 ,2
,2DNA methylation in the medial prefrontal cortex regulates alcohol-related behavior in rats
Hengzhen Cui1, Mizhu Sun2, Runzhi Wang2, Chenyu Li3, Yuxuan Huang3, Qiuju Huang3, Xiaomeng Qiao ,2
,2通讯作者: 乔晓孟,博士,讲师,研究方向:酒精滥用的神经生物学机制。E-mail:xiaomeng416520@126.com
编委: 吴强
收稿日期:2019-09-2修回日期:2019-11-21网络出版日期:2020-01-20
| 基金资助: |
Editorial board:
Received:2019-09-2Revised:2019-11-21Online:2020-01-20
| Fund supported: |
作者简介 About authors
崔亨贞,本科生,专业方向:基础医学。E-mail:chzzzu@163.com。

摘要
酒精滥用不仅导致组织器官损伤,还易诱发神经精神疾病。研究表明,DNA甲基化在酒精诱导基因表达和行为改变中发挥重要作用,但具体的神经生物学机制尚未被阐明。为了探索DNA甲基化在酒精滥用中的作用机制,本研究选取健康成年雄性 SD大鼠(Rattus norvegicus) 32只,随机分为饮水对照组(n=16)和慢性酒精暴露组(n=16),运用双瓶选择实验(two bottle choice test, TBCT)评估大鼠酒精偏爱率(alcohol preference),通过旷场行为(open field test, OFT)评估活动状态并检测血酒精浓度。分离两组大鼠内侧前额叶皮质(medial prefrontal cortex, mPFC),提取总DNA,利用简化代表性重亚硫酸盐测序技术(reduced representation bisulfite sequencing, RRBS)构建mPFC甲基化谱,对差异基因进行功能富集和通路分析,筛选与酒精滥用密切相关的甲基化差异基因,运用qRT-PCR技术检测差异基因的表达,验证DNA甲基化对基因的表达调控;利用qRT-PCR和Western blot检测甲基转移酶(DNA methyltransferases, DNMTs)和甲基化CpG 位点结合蛋白2 (methyl CpG binding protein 2, MeCP2)的表达;同时,还检测了短期酒精暴露(7 d)对大鼠mPFC内DNMTs和MeCP2的影响(n=8/组)。结果表明,慢性酒精暴露大鼠mPFC内基因启动子区甲基化水平显著升高。与酒精滥用密切相关的差异基因中,慢性酒精暴露组Ntf3和Ppm1G启动子区甲基化水平升高,mRNA表达降低;Hap1和DUSP1启动子区甲基化水平降低,mRNA表达升高。慢性酒精暴露使DNMT3B和MeCP2 mRNA和蛋白表达升高,而短期内酒精暴露不影响它们的表达。本研究初步证实DNA甲基化与酒精滥用的发展相关,可能受DNMT3B和MeCP2分子的调控,并发现了与酒精滥用相关的靶基因Ntf3、Ppm1G、Hap1和DUSP1,为研究酒精滥用的神经生物学机制提供了新见解,同时为酒精滥用治疗提供了可能的药理学靶点。
关键词:
Abstract
Alcohol abuse causes tissue and organ damage, and may participate neuropsychiatric diseases. Studies have shown that DNA methylation plays an important role in gene expression and behavioral changes induced by alcohol, however the causative neurobiological mechanisms have not been clarified. In this study, 32 healthy adult male SD rats were randomly divided into a drinking water control group (n=16) and a chronic alcohol exposure group (n=16). The alcohol preference and locomotor activity of rats were evaluated by two-bottle choice test (TBCT) and open-field test (OFT). DNA methylation in the medial prefrontal cortex (mPFC) tissue was detected by the reduced representative bisulfite sequencing (RRBS) technology. The methylation differential genes closely related to alcohol abuse were screened. qRT-PCR was used to verify the mRNA expression patterns of differential genes. qRT-PCR and Western blot were used to detect the expression of DNA methyltransferases (DNMTs) and methyl CpG binding protein 2 (MeCP2). Furthermore, the effect of short-term alcohol exposure (7 days) on DNMTs and MeCP2 in the mPFC of rats was tested (n=8/group). The results indicated that the methylation level of promoter region in the mPFC of rats exposed to chronic alcohol was significantly increased. In addition, the increased methylation levels in the promoter of Ntf3 and Ppm1G were accompanied by down-regulated mRNA levels in the chronic alcohol exposure group. The decreased methylation levels in the promoter of Hap1 and DUSP1 were accompanied by up-regulated mRNA levels. Furthermore, chronic alcohol exposure increased the mRNA and protein levels of DNMT3B and MeCP2. However, short term alcohol exposure did not affect their expression. This present study provides evidence that DNA methylation is associated with the development of alcohol abuse, which may be regulated by DNMT3B and MeCP2. The target genes Ntf3, Ppm1G, Hap1, and DUSP1 related to alcohol abuse were discovered as well, providing new insights into the neurobiological mechanism of alcohol abuse and the potential pharmacological targets for the treatment of alcohol abuse.
Keywords:
PDF (1502KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
崔亨贞, 孙蜜烛, 王润芝, 李辰雨, 黄予暄, 黄秋菊, 乔晓孟. 内侧前额叶皮质DNA甲基化调控大鼠酒精相关行为. 遗传[J], 2020, 42(1): 112-125 doi:10.16288/j.yczz.19-261
Hengzhen Cui.
酒精是世界公认的成瘾性精神活性物质,长期过量饮酒会引起多种危害。在发达国家,酒精引起的疾病负担(约10%)仅次于烟草和高血压,而在一些发展中国家,酒精甚至是致病致残的首要危险因素。《2017年全球酒精、烟草和非法药物使用状况报告》指出,2015年在成年人群中,重度饮酒的患病率为18.4%,其中每10万人中就有843.2人患有酒精依赖[1]。全世界因酒精滥用造成的死亡达330万例(占所有死亡人数的5.9%),并产生5.1%的疾病负担,造成2500亿的经济损失[2]。中国的酒文化源远流长,带来的各种问题也显而易见。2015年我国重度饮酒的患病率在20%~25%[1],青少年酗酒率呈逐年上升趋势[3]。酒精滥用对个人的身心健康有着极大威胁。摄入过量酒精可以诱发肝硬化,增加肝癌的发生率,并造成胃肠道功能紊乱及心血管疾病;长期饮酒可诱发神经精神疾病,包括胎儿酒精谱系障碍和依赖,导致焦虑、抑郁甚至认知功能障碍[4]。因此,对酒精滥用的病因机制进行研究具有十分重要的生物医学及社会学意义。
酒精反复摄入会引起相关基因的表达变化[5,6],即使停止饮酒后这种改变仍可持续,并且可以遗传。这种基因表达改变的内在机制可能由表观遗传介导[7]。近年来,越来越多的研究聚焦酒精滥用相关基因表达改变的表观遗传机制,酒精可能使基因的甲基化或组蛋白乙酰化发生改变,从而增强或抑制转录,引起基因表达持久性改变。Zhu等[8]通过对1465个健康受试者外周血全基因组甲基化进行分析,发现饮酒量与基因组整体甲基化水平呈负相关。Mg2+/Mn2+依赖的蛋白质磷酸酶1G (protein phosphatase-Mg2+/Mn2+- dependent-1G, Ppm1G)被发现与酒精使用障碍相关,在酒精使用障碍患者的外周血基因组中,Ppm1G启动子区甲基化水平升高,mRNA表达降低,而且其与499名酗酒青少年的冲动性行为增加显著相关[9]。在对啮齿类动物的研究中发现,慢性酒精暴露改变了小鼠(Mus musculus)不同脑区5-羟色胺受体3a (serotonin receptor 3a, Htr3a)启动子区的甲基化,Hip中Htr3a启动子区甲基化水平升高,而背内侧纹状体(dorsomedial striatum, DMSTR)和dmPFC内Htr3a启动子区甲基化水平降低[10]。有研究表明,低甲基化药物(如甲基转移酶抑制剂)能减少慢性酒精暴露大鼠的饮酒量[11,12]。由此可见,DNA选择性地响应环境因素,进而改变转录机制,参与细胞生长发育、周期调节、信号转导等多种生命过程,导致大脑中生理和病理过程的改变。
本研究通过建立大鼠慢性酒精暴露模型,提取内侧前额叶皮质(medial prefrontal cortex, mPFC) DNA,运用简化代表性重亚硫酸盐测序(reduced representation bisulfite sequencing,RRBS)技术检测mPFC全基因组甲基化水平,筛选出与酒精滥用相关的甲基化差异基因;进而对差异基因进行GO功能富集和KEGG通路分析;运用实时荧光定量PCR技术 (qRT-PCR) 对甲基化差异基因进行扩大样本量验证;运用Western blot技术对调控DNA甲基化的关键酶及分子进行蛋白表达检测,探索DNA甲基化在酒精滥用发展过程中的作用。
1 材料与方法
1.1 实验动物
健康成年雄性 SD大鼠48只,6~8周龄,体重180~220 g。实验动物均由郑州大学实验动物中心提供,在无特定病原体(specific pathogen free, SPF)的动物标准环境饲养。饲养室环境安静,温度区间为21~25℃,湿度区间为40%~60%,保持12 h明暗周期(7:00 AM灯亮,7:00 PM灯暗),实验均在明周期进行。该项目经郑州大学实验动物管理与使用委员会批准,并严格遵守中华人民共和国科技部《关于善待实验动物的指导意见》。所有大鼠均未经过任何行为学训练,单只单笼饲养,自由摄食饮水,并于实验前一周时间提前适应新环境。1.2 慢性酒精暴露
所有动物被随机分为两组:慢性酒精暴露组(n=16)和饮水对照组(n=16),其中每组各12只用于行为学检测及分子生物学实验,剩余4只用于RRBS测序。慢性酒精暴露模型建立之前,酒精暴露组大鼠首先接受连续5 d的双瓶选择实验(two bottle choice test, TBCT),以判断大鼠的基础酒精偏爱率(alcohol preference)。同一饲养笼左右两边各一瓶6% (V/V)的酒精水溶液和自来水,供大鼠自由选择。为防止大鼠对位置的偏爱,需每天交换酒瓶和水瓶的位置。每天17:00 PM定时称量酒瓶和水瓶的重量,计算酒精溶液和水溶液的消耗体积,酒精偏爱率=酒精溶液消耗体积/液体总摄入体积×100%。双瓶选择实验结束后,开始建立慢性酒精暴露模型。实验组大鼠自由饮用6%的酒精水溶液,对照组大鼠自由饮水,每日7:00 AM更换新的酒和水,连续35 d。35 d内每日5:00 PM称量大鼠体重并记录饮酒量,第7、14、21、28、35 d分别进行旷场行为(open field test, OFT)和血酒精浓度检测。第36 d起重新进行一次双瓶选择实验,以判定模型是否建立成功。随后以颈椎脱臼法处死大鼠,分离mPFC核团,保存于-80℃冰箱中备用。对于短期酒精暴露模型(n=8/组),大鼠自由饮用6%的酒精水溶液连续7 d,第7 d早上处死大鼠,分离mPFC核团,一半核团提取mRNA,一半提取蛋白质用于后续分析。1.3 旷场行为
黑色正方形敞口实验箱(100 cm×100 cm×45 cm),中心区域占箱底总面积的60%,上方2 m处悬挂30 W照明灯。在酒精暴露的第7、14、21、28、35 d早上7:30,将所有大鼠逐只转运至行为学实验室适应30 min,随后将大鼠放至箱子角落进行15 min的自由探索,由摄像装置记录并分析15 min内的数据,包括:(1)总活动距离(total distance):大鼠在实验箱的总活动路程,反应大鼠的水平动机;(2)中心区域停留时间比(pencentage of time in the center):反应大鼠的探索行为和焦虑样情绪。1.4 血酒精浓度(blood alcohol level, BALs)检测
旷场实验结束后,立即对酒精暴露组大鼠进行鼠尾静脉采血,每只抽取尾静脉血约0.2 mL。随后采用顶空-气相色谱法测定BALs。实验前平衡温度20 min,从冰箱内取出血样100 μL于5 mL顶空气瓶中,加入0.1 g/L叔丁醇内标溶液500 μL,用钳口器将空瓶用硅胶垫密封后放入自动顶空恒温炉中,吹洗2 min,进行预热,听到响声后代表预热完成,开始进样,进样口温度为150℃,再听到一次响声时点击“START”按钮,开始测定,并进行样品记录。1.5 简化代表性重亚硫酸盐测序
两组大鼠分别选取3只,提取mPFC基因组DNA。限制性内切酶Msp Ⅰ将DNA切割为40~220 bp大小的片段,在DNA分子两端引入接头序列及索引序列,割胶后回收。按照标准流程使用QIAquick Gel Extraction kit进行纯化,洗脱,将DNA保存在-20℃冰箱备用。按照标准流程使用EpiTect Bisulfite Kit对纯化产物进行重亚硫酸氢盐处理,洗脱DNA。通过PCR扩增完成文库构建。PCR扩增引物为:F: 5′-AATGATACGGCGACCACCGA-3′;
R: 5′-CAAGCAGAAGACGGCATACGA-3′。
PCR扩增条件:95℃ 5 min,98℃ 30 s;98℃ 10 s,60℃ 30 s,72℃ 30 s,15个循环;最后再72℃ 5 min;4℃保存。
将构建好的文库等摩尔浓度混合,对文库进行定量后,利用Illumina Nextseq 500 Desktop Sequencer上机测序。得到原始测序数据后,对其进行质量控制以获得高质量数据进行后续分析。将测序序列比对到基因组,得到不同基因组环境的甲基化情况,并基于二项分布检验和1% FDR约束对甲基化位点进行筛选。统计甲基化程度的分布情况,比较样本之间甲基化程度的一致性。最后,运用GOseq对差异甲基化区域最近邻基因进行功能注释,用KEGG pathway对甲基化差异基因进行通路分析。
1.6 实时荧光定量PCR
采用MiniBEST Universal RNA Extraction 试剂盒(日本TaKaRa公司)提取两组大鼠(n=12/组)一侧mPFC的RNA,并测定RNA浓度。随后以RNA为模板合成互补的cDNA。以cDNA产物为模板,进行荧光实时定量PCR反应。目的基因的引物序列见表1。Table 1
表1
表1大鼠目的基因扩增引物序列信息
Table 1
| 基因名称 | 引物序列(5′→3′) |
|---|---|
| Ntf3 | F: GGCAACAGAGACGCACAAT |
| R: TCCTCCGTGGTGATGTTCTA | |
| Pdp1 | F: CTCAATTACCTGGTGCTTCG |
| R: GCATTGTGGTCATTAGAGAGC | |
| Ppm1G | F: TCATTGCTTGTGACGGCATC |
| R: ATGACAATAACCGAAGCTCCC | |
| FLT1 | F: TAACGCAATCTTCTTAACGTC |
| R: TTAATCCTCACAAACGTCCC | |
| N1g2 | F: CAGAAGAGACAGAGCAGGCATC |
| R: GCAGAGACCACCACATTCATC | |
| Hap1 | F: CAAGGATGAATGTGGTGGTCTC |
| R: CTGCTGCCGCTTAGAATGG | |
| NFκB-p100 | F: TCGACCTCCACCGGATCTTT |
| R: CTGTGTCTTTTGGCTTCGGC | |
| RTKN2 | F: CCATAGGATGCTAATCTCCGTTT |
| R: AAGATTACTGCGGTCAACCTT | |
| DUSP1 | F: CATTCCCGATGACATACACGTT |
| R: CATTCCCGATGACATACACGTT | |
| DUSP8 | F: TCACTGGTCTGGCTGGCATTTC |
| R: AGTTGAAGTTGGGCGAGATGG | |
| DNMT1 | F: GTTCCTTGTAGGCGAGTGTG |
| R: TTGCGTAGTCCTGGCTGTAC | |
| DNMT3A | F: GCA AAGTGAGGACCATTACC |
| R: GCCAAACACCCTTTCCAT | |
| DNMT3B | F: GTGCGTCGTTCAGGCAGTAG |
| R: GTTCTCGGCTCTCCTCATCT | |
| MeCP2 | F: GCCTTCAGCCCACCATTCTG |
| R: CGGTCACGAATGATGGAACG | |
| GAPDH | F: ATGGGGAAGGTGAAGGTCG |
| R: GGGGTCATTGATGGCAACAA |
新窗口打开|下载CSV
1.7 Western blot检测
提取两组大鼠一侧mPFC的蛋白质,用二喹啉甲酸(bicinchoninic acid, BCA)法进行蛋白定量。为避免电泳时由于蛋白浓度的差异造成的条带深浅不一,根据测得的蛋白浓度(均高于3 μg/μL),将所有样本的总蛋白浓度均调整为3 μg/μL,用RIPA裂解液和5×的蛋白上样缓冲液稀释所提取的蛋白。随后进行聚丙烯酰胺凝胶电泳(SDS-PAGE)、转膜并孵育抗体。一抗分别为抗DNMT1兔多克隆抗体(美国Epigentek公司)、抗DNMT3A兔多克隆抗体(美国Epigentek公司)、抗DNMT3B兔多克隆抗体(美国Epigentek公司)、抗MeCP2小鼠单克隆抗体(英国Abcam Technology公司)。二抗抗体孵育完成并洗膜后进行化学发光检测。采用美国Bio-rad公司的Image Lab软件进行分析,采集灰度值,计算目的蛋白条带的平均光密度,进行相对定量分析。用目的蛋白与内参蛋白(β-actin)条带的光密度比值来表示检测蛋白的相对表达水平。1.8 数据处理与统计学分析
采用SPSS 20.0和Graphpad Prism 6.0软件进行统计学分析,以均值 ± 标准误(mean ± S.E.M.)表示实验数据。对于模型建立期间两组大鼠的体重变化和每周一次的旷场行为测试结果,以时间为组内因素,饮酒或饮水为组间因素进行重复测量的双因素方差分析(Two-way ANOVA with repeated measurements, RM two-way ANOVA);两个因素有交互作用时,进行Sidak’s post hoc多组之间的两两比较。对于35 d内慢性酒精暴露组大鼠的酒精摄入量(g/kg/d)、血酒精浓度和酒精偏爱率,进行重复测量的单因素方差分析(One-way ANOVA with repeated measurements, RM one-way ANOVA);并进行Sidak’s post hoc不同时间点的两两比较。对于两组大鼠mPFC内相关分子的mRNA和蛋白表达,以相对于饮水组的相对倍数表示,采用非配对t检验进行比较。以P<0.05为显著性检验水准。2 结果与分析
2.1 大鼠体重、酒精摄入量、血酒精浓度及酒精偏爱率的变化
2.1.1 35 d模型建立期间大鼠体重和酒精摄入量变化为观测慢性酒精暴露对大鼠基础新陈代谢的影响,每日5:00 PM称量大鼠体重。重复测量的双因素方差分析结果表明,慢性酒精暴露模型建立35 d内,饮用6%酒精水溶液对大鼠体重未产生显著性影响[FAlcohol(1,22)=0.649, P=0.429]。在整个实验期间,两组大鼠的体重表现出同步增长[FTime(5,110)=932.300, P<0.0001] (图1A)。结果表明,饮用6%酒精水溶液35 d对大鼠体重不产生影响,基本不影响大鼠的新陈代谢。
对于酒精摄入量,重复测量的单因素方差分析表明,整个实验过程中大鼠饮酒量保持稳定[F(5,66)= 2.268, P=0.121],平均摄入量为6.47±1.08 g/kg/d (图1B)。随着饮用时间增长,大鼠饮酒量并没有增加,始终保持稳定,说明大鼠没有产生酒精耐受。
2.1.2 慢性酒精暴露大鼠的血酒精浓度
在酒精暴露第7、14、21、28、35 d,对酒精暴露组大鼠的尾静脉进行采血,检测大鼠BALs,以了解酒精在大鼠体内的维持状况。整个实验过程中,酒精暴露组大鼠的BALs平均为83.54±1.24 mg/dL,重复测量的单因素方差分析表明,酒精暴露组大鼠BALs保持稳定[F(4,55)=0.4265, P=0.936] (图1C)。结果表明,6% (V/V)酒精水溶液的慢性暴露可以让大鼠体内维持一定的BALs,大鼠相关行为的异常是酒精作用的结果。
2.1.3 模型建立前后大鼠酒精偏爱率
慢性酒精暴露开始前和结束后,对酒精暴露组大鼠进行5 d的双瓶选择实验。在模型建立之前,重复测量的单因素方差分析表明,酒精暴露组大鼠的酒精偏爱率保持较低的稳定水平[F(4,55)=0.467, P=0.721],平均偏爱率为17.12±1.50%。而经过35 d的酒精暴露之后,慢性酒精暴露组大鼠的酒精偏爱率明显升高,重复测量的单因素方差分析显示,时间因素影响了大鼠的酒精偏爱率[F(4,55)=3.905, P< 0.05],测试第5 d,慢性酒精暴露组大鼠的酒精偏爱率显著高于第1 d (P<0.05)。5 d内大鼠酒精偏爱率的最低值为43.10±5.54%,远高于模型建立前大鼠的酒精偏爱率(图1D)。
图1
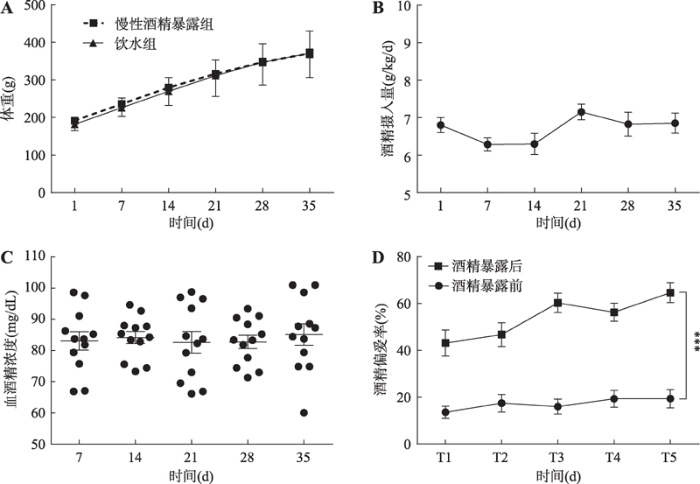 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1大鼠体重、饮酒量、血酒精浓度和酒精偏爱率的变化
A:慢性酒精暴露期间大鼠体重变化;B:大鼠每天酒精摄入量;C:血酒精浓度;D:模型建立前后大鼠酒精偏爱率。与酒精暴露前相比,***: P<0.001表示差异显著,n=12/组。
Fig. 1Changes of body weight, alcohol intake, BALs, and alcohol preference in rats
2.2 酒精对大鼠旷场行为的影响
为研究连续的酒精作用对大鼠自主活动量和焦虑样行为的影响,在酒精暴露第7、14、21、28和35 d对两组大鼠进行旷场实验。结果发现,大鼠偏向于在旷场外围活动,这与啮齿类动物的趋壁性习性相吻合(图2A)。重复测量的双因素方差分析显示,在35 d的慢性酒精暴露过程中,大鼠的水平活动量随时间有所变化[FTime(4,88)=24.150, P<0.0001],酒精因素所起的作用不大[FAlcohol(1,22)=2.868, P=0.113]。因酒精和时间因素没有显著的交互作用,本研究无法进行多重比较分析,为了使结果更直观,本研究展示了具体的总活动距离(mean±S.E.M.)。慢性酒精暴露组和饮水对照组相比,在饮酒第7 d大鼠水平活动量有增加趋势(饮水组:5457.75±192.75 cm vs. 酒精暴露组:7017.95±213.73 cm),表明短期酒精暴露有刺激运动的作用;随后这种差异在第14、21 d逐渐消失,饮水组和酒精暴露组总活动距离一致,表明神经系统对酒精的适应性;而在饮酒第28 d和第35 d大鼠水平活动量有减少趋势:第28 d,饮水组:5824.94± 144.86 cm vs. 酒精暴露组:4494.54±180.74 cm;第35 d,饮水组:5602.01±222.11cm vs. 酒精暴露组:3635.22±208.12 cm (图2B),反映了慢性酒精暴露的抑制性作用。
图2
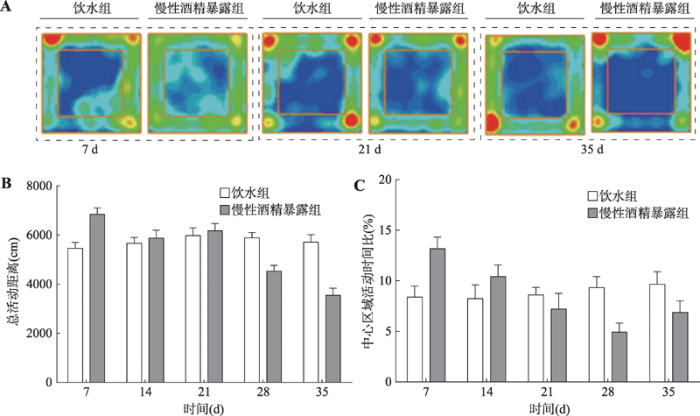 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2酒精暴露期间大鼠的旷场行为
A:慢性酒精暴露第7 d、21 d、35 d大鼠旷场行为热图;B:慢性酒精暴露第7 d、14 d、21 d、28 d、35 d大鼠总活动距离;C:大鼠在中心区域停留时间比。
Fig. 2The open-field behavior in rats during alcohol exposure
同样,本研究也检测了中心区域探索的时间百分比。重复测量的双因素方差分析显示,时间因素显著影响了大鼠对中心区域的探索[FTime(4,88)=4.607, P<0.05],而酒精因素所起的作用不大[FAlcohol(1,22)= 0.105, P=0.751] (图2C)。与总活动距离结果一致,在饮酒第7 d和第14 d大鼠向中心区域探索的时间有增加趋势:第7 d,饮水组:8.38±1.10% vs. 酒精暴露组:13.18±1.13%;第14 d,饮水组:8.24±1.35% vs. 酒精暴露组:10.40±1.16%。随后这种差异在第21 d消失,而在饮酒第28 d和35 d酒精暴露组大鼠向中心区域探索的时间有减少趋势:第28 d,饮水组:9.32±1.10% vs. 酒精暴露组:4.93±0.89%;第14 d,饮水组:9.64±1.25% vs. 酒精暴露组:6.87± 1.13%。
2.3 慢性酒精暴露大鼠mPFC内DNA甲基化谱
双瓶选择实验之后,分别选取饮水对照组和慢性酒精暴露组大鼠各4只,立即将其处死,分离mPFC核团,提取mPFC总DNA,浓度和质量测定后分别选取每组3个样本进行RRBS测序。从图3A可以看出,两组样品中甲基化的胞嘧啶CpG类型的比例最高,这是因为哺乳动物的胞嘧啶甲基化一般发生在CpG位点。其余类型的甲基化均较低,对总甲基化水平贡献不大。对于不同胞嘧啶类型的甲基化水平,尽管慢性酒精暴露组均略高于饮水对照组,但总体上两组甲基化水平没有差异。为了更进一步分析CpG的甲基化变化,本研究对两组样本的CpG进行了不同特征区域的注释(图3B)。结果表明,甲基化差异区域有66%存在于基因连接区,23%存在于内含子区,9%存在于外显子区,2%存在于启动子区。在所有甲基化存在差异的CpG位点,仅有2%存在于启动子区的CpG岛,4%存在于CpG岛岸。
本研究进一步对甲基化水平差异最大的基因进行GO功能富集(Top 20)和KEGG通路分析(Top 30)。GO结果显示,差异基因多为突触相关基因、谷氨酸受体、跨膜转运体和离子通道,这些基因与神经系统功能密切相关(图3C)。通过对差异基因进行KEGG通路分析,发现甲基化差异基因与mTOR信号通路、可卡因成瘾、Ras信号通路、cAMP信号通路、FoxO信号通路、安非他命成瘾和Rap1信号通路等密切相关(图3D)。
图3
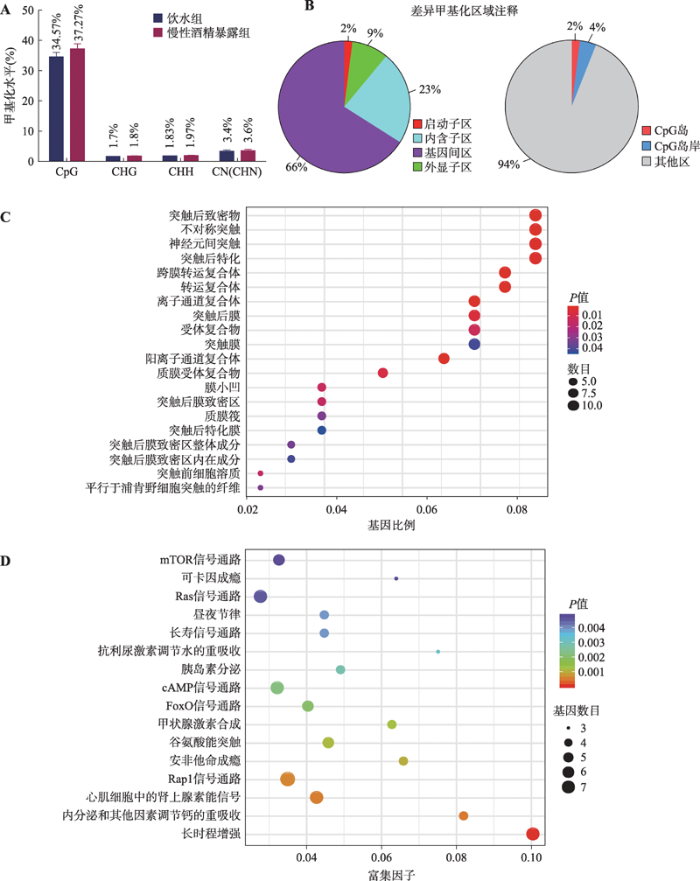 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3RRBS测序结果
A:两组样品中不同胞嘧啶类型的平均甲基化水平;B:差异甲基化区域在功能元件上的注释;C:甲基化基因的GO功能富集(Top 20);D:甲基化差异基因的KEGG通路分析(Top 30)。不同胞嘧啶类型包括CpG、CHG、CHH、CN,H代表除了G以外的A、T、C碱基。
Fig. 3Results of RRBS sequencing
2.4 慢性酒精暴露大鼠mPFC内基因启动子区甲基化水平及酒精滥用相关基因的筛选
哺乳动物启动子区的CpG位点的甲基化水平与转录调节密切相关,因此本研究着重分析了启动子区CpG差异位点。为了更直观地表现出慢性酒精暴露组与饮水对照组大鼠mPFC内基因启动子区甲基化的差异,本研究对位于启动子区的340个差异位点进行了热图分析(图4A)。热图中每一行为一个位点,每一列代表一个样本,左边3列为酒精暴露组,右边3列为饮水组。最上边的树状结构表示不同样本间的聚类,本图中慢性酒精暴露组聚在一起,饮水对照组聚在一起,说明组内样本亲缘关系更近,样本聚类良好。左边的树状结构为差异位点的聚类关系。尽管两组样本mPFC内整体甲基化水平无显著差异,但在启动子区,慢性酒精暴露组的甲基化水平显著高于饮水对照组。其中与对照组相比,慢性酒精暴露组大鼠mPFC内存在67个低甲基化位点和273个高甲基化位点,这些位点涉及到241个已知基因。通过滥用与成瘾数据库,并查阅相关文献,筛选出与药物依赖及酒精滥用密切相关的启动子区甲基化差异基因共10个。与饮水对照组大鼠相比,慢性酒精暴露组大鼠mPFC内基因启动子区甲基化升高的基因包括神经营养因子3 (neurotrophin factor-3, Ntf3) (P<0.001)、丙酮酸脱氢酶磷酸酶催化亚单元1 (pyruvate dehyrogenase phosphatase catalytic subunit 1, Pdp1) (P<0.01)、Mg2+/Mn2+依赖的蛋白质磷酸酶1G (protein phosphatase-Mg2+/Mn2+-dependent-1G, Ppm1G) (P<0.05)、FMS样酪氨酸激酶1 (FMS-like tyrosine kinase 1, FLT1) (P<0.0001)和神经连接蛋白2 (neuroligin 2, Nlg2) (P<0.05),启动子区甲基化降低的基因包括亨廷顿相关蛋白1 (huntingtin-associated protein 1, Hap1) (P<0.05)、核因子κB (nuclear factor kappa B p100, NFκB-p100) (P<0.05)、RTKN蛋白2 (rhotekin 2, RTKN2) (P<0.01)、双特异性磷酸酶1 (dual specificity phosphatase 1, DUSP1) (P<0.05)和双特异性磷酸酶8 (dual specificity phosphatase 8, DUSP8) (P<0.05) (图4B)。
图4
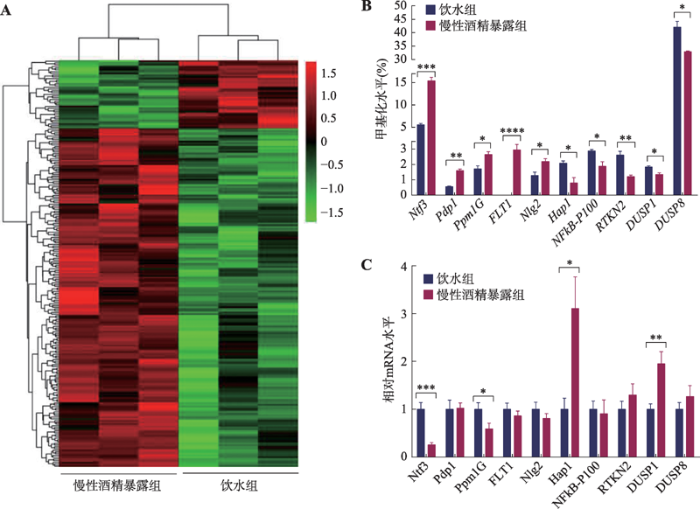 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4启动子区甲基化水平及甲基化差异基因转录验证
A:启动子区甲基化差异位点聚类图。红、黑、绿信号代表甲基化程度的高低,红色信号越强,代表甲基化程度越高,反之则代表甲基化程度越低。B:慢性酒精暴露组和饮水对照组mPFC内10个基因启动子区的甲基化程度。C:慢性酒精暴露组和饮水对照组mPFC内10个甲基化差异基因的mRNA表达水平。与饮水组相比,*: P<0.05表示有差异,**: P<0.01表示有明显差异,***: P<0.001表示差异显著,****: P<0.0001表示差异极显著。
Fig. 4The methylation levels of promoter region and transcription verification of methylation differential genes
本研究进一步利用qRT-PCR对这10个甲基化差异基因进行转录水平的验证(图4C)。结果表明,慢性酒精暴露后,大鼠mPFC内Ntf3、Ppm1G启动子区CpG位点甲基化水平升高,抑制转录,导致其mRNA水平降低;而Hap1和DUSP1启动子区CpG位点甲基化水平降低,解除或削弱了转录抑制作用,导致其mRNA水平升高。
2.5 慢性酒精暴露大鼠mPFC内甲基化调控酶的表达
经过35 d的慢性酒精暴露和5 d的双瓶选择,本研究检测了大鼠mPFC内DNMTs和MeCP2蛋白和mRNA表达改变(图5)。非配对t检验显示,相较于饮水对照组,酒精暴露组大鼠mPFC内的DNMT1 (蛋白:t=0.824, P=0.424; mRNA: t=0.885, P=0.391)和DNMT3A (蛋白: t=1.510, P=0.153; mRNA: t= 0.064, P=0.950)表达无明显改变,而DNMT3B (蛋白: t=9.800, P<0.0001; mRNA: t=5.364, P<0.0001)和MeCP2 (蛋白: t=13.94, P<0.0001; mRNA: t=4.652, P<0.001)表达显著升高。图5
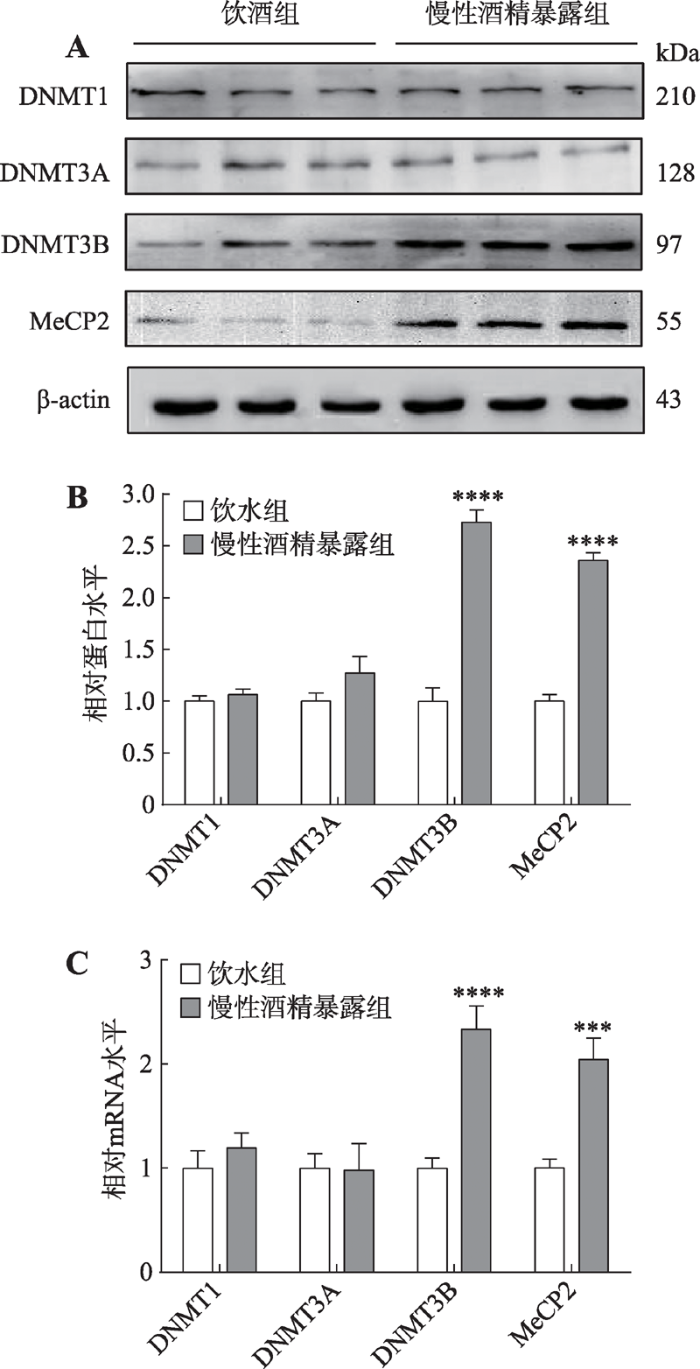 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5慢性酒精暴露后大鼠mPFC内DNMT1、DNMT3A、DNMT3B和MeCP2蛋白及mRNA表达改变
A:慢性酒精暴露后大鼠mPFC内DNMT1、DNMT3A、DNMT3B和MeCP2蛋白条带;B:慢性酒精暴露后大鼠mPFC内DNMT1、DNMT3A、DNMT3B和MeCP2蛋白表达;C:慢性酒精暴露后大鼠mPFC内DNMT1、DNMT3A、DNMT3B和MeCP2的mRNA表达。与饮水组比较,***: P<0.001表示差异显著,****: P< 0.0001表示差异极显著。
Fig. 5Changes in DNMT1, DNMT3A, DNMT3B and MeCP2 protein and mRNA expression in rats mPFC tissue after chronic alcohol exposure
2.6 短期酒精暴露对大鼠mPFC内甲基化调控酶的影响
为了明确短期内酒精暴露对甲基化调控酶的作用,让大鼠自由饮酒7 d后,本研究检测了mPFC内DNMTs和MeCP2的蛋白和mRNA表达改变(图6)。非配对t检验显示,短期酒精暴露不影响DNMT1 (蛋白: t=0.092, P=0.928; mRNA: t=0.440, P=0.670)、DNMT3A (蛋白: t=0.153, P=0.880; mRNA: t=0.209, P=0.739)、DNMT3B (蛋白: t=0.198, P=0.846; mRNA: t=0.182, P=0.859)和MeCP2 (蛋白: t=0.060, P=0.953; mRNA: t=0.394, P=0.653)的表达。图6
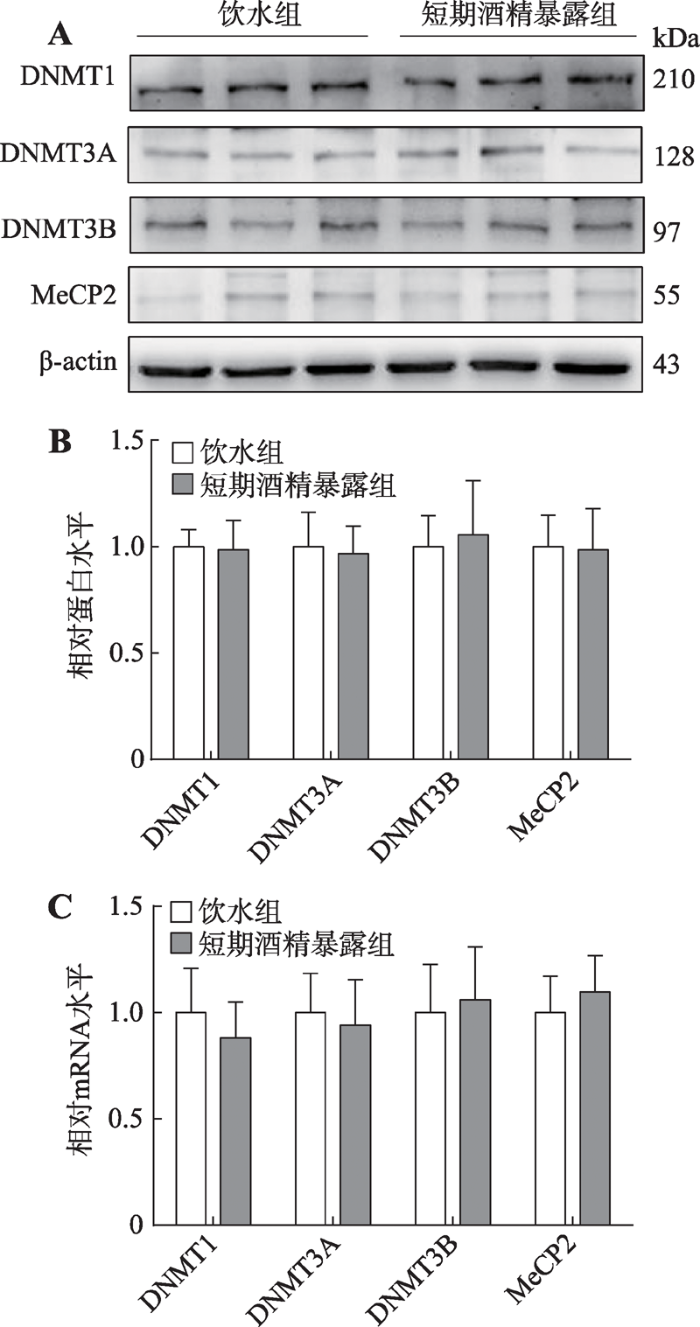 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6短期酒精暴露后大鼠mPFC内DNMT1、DNMT3A、DNMT3B和MeCP2蛋白和mRNA表达改变
A:短期酒精暴露后大鼠mPFC内DNMT1、DNMT3A、DNMT3B和MeCP2蛋白条带;B:短期酒精暴露后大鼠mPFC内DNMT1、DNMT3A、DNMT3B和MeCP2的蛋白表达;C:短期酒精暴露后大鼠mPFC内DNMT1、DNMT3A、DNMT3B和MeCP2的mRNA表达。
Fig. 6Changes of DNMT1, DNMT3A, DNMT3B, and MeCP2 protein and mRNA levels in the rats mPFC tissue after short-term alcohol exposure
3 讨论
酒精滥用是一种由遗传因素和环境因素共同作用的复杂疾病,涉及奖赏、动机、学习记忆、情绪和执行功能等多种神经生物学过程,尽管科研人员对其神经生物学机制进行了深入研究,但尚未研发出有效的药物。前额叶皮质参与调控认知、情感、情绪、动机、决策等生物学进程,其中mPFC与酒精滥用引起的神经适应性改变密切相关。DNA甲基化在长时程记忆的形成以及药物成瘾中都具有重要作用,DNMTs可通过催化5?-CpG-3?上的胞嘧啶发生甲基化来调节下游基因的表达,从而产生持久的神经生物学改变并最终影响个体行为调控,然而在mPFC内DNMTs的活性对于酒精相关行为的调控作用仍不清楚。本研究建立大鼠慢性酒精暴露模型,检测了大鼠的自主活动和焦虑样行为,发现在饮酒第7 d酒精暴露组大鼠总活动距离和中心区域探索时间比饮水对照组高,表明短期内低浓度饮酒,可以增加大鼠的自主活动量,并在一定程度上减轻焦虑样情绪。随着饮酒时间的增加,在连续饮酒21 d后酒精暴露组与饮水对照组大鼠之间活动总距离和中心区探索时间差异逐渐缩小,反映了长期饮酒后大鼠对酒精的适应性。而饮酒28 d后,大鼠总活动距离和中心区探索时间又减少并低于饮水对照组,表明慢性低浓度饮酒给大鼠带来了不适感,减少了大鼠的自主活动量,并加重了大鼠的焦虑样行为。与本研究结果一致的是,Boerngen-Lacerda等[13]发现2 g/kg的酒精对小鼠急性灌胃或灌胃7 d,均增加了小鼠对高架十字迷宫开臂的探索时间和旷场实验的自主活动距离。出生4~9 d的雄性大鼠幼崽暴露在5.25 g/kg/d的酒精环境下连续一个月,活动性和探索行为均降低,并且社交行为变少[14]。大量的动物实验和临床资料表明,焦虑与酒精滥用两者之间存在复杂的交互联系[15,16]。酒精滥用和依赖作为慢性复发性疾病,在发展过程中焦虑起了促进作用[17]。短期内饮酒降低短时程焦虑,而长期饮酒反而增加长时程焦虑。多数慢性饮酒或酒精戒断能够通过生理-心理-社会机制引起显著的焦虑症状。个体饮用一定浓度酒精后,酒精的心理或药理作用可以缓解焦虑症状,在此过程中,酒精通过负强化作用促进焦虑个体提高饮酒量和饮酒频率[18]。
尽管DNA甲基化异常可以增加很多疾病的患病风险,酒精滥用的甲基化研究尚处于早期阶段。目前的研究偏向于饮酒个体外周血的基因组甲基化或酒精诱导的某个基因甲基化的异常。有研究表明酒精滥用患者外周血DNA甲基化较健康对照组显著升高,伴随DNMT3A和DNMT3B mRNA水平的降低[19,20]。利用甲基化芯片对台湾地区酒依赖患者外周血全基因组DNA进行检测,发现酒依赖组整体DNA甲基化程度较对照组明显升高[21]。与血液基因组相比,来自大脑基因组的甲基化差异更能说明酒精相关行为的中枢控制机制。本研究通过提取慢性酒精暴露大鼠mPFC基因组DNA,进行RRBS测序分析,发现两组大鼠mPFC内整体基因组甲基化无显著差异,但仍有911个高甲基化差异甲基化区域(differentially methylated region, DMR)和560个低甲基化DMR,进一步分析启动子发现,慢性酒精暴露组大鼠mPFC启动子区甲基化水平显著高于饮水对照组。通过对差异基因的功能富集分析和通路分析,本研究筛选出与酒精滥用密切相关的甲基化差异基因Ntf3、Ppm1G、Hap1和DUSP1,在mRNA水平验证了其表达。其中,Ntf3在神经元分化、突触发育和可塑性起关键的调控作用[22,23],本实验室前期研究也证实Ntf3是酒精滥用的靶基因[12]。Ruggeri[9]等对18对患有酒精使用障碍的单卵双胞胎外周血DNA进行了全基因组甲基化分析,证明Ppm1G与酒精使用障碍和行为控制期间的大脑活动有关。此外,Hap通过影响钙信号通路从而调控神经元兴奋性[24,25]。同时,DUSP1可以使丝裂原活化蛋白激酶(mitogen activated protein kinases,MAPK)苏氨酸/酪氨酸残基特异性去磷酸化,从而调控细胞的生长、分化、增殖及凋亡[26]。以上甲基化差异基因的筛选为酒精滥用的分子机制提供了可能靶点。
DNA甲基化是由甲基转移酶(DNA methyltransferase, DNMTs)调控的。DNMTs大致分为两类:(1)从头甲基化酶,包括DNMT3A和DNMT3B,在未经甲基化的DNA上建立初始甲基化模式;(2)维持甲基化酶DNMT1,在DNA复制过程中将甲基化模式从亲本DNA链上复制到子代链[27,28]。DNMTs的这些独特功能可以确保DNA甲基化模式在不同个体中以组织特异性方式维持和保存。此外,DNMTs、甲基化CpG位点结合蛋白-2(MeCP2)、HMTs、HDACs可能通过复杂的交互作用影响DNA的甲基化水平,调节相关基因的表达,从而对酒精相关表型发挥作用[29]。DNMT1和DNMT3A基因双敲除小鼠Hip神经元体积减小,学习和记忆能力受损[30]。大鼠伏隔核(nucleus accumbens, NAc)中DNMT3A过表达能够增加相应神经元的树突棘密度,同时减弱可卡因诱导的奖赏效应[31]。MeCP2基因敲除小鼠比野生型小鼠对酒精有更强的敏感性和戒断效应[32]。本研究也发现,慢性酒精暴露后,大鼠mPFC内DNMT3B和MeCP2表达升高。由此看来,动态的DNA甲基化调节对酒精相关表型的改变和维持至关重要,而DNMTs和MeCP2作为DNA甲基化的调控者,所起的重要作用不言而喻。然而,大鼠饮酒7 d,其mPFC内DNMTs和MeCP2未发生显著改变。由于表观遗传修饰是环境因素长期刺激的结果,并可以遗传给后代[33],猜测短期酒精作用不足以引起甲基化调控酶及DNA甲基化谱的改变。
本研究成功建立了大鼠慢性酒精暴露模型,获得慢性酒精暴露大鼠mPFC内DNA甲基化谱,并筛选出与酒精滥用相关的基因Ntf3、Ppm1G、Hap1和DUSP1,且明晰了调控DNA甲基化的关键酶,为未来表观遗传学在酒精滥用中的机制研究提供了基础,将来尚需从以下两个方面进行深入研究:(1)所得甲基化谱样本量太小,应该针对与酒精滥用相关的甲基化差异基因,采用BSP挑克隆测序进行扩大样本量验证,使筛选出的差异基因更可靠。(2)本研究发现DNMT3B和MeCP2调控酒精诱导的DNA甲基化的改变,接下来应运用甲基转移酶特异性干预剂验证它们在酒精滥用中的作用。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1111/add.14234URLPMID:29749059 [本文引用: 2]

This review provides an up-to-date curated source of information on alcohol, tobacco and illicit drug use and their associated mortality and burden of disease. Limitations in the data are also discussed, including how these can be addressed in the future.
DOI:10.1016/j.abrep.2019.100216URLPMID:31692685 [本文引用: 1]

High-level alcohol consumption is common in, and central to, the student community. Among adults, high-level alcohol consumption, and sometimes also low, has been associated with poorer social integration and mental health. We aimed to investigate how alcohol consumption relates to life satisfaction and mental health among students in higher education.
DOI:10.3390/ijerph120202037URLPMID:25685952 [本文引用: 1]

The current study was designed to investigate the drinking patterns and association between socio-demographic factors and adolescents' alcohol use among high school students from China's three metropolises, Beijing, Shanghai, and Guangzhou. Using a self-administered questionnaire, we conducted a cross-sectional survey among 13,811 high school students from 136 schools between May and June 2013. A two-stage stratified sampling method was used for subject selection. The prevalence of lifetime drinking was 52.5%; in addition, 38.5% of the students were past-year drinkers, while 20.1% of them had consumed alcohol in the past 30 days. During the past year, 29.7% of the students reported that they drank once per month or less, and 22.0% of the students drank less than one standard drink (SD) per occasion. For the students who were not living with their mothers, as well as the students in higher socioeconomic status (SES), the adjusted odds of past and current drinking were significantly higher, compared with those who lived with both parents and low SES. Due to the high prevalence of alcohol consumption among junior and senior high school students in metropolises, attention should be paid by parents, school administrators, educational and public health agencies for making efforts collectively to reduce alcohol availability and drinking among adolescents.
DOI:10.1080/14740338.2018.1404025URLPMID:29120249 [本文引用: 1]

Alcohol Use Disorders (AUD) is a leading cause of mortality and morbidity worldwide. At present disulfiram, naltrexone and acamprosate are approved for the treatment of AUD in U.S. and Europe. Nalmefene is approved in Europe and sodium oxybate is approved in Italy and Austria only. Baclofen received a 'temporary recommendation for use' in France.
DOI:10.3724/SP.J.1005.2014.00011URL [本文引用: 1]
DOI:10.3724/SP.J.1005.2014.00011URL [本文引用: 1]
DOI:10.3724/SP.J.1005.2008.00413URL [本文引用: 1]
DOI:10.3724/SP.J.1005.2008.00413URL [本文引用: 1]
DOI:10.1016/j.neuropharm.2013.04.004URL [本文引用: 1]

Drug addiction involves potentially life-long behavioral abnormalities that are caused in vulnerable individuals by repeated exposure to a drug of abuse. The persistence of these behavioral changes suggests that long-lasting changes in gene expression, within particular regions of the brain, may contribute importantly to the addiction phenotype. Work over the past decade has demonstrated a crucial role for epigenetic mechanisms in driving lasting changes in gene expression in diverse tissues, including brain. This has prompted recent research aimed at characterizing the influence of epigenetic regulatory events in mediating the lasting effects of drugs of abuse on the brain in animal models of drug addiction. This review provides a progress report of this still early work in the field. As will be seen, there is robust evidence that repeated exposure to drugs of abuse induces changes within the brain's reward regions in three major modes of epigenetic regulation histone modifications such as acetylation and methylation, DNA methylation, and non-coding RNAs. In several instances, it has been possible to demonstrate directly the contribution of such epigenetic changes to addiction-related behavioral abnormalities. Studies of epigenetic mechanisms of addiction are also providing an unprecedented view of the range of genes and non-genic regions that are affected by repeated drug exposure and the precise molecular basis of that regulation. Work is now needed to validate key aspects of this work in human addiction and evaluate the possibility of mining this information to develop new diagnostic tests and more effective treatments for addiction syndromes. (C) 2013 Elsevier Ltd.
DOI:10.1093/ije/dyq154URL [本文引用: 1]

Background Estimates of global DNA methylation from repetitive DNA elements, such as Alu and LINE-1, have been increasingly used in epidemiological investigations because of their relative low-cost, high-throughput and quantitative results. Nevertheless, determinants of these methylation measures in healthy individuals are still largely unknown. The aim of this study was to examine whether age, gender, smoking habits, alcohol drinking and body mass index (BMI) are associated with Alu or LINE-1 methylation levels in blood leucocyte DNA of healthy individuals.
Methods Individual data from five studies including a total of 1465 healthy subjects were combined. DNA methylation was quantified by PCR-pyrosequencing.
Results Age [beta = -0.011% of 5-methyl-cytosine (% 5mC)/year, 95% confidence interval (CI) -0.020 to -0.001% 5mC/year] and alcohol drinking (beta = -0.214, 95% CI -0.415 to -0.013) were inversely associated with Alu methylation. Compared with females, males had lower Alu methylation (beta = -0.385, 95% CI -0.665 to -0.104) and higher LINE-1 methylation (-0.796, 95% CI 0.261 to 1.330). No associations were found with smoking or BMI. Percent neutrophils and lymphocytes in blood counts exhibited a positive (beta=0.036, 95% CI 0.010 to 0.061) and negative (beta = -0.038, 95% CI -0.065 to -0.012) association with LINE-1 methylation, respectively.
Conclusions Global methylation measures in blood DNA vary in relation with certain host and lifestyle characteristics, including age, gender, alcohol drinking and white blood cell counts. These findings need to be considered in designing epidemiological investigations aimed at identifying associations between DNA methylation and health outcomes.
DOI:10.1176/appi.ajp.2014.14030382URLPMID:25982659 [本文引用: 2]

The genetic component of alcohol use disorder is substantial, but monozygotic twin discordance indicates a role for nonheritable differences that could be mediated by epigenetics. Despite growing evidence associating epigenetics and psychiatric disorders, it is unclear how epigenetics, particularly DNA methylation, relate to brain function and behavior, including drinking behavior.
DOI:10.1111/j.1530-0277.2012.01906.xURLPMID:22834954 [本文引用: 1]

Abnormal DNA methylation has been observed in promoter regions of a number of genes in human alcoholics. It is unclear whether DNA methylation changes in alcoholics result directly from alcohol consumption or predated the occurrence of alcohol abuse or dependence and whether altered DNA methylation influences gene expression.
.
DOI:10.1523/JNEUROSCI.4571-14.2015URLPMID:25878287 [本文引用: 1]

Recent studies have suggested an association between alcoholism and DNA methylation, a mechanism that can mediate long-lasting changes in gene transcription. Here, we examined the contribution of DNA methylation to the long-term behavioral and molecular changes induced by a history of alcohol dependence. In search of mechanisms underlying persistent rather than acute dependence-induced neuroadaptations, we studied the role of DNA methylation regulating medial prefrontal cortex (mPFC) gene expression and alcohol-related behaviors in rats 3 weeks into abstinence following alcohol dependence. Postdependent rats showed escalated alcohol intake, which was associated with increased DNA methylation as well as decreased expression of genes encoding synaptic proteins involved in neurotransmitter release in the mPFC. Infusion of the DNA methyltransferase inhibitor RG108 prevented both escalation of alcohol consumption and dependence-induced downregulation of 4 of the 7 transcripts modified in postdependent rats. Specifically, RG108 treatment directly reversed both downregulation of synaptotagmin 2 (Syt2) gene expression and hypermethylation on CpG#5 of its first exon. Lentiviral inhibition of Syt2 expression in the mPFC increased aversion-resistant alcohol drinking, supporting a mechanistic role of Syt2 in compulsive-like behavior. Our findings identified a functional role of DNA methylation in alcohol dependence-like behavioral phenotypes and a candidate gene network that may mediate its effects. Together, these data provide novel evidence for DNA methyltransferases as potential therapeutic targets in alcoholism.
DOI:10.1371/journal.pone.0179469URLPMID:28614398 [本文引用: 2]

Recent studies have indicated that DNA methylation plays an important role in the development of alcohol abuse. 5-Aza-2'-deoxycytidine (5-Aza-dc), an inhibitor of DNA methyltransferases, was FDA approved for myelodysplastic syndrome treatment. However, it is unclear whether 5-Aza-dc is involved in alcohol abuse. In this study, using a chronic alcohol exposure model in rats, 5-Aza-dc was injected into the medial prefrontal cortex (mPFC). Alcohol-drinking behavior and the anxiety related behavior were evaluated by two-bottle choice and open field test. We found that 5-Aza-dc injection into the mPFC significantly decreased alcohol consumption and alcohol preference in alcohol-exposure rats, corresponding to the reduced blood alcohol levels. Although 5-Aza-dc potentiated the anxiety-like behavior of alcohol-exposure rats, it had no effect on the locomotor activity. Moreover, both of the mRNA and protein levels of DNA Methyltransferase 3A (DNMT3A) and DNMT3B in the mPFC were upregulated after 35 days of alcohol exposure and this upregulation could be reversed by 5-Aza-dc treatment. Additionally, 5-Aza-dc reversed the alcohol-induced downregulation of neurotrophin-3 (Ntf3), correspondingly the expression of its receptor-TrkC was reduced. These findings identified a functional role of 5-Aza-dc in alcohol-related behavioral phenotypes and one of the potential target genes, Ntf3. We also provide novel evidence for DNA methyltransferases as potential therapeutic targets in alcohol abuse.
DOI:10.1016/s0091-3057(00)00360-9URLPMID:11124385 [本文引用: 1]

The responses of mice to low doses of acutely and chronically administered ethanol (2.0 g/kg) and diazepam (2.0 mg/kg) were observed in activity cages, the open field and the elevated plus-maze. After prolonged administration, ethanol significantly increased locomotion in the activity cages and the plus-maze. In the open field, an increase was only observed in the tests performed after 7 and 14 days of treatment. Ethanol increased the open-arm time in the plus-maze in all the tests, including after acute administration, suggesting an anxiolytic effect. Diazepam induced an anxiolytic effect after 14 days of daily injections but had no stimulant effect on locomotion. Moreover, after prolonged administration sensitization to the anxiolytic, but not to the stimulant effect, was observed. In short, the present paper's data support the hypothesis that the stimulant and anxiolytic effects of ethanol are probably being mediated by distinct mechanisms. Furthermore, these data support the hypothesis that drugs that lead to abusive use, such as ethanol, may act both as positive and negative reinforcement.
DOI:10.1016/j.alcohol.2014.07.005URL [本文引用: 1]

Environmental complexity (EC) is a powerful, stimulating paradigm that engages animals through a variety of sensory and motor pathways. Exposure to EC (30 days) following 12 days of wheel running preserves hippocampal neuroplasticity in male rats neonatally. exposed to alcohol during the third-trimester equivalent (binge-like exposure on postnatal days [PD] 4-9). The current experiment investigates the importance of various components of EC (physical activity, exploration, social interaction, novelty) and examines whether neonatal alcohol exposure affects how male rats interact with their environment and other male rats. Male pups were assigned to I of 3 neonatal conditions from PD 4-9: suckle control (SC), sham-intubated (SI), or alcohol-exposed (AE, 5.25 g/kg/day). From PD 30-42 animals were housed with 24-h access to a voluntary running wheel. The animals were then placed in EC from PD 42-72 (9 animals/cage, counterbalanced by neonatal condition). During EC, the animals were filmed for five 30-min sessions (PD 42, 48, 56, 64, 68). For the first experiment, the videos were coded for distance traveled in the cage, overall locomotor activity, time spent near other animals, and interaction with toys. For the second experiment, the videos were analyzed for wrestling, mounting, boxing, grooming, sniffing, and crawling over/under. AE animals were found to be less active and exploratory and engaged in fewer mounting behaviors compared to control animals. Results suggest that after exposure to wheel running, AE animals still have deficits in activity and social behaviors while housed in EC compared to control animals with the same experience. (C) 2014 Elsevier Inc.
DOI:10.1016/j.addbeh.2016.02.006URLPMID:26894561 [本文引用: 1]

Social anxiety disorder more than quadruples the risk of developing an alcohol use disorder, yet it is inconsistently linked to drinking frequency. Inconsistent findings may be at least partially due to lack of attention to drinking context - it may be that socially anxious individuals are especially vulnerable to drinking more often in specific contexts that increase their risk for alcohol-related problems. For instance, socially anxious persons may drink more often while alone, before social situations for &quot;liquid courage&quot; and/or after social situations to manage negative thoughts about their performance. Among current (past-month) drinkers (N=776), social anxiety was significantly, positively related to solitary drinking frequency and was negatively related to social drinking frequency. Social anxiety was indirectly (via solitary drinking frequency) related to greater past-month drinking frequency and more drinking-related problems. Social anxiety was also indirectly (via social drinking frequency) negatively related to past-month drinking frequency and drinking-related problems. Findings suggest that socially anxious persons may be vulnerable to more frequent drinking in particular contexts (in this case alone) and that this context-specific drinking may play an important role in drinking problems among these high-risk individuals.
DOI:10.1016/j.alcohol.2017.04.002URLPMID:28847379 [本文引用: 1]

Previous studies suggested that exposure of Sardinian alcohol-preferring (sP) rats to daily drinking sessions of 1?h, during the dark phase of the light/dark cycle, with multiple alcohol concentrations, and unpredictable access to alcohol, resulted in exceptionally high intakes of alcohol when the drinking session occurred over the last hours of the dark phase. Additionally, higher levels of anxiety-related behaviors were observed at the 12th, rather than 1st, hour of the dark phase, suggesting that uncertainty of time of alcohol access and expectation of alcohol availability produced an emotional &quot;distress&quot;. The present study was designed to provide pharmacological support to the hypothesis that high alcohol intake under this drinking procedure is secondary to exacerbation of the anxiety-like state of sP rats. To this end, sP rats were initially exposed to daily 1-h drinking sessions during the dark phase and with multiple alcohol concentrations (0%, 10%, 20%, and 30%; v/v); time of alcohol exposure was changed each day and was unpredictable to rats. Rats were then treated acutely with non-sedative doses of diazepam (0, 1, 2, and 3?mg/kg; intraperitoneally [i.p.]) before two drinking sessions occurring at the 1st and 12th hour of the dark phase, respectively. Treatment with diazepam was ineffective at the 1st hour; conversely, it selectively reduced alcohol intake (up to 50% at the dose of 3?mg/kg) at the 12th hour. The preferential effectiveness of diazepam in reducing alcohol intake when the drinking session occurred at the 12th hour of the dark phase is consistent with the hypothesis that uncertainty of time of alcohol access and expectation of alcohol availability generated an emotional &quot;distress&quot; that rats counterbalanced with high alcohol drinking; the results of the present study are interpreted as the anxiolytic effects of diazepam substituting for those of alcohol, resulting in the observed reduction in alcohol intake.
DOI:10.1080/10826084.2019.1650773URLPMID:31429362 [本文引用: 1]

Background: Alexithymia is a personality trait associated with emotion regulation difficulties. Up to 67% of alcohol-dependent patients in treatment have alexithymia. Objectives: The objective of this study was to investigate the direct and indirect effects of alexithymia, negative mood (stress, anxiety, and depression) and alcohol craving on alcohol dependence severity. Methods: Three hundred and fifty-five outpatients (mean age = 38.70, SD = 11.00, 244 males, range 18-71?years) undergoing Cognitive-Behavioral Therapy for alcohol dependence completed the Toronto Alexithymia Scale (TAS-20), Depression Anxiety Stress Scales (DASS-21), Obsessive Compulsive Drinking Scale (OCDS), and Alcohol Use Disorders Identification Test (AUDIT) prior to the first treatment session. Results: Alexithymia had an indirect effect on alcohol dependence severity, via both negative mood and alcohol craving (b?=?0.03, seb?=?0.008, 95% CI: 0.02-0.05). An indirect effect of negative mood on alcohol dependence via alcohol craving was also observed (b?=?0.12, seb?=?0.03, 95% CI: 0.07-0.16). Conclusions/importance: Alexithymia worked through negative mood and alcohol craving leading to increased alcohol dependence severity, indicating that craving had an indirect effect on the relationship between alexithymia and alcohol dependence severity. Targeting alcohol craving and negative mood for alcohol-dependent patients with alexithymia seems warranted.
DOI:10.1111/adb.12414URLPMID:27353220 [本文引用: 1]

Based on the knowledge that alcohol misuse causes a multitude of diseases and increased mortality, this systematic review examines whether a reduction of the individual alcohol consumption can contribute to a minimization of health risks within a harm reduction approach. In fact, the reviewed 63 studies indicate that interventions aiming at alcohol reduction (including total abstinence as one possible therapeutic aim) indeed resulted in or were associated with positive effects in harmful, hazardous or alcohol-dependent drinkers. Major benefits were observed for reducing alcohol-associated injuries, recovery of ventricular heart function in alcoholic cardiomyopathy, blood pressure lowering, normalization of biochemical parameter, body weight reduction, histological improvement in pre-cirrhotic alcohol-related liver disease and slowed progression of an already existing alcohol-attributable liver fibrosis. Furthermore, reduced withdrawal symptoms, prevalence of psychiatric episodes and duration of in-patient hospital days, improvement of anxiety and depression symptoms, self-confidence, physical and mental quality of life, fewer alcohol-related adverse consequences as well as lower psychosocial stress levels and better social functioning can result from reduced alcohol intake. The reviewed literature demonstrated remarkable socioeconomic cost benefits in areas such as the medical health-care system or workforce productivity. Individuals with heightened vulnerability further benefit significantly from alcohol reduction (e.g. hypertension, hepatitis C, psychiatric co-morbidities, pregnancy, but also among adolescents and young adults). Concluding, the reviewed studies strongly support and emphasize the importance and benefits of early initial screening for problematic alcohol use followed by brief and other interventions in first contact medical health-care facilities to reduce alcohol intake.
.
DOI:10.1007/s00702-005-0413-2URLPMID:16463117 [本文引用: 1]

DNA methyltransferases (DNMTs) are involved within the epigenetic control of DNA methylation processes. Recently, it has been shown that the genomic DNA methylation in patients with alcoholism is increased. In the present controlled study we observed a significant decrease of mRNA expression of DNMT-3a and DNMT-3b when comparing alcoholic patients (n = 59) with healthy controls (n = 66): DNMT-3a (t = -2.38, p = 0.019), DNMT-3b (t = -2.65, p = 0.008). No significant differences were seen for DNMT-1 and Mbd-2 (Methyl-CpG-Binding-Domain protein 2) expression. Additionally, we observed a significant negative correlation between DNMT-3b expression and the blood alcohol concentration (r = -0.45, p = 0.003) which might explain the decrease of DNMT-3b mRNA expression in alcoholic patients. Using a multivariate model we observed that the increase (10%) of genomic DNA methylation in patients with alcoholism was significantly associated with their lowered DNMT-3b mRNA expression (multiple linear regression, p = 0.014). Since methylation of DNA is an important epigenetic factor in regulation of gene expression these findings may have important implications for a possible subsequent derangement of epigenetic control in these patients.
.
DOI:10.1007/s00702-004-0232-xURLPMID:15565495 [本文引用: 1]

Higher plasma homocysteine concentrations can influence genomic DNA methylation in peripheral blood cells. In the present controlled study we observed a significant increase (10%) of genomic DNA methylation in patients with alcoholism (t = -3.16, df = 158, p = 0.002) which was significantly associated with their elevated homocysteine levels (multiple linear regression, p &lt; 0.001). Since methylation of DNA is an important epigenetic factor in regulation of gene expression these findings may have important implications for a possible subsequent derangement of epigenetic control these patients.
DOI:10.1016/j.compbiomed.2014.12.003URLPMID:25555412 [本文引用: 1]

Alcoholism has always been a major public health concern in Taiwan, especially in the aboriginal communities. Emerging evidence supports the association between DNA methylation and alcoholism, though very few studies have examined the effect of chronic alcohol consumption on the epignome. Since 1986, we have been following up on the mental health conditions of four major aboriginal peoples of Taiwan. The 993 aboriginal people who underwent the phase 1 (1986) clinical interviews were followed up through phase 2 (1990-1992), and phase 3 (2003-2009). Selected individuals for the current study included 10 males from the phase 1 normal cohort who remained normal at phase 2 and became dependent on alcohol by phase 3 and 10 control subjects who have not had any drinking problems throughout the study. We profiled the DNA methylation changes in the blood samples collected at phases 2 and 3. Enrichment analyses have identified several biological processes related to immune system responses and aging in the control group. In contrast, differentially methylated genes in the case group were mostly associated with susceptibility to infections, as well as pathways related to muscular contraction and neural degeneration. The methylation levels of six genes were found to correlate with alcohol consumption. These include genes involved in neurogenesis (NPDC1) and inflammation (HERC5), as well as alcoholism-associated genes ADCY9, CKM, and PHOX2A. Given the limited sample size, our approach uncovered genes and disease pathways associated with chronic alcohol consumption at the epigenetic level. The results offer a preliminary methylome map that enhances our understanding of alcohol-induced damages and offers new targets for alcohol injury research.
DOI:10.1242/dev.069997URLPMID:22318233 [本文引用: 1]

Sensory neurons possess the central and peripheral branches and they form unique spinal neural circuits with motoneurons during development. Peripheral branches of sensory axons fasciculate with the motor axons that extend toward the peripheral muscles from the central nervous system (CNS), whereas the central branches of proprioceptive sensory neurons directly innervate motoneurons. Although anatomically well documented, the molecular mechanism underlying sensory-motor interaction during neural circuit formation is not fully understood. To investigate the role of motoneuron on sensory neuron development, we analyzed sensory neuron phenotypes in the dorsal root ganglia (DRG) of Olig2 knockout (KO) mouse embryos, which lack motoneurons. We found an increased number of apoptotic cells in the DRG of Olig2 KO embryos at embryonic day (E) 10.5. Furthermore, abnormal axonal projections of sensory neurons were observed in both the peripheral branches at E10.5 and central branches at E15.5. To understand the motoneuron-derived factor that regulates sensory neuron development, we focused on neurotrophin 3 (Ntf3; NT-3), because Ntf3 and its receptors (Trk) are strongly expressed in motoneurons and sensory neurons, respectively. The significance of motoneuron-derived Ntf3 was analyzed using Ntf3 conditional knockout (cKO) embryos, in which we observed increased apoptosis and abnormal projection of the central branch innervating motoneuron, the phenotypes being apparently comparable with that of Olig2 KO embryos. Taken together, we show that the motoneuron is a functional source of Ntf3 and motoneuron-derived Ntf3 is an essential pre-target neurotrophin for survival and axonal projection of sensory neurons.
.
DOI:10.1523/JNEUROSCI.4024-15.2016URLPMID:27122038 [本文引用: 1]

Neurotrophin-3 (NT-3) is a secreted neurotrophic factor that binds neurotrophin receptor tyrosine kinase C (TrkC), which in turn binds to presynaptic protein tyrosine phosphatase σ (PTPσ) to govern excitatory synapse development. However, whether and how NT-3 cooperates with the TrkC-PTPσ synaptic adhesion pathway and TrkC-mediated intracellular signaling pathways in rat cultured neurons has remained unclear. Here, we report that NT-3 enhances TrkC binding affinity for PTPσ. Strikingly, NT-3 treatment bidirectionally regulates the synaptogenic activity of TrkC: at concentrations of 10-25 ng/ml, NT-3 further enhanced the increase in synapse density induced by TrkC overexpression, whereas at higher concentrations, NT-3 abrogated TrkC-induced increases in synapse density. Semiquantitative immunoblotting and optogenetics-based imaging showed that 25 ng/ml NT-3 or light stimulation at a power that produced a comparable level of NT-3 (6.25 μW) activated only extracellular signal-regulated kinase (ERK) and Akt, whereas 100 ng/ml NT-3 (light intensity, 25 μW) further triggered the activation of phospholipase C-γ1 and CREB independently of PTPσ. Notably, disruption of TrkC intracellular signaling pathways, extracellular ligand binding, or kinase activity by point mutations compromised TrkC-induced increases in synapse density. Furthermore, only sparse, but not global, TrkC knock-down in cultured rat neurons significantly decreased synapse density, suggesting that intercellular differences in TrkC expression level are critical for its synapse-promoting action. Together, our data demonstrate that NT-3 is a key factor in excitatory synapse development that may direct higher-order assembly of the TrkC/PTPσ complex and activate distinct intracellular signaling cascades in a concentration-dependent manner to promote competition-based synapse development processes.
DOI:10.1016/s0896-6273(03)00366-0URLPMID:12873381 [本文引用: 1]

Huntington's disease (HD) is caused by polyglutamine expansion (exp) in huntingtin (Htt). The type 1 inositol (1,4,5)-triphosphate receptor (InsP3R1) is an intracellular calcium (Ca2+) release channel that plays an important role in neuronal function. In a yeast two-hybrid screen with the InsP3R1 carboxy terminus, we isolated Htt-associated protein-1A (HAP1A). We show that an InsP3R1-HAP1A-Htt ternary complex is formed in vitro and in vivo. In planar lipid bilayer reconstitution experiments, InsP3R1 activation by InsP3 is sensitized by Httexp, but not by normal Htt. Transfection of full-length Httexp or caspase-resistant Httexp, but not normal Htt, into medium spiny striatal neurons faciliates Ca2+ release in response to threshold concentrations of the selective mGluR1/5 agonist 3,5-DHPG. Our findings identify a novel molecular link between Htt and InsP3R1-mediated neuronal Ca2+ signaling and provide an explanation for the derangement of cytosolic Ca2+ signaling in HD patients and mouse models.
DOI:10.3389/fncel.2018.00381URLPMID:30455632 [本文引用: 1]

Huntington's disease (HD) is a hereditary neurodegenerative disease that is caused by polyglutamine expansion within the huntingtin (HTT) gene. One of the cellular activities that is dysregulated in HD is store-operated calcium entry (SOCE), a process by which Ca2+ release from the endoplasmic reticulum (ER) induces Ca2+ influx from the extracellular space. HTT-associated protein-1 (HAP1) is a binding partner of HTT. The aim of the present study was to examine the role of HAP1A protein in regulating SOCE in YAC128 mice, a transgenic model of HD. After Ca2+ depletion from the ER by the activation of inositol-(1,4,5)triphosphate receptor type 1 (IP3R1), we detected an increase in the activity of SOC channels when HAP1 protein isoform HAP1A was overexpressed in medium spiny neurons (MSNs) from YAC128 mice. A decrease in the activity of SOC channels in YAC128 MSNs was observed when HAP1 protein was silenced. In YAC128 MSNs that overexpressed HAP1A, an increase in activity of IP3R1 was detected while the ionomycin-sensitive ER Ca2+ pool decreased. 6-Bromo-N-(2-phenylethyl)-2,3,4,9-tetrahydro-1H-carbazol-1-amine hydrochloride (C20H22BrClN2), identified in our previous studies as a SOCE inhibitor, restored the elevation of SOCE in YAC128 MSN cultures that overexpressed HAP1A. The IP3 sponge also restored the elevation of SOCE and increased the release of Ca2+ from the ER in YAC128 MSN cultures that overexpressed HAP1A. The overexpression of HAP1A in the human neuroblastoma cell line SK-N-SH (i.e., a cellular model of HD (SK-N-SH HTT138Q)) led to the appearance of a pool of constitutively active SOC channels and an increase in the expression of STIM2 protein. Our results showed that HAP1A causes the activation of SOC channels in HD models by affecting IP3R1 activity.
DOI:10.1007/s00213-013-3165-yURLPMID:23771439 [本文引用: 1]

Clozapine affects the extracellular signal-regulated kinase 1/2 (ERK1/2) pathway in the brain, which plays an important role in its antipsychotic action. However, previous findings are inconsistent, and related molecular mechanisms require further clarification.
URLPMID:16341272 [本文引用: 1]

The term epigenetic modification denotes reversible traits of gene expression that do not include alterations to the DNA sequence. These epigenetic alterations are responsible for chromatin structure stability, genome integrity, modulation of tissue-specific gene expression, embryonic development, genomic imprinting and X-chromosome inactivation in females. Epigenetic changes include reversible DNA methylation and histone acetylation or methylation. The modification of mammalian genomic DNA includes the methylation at the 5-position of the cytosine (C) residue within cytosine-guanine dinucleotides (CpG), resulting in the formation of 5-methylcytosine (m5C). Regulatory DNA sequences in vertebrates often have little or no methylation. The methylation of mammalian genomic DNA is catalyzed by DNA methyltransferases (DNMTs), which play a special role in the initiation of chromatin remodeling and gene expression regulation. The mammalian DNMTs are DNMT1, DNMT3A and DNMT3B, which together with accessory proteins, like DNMT3L, are responsible for methylation pattern acquisition during gametogenesis, embryogenesis and somatic tissue development. Reversible epigenetic alterations lead to selective utilization of genome information through the activation or inactivation of transcription of functional genes during gametogenesis, embryogenesis and cell differentiation. Recently, several disparate isoforms of DNMT1 were identified in human somatic and female and male germ cells. Recent advances in the investigation of DNMT function in epigenetic DNA changes have formed the basis of the understanding of various disorder etiopathogeneses, and as a result, have facilitated and enabled new therapies with respect to these diseases.
URLPMID:24846989 [本文引用: 1]

DNA methylation plays important roles in cell differentiation, embryonic development, host adaptations to environmental factors, and pathogenesis through regulation of gene transcription and imprinting, X-inactivation, and defense of foreign genetic material invasion, is currently one of the hottest research fields on epigenetics. In the past few years, a number of important findings on DNA methylation have been achieved. These findings include discovery of TETs-catalyzed cytosine hydroxymethylation and its functions in the early embryonic development; the relationship between active and passive DNA demethylation; establishment and maintenance of DNA methylation patterns and their associations with histone modifications, chromatin configuration, polycomb group proteins and non-coding RNA bindings. DNA methylation has become a new potential biomarker and therapy target.
URLPMID:24846989 [本文引用: 1]

DNA methylation plays important roles in cell differentiation, embryonic development, host adaptations to environmental factors, and pathogenesis through regulation of gene transcription and imprinting, X-inactivation, and defense of foreign genetic material invasion, is currently one of the hottest research fields on epigenetics. In the past few years, a number of important findings on DNA methylation have been achieved. These findings include discovery of TETs-catalyzed cytosine hydroxymethylation and its functions in the early embryonic development; the relationship between active and passive DNA demethylation; establishment and maintenance of DNA methylation patterns and their associations with histone modifications, chromatin configuration, polycomb group proteins and non-coding RNA bindings. DNA methylation has become a new potential biomarker and therapy target.
DOI:10.1177/1073858415579635URLPMID:25832671 [本文引用: 1]

The establishment of synaptic plasticity and long-term memory requires lasting cellular and molecular modifications that, as a whole, must endure despite the rapid turnover of their constituent parts. Such a molecular feat must be mediated by a stable, self-perpetuating, cellular information storage mechanism. DNA methylation, being the archetypal cellular information storage mechanism, has been heavily implicated as being necessary for stable activity-dependent transcriptional alterations within the CNS. This review details the foundational discoveries from both gene-targeted and whole-genome sequencing studies that have brought DNA methylation to our attention as a chief regulator of activity- and experience-dependent transcriptional alterations within the CNS. We present a hypothetical framework to resolve disparate experimental findings regarding distinct manipulations of DNA methylation and their effect on memory, taking into account the unique impact activity-dependent alterations in DNA methylation potentially have on both memory-promoting and memory-suppressing gene expression. And last, we discuss potential avenues for future inquiry into the role of DNA methylation during remote memory formation.
DOI:10.1038/nn.2514URLPMID:20228804 [本文引用: 1]

Dnmt1 and Dnmt3a are important DNA methyltransferases that are expressed in postmitotic neurons, but their function in the CNS is unclear. We generated conditional mutant mice that lack Dnmt1, Dnmt3a or both exclusively in forebrain excitatory neurons and found that only double knockout (DKO) mice showed abnormal long-term plasticity in the hippocampal CA1 region together with deficits in learning and memory. Although we found no neuronal loss, hippocampal neurons in DKO mice were smaller than in the wild type; furthermore, DKO neurons showed deregulated expression of genes, including the class I MHC genes and Stat1, that are known to contribute to synaptic plasticity. In addition, we observed a significant decrease in DNA methylation in DKO neurons. We conclude that Dnmt1 and Dnmt3a are required for synaptic plasticity, learning and memory through their overlapping roles in maintaining DNA methylation and modulating neuronal gene expression in adult CNS neurons.
DOI:10.1038/nn.2619URLPMID:20729844 [本文引用: 1]

Despite abundant expression of DNA methyltransferases (Dnmts) in brain, the regulation and behavioral role of DNA methylation remain poorly understood. We found that Dnmt3a expression was regulated in mouse nucleus accumbens (NAc) by chronic cocaine use and chronic social defeat stress. Moreover, NAc-specific manipulations that block DNA methylation potentiated cocaine reward and exerted antidepressant-like effects, whereas NAc-specific Dnmt3a overexpression attenuated cocaine reward and was pro-depressant. On a cellular level, we found that chronic cocaine use selectively increased thin dendritic spines on NAc neurons and that DNA methylation was both necessary and sufficient to mediate these effects. These data establish the importance of Dnmt3a in the NAc in regulating cellular and behavioral plasticity to emotional stimuli.
DOI:10.1111/adb.12047URL [本文引用: 1]

We have investigated the expression of chromatin-regulating genes in the prefrontal cortex and in the shell subdivision of the nucleus accumbens during protracted withdrawal in mice with increased ethanol drinking after chronic intermittent ethanol (CIE) vapor exposure and in mice with a history of non-dependent drinking. We observed that the methyl-CpG binding protein 2 (MeCP2) was one of the few chromatin-regulating genes to be differentially regulated by a history of dependence. As MeCP2 has the potential of acting as a broad gene regulator, we investigated sensitivity to ethanol and ethanol drinking in MeCP2(308/Y) mice, which harbor a truncated MeCP2 allele but have a milder phenotype than MeCP2 null mice. We observed that MeCP2(308/Y) mice were more sensitive to ethanol's stimulatory and sedative effects than wild-type (WT) mice, drank less ethanol in a limited access 2 bottle choice paradigm and did not show increased drinking after induction of dependence with exposure to CIE vapors. Alcohol metabolism did not differ in MeCP2(308/Y) and WT mice. Additionally, MeCP2(308/Y) mice did not differ from WT mice in ethanol preference in a 24-hour paradigm nor in their intake of graded solutions of saccharin or quinine, suggesting that the MeCP2(308/Y) mutation did not alter taste function. Lastly, using the Gene Set Enrichment Analysis algorithm, we found a significant overlap in the genes regulated by alcohol and by MeCP2. Together, these results suggest that MeCP2 contributes to the regulation of ethanol sensitivity and drinking.
DOI:10.1111/ajad.12465URLPMID:27759945 [本文引用: 1]

Risk for alcohol use disorders (AUDs) is influenced by gene-environment interactions. Environmental factors can affect gene expression through epigenetic mechanisms such as DNA methylation. This review outlines the findings regarding the association of DNA methylation and AUDs.
