达水平的变化,通过染色质免疫沉淀(chromatin immunoprecipitation, ChIP)实验结合深度测序,探寻敲除RFX5对于染色质空间架构蛋白CTCF和cohesin蛋白复合体在DNA结合方面的影响,最后通过定量高分辨率染色体构象捕获(quantitative high-resolution chromosome conformation capture copy, QHR-4C)实验探究敲除RFX5对原钙粘蛋白α基因簇启动子与增强子之间三维空间结构的影响,发现RFX5能够抑制染色质远距离环化,对进一步了解RFX5蛋白的功能有重要意义。
1 材料与方法
1.1 实验材料
pGL3-U6-sgRNA-Puromycin-BsaI质粒由南京大学黄行许教授提供;人源化pST374-Cas9-ZF-NLS质粒由北京大学席建忠教授提供;人子宫内膜癌细胞系HEC-1-B来自于美国菌种保藏中心(ATCC);MEM培养基购自美国HyClone公司;胰酶、青霉素/链霉素双抗、谷氨酰胺和丙酮酸钠均购自美国Gibco公司;胎牛血清购自南美ExCell公司;sgRNA、测序引物由北京擎科新业生物技术有限公司、上海生工生物工程(上海)股份有限公司合成;质粒小量提取试剂盒、DNA凝胶回收试剂盒均购自美国AXYGEN公司;MinElute Gel Extraction kit购自德国QIAGEN公司;Lipo3000购自美国Invitrogen公司;Bsa I限制性内切酶、NEB Buffer 2、T4 DNA连接酶、T4 DNA连接酶缓冲液、Poly (A) mRNA Magnetic Isolation Module、RNA文库制备试剂盒、末端修复酶、末端修复酶缓冲液、Klenow Fragment (3?→5? exo-)、dA-Tailing反应缓冲液、Blunt/TA连接酶混合液、NEB Buffer 2.1、T4 DNA聚合酶、Lambda核酸外切酶、RecJf核酸外切酶、蛋白酶K、Q5? Hot Start HiFi PCR Master Mix等均购自美国NEB公司;TRIzol购自美国Ambion公司;AMPure? XP Beads购自美国Beckman Coulter公司;Green Taq Mix、dNTPs购自南京诺唯赞生物科技有限公司;Protein G beads、甲醛溶液、RNase A、糖原、Fast Digest BamH I限制性内切酶均购自美国Thermo公司;CirLigase? ssDNA Ligase试剂盒购自美国Epicentre公司;Rad21抗体、CTCF抗体均购自英国Abcam公司。1.2 构建pGL3-U6-sgRNA质粒
设计合成sgRNA的寡核苷酸序列如表1所示。购买的单链sgRNA首先用ddH2O稀释为终浓度20 μmol/L,接着进行引物退火步骤,反应体系如下(总体积为20 μL):2 μL sgRNA-F,2 μL sgRNA-R,2 μL 10 × NEB Buffer 2,14 μL ddH2O,经微量移液器吹打混匀后放入PCR仪中。反应条件为:95℃预变性5 min,缓慢降温95℃ 20 s 351个循环(自第二个循环开始逐步降温,每个循环降温0.2℃),最终产物4℃保存。环形pGL3-U6-sgRNA质粒通过BsaI酶切后,再经琼脂糖凝胶电泳的分离,切胶纯化得到线性pGL3-U6-sgRNA载体。将该线性化载体与退火产物进行连接反应,体系如下(总体积为5 μL):0.5 μL T4 DNA连接酶,0.5 μL T4 DNA连接酶缓冲液,0.5 μL线性pGL3-U6-sgRNA载体,0.5 μL退火产物,3 μL ddH2O,混匀后室温静置30 min;将上述反应体系加入含感受态细胞的1.5 mL离心管中,冰上静置30 min后放入42℃水浴锅,热激45 s立即拿出,冰上静置2 min;加入500 μL LB无抗培养基,并置于37℃摇床中培养45 min,均匀地涂在含有氨苄抗性的LB固体培养基上,37℃培养16~17 h;挑取2~3个单菌落摇菌培养,用质粒小量提取试剂盒提取质粒,并测序。
Table 1
表1
表1引物序列
Table 1
| 类型 | 引物名称 | 序列(5′→3′) |
|---|---|---|
| PCR | F1 | GGGAAGTCGTGGCGAGATTA |
| F2 | GTGCCCTGAAAGTGGCTACA | |
| R1 | CTGGGTGACTCAGCTGTCTG | |
| R2 | GCTGGAGGTCACACACAAGA | |
| R3 | AGAGTAGCCTGGTGTTAGCG | |
| sgRNA | sgRNA1F | ACCGACCCTTCTTCAGAGGCTCCG |
| sgRNA1R | AAACCGGAGCCTCTGAAGAAGGGT | |
| sgRNA2F | ACCGAAGCAACACCCCCATGATAC | |
| sgRNA2R | AAACGTATCATGGGGGTGTTGCTT | |
| Index | P5-HS5-1-F2 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTGTTTTGGCGGCGACAAATTCG |
| P5-α12-F3 | AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTAGTCCAATCATTCACGGAATAGGATC | |
| P7-index-1 | CAAGCAGAAGACGGCATACGAGATCGAGTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-2 | CAAGCAGAAGACGGCATACGAGATTCTCCGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-3 | CAAGCAGAAGACGGCATACGAGATAATGAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-4 | CAAGCAGAAGACGGCATACGAGATGGAATCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-5 | CAAGCAGAAGACGGCATACGAGATAGCTTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-6 | CAAGCAGAAGACGGCATACGAGATGCGCATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-7 | CAAGCAGAAGACGGCATACGAGATGCGCGAGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-8 | CAAGCAGAAGACGGCATACGAGATAGAGTACTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-9 | CAAGCAGAAGACGGCATACGAGATGCTCCGTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-10 | CAAGCAGAAGACGGCATACGAGATCATGAGAGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-11 | CAAGCAGAAGACGGCATACGAGATTGAATCGCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-12 | CAAGCAGAAGACGGCATACGAGATGTCGCGTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-13 | CAAGCAGAAGACGGCATACGAGATCCGCATGAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-14 | CAAGCAGAAGACGGCATACGAGATCCACAATCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-15 | CAAGCAGAAGACGGCATACGAGATTTCATCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-16 | CAAGCAGAAGACGGCATACGAGATTAGAGTGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-17 | CAAGCAGAAGACGGCATACGAGATTAACTCGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-18 | CAAGCAGAAGACGGCATACGAGATTATACGGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-19 | CAAGCAGAAGACGGCATACGAGATGAGTCAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-20 | CAAGCAGAAGACGGCATACGAGATGACATAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-21 | CAAGCAGAAGACGGCATACGAGATGTATTAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-22 | CAAGCAGAAGACGGCATACGAGATGACACAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-23 | CAAGCAGAAGACGGCATACGAGATGCGATAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-24 | CAAGCAGAAGACGGCATACGAGATCTCTATGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT | |
| P7-index-25 | CAAGCAGAAGACGGCATACGAGATTACGCAGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT |
新窗口打开|下载CSV
1.3 细胞培养
HEC-1-B细胞培养在含5% CO2浓度的37℃恒温培养箱中。培养基为加入10%胎牛血清、2 mmol/L谷氨酰胺、1%青霉素/链霉素和1 mmol/L丙酮酸钠的MEM完全培养基。1.4 转染与单细胞克隆株的筛选
将处于对数生长期的约2×105/mL的HEC-1-B细胞传代至12孔板培养,待细胞生长密度为70%~ 90%时转染:先用MEM培养基稀释转染试剂Lipo3000,混匀后室温静置5 min;另取一管MEM培养基稀释sgRNA质粒、Cas9质粒和转染试剂P3000,并将Lipo3000加入其中,室温孵育20 min后全部加至12孔板的培养基中。培养48 h后,将培养基换为含有2 ng/μL嘌呤霉素的MEM完全培养基进行药物筛选。药筛4 d后,更换为普通的MEM完全培养基继续培养。待细胞量足够时,经过胰酶消化后,取1/10细胞置于PCR管中,室温11,200 r/min离心3 min,弃去上清。向PCR管中加入10 μL碱裂解液,吹打混匀后放入PCR仪中,98℃ 20 min裂解细胞释放DNA;再加入等体积的中和液,吹打混匀于-20℃冰箱中保存待用。通过设计PCR引物鉴定上述转染细胞中是否含有片段删除,若存在,则将12孔板中剩余的细胞进行单克隆铺板。一周后在显微镜下观察孔内细胞是否为单一团状并做标记,两周后对单细胞克隆株进行基因型鉴定。
1.5 单细胞克隆株的鉴定
单细胞克隆经胰酶消化后,取部分细胞经上述碱裂解法进行裂解,获得的细胞基因组作为模板,保存至-20℃冰箱待用。以特异性引物通过PCR扩增反应得到目的片段,扩增产物经琼脂糖凝胶电泳分离,并回收测序从而鉴定单细胞克隆株的基因型。若出现双峰的情况,则通过TA克隆进一步鉴定。鉴定单细胞克隆株的引物序列如表1所示。PCR反应体系如下(总体积为10 μL):1 μL模板DNA,0.5 μL 10 μmol/L正向引物,0.5 μL 10 μmol/L反向引物,5 μL 2 × Green Taq Mix,3 μL ddH2O。上述反应体系经微量移液器吹打混匀后放入PCR仪中。反应条件为:94℃预变性2 min;94℃变性30 s,55℃退火30 s (退火温度根据引物的GC含量而定),72℃延伸1 min (延伸时间根据PCR产物的长度改变,Taq酶延伸速度约为1 kb/min),35~37个循环;72℃ 2 min完全延伸,最终产物4℃保存。
TA克隆:PCR产物与T载体室温连接30 min;连接产物加入含感受态细胞的1.5 mL离心管中,冰上静置30 min;42℃热激45 s;冰上2 min;向离心管加入500 μL LB无抗培养基,37℃摇床中培养45 min;均匀涂在含有氨苄抗性的LB固体培养基上,37℃培养箱中培养16~17 h;挑取8~10个单菌落摇菌培养,用质粒小量提取试剂盒提取质粒,并送测序。
1.6 单细胞克隆株总RNA的提取
将处于对数生长期的细胞传代至12孔板培养,待细胞生长密度为80%~90%时收集细胞提取RNA:丢弃原孔中的培养基,用1×PBS清洗2~3次并弃去上清;加入500 μL/孔TRIzol溶液,室温静置15 min;吹打混匀后转至1.5 mL离心管中;加入100 μL三氯甲烷,震荡仪剧烈震荡15 s,室温孵育2~3 min;放入4℃离心机中12,000 r/min离心15 min;转移上清(约200 μL左右)至新的1.5 mL离心管中;加入250 μL异丙醇,上下颠倒混匀,室温孵育10 min;放入4℃离心机中12,000 r/min离心10 min,弃去上清;加入500 μL 75%乙醇,上下颠倒10次混匀,放入4℃离心机中12,000 r/min离心5 min,弃去上清(勿动沉淀);短暂离心以除去管壁上残留的液滴,室温开盖干燥10 min;加入20 μL无核酸酶的水溶解沉淀,震荡混匀,用NanoDrop2000测浓度后于-80℃冰箱保存。1.7 RNA-seq建库和高通量测序
单细胞克隆株总RNA抽提后,使用Poly (A) mRNA Magnetic Isolation Module试剂盒提取成熟的mRNA,合成并纯化获得cDNA;使用RNA文库制备试剂盒经过末端修复、加接头、PCR扩增、纯化等步骤构建文库,经Qubit3 Fluorometer测量浓度后于-20℃冰箱保存,以备高通量测序。文库经苏州金唯智生物科技有限公司的Illumina HiSeq平台进行测序,利用特异性index序列对测序结果进行拆分,通过Cufflinks、STAR等软件进行数据分析[48]。所用P7-index序列见表1。1.8 RFX5与CTCF蛋白结合位点分析
ChIP-seq数据来自GEO,所分析的细胞系为人宫颈癌细胞系HeLa-S3,RFX5蛋白的ChIP-seq数据编号为GSM935560,CTCF蛋白的ChIP-seq数据编号为GSM749739。利用Bedtools分析结合峰,并画出韦恩图。Pcdhα12启动子处RFX5和CTCF蛋白的ChIP-seq数据来自人神经母细胞瘤细胞系SK-N-SH,数据编号分别为GSM1003629和GSM1003633。1.9 单细胞克隆株ChIP-nexus建库与高通量测序
将处于对数生长期的细胞系传代至10 cm培养皿培养,待细胞生长密度为80%~90%时收集细胞:丢弃原10 cm培养皿中的培养基,用1 × PBS清洗1次并弃去上清;加入1 mL/皿胰酶溶液,置于37℃培养箱中孵育2~3 min;加入1 mL新鲜的MEM培养基终止胰酶消化,吹打混匀后转至15 mL离心管中;放入离心机中室温2000 r/min离心5 min,弃去上清(勿动下层细胞沉淀);加入10 mL培养基重悬沉淀;加入670 μL 16%多聚甲醛溶液,置于旋转仪中,室温颠倒混匀10 min;加入712.5 μL 2 mol/L的甘氨酸溶液,置于旋转仪中,室温颠倒混匀5 min;放入4℃离心机中3300 r/min离心10 min,弃去上清;用预冷的1×PBS重悬细胞沉淀,放入4℃离心机中3300 r/min离心5 min,弃去上清;短暂离心以除去管壁上的残留液,用微量移液器吸取上清并丢弃;立即用液氮速冻后放入-80℃冰箱保存待用或直接进行裂解超声。超声后,放入4℃离心机中以最大转速离心10 min;转移上清至新的无菌1.5 mL离心管中,加入适量抗体,置于4℃冰箱旋转仪中缓慢旋转孵育过夜;加入适量Protein G beads孵育;接着进行末端修复、3?末端添加一个腺嘌呤脱氧核苷酸、连接接头、接头填补和修剪、Lambda和RecJf核酸外切酶的酶切反应、DNA洗脱与纯化、单链环化等步骤构建文库[49],经过琼脂糖凝胶电泳分离,切胶回收。用Qubit3 Fluorometer测浓度后于-20℃冰箱保存,以备高通量测序。文库经苏州金唯智生物科技有限公司的Illumina HiSeq平台进行测序,根据index序列对测序结果进行拆分,通过Bowtie2比对到GRCh37/hg19基因组,用BamCoverage进行归一化处理,标准化到RPKM (reads per kilobase per million mapped reads)。所用P7-index序列见表1。
1.10 单细胞克隆株QHR-4C建库与高通量测序
将生长旺盛的细胞系传代至10 cm培养皿培养,待细胞生长密度为80%~90%时收集细胞。接着进行酶切、连接、DNA纯化、超声、线性PCR扩增、加接头、PCR扩增、纯化等步骤构建文库[15,50]。用Qubit3 Fluorometer测量浓度后置于-20℃冰箱保存,以备高通量测序。文库经苏州金唯智生物科技有限公司的Illumina HiSeq平台进行测序,根据index序列对测序结果进行拆分,去除重复的Reads,通过Bowtie2比对到GRCh37/hg19基因组,用Samtools将比对文件进行转换,随即用R3Cseq计算每个DpnII酶切片段的Reads数,将总Reads标准化到一百万,最后根据幂律分布对观测点附近进行反向累积拟合得到染色质互作图。由于超声产生DNA末端的随机性,将RFX5敲除细胞与野生型细胞数据进行相减得到差异性数据[15,50]。所用P7-index序列见表1。2 结果与分析
2.1 使用CRISPR/Cas9大片段编辑技术建立RFX5基因敲除的单细胞克隆株
RFX5位于人类1号染色体,包含11个外显子,起始密码子位于第3个外显子,终止密码子位于第11个外显子(图1B)。分析宫颈癌细胞系HeLa-S3中RFX5与CTCF蛋白的ChIP-seq数据,可观察到两者有许多重叠峰(图1C),因此猜测RFX5可能与CTCF有相似的功能,互相协作共同调控染色体的三维空间构象。在原钙粘蛋白α基因簇中,与CTCF蛋白在启动子和HS5-1增强子中各有两个结合位点的情况类似,RFX5在启动子和增强子中也分别有两个峰,并且与CTCF峰有部分重叠。例如在启动子Pcdhα12及增强子HS5-1b位点处,RFX5与CTCF蛋白结合峰重叠(图1D)。为了研究RFX5在原钙粘蛋白α基因簇表达调控中的功能,本研究选用表达原钙粘蛋白α基因的HEC-1-B细胞系,应用CRISPR/Cas9技术,通过两个sgRNA,将Cas9蛋白定位至目的基因。Cas9蛋白在目的DNA片段互补链PAM (protospacer adjacent motif)序列上游3 bp处进行切割,非互补链PAM序列上游3~6 bp处进行切割,造成DNA双链断裂[42,43,44],经过细胞修复导致基因片段删除。通过筛选,最终得到RFX5基因敲除的单细胞克隆株。图1
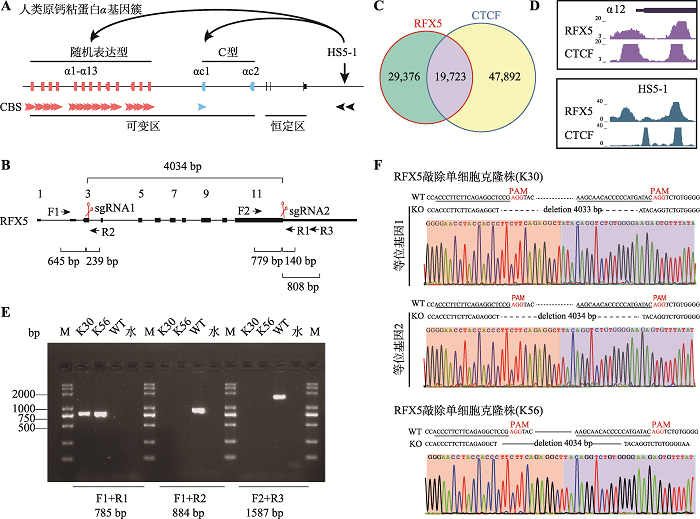 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1使用CRISPR/Cas9技术建立RFX5基因敲除的单细胞克隆株
Fig. 1Knockout of the RFX5 gene using CRISPR DNA-fragment editing by Cas9 with two sgRNAs
A:人类原钙粘蛋白α (Pcdhα)基因簇示意图。人类原钙粘蛋白α基因簇包含15个可变区外显子(13个随机表达型及2个C型)和3个恒定区外显子。每1个随机表达型外显子的启动子上都有2个正向的CTCF结合位点(CBS),C型外显子Pcdhαc1的启动子上有1个正向的CBS,而Pcdhαc2的启动子上没有CBS。原钙粘蛋白α基因簇下游的增强子HS5-1上含有两个反向的CBS。在CTCF和cohesin蛋白复合体的作用下,HS5-1增强子与原钙粘蛋白α基因启动子之间(Pcdhαc2除外)形成染色质环调控这些基因的表达。B:敲除RFX5基因所使用的一对sgRNA (sgRNA1和sgRNA2)以及各鉴定引物的位置示意图。C:分析比较HeLa-S3细胞中RFX5蛋白和CTCF蛋白结合峰的重叠情况。D:RFX5在Pcdhα12启动子和HS5-1增强子中分别有两个结合峰,并且部分与CTCF结合峰有重叠。E:RFX5基因敲除的单细胞克隆株K30和K56的基因型鉴定结果。以野生型细胞(WT)作为对照;用F1/R2或F2/R3引物,在sgRNA切点处,WT可扩增出884 bp或1587 bp片段,K30和K56中未检测到以上2个片段;用引物F1/R1,在K30和K56中扩增出785 bp的条带,而WT无条带。F:F1/R1引物在K30和K56单细胞克隆株中扩增产物的测序结果图。
设计两个sgRNA (sgRNA1和sgRNA2),分别定位在启动子下游101 bp和终止密码子处(PAM上游3 bp),二者相距4034 bp (图1B)。利用这两个sgRNA与Cas9切割基因,能造成RFX5基因绝大部分编码序列的删除。在切口位置上下游设计引物,检测编辑后细胞的基因型。引物F1设计在sgRNA1切口上游645 bp,引物R1设计在sgRNA2切口下游140 bp。使用F1和R1鉴定片段删除的单细胞克隆株,经过PCR反应可获得长度为785 bp的产物,而野生型细胞无条带(图1E)。为排除杂合子,对有片段删除的单细胞克隆株进一步在sgRNA切口上下游进行PCR扩增鉴定。对于sgRNA1切口设计引物R2,位于切口下游239 bp。sgRNA1切口处使用引物F1和R2进行PCR,野生型或杂合子可获得片段长度为884 bp的产物,而纯合子敲除无条带(图1E)。sgRNA2切口处设计引物F2,位于sgRNA2上游779 bp,引物R3设计在sgRNA2下游808 bp (图1B)。使用引物F2和R3进行PCR,野生型或杂合子可获得片段长度为1587 bp的产物,而纯合子敲除无条带(图1E)。经过上述PCR鉴定,从56个单细胞克隆株筛选到两个RFX5敲除纯合子单细胞克隆株:K30和K56。PCR产物测序显示K30存在两种不同的染色体基因型,一种为Cas9精确地切割在PAM上游3 bp处后的连接产物,另一种加入一个T碱基,可能为Cas9切割在sgRNA2非互补链PAM上游4 bp处所致。K56只存在一种基因型,即Cas9精确地切割在PAM上游3 bp处后的连接产物(图1F)。
2.2 敲除RFX5基因对原钙粘蛋白α基因簇表达的影响
为了研究RFX5敲除对基因表达产生的影响,对获得的两个单细胞克隆株分别进行RNA-seq实验。首先分析RFX5基因的表达,结果显示两个克隆中均检测不到RFX5的成熟mRNA,进一步证明了单细胞克隆株K30和K56已成功敲除RFX5基因(图2,A和B)。进一步分析了RFX5基因敲除对原钙粘蛋白α基因簇转录水平的影响,结果发现,在RFX5敲除的K30和K56单细胞克隆株中,相比于野生型细胞,原钙粘蛋白α基因簇基因总体转录水平升高(图2C)。野生型细胞主要表达Pcdhα6、Pcdhα12、Pcdhαc1以及Pcdhαc2,其他Pcdhα基因不表达。在RFX5敲除的K30单细胞克隆株中,相比于野生型细胞,表达基因的个数没有变化,但表达水平升高,其中Pcdhα6表达升高1.55倍,Pcdhα12表达升高1.34倍,Pcdhαc2表达升高1.77倍(图2D)。在K56单细胞克隆株中,与K30单细胞克隆株一致,上述3个基因表达均升高,其中Pcdhα6表达升高1.56倍,Pcdhα12表达升高1.86倍,Pcdhαc2表达升高1.9倍(图2E)。这些结果表明,RFX5基因敲除显著提高了原钙粘蛋白α基因簇的基因转录水平。图2
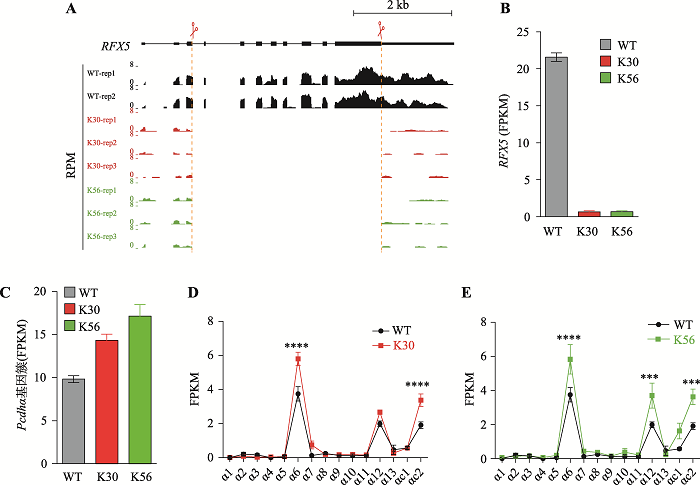 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2RFX5基因敲除对原钙粘蛋白α基因簇转录水平的影响
A:RNA-seq数据分析比较WT、K30、K56单细胞克隆株中RFX5基因外显子转录水平。B:RNA-seq数据分析比较WT、K30、K56单细胞克隆株中RFX5基因转录水平。C:RNA-seq数据分析比较WT、K30、K56单细胞克隆株中原钙粘蛋白α基因簇基因总体转录水平。D:RNA-seq数据分析比较WT和K30细胞中原钙粘蛋白α基因簇各基因转录水平。E:RNA-seq数据分析比较WT和K56细胞中原钙粘蛋白α基因簇各基因转录水平。***:P < 0.001;****:P < 0.0001;FPKM:fragments per kilobase of transcript per million mapped reads;RPM:reads of transcript per million mapped reads。
Fig. 2Effects of RFX5 deletion on the expression of the clustered Pcdhα genes
2.3 敲除RFX5基因增加CTCF和cohesin蛋白在原钙粘蛋白α基因簇的结合
为了探寻RFX5基因如何调控原钙粘蛋白α基因簇的转录,首先对CTCF蛋白在原钙粘蛋白α基因簇的结合情况进行研究。CTCF蛋白是一种转录因子,包含11个锌指结构域,可方向性地与DNA结合,调控基因的表达[51]。此外,CTCF可作为绝缘子,与cohesin蛋白复合体相互作用,协同决定染色质环形成的方向性[13,14,52,53]。采用ChIP-nexus实验,发现在RFX5基因敲除的两个单细胞克隆株K30和K56中,CTCF在转录升高的Pcdhα6和Pcdhα12基因启动子上的结合均高于野生型克隆,在增强子HS5-1处的结合也同时升高(图3A)。在不表达的Pcdhα8和Pcdhα10基因启动子处,CTCF的结合水平也高于野生型。总体而言,RFX5基因的敲除显著提高了CTCF蛋白在原钙粘蛋白α基因簇位点的结合。结合在正向-反向CBS上的一对CTCF蛋白能够阻断cohesin蛋白复合体在DNA上的滑动,从而形成染色质环。因此,绝大多数CBS上都有cohesin蛋白复合体的结合[13,14,54,55]。为了研究RFX5的敲除是否影响cohesin蛋白复合体的结合,对cohesin蛋白复合体亚基RAD21也进行了ChIP-nexus实验。类似于CTCF,结果发现RAD21在Pcdhα6、Pcdhα8、Pcdhα10、Pcdhα12以及HS5-1处的结合也高于野生型克隆(图3B)。
图3
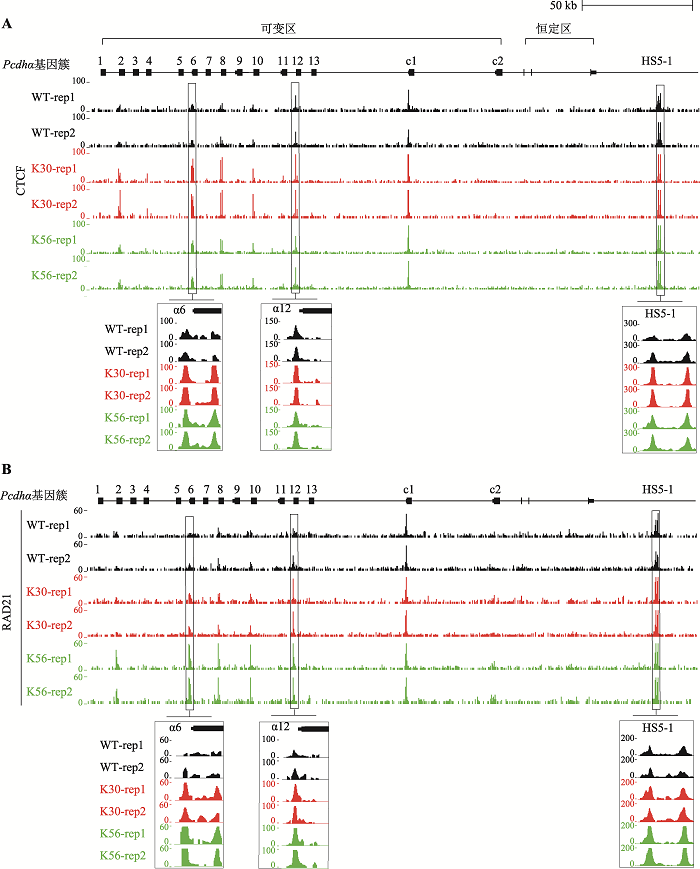 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3RFX5敲除促进CTCF和RAD21在原钙粘蛋白α基因簇位点的结合
A:ChIP-nexus数据分析比较WT、K30和K56单细胞克隆株中,CTCF蛋白在原钙粘蛋白α基因簇的结合情况。B:ChIP-nexus数据分析比较WT、K30和K56单细胞克隆株中,cohesin蛋白复合体亚基RAD21蛋白在原钙粘蛋白α基因簇的结合情况。对每个克隆分别进行了两次独立的重复实验(rep)。
Fig. 3RFX5 deletion results in the increased binding of CTCF and RAD21 in the Pcdhα gene cluster
2.4 RFX5影响原钙粘蛋白α基因簇的三维空间构象
基于RFX5对CTCF和cohesin复合体在原钙粘蛋白α基因簇位点结合水平的影响,推测RFX5可能通过影响染色质三维空间构象从而调控原钙粘蛋白α基因簇的表达,因此对原钙粘蛋白α基因簇进行可定量的高分辨率染色质构象捕获(QHR-4C)实验。该实验能够研究某个观测点(viewpoint, VP)与全基因组其他位点之间的空间关系[15]。本研究首先以增强子HS5-1为观测点,发现在野生型细胞中,HS5-1与Pcdhαc1以及Pcdhα6-Pcdhα12之间形成远距离的染色质环(图4A)。在RFX5敲除的两个单细胞克隆株K30和K56中,HS5-1与这些基因之间的远距离相互作用增加(图4A)。为了进一步确定这些基因组位点间空间结构的变化,选取Pcdhα12启动子为观测点,发现在RFX5基因敲除的两个单细胞克隆株中,Pcdhα12与增强子HS5-1之间的远距离相互作用也同样增加(图4B)。这些结果说明,敲除RFX5基因后,原钙粘蛋白α基因簇启动子与远端增强子HS5-1之间的染色质相互作用增强,这可能是Pcdhα基因表达上调的原因。综上所述,本研究发现敲除RFX5基因导致原钙粘蛋白α基因簇表达升高,并且RFX5会阻碍CTCF蛋白和cohesin蛋白复合体在原钙粘蛋白α基因簇的结合。此外,RFX5参与调控染色质环的形成,进而影响染色质三维空间结构。
图4
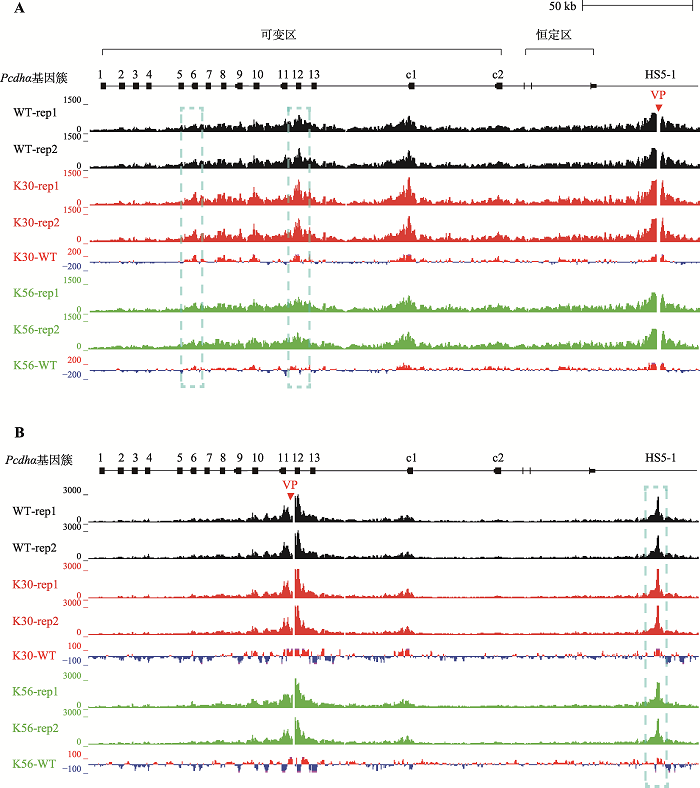 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4RFX5敲除增加原钙粘蛋白α基因簇增强子与启动子的远距离空间互作
A:RFX5敲除导致增强子与启动子间相互作用增加。在染色质构象捕获QHR-4C实验中,以HS5-1为观测点(viewpoint, VP),显示K30和K56单细胞克隆株中增强子HS5-1与原钙粘蛋白α基因簇启动子之间的染色质相互作用。K30和K56分别与WT相减(K30-WT, K56-WT)发现RFX5敲除后HS5-1与Pcdhα启动子之间的染色质相互作用高于WT克隆。B:RFX5敲除导致启动子与增强子间相互作用增加。以Pcdhα12为观测点的染色质构象捕获实验,显示K30和K56单细胞克隆株中Pcdhα12启动子与增强子HS5-1之间的染色质相互作用。K30和K56分别与WT相减(K30-WT, K56-WT)发现RFX5敲除后Pcdhα12启动子与HS5-1之间的染色质相互作用高于WT克隆。
Fig. 4RFX5 deletion results in increased long-range chromatin interactions between enhancers and promoters in the Pcdhα gene cluster
3 讨论
基因组空间构象的变化在很大程度上影响基因的表达,因此对基因组空间构象调控机制的研究具有重要意义。CTCF通过与DNA的结合,阻碍cohesin蛋白复合体的滑动从而使DNA成环,造成远距离的启动子、增强子彼此靠近,以此调控基因的表达[16,54]。以原钙粘蛋白基因簇为例,基因簇下游的增强子与上游基因启动子之间形成远距离染色质环,调控上游基因的表达[12,56]。本研究主要集中在RFX5蛋白对原钙粘蛋白α基因簇的表达调控,以及探索其对于原钙粘蛋白α基因簇三维空间结构的影响。本研究使用CRISPR/Cas9技术,通过导入两个sgRNA,将Cas9蛋白定位在目的片段处进行切割,造成DNA双链断裂,经细胞修复机制得到RFX5基因敲除的单细胞克隆株K30和K56。K30的两条染色体有一个碱基不同,而K56两条染色体基因型相同。本研究对这两个单细胞克隆株的RNA-seq、ChIP-nexus、以及染色质构象捕获实验发现,RFX5敲除后,Pcdhα6、Pcdhα12、Pcdhαc2转录升高。推测这些转录水平变化是由于RFX5敲除后,蛋白CTCF和cohesin在原钙粘蛋白α基因簇的结合升高,使得增强子与启动子之间的染色质相互作用增加而导致的(图5)。该现象表明RFX5蛋白通过抑制CTCF和cohesin与DNA的结合调控染色质高级结构,从而参与原钙粘蛋白α基因簇的转录调控(图5)。然而进一步分析RFX5敲除后基因组范围的转录变化时,发现转录水平降低的基因数量多于转录水平升高的基因,说明RFX5蛋白在不同的基因位点可能扮演的角色不同。
图5
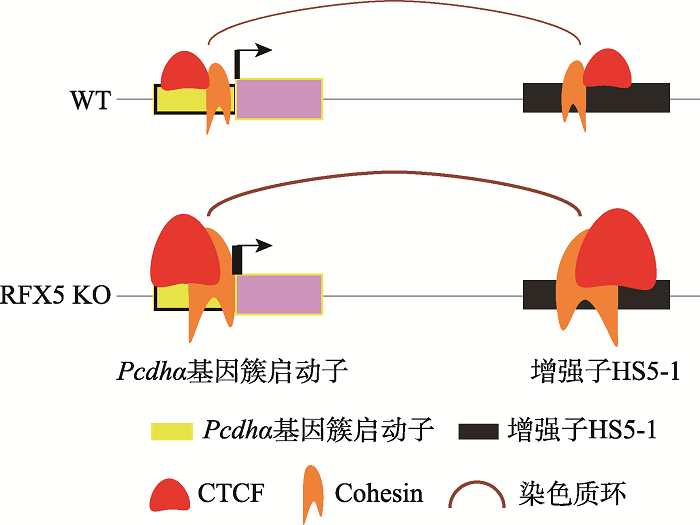 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5RFX5蛋白调控原钙粘蛋白α基因簇表达的模式图
A:野生型细胞(WT)中原钙粘蛋白α基因簇基因调控示意图。在野生型细胞中,RFX5、CTCF蛋白和cohesin复合体结合在原钙粘蛋白α基因簇启动子和增强子上,通过CTCF/cohesin介导的染色质环,增强子调控启动子的活性。B:RFX5敲除细胞(KO)中原钙粘蛋白α基因簇基因调控示意图。敲除RFX5基因后,CTCF蛋白和cohesin复合体在启动子、增强子的结合增加,使得增强子与启动子之间的染色质环增多,启动子的活性增加。
Fig. 5A model for the RFX5 regulation of gene expression of the Pcdhα gene cluster through chromatin looping
通过ChIP-nexus和染色质构象捕获QHR-4C实验,发现敲除RFX5基因后,原钙粘蛋白α基因簇中启动子以及增强子HS5-1处的CTCF和cohesin的结合水平升高。在RFX5敲除的单细胞克隆株中,Pcdhα6和Pcdhα12启动子处的CTCF/cohesin结合增加,以及这两个启动子与增强子HS5-1之间的染色质相互作用增加,这些均能解释Pcdhα6和Pcdhα12这两个基因转录水平升高的现象。但是,RFX5的敲除如何上调Pcdhαc2的转录水平仍是未解决的问题。Pcdhαc2是原钙粘蛋白α基因簇的特殊一员,它的启动子上没有CTCF蛋白的结合,并且它的转录不受增强子HS5-1的调控,而是由另外一个增强子HS7调控[12,57,58]。本研究在Pcdhαc2的启动子上没有发现RFX5的结合,但在增强子HS7上发现有RFX5的强结合,说明RFX5可能通过结合增强子HS7调控Pcdhαc2的转录。
综上所述,本研究发现RFX5蛋白参与调控原钙粘蛋白α基因簇的转录,敲除RFX5导致该基因簇的转录水平升高。进一步研究发现RFX5抑制染色质环的形成,在三维基因组空间构象中发挥作用。本研究工作将为后续对RFX5蛋白更深层次的研究提供参考。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1146/annurev-genom-091212-153515URLPMID:23875797 [本文引用: 1]

In vivo, the human genome functions as a complex, folded, three-dimensional chromatin polymer. Understanding how the human genome is spatially organized and folded inside the cell nucleus is therefore central to understanding how genes are regulated in normal development and dysregulated in disease. Established light microscopy-based approaches and more recent molecular chromosome conformation capture methods are now combining to give us unprecedented insight into this fascinating aspect of human genomics.
DOI:10.1016/s0092-8674(00)80789-8URLPMID:10380929 [本文引用: 2]

We have identified 52 novel human cadherin-like genes organized into three closely linked clusters. Comparison of the genomic DNA sequences with those of representative cDNAs reveals a striking genomic organization similar to that of immunoglobulin and T cell receptor gene clusters. The N-terminal extracellular and transmembrane domains of each cadherin protein are encoded by a distinct and unusually large exon. These exons are organized in a tandem array. By contrast, the C-terminal cytoplasmic domain of each protein is identical and is encoded by three small exons located downstream from the cluster of N-terminal exons. This unusual organization has interesting implications regarding the molecular code required to establish complex networks of neuronal connections in the brain and the mechanisms of cell-specific cadherin-like gene expression.
DOI:10.1534/genetics.104.037606URLPMID:15744052 [本文引用: 1]

To explain the mechanism for specifying diverse neuronal connections in the brain, Sperry proposed that individual cells carry chemoaffinity tags on their surfaces. The enormous complexity of these connections requires a tremendous diversity of cell-surface proteins. A large number of neural transmembrane protocadherin (Pcdh) proteins is encoded by three closely linked human and mouse gene clusters (alpha, beta, and gamma). To gain insight into Pcdh evolution, I performed comprehensive comparative cDNA and genomic DNA analyses for the three clusters in the chimpanzee, rat, and zebrafish genomes. I found that there are species-specific duplications in vertebrate Pcdh genes and that additional diversity is generated through alternative splicing within the zebrafish
DOI:10.1093/jmcb/mjs034URL [本文引用: 1]

Dendritic patterning and spine morphogenesis are crucial for the assembly of neuronal circuitry to ensure normal brain development and synaptic connectivity as well as for understanding underlying mechanisms of neuropsychiatric diseases and cognitive impairments. The Rho GTPase family is essential for neuronal morphogenesis and synaptic plasticity by modulating and reorganizing the cytoskeleton. Here, we report that protocadherin (Pcdh) clusters and cell adhesion kinases (CAKs) play important roles in dendritic development and spine elaboration. The knockout of the entire Pcdh cluster results in the dendritic simplification and spine loss in CA1 pyramidal neurons in vivo and in cultured primary hippocampal neurons in vitro. The knockdown of the whole Pcdh cluster or in combination with the Pcdh knockout results in similar dendritic and spine defects in vitro. The overexpression of proline-rich tyrosine kinase 2 (Pyk2, also known as CAK, RAFTK, FAK2, and CADTK) recapitulates these defects and its knockdown rescues the phenotype. Moreover, the genetic deletion of the Pcdh cluster results in phosphorylation and activation of Pyk2 and focal adhesion kinase (Fak) and the inhibition of Rho GTPases in vivo. Finally, the overexpression of Pyk2 leads to inactivation of Rac1 and, conversely, the constitutive active Rac1 rescues the dendritic and spine morphogenesis defects caused by the knockout of the Pcdh cluster and the knockdown of the Pcdh cluster. Thus, the involvement of the Pcdh-CAK-Rho GTPase pathway in the dendritic development and spine morphogenesis has interesting implications for proper assembly of neuronal connections in the brain.
DOI:10.1073/pnas.060027397URLPMID:10716726 [本文引用: 1]

Recent studies revealed a striking difference in the genomic organization of classic cadherin genes and one family of
DOI:10.1101/gr.167301URLPMID:11230163 [本文引用: 2]

The genomic organization of the human protocadherin alpha, beta, and gamma gene clusters (designated Pcdh alpha [gene symbol PCDHA], Pcdh beta [PCDHB], and Pcdh gamma [PCDHG]) is remarkably similar to that of immunoglobulin and T-cell receptor genes. The extracellular and transmembrane domains of each protocadherin protein are encoded by an unusually large
DOI:10.1016/j.neuroscience.2006.10.011URLPMID:17110050 [本文引用: 1]

Three closely-linked clusters of protocadherin (Pcdh) genes (alpha, beta, and gamma) encoding more than 50 distinct mRNAs have been identified in humans and mice, and proposed to play important roles in neuronal connectivity in the CNS. The human and mouse Pcdh alpha and gamma clusters each span a region of about 300 kb genomic DNA, and are each organized into a tandem array of more than a dozen highly-similar
DOI:10.1101/gr.1225204URLPMID:14672974 [本文引用: 1]

A large family of neural protocadherin (Pcdh) proteins is encoded by three closely linked mammalian gene clusters (alpha, beta, and gamma). Pcdh alpha and gamma clusters have a striking genomic organization. Specifically, each
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/s1097-2765(02)00578-6URLPMID:12150904 [本文引用: 1]

A family of mammalian protocadherin (Pcdh) proteins is encoded by three closely linked gene clusters (alpha, beta, and gamma). Multiple alpha and gamma Pcdh mRNAs are expressed in distinct patterns in the nervous system and are generated by alternative pre-mRNA splicing between different
DOI:10.1242/dev.090621URL [本文引用: 1]

The majority of vertebrate protocadherin (Pcdh) genes are clustered in a single genomic locus, and this remarkable genomic organization is highly conserved from teleosts to humans. These clustered Pcdhs are differentially expressed in individual neurons, they engage in homophilic trans-interactions as multimers and they are required for diverse neurodevelopmental processes, including neurite self-avoidance. Here, we provide a concise overview of the molecular and cellular biology of clustered Pcdhs, highlighting how they generate single cell diversity in the vertebrate nervous system and how such diversity may be used in neural circuit assembly.
DOI:10.1073/pnas.0609445104URLPMID:17172445 [本文引用: 3]

The clustered protocadherins (Pcdh) are encoded by three closely linked gene clusters (Pcdh-alpha, -beta, and -gamma) that span nearly 1 million base pairs of DNA. The Pcdh-alpha gene cluster encodes a family of 14 distinct cadherin-like cell surface proteins that are expressed in neurons and are present at synaptic junctions. Individual Pcdh-alpha mRNAs are assembled from one of 14
URLPMID:23204437 [本文引用: 4]
DOI:10.1016/j.cell.2015.07.038URLPMID:26276636 [本文引用: 4]

CTCF and the associated cohesin complex play a central role in insulator function and higher-order chromatin organization of mammalian genomes. Recent studies identified a correlation between the orientation of CTCF-binding sites (CBSs) and chromatin loops. To test the functional significance of this observation, we combined CRISPR/Cas9-based genomic-DNA-fragment editing with chromosome-conformation-capture experiments to show that the location and relative orientations of CBSs determine the specificity of long-range chromatin looping in mammalian genomes, using protocadherin (Pcdh) and beta-globin as model genes. Inversion of CBS elements within the Pcdh enhancer reconfigures the topology of chromatin loops between the distal enhancer and target promoters and alters gene-expression patterns. Thus, although enhancers can function in an orientation-independent manner in reporter assays, in the native chromosome context, the orientation of at least some enhancers carrying CBSs can determine both the architecture of topological chromatin domains and enhancer/promoter specificity. These findings reveal how 3D chromosome architecture can be encoded by linear genome sequences.
DOI:10.1186/s13059-020-01984-7URLPMID:32293525 [本文引用: 4]

BACKGROUND: CTCF is a key insulator-binding protein, and mammalian genomes contain numerous CTCF sites, many of which are organized in tandem. RESULTS: Using CRISPR DNA-fragment editing, in conjunction with chromosome conformation capture, we find that CTCF sites, if located between enhancers and promoters in the protocadherin (Pcdh) and beta-globin clusters, function as an enhancer-blocking insulator by forming distinct directional chromatin loops, regardless whether enhancers contain CTCF sites or not. Moreover, computational simulation in silico and genetic deletions in vivo as well as dCas9 blocking in vitro revealed balanced promoter usage in cell populations and stochastic monoallelic expression in single cells by large arrays of tandem CTCF sites in the Pcdh and immunoglobulin heavy chain (Igh) clusters. Furthermore, CTCF insulators promote, counter-intuitively, long-range chromatin interactions with distal directional CTCF sites, consistent with the cohesin
DOI:10.1038/nature09380URLPMID:20720539 [本文引用: 2]

Transcription factors control cell-specific gene expression programs through interactions with diverse coactivators and the transcription apparatus. Gene activation may involve DNA loop formation between enhancer-bound transcription factors and the transcription apparatus at the core promoter, but this process is not well understood. Here we report that mediator and cohesin physically and functionally connect the enhancers and core promoters of active genes in murine embryonic stem cells. Mediator, a transcriptional coactivator, forms a complex with cohesin, which can form rings that connect two DNA segments. The cohesin-loading factor Nipbl is associated with mediator-cohesin complexes, providing a means to load cohesin at promoters. DNA looping is observed between the enhancers and promoters occupied by mediator and cohesin. Mediator and cohesin co-occupy different promoters in different cells, thus generating cell-type-specific DNA loops linked to the gene expression program of each cell.
DOI:10.1093/nar/24.5.803URLPMID:8600444 [本文引用: 1]

Until recently, the RFX family of DNA binding proteins consisted exclusively of four mammalian members (RFX1-RFX4) characterized by a novel highly conserved DNA binding domain. Strong conservation of this DNA binding domain precluded a precise definition of the motif required for DNA binding. In addition, the biological systems in which these RFX proteins are implicated remained obscure. The recent identification of four new RFX genes has now shed light on the evolutionary conservation of the RFX family, contributed greatly to a detailed characterization of the RFX DNA binding motif, and provided clear evidence for the function of some of the RFX proteins. RFX proteins have been conserved throughout evolution in a wide variety of species, including Saccharomyces cerevisiae, Schizosaccharomyces pombe, Caenorhabditis elegans, mouse and man. The characteristic RFX DNA binding motif has been recruited into otherwise very divergent regulatory factors functioning in a diverse spectrum of unrelated systems, including regulation of the mitotic cell cycle in fission yeast, the control of the immune response in mammals, and infection by human hepatitis B virus.
DOI:10.1038/35002634URLPMID:10706293 [本文引用: 1]

Regulatory factor X (RFX) proteins are transcriptional activators that recognize X-boxes (DNA of the sequence 5'-GTNRCC(0-3N)RGYAAC-3', where N is any nucleotide, R is a purine and Y is a pyrimidine) using a highly conserved 76-residue DNA-binding domain (DBD). DNA-binding defects in the protein RFX5 cause bare lymphocyte syndrome or major histocompatibility antigen class II deficiency. RFX1, -2 and -3 regulate expression of other medically important gene products (for example, interleukin-5 receptor alpha chain, IL-5R alpha). Fusions of the ligand-binding domain of the oestrogen receptor with the DBD of RFX4 occur in some human breast tumours. Here we present a 1.5 A-resolution structure of two copies of the DBD of human RFX1 (hRFX1) binding cooperatively to a symmetrical X-box. hRFX1 is an unusual member of the winged-helix subfamily of helix-turn-helix proteins because it uses a beta-hairpin (or wing) to recognize DNA instead of the recognition helix typical of helix-turn-helix proteins. A new model for interactions between linker histones and DNA is proposed.
DOI:10.1146/annurev.immunol.19.1.331URLPMID:11244040 [本文引用: 1]

The bare lymphocyte syndrome (BLS) is a hereditary immunodeficiency resulting from the absence of major histocompatibility complex class II (MHCII) expression. Considering the central role of MHCII molecules in the development and activation of CD4(+) T cells, it is not surprising that the immune system of the patients is severely impaired. BLS is the prototype of a
DOI:10.1146/annurev.immunol.14.1.301URLPMID:8717517 [本文引用: 1]

Precise regulation of major histocompatibility complex class II (MHC-II) gene expression plays a crucial role in the control of the immune response. A major breakthrough in the elucidation of the molecular mechanisms involved in MHC-II regulation has recently come from the study of patients that suffer from a primary immunodeficiency resulting from regulatory defects in MHC-II expression. A genetic complementation cloning approach has led to the isolation of CIITA and RFX5, two essential MHC-II gene transactivators. CIITA and RFX5 are mutated in these patients, and the wild-type genes are capable of correcting their defect in MHC-II expression. The identification of these regulatory factors has furthered our understanding of the molecular mechanisms that regulate MHC-II genes. CIITA was found to be a non-DNA binding transactivator that functions as a molecular switch controlling both constitutive and inducible MHC-II expression. The finding that RFX5 is a subunit of the nuclear RFX-complex has confirmed that a deficiency in the binding of this complex is indeed the molecular basis for MHC-II deficiency in the majority of patients. Furthermore, the study of RFX has demonstrated that MHC-II promoter activity is dependent on the binding of higher-order complexes that are formed by highly specific cooperative binding interactions between certain MHC-II promoter-binding proteins. Two of these proteins belong to families of which the other members, although capable of binding to the same DNA motifs, are probably not directly involved in the control of MHC-II expression. Finally, the facts that CIITA and RFX5 are both essential and highly specific for MHC-II genes make possible novel strategies designed to achieve immunomodulation via transcriptional intervention.
DOI:10.1016/s0022-3476(05)80388-9URLPMID:8229525 [本文引用: 1]

Major histocompatibility complex class II deficiency (bare lymphocyte syndrome) is a rare primary immunodeficiency disorder characterized by profound defects in human leukocyte antigen class II expression, inconsistent and incomplete expression of human leukocyte antigen class I molecules, and a complete lack of cellular and humoral immune responses to foreign antigens. To define the clinical and immunologic characteristics, outcome, and natural history of major histocompatibility complex class II deficiency, we retrospectively analyzed 30 consecutive patients. Clinical onset occurred in the first year of life, usually involving recurrent bronchopulmonary infections and chronic diarrhea. The clinical course was complicated by viral meningoencephalitis, hepatitis, cholangitis, and various autoimmune phenomena. Prognosis was very poor: the mean age at the time of death was 4 years. The main cause of death was overwhelming viral infection. Recent advances in bone marrow transplantation have raised hopes of curative treatment: 6 of 14 patients who underwent bone marrow transplantation were cured. Long-term survival after human leukocyte antigen-identical and haploidentical bone marrow transplantation seemed to depend primarily on the presence of preexisting viral infections.
DOI:10.1097/00005792-200111000-00006URL [本文引用: 1]
URLPMID:2517209 [本文引用: 1]

MHC class II deficiency is an inherited immunodeficiency disease characterized by the presence of a normal number of T and B lymphocytes and profound anomaly of cellular and humoral responses to foreign antigens. All bone-marrow-derived cells (including B lymphocytes, monocytes and activated T lymphocytes) and also enterocytes and endothelial cells do not express all HLA class II (DR, DQ and DP) molecules on their membrane. It is known that the proper recognition of foreign antigens depends on their presentation, together with HLA class II molecules, on the membrane of antigen-presenting cells. MHC class II deficient combined immunodeficiency confirms the important role of MHC gene products in immune-defence mechanisms. Patients suffer from repeated and severe infections that are frequently the cause of death. The defect in HLA class II expression is the consequence of a lack of synthesis of HLA class II alpha and beta chains in patients' cells. Studies performed at DNA and RNA levels showed that there was no gross abnormality of MHC class II genes and that mRNA for all HLA molecules was not detected in patients' cells. These results, together with segregation studies performed in several families, suggested that the defect in HLA class II gene expression involves a transacting regulatory factor. Direct transcription assays showed that the disease is characterized by an absence of HLA class II gene transcription. An analysis of the specific binding of nuclear proteins from patients' cell lines to HLA class II promotor showed that a specific protein, RF-X, which normally binds to a regulatory sequence common to HLA class II promotors, is affected in MHC class II combined immunodeficiency.
DOI:10.1101/gad.9.9.1021URLPMID:7744245 [本文引用: 1]

Regulation of MHC class II gene expression is an essential aspect of the control of the immune response. Primary MHC class II deficiency is a genetically heterogeneous disease of gene regulation that offers the unique opportunity of a genetic approach for the identification of the functionally relevant regulatory genes and factors. Most patients exhibit a characteristic defect in the binding of a nuclear complex, RFX, to the X box motif of MHC class II promoters. Genetic complementation of a B-lymphocyte cell line from such a patient with a cDNA expression library has allowed us to isolate RFX5, the regulatory gene responsible for the MHC class II deficiency. This gene encodes a novel DNA-binding protein that is indeed a subunit of the RFX complex. Mutations in the RFX5 gene have been characterized in two patients. Transfection of the patient's cells with the RFX5 cDNA repairs the binding defect and fully restores expression of all the endogenous MHC class II genes in vivo.
URLPMID:8110378 [本文引用: 1]

The major histocompatibility complex (MHC) class II genes encode surface molecules that are required for presentation of antigenic peptides to helper T-cells. The concentration of these proteins on the surface of effector cells (antigen-presenting cells such as B-cells and macrophage) is one of the parameters affecting the intensity of the immune response. Many studies have thus focused their attention on the mechanisms that control the expression of class II genes, particularly in B-cells. The anatomy of MHC class II promoters has been dissected in detail, and many trans-acting factors and their cognate DNA regulatory elements have been identified and characterized, thus helping to elucidate the molecular circuitry which determines tissue-specific, coordinate expression of these genes. In most cases, regulation has been investigated at the level of mRNA transcription. MHC class II gene expression has been observed as well, under physiological conditions, in many other tissues and organs such as brain, thyroid, thymus, and intestine, thus implying that class II molecules may be involved, whether directly or indirectly, in the modulation of other important biological responses in addition to the control of the immune reaction against soluble antigens. Spurious MHC class II activity is also detected in tumor cells and in other pathological conditions such as those found in autoimmune, inflammatory, and infectious diseases. In autoimmunity, cells that express class II molecules may present tissue-specific antigens, thus triggering a mechanism of self-destruction. In tumors, instead, unscheduled MHC class II expression may be part of a mechanism that prevents tumor progression. Comprehension of the regulatory functions operating in pathological conditions as compared to those active in B-cells and in macrophages is still rudimentary. Because of the possible pathogenetic importance of aberrant class II expression, knowledge of the cis- and trans-acting elements controlling gene expression at either the transcriptional or posttranscriptional level may allow the development of strategies for immunointervention against these diseases.
DOI:10.1016/j.jaci.2014.06.001URLPMID:25001848 [本文引用: 2]

Deficiencies of MHC complex class I or II are rare primary immunodeficiencies, both of which are inherited in an autosomal recessive pattern. MHC class II deficiency is a prototype of a disease of gene regulation. Defects in transacting regulatory factors required for expression of MHC class II genes, rather than the genes themselves, are responsible for the disease phenotype. The affected genes are known to encode 4 distinct regulatory factors controlling transcription of MHC class II genes. These transacting factors are the class II transactivator and 3 subunits of regulatory factor X (RFX): RFX containing ankyrin repeats (RFXANK), the fifth member of the RFX family (RFX5), and RFX-associated protein (RFXAP). Mutations in one of each define 4 distinct complementation groups termed A, B, C, and D, respectively. MHC class I deficiency is extremely rare and has been reported in less than 30 patients worldwide. Here we review the clinical, genetic, and molecular features that characterize these primary immunodeficiencies and discuss therapy options. Beyond the description of MHC class I and II deficiencies, their discovery has fascinated scientists and clinicians because of their ability to reveal the molecular basis of MCH regulation.
DOI:10.1146/annurev.iy.12.040194.001355URLPMID:8011283 [本文引用: 1]

MHC class II molecules assemble in the endoplasmic reticulum in a chaperone-mediated fashion to form a nine-chain structure consisting of three alpha beta dimers associated with an invariant chain trimer. This complex is transported through the Golgi apparatus and into the endosomal system. The signal for endosomal targeting resides in the cytoplasmic tail of the invariant chain. Current evidence argues that the segregation of the class II-invariant chain complex from the constitutive pathway of membrane protein transport occurs in the trans-Golgi network. However, class II-invariant chain complexes that reach the cell surface are also rapidly internalized into endosomes. Within the endosomal system, probably in a late endosome/pre-lysosome, the invariant chain is degraded, releasing alpha beta dimers that bind peptides predominantly derived from endocytosed proteins. Evidence suggests that many of these peptides are actually generated in lysosomes. The precise mechanisms involved in forming class II-peptide complexes are unclear, although the existence of antigen-processing mutants argues that additional gene products, at least one of which is encoded in the MHC, are involved. After binding peptides, class II molecules are transported by an unknown route to the cell surface, where their primary function of presenting antigenic peptides to CD4+ T cells is carried out.
DOI:10.1002/(SICI)1098-1004(1997)10:6<430::AID-HUMU3>3.0.CO;2-HURLPMID:9401005 [本文引用: 1]

MHC class II deficiency is a severe primary immunodeficiency characterised by the absence of major histocompatibility complex class II (MHC-II) gene expression. It is genetically heterogeneous and can result from defects in at least four different trans-acting regulatory genes required for transcription of MHC-II genes. One of these genes has recently been shown to encode a novel DNA binding protein called RFX5, which is one subunit of a heteromeric protein complex (RFX) that binds to the promoters of MHC-II genes. We have characterised the mutations in all four patients known to harbour a defect in the RFX5 gene and have mapped this new human disease gene to chromosome 1 band q21, a region frequently exhibiting chromosomal aberrations in a variety of preneoplastic and neoplastic diseases.
DOI:10.1016/s1286-4579(99)00234-8URLPMID:10614001 [本文引用: 1]
DOI:10.1016/s1074-7613(00)80467-7URLPMID:9491996 [本文引用: 1]

Patients with major histocompatibility complex class II (MHC-II) deficiency are known to carry mutations in either the RFX complex or the trans-activator CIITA. While the pivotal role of CIITA for MHC-II gene transcription is supported by the essential absence of MHC-II molecules in CIITA-deficient mice, we demonstrate here that RFX5-/- mice retain expression of MHC-II in thymic medulla, mature dendritic cells, and activated B cells. Nevertheless, RFX5-/- mice develop a severe immunodeficiency due to the lack of MHC-II in thymic cortex, failure of positive selection of CD4+ T cells, and absence of MHC-II on resting B cells and resident or IFNgamma-activated macrophages. This differential requirement for CIITA and RFX5 in subsets of antigen-presenting cells may be specific for the mouse; it may, however, also exist in humans without having been noticed so far.
DOI:10.1128/mcb.20.10.3364-3376.2000URLPMID:10779326 [本文引用: 1]

Major histocompatibility complex class II (MHC-II) molecules occupy a pivotal position in the adaptive immune system, and correct regulation of their expression is therefore of critical importance for the control of the immune response. Several regulatory factors essential for the transcription of MHC-II genes have been identified by elucidation of the molecular defects responsible for MHC-II deficiency, a hereditary immunodeficiency disease characterized by regulatory defects abrogating MHC-II expression. Three of these factors, RFX5, RFXAP, and RFXANK, combine to form the RFX complex, a regulatory protein that binds to the X box DNA sequence present in all MHC-II promoters. In this study we have undertaken a dissection of the structure and function of RFX5, the largest subunit of the RFX complex. The results define two distinct domains serving two different essential functions. A highly conserved N-terminal region of RFX5 is required for its association with RFXANK and RFXAP, for assembly of the RFX complex in vivo and in vitro, and for binding of this complex to its X box target site in the MHC-II promoter. This N-terminal region is, however, not sufficient for activation of MHC-II expression. This requires an additional domain within the C-terminal region of RFX5. This C-terminal domain mediates cooperative binding between the RFX complex and NF-Y, a transcription factor binding to the Y box sequence of MHC-II promoters. This provides direct evidence that RFX5-mediated cooperative binding between RFX and NF-Y plays an essential role in the transcriptional activation of MHC-II genes.
DOI:10.1016/j.molimm.2013.05.235URL [本文引用: 1]

Major histocompatibility complexes class II are responsible for the antigen presentation that shapes the repertoire of the adaptive immune responses. All members of the MHCII family of genes are controlled by the same set of conserved transcription factors and promoter elements, resulting in coordinated transcription. We report the role of a previously unidentified AT-hook motif of the MHCII regulatory factor RFX5, and show that this is involved in regulating the transcription of the HLA-DQ, but not HLA-DR, MHCII isotype. Furthermore, PRMT6, an arginine methyltransferase known to methylate AT-hook motifs, down-regulates the expression of HLA-DQ, but not HLA-DR, in an AT-hook-dependent manner. This can provide a fine-tuning mechanism for isotype-specific transcriptional regulation, where a post-translational modification modulates the relative levels of the MHCII isotypes. (C) 2013 Elsevier Ltd.
DOI:10.1021/bi6023868URLPMID:17279624 [本文引用: 2]

Major histocompatability complex class II (MHCII) molecules are an essential component of the mammalian adaptive immune response. The expression of MHCII genes is regulated by a cell-specific multiprotein complex, termed the MHCII enhanceosome. The heterotrimeric RFX complex is the key DNA-binding component of the MHCII enhanceosome. The RFX complex is comprised of three proteins, RFXB, RFXAP, and RFX5, all of which are required for DNA binding and activation of MHCII gene expression. Static light scattering and chemical cross-linking of the three RFX proteins show that RFXB and RFXAP are monomers and that RFX5 dimerizes through two separate domains. One of these domains, the oligomerization domain, promotes formation of a dimer of dimers of RFX5. In addition, we show that the RFX complex forms a 2:1:1 complex of RFX5.RFXAP.RFXB, which can associate with a further dimer of RFX5 to form a 4:1:1 complex through the oligomerization domain of RFX5. On the basis of these studies, we propose DNA-binding models for the interaction between the RFX complex and the MHCII promoter including a DNA looping model. We also provide direct evidence that the RFX5(L66A) point mutation prevents dimerization of the RFX complexes and propose a model for how this results in a loss of MHCII gene expression.
DOI:10.1128/mcb.20.17.6587-6599.2000URLPMID:10938133 [本文引用: 1]

The bare lymphocyte syndrome, a severe combined immunodeficiency due to loss of major histocompatibility complex (MHC) class II gene expression, is caused by inherited mutations in the genes encoding the heterotrimeric transcription factor RFX (RFX-B, RFX5, and RFXAP) and the class II transactivator CIITA. Mutagenesis of the RFX genes was performed, and the properties of the proteins were analyzed with regard to transactivation, DNA binding, and protein-protein interactions. The results identified specific domains within each of the three RFX subunits that were necessary for RFX complex formation, including the ankyrin repeats of RFX-B. DNA binding was dependent on RFX complex formation, and transactivation was dependent on a region of RFX5. RFX5 was found to interact with CIITA, and this interaction was dependent on a proline-rich domain within RFX5. Thus, these studies have defined the protein domains required for the functional regulation of MHC class II genes.
DOI:10.1016/j.jmb.2010.08.025URLPMID:20732328 [本文引用: 1]

The mammalian immune response is mediated by a heterotetrameric transcriptional control complex, called regulatory factor X (RFX), that regulates the expression of major histocompatibility complex class II genes. RFX comprises three proteins: RFX5 (two copies), RFXAP, and RFXB, and mutations and deletions that prevent the assembly of the RFX complex have been linked to a severe immunodeficiency disorder. Two RFX5 molecules and one RFXAP molecule assemble in the cytoplasm prior to nuclear localization, a process mediated by an N-terminal
DOI:10.4049/jimmunol.173.1.410URLPMID:15210800 [本文引用: 1]

Comprised of RFX5, RFXAP, and RFX-B/ANK, the regulatory factor X (RFX) complex is an obligate transcription factor required for the expression of MHC class II genes. RFX functions by binding to the conserved X1 box sequence located upstream of all MHC class II genes. Using a mutagenesis scheme and a yeast heterologous reporter system, the mechanism by which the RFX complex is transported into the nucleus was examined. The results have identified specific nuclear localization signals (NLS) in both RFX5 and RFXAP that direct the nuclear translocation and expression of MHC class II genes. Additionally, a nuclear export signal was identified in the N terminus of RFXAP. RFX-B was poorly localized to the nucleus, and no specific NLS was identified. Whereas RFX5 could import an RFXAP NLS mutant into the nucleus, it had no effect on the import of RFX-B. The results suggest that although RFX5 and RFXAP could assemble before nuclear import, RFX-B association with the complex does not take place until after the subunits enter the nucleus. The identification of nuclear import and export sites on RFX molecules provides potential targets to modulate MHC class II expression.
DOI:10.1074/jbc.M111712200URLPMID:11986307

The transcription start site of the collagen alpha2(1) gene (COL1A2) has a sequence-specific binding site for a DNA methylation-responsive binding protein called regulatory factor for X-box 1 (RFX1) (Sengupta, P. K., Erhlich, M., and Smith, B. D. (1999) J. Biol. Chem. 274, 36649-36655). In this report, we demonstrate that RFX1 forms homodimers as well as heterodimers with RFX2 spanning the collagen transcription start site. Methylation at +7 on the coding strand increases RFX1 complex formation in gel shift assays. Methylation on the template strand, however, does not increase RFX1 complex formation. DNA from human fibroblasts contains minimal methylation on the coding strand (<4%) with variable methylation on the template strand. RFX1 acts as a repressor of collagen transcription as judged by in vitro transcription and co-transfection assays with an unmethylated collagen promoter-reporter construct. In addition, an RFX5 complex present in human fibroblasts interacts with the collagen RFX site, which is not sensitive to methylation. This is the first demonstration of RFX5 complex formation on a gene other than major histocompatibility complex (MHC) promoters. Also, RFX5 represses transcription of a collagen promoter-reporter construct in rat fibroblasts that have no detectable RFX5 complex formation or protein. RFX5 complex activates MHC II transcription by interacting with an interferon-gamma (IFN-gamma)-inducible protein, major histocompatibility class II trans-activator (CIITA). Collagen transcription is repressed by IFN-gamma in a dose-dependent manner in human but not in rat fibroblasts. IFN-gamma enhances RFX5 binding activity, and CIITA is present in the RFX5 complex of IFN-gamma-treated human fibroblasts. CIITA repressed collagen gene transcription more effectively in human fibroblasts than in rat fibroblasts, suggesting that the RFX5 complex may, in part, recruit CIITA protein to the collagen transcription start site. Thus the RFX family may be important repressors of collagen gene transcription through a RFX binding site spanning the transcription start site.
DOI:10.1002/ijc.25106URLPMID:20013806

Colorectal cancers (CRCs) develop on the basis of a deficient DNA mismatch repair (MMR) system in about 15% of cases. MMR-deficient CRC lesions show high-level microsatellite instability (MSI-H) and accumulate numerous mutations located at coding microsatellite loci that lead to the generation of immunogenic neopeptides. Consequently, the host's antitumoral immune response is of high importance for the course of the disease in MSI-H CRC patients. Accordingly, immune evasion mediated by impairment of HLA class I antigen presentation is frequently observed in these cancers. In this study, we aimed at a systematic analysis of alterations affecting HLA class II antigen expression in MSI-H CRC. HLA class II antigens are expressed by only two-thirds of MSI-H CRCs. The mechanisms underlying the lack of HLA class II antigens in a subset of MSI-H CRCs remain unknown. We here screened HLA class II regulatory genes for the presence of coding microsatellites and identified mutations of the essential regulator genes RFX5 in 9 (26.9%) out of 34 and CIITA in 1 (2.9%) out of 34 MSI-H CRCs. RFX5 mutations were related to lack of or faint HLA class II antigen expression (p = 0.006, Fisher's exact test). Transfection with wild-type RFX5 was sufficient to restore interferon gamma-inducible HLA class II antigen expression in the RFX5-mutant cell line HDC108. We conclude that somatic mutations of the RFX5 gene represent a novel mechanism of loss of HLA class II antigen expression in tumor cells, potentially contributing to immune evasion in MSI-H CRCs.
DOI:10.3892/or.2016.5240URLPMID:27840983 [本文引用: 1]

Regulatory factor X-5 (RFX5) was previously characterized as an essential and highly specific regulator of major histocompatibility class II (MHCII) gene expression in the immune system. We found that RFX5 is significantly upregulated in hepatocellular carcinoma (HCC) tumors and cell lines compared with non-tumor tissues in mRNA expression levels, but it fails to induce the expression of MHCII. However, RFX5 can strongly bind to the tripeptidyl peptidase 1 (TPP1) promoter region and then increase its transcriptional activity. We also found that manipulation the expression of RFX5 can significantly affect the expression of TPP1 in HepG2, which suggested that RFX5 can transcriptionally activate TPP1 in HCC. Moreover, TPP1 is overexpressed in HCC tissues and significantly correlated with poor prognosis of HCC patients, suggesting that it may have potential biological implications in HCC.
DOI:10.1126/science.1225829URLPMID:22745249 [本文引用: 1]

Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems provide bacteria and archaea with adaptive immunity against viruses and plasmids by using CRISPR RNAs (crRNAs) to guide the silencing of invading nucleic acids. We show here that in a subset of these systems, the mature crRNA that is base-paired to trans-activating crRNA (tracrRNA) forms a two-RNA structure that directs the CRISPR-associated protein Cas9 to introduce double-stranded (ds) breaks in target DNA. At sites complementary to the crRNA-guide sequence, the Cas9 HNH nuclease domain cleaves the complementary strand, whereas the Cas9 RuvC-like domain cleaves the noncomplementary strand. The dual-tracrRNA:crRNA, when engineered as a single RNA chimera, also directs sequence-specific Cas9 dsDNA cleavage. Our study reveals a family of endonucleases that use dual-RNAs for site-specific DNA cleavage and highlights the potential to exploit the system for RNA-programmable genome editing.
DOI:10.1038/nature09523URLPMID:21048762 [本文引用: 1]

Bacteria and Archaea have developed several defence strategies against foreign nucleic acids such as viral genomes and plasmids. Among them, clustered regularly interspaced short palindromic repeats (CRISPR) loci together with cas (CRISPR-associated) genes form the CRISPR/Cas immune system, which involves partially palindromic repeats separated by short stretches of DNA called spacers, acquired from extrachromosomal elements. It was recently demonstrated that these variable loci can incorporate spacers from infecting bacteriophages and then provide immunity against subsequent bacteriophage infections in a sequence-specific manner. Here we show that the Streptococcus thermophilus CRISPR1/Cas system can also naturally acquire spacers from a self-replicating plasmid containing an antibiotic-resistance gene, leading to plasmid loss. Acquired spacers that match antibiotic-resistance genes provide a novel means to naturally select bacteria that cannot uptake and disseminate such genes. We also provide in vivo evidence that the CRISPR1/Cas system specifically cleaves plasmid and bacteriophage double-stranded DNA within the proto-spacer, at specific sites. Our data show that the CRISPR/Cas immune system is remarkably adapted to cleave invading DNA rapidly and has the potential for exploitation to generate safer microbial strains.
DOI:10.1093/jmcb/mjv016URLPMID:25757625 [本文引用: 3]

The human genome contains millions of DNA regulatory elements and a large number of gene clusters, most of which have not been tested experimentally. The clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated nuclease 9 (Cas9) programed with a synthetic single-guide RNA (sgRNA) emerges as a method for genome editing in virtually any organisms. Here we report that targeted DNA fragment inversions and duplications could easily be achieved in human and mouse genomes by CRISPR with two sgRNAs. Specifically, we found that, in cultured human cells and mice, efficient precise inversions of DNA fragments ranging in size from a few tens of bp to hundreds of kb could be generated. In addition, DNA fragment duplications and deletions could also be generated by CRISPR through trans-allelic recombination between the Cas9-induced double-strand breaks (DSBs) on two homologous chromosomes (chromatids). Moreover, junctions of combinatorial inversions and duplications of the protocadherin (Pcdh) gene clusters induced by Cas9 with four sgRNAs could be detected. In mice, we obtained founders with alleles of precise inversions, duplications, and deletions of DNA fragments of variable sizes by CRISPR. Interestingly, we found that very efficient inversions were mediated by microhomology-mediated end joining (MMEJ) through short inverted repeats. We showed for the first time that DNA fragment inversions could be transmitted through germlines in mice. Finally, we applied this CRISPR method to a regulatory element of the Pcdhalpha cluster and found a new role in the regulation of members of the Pcdhgamma cluster. This simple and efficient method should be useful in manipulating mammalian genomes to study millions of regulatory DNA elements as well as vast numbers of gene clusters.
DOI:10.1016/j.molcel.2018.06.021URLPMID:30033371 [本文引用: 2]

Chromosomal rearrangements including large DNA-fragment inversions, deletions, and duplications by Cas9 with paired sgRNAs are important to investigate genome structural variations and developmental gene regulation, but little is known about the underlying mechanisms. Here, we report that disrupting CtIP or FANCD2, which have roles in alternative non-homologous end joining, enhances precise DNA-fragment deletion. By analyzing the inserted nucleotides at the junctions of DNA-fragment editing of deletions, inversions, and duplications and characterizing the cleaved products, we find that Cas9 endonucleolytically cleaves the noncomplementary strand with a flexible scissile profile upstream of the -3 position of the PAM site in vivo and in vitro, generating double-strand break ends with 5' overhangs of 1-3 nucleotides. Moreover, we find that engineered Cas9 nucleases have distinct cleavage profiles. Finally, Cas9-mediated nucleotide insertions are nonrandom and are equal to the combined sequences upstream of both PAM sites with predicted frequencies. Thus, precise and predictable DNA-fragment editing could be achieved by perturbing DNA repair genes and using appropriate PAM configurations.
DOI:10.1038/s41421-019-0120-zURLPMID:31636963 [本文引用: 2]
DOI:10.1126/science.1229223URL [本文引用: 1]

Allostery is well documented for proteins but less recognized for DNA-protein interactions. Here, we report that specific binding of a protein on DNA is substantially stabilized or destabilized by another protein bound nearby. The ternary complex's free energy oscillates as a function of the separation between the two proteins with a periodicity of similar to 10 base pairs, the helical pitch of B-form DNA, and a decay length of similar to 15 base pairs. The binding affinity of a protein near a DNA hairpin is similarly dependent on their separation, which-together with molecular dynamics simulations-suggests that deformation of the double-helical structure is the origin of DNA allostery. The physiological relevance of this phenomenon is illustrated by its effect on gene expression in live bacteria and on a transcription factor's affinity near nucleosomes.
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/nprot.2012.016URL [本文引用: 1]

Recent advances in high-throughput cDNA sequencing (RNA-seq) can reveal new genes and splice variants and quantify expression genome-wide in a single assay. The volume and complexity of data from RNA-seq experiments necessitate scalable, fast and mathematically principled analysis software. TopHat and Cufflinks are free, open-source software tools for gene discovery and comprehensive expression analysis of high-throughput mRNA sequencing (RNA-seq) data. Together, they allow biologists to identify new genes and new splice variants of known ones, as well as compare gene and transcript expression under two or more conditions. This protocol describes in detail how to use TopHat and Cufflinks to perform such analyses. It also covers several accessory tools and utilities that aid in managing data, including CummeRbund, a tool for visualizing RNA-seq analysis results. Although the procedure assumes basic informatics skills, these tools assume little to no background with RNA-seq analysis and are meant for novices and experts alike. The protocol begins with raw sequencing reads and produces a transcriptome assembly, lists of differentially expressed and regulated genes and transcripts, and publication-quality visualizations of analysis results. The protocol's execution time depends on the volume of transcriptome sequencing data and available computing resources but takes less than 1 d of computer time for typical experiments and similar to 1 h of hands-on time.
DOI:10.1038/nbt.3121URLPMID:25751057 [本文引用: 1]

Understanding how eukaryotic enhancers are bound and regulated by specific combinations of transcription factors is still a major challenge. To better map transcription factor binding genome-wide at nucleotide resolution in vivo, we have developed a robust ChIP-exo protocol called ChIP-nexus (chromatin immunoprecipitation experiments with nucleotide resolution through exonuclease, unique barcode and single ligation), which utilizes an efficient DNA self-circularization step during library preparation. Application of ChIP-nexus to four proteins--human TBP and Drosophila NFkB, Twist and Max--shows that it outperforms existing ChIP protocols in resolution and specificity, pinpoints relevant binding sites within enhancers containing multiple binding motifs, and allows for the analysis of in vivo binding specificities. Notably, we show that Max frequently interacts with DNA sequences next to its motif, and that this binding pattern correlates with local DNA-sequence features such as DNA shape. ChIP-nexus will be broadly applicable to the study of in vivo transcription factor binding specificity and its relationship to cis-regulatory changes in humans and model organisms.
DOI:10.1038/nprot.2016.043URLPMID:27031497 [本文引用: 2]

Unbiased, high-throughput assays for detecting and quantifying DNA double-stranded breaks (DSBs) across the genome in mammalian cells will facilitate basic studies of the mechanisms that generate and repair endogenous DSBs. They will also enable more applied studies, such as those to evaluate the on- and off-target activities of engineered nucleases. Here we describe a linear amplification-mediated high-throughput genome-wide sequencing (LAM-HTGTS) method for the detection of genome-wide 'prey' DSBs via their translocation in cultured mammalian cells to a fixed 'bait' DSB. Bait-prey junctions are cloned directly from isolated genomic DNA using LAM-PCR and unidirectionally ligated to bridge adapters; subsequent PCR steps amplify the single-stranded DNA junction library in preparation for Illumina Miseq paired-end sequencing. A custom bioinformatics pipeline identifies prey sequences that contribute to junctions and maps them across the genome. LAM-HTGTS differs from related approaches because it detects a wide range of broken end structures with nucleotide-level resolution. Familiarity with nucleic acid methods and next-generation sequencing analysis is necessary for library generation and data interpretation. LAM-HTGTS assays are sensitive, reproducible, relatively inexpensive, scalable and straightforward to implement with a turnaround time of <1 week.
DOI:10.1128/mcb.16.6.2802URLPMID:8649389 [本文引用: 1]

We have isolated and analyzed human CTCF cDNA clones and show here that the ubiquitously expressed 11-zinc-finger factor CTCF is an exceptionally highly conserved protein displaying 93% identity between avian and human amino acid sequences. It binds specifically to regulatory sequences in the promoter-proximal regions of chicken, mouse, and human c-myc oncogenes. CTCF contains two transcription repressor domains transferable to a heterologous DNA binding domain. One CTCF binding site, conserved in mouse and human c-myc genes, is found immediately downstream of the major P2 promoter at a sequence which maps precisely within the region of RNA polymerase II pausing and release. Gel shift assays of nuclear extracts from mouse and human cells show that CTCF is the predominant factor binding to this sequence. Mutational analysis of the P2-proximal CTCF binding site and transient-cotransfection experiments demonstrate that CTCF is a transcriptional repressor of the human c-myc gene. Although there is 100% sequence identity in the DNA binding domains of the avian and human CTCF proteins, the regulatory sequences recognized by CTCF in chicken and human c-myc promoters are clearly diverged. Mutating the contact nucleotides confirms that CTCF binding to the human c-myc P2 promoter requires a number of unique contact DNA bases that are absent in the chicken c-myc CTCF binding site. Moreover, proteolytic-protection assays indicate that several more CTCF Zn fingers are involved in contacting the human CTCF binding site than the chicken site. Gel shift assays utilizing successively deleted Zn finger domains indicate that CTCF Zn fingers 2 to 7 are involved in binding to the chicken c-myc promoter, while fingers 3 to 11 mediate CTCF binding to the human promoter. This flexibility in Zn finger usage reveals CTCF to be a unique
DOI:10.1038/nature08079URLPMID:19458616 [本文引用: 1]

Cohesin-mediated sister chromatid cohesion is essential for chromosome segregation and post-replicative DNA repair. In addition, evidence from model organisms and from human genetics suggests that cohesin is involved in the control of gene expression. This non-canonical role has recently been rationalized by the findings that mammalian cohesin complexes are recruited to a subset of DNase I hypersensitive sites and to conserved noncoding sequences by the DNA-binding protein CTCF. CTCF functions at insulators (which control interactions between enhancers and promoters) and at boundary elements (which demarcate regions of distinct chromatin structure), and cohesin contributes to its enhancer-blocking activity. The underlying mechanisms remain unknown, and the full spectrum of cohesin functions remains to be determined. Here we show that cohesin forms the topological and mechanistic basis for cell-type-specific long-range chromosomal interactions in cis at the developmentally regulated cytokine locus IFNG. Hence, the ability of cohesin to constrain chromosome topology is used not only for the purpose of sister chromatid cohesion, but also to dynamically define the spatial conformation of specific loci. This new aspect of cohesin function is probably important for normal development and disease.
DOI:10.1038/nature06634URLPMID:18235444 [本文引用: 1]

Cohesin complexes mediate sister-chromatid cohesion in dividing cells but may also contribute to gene regulation in postmitotic cells. How cohesin regulates gene expression is not known. Here we describe cohesin-binding sites in the human genome and show that most of these are associated with the CCCTC-binding factor (CTCF), a zinc-finger protein required for transcriptional insulation. CTCF is dispensable for cohesin loading onto DNA, but is needed to enrich cohesin at specific binding sites. Cohesin enables CTCF to insulate promoters from distant enhancers and controls transcription at the H19/IGF2 (insulin-like growth factor 2) locus. This role of cohesin seems to be independent of its role in cohesion. We propose that cohesin functions as a transcriptional insulator, and speculate that subtle deficiencies in this function contribute to 'cohesinopathies' such as Cornelia de Lange syndrome.
DOI:10.1126/science.abb0981URLPMID:32409525 [本文引用: 2]

As a ring-shaped adenosine triphosphatase (ATPase) machine, cohesin organizes the eukaryotic genome by extruding DNA loops and mediates sister chromatid cohesion by topologically entrapping DNA. How cohesin executes these fundamental DNA transactions is not understood. Using cryo-electron microscopy (cryo-EM), we determined the structure of human cohesin bound to its loader NIPBL and DNA at medium resolution. Cohesin and NIPBL interact extensively and together form a central tunnel to entrap a 72-base pair DNA. NIPBL and DNA promote the engagement of cohesin's ATPase head domains and ATP binding. The hinge domains of cohesin adopt an
DOI:10.1073/pnas.1911708117URLPMID:31937660 [本文引用: 1]

The DNA-binding protein CCCTC-binding factor (CTCF) and the cohesin complex function together to shape chromatin architecture in mammalian cells, but the molecular details of this process remain unclear. Here, we demonstrate that a 79-aa region within the CTCF N terminus is essential for cohesin positioning at CTCF binding sites and chromatin loop formation. However, the N terminus of CTCF fused to artificial zinc fingers was not sufficient to redirect cohesin to non-CTCF binding sites, indicating a lack of an autonomously functioning domain in CTCF responsible for cohesin positioning. BORIS (CTCFL), a germline-specific paralog of CTCF, was unable to anchor cohesin to CTCF DNA binding sites. Furthermore, CTCF-BORIS chimeric constructs provided evidence that, besides the N terminus of CTCF, the first two CTCF zinc fingers, and likely the 3D geometry of CTCF-DNA complexes, are also involved in cohesin retention. Based on this knowledge, we were able to convert BORIS into CTCF with respect to cohesin positioning, thus providing additional molecular details of the ability of CTCF to retain cohesin. Taken together, our data provide insight into the process by which DNA-bound CTCF constrains cohesin movement to shape spatiotemporal genome organization.
[本文引用: 1]
[本文引用: 1]
DOI:10.1073/pnas.1114357108URLPMID:21949399 [本文引用: 1]

The mouse protocadherin (Pcdh) -alpha, -beta, and -gamma gene clusters encode more than 50 protein isoforms, the combinatorial expression of which generates vast single-cell diversity in the brain. At present, the mechanisms by which this diversity is expressed are not understood. Here we show that two transcriptional enhancer elements, HS5-1 and HS7, play a critical role in Pcdhalpha gene expression in mice. We show that the HS5-1 element functions as an enhancer in neurons and a silencer in nonneuronal cells. The enhancer activity correlates with the binding of zinc finger DNA binding protein CTCF to the target promoters, and the silencer activity requires the binding of the REST/NRSF repressor complex in nonneuronal cells. Thus, the HS5-1 element functions as a neuron-specific enhancer and nonneuronal cell repressor. In contrast, the HS7 element functions as a Pcdhalpha cluster-wide transcription enhancer element. These studies reveal a complex organization of regulatory elements required for generating single cell Pcdh diversity.
DOI:10.1073/pnas.1205074109URLPMID:22550178 [本文引用: 1]

Extraordinary single-cell diversity is generated in the vertebrate nervous system by the combinatorial expression of the clustered protocadherin genes (Pcdhalpha, -beta, and -gamma). This diversity is generated by a combination of stochastic promoter choice and alternative pre-mRNA splicing. Here we show that both the insulator-binding protein CTCF and the cohesin complex subunit Rad21 bind to two highly conserved DNA sequences, the first within and the second downstream of transcriptionally active Pcdhalpha promoters. Both CTCF and Rad21 bind to these sites in vitro and in vivo, this binding directly correlates with alternative isoform expression, and knocking down CTCF expression reduces alternative isoform expression. Remarkably, a similarly spaced pair of CTCF/Rad21 binding sites was identified within a distant enhancer element (HS5-1), which is required for normal levels of alternative isoform expression. We also identify an additional, unique regulatory role for cohesin, as Rad21 binds to another enhancer (HS7) independently of CTCF, and knockdown of Rad21 reduces expression of the constitutive, biallelically expressed Pcdhalpha isoforms alphac1 and alphac2. We propose that CTCF and the cohesin complex initiate and maintain Pcdhalpha promoter choice by mediating interactions between Pcdhalpha promoters and enhancers.
