 ,1,2, 黄冰艳1, 苗利娟1, 徐静1, 石磊1, 张忠信1, 孙子淇1, 刘华1, 齐飞艳1, 董文召1, 郑峥1, 张新友
,1,2, 黄冰艳1, 苗利娟1, 徐静1, 石磊1, 张忠信1, 孙子淇1, 刘华1, 齐飞艳1, 董文召1, 郑峥1, 张新友 ,1
,1Genetic Analysis of Peanut Kernel Traits in a Nested-crossing Population by Major Gene Plus Polygenes Mixed Model
ZHANG MaoNing ,1,2, HUANG BingYan1, MIAO LiJuan1, XU Jing1, SHI Lei1, ZHANG ZhongXin1, SUN ZiQi1, LIU Hua1, QI FeiYan1, DONG WenZhao1, ZHENG Zheng1, ZHANG XinYou
,1,2, HUANG BingYan1, MIAO LiJuan1, XU Jing1, SHI Lei1, ZHANG ZhongXin1, SUN ZiQi1, LIU Hua1, QI FeiYan1, DONG WenZhao1, ZHENG Zheng1, ZHANG XinYou ,1
,1通讯作者:
责任编辑: 李莉
收稿日期:2020-12-5修回日期:2021-03-3网络出版日期:2021-07-01
| 基金资助: |
Received:2020-12-5Revised:2021-03-3Online:2021-07-01
作者简介 About authors
张毛宁,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (7697KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张毛宁, 黄冰艳, 苗利娟, 徐静, 石磊, 张忠信, 孙子淇, 刘华, 齐飞艳, 董文召, 郑峥, 张新友. 巢式杂交分离群体的花生籽仁性状的主基因+多基因混合遗传模型分析[J]. 中国农业科学, 2021, 54(13): 2916-2930 doi:10.3864/j.issn.0578-1752.2021.13.019
ZHANG MaoNing, HUANG BingYan, MIAO LiJuan, XU Jing, SHI Lei, ZHANG ZhongXin, SUN ZiQi, LIU Hua, QI FeiYan, DONG WenZhao, ZHENG Zheng, ZHANG XinYou.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】花生(Arachis hypogaea L.)作为原产自南美洲的一种重要的经济作物和油料作物[1],其市场价值在中国所有作物中位居第四,仅次于水稻、小麦和玉米[2]。同时花生种子的油脂含量约50%,蛋白质含量约26%,是世界重要植物油脂和植物蛋白的来源。花生籽仁含有丰富的油酸、亚油酸、白藜芦醇等营养物质,有益于人体的健康[3]。作为世界上最大的花生生产国,中国虽然花生总产量及单产都在稳步提高,但仍不能满足国内市场对花生的需求,花生进口快速增长,而且中国花生进口来源潜力有限[4]。因此,高产、优质和专用型花生新品种选育不仅能提高中国农业产值,同时也能稳定中国植物油脂及植物蛋白的安全供给。花生产量性状是数量性状,受多基因调控[5]。花生籽仁表型性状包括籽仁长、宽、长宽比、表面积、表面周长及百仁重等不仅影响花生外观及花生加工品质[6],同时也直接影响花生产量。对花生籽仁表型性状开展遗传机理研究,将为高产优质的专用型花生新品种培育奠定理论基础和提供实践依据。【前人研究进展】花生产量性状相关性状,如:荚果的长、宽、长宽比、百果重[7,8,9]、出仁率[10]以及籽仁的长、宽、长宽比、百仁重[11]等大多数性状均属于数量性状。近年来,植物数量性状的遗传研究方法得到了长足发展,利用数量性状的遗传模型分析方法,解析重要农艺性状的遗传规律,为现代植物育种提供了重要的参考。利用双亲杂交构建的遗传群体进行表型分析,GAI等[12]提出的主基因+多基因遗传模型分析体系已广泛应用于小麦穗部性状[13]、不结球白菜叶片数和分蘖数[14]、玉米粒长[15]、黄瓜单性结实性[16]、甜瓜果抗裂性[17]等多种作物的重要农艺性状解析。该分析方法也在花生籽仁的蛋白质含量[18]、油酸和亚油酸含量[19]、白藜芦醇含量[20]等重要性状研究上得到应用。籽仁性状的相关研究表明,百仁重的遗传符合多对主基因+多基因加性上位性模型,籽仁的长宽比受到2对主基因控制,籽仁长符合多对主基因控制的加性-上位性遗传模型,籽仁宽符合加性或显性上位性主基因+多基因混合遗传模型[21,22,23]。前人研究采用的材料、种植环境不同,遗传规律存在一定差异,且所用群体为双亲杂交的分离或RIL群体,存在一定的局限性。【本研究切入点】近年来,多亲杂交群体逐渐被复杂数量性状的遗传研究所采用,但采用巢式杂交群体分析花生籽仁性状的遗传模式鲜有报道。【拟解决的关键问题】本研究将花生籽仁性状细分为籽仁长、宽、长宽比、表面积、表面周长和单仁重等6个性状,以豫花15共同亲本与其他基础亲本远杂9102、中花6号、粤油20、伏花生、NC94022,配置5个杂交组合,获得含有1 812个F2单株的巢式杂交F2:3家系群体,确定花生籽仁的6个表型性状的遗传模式,分析该6个性状的调控主基因数及加性、上位性效应,探究花生籽仁性状的遗传模式,为花生高产优质专用型新品种培育奠定理论基础。1 材料与方法
1.1 试验材料
以豫花15(YH15)为巢式杂交群体的共同亲本,远杂9102(YZ9102)、中花6号(ZH6)、粤油20(YY20)、伏花生(FHS)及NC94022为基础亲本。其中YH15和YZ9102为河南省农业科学院选育的通过国家鉴定的花生品种,分别为中间型大果品种和珍珠豆型高产品种;ZH6和YY20为中国农业科学院油料作物研究所和广东省农业科学院选育的珍珠豆型高产品种;FHS是山东省选育的珍珠豆型早熟品种;NC94022是美国普通型小果(兰娜型)品种。5个杂交组合远杂9102×豫花15(YZ9102×YH15)、中花6号×豫花15(ZH6×YH15)、豫花15×粤油20(YH15×YY20)、豫花15×伏花生(YH15×FHS)、豫花15×NC94022(YH15×NC94022)的F2单株收获,分别获得472、392、286、393和269个F2:3家系的籽仁,构成一个含1 812个F2单株的巢式杂交分离群体。
1.2 表型测定
单株收获F2的荚果经晾晒风干后,选剥均匀一致的成熟饱满荚果,挑选10个饱满且整齐的F2:3籽仁,依据《花生种质资源描述规范和数据标准》[24],利用万深SC-G型自动考种分析及千粒重仪,获得籽仁的6个表型性状:籽仁长(length of kernel,KL)、籽仁宽(width of kernel,KW)、籽仁长宽比(ratio of kernel length to kernel width,KL/KW)、籽仁表面积(kernel surface area,KSA)、籽仁表面周长(kernel surface perimeter,KSP)和单仁重(average- kernel weight,AKW)。1.3 数据分析
运用Excel软件进行表型数据、偏度、峰度等系数估计值基本统计;运用Origin和Graphpad prism7.0等软件对试验数据进行可视化处理;运用R软件包绘制频率分布直方图、邦弗朗尼(Bonferroni)相关性及线性拟合线。运用SEAv1.0软件,使用程序SEA-F2:3[25]对表型数据进行分析。以AIC(Akaike’s Information Criterion)最小值或较小值的模型作为备选模型,再进行适合性检验包括:均匀性检验(U12、U22、U32)、Smirnov检验(Wn2)和Kolmogorov检验(Dn),从备选的模型中选择适合性检验参数未达显著水平的模型,通过对比各模型的遗传参数,确定最佳遗传模型,再对最佳模型进行遗传参数的分析。
2 结果
2.1 花生籽仁表型性状特征及次数分布
通过对花生籽仁表型性状进行统计(图1),6个亲本的籽仁不同的表型性状均存在差异,豫花15、伏花生的籽仁长、宽、表面积、表面周长和单仁重较其他亲本更大,分别为17.17 mm、9.32 mm、126.62 mm2、44.93 mm、0.85 g和16.93 mm、9.93 mm、130.38 mm2、45.53 mm、0.865 g,NC94022的长宽比值为2.12,居所有亲本之首。图1
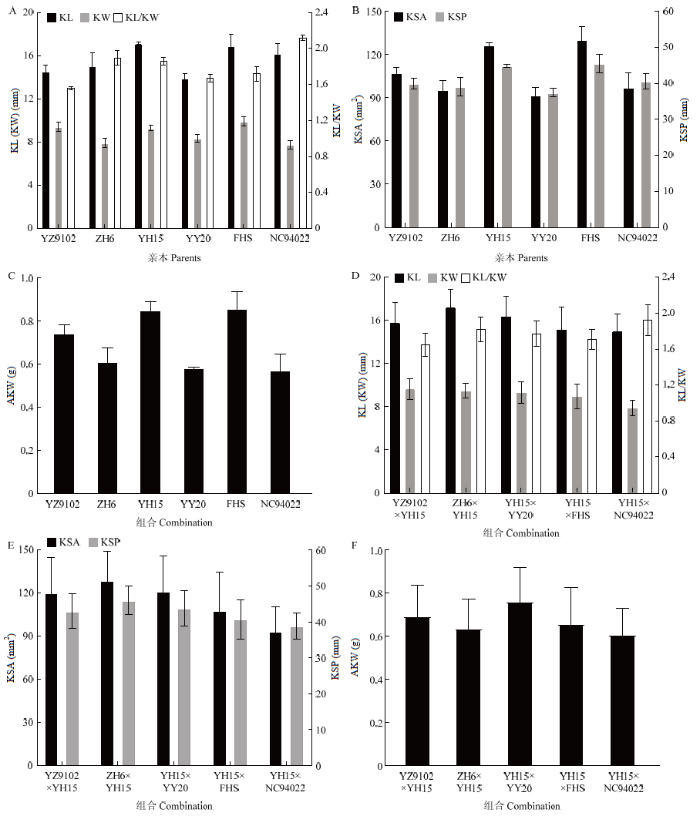 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1亲本及F2:3籽仁表型性状的群体分布
A:亲本籽仁长、宽、长宽比;B:亲本的籽仁表面积及表面周长;C:亲本的单仁重;D:F2:3的籽仁长、宽、长宽比;E:F2:3的表面积及表面周长;F:F2:3的单仁重。KL:籽仁长;KW:籽仁宽;KL/KW:籽仁长宽比;KSA:籽仁表面积;KSP:籽仁表面周长。下同
Fig. 1Population distribution of kernel phenotypic traits in parents and F2:3
A: KL, KW and KL/KW of parents; B: Kernel surface area and perimeter of parents; C: Average-kernel weight of parents; D: KL, KW and KL/KW of F2:3; E: Kernel surface area and perimeter of F2:3; F: Average-kernel weight of F2:3. KL: Kernel length; KW: Kernel width; KL/KW: Ratio of kernel length to kernel width; KSA: Kernel surface area; KSP: Kernel surface perimeter. The same as below
在巢式杂交群体的5个组合中,籽仁长平均值最大的组合为ZH6×YH15,平均值达17.20 mm;籽仁宽值最大为9.63 mm,组合为YZ9102×YH15;籽仁长宽比值最大为1.92,组合为YH15×NC94022,籽仁表面积最大值为128.97 mm2,组合为ZH6×YH15;籽仁表面周长最大值为46.05 mm,组合为ZH6×YH15;籽仁单仁重最大值为0.765 g,组合为YH15×YY20。籽仁长、宽、长宽比、表面积、表面周长、单仁重的变异幅度分别为10.01—21.54 mm、5.89—12.74 mm、1.28—2.49、50.97—197.60 mm2、27.83 mm—56.50 mm、0.22—1.21 g。籽仁长宽比变异系数较其他性状小;籽仁表面积、单仁重变异系数值较其他性状大,且较其他性状变异表现更为突出且丰富(图1-D—图1-F,表1)。
Table 1
表1
表1巢式杂交群体籽仁表型性状变异
Table 1
| 组合 Combination | 性状 Trait | 均值 Mean | 标准差 SD | 变异系数 CV (%) | 最大值 Max | 最小值 Min | 峰度 Kurt | 偏度 Skew |
|---|---|---|---|---|---|---|---|---|
| YZ9102×YH15 | 籽仁长 KL (mm) | 15.80 | 1.85 | 11.71 | 21.43 | 10.43 | -0.13 | 0.13 |
| 籽仁宽 KW (mm) | 9.63 | 0.98 | 10.14 | 12.74 | 7.09 | -0.26 | 0.14 | |
| 籽仁长宽比KL/KW | 1.65 | 0.13 | 7.94 | 2.01 | 1.28 | -0.19 | 0.08 | |
| 籽仁表面积KSA (mm2) | 120.28 | 24.52 | 20.39 | 197.60 | 60.67 | -0.15 | 0.38 | |
| 籽仁表面周长 KSP (mm) | 42.96 | 4.81 | 11.20 | 56.43 | 29.45 | -0.31 | 0.14 | |
| 单仁重 AKW (g) | 0.70 | 0.14 | 20.48 | 1.14 | 0.32 | 0.05 | 0.03 | |
| ZH6×YH15 | 籽仁长 KL (mm) | 17.20 | 1.65 | 9.61 | 21.31 | 12.18 | -0.32 | -0.02 |
| 籽仁宽 KW (mm) | 9.48 | 0.71 | 7.46 | 11.39 | 7.37 | -0.10 | 0.09 | |
| 籽仁长宽比KL/KW | 1.82 | 0.14 | 7.69 | 2.33 | 1.42 | 0.26 | 0.10 | |
| 籽仁表面积 KSA (mm2) | 128.97 | 20.22 | 15.68 | 186.21 | 69.80 | -0.20 | 0.18 | |
| 籽仁表面周长 KSP (mm) | 46.05 | 3.98 | 8.64 | 56.50 | 33.15 | -0.27 | -0.02 | |
| 单仁重 AKW (g) | 0.64 | 0.13 | 21.31 | 1.16 | 0.29 | 0.30 | 0.33 | |
| YH15×YY20 | 籽仁长 KL (mm) | 16.41 | 1.85 | 11.29 | 21.54 | 11.89 | -0.42 | 0.09 |
| 籽仁宽 KW (mm) | 9.31 | 1.01 | 10.81 | 11.67 | 7.00 | -0.73 | 0.11 | |
| 籽仁长宽比KL/KW | 1.77 | 0.14 | 7.80 | 2.12 | 1.40 | -0.25 | -0.01 | |
| 籽仁表面积 KSA (mm2) | 121.21 | 24.55 | 20.25 | 185.38 | 69.52 | -0.54 | 0.25 | |
| 籽仁表面周长 KSP (mm) | 43.82 | 4.95 | 11.31 | 55.96 | 32.62 | -0.65 | 0.11 | |
| 单仁重 AKW (g) | 0.76 | 0.16 | 20.77 | 1.21 | 0.36 | 1.23 | -0.27 | |
| YH15×FHS | 籽仁长 KL (mm) | 15.17 | 2.07 | 13.65 | 21.18 | 10.01 | -0.27 | 0.22 |
| 籽仁宽 KW (mm) | 8.94 | 1.13 | 12.66 | 12.10 | 6.28 | -0.62 | 0.15 | |
| 籽仁长宽比KL/KW | 1.71 | 0.11 | 6.73 | 2.06 | 1.36 | 0.32 | 0.25 | |
| 籽仁表面积 KSA (mm2) | 107.71 | 27.25 | 25.30 | 194.25 | 50.97 | -0.19 | 0.47 | |
| 籽仁表面周长 KSP (mm) | 40.73 | 5.53 | 13.57 | 56.30 | 27.83 | -0.44 | 0.21 | |
| 单仁重 AKW (g) | 0.65 | 0.17 | 25.77 | 1.16 | 0.22 | -0.07 | 0.25 | |
| YH15×NC94022 | 籽仁长 KL (mm) | 15.03 | 1.57 | 10.41 | 20.21 | 11.12 | 0.27 | 0.18 |
| 籽仁宽 KW (mm) | 7.86 | 0.73 | 9.32 | 10.52 | 5.89 | 1.16 | 0.16 | |
| 籽仁长宽比KL/KW | 1.92 | 0.17 | 8.72 | 2.49 | 1.54 | 1.92 | 0.76 | |
| 籽仁表面积 KSA (mm2) | 93.62 | 16.90 | 18.05 | 167.13 | 51.71 | 1.56 | 0.64 | |
| 籽仁表面周长 KSP (mm) | 38.82 | 3.72 | 9.59 | 52.61 | 29.22 | 0.70 | 0.28 | |
| 单仁重 AKW (g) | 0.61 | 0.12 | 20.22 | 0.94 | 0.31 | -0.43 | 0.05 |
新窗口打开|下载CSV
图2表明巢式杂交群体中F2:3家系的籽仁性状,籽仁长、宽、长宽比、表面积、表面周长、单仁重呈现出连续的正态分布,具有连续多峰分布特点,符合主基因加多基因的特征,表明可能存在主基因。
图2
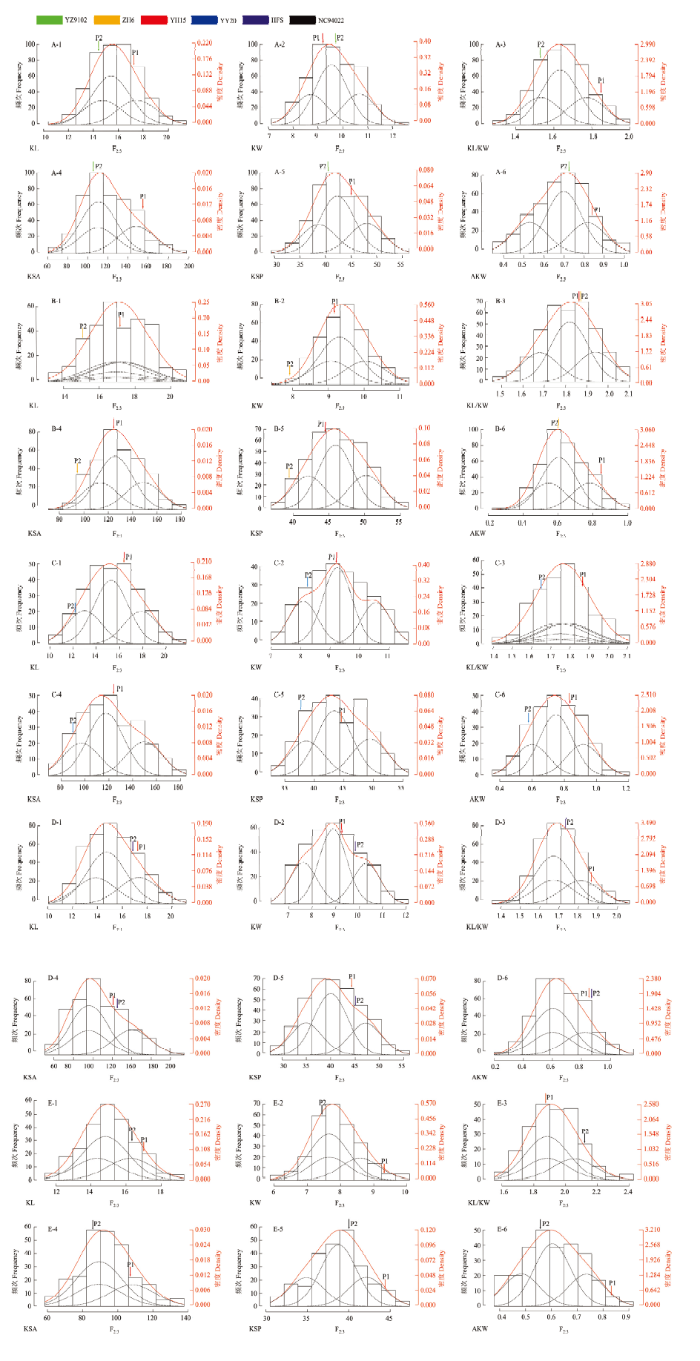 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2巢式杂交群体籽仁的6个表型性状的次数分布(柱形)、拟混合分布(红线)与成分(黑线)分布
A、B、C、D、E分别表示巢式杂交群体中的组合YZ9102×YH15、ZH6×YH15、YH15×YY20、YH15×FHS、YH15×NC94022
Fig. 2Frequent (column), mixed (red line, theoretical), and component (black line) distributions for 6 phenotypic characters of kernel in nested-crossing population
A, B, C, D, and E represent YZ9102×YH15, ZH6×YH15, YH15×YY20, YH15×FHS and YH15×NC94022 combinations in nested-crossing population, respectively
2.2 花生籽仁表型性状的遗传相关性
对巢式杂交群体的籽仁表型性状(籽仁长、宽、长宽比、表面积、表面周长、单仁重)进行邦弗朗尼(Bonferroni)相关性及线性拟合线的分析,建立了花生F2:3家系的线性拟合散点图矩阵(图3)。分析巢式杂交群体中5个组合的籽仁性状的频率分布,每个变量的分布显示在对角线上。相关和拟合曲线分析表明,所研究的多数性状存在一定的相关性,部分性状之间存在较强的相关性。图3
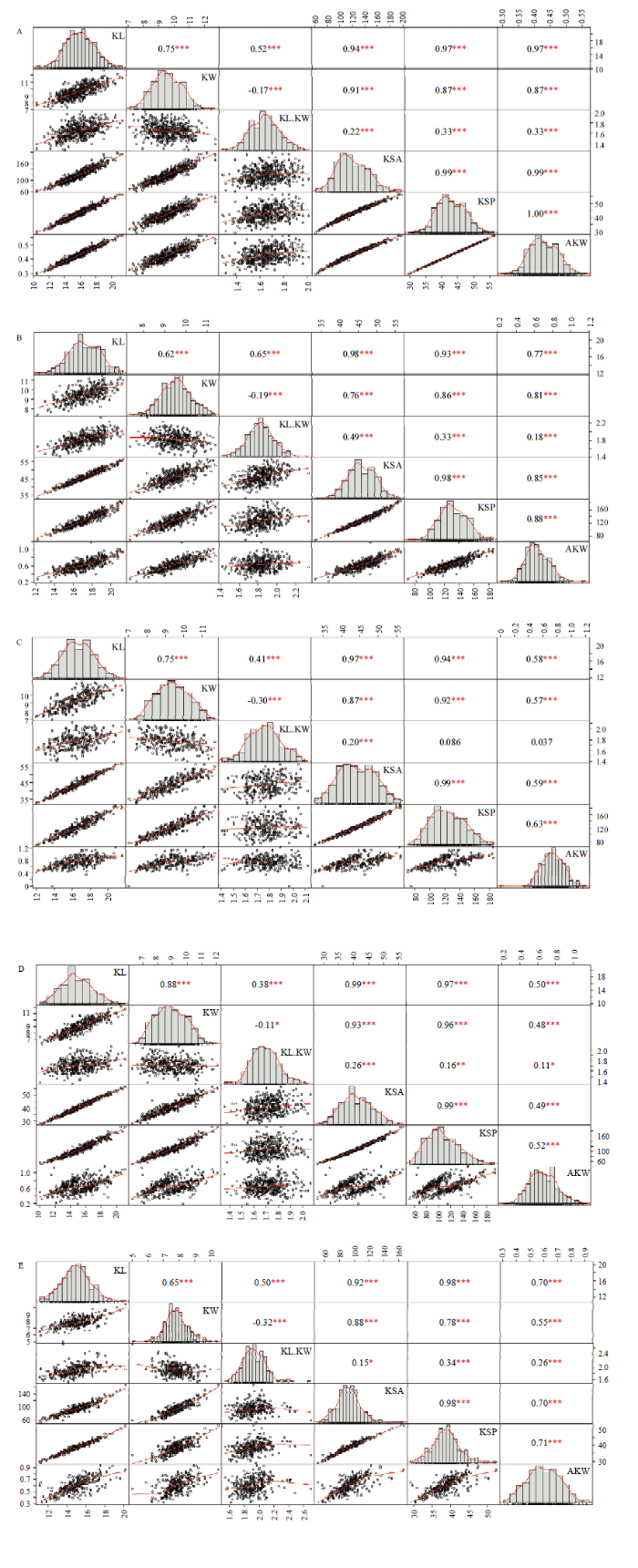 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3巢式杂交群体荚果的6个表型性状的频率分布?相关性和拟合曲线
A、B、C、D、E分别表示巢式杂交群体中的组合YZ9102×YH15、ZH6×YH15、YH15×YY20、YH15×FHS、YH15×NC94022。图中的上三角形显示了2个籽仁性状之间的相关系数以及显著性水平,其中,*:P<0.05,**:P<0.01,***:P<0.001;下三角形用散点图显示2个籽仁性状之间拟合度。对角线是6个籽仁性状的直方图
Fig. 3Frequency distribution, correlation and fit curve of six phenotypic characters in nested-crossing population
A, B, C, D, and E represent YZ9102×YH15, ZH6×YH15, YH15×YY20, YH15×FHS and YH15×NC94022 combinations in nested-crossing population, respectively. The upper triangle in the figure shows the correlation coefficient and significance level between the two kernel traits. Where, *:P<0.05, **:P<0.01, ***:P<0.001; the lower triangle shows the fitness between the two kernels traits with scatter plots. Diagonal is a histogram of the six kernels traits
2.3 花生籽仁表型性状的遗传模型的建立
应用SEA分析软件,选择F2:3群体类型,分别对巢式杂交群体的各组合的籽仁表型性状进行主基因+多基因模型分析,5个组合的籽仁的6个不同的表型性状均获得3类共11种遗传模型(电子附表1),并对其进行U12、U22、U32、nW2、Dn等5种适合性检验(电子附表1)。在该巢式群体中的6个表型性状,除了组合YZ9102×YH15的籽仁表面积(KSA)的模型2MG-A和组合YH15×FHS的籽仁表面积(KSA)的0MG、2MG-EA、1MG-A、2MG-EAD、2MG-A、1MG-EAD、2MG-CD、2MG-ADI模型出现适应性达显著外,其余组合性状的模型均未达到显著水平。Supplement table 1
附表1
附表1籽仁表型性状遗传模式分析
Supplement table 1
| 组合 Combination | 籽仁长 KL | 籽仁宽 KW | 籽仁长宽比 KL/KW | 籽仁表面积 KSA | 籽仁表面周长 KSP | 平均单仁重 SKW | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 模型 Models | 最大似然值Maximum likelihod value | AIC值 AIC value | 模型 Models | 最大似然值Maximum likelihod value | AIC值 AIC value | 模型 Models | 最大似然值Maximum likelihod value | AIC值 AIC value | 模型 Models | 最大似然值Maximum likelihod value | AIC值 AIC value | 模型 Models | 最大似然值Maximum likelihod value | AIC值 AIC value | 模型 Models | 最大似然值Maximum likelihod value | AIC值 AIC value | ||
| YZ9102×YH15 | 0MG | -959.99 | 1923.97 | 1MG-AD | -656.46 | 1320.93 | 0MG | 289.99 | -575.98 | 1MG-AD | -2163.74 | 4335.49 | 1MG-AD | -1408.02 | 2824.04 | 1MG-AD | -1879.32 | 3766.63 | |
| 1MG-NCD | -958.76 | 1925.52 | 0MG | -658.57 | 1321.14 | 1MG-AD | 291.02 | -574.03 | 1MG-NCD | -2163.98 | 4335.95 | 1MG-NCD | -1408.58 | 2825.15 | 2MG-EAD | -1880.98 | 3767.95 | ||
| 1MG-AD | -958.85 | 1925.70 | 1MG-NCD | -657.14 | 1322.29 | 1MG-A | 289.99 | -573.98 | 2MG-AD | -2163.74 | 4339.48 | 0MG | -1411.09 | 2826.17 | 1MG-EAD | -1880.14 | 3768.29 | ||
| 1MG-A | -959.98 | 1925.97 | 1MG-A | -658.57 | 1323.13 | 2MG-EA | 289.99 | -573.98 | 0MG | -2172.86 | 4349.72 | 2MG-AD | -1408.02 | 2828.04 | 2MG-CD | -1880.15 | 3768.29 | ||
| 2MG-EA | -959.98 | 1925.97 | 2MG-EA | -658.57 | 1323.13 | 2MG-EAD | 289.98 | -573.96 | 2MG-EA | -2172.85 | 4351.71 | 1MG-A | -1411.09 | 2828.17 | 0MG | -1882.81 | 3769.62 | ||
| 2MG-EAD | -959.99 | 1925.99 | 2MG-EAD | -658.58 | 1323.15 | 1MG-NCD | 290.76 | -573.53 | 1MG-A | -2172.86 | 4351.72 | 2MG-EA | -1411.09 | 2828.17 | 2MG-AD | -1879.32 | 3770.63 | ||
| 2MG-A | -959.98 | 1927.97 | 2MG-AD | -656.46 | 1324.92 | 2MG-A | 289.99 | -571.98 | 2MG-EAD | -2172.86 | 4351.73 | 2MG-EAD | -1411.09 | 2828.19 | 1MG-A | -1882.81 | 3771.62 | ||
| 1MG-EAD | -959.99 | 1927.98 | 2MG-A | -658.57 | 1325.13 | 1MG-EAD | 289.98 | -571.96 | 2MG-A | -2172.86 | 4353.72 | 2MG-A | -1411.08 | 2830.17 | 2MG-EA | -1882.81 | 3771.62 | ||
| 2MG-CD | -959.99 | 1927.99 | 1MG-EAD | -658.57 | 1325.15 | 2MG-CD | 289.98 | -571.96 | 1MG-EAD | -2172.86 | 4353.73 | 1MG-EAD | -1411.09 | 2830.19 | 2MG-A | -1882.81 | 3773.61 | ||
| 2MG-AD | -958.69 | 1929.37 | 2MG-CD | -658.57 | 1325.15 | 2MG-AD | 291.02 | -570.03 | 2MG-CD | -2172.86 | 4353.73 | 2MG-CD | -1411.09 | 2830.19 | 1MG-NCD | -1882.82 | 3773.63 | ||
| 2MG-ADI | -959.96 | 1939.91 | 2MG-ADI | -658.54 | 1337.07 | 2MG-ADI | 290.00 | -560.00 | 2MG-ADI | -2172.01 | 4364.02 | 2MG-ADI | -1411.05 | 2842.11 | 2MG-ADI | -1882.78 | 3785.55 | ||
| ZH6×YH15 | 2MG-EAD | -696.02 | 1398.03 | 0MG | -420.43 | 844.852 | 0MG | 244.26 | -484.53 | 1MG-AD | -1732.76 | 3473.521 | 1MG-AD | -1081.861 | 2171.72 | 1MG-NCD | -1562.88 | 3133.77 | |
| 1MG-AD | -696.24 | 1400.48 | 1MG-A | -420.42 | 846.85 | 1MG-AD | 245.38 | -482.76 | 0MG | -1734.78 | 3473.554 | 0MG | -1084.278 | 2172.56 | 1MG-AD | -1562.99 | 3133.98 | ||
| 2MG-AD | -696.24 | 1404.48 | 2MG-EA | -420.42 | 846.85 | 2MG-EAD | 244.29 | -482.57 | 1MG-NCD | -1732.85 | 3473.693 | 1MG-A | -1084.274 | 2174.55 | 0MG | -1566.34 | 3136.69 | ||
| 2MG-CD | -699.82 | 1407.63 | 2MG-EAD | -420.43 | 846.869 | 2MG-EA | 244.27 | -482.53 | 2MG-EA | -1734.78 | 3475.549 | 2MG-EA | -1084.275 | 2174.55 | 2MG-AD | -1562.99 | 3137.98 | ||
| 1MG-EAD | -699.82 | 1407.63 | 1MG-NCD | -419.77 | 847.546 | 1MG-A | 244.27 | -482.53 | 1MG-A | -1734.78 | 3475.55 | 2MG-EAD | -1084.286 | 2174.57 | 2MG-EA | -1566.34 | 3138.68 | ||
| 0MG | -701.89 | 1407.77 | 1MG-AD | -419.80 | 847.609 | 2MG-CD | 244.94 | -481.88 | 2MG-EAD | -1734.78 | 3475.566 | 1MG-NCD | -1083.612 | 2175.22 | 1MG-A | -1566.34 | 3138.68 | ||
| 1MG-A | -701.88 | 1409.76 | 2MG-A | -420.42 | 848.848 | 1MG-EAD | 244.94 | -481.88 | 2MG-AD | -1732.76 | 3477.52 | 2MG-AD | -1081.86 | 2175.72 | 2MG-EAD | -1566.35 | 3138.70 | ||
| 2MG-EA | -701.88 | 1409.76 | 1MG-EAD | -420.43 | 848.866 | 2MG-A | 244.27 | -480.53 | 2MG-A | -1734.77 | 3477.548 | 2MG-A | -1084.271 | 2176.54 | 2MG-A | -1566.34 | 3140.68 | ||
| 2MG-A | -701.88 | 1411.76 | 2MG-CD | -420.43 | 848.867 | 1MG-NCD | 244.26 | -480.51 | 1MG-EAD | -1734.78 | 3477.564 | 1MG-EAD | -1084.284 | 2176.57 | 1MG-EAD | -1566.35 | 3140.69 | ||
| 1MG-NCD | -701.89 | 1411.78 | 2MG-AD | -419.80 | 851.609 | 2MG-AD | 245.38 | -478.77 | 2MG-CD | -1734.78 | 3477.564 | 2MG-CD | -1084.289 | 2176.58 | 2MG-CD | -1566.35 | 3140.70 | ||
| 2MG-ADI | -701.83 | 1423.66 | 2MG-ADI | -420.41 | 860.817 | 2MG-ADI | 244.29 | -468.58 | 2MG-ADI | -1734.70 | 3489.398 | 2MG-ADI | -1084.263 | 2188.53 | 2MG-ADI | -1566.21 | 3152.42 | ||
| YH15×YY20 | 0MG | -584.03 | 1172.05 | 1MG-AD | -403.72 | 815.444 | 0MG | 159.67 | -315.35 | 1MG-AD | -1257.01 | 2522.023 | 1MG-AD | -820.9228 | 1649.85 | 0MG | -1173.89 | 2351.78 | |
| 1MG-AD | -582.14 | 1172.28 | 2MG-AD | -403.72 | 819.444 | 1MG-A | 159.68 | -313.35 | 1MG-NCD | -1257.41 | 2522.813 | 1MG-NCD | -822.6339 | 1653.27 | 1MG-AD | -1172.70 | 2353.40 | ||
| 1MG-A | -584.02 | 1174.04 | 1MG-NCD | -409.22 | 826.442 | 2MG-EA | 159.68 | -313.35 | 2MG-AD | -1257.01 | 2526.018 | 0MG | -824.8229 | 1653.65 | 1MG-A | -1173.89 | 2353.78 | ||
| 2MG-EA | -584.02 | 1174.04 | 0MG | -412.19 | 828.389 | 2MG-EAD | 159.67 | -313.33 | 0MG | -1261.74 | 2527.484 | 2MG-AD | -820.9221 | 1653.84 | 2MG-EA | -1173.89 | 2353.78 | ||
| 2MG-EAD | -584.03 | 1174.06 | 1MG-A | -412.19 | 830.374 | 1MG-NCD | 159.68 | -311.37 | 2MG-EA | -1261.74 | 2529.47 | 1MG-A | -824.8159 | 1655.63 | 2MG-EAD | -1173.90 | 2353.80 | ||
| 1MG-NCD | -583.11 | 1174.22 | 2MG-EA | -412.19 | 830.377 | 2MG-A | 159.68 | -311.36 | 1MG-A | -1261.74 | 2529.472 | 2MG-EA | -824.8174 | 1655.64 | 1MG-NCD | -1173.40 | 2354.79 | ||
| 2MG-A | -584.02 | 1176.03 | 2MG-EAD | -412.21 | 830.419 | 1MG-AD | 159.68 | -311.35 | 2MG-EAD | -1261.75 | 2529.492 | 2MG-EAD | -824.8306 | 1655.66 | 2MG-A | -1173.89 | 2355.77 | ||
| 1MG-EAD | -584.03 | 1176.06 | 2MG-A | -412.18 | 832.353 | 1MG-EAD | 159.67 | -311.33 | 2MG-A | -1261.73 | 2531.464 | 2MG-A | -824.8067 | 1657.61 | 2MG-CD | -1173.90 | 2355.80 | ||
| 2MG-CD | -584.03 | 1176.06 | 1MG-EAD | -412.21 | 832.411 | 2MG-CD | 159.66 | -311.33 | 1MG-EAD | -1261.75 | 2531.491 | 2MG-CD | -824.8297 | 1657.66 | 1MG-EAD | -1173.90 | 2355.81 | ||
| 2MG-AD | -582.14 | 1176.28 | 2MG-CD | -412.21 | 832.427 | 2MG-AD | 160.70 | -309.41 | 2MG-CD | -1261.75 | 2531.492 | 1MG-EAD | -824.8359 | 1657.67 | 2MG-AD | -1172.70 | 2357.39 | ||
| 2MG-ADI | -583.98 | 1187.96 | 2MG-ADI | -412.14 | 844.289 | 2MG-ADI | 159.69 | -299.37 | 2MG-ADI | -1261.47 | 2542.949 | 2MG-ADI | -824.7699 | 1669.54 | 2MG-ADI | -1173.86 | 2367.73 | ||
| YH15×FHS | 1MG-NCD | -840.29 | 1688.58 | 1MG-AD | -597.78 | 1203.56 | 0MG | 293.68 | -583.36 | 1MG-AD | -1831.51 | 3671.015 | 1MG-AD | -1215.35 | 2438.70 | 1MG-AD | -1657.61 | 3323.22 | |
| 1MG-AD | -840.41 | 1688.81 | 2MG-AD | -597.78 | 1207.56 | 1MG-AD | 295.35 | -582.71 | 1MG-NCD | -1831.90 | 3671.808 | 1MG-NCD | -1216.119 | 2440.24 | 1MG-NCD | -1657.70 | 3323.41 | ||
| 0MG | -843.13 | 1690.27 | 1MG-NCD | -602.16 | 1212.32 | 1MG-NCD | 295.11 | -582.22 | 2MG-AD | -1831.51 | 3675.013 | 2MG-AD | -1215.35 | 2442.70 | 0MG | -1660.06 | 3324.12 | ||
| 2MG-EA | -843.13 | 1692.26 | 0MG | -605.49 | 1214.98 | 2MG-EA | 293.68 | -581.36 | 0MG | -1842.40 | 3688.807 | 0MG | -1220.225 | 2444.45 | 2MG-EA | -1660.06 | 3326.11 | ||
| 1MG-A | -843.13 | 1692.26 | 1MG-A | -605.49 | 1216.97 | 1MG-A | 293.68 | -581.36 | 2MG-EA | -1842.40 | 3690.789 | 2MG-EA | -1220.221 | 2446.44 | 1MG-A | -1660.06 | 3326.12 | ||
| 2MG-EAD | -843.14 | 1692.28 | 2MG-EA | -605.49 | 1216.97 | 2MG-EAD | 293.68 | -581.35 | 1MG-A | -1842.40 | 3690.803 | 1MG-A | -1220.221 | 2446.44 | 2MG-EAD | -1660.06 | 3326.13 | ||
| 2MG-AD | -840.19 | 1692.38 | 2MG-EAD | -605.50 | 1216.99 | 1MG-EAD | 293.68 | -579.35 | 2MG-EAD | -1842.41 | 3690.814 | 2MG-EAD | -1220.23 | 2446.46 | 2MG-AD | -1657.61 | 3327.22 | ||
| 2MG-A | -843.13 | 1694.26 | 2MG-A | -605.48 | 1218.96 | 2MG-A | 293.68 | -579.35 | 2MG-A | -1842.40 | 3692.806 | 2MG-A | -1220.219 | 2448.44 | 2MG-A | -1660.06 | 3328.12 | ||
| 1MG-EAD | -843.14 | 1694.27 | 1MG-EAD | -605.50 | 1218.99 | 2MG-CD | 293.68 | -579.35 | 1MG-EAD | -1842.41 | 3692.812 | 1MG-EAD | -1220.229 | 2448.46 | 1MG-EAD | -1660.06 | 3328.13 | ||
| 2MG-CD | -843.14 | 1694.27 | 2MG-CD | -605.50 | 1218.99 | 2MG-AD | 295.35 | -578.71 | 2MG-CD | -1842.41 | 3692.813 | 2MG-CD | -1220.229 | 2448.46 | 2MG-CD | -1660.06 | 3328.13 | ||
| 2MG-ADI | -843.02 | 1706.03 | 2MG-ADI | -605.44 | 1230.87 | 2MG-ADI | 293.90 | -567.81 | 2MG-ADI | -1839.82 | 3699.64 | 2MG-ADI | -1220.108 | 2460.22 | 2MG-ADI | -1659.86 | 3339.72 | ||
| YH15×NC94022 | 0MG | -478.24 | 960.48 | 0MG | -286.25 | 576.491 | 0MG | 119.22 | -234.45 | 0MG | -1097.35 | 2198.689 | 0MG | -680.8403 | 1365.68 | 1MG-AD | -1031.72 | 2071.45 | |
| 1MG-A | -478.24 | 962.48 | 1MG-AD | -284.66 | 577.311 | 2MG-EA | 119.23 | -232.45 | 1MG-AD | -1096.25 | 2200.493 | 2MG-EAD | -680.6691 | 1367.34 | 0MG | -1033.78 | 2071.55 | ||
| 2MG-EA | -478.24 | 962.48 | 1MG-NCD | -284.89 | 577.789 | 1MG-A | 119.22 | -232.44 | 2MG-EA | -1097.34 | 2200.683 | 1MG-A | -680.8372 | 1367.67 | 1MG-A | -1033.77 | 2073.54 | ||
| 2MG-EAD | -478.25 | 962.50 | 2MG-EA | -286.24 | 578.485 | 2MG-EAD | 119.22 | -232.44 | 1MG-A | -1097.35 | 2200.69 | 2MG-EA | -680.8376 | 1367.68 | 2MG-EA | -1033.77 | 2073.54 | ||
| 1MG-NCD | -477.98 | 963.95 | 1MG-A | -286.25 | 578.495 | 1MG-AD | 120.02 | -232.05 | 2MG-EAD | -1097.35 | 2200.697 | 1MG-AD | -679.9577 | 1367.92 | 2MG-EAD | -1033.78 | 2073.56 | ||
| 1MG-AD | -477.99 | 963.98 | 2MG-EAD | -286.25 | 578.498 | 1MG-NCD | 119.88 | -231.77 | 1MG-NCD | -1096.40 | 2200.801 | 2MG-CD | -680.8213 | 1369.64 | 1MG-NCD | -1032.98 | 2073.96 | ||
| 2MG-A | -478.24 | 964.48 | 1MG-EAD | -286.25 | 580.496 | 1MG-EAD | 119.22 | -230.44 | 2MG-A | -1097.35 | 2202.694 | 1MG-EAD | -680.8291 | 1369.66 | 2MG-AD | -1031.72 | 2075.44 | ||
| 1MG-EAD | -478.25 | 964.50 | 2MG-CD | -286.25 | 580.497 | 2MG-CD | 119.22 | -230.44 | 1MG-EAD | -1097.35 | 2202.695 | 2MG-A | -680.8352 | 1369.67 | 2MG-A | -1033.77 | 2075.53 | ||
| 2MG-CD | -478.25 | 964.50 | 2MG-A | -286.25 | 580.505 | 2MG-A | 119.22 | -230.44 | 2MG-CD | -1097.35 | 2202.696 | 1MG-NCD | -680.8468 | 1369.69 | 1MG-EAD | -1033.78 | 2075.56 | ||
| 2MG-AD | -477.99 | 967.97 | 2MG-AD | -284.66 | 581.312 | 2MG-AD | 120.59 | -229.18 | 2MG-AD | -1096.25 | 2204.493 | 2MG-AD | -679.9576 | 1371.92 | 2MG-CD | -1033.78 | 2075.56 | ||
| 2MG-ADI | -478.22 | 976.44 | 2MG-ADI | -285.29 | 590.582 | 2MG-ADI | 120.09 | -220.19 | 2MG-ADI | -1096.95 | 2213.901 | 2MG-ADI | -680.8271 | 1381.65 | 2MG-ADI | -1033.73 | 2087.45 | ||
新窗口打开|下载CSV
Supplement table 2
附表2
附表2遗传模式的适合性测验
Supplement table 2
| 组合 Combination | 性状 Trail | 模型 Models | U12 | PU12 | U22 | PU22 | U32 | PU32 | nW2 | PnW2 | Dn | PDn |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| YZ9102×YH15 | 籽仁长 KL | 0MG | 0.01 | 0.77 | 0.00 | 0.86 | 0.04 | 0.65 | 0.00 | 0.76 | 0.03 | 0.77 |
| 1MG-NCD | 0.00 | 0.99 | 0.00 | 0.93 | 0.01 | 0.76 | 0.00 | 0.98 | 0.02 | 0.99 | ||
| 1MG-AD | 0.00 | 0.99 | 0.00 | 0.92 | 0.02 | 0.71 | 0.00 | 0.97 | 0.02 | 0.98 | ||
| 1MG-A | 0.01 | 0.77 | 0.00 | 0.87 | 0.06 | 0.63 | 0.00 | 0.76 | 0.03 | 0.77 | ||
| 2MG-EA | 0.01 | 0.77 | 0.00 | 0.87 | 0.06 | 0.63 | 0.00 | 0.76 | 0.03 | 0.77 | ||
| 2MG-EAD | 0.01 | 0.77 | 0.00 | 0.87 | 0.06 | 0.62 | 0.00 | 0.76 | 0.03 | 0.76 | ||
| 2MG-A | 0.01 | 0.76 | 0.00 | 0.86 | 0.06 | 0.63 | 0.00 | 0.76 | 0.03 | 0.76 | ||
| 1MG-EAD | 0.01 | 0.77 | 0.00 | 0.87 | 0.06 | 0.63 | 0.00 | 0.76 | 0.03 | 0.76 | ||
| 2MG-CD | 0.01 | 0.77 | 0.00 | 0.87 | 0.06 | 0.63 | 0.00 | 0.76 | 0.03 | 0.76 | ||
| 2MG-AD | 0.00 | 0.99 | 0.00 | 0.97 | 0.00 | 0.84 | 0.00 | 0.99 | 0.02 | 1.00 | ||
| 2MG-ADI | 0.01 | 0.77 | 0.00 | 0.87 | 0.06 | 0.63 | 0.00 | 0.77 | 0.03 | 0.77 | ||
| 籽仁宽 KW | 1MG-AD | 0.00 | 0.99 | 0.00 | 0.97 | 0.00 | 0.85 | 0.00 | 0.97 | 0.03 | 0.87 | |
| 0MG | 0.00 | 0.79 | 0.00 | 0.97 | 0.52 | 0.40 | 0.01 | 0.56 | 0.04 | 0.60 | ||
| 1MG-NCD | 0.00 | 0.95 | 0.00 | 0.83 | 0.15 | 0.53 | 0.00 | 0.87 | 0.03 | 0.62 | ||
| 1MG-A | 0.00 | 0.79 | 0.00 | 0.97 | 0.59 | 0.38 | 0.01 | 0.55 | 0.04 | 0.59 | ||
| 2MG-EA | 0.00 | 0.79 | 0.00 | 0.97 | 0.59 | 0.38 | 0.01 | 0.55 | 0.04 | 0.59 | ||
| 2MG-EAD | 0.00 | 0.79 | 0.00 | 0.97 | 0.59 | 0.38 | 0.01 | 0.55 | 0.04 | 0.59 | ||
| 2MG-AD | 0.00 | 0.99 | 0.00 | 0.97 | 0.00 | 0.85 | 0.00 | 0.97 | 0.03 | 0.87 | ||
| 2MG-A | 0.01 | 0.79 | 0.00 | 0.97 | 0.59 | 0.38 | 0.01 | 0.55 | 0.04 | 0.59 | ||
| 1MG-EAD | 0.00 | 0.79 | 0.00 | 0.97 | 0.59 | 0.38 | 0.01 | 0.55 | 0.04 | 0.59 | ||
| 2MG-CD | 0.00 | 0.79 | 0.00 | 0.97 | 0.59 | 0.38 | 0.01 | 0.55 | 0.04 | 0.59 | ||
| 2MG-ADI | 0.00 | 0.80 | 0.00 | 0.97 | 0.59 | 0.38 | 0.01 | 0.56 | 0.04 | 0.59 | ||
| 籽仁长宽比KL/KW | 2MG-ADI | 0.00 | 0.84 | 0.00 | 0.89 | 0.00 | 0.82 | 0.01 | 0.75 | 0.03 | 0.78 | |
| 0MG | 0.00 | 0.84 | 0.00 | 0.88 | 0.00 | 0.84 | 0.01 | 0.75 | 0.03 | 0.79 | ||
| 1MG-AD | 0.00 | 0.99 | 0.00 | 0.99 | 0.00 | 0.91 | 0.00 | 0.78 | 0.04 | 0.43 | ||
| 1MG-A | 0.00 | 0.83 | 0.00 | 0.89 | 0.00 | 0.82 | 0.01 | 0.75 | 0.03 | 0.78 | ||
| 2MG-EA | 0.00 | 0.84 | 0.00 | 0.89 | 0.00 | 0.82 | 0.01 | 0.75 | 0.03 | 0.78 | ||
| 2MG-EAD | 0.00 | 0.83 | 0.00 | 0.88 | 0.00 | 0.82 | 0.01 | 0.75 | 0.03 | 0.78 | ||
| 1MG-NCD | 0.00 | 0.95 | 0.00 | 0.93 | 0.00 | 0.90 | 0.00 | 0.78 | 0.04 | 0.44 | ||
| 2MG-A | 0.00 | 0.83 | 0.00 | 0.88 | 0.00 | 0.82 | 0.01 | 0.75 | 0.03 | 0.78 | ||
| 1MG-EAD | 0.00 | 0.83 | 0.00 | 0.88 | 0.00 | 0.82 | 0.01 | 0.75 | 0.03 | 0.78 | ||
| 2MG-CD | 0.00 | 0.83 | 0.00 | 0.88 | 0.00 | 0.82 | 0.01 | 0.75 | 0.03 | 0.78 | ||
| 2MG-AD | 0.00 | 0.99 | 0.00 | 0.99 | 0.00 | 0.90 | 0.00 | 0.78 | 0.04 | 0.44 | ||
| 籽仁表面积KSA | 1MG-AD | 0.00 | 0.87 | 0.00 | 0.97 | 0.04 | 0.65 | 0.01 | 0.67 | 0.03 | 0.79 | |
| 1MG-NCD | 0.00 | 0.84 | 0.00 | 0.90 | 0.01 | 0.76 | 0.01 | 0.70 | 0.03 | 0.81 | ||
| 2MG-AD | 0.00 | 0.87 | 0.00 | 0.97 | 0.04 | 0.65 | 0.01 | 0.67 | 0.03 | 0.79 | ||
| 0MG | 0.51 | 0.40 | 0.12 | 0.55 | 0.66 | 0.37 | 0.17 | 0.07 | 0.06 | 0.05 | ||
| 2MG-EA | 0.51 | 0.40 | 0.12 | 0.56 | 0.75 | 0.35 | 0.17 | 0.07 | 0.06 | 0.05 | ||
| 1MG-A | 0.51 | 0.40 | 0.12 | 0.56 | 0.75 | 0.35 | 0.17 | 0.07 | 0.06 | 0.05 | ||
| 2MG-EAD | 0.51 | 0.40 | 0.12 | 0.56 | 0.75 | 0.35 | 0.17 | 0.07 | 0.06 | 0.05 | ||
| 2MG-A | 0.56 | 0.39 | 0.13 | 0.55 | 0.76 | 0.35 | 0.17 | 0.07 | 0.06 | 0.04 | ||
| 1MG-EAD | 0.51 | 0.40 | 0.12 | 0.56 | 0.75 | 0.35 | 0.17 | 0.07 | 0.06 | 0.05 | ||
| 2MG-CD | 0.51 | 0.40 | 0.12 | 0.56 | 0.75 | 0.35 | 0.17 | 0.07 | 0.06 | 0.05 | ||
| 2MG-ADI | 0.41 | 0.42 | 0.09 | 0.59 | 0.77 | 0.35 | 0.15 | 0.08 | 0.06 | 0.06 | ||
| 籽仁表面周长KSP | 1MG-AD | 0.00 | 0.98 | 0.00 | 0.94 | 0.02 | 0.70 | 0.01 | 0.72 | 0.04 | 0.60 | |
| 1MG-NCD | 0.00 | 0.96 | 0.00 | 0.83 | 0.20 | 0.50 | 0.01 | 0.63 | 0.04 | 0.43 | ||
| 0MG | 0.01 | 0.73 | 0.00 | 0.94 | 1.16 | 0.30 | 0.04 | 0.25 | 0.04 | 0.43 | ||
| 2MG-AD | 0.00 | 0.98 | 0.00 | 0.94 | 0.02 | 0.70 | 0.01 | 0.72 | 0.04 | 0.60 | ||
| 1MG-A | 0.01 | 0.73 | 0.00 | 0.94 | 1.30 | 0.29 | 0.05 | 0.25 | 0.04 | 0.43 | ||
| 2MG-EA | 0.01 | 0.73 | 0.00 | 0.94 | 1.30 | 0.29 | 0.05 | 0.25 | 0.04 | 0.42 | ||
| 2MG-EAD | 0.02 | 0.73 | 0.00 | 0.94 | 1.31 | 0.29 | 0.05 | 0.25 | 0.04 | 0.42 | ||
| 2MG-A | 0.02 | 0.72 | 0.00 | 0.94 | 1.30 | 0.29 | 0.05 | 0.25 | 0.04 | 0.42 | ||
| 1MG-EAD | 0.02 | 0.73 | 0.00 | 0.94 | 1.31 | 0.29 | 0.05 | 0.25 | 0.04 | 0.42 | ||
| 2MG-CD | 0.02 | 0.73 | 0.00 | 0.94 | 1.31 | 0.29 | 0.05 | 0.25 | 0.04 | 0.42 | ||
| 2MG-ADI | 0.01 | 0.73 | 0.00 | 0.95 | 1.30 | 0.29 | 0.04 | 0.25 | 0.04 | 0.43 | ||
| 平均单仁重 AKW | 1MG-AD | 0.00 | 0.98 | 0.00 | 0.96 | 0.00 | 0.92 | 0.00 | 1.00 | 0.02 | 1.00 | |
| 2MG-EAD | 0.00 | 0.98 | 0.00 | 0.95 | 0.01 | 0.76 | 0.00 | 0.99 | 0.02 | 1.00 | ||
| 1MG-EAD | 0.00 | 0.91 | 0.00 | 0.96 | 0.00 | 0.79 | 0.00 | 1.00 | 0.02 | 1.00 | ||
| 2MG-CD | 0.00 | 0.91 | 0.00 | 0.96 | 0.00 | 0.79 | 0.00 | 1.00 | 0.02 | 1.00 | ||
| 0MG | 0.02 | 0.72 | 0.06 | 0.63 | 0.10 | 0.57 | 0.01 | 0.61 | 0.03 | 0.76 | ||
| 2MG-AD | 0.00 | 0.98 | 0.00 | 0.96 | 0.00 | 0.92 | 0.00 | 1.00 | 0.02 | 1.00 | ||
| 1MG-A | 0.02 | 0.72 | 0.06 | 0.62 | 0.12 | 0.56 | 0.01 | 0.60 | 0.03 | 0.76 | ||
| 2MG-EA | 0.02 | 0.72 | 0.06 | 0.62 | 0.12 | 0.55 | 0.01 | 0.60 | 0.03 | 0.76 | ||
| 2MG-A | 0.02 | 0.72 | 0.06 | 0.62 | 0.12 | 0.55 | 0.01 | 0.60 | 0.03 | 0.76 | ||
| 1MG-NCD | 0.02 | 0.72 | 0.06 | 0.62 | 0.12 | 0.55 | 0.01 | 0.60 | 0.03 | 0.76 | ||
| 2MG-ADI | 0.02 | 0.72 | 0.06 | 0.62 | 0.12 | 0.56 | 0.01 | 0.61 | 0.03 | 0.77 | ||
| ZH6×YH15 | 籽仁长 KL | 2MG-EAD | 0.00 | 0.89 | 0.00 | 0.92 | 0.88 | 0.33 | 0.01 | 0.51 | 0.04 | 0.46 |
| 1MG-AD | 0.00 | 0.89 | 0.00 | 0.82 | 0.02 | 0.69 | 0.01 | 0.72 | 0.03 | 0.77 | ||
| 2MG-AD | 0.00 | 0.89 | 0.00 | 0.82 | 0.02 | 0.69 | 0.01 | 0.72 | 0.03 | 0.77 | ||
| 2MG-CD | 0.00 | 0.97 | 0.01 | 0.75 | 4.00 | 0.16 | 0.03 | 0.30 | 0.04 | 0.46 | ||
| 1MG-EAD | 0.00 | 0.97 | 0.01 | 0.75 | 3.99 | 0.16 | 0.03 | 0.30 | 0.04 | 0.46 | ||
| 0MG | 0.00 | 0.81 | 0.14 | 0.54 | 5.27 | 0.13 | 0.03 | 0.34 | 0.05 | 0.38 | ||
| 1MG-A | 0.00 | 0.81 | 0.15 | 0.53 | 5.77 | 0.12 | 0.03 | 0.34 | 0.05 | 0.37 | ||
| 2MG-EA | 0.00 | 0.81 | 0.15 | 0.53 | 5.77 | 0.12 | 0.03 | 0.34 | 0.05 | 0.37 | ||
| 2MG-A | 0.00 | 0.80 | 0.15 | 0.53 | 5.76 | 0.12 | 0.03 | 0.34 | 0.05 | 0.37 | ||
| 1MG-NCD | 0.00 | 0.81 | 0.15 | 0.53 | 5.77 | 0.12 | 0.03 | 0.34 | 0.05 | 0.37 | ||
| 2MG-ADI | 0.00 | 0.81 | 0.14 | 0.54 | 5.77 | 0.12 | 0.03 | 0.34 | 0.05 | 0.38 | ||
| 籽仁宽 KW | 0MG | 0.00 | 0.83 | 0.00 | 0.80 | 0.00 | 0.87 | 0.00 | 0.90 | 0.03 | 0.91 | |
| 1MG-A | 0.00 | 0.83 | 0.00 | 0.81 | 0.00 | 0.89 | 0.00 | 0.90 | 0.03 | 0.91 | ||
| 2MG-EA | 0.00 | 0.83 | 0.00 | 0.81 | 0.00 | 0.89 | 0.00 | 0.90 | 0.03 | 0.91 | ||
| 2MG-EAD | 0.00 | 0.83 | 0.00 | 0.81 | 0.00 | 0.89 | 0.00 | 0.90 | 0.03 | 0.91 | ||
| 1MG-NCD | 0.00 | 0.96 | 0.00 | 0.99 | 0.00 | 0.81 | 0.00 | 0.95 | 0.02 | 0.98 | ||
| 1MG-AD | 0.00 | 0.96 | 0.00 | 1.00 | 0.00 | 0.84 | 0.00 | 0.96 | 0.02 | 0.98 | ||
| 2MG-A | 0.00 | 0.83 | 0.00 | 0.80 | 0.00 | 0.89 | 0.00 | 0.90 | 0.03 | 0.91 | ||
| 1MG-EAD | 0.00 | 0.83 | 0.00 | 0.81 | 0.00 | 0.89 | 0.00 | 0.90 | 0.03 | 0.91 | ||
| 2MG-CD | 0.00 | 0.83 | 0.00 | 0.81 | 0.00 | 0.89 | 0.00 | 0.90 | 0.03 | 0.91 | ||
| 2MG-AD | 0.00 | 0.96 | 0.00 | 1.00 | 0.00 | 0.84 | 0.00 | 0.96 | 0.02 | 0.98 | ||
| 2MG-ADI | 0.00 | 0.83 | 0.00 | 0.81 | 0.00 | 0.89 | 0.00 | 0.90 | 0.03 | 0.92 | ||
| 籽仁长宽比KL/KW | 0MG | 0.00 | 0.87 | 0.00 | 0.80 | 0.02 | 0.70 | 0.00 | 0.99 | 0.03 | 0.93 | |
| 1MG-AD | 0.00 | 0.96 | 0.00 | 0.98 | 0.00 | 0.93 | 0.00 | 1.00 | 0.02 | 1.00 | ||
| 2MG-EAD | 0.00 | 0.87 | 0.00 | 0.79 | 0.03 | 0.67 | 0.00 | 0.99 | 0.03 | 0.93 | ||
| 2MG-EA | 0.00 | 0.87 | 0.01 | 0.79 | 0.03 | 0.67 | 0.00 | 0.98 | 0.03 | 0.92 | ||
| 1MG-A | 0.00 | 0.87 | 0.01 | 0.79 | 0.03 | 0.67 | 0.00 | 0.98 | 0.03 | 0.92 | ||
| 2MG-CD | 0.00 | 0.96 | 0.00 | 0.97 | 0.01 | 0.74 | 0.00 | 0.99 | 0.02 | 0.97 | ||
| 1MG-EAD | 0.00 | 0.96 | 0.00 | 0.97 | 0.01 | 0.74 | 0.00 | 0.99 | 0.02 | 0.97 | ||
| 2MG-A | 0.00 | 0.86 | 0.01 | 0.79 | 0.03 | 0.67 | 0.00 | 0.98 | 0.03 | 0.92 | ||
| 1MG-NCD | 0.00 | 0.87 | 0.01 | 0.79 | 0.03 | 0.67 | 0.00 | 0.98 | 0.03 | 0.92 | ||
| 2MG-AD | 0.00 | 0.96 | 0.00 | 0.98 | 0.00 | 0.93 | 0.00 | 1.00 | 0.02 | 1.00 | ||
| 2MG-ADI | 0.00 | 0.87 | 0.00 | 0.79 | 0.03 | 0.67 | 0.00 | 0.99 | 0.03 | 0.93 | ||
| 籽仁表面积KSA | 1MG-AD | 0.00 | 0.98 | 0.00 | 0.97 | 0.00 | 0.79 | 0.00 | 0.97 | 0.02 | 0.98 | |
| 0MG | 0.02 | 0.72 | 0.00 | 0.85 | 0.14 | 0.54 | 0.01 | 0.58 | 0.04 | 0.56 | ||
| 1MG-NCD | 0.00 | 1.00 | 0.00 | 0.92 | 0.02 | 0.70 | 0.00 | 0.95 | 0.03 | 0.96 | ||
| 2MG-EA | 0.02 | 0.72 | 0.00 | 0.85 | 0.17 | 0.52 | 0.01 | 0.57 | 0.04 | 0.56 | ||
| 1MG-A | 0.02 | 0.72 | 0.00 | 0.85 | 0.17 | 0.52 | 0.01 | 0.57 | 0.04 | 0.56 | ||
| 2MG-EAD | 0.02 | 0.72 | 0.00 | 0.85 | 0.17 | 0.52 | 0.01 | 0.57 | 0.04 | 0.56 | ||
| 2MG-AD | 0.00 | 0.98 | 0.00 | 0.97 | 0.00 | 0.79 | 0.00 | 0.97 | 0.02 | 0.98 | ||
| 2MG-A | 0.02 | 0.71 | 0.00 | 0.85 | 0.17 | 0.52 | 0.01 | 0.57 | 0.04 | 0.55 | ||
| 1MG-EAD | 0.02 | 0.72 | 0.00 | 0.85 | 0.17 | 0.52 | 0.01 | 0.57 | 0.04 | 0.56 | ||
| 2MG-CD | 0.02 | 0.72 | 0.00 | 0.85 | 0.17 | 0.52 | 0.01 | 0.57 | 0.04 | 0.56 | ||
| 2MG-ADI | 0.01 | 0.73 | 0.00 | 0.86 | 0.17 | 0.52 | 0.01 | 0.59 | 0.04 | 0.57 | ||
| 籽仁表面周长KSP | 1MG-AD | 0.00 | 0.98 | 0.00 | 0.95 | 0.01 | 0.73 | 0.00 | 0.82 | 0.03 | 0.89 | |
| 0MG | 0.00 | 0.91 | 0.00 | 0.87 | 1.50 | 0.27 | 0.01 | 0.50 | 0.04 | 0.62 | ||
| 1MG-A | 0.00 | 0.91 | 0.00 | 0.86 | 1.69 | 0.25 | 0.01 | 0.50 | 0.04 | 0.61 | ||
| 2MG-EA | 0.00 | 0.91 | 0.00 | 0.86 | 1.69 | 0.25 | 0.01 | 0.50 | 0.04 | 0.61 | ||
| 2MG-EAD | 0.00 | 0.91 | 0.00 | 0.86 | 1.69 | 0.25 | 0.01 | 0.49 | 0.04 | 0.61 | ||
| 1MG-NCD | 0.00 | 0.92 | 0.01 | 0.73 | 0.90 | 0.33 | 0.01 | 0.61 | 0.04 | 0.56 | ||
| 2MG-AD | 0.00 | 0.98 | 0.00 | 0.95 | 0.01 | 0.73 | 0.00 | 0.82 | 0.03 | 0.89 | ||
| 2MG-A | 0.00 | 0.91 | 0.00 | 0.86 | 1.68 | 0.26 | 0.01 | 0.50 | 0.04 | 0.62 | ||
| 1MG-EAD | 0.00 | 0.91 | 0.00 | 0.86 | 1.69 | 0.25 | 0.01 | 0.49 | 0.04 | 0.61 | ||
| 2MG-CD | 0.00 | 0.91 | 0.00 | 0.86 | 1.69 | 0.25 | 0.01 | 0.49 | 0.04 | 0.61 | ||
| 2MG-ADI | 0.00 | 0.91 | 0.00 | 0.86 | 1.68 | 0.26 | 0.01 | 0.50 | 0.04 | 0.61 | ||
| 平均单仁重 AKW | 1MG-NCD | 0.00 | 0.98 | 0.00 | 0.99 | 0.00 | 0.86 | 0.00 | 0.99 | 0.02 | 0.99 | |
| 1MG-AD | 0.00 | 0.97 | 0.00 | 0.98 | 0.00 | 0.94 | 0.00 | 0.99 | 0.02 | 0.99 | ||
| 0MG | 0.07 | 0.61 | 0.04 | 0.65 | 0.00 | 0.87 | 0.03 | 0.35 | 0.05 | 0.24 | ||
| 2MG-AD | 0.00 | 0.97 | 0.00 | 0.98 | 0.00 | 0.94 | 0.00 | 0.99 | 0.02 | 0.99 | ||
| 2MG-EA | 0.07 | 0.61 | 0.04 | 0.66 | 0.00 | 0.85 | 0.03 | 0.35 | 0.05 | 0.24 | ||
| 1MG-A | 0.07 | 0.61 | 0.04 | 0.66 | 0.00 | 0.85 | 0.03 | 0.35 | 0.05 | 0.24 | ||
| 2MG-EAD | 0.07 | 0.61 | 0.04 | 0.66 | 0.00 | 0.85 | 0.03 | 0.35 | 0.05 | 0.24 | ||
| 2MG-A | 0.07 | 0.60 | 0.04 | 0.65 | 0.00 | 0.85 | 0.03 | 0.34 | 0.05 | 0.24 | ||
| 1MG-EAD | 0.07 | 0.61 | 0.04 | 0.66 | 0.00 | 0.85 | 0.03 | 0.35 | 0.05 | 0.24 | ||
| 2MG-CD | 0.07 | 0.61 | 0.04 | 0.66 | 0.00 | 0.85 | 0.03 | 0.35 | 0.05 | 0.24 | ||
| 2MG-ADI | 0.06 | 0.62 | 0.03 | 0.67 | 0.00 | 0.85 | 0.03 | 0.36 | 0.05 | 0.25 | ||
| YH15×YY20 | 籽仁长 KL | 0MG | 0.00 | 0.87 | 0.00 | 0.94 | 0.83 | 0.34 | 0.01 | 0.75 | 0.04 | 0.84 |
| 1MG-AD | 0.00 | 0.95 | 0.00 | 0.98 | 0.02 | 0.72 | 0.00 | 0.95 | 0.04 | 0.85 | ||
| 1MG-A | 0.00 | 0.87 | 0.00 | 0.93 | 0.97 | 0.32 | 0.01 | 0.74 | 0.04 | 0.84 | ||
| 2MG-EA | 0.00 | 0.87 | 0.00 | 0.93 | 0.97 | 0.32 | 0.01 | 0.74 | 0.04 | 0.83 | ||
| 2MG-EAD | 0.00 | 0.87 | 0.00 | 0.93 | 0.97 | 0.32 | 0.01 | 0.74 | 0.04 | 0.84 | ||
| 1MG-NCD | 0.00 | 0.95 | 0.01 | 0.79 | 0.55 | 0.39 | 0.00 | 0.78 | 0.05 | 0.57 | ||
| 2MG-A | 0.00 | 0.86 | 0.00 | 0.94 | 0.96 | 0.32 | 0.01 | 0.74 | 0.04 | 0.84 | ||
| 1MG-EAD | 0.00 | 0.87 | 0.00 | 0.93 | 0.97 | 0.32 | 0.01 | 0.74 | 0.04 | 0.84 | ||
| 2MG-CD | 0.00 | 0.87 | 0.00 | 0.93 | 0.97 | 0.32 | 0.01 | 0.74 | 0.04 | 0.84 | ||
| 2MG-AD | 0.00 | 0.95 | 0.00 | 0.98 | 0.02 | 0.72 | 0.00 | 0.95 | 0.04 | 0.85 | ||
| 2MG-ADI | 0.00 | 0.88 | 0.00 | 0.92 | 0.96 | 0.32 | 0.01 | 0.74 | 0.04 | 0.83 | ||
| 籽仁宽 KW | 1MG-AD | 0.00 | 0.98 | 0.00 | 0.96 | 0.01 | 0.79 | 0.00 | 1.00 | 0.03 | 0.99 | |
| 2MG-AD | 0.00 | 0.98 | 0.00 | 0.96 | 0.01 | 0.79 | 0.00 | 1.00 | 0.03 | 0.99 | ||
| 1MG-NCD | 0.00 | 0.87 | 0.04 | 0.66 | 1.45 | 0.27 | 0.01 | 0.67 | 0.04 | 0.68 | ||
| 0MG | 0.00 | 0.84 | 0.00 | 0.87 | 4.14 | 0.15 | 0.01 | 0.57 | 0.04 | 0.71 | ||
| 1MG-A | 0.00 | 0.84 | 0.00 | 0.86 | 4.61 | 0.14 | 0.01 | 0.56 | 0.04 | 0.70 | ||
| 2MG-EA | 0.00 | 0.84 | 0.00 | 0.86 | 4.62 | 0.14 | 0.01 | 0.55 | 0.04 | 0.70 | ||
| 2MG-EAD | 0.00 | 0.84 | 0.00 | 0.87 | 4.62 | 0.14 | 0.01 | 0.55 | 0.04 | 0.70 | ||
| 2MG-A | 0.00 | 0.83 | 0.00 | 0.87 | 4.57 | 0.14 | 0.01 | 0.55 | 0.04 | 0.70 | ||
| 1MG-EAD | 0.00 | 0.84 | 0.00 | 0.87 | 4.62 | 0.14 | 0.01 | 0.55 | 0.04 | 0.69 | ||
| 2MG-CD | 0.00 | 0.84 | 0.00 | 0.87 | 4.61 | 0.14 | 0.01 | 0.55 | 0.04 | 0.70 | ||
| 2MG-ADI | 0.00 | 0.85 | 0.00 | 0.86 | 4.56 | 0.14 | 0.01 | 0.56 | 0.04 | 0.70 | ||
| 籽仁长宽比KL/KW | 0MG | 0.00 | 0.88 | 0.00 | 0.93 | 0.00 | 0.83 | 0.00 | 0.93 | 0.03 | 0.93 | |
| 1MG-A | 0.00 | 0.88 | 0.00 | 0.94 | 0.00 | 0.80 | 0.00 | 0.93 | 0.03 | 0.93 | ||
| 2MG-EA | 0.00 | 0.88 | 0.00 | 0.94 | 0.00 | 0.80 | 0.00 | 0.93 | 0.03 | 0.93 | ||
| 2MG-EAD | 0.00 | 0.88 | 0.00 | 0.93 | 0.00 | 0.80 | 0.00 | 0.93 | 0.03 | 0.93 | ||
| 1MG-NCD | 0.00 | 0.89 | 0.00 | 0.94 | 0.00 | 0.80 | 0.00 | 0.93 | 0.03 | 0.93 | ||
| 2MG-A | 0.00 | 0.88 | 0.00 | 0.93 | 0.00 | 0.80 | 0.00 | 0.93 | 0.03 | 0.93 | ||
| 1MG-AD | 0.00 | 0.88 | 0.00 | 0.94 | 0.00 | 0.80 | 0.00 | 0.93 | 0.03 | 0.93 | ||
| 1MG-EAD | 0.00 | 0.88 | 0.00 | 0.93 | 0.00 | 0.80 | 0.00 | 0.93 | 0.03 | 0.93 | ||
| 2MG-CD | 0.00 | 0.88 | 0.00 | 0.93 | 0.00 | 0.80 | 0.00 | 0.93 | 0.03 | 0.93 | ||
| 2MG-AD | 0.00 | 0.97 | 0.00 | 1.00 | 0.00 | 0.86 | 0.00 | 0.95 | 0.03 | 0.98 | ||
| 2MG-ADI | 0.00 | 0.88 | 0.00 | 0.94 | 0.00 | 0.80 | 0.00 | 0.93 | 0.03 | 0.93 | ||
| 籽仁表面积KSA | 1MG-AD | 0.00 | 0.92 | 0.00 | 0.98 | 0.07 | 0.61 | 0.00 | 0.90 | 0.04 | 0.72 | |
| 1MG-NCD | 0.00 | 0.99 | 0.00 | 0.85 | 0.43 | 0.42 | 0.00 | 0.79 | 0.04 | 0.76 | ||
| 2MG-AD | 0.00 | 0.92 | 0.00 | 0.98 | 0.07 | 0.61 | 0.00 | 0.90 | 0.04 | 0.72 | ||
| 0MG | 0.03 | 0.67 | 0.00 | 0.91 | 2.08 | 0.23 | 0.03 | 0.36 | 0.06 | 0.21 | ||
| 2MG-EA | 0.03 | 0.67 | 0.00 | 0.92 | 2.37 | 0.21 | 0.03 | 0.35 | 0.06 | 0.20 | ||
| 1MG-A | 0.03 | 0.67 | 0.00 | 0.92 | 2.37 | 0.21 | 0.03 | 0.35 | 0.06 | 0.20 | ||
| 2MG-EAD | 0.03 | 0.67 | 0.00 | 0.92 | 2.38 | 0.21 | 0.03 | 0.35 | 0.06 | 0.20 | ||
| 2MG-A | 0.04 | 0.66 | 0.00 | 0.90 | 2.36 | 0.22 | 0.03 | 0.35 | 0.06 | 0.20 | ||
| 1MG-EAD | 0.03 | 0.67 | 0.00 | 0.92 | 2.38 | 0.21 | 0.03 | 0.35 | 0.06 | 0.20 | ||
| 2MG-CD | 0.03 | 0.67 | 0.00 | 0.92 | 2.38 | 0.21 | 0.03 | 0.35 | 0.06 | 0.20 | ||
| 2MG-ADI | 0.03 | 0.69 | 0.00 | 0.94 | 2.35 | 0.22 | 0.02 | 0.37 | 0.06 | 0.22 | ||
| 籽仁表面周长KSP | 1MG-AD | 0.00 | 0.96 | 0.00 | 0.92 | 0.16 | 0.53 | 0.01 | 0.71 | 0.04 | 0.68 | |
| 1MG-NCD | 0.00 | 0.93 | 0.02 | 0.71 | 1.67 | 0.26 | 0.01 | 0.50 | 0.05 | 0.38 | ||
| 0MG | 0.00 | 0.84 | 0.00 | 0.86 | 4.83 | 0.14 | 0.03 | 0.31 | 0.05 | 0.43 | ||
| 2MG-AD | 0.00 | 0.96 | 0.00 | 0.92 | 0.16 | 0.53 | 0.01 | 0.71 | 0.04 | 0.68 | ||
| 1MG-A | 0.00 | 0.84 | 0.00 | 0.85 | 5.38 | 0.13 | 0.03 | 0.30 | 0.05 | 0.42 | ||
| 2MG-EA | 0.00 | 0.84 | 0.00 | 0.85 | 5.39 | 0.13 | 0.03 | 0.30 | 0.05 | 0.42 | ||
| 2MG-EAD | 0.00 | 0.84 | 0.00 | 0.85 | 5.39 | 0.13 | 0.03 | 0.30 | 0.05 | 0.42 | ||
| 2MG-A | 0.00 | 0.83 | 0.00 | 0.86 | 5.33 | 0.13 | 0.03 | 0.30 | 0.05 | 0.41 | ||
| 2MG-CD | 0.00 | 0.84 | 0.00 | 0.85 | 5.39 | 0.13 | 0.03 | 0.30 | 0.05 | 0.42 | ||
| 1MG-EAD | 0.00 | 0.84 | 0.00 | 0.85 | 5.40 | 0.13 | 0.03 | 0.30 | 0.05 | 0.42 | ||
| 2MG-ADI | 0.00 | 0.85 | 0.00 | 0.85 | 5.32 | 0.13 | 0.03 | 0.30 | 0.05 | 0.42 | ||
| 平均单仁重 AKW | 0MG | 0.00 | 0.90 | 0.00 | 0.93 | 0.44 | 0.42 | 0.00 | 0.84 | 0.03 | 0.90 | |
| 1MG-AD | 0.00 | 0.97 | 0.00 | 0.97 | 0.01 | 0.77 | 0.00 | 0.99 | 0.03 | 0.99 | ||
| 1MG-A | 0.00 | 0.90 | 0.00 | 0.92 | 0.52 | 0.39 | 0.00 | 0.83 | 0.03 | 0.90 | ||
| 2MG-EA | 0.00 | 0.90 | 0.00 | 0.92 | 0.53 | 0.39 | 0.00 | 0.83 | 0.03 | 0.90 | ||
| 2MG-EAD | 0.00 | 0.90 | 0.00 | 0.93 | 0.53 | 0.39 | 0.00 | 0.83 | 0.03 | 0.89 | ||
| 1MG-NCD | 0.00 | 0.96 | 0.00 | 0.81 | 0.30 | 0.46 | 0.00 | 0.89 | 0.03 | 0.88 | ||
| 2MG-A | 0.00 | 0.90 | 0.00 | 0.93 | 0.52 | 0.40 | 0.00 | 0.83 | 0.03 | 0.89 | ||
| 2MG-CD | 0.00 | 0.90 | 0.00 | 0.93 | 0.52 | 0.39 | 0.00 | 0.83 | 0.03 | 0.89 | ||
| 1MG-EAD | 0.00 | 0.90 | 0.00 | 0.93 | 0.53 | 0.39 | 0.00 | 0.83 | 0.03 | 0.89 | ||
| 2MG-AD | 0.00 | 0.97 | 0.00 | 0.97 | 0.01 | 0.77 | 0.00 | 0.99 | 0.03 | 0.99 | ||
| 2MG-ADI | 0.00 | 0.91 | 0.00 | 0.92 | 0.52 | 0.40 | 0.00 | 0.84 | 0.03 | 0.90 | ||
| YH15×FHS | 籽仁长 KL | 1MG-NCD | 0.00 | 0.98 | 0.00 | 0.93 | 0.00 | 0.79 | 0.00 | 0.83 | 0.03 | 0.81 |
| 1MG-AD | 0.00 | 0.97 | 0.00 | 0.90 | 0.02 | 0.72 | 0.00 | 0.82 | 0.03 | 0.83 | ||
| 0MG | 0.03 | 0.68 | 0.01 | 0.78 | 0.06 | 0.62 | 0.02 | 0.49 | 0.05 | 0.26 | ||
| 2MG-EA | 0.03 | 0.68 | 0.01 | 0.78 | 0.07 | 0.60 | 0.02 | 0.49 | 0.05 | 0.26 | ||
| 1MG-A | 0.03 | 0.68 | 0.01 | 0.78 | 0.07 | 0.60 | 0.02 | 0.49 | 0.05 | 0.26 | ||
| 2MG-EAD | 0.03 | 0.68 | 0.01 | 0.78 | 0.07 | 0.60 | 0.02 | 0.49 | 0.05 | 0.26 | ||
| 2MG-AD | 0.00 | 0.99 | 0.00 | 0.99 | 0.00 | 0.91 | 0.00 | 0.85 | 0.03 | 0.80 | ||
| 2MG-A | 0.03 | 0.67 | 0.01 | 0.78 | 0.07 | 0.60 | 0.02 | 0.48 | 0.05 | 0.26 | ||
| 1MG-EAD | 0.03 | 0.68 | 0.01 | 0.78 | 0.07 | 0.60 | 0.02 | 0.49 | 0.05 | 0.26 | ||
| 2MG-CD | 0.03 | 0.68 | 0.01 | 0.78 | 0.07 | 0.60 | 0.02 | 0.49 | 0.05 | 0.26 | ||
| 2MG-ADI | 0.03 | 0.69 | 0.00 | 0.79 | 0.07 | 0.60 | 0.01 | 0.50 | 0.05 | 0.27 | ||
| 籽仁宽 KW | 1MG-AD | 0.00 | 0.91 | 0.00 | 0.97 | 0.00 | 0.79 | 0.00 | 0.97 | 0.03 | 0.96 | |
| 2MG-AD | 0.00 | 0.91 | 0.00 | 0.97 | 0.00 | 0.79 | 0.00 | 0.97 | 0.03 | 0.96 | ||
| 1MG-NCD | 0.00 | 0.91 | 0.03 | 0.68 | 2.20 | 0.22 | 0.01 | 0.56 | 0.04 | 0.48 | ||
| 0MG | 0.01 | 0.78 | 0.00 | 0.90 | 5.83 | 0.12 | 0.02 | 0.40 | 0.04 | 0.47 | ||
| 1MG-A | 0.01 | 0.78 | 0.00 | 0.90 | 6.35 | 0.11 | 0.02 | 0.39 | 0.04 | 0.46 | ||
| 2MG-EA | 0.01 | 0.78 | 0.00 | 0.90 | 6.36 | 0.11 | 0.02 | 0.39 | 0.04 | 0.46 | ||
| 2MG-EAD | 0.01 | 0.78 | 0.00 | 0.90 | 6.37 | 0.11 | 0.02 | 0.39 | 0.04 | 0.46 | ||
| 2MG-A | 0.01 | 0.77 | 0.00 | 0.90 | 6.34 | 0.11 | 0.02 | 0.39 | 0.04 | 0.46 | ||
| 1MG-EAD | 0.01 | 0.78 | 0.00 | 0.90 | 6.36 | 0.11 | 0.02 | 0.39 | 0.04 | 0.46 | ||
| 2MG-CD | 0.01 | 0.78 | 0.00 | 0.90 | 6.36 | 0.11 | 0.02 | 0.39 | 0.04 | 0.46 | ||
| 2MG-ADI | 0.01 | 0.79 | 0.00 | 0.89 | 6.33 | 0.11 | 0.02 | 0.40 | 0.04 | 0.46 | ||
| 籽仁长宽比KL/KW | 0MG | 0.04 | 0.65 | 0.12 | 0.55 | 0.11 | 0.56 | 0.01 | 0.68 | 0.03 | 0.81 | |
| 1MG-AD | 0.00 | 0.90 | 0.01 | 0.79 | 0.14 | 0.54 | 0.00 | 0.94 | 0.03 | 0.91 | ||
| 1MG-NCD | 0.00 | 0.87 | 0.01 | 0.75 | 0.19 | 0.51 | 0.00 | 0.92 | 0.03 | 0.90 | ||
| 2MG-EA | 0.05 | 0.64 | 0.12 | 0.56 | 0.09 | 0.58 | 0.01 | 0.68 | 0.03 | 0.81 | ||
| 1MG-A | 0.05 | 0.64 | 0.12 | 0.56 | 0.09 | 0.58 | 0.01 | 0.68 | 0.03 | 0.81 | ||
| 2MG-EAD | 0.05 | 0.64 | 0.12 | 0.56 | 0.09 | 0.58 | 0.01 | 0.68 | 0.03 | 0.81 | ||
| 1MG-EAD | 0.05 | 0.64 | 0.12 | 0.56 | 0.09 | 0.58 | 0.01 | 0.68 | 0.03 | 0.81 | ||
| 2MG-A | 0.05 | 0.64 | 0.12 | 0.55 | 0.09 | 0.58 | 0.01 | 0.67 | 0.03 | 0.80 | ||
| 2MG-CD | 0.05 | 0.64 | 0.12 | 0.56 | 0.09 | 0.58 | 0.01 | 0.68 | 0.03 | 0.81 | ||
| 2MG-AD | 0.00 | 0.90 | 0.01 | 0.79 | 0.14 | 0.54 | 0.00 | 0.94 | 0.03 | 0.91 | ||
| 2MG-ADI | 0.04 | 0.66 | 0.10 | 0.57 | 0.09 | 0.58 | 0.01 | 0.71 | 0.03 | 0.83 | ||
| 籽仁表面积KSA | 1MG-AD | 0.00 | 0.90 | 0.00 | 0.98 | 0.02 | 0.69 | 0.00 | 0.87 | 0.03 | 0.87 | |
| 1MG-NCD | 0.00 | 0.86 | 0.00 | 0.92 | 0.01 | 0.79 | 0.00 | 0.89 | 0.03 | 0.87 | ||
| 2MG-AD | 0.00 | 0.90 | 0.00 | 0.99 | 0.02 | 0.69 | 0.00 | 0.87 | 0.03 | 0.87 | ||
| 0MG | 0.67 | 0.37 | 0.25 | 0.48 | 0.20 | 0.50 | 0.12 | 0.10 | 0.08 | 0.02 | ||
| 2MG-EA | 0.67 | 0.37 | 0.24 | 0.48 | 0.24 | 0.48 | 0.13 | 0.10 | 0.08 | 0.02 | ||
| 1MG-A | 0.67 | 0.36 | 0.24 | 0.48 | 0.24 | 0.48 | 0.13 | 0.10 | 0.08 | 0.02 | ||
| 2MG-EAD | 0.68 | 0.36 | 0.24 | 0.48 | 0.24 | 0.48 | 0.13 | 0.10 | 0.08 | 0.02 | ||
| 2MG-A | 0.74 | 0.35 | 0.27 | 0.47 | 0.25 | 0.48 | 0.13 | 0.10 | 0.08 | 0.02 | ||
| 1MG-EAD | 0.67 | 0.36 | 0.24 | 0.48 | 0.24 | 0.48 | 0.13 | 0.10 | 0.08 | 0.02 | ||
| 2MG-CD | 0.68 | 0.36 | 0.24 | 0.48 | 0.24 | 0.48 | 0.13 | 0.10 | 0.08 | 0.02 | ||
| 2MG-ADI | 0.34 | 0.45 | 0.09 | 0.59 | 0.38 | 0.43 | 0.08 | 0.16 | 0.07 | 0.04 | ||
| 籽仁表面周长KSP | 1MG-AD | 0.00 | 0.95 | 0.00 | 0.99 | 0.00 | 0.83 | 0.00 | 0.90 | 0.03 | 0.89 | |
| 1MG-NCD | 0.00 | 0.96 | 0.00 | 0.84 | 0.14 | 0.54 | 0.00 | 0.81 | 0.03 | 0.85 | ||
| 2MG-AD | 0.00 | 0.95 | 0.00 | 0.99 | 0.00 | 0.83 | 0.00 | 0.90 | 0.03 | 0.89 | ||
| 0MG | 0.03 | 0.67 | 0.00 | 0.86 | 0.79 | 0.35 | 0.02 | 0.39 | 0.05 | 0.20 | ||
| 2MG-EA | 0.04 | 0.67 | 0.00 | 0.86 | 0.91 | 0.33 | 0.02 | 0.39 | 0.05 | 0.20 | ||
| 1MG-A | 0.04 | 0.67 | 0.00 | 0.86 | 0.91 | 0.33 | 0.02 | 0.39 | 0.05 | 0.20 | ||
| 2MG-EAD | 0.04 | 0.66 | 0.00 | 0.86 | 0.91 | 0.33 | 0.02 | 0.39 | 0.05 | 0.20 | ||
| 2MG-A | 0.04 | 0.66 | 0.00 | 0.85 | 0.91 | 0.33 | 0.02 | 0.38 | 0.05 | 0.19 | ||
| 1MG-EAD | 0.04 | 0.66 | 0.00 | 0.86 | 0.91 | 0.33 | 0.02 | 0.39 | 0.05 | 0.20 | ||
| 2MG-CD | 0.04 | 0.66 | 0.00 | 0.86 | 0.91 | 0.33 | 0.02 | 0.39 | 0.05 | 0.20 | ||
| 2MG-ADI | 0.03 | 0.67 | 0.00 | 0.87 | 0.90 | 0.33 | 0.02 | 0.40 | 0.05 | 0.21 | ||
| 平均单仁重 AKW | 1MG-AD | 0.00 | 0.95 | 0.00 | 0.98 | 0.00 | 0.88 | 0.00 | 0.83 | 0.03 | 0.81 | |
| 1MG-NCD | 0.00 | 0.92 | 0.00 | 0.94 | 0.00 | 0.94 | 0.00 | 0.84 | 0.03 | 0.80 | ||
| 0MG | 0.05 | 0.64 | 0.02 | 0.70 | 0.00 | 0.82 | 0.01 | 0.50 | 0.04 | 0.63 | ||
| 2MG-EA | 0.05 | 0.64 | 0.02 | 0.70 | 0.00 | 0.79 | 0.01 | 0.49 | 0.04 | 0.62 | ||
| 1MG-A | 0.05 | 0.64 | 0.02 | 0.70 | 0.00 | 0.79 | 0.01 | 0.49 | 0.04 | 0.62 | ||
| 2MG-EAD | 0.05 | 0.64 | 0.02 | 0.70 | 0.00 | 0.79 | 0.01 | 0.49 | 0.04 | 0.62 | ||
| 2MG-AD | 0.00 | 0.95 | 0.00 | 0.98 | 0.00 | 0.88 | 0.00 | 0.83 | 0.03 | 0.81 | ||
| 2MG-A | 0.05 | 0.63 | 0.02 | 0.69 | 0.00 | 0.79 | 0.02 | 0.49 | 0.04 | 0.61 | ||
| 1MG-EAD | 0.05 | 0.64 | 0.02 | 0.70 | 0.00 | 0.79 | 0.01 | 0.49 | 0.04 | 0.62 | ||
| 2MG-CD | 0.05 | 0.64 | 0.02 | 0.70 | 0.00 | 0.79 | 0.01 | 0.49 | 0.04 | 0.62 | ||
| 2MG-ADI | 0.04 | 0.66 | 0.02 | 0.72 | 0.00 | 0.79 | 0.01 | 0.51 | 0.04 | 0.65 | ||
| YH15×NC94022 | 籽仁长 KL | 0MG | 0.00 | 0.91 | 0.00 | 0.90 | 0.00 | 0.95 | 0.00 | 1.00 | 0.02 | 1.00 |
| 1MG-A | 0.00 | 0.91 | 0.00 | 0.91 | 0.00 | 0.98 | 0.00 | 1.00 | 0.02 | 1.00 | ||
| 2MG-EA | 0.00 | 0.91 | 0.00 | 0.91 | 0.00 | 0.98 | 0.00 | 1.00 | 0.03 | 1.00 | ||
| 2MG-EAD | 0.00 | 0.91 | 0.00 | 0.90 | 0.00 | 0.98 | 0.00 | 1.00 | 0.03 | 1.00 | ||
| 1MG-NCD | 0.00 | 0.97 | 0.00 | 0.99 | 0.00 | 0.93 | 0.00 | 1.00 | 0.02 | 1.00 | ||
| 1MG-AD | 0.00 | 0.97 | 0.00 | 0.99 | 0.00 | 0.95 | 0.00 | 1.00 | 0.02 | 1.00 | ||
| 2MG-A | 0.00 | 0.90 | 0.00 | 0.90 | 0.00 | 0.98 | 0.00 | 1.00 | 0.02 | 1.00 | ||
| 1MG-EAD | 0.00 | 0.91 | 0.00 | 0.90 | 0.00 | 0.98 | 0.00 | 1.00 | 0.03 | 1.00 | ||
| 2MG-CD | 0.00 | 0.91 | 0.00 | 0.90 | 0.00 | 0.98 | 0.00 | 1.00 | 0.03 | 1.00 | ||
| 2MG-AD | 0.00 | 0.97 | 0.00 | 0.99 | 0.00 | 0.95 | 0.00 | 1.00 | 0.02 | 1.00 | ||
| 2MG-ADI | 0.00 | 0.91 | 0.00 | 0.91 | 0.00 | 0.98 | 0.00 | 1.00 | 0.02 | 1.00 | ||
| 籽仁宽 KW | 0MG | 0.04 | 0.66 | 0.17 | 0.52 | 0.58 | 0.38 | 0.02 | 0.45 | 0.06 | 0.37 | |
| 1MG-AD | 0.00 | 0.95 | 0.01 | 0.77 | 0.80 | 0.34 | 0.00 | 0.77 | 0.05 | 0.62 | ||
| 1MG-NCD | 0.00 | 0.92 | 0.01 | 0.73 | 0.94 | 0.32 | 0.01 | 0.73 | 0.05 | 0.56 | ||
| 2MG-EA | 0.04 | 0.66 | 0.16 | 0.53 | 0.49 | 0.40 | 0.02 | 0.45 | 0.06 | 0.37 | ||
| 1MG-A | 0.04 | 0.66 | 0.16 | 0.53 | 0.50 | 0.40 | 0.02 | 0.45 | 0.06 | 0.37 | ||
| 2MG-EAD | 0.04 | 0.66 | 0.16 | 0.53 | 0.49 | 0.40 | 0.02 | 0.45 | 0.06 | 0.37 | ||
| 1MG-EAD | 0.04 | 0.66 | 0.16 | 0.53 | 0.49 | 0.40 | 0.02 | 0.45 | 0.06 | 0.37 | ||
| 2MG-CD | 0.04 | 0.66 | 0.16 | 0.53 | 0.49 | 0.40 | 0.02 | 0.45 | 0.06 | 0.37 | ||
| 2MG-A | 0.04 | 0.65 | 0.18 | 0.52 | 0.50 | 0.40 | 0.02 | 0.45 | 0.06 | 0.36 | ||
| 2MG-AD | 0.00 | 0.95 | 0.01 | 0.77 | 0.80 | 0.34 | 0.00 | 0.77 | 0.05 | 0.62 | ||
| 2MG-ADI | 0.01 | 0.73 | 0.08 | 0.60 | 0.40 | 0.43 | 0.01 | 0.57 | 0.05 | 0.46 | ||
| 籽仁长宽比KL/KW | 0MG | 0.01 | 0.77 | 0.01 | 0.74 | 0.00 | 0.86 | 0.00 | 0.87 | 0.04 | 0.70 | |
| 2MG-EA | 0.01 | 0.77 | 0.01 | 0.75 | 0.00 | 0.89 | 0.00 | 0.87 | 0.04 | 0.70 | ||
| 1MG-A | 0.01 | 0.77 | 0.01 | 0.75 | 0.00 | 0.89 | 0.00 | 0.87 | 0.04 | 0.70 | ||
| 2MG-EAD | 0.01 | 0.77 | 0.01 | 0.75 | 0.00 | 0.89 | 0.00 | 0.87 | 0.04 | 0.70 | ||
| 1MG-AD | 0.00 | 0.91 | 0.00 | 0.88 | 0.00 | 0.85 | 0.00 | 0.95 | 0.03 | 0.93 | ||
| 1MG-NCD | 0.00 | 0.89 | 0.00 | 0.85 | 0.00 | 0.83 | 0.00 | 0.95 | 0.03 | 0.90 | ||
| 1MG-EAD | 0.01 | 0.77 | 0.01 | 0.75 | 0.00 | 0.89 | 0.00 | 0.87 | 0.04 | 0.70 | ||
| 2MG-CD | 0.01 | 0.77 | 0.01 | 0.75 | 0.00 | 0.89 | 0.00 | 0.87 | 0.04 | 0.70 | ||
| 2MG-A | 0.01 | 0.75 | 0.01 | 0.73 | 0.00 | 0.88 | 0.00 | 0.86 | 0.04 | 0.69 | ||
| 2MG-AD | 0.00 | 0.98 | 0.00 | 0.96 | 0.01 | 0.79 | 0.00 | 0.94 | 0.03 | 0.98 | ||
| 2MG-ADI | 0.00 | 0.83 | 0.00 | 0.82 | 0.00 | 0.94 | 0.00 | 0.92 | 0.04 | 0.82 | ||
| 籽仁表面积KSA | 0MG | 0.01 | 0.75 | 0.05 | 0.64 | 0.16 | 0.53 | 0.01 | 0.75 | 0.04 | 0.76 | |
| 1MG-AD | 0.00 | 0.97 | 0.00 | 0.85 | 0.17 | 0.52 | 0.00 | 0.86 | 0.04 | 0.82 | ||
| 2MG-EA | 0.01 | 0.75 | 0.05 | 0.64 | 0.13 | 0.55 | 0.00 | 0.76 | 0.04 | 0.77 | ||
| 1MG-A | 0.01 | 0.75 | 0.05 | 0.64 | 0.13 | 0.55 | 0.00 | 0.76 | 0.04 | 0.77 | ||
| 2MG-EAD | 0.01 | 0.75 | 0.05 | 0.64 | 0.13 | 0.55 | 0.00 | 0.76 | 0.04 | 0.77 | ||
| 1MG-NCD | 0.00 | 0.94 | 0.00 | 0.81 | 0.21 | 0.50 | 0.00 | 0.84 | 0.04 | 0.82 | ||
| 2MG-A | 0.01 | 0.74 | 0.05 | 0.63 | 0.13 | 0.55 | 0.01 | 0.75 | 0.04 | 0.76 | ||
| 1MG-EAD | 0.01 | 0.75 | 0.05 | 0.64 | 0.13 | 0.55 | 0.00 | 0.76 | 0.04 | 0.77 | ||
| 2MG-CD | 0.01 | 0.75 | 0.05 | 0.64 | 0.13 | 0.55 | 0.00 | 0.76 | 0.04 | 0.77 | ||
| 2MG-AD | 0.00 | 0.97 | 0.00 | 0.85 | 0.17 | 0.52 | 0.00 | 0.86 | 0.04 | 0.82 | ||
| 2MG-ADI | 0.01 | 0.78 | 0.03 | 0.68 | 0.11 | 0.56 | 0.00 | 0.80 | 0.04 | 0.81 | ||
| 籽仁表面周长KSP | 0MG | 0.00 | 0.91 | 0.00 | 0.92 | 0.00 | 0.96 | 0.00 | 0.97 | 0.03 | 0.99 | |
| 2MG-EAD | 0.00 | 1.00 | 0.00 | 0.99 | 0.00 | 0.95 | 0.00 | 0.98 | 0.03 | 0.97 | ||
| 1MG-A | 0.00 | 0.91 | 0.00 | 0.91 | 0.00 | 0.99 | 0.00 | 0.97 | 0.03 | 0.99 | ||
| 2MG-EA | 0.00 | 0.91 | 0.00 | 0.91 | 0.00 | 0.99 | 0.00 | 0.97 | 0.03 | 0.99 | ||
| 1MG-AD | 0.00 | 0.98 | 0.00 | 0.89 | 0.04 | 0.65 | 0.00 | 0.93 | 0.04 | 0.89 | ||
| 2MG-CD | 0.00 | 0.91 | 0.00 | 0.92 | 0.00 | 0.99 | 0.00 | 0.97 | 0.03 | 0.99 | ||
| 1MG-EAD | 0.00 | 0.91 | 0.00 | 0.92 | 0.00 | 0.99 | 0.00 | 0.97 | 0.03 | 0.99 | ||
| 2MG-A | 0.00 | 0.91 | 0.00 | 0.91 | 0.00 | 0.99 | 0.00 | 0.97 | 0.03 | 0.99 | ||
| 1MG-NCD | 0.00 | 0.90 | 0.00 | 0.91 | 0.00 | 0.99 | 0.00 | 0.97 | 0.03 | 0.99 | ||
| 2MG-AD | 0.00 | 0.98 | 0.00 | 0.89 | 0.04 | 0.65 | 0.00 | 0.93 | 0.04 | 0.89 | ||
| 2MG-ADI | 0.00 | 0.91 | 0.00 | 0.92 | 0.00 | 0.99 | 0.00 | 0.97 | 0.03 | 0.99 | ||
| 平均单仁重 AKW | 1MG-AD | 0.00 | 0.95 | 0.00 | 0.99 | 0.01 | 0.76 | 0.00 | 0.96 | 0.03 | 0.97 | |
| 0MG | 0.00 | 0.89 | 0.00 | 0.92 | 0.84 | 0.34 | 0.00 | 0.76 | 0.04 | 0.80 | ||
| 1MG-A | 0.00 | 0.89 | 0.00 | 0.91 | 0.98 | 0.32 | 0.01 | 0.74 | 0.04 | 0.79 | ||
| 2MG-EA | 0.00 | 0.89 | 0.00 | 0.91 | 0.99 | 0.32 | 0.01 | 0.74 | 0.04 | 0.79 | ||
| 2MG-EAD | 0.00 | 0.89 | 0.00 | 0.91 | 0.99 | 0.32 | 0.01 | 0.74 | 0.04 | 0.79 | ||
| 1MG-NCD | 0.00 | 0.95 | 0.01 | 0.78 | 0.54 | 0.39 | 0.00 | 0.80 | 0.04 | 0.78 | ||
| 2MG-AD | 0.00 | 0.95 | 0.00 | 0.99 | 0.01 | 0.76 | 0.00 | 0.96 | 0.03 | 0.97 | ||
| 2MG-A | 0.00 | 0.88 | 0.00 | 0.92 | 0.97 | 0.32 | 0.01 | 0.74 | 0.04 | 0.79 | ||
| 1MG-EAD | 0.00 | 0.89 | 0.00 | 0.91 | 0.99 | 0.32 | 0.01 | 0.74 | 0.04 | 0.79 | ||
| 2MG-CD | 0.00 | 0.89 | 0.00 | 0.91 | 0.99 | 0.32 | 0.01 | 0.74 | 0.04 | 0.79 | ||
| 2MG-ADI | 0.00 | 0.90 | 0.00 | 0.90 | 0.98 | 0.32 | 0.01 | 0.75 | 0.04 | 0.80 |
新窗口打开|下载CSV
2.4 花生籽仁表型性状的遗传参数的估计
基于AIC值对通过适合性检验的模型进行升序排序,选择前5个模型作为备选模型(表2),并对具有统计学意义备选遗传模型进行遗传参数估计,基于AIC最小值或较小值选择遗传力较大的模型作为最佳遗传模型,巢式杂交群体中不同组合的不同籽仁性状的最适遗传模型存在差异,籽仁长(KL)在巢式杂交群体中有3个组合符合1对主基因负向完全显性模型(1MG-NCD)模型,分别是组合YZ9102×YH15、YH15×FHS、YH15×NC94022,主基因遗传率分别为31.46%、40.85%和22.95%,组合ZH6×YH15符合2对主基因加性-等加性显性(2MG-EAD)模型,主基因遗传率为68.33%,组合YH15×YY20符合1对主基因加性-显性模型(1MG-AD)模型,主基因遗传率为58.54%;籽仁宽(KW)在巢式杂交群体中有4个组合符合1MG-AD模型,分别是组合YZ9102×YH15、YH15×YY20、YH15×FHS、YH15×NC94022,主基因遗传率分别为53.68%、77.06%、73.91%和31.81%,组合ZH6×YH15符合1MG-NCD模型,主基因遗传率分别为28.26%;籽仁长宽比(KL/KW)在巢式杂交群体中有4个组合符合1MG-AD模型,分别是组合YZ9102×YH15、ZH6×YH15、YH15×FHS和YH15×NC94022,主基因遗传率分别为43.64%、52.89%、28.02%和22.99%,组合YH15×YY20符合2MG-EAD模型,主基因遗传率为3.80%;籽仁表面积(KSA)和籽仁表面周长(KSP)在巢式杂交群体中5个组合均符合1MG-AD模型,主基因遗传率分别为46.81%、43.61%、62.13%、62.38%、27.85%和50.90%、59.09%、63.12%、62.38%、58.14%;单仁重(AKW)在巢式杂交群体中有4个组合符合1MG-AD模型,分别是组合YZ9102×YH15、YH15×YY20、YH15×FHS和YH15×NC94022,主基因遗传率分别为57.08%、55.03%、32.94%和62.02%,组合ZH6×YH15符合1MG-NCD模型,主基因遗传率分别为44.40%(表3)。Table 2
表2
表2籽仁表型性状遗传模式分析
Table 2
| 组合 Combination | 籽仁长KL | 籽仁宽KW | 籽仁长宽比KL/KW | 籽仁表面积KSA | 籽仁表面周长KSP | 单仁重 SKW | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 模型 Models | 最大似 然值 Maximum likelihood value | AIC值 AIC value | 模型 Models | 最大似 然值 Maximum likelihood value | AIC值 AIC value | 模型 Models | 最大似 然值 Maximum likelihood value | AIC值 AIC value | 模型 Models | 最大似 然值 Maximum likelihood value | AIC值 AIC value | 模型 Models | 最大似 然值 Maximum likelihood value | AIC值 AIC value | 模型 Models | 最大似 然值 Maximum likelihood value | AIC值 AIC value | |
| YZ9102× YH15 | 0MG | -959.99 | 1923.97 | 1MG-AD | -656.46 | 1320.93 | 0MG | 289.99 | -575.98 | 1MG-AD | -2163.74 | 4335.49 | 1MG-AD | -1408.02 | 2824.04 | 1MG-AD | -1879.32 | 3766.63 |
| 1MG-NCD | -958.76 | 1925.52 | 0MG | -658.57 | 1321.14 | 1MG-AD | 291.02 | -574.03 | 1MG-NCD | -2163.98 | 4335.95 | 1MG-NCD | -1408.58 | 2825.15 | 2MG-EAD | -1880.98 | 3767.95 | |
| 1MG-AD | -958.85 | 1925.70 | 1MG-NCD | -657.14 | 1322.29 | 1MG-A | 289.99 | -573.98 | 2MG-AD | -2163.74 | 4339.48 | 0MG | -1411.09 | 2826.17 | 1MG-EAD | -1880.14 | 3768.29 | |
| 1MG-A | -959.98 | 1925.97 | 1MG-A | -658.57 | 1323.13 | 2MG-EA | 289.99 | -573.98 | 0MG | -2172.86 | 4349.72 | 2MG-AD | -1408.02 | 2828.04 | 2MG-CD | -1880.15 | 3768.29 | |
| 2MG-EA | -959.98 | 1925.97 | 2MG-EA | -658.57 | 1323.13 | 2MG-EAD | 289.98 | -573.96 | 2MG-EA | -2172.85 | 4351.71 | 1MG-A | -1411.09 | 2828.17 | 0MG | -1882.81 | 3769.62 | |
| ZH6× YH15 | 2MG-EAD | -696.02 | 1398.03 | 0MG | -420.43 | 844.85 | 0MG | 244.26 | -484.53 | 1MG-AD | -1732.76 | 3473.52 | 1MG-AD | -1081.86 | 2171.72 | 1MG-NCD | -1562.88 | 3133.77 |
| 1MG-AD | -696.24 | 1400.48 | 1MG-A | -420.42 | 846.85 | 1MG-AD | 245.38 | -482.76 | 0MG | -1734.78 | 3473.55 | 0MG | -1084.28 | 2172.56 | 1MG-AD | -1562.99 | 3133.98 | |
| 2MG-AD | -696.24 | 1404.48 | 2MG-EA | -420.42 | 846.85 | 2MG-EAD | 244.29 | -482.57 | 1MG-NCD | -1732.85 | 3473.69 | 1MG-A | -1084.27 | 2174.55 | 0MG | -1566.34 | 3136.69 | |
| 2MG-CD | -699.82 | 1407.63 | 2MG-EAD | -420.43 | 846.87 | 2MG-EA | 244.27 | -482.53 | 2MG-EA | -1734.78 | 3475.55 | 2MG-EA | -1084.28 | 2174.55 | 2MG-AD | -1562.99 | 3137.98 | |
| 1MG-EAD | -699.82 | 1407.63 | 1MG-NCD | -419.77 | 847.55 | 1MG-A | 244.27 | -482.53 | 1MG-A | -1734.78 | 3475.55 | 2MG-EAD | -1084.29 | 2174.57 | 2MG-EA | -1566.34 | 3138.68 | |
| YH15× YY20 | 0MG | -584.03 | 1172.05 | 1MG-AD | -403.72 | 815.44 | 0MG | 159.67 | -315.35 | 1MG-AD | -1257.01 | 2522.02 | 1MG-AD | -820.92 | 1649.85 | 0MG | -1173.89 | 2351.78 |
| 1MG-AD | -582.14 | 1172.28 | 2MG-AD | -403.72 | 819.44 | 1MG-A | 159.68 | -313.35 | 1MG-NCD | -1257.41 | 2522.81 | 1MG-NCD | -822.63 | 1653.27 | 1MG-AD | -1172.70 | 2353.40 | |
| 1MG-A | -584.02 | 1174.04 | 1MG-NCD | -409.22 | 826.44 | 2MG-EA | 159.68 | -313.35 | 2MG-AD | -1257.01 | 2526.02 | 0MG | -824.82 | 1653.65 | 1MG-A | -1173.89 | 2353.78 | |
| 2MG-EA | -584.02 | 1174.04 | 0MG | -412.19 | 828.39 | 2MG-EAD | 159.67 | -313.33 | 0MG | -1261.74 | 2527.48 | 2MG-AD | -820.92 | 1653.84 | 2MG-EA | -1173.89 | 2353.78 | |
| 2MG-EAD | -584.03 | 1174.06 | 1MG-A | -412.19 | 830.37 | 1MG-NCD | 159.68 | -311.37 | 2MG-EA | -1261.74 | 2529.47 | 1MG-A | -824.82 | 1655.63 | 2MG-EAD | -1173.90 | 2353.80 | |
| YH15× FHS | 1MG-NCD | -840.29 | 1688.58 | 1MG-AD | -597.78 | 1203.56 | 0MG | 293.68 | -583.36 | 1MG-AD | -1831.51 | 3671.02 | 1MG-AD | -1215.35 | 2438.70 | 1MG-AD | -1657.61 | 3323.22 |
| 1MG-AD | -840.41 | 1688.81 | 2MG-AD | -597.78 | 1207.56 | 1MG-AD | 295.35 | -582.71 | 1MG-NCD | -1831.90 | 3671.81 | 1MG-NCD | -1216.12 | 2440.24 | 1MG-NCD | -1657.70 | 3323.41 | |
| 0MG | -843.13 | 1690.27 | 1MG-NCD | -602.16 | 1212.32 | 1MG-NCD | 295.11 | -582.22 | 2MG-AD | -1831.51 | 3675.01 | 2MG-AD | -1215.35 | 2442.70 | 0MG | -1660.06 | 3324.12 | |
| 2MG-EA | -843.13 | 1692.26 | 0MG | -605.49 | 1214.98 | 2MG-EA | 293.68 | -581.36 | 0MG | -1842.40 | 3688.81 | 0MG | -1220.23 | 2444.45 | 2MG-EA | -1660.06 | 3326.11 | |
| 1MG-A | -843.13 | 1692.26 | 1MG-A | -605.49 | 1216.97 | 1MG-A | 293.68 | -581.36 | 2MG-EA | -1842.40 | 3690.79 | 2MG-EA | -1220.22 | 2446.44 | 1MG-A | -1660.06 | 3326.12 | |
| YH15× NC94022 | 0MG | -478.24 | 960.48 | 0MG | -286.25 | 576.49 | 0MG | 119.22 | -234.45 | 0MG | -1097.35 | 2198.69 | 0MG | -680.84 | 1365.68 | 1MG-AD | -1031.72 | 2071.45 |
| 1MG-A | -478.24 | 962.48 | 1MG-AD | -284.66 | 577.31 | 2MG-EA | 119.23 | -232.45 | 1MG-AD | -1096.25 | 2200.49 | 2MG-EAD | -680.67 | 1367.34 | 0MG | -1033.78 | 2071.55 | |
| 2MG-EA | -478.24 | 962.48 | 1MG-NCD | -284.89 | 577.79 | 1MG-A | 119.22 | -232.44 | 2MG-EA | -1097.34 | 2200.68 | 1MG-A | -680.84 | 1367.67 | 1MG-A | -1033.77 | 2073.54 | |
| 2MG-EAD | -478.25 | 962.50 | 2MG-EA | -286.24 | 578.49 | 2MG-EAD | 119.22 | -232.44 | 1MG-A | -1097.35 | 2200.69 | 2MG-EA | -680.84 | 1367.68 | 2MG-EA | -1033.77 | 2073.54 | |
| 1MG-NCD | -477.98 | 963.95 | 1MG-A | -286.25 | 578.50 | 1MG-AD | 120.02 | -232.05 | 2MG-EAD | -1097.35 | 2200.70 | 1MG-AD | -679.96 | 1367.92 | 2MG-EAD | -1033.78 | 2073.56 | |
新窗口打开|下载CSV
Table 3
表3
表3巢式杂交群体籽仁性状的二阶参数估计值
Table 3
| 性状 Trait | YZ9102×YH15 | ZH6×YH15 | YH15×YY20 | YH15×FHS | YH15×NC94022 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 模型 Models | 二阶参数 Second order genetic parameters | 模型 Models | 二阶参数 Second order genetic parameters | 模型 Models | 二阶参数 Second order genetic parameters | 模型 Models | 二阶参数 Second order genetic parameters | 模型 Models | 二阶参数 Second order genetic parameters | ||||||
| σ2mg | h2pg (%) | σ2mg | h2pg (%) | σ2mg | h2pg (%) | σ2mg | h2pg (%) | σ2mg | h2pg (%) | ||||||
| 籽仁长 KL | 0MG | 2MG-EAD | 1.61 | 68.33 | 0MG | 1MG-NCD | 1.75 | 40.85 | 0MG | ||||||
| 1MG-NCD | 1.08 | 31.46 | 1MG-AD | 1.64 | 69.52 | 1MG-AD | 2.04 | 58.54 | 1MG-AD | 1.55 | 36.11 | 1MG-A | 0.07 | 3.23 | |
| 1MG-AD | 0.93 | 27.09 | 2MG-AD | 1.64 | 69.59 | 1MG-A | 0.11 | 3.01 | 0MG | 2MG-EA | 0.08 | 3.60 | |||
| 1MG-A | 0.06 | 1.85 | 2MG-CD | 0.89 | 37.49 | 2MG-EA | 0.12 | 3.37 | 2MG-EA | 0.10 | 2.44 | 2MG-EAD | 0.06 | 2.72 | |
| 2MG-EA | 0.07 | 2.06 | 1MG-EAD | 0.88 | 37.25 | 2MG-EAD | 0.07 | 2.12 | 1MG-A | 0.09 | 2.21 | 1MG-NCD | 0.51 | 22.95 | |
| 籽仁宽 KW | 1MG-AD | 0.51 | 53.68 | 0MG | 1MG-AD | 0.81 | 77.06 | 1MG-AD | 0.95 | 73.91 | 0MG | ||||
| 0MG | 1MG-A | 0.01 | 2.21 | 2MG-AD | 0.81 | 77.08 | 2MG-AD | 0.95 | 73.96 | 1MG-AD | 0.16 | 31.81 | |||
| 1MG-NCD | 0.33 | 34.01 | 2MG-EA | 0.01 | 2.47 | 1MG-NCD | 0.50 | 48.00 | 1MG-NCD | 0.57 | 44.57 | 1MG-NCD | 0.17 | 34.06 | |
| 1MG-A | 0.02 | 1.85 | 2MG-EAD | 0.01 | 2.07 | 0MG | 0MG | 2MG-EA | 0.02 | 3.38 | |||||
| 2MG-EA | 0.02 | 2.07 | 1MG-NCD | 0.14 | 28.26 | 1MG-A | 0.03 | 3.02 | 1MG-A | 0.03 | 2.22 | 1MG-A | 0.02 | 3.16 | |
| 籽仁长 宽比 KL/KW | 2MG-ADI | 0.00 | 2.92 | 0MG | 0MG | 0MG | 0MG | ||||||||
| 0MG | 1MG-AD | 0.01 | 52.89 | 1MG-A | 0.00 | 3.00 | 1MG-AD | 0.00 | 28.02 | 2MG-EA | 0.00 | 3.40 | |||
| 1MG-AD | 0.01 | 43.64 | 2MG-EAD | 0.00 | 2.29 | 2MG-EA | 0.00 | 3.37 | 1MG-NCD | 0.00 | 30.07 | 1MG-A | 0.00 | 3.18 | |
| 1MG-A | 0.00 | 1.85 | 2MG-EA | 0.00 | 27.08 | 2MG-EAD | 0.00 | 3.80 | 2MG-EA | 0.00 | 2.38 | 2MG-EAD | 0.00 | 0.99 | |
| 2MG-EA | 0.00 | 2.08 | 1MG-A | 0.00 | 1.38 | 1MG-NCD | 0.00 | 2.73 | 1MG-A | 0.00 | 2.20 | 1MG-AD | 0.01 | 22.99 | |
| 籽仁表 面积 KSA | 1MG-AD | 279.12 | 46.81 | 1MG-AD | 178.68 | 43.61 | 1MG-AD | 377.44 | 62.13 | 1MG-AD | 377.37 | 50.68 | 0MG | ||
| 1MG-NCD | 305.43 | 51.22 | 0MG | 1MG-NCD | 307.74 | 50.66 | 1MG-NCD | 410.19 | 55.09 | 1MG-AD | 66.74 | 27.85 | |||
| 2MG-AD | 279.92 | 46.94 | 1MG-NCD | 153.10 | 37.36 | 2MG-AD | 379.03 | 62.39 | 2MG-AD | 378.30 | 50.80 | 2MG-EA | 8.31 | 3.47 | |
| 0MG | 2MG-EA | 10.04 | 2.45 | 0MG | 1MG-A | 7.71 | 3.22 | ||||||||
| 2MG-EA | 12.03 | 2.02 | 1MG-A | 9.08 | 2.21 | 2MG-EA | 21.28 | 3.50 | 2MG-EAD | 2.69 | 1.12 | ||||
| 籽仁表 面周长 KSP | 1MG-AD | 11.80 | 50.90 | 1MG-AD | 9.02 | 59.09 | 1MG-AD | 15.62 | 63.12 | 1MG-AD | 19.11 | 62.38 | 0MG | ||
| 1MG-NCD | 9.24 | 39.85 | 0MG | 1MG-NCD | 10.84 | 43.81 | 1MG-NCD | 14.10 | 46.02 | 2MG-EAD | 2.90 | 26.26 | |||
| 0MG | 1MG-A | 0.34 | 2.23 | 0MG | 2MG-AD | 19.14 | 62.46 | 1MG-A | 0.36 | 3.28 | |||||
| 2MG-AD | 11.82 | 50.98 | 2MG-EA | 0.39 | 2.52 | 2MG-AD | 15.64 | 63.23 | 0MG | 2MG-EA | 0.42 | 3.77 | |||
| 1MG-A | 0.43 | 1.85 | 2MG-EAD | 0.43 | 2.83 | 1MG-A | 0.78 | 3.15 | 2MG-EA | 0.76 | 2.48 | 1MG-AD | 6.43 | 58.14 | |
| 单仁重 AKW | 1MG-AD | 106.38 | 57.08 | 1MG-NCD | 78.63 | 44.40 | 0MG | 1MG-AD | 94.20 | 32.94 | 1MG-AD | 89.15 | 62.02 | ||
| 2MG-EAD | 86.42 | 46.37 | 1MG-AD | 69.60 | 39.30 | 1MG-AD | 129.58 | 55.03 | 1MG-NCD | 104.52 | 36.54 | 0MG | |||
| 1MG-EAD | 76.08 | 40.82 | 0MG | 1MG-A | 7.15 | 3.03 | 0MG | 1MG-A | 4.65 | 3.24 | |||||
| 2MG-CD | 76.29 | 40.94 | 2MG-AD | 69.89 | 39.47 | 2MG-EA | 8.02 | 3.40 | 2MG-EA | 6.94 | 2.43 | 2MG-EA | 5.21 | 3.63 | |
| 0MG | 2MG-EA | 4.31 | 2.44 | 2MG-EAD | 6.30 | 2.68 | 1MG-A | 6.34 | 2.22 | 2MG-EAD | 3.20 | 2.22 | |||
新窗口打开|下载CSV
3 讨论
3.1 花生籽仁性状的遗传模型
花生籽仁性状包括多个复杂的数量遗传性状,如籽仁长、宽、长宽比、表面积、表面周长、百仁重等,由多个遗传效应不同的基因控制。殷冬梅等[26]曾利用双列杂交F2分析了花生分离世代百仁重的遗传效应,得出百仁重性状的遗传具有加性和显性效应的结论。张新友[27]、刘华[28]针对花生RIL群体研究结果均表明花生百仁重受2对主基因加多基因控制。而本研究中,在巢式杂交群体中有4个群体的单仁重的遗传模型符合1对主基因加性-显性模型,1个群体的单仁重遗传模型符合1对主基因负向完全显性模型,与前人研究存在差异。张胜忠等[22]分析种植在不同环境的花生RIL群体的种子长宽比表型数据,结果表明,花生种子长宽比的遗传受2对主基因控制。本研究结果显示籽仁长宽比在巢式杂交群体中有4个组合符合1对主基因加性-显性模型,1个组合符合2对主基因加性-等加性显性模型,与前人研究有一定差异。本研究结果籽仁长和宽在巢式杂交群体中分别有3个组合符合1对主基因负向完全显性模型、1个组合符合1对主基因加性-显性模型、1个组合符合2对主基因加性-等加性显性模型和4个组合符合1对主基因加性-显性模型、1个组合符合1对主基因负向完全显性模型。结果与张晓霞等[23]利用正反交的2个F2群体获得籽仁长符合3对主基因控制的加性-上位性遗传模型、宽符合1对具有加性效应的主基因+多基因混合遗传模型或2对具有显性上位效应的主基因+多基因混合遗传模型的结果,存在一定差异。本研究所采用的亲本种类、亲本数量、群体类型、种植环境等均与前人研究存在较明显的差异,结果中符合1对主基因加性-显性模型的性状及组合最多,其中籽仁表面积、表面周长在所有组合中均符合1MG-AM模型,籽仁宽、籽仁长宽比、单仁重均有4个组合符合1MG-AD模型,籽仁长有1个组合符合1MG-AD模型;其次是1对主基因负向完全显性模型,籽仁长有3个组合符合1MG-NCD模型,籽仁宽、单仁重各有1个组合符合1MG-NCD模型;2对主基因加性-等加性显性模型也在少量性状中有所表现,籽仁长、长宽比均有1个组合符合2MG-EAD模型,主基因遗传力3.80%—77.06%。不同组合的加性、显性和上位性基因效应值各不相同,遗传背景及种植环境对确定数量性状的遗传模型分析具有一定影响。
3.2 自然群体及双亲群体的局限性
全基因组关联研究(genome-wide association study,GWAS)、数量性状基因定位(quantitative trait locusmapping,QTL定位)等多种定位手段被应用于自然群体或各种双亲构建的不同连锁作图群体,用来定位如稻粒直径、粒重[29],小麦粒重、粒长[30],玉米粒重[31]等复杂的数量性状。CHEN等[32]针对花生的2个F2:3群体分别对种子长、宽进行QTL分析,鉴定了新的调控籽仁长、籽仁宽的QTL数量分别为5个和1个及0个和2个。宋延滨[33]和曾新颖[11]利用花生RIL群体进行花生籽仁形态相关性状QTL定位,二者结果存在差异,籽仁长、宽、长宽比、百仁重在多年重复检测到的QTL数量分别有7、4、9和9个及18、16、18和14个。PANDEY等[34]对来自不同国家共48个半干旱区域的300个基因型进行全基因组关联分析,发现调控花生籽仁的长、宽、重的性状关联标记(markers- trait association,MATs)分别是9、3和5个。由于环境、群体亲本等诸多因素导致利用不同的RIL群体定位籽仁相关性状的结果不一致,且采用GWAS对自然群体的数量性状定位结果与双亲群体的QTL定位结果也存在较大差异。对于花生籽仁性状的调控位点挖掘,目前,利用单一RIL群体的QTL定位区间较大;而GWAS难以定位到稀有变异,且由于自然群体的结构和分层现象,影响GWAS结果的准确性。因此,有必要构建新的数量性状定位群体。
3.3 巢式群体挖掘数量性状位点的潜力
NAM(nested-association mapping)群体,是由一个共同亲本与多个其他不同亲本配合而成的含有多个RIL组合的群体,具有丰富的遗传变异类型,同时其具有更多的基因重组机会,弥补了自然群体和双亲群体的缺陷,提高了等位基因的多样性和QTL定位能力,现已被运用到小麦的开花期[35]、玉米籽仁大小及百仁重[36]、大豆种子的耐渍性[37]等各种作物的不同数量性状的QTL定位中。NAM群体在油料作物的产量相关性状的QTL定位方面已得到初步应用并展示出良好的应用前景。XAVIER等[38]分析了大豆NAM群体的产量稳定性,发现第4、6、9、13、15、18染色体上存在环境互作QTL,其中第18染色体上存在与大豆产量性状显著关联的QTL。SONG等[39]利用油菜NAM群体检测到控制角果长、粒重的SNP位于目标基因BnaA9.CYP78A9非编码区。GANGURDE等[40]采用花生的NAM群体进行了百果重和百仁重的QTL定位,结果表明,不同的NAM群体其百果重、百仁重的QTL数量不同,分别是12和8个以及13和11个,主要分布于A05、A06、B05和B06染色体上。本研究采用的材料是以豫花15为共同亲本,远杂9102、中花6号、粤油20、伏花生和NC94022为基础亲本,配成包括远杂9102×豫花15、中花6号×豫花15、豫花15×粤油20、豫花15×伏花生和豫花15×NC94022共5个组合的巢式杂交群体。该群体的6个亲本包含了普通型、珍珠豆型、中间型等生产上广泛应用的品种类型。且将花生的籽仁形态细分为6个不同的性状,该6个性状在6个亲本中均表现较为明显的差异,在F2:3中分离明显,遗传变异类型丰富,为进一步进行QTL定位解析花生籽仁性状遗传机制提供了材料基础。
4 结论
在巢式杂交群体中,籽仁表型性状的遗传表现出不同的模型,多数组合的籽仁宽、长宽比、表面积、表面周长、单仁重符合1MG-AD模型,籽仁长有3个组合符合1MG-NCD模型,个别组合的个别性状符合2MG-EAD模型,各主基因遗传力、遗传效应均存在差异,表明该巢式杂交群体中不同组合的籽仁性状的调控基因有差异。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1038/ng.3517URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1186/s12864-018-5288-3URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00122-017-3000-7URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00122-017-2915-3URL [本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.1007/s11703-007-0001-3URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.scienta.2016.03.018URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.scienta.2015.04.004URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1016/j.molp.2018.03.005URL [本文引用: 1]
DOI:10.3389/fpls.2018.01045URL [本文引用: 1]
DOI:10.1007/s00438-019-01631-2URL [本文引用: 1]
DOI:10.1186/s12863-016-0337-xURL [本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1371/journal.pone.0105228URL [本文引用: 1]
DOI:10.1111/pbi.2019.17.issue-7URL [本文引用: 1]
DOI:10.1007/s00122-019-03426-wURL [本文引用: 1]
DOI:10.1016/j.cj.2020.06.008URL [本文引用: 1]
[本文引用: 1]
DOI:10.1038/s41477-019-0577-7URL [本文引用: 1]
DOI:10.1111/pbi.v18.6URL [本文引用: 1]
