 ,*华中农业大学, 作物遗传改良国家重点实验室, 武汉 430070
,*华中农业大学, 作物遗传改良国家重点实验室, 武汉 430070Chinese Scientists Reveal Genome-wide Off-targeted Editing of Cytosine Base Editor
Kabin Xie ,*National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China
,*National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, China通讯作者:
收稿日期:2019-01-21接受日期:2019-01-22网络出版日期:2019-07-01
Corresponding authors:
Received:2019-01-21Accepted:2019-01-22Online:2019-07-01

摘要
关键词:
Abstract
Keywords:
PDF (868KB)摘要页面多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
引用本文
谢卡斌. 中国科学家发现胞嘧啶单碱基编辑工具存在 基因组范围的脱靶. 植物学报, 2019, 54(3): 296-299 doi:10.11983/CBB19033
Xie Kabin.
基于CRISPR系统的基因组编辑技术是近年来的革命性技术, 现已广泛应用于基础研究、疾病治疗和作物遗传改良等各个方面。与此同时, 对CRISPR的改进和探索也在不断进行。近几年, 多种CRISPR系统(如Cas12a和Cas13)被发掘并改造成基因编辑工具; 此外, 最常用的源自酿脓链球菌(Streptococus pyogenes)的Cas9 (Streptococcus pyogenes Cas9, 简称Cas9)蛋白也被定向改造出多个变体, 包括识别不同PAM (protospacer adjacent motif)的Cas9变体和高保真的Cas9 (High-fidelity Cas9, HF-Cas9)等。随着CRISPR技术的日臻完善也出现了许多新型的应用, 尤其是基于CRISPR-Cas的单碱基编辑技术, 能够精确地将生物体内DNA靶位点上的单个碱基进行转换, 为遗传信息的精准编辑提供了一种新的策略。单碱基编辑技术被Science杂志选为2017年度的十大科学突破之一(
目前的单碱基编辑技术依赖简单的生物化学原理, 但通过非常巧妙的设计来实现(图1A): 利用脱氨酶将胞嘧啶(C)或腺嘌呤(A)上的氨基去掉, 分别转换为尿嘧啶(U)或次黄嘌呤(I), 然后在DNA复制或修复时进一步将两者转换为胸腺嘧啶(T)或鸟嘌呤(G), 从而实现生物体DNA序列的靶向碱基转换。因此, 单碱基编辑技术依赖核苷酸特异的脱氨酶。根据靶向碱基的不同, 单碱基编辑技术可以分为胞嘧啶单碱基编辑(cytosine base editor, CBE)和腺嘌呤单碱基编辑(adenine base editor, ABE)。2016年4月, 哈佛大学的David R. Liu实验室首次成功开发了靶向编辑胞嘧啶的工具BE3 (base editor 3) (图1B), 在生物体内实现了靶位点内C?G碱基对到T?A碱基对的转换。BE3编辑器是将具有切口酶活性的nCas9 (Cas9 nickase) 蛋白融合1个来自大鼠(Rattus norvegicus)的胞嘧啶脱氨酶(APOBEC1), 此外还包含1个尿嘧啶糖基化酶抑制子(Uracil DNA glycosylase inhibitor, UGI), 用来抑制细胞的DNA修复系统(将U?G复原为C?G的修复)。在sgRNA (single guide RNA)的引导下, 该BE3可在细胞内高效率地将靶DNA编辑窗口序列中的C转换为T (Komor et al., 2016)。此外, 日本神户大学Akihiko Kondo和上海交通大学常兴实验室分别利用来源于七鳃鳗(Petromyzon marinus)和经修改的人(Homo sapiens)源的激活诱导性胞嘧啶脱氨酶(activation-induced deaminase, AID)开发出类似的CBE工具(Ma et al., 2016; Nishida et al., 2016)。相比CBE编辑工具, ABE编辑工具的开发更具挑战性。目前, 在生物体内尚未发现能够作用于单链DNA的腺嘌呤脱氨酶, 因此需要通过遗传工程来创造能作用于DNA的腺嘌呤脱氨酶才能构建ABE编辑器。2017年, David R. Liu实验室进一步通过分子进化技术, 在大肠杆菌(Escherichia coli) tRNA脱氨酶(TadA)的基础上改造出能作用于单链DNA底物的腺嘌呤脱氨酶(标记为TadA*), 将TadA和TadA*同时与nCas9融合(图1B), 得到能高效率实现A?T到G?C转换的ABE编辑器(Gaudelli et al., 2017)。
图1
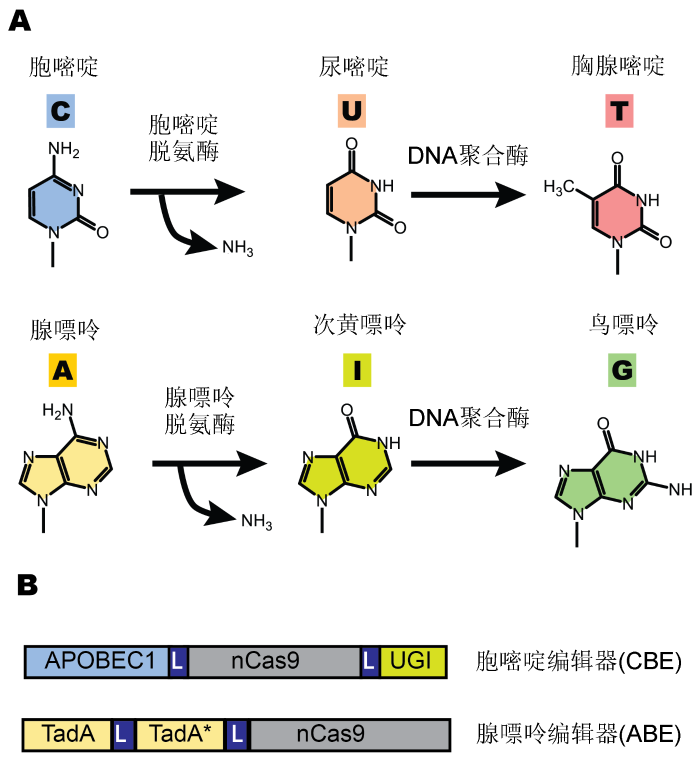 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1基于CRISPR-Cas9的单碱基编辑
(A) 利用脱氨酶进行DNA碱基转换的原理; (B) 胞嘧啶编辑器(CBE, 以BE3为例)和腺嘌呤编辑器(ABE)的结构示意图。L: 连接肽
Figure 1CRISRP-Cas9 base editing
(A) Deaminase-mediated base editing reactions; (B) The schematic structure of cytosine (CBE, take BE3 as an example) and adenine (ABE) base editors. L: Peptide linker
CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(May, 2017)。在植物中, 多个研究小组均证实CBE单碱基编辑工具(Li et al., 2017; Lu and Zhu, 2017; Shimatani et al., 2017; Zong et al., 2017; Ren et al., 2018)和后来的ABE单碱基编辑工具(Hua et al., 2018; Kang et al., 2018; Li et al., 2018; Yan et al., 2018)能够高效地在sgRNA靶位点对目标碱基进行转换。与动物中的研究结果一致, 单碱基编辑工具的作用范围会限制在靶位点的一定区域内(“编辑窗口”)。同时, 对单碱基编辑工具的改进也没有停止。例如, 利用高保真的Cas9变体改造BE3 (HF1-BE3)以降低Cas9脱靶的风险, 而利用更高活性的APOBEC3A或AID超活性突变体可以提高CBE编辑工具的效率(Rees and Liu, 2018)。此外, 在全基因组水平检测单碱基编辑工具的脱靶也尤为重要。
最近, 中国科学院遗传与发育生物学研究所高彩霞实验室对ABE和CBE两个单碱基编辑工具进行深入研究, 发现利用APOBEC1和UGI构建的BE3和HF1-BE3两个CBE编辑工具存在全基因组范围不可预测的脱靶现象, 但利用ABE编辑工具则没有检测到脱靶(Jin et al., 2019)。Jin等(2019)在水稻(Oryza sativa)中对ABE、BE3和HF1-BE3单碱基编辑器进行了脱靶编辑分析。他们通过60×深度的基因组测序比较了同时转化的愈伤组织再生植株和转化单碱基编辑器的水稻材料, 其中包括装载了gRNA的载体和空载体。除了检测到预期的靶位点编辑之外, 他们还发现转化BE3和HF1-BE3的水稻中SNVs (single nucleotide variants)的频率显著高于对照。进一步分析发现, 这些植株的C>T (G>A)转换突变的数目高于对照。在转化ABE的水稻植株中, 各种类型的SNVs频率与对照相比均无显著差异。以上结果表明, BE3和HF1-BE3这两个CBE胞嘧啶编辑工具存在脱靶现象, 而在ABE系统中则没有可检测到的脱靶现象。此外, 研究表明, BE3和HF1-BE3植株中所检测到C>T脱靶编辑位点的序列与sgRNA的引导序列无相似性, 且不能够通过目前的Cas9脱靶预测软件来预测。作者推断这种全基因组范围的脱靶编辑很可能是所用到的胞嘧啶脱氨酶或者UGI导致的, 而不是Cas9和gRNA脱靶。作者也特别注意到, C>T脱靶碱基转换的分布在染色体间没有显著差异, 但偏向于分布在转录活跃的基因编码区。
Jin等(2019)的研究结果表明, ABE编辑器能够精准实现单碱基编辑, 但BE3和HF1-BE3的胞嘧啶编辑器在全基因组范围都有脱靶编辑, 而脱靶的原因很可能是所用的胞嘧啶脱氨酶或UGI引起基因组随机变异。该研究对单碱基编辑工具的应用和改造具有重要指导意义。值得注意的是, 动物中单碱基编辑工具的脱靶分析也发现了同样的现象(Zuo et al., 2019)。虽然现在还不确定利用其它胞嘧啶脱氨酶(如AID和APOBEC3A)构建的CBE是否也存在基因组范围的脱靶, 但相信很快就会有答案。如何降低或消除胞嘧啶单碱基编辑工具的脱靶, 将是基因编辑技术优化的一个重要方向。此外, 降低DNA结合活性的胞嘧啶脱氨酶变体(Kim et al., 2017), 可能为降低全基因组范围脱靶提供新的解决思路。
(责任编辑: 朱亚娜)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1038/s41586-018-0070-xURLPMID:29160308 [本文引用: 1]

Abstract The spontaneous deamination of cytosine is a major source of transitions from C090004G to T090004A base pairs, which account for half of known pathogenic point mutations in humans. The ability to efficiently convert targeted A090004T base pairs to G090004C could therefore advance the study and treatment of genetic diseases. The deamination of adenine yields inosine, which is treated as guanine by polymerases, but no enzymes are known to deaminate adenine in DNA. Here we describe adenine base editors (ABEs) that mediate the conversion of A090004T to G090004C in genomic DNA. We evolved a transfer RNA adenosine deaminase to operate on DNA when fused to a catalytically impaired CRISPR-Cas9 mutant. Extensive directed evolution and protein engineering resulted in seventh-generation ABEs that convert targeted A090004T base pairs efficiently to G090004C (approximately 50% efficiency in human cells) with high product purity (typically at least 99.9%) and low rates of indels (typically no more than 0.1%). ABEs introduce point mutations more efficiently and cleanly, and with less off-target genome modification, than a current Cas9 nuclease-based method, and can install disease-correcting or disease-suppressing mutations in human cells. Together with previous base editors, ABEs enable the direct, programmable introduction of all four transition mutations without double-stranded DNA cleavage.
DOI:10.1016/j.molp.2018.02.007URL [本文引用: 1]
[本文引用: 3]
DOI:10.1038/s41477-018-0178-xURL [本文引用: 1]

CRISPR-Cas9 system is now widely used to edit a target genome in animals and plants. Cas9 protein derived from Streptococcus pyogenes (SpCas9) cleaves double-stranded DNA targeted by a chimeric single-guide RNA (sgRNA). For plant genome editing, Agrobacterium-mediated T-DNA transformation has been broadly used to express Cas9 proteins and sgRNAs under the control of CaMV 35S and U6/U3... [Show full abstract]
DOI:10.1038/nbt.3803URLPMID:28191901 [本文引用: 1]

Base editing is a recently developed approach to genome editing that uses a fusion protein containing a catalytically defectiveStreptococcus pyogenesCas9, a cytidine deaminase, and an inhibitor of base excision repair to induce programmable, single-nucleotide changes in the DNA of living cells without generating double-strand DNA breaks, without requiring a donor DNA template, and without inducing an excess of stochastic insertions and deletions1. Here we report the development of five new C鈫扵 (or G鈫扐) base editors that use natural and engineered Cas9 variants with different protospacer-adjacent motif (PAM) specificities to expand the number of sites that can be targeted by base editing by 2.5-fold. Additionally, we engineered new base editors containing mutated cytidine deaminase domains that narrow the width of the apparent editing window from approximately 5 nucleotides to as little as 1 to 2 nucleotides, enabling the discrimination of neighboring C nucleotides that would previously be edited with comparable efficiency, thereby doubling the number of disease-associated target Cs that can be corrected preferentially over nearby non-target Cs. Collectively, these developments substantially increase the targeting scope of base editing and establish the modular nature of base editors.
DOI:10.1038/nature17946URLPMID:27096365 [本文引用: 1]

Current genome-editing technologies introduce double-stranded (ds) DNA breaks at a target locus as the first step to gene correction.1,2Although most genetic diseases arise from point mutations, current approaches to point mutation correction are inefficient and typically induce an abundance of random insertions and deletions (indels) at the target locus from the cellular response to dsDNA breaks.1,2Here we report the development of base editing, a new approach to genome editing that enables the direct, irreversible conversion of one target DNA base into another in a programmable manner, without requiring dsDNA backbone cleavage or a donor template. We engineered fusions of CRISPR/Cas9 and a cytidine deaminase enzyme that retain the ability to be programmed with a guide RNA, do not induce dsDNA breaks, and mediate the direct conversion of cytidine to uridine, thereby effecting a C→T (or G→A) substitution. The resulting “base editors” convert cytidines within a window of approximately five nucleotides (nt), and can efficiently correct a variety of point mutations relevant to human disease. In four transformed human and murine cell lines, second- and third-generation base editors that fuse uracil glycosylase inhibitor (UGI), and that use a Cas9 nickase targeting the non-edited strand, manipulate the cellular DNA repair response to favor desired base-editing outcomes, resulting in permanent correction of 6515-75% of total cellular DNA with minimal (typically ≤ 1%) indel formation. Base editing expands the scope and efficiency of genome editing of point mutations.
DOI:10.1186/s13059-018-1443-zURL [本文引用: 1]

Nucleotide base editors in plants have been limited to conversion of cytosine to thymine. Here, we describe a new plant adenine base editor based on an evolved tRNA adenosine deaminase fused to the nickase CRISPR/Cas9, enabling A61T to G61C conversion at frequencies up to 7.5% in protoplasts and 59.1% in regenerated rice and wheat plants. An endogenous gene is also successfully modified through introducing a gain-of-function point mutation to directly produce an herbicide-tolerant rice plant. With this new adenine base editing system, it is now possible to precisely edit all base pairs, thus expanding the toolset for precise editing in plants. The online version of this article (10.1186/s13059-018-1443-z) contains supplementary material, which is available to authorized users.
DOI:10.1016/j.molp.2016.12.001URLPMID:27940306 [本文引用: 1]
DOI:10.1016/j.molp.2016.11.013URLPMID:27932049 [本文引用: 1]
DOI:10.1038/nmeth.4027URLPMID:27723754 [本文引用: 1]

Abstract A large number of genetic variants have been associated with human diseases. However, the lack of a genetic diversification approach has impeded our ability to interrogate functions of genetic variants in mammalian cells. Current screening methods can only be used to disrupt a gene or alter its expression. Here we report the fusion of activation-induced cytidine deaminase (AID) with nuclease-inactive clustered regularly interspaced short palindromic repeats (CRISPR)-associated protein 9 (dCas9) for efficient genetic diversification, which enabled high-throughput screening of functional variants. Guided by single guide (sg)RNAs, dCas9-AID-P182X (AIDx) directly changed cytidines or guanines to the other three bases independent of AID hotspot motifs, generating a large repertoire of variants at desired loci. Coupled with a uracil-DNA glycosylase inhibitor, dCas9-AIDx converted targeted cytidines specifically to thymines, creating specific point mutations. By targeting BCR-ABL with dCas9-AIDx, we efficiently identified known and new mutations conferring imatinib resistance in chronic myeloid leukemia cells. Thus, targeted AID-mediated mutagenesis (TAM) provides a forward genetic tool to screen for gain-of-function variants at base resolution.
DOI:10.1038/nbt.3871URLPMID:28486457 [本文引用: 1]

Research on base editing leads to new enzymes, genome-wide specificity measurements, and edited plants and animals.
DOI:10.1126/science.aaf8729URLPMID:27492474 [本文引用: 1]

The generation of genetic variation (somatic hypermutation) is an essential process for the adaptive immune system in vertebrates. We demonstrate the targeted single-nucleotide substitution of DNA using hybrid vertebrate and bacterial immune systems components. Nuclease-deficient type II CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR-associated) and the activation-induced cytidine deaminase (AID) ortholog PmCDA1 were engineered to form a synthetic complex (Target-AID) that performs highly efficient target-specific mutagenesis. Specific point mutation was induced primarily at cytidines within the target range of five bases. The toxicity associated with the nuclease-based CRISPR/Cas9 system was greatly reduced. Although combination of nickase Cas9(D10A) and the deaminase was highly effective in yeasts, it also induced insertion and deletion (indel) in mammalian cells. Use of uracil DNA glycosylase inhibitor suppressed the indel formation and improved the efficiency.
DOI:10.1038/s41576-018-0059-1 [本文引用: 1]
DOI:10.1016/j.molp.2018.01.005URLPMID:29382569 [本文引用: 1]

ACCEPTED MANUSCRIPT codon-optimized for expression in rice (Figure S1) and attached to 5使 terminal end of Cas9n-NLS using XTEN linker (Figure 1A and 1B). Therefore, it is likely that the editing window of hAID*鈭 would be restricted to the sgRNA-targeting site similar to the case of APOBEC1. To be noted, UGI was excluded at this time since more types of nucleotide conversions, produced in uracil N-glycosylase (UDG)-initiated base excision repair (BER), would be anticipated.The hAID*鈭-XTEN-Cas9n-NLS chimeric gene (termed rBE5) were first tested in rice leaf sheath protoplast, with the sgRNAs targeting a GCAC-containing ApaLI restriction site in OsRLCK185 and a TCC-containing BamHI restriction site in OsCERK1, respectively. Sequencing data showed that distinct mutations with high frequency of C>T conversions occurred in the editing window (Figure S2A-S2H), suggesting rBE5 functions on GC, AC, TC as well as CC in rice cells.As a follow-up to our previous study of the rBE3 system (Ren et al., 2017), rBE3 gene in the binary vector pUbi:rBE3 was replaced by rBE5 gene, namely pUbi:rBE5, and then further tested for efficiency in stable transgenic rice plants through Agrobacterium-mediated transformation. Pi-d2, an agriculturally important rice blast R gene in which a GC-containing region was resistant to rBE3-mediated editing in our earlier experiment, was first chosen for targeting (Figure 1C). As reported previously, a single amino acid substitution at position 441 in the recessive allele of Pi-d2 gene results in loss of resistance to Magnaporthe oryzae (Chen et al., 2006). Therefore, we assumed that introducing a G>A mutation (M441I) in endogenous pi-d2kit using rBE5 could recover its biological function. The same protospacer for rBE3 was used in this experiment, totally 26 independent transgenic lines of Kitaake were obtained after transformation and genotyped subsequently by Sanger sequencing. 8 heterozygous lines (30.8% efficiency) carrying a desired single G to A conversion at position -17 upstream of PAM were identified (Figure 1D-1E, Figure S3A), suggesting that rBE5 is much more efficient on target C that immediately follow a G than rBE3 does. We assumed that the successful gene correction of pi-d2 would be attributable to both the specificity and the improved activity of hAID*鈭. Remarkably, pi-d2 alleles with Indel mutations were also detected in 5 lines (Figure S3B-S3C).
DOI:10.1038/nbt.3833URLPMID:28346401 [本文引用: 1]

Targeted editing of single base pairs is achieved in monocot rice and dicot tomato using Target-AID (Cas9 activation-induced cytidine deaminase fusion).
DOI:10.1016/j.molp.2018.02.008URL [本文引用: 1]

Dear Editor, The newly developed CRISPR/Cas9-mediated base editing technology with cytosine deaminase is capable of precisely and efficiently introducing point mutations at the target genomic locus,which does not require double-stranded DNA breaks or any donor templates and thus exhibit a great potential for gene correction and genetic diversification in yeasts,plants,and mammalian and human cells (Komor et al.,2016;Nishida et al.,2016;Lu and Zhu,2017;Ren et al.,2017).However,compared with AID/APOBEC1 members of the cytosine deaminase family that are widely utilized in base editing to induce cytidine (C) to thymine (T) conversion,adenosine deaminase is far from being applicable since TadA/ADAR members act strictly on duplex RNA,or DNA/RNA hybrids with mismatches,instead of single-stranded DNA (Zheng et al.,2017).To address this problem,great efforts have been invested recently in identifying Escherichia coli TadA variants that accept DNA as a substrate through rounds of protein evolution and engineering,ultimately leading to a number of adenine base editors (ABEs) with great efficiencies and broadened sequence compatibility in inducing nucleotide changes at a wide range of target genomic loci in human cells (Gaudelli et al.,2017).In these ABE systems,the TadA:TadA* heterodimer is guided by the Cas9n/single guide RNA (sgRNA) complex to the target site,and the engineered TadA*,but not the wild-type TadA,functions as the active tRNA adenosine deaminase that turns adenine (A) to inosine (鈪) in singlestranded genomic DNA,subsequently resulting in A to guanine (G) mutation in genome during DNA repair or DNA replication (Gaudelli et al.,2017).These tools,together with previous base editors,enable programmable introduction of all four transitions (C to T,G to A,A to G,and T to C) at the target loci in the genome,greatly expanding the capabilities of base editing.Here,we report the development of fluorescencetracking base editing systems with E.coil TadA variants and Cas9 variants in rice.
DOI:10.1038/nbt.3811URLPMID:28244994 [本文引用: 1]

Single DNA base pairs are edited in wheat, rice and maize using a Cas9 nickase fusion protein.
[本文引用: 1]
Programmable base editing of A·T to G·C in genomic DNA without DNA cleavage
1
2017
... 目前的单碱基编辑技术依赖简单的生物化学原理, 但通过非常巧妙的设计来实现(
Precise A·T to G·C base editing in the rice genome
1
2018
... CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(
Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice
3
2019
... 最近, 中国科学院遗传与发育生物学研究所高彩霞实验室对ABE和CBE两个单碱基编辑工具进行深入研究, 发现利用APOBEC1和UGI构建的BE3和HF1-BE3两个CBE编辑工具存在全基因组范围不可预测的脱靶现象, 但利用ABE编辑工具则没有检测到脱靶(
... ).
...
Precision genome engineering th- rough adenine base editing in plants
1
2018
... CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(
Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions
1
2017
...
Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage
1
2016
... 目前的单碱基编辑技术依赖简单的生物化学原理, 但通过非常巧妙的设计来实现(
Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion
1
2018
... CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(
Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system
1
2017
... CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(
Precise editing of a target base in the rice genome using a modified CRISPR/Cas9 system
1
2017
... CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(
Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells
1
2016
... 目前的单碱基编辑技术依赖简单的生物化学原理, 但通过非常巧妙的设计来实现(
Base editing on the rise
1
2017
... CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(
Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems
1
2016
... 目前的单碱基编辑技术依赖简单的生物化学原理, 但通过非常巧妙的设计来实现(
Base editing: precision chemistry on the genome and transcriptome of living cells
1
2018
... CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(
Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant
1
2018
... CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(
Targeted base editing in rice and tomato using a CRISPR-1)Cas9 cytidine deaminase fusion
1
2017
... CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(
Highly efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice
1
2018
... CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(
Precise base editing in rice, wheat and maize with a Cas9-cytidine deaminase fusion
1
2017
... CBE和ABE编辑器的效率很快就在多个动物和植物物种中得到了验证和优化(
Cytosine base editor generates substantial off-target single-nucleotide variants in mouse embryos
1
2019
...
备案号: 京ICP备16067583号-21
版权所有 © 2021 《植物学报》编辑部
地址:北京香山南辛村20号 邮编:100093
电话:010-62836135 010-62836131 E-mail:cbb@ibcas.ac.cn
本系统由北京玛格泰克科技发展有限公司设计开发
