 ,, 王静
,, 王静 ,石河子大学动物科技学院,石河子 832000
,石河子大学动物科技学院,石河子 832000Exploration and characterization of a universal piggyBac transposon vector for efficient transgene studies in sheep
Guangdong Hu, Kexing Hao, Tao Huang, Weibin Zeng, Xinli Gu ,, Jing Wang
,, Jing Wang ,College of Animal Science and Technology, Shihezi University, Shihezi 832000, China
,College of Animal Science and Technology, Shihezi University, Shihezi 832000, China通讯作者:
编委: 蒋思文
收稿日期:2017-12-25修回日期:2018-03-20网络出版日期:2018-08-16
| 基金资助: |
Received:2017-12-25Revised:2018-03-20Online:2018-08-16
| Fund supported: |
作者简介 About authors
胡广东,博士,副教授,研究方向:动物胚胎工程,动物生物技术E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1012KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
胡广东, 郝科兴, 黄涛, 曾维斌, 谷新利, 王静. 绵羊高效转基因通用型piggyBac转座子载体构建及功能验证[J]. 遗传, 2018, 40(8): 647-656 doi:10.16288/j.yczz.17-421
Guangdong Hu, Kexing Hao, Tao Huang, Weibin Zeng, Xinli Gu, Jing Wang.
piggyBac (PB)转座子最初是由Malcolm J. Fraser在粉纹夜蛾(Trichoplusia ni)中发现并将其应用于转基因研究,该转座子在多个宿主中具有转座活性[1]。PB转座子全长2472 bp,中间包含一个长2.1 kb的转录区域,该转录区域由一个启动子和一个长1.8 kb的外显子组成,编码大小约为64 kDa的转座酶[2]。在转基因相关操作中,PB转座子被改造成由供体质粒和辅助质粒组成的双质粒系统,供体质粒包含PB转座子必须的两个重复末端—5°TR和3°TR,可携带外源基因;辅助质粒包含一个表达转座酶的表达框,两个质粒共转染细胞,在转座酶的作用下使供体质粒特异性整合到宿主基因组的TTAA位点,进而实现导入外源基因的目的[3]。
自Ding等[4]证实PB转座子能在人(Homo sapiens)和小鼠(Mus musculus)细胞进行高效转座以来,该系统也被应用在牛(Bos taurus)[5, 6]、山羊(Capra hircus)[7]和猪(Sus scrofa)[8, 9]等哺乳动物中进行转基因操作,是目前常用的一种高效基因转移工具。基因密码子偏好性对蛋白表达有重要影响[10],优化PBase基因密码子是维持PBase稳定表达和保障PB转座发生的重要改进手段。本文在已有研究的基础上,将PBase基因进行偏好绵羊密码子优化后,成功构建了一种能在绵羊体细胞中具有功能的PB转座子载体,为绵羊转基因相关研究提供了一种高效工具。
1 材料和方法
1.1 材料
PBase CDS原始序列(GenBank登录号:EF591492.1)由南京金斯瑞生物公司进行偏好绵羊密码子优化、合成(两端含有SacⅡ、BamHⅠ酶切位点),并克隆到pUC57 Simple-T载体中,将该质粒命名为pUC57-SPBase。PBase表达载体pBNW-TD2、PB供体质粒载体pBNW-TP1均由本实验室保存。1.2 引物设计与合成
为扩增pTK-SPBase-ployA表达框,利用primer 5.0软件设计引物PPP F和PPP R,上下游引物分别引入XhoⅠ和NotⅠ酶切位点,以PB转座子5′TR端 313 bp大小核苷酸序列为模板(GenBank登录号:AB593377.1),设计3条特异性引物,分别命名为SP1、SP2和SP3,用于热不对称PCR (thermal asymmetric interlaced PCR, Tail-PCR)检测。上述所有引物均由上海生工生物工程技术服务有限公司合成(表1)。Table 1
表1
表1 本文所用的引物序列信息
Table 1
| 名称 | 引物序列(3′→5′) | 用途 | 片段大小 |
|---|---|---|---|
| PPP F | CTCGCTCGAGTAAAACGACGGCCAGTGAAT | pTK-SPBase-ployA 表达框PCR扩增 | 2.2 kb |
| PPP R | ATCGGCGGCCGCCGCTTACAATTTACGCGTTAAC | ||
| SP1 | TGACTCACGCGGTCGTTATAGTTCA | Tail-PCR检测 | |
| SP2 | GTGACACTTACCGCATTGACAAGCA | ||
| SP3 | CGGATTCGCGCTATTTAGAAAGAG |
新窗口打开|下载CSV
1.3 PB转座子单质粒通用型载体构建
以质粒pUC57-SPBase和pBNW-TD2[11]为模板,利用SacⅡ、BamHⅠ(NEB公司,美国)进行双酶切,回收优化后的PBase目的片段和pBNW-TD2骨架载体,加入T4 DNA连接酶(TaKaRa (大连)有限公司)于4℃过夜连接。将连接产物转入感受态大肠杆菌DH5α中,过夜培养,挑取单菌落,扩大培养后提取质粒。以该质粒为模板,利用PPP F和PPP R引物进行PCR,扩增pTK-SPBase-ployA表达框基因。扩增体系:5×PS buffer 10 μL,2.5 U dNTP mixture 5 μL,上下游引物各1 μL,模板150 ng,0.5 μL PrimeSTAR HS DNA Polymerase (TaKaRa (大连)有限公司) 0.5 μL,31.5 μL ddH2O,共50 μL。扩增条件:95℃ 5 min;95℃ 30 s,58℃ 30 s,72℃ 2.5 min,30个循环;最后再72℃延伸 10 min。PCR扩增结束后,取5 μL扩增产物在1 %琼脂糖凝胶上进行电泳检测。利用胶回收试剂盒(上海生工生物工程技术服务有限公司)胶回收上述目的基因片段,并将其与pBNW-TP1载体分别用NotⅠ和XhoⅠ(NEB公司,美国)双酶切,回收目的基因片段和载体片段,加入T4 DNA连接酶于4℃过夜连接,将连接产物转化入感受态大肠杆菌DH 5α (北京天根生化科技有限公司)中,过夜培养挑取单菌落,扩大培养后提取质粒,该载体命名为pBNW-TP2 (图1),用NotⅠ和XhoⅠ双酶切进行鉴定。图1
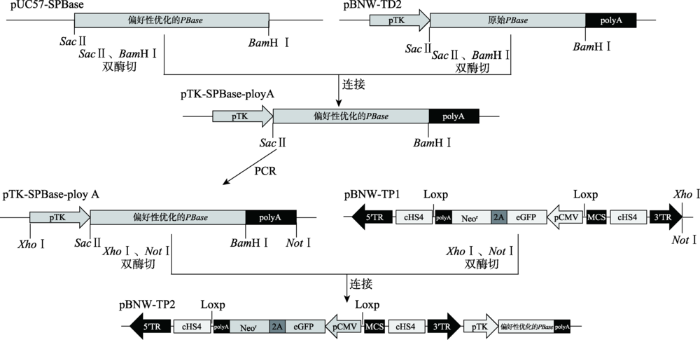 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1基于绵羊PB转座子通用型载体pBNW-TP2构建流程图
Fig. 1The flow chart of the universal PB transposon vector pBNW-TP2 for sheep
1.4 绵羊成纤维细胞和乳腺上皮细胞的培养、转染和筛选
利用组织块分离法分离绵羊成纤维细胞和乳腺上皮原代细胞,在含10%胎牛血清(Gibco公司,美国)、100 U/mL青霉素、100 mg/L链霉素的DMEM培养基(Gibco公司,美国)中培养,置于37℃、5%CO2和90%相对湿度的细胞培养箱中培养。利用电穿孔的方式对细胞进行质粒转染。电转液成分包括:120 mmol/L KCl,0.15 mmol/L CaCl2,10 mmol/L K2HPO4,5 mmol/L MgCl2。电穿孔转染具体程序如下:待原代细胞在60 mm平皿中生长至90%左右时,弃去培养基,PBS洗一遍,胰酶消化细胞,将细胞悬液转移至4 mL离心管中,1000 r/min 离心5 min。弃上清,将细胞用Opti-MEM充悬清洗两次转移至1.5 mL EP管中,随后利用100 μL电转工作液重悬细胞并吹打均匀,同时在另一个1.5 mL EP管中将转染的质粒用100 μL的电转液预先混合。转染细胞时,加入5 μg pBNW-TP2质粒,随后将质粒混合液和细胞悬液混合,并加入电转杯中进行电击。转染成纤维细胞电击参数:电压510 V,脉冲时间2 ms,电击1次。转染乳腺上皮细胞电击参数除电击次数为3次外,其余参数均不变。电击后将电转杯转移至冰上放置10 min,最后将每个电转杯中的液体接种至2个90 mm平皿中,加入含10%胎牛血清的完全培养基DMEM,放置于细胞培养箱,48 h后在细胞中加入包含800 μg/mL浓度的G418选择性培养基进行细胞筛选。3 d换液一次,持续筛选15 d后观察阳性细胞克隆,挑取阳性细胞克隆并扩大培养,保存备用。1.5 PB转座整合位点检测
利用基因组提取试剂盒(北京天根生化科技有限公司)提取扩大培养后的阳性细胞基因组DNA,参考文献[12],利用Tail-PCR检测PB转座子5′TR插入位点基因组DNA片段,其中特异性引物序列见表1。以提取的阳性细胞基因组DNA为模板,严格按照染色体步移试剂盒(TaKaRa (大连)有限公司)说明书中Tail-PCR体系和步骤,经三轮巢式PCR进行DNA片段扩增。将第3轮扩增出的所有PCR特异性条带经胶回收后连接至pMD18-T载体(TaKaRa (大连)有限公司),由南京金斯瑞生物科技有限公司测序。测序结果输入美国加州大学基因组生物信息学网站(http://genome.ucsc.edu/),确定整合染色体位点并分析其整合位置特征。1.6 转座效率检测
绵羊成纤维细胞和乳腺上皮细胞按照1.4中方法转染后移至90 mm平皿,进行G418筛选,15 d后观察细胞克隆。用4%的多聚甲醛固定,0.2%亚甲蓝溶液染色30 min,去离子水清洗后,统计细胞克隆数。本操作同等条件下重复3次,统计结果以平均数±标准误表示。利用非配对t检验(Student’s t-test)对细胞克隆数进行统计学分析,转染相同量的pBNW-TP1+CDNA4/HisMaxA (Invitrogen公司,美国)载体混合物为对照组。2 结果与分析
2.1 PBase密码子偏好性优化结果
利用OptimumGeneTM软件计算PBase密码子在绵羊中出现的频率,使其密码子适应指数(codon adaptation index, CAI)提高至0.97 (图2A),通过对PBase原有氨基酸密码子进行计算,结果显示98个氨基酸密码子在绵羊中出现频率较低,对其进行修正(图2B),对GC含量进行平衡,使整体GC含量达到56.36% (图2C)。将优化后的核酸序列TA克隆至pUC57 Simple-T载体,将其命名为pUC57-SPBase,经BamHⅠ酶切,琼脂糖凝胶电泳显示有4.4 kb大小的条带(图3),表明该载体构建正确,-20℃保存备用。图2
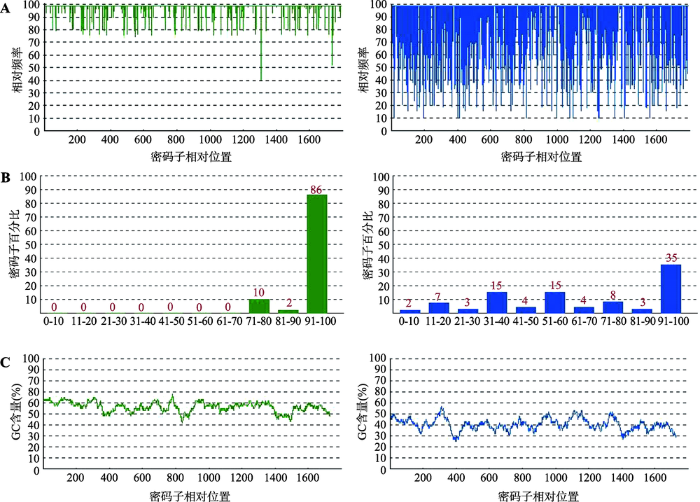 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2PBase密码子偏好性优化结果
A:PBase基因偏好性优化CAI (0.97)指数调整结果。优化后CAI指数为0.97 (绿),优化前CAI指数为0.58 (蓝)。B:PBase基因密码子偏好性调整结果。对98个氨基酸密码子进行优化(绿)。C:PBase基因GC含量调整结果。优化后GC含量为56.36% (绿),优化前为40.3% (蓝) 。
Fig. 2Optimization results of the PBase codon bias
图3
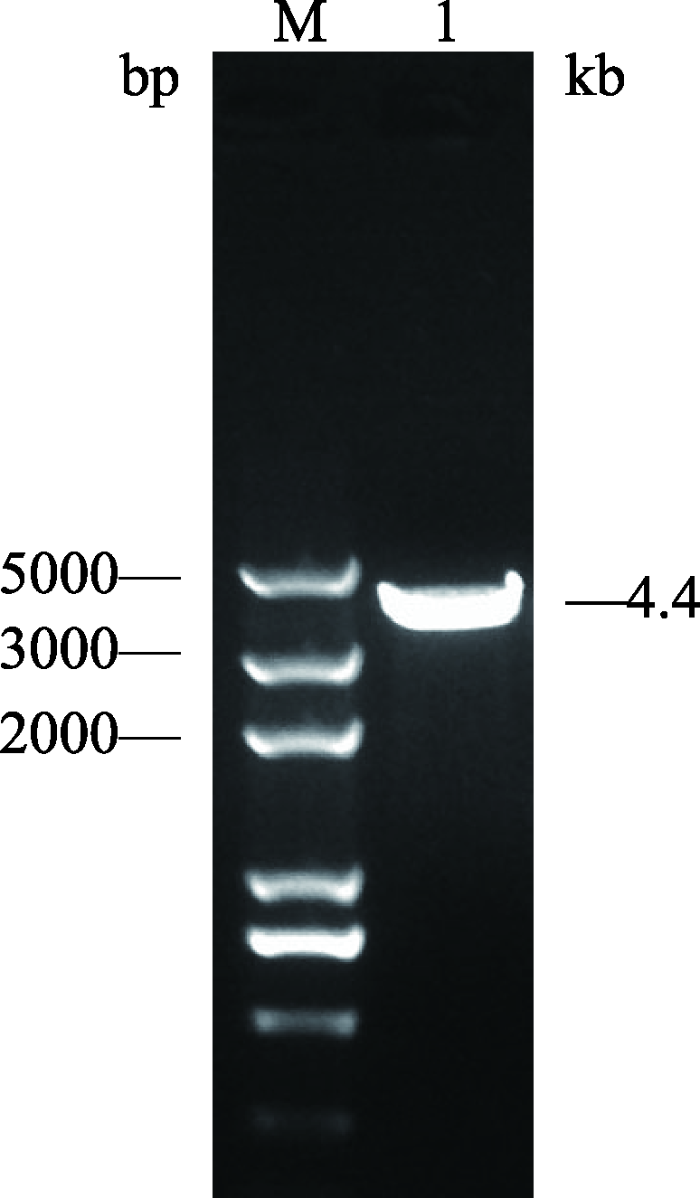 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3pUC57-PBase单酶切结果
M:Trans 2K Plus DNA marker;1:pUC57-PBase单酶切结果(4.4 kb)。
Fig. 3Single digestion results of pUC57-PBase
2.2 重组质粒酶切鉴定结果
PCR扩增pTK-SPBase-ployA表达框,两端分别携带XhoⅠ和NotⅠ酶切位点,琼脂糖凝胶电泳结果显示,pTK-SPBase-ployA表达框扩增成功(2.2 kb) (图4)。该片段经酶切,克隆至pBNW-TP1载体,成功构建了基于绵羊PB单质粒转座系统pBNW-TP2 (图1),XhoⅠ和NotⅠ双酶切鉴定结果显示,片段大小为2.2 kb,与目的片段相符,表明该载体构建成功(图5)。图4
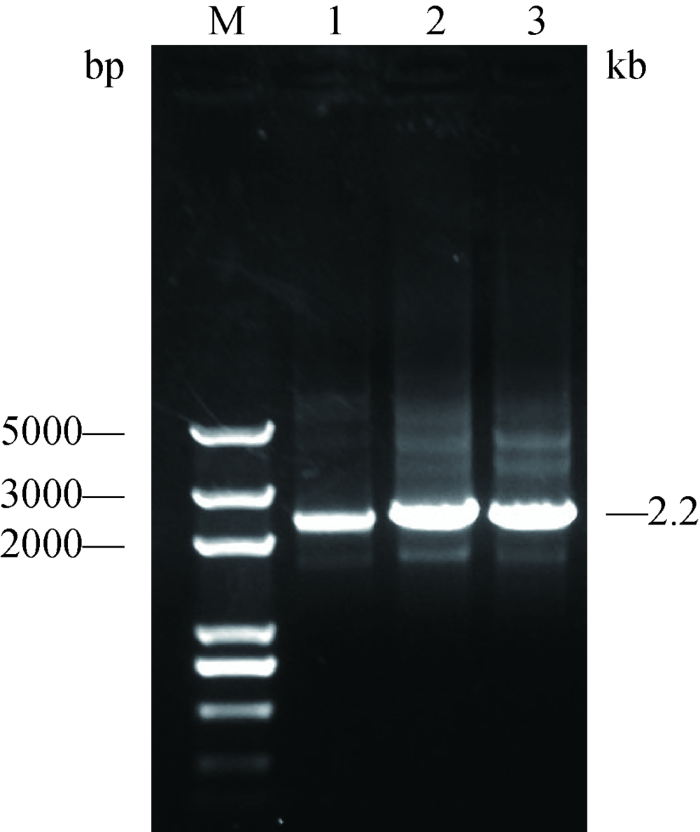 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4pTK-SPBase PCR扩增结果
M:Trans 2K Plus DNA marker;1~3:pTK-SPBase-ployA表达框PCR扩增结果。
Fig. 4PCR amplification results of pTK-SPBase
图5
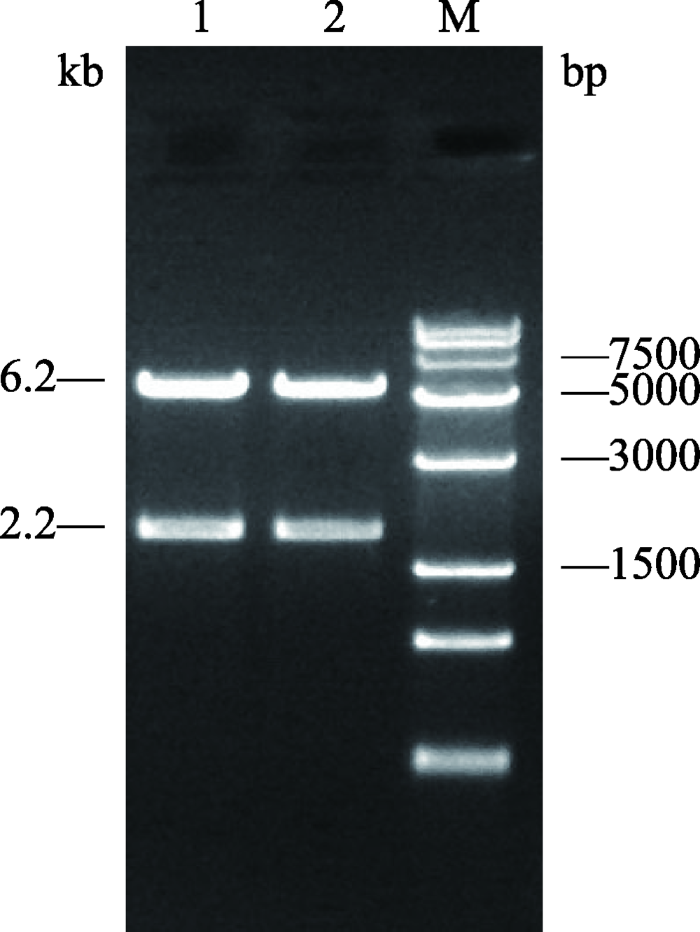 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5pBNW-TP2双酶切结果
M:Trans 15K DNA marker;1,2:pBNW-TP2载体XhoⅠ和NotⅠ双酶切鉴定结果。2.2 kb为pTK-SPBase-ployA表达框,6.2 kb为载体骨架。
Fig. 5Dual digestion results of pBNW-TP2
2.3 pBNW-TP2载体介导绵羊成纤维细胞和乳腺上皮细胞转基因细胞株特征观察
利用组织块分离法分离纯化绵羊成纤维和乳腺上皮细胞,成纤维细胞呈梭状密集排列(图6 A),乳腺上皮细胞外形呈鹅卵石状,细胞成簇生长形成岛状结构(图6 B),提示绵羊胎儿成纤维原代细胞和乳腺上皮原代细胞分离成功。图6
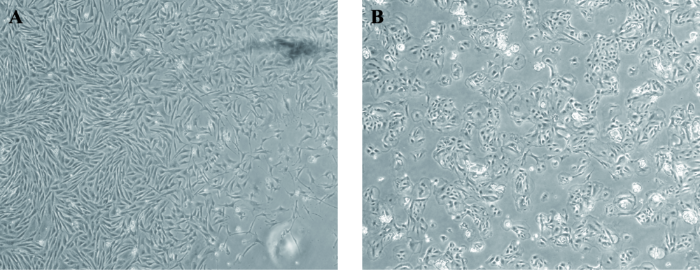 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6绵羊成纤维原代细胞和乳腺上皮细胞形态观察(40×)
A:绵羊成纤维细胞;B:绵羊乳腺上皮细胞。
Fig. 6Morphological observation of ovine primary fibroblasts and mammary epithelial cells (40×)
细胞传至第三代后,将pBNW-TP2质粒利用电转染方式转染至绵羊成纤维细胞和乳腺上皮细胞中, 转染后的细胞置于90皿,按照1.4的方法持续筛选15 d,观察阳性细胞克隆,绵羊成纤维细胞克隆明场下呈簇状克隆样(图7 A),暗场可见绿色荧光(图7 B),绵羊乳腺上皮细胞明场中可见鹅卵石状细胞呈岛状生长(图7 C),暗场下形态特征与明场相同并可见绿色荧光(图7 D)。
图7
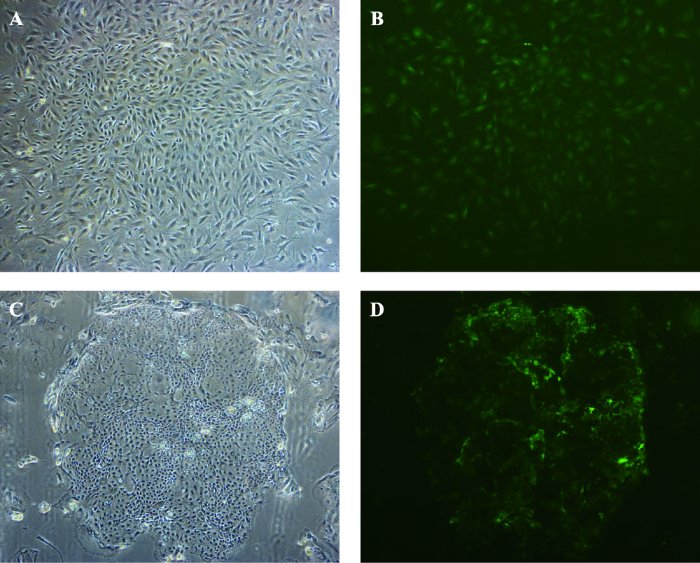 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7绵羊成纤维细胞和乳腺上皮细胞单克隆特征观察结果(40×)
A:绵羊成纤维细胞单克隆(明场);B:绵羊成纤维细胞单克隆(暗场);C:绵羊乳腺上皮细胞单克隆(明场);D:绵羊乳腺上皮细胞单克隆(暗场)
Fig. 7Morphological observation of stably selected ovine fibroblasts and mammary epithelial cells (40×)
2.4 PB整合位点检测结果
利用G418筛选获取绵羊成纤维和乳腺上皮阳性细胞,利用Tail-PCR对PB转座整合位点侧翼基因组序列进行检测并分析其特点。结果显示,11个成纤维细胞克隆和9个乳腺上皮细胞克隆中发生了PB转座子TTAA位点特异性整合,其整合位置分别位于绵羊不同染色体上,其中成纤维细胞中10个整合位点位于功能基因间,1个整合位点未检测到相关信息,乳腺上皮细胞9个整合位点均位于功能基因间(表2)。Table 2
表2
表2 PB转座系统在两种绵羊体细胞中整合位点的侧翼序列
Table 2
| 编号 | 5′重复末端 | 侧翼序列 | 位置 |
|---|---|---|---|
| 绵羊成纤维细胞 | |||
| #1 | TAGGGTTAA | CCGGTCGAGCGTGGCGTGTTGTGATTAATGAGAGCACCTCCCACT | Chr.1 — |
| #2 | TAGGGTTAA | AGCACCGCTGGCATCGCTATCTCCAGGTGCCTGGATCAAGGGGGG | Chr.1 基因间 |
| #3 | TAGGGTTAA | TATGATAGTTCTATTTGCTGCTGGCACATATGGAATTGTCACAGCTT | Chr.2 基因间 |
| #4 | TAGGGTTAA | GAATTTTCCAGAGTTTGTTATGATCCACACAAAGGCTTTGGCATAG | Chr.23 基因间 |
| #5 | TAGGGTTAA | AGGCTCAAGAGGGAAGGTGGAATTTCTCTCGAGACACCGCAGGG | Chr.9 基因间 |
| #6 | TAGGGTTAA | ACTAGAGAGGAACTCCAGGGATCATGCCCCTATTCCAAAAGACCC | Chr.15 基因间 |
| #7 | TAGGGTTAA | TCTGTCCCCAGGTGGGTATGGAGGCTAGTTGAGTTGACTGAGCTG | Chr.6 基因间 |
| #8 | TAGGGTTAA | CACATAGGGGTCTTTTGGAATGGTGGCAAGAACCCTGGAGCTCCT | Chr.6 基因间 |
| #9 | TAGGGTTAA | AAAAGTGCTGGAATGAAAACGTTGTTCATGTAATCACAGCCACCT | Chr.X 基因间 |
| #10 | TAGGGTTAA | AAAGGGAACCTTCCTGTTACATTATAGTTTCCTTAATTGCAAGAATC | Chr.13 基因间 |
| #11 | TAGGGTTAA | GAATAGCTTTCCCTTATGGTGGTCCTAAATTTCAAGGTAAGACATAC | Chr.18 基因间 |
| 绵羊乳腺上皮细胞 | |||
| #1 | TAGGGTTAA | TATCCCCTAATAATTTCTGGAAGTCTGACACAGGATACAGCATGCT | Chr.11 基因间 |
| #2 | TAGGGTTAA | GTTGCCTCAAGGGTGTCAAGGACCCTTTCGAGGATCAAGAAGGAA | Chr.11 基因间 |
| #3 | TAGGGTTAA | GTCGGTCCTCGGGGTGATCGTCGAGCCAGTGCAGGGGCGTCAGGT | Chr.21 基因间 |
| #4 | TAGGGTTAA | GGGTTTTTTGGTCCACCTTATAGTTTGTCTTCGATTAGTACTTTAATA | Chr.X 基因间 |
| #5 | TAGGGTTAA | TCTGTCCCCAGGTGGGTATGGAGGCTAGTTGAGTTGACTGAGCTG | Chr.6 基因间 |
| #6 | TAGGGTTAA | TGTAGAAATACAAGATTAGATTAGGGAGAAATAGTGTAGTAGGAAA | Chr.9 基因间 |
| #7 | TAGGGTTAA | AATATCTCCAACCTGAGTCACGGCGGAAACCGCCAAACTGACACT | Chr.1 基因间 |
| #8 | TAGGGTTAA | AAGCAAGCCGTATTACAGGTGATACATCTCACACACGGACGTGAC | Chr.1 基因间 |
| #9 | TAGGGTTAA | AATCAAAGCCCATGAAACCGTGCTGCACACCAGGAATTGTTAGAG | Chr.2 基因间 |
新窗口打开|下载CSV
2.5 PB转座效率检测结果
将pBNW-TP2转染到绵羊成纤维细胞和乳腺上皮细胞中,G418筛选细胞阳性克隆,进行质粒转染,对其进行PB转座效率检测,3次细胞克隆计数结果经非配对t检验(Student’s t-test)统计。结果显示,由pBNW-TP2载体介导的绵羊成纤维细胞克隆数与对照组差异极其显著(图8A,P<0.01),绵羊乳腺上皮细胞克隆数与对照组差异极其显著(图8B,P<0.01),外源基因在上述两种绵羊细胞中整合效率显著提升,提示pBNW-TP2载体以PB转座的方式高效介导了转基因过程。图8
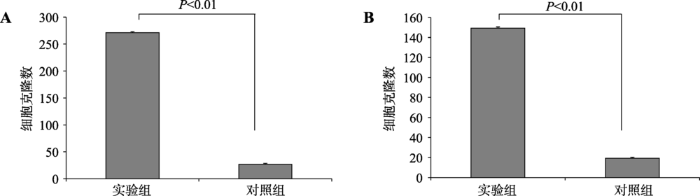 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8绵羊成纤维细胞和乳腺上皮细胞中PB转座效率统计分析
Fig. 8The statistical analysis of PB transposon integration efficiency in two ovine somatic cells
A:绵羊成纤维细胞PB转座效率统计结果;B:绵羊成纤维细胞PB转座效率统计结果。转染pBNW-TP2质粒为实验组,pBNW-TP1+empty vector(pCDNA4/HisMaxA)为对照组,P<0.01表示差异极显著。
3 讨 论
密码子优化和降低稀有密码子出现频率是基因异源高效表达的常用方法。在此过程中,需避免蛋白N端成串出现稀有密码子,这是提升蛋白翻译效率的必要手段[13]。此外,对宿主进行密码子偏好性分析,并据此对外源基因CDS区重新合成,使其密码子组成频率更接近宿主[14],可显著提升外源基因的表达效率。PB转座酶来自粉纹夜蛾,其密码子偏好性与绵羊差别较大,本研究中通过对绵羊常见密码子出现频率进行分析,将PBase原始序列中的98个氨基酸密码子进行修正(图2:A和B),使其密码子组成特征与绵羊相似。同时,为了维持PBasemRNA在细胞中的稳定性,将其GC含量调整至56.36%(图2C),从而为PBase基因在绵羊细胞内的高效稳定表达提供了保障,PB转座效率统计分析结果显示(图8),本研究构建的通用型载体pBNW- TP2可在绵羊成纤维细胞和乳腺上皮细胞中高效转座,表明针对PBase进行绵羊密码子偏好性优化,可有效介导绵羊体细胞中PB转座的发生。哺乳动物中具有活性的DNA转座子主要有SB (sleeping beauty)、Tol2和PB转座子[15],相比于另外两种转座子,PB转座子由于其外源基因负载容量大(100 kb)[16]、整合效率高[4]以及极强的可塑性等优点而被广泛应用于功能基因筛选[17]、基因治疗[18, 19, 20]、转基因动物生产[7, 21]、IPS诱导[22, 23]及RNA干扰[24]等转基因操作中。为了使该系统应用更加方便,PB转座子被改造成由一个供体质粒(包含5′TR和3′TR)和一个辅助质粒(表达PBase)组成的双质粒系统,且供体质粒和辅助质粒比例达到1:1可介导PB转座事件发生[25],而为了保证PB转座的有效进行,在实验中辅助质粒与供体质粒的比例通常高于1:1,从而引发PB转座酶持续过量,进而导致PB转座子供体质粒在宿主基因组中不断发生转座[26],造成潜在的遗传风险。研究表明,将供体质粒和辅助质粒整合为单质粒载体,在保证PB转座活性的同时可降低PB转座子在基因组中不断转座的概率[27],pBNW- TP2载体(图1)正是基于上述原因而构建,该载体作为一种PB转座子单质粒系统可以在绵羊成纤维和乳腺上皮细胞中高效介导转基因过程,并特异性整合至TTAA位点(表2,图8),具备PB转座子特性且提升了实验操作便捷性,是对传统的PB转座子双质粒系统的显著改进。
研究表明,PB转座子能在牛[28, 29]和山羊[30]等反刍动物成纤维细胞中发生转座,其整合位点多位于基因内含子区或基因间。在人HEK293细胞和Hela细胞中,PB转座子倾向于整合入基因的转录起始位点和长末端重复元件区域[31],在人T细胞中,PB转座子偏好整合于转录单位、CpG岛和转录起始位点区域[32]。本研究在绵羊成纤维细胞和乳腺上皮细胞中共获得20个PB转座子整合位点,其插入位置分别位于1号、2号、9号、15号、6号、11号、13号、18号、21号、23号和X染色体上(表2),核苷酸序列特征分析表明,19个整合位点位于功能基因组间,与牛和山羊细胞整合位点特征相似,提示PB转座子可能在反刍动物基因组中整合位点特征趋于一致。由于本研究在绵羊基因组中仅获得20个转座位点,其是否位于转录单位、CpG 岛和转录起始位点区域,则需要获取更多的绵羊PB转座位点进行深入研究。综上所述,PB转座子在绵羊体细胞中的整合位置倾向于功能基因间,对功能基因表达产生重大影响的可能性较小,安全性较高。
在绵羊体细胞中对PB转座子的相关特征进行分析目前尚未见报道,本研究证实PB转座子能在绵羊成纤维细胞和乳腺上皮细胞中以TTAA靶向性整合的方式介导外源基因插入(图7,图8),是对PB转座子在动物体细胞中介导转基因的一个有效补充。此外,我们在实际研究中发现,乳腺上皮细胞质粒转染较困难,转染后外源基因稳定整合至宿主基因组的效率不高,本研究发现PB转座子可携带外源基因在绵羊乳腺上皮细胞中发生PB转座,为绵羊乳腺细胞转基因相关操作提供了一种有效工具。
本研究通过对已有PB转座单质粒中PBase进行偏好绵羊密码子优化,构建了一种能在绵羊体细胞中进行高效转基因的通用性载体pBNW-TP2,通过对其转座特征进行分析,证实该载体可在绵羊成纤维和乳腺上皮细胞中以TTAA特异性整合的方式高效介导转基因过程,为PB转座子介导的绵羊体细胞转基因相关研究提供了理论参考和技术基础。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URL [本文引用: 1]
.
URL [本文引用: 1]
.
URL [本文引用: 1]
.
URLPMID:16096065 [本文引用: 2]

Transposable elements have been routinely used for genetic manipulation in lower organisms, including generating transgenic animals and insertional mutagenesis. In contrast, the usage of transposons in mice and other vertebrate systems is still limited due to the lack of an efficient transposition system. We have tested the ability of piggyBac ( PB), a DNA transposon from the cabbage looper moth Trichoplusia ni, to transpose in mammalian systems. We show that PB elements carrying multiple genes can efficiently transpose in human and mouse cell lines and also in mice. PB permits the expression of the marker genes it carried. During germline transposition, PB could excise precisely from original insertion sites and transpose into the mouse genome at diverse locations, preferably transcription units. These data provide a first and critical step toward a highly efficient transposon system for a variety of genetic manipulations including transgenesis and insertional mutagenesis in mice and other vertebrates.
.
URLPMID:21747215 [本文引用: 1]

Abstract Transgenic research on cattle embryos has been developed to date using viral or plasmid DNA delivery systems. In this study, a different gene delivery system, piggybac transposition, was employed to investigate if it can be applied for producing transgenic cattle embryos. Green or red fluorescent proteins (GFP or RFP) were transfected into donor fibroblasts, and then transfected donor cells were reprogrammed in enucleated oocytes through SCNT and developed into pre-implantation stage embryos. GFP was expressed in donor cells and in cloned embryos without any mosaicism. Induction of RFP expression was regulated by doxycycline treatment in donor fibroblasts and pre-implantational stage embryos. In conclusion, this study demonstrated that piggybac transposition could be a mean to deliver genes into bovine somatic cells or embryos for transgenic research.
URLPMID:21747215 [本文引用: 1]

Abstract Transgenic research on cattle embryos has been developed to date using viral or plasmid DNA delivery systems. In this study, a different gene delivery system, piggybac transposition, was employed to investigate if it can be applied for producing transgenic cattle embryos. Green or red fluorescent proteins (GFP or RFP) were transfected into donor fibroblasts, and then transfected donor cells were reprogrammed in enucleated oocytes through SCNT and developed into pre-implantation stage embryos. GFP was expressed in donor cells and in cloned embryos without any mosaicism. Induction of RFP expression was regulated by doxycycline treatment in donor fibroblasts and pre-implantational stage embryos. In conclusion, this study demonstrated that piggybac transposition could be a mean to deliver genes into bovine somatic cells or embryos for transgenic research.
.
URLPMID:22738962 [本文引用: 2]

PiggyBac (PB) has recently been found to be functional in various organisms. To verify and exploit its application in the cashmere goat, a PB transposon system including donor and helper vector of was developed, in which the EGFP gene in donor of vector was used as reporter. Cashmere goat fetal fibroblasts cells (GFFs) were transfected with the PB transposon system and the efficiency of gene transfer was determined. Compared with random integration, PB-mediated EGFP expression levels increased 7.78-fold in the GFFs, confirming that the PB transposon system constructed successfully mediated efficient foreign gene integration in the GFFs. To further investigate the characteristics of PB-mediated integration instance, PB integration site distribution in the goat genome was examined. The results showed that PB had a preference for AT rich regions of the goat genome. Thus this study confirms the function of PB transposon in GFFs and provides a potential genetic tool for producing transgenic goats.
.
URL [本文引用: 1]
.
[本文引用: 1]
.
URL [本文引用: 1]
.
URL [本文引用: 1]
URL [本文引用: 1]
.
URL [本文引用: 1]
.
URLPMID:3503753 [本文引用: 1]

Background Sustainable utilization of plant biomass as renewable source for fuels and chemical building blocks requires a complex mixture of diverse enzymes, including hydrolases which comprise the largest class of lignocellulolytic enzymes. These enzymes need to be available in large amounts at a low price to allow sustainable and economic biotechnological processes. Over the past years Pichia pastoris has become an attractive host for the cost-efficient production and engineering of heterologous (eukaryotic) proteins due to several advantages. Results In this paper codon optimized genes and synthetic alcohol oxidase 1 promoter variants were used to generate Pichia pastoris strains which individually expressed cellobiohydrolase 1, cellobiohydrolase 2 and beta-mannanase from Trichoderma reesei and xylanase A from Thermomyces lanuginosus. For three of these enzymes we could develop strains capable of secreting gram quantities of enzyme per liter in fed-batch cultivations. Additionally, we compared our achieved yields of secreted enzymes and the corresponding activities to literature data. Conclusion In our experiments we could clearly show the importance of gene optimization and strain characterization for successfully improving secretion levels. We also present a basic guideline how to correctly interpret the interplay of promoter strength and gene dosage for a successful improvement of the secretory production of lignocellulolytic enzymes in Pichia pastoris.
Magsci [本文引用: 1]

近年来, 转座子介导的插入突变在哺乳动物的分子遗传学研究中得到了广泛的应用。转座子作为一种简便高效的遗传学操作工具, 在构建转基因动物模型、基因治疗、细胞水平和动物整体水平的基因功能研究等方面发挥了重要的作用。文章重点介绍DNA转座子的结构、功能及其应用于小鼠分子遗传学领域的最新研究进展。
Magsci [本文引用: 1]

近年来, 转座子介导的插入突变在哺乳动物的分子遗传学研究中得到了广泛的应用。转座子作为一种简便高效的遗传学操作工具, 在构建转基因动物模型、基因治疗、细胞水平和动物整体水平的基因功能研究等方面发挥了重要的作用。文章重点介绍DNA转座子的结构、功能及其应用于小鼠分子遗传学领域的最新研究进展。
.
URLPMID:21948799 [本文引用: 1]

Abstract The development of technologies that allow the stable delivery of large genomic DNA fragments in mammalian systems is important for genetic studies as well as for applications in gene therapy. DNA transposons have emerged as flexible and efficient molecular vehicles to mediate stable cargo transfer. However, the ability to carry DNA fragments >10 kb is limited in most DNA transposons. Here, we show that the DNA transposon piggyBac can mobilize 100-kb DNA fragments in mouse embryonic stem (ES) cells, making it the only known transposon with such a large cargo capacity. The integrity of the cargo is maintained during transposition, the copy number can be controlled and the inserted giant transposons express the genomic cargo. Furthermore, these 100-kb transposons can also be excised from the genome without leaving a footprint. The development of piggyBac as a large cargo vector will facilitate a wider range of genetic and genomic applications. The Author(s) 2011. Published by Oxford University Press.
.
URLPMID:21185377 [本文引用: 1]

Transposons are an attractive system to use in genetic screens as they are molecularly tractable and the disrupted loci that give rise to the desired phenotype are easily mapped. We consider herein the characteristics of the piggyBac transposon system in complementing existing mammalian screen strategies, including the Sleeping Beauty transposon system. We also describe the design of the piggyBac resources that we have developed for both forward and reverse genetic screens, and the protocols we use in these experiments.
.
URLPMID:26742580 [本文引用: 1]

Abstract Selection of suitable delivery system is one of the crucial aspects in gene therapy that determines the efficiency of gene therapy. The past two decades have witnessed extensive efforts for finding safe and efficient vectors to overcome the limitations of viral vectors. The utilization of DNA transposon-based vectors for gene therapy has emerged as a promising non-viral alternative. DNA 'cut-and-paste' is one of the main mechanisms of genome engineering by transposon elements. However, the lack of an efficient transposition system has limited the utilization of transposon vectors in mice and mammalian systems. PiggyBac (PB) is known as a highly efficient DNA transposon originally isolated from Trichoplusia ni as an alternative to Sleeping Beauty (SB). It has been shown that PB can be functional in various species including mammalian systems. This vector could overcome some limitations of other vectors in cancer gene therapy. Some advantages of PB include the capacity for integration into the genome and providing a stable expression, capacity to harbor 10 and 9.1 b of foreign DNA into the host genome, without a significant reduction in their transposition activity and display non-overlapping targeting preferences. However, to advance PB to clinical applications, some obstacles still require to be overcome to improve its safety and efficiency. Hence, it seems that this vector could open new horizons in gene and cancer therapy.Cancer Gene Therapy advance online publication, 8 January 2016; doi:10.1038/cgt.2015.68.
.
URL [本文引用: 1]
.
URL [本文引用: 1]
.
URL [本文引用: 1]
.
URLPMID:19252478 [本文引用: 1]

Abstract Transgenic expression of just four defined transcription factors (c-Myc, Klf4, Oct4 and Sox2) is sufficient to reprogram somatic cells to a pluripotent state. The resulting induced pluripotent stem (iPS) cells resemble embryonic stem cells in their properties and potential to differentiate into a spectrum of adult cell types. Current reprogramming strategies involve retroviral, lentiviral, adenoviral and plasmid transfection to deliver reprogramming factor transgenes. Although the latter two methods are transient and minimize the potential for insertion mutagenesis, they are currently limited by diminished reprogramming efficiencies. piggyBac (PB) transposition is host-factor independent, and has recently been demonstrated to be functional in various human and mouse cell lines. The PB transposon/transposase system requires only the inverted terminal repeats flanking a transgene and transient expression of the transposase enzyme to catalyse insertion or excision events. Here we demonstrate successful and efficient reprogramming of murine and human embryonic fibroblasts using doxycycline-inducible transcription factors delivered by PB transposition. Stable iPS cells thus generated express characteristic pluripotency markers and succeed in a series of rigorous differentiation assays. By taking advantage of the natural propensity of the PB system for seamless excision, we show that the individual PB insertions can be removed from established iPS cell lines, providing an invaluable tool for discovery. In addition, we have demonstrated the traceless removal of reprogramming factors joined with viral 2A sequences delivered by a single transposon from murine iPS lines. We anticipate that the unique properties of this virus-independent simplification of iPS cell production will accelerate this field further towards full exploration of the reprogramming process and future cell-based therapies.
.
URLPMID:2677165 [本文引用: 1]

Abstract Induced pluripotent stem cells (iPSCs) have been generated from somatic cells by transgenic expression of Oct4 (Pou5f1), Sox2, Klf4 and Myc. A major difficulty in the application of this technology for regenerative medicine, however, is the delivery of reprogramming factors. Whereas retroviral transduction increases the risk of tumorigenicity, transient expression methods have considerably lower reprogramming efficiencies. Here we describe an efficient piggyBac transposon-based approach to generate integration-free iPSCs. Transposons carrying 2A peptide-linked reprogramming factors induced reprogramming of mouse embryonic fibroblasts with equivalent efficiencies to retroviral transduction. We removed transposons from these primary iPSCs by re-expressing transposase. Transgene-free iPSCs could be identified by negative selection. piggyBac excised without a footprint, leaving the iPSC genome without any genetic alteration. iPSCs fulfilled all criteria of pluripotency, such as pluripotency gene expression, teratoma formation and contribution to chimeras. piggyBac transposon-based reprogramming may be used to generate therapeutically applicable iPSCs.
.
URL [本文引用: 1]
.
URLPMID:18579772 [本文引用: 1]

Transposon systems are widely used for generating mutations in various model organisms. PiggyBac (PB) has recently been shown to transpose efficiently in the mouse germ line and other mammalian cell lines. To facilitate PB's application in mammalian genetics, we characterized the properties of the PB transposon in mouse embryonic stem (ES) cells. We first measured the transposition efficiencies of PB transposon in mouse embryonic stem cells. We next constructed a PB/SB hybrid transposon to compare PB and Sleeping Beauty (SB) transposon systems and demonstrated that PB transposition was inhibited by DNA methylation. The excision and reintegration rates of a single PB from two independent genomic loci were measured and its ability to mutate genes with gene trap cassettes was tested. We examined PB's integration site distribution in the mouse genome and found that PB transposition exhibited local hopping. The comprehensive information from this study should facilitate further exploration of the potential of PB and SB DNA transposons in mammalian genetics.
.
URL [本文引用: 1]
.
URL [本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
Magsci [本文引用: 1]

piggyBac(PB)转座子作为一种遗传工具被广泛应用于多个物种的转基因及插入突变研究, 目前PB转座子在牛中的相关研究还较少。为了获得PB转座子在牛基因组中的整合位点, 总结其转座特征, 文章构建了PB[CMV-EGFP]和pcDNA-PBase二元转座系统, 利用细胞核电转技术共转染牛耳组织成纤维细胞, 经G-418筛选, 获得了稳定转染EGFP的转基因细胞系; 提取细胞基因组DNA, 利用基因组步移技术扩增PB转座子5′ Bac区插入位置的DNA序列; 通过与牛基因组序列进行BLAST比对, 得到PB转座子在牛基因组中的插入位点。文章共获得了8个有效的整合位点, 但仅有5个位点定位到染色体1、2、11和X染色体上。序列分析表明:在牛基因组中, PB转座子可特异性的插入到“TTAA”位置, 并整合到基因间的非调控区; 分析整合位点“TTAA”相邻一侧的5个碱基组成, 发现PB转座子5′端倾向于插入到GC(62.5%)碱基富集区。该研究表明, PB转座子可以在牛基因组中发生转座, 获得的整合位点信息为利用PB转座子在牛上开展遗传学研究提供了理论参考。
Magsci [本文引用: 1]

piggyBac(PB)转座子作为一种遗传工具被广泛应用于多个物种的转基因及插入突变研究, 目前PB转座子在牛中的相关研究还较少。为了获得PB转座子在牛基因组中的整合位点, 总结其转座特征, 文章构建了PB[CMV-EGFP]和pcDNA-PBase二元转座系统, 利用细胞核电转技术共转染牛耳组织成纤维细胞, 经G-418筛选, 获得了稳定转染EGFP的转基因细胞系; 提取细胞基因组DNA, 利用基因组步移技术扩增PB转座子5′ Bac区插入位置的DNA序列; 通过与牛基因组序列进行BLAST比对, 得到PB转座子在牛基因组中的插入位点。文章共获得了8个有效的整合位点, 但仅有5个位点定位到染色体1、2、11和X染色体上。序列分析表明:在牛基因组中, PB转座子可特异性的插入到“TTAA”位置, 并整合到基因间的非调控区; 分析整合位点“TTAA”相邻一侧的5个碱基组成, 发现PB转座子5′端倾向于插入到GC(62.5%)碱基富集区。该研究表明, PB转座子可以在牛基因组中发生转座, 获得的整合位点信息为利用PB转座子在牛上开展遗传学研究提供了理论参考。
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URLPMID:19752750 [本文引用: 1]

Abstract The piggyBac transposon system represents a promising nonviral tool for gene delivery and discovery, and may also be of value for clinical gene therapy. PiggyBac is a highly efficient integrating vector that stably transfects (approximately 40%) of primary human T cells for potential adoptive immunotherapy applications. To evaluate the potential genotoxicity of piggyBac, we compared 228 integration sites in primary human T cells to integrations in 2 other human-derived cell lines (HEK293 and HeLa) and randomly simulated integrations into the human genome. Our results revealed distinct differences between cell types. PiggyBac had a nonrandom integration profile and a preference for transcriptional units (approximately 50% into RefSeq genes in all cell types), CpG islands (18% in T cells and 8% in other human cells), and transcriptional start sites (<5 kb, 16% to 20% in all cell types). PiggyBac also preferred TTAA but not AT-rich regions of the human genome. We evaluated the expression of mapped genes into which piggyBac integrated, and found selection of more active genes in primary human T cells compared with other human cell types, possibly due to concomitant T-cell activation during transposition. Importantly, we found that in comparison to what has been reported for gammaretroviral and human lenitviral vectors, piggyBac had decreased integration frequency into or within 50 kb of the transcriptional start sites of known proto-oncogenes. Hence the piggyBac nonviral gene delivery system seems to represent a promising gene transfer system for clinical applications using human T lymphocytes.
