 ,1, 杜伟1, 侯思宇1, 王东航1, 冯红梅1, 韩渊怀1, 周美亮2, 张凯旋2, 刘龙龙3, 王俊珍4, 李红英1, 孙朝霞
,1, 杜伟1, 侯思宇1, 王东航1, 冯红梅1, 韩渊怀1, 周美亮2, 张凯旋2, 刘龙龙3, 王俊珍4, 李红英1, 孙朝霞 ,1
,1Identification of ARF Gene Family and Expression Pattern Induced by Auxin in Fagopyrum tataricum
HAO YanRong ,1, DU Wei1, HOU SiYu1, WANG DongHang1, FENG HongMei1, HAN YuanHuai1, ZHOU MeiLiang2, ZHANG KaiXuan2, LIU LongLong3, WANG JunZhen4, LI HongYing1, SUN ZhaoXia
,1, DU Wei1, HOU SiYu1, WANG DongHang1, FENG HongMei1, HAN YuanHuai1, ZHOU MeiLiang2, ZHANG KaiXuan2, LIU LongLong3, WANG JunZhen4, LI HongYing1, SUN ZhaoXia ,1
,1通讯作者:
责任编辑: 李莉
收稿日期:2020-03-25接受日期:2020-06-22网络出版日期:2020-12-01
| 基金资助: |
Received:2020-03-25Accepted:2020-06-22Online:2020-12-01
作者简介 About authors
郝彦蓉,Tel:19834543980;E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (4953KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
郝彦蓉, 杜伟, 侯思宇, 王东航, 冯红梅, 韩渊怀, 周美亮, 张凯旋, 刘龙龙, 王俊珍, 李红英, 孙朝霞. 苦荞ARF基因家族的鉴定及生长素诱导下的表达模式[J]. 中国农业科学, 2020, 53(23): 4738-4749 doi:10.3864/j.issn.0578-1752.2020.23.002
HAO YanRong, DU Wei, HOU SiYu, WANG DongHang, FENG HongMei, HAN YuanHuai, ZHOU MeiLiang, ZHANG KaiXuan, LIU LongLong, WANG JunZhen, LI HongYing, SUN ZhaoXia.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】荞麦(Fagopyrum)是起源于中国的蓼科双子叶杂粮作物,距今已有2 000多年的栽培史[1]。苦荞(Fagopyrum tataricum)是重要的荞麦栽培种,其适应性广,营养价值高,已成为备受关注的“药食同源”作物[2]。苦荞中氨基酸含量均衡,富含人体必需氨基酸,特别是主粮作物较为缺乏的赖氨酸,特有的黄酮类物质——芦丁,更使其在营养保健方面功效显著[3]。但由于苦荞自身的结构特点,植株较高、茎秆中空、脆性强,使得苦荞在生长发育过程中容易受到外界自然环境的影响,发生茎秆倒伏和弯曲,造成收获困难,制约苦荞产量的提升[4,5]。因此,探讨苦荞株高影响因素,可在一定程度上提高苦荞产量,提升苦荞收获指数,促进农民种植积极性。【前人研究进展】苦荞株高由基因型决定,随栽培条件(包括水肥、土壤等条件)、地理环境而变化。植物节间缩短、节间数减少或细胞伸长异常等都与植物激素调控相关[6],研究发现,GA、BR、SLs、IAA等植物激素的合成或信号转导都与植物株高有关[7,8,9,10]。生长素(auxin)作为最早被人们发现的植物激素,具有调节茎的生长速率、抑制侧芽、促进生根等作用[11],参与植物生长调控的大部分过程。而植物对生长素的反应受许多基因的调控[12],生长素响应因子(auxin response factor,ARF)则是其中一类较为重要的转录因子,它能够特异地与生长素响应基因启动子区域的生长素响应元件(auxin response element,AuxRE)TGTCTC结合,激活或抑制基因的表达,从而调控植物的生长发育[13]。ARF基因已在不同物种的基因组中被鉴定出来,最早是在模式植物拟南芥中被发现的,目前已有23个ARF成员[14],随后在水稻[15]、杨树[16]、玉米[17]、番茄[18]、大豆[19]、葡萄[20]等植物中也鉴定出ARF基因家族。WU等[21]研究发现在拟南芥中过表达芒果MiARF2抑制了根和下胚轴的生长。JUNG等[22]研究发现拟南芥突变体dlf-1D叶片变小,生育能力降低,表现出植株矮化,在该突变体中AtARF8的转录水平显著升高。HUANG等[23]研究发现水稻过表达OsmiR160抗性转基因植株的OsARF18,在生长发育中表现出多种缺陷,包括植株矮化、叶片卷曲和种子变小等。【本研究切入点】苦荞基因组测序的完成[24],更有利于苦荞基因的鉴定及功能分析,尽管LIU等[25]对苦荞ARF基因家族已有报道,但其调控下胚轴伸长的机理尚不明确。【拟解决的关键问题】本研究从转录组数据入手,鉴定苦荞ARF基因家族成员并分析其结构特征,分析其在不同组织器官中的表达规律以及研究其在生长素信号途径中的作用,将有助于解析该家族基因的作用机制,也为进一步解析苦荞ARF基因与株高的关系,解决生产上苦荞易倒伏、产量降低的问题提供理论依据。1 材料与方法
1.1 植物材料
以黑丰一号苦荞为试验材料,2019年种植于山西农业大学农作站。大田种植取生长状态良好植株的幼嫩根、茎、新叶、开放花、未成熟籽粒和成熟籽粒于1.5 mL离心管中,液氮速冻后储存于-80℃备用。选取4份苦荞资源PI658429、PI647612、ZNQ189、PI673849进行外源生长素处理(所有材料均来源于山西农业大学)。室内试验在中国农业科学院荞麦基因资源创新研究组进行。1.2 苦荞转录组测序及FtARFs基因筛选
利用RNA-seq对苦荞黑丰一号根、茎、叶、花、未成熟籽粒和成熟籽粒6个组织(每个组织3份重复),共计18个样品进行转录组分析。以“auxin response factor”为关键词,对上述转录组测序结果中NR注释信息进行筛选,挑选出生长素响应因子(ARFs)家族成员,结合pfam注释信息,筛选含有Auxin_resp结构域的基因。以已公布的pinku基因组为参考基因组(
1.3 苦荞ARF基因的结构、理化性质、染色体定位和启动子序列分析
使用NCBI CDD(1.4 系统进化树的构建
利用String网站(1.5 外源激素处理及FtARFs表达量分析
从苦荞转录组数据中获得FtARFs在根、茎、叶、花、未成熟和成熟籽粒中的FPKM值,利用TBtools软件绘制基因表达热图,分析在不同组织中特异表达的基因,为了检测在苦荞茎秆发育过程中,FtARFs的表达是否受到生长素的调控,对苦荞幼苗进行外源生长素处理。田间调查100份苦荞资源的株高与主茎节数,筛选2017—2019年株高较稳定资源4份(高秆2份:ZNQ189与PI673849,矮秆2份:PI658429与PI647612),于MS固体培养基培养7 d后,取3株长势一致的无菌苗于液体MS培养基中,室温震荡(120 r/min)1 d后,加入0.5 mg·L-1 IAA(IAA溶于二甲基亚砜(DMSO)中),于不同时间(0、0.5、1、6、12、24和48 h)对下胚轴取样,对照组添加同体积的二甲基亚砜。取50—100 mg样品加液氮充分研磨后,利用多糖多酚植物总RNA提取试剂盒(天根生化科技北京有限公司)提取上述样品的RNA,使用HiScript? III 1st Strand cDNA Synthesis Kit(+gDNA wiper)试剂盒(南京诺唯赞生物技术有限公司)进行反转录获得cDNA第一链,保存于-20℃备用。以FtHis为内参基因,同时设计FtARFs基因特异引物(表1),以上述cDNA为模板,使用南京诺唯赞生物技术有限公司试剂盒进行qRT-PCR检测,设置生物学重复3次,使用2-ΔΔCt法计算基因的相对表达量。
Table 1
表1
表1引物序列
Table 1
| 基因名称 Gene name | 正向引物 Forward primers (5′-3′) | 反向引物 Reverse primers (5′-3′) |
|---|---|---|
| FtARF1 | AACCCGAGGACACACCTTTC | GTTGCCTTGAATAAGCGGGC |
| FtARF2 | GCTTGTCGGAGATGACCCTT | CTGCACTCCTTCCTTGCTCA |
| FtARF4 | CGGGCTGATGTTACCCATGA | CCTGCACTCGGCAGTTCATA |
| FtARF14 | GCAAATGGGCTTCAACCGAG | CCAAGCCATCCATACCAGCA |
| FtARF16 | CCAACCTTTGTCTCCGCAAG | CGAGGAACAGAAAAACCCCC |
| FtARF17 | CGAGGATTGCTTCCCTCCTT | CTCCATCCTGTCGTGAGCAA |
| FtARF18 | ATAGCATGCACATCGGCCTT | ACTTTGCCAGAGGAACGACA |
| FtARF22 | CCACGATGTGGTAAACGGGA | TTGAGTCCGCACAGAACCTC |
| FtARF23 | ACTGAATGGAAGTTCCGCCA | AACCGCATCCCCTGAAACAA |
| FtHis | ATTCCAGAGGCTTGTTCGTG | CATAATGGTGACACGCTTGG |
新窗口打开|下载CSV
1.6 苦荞下胚轴石蜡切片观察
含有0.5 mg·L-1 IAA的MS固体培养基培养4份材料7 d后,各取10株无菌苗统计下胚轴长度;同时,取1 cm左右下胚轴鲜样,用FAA固定液(甲醛5 mL+冰醋酸5 mL+70%乙醇90 mL)固定24 h以上,参照王茜茹等[26]方法进行石蜡切片,经番红固绿染色后,于LEICA DM500光学显微镜下进行观察。1.7 数据分析
使用Excel 2019办公软件进行数据分析。利用欧氏层次聚类方法,使用TBtools绘制热图。使用GraphPad Prism 5(San Diego California USA,www.graphpad.com)进行单因素方差分析。2 结果
2.1 FtARFs筛选及蛋白理化性质分析
通过对转录组数据中NR注释信息的筛选,共筛选出41个苦荞ARF候选基因,对上述候选基因pfam注释信息进行分析,剔除不含有典型ARF结构域的冗余蛋白后,最终得到26个苦荞ARF基因,并绘制出FtARFs基因在8条染色体上的分布图(图1),结果显示,除Chr.4(第4染色体)外,均有ARF基因分布。其中在Chr.3分布最多,包含5个基因,其次为Chr.1、Chr.2、Chr.5和Chr.8上各有4个基因分布,而在Chr.6和Chr.7仅有2—3个基因分布。根据苦荞ARF基因在染色体上的物理位置分布,按顺序命名为FtARF1—FtARF26。理化性质分析表明,蛋白长度为331—1 083 aa,CDS长度为720—3 252 bp,分子量为37.18—120.49 kD,理论等电点为5.34—8.63。图1
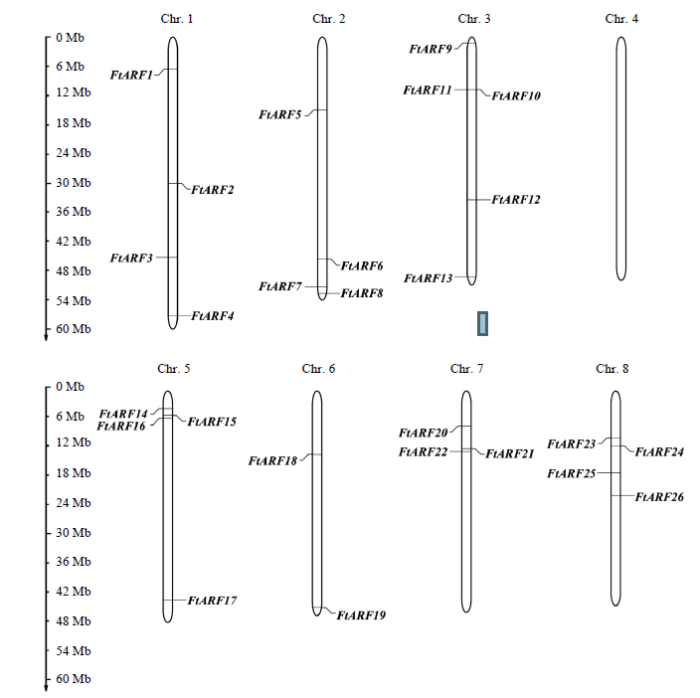 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1FtARFs基因染色体分布图
Fig. 1The distribution of FtARFs on chromosomes
2.2 苦荞ARF系统进化及蛋白结构域、蛋白基序和基因结构分析
系统进化树分析可将苦荞ARF蛋白分为4组,ClassⅠ与ClassⅢ蛋白结构域较完整,除FtARF1、FtARF11、FtARF12、FtARF16和FtARF24缺少C端的Aux/IAA结构域外,其余蛋白均包含3个结构域。ClassⅡ中FtARF23蛋白结构域完整,FtARF9和FtARF17含有2个结构域。ClassⅣ中,所有蛋白均只含有B3结构域和Auxin_resp结构域。进一步对苦荞ARF蛋白保守基序分析,共鉴定了10个motif,其中FtARF19含有6个基序,FtARF11、FtARF12、FtARF16和FtARF22含有7个基序,FtARF9和FtARF15含有8个基序,FtARF24和FtARF26包含9个基序,其余蛋白均含有10个保守基序(图2)。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2苦荞ARF家族的系统发育树、保守结构域和基因结构
Fig. 2The phylogenetic tree, conserved domains and gene structures of ARF family in tartary buckwheat
基因结构分析显示,26个FtARFs基因均含有外显子和内含子,不同苦荞ARF所含外显子数量差异很大,ClassⅠ成员的外显子数量差异较大,有10—15个,ClassⅡ成员的外显子数量为10—12个,ClassⅢ除个别基因含有7和15个外显子外,其余基因均含有14个外显子,ClassⅣ成员的外显子数量最少,为2—4个。这些结果暗示着FtARFs基因的结构差异可能影响着其功能的发挥,因此,有必要对该类基因进行进一步的功能分析。
2.3 不同植物ARF蛋白进化关系
为了充分揭示苦荞ARF基因家族的进化关系,选取拟南芥、水稻、甜荞、甜菜和大豆,6个物种共114个ARF蛋白构建系统进化树(图3)。聚类分析显示,共分为4个类群(GroupⅠ—GroupⅣ),分别含有38、15、38和23个成员,然而不同类群的蛋白在功能上可能存在一定差异,其中,苦荞FtARFs在4个类群中均有分布,如在GroupⅠ中包含8个成员(FtARF1、FtARF3、FtARF5、FtARF10、FtARF11、FtARF12、FtARF20和FtARF22),Group Ⅱ中包含3个成员(FtARF9、FtARF17和FtARF23),Group Ⅲ中包含10个成员(FtARF2、FtARF4、FtARF6、FtARF8、FtARF13、FtARF14、FtARF16、FtARF18、FtARF24和FtARF25),Group Ⅳ中包含5个成员(FtARF7、FtARF15、FtARF19、FtARF22和FtARF26)。同时发现有18个苦荞ARF成员与甜荞ARF高度同源。在Group Ⅰ中,拟南芥AtARF12—AtARF15、AtARF20— AtARF23成员单独聚在一个分支上,在其他5个物种中,均未发现与其高度同源的序列。图3
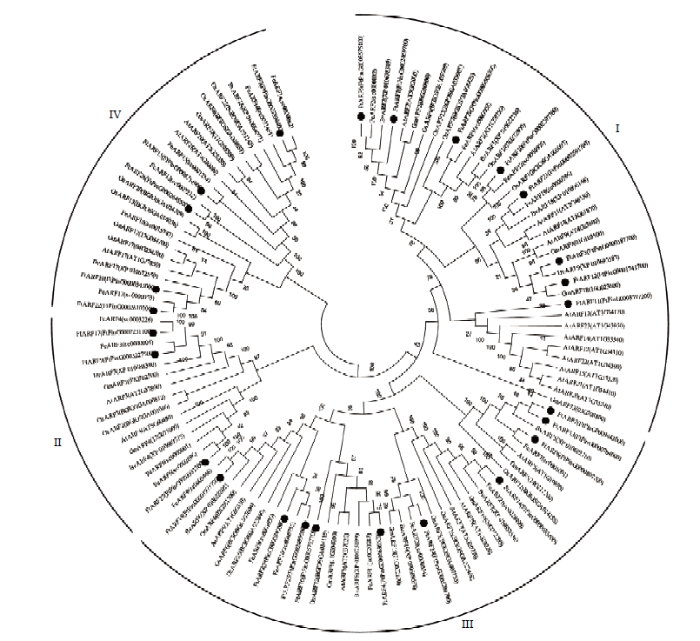 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3苦荞与其他物种ARF蛋白系统进化树
Fig. 3The phylogenetic tree of ARF protein in tartary buckwheat and other species
2.4 苦荞ARF基因的表达分析
为了解苦荞生长发育过程中ARF基因的表达变化,利用6个组织转录组数据的FPKM值绘制热图(图4),发现FtARFs基因具有组织特异性表达。其中,FtARF10在成熟籽粒中特异表达,FtARF19和FtARF26在未成熟籽粒中表达量高,在成熟籽粒中表达量降低,且FtARF10、FtARF19和FtARF26在聚类过程中高度同源,表明其可能在种子成熟发育过程中起重要作用。而另有7个基因在根中呈现高表达水平,这些基因可能参与根系发育及细胞伸长。但在叶中仅有3个基因特异表达,其余基因表达量都较低,可能是因为展开的叶片在促进细胞伸长方面要求较低。同时发现在花中基因表达量较高,有4个基因呈现表达差异。值得注意的是,FtARF1、FtARF2、FtARF4、FtARF14、FtARF16、FtARF17、FtARF18、FtARF22和FtARF23在下胚轴中特异表达,暗示其可能与下胚轴伸长有关。图4
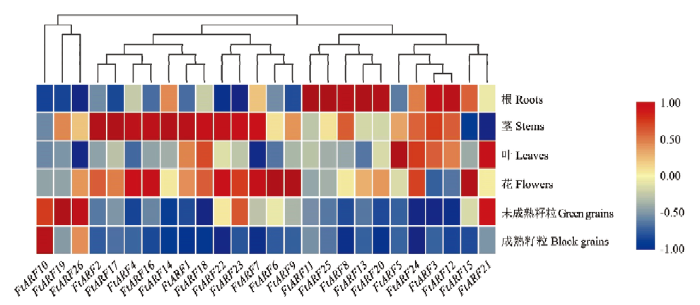 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4苦荞ARF基因组织特异性表达
Fig. 4The tissue specific expression of ARF genes in tartary buckwheat
2.5 苦荞ARF基因家族启动子序列分析
利用在线网站PlantCARE预测9个下胚轴特异表达的FtARFs基因上游2 000 bp序列的顺式作用元件位点(图5),发现其具有多个激素响应元件,包括生长素响应元件、脱落酸响应元件、茉莉酸响应元件、水杨酸响应元件以及赤霉素响应元件。此外,该区域还发现一些其他胁迫响应元件,如光响应元件、干旱响应元件以及低温响应元件等。其中发现5个基因(FtARF1、FtARF2、FtARF4、FtARF16和FtARF18)具有生长素响应元件TGA-element,而4个基因(FtARF14、FtARF17、FtARF22和FtARF23)启动子区域没有生长素响应元件。图5
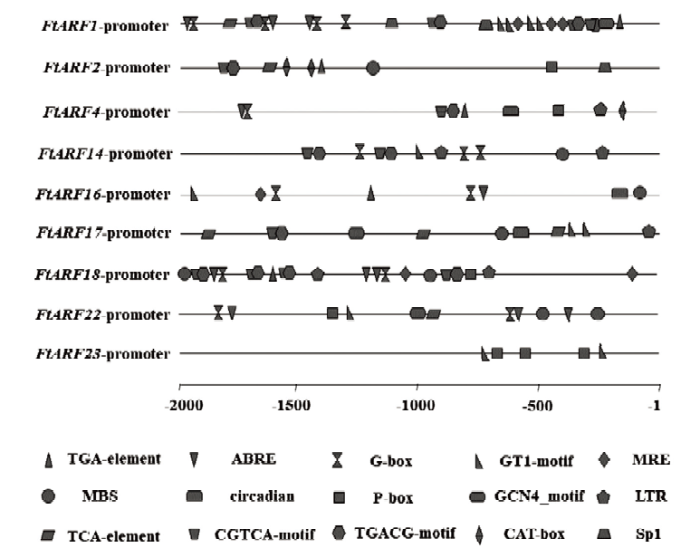 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5苦荞ARF基因启动子元件分析
Fig. 5The analysis of ARF gene promoter elements in tartary buckwheat
2.6 IAA诱导下胚轴长度变异与FtARFs基因表达分析
FtARFs基因的启动子顺式作用元件预测结果表明多个FtARFs基因受生长素诱导。选取株高较高和较低材料各2份,经IAA处理后,2份矮秆材料的下胚轴1个被抑制(PI658429),1个被促进(PI647612),2份高秆材料下胚轴均伸长,但伸长趋势不一,ZNQ189伸长不明显(图6-A和图6-C)。对其下胚轴进行纵向切片并测量其细胞长度(图6-B和图6-D),结果显示,PI658429在生长素处理后细胞长度缩短0.4倍左右,其余3份材料在生长素处理后,细胞长度均伸长0.5倍以上,与幼苗表型变化相一致。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6外源激素IAA处理7 d后苦荞幼苗的生长情况及细胞学观察
A:生长素处理前后幼苗生长情况;B:生长素处理前后下胚轴细胞纵切图,黑色方框代表单个细胞大小;C:幼苗下胚轴长度变化;D:下胚轴细胞长度变化;图中*代表差异显著(P<0.05),**代表差异极显著(P<0.01)
Fig. 6The growth and cytological observation of buckwheat seedlings after 7 days treatment with IAA
A: Growth of seedlings before and after IAA treatment; B: Longitudinal section of hypocotyl cells before and after IAA treatment, and the black box represents the size of a single cell; C: The change in stem length of seedlings; D: The change in length of hypocotyl cells; *above bars indicated significant difference (P<0.05), ** indicated very significant difference (P<0.01)
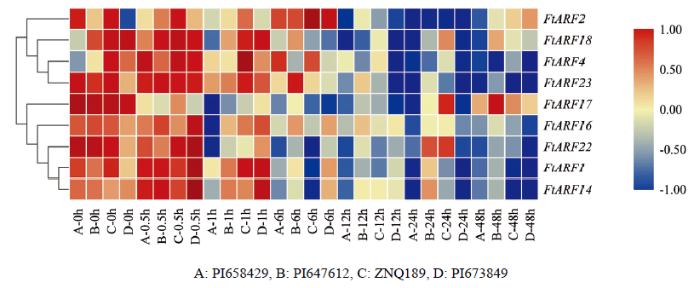
qRT-PCR检测IAA诱导下9个茎秆特异表达的FtARFs基因的相对表达量。结果显示(图7),在IAA处理前期(0.5—1 h),除FtARF2、FtARF17与FtARF22外,其余基因表达水平均升高。FtARF17在未经处理的材料中表达水平较高,而经IAA处理后,表达水平大幅下降,这可能是受到IAA抑制所引起的,并且在启动子元件分析中,该基因缺乏IAA响应元件也有印证。与之规律类似的还有FtARF22,该基因在0.5 h处理点极短响应,随后即出现表达水平大幅降低,推测与缺乏生长素响应元件有关。
图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7下胚轴伸长相关的FtARFs基因受IAA诱导的相对表达量
Fig. 7The IAA induced relative expression of FtARFs related to hypocotyl elongation
值得注意的是,PI658429在IAA处理后,下胚轴被明显抑制;相应地,所检测的FtARFs基因在处理0.5 h后都表现为表达量降低,推测可能是由于该品种无法响应持续的激素信号所引起的。PI647612和PI673849在IAA处理后,下胚轴呈现增长趋势(增幅为1 cm左右),在处理0.5 h后,分别有6个和7个基因表达量提高,其中PI673849材料中FtARF2初期表达水平较低,随后表达水平持续升高,在6 h达到最高值,可部分解释该材料经IAA处理后呈现的下胚轴明显伸长的趋势。ZNQ189在IAA处理后,下胚轴增长但不明显,处理0.5 h时,基因表达差异不显著,在处理1和6 h时,FtARF4和FtARF2表达量都有一定的提高,说明这两个基因响应IAA的诱导。总之,不同材料对生长素响应不同,且大多数FtARFs基因受到IAA诱导,在处理初期(0.5—1 h)表达升高,处理后期表达降低。
3 讨论
近年来,从全基因组水平研究基因家族的分类、序列特点、进化特征和功能预测等已成为生物学分析的一种重要手段。ARF基因家族已被证明调控植物生长发育多个过程,具有重要的生物学功能[27]。本研究鉴定的苦荞ARF蛋白多含有3个保守结构域,结构域之间的关联决定其功能。如,B3结构域可以与ARF基因启动子上的作用元件结合,通过Auxin_resp结构域激活或抑制相关基因的表达,ARF激活子可与Aux /IAA抑制子通过CTD结构域二聚化来抑制基因的表达[28],而缺失的Aux/IAA结构域可能造成功能的多样化。系统进化是研究基因家族功能,预测基因功能的重要工具。由于苦荞分子生物学研究还处在起步期,因此,利用系统进化树对基因功能进行分类就显得尤为重要。本研究中的ARF蛋白聚为4类,该结果与梨[29]和葡萄[20]等研究结果一致,其中Class Ⅳ外显子数量明显少于其他类群,在苹果[30]、梨[29]等物种中也发现了类似现象,且FtARFs基因家族成员外显子数量为2—15个,与拟南芥[14]、葡萄[20]和番茄[18]相似。由此可见,ARF基因家族在结构进化上具有一定的保守性和一致性,这为其功能研究提供了参考依据。同时,对114个ARF蛋白的聚类结果深入分析发现,FtARF20与拟南芥AtARF1,FtARF1和FtARF5与AtARF2遗传距离接近,ELLIS等[31]研究发现AtARF1和AtARF2主要调控叶片衰老、开花时间和种子大小等,预测苦荞中该类基因编码的蛋白可能具有类似功能。FENG等[32]研究发现,拟南芥AtARF11促进侧根形成,在激素信号途径中起关键作用,而FtARF12恰与其高度同源。同样,发现FtARF2、FtARF6、FtARF13与AtARF5聚为一类,预测其与极性生长及细胞扩增有关[33]。FtARF8、FtARF18、FtARF25与AtARF6聚为一类,WU等[34]发现拟南芥AtARF6调控种子心皮发育,促进开花和果实成熟。FtARF14、FtARF24与AtARF19同源性较高,预测其能促进侧根的形成,在激素信号通路中发挥重要作用[35]。值得关注的是,AtARF8能调控下胚轴的伸长,并且是果实起始发育的负调控子[36],本研究发现其同源基因FtARF4和FtARF16为下胚轴特异表达基因,表明极有可能调控下胚轴伸长,值得进一步研究。
作为最广泛的植物激素,生长素对植株的生长发育具有重要作用。TIWARI等[37,38,39]研究发现,生长素以浓度依赖的方式调节植物的生长发育。当生长素浓度低时,Aux/IAA抑制子与ARF转录因子相结合,抑制ARF的活性,当生长素浓度提高时,Aux/IAA被泛素化,ARF转录因子变为有活性的形式,激活或抑制下游基因的表达。CHENG等[40]发现拟南芥特定组织中生长素合成缺陷会造成突变体顶端优势丧失、株高下降、叶片扭曲等发育缺陷,对植株形态产生重要影响。同时,拟南芥AtARF19受到外源生长素和乙烯的诱导,外源生长素浓度会影响miR390的表达,从而影响AtARF4介导的侧根形成[41]。可见,生长素的含量和分布对植株形态建成具有重要影响,因此,研究与生长素含量变化相关的基因,对解析植物生长发育具有指导作用。本研究使用0.5 mg·L-1 IAA处理苦荞幼苗,不同的材料表现出不同的生长状态,PI658429下胚轴变短,其余材料下胚轴呈现出增长趋势,王红飞等[42]研究发现下胚轴伸长与细胞伸长直接相关,本研究对其下胚轴纵向切片发现,PI658429生长素处理后,细胞长度变短,其余材料细胞均伸长,因此推测4份材料在生长素处理后表现出不同的表型变化主要是由于细胞长度变化引起的。在生长素处理0.5 h时,大多数下胚轴特异表达的FtARFs基因表达量上调,在处理前期(0.5—1 h)表达量升高,处理后期表达量降低,盛慧等[43]使用生长素处理黄瓜幼苗发现,ARF基因的表达受到IAA的正调控。LIU等[25]对苦荞ARF研究发现在不同的组织和器官中,FtARFs基因的转录丰度变化很大,且具有组织特异性表达,外源NAA处理试验表明FtARFs基因在果实发育过程中对生长素正响应。同时WALLER等[44]也研究发现外源生长素在15—30 min内上调OsARF1 mRNA的稳态水平。因此,推测FtARFs基因在响应IAA信号诱导苦荞下胚轴伸长中发挥作用。但是发现虽然大多数FtARFs基因对外源生长素处理有反应,但生长素响应元件仅存在于其中5个FtARFs基因的启动子区,因此,FtARFs基因受生长素诱导的机制有待进一步阐明。
4 结论
鉴定出26个苦荞ARF家族基因,其具有组间多样性和组内保守性的特征,且FtARFs具有组织特异性表达,下胚轴长度变化与细胞长度变化相关,推测IAA可能调控苦荞下胚轴的伸长。(责任编辑 李莉)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1023/A:1008741513277URL [本文引用: 1]

The phylogenetic relationships among cultivated landraces and natural populations of wild subspecies of Tatary buckwheat were investigated at the individual level by constructing a phylogenetic tree based on RAPD markers. As the PCR templates, DNA of individuals rather than bulked samples, was used. Ten individuals from 10 cultivated landraces, 71 individuals from 21 natural populations of wild subspecies, and 7 individuals from 3 weedy Tatary buckwheat were provided for RAPD analyses. Three groups were recognized: (1) all cultivated landraces and wild subspecies from central Tibet and northern Pakistan; (2) 10 individuals of wild subspecies from northwestern Yunnan; (3) the remaining individuals of wild subspecies from northwestern Yunnan and all individuals of wild subspecies from Sichuan. Group (2) was phylogenetically closely related to group (1). The origin of cultivated Tatary buckwheat, the hybrid origin of weedy Tatary buckwheat and of the wild populations from central Tibet and northern Pakistan are discussed. We arrive at the conclusion that cultivated Tatary buckwheat probably originated in northwestern Yunnan in China.
DOI:10.1007/s00425-018-03080-4URLPMID:30623242 [本文引用: 1]

MAIN CONCLUSION: Emerging insights in buckwheat molecular genetics allow the integration of genomics driven breeding to revive this ancient crop of immense nutraceutical potential from Asia. Out of several thousand known edible plant species, only four crops-rice, wheat, maize and potato provide the largest proportion of daily nutrition to billions of people. While these crops are the primary supplier of carbohydrates, they lack essential amino acids and minerals for a balanced nutrition. The overdependence on only few crops makes the future cropping systems vulnerable to the predicted climate change. Diversifying food resources through incorporation of orphan or minor crops in modern cropping systems is one potential strategy to improve the nutritional security and mitigate the hostile weather patterns. One such crop is buckwheat, which can contribute to the agricultural sustainability as it grows in a wide range of environments, requires relatively low inputs and possess balanced amino acid and micronutrient profiles. Additionally, gluten-free nature of protein and nutraceutical properties of secondary metabolites make the crop a healthy alternative of wheat-based diet in developed countries. Despite enormous potential, efforts for the genetic improvement of buckwheat are considerably lagged behind the conventional cereal crops. With the draft genome sequences in hand, there is a great scope to speed up the progress of genetic improvement of buckwheat. This article outlines the state of the art in buckwheat research and provides concrete perspectives how modern breeding approaches can be implemented to accelerate the genetic gain. Our suggestions are transferable to many minor and underutilized crops to address the issue of limited genetic gain and low productivity.
DOI:10.1016/j.biotechadv.2020.107650URLPMID:33091484 [本文引用: 1]

Biotechnological application of microalgae cultures at large scale has significant potential in the various fields of biofuels, food and feed, cosmetic, pharmaceutic, environmental remediation and water treatment. Despite this great potential application, industrialisation of microalgae culture and valorisation is still faced with serious remaining challenges in culture scale-up, harvesting and extraction of target molecules. This review presents a general summary of current techniques for harvesting and extraction of biomolecules from microalgae, their relative merits and potential for industrial application. The cell wall composition and its impact on microalgae cell disruption is discussed. Additionally, more recent progress and promising experimental methods and studies are summarised that would allow the reader to further investigate the state of the art. A final survey of energetic assessments of the different techniques is also made. Bead milling and high-pressure homogenisation seem to give clear advantages in terms of target high value compounds extraction from microalgae, with enzyme hydrolysis as a promising emerging technique. Future industrialisation of microalgae for high scale biotechnological processing will require the establishment of universal comparison-standards that would enable easy assessment of one technique against another.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1104/pp.18.00023URLPMID:29545269 [本文引用: 1]

Semidwarfing genes have improved crop yield by reducing height, improving lodging resistance, and allowing plants to allocate more assimilates to grain growth. In wheat (Triticum aestivum), the Rht18 semidwarfing gene was identified and deployed in durum wheat before it was transferred into bread wheat, where it was shown to have agronomic potential. Rht18, a dominant and gibberellin (GA) responsive mutant, is genetically and functionally distinct from the widely used GA-insensitive semidwarfing genes Rht-B1b and Rht-D1b In this study, the Rht18 gene was identified by mutagenizing the semidwarf durum cultivar Icaro (Rht18) and generating mutants with a range of tall phenotypes. Isolating and sequencing chromosome 6A of these
DOI:10.1101/gad.243675.114URLPMID:25085420 [本文引用: 1]

Signaling by the hormones brassinosteroid (BR) and gibberellin (GA) is critical to normal plant growth and development and is required for hypocotyl elongation in response to dark and elevated temperatures. Active BR signaling is essential for GA promotion of hypocotyl growth and suppresses the dwarf phenotype of GA mutants. Cross-talk between these hormones occurs downstream from the DELLAs, as GA-induced destabilization of these GA signaling repressors is not affected by BRs. Here we show that the light-regulated PIF4 (phytochrome-interacting factor 4) factor is a phosphorylation target of the BR signaling kinase BRASSINOSTEROID-INSENSITIVE 2 (BIN2), which marks this transcriptional regulator for proteasome degradation. Expression of a mutated PIF41A protein lacking a conserved BIN2 phosphorylation consensus causes a severe elongated phenotype and strongly up-regulated expression of the gene targets. However, PIF41A is not able to suppress the dwarf phenotype of the bin2-1 mutant with constitutive activation of this kinase. PIFs were shown to be required for the constitutive BR response of bes1-D and bzr1-1D mutants, these factors acting in an interdependent manner to promote cell elongation. Here, we show that bes1-D seedlings are still repressed by the inhibitor BRZ in the light and that expression of the nonphosphorylatable PIF41A protein makes this mutant fully insensitive to brassinazole (BRZ). PIF41A is preferentially stabilized at dawn, coinciding with the diurnal time of maximal growth. These results uncover a main role of BRs in antagonizing light signaling by inhibiting BIN2-mediated destabilization of the PIF4 factor. This regulation plays a prevalent role in timing hypocotyl elongation to late night, before light activation of phytochrome B (PHYB) and accumulation of DELLAs restricts PIF4 transcriptional activity.
DOI:10.1093/pcp/pcq075URLPMID:20498118 [本文引用: 1]

Strigolactones (SLs) are newly discovered plant hormones that regulate plant growth and development including shoot branching. They also stimulate symbiosis with arbuscular mycorrhizal fungi. Rice has at least three genes that are involved in SL synthesis (D10, D17/HTD1 and D27) and at least two genes that are involved in SL signaling (D3) and SL signaling or downstream metabolism (D14/D88/HTD2). We observed that mesocotyl elongation in darkness was greater in rice mutants defective in these genes than in the wild type. Exogenous application of a synthetic SL analog, GR24, rescued the phenotype of mesocotyl elongation in the SL-deficient mutants, d10-1, d17-1 and d27-1, in a dose-dependent manner, but did not affect mesocotyl lengths of the SL-insensitive mutants, d3-1 and d14-1. No significant differences in cell length were found between the d mutants and the wild type, except for some cells on the lower half of the d3-1 mesocotyl that were shortened. On the other hand, the number of cells in the mesocotyls was 3- to 6-fold greater in the d mutants than in the wild type. Treatment with GR24 reduced the number of cells in the d10-1 mesocotyl to the wild-type level, but did not affect the number of cells in the d3-1 and d14-1 mesocotyls. These findings indicate that SLs negatively regulate cell division, but not cell elongation, in the mesocotyl during germination and growth of rice in darkness.
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/s41467-020-14395-wURLPMID:32015349 [本文引用: 1]

Auxin determines the developmental fate of plant tissues, and local auxin concentration is precisely controlled. The role of auxin transport in modulating local auxin concentration has been widely studied but the regulation of local auxin biosynthesis is less well understood. Here, we show that TRYPTOPHAN AMINOTRANSFERASE OF ARABIDOPSIS (TAA1), a key enzyme in the auxin biosynthesis pathway in Arabidopsis thaliana is phosphorylated at Threonine 101 (T101). T101 phosphorylation status can act as an on/off switch to control TAA1-dependent auxin biosynthesis and is required for proper regulation of root meristem size and root hair development. This phosphosite is evolutionarily conserved suggesting post-translational regulation of auxin biosynthesis may be a general phenomenon. In addition, we show that auxin itself, in part via TRANS-MEMBRANE KINASE 4 (TMK4), can induce T101 phosphorylation of TAA1 suggesting a self-regulatory loop whereby local auxin signalling can suppress biosynthesis. We conclude that phosphorylation-dependent control of TAA1 enzymatic activity may contribute to regulation of auxin concentration in response to endogenous and/or external cues.
DOI:10.1093/jxb/erx237URLPMID:28992135 [本文引用: 1]

The phytohormone auxin is involved in almost all developmental processes in land plants. Most, if not all, of these processes are mediated by changes in gene expression. Auxin acts on gene expression through a short nuclear pathway that converges upon the activation of a family of DNA-binding transcription factors. These AUXIN RESPONSE FACTORS (ARFs) are thus the effector of auxin response and translate the chemical signal into the regulation of a defined set of genes. Given the limited number of dedicated components in auxin signaling, distinct properties among the ARF family probably contribute to the establishment of multiple unique auxin responses in plant development. In the two decades following the identification of the first ARF in Arabidopsis, much has been learnt about how these transcription factors act, and how they generate unique auxin responses. Progress in genetics, biochemistry, genomics, and structural biology has helped to develop mechanistic models for ARF action. However, despite intensive efforts, many central questions are yet to be addressed. In this review, we highlight what has been learnt about ARF transcription factors, and identify outstanding questions and challenges for the near future.
DOI:10.1016/j.pbi.2007.08.014URL [本文引用: 1]

Auxin signaling is key to many plant growth and developmental processes from embryogenesis to senescence. Most, if not all, of these processes are initiated and/or mediated through auxin-regulated gene expression. Two types of transcription factor families are required for controlling expression of auxin response genes. One of these, the auxin response factor (ARF) family, functions by binding to auxin response elements (AuxREs) on promoters of auxin response genes, activating or repressing the auxin response genes, and recruiting a second family of transcription factors, the Aux/IAA repressors, that confer an auxin response to the genes. Recent advances have provided information on regulation of ARF gene expression, ARF roles in growth and developmental processes, and target genes regulated by ARFs.
DOI:10.1105/tpc.104.028316URLPMID:15659631 [本文引用: 2]

The AUXIN RESPONSE FACTOR (ARF) gene family products, together with the AUXIN/INDOLE-3-ACETIC ACID proteins, regulate auxin-mediated transcriptional activation/repression. The biological function(s) of most ARFs is poorly understood. Here, we report the identification and characterization of T-DNA insertion lines for 18 of the 23 ARF gene family members in Arabidopsis thaliana. Most of the lines fail to show an obvious growth phenotype except of the previously identified arf2/hss, arf3/ett, arf5/mp, and arf7/nph4 mutants, suggesting that there are functional redundancies among the ARF proteins. Subsequently, we generated double mutants. arf7 arf19 has a strong auxin-related phenotype not observed in the arf7 and arf19 single mutants, including severely impaired lateral root formation and abnormal gravitropism in both hypocotyl and root. Global gene expression analysis revealed that auxin-induced gene expression is severely impaired in the arf7 single and arf7 arf19 double mutants. For example, the expression of several genes, such as those encoding members of LATERAL ORGAN BOUNDARIES domain proteins and AUXIN-REGULATED GENE INVOLVED IN ORGAN SIZE, are disrupted in the double mutant. The data suggest that the ARF7 and ARF19 proteins play essential roles in auxin-mediated plant development by regulating both unique and partially overlapping sets of target genes. These observations provide molecular insight into the unique and overlapping functions of ARF gene family members in Arabidopsis.
DOI:10.1016/j.gene.2007.01.006URL [本文引用: 1]
DOI:10.1186/1471-2229-7-59URL [本文引用: 1]
DOI:10.1186/1471-2164-12-178URL [本文引用: 1]
DOI:10.1007/s00299-011-1113-zURL [本文引用: 2]

Auxin response factors (ARFs) encode transcriptional factors that bind specifically to the TGTCTC-containing auxin response elements found in the promoters of primary/early auxin response genes that regulate plant development. In this study, investigation of the tomato genome revealed 21 putative functional ARF genes (SlARFs), a number comparable to that found in Arabidopsis (23) and rice (25). The full cDNA sequences of 15 novel SlARFs were isolated and delineated by sequencing of PCR products. A comprehensive genome-wide analysis of this gene family is presented, including the gene structures, chromosome locations, phylogeny, and conserved motifs. In addition, a comparative analysis between ARF family genes in tomato and maize was performed. A phylogenetic tree generated from alignments of the full-length protein sequences of 21 OsARFs, 23 AtARFs, 31 ZmARFs, and 21 SlARFs revealed that these ARFs were clustered into four major groups. However, we could not find homologous genes in rice, maize, or tomato with AtARF12-15 and AtARF20-23. The expression patterns of tomato ARF genes were analyzed by quantitative real-time PCR. Our comparative analysis will help to define possible functions for many of these newly isolated ARF-family genes in plant development.
DOI:10.1093/dnares/dst027URL [本文引用: 1]

In plants, the auxin response factor (ARF) transcription factors play important roles in regulating diverse biological processes, including development, growth, cell division and responses to environmental stimuli. An exhaustive search of soybean genome revealed 51 GmARFs, many of which were formed by genome duplications. The typical GmARFs (43 members) contain a DNA-binding domain, an ARF domain and an auxin/indole acetic acid (AUX/IAA) dimerization domain, whereas the remaining eight members lack the dimerization domain. Phylogenetic analysis of the ARFs from soybean and Arabidopsis revealed both similarity and divergence between the two ARF families, as well as enabled us to predict the functions of the GmARFs. Using quantitative real-time polymerase chain reaction (qRT-PCR) and available soybean Affymetrix array and Illumina transcriptome sequence data, a comprehensive expression atlas of GmARF genes was obtained in various organs and tissues, providing useful information about their involvement in defining the precise nature of individual tissues. Furthermore, expression profiling using qRT-PCR and microarray data revealed many water stress-responsive GmARFs in soybean, albeit with different patterns depending on types of tissues and/or developmental stages. Our systematic analysis has identified excellent tissue-specific and/or stress-responsive candidate GmARF genes for in-depth in planta functional analyses, which would lead to potential applications in the development of genetically modified soybean cultivars with enhanced drought tolerance.
DOI:10.1007/s00299-014-1622-7URL [本文引用: 3]

Our study has identified and analyzed the VvARF gene family that may be associated with the development of grape berry and other tissues.
Auxin response factors (ARFs) are transcription factors that regulate the expression of auxin responsive genes through specific binding to auxin response elements (AuxREs). The ARF genes are represented by a large multigene family in plants. Until now, many ARF families have been characterized based on genome resources. However, there is no specialized research about ARF genes in grapevine (Vitis vinifera). In this study, a comprehensive bioinformatics analysis of the grapevine ARF gene family is presented, including chromosomal locations, phylogenetic relationships, gene structures, conserved domains and expression profiles. Nineteen VvARF genes were identified and categorized into four groups (Classes 1, 2, 3 and 4). Most of VvARF proteins contain B3, AUX_RESP and AUX_IAA domains. The VvARF genes were widely expressed in a range of grape tissues, and fruit had higher transcript levels for most VvARFs detected in the EST sources. Furthermore, analysis of expression profiles indicated some VvARF genes may play important roles in the regulation of grape berry maturation processes. This study which provided basic genomic information for the grapevine ARF gene family will be useful in selecting candidate genes related to tissue development in grapevine and pave the way for further functional verification of these VvARF genes.
DOI:10.1007/s11033-010-9990-8URL [本文引用: 1]

An auxin response factor 2 gene, MiARF2, was cloned in our previous study [1] from the cotyledon section of mango (Mangifera indica L. cv. Zihua) during adventitious root formation, which shares an 84% amino acid sequence similarity to Arabidopsis ARF2. This study was to examine the effects of over-expression of the full-length MiARF2 open reading frame on the root and hypocotyl growth in Arabidopsis. Phenotype analysis showed that the T(3) transgenic lines had about 20-30% reduction in the length of hypocotyls and roots of the seedlings in comparison with the wild-type. The transcription levels of ANT and ARGOS genes which play a role in controlling organ size and cell proliferation in the transgenic seedlings also decreased. Therefore, the inhibited root and hypocotyl growth in the transgenic seedlings may be associated with the down-regulated transcription of ANT and ARGOS by the over-expression of MiARF2. This study also suggests that although MiARF2 only has a single DNA-binding domain (DBD), it can function as other ARF-like proteins containing complete DBD, middle region (MR) and carboxy-terminal dimerization domain (CTD).
DOI:10.1007/s10059-010-0055-6URL [本文引用: 1]

Auxin and brassinosteroid (BR) play essential roles in diverse aspects of growth and developmental processes in plants mainly through coordinate regulation of cell division, elongation, and differentiation. Consistent with the overlapped roles, accumulating evidence indicates that the two growth hormones act in a synergistic as well as in an interdependent manner in many cases, although the underlying molecular mechanisms are not fully understood. Here, we demonstrate that auxin and BR signaling pathways are interconnected at the transcriptional level via a negative feedback loop. An Arabidopsis activating tagging mutant dlf-1D exhibited dwarfed growth with small, dark-green leaves and reduced fertility. Hormone feeding assays revealed that the mutant phenotype is caused by the reduction of endogenous BR level. Consistent with this, a gene encoding the CYP72C1 enzyme that catabolizes BR was up-regulated. Notably, the transcript level of the ARF8 transcription factor gene, which modulates the expression of auxin-responsive genes, was significantly elevated in the mutant. In addition, the ARF8 gene expression was significantly reduced by BR but induced by brassinazole, a BR biosynthetic inhibitor. On the other hand, two BR catabolic pathway genes, DLF (CYP72C1) and BAS1, were induced by auxin. Our observations indicate that at least part of auxin and BR signaling pathways are unified through a transcriptional feedback control of the DLF and ARF8 genes.
DOI:10.1038/srep29938URL [本文引用: 1]
DOI:10.1016/j.molp.2017.08.013URLPMID:28866080 [本文引用: 1]

Tartary buckwheat (Fagopyrum tataricum) is an important pseudocereal crop that is strongly adapted to growth in adverse environments. Its gluten-free grain contains complete proteins with a well-balanced composition of essential amino acids and is a rich source of beneficial phytochemicals that provide significant health benefits. Here, we report a high-quality, chromosome-scale Tartary buckwheat genome sequence of 489.3 Mb that is assembled by combining whole-genome shotgun sequencing of both Illumina short reads and single-molecule real-time long reads, sequence tags of a large DNA insert fosmid library, Hi-C sequencing data, and BioNano genome maps. We annotated 33 366 high-confidence protein-coding genes based on expression evidence. Comparisons of the intra-genome with the sugar beet genome revealed an independent whole-genome duplication that occurred in the buckwheat lineage after they diverged from the common ancestor, which was not shared with rosids or asterids. The reference genome facilitated the identification of many new genes predicted to be involved in rutin biosynthesis and regulation, aluminum stress resistance, and in drought and cold stress responses. Our data suggest that Tartary buckwheat's ability to tolerate high levels of abiotic stress is attributed to the expansion of several gene families involved in signal transduction, gene regulation, and membrane transport. The availability of these genomic resources will facilitate the discovery of agronomically and nutritionally important genes and genetic improvement of Tartary buckwheat.
DOI:10.3390/ijms19113526URL [本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1105/tpc.108.060418URLPMID:18647826 [本文引用: 1]
DOI:10.3864/j.issn.0578-1752.2018.02.012URL [本文引用: 2]

【Objective】The objectives of this research are to identify the auxin response factor (ARF) family genes from pear (Pyrus bretschneideri) genome, to know the profile of ARF family such as gene number, gene structure and tissue expression in pear, and to provide theoretical basis for revealing what roles the ARF play in auxin signaling pathway and in growth and development of dwarf pear.【Method】ARF genes in pear genome were identified by BLAST software based on ARF genes from apple and Arabidopsis. SMART, PROSITE, WebLogo 3, DNAMAN 5, MEME, GSDS 2 and MEGA 5.1 software were used for bioinformatics analysis of ARF protein and gene sequences. The qPCR method was used to detect the relative expression of ARF genes in different tissues of dwarf pear ‘Zhongai 1’ and in xylem and phloem of 3 pear cultivars with different growth vigor.【Result】Total of 31 ARF genes were identified from pear genome. All the PbARFs contain two domains of Auxin_resp and B3, and except for PbARF11, 12, 24, 25 and 26, the rest also contain a PB1 domain. Conservative motif analysis result showed that there are 15 motifs in PbARFs, but not every PbARF protein contains all the motifs. The PbARFs were divided into four classes based on phylogenetic analysis. Gene structure analysis result showed that there are 2-15 exons in PbARFs, The gene structure of PbARF is high conservative. The qPCR result showed that all the PbARF genes were expressed in the root, phloem, xylem, leaf, flower and fruit of ‘Zhongai 1’ pear and the expression pattern was various. The relative expression of PbARF29 in the phloem of 3 pear cultivars showed that the more dwarf the plant, the higher the expression, and the relative expression of PbARF16, 17, 18, 27 in the xylem of 3 pear cultivars showed that the more dwarf the plant, the lower the expression.【Conclusion】Auxin response factor family in pear contains 31 genes. All the 31 PbARF proteins contain both of Auxin_resp and B3 conservative domains, and were divided into four classes. The gene structure of PbARF is high conservative. All the 31 PbARFs were expressed in different tissues of ‘Zhongai 1’, root of rootstock Pyrus betulifolia and in phloem and xylem of 3 cultivars. Thereinto, the expression of PbARF29, 16, 17, 18, 27 may be relevant to pear plant height.
DOI:10.3864/j.issn.0578-1752.2018.02.012URL [本文引用: 2]

【Objective】The objectives of this research are to identify the auxin response factor (ARF) family genes from pear (Pyrus bretschneideri) genome, to know the profile of ARF family such as gene number, gene structure and tissue expression in pear, and to provide theoretical basis for revealing what roles the ARF play in auxin signaling pathway and in growth and development of dwarf pear.【Method】ARF genes in pear genome were identified by BLAST software based on ARF genes from apple and Arabidopsis. SMART, PROSITE, WebLogo 3, DNAMAN 5, MEME, GSDS 2 and MEGA 5.1 software were used for bioinformatics analysis of ARF protein and gene sequences. The qPCR method was used to detect the relative expression of ARF genes in different tissues of dwarf pear ‘Zhongai 1’ and in xylem and phloem of 3 pear cultivars with different growth vigor.【Result】Total of 31 ARF genes were identified from pear genome. All the PbARFs contain two domains of Auxin_resp and B3, and except for PbARF11, 12, 24, 25 and 26, the rest also contain a PB1 domain. Conservative motif analysis result showed that there are 15 motifs in PbARFs, but not every PbARF protein contains all the motifs. The PbARFs were divided into four classes based on phylogenetic analysis. Gene structure analysis result showed that there are 2-15 exons in PbARFs, The gene structure of PbARF is high conservative. The qPCR result showed that all the PbARF genes were expressed in the root, phloem, xylem, leaf, flower and fruit of ‘Zhongai 1’ pear and the expression pattern was various. The relative expression of PbARF29 in the phloem of 3 pear cultivars showed that the more dwarf the plant, the higher the expression, and the relative expression of PbARF16, 17, 18, 27 in the xylem of 3 pear cultivars showed that the more dwarf the plant, the lower the expression.【Conclusion】Auxin response factor family in pear contains 31 genes. All the 31 PbARF proteins contain both of Auxin_resp and B3 conservative domains, and were divided into four classes. The gene structure of PbARF is high conservative. All the 31 PbARFs were expressed in different tissues of ‘Zhongai 1’, root of rootstock Pyrus betulifolia and in phloem and xylem of 3 cultivars. Thereinto, the expression of PbARF29, 16, 17, 18, 27 may be relevant to pear plant height.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1242/dev.00925URLPMID:14973283 [本文引用: 1]

Transcription factors of the auxin response factor (ARF) family have been implicated in auxin-dependent gene regulation, but little is known about the functions of individual ARFs in plants. Here, interaction assays, expression studies and combinations of multiple loss- and gain-of-function mutants were used to assess the roles of two ARFs, NONPHOTOTROPIC HYPOCOTYL 4 (NPH4/ARF7) and MONOPTEROS (MP/ARF5), in Arabidopsis development. Both MP and NPH4 interact strongly and selectively with themselves and with each other, and are expressed in vastly overlapping domains. We show that the regulatory properties of both genes are far more related than suggested by their single mutant phenotypes. NPH4 and MP are capable of controlling both axis formation in the embryo and auxin-dependent cell expansion. Interaction of MP and NPH4 in Arabidopsis plants is indicated by their joint requirement in a number of auxin responses and by synergistic effects associated with the co-overexpression of both genes. Finally, we demonstrate antagonistic interaction between ARF and Aux/IAA gene functions in Arabidopsis development. Overexpression of MP suppresses numerous defects associated with a gain-of-function mutation in BODENLOS (BDL)/IAA12. Together these results provide evidence for the biological relevance of ARF-ARF and ARF-Aux/IAA interaction in Arabidopsis plants and demonstrate that an individual ARF can act in both invariantly programmed pattern formation as well as in conditional responses to external signals.
DOI:10.1242/dev.02602URLPMID:17021043 [本文引用: 1]

In flowering plants, diploid sporophytic tissues in ovules and anthers support meiosis and subsequent haploid gametophyte development. These analogous reproductive functions suggest that common mechanisms may regulate ovule and anther development. Two Arabidopsis Auxin Response Factors, ARF6 and ARF8, regulate gynoecium and stamen development in immature flowers. Wild-type pollen grew poorly in arf6 arf8 gynoecia, correlating with ARF6 and ARF8 expression in style and transmitting tract. ARF6 and ARF8 transcripts are cleavage targets of the microRNA miR167, and overexpressing miR167 mimicked arf6 arf8 phenotypes. Mutations in the miR167 target sites of ARF6 or ARF8 caused ectopic expression of these genes in domains of both ovules and anthers where miR167 was normally present. As a result, ovule integuments had arrested growth, and anthers grew abnormally and failed to release pollen. Thus, miR167 is essential for correct patterning of gene expression, and for fertility of both ovules and anthers. The essential patterning function of miR167 contrasts with cases from animals in which miRNAs reinforce or maintain transcriptionally established gene expression patterns.
DOI:10.1002/dvg.22729URL [本文引用: 1]

Patterning of numerous features of plants depends on transduction of the auxin signal. Auxin signaling is mediated by several pathways, the best understood of which relies on the function of the MONOPTEROS (MP) gene. Seven mp mutant alleles have been described in the widely used Columbia background of Arabidopsis: two extensively characterized and five only partially characterized. One of these five mp alleles appears to be extinct and thus unavailable for analysis. We show that two of the four remaining, partially characterized mp alleles reported to be in the Columbia background are in fact not in this background. We extend characterization of the remaining two Columbia alleles of mp, and we identify and characterize four new alleles of mp in the Columbia background, among which the first low-expression allele of mp and the strongest Columbia allele of mp. These genetic resources provide the research community with new experimental opportunities for insight into the function of MP-dependent auxin signaling in plant development. genesis 00:1-9. (c) 2013 Wiley Periodicals, Inc.
DOI:10.1111/j.1365-313X.2004.02220.xURLPMID:15469491 [本文引用: 1]

Auxin response factor (ARF) family genes play a central role in controlling sensitivity to the plant hormone auxin. We characterized the function of ARF8 in Arabidopsis by investigating a T-DNA insertion line (arf8-1) and overexpression lines (ARF8 OX) of ARF8. arf8-1 showed a long-hypocotyl phenotype in either white, blue, red or far-red light conditions, in contrast to ARF8 OX that displayed short hypocotyls in the light. Stronger and weaker apical dominance, and promotion and inhibition of lateral root formation were observed in arf8-1 and ARF8 OX respectively. Sensitivity to auxin was unaltered in arf8-1 hypocotyls with respect to growth inhibition caused by exogenously applied auxin and growth promotion induced by higher temperatures. ARF8 expression was observed constitutively in shoot and root apexes, and was induced in the light condition in hypocotyls. Free IAA contents were approximately 30% reduced in light-grown hypocotyls of ARF8 OX, but were similar between those of arf8-1 and wild type. Expression of the three GH3 genes was reduced in arf8-1 and increased in ARF8 OX, indicating that they are targets of ARF8 transcriptional control. Because the three GH3 proteins may be involved in the conjugation of IAA as suggested by Staswick et al. (2002), and because two of the three GH3 genes are auxin inducible, ARF8 may control the free IAA level in a negative feedback fashion by regulating GH3 gene expression. ARF family genes seem to control both auxin sensitivity and homeostasis in Arabidopsis.
DOI:10.1105/tpc.008417URLPMID:12566590 [本文引用: 1]

Auxin response factors (ARFs) are transcription factors that bind to TGTCTC auxin response elements in promoters of early auxin response genes. ARFs have a conserved N-terminal DNA binding domain (DBD) and in most cases a conserved C-terminal dimerization domain (CTD). The ARF CTD is related in amino acid sequence to motifs III and IV found in Aux/IAA proteins. Just C terminal to the DBD, ARFs contain a nonconserved region referred to as the middle region (MR), which has been proposed to function as a transcriptional repression or activation domain. Results with transfected protoplasts reported here show that ARFs with Q-rich MRs function as activators, whereas most, if not all other ARFs, function as repressors. ARF DBDs alone are sufficient to recruit ARFs to their DNA target sites, and auxin does not influence this recruitment. ARF MRs alone function as activation or repression domains when targeted to reporter genes via a yeast Gal4 DBD, and auxin does not influence the potency of activation or repression. ARF CTDs, along with a Q-rich MR, are required for an auxin response whether the MRs plus CTDs are recruited to a promoter by an ARF DBD or by a Gal4 DBD. The auxin response is mediated by the recruitment of Aux/IAA proteins to promoters that contain a DNA binding protein with a Q-rich MR and an attached CTD.
DOI:10.1105/tpc.105.036236URLPMID:16141189 [本文引用: 1]
DOI:10.1038/446621aURLPMID:17410164 [本文引用: 1]
DOI:10.1101/gad.1415106URLPMID:16818609 [本文引用: 1]

Auxin biosynthesis in plants has remained obscure although auxin has been known for decades as a key regulator for plant growth and development. Here we define the YUC gene family and show unequivocally that four of the 11 predicted YUC flavin monooxygenases (YUC1, YUC2, YUC4, and YUC6) play essential roles in auxin biosynthesis and plant development. The YUC genes are mainly expressed in meristems, young primordia, vascular tissues, and reproductive organs. Overexpression of each YUC gene leads to auxin overproduction, whereas disruption of a single YUC gene causes no obvious developmental defects. However, yuc1yuc4, yuc2yuc6, all of the triple and quadruple mutants of the four YUC genes, display severe defects in floral patterning, vascular formation, and other developmental processes. Furthermore, inactivation of the YUC genes leads to dramatically reduced expression of the auxin reporter DR5-GUS in tissues where the YUC genes are expressed. Moreover, the developmental defects of yuc1yuc4 and yuc1yuc2yuc6 are rescued by tissue-specific expression of the bacterial auxin biosynthesis gene iaaM, but not by exogenous auxin, demonstrating that spatially and temporally regulated auxin biosynthesis by the YUC genes is essential for the formation of floral organs and vascular tissues.
DOI:10.1104/pp.105.070987URLPMID:16461383 [本文引用: 1]

Although auxin response factors (ARFs) are the first well-characterized proteins that bind to the auxin response elements, elucidation of the roles of each ARF gene in auxin responses and plant development has been challenging. Here we show that ARF19 and ARF7 not only participate in auxin signaling, but also play a critical role in ethylene responses in Arabidopsis (Arabidopsis thaliana) roots, indicating that the ARFs serve as a cross talk point between the two hormones. Both arf19 and arf7 mutants isolated from our forward genetic screens are auxin resistant and the arf19arf7 double mutant had stronger auxin resistance than the single mutants and displayed phenotypes not seen in the single mutants. Furthermore, we show that a genomic fragment of ARF19 not only complements arf19, but also rescues arf7. We conclude that ARF19 complements ARF7 at the protein level and that the ARF7 target sequences are also recognized by ARF19. Therefore, it is the differences in expression level/pattern and not the differences in protein sequences between the two ARFs that determines the relative contribution of the two ARFs in auxin signaling and plant development. In addition to being auxin resistant, arf19 has also ethylene-insensitive roots and ARF19 expression is induced by ethylene treatment. This work provides a sensitive genetic screen for uncovering auxin-resistant mutants including the described arf mutants. This study also provides a likely mechanism for coordination and integration of hormonal signals to regulate plant growth and development.
DOI:10.11983/CBB17065URL [本文引用: 1]

作为一种正常的生命现象, 植物下胚轴伸长是长期自然选择的结果, 也是植物进行光合作用、实现自养的必要前提。但下胚轴过度伸长容易造成幼苗徒长, 使植株长势弱, 抗逆能力差, 不利于产量提高和品质改良。该文综述了被子植物下胚轴的发育过程、下胚轴伸长的细胞学机制、植物激素及环境信号调控下胚轴细胞伸长分子机制的最新研究进展, 并展望了未来的研究方向。
DOI:10.11983/CBB17065URL [本文引用: 1]

作为一种正常的生命现象, 植物下胚轴伸长是长期自然选择的结果, 也是植物进行光合作用、实现自养的必要前提。但下胚轴过度伸长容易造成幼苗徒长, 使植株长势弱, 抗逆能力差, 不利于产量提高和品质改良。该文综述了被子植物下胚轴的发育过程、下胚轴伸长的细胞学机制、植物激素及环境信号调控下胚轴细胞伸长分子机制的最新研究进展, 并展望了未来的研究方向。
DOI:10.3864/j.issn.0578-1752.2014.10.012URL [本文引用: 1]

【Objective】 The objective of this study is to identify the cucumber auxin response factor (ARF), forecast the small RNAs and verify the relationship between ARF with small RNAs and auxin. The expression pattern of ARF during seed germination was analyzed and the effect of ARF on the process of parthenocarpy and seed germination was deduced. 【Method】 The Arabidopsis and rice ARF proteins were used to search the cucumber genome database. Then, the retrieved cucumber ARF family structure was analyzed and the small RNAs were predicted. The predicted gma-MIR160o precursur was built into the pCAMBIA2301. By Agrobacterium-mediated method, it was imported into parthenocarpy cucumber varieties. Transgenic plants were validated by RT-PCR. Using real-time RT-PCR method, the expression patterns of ARF were analyzed in auxin treatment, flowing and seed germination. 【Result】 By comparison with the Arabidopsis and rice ARF protein sequences, there are 18 cucumber ARF protein sequences. ARF proteins were divided into four classes. The number of exons varied from 2 to 18, and the structure was similar among the same class. Phylogenetic tree showed that the similarity of 18 genes was not high. Small RNAs corresponds to ARF genes in cucumber were found. Csa010564, Csa011935, Csa015176, Csa020560 and Csa022361 were miR160 targets. Transgenic test further exhibited mRNA abundance of Csa010564, Csa011935 and Csa015176 decreased, while Csa020560 and Csa022361 rose slightly, indicating that Csa010564, Csa011935 and Csa015176 were miR160 targets. The results of real-time RT-PCR exhibited that mRNA abundance of Csa007296, Csa011935 and Csa015176 in roots, stems and leaves were higher than wild type, indicating the expressions of these genes were positively regulated by auxin. Meanwhile, the expressions in leaves and female flowers were decreased on the second flowering day, while the expression of the ovary was the opposite, especially Csa011935 and Csa015176, indicating that ARF genes play a crucial role in the development of the ovary. In the experiment of the auxin treatment, the expressions in different tissues were up-regulated. The expression of ARF genes in the seed germination was analyzed. The results showed that the expression peak of most genes were 12 h and 48 h in imbibition. 【Conclusion】 The ARF genes were regulated by auxin and the corresponding small RNAs. ARF genes may play a key role in the parthenocarpy and seed germination of cucumber .
DOI:10.3864/j.issn.0578-1752.2014.10.012URL [本文引用: 1]

【Objective】 The objective of this study is to identify the cucumber auxin response factor (ARF), forecast the small RNAs and verify the relationship between ARF with small RNAs and auxin. The expression pattern of ARF during seed germination was analyzed and the effect of ARF on the process of parthenocarpy and seed germination was deduced. 【Method】 The Arabidopsis and rice ARF proteins were used to search the cucumber genome database. Then, the retrieved cucumber ARF family structure was analyzed and the small RNAs were predicted. The predicted gma-MIR160o precursur was built into the pCAMBIA2301. By Agrobacterium-mediated method, it was imported into parthenocarpy cucumber varieties. Transgenic plants were validated by RT-PCR. Using real-time RT-PCR method, the expression patterns of ARF were analyzed in auxin treatment, flowing and seed germination. 【Result】 By comparison with the Arabidopsis and rice ARF protein sequences, there are 18 cucumber ARF protein sequences. ARF proteins were divided into four classes. The number of exons varied from 2 to 18, and the structure was similar among the same class. Phylogenetic tree showed that the similarity of 18 genes was not high. Small RNAs corresponds to ARF genes in cucumber were found. Csa010564, Csa011935, Csa015176, Csa020560 and Csa022361 were miR160 targets. Transgenic test further exhibited mRNA abundance of Csa010564, Csa011935 and Csa015176 decreased, while Csa020560 and Csa022361 rose slightly, indicating that Csa010564, Csa011935 and Csa015176 were miR160 targets. The results of real-time RT-PCR exhibited that mRNA abundance of Csa007296, Csa011935 and Csa015176 in roots, stems and leaves were higher than wild type, indicating the expressions of these genes were positively regulated by auxin. Meanwhile, the expressions in leaves and female flowers were decreased on the second flowering day, while the expression of the ovary was the opposite, especially Csa011935 and Csa015176, indicating that ARF genes play a crucial role in the development of the ovary. In the experiment of the auxin treatment, the expressions in different tissues were up-regulated. The expression of ARF genes in the seed germination was analyzed. The results showed that the expression peak of most genes were 12 h and 48 h in imbibition. 【Conclusion】 The ARF genes were regulated by auxin and the corresponding small RNAs. ARF genes may play a key role in the parthenocarpy and seed germination of cucumber .
DOI:10.1023/A:1019818110761URL [本文引用: 1]

We screened for auxin-induced genes with an expression correlated to the auxin-induced growth response from rice coleoptiles by fluorescent differential display. A rice homologue of the auxin response factor (ARF) family of transcriptional regulators, OsARF1, was identified. An OsARF1:GFP fusion protein was localized to the nucleus. Steady-state levels of OsARF1 mRNA correlated positively with auxin-dependent differential growth: gravitropic stimulation enhanced the amount of OsARF1 transcript in the lower, faster-growing flank accompanied by a decrease in the upper flank of gravitropically stimulated rice coleoptiles. Exogenous auxin up-regulated the steady-state level of OsARF1 mRNA within 15–30min. This up-regulation is independent of de novo protein synthesis. Thus, OsARF1 is the first ARF that classifies as an early auxin-responsive gene. The observed auxin-dependent regulation comprises a new level of regulation in auxin-induced gene expression and is discussed as a possible feedback mechanism in plant growth control.
