 ,1, 韩占品1, 贺丽霞
,1, 韩占品1, 贺丽霞 ,2, 杨亚苓1, 尤书燕2, 邓琳2, 王春国2
,2, 杨亚苓1, 尤书燕2, 邓琳2, 王春国2Cloning and Functional Analysis of BraERF023a Under Salt and Drought Stresses in Cauliflower (Brassica oleracea L. var. botrytis)
LI Hui ,1, HAN ZhanPin1, HE LiXia
,1, HAN ZhanPin1, HE LiXia ,2, YANG YaLing1, YOU ShuYan2, DENG Lin2, WANG ChunGuo2
,2, YANG YaLing1, YOU ShuYan2, DENG Lin2, WANG ChunGuo2通讯作者:
责任编辑: 赵伶俐
收稿日期:2020-04-12接受日期:2020-06-29网络出版日期:2021-01-01
| 基金资助: |
Received:2020-04-12Accepted:2020-06-29Online:2021-01-01
作者简介 About authors
李慧,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2728KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
李慧, 韩占品, 贺丽霞, 杨亚苓, 尤书燕, 邓琳, 王春国. 花椰菜BraERF023a的克隆及在响应盐和干旱胁迫中的功能[J]. 中国农业科学, 2021, 54(1): 152-163 doi:10.3864/j.issn.0578-1752.2021.01.011
LI Hui, HAN ZhanPin, HE LiXia, YANG YaLing, YOU ShuYan, DENG Lin, WANG ChunGuo.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】ERF转录因子隶属于AP2/ERF转录因子超家族,在植物响应逆境胁迫中发挥重要作用。花椰菜是重要的蔬菜作物,土壤盐渍及干旱是造成花椰菜种植减产的主要环境灾害。但对花椰菜响应盐及干旱胁迫的分子基础认识十分有限,这已成为采用分子育种等手段创制花椰菜抗逆新种质及培育新品种的重要瓶颈。因此,克隆ERF等逆境应答相关转录因子或基因并阐释其功能,对开展抗逆花椰菜分子育种及基因靶向育种具有重要意义。【前人研究进展】植物在生长发育过程中需要不断应对持续变化的不利环境,如动物啃食、病原体侵染等生物胁迫和干旱、高温、土壤盐渍等非生物胁迫。这其中干旱、盐害及温度胁迫是影响植物地理分布,造成农作物减产甚至绝收的主要环境灾害[1]。因此,阐释植物响应逆境胁迫的分子基础及调控机制,进而在作物中开展抗逆分子育种一直是植物学研究的前沿及热点领域。已有研究表明,WRKY、MYB、bZIP、NAC和AP2/ERF等转录因子家族成员在植物生长发育及响应各类逆境胁迫中发挥重要作用[2,3],其中AP2/ERF转录因子家族为植物所特有。该家族成员具有参与DNA结合的AP2保守结构域,该结构域约由60个氨基酸残基按照3个β折叠和1个α螺旋的方式构成一个典型三维结构[4,5,6]。根据AP2结构域的数量和识别序列的不同,AP2/ERF转录因子家族可以分为AP2、ERF、DREB、RAV和Soloist 5个亚家族[7,8]。其中ERF亚家族只含有1个AP2结构域,被证实主要通过识别并特异性结合各类胁迫响应相关基因顺式作用元件,而在植物抵抗各种逆境胁迫过程中发挥重要的调控作用[6,9]。水稻中ERF亚家族成员SNORKEL1(SK1)和SNORKEL2(SK2)可以促进水稻节间伸长,从而使植株可以在深水环境中存活[10]。在拟南芥中过表达菊花CmERF053的研究表明,CmERF053可能参与细胞分裂相关的侧枝形成调控途径,同时在植物抵御干旱胁迫中发挥作用[11]。大豆中GmERF6能够被干旱诱导表达,将GmERF6在拟南芥中过表达可显著提高转化株的抗旱性[12]。在拟南芥中过表达甜薯IbRAP2-12和芜菁BrERF4同样也可提高转化株对盐和干旱胁迫的耐受性[13,14]。PARK等[15]研究发现拟南芥AtERF71/HRE2功能缺失可使突变株表现为盐敏感,而将AtERF71/HRE2转入突变体中,植株表现出耐盐性,可成功抵抗渗透胁迫,且提高转化株对低氧胁迫的耐受能力。此外,研究证实,辣椒的CaERFLP1[16]和CaPF1[17],番茄的TERF1[18]和JERF1[19]以及小麦中TaERF1[20]的过表达均可显著提高转化株对盐及低温等逆境的适应能力。综上所述,ERF转录因子已被证实在植物响应逆境胁迫,特别是非生物逆境胁迫中发挥重要作用。【本研究切入点】虽然在不同植物中已有多个ERF转录因子被克隆并证实其在逆境应答中发挥重要作用,但对ERF转录因子在花椰菜逆境应答中的功能及调控机制仍知之甚少。笔者实验室前期对花椰菜中的AP2/ERF家族成员采用高通量转录组测序方法进行了分析,从中筛选、鉴定到146个ERF转录因子,并证实包括BraERF025a等在内的多个ERF亚家族成员在盐及干旱胁迫前后呈现显著差异表达特征[21]。进一步的分子进化树分析结果表明BraERF023a是与BraERF025a同属于一个进化分支的ERF亚家族成员,二者具有较近的亲缘关系[21]。但BraERF023a是否也响应盐及干旱胁迫,且其是否在花椰菜抵抗盐及干旱胁迫中发挥作用仍未知。【拟解决的关键问题】对花椰菜中的BraERF023a进行克隆,揭示其序列特征,以及在盐及干旱胁迫条件下的表达特征,并通过过表达分析阐释BraERF023a在盐及干旱胁迫应答中的功能。1 材料与方法
试验于2017—2019年在南开大学生命科学学院植物细胞及分子遗传学实验室进行。1.1 试验材料
供试花椰菜‘津品70’种子由天津市科润蔬菜研究所孙德岭研究员惠赠。拟南芥为哥伦比亚生态型。克隆载体pEASY-T1购自北京全式金生物技术有限公司。植物表达载体pCAMBIA3301、大肠杆菌DH5α、农杆菌菌株LBA4404由笔者实验室保存。1.2 DNA提取
野生型和过表达BraERF023a转基因拟南芥、花椰菜幼苗新鲜组织,于研钵中加入液氮快速研磨成粉末状,采用经典CTAB法提取DNA。ddH2O于4℃过夜溶解DNA沉淀。在溶解液中RNase A于37℃金属浴中反应2 h除去RNA。NanoDrop?ND-1000检测DNA浓度和质量。高质量的DNA于-20℃保存备用。1.3 RNA提取及反转录
参照植物总RNA提取试剂盒的操作说明提取花椰菜及拟南芥叶片总RNA。NanoDrop?ND-1000检测提取RNA的浓度,2%琼脂糖凝胶电泳检测RNA完整性。采用M-MLV反转录酶合成cDNA第一链(具体操作参照Promega公司的M-MLV反转录说明书进行)。1.4 花椰菜不同逆境胁迫处理
待花椰菜幼苗长至四片真叶时,将幼苗从营养土中取出,自来水缓缓冲洗根部,注意不要伤害到根系,随后将幼苗分为两组,每组至少16个单株。将其中一组幼苗根部直接浸入含有200 mmol?L-1 NaCl的水溶液进行盐胁迫处理;另外一组幼苗根部浸入含20% PEG6000的水溶液中进行模拟干旱处理。分别在0、4、8、12和24 h时摘取叶片并进行总RNA提取。提取的RNA反转录合成cDNA第一链,-80℃保存备用。1.5 不同胁迫处理条件下BraERF023a表达特征的定量表达分析
采用qRT-PCR方法对花椰菜不同胁迫处理下BraERF023a的表达特征进行分析。反应体系为20 μL,其中含有10 μL 2×SYBR Green反应mix(罗氏公司)、1 μL BraERF023a定量引物(表1)。以花椰菜BraActin为内参,采用2-??CT法进行定量数据分析。Table 1
表1
表1 BraERF023a克隆、载体构建和定量RT-PCR分析引物序列
Table 1
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|
| BraERF023a-F | ATGCTGAGTACAGTGAGTGAAACC |
| BraERF023a-R | TCACAGACACGCCATGAACTG |
| Nco I-BraERF023a-F | CCATGGATGCTGAGTACAGTGAGTG |
| BstE II-BraERF023a-R | GGTCACCTCACAGACACGCCAT |
| qBraERF023a-F | CCACCACTTACCAACGAAC |
| qBraERF023a-R | AAGATCCGAGCCAAATCCG |
| 35S-F | AACAGAACTCGCCGTAAAG |
| BraActin-F | GCTCCTCTTAACCCAAAGGC |
| BraActin-R | CACACCATCACCAGAATCCAGC |
| AtActin-F | GGTAACATTGTGCTCAGTGGTGG |
| AtActin-R | AACGACCTTAATCTTCATGCTGC |
新窗口打开|下载CSV
1.6 BraERF023a植物过表达载体构建
以花椰菜cDNA为模板扩增具有Nco I和BstE II酶切位点的BraERF023a编码区cDNA。将BraERF023a的PCR扩增产物与pEASY-T1克隆载体连接(参照说明书),采用热激法将重组载体转化大肠杆菌DH5α。利用Nco I-BraERF023a-F/BstE II-BraERF023a-R引物组合对阳性克隆进行鉴定(表1),并进一步对阳性克隆进行测序分析。对含有BraERF023a正确序列的阳性克隆及pCAMBIA3301表达载体分别进行Nco I和BstE II双酶切,回收酶切片段,利用T4 DNA连接酶将BraERF023a连接至pCAMBIA3301载体的CaMV 35S启动子下游,筛选获得35S::BraERF023a重组表达载体。采用热激法转化农杆菌LBA4404,利用BraERF023a特异引物和组合引物(35S-F/ BraERF023a-R)对农杆菌阳性转化菌株进行分子鉴定(表1)。将含有35S::BraERF023a重组质粒的农杆菌采用浸花法遗传转化拟南芥。1.7 农杆菌介导BraERF023a过表达载体遗传转化拟南芥
于250 mL液体YEB培养基中,扩大培养含有35S::BraERF023a重组质粒的阳性农杆菌,使其OD600值约为1.2—1.6,5 500 r/min离心10 min收集菌体,用500 mL 5%的蔗糖溶液(含终浓度为0.05%的silwet L-77)重悬菌体,配成转化液。于盛花期将拟南芥花序浸入转化液中浸染30 s,用保鲜膜包好植株,平置于托盘中避光培养24 h,7 d后再次转化,两次转化完成后将拟南芥置于培养室中继续培养至种子成熟,收集种子,记为T0代。1.8 BraERF023a过表达拟南芥的抗性筛选及鉴定
将春化后的BraERF023a转基因T0代拟南芥种子种于营养土中,待其长出两片真叶后,喷洒稀释10 000倍的Basta溶液,对转化株进行抗性筛选。对筛选获得的T0、T1、T2和T3代抗性转化株进一步提取其DNA,利用特异引物(BraERF023a-F/BraERF023a- R)和组合引物(35S-F/BraERF023a-R)进行PCR扩增(表1),检测BraERF023a是否已整合到拟南芥基因组中。进一步提取T3代纯合抗性转化株RNA,以qBraERF023a-F/qBraERF023a-R为引物,采用qRT-PCR方法检测BraERF023a在拟南芥转化株中的表达特征,收获纯合的T3代转化株种子用于后续分析。1.9 BraERF023a过表达拟南芥的表型及抗性分析
将 T3代过表达BraERF023a拟南芥转化株系和野生型对照种子同时播种,在相同的环境条件下进行培养,并对其株高、茎粗、抽薹时间、分枝数目、果荚长短等表型进行观察。此外,待过表达BraERF023a拟南芥转化株系和野生型对照长至3周时,在一次性浇足水后7 d,改为浇灌200 mmol?L-1 NaCl溶液进行为期3周的盐胁迫处理,3周后观察二者在盐胁迫下的表型变化。对长至3周的过表达BraERF023a拟南芥转化株系和野生型对照在一次性浇足水后,停止浇水并进行为期20 d的干旱胁迫处理,之后复水,观察复水10 d后二者的干旱胁迫表型并统计其存活率。2 结果
2.1 BraERF023a的克隆及序列分析
分别以花椰菜DNA、cDNA为模板,利用特异性引物组合BraERF023a-F/BraERF023a-R扩增BraERF023a的全长编码区(表1)。扩增产物的电泳检测结果表明,无论是以DNA还是cDNA为模板均能扩增出一条约600 bp的特异条带,其长度与前期转录组测序获得的BraERF023a序列长度一致,证实花椰菜BraERF023a序列中无内含子。进一步对扩增产物进行TA克隆及测序,测序结果显示,BraERF023a编码区全长597 bp,预期编码一个含有198个氨基酸的蛋白质(图1)。序列分析发现,BraERF023a包含一个AP2保守结构域,该结构域包含3个β折叠和1个α螺旋,且保守结构域中第14和19位氨基酸分别为丙氨酸和天冬氨酸,均符合ERF转录因子的典型结构特征(图1)。进一步利用ProtFun 2.2 Server(http://www. cbs.dtu.dk/services/ProtFun/)对BraERF023a的蛋白功能进行预测分析。结果显示,BraERF023a可能主要在植物生长发育及转录调控两个过程中发挥作用。此外,序列比对及聚类分析结果显示,BraERF023a与B. napus、B. oleracea、B. rapa等十字花科芸薹属其他植物中的ERF023序列具有较高相似性(大于90%),而与水稻、玉米、高粱等禾本科植物中的ERF023相似性较低,表明ERF023在十字花科芸薹属作物中具有较高保守性,其可能具有相似的功能。图1
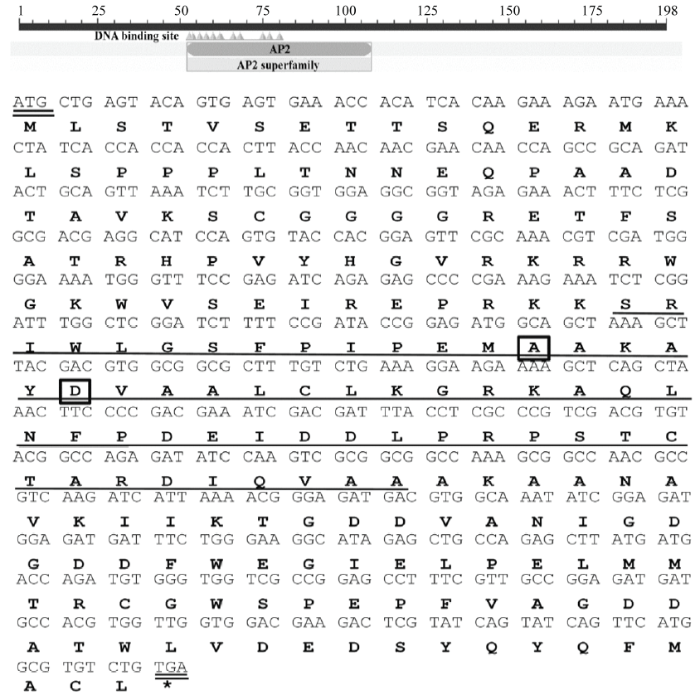 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1花椰菜BraERF023a序列分析
横线显示AP2保守结构域,方框显示ERF转录因子AP2结构域内保守的第14位和19位氨基酸
Fig. 1Sequence analysis of BraERF023a in cauliflower
Sequences marked by the black lines indicated the AP2 domain, and the boxes indicated the conserved fourteenth and nineteenth amino acids in the AP2 domain of ERFs, respectively
2.2 BraERF023a在盐胁迫和模拟干旱处理条件下的表达特征
200 mmol?L-1 NaCl盐处理条件下,BraERF023a的表达量随着处理时间的延长显著升高,表明盐胁迫处理可以显著诱导BraERF023a的表达(图2-a)。与其相似,在20%的PEG6000模拟干旱胁迫处理条件下,花椰菜中的BraERF023a在胁迫处理后不同时间段的表达量均显著高于处理前(图2-b),表明BraERF023a不但正向响应盐胁迫,也受干旱胁迫的诱导表达。图2
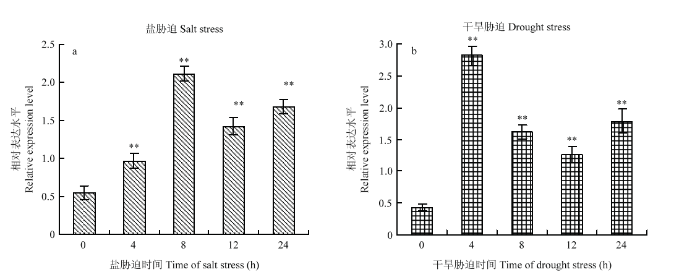 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2花椰菜盐(a)和干旱(b)胁迫条件下BraERF023a表达特征的 qRT-PCR分析结果
**表示差异极显著(P<0.01)。下同
Fig. 2Expression profiles of BraERF023a under salt (a) and drought (b) stresses detected by qRT-PCR
** indicated the difference is significant at the 0.01 level. The same as below
2.3 BraERF023a表达载体构建及鉴定
分别采用基因特异引物(BraERF023a-F/BraERF023a- R)和组合引物(35S-F/BraERF023a-R)进行菌液PCR,鉴定重组载体是否已成功转入农杆菌中。随机挑取6个阳性克隆进行检测,PCR产物的电泳结果显示在6个阳性克隆中均可检测到长度约为600 bp和1 100 bp的特异性条带(图3-a),条带大小均符合预期,表明35S::BraERF023a过表达载体已成功转入农杆菌中。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3农杆菌及拟南芥转化株中BraERF023a的分子鉴定
a:分别采用BraERF023a特异扩增引物及BraERF023a与载体序列组合引物对农杆菌阳性克隆进行菌液PCR的检测结果,M:DNA marker,1—6:6个阳性克隆特异引物PCR检测结果,1′—6′:6个阳性克隆组合引物PCR检测结果。b:分别采用BraERF023a特异扩增引物及BraERF023a与载体序列组合引物对拟南芥转化株的单株PCR检测结果,M:DNA marker,1—10:10个单株特异引物PCR检测结果,1′—10′:10个单株组合引物PCR检测结果。c:不同BraERF023a过表达拟南芥转化株系中BraERF023a表达水平的qRT-PCR检测结果,CK:对照组,L3/L4/L6/L8/L9:BraERF023a过表达拟南芥株系
Fig. 3Identification of BraERF023a in Agrobacterium tumefaciens and transgenic lines in Arabidopsis
a: Identification of Agrobacterium tumefaciens with BraERF023a by PCR used the specific primers and combined primers, respectively. M: DNA marker, 1-6 indicated PCR identification used the specific primers, 1′-6′ indicated PCR identification used the combined primers. b: Identification of individual Arabidopsis plant with BraERF023a by PCR used the specific primers and combined primers, respectively. M: DNA marker, 1-10 indicated PCR identification of individual plants used the specific primers, 1′-10′ indicated PCR identification of individual plant used the combined primers. c: Expression levels of BraERF023a in differential overexpression BraERF023a transgenic Arabidopsis lines detected by qRT-PCR, CK: Wild-type control, L3/L4/L6/L8/L9 indicated the five independent transgenic lines
2.4 BraERF023a过表达拟南芥转化株系筛选及分子鉴定
野生型拟南芥于盛花期经农杆菌浸花法侵染后培养至种子成熟,记为T0代,T0代种子萌发后进行Basta抗性筛选。筛选结果显示,绝大部分幼苗叶片逐渐黄化,26个植株生长发育状况正常,初步鉴定为抗性转化株系。随机选取其中10个抗性转化株系分别进行DNA水平及转录水平的分子鉴定。以DNA为模板,采用BraERF023a特异引物及BraERF023a与载体序列组合引物分别进行PCR检测,在其中5个株系中可以分别扩增出与预期大小一致的约600 bp和1 100 bp的特异性条带,表明BraERF023a已成功整合到这些株系的基因组中(图3-b)。进一步采用qRT-PCR方法对BraERF023a在这5个阳性株系中的表达特征进行分析。结果显示,BraERF023a在这5个株系中的表达水平显著高于野生型对照(图3-c),证实BraERF023a不但成功整合到转化株基因组内,并且实现在转化株的高表达。将这5个T0代植株进行多代自交纯合,T3代株系用于后续分析。2.5 BraERF023a过表达转基因拟南芥表型分析
为揭示BraERF023a在拟南芥中过表达是否影响转化株的生长发育,对过表达BraERF023a转基因拟南芥在正常生长条件下不同生长阶段的表型特征进行观察、分析。结果显示,幼苗生长初期(3周以前),过表达BraERF023a转基因拟南芥株系与对照相比无明显差异,但在幼苗生长3周后,过表达BraERF023a转化株系的长势明显优于对照(图4)。待植株进入成熟期,对其株高、分枝数目等表型特征进行统计分析。结果显示,过表达BraERF023a转化株系相对于对照植株表现为植株更高、分枝数目更多、花茎更粗,整体生物量增加。但二者花序大小及果荚长度无明显差异。在抽薹时间方面,过表达BraERF023a转化株系比对照滞后约一周(表2)。上述结果表明,在拟南芥中过表达BraERF023a对转化株的生长发育有一定影响,其可以促进转化株的生长发育,延长其抽薹期,但对转化株育性、结种量无显著影响。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4BraERF023a过表达拟南芥不同生长发育时期的表型
a、b和c分别显示生长3周、4周及8周的BraERF023a过表达转化株系及对照的表型。CK:对照组,L4/L8/L9:过表达BraERF023a转化株系
Fig. 4Phenotypes of overexpression BraERF023a transgenic Arabidopsis at different developmental stages
a, b and c indicated the phenotypes of 3-week-old, 4-week-old and 8-week-old overexpression BraERF023a transgenic Arabidopsis and the control (CK). L4/L8/L9 indicated three independent overexpression BraERF023a transgenic lines
Table 2
表2
表2过表达BraERF023a转基因拟南芥表型分析
Table 2
| 表型Phenotype | 对照CK | L4 | L8 | L9 |
|---|---|---|---|---|
| 株高 Stem length (cm) | 43.125±2.780c | 56.750±3. 862a | 51.125±1.0307ab | 49.625±3.0923b |
| 抽薹时间 Bolting time (d) | 26.00±2.4495b | 32.00±2.4495a | 29±1.9148a | 31.00±1.2583a |
| 茎粗 Stem diameter (mm) | 1.2375±0.0727b | 1.7800±0.2191a | 1.6300±0.1551a | 1.7125±0.1059a |
| 主茎 Stem (number) | 5.75±0.9574c | 6.75±1.0708ab | 6.25±0.5000b | 7.775±0.5774a |
| 一级分枝 Primary branch (number) | 13.75±2.630b | 15.00±2.449ab | 16.00±3.742a | 13.75±5.132b |
| 二级分枝 Secondary branch (number) | 5.00±0.816a | 5.00±1.155a | 5.75±2.602a | 6.00±1.15a |
| 果荚长 Pod length (mm) | 18.000±1.0475a | 17.153±1.3553a | 17.600±1.2035a | 18.028±0.8587a |
新窗口打开|下载CSV
2.6 BraERF023a过表达转基因拟南芥盐胁迫分析
BraERF023a过表达转基因拟南芥和野生型对照经盐胁迫后生长均受到一定抑制。但对照组拟南芥叶片萎蔫黄化严重甚至全部枯萎,植株存活率低;而过表达BraERF023a转化株系叶片虽也发生黄化,但程度显著低于对照(图5),且尽管受到盐胁迫,BraERF023a过表达转化株植株最终的存活率(86%)显著高于对照组(41%)。表明在拟南芥中过表达BraERF023a可以提高转化株抵御盐胁迫的能力。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5过表达BraERF023a转基因拟南芥盐胁迫表型分析
a:过表达BraERF023a拟南芥转化株系(L4、L8、L9)及对照组(CK)200 mmol·L-1 NaCl 盐胁迫处理前表型;b:过表达BraERF023a拟南芥转化株系(L4、L8、L9)及对照组(CK)200 mmol·L-1 NaCl 盐胁迫处理3周后表型
Fig. 5Phenotypes of overexpression BraERF023a transgenic Arabidopsis under salt stress
a: The phenotypes of overexpression BraERF023a transgenic Arabidopsis (L4, L8, L9) and control before salt stress (200 mmol·L-1 NaCl); b: The phenotypes of overexpression BraERF023a transgenic Arabidopsis (L4, L8, L9) and control under salt stress (200 mmol?L-1 NaCl) for three weeks
2.7 BraERF023a过表达转基因拟南芥干旱胁迫分析
干旱胁迫处理20 d后,BraERF023a过表达转化株系和野生型植株均萎蔫严重,但野生型植株基本已全部萎蔫黄化至死亡,而BraERF023a过表达转化株系萎蔫黄化程度相对野生型较轻,部分转化株系叶片仍保持绿色(图6)。对所有干旱处理植株进行复水,复水10 d后,其中2个过表达BraERF023a转化株系中(L8、L9)的大部分植株恢复生长,而对照组绝大部分已枯萎(图6)。且过表达BraERF023a转化株系的存活率显著高于对照组(图7)。表明在拟南芥中过表达BraERF023a可以显著提高转化株的耐旱性。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6过表达BraERF023a转基因拟南芥干旱胁迫表型分析
L4、L8、L9显示3个独立的过表达BraERF023a拟南芥转化株系;CK为对照
Fig. 6Phenotypes of overexpression BraERF023a transgenic Arabidopsis under drought stress
L4, L8, L9 indicated three independent overexpression BraERF023a transgenic lines; CK indicated wild-type control
图7
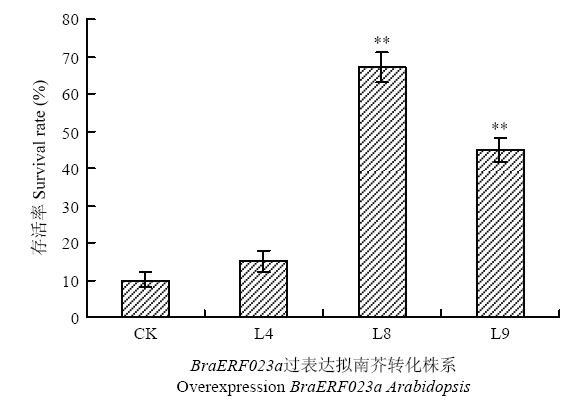 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7过表达BraERF023a转基因拟南芥干旱胁迫后存活率分析
L4、L8、L9显示3个独立的过表达BraERF023a拟南芥转化株系;CK为对照
Fig. 7Survival rate of overexpression BraERF023a transgenic Arabidopsis under drought stress
L4, L8 and L9 indicated three independent overexpression BraERF023a transgenic lines; CK indicated wild-type control
3 讨论
3.1 BraERF023a在植物中的分子进化特征分析
AP2/ERF转录因子最初在植物中被发现。1994年JOFUKU等[4]从模式植物拟南芥中分离到第一个AP2转录因子APETALA2,该转录因子与花发育和种子萌发有关。随后AP2/ERF转录因子家族成员陆续从不同植物中被鉴定、克隆,并证实其为植物所特有。基因结构分析表明AP2/ERF家族成员均含有保守的AP2结构域[5]。根据AP2结构域的数目及序列特征,AP2/ERF家族中的ERF亚族和DREB亚族均含有单个的AP2结构域[6],但是AP2结构域第14位和第19位氨基酸残基在两者中不同。ERF亚族成员AP2结构域的第14位和19位分别为丙氨酸和天冬氨酸,而DREB亚族成员该结构域第14位和19位分别为缬氨酸和谷氨酸[7]。通过对BraERF023a的序列分析发现,其具有1个AP2结构域,并且保守结构域第14位和19位分别为丙氨酸和天冬氨酸,符合ERF亚族成员的典型结构特征,证实其为ERF转录因子。已有研究证实ERF转录因子可与GCC盒(AGCCGCC)等下游靶基因的顺式作用元件结合,从而在基因转录调控中发挥作用[9,22]。进一步的序列比对发现,BraERF023a与双子叶植物,特别是与花椰菜同属于十字花科芸薹属的其他植物中的ERF023在进化上具有较高保守性,而与单子叶植物中该转录因子的序列同源性较低,暗示BraERF023a在祖先植物进化形成单子叶和双子叶植物时,序列已发生分化,但其功能是否也发生了亚功能化,有待进一步探究。在克隆并揭示BraERF023a序列及表达特征基础上,通过遗传转化策略将其在花椰菜中进行过表达,并分析过表达BraERF023a花椰菜转化株系生长发育及在逆境胁迫条件下的表型及生理特征,对于阐释BraERF023a的功能具有重要价值。但花椰菜遗传转化效率受材料基因型、遗传背景等因素影响很大,不同基因型材料遗传转化效率普遍不高,很难在短期内获得转化株系,即使获得了转化株系,很多转化株系也不能正常结籽。因此,转化效率低及结籽困难一直是花椰菜基础研究进展缓慢的一个重要瓶颈。基于此,本研究为探究花椰菜BraERF023a的功能,首先将其转入到易于转化的模式植物拟南芥中进行功能分析。BraERF023a在花椰菜中过表达及采用CRISPR/Cas9基因编辑技术敲除的工作正在进行中。3.2 BraERF023a响应盐和干旱胁迫分析
已有研究表明,ERF转录因子亚家族成员主要在植物逆境响应中发挥作用。但不同植物来源的ERF成员或同一植物中不同的ERF成员对逆境胁迫的响应特征不尽相同。JERF1在烟草中过表达可增强转化株对低温及高盐的耐受[23]。拟南芥AtERF1的表达受ABA负调控,其过表达导致ABA积累和脯氨酸增加,进而增强转化株对干旱、高盐和热胁迫的耐受性[24]。过量表达 MdERF72的苹果愈伤组织在高盐和低温胁迫处理下,生长势明显强于野生型对照,表明MdERF72在响应高盐、低温胁迫过程中发挥着重要的正调控作用[25]。苜蓿中ERF转录因子MfERF1过表达可促进转化株多胺的合成与转化,提高植株抗氧化能力和脯氨酸积累,进而提高转化株对低温的耐受[26]。BOLT等[27]研究发现ERF105在拟南芥对冻害的耐受性中发挥重要作用,是拟南芥冷驯化中所必需的转录调控因子。ERF102和ERF103也被证实在拟南芥冷驯化中扮演重要角色[28]。有关花椰菜中ERF转录因子的报道很少。笔者课题组前期的研究表明,在花椰菜中至少存在146个ERF成员,这其中绝大部分成员的功能未知[21]。本研究结果表明,盐及干旱逆境胁迫可显著诱导花椰菜中BraERF023a的表达,暗示BraERF023a是一个盐及干旱正向响应应答因子。进一步的过表达功能分析表明,在拟南芥中过表达BraERF023a可显著提高转化株对盐及干旱胁迫的耐受。该结果进一步证实,BraERF023a在植物耐盐及抗旱应答中发挥重要的正向调控作用。此外,前期研究结果表明花椰菜中除BraERF023a外,还具有一个BraERF023a的等位变异,命名为BraERF023b,其编码区长度为567 bp,比BraERF023a少30 bp[21]。在花椰菜中BraERF023b是功能性冗余还是具有其他功能有待进一步探究。目前,有关不同植物中ERF023的研究很少。在杏桃干旱胁迫响应的研究中发现,干旱胁迫可诱导其内PpERF023的表达,暗示PpERF023在干旱胁迫应答中发挥重要作用[29]。杏桃ERF023在拟南芥中的同源基因AtHARDY被证实在提高植株的水分利用效率,提高对干旱胁迫的适应能力过程中发挥重要作用[30]。这暗示不同植物来源的ERF023在干旱胁迫应答中可能发挥重要作用,但其是否也都参与盐胁迫应答并发挥作用有待进一步探究。在葡萄中的研究表明,ERF023可能参与葡萄对病原菌的防御,其过表达在提高葡萄对灰霉病的抗性方面有一定作用[31]。暗示ERF023不但在非生物逆境胁迫中发挥作用,也响应生物胁迫。因此,进一步克隆不同植物中的ERF023,并揭示其对不同逆境胁迫响应的分子机制,对于阐释ERF类转录因子的功能具有重要意义。3.3 BraERF023a在生长发育中的功能分析
尽管大部分研究表明ERF类转录因子主要在植物响应逆境胁迫中发挥作用。但也有研究表明一些ERF转录因子参与植物生长发育调控。如拟南芥中ERF转录因子家族成员TINY可抑制植物激素BR调控的生长发育过程,同时也调控植株对干旱的胁迫反应。TINY 过表达可导致植物生长发育迟缓[32]。在水稻中ERF转录因子OsEATB可通过下调赤霉素生物合成基因限制节间伸长,导致植株矮化[33]。NAKANO等[34]研究表明,番茄SlERF52在花梗离区特异表达,其在花梗脱落中发挥重要作用。过量表达拟南芥AtERF019不但可以提高植物的耐旱性,还可以延迟植株生长和衰老[35]。在黄瓜中鉴定出138个ERF家族成员,其中多个成员被认为在雌花发育过程中发挥调控作用[36]。本研究结果表明,正常生长条件下,在拟南芥中过表达BraERF023a可使转化株表现出显著不同于野生型对照的生长发育特征,如相对于野生型对照,过表达BraERF023a转基因拟南芥植株更高,主茎加粗,莲座和一级分枝数增多、生物量提高等,并且过表达转化株的抽薹时间延长,致使其生长期延长(表2、图4)。该结果表明在拟南芥中过表达BraERF023a可以促进转化株的生长发育,暗示BraERF023a不但是一个盐及干旱胁迫响应的正向调控因子,其在植物生长发育调控过程中也可能发挥重要作用,有待对其调控机制进行深入探究。4 结论
花椰菜BraERF023a含有一个保守的AP2结构域,其与十字花科芸薹属植物中的ERF023具有较高序列同源性。转录表达特征分析证实盐及干旱胁迫可显著诱导BraERF023a的表达。在拟南芥中过表达BraERF023a可促进转化株的生长发育,并显著提高转化株对盐及干旱胁迫的耐受力。结果证实,BraERF023a在植物响应盐及干旱胁迫中发挥重要的正向调控作用,是一个优质耐盐抗旱基因,在花椰菜抗逆分子育种中具有重要应用价值。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1126/science.1186834URLPMID:20150494 [本文引用: 1]

Population growth, arable land and fresh water limits, and climate change have profound implications for the ability of agriculture to meet this century's demands for food, feed, fiber, and fuel while reducing the environmental impact of their production. Success depends on the acceptance and use of contemporary molecular techniques, as well as the increasing development of farming systems that use saline water and integrate nutrient flows.
DOI:10.1111/pbr.12645URLPMID:31598026 [本文引用: 1]

Groundnut is an important food and oil crop in the semiarid tropics, contributing to household food consumption and cash income. In Asia and Africa, yields are low attributed to various production constraints. This review paper highlights advances in genetics, genomics and breeding to improve the productivity of groundnut. Genetic studies concerning inheritance, genetic variability and heritability, combining ability and trait correlations have provided a better understanding of the crop's genetics to develop appropriate breeding strategies for target traits. Several improved lines and sources of variability have been identified or developed for various economically important traits through conventional breeding. Significant advances have also been made in groundnut genomics including genome sequencing, marker development and genetic and trait mapping. These advances have led to a better understanding of the groundnut genome, discovery of genes/variants for traits of interest and integration of marker-assisted breeding for selected traits. The integration of genomic tools into the breeding process accompanied with increased precision of yield trialing and phenotyping will increase the efficiency and enhance the genetic gain for release of improved groundnut varieties.
DOI:10.1111/pbr.12645URLPMID:31598026 [本文引用: 1]

Groundnut is an important food and oil crop in the semiarid tropics, contributing to household food consumption and cash income. In Asia and Africa, yields are low attributed to various production constraints. This review paper highlights advances in genetics, genomics and breeding to improve the productivity of groundnut. Genetic studies concerning inheritance, genetic variability and heritability, combining ability and trait correlations have provided a better understanding of the crop's genetics to develop appropriate breeding strategies for target traits. Several improved lines and sources of variability have been identified or developed for various economically important traits through conventional breeding. Significant advances have also been made in groundnut genomics including genome sequencing, marker development and genetic and trait mapping. These advances have led to a better understanding of the groundnut genome, discovery of genes/variants for traits of interest and integration of marker-assisted breeding for selected traits. The integration of genomic tools into the breeding process accompanied with increased precision of yield trialing and phenotyping will increase the efficiency and enhance the genetic gain for release of improved groundnut varieties.
DOI:10.1016/S1369-5266(02)00289-3URL [本文引用: 1]

Abstract
Transcriptional control of the expression of stress-responsive genes is a crucial part of the plant response to a range of abiotic and biotic stresses. Research carried out in the past few years has been productive in identifying transcription factors that are important for regulating plant responses to these stresses. These studies have also revealed some of the complexity and overlap in the responses to different stresses, and are likely to lead to new ways to enhance crop tolerance to disease and environmental stress.DOI:10.1105/tpc.6.9.1211URLPMID:7919989 [本文引用: 2]

APETALA2 (AP2) plays a central role in the establishment of the floral meristem, the specification of floral organ identity, and the regulation of floral homeotic gene expression in Arabidopsis. We show here that in addition to its functions during flower development, AP2 activity is also required during seed development. We isolated the AP2 gene and found that it encodes a putative nuclear protein that is distinguished by an essential 68-amino acid repeated motif, the AP2 domain. Consistent with its genetic functions, we determined that AP2 is expressed at the RNA level in all four types of floral organs--sepals, petals, stamens, and carpels--and in developing ovules. Thus, AP2 gene transcription does not appear to be spatially restricted by the floral homeotic gene AGAMOUS as predicted by previous studies. We also found that AP2 is expressed at the RNA level in the inflorescence meristem and in nonfloral organs, including leaf and stem. Taken together, our results suggest that AP2 represents a new class of plant regulatory proteins that may play a general role in the control of Arabidopsis development.
DOI:10.1093/emboj/17.18.5484URLPMID:9736626 [本文引用: 2]

The 3D solution structure of the GCC-box binding domain of a protein from Arabidopsis thaliana in complex with its target DNA fragment has been determined by heteronuclear multidimensional NMR in combination with simulated annealing and restrained molecular dynamic calculation. The domain consists of a three-stranded anti-parallel beta-sheet and an alpha-helix packed approximately parallel to the beta-sheet. Arginine and tryptophan residues in the beta-sheet are identified to contact eight of the nine consecutive base pairs in the major groove, and at the same time bind to the sugar phosphate backbones. The target DNA bends slightly at the central CG step, thereby allowing the DNA to follow the curvature of the beta-sheet.
DOI:10.1104/pp.105.073783URLPMID:16407444 [本文引用: 3]

Genes in the ERF family encode transcriptional regulators with a variety of functions involved in the developmental and physiological processes in plants. In this study, a comprehensive computational analysis identified 122 and 139 ERF family genes in Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa L. subsp. japonica), respectively. A complete overview of this gene family in Arabidopsis is presented, including the gene structures, phylogeny, chromosome locations, and conserved motifs. In addition, a comparative analysis between these genes in Arabidopsis and rice was performed. As a result of these analyses, the ERF families in Arabidopsis and rice were divided into 12 and 15 groups, respectively, and several of these groups were further divided into subgroups. Based on the observation that 11 of these groups were present in both Arabidopsis and rice, it was concluded that the major functional diversification within the ERF family predated the monocot/dicot divergence. In contrast, some groups/subgroups are species specific. We discuss the relationship between the structure and function of the ERF family proteins based on these results and published information. It was further concluded that the expansion of the ERF family in plants might have been due to chromosomal/segmental duplication and tandem duplication, as well as more ancient transposition and homing. These results will be useful for future functional analyses of the ERF family genes.
DOI:10.1006/bbrc.2001.6299URLPMID:11798174 [本文引用: 2]

DRE/CRT is a cis-acting element that is involved in gene expression responsive to drought and low-temperature stress in higher plants. DREB1A/CBF3 and DREB2A are transcription factors that specifically bind to DRE/CRT in Arabidopsis. We precisely analyzed the DNA-binding specificity of DREBs. Both DREBs specifically bound to six nucleotides (A/GCCGAC) of DRE. However, these proteins had different binding specificities to the second or third nucleotides of DRE. Gel mobility shift assay using mutant DREB proteins showed that the two amino acids, valine and glutamic acid conserved in the ERF/AP2 domains, especially valine, have important roles in DNA-binding specificity. In the Arabidopsis genome, 145 DREB/ERF-related proteins are encoded. These proteins were classified into five groups-AP-2 subfamily, RAV subfamily, DREB subfamily, ERF subfamily, and others. The DREB subfamily included three novel DREB1A- and six DREB2A-related proteins. We analyzed expression of novel genes for these proteins and discuss their roles in stress-responsive gene expression.
DOI:10.1093/nar/gkg292URLPMID:12655002 [本文引用: 1]

The Arabidopsis protein AINTEGUMENTA (ANT) is an important regulator of organ growth during flower development. ANT is a member of the AP2 subclass of the AP2/ERF family of plant-specific transcription factors. These proteins contain either one or two copies of a DNA-binding domain called the AP2 domain. Here, it is shown that ANT can act as a transcriptional activator in yeast through binding to a consensus ANT-binding site. This activity was used as the basis for a genetic screen to identify amino acids that are critical for the DNA binding ability of ANT. Mutants that showed reduced or no activation of a reporter gene under the control of ANT-binding sites were identified. The mutations identified in the screen as well as additional site-directed mutations suggest that the mode of DNA recognition by members of the AP2 subfamily is distinct from that of ERF proteins. Surprisingly, it appears that each AP2 domain of ANT uses different amino acids to contact DNA. Identification of several linker mutations argues that this sequence acts in the positioning of each AP2 domain on the DNA or makes direct DNA contacts.
DOI:10.1146/annurev.arplant.57.032905.105444URL [本文引用: 2]
DOI:10.1038/nature08258URLPMID:19693083 [本文引用: 1]

Living organisms must acquire new biological functions to adapt to changing and hostile environments. Deepwater rice has evolved and adapted to flooding by acquiring the ability to significantly elongate its internodes, which have hollow structures and function as snorkels to allow gas exchange with the atmosphere, and thus prevent drowning. Many physiological studies have shown that the phytohormones ethylene, gibberellin and abscisic acid are involved in this response, but the gene(s) responsible for this trait has not been identified. Here we show the molecular mechanism of deepwater response through the identification of the genes SNORKEL1 and SNORKEL2, which trigger deepwater response by encoding ethylene response factors involved in ethylene signalling. Under deepwater conditions, ethylene accumulates in the plant and induces expression of these two genes. The products of SNORKEL1 and SNORKEL2 then trigger remarkable internode elongation via gibberellin. We also demonstrate that the introduction of three quantitative trait loci from deepwater rice into non-deepwater rice enabled the latter to become deepwater rice. This discovery will contribute to rice breeding in lowland areas that are frequently flooded during the rainy season.
DOI:10.1007/s00299-018-2290-9URLPMID:29687169 [本文引用: 1]

KEY MESSAGE: We find that the DREB subfamily transcription factor, CmERF053, has a novel function to regulate the development of shoot branching and lateral root in addition to affecting abiotic stress. Dehydration-responsive element binding proteins (DREBs) are important plant transcription factors that regulate various abiotic stresses. Here, we isolated an APETALA2/ethylene-responsive factor (AP2/ERF) transcription factor from chrysanthemum (Chrysanthemum morifolium 'Jinba'), CmERF053, the expression of which was rapidly up-regulated by main stem decapitation. Phylogenetic analysis indicated that it belongs to the A-6 group of the DREB subfamily, and the subcellular localization assay confirmed that CmERF053 was a nuclear protein. Overexpression of CmERF053 in Arabidopsis exhibited positive effects of plant lateral organs, which had more shoot branching and lateral roots than did the wild type. We also found that the expression of CmERF053 in axillary buds was induced by exogenous cytokinins. These results suggested that CmERF053 may be involved in cytokinins-related shoot branching pathway. In this study, an altered auxin distribution was observed during root elongation in the seedlings of the overexpression plants. Furthermore, overexpress CmERF053 gene could enhance drought tolerance. Together, these findings indicated that CmERF053 plays crucial roles in regulating shoot branching, lateral root, and drought stress in plant. Moreover, our study provides potential application value for improving plant productivity, ornamental traits, and drought tolerance.
URL [本文引用: 1]

GmERF6 is a new ERF transcription repressor isolated from soybean. The function of GmERF6 was further analyzed in this research. GmERF6 protein localized to the nucleus when transiently expressed in onion epidermal cells. Furthermore, EMSA showed GmERF6 protein binding to the GCC-box element in vitro . The seed germination rates of GmERF6 transgenic Arabidopsis were significantly higher than that of wild type Arabidopsis under drought stress. GmERF6 transgenic Arabidopsis grew much healthier than wild-type Arabidopsis by removing water for 18 d. These data indicated that GmERF6 enhanced drought tolerance in transgenic Arabidopsis . These results suggest GmERF6 is likely to be a useful tool for improving drought tolerance of plants.
URL [本文引用: 1]

GmERF6 is a new ERF transcription repressor isolated from soybean. The function of GmERF6 was further analyzed in this research. GmERF6 protein localized to the nucleus when transiently expressed in onion epidermal cells. Furthermore, EMSA showed GmERF6 protein binding to the GCC-box element in vitro . The seed germination rates of GmERF6 transgenic Arabidopsis were significantly higher than that of wild type Arabidopsis under drought stress. GmERF6 transgenic Arabidopsis grew much healthier than wild-type Arabidopsis by removing water for 18 d. These data indicated that GmERF6 enhanced drought tolerance in transgenic Arabidopsis . These results suggest GmERF6 is likely to be a useful tool for improving drought tolerance of plants.
DOI:10.1016/j.plantsci.2019.01.009URLPMID:30824052 [本文引用: 1]

The manipulation of APETALA2/ethylene responsive factor (AP2/ERF) genes in plants makes great contributions on resistance to abiotic stresses. Here, we cloned an AP2/ERF gene from the salt-tolerant sweetpotato line ND98 and named IbRAP2-12. IbRAP2-12 protein expressed in nuclear revealed by transient expression in tobacco epidermal cells, and IbRAP2-12 exhibited transcriptional activation using heterologous expression assays in yeast. IbRAP2-12 was induced by NaCl (200 mM), 20% polyethylene glycol (PEG) 6000, 100 muM abscisic acid (ABA), 100 muM ethephon and 100 muM methyl jasmonate (MeJA). IbRAP2-12-overexpressing Arabidopsis lines were more tolerant to salt and drought stresses than wild type plants. Transcriptome analysis showed that genes involved in the ABA signalling, JA signalling, proline biosynthesis and reactive oxygen species (ROS) scavenging processes were up-regulated in IbRAP2-12 overexpression lines under salt and drought stresses. In comparing with WT, the contents of ABA, JA and proline were significantly increased, while hydrogen peroxide (H2O2) and the rate of water loss were significantly reduced in transgenic lines under salt and drought stresses. All these results demonstrated the roles of IbRAP2-12 in enhancing salt and drought tolerance in transgenic Arabidopsis lines. Thus, this IbRAP2-12 gene can be used to increase the tolerance ability during abiotic stresses in plants.
DOI:10.1007/s10059-010-0114-zURL [本文引用: 1]
DOI:10.1016/j.bbrc.2011.09.039URL [本文引用: 1]

Various transcription factors are involved in the response to environmental stresses in plants. In this study, we characterized AtERF71/HRE2, a member of the Arabidopsis AP2/ERF family, as an important regulator of the osmotic and hypoxic stress responses in plants. Transcript level of AtERF71/HRE2 was highly increased by anoxia, NaCl, mannitol, ABA, and MV treatments. aterf71/hre2 loss-of-function mutants displayed higher sensitivity to osmotic stress such as high salt and mannitol, accumulating higher levels of ROS under high salt treatment. In contrast, AtERF71/HRE2-overexpressing transgenic plants showed tolerance to salt and mannitol as well as flooding and MV stresses, exhibiting lower levels of ROS under high salt treatment. AtERF71/HRE2 protein was localized in the nucleus, and the C-terminal region of AtERF71/HRE2 was required for transcription activation activity. Taken together, our results suggest that AtERF71/HRE2 might function as a transcription factor involved in the response to osmotic stress as well as hypoxia. (C) 2011 Elsevier Inc.
DOI:10.1007/s11103-004-0417-6URL [本文引用: 1]

From a pathogen-inoculated hot pepper (Capsicum annuumL. cv. Pukang) leaf EST, we identified a cDNA clone, pCaERFLP1, encoding a putative transcription factor that contains a single ERF/AP2 DNA binding domain. CaERFLP1 was most closely related to tomato LeERF2 (73%), both of which belong to the novel ERF class IV typified by the N-terminal MCGGAIL signature sequence, while it had a limited sequence identity (25–30%) with Arabidopsis AtERFs and tobacco NtERFs. Quantitative gel retardation assays revealed that bacterially expressed full-length CaERFLP1 was able to form a specific complex with both the GCC box and DRE/CRT motif, with its binding affinity for GCC being stronger than for DRE/CRT. When fused to the GAL4 DNA binding domain, the N-terminal CaERFLP11–37 and C-terminal CaERFLP1198–264 mutant polypeptides could function individually as transactivators in yeast. This suggests that two separate domains of CaERFLP1 may play distinct roles in transcription activation. In particle co-bombardment experiments, CaERFLP1 activated the transcription of reporter genes containing the 4X[GCC] element in tobacco cells. In hot pepper plants, the steady-state level of CaERFLP1mRNA was markedly induced by multiple environmental factors, such as pathogen infection, ethylene, mechanical wounding and high salinity. Furthermore, ectopic expression of CaERFLP1 in transgenic tobacco plants resulted in partially improved tolerance against the bacterial pathogen Pseudomonas syringae and salt stress (100mM NaCl). Consistently, various defense-related genes, including GCC box-containing PR genes and the DRE/CRT-containing LTI45 (ERD10) gene, were constitutively expressed in 35S::CaERFLP1 tobacco plants. Thus, it appears that CaERFLP1 is functional in tobacco cells, where it induces the transactivation of some GCC- and DRE/CRT-genes to trigger a subset of stress response. Here, the possible biological role(s) of CaERFLP1 is discussed, especially with regard to the possibility that CaERFLP1 has multiple functions in the regulation of GCC- and DRE/CRT-mediated gene expression in hot pepper plants.
DOI:10.1104/pp.104.042903URLPMID:15347795 [本文引用: 1]

An ERF/AP2-type transcription factor (CaPF1) was isolated by differential-display reverse transcription-PCR, following inoculation of the soybean pustule pathogen Xanthomonas axonopodis pv glycines 8ra, which induces hypersensitive response in pepper (Capsicum annuum) leaves. CaPF1 mRNA was induced under conditions of biotic and abiotic stress. Higher levels of CaPF1 transcripts were observed in disease-resistant tissue compared with susceptible tissue. CaPF1 expression was additionally induced using various treatment regimes, including ethephon, methyl jasmonate, and cold stress. To determine the role of CaPF1 in plants, transgenic Arabidopsis and tobacco (Nicotiana tabacum) plants expressing higher levels of CaPF1 were generated. Gene expression analyses of transgenic Arabidopsis and tobacco revealed that the CaPF1 level in transgenic plants affects expression of genes that contain either a GCC or a CRT/DRE box in their promoter regions. Furthermore, transgenic Arabidopsis plants expressing CaPF1 displayed tolerance against freezing temperatures and enhanced resistance to Pseudomonas syringae pv tomato DC3000. Disease tolerance was additionally observed in CaPF1 transgenic tobacco plants. The results collectively indicate that CaPF1 is an ERF/AP2 transcription factor in hot pepper plants that may play dual roles in response to biotic and abiotic stress in plants.
DOI:10.1016/j.febslet.2004.07.064URL [本文引用: 1]
DOI:10.1007/s00425-004-1347-xURL [本文引用: 1]

Ethylene responsive factors (ERFs) are important plant-specific transcription factors, some of which have been demonstrated to interact with the ethylene-responsive GCC box and the dehydration-responsive element (DRE); however, data on the roles of ERF proteins in connection with various signaling pathways are limited. In this research, we used the GCC box, an essential cis-acting element responsive to ethylene and methyl jasmonate (MeJA), as bait in a yeast one-hybrid system to isolate transcription factors from tomato (Lycopersicon esculentum Mill.). One of the cDNAs, which was designated Jasmonate and Ethylene Response Factor1 (JERF1), encodes an ERF protein, containing a conserved ERF DNA-binding motif and functioning as a transcriptional activator in yeast through targeting to the nucleus in onion (Allium cepa L.) epidermal cells. Biochemical analysis revealed that JERF1 bound not only to the GCC box but also to the DRE sequence. Expression of the JERF1 gene in tomato was induced by ethylene, MeJA, abscisic acid (ABA) and salt treatment, indicating that JERF1 might act as a connector among different signal transduction pathways. Further research with transgenic JERF1 tobacco (Nicotiana tabacum L.) plants indicated that overexpressing JERF1 activated expression of GCC box-containing genes such as osmotin, GLA, Prb-1b and CHN50 under normal growth conditions, and subsequently resulted in enhanced tolerance to salt stress, suggesting that JERF1 modulates osmotic tolerance by activation of downstream gene expression through interaction with the GCC box or DRE.
[本文引用: 1]
DOI:10.3389/fpls.2017.00946URLPMID:28642765 [本文引用: 4]

The AP2/ERF transcription factors (TFs) comprise one of the largest gene superfamilies in plants. These TFs perform vital roles in plant growth, development, and responses to biotic and abiotic stresses. In this study, 171 AP2/ERF TFs were identified in cauliflower (Brassica oleracea L. var. botrytis), one of the most important horticultural crops in Brassica. Among these TFs, 15, 9, and 1 TFs were classified into the AP2, RAV, and Soloist family, respectively. The other 146 TFs belong to ERF family, which were further divided into the ERF and DREB subfamilies. The ERF subfamily contained 91 TFs, while the DREB subfamily contained 55 TFs. Phylogenetic analysis results indicated that the AP2/ERF TFs can be classified into 13 groups, in which 25 conserved motifs were confirmed. Some motifs were group- or subgroup- specific, implying that they are significant to the functions of the AP2/ERF TFs of these clades. In addition, 35 AP2/ERF TFs from the 13 groups were selected randomly and then used for expression pattern analysis under salt and drought stresses. The majority of these AP2/ERF TFs exhibited positive responses to these stress conditions. In specific, Bra-botrytis-ERF054a, Bra-botrytis-ERF056, and Bra-botrytis-CRF2a demonstrated rapid responses. By contrast, six AP2/ERF TFs were showed to delay responses to both stresses. The AP2/ERF TFs exhibiting specific expression patterns under salt or drought stresses were also confirmed. Further functional analysis indicated that ectopic overexpression of Bra-botrytis-ERF056 could increase tolerance to both salt and drought treatments. These findings provide new insights into the AP2/ERF TFs present in cauliflower, and offer candidate AP2/ERF TFs for further studies on their roles in salt and drought stress tolerance.
[本文引用: 1]
DOI:10.1007/s00425-007-0528-9URL [本文引用: 1]

Increasing evidences indicate that ethylene responsive factor (ERF) proteins regulate a variety of biotic and abiotic stress responses, and plant development as well. Previously we demonstrated that JERF1, encoding an ERF transcriptional activator, is inducible by ethylene, MeJA, ABA, and NaCl, suggesting its possible regulation in multiple stress responses. In the present paper, we report that expressing JERF1 in tobacco increases the seed germination under mannitol treatment, and enhances the tolerance to high salinity and low temperature, through accumulating sodium in vacuole of leaves and stabilizing the plasma membrane, respectively, and significantly increases the growth of tobacco roots and leaves under salinity and low temperature through an unknown mechanism. The evidence that JERF1 interacts with multiple cis-acting elements, such as GCC-box, DRE, and CE1, to activate the expression of stress-related genes, supports the possible involvement of JERF1 in multiple plant stress responses with ABA-dependent and ABA-independent manner. More importantly, we reveal that expressing JERF1 in tobacco transcriptionally regulates the expression of ABA biosynthesis-related gene NtSDR, resulting in the increase of the ABA content. Together, our results indicate that JERF1 interacts with multiple cis-acting elements and activates the expression of stress responsive and ABA biosynthesis-related genes, consequently causing ABA biosynthesis, and ultimately enhancing tobacco tolerance and growth under high salinity and low temperature.
DOI:10.1104/pp.113.221911URL [本文引用: 1]
DOI:10.3864/j.issn.0578-1752.2019.23.017URL [本文引用: 1]

【Objective】Ethylene response factor (ERF) , a plant-specific transcription factor, is involved in the growth and development of root formation, hypocotyl elongation, fruit ripening, and organ senescence. It also plays a vital role in regulating responses of plant biological and abiotic stress, as well as fruit qualities. In this study, we cloned the apple ethylene response factor MdERF72. Subsequently, a series of expression analysis and functional identification of transgenic apple calli were performed to study its role in abiotic stress responses. These results provided a theoretical basis for exploring the functions of MdERF72 in plant growth and development.【Method】Using Orin apple calli (Malus calli stica Borkh.) as the test material, the MdERF72 was cloned from apple fruits by RT-PCR assay. Bioinformatics methods were used to analyze its amino acid sequence, physicochemical properties genetic relationship, and spatial structure. MEGA5.0 was used to construct the phylogenetic tree for analyzing the homology of its protein sequence with ERF-B2 subfamily in Arabidopsis. The real-time fluorescence PCR (qRT-PCR) assays were performed to analyse the expression of MdERF72 in different organs and tissues of apple, as well as in apple fruits during different developmental stages. Meanwhile, the expression of MdERF72 in Gala apple tissue culture seedlings treated with ACC, NaCl and low-temperature was detected by qRT-PCR assay. We also constructed its overexpression vector and obtained stable overexpression apple calli through Agrobacterium-mediated genetic transformation. The fresh weight, malondialdehyde content, electrical conductivity, hydrogen peroxide content and superoxide anion content of the wild type and transgenic apple calli were detected after NaCl and low temperature treatment. 【Result】 MdERF72 was located on chromosome 13 in apple genome, which had an AP2/ERF domain, unique to ERF family. Phylogenetic tree analyses indicated that the apple MdERF72 exhibited the highest sequence similarity to Arabidopsis AtERF72, and belonged to the B2 subfamily of ERF family. Analysis of amino acid physicochemical properties indicated that MdERF72 encodes 253 amino acids, and its protein molecular weight was predicted as 27.61 kD, the isoelectric point (pI) was 5.10. In addition, the pro-hydrophobic prediction showed that the hydrophobic portion of MdERF72 was larger than the hydrophilic portion, indicating that it belonged to a hydrophobic protein. Phosphorylation site analysis revealed that MdERF72 had only one threonine phosphorylation site, suggesting that the protein might be regulated by phosphorylation. The results revealed that the MdERF72 promoter sequence contains cis-acting elements associated with jasmonic acid (JA), auxin and drought signals. qRT-PCR analysis showed that MdERF72 was a positive regulatory transcription factor of ethylene, which was expressed in all tissues of apple. Its expression in fruits and stems was relatively high, and gradually increased with the fruit ripening. The expression of MdERF72 in apple tissue culture seedlings was significantly induced by high salt and low temperature. Under the treatment of high salt and low temperature stresses, the MdERF72- overexpressing apple calli had stronger growth potential than the wild type control, and the conductivity, malondialdehyde, hydrogen peroxide and superoxide anion content were lower than the wild type control, indicating that MdERF72 increased the resistance to salt and low temperature stresses.【Conclusion】MdERF72 played an important role in the regulation of high salt and low temperature stresses. Overexpression of MdERF72 could increase the resistance of apple calli to high salt and low temperature stresses.
DOI:10.3864/j.issn.0578-1752.2019.23.017URL [本文引用: 1]

【Objective】Ethylene response factor (ERF) , a plant-specific transcription factor, is involved in the growth and development of root formation, hypocotyl elongation, fruit ripening, and organ senescence. It also plays a vital role in regulating responses of plant biological and abiotic stress, as well as fruit qualities. In this study, we cloned the apple ethylene response factor MdERF72. Subsequently, a series of expression analysis and functional identification of transgenic apple calli were performed to study its role in abiotic stress responses. These results provided a theoretical basis for exploring the functions of MdERF72 in plant growth and development.【Method】Using Orin apple calli (Malus calli stica Borkh.) as the test material, the MdERF72 was cloned from apple fruits by RT-PCR assay. Bioinformatics methods were used to analyze its amino acid sequence, physicochemical properties genetic relationship, and spatial structure. MEGA5.0 was used to construct the phylogenetic tree for analyzing the homology of its protein sequence with ERF-B2 subfamily in Arabidopsis. The real-time fluorescence PCR (qRT-PCR) assays were performed to analyse the expression of MdERF72 in different organs and tissues of apple, as well as in apple fruits during different developmental stages. Meanwhile, the expression of MdERF72 in Gala apple tissue culture seedlings treated with ACC, NaCl and low-temperature was detected by qRT-PCR assay. We also constructed its overexpression vector and obtained stable overexpression apple calli through Agrobacterium-mediated genetic transformation. The fresh weight, malondialdehyde content, electrical conductivity, hydrogen peroxide content and superoxide anion content of the wild type and transgenic apple calli were detected after NaCl and low temperature treatment. 【Result】 MdERF72 was located on chromosome 13 in apple genome, which had an AP2/ERF domain, unique to ERF family. Phylogenetic tree analyses indicated that the apple MdERF72 exhibited the highest sequence similarity to Arabidopsis AtERF72, and belonged to the B2 subfamily of ERF family. Analysis of amino acid physicochemical properties indicated that MdERF72 encodes 253 amino acids, and its protein molecular weight was predicted as 27.61 kD, the isoelectric point (pI) was 5.10. In addition, the pro-hydrophobic prediction showed that the hydrophobic portion of MdERF72 was larger than the hydrophilic portion, indicating that it belonged to a hydrophobic protein. Phosphorylation site analysis revealed that MdERF72 had only one threonine phosphorylation site, suggesting that the protein might be regulated by phosphorylation. The results revealed that the MdERF72 promoter sequence contains cis-acting elements associated with jasmonic acid (JA), auxin and drought signals. qRT-PCR analysis showed that MdERF72 was a positive regulatory transcription factor of ethylene, which was expressed in all tissues of apple. Its expression in fruits and stems was relatively high, and gradually increased with the fruit ripening. The expression of MdERF72 in apple tissue culture seedlings was significantly induced by high salt and low temperature. Under the treatment of high salt and low temperature stresses, the MdERF72- overexpressing apple calli had stronger growth potential than the wild type control, and the conductivity, malondialdehyde, hydrogen peroxide and superoxide anion content were lower than the wild type control, indicating that MdERF72 increased the resistance to salt and low temperature stresses.【Conclusion】MdERF72 played an important role in the regulation of high salt and low temperature stresses. Overexpression of MdERF72 could increase the resistance of apple calli to high salt and low temperature stresses.
[本文引用: 1]
DOI:10.1111/pce.12838URL [本文引用: 1]
[本文引用: 1]
DOI:10.1371/journal.pone.0205493URLPMID:30308016 [本文引用: 1]

Drought is one of the main abiotic stresses with far-reaching ecological and socioeconomic impacts, especially in perennial food crops such as Prunus. There is an urgent need to identify drought resilient rootstocks that can adapt to changes in water availability. In this study, we tested the hypothesis that PEG-induced water limitation stress will simulate drought conditions and drought-related genes, including transcription factors (TFs), will be differentially expressed in response to this stress. 'Garnem' genotype, an almond x peach hybrid [P. amygdalus Batsch, syn P. dulcis (Mill.) x P. persica (L.) Batsch] was exposed to PEG-6000 solution, and a time-course transcriptome analysis of drought-stressed roots was performed at 0, 2 and 24 h time points post-stress. Transcriptome analysis resulted in the identification of 12,693 unique differentially expressed contigs (DECs) at the 2 h time point, and 7,705 unique DECs at the 24 h time point after initiation of the drought treatment. Interestingly, three drought-induced genes, directly related to water use efficiency (WUE) namely, ERF023 TF; LRR receptor-like serine/threonine-kinase ERECTA; and NF-YB3 TF, were found induced under stress. The RNAseq results were validated with quantitative RT-PCR analysis of eighteen randomly selected differentially expressed contigs (DECs). Pathway analysis in the present study provides valuable information regarding metabolic events that occur during stress-induced signalling in 'Garnem' roots. This information is expected to be useful in understanding the potential mechanisms underlying drought stress responses and drought adaptation strategies in Prunus species.
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1105/tpc.18.00918URLPMID:31126980 [本文引用: 1]

APETALA2/ETHYLENE RESPONSIVE FACTOR (AP2/ERF) family transcription factors have well-documented functions in stress responses, but their roles in brassinosteroid (BR)-regulated growth and stress responses have not been established. Here, we show that the Arabidopsis (Arabidopsis thaliana) stress-inducible AP2/ERF transcription factor TINY inhibits BR-regulated growth while promoting drought responses. TINY-overexpressing plants have stunted growth, increased sensitivity to BR biosynthesis inhibitors, and compromised BR-responsive gene expression. By contrast, tiny tiny2 tiny3 triple mutants have increased BR-regulated growth and BR-responsive gene expression. TINY positively regulates drought responses by activating drought-responsive genes and promoting abscisic acid-mediated stomatal closure. Global gene expression studies revealed that TINY and BRs have opposite effects on plant growth and stress response genes. TINY interacts with and antagonizes BRASSINOSTERIOID INSENSITIVE1-ETHYL METHANESULFONATE SUPRESSOR1 (BES1) in the regulation of these genes. Glycogen synthase kinase 3-like protein kinase BR-INSENSITIVE2 (BIN2), a negative regulator in the BR pathway, phosphorylates and stabilizes TINY, providing a mechanism for BR-mediated downregulation of TINY to prevent activation of stress responses under optimal growth conditions. Taken together, our results demonstrate that BR signaling negatively regulates TINY through BIN2 phosphorylation and TINY positively regulates drought responses, as well as inhibiting BR-mediated growth through TINY-BES1 antagonistic interactions. Our results thus provide insight into the coordination of BR-regulated growth and drought responses.
DOI:10.1104/pp.111.179945URL [本文引用: 1]

Plant height is a decisive factor in plant architecture. Rice (Oryza sativa) plants have the potential for rapid internodal elongation, which determines plant height. A large body of physiological research has shown that ethylene and gibberellin are involved in this process. The APETALA2 (AP2)/Ethylene-Responsive Element Binding Factor (ERF) family of transcriptional factors is only present in the plant kingdom. This family has various developmental and physiological functions. A rice AP2/ERF gene, OsEATB (for ERF protein associated with tillering and panicle branching) was cloned from indica rice variety 9311. Bioinformatic analysis suggested that this ERF has a potential new function. Ectopic expression of OsEATB showed that the cross talk between ethylene and gibberellin, which is mediated by OsEATB, might underlie differences in rice internode elongation. Analyses of gene expression demonstrated that OsEATB restricts ethylene-induced enhancement of gibberellin responsiveness during the internode elongation process by down-regulating the gibberellin biosynthetic gene, ent-kaurene synthase A. Plant height is negatively correlated with tiller number, and higher yields are typically obtained from dwarf crops. OsEATB reduces rice plant height and panicle length at maturity, promoting the branching potential of both tillers and spikelets. These are useful traits for breeding high-yielding crops.
DOI:10.1093/jxb/eru154URL [本文引用: 1]

In plants, abscission removes senescent, injured, infected, or dispensable organs. Induced by auxin depletion and an ethylene burst, abscission requires pronounced changes in gene expression, including genes for cell separation enzymes and regulators of signal transduction and transcription. However, the understanding of the molecular basis of this regulation remains incomplete. To examine gene regulation in abscission, this study examined an ERF family transcription factor, tomato (Solanum lycopersicum) ETHYLENE-RESPONSIVE FACTOR 52 (SlERF52). SlERF52 is specifically expressed in pedicel abscission zones (AZs) and SlERF52 expression is suppressed in plants with impaired function of MACROCALYX and JOINTLESS, which regulate pedicel AZ development. RNA interference was used to knock down SlERF52 expression to show that SlERF52 functions in flower pedicel abscission. When treated with an abscission-inducing stimulus, the SlERF52-suppressed plants showed a significant delay in flower abscission compared with wild type. They also showed reduced upregulation of the genes for the abscission-associated enzymes cellulase and polygalacturonase. SlERF52 suppression also affected gene expression before the abscission stimulus, inhibiting the expression of pedicel AZ-specific transcription factor genes, such as the tomato WUSCHEL homologue, GOBLET, and Lateral suppressor, which may regulate meristematic activities in pedicel AZs. These results suggest that SlERF52 plays a pivotal role in transcriptional regulation in pedicel AZs at both pre-abscission and abscission stages.
DOI:10.1093/jxb/erw429URLPMID:28204526 [本文引用: 1]

The transcription factor superfamily, APETALA2/ethylene response factor, is involved in plant growth and development, as well as in environmental stress responses. Here, an uncharacterized gene of this family, AtERF019, was studied in Arabidopsis thaliana under abiotic stress situations. Arabidopsis plants overexpressing AtERF019 showed a delay in flowering time of 7 days and a delay in senescence of 2 weeks when comparison with wild type plants. These plants also showed increased tolerance to water deficiency that could be explained by a lower transpiration rate, owing to their smaller stomata aperture and lower cuticle and cell wall permeability. Furthermore, using a bottom-up proteomic approach, proteins produced in response to stress, namely branched-chain-amino-acid aminotransferase 3 (BCAT3) and the zinc finger transcription factor oxidative stress 2, were only identified in plants overexpressing AtERF019. Additionally, a BCAT3 mutant was more sensitive to water-deficit stress than wild type plants. Predicted gene targets of AtERF019 were oxidative stress 2 and genes related to cell wall metabolism. These data suggest that AtERF019 could play a primary role in plant growth and development that causes an increased tolerance to water deprivation, so strengthening their chances of reproductive success.
DOI:10.3864/j.issn.0578-1752.2020.01.013URL [本文引用: 1]

【Objective】The objectives of this research were to identify the Ethylene Response Factor (ERF) family genes from cucumber (Cucumis sativus L.) genome and to know the profile of ERF family such as gene number, gene structure and expression characters in cucumber, so as to provide a theoretical basis for exploring what roles the ERF transcription factors played in female flowers development. 【Method】 The ERF genes in cucumber genome were identified by BLAST software in the 9930_V2 genome database based on Arabidopsis ERF genes. EMBOSS, MEME, TBtools, ExPASy and MEGA 7.0 software were used for carrying out various bioinformatics analysis of ERF. The qRT-PCR was used to detect the expression of cucumber ERF gene family in female buds’ differentiation. 【Result】 Total of 138 ERF genes were identified from cucumber genome, which could be divided into 10 classes. These 138 ERF genes were named from CsERF1 to CsERF138, and the number of amino acids of these ERF genes was between 126 and 745. Multiple sequence alignments and Motif analysis showed that the CsERF gene family had two conserved domains, namely AP2/ERF domain and B3 domain. There were 19 ERF genes showed different expression between FFMMAA and ffMMAA genotype, 9 of them up-regulated in FFMMAA genotype and the other 10 up-regulated in ffMMAA genotype. The expression trend analysis showed 31 ERF genes up-regulated and 30 genes down-regulated in the initial-phase of female buds’ differentiation. Furthermore, it was demonstrated that CsERF9 and CsERF31 could bind the GCC-box. 【Conclusion】 138 ERF gene family members were identified from the cucumber 9930_V2 genome. These entire ERF gene shared AP2/ERF domain. Some of the ERF family members were related to sex determination and female flower development and could bind GCC-box to regulate downstream genes expression.
DOI:10.3864/j.issn.0578-1752.2020.01.013URL [本文引用: 1]

【Objective】The objectives of this research were to identify the Ethylene Response Factor (ERF) family genes from cucumber (Cucumis sativus L.) genome and to know the profile of ERF family such as gene number, gene structure and expression characters in cucumber, so as to provide a theoretical basis for exploring what roles the ERF transcription factors played in female flowers development. 【Method】 The ERF genes in cucumber genome were identified by BLAST software in the 9930_V2 genome database based on Arabidopsis ERF genes. EMBOSS, MEME, TBtools, ExPASy and MEGA 7.0 software were used for carrying out various bioinformatics analysis of ERF. The qRT-PCR was used to detect the expression of cucumber ERF gene family in female buds’ differentiation. 【Result】 Total of 138 ERF genes were identified from cucumber genome, which could be divided into 10 classes. These 138 ERF genes were named from CsERF1 to CsERF138, and the number of amino acids of these ERF genes was between 126 and 745. Multiple sequence alignments and Motif analysis showed that the CsERF gene family had two conserved domains, namely AP2/ERF domain and B3 domain. There were 19 ERF genes showed different expression between FFMMAA and ffMMAA genotype, 9 of them up-regulated in FFMMAA genotype and the other 10 up-regulated in ffMMAA genotype. The expression trend analysis showed 31 ERF genes up-regulated and 30 genes down-regulated in the initial-phase of female buds’ differentiation. Furthermore, it was demonstrated that CsERF9 and CsERF31 could bind the GCC-box. 【Conclusion】 138 ERF gene family members were identified from the cucumber 9930_V2 genome. These entire ERF gene shared AP2/ERF domain. Some of the ERF family members were related to sex determination and female flower development and could bind GCC-box to regulate downstream genes expression.
