 ,*山东农业大学农学院, 泰安 271018
,*山东农业大学农学院, 泰安 271018Recent Progress in Evolutionary Technology of CRISPR/Cas9 System for Plant Genome Editing
Yuekai Su, Jingren Qiu, Han Zhang, Zhenqiao Song, Jianhua Wang ,*College of Agronomy, Shandong Agricultural University, Tai’an 271018, China
,*College of Agronomy, Shandong Agricultural University, Tai’an 271018, China通讯作者:
收稿日期:2018-07-2接受日期:2018-08-9网络出版日期:2019-07-01
| 基金资助: |
Corresponding authors:
Received:2018-07-2Accepted:2018-08-9Online:2019-07-01

摘要
关键词:
Abstract
Keywords:
PDF (1090KB)摘要页面多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
引用本文
苏钺凯, 邱镜仁, 张晗, 宋振巧, 王建华. CRISPR/Cas9系统在植物基因组编辑中技术改进与创新的研究进展. 植物学报, 2019, 54(3): 385-395 doi:10.11983/CBB18151
Su Yuekai, Qiu Jingren, Zhang Han, Song Zhenqiao, Wang Jianhua.
在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(Chamberlain et al., 2004)。该技术可以使研究人员在极短的时间内模拟基因的自然突变, 甚至能够完成自然选择中无法完成的基因演变。从最初的归巢核酸内切酶(meganucleases) (Puchta et al., 1993; Redondo et al., 2008)到第1代人工核酸内切酶——锌指核酸酶(zinc finger nucleases, ZFNs) (Durai et al., 2005), 再到第2代人工核酸内切酶——类转录激活效应因子核酸酶(transcription activator-like effector nucleases, TALENs) (Miller et al., 2011; Li et al., 2012), 以及近几年兴起的第3代人工核酸内切酶——CRISPR/Cas9系统(Cong et al., 2013; Jinek et al., 2013), 基因组编辑技术已经逐渐成为基因改造及基因功能研究中不可或缺的技术手段(Shan and Gao, 2015)。尤其是 CRISPR/Cas9技术, 凭借其效率高、成本低和操作简单等优势(Gaj et al., 2013; Musso-lino and Cathomen, 2013), 自创始之初便展现出非常广阔的应用前景。2013年8月, 3家实验室同时发表了CRISPR/Cas9技术在植物基因组编辑中的应用(Shan et al., 2013; Nekrasov et al., 2013; Li et al., 2013), 由此拉开了CRISPR/Cas9技术在植物中应用的序幕。本文简要介绍CRISPR系统的发现与原理, 重点阐述近几年在植物领域对CRISPR/Cas系统进行的优化与改造以及基因组编辑技术在植物领域面临的挑战。
1 CRISPR/Cas9基因组编辑技术
1.1 CRISPR/Cas系统的起源
CRISPR/Cas系统最早是由日本大阪大学的一个研究团队在1987年对大肠杆菌碱性磷酸酶基因进行研究时发现的(Ishino et al., 1987), 只是当时这一段特殊的重复序列并未引起普遍关注。随后研究人员发现在细菌和古细菌的基因组中广泛存在这种带有间隔序列的多个重复序列回文结构。2002年, 科学家最终将这种重复序列正式命名为CRISPR (clustered re- gularly interspaced short palindromic repeats)序列(Jansen et al., 2002)。虽然人们猜测该序列可能与细菌的自身免疫功能有关, 但直到2007年, Barrangou等(2007)才通过实验首次发现并证明嗜热链球菌利用CRISPR/Cas系统抵御噬菌体的入侵。2008年, 研究人员在表皮葡萄球菌(Staphylococcus epidermi- dis) (Marraffini and Sontheimer, 2008)和大肠杆菌(Escherichia coli) (Brouns et al., 2008)中均发现成熟的crRNAs (CRISPR RNAs)可以与Cas9蛋白形成复合体, 并通过实验证明该复合体可以干扰噬菌体的增殖。这些研究为CRISPR系统此后突飞猛进地发展奠定了基础, 科学家由此逐步揭开了CRISPR系统作用机理的神秘面纱。图1
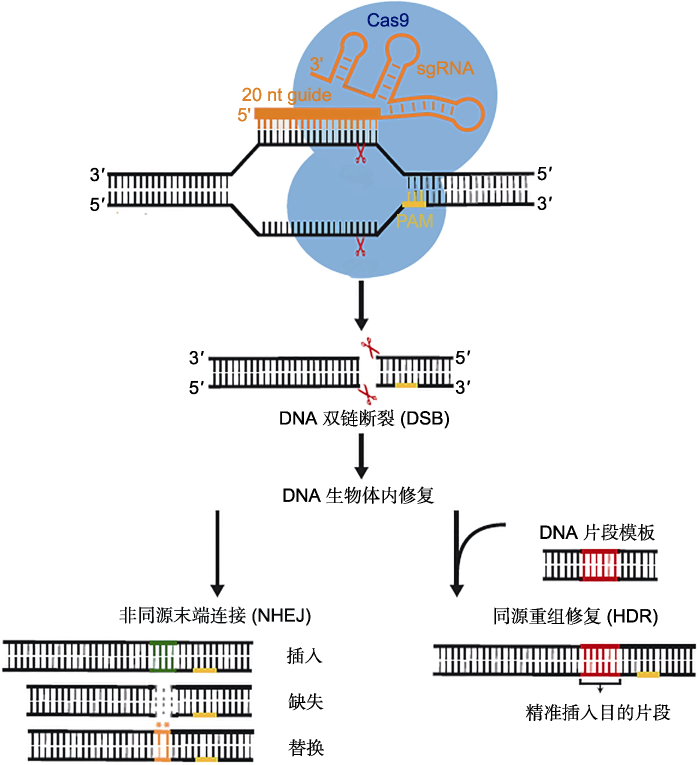 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1CRISPR/Cas9基因组编辑原理(Jiang and Doudna, 2017)
来自酿脓链球菌(Streptococcus pyogenes)的Cas9蛋白(SpCas9)与包含20 nt识别序列的向导RNA (sgRNA)结合, 然后定向切割位于原型间隔序列毗邻基序(PAM)区域上游的靶序列造成双链断裂(DSB), 最终借助生物体内自身修复实现基因组编辑。
Figure 1The schematic diagram of CRISPR/Cas9 genome editing (Jiang and Doudna, 2017)
The Cas9 protein from Streptococcus pyogenes (SpCas9) and associated guide RNA (sgRNA), containing a 20 nt recognition sequence, will cleave a target sequence located upstream of the protospacer adjacent motif (PAM) region, resulting in double-strand break (DSB). Ultimately use the repair in the organism to achieve genome editing.
1.2 CRISPR/Cas9基因组编辑技术的原理
目前, 应用于基因组编辑的CRISPR系统主要以来自酿脓链球菌(Streptococcus pyogenes)的CRISPR/ Cas9系统应用最广, 且研究得最为透彻。该系统由Cas9蛋白、crRNA和tracrRNA (trans-activating CRISPR RNA) 3部分组成。其中, crRNA是由pre- crRNA (precursor crRNA)经tracrRNA和RNase III加工后得到, 主要负责引导CRISPR/Cas系统对特异靶位点的识别(Deltcheva et al., 2011)。每个crRNA只有几十个碱基, 与前2代基因组编辑技术ZFNs和TALENs相比, 载体变得更小, 更加容易构建, 且这个系统识别的PAM (protospacer adjacent motif)序列NGG广泛存在于各物种的基因组序列中(Gasiunas et al., 2012)。Jinek等(2012)揭示了II型CRISPR系统切割DNA双链的分子机制并完成对CRISPR/ Cas9系统的改造和优化, 他们开创性地将原本分开作用的crRNA和tracrRNA整合形成1条sgRNA (single guide RNA), 并成功地在体外定点切割线性寡聚核苷酸片段和环状质粒DNA。他们还对Cas9不同结构域的作用以及PAM序列的重要性等多个问题进行研究, 这些研究成为CRISPR/Cas9基因组编辑系统最终能广泛应用于生物体的重要基础(Cong et al., 2013)。至此, CRISPR/Cas9基因组编辑体系已初步成型。简化后的CRISPR/Cas9系统只由两部分组成: sgRNA和Cas9蛋白。首先, 成功表达的sgRNA通过自身的Cas9把手(Cas9 handle)与Cas9蛋白形成复合体; 然后, 复合体开始搜寻并匹配PAM序列, 随后sgRNA的碱基互补配对区序列通过碱基互补配对原则识别目标基因的靶序列; 最后, Cas9蛋白利用自身的核酸内切酶活性对靶序列进行切割, 形成DNA双链断裂(double-strand break, DSB)(图1)。
真核生物细胞内有2种修复DSB的方式: 一种是非同源末端连接(nonhomologous end-joining, NHEJ), 另一种是同源重组修复(homology-directed repair, HDR)。在自然条件下, 同源重组修复出现的概率极低, 因此细胞中DSB的修复方式主要为NHEJ (Sy-mington and Gautier, 2011)。在NHEJ修复过程中, 断裂链的末端会直接随机连接, 修复后的DNA双链容易出现个别碱基的缺失或插入, 使靶基因的核苷酸序列发生移位, 从而导致基因功能的改变, 正是利用这一点可以使经过CRISPR/Cas9系统编辑后的基因被敲除; 在提供同源序列的情况下, HDR途径会以同源序列为模板进行合成修复, 从而对Cas9切割的目标DNA序列进行精确编辑, 如特定核苷酸序列的定点替换或者插入突变体。
2 CRISPR/Cas9基因组编辑技术在植物领域中的改进与创新
目前, 在医学(Liang et al., 2015)、农业(Xing et al., 2016)和生物制药(Peng et al., 2015)等领域的研究中, CRISPR/Cas9基因组编辑技术都发挥了不可替代的作用。相比动物和微生物, CRISPR/Cas9在植物中的应用相对滞后, 但近几年在科学家的不懈努力下, CRISPR/Cas9系统在植物领域中得到了不断的改进与优化, 研究成果层出不穷, 主要集中在以下几个方面。2.1 CRISPR/Cas9技术的编辑效率
2.1.1 CRISPR系统元件对基因组编辑效率的影响研究人员发现靶位点的核酸组成(即GC含量)、Cas9的表达水平以及靶区域的表观遗传等都会对基因组编辑效率产生影响。Ma等(2015)研究发现高GC碱基(50%-70%)含量在水稻(Oryza sativa)目标基因组中的编辑效率相对更高, 同时还发现sgRNA中目标配对序列茎环结构的形成往往会降低基因组编辑效率。Xu等(2014)为了比较未优化和优化后的Cas9基因编辑效率, 分别使用经过水稻密码子优化和人类(Homo sapiens)细胞密码子优化后的Cas9基因(p Spcas9和Spcas9)靶向编辑2个相同的位点, 结果表明p Spcas9比Spcas9突变效率显著提高。然而在双子叶植物中, 使用在人类细胞中进行优化后的密码子同样可以进行高效的基因组编辑, Cas9基因密码子的优化可能并不是必需的。此外, 通过使用化学修饰的sgRNA和Cas9进行基因组编辑也可以提高基因组编辑效率。Hendel等(2015)通过核苷亚磷酰胺对sgRNA的稳定作用进行深度测序, 结果显示, 在3'和5'末端对3个末端核苷酸的修饰改善了靶位点的编辑效率, 同时脱靶位效应有所下降。
2018年, 中国科学院遗传与发育生物学研究所韩方普团队使用玉米(Zea mays) DMC1基因启动子, 构建DPC (DMC1 promoter-controlled) CRISPR-Cas9载体系统, 并用其转化玉米幼胚, 结果发现抗性愈伤组织的基因编辑效率高达100%, 并在T0代植株中就出现了60%-70%的纯合或双等位突变体。 经全基因组测序分析, 在预测的1 000多个潜在脱靶位点处并没有发现脱靶突变(Feng et al., 2018)。
2.1.2 环境因素对基因编辑效率的影响
Le Blanc等(2017)的研究结果表明, 与在标准温度(22°C)下连续生长的植物相比, 在37°C经历热激的拟南芥(Arabidopsis thaliana)显示出更高的基因编辑效率。依赖于GFP报告基因的定量分析表明, 拟南芥在37°C处理后体细胞基因表达量可以提高5倍, 生殖细胞基因表达量可以提高100倍以上。该团队还发现温度对突变率的影响不限于拟南芥, 在37°C热激的柑橘(Citrus reticulata)中CRISPR/Cas9的靶向突变效率也有类似的增加。体外实验证明, 来自化脓链球菌的Cas9 (SpCas9)在37°C比在22°C下活性更高, 更易产生双链DNA断裂, 由此表明温度对CRISPR/Cas9的体内作用机制具有潜在影响。
2.2 CRISPR/Cas9技术的编辑范围
除应用最广的SpCas9外, 其它不同来源的Cas9也具有基因编辑能力。研究人员从金黄色葡萄球菌(Staphylococcus aureus)中找到1种较小的Cas9核酸酶(SaCas9) (Ran et al., 2015), 它比SpCas9小25%, PAM识别序列为NNGRRT。此外, 来自嗜热乳链球菌(Streptococcus thermophilus)的Cas9核酸酶(St1Cas9)可以识别NNAGAA的PAM序列(Deveau et al., 2008; Horvath, 2008)。Steinert等(2015)将经密码子优化的SaCas9和St1Cas9两种Cas9蛋白成功应用于拟南芥中, 并获得了较高的基因组编辑效率。这些SpCas9同源蛋白的发现扩大了基因组编辑的范围, 为研究人员提供了更多的选择。Meng等(2018)发现CRISPR/Cas9系统能够在水稻中高效识别和编辑NAG位点, 相比传统的NGG位点, NAG邻近基序靶位点的容错能力更低, 表现出更低的脱靶效应。这项研究成果不仅拓展了CRISPR/ Cas9系统在水稻基因组中的可编辑范围, 而且也表明在进行基因组编辑时需更多考虑到NAG邻近基序处的潜在脱靶效应。中国水稻研究所王克剑课题组(Hu et al., 2016)通过定点突变的方法对SpCas9蛋白进行改造, 获得了2种SpCas9变体(VQR和VRER), 并通过一系列的转基因实验证明, 在水稻中VQR可以高效识别NGA的PAM序列, 而VRER则可以识别NGCG的PAM序列。这2种突变体使得CRISPR/Cas9系统在水稻中的可编辑范围拓展到原有的2倍以上。随后, 他们又通过修饰sgRNA的结构以及使用强内源启动子, 显著提高了VQR变体的编辑效率(Hu et al., 2017a)。哈佛大学David Liu研究团队利用噬菌体辅助持续进化(PACE)方法获得SpCas9最新突变体xCas9, xCas9可识别的PAM位点为NG、GAA和GAT, 并且成功应用于转录激活、基因敲除和单碱基编辑。研究表明, xCas9不仅应用范围更大, 而且特异性要高于SpCas9 (Xing et al., 2014)。
2.3 多基因同时编辑与单碱基精准编辑
在后基因组时代, 功能基因组研究的重心逐渐由单一基因转向多基因功能研究。CRISPR/Cas9技术凭借其简单、高效等优点使多基因同时敲除变得可行。Xing等(2014)构建了1套适合进行植物多基因编辑的CRISPR/Cas9载体系统, 在拟南芥的T1代中实现了三基因同步敲除。Xie等(2015)利用内源的转运RNA (transfer RNA, tRNA)加工系统提高了CRISPR/Cas9进行多基因编辑的能力, 他们将多个tRNA-ssgRNA结构串联排列, 并通过单个多顺反子转录、加工成多个ssgRNA。利用这一策略在水稻中实现了多达8个位点同时突变, 其中个别位点效率高达100%。由于tRNA及其加工系统在所有生物中都非常保守, 因此这一策略有望在其它物种的多基因编辑中广泛应用。Yu等(2018)则发现在敲除多基因时, 通常做法是针对1个基因设计1条sgRNA, 这种设计方法虽然提高了敲除效率, 但每多加1条sgRNA, 脱靶风险将呈指数级增加。为了解决这一问题, 该研究组以编码拟南芥核糖体大亚基AtRPL10亚家族的3个同源基因AtRPL10A (AT1G14320)、AtRPL10B (AT1G26910)及AtRPL10C (AT1G66580)为切入点, 通过序列分析找到同时满足编辑3个基因的1条sgRNA, 利用Cas9同时编辑3个基因。经单克隆测序分析统计, 显示在Line9植株中, AtRPL10A、AtRPL10B以及AtRPL10C的编辑效率分别为8.8%、3.8%及23.6%。2016年, 在哺乳动物细胞中成功对靶标基因实现了单碱基编辑(C-T)。随后, 单碱基编辑技术在水稻、小麦(Triticum aestivum)、玉米、拟南芥和番茄(Solanum lycopersicum)等多种植物中得到应用, 然而所采用的APOBEC1胞嘧啶脱氨酶表现出对TC的偏好性, 对序列为GC的编辑效率较低。鉴于此, Ren等(2018)采用人的AID胞嘧啶脱氨酶替换APOBEC1的方法, 在水稻中解决了TC偏好性问题, 对GC和TC都有较高的编辑效率。
大肠杆菌腺嘌呤脱氨酶(ecTadA)能够对正常DNA上的腺嘌呤进行脱氨, 将腺嘌呤A转变为次黄嘌呤I, 损伤DNA在重新复制过程中被聚合酶作用, 导致A-T配对转换为G-C配对(Gaudelli et al., 2017)。2018年4月, 中国农业科学院植物保护研究所Yan等(2018)引入TadA及其2个突变体TadA*7.10和TadA*7.8, 并将其密码子优化后, 分别与CRISPR/Cas9系统融合, 开发了rBE14、rBE15、rBE17和rBE18多套碱基编辑器, 对OsMPK6、OsMPK13、OsSERK2及OsWRKY45四个水稻内源基因靶位点进行编辑, 成功获得预期的水稻突变体材料, 效率高达62%。同时, 该套系统引入了全新的紫外激活GFP检测系统, 可以通过手持式紫外灯直接照射水稻组织, 使后代群体中的转基因分离检测十分便利。Molecular Plant同期还发表了中国科学院上海植物逆境生物学研究中心朱健康研究团队关于单碱基编辑的研究, 该文报道, 通过融合SpCas9 (D10A)和ecTadA*7.10的方法实现了对IPA1等基因的编辑, 编辑效率高达45.2% (Kai et al., 2018)。
中国科学院遗传与发育生物学研究所高彩霞研究组利用Cas9变体(nCas9-D10A)融合ecTadA和ecTadA*二聚体(Li et al., 2018), 建立并优化了高效精确的植物ABE (Adenine Base Editor)单碱基编辑系统, 在植物中实现了高效的A·T转换为G·C的碱基替换, 为植物基因组功能解析和作物遗传改良及新品种培育提供了重要技术支撑。
2.4 CRISPR/Cas9技术的安全性
人们对CRISPR/Cas9技术安全性的担心主要围绕2个问题, 一是基因组编辑系统的脱靶效应, 二是基因组编辑过程中外源基因的插入。2.4.1 降低脱靶效应
目前, 所有的基因组编辑技术都存在脱靶问题(王影等, 2018)。在已完成全基因组测序的情况下, 可以使用在线工具预测脱靶位点, 从而选择脱靶概率相对较低的sgRNA进行实验。然而, 许多植物的基因组还没有完全测序, 且在许多作物中, 相同物种的不同品种在可变基因座处具有多态性(Mikami et al., 2016)。相比哺乳动物系统中大量的信息(Corrigan-Curay et al., 2015), 目前描述植物中CRISPR/Cas9基因组编辑的脱靶效应的数据还较少, 植物系统中可能影响靶点特异性的潜在因素尚不清楚。
从生物信息学角度来看, 较长的DNA靶序列会降低脱靶错配的可能性, 但Fu等(2014)发现长度为17-19 nt的sgRNA显示出比原先20 nt的sgRNA更好的靶向活性, 同时脱靶率更低; 然而, 将sgRNA进一步缩短至15 nt则导致其活性丧失。对此可解释为: 在错配位点缩短的sgRNA与DNA的结合能力降低, 从而保持较高的靶点编辑效率以及更低的脱靶效应。
Endo等(2015)研究发现在植物中进行基因编辑时, 通常需要同时修饰多个同源基因, 即具有相似但不相同的DNA序列的基因, 以获得期望的表型, 于是他们利用CRISPR/Cas9的这种脱靶效应来诱导多种同源基因的敲除。他们针对细胞周期蛋白依赖性激酶B2 (CDKB2)设计了一段20 bp的序列作为sgRNA, 这20 bp与其它水稻CDK基因(CDKA1、CDKA2和CDKB1)具有不同数目的错配, 并在水稻中验证了该方法的可行性。Kweon等(2017)开发出一种融合向导RNA (fgRNA), 通过与Cas9和Cpf1蛋白共同发挥功能来降低脱靶效应, 并且不会降低其特异性和编辑效率。
2.4.2 消除外源基因插入
近年来, 转基因作物的安全性一直处于社会舆论的风口浪尖, 基因组编辑技术的出现为植物分子育种提供了前所未有的新机遇。传统转基因技术插入位点随机, 常导致许多不利结果, 如内源基因被破坏及外源基因沉默; 而基因组编辑的插入位点与编辑位点通常相隔较远, 通过自交得到的F1代中, 可以比较容易地分离得到不含有外源转基因成分的植株。
Woo等(2015)将纯化的Cas9蛋白与sgRNA预组装成蛋白质复合物溶液, 之后利用生成的蛋白质复合物溶液转染到拟南芥、烟草(Nicotiana tabacum)、莴苣(Lactuca sativa)和水稻的原生质体中, 在再生植物中实现了46%的靶向诱变, 且靶向位点的突变如同自然突变一样, 可以通过生殖细胞遗传给下一代。该方法在不将外源DNA导入细胞的情况下编辑植物基因组, 避免了使用可能会导致外源DNA介入的农杆菌介导法, 使安全性显著提高。
Liang等(2018)通过基因枪将CRISPR/Cas9 IVT (in vitro transcripts)和RNP (核糖核蛋白复合体)导入小麦未成熟幼胚, 建立了基因组定点修饰的DNA-free 基因组编辑体系。该方法只需要9-11周即可在T0代获得不含外源DNA的靶向修饰小麦突变体。由于IVT和RNP是以瞬时表达方式发挥作用, 故可成功避免外源DNA整合到宿主基因组中且大大降低了脱靶效应。
水稻中的CYP81A6编码P450细胞色素蛋白, 该蛋白失活后水稻对除草剂苯达松表现敏感。基因编辑后的个体中如果有CRISPR/Cas9存在且正常表达就会对苯达松表现敏感。浙江大学舒庆尧研究团队将靶标为CYP81A6的RNAi干涉片段与CRISPR/Cas9编辑元件串联整合,得到96个独立转化事件, 其中29个事件对苯达松表现敏感(CRISPR/Cas9正常表达)。后续通过在T1代喷施苯达松, 就可筛选得到不含外源基因的植株(Lu et al., 2017)。
华中农业大学赵云德研究团队通过利用自杀基因与CRISPR载体融合, 开发出高效去除含有Cas9转化事件的新技术, 通过基因型鉴定发现所有的T1代植株都不含有Cas9 (He et al., 2018)。这些DNA-free基因组编辑体系的建立将有助于进一步完善作物基因组编辑技术, 推进基因组编辑育种产业化进程。
2017年, 美国农业部(USDA)宣布将不会对采用基因组编辑工具CRISPR/Cas9进行遗传工程改造的双孢蘑菇(Agaricus bisporus)实施管控。这种蘑菇是由宾夕法尼亚州立大学的杨亦农(Yinong Yang)课题组改造的一种抗褐变白蘑菇(Waltz, 2016), 也是全球第一例获得商业化许可的基因编辑食品。
2.5 基因片段的插入与替换
目前, CRISPR/Cas9系统在植物领域中的应用大部分都通过NHEJ修复方式形成错配, 从而造成基因靶向敲除; 而在植物染色体上定点插入或替换一段序列一直以来都是困扰科学家的一个难题, 必须要借助HDR修复方式才能解决。Schiml等(2015)首次报道了通过CRISPR/Cas9系统利用HDR介导的修复机制在拟南芥ADH1基因位点上实现卡那抗性片段的导入。Sun等(2016)通过HDR修复方式修饰了水稻内源性ALS基因, 从而使其具有抗除草剂特性, 并发现当质粒和自由双链DNA形态同时存在时, 提供HDR供体能够有效提高植物同源重组修复效率。同年, 高彩霞团队与李家洋团队合作, 利用Cas9和1对sgRNA从DNA供体上剪切下一段序列, 之后再插入到水稻基因组的靶位点上, 从而实现基因的靶向敲入(Li et al., 2016)。据报道, 以小麦矮缩病毒WDV (wheat dwarf virus)作为载体, 通过提高供体DNA的拷贝数, 可以大幅提升敲入效率, 并在水稻中获得成功, 敲入效率最高可达19.4% (Wang et al., 2017)。朱健康研究团队报道了一种在拟南芥中基于二代转化策略与CRISPR/Cas9系统的超长基因片段高效精准敲入以及替换技术(Miki et al., 2018)。它无需任何标记辅助筛选就可以在拟南芥基因组精准定向地实现单氨基酸或大片段的插入或替换, 效率高达5%- 10%, 极大地推动了高等植物基因编辑技术的发展。
2.6 靶向基因转录调控
将不同的功能蛋白(转录抑制、转录激活的蛋白元件)与dCas9 (Dead Cas9, 无DNA切割活性的Cas9)融合表达, 利用sgRNA的特异性引导功能, 可以实现转录抑制或转录激活等不同的遗传操作。Baazim (2014)采用CRISPR/dCas9系统在烟草的瞬时转化实验中成功实现了对目标基因表达的激活与干扰。 在dCas9的C端融合转录激活因子EDLL或TAD (TAL effector激活结构域), 通过半定量PCR实验, 结果表明GUS基因或PDS基因可以被激活; 在dCas9蛋白的C端融合SRDX转录抑制结构域, 结果表明dCas9-SRDX可以靶向性干扰烟草PDS基因的表达。Cheng等(2016)构建出一种全新的基因调控系统, 称为Casilio系统。该系统包括dCas9蛋白, 附带有多个Pumilio/FBF结合位点的sgRNA以及与Pumilio/FBF结构域融合的效应蛋白, 将CRISPR/Cas9系统和Pumilio RNA结合蛋白结合在一起, 可实现对基因的高效调控。美国马里兰大学戚益平实验室与中国电子科技大学张勇实验室合作开发的CRISPR-Act 2.0比该实验室报道的第1代dCas9-VP64系统更具转录激活效应, 已在水稻细胞中成功进行多基因激活, 表明该系统在植物基因调控中具有很好的应用前景(Lowder et al., 2018)。3 展望
与前2代基因编辑技术相比, CRISPR/Cas9技术的出现为广大科研人员提供了一个简单、廉价且高效的遗传操作平台, 不仅能为基础研究提供强大支持, 还能将功能基因组学的研究成果加速向产品转化(李红和谢卡斌, 2017)。尤其在植物研究中, 基因组编辑技术独有以下优势: 其一不涉及伦理问题; 其二脱靶效应对植物的影响较小, 即使在T0代有脱靶现象发生, 在后代分离中也可以把脱靶位点分离出去。随着CRISPR/Cas9基因组编辑技术的不断改进和创新, 人们对CRISPR/Cas9系统运行机理的了解进一步加深, 原本一些困扰研究人员的问题已经迎刃而解。 例如, PAM限制基因组编辑范围和单碱基的精准编辑。然而, 植物基因组编辑也依然存在着一些亟待解决的问题。例如, 如何更高效地将基因组编辑材料运送到目的植物中, 随后产生定点编辑的植物(Altpeter et al., 2016)。目前, 虽然有多种可用的遗传转化方法, 但往往仅局限于特定的基因型、组织和培养类型, 并不能应用于所有的植物品种。另外, 目前大多数的基因组编辑依旧聚焦于创制DSB进而破坏基因功能, 仅有少数报道是运用HDR修复方式对内源基因进行序列插入和替换, 其主要原因是由于缺乏对基因组编辑的分子机理研究, 同时还缺少转化多种基因组编辑材料的有效手段与工具(冉毅东等, 2017)。
近年来, 一些新的基因组编辑系统不断问世。2015年, 麻省理工学院张峰团队报道了一种新的CRISPR效应蛋白Cas12a (Cpf1) (Zetsche et al., 2015), 并先后在动物(Kim et al., 2016; Hur et al., 2016)、微生物(Ungerer and Pakrasi, 2016)以及植物(Xu et al., 2017; Hu et al., 2017b)的基因组编辑中成功应用。Cas13a (C2c2)是能够靶向和切割噬菌体基因组的单链RNA (ssRNA)分子。据Aman等(2018)报道, Jesser等使用CRISPR/Cas13a干扰植物中的RNA病毒——芜菁花叶病毒(TuMV), 并取得了成功。以色列的科学家在细菌体内发现了10种新的免疫防御系统, 这些新系统将为开发新的基因组编辑工具带来希望(Doron et al., 2018)。相信在不久的将来, 不断改进优化的CRISPR/Cas9系统以及更多新型的基因组编辑技术必将在作物遗传改良和品种选育方面发挥更大的作用。
(责任编辑: 白羽红)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
DOI:10.1016/j.molp.2017.11.010URL [本文引用: 1]

User-friendly tools for robust transcriptional activation of endogenous genes are highly demanded in plants. We previously showed that a dCas9-VP64 system consisting of the deactivated CRISPR- associated protein 9 (dCasg) fused with four tandem repeats of the transcriptional activator VP16 0/1=64) could be used for transcriptional activation of endogenous genes in plants. In this study, we developed a second generation of vector systems for enhanced transcriptional activation in plants. We tested multiple strategies for dCasg-based transcriptional activation, and found that simultaneous recruitment of VP64 by dCas9 and a modified guide RNA scaffold gRNA2.0 (designated CRISPR-Act2.0) yielded stronger transcrip- tional activation than the dCas9-VP64 system. Moreover, we developed a multiplex transcription activator- likeeffector activation (mTALE-Act) system for simultaneous activation of up to four genes in plants. Our results suggest that mTALE-Act is even more effective than CRISPR-Act2.0 in most cases tested. In addition, we explored tissue-specific gene activation using positive feedback loops. Interestingly, our study revealed that certain endogenous genes are more amenable than others to transcriptional activation, and tightly regulated genes may cause target gene silencing when perturbed by activation probes. Hence, these new tools could be used to investigate gene regulatory networks and their control mechanisms. Assembly of multiplex CRISPR-Act2.0 and mTALE-Act systems are both based on streamlined and PCR-independent Golden Gate and Gateway cloning strategies, which will facilitate transcriptional activation applications in both dicots and monocots.
DOI:10.1111/pbi.12788URLPMID:5633759

To produce agronomically competitive rice with nutritionally superior, environmentally safe phytic acid (PA) levels, hairpin RNA (hpRNA)- and artificial microRNA (amiRNA)-mediated gene silencing approaches were explored to reduce both myo-inositol kinase gene (OsMIK) expression and PA accumulation in rice seeds. hpRNA and amiRNA sequences targeted to OsMIK (hpMIK and amiMIK), under the control... [Show full abstract]
DOI:10.1016/j.molp.2015.04.007URLPMID:25917172 [本文引用: 2]

A聽robust CRISPR/Cas9 vector system for multiplex genome editing in monocot and dicot plants was developed. Multiple sgRNA expression cassettes can be assembled into the binary CRISPR/Cas9 vectors in one round of cloning. This system can uniformly, efficiently, and simultaneously produce multiple heritable mutations in rice and Arabidopsis by targeting multiple genes or genomic sites via single transformation events.
DOI:10.1007/s11427-017-9247-9URL [本文引用: 2]

正Dear Editor,The CRISPR-Cas9(clustered regularly interspaced short palindromic repeats/Cas9)system has been widely used for a variety of applications,including targeted gene knockout,gene insertion,gene replacement and base editing.Despite its wide use,the genome editing using CRISPR-Cas9 is performed almost exclusively at sites containing canonical NGG protospacer adjacent motifs(PAMs).To overcome the PAM constraint of the CRISPR-Cas9 system,many attempts have been made to develop various Cas9 orthologs and
DOI:10.1093/pcp/pcw049URLPMID:26936792 [本文引用: 1]

Recent reports of CRISPR- (clustered regularly interspaced short palindromic repeats)/Cas9 (CRISPR-associated protein 9) mediated heritable mutagenesis in plants highlight the need for accuracy of the mutagenesis directed by this system. Off-target mutations are an important issue when considering functional gene analysis, as well as the molecular breeding of crop plants with large genome size, i.e. with many duplicated genes, and where the whole-genome sequence is still lacking. In mammals, off-target mutations can be suppressed by using Cas9 paired nickases together with paired guide RNAs (gRNAs). However, the performance of Cas9 paired nickases has not yet been fully assessed in plants. Here, we analyzed on- and off-target mutation frequency in rice calli and regenerated plants using Cas9 nuclease or Cas9 nickase with paired gRNAs. When Cas9 paired nickases were used, off-target mutations were fully suppressed in rice calli and regenerated plants. However, on-target mutation frequency also decreased compared with that induced by the Cas9 paired nucleases system. Since the gRNA sequence determines specific binding of Cas9 protein鈥揼RNA ribonucleoproteins at the targeted sequence, the on-target mutation frequency of Cas9 paired nickases depends on the design of paired gRNAs. Our results suggest that a combination of gRNAs that can induce mutations at high efficiency with Cas9 nuclease should be used together with Cas9 nickase. Furthermore, we confirmed that a combination of gRNAs containing a one nucleotide (1 nt) mismatch toward the target sequence could not induce mutations when expressed with Cas9 nickase. Our results clearly show the effectiveness of Cas9 paired nickases in delivering on-target specific mutations.
[本文引用: 1]
DOI:10.1038/nbt.1755
DOI:10.1038/nbt.2527 [本文引用: 2]
[本文引用: 1]
DOI:10.1038/srep16705URLPMID:4642324 [本文引用: 1]

Precise genome modification in large domesticated animals is desirable under many circumstances. In the past it is only possible through lengthy and burdensome cloning procedures. Here we attempted to achieve that goal through the use of the newest genome-modifying tool CRISPR/Cas9. We set out to knockin human albumin cDNA into pigAlblocus for the production of recombinant human serum albumin (rHSA). HSA is a widely used human blood product and is in high demand. We show that homologous recombination can occur highly efficiently in swine zygotes. All 16 piglets born from the manipulated zygotes carry the expected knockin allele and we demonstrated the presence of human albumin in the blood of these piglets. Furthermore, the knockin allele was successfully transmitted through germline. This success in precision genomic engineering is expected to spur exploration of pigs and other large domesticated animals to be used as bioreactors for the production of biomedical products or creation of livestock strains with more desirable traits.
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/nature07343URLPMID:18987743 [本文引用: 1]

Abstract Xeroderma pigmentosum is a monogenic disease characterized by hypersensitivity to ultraviolet light. The cells of xeroderma pigmentosum patients are defective in nucleotide excision repair, limiting their capacity to eliminate ultraviolet-induced DNA damage, and resulting in a strong predisposition to develop skin cancers. The use of rare cutting DNA endonucleases-such as homing endonucleases, also known as meganucleases-constitutes one possible strategy for repairing DNA lesions. Homing endonucleases have emerged as highly specific molecular scalpels that recognize and cleave DNA sites, promoting efficient homologous gene targeting through double-strand-break-induced homologous recombination. Here we describe two engineered heterodimeric derivatives of the homing endonuclease I-CreI, produced by a semi-rational approach. These two molecules-Amel3-Amel4 and Ini3-Ini4-cleave DNA from the human XPC gene (xeroderma pigmentosum group C), in vitro and in vivo. Crystal structures of the I-CreI variants complexed with intact and cleaved XPC target DNA suggest that the mechanism of DNA recognition and cleavage by the engineered homing endonucleases is similar to that of the wild-type I-CreI. Furthermore, these derivatives induced high levels of specific gene targeting in mammalian cells while displaying no obvious genotoxicity. Thus, homing endonucleases can be designed to recognize and cleave the DNA sequences of specific genes, opening up new possibilities for genome engineering and gene therapy in xeroderma pigmentosum patients whose illness can be treated ex vivo.
DOI:10.1016/j.molp.2018.01.005URLPMID:29382569

ACCEPTED MANUSCRIPT codon-optimized for expression in rice (Figure S1) and attached to 5使 terminal end of Cas9n-NLS using XTEN linker (Figure 1A and 1B). Therefore, it is likely that the editing window of hAID*鈭 would be restricted to the sgRNA-targeting site similar to the case of APOBEC1. To be noted, UGI was excluded at this time since more types of nucleotide conversions, produced in uracil N-glycosylase (UDG)-initiated base excision repair (BER), would be anticipated.The hAID*鈭-XTEN-Cas9n-NLS chimeric gene (termed rBE5) were first tested in rice leaf sheath protoplast, with the sgRNAs targeting a GCAC-containing ApaLI restriction site in OsRLCK185 and a TCC-containing BamHI restriction site in OsCERK1, respectively. Sequencing data showed that distinct mutations with high frequency of C>T conversions occurred in the editing window (Figure S2A-S2H), suggesting rBE5 functions on GC, AC, TC as well as CC in rice cells.As a follow-up to our previous study of the rBE3 system (Ren et al., 2017), rBE3 gene in the binary vector pUbi:rBE3 was replaced by rBE5 gene, namely pUbi:rBE5, and then further tested for efficiency in stable transgenic rice plants through Agrobacterium-mediated transformation. Pi-d2, an agriculturally important rice blast R gene in which a GC-containing region was resistant to rBE3-mediated editing in our earlier experiment, was first chosen for targeting (Figure 1C). As reported previously, a single amino acid substitution at position 441 in the recessive allele of Pi-d2 gene results in loss of resistance to Magnaporthe oryzae (Chen et al., 2006). Therefore, we assumed that introducing a G>A mutation (M441I) in endogenous pi-d2kit using rBE5 could recover its biological function. The same protospacer for rBE3 was used in this experiment, totally 26 independent transgenic lines of Kitaake were obtained after transformation and genotyped subsequently by Sanger sequencing. 8 heterozygous lines (30.8% efficiency) carrying a desired single G to A conversion at position -17 upstream of PAM were identified (Figure 1D-1E, Figure S3A), suggesting that rBE5 is much more efficient on target C that immediately follow a G than rBE3 does. We assumed that the successful gene correction of pi-d2 would be attributable to both the specificity and the improved activity of hAID*鈭. Remarkably, pi-d2 alleles with Indel mutations were also detected in 5 lines (Figure S3B-S3C).
DOI:10.1111/tpj.12704URLPMID:25327456 [本文引用: 2]

Summary The CRISPR/Cas nuclease is becoming a major tool for targeted mutagenesis in eukaryotes by inducing double-strand breaks (DSBs) at pre-selected genomic sites that are repaired by non-homologous end joining (NHEJ) in an error-prone way. In plants, it could be demonstrated that the Cas9 nuclease is able to induce heritable mutations in Arabidopsis thaliana and rice. Gene targeting (GT) by homologous recombination (HR) can also be induced by DSBs. Using a natural nuclease and marker genes, we previously developed an in planta GT strategy in which both a targeting vector and targeting locus are activated simultaneously via DSB induction during plant development. Here, we demonstrate that this strategy can be used for natural genes by CRISPR/Cas-mediated DSB induction. We were able to integrate a resistance cassette into the ADH1 locus of A.聽thaliana via HR. Heritable events were identified using a PCR-based genotyping approach, characterised by Southern blotting and confirmed on the sequence level. A major concern is the specificity of the CRISPR/Cas nucleases. Off-target effects might be avoided using two adjacent sgRNA target sequences to guide the Cas9 nickase to each of the two DNA strands, resulting in the formation of a DSB. By amplicon deep sequencing, we demonstrate that this Cas9 paired nickase strategy has a mutagenic potential comparable with that of the nuclease, while the resulting mutations are mostly deletions. We also demonstrate the stable inheritance of such mutations in A.聽thaliana .
DOI:10.16288/j.yczz.15-156URLPMID:26496748 [本文引用: 1]

Genome editing technologies using engineered nucleases have been widely used in many model organisms. Genome editing with sequence-specific nuclease(SSN) creates DNA double-strand breaks(DSBs) in the genomic target sites that are primarily repaired by the non-homologous end joining(NHEJ) or homologous recombi- nation(HR) pathways, which can be employed to achieve targeted genome modifications such as gene mutations, insertions, replacements or chromosome rearrangements. There are three major SSNs鈥攝inc finger nuclease(ZFN), transcription activator-like effector nuclease(TALEN) and clustered regularly interspaced short palindromic repeats/CRISPR-associated 9(CRISPR/Cas9) system. In contrast to ZFN and TALEN, which require substantial protein engineering to each DNA target, the CRISPR/Cas9 system requires only a change in the guide RNA. For this reason, the CRISPR/Cas9 system is a simple, inexpensive and versatile tool for genome engineering. Furthermore, a modified version of the CRISPR/Cas9 system has been developed to recruit heterologous domains that can regulate endogenous gene expression, such as activation, depression and epigenetic regulation. In this review, we summarize the development and applications of genome editing technologies for basic research and biotechnology, as well as highlight challenges and future directions, with particular emphasis on plants.
DOI:10.1038/nbt.2650URLPMID:23929338

The article offers information on genome modification of crop plants using a CRISPR-Cas system. It states that genome editing technologies using zinc finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) can also generate genome modifications. Photographs related to genome editing in rice and wheat using an engineered type II CRISPR-Cas system are also presented.
[本文引用: 1]
DOI:10.1016/j.molp.2016.01.001URLPMID:26768120 [本文引用: 1]
DOI:10.1146/annurev-genet-110410-132435URLPMID:21910633 [本文引用: 2]

DNA double-strand breaks (DSBs) are cytotoxic lesions that can result in mutagenic events or cell death if left unrepaired or repaired inappropriately. Cells use two major pathways for DSB repair: nonhomologous end joining (NHEJ) and homologous recombination (HR). The choice between these pathways depends on the phase of the cell cycle and the nature of the DSB ends. A critical determinant of repair pathway choice is the initiation of 5'-3' resection of DNA ends, which commits cells to homology-dependent repair, and prevents repair by classical NHEJ. Here, we review the components of the end resection machinery, the role of end structure, and the cell-cycle phase on resection and the interplay of end processing with NHEJ.
DOI:10.1038/srep39681URLPMID:28000776 [本文引用: 1]

Cyanobacteria are the ideal organisms for the production of a wide range of bioproducts as they can convert CO2directly into the desired end product using solar energy. Unfortunately, the engineering of cyanobacteria to create efficient cell factories has been impaired by the cumbersome genetic tools that are currently available for these organisms; especially when trying to accumulate multiple modifications. We sought to construct an efficient and precise tool for generating numerous markerless modifications in cyanobacteria using CRISPR technology and the alternative nuclease, Cpf1. In this study we demonstrate rapid engineering of markerless knock-ins, knock-outs and point mutations in each of three model cyanobacteria;Synechococcus, SynechocystisandAnabaena. The markerless nature ofcpf1genome editing will allow for complex genome modification that was not possible with previously existing technology while facilitating the development of cyanobacteria as highly modified biofactories.
DOI:10.1038/nature.2016.19754URLPMID:27111611 [本文引用: 1]

A fungus engineered with the CRISPR–Cas9 technique can be cultivated and sold without further oversight.
DOI:10.1016/j.molp.2017.03.002URLPMID:28315751 [本文引用: 1]
DOI:10.1038/nbt.3389URLPMID:26479191

Abstract Editing plant genomes without introducing foreign DNA into cells may alleviate regulatory concerns related to genetically modified plants. We transfected preassembled complexes of purified Cas9 protein and guide RNA into plant protoplasts of Arabidopsis thaliana, tobacco, lettuce and rice and achieved targeted mutagenesis in regenerated plants at frequencies of up to 46%. The targeted sites contained germline-transmissible small insertions or deletions that are indistinguishable from naturally occurring genetic variation.
DOI:10.1073/pnas.1420294112URLPMID:25733849 [本文引用: 2]

The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein 9 nuclease (Cas9) system is being harnessed as a powerful tool for genome engineering in basic research, molecular therapy, and crop improvement. This system uses a small guide RNA (gRNA) to direct Cas9 endonuclease to a specific DNA site; thus, its...
DOI:10.1186/s12870-014-0327-yURLPMID:4262988 [本文引用: 2]

Background To accelerate the application of the CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/ CRISPR-associated protein 9) system to a variety of plant species, a toolkit with additional plant selectable markers, more gRNA modules, and easier methods for the assembly of one or more gRNA expression cassettes is required. Results We developed a CRISPR/Cas9 binary vector set based on the pGreen or pCAMBIA backbone, as well as a gRNA (guide RNA) module vector set, as a toolkit for multiplex genome editing in plants. This toolkit requires no restriction enzymes besides BsaI to generate final constructs harboring maize-codon optimized Cas9 and one or more gRNAs with high efficiency in as little as one cloning step. The toolkit was validated using maize protoplasts, transgenic maize lines, and transgenic Arabidopsis lines and was shown to exhibit high efficiency and specificity. More importantly, using this toolkit, targeted mutations of three Arabidopsis genes were detected in transgenic seedlings of the T1 generation. Moreover, the multiple-gene mutations could be inherited by the next generation. Conclusions We developed a toolkit that facilitates transient or stable expression of the CRISPR/Cas9 system in a variety of plant species, which will facilitate plant research, as it enables high efficiency generation of mutants bearing multiple gene mutations.
DOI:10.16288/j.yczz.15-398URLPMID:27001476

CRISPR(Clustered regularly interspaced short palindromic repeats)/Cas(CRISPR associated proteins) is an acquired immune system found in bacteria and archaea that fight against invasion of viruses or plasmids. CRISPR/Cas systems are currently classified into three main types: I, II and III, of which type II has relatively simple components. The CRISPR/Cas9 technology modified from type II CRISPR/Cas system has been developed as an efficient genome editing tool. Since the initial application of the CRISPR/Cas9 technology in mammals in 2013, the reports of this system for genomic editing has skyrocketed. Farm animals are not only economically important animals, but also ideal animal models for human diseases and biomedical studies. In this review, we summarize the applications of CRISPR/Cas9 in farm animals, briefly describe the off-target effects and the main solutions, and finally highlight the future perspectives of this technology.
[本文引用: 2]
DOI:10.1111/pbi.12669URLPMID:27875019

CRISPR‐Cpf1 is a newly identifiedCRISPR‐Cas system, and Cpf1 was recently engineered as a molecular tool for targeted genome editing in mammalian cells. To test whether the engineeredCRISPR‐Cpf1 system could induce the production of rice mutants, we selected two genome targets in theOsPDSandOsBELgenes. Our results show that both targets could be efficiently mutated in transgenic rice plants usingCRISPR‐Cpf1. We found that pre‐crRNAs with a full‐length direct repeat sequence exhibited considerably increased efficiencies compared with mature crRNAs. In addition, the specificity and transmission of the mutation were investigated, and the behaviours of crRNA‐Cpf1‐induced plant targeted genome mutagenesis were assessed. Taken together, our results indicate thatCRISPR‐Cpf1 expression via stable transformation can efficiently generate specific and heritable targeted mutations in rice and thereby constitutes a novel and important approach to specific and precise plant genome editing.
DOI:10.1016/j.molp.2018.02.008URL [本文引用: 2]

Dear Editor, The newly developed CRISPR/Cas9-mediated base editing technology with cytosine deaminase is capable of precisely and efficiently introducing point mutations at the target genomic locus,which does not require double-stranded DNA breaks or any donor templates and thus exhibit a great potential for gene correction and genetic diversification in yeasts,plants,and mammalian and human cells (Komor et al.,2016;Nishida et al.,2016;Lu and Zhu,2017;Ren et al.,2017).However,compared with AID/APOBEC1 members of the cytosine deaminase family that are widely utilized in base editing to induce cytidine (C) to thymine (T) conversion,adenosine deaminase is far from being applicable since TadA/ADAR members act strictly on duplex RNA,or DNA/RNA hybrids with mismatches,instead of single-stranded DNA (Zheng et al.,2017).To address this problem,great efforts have been invested recently in identifying Escherichia coli TadA variants that accept DNA as a substrate through rounds of protein evolution and engineering,ultimately leading to a number of adenine base editors (ABEs) with great efficiencies and broadened sequence compatibility in inducing nucleotide changes at a wide range of target genomic loci in human cells (Gaudelli et al.,2017).In these ABE systems,the TadA:TadA* heterodimer is guided by the Cas9n/single guide RNA (sgRNA) complex to the target site,and the engineered TadA*,but not the wild-type TadA,functions as the active tRNA adenosine deaminase that turns adenine (A) to inosine (鈪) in singlestranded genomic DNA,subsequently resulting in A to guanine (G) mutation in genome during DNA repair or DNA replication (Gaudelli et al.,2017).These tools,together with previous base editors,enable programmable introduction of all four transitions (C to T,G to A,A to G,and T to C) at the target loci in the genome,greatly expanding the capabilities of base editing.Here,we report the development of fluorescencetracking base editing systems with E.coil TadA variants and Cas9 variants in rice.
DOI:10.1111/jipb.12622URLPMID:29226588 [本文引用: 1]

We report that a solo single-guide RNA(sg RNA) seed is capable of guiding Clustered Regularly Interspaced Short Palindromic Repeats(CRISPR)/CRISPR脿associated 9(CRISRP/Cas_9) to simultaneously edit multiple genes At RPL_(10)A, At RPL_(10)B and At RPL_(10)C in Arabidopsis. Our results also demonstrate that it is possible to use CRISPR/Cas_9 technology to create At RPL_(10) triple mutants which otherwise cannot be generated by conventional genetic crossing. Compared to other conventional multiplex CRISPR/Cas systems, a single sg RNA seed has the advantage of reducing off-target gene-editing. Such a gene editing system might be also applicable to modify other homologous genes, or even less-homologous sequences for multiple gene-editing in plants and other organisms.
DOI:10.13345/j.cjb.170171URL [本文引用: 1]

在过去的4年中,CRISPR/Cas9基因组编辑技术成为生命科学领域的革命性工具,为植物学基础研究和农作物遗传改良提供了高效、快速而又廉价的遗传操作工具.利用CRISPR/Cas9系统可以实现精准的knock-out和knock-in等遗传操作,也可用于靶向激活或抑制基因的表达.在CRISPR/Cas9被广泛地用于基因组编辑的同时,它的编辑能力、效率和精确度也在不断地改进和完善,特别是CRISPR/Cpf1系统的发掘和单碱基编辑技术的创建,使CRISPR系统正逐步成为一个理想的遗传工程技术平台.此外,利用CRISPR/Cas9技术改良的农作物品种也已经涌现,这必将推动精准基因组编辑技术在农作物遗传改良中的应用和发展.
URL [本文引用: 1]

目前科研人员已经研发了许多基因组编辑工具,新的基因组编辑工具也在不断地探索中,其中有一些编辑工具,如CRISPR/Cas9系统已经被广泛地应用于植物遗传学、生物技术和育种等各方面.基因组编辑技术可以对农艺性状相关基因的特异序列进行精准编辑,从而开启了作物改良的新时代.本文聚焦于植物基因组编辑试剂材料导入及转化的最新方法及进展,比较了各种导入及转化系统的优缺点,最后讨论了植物基因组编辑试剂材料导入及转化未来可能的发展方向.
DOI:10.11983/CBB18004URL [本文引用: 1]

近年来,CRISPR定点编辑技术发展迅猛,在动物、植物和微生物中均得到广泛应用.其中,备受关注的脱靶现象也是研究的热点,迄今己取得了重要进展.该文介绍了脱靶现象的产生原理及体内和体外检测脱靶现象的方法,评价了通过改进sgRNA设计和优化CRISPR系统等来降低脱靶率的方法.在植物基因组定点编辑过程中,应适时检测脱靶现象,提高脱靶检测的精确度和准确度.
DOI:10.1105/tpc.16.00196URLPMID:27335450 [本文引用: 1]

Abstract Plant transformation has enabled fundamental insights into plant biology and revolutionized commercial agriculture. Unfortunately, for most crops, transformation and regeneration remain arduous even after more than 30 years of technological advances. Genome editing provides novel opportunities to enhance crop productivity but relies on genetic transformation and plant regeneration, which are bottlenecks in the process. Here, we review the state of plant transformation and point to innovations needed to enable genome editing in crops. Plant tissue culture methods need optimization and simplification for efficiency and minimization of time in culture. Currently, specialized facilities exist for crop transformation. Single-cell and robotic techniques should be developed for high-throughput genomic screens. Plant genes involved in developmental reprogramming, wound response, and/or homologous recombination should be used to boost the recovery of transformed plants. Engineering universal Agrobacterium tumefaciens strains and recruiting other microbes, such as Ensifer or Rhizobium, could facilitate delivery of DNA and proteins into plant cells. Synthetic biology should be employed for de novo design of transformation systems. Genome editing is a potential game-changer in crop genetics when plant transformation systems are optimized. 2016 American Society of Plant Biologists. All rights reserved.
DOI:10.1186/s13059-017-1381-1URL

CRISPR/Cas systems confer immunity against invading nucleic acids and phages in bacteria and archaea. CRISPR/Cas13a (known previously as C2c2) is a class 2 type VI-A ribonuclease capable of targeting and cleaving single-stranded RNA (ssRNA) molecules of the phage genome. Here, we employ CRISPR/Cas13a to engineer interference with an RNA virus, Turnip Mosaic Virus (TuMV), in plants. CRISPR/Cas13a produces interference against green fluorescent protein (GFP)-expressing TuMV in transient assays and stable overexpression lines of Nicotiana benthamiana. CRISPR RNA (crRNAs) targeting the HC-Pro and GFP sequences exhibit better interference than those targeting other regions such as coat protein (CP) sequence. Cas13a can also process pre-crRNAs into functional crRNAs. Our data indicate that CRISPR/Cas13a can be used for engineering interference against RNA viruses, providing a potential novel mechanism for RNA-guided immunity against RNA viruses and for other RNA manipulations in plants. The online version of this article (doi:10.1186/s13059-017-1381-1) contains supplementary material, which is available to authorized users.
[本文引用: 1]
DOI:10.1126/science.1138140URLPMID:17379808 [本文引用: 1]

Clustered regularly interspaced short palindromic repeats (CRISPR) are a distinctive feature of the genomes of most Bacteria and Archaea and are thought to be involved in resistance to bacteriophages. We found that, after viral challenge, bacteria integrated new spacers derived from phage genomic sequences. Removal or addition of particular spacers modified the phage-resistance phenotype of the cell. Thus, CRISPR, together with associated cas genes, provided resistance against phages, and resistance specificity is determined by spacer-phage sequence similarity.
DOI:10.1126/science.1159689URL [本文引用: 1]
[本文引用: 1]
DOI:10.1038/cr.2016.3URLPMID:4746610

Cell death and differentiation is a monthly research journal focused on the exciting field of programmed cell death and apoptosis. It provides a single accessible source of information for both scientists and clinicians, keeping them up-to-date with advances in the field. It encompasses programmed cell death, cell death induced by toxic agents, differentiation and the interrelation of these with cell proliferation.
DOI:10.1007/978-1-4939-1862-1_10URLPMID:23287718 [本文引用: 2]

The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-Cas9 system is an adaptive immune system that exists in a variety of microbes. It could be engineered to function in eukaryotic c
DOI:10.1038/mt.2015.54URLPMID:25943494 [本文引用: 1]

http://www.nature.com/doifinder/10.1038/mt.2015.54
DOI:10.1038/nature09886URLPMID:3070239 [本文引用: 1]

Abstract CRISPR/Cas systems constitute a widespread class of immunity systems that protect bacteria and archaea against phages and plasmids, and commonly use repeat/spacer-derived short crRNAs to silence foreign nucleic acids in a sequence-specific manner. Although the maturation of crRNAs represents a key event in CRISPR activation, the responsible endoribonucleases (CasE, Cas6, Csy4) are missing in many CRISPR/Cas subtypes. Here, differential RNA sequencing of the human pathogen Streptococcus pyogenes uncovered tracrRNA, a trans-encoded small RNA with 24-nucleotide complementarity to the repeat regions of crRNA precursor transcripts. We show that tracrRNA directs the maturation of crRNAs by the activities of the widely conserved endogenous RNase III and the CRISPR-associated Csn1 protein; all these components are essential to protect S. pyogenes against prophage-derived DNA. Our study reveals a novel pathway of small guide RNA maturation and the first example of a host factor (RNase III) required for bacterial RNA-mediated immunity against invaders.
[本文引用: 1]
DOI:10.1126/science.aar4120URLPMID:29371424 [本文引用: 1]

The arms race between bacteria and phages led to the development of sophisticated antiphage defense systems, including CRISPR-Cas and restriction-modification systems. Evidence suggests that unknown defense systems are located in “defense islands” in microbial genomes. We comprehensively characterized the bacterial defensive arsenal by examining gene families that are clustered next to known defense genes in prokaryotic genomes. Candidate defense systems were systematically engineered and validated in model bacteria for their antiphage activities. We report nine previously unknown antiphage systems and one antiplasmid system that are widespread in microbes and strongly protect against foreign invaders. These include systems that adopted components of the bacterial flagella and condensin complexes. Our data also suggest a common, ancient ancestry of innate immunity components shared between animals, plants, and bacteria.
DOI:10.1093/nar/gki912URLPMID:1270952 [本文引用: 1]

Custom-designed zinc finger nucleases (ZFNs), proteins designed to cut at specific DNA sequences, are becoming powerful tools in gene targeting--the process of replacing a gene within a genome by homologous recombination (HR). ZFNs that combine the non-specific cleavage domain (N) of FokI endonuclease with zinc finger proteins (ZFPs) offer a general way to deliver a site-specific double-strand break (DSB) to the genome. The development of ZFN-mediated gene targeting provides molecular biologists with the ability to site-specifically and permanently modify plant and mammalian genomes including the human genome via homology-directed repair of a targeted genomic DSB. The creation of designer ZFNs that cleave DNA at a pre-determined site depends on the reliable creation of ZFPs that can specifically recognize the chosen target site within a genome. The (Cys2His2) ZFPs offer the best framework for developing custom ZFN molecules with new sequence-specificities. Here, we explore the different approaches for generating the desired custom ZFNs with high sequence-specificity and affinity. We also discuss the potential of ZFN-mediated gene targeting for 'directed mutagenesis' and targeted 'gene editing' of the plant and mammalian genome as well as the potential of ZFN-based strategies as a form of gene therapy for human therapeutics in the future.
DOI:10.1093/pcp/pcu154URLPMID:4301742 [本文引用: 1]

The clustered regularly interspaced short palindromic repeat (CRISPR)-associated endonuclease 9 (CRISPR/Cas9) system has been demonstrated to be a robust genome engineering tool in a variety of organisms including plants. However, it has been shown that the CRISPR/Cas9 system cleaves genomic DNA sequences containing mismatches to the guide RNA strand. We expected that this low specificity could be exploited to induce multihomeologous and multiparalogous gene knockouts. In the case of polyploid plants, simultaneous modification of multiple homeologous genes, i.e. genes with similar but not identical DNA sequences, is often needed to obtain a desired phenotype. Even in diploid plants, disruption of multiparalogous genes, which have functional redundancy, is often needed. To validate the applicability of the CRISPR/Cas9 system to target mutagenesis of paralogous genes in rice, we designed a single-guide RNA (sgRNA) that recognized 20 bp sequences of cyclin-dependent kinase B2 (CDKB2) as an on-target locus. These 20 bp possess similarity to other rice CDK genes (CDKA1, CDKA2 and CDKB1) with different numbers of mismatches. We analyzed mutations in these four CDK genes in plants regenerated from Cas9/sgRNA-transformed calli and revealed that single, double and triple mutants of CDKA2, CDKB1 and CDKB2 can be created by a single sgRNA.
DOI:10.1111/pbi.12920URLPMID:29569825 [本文引用: 1]

Recently, engineered minichromosomes have been produced using a telomere-mediated truncation technique in some plants. However, the study on transferring genes to minichromosomes is very limited. Here, telomere-mediated truncation was successfully performed in common wheat (Triticum aestivum) to generate stable truncated chromosomes accompanied by a relatively high frequency of chromosomal... [Show full abstract]
DOI:10.1038/nbt.2808URLPMID:24463574 [本文引用: 1]

Abstract Clustered, regularly interspaced, short palindromic repeat (CRISPR) RNA-guided nucleases (RGNs) are highly efficient genome editing tools. CRISPR-associated 9 (Cas9) RGNs are directed to genomic loci by guide RNAs (gRNAs) containing 20 nucleotides that are complementary to a target DNA sequence. However, RGNs can induce mutations at sites that differ by as many as five nucleotides from the intended target. Here we report that truncated gRNAs, with shorter regions of target complementarity <20 nucleotides in length, can decrease undesired mutagenesis at some off-target sites by 5,000-fold or more without sacrificing on-target genome editing efficiencies. In addition, use of truncated gRNAs can further reduce off-target effects induced by pairs of Cas9 variants that nick DNA (paired nickases). Our results delineate a simple, effective strategy to improve the specificities of Cas9 nucleases or paired nickases.
[本文引用: 1]
[本文引用: 1]
DOI:10.1073/pnas.1208507109URL [本文引用: 1]

Clustered, regularly interspaced, short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems provide adaptive immunity against viruses and plasmids in bacteria and archaea. The silencing of invading nucleic acids is executed by ribonucleoprotein complexes preloaded with small, interfering CRISPR RNAs (crRNAs) that act as guides for targeting and degradation of foreign nucleic acid. Here, we demonstrate that the Cas9-crRNA complex of the Streptococcus thermophilus CRISPR3/Cas system introduces in vitro a double-strand break at a specific site in DNA containing a sequence complementary to crRNA. DNA cleavage is executed by Cas9, which uses two distinct active sites, RuvC and HNH, to generate site-specific nicks on opposite DNA strands. Results demonstrate that the Cas9-crRNA complex functions as an RNA-guided endonuclease with RNA-directed target sequence recognition and protein-mediated DNA cleavage. These findings pave the way for engineering of universal programmable RNA-guided DNA endonucleases.
DOI:10.1038/s41586-018-0070-xURLPMID:29160308 [本文引用: 1]

Abstract The spontaneous deamination of cytosine is a major source of transitions from C090004G to T090004A base pairs, which account for half of known pathogenic point mutations in humans. The ability to efficiently convert targeted A090004T base pairs to G090004C could therefore advance the study and treatment of genetic diseases. The deamination of adenine yields inosine, which is treated as guanine by polymerases, but no enzymes are known to deaminate adenine in DNA. Here we describe adenine base editors (ABEs) that mediate the conversion of A090004T to G090004C in genomic DNA. We evolved a transfer RNA adenosine deaminase to operate on DNA when fused to a catalytically impaired CRISPR-Cas9 mutant. Extensive directed evolution and protein engineering resulted in seventh-generation ABEs that convert targeted A090004T base pairs efficiently to G090004C (approximately 50% efficiency in human cells) with high product purity (typically at least 99.9%) and low rates of indels (typically no more than 0.1%). ABEs introduce point mutations more efficiently and cleanly, and with less off-target genome modification, than a current Cas9 nuclease-based method, and can install disease-correcting or disease-suppressing mutations in human cells. Together with previous base editors, ABEs enable the direct, programmable introduction of all four transition mutations without double-stranded DNA cleavage.
DOI:10.1016/j.molp.2018.05.005URLPMID:29857174
DOI:10.1038/nbt.3290URLPMID:4729442 [本文引用: 2]

CRISPR-Cas-mediated genome editing relies on guide RNAs that direct site-specific DNA cleavage facilitated by the Cas endonuclease. Here we report that chemical alterations to synthesized single guide RNAs (sgRNAs) enhance genome editing efficiency in human primary T cells and CD34(+) hematopoietic stem and progenitor cells. Co-delivering chemically modified sgRNAs with Cas9 mRNA or protein is an efficient RNA- or ribonucleoprotein (RNP)-based delivery method for the CRISPR-Cas system, without the toxicity associated with DNA delivery. This approach is a simple and effective way to streamline the development of genome editing with the potential to accelerate a wide array of biotechnological and therapeutic applications of the CRISPR-Cas technology.
DOI:10.1038/nature26155URLPMID:29512652

Programmable DNA nucleases have provided scientists with the unprecedented ability to probe, regulate, and manipulate the human genome. Zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and the clustered regularly interspaced short palindromic repeat-Cas9 system (CRISPR-Cas9) represent a powerful array of tools that can bind to and cleave a specified DNA... [Show full abstract]
DOI:10.1111/pbi.12771URLPMID:28605576 [本文引用: 2]

Abstract Clustered regularly interspaced short palindromic repeats-associated protein 9 (CRISPR-Cas9) is a revolutionary technology that enables efficient genomic modification in many organisms. Currently, the wide use of Streptococcus pyogenes Cas9 (SpCas9) primarily recognises sites harbouring a canonical NGG protospacer adjacent motif (PAM). The newly developed VQR (D1135V/R1335Q/T1337R) variant of Cas9 has been shown to cleave sites containing NGA PAM in rice, which greatly expanded the range of genome editing. However, the low editing efficiency of the VQR variant remains, which limits its wide application in genome editing. In this study, by modifying the single guide RNA (sgRNA) structure and using strong endogenous promoters, we significantly increased the editing efficiency of the VQR variant. The modified CRISPR-Cas9-VQR system provides a robust toolbox for multiplex genome editing at sites containing non-canonical NGA PAM. This article is protected by copyright. All rights reserved. This article is protected by copyright. All rights reserved.
DOI:10.1016/j.molp.2016.03.003URLPMID:26995294

Dear Editor, The CRISPR/Cas9 system has emerged as a versatile molecular tool for genome editing in various organisms,including plants.In this system,the specificity of Cas9-directed DNA cleavage strictly requires the presence of a chimeric single guide RNA (sgRNA) and a short trinucleotide protospacer adjacent motif (PAM) in the genome (Anders et al.,2014;Sternberg et al.,2014).To date,the Cas9 used in plants has only been shown to recognize PAM sequences in the canonical form NGG (Li et al.,2013;Miao et al.,2013;Shan et al.,2013;Ma et al.,2015b).As such,the range of sequences for genome editing in plants is limited to sites containing an NGG motif.Many attempts have been made to overcome this constraint,and recent work has revealed that Cas9 can be modified to recognize alternative PAM sequences in zebrafish and human cells (Kleinstiver et al.,2015).However,the targeting range limitations of the CRISPR/Cas9 systems in plants have yet to be resolved.The widely cultivated plant rice is not only an important food crop but also a model crop plant because of its relatively small genome and relative ease of transformation.To expand the range of genome editing in rice,we generated two Cas9 variants as reported (Kleinstiver et al.,2015) and investigated their genome editing performance in rice.We demonstrate that Cas9 can be engineered to target sites containing alternative non-canonical PAMs in rice,which significantly broadens the range of genome editing.
DOI:10.1016/j.jgg.2016.12.001URLPMID:28043782 [本文引用: 2]

正Cpfl is a class 2/type V CRISPR effector that has been recently harnessed for genome editing(Zetsche et al.,2015;Hur et al.,2016;Kim et al.,2016).Cpfl recognizes thymidine-rich sequence as the protospacer-adjacent motif(PAM)at the 5'end of target sequences.In addition,Cpfl requires only a single shorter crRNA and
DOI:10.1038/nbt.3596URLPMID:27272385 [本文引用: 1]

[No abstract available]
[本文引用: 1]
DOI:10.1089/15362310252780816URLPMID:11883425 [本文引用: 2]

The rapid increase in genomic sequences provides new opportunities for comparative genomics. In this report, we describe a novel family of repeat sequences that is present in Bacteria and Archaea but not in Eukarya. The repeat loci typically consisted of repetitive stretches of nucleotides with a length of 25 to 37 bp alternated by nonrepetitive DNA spacers of approximately equal size as the repeats. The nucleotide sequences and the size of the repeats were highly conserved within a species, but between species the sequences showed no similarity. Due to their characteristic structure, we have designated this family of repeat loci as SPacers Interspersed Direct Repeats (SPIDR). The SPIDR loci were identified in more than forty different prokaryotic species. Individual species such as Mycobacterium tuberculosis contain one SPIDR locus, while other species such as Methanococcus jannaschii contained up to 20 different loci. The number of repeats in a locus varies greatly from two repeats to several dozens of repeats. The SPIDR loci were flanked by a common 300-500-bp leader sequence, which appeared to be conserved within a species but not between species. The SPIDR locus of M. tuberculosis is extensively used for strain typing. The finding of SPIDR loci in other prokaryotes, including the pathogens Salmonella, Campylobacter, and Pasteurella may extend this surveillance to other species.
DOI:10.1146/annurev-biophys-062215-010822URLPMID:28375731

Abstract Many bacterial clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated (Cas) systems employ the dual RNA-guided DNA endonuclease Cas9 to defend against invading phages and conjugative plasmids by introducing site-specific double-stranded breaks in target DNA. Target recognition strictly requires the presence of a short protospacer adjacent motif (PAM) flanking the target site, and subsequent R-loop formation and strand scission are driven by complementary base pairing between the guide RNA and target DNA, Cas9-DNA interactions, and associated conformational changes. The use of CRISPR-Cas9 as an RNA-programmable DNA targeting and editing platform is simplified by a synthetic single-guide RNA (sgRNA) mimicking the natural dual trans-activating CRISPR RNA (tracrRNA)-CRISPR RNA (crRNA) structure. This review aims to provide an in-depth mechanistic and structural understanding of Cas9-mediated RNA-guided DNA targeting and cleavage. Molecular insights from biochemical and structural studies provide a framework for rational engineering aimed at altering catalytic function, guide RNA specificity, and PAM requirements and reducing off-target activity for the development of Cas9-based therapies against genetic diseases.
DOI:10.1126/science.1225829URL [本文引用: 2]
DOI:10.7554/eLife.00471URLPMID:23386978 [本文引用: 1]

10.7554/eLife.00471.001Type II CRISPR immune systems in bacteria use a dual RNA-guided DNA endonuclease, Cas9, to cleave foreign DNA at specific sites. We show here that Cas9 assembles with hybrid guide RNAs in human cells and can induce the formation of double-strand DNA breaks (DSBs) at a site complementary to the guide RNA sequence in genomic DNA. This cleavage activity requires both Cas9 and the complementary binding of the guide RNA. Experiments using extracts from transfected cells show that RNA expression and/or assembly into Cas9 is the limiting factor for Cas9-mediated DNA cleavage. In addition, we find that extension of the RNA sequence at the 3??? end enhances DNA targeting activity in vivo. These results show that RNA-programmed genome editing is a facile strategy for introducing site-specific genetic changes in human cells.DOI: http://dx.doi.org/10.7554/eLife.00471.001
DOI:10.1016/j.molp.2018.02.007URL [本文引用: 1]
DOI:10.1038/nbt.3614URLPMID:27272387

react-text: 457 Decreasing the acquisition time in bioluminescence imaging (BLI) and bioluminescence tomography (BLT) will enable animals to be imaged within the window of stable emission of the bioluminescent source, a higher imaging throughput and minimisation of the time which an animal is anaesthetised. This work investigates, through simulation using a heterogeneous mouse model, two methods of decreasing... /react-text react-text: 458 /react-text [Show full abstract]
DOI:10.1038/s41467-017-01650-wURLPMID:29167440 [本文引用: 1]

The bacteria-derived clustered regularly interspaced short palindromic repeat (CRISPR)–Cas systems are powerful tools for genome engineering. Recently, in addition to Cas protein engineering, the improvement of guide RNAs are also performed, contributing to broadening the research area of CRISPR–Cas9 systems. Here we develop a fusion guide RNA (fgRNA) that functions with both Cas9 and Cpf1 proteins to induce mutations in human cells. Furthermore, we demonstrate that fgRNAs can be used in multiplex genome editing and orthogonal genome manipulation with two types of Cas proteins. Our results show that fgRNAs can be used as a tool for performing multiple gene manipulations. Cas9 and Cpf1 have both been adapted for genome engineering, editing and gene expression regulation. Here the authors design a fusion guide RNA that can interact with both proteins for multiple and orthogonal genome manipulation.
DOI:10.1111/tpj.13782URLPMID:29161464 [本文引用: 2]

Abstract The CRISPR/Cas9 system has greatly improved our ability to engineer targeted mutations in eukaryotic genomes. While CRISPR/Cas9 appears to work universally, the efficiency of targeted mutagenesis and the adverse generation of off-target mutations vary greatly between different organisms. In this study, we report that Arabidopsis plants subjected to heat stress at 3700°C show much higher frequencies of CRISPR-induced mutations compared to plants grown continuously at the standard temperature (2200°C). Using quantitative assays relying on GFP reporter genes, we found that targeted mutagenesis by CRISPR/Cas9 in Arabidopsis is increased by ~5-fold in somatic tissues and up to 100-fold in the germline upon heat treatment. This effect of temperature on the mutation rate is not limited to Arabidopsis, as we observed a similar increase in targeted mutations by CRISPR/Cas9 in Citrus plants exposed to heat stress at 3700°C. In vitro assays demonstrate that Cas9 from Streptococcus pyogenes (SpCas9) is more active in creating double-stranded DNA breaks at 3700°C than at 2200°C, thus indicating a potential contributing mechanism for the in vivo effect of temperature on CRISPR/Cas9. This study reveals the importance of temperature in modulating SpCas9 activity in eukaryotes, and provides a simple method to increase on-target mutagenesis in plants using CRISPR/Cas9. This article is protected by copyright. All rights reserved. This article is protected by copyright. All rights reserved.
DOI:10.1186/s13059-018-1443-zURL [本文引用: 1]

Nucleotide base editors in plants have been limited to conversion of cytosine to thymine. Here, we describe a new plant adenine base editor based on an evolved tRNA adenosine deaminase fused to the nickase CRISPR/Cas9, enabling A61T to G61C conversion at frequencies up to 7.5% in protoplasts and 59.1% in regenerated rice and wheat plants. An endogenous gene is also successfully modified through introducing a gain-of-function point mutation to directly produce an herbicide-tolerant rice plant. With this new adenine base editing system, it is now possible to precisely edit all base pairs, thus expanding the toolset for precise editing in plants. The online version of this article (10.1186/s13059-018-1443-z) contains supplementary material, which is available to authorized users.
DOI:10.1038/nplants.2016.139URLPMID:27618611 [本文引用: 1]

Abstract Sequence-specific nucleases have been exploited to create targeted gene knockouts in various plants(1), but replacing a fragment and even obtaining gene insertions at specific loci in plant genomes remain a serious challenge. Here, we report efficient intron-mediated site-specific gene replacement and insertion approaches that generate mutations using the non-homologous end joining (NHEJ) pathway using the clustered regularly interspaced short palindromic repeats (CRISPR)-CRISPR-associated protein 9 (Cas9) system. Using a pair of single guide RNAs (sgRNAs) targeting adjacent introns and a donor DNA template including the same pair of sgRNA sites, we achieved gene replacements in the rice endogenous gene 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) at a frequency of 2.0%. We also obtained targeted gene insertions at a frequency of 2.2% using a sgRNA targeting one intron and a donor DNA template including the same sgRNA site. Rice plants harbouring the OsEPSPS gene with the intended substitutions were glyphosate-resistant. Furthermore, the site-specific gene replacements and insertions were faithfully transmitted to the next generation. These newly developed approaches can be generally used to replace targeted gene fragments and to insert exogenous DNA sequences into specific genomic sites in rice and other plants.
[本文引用: 1]
DOI:10.1038/nbt.2199URLPMID:22565958 [本文引用: 1]

The article focuses on a study which examined the use of transcriptor activator-like (TAL) effector nucleases (TALEN) in editing a specific S gene in rice to prevent the virulence strategy of Xanthomonas oryzae. For TALEN-based disruption, the rice bacterial blight susceptibility gene was targeted. Results of the study showed that bacterial infection assays were resistant to infection by pathogenic Xoo.
DOI:10.1016/j.cell.2015.09.038URLPMID:26422227

Cpf1 is a RNA-guided DNA nuclease that provides immunity in bacteria and can be adapted for genome editing in mammalian cells.
DOI:10.1007/s13238-015-0153-5URLPMID:25894090 [本文引用: 1]

Abstract Genome editing tools such as the clustered regularly interspaced short palindromic repeat (CRISPR)-associated system (Cas) have been widely used to modify genes in model systems including animal zygotes and human cells, and hold tremendous promise for both basic research and clinical applications. To date, a serious knowledge gap remains in our understanding of DNA repair mechanisms in human early embryos, and in the efficiency and potential off-target effects of using technologies such as CRISPR/Cas9 in human pre-implantation embryos. In this report, we used tripronuclear (3PN) zygotes to further investigate CRISPR/Cas9-mediated gene editing in human cells. We found that CRISPR/Cas9 could effectively cleave the endogenous 脦虏-globin gene (HBB). However, the efficiency of homologous recombination directed repair (HDR) of HBB was low and the edited embryos were mosaic. Off-target cleavage was also apparent in these 3PN zygotes as revealed by the T7E1 assay and whole-exome sequencing. Furthermore, the endogenous delta-globin gene (HBD), which is homologous to HBB, competed with exogenous donor oligos to act as the repair template, leading to untoward mutations. Our data also indicated that repair of the HBB locus in these embryos occurred preferentially through the non-crossover HDR pathway. Taken together, our work highlights the pressing need to further improve the fidelity and specificity of the CRISPR/Cas9 platform, a prerequisite for any clinical applications of CRSIPR/Cas9-mediated editing.
Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9
1
2018
... 将不同的功能蛋白(转录抑制、转录激活的蛋白元件)与dCas9 (Dead Cas9, 无DNA切割活性的Cas9)融合表达, 利用sgRNA的特异性引导功能, 可以实现转录抑制或转录激活等不同的遗传操作.
Robust transcriptional activation in plants using multiplexed CRISPR-Act 2.0 and mTALE-Act systems
1
2018
... 水稻中的CYP81A6编码P450细胞色素蛋白, 该蛋白失活后水稻对除草剂苯达松表现敏感.基因编辑后的个体中如果有CRISPR/Cas9存在且正常表达就会对苯达松表现敏感.浙江大学舒庆尧研究团队将靶标为CYP81A6的RNAi干涉片段与CRISPR/Cas9编辑元件串联整合,得到96个独立转化事件, 其中29个事件对苯达松表现敏感(CRISPR/Cas9正常表达).后续通过在T1代喷施苯达松, 就可筛选得到不含外源基因的植株(
CRISPR-S: an active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants
0
2017
A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants
2
2015
... CRISPR/Cas系统最早是由日本大阪大学的一个研究团队在1987年对大肠杆菌碱性磷酸酶基因进行研究时发现的(
... 研究人员发现靶位点的核酸组成(即GC含量)、Cas9的表达水平以及靶区域的表观遗传等都会对基因组编辑效率产生影响.
CRISPR interference limits horizontal gene transfer in
0
2008
Robust genome editing of CRISPR-Cas9 at NAG PAMs in rice
2
2018
...
... 目前, 所有的基因组编辑技术都存在脱靶问题(
Precision targeted mutagenesis via Cas9 paired nickases in rice
1
2016
... 朱健康研究团队报道了一种在拟南芥中基于二代转化策略与CRISPR/Cas9系统的超长基因片段高效精准敲入以及替换技术(
CRISPR/Cas9-mediated gene targeting in
1
2018
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
A tale nuclease architecture for efficient genome editing
0
2011
RNA guides genome engineering
2
2013
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
... ;
Targeted mutagenesis in the model plant
1
2013
... 目前, 在医学(
Production of human albumin in pigs through CRISPR/Cas9-mediated knockin of human cDNA into swine albumin locus in the zygotes
1
2015
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
Homologous recombination in plant cells is enhanced by
1
1993
... 除应用最广的SpCas9外, 其它不同来源的Cas9也具有基因编辑能力.研究人员从金黄色葡萄球菌(Staphylococcus aureus)中找到1种较小的Cas9核酸酶(SaCas9) (
In vivo genome editing using
1
2015
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
Molecular basis of xeroderma pigmentosum group C DNA recognition by engineered meganucleases
1
2008
... 2016年, 在哺乳动物细胞中成功对靶标基因实现了单碱基编辑(C-T).随后, 单碱基编辑技术在水稻、小麦(Triticum aestivum)、玉米、拟南芥和番茄(Solanum lycopersicum)等多种植物中得到应用, 然而所采用的APOBEC1胞嘧啶脱氨酶表现出对TC的偏好性, 对序列为GC的编辑效率较低.鉴于此,
Improved base editor for efficiently inducing genetic variations in rice with CRISPR/Cas9-guided hyperactive hAID mutant
0
2018
The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in
2
2015
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
... 目前, CRISPR/Cas9系统在植物领域中的应用大部分都通过NHEJ修复方式形成错配, 从而造成基因靶向敲除; 而在植物染色体上定点插入或替换一段序列一直以来都是困扰科学家的一个难题, 必须要借助HDR修复方式才能解决.
Research progress of genome editing and derivative technologies in plants
1
2015
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
Targeted genome modification of crop plants using a CRISPR-Cas system
0
2013
Highly efficient heritable plant genome engineering using Cas9 orthologues from
1
2015
... 除应用最广的SpCas9外, 其它不同来源的Cas9也具有基因编辑能力.研究人员从金黄色葡萄球菌(Staphylococcus aureus)中找到1种较小的Cas9核酸酶(SaCas9) (
Engineering herbicide-resistant rice plants through CRISPR/Cas9-mediated homologous recombination of acetolactate synthase
1
2016
... 目前, CRISPR/Cas9系统在植物领域中的应用大部分都通过NHEJ修复方式形成错配, 从而造成基因靶向敲除; 而在植物染色体上定点插入或替换一段序列一直以来都是困扰科学家的一个难题, 必须要借助HDR修复方式才能解决.
Double-strand break end resection and repair pathway choice
2
2011
... 真核生物细胞内有2种修复DSB的方式: 一种是非同源末端连接(nonhomologous end-joining, NHEJ), 另一种是同源重组修复(homology-directed repair, HDR).在自然条件下, 同源重组修复出现的概率极低, 因此细胞中DSB的修复方式主要为NHEJ (
... 近年来, 一些新的基因组编辑系统不断问世.2015年, 麻省理工学院张峰团队报道了一种新的CRISPR效应蛋白Cas12a (Cpf1) (
Cpf1 is a versatile tool for CRISPR genome editing across diverse species of cyanobacteria
1
2016
... 2017年, 美国农业部(USDA)宣布将不会对采用基因组编辑工具CRISPR/Cas9进行遗传工程改造的双孢蘑菇(Agaricus bisporus)实施管控.这种蘑菇是由宾夕法尼亚州立大学的杨亦农(Yinong Yang)课题组改造的一种抗褐变白蘑菇(
Gene-edited CRISPR mushroom escapes US regulation
1
2016
... 目前, CRISPR/Cas9系统在植物领域中的应用大部分都通过NHEJ修复方式形成错配, 从而造成基因靶向敲除; 而在植物染色体上定点插入或替换一段序列一直以来都是困扰科学家的一个难题, 必须要借助HDR修复方式才能解决.
Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system
1
2017
...
DNA-free genome editing in plants with preassembled CRISPRCas9 ribonucleoproteins
0
2015
Boosting CRISPR/ Cas9 multiplex editing capability with the endogenous tRNA-processing system
2
2015
...
... 在后基因组时代, 功能基因组研究的重心逐渐由单一基因转向多基因功能研究.CRISPR/Cas9技术凭借其简单、高效等优点使多基因同时敲除变得可行.
A CRISPR/Cas9 toolkit for multiplex genome editing in plants
2
2014
... 目前, 在医学(
... 在后基因组时代, 功能基因组研究的重心逐渐由单一基因转向多基因功能研究.CRISPR/Cas9技术凭借其简单、高效等优点使多基因同时敲除变得可行.
Application of CRISPR/ Cas9 mediated genome editing in farm animals
0
2016
Gene targeting using the
2
2014
... 研究人员发现靶位点的核酸组成(即GC含量)、Cas9的表达水平以及靶区域的表观遗传等都会对基因组编辑效率产生影响.
... 近年来, 一些新的基因组编辑系统不断问世.2015年, 麻省理工学院张峰团队报道了一种新的CRISPR效应蛋白Cas12a (Cpf1) (
0
Generation of targeted mutant rice using a CRISPR-Cpf1 system
0
2017
High-efficient A·T to G·C base editing by Cas9n-guided tRNA adenosine deaminase in rice
2
2018
... 大肠杆菌腺嘌呤脱氨酶(ecTadA)能够对正常DNA上的腺嘌呤进行脱氨, 将腺嘌呤A转变为次黄嘌呤I, 损伤DNA在重新复制过程中被聚合酶作用, 导致A-T配对转换为G-C配对(
... 近年来, 一些新的基因组编辑系统不断问世.2015年, 麻省理工学院张峰团队报道了一种新的CRISPR效应蛋白Cas12a (Cpf1) (
Multigene editing via CRISPR/Cas9 guided by a single-sgRNA seed in
1
2018
... 在后基因组时代, 功能基因组研究的重心逐渐由单一基因转向多基因功能研究.CRISPR/Cas9技术凭借其简单、高效等优点使多基因同时敲除变得可行.
植物CRISPR基因组编辑技术的新进展
1
2017
... 与前2代基因编辑技术相比, CRISPR/Cas9技术的出现为广大科研人员提供了一个简单、廉价且高效的遗传操作平台, 不仅能为基础研究提供强大支持, 还能将功能基因组学的研究成果加速向产品转化(
植物基因组编辑试剂材料的导入及转化系统的研究现状及前景
1
2017
... 随着CRISPR/Cas9基因组编辑技术的不断改进和创新, 人们对CRISPR/Cas9系统运行机理的了解进一步加深, 原本一些困扰研究人员的问题已经迎刃而解. 例如, PAM限制基因组编辑范围和单碱基的精准编辑.然而, 植物基因组编辑也依然存在着一些亟待解决的问题.例如, 如何更高效地将基因组编辑材料运送到目的植物中, 随后产生定点编辑的植物(
CRISPR/Cas9基因组定点编辑中脱靶现象的研究进展
1
2018
... 目前, 所有的基因组编辑技术都存在脱靶问题(
Advancing crop transformation in the era of genome editing
1
2016
... 随着CRISPR/Cas9基因组编辑技术的不断改进和创新, 人们对CRISPR/Cas9系统运行机理的了解进一步加深, 原本一些困扰研究人员的问题已经迎刃而解. 例如, PAM限制基因组编辑范围和单碱基的精准编辑.然而, 植物基因组编辑也依然存在着一些亟待解决的问题.例如, 如何更高效地将基因组编辑材料运送到目的植物中, 随后产生定点编辑的植物(
RNA virus interference via CRISPR/Cas13a system in plants
0
2018
RNA-guided transcriptional regulation in plants via dCas9 chimeric proteins. Ph.D. thesis. Thuwal, Kingdom of Saudi Arabia: King Abdullah University of Science and
1
2014
... 将不同的功能蛋白(转录抑制、转录激活的蛋白元件)与dCas9 (Dead Cas9, 无DNA切割活性的Cas9)融合表达, 利用sgRNA的特异性引导功能, 可以实现转录抑制或转录激活等不同的遗传操作.
Crispr provides acquired resistance against viruses in prokaryotes
1
2007
... CRISPR/Cas系统最早是由日本大阪大学的一个研究团队在1987年对大肠杆菌碱性磷酸酶基因进行研究时发现的(
Small CRISPR RNAs guide antiviral defense in prokaryotes
1
2008
... CRISPR/Cas系统最早是由日本大阪大学的一个研究团队在1987年对大肠杆菌碱性磷酸酶基因进行研究时发现的(
Gene targeting in stem cells from individuals with
1
2004
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
Casilio: a versatile CRISPR-Cas9-Pumilio hybrid for gene regulation and genomic labelling
0
2016
Multiplex genome engineering using CRISPR/Cas systems
2
2013
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
... 系统切割DNA双链的分子机制并完成对CRISPR/ Cas9系统的改造和优化, 他们开创性地将原本分开作用的crRNA和tracrRNA整合形成1条sgRNA (single guide RNA), 并成功地在体外定点切割线性寡聚核苷酸片段和环状质粒DNA.他们还对Cas9不同结构域的作用以及PAM序列的重要性等多个问题进行研究, 这些研究成为CRISPR/Cas9基因组编辑系统最终能广泛应用于生物体的重要基础(
Genome editing technologies: defining a path to clinic: genomic editing: establishing preclinical toxicology standards, Bethesda, Maryland 10 June 2014
1
2015
... 目前, 所有的基因组编辑技术都存在脱靶问题(
CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III
1
2011
... 目前, 应用于基因组编辑的CRISPR系统主要以来自酿脓链球菌(Streptococcus pyogenes)的CRISPR/ Cas9系统应用最广, 且研究得最为透彻.该系统由Cas9蛋白、crRNA和tracrRNA (trans-activating CRISPR RNA) 3部分组成.其中, crRNA是由pre- crRNA (precursor crRNA)经tracrRNA和RNase III加工后得到, 主要负责引导CRISPR/Cas系统对特异靶位点的识别(
Phage response to CRISPR-encoded resistance in
1
2008
... 除应用最广的SpCas9外, 其它不同来源的Cas9也具有基因编辑能力.研究人员从金黄色葡萄球菌(Staphylococcus aureus)中找到1种较小的Cas9核酸酶(SaCas9) (
Systematic discovery of antiphage defense systems in the microbial pangenome
1
2018
... 近年来, 一些新的基因组编辑系统不断问世.2015年, 麻省理工学院张峰团队报道了一种新的CRISPR效应蛋白Cas12a (Cpf1) (
Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells
1
2005
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice
1
2015
...
High efficiency genome editing using a dmc1 promoter-controlled CRISPR/Cas9 system in maize
1
2018
... 2018年, 中国科学院遗传与发育生物学研究所韩方普团队使用玉米(Zea mays) DMC1基因启动子, 构建DPC (DMC1 promoter-controlled) CRISPR-Cas9载体系统, 并用其转化玉米幼胚, 结果发现抗性愈伤组织的基因编辑效率高达100%, 并在T0代植株中就出现了60%-70%的纯合或双等位突变体. 经全基因组测序分析, 在预测的1 000多个潜在脱靶位点处并没有发现脱靶突变(
Improving CRISPR-Cas nuclease specificity using truncated guide RNAs
1
2014
... 从生物信息学角度来看, 较长的DNA靶序列会降低脱靶错配的可能性, 但
ZFN, TALEN, and
1
2013
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
1
... 目前, 应用于基因组编辑的CRISPR系统主要以来自酿脓链球菌(Streptococcus pyogenes)的CRISPR/ Cas9系统应用最广, 且研究得最为透彻.该系统由Cas9蛋白、crRNA和tracrRNA (trans-activating CRISPR RNA) 3部分组成.其中, crRNA是由pre- crRNA (precursor crRNA)经tracrRNA和RNase III加工后得到, 主要负责引导CRISPR/Cas系统对特异靶位点的识别(
Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria
1
2012
... 大肠杆菌腺嘌呤脱氨酶(ecTadA)能够对正常DNA上的腺嘌呤进行脱氨, 将腺嘌呤A转变为次黄嘌呤I, 损伤DNA在重新复制过程中被聚合酶作用, 导致A-T配对转换为G-C配对(
Programmable base editing of A?T to G?C in genomic DNA without DNA cleavage
1
2017
... 华中农业大学赵云德研究团队通过利用自杀基因与CRISPR载体融合, 开发出高效去除含有Cas9转化事件的新技术, 通过基因型鉴定发现所有的T1代植株都不含有Cas9 (
Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants
0
2018
Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells
2
2015
... 研究人员发现靶位点的核酸组成(即GC含量)、Cas9的表达水平以及靶区域的表观遗传等都会对基因组编辑效率产生影响.
... 除应用最广的SpCas9外, 其它不同来源的Cas9也具有基因编辑能力.研究人员从金黄色葡萄球菌(Staphylococcus aureus)中找到1种较小的Cas9核酸酶(SaCas9) (
Diversity, activity, and evolution of CRISPR loci in
0
2008
Evolved Cas9 variants with broad PAM compatibility and high DNA specificity
0
2018
a). Increasing the efficiency of CRISPR-Cas9-VQR precise genome editing in rice
2
2017
...
... )通过定点突变的方法对SpCas9蛋白进行改造, 获得了2种SpCas9变体(VQR和VRER), 并通过一系列的转基因实验证明, 在水稻中VQR可以高效识别NGA的PAM序列, 而VRER则可以识别NGCG的PAM序列.这2种突变体使得CRISPR/Cas9系统在水稻中的可编辑范围拓展到原有的2倍以上.随后, 他们又通过修饰sgRNA的结构以及使用强内源启动子, 显著提高了VQR变体的编辑效率(
Expanding the range of CRISPR/Cas9 genome editing in rice
0
2016
b). Targeted mutagenesis in rice using CRISPR-Cpf1 system
2
2017
... 近年来, 一些新的基因组编辑系统不断问世.2015年, 麻省理工学院张峰团队报道了一种新的CRISPR效应蛋白Cas12a (Cpf1) (
... ;
Targeted mutagenesis in mice by electroporation of Cpf1 ribonucleo proteins
1
2016
... CRISPR/Cas系统最早是由日本大阪大学的一个研究团队在1987年对大肠杆菌碱性磷酸酶基因进行研究时发现的(
Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in
1
1987
... CRISPR/Cas系统最早是由日本大阪大学的一个研究团队在1987年对大肠杆菌碱性磷酸酶基因进行研究时发现的(
Identification of a novel family of sequence repeats among prokaryotes
2
2002
... CRISPR/Cas系统最早是由日本大阪大学的一个研究团队在1987年对大肠杆菌碱性磷酸酶基因进行研究时发现的(
... 来自酿脓链球菌(Streptococcus pyogenes)的Cas9蛋白(SpCas9)与包含20 nt识别序列的向导RNA (sgRNA)结合, 然后定向切割位于原型间隔序列毗邻基序(PAM)区域上游的靶序列造成双链断裂(DSB), 最终借助生物体内自身修复实现基因组编辑.
CRISPR-Cas9 structures and mechanisms
0
2017
A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity
2
2012
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
... 目前, 应用于基因组编辑的CRISPR系统主要以来自酿脓链球菌(Streptococcus pyogenes)的CRISPR/ Cas9系统应用最广, 且研究得最为透彻.该系统由Cas9蛋白、crRNA和tracrRNA (trans-activating CRISPR RNA) 3部分组成.其中, crRNA是由pre- crRNA (precursor crRNA)经tracrRNA和RNase III加工后得到, 主要负责引导CRISPR/Cas系统对特异靶位点的识别(
RNA-programmed genome editing in human cells
1
2013
... 大肠杆菌腺嘌呤脱氨酶(ecTadA)能够对正常DNA上的腺嘌呤进行脱氨, 将腺嘌呤A转变为次黄嘌呤I, 损伤DNA在重新复制过程中被聚合酶作用, 导致A-T配对转换为G-C配对(
Precise A·T to G·C base editing in the rice genome
1
2018
... 近年来, 一些新的基因组编辑系统不断问世.2015年, 麻省理工学院张峰团队报道了一种新的CRISPR效应蛋白Cas12a (Cpf1) (
Generation of knockout mice by Cpf1-mediated gene targeting
0
2016
Fusion guide RNAs for orthogonal gene manipulation with Cas9 and Cpf1
1
2017
...
Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress
2
2017
...
... 中国科学院遗传与发育生物学研究所高彩霞研究组利用Cas9变体(nCas9-D10A)融合ecTadA和ecTadA*二聚体(
Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion
1
2018
... 目前, CRISPR/Cas9系统在植物领域中的应用大部分都通过NHEJ修复方式形成错配, 从而造成基因靶向敲除; 而在植物染色体上定点插入或替换一段序列一直以来都是困扰科学家的一个难题, 必须要借助HDR修复方式才能解决.
Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9
1
2016
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
Multiplex and homologous recombination-mediated genome editing in
1
2013
... 在现代生物学研究中, 基因组编辑技术(genome editing)是一种通过人工设计和改造核酸酶达到对基因组进行定点修饰的技术(
High-efficiency talen-based gene editing produces disease-resistant rice
1
2012
... 目前, 在医学(
Cpf1 is a single RNA-guided Endonuclease of a Class 2 CRISPR- Cas system
0
2015
CRISPR/Cas9- mediated gene editing in human tripronuclear zygotes
1
2015
...
备案号: 京ICP备16067583号-21
版权所有 © 2021 《植物学报》编辑部
地址:北京香山南辛村20号 邮编:100093
电话:010-62836135 010-62836131 E-mail:cbb@ibcas.ac.cn
本系统由北京玛格泰克科技发展有限公司设计开发
