 ,, �����, ��ʤ��, �Ƽ���, ����, ��ӱ, �����, ����, ������, �����, ������
,, �����, ��ʤ��, �Ƽ���, ����, ��ӱ, �����, ����, ������, �����, ������ ,����ũҵ��ѧ��ѧԺ/��ʩ��ʡ�������������ص�ʵ����/��������ʩ�����Ӧ�ü������ҵط����Ϲ����о����ģ�������,���� 110866
,����ũҵ��ѧ��ѧԺ/��ʩ��ʡ�������������ص�ʵ����/��������ʩ�����Ӧ�ü������ҵط����Ϲ����о����ģ�������,���� 110866Light Regulation of Anthocyanin Biosynthesis in Horticultural Crops
WANG Feng ,, WANG XiuJie, ZHAO ShengNan, YAN JiaRong, BU Xin, ZHANG Ying, LIU YuFeng, XU Tao, QI MingFang, QI HongYan, LI TianLai
,, WANG XiuJie, ZHAO ShengNan, YAN JiaRong, BU Xin, ZHANG Ying, LIU YuFeng, XU Tao, QI MingFang, QI HongYan, LI TianLai ,College of Horticulture, Shenyang Agricultural University/The State Education Ministry and Liaoning Provincial Key Laboratory of Protected Horticulture/National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Shenyang 110866
,College of Horticulture, Shenyang Agricultural University/The State Education Ministry and Liaoning Provincial Key Laboratory of Protected Horticulture/National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Shenyang 110866ͨѶ����:
���α༭: ������
�ո�����:2020-04-6��������:2020-06-30�����������:2020-12-01
| ��������: |
Received:2020-04-6Accepted:2020-06-30Online:2020-12-01
����� About authors
����,E-mail��

ժҪ
�ؼ��ʣ�
Abstract
Keywords��
PDF (743KB)Ԫ������ά�����������������EndNote|Ris|Bibtex�ղر���
�������ø�ʽ
����, �����, ��ʤ��, �Ƽ���, ����, ��ӱ, �����, ����, ������, �����, ������. �����ֲ�ﻨ��������ϳɵĵ�������[J]. �й�ũҵ��ѧ, 2020, 53(23): 4904-4917 doi:10.3864/j.issn.0578-1752.2020.23.015
WANG Feng, WANG XiuJie, ZHAO ShengNan, YAN JiaRong, BU Xin, ZHANG Ying, LIU YuFeng, XU Tao, QI MingFang, QI HongYan, LI TianLai.
���ſ�ѧ����Դ����ʶ�루OSID����

����������Ȼ���й㷺������ֲ���е�ˮ������Ȼɫ��,��ֲ��Ļ���ҶƬ��ʵ�������о��л��ۡ������ؿ���ͨ����������ɫ�ʰ���ֲ����������ͷ����ý��,����ֲ�ﴫ�ۺ�ɢ�����ӡ�����,�����ؾ��н�ǿ�Ŀ���������,���Լ���ֲ����֯���ܻ�������ROS����в�ȵ��˺�,ͬʱ,�ڿ�˥��[1]��������Ѫ�ܼ���[2]�ȷ��������Ҳ�б�����Ч�������ΪӰ��ֲ�ﻨ���غϳɵ���Ҫ��������,�ܵ�Խ��Խ��Ĺ�ע,������Ի���������ϳɵĵ�������,�Ծ�������ֲ�ﲻͬ���ٻ����صĺ���������ղ�ƷӪ���ɷּ����屣���ȷ��������Ҫ���о���Ӧ�ü�ֵ��
1 �����ص�����ϳɴ�л
����������ϳ�;����ֲ�����ͪ;����һ����֧;��,��Ҫ��������������С�һ����˵,��������3����������øA���Ӻ�һ��4-�㶹����øA������ϸ�����н��,���ɲ��ͪ��ø��CHS�����ò������ͪ��ͼ1�������ͪ�����ͪ�칹ø��CHI�����γɻ���ͪ��Ƥ��,����ͪ-3-�ǻ�ø��F3H��������ͪ��Ƥ���γɶ����ͪ����DHK����DHKһ����ֱ�ӱ������ͪ����ԭø��DFR�����γ���ɫ���ÿ���;��һ����,DHK���ɶ����ͪ��-3��-�ǻ�ø��F3��H�������γɶ�����Ƥ�أ�DHQ��,DHQ��DHK�ڶ����ͪ��-3��, 5��-�ǻ�ø��F3��5��H���������γɶ�����÷�أ�DHM����DHQ��DHM��DFR����ֱ��γ���ɫʸ�����غ���ɫ������ء�3����ɫ�������ڻ����غϳ�ø��ANS�������γ����ÿ��ء�ʸ�����غͷ�����ء����ȶ��Ļ�����ͨ�����ͪ-3-O-�����ǻ�ת��ø��UFGT�����γ��ȶ��Ļ����ա�ʸ�������վ��ɼ�ת��ø��OMT�����γ���ҩ����,��������վ���OMT���γ�ǣţ�����պͽ������գ�ͼ1����ͼ1
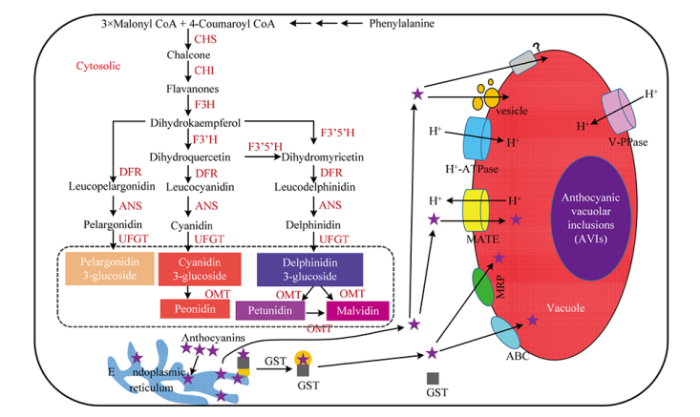 �´��ڴ�|����ԭͼZIP|����PPT
�´��ڴ�|����ԭͼZIP|����PPTͼ1��ֲ�ﻨ��������ϳ�;������ת����ʾ��ͼ
Fig. 1Models for anthocyanin biosynthesis and sequestration of anthocyanin into vacuole in horticultural plants
CHS�����ͪ��ø Chalcone synthase;CHI�����ͪ�칹ø Chalcone isomerase;F3H������ͪ-3��-�ǻ�ø Flavanone-3��-hydroxylase;F3��H�������ͪ��-3��-�ǻ�ø Dihydroflavonoid-3'-hydroxylase;F3��5��H�������ͪ��-3��, 5��-�ǻ�ø Dihydroflavonoid-3', 5'-hydroxylase;DFR�������ͪ����ԭø Dihydroflavonol reductase;ANS�������غϳ�ø Anthocyanin synthase;UFGT�����ն�����-������-���ͪ-3-���ǻ�ת��ø UDP-glucose flavonoid 3-glucosyltransferase;OMT����ת��ø O-methyltransferases;GST��������ת��ø Glutathione S-transferase;MRP����ҩ��ҩ��ص��� Multidrug resistance-associated protein;MATE����ҩ���ж��������ų����嵰�� Multidrug and toxic compound extrusion;ABC��C�͵�ATP��ϵ��� C type of ATP-binding cassette;AVIs����������Һ���ں��� Anthocyanic vacuolar inclusions;Phenylalanine������������;3��Malonyl CoA��3����������øA;4-Coumaroyl CoA��4-�㶹����øA;Chalcone�����ͪ;Flavanones������ͪ;Dihydrokaempferol��DHK�����������ͪ;Dihydroquercetin��DHQ���������ݾ�;Dihydromyricetin��DHM����������÷ͪ;Leucopelargonidin����ɫ���ÿ���;Leucocyanidin����ɫʸ������;Leucodelphinidin����ɫ�������;Pelargonidin�����ÿ���;Cyanidin��ʸ������;Delphinidin���������;Pelargonidin 3-glucoside�����ÿ�����;Cyanidin 3-glucoside��ʸ��������;Delphinidin 3-glucoside�����������;Peonidin����ҩ��������;Petunidin��ǣţ������;Malvidin����������;Anthocyanins��������;Cytosolic��ϸ����;Endoplasmic reticulum��������
�������պϳɺ���Ҫ��ת�˵���ת�����ݵ�Э������Ч����Һ����ת�˲����أ�ͼ1��,�Ӷ���ֹ���������������������Լ���ϸ����ɶ��������ڻ�����������������Һ�ݻ�����Ӱ�컨�����յij�ɫ,�����Ի�����ʹ����ɫ����,�����Ի�����ʹ����ɫ����[3],���,��ȷ�������յ�Һ����ת���Ƽ�����Ҫ���о�����,��������Һ��ת�˵���Ҫ��ʽΪ����GSTЭ���±�����λ��Һ�ݸ���,Һ��Ĥ�ϵ�MRP��ת�˵���ʶ������Ĥת����Һ��[4];��Һ��Ĥ�ϵ�MATE��ת�˵������Ĥת�˵�Һ����,���������Ҫ��H+-ATPase��H+-pyrophosphatase��V-PPase�����ӱý��������䵽Һ���ж�Һ���ữ[5];�����ݰ���,ϸ��Ĥ������������Ĥ�ںϵķ�ʽֱ�ӽ����������䵽����Һ����,������̲�����GST��MRP3����Э��[4]������,ABC��ת����Ҳ���뻨�����յ�Һ�����պ����Ź���[6]��
2 ����������ϳɵ�ת¼����
����������ϳ�;������Ҫ������ת¼ˮƽ���ܶ���ת¼���ӵĵ���,��Ҫ����MYB��bHLH��WDRת¼����,��3��ת¼���ӿ����γ�MBW������[7]�����Ͻ��л����غϳɻ����Ϊ���ںϳɻ���early biosynthetic genes,EBGs���ͺ��ںϳɻ���later biosynthetic genes,LBGs��,EBGs����CHS��CHI��F3H��F3��H,LBGs����DFR��ANS��UFGT[8]��EBGs�ı��ﲻ��MBW���������,��LBGs�ı�����MBW���������[9]��2.1 MYBת¼����
MYB������N�˺��п���DNA��ϵı���MYB�ṹ��,C���ڲ�ͬ���ּ����б���ϴ�,������ת¼��������ƽṹ�������[10]��MYB�ṹ����1-4��R����R repeat�����,���ݺ���R�����������Ϊ1R-MYB��R2R3-MYB��3R-MYB��4R-MYB��������2��R�����R2R3-MYB�������MYBת¼����,Ҳ�Dz���������ͪ��л�ͻ���������ϳɵ���Ҫת¼���ӡ�MYB�������쳣��л�����������MYBʶ������[MYB-recognizing elements,MRE;ʶ��λ��ANCNN(C/A)C]�쳣,�����ܵ��»����ص�����ϳɲ�����[11]��������C1��Colorless-1����ֲ�����������ֵ�R2R3-MYBת¼����,��������������ZmCHS�� ZmDFR�ı���,�Ӷ��ٽ��������صĺϳ�[12]����ǣţ��MYB��ת¼����PhAN2��anthocyanin 2����C�˾���������C1���ƵĽṹ��,��PhAN4��������PhDFR�ı���,�Ӷ��ٽ���ǣţ�����صĺϳ�[13]���밫ǣţPhAN2ͬԴ�������ӽ��ٺϵ�LhMYB6��LhMYB12ͬ�����Դٽ��װٺϻ����صĻ���[14]������о�����,Ұ������SlAN2-likeת¼���ӿ��Լ���SlAN1�ı���,�յ����ѹ�Ƥ�л����صĻ���[15],��SlAN2-likeת¼�����SlMYBATV�ᷴ������SlAN2-like�ı���[16];����,SlAN2-like��ѡ���Լ��жԷ��ѻ����صĻ���Ҳ��Ӱ��[17]���о�����,���Ͻ�MYBת¼����AtPAP1��production of anthocyanin pigment 1������������AtCHS��AtCHI��AtANS�Ȼ���ı���,���Ҵٽ��ǻ�ת��ø����AtUGT78D2��AtUGT75C1�ı���,�յ�ʸ�����ش�������[18]������ֲ���ϻ���ޣ������ޣ�Ͱ���Ҷ����AtPAP1ͬԴ��LAP1��legume anthocyanin production 1��ͬ���ܹ���Ч�شٽ�ʸ�����ػ���[19]��ƻ��MdMYB10��AtPAP1�нϸ�ͬԴ��[20],MdMYB10��ϵ���������������23 bp�Ĵ����ظ�������,��������ת¼,�����ٽ�MdDFR�ı���,ʹƻ�����⻨���ػ���,����ɫ����ƻ����MdMYB10����������ȱ���������,����MdMYB10��ת¼ˮƽ�ϵ�,�ƻ��˻����صĻ���[21]���о�����,MYB10�ڹ�ʵ�������γ����DZ��ص�,��ݮ[22]����ӣ��[23]������[24]����[25]�ȹ�ʵ�������γɾ���MYB10��ء�����,ֲ����һЩ�����MYBҲ�ܹ�Ӱ�컨���صĻ��ۡ������Ͻ�AtMYB113��AtMYB114[9],�����е�GtMYB3[26],�ջ��е�CmMYB6[27],�����е�VvMYBA��VvMYB5a��VvMYB5b[28],ƻ����MdMYB9��MdMYB11��MdMYB110[29,30],�Լ�����SlMYB75[31]��
Ϊ��֤��������ֲ�����ڵĴ�лƽ��,һЩMYB�ڻ���������ϳ������Ÿ��������ӵ����á������Ͻ�AtMYB2������AtF3H��AtDFR��AtANS�ı���[32];����VvMYB4-like����VvDFR��VvANS�ı���[33];ƻ��MdMYB6�����������MdDFR��MdCHS�Ȼ���ı���[34];������ݮFcMYB1������FcANS�ı���[35]����Щ������MYBһ����ֱ��ת¼���ƻ����غϳ�;����ػ���ı���;��һ����ͨ������ػ����غϳɵ����������ӽ��,�������ǵı�����ƻ�MBW��������γɡ�����ж���TLLLFR���ƽṹ���AtMYBL2ת¼����,ͨ����AtTT8��Ͻ�������AtTT8��AtDFR�ı���[36];AtSPL9��squamosa promoter binding protein-like 9��ת¼����ͨ���ƻ�MBW��������ȶ�������AtDFR�ı���[37];�����е�VvMYBC2-L1��VvMYBC2-L3��VvAN1���,�ƻ�MBW��������ȶ���,���ƻ����صĻ���[38]���о�����,��ǣţPhMYB27һ����ͨ����PhAN1����,������Cĩ��EAR��������PhAN1�ı���[39];��һ����ͨ����bHLH��ת¼����PhJAF13��PhAN11��ϻ���,�ƻ�MBW�������γ�,ʹMBW������Ӽ���״̬��Ϊ����״̬,�������ƻ����غϳ�[40]��AtCPC��CAPRICE����PhMYB27��ͬ,����Cĩ�˲������ƻ����R3-MYB,����ֱ����MBW��������,���Ǽ�Ӽ���MBW������Ļ���,�Ӷ����ƻ����ص�����ϳ�[41]��
2.2 bHLHת¼����
bHLH��basic helix-loop-helix��ת¼���Ӿ�����DNA��ϵĸ߶ȱ��ص�bHLH�ṹ��,����ṹ���N����13��18����ˮ������л���ɼ�������basic��,������DNA�Ӵ�,C��ΪHLH��,��Ҫ����ͬԴ����Դ��������γ�,���еĻ�״����Loop,L��������2��������helix,H����bHLHת¼���Ӿ�����MBW������ʶ��л����������ϵ�ת¼���λ�㼰����л���ת¼��������[42]���о�����,��������ص�bHLH�л������Чʶ�����У�bHLH-recognizing elements,BRE��ΪCACN(A/C/T)(G/T) [11]��bHLH�������쳣��л�����������BRE�쳣,�����ܵ��»����غϳ��쳣�����緢�ֵ�һ�����ػ����غϳɵ�bHLH����ת¼�����������е�R1��B1��Lc��Leaf color����Sn[43,44]��ZmLc�ܹ�����ZmCHS��ZmDFR�ı���,�ٽ������ص�����ϳ�[13]��������RͬԴ�����Ͻ�bHLHת¼����ΪAtTT8��transparent testa 8����AtGL3��glabra 3����AtEGL3��enhancer of glabra 3��,����ͨ������AtDFR�ı������ٽ������ص�����ϳ�[45]������ޣMtTT8�������Ͻ�AtTT8ͬԴ��bHLH��ת¼����,������MtLAP1��MtWD40-1�����γ�MBW������,�ٽ������صĺϳ�[46]���о�����,��ǣţ��PhJAF13��PhAN1�ǵ��ػ���������ϳɵ���ҪbHLH���������,���Ǿ�����PhAN2��������DFR�ı���,��PhAN1������ֱ�Ӽ���DFR�ı���[42]��ͬ��,�̲��е�NtAN1a��NtAN2��������NtDFR��NtCHS�ı���[47]������bHLH��������GMYC1��MYBת¼����AN2��GMYB10����,������ڻ��ں���Ƥ��λ�ٽ�DFR�ı���[48];��ɫ�����bHLH�ͻ���GbMYC1��MYBת¼����GbMYB1��ͬ�����ܼ���GbDFR��GbANS������,�յ������غϳ�[49]������,ƻ��MdbHLH3��MdbHLH33��ͬ��MdMYB10ת¼���ӻ���,�ٽ���ʵ��ɫ���[20];��֦LcbHLH1 ��LcbHLH3 ת¼������LcMYB1��ͬ����,�ٽ������ػ���[50];����GtbHLH1��������GtMYB3����,�ٽ��������껨���غϳ�[26];���ްٺ�LhbHLH2����LhMYB6��LhMYB12�����,����LhDFR��LhCHSa��LhCHSb�ı���ٽ������صĺϳ�[14]�����Ͻ������,bHLHת¼���ӳ���ֱ�ӵ���DFR�ı�����,��������MYBת¼���ӻ���,���ػ����صĺϳɡ�
2.3 WDR������
WDR��WD40 repeat proteins,WDR����������б��ض�����Ķ����ظ�����,ÿ���ظ���WD��Ԫ�����40��60��������л����,WDR�ڵ����ʻ���ʱ��Ϊ֧����̶����á���ǣţ��PhAN11�ǵ�1�������ֵ��ػ���������ϳɵ�WDR����,��������PhAN2������,ͨ������PhAN2�ı�����ػ����صĺϳ�[51]�����Ͻ�AtTTG1��transparent testa glabra 1���밫ǣţPhAN11ͬԴ,��һ������bHLH��ת¼����AtGL3������,ֱ�Ӵٽ�AtDFR��AtANS�ı���;��һ����,AtTTG1��AtPAP1��AtGL3/AtEGL3����,�γ�MYB/bHLH/TTG1ת¼������,�������ػ����ص�����ϳ�[9]��������Լ��20������Ӱ���仨���صIJ���,����ZmPAC1��pale aleurone color 1���������ڰ�ǣţPhAN11�����Ͻ�AtTTGl��WDR����,Zmpac1��ȱʧ���»����غ�������[52]���о�����,��÷MrWD40-1��MrMYB1��MrbHLH1�����,������÷�����صĻ���[53];ƻ��MdTTG1��bHLHת¼���Ӻ�MYB ת¼����������γɸ�����,������غϳɻ���MdDFR��MdUFGT��ת¼[54];��ݮWD40ת¼����VcWDL2��VcMYBL1��VcbHLHL1����,���������ݮ��ʵ�����صĺϳ�[55];����,������ʵ�е�CaWD40��Ĭ��,�����Ļ����غ������Լ���[56]������WDR���ڹ����Ա��ﵰ�ס���֯�����Խ���,����һ���������,��������ͻ���Ӱ��MBW��������γɻ���Ķ�λ���źŴ���,�����ƻ���MBW��������ڶ�л���ı�����ء�2.4 �������͵�������
��MYB��bHLH��WDR��,miRNA��JAZ���ס�bZIP����Ҳ������ֲ�ﻨ��������ϳɵĵ��ء��о�����,miR828/TAS4-siR81(-)���뻨��������������PAP1��PAP2��MYB113���,�������ǵı���,�������������Ͻ滨���صĻ���[57]��miR156����λ��SPL9ת¼���ӿ�ʹMBWת¼�������ò��ȶ�,���ո��������Ͻ滨���ص�����ϳ�[37]������,����miR858��������R2R3-MYB�ı���,���ٻ����صĺϳ�[58]�����Ͻ��������ź�ͨ·�е�AtJAZ���ײ���������bHLHs��AtGL3��AtEGL3��AtTT8����R2R3-MYBs��AtPAP1��AtPAP2�������,�谭MBW��������γ�,���ƻ����ص�����ϳ�,���ҿ�����Ϊ���������Ӳ���JA�յ��Ļ����ػ���[59]������,bZIP��ת¼����,��HY5��������ֱ�ӵ���F3H��CHS�Ȼ����ؽṹ����ı���[60],���ҿ��Խ�ϵ�PAP1��MYBL2��MYB10��ת¼������������Ӱ�����ǵı���,ͬʱ������ͨ������miR858�ı���,��ӵ��ػ�������ػ���ı���[61,62,63]������,HY5������PIF3��phytochrome interacting factor 3����bHLH������,��ͬ���ػ����غϳ�;�������ΰл���,�Ӷ��ٽ������صĺϳ�[64]��3 �����ֲ�ﻨ���ص�����ϳ�
����Ӱ��ֲ�ﻨ���غϳɵ�����Ҫ��������֮һ����ֲ����ɫ��֯��ϸ����,��ͨ�������弰��ϵ��Ӵ��ݵ���ֲ�ﻨ���صĺϳ������,�Ӷ�����ֲ����֯���ܻ�������ROS���ȵ�в�ȼ�����ֲ��ɫ����γɡ����,���ĶԹ��źŵ���ֲ�ﻨ���غϳɼ���л�����ܽᣨͼ2��,�����������¡�ͼ2
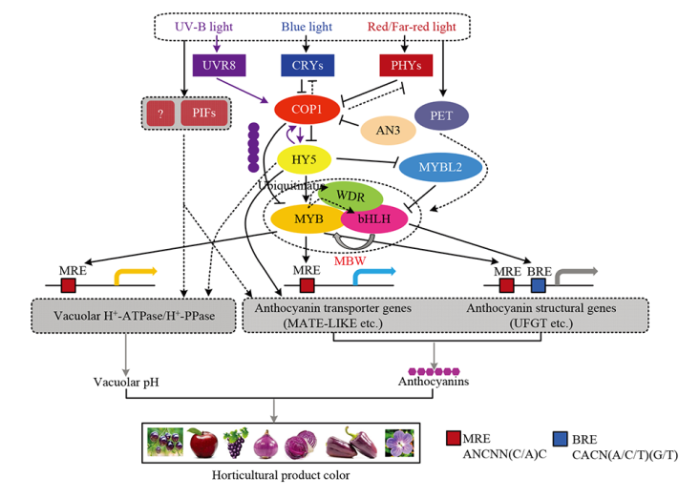 �´��ڴ�|����ԭͼZIP|����PPT
�´��ڴ�|����ԭͼZIP|����PPTͼ2���źŲ�����ֲ�ﻨ���ش�л�ĵ�������
UVR8��UV-B������;CRYs������ɫ��;PHYs������ɫ��;MBW��MYB-bHLH-WDR���ɵĸ�����;COP1������̬�����е�E3��������ø����;HY5�����ź�ת¼����;PIFs������ɫ�ػ�������;AN3�������غϳɵ�������;PET����ϵ��Ӵ���;MRE��MYBʶ������;BRE��bHLHʶ������
Fig. 2Models for light signaling regulation of anthocyanin biosynthesis in horticultural plants
UVR8: UV resistance locus 8; CRYs: crytochrome; PHYs: phytochromes; MBW: MYB-bHLH-WDR complex; COP1: constitutively photomorphogenic 1; HY5: elongated hypocotyl 5; PIFs: phytochrome interacting factors; AN3: ANGUSTIFOLIA3; PET: photosynthetic electron transport; MRE: MYB-recognizing elements; BRE: bHLH-recognizing elements
3.1 ��ǿ����ֲ�ﻨ���ص�����ϳ�
ǿ����Դ̼�����ֲ�ﻨ���ص��γ������[65]�����ѣ�Aft/Aft atv/atv����ʵ��¶�ڹ��²��ֱ��������ֵĻ����غ�����[66];���ջ�����кڰ�������,�仨���ر�����[67]����Ҫԭ���ǹ������߱�������ⰱø��PAL�������ͪ��ø��CHS���������ͪ��4-��ԭø��DFR�������ͪ��������ת��ø��UFGT���Ȼ����غϳ�;���йؼ��ϳ�ø�Ļ���,�����ٽ������صĺϳɺͻ��ۡ��о�����,ǿ����Դٽ�����������ϳ�;���ṹ����͵��ڻ���ı����ǿ��ٽ����Ͻ滨���غϳɽṹ����AtCHS��AtF3H��AtDFR�����ڻ���AtPAP1��AtPAP2�ı���,�Ӷ��ٽ�ֲ�ﻨ���صĺϳ������[68]��ǿ��ٽ���ǣţPhCHS��PhCHI��PhFLS�ı���,�������ڰ���ʹ��ǣţ�����յ�ֲ��Ļ����ؽṹ����������µ�����������,ʹֲ����ְ���dzɫ��[69,70]����ݮ��ʵ��FaCHS��FaCHI��FaF3H��FaDFR��FaANS��FaUFGT�Ƚṹ����ת¼����FaMYB10��FaMYB1�ı����������Ź���ǿ�ȵĽ��Ͷ�����,�����ʹ��ݮ��ɫ�����������غ�������[71]��ǿ������յ����Ѻ�������ֲ��R2R3-MYBת¼����SlAN2��CaMYBa�ı���,���ƺ����ƻ���İ�ǣţPhMYB27�ı���,�Ӷ��ٽ������صĻ���[39]������,ǿ����Դٽ�����SlAN11��SlTT8��SlAN2�����,�γ�MBW������,�����ٽ����ѻ����ؽṹ����ı��P�����صĻ���,��SlMYBL2�������ƻ����غϳɻ���ı���[72]��ǿ���������ƻ��MdMYB1�ı���,�Ӷ��յ������λ���ı���,�ٽ���Ƥ�л����صĻ���[73]������,ǿ�����ͨ����������CRY1��crytochrome 1���յ�HY5�ı���,�����ٽ�ֲ�ﻨ���صĻ���[74]������,��ϵ��Ӵ��ݿ�����ֲ�ﻨ���غϳ���Ҳ������Ҫ������[75],���,���źŶ�ֲ�ﻨ���غϳɵĵ����Ƕ�ͨ·Э���ĸ��ӹ��̡�3.2 ���ʵ���ֲ�ﻨ���ص�����ϳ�
3.2.1 ����⣨UV�������� ��ֲ�ﻨ���غϳɺͻ��۵Ĺ�����,��ͬ���ʶ�ֲ�ﻨ�����γɵĵ������ò�ͬ�����ڴ����ֲ��,�����ߣ�UV���ǻ����ɫ�������ػ��۵���Ҫ����[76]��UV�������ɶ̵������η�ΪUV-C��UV-B��UV-A���о�����,UV-C���Դٽ��ϸ�������������ת��ø����BoSCPL�Լ�R2R3-MYB����ת¼����MYB114��PAP1�ı���,�����ٽ������غϳ�[77]������,UV-C���������÷��ʵ�е�PAL��CHI��4-�㶹����øA����ø��4CL����������⻯ø��C4H���Ļ���,�Ӷ����ӻ����غ����ͪ���ʵĺϳ������[78]���о�����,һ���̶ȵ�UV-B����ͨ����ǿ��ݮ��CHIø��������仨���صĺ���[79]��UV-B��ֲ��������Ҫͨ��������ʽ�յ������صĻ��ۡ�ֲ��UVR8��UV resistance locus 8����֪UV-B�����,�ɶ�������ʽת��Ϊ������ʽ[80]��������ʽUVR8��COP1��SPA�ۺϲ���UVR8-SPA- COP1������[81],���ۼ���ϸ�����С����ָ�����һ��������MYB��bHLH��WDR 3��ת¼������Ӧ,�ٽ�MBW�������γ�,ֱ�ӻ��Ӵٽ������غϳ�;���и�����ı���[82]����UV-B�����յ�ݫ��ҶƬ��CHS�ı���[83],�Լ�����DFR��F3H�ı���[84],�������UVR8��MYB13�Ļ������[85]����һ����,UVR8-SPA-COP1��������ȶ�HY5����[86]��ͼ2��,HY5ת¼���ӿ��Լ���R2R3-MYBת¼����,�ٽ�ֲ�ﻨ���غϳɻ���ı���,�Ӷ��յ������صĻ���[61]����UV-B��,ƻ����MdHY5��MdMYBA�������ӽ��,������ת¼,�����ٽ�ƻ�������صĺϳ�[64]���о�����,��������������������UV�������,��ʵ��ɫ����,�������ӽ���UV-A�����,������ɫ����[87]���о�����,������,UV-A������߷��������ʵ�л����صĻ���[88],ͬʱ,UV-A�����յ���ݼF3H��ANS��CHS��DFR�ı���[89]��ֲ��ͨ������ɫ��CRY����UV-A������[90]���о�����,���Ͻ�cry1ͻ������AtCHS�����µ�,�����ػ��ۼ���[76]��������,CRY1��CRY2������COP1��WD40�����,���������û��Ʋ�ͬ��CRY1��SPA1��suppressor of phyA1������,��SPA1��COP1����,�Ӷ��谭COP1-SPA1����������γ�[91];��CRY2-SPA1��������ǿ��CRY2-COP1�Ļ���,CRY1/2-SPA1����þ�������COP1��E3��������ø����,�Ӷ�ʹCOP1����ת¼���ӵĵ��״����ȶ�״̬[92]���о�����,CRY1��CRY2���Լ�����ź�ת¼����HY5��COL5,�����ٽ�����������ϳɽṹ����MYB10�ı���,�ٽ������صĻ���[86]���о�����,����SmCRYs�ڹ�������SmCOP1�Ļ���,��ʹSmHY5��SmMYB1��ϵ�SmCHS��SmDFR���������ϼ������ǵı���,�����ٽ����ӻ����صĻ���;���ڰ���,SmCRYs��������SmCOP1�Ļ���,SmHY5��SmMYB1��SmCOP1�ж���ͨ��26S���ص���ø��;������,��ֹ������SmMYB1����Ļ����غϳ�;��[93]������о�����,���Ͻ�ת¼����������AtAN3��angustifolia 3�����Խ�ϵ�COP1����������,������COP1�ı���;ȱʧAtAN3��,ֲ��Ļ����غ������Խ���[94]��ͼ2��,��AtAN3�Ƿ�ͨ��������Ӱ��ֲ�ﻨ���صĺϳ��в�������о�����,��������ı���������߷�������Ļ����غ���[95]������ٽ���÷��ʵMrMYB1�������غϳ���ؽṹ����MrCHI��MrF3H��MrF3��H��MrDFR1��MrDFR2��MrANS�ı���,�Ӷ��ٽ���ʵ�л����صĺϳ�[96]��ӣ��������Ͱ�-��-�̹��������������PALø�Ļ���,ʹ��ʵ�еĻ����غ�������[97]������,�������ͨ��PHOT��CRY�������Ӳ�ݮ�л����صĻ���[98,99]�����Ͻ������,�̲�����UV���������ͨ��UVR8��CRY,��ת¼��ת¼����Ӱ��MBW�������������ι��ź�ת¼���ӵı���,����Ӱ��ֲ�ﻨ���ص�����ϳ�,�������û��ƻ�����һ�������о���
3.2.2 ����Զ��� ����Զ�����ֲ�ﻨ���ص�����ϳ���Ҳ������Ҫ���á��о�����,����յ��ѿ��߲���CHS�ı��P�����ӹ�ʵ�л�ͪ�����ʵĺϳ�,�����ٽ������صĺϳ�[100]������,�ߺ��/Զ�������������ķ���,�仨���������ϳ�,���ͺ��/Զ���������,���ѻ����صĺϳ�ȴ�ܵ���������[101,102],����Ҫ�Ǹ߱����ĺ����Դٽ�DFRø�ͻ�ɫ��˫��ø��LDOX������,�����ٽ������صĻ���,��Զ������ࣨ�ͱ����ĺ��/Զ��⣩����DFRø��LDOXø�Ĵ�����[101]������ɫ���Ǹ��ܺ���Զ������Ҫ�����塣�о�����,�������phyB��Ҫͨ�����ַ�ʽ�������ã�ͼ2�������ʹphyB�Ի��Ե�Pfr��ʽ��ϸ���ʽ���ϸ����,һ����ͨ������ɫ��-ת¼����;��ֱ�ӵ��ػ������;��һ����ͨ������ɫ��-COP1;����ӵ��ػ������[103]������Զ����źŵ��ػ����غϳɵĻ����в����,��ΪԶ����������ɫ��A��phyA���ͺ������phyB���ܹ�����COP1�Ļ���,��COP1�����ػ����صĺϳ�[104],������Զ���ı������ͺ��/Զ��������ȴ���ͻ����صĻ���,�����������Զ����phyB�Ķۻ����ô���Զ����phyA�ļ������á�
3.3 �����ڣ�����ʱ������ֲ�ﻨ��������ϳɵĵ���
������Ҳ��Ӱ��ֲ�ﻨ���ص�����ϳɡ��о�����,���ò��⼼���ӳ�����ʱ�����������ֲ�ﻨ���صĻ���[105]�����տ�������̲ݻ���������ϳɻ���ı�����,���ӻ����صĺϳ�,���ڰ��������෴[8]������ݼ���¸����й��մ���ʱ,CHS���������Ź���ʱ����ӳ�������[106]������,�ӳ�����ʱ��������ǿ��Ҷ��ҶƬ��PALø�Ļ���,�ٽ������ص�����ϳ�,ʹ��Ҷɫ���[107]���ڰ���ֲ�ﻨ���غ����ϵͿ�����COP1�й�,��Ϊcop1ͻ�����ںڰ����ܲ���������[104]���ڰ���,COP1��ϸ�����д�������,������ͨ��E3����;���������ι��źż�������������ת¼���ӽ���[92]������,�ڰ���,SPA�µ�PAP1��PAP2ת¼ˮƽ[108]�����,COP1-SPA�������ǹ��ջ�ڰ�����ֲ�ﻨ��������ϳɵ����ĵ�������[104]��Ȼ��,���ǹ���ʱ��Խ��,�����ػ���Խ�ࡣ�������ӽ��ٺϡ�Vivaldi���ںڰ�����������ʱ,�����غ����ܵ�,���պ�Ѹ������,������ʱ�������,�����غ���������[14]�����,��ȷ����ʱ����ֲ�ﻨ���ػ��۵�Ӱ��,�Ժ�������LED����������ֲ�ﻨ���صĻ�������Ҫ���塣����������壨UVR8��CRYs��PHOTs��PHYs����Ҫͨ��COP1��HY5��PIFs�ȹ��ź�����������ֲ�ﻨ���صĺϳ�,��Щ���ź�����һ����ֱ�ӽ�ϵ����ػ����غϳɵ�MYB��bHLH��WDR������ת¼�����ϣ�ͼ2��,ת¼������������ǵı���[61,62,63];��һ����,��Щ���ź�����ͨ����MYB��bHLH��WDR������ת¼���ӵ�����,Ӱ�������γɵ�MBW��������ȶ���[93,104],�������ػ����صĺϳɡ�����,��Щ���ź����ӻ�����ͨ��������MBW��ͨ·���ػ����صĺϳ�[64,65],��HY5ͨ������miR858Ӱ�컨���صĺϳ�[58,60];����,һЩδ֪�Ĺ���Ӧ���ӿ����Բ�����MBWͨ·�ķ�ʽֱ�ӻ��ӵص��ػ����غϳɻ����Һ��Ĥ�ϵ���ת����,�ı�Һ���ữ,���ڻ����صĺϳ�[5,6]��
4 ��������﹤�̸�����ֲ��Ļ�����
4.1 ͨ�����ع��������ֲ�ﻨ����
�����ؼ�Ӱ��ֲ���ɫ��,Ҳ��Ϊ���������ʶ����彡������,���,��������ﻨ���غ�����Ϊ���ƹ�ʵӪ�����ȵ㡣��ǿ�����ʡ�����ʱ���ȹ��,���Ըı���ֲ�ﻨ���صĺ����������ӹ��տ��Դٽ�ƻ����Ƥ���ۻ�����,ʹ��Ƥ���[109]�����ѣ�Aft/Aft atv/atv����ʵ��¶�ڹ��²��ֱ��������ֵĻ����غ�����[66]��ǿ���յ����Ѻ�������SlAN2��CaMYBa����,�ٽ����Ѻ����������صĻ���[39]���������ӽ���UV-A�����,������ɫ����[87]�����ӹ��������ı������Դٽ������л����صĺ���[95]���о�����,����ĵ����Ǹ��������ﻨ���ص���Ҫ�ֶ�,���,LED���⼼����Ϊ����������ɫ��Ĺؼ�������4.2 ͨ�����عؼ�����������﹤�̸���
��ͳ�ӽ����ձ�Ϊ�������ּ�������ֲ�ﻨ���صĸ������ڳ���ͨ���ı���ػ����عؼ�����ı���ˮƽ�����ı仨���ػ���,�ǻ��������﹤�̺ʹ�л���̵�һ����Ҫ���ԡ�������DELILA��ROSEA1ת�뷬�Ѻ�,ת�����ѵĹ�Ƥ�����������ɫ,�����غ���Զ������ݮ[110],�����ķ��Ѷ�������忹��������������Ҫ�Ĺ�Ч��õ�塢����ܰ�;ջ��й�������F3��5��H,����ϳɷ�����س�������ɫ,����˴�ͳ���ּ������������������������ɫ���������[111,112]����PAP1ת���̲��пɲ���1�־���ҩ�ü�ֵ�Ļ�����Cyanidin 3-O-rutinoside,�����ֳɷ�ռ�ܻ����غ�����98%[113]����һ�о�ʹ��ҵ����ȡ��������ҩ�ù��ܵĻ����س�Ϊ����,ͬʱ���̲���Ϊԭ�Ͻ����������ɱ�,����شٽ��˻����ش�л���̺����﹤�̵ķ�չ��5 չ��
������,������������ӹ�ע��������ֲ��ɫ���γɡ������������屣���ȷ��������,������Ϊ�����غϳɵ���Ҫ����Ҳ�ܵ�Խ��Խ��Ĺ�ע��������Ի���������ϳɵĵ�������Ծ�������ֲ�ﲻͬ���ٻ����صĺ�������Ҫ������ѧ���塣������������ģʽֲ���ǣţ�����Ͻ棩�Ի����ص�����ϳ�;�����˽��������,����½����¡�ͼ�����һЩ�������ֻ����غϳɺ���ת;���Ļ���,�����ڹ��źŶ�ֲ�ﻨ��������ϳɼ���ת���ػ��Ƶ��о��Ÿո���,������������ֵ�ý�һ��˼������1����ͬ��������MBW������Ļ������ع�ϵ�в�������Ŀǰ����HY5�Ի����ػ��۵ĵ��ػ���ȡ��һ����չ��,��MBW�����廥�����������ź����Ӽ��书�ܵĽ���,�Թ������źŵ��ػ����ص��ź����������ش�;��2�������Ѿ�������������ػ���������ϳɵ�����������,���为���������о���Խ���,�����ǹ����Щ���������ӵ����û��������ѷ�;��3���������ǻ������ǻ������������ȴ����öԻ����ص��γɺ���������Ҫ����,�����ź��Ƿ�Ӱ����Щ���ι����в����;��4����ֲ���ʵ�л����صĻ���ͨ���ܹ��ա��¶ȵȻ������ع�ͬ���õ�Ӱ��,̽��������������������ڵ��ػ������еĻ�������,�Ժ���������ʩ���������ֶ�,��������ֲ�ﻨ���ص��γ�,������������ֲ�ﻨɫ������ղ�Ʒ��Ӫ����ֵ����Ҫ��ָ�����塣�����α༭ ��������
�ο����� ԭ��˳��
������ȵ���
������������
�����ڿ�Ӱ������
DOI:10.3892/ijmm.2018.3378URLPMID:29328429 [��������: 1]

Oxidative stress is an important contributory factor resulting the development of kidney injury in patients with diabetes. Numerous in vitro and in vivo studies have suggested that anthocyanins, natural phenols commonly existing in numerous fruits and vegetables, exhibit important antioxidative, antiin fl ammatory and antihyperlipidemic effects; however, their effects and underlying mechanisms on diabetic nephropathy (DN) have not yet been fully determined. In the present study, the regulation of apoptosis metabolism and antioxidative effects exhibited by anthocyanins [grape seed procyanidin (GSPE) and cyanidin3Obetaglucoside chloride (C3G)] were investigated, and the molecular mechanism underlying this process was investigated in vivo and in vitro. GSPE administration was revealed to suppress renal cell apoptosis, as well as suppress the expression of Bcl2 in diabetic mouse kidneys. Furthermore, GSPE administration was demonstrated to suppress the expression of thioredoxin interacting protein (TXNIP), in addition to enhancing p38 mitogenactivation protein kinase (MAPK) and extracellular signalregulated kinase 1/2 (ERK1/2) oxidase activity in diabetic mouse kidneys. In vitro experiments using HK2 cells revealed that C3G suppressed the generation of HGmediated reactive oxygen species, cellular apoptosis, the expression of cleaved caspase3 and the Bax/Bcl2 ratio; and enhanced the expression of cytochrome c released from mitochondria. Furthermore, treatment with C3G was revealed to suppress the expression of TXNIP, in addition to the phosphorylation of p38 MAPK and ERK1/2 oxidase activity in HK2 cells under HG conditions. In addition, treatment with C3G was revealed to attenuate the HGinduced suppression of the biological activity of thioredoxin, and to enhance the expression of thioredoxin 2 in HK2 cells under HG conditions. In conclusion, the present study demonstrated that anthocyanins may exhibit protective effects against HGinduced renal injury in DN via antioxidant activity.
DOI:10.1139/cjpp-2016-0667URLPMID:28384410 [��������: 1]

Lingonberry grown in northern Manitoba, Canada, contains exceptionally high levels of anthocyanins and other polyphenols. Previous studies from our lab have shown that lingonberry anthocyanins can protect H9c2 cells from ischemia-reperfusion injury and anthocyanin-rich diets have been shown to be associated with decreased cardiovascular disease and mortality. Oxidative stress can impair function and trigger apoptosis in cardiomyocytes. This study investigated the protective effects of physiologically relevant doses of lingonberry extracts and pure anthocyanins against hydrogen-peroxide-induced cell death. Apoptosis and necrosis were detected in H9c2 cells after hydrogen peroxide treatment via flow cytometry using FLICA 660 caspase 3/7 combined with YO-PRO-1 and then confirmed with Hoechst staining and fluorescence microscopy. Each of the 3 major anthocyanins found in lingonberry (cyanidin-3-galactoside, cyanidin-3-glucoside, and cyanidin-3-arabinoside) was protective against hydrogen-peroxide-induced apoptosis in H9c2 cells at 10 ng.mL(-1) (20 nmol.L(-1)) and restored the number of viable cells to match the control group. A combination of the 3 anthocyanins was also protective and a lingonberry extract tested at 3 concentrations produced a dose-dependent protective effect. Lingonberry anthocyanins protected cardiac cells from oxidative-stress-induced apoptosis and may have cardioprotective effects as a dietary modification.
DOI:10.1038/373291a0URL [��������: 1]
DOI:10.1104/pp.107.105064URLPMID:17921343 [��������: 2]

Plants produce a very large number of specialized compounds that must be transported from their site of synthesis to the sites of storage or disposal. Anthocyanin accumulation has provided a powerful system to elucidate the molecular and cellular mechanisms associated with the intracellular trafficking of phytochemicals. Benefiting from the unique fluorescent properties of anthocyanins, we show here that in Arabidopsis (Arabidopsis thaliana), one route for anthocyanin transport to the vacuole involves vesicle-like structures shared with components of the secretory pathway. By colocalizing the red fluorescence of the anthocyanins with green fluorescent protein markers of the endomembrane system in Arabidopsis seedlings, we show that anthocyanins are also sequestered to the endoplasmic reticulum and to endoplasmic reticulum-derived vesicle-like structures targeted directly to the protein storage vacuole in a Golgi-independent manner. Moreover, our results indicate that vacuolar accumulation of anthocyanins does not depend solely on glutathione S-transferase activity or ATP-dependent transport mechanisms. Indeed, we observed a dramatic increase of anthocyanin-filled subvacuolar structures, without a significant effect on total anthocyanin levels, when we inhibited glutathione S-transferase activity, or the ATP-dependent transporters with vanadate, a general ATPase inhibitor. Taken together, these results provide evidence for an alternative novel mechanism of vesicular transport and vacuolar sequestration of anthocyanins in Arabidopsis.
DOI:10.1104/pp.15.01333URLPMID:26637549 [��������: 2]

Tonoplast transporters, including proton pumps and secondary transporters, are essential for plant cell function and for quality formation of fleshy fruits and ornamentals. Vacuolar transport of anthocyanins, malate, and other metabolites is directly or indirectly dependent on the H(+)-pumping activities of vacuolar H(+)-ATPase (VHA) and/or vacuolar H(+)-pyrophosphatase, but how these proton pumps are regulated in modulating vacuolar transport is largely unknown. Here, we report a transcription factor, MdMYB1, in apples that binds to the promoters of two genes encoding the B subunits of VHA, MdVHA-B1 and MdVHA-B2, to transcriptionally activate its expression, thereby enhancing VHA activity. A series of transgenic analyses in apples demonstrates that MdMYB1/10 controls cell pH and anthocyanin accumulation partially by regulating MdVHA-B1 and MdVHA-B2. Furthermore, several other direct target genes of MdMYB10 are identified, including MdVHA-E2, MdVHP1, MdMATE-LIKE1, and MdtDT, which are involved in H(+)-pumping or in the transport of anthocyanins and malates into vacuoles. Finally, we show that the mechanism by which MYB controls malate and anthocyanin accumulation in apples also operates in Arabidopsis (Arabidopsis thaliana). These findings provide novel insights into how MYB transcription factors directly modulate the vacuolar transport system in addition to anthocyanin biosynthesis, consequently controlling organ coloration and cell pH in plants.
DOI:10.1016/B978-0-12-407695-2.00009-3URLPMID:23890387 [��������: 2]

The vacuole is the largest compartment in plant cells, often occupying more than 80% of the total cell volume. This organelle accumulates a large variety of endogenous ions, metabolites, and xenobiotics. The compartmentation of divergent substances is relevant for a wide range of biological processes, such as the regulation of stomata movement, defense mechanisms against herbivores, flower coloration, etc. Progress in molecular and cellular biology has revealed that a large number of transporters and channels exist at the tonoplast. In recent years, various biochemical and physiological functions of these proteins have been characterized in detail. Some are involved in maintaining the homeostasis of ions and metabolites, whereas others are related to defense mechanisms against biotic and abiotic stresses. In this review, we provide an updated inventory of vacuolar transport mechanisms and a comprehensive summary of their physiological functions.
DOI:10.1016/j.tplants.2004.12.011URLPMID:15708343 [��������: 1]

A protein complex composed of MYB and bHLH transcription factors associated with a WD40 repeat protein initiates multiple cellular differentiation pathways in a range of plants. Recent reports have provided the first coherent models of the network of interactions that lead to diverse cell fates through the activity of this protein complex. The resulting flexibility in plant morphology is likely to have played a major role in angiosperm evolution and success. The complex appears to have arisen in the land plant lineage, although its component parts are considerably more ancient. Here, we review the evolutionary history of the MYB-bHLH-WD40 protein complex and its role in generating plant epidermal cellular diversity.
DOI:10.3389/fchem.2018.00052URLPMID:29594099 [��������: 2]

Anthocyanins are a group of polyphenolic pigments that are ubiquitously found in the plant kingdom. In plants, anthocyanins play a role not only in reproduction, by attracting pollinators and seed dispersers, but also in protection against various abiotic and biotic stresses. There is accumulating evidence that anthocyanins have health-promoting properties, which makes anthocyanin metabolism an interesting target for breeders and researchers. In this review, the state of the art knowledge concerning anthocyanins in the Solanaceous vegetables, i.e., pepper, tomato, eggplant, and potato, is discussed, including biochemistry and biological function of anthocyanins, as well as their genetic and environmental regulation. Anthocyanin accumulation is determined by the balance between biosynthesis and degradation. Although the anthocyanin biosynthetic pathway has been well-studied in Solanaceous vegetables, more research is needed on the inhibition of biosynthesis and, in particular, the anthocyanin degradation mechanisms if we want to control anthocyanin content of Solanaceous vegetables. In addition, anthocyanin metabolism is distinctly affected by environmental conditions, but the molecular regulation of these effects is poorly understood. Existing knowledge is summarized and current gaps in our understanding are highlighted and discussed, to create opportunities for the development of anthocyanin-rich crops through breeding and environmental management.
DOI:10.1111/j.1365-313X.2007.03373.xURLPMID:18036197 [��������: 3]

In all higher plants studied to date, the anthocyanin pigment pathway is regulated by a suite of transcription factors that include Myb, bHLH and WD-repeat proteins. However, in Arabidopsis thaliana, the Myb regulators remain to be conclusively identified, and little is known about anthocyanin pathway regulation by TTG1-dependent transcriptional complexes. Previous overexpression of the PAP1 Myb suggested that genes from the entire phenylpropanoid pathway are targets of regulation by Myb/bHLH/WD-repeat complexes in Arabidopsis, in contrast to other plants. Here we demonstrate that overexpression of Myb113 or Myb114 results in substantial increases in pigment production similar to those previously seen as a result of over-expression of PAP1, and pigment production in these overexpressors remains TTG1- and bHLH-dependent. Also, plants harboring an RNAi construct targeting PAP1 and three Myb candidates (PAP2, Myb113 and Myb114) showed downregulated Myb gene expression and obvious anthocyanin deficiencies. Correlated with these anthocyanin deficiencies is downregulation of the same late anthocyanin structural genes that are downregulated in ttg1 and bHLH anthocyanin mutants. Expression studies using GL3:GR and TTG1:GR fusions revealed direct regulation of the late biosynthetic genes only. Functional diversification between GL3 and EGL3 with regard to activation of gene targets was revealed by GL3:GR studies in single and double bHLH mutant seedlings. Expression profiles for Myb and bHLH regulators are also presented in the context of pigment production in young seedlings.
DOI:10.1016/j.tplants.2010.06.005URL [��������: 1]

The MYB family of proteins is large, functionally diverse and represented in all eukaryotes. Most MYB proteins function as transcription factors with varying numbers of MYB domain repeats conferring their ability to bind DNA. In plants, the MYB family has selectively expanded, particularly through the large family of R2R3-MYB. Members of this family function in a variety of plant-specific processes, as evidenced by their extensive functional characterization in Arabidopsis (Arabidopsis thaliana). MYB proteins are key factors in regulatory networks controlling development, metabolism and responses to biotic and abiotic stresses. The elucidation of MYB protein function and regulation that is possible in Arabidopsis will provide the foundation for predicting the contributions of MYB proteins to the biology of plants in general.
DOI:10.1093/jxb/erv173URLPMID:25911741 [��������: 2]

Cellular activities such as compound synthesis often require the transcriptional activation of an entire pathway; however, the molecular mechanisms underlying pathway activation have rarely been explained. Here, the cis regulatory architecture of the anthocyanin pathway genes targeted by the transcription factor (TF) complex including MYB, bHLH, and WDR was systematically analysed in one species and the findings extended to others. In Ipomoea purpurea, the IpMYB1-IpbHLH2-IpWDR1 (IpMBW) complex was found to be orthologous to the PAP1-GL3-TTG1 (AtPGT) complex of Arabidopsis thaliana, and interacted with a 7-bp MYB-recognizing element (MRE) and a 6-bp bHLH-recognizing element (BRE) at the proximal promoter region of the pathway genes. There was little transcription of the gene in the absence of the MRE or BRE. The cis elements identified experimentally converged on two syntaxes, ANCNNCC for MREs and CACN(A/C/T)(G/T) for BREs, and our bioinformatic analysis showed that these were present within anthocyanin gene promoters in at least 35 species, including both gymnosperms and angiosperms. For the anthocyanin pathway, IpMBW and AtPGT recognized the interspecific promoters of both early and later genes. In A. thaliana, the seed-specific TF complex (TT2, TT8, and TTG1) may regulate all the anthocyanin pathway genes, in addition to the proanthocyanidin-specific BAN. When multiple TF complexes in the anthocyanin pathway were compared, the cis architecture played a role larger than the TF complex in determining the variation in promoter activity. Collectively, a cis logic common to the pathway gene promoters was found, and this logic is essential for the trans factors to regulate the pathway.
URLPMID:3428265 [��������: 1]

The structure of the wild-type c1 locus of Zea mays was determined by sequence analysis of one genomic and two cDNA clones. The coding region is composed of three exons (150 bp, 129 bp and one, at least 720 bp) and two small introns (88 bp and 145 bp). Transcription of the mRNAs corresponding to the two cDNA clones cLC6 (1.1 kb) and cLC28 (2.1 kb) starts from the same promoter. Both cDNAs are identical except that cLC28 extends further at its 3' end. A putative protein, 273 amino acids in length was deduced from the sequence of both transcripts. It contains two domains, one basic and the other acidic and might function as a transcriptional activator. The basic domain of this c1-encoded protein shows 40% sequence homology to the protein products of animal myb proto-oncogenes.
DOI:10.1105/tpc.5.11.1497URLPMID:12271045 [��������: 2]

In this study, we demonstrate that in petunia at least four regulatory genes (anthocyanin-1 [an1], an2, an4, and an11) control transcription of a subset of structural genes from the anthocyanin pathway by using a combination of RNA gel blot analysis, transcription run-on assays, and transient expression assays. an2- and an11- mutants could be transiently complemented by the maize regulatory genes Leaf color (Lc) or Colorless-1 (C1), respectively, whereas an1- mutants only by Lc and C1 together. In addition, the combination of Lc and C1 induces pigment accumulation in young leaves. This indicates that Lc and C1 are both necessary and sufficient to produce pigmentation in leaf cells. Regulatory pigmentation genes in maize and petunia control different sets of structural genes. The maize Lc and C1 genes expressed in petunia differentially activate the promoters of the chalcone synthase genes chsA and chsJ in the same way that the homologous petunia genes do. This suggests that the regulatory proteins in both species are functionally similar and that the choice of target genes is determined by their promoter sequences. We present an evolutionary model that explains the differences in regulation of pigmentation pathways of maize, petunia, and snapdragon.
DOI:10.1093/pcp/pcq011URLPMID:20118109 [��������: 3]

Anthocyanins are secondary metabolites that contribute to colors of flowers, fruits and leaves. Asiatic hybrid lily (Lilium spp.) accumulates cyanidin anthocyanins in flower tepals, tepal spots and leaves of juvenile shoots. To clarify their mechanisms of regulation of anthocyanin pigmentation, two full-length cDNAs of R2R3-MYB (LhMYB6 and LhMYB12) were isolated from the anthocyanin-accumulating tepals of cultivar 'Montreux'. Analysis of the deduced amino acid sequences indicated they have homology with petunia AN2, homologous sequences of which had not been isolated in species of monocots. Yeast two-hybrid analysis showed that LhMYB6 and LhMYB12 interacted with the Lilium hybrid basic helix-loop-helix 2 (LhbHLH2) protein. Transient expression analysis indicated that co-expression of LhMYB6 and LhbHLH2 or LhMYB12 and LhbHLH2, introduced by a microprojectile, activated the transcription of anthocyanin biosynthesis genes in lily bulbscales. Spatial and temporal transcription of LhMYB6 and LhMYB12 was analyzed. The expression of LhMYB12 corresponded well with anthocyanin pigmentation in tepals, filaments and styles, and that of LhMYB6 correlated with anthocyanin spots in tepals and light-induced pigmentation in leaves. These results indicate that LhMYB6 and LhMYB12 positively regulate anthocyanin biosynthesis and determine organ- and tissue-specific accumulation of anthocyanin.
DOI:10.1016/j.molp.2019.10.010URLPMID:31678614 [��������: 1]

Dietary anthocyanins are important health-promoting antioxidants that make a major contribution to the quality of fruits. It is intriguing that most tomato cultivars do not produce anthocyanins in fruit. However, the purple tomato variety Indigo Rose, which has the dominant Aft locus combined with the recessive atv locus from wild tomato species, exhibits light-dependent anthocyanin accumulation in the fruit skin. Here, we report that Aft encodes a functional anthocyanin activator named SlAN2-like, while atv encodes a nonfunctional version of the anthocyanin repressor SlMYBATV. The expression of SlAN2-like is responsive to light, and the functional SlAN2-like can activate the expression of both anthocyanin biosynthetic genes and their regulatory genes, suggesting that SlAN2-like acts as a master regulator in the activation of anthocyanin biosynthesis. We further showed that cultivated tomatoes contain nonfunctional alleles of SlAN2-like and therefore fail to produce anthocyanins. Consistently, expression of a functional SlAN2-like gene driven by the fruit-specific promoter in a tomato cultivar led to the activation of the entire anthocyanin biosynthesis pathway and high-level accumulation of anthocyanins in both the peel and flesh. Taken together, our study exemplifies that efficient engineering of complex metabolic pathways could be achieved through tissue-specific expression of master transcriptional regulators.
DOI:10.1111/nph.16272URLPMID:31625612 [��������: 1]

Anthocyanin fruit (Aft) and atroviolacea (atv) were characterized in wild tomato and can enhance anthocyanin content in tomato fruit. However, the gene underlying the Aft locus and the mechanism by which Aft and atv act remain largely unknown. In this study, the Aft locus was fine-mapped to an approximately 145-kb interval on chromosome 10, excluding SlAN2 (Solyc10g086250), SlANT1 (Solyc10g086260) and SlANT1-like (Solyc10g086270), which have previously been suggested as candidates. Thus, the R2R3-MYB transcription factor SlAN2-like (Solyc10g086290) was considered the best candidate gene for Aft. The CRISPR/Cas9-mediated SlAN2-like mutants show a much lower accumulation of anthocyanins associated with the downregulation of multiple anthocyanin-related genes compared to the wild-type tomato, indicating that SlAN2-like is responsible for the Aft phenotype. The repressive function of SlMYBATV also was confirmed through the CRISPR/Cas9 approach. A yeast-two-hybrid assay revealed that SlMYBATV interacts with the bHLH protein SlJAF13. Furthermore, yeast-one-hybrid and dual-luciferase transient expression assays showed that Aft directly binds to the SlMYBATV promoter and activates its expression. The results herein provide candidate genes to enhance anthocyanin content in tomato fruit. This research also provides insight into a mechanism involving the Aft-SlMYBATV pathway that fine-tunes anthocyanin accumulation in tomato fruit.
DOI:10.1016/j.xplc.2019.100006URL [��������: 1]
DOI:10.1111/j.1365-313X.2005.02371.xURLPMID:15807784 [��������: 1]

The integration of metabolomics and transcriptomics can provide precise information on gene-to-metabolite networks for identifying the function of unknown genes unless there has been a post-transcriptional modification. Here, we report a comprehensive analysis of the metabolome and transcriptome of Arabidopsis thaliana over-expressing the PAP1 gene encoding an MYB transcription factor, for the identification of novel gene functions involved in flavonoid biosynthesis. For metabolome analysis, we performed flavonoid-targeted analysis by high-performance liquid chromatography-mass spectrometry and non-targeted analysis by Fourier-transform ion-cyclotron mass spectrometry with an ultrahigh-resolution capacity. This combined analysis revealed the specific accumulation of cyanidin and quercetin derivatives, and identified eight novel anthocyanins from an array of putative 1800 metabolites in PAP1 over-expressing plants. The transcriptome analysis of 22,810 genes on a DNA microarray revealed the induction of 38 genes by ectopic PAP1 over-expression. In addition to well-known genes involved in anthocyanin production, several genes with unidentified functions or annotated with putative functions, encoding putative glycosyltransferase, acyltransferase, glutathione S-transferase, sugar transporters and transcription factors, were induced by PAP1. Two putative glycosyltransferase genes (At5g17050 and At4g14090) induced by PAP1 expression were confirmed to encode flavonoid 3-O-glucosyltransferase and anthocyanin 5-O-glucosyltransferase, respectively, from the enzymatic activity of their recombinant proteins in vitro and results of the analysis of anthocyanins in the respective T-DNA-inserted mutants. The functional genomics approach through the integration of metabolomics and transcriptomics presented here provides an innovative means of identifying novel gene functions involved in plant metabolism.
DOI:10.1111/j.1365-313X.2009.03885.xURLPMID:19368693 [��������: 1]

MYB transcription factors help to control anthocyanin biosynthesis in plants, and ectopic expression of the Arabidopsis Production of Anthocyanin Pigment 1 (PAP1) transcription factor activates the anthocyanin pathway in tobacco, suggesting the general utility of such factors for metabolic engineering of anthocyanins and anthocyanin-derived compounds such as proanthocyanidins (condensed tannins). However, PAP1 does not activate anthocyanin biosynthesis in the model legume Medicago truncatula or in alfalfa (Medicago sativa). A related Legume Anthocyanin Production 1 (LAP1) gene was identified from the genome of M. truncatula. When constitutively expressed in transgenic alfalfa, M. truncatula or white clover, LAP1 induced massive accumulation of anthocyanin pigments comprising multiple glycosidic conjugates of cyanidin. Oligomeric/polymeric compounds with some diagnostic characteristics of proanthocyanidins also accumulated in LAP1-expressing plants, but these compounds were not composed of (epi)catechin units. Over 260 and 70 genes were up-regulated in leaves of alfalfa or M. truncatula, respectively, in response to constitutive expression of LAP1, many of which are involved in anthocyanin biosynthesis. In particular, the glucosyltransferase UGT78G1, previously identified as showing preference for isoflavonoid substrates in vitro, was strongly up-regulated by LAP1, and appears to function as an anthocyanin glycosyltransferase in vivo. Over-expression of UGT78G1 in transgenic alfalfa resulted in increased anthocyanin accumulation when plants were exposed to abiotic stress.
DOI:10.1111/j.1365-313X.2006.02964.xURLPMID:17181777 [��������: 2]

Anthocyanin concentration is an important determinant of the colour of many fruits. In apple (Malus x domestica), centuries of breeding have produced numerous varieties in which levels of anthocyanin pigment vary widely and change in response to environmental and developmental stimuli. The apple fruit cortex is usually colourless, although germplasm does exist where the cortex is highly pigmented due to the accumulation of either anthocyanins or carotenoids. From studies in a diverse array of plant species, it is apparent that anthocyanin biosynthesis is controlled at the level of transcription. Here we report the transcript levels of the anthocyanin biosynthetic genes in a red-fleshed apple compared with a white-fleshed cultivar. We also describe an apple MYB transcription factor, MdMYB10, that is similar in sequence to known anthocyanin regulators in other species. We further show that this transcription factor can induce anthocyanin accumulation in both heterologous and homologous systems, generating pigmented patches in transient assays in tobacco leaves and highly pigmented apple plants following stable transformation with constitutively expressed MdMYB10. Efficient induction of anthocyanin biosynthesis in transient assays by MdMYB10 was dependent on the co-expression of two distinct bHLH proteins from apple, MdbHLH3 and MdbHLH33. The strong correlation between the expression of MdMYB10 and apple anthocyanin levels during fruit development suggests that this transcription factor is responsible for controlling anthocyanin biosynthesis in apple fruit; in the red-fleshed cultivar and in the skin of other varieties, there is an induction of MdMYB10 expression concurrent with colour formation during development. Characterization of MdMYB10 has implications for the development of new varieties through classical breeding or a biotechnological approach.
DOI:10.1105/tpc.108.059329URLPMID:19151225 [��������: 1]

Mutations in the genes encoding for either the biosynthetic or transcriptional regulation of the anthocyanin pathway have been linked to color phenotypes. Generally, this is a loss of function resulting in a reduction or a change in the distribution of anthocyanin. Here, we describe a rearrangement in the upstream regulatory region of the gene encoding an apple (Malus x domestica) anthocyanin-regulating transcription factor, MYB10. We show that this modification is responsible for increasing the level of anthocyanin throughout the plant to produce a striking phenotype that includes red foliage and red fruit flesh. This rearrangement is a series of multiple repeats, forming a minisatellite-like structure that comprises five direct tandem repeats of a 23-bp sequence. This MYB10 rearrangement is present in all the red foliage apple varieties and species tested but in none of the white fleshed varieties. Transient assays demonstrated that the 23-bp sequence motif is a target of the MYB10 protein itself, and the number of repeat units correlates with an increase in transactivation by MYB10 protein. We show that the repeat motif is capable of binding MYB10 protein in electrophoretic mobility shift assays. Taken together, these results indicate that an allelic rearrangement in the promoter of MYB10 has generated an autoregulatory locus, and this autoregulation is sufficient to account for the increase in MYB10 transcript levels and subsequent ectopic accumulation of anthocyanins throughout the plant.
DOI:10.1093/jxb/ert377URL [��������: 1]

This work characterized the role of the R2R3-MYB10 transcription factor (TF) in strawberry fruit ripening. The expression of this TF takes place mainly in the fruit receptacle and is repressed by auxins and activated by abscisic acid (ABA), in parallel to the ripening process. Anthocyanin was not produced when FaMYB10 expression was transiently silenced in fruit receptacles. An increase in FaMYB10 expression was observed in water-stressed fruits, which was accompanied by an increase in both ABA and anthocyanin content. High-throughput transcriptomic analyses performed in fruits with downregulated FaMYB10 expression indicated that this TF regulates the expression of most of the Early-regulated Biosynthesis Genes (EBGs) and the Late-regulated Biosynthesis Genes (LBGs) genes involved in anthocyanin production in ripened fruit receptacles. Besides, the expression of FaMYB10 was not regulated by FaMYB1 and vice versa. Taken together, all these data clearly indicate that the Fragaria ananassa MYB10 TF plays a general regulatory role in the flavonoid/phenylpropanoid pathway during the ripening of strawberry.
DOI:10.1371/journal.pone.0126991URLPMID:25978735 [��������: 1]

Anthocyanins are essential contributors to fruit coloration, an important quality feature and a breed determining trait of a sweet cherry fruit. It is well established that the biosynthesis of anthocyanins is regulated by an interplay of specific transcription factors belonging to MYB and bHLH families accompanied by a WD40 protein. In this study, we isolated and analyzed PaWD40, PabHLH3, PabHLH33, and several closely related MYB10 gene variants from different cultivars of sweet cherry, analyzed their expression in fruits with different anthocyanin levels at several developmental stages, and determined their capabilities to modulate anthocyanin synthesis in leaves of two Nicotiana species. Our results indicate that transcription level of variant PaMYB10.1-1 correlates with fruit coloration, but anthocyanin synthesis in Nicotiana was induced by another variant, PaMYB10.1-3, which is moderately expressed in fruits. The analysis of two fruit-expressed bHLH genes revealed that PabHLH3 enhances MYB-induced anthocyanin synthesis, whereas PabHLH33 has strong inhibitory properties.
DOI:10.1186/1471-2229-13-68URLPMID:23617716 [��������: 1]

BACKGROUND: Flavonoids such as anthocyanins, flavonols and proanthocyanidins, play a central role in fruit colour, flavour and health attributes. In peach and nectarine (Prunus persica) these compounds vary during fruit growth and ripening. Flavonoids are produced by a well studied pathway which is transcriptionally regulated by members of the MYB and bHLH transcription factor families. We have isolated nectarine flavonoid regulating genes and examined their expression patterns, which suggests a critical role in the regulation of flavonoid biosynthesis. RESULTS: In nectarine, expression of the genes encoding enzymes of the flavonoid pathway correlated with the concentration of proanthocyanidins, which strongly increases at mid-development. In contrast, the only gene which showed a similar pattern to anthocyanin concentration was UDP-glucose-flavonoid-3-O-glucosyltransferase (UFGT), which was high at the beginning and end of fruit growth, remaining low during the other developmental stages. Expression of flavonol synthase (FLS1) correlated with flavonol levels, both temporally and in a tissue specific manner. The pattern of UFGT gene expression may be explained by the involvement of different transcription factors, which up-regulate flavonoid biosynthesis (MYB10, MYB123, and bHLH3), or repress (MYB111 and MYB16) the transcription of the biosynthetic genes. The expression of a potential proanthocyanidin-regulating transcription factor, MYBPA1, corresponded with proanthocyanidin levels. Functional assays of these transcription factors were used to test the specificity for flavonoid regulation. CONCLUSIONS: MYB10 positively regulates the promoters of UFGT and dihydroflavonol 4-reductase (DFR) but not leucoanthocyanidin reductase (LAR). In contrast, MYBPA1 trans-activates the promoters of DFR and LAR, but not UFGT. This suggests exclusive roles of anthocyanin regulation by MYB10 and proanthocyanidin regulation by MYBPA1. Further, these transcription factors appeared to be responsive to both developmental and environmental stimuli.
DOI:10.1104/pp.113.214700URL [��������: 1]

Varieties of the European pear (Pyrus communis) can produce trees with both red- and green-skinned fruits, such as the Max Red Bartlett (MRB) variety, although little is known about the mechanism behind this differential pigmentation. In this study, we investigated the pigmentation of MRB and its green-skinned sport (MRB-G). The results suggest that a reduction in anthocyanin concentration causes the MRB-G sport. Transcript levels of PcUFGT (for UDP-glucose:flavonoid 3-O-glucosyltransferase), the key structural gene in anthocyanin biosynthesis, paralleled the change of anthocyanin concentration in both MRB and MRB-G fruit. We cloned the PcMYB10 gene, a transcription factor associated with the promoter of PcUFGT. An investigation of the 2-kb region upstream of the ATG translation start site of PcMYB10 showed the regions 2604 to 2911 bp and 21,218 to 21,649 bp to be highly methylated. A comparison of the PcMYB10 promoter methylation level between the MRB and MRB-G forms indicated a correlation between hypermethylation and the green-skin phenotype. An Agrobacterium tumefaciens infiltration assay was conducted on young MRB fruits by using a plasmid constructed to silence endogenous PcMYB10 via DNA methylation. The infiltrated fruits showed blocked anthocyanin biosynthesis, higher methylation of the PcMYB10 promoter, and lower expression of PcMYB10 and PcUFGT. We suggest that the methylation level of PcMYB10 is associated with the formation of the green-skinned sport in the MRB pear. The potential mechanism behind the regulation of anthocyanin biosynthesis is discussed.
DOI:10.1093/pcp/pcn163URLPMID:18974195 [��������: 2]

Gentian plants have vivid blue-colored flowers, caused by accumulation of a polyacylated anthocyanin 'gentiodelphin'. We previously performed expression analysis of gentiodelphin biosynthetic genes, and hypothesized that the white-flowered gentian cultivar 'Polarno White' might have resulted from the mutation of certain regulatory factors responsible for anthocyanin biosynthesis in flower petals. In this study, we isolated 26 R2R3-MYB gene fragments including four full-length cDNAs (GtMYB2a, GtMYB2b, GtMYB3 and GtMYB4) and one basic helix-loop-helix (bHLH) gene (GtbHLH1) from blue-flowered gentian by degenerate PCR and rapid amplification of cDNA ends (RACE). Phylogenetic tree analysis showed that GtMYB3 was categorized into a clade involved in anthocyanin biosynthesis including petunia AN2 and Arabidopsis PAP1. On the other hand, GtbHLH1 exhibited high identity with petunia AN1 based on both phylogenetic and genomic structural analyses. Temporal profiles of GtMYB3 and GtbHLH1 transcript levels corresponded well with those of gentiodelphin accumulation and their biosynthetic genes in petals. Yeast two-hybrid analysis showed that GtbHLH1 interacted with GtMYB3. Moreover, transient expression analysis indicated that the co-expression of GtMYB3 and GtbHLH1 could enhance the promoter activities of late anthocyanin biosynthetic genes in tobacco BY2 cells. We also revealed that in cv. 'Polarno White' the GtMYB3 genes were mutated by insertions of transposable elements or uncharacterized sequences, indicating that the white coloration was caused by GtMYB3 mutation. These results strongly suggested that GtMYB3 and GtbHLH1 are involved in the regulation of gentiodelphin biosynthesis in gentian flowers.
DOI:10.1016/j.scienta.2015.08.018URL [��������: 1]
DOI:10.1093/jxb/erv159URLPMID:26071528 [��������: 1]

In the last decade, great progress has been made in clarifying the main determinants of anthocyanin accumulation in grape berry skin. However, the molecular details of the fine variation among cultivars, which ultimately contributes to wine typicity, are still not completely understood. To shed light on this issue, the grapes of 170 F1 progeny from the cross 'Syrah'x'Pinot Noir' were characterized at the mature stage for the content of 15 anthocyanins during four growing seasons. This huge data set was used in combination with a dense genetic map to detect genomic regions controlling the anthocyanin pathway both at key enzymatic points and at particular branches. Genes putatively involved in fine tuning the global regulation of anthocyanin biosynthesis were identified by exploring the gene predictions in the QTL (quantitative trait locus) confidence intervals and their expression profile during berry development in offspring with contrasting anthocyanin accumulation. New information on some aspects which had scarcely been investigated so far, such as anthocyanin transport into the vacuole, or completely neglected, such as acylation, is provided. These genes represent a valuable resource in grapevine molecular-based breeding programmes to improve both fruit and wine quality and to tailor wine sensory properties according to consumer demand.
DOI:10.1104/pp.112.206771URL [��������: 1]

Anthocyanin accumulation is coordinated in plants by a number of conserved transcription factors. In apple (Malus 3 domestica), an R2R3 MYB transcription factor has been shown to control fruit flesh and foliage anthocyanin pigmentation (MYB10) and fruit skin color (MYB1). However, the pattern of expression and allelic variation at these loci does not explain all anthocyanin-related apple phenotypes. One such example is an open-pollinated seedling of cv Sangrado that has green foliage and develops red flesh in the fruit cortex late in maturity. We used methods that combine plant breeding, molecular biology, and genomics to identify duplicated MYB transcription factors that could control this phenotype. We then demonstrated that the red-flesh cortex phenotype is associated with enhanced expression of MYB110a, a paralog of MYB10. Functional characterization of MYB110a showed that it was able to up-regulate anthocyanin biosynthesis in tobacco (Nicotiana tabacum). The chromosomal location of MYB110a is consistent with a whole-genome duplication event that occurred during the evolution of apple within the Maloideae. Both MYB10 and MYB110a have conserved function in some cultivars, but they differ in their expression pattern and response to fruit maturity.
DOI:10.1093/pcp/pcu205URLPMID:25527830 [��������: 1]

Anthocyanin and proanthocyanidin (PA) are important secondary metabolites and beneficial to human health. Their biosynthesis is induced by jasmonate (JA) treatment and regulated by MYB transcription factors (TFs). However, which and how MYB TFs regulate this process is largely unknown in apple. In this study, MdMYB9 and MdMYB11 which were induced by methyl jasmonate (MeJA) were functionally characterized. Overexpression of MdMYB9 or MdMYB11 promoted not only anthocyanin but also PA accumulation in apple calluses, and the accumulation was further enhanced by MeJA. Subsequently, yeast two-hybrid, pull-down and bimolecular fluorescence complementation assays showed that both MYB proteins interact with MdbHLH3. Moreover, Jasmonate ZIM-domain (MdJAZ) proteins interact with MdbHLH3. Furthermore, chromatin immunoprecipitation-quantitative PCR and yeast one-hybrid assays demonstrated that both MdMYB9 and MdMYB11 bind to the promoters of ANS, ANR and LAR, whereas MdbHLH3 is recruited to the promoters of MdMYB9 and MdMYB11 and regulates their transcription. In addition, transient expression assays indicated that overexpression of MdJAZ2 inhibits the recruitment of MdbHLH3 to the promoters of MdMYB9 and MdMYB11. Our findings provide new insight into the mechanism of how MeJA regulates anthocyanin and PA accumulation in apple.
DOI:10.1038/s41438-018-0098-yURLPMID:30729012 [��������: 1]

Genetic manipulation of genes to upregulate specific branches of metabolic pathways is a method that is commonly used to improve fruit quality. However, the use of a single gene to impact several metabolic pathways is difficult. Here, we show that overexpression of the single gene SlMYB75 (SlMYB75-OE) is effective at improving multiple fruit quality traits. In these engineered fruits, the anthocyanin content reached 1.86 mg g(-1) fresh weight at the red-ripe stage, and these SlMYB75-OE tomatoes displayed a series of physiological changes, including delayed ripening and increased ethylene production. In addition to anthocyanin, the total contents of phenolics, flavonoids and soluble solids in SlMYB75-OE fruits were enhanced by 2.6, 4, and 1.2 times, respectively, compared to those of wild-type (WT) fruits. Interestingly, a number of aroma volatiles, such as aldehyde, phenylpropanoid-derived and terpene volatiles, were significantly increased in SlMYB75-OE fruits, with some terpene volatiles showing more than 10 times higher levels than those in WT fruits. Consistent with the metabolic assessment, transcriptomic profiling indicated that the genes involved in the ethylene signaling, phenylpropanoid and isoprenoid pathways were greatly upregulated in SlMYB75-OE fruits. Yeast one-hybrid and transactivation assays revealed that SlMYB75 is able to directly bind to the MYBPLANT and MYBPZM cis-regulatory elements and to activate the promoters of the LOXC, AADC2 and TPS genes. The identification of SlMYB75 as a key regulator of fruit quality attributes through the transcriptional regulation of downstream genes involved in several metabolic pathways opens new avenues towards engineering fruits with a higher sensory and nutritional quality.
DOI:10.1111/j.1365-313X.2008.03564.xURLPMID:18532978 [��������: 1]

SUMMARY: In Arabidopsis thaliana, several MYB and basic helix-loop-helix (BHLH) proteins form ternary complexes with TTG1 (WD-Repeats) and regulate the transcription of genes involved in anthocyanin and proanthocyanidin (PA) biosynthesis. Similar MYB-BHLH-WDR (MBW) complexes control epidermal patterning and cell fates. A family of small MYB proteins (R3-MYB) has been shown to play an important role in the regulation of epidermal cell fates, acting as inhibitors of the MBW complexes. However, so far none of these small MYB proteins have been demonstrated to regulate flavonoid biosynthesis. The genetic and molecular analyses presented here demonstrated that Arabidopsis MYBL2, which encodes a R3-MYB-related protein, is involved in the regulation of flavonoid biosynthesis. The loss of MYBL2 activity in the seedlings of two independent T-DNA insertion mutants led to a dramatic increase in the accumulation of anthocyanin. In addition, overexpression of MYBL2 in seeds inhibited the biosynthesis of PAs. These changes in flavonoid content correlate well with the increased level of mRNA of several structural and regulatory anthocyanin biosynthesis genes. Interestingly, transient expression analyses in A. thaliana cells suggested that MYBL2 interacts with MBW complexes in planta and directly modulates the expression of flavonoid target genes. These results are fully consistent with the molecular interaction of MYBL2 with BHLH proteins observed in yeast. Finally, MYBL2 expression studies, including its inhibition by light-induced stress, allowed us to hypothesise a physiological role for MYBL2. Taken together, these results bring new insights into the transcriptional regulation of flavonoid biosynthesis and provide new clues and tools for further investigation of its developmental and environmental regulation.
DOI:10.1007/s11103-015-0394-yURLPMID:26497001 [��������: 1]

In grapevine, anthocyanins and proanthocyanidins are the main flavonoids in berries, which are associated to organoleptic properties in red wine such as color and astringency. Flavonoid pathway is specifically regulated at transcriptional level and several R2R3-MYB proteins have shown to act as positive regulators. However, some members of this family have shown to repress the flavonoid biosynthesis. In this work, we present the characterization of VvMYB4-like gene, which encodes a putative transcriptional factor highly expressed in the skin of berries at the pre veraison stage in grapevine. Its over-expression in tobacco resulted in the loss of pigmentation in flowers due a decrease in anthocyanin accumulation. Severity in anthocyanin suppression observed in petals could be associated with the expression level of the VvMYB4-like transgene. Expression analysis of flavonoid structural genes revealed the strong down-regulation of the flavonoid-related genes anthocyanidin synthase (ANS) and dihydroflavonol reductase (DFR) genes and also the reduction of the anthocyanin-related gene UDP glucose:flavonoid 3-O-glucosyl transferase (UFGT), which was dependent of the transgene expression. In addition, expression of VvMYB4-like in the model plant Arabidopsis showed similar results, with the higher down-regulation observed in the AtDFR and AtLDOX genes. These results suggest that VvMYB4-like may play an important role in regulation of anthocyanin biosynthesis in grapevine acting as a transcriptional repressor of flavonoid structural genes.
DOI:10.1007/s11240-010-9912-4URL [��������: 1]
DOI:10.1016/j.phytochem.2013.02.016URLPMID:23522932 [��������: 1]

Anthocyanins and proanthocyanidins (PAs), flavonoid-derived metabolites with different physiological roles, are produced by plants in a coordinated manner during fruit development by the action of transcription factors (TFs). These regulatory proteins have either an activating or repressing effect over structural genes from the biosynthetic pathway under their control. FaMYB1, a TF belonging to the R2R3-MYB family and isolated from commercial strawberry fruit (Fragariaxananassa), was reported as a transcriptional repressor and its heterologous over-expression in tobacco flowers suppressed flavonoid-derived compound accumulation. FcMYB1, an ortholog of FaMYB1 isolated from the white Chilean strawberry (Fragaria chiloensis ssp. chiloensis f. chiloensis), showed higher transcript levels in white (F. chiloensis) than in red (F.xananassa cv. Camarosa) fruits. In order to assess its contribution to the discolored phenotype in F. chiloensis, FcMYB1 was transiently down-regulated in planta using an RNAi-based approach. Quantitative real-time PCR on FcMYB1 down-regulated fruits resulted an up-regulation of anthocyanidin synthase (ANS) and a strong repression of anthocyanidin reductase (ANR) and leucoanthocyanidin reductase (LAR) transcript accumulation. In addition, these fruits showed increased concentrations of anthocyanins and undetectable levels of flavan 3-ols. Altogether, these results indicate a role for FcMYB1 in regulation of the branching-point of the anthocyanin/PA biosynthesis determining the discolored phenotype of the white Chilean strawberry fruit.
DOI:10.1111/j.1365-313X.2008.03565.xURLPMID:18532977 [��������: 1]

SUMMARY: In Arabidopsis, MYB transcription factors regulate flavonoid biosynthesis via the formation of protein complexes with a basic helix-loop-helix (bHLH) transcription factor and a WD40 repeat protein. Several R3-type single-MYB proteins (R3-MYB), such as CPC and TRY, act as negative regulators of the development of epidermal cells. However, such regulators of flavonoid biosynthesis have not yet been reported, to our knowledge. We show here that an R3-MYB protein, AtMYBL2, acts as a transcriptional repressor and negatively regulates the biosynthesis of anthocyanin in Arabidopsis. In an AtMYBL2 knockout line (mybl2), the expression of the DFR and TT8 genes was enhanced and resulted in the ectopic accumulation of anthocyanin, while ectopic expression of AtMYBL2 or of a chimeric repressor that is a dominant negative form of AtMYBL2 suppressed the expression of DFR and TT8, and the biosynthesis of anthocyanin. The expression of AtMYBL2 was detected in various tissues but not in those in which anthocyanin accumulated or TT8 was expressed. The minimal repression domain of AtMYBL2 was found to be the six amino acids (TLLLFR) at the carboxyl terminus, and TLLLFR appears to be a novel repression motif that is different from the ERF-associated amphiphilic repression (EAR) motif. The defective phenotype of mybl2 mutants was complemented by 35S:AtMYBL2 but enhanced by a truncated form of AtMYBL2 from which the repression domain had been deleted. AtMYBL2 bound directly to TT8 protein and this complex suppressed the expression of DFR and TT8. The repression activity of AtMYBL2 appears to play a critical role in the regulation of anthocyanin biosynthesis.
DOI:10.1105/tpc.111.084525URLPMID:21487097 [��������: 2]

Flavonoids are synthesized through an important metabolic pathway that leads to the production of diverse secondary metabolites, including anthocyanins, flavonols, flavones, and proanthocyanidins. Anthocyanins and flavonols are derived from Phe and share common precursors, dihydroflavonols, which are substrates for both flavonol synthase and dihydroflavonol 4-reductase. In the stems of Arabidopsis thaliana, anthocyanins accumulate in an acropetal manner, with the highest level at the junction between rosette and stem. We show here that this accumulation pattern is under the regulation of miR156-targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes, which are deeply conserved and known to have important roles in regulating phase change and flowering. Increased miR156 activity promotes accumulation of anthocyanins, whereas reduced miR156 activity results in high levels of flavonols. We further provide evidence that at least one of the miR156 targets, SPL9, negatively regulates anthocyanin accumulation by directly preventing expression of anthocyanin biosynthetic genes through destabilization of a MYB-bHLH-WD40 transcriptional activation complex. Our results reveal a direct link between the transition to flowering and secondary metabolism and provide a potential target for manipulation of anthocyanin and flavonol content in plants.
DOI:10.1104/pp.114.256172URLPMID:25659381 [��������: 1]

Because of the vast range of functions that phenylpropanoids possess, their synthesis requires precise spatiotemporal coordination throughout plant development and in response to the environment. The accumulation of these secondary metabolites is transcriptionally controlled by positive and negative regulators from the MYB and basic helix-loop-helix protein families. We characterized four grapevine (Vitis vinifera) R2R3-MYB proteins from the C2 repressor motif clade, all of which harbor the ethylene response factor-associated amphiphilic repression domain but differ in the presence of an additional TLLLFR repression motif found in the strong flavonoid repressor Arabidopsis (Arabidopsis thaliana) AtMYBL2. Constitutive expression of VvMYB4a and VvMYB4b in petunia (Petunia hybrida) repressed general phenylpropanoid biosynthetic genes and selectively reduced the amount of small-weight phenolic compounds. Conversely, transgenic petunia lines expressing VvMYBC2-L1 and VvMYBC2-L3 showed a severe reduction in petal anthocyanins and seed proanthocyanidins together with a higher pH of crude petal extracts. The distinct function of these regulators was further confirmed by transient expression in tobacco (Nicotiana benthamiana) leaves and grapevine plantlets. Finally, VvMYBC2-L3 was ectopically expressed in grapevine hairy roots, showing a reduction in proanthocyanidin content together with the down-regulation of structural and regulatory genes of the flavonoid pathway as revealed by a transcriptomic analysis. The physiological role of these repressors was inferred by combining the results of the functional analyses and their expression patterns in grapevine during development and in response to ultraviolet B radiation. Our results indicate that VvMYB4a and VvMYB4b may play a key role in negatively regulating the synthesis of small-weight phenolic compounds, whereas VvMYBC2-L1 and VvMYBC2-L3 may additionally fine tune flavonoid levels, balancing the inductive effects of transcriptional activators.
DOI:10.1111/j.1365-313X.2010.04465.xURLPMID:21235651 [��������: 3]

We present an investigation of anthocyanin regulation over the entire petunia plant, determining the mechanisms governing complex floral pigmentation patterning and environmentally induced vegetative anthocyanin synthesis. DEEP PURPLE (DPL) and PURPLE HAZE (PHZ) encode members of the R2R3-MYB transcription factor family that regulate anthocyanin synthesis in petunia, and control anthocyanin production in vegetative tissues and contribute to floral pigmentation. In addition to these two MYB factors, the basic helix-loop-helix (bHLH) factor ANTHOCYANIN1 (AN1) and WD-repeat protein AN11, are also essential for vegetative pigmentation. The induction of anthocyanins in vegetative tissues by high light was tightly correlated to the induction of transcripts for PHZ and AN1. Interestingly, transcripts for PhMYB27, a putative R2R3-MYB active repressor, were highly expressed during non-inductive shade conditions and repressed during high light. The competitive inhibitor PhMYBx (R3-MYB) was expressed under high light, which may provide feedback repression. In floral tissues DPL regulates vein-associated anthocyanin pigmentation in the flower tube, while PHZ determines light-induced anthocyanin accumulation on exposed petal surfaces (bud-blush). A model is presented suggesting how complex floral and vegetative pigmentation patterns are derived in petunia in terms of MYB, bHLH and WDR co-regulators.
DOI:10.1105/tpc.113.122069URLPMID:24642943 [��������: 1]

Plants require sophisticated regulatory mechanisms to ensure the degree of anthocyanin pigmentation is appropriate to myriad developmental and environmental signals. Central to this process are the activity of MYB-bHLH-WD repeat (MBW) complexes that regulate the transcription of anthocyanin genes. In this study, the gene regulatory network that regulates anthocyanin synthesis in petunia (Petunia hybrida) has been characterized. Genetic and molecular evidence show that the R2R3-MYB, MYB27, is an anthocyanin repressor that functions as part of the MBW complex and represses transcription through its C-terminal EAR motif. MYB27 targets both the anthocyanin pathway genes and basic-helix-loop-helix (bHLH) ANTHOCYANIN1 (AN1), itself an essential component of the MBW activation complex for pigmentation. Other features of the regulatory network identified include inhibition of AN1 activity by the competitive R3-MYB repressor MYBx and the activation of AN1, MYB27, and MYBx by the MBW activation complex, providing for both reinforcement and feedback regulation. We also demonstrate the intercellular movement of the WDR protein (AN11) and R3-repressor (MYBx), which may facilitate anthocyanin pigment pattern formation. The fundamental features of this regulatory network in the Asterid model of petunia are similar to those in the Rosid model of Arabidopsis thaliana and are thus likely to be widespread in the Eudicots.
DOI:10.1093/mp/ssp030URLPMID:19825656 [��������: 1]

Single-repeat R3 MYB transcription factors like CPC (CAPRICE) are known to play roles in developmental processes such as root hair differentiation and trichome initiation. However, none of the six Arabidopsis single-repeat R3 MYB members has been reported to regulate flavonoid biosynthesis. We show here that CPC is a negative regulator of anthocyanin biosynthesis. In the process of using CPC to test GAL4-dependent driver lines, we observed a repression of anthocyanin synthesis upon GAL4-mediated CPC overexpression. We demonstrated that this is not due to an increase in nutrient uptake because of more root hairs. Rather, CPC expression level tightly controls anthocyanin accumulation. Microarray analysis on the whole genome showed that, of 37 000 features tested, 85 genes are repressed greater than three-fold by CPC overexpression. Of these 85, seven are late anthocyanin biosynthesis genes. Also, anthocyanin synthesis genes were shown to be down-regulated in 35S::CPC overexpression plants. Transient expression results suggest that CPC competes with the R2R3-MYB transcription factor PAP1/2, which is an activator of anthocyanin biosynthesis genes. This report adds anthocyanin biosynthesis to the set of programs that are under CPC control, indicating that this regulator is not only for developmental programs (e.g. root hairs, trichomes), but can influence anthocyanin pigment synthesis.
DOI:10.1105/tpc.12.9.1619URLPMID:11006336 [��������: 2]

The petunia loci anthocyanin1 (an1), an2, an4, and an11 are required for the transcription of anthocyanin biosynthetic genes in floral organs. The an2 and an11 loci were recently cloned and shown to encode a MYB-domain transcriptional activator and a cytosolic WD40 protein, respectively. Here, we report the isolation of an1 by transposon tagging. an1 encodes a new member of the basic helix-loop-helix family of transcription factors that is functionally and evolutionarily distinct from JAF13, the apparent petunia ortholog of maize RED1 and snapdragon DELILA. We provide genetic evidence that the transcription factors encoded by an1, an2, and an4 operate in an unexpectedly complex regulatory hierarchy. In leaves, ectopic expression of AN2 induces an1 expression, whereas in anthers, an1 expression depends on an4, encoding (or controlling) a MYB protein that is paralogous to AN2. Experiments with transgenic plants expressing a post-translationally controlled AN1-GLUCOCORTICOID RECEPTOR fusion protein indicated that independent of protein synthesis, AN1 directly activates the expression of the dfrA gene encoding the enzyme dihydroflavonol 4-reductase and of Pmyb27 encoding a MYB-domain protein of unknown function.
DOI:10.1105/tpc.1.12.1175URLPMID:2535537 [��������: 1]

Genetic studies in maize have identified several regulatory genes that control the tissue-specific synthesis of the purple anthocyanin pigments during development. Two such genes, R and B, exhibit extensive allelic diversity with respect to the tissue specificity and developmental timing of anthocyanin synthesis. Previous genetic studies demonstrated that certain B alleles can substitute for R function, and in these cases only one functional allele at either locus is required for pigment synthesis in the aleurone. In addition, biochemical studies have shown that both genes act on the same biosynthetic pathway, suggesting that the genes are functionally duplicate. In this report we describe DNA hybridization experiments that demonstrate that the functionally duplicate nature of B and R is reflected in DNA sequence similarity between the two genes. We took advantage of this homology and used the R genomic sequences to clone B. Two different strategies were pursued and two genomic clones isolated, a 2.5-kilobase BgIII fragment linked to the b allele in W23 inbred stocks and a 1.0-kilobase HindIII fragment linked to the B allele in CM37 stocks. Examination of several independent transposable element insertion mutations in B and revertant derivatives demonstrated that our clones recognize the functional B gene. Genomic clones representing the entire B-Peru allele were isolated, and a detailed restriction map was prepared. Using these clones we have identified a 2.2-kilobase mRNA in husks from plants containing either B-I or B-Peru alleles, but no B mRNA was detected in plants containing a b allele. The transcript is at least 100 times more abundant in strongly pigmented B-I husks than in weakly pigmented B-Peru husk tissue. Expression of functional B alleles in husk tissue correlates with the coordinate increase in mRNA levels of two structural genes of the pathway, A1 and Bz1, consistent with the postulated role of B as a regulatory gene.
DOI:10.1093/nar/20.2.373URLPMID:1741268 [��������: 1]
DOI:10.1111/j.1365-313X.2004.02138.xURLPMID:15255866 [��������: 1]

Genetic analyses have demonstrated that together with TTG1, a WD-repeat (WDR) protein, TT2 (MYB), and TT8 (bHLH) are necessary for the correct expression of BANYULS (BAN). This gene codes for the core enzyme of proanthocyanidin biosynthesis in Arabidopsis thaliana seed coat. The interplays of TT2, TT8, and their closest MYB/bHLH relatives, with TTG1 and the BAN promoter have been investigated using a combination of genetic and molecular approaches, both in yeast and in planta. The results obtained using glucocorticoid receptor fusion proteins in planta strongly suggest that TT2, TT8, and TTG1 can directly activate BAN expression. Experiments using yeast two- and three-hybrid clearly demonstrated that TT2, TT8, and TTG1 can form a stable ternary complex. Furthermore, although TT2 and TT8 were able to bind to the BAN promoter when simultaneously expressed in yeast, the activity of the complex correlated with the level of TTG1 expression in A. thaliana protoplasts. In addition, transient expression experiments revealed that TTG1 acts mainly through the bHLH partner (i.e. TT8 or related proteins) and that TT2 cannot be replaced by any other related A. thaliana MYB proteins to activate BAN. Finally and consistent with these results, the ectopic expression of TT2 was sufficient to trigger BAN activation in vegetative parts, but only where TTG1 was expressed. Taken together, these results indicate that TT2, TT8, and TTG1 can form a ternary complex directly regulating BAN expression in planta.
DOI:10.1111/nph.13816URLPMID:26725247 [��������: 1]

The MYB- basic helix-loop-helix (bHLH)-WD40 complexes regulating anthocyanin and proanthocyanidin (PA) biosynthesis in plants are not fully understood. Here Medicago truncatula bHLH MtTT8 was characterized as a central component of these ternary complexes that control anthocyanin and PA biosynthesis. Mttt8 mutant seeds have a transparent testa phenotype with reduced PAs and anthocyanins. MtTT8 restores PA and anthocyanin productions in Arabidopsis tt8 mutant. Ectopic expression of MtTT8 restores anthocyanins and PAs in mttt8 plant and hairy roots and further enhances both productions in wild-type hairy roots. Transcriptomic analyses and metabolite profiling of mttt8 mutant seeds and M. truncatula hairy roots (mttt8 mutant, mttt8 mutant complemented with MtTT8, or MtTT8 overexpression lines) indicate that MtTT8 regulates a subset of genes involved in PA and anthocyanin biosynthesis. MtTT8 is genetically regulated by MtLAP1, MtPAR and MtWD40-1. Combinations of MtPAR, MtLAP1, MtTT8 and MtWD40-1 activate MtTT8 promoter in yeast assay. MtTT8 interacts with these transcription factors to form regulatory complexes. MtTT8, MtWD40-1 and an MYB factor, MtPAR or MtLAP1, interacted and activated promoters of anthocyanidin reductase and anthocyanidin synthase to regulate PA and anthocyanin biosynthesis, respectively. Our results provide new insights into the complex regulation of PA and anthocyanin biosynthesis in M. truncatula.
DOI:10.1007/s00425-011-1407-yURL [��������: 1]

The basic helix-loop-helix (bHLH) transcription factors (TFs) comprise one of the largest families of TFs involved in developmental and physiological processes in plants. Here, we describe the functional characterization of two bHLH TFs (NtAn1a and NtAn1b) isolated from tobacco (Nicotiana tabacum) flowers. NtAn1a and NtAn1b originate from two ancestors of tobacco, N. sylvestris and N. tomentosiformis, respectively. NtAn1a and NtAn1b share high sequence similarity with other known flavonoid-related bHLH TFs and are predominantly expressed in flowers. GUS expression driven by the NtAn1a promoter is consistent with NtAn1 transcript profile in tobacco flowers. Both NtAn1a and NtAn1b are transcriptional activators as demonstrated by transactivation assays using yeast cells and tobacco protoplasts. Ectopic expression of NtAn1a or NtAn1b enhances anthocyanin accumulation in tobacco flowers. In transgenic tobacco expressing NtAn1a or NtAn1b, both subsets of early and late flavonoid pathway genes were up-regulated. Yeast two-hybrid assays showed that NtAn1 proteins interact with the previously characterized R2R3-MYB TF, NtAn2. The NtAn1-NtAn2 complex activated the promoters of two key anthocyanin pathway genes, dihydroflavonol reductase and chalcone synthase. The promoter activation is severely repressed by dominant repressive forms of either NtAn1a or NtAn2, created by fusing the SRDX repressor domain to the TFs. Our results show that NtAn1 and NtAn2 act in concert to regulate the anthocyanin pathway in tobacco flowers and NtAn2 up-regulates NtAn1 gene expression.
DOI:10.1104/pp.103.026039URLPMID:14605235 [��������: 1]

We have identified an R2R3-type MYB factor, GMYB10, from Gerbera hybrida (Asteraceae) that shares high sequence homology to and is phylogenetically grouped together with the previously characterized regulators of anthocyanin pigmentation in petunia (Petunia hybrida) and Arabidopsis. GMYB10 is able to induce anthocyanin pigmentation in transgenic tobacco (Nicotiana tabacum), especially in vegetative parts and anthers. In G. hybrida, GMYB10 is involved in activation of anthocyanin biosynthesis in leaves, floral stems, and flowers. In flowers, its expression is restricted to petal epidermal cell layers in correlation with the anthocyanin accumulation pattern. We have shown, using yeast (Saccharomyces cerevisiae) two-hybrid assay, that GMYB10 interacts with the previously isolated bHLH factor GMYC1. Particle bombardment analysis was used to show that GMYB10 is required for activation of a late anthocyanin biosynthetic gene promoter, PGDFR2. cis-Analysis of the target PGDFR2 revealed a sequence element with a key role in activation by GMYB10/GMYC1. This element shares high homology with the anthocyanin regulatory elements characterized in maize (Zea mays) anthocyanin promoters, suggesting that the regulatory mechanisms involved in activation of anthocyanin biosynthesis have been conserved for over 125 million years not only at the level of transcriptional regulators but also at the level of the biosynthetic gene promoters.
DOI:10.1016/j.plaphy.2010.11.006URL [��������: 1]

Gynura bicolor DC. is a traditional vegetable in Japan. G. bicolor grown in the field has adaxial sides of leaves that are green and abaxial sides that are reddish purple. It has been reported that the responsible reddish purple pigments are anthocyanins, which are acylated and highly stable. We have reported that cultured G. bicolor plantlets treated with methyl jasmonate (MJ) exhibited anthocyanin accumulation in roots, and this was affected by light irradiation. In the present study, to clarify this accumulation induced by MJ treatment, we isolated anthocyanin biosynthesis and regulatory genes from G. bicolor. Expression analysis revealed up-regulated expression of flavonoid biosynthesis genes, GbCHS, GbCHI, GbDFR and GbANS. Furthermore, it was shown that isolated regulatory genes, GbMYB1 and GbMYC1, were also upregulated by MJ treatment. In addition, it was shown that co-expression of GbMYB1 and GbMYC1 could activate GbDFR and GbANS gene promoters in transient assays with tobacco protoplasts. These results strongly indicate that GbMYB1 and GbMYC1 coordinately regulate flavonoid biosynthetic genes induced by MJ treatment, and thereby cause anthocyanin accumulation in roots. (C) 2010 Elsevier Masson SAS.
DOI:10.3389/fpls.2016.00166URLPMID:26925082 [��������: 1]

Anthocyanin biosynthesis requires the MYB-bHLH-WD40 protein complex to activate the late biosynthetic genes. LcMYB1 was thought to act as key regulator in anthocyanin biosynthesis of litchi. However, basic helix-loop-helix proteins (bHLHs) as partners have not been identified yet. The present study describes the functional characterization of three litchi bHLH candidate anthocyanin regulators, LcbHLH1, LcbHLH2, and LcbHLH3. Although these three litchi bHLHs phylogenetically clustered with bHLH proteins involved in anthcoyanin biosynthesis in other plant, only LcbHLH1 and LcbHLH3 were found to localize in the nucleus and physically interact with LcMYB1. The transcription levels of all these bHLHs were not coordinated with anthocyanin accumulation in different tissues and during development. However, when co-infiltrated with LcMYB1, both LcbHLH1 and LcbHLH3 enhanced anthocyanin accumulation in tobacco leaves with LcbHLH3 being the best inducer. Significant accumulation of anthocyanins in leaves transformed with the combination of LcMYB1 and LcbHLH3 were noticed, and this was associated with the up-regulation of two tobacco endogenous bHLH regulators, NtAn1a and NtAn1b, and late structural genes, like NtDFR and NtANS. Significant activity of the ANS promoter was observed in transient expression assays either with LcMYB1-LcbHLH1 or LcMYB1-LcbHLH3, while only minute activity was detected after transformation with only LcMYB1. In contrast, no activity was measured after induction with the combination of LcbHLH2 and LcMYB1. Higher DFR expression was also oberseved in paralleling with higher anthocyanins in co-transformed lines. LcbHLH1 and LcbHLH3 are essential partner of LcMYB1 in regulating the anthocyanin production in tobacco and probably also in litchi. The LcMYB1-LcbHLH complex enhanced anthocyanin accumulation may associate with activating the transcription of DFR and ANS.
DOI:10.1101/gad.11.11.1422URLPMID:9192870 [��������: 1]

In petunia flowers, the loci an1, an2, and an11 control the pigmentation of the flower by stimulating the transcription of anthocyanin biosynthetic genes. The an1 and an2 locus were recently cloned and encode a basic helix-loop-helix (bHLH) and MYB-domain transcriptional activator, respectively. Here, we report the isolation of the an11 locus by transposon tagging. RNA gel blot experiments show that an11 is expressed independently from an1 and an2 throughout plant development, as well as in tissues that do not express the anthocyanin pathway. It encodes a novel WD-repeat protein that is highly conserved even in species that do not produce anthocyanins such as yeast, nematodes, and mammals. The observation that the human an11 homolog partially complements the an11 petunia mutant in transient assays shows that sequence similarity reflects functional conservation. Overexpression of an2 in an11- petals restored the activity of a structural anthocyanin gene in transient assays, indicating that AN11 acts upstream of AN2. Cell fractionation experiments show that the bulk of the AN11 protein is localized in the cytoplasm. Taken together, this indicates that AN11 is a cytoplasmic component of a conserved signal transduction cascade that modulates AN2 function in petunia, thereby linking cellular signals with transcriptional activation.
DOI:10.1105/tpc.018796URLPMID:14742877 [��������: 1]

The pale aleurone color1 (pac1) locus, required for anthocyanin pigment in the aleurone and scutellum of the Zea mays (maize) seed, was cloned using Mutator transposon tagging. pac1 encodes a WD40 repeat protein closely related to anthocyanin regulatory proteins ANTHOCYANIN11 (AN11) (Petunia hybrida [petunia]) and TRANSPARENT TESTA GLABRA1 (TTG1) (Arabidopsis thaliana). Introduction of a 35S-Pac1 transgene into A. thaliana complemented multiple ttg1 mutant phenotypes, including ones nonexistent in Z. mays. Hybridization of Z. mays genomic BAC clones with the pac1 sequence identified an additional related gene, mp1. PAC1 and MP1 deduced protein sequences were used as queries to build a phylogenetic tree of homologous WD40 repeat proteins, revealing an ancestral gene duplication leading to two clades in plants, the PAC1 clade and the MP1 clade. Subsequent duplications within each clade have led to additional WD40 repeat proteins in particular species, with all mutants defective in anthocyanin expression contained in the PAC1 clade. Substantial differences in pac1, an11, and ttg1 mutant phenotypes suggest the evolutionary divergence of regulatory mechanisms for several traits that cannot be ascribed solely to divergence of the dicot and monocot protein sequences.
DOI:10.1007/s11240-013-0361-8URL [��������: 1]
DOI:10.1016/j.jplph.2012.01.015URL [��������: 1]

The abundance of anthocyanins and proanthocyanins in apples is tightly regulated by three classes of regulatory factors, MYB, bHLH and WD40 proteins, only some of which have been previously identified. In this study, we identified an apple WD40 protein (MdTTG1) that promotes the accumulation of anthocyanins. The biosynthetic genes required downstream in the flavonoid pathway were up-regulated when MdTTG1 was over-expressed in Arabidopsis. Consistent with its role as a transcriptional regulator, an MdTTG1-GFP fusion protein was observed only in the nucleus. We assayed the expression patterns of this gene in different organs and found that they were positively correlated with anthocyanin accumulation in the apple. Yeast two-hybrid and bimolecular fluorescence complementation assays demonstrated that MdTTG1 interacted with bHLH transcription factors (TFs) but not MYB protein, whereas bHLH was known to interact with MYB in apples. However, based on a ChIP assay, MdTTG1 does not appear to bind to the promoter of the anthocyanin biosynthetic genes MdDFR and MdUFGT. Taken together, these results suggest that the apple WD40 protein MdTTG1 interacts with bHLH but not MYB proteins to regulate anthocyanin accumulation. (C) 2012 Elsevier GmbH.
DOI:10.3390/genes10070496URL [��������: 1]
DOI:10.1007/s10535-014-0427-4URL [��������: 1]

The MYB, MYC, and WD40 transcription factors (TFs) are known to regulate the expression of structural biosynthetic genes at different steps depending on the plant species. In this work, we used an agroinfiltration protocol with Tobacco rattle virus (TRV) constructs containing partial sequences from MYB or WD40 for virus-induced gene silencing (VIGS) to demonstrate their participation in the regulation of anthocyanin biosynthesis in chilli pepper (Capsicum eximium) fruits. The accumulation of anthocyanins in chilli pepper fruits of plants transformed with TRV2-MYB and TRV2-WD40 constructs was significantly reduced compared to the control or empty TRV2-transformed plants. A significant reduction in gene expression of both TFs was also detected. The expressions of the chalcone synthase (CHS), chalcone isomerase (CHI), flavonoid 3',5'-hydroxylase (F3'5'H), dihydroflavonol 4-reductase (DFR), and UDP-glucose:flavonoid 3-O-glucosyltransferase (3GT) genes were decreased in the plants transformed with the TRV2-MYB construct but not the transcription of flavanone 3-hydroxylase (F3H). When chilli pepper plants were infected with the TRV2-WD40 construct, a significant reduction in CHS, F3H, F3'5'H, DFR and 3GT expression, but not in CHI in the fruits was observed.
DOI:10.1007/s11240-013-0349-4URL [��������: 1]
DOI:10.1007/s00425-015-2305-5URLPMID:25916310 [��������: 2]

MAIN CONCLUSION: Our work strongly suggests that microRNA858 regulates anthocyanin biosynthesis in tomato by modulating the expression of two R2R3 MYB transcription factors, underscoring the importance of microRNAs in the gene regulatory network controlling specialized metabolism in plants. The biological functions of microRNA858 (miR858), a recently identified small RNA, are not well understood. Here, we identified miR858 as a negative regulator of anthocyanin biosynthesis in tomato (Solanum lycopersicum). RNA ligase-mediated 5'RACE cleavage assay showed that miR858 mediates the cleavage of SlMYB7-like and SlMYB48-like transcripts in tomato. Expression analysis revealed an inverse correlation between the accumulation of miR858 and its target SlMYB7-like mRNA, in different tissues of tomato. Subsequently, a small tandem target mimic construct for blocking miR858 (STTM858) was generated and transformed into tomato. The majority of endogenous miR858 was blocked in STTM858 over-expressing tomato plants, whereas SlMYB7-like transcripts increased significantly. Concomitantly, upregulated expression was detected for several anthocyanin biosynthetic genes, including PAL, CHS, DFR, ANS and 3GT. As a result, anthocyanins were highly accumulated in young seedlings, leaves, stems and leaf buds of the transgenic plants under normal growth conditions. In addition, over-expression of STTM858 in tomato activated another MYB transcription factor, SlMYB48, implicating the possible involvement of SlMYB48 in anthocyanin biosynthesis.
DOI:10.1105/tpc.111.083261URLPMID:21551388 [��������: 1]

Jasmonates (JAs) mediate plant responses to insect attack, wounding, pathogen infection, stress, and UV damage and regulate plant fertility, anthocyanin accumulation, trichome formation, and many other plant developmental processes. Arabidopsis thaliana Jasmonate ZIM-domain (JAZ) proteins, substrates of the CORONATINE INSENSITIVE1 (COI1)-based SCF(COI1) complex, negatively regulate these plant responses. Little is known about the molecular mechanism for JA regulation of anthocyanin accumulation and trichome initiation. In this study, we revealed that JAZ proteins interact with bHLH (Transparent Testa8, Glabra3 [GL3], and Enhancer of Glabra3 [EGL3]) and R2R3 MYB transcription factors (MYB75 and Glabra1), essential components of WD-repeat/bHLH/MYB transcriptional complexes, to repress JA-regulated anthocyanin accumulation and trichome initiation. Genetic and physiological evidence showed that JA regulates WD-repeat/bHLH/MYB complex-mediated anthocyanin accumulation and trichome initiation in a COI1-dependent manner. Overexpression of the MYB transcription factor MYB75 and bHLH factors (GL3 and EGL3) restored anthocyanin accumulation and trichome initiation in the coi1 mutant, respectively. We speculate that the JA-induced degradation of JAZ proteins abolishes the interactions of JAZ proteins with bHLH and MYB factors, allowing the transcriptional function of WD-repeat/bHLH/MYB complexes, which subsequently activate respective downstream signal cascades to modulate anthocyanin accumulation and trichome initiation.
DOI:10.1016/j.molp.2016.07.003URLPMID:27450422 [��������: 2]

Extensive studies in various plants show that the anthocyanin biosynthetic process is affected by environmental factors and regulated by many transcription factors through sophisticated regulatory networks. However, it remains largely unclear about the roles of microRNA in this process. Here, we demonstrate that miR858a is a positive regulator of anthocyanin biosynthesis in Arabidopsis seedlings. Overexpression of miR858a enhances the accumulation of anthocyanins, whereas the reduced miR858a activity results in low levels of anthocyanins in STTM858 transgenic plants. We found that miR858a inhibits the expression of MYBL2, a key negative regulator of anthocyanin biosynthesis, by translational repression. In addition, ELONGATED HYPOCOTYL 5 (HY5) was shown to directly bind the MYBL2 promoter and represses its expression via specific histone modifications. Interestingly, we found that miR858a exhibits light-responsive expression in an HY5-dependent manner. Together, these results delineate the HY5-MIR858a-MYBL2 loop as a cellular mechanism for modulating anthocyanin biosynthesis, suggesting that integration of transcriptional and posttranscriptional regulation is critical for governing proper anthocyanin accumulation in response to light and other environmental factors.
DOI:10.1016/j.febslet.2013.03.037URL [��������: 3]

Several positive transcription factors regulate Arabidopsis anthocyanin biosynthesis. HY5, a component of light-signaling pathways, and PAP1, an R2R3-MYB transcription factor, share common regulatory targets on anthocyanin biosynthesis genes. The epistatic interactions between the two transcription factors are currently unknown. To address this problem, we analyzed crosses between hy5 and pap1 mutants (hy5pap1) or pap1D overexpressors (hy5pap1D), performed chromatin immunoprecipitation-qPCR, and determined the PAP1 promoter region through deletion analysis. The results show that HY5 regulates PAP1 expression via direct binding to G- and ACE-boxes in the promoter region, which suggests bifurcate regulation of anthocyanin biosynthesis by HY5 via transcriptional activation of PAP1. (C) 2013 Federation of European Biochemical Societies. Published by Elsevier B. V.
DOI:10.1111/tpj.13077URLPMID:26576746 [��������: 2]

Photomorphogenesis is an essential program in plant development. This process is effected by the balanced cooperation of many factors under light and dark conditions. In a previous study, we showed that MYB hypocotyl elongation-related (MYBH) is involved in cell elongation. To expand our understanding of MYBH function, we performed a yeast two-hybrid assay and identified an MYB-like Domain transcription factor (MYBD). In this study, we investigated the function of MYBD, which is an MYBH homolog involved in anthocyanin accumulation. MYBD expression increased in response to light or cytokinin, and MYBD enhanced anthocyanin biosynthesis via repression of MYBL2, which encodes a transcription factor that has a negative effect on this process. In addition, MYBD binding in vivo to the MYBL2 promoter and the lower level of histone H3K9 acetylation at the upstream region of MYBL2 in MYBD over-expressing plants in comparison with wild-type plants imply that MYBD represses MYBL2 expression via an epigenetic mechanism. HY5 directly binds to the MYBD promoter, which indicates that MYBD acts on HY5-downstream in light- or cytokinin-triggered signaling pathways, leading to anthocyanin accumulation. Our results suggest that, although MYBD and MYBH are homologs, they act in opposite ways during plant photomorphogenesis, and these functions should be examined in further studies.
DOI:10.1038/hortres.2017.56URLPMID:29114391 [��������: 2]

[This corrects the article DOI: 10.1038/hortres.2017.23.].
DOI:10.1111/j.1365-313X.2006.03021.xURLPMID:17319847 [��������: 3]

Phytochromes are red/far-red light receptors that regulate various light responses by initiating the transcriptional cascades responsible for changing the expression patterns of 10-30% of the entire plant transcriptome. Several transcription factors that are thought to participate in this process have been identified, but the functional relationships among them have not yet been fully elucidated. Here we investigated the functional relationship between two such transcription factors, PIF3 and HY5, and their effects on anthocyanin biosynthesis. Our results revealed that PIF3 and HY5 do not regulate each other at either the transcriptional or the protein levels in continuous light conditions, suggesting that they are not directly linked within phytochrome-mediated signaling. We found that both PIF3 and HY5 positively regulate anthocyanin biosynthesis by activating the transcription of the same anthocyanin biosynthetic genes, but the positive effects of PIF3 required functional HY5. Chromatin immunoprecipitation analyses indicated that both PIF3 and HY5 regulate anthocyanin biosynthetic gene expression by directly binding to different regions of the gene promoters in vivo. Additional experiments revealed that PIF3 bound the promoters regardless of light and HY5. Collectively, these data show that PIF3 and HY5 regulate anthocyanin biosynthesis by simultaneously binding anthocyanin biosynthetic gene promoters at separate sequence elements.
DOI:10.4161/15592316.2014.970440URLPMID:25482806 [��������: 2]

In Arabidopsis and many other plant species, anthocyanin pigments accumulate only after light exposure and not in darkness. Excess light of very high fluence rates leads to a further, very strong increase in anthocyanin levels. How excess light is sensed is not well understood. Here, we show that mutations in the key repressor of light signaling, the COP1/SPA complex, cause a strong hyperaccumulation of anthocyanins not only under normal light but also under excess, high light conditions. Hence, normal light signaling via COP1/SPA is required to prevent hyperaccumulation of anthocyanins under these high light conditions. However, since cop1 and spa mutants show a similar high-light responsiveness of anthocyanin accumulation as the wild type it remains to be resolved whether COP1/SPA is directly involved in the high-light response itself.
DOI:10.1016/j.plaphy.2013.05.012URL [��������: 2]

The production of anthocyanins in the tomato (Solanum lycopersicum L.) fruit is normally absent or poor, but a number of mutants or introgression lines are known to increase anthocyanin levels in vegetative and reproductive tissues. Through conventional breeding, a genetic combination was obtained with the remarkable phenotype of a deep purple fruit pigmentation, due to an accumulation of anthocyanins on the peel. Such a genotype was named Sun Black (SB) as a consequence of its sensitivity to light induction. When characterized for morpho-agronomic traits, SB plants showed increased fertility. Purple fruits displayed an arrangement of the epicarp cells different from normal tomatoes, a feature that could account for different mechanical properties and shelf-life potential. The SB genotype and, to a lesser extent, its single mutant parents showed the capacity to accumulate anthocyanins in the seedling root when grown under light. This phenotype, which was greatly improved by the addition of sucrose to the germination medium, proved to be useful as selection index and gave new insights for in vitro production of anthocyanin extracts. To assess the nutraceutical potential of purple tomatoes, we tested the activity of SB skin extracts on the proliferation of two human cancer cells lines. Cell proliferation was significantly inhibited by SB extract in a dose-dependent manner. When the bioactivity of SB extracts was compared with that of other anthocyanin-containing fruits or vegetables, a significant "Extract*Line" interaction was evidenced, suggesting a crucial role for the extract composition in terms of anthocyanidins and other eventual cell growth-inhibiting compounds. (C) 2013 Elsevier Masson SAS.
DOI:10.1080/14620316.2004.11511725URL [��������: 1]
DOI:10.1016/j.jplph.2007.06.010URLPMID:17766004 [��������: 1]

In this work we analysed, at the transcript level, the response of Arabidopsis anthocyanin regulatory genes of the MYB (PAP1 and PAP2), bHLH (TT8, EGL3 and GL3) and WD40 (TTG1) families to white light in seedlings and to different light qualities in rosette leaves. Our experiments showed strong light induction of the MYB genes PAP1 and PAP2. In particular, the kinetics of PAP1 expression preceded those of PAP2 and all of the structural genes (CHS, DFR, F3H, LDOX), consistent with the hypothesis that it has a key role in light induction of anthocyanin biosynthesis. All bHLH genes analysed showed light induction, and in seedlings their expression preceded that of the late structural genes, suggesting their possible role in light regulation of these structural genes. TTG1 expression is essentially constitutive in both systems. Experiments with transgenic lines over-expressing the MYB factors show that PAP1, but not PAP2, strongly stimulates expression of the anthocyanin structural gene encoding dihydroflavonol reductase, but neither factor affected expression of the early flavonoid biosynthesis gene encoding chalcone synthase. Consistent with these findings, PAP1, but not PAP2, stimulated light induction of anthocyanin biosynthesis in seedlings. We conclude that specific members of the MYB and bHLH families play important roles in regulating anthocyanin biosynthesis in response to different light qualities in Arabidopsis.
DOI:10.1105/tpc.105.034041URLPMID:16603655 [��������: 1]

The Petunia hybrida genes ANTHOCYANIN1 (AN1) and AN2 encode transcription factors with a basic-helix-loop-helix (BHLH) and a MYB domain, respectively, that are required for anthocyanin synthesis and acidification of the vacuole in petal cells. Mutation of PH4 results in a bluer flower color, increased pH of petal extracts, and, in certain genetic backgrounds, the disappearance of anthocyanins and fading of the flower color. PH4 encodes a MYB domain protein that is expressed in the petal epidermis and that can interact, like AN2, with AN1 and the related BHLH protein JAF13 in yeast two-hybrid assays. Mutation of PH4 has little or no effect on the expression of structural anthocyanin genes but strongly downregulates the expression of CAC16.5, encoding a protease-like protein of unknown biological function. Constitutive expression of PH4 and AN1 in transgenic plants is sufficient to activate CAC16.5 ectopically. Together with the previous finding that AN1 domains required for anthocyanin synthesis and vacuolar acidification can be partially separated, this suggests that AN1 activates different pathways through interactions with distinct MYB proteins.
DOI:10.1093/jxb/erp097URLPMID:19380423 [��������: 1]

The Lc petunia system, which displays enhanced, light-induced vegetative pigmentation, was used to investigate how high light affects anthocyanin biosynthesis, and to assess the effects of anthocyanin pigmentation upon photosynthesis. Lc petunia plants displayed intense purple anthocyanin pigmentation throughout the leaves and stems when grown under high-light conditions, yet remain acyanic when grown under shade conditions. The coloured phenotypes matched with an accumulation of anthocyanins and flavonols, as well as the activation of the early and late flavonoid biosynthetic genes required for flavonol and anthocyanin production. Pigmentation in Lc petunia only occurred under conditions which normally induce a modest amount of anthocyanin to accumulate in wild-type Mitchell petunia [Petunia axillaris x (Petunia axillaris x Petunia hybrida cv. 'Rose of Heaven')]. Anthocyanin pigmentation in Lc petunia leaves appears to screen underlying photosynthetic tissues, increasing light saturation and light compensation points, without reducing the maximal photosynthetic assimilation rate (A(max)). In the Lc petunia system, where the bHLH factor Leaf colour is constitutively expressed, expression of the bHLH (Lc) and WD40 (An11) components of the anthocyanin regulatory system were not limited, suggesting that the high-light-induced anthocyanin pigmentation is regulated by endogenous MYB transcription factors.
[��������: 1]
[��������: 1]
DOI:10.1038/s41438-019-0138-2URLPMID:31098031 [��������: 1]

Pigment intensity and patterns are important factors that determine the nutritional and market values of tomato fruits. The acropetal manner of light-dependent anthocyanin accumulation with the highest levels at the stem end of the fruit makes Pro35S:BrTT8 tomato plants an ideal system for investigating the effects of light intensity on anthocyanin biosynthesis. Extensive transcript analyses indicate that anthocyanin pigmentation in Pro35S:BrTT8 plants under high light might be coordinately regulated by the exogenous protein BrTT8 and endogenous proteins SlAN2 and SlMYBL2. Furthermore, yeast two-hybrid assays showed that BrTT8 could interact efficiently with SlAN2, SlMYBL2, and SlAN11. Moreover, the physical interaction between BrTT8 and SlAN2 was validated by FRET. Simultaneous overexpression of SlAN2 and BrTT8 activated significant anthocyanin biosynthesis in infiltrated tobacco leaves. In addition, the ability of SlMYBL2 to suppress anthocyanin accumulation was also demonstrated in infiltrated tobacco leaves. Altogether, these results prove that tissue-specific assemblage of the heterogeneous MYB-bHLH-WD40 complex consisting of SlAN2, BrTT8 and SlAN11 triggers nonuniform anthocyanin accumulation in tomato fruit under high light. Additionally, it is proposed that a negative-feedback loop fulfilled by SlMYBL2 also participates in the regulation of anthocyanin production.
DOI:10.1016/j.scienta.2019.01.034URL [��������: 1]
DOI:10.1104/pp.107.098293URLPMID:17478635 [��������: 1]

Exposure to high irradiance results in dramatic changes in nuclear gene expression in plants. However, little is known about the mechanisms by which changes in irradiance are sensed and how the information is transduced to the nucleus to initiate the genetic response. To investigate whether the photoreceptors are involved in the response to high irradiance, we analyzed expression of EARLY LIGHT-INDUCIBLE PROTEIN1 (ELIP1), ELIP2, ASCORBATE PEROXIDASE2 (APX2), and LIGHT-HARVESTING CHLOROPHYLL A/B-BINDING PROTEIN2.4 (LHCB2.4) in the phytochrome A (phyA), phyB, cryptochrome1 (cry1), and cry2 photoreceptor mutants and long hypocotyl5 (hy5) and HY5 homolog (hyh) transcription factor mutants. Following exposure to high intensity white light for 3 h (1,000 mumol quanta m(-2) s(-1)) expression of ELIP1/2 and APX2 was strongly induced and LHCB2.4 expression repressed in wild type. The cry1 and hy5 mutants showed specific misregulation of ELIP1/2, and we show that the induction of ELIP1/2 expression is mediated via CRY1 in a blue light intensity-dependent manner. Furthermore, using the Affymetrix Arabidopsis (Arabidopsis thaliana) 24 K Gene-Chip, we showed that 77 of the high light-responsive genes are regulated via CRY1, and 26 of those genes were also HY5 dependent. As a consequence of the misregulation of these genes, the cry1 mutant displayed a high irradiance-sensitive phenotype with significant photoinactivation of photosystem II, indicated by reduced maximal fluorescence ratio. Thus, we describe a novel function of CRY1 in mediating plant responses to high irradiances that is essential to the induction of photoprotective mechanisms. This indicates that high irradiance can be sensed in a chloroplast-independent manner by a cytosolic/nucleic component.
DOI:10.4161/psb.6.1.14082URLPMID:21248473 [��������: 1]

Light is the ultimate energy source for photo-autotrophs on earth. For green plants, however, it can also be toxic under certain stressful environmental conditions and at critical developmental stages. Anthocyanins, a class of flavonoids, act as an effective screening mechanism that allows plant survival and proliferation under occasional periods of harmful irradiation through modulation of light absorption. Apart from light-sensing through photoreceptors such as phytochrome and cryptochrome, plants use the photosynthetic electron transfer (PET) chain to integrate light information. The redox status of the plastoquinone (PQ) pool of the PET chain regulates anthocyanin biosynthesis genes, together with the plant hormone ethylene and plant hormone-like sugars. A complex signaling apparatus in acyanic cells appears to transduce information to cyanic cells to regulate anthocyanin production through an intercellular signaling pathway that remains largely uncharacterized. This review will highlight recent advances in this field and their implications for the regulation of anthocyanin pigmentation.
DOI:10.3969/j.issn.1674-3466.2010.03.002URL [��������: 2]

Anthocyanin is one of the most important plant pigments for the color of flowers, fruits and seedlings. Anthocyanin synthesis and accumulation are closely related to plant growth and development and are subject to internal and external factors. Activation of the anthocyanin pathway and accumulation of the pigment require many environmental signals. Many studies have shown that environmental factors induce anthocyanin accumulation via the activation of anthocyanin biosynthetic genes. This paper reviews the effect of major environmental factors on gene expression patterns of anthocyanin synthesis and regulation of anthocyanin accumulation and stability. Light is one of the most important stimulators, and light quality is more important than light intensity. Low temperatures increase and high temperatures decrease anthocyanin concentration. Most of the structural and regulation genes involved in anthocyanin biosynthesis can be regulated by different sugars. Three aspects demand further research: the relationship between flower development and flower pigmentation, the response of flower color to environmental factors, and the resistance mechanism of anthocyanin to stress. Controlling flower color by environmental factors will greatly improve the quality of ornamentals.
DOI:10.3969/j.issn.1674-3466.2010.03.002URL [��������: 2]

Anthocyanin is one of the most important plant pigments for the color of flowers, fruits and seedlings. Anthocyanin synthesis and accumulation are closely related to plant growth and development and are subject to internal and external factors. Activation of the anthocyanin pathway and accumulation of the pigment require many environmental signals. Many studies have shown that environmental factors induce anthocyanin accumulation via the activation of anthocyanin biosynthetic genes. This paper reviews the effect of major environmental factors on gene expression patterns of anthocyanin synthesis and regulation of anthocyanin accumulation and stability. Light is one of the most important stimulators, and light quality is more important than light intensity. Low temperatures increase and high temperatures decrease anthocyanin concentration. Most of the structural and regulation genes involved in anthocyanin biosynthesis can be regulated by different sugars. Three aspects demand further research: the relationship between flower development and flower pigmentation, the response of flower color to environmental factors, and the resistance mechanism of anthocyanin to stress. Controlling flower color by environmental factors will greatly improve the quality of ornamentals.
[D].
[��������: 1]
[D].
[��������: 1]
DOI:10.7506/spkx1002-6630-201512048URL [��������: 1]

The effects of UV-C treatment at different dosages on postharvest quality, antioxidant activity, phenylpropanemetabolite contents, and related enzymes activities in Chinese bayberry fruit (cv. Dongkui)were investigated. The fruit weretreated with 0, 1.5, 3.0, 4.5 and 6.0 kJ/m2 of UV-C and then stored at 5 �� for up to 12 days. The results indicated that UV-Ctreatment at 3.0 kJ/m2 had the most significant inhibitory effect on fruit decay. The decreases of fruit firmness and vitamin Ccontent were retarded and thus fruit quality was maintained. Meanwhile, this UV-C treatment significantly enhanced theactivities of phenyalanine ammonialyase, 4-coumarate-CoA ligase, chalcone isomerase and cinnamate 4-hydroxylase, andmaintained high levels of total phenolics, anthocyanin, carotenoids and flavonoid and high antioxidant activity during thestorage. These results suggest that UV-C treatment has a promising application prospect in quality maintenance of harvestedChinese bayberry fruit.
DOI:10.7506/spkx1002-6630-201512048URL [��������: 1]

The effects of UV-C treatment at different dosages on postharvest quality, antioxidant activity, phenylpropanemetabolite contents, and related enzymes activities in Chinese bayberry fruit (cv. Dongkui)were investigated. The fruit weretreated with 0, 1.5, 3.0, 4.5 and 6.0 kJ/m2 of UV-C and then stored at 5 �� for up to 12 days. The results indicated that UV-Ctreatment at 3.0 kJ/m2 had the most significant inhibitory effect on fruit decay. The decreases of fruit firmness and vitamin Ccontent were retarded and thus fruit quality was maintained. Meanwhile, this UV-C treatment significantly enhanced theactivities of phenyalanine ammonialyase, 4-coumarate-CoA ligase, chalcone isomerase and cinnamate 4-hydroxylase, andmaintained high levels of total phenolics, anthocyanin, carotenoids and flavonoid and high antioxidant activity during thestorage. These results suggest that UV-C treatment has a promising application prospect in quality maintenance of harvestedChinese bayberry fruit.
[��������: 1]
[��������: 1]
DOI:10.1073/pnas.1316622110URL [��������: 1]

The evolutionarily conserved CONSTITUTIVE PHOTOMORPHOGENESIS 1 (COP1) is a RING and WD40 protein that functions as a substrate receptor of CULLIN4-DAMAGED DNA BINDING PROTEIN 1 (CUL4-DDB1)-based E3 ubiquitin ligases in both plants and animals. In Arabidopsis, COP1 is a central repressor of photomorphogenesis in the form of COP1-SUPPRESSOR OF PHYA (SPA) complex(es). CUL4-DDB1-COP1-SPA suppresses the photomorphogenic program by targeting the transcription factor ELONGATED HYPOCOTYL 5 for degradation. Intriguingly, under photomorphogenic UV-B light, COP1 reverses its repressive role and promotes photomorphogenesis. However, the mechanism by which COP1 is functionally switched is still obscure. Here, we demonstrate that UV-B triggers the physical and functional disassociation of the COP1-SPA core complex(es) from CUL4-DDB1 and the formation of a unique complex(es) containing the UV-B receptor UV RESISTANCE LOCUS 8 (UVR8). The establishment of this UV-B-dependent COP1 complex(es) is associated with its positive modulation of ELONGATED HYPOCOTYL 5 stability and activity, which sheds light on the mechanism of COP1's promotive action in UV-B-induced photomorphogenesis.
DOI:10.3389/fpls.2016.00153URLPMID:26909096 [��������: 1]

In the last decade plant biotechnologists and breeders have made several attempt to improve the antioxidant content of plant-derived food. Most efforts concentrated on increasing the synthesis of antioxidants, in particular anthocyanins, by inducing the transcription of genes encoding the synthesizing enzymes. We present here an overview of economically interesting plant species, both food crops and ornamentals, in which anthocyanin content was improved by traditional breeding or transgenesis. Old genetic studies in petunia and more recent biochemical work in brunfelsia, have shown that after synthesis and compartmentalization in the vacuole, anthocyanins need to be stabilized to preserve the color of the plant tissue over time. The final yield of antioxidant molecules is the result of the balance between synthesis and degradation. Therefore the understanding of the mechanism that determine molecule stabilization in the vacuolar lumen is the next step that needs to be taken to further improve the anthocyanin content in food. In several species a phenomenon known as fading is responsible for the disappearance of pigmentation which in some case can be nearly complete. We discuss the present knowledge about the genetic and biochemical factors involved in pigment preservation/destabilization in plant cells. The improvement of our understanding of the fading process will supply new tools for both biotechnological approaches and marker-assisted breeding.
DOI:10.1104/pp.112.199703URLPMID:22855936 [��������: 1]

MdMYB1 is a crucial regulator of light-induced anthocyanin biosynthesis and fruit coloration in apple (Malus domestica). In this study, it was found that MdMYB1 protein accumulated in the light but degraded via a ubiquitin-dependent pathway in the dark. Subsequently, the MdCOP1-1 and MdCOP1-2 genes were isolated from apple fruit peel and were functionally characterized in the Arabidopsis (Arabidopsis thaliana) cop1-4 mutant. Yeast (Saccharomyces cerevisiae) two-hybrid, bimolecular fluorescence complementation, and coimmunoprecipitation assays showed that MdMYB1 interacts with the MdCOP1 proteins. Furthermore, in vitro and in vivo experiments indicated that MdCOP1s are necessary for the ubiquitination and degradation of MdMYB1 protein in the dark and are therefore involved in the light-controlled stability of the MdMYB1 protein. Finally, a viral vector-based transformation approach demonstrated that MdCOP1s negatively regulate the peel coloration of apple fruits by modulating the degradation of the MdMYB1 protein. Our findings provide new insight into the mechanism by which light controls anthocyanin accumulation and red fruit coloration in apple and even other plant species.
DOI:10.1007/s00299-006-0255-xURL [��������: 1]

Molecular analysis of gene expression differences between green and red lettuce leaves was performed using the SSH method. BlastX comparisons of subtractive expressed sequence tags (ESTs) indicated that 7.6% of clones encoded enzymes involved in secondary metabolism. Such clones had a particularly high abundance of flavonoid-metabolism proteins (6.5%). Following SSH, 566 clones were rescreened for differential gene expression using dot-blot hybridization. Of these, 53 were found to overexpressed during red coloration. The up-regulated expression of six genes was confirmed by Northern blot analyses. The expression of chalcone synthase (CHS), flavanone 3-hydroxylase (F3H), and dihydroflavonol 4-reductase (DFR) genes showed a positive correlation with anthocyanin accumulation in UV-B-irradiated lettuce leaves; flavonoid 3′,5′-hydroxylase (F3′,5′H) and anthocyanidin synthase (ANS) were expressed continuously in both samples. These results indicated that the genes CHS, F3H, and DFR coincided with increases in anthocyanin accumulation during the red coloration of lettuce leaves. This study show a relationship between red coloration and the expression of up-regulated genes in lettuce. The subtractive cDNA library and EST database described in this study represent a valuable resource for further research for secondary metabolism in the vegetable crops.
[��������: 1]
[��������: 1]
DOI:10.1016/j.molp.2020.10.009URLPMID:33164768 [��������: 1]
[��������: 2]
DOI:10.2525/ecb1963.9.3-4_9URL [��������: 2]
DOI:10.1007/s10725-010-9472-yURL [��������: 1]

Ultraviolet A (UV-A) is an environmental stimulus, and UV-A-induced anthocyanin biosynthesis has been previously investigated in the tomato plant (Solanum lycopersicum L.). UV-A induced anthocyanin biosynthesis in tomato seedlings and co-irradiation with visible light and UV-A did not influence the content of UV-A-induced anthocyanin accumulation. UV-A irradiation induced significant accumulations of anthocyanin in both the cotyledons and hypocotyls of tomato seedlings. Anthocyanin production increased gradually in the tomato hypocotyls after exposure to UV-A and reached maximum levels at 12h. In the cotyledons, anthocyanin accumulation was significantly increased at 1h after UV-A exposure and was reduced afterward; however, it increased again beginning at 3h. The expression of the phenylalanine ammonia-lyase (SlPAL5) gene was shown to be increased after UV-A exposure in a time-dependent manner. UV-A irradiation was also shown to induce anthocyanin accumulation in the epidermis of the tomato fruit; however, SlPAL5 transcripts were detected only at 3 and 24h after UV-A treatment. After a 1h pulse of UV-A, SlPAL5 transcripts were increased significantly in tomato cotyledons and hypocotyls after transfer to dark conditions for a short time.
DOI:10.1093/jxb/erm036URLPMID:17426056 [��������: 1]

Ultraviolet A (UV-A)-mediated regulation of anthocyanin biosynthesis was investigated in swollen hypocotyls of the red turnip 'Tsuda'. The shaded swollen hypocotyls which contained negligible anthocyanin were exposed to artificial light sources including low fluence UV-B, UV-A, blue, red, far-red, red plus UV-A, far-red plus UV-A, and blue plus red. Among these lights, only UV-A induced anthocyanin biosynthesis and co-irradiation of red or far-red with UV-A did not affect the extent of UV-A-induced anthocyanin accumulation. The expression of phenylalanine ammonia lyase (PAL; EC 4.3.1.5), chalcone synthase (CHS; EC 2.3.1.74), flavanone 3-hydroxylase (F3H; EC 1.14.11.9), dihydroflavonol 4-reductase (DFR; EC 1.1.1.219), and anthocyanidin synthase (ANS; EC 1.14.11.19) genes was increased with time during a 24 h exposure to UV-A. In contrast, irradiation with red, blue, UV-B, and a combination of blue with red failed to induce CHS expression. Microarray analysis showed that only a few genes, including CHS and F3H, were induced significantly by UV-A, while a separate set of many genes was induced by low fluence UV-B. The UV-A-specific induction of anthocyanin biosynthesis and the unique gene expression profile upon UV-A irradiation as compared with blue and UV-B demonstrated that the observed induction of anthocyanin biosynthesis in red turnips was mediated by a distinct UV-A-specific photoreceptor, but not by phytochromes, UV-A/blue photoreceptors, or UV-B photoreceptors.
DOI:10.1038/nrg2049URLPMID:17304247 [��������: 1]

Plants have evolved complex and sophisticated transcriptional networks that mediate developmental changes in response to light. These light-regulated processes include seedling photomorphogenesis, seed germination and the shade-avoidance and photoperiod responses. Understanding the components and hierarchical structure of the transcriptional networks that are activated during these processes has long been of great interest to plant scientists. Traditional genetic and molecular approaches have proved powerful in identifying key regulatory factors and their positions within these networks. Recent genomic studies have further revealed that light induces massive reprogramming of the plant transcriptome, and that the early light-responsive genes are enriched in transcription factors. These combined approaches provide new insights into light-regulated transcriptional networks.
DOI:10.1016/j.tplants.2012.05.004URL [��������: 1]

COP1 and DET1 are among the first repressors of photomorphogenesis to be identified, more than 20 years ago. Discovery of these repressors as conserved regulators of the ubiquitin-proteasome system has established protein degradation as a central theme in light signal transduction. COP1 is a RING E3 ubiquitin ligase that targets key regulators for degradation, and DET1 complexes with COP10 and DDB1, which is proposed to aid in COP1-mediated degradation. Recent studies have strengthened the role of COP1 as a major signaling center. DET1 is also emerging as a chromatin regulator in repressing gene expression. Here, we review current understanding on COP1 and DET1, with a focus on their role as part of two distinct, multimeric CUL4-based E3 ligases.
DOI:10.1101/gad.2025011URLPMID:21511871 [��������: 2]

Plant photoreceptors mediate light suppression of the E3 ubiquitin ligase COP1 (CONSTITUTIVE PHOTOMORPHOGENIC 1) to affect gene expression and photomorphogenesis. However, how photoreceptors mediate light regulation of COP1 activity remains unknown. We report here that Arabidopsis blue-light receptor cryptochrome 1 (CRY1) undergoes blue-light-dependent interaction with the COP1-interacting protein SPA1 (SUPPRESSOR OF PHYTOCHROME A). We further show that the CRY1-SPA1 interaction suppresses the SPA1-COP1 interaction and COP1-dependent degradation of the transcription factor HY5. These results are consistent with a hypothesis that photoexcited CRY1 interacts with SPA1 to modulate COP1 activity and plant development.
DOI:10.1007/s11418-015-0935-3URLPMID:26481011 [��������: 2]

Alcoholic liver disease (ALD) is a serious and challenging health issue. In the past decade, natural components possessing hepatoprotective properties have gained more attention for ALD intervention. In this study, the phytochemical components of anthocyanins from purple potato were assessed using UPLC-MS/MS, and the hepatoprotective effects of purple potato anthocyanins (PPAs) were investigated in the ALD mouse model. Serum and liver biochemical parameters were determined, along with histopathological changes in liver tissue. In addition, the major contributors to alcohol-induced oxidative stress were assessed. The results indicated that the levels of aspartate transaminase and alanine transaminase were lower in the serum of the PPA-treated group than the alcohol-treated group. PPAs significantly inhibited the reduction of total cholesterol and triglycerides. Higher levels of superoxide dismutase and reduced glutathione enzymes as well as a reduction in the formation of malondialdehyde occurred in mice fed with PPAs. In addition, PPAs protected against increased alcohol-induced levels and activity of cytochrome P450 2E1 (CYP2E1), which demonstrates the effects of PPAs against alcohol-induced oxidative stress and liver injury. This study suggests that PPAs could be an effective therapeutic agent in alcohol-induced liver injuries by inhibiting CYP2E1 expression and thereby strengthening antioxidant defenses.
DOI:10.1111/pce.12456URLPMID:25256341 [��������: 1]

ANGUSTIFOLIA3 (AN3), a transcription coactivator, is implicated in modulating cell proliferation. In this study, I found that AN3 is a novel regulator of anthocyanin biosynthesis and light-induced root elongation. Seedlings and seeds lacking AN3 activity presented significantly reduced anthocyanin accumulation and light-induced root elongation, whereas those of transgenic plants harbouring the 35S:AN3 construct exhibited increased anthocyanin accumulation. AN3 is required for the proper expression of other genes that affect anthocyanin accumulation and light-induced root elongation, Constitutive Photomorphogenic1 (COP1), encoding a RING motif - containing E3 ubiquitin ligase. AN3 was associated with COP1 promoter in vivo. Thus, AN3 may act with other proteins that bind to COP1 promoter to promote anthocyanin accumulation and inhibit light-induced root elongation.
DOI:10.1016/j.scienta.2016.11.005URL [��������: 2]
DOI:10.1016/j.scienta.2014.09.022URL [��������: 1]

Effect of blue light-emitting diode light (470 nm) treatment on anthocyanin accumulation and expression of related genes in Chinese bayberry fruit was investigated. Bayberries hand-harvested when nearly 3/4 of the fruit turned red were exposed to blue light for 8 days at 10 degrees C. Anthocyanin accumulated in bayberries during storage, which can be drastically enhanced by blue light treatment. The blue light treatment also increased the expression of MrMYB I and structural genes involved in anthocyanin biosynthesis such as MrCHI, MrF3H, MrF3'H, MrDFR1, MrDFR2 and MrANS. These findings suggested that the induction of anthocyanin accumulation by blue light was associated with the increased expression of anthocyanin biosynthetic and regulatory genes. Our results suggested that blue light treatment may be a useful technique to enhance commercial and nutritional value of Chinese bayberry fruit. (C) 2014 Elsevier B.V.
DOI:10.1016/j.postharvbio.2018.11.011URL [��������: 1]
DOI:10.1007/s10265-013-0582-2URL [��������: 1]

Anthocyanins are widespread, essential secondary metabolites in higher plants during color development in certain flowers and fruits. In strawberries, anthocyanins are also key contributors to fruit antioxidant capacity and nutritional value. However, the effects of different light qualities on anthocyanin accumulation in strawberry (Fragaria x ananassa, cv. Sachinoka) fruits remain elusive. In the present study, we showed the most efficient increase in anthocyanin content occurred by blue light irradiation. Light sensing at the molecular level was investigated by isolation of two phototropin (FaPHOT1 and FaPHOT2), two cryptochrome (FaCRY1 and FaCRY2), and two phytochrome (FaPHYA and FaPHYB) homologs. Expression analysis revealed only FaPHOT2 transcripts markedly increased depending on fruit developmental stage, and a corresponding increase in anthocyanin content was detected. FaPHOT2 knockdown resulted in decreased anthocyanin content; however, overexpression increased anthocyanin content. These findings suggested blue light induced anthocyanin accumulation, and FaPHOT2 may play a role in sensing blue light, and mediating anthocyanin biosynthesis in strawberry fruits. This is the first report to find a relationship between visible light sensing, and color development in strawberry fruits.
DOI:10.1021/jf501120uURL [��������: 1]

Blue light irradiation was applied to postharvest strawberry fruit to explore its influence on anthocyanin content and anthocyanin biosynthetic enzyme activities. Strawberry fruit was irradiated with blue light at 40 mu mol m(-2) s(-1) for 12 days at 5 degrees C. The results indicated that blue light treatment improved total anthocyanin content in strawberry fruit during storage. Meanwhile, the treatment increased the activities of glucose-6-phosphate, shikimate dehydrogenase, tyrosine ammonia-lyase, phenylalanine ammonia-lyase, cinnamate-4-hydroxylase, 4-coumarate/coenzyme A ligase, dihydroflavonol-4-reductase, chalcone synthase, flavanone-3-beta-hydroxylase, anthocyanin synthase, and UDP-glycose flavonoid-3-O-glycosyltranferase, which suggested that the enhancement of anthocyanin concentration by blue light might result from the activation of its related enzymes. Blue light might be proposed as a supplemental light source in the storage of strawberry fruit to improve its anthocyanin content.
DOI:10.1080/07929978.1999.10676777URL [��������: 1]
[��������: 2]
[��������: 2]
DOI:10.1016/j.plantsci.2015.06.001URLPMID:26259175 [��������: 1]

Light is an important environmental factor inducing anthocyanin accumulation in plants. Phytochrome-interacting factors (PIFs) have been shown to be a family of bHLH transcription factors involved in light signaling in Arabidopsis. Red light effectively increased anthocyanin accumulation in wild-type Col-0, whereas the effects were enhanced in pif4 and pif5 mutants but impaired in overexpression lines PIF4OX and PIF5OX, indicating that PIF4 and PIF5 are both negative regulators for red light-induced anthocyanin accumulation. Consistently, transcript levels of several genes involved in anthocyanin biosynthesis and regulatory pathway, including CHS, F3'H, DFR, LDOX, PAP1 and TT8, were significantly enhanced in mutants pif4 and pif5 but decreased in PIF4OX and PIF5OX compared to in Col-0, indicating that PIF4 and PIF5 are transcriptional repressor of these gene. Transient expression assays revealed that PIF4 and PIF5 could repress red light-induced promoter activities of F3'H and DFR in Arabidopsis protoplasts. Furthermore, chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) test and electrophoretic mobility shift assay (EMSA) showed that PIF5 could directly bind to G-box motifs present in the promoter of DFR. Taken together, these results suggest that PIF4 and PIF5 negatively regulate red light-induced anthocyanin accumulation through transcriptional repression of the anthocyanin biosynthetic genes in Arabidopsis.
DOI:10.1073/pnas.1120764109URL [��������: 1]

Phytochromes (phy) are red/far-red-absorbing photoreceptors that regulate the adaption of plant growth and development to changes in ambient light conditions. The nuclear transport of the phytochromes upon light activation is regarded as a key step in phytochrome signaling. Although nuclear import of phyA is regulated by the transport facilitators far red elongated hypocotyl 1 (FHY1) and fhy1-like, an intrinsic nuclear localization signal was proposed to be involved in the nuclear accumulation of phyB. We recently showed that nuclear import of phytochromes can be analyzed in a cell-free system consisting of isolated nuclei of the unicellular green algae Acetabularia acetabulum. We now show that this system is also versatile to elucidate the mechanism of the nuclear transport of phyB. We tested the nuclear transport characteristics of full-length phyB as well as N-and C-terminal phyB fragments in vitro and showed that the nuclear import of phyB can be facilitated by phytochrome-interacting factor 3 (PIF3). In vivo measurements of phyB nuclear accumulation in the absence of PIF1, -3, -4, and -5 indicate that these PIFs are the major transport facilitators during the first hours of deetiolation. Under prolonged irradiations additional factors might be responsible for phyB nuclear transport in the plant.
DOI:10.1105/tpc.6.4.487URLPMID:8205001 [��������: 4]

The Arabidopsis protein COP1, encoded by the constitutive photomorphogenic locus 1, is an essential regulatory molecule that plays a role in the repression of photomorphogenic development in darkness and in the ability of light-grown plants to respond to photoperiod, end-of-day far-red treatment, and ratio of red/far-red light. The COP1 protein contains three recognizable structural domains: starting from the N terminus, they are the zinc binding motif, the putative coiled-coil region, and the domain with multiple WD-40 repeats homologous to the beta subunit of trimeric G-proteins (G beta). To understand the functional implications of these structural motifs, 17 recessive mutations of the COP1 gene have been isolated based on their constitutive photomorphogenic seedling development in darkness. These mutations define three phenotypic classes: weak, strong, and lethal. The mutations that fall into the lethal class are possible null mutations of COP1. Molecular analysis of the nine mutant alleles that accumulated mutated forms of COP1 protein revealed that disruption of the G beta-protein homology domain or removal of the very C-terminal 56 amino acids are both deleterious to COP1 function. In-frame deletions or insertions of short amino acid stretches between the putative coiled-coil and G beta-protein homology domains strongly compromised COP1 function. However, a mutation resulting in a COP1 protein with only the N-terminal 282 amino acids, including both the zinc binding and the coiled-coil domains, produced a weak phenotypic defect. These results indicated that the N-terminal half of COP1 alone retains some activity and a disrupted C-terminal domain masks this remaining activity.
DOI:10.12677/BR.2019.82016URL [��������: 1]
DOI:10.12677/BR.2019.82016URL [��������: 1]
[D].
[��������: 1]
[D].
[��������: 1]
[D].
[��������: 1]
[D].
[��������: 1]
DOI:10.1111/tpj.12153URLPMID:23425305 [��������: 1]

Anthocyanins are natural pigments that accumulate only in light-grown and not in dark-grown Arabidopsis plants. Repression of anthocyanin accumulation in darkness requires the CONSTITUTIVELY PHOTOMORPHOGENIC1/SUPPRESSOR OF PHYA-105 (COP1/SPA) ubiquitin ligase, as cop1 and spa mutants produce anthocyanins also in the dark. Here, we show that COP1 and SPA proteins interact with the myeloblastosis (MYB) transcription factors PRODUCTION OF ANTHOCYANIN PIGMENT1 (PAP)1 and PAP2, two members of a small protein family that is required for anthocyanin accumulation and for the expression of structural genes in the anthocyanin biosynthesis pathway. The increased anthocyanin levels in cop1 mutants requires the PAP1 gene family, indicating that COP1 functions upstream of the PAP1 gene family. PAP1 and PAP2 proteins are degraded in the dark and this degradation is dependent on the proteasome and on COP1. Hence, the light requirement for anthocyanin biosynthesis results, at least in part, from the light-mediated stabilization of PAP1 and PAP2. Consistent with this conclusion, moderate overexpression of PAP1 leads to an increase in anthocyanin levels only in the light and not in darkness. Here we show that SPA genes are also required for reducing PAP1 and PAP2 transcript levels in dark-grown seedlings. Taken together, these results indicate that the COP1/SPA complex affects PAP1 and PAP2 both transcriptionally and post-translationally. Thus, our findings have identified mechanisms via which the COP1/SPA complex controls anthocyanin levels in Arabidopsis that may be useful for applications in biotechnology directed towards increasing anthocyanin content in plants.
DOI:10.1104/pp.106.088104URLPMID:17012405 [��������: 1]

Anthocyanins are secondary metabolites found in higher plants that contribute to the colors of flowers and fruits. In apples (Malus domestica Borkh.), several steps of the anthocyanin pathway are coordinately regulated, suggesting control by common transcription factors. A gene encoding an R2R3 MYB transcription factor was isolated from apple (cv Cripps' Pink) and designated MdMYB1. Analysis of the deduced amino acid sequence suggests that this gene encodes an ortholog of anthocyanin regulators in other plants. The expression of MdMYB1 in both Arabidopsis (Arabidopsis thaliana) plants and cultured grape cells induced the ectopic synthesis of anthocyanin. In the grape (Vitis vinifera) cells MdMYB1 stimulated transcription from the promoters of two apple genes encoding anthocyanin biosynthetic enzymes. In ripening apple fruit the transcription of MdMYB1 was correlated with anthocyanin synthesis in red skin sectors of fruit. When dark-grown fruit were exposed to sunlight, MdMYB1 transcript levels increased over several days, correlating with anthocyanin synthesis in the skin. MdMYB1 gene transcripts were more abundant in red skin apple cultivars compared to non-red skin cultivars. Several polymorphisms were identified in the promoter of MdMYB1. A derived cleaved amplified polymorphic sequence marker designed to one of these polymorphisms segregated with the inheritance of skin color in progeny from a cross of an unnamed red skin selection (a sibling of Cripps' Pink) and the non-red skin cultivar Golden Delicious. We conclude that MdMYB1 coordinately regulates genes in the anthocyanin pathway and the expression level of this regulator is the genetic basis for apple skin color.
DOI:10.1038/nbt.1506URLPMID:18953354 [��������: 1]

Dietary consumption of anthocyanins, a class of pigments produced by higher plants, has been associated with protection against a broad range of human diseases. However, anthocyanin levels in the most commonly eaten fruits and vegetables may be inadequate to confer optimal benefits. When we expressed two transcription factors from snapdragon in tomato, the fruit of the plants accumulated anthocyanins at levels substantially higher than previously reported for efforts to engineer anthocyanin accumulation in tomato and at concentrations comparable to the anthocyanin levels found in blackberries and blueberries. Expression of the two transgenes enhanced the hydrophilic antioxidant capacity of tomato fruit threefold and resulted in fruit with intense purple coloration in both peel and flesh. In a pilot test, cancer-susceptible Trp53(-/-) mice fed a diet supplemented with the high-anthocyanin tomatoes showed a significant extension of life span.
DOI:10.1093/pcp/pcm131URLPMID:17925311 [��������: 1]

Flower color is mainly determined by anthocyanins. Rosa hybrida lacks violet to blue flower varieties due to the absence of delphinidin-based anthocyanins, usually the major constituents of violet and blue flowers, because roses do not possess flavonoid 3',5'-hydoxylase (F3'5'H), a key enzyme for delphinidin biosynthesis. Other factors such as the presence of co-pigments and the vacuolar pH also affect flower color. We analyzed the flavonoid composition of hundreds of rose cultivars and measured the pH of their petal juice in order to select hosts of genetic transformation that would be suitable for the exclusive accumulation of delphinidin and the resulting color change toward blue. Expression of the viola F3'5'H gene in some of the selected cultivars resulted in the accumulation of a high percentage of delphinidin (up to 95%) and a novel bluish flower color. For more exclusive and dominant accumulation of delphinidin irrespective of the hosts, we down-regulated the endogenous dihydroflavonol 4-reductase (DFR) gene and overexpressed the Irisxhollandica DFR gene in addition to the viola F3'5'H gene in a rose cultivar. The resultant roses exclusively accumulated delphinidin in the petals, and the flowers had blue hues not achieved by hybridization breeding. Moreover, the ability for exclusive accumulation of delphinidin was inherited by the next generations.
DOI:10.1093/pcp/pct110URLPMID:23926066 [��������: 1]

Chrysanthemums (Chrysanthemumxmorifolium Ramat.) are an important cut-flower and potted plant crop in the horticultural industry world wide. Chrysanthemums express the flavonoid 3'-hydroxylase (F3'H) gene and thus accumulate anthocyanins derived from cyanidin in their inflorescences which appear pink/red. Delphinidin-based anthocyanins are lacking due to the deficiency of a flavonoid 3', 5'-hydroxylase (F3'5'H), and so violet/blue chrysanthemum flower colors are not found. In this study, together with optimization of transgene expression and selection of the host cultivars and gene source, F3'5'H genes have been successfully utilized to produce transgenic bluish chrysanthemums that accumulate delphinidin-based anthocyanins. HPLC analysis and feeding experiments with a delphinidin precursor identified 16 cultivars of chrysanthemums out of 75 that were predicted to turn bluish upon delphinidin accumulation. A selection of eight cultivars were successfully transformed with F3'5'H genes under the control of different promoters. A pansy F3'5'H gene under the control of a chalcone synthase promoter fragment from rose resulted in the effective diversion of the anthocyanin pathway to produce delphinidin in transgenic chrysanthemum flower petals. The resultant petal color was bluish, with 40% of total anthocyanidins attributed to delphinidin. Increased delphinidin levels (up to 80%) were further achieved by hairpin RNA interference-mediated silencing of the endogenous F3'H gene. The resulting petal colors were novel bluish hues, not possible by hybridization breeding. This is the first report of the production of anthocyanins derived from delphinidin in chrysanthemum petals leading to novel flower color.
DOI:10.1111/ppl.12475URLPMID:27229540 [��������: 1]

In this study, we investigate the metabolic engineering of anthocyanins in two dark tobacco crops (Narrow Leaf Madole and KY171) and evaluate the effects on physiological features of plant photosynthesis. Arabidopsis PAP1 (production of anthocyanin pigment 1) gene (AtPAP1) encodes a R2R3-type MYB transcript factor that is a master component of regulatory complexes controlling anthocyanin biosynthesis. AtPAP1 was introduced to Narrow Leaf Madole and KY171 plants. Multiple transgenic plants developed red/purple pigmentation in different tissues. Quantitative real-time polymerase chain reaction (qRT-PCR) analysis showed that the expression levels of six pathway genes were increased two- to eight-fold in AtPAP1 transgenic plants compared with vector control plants. Dihydroflavonol reductase and anthocyanidin synthase genes that were not expressed in wild-type plants were activated. Spectrophotometric measurement showed that the amount of anthocyanins in AtPAP1 transgenic plants were 400-800 microg g(-1) fresh weight (FW). High-performance liquid chromatography (HPLC) analysis showed that one main anthocyanin molecule accounted for approximately 98% of the total anthocyanins. Tandem MS/MS analysis using HPLC coupled to electrospray ionization and quadrupole time-of-flight mass spectrometry identified the main anthocyanin as cyanidin 3-O-rutinoside, an important medicinal anthocyanin. Analysis of photosynthesis rate, chlorophylls and carotenoids contents showed no differences between red/purple transgenic and control plants, indicating that this metabolic engineering did not alter photosynthetic physiological traits. This study shows that AtPAP1 is of significance for metabolic engineering of anthocyanins in crop plants for value-added traits.
