 ,1,2,3
,1,2,3Post-transcriptional regulation of chicken PPARγ transcript variant 3 by upstream open reading frame
Yankai Chu1,2,3, Yanfei Jin1,2,3, Tianyu Xing1,2,3, Guangwei Ma1,2,3, Tingting Cui1,2,3, Xiaohong Yan1,2,3, Hui Li1,2,3, Ning Wang ,1,2,3
,1,2,3通讯作者:
编委: 赵要风
收稿日期:2018-02-7修回日期:2018-05-31网络出版日期:2018-08-16
| 基金资助: |
Received:2018-02-7Revised:2018-05-31Online:2018-08-16
| Fund supported: |
作者简介 About authors
褚衍凯,硕士研究生,专业方向:动物遗传育种与繁殖E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (796KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
褚衍凯, 靳艳飞, 邢天宇, 马广伟, 崔婷婷, 闫晓红, 李辉, 王宁. 鸡PPARγ基因转录本3上游开放阅读框转录后的调控作用[J]. 遗传, 2018, 40(8): 657-667 doi:10.16288/j.yczz.18-038
Yankai Chu, Yanfei Jin, Tianyu Xing, Guangwei Ma, Tingting Cui, Xiaohong Yan, Hui Li, Ning Wang.
过氧化物酶体增殖物激活受体γ (peroxisome proliferator-activated receptor gamma, PPARγ)是一个配体依赖型的转录因子,属于核内激素受体超家族成员。PPARγ被配体激活后可以与视黄醛X受体α(retinoid X receptor alpha, RXRα)形成异二聚体,结合于靶基因启动子区的过氧化物酶体增殖物反应元件(PPAR response elements, PPREs)来调控基因表 达[1, 2]。PPARγ是调控脂肪生成最关键的转录调控因子,也是许多脂类代谢基因的转录调控因子,与肥胖、Ⅱ型糖尿病和高血压等疾病密切相关[3,4,5]。研究发现,PPARγ基因是一个受多个启动子调控的基因,人类(Homo sapiens)和小鼠(Mus musculus)的PPARγ基因分别有4个和2个启动子[6, 7]。由于选择性剪接,人的PPARγ基因可以产生4种转录本—hPPARγs 1-4,小鼠的PPARγ基因可以产生2种转录本—mPPARγs 1-2,这些PPARγ转录异构体之间的差别主要是5′端非翻译区(5′ untranslated region, 5′UTR)序列和长度不同[6, 7]。本实验室前期研究发现,鸡(Gallus domesticus)PPARγ基因有3个启动子,由于使用不同启动子和选择性拼接,鸡可以产生5种不同的PPARγ转录本(cPPARγs 1-5),这5种转录本的差别也在于5′UTR序列和长度不同[8]。序列分析发现,鸡PPARγ基因5个转录本的5′UTR与哺乳动物PPARγ基因各转录本的5′UTR序列和长度均有较大差异[9],提示鸡PPARγ基因5′UTR的调控作用和机制可能与哺乳动物不同。
真核生物mRNA的5′UTR对基因表达具有重要的调控作用,既可以促进基因表达也可以抑制基因表达[10]。目前在真核生物5′UTR区已鉴定出大量的顺式调控元件,如5′帽结构、翻译起始基序、核糖体内部进入位点(internal ribosome entry sites, IRESs)、5′-末端寡嘧啶区(5′ terminal oligo-pyrimidine, 5′TOPs)、G-四链体(G-quadruplexes)、5′UTR的二级结构、上游AUG密码子(upstream AUG codons, uAUGs)和上游开放阅读框(upstream open reading frame, uORF)等[11, 12],这些调控元件可以在转录后以及翻译水平来调控基因表达[13]。其中,uORF是一个重要的顺式调控元件,哺乳动物uORF的转录后调控研究已经较为深入[14]。
uORF是由上游起始密码子(upstream ATG, uATG)和上游终止密码子(upstream stop codon, uSTOP)构成的一类短小的开放阅读框,一般位于5′UTR区。生物信息学分析发现,人类约49%的转录本含有uORF,小鼠约44%的转录本含有uORF,并且许多uORF在物种间是高度保守的,提示这些uORF序列是选择进化的结果[15, 16]。对哺乳动物的研究发现,在正常情况下,uORF可以通过降低下游编码区的翻译效率或引起mRNA降解来抑制基因表达。然而,当细胞处于应激状态时,uORF可以促进某些应激相关基因的mRNA表达[17]。
目前,对PPARγ基因的调控研究主要集中在哺乳动物上,研究内容主要包括PPARγ基因的转录调控和表观遗传调控等。但对PPARγ基因5′UTR转录后调控的研究报道还很少。本实验室前期通过生物信息学分析发现,与人和小鼠等哺乳动物PPARγ基因的转录本不同,鸡PPARγ基因的多个转录本5′UTR都存在uORF,但其调控作用还不清楚。为了揭示uORF在鸡PPARγ基因中的调控作用,本研究选择鸡PPARγ转录本3(cPPARγ3),采用基因定点突变和报告基因等技术对uORF的转录后调控作用进行 分析,证实uORF抑制鸡PPARγ基因的翻译,研究结果为揭示鸡PPARγ基因的转录后调控机制奠定基础。
1 材料和方法
1.1 材料
psiCHECKTM-2报告基因载体购自Promega公司(美国),pcDNA3.1(+)真核表达载体购自Invitrogen公司(美国)。鸡胚成纤维细胞系DF1购自上海生命科学研究院细胞资源中心。鸡永生化前脂肪细胞系(immortalized chicken pre-adipocytes, ICPA)为本实验室保存[17]。1.2 生物信息学分析
利用StarORF (http://star.mit.edu/index.html)、UTRscan (http://itbtools.ba.itb.cnr.it/utrscan)和RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw)软件预测鸡cPPARγ3的5′UTR区潜在的顺式调控元件。利用RNAFOLD (http://rna.tbi.univie.ac.at)、MFOLD (http:// mfold.rna.albany.edu)和GEEBEE (http://www.genebee. msu.su)软件预测鸡PPARγ基因5′UTR的高级结构。1.3 引物设计及合成
根据本实验室前期得到的鸡cPPARγ3的5′端mRNA序列(GenBank登录号:KP736528)设计qRT- PCR表达检测引物cPPARγ3-F1/cPPARγ3-R1;根据在Promega公司官方网站(www.promega.com/vectors/)中获取的psiCHECK?-2载体全长序列,设计hRluc基因的点突变引物hRluc-F1/hRluc-R1和qRT-PCR表达检测引物hRluc-F2/hRluc-R2,选择NONO基因(NM_001031532)作为内参基因。根据鸡PPARγ基因(NM_001001460.1)的mRNA序列,设计CDS区克隆引物cPPARγ-F/cPPARγ-R。所有引物均利用Primer Premier 5.0软件设计,由上海英骏生物技术有限公司合成。引物信息见表1。Table 1
表1
表1 本研究所使用的引物信息
Table 1
| 引物 | 序列(5′→3′) | 片段大小(bp) | 用途 |
|---|---|---|---|
| cPPARγ3-F1 | GAGCCTGCAACAAATTACAATGG | 166 | qRT-PCR |
| cPPARγ3-R1 | AGAAAAATAGGGAGGAGAAGGAGG | ||
| hRluc-F2 | ACCAAGACAAGATCAAGGCC | 195 | qRT-PCR |
| hRluc-R2 | GAACTCCTCAGGCTCCAGT | ||
| NONO-F | AGAAGCAGCAGCAAGAAC | 115 | qRT-PCR |
| NONO-R | TCCTCCATCCTCCTCAGT | ||
| cPPAR γ-F | GAATTCATGGTTGACACAGAAATGCCGT | 1451 | CDS区克隆 |
| cPPARγ -R | CCTCGAGGAGGATAAGAACTACTATCGCC | ||
| hRluc-F1 | ACTATAGGCTAGCCACCTTGGCTTCCAAGG | 6273 | hRluc点突变 |
| hRluc-R1 | AGGTGGCTAGCCTATAGTCATGAATTATGCTG |
新窗口打开|下载CSV
1.4 载体构建
1.4.1 分析5′UTR转录后调控的报告基因载体构建利用点突变试剂盒(全式金,北京)将psiCHECK?-2载体的hRluc基因的起始密码子ATG突变为TTG,突变后的载体命名为psiCHECK2M[18],测序验证无误后用于后续研究。
1.4.2 鸡cPPARγ3-5′UTR报告基因载体构建
将带有PPARγ基因起始密码子ATG的野生型cPPARγ3的5′UTR序列(WT)和uORF突变型的5′UTR序列(Mut,uORF中的uATG突变为终止密码子TGA)分别插入psiCHECK2M载体的hRluc基 因上游的NheⅠ酶切位点处,获得野生型和uORF突变型的5′UTR报告基因载体(psiCHECK2M- cPPARγ3-5′UTR-WT和psiCHECK2M-cPPARγ3- 5′UTR-Mut),测序验证无误后用于后续研究。
1.4.3 鸡cPPARγ3真核表达载体构建
利用引物cPPARγ-F/cPPARγ-R,以ICPA细胞的cDNA为模板,PCR扩增得到鸡PPARγ基因CDS片段(两端携带EcoRⅠ和XhoⅠ酶切位点)。将PCR产物和pcDNA3.1质粒分别用EcoRⅠ和XhoⅠ进行双酶切,回收纯化得到目的片段,使用T4连接酶连接得到重组质粒pcDNA3.1-CDS。人工合成鸡cPPARγ3野生型5′UTR片段和uORF突变的5′UTR片段(两端均携带NheⅠ和XhoⅠ酶切位点)克隆至pUC57载体中,分别命名为pUC57-WT-1和pUC57- Mut-1。用NheⅠ和XhoⅠ双酶切重组质粒pcDNA3.1- CDS,同时用NheⅠ和XhoⅠ分别双酶切pUC57- WT-1和pUC57-Mut-1,回收纯化获得目的片段,T4连接酶将目的片段与线性化质粒pcDNA3.1-CDS连接得到重组质粒pcDNA3.1-cPPARγ3-WT和pcDNA3.1-cPPARγ3-Mut,测序验证无误后用于后续研究。
1.4.4 鸡cPPARγ3 5′UTR启动子报告基因载体的构建
用XhoⅠ和HindⅢ双酶切pGL3-basic报告基因质粒,琼脂糖凝胶电泳后,胶回收得到线性化质粒。将人工合成的鸡cPPARγ3野生型5′UTR片段和uORF突变的5′UTR片段(两端均携带XhoⅠ和HindⅢ酶切位点)克隆至pUC57载体中,分别命名为pUC57-WT-2和pUC57-Mut-2。用XhoⅠ和HindⅢ
分别双酶切pUC57-WT-2和pUC57-Mut-2质粒,回收纯化后获得目的片段。使用T4连接酶将目的片段与线性化的pGL3-basic质粒连接,获得鸡cPPARγ3 5′UTR启动子活性分析用报告基因载体pGL3-basic- cPPARγ3-WT和pGL3-basic-cPPARγ3-Mut,测序验证无误后用于后续研究。
1.5 细胞培养
DF1细胞和ICPA细胞均培养在添加了10%胎牛血清及终浓度为100 U/mL青霉素、100 mg/L链霉素的DMEM培养基中,置于37℃含5% CO2和90%相对湿度的细胞培养箱中传代培养,24~48 h传代一次。1.6 双荧光素酶报告基因活性检测
将DF1细胞和ICPA细胞分别接种到24孔板中(2×105个/孔),待细胞汇合度达到50%~70%时,参考Lipofectamine®2000(Invitrogen, 美国)说明书,将报告基因质粒分别转染细胞,转染48 h后收集细胞,按双荧光素酶检测试剂盒(Promega, 美国)说明书操作,分别检测各组双荧光素酶报告基因活性。1.7 RNA提取及实时荧光定量PCR (qRT-PCR)
将重组质粒pcDNA3.1-cPPARγ3-WT和pcDNA3.1-cPPARγ3-Mut分别转染DF1细胞和ICPA细胞,转染48 h后,收集细胞,按照试剂盒RNeasy Plus Mini Kit(QIAGEN,德国)说明书提取细胞总RNA。取等量的RNA (1 μg)按照反转录试剂盒PrimeScript?RT reagent Kit with gDNA Erase(宝生物工程(大连)有限公司)说明书进行反转录,然后采用qRT-PCR方法检测cPPARγ3 mRNA表达。将psiCHECK2-cPPARγ3-5′UTR-WT和psiCHECK2- cPPARγ3-5′UTR-Mut分别转染DF1细胞和ICPA细胞,提取细胞总RNA,反转录后采用qRT-PCR检测hRluc基因的mRNA表达。qRT-PCR反应在ABI 7500荧光定量PCR仪上进行。反应体系为:FastStart Universal SYBR Green Master (ROX) (Roche,德国) 5 μL,cDNA模板1 μL,上、下游引物(10 μmol/L)各0.2 μL,水3.6 μL,总体积为10 μL。反应条件:95℃预变性10 min;95℃变性15 s,60℃复性延伸60 s,共40个循环。以NONO基因为内参基因,采用2-ΔΔCt法计算mRNA的相对表达量[19]。
1.8 Western blot分析
将重组质粒pcDNA3.1-cPPARγ3-WT和pcDNA3.1-cPPARγ3-Mut分别转染ICPA细胞,转染48 h后,用PBS洗细胞3遍,加入含有1 mmol/L PMSF的细胞裂解液,振荡混匀后冰上静置30 min,12 000 r/min离心5 min,取上清,用BCA方法测定蛋白浓度。蛋白样经过12% SDS-PAGE电泳分离后,电转至NC膜上,5%脱脂奶粉室温封闭1 h;PBST洗涤后加入1:1000稀释的鸡PPARγ一抗(实验室保存)室温振荡孵育2 h;PBST洗涤后加入1:5000稀释的辣根过氧化物酶标记的山羊抗兔二抗(北京中杉金桥生物技术有限公司)室温振荡孵育1 h,洗膜后进行ECL显色。内参对照为β-actin。1.9 数据分析
运用SAS 9.2软件(SAS Institute Inc.)分析实验数据,数据结果表示为平均数±标准误(x±SEM),采用t-test检验,n.s.表示差异不显著,*P<0.05为差异显著,**P<0.01,***P<0.001,****P<0.0001为差异极显著。2 结果与分析
2.1 鸡cPPARγ3 5′UTR区的生物信息学分析
鸡cPPARγ3的5′UTR区全长300 bp,利用StarORF (http://star.mit.edu/index.html)和UTRscan (http://itbtools.ba.itb.cnr.it/utrscan)软件分析鸡cPPARγ3 mRNA的5′UTR序列。结果显示,在-39/-28 (起始密码ATG的A为+1)区域存在一个uORF,长度为12 bp (图1A)。对uORF中的上游起始密码子uATG的序列环境分析发现,uATG和编码区ATG(main ATG, mATG)的序列环境相似,基本符合Kozak规则[20, 21](图1B)。图1
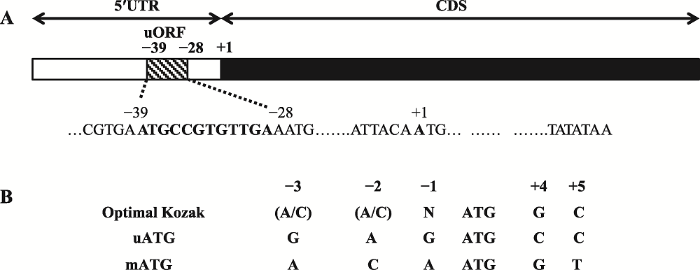 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1鸡PPARγ基因转录本3(cPPARγ3)5′UTR区的生物信息学分析
A:鸡PPARγ基因转录本3(cPPARγ3)5′UTR区uORF的预测分析;B:uORF的起始密码子(uATG)和编码区起始密码子(mATG)序列分析。
Fig. 1Bioinformatics analysis of the 5′UTR of chicken PPARγ transcript variant 3 (cPPARγ3)
2.2 鸡cPPARγ3的uORF对报告基因的转录后调控分析
为研究鸡cPPARγ3 5′UTR区uORF的转录后调控作用,本文首先构建了5′UTR转录后调控分析用的报告基因载体。将psiCHECK-2载体中hRluc基因的起始密码子ATG突变为TTG,使其不能发挥作用,将这一报告基因载体命名为psiCHECK2M。将3′ 末端携带起始密码子的5′UTR片段插入到hRluc基因上游的NheⅠ酶切位点,所插入的5′UTR片段作为hRluc基因的5′UTR,并使用5′UTR片段3′末端所携带的起始密码子ATG来起始翻译。利用这一构建的5′UTR报告基因载体可以分析5′UTR的转录后调控作用[18]。双荧光素酶报告基因结果显示,与野生型载体(psiCHECK-2)相比,hRluc基因的ATG突变后报告基因活性在DF1细胞和ICPA细胞中均极显著下降(P<0.001,图2A),表明本研究成功构建了用于分析5′UTR转录后调控作用的报告基因载体(psiCHECK2M),可以用于后续相关实验。图2
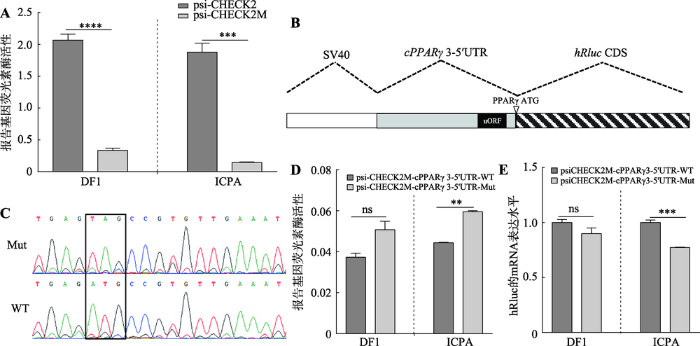 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2鸡cPPARγ3 5′UTR区uORF突变对报告基因活性和表达的影响
A:psiCHECK-2M报告基因载体的验证分析;B:cPPARγ3的5′UTR报告基因载体结构示意图;C:5′UTR野生型报告基因载体(psiCHECK2M-cPPARγ3-5′UTR-WT)和uORF突变的5′UTR报告基因载体(psiCHECK2M-cPPARγ3-5′UTR-Mut)测序比对(黑色方框位置为uORF的突变位点);D:双荧光素酶报告基因技术检测uORF突变对hRluc报告基因活性的影响;E:qRT-PCR检测uORF突变对hRluc基因 mRNA表达的影响。**:P<0.01;***:P<0.001;****:P<0.0001;ns表示差异不显著。
Fig. 2The effects of uORF mutaiton in cPPARγ3 5′UTR on reporter activity and mRNA expression
将cPPARγ3野生型的5′UTR序列(WT)和uORF突变型的5′UTR序列(Mut)别插入到psiCHECK2M载体hRluc基因上游的NheⅠ酶切位点(图2,B和C)。双荧光素酶报告基因检测结果表明:在ICPA细胞中,psiCHECK2M-cPPARγ3-5′UTR-Mut的报告基因活性极显著高于野生型(psiCHECK2M-cPPARγ3- 5′UTR-WT) (P<0.01,图2D);在DF1细胞中,psiCHECK2M-cPPARγ3-5′UTR-Mut的hRluc报告基因活性
高于野生型(psiCHECK2M-cPPARγ3-5′UTR-WT),但没有达到统计学差异显著(P>0.05,图2D)。为了进一步分析uORF的调控机制,本研究采用qRT-PCR方法分别检测了psiCHECK2M-cPPARγ3-5′UTR-WT和psiCHECK2M-cPPARγ3-5′UTR-Mut转染细胞中hRluc基因的mRNA表达情况。结果显示:在ICPA细胞中,uORF突变后,hRluc基因mRNA表达水平极显著降低(P<0.001,图2E);在DF1细胞中,uORF突变后,hRluc基因的mRNA表达水平低于野生型,但是差异不显著(P>0.05,图2E)。上述数据表明,uORF可能通过抑制下游基因的翻译来调控基因表达。
2.3 鸡cPPARγ3 uORF对PPARγ基因的转录后调控分析
本文进一步分析了5′UTR区的uORF对cPPARγ3的转录后调控作用,将构建的重组质粒pcDNA3.1- cPPARγ3-WT和pcDNA3.1-cPPARγ3-Mut分别转染DF1细胞和ICPA细胞,检测鸡cPPARγ3的mRNA和蛋白表达。qRT-PCR检测结果显示,在这两种细胞中,与pcDNA3.1-cPPARγ3-WT转染细胞相比,pcDNA3.1-cPPARγ3-Mut转染细胞后,cPPARγ3的mRNA表达显著降低(P<0.05) (图3A)。Western blot结果显示,与pcDNA3.1-cPPARγ3-WT转染细胞相比,pcDNA3.1-cPPARγ3-Mut转染细胞的PPARγ蛋白表达显著升高,大约上调2.3倍(P<0.0001) (图3,B和C)。上述结果表明,uORF通过抑制鸡PPARγ基因的翻译来调控其表达。图3
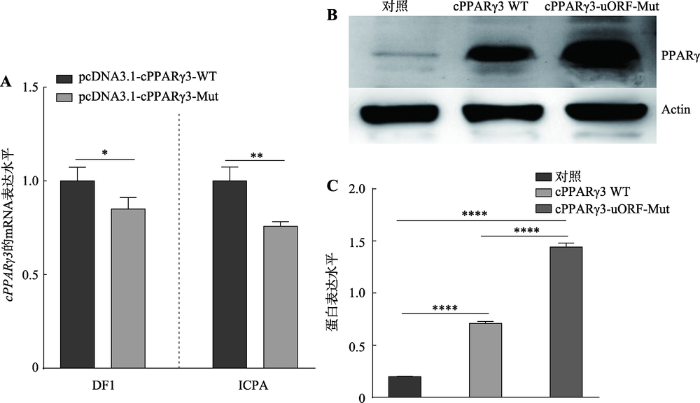 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图35′UTR区uORF突变对cPPARγ3的mRNA和蛋白表达的影响
A:qRT-PCR检测5′UTR区uORF突变对鸡cPPARγ3 mRNA表达的影响;B:Western blot检测5′UTR区uORF突变对鸡PPARγ蛋白表达的影响;C:uORF突变对鸡PPARγ蛋白表达影响的定量分析。*:P<0.05;**:P<0.01;****:P<0.0001。
Fig. 3The effects of 5′UTR uORF mutaiton on cPPARγ3 mRNA and protein levels
2.4 鸡cPPARγ3 5′UTR区的启动子活性检测
qRT-PCR检测发现,uORF突变会导致hRluc基因和鸡cPPARγ3的mRNA表达水平下降(图2E,图3A),这提示5′UTR相应的DNA序列可能具有启动子活性,uORF的突变可能影响5′UTR的启动子活性,从而引起下游基因mRNA表达变化。为确定鸡cPPARγ3-5′UTR区是否具有启动子活性,本文将构建的野生型(pGL3-basic-cPPARγ3-WT)和uORF突变的5′UTR启动子报告基因载体(pGL3-basic- cPPARγ3-Mut)分别转染ICPA细胞和DF1细胞。双荧光素酶报告基因检测结果显示,在ICPA细胞和DF1细胞中,野生型和uORF突变的启动子报告基因载体的活性均极显著的高于pGL3-basic空载体,活性约是pGL3-basic空载体的20倍(图4,A和B)。与野生型报告基因载体相比,uORF突变后,启动子活性在两种细胞中均极显著下降,分别下降约21%和13%(P<0.01)(图4,A和B)。这些结果表明,鸡cPPARγ3 5′UTR区序列具有启动子活性,uORF突变会导致启动子活性显著降低,这与上述hRluc基因和cPPARγ3的mRNA表达变化相一致。图4
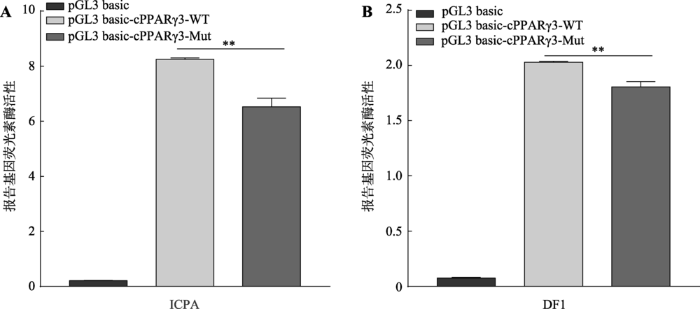 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4鸡cPPARγ3 5′UTR区的启动子活性检测
A:cPPARγ3 5°UTR区在ICPA细胞中的启动子活性检测分析;B:cPPARγ3 5°UTR区在DF1细胞中的启动子活性检测分析。**:P<0.01。
Fig. 4The promoter activity analysis of the 5′UTR of cPPARγ3
3 讨 论
uORF是一种重要的顺式调控元件,通常可以抑制目的基因的翻译[22]。如癌基因mdm2在正常细胞中主要表达含有2个uORFs的L-mdm2转录本,这些uORFs抑制了mdm2基因的翻译,mdm2蛋白表达量很低,因此不致癌;癌细胞中mdm2基因主要表达缺少uORF的S-mdm2转录本,mdm2蛋白表达量比正常细胞高20倍以上[23]。当机体处于应激状态时,uORF可以促进ATF4、CAT-1等应激相关基因的翻译[24]。此外,有研究发现一些uORF对蛋白的翻译没有影响[25, 26]。本研究发现鸡cPPARγ3 5′UTR区的uORF突变后,cPPARγ3的mRNA表达水平显著下降(图3A),但是鸡cPPARγ3蛋白表达水平极显著升高(图3,B和C),说明该uORF抑制鸡cPPARγ3的翻译。真核生物mRNA通常以“核糖体扫描”机制进行翻译[27],由于5′UTR区的uORF位于基因编码区阅读框(main open reading frame, mORF)的上游,会优先被扫描,所以uORF的序列组成、二级结构等都会影响基因下游编码区的翻译。研究发现,uORF可以通过多种方式调控基因表达:(1)核糖体在扫描过程中将uORF当做为编码区ORF (mORF)进行翻译,导致mORF的翻译受到抑制[28];(2)核糖体扫描并翻译uORF,在翻译的延伸期或终止期形成“路障”,阻碍核糖体继续向3′端扫描或引起翻译起始复合物的解体从而抑制蛋白翻译;(3) uORF可以引起无义介导的mRNA降解(nonsense-mediated mRNA decay, NMD),使模板mRNA的降解,无法翻译下游的编码区[29]。本研究发现,cPPARγ3的uATG基本符合Kozak规则,提示该uATG在翻译起始阶段可能被核糖体识别并起始翻译,从而降低了下游编码区的翻译效率。本研究证实cPPARγ3的uORF显著抑制鸡PPARγ的翻译(图3,B和C),但uORF抑制cPPARγ3蛋白翻译的具体作用机制还有待于进一步探究。
目前已发现许多基因的5′UTR所对应的DNA序列也是其启动子的一部分,具有启动子活性。例如,研究发现CCN1、CCN2、CCN4基因的5′UTR区都有启动子活性[30,31]。本研究发现,鸡cPPARγ3 5′UTR对应的DNA序列也具有启动子活性,且uORF中的uATG突变会导致启动子活性显著降低 (图4,A和B),这与hRluc基因和cPPARγ3的mRNA表达变化相一致(图2E,图3A)。本研究构建的5′UTR报告基因载体和cPPARγ3真核表达载体中,5′UTR区对应DNA序列在转录时可以作为转录调控元件调控下游基因的转录;当被转录为mRNA的5′UTR后,它又能作为转录后调控元件发挥转录后调控作用。这是uORF突变导致下游基因(hRluc和cPPARγ3)的蛋白和mRNA表达不一致的原因。
在uORF转录后调控分析中,与野生型psiCHECK2M-cPPARγ3-5′UTR-WT相比,在ICPA细胞中psiCHECK2M-cPPARγ3-5′UTR-Mut的报告基因活性极显著升高,hRluc基因的mRNA表达显著下降;但是在DF1细胞中,尽管变化趋势与ICPA细胞中的结果相似,但psiCHECK2M-cPPARγ3-5′UTR-Mut的报告基因活性和报告基因hRluc的mRNA变化幅度均未达到统计学显著水平(图2,D和E)。与本研究情况类似,人PTCH1b基因5′UTR的uORF在HEK293T和MCF7细胞中的调控强度也存在差 异[32]。我们推测,cPPARγ3的uORF发挥作用需要RNA结合蛋白、非编码RNA以及下游基因mRNA等相互作用,而ICPA和DF1细胞所表达的RNA结合蛋白和非编码RNA的种类和含量可能存在一定的差异,导致uORF在这两种细胞中的作用强度 不同。
研究发现,不同5′末端序列的PPARγ转录异构体有不同的组织表达模式,如人和小鼠的转录异构体PPARγ1呈现广泛的组织表达,而PPARγ2的表达主要限于脂肪组织,人的转录异构体hPPARγ3主要在白色脂肪组织、结肠和巨噬细胞中表达[33]。同样,本实验室前期研究发现,鸡PPARγ基因不同转录本的组织表达谱也不同[9],这提示5′UTR对各个转录本的调控作用可能不同。人PPARγ基因各转录本的5′UTR区由于其二级结构和包含的调控元件存在差异致使5′UTR对PPARγ的翻译有不同的调控作 用[34]。鸡PPARγ基因的5′UTR与人和鼠的同源性很低,与人和鼠的PPARγ基因的5′UTR区不同,鸡PPARγ基因的5′UTR存在uORF,提示PPARγ基因在鸡和哺乳动物中的转录后调控机制存在差异。PPARγ在脂肪生成、糖脂代谢等方面都发挥重要的调控作用,而鸟类的这些生物学过程均与哺乳动物有较大的区别,如鸡的脂肪组织对胰岛素不敏感,鸡血清中的葡萄糖浓度是哺乳动物的2倍[35,36],鸡PPARγ基因的uORF可能是导致这些差异的原因之一。研究发现,人类许多基因由于碱基突变可以产生新的uORF,这些uORF会导致基因表达异常和疾病的发生。例如,LDLR基因一个C碱基的缺失突变,产生一个新的uORF,抑制了LDLR基因的翻译,并最终引起人类家族性高胆固醇血症[37];KCNJ11基因中-34位的C/T突变产生了一个新的uORF,该uORF抑制了KCNJ11基因的翻译,从而增加了先天性高胰岛素发生的风险[38]。未来有必要进一步深入探究鸡PPARγ基因中uORF在脂肪生物学中的作用和作用机制。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URL [本文引用: 1]
Magsci [本文引用: 1]

为分析鸡<em>PPARγ</em>基因的组织表达特性及其在脂肪细胞增殖和分化过程中的功能, 文章以东北农业大学高、低腹脂双向选择品系肉鸡为实验材料, 利用Western blotting方法, 检测<em>PPARγ</em>基因的组织表达特性及其在高、低脂系肉鸡腹部脂肪组织间的表达差异; 采用RNAi技术, 在鸡原代脂肪细胞中抑制<em>PPARγ</em>基因的表达后, 通过MTT和油红O提取比色的方法, 研究鸡<em>PPARγ</em>基因对脂肪细胞增殖和分化的调控作用; 利用Real-time PCR和Western blotting技术, 分析<em>PPARγ</em>基因表达下调后, 其他脂肪细胞分化转录因子以及与脂肪细胞分化相关的重要基因的表达变化情况。结果表明, <em>PPARγ</em>基因在7周龄高脂系肉鸡腹部脂肪组织、肌胃、脾脏、肾脏组织中表达量较高, 在心脏中表达量较低, 在肝脏、胸肌、腿肌、十二指肠中未检测到表达信号; 与高脂系相比, <em>PPARγ</em>基因在5和7周龄低脂系肉鸡腹部脂肪组织中的表达量较低(<em>P</em><0.05); <em>PPARγ</em>基因的表达量下降后, 鸡脂肪细胞的增殖能力增强, 分化能力减弱; 同时, <em>C/EBPα、SREBP1、A-FABP、Perilipin1、LPL、IGFBP-2</em>基因的表达量均下降(<em>P</em><0.05)。由此可见, PPARγ基因的表达可能与肉鸡腹部脂肪的沉积有一定的关系, 该基因可能是调控鸡脂肪细胞增殖与分化的关键因子。
Magsci [本文引用: 1]

为分析鸡<em>PPARγ</em>基因的组织表达特性及其在脂肪细胞增殖和分化过程中的功能, 文章以东北农业大学高、低腹脂双向选择品系肉鸡为实验材料, 利用Western blotting方法, 检测<em>PPARγ</em>基因的组织表达特性及其在高、低脂系肉鸡腹部脂肪组织间的表达差异; 采用RNAi技术, 在鸡原代脂肪细胞中抑制<em>PPARγ</em>基因的表达后, 通过MTT和油红O提取比色的方法, 研究鸡<em>PPARγ</em>基因对脂肪细胞增殖和分化的调控作用; 利用Real-time PCR和Western blotting技术, 分析<em>PPARγ</em>基因表达下调后, 其他脂肪细胞分化转录因子以及与脂肪细胞分化相关的重要基因的表达变化情况。结果表明, <em>PPARγ</em>基因在7周龄高脂系肉鸡腹部脂肪组织、肌胃、脾脏、肾脏组织中表达量较高, 在心脏中表达量较低, 在肝脏、胸肌、腿肌、十二指肠中未检测到表达信号; 与高脂系相比, <em>PPARγ</em>基因在5和7周龄低脂系肉鸡腹部脂肪组织中的表达量较低(<em>P</em><0.05); <em>PPARγ</em>基因的表达量下降后, 鸡脂肪细胞的增殖能力增强, 分化能力减弱; 同时, <em>C/EBPα、SREBP1、A-FABP、Perilipin1、LPL、IGFBP-2</em>基因的表达量均下降(<em>P</em><0.05)。由此可见, PPARγ基因的表达可能与肉鸡腹部脂肪的沉积有一定的关系, 该基因可能是调控鸡脂肪细胞增殖与分化的关键因子。
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:25450819 [本文引用: 1]

Peroxisome proliferator-activated receptor gamma (PPARγ) has been the focus of intense research because ligands for this receptor have emerged as potent insulin sensitizers used in the treatment of type 2 diabetes. There have been described three PPAR isotypes α, δ and γ which have an integrated role in controlling the expression of genes playing key roles in the storage and mobilization of lipids, in glucose metabolism, in morphogenesis and inflammatory response. Recent advances include the discovery of novel genes that are regulated by PPARγ, which helps to explain how activation of this adipocyte predominant transcription factor regulates glucose and lipid homeostasis. Increased levels of circulating free fatty acids and lipid accumulation in non-adipose tissue have been implicated in the development of insulin resistance. This situation is improved by PPARγ ligands, which promotes fatty acid storage in fat deposits and regulates the expression of adipocyte-secreted hormones that impacts on glucose homeostasis. So the net result of the pleiotropic effects of PPARγ ligands is improvement of insulin sensitivity. This review highlights the roles that PPAR gamma play in the regulation of gene expression of multiple diseases including obesity, diabetes and cancer and highlights the gene isolation transformation role. Further studies are needed for the transformation of PPAR gamma gene in plants and evaluate in animals for the treatment of type 2 diabetes.
URL [本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 2]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:26896445 [本文引用: 1]

Regulation of gene expression is fundamental in establishing cellular diversity and a target of natural selection. Untranslated mRNA regions (UTRs) are key mediators of post-transcriptional regulation. Previous studies have predicted thousands of ORFs in 5 UTRs, the vast majority of which have unknown function. Here, we present a systematic analysis of the translation and function of upstream open reading frames (uORFs) across vertebrates. Using high-resolution ribosome footprinting, we find that (i) uORFs are prevalent within vertebrate transcriptomes, (ii) the majority show signatures of active translation, and (iii) uORFs act as potent regulators of translation and RNA levels, with a similar magnitude to miRNAs. Reporter experiments reveal clear repression of downstream translation by uORFs/oORFs. uORF number, intercistronic distance, overlap with the CDS, and initiation context most strongly influence translation. Evolution has targeted these features to favor uORFs amenable to regulation over constitutively repressive uORFs/oORFs. Finally, we observe that the regulatory potential of uORFs on individual genes is conserved across species. These results provide insight into the regulatory code within mRNA leader sequences and their capacity to modulate translation across vertebrates. Synopsis
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:20965418 [本文引用: 2]

A number of stresses, including nutrient stress, temperature shock, DNA damage, and hypoxia, can lead to changes in gene expression patterns caused by a general shutdown and reprogramming of protein synthesis. Each of these stress conditions results in selective recruitment of ribosomes to mRNAs whose protein products are required for responding to stress. This recruitment is regulated by elements within the 5′ and 3′ untranslated regions of mRNAs, including internal ribosome entry segments, upstream open reading frames, and microRNA target sites. These elements can act singly or in combination and are themselves regulated by trans-acting factors. Translational reprogramming can result in increased life span, and conversely, deregulation of these translation pathways is associated with disease including cancer and diabetes.
[本文引用: 2]
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:16213112 [本文引用: 1]

The mechanism of initiation of translation differs between prokaryotes and eukaryotes, and the strategies used for regulation differ accordingly. Translation in prokaryotes is usually regulated by blocking access to the initiation site. This is accomplished via base-paired structures (within the mRNA itself, or between the mRNA and a small trans-acting RNA) or via mRNA-binding proteins. Classic examples of each mechanism are described. The polycistronic structure of mRNAs is an important aspect of translational control in prokaryotes, but polycistronic mRNAs are not usable (and usually not produced) in eukaryotes. Four structural elements in eukaryotic mRNAs are important for regulating translation: (i) the m7G cap; (ii) sequences flanking the AUG start codon; (iii) the position of the AUG codon relative to the 5 end of the mRNA; and (iv) secondary structure within the mRNA leader sequence. The scanning model provides a framework for understanding these effects. The scanning mechanism also explains how small open reading frames near the 5 end of the mRNA can down-regulate translation. This constraint is sometimes abrogated by changing the structure of the mRNA, sometimes with clinical consequences. Examples are described. Some mistaken ideas about regulation of translation that have found their way into textbooks are pointed out and corrected.
URLPMID:1008439 [本文引用: 1]

ABSTRACT Continuing discoveries of new and surprising mechanisms of gene regulation suggest that our understanding of this complex and ubiquitous biological process remains incomplete. Emerg- ing examples illustrate that many and perhaps all genes are regulated at multiple steps including transcription, posttran- scriptional processing, nuclear export and localization, stabil- ity, and translation of mature mRNA molecules. Translation itself is regulated by a diverse collection of mechanisms that act not only at the initiation step but also during elongation and termination and even after termination. Among the various cis elements in mRNAs (43) that partic- ipate in regulating translation are AUG codons within tran- script leaders (upstream AUGs (uAUGs)) and, in some cases, associated upstream open reading frames (uORFs). Based on a 1987 survey, less than 10% of eukaryotic mRNAs contain AUG codons within their transcript leader regions (often er- roneously referred to as 59 untranslated regions). However, uAUGs are conspicuously common in certain classes of genes, including two-thirds of oncogenes and many other genes in- volved in the control of cellular growth and differentiation (29, 31, 42). Despite the wealth of sequence data being generated by large-scale sequencing projects, extracting an up-to-date, comprehensive, and accurate estimate of the number of genes with uORFs is a formidable task. Only a minority of database entries are based on careful mRNA mapping data with anno- tations that identify the precise start of the transcript leader. Moreover, the use of alternative transcriptional start sites, alternative RNA processing, and alternative initiation codons complicates the determination of what exactly constitutes the transcript leader. Nonetheless, it is clear that uAUGs are not uncommon in genes with critical cellular roles, and identifying when and how they function is necessary if we are to achieve a comprehensive understanding of the interesting genes that contain these elements and of eukaryotic gene regulation in general. Some of the general principles by which uORFs participate in translational control are beginning to be understood. In this article, we first review these principles, which include the pro- cess of recognition of uORFs, regulation of reinitiation at downstream cistrons after translation of uORFs, and regula- tory effects of peptides encoded by uORFs. We then illustrate how these principles are applied by reviewing several specific examples where the roles of uORFs in translational control have been well characterized.
[本文引用: 1]
URL [本文引用: 1]

ABSTRACT Transport of the essential amino acids arginine and lysine is critical for the survival of mammalian cells. The adaptive response to nutritional stress involves increased translation of the arginine/lysine transporter (cat-1) mRNA via an internal ribosome entry site (IRES) within the mRNA leader. Induction of cat-1 IRES activity requires both translation of a small upstream open reading frame (uORF) within the IRES and phosphorylation of the translation initiation factor eIF2 . We show here that translation of the upstream ORF unfolds an inhibitory structure in the mRNA leader, eliciting a conformational change that yields an active IRES. The IRES, whose activity is induced by amino acid starvation, is created by RNA-RNA interactions between the 5 end of the leader and downstream sequences. This study suggests that the structure of the IRES is dynamic and regulation of this RNA structure is a mechanism of translational control.
URL [本文引用: 1]
URLPMID:1516788842 [本文引用: 1]

Abstract The aspartyl protease BACE1 has a pivotal role in the pathogenesis of Alzheimer's disease. Recently, it was shown that in Alzheimer's disease patients, BACE1 levels were elevated although mRNA levels were not changed compared with controls. Here, we demonstrate that the 5'-untranslated region (5'UTR) of BACE1 controls the rate of BACE1 translation. In the presence of the 5'UTR, we observed more than 90% reduction of BACE1 protein levels in HEK293, COS7 and H4 cells, and a similar reduction of BACE1 activity in vitro. mRNA levels were not affected, demonstrating that the 5'UTR repressed the translation but not the transcription of BACE1. The 3'UTR did not affect BACE1 expression. An extensive mutagenesis analysis predicts that the GC-rich region of the 5'UTR forms a constitutive translation barrier, which may prevent the ribosome from efficiently translating the BACE1 mRNA. Our data therefore demonstrate translational repression as a new mechanism controlling BACE1 expression.
URLPMID:27313038 [本文引用: 1]

The eukaryotic 5' untranslated region (UTR) is critical for ribosome recruitment to the messenger RNA (mRNA) and start codon choice and plays a major role in the control of translation efficiency and shaping the cellular proteome. The ribosomal initiation complex is assembled on the mRNA via a cap-dependent or cap-independent mechanism. We describe various mechanisms controlling ribosome scanning and initiation codon selection by 5' upstream open reading frames, translation initiation factors, and primary and secondary structures of the 5'UTR, including particular sequence motifs. We also discuss translational control via phosphorylation of eukaryotic initiation factor 2, which is implicated in learning and memory, neurodegenerative diseases, and cancer.
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URLPMID:9821958 [本文引用: 1]

Abstract PPARgamma is a member of the peroxisome proliferator activated receptors (PPAR) subfamily of nuclear receptors. So far two PPARgamma isoforms, PPARgamma1 and PPARgamma2, were known in mammals. We describe the structure of a novel human PPARgamma subtype, PPARgamma3. The PPARgamma3 mRNA is transcribed from a novel promoter localized 5' of exon A2. PPARgamma3 mRNA expression was restricted to adipose tissue and large intestine. Similar to human PPARgamma1 and -2, PPARy3 is activated by thiazolidinediones and prostaglandin J derivatives and binds with high affinity to a PPRE.
[本文引用: 1]
URL [本文引用: 1]
URLPMID:25015647 [本文引用: 1]

Abstract Gene loss is one of the main drivers in the evolution of genomes and species. The demonstration that a gene has been lost by pseudogenization is truly complete when one finds the pseudogene in the orthologous genomic region with respect to active genes in other species. In some cases, the identification of such orthologous loci is not possible because of chromosomal rearrangements or if the gene of interest has not yet been sequenced. This question is particularly important in the case of birds because the genomes of avian species possess only about 15,000 predicted genes, in comparison with 20,000 in mammals. Yet, gene loss raises the question of which functions are affected by the changes in gene counts. We describe a systematic approach that makes it possible to demonstrate gene loss in the chicken genome even if a pseudogene has not been found. By using phylogenetic and synteny analysis in vertebrates, genome-wide comparisons between the chicken genome and expressed sequence tags, RNAseq data analysis, statistical analysis of the chicken genome, and radiation hybrid mapping, we show that resistin, TNF , and PAI-1 (SERPINE1), three genes encoding adipokines inhibiting insulin sensitivity, have been lost in chicken and zebra finch genomes. Moreover, omentin, a gene encoding an adipokine that enhances insulin sensitivity, has also been lost in the chicken genome. Overall, only one adipokine inhibiting insulin sensitivity and five adipokines enhancing insulin sensitivity are still present in the chicken genome. These genetic differences between mammals and chicken, given the functions of the genes in mammals, would have dramatic consequences on chicken endocrinology, leading to novel equilibriums especially in the regulation of energy metabolism, insulin sensitivity, as well as appetite and reproduction. The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
URLPMID:15823276 [本文引用: 1]

Familial hypercholesterolaemia (FH) is an autosomal dominant disorder of lipoprotein metabolism. In the majority of patients FH is caused by mutations in the gene for the low-density lipoprotein receptor (LDLR), and to date more than 700 mutations have been reported worldwide. In this study, 36 paediatric patients with a clinical diagnosis of FH (20 homozygous and 16 heterozygotes) were screened for mutations in the LDLR gene. Each exon, with intron xon junctions, was screened by capillary fluorescent SSCP (F-SSCP) and heteroduplex analysis. Samples showing different band patterns were sequenced. Ten novel (including three frame shift small deletions or insertions) and seven known mutations were detected. A total of 37 out of the predicted 56 FH-causing alleles were identified (66.1%). No patients with the R3500Q mutation in the APOB gene were found. W556R was the most common mutation, explaining 21.4% of the predicted defective LDLR alleles. The novel sequence changes were deemed to be pathogenic if they altered a conserved amino acid (L143P, D147E, Q233H-C234G, C347G) or occurred in or close to a splice site (IVS 16 + 5) and were absent in DNA from 50 healthy Turkish subjects. These data confirm the genetic heterogeneity of FH in Turkey, and demonstrate the usefulness of F-SSCP for mutation detection.
URLPMID:12364426 [本文引用: 1]

Mutations in genes encoding the two subunits of the beta-cell ATP-sensitive potassium channel (K(ATP)) channel (SUR1 and Kir6.2) are the major cause of congenital hyperinsulinism (CHI). In this study, the K(ATP) channel genes were screened in a population-based study that included all verified Finnish CHI patients (n = 43) in a 27-yr period. Seven different mutations were identified, which accounted for 60% of all cases. The functional consequences of the major missense mutations were studied in vivo by determining acute (1-3 min) plasma insulin and C-peptide responses to calcium (n = 18), glucose (n = 12), and tolbutamide (n = 11) in those CHI patients who were able to take part in these studies. C-peptide and insulin responses to calcium were significantly higher in the patients with SUR1-E1506K mutation, compared with patients without K(ATP) channel mutations. The patients with SUR1-V187D mutation showed a reduced response to tolbutamide but unexpectedly did not show any response to calcium stimulation. A compound heterozygous patient with Kir6.2-(-54)/K67N mutations responded to calcium but also to tolbutamide. In conclusion, our results show that a positive response in the calcium test is indicative of a K(ATP) channel mutation, but all mutations cannot be identified with this method. The insulin response to tolbutamide in patients with SUR1 mutations is impaired to different extents, depending on the genotype. The combination of calcium and tolbutamide tests is a useful tool for the detection of CHI patients with K(ATP) channel dysfunction. Our results, however, also demonstrate the complexity of these responses and the difficulties in their interpretation.
