 ,1,2
,1,2Three-dimensional structure and function of chromatin regulated by “liquid-liquid phase separation” of biological macromolecules.
Xiaomeng Gao1,2, Zhihua Zhang ,1,2
,1,2通讯作者: 张治华,博士,研究员,研究方向:三维基因组结构研究。E-mail:zhangzhihua@big.ac.cn
编委: 史岸冰
收稿日期:2019-09-4修回日期:2019-11-4网络出版日期:2020-01-20
| 基金资助: |
Editorial board:
Received:2019-09-4Revised:2019-11-4Online:2020-01-20
| Fund supported: |
作者简介 About authors
高晓萌,硕士研究生,专业方向:三维基因组结构研究。E-mail:gaoxiaomeng2018m@big.ac.cn。

摘要
生物大分子的相分离聚集(简称相分离)是驱动细胞内无膜细胞器形成的主要机制,参与众多生物学过程并和多种人类疾病密切相关,如神经退行性疾病等。近年来,研究人员围绕相分离现象的分子机制和生物学功能,发现了相分离与信号传导、染色质结构、基因表达、转录调控等一系列生物学过程存在紧密关联,为理解细胞命运决定和疾病发生提供了新的视角,为疾病治疗和新药研发开辟了新的可能途径。本文在回顾了相分离研究的发展过程、相分离现象在生物学中的应用,以及相分离与疾病的关系的基础上,重点分析了近年来相分离与染色质结构关联方面的研究突破,包括相分离如何感知并重塑染色质结构、超级增强子如何通过相分离调节基因表达、共转录激活因子如何通过相分离参与基因表达调控等,以期为进一步理解相分离与染色质空间结构的关系提供参考。
关键词:
Abstract
Phase separation drives biomacromolecule condensation (phase separation) and is the main mechanism for the formation of membrane-less organelles in cells. Phase separation is involved in many biological processes and is closely associated to various human diseases, e.g., neurodegenerative diseases. Focusing on the molecular mechanism and functions, researchers have recently revealed close associations s between phase separation and various biological functions, such as signal transduction, chromosome structure, gene expression, and transcriptional regulation. These findings have provided new perspectives in understanding cell fate decisions and disease processes, thereby offering novel approaches for future drug discovery and development of disease treatments in medicine. In this review, we summarized the current progress in the field of phase separation research. We focused on its application on understanding how phase separation remodels the chromatin structure, assembles co-activators and super-enhancers in regulation of gene expression, in order to further understand the relationship between phase separation and chromatin spatial structures. Finally, we also outline the challenges in reference to future research directions in the field.
Keywords:
PDF (468KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
高晓萌, 张治华. 生物大分子“液-液相分离”调控染色质三维空间结构和功能. 遗传[J], 2020, 42(1): 45-56 doi:10.16288/j.yczz.19-266
Xiaomeng Gao.
细胞发达的内膜系统将细胞内部划分成许多功能区块,形成具有各种特定功能的细胞器。除了这类具膜细胞器之外,细胞内还存在许多无膜细胞器,如中心体(centriole)、应力颗粒(stress granules)、核小斑点(nuclear speckles)等[1]。研究表明,生物大分子的相分离(phase separation)凝聚是驱动无膜细胞器形成的主要机制[2]。物质的相分离是在自然界中普遍存在的物理现象,通常指的是在均匀的混合液/气态物质中自发形成的不同相。在体外,相分离是可逆的,受到诸如温度、离子强度、翻译后修饰等物理或化学因素的影响,从而形成或者解离液滴[3]。在体内相分离通常是一个动态的过程,并且通常指的是“液-液相分离”(liquid-liquid phase separation),在不引起歧义的情况下,以下简称相分离。
2009年,德国马普研究所Tony Hyman教授与其博士后研究员Cliff Brangwynne发现线虫(Caenorhabditis elegans)胚胎中的RNA和包含蛋白质的结合体P颗粒(P granules)实际上是由蛋白质通过相分离形成的凝聚小球[4]。体外实验显示,在过饱和溶液中会自发地分成一种浓相和一种稀相并且稳定地共存。2011年,该实验室对核仁进行了类似的研究,证实了相分离是核仁发生的基础[5]。此后,研究人员开始对细胞中的相分离现象进行广泛而深入的研究。细胞内的相分离是由弱多价相互作用引起的,通常由内在无序蛋白质、内在无序区域或者低序列复杂结构域介导[6,7,8]。基因组的活性与相分离现象有关,相分离发生部位通常与染色质密度较低区域相关联。越来越多的研究表明相分离现象对细胞核的染色质空间结构的组织调控非常重要。而染色质的组织结构是细胞核内生命活动,包括基因表达、DNA复制、损伤修复、基因组稳定性以及转座子沉默等的基本物理平台[9],因此,探究相分离现象与染色质的结构和功能的关系具有重要的意义。本文从相分离的形成、生物学意义以及相分离与染色质空间结构和转录调控的关系入手,结合最前沿的研究进展,介绍了相分离研究在当前生物学研究中的重大突破,为全面了解相分离研究领域提供参考。
1 相分离现象及实验检测
1.1 相分离的形成
2009年,Brangwynne 等[4]在研究秀丽隐杆线虫中无膜P颗粒的形成过程时,发现其具备以下特征:P颗粒游离状态下为球形,附着在细胞核表面时为非球形,颗粒间有明显的边界;在压力状态下可以流动,并可由小液滴融合形成大的液滴;P颗粒具有很强的黏性和表面张力;光脱色荧光恢复实验表明P颗粒内部分子可与外部溶液中的分子快速交换。结合其他生物分子凝聚体与P颗粒的相似特征,推测这是一种相分离现象。生物大分子可以通过多价相互作用形成液-液相分离(图1)[8]。多价相互作用是指一个多价配体与一个或多个功能亲和(表观亲和)增强的受体结合并协同作用的过程。细胞内生物大分子的多价相互作用形成方式主要包括线性重复功能性的结构域、寡聚化蛋白和多个位点蛋白的表观修饰(磷酸化、甲基化、乙酰化和泛素化等)[10]。内在无序蛋白(intrinsically disordered proteins)或内在无序区域(intrinsically disordered regions)是一种重要的多价相互作用来源。内在无序蛋白和内在无序区域与相分离的形成密切相关,其磷酸化会导致凝聚物的降解。很多内在无序蛋白和内在无序区域都含有低序列复杂结构域(low-complexity domains)。低序列复杂结构域通常仅富集Gly、Ser、Tyr和Gln等残基类型,即具有强氨基酸偏好性和自我维持聚集潜力[11,12]。这种序列特性赋予其特殊的生物化学与生物物理性质。例如,朊病毒样结构域是一种富含不带电极性氨基酸的低序列复杂结构域,具有自组装和形成凝聚物的趋势[13]。朊病毒样结构域富含参与生理相分离和病理蛋白聚集的蛋白。与其他蛋白质相比,内在无序蛋白和内在无序区域缺乏明确的蛋白质折叠构象,具有高度的灵活性,从而违背了经典的结构-功能范式,这种结构可塑性使它们能够动态地采用不同的构象,并经历混杂的多价形式和异型相互作用[8]。在体外实验中,低序列复杂结构域当与它们的多价同源配体混合时,可以发生寡聚和相分离,如低序列复杂结构域中的RNA结合域会与重复RNAs结合发生寡聚作用[14,15]。内在无序区域还可以通过静电、极性和疏水作用以及产生淀粉样纤维和其他潜在玻璃结构相互作用等模式来实现自缔结[7]。
图1
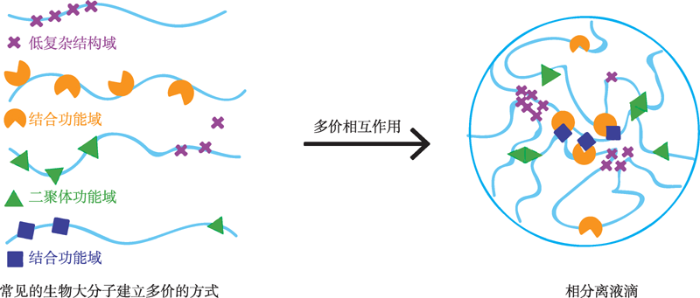 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1相分离现象形成过程
Fig. 1The formation of phase separation
1.2 相分离的实验检测技术
体外的相分离现象可以使用普通的光学显微镜观察。发生相分离的体系具有溶液从澄清变浑浊的特点,在镜检中可见油滴状颗粒。目前被广为接受的相分离检测标准是:形成球状液滴结构的生物分子凝聚体;液滴具有流动性,可以发生融合现象;使用光脱色荧光恢复技术(fluorescence recovery after photobleaching),可见荧光漂白恢复。建立可操控的体内相分离体系是相关研究的基础。CasDrop系统(图2)是由普林斯顿大学Brangwynne实验室开发的基于Cas9基因编辑技术的细胞内相分离系统[16]。该系统由3部分组成:(1) SunTag标记的dCas9蛋白, 用于将体系锚定到特定的基因位点;(2) scFv标记的ST同源物、sfGFP和光发生二聚蛋白iLID,用于构建光敏的多聚体。前两种结构可以自组装成一个多聚体蛋白复合物(dCas9-ST-GFP-iLID),从而实现位点的可视化,同时为招募各种蛋白复合物(包括包含内在无序区域的蛋白)提供了光诱导的结合支架; (3) sspB标记的目的蛋白内在无序区域区域,用于检测相变行为。当sgRNA表达时,scFv-GFP-iLID蛋白被SunTag募集到SunTag-dCas9结合的基因位点处,形成dCas9-ST-GFP-iLID蛋白的多聚体;当sgRNA不表达时,dCas9-ST-GFP-iLID蛋白在胞内则成弥散状。在蓝光刺激下,sspB结合到iLID上,目的蛋白内在无序区域相分离使得液滴快速凝结,频繁合成。该体系可以定量、定位地研究多种蛋白的相分离现象。图2
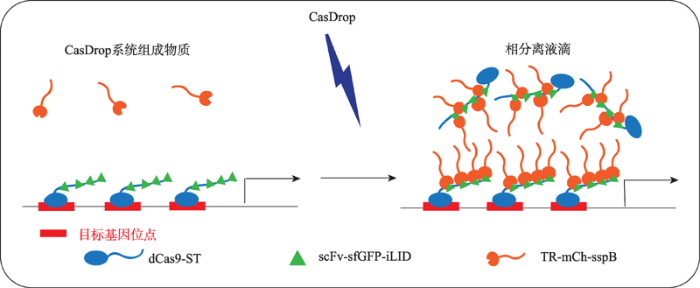 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2CasDrop相分离检测系统 根据文献[16]修改绘制。
Fig. 2The phase separation detection system—CasDrop
2 生命体内的相分离及生物意义
2.1 相分离调节信号转导
由于相分离形成蛋白质分子的聚集,使得它在信号传导过程中起到重要作用。例如,Ras蛋白是在真核细胞中对细胞生长和信号传导发挥着重要作用的因子,T细胞使用Ras作为发起保护性反应的入侵者报警通道的分子开关。Ras蛋白的激活需要通过相分离形成的凝聚液滴维持充分的反应时间[17]。T细胞与Ras的相互作用依赖于LAT、GRB2和SOS三种中间蛋白而起作用[18]。其中SOS是一种重要的Ras激活剂,它通过GRB2蛋白招募并激活膜上受体蛋白,随后激活Ras蛋白[19]。膜信号受体的激活促进LAT磷酸化,生成的多价磷酸化酪氨酸位点作为GRB2的结合位点招募GRB2[20]。LAT-GRB2-SOS复合体通过相分离在膜上形成二维生物分子凝聚液滴或凝胶[21,22],从而增加了SOS在细胞膜上停留时间。研究人员通过设计体外重组试验测量单SOS分子的膜招募动力学和Ras激活动力学,使用显微镜来观察膜上的T细胞受体请求单个SOS分子活化时,发现SOS不会立即响应,而是等待10~30 s才进入活性状态并发挥作用,因此由于相分离现象导致的SOS膜结合时间增加可以促进Ras蛋白完全激活[17]。2.2 相分离帮助细胞适应环境
相分离对生理环境中的微小变化(pH改变等)非常敏感,因此相分离为细胞感知环境变化提供了一种高效的方式[3]。例如,朊蛋白样结构域可以感知环境条件的变化,通过改变相分离的状态,保护细胞内的生物活动[6,23]。出芽酵母的翻译终止因子Sup35是一种典型的朊病毒蛋白。细胞内一组带负电荷的氨基酸可感知环境的pH变化,通过pH诱导,Sup35经过液相分离形成一种保护性蛋白凝胶。相分离受到相邻两个应激传感器(即朊蛋白样结构域)的调节。这种朊蛋白样结构域之间的协同作用使得翻译终止因子Sup35在应激期间快速地形成保护性凝聚物。缺乏朊病毒样结构域的细胞在从应激状态恢复时表现出翻译活性受损和生长缺陷。朊蛋白样结构域改变了Sup35的鸟苷三磷酸酶结构域的不可逆聚集状态,从而确保这种翻译终止因子在恶劣的环境条件下保持功能[24]。总之,Sup35的朊蛋白样结构域是一种受到高度调节的分子装置,具有感知和响应细胞内物理化学变化的能力,允许细胞对特定的环境条件作出反应。2.3 相分离与疾病
相分离和多种疾病相关。细胞内相分离倾向于凝聚蛋白质,该过程伴随着潜在的危险。蛋白质浓度过高往往会引起聚合反应或发生堵塞,形成固体凝胶甚至晶体,不再为细胞内化学反应提供必要的环境。由于在神经退行性疾病中经常发生病变的几个关键蛋白是无膜细胞器的组成部分,因此,这些装配体在形成、维持或清除方面的失调就可能导致疾病的发生[11,25]。例如,许多神经退行性疾病的特征都包含大量致病蛋白的聚集,比如阿尔茨海默症(Alzheimer’s disease, AD)中的淀粉样蛋白和tau蛋白的聚集[26],帕金森综合征(Parkinson’s disease, PD)中的突触核蛋白斑块[27],或者肌萎缩侧索硬化症(amyotrophic lateral sclerosis, ALS)中的斑块[28]。基因的相关突变可以促进这种液体到固体的相分离。例如,FUS的朊病毒样结构域内的G156E突变最近被证明可以加速从液滴到蛋白质捕获纤维水凝胶和神经毒性蛋白聚集物的转变[29,30],而hnRNPA2B和hnRNPA1的类朊病毒样结构域发生致病突变,加剧了蛋白内在聚集趋势,形成自种原纤维,从而促进细胞聚集体的形成[31]。当这些突变存在于内在无序区的β拉链结构域时,其更加倾向于折叠为稳定的类淀粉蛋白结构[32]。致病突变还可以通过产生异常的蛋白质或RNA来影响相分离。例如,由FUS (TLS)、EWS (EWSR1)和TAF15组成的FET蛋白家族,参与RNA存储位点的相变[33],并通过RNA刺激的过程组装成高阶结构[34,35],这些功能在人类受损从而造成疾病。RNA重复序列扩增也会导致功能异常,重复的RNA位点可以捕获RNA结合蛋白导致其功能丧失。研究人员已经发现一些RNA重复序列可以进行翻译过程[36],如肌萎缩侧索硬化症致病重复序列GGGGCC可以产生不同的二肽重复,其中的甘氨酸-精氨酸、脯氨酸-精氨酸二肽重复序列定位于不同的无膜细胞器[37]。线粒体功能紊乱也会促进衰老和神经退化性疾病的发生,它可能导致ATP水平下降[38]。有证据表明,细胞ATP可以作为化学助溶物直接阻止相分离和聚合。因此在老化和疾病中,线粒体呼吸功能的缺陷可能促进蛋白质聚集。近年来,很多研究人员开始探讨癌症与相分离之间的关系。肿瘤抑制因子SPOP突变可导致前列腺、乳腺和其他实体肿瘤[39]。SPOP是cullin3环泛素连接酶的底物的配体,定位于核斑点处[40]。癌症相关的SPOP突变会干扰连接酶底物募集,底物在体外可触发SPOP的相分离和细胞内无膜细胞器的共定位。酶活性与细胞共定位和体外中尺度的SPOP-底物组装形成有关。疾病相关的SPOP突变可导致原癌蛋白的积累,从而干扰无膜细胞器的相分离和共定位,这表明该E3连接酶的底物相分离是泛素依赖蛋白稳定的再泛素化的基础[41]。在某些癌症中[42],易位会破坏FET蛋白的低复杂度区域,从而改变融合蛋白的核酸结合特性。
以上简单介绍了相分离与疾病之间的重要关系,但是相分离的很多具体的致病机理还不是非常明确。研究人员发现染色质三维结构与相分离有关,研究相分离与基因组结构之间的关系有助于人们进一步揭示相分离的致病机理。
2.4 相分离的其他生物学功能
相分离因为将参与相变的分子进行了局部浓缩,提高了细胞内关键酶或蛋白复合物的局部浓度,所以在细胞内,相分离还具有促进信号传导[5]等生化反应过程、介导蛋白质定位和隔离分子防止反应失活等诸多作用[43]。随着研究手段的进步,更多的相分离生物学功能正在逐渐被解释出来。3 相分离与染色质结构
3.1 染色质结构
染色质是细胞核内生命活动的物质环境,染色质的空间结构和细胞核内的诸多生命过程息息相关。在高通量基因组测序技术高速发展的推动下,以染色质免疫共沉淀测序(chromatin immunoprecipitation sequencing, ChIP-seq)[44,45]、高通量染色质构象捕获测序(high input chromosome conformation capture, Hi-C)[46,47]和转座酶可接近性核染色质序列测序(assay for transposase accessible chromatin, ATAC-seq)[48,49]等技术为代表的一系列技术推动了染色质三维结构的研究迅速发展。近年来,研究人员发现,细胞核内不仅有大量相分离现象的存在,并且相分离现象与染色质的空间结构具有重要的联系。2019年,Gibson等[50]甚至发现,在体外纯化的串联核小体可以通过其核心蛋白内的无序区域间的多价相互作用形成相分离凝聚,并且在组蛋白H1的帮助下,这一相分离凝聚可以更加高效的发生。由于核小体的串联是染色质的最基本结构单元,这一发现为细胞核中染色质的整体结构形成提供了一个“基态”。也就是说,更精细的染色质结构形成很可能是在这一相分离凝聚体之上发生的[50]。这一发现同时对常见的染色质多聚体物理模型提出了挑战[51]。3.2 相分离感知并重塑染色质结构
相分离感知并重塑染色质结构的研究工作目前主要是Brangwynne实验室利用其所建立的CasDrop系统通过对FUS、BRD4和TAF15蛋白的研究做出的。他们的研究发现,内在无序区域组装而成的凝聚体具有广泛的序列特征,它们不仅在低密度染色质区域优先成凝聚核,而且在成核和生长过程中会排斥该区域的染色质[52]。首先,3种蛋白均可在CasDrop系统诱导下发生“液-液相分离”现象,其中BRD4中的一个长内在无序区域BRD4ΔN形成液滴的能力最强。其次,以H2B-miRFP670荧光强度作为测定染色质密度的指标,测定相分离液滴在液滴凝聚前后以及整个细胞核中染色质的密度分布。通过比较发现,在液滴区域,H2B强度的分布明显低于整个核中H2B的分布,并且H2B-miRFP670在相变处的荧光强度随时间延长而逐步下降。这暗示着,随着液滴的增长,液滴所在区域的染色质会被推出该区域。与此相一致的是,细胞核内的无膜结构,像核仁、Cajal体和PML体等也倾向于在染色质低密度区域形成[53]。为进一步研究为什么倾向于排斥染色质的液滴会在低染色质密度区域优先生成,Shin等[16]建立了凝聚物与可变形染色质网络的力学相互作用的数学模型,液滴成核时的自由能损耗?F可通过下式描绘:
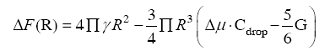
其中R、γ、?μ、Cdrop和G分别代表液滴半径、液滴表面张力、过饱和溶液中分子与液滴相的化学势差、液滴内分子的饱和浓度、形变区域的杨氏(Young’s)弹性模量。
该模型认为较小液滴在所有染色质密度区域都可以形成,当液滴增大到光学显微镜可分辨大小时则倾向于在染色质密度低的区域形成。在染色质密度足够的区域,机械变形能阻止液滴的生长超过临界尺寸。利用CasDrop体系观察了BRD4在微卫星序列区、端粒和致密异染色质区的相分离现象,发现BRD4在染色质低密度区域能够被诱导形成液滴,而在染色质高密度区,BRD4的液滴则仅在其周围形成液滴。当CasDrop体系将BRD4的液滴锚定在异染色质区域内HP1α密度相对较低的区域时,随着液滴的增大,HP1α被逐步剔除出该区域;而部分增大的BRD4液滴则可以扰乱异染色质的结构,使得该区域形成染色质密度下降。这些发现表明在相变发生的过程中,其周围的染色质提供了一定的机械阻力。
相分离形成的液滴对染色质具有排斥作用,导致染色质结构重塑;同时,染色质结构对相分离现象也有一定的影响,相分离液滴倾向于在染色质结构疏松、低密度区域形成,周围染色质对液滴的形成具有一定的机械阻力。相分离的这些性质被总结为一个染色质过滤模型,即结合在特定基因位点的蛋白质的相分离能够将距离较远的基因拉近,且排斥不含结合位点的基因区域。这保证了超级转录因子在染色质结构疏松的区域的聚积、对染色质的折叠、和促进活跃基因的表达。
3.3 相分离驱动异染色质区域的形成
组成型异染色质是真核生物基因组的重要组成部分,在细胞核结构、DNA修复、基因组稳定性、转座子沉默以及基因表达中起着重要作用[9]。一个异染色质形成模型认为HP1α蛋白发生液-液相分离,形成容纳染色质而排斥RNA聚合酶等分子的液相稳定区室,是异染色质形成的第一步,并通过对果蝇、小鼠的体内体外实验进行验证。果蝇HP1α蛋白尾部富集内在无序域和低复杂结构域。为确定HP1α蛋白是否可以在体外发生相分离,研究人员纯化了果蝇HP1α蛋白,发现在22℃、高蛋白浓度和低盐状态下,HP1α的水溶液自动分解形成液滴并在37℃时溶解。为探究相分离现象与体内异染色质现象的相关性,研究人员分析了早期果蝇胚胎异染色质形成的第一阶段,发现异染色质在受精后第11~13个核周期开始形成,但直到第14个周期才成熟为稳定的结构域。在每一个周期中GFP-HP1α都表现出与相分离的液相区相关联的成核、生长和聚变相关的动态变化。在第13~14个周期早期胚胎中,HP1α液滴在融合后聚集并保持圆形。随着第14个周期的进展,其圆度较低。通过光漂白后荧光恢复实验测量10~14个周期和原肠胚形成后的HP1α蛋白活动和静止的组分比率,表明液滴圆度的丧失伴随着HP1α静止组分的显著增加,由此推测这可能是HP1α与染色质聚合物的结合程度增多的结果[52]。大分子间弱疏水相互作用可以促进相分离。例如,在果蝇S2和小鼠NIH3T3细胞中加入1,6-己二醇(特异性破坏疏水相互作用力),培养2 min后,HP1在异染色质区明显但不完全分散;去除1,6-己二醇区域内的HP1富集部分恢复。与野生型对照组相比,含有破坏二聚作用(1191E)或非组蛋白结合体(W200A)的点突变的GFP-HP1α蛋白的迁移率显著增加。这一体外实验说明,成熟异染色质结构域的完整性依赖于弱疏水相互作用,而二聚作用和与非组蛋白结合体的相互作用有助于HP1的固定,由此推断成熟异染色质形成与相分离有关并且包括固定(静态)和移动(液态) HP1α区域。为进一步验证通过相分离产生异染色质结构域的假设,研究人员详细分析了这些结构域内外的HP1α动态变化。数据表明HP1α和其他异染色质蛋白在相界面上协调运动。在果蝇和哺乳动物细胞中,HP1α的动态变化与包含异染色质蛋白的相分离成分动态变化一致。通过果蝇S2细胞HP1α去除实验发现GFP-HP1α失去协调运动并在细胞内呈分散状态。
基于一系列体内和体外数据的组合,研究人员提出一个由相分离驱动异染色质结构域形成的模型,从整体上将异染色质结构域的涌现特性与局部分子相互作用联系起来。该模型认为异染色质的形成始于相分离的成核。起初,所有的HP1α都是可自由移动的,通过多个HP1分子与其他异染色质组分之间形成多价、弱疏水相互作用,开始形成原始液滴。随着时间的推移,这些液滴发生了生长。在此期间,球型液滴通过招募更多的HP1α和变粗而变得更大,一些HP1α移动性丧失。HP1α液滴圆度的丧失随着固定的、染色质结合的HP1α的增加而降低,从而形成由液体和稳定的腔室组成的成熟异染色质区域[52]。
3.4 形成染色质亚区室(subcompartement)的相分离机制
真核细胞中染色质可被划分为生物学功能不同的亚区室,关于亚区室形成方式有两种不同的基于相分离的解释。一个是交联不同染色质区段的染色质相关蛋白可以诱导聚合物-聚合物相分离,它产生一个有序的折叠小球[54];另一个是染色质相关蛋白与蛋白通过多价相互作用促进液-液相分离的发生[3]。聚合物-聚合物相分离机制基于可溶性桥连因子的结合,这些桥因子组装成类似于一个塌陷的聚合物球的动态交联的染色质支架。在分子水平上,桥接和染色质环的形成可以通过核小体间相互作用介导。由于染色质相关蛋白和转录因子含有两个或多个染色质、DNA识别结构域,因此具有桥联不同核小体的潜力[55]。桥接可以驱动相应染色质区域与不同类型的桥接因子相互作用的相分离。
液-液相分离驱动相分离主要通过多价相互作用形成液滴,并围绕染色质上的各个结合位点组装成亚区室。聚合物-聚合物相分离中的桥接作用可能间接地产生于多价结合染色质之间的相互作用。因此,尽管在不同的分子基础上,聚合物-聚合物相分离和液-液相分离原则上都可以促进紧凑染色质组分的形成,通过聚合物-聚合物相分离形成的亚区室严格依赖于染色质支架,在没有染色质的情况下会进行拆卸,而通过液-液相分离形成的亚区可以独立于染色质支架而存在。
4 相分离和基因表达调控
4.1 超级增强子通过相分离调节基因表达
超级增强子(super enhancer)由一组连续排列的增强子组成,具有比典型增强子更高水平的调控基因转录的能力[56],然而其调控基因表达的具体分子机制还不是很清楚。2017年,Richard Young实验室提出一个解释超级增强子通过相分离参与基因调控的模型[57]。在该模型中,增强子的蛋白质和核酸成分被表示为链状分子,每个分子都含有一组残基,这些残基可能与其他链发生相互作用。这些残基代表那些链状分子中可以进行可逆化学修饰的位点,其上的化学修饰与它们形成链之间非共价交联的能力相关。例如,RNA Pol Ⅱ的C端结构域(C-terminal domain, CTD)七肽重复序列,以及转录因子、共作用因子都有可能是这样的长链分子。这些分子上的磷酸化或去磷酸化修饰,或者组蛋白和其他蛋白质上的乙酰化、甲基化或其他类型的化学修饰,都会影响大分子之间的相互作用,多个链分子相互作用,交联形成网状聚集体。2018年,该实验室通过实验证明了上述的超级增强子基因调控模型[58]。他们发现胚胎干细胞中的转录共激活因子MED1和BRD4可以在超级增强子处形成相分离的凝聚体,并将转录物质富集在超级增强子周围,形成转录过程中的区室化[58]。转录共激活因子MED1和BRD4是超级增强子的重要组成蛋白,超级增强子与MED1和BRD4的凝聚物相关联。在稳定表达mEGFP-BRD4/mEGFP-MED1的mESC细胞系中,MED1和BRD4在细胞核中呈现亮斑状且细胞核中的超级增强子与MED1和BRD4共定位,说明MED1与BRD4与超级增强子的功能有关。通过对BRD4与MED1形成的亮斑进行荧光漂白恢复实验,发现BRD4和MED1可以迅速恢复荧光性,当细胞内ATP被消耗时,BRD4和MED1的荧光恢复效果显著降低;当用3% 1,6-己二醇处理细胞后,细胞内的BRD4和MED1亮斑数目明显减少。这些现象说明BRD4和MED1亮斑具有相分离凝聚物的性质。通过ChIP-seq实验发现,1,6-己二醇处理后的细胞中,BRD4和MED1在超级增强子处的结合能力显著下降,同时由超级增强子调控的基因表达水平也显著降低。说明转录共激活因子BRD4与MED1在细胞中通过相分离形式在超级增强子处成簇分布,并且起到调控基因表达的作用。BRD4和MED1中均检测到具有长的内在无序区域。通对BRD4与MED1的内在无序区域进行纯化,发现它们在体外可自发组装形成液滴。这些液滴表现出弱蛋白-蛋白相互作用网络凝聚物的特征。液滴的大小与蛋白浓度、缓冲体系的盐浓度及疏水性等因素有关。在体内,利用蓝光报告系统OptoDroplet证明内在无序区域可以发生相分离,在蓝光刺激的情况下可以观察到细胞内亮斑的形成,并且亮斑之间有融合的现象。这些现象说明内在无序区域参与了与超级增强子共定位的BRD4和MED1的相分离现象,且BRD4和MED1相分离凝聚物具有普通相分离液滴的性质。这一系列的实验显示,BRD4和MED1形成了相分离液体,参与了超级增强子的基因表达调控。
4.2 转录因子通过相分离参与基因表达调控
转录因子由DNA结合域(DNA-binding domains)和激活域(activation domains)组成[59]。对转录因子的DNA结合域已经有比较深入的研究[60],但对激活域行使功能的机制,到目前为止仍然了解甚少。研究发现激活域可以与介质共激活剂共同作用发生相分离,激活基因表达[61]。OCT4是对胚胎干细胞的多能状态维持至关重要的主要转录因子,也是胚胎干细胞超级增强子中的一个转录因子[56]。OCT4可以与转录中介体复合物(mediator)的亚基MED1相互作用,在胚胎干细胞中的超级增强子上形成相分离凝聚物[62]。新生RNA荧光原位杂交的免疫荧光成像显示,在关键的多能性基因Esrrb、Nanog、Trim28和Mir290的超级增强子处存在不连续的OCT4斑点。ChIP-seq结果显示OCT4和MED1在超级增强子附近共定位。以Nanog基因为研究对象,当OCT4暴露于小分子dTag中时,OCT4发生迅速泛素化并降解,凝聚在超级增强子处的MED1和Nanog的基因表达也随之下降。在胚胎干细胞的分化过程中,Mir290超级增强子附近的OCT4的丢失,MED1和Mir290的基因表达也显著降低。这些结果说明MED1在超级增强子处的凝聚依赖于OCT4,且OCT4和MED1与基因转录水平有关。
转录因子OCT4的激活域通过与MED1-IDR 相互作用激活基因表达。OCT4-GFP自身不形成相分离液滴,而MED1-IDR-GFP可以形成相分离液滴,且液滴大小与溶液浓度相关。当将两种蛋白混合时,OCT4-GFP蛋白可以被招募进入MED1-IDR-GFP液滴中,并显示出液滴的性质。通过分析OCT4中氨基酸频率和电荷偏好性发现OCT4内在无序区域中富含脯氨酸和甘氨酸,并且具有全部的酸性氨基酸。由于转录因子的激活域中富含酸性氨基酸和脯氨酸,作者推测激活域中的脯氨酸或酸性氨基酸可能促进与相分离的MED1-IDR液滴的相互作用。通过设计荧光标记实验发现具有酸性氨基酸基序的多肽容易进入MED1液滴中,且MED1-IDR和转录中介体复合物招募OCT4要依赖于激活域酸性氨基酸提供的静电作用力。OCT4 激活域的酸性氨基酸基序对于其进入MED1的液滴以及基因激活功能都是必需的,它通过与MED1聚集形成液滴发挥其基因激活功能。这个例子展示了转录因子可以和转录中介体复合物发生液-液相分离从而激活基因表达。
4.3 一种新的相分离调控基因转录模型
最近研究人员提出了一种新的基因转录模型解释RNA聚合酶Ⅱ(Pol Ⅱ)在起始和延伸阶段的调控机制[63]。该模型认为基因的转录受控于两个相分离凝聚体。一个是在启动子附近形成的启动子动态凝聚体,由包含转录因子、共激活因子、非磷酸化的Pol Ⅱ和启动因子组成。另一个是在基因体中的瞬时凝聚体,由磷酸化Pol Ⅱ、新生RNA、延伸因子、RNA加工因子以及特异性延伸共激活因子组成。启动子凝析体促进转录起始、RNA合成和Pol Ⅱ磷酸化。Pol Ⅱ的C端非结构区域的磷酸化驱使Pol Ⅱ从启动子凝聚体中析出,进入基因体凝聚体中。当Pol Ⅱ到达基因的末端时,C端非结构区域脱磷使得Pol Ⅱ从基因体凝析液中释放出来,并转移回启动子凝聚物。简单的说,就是Pol Ⅱ通过C端非结构区域的磷酸化调控其在不同功能聚集体之间的穿梭,完成高效并且可调控的转录。该模型假说提出后不久,Richard Young实验室发表了跟上述模型一致的实验结果[64]。在Young的结果中,更明确的指出了剪接复合物在基因体凝聚体中的关键性作用,即Pol Ⅱ的C端非结构区域的磷酸化驱动从转录起始复合物到RNA加工复合物的转换。这也进一步强调了C端非结构区域磷酸化作为调节Pol Ⅱ结合不同复合物偏好性的机制。5 结论与展望
从发现相分离现象与无膜细胞器形成之间的联系之后,研究人员一直致力于相分离的形成和功能机制的研究,在相分离与染色质结构、基因表达、转录调控、信号传导等方面取得了重要突破。相分离现象与神经退行性疾病、癌症等复杂疾病密切关联的研究结果有助于研究人员进一步了解疾病的致病机理,进而促进新的药物开发。但是相分离研究中仍然存在诸多困难与挑战。首先,目前并没有广泛接受的标准来定义发生相分离的染色质结构;其次,简单的光学观测还不足以区分相分离。例如,体外过表达一种蛋白质并看到一个大的球形液滴是常见的,内源性表达的蛋白质也可能存在类似的液滴。在实验方法方面,细胞内相分离物质纯化比较困难,很多体外验证的实验难以在体内进行开展。在未来的研究中,除了对新的实验方法以解决细胞内实验困难有迫切需求之外,新的影像分析识别算法,建立分子层面的组学和宏观的相分离之间的联系,也是一个重要的待解决问题。另外,对于相分离在染色质构象形成,相分离对细胞核内的生命过程的功能性作用是不是具有普遍性等一些基本的问题,在学界仍然存在很多不同的声音。这一方面说明相分离的确是一个重要的,受到广泛关注的生理现象。而另一方面,也说明相分离的研究还需要被更深入全面的检视。在可预见的未来,相分离的研究将在实验技术和计算技术的双重推动下,揭示更深入的生命科学问题。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1186/s12964-015-0125-7URLPMID:26727894 [本文引用: 1]

Inside eukaryotic cells, macromolecules are partitioned into membrane-bounded compartments and, within these, some are further organized into non-membrane-bounded structures termed membrane-less organelles. The latter structures are comprised of heterogeneous mixtures of proteins and nucleic acids and assemble through a phase separation phenomenon similar to polymer condensation. Membrane-less organelles are dynamic structures maintained through multivalent interactions that mediate diverse biological processes, many involved in RNA metabolism. They rapidly exchange components with the cellular milieu and their properties are readily altered in response to environmental cues, often implicating membrane-less organelles in responses to stress signaling. In this review, we discuss: (1) the functional roles of membrane-less organelles, (2) unifying structural and mechanistic principles that underlie their assembly and disassembly, and (3) established and emerging methods used in structural investigations of membrane-less organelles.
.
DOI:10.1016/j.sbi.2016.10.015URLPMID:27838525 [本文引用: 1]

Although the cellular interior is crowded with various biological macromolecules, the distribution of these macromolecules is highly inhomogeneous. Eukaryotic cells contain numerous proteinaceous membrane-less organelles (PMLOs), which are condensed liquid droplets formed as a result of the reversible and highly controlled liquid-liquid phase transitions. The interior of these cellular bodies represents an overcrowded milieu, since their protein concentrations are noticeably higher than those of the crowded cytoplasm and nucleoplasm. PMLOs are different in size, shape, and composition, and almost invariantly contain intrinsically disordered proteins (e.g., eIF4B and TDP43 in stress granules, TTP in P-bodies, RDE-12 in nuage, RNG105 in RNA granules, centrins in centrosomes, NOPP140 in nucleoli, SRSF4 in nuclear speckles, Saf-B in nuclear stress bodies, NOLC1 in Cajal bodies, CBP in PML nuclear bodies, SOX9 in paraspeckles, KSRP in perinucleolar compartment, and hnRNPG and Sam68 in Sam68 nuclear body, to name a few), which indicates that the formation of these phase-separated droplets is crucially dependent on intrinsic disorder. The goal of this review is to show the roles of intrinsic disorder in the magic behind biological liquid-liquid phase transitions that lead to the formation of PMLOs.
.
DOI:10.1038/nrm.2017.7URLPMID:28225081 [本文引用: 3]

Biomolecular condensates are micron-scale compartments in eukaryotic cells that lack surrounding membranes but function to concentrate proteins and nucleic acids. These condensates are involved in diverse processes, including RNA metabolism, ribosome biogenesis, the DNA damage response and signal transduction. Recent studies have shown that liquid-liquid phase separation driven by multivalent macromolecular interactions is an important organizing principle for biomolecular condensates. With this physical framework, it is now possible to explain how the assembly, composition, physical properties and biochemical and cellular functions of these important structures are regulated.
.
DOI:10.1126/science.1172046URLPMID:19460965 [本文引用: 2]

In sexually reproducing organisms, embryos specify germ cells, which ultimately generate sperm and eggs. In Caenorhabditis elegans, the first germ cell is established when RNA and protein-rich P granules localize to the posterior of the one-cell embryo. Localization of P granules and their physical nature remain poorly understood. Here we show that P granules exhibit liquid-like behaviors, including fusion, dripping, and wetting, which we used to estimate their viscosity and surface tension. As with other liquids, P granules rapidly dissolved and condensed. Localization occurred by a biased increase in P granule condensation at the posterior. This process reflects a classic phase transition, in which polarity proteins vary the condensation point across the cell. Such phase transitions may represent a fundamental physicochemical mechanism for structuring the cytoplasm.
.
DOI:10.1073/pnas.1017150108URLPMID:21368180 [本文引用: 2]

For most intracellular structures with larger than molecular dimensions, little is known about the connection between underlying molecular activities and higher order organization such as size and shape. Here, we show that both the size and shape of the amphibian oocyte nucleolus ultimately arise because nucleoli behave as liquid-like droplets of RNA and protein, exhibiting characteristic viscous fluid dynamics even on timescales of < 1 min. We use these dynamics to determine an apparent nucleolar viscosity, and we show that this viscosity is ATP-dependent, suggesting a role for active processes in fluidizing internal contents. Nucleolar surface tension and fluidity cause their restructuring into spherical droplets upon imposed mechanical deformations. Nucleoli exhibit a broad distribution of sizes with a characteristic power law, which we show is a consequence of spontaneous coalescence events. These results have implications for the function of nucleoli in ribosome subunit processing and provide a physical link between activity within a macromolecular assembly and its physical properties on larger length scales.
.
DOI:10.1016/j.cell.2012.04.017URL [本文引用: 2]

Eukaryotic cells contain assemblies of RNAs and proteins termed RNA granules. Many proteins within these bodies contain KH or RRM RNA-binding domains as well as low complexity (LC) sequences of unknown function. We discovered that exposure of cell or tissue lysates to a biotinylated isoxazole (b-isox) chemical precipitated hundreds of RNA-binding proteins with significant overlap to the constituents of RNA granules. The LC sequences within these proteins are both necessary and sufficient for b-isox-mediated aggregation, and these domains can undergo a concentration-dependent phase transition to a hydrogel-like state in the absence of the chemical. X-ray diffraction and EM studies revealed the hydrogels to be composed of uniformly polymerized amyloid-like fibers. Unlike pathogenic fibers, the LC sequence-based polymers described here are dynamic and accommodate heterotypic polymerization. These observations offer a framework for understanding the function of LC sequences as well as an organizing principle for cellular structures that are not membrane bound.
.
DOI:10.1016/j.molcel.2015.01.013URLPMID:25747659 [本文引用: 2]

Cells chemically isolate molecules in compartments to both facilitate and regulate their interactions. In addition to membrane-encapsulated compartments, cells can form proteinaceous and membraneless organelles, including nucleoli, Cajal and PML bodies, and stress granules. The principles that determine when and why these structures form have remained elusive. Here, we demonstrate that the disordered tails of Ddx4, a primary constituent of nuage or germ granules, form phase-separated organelles both in live cells and in vitro. These bodies are stabilized by patterned electrostatic interactions that are highly sensitive to temperature, ionic strength, arginine methylation, and splicing. Sequence determinants are used to identify proteins found in both membraneless organelles and cell adhesion. Moreover, the bodies provide an alternative solvent environment that can concentrate single-stranded DNA but largely exclude double-stranded DNA. We propose that phase separation of disordered proteins containing weakly interacting blocks is a general mechanism for forming regulated, membraneless organelles.
.
DOI:10.1038/nature10879URL [本文引用: 3]

Cells are organized on length scales ranging from angstrom to micrometres. However, the mechanisms by which angstrom-scale molecular properties are translated to micrometre-scale macroscopic properties are not well understood. Here we show that interactions between diverse synthetic, multivalent macromolecules (including multi-domain proteins and RNA) produce sharp liquid-liquid demixing phase separations, generating micrometre-sized liquid droplets in aqueous solution. This macroscopic transition corresponds to a molecular transition between small complexes and large, dynamic supramolecular polymers. The concentrations needed for phase transition are directly related to the valency of the interacting species. In the case of the actin-regulatory protein called neural Wiskott-Aldrich syndrome protein (N-WASP) interacting with its established biological partners NCK and phosphorylated nephrin(1), the phase transition corresponds to a sharp increase in activity towards an actin nucleation factor, the Arp2/3 complex. The transition is governed by the degree of phosphorylation of nephrin, explaining how this property of the system can be controlled to regulatory effect by kinases. The widespread occurrence of multivalent systems suggests that phase transitions may be used to spatially organize and biochemically regulate information throughout biology.
.
DOI:10.1353/pbm.2015.0024URLPMID:26750599 [本文引用: 2]

The concept of chromatin as a complex of nucleic acid and proteins in the cell nucleus was developed by cytologists and biochemists in the late 19th century. It was the starting point for biochemical research on DNA and nuclear proteins. Although interest in chromatin declined rapidly at the beginning of the 20th century, a few decades later a new focus on chromatin emerged, which was not only related to its structure, but also to its function in gene regulatory processes in the development of higher organisms. Since the late 20th century, research on chromatin modifications has also been conducted under the label of epigenetics. This article highlights the major phases of chromatin research until the present time and introduces major investigators and their scientific and philosophical outlooks.
DOI:10.1360/N972019-00281URL [本文引用: 1]
DOI:10.1360/N972019-00281URL [本文引用: 1]
.
DOI:10.1016/j.cell.2013.07.038URL [本文引用: 2]

The molecular processes that contribute to degenerative diseases are not well understood. Recent observations suggest that some degenerative diseases are promoted by the accumulation of nuclear or cytoplasmic RNA-protein (RNP) aggregates, which can be related to endogenous RNP granules. RNP aggregates arise commonly in degenerative diseases because RNA-binding proteins commonly self-assemble, in part through prion-like domains, which can form self-propagating amyloids. RNP aggregates may be toxic due to multiple perturbations of posttranscriptional control, thereby disrupting the normal "ribostasis" of the cell. This suggests that understanding and modulating RNP assembly or clearance may be effective approaches to developing therapies for these diseases.
.
DOI:10.1016/j.brainres.2012.01.016URL [本文引用: 1]

Prions are self-templating protein conformers that are naturally transmitted between individuals and promote phenotypic change. In yeast, prion-encoded phenotypes can be beneficial, neutral or deleterious depending upon genetic background and environmental conditions. A distinctive and portable 'prion domain' enriched in asparagine, glutamine, tyrosine and glycine residues unifies the majority of yeast prion proteins. Deletion of this domain precludes prionogenesis and appending this domain to reporter proteins can confer prionogenicity. An algorithm designed to detect prion domains has successfully identified 19 domains that can confer prion behavior. Scouring the human genome with this algorithm enriches a select group of RNA-binding proteins harboring a canonical RNA recognition motif (RRM) and a putative prion domain. Indeed, of 210 human RRM-bearing proteins, 29 have a putative prion domain, and 12 of these are in the top 60 prion candidates in the entire genome. Startlingly, these RNA-binding prion candidates are inexorably emerging, one by one, in the pathology and genetics of devastating neurodegenerative disorders, including amyotrophic lateral sclerosis (ALS), frontotemporal lobar degeneration with ubiquitin-positive inclusions (FTLD-U), Alzheimer's disease and Huntington's disease. For example, FUS and TDP-43, which rank 1st and 10th among RRM-bearing prion candidates, form cytoplasmic inclusions in the degenerating motor neurons of ALS patients and mutations in TDP-43 and FUS cause familial ALS. Recently, perturbed RNA-binding proteostasis of TAF15, which is the 2nd ranked RRM-bearing prion candidate, has been connected with ALS and FTLD-U. We strongly suspect that we have now merely reached the tip of the iceberg. We predict that additional RNA-binding prion candidates identified by our algorithm will soon surface as genetic modifiers or causes of diverse neurodegenerative conditions. Indeed, simple prion-like transfer mechanisms involving the prion domains of RNA-binding proteins could underlie the classical non-cell-autonomous emanation of neurodegenerative pathology from originating epicenters to neighboring portions of the nervous system.
This article is part of a Special Issue entitled RNA-Binding Proteins. (c) 2012 Elsevier B.V.
.
DOI:10.1016/j.cell.2009.02.044URLPMID:19345193 [本文引用: 1]

Prions are proteins that convert between structurally and functionally distinct states, one or more of which is transmissible. In yeast, this ability allows them to act as non-Mendelian elements of phenotypic inheritance. To further our understanding of prion biology, we conducted a bioinformatic proteome-wide survey for prionogenic proteins in S. cerevisiae, followed by experimental investigations of 100 prion candidates. We found an unexpected amino acid bias in aggregation-prone candidates and discovered that 19 of these could also form prions. At least one of these prion proteins, Mot3, produces a bona fide prion in its natural context that increases population-level phenotypic heterogeneity. The self-perpetuating states of these proteins present a vast source of heritable phenotypic variation that increases the adaptability of yeast populations to diverse environments.
.
DOI:10.1002/anie.201402885URLPMID:24862735 [本文引用: 1]

In eukaryotic cells, components of the 5' to 3' mRNA degradation machinery can undergo a rapid phase transition. The resulting cytoplasmic foci are referred to as processing bodies (P-bodies). The molecular details of the self-aggregation process are, however, largely undetermined. Herein, we use a bottom-up approach that combines NMR spectroscopy, isothermal titration calorimetry, X-ray crystallography, and fluorescence microscopy to probe if mRNA degradation factors can undergo phase transitions in?vitro. We show that the Schizosaccharomyces pombe Dcp2 mRNA decapping enzyme, its prime activator Dcp1, and the scaffolding proteins Edc3 and Pdc1 are sufficient to reconstitute a phase-separation process. Intermolecular interactions between the Edc3?LSm domain and at least 10?helical leucine-rich motifs in Dcp2 and Pdc1 build the core of the interaction network. We show that blocking of these interactions interferes with the clustering behavior, both in?vitro and in?vivo.
DOI:10.1016/j.cell.2016.07.008URLPMID:27565345 [本文引用: 1]

Postsynaptic densities (PSDs) are membrane semi-enclosed, submicron protein-enriched cellular compartments beneath postsynaptic membranes, which constantly exchange their components with bulk aqueous cytoplasm in synaptic spines. Formation and activity-dependent modulation of PSDs is considered as one of the most basic molecular events governing synaptic plasticity in the nervous system. In this study, we discover that SynGAP, one of the most abundant PSD proteins and a Ras/Rap GTPase activator, forms a homo-trimer and binds to multiple copies of PSD-95. Binding of SynGAP to PSD-95 induces phase separation of the complex, forming highly concentrated liquid-like droplets reminiscent of the PSD. The multivalent nature of the SynGAP/PSD-95 complex is critical for the phase separation to occur and for proper activity-dependent SynGAP dispersions from the PSD. In addition to revealing a dynamic anchoring mechanism of SynGAP at the PSD, our results also suggest a model for phase-transition-mediated formation of PSD.
DOI:10.1016/j.cell.2018.10.057URLPMID:30500535 [本文引用: 3]

Phase transitions involving biomolecular liquids are a fundamental mechanism underlying intracellular organization. In the cell nucleus, liquid-liquid phase separation of intrinsically disordered proteins (IDPs) is implicated in assembly of the nucleolus, as well as transcriptional clusters, and other nuclear bodies. However, it remains unclear whether and how physical forces associated with nucleation, growth, and wetting of liquid condensates can directly restructure chromatin. Here, we use CasDrop, a novel CRISPR-Cas9-based optogenetic technology, to show that various IDPs phase separate into liquid condensates that mechanically exclude chromatin as they grow and preferentially form in low-density, largely euchromatic regions. A minimal physical model explains how this stiffness sensitivity arises from lower mechanical energy associated with deforming softer genomic regions. Targeted genomic loci can nonetheless be mechanically pulled together through surface tension-driven coalescence. Nuclear condensates may thus function as mechano-active chromatin filters, physically pulling in targeted genomic loci while pushing out non-targeted regions of the neighboring genome. VIDEO ABSTRACT.
.
DOI:10.1126/science.aau5721URLPMID:30846600 [本文引用: 2]

The guanine nucleotide exchange factor (GEF) Son of Sevenless (SOS) is a key Ras activator that is autoinhibited in the cytosol and activates upon membrane recruitment. Autoinhibition release involves structural rearrangements of the protein at the membrane and thus introduces a delay between initial recruitment and activation. In this study, we designed a single-molecule assay to resolve the time between initial receptor-mediated membrane recruitment and the initiation of GEF activity of individual SOS molecules on microarrays of Ras-functionalized supported membranes. The rise-and-fall shape of the measured SOS activation time distribution and the long mean time scale to activation (~50 seconds) establish a basis for kinetic proofreading in the receptor-mediated activation of Ras. We further demonstrate that this kinetic proofreading is modulated by the LAT (linker for activation of T cells)-Grb2-SOS phosphotyrosine-driven phase transition at the membrane.
.
DOI:10.1038/nsmb1133URLPMID:16906159 [本文引用: 1]

Receptor oligomerization is vital for activating intracellular signaling, in part by initiating events that recruit effector and adaptor proteins to sites of active signaling. Whether these distal molecules themselves oligomerize is not well appreciated. In this study, we examined the molecular interactions of the adaptor protein GRB2. In T cells, the SH2 domain of GRB2 binds phosphorylated tyrosines on the adaptor protein LAT and the GRB2 SH3 domains associate with the proline-rich regions of SOS1 and CBL. Using biochemical and biophysical techniques in conjunction with confocal microscopy, we observed that the simultaneous association of GRB2, via its SH2 and SH3 domains, with multivalent ligands led to the oligomerization of these ligands, which affected signaling. These data suggest that multipoint binding of distal adaptor proteins mediates the formation of oligomeric signaling clusters vital for intracellular signaling.
.
DOI:10.1101/cshperspect.a031534URLPMID:29610148 [本文引用: 1]

The guanine-nucleotide exchange factor (GEF) Son-of-Sevenless (SOS) plays a critical role in metazoan signaling by converting Ras?GDP (guanosine diphosphate) to Ras?GTP (guanosine triphosphate) in response to tyrosine kinase activation. Structural studies have shown that SOS differs from other Ras-specific GEFs in that SOS is itself activated by Ras?GTP binding to an allosteric site, distal to the site of nucleotide exchange. The activation of SOS involves membrane recruitment and conformational changes, triggered by lipid binding, that open the allosteric binding site for Ras?GTP. This is in contrast to other Ras-specific GEFs, which are activated by second messengers that more directly affect the active site. Allosteric Ras?GTP binding stabilizes SOS at the membrane, where it can turn over other Ras molecules processively, leading to an ultrasensitive response that is distinct from that of other Ras-specific GEFs.
.
DOI:10.1016/j.bpj.2009.01.019URLPMID:19348745 [本文引用: 1]

Ligand-induced receptor aggregation is a well-known mechanism for initiating intracellular signals but oligomerization of distal signaling molecules may also be required for signal propagation. Formation of complexes containing oligomers of the transmembrane adaptor protein, linker for the activation of T cells (LAT), has been identified as critical in mast cell and T cell activation mediated by immune response receptors. Cross-linking of LAT arises from the formation of a 2:1 complex between the adaptor Grb2 and the nucleotide exchange factor SOS1, which bridges two LAT molecules through the interaction of the Grb2 SH2 domain with a phosphotyrosine on LAT. We model this oligomerization and find that the valence of LAT for Grb2, which ranges from zero to three, is critical in determining the nature and extent of aggregation. A dramatic rise in oligomerization can occur when the valence switches from two to three. For valence three, an equilibrium theory predicts the possibility of forming a gel-like phase. This prediction is confirmed by stochastic simulations, which make additional predictions about the size of the gel and the kinetics of LAT oligomerization. We discuss the model predictions in light of recent experiments on RBL-2H3 and Jurkat E6.1 cells and suggest that the gel phase has been observed in activated mast cells.
. .
DOI:10.1021/jacs.7b09387URLPMID:29182244 [本文引用: 1]

Tyrosine phosphorylation of membrane receptors and scaffold proteins followed by recruitment of SH2 domain-containing adaptor proteins constitutes a central mechanism of intracellular signal transduction. During early T-cell receptor (TCR) activation, phosphorylation of linker for activation of T cells (LAT) leading to recruitment of adaptor proteins, including Grb2, is one prototypical example. LAT contains multiple modifiable sites, and this multivalency may provide additional layers of regulation, although this is not well understood. Here, we quantitatively analyze the effects of multivalent phosphorylation of LAT by reconstituting the initial reactions of the TCR signaling pathway on supported membranes. Results from a series of LAT constructs with combinatorial mutations of tyrosine residues reveal a previously unidentified allosteric mechanism in which the binding affinity of LAT:Grb2 depends on the phosphorylation at remote tyrosine sites. Additionally, we find that LAT:Grb2 binding affinity is altered by membrane localization. This allostery mainly regulates the kinetic on-rate, not off-rate, of LAT:Grb2 interactions. LAT is an intrinsically disordered protein, and these data suggest that phosphorylation changes the overall ensemble of configurations to modulate the accessibility of other phosphorylated sites to Grb2. Using Grb2 as a phosphorylation reporter, we further monitored LAT phosphorylation by TCR ζ chain-recruited ZAP-70, which suggests a weakly processive catalysis on membranes. Taken together, these results suggest that signal transmission through LAT is strongly gated and requires multiple phosphorylation events before efficient signal transmission is achieved.
.
DOI:10.1016/j.bpj.2017.08.024URLPMID:29045874 [本文引用: 1]

Biochemical signaling pathways often involve proteins with multiple, modular interaction domains. Signaling activates binding sites, such as by tyrosine phosphorylation, which enables protein recruitment and growth of networked protein assemblies. Although widely observed, the physical properties of the assemblies, as well as the mechanisms by which they function, remain largely unknown. Here we examine molecular mobility within LAT:Grb2:SOS assemblies on supported membranes by single-molecule tracking. Trajectory analysis reveals a discrete temporal transition to subdiffusive motion below a characteristic timescale, indicating that the LAT:Grb2:SOS assembly has the dynamical structure of a loosely entangled polymer. Such dynamical analysis is also applicable in living cells, where it offers another dimension on the characteristics of cellular signaling assemblies.
DOI:10.1016/j.cell.2017.02.027URLPMID:28283059 [本文引用: 1]

In eukaryotic cells, diverse stresses trigger coalescence of RNA-binding proteins into stress granules.?In?vitro, stress-granule-associated proteins can demix to form liquids, hydrogels, and other assemblies lacking fixed stoichiometry. Observing these phenomena has generally required conditions far removed from physiological stresses. We show that poly(A)-binding protein (Pab1 in yeast), a defining marker of stress granules, phase separates and forms hydrogels in?vitro upon exposure to physiological stress conditions. Other RNA-binding proteins depend upon low-complexity regions (LCRs) or RNA for phase separation, whereas Pab1's LCR is not required for demixing, and RNA inhibits it. Based on unique evolutionary patterns, we create LCR mutations, which systematically tune its biophysical properties and Pab1 phase separation in?vitro and in?vivo. Mutations that impede phase separation reduce organism fitness during prolonged stress. Poly(A)-binding protein thus acts as a physiological stress sensor, exploiting phase separation to precisely mark stress onset, a broadly generalizable mechanism.
DOI:10.1126/science.359.6371.126URLPMID:29302016 [本文引用: 1]

Innate lymphoid cells (ILCs) are innate counterparts of adaptive T lymphocytes, contributing to host defense, tissue repair, metabolic homeostasis, and inflammatory diseases. ILCs have been considered to be tissue-resident cells, but whether ILCs move between tissue sites during infection has been unclear. We show here that interleukin-25- or helminth-induced inflammatory ILC2s are circulating cells that arise from resting ILC2s residing in intestinal lamina propria. They migrate to diverse tissues based on sphingosine 1-phosphate (S1P)-mediated chemotaxis that promotes lymphatic entry, blood circulation, and accumulation in peripheral sites, including the lung, where they contribute to anti-helminth defense and tissue repair. This ILC2 expansion and migration is a behavioral parallel to the antigen-driven proliferation and migration of adaptive lymphocytes to effector sites and indicates that ILCs complement adaptive immunity by providing both local and distant tissue protection during infection.
.
DOI:10.1083/jcb.201302044URLPMID:23629963 [本文引用: 1]

Amyotrophic lateral sclerosis (ALS) is a fatal human neurodegenerative disease affecting primarily motor neurons. Two RNA-binding proteins, TDP-43 and FUS, aggregate in the degenerating motor neurons of ALS patients, and mutations in the genes encoding these proteins cause some forms of ALS. TDP-43 and FUS and several related RNA-binding proteins harbor aggregation-promoting prion-like domains that allow them to rapidly self-associate. This property is critical for the formation and dynamics of cellular ribonucleoprotein granules, the crucibles of RNA metabolism and homeostasis. Recent work connecting TDP-43 and FUS to stress granules has suggested how this cellular pathway, which involves protein aggregation as part of its normal function, might be coopted during disease pathogenesis.
.
DOI:10.1038/nrm2873URLPMID:20308987 [本文引用: 1]

Neurodegenerative diseases are commonly associated with the accumulation of intracellular or extracellular protein aggregates. Recent studies suggest that these aggregates are capable of crossing cellular membranes and can directly contribute to the propagation of neurodegenerative disease pathogenesis. We propose that, once initiated, neuropathological changes might spread in a 'prion-like' manner and that disease progression is associated with the intercellular transfer of pathogenic proteins. The transfer of naked infectious particles between cells could therefore be a target for new disease-modifying therapies.
.
DOI:10.1146/annurev-pathol-011110-130242URL [本文引用: 1]
.
DOI:10.1038/nrn3430URL [本文引用: 1]

Several recent breakthroughs have provided notable insights into the pathogenesis of amyotrophic lateral sclerosis (ALS), with some even shifting our thinking about this neurodegenerative disease and raising the question as to whether this disorder is a proteinopathy, a ribonucteopathy or both. In addition, these breakthroughs have revealed mechanistic links between ALS and frontotemporal dementia, as well as between ALS and other neurodegenerative diseases, such as the cerebellar atrophies, nnyotonic dystrophy and inclusion body myositis. Here, we summarize the new findings in ALS research, discuss what they have taught us about this disease and examine issues that are still outstanding.
.
DOI:10.1016/j.cell.2015.07.047URLPMID:26317470 [本文引用: 1]

Many proteins contain disordered regions of low-sequence complexity, which cause aging-associated diseases because they are prone to aggregate. Here, we study FUS, a prion-like protein containing intrinsically disordered domains associated with the neurodegenerative disease ALS. We show that, in cells, FUS forms liquid compartments at sites of DNA damage and in the cytoplasm upon stress. We confirm this by reconstituting liquid FUS compartments in vitro. Using an in vitro "aging" experiment, we demonstrate that liquid droplets of FUS protein convert with time from a liquid to an aggregated state, and this conversion is accelerated by patient-derived mutations. We conclude that the physiological role of FUS requires forming dynamic liquid-like compartments. We propose that liquid-like compartments carry the trade-off between functionality and risk of aggregation and that aberrant phase transitions within liquid-like compartments lie at the heart of ALS and, presumably, other age-related diseases.
.
DOI:10.1038/nature11922URL [本文引用: 1]

Algorithms designed to identify canonical yeast prions predict that around 250 human proteins, including several RNA-binding proteins associated with neurodegenerative disease, harbour a distinctive prion-like domain (PrLD) enriched in uncharged polar amino acids and glycine. PrLDs in RNA-binding proteins are essential for the assembly of ribonucleoprotein granules. However, the interplay between human PrLD function and disease is not understood. Here we define pathogenic mutations in PrLDs of heterogeneous nuclear ribonucleoproteins (hnRNPs) A2B1 and A1 in families with inherited degeneration affecting muscle, brain, motor neuron and bone, and in one case of familial amyotrophic lateral sclerosis. Wild-type hnRNPA2 (the most abundant isoform of hnRNPA2B1) and hnRNPA1 show an intrinsic tendency to assemble into self-seeding fibrils, which is exacerbated by the disease mutations. Indeed, the pathogenic mutations strengthen a 'steric zipper' motif in the PrLD, which accelerates the formation of self-seeding fibrils that cross-seed polymerization of wild-type hnRNP. Notably, the disease mutations promote excess incorporation of hnRNPA2 and hnRNPA1 into stress granules and drive the formation of cytoplasmic inclusions in animal models that recapitulate the human pathology. Thus, dysregulated polymerization caused by a potent mutant steric zipper motif in a PrLD can initiate degenerative disease. Related proteins with PrLDs should therefore be considered candidates for initiating and perhaps propagating proteinopathies of muscle, brain, motor neuron and bone.
.
DOI:10.1016/j.neuron.2015.10.030URLPMID:26526393 [本文引用: 1]

The mechanisms by which mutations in FUS and other RNA binding proteins cause ALS and FTD remain controversial. We propose a model in which low-complexity (LC) domains of FUS drive its physiologically reversible assembly into membrane-free, liquid droplet and hydrogel-like structures. ALS/FTD mutations in LC or non-LC domains induce further phase transition into poorly soluble fibrillar hydrogels distinct from conventional amyloids. These assemblies are necessary and sufficient for neurotoxicity in a C. elegans model of FUS-dependent neurodegeneration. They trap other ribonucleoprotein (RNP) granule components and disrupt RNP granule function. One consequence is impairment of new protein synthesis by cytoplasmic RNP granules in axon terminals, where RNP granules regulate local RNA metabolism and translation. Nuclear FUS granules may be similarly affected. Inhibiting formation of these fibrillar hydrogel assemblies mitigates neurotoxicity and suggests a potential therapeutic strategy that may also be applicable to ALS/FTD associated with mutations in other RNA binding proteins.
.
DOI:10.1073/pnas.0511295103URLPMID:16537487 [本文引用: 1]

Based on the crystal structure of the cross-beta spine formed by the peptide NNQQNY, we have developed a computational approach for identifying those segments of amyloidogenic proteins that themselves can form amyloid-like fibrils. The approach builds on experiments showing that hexapeptides are sufficient for forming amyloid-like fibrils. Each six-residue peptide of a protein of interest is mapped onto an ensemble of templates, or 3D profile, generated from the crystal structure of the peptide NNQQNY by small displacements of one of the two intermeshed beta-sheets relative to the other. The energy of each mapping of a sequence to the profile is evaluated by using ROSETTADESIGN, and the lowest energy match for a given peptide to the template library is taken as the putative prediction. If the energy of the putative prediction is lower than a threshold value, a prediction of fibril formation is made. This method can reach an accuracy of approximately 80% with a P value of approximately 10(-12) when a conservative energy threshold is used to separate peptides that form fibrils from those that do not. We see enrichment for positive predictions in a set of fibril-forming segments of amyloid proteins, and we illustrate the method with applications to proteins of interest in amyloid research.
.
DOI:10.1016/j.cell.2012.04.016URL [本文引用: 1]

Cellular granules lacking boundary membranes harbor RNAs and their associated proteins and play diverse roles controlling the timing and location of protein synthesis. Formation of such granules was emulated by treatment of mouse brain extracts and human cell lysates with a biotinylated isoxazole (b-isox) chemical. Deep sequencing of the associated RNAs revealed an enrichment for mRNAs known to be recruited to neuronal granules used for dendritic transport and localized translation at synapses. Precipitated mRNAs contain extended 30 UTR sequences and an enrichment in binding sites for known granule-associated proteins. Hydrogels composed of the low complexity (LC) sequence domain of FUS recruited and retained the same mRNAs as were selectively precipitated by the b-isox chemical. Phosphorylation of the LC domain of FUS prevented hydrogel retention, offering a conceptual means of dynamic, signal-dependent control of RNA granule assembly.
.
DOI:10.1016/j.celrep.2013.11.017URLPMID:24268778 [本文引用: 1]

The abundant nuclear RNA binding protein FUS binds the C-terminal domain (CTD) of RNA polymerase II in an RNA-dependent manner, affecting Ser2 phosphorylation and transcription. Here, we examine the mechanism of this process and find that RNA binding nucleates the formation of higher-order FUS ribonucleoprotein assemblies that bind the CTD. Both the low-complexity domain and the arginine-glycine rich domain of FUS contribute to assembly. The assemblies appear fibrous by electron microscopy and have characteristics of β zipper structures. These results support the emerging view that the pathologic protein aggregation seen in neurodegenerative diseases such as amyotrophic lateral sclerosis may occur via the exaggeration of functionally important assemblies of RNA binding proteins.
.
DOI:10.1146/annurev-biochem-060614-034325URLPMID:25494299 [本文引用: 1]

Members of the FET protein family, consisting of FUS, EWSR1, and TAF15, bind to RNA and contribute to the control of transcription, RNA processing, and the cytoplasmic fates of messenger RNAs in metazoa. FET proteins can also bind DNA, which may be important in transcription and DNA damage responses. FET proteins are of medical interest because chromosomal rearrangements of their genes promote various sarcomas and because point mutations in FUS or TAF15 can cause neurodegenerative diseases such as amyotrophic lateral sclerosis and frontotemporal lobar dementia. Recent results suggest that both the normal and pathological effects of FET proteins are modulated by low-complexity or prion-like domains, which can form higher-order assemblies with novel interaction properties. Herein, we review FET proteins with an emphasis on how the biochemical properties of FET proteins may relate to their biological functions and to pathogenesis.
.
DOI:10.1093/hmg/ddt371URLPMID:23918658 [本文引用: 1]

Well-established rules of translational initiation have been used as a cornerstone in molecular biology to understand gene expression and to frame fundamental questions on what proteins a cell synthesizes, how proteins work and to predict the consequences of mutations. For a group of neurological diseases caused by the abnormal expansion of short segments of DNA (e.g. CAG?CTG repeats), mutations within or outside of predicted coding and non-coding regions are thought to cause disease by protein gain- or loss-of-function or RNA gain-of-function mechanisms. In contrast to these predictions, the recent discovery of repeat-associated non-ATG (RAN) translation showed expansion mutations can express homopolymeric expansion proteins in all three reading frames without an AUG start codon. This unanticipated, non-canonical type of protein translation is length-and hairpin-dependent, takes place without frameshifting or RNA editing and occurs across a variety of repeat motifs. To date, RAN proteins have been reported in spinocerebellar ataxia type 8 (SCA8), myotonic dystrophy type 1 (DM1), fragile X tremor ataxia syndrome (FXTAS) and C9ORF72 amyotrophic lateral sclerosis/frontotemporal dementia (ALS/FTD). In this article, we review what is currently known about RAN translation and recent progress toward understanding its contribution to disease.
.
DOI:10.1126/science.1232927URL [本文引用: 1]

Expansion of a GGGGCC hexanucleotide repeat upstream of the C9orf72 coding region is the most common cause of familial frontotemporal lobar degeneration and amyotrophic lateral sclerosis (FTLD/ALS), but the pathomechanisms involved are unknown. As in other FTLD/ALS variants, characteristic intracellular inclusions of misfolded proteins define C9orf72 pathology, but the core proteins of the majority of inclusions are still unknown. Here, we found that most of these characteristic inclusions contain poly-(Gly-Ala) and, to a lesser extent, poly-(Gly-Pro) and poly-(Gly-Arg) dipeptide-repeat proteins presumably generated by non-ATG-initiated translation from the expanded GGGGCC repeat in three reading frames. These findings directly link the FTLD/ALS-associated genetic mutation to the predominant pathology in patients with C9orf72 hexanucleotide expansion.
.
DOI:10.1038/nature05292URLPMID:17051205 [本文引用: 1]

Many lines of evidence suggest that mitochondria have a central role in ageing-related neurodegenerative diseases. Mitochondria are critical regulators of cell death, a key feature of neurodegeneration. Mutations in mitochondrial DNA and oxidative stress both contribute to ageing, which is the greatest risk factor for neurodegenerative diseases. In all major examples of these diseases there is strong evidence that mitochondrial dysfunction occurs early and acts causally in disease pathogenesis. Moreover, an impressive number of disease-specific proteins interact with mitochondria. Thus, therapies targeting basic mitochondrial processes, such as energy metabolism or free-radical generation, or specific interactions of disease-related proteins with mitochondria, hold great promise.
.
DOI:10.1158/2159-8290.CD-12-0095URL [本文引用: 1]

The cBio Cancer Genomics Portal (http://cbioportal.org) is an open-access resource for interactive exploration of multidimensional cancer genomics data sets, currently providing access to data from more than 5,000 tumor samples from 20 cancer studies. The cBio Cancer Genomics Portal significantly lowers the barriers between complex genomic data and cancer researchers who want rapid, intuitive, and high-quality access to molecular profiles and clinical attributes from large-scale cancer genomics projects and empowers researchers to translate these rich data sets into biologic insights and clinical applications. Cancer Discov; 2(5); 401-4. (c) 2012 AACR.
.
DOI:10.1128/MCB.01297-06URLPMID:17189428 [本文引用: 1]

Androgens have key roles in normal physiology and in male sexual differentiation as well as in pathological conditions such as prostate cancer. Androgens act through the androgen receptor (AR), which is a ligand-modulated transcription factor. Antiandrogens block AR function and are widely used in disease states, but little is known about their mechanism of action in vivo. Here, we describe a rapid differential interaction of AR with target genomic sites in living cells in the presence of agonists which coincides with the recruitment of BRM ATPase complex and chromatin remodeling, resulting in transcriptional activation. In contrast, the interaction of antagonist-bound or mutant AR with its target was found to be kinetically different: it was dramatically faster, occurred without chromatin remodeling, and resulted in the lack of transcriptional inhibition. Fluorescent resonance energy transfer analysis of wild-type AR and a transcriptionally compromised mutant at the hormone response element showed that intramolecular interactions between the N and C termini of AR play a key functional role in vivo compared to intermolecular interactions between two neighboring ARs. These data provide a kinetic and mechanistic basis for regulation of gene expression by androgens and antiandrogens in living cells.
[本文引用: 1]
.
DOI:10.1093/hmg/ddr299URL [本文引用: 1]

Discrete and punctate nuclear RNA foci are characteristic molecular hallmarks of pathogenesis in myotonic dystrophy type 1 and type 2. Intranuclear RNA inclusions of distinct morphology have also been found in fragile X-associated tremor ataxia syndrome, Huntington's disease-like 2, spinocerebellar ataxias type 8, type 10 and type 31. These neurological diseases are associated with the presence of abnormally long simple repeat expansions in their respective genes whose expression leads to the formation of flawed transcripts with altered metabolisms. Expanded CUG, CCUG, CGG, CAG, AUUCU and UGGAA repeats are associated with the diseases and accumulate in nuclear foci, as demonstrated in variety of cells and tissues of human and model organisms. These repeat RNA foci differ in size, shape, cellular abundance and protein composition and their formation has a negative impact on cellular functions. This review summarizes the efforts of many laboratories over the past 15 years to characterize nuclear RNA foci that are recognized as important triggers in the mutant repeat RNA toxic gain-of-function mechanisms of pathogenesis in neurological disorders.
.
DOI:10.1016/j.cell.2016.04.047URLPMID:27212236 [本文引用: 1]

The nucleolus and other ribonucleoprotein (RNP) bodies are membrane-less organelles that appear to assemble through phase separation of their molecular components. However, many such RNP bodies contain internal subcompartments, and the mechanism of their formation remains unclear. Here, we combine in?vivo and in?vitro studies, together with computational modeling, to show that subcompartments within the nucleolus represent distinct, coexisting liquid phases. Consistent with their in?vivo immiscibility, purified nucleolar proteins phase separate into droplets containing distinct non-coalescing phases that are remarkably similar to nucleoli in?vivo. This layered droplet organization is caused by differences in the biophysical properties of the phases-particularly droplet surface tension-which arises from sequence-encoded features of their macromolecular components. These results suggest that phase separation can give rise to multilayered liquids that may facilitate sequential RNA processing reactions in a variety of RNP bodies. PAPERCLIP.
.
DOI:10.1038/nbt.3383URLPMID:26458175 [本文引用: 1]

Chromatin profiling provides a versatile means to investigate functional genomic elements and their regulation. However, current methods yield ensemble profiles that are insensitive to cell-to-cell variation. Here we combine microfluidics, DNA barcoding and sequencing to collect chromatin data at single-cell resolution. We demonstrate the utility of the technology by assaying thousands of individual cells and using the data to deconvolute a mixture of ES cells, fibroblasts and hematopoietic progenitors into high-quality chromatin state maps for each cell type. The data from each single cell are sparse, comprising on the order of 1,000 unique reads. However, by assaying thousands of ES cells, we identify a spectrum of subpopulations defined by differences in chromatin signatures of pluripotency and differentiation priming. We corroborate these findings by comparison to orthogonal single-cell gene expression data. Our method for single-cell analysis reveals aspects of epigenetic heterogeneity not captured by transcriptional analysis alone.
.
DOI:10.1038/nmeth.1626URLPMID:21642965 [本文引用: 1]

Genome-wide profiling of transcription factors based on massive parallel sequencing of immunoprecipitated chromatin (ChIP-seq) requires nanogram amounts of DNA. Here we describe a high-fidelity, single-tube linear DNA amplification method (LinDA) for ChIP-seq and reChIP-seq with picogram DNA amounts obtained from a few thousand cells. This amplification technology will facilitate global analyses of transcription-factor binding and chromatin with very small cell populations, such as stem or cancer-initiating cells.
.
DOI:10.1126/science.1181369URLPMID:19815776 [本文引用: 1]

We describe Hi-C, a method that probes the three-dimensional architecture of whole genomes by coupling proximity-based ligation with massively parallel sequencing. We constructed spatial proximity maps of the human genome with Hi-C at a resolution of 1 megabase. These maps confirm the presence of chromosome territories and the spatial proximity of small, gene-rich chromosomes. We identified an additional level of genome organization that is characterized by the spatial segregation of open and closed chromatin to form two genome-wide compartments. At the megabase scale, the chromatin conformation is consistent with a fractal globule, a knot-free, polymer conformation that enables maximally dense packing while preserving the ability to easily fold and unfold any genomic locus. The fractal globule is distinct from the more commonly used globular equilibrium model. Our results demonstrate the power of Hi-C to map the dynamic conformations of whole genomes.
.
DOI:10.1016/j.cell.2014.11.021URL [本文引用: 1]

We use in situ Hi-C to probe the 3D architecture of genomes, constructing haploid and diploid maps of nine cell types. The densest, in human lymphoblastoid cells, contains 4.9 billion contacts, achieving 1 kb resolution. We find that genomes are partitioned into contact domains (median length, 185 kb), which are associated with distinct patterns of histone marks and segregate into six subcompartments. We identify similar to 10,000 loops. These loops frequently link promoters and enhancers, correlate with gene activation, and show conservation across cell types and species. Loop anchors typically occur at domain boundaries and bind CTCF. CTCF sites at loop anchors occur predominantly (>90%) in a convergent orientation, with the asymmetric motifs "facing'' one another. The inactive X chromosome splits into two massive domains and contains large loops anchored at CTCF-binding repeats.
DOI:10.1186/s12864-018-4559-3URLPMID:29490630 [本文引用: 1]

ATAC-seq (Assays for Transposase-Accessible Chromatin using sequencing) is a recently developed technique for genome-wide analysis of chromatin accessibility. Compared to earlier methods for assaying chromatin accessibility, ATAC-seq is faster and easier to perform, does not require cross-linking, has higher signal to noise ratio, and can be performed on small cell numbers. However, to ensure a successful ATAC-seq experiment, step-by-step quality assurance processes, including both wet lab quality control and in silico quality assessment, are essential. While several tools have been developed or adopted for assessing read quality, identifying nucleosome occupancy and accessible regions from ATAC-seq data, none of the tools provide a comprehensive set of functionalities for preprocessing and quality assessment of aligned ATAC-seq datasets.
.
DOI:10.1038/NMETH.2688URL [本文引用: 1]

We describe an assay for transposase-accessible chromatin using sequencing (ATAC-seq), based on direct in vitro transposition of sequencing adaptors into native chromatin, as a rapid and sensitive method for integrative epigenomic analysis. ATAC-seq captures open chromatin sites using a simple two-step protocol with 500-50,000 cells and reveals the interplay between genomic locations of open chromatin, DNA-binding proteins, individual nucleosomes and chromatin compaction at nucleotide resolution. We discovered classes of DNA-binding factors that strictly avoided, could tolerate or tended to overlap with nucleosomes. Using ATAC-seq maps of human CD4(+) T cells from a proband obtained on consecutive days, we demonstrated the feasibility of analyzing an individual's epigenome on a timescale compatible with clinical decision-making.
DOI:10.1016/j.cell.2019.08.037URLPMID:31543265 [本文引用: 2]

Eukaryotic chromatin is highly condensed but dynamically accessible to regulation and organized into subdomains. We demonstrate that reconstituted chromatin undergoes histone tail-driven liquid-liquid phase separation (LLPS) in physiologic salt and when microinjected into cell nuclei, producing dense and dynamic droplets. Linker histone H1 and internucleosome linker lengths shared across eukaryotes promote phase separation of chromatin, tune droplet properties, and coordinate to form condensates of consistent density in manners that parallel chromatin behavior in cells. Histone acetylation by p300 antagonizes chromatin phase separation, dissolving droplets in?vitro and decreasing droplet formation in nuclei. In the presence of multi-bromodomain proteins, such as BRD4, highly acetylated chromatin forms a new phase-separated state with droplets of distinct physical properties, which can be immiscible with unmodified chromatin droplets, mimicking nuclear chromatin subdomains. Our data suggest a framework, based on intrinsic phase separation of the chromatin polymer, for understanding the organization and regulation of eukaryotic genomes.
.
DOI:10.1007/s40484-016-0082-1URL [本文引用: 1]
.
DOI:10.1038/nature22989URLPMID:28636597 [本文引用: 3]

Constitutive heterochromatin is an important component of eukaryotic genomes that has essential roles in nuclear architecture, DNA repair and genome stability, and silencing of transposon and gene expression. Heterochromatin is highly enriched for repetitive sequences, and is defined epigenetically by methylation of histone H3 at lysine 9 and recruitment of its binding partner heterochromatin protein 1 (HP1). A prevalent view of heterochromatic silencing is that these and associated factors lead to chromatin compaction, resulting in steric exclusion of regulatory proteins such as RNA polymerase from the underlying DNA. However, compaction alone does not account for the formation of distinct, multi-chromosomal, membrane-less heterochromatin domains within the nucleus, fast diffusion of proteins inside the domain, and other dynamic features of heterochromatin. Here we present data that support an alternative hypothesis: that the formation of heterochromatin domains is mediated by phase separation, a phenomenon that gives rise to diverse non-membrane-bound nuclear, cytoplasmic and extracellular compartments. We show that Drosophila HP1a protein undergoes liquid-liquid demixing in vitro, and nucleates into foci that display liquid properties during the first stages of heterochromatin domain formation in early Drosophila embryos. Furthermore, in both Drosophila and mammalian cells, heterochromatin domains exhibit dynamics that are characteristic of liquid phase-separation, including sensitivity to the disruption of weak hydrophobic interactions, and reduced diffusion, increased coordinated movement and inert probe exclusion at the domain boundary. We conclude that heterochromatic domains form via phase separation, and mature into a structure that includes liquid and stable compartments. We propose that emergent biophysical properties associated with phase-separated systems are critical to understanding the unusual behaviours of heterochromatin, and how chromatin domains in general regulate essential nuclear functions.
.
DOI:10.1016/j.tig.2011.05.006URL [本文引用: 1]

Nuclear bodies including nucleoli, Cajal bodies, nuclear speckles, Polycomb bodies, and paraspeckles are membraneless subnuclear organelles. They are present at steady-state and dynamically respond to basic physiological processes as well as to various forms of stress, altered metabolic conditions and alterations in cellular signaling. The formation of a specific nuclear body has been suggested to follow a stochastic or ordered assembly model. In addition, a seeding mechanism has been proposed to assemble, maintain, and regulate particular nuclear bodies. In coordination with noncoding RNAs, chromatin modifiers and other machineries, various nuclear bodies have been shown to sequester and modify proteins, process RNAs and assemble ribonucleoprotein complexes, as well as epigenetically regulate gene expression. Understanding the functional relationships between the 3D organization of the genome and nuclear bodies is essential to fully uncover the regulation of gene expression and its implications for human disease.
.
DOI:10.1016/j.bpj.2018.03.011URLPMID:29628210 [本文引用: 1]

Chromatin is partitioned on multiple length scales into subcompartments that differ from each other with respect to their molecular composition and biological function. It is a key question how these compartments can form even though diffusion constantly mixes the nuclear interior and rapidly balances concentration gradients of soluble nuclear components. Different biophysical concepts are currently used to explain the formation of "chromatin bodies" in a self-organizing manner and without consuming energy. They rationalize how soluble protein factors that are dissolved in the liquid nuclear phase, the nucleoplasm, bind and organize transcriptionally active or silenced chromatin domains. In addition to cooperative binding of proteins to a preformed chromatin structure, two different mechanisms for the formation of phase-separated chromatin subcompartments have been proposed. One is based on bridging proteins that cross-link polymer segments with particular properties. Bridging can induce a collapse of the nucleosome chain and associated factors into an ordered globular phase. The other mechanism is based on multivalent interactions among soluble molecules that bind to chromatin. These interactions can induce liquid-liquid phase separation, which drives the assembly of liquid-like nuclear bodies around the respective binding sites on chromatin. Both phase separation mechanisms can explain that chromatin bodies are dynamic spherical structures, which can coalesce and are in constant and rapid exchange with the surrounding nucleoplasm. However, they make distinct predictions about how the size, density, and stability of chromatin bodies depends on the concentration and interaction behavior of the molecules involved. Here, we compare the different biophysical mechanisms for the assembly of chromatin bodies and discuss experimental strategies to distinguish them from each other. Furthermore, we outline the implications for the establishment and memory of functional chromatin state patterns.
.
DOI:10.1038/nrm2298URLPMID:18037899 [本文引用: 1]

Various chemical modifications on histones and regions of associated DNA play crucial roles in genome management by binding specific factors that, in turn, serve to alter the structural properties of chromatin. These so-called effector proteins have typically been studied with the biochemist's paring knife--the capacity to recognize specific chromatin modifications has been mapped to an increasing number of domains that frequently appear in the nuclear subset of the proteome, often present in large, multisubunit complexes that bristle with modification-dependent binding potential. We propose that multivalent interactions on a single histone tail and beyond may have a significant, if not dominant, role in chromatin transactions.
.
DOI:10.1016/j.cell.2013.03.035URL [本文引用: 2]

Master transcription factors Oct4, Sox2, and Nanog bind enhancer elements and recruit Mediator to activate much of the gene expression program of pluripotent embryonic stem cells (ESCs). We report here that the ESC master transcription factors form unusual enhancer domains at most genes that control the pluripotent state. These domains, which we call super-enhancers, consist of clusters of enhancers that are densely occupied by the master regulators and Mediator. Super-enhancers differ from typical enhancers in size, transcription factor density and content, ability to activate transcription, and sensitivity to perturbation. Reduced levels of Oct4 or Mediator cause preferential loss of expression of super-enhancer-associated genes relative to other genes, suggesting how changes in gene expression programs might be accomplished during development. In other more differentiated cells, super-enhancers containing cell-type-specific master transcription factors are also found at genes that define cell identity. Super-enhancers thus play key roles in the control of mammalian cell identity.
.
DOI:10.1016/j.cell.2017.02.007URLPMID:28340338 [本文引用: 1]

Phase-separated multi-molecular assemblies provide a general regulatory mechanism to compartmentalize biochemical reactions within cells. We propose that a phase separation model explains established and recently described features of transcriptional control. These features include the formation of super-enhancers, the sensitivity of super-enhancers to perturbation, the transcriptional bursting patterns of enhancers, and the ability of an enhancer to produce simultaneous activation at multiple genes. This model provides a conceptual framework to further explore principles of gene control in mammals.
DOI:10.1126/science.361.6409.1410URLPMID:30262504 [本文引用: 2]

Hox genes encode conserved developmental transcription factors that govern anterior-posterior (A-P) pattering in diverse bilaterian animals, which display bilateral symmetry. Although Hox genes are also present within Cnidaria, these simple animals lack a definitive A-P axis, leaving it unclear how and when a functionally integrated Hox code arose during evolution. We used short hairpin RNA (shRNA)-mediated knockdown and CRISPR-Cas9 mutagenesis to demonstrate that a Hox-Gbx network controls radial segmentation of the larval endoderm during development of the sea anemone Nematostella vectensis. Loss of Hox-Gbx activity also elicits marked defects in tentacle patterning along the directive (orthogonal) axis of primary polyps. On the basis of our results, we propose that an axial Hox code may have controlled body patterning and tissue segmentation before the evolution of the bilaterian A-P axis.
.
DOI:10.1016/0092-8674(85)90246-6URLPMID:3907859 [本文引用: 1]

We describe a new protein that binds to DNA and activates gene transcription in yeast. This protein, LexA-GAL4, is a hybrid of LexA, an Escherichia coli repressor protein, and GAL4, a Saccharomyces cerevisiae transcriptional activator. The hybrid protein, synthesized in yeast, activates transcription of a gene if and only if a lexA operator is present near the transcription start site. Thus, the DNA binding function of GAL4 can be replaced with that of a prokaryotic repressor without loss of the transcriptional activation function. These results suggest that DNA-bound LexA-GAL4 and DNA-bound GAL4 activate transcription by contacting other proteins.
.
DOI:10.1186/gb-2009-10-3-r29URLPMID:19284633 [本文引用: 1]

Unravelling regulatory programs governed by transcription factors (TFs) is fundamental to understanding biological systems. TFCat is a catalog of mouse and human TFs based on a reliable core collection of annotations obtained by expert review of the scientific literature. The collection, including proven and homology-based candidate TFs, is annotated within a function-based taxonomy and DNA-binding proteins are organized within a classification system. All data and user-feedback mechanisms are available at the TFCat portal (http://www.tfcat.ca).
DOI:10.1016/j.cell.2018.10.042URLPMID:30449618 [本文引用: 1]

Gene expression is controlled by transcription factors (TFs) that consist of DNA-binding domains (DBDs) and activation domains (ADs). The DBDs have been well characterized, but little is known about the mechanisms by which ADs effect gene activation. Here, we report that diverse ADs form phase-separated condensates with the Mediator coactivator. For the OCT4 and GCN4 TFs, we show that the ability to form phase-separated droplets with Mediator in?vitro and the ability to activate genes?in?vivo are dependent on the same amino acid residues. For the estrogen receptor (ER), a ligand-dependent activator, we show that estrogen enhances phase separation with Mediator, again linking phase separation with gene activation. These results suggest that diverse TFs can interact with Mediator through the phase-separating capacity of their ADs and that formation of condensates with Mediator is involved in gene activation.
.
DOI:10.1016/j.stem.2013.04.013URL [本文引用: 1]

The chromatin state of pluripotency genes has been studied extensively in embryonic stem cells (ESCs) and differentiated cells, but their potential interactions with other parts of the genome remain largely unexplored. Here, we identified a genome-wide, pluripotency-specific interaction network around the Nanog promoter by adapting circular chromosome conformation capture sequencing. This network was rearranged during differentiation and restored in induced pluripotent stem cells. A large fraction of Nanog-interacting loci were bound by Mediator or cohesin in pluripotent cells. Depletion of these proteins from ESCs resulted in a disruption of contacts and the acquisition of a differentiation-specific interaction pattern prior to obvious transcriptional and phenotypic changes. Similarly, the establishment of Nanog interactions during reprogramming often preceded transcriptional upregulation of associated genes, suggesting a causative link. Our results document a complex, pluripotency-specific chromatin "interactome'' for Nanog and suggest a functional role for long-range genomic interactions in the maintenance and induction of pluripotency.
.
DOI:10.1038/s41586-019-1517-4URLPMID:31462772 [本文引用: 1]

The regulated transcription of genes determines cell identity and function. Recent structural studies have elucidated mechanisms that govern the regulation of transcription by RNA polymerases during the initiation and elongation phases. Microscopy studies have revealed that transcription involves the condensation of factors in the cell nucleus. A model is emerging for the transcription of protein-coding genes in which distinct transient condensates form at gene promoters and in gene bodies to concentrate the factors required for transcription initiation and elongation, respectively. The transcribing enzyme RNA polymerase?II may shuttle between these condensates in a phosphorylation-dependent manner. Molecular principles are being defined?that rationalize transcriptional organization and regulation, and that will guide future investigations.
.
DOI:10.1038/s41586-019-1464-0URLPMID:31391587 [本文引用: 1]

The synthesis of pre-mRNA by RNA polymerase II (Pol?II) involves the formation of a transcription initiation complex, and a transition to an elongation complex1-4. The large subunit of Pol?II contains an intrinsically disordered C-terminal domain that is phosphorylated by cyclin-dependent kinases during the transition from initiation to elongation, thus influencing the interaction of the C-terminal domain with different components of the initiation or the RNA-splicing apparatus5,6. Recent observations suggest that this model provides only a partial picture of the effects of phosphorylation of the C-terminal domain7-12. Both the transcription-initiation machinery and the splicing machinery can form phase-separated condensates that contain large numbers of component molecules: hundreds of molecules of Pol?II and mediator are concentrated in condensates at super-enhancers7,8, and large numbers of splicing factors are concentrated in nuclear speckles, some of which occur at highly active transcription sites9-12. Here we investigate whether the phosphorylation of the Pol?II C-terminal domain regulates the incorporation of Pol II into phase-separated condensates that are associated with transcription initiation and splicing. We find that the hypophosphorylated C-terminal domain of Pol?II is incorporated into mediator condensates and that phosphorylation by regulatory cyclin-dependent kinases reduces this incorporation. We also find that the hyperphosphorylated C-terminal domain is preferentially incorporated into condensates that are formed by splicing factors. These results suggest that phosphorylation of the Pol?II C-terminal domain drives an exchange from condensates that are involved in transcription initiation to those that are involved in RNA processing, and implicates phosphorylation as a mechanism that regulates condensate preference.
