牛煦然, 尹树明, 陈曦, 邵婷婷, 李大力
, 上海市调控生物学重点实验室,华东师范大学生命科学学院,上海 200241
Gene editing technology and its recent progress in disease therapy Xuran Niu, Shuming Yin, Xi Chen, Tingting Shao, Dali Li
, Shanghai Key Laboratory of Regulatory Biology, School of Life Sciences, East China Normal University, Shanghai 200241, China
通讯作者: 李大力,博士,研究员,研究方向:基因编辑。E-mail: dlli@bio.ecnu.edu.cn 编委: 谷峰
收稿日期: 2019-04-16
修回日期: 2019-05-29
网络出版日期: 2019-07-20
基金资助: 国家自然科学基金项目资助 .81670470 国家自然科学基金项目资助 .81873685
Received: 2019-04-16
Revised: 2019-05-29
Online: 2019-07-20
Fund supported: Supported by the National Natural Science Foundation of China .81670470 Supported by the National Natural Science Foundation of China .81873685
作者简介 About authors
牛煦然,本科在读,专业方向:生物科学E-mail:574569231@qq.com。
摘要 基因编辑是一种基于人工核酸酶的遗传操作技术,能精确地对DNA或RNA进行高效改造。基因编辑除了在基础研究、生物育种和药物筛选等方面展现了巨大前景之外,在疾病治疗(特别是基因遗传病)领域的前景与进展尤为引人注目。本文在介绍基因编辑技术的发展及其在疾病治疗中不同策略的基础上,重点围绕遗传疾病的基因治疗研究,综述了基因编辑技术(包括单碱基编辑和表观调控等技术)在血液系统、肝脏、肌肉和神经系统等疾病治疗的研究进展,并对基因编辑治疗疾病的未来发展进行了展望。 关键词: 基因编辑 ;
基因治疗 ;
遗传病 Abstract Gene editing is a genetic manipulation technology which utilizes bacterial nucleases to accurately and efficiently modify DNA or RNA. Gene editing has broad applications in basic research, breeding, and drug screening, and it is gaining validity and applicability to the therapy of many diseases especially genetic-based disease. In this review, we summarize the development of gene editing technology, its different strategies and applications in the treatment of disease, and the research of gene editing therapy for genetic diseases (including base editor and epigenetic regulation) in the treatment of disorders and diseases of the blood system, liver, muscle and nervous system. Finally, we discuss the future development prospects of gene editing therapy. Keywords: gene editing ;
gene therapy ;
genetic diseases PDF (713KB) 元数据 多维度评价 相关文章 导出 EndNote |
Ris |
Bibtex 收藏本文 本文引用格式 牛煦然, 尹树明, 陈曦, 邵婷婷, 李大力. 基因编辑技术及其在疾病治疗中的研究进展[J].
遗传 , 2019, 41(7): 582-598 doi:10.16288/j.yczz.19-102
Xuran Niu, Shuming Yin, Xi Chen, Tingting Shao, Dali Li.
Gene editing technology and its recent progress in disease therapy [J].
Hereditas(Beijing) , 2019, 41(7): 582-598 doi:10.16288/j.yczz.19-102
1953年,Watson与Crick提出的DNA双螺旋结构拉开了现代分子生物学的序幕,也打开了人类从DNA分子层面深入认识遗传疾病的大门。随着测序和分析技术的不断发展,目前已知的人类遗传病超过6000种,其中3200种为单基因遗传病
[1 ] 。虽然人们对遗传疾病的了解和诊断有了长足的发展,但是对于这些疾病的治疗仍然举步维艰:超过95%的遗传疾病都没有有效的治疗方法,更谈不上治愈
[1 ] 。相对于传统的小分子或者酶替代疗法需要不断给药持续治疗,基因治疗有望一次治疗达到长期疗效,是遗传疾病治愈的希望。近几年,基因治疗领域蓬勃发展,目前已经有Luxturna、Glybera Strimvelis等3个治疗罕见病的药物获批上市
[2 ,3 ,4 ] ,预计在近年将有更多的药物获得批准,应用于临床。然而,传统的基因治疗方法存在着潜在的致瘤风险,治疗的时效性还有待更长时间的考验。基因编辑技术的出现,将为基因治疗增添新的升级工具,弥补传统基因疗法的不足,有望真正实现一次治疗终身治愈的目标,成为目前研究的热点。在未来,采用基因编辑技术进行基因治疗,也必将会在生物医学与临床治疗领域带来更为深刻的变革。本文即从基因编辑技术的发展沿革入手,对近年来基因编辑在疾病治疗中的研究进展进行总结。
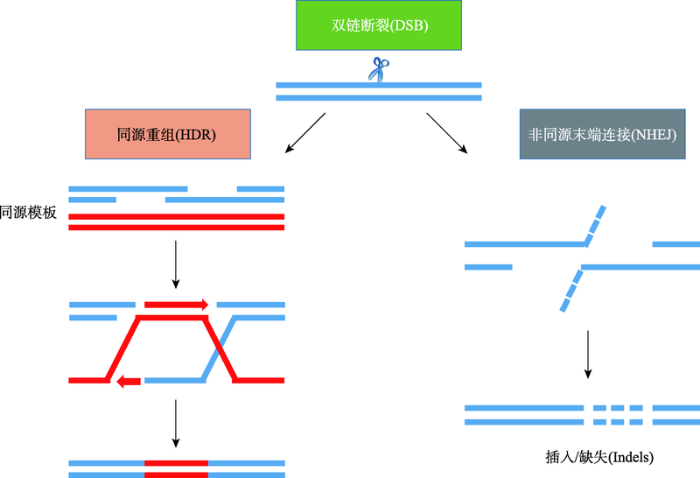
1 基因编辑技术的发展 近年来,基于多种高效核酸酶的发现,基因编辑技术得到了快速发展和广泛应用。其中以人工核酸内切酶介导的基因组编辑技术发展最为迅速
[5 ] ,主要包括4种:大范围核酸酶技术(meganuclease)、锌指核酸酶技术(zinc finger nuclease, ZFN)、转录激活因子样效应物核酸酶技术(transcription activator-like effector nuclease, TALEN)和成簇的规律间隔的短回文重复序列(clustered regularly interspaced short palindromic repeats, CRISPR)/CRISPR相关蛋白(CRISPR-associated proteins, Cas)系统。通过人工核酸酶可以精确靶向诱导双链DNA断裂(double strand break, DSB),在DSB产生之后,细胞内将启动两种主要的修复机制
[6 ] :在通常情况下,细胞主要通过非同源末端连接(non-homologous end joining, NHEJ)的方式进行修复,NHEJ可以在DSB位点有效的产生不同长度片段的插入或缺失(insertions and/or deletions, indel),通常导致基因功能失活;在存在同源序列的DNA模板的情况下,细胞还会采取同源重组(homology-directed repair, HDR)的方式进行修复,可以实现特定位点的精确插入、缺失或者碱基置换
[7 ] (
图1 )。然而,相对于ZFN技术、TALEN技术和CRISPR/Cas9技术,2016年诞生的单碱基编辑技术(base editor, BE),可以在不需要同源模板的情况下实现精确的碱基替换,同时不会产生双链断裂,具有更安全、高效的特点,应用潜力大
[8 ,9 ,10 ] 。与此同时,随着科学家对基因编辑工具认识的加深和不断改造,基因编辑工具的应用领域也从基因组编辑发展到了表观基因组编辑和转录组调控领域,开发出一系列表观基因组编辑工具和转录调控相关工具。这些工具展示了基因编辑技术的超强可塑性,增加了更多的应用场景,也在基因治疗中展现了优势。
图1 新窗口打开 |
下载原图ZIP |
生成PPT 图1基因编辑原理 Fig. 1Schematic of gene editing 1.1 大范围核酸酶技术 大范围核酸酶是早期的基因编辑工具,为一种具有较大切割位点(通常为12~45 bp)的序列特异性核酸内切酶
[11 ] 。在自然界中,大范围核酸酶以归巢内切核酸酶(homing endonuclease, HE)代表
[12 ] 。目前,大范围核酸酶已经用于在细胞系和植物中诱导靶序列附近产生DSB,以达到在基因组中敲除内源基因或敲入外源基因的目的,或者是矫正与单基因疾病相关的突变
[13 ] 。在诸多核酸酶之中,它的编辑精度更高,自身编码基因也比较小(只有1 kb左右),能容易包装入AAV载体中。2018年,
Nature Biotechnology 杂志发表了一项用大范围核酸内切酶I-CreI 靶向敲除非人类灵长类动物的
PCSK9 基因,成功降低了体内胆固醇的水平的研究
[14 ] 。但是,对大范围核酸酶诱导重组的应用长期以来受到天然大范围核酸酶种类的限制,以及大部分大范围核酸酶很难在人基因组上找到合适的位点,这都使得大范围核酸酶的广泛应用较为困难。
1.2 锌指核酸酶技术 锌指蛋白最早在1984年由科学家们从非洲爪蟾的转录因子中发现,后来逐步发展为锌指核酸酶技术
[15 ] 。ZFN主要包括用于识别和结合特定的DNA序列重复的锌指蛋白(zinc finger protein, ZFP)和来源于海床黄杆菌(
Flavobacterium okeanokoites )的一种可以通过二聚体化非特异地切割DNA的核酸内切酶
Fok I
[16 ,17 ] 。利用一对串联的锌指结合特异DNA,将
Fok I的两个亚基带到特定基因组位点,对DNA进行切割产生DSB。识别特定DNA的锌指序列需要通过文库筛选来确定,非常耗时耗力,也需要丰富的经验,一定程度上限制了其发展。此外,因为锌指核酸酶存在上下游DNA序列的依赖效应
[18 ,19 ,20 ] ,这进一步加大了筛选的难度,也由此可能产生更多的脱靶效应,引发细胞毒性,故而对ZFN的应用推广也产生了一定的阻碍。不可否认,ZFN是最早开发的开创性技术,也是往临床推进最为深入的技术,为后续基因编辑技术的快速发展奠定了基础。
1.3 转录激活子样效应物技术 1989年,科学家们从植物病原体黄单胞菌属(
Xanthomonas )中克隆出一种avrBs3蛋白,该蛋白质可以结合植物宿主基因组并激活转录
[21 ] 。2007年,这种特殊的分泌蛋白质被命名为转录激活因子样效应物TALE
[22 ,23 ] 。TALE如何结合DNA的机制在2009年逐步被科学家们成功解析
[24 ,25 ] 。2012年,TALEN技术正式出现,ZFN技术随之被渐渐取代
[26 ] 。与锌指核酸酶技术相似,TALEN也由两部分组成,一部分是TALE蛋白所在的DNA的特异性识别和结合区域;另一部分是与ZFN相同的
Fok I核酸酶
[27 ,28 ] 。相比于ZFN而言,TALEN的设计比较简单,只需要设计1个TALE分子就可以识别一个碱基,理论上可以靶向基因组任何区域
[29 ] 。TALEN技术的出现,在一定程度上解决了ZFN技术存在的脱靶问题,具有设计简单,特异性和活性更高的优点,成功地应用在小鼠、大鼠、果蝇和拟南芥等模式生物上,成为基因功能研究和基因治疗研究中有力的工具
[30 ,31 ,32 ,33 ] 。
1.4 CRISPR/Cas技术 1987年,Nakata研究组在分析大肠杆菌时偶然地发现在其基因组上的
iap 基因存在含有29个碱基的高度同源序列重复性,且这些重复序列被含32个碱基的序列间隔开
[34 ] 。此后,类似的重复序列也不断在其他微生物中被发现。2002年。这种重复序列被正式命名为CRISPR,并且在重复序列附近发现了一系列保守的CRISPR相关基因(Cas)
[35 ] 。2012年,Gasiunas等
[36 ] 发现,使用单一引导RNA(single-guide RNA, sgRNA)能和天然的crRNA和tracrRNA一样地有效介导Cas9靶向切割DNA片段,并逐步将CRISPR/Cas系统优化为更容易进行操作的Cas9/ sgRNA系统。
最早发现具有基因编辑功能和最广泛使用的CRISPR/Cas9是一种来源于酿脓链球菌(
Streptococcus pyogenes )的Cas9(SpCas9)蛋白
[37 ] ,由RuvC和HNH这两个重要结构域组成。RuvC结构域负责切割靶向链,而HNH负责切割与sgRNA互补配对的非靶向链,切割位点通常位于PAM (protospacer- adjacent motifs)序列(通常为NGG)上游3 nt处。相比于传统的ZFN和 TALEN技术,CRISPR/Cas9技术更为简单,只需要构建针对特定位点的sgRNA,而且效率也比前面几种技术更高,在模型的构建和疾病治疗研究中发挥越来越重要的作用。当然,CRISPR/Cas9系统仍然存在着一定的局限性,这种局限性主要体现在功能发挥时系统对DNA上PAM序列的依赖性以及切割时潜在的脱靶效应
[38 ,39 ,40 ] 。在CRISPR/Cas9系统中,Cas9只在具有PAM序列且可以与sgRNA互补配对的靶序列发生切割。基于这些需要改善的方面,研究人员开创性地发现了一些识别更多样PAM的CRISPR/Cas系统,如:识别NNGRRT(R代表嘌呤碱基)的SaCas9
[41 ] ,识别多A/T的Cas12(Cpf1)系统
[42 ] ,同时也对已有的SpCas9进行结构优化和改造,开发出了以NG为PAM的xCas9
[43 ] 和SpCas9-NG
[44 ] 等。对于脱靶的问题,科学家们根据Cas9结构特点设计出一系列精确性更高的Cas9突变体(如eCas91.0、Cas9-HF1、HypaCas9等)
[45 ,46 ] 。研究证明,这些优化或改造的Cas9可降低50~1500倍的脱靶率,极大地解决了脱靶问题
[47 ] 。
1.5 单碱基编辑技术 单碱基编辑技术(base editor, BE)是一种基于脱氨酶与CRISPR/Cas9系统融合形成的技术。2016年哈佛大学David Liu实验室首次报道,通过将SpCas9与胞嘧啶脱氨酶(cytidine deaminase, CyD, 如APOBEC1)融合,可以在一定的突变窗口内实现胞嘧啶(cytosine, C)到胸腺嘧啶(thymine, T)的单碱基转换(cytosine base editor, CBE)
[48 ] 。随后多个实验室也发表了类似的工具,并在这些工具的基础上进行了更为深入的改造与优化。Nishida等
[49 ] 将来源于七鳃鳗的胞嘧啶脱氨酶与CRISPR/Cas9和尿嘧啶糖苷酶抑制蛋白(uracil DNA glycosylase inhibitor, UGI)融合,可在哺乳动物细胞中实现15%~55%靶向突变。中国科学院上海生命科学研究院常兴课题组将人源胞嘧啶脱氨酶融合dCas9的C端(dCas9-AIDx)形成的TAM (targeted AID-mediated mutagenesis)可通过诱变产生局部序列的多样化
[50 ] 。2017年10月底,David Liu实验室在原有BE的基础上实现了腺嘌呤(adenine, A)到鸟嘌呤(guanine, G)的精确转换(adenine base editor, ABE)
[51 ] ,为基因编辑提供了新的工具。相比于CRISPR/Cas9技术,BE技术可以既不引入DNA双链断裂,又不需要重组修复模板,而且其效率远远高于由发生双链断裂引起的同源重组修复方式,对于许多点突变造成的遗传疾病具有很大的应用潜能
[52 ] 。
1.6 表观基因组编辑和转录调控 基因编辑工具的迅速发展使得在天然染色质环境中有针对性地进行表观基因组编辑和转录调控成为可能。目前,表观遗传修饰和转录调控的设计主要基于核酸酶的结合运用,其中以工程化缺陷型核酸酶(dCas9)的结合最为有效,其设计原理在于把各种表观调控效应器融合到dCas9上,利用核酸酶可以靶向识别并结合DNA的特性,从而在特定位点上实现表观基因组的编辑。在常用的表观遗传编辑工具中,dCas9-KRAB
[53 ] 和dCas9-VP64
[54 ,55 ] 分别是用于抑制和激活基因表达的调节因子。此外,dCas9- VPR
[56 ] 和dCas9-TV
[57 ] 是比dCas9-VP64更有效的转录激活剂。这些反式调控结构域和蛋白是通过向启动子区域的dCas9靶向位点阻碍RNA聚合酶的结合或募集内源性转录复合物来发挥作用的。另一方面,Cas9还可以与表观遗传修饰酶来进行融合,直接催化DNA或者组蛋白的表观修饰,调控基因的表达。dCas9-DNMT3A已被证实可使靶向启动子上的DNA瞬时甲基化,从而抑制基因的表达
[58 ] 。相比之下,使用dCas9-TET1则可实现启动子的快速和瞬时DNA去甲基化,诱导靶基因的表达上调
[58 ,59 ] 。此外,通过dCas9-PRDM9和dCas9 -DOT1L可以实现组蛋白稳定的甲基化,以重新启动表观遗传相关沉默基因的表达
[60 ] 。目前也有研究报道通过dCas9- LSD1诱导增强子的组蛋白脱甲基化,可下调靶基因的表达
[61 ] 。此外,通过使用dCas9-P300
[62 ] 和dCas9- HDAC3
[63 ] ,在靶向增强子和启动子区域可实现组蛋白瞬时高效的乙酰化和脱乙酰化,调控基因的表达。各种调控效应器同dCas9的结合极大地扩展了CRISPR在转录调控和表观基因组编辑的应用范围,而相关研究也表明核酸酶的表观基因组编辑具有极高的安全性和特异性,并且没有明显的脱靶效应
[52 ,64 ] ,为体内表观基因修饰和治疗奠定了基础。
图2 新窗口打开 |
下载原图ZIP |
生成PPT 图2不同类型的基因编辑技术 A:大范围核酸酶技术(图中蓝色部分为核酸酶蛋白);B:锌指核酸酶技术,重复的锌指蛋白作为基因组识别原件,
Fok I作为切割原件;C:TALEN技术,TALE重复结构域作为基因组识别原件,
Fok I作为切割原件;D:CRISPR/Cas9技术(蓝色部分为Cas9蛋白,红色部分为Guide RNA); E:单碱基编辑与转录调控系统,利用只具有识别活性的dCas9蛋白融合酶或调控原件,对基因组进行编辑与操作。
Fig. 2Different types of gene editing techniques 2 基因治疗的策略及其载体系统 基因疗法(gene therapeutics)的概念最早由美国科学家Michael Blease于1968年提出。直到1989年,美国的联邦食品与药品管理局(Food and Drug Administration, FDA)才首次同意了将载体导入的基因治疗方法用于晚期恶性黑色素瘤患者,标志着人类基因治疗(gene therapy)临床试验的开启。FDA对基因治疗的定义是“通过各种手段修复缺陷基因,以实现减缓或者治愈疾病目的的技术”。可以看到,基因治疗主要是指将外源正常基因导入靶细胞,以纠正或补偿缺陷和异常基因引起的疾病,从而达到治疗目的。至今已有超过2600多项基因治疗的临床试验获批开展。2012年,欧洲批准了格利贝拉(Glybera)作为第一个上市的基因药物,用于治疗罕见性遗传病——脂蛋白脂肪酶缺乏症(lipoprotein lipase deficiency, LPLD)。虽然该药物最后因为定价太高和需求不足而退出市场,但作为一个标志性的产品,代表着基因药物正式作为疾病治疗的新方法应用于临床。最近几年,基因治疗更是取得蓬勃发展。2016年,基因治疗药物Strimvelis被欧洲药品管理局(European Medicines Agency, EMA)批准,用于治疗儿童腺苷脱氨酶(adenosine deaminase, ADA)缺乏性重度联合免疫缺陷症(ADA-severe combined immunodeficiency, ADA-SCID)。2017年,来自美国Spark公司的具有跨时代意义的眼部罕见病基因治疗产品Luxturna获批上市,这也是FDA批准的首个基因治疗药物。另外,对于血友病、β地中海贫血、杜氏肌营养不良和1型脊髓性肌萎缩症(spinal muscular atrophy type 1, SMA1)等疾病而言,相应的基因治疗药物都在临床实验中取得了令人振奋的治疗效果,有望在近年获批上市,为相应疾病的治疗提供新的选择。
2.1 基因治疗的策略 长期以来,在科研和临床实践中,基因治疗的基本策略主要包括两类:一种是利用较为直接的方法对局部或全身注射含有修复基因片段的病毒或非病毒载体(
in vivo therapy);另一种是将患者的体细胞(如造血干细胞)从体内分离,在体外进行培养并使用携带有正常基因片段的载体修复突变基因,最后通过注射的方法将改良后的细胞回输入患者体内(
ex vivo therapy)以进行疾病的治疗
[65 ] (
图3 )。两种策略在不同组织器官的疾病治疗中各有侧重,为多种疾病的治疗提供了方法学支持。
ex vivo 方法对于目的细胞(如造血干细胞)的遗传改造是在实验室中进行的,即使存在编辑效率较低的可能,但仍然可以通过实验筛选出正确改造的细胞,之后只需要进行扩大培养再回输回患者体内即可。而
in vivo 方法的整个编辑过程都是在体内进行,这就需要编辑方法和工具有较高活性;此外,在
in vivo 方法中,病毒载体注入人体之后,安全性是最关键的指标。关于治疗策略的安全性和有效性等问题将在下文做进一步讨论。
图3 新窗口打开 |
下载原图ZIP |
生成PPT 图3基因治疗策略 Fig.3Strategies of gene therapy 2.2 基因治疗的载体系统 由于基因治疗需要将相应的基因片段递送入人体或动物细胞中,因此想要进行基因治疗,必须要有好的载体系统。所谓载体系统,主要是指递送外源基因片段的运输工具。目前基因治疗过程中用到的载体系统主要分为两类:一类是病毒类载体,另一类是非病毒类的载体。
2.2.1 病毒载体
病毒是一类能对人体细胞及其它动物细胞进行有效感染的微生物。由于其具有良好的基因递送效能,故而被科学家们改造成为进行基因治疗的主要递送工具。按照时间的发展,在基因治疗中使用最广泛的病毒载体包括逆转录病毒(retro virus, RV)和慢病毒(lentivirus, LV))、腺病毒(adeno virus, Adv)以及腺相关病毒(adeno associated virus, AAV)等载体。
逆转录病毒是最早应用于基因治疗的一类载体,是一种RNA病毒,其中以RV和LV最为常用。逆转录病毒可以把目的基因随机整合入宿主基因组,从而长期稳定表达目的基因。但由于逆转录病毒的随机整合特点,容易干扰宿主基因的正常表达,具有潜在的致瘤风险。不过在重度联合免疫缺陷症与地中海贫血疾病的治疗中,逆转录病毒仍然表现出很好的治疗效果,且未报道出相应的副作用
[66 ,67 ] 。
Adv是一类无包膜的双链DNA病毒。在临床和科研中,主要使用的是Adv5型腺病毒载体。与上面所述的逆转录病毒不同,Adv不会将基因片段整合入宿主基因组,属于非整合型的病毒载体。不过,Adv在使用中容易引发较强烈的免疫反应,故而在使用中存在一定的限制
[68 ] 。
AAV是目前应用前景比较广泛的一类基因转运工具。与Adv相同,AAV仍然属于非整合型的载体,递送入细胞的基因片段可以在染色体外稳定表达
[69 ] 。不过,由于其导入基因以游离状态存在,故而在细胞分裂中容易丢失。且其衣壳蛋白对应的不同血清型所导致的免疫原性也可能使AAV介导的治疗效果大打折扣。
2.2.2 非病毒载体
非病毒载体的使用原理是采用人工合成的载体材料的物理化学性质来介导基因的转移。与病毒载体相比较,非病毒载体具有成本低、制备简单、便于大规模生产、安全性高、外源基因长度不受限制等优点,在表达反义寡核苷酸等特殊外源基因片段中,有着传统病毒载体不可替代的作用。正在研究的非病毒载体主要包括脂质体、分子偶联受体、聚合物(聚-L-赖氨酸、聚乙烯亚胺等)、复合载体以及纳米粒子载体等
[70 ] 。常用的基因转移脂质体包括阳离子、中性和阴离子脂质体,其中阳离子脂质体研究的最为广泛;而应用于基因转移的无机纳米粒子主要包括硅、碳纳米管、铁氧化物等,它们主要通过穿过细胞膜将药物或生物分子转运到生物体中而起到治疗疾病的作用
[70 ] 。虽然它们仍存在转染效率较低等问题,但也在基因治疗领域扮演着越来越重要的角色。
3 基因编辑技术在基因治疗中的应用 3.1 基因敲除在基因治疗中的应用 基因敲除(knock out, KO)是基因编辑工具的一个重要功能,可以实现基因组DNA特异片段的删除,进而导致目标基因的失活或激活等。在众多的遗传疾病中,有不少疾病便是与基因的异常表达相关。通过基因编辑工具敲除异常表达的基因或删除突变基因的特定区域,是遗传疾病基因治疗的一大策略。从基因敲除的整体情况上看,基因敲除型基因治疗可以分为3种类型:第一类是基因的完全敲除;第二类是基因部分功能区域的敲除(如增强子区域);第三类是杂合子中显性突变基因的敲除。这三类敲除在不同疾病研究中各有应用,为基因治疗提供了丰富的策略。
基因完全敲除策略在代谢类疾病治疗中最为典型(如家族性高胆固醇血症和I型酪氨酸血症血症),通过完全敲除代谢过程中的负调控蛋白或阻断上流代谢通路,间接地治疗疾病。PCSK9蛋白能促进低密度脂蛋白受体(low-density lipoprotein receptor, LDLR)的降解而提高血浆低密度脂蛋白胆固醇(low-density lipoprotein-cholesterol, LDL-C)的水平,是降低胆固醇水平和治疗心血管疾病的重要靶点。研究表明利用Cas9敲除肝脏中
PCSK9 可使血液中低密度脂蛋白胆固醇水平下降35%~40%,对家族性高胆固醇血症起到了良好的治疗效果
[71 ] 。酪氨酸血症是一种因延胡索酰乙酰乙酸水解酶(fuarylacetoacetate hydrolase, Fah)缺乏引起酪氨酸代谢异常,并引起一系列并发症的疾病。4-羟基苯丙酮酸双氧酶(4-hydroxyphenylpyruvate dioxygenase, Hpd)是酪氨酸分解途径中位于上游的代谢酶,可将4-羟基苯丙酮酸代谢为尿黑酸,而尿黑酸可进一步代谢生成延胡索酰乙酰乙酸。研究者们利用Cas9敲除肝细胞中的
Hpd ,从而降低进入酪氨酸分解下游代谢途径的尿黑酸,可将严重的I型酪氨酸血症转化为良性的III型酪氨酸血症,起到疾病的治疗作用
[72 ] 。
基因部分功能区域敲除策略被经常应用于血液疾病和肌营养不良症(duchenne muscular dystrophy, DMD)的治疗。β地中海贫血是临床上常见的贫血疾病。地中海贫血又称珠蛋白生成障碍性贫血,是由于构成β珠蛋白的基因缺失或突变,导致构成血红蛋白的α链和β链珠蛋白的合成比例失衡,而造成红细胞寿命缩短的一种溶血性贫血。β地中海贫血的独特之处在于,β珠蛋白功能或表达上的缺陷可以通过诱导γ珠蛋白的上调来进行补偿
[73 ] 。γ珠蛋白是一种在胎儿发育期间表达但在出生后沉默的特殊珠蛋白。γ珠蛋白在成人体内表达下降的一大原因是细胞内存在一种些转录抑制因子,它可以抑制γ珠蛋白表达,其中以BCL11A分子研究最为透彻
[74 ] 。因此,研究者们创新性地提出敲除
BCL11A 基因作为治疗β地中海贫血的方法。然而,后续研究观察到在所有造血细胞系中如果不存在BCL11A对于非红细胞样细胞是有害的
[75 ] 。有趣的是,科学家们在
BCL11A 的基因座上发现了一种增强子元件,可特异性地增强红细胞中的BCL11A的表达
[76 ,77 ] 。因此,研究人员通过基因编辑技术敲除增强子元件从而起到抑制
BCL11A 表达的作用,能够在红细胞谱系细胞中实现γ-珠蛋白的上调
[76 ,77 ,78 ] 。同样,研究人员也通过敲除BCL11A的结合位点来解除BCL11A对γ-珠蛋白表达的抑制作用,从而提高γ-珠蛋白的表达
[79 ] 。肌营养不良症由肌营养不良蛋白基因(
Dmd )发生移码突变引起,并编码不正常的蛋白。不过,科学家们使用基因编辑工具敲除功能突变的一个或多个外显子,产生片段缩短的mRNA,该mRNA仍可翻译成有正常功能的蛋白质
[80 ,81 ,82 ] ,该方案有望用以解决大部分DMD患者群体的疾病治疗问题。
功能获得型突变(gain-of-function mutations)是许多显性遗传病的病因。对于杂合个体而言,失活一条染色体上的显性基因即可起到疾病治疗的效果。以亨廷顿舞蹈症(Huntington's disease)为例,美国哈佛医学院Jong-Min Lee实验室利用Cas9失活突变基因的研究对于这类显性遗传病的治疗而言具有相当重要的指导意义,由于很多基因在序列上存在多个突变位点,所以治疗前对患者基因突变位点的单核苷酸多态(single nucleotide polymorphism, SNP)检测也为这类疾病的精准治疗提供了前提
[83 ] 。2018年,David Liu实验室利用脂质体递送一种Cas9-gRNA复合体的核糖核蛋白(ribonucleotide protein, RNP)用以失活发生显性突变的
Tmc1 (transmembrane channel- like gene family 1)基因,对贝多芬模型小鼠的耳聋疾病进行了治疗,缓解了小鼠失聪的症状
[84 ] 。
3.2 基因敲入在基因治疗中的应用 与基因敲除相对应,基因敲入(knock in, KI)是基因治疗的另一种策略,应用于因关键基因突变失活而导致的遗传疾病的治疗。
利用同源重组的方式进行基因敲入是使用最为广泛的一种传统策略。通过同源重组方式进行基因治疗可分为两类:点突变修复和大片段(cDNA)敲入。根据临床病例及相关测序结果统计,由于点突变导致的疾病占人类遗传疾病总数的2/3
[52 ] ,因此通过精准的基因编辑工具进行点突变修复来治疗遗传疾病最为有益。以肝脏疾病为例,研究人员在小鼠模型中分别通过质粒、AAV和纳米材料递送CRISPR/Cas9系统和修复模板进行体内基因编辑治疗I型遗传性酪氨酸血症,结果显示都能完全治愈患病小鼠
[85 ] 。本实验室在大鼠模型上通过递送nCas9进行在体治疗I型遗传性酪氨酸血症,相比于Cas9,nCas9不仅可以治愈患病大鼠,同时能够有效地降低脱靶
[86 ] 。虽然对于I型遗传性酪氨酸血症来说,修复的细胞具有扩增优势,因此只需要修复不到1%的细胞就可以治愈疾病,而对于大多数肝脏类疾病并不适用。但是该类研究是证明在肝脏中用CRISPR/Cas9可以有效进行体内基因编辑的一个里程碑。而对肝细胞没有生长优势的疾病(如血友病),本实验室也在B型血友病小鼠中利用CRISPR/Cas9技术修复突变的
FIX 基因,结果表明只需修复肝细胞中0.6%的
FIX 基因突变就有显著疗效
[87 ] 。同时,
Nature Biotechnology 报道通过两个AAV分别递送SaCas9和修复模板治疗因鸟氨酸转氨甲酰酶(ornithine transaminase, OTC)缺乏的高血氨症,在幼鼠的肝细胞中可以达到10%的修复效率,能够有效治愈疾病,但是在成年鼠中重组效率只有不到2%,没有治疗效果
[88 ] 。这也提示成年动物肝细胞同源重组效率较低,还有很大改进的空间。
然而,事实上还有许多病人并不是由于单碱基突变导致的疾病,往往由于不同位置的多个突变或大片段缺失或插入,甚至是基因结构改变等原因导致疾病产生。而对于患这类疾病的病人,大片段(cDNA)敲入的策略毫无疑问是最好的方法,尤其是在基因组的一些安全位点或高表达基因的3° UTR处进行敲入。2011年,Li等
[89 ] 首先利用ZFN结合AAV载体,将FIX cDNA通过尾静脉注射导入到小鼠体内并实现了有效的整合,这一研究现已经进入临床试验阶段,用于B型血友病新生儿的治疗。但是无论是通过点突变修复,还是大片段敲入进行基因治疗,都依赖于细胞的同源重组机制,而同源重组仅只发生于细胞周期的S/G
2 期,故只能在分裂细胞或器官中实现有效的疾病基因治疗,此外,由于同源重组效率较低,这都成了限制同源重组进行基因治疗的主要障碍
[90 ] 。
因此,科学家在想办法提高同源重组效率的同时,也在思考如何利用不依赖细胞周期的非同末端连接的机制来进行基因治疗。2016年,科学家新发现了一种非同源末端连接优化后的非同源依赖的靶向整合(homology independent targeted integration, HITI)技术
[91 ] ,相比于HDR,HITI第一次在非分裂细胞中成功实现了高效的外源基因靶向敲入,并用于视网膜色素变性大鼠模型的修复。结果表明,HITI介导的体内基因敲入效率可达10%,对视网膜色素变性起到了良好的治疗效果
[91 ] 。2017年,中国科学院神经科学研究所杨辉实验室设计出一种长同源序列末端连接(homology-mediated end-joining, HMEJ)介导的定点基因整合方式,这是一种基于CRISPR/ Cas9系统,利用sgRNA靶向位点和长同源臂(约800 bp)的供体载体来实现高效的精确整合的技术
[92 ] ,并利用其导入
Fah 基因,进行酪氨酸血症的治疗研究,具有一定的疗效
[93 ] 。
3.3 单碱基编辑系统在基因治疗中的应用 自BE系统出现之后,对于它的应用拓展一直以来就是研究者们关注的方向。由于BE系统在不造成双链断裂的前提下能引入精确的碱基转换,避免了DSB造成的靶点和非靶点序列的缺失、插入等突变,这对于基因治疗而言无疑是一个绝佳的工具。
因为BE系统可以实现其窗口内C到T或者A到G的定点突变,故而很容易实现在开放阅读框内遗传密码子的改变(错义突变)或提前产生终止密码子。如果使得基因上提前出现终止密码子(TAA、TGA或TAG),即可实现相应基因的敲除
[94 ,95 ] 。2018年10月,
Nature Medicine 在线发表通过单碱基编辑技术来治疗肝脏类代谢疾病的研究
[96 ] ,研究人员分别通过AAV递送CBE工具靶向肝细胞中
PCSK 9基因,提前产生终止码子,从而降低肝细胞的PCSK9表达,成功在小鼠模型中降低了胆固醇含量;同样通过靶向提前终止Fah的上游基因
Hpd 的表达,阻止酪氨酸代谢过程中有毒物质的产生,成功治愈了I型遗传性酪氨酸血症。
除了通过产生终止密码子进行基因的敲除,BE技术还可以用以进行突变基因的修复,从而达到疾病治疗的目的。韩国首尔国立大学Jin-Soo Kim实验室将利用反式剪接腺相关病毒载体将ABE系统导入到成年DMD小鼠的肌肉中,纠正了
Dmd 基因20号外显子上的一个无义突变,恢复了17%的肌营养不良蛋白表达,明显改善了患病小鼠的肌肉功能
[97 ] 。而2018年,
Nature Medicine 发表的另一项研究表明CBE工具可以治愈患苯丙酮尿症的成年小鼠模型,并且修复效率在RNA水平上达到63%
[98 ] 。这样振奋人心的结果对此类没有肝细胞生长优势的疾病的治疗带来了希望,同时也说明单碱基编辑技术在成体疾病的治疗上具有极大临床应用潜能。
3.4 表观遗传调控在基因治疗中的应用 基因组表观遗传修饰的异常与许多疾病的发生发展密切相关,如代谢紊乱、心血管疾病和癌症等等。基于蛋白编码的基因研究是不足以解释这一类疾病的发生机理,这些疾病的发病机制主要是基因和环境共同作用的结果。转录调控和表观遗传修饰改变的可逆性为治疗这类疾病提供了新的希望。基因定点修饰技术的发展更为其精准治疗增加了可能。
目前利用转录调控和表观遗传定点修饰技术在体内治疗表观遗传异常引起疾病的研究还屈指可数。2017年,Liao等
[99 ] 采用了一种gRNA融合MS2- P65-HSF1 (MPH)转录激活复合物的策略,将截短的gRNA (dgRNA)和MPH包装到两个单独的AAV载体中,共同递送到表达dCas9的转基因小鼠中。通过靶向激活相应的功能性基因成功地修复了I型糖尿病、急性肾损伤和肌营养不良的疾病表型。同时另一项突破性的工作则使用一种SunTag (dCas9- 10xGCN4)系统融合多个拷贝的转录激活蛋白(p65- HSF1),构建了一种Cre依赖性的SunTag-p65-HSF1 (SPH)转基因小鼠模型。使用AAV8将Cre和sgRNAs递送到SPH转基因小鼠中,通过激活内源性神经源性转录因子的表达,在小鼠体内成功地实现了星形胶质细胞直接转化为功能性的神经元
[100 ] 。这一研究表明,在不需要使用外源重编程因子或转录因子的前提下,通过转录调控的CRISPR系统可以实现特定细胞的重编程或不同细胞类型之间的转分化,为体外细胞基因治疗遗传疾病提供了新的策略。
2018年12月,
Science 在线发表了一篇使用CRISPRa系统在小鼠中成功修复一种因单倍剂量不足引起的肥胖
[101 ] 。研究人员通过AAV在SIM1基因或MC4R基因部分功能丧失的小鼠脑部递送dCas9-vp64和sgRNA的方式成功激活了SIM1或MC4R蛋白的表达,成功抑制了肥胖的表型。这一策略给罹患单倍体剂量不足引起的疾病患者带来转机。
基因中CpG岛中的5' C经常突变引起高甲基化、羟甲基化等修饰,研究表明这些异常修饰会影响基因的表达调控,最终引起疾病
[102 ] 。脆性X综合征(fragile X syndrome, FXS)即是一种由
FMR1 基因5° UTR区中CGG三核甘酸重复序列扩增突变并高甲基化,使
FMR 基因沉默而导致的疾病。最近的一项研究通过利用dCas9融合Tet甲基胞嘧啶双加氧酶1(Tet methylcytosine dioxygenase 1, Tet1)转染FXS iPSCs细胞系,成功靶向诱导
FMR 基因5' UTR CpG岛去甲基化,为这些因异常甲基化引起的疾病的治疗奠定了基础
[103 ] 。
基于CRISPR的基因组转录调控和表观遗传修饰的优点在于能够调控基因的表达而不会造成永久性的DNA损伤,因而不会产生有害突变和脱靶效应,并且表观遗传疗法可以通过同时调节多个基因活性来提供更好的治疗效果,进一步的研究可弥补基因治疗存在的不足,是基因治疗研究的新热点。
4 问题与展望 4.1 基因编辑治疗所面临的问题 虽然采用基因编辑技术进行基因治疗在基础研究领域已经取得了不少突破性的进展,可是目前在临床应用方面仍然存在几大困难与挑战。
首先是治疗的安全性。进行外源基因递送时采用的整合型载体(如LV载体)产生的可随细胞分裂而保留的插入突变和病毒载体本身所产生的免疫反应(如AAV衣壳蛋白引起的免疫排斥)是基因治疗在发展过程中备受讨论的风险问题,同时基于各种基因编辑技术的基因治疗存在脱靶问题也使受众产生担忧。除此之外,2018年6月发表在
Nature Medicine 的两篇文章指出
[104 ,105 ] ,在某些情况下,CRISPR介导的基因组编辑可以导致p53介导的应激反应和细胞周期停滞现象,强调人们有必要从更多角度去理解影响体内和体外可以有效进行基因组编辑的因素,也侧面显示出基因编辑技术仍存在一些需要人们去深入思考的潜在问题(如是否存在潜在的致瘤风险)。2019年3月,中国科学家杨辉与高彩霞也在
Science 上“背靠背”发文,发现CBE系统分别在小鼠胚胎和水稻中导致较多的脱靶性单核苷酸突变
[106 ,107 ] ,这也启示人们在利用BE系统展开相关应用尤其是临床应用之前必须做好足够的安全性评估,以避免大量脱靶导致的严重安全性问题。
其次,就治疗的有效性而言,载体的递送效率直接影响到治疗本身的效率。这种递送效率包括载体对外源基因本身携带的能力,对细胞的亲和性以及最终的感染效率等3个方面。在进行基因编辑治疗时靶点的活性和编辑的效率对于治疗的有效性也不容忽视。
最后,基因治疗的伦理也是不容忽视的问题
[108 ] 。争议颇多的生殖细胞的编辑长期以来就受到科学家和社会学家的广泛反对,而在现有的科技与社会背景下,对于胚胎的基因编辑仍然也饱受争议。少数研究者认为对于胚胎DNA的修饰或编辑可以加深人们对于人类早期发育的理解,研究人员同时希望在较长时间之后,对于胚胎的实验研究可以在临床中用于修复致病的基因突变,进而避免将遗传性疾病传给下一代。但是大部分伦理学家和许多研究者仍然担心这些技术会被用于非医学目的。包括我国在内的许多国家禁止对胚胎的编辑研究,只允许在非生殖性的体细胞中进行编辑
[109 ] 。但是,对于体细胞的编辑也应当小心谨慎,避免可能的潜在问题(如给治疗者带来严重的附加疾病)出现
[110 ,111 ,112 ] 。
4.2 基因编辑治疗的发展前景 基因治疗从诞生到现在,已经逐步发展成为临床领域的重要治疗方式,且在治疗方法和药物开发方面取得了重大进展。3个治疗罕见病的药物已经走入了遗传病患者的生活之中,且还有不少药物正在临床试验中接受着检验,可以预想到,随着社会需要的增强和相关药企的愈发重视,在未来还会有更多的药物获得批准,走向临床。目前,在新兴技术的驱动下,国际上已经出现了以CRISPR/Cas9技术为核心的三大基因编辑公司:Editas Medicine、CRISPR Therapeutics和Intellia Therapeutics。这三大基因编辑公司均致力于重大遗传疾病和肿瘤的基因治疗药物的研发,旨在攻克一系列人类重大疾病。2017年底,患有亨特氏综合征的美国男子布莱恩·马德(Brian Madeux)在加州大学旧金山分校贝尼奥夫儿童医院接受了一次大胆的治疗:通过AAV递送基因编辑工具ZFN进行体内基因编辑来治疗疾病,这是世界上首次通过体内基因编辑来治疗遗传疾病的报道,更加说明基因编辑对于遗传疾病,尤其是肝脏类代谢疾病的治疗具有极其重要的临床应用潜能
[113 ] 。基因编辑技术在近年来也和肿瘤的免疫治疗相结合
[114 ,115 ,116 ] ,为人类的疾病治疗提供了更多更新的选择。再回到技术本身,虽然CBE系统在研究中出现了较高的脱靶性,但是ABE系统仍然被证明具有较好的保真性
[106 ,107 ] 。此外,碱基编辑器由于不产生DSB,故而也跳过了p53等风险因素,可能具有更好的安全性。在未来的基础医学实践中,碱基编辑器或许会发挥其特有的优势,创造更多的成果。
截止目前,基因编辑技术已经在基础科研和临床应用研究中取得了重大突破与进展,并不断创造着社会经济价值。随着现代医学的进步和个体化治疗的推进,基因编辑治疗的广泛化、个体化与最优化也将进一步发展,并在未来更多地走入患者的治疗与生活中。尽管目前对于基因治疗还有许多技术难题有待解决,但可以相信随着人类基因表达调控机制的阐明,以及转基因技术的发展和转基因方法的完善,采用基因编辑技术进行的基因治疗方式必将成为21世纪人类攻克疑难病症的一种常规治疗手段,为维护人类健康做出重要贡献。
[1] Chu SY Weng CY Hu Li Za Zhi 2017 ,64(5 ):11 -17 . [本文引用: 2] 褚思义 , 翁纯英 . 遗传/罕见病简介及遗传咨询的应用护理杂志 , 2017 ,64(5 ):11 -17 . [本文引用: 2] [2] Darrow JJ Drug Discov Today 2019 ,24(4 ):949 -954 . URL [本文引用: 1] [3] Han X Ni W Value Health 2015 ,18(7 ):A756 . [本文引用: 1] [4] Schimmer J Breazzano S Hum Gene Ther Clin Dev 2016 ,27(2 ):57 -61 . URL [本文引用: 1] [5] Gupta SK Shukla P Crit Rev Biotechnol 2017 ,37(5 ):672 -684 . URL [本文引用: 1] [6] Takata M Sasaki MS Sonoda E Morrison C Hashimoto M Utsumi H Yamaguchi-Iwai Y Shinohara A Takeda S EMBO J 1998 ,17(18 ):5497 -5508 . URL [本文引用: 1] [7] Lieber MR Ma Y Pannicke U Schwarz K Nat Rev Mol Cell Bio 2003 ,4(9 ):712 -720 . [本文引用: 1] [8] Joung JK Sander JD Nat Rev Mol Cell Bio 2012 ,14(1 ):49 -55 . [本文引用: 1] [9] Sander JD Joung JK Nat Biotechnol 2014 ,32(4 ):347 -355 . [本文引用: 1] [10] Komor AC Badran AH Liu DR Cell 2017 ,168(1-2 ):20 -36 . URL [本文引用: 1] [11] Marcaida MJ Prieto J Redondo P Proc Natl Acad Sci USA 2008 ,105(44 ):16888 -16893 . URL [本文引用: 1] [12] Smith J Grizot S Arnould S Duclert A Epinat JC Chames P Prieto J Redondo P Blanco FJ Bravo J Montoya G Paques F Duchateau P Nucleic Acids Res 2006 ,34(22 ):e149 . URL [本文引用: 1] [13] Silva G Poirot L Galetto R Smith J Montoya G Duchateau P Paques F Curr Gene Ther 2011 ,11(1 ):11 -27 . URL [本文引用: 1] [14] Wang L Smith J Breton C Clark P Zhang J Ying L Che Y Lape J Bell P Calcedo R Buza EL Saveliev A Bartsevich VV He Z White J Li M Jantz D Wilson JM Nat Biotechnol 2018 ,36(8 ):717 -725 . [本文引用: 1] [15] Mani M Kandavelou K Dy FJ Durai S Chandrasegaran S Biochem Biophys Res Commun 2005 ,335(2 ):447 -457 . URL [本文引用: 1] [16] Carroll D Genetics 2011 ,188(4 ):773 -782 . URL [本文引用: 1] [17] Kim YG Cha J Chandrasegaran S Proc Natl Acad Sci USA 1996 ,93(3 ):1156 -1160 . URL [本文引用: 1] [18] Ramirez CL Foley JE Wright DA Müller-Lerch F Rahman SH Cornu TI Winfrey RJ Sander JD Fu F Townsend JA Cathomen T Voytas DF Joung JK Nat Methods 2008 ,5(5 ):374 -375 . [本文引用: 1] [19] Lam KN van Bakel H Cote AG van der Ven A Hughes TR Nucleic Acids Res 2011 ,39(11 ):4680 -4690 . URL [本文引用: 1] [20] Ul Ain Q Chung JY Kim YH J Control Release 2014 ,205:120 -127 . [本文引用: 1] [21] Bonas U Stall RE Staskawicz B Mol Gen Genet 1989 ,218(1 ):127 -136 . URL [本文引用: 1] [22] Kay S Hahn S Marois E Hause G Bonas U Science 2007 ,318(5850 ):648 -651 . URL [本文引用: 1] [23] Sugio A Yang B Zhu T White FF Proc Natl Acad Sci USA 2007 ,104(25 ):10720 -10725 . URL [本文引用: 1] [24] Boch J Scholze H Schornack S Landgraf A Hahn S Kay S Lahaye T Nickstadt A Bonas U Science 2009 ,326(5959 ):1509 -1512 . URL [本文引用: 1] [25] Moscou MJ Bogdanove AJ Science 2009 ,326(5959 ):1501 . URL [本文引用: 1] [26] Bedell VM Wang Y Campbell JM Poshusta TL Starker CG Krug RG 2nd Tan W Penheiter SG Ma AC Leung AY Fahrenkrug SC Carlson DF Voytas DF Clark KJ Essner J Ekker SC Nature 2012 ,491(7422 ):114 -118 . [本文引用: 1] [27] Tan WS Carlson DF Walton MW Fahrenkrug SC Hackett PB Adv Genet 2012 ,80:37 -97 . URL [本文引用: 1] [28] Joung JK Sander JD Nat Rev Mol Cell Biol 2013 ,14(1 ):49 -55 . [本文引用: 1] [29] Lee HB Sundberg BN Sigafoos AN Clark KJ Front Genet 2016 ,7:47 . [本文引用: 1] [30] Qiu Z Liu M Chen Z Shao Y Pan H Wei G Yu C Zhang L Li X Wang P Fan HY Du B Liu B Liu M Li D Nucleic Acids Res 2013 ,41(11 ):e120 . URL [本文引用: 1] [31] Tong C Huang G Ashton C Wu H Yan H Ying QL J Genet Genomics 2012 ,39(6 ):275 -280 . URL [本文引用: 1] [32] Liu J Li C Yu Z Huang P Wu H Wei C Zhu N Shen Y Chen Y Zhang B Deng WM Jiao R J Genet Genomics 2012 ,39(5 ):209 -215 . URL [本文引用: 1] [33] Christian M Qi Y Zhang Y Voytas DF G3 (Bethesda) 2013 ,3(10 ):1697 -1705 . URL [本文引用: 1] [34] Ishino Y Shinagawa H Makino K Amemura M Nakata A J Bacteriol 1987 ,169(12 ):5429 -5433 . URL [本文引用: 1] [35] Jansen R Embden JD Gaastra W Schouls LM Mol Microbiol 2002 ,43(6 ):1565 -1575 . URL [本文引用: 1] [36] Gasiunas G Barrangou R Horvath P Siksnys V Proc Natl Acad Sci USA 2012 ,109(39 ):E2579 -2586 . URL [本文引用: 1] [37] Fonfara I Le Rhun A Chylinski K Makarova KS Lécrivain AL Bzdrenga J Koonin EV Charpentier E Nucleic Acids Res 2014 ,42(4 ):2577 -2590 . URL [本文引用: 1] [38] Hsu PD Scott DA Weinstein JA Ran FA Konermann S Agarwala V Li Y Fine EJ Wu X Shalem O Cradick TJ Marraffini LA Bao G Zhang F Nat Biotechnol 2013 ,31(9 ):827 -832 . [本文引用: 1] [39] Pattanayak V Lin S Guilinger JP Ma E Doudna JA Liu DR Nat Biotechnol 2013 ,31(9 ):839 -843 . [本文引用: 1] [40] Fu Y Foden JA Khayter C Maeder ML Reyon D Joung JK Sander JD Nat Biotechnol 2013 ,31(9 ):822 -826 . [本文引用: 1] [41] Ran FA Cong L Yan WX Scott DA Gootenberg JS Kriz AJ Zetsche B Shalem O Wu X Makarova KS Koonin EV Sharp PA Zhang F Nature 2015 ,520(7546 ):186 -191 . [本文引用: 1] [42] Zetsche B Gootenberg JS Abudayyeh OO Slaymaker IM Makarova KS Essletzbichler P Volz SE Joung J van der Oost J Regev A Koonin EV Zhang F Cell 2015 ,163(3 ):759 -771 . URL [本文引用: 1] [43] Hu JH Miller SM Geurts MH Tang W Chen L Sun N Zeina CM Gao X Rees HA Lin Z Liu DR Nature 2018 ,556(7699 ):57 -63 . [本文引用: 1] [44] Nishimasu H Shi X Ishiguro S Gao L Hirano S Okazaki S Noda T Abudayyeh OO Gootenberg JS Mori H Oura S Holmes B Tanaka M Seki M Hirano H Aburatani H Ishitani R Ikawa M Yachie N Zhang F Nureki O Science 2018 ,361(6408 ):1259 -1262 . URL [本文引用: 1] [45] Singh D Wang Y Mallon J Yang O Fei J Poddar A Ceylan D Bailey S Ha T Nat Struct Mol Biol 2018 ,25(4 ):347 -354 . [本文引用: 1] [46] Chen JS Dagdas YS Kleinstiver BP Welch MM Sousa AA Harrington LB Sternberg SH Joung JK Yildiz A Doudna JA Nature 2017 ,550(7676 ):407 -410 . [本文引用: 1] [47] Ran FA Hsu PD Lin CY Gootenberg JS Konermann S Trevino AE Scott DA Inoue A Matoba S Zhang Y Zhang F Cell 2013 ,154(6 ):1380 -1389 . URL [本文引用: 1] [48] Komor AC Kim YB Packer MS Zuris JA Liu DR Nature 2016 ,533(7603 ):420 -424 . [本文引用: 1] [49] Nishida K Arazoe T Yachie N Banno S Kakimoto M Tabata M Mochizuki M Miyabe A Araki M Hara KY Shimatani Z Kondo A Science 2016 , 353(6305 ): .aaf8729 . URL [本文引用: 1] [50] Ma Y Zhang J Yin W Zhang Z Song Y Chang X Nat Methods 2016 ,13(12 ):1029 -1035 . [本文引用: 1] [51] Gaudelli NM Komor AC Rees HA Packer MS Badran AH Bryson DI Liu DR Nature 2017 ,551(7681 ):464 -471 . [本文引用: 1] [52] Wei Y Zhang XH Li DL Hereditas (Beijing) 2017 ,39(12 ):1115 -1121 . [本文引用: 3] 魏瑜 , 张晓辉 , 李大力 . 基因编辑之“新宠”—单碱基基因组编辑系统遗传 , 2017 ,39(12 ):1115 -1121 . [本文引用: 3] [53] Gilbert LA Larson MH Morsut L Liu Z Brar GA Torres SE Stern-Ginossar N Brandman O Whitehead EH Doudna JA Lim WA Weissman JS Qi LS Cell 2013 ,154(2 ):442 -451 . URL [本文引用: 1] [54] Maeder ML Linder SJ Cascio VM Fu Y Ho QH Joung JK Nat Methods 2013 ,10(10 ):977 -979 . [本文引用: 1] [55] Perez-Pinera P Kocak DD Vockley CM Adler AF Kabadi AM Polstein LR Thakore PI Glass KA Ousterout DG Leong KW Guilak F Crawford GE Reddy TE Gersbach CA Nat Methods 2013 ,10(10 ):973 -976 . [本文引用: 1] [56] Chavez A Scheiman J Vora S Pruitt BW Tuttle M P R Iyer E Lin S Kiani S Guzman CD Wiegand DJ Ter-Ovanesyan D Braff JL Davidsohn N Housden BE Perrimon N Weiss R Aach J Collins JJ Church GM Nat Methods 2015 ,12(4 ):326 -328 . [本文引用: 1] [57] Li Z Zhang D Xiong X Yan B Xie W Sheen J Li JF Nat Plants 2017 ,3(12 ):930 -936 . URL [本文引用: 1] [58] Liu XS Wu H Ji X Stelzer Y Wu X Czauderna S Shu J Dadon D Young RA Jaenisch R Cell 2016 ,167(1 ):233 -247 . URL [本文引用: 2] [59] Liu P Chen M Liu Y Qi LS Ding S Cell Stem Cell 2018 ,22(2 ):252 -261 . URL [本文引用: 1] [60] Cano-Rodriguez D Gjaltema RA Jilderda LJ Jellema P Dokter-Fokkens J Ruiters MH Rots MG Nat Commun 2016 ,7:12284 . [本文引用: 1] [61] Kearns NA Pham H Tabak B Genga RM Silverstein NJ Garber M Maehr R Nat Methods 2015 ,12(5 ):401 -403 . [本文引用: 1] [62] Hilton IB D'Ippolito AM Vockley CM Thakore PI Crawford GE Reddy TE Gersbach CA Nat Biotechnol 2015 ,33(5 ):510 -517 . [本文引用: 1] [63] Kwon DY Zhao YT Lamonica JM Zhou Z Nat Commun 2017 ,8:15315 . [本文引用: 1] [64] Mendenhall EM Williamson KE Reyon D Zou JY Ram O Joung JK Bernstein BE Nat Biotechnol 2013 ,31(12 ):1133 -1136 . [本文引用: 1] [65] Naldini L Nature 2015 ,526(7573 ):351 -360 . [本文引用: 1] [66] Sinn PL Sauter SL McCray PB Gene Ther 2005 ,12(14 ):1089 -1098 . [本文引用: 1] [67] Cavazzana-Calvo M Payen E Negre O Wang G Hehir K Fusil F Down J Denaro M Brady T Westerman K Cavallesco R Gillet-Legrand B Caccavelli L Sgarra R Maouche-Chrétien L Bernaudin F Girot R Dorazio R Mulder GJ Polack A Bank A Soulier J Larghero J Kabbara N Dalle B Gourmel B Socie G Chrétien S Cartier N Aubourg P Fischer A Cornetta K Galacteros F Beuzard Y Gluckman E Bushman F Hacein-Bey- Abina S Leboulch P Nature 2010 ,467(7313 ):318 -322 . [本文引用: 1] [68] Wickham TJ Gene Ther 2000 ,7(2 ):110 -114 . [本文引用: 1] [69] Kotterman MA Schaffer DV Nat Rev Genet 2014 ,15(7 ):445 -451 . [本文引用: 1] [70] Jin S Ye K Biotechnol Prog 2007 ,23(1 ):32 -41 . URL [本文引用: 2] [71] Ding Q Strong A Patel KM Ng SL Gosis BS Regan SN Cowan CA Rader DJ Musunuru K Circ Res 2014 ,115(5 ):488 -492 . URL [本文引用: 1] [72] Pankowicz FP Barzi M Legras X Hubert L Mi T Tomolonis JA Ravishankar M Sun Q Yang D Borowiak M Sumazin P Elsea SH Bissig-Choisat B Bissig KD Nat Commun 2016 ,7:12642 . [本文引用: 1] [73] Forget BG Ann Ny Acad Sci 1998 ,850:38 -44 . URL [本文引用: 1] [74] Thein SL Blood Cell Mol Dis 2017 ,70:54 -65 . [本文引用: 1] [75] Tsang JC Yu Y Burke S Buettner F Wang C Kolodziejczyk AA Teichmann SA Lu L Liu P Genome Biol 2015 ,16:178 . URL [本文引用: 1] [76] Bauer DE Kamran SC Lessard S Xu J Fujiwara Y Lin C Shao Z Canver MC Smith EC Pinello L Sabo PJ Vierstra J Voit RA Yuan GC Porteus MH Stamatoyannopoulos JA Lettre G Orkin SH Science 2013 ,342(6155 ):253 -257 . URL [本文引用: 2] [77] Canver MC Smith EC Sher F Pinello L Sanjana NE Shalem O Chen DD Schupp PG Vinjamur DS Garcia SP Luc S Kurita R Nakamura Y Fujiwara Y Maeda T Yuan GC Zhang F Orkin SH Bauer DE Nature 2015 ,527(7577 ):192 -197 . [本文引用: 2] [78] Vierstra J Reik A Chang KH Stehling-Sun S Zhou Y Hinkley SJ Paschon DE Zhang L Psatha N Bendana YR O'Neil CM Song AH Mich AK Liu PQ Lee G Bauer DE Holmes MC Orkin SH Papayannopoulou T Stamatoyannopoulos G Rebar EJ Gregory PD Urnov FD Stamatoyannopoulos JA Nat Methods 2015 ,12(10 ):927 -930 . [本文引用: 1] [79] Martyn GE Wienert B Yang L Shah M Norton LJ Burdach J Kurita R Nakamura Y Pearson RCM Funnell APW Quinlan KGR Crossley M Nat Genet 2018 ,50(4 ):498 -503 . [本文引用: 1] [80] Ousterout DG Kabadi AM Thakore PI Perez-Pinera P Brown MT Majoros WH Reddy TE Gersbach CA Mol Ther 2015 ,23(3 ):523 -532 . URL [本文引用: 1] [81] Ousterout DG Kabadi AM Thakore PI Majoros WH Reddy TE Gersbach CA Nat Commun 2015 ,6:6244 . [本文引用: 1] [82] Li HL Fujimoto N Sasakawa N Shirai S Ohkame T Sakuma T Tanaka M Amano N Watanabe A Sakurai H Yamamoto T Yamanaka S Hotta A Stem Cell Reports 2015 ,4(1 ):143 -154 . URL [本文引用: 1] [83] Shin JW Kim KH Chao MJ Atwal RS Gillis T MacDonald ME Gusella JF Lee JM Hum Mol Genet 2016 ,25(20 ):4566 -4576 . [本文引用: 1] [84] Gao X Tao Y Lamas V Huang M Yeh WH Pan B Hu YJ Hu JH Thompson DB Shu Y Li Y Wang H Yang S Xu Q Polley DB Liberman MC Kong WJ Holt JR Chen ZY Liu DR Nature 2018 ,553(7687 ):217 -221 . [本文引用: 1] [85] Yin H Xue W Chen S Bogorad RL Benedetti E Grompe M Koteliansky V Sharp PA Jacks T Anderson DG Nat Biotechnol 2014 ,32(6 ):551 -553 . [本文引用: 1] [86] Shao Y Wang L Guo N Wang S Yang L Li Y Wang M Yin S Han H Zeng L Zhang L Hui L Ding Q Zhang J Geng H Liu M Li D J Biol Chem 2018 ,293(18 ):6883 -6892 . URL [本文引用: 1] [87] Guan Y Ma Y Li Q Sun Z Ma L Wu L Wang L Zeng L Shao Y Chen Y Ma N Lu W Hu K Han H Yu Y Huang Y Liu M Li D EMBO Mol Med 2016 ,8(5 ):477 -488 . URL [本文引用: 1] [88] Yang Y Wang L Bell P McMenamin D He Z White J Yu H Xu C Morizono H Musunuru K Batshaw ML Wilson JM Nat Biotechnol 2016 ,34(3 ):334 -338 . [本文引用: 1] [89] Li H Haurigot V Doyon Y Li T Wong SY Bhagwat AS Malani N Anguela XM Sharma R Ivanciu L Murphy SL Finn JD Khazi FR Zhou S Paschon DE Rebar EJ Bushman FD Gregory PD Holmes MC High KA Nature 2011 ,475(7355 ):217 -221 . [本文引用: 1] [90] Cong L Ran FA Cox D Lin S Barretto R Habib N Hsu PD Wu X Jiang W Marraffini LA Zhang F Science 2013 ,339(6121 ):819 -823 . URL [本文引用: 1] [91] Suzuki K Tsunekawa Y Hernandez-Benitez R Wu J Zhu J Kim EJ Hatanaka F Yamamoto M Araoka T Li Z Kurita M Hishida T Li M Aizawa E Guo S Chen S Goebl A Soligalla RD Qu J Jiang T Fu X Jafari M Esteban CR Berggren WT Lajara J Nuñez-Delicado E Guillen P Campistol JM Matsuzaki F Liu GH Magistretti P Zhang K Callaway EM Zhang K Belmonte JC Nature 2016 ,540(7631 ):144 -149 . [本文引用: 2] [92] Yao X Wang X Hu X Liu Z Liu J Zhou H Shen X Wei Y Huang Z Ying W Wang Y Nie YH Zhang CC Li S Cheng L Wang Q Wu Y Huang P Sun Q Shi L Yang H Cell Res 2017 ,27(6 ):801 -814 . [本文引用: 1] [93] Yao X Wang X Liu J Shi L Huang P Yang H J Vis Exp 2018 (133 ). [本文引用: 1] [94] Komor AC Kim YB Packer MS Zuris JA Liu DR Nature 2016 ,533(7603 ):420 -424 . [本文引用: 1] [95] Billon P Bryant EE Joseph SA Nambiar TS Hayward SB Rothstein R Ciccia A Mol Cell 2017 , 67(6 ): 1068 -1079 . e4 . URL [本文引用: 1] [96] Rossidis AC Stratigis JD Chadwick AC Hartman HA Ahn NJ Li H Singh K Coons BE Li L Lv W Zoltick PW Alapati D Zacharias W Jain R Morrisey EE Musunuru K Peranteau WH Nat Med 2018 ,24(10 ):1513 -1518 . [本文引用: 1] [97] Ryu SM Koo T Kim K Lim K Baek G Kim ST Kim HS Kim DE Lee H Chung E Kim JS Nat Biotechnol 2018 ,36(6 ):536 -539 . [本文引用: 1] [98] Villiger L Grisch-Chan HM Lindsay H Ringnalda F Pogliano CB Allegri G Fingerhut R Häberle J Matos J Robinson MD Thöny B Schwank G Nat Med 2018 ,24(10 ):1519 -1525 . [本文引用: 1] [99] Liao HK Hatanaka F Araoka T Reddy P Wu MZ Sui Y Yamauchi T Sakurai M O'Keefe DD Núñez- Delicado E Guillen P Campistol JM Wu CJ Lu LF Esteban CR Izpisua Belmonte JC Cell 2017 , 171(7 ): 1495 -11507 . e15 . URL [本文引用: 1] [100] Zhou H Liu J Zhou C Gao N Rao Z Li H Hu X Li C Yao X Shen X Sun Y Wei Y Liu F Ying W Zhang J Tang C Zhang X Xu H Shi L Cheng L Huang P Yang H Nat Neurosci 2018 ,21(3 ):440 -446 . [本文引用: 1] [101] Matharu N Rattanasopha S Tamura S Maliskova L Wang Y Bernard A Hardin A Eckalbar WL Vaisse C Ahituv N Science 2018 ,363(6424 ):231 -243 . [本文引用: 1] [102] Xu Y Wu F Tan L Kong L Xiong L Deng J Barbera AJ Zheng L Zhang H Huang S Min J Nicholson T Chen T Xu G Shi Y Zhang K Shi YG Mol Cell 2011 ,42(4 ):451 -464 . URL [本文引用: 1] [103] Liu XS Wu H Krzisch M Wu X Graef J Muffat J Hnisz D Li CH Yuan B Xu C Li Y Vershkov D Cacace A Young RA Jaenisch R Cell 2018 , 172(5 ): 979 -992 . e6 . URL [本文引用: 1] [104] Haapaniemi E Botla S Persson J Schmierer B Taipale J Nat Med 2018 ,24(7 ):927 -930 . [本文引用: 1] [105] Ihry RJ Worringer KA Salick MR Frias E Ho D Theriault K Kommineni S Chen J Sondey M Ye C Randhawa R Kulkarni T Yang Z McAllister G Russ C Reece-Hoyes J Forrester W Hoffman GR Dolmetsch R Kaykas A Nat Med 2018 ,24(7 ):939 -946 . [本文引用: 1] [106] Zuo E Sun Y Wei W Yuan T Ying W Sun H Yuan L Steinmetz LM Li Y Yang H Science 2019 ,364(6437 ):289 -292 . [本文引用: 2] [107] Jin S Zong Y Gao Q Zhu Z Wang Y Qin P Liang C Wang D Qiu JL Zhang F Gao C Science 2019 ,364(6437 ):292 -295 . [本文引用: 2] [108] Chu JY Hereditas(Beijing) 2019 ,41(5 ):447 -450 . [本文引用: 1] 褚嘉祐 . 遗传学研究领域发展过程中不可忽视的伦理学问题遗传 , 2019 ,41(5 ):447 -450 . [本文引用: 1] [109] David C Nature, news 2018 -10-03. [本文引用: 1] [110] Woods NB Bottero V Schmidt M von Kalle C Verma IM Nature 2006 ,440(7088 ):1123 . [本文引用: 1] [111] Mavilio F Ferrari G EMBO Rep 2008 ,9(1S ):S64 -S69 . URL [本文引用: 1] [112] Lappé M J Med Philos 1991 ,16(6 ):621 -639 . URL [本文引用: 1] [113] Jocelyn K Here’s what you need to know. Science,news 2017 -11-15. [本文引用: 1] [114] Boissel S Jarjour J Astrakhan A Adey A Gouble A Duchateau P Shendure J Stoddard BL Certo MT Baker D Scharenberg AM Nucleic Acids Res 2013 ,42(4 ):2591 -2601 . [本文引用: 1] [115] Beane JD Lee G Zheng Z Mendel M Abate-Daga D Bharathan M Black M Gandhi N Yu Z Chandran S Giedlin M Ando D Miller J Paschon D Guschin D Rebar EJ Reik A Holmes MC Gregory PD Restifo NP Rosenberg SA Morgan RA Feldman SA Mol Ther 2015 ,23(8 ):1380 -1390 . URL [本文引用: 1] [116] Reik A Holmes MC Zhou Y Mendel M Liu PQ Lee G Paschon D Rebar E Ando D DiGiusto D Gregory PD Jensen MC Blood 2007 ,110:2597 . [本文引用: 1]  ,上海市调控生物学重点实验室,华东师范大学生命科学学院,上海 200241
,上海市调控生物学重点实验室,华东师范大学生命科学学院,上海 200241 ,Shanghai Key Laboratory of Regulatory Biology, School of Life Sciences, East China Normal University, Shanghai 200241, China
,Shanghai Key Laboratory of Regulatory Biology, School of Life Sciences, East China Normal University, Shanghai 200241, China
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT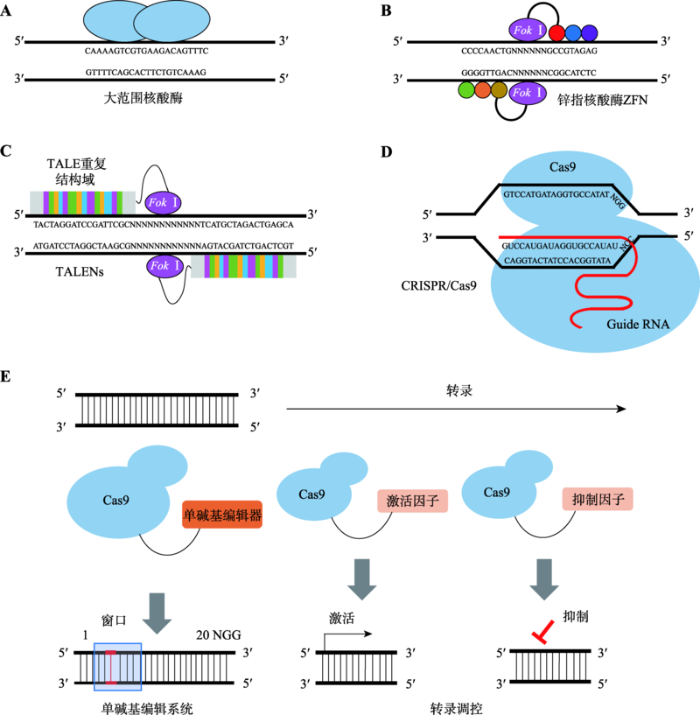 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT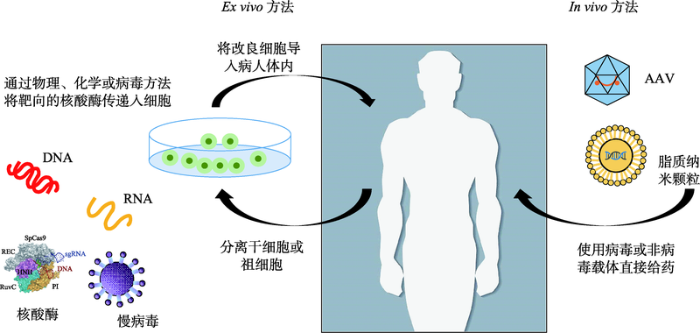 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT