 ,上海交通大学农业与生物学院单细胞生物学联合研究中心,上海 200240
,上海交通大学农业与生物学院单细胞生物学联合研究中心,上海 200240Progresses on the structure and function of cohesin
Yu Zhang, Yuda Fang ,Joint Center for Single Cell Biology, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China
,Joint Center for Single Cell Biology, School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China通讯作者: 方玉达,博士,****,博士生导师,研究方向:植物细胞核与染色质的结构和功能。E-mail:yuda.fang@sjtu.edu.cn
编委: 史庆华
收稿日期:2019-11-7修回日期:2019-12-10网络出版日期:2020-01-20
| 基金资助: |
Editorial board:
Received:2019-11-7Revised:2019-12-10Online:2020-01-20
| Fund supported: |
作者简介 About authors
张雨,博士后,研究方向:染色质结构与功能。E-mail:zhangyu2065@sjtu.edu.cn。

摘要
Cohesin是一类在真核生物进化过程中保守的蛋白复合体,由4个重要亚基相互作用形成环状结构,在细胞分裂过程中参与维持染色体的有序排布。在动物中研究发现cohesin还可以作为分子间的连结器介导绝缘子/增强子-启动子间长距离交互,导致基因表达增强或者抑制,但在植物中关于cohesin在调控基因表达和维持染色体构象方面的研究却相对滞后。本文介绍了cohesin的结构特点和主要组成亚基,对调控cohesin在染色质上动态变化的相关因子进行了总结,并结合近年来植物中cohesin的功能研究和动物中cohesin在三维基因组及转录调控中的重要作用,展望了植物中cohesin在转录调控中的潜在功能。
关键词:
Abstract
Cohesin is an evolutionarily conserved protein complex in eukaryotes. The four subunits of cohesin form a ring structure that plays an important role in maintaining the orderly arrangement of chromatin during cell division. In addition, metazoan cohesin was found to act as an intermolecular linker, which regulates insulator/enhancer-promoter interactions, leading to either enhancement or inhibition of gene expressions. However, little is known about the role of cohesin in the transcriptional regulation in plants. In the review, we introduce the structure and core subunits of cohesin, and summarize the factors that regulate its dynamic changes on chromatin. Based on the functional study of plant cohesin in recent years and researches in animals about the roles of cohesin in the three-dimensional genome organization and transcriptional regulation, we prospect the potential functions of plant cohesin in regulating transcription.
Keywords:
PDF (551KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张雨, 方玉达. Cohesin结构及功能研究进展. 遗传[J], 2020, 42(1): 57-72 doi:10.16288/j.yczz.19-288
Yu Zhang.
细胞核是细胞遗传与代谢的调控中心,遗传物质DNA有序且密集地分布其中。人类细胞核基因组的物理长度约102 cm,即使基因组较小的拟南芥(Arabidopsis thaliana),其细胞核基因组也有3.8 cm,而这些DNA通过折叠浓缩后储存在仅有几微米的细胞核内。染色体经过折叠形成有序的三维结构,这一过程很大程度上依赖染色体结构维持蛋白(structural maintenance of chromosomes, SMC)的调控[1,2,3]。SMC复合体从真菌、植物到人类都非常保守,包括cohesin、condensin和SMC5/6三大类。Condensin的功能主要与染色体内部的凝聚相关,当人类细胞敲除condensin后,导致染色体不能凝聚,不能形成正常姐妹染色单体,在分裂后期姐妹染色单体也不能正常分离。SMC5/6功能主要与DNA的损伤修复相关。关于cohesin的功能,早期人们研究发现其在酵母细胞有丝分裂和减数分裂过程中都发挥重要功能。在分裂过程中,cohesin可以维持染色体的正常形态,保证姐妹染色单体及同源染色体在细胞的不同分裂时期正确分布[4,5,6]。而在间期,cohesin维持染色质形成不同的空间结构,调控基因表达,还与DNA复制、DNA损伤修复相关[5,7~9]。最近的研究还发现cohesin介导的染色质环挤出动态过程对RAG (recombination-activating gene)扫描损伤位点起到促进作用,并在数量众多的V(D)J (variable- diversity-joining)重排和交错转化重组(cross switch recombination, CSR)过程中发挥重要作用[10]。
Cohesin在维持染色质构象及调控转录方面的研究也成为三维基因组学和表观遗传学研究的热点。本文在介绍cohesin结构特点、主要组成亚基及其功能的基础上,对cohesin在染色质上从招募到稳定结合,再到解离过程中调控其动态变化的作用因子进行了总结,并结合近年来在哺乳动物及酵母中的相关研究,讨论了cohesin在植物与动物中功能的保守程度,对植物cohesin在基因表达调控中的潜在功能进行了展望。
1 Cohesin结构
1.1 SMC蛋白复合体的结构特点
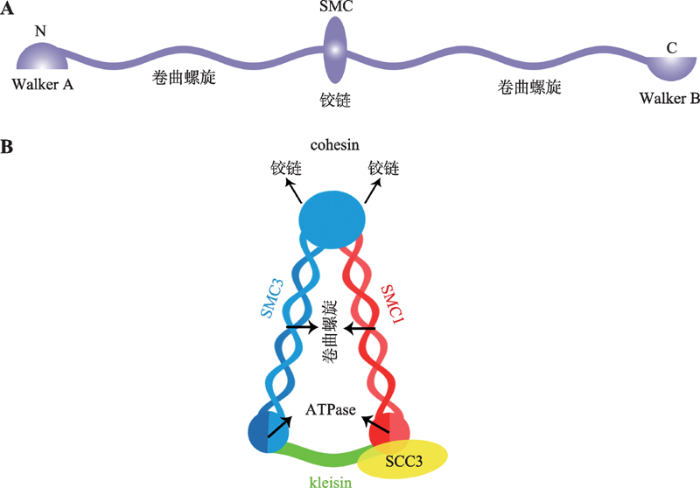
真核生物的SMC复合体都是在两个SMC蛋白组成的异源二聚体基础上形成的[11,12,13]。每个SMC 蛋白由1000~1500个氨基酸组成,中间是球状的铰链(hinge)结构域,铰链结构域两侧延伸形成卷曲螺旋(coiled-coils)结构域[14],卷曲螺旋结构域终端分别为Walker A和Walker B结构域,即SMC蛋白N端的Walker A和C端的Walker B结构域。Walker A含有核苷酸结合结构域(nucleotide-binding domain, NBD),Walker B含有与典型ATP酶同源的ATP结合结构域(ATP-binding cassette, ABC)。单个SMC蛋白以hinge结构为中心,两侧的coiled-coils结构域反向平行相互作用在一起,这使得SMC的N端Walker A和C端Walker B结构域相互靠近在一起,形成有功能ATP酶(ATPase)结构域(图1,A和B)[15,16,17]。SMC的ATPase位点对于整个SMC蛋白复合体在DNA上的结合和解离至关重要[18]。1.2 Cohesin主要亚基
Cohesin是SMC复合体中的一类,由SMC1、SMC3和SCC3 (在动物中是Rad21)以及kleisin亚基组成的环状套索结构[14,19]。其中,SMC1与SMC3是典型的SMC 蛋白,SMC1和SMC3的hinge结构域相互作用形成V形的异源二聚体,底部由kleisin亚基将两个SMC蛋白的ATP酶结构域连接形成闭合环状V形复合体(图1B)[20,21]。Kleisin亚基与SCC3亚基相互作用,进而招募SCC3形成完整的cohesin蛋白复合体[22]。酵母中发现SCC3 (SA2)的C端与kleisin相结合。蛋白结构分析发现,SCC3内部凹面可以与kleisin (Rad21/Scc1-M)亚基中间很大一段相互作用[23]。目前在植物中还没有关于cohesin各亚基间相互作用的报道。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1SMC类蛋白及cohesin的结构示意图
A:SMC类蛋白保守结构域示意图;B:Cohesin结构示意图。根据参考文献[18]绘制。
Fig. 1The diagram of SMC proteins and the cohesin complex
拟南芥cohesin的AtSMC1和AtSMC3亚基与酵母和哺乳动物SMC家族相比蛋白同源性很高。拟南芥atsmc1和atsmc3单突纯合突变体种子在发育过程中胚和胚乳都存在严重缺陷[24,25,26],胚胎发育早期就死亡,由此可见cohesin在胚胎发育早期即已经发挥着重要作用。拟南芥中AtSCC3不存在基因冗余现象,与酵母中SCC3蛋白有40%的同源性。动物中SCC3亚基含有HEAT-repeat (Huntingtin, elongation factor 3, protein phosphatase 2A)结构域[27],而拟南芥中AtSCC3却不含有HEAT-repeat结构域。拟南芥atscc3纯合突变体在胚胎发育早期缺陷致死,Chelysheva等[28]发现Ws(Wassileskija)拟南芥弱的atscc3-1突变体植株与野生型相比表现出矮小、晚花、育性降低、有丝分裂及减数分裂均发生异常。
Kleisin亚基在拟南芥、水稻(Oryza sativa L.)和玉米(Zea mays L.)中均有研究。在拟南芥和水稻中kleisin 亚基的4个同源蛋白相对保守,玉米中仅有AFD1一个同源蛋白,酿酒酵母(Saccharomyces cerevisiae)及脊椎动物中kleisin 亚基有RAD21和REC8两个同源蛋白(表1)[29]。拟南芥中Kleisin亚基的4个同源蛋白分别为:AtSYN1、AtSYN2、AtSYN3和AtSYN4[30,31,32,33]。Atsyn1突变体雌雄配子不育,但其营养生长等生长发育过程均正常[34,35],表明AtSYN1蛋白主要在减数分裂形成配子过程中发挥重要功能[34,36]。最近研究发现,拟南芥第一次减数分裂过程中cohesin维持在着丝粒区域依赖两个蛋白磷酸化酶对AtSYN1的去磷酸化作用[37,38]。AtSYN3主要定位在核仁,与rDNA结构维持及rRNA转录和加工成熟有关[33]。AtSYN2和AtSYN4在有丝分裂中发挥重要功能。AtSYN2与种子萌发过程中DNA损伤后修复相关[35],而AtSYN4与苗期体细胞DNA损伤修复相关[39]。酵母双杂交实验证明,AtSYN4 也可以与磷酸化酶PP2A B'α、PP2AB'β和PP2AB'ζ相互作用,磷酸化酶与AtSYN4的相互作用可能与有丝分裂过程中cohesin在着丝粒上的维持相关[37,38]。
水稻中RAD21-4/OsREC8是酵母中REC8的同源蛋白,OsREC8在减数分裂过程中保证同源染色体正确的配对及联会(表1)。Osrec8突变体及OsRAD21- 4RNAi植株在营养生长阶段与野生型相比无明显差异,但育性均显著降低[40,41]。OsRAD21-1在水稻各组织中均有表达,但在花和芽中表达明显高于叶和根。OsRAD21-3在花中高表达。OsRAD21-3RNAi植株的花粉有丝分裂异常,染色体不能正常分离,且花粉活力严重降低,但花粉的减数分裂并无异常,表明OsRAD21-3在花粉减数分裂后的有丝分裂过程中发挥作用[42]。通过原位杂交发现OsRAD21-2 多在细胞分裂旺盛的组织中高表达,且异位表达OsRAD21-2后水稻细胞生长迟缓、植株发育异常[43]。
玉米AFD1是REC8的同源蛋白,其功能与同源染色体配对、联会复合体的形成及RAD51在染色体上的分布有关。AFD1会影响染色体在细线期及偶线期的分布。Afd1突变体中染色体在偶线期不能呈“花束”形态(bouquet formation)分布,减数分裂发生异常[44]。
2 调节cohesin在染色质上动态变化的因子
Cohesin在细胞分裂过程中重要的功能是维持姐妹染色单体有序地分布。在显微镜下可以观察到,在细胞分裂前期到中期cohesin都结合在染色体臂以及着丝粒区域,维持两条姐妹染色单体粘连在一起,在分裂中后期cohesin从染色体臂上解离下来,末期着丝粒上的cohesin也解离下来,姐妹染色单体得以正常分离。在转录过程中cohesin也随着RNA聚合酶及转录因子从转录起始位点向转录终止位点移动[45,46]。可见cohesin在染色质上的结合是动态变化的。Cohesin在染色质上的动态变化在拟南芥,酵母,线虫(Caenorhabditis elegans)和人类(Homo sapiens)中均有相关研究[4~6,47,48]。Cohesin的动态变化依赖很多蛋白,如:SCC2负责在DNA上招募cohesin,而cohesin在染色质上的维持依赖CTF7/ECO1 (chromosome transmission fidelity/establishment of cohesion 1)。另外,WAPL (wings apart-like protein)和PDS5 (precocious dissociation of sisters protein 5)因子与cohesin从DNA上解离有关(表1)。Table 1
表1
表1Cohesin亚基及相关调控因子
Table 1
| 类型 | 酿酒酵母 | 线虫 | 脊椎动物 | 拟南芥 | 水稻 | 玉米 |
|---|---|---|---|---|---|---|
| cohesin 亚基 | PSM1 | HIM-1 | SMC1α SMC1β | SMC1 | SMC1 | |
| PSM3 | SMC-3 | SMC3 | SMC3 | SMC3 | ||
| RAD21 REC8 | SCC-1/ COH-2 COH-1 COH-3 REC8 | RAD21 RAD21L REC8 | SYN2/ RAD21.1 SYN4/ RAD21.3 SYN3/ RAD21.2 SYN1/DIF1 | RAD21-1 RAD21-2 RAD21-3 RAD21-4 /OsREC8 | AFD1 | |
| PSC3 REC11 | SCC3 | SA1 SA2 STAG3 | SCC3 | SCC3* | ||
| 加载因子 | MIS4 | NIPBL | NIPBL | SCC2 | OsJ_22834* OsI_24633* | |
| SSL3 | Mau-2 | SCC4/MAU2 | SCC4 | DEK15 | ||
| 维持和解离因子 | CTF7/ ECO1 | ESCO1 ESCO2 | CTF7 | OsI_19739* | ESC01* | |
| PDS5 | ELV-14 | PDS5A PDS5B | PDS5E PDS5B PDS5D PDS5C PDS5A | |||
| WPL1 | WAPL-1 | WAPAL | WAPL1 WAPL2 | OsI_34181* | ACL54412* | |
| CUT2 | IFY-1 | Securin/ PTTG | ||||
| CUT1 | SEP-1 | Separase/ ESP1 | AtESP1 | OsI_09098* OsI_08535* | ||
| PLO1 | POLO | PLK1 | ||||
| SGO2 SGO1 | SGO1 | SGO2 SGO1 | SGO1 SGO2 | OsSGO1 OsSGO2* | ZMSGO2 ZMSGO1 |
新窗口打开|下载CSV
2.1 Cohesin在染色质上的加载
在DNA复制开始前,cohesin的加载因子SCC2和SCC4先在染色质上结合,进而招募cohesin在染色质上结合[6,47,49,50]。在酿酒酵母中,SCC4可以稳定SCC2在染色质上的结合[49,51],两者作用在一起形成cohesin的加载因子,从而招募cohesin[6,52]。Cohesin与SCC2在DNA上的结合位点并不是随机的,两者染色质免疫共沉淀-高通量测序(ChIP-seq)的分析结果发现它们各自结合位点可能没有重叠[53,54],这可能由于cohesin最初是依赖SCC2与DNA的结合,但cohesin在染色质上的结合位置是动态变化的,cohesin会在其他因子的作用下移动,如间期cohesin随着转录过程中RNA聚合酶在染色质上移动。Cohesin多分布在转录相对活跃的地方,且随转录过程在转录终止区域富集[55,56,57]。SCC2与cohesin结合的程度会影响cohesin在染色质上的移动。当SCC2突变后,cohesin在转录起始位点的结合能力也降低[58]。SCC2和SCC4影响cohesin在染色质上结合的具体机制还不完全清楚。酵母研究发现,SCC2和SCC4突变后,完整的cohesin环可以形成,但不能与DNA结合。Scc2和scc4突变体的表型与SMC1和SMC3的ATPase结构域突变后的表型类似,即cohesin可以形成完整的环状复合体,但也不能结合DNA。据此推测,SCC2可以加强SMC3和SMC1的ATPase活性以及催化DNA形成容易被cohesin有效结合的拓扑异构结构,进而影响cohesin在染色质上的结合[14,21,59~61]。拟南芥中cohesin加载因子的同源蛋白为AtSCC2和AtSCC4。AtSCC2与动物中同源蛋白有20%的同源性,除了有动物中共有的HEAT-repeat结构域外,AtSCC2还有植物中特有的植物同源结构域(plant homeodomain, PHD)。PHD结构域与组蛋白表观修饰以及基因表达调控相关[62,63]。在植物中,SCC2也是非常重要的蛋白。拟南芥atscc2纯合突变体在种子形成过程中胚乳过度增生分裂、发育异常、胚胎早期致死[62]。拟南芥atscc4纯合突变体胚胎在心形胚形成阶段不能对称分裂,胚柄处过度增生[64]。拟南芥atSCC2 RNAi植株中可观察到减数分裂过程中染色体分离紊乱,同时结合在染色质上的AtSCC3蛋白也减少,并出现姐妹染色单体黏连,染色体桥及分裂后细胞中染色体数目异常的现象[62]。在atscc2atscc4双突变体背景下,生长素报告基因pDR5rev::3xVENUS-N7被限制在胚柄底部细胞中表达,而野生型中报告基因在胚柄顶部细胞中表达。这表明AtSCC2和AtSCC4的缺失会导致胚胎发育过程中胚柄细胞胚胎潜能的改变。植物和酵母中都发现,SCC4可以与SCC2的N端稳定地相互作用在一起,但植物AtSCC4与AtSCC2之间的相互作用不会影响AtSCC4的定位。拟南芥AtSCC2的突变并没有改变植物体细胞核中AtSCC4的定位[54]。此外,有丝分裂间期AtSCC4与kleisin亚基AtSYN4共定位[54],而AtSCC2的主要功能被认为在减数分裂过程中影响cohesin的定位[62],这表明在拟南芥中AtSCC4与AtSCC2功能存在特异性。最近研究发现,玉米中DEK15是SCC4的同源蛋白。在dek15突变体中,姐妹染色单体形态异常,非整倍数细胞增多,且种子胚乳发育异常,胚胎早期死亡率增加。玉米DEK15对于染色体精确的分离非常重要,且可以协同染色质重塑因子促进cohesin在染色质上的结合[65]。
2.2 Cohesin在染色质上的维持
在有丝分裂S期前,cohesin在SCC2和SCC4的招募下与DNA结合。从S期到分裂中期,cohesin一直结合在染色体臂及着丝粒上,维持姐妹染色单体连接在一起,直至后期cohesin从染色体上解离下来。在这个过程中,cohesin复合体在染色质上的维持依赖几个关键蛋白:ECO1 (establishment of cohesion 1)又称为CTF7 (chromosome transmission fidelity 7),以及sororin因子。酵母中CTF7/ECO1是乙酰转移酶,在S期可以对SMC3的head结构域的两个赖氨酸残基进行乙酰化修饰[66,67,68,69]。SMC3的ATPase位点K112和K113位被乙酰化后,ATPase结构域关闭,使kleisin亚基与SMC亚基结合紧密,进而使cohesin环状结构稳定[68,69,70]。SMC3的这两个赖氨酸残基位点在多种生物中都是非常保守的,在人体细胞中,ESCO1 (establishment of sister chromatid cohesion N-acetyltransferase 1)和ESCO2两个乙酰化酶同样可以乙酰化SMC3[69,70]。酵母CTF7缺失会造成染色质状态混乱,导致cohesin在染色体臂及着丝粒上分布异常,以及细胞周期异常[71,72]。酵母CTF7/ECO1与增值细胞核抗原(proliferating cell nuclear antigen, PCNA)和复制因子C (replication factor C, RFC)复合体直接相互作用,这表明在姐妹染色单体形成过程中,DNA的复制和cohesin作用下的姐妹染色单体粘连是同时进行的[73,74]。
在脊椎动物中,还存在另外一个对cohesin与染色质的稳定结合起到重要作用的sororin因子。由于一些解离因子的存在,仅仅乙酰化的SMC3不足以让cohesin在复制过程中稳定地结合在染色质上,还需要乙酰化结合蛋白sororin来维持整个复合体的稳定。Sororin含有FGF结合序列,可以结合在PDS5 (precocious dissociation of sisters 5)蛋白上,进而起到稳定cohesin-DNA的作用[75,76,77]。在裂殖酵母(Schizosaccharomyces pombe)中,PDS5可以加强SMC3的乙酰化[78]。PDS5在间期与sororin相互作用,有协助cohesin结合DNA,并有维持cohesin与DNA稳定结合的功能。在后期,PDS5与解离因子相互作用,促进cohesin从DNA上解离下来,可见PDS5与不同因子相互作用发挥的功能也不同[79,80,81]。
拟南芥AtCTF7可以互补酵母ctf7突变体表型[82,83,84],这表明cohesin在细胞分裂过程中的功能在拟南芥和酵母中是非常保守的。AtCTF7包含PIP-BOX (PCNA-interacting protein BOX)、一个C2H2锌指蛋白结构域和一个乙酰转移酶结构域[82]。与其他生物相同,AtCTF7功能也有剂量效应,atctf7+/-杂合体雄配子异常,小孢子母细胞发育正常,植物营养生长无明显异常,但育性降低。完全缺失AtCTF7的突变体拟南芥表现出严重的生长缺陷表型:胚胎在发育到球形胚阶段就严重畸形,仅能获得少数纯合植株,表现出极矮小、不育的表型,同时cohesin在染色质上的结合明显减少[82,83]。过表达CTF7也会导致拟南芥胚珠在发育早期死亡[85]。
2.3 Cohesin从染色质上解离
WAPL是调控cohesin从染色质上的解离下来的关键因子。有丝分裂中后期,cohesin开始逐渐从 染色体臂上解离下来,仅保留在着丝粒区域。起始cohesin从染色体臂上解离下来的过程与SCC3亚基的磷酸化相关,这个磷酸化过程依赖于WAPL解离因子[86]。有丝分裂后期,SCC3与sororin被磷酸化,磷酸化后的sororin不再与PDS5相互作用,PDS5与解离因子WAPL相互作用,PDS5-WAPL复合体促进cohesin从染色体壁上解离下来。Cohesin从着丝粒上解离下来的过程依赖蛋白酶对kleisin亚基的水解,整个过程WAPL-PDS5-SCC3协同发挥作用[79,87,88]。拟南芥中PDS5有5个同源基因,在不同器官中检测AtPDS5表达量,发现在种子成熟过程中其表达量明显下降。当植株被γ射线照射后,AtPDS5表达上升。敲除AtPDS5后,减数分裂只轻微受到影响,但是DNA的同源重组修复能力明显减弱[89]。拟南芥中WAPL有两个同源基因AtWAPL1和AtWAPL2[90],而AtCTF7仅有一个拷贝[82],分子及遗传学实验证明AtWAPL和AtCTF7二者功能拮抗[91]。AtWAPL1和AtWAPL2 T-DNA插入突变体在植物生长发育以及育性方面都没有异常[90],Atwapl1-1 atwapl2纯合双突变体在营养生长阶段与野生型相比没有差异,但雌配子雄配子活性下降,植株育性降低。在减数分裂方面,双突变体的同源染色体配对异常,纺锤体形成异常,且cohesin在染色体臂上滞留,出现黏连在一起的姐妹染色单体,在后期不能正常分离[90]。WAPL在许多生物有丝分裂过程中发挥重要功能,减数分裂中的研究较少。对拟南芥AtWAPL的研究发现,其在植物减数分裂中同样发挥重要功能。拟南芥atctf7+/-杂合子突变体植株育性降低,纯合突变体植株生长发育严重缺陷,并且不育[83]。Kuntal De等[91]在研究AtCTF7和AtWAPL功能时发现,将atwapl1-1 wapl2 纯合突变体与atctf7+/-突变体杂交,获得atwapl1-1 wapl2 ctf7三突纯合突变体,其生长发育与野生型无明显差异,但育性比atwapl1-1 wapl2和atctf7+/-低,可见AtWAPL蛋白缺失可以抵消atctf7突变体在有丝分裂过程中cohesin不能结合到染色体上的缺陷。同时表明作为调控cohesin动态变化的因子,AtWAPL和AtCTF7在功能上相互拮抗。
3 Cohesin功能
早期关于cohesin的研究大多集中在细胞分裂过程中,其中在有丝分裂和减数分裂过程中cohesin对于姐妹染色单体间有序的凝聚在一起发挥着重要功能。近期研究表明cohesin还可以在分子间起到连接的作用,在长距离范围内影响DNA的交互,进而调控转录。另外,cohesin在DNA损伤修复方面也发挥重要功能,Scc1亚基就是在酵母中筛选易发生DNA损伤突变体时发现的[92,93]。对非洲爪蟾(Xenopus laevis)和鸡(Gallus gallus)的细胞进行持续γ射线照射会导致染色体的断裂,此过程伴随着cohesin在DNA上的结合增多,以及cohesin动态变化会更加活跃[94]。3.1 Cohesin在细胞分裂中的功能
一个细胞在分裂成两个不同细胞的过程中需要很多蛋白协同发挥作用,并要经历几个重要时期以确保正常的细胞能继续完成整个细胞周期,阻止异常的细胞进行分裂。有丝分裂过程中,G1期需要完成细胞健康与否的分拣,正常的细胞进入S期,异常的细胞不再进行分裂。G2期确保细胞完成了正确的DNA复制过程,才能进入分裂期。在S期染色体经历了复制过程,产生两个一样的姐妹染色单体。从S期DNA开始复制起cohesin就将两个姐妹染色单体有序地黏连在一起,直到分裂后期才完全从染色体上解离下来。这个机制在所有真核生物中都是非常保守的[95,96,97,98,99]。体细胞进行有丝分裂的过程中,G1期SCC2和SCC4招募cohesin与DNA结合,这个过程也依赖SMC蛋白ATP水解酶活性。SMC1和SMC3形成的hinge结构是DNA链进入cohesin环的“入口”[60]。Cohesin与DNA结合后,从间期到中期,在染色体上的维持依赖于ECO1/CTF7这个乙酰转移酶对SMC3亚基的乙酰化作用,以及sororin-PDS5蛋白的结合抑制了WAPL蛋白打开cohesin环的作用[77,100,101]。在S期,cohesin在DNA上的加载与DNA的复制过程协同进行[102]。在前期-中期转换的阶段,染色体臂上的cohesin开始解离下来,这个过程依赖一些有丝分裂激酶的作用。以哺乳动物为例,cohesin的SA (SCC3)亚基被Plk1磷酸化以及sororin蛋白被Cdk1和Aurora B磷酸化都与cohesin从染色体臂上的解离相关,其中WAPL也发挥重要作用[103,104]。但在有丝分裂后期姐妹染色单体分离之前,cohesin会一直结合在着丝粒上,此时SGO1以及PP2A会保护SA及sororin不被磷酸化,从而使cohesin维持在着丝粒上[103,105]。中后期纺锤体上的微管向细胞两极牵引,此时着丝粒上的cohesin产生的内聚力可以抵消掉部分纺锤体的牵引力。在中期赤道板上的姐妹染色单体有了分别向两极移动的重新定向,确保染色体可以正常移动到两极后,才进行后期着丝粒解凝聚。这时cohesin的kleisin亚基在蛋白水解酶作用下水解,致使cohesin从着丝粒上解离下来,姐妹染色单体向两极移动[12,106]。
在减数分裂过程中cohesin同样发挥着重要作用。在哺乳动物生殖细胞中,与体细胞相比cohesin的SMC1α亚基及SA1和SA2亚基绝大多数被SMCβ及STAG3/SA3代替,SMCβ及STAG3/SA3是减数分裂特异的cohesin亚基[107,108,109]。生殖细胞中的kleisin亚基为REC8和RAD21L,这也是哺乳动物中减数分裂特异的亚基(表2)。减数分裂过程中cohesin在DNA上的结合和维持过程同样是依赖SCC2和SCC4、PDS5以及sororin,且这些调控因子在减数分裂和有丝分裂中的功能保守[110,111,112]。在减数分裂过程中,kleisin亚基与cohesin在染色质上的时空分布相关。在哺乳动物减数分裂前期,REC8类cohesin在DNA复制前结合到染色质上,大量REC8类cohesin与DNA的结合会贯穿整个减数分裂过程,直到第二次减数分裂中期。而RAD21L类cohesin大多是在DNA复制完成之后与染色体结合,且在第一次减数分裂的粗线期后期就从染色体上解离下来[113,114,115]。减数分裂过程中cohesin从染色体上的解离过程同样依赖WAPL[116],其机制也与有丝分裂相同。在酿酒酵母中,减数分裂SMC亚基与有丝分裂亚基相同,都为PSM1和PSM2。Klesin亚基与有丝分裂不同,为减数分裂特异的REC8,有丝分裂中的PSC3亚基在减数分裂中为REC11。目前已知拟南芥cohesin亚基中只有SYN1是减数分裂特有的(表2)。
Table 2
表2
表2Cohesin亚基在有丝分裂及减数分裂中的比较
Table 2
| cohesin 亚基 | ||
|---|---|---|
| 有丝分裂 | 减数分裂 | |
| 酿酒酵母 | PSM1 | PSM1 |
| PSM3 | PSM3 | |
| RAD21 | REC8 | |
| PSC3 | REC11 | |
| 脊椎动物 | SMC1α | SMC1β |
| SMC3 | SMC3 | |
| RAD21 | RAD21L REC8 | |
| SA1 SA2 | STAG3 | |
| 拟南芥 | SMC1 | SMC1 |
| SMC3 | SMC3 | |
| SYN2 SYN4 SYN3 | SYN1 | |
| SCC3 | SCC3 | |
新窗口打开|下载CSV
无论是有丝分裂还是减数分裂,cohesin对维持姐妹染色单体凝聚在一起发挥着重要功能,这种凝聚力从间期DNA复制开始一直持续到中后期姐妹染色单体分开。如果缺少了分子间的凝聚力,会导致基因组不稳定、非整倍体细胞增多、DNA修复力下降、染色体异位等异常[117,118,119]。
3.2 Cohesin在维持染色质构像及基因表达调控中的功能
最早是在果蝇(Drosophila melanogaster)中发现cohesin具有转录调控的功能。Cohesin的加载因子 Nipped-B(SCC2)发生突变后,Homeo box基因的表达受到抑制,Nipped-B可以介导Homeo box基因区域增强子-启动子的相互作用。如果cohesin不能结合到Homeo box基因上,启动子不能与增强子互作,基因转录水平降低[120]。同样,当人缺失了cohesin 加载因子CdLS(SCC2)会造成科妮莉亚·德·兰格发育综合征(Cornelia de Lange syndrome),这是一种引起上肢发育畸形、智力缺陷的疾病,其致病原因是由于CdLS的缺失导致下游基因转录调控异常[121,122]。CTCF(CCCTC-binding factor)是协同cohesin维持染色质三维结构及调控转录的关键因子。染色质在细胞核内相互作用形成拓扑异构相关结构域(topologically associating domain, TAD),TADs是与染色质三维结构功能相关的重要区域,TADs内部染色质交互密集,TADs之间染色质交互频率低[123]。有研究提出TAD的主要作用是限制启动子和增强子间的相互作用[124,125]。不同TAD之间被边界区域(boundary)隔开,边界区域富集CTCF和cohesin(图2)[126],且多富集转录相对活跃的管家基因[127,128,129,130]。边界区域基因表达相对活跃,与染色质结构相对松散,以及富集着一些与活跃染色质相关的组蛋白修饰标记(H3K4me3和H3K36me3)相关。
图2
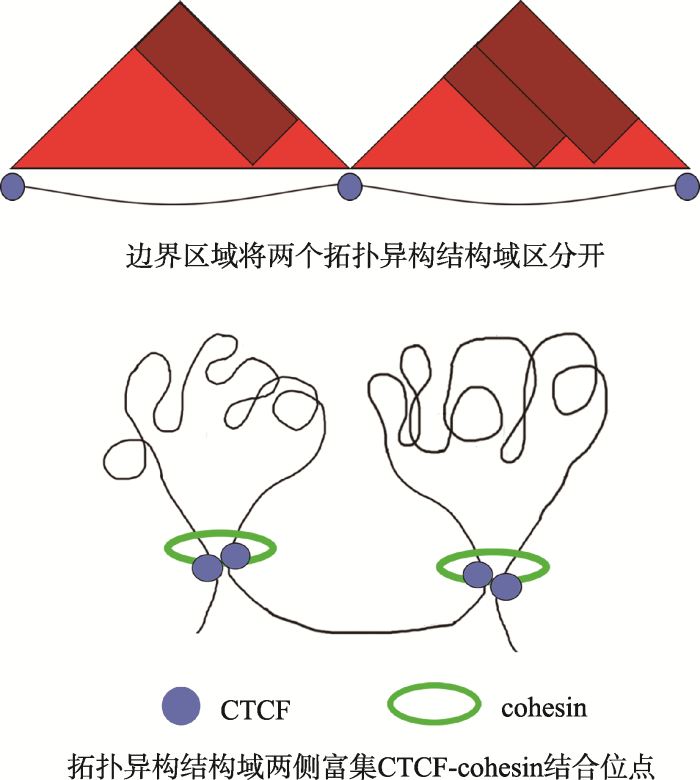 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2拓扑异构结构域的二维结构示意图
根据参考文献[126]绘制。
Fig. 2The two-dimensional structure diagram of TADs
拟南芥中,染色体组织形态上没有明显的TAD。同时,拟南芥中也缺少动物中经典的CTCF绝缘蛋白,这与拟南芥中缺少典型的TAD存在相关性。仅有很少的可信证据表明在拟南芥中存在类似于绝缘元件的DNA (insulator-like DNA)序列。然而,在对拟南芥进行高分辨率的全基因组染色质构象捕获(Hi-C)后发现超过1000个类似TAD(TAD-like)的区域[131]。拟南芥中这些区域和动物中的TAD有着相似的特性:在TAD内部,染色质交互密集;在TAD之间,染色质的交互受到限制。同样它们在染色体松散的地方以及基因表达活跃的地方富集[131,132]。但植物中还没有关于cohesin与三维基因组的相关报道。
研究发现cohesin加载因子、SMC和kleisin不同亚基在全基因组上的结合位点与CTCF有显著重叠,并且cohesin与CTCF共同对这些基因转录起到抑制的作用。尽管CTCF和cohesin在很多环状DNA结构处共同结合,但是它们在维持染色质构象上的功能不尽相同。CTCF与转录抑制相关,而cohesin除了与CTCF共同作用的位点外,还在很多基因位点与转录激活相关[127]。根据染色质包装紧密程度可以将cohesin的结合位点分类:在包装紧密的DNA结合位点,通常是cohesin与CTCF共同结合的位点;染色质包装松散的DNA结合位点,通常没有CTCF结合,这些区域大多为启动子或增强子区[133,134,135,136,137,138,139]。Cohesin还和一些其它的调控因子如调控蛋白复合体(mediator complex)相互作用发挥转录激活作用[133,134,135,136,137]。可见cohesin作为分子间桥梁,通过影响长距离范围内DNA上调控元件如:绝缘子/增强子-启动子(insulator/enhancer-promoter)之间的染色质交互来调控转录。Cohesin将增强子-启动子拉近在一起时,可以起到转录激活作用,此时cohesin多与转录因子或mediator共同起作用;当cohesin将绝缘子-启动子拉近在一起时,可以起到转录抑制功能(图3)[18,140],此时cohesin多与CTCF共同发挥作用。
图3
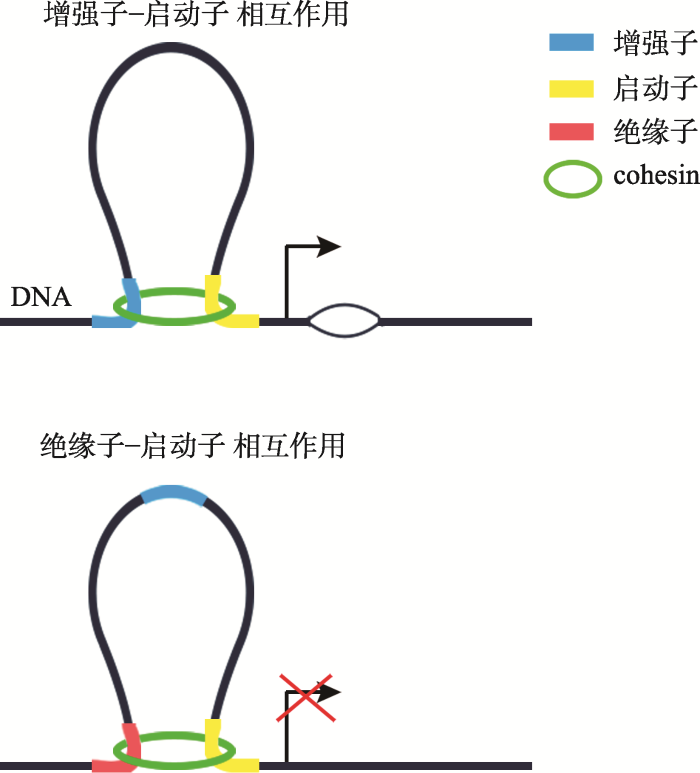 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3Cohesin在基因表达调控中的功能
根据参考文献[18]绘制。
Fig. 3Models of the functions of cohesin in transcriptional regulation
在复制过程中关于cohesin在染色质上结合机制的研究相对较多,最近Murayama等[141]人用详尽巧妙的体外实验探索了cohesin在DNA复制过程中动态结合DNA的机制:cohesin在加载因子及ATP的存在下可以结合在双链DNA上,当cohesin先结合一分子DNA双链上后,仅能再结合一分子单链DNA,这个过程也是依赖加载因子和ATP的作用。Cohesin结合了单链DNA后,体外再给予单链DNA、DNA聚合酶和dNTP等条件会稳定整个DNA-cohesin结构。这个过程成功模拟了复制叉形成及推进过程中cohesin动态结合DNA的过程[142,143]。而在转录过程中,是否会形成DNA-cohesin-RNA复合体,目前还不是很清楚,还没有直接的证据表明cohesin能在转录过程中可以沿着DNA移动。最近Peters和他的同事利用遗传学结合基因组学方法研究发现在转录过程中cohesin在转录复合体的作用下可以随转录进程移动。在ctcf突变体中,cohesin在转录起始位点结合增加30%。在ctf7 wapl双突变体中,cohesin在转录终止位点下游滞留。基因的转录程度不同,cohesin的分布也随之不均一,转录活跃区域的基因上的cohesin会被推到转录不活跃区域[144]。这些结果暗示转录过程中cohesin可能在PolII-TFs的推动下从转录起始位点向转录终止位点移动。全基因组范围内PolII-ChIP-seq,RAD21-ChIP-seq也表明cohesin不仅可以通过控制长距离范围内DNA上转录元件的结合调控转录,也可以直接与转录复合体等相关因子在转录起始位点发挥作用[144]。
4 结语与展望
对cohesin复合体研究至今已有30多年的时间,除了其主要组成亚基SMC1、SMC3、kleisin和SCC3以外,许多与其功能相关的因子也被发现,包括cohesin加载因子SCC2、SCC4和解离因子WAPL等。这些研究使得cohesin在细胞分裂过程中的功能及机制逐渐清晰。在细胞周期中,cohesin对于维持染色体的正常形态和有序排布是至关重要的。在拟南芥和水稻中,cohesin在有丝分裂及减数分裂中的功能与酵母,哺乳动物高度保守。此外,cohesin在植物中对于胚胎发育、育性及DNA损伤修复也发挥重要作用。在cohesin的作用下,两条姐妹染色单体靠凝聚力联系在一起,这对于染色体在整个细胞周期中正确的动态变化和正确的分布是至关重要的。在真核生物有丝分裂过程中,DNA复制同时cohesin就已经开始发挥作用。在S期DNA成功复制后就形成了联系在一起的姐妹染色单体,直到末期cohesin从染色体上解离下来,新的子细胞形成。在第一次减数分裂中期,cohesin确保联会合复合体形成,保证第一次减数分裂后期同源染色体间可以正常交换和分离。当植物缺失cohesin的SMC亚基后在胚胎发育早期就死亡;一些敲低cohesin表达的植物有丝分裂,减数分裂染色体形态分布严重异常,育性明显降低。这都表明,cohesin对于一个物种的生存和繁衍有着重要的影响。
近些年,ChIP-seq技术及Hi-C技术的应用,为cohesin调控染色质间相互作用、影响基因表达提供了很多证据。在动物中,cohesin是一个研究染色质长距离交互、三维基因组与转录调控关系的重要蛋白复合体,在植物中却缺少相关研究。在哺乳动物中发现cohesin与染色质构像及转录调控相关功能与CTCF这个关键因子紧密联系,但在拟南芥中并不存在CTCF的同源蛋白。另外,拟南芥染色体组织形态上没有明显的TADs,但有超过1000个类似TAD的区域,并且这些TAD-like的区域性质与动物中TAD的特性相类似。拟南芥基因组中没有典型的TAD结构域,这可能与缺少CTCF相关,但TAD-like区域与TAD性质相似,推测拟南芥cohesin在维持三维基因组结构及转录调控中可能同样会发挥功能。动物细胞中发现cohesin与一些中介因子(mediator)、转录因子及转录复合体相互作用,并且它们在基因组上有显著共同结合位点。在植物缺少CTCF的情况下,cohesin是否能与一些其他类型转录因子相互作用来调控基因表达及是否参与植物三维基因组产生和维持是值得进一步研究的。拟南芥中有研究发现在AtSYN3 RNAi植株中,与同源染色体联合及染色体同源重组相关基因表达水平发生变化[145],以及在Atctf7突变体中MU1、COPIA28等基因的转录水平也发生了变化[83,85]。这些基因转录水平的变化是否直接由cohesin引起的并不清楚,其中的机制也没有研究,有待进一步探索。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/j.tig.2018.03.003URLPMID:29606284 [本文引用: 1]

What drives the formation of chromatin loops has been a long-standing question in chromosome biology. Recent work provides major insight into the basic principles behind loop formation. Structural maintenance of chromosomes (SMC) complexes, that are conserved from bacteria to humans, are key to this process. The SMC family includes condensin and cohesin, which structure chromosomes to enable mitosis and long-range gene regulation. We discuss novel insights into the mechanism of loop formation and the implications for how these complexes ultimately shape chromosomes. A picture is emerging in which these complexes form small loops that they then processively enlarge. It appears that SMC complexes act by family-wide basic principles, with complex-specific levels of control.
DOI:10.1016/j.cell.2016.01.033URLPMID:26919425 [本文引用: 1]

Condensins are large protein complexes that play a central role in chromosome organization and segregation in the three domains of life. They display highly characteristic, rod-shaped structures with SMC (structural maintenance of chromosomes) ATPases as their core subunits and organize large-scale chromosome structure through active mechanisms. Most eukaryotic species have two distinct condensin complexes whose balanced usage is adapted flexibly to different organisms and cell types. Studies of bacterial condensins provide deep insights into the fundamental mechanisms of chromosome segregation. This Review surveys both conserved features and rich variations of condensin-based chromosome organization and discusses their evolutionary implications.
DOI:10.1016/j.cub.2018.08.034URLPMID:30399354 [本文引用: 1]

Protein complexes built of structural maintenance of chromosomes (SMC) and kleisin subunits, including cohesin, condensin and the Smc5/6 complex, are master organizers of genome architecture in all kingdoms of life. How these large ring-shaped molecular machines use the energy of ATP hydrolysis to change the topology of chromatin fibers has remained a central unresolved question of chromosome biology. A currently emerging concept suggests that the common principle that underlies the essential functions of SMC protein complexes in the control of gene expression, chromosome segregation or DNA damage repair is their ability to expand DNA into large loop structures. Here, we review the current knowledge about the biochemical and structural properties of SMC protein complexes that might enable them to extrude DNA loops and compare their action to other motor proteins and nucleic acid translocases. We evaluate the currently predominant models of active loop extrusion and propose a detailed version of a 'scrunching' model, which reconciles much of the available mechanistic data and provides an elegant explanation for how SMC protein complexes fulfill an array of seemingly diverse tasks during the organization of genomes.
DOI:10.1038/22774URLPMID:10440376 [本文引用: 2]

When cells exit from mitotic cell division, their sister chromatids lose cohesion and separate to opposite poles of the dividing cell, resulting in equational chromosome segregation. In contrast, the reductional segregation of the first stage of meiotic cell division (meiosis I) requires that sister chromatids remain associated through their centromeres and move together to the same pole. Centromeric cohesion is lost as cells exit from meiosis II and sister chromatids can then separate. The fission yeast cohesin protein Rec8 is specific to and required for meiosis. Here we show that Rec8 appears in the centromeres and adjacent chromosome arms during the pre-meiotic S phase. Centromeric Rec8 persists throughout meiosis I and disappears at anaphase of meiosis II. When the rec8 gene is deleted, sister chromatids separate at meiosis I, resulting in equational rather than reductional chromosome segregation. We propose that the persistence of Rec8 at centromeres during meiosis I maintains sister-chromatid cohesion, and that its presence in the centromere-adjacent regions orients the kinetochores so that sister chromatids move to the same pole. This results in the reductional pattern of chromosome segregation necessary to reduce a diploid zygote to haploid gametes.
DOI:10.1016/S0092-8674(00)80609-1URLPMID:10412984 [本文引用: 2]

A multisubunit complex, called cohesin, containing Smc1p, Smc3p, Scc1p, and Scc3p, is required for sister chromatid cohesion in mitotic cells. We show here that Smc3p and a meiotic version of Scc1p called Rec8p are required for cohesion between sister chromatids, for formation of axial elements, for reciprocal recombination, and for preventing hyperresection of double-strand breaks during meiosis. Both Rec8p and Smc3p colocalize with chromosome cores independently of synapsis during prophase I and largely disappear from chromosome arms after pachytene but persist in the neighborhood of centromeres until the onset of anaphase II. The eukaryotic cell's cohesion apparatus is required both for the repair of recombinogenic lesions and for chromosome segregation and therefore appears to lie at the heart of the meiotic process.
DOI:10.1016/s1097-2765(00)80420-7URLPMID:10882066 [本文引用: 4]

Cohesion between sister chromatids depends on a multisubunit cohesin complex that binds to chromosomes around DNA replication and dissociates from them at the onset of anaphase. Scc2p, though not a cohesin subunit, is also required for sister chromatid cohesion. We show here that Scc2p forms a complex with a novel protein, Scc4p, which is also necessary for sister cohesion. In scc2 or scc4 mutants, cohesin complexes form normally but fail to bind both to centromeres and to chromosome arms. Our data suggest that a major role for the Scc2p/Scc4p complex is to facilitate the loading of cohesin complexes onto chromosomes.
DOI:10.1016/j.yexcr.2006.06.024URLPMID:16876157 [本文引用: 1]

Replicated DNA molecules are physically connected by cohesin complexes from the time of their synthesis in S-phase until they are segregated during anaphase of the subsequent mitosis or meiosis. This sister chromatid cohesion is essential for the biorientation of chromosomes on the mitotic or meiotic spindle. In addition, cohesion is also essential during G2-phase of the cell cycle to allow repair of DNA double-strand breaks by homologous recombination. Although cohesion can normally only be established during S-phase, recent work in yeast has shown that DNA double-strand breaks induce the recruitment of cohesin to the damage site and lead to the de novo formation of cohesion at this site. It is unknown if similar mechanisms operate in higher eukaryotes, but in mammalian cells phosphorylation of the cohesin subunit Smc1 by the protein kinase Atm has been shown to be important for DNA repair. We discuss how cohesin and sister chromatid cohesion might facilitate the repair of damaged DNA.
DOI:10.1038/s41594-019-0187-0URLPMID:30778236

In meiotic prophase, chromosomes are organized into compacted loop arrays to promote homolog pairing and recombination. Here, we probe the architecture of the mouse spermatocyte genome in early and late meiotic prophase using chromosome conformation capture (Hi-C). Our data support the established loop array model of meiotic chromosomes, and infer loops averaging 0.8-1.0?megabase pairs (Mb) in early prophase and extending to 1.5-2.0?Mb in late prophase as chromosomes compact and homologs undergo synapsis. Topologically associating domains (TADs) are lost in meiotic prophase, suggesting that assembly of the meiotic chromosome axis alters the activity of chromosome-associated cohesin complexes. While TADs are lost, physically separated A and B compartments are maintained in meiotic prophase. Moreover, meiotic DNA breaks and interhomolog crossovers preferentially form in the gene-dense A compartment, revealing a role for chromatin organization in meiotic recombination. Finally, direct detection of interhomolog contacts genome-wide reveals the structural basis for homolog alignment and juxtaposition by the synaptonemal complex.
URL [本文引用: 1]
URL [本文引用: 1]
DOI:10.1038/s41586-019-1547-yURLPMID:31511698 [本文引用: 1]

The RAG endonuclease initiates Igh V(D)J assembly in B cell progenitors by joining D segments to JH segments, before joining upstream VH segments to DJH intermediates1. In mouse progenitor B cells, the CTCF-binding element (CBE)-anchored chromatin loop domain2 at the 3' end of Igh contains an internal subdomain that spans the 5' CBE anchor (IGCR1)3, the DH segments, and a RAG-bound recombination centre (RC)4. The RC comprises the JH-proximal D segment (DQ52), four JH segments, and the intronic enhancer (iEμ)5. Robust RAG-mediated cleavage is restricted to paired V(D)J segments flanked by complementary recombination signal sequences (12RSS and 23RSS)6. D segments are flanked downstream and upstream by 12RSSs that mediate deletional joining with convergently oriented JH-23RSSs and VH-23RSSs, respectively6. Despite 12/23 compatibility, inversional D-to-JH joining via upstream D-12RSSs is rare7,8. Plasmid-based assays have attributed the lack of inversional D-to-JH joining to sequence-based preference for downstream D-12RSSs9, as opposed to putative linear scanning mechanisms10,11. As RAG linearly scans convergent CBE-anchored chromatin loops4,12-14, potentially formed by cohesin-mediated loop extrusion15-18, we revisited its scanning role. Here we show that the chromosomal orientation of JH-23RSS programs RC-bound RAG to linearly scan upstream chromatin in the 3' Igh subdomain for convergently oriented D-12RSSs and, thereby, to mediate deletional joining of all D segments except RC-based DQ52, which joins by a diffusion-related mechanism. In a DQ52-based RC, formed in the absence of JH segments, RAG bound by the downstream DQ52-RSS scans the downstream constant region exon-containing 3' Igh subdomain, in which scanning can be impeded by targeted binding of nuclease-dead Cas9, by transcription through repetitive Igh switch sequences, and by the 3' Igh CBE-based loop anchor. Each scanning impediment focally increases RAG activity on potential substrate sequences within the impeded region. High-resolution mapping of chromatin interactions in the RC reveals that such focal RAG targeting is associated with corresponding impediments to the loop extrusion process that drives chromatin past RC-bound RAG.
DOI:10.1016/s0092-8674(00)80233-0URLPMID:9160743 [本文引用: 1]

We report here purification and characterization of chromosome condensation protein complexes (termed condensins) containing XCAP-C and XCAP-E, two Xenopus members of the SMC family. Sucrose density gradient centrifugation reveals two major forms of condensins. The 8S form is a heterodimer of XCAP-C and XCAP-E, whereas the 13S form contains three additional subunits. One of them is identified as a homolog of the Drosophila Barren protein whose mutation shows a defect in chromosome segregation. Chromosomal targeting of condensins is mitosis-specific and is independent of topoisomerase IIalpha. 13S condensin is required for condensation, as demonstrated by immunodepletion and rescue experiments. Our results suggest that the condensin complexes represent the most abundant structural components of mitotic chromosomes and play a central role in driving chromosome condensation.
DOI:10.1038/21831URLPMID:10403247 [本文引用: 2]

Cohesion between sister chromatids is established during DNA replication and depends on a multiprotein complex called cohesin. Attachment of sister kinetochores to the mitotic spindle during mitosis generates forces that would immediately split sister chromatids were it not opposed by cohesion. Cohesion is essential for the alignment of chromosomes in metaphase but must be abolished for sister separation to start during anaphase. In the budding yeast Saccharomyces cerevisiae, loss of sister-chromatid cohesion depends on a separating protein (separin) called Esp1 and is accompanied by dissociation from the chromosomes of the cohesion subunit Scc1. Here we show that Esp1 causes the dissociation of Scc1 from chromosomes by stimulating its cleavage by proteolysis. A mutant Scc1 is described that is resistant to Esp1-dependent cleavage and which blocks both sister-chromatid separation and the dissociation of Scc1 from chromosomes. The evolutionary conservation of separins indicates that the proteolytic cleavage of cohesion proteins might be a general mechanism for triggering anaphase.
DOI:10.1128/MCB.25.1.172-184.2005URLPMID:15601840 [本文引用: 1]

The rad18 gene of Schizosaccharomyces pombe is an essential gene that is involved in several different DNA repair processes. Rad18 (Smc6) is a member of the structural maintenance of chromosomes (SMC) family and, together with its SMC partner Spr18 (Smc5), forms the core of a high-molecular-weight complex. We show here that both S. pombe and human Smc5 and -6 interact through their hinge domains and that four independent temperature-sensitive mutants of Rad18 (Smc6) are all mutated at the same glycine residue in the hinge region. This mutation abolishes the interactions between the hinge regions of Rad18 (Smc6) and Spr18 (Smc5), as does mutation of a conserved glycine in the hinge region of Spr18 (Smc5). We purified the Smc5-6 complex from S. pombe and identified four non-SMC components, Nse1, Nse2, Nse3, and Rad62. Nse3 is a novel protein which is related to the mammalian MAGE protein family, many members of which are specifically expressed in cancer tissue. In initial steps to understand the architecture of the complex, we identified two subcomplexes containing Rad18-Spr18-Nse2 and Nse1-Nse3-Rad62. The subcomplexes are probably bridged by a weaker interaction between Nse2 and Nse3.
DOI:10.1016/s1097-2765(02)00515-4URLPMID:11983169 [本文引用: 3]

Sister chromatids are held together by the multisubunit cohesin complex, which contains two SMC (Smc1 and Smc3) and two non-SMC (Scc1 and Scc3) proteins. The crystal structure of a bacterial SMC "hinge" region along with EM studies and biochemical experiments on yeast Smc1 and Smc3 proteins show that SMC protamers fold up individually into rod-shaped molecules. A 45 nm long intramolecular coiled coil separates the hinge region from the ATPase-containing "head" domain. Smc1 and Smc3 bind to each other via heterotypic interactions between their hinges to form a V-shaped heterodimer. The two heads of the V-shaped dimer are connected by different ends of the cleavable Scc1 subunit. Cohesin therefore forms a large proteinaceous loop within which sister chromatids might be entrapped after DNA replication.
DOI:10.1002/prot.20795URLPMID:16437548 [本文引用: 1]

The SMC (structural maintenance of chromosomes) proteins are a highly conserved and ubiquitous family of ATPases, found in nearly all living organisms examined, where they play crucial roles in transmission of the hereditary material. However, the extent to which efficient ATP hydrolysis is required for SMC function has been a matter of some debate. Here we investigate the potential functional significance of ATP binding and hydrolysis in different eukaryotic SMC proteins, both by comparing the conservation of conserved ATPase motifs and by exploring potential coevolution between associated domains. In this way, we have been able to account for the reduced requirement for ATPase activity in cohesin's SMC3 and demonstrate the greater apparent conservation requirements for such activity in condensin SMC proteins. Finally, we explore possible interactions between the SMC and non-SMC components of the condensin complex that are required for full condensin activity and may modulate ATPase activity in the holocomplex.
DOI:10.1096/fasebj.6.9.1377140URLPMID:1377140 [本文引用: 1]

The traffic ATPases superfamily includes known transporters, both prokaryotic and eukaryotic, including the medically important proteins, P-glycoprotein, and the cystic fibrosis gene product (CFTR), which is known to be a Cl- channel. The structure and mechanism of action of the best-studied members of the superfamily, the periplasmic permeases, are described and related to that of CFTR and eukaryotic traffic ATPases in general. The contention is put forward that the distinction between the architecture and mechanisms of action of channels and transporters is blurred.
DOI:10.1016/s0092-8674(00)80890-9URLPMID:10892749 [本文引用: 1]

To clarify the key role of Rad50 in DNA double-strand break repair (DSBR), we biochemically and structurally characterized ATP-bound and ATP-free Rad50 catalytic domain (Rad50cd) from Pyrococcus furiosus. Rad50cd displays ATPase activity plus ATP-controlled dimerization and DNA binding activities. Rad50cd crystal structures identify probable protein and DNA interfaces and reveal an ABC-ATPase fold, linking Rad50 molecular mechanisms to ABC transporters, including P glycoprotein and cystic fibrosis transmembrane conductance regulator. Binding of ATP gamma-phosphates to conserved signature motifs in two opposing Rad50cd molecules promotes dimerization that likely couples ATP hydrolysis to dimer dissociation and DNA release. These results, validated by mutations, suggest unified molecular mechanisms for ATP-driven cooperativity and allosteric control of ABC-ATPases in DSBR, membrane transport, and chromosome condensation by SMC proteins.
DOI:10.1038/nrm3857URLPMID:25145851 [本文引用: 4]

Structural maintenance of chromosomes (SMC) complexes, which in eukaryotic cells include cohesin, condensin and the Smc5/6 complex, are central regulators of chromosome dynamics and control sister chromatid cohesion, chromosome condensation, DNA replication, DNA repair and transcription. Even though the molecular mechanisms that lead to this large range of functions are still unclear, it has been established that the complexes execute their functions through their association with chromosomal DNA. A large set of data also indicates that SMC complexes work as intermolecular and intramolecular linkers of DNA. When combining these insights with results from ongoing analyses of their chromosomal binding, and how this interaction influences the structure and dynamics of chromosomes, a picture of how SMC complexes carry out their many functions starts to emerge.
DOI:10.1128/mcb.23.16.5638-5650.2003URLPMID:12897137 [本文引用: 1]

We show that Bacillus subtilis SMC (structural maintenance of chromosome protein) localizes to discrete foci in a cell cycle-dependent manner. Early in the cell cycle, SMC moves from the middle of the cell toward opposite cell poles in a rapid and dynamic manner and appears to interact with different regions on the chromosomes during the cell cycle. SMC colocalizes with its interacting partners, ScpA and ScpB, and the specific localization of SMC depends on both Scp proteins, showing that all three components of the SMC complex are required for proper localization. Cytological and biochemical experiments showed that dimeric ScpB stabilized the binding of ScpA to the SMC head domains. Purified SMC showed nonspecific binding to double-stranded DNA, independent of Scp proteins or ATP, and was retained on DNA after binding to closed DNA but not to linear DNA. The SMC head domains and hinge region did not show strong DNA binding activity, suggesting that the coiled-coil regions in SMC mediate an association with DNA and that SMC binds to DNA as a ring-like structure. The overproduction of SMC resulted in global chromosome compaction, while SMC was largely retained in bipolar foci, suggesting that the SMC complex forms condensation centers that actively affect global chromosome compaction from a defined position on the nucleoid.
DOI:10.1016/s1097-2765(03)00108-4URLPMID:12667442 [本文引用: 1]

We describe a superfamily of eukaryotic and prokaryotic proteins (kleisins) that includes ScpA, Scc1, Rec8, and Barren. Scc1 interacts with SMC proteins through N- and C-terminal domains to form a ring-like structure. Since these are the only domains conserved among kleisins, we suggest that ring formation with SMC proteins may define this family.
DOI:10.1016/s0092-8674(03)00162-4URLPMID:12654244 [本文引用: 2]

The cohesin complex is essential for sister chromatid cohesion during mitosis. Its Smc1 and Smc3 subunits are rod-shaped molecules with globular ABC-like ATPases at one end and dimerization domains at the other connected by long coiled coils. Smc1 and Smc3 associate to form V-shaped heterodimers. Their ATPase heads are thought to be bridged by a third subunit, Scc1, creating a huge triangular ring that could trap sister DNA molecules. We address here whether cohesin forms such rings in vivo. Proteolytic cleavage of Scc1 by separase at the onset of anaphase triggers its dissociation from chromosomes. We show that N- and C-terminal Scc1 cleavage fragments remain connected due to their association with different heads of a single Smc1/Smc3 heterodimer. Cleavage of the Smc3 coiled coil is sufficient to trigger cohesin release from chromosomes and loss of sister cohesion, consistent with a topological association with chromatin.
DOI:10.1146/annurev.biochem.74.082803.133219URLPMID:15952899 [本文引用: 1]

Protein complexes consisting of structural maintenance of chromosomes (SMC) and kleisin subunits are crucial for the faithful segregation of chromosomes during cell proliferation in prokaryotes and eukaryotes. Two of the best-studied SMC complexes are cohesin and condensin. Cohesin is required to hold sister chromatids together, which allows their bio-orientation on the mitotic spindle. Cleavage of cohesin's kleisin subunit by the separase protease then triggers the movement of sister chromatids into opposite halves of the cell during anaphase. Condensin is required to organize mitotic chromosomes into coherent structures that prevent them from getting tangled up during segregation. Here we describe the discovery of SMC complexes and discuss recent advances in determining how members of this ancient protein family may function at a mechanistic level.
DOI:10.1016/j.tcb.2016.04.002URLPMID:27134029 [本文引用: 1]

Cohesin facilitates sister chromatid cohesion through the formation of a large ring structure that encircles DNA. Its function relies on two structural maintenance of chromosomes (Smc) proteins, which are found in almost all organisms tested, from bacteria to humans. In accordance with their ubiquity, Smc complexes, such as cohesin, condensin, Smc5-6, and the dosage compensation complex, affect almost all processes of DNA homeostasis. Although their precise molecular mechanism remains enigmatic, here we provide an overview of the architecture of eukaryotic Smc complexes with a particular focus on cohesin, which has seen the most progress recently. Given the evident conservation of many structural features between Smc complexes, it is expected that architecture and topology will have a significant role when deciphering their precise molecular mechanisms.
DOI:10.1046/j.1365-313x.2002.01224.xURLPMID:11846874 [本文引用: 1]

The titan (ttn) mutants of Arabidopsis exhibit striking alterations in chromosome dynamics and cell division during seed development. Endosperm defects include aberrant mitoses and giant polyploid nuclei. Mutant embryos differ in cell size, morphology and viability, depending on the locus involved. Here we demonstrate that three TTN genes encode chromosome scaffold proteins of the condensin (SMC2) and cohesin (SMC1 and SMC3) classes. These proteins have been studied extensively in yeast and animal systems, where they modulate chromosome condensation, chromatid separation, and dosage compensation. Arabidopsis contains single copies of SMC1 and SMC3 cohesins. We used forward genetics to identify duplicate T-DNA insertions in each gene. These mutants (ttn7 and ttn8) have similar titan phenotypes: giant endosperm nuclei and arrested embryos with a few small cells. A single SMC2 knockout (ttn3) was identified and confirmed by molecular complementation. The weak embryo phenotype observed in this mutant may result from expression of a related gene (AtSMC2) with overlapping functions. Further analysis of titan mutants and the SMC gene family in Arabidopsis should provide clues to chromosome mechanics in plants and insights into the regulation of nuclear activity during endosperm development.
DOI:10.1046/j.1365-313x.1998.00268.xURLPMID:9807824 [本文引用: 1]

We describe in this report a novel class of mutants that should facilitate the identification of genes required for progression through the mitotic cell cycle during seed development in angiosperms. Three non-allelic titan (ttn) mutants with related but distinct phenotypes are characterized. The common feature among these mutants is that endosperm nuclei become greatly enlarged and highly polyploid. The mutant embryo is composed of a few giant cells in ttn1, several small cells in ttn2, and produces a normal plant in ttn3. Condensed chromosomes arrested at prophase of mitosis are found in the free nuclear endosperm of ttn1 and ttn2 seeds. Large mitotic figures with excessive numbers of chromosomes are visible in ttn3 endosperm. The ttn1 mutation appears to disrupt cytoskeletal organization because endosperm nuclei fail to migrate to the chalazal end of the seed. How double fertilization leads to the establishment of distinct patterns of mitosis and cytokinesis in the embryo and endosperm is a central question in plant reproductive biology. Molecular isolation of TITAN genes should help to answer this question, as well as related issues concerning cell cycle regulation, chromosome movement and endosperm identity in angiosperms.
DOI:10.1242/dev.00542URLPMID:12783798 [本文引用: 1]

Proper chromatin condensation and sister chromatid resolution are essential for the maintenance of chromosomal integrity during cell division, and is in part mediated by a conserved multisubunit apparatus termed the condensin complex. The core subunits of the complex are members of the SMC2 (Structural Maintenance of Chromosomes) and SMC4 gene families. We have cloned an Arabidopsis gene, AtCAP-E1, which is a functional ortholog of the yeast SMC2 gene. A second, highly homologous SMC2 gene, AtCAPE-2, was identified by the Arabidopsis genome project. SMC2 gene expression in Arabidopsis was correlated with the mitotic activity of tissues, with high level expression observed in meristematic cells. The two genes are differentially expressed with AtCAP-E1 accounting for more than 85% of the total SMC2 transcript pool. The titan3 mutant is the result of a T-DNA insertion into AtCAP-E1, but other than subtle endosperm defects, titan3 is viable and fecund. We identified a T-DNA insertion mutant of AtCAP-E2, which showed no obvious mutant phenotype, indicating that the two genes are functionally redundant. Genetic crosses were employed to examine the consequences of reduced SMC2 levels. Both male and female gametogenesis were compromised in double mutant spores. Embryo lethality was observed for both double homozygous and AtCAP-E1(-/-), AtCAP-E2(+/-) plants; arrest occurred at or before the globular stage and was associated with altered planes of cell division in both the suspensor and the embryo. Down regulation of both genes by antisense technology, as well as in AtCAP-E1(+/-), AtCAP-E2(-/-) plants results in meristem disorganization and fasciation. Our data are consistent with the interpretation that threshold levels of SMC2 proteins are required for normal development and that AtCAP-E2 may have a higher affinity for its target than AtCAP-E1.
.
DOI:10.1083/jcb.151.4.749URLPMID:11076961 [本文引用: 1]

In eukaryotes, sister chromatids remain connected from the time of their synthesis until they are separated in anaphase. This cohesion depends on a complex of proteins called cohesins. In budding yeast, the anaphase-promoting complex (APC) pathway initiates anaphase by removing cohesins from chromosomes. In vertebrates, cohesins dissociate from chromosomes already in prophase. To study their mitotic regulation we have purified two 14S cohesin complexes from human cells. Both complexes contain SMC1, SMC3, SCC1, and either one of the yeast Scc3p orthologs SA1 and SA2. SA1 is also a subunit of 14S cohesin in Xenopus. These complexes interact with PDS5, a protein whose fungal orthologs have been implicated in chromosome cohesion, condensation, and recombination. The bulk of SA1- and SA2-containing complexes and PDS5 are chromatin-associated until they become soluble from prophase to telophase. Reconstitution of this process in mitotic Xenopus extracts shows that cohesin dissociation does neither depend on cyclin B proteolysis nor on the presence of the APC. Cohesins can also dissociate from chromatin in the absence of cyclin-dependent kinase 1 activity. These results suggest that vertebrate cohesins are regulated by a novel prophase pathway which is distinct from the APC pathway that controls cohesins in yeast.
.
DOI:10.1242/jcs.02583URLPMID:16176934 [本文引用: 1]

The success of the first meiotic division relies (among other factors) on the formation of bivalents between homologous chromosomes, the monopolar orientation of the sister kinetochores at metaphase I and the maintenance of centromeric cohesion until the onset of anaphase II. The meiotic cohesin subunit, Rec8 has been reported to be one of the key players in these processes, but its precise role in kinetochore orientation is still under debate. By contrast, much less is known about the other non-SMC cohesin subunit, Scc3. We report the identification and the characterisation of AtSCC3, the sole Arabidopsis homologue of Scc3. The detection of AtSCC3 in mitotic cells, the embryo lethality of a null allele Atscc3-2, and the mitotic defects of the weak allele Atscc3-1 suggest that AtSCC3 is required for mitosis. AtSCC3 was also detected in meiotic nuclei as early as interphase, and bound to the chromosome axis from early leptotene through to anaphase I. We show here that both AtREC8 and AtSCC3 are necessary not only to maintain centromere cohesion at anaphase I, but also for the monopolar orientation of the kinetochores during the first meiotic division. We also found that AtREC8 is involved in chromosome axis formation in an AtSPO11-1-independent manner. Finally, we provide evidence for a role of AtSPO11-1 in the stability of the cohesin complex.
DOI:10.2174/138920311795684904URLPMID:21348848 [本文引用: 2]

Cohesin complexes are critical for holding sister chromatids together during nuclear division. They also play important roles in the compaction of chromosomes and their bipolar attachment to the spindle, DNA double strand break repair, and the regulation of gene expression. Studies on sister chromatid cohesion in a wide range of organisms have shown that the proteins involved, and the general events of this important process are conserved between yeast, plants and animals. However, species-specific differences have been identified. In this review a general overview of cohesins, their roles and mechanisms of action is presented, followed by a review of our current state of knowledge on plant cohesins. While plants utilize the same general set of cohesin proteins and similar processes to establish and release sister chromatid cohesion, they also exhibit a number of unique features that are likely to provide interesting new insights into the roles of these important proteins.
DOI:10.1046/j.1365-313x.1997.11040659.xURLPMID:9161029 [本文引用: 1]

Fluorescence microscopy was used to study meiosis in microsporocytes from wild-type Arabidopsis thaliana and a T-DNA-tagged meiotic mutant. Techniques for visualizing chromosomes and beta-tubulin in other plant species were evaluated and modified in order to develop a method for analyzing meiosis in A. thaliana anthers. Like most dicots, A. thaliana microsporocytes undergo simultaneous cytokinesis in which both meiotic divisions are completed prior to cytokinesis. However, two unique events were observed in wild-type A. thaliana that have not been reported in other angiosperms: (1) polarization of the microsporocyte cytoskeleton during prophase I prior to nuclear envelope breakdown, and (2) extensive depolymerization of microtubules just prior to metaphase II. The first observation could have implications regarding a previously uncharacterized mechanism for determining the axis of the metaphase I spindle during microsporogenesis. The second observation is peculiar since microtubules are known to be involved in chromosome alignment in other species; possible explanations will be discussed. A T-DNA-tagged meiotic mutant of A. thaliana (syn1), which had previously been shown to produce abnormal microspores with variable DNA content, was also cytologically characterized. The first observable defect occurs in microsporocytes at telophase I, where some chromosomes are scattered throughout the cytoplasm, usually attached to stray microtubules. Subsequent development stages are affected, leading to complete male sterility. Based on similarities to synaptic mutants that have been described in other species, it is suggested that this mutant is defective in synaptonemal complex formation and/or cohesion between sister chromatids.
DOI:10.1242/jcs.00601URLPMID:12783989 [本文引用: 1]

The faithful transmission of chromosomes during mitosis and meiosis requires the establishment and subsequent release of cohesion between replicated chromosomes. Sister chromatid cohesion is mediated, in large part, by the cohesin complex, which consists of four highly conserved proteins: SMC1, SMC3, SCC1/REC8 and SCC3. Mitotic cohesin complexes contain SSC1, whereas meiotic cohesin complexes contain the related REC8 protein. As part of studies to identify and characterize proteins required for meiosis in plants, we previously identified a putative Arabidopsis REC8 homolog, referred to as syn1. Preliminary cytological studies indicated that syn1 plants exhibit defects in meiotic chromosome cohesion and condensation that result in fragmentation of the chromosomes and the formation of polyads. In the experiments presented here we show that SYN1 encodes a protein that localizes to arms of meiotic chromosomes from approximately meiotic interphase to anaphase I. The protein is not detected at the centromeres or after metaphase I. Furthermore, fluorescence in situ hybridization experiments on microsporocytes from syn1 plants demonstrate that the mutation eliminates arm cohesion as early as interphase, whereas centromere cohesion is maintained until approximately anaphase I. These results indicate that although the main role of SYN1 is in chromosome arm cohesion, it is also important for maintaining cohesion at the centromeres during late stages of meiosis I.
DOI:10.1016/s0378-1119(01)00499-1URLPMID:11410371 [本文引用: 1]

Sister chromatid cohesion is required for proper chromosome segregation during cell division. One group of proteins that is essential for sister chromatid cohesion during mitosis and meiosis is the RAD21/REC8 family of cohesin proteins. Two cohesin proteins are found in yeast; one that functions mainly in mitosis while the other participates in meiosis. In contrast, only one cohesin gene appears to be present in Drosophila. In previous studies we identified an Arabidopsis cohesin protein that is required for meiosis. In this report we describe the isolation and characterization of two additional Arabidopsis cohesin genes. The structure of the genes suggests that they arose via a gene duplication event followed by extensive sequence evolution. Transcripts for the two genes are present throughout the plant and are highest in regions of active cell division, suggesting that the proteins may participate in chromosome cohesion during mitosis.
DOI:10.1111/j.1365-313X.2007.03106.xURLPMID:17488242 [本文引用: 2]

Alpha-kleisins are core components of meiotic and mitotic cohesin complexes. Arabidopsis contains genes for four alpha-kleisin proteins encoded by SYN genes. SYN1, a REC8 ortholog, is essential for meiosis, while SYN2 and SYN4 appear to be SCC1 orthologs and function in mitosis. Our analysis of AtSYN3 shows that it localizes primarily in the nucleolus of both meiotic and mitotic cells. Furthermore, analysis of plants containing an AtSYN3 T-DNA knockout mutation demonstrated that it is essential for megagametogenesis and plays an important role in pollen. These results suggest that SYN3 may not function as part of a typical cohesin complex; rather it may have evolved a specialized role in controlling rDNA structure, transcription or rRNA processing.
DOI:10.1046/j.1365-313x.1999.00548.xURLPMID:10504568 [本文引用: 2]

Cohesins are a group of conserved proteins responsible for cohesion between replicated sister chromatids during mitosis and meiosis and which are implicated in double-strand break repair and meiotic recombination. We describe here the identification and characterisation of an Arabidopsis gene - DETERMINATE, INFERTILE1 (DIF1), which is a homolog of the Schizosaccharomyces pombe REC8/RAD21 cohesin genes, and is essential for meiotic chromosome segregation. Five independent alleles of the DIF1 gene were isolated by transposon mutagenesis, and the mutants show complete male and female sterility. Pollen mother cells (PMCs) of dif1 mutants show multiple meiotic defects which are represented by univalent chromosomes and chromosome fragmentation at metaphase I, and acentric fragments and chromatin bridges in meiosis I and II. Consequently, chromosome segregation is strongly affected, resulting in meiotic products of uneven size, shape and of variable ploidy. The similarities in phenotype, and the sequence homology between DIF1 and the REC8/RAD21 cohesins suggests that cohesin function is largely conserved between eukaryotes and highlights the essential role cohesins play in plant meiosis.
DOI:10.1093/jxb/erj083URLPMID:16488915 [本文引用: 2]

The RAD21/REC8 gene family has been implicated in sister chromatid cohesion and DNA repair in several organisms. Unlike most eukaryotes, Arabidopsis thaliana has three RAD21 gene homologues, and their cloning and characterization are reported here. All three genes, AtRAD21.1, AtRAD21.2, and AtRAD21.3, are expressed in tissues rich in cells undergoing cell division, and AtRAD21.3 shows the highest relative level of expression. An increase in steady-state levels of AtRAD21.1 transcript was also observed, specifically after the induction of DNA damage. Phenotypic analysis of the atrad21.1 and atrad21.3 mutants revealed that neither of the single mutants was lethal, probably due to the redundancy in function of the AtRAD21 genes. However, AtRAD21.1 plays a critical role in recovery from DNA damage during seed imbibition, prior to germination, as atrad21.1 mutant seeds are hypersensitive to radiation damage.
DOI:10.1105/tpc.11.3.417URLPMID:10072401 [本文引用: 1]

The proper pairing, recombination, and segregation of chromosomes are central to meiosis and sexual reproduction. The syn1 mutation was previously identified as a synaptic mutant in a T-DNA-tagged population of plants. SYN1 has been isolated and found to exhibit similarity to Schizosaccharomyces pombe RAD21 and RAD21-like proteins, which are required for chromosome condensation and sister chromatid cohesion during mitosis. Plants homozygous for syn1 are male and female sterile and show defects in chromosome condensation and pairing beginning at leptonema of meiosis I. Fragmentation of the chromosomes was observed at metaphase I. Alternative promoters produced two SYN1 transcripts. One transcript was expressed at low levels in most tissues, whereas the other was expressed only in prebolting buds. DNA blot analyses suggest that Arabidopsis contains a small RAD21 gene family. Consistent with the DNA blot data, a second Arabidopsis RAD21-like gene has been identified. These results suggest that different RAD21-like proteins play essential roles in chromosome condensation and pairing during both meiosis and mitosis.
DOI:10.1104/pp.18.01320URLPMID:30705069 [本文引用: 2]

During meiosis, the stepwise release of sister chromatid cohesion is crucial for the equal distribution of genetic material to daughter cells, enabling generation of fertile gametophytes. However, the molecular mechanism that protects centromeric cohesion from release at meiosis I is unclear in Arabidopsis (Arabidopsis thaliana). Here, we report that the protein phosphatase 2A regulatory subunits B'α and B'β participate in the control of sister chromatid separation. The double mutant b'αβ exhibited severe male and female sterility, caused by the lack of a nucleus or presence of an abnormal nucleus in mature microspores and embryo sacs. 4',6-Diamidino-2-phenylindole staining revealed unequal amounts of DNA in the mononuclear microspores. Transverse sections of the anthers revealed unevenly sized tetrads with or without a nucleus, suggesting a defect in meiocyte meiosis. An analysis of chromosome spreads showed that the sister chromatids separated prematurely at anaphase I in b'αβ Immunoblotting showed that AtRECOMBINATION DEFECTIVE8 (AtREC8), a key member of the cohesin complex, was hyperphosphorylated in b'αβ anthers and pistils during meiosis but hypophosphorylated in the wild type. Furthermore, yeast two-hybrid and bimolecular fluorescence complementation assays showed that B'α and B'β interact specifically with AtREC8, AtSHUGOSHIN1 (AtSGO1), AtSGO2, and PATRONUS1. Given that B'α was reported to localize to the centromere in meiotic cells, we propose that protein phosphatase 2A B'α and B'β are recruited by AtSGO1/2 and PATRONUS1 to dephosphorylate AtREC8 at the site of centromere cohesion to shield it from cleavage until anaphase II, contributing to the balanced separation of sister chromatids at meiosis.
DOI:10.1104/pp.18.00281URLPMID:30061120 [本文引用: 2]

The correct separation of homologous chromosomes during meiosis I, and sister chromatids during meiosis II, relies on the tight control of the cohesion complex. The phosphorylation and subsequent cleavage of the meiotic recombination protein REC8 (REC8-like family protein [SYN1] in Arabidopsis [Arabidopsis thaliana]), the α-kleisin subunit of the cohesion ring, along the chromosome arms at meiosis I allows crossovers and separation of homologous chromosomes without chromatid dissociation. REC8 continues to localize and function at the centromeres up to metaphase II and, in yeast and vertebrates, is protected from cleavage by means of protein phosphatase 2A (PP2A)-mediated dephosphorylation. Here, we show that, in plants, centromeric sister chromatid cohesion until meiosis II also requires the activity of a PP2A-type phosphatase complex. The combined absence of the regulatory subunits PP2AB'α and PP2AB'β leads to the premature loss of chromosome cohesion in meiosis I. Male meiocytes of the pp2ab'αβ double mutant display premature depletion of SYN1. The PP2AA1 structural and B'α regulatory subunit localize specifically to centromeres until metaphase II, supporting a role for the PP2A complex in the SYN1-mediated maintenance of centromeric cohesion in plant meiosis.
DOI:10.1186/s12870-014-0353-9URLPMID:25511710 [本文引用: 1]

The RAD21 cohesin plays, besides its well-recognised role in chromatid cohesion, a role in DNA double strand break (dsb) repair. In Arabidopsis there are three RAD21 paralog genes (AtRAD21.1, AtRAD21.2 and AtRAD21.3), yet only AtRAD21.1 has been shown to be required for DNA dsb damage repair. Further investigation of the role of cohesins in DNA dsb repair was carried out and is here reported.
DOI:10.1104/pp.111.177428URL [本文引用: 1]

The successful transmission of chromosomes during mitosis and meiosis relies on the establishment and subsequent release of cohesion between replicated chromatids. Cohesion is mediated by a four-subunit structural maintenance of chromosome complex, called cohesins. REC8 is a key component of this meiotic cohesion complex in most model organisms studied to date. Here, we isolated and dissected the functions of OsREC8, a rice (Oryza sativa) REC8 homolog, using two null Osrec8 mutants. We showed that OsREC8 encodes a protein that localized to meiotic chromosomes from approximately meiotic interphase to metaphase I. Homologous pairing and telomere bouquet formation were abnormal in Osrec8 meiocytes. Furthermore, fluorescent in situ hybridization experiments on Osrec8 meiocytes demonstrated that the mutation eliminated meiotic centromeric cohesion completely during prophase I and also led to the bipolar orientation of the kinetochores during the first meiotic division and accordingly resulted in premature separation of sister chromatid during meiosis I. Immunolocalization analyses revealed that the loading of PAIR2, PAIR3, OsMER3, and ZEP1 all depended on OsREC8. By contrast, the presence of the OsREC8 signal in pair2, pair3, Osmer3, and zep1 mutants indicated that the loading of OsREC8 did not rely on these four proteins. These results suggest that OsREC8 has several essential roles in the meiotic processes.
DOI:10.1007/s11103-005-4922-zURL [本文引用: 1]

In yeast, Rad21/Scc1 and its meiotic variant Rec8 are key players in the establishment and subsequent dissolution of sister chromatid cohesion for mitosis and meiosis, respectively, which are essential for chromosome segregation. Unlike yeast, our identification revealed that the rice genome has 4 RAD21-like genes that share lower than 21% identity at polypeptide levels, and each is present as a single copy in this genome. Here we describe our analysis of the function of OsRAD21-4 by RNAi. Western blot analyses indicated that the protein was most abundant in young flowers and less in leaves and buds but absent in roots. In flowers, the expression was further defined to premeiotic pollen mother cells (PMCs) and meiotic PMCs of anthers. Meiotic chromosome behaviors were monitored from male meiocytes of OsRAD21-4-deficient lines mediated by RNAi. The male meiocytes showed multiple aberrant events at meiotic prophase I, including over-condensation of chromosomes, precocious segregation of homologues and chromosome fragmentation. Fluorescence in situ hybridization experiments revealed that the deficient lines were defective in homologous pairing and cohesion at sister chromatid arms. These defects resulted in unequal chromosome segregation and aberrant spore generation. These observations suggest that OsRad21-4 is essential for efficient meiosis.
DOI:10.1111/j.1365-313X.2007.03190.xURLPMID:17617177 [本文引用: 1]

In contrast to animals, in which products of meiosis differentiate directly into sperm, flowering plants employ a specific mechanism to give rise to functional sperm cells, the specifics of which remain largely unknown. A previous study revealed that, compared to yeast and vertebrates, which have two proteins (Rad21 and its meiosis-specific variant Rec8) that play a vital role in sister chromatid cohesion and segregation for mitosis and meiosis, respectively, the rice genome encodes four Rad21/Rec8 proteins (OsRad21s). In this paper, phylogenetic and immunostaining analyses reveal that OsRad21-3 is an orthologue of yeast Rad21. OsRAD21-3 transcript and protein accumulated preferentially in flowers, with low levels in vegetative tissues. In flowers, they persisted from the stamen and carpel primordia stages until the mature pollen stage. OsRAD21-3-deficient RNAi lines showed arrested pollen mitosis, aberrant pollen chromosome segregation and aborted pollen grains, which led to disrupted pollen viability. However, male meiosis in these RNAi lines did not appear to be severely disrupted, which suggests that the main involvement of OsRAD21-3 is in post-meiotic pollen development by affecting pollen mitosis. Furthermore, of the four OsRAD21 genes in the rice genome, only OsRAD21-3 was expressed in pollen grains. Given that the mechanism involving generation of sperm cells differs between flowering plants and metozoans, this study shows, in part, why flowering plants of rice and Arabidopsis have four Rad21/Rec8 proteins, as compared with two in yeast and metozoans, and gives some clues to the functional differentiation of Rad21/Rec8 proteins during evolution.
.
DOI:10.1111/j.1744-7909.2010.01009.xURLPMID:21205177 [本文引用: 1]

Rad21 and its meiotic counterpart Rec8, the key components of the cohesin complex, are essential for sister chromatid cohesion and chromosome segregation in mitosis and meiosis, respectively. In contrast to yeast and vertebrates, which have only two RAD21/REC8 genes, the rice genome encodes four Rad21/Rec8 proteins. Here, we report on the cloning and characterization of OsRAD21-2 from rice (Oryza sativa L.). Phylogenetic analysis of the full-length amino acids showed that OsRad21-2 was grouped into the plant-specific Rad21 subfamily. Semi-quantitative reverse transcription-polymerase chain reaction revealed OsRAD21-2 preferentially expressed in premeiotic flowers. Further RNA in situ hybridization analysis and promoter::β-glucuronidase staining indicated that OsRAD21-2 was mainly expressed in actively dividing tissues including premeiotic stamen, stem intercalary meristem, leaf meristem, and root pericycle. Ectopic expression of OsRAD21-2 in fission yeast resulted in cell growth delay and morphological abnormality. Flow cytometric analysis revealed that the OsRAD21-2-expressed cells were arrested in G2 phase. Our results suggest that OsRad21-2 functions in regulation of cell division and growth.
.
DOI:10.1242/jcs.03054URLPMID:16868028 [本文引用: 1]

REC8 is a master regulator of chromatin structure and function during meiosis. Here, we dissected the functions of absence of first division (afd1), a maize rec8/alpha-kleisin homolog, using a unique afd1 allelic series. The first observable defect in afd1 mutants is the inability to make a leptotene chromosome. AFD1 protein is required for elongation of axial elements but not for their initial recruitment, thus showing that AFD1 acts downstream of ASY1/HOP1. AFD1 is associated with the axial and later the lateral elements of the synaptonemal complex. Rescuing 50% of axial element elongation in the weakest afd1 allele restored bouquet formation demonstrating that extent of telomere clustering depends on axial element elongation. However, rescuing bouquet formation was not sufficient for either proper RAD51 distribution or homologous pairing. It provides the basis for a model in which AFD1/REC8 controls homologous pairing through its role in axial element elongation and the subsequent distribution of the recombination machinery independent of bouquet formation.
DOI:10.1038/nature08550URLPMID:19907496 [本文引用: 1]

Cohesin not only links sister chromatids but also inhibits the transcriptional machinery's interaction with and movement along chromatin. In contrast, replication forks must traverse such cohesin-associated obstructions to duplicate the entire genome in S phase. How this occurs is unknown. Through single-molecule analysis, we demonstrate that the replication factor C (RFC)-CTF18 clamp loader (RFC(CTF18)) controls the velocity, spacing and restart activity of replication forks in human cells and is required for robust acetylation of cohesin's SMC3 subunit and sister chromatid cohesion. Unexpectedly, we discovered that cohesin acetylation itself is a central determinant of fork processivity, as slow-moving replication forks were found in cells lacking the Eco1-related acetyltransferases ESCO1 or ESCO2 (refs 8-10) (including those derived from Roberts' syndrome patients, in whom ESCO2 is biallelically mutated) and in cells expressing a form of SMC3 that cannot be acetylated. This defect was a consequence of cohesin's hyperstable interaction with two regulatory cofactors, WAPL and PDS5A (refs 12, 13); removal of either cofactor allowed forks to progress rapidly without ESCO1, ESCO2, or RFC(CTF18). Our results show a novel mechanism for clamp-loader-dependent fork progression, mediated by the post-translational modification and structural remodelling of the cohesin ring. Loss of this regulatory mechanism leads to the spontaneous accrual of DNA damage and may contribute to the abnormalities of the Roberts' syndrome cohesinopathy.
DOI:10.1016/j.cub.2011.08.036URL [本文引用: 1]

Background: The cohesin complex mediates sister chromatid cohesion and regulates gene transcription. Prior studies show that cohesin preferentially binds and regulates genes that control growth and differentiation and that even mild disruption of cohesin function alters development. Here we investigate how cohesin specifically recognizes and regulates genes that control development in Drosophila.
Results: Genome-wide analyses show that cohesin selectively binds genes in which RNA polymerase II (Pol II) pauses just downstream of the transcription start site. These genes often have GAGA factor (GAF) binding sites 100 base pairs (bp) upstream of the start site, and GT dinucleotide repeats 50 to 800 bp downstream in the plus strand. They have low levels of histone H3 lysine 36 trimethylation (H3K36me3) associated with transcriptional elongation, even when highly transcribed. Cohesin depletion does not reduce polymerase pausing, in contrast to depletion of the NELF (negative elongation factor) pausing complex. Cohesin, NELF, and Spt5 pausing and elongation factor knockdown experiments indicate that cohesin does not inhibit binding of polymerase to promoters or physically block transcriptional elongation, but at genes that it strongly represses, it hinders transition of paused polymerase to elongation at a step distinct from those controlled by Spt5 and NELF.
Conclusions: Our findings argue that cohesin and pausing factors are recruited independently to the same genes, perhaps by GAF and the GT repeats, and that their combined action determines the level of actively elongating RNA polymerase.
DOI:10.1016/j.cub.2004.07.053URL [本文引用: 2]

Abstract
The cohesin complex is a central player in sister chromatid cohesion, a process that ensures the faithful segregation of chromosomes in mitosis and meiosis [1] and [2]. Previous genetic studies in yeast show that Scc2/Mis4, a HEAT-repeat-containing protein, is required for the loading of cohesin onto chromatin [3] and [4]. In this study, we have identified two isoforms of Scc2 in humans and Xenopus (termed Scc2A and Scc2B), which are encoded by a single gene but have different carboxyl termini created by alternative splicing. Both Scc2A and Scc2B bind to chromatin concomitant with cohesin during DNA replication in Xenopus egg extracts. Simultaneous immunodepletion of Scc2A and Scc2B from the extracts impairs the association of cohesin with chromatin, leading to severe defects in sister chromatid pairing in the subsequent mitosis. The loading of Scc2 onto chromatin is inhibited in extracts treated with geminin but not with p21CIP1, suggesting that this step depends on replication licensing but not on the initiation of DNA replication. Upon mitotic entry, Scc2 is removed from chromatin through a mechanism that requires cdc2 but not aurora B or polo-like kinase. Our results suggest that vertebrate Scc2 couples replication licensing to sister chromatid cohesion by facilitating the loading of cohesin onto chromatin.DOI:10.1101/gad.275203URLPMID:14563680 [本文引用: 1]

Chromosomal processes related to formation and function of meiotic chiasmata have been analyzed in Sordaria macrospora. Double-strand breaks (DSBs), programmed or gamma-rays-induced, are found to promote four major events beyond recombination and accompanying synaptonemal complex formation: (1) juxtaposition of homologs from long-distance interactions to close presynaptic coalignment at midleptotene; (2) structural destabilization of chromosomes at leptotene/zygotene, including sister axis separation and fracturing, as revealed in a mutant altered in the conserved, axis-associated cohesin-related protein Spo76/Pds5p; (3) exit from the bouquet stage, with accompanying global chromosome movements, at zygotene/pachytene (bouquet stage exit is further found to be a cell-wide regulatory transition and DSB transesterase Spo11p is suggested to have a new noncatalytic role in this transition); (4) normal occurrence of both meiotic divisions, including normal sister separation. Functional interactions between DSBs and the spo76-1 mutation suggest that Spo76/Pds5p opposes local destabilization of axes at developing chiasma sites and raise the possibility of a regulatory mechanism that directly monitors the presence of chiasmata at metaphase I. Local chromosome remodeling at DSB sites appears to trigger an entire cascade of chromosome movements, morphogenetic changes, and regulatory effects that are superimposed upon a foundation of DSB-independent processes.
DOI:10.1016/j.cub.2006.03.049URL [本文引用: 2]

Summary
Background
Sister-chromatid cohesion depends on the cohesin complex whose association with chromatin is mediated by Scc2 and Scc4 in budding yeast. Both cohesin and Scc2 have been conserved from yeast to humans, but no Scc4 orthologs have been identified. Mutation of Scc2 orthologs causes defects in cohesion, transcription, and development, resulting in Cornelia de Lange syndrome in humans.Results
We have identified a family of tetratricopeptide repeat proteins that share weak sequence similarities with yeast Scc4. This family includes MAU-2, which is required for development of the nervous system in Caenorhabditis elegans. We show that the human member of this family is associated with Scc2, is bound to chromatin from telophase until prophase, and is required for association of cohesin with chromatin during interphase. Cells lacking Scc4 lose sister-chromatid cohesion precociously and arrest in prometaphase. Mitotic chromosomes in Scc4-depleted cells lack cohesin, even though the cohesin-protecting proteins Sgo1 and Bub1 are normally enriched at centromeres and separase does not seem to be active.Conclusion
Our data indicate that human Scc4 is required for the association of cohesin with chromatin, which is a prerequisite for the establishment of sister-chromatid cohesion and for chromosome biorientation in mitosis. The proteinaceous machinery that is required for loading of cohesin onto chromatin is therefore conserved from yeast to humans. The finding that Caenorhabditis elegans MAU-2 is an ortholog of Scc4 further supports the notion that the Scc2-Scc4 complex is required for developmental processes in metazoans.DOI:10.1146/annurev.cellbio.24.110707.175350URLPMID:18616427 [本文引用: 1]

In eukaryotes, the process of sister chromatid cohesion holds the two sister chromatids (the replicated chromosomes) together from DNA replication to the onset of chromosome segregation. Cohesion is mediated by cohesin, a four-subunit SMC (structural maintenance of chromosome) complex. Cohesin and cohesion are required for proper chromosome segregation, DNA repair, and gene expression. To carry out these functions, cohesion is regulated by elaborate mechanisms involving a growing list of cohesin auxiliary factors. These factors control the timing and position of cohesin binding to chromatin, activate chromatin-bound cohesin to become cohesive, and orchestrate the orderly dissolution of cohesion. The 45-nm ringlike architecture of soluble cohesin is compatible with dramatically different mechanisms for both chromatin binding and cohesion generation. Solving the mechanism of cohesion and its complex regulation presents significant challenges but offers the potential to provide important insights into higher-order chromosome organization and chromosome biology.
DOI:10.7554/eLife.06057URLPMID:26038942 [本文引用: 1]

The cohesin ring holds newly replicated sister chromatids together until their separation at anaphase. Initiation of sister chromatid cohesion depends on a separate complex, Scc2(NIPBL)/Scc4(Mau2) (Scc2/4), which loads cohesin onto DNA and determines its localization across the genome. Proper cohesin loading is essential for cell division, and partial defects cause chromosome missegregation and aberrant transcriptional regulation, leading to severe developmental defects in multicellular organisms. We present here a crystal structure showing the interaction between Scc2 and Scc4. Scc4 is a TPR array that envelops an extended Scc2 peptide. Using budding yeast, we demonstrate that a conserved patch on the surface of Scc4 is required to recruit Scc2/4 to centromeres and to build pericentromeric cohesion. These findings reveal the role of Scc4 in determining the localization of cohesin loading and establish a molecular basis for Scc2/4 recruitment to centromeres.
DOI:10.1016/j.cell.2017.08.017URLPMID:28938124 [本文引用: 1]

The ring-shaped cohesin complex brings together distant DNA domains to maintain, express, and segregate the genome. Establishing specific chromosomal linkages depends on cohesin recruitment to defined loci. One such locus is the budding yeast centromere, which is a paradigm for targeted cohesin loading. The kinetochore, a multiprotein complex that connects centromeres to microtubules, drives the recruitment of high levels of cohesin to link sister chromatids together. We have exploited this system to determine the mechanism of specific cohesin recruitment. We show that phosphorylation of the Ctf19 kinetochore protein by a conserved kinase, DDK, provides a binding site for the Scc2/4 cohesin loading complex, thereby directing cohesin loading to centromeres. A similar mechanism targets cohesin to chromosomes in vertebrates. These findings represent a complete molecular description of targeted cohesin loading, a phenomenon with wide-ranging importance in chromosome segregation and, in multicellular organisms, transcription regulation.
DOI:10.1101/gad.1675708URLPMID:18708580 [本文引用: 1]

Eukaryotic chromosomes reach their stable rod-shaped appearance in mitosis in a reaction dependent on the evolutionarily conserved condensin complex. Little is known about how and where condensin associates with chromosomes. Here, we analyze condensin binding to budding yeast chromosomes using high-resolution oligonucleotide tiling arrays. Condensin-binding sites coincide with those of the loading factor Scc2/4 of the related cohesin complex. The sites map to tRNA and other genes bound by the RNA polymerase III transcription factor TFIIIC, and ribosomal protein and SNR genes. An ectopic B-box element, recognized by TFIIIC, constitutes a minimal condensin-binding site, and TFIIIC and the Scc2/4 complex promote functional condensin association with chromosomes. A similar pattern of condensin binding is conserved along fission yeast chromosomes. This reveals that TFIIIC-binding sites, including tRNA genes, constitute a hitherto unknown chromosomal feature with important implications for chromosome architecture during both interphase and mitosis.
DOI:10.1038/nature24281URLPMID:29094699 [本文引用: 3]

Imaging and chromosome conformation capture studies have revealed several layers of chromosome organization, including segregation into megabase-sized active and inactive compartments, and partitioning into sub-megabase domains (TADs). It remains unclear, however, how these layers of organization form, interact with one another and influence genome function. Here we show that deletion of the cohesin-loading factor Nipbl in mouse liver leads to a marked reorganization of chromosomal folding. TADs and associated Hi-C peaks vanish globally, even in the absence of transcriptional changes. By contrast, compartmental segregation is preserved and even reinforced. Strikingly, the disappearance of TADs unmasks a finer compartment structure that accurately reflects the underlying epigenetic landscape. These observations demonstrate that the three-dimensional organization of the genome results from the interplay of two independent mechanisms: cohesin-independent segregation of the genome into fine-scale compartments, defined by chromatin state; and cohesin-dependent formation of TADs, possibly by loop extrusion, which helps to guide distant enhancers to their target genes.
DOI:10.1038/nature02742URLPMID:15229615 [本文引用: 1]

Sister chromatids, the products of eukaryotic DNA replication, are held together by the chromosomal cohesin complex after their synthesis. This allows the spindle in mitosis to recognize pairs of replication products for segregation into opposite directions. Cohesin forms large protein rings that may bind DNA strands by encircling them, but the characterization of cohesin binding to chromosomes in vivo has remained vague. We have performed high resolution analysis of cohesin association along budding yeast chromosomes III-VI. Cohesin localizes almost exclusively between genes that are transcribed in converging directions. We find that active transcription positions cohesin at these sites, not the underlying DNA sequence. Cohesin is initially loaded onto chromosomes at separate places, marked by the Scc2/Scc4 cohesin loading complex, from where it appears to slide to its more permanent locations. But even after sister chromatid cohesion is established, changes in transcription lead to repositioning of cohesin. Thus the sites of cohesin binding and therefore probably sister chromatid cohesion, a key architectural feature of mitotic chromosomes, display surprising flexibility. Cohesin localization to places of convergent transcription is conserved in fission yeast, suggesting that it is a common feature of eukaryotic chromosomes.
DOI:10.1016/j.cub.2010.12.004URL [本文引用: 1]

Background: The Cohesin complex that holds sister chromatins together until anaphase is comprised of three core subunits: Smc1 and Smc3, two long-rod-shaped proteins with an ABC-like ATPase head (nucleotide-binding domain [NBD]) and a dimerization domain linked by a 50 nm long intramolecular antiparallel coiled-coil, and Scc1, an alpha-kleisin subunit interconnecting the NBD domains of Smc1 and Smc3. Cohesin's stable association with chromosomes is thought to involve entrapment of chromatin fibers by its tripartite Smc1-Smc3-Scc1 ring via a poorly understood mechanism dependent on a separate Scc2/4 loading complex. A key issue concerns where entrapment initially takes place: at sites where cohesin is found stably associated or at distinct "loading" sites from which it translocates.
Results: In this study, we find transition state mutant versions (Smc1E1158Q and SmcE1155Q) defective in disengagement of their nucleotide binding domains (NBDs), unlike functional cohesin, colocalize with Scc2/4 at core centromeres, sites that catalyze wild-type cohesin's recruitment to sequences 20 kb or more away. In addition to Scc2/4, the unstable association of transition state complexes with core centromeres requires Scc1's association with Smc1 and Smc3 NBDs, ATP-driven NBD engagement, cohesin's Scc3 subunit, and its hinge domain.
Conclusion: We propose that cohesin's association with chromosomes is driven by two key events. NBD engagement driven by ATP binding produces an unstable association with specific loading sites like core centromeres, whereas subsequent ATP hydrolysis triggers DNA entrapment, which permits translocation along chromatin fibers.
DOI:10.1016/j.cub.2013.02.022URL [本文引用: 1]

Cohesin is a conserved ring-shaped multiprotein complex that participates in chromosome segregation, DNA repair, and transcriptional regulation [1, 2]. Cohesin loading onto chromosomes universally requires the Scc2/4 "loader" complex (also called NippedBL/Mau2), mutations in which cause the developmental disorder Cornelia de Lange syndrome in humans [1-9]. Cohesin is most concentrated in the pericentromere, the region surrounding the centromere [10-15]. Enriched pericentromeric cohesin requires the Ctf19 kinetochore subcomplex in budding yeast [16-18]. Here, we uncover the spatial and temporal determinants for Scc2/4 centromere association. We demonstrate that the critical role of the Ctf19 complex is to enable Scc2/4 association with centromeres, through which cohesin loads and spreads onto the adjacent pericentromere. We show that, unexpectedly, Scc2 association with centromeres depends on cohesin itself. The absence of the Scc1/Mcd1/Rad21 cohesin subunit precludes Scc2 association with centromeres from anaphase until late G1. Expression of SCC1 is both necessary and sufficient for the binding of cohesin to its loader, the association of Scc2 with centromeres, and cohesin loading. We propose that cohesin triggers its own loading by enabling Scc2/4 to connect with chromosomal landmarks, which at centromeres are specified by the Ctf19 complex. Overall, our findings provide a paradigm for the spatial and temporal control of cohesin loading.
DOI:10.7554/eLife.30000URLPMID:28914604 [本文引用: 1]

The cohesin complex mediates DNA-DNA interactions both between (sister chromatid cohesion) and within chromosomes (DNA looping). It has been suggested that intra-chromosome loops are generated by extrusion of DNAs through the lumen of cohesin's ring. Scc2 (Nipbl) stimulates cohesin's ABC-like ATPase and is essential for loading cohesin onto chromosomes. However, it is possible that the stimulation of cohesin's ATPase by Scc2 also has a post-loading function, for example driving loop extrusion. Using fluorescence recovery after photobleaching (FRAP) and single-molecule tracking in human cells, we show that Scc2 binds dynamically to chromatin, principally through an association with cohesin. Scc2's movement within chromatin is consistent with a 'stop-and-go' or 'hopping' motion. We suggest that a low diffusion coefficient, a low stoichiometry relative to cohesin, and a high affinity for chromosomal cohesin enables Scc2 to move rapidly from one chromosomal cohesin complex to another, performing a function distinct from loading.
DOI:10.1016/j.cub.2003.10.036URL [本文引用: 1]

Abstract
Background: A multi-subunit protein complex called cohesin is involved in holding sister chromatids together after DNA replication. Cohesin contains four core subunits: Smc1, Smc3, Scc1, and Scc3. Biochemical studies suggest that Smc1 and Smc3 each form 50 nm-long antiparallel coiled coils (arms) and bind to each other to form V-shaped heterodimers with globular ABC-like ATPases (created by the juxtaposition of N- and C-terminal domains) at their apices. These Smc “heads” are connected by Scc1, creating a tripartite proteinaceous ring.Results: To investigate the role of Smc1 and Smc3's ATPase domains, we engineered smc1 and smc3 mutations predicted to abolish either ATP binding or hydrolysis. All mutations abolished Smc protein function. The binding of ATP to Smc1, but not Smc3, was essential for Scc1's association with Smc1/3 heterodimers. In contrast, mutations predicted to prevent hydrolysis of ATP bound to either head abolished cohesin's association with chromatin but not Scc1's ability to connect Smc1's head with that of Smc3. Inactivation of the Scc2/4 complex had a similar if not identical effect; namely, the production of tripartite cohesin rings that cannot associate with chromosomes.
Conclusions: Cohesin complexes whose heads have been connected by Scc1 must hydrolyze ATP in order to associate stably with chromosomes. If chromosomal association is mediated by the topological entrapment of DNA inside cohesin's ring, then ATP hydrolysis may be responsible for creating a gate through which DNA can enter. We suggest that ATP hydrolysis drives the temporary disconnection of Scc1 from Smc heads that are needed for DNA entrapment and that this process is promoted by Scc2/4.
DOI:10.1016/j.cell.2006.08.048URLPMID:17081975 [本文引用: 1]

Cohesin is a multisubunit complex that mediates sister-chromatid cohesion. Its Smc1 and Smc3 subunits possess ABC-like ATPases at one end of 50 nm long coiled coils. At the other ends are pseudosymmetrical hinge domains that interact to create V-shaped Smc1/Smc3 heterodimers. N- and C-terminal domains within cohesin's kleisin subunit Scc1 bind to Smc3 and Smc1 ATPase heads respectively, thereby creating a huge tripartite ring. It has been suggested that cohesin associates with chromosomes by trapping DNA within its ring. Opening of the ring due to cleavage of Scc1 by separase destroys sister-chromatid cohesion and triggers anaphase. We show that cohesin's hinges are not merely dimerization domains. They are essential for cohesin's association with chromosomes, which is blocked by artificially holding hinge domains together but not by preventing Scc1's dissociation from SMC ATPase heads. Our results suggest that entry of DNA into cohesin's ring requires transient dissociation of Smc1 and Smc3 hinge domains.
DOI:10.1371/journal.pbio.0040242URLPMID:16802858 [本文引用: 1]

Saccharomyces cerevisiae Scc2 binds Scc4 to form an essential complex that loads cohesin onto chromosomes. The prevalence of Scc2 orthologs in eukaryotes emphasizes a conserved role in regulating sister chromatid cohesion, but homologs of Scc4 have not hitherto been identified outside certain fungi. Some metazoan orthologs of Scc2 were initially identified as developmental gene regulators, such as Drosophila Nipped-B, a regulator of cut and Ultrabithorax, and delangin, a protein mutant in Cornelia de Lange syndrome. We show that delangin and Nipped-B bind previously unstudied human and fly orthologs of Caenorhabditis elegans MAU-2, a non-axis-specific guidance factor for migrating cells and axons. PSI-BLAST shows that Scc4 is evolutionarily related to metazoan MAU-2 sequences, with the greatest homology evident in a short N-terminal domain, and protein-protein interaction studies map the site of interaction between delangin and human MAU-2 to the N-terminal regions of both proteins. Short interfering RNA knockdown of human MAU-2 in HeLa cells resulted in precocious sister chromatid separation and in impaired loading of cohesin onto chromatin, indicating that it is functionally related to Scc4, and RNAi analyses show that MAU-2 regulates chromosome segregation in C. elegans embryos. Using antisense morpholino oligonucleotides to knock down Xenopus tropicalis delangin or MAU-2 in early embryos produced similar patterns of retarded growth and developmental defects. Our data show that sister chromatid cohesion in metazoans involves the formation of a complex similar to the Scc2-Scc4 interaction in the budding yeast. The very high degree of sequence conservation between Scc4 homologs in complex metazoans is consistent with increased selection pressure to conserve additional essential functions, such as regulation of cell and axon migration during development.
DOI:10.1111/j.1365-313X.2009.03845.xURLPMID:19228337 [本文引用: 4]

Adherin plays an important role in loading the cohesin complex onto chromosomes, and is essential for the establishment of sister-chromatid cohesion. We have identified and analyzed the Arabidopsis adherin homolog AtSCC2. Interestingly, the sequence analysis of AtSCC2 and of other putative plant adherin homologs revealed the presence of a PHD finger, which is not found in their fungal and animal counterparts. AtSCC2 is identical to EMB2773, and mutants show early embryo lethality and formation of giant endosperm nuclei. A role for AtSCC2 in sister-chromatid cohesion was established by using conditional RNAi and examining meiotic chromosome organization. AtSCC2-RNAi lines showed sterility, arising from the following defects in meiotic chromosome organization: failure of homologous pairing, loss of sister-chromatid cohesion, mixed segregation of chromosomes and chromosome fragmentation. The mutant phenotype, which included defects in chromosome organization and cohesion in prophase I, is distinct from that of the Arabidopsis cohesin mutant Atrec8, which retains centromere cohesion up to anaphase I. Immunostaining experiments revealed the aberrant distribution of the cohesin subunit AtSCC3 on chromosomes, and defects in chromosomal axis formation, in the meiocytes of AtSCC2-RNAi lines. These results demonstrate a role for AtSCC2 in sister-chromatid cohesion and centromere organization, and show that the machinery responsible for the establishment of cohesion is conserved in plants.
DOI:10.3724/SP.J.1005.2014.0208URL [本文引用: 1]

Histone modification is one important sort of the epigenetic modifications, including acetylation, formylation, methylation, phosphorylation, ubiquitination and SUMOylation. By forming a complicated network, these modifications control the expression of genes. Histone methylation occurs mainly on the lysine residues, and plays a key role during flowering and stress response of plants, through changing the methylation status of lysine residues and the ratio of methylation. Triple-methylation of H3K4 promotes FLC expression but triple-methylation of H3K27 inhibits its expression. H3K4me3 activates the expression of PtdIns5P gene to initiate lipid synthesis signal pathway in response to drought stress. On the contrary, the low levels of H3K27me3 induce the expression of COR15A and ATGOLS3, which encode for low temperature protective proteins of chloroplast (Cor15am) and Galactional Synthase (GOLS), in order to resist cold stress. In this review, we summarize the molecular mechanisms of histone lysine methylation involved in DNA methylation, plant flowering and stress response.
DOI:10.3724/SP.J.1005.2014.0208URL [本文引用: 1]

Histone modification is one important sort of the epigenetic modifications, including acetylation, formylation, methylation, phosphorylation, ubiquitination and SUMOylation. By forming a complicated network, these modifications control the expression of genes. Histone methylation occurs mainly on the lysine residues, and plays a key role during flowering and stress response of plants, through changing the methylation status of lysine residues and the ratio of methylation. Triple-methylation of H3K4 promotes FLC expression but triple-methylation of H3K27 inhibits its expression. H3K4me3 activates the expression of PtdIns5P gene to initiate lipid synthesis signal pathway in response to drought stress. On the contrary, the low levels of H3K27me3 induce the expression of COR15A and ATGOLS3, which encode for low temperature protective proteins of chloroplast (Cor15am) and Galactional Synthase (GOLS), in order to resist cold stress. In this review, we summarize the molecular mechanisms of histone lysine methylation involved in DNA methylation, plant flowering and stress response.
DOI:10.1242/jcs.196865URLPMID:28137757 [本文引用: 1]

Factors regulating dynamics of chromatin structure have direct impact on expression of genetic information. Cohesin is a multi-subunit protein complex that is crucial for pairing sister chromatids during cell division, DNA repair and regulation of gene transcription and silencing. In non-plant species, cohesin is loaded on chromatin by the Scc2-Scc4 complex (also known as the NIBPL-MAU2 complex). Here, we identify the Arabidopsis homolog of Scc4, which we denote Arabidopsis thaliana (At)SCC4, and show that it forms a functional complex with AtSCC2, the homolog of Scc2. We demonstrate that AtSCC2 and AtSCC4 act in the same pathway, and that both proteins are indispensable for cell fate determination during early stages of embryo development. Mutant embryos lacking either of these proteins develop only up to the globular stage, and show the suspensor overproliferation phenotype preceded by ectopic auxin maxima distribution. We further establish a new assay to reveal the AtSCC4-dependent dynamics of cohesin loading on chromatin in vivo Our findings define the Scc2-Scc4 complex as an evolutionary conserved machinery controlling cohesin loading and chromatin structure maintenance, and provide new insight into the plant-specific role of this complex in controlling cell fate during embryogenesis.
DOI:10.1105/tpc.18.00921URLPMID:30705131 [本文引用: 1]

Cohesin complexes maintain sister chromatid cohesion to ensure proper chromosome segregation during mitosis and meiosis. In plants, the exact components and functions of the cohesin complex remain poorly understood. Here, we positionally cloned the classic maize (Zea mays) mutant defective kernel 15 (dek15), revealing that it encodes a homolog of SISTER CHROMATID COHESION PROTEIN 4 (SCC4), a loader subunit of the cohesin ring. Developing dek15 kernels contained fewer cells than the wild type, but had a highly variable cell size. The dek15 mutation was found to disrupt the mitotic cell cycle and endoreduplication, resulting in a reduced endosperm and embryo lethality. The cells in the dek15 endosperm and embryo exhibited precocious sister chromatid separation and other chromosome segregation errors, including misaligned chromosomes, lagging chromosomes, and micronuclei, resulting in a high percentage of aneuploid cells. The loss of Dek15/Scc4 function upregulated the expression of genes involved in cell cycle progression and stress responses, and downregulated key genes involved in organic synthesis during maize endosperm development. Our yeast two-hybrid screen identified the chromatin remodeling proteins chromatin remodeling factor 4, chromatin remodeling complex subunit B (CHB)102, CHB105, and CHB106 as SCC4-interacting proteins, suggesting a possible mechanism by which the cohesin ring is loaded onto chromatin in plant cells. This study revealed biological functions for DEK15/SCC4 in mitotic chromosome segregation and kernel development in maize.
DOI:10.1126/science.1157774URLPMID:18653893 [本文引用: 1]

Replicated chromosomes are held together by the chromosomal cohesin complex from the time of their synthesis in S phase onward. This requires the replication fork-associated acetyl transferase Eco1, but Eco1's mechanism of action is not known. We identified spontaneous suppressors of the thermosensitive eco1-1 allele in budding yeast. An acetylation-mimicking mutation of a conserved lysine in cohesin's Smc3 subunit makes Eco1 dispensable for cell growth, and we show that Smc3 is acetylated in an Eco1-dependent manner during DNA replication to promote sister chromatid cohesion. A second set of eco1-1 suppressors inactivate the budding yeast ortholog of the cohesin destabilizer Wapl. Our results indicate that Eco1 modifies cohesin to stabilize sister chromatid cohesion in parallel with a cohesion establishment reaction that is in principle Eco1-independent.
DOI:10.1126/science.1157880URLPMID:18653894 [本文引用: 1]

Chromosome segregation, transcriptional regulation, and repair of DNA double-strand breaks require the cohesin protein complex. Cohesin holds the replicated chromosomes (sister chromatids) together to mediate sister chromatid cohesion. The mechanism of how cohesion is established is unknown. We found that in budding yeast, the head domain of the Smc3p subunit of cohesin is acetylated by the Eco1p acetyltransferase at two evolutionarily conserved residues, promoting the chromatin-bound cohesin to tether sister chromatids. Smc3p acetylation is induced in S phase after the chromatin loading of cohesin and is suppressed in G(1) and G(2)/M. Smc3 head acetylation and its cell cycle regulation provide important insights into the biology and mechanism of cohesion establishment.
DOI:10.1016/j.molcel.2009.02.028URLPMID:19328069 [本文引用: 2]

Cohesin's Smc1, Smc3, and Scc1 subunits form a tripartite ring that entraps sister DNAs. Scc3, Pds5, and Rad61 (Wapl) are regulatory subunits that control this process. We describe here smc3, scc3, pds5, and rad61 mutations that permit yeast cell proliferation and entrapment of sister DNAs by cohesin rings in the absence of Eco1, an acetyl transferase normally essential for establishing sister chromatid cohesion. The smc3 mutations cluster around and include a highly conserved lysine (K113) close to Smc3's ATP-binding pocket, which, together with K112, is acetylated by Eco1. Lethality caused by mutating both residues to arginine is suppressed by the scc3, pds5, and rad61 mutants. Scc3, Pds5, and Rad61 form a complex and inhibit entrapment of sister DNAs by a process involving the "K112/K113" surface on Smc3's ATPase. According to this model, Eco1 promotes sister DNA entrapment partly by relieving an antiestablishment activity associated with Scc3, Pds5, and Rad61.
DOI:10.1016/j.molcel.2008.06.006URLPMID:18614053 [本文引用: 3]

Sister chromatid cohesion is normally established in S phase in a process that depends on the cohesion establishment factor Eco1, a conserved acetyltransferase. However, due to the lack of known in vivo substrates, how Eco1 regulates cohesion is not understood. Here we report that yeast Eco1 and its human ortholog, ESCO1, both acetylate Smc3, a component of the cohesin complex that physically holds the sister chromatid together, at two conserved lysine residues. Mutating these lysine residues to a nonacetylatable form leads to increased loss of sister chromatid cohesion and genome instability in both yeast and human. In addition, we clarified that the acetyltransferase activity of Eco1 is essential for its function. Our study thus identified a molecular target for the acetyltransferase Eco1 and revealed that Smc3 acetylation is a conserved mechanism in regulating sister chromatid cohesion.
DOI:10.1091/mbc.e04-12-1063URLPMID:15958495 [本文引用: 2]

Genetic studies in yeast and Drosophila have uncovered a conserved acetyltransferase involved in sister-chromatid cohesion. Here, we described the two human orthologues, previously named EFO1/ESCO1 and EFO2/ESCO2. Similar to their yeast (Eco1/Ctf7 and Eso1) and fly (deco) counterparts, both proteins feature a conserved C-terminal domain consisting of a H2C2 zinc finger motif and an acetyltransferase domain that is able to catalyze autoacetylation reaction in vitro. However, no similarity can be detected outside of the conserved domain. RNA interference depletion experiment revealed that EFO1/ESCO1 and EFO2/ESCO2 were not redundant and that both were required for proper sister-chromatid cohesion. The difference between EFO1 and EFO2 also is reflected in their cell cycle regulation. In mitosis, EFO1 is phosphorylated, whereas EFO2 is degraded. Furthermore, both proteins associate with chromosomes, and the chromosome binding depends on the diverse N-terminal domains. We propose that EFO1 and EFO2 are targeted to different chromosome structures to help establish or maintain sister-chromatid cohesion.
DOI:10.1101/gad.13.3.307URLPMID:9990855 [本文引用: 1]

CTF7 (chromosome transmission fidelity) gene in budding yeast encodes an essential protein that is required for high-fidelity chromosome transmission and contains regions of identity conserved from yeast to man. ctf7 mutant cells arrested prior to anaphase onset contain separated sister chromatids. Thus, Ctf7p is essential for cohesion. Cohesion is established during S phase and then maintained until mitosis. However, Ctf7p activity is required only during S phase, suggesting that Ctf7p functions in the establishment of cohesion. In addition, ctf7 genetically interacts with DNA metabolism mutations pol30 (PCNA) and ctf18 (an RF-C like protein) and ctf7 temperature sensitivity and chromosome loss are rescued by high levels of POL30. These findings provide the first evidence that links the establishment of sister chromatid cohesion to the DNA replication machinery and suggest that the assembly of cohesion (and possibly condensation) complexes are coupled to PCNA-dependent DNA replication. The analysis of Ctf7p also reveals an important connection between sister chromatid cohesion, spindle integrity and the spindle assembly checkpoint.
DOI:10.1371/journal.pgen.0030012URLPMID:17238288 [本文引用: 1]

The cohesion of sister chromatids is mediated by cohesin, a protein complex containing members of the structural maintenance of chromosome (Smc) family. How cohesins tether sister chromatids is not yet understood. Here, we mutate SMC1, the gene encoding a cohesin subunit of budding yeast, by random insertion dominant negative mutagenesis to generate alleles that are highly informative for cohesin assembly and function. Cohesins mutated in the Hinge or Loop1 regions of Smc1 bind chromatin by a mechanism similar to wild-type cohesin, but fail to enrich at cohesin-associated regions (CARs) and pericentric regions. Hence, the Hinge and Loop1 regions of Smc1 are essential for the specific chromatin binding of cohesin. This specific binding and a subsequent Ctf7/Eco1-dependent step are both required for the establishment of cohesion. We propose that a cohesin or cohesin oligomer tethers the sister chromatids through two chromatin-binding events that are regulated spatially by CAR binding and temporally by Ctf7 activation, to ensure cohesins crosslink only sister chromatids.
DOI:10.1128/mcb.23.8.2999-3007.2003URLPMID:12665596 [本文引用: 1]

CTF7/ECO1 is an essential yeast gene required for the establishment of sister chromatid cohesion. The findings that CTF7/ECO1, POL30 (PCNA), and CHL12/CTF18 (a replication factor C [RFC] homolog) genetically interact provided the first evidence that the processes of cohesion establishment and DNA replication are intimately coupled-a link now confirmed by other studies. To date, however, it is unknown how Ctf7p/Eco1p function is coupled to DNA replication or whether Ctf7p/Eco1p physically associates with any components of the DNA replication machinery. Here, we report that Ctf7p/Eco1p associates with proteins that perform partially redundant functions in DNA replication. Chl12p/Ctf18p combines with Rfc2p to Rfc5p to form one of three independent RFC complexes. By chromatographic methods, Ctf7p/Eco1p was found to associate with Chl12/Ctf18p and with Rfc2p, Rfc3p, Rfc4p, and Rfc5p. The association between Ctf7p/Eco1p and this RFC complex is biologically relevant in that (i) Ctf7p/Eco1p cosediments with Chl12p/Ctf18p in vivo and (ii) rfc5-1 mutant cells exhibit precocious sister separation. Previous studies revealed that Rfc1p or Rad24p associates with Rfc2p to Rfc5p to form two other RFC complexes independent of Ctf18p-RFC complexes. These Rfc1p-RFC and Rad24p-RFC complexes function in DNA replication or repair and DNA damage checkpoint pathways. Importantly, Ctf7p/Eco1p also associates with Rfc1p and Rad24p, suggesting that these RFC complexes also play critical roles in cohesion establishment. The associations between Ctf7p/Eco1p and RFC subunits provide novel evidence regarding the physical linkage between cohesion establishment and DNA replication. Furthermore, the association of Ctf7p/Eco1p with each of three RFC complexes supplies new insights into the functional redundancy of RFC complexes in cohesion establishment.
DOI:10.1016/j.molcel.2006.08.018URLPMID:16962805 [本文引用: 1]

Two identical sister copies of eukaryotic chromosomes are synthesized during S phase. To facilitate their recognition as pairs for segregation in mitosis, sister chromatids are held together from their synthesis onward by the chromosomal cohesin complex. Replication fork progression is thought to be coupled to establishment of sister chromatid cohesion, facilitating identification of replication products, but evidence for this has remained circumstantial. Here we show that three proteins required for sister chromatid cohesion, Eco1, Ctf4, and Ctf18, are found at, and Ctf4 travels along chromosomes with, replication forks. The ring-shaped cohesin complex is loaded onto chromosomes before S phase in an ATP hydrolysis-dependent reaction. Cohesion establishment during DNA replication follows without further cohesin recruitment and without need for cohesin to re-engage an ATP hydrolysis motif that is critical for its initial DNA binding. This provides evidence for cohesion establishment in the context of replication forks and imposes constraints on the mechanism involved.
DOI:10.1016/j.cub.2007.02.029URL [本文引用: 1]

Summary
Sister chromatid cohesion depends on cohesin [1], [2] and [3]. Cohesin associates with chromatin dynamically throughout interphase [4]. During DNA replication, cohesin establishes cohesion [5], and this process coincides with the generation of a cohesin subpopulation that is more stably bound to chromatin [4]. In mitosis, cohesin is removed from chromosomes, enabling sister chromatid separation [6]. How cohesin associates with chromatin and establishes cohesion is poorly understood. By searching for proteins that are associated with chromatin-bound cohesin, we have identified sororin, a protein that was known to be required for cohesion [7]. To obtain further insight into sororin's function, we have addressed when during the cell cycle sororin is required for cohesion. We show that sororin is dispensable for the association ofcohesin with chromatin but that sororin is essential for proper cohesion during G2 phase. Like cohesin, sororin is also needed for efficient repair of DNA double-strand breaks in G2. Finally, sororin is required for the presence of normal amounts of the stably chromatin-bound cohesin population in G2. Our data indicate that sororin interacts with chromatin-bound cohesin and functions during the establishment or maintenance of cohesion in S or G2 phase, respectively.DOI:10.1128/MCB.01284-10URL [本文引用: 1]

In budding yeast and humans, cohesion establishment during S phase requires the acetyltransferase Eco1/Esco1-2, which acetylates the cohesin subunit Smc3 on two conserved lysine residues. Whether Smc3 is the sole Eco1/Esco1-2 effector and how Smc3 acetylation promotes cohesion are unknown. In fission yeast (Schizosaccharomyces pombe), as in humans, cohesin binding to G(1) chromosomes is dynamic and the unloading reaction is stimulated by Wpl1 (human ortholog, Wapl). During S phase, a subpopulation of cohesin becomes stably bound to chromatin in an Eso1 (fission yeast Eco1/Esco1-2)-dependent manner. Cohesin stabilization occurs unevenly along chromosomes. Cohesin remains largely labile at the rDNA repeats but binds mostly in the stable mode to pericentromere regions. This pattern is largely unchanged in eso1 Delta wpl1 Delta cells, and cohesion is unaffected, indicating that the main Eso1 role is counteracting Wpl1. A mutant of Psm3 (fission yeast Smc3) that mimics its acetylated state renders cohesin less sensitive to Wpl1-dependent unloading and partially bypasses the Eso1 requirement but cannot generate the stable mode of cohesin binding in the absence of Eso1. Conversely, nonacetylatable Psm3 reduces the stable cohesin fraction and affects cohesion in a Wpl1-dependent manner, but cells are viable. We propose that Psm3 acetylation contributes to Eso1 counteracting of Wpl1 to secure stable cohesin interaction with postreplicative chromosomes but that it is not the sole molecular event by which this occurs.
DOI:10.1016/j.cell.2010.10.031URLPMID:21111234 [本文引用: 2]

Sister chromatid cohesion is essential for chromosome segregation and is mediated by cohesin bound to DNA. Cohesin-DNA interactions can be reversed by the cohesion-associated protein Wapl, whereas a stably DNA-bound form of cohesin is thought to mediate cohesion. In vertebrates, Sororin is essential for cohesion and stable cohesin-DNA interactions, but how Sororin performs these functions is unknown. We show that DNA replication and cohesin acetylation promote binding of Sororin to cohesin, and that Sororin displaces Wapl from its binding partner Pds5. In the absence of Wapl, Sororin becomes dispensable for cohesion. We propose that Sororin maintains cohesion by inhibiting Wapl's ability to dissociate cohesin from DNA. Sororin has only been identified in vertebrates, but we show that many invertebrate species contain Sororin-related proteins, and that one of these, Dalmatian, is essential for cohesion in Drosophila. The mechanism we describe here may therefore be widely conserved among different species.
DOI:10.1038/embor.2012.72URL [本文引用: 1]

Pds5 and Wpl1 act as anti-establishment factors preventing sister-chromatid cohesion until counteracted in S-phase by the cohesin acetyl-transferase Eso1. However, Pds5 is also required to maintain sister-chromatid cohesion in G2. Here, we show that Pds5 is essential for cohesin acetylation by Eso1 and ensures the maintenance of cohesion by promoting a stable cohesin interaction with replicated chromosomes. The latter requires Eso1 only in the presence of Wapl, indicating that cohesin stabilization relies on Eso1 only to neutralize the anti-establishment activity. We suggest that Eso1 requires Pds5 to counteract anti-establishment. This allows both cohesion establishment and Pds5-dependent stable cohesin binding to chromosomes.
DOI:10.1016/j.cell.2006.09.040URLPMID:17113138 [本文引用: 2]

Cohesin establishes sister-chromatid cohesion from S phase until mitosis or meiosis. To allow chromosome segregation, cohesion has to be dissolved. In vertebrate cells, this process is mediated in part by the protease separase, which destroys a small amount of cohesin, but most cohesin is removed from chromosomes without proteolysis. How this is achieved is poorly understood. Here, we show that the interaction between cohesin and chromatin is controlled by Wapl, a protein implicated in heterochromatin formation and tumorigenesis. Wapl is associated with cohesin throughout the cell cycle, and its depletion blocks cohesin dissociation from chromosomes during the early stages of mitosis and prevents the resolution of sister chromatids until anaphase, which occurs after a delay. Wapl depletion also increases the residence time of cohesin on chromatin in interphase. Our data indicate that Wapl is required to unlock cohesin from a particular state in which it is stably bound to chromatin.
DOI:10.1016/j.cub.2009.01.062URL [本文引用: 1]

Summary
Sister chromatid cohesion, which is mediated by the cohesin complex, is vital for faithful segregation of chromosomes in mitosis and meiosis (reviewed in [1]). Cohesion is established during S phase, and this process requires the function of the acetyltransferase Eco1/Ctf7 [2], [3], [4] and [5]. The mechanism of the cohesion establishment is, however, still unclear. Here, we describe isolation and identification of genetic suppressors of budding yeast eco1-1 temperature-sensitive mutant. By using a recently described microarray-based method [6], we successfully mapped 11 intergenic suppressor mutations in two genes, wpl1 (also known as rad61) and pds5. Pds5 is a known accessory factor of cohesin complex [7], [8], [9], [10] and [11], and we show that Wpl1/Rad61 protein forms a complex with Pds5 and colocalizes with cohesin on chromosomes, as its presumed human homolog Wapl [12] and [13]. Impaired function of Wpl1-Pds5 complex makes Eco1 dispensable for cell survival. We also provide evidence that Wpl1 is required for efficient association of cohesin with G2 phase chromosomes and that Eco1 promotes dissociation of Wpl1-Pds5 from cohesin via acetylation of Smc3, a cohesin subunit. Taken together, the presented data suggest that Wpl1-Pds5 complex is inhibitory for cohesion establishment and that Eco1 establishes cohesion by hindering the function of Wpl1-Pds5 temporally in S phase.DOI:10.1016/j.cell.2012.07.028URL [本文引用: 1]

Sister chromatid cohesion is mediated by entrapment of sister DNAs by a tripartite ring composed of cohesin's Smc1, Smc3, and alpha-kleisin subunits. Cohesion requires acetylation of Smc3 by Eco1, whose role is to counteract an inhibitory (antiestablishment) activity associated with cohesin's Wapl subunit. We show that mutations abrogating antiestablishment activity also reduce turnover of cohesin on pericentric chromatin. Our results reveal a "releasing" activity inherent to cohesin complexes transiently associated with Wapl that catalyzes their dissociation from chromosomes. Fusion of Smc3's nucleotide binding domain to alpha-kleisin's N-terminal domain also reduces cohesin turnover within pericentric chromatin and permits establishment of Wapl-resistant cohesion in the absence of Eco1. We suggest that releasing activity opens the Smc3/alpha-kleisin interface, creating a DNA exit gate distinct from its proposed entry gate at the Smc1/3 interface. According to this notion, the function of Smc3 acetylation is to block its dissociation from alpha-kleisin. The functional implications of regulated ring opening are discussed.
DOI:10.1104/pp.110.157560URLPMID:20671110 [本文引用: 4]

CTF7 is an essential gene in yeast that is required for the formation of sister chromatid cohesion. While recent studies have provided insights into how sister chromatid cohesion is established, less is known about how specifically CTF7 facilitates the formation of cohesion, and essentially nothing is known about how sister chromatid cohesion is established in plants. In this report, we describe the isolation and characterization of CTF7 from Arabidopsis (Arabidopsis thaliana). Arabidopsis CTF7 is similar to Saccharomyces cerevisiae CTF7 in that it lacks an amino-terminal extension, exhibits acetyltransferase activity, and can complement a yeast ctf7 temperature-sensitive mutation. CTF7 transcripts are found throughout the plant, with the highest levels present in buds. Seeds containing T-DNA insertions in CTF7 exhibit mitotic defects in the zygote. Interestingly, the endosperm developed normally in ctf7 seeds, suggesting that CTF7 is not essential for mitosis in endosperm nuclei. Minor defects were observed in female gametophytes of ctf7(+/-) plants, and plants that overexpress CTF7 exhibited female gametophyte lethality. Pollen development appeared normal in both CTF7 knockout and overexpression plants. Therefore, proper levels of CTF7 are critical for female gametophyte and embryo development but not for the establishment of mitotic cohesion during microgametogenesis or during endosperm development.
DOI:10.1111/tpj.12261URL [本文引用: 4]

The proper transmission of DNA in dividing cells is crucial for the survival of eukaryotic organisms. During cell division, faithful segregation of replicated chromosomes requires their tight attachment, known as sister chromatid cohesion, until anaphase. Sister chromatid cohesion is established during S-phase in a process requiring an acetyltransferase that in yeast is known as Establishment of cohesion 1 (Eco1). Inactivation of Eco1 typically disrupts chromosome segregation and homologous recombination-dependent DNA repair in dividing cells, ultimately resulting in lethality. We report here the isolation and detailed characterization of two homozygous T-DNA insertion mutants for the Arabidopsis thaliana Eco1 homolog, CHROMOSOME TRANSMISSION FIDELITY 7/ESTABLISHMENT OF COHESION 1 (CTF7/ECO1), called ctf7-1 and ctf7-2. Mutants exhibited dwarfism, poor anther development and sterility. Analysis of somatic tissues by flow cytometry, scanning electron microscopy and quantitative real-time PCR identified defects in DNA repair and cell division, including an increase in the area of leaf epidermal cells, an increase in DNA content and the upregulation of genes involved in DNA repair including BRCA1 and PARP2. No significant change was observed in the expression of genes that influence entry into the endocycle. Analysis of meiocytes identified changes in chromosome morphology and defective segregation; the abundance of chromosomal-bound cohesion subunits was also reduced. Transcript levels for several meiotic genes, including the recombinase genes DMC1 and RAD51C and the S-phase licensing factor CDC45 were elevated in mutant anthers. Taken together our results demonstrate that Arabidopsis CTF7/ECO1 plays important roles in the preservation of genome integrity and meiosis.
DOI:10.1186/1471-2229-13-117URLPMID:23941555 [本文引用: 1]

The establishment of sister chromatid cohesion followed by its controlled release at the metaphase to anaphase transition is necessary for faithful segregation of chromosomes in mitosis and meiosis. Cohesion is established by the action of Ctf7/Eco1 on the cohesin complex during DNA replication following loading of cohesin onto chromatin by the Scc2-Scc4 complex. Ctf7 is also required for sister chromatid cohesion during repair of DNA double strand breaks. Ctf7 contains an acetyltransferase domain and a zinc finger motif and acetylates conserved lysine residues in the Smc3 subunit of cohesin. In Arabidopsis CTF7 is encoded by a single gene and mutations in AtCTF7 cause embryo lethality indicating that the gene is essential.
DOI:10.1186/s12870-015-0452-2URLPMID:25848842 [本文引用: 2]

Eco1/Ctf7 is essential for the establishment of sister chromatid cohesion during S phase of the cell cycle. Inactivation of Ctf7/Eco1 leads to a lethal phenotype in most organisms. Altering Eco1/Ctf7 levels or point mutations in the gene can lead to alterations in nuclear division as well as a wide range of developmental defects. Inactivation of Arabidopsis CTF7 (AtCTF7) results in severe defects in reproduction and vegetative growth.
DOI:10.1371/journal.pbio.0030069URLPMID:15737063 [本文引用: 1]

Cohesin is a protein complex that is required to hold sister chromatids together. Cleavage of the Scc1 subunit of cohesin by the protease separase releases the complex from chromosomes and thereby enables the separation of sister chromatids in anaphase. In vertebrate cells, the bulk of cohesin dissociates from chromosome arms already during prophase and prometaphase without cleavage of Scc1. Polo-like kinase 1 (Plk1) and Aurora-B are required for this dissociation process, and Plk1 can phosphorylate the cohesin subunits Scc1 and SA2 in vitro, consistent with the possibility that cohesin phosphorylation by Plk1 triggers the dissociation of cohesin from chromosome arms. However, this hypothesis has not been tested yet, and in budding yeast it has been found that phosphorylation of Scc1 by the Polo-like kinase Cdc5 enhances the cleavability of cohesin, but does not lead to separase-independent dissociation of cohesin from chromosomes. To address the functional significance of cohesin phosphorylation in human cells, we have searched for phosphorylation sites on all four subunits of cohesin by mass spectrometry. We have identified numerous mitosis-specific sites on Scc1 and SA2, mutated them, and expressed nonphosphorylatable forms of both proteins stably at physiological levels in human cells. The analysis of these cells lines, in conjunction with biochemical experiments in vitro, indicate that Scc1 phosphorylation is dispensable for cohesin dissociation from chromosomes in early mitosis but enhances the cleavability of Scc1 by separase. In contrast, our data reveal that phosphorylation of SA2 is essential for cohesin dissociation during prophase and prometaphase, but is not required for cohesin cleavage by separase. The similarity of the phenotype obtained after expression of nonphosphorylatable SA2 in human cells to that seen after the depletion of Plk1 suggests that SA2 is the critical target of Plk1 in the cohesin dissociation pathway.
DOI:10.1073/pnas.1304594110URLPMID:23776203 [本文引用: 1]

Cohesin, along with positive regulators, establishes sister-chromatid cohesion by forming a ring to circle chromatin. The wings apart-like protein (Wapl) is a key negative regulator of cohesin and forms a complex with precocious dissociation of sisters protein 5 (Pds5) to promote cohesin release from chromatin. Here we report the crystal structure and functional characterization of human Wapl. Wapl contains a flexible, variable N-terminal region (Wapl-N) and a conserved C-terminal domain (Wapl-C) consisting of eight HEAT (Huntingtin, Elongation factor 3, A subunit, and target of rapamycin) repeats. Wapl-C folds into an elongated structure with two lobes. Structure-based mutagenesis maps the functional surface of Wapl-C to two distinct patches (I and II) on the N lobe and a localized patch (III) on the C lobe. Mutating critical patch I residues weaken Wapl binding to cohesin and diminish sister-chromatid resolution and cohesin release from mitotic chromosomes in human cells and Xenopus egg extracts. Surprisingly, patch III on the C lobe does not contribute to Wapl binding to cohesin or its known regulators. Although patch I mutations reduce Wapl binding to intact cohesin, they do not affect Wapl-Pds5 binding to the cohesin subcomplex of sister chromatid cohesion protein 1 (Scc1) and stromal antigen 2 (SA2) in vitro, which is instead mediated by Wapl-N. Thus, Wapl-N forms extensive interactions with Pds5 and Scc1-SA2. Wapl-C interacts with other cohesin subunits and possibly unknown effectors to trigger cohesin release from chromatin.
DOI:10.1016/j.cub.2006.10.061URL [本文引用: 1]

Summary
Background
The linkage between duplicated chromosomes (sister chromatids) is established during S phase by the action of cohesin, a multisubunit complex conserved from yeast to humans. Most cohesin dissociates from chromosome arms when the cell enters mitotic prophase, leading to the formation of metaphase chromosomes with two cytologically discernible chromatids. This process is known as sister-chromatid resolution. Although two mitotic kinases have been implicated inthis process, it remains unknown exactly how the cohesin-mediated linkage is destabilized at a mechanistic level.Results
The wings apart-like (Wapl) protein was originally identified as a gene product that potentially regulates heterochromatin organization in Drosophila melanogaster. We show that the human ortholog of Wapl isa cohesin-binding protein that facilitates cohesin's timely release from chromosome arms during prophase. Depletion of Wapl from HeLa cells causes transient accumulation of prometaphase-like cells with chromosomes that display poorly resolved sister chromatids with a high level of cohesin. Reduction of cohesin relieves the Wapl-depletion phenotype, and depletion of Wapl rescues premature sister separation observed inSgo1-depleted or Esco2-depleted cells. Conversely, overexpression of Wapl causes premature separationof sister chromatids. Wapl physically associates with cohesin in HeLa-cell nuclear extracts. Remarkably, in vitro reconstitution experiments demonstrate that Wapl formsa stoichiometric, ternary complex with two regulatory subunits of cohesin, implicating its noncatalytic function in inactivating cohesin's ability to interact with chromatin.Conclusions
Wapl is a new regulator of sister chromatid resolution and promotes release of cohesin from chromosomes by directly interacting with its regulatory subunits.DOI:10.3389/fpls.2015.01034URLPMID:26648949 [本文引用: 1]

Maintenance and precise regulation of sister chromatid cohesion is essential for faithful chromosome segregation during mitosis and meiosis. Cohesin cofactors contribute to cohesin dynamics and interact with cohesin complexes during cell cycle. One of these, PDS5, also known as SPO76, is essential during mitosis and meiosis in several organisms and also plays a role in DNA repair. In yeast, the complex Wapl-Pds5 controls cohesion maintenance and colocalizes with cohesin complexes into chromosomes. In Arabidopsis, AtWAPL proteins are essential during meiosis, however, the role of AtPDS5 remains to be ascertained. Here we have isolated mutants for each of the five AtPDS5 genes (A-E) and obtained, after different crosses between them, double, triple, and even quadruple mutants (Atpds5a Atpds5b Atpds5c Atpds5e). Depletion of AtPDS5 proteins has a weak impact on meiosis, but leads to severe effects on development, fertility, somatic homologous recombination (HR) and DNA repair. Furthermore, this cohesin cofactor could be important for the function of the AtSMC5/AtSMC6 complex. Contrarily to its function in other species, our results suggest that AtPDS5 is dispensable during the meiotic division of Arabidopsis, although it plays an important role in DNA repair by HR.
DOI:10.1371/journal.pgen.1004497URLPMID:25033056 [本文引用: 3]

Sister chromatid cohesion, which is mediated by the cohesin complex, is essential for the proper segregation of chromosomes in mitosis and meiosis. The establishment of stable sister chromatid cohesion occurs during DNA replication and involves acetylation of the complex by the acetyltransferase CTF7. In higher eukaryotes, the majority of cohesin complexes are removed from chromosomes during prophase. Studies in fly and human have shown that this process involves the WAPL mediated opening of the cohesin ring at the junction between the SMC3 ATPase domain and the N-terminal domain of cohesin's α-kleisin subunit. We report here the isolation and detailed characterization of WAPL in Arabidopsis thaliana. We show that Arabidopsis contains two WAPL genes, which share overlapping functions. Plants in which both WAPL genes contain T-DNA insertions show relatively normal growth and development but exhibit a significant reduction in male and female fertility. The removal of cohesin from chromosomes during meiotic prophase is blocked in Atwapl mutants resulting in chromosome bridges, broken chromosomes and uneven chromosome segregation. In contrast, while subtle mitotic alterations are observed in some somatic cells, cohesin complexes appear to be removed normally. Finally, we show that mutations in AtWAPL suppress the lethality associated with inactivation of AtCTF7. Taken together our results demonstrate that WAPL plays a critical role in meiosis and raises the possibility that mechanisms involved in the prophase removal of cohesin may vary between mitosis and meiosis in plants.
DOI:10.1105/tpc.15.00781URLPMID:26813623 [本文引用: 2]

Sister chromatid cohesion, which is mediated by the cohesin complex, is essential for the proper segregation of chromosomes during mitosis and meiosis. Stable binding of cohesin with chromosomes is regulated in part by the opposing actions of CTF7 (CHROMOSOME TRANSMISSION FIDELITY7) and WAPL (WINGS APART-LIKE). In this study, we characterized the interaction between Arabidopsis thaliana CTF7 and WAPL by conducting a detailed analysis of wapl1-1 wapl2 ctf7 plants. ctf7 plants exhibit major defects in vegetative growth and development and are completely sterile. Inactivation of WAPL restores normal growth, mitosis, and some fertility to ctf7 plants. This shows that the CTF7/WAPL cohesin system is not essential for mitosis in vegetative cells and suggests that plants may contain a second mechanism to regulate mitotic cohesin. WAPL inactivation restores cohesin binding and suppresses ctf7-associated meiotic cohesion defects, demonstrating that WAPL and CTF7 function as antagonists to regulate meiotic sister chromatid cohesion. The ctf7 mutation only had a minor effect on wapl-associated defects in chromosome condensation and centromere association. These results demonstrate that WAPL has additional roles that are independent of its role in regulating chromatin-bound cohesin.
DOI:10.1093/nar/20.24.6605URLPMID:1480481 [本文引用: 1]

Analysis of the Schizosaccharomyces pombe chromosomes by pulsed field gel electrophoresis showed that the fission yeast has a very efficient DNA double-strand-break (dsb) repair system, which properly restores the three chromosomes after they are degraded by gamma-irradiation. The radiation-sensitive mutant rad21-45 is deficient in this repair pathway but is capable of cell-cycle arrest in G2 following DNA damage. We cloned the rad21 gene by complementing the radiation sensitivity of the rad21-45 mutant. The plasmid-borne gene completely reestablished the DNA dsb repair pathway. The rad21 gene was localized to chromosome III by hybridization. The transcript is 2.5 kb long and expressed at a moderate level. The 1884-bp open reading frame encodes a 628 amino acid, very acidic peptide with a calculated molecular mass of 67,854 D. The rad21 gene shows no significant homology to other known nucleotide or peptide sequences. The inability of the mutant to perform efficient DNA repair is caused by a single base substitution, which changes wild-type isoleucine67 into threonine in the mutant. Deletion of the genomic rad21 gene showed that it is essential for mitotic growth of S.pombe.
DOI:10.1007/s004380050634URLPMID:9491073 [本文引用: 1]

The Saccharomyces cerevisiae gene RHC21 is a homologue of the fission yeast rad21+ gene, which affects the sensitivity of cells to gamma-irradiation and is essential for cell growth in S. pombe. Disruption of the RHC21 gene showed that it is also essential in S. cerevisiae. To examine its function in cell growth further, we have isolated temperature-sensitive mutants for the RHC21 gene and characterized one of them, termed rhc21-sk16. When this mutant was incubated at 36 degrees C, the percentage of large-budded cells was increased. Most of the large-budded cells had aberrant nuclear structures, such as unequally extended nuclear DNA with incompletely elongated spindles across the mother-daughter neck or only in a mother cell. Furthermore, a circular minichromosome is more unstable in the mutant than in the wild-type, even at 25 degrees C. Flow cytometry showed that the bulk of DNA replication takes place normally at the restrictive temperature in the mutant. These results indicated that the RHC21 gene is required for proper segregation of the chromosomes. In addition, we found that the mutant is sensitive not only to UV radiation and gamma-rays but also to the antimicrotubule agent nocodazole at 25 degrees C. This suggests that the RHC21 gene is involved in the microtubule function. We discuss how the RHC21 gene product may be involved in chromosome segregation and microtubule function.
DOI:10.1016/s1534-5807(01)00088-0URLPMID:11740938 [本文引用: 1]

Proteolytic cleavage of the cohesin subunit Scc1 is a consistent feature of anaphase onset, although temporal differences exist between eukaryotes in cohesin loss from chromosome arms, as distinct from centromeres. We describe the effects of genetic deletion of Scc1 in chicken DT40 cells. Scc1 loss caused premature sister chromatid separation but did not disrupt chromosome condensation. Scc1 mutants showed defective repair of spontaneous and induced DNA damage. Scc1-deficient cells frequently failed to complete metaphase chromosome alignment and showed chromosome segregation defects, suggesting aberrant kinetochore function. Notably, the chromosome passenger INCENP did not localize normally to centromeres, while the constitutive kinetochore proteins CENP-C and CENP-H behaved normally. These results suggest a role for Scc1 in mitotic regulation, along with cohesion.
.
DOI:10.1083/jcb.117.5.921URLPMID:1315785 [本文引用: 1]

We have produced metaphase spindles and induced them to enter anaphase in vitro. Sperm nuclei were added to frog egg extracts, allowed to replicate their DNA, and driven into metaphase by the addition of cytoplasm containing active maturation promoting factor (MPF) and cytostatic factor (CSF), an activity that stabilizes MPF. Addition of calcium induces the inactivation of MPF, sister chromatid separation and anaphase chromosome movement. DNA topoisomerase II inhibitors prevent chromosome segregation at anaphase, demonstrating that the chromatids are catenated at metaphase and that decatenation occurs at the start of anaphase. Topoisomerase II activity towards exogenous substrates does not increase at the metaphase to anaphase transition, showing that chromosome separation at anaphase is not triggered by a bulk activation of topoisomerase II.
DOI:10.1007/s00412-007-0131-7URL [本文引用: 1]

PICH (Plk1-interacting checkpoint helicase) was recently identified as an essential component of the spindle assembly checkpoint and shown to localize to kinetochores, inner centromeres, and thin threads connecting separating chromosomes even during anaphase. In this paper, we have used immuno-fiber fluorescence in situ hybridization and chromatin-immunoprecipitation to demonstrate that PICH associates with centromeric chromatin during anaphase. Furthermore, by careful analysis of PICH-positive anaphase threads through FISH as well as bromo-deoxyurdine and CREST labeling, we strengthen the evidence that these threads comprise mainly alphoid centromere deoxyribonucleic acid. Finally, by timing the addition of ICRF-193 (a specific inhibitor of topoisomerase-II alpha) to cells synchronized in anaphase, we demonstrate that topoisomerase activity is required specifically to resolve PICH-positive threads during anaphase (as opposed to being required to prevent the formation of such threads during earlier cell cycle stages). These data indicate that PICH associates with centromeres during anaphase and that most PICH-positive threads evolve from inner centromeres as these stretch in response to tension. Moreover, they show that topoisomerase activity is required during anaphase for the resolution of PICH-positive threads, implying that the complete separation of sister chromatids occurs later than previously assumed.
DOI:10.1038/ncb2018URLPMID:20081838 [本文引用: 1]

The metaphase-anaphase transition is orchestrated through proteolysis of numerous proteins by a ubiquitin protein ligase called the anaphase-promoting complex or cyclosome (APC/C). A crucial aspect of this process is sister chromatid separation, which is thought to be mediated by separase, a thiol protease activated by the APC/C. Separase cleaves cohesin, a ring-shaped complex that entraps sister DNAs. It is a matter of debate whether cohesin-independent forces also contribute to sister chromatid cohesion. Using 4D live-cell imaging of Drosophila melanogaster syncytial embryos blocked in metaphase (via APC/C inhibition), we show that artificial cohesin cleavage is sufficient to trigger chromosome disjunction. This is nevertheless insufficient for correct chromosome segregation. Kinetochore-microtubule attachments are rapidly destabilized by the loss of tension caused by cohesin cleavage in the presence of high Cdk1 (cyclin-dependent kinase 1) activity, as occurs when the APC/C cannot destroy mitotic cyclins. Metaphase chromosomes undergo a bona fide anaphase when cohesin cleavage is combined with Cdk1 inhibition. We conclude that only two key events, opening of cohesin rings and downregulation of Cdk1, are sufficient to drive proper segregation of chromosomes in anaphase.
DOI:10.1091/mbc.e05-11-1089URLPMID:16510521 [本文引用: 1]

Cohesin maintains sister chromatid cohesion until its Rad21/Scc1/Mcd1 is cleaved by separase during anaphase. DNA topoisomerase II (topo II) maintains the proper topology of chromatid DNAs and is essential for chromosome segregation. Here we report direct observations of mitotic progression in individual HeLa cells after functional disruptions of hRad21, NIPBL, a loading factor for hRad21, and topo II alpha,beta by RNAi and a topo II inhibitor, ICRF-193. Mitosis is delayed in a Mad2-dependent manner after disruption of either or both cohesin and topo II. In hRad21 depletion, interphase pericentric architecture becomes aberrant, and anaphase is virtually permanently delayed as preseparated chromosomes are misaligned on the metaphase spindle. Topo II disruption perturbs centromere organization leading to intense Bub1, but no Mad2, on kinetochores and sustains a Mad2-dependent delay in anaphase onset with persisting securin. Thus topo II impinges upon centromere/kinetochore function. Disruption of topo II by RNAi or ICRF-193 overrides the mitotic delay induced by cohesin depletion: sister centromeres are aligned and anaphase spindle movements occur. The ensuing accumulation of catenations in preseparated sister chromatids may overcome the reduced tension arising from cohesin depletion, causing the override. Cohesin and topo II have distinct, yet coordinated functions in metaphase alignment.
URL [本文引用: 1]
URL [本文引用: 1]
DOI:10.1073/pnas.1708291114URLPMID:28847955 [本文引用: 1]

Sister chromatids are tethered together by the cohesin complex from the time they are made until their separation at anaphase. The ability of cohesin to tether sister chromatids together depends on acetylation of its Smc3 subunit by members of the Eco1 family of cohesin acetyltransferases. Vertebrates express two orthologs of Eco1, called Esco1 and Esco2, both of which are capable of modifying Smc3, but their relative contributions to sister chromatid cohesion are unknown. We therefore set out to determine the precise contributions of Esco1 and Esco2 to cohesion in vertebrate cells. Here we show that cohesion establishment is critically dependent upon Esco2. Although most Smc3 acetylation is Esco1 dependent, inactivation of the ESCO1 gene has little effect on mitotic cohesion. The unique ability of Esco2 to promote cohesion is mediated by sequences in the N terminus of the protein. We propose that Esco1-dependent modification of Smc3 regulates almost exclusively the noncohesive activities of cohesin, such as DNA repair, transcriptional control, chromosome loop formation, and/or stabilization. Collectively, our data indicate that Esco1 and Esco2 contribute to distinct and separable activities of cohesin in vertebrate cells.
DOI:10.1038/emboj.2013.230URLPMID:24141881 [本文引用: 1]

Cohesin mediates sister chromatid cohesion and contributes to the organization of interphase chromatin through DNA looping. In vertebrate somatic cells, cohesin consists of Smc1, Smc3, Rad21, and either SA1 or SA2. Three additional factors Pds5, Wapl, and Sororin bind to cohesin and modulate its dynamic association with chromatin. There are two Pds5 proteins in vertebrates, Pds5A and Pds5B, but their functional specificity remains unclear. Here, we demonstrate that Pds5 proteins are essential for cohesion establishment by allowing Smc3 acetylation by the cohesin acetyl transferases (CoATs) Esco1/2 and binding of Sororin. While both proteins contribute to telomere and arm cohesion, Pds5B is specifically required for centromeric cohesion. Furthermore, reduced accumulation of Aurora B at the inner centromere region in cells lacking Pds5B impairs its error correction function, promoting chromosome mis-segregation and aneuploidy. Our work supports a model in which the composition and function of cohesin complexes differs between different chromosomal regions.
DOI:10.1016/j.celrep.2017.08.092URLPMID:28930671 [本文引用: 1]

To ensure disjunction to opposite poles during anaphase, sister chromatids must be held together following DNA replication. This is mediated by cohesin, which is thought to entrap sister DNAs inside a tripartite ring composed of its Smc and kleisin (Scc1) subunits. How such structures are created during S phase is poorly understood, in particular whether they are derived from complexes that had entrapped DNAs prior to replication. To address this, we used selective photobleaching to determine whether cohesin associated with chromatin in G1 persists in?situ after replication. We developed a non-fluorescent HaloTag ligand to discriminate the fluorescence recovery signal from labeling of newly synthesized Halo-tagged Scc1 protein (pulse-chase or pcFRAP). In cells where cohesin turnover is inactivated by deletion of WAPL, Scc1 can remain associated with chromatin throughout S phase. These findings suggest that cohesion might be generated by cohesin that is already bound to un-replicated DNA.
DOI:10.1073/pnas.1305020110URLPMID:23901111 [本文引用: 2]

Sister chromatid cohesion depends on Sororin, a protein that stabilizes acetylated cohesin complexes on DNA by antagonizing the cohesin release factor Wings-apart like protein (Wapl). Cohesion is essential for chromosome biorientation but has to be dissolved to enable sister chromatid separation. To achieve this, the majority of cohesin is removed from chromosome arms in prophase and prometaphase in a manner that depends on Wapl and phosphorylation of cohesin's subunit stromal antigen 2 (SA2), whereas centromeric cohesin is cleaved in metaphase by the protease separase. Here we show that the mitotic kinases Aurora B and Cyclin-dependent kinase 1 (Cdk1) destabilize interactions between Sororin and the cohesin subunit precocious dissociation of sisters protein 5 (Pds5) by phosphorylating Sororin, leading to release of acetylated cohesin from chromosome arms and loss of cohesion. At centromeres, the cohesin protector shugoshin (Sgo1)-protein phosphatase 2A (PP2A) antagonizes Aurora B and Cdk1 partly by dephosphorylating Sororin and thus maintains cohesion until metaphase. We propose that the stepwise loss of cohesion between chromosome arms and centromeres is caused by local regulation of Wapl activity, which is controlled by the phosphorylation state of Sororin.
DOI:10.1038/ncb2637URLPMID:23242214 [本文引用: 1]

Timely dissolution of sister-chromatid cohesion in mitosis ensures accurate chromosome segregation to guard against aneuploidy and tumorigenesis. The complex of shugoshin and protein phosphatase 2A (SGO1-PP2A) protects cohesin at centromeres from premature removal by mitotic kinases and WAPL in prophase. Here we address the regulation and mechanism of human SGO1 in centromeric cohesion protection, and show that cyclin-dependent kinase (CDK)-mediated, mitosis-specific phosphorylation of SGO1 activates its cohesion-protection function and enables its direct binding to cohesin. The phospho-SGO1-bound cohesin complex contains PP2A, PDS5 and hypophosphorylated sororin, but lacks WAPL. Expression of non-phosphorylatable sororin bypasses the requirement for SGO1-PP2A in centromeric cohesion. Thus, mitotic phosphorylation of SGO1 targets SGO1-PP2A to cohesin, promotes dephosphorylation of PDS5-bound sororin and protects centromeric cohesin from WAPL. PP2A-orchestrated, site-selective dephosphorylation of cohesin and its regulators underlies centromeric cohesion protection.
.
DOI:10.1242/jcs.212100URLPMID:29724914 [本文引用: 1]

The DNA-embracing, ring-shaped multiprotein complex cohesin mediates sister chromatid cohesion and is stepwise displaced in mitosis by Wapl and separase (also known as ESPL1) to facilitate anaphase. Proper regulation of chromosome cohesion throughout meiosis is critical for preventing formation of aneuploid gametes, which are associated with trisomies and infertility in humans. Studying cohesion in meiocytes is complicated by their difficult experimental amenability and the absence of cohesin turnover. Here, we use cultured somatic cells to unravel fundamental aspects of meiotic cohesin. When expressed in Hek293 cells, the kleisin Rec8 displays no affinity for the peripheral cohesin subunits Stag1 or Stag2 and remains cytoplasmic. However, co-expression of Stag3 is sufficient for Rec8 to enter the nucleus, load onto chromatin, and functionally replace its mitotic counterpart Scc1 (also known as RAD21) during sister chromatid cohesion and dissolution. Rec8-Stag3 cohesin physically interacts with Pds5, Wapl and sororin (also known as CDCA5). Importantly, Rec8-Stag3 cohesin is shown to be susceptible to Wapl-dependent ring opening and sororin-mediated protection. These findings exemplify that our model system is suitable to rapidly generate testable predictions for important unresolved issues of meiotic cohesion regulation.
DOI:10.1038/35019529URLPMID:10934469 [本文引用: 1]

The multisubunit protein complex cohesin is required to establish cohesion between sister chromatids during S phase and to maintain it during G2 and M phases. Cohesin is essential for mitosis, and even partial defects cause very high rates of chromosome loss. In budding yeast, cohesin associates with specific sites which are distributed along the entire length of a chromosome but are more dense in the vicinity of the centromere. Real-time imaging of individual centromeres tagged with green fluorescent protein suggests that cohesin bound to centromeres is important for bipolar attachment to microtubules. This cohesin is, however, incapable of resisting the consequent force, which leads to sister centromere splitting and chromosome stretching. Meanwhile, cohesin bound to sequences flanking the centromeres prevents sister chromatids from completely unzipping and is required to pull back together sister centromeres that have already split. Cohesin therefore has a central role in generating a dynamic tension between microtubules and sister chromatid cohesion at centromeres, which lasts until chromosome segregation is finally promoted by separin-dependent cleavage of the cohesin subunit Scc1p.
DOI:10.1128/MCB.21.20.6984-6998.2001URLPMID:11564881 [本文引用: 1]

Structural maintenance of chromosomes (SMC) proteins fulfill pivotal roles in chromosome dynamics. In yeast, the SMC1-SMC3 heterodimer is required for meiotic sister chromatid cohesion and DNA recombination. Little is known, however, about mammalian SMC proteins in meiotic cells. We have identified a novel SMC protein (SMC1beta), which-except for a unique, basic, DNA binding C-terminal motif-is highly homologous to SMC1 (which may now be called SMC1alpha) and is not present in the yeast genome. SMC1beta is specifically expressed in testes and coimmunoprecipitates with SMC3 from testis nuclear extracts, but not from a variety of somatic cells. This establishes for mammalian cells the concept of cell-type- and tissue-specific SMC protein isoforms. Analysis of testis sections and chromosome spreads of various stages of meiosis revealed localization of SMC1beta along the axial elements of synaptonemal complexes in prophase I. Most SMC1beta dissociates from the chromosome arms in late-pachytene-diplotene cells. However, SMC1beta, but not SMC1alpha, remains chromatin associated at the centromeres up to metaphase II. Thus, SMC1beta and not SMC1alpha is likely involved in maintaining cohesion between sister centromeres until anaphase II.
DOI:10.1038/ncb1135URLPMID:15146193 [本文引用: 1]

Sister chromatid cohesion ensures the faithful segregation of chromosomes in mitosis and in both meiotic divisions. Meiosis-specific components of the cohesin complex, including the recently described SMC1 isoform SMC1 beta, were suggested to be required for meiotic sister chromatid cohesion and DNA recombination. Here we show that SMC1 beta-deficient mice of both sexes are sterile. Male meiosis is blocked in pachytene; female meiosis is highly error-prone but continues until metaphase II. Prophase axial elements (AEs) are markedly shortened, chromatin extends further from the AEs, chromosome synapsis is incomplete, and sister chromatid cohesion in chromosome arms and at centromeres is lost prematurely. In addition, crossover-associated recombination foci are absent or reduced, and meiosis-specific perinuclear telomere arrangements are impaired. Thus, SMC1 beta has a key role in meiotic cohesion, the assembly of AEs, synapsis, recombination, and chromosome movements.
DOI:10.1002/mrd.1104URLPMID:11599053 [本文引用: 1]

Affected males (as/as) from the mutant TT rat strain are sterile due to spermatogenesis impairment with meiotic arrest at the pachytene stage. The as locus is on rat chromosome 12, in a region that shows conserved synteny to cM 74-94 on mouse chromosome 5. Stag3, a new member of the stromalin protein family, is expressed specifically in testis and associates to the synaptonemal complex. Mouse Stag3 gene has been assigned to cM 78 on chromosome 5. In this study, we have characterized the rat Stag3 gene and examined it as a candidate for male infertility in as/as rats. The rat Stag3 cDNA is 4181 nucleotides long, contains a highly polymorphic hexanucleotide repeat in the coding region, and encodes a 1256 amino acid protein with 93 and 77% sequence identity to mouse and human Stag3 proteins, respectively. No mutations or differences in size or abundance of Stag3 mRNA were detected between as/as and control rats, suggesting that Stag3 is not responsible for the aspermic phenotype. In addition, immunohistochemistry with antibodies against SCP1 and SPC3 proteins suggest that the synaptonemal complex structures are not primarily affected in these rats.
DOI:10.3390/genes1030484URLPMID:24710098 [本文引用: 1]

During the first meiotic prophase, the cohesin complex is localized to the chromosome axis and contributes to chromosome organization, pairing, synapsis, and recombination. The PDS5 protein, an accessory factor of the cohesin complex, is known to be a component of meiotic chromosome cores in fungi and to be implicated in meiotic chromosome structure and function. We found by immunoblotting experiments that a mammalian PDS5 protein, PDS5B, is abundantly expressed in mouse testis compared to other tissues. Immunofluorescence labeling experiments revealed that PDS5B is highly expressed in spermatogonia and that most PDS5B is depleted from chromatin as cells enter meiosis. During the first meiotic prophase, PDS5B associates with the axial cores of chromosomes. The axial association of PDS5B was observed also in the absence of synaptonemal complex proteins, such as SYCP1 and SYCP3, suggesting that PDS5B is an integral part of the chromosome axis as defined by the cohesin complex. These results suggest that PDS5B modulates cohesin functions in spermatocytes as well as in spermatogonia, contributing to meiotic chromosome structure and function.
DOI:10.15252/embr.201541060URLPMID:26951638 [本文引用: 1]

The distribution and regulation of the cohesin complexes have been extensively studied during mitosis. However, the dynamics of their different regulators in vertebrate meiosis is largely unknown. In this work, we have analyzed the distribution of the regulatory factor Sororin during male mouse meiosis. Sororin is detected at the central region of the synaptonemal complex during prophase I, in contrast with the previously reported localization of other cohesin components in the lateral elements. This localization of Sororin depends on the transverse filaments protein SYCP1, but not on meiosis-specific cohesin subunits REC8 and SMC1β. By late prophase I, Sororin accumulates at centromeres and remains there up to anaphase II The phosphatase activity of PP2A seems to be required for this accumulation. We hypothesize that Sororin function at the central region of the synaptonemal complex could be independent on meiotic cohesin complexes. In addition, we suggest that Sororin participates in the regulation of centromeric cohesion during meiosis in collaboration with SGO2-PP2A.
DOI:10.1007/s10577-016-9542-8URLPMID:28050734 [本文引用: 1]

During meiotic prophase, cohesin complexes mediate cohesion between sister chromatids and promote pairing and synapsis of homologous chromosomes. Precisely how the activity of cohesin is controlled to promote these events is not fully understood. In metazoans, cohesion establishment between sister chromatids during mitotic divisions is accompanied by recruitment of the cohesion-stabilizing protein Sororin. During somatic cell division cycles, Sororin is recruited in response to DNA replication-dependent modification of the cohesin complex by ESCO acetyltransferases. How Sororin is recruited and acts in meiosis is less clear. Here, we have surveyed the chromosomal localization of Sororin and its relationship to the meiotic cohesins and other chromatin modifiers with the objective of determining how Sororin contributes to meiotic chromosome dynamics. We show that Sororin localizes to the cores of meiotic chromosomes in a manner that is dependent on synapsis and the synaptonemal complex protein SYCP1. In contrast, cohesin, with which Sororin interacts in mitotic cells, shows axial enrichment on meiotic chromosomes even in the absence of synapsis between homologs. Using high-resolution microscopy, we show that Sororin is localized to the central region of the synaptonemal complex. These results indicate that Sororin regulation during meiosis is distinct from its regulation in mitotic cells and may suggest that it interacts with a distinctly different partner to ensure proper chromosome dynamics in meiosis.
DOI:10.1101/gad.237313.113URL [本文引用: 1]

During meiosis, homologous chromosome (homolog) pairing is promoted by several layers of regulation that include dynamic chromosome movement and meiotic recombination. However, the way in which homologs recognize each other remains a fundamental issue in chromosome biology. Here, we show that homolog recognition or association initiates upon entry into meiotic prophase before axis assembly and double-strand break (DSB) formation. This homolog association develops into tight pairing only during or after axis formation. Intriguingly, the ability to recognize homologs is retained in Sun1 knockout spermatocytes, in which telomere-directed chromosome movement is abolished, and this is the case even in Spo11 knockout spermatocytes, in which DSB-dependent DNA homology search is absent. Disruption of meiosis-specific cohesin RAD21L precludes the initial association of homologs as well as the subsequent pairing in spermatocytes. These findings suggest the intriguing possibility that homolog recognition is achieved primarily by searching for homology in the chromosome architecture as defined by meiosis-specific cohesin rather than in the DNA sequence itself.
DOI:10.1038/embor.2011.2URL [本文引用: 1]

We identify a new mammalian cohesin subunit, RAD21-like protein (RAD21L), with sequence similarity to RAD21 and REC8. RAD21L localizes along axial elements in early meiotic prophase, in a manner that is spatiotemporally different to either REC8 or RAD21. Remarkably, RAD21L and REC8 have symmetrical, mutually exclusive localization on the not-yet-synapsed homologues, implying that the cohesin patterning could provide a code for homologue recognition. RAD21 transiently localizes to axial elements after the dissociation of RAD21L and REC8 in late pachytene, a period of recombination repair. Further, we show that the removal of cohesins and synaptonemal complex during late meiotic prophase is promoted by Polo-like kinase 1, which is similar to the mitotic prophase pathway.
.
DOI:10.1083/jcb.201008005URLPMID:21242291 [本文引用: 1]

Cohesins are multi-subunit protein complexes that regulate sister chromatid cohesion during mitosis and meiosis. Here we identified a novel kleisin subunit of cohesins, RAD21L, which is conserved among vertebrates. In mice, RAD21L is expressed exclusively in early meiosis: it apparently replaces RAD21 in premeiotic S phase, becomes detectable on the axial elements in leptotene, and stays on the axial/lateral elements until mid pachytene. RAD21L then disappears, and is replaced with RAD21. This behavior of RAD21L is unique and distinct from that of REC8, another meiosis-specific kleisin subunit. Remarkably, the disappearance of RAD21L at mid pachytene correlates with the completion of DNA double-strand break repair and the formation of crossovers as judged by colabeling with molecular markers, γ-H2AX, MSH4, and MLH1. RAD21L associates with SMC3, STAG3, and either SMC1α or SMC1β. Our results suggest that cohesin complexes containing RAD21L may be involved in synapsis initiation and crossover recombination between homologous chromosomes.
DOI:10.1016/j.celrep.2016.09.059URLPMID:27760328 [本文引用: 1]

Mammalian NIMA-like kinase-1 (NEK1) is a dual-specificity kinase highly expressed in mouse germ cells during prophase I of meiosis. Loss of NEK1 induces retention of cohesin on chromosomes at meiotic prophase I. Timely deposition and removal of cohesin is essential for accurate chromosome segregation. Two processes regulate cohesin removal: a non-proteolytic mechanism involving WAPL, sororin, and PDS5B and direct cleavage by separase. Here, we demonstrate a role for NEK1 in the regulation of WAPL loading during meiotic prophase I, via an interaction between NEK1 and PDS5B. This regulation of WAPL by NEK1-PDS5B is mediated by protein phosphatase 1 gamma (PP1γ), which both interacts with and is a phosphotarget of NEK1. Taken together, our results reveal that NEK1 phosphorylates PP1γ, leading to the dephosphorylation of WAPL, which, in turn, results?in?its retention on chromosome cores to promote loss of cohesion at the end of prophase I in mammals.
.
DOI:10.1083/jcb.201201100URLPMID:22711701 [本文引用: 1]

Cohesin is a conserved multisubunit protein complex that participates in chromosome segregation, DNA damage repair, chromatin regulation, and synaptonemal complex (SC) formation. Yeast, but not mice, depleted of the cohesin subunit Rec8 are defective in the formation of the axial elements (AEs) of the SC, suggesting that, in mammals, this function is not conserved. In this paper, we show that spermatocytes from mice lacking the two meiosis-specific cohesin subunits RAD21L and REC8 were unable to initiate RAD51- but not DMC1-mediated double-strand break repair, were not able to assemble their AEs, and arrested as early as the leptotene stage of prophase I, demonstrating that cohesin plays an essential role in AE assembly that is conserved from yeast to mammals.
DOI:10.1016/j.cell.2011.07.003URL [本文引用: 1]

Meiotic recombination between homologous chromosomes initiates via programmed DNA double-strand breaks (DSBs), generated by complexes comprising Spo11 transesterase plus accessory proteins. DSBs arise concomitantly with the development of axial chromosome structures, where the coalescence of axis sites produces linear arrays of chromatin loops. Recombining DNA sequences map to loops, but are ultimately tethered to the underlying axis. How and when such tethering occurs is currently unclear. Using ChIPchip in yeast, we show that Spo11-accessory proteins Rec114, Mer2, and Mei4 stably interact with chromosome axis sequences, upon phosphorylation of Mer2 by S phase Cdk. This axis tethering requires meiotic axis components (Red1/Hop1) and is modulated in a domain-specific fashion by cohesin. Loss of Rec114, Mer2, and Mei4 binding correlates with loss of DSBs. Our results strongly suggest that hot-spot sequences become tethered to axis sites by the DSB machinery prior to DSB formation.
DOI:10.1016/j.cub.2011.07.007URL [本文引用: 1]

Background: Chromosome segregation and the repair of DNA double-strand breaks (DSBs) by homologous recombination require cohesin, the protein complex that mediates sister chromatid cohesion (SCC). In addition, cohesin is also required for the integrity of DNA damage checkpoints in somatic cells, where cohesin loading depends on a conserved complex containing the Scc2/Nipbl protein. Although cohesin is required for the completion of meiotic recombination, little is known about how cohesin promotes the repair of meiotic DSBs and about the factors that promote loading of cohesin during meiosis.
Results: Here we show that during Caenorhabditis elegans meiosis, loading of cohesin requires SCC-2, whereas the cohesin-related complexes condensin and SMC-5/6 can be loaded by mechanisms independent of both SCC-2 and cohesin. Although the lack of cohesin in scc-2 mutants impairs the repair of meiotic DSBs, surprisingly, the persistent DNA damage fails to trigger an apoptotic response of the conserved pachytene DNA damage checkpoint. Mutants carrying an scc-3 allele that abrogates loading of meiotic cohesin are also deficient in the apoptotic response of the pachytene checkpoint, and both scc-2 and scc-3 mutants fail to recruit the DNA damage sensor 9-1-1 complex onto persistent damage sites during meiosis. Furthermore, we show that meiotic cohesin is also required for the timely loading of the RAD-51 recombinase to irradiation-induced DSBs.
Conclusions: We propose that meiotic cohesin promotes DSB processing and recruitment of DNA damage checkpoint proteins, thus implicating cohesin in the earliest steps of the DNA damage response during meiosis.
URLPMID:10353901 [本文引用: 1]

How enhancers are able to activate promoters located several kilobases away is unknown. Activation by the wing margin enhancer in the cut gene, located 85 kb from the promoter, requires several genes that participate in the Notch receptor pathway in the wing margin, including scalloped, vestigial, mastermind, Chip, and the Nipped locus. Here we show that Nipped mutations disrupt one or more of four essential complementation groups: l(2)41Ae, l(2)41Af, Nipped-A, and Nipped-B. Heterozygous Nipped mutations modify Notch mutant phenotypes in the wing margin and other tissues, and magnify the effects that mutations in the cis regulatory region of cut have on cut expression. Nipped-A and l(2)41Af mutations further diminish activation by a wing margin enhancer partly impaired by a small deletion. In contrast, Nipped-B mutations do not diminish activation by the impaired enhancer, but increase the inhibitory effect of a gypsy transposon insertion between the enhancer and promoter. Nipped-B mutations also magnify the effect of a gypsy insertion in the Ultrabithorax gene. Gypsy binds the Suppressor of Hairy-wing insulator protein [Su(Hw)] that blocks enhancer-promoter communication. Increased insulation by Su(Hw) in Nipped-B mutants suggests that Nipped-B products structurally facilitate enhancer-promoter communication. Compatible with this idea, Nipped-B protein is homologous to a family of chromosomal adherins with broad roles in sister chromatid cohesion, chromosome condensation, and DNA repair.
DOI:10.1038/ng1364URLPMID:15146186 [本文引用: 1]

Cornelia de Lange syndrome (CdLS; OMIM 122470) is a dominantly inherited multisystem developmental disorder characterized by growth and cognitive retardation; abnormalities of the upper limbs; gastroesophageal dysfunction; cardiac, ophthalmologic and genitourinary anomalies; hirsutism; and characteristic facial features. Genital anomalies, pyloric stenosis, congenital diaphragmatic hernias, cardiac septal defects, hearing loss and autistic and self-injurious tendencies also frequently occur. Prevalence is estimated to be as high as 1 in 10,000 (ref. 4). We carried out genome-wide linkage exclusion analysis in 12 families with CdLS and identified four candidate regions, of which chromosome 5p13.1 gave the highest multipoint lod score of 2.7. This information, together with the previous identification of a child with CdLS with a de novo t(5;13)(p13.1;q12.1) translocation, allowed delineation of a 1.1-Mb critical region on chromosome 5 for the gene mutated in CdLS. We identified mutations in one gene in this region, which we named NIPBL, in four sporadic and two familial cases of CdLS. We characterized the genomic structure of NIPBL and found that it is widely expressed in fetal and adult tissues. The fly homolog of NIPBL, Nipped-B, facilitates enhancer-promoter communication and regulates Notch signaling and other developmental pathways in Drosophila melanogaster.
DOI:10.1038/ng1363URLPMID:15146185 [本文引用: 1]

Cornelia de Lange syndrome (CdLS) is a multiple malformation disorder characterized by dysmorphic facial features, mental retardation, growth delay and limb reduction defects. We indentified and characterized a new gene, NIPBL, that is mutated in individuals with CdLS and determined its structure and the structures of mouse, rat and zebrafish homologs. We named its protein product delangin. Vertebrate delangins have substantial homology to orthologs in flies, worms, plants and fungi, including Scc2-type sister chromatid cohesion proteins, and D. melanogaster Nipped-B. We propose that perturbed delangin function may inappropriately activate DLX genes, thereby contributing to the proximodistal limb patterning defects in CdLS. Genome analyses typically identify individual delangin or Nipped-B-like orthologs in diploid animal and plant genomes. The evolution of an ancestral sister chromatid cohesion protein to acquire an additional role in developmental gene regulation suggests that there are parallels between CdLS and Roberts syndrome.
DOI:10.1016/j.molcel.2019.03.037URLPMID:31226277 [本文引用: 1]

The condensin protein complex plays a key role in the structural organization of genomes. How the ATPase activity of its SMC subunits drives large-scale changes in chromosome topology has remained unknown. Here we reconstruct, at near-atomic resolution, the sequence of events that take place during the condensin ATPase cycle. We show that ATP binding induces a conformational switch in the Smc4 head domain that releases its hitherto undescribed interaction with the Ycs4 HEAT-repeat subunit and promotes its engagement with the Smc2 head into an asymmetric heterodimer. SMC head dimerization subsequently enables nucleotide binding at the second active site and disengages the Brn1 kleisin subunit from the Smc2 coiled coil to open the condensin ring. These large-scale transitions in the condensin architecture lay out a mechanistic path for its ability to extrude DNA helices into large loop structures.
DOI:10.1101/gr.163519.113URL [本文引用: 1]

Long-range regulatory interactions play an important role in shaping gene-expression programs. However, the genomic features that organize these activities are still poorly characterized. We conducted a large operational analysis to chart the distribution of gene regulatory activities along the mouse genome, using hundreds of insertions of a regulatory sensor. We found that enhancers distribute their activities along broad regions and not in a gene-centric manner, defining large regulatory domains. Remarkably, these domains correlate strongly with the recently described TADs, which partition the genome into distinct self-interacting blocks. Different features, including specific repeats and CTCF-binding sites, correlate with the transition zones separating regulatory domains, and may help to further organize promiscuously distributed regulatory influences within large domains. These findings support a model of genomic organization where TADs confine regulatory activities to specific but large regulatory domains, contributing to the establishment of specific gene expression profiles.
[本文引用: 1]
DOI:10.1038/s41576-018-0060-8URLPMID:30367165 [本文引用: 2]

Studies of 3D chromatin organization have suggested that chromosomes are hierarchically organized into large compartments composed of smaller domains called topologically associating domains (TADs). Recent evidence suggests that compartments are smaller than previously thought and that the?transcriptional or chromatin state is responsible for interactions leading to the formation of small compartmental domains in all organisms. In vertebrates, CTCF forms loop domains, probably via an extrusion process involving cohesin. CTCF loops cooperate with compartmental domains to establish the 3D organization of the genome. The continuous extrusion of the chromatin fibre by cohesin may also be responsible for the establishment of enhancer-promoter interactions and stochastic aspects of the transcription process. These observations suggest that the 3D organization of the genome is an emergent property of chromatin and its components, and thus may not be only a determinant but also a consequence of its function.
DOI:10.1073/pnas.1317788111URLPMID:24335803 [本文引用: 2]

Recent studies of genome-wide chromatin interactions have revealed that the human genome is partitioned into many self-associating topological domains. The boundary sequences between domains are enriched for binding sites of CTCC-binding factor (CTCF) and the cohesin complex, implicating these two factors in the establishment or maintenance of topological domains. To determine the role of cohesin and CTCF in higher-order chromatin architecture in human cells, we depleted the cohesin complex or CTCF and examined the consequences of loss of these factors on higher-order chromatin organization, as well as the transcriptome. We observed a general loss of local chromatin interactions upon disruption of cohesin, but the topological domains remain intact. However, we found that depletion of CTCF not only reduced intradomain interactions but also increased interdomain interactions. Furthermore, distinct groups of genes become misregulated upon depletion of cohesin and CTCF. Taken together, these observations suggest that CTCF and cohesin contribute differentially to chromatin organization and gene regulation.
DOI:10.1038/nature11082URL [本文引用: 1]

The spatial organization of the genome is intimately linked to its biological function, yet our understanding of higher order genomic structure is coarse, fragmented and incomplete. In the nucleus of eukaryotic cells, interphase chromosomes occupy distinct chromosome territories, and numerous models have been proposed for how chromosomes fold within chromosome territories(1). These models, however, provide only few mechanistic details about the relationship between higher order chromatin structure and genome function. Recent advances in genomic technologies have led to rapid advances in the study of three-dimensional genome organization. In particular, Hi-C has been introduced as a method for identifying higher order chromatin interactions genome wide(2). Here we investigate the three-dimensional organization of the human and mouse genomes in embryonic stem cells and terminally differentiated cell types at unprecedented resolution. We identify large, megabase-sized local chromatin interaction domains, which we term 'topological domains', as a pervasive structural feature of the genome organization. These domains correlate with regions of the genome that constrain the spread of heterochromatin. The domains are stable across different cell types and highly conserved across species, indicating that topological domains are an inherent property of mammalian genomes. Finally, we find that the boundaries of topological domains are enriched for the insulator binding protein CTCF, housekeeping genes, transfer RNAs and short interspersed element (SINE) retrotransposons, indicating that these factors may have a role in establishing the topological domain structure of the genome.
DOI:10.1016/j.cell.2014.11.021URL [本文引用: 1]

We use in situ Hi-C to probe the 3D architecture of genomes, constructing haploid and diploid maps of nine cell types. The densest, in human lymphoblastoid cells, contains 4.9 billion contacts, achieving 1 kb resolution. We find that genomes are partitioned into contact domains (median length, 185 kb), which are associated with distinct patterns of histone marks and segregate into six subcompartments. We identify similar to 10,000 loops. These loops frequently link promoters and enhancers, correlate with gene activation, and show conservation across cell types and species. Loop anchors typically occur at domain boundaries and bind CTCF. CTCF sites at loop anchors occur predominantly (>90%) in a convergent orientation, with the asymmetric motifs "facing'' one another. The inactive X chromosome splits into two massive domains and contains large loops anchored at CTCF-binding repeats.
DOI:10.1038/s41580-019-0132-4URLPMID:31197269 [本文引用: 1]

In eukaryotes, the genome does not exist as a linear molecule but instead is hierarchically packaged inside the nucleus. This complex genome organization includes multiscale structural units of chromosome territories, compartments, topologically associating domains, which are often demarcated by architectural proteins such as CTCF and cohesin, and chromatin loops. The 3D organization of chromatin modulates biological processes such as transcription, DNA replication, cell division and meiosis, which are crucial for cell differentiation and animal development. In this Review, we discuss recent progress in our understanding of the general principles of chromatin folding, its regulation and its functions in mammalian development. Specifically, we discuss the dynamics of 3D chromatin and genome organization during gametogenesis, embryonic development, lineage commitment and stem cell differentiation, and focus on the functions of chromatin architecture in transcription regulation. Finally, we discuss the role of 3D genome alterations in the aetiology of developmental disorders and human diseases.
DOI:10.1101/gr.170332.113URLPMID:25367294 [本文引用: 2]

The spatial arrangement of interphase chromosomes in the nucleus is important for gene expression and genome function in animals and in plants. The recently developed Hi-C technology is an efficacious method to investigate genome packing. Here we present a detailed Hi-C map of the three-dimensional genome organization of the plant Arabidopsis thaliana. We find that local chromatin packing differs from the patterns seen in animals, with kilobasepair-sized segments that have much higher intrachromosome interaction rates than neighboring regions, representing a dominant local structural feature of genome conformation in A. thaliana. These regions, which appear as positive strips on two-dimensional representations of chromatin interaction, are enriched in epigenetic marks H3K27me3, H3.1, and H3.3. We also identify more than 400 insulator-like regions. Furthermore, although topologically associating domains (TADs), which are prominent in animals, are not an obvious feature of A. thaliana genome packing, we found more than 1000 regions that have properties of TAD boundaries, and a similar number of regions analogous to the interior of TADs. The insulator-like, TAD-boundary-like, and TAD-interior-like regions are each enriched for distinct epigenetic marks and are each correlated with different gene expression levels. We conclude that epigenetic modifications, gene density, and transcriptional activity combine to shape the local packing of the A. thaliana nuclear genome.
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.cell.2008.01.011URLPMID:18237772 [本文引用: 2]

Cohesins mediate sister chromatid cohesion, which is essential for chromosome segregation and postreplicative DNA repair. In addition, cohesins appear to regulate gene expression and enhancer-promoter interactions. These noncanonical functions remained unexplained because knowledge of cohesin-binding sites and functional interactors in metazoans was lacking. We show that the distribution of cohesins on mammalian chromosome arms is not driven by transcriptional activity, in contrast to S. cerevisiae. Instead, mammalian cohesins occupy a subset of DNase I hypersensitive sites, many of which contain sequence motifs resembling the consensus for CTCF, a DNA-binding protein with enhancer blocking function and boundary-element activity. We find cohesins at most CTCF sites and show that CTCF is required for cohesin localization to these sites. Recruitment by CTCF suggests a rationale for noncanonical cohesin functions and, because CTCF binding is sensitive to DNA methylation, allows cohesin positioning to integrate DNA sequence and epigenetic state.
DOI:10.1073/pnas.0801273105URLPMID:18550811 [本文引用: 2]

Cohesin is required to prevent premature dissociation of sister chromatids after DNA replication. Although its role in chromatid cohesion is well established, the functional significance of cohesin's association with interphase chromatin is not clear. Using a quantitative proteomics approach, we show that the STAG1 (Scc3/SA1) subunit of cohesin interacts with the CCTC-binding factor CTCF bound to the c-myc insulator element. Both allele-specific binding of CTCF and Scc3/SA1 at the imprinted IGF2/H19 gene locus and our analyses of human DM1 alleles containing base substitutions at CTCF-binding motifs indicate that cohesin recruitment to chromosomal sites depends on the presence of CTCF. A large-scale genomic survey using ChIP-Chip demonstrates that Scc3/SA1 binding strongly correlates with the CTCF-binding site distribution in chromosomal arms. However, some chromosomal sites interact exclusively with CTCF, whereas others interact with Scc3/SA1 only. Furthermore, immunofluorescence microscopy and ChIP-Chip experiments demonstrate that CTCF associates with both centromeres and chromosomal arms during metaphase. These results link cohesin to gene regulatory functions and suggest an essential role for CTCF during sister chromatid cohesion. These results have implications for the functional role of cohesin subunits in the pathogenesis of Cornelia de Lange syndrome and Roberts syndromes.
DOI:10.1038/emboj.2008.1URLPMID:18219272 [本文引用: 2]

Cohesins, which mediate sister chromatin cohesion, and CTCF, which functions at chromatin boundaries, play key roles in the structural and functional organization of chromosomes. We examined the binding of these two factors on the Kaposi's sarcoma-associated herpesvirus (KSHV) episome during latent infection and found a striking colocalization within the control region of the major latency transcript responsible for expressing LANA (ORF73), vCyclin (ORF72), vFLIP (ORF71), and vmiRNAs. Deletion of the CTCF-binding site from the viral genome disrupted cohesin binding, and crippled colony formation in 293 cells. Clonal instability correlated with elevated expression of lytic cycle gene products, notably the neighbouring promoter for K14 and vGPCR (ORF74). siRNA depletion of RAD21 from latently infected cells caused an increase in K14 and ORF74, and lytic inducers caused a rapid dissociation of RAD21 from the viral genome. RAD21 and SMC1 also associate with the cellular CTCF sites at mammalian c-myc promoter and H19/Igf2 imprinting control region. We conclude that cohesin subunits associate with viral and cellular CTCF sites involved in complex gene regulation and chromatin organization.
DOI:10.1038/nature06634URLPMID:18235444 [本文引用: 2]

Cohesin complexes mediate sister-chromatid cohesion in dividing cells but may also contribute to gene regulation in postmitotic cells. How cohesin regulates gene expression is not known. Here we describe cohesin-binding sites in the human genome and show that most of these are associated with the CCCTC-binding factor (CTCF), a zinc-finger protein required for transcriptional insulation. CTCF is dispensable for cohesin loading onto DNA, but is needed to enrich cohesin at specific binding sites. Cohesin enables CTCF to insulate promoters from distant enhancers and controls transcription at the H19/IGF2 (insulin-like growth factor 2) locus. This role of cohesin seems to be independent of its role in cohesion. We propose that cohesin functions as a transcriptional insulator, and speculate that subtle deficiencies in this function contribute to 'cohesinopathies' such as Cornelia de Lange syndrome.
DOI:10.1038/nature09380URLPMID:20720539 [本文引用: 2]

Transcription factors control cell-specific gene expression programs through interactions with diverse coactivators and the transcription apparatus. Gene activation may involve DNA loop formation between enhancer-bound transcription factors and the transcription apparatus at the core promoter, but this process is not well understood. Here we report that mediator and cohesin physically and functionally connect the enhancers and core promoters of active genes in murine embryonic stem cells. Mediator, a transcriptional coactivator, forms a complex with cohesin, which can form rings that connect two DNA segments. The cohesin-loading factor Nipbl is associated with mediator-cohesin complexes, providing a means to load cohesin at promoters. DNA looping is observed between the enhancers and promoters occupied by mediator and cohesin. Mediator and cohesin co-occupy different promoters in different cells, thus generating cell-type-specific DNA loops linked to the gene expression program of each cell.
DOI:10.1038/nature11243URL [本文引用: 1]

The laboratory mouse is the most widely used mammalian model organism in biomedical research. The 2.6 x 10(9) bases of the mouse genome possess a high degree of conservation with the human genome(1), so a thorough annotation of the mouse genome will be of significant value to understanding the function of the human genome. So far, most of the functional sequences in the mouse genome have yet to be found, and the cis-regulatory sequences in particular are still poorly annotated. Comparative genomics has been a powerful tool for the discovery of these sequences(2), but on its own it cannot resolve their temporal and spatial functions. Recently, ChIP-Seq has been developed to identify cis-regulatory elements in the genomes of several organisms including humans, Drosophila melanogaster and Caenorhabditis elegans(3-5). Here we apply the same experimental approach to a diverse set of 19 tissues and cell types in the mouse to produce a map of nearly 300,000 murine cis-regulatory sequences. The annotated sequences add up to 11% of the mouse;[GRAPHICS];genome, and include more than 70% of conserved non-coding sequences. We define tissue-specific enhancers and identify potential transcription factors regulating gene expression in each tissue or cell type. Finally, we show that much of the mouse genome is organized into domains of coordinately regulated enhancers and promoters. Our results provide a resource for the annotation of functional elements in the mammalian genome and for the study of mechanisms regulating tissue-specific gene expression.
DOI:10.1101/gr.100479.109URLPMID:20219941 [本文引用: 1]

The cohesin protein complex holds sister chromatids in dividing cells together and is essential for chromosome segregation. Recently, cohesin has been implicated in mediating transcriptional insulation, via its interactions with CTCF. Here, we show in different cell types that cohesin functionally behaves as a tissue-specific transcriptional regulator, independent of CTCF binding. By performing matched genome-wide binding assays (ChIP-seq) in human breast cancer cells (MCF-7), we discovered thousands of genomic sites that share cohesin and estrogen receptor alpha (ER) yet lack CTCF binding. By use of human hepatocellular carcinoma cells (HepG2), we found that liver-specific transcription factors colocalize with cohesin independently of CTCF at liver-specific targets that are distinct from those found in breast cancer cells. Furthermore, estrogen-regulated genes are preferentially bound by both ER and cohesin, and functionally, the silencing of cohesin caused aberrant re-entry of breast cancer cells into cell cycle after hormone treatment. We combined chromosomal interaction data in MCF-7 cells with our cohesin binding data to show that cohesin is highly enriched at ER-bound regions that capture inter-chromosomal loop anchors. Together, our data show that cohesin cobinds across the genome with transcription factors independently of CTCF, plays a functional role in estrogen-regulated transcription, and may help to mediate tissue-specific transcriptional responses via long-range chromosomal interactions.
DOI:10.1038/nrm.2016.30URLPMID:27075410 [本文引用: 1]

SMC (structural maintenance of chromosomes) complexes - which include condensin, cohesin and the SMC5-SMC6 complex - are major components of chromosomes in all living organisms, from bacteria to humans. These ring-shaped protein machines, which are powered by ATP hydrolysis, topologically encircle DNA. With their ability to hold more than one strand of DNA together, SMC complexes control a plethora of chromosomal activities. Notable among these are chromosome condensation and sister chromatid cohesion. Moreover, SMC complexes have an important role in DNA repair. Recent mechanistic insight into the function and regulation of these universal chromosomal machines enables us to propose molecular models of chromosome structure, dynamics and function, illuminating one of the fundamental entities in biology.
DOI:10.1016/j.cell.2017.12.021URLPMID:29358048 [本文引用: 1]

The ring-shaped structural maintenance of chromosome (SMC) complexes are multi-subunit ATPases that topologically encircle DNA. SMC rings make vital contributions to numerous chromosomal functions, including mitotic chromosome condensation, sister chromatid cohesion, DNA repair, and transcriptional regulation. They are thought to do so by establishing interactions between more than one DNA. Here, we demonstrate DNA-DNA tethering by the purified fission yeast cohesin complex. DNA-bound cohesin efficiently and topologically captures a second DNA, but only if that is single-stranded DNA (ssDNA). Like initial double-stranded DNA (dsDNA) embrace, second ssDNA capture is ATP-dependent, and it strictly requires the cohesin loader complex. Second-ssDNA capture is relatively labile but is converted into stable dsDNA-dsDNA cohesion through DNA synthesis. Our study illustrates second-DNA capture by an SMC complex and provides a molecular model for the establishment of sister chromatid cohesion.
DOI:10.1016/j.cell.2018.11.036URLPMID:30595451 [本文引用: 1]

The temporal order of DNA replication (replication timing [RT]) is highly coupled with genome architecture, but cis-elements regulating either remain elusive. We created a series of CRISPR-mediated deletions and inversions of a pluripotency-associated topologically associating domain (TAD) in mouse ESCs. CTCF-associated domain boundaries were dispensable for RT. CTCF protein depletion weakened most TAD boundaries but had no effect on RT or A/B compartmentalization genome-wide. By contrast, deletion of three intra-TAD CTCF-independent 3D contact sites caused a domain-wide early-to-late RT shift, an A-to-B compartment switch, weakening of TAD architecture, and loss of transcription. The dispensability of TAD boundaries and the?necessity of these "early replication control elements" (ERCEs) was validated by deletions and inversions at additional domains. Our results demonstrate that discrete cis-regulatory elements orchestrate domain-wide RT, A/B compartmentalization, TAD architecture, and transcription, revealing fundamental principles linking genome structure and function.
DOI:10.1007/s00294-018-0824-xURLPMID:29549581 [本文引用: 1]

Proliferating cells need to accurately duplicate and pass their genetic material on to daughter cells. Problems during replication and partition challenge the structural and numerical integrity of chromosomes. Diverse mechanisms, as the DNA replication checkpoint, survey the correct progression of replication and couple it with other cell cycle events to preserve genome integrity. The structural maintenance of chromosomes (SMC) cohesin complex primarily contributes to chromosome duplication by mediating the tethering of newly replicated sister chromatids, thus assisting their equal segregation in mitosis. In addition, cohesin exerts important functions in genome organization, gene expression and DNA repair. These are determined by cohesin's ability to bring together different DNA segments and, hence, by the fashion and dynamics of its interaction with chromatin. It recently emerged that cohesin contributes to the protection of stalled replication forks through a mechanism requiring its timely mobilization from unreplicated DNA and relocation to nascent strands. This mechanism relies on DNA replication checkpoint-dependent cohesin ubiquitylation and promotes nascent sister chromatid entrapment, likely contributing to preserve stalled replisome-fork architectural integrity. Here we review how cohesin dynamic association to chromatin is controlled through post-translational modifications to dictate its functions during chromosome duplication. We also discuss recent insights on the mechanism that mediates interfacing of replisome components with chromatin-bound cohesin and its contribution to the establishment of sister chromatid cohesion and the protection of stalled replication forks.
DOI:10.1038/nature22063URLPMID:28424523 [本文引用: 2]

Mammalian genomes are spatially organized by CCCTC-binding factor (CTCF) and cohesin into chromatin loops and topologically associated domains, which have important roles in gene regulation and recombination. By binding to specific sequences, CTCF defines contact points for cohesin-mediated long-range chromosomal cis-interactions. Cohesin is also present at these sites, but has been proposed to be loaded onto DNA elsewhere and to extrude chromatin loops until it encounters CTCF bound to DNA. How cohesin is recruited to CTCF sites, according to this or other models, is unknown. Here we show that the distribution of cohesin in the mouse genome depends on transcription, CTCF and the cohesin release factor Wings apart-like (Wapl). In CTCF-depleted fibroblasts, cohesin cannot be properly recruited to CTCF sites but instead accumulates at transcription start sites of active genes, where the cohesin-loading complex is located. In the absence of both CTCF and Wapl, cohesin accumulates in up to 70 kilobase-long regions at 3'-ends of active genes, in particular if these converge on each other. Changing gene expression modulates the position of these 'cohesin islands'. These findings indicate that transcription can relocate mammalian cohesin over long distances on DNA, as previously reported for yeast cohesin, that this translocation contributes to positioning cohesin at CTCF sites, and that active genes can be freed from cohesin either by transcription-mediated translocation or by Wapl-mediated release.
DOI:10.1111/j.1365-313X.2012.04979.xURL [本文引用: 1]

a-Kleisins are core components of meiotic and mitotic cohesin complexes. Arabidopsis contains four genes that encode a-kleisin proteins: SYN1, SYN2, SYN3 and SYN4. SYN1, a REC8 ortholog, is essential for meiosis, while SYN2 and SYN4 appear to be SCC1 orthologs and function in mitosis. SYN3 is essential for megagametogenesis and is enriched in the nucleolus of meiotic and mitotic cells. In this study the role of SYN3 during meiosis was investigated by characterization of plants that express SYN3-RNAi constructs from either meiotic DMC1, native SYN3, or inducible PX7 promoters. Reduction of SYN3 caused defects in homologous chromosome synapsis and synaptonemal complex (SC) formation during male and female meiosis. Consistent with this observation, relatively little signal for the SC component ZYP1 was detected on the chromosomes of SYN3-RNAi plants. ZYP1 transcript levels were relatively normal, but several transcripts for genes that encode proteins involved in meiotic recombination were altered, which suggested that a reduction in SYN3 may inhibit meiotic progression by alteration of meiotic gene expression.
