 ,1,2, 杨勇1,2, 魏唯2, 林辰涛2, 马留银
,1,2, 杨勇1,2, 魏唯2, 林辰涛2, 马留银 ,2
,2Identification and expression analyses of the NAC transcription factor family in Spartina alterniflora
Taotao Wang ,1,2, Yong Yang1,2, Wei Wei2, Chentao Lin2, Liuyin Ma
,1,2, Yong Yang1,2, Wei Wei2, Chentao Lin2, Liuyin Ma ,2
,2通讯作者: 马留银,博士,副教授,博士生导师,研究方向:植物转录后调控。E-mail:lma223@163.com
第一联系人:
编委: 赵方庆
收稿日期:2019-08-27修回日期:2019-12-26网络出版日期:2020-02-21
| 基金资助: |
Received:2019-08-27Revised:2019-12-26Online:2020-02-21
| Fund supported: |

摘要
互花米草(Spartina alterniflora)作为一种海岸带盐生植物,高度耐盐胁迫,但因为缺少参考基因组,其耐盐的分子机制却尚未见报道。NAC家族蛋白是植物特有的转录因子,调控植物的生长发育和胁迫应答。为了鉴定互花米草NAC蛋白(SaNAC)并探究它们与互花米草生长发育及胁迫响应之间的关系,本研究以互花米草三代全长转录组数据为参考,通过与水稻(Oryza sativa)、拟南芥(Arabidopsis thaliana)和玉米(Zea mays)NAC蛋白序列进行比对,并结合保守功能域进一步筛选,最终找到62个SaNAC蛋白。从蛋白序列比对、进化、motif预测、同源性比较、亚细胞定位、组织表达以及非生物胁迫下的基因差异表达等方面分别对互花米草NAC家族成员进行分析,结果发现SaNAC蛋白均含有保守的NAM结构域,且在进化上与水稻NAC家族具有一定的相似性;SaNAC家族中的两个蛋白SaNAC9和SaNAC49在细胞核表达;另外,本研究还发现互花米草SaNAC基因表达具有高度组织和胁迫应答差异性。这些结果表明互花米草NAC转录因子家族不仅具有保守的功能域,而且在调控互花米草的生长发育和非生物胁迫响应过程中具有重要的作用。
关键词:
Abstract
As a coastal halophyte, Spartina alterniflora has high salt tolerance. However, the mechanism at the molecular level has not been widely studied due to the absence of a reference genome. The proteins of NAC families are plant-specific transcription factors that regulate the growth, development and stress response in plants. To identify the NAC family and explore the relationship between NAC proteins and the growth, development and stress response of Spatina alterniflora, full-length transcriptome data of Spartina alterniflora by the third generation sequencing technology was used as reference sequences in this study to blast with the NAC protein sequences from Oryza sativa, Arabidopsis thaliana and Zea mays. Finally, 62 SaNAC proteins were found in Spartina alterniflora by deep analysis on conserved domains. Then we analyzed sequence alignment, evolution, motif prediction, homology comparison, subcellular localization, tissue and abiotic stress-induced gene differential expression profile on the NAC family members in Spartina alterniflora. As a result, all SaNAC proteins were found containing a conserved NAM domain and having certain evolutionary similarity with rice; two family proteins, SaNAC9 and SaNAC49, were expressed in the nucleus; moreover, SaNAC genes were identified to have distinct expressional profiles in different tissues and stress response of Spartina alterniflora. These results indicated the SaNAC transcription factor family not only had conserved functional domains but also played important role in the regulation of growth, development and abiotic stress response.
Keywords:
PDF (3469KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王涛涛, 杨勇, 魏唯, 林辰涛, 马留银. 互花米草NAC转录因子家族的鉴定与表达分析. 遗传[J], 2020, 42(2): 194-211 doi:10.16288/j.yczz.19-250
Taotao Wang.
由于固定生长的特点,植物极度依赖其所在的生存环境,因此环境因素对植物生长发育有着深远的影响[1]。其中,土壤盐渍化是制约植物生产的主要环境压力之一,高盐胁迫诱使细胞膜解体、产生活性氧、削弱代谢过程、抑制光合作用和减弱营养吸收,进而延缓植物生长和发育、降低农林作物品质[2]。因此,研究植物高盐耐受性的调控机理具有十分重要的理论与实践价值。
转录因子是一类重要的调控子,一般通过结合在胁迫相关基因的顺式作用元件,参与植物对干旱、高盐、低温等非生物胁迫的响应[3]。NAC (NAM、ATAF1/2和CUC2)家族是植物特有的且最大的转录因子家族之一。NAC家族蛋白在N端拥有一个大约160个氨基酸残基大小的保守区域,该区域被进一步分为5个亚结构域(A~E)[4,5]。在植物中,NAC转录因子调控大量的生长过程,包括花形态建成[6]、根部发育[7]、叶片衰老[8]和种子萌发[9]等。同时NAC转录因子也参与植物各类胁迫应答过程[10],例如小麦(Triticum aestivum) NAC家族因子TaNAC2、TaNAC47和TaNAC67在促进自身耐多种胁迫压力过程中具有重要的作用[11,12,13],林木类植物刚毛柽柳(Tamarix hispida) ThNAC13因子能促进柽柳和拟南芥(Arabidopsis thaliana)更加耐盐及渗透胁迫[14],表明NAC转录因子在促进农林物种耐胁迫改良方面均具有重要的利用价值。
随着高通量测序技术的发展,越来越多的植物NAC家族被发掘和鉴定,大大加速了NAC蛋白在调控植物生长发育和胁迫应答方面的研究。研究人员不仅在拟南芥和水稻(Oryza sativa)中发现117和151个NAC蛋白[15],在大豆(Glycine max)和乌拉尔图小麦(T.urartu)中也发现含有152和87个NAC蛋白[16,17]。同时,在一些林木植物如果梅(Prunus mume)、白梨(Pyrus bretschneideri)和海岸松(Pinus pinaster)中也分别识别出113、146和37个NAC因子[18,19,20]。分析NAC家族因子有助于探究植物在非生物胁迫下的应答机理,然而大多数植物因为缺少高质量的参考基因组,NAC家族还没有被系统地鉴定和研究。第三代转录组测序(Pacbio单分子测序)技术的诞生为很多缺少基因组的物种提供了可参考的全长转录组序列[21],在一定程度上解决了这些物种家族分析研究上的难题。
互花米草(Spartina alterniflora)作为耐盐性良好的盐生植物,自1979年引入我国以后,广泛分布于沿海地区的潮滩及河口湾地带,根系发达,茎秆粗壮,能够忍受恶劣的气候和环境,尤其是高盐胁迫。本研究以互花米草为对象,将全长转录组序列作为参考,通过与拟南芥、水稻和玉米NAC蛋白序列进行比对,并经过功能域分析和筛选,获得62个SaNAC蛋白,随后从序列比对、进化比较、蛋白定位、组织表达以及非生物胁迫下的基因差异表达等角度对SaNAC家族进行系统的分析,完成了NAC转录因子家族在互花米草转录组水平上的初步鉴定以及在生长发育和胁迫响应过程中的表达分析,为进一步探究互花米草NAC蛋白功能及遗传应用提供理论依据。
1 材料与方法
1.1 材料
互花米草种子和植株采自江苏省赣榆市(E119°16?, N34°46?),把种子浸泡在1.5%盐水中并且储存于4℃以备长期使用。幼苗种植:将互花米草种子放在含1/2 Hoaglang营养液的滤纸上萌发7 d,然后转移至含有营养液的生长盒中继续生长3周,期间每隔3 d更换一次营养液。盐处理:将互花米草幼苗分别培养在正常营养液以及含350、500和800 mmol/L NaCl的营养液中24 h,然后进行整株取样并立即置于液氮保存。干旱处理:将4周大的互花米草幼苗放在干燥的滤纸上,分别在第0 h、1 h、8 h和24 h取样并立即置于液氮保存。ABA处理:将4周大的互花米草幼苗放入含有100 μmol/L ABA[22]的营养液里培养,分别在第0 h、1 h、8 h和24 h取样并冻存。1.2 互花米草NAC家族的鉴定
拟南芥、水稻以及玉米NAC家族蛋白序列从植物转录因子数据库上(1.3 序列比较、同源进化树分析和motif预测
利用DNAMAN软件进行复合序列比对,保守的结构域参照拟南芥和水稻的NAC家族进行划分[5]。利用MEGA7软件构建系统进化树,选择Neighbour- Joining方法,并设置自检次数为1000次。利用MEME (1.4 烟草瞬时表达
利用基因特异性引物(表1)从互花米草cDNA中扩增SaNAC9和SaNAC49基因全长CDS序列,并连接到植物表达载体pCambia3301上GFP标签的N端,随后将载体借助农杆菌AGL0瞬时转入烟草(Nicotiana tabacum L.)叶片中。烟草浸染实验步骤:首先从转化目的载体的培养板上挑取单克隆菌落培养并进行PCR鉴定,然后取200 μL阳性菌液接入50 mL含有10 mmol/L MES、20 μmol/L 乙酰丁香酮以及相应抗生素的LB液体培养基中培养过夜。第二天,将菌液离心(4000 r/min,15 min),并用适量体积重悬液(10 mmol/L MES、150 μmol/L乙酰丁香酮、10 mmol/L MgCl2)将沉淀混匀至OD值为1.0~1.5之间,重悬的菌液放置室温2 h后用无菌注射器轻轻注射入新鲜烟草的叶片中,并对注射位置进行标记。将烟草放在暗室过夜培养后继续放置温室生长1~2 d。进行显微观察时,取小块注射位置的叶片浸泡在1 mg/L的DAPI染液中30 min,分别在倒置显微镜(型号Zeiss Axio Observer.A1)下观察SaNAC9、SaNAC49以及对照蛋白在GFP和DAPI通道下的荧光表达,找到同时表达的核定位信号后与明场图片进行重叠。1.5 SaNAC基因家族表达模式分析与验证
1.5.1 RNA-seq数据分析表达模式分析参考互花米草转录组测序结果[23],RNA-seq数据从Bioproject(PRJNA413596)下载。使用bowtie2 (v2.2.9)[24]将测序数据与互花米草全长转录组数据进行比对,然后对数据进行均一化处理,最后使用RSME[25]对处理后的数据进行计算,得到基因标准化后的表达量FPKM值。
Table 1
表1
表1本研究使用的引物序列
Table 1
| 基因名称 | 引物序列(5′→3′) | 引物用途 |
|---|---|---|
| SaNAC1 | F: AACACACTATCCTGCCTGCT | qRT-PCR |
| R: AGCTAGGTTCAAAGGACGCT | ||
| SaNAC5 | F: TCCATCCTTCTGACGCTGAA | qRT-PCR |
| R: TGTTGCCCTGTTTGATCTGC | ||
| SaNAC9 | F: CGAGGAGCTCATCACGTACT | qRT-PCR |
| R: TTAGTGGCACGGTTTGTTCG | ||
| SaNAC11 | F: ACTGCCACCACAAAATCGAC | qRT-PCR/RT-PCR |
| R: TAACATATGCCGTCCTCCCC | ||
| SaNAC15 | F: CAAGAAGGTGGTCAACGAGC | qRT-PCR |
| R: TCGCCTTCCAGTATCCAGTC | ||
| SaNAC17 | F: CCTCTACAAGTTCGACCCGT | qRT-PCR/RT-PCR |
| R: GACGAGCGCCTTCTTGATG | ||
| SaNAC18 | F: CTTGGTTCCATACAGCAGCC | qRT-PCR |
| R: GCTCTTCGCCTTGACATCTG | ||
| SaNAC19 | F: ATCATGCACGAGTACAGGCT | qRT-PCR |
| R: GCGCGTTCTTGTTGTTCTTG | ||
| SaNAC22 | F: AACTGGGTCATGCACGAGTA | qRT-PCR/RT-PCR |
| R: TCATCCTCCTCCTCTTCCCA | ||
| SaNAC24 | F: GGCGAGAAGGAGTGGTACTT | qRT-PCR |
| R: CTCGTGCATGATCCAGTTGG | ||
| SaNAC25 | F: TCAAGGTTCGAACGAGACCA | qRT-PCR |
| R: TTCATAGTGCCATCCCGACA | ||
| SaNAC26 | F: ATCCACATACCCCACCCAAG | qRT-PCR |
| R: CCGGAAGAAGACGACGAGTA | ||
| SaNAC28 | F: GGTGAGGAGGAACAGAACGA | qRT-PCR/RT-PCR |
| R: CCTGCCCTTGTAGTACACCA | ||
| SaNAC30 | F: AGTGGTACTTCTTCTCGCCG | qRT-PCR |
| R: CTCGTGCATGATCCAGTTGG | ||
| SaNAC31 | F: GATCGTCTCGCACTACCTCA | qRT-PCR/RT-PCR |
| R: TTGTCCTTTCCGGTAGCCTT | ||
| SaNAC37 | F: AAGAACGAGTGGGAGAAGGC | qRT-PCR/RT-PCR |
| R: TAGCTGAGGTCGACGAACAG | ||
| SaNAC38 | F: AGTCTCTCCGTGCTTCAACA | qRT-PCR |
| R: CTCTAGAAGCTCCTGGTCCG | ||
| SaNAC43 | F: CTCCTCCTGGCTAACTCGAC | qRT-PCR/RT-PCR |
| R: TCCCCACGTTAGGATGATGG | ||
| SaNAC45 | F: GAGGAGCTCATCACGCACTA | qRT-PCR |
| R: AAGATCTCCCTGTCCTTGCC |
新窗口打开|下载CSV
Table 1 (Continued)
续表1
续表1本研究使用的引物序列
Table 1 (Continued)
| 基因名称 | 引物序列(5′→3′) | 引物用途 |
|---|---|---|
| SaNAC46 | F: GGCGAGAAGGAGTGGTACTT | qRT-PCR/RT-PCR |
| R: CTCGTGCATGATCCAGTTGG | ||
| SaNAC51 | F: GCTCGTCAAATCCTACCTGC | qRT-PCR/RT-PCR |
| R: TTGGATTTGGCCTCGTTGTG | ||
| SaNAC56 | F: TCGACATGACCACCTCCTAC | qRT-PCR/RT-PCR |
| R: ATGCTCTGGATGTCGTCGAA | ||
| SaNAC57 | F: GAAGAGCTGGTGGTGCAGTA | qRT-PCR |
| R: CCGGATCGCGAAGAAGTACT | ||
| SaNAC59 | F: CATGATGTTGGACTGGGTGC | qRT-PCR/RT-PCR |
| R: ATGGAACTGGTGGTGATCGT | ||
| SaNAC60 | F: AGGGCGAGTGGTACTTCTTC | qRT-PCR/RT-PCR |
| R: CTTCTTGACGCCGATCATGG | ||
| SaACTIN | F: AGGGCAGTTTTCCCTAGCAT | qRT-PCR/RT-PCR |
| R: CTCTCTTGGACTGTGCCTCA | ||
| SaNAC9 | F: TGACCTCGAGACTAGTATGAGTACGGAAGGGTCAGG | 亚细胞定位载体构建 |
| R: AGGTGGAGGTCCCCCGGGCACCTGGTAACCAGCAGCA | ||
| SaNAC49 | F: TGACCTCGAGACTAGTATGGAGATGGAGCAGGATCTC | 亚细胞定位载体构建 |
| R: AGGTGGAGGTCCCCCGGGGTAGAGCAGATTGGCCAGGGT |
新窗口打开|下载CSV
1.5.2 RT-PCR验证
用试剂盒(广州美基生物公司,R4151-02)提取不同组织(叶、茎、根和地下茎)以及胁迫处理的互花米草幼苗总RNA,具体操作参照说明书进行,对RNA进行定性定量检测确保其质量满足后续实验。取1 μg RNA,利用MonScript? RTIII All-in-One Mix反转录试剂盒(武汉莫纳生物公司,RN05004M)合成第一链cDNA,具体操作参照说明书进行,将cDNA的浓度稀释至5 ng/μL后备用。挑选SaNAC11、SaNAC17、SaNAC22、SaNAC28、SaNAC31、SaNAC37、SaNAC43、SaNAC46、SaNAC51、SaNAC56、SaNAC59和SaNAC60基因进行RT-PCR验证,引物序列如表1所示。扩增前先用内参基因SaACTIN对模板进行均一化,然后取合适体积的模板进行PCR。PCR反应体系:10 μL MonAmp? 2× Taq Mix (武汉莫纳生物公司,MP05001),0.5 μL正向引物,0.5 μL反向引物,适当体积的模板cDNA,适量体积RNAse-free水,总体系20 μL。反应程序:94℃ 3 min;94℃ 30 s,55℃ 30 s,72℃ 30 s,35个循环;72℃ 5 min。
1.5.3 qRT-PCR验证
通过qRT-PCR验证SaNAC1、SaNAC5、SaNAC9、SaNAC15、SaNAC18、SaNAC19、SaNAC22、SaNAC24、SaNAC25、SaNAC26、SaNAC30、SaNAC31、SaNAC38、SaNAC45、SaNAC51和SaNAC57基因在互花米草叶、茎、地下茎和根部的表达量,同时验证SaNAC11、SaNAC18、SaNAC22、SaNAC25、SaNAC28、SaNAC31、SaNAC37、SaNAC43、SaNAC45、SaNAC46、SaNAC56和SaNAC59基因在ABA及干旱处理下的表达量。引物在Primer3 (http://primer3.ut.ee/)网站上设计,GC含量在40%~60%且Tm值在55℃~65℃之间,序列如表1所示。每个qRT-PCR反应体系包含1 μL正反向引物(10 μmol/L)、1 μL cDNA模板、7 μL RNAse-free水和10 μL SYBRGreen Master Mix(莫纳生物,RN04002M)。反应程序为95℃ 5 min;95℃ 10 s,60℃ 10 s,72℃ 30 s,40个循环。样本之间的表达量差异使用t-test进行计算,P值小于0.01定义为具有显著性差异。
2 结果与分析
2.1 互花米草NAC转录因子家族鉴定
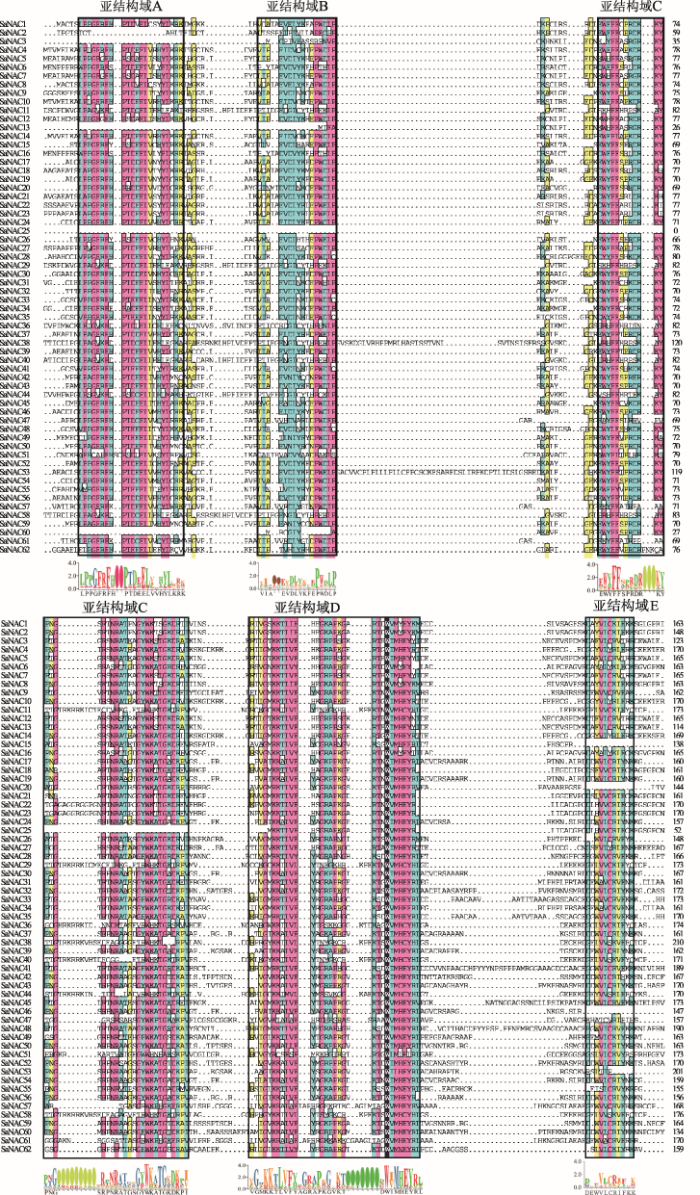
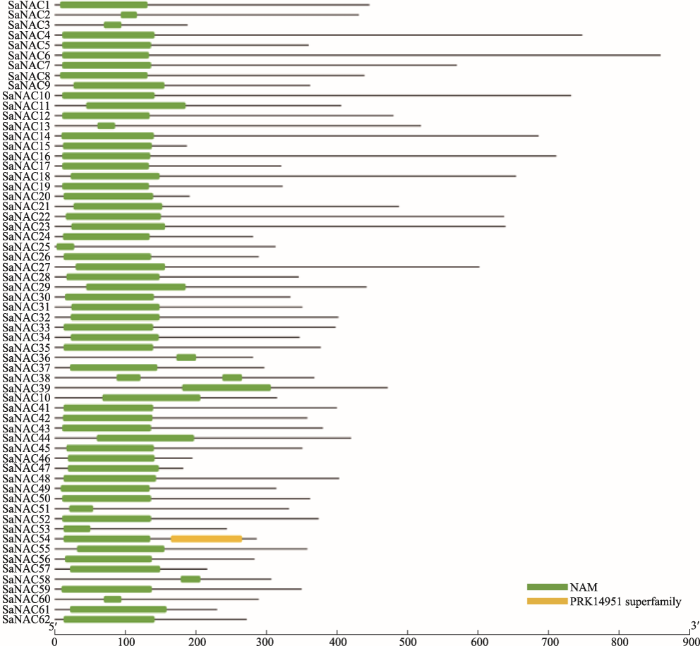
为了从转录组数据库中鉴定出含有功能域的NAC转录因子,本研究首先对复合序列比对产生的所有可能NAC蛋白进行保守功能域分析。CDD (Conserved Domain Database)预测结果显示,62个SaNAC蛋白在N端均含有NAM结构域(附图1)。NAM是NAC家族蛋白特有的结构域(NAM、ATAF1/2和CUC2)之一,也是NAC转录因子能够发挥结合功能的关键区域,预测结果表明62个SaNAC蛋白均是具有潜在NAC功能的转录因子。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1互花米草NAC蛋白序列比对
黑色线框圈出的位置代表不同的亚结构域A~E,不同颜色代表序列之间的相似性程度(黑色>红色>绿色>黄色),线框下方的Logo代表对应亚结构域的保守序列。
Fig. 1Sequence alignment of Spartina alterniflora NAC proteins
根据三代全长转录组数据对所有互花米草基因的命名[23],按从小到大的顺序将互花米草NAC蛋白命名为SaNAC1~SaNAC62,然后根据氨基酸序列对所有SaNAC蛋白的基础特征包括氨基酸数量、蛋白分子重量、等电点以及蛋白定位进行预测(表2)。通过分析发现,SaNAC蛋白大小介于186~ 858个氨基酸之间,蛋白分子量则介于21.2~93.2 kDa之间,其中SaNAC6和SaNAC15分别对应最大和最小的蛋白。所有SaNAC蛋白的等电点在4.3~ 10.7之间,SaNAC47和SaNAC2蛋白分别具有最大和最小的等电点。蛋白定位预测结果显示大多数SaNAC蛋白定位在细胞核,少数蛋白定位在细胞外(表2)。
Table 2
表2
表2互花米草NAC蛋白基本信息
Table 2
| 转录本序列号 | 蛋白名称 | 氨基酸数量(aa) | 分子量(kDa) | 等电点 | 预测的蛋白定位 |
|---|---|---|---|---|---|
| Cluster275-001 | SaNAC1 | 445 | 49.6 | 4.56 | 细胞核 |
| Cluster1229-001 | SaNAC2 | 430 | 61.2 | 4.3 | 细胞外 |
| Cluster1867-001 | SaNAC3 | 187 | 21.4 | 9.91 | 细胞核 |
| Cluster2112-001 | SaNAC4 | 747 | 82.7 | 4.71 | 细胞外 |
| Cluster4261-001 | SaNAC5 | 359 | 39.3 | 5.31 | 细胞核 |
| Cluster4343-001 | SaNAC6 | 858 | 93.2 | 4.47 | 细胞外 |
| Cluster5577-001 | SaNAC7 | 569 | 62.1 | 5.62 | 细胞核 |
| Cluster6349-001 | SaNAC8 | 438 | 48.9 | 4.59 | 细胞核 |
| Cluster7422-001 | SaNAC9 | 361 | 39.1 | 8.93 | 细胞核 |
| Cluster9525-001 | SaNAC10 | 731 | 80.7 | 4.86 | 细胞外 |
| Cluster10144-001 | SaNAC11 | 405 | 45.6 | 8.48 | 细胞核 |
| Cluster12101-001 | SaNAC12 | 479 | 52.8 | 5.8 | 细胞核 |
| Cluster13102-001 | SaNAC13 | 518 | 56 | 5.55 | 细胞核 |
| Cluster14225-001 | SaNAC14 | 685 | 75.8 | 4.85 | 细胞外 |
| Cluster15400-004 | SaNAC15 | 186 | 21.2 | 9.57 | 细胞核 |
| Cluster15711-001 | SaNAC16 | 710 | 77.7 | 4.61 | 细胞外 |
| Cluster16418-001 | SaNAC17 | 320 | 34.8 | 5.92 | 细胞核 |
| Cluster17526-001 | SaNAC18 | 653 | 71.4 | 4.73 | 细胞外 |
| Cluster17869-001 | SaNAC19 | 322 | 35.2 | 6.01 | 细胞核 |
| Cluster18322-001 | SaNAC20 | 190 | 21.5 | 9.94 | 细胞核 |
| Cluster18506-001 | SaNAC21 | 487 | 53.4 | 4.46 | 细胞核 |
| Cluster18550-001 | SaNAC22 | 636 | 69.1 | 4.53 | 细胞外 |
| Cluster18579-001 | SaNAC23 | 638 | 69.4 | 4.5 | 细胞外 |
| Cluster19615-001 | SaNAC24 | 280 | 31.2 | 8.74 | 细胞核 |
| Cluster20092-001 | SaNAC25 | 312 | 34.2 | 4 | 细胞外 |
| Cluster20280-001 | SaNAC26 | 288 | 31.9 | 6.92 | 细胞核 |
| Cluster21221-001 | SaNAC27 | 601 | 67 | 5.76 | 细胞外 |
| Cluster23463-001 | SaNAC28 | 345 | 39.8 | 6.25 | 细胞核 |
新窗口打开|下载CSV
Table 2 (Continued)
续表2
续表2互花米草NAC蛋白基本信息
Table 2 (Continued)
| 转录本序列号 | 蛋白名称 | 氨基酸数量(aa) | 分子量(kDa) | 等电点 | 预测的蛋白定位 |
|---|---|---|---|---|---|
| Cluster24200-001 | SaNAC29 | 441 | 49.8 | 7.43 | 细胞外 |
| Cluster24596-001 | SaNAC30 | 333 | 36.5 | 5.75 | 细胞核 |
| Cluster24844-001 | SaNAC31 | 350 | 38.6 | 5.97 | 细胞核 |
| Cluster24971-001 | SaNAC32 | 401 | 43.7 | 6.2 | 细胞核 |
| Cluster25024-001 | SaNAC33 | 397 | 42.6 | 6.39 | 细胞核 |
| Cluster25479-001 | SaNAC34 | 346 | 38.2 | 6.03 | 细胞核 |
| Cluster25519-001 | SaNAC35 | 376 | 40.4 | 6.27 | 细胞核 |
| Cluster25968-001 | SaNAC36 | 280 | 31.7 | 7.62 | 细胞核 |
| Cluster26335-001 | SaNAC37 | 296 | 33.3 | 5.84 | 细胞核 |
| Cluster26479-001 | SaNAC38 | 367 | 40.3 | 8.97 | 细胞核 |
| Cluster26810-001 | SaNAC39 | 471 | 53 | 8.25 | 细胞核 |
| Cluster26901-001 | SaNAC40 | 314 | 34.7 | 8.68 | 细胞核 |
| Cluster27315-001 | SaNAC41 | 399 | 43.3 | 6.59 | 细胞核 |
| Cluster27759-001 | SaNAC42 | 357 | 40 | 5.59 | 细胞核 |
| Cluster27876-001 | SaNAC43 | 379 | 41 | 8.26 | 细胞核 |
| Cluster27938-001 | SaNAC44 | 419 | 46.8 | 5.18 | 细胞核 |
| Cluster28031-001 | SaNAC45 | 350 | 37.8 | 6.4 | 细胞核 |
| Cluster28253-001 | SaNAC46 | 194 | 21.7 | 9.97 | 细胞核 |
| Cluster28923-001 | SaNAC47 | 181 | 20.1 | 10.4 | 细胞核 |
| Cluster28941-001 | SaNAC48 | 402 | 44.1 | 6.12 | 细胞核 |
| Cluster28982-001 | SaNAC49 | 313 | 35.2 | 6.72 | 细胞核 |
| Cluster29067-001 | SaNAC50 | 361 | 40.2 | 6.79 | 细胞核 |
| Cluster29514-001 | SaNAC51 | 331 | 35.6 | 6.12 | 细胞外 |
| Cluster29630-001 | SaNAC52 | 373 | 40.5 | 8.26 | 细胞核 |
| Cluster29846-001 | SaNAC53 | 243 | 26.8 | 9.78 | 细胞核 |
| Cluster30123-001 | SaNAC54 | 285 | 31.3 | 8.43 | 细胞核 |
| Cluster30128-001 | SaNAC55 | 357 | 39.5 | 9.36 | 细胞核 |
| Cluster30156-001 | SaNAC56 | 282 | 31.5 | 8.69 | 细胞核 |
| Cluster30326-001 | SaNAC57 | 215 | 23.2 | 9.96 | 细胞核 |
| Cluster30465-001 | SaNAC58 | 306 | 33.9 | 8.63 | 细胞核 |
| Cluster30667-001 | SaNAC59 | 349 | 38.8 | 6.35 | 细胞核 |
| Cluster30676-001 | SaNAC60 | 288 | 31.7 | 8.92 | 细胞核 |
| Cluster31570-001 | SaNAC61 | 229 | 24.4 | 10.15 | 细胞核 |
| Cluster32036-001 | SaNAC62 | 271 | 30.6 | 7.03 | 细胞核 |
新窗口打开|下载CSV
2.2 蛋白序列比对及功能域分析
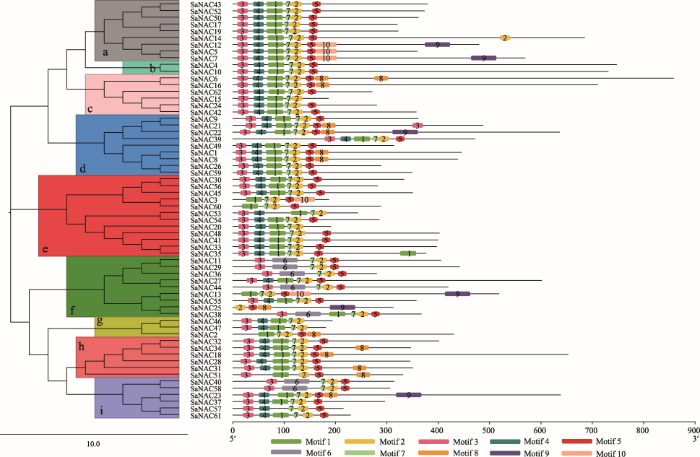
复合序列比对分析结果显示,所有的互花米草NAC蛋白在N端都具有保守的NAC结构域(图1),这与拟南芥和水稻的报道结果相一致[5]。除了少数基因具有不完整的NAC结构域外,大多数基因具有5个保守的亚结构域(A~E)。其中,蛋白SaNAC2、SaNAC3、SaNAC13和SaNAC60缺少A、B结构域;SaNAC58缺少B、C结构域;SaNAC25缺少A、B和C结构域;SaNAC46、47、51和53缺少E结构域。蛋白序列比对结果进一步证明了SaNAC家族蛋白在N端具有保守的NAM功能域(A~D),表明SaNAC转录因子可能与其他植物NAC蛋白功能相似。为了研究互花米草NAC转录因子之间的进化关系,本研究利用62个SaNAC蛋白序列构建NJ进化树。通过进化树分析将互花米草NAC家族分成9个小组,分别命名为a~I (图2)。其中小组e中包含最多的SaNAC蛋白,有12个,小组b中的SaNAC蛋白最少,只有2个,分别是SaNAC4和SaNAC10。为了进一步探究NAC蛋白功能域之间的关系,对所有的SaNAC蛋白序列进行motif分析(MEME),通过预测最终获得10个保守的motif (motif 1~10) (图2)。分析motif与蛋白结构域,发现motif 3、4、1、7、2和5分别对应SaNAC转录因子N端5个保守的亚结构A~D,其中motif 2和motif 7共同组成结构域D。另外,SaNAC11、29、36、38、40和58蛋白缺少motif 1和4,但是都拥有motif 6。Motif 8则主要存在于小组d和h中,而motif 9则出现在一些SaNAC蛋白的C端。结合小组分类分析motif,发现小组g中的全部NAC蛋白均具有不完整的NAC亚结构,而小组a、b、d里的NAC蛋白亚结构最为完整,其他小组里都有个别基因缺少某些亚结构(图2)。
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2互花米草NAC家族进化及MEME分析
左侧为SaNAC家族NJ进化树,a~i代表进化树不同的小组;右侧为SaNAC蛋白MEME预测结果,1~10代表不同的motif。
Fig. 2Evolutionary and MEME analysis of the Spartina alterniflora NAC family
2.3 互花米草与水稻NAC家族进化分析
为了探究互花米草与水稻NAC家族成员之间的进化关系,本研究利用MEGA7软件构建互花米草与水稻NAC蛋白NJ进化树(图3)。结果显示,所有NAC蛋白被分为12个小组(Ⅰ~Ⅻ),其中小组Ⅰ中蛋白数量最多,包含11个SaNAC和18个OsNAC蛋白,小组Ⅲ中蛋白数量最少,不包含SaNAC蛋白。小组Ⅷ和Ⅺ中也只包含1个(SaNAC9)及2个(SaNAC36和SaNAC44) SaNAC蛋白,说明互花米草基因组在长期的进化过程中可能丢失了一些NAC基因。但是,几乎所有小组都同时包含互花米草和水稻NAC蛋白,表明两个物种的NAC家族在进化水平上具有一定的相似性。图3
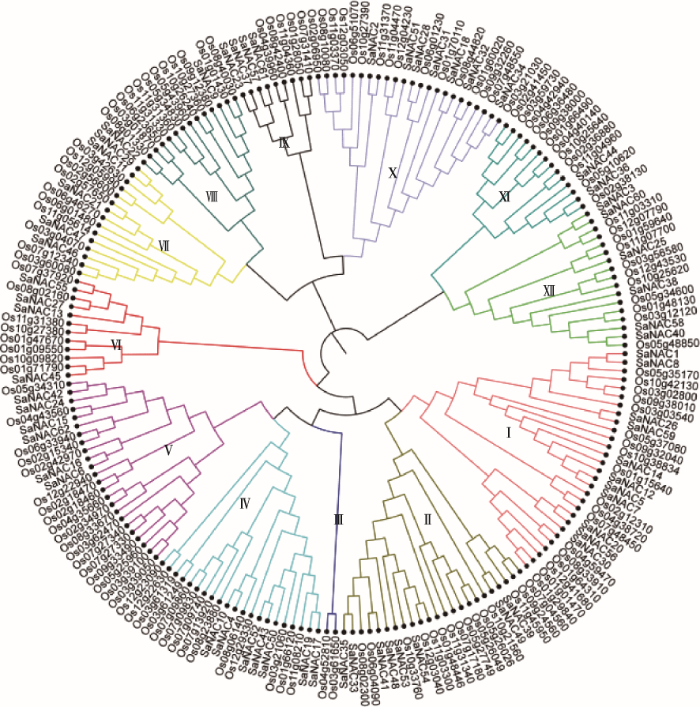 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3互花米草与水稻NAC家族系统进化树
Ⅰ~Ⅻ分别代表进化树的不同小组。
Fig. 3Phylogenetic tree of Spatina alterniflora and Oryza sativa NAC families
2.4 互花米草NAC蛋白亚细胞定位
蛋白定位预测结果表明大多数互花米草NAC转录因子定位在细胞核中(表1)。为了进一步验证SaNAC蛋白的表达定位,本研究将SaNAC9和SaNAC49基因全长CDS序列克隆到植物表达载体pCambia3301中,并利用烟草瞬时转化系统将目的载体和空载体对照同时转入烟草叶片表皮中。通过对SaNAC蛋白表达GFP信号的观察以及烟草叶片细胞核的染色,可以清楚的发现SaNAC9和SaNAC49蛋白在烟草表皮细胞的细胞核中表达(图4)。图4
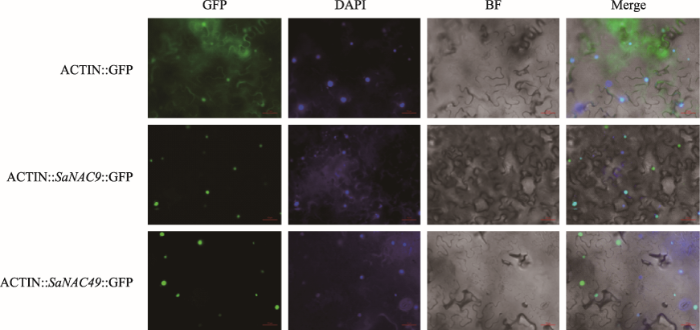 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4互花米草NAC蛋白亚细胞定位
BF代表明场通道下的细胞,GFP和DAPI分别代表绿色荧光和细胞核信号,Merge代表BF、GFP和DAPI图像的重叠。所有图像在显微镜下放大倍数:20 ×。
Fig. 4Subcellular localization of Spatina alterniflora NAC proteins
2.5 互花米草NAC基因在不同组织中的表达特征
为了研究SaNAC基因在互花米草不同组织中的表达情况,本研究随机挑选16个SaNAC基因,并通过qRT-PCR检测它们在互花米草的叶、茎、根以及地下茎中的表达量。结果显示,这些SaNAC基因在互花米草不同组织中的表达量各不相同,其中SaNAC15、SaNAC18、SaNAC19、SaNAC30、SaNAC31、SaNAC38、SaNAC51和SaNAC71在叶片表达量最高,而SaNAC1、SaNAC5、SaNAC22、SaNAC24和SaNAC25则在地下茎中有较高的表达量(图5)。另外,SaNAC9、SaNAC26和SaNAC45在根部有较高的表达量,这些基因在茎中的表达量都相对较低(图5)。结果显示,SaNAC基因的表达具有一定的组织差异性,表明SaNAC基因可能与互花米草的生长发育有关。图5
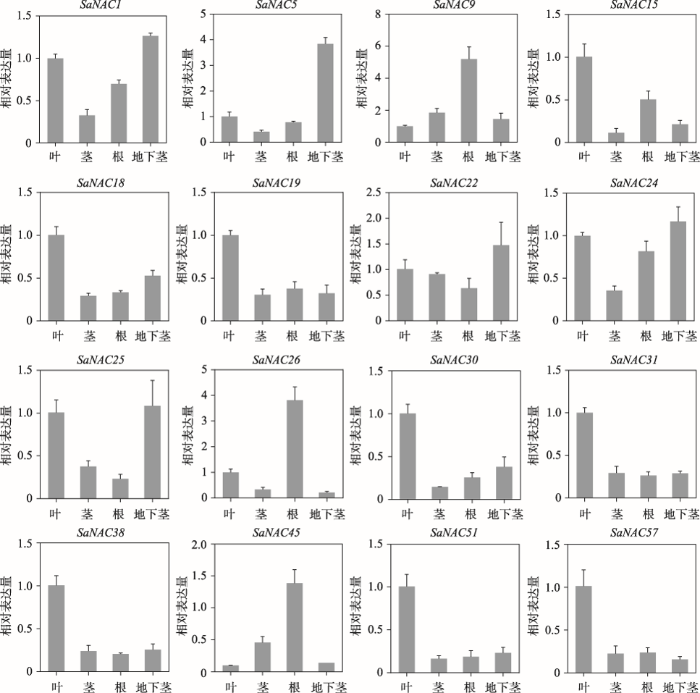 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图516个SaNAC基因组织表达特征
每个样品取3个生物学重复,叶片的表达量被均一化成1。
Fig. 5Tissue expression pattern of 16 SaNAC genes
2.6 互花米草NAC基因在盐胁迫下的表达特征
为了探究SaNAC基因在盐胁迫下的表达特征,结合不同浓度(0、350、500以及800 mmol/L)NaCl处理下的互花米草RNA-seq测序数据[23],对所有SaNAC基因的表达水平进行了系统分析。通过不同盐浓度处理下的表达热图可以看出,大部分SaNAC基因在盐处理下发生了差异表达(图6A),说明盐胁迫可以调控SaNAC基因的表达。其中SaNAC6、SaNAC11、SaNAC23、SaNAC28、SaNAC29、SaNAC33、SaNAC38和SaNAC58随着盐浓度的升高表达量持续降低(图6A),表明这些基因可能负向调控互花米草的盐胁迫响应。在800 mmol/L高盐处理时,与对照组相比,SaNAC3、SaNAC17、SaNAC18以及SaNAC22等25个基因表达量显著增加,SaNAC31基因的表达量上调最多,增加近60倍,表明高盐对SaNAC基因具有显著调控作用。其中SaNAC17、SaNAC32、SaNAC45以及SaNAC46等10个基因随着盐浓度的增加,表达量呈逐渐上升趋势(图6A)。另外,一些SaNAC基因随着盐浓度的增加,表达量呈先升高后降低的趋势,例如SaNAC8、SaNAC14、SaNAC20以及SaNAC26等(图6A),表明这些基因虽然受盐胁迫的调控,但具有一定的浓度范围,一旦超过最适的浓度,则开始发生负向调控。为了证明RNA-seq数据分析的可信性,本研究挑选了SaNAC11、SaNAC17、SaNAC22、SaNAC28、SaNAC31、SaNAC37、SaNAC43、SaNAC46、SaNAC51、SaNAC56、SaNAC59和SaNAC60基因进行RT-PCR验证(图6B),实验结果与RNA-seq分析结果基本一致,进一步证明盐胁迫调控互花米草NAC家族基因的表达。图6
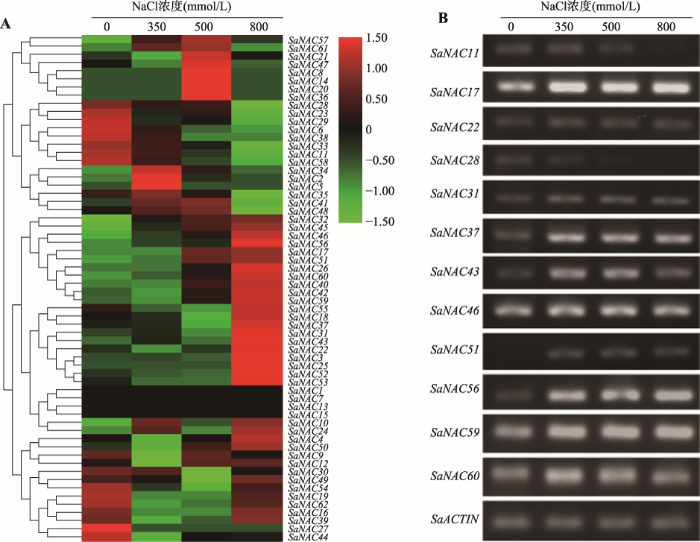 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6SaNAC基因在不同盐浓度下的表达特征
A:不同浓度盐胁迫下所有SaNAC基因的表达热图。表达量的值经过均一化后呈现在热图的右边,红色代表高表达,绿色代表低表达。B:12个盐胁迫应答基因的RT-PCR验证SaACTIN作为RT-PCR扩增均一化的内参基因。
Fig. 6Expression profiles of SaNAC genes under different concentration of salinity
2.7 互花米草NAC基因在ABA处理下的表达特征
NAC家族除了调控植物响应盐胁迫,还参与多种胁迫的调控过程[12,13]。脱落酸(abscisic acid, ABA)信号通路已被确定为植物非生物胁迫反应的中枢调节因子,可以引起植物生理反应和基因的表达[26],为了探究互花米草NAC基因在ABA胁迫下的表达变化,对12个随机的SaNAC基因在ABA处理下的表达量进行检测。qRT-PCR结果显示,在ABA处理下,大部分检测的SaNAC基因发生了表达量的改变(图7)。其中,SaNAC11、SaNAC25、SaNAC43及SaNAC59的表达随着ABA处理时间的增加逐渐降低,SaNAC18、SaNAC22、SaNAC31、SaNAC37、SaNAC45、SaNAC46及SaNAC56则在ABA处理下呈现显著上调的趋势,这些基因中只有SaNAC28的表达水平没有受到ABA的影响(图7),表明SaNAC基因的表达受到ABA的调控。图7
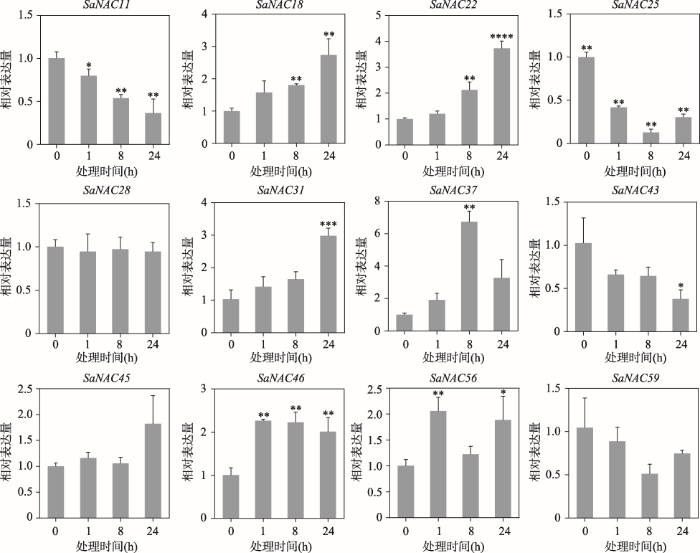 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7ABA处理下12个SaNAC基因的表达特征
每个时间点的样本取3个生物学重复,处理0 h的样本表达量被均一化为1,星号代表ABA处理1 h、8 h或者24 h的样本与0 h样本之间具有差异。*表示P<0.05,样本之间具有统计学差异;**表示P<0.01,样本之间具有显著性差异;***表示P<0.001,****表示P<0.0001,均代表样本之间具有极显著差异。
Fig. 7Expression pattern of 12 SaNAC genes under ABA treatment
2.8 互花米草NAC基因在干旱处理下的表达特征
与ABA胁迫类似,植物对干旱胁迫的应答同样是一个复杂的调控过程[27]。研究表明水稻ONAC066和ONAC095蛋白都参与干旱胁迫的调控[28,29],大豆NAC家族因子在抗旱过程中也具有重要作用[30],表明NAC蛋白是植物干旱胁迫响应过程中的重要调控子。本研究对12个SaNAC基因在干旱胁迫下的基因表达进行qRT-PCR验证,结果显示,与ABA胁迫类似,干旱胁迫促进大多数SaNAC基因发生差异表达(图8)。其中SaNAC11、SaNAC18、SaNAC22、SaNAC31、SaNAC37、SaNAC45和SaNAC46在干旱处理时表达量均呈显著上调趋势,且除SaNAC37外,其他基因的表达量都表现出先上升后下降的趋势。SaNAC25、SaNAC28、SaNAC43、SaNAC56和SaNAC59在干旱胁迫下表现出显著下调的趋势,其中SaNAC25和SaNAC28基因的表达量在1 h干旱处理时先降至最低,然后随着处理时间的增加开始逐渐增加(图8)。这些结果表明干旱胁迫同样可以调控SaNAC基因的表达。图8
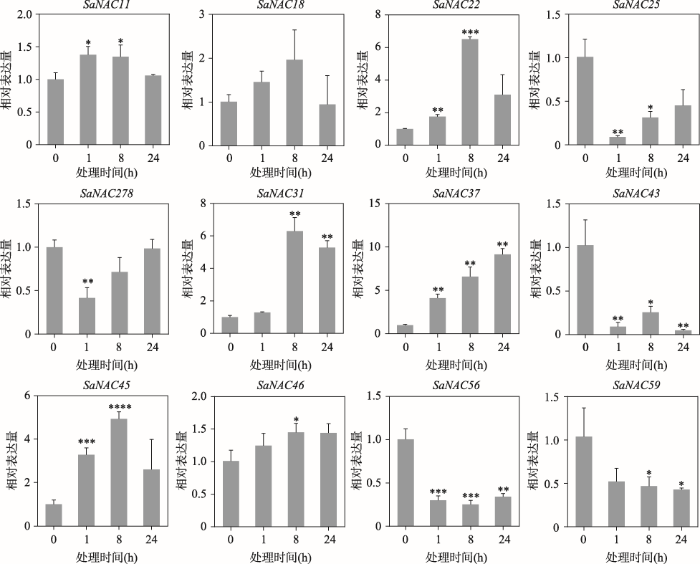 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8干旱处理下12个SaNAC基因的表达特征
每个时间点的样本取3个生物学重复,处理0 h的样本表达量被均一化为1,星号代表干旱处理1 h、8 h或者24 h的样本与0 h样本之间具有显著性差异。*表示P<0.05,样本之间具有统计学差异;**表示P<0.01,样本之间具有显著性差异;***表示P<0.001,****表示P<0.0001,均代表样本之间具有极显著差异。
Fig. 8Expression pattern of 12 SaNAC genes under drought treatment
3 讨论
互花米草作为高度耐盐的盐生植物,在沿海地带具有良好的生长优势。NAC家族蛋白具有保守的结构域和功能域,对植物的生长发育以及非生物胁迫应答具有重要的作用[16,18,20]。本研究从互花米草全长转录组数据中鉴定出62个完整的NAC转录因子,明显少于拟南芥和水稻基因组中的数目[5]。三代测序技术虽然可以得到较长的基因序列,但受到文库构建和机器读取不确定性的影响,相比基因组测序存在基因测序不全的缺点,因此互花米草NAC家族成员的完整性还有待进一步确定和完善。蛋白序列比对结果显示SaNAC家族蛋白在N端含有基本的NAC结构域,除了SaNAC2、SaNAC3和SaNAC13等一些蛋白缺少个别亚结构域(A~E)外,绝大多数SaNAC蛋白都含有完整的NAC结构。NAC蛋白结构可以分为两个部分:N端保守的DNA结合区域(BD)和C端变化的转录激活区域(TR)[5],其中BD包含150~160个氨基酸残基,主要被分为A~E五个亚结构域,与DNA结合、同源或异源二聚体的形成以及核定位具有密切联系。保守功能域预测结果显示SaNAC蛋白均含有NAM功能域, NAM区域虽然只包含A~D共4个NAC亚结构域,但结构域E却是AtNAM蛋白一个重要的DNA结合区域[31],因此可以用NAM结构域来替代分析NAC结构域。同时,亚细胞定位实验也证明SaNAC9和SaNAC49在细胞核里表达,这些分析结果共同表明SaNAC蛋白可能发挥植物NAC转录因子的功能。
在进化分组上共同位于某一小组的蛋白一般具有相似程度较高的序列,因此可能发挥相似的蛋白功能。互花米草与水稻同为单子叶植物,在结构上具有一定的相似之处,而且研究表明互花米草基因与水稻具有90%以上的相似度[32],因此分析它们NAC家族蛋白之间的进化保守性(图3),有助于更好的研究互花米草NAC蛋白的功能。系统进化树分析结果显示互花米草与水稻NAC蛋白相似程度较高,因此可以通过分析水稻NAC蛋白来预测互花米草NAC同源蛋白的功能。水稻ONAC095 (Os06g51070)被证明参与干旱和冷胁迫的响应,在干旱和ABA处理下,ONAC095基因表达量增加,通过显性嵌合抑制因子抑制ONAC095基因的表达后减少了水稻体内水分的丢失并且增加了脯氨酸和可溶性糖的含量,同时使一些干旱相关基因的表达发生上调,从而促进水稻抗干旱胁迫[29]。互花米草SaNAC2、SaNAC18、SaNAC28、SaNAC31、SaNAC32、SaNAC34及SaNAC51与OsNAC095蛋白共同位于X小组, 研究发现SaNAC18、SaNAC28和SaNAC31基因在干旱处理下的表达量同样增加,因此猜测这些蛋白可能同样参与互花米草对干旱胁迫的调控。
组织表达差异是基因选择性表达的结果,实验结果表明SaNAC基因在互花米草叶、茎、根以及地下茎中的表达具有明显差异。SaNAC22基因在拟南芥里的同源基因ANAC053编码一种转录激活因子,在干旱诱导的叶片衰老过程中,通过与ROS生物合成酶基因的启动子直接结合来促进ROS的产生[33]。互花米草能够生长在潮水涨落频繁的海岸带,主要原因之一是具有发达的地下根茎系统,地下茎的横向扩张也是互花米草繁殖的主要途径之一。本研究结果显示SaNAC22基因在互花米草地下茎中的表达量相对较高,同时在干旱处理下表达量也显著上调,因此猜测SaNAC22基因参与的干旱应答过程可能在互花米草地下茎中进行。SaNAC18在拟南芥的同源基因ATNAC2,参与调控拟南芥韧皮部薄壁转移细胞壁的生长[34],SaNAC38的同源基因ANAC073,也影响拟南芥纤维次生细胞壁发育以及增加纤维细胞面积相关基因的表达[35],SaNAC18和SaNAC38基因在互花米草叶片中具有较高的表达,却在茎秆部分表达量较低,表明它们很可能同样参与调控互花米草器官的生长发育。另外,叶片中含有盐腺是互花米草耐盐的特点之一,根部吸收的盐大多可以由盐腺排出体外,因此叶片在互花米草的耐盐过程中也极为重要[36]。在研究红树林的盐腺时,发现相比叶肉组织,在富含盐腺的组织中红树林AoNAC32基因具有更高的表达量[37],说明NAC家族因子可能参与调控盐腺器官的发育。因此,大量SaNAC基因在互花米草叶片中的高度表达,可能与盐腺的发育以及耐盐的调控有关。
为了系统的研究非生物胁迫对SaNAC基因表达的调控,本研究同时分析了盐、ABA以及干旱胁迫对SaNAC基因表达的影响。耐盐是互花米草最显著的特征,找出耐盐相关基因对研究其机理具有重要意义。非生物胁迫的控制机制依赖于大量胁迫相关基因的激活和调控[38],通过RNA-seq数据和qRT- PCR分析发现SaNAC18、SaNAC22、SaNAC31、SaNAC37、SaNAC45和SaNAC46基因同时受到高盐、ABA和干旱胁迫的调控,基因的表达量在3种胁迫下均显著增加,表明这些基因都是潜在的胁迫相关基因,在非生物胁迫的调控过程中可能扮演重要的角色。其中,SaNAC22的同源基因ANAC053参与调控拟南芥对干旱胁迫的应答[33],SaNAC37的同源基因ANAC081调控拟南芥细胞的衰老[39],而SaNAC46同源基因ANAC2则可以负向调控ABA信号通路[40],充分显示SaNAC基因在互花米草组织发育和对非生物胁迫应答方面的重要作用,同时也表明互花米草对非生物胁迫的应答是一个多基因复杂调控的过程。本研究丰富了互花米草NAC家族转录因子的信息,不仅为更好的研究互花米草生长发育和胁迫响应机制提供一定的参考,同时也为农林作物耐盐改良提供良好的基因资源。
附录
附图1详见文章电子版 www.chinagene.cn。附图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图1SaNAC蛋白保守功能域
Supplementle Fig. 1Conserved functional domains of SaNAC proteins
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/j.cell.2016.08.029URLPMID:27716505 [本文引用: 1]

As sessile organisms, plants must cope with abiotic stress such as soil salinity, drought, and extreme temperatures. Core stress-signaling pathways involve protein kinases related to the yeast SNF1 and mammalian AMPK, suggesting that stress signaling in plants evolved from energy sensing. Stress signaling regulates proteins critical for ion and water transport and for metabolic and gene-expression reprogramming to bring about ionic and water homeostasis and cellular stability under stress conditions. Understanding stress signaling and responses will increase our ability to improve stress resistance in crops to achieve agricultural sustainability and food security for a growing world population.
[本文引用: 1]
DOI:10.3390/ijms19061634URLPMID:29857524 [本文引用: 1]

Owing to diverse abiotic stresses and global climate deterioration, the agricultural production worldwide is suffering serious losses. Breeding stress-resilient crops with higher quality and yield against multiple environmental stresses via application of transgenic technologies is currently the most promising approach. Deciphering molecular principles and mining stress-associate genes that govern plant responses against abiotic stresses is one of the prerequisites to develop stress-resistant crop varieties. As molecular switches in controlling stress-responsive genes expression, transcription factors (TFs) play crucial roles in regulating various abiotic stress responses. Hence, functional analysis of TFs and their interaction partners during abiotic stresses is crucial to perceive their role in diverse signaling cascades that many researchers have continued to undertake. Here, we review current developments in understanding TFs, with particular emphasis on their functions in orchestrating plant abiotic stress responses. Further, we discuss novel molecular mechanisms of their action under abiotic stress conditions. This will provide valuable information for understanding regulatory mechanisms to engineer stress-tolerant crops.
DOI:10.1105/tpc.9.6.841URLPMID:9212461 [本文引用: 1]

Mutations in CUC1 and CUC2 (for CUP-SHAPED COTYLEDON), which are newly identified genes of Arabidopsis, caused defects in the separation of cotyledons (embryonic organs), sepals, and stamens (floral organs) as well as in the formation of shoot apical meristems. These defects were most apparent in the double mutant. Phenotypes of the mutants suggest a common mechanism for separating adjacent organs within the same whorl in both embryos and flowers. We cloned the CUC2 gene and found that the encoded protein was homologous to the petunia NO APICAL MERISTEM (NAM) protein, which is thought to act in the development of embryos and flowers.
DOI:10.1093/dnares/10.6.239URLPMID:15029955 [本文引用: 5]

The NAC domain was originally characterized from consensus sequences from petunia NAM and from Arabidopsis ATAF1, ATAF2, and CUC2. Genes containing the NAC domain (NAC family genes) are plant-specific transcriptional regulators and are expressed in various developmental stages and tissues. We performed a comprehensive analysis of NAC family genes in Oryza sativa (a monocot) and Arabidopsis thaliana (a dicot). We found 75 predicted NAC proteins in full-length cDNA data sets of O. sativa (28,469 clones) and 105 in putative genes (28,581 sequences) from the A. thaliana genome. NAC domains from both predicted and known NAC family proteins were classified into two groups and 18 subgroups by sequence similarity. There were a few differences in amino acid sequences in the NAC domains between O. sativa and A. thaliana. In addition, we found 13 common sequence motifs from transcriptional activation regions in the C-terminal regions of predicted NAC proteins. These motifs probably diverged having correlations with NAC domain structures. We discuss the relationship between the structure and function of the NAC family proteins in light of our results and the published data. Our results will aid further functional analysis of NAC family genes.
DOI:10.1016/s0092-8674(00)80902-2URLPMID:9489703 [本文引用: 1]

To understand how homeotic genes affect morphogenesis and differentiation, their target genes must be identified. In Arabidopsis flowers, the homeotic protein heterodimer APETALA3/PISTILLATA is necessary for petal and stamen formation. Here, AP3/PI function was put under posttranslational control to analyze its immediate effect on the floral mRNA population, with indirect effects blocked by cycloheximide. Using differential display, a target gene of AP3/PI was identified (NAP:NAC-LIKE, ACTIVATED BY AP3/PI), which is homologous to genes required for meristem establishment and separation of floral organs. The expression pattern of NAP and the phenotypes caused by its misexpression suggest that it functions in the transition between growth by cell division and cell expansion in stamens and petals.
DOI:10.1186/s12870-015-0530-5URLPMID:26048392 [本文引用: 1]

Long non-coding RNAs (lncRNAs) have been shown to play crucially regulatory roles in diverse biological processes involving complex mechanisms. However, information regarding the number, sequences, characteristics and potential functions of lncRNAs in plants is so far overly limited.
DOI:10.1016/j.pbi.2016.06.002URLPMID:27314623 [本文引用: 1]

Leaf senescence is finely tuned by many regulatory factors such as NAC (NAM/ATAF/CUC) transcription factors (TFs). NACs comprise one of the largest families of TFs in plants, many of which are differentially regulated during leaf senescence and play a major role in leaf senescence. Recent studies advanced our understanding on the structural and functional features of NAC TFs including target binding specificities of the N-terminal DNA binding domain and dynamic interaction of the C-terminal intrinsically disordered domain with other proteins. NAC TFs control other NACs and also interact with NACs or other TFs to fine-tune the expression of target genes. These studies clearly demonstrated the highly complex characteristics of NAC regulatory networks, which are dynamically regulated temporally and spatially and effectively integrate multiple developmental and environmental signals.
DOI:10.1104/pp.111.177071URL [本文引用: 1]

Seed germination is regulated through elaborately interacting signaling networks that integrate diverse environmental cues into hormonal signaling pathways. Roles of gibberellic acid and abscisic acid in germination have been studied extensively using Arabidopsis (Arabidopsis thaliana) mutants having alterations in seed germination. Auxin has also been implicated in seed germination. However, how auxin influences germination is largely unknown. Here, we demonstrate that auxin is linked via the IAA30 gene with a salt signaling cascade mediated by the NAM-ATAF1/2-CUC2 transcription factor NTM2/Arabidopsis NAC domain-containing protein 69 (for NAC with Transmembrane Motif1) during seed germination. Germination of the NTM2-deficient ntm2-1 mutant seeds exhibited enhanced resistance to high salinity. However, the salt resistance disappeared in the ntm2-1 mutant overexpressing the IAA30 gene, which was induced by salt in a NTM2-dependent manner. Auxin exhibited no discernible effects on germination under normal growth conditions. Under high salinity, however, whereas exogenous application of auxin further suppressed the germination of control seeds, the auxin effects were reduced in the ntm2-1 mutant. Consistent with the inhibitory effects of auxin on germination, germination of YUCCA 3-overexpressing plants containing elevated levels of active auxin was more severely influenced by salt. These observations indicate that auxin delays seed germination under high salinity through cross talk with the NTM2-mediated salt signaling in Arabidopsis.
DOI:10.3724/SP.J.1005.2012.00993URL [本文引用: 1]

NAC transcription factors belong to a unique class of transcription factors in plants. The common characteristics of the NAC proteins are the presence of a conserved NAC domain, comprising of about 150 amino acids in N-terminals and a highly variable transcriptional regulation region in C-terminals. Extensive studies have revealed that NAC transcription factors not only play important roles in plant growth and development, but also have functions in regulation of responses to biotic and abiotic stresses. In this minireview, we summarized the functions and mechanisms of the NAC transcriptional factors in plant abiotic and biotic stress responses. We also discussed future directions towards understanding the biological functions of the members of the NAC transcriptional factors in plants.
DOI:10.3724/SP.J.1005.2012.00993URL [本文引用: 1]

NAC transcription factors belong to a unique class of transcription factors in plants. The common characteristics of the NAC proteins are the presence of a conserved NAC domain, comprising of about 150 amino acids in N-terminals and a highly variable transcriptional regulation region in C-terminals. Extensive studies have revealed that NAC transcription factors not only play important roles in plant growth and development, but also have functions in regulation of responses to biotic and abiotic stresses. In this minireview, we summarized the functions and mechanisms of the NAC transcriptional factors in plant abiotic and biotic stress responses. We also discussed future directions towards understanding the biological functions of the members of the NAC transcriptional factors in plants.
DOI:10.1371/journal.pone.0084359URLPMID:24427285 [本文引用: 1]

Abiotic stresses are major environmental factors that affect agricultural productivity worldwide. NAC transcription factors play pivotal roles in abiotic stress signaling in plants. As a staple crop, wheat production is severely constrained by abiotic stresses whereas only a few NAC transcription factors have been characterized functionally. To promote the application of NAC genes in wheat improvement by biotechnology, a novel NAC gene designated TaNAC67 was characterized in common wheat. To determine its role, transgenic Arabidopsis overexpressing TaNAC67-GFP controlled by the CaMV-35S promoter was generated and subjected to various abiotic stresses for morphological and physiological assays. Gene expression showed that TaNAC67 was involved in response to drought, salt, cold and ABA treatments. Localization assays revealed that TaNAC67 localized in the nucleus. Morphological analysis indicated the transgenics had enhanced tolerances to drought, salt and freezing stresses, simultaneously supported by enhanced expression of multiple abiotic stress responsive genes and improved physiological traits, including strengthened cell membrane stability, retention of higher chlorophyll contents and Na(+) efflux rates, improved photosynthetic potential, and enhanced water retention capability. Overexpression of TaNAC67 resulted in pronounced enhanced tolerances to drought, salt and freezing stresses, therefore it has potential for utilization in transgenic breeding to improve abiotic stress tolerance in crops.
DOI:10.1093/jxb/err462URL [本文引用: 2]

Environmental stresses such as drought, salinity, and cold are major factors that significantly limit agricultural productivity. NAC transcription factors play essential roles in response to various abiotic stresses. However, the paucity of wheat NAC members functionally characterized to date does not match the importance of this plant as a world staple crop. Here, the function of TaNAC2 was characterized in Arabidopsis thaliana. A fragment of TaNAC2 was obtained from suppression subtractive cDNA libraries of wheat treated with polyethylene glycol, and its full-length cDNA was obtained by searching a full-length wheat cDNA library. Gene expression profiles indicated that TaNAC2 was involved in response to drought, salt, cold, and abscisic acid treatment. To test its function, transgenic Arabidopsis lines overexpressing TaNAC2-GFP controlled by the cauliflower mosaic virus 35S promoter were generated. Overexpression of TaNAC2 resulted in enhanced tolerances to drought, salt, and freezing stresses in Arabidopsis, which were simultaneously demonstrated by enhanced expression of abiotic stress-response genes and several physiological indices. Therefore, TaNAC2 has potential for utilization in transgenic breeding to improve abiotic stress tolerances in crops.
DOI:10.3389/fpls.2015.01174URLPMID:26834757 [本文引用: 2]

NAC transcription factors play diverse roles in plant development and responses to abiotic stresses. However, the biological roles of NAC family members in wheat are not well understood. Here, we reported the isolation and functional characterization of a novel wheat TaNAC47 gene. TaNAC47 encoded protein, localizing in the nucleus, is able to bind to the ABRE cis-element and transactivate transcription in yeast, suggesting that it likely functions as a transcriptional activator. We also showed that TaNAC47 is differentially expressed in different tissues, and its expression was induced by the stress treatments of salt, cold, polyethylene glycol and exogenous abscisic acid. Furthermore, overexpression of TaNAC47 in Arabidopsis resulted in ABA hypersensitivity and enhancing tolerance of transgenic plants to drought, salt, and freezing stresses. Strikingly, overexpression of TaNAC47 was found to activate the expression of downstream genes and change several physiological indices that may enable transgenic plants to overcome unfavorable environments. Taken together, these results uncovered an important role of wheat TaNAC47 gene in response to ABA and abiotic stresses.
DOI:10.3389/fpls.2017.00635URLPMID:28491072 [本文引用: 1]

NAC (NAM, ATAF1/2, and CUC2) proteins play critical roles in many plant biological processes and environmental stress. However, NAC proteins from Tamarix hispida have not been functionally characterized. Here, we studied a NAC gene from T. hispida, ThNAC13, in response to salt and osmotic stresses. ThNAC13 is a nuclear protein with a C-terminal transactivation domain. ThNAC13 can bind to NAC recognized sites and calmodulin-binding NAC (CBNAC) binding element. Overexpression of ThNAC13 in Arabidopsis improved seed germination rate and increased root growth and fresh weight gain under salt or osmotic stress. Transgenic T. hispida plants transiently overexpressing ThNAC13 and with RNAi-silenced ThNAC13 were generated for gain- and loss-of-function experiments. Following exposure to salt or osmotic stress, overexpression of ThNAC13 induced superoxide dismutase (SOD) and peroxidase (POD) activities, chlorophyll and proline contents; decreased the reactive oxygen species (ROS) and malondialdehyde levels; and reduced electrolyte leakage rates in both transgenic Tamarix and Arabidopsis plants. In contrast, RNAi-silenced ThNAC13 showed the opposite results in transgenic Tamarix. Furthermore, ThNAC13 induced the expression of SODs and PODs in transgenic Arabidopsis. These results suggest that ThNAC13 improves salt and osmotic tolerance by enhancing the ROS-scavenging capability and adjusting osmotic potential.
DOI:10.1016/j.gene.2010.06.008URLPMID:20600702 [本文引用: 1]

We investigated 151 non-redundant NAC genes in rice and 117 in Arabidopsis. A complete overview of this gene family in rice is presented, including gene structures, phylogenies, genome localizations, and expression profiles. We also performed a comparative analysis of these genes in rice and Arabidopsis. Conserved amino acid residues and phylogeny construction using the NAC conserved domain sequence suggest that OsNAC gene family was classified broadly into two major groups (A and B) and sixteen subgroups in rice. We presented more specific phylogenetic analysis of OsNAC proteins based on the DNA-binding domain and known gene function, respectively. Loss of introns was observed in the segmental duplication. Homologous, paralogous, and orthologous searches of rice and Arabidopsis revealed that the major functional diversification within the NAC gene family predated the divergence of monocots and dicots. The chromosomal localizations of OsNAC genes indicated nine segmental duplication events involving 18 genes; 32 non-redundant OsNAC genes were involved in tandem duplications. Expression levels of this gene family were checked under various abiotic stresses (cold, drought, submergence, laid-down submergence, osmotic, salinity and hormone) and biotic stresses [infection with rice viruses such as RSV (rice stripe virus) and RTSV (rice tungro spherical virus)]. Biotic stresses are novel work and increase the possibilities for finding the best candidate genes. A preliminary search based on our microarray (22K and 44K) data suggested that more than 45 and 26 non-redundant genes in this family were upregulated in response to abiotic and biotic stresses, respectively. All of the genes were further investigated for their stress responsiveness by RT-PCR analysis. Six genes showed preferential expression under both biotic RSV and RTSV stress. Eleven genes were upregulated by at least three abiotic treatments. Our study provides a very useful reference for cloning and functional analysis of members of this gene family in rice.
DOI:10.1186/s12870-018-1367-5URLPMID:30041622 [本文引用: 2]

Transcription factors operate as important switches of transcription networks, and NAC (NAM, ATAF, and CUC) transcription factors are a plant-specific family involved in multiple biological processes. However, this gene family has not been systematically characterized in cotton.
[本文引用: 1]
[本文引用: 1]
DOI:10.1186/s12870-019-1760-8URLPMID:31023218 [本文引用: 2]

Although the genome of Chinese white pear ('Dangshansuli') has been released, little is known about the functions, evolutionary history and expression patterns of NAC families in this species to date.
DOI:10.1186/s12870-015-0640-0URLPMID:26500018 [本文引用: 1]

NAC transcription factors comprise a large plant-specific gene family involved in the regulation of diverse biological processes. Despite the growing number of studies on NAC transcription factors in various species, little information is available about this family in conifers. The goal of this study was to identify the NAC transcription family in maritime pine (Pinus pinaster), to characterize ATAF-like genes in response to various stresses and to study their molecular regulation.
DOI:10.3390/genes9100494URLPMID:30322087 [本文引用: 2]

NAC transcription factors (TFs) participate in multiple biological processes, including biotic and abiotic stress responses, signal transduction and development. Cold stress can adversely impact plant growth and development, thereby limiting agricultural productivity. Prunus mume, an excellent horticultural crop, is widely cultivated in Asian countries. Its flower can tolerate freezing-stress in the early spring. To investigate the putative NAC genes responsible for cold-stress, we identified and analyzed 113 high-confidence PmNAC genes and characterized them by bioinformatics tools and expression profiles. These PmNACs were clustered into 14 sub-families and distributed on eight chromosomes and scaffolds, with the highest number located on chromosome 3. Duplicated events resulted in a large gene family; 15 and 8 pairs of PmNACs were the result of tandem and segmental duplicates, respectively. Moreover, three membrane-bound proteins (PmNAC59/66/73) and three miRNA-targeted genes (PmNAC40/41/83) were identified. Most PmNAC genes presented tissue-specific and time-specific expression patterns. Sixteen PmNACs (PmNAC11/19/20/23/41/48/58/74/75/76/78/79/85/86/103/111) exhibited down-regulation during flower bud opening and are, therefore, putative candidates for dormancy and cold-tolerance. Seventeen genes (PmNAC11/12/17/21/29/42/30/48/59/66/73/75/85/86/93/99/111) were highly expressed in stem during winter and are putative candidates for freezing resistance. The cold-stress response pattern of 15 putative PmNACs was observed under 4 °C at different treatment times. The expression of 10 genes (PmNAC11/20/23/40/42/48/57/60/66/86) was upregulated, while 5 genes (PmNAC59/61/82/85/107) were significantly inhibited. The putative candidates, thus identified, have the potential for breeding the cold-tolerant horticultural plants. This study increases our understanding of functions of the NAC gene family in cold tolerance, thereby potentially intensifying the molecular breeding programs of woody plants.
DOI:10.1016/j.gpb.2015.08.002URLPMID:26542840 [本文引用: 1]

Single-molecule, real-time sequencing developed by Pacific BioSciences offers longer read lengths than the second-generation sequencing (SGS) technologies, making it well-suited for unsolved problems in genome, transcriptome, and epigenetics research. The highly-contiguous de novo assemblies using PacBio sequencing can close gaps in current reference assemblies and characterize structural variation (SV) in personal genomes. With longer reads, we can sequence through extended repetitive regions and detect mutations, many of which are associated with diseases. Moreover, PacBio transcriptome sequencing is advantageous for the identification of gene isoforms and facilitates reliable discoveries of novel genes and novel isoforms of annotated genes, due to its ability to sequence full-length transcripts or fragments with significant lengths. Additionally, PacBio's sequencing technique provides information that is useful for the direct detection of base modifications, such as methylation. In addition to using PacBio sequencing alone, many hybrid sequencing strategies have been developed to make use of more accurate short reads in conjunction with PacBio long reads. In general, hybrid sequencing strategies are more affordable and scalable especially for small-size laboratories than using PacBio Sequencing alone. The advent of PacBio sequencing has made available much information that could not be obtained via SGS alone.
DOI:10.1007/s00299-013-1537-8URLPMID:24247851 [本文引用: 1]

Adenosine diphosphate-ribosylation factors (ARFs) are small guanine nucleotide-binding proteins that play an important role in intracellular protein trafficking necessary for undertaking multiple physiological functions in plant growth and developmental processes. However, little is known about the mechanism of ARF functioning at the molecular level, as well as its involvement in abiotic stress tolerance. In this study, we demonstrated the direct involvement of an ARF gene SaARF from a grass halophyte Spartina alterniflora in abiotic stress adaptation for the first time. SaARF, which encodes a protein with predicted molecular mass of 21 kDa, revealed highest identity with ARF of Oryza sativa. The SaARF gene is transcriptionally regulated by salt, drought, cold, and ABA in the leaves and roots of S. alterniflora. Arabidopsis plants overexpressing SaARF showed improved seed germination and survival of seedlings under salinity stress. Similarly, SaARF transgenic Arabidopsis plants were more tolerant to drought stress, compared to wild-type plants, by maintaining chlorophyll synthesis, increasing osmolyte synthesis, and stabilizing membrane integrity. Oxidative damage due to moisture stress in transgenic Arabidopsis was also reduced possibly by activating antioxidant genes, AtSOD1 and AtCAT. Our results suggest that enhanced drought and salinity tolerance conferred by the SaARF gene may be due to its role in mediating multiple abiotic stress tolerance mechanisms.
URLPMID:32044993 [本文引用: 4]

Spartina alterniflora (Spartina) is the only halophyte in the salt marsh. However, the molecular basis of its high salt tolerance remains elusive. In this study, we used PacBio full-length single molecule long-read sequencing and RNA-seq to elucidate the transcriptome dynamics of high salt tolerance in Spartina by salt-gradient experiments. High quality unigenes, transcription factors, non-coding RNA and Spartina specific transcripts were identified. Co-expression network analysis found that protein kinases-encoding genes (SaOST1, SaCIPK10 and SaLRRs) are hub genes in the salt tolerance regulatory network. High salt stress induced expression of transcription factors but repressed expression of long non-coding RNAs. The Spartina transcriptome is closer to rice than Arabidopsis, and a higher proportion of transporter and transcription factor-encoding transcripts have been found in Spartina. Transcriptome analysis showed that high salt stress induced the expression of carbohydrate metabolism, especially cell wall biosynthesis-related genes in Spartina, while repressed its expression in rice. Compared with rice, high salt stress highly induced the expression of stress response, protein modification and redox-related gene expression, and greatly inhibited translation in Spartina. High salt stress also induced alternative splicing in Spartina, while differentially expressed alternative splicing events associated with photosynthesis were over-represented in Spartina but not in rice. Finally, we built the SAPacBio website for visualizing full-length transcriptome sequences, transcription factors, ncRNAs, salt-tolerant genes, and alternative splicing events in Spartina. Overall, this study suggests that salt tolerance mechanism in Spartina is different from rice from many aspects and is far more complex than expected.
DOI:10.1038/nmeth.1923URLPMID:22388286 [本文引用: 1]

As the rate of sequencing increases, greater throughput is demanded from read aligners. The full-text minute index is often used to make alignment very fast and memory-efficient, but the approach is ill-suited to finding longer, gapped alignments. Bowtie 2 combines the strengths of the full-text minute index with the flexibility and speed of hardware-accelerated dynamic programming algorithms to achieve a combination of high speed, sensitivity and accuracy.
DOI:10.1186/1471-2105-12-323URLPMID:21816040 [本文引用: 1]

RNA-Seq is revolutionizing the way transcript abundances are measured. A key challenge in transcript quantification from RNA-Seq data is the handling of reads that map to multiple genes or isoforms. This issue is particularly important for quantification with de novo transcriptome assemblies in the absence of sequenced genomes, as it is difficult to determine which transcripts are isoforms of the same gene. A second significant issue is the design of RNA-Seq experiments, in terms of the number of reads, read length, and whether reads come from one or both ends of cDNA fragments.
DOI:10.1199/tab.0166URLPMID:24273463 [本文引用: 1]

Abscisic acid (ABA) is one of the &quot;classical&quot; plant hormones, i.e. discovered at least 50 years ago, that regulates many aspects of plant growth and development. This chapter reviews our current understanding of ABA synthesis, metabolism, transport, and signal transduction, emphasizing knowledge gained from studies of Arabidopsis. A combination of genetic, molecular and biochemical studies has identified nearly all of the enzymes involved in ABA metabolism, almost 200 loci regulating ABA response, and thousands of genes regulated by ABA in various contexts. Some of these regulators are implicated in cross-talk with other developmental, environmental or hormonal signals. Specific details of the ABA signaling mechanisms vary among tissues or developmental stages; these are discussed in the context of ABA effects on seed maturation, germination, seedling growth, vegetative stress responses, stomatal regulation, pathogen response, flowering, and senescence.
DOI:10.3389/fpls.2015.00084URLPMID:25741357 [本文引用: 1]

Advances have been made in the development of drought-tolerant transgenic plants, including cereals. Rice, one of the most important cereals, is considered to be a critical target for improving drought tolerance, as present-day rice cultivation requires large quantities of water and as drought-tolerant rice plants should be able to grow in small amounts of water. Numerous transgenic rice plants showing enhanced drought tolerance have been developed to date. Such genetically engineered plants have generally been developed using genes encoding proteins that control drought regulatory networks. These proteins include transcription factors, protein kinases, receptor-like kinases, enzymes related to osmoprotectant or plant hormone synthesis, and other regulatory or functional proteins. Of the drought-tolerant transgenic rice plants described in this review, approximately one-third show decreased plant height under non-stressed conditions or in response to abscisic acid treatment. In cereal crops, plant height is a very important agronomic trait directly affecting yield, although the improvement of lodging resistance should also be taken into consideration. Understanding the regulatory mechanisms of plant growth reduction under drought stress conditions holds promise for developing transgenic plants that produce high yields under drought stress conditions. Plant growth rates are reduced more rapidly than photosynthetic activity under drought conditions, implying that plants actively reduce growth in response to drought stress. In this review, we summarize studies on molecular regulatory networks involved in response to drought stress. In a separate section, we highlight progress in the development of transgenic drought-tolerant rice plants, with special attention paid to field trial investigations.
DOI:10.1186/s12870-019-1883-yURLPMID:31238869 [本文引用: 1]

NAC (NAM, ATAF and CUC) transcriptional factors constitute a large family with more than 150 members in rice and several members of this family have been demonstrated to play crucial roles in rice abiotic stress response. In the present study, we report the function of a novel stress-responsive NAC gene, ONAC066, in rice drought and oxidative stress tolerance.
DOI:10.1186/s12870-016-0897-yURLPMID:27646344 [本文引用: 2]

The NAC (NAM, ATAF and CUC) transcriptional factors constitute a large family with more than 150 members in rice and some of them have been demonstrated to play crucial roles in plant abiotic stress response. Here, we report the characterization of a rice stress-responsive NAC gene, ONAC095, and the exploration of its function in drought and cold stress tolerance.
DOI:10.1186/s12870-017-1001-yURLPMID:28241800 [本文引用: 1]

The NAC gene family is notable due to its large size, as well as its relevance in crop cultivation - particularly in terms of enhancing stress tolerance of plants. These plant-specific proteins contain NAC domain(s) that are named after Petunia NAM and Arabidopsis ATAF1/2 and CUC2 transcription factors based on the consensus sequence they have. Despite the knowledge available regarding NAC protein function, an extensive study on the possible use of GmNACs in developing soybean cultivars with superior drought tolerance is yet to be done.
DOI:10.1023/a:1016028530943URLPMID:12175016 [本文引用: 1]

The petunia NAM and ArabidopsisATAF1 and CUC2 genes define the conserved NAC domain. In petunia, loss-of-function nam mutants result in embryos that fail to elaborate shoot apical meristems (SAM), and nam seedlings do not develop shoots and leaves. We have isolated a NAC domain gene, AtNAM, from an Arabidopsis developing seed cDNA library. Expression of AtNAM mRNA is restricted primarily to the region of the embryo including the SAM. The AtNAM gene contains three exons and is located on Chromosome 1. In vivo assays in yeast demonstrate that AtNAM encodes a transcription factor and that the NAC domain includes a specific DNA binding domain (DBD). The AtNAM DBD is contained within a 60 amino acid region which potentially folds into a helix-turn-helix motif that specifically binds to the CaMV 35S promoter. The putative transcriptional activation domain is located in the C-terminal region of the protein, a highly divergent region among NAC domain-containing genes. The Arabidopsis genome contains 90 predicted NAC domain genes; we refer to these collectively as the AtNAC superfamily. The first two exons of all members of this superfamily encode the NAC domain. Most AtNAC genes contain three exons with the last exon encoding an activation domain. A subfamily of AtNAC genes contains additional terminal exons coding for protein domains whose functions are unknown.
DOI:10.1186/s12864-016-3017-3URLPMID:27542721 [本文引用: 1]

Soil salinity affects growth and yield of crop plants. Plants respond to salinity by physiological and biochemical adjustments through a coordinated regulation and expression of a cascade of genes. Recently, halophytes have attracted attention of the biologists to understand their salt adaptation mechanisms. Spartina alterniflora (smooth cordgrass) is a Louisiana native monocot halophyte that can withstand salinity up to double the strength of sea water. To dissect the molecular mechanisms underlying its salinity adaptation, leaf and root transcriptome of S. alterniflora was sequenced using 454/GS-FLX.
DOI:10.1111/j.1365-313X.2012.04932.xURL [本文引用: 2]

Reactive oxygen species (ROS) are produced in plant cells primarily as by-products of aerobic energy metabolism. They are also generated during plant adaptation responses to environmental stresses, such as drought and high salinity. Therefore, plants have evolved ROS-detoxifying enzymes and antioxidants to cope with ROS accumulation. However, if stress conditions are prolonged, the level of ROS will surpass the capacity of the detoxifying machinery, causing oxidative damage to cellular constituents. It is known that ROS act in abscisic acid-mediated stress responses to sustain plant survival under adverse growth conditions. However, it is largely unknown how ROS metabolism is linked to stress responses. Here, we show that a drought-responsive NAC transcription factor NTL4 promotes ROS production by binding directly to the promoters of genes encoding ROS biosynthetic enzymes during drought-induced leaf senescence. Leaf senescence was accelerated in 35S:4?C transgenic plants over-expressing an active form of NTL4 under drought conditions. The 35S:4?C transgenic plants were hypersensitive to drought, and ROS accumulated in the leaves. In contrast, ROS levels were reduced in NTL4-deficient ntl4 mutants, which exhibited delayed leaf senescence and enhanced drought resistance. These observations indicate that NTL4 acts as a molecular switch that couples ROS metabolism to drought-induced leaf senescence in Arabidopsis.
DOI:10.3389/fpls.2018.00341URLPMID:29599795 [本文引用: 1]

Transfer cells (TCs) play important roles in facilitating enhanced rates of nutrient transport at key apoplasmic/symplasmic junctions along the nutrient acquisition and transport pathways in plants. TCs achieve this capacity by developing elaborate wall ingrowth networks which serve to increase plasma membrane surface area thus increasing the cell's surface area-to-volume ratio to achieve increased flux of nutrients across the plasma membrane. Phloem parenchyma (PP) cells of Arabidopsis leaf veins trans-differentiate to become PP TCs which likely function in a two-step phloem loading mechanism by facilitating unloading of photoassimilates into the apoplasm for subsequent energy-dependent uptake into the sieve element/companion cell (SE/CC) complex. We are using PP TCs in Arabidopsis as a genetic model to identify transcription factors involved in coordinating deposition of the wall ingrowth network. Confocal imaging of pseudo-Schiff propidium iodide-stained tissue revealed different profiles of temporal development of wall ingrowth deposition across maturing cotyledons and juvenile leaves, and a basipetal gradient of deposition across mature adult leaves. RNA-Seq analysis was undertaken to identify differentially expressed genes common to these three different profiles of wall ingrowth deposition. This analysis identified 68 transcription factors up-regulated two-fold or more in at least two of the three experimental comparisons, with six of these transcription factors belonging to Clade III of the NAC-domain family. Phenotypic analysis of these NAC genes using insertional mutants revealed significant reductions in levels of wall ingrowth deposition, particularly in a double mutant of NAC056 and NAC018, as well as compromised sucrose-dependent root growth, indicating impaired capacity for phloem loading. Collectively, these results support the proposition that Clade III members of the NAC-domain family in Arabidopsis play important roles in regulating wall ingrowth deposition in PP TCs.
DOI:10.1186/1471-2229-11-173URLPMID:22133261 [本文引用: 1]

NAC domain transcription factors initiate secondary cell wall biosynthesis in Arabidopsis fibres and vessels by activating numerous transcriptional regulators and biosynthetic genes. NAC family member SND2 is an indirect target of a principal regulator of fibre secondary cell wall formation, SND1. A previous study showed that overexpression of SND2 produced a fibre cell-specific increase in secondary cell wall thickness in Arabidopsis stems, and that the protein was able to transactivate the cellulose synthase8 (CesA8) promoter. However, the full repertoire of genes regulated by SND2 is unknown, and the effect of its overexpression on cell wall chemistry remains unexplored.
[本文引用: 1]
DOI:10.1186/s12870-014-0291-6URLPMID:25404140 [本文引用: 1]

Salt stress is a major challenge for growth and development of plants. The mangrove tree Avicennia officinalis has evolved salt tolerance mechanisms such as salt secretion through specialized glands on its leaves. Although a number of structural studies on salt glands have been done, the molecular mechanism of salt secretion is not clearly understood. Also, studies to identify salt gland-specific genes in mangroves have been scarce.
DOI:10.3389/fpls.2017.01564URLPMID:29033955 [本文引用: 1]

The sessile lifestyle of plants requires them to cope with stresses in situ. Plants overcome abiotic stresses by altering structure/morphology, and in some extreme conditions, by compressing the life cycle to survive the stresses in the form of seeds. Genetic and molecular studies have uncovered complex regulatory processes that coordinate stress adaptation and tolerance in plants, which are integrated at various levels. Investigating natural variation in stress responses has provided important insights into the evolutionary processes that shape the integrated regulation of adaptation and tolerance. This review primarily focuses on the current understanding of how transcriptional, post-transcriptional, post-translational, and epigenetic processes along with genetic variation orchestrate stress responses in plants. We also discuss the current and future development of computational tools to identify biologically meaningful factors from high dimensional, genome-scale data and construct the signaling networks consisting of these components.
DOI:10.1111/tpj.13067URLPMID:26518251 [本文引用: 1]

Leaf senescence is the terminal phenotype of plant leaf development, and ethylene is a major plant hormone inducing leaf senescence. Recent studies have shown that abscisic acid (ABA) also induces leaf senescence. However, the detailed mechanisms of ABA-induced leaf senescence remain unclear. We focused on the A subfamily of stress-responsive NAC (SNAC-A) transcription factors, the expression of which is induced by abiotic stresses, particularly ABA. Gene expression analysis revealed that seven SNAC-A genes including ANAC055, ANAC019, ANAC072/RD26, ANAC002/ATAF1, ANAC081/ATAF2, ANAC102 and ANAC032 were induced by long-term treatment with ABA and/or during age-dependent senescence. The SNAC-A septuple mutant clearly showed retardation of ABA-inducible leaf senescence. Microarray analysis indicated that SNAC-As induce ABA- and senescence-inducible genes. In addition, comparison of the expression profiles of the downstream genes of SNAC-As and ABA-responsive element (ABRE)-binding protein (AREB)/ABRE-binding factor (ABF) (AREB/ABFs) indicates that SNAC-As induce a different set of ABA-inducible genes from those mediated by AREB/ABFs. These results suggest that SNAC-As play crucial roles in ABA-induced leaf senescence signaling. We also discuss the function of SNAC-As in the transcriptional change of leaf senescence as well as in ABA response under abiotic stress conditions.
DOI:10.1007/s10265-016-0833-0URLPMID:27216423 [本文引用: 1]

NAC (NAM, ATAF1/2, CUC2) transcription factors are plant-specific and have diverse functions in many plant developmental processes and responses to stress. In our previous study, we found that the expression of ATAF1, an Arabidopsis NAC gene, was obviously induced by high-salinity and abscisic acid (ABA). The overexpression of ATAF1 in Arabidopsis increased plant sensitivity to ABA and salt. To investigate whether ATAF1 affects the sensitivity of monocotyledon plant to salt and ABA, ATAF1 transgenic rice were generated. Transgenic rice exhibited significantly improved salt tolerance and insensitivity to ABA. The results of real-time PCR showed that ATAF1 overexpression in rice elevated the transcription of OsLEA3, OsSalT1 and OsPM1, which are stress-associated genes. Our results indicate that ATAF1 plays an important role in response to salt stress and may be utilized to improve the salt tolerance of rice.
