 ,1, 吴凯伸2, 惠建1, 白海波1, 马斯霜1, 李靖宇2
,1, 吴凯伸2, 惠建1, 白海波1, 马斯霜1, 李靖宇2Production of High Linoleic Acid Rice by Genome Editing
CHEN XiaoJun ,1, WU KaiShen2, HUI Jian1, BAI HaiBo1, MA SiShuang1, LI JingYu2
,1, WU KaiShen2, HUI Jian1, BAI HaiBo1, MA SiShuang1, LI JingYu2责任编辑: 李莉
收稿日期:2020-11-2接受日期:2021-01-29
| 基金资助: |
Received:2020-11-2Accepted:2021-01-29
作者简介 About authors
陈晓军,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2753KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
陈晓军, 吴凯伸, 惠建, 白海波, 马斯霜, 李靖宇. 基于基因编辑技术创制高亚油酸水稻材料[J]. 中国农业科学, 2021, 54(14): 2931-2940 doi:10.3864/j.issn.0578-1752.2021.14.001
CHEN XiaoJun, WU KaiShen, HUI Jian, BAI HaiBo, MA SiShuang, LI JingYu.
开放科学(资源服务)标识码(OSID):

0 引言
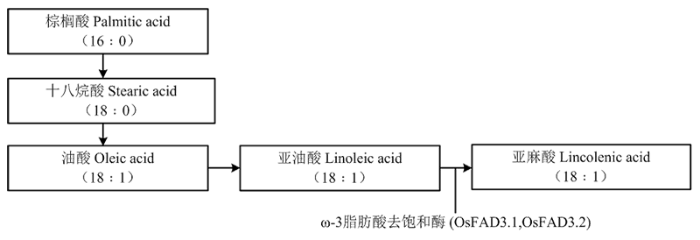
【研究意义】亚油酸(十八碳二烯酸)是一种人体必需的脂肪酸,具有保护心脑血管的功效,通过加快体内脂肪代谢与分解,减少胆固醇在血管壁上形成积存,有效预防动脉硬化的发生。亚油酸还具有提高人体免疫力、促进骨骼发育、提高记忆力、预防脑部功能退化[1,2]等功能。米糠是稻谷加工过程中的副产品,可使用压榨法或浸出法从中制取米糠油,米糠油是一种营养价值很高的食用油品种。中国米糠油原料资源丰富,但米糠油的生产和消费还处在起步阶段。据统计,中国水稻总产量约2亿t,米糠产量占7%,米糠出油率约15%,按此粗算,如完全利用,可产200万t米糠油,约等于中国第二大油料作物——花生的产油量,相当于1 200万t大豆的含油量,因此,提高水稻中米糠油的含量,可有效缓解食用油供求失衡的压力[3]。【前人研究进展】米糠油中亚油酸和油酸含量分别为38%和42%。水稻中亚油酸转化亚麻酸主要由ω-3脂肪酸去饱和酶(FAD3.1和FAD3.2)催化合成(图1)[4]。ABE等[4]利用基因编辑技术敲除了水稻控制油酸转化为亚油酸的关键酶基因——OsFAD2-1,结果表明,油酸含量较野生型增加2倍。随着基因编辑技术的迅速兴起,各种Cas9突变体[5,6]、基因定点突变[7,8,9,10]、单碱基替换[11,12,13,14]等基因编辑手段和方法对植物合成途径的精准控制成为可能。【本研究切入点】基于前人研究,米糠油中高油酸含量已经实现,但有关高亚油酸米糠油的研究鲜见报道。水稻中亚油酸到亚麻酸的合成由2个冗余基因控制。因此,同时敲除这两个基因,成为改良水稻含油量的难点。【拟解决的关键问题】本研究基于CRISPR-Cas9编辑技术原理,构建、转化一个植物基因编辑载体同时敲除ω-3脂肪酸去饱和酶的2个基因(OsFAD3.1和OsFAD3.2),阻断亚油酸转化亚麻酸的途径,为创制富集亚油酸水稻材料提供依据。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1水稻中脂肪酸合成路径[4]
Fig. 1The pathway of fatty acid synthesis in rice[4]
1 材料与方法
1.1 材料
水稻转化受体材料富源4号由宁夏农林科学院农作物研究所提供。基因编辑载体AarI-Cas9- PC1300由中国水稻研究所王克剑研究员惠赠。试验于2019年完成载体构建、基因转化,2020年完成籽粒农艺性状调查,均在宁夏农业生物技术重点实验室完成;2020年盆栽试验在宁永王太堡网室中进行。1.2 靶位点设计
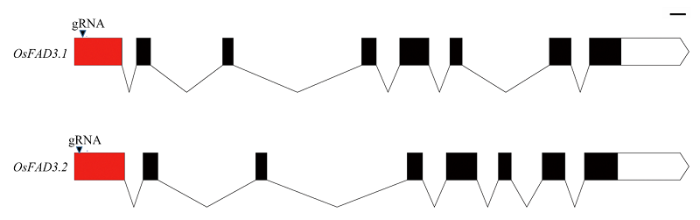
ω-3脂肪酸去饱和酶基因OsFAD3.1(LOC_ Os11g01340)和OsFAD3.2(LOC_Os12g01370)均由8个外显子和7个内含子组成,分别分布在水稻第11和第12染色体(图2)。二者cDNA同源性达97.32%,且第一外显子同源性达93.14%。2个基因编码区的同源性高度一致,为由1个gRNA同时编辑2个基因提供了必要条件。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2OsFAD3.1和OsFAD3.2的结构比较
bar=100 bp
Fig. 2Structure comparison of OsFAD3.1 and OsFAD3.2
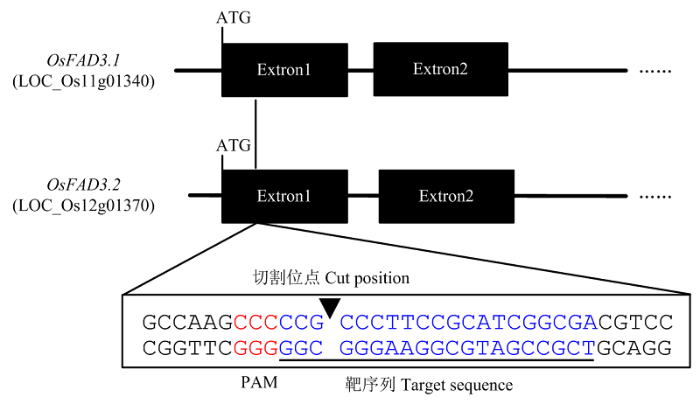
基因编辑的特异性取决于gRNA的特异性。选取2个基因编码区序列一致的区域,寻找基因编辑所必备的PAM结构(图3),设计2个靶位点gRNA1和gRNA2的引物序列(FAD3-cas9-SE1F:5′-ggcaTCGCC GATGCGGAAGGGCGG-3′,FAD3-cas9-SE1R:5′-aaac CCGCCCTTCCGCATCGGCGA-3′;FAD3-cas9-SE2F:5′-ggcaTCTTGCGCCAGCAGTGGGCG-3′,FAD3-cas9- SE2R:5′-aaacCGCCCACTGCTGGCGCAAGA-3′。下划线为与基因编辑载体AarI-Cas-9-PC1300相连接所需的黏性末端),引物由捷瑞生物北京分公司合成。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3gRNA1靶位点设计示意图
Fig. 3Design of gRNA1 target site
1.3 基因编辑载体的构建
gRNA1和gRNA2引物由捷瑞生物公司以脱盐方式合成4 OD,依据合成量,将合成好的上下游引物分别加入去离子ddH2O,并稀释至100 μmol·L-1。分别吸取上述引物(FAD3-cas9-SE1F/FAD3-cas9- SE1R和FAD3-cas9-SE2F/FAD3-cas9-SE2R)的母液各10 μL加入同一PCR管中进行退火。退火程序为98℃ 5 min,自然冷却至室温。gRNA1和gRNA2黏性末端片段制备完成,-20℃保存备用。将AarI-Cas9-PC1300载体经AarⅠ 37℃酶切,酶切体系为载体质粒10 μL、AarⅠ 2 μL、10×Buffer 5 μL、50×oligonucleotide(0.025 mmol·L-1)1 μL,补充ddH2O至50 μL。酶切产物回收、纯化,用30 μL ddH2O洗脱,-20℃保存备用。
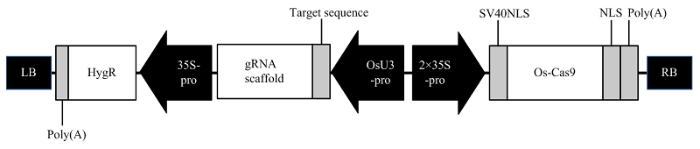
将gRNA1和gRNA2片段分别与AarI-Cas9- PC1300载体连接(图4)。连接体系为T4 DNA Ligase1 μL、5×Rapid Buffer 2 μL、退火产物片段6 μL和AarI- Cas9-PC1300载体1 μL。22℃温浴20 min。将连接产物转化DH5α,过夜培养,筛选阳性克隆测序,检测这两个载体是否构建成功,并命名为FAD3-S1-PC1300和FAD3-S2-PC1300。
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4植物基因编辑载体示意图
Fig. 4Plant gene editing vector
1.4 农杆菌介导的基因编辑
参考TOKI等[15]方法将植物基因编辑载体FAD3- S1-PC1300和FAD3-S2-PC1300进行基因转化。1.5 基因编辑植株的靶位点基因型鉴定
为了精确检测编辑靶位点的突变类型,将对T1代植株进行基因型鉴定。按照陈晓军等[16]方法提取植株叶片DNA。在靶位点上、下游分别设计引物(OsFAD3.1-dect-F:5′-CCTTCCTTCCTTCCTTCCTG GG-3′,OsFAD3.1-dect-R:5′-GAAATACCAGTCGTGG CCGAG-3′;OsFAD3.2-dect-F:5′-GGAGGAGCTTGGT TTCTGGG-3′,OsFAD3.2-dect-R:5′-GGCCAGGCCGC CCAGGCGA-3′),OsFAD3.1扩增片段508 bp,OsFAD3.2扩增片段447 bp。一个转化植株同时对2个靶位点进行突变类型鉴定。PCR反应体系为模板DNA 2 μL、2×Mixture 20 μL、上下引物各1 μL,补充ddH2O至40 μL。PCR反应程序为93℃ 3 min;93℃ 30 s,55℃ 30 s,72℃ 30 s,35个循环;72℃ 10 min。PCR产物经1%琼脂糖凝胶电泳检测,其余产物Sanger测序留用。1.6 基因编辑植株的籽粒农艺性状测定
利用万深SC-A1型种子自动考种及千粒重仪对受体材料和基因编辑材料进行考种。测定平均粒长(average grain length,AGL)、平均粒宽(average grain width,AGW)和千粒重(thousand grain weight,TGW)。1.7 基因编辑植株的脂肪酸测定
采用国家标准GB5009.168-2016第一法内标法,利用气质联用对3份受体材料和3份基因编辑材料进行37种脂肪酸测定。所用水稻材料去内外稃后备用。测定的脂肪酸包括丁酸C4:0、己酸C6:0、辛酸C8:0、癸酸C10:0、十一烷酸C11:0、月桂酸C12:0、十三烷酸C13:0、肉豆蔻酸C14:0、肉豆蔻烯酸C14、十五烷酸C15:0、十五碳烯酸C15:1、棕榈酸C16:0、棕榈油酸C16:1、十七烷酸C17:0、十七碳烯酸C17:1、十八烷酸C18:0、油酸C18:1、反油酸C18:1、亚油酸C18:2、反亚油酸C18:2TT,trans-9,12、γ-亚麻酸C18:3,cis- 6,9,12、亚麻酸C18:3,cis-9,12,15、花生酸C20:0、二十碳烯酸C20:1,cis-11、二十碳三烯酸C20:3,cis-11, 14,17、二十碳三烯酸C20:3,cis-8,11,14、花生四烯酸C20:4,cis-5,8,11,14、二十碳五烯酸(EPA-M)C20:5,cis-5,8,11,14,17、二十一烷酸C21:0、二十碳二烯酸C20:2,cis-11,14、二十二碳酸C22:0、芥酸C22:1,cis-13、二十二碳二烯酸C22:2,cis-13,16、二十二碳六烯酸(DHA-M)C22:6,cis-4,7,10,13,16,19、二十三烷酸C23:0、二十四烷酸C24:0和二十四碳烯酸C24:1,cis-15。2 结果
2.1 植物编辑载体的鉴定
通过对所构建的植物基因编辑载体进行测序鉴定,靶位点gRNA1的20个碱基正确连入编辑载体FAD3-S1-PC1300(图5-A);靶位点gRNA2的20个碱基也正确连入编辑载体FAD3-S2-PC1300(图5-B)。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5植物基因编辑载体测序
A:FAD3-S1-PC1300载体测序;B:FAD3-S2-PC1300载体测序 A: Sequence of FAD3-S1-PC1300; B: Sequence of FAD3-S2-PC1300
Fig. 5Sequence gene editing vector
2.2 OsFAD3突变体的鉴定
在T1代测序中,可以观察到同一植株2个靶位点OsFAD3.1和OsFAD3.2都有纯合突变,即在第二代中获得2个位点的纯合突变体(图6)。但2个载体存在编辑效率的差异。FAD3-S1-PC1300在T1代中出现了2个靶位点均纯合的植株,而FAD3-S2-PC1300在T1代中出现2个靶位点杂合的植株较多(图6)。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6OsFAD3.1与OsFAD3.2同时编辑的植株测序
A、B:FAD3-S1-PC1300基因编辑载体转化植株测序;C、D:FAD3-S2-PC1300基因编辑载体转化植株测序;A、C:OsFAD3.1靶位点;B、D:OsFAD3.2靶位点;箭头所示位置为编辑位点
Fig. 6Simultaneous editing of OsFAD3.1 and OsFAD3.2
A, B: FAD3-S1-PC1300 editing plant; C, D: FAD3-S2-PC1300 editing plant. A, C: The target of OsFAD3.1; B, D: The target of OsFAD3.2.Arrows indicate editing position
在FAD3-S1-PC1300载体的基因编辑后代纯合个体中,大多数突变发生在PAM结构前3个碱基,符合增加或减少1个碱基的预期。但也有大片段缺失(-40 bp)(图7-A),可见植物体发生基因编辑的机制还很复杂,可能与染色体DNA的高级结构相关。
图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7编辑植株测序
A:FAD3-S1-PC1300编辑植株;B:FAD3-S2-PC1300编辑植株
Fig. 7The sequence of editing plants
A: The sequence of editing plants with FAD3-S1-PC1300; B: The sequence of editing plants with FAD3-S2-PC1300
在FAD3-S2-PC1300载体的基因编辑后代纯合个体中,2个靶位点同时出现纯合突变的植株较少。在T1代测序后代株系中有2个靶位点杂合的植株,表明2个靶位点同时发生了基因编辑(图7-B)。
在所有2个载体的纯合突变体中,同一株系后代只有2种突变形式,表明T0代嵌合体的数目较少。
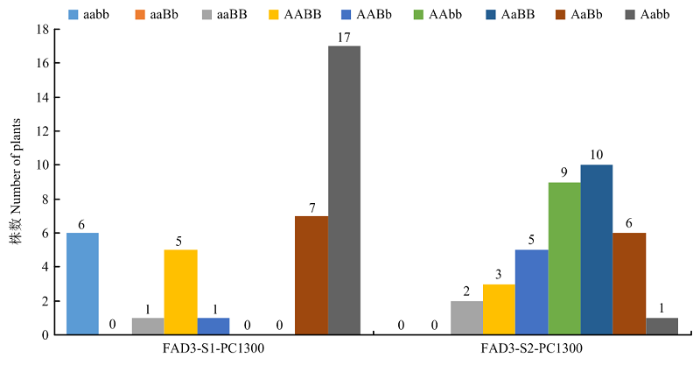
对FAD3-S1-PC1300载体的基因编辑后代4个株系共37株(n=15,n=3,n=9,n=10)植株进行测序,对FAD3-S2-PC1300载体的基因编辑后代4个株系共36株(n=14,n=3,n=10,n=9)植株进行测序,统计OsFAD3.1(AA)和OsFAD3.2(BB)基因型(图9)。2个载体转化后代均没有aaBb基因型,FAD3-S1- PC1300载体中没有AAbb和AaBB;FAD3-S2-PC1300载体中没有aabb和aaBb。
图9
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图99种基因型在基因编辑后代中的分布
Fig. 9Distribution of nine genotypes in the progeny of gene editing
2.3 OsFAD3双突变体的籽粒农艺性状测定
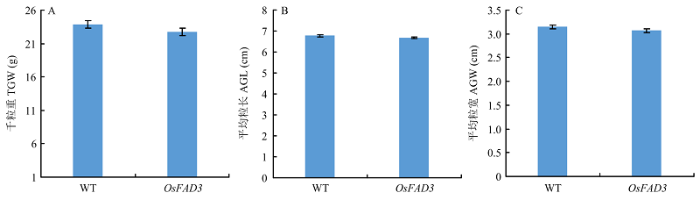
在受体材料和基因编辑的OsFAD3双突变体稻米材料中,营养生长期和生殖生长期在株高、叶片形态和颜色、穗型上均无显著改变。此外,还重点调查了其籽粒性状,它们的千粒重、平均粒长、平均粒宽均无显著变化,受体材料千粒重(TGW)、平均粒长(AGL)、平均粒宽(AGW)分别为23.84 g、6.77 cm和3.14 cm;基因编辑双位点纯合突变体材料的千粒重、平均粒长、平均粒宽分别为22.78 g、6.67 cm和3.07 cm(图10)。图10
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图10OsFAD3双突变体的籽粒农艺性状测定
A:千粒重;B:平均粒长;C:平均粒宽
Fig. 10Agronomic trait of grain in OsFAD3 double mutant
A: Thousand grain weight; B: Average grain length; C: Average grain width
2.4 OsFAD3双突变体的脂肪酸含量测定
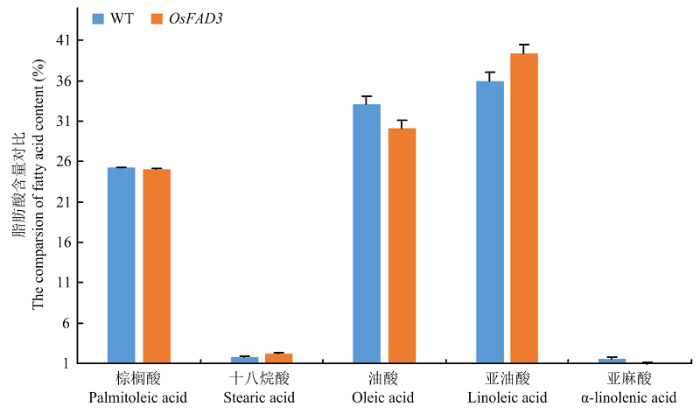
在受体材料和基因编辑OsFAD3双突变体稻米材料中,检测到10种脂肪酸,包括肉豆蔻酸、棕榈酸、十八烷酸、油酸、亚油酸、花生酸、二十碳烯酸、亚麻酸、芥酸和二十四烷酸(图11)。受体稻米材料中脂肪酸相对含量分别为棕榈酸(25.23±0.06)%、十八烷酸(1.77±0.13)%、油酸(33.12±0.95)%、亚油酸(36.01±1.07)%和亚麻酸(1.59±0.22)%。基因编辑稻米材料中脂肪酸相对含量分别为棕榈酸(25.05±0.41)%、十八烷酸(2.19±0.15)%、油酸(30.15±0.31)%、亚油酸(39.36±0.72)%和亚麻酸(0.89±0.14)%。基因编辑稻米亚油酸相对含量提高3.36个百分点。亚麻酸相对含量降低0.7个百分点。结果表明,OsFAD3双突变对棕榈酸、十八烷酸的相对含量影响不大,但对亚油酸的含量显著提高。图11
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图11脂肪酸含量对比
Fig. 11Comparison of fatty acid content
3 讨论
据生物信息分析,在水稻中共有20个脂肪酸去饱和酶基因[17],其表达产物分布在细胞的内质网、叶绿体、线粒体和质膜上,其中,OsFAD3-1和OsFAD3-2定位在内质网上。植物体内,根据电子供体不同可将ω-3脂肪酸去饱和酶分为两类,一类定位于植物细胞的内质网,以细胞色素b5为电子供体,作用于磷脂酞甘油(phosphatidylglyeerol,PG)或其他磷脂;另一类定位于植物细胞质体,以铁氧还蛋白为电子供体, 作用于磷脂酞甘油,硫脂和半乳糖脂。植物体内有2条多聚不饱和脂肪酸的生物合成途径。其中一条在微体中合成,另一条在质体膜上。自从克隆了拟南芥内质网ω-3脂肪酸去饱和酶基因[18]以来,又陆续克隆了核桃[19]、西班牙鼠尾草[20]、紫苏[20,21]、油茶[22]、油菜[23]、高粱[24]、大豆[25]等的不饱和脂肪酸的合成基因。植物中的不饱和脂肪酸由脂肪酸去饱和酶合成,它在植物的生长发育以及植物非生物胁迫方面起着重要的作用。水稻叶中OsFAD2、OsFAD6、OsFAD3和OsFAD7的mRNA表达具有昼夜节律性,在光照下表达量低,而在随后的黑暗中表达量高,OsFAD6和OsFAD7的mRNA表达的昼夜节律性可能与水稻幼苗叶片中NADPH量的改变有关[26]。
OsFAD3在根组织中含量丰富,而在叶片中几乎检测不到。根组织中,在15℃和20℃条件下OsFAD3高水平表达,在低于10℃条件下其水平显著降低。在正常生长温度和低温条件下,OsFAD3在叶组织中的积累保持在相当低的水平。OsFAD3在耐冷性和不耐冷性水稻品种中表现出相似的温度响应[27]。
YIN等[28]从水稻和大豆中克隆了2个ω3/Δ15脂肪酸去饱和酶基因,并在胚特异启动子REG驱动下将它们导入水稻中。转基因株系中,胚和米糠中的α-亚麻酸(α-linolenic acid,ALA)含量分别增加到6.09和5.86 mg·g-1,这比非转基因对照的0.24和0.21 mg·g-1提高了25.4和27.9倍。而本试验中,亚麻酸相对含量则由(1.59±0.22)%降低到(0.89±0.14)%。说明完全缺失该基因确实关闭了下游亚麻酸的合成。
LIU等[29]过量表达定位于内质网的GmFAD3-1和OsFAD3的转基因株系,种子中ALA含量从0.36 mg·g-1分别提升到8.57和10.06 mg·g-1,是非转基因植株的23.8和27.9倍。在水稻种子中OsFAD3催化亚油酸(linoleic acid,LA)向ALA转化的活性强于GmFAD3-1。过量表达定位于内质网的GmFAD3-2/ GmFAD3-3以及叶绿体定位的OsFAD7/OsFAD8对于提高水稻种子ALA含量的作用较小。
本研究表明,获得的ω-3脂肪酸去饱和酶2个基因的突变材料能够增加亚油酸的含量3.36个百分点。
由于冗余功能基因的存在,多拷贝基因的生物学功能一直是研究难点。一个gRNA打靶2个同源基因这一策略客观上要求在基因编辑后代中能够利用引物的特异性把2个靶基因分开。只有这样才能同时对2个基因进行靶位点的鉴定。本研究通过获得了一系列基因编辑材料,对研究功能基因的剂量效应[30]也有一定帮助。另外,同时敲除3个或以上的高拷贝基因可能也达到预计的目的。
4 结论
根据基因编辑原理,达到了一个载体同敲除2个同源基因的目的,并且易获得2个位点的纯合编辑材料,同时亚油酸含量提高了3.36个百分点。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.plaphy.2018.04.033URL [本文引用: 4]
DOI:10.1038/s41587-020-0490-7URL [本文引用: 1]
DOI:10.1038/s41477-020-0614-6URL [本文引用: 1]
DOI:10.1111/pbi.2019.17.issue-1URL [本文引用: 1]
[本文引用: 1]
DOI:10.1038/nbt.4273URL [本文引用: 1]
DOI:10.1111/pbi.2018.16.issue-4URL [本文引用: 1]
DOI:10.1038/s41587-020-0455-xURL [本文引用: 1]
DOI:10.1111/pbi.v18.11URL [本文引用: 1]
DOI:10.1016/j.molp.2020.03.011URL [本文引用: 1]
DOI:10.1016/j.molp.2020.03.010URL [本文引用: 1]
DOI:10.1111/tpj.2006.47.issue-6URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/s41598-019-55648-zURL [本文引用: 1]
DOI:10.1126/science.1455229URL [本文引用: 1]
DOI:10.1186/s12864-020-6692-zURL [本文引用: 1]
DOI:10.1371/journal.pone.0191432URL [本文引用: 2]
DOI:10.1007/s00299-016-2053-4URL [本文引用: 1]
DOI:10.3389/fpls.2020.00011URL [本文引用: 1]
DOI:10.1111/pbi.v17.12URL [本文引用: 1]
DOI:10.3390/ijms20215359URL [本文引用: 1]
DOI:10.3389/fpls.2019.01812URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s11032-013-0014-yURL [本文引用: 1]
DOI:10.1093/jxb/ers051URL [本文引用: 1]
DOI:10.1111/pbi.v18.2URL [本文引用: 1]
