 ,1, 李铃仙1, 曹阳2, 张玮1, 彭军波1, 燕继晔1, 李兴红
,1, 李铃仙1, 曹阳2, 张玮1, 彭军波1, 燕继晔1, 李兴红 ,1
,1Prediction and Analysis of Candidate Secreted Proteins from the Genome of Lasiodiplodia theobromae
XING QiKai ,1, LI LingXian1, CAO Yang2, ZHANG Wei1, PENG JunBo1, YAN JiYe1, LI XingHong
,1, LI LingXian1, CAO Yang2, ZHANG Wei1, PENG JunBo1, YAN JiYe1, LI XingHong ,1
,1通讯作者:
责任编辑: 岳梅
收稿日期:2020-02-14接受日期:2020-03-20网络出版日期:2020-12-16
| 基金资助: |
Received:2020-02-14Accepted:2020-03-20Online:2020-12-16
作者简介 About authors
邢启凯,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1530KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
邢启凯, 李铃仙, 曹阳, 张玮, 彭军波, 燕继晔, 李兴红. 可可毛色二孢全基因组分泌蛋白的预测及分析[J]. 中国农业科学, 2020, 53(24): 5027-5038 doi:10.3864/j.issn.0578-1752.2020.24.006
XING QiKai, LI LingXian, CAO Yang, ZHANG Wei, PENG JunBo, YAN JiYe, LI XingHong.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】葡萄溃疡病(Botryosphaeria dieback)是葡萄(Vitis vinifera)生产上最重要的枝干病害之一,在世界葡萄主产区造成巨大的经济损失[1]。通常认为,该类型病原真菌主要通过自然孔口或修剪伤口侵入,感病果木出现枝干溃疡、果梗干枯、果实干缩、掉粒以及树势减弱等症状,更严重的会引起根腐,造成整株果木枯死[2]。至今,在我国葡萄产区共分离获得6种葡萄座腔菌科(Botryosphaeriaceae)真菌可引起葡萄溃疡病,其中优势种群为可可毛色二孢(Lasiodiplodia theobromae)和葡萄座腔菌(Botryosphaeria dothidea),而致病力最强的为可可毛色二孢[3,4,5]。目前普遍使用化学药剂防治葡萄溃疡病,但至今没有获得低毒环保并能够高效防治该病害的有效药剂。病原真菌分泌蛋白(secreted protein)在病原真菌与宿主植物的互作中起着至关重要的作用,其直接影响病原菌的侵入、扩展、定殖以及病害发生[6,7]。可可毛色二孢全基因组范围内分泌蛋白的筛选鉴定,将对揭示其致病机理,进一步制定葡萄溃疡病的防治新策略具有重要意义。【前人研究进展】分泌蛋白是指在病原菌体内合成后,通过内质网/高尔基体等分泌途径转运到宿主细胞质膜外空间或细胞内发挥功能的蛋白分子[8,9]。经典的分泌蛋白具有以下特征[10]:(1)氨基酸N端含有信号肽序列;(2)无糖基磷脂酰肌醇(glycosylphosphatidylinositol,GPI)锚定位点;(3)没有跨膜结构域;(4)没有将蛋白输送至叶绿体、线粒体等胞内细胞器的定位信号。研究表明,在病原真菌的侵入、扩展和定殖等致病过程中,往往有大量的分泌蛋白参与其中,扮演着重要角色[11,12]。例如,在侵入宿主植物期间,病原真菌可以分泌大量的植物细胞壁降解酶、蛋白水解酶和代谢相关酶等酶类以及激发子蛋白,进而降解植物细胞壁以及植物细胞内复杂的碳氮化合物,一方面获得营养物质供病原菌生长,促使其在宿主中的侵入、定殖和扩散;另一方面阻碍或抑制宿主植物的免疫系统,从而完成其致病过程[9,13]。病原物来源的分泌蛋白与宿主植物受体蛋白的识别和信号传导研究是揭示病原物与宿主互作机制的关键点[14,15],因而研究者完成了多种病原菌全基因组水平的分泌蛋白预测研究[16,17,18,19,20,21],为其他病原菌分泌蛋白的预测及分析提供了参考。【本研究切入点】目前,关于葡萄溃疡病的研究主要集中在病原真菌的种类鉴定、种群结构、侵染过程以及毒素分泌等方面,关于其病原菌分泌蛋白的研究尚未报道。实验前期首先应用SOAPdenovo组装软件对可可毛色二孢菌株CSS-01s进行了de novo测序组装分析[13],其全基因组大小为43.3 M,测序深度为90X,GC含量为54.77%[22]。通过组装拼接共获得60条contigs和29条scaffolds,并且通过基因软件预测并与各数据库比对共注释获得12 902个蛋白序列[22]。【拟解决的关键问题】在已公布的可可毛色二孢菌株CSS-01s全基因组信息的基础上[22],根据分泌蛋白典型特征,利用生物信息学在线程序进行预测分析,以期获得所有可编码经典分泌蛋白的相关基因,为揭示葡萄与可可毛色二孢等葡萄座腔菌科真菌互作的分子机制,并进一步持续有效地开展葡萄溃疡病的防控打下基础。1 材料与方法
试验于2018—2019年在北京市农林科学院完成。1.1 供试材料
供试可可毛色二孢菌株CSS-01s为课题组分离、单孢纯化、鉴定并保存。可可毛色二孢12 902个蛋白序列来源于PRJNA339237(accession number SRP107819)[22]。一年生葡萄‘夏黑’(Summer Black)绿枝条由北京市林业果树科学研究院提供。酵母信号序列诱捕载体pSUC2T7M13ORI(pSUC2)、对照载体和酵母菌株YTK12(suc2Δ9trp1Δade2-101 ura3-52)由中国农业大学植物保护学院孙文献教授提供。1.2 分泌蛋白筛选程序
应用SignalP v5.0[23](1.3 分泌蛋白信号肽功能分析
1.3.1 载体构建 参照JACOBS等[30,31]方法,用蔗糖酶缺陷的酵母分泌系统对预测的分泌蛋白信号肽的功能进行分析验证。根据可可毛色二孢效应子序列信息,用生物信息学在线程序SignalP v5.0进行预测分析所选效应子的信号肽区域,信号肽通常是位于蛋白质N端、大小为40个氨基酸的多肽。依据该目的区段,利用Primer5.0在线程序设计载体构建所需的特异性引物,并在引物5′和3′端分别添加EcoR I和Xho I酶切位点,保证目的片段可单方向插入功能质粒。Trizol(Invitrogen)法提取可可毛色二孢的RNA,反转录得到cDNA。以此为模板进行PCR扩增,对表1所选的9个候选分泌蛋白信号肽片段进行扩增,反应程序:95℃预变性3 min;进入95℃变性30 s,60℃退火30 s,72℃延伸20 s,共32个循环;72℃延伸10 min。所用引物见表1,引物由上海生工生物工程有限公司合成。用EcoR I和Xho I酶切PCR片段,将回收产物连接到pSUC2相同的酶切位点间,随后对克隆进行PCR筛选鉴定,最后将重组质粒送北京擎科新业生物技术有限公司进行测序验证。Table 1
表1
表1分泌蛋白信号肽活性测定载体构建引物序列
Table 1
| 基因 Gene | 正向引物序列 Forward primer (5′-3′) | 反向引物序列 Reverse primer (5′-3′) |
|---|---|---|
| LT_159 | TTTATGAATTCATGGTCAAGGCTTCCACC | TAATACTCGAGGGCATCGGTGAAGGTGCAG |
| LT_188 | TTTATGAATTCATGCGTGTTTCGACTCTTC | TAATACTCGAGAAAGAAGGTGAAGGTAGAAG |
| LT_233 | TTTATGAATTCATGGTCAAGGTTTCCACC | TAATACTCGAGGGTGAAAGTGCAGCTGG |
| LT_359 | TTTATGAATTCATGCCTTCCCTCAAGTC | TAATACTCGAGGTTTTCGGCGGCCTGGG |
| LT_595 | TACAGGAATTCATGCGTTCCTCTGCTC | GACTGCTCGAGCACGATGTCGAGATCAG |
| LT_62 | CCGGAATTCATGGGCTGGTTTTGGTTC | CCGCTCGAGCACGACGGTGATCGTCG |
| LT_936 | TACTAGAATTCATGAAGGCTTCCGGTC | CACTTCTCGAGACCGTTGACAGCCTGAC |
| LT_1541 | TTTATGAATTCATGGTGTCCTTCCGCTCTC | TAATACTCGAGGAGAGACTGCCTGGCAATC |
| LT_1698 | TTTATGAATTCATGAAGTTCTCTACCACC | TAATACTCGAGGTCCTCGGTGACCTCGCC |
新窗口打开|下载CSV
1.3.2 酵母感受态的制备与载体转化 依照Frozen- EZ Yeast Transformation II KitTM(Zymo Research,Orange,CA,USA)提供的方法制备蔗糖酶缺陷型酵母YTK12菌株的感受态,具体方法参照试剂盒说明书。将酵母YTK12菌株在YPDA平板上划线,30℃培养3—5 d;挑单斑至YPDA液体培养基摇培,直到OD600=0.7—0.8,收集菌液,用E1悬浮;再次离心倒掉上清,加入500 μL的E2,即为制备好的YTK18感受态,-80℃保存备用。
将构建好的载体、阳性对照载体pSUC2-Avr1b以及阴性对照载体pSUC2-Mg87分别转化到酵母YTK18感受态中,涂到CMD-W培养基(0.67%不含氨基酸的酵母氮源、0.075%色氨酸一缺培养基、0.1%葡萄糖、2%蔗糖和2%琼脂)上培养3—5 d。将筛选获得的转化子划线到YPRAA培养基(1%酵母提取物、2%蛋白胨、2 μg·L-1的抗霉素A、2%棉籽糖和2%琼脂)上培养3—5 d,通过观察酵母菌在YPRAA上的生长状况来判断所选分泌蛋白的信号肽是否具有分泌功能。
1.4 可可毛色二孢侵染下分泌蛋白基因的表达分析
参考YAN等[3,22]接种葡萄座腔病菌的方法,用可可毛色二孢CSS-01s菌株接种一年生葡萄‘夏黑’绿枝条,于接种后0、6和12 h取接种点0.5—2 cm葡萄枝条表皮,液氮冻存后于-80℃冰箱保存备用。组织经液氮充分研磨后,用EASYspin Plus多糖多酚复杂植物RNA提取试剂盒(艾德莱生物)提取总RNA,经DNase I(TaKaRa)处理后,用SuperScript III反转录酶(Invitrogen)反转录得到第一链cDNA,-20℃保存,用于后续的实时荧光定量PCR分析(qRT-PCR)。依据全基因组预测的分泌蛋白序列设计特异性引物(表2),以LtActin(KAB2581229.1)作为内参基因,进行qRT-PCR分析。采用SYBR Green I荧光染料法,试验过程参照TaKaRa说明书。20 μL反应体系:2×TB Green Premix Ex Taq II(Tli RNaseH Plus)10 μL,正反引物(0.5 μmol·L-1)3.5 μL,cDNA 0.5 μL,ROX Reference Dye II 0.5 μL,补水至总体积为20 μL。qRT-PCR在ABI7500仪器上进行。反应程序:95℃ 5 min;进入95℃ 30 s,60℃ 30 s,共45个循环。每个反应得到相应的Ct值。每个样品进行3个平行反应,取均值后,用2-ΔΔCt方法计算基因的相对表达量。Table 2
表2
表2实时荧光定量PCR所用引物序列
Table 2
| 基因 Gene | 正向引物序列 Forward primer (5′-3′) | 反向引物序列 Reverse primer (5′-3′) |
|---|---|---|
| LT_159 | CCAGCAGGACTACAAGAA | CCAGAGGTAGACCAGTTC |
| LT_188 | CTACCTTGCCGACCTTAA | GATGATGTTGCCGTTGAA |
| LT_233 | GAGCAGGACTACGAGAAC | CGCAGAGGATGTAGATGT |
| LT_359 | CAAGTCTTCCTCCATCCA | GATCTGAGCCGAGTTGTA |
| LT_595 | AGATGGTCTGGAAGAACTC | CGTACTCGTCAAGGATGT |
| LT_62 | GGAATCAACGACGACTCT | CGCACTGTGTTGGTTATG |
| LT_936 | CTACAACGAAATCAGCGAAT | ATGGTGGTGGTCTTCTTC |
| LT_1541 | CAACGGCTACTACTACTCTT | TTGATGTTCCTGGCACTG |
| LT_1698 | AATGGTGCTCAGTTCTACA | AGATGTTGATGAGGAGACC |
| LT_Actin | TCTTCGCTCGAGAAGTCGTA | ACAATGGAAGGTCCGCTCTC |
新窗口打开|下载CSV
2 结果
2.1 可可毛色二孢全基因组水平的分泌蛋白筛选鉴定
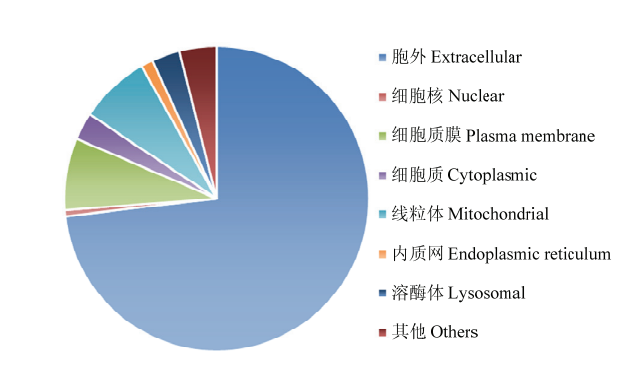
基于可可毛色二孢全基因组测序数据[22],对12 902条蛋白序列,通过SignalP v5.0[23]预测得到937个在氨基端含有典型信号肽序列的蛋白,占全基因组蛋白序列总数的7.26%。随后以ProtComp v9.0[24]在线程序对这937个含有信号肽的蛋白序列的细胞定位进行预测分析,结果如图1所示,共有685个蛋白可转运至细胞外,是胞外分泌蛋白类型,占比73%;另有252个蛋白未分泌到细胞外,其中最多的72个蛋白序列转运至细胞质膜(7.7%),其次是转运至线粒体(7.5%)、溶酶体(3%)和细胞质(2.9%);另外转运至过氧化酶体、细胞核、内质网和液泡的共有39个,占比4.2%。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1可可毛色二孢中937个具有信号肽蛋白的亚细胞定位
Fig. 1Subcellular localization of 937 proteins with signal peptide in L. theobromae
为了排除含有跨膜结构域的蛋白序列,通过TMHMM v2.0在线软件[25]对氨基端含有典型信号肽的蛋白序列进行跨膜螺旋结构分析,结果如图2所示,该685个蛋白序列中,有576个蛋白序列不含跨膜结构域,占比84.1%;有98个蛋白序列只含有1个跨膜结构域,占比14.3%;其余11个蛋白序列含有 2—3个跨膜结构域,占比1.6%。
图2
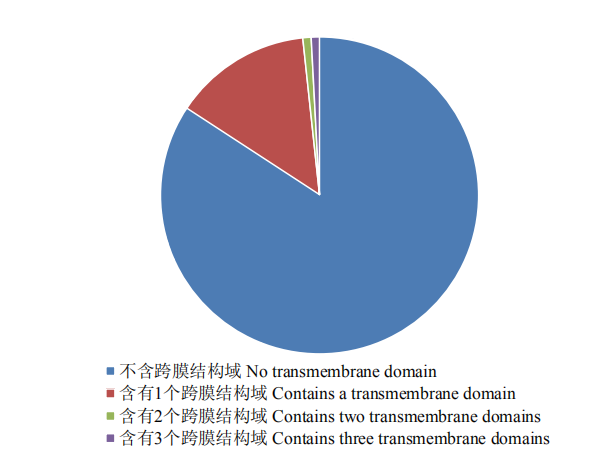 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2可可毛色二孢中685个外泌蛋白的跨膜结构域分析
Fig. 2The transmembrane domain analysis of 685 proteins transported to the extracellular space in L. theobromae
为了在预测蛋白中将这一部分蛋白去除,对576个蛋白序列进一步应用big-PI Fungal predictor[26]在线软件进行GPI锚定位点预测分析,结果发现552个蛋白序列不具有GPI锚定位点,而24个蛋白含有GPI锚定位点。通过对以上552个非GPI锚定蛋白序列测试发现,这552个测试蛋白序列均具有胞外分泌途径信号肽(SP)。
综合运用上述5种生物信息学算法程序,对可可毛色二孢全基因组12 902个蛋白序列进行预测筛选,最终获得552个具有信号肽、不含有跨膜结构域和GPI锚定位点并可外泌到细胞外的具有典型特征的经典分泌蛋白,占全基因组预测蛋白总数的4.3%。
2.2 可可毛色二孢中分泌蛋白的信号肽特征
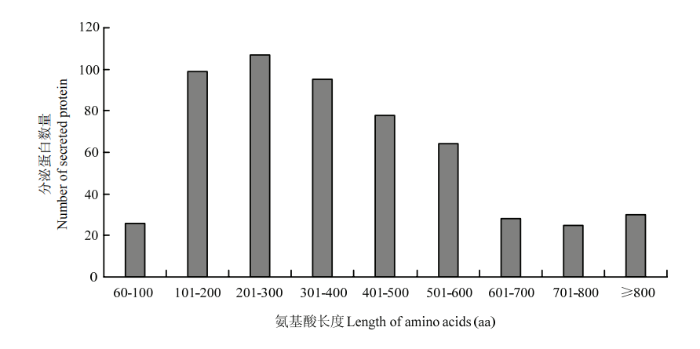
在552个具有分泌蛋白信号肽的蛋白序列中,蛋白序列长度最小的是66 aa,最大的长度是2 269 aa,大多集中于101—400 aa,占蛋白序列总数的54.5%(图3),上述结果表明,可可毛色二孢中所预测的典型分泌蛋白多属于小型蛋白。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3可可毛色二孢典型分泌蛋白的序列长度
Fig. 3Length of typical secreted protein sequences in L. theobromae
信号肽氨基酸长度统计结果如图4所示,候选分泌蛋白的信号肽长度多集中在18—20 aa,有253个,占预测分泌蛋白总数的45.8%;其中,含信号肽长度为20 aa的蛋白数量最多,有91个,占比16.5%。关于候选分泌蛋白的信号肽酶识别位点,利用LipoP v1.0[28]在线程序进行预测分析,结果表明含有Sp I型信号肽识别位点的分泌蛋白序列有522个,占比94.6%;另有10个分泌蛋白含有Sp II型信号肽识别位点,2个含有CYT型信号肽识别位点。以上结果说明大部分的可可毛色二孢候选分泌蛋白是通过Sp I型信号肽酶识别并切割掉信号肽。
图4
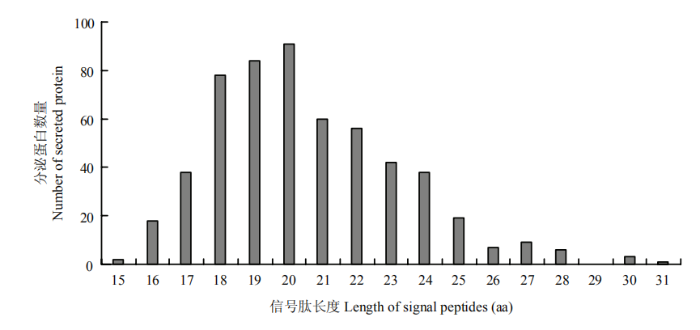 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4可可毛色二孢分泌蛋白的信号肽长度分布
Fig. 4Length distribution of signal peptides of secreted protein in L. theobromae
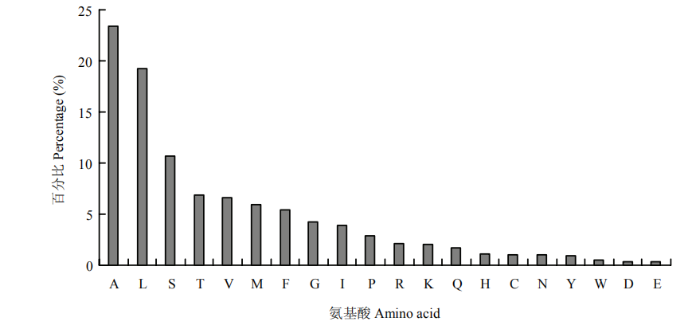
进一步对20种氨基酸在候选分泌蛋白信号肽区段中的使用频率进行统计分析,结果如图5所示,在组成信号肽的20种氨基酸中,有129个丙氨酸(A),数量最多,占分泌蛋白总数的23.4%;其次为亮氨酸(L),数量为106个,占全部的19.2%;非极性、疏水的氨基酸使用频率最高(A、G、I、L、P和V),占蛋白总数的60.2%;其次是极性、不带电荷的氨基酸(C、M、N、Q、S和T),占全部的27.2%;带正电荷的碱性氨基酸(K、R和H)所占比例为5.2%;带负电荷的酸性氨基酸(A和E)占23.7%;芳香族氨基酸(W、F和Y)占6.8%。
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5可可毛色二孢分泌蛋白氨基酸使用频率
Fig. 5The frequency of amino acids of secreted protein in L. theobromae
对可可毛色二孢候选分泌蛋白信号肽切割位点进行统计分析,结果如表3所示,在信号肽和成熟分泌蛋白切割位点-3、-2、-1、1、2、3位最多的氨基酸分别为A、L、A、A、P、T,所占比例分别为47.0%、16.4%、73.0%、27.3%、27.9%、14.0%。位于信号肽切割位点之前的-3、-2、-1位的氨基酸组成为A-S-A,属于比较典型的A-X-A类型,可被Sp I型信号肽酶识别并切割,这与LipoP v1.0预测的结果相一致。
Table 3
表3
表3可可毛色二孢分泌蛋白信号肽切割位点的氨基酸组成分布
Table 3
| 氨基酸类型 Type of amino acids | 信号肽切割位点-3到3位的氨基酸组成 Frequency of amino acids from -3 to +3 at signal peptide cleavage site of secreted proteins | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -3 | -2 | -1 | 1 | 2 | 3 | |||||||
| 数量 Amount | 百分比 Percentage (%) | 数量 Amount | 百分比 Percentage (%) | 数量 Amount | 百分比 Percentage (%) | 数量 Amount | 百分比 Percentage (%) | 数量 Amount | 百分比 Percentage (%) | 数量 Amount | 百分比 Percentage (%) | |
| A | 249 | 47.0 | 60 | 11.3 | 387 | 73.0 | 145 | 27.3 | 26 | 4.9 | 35 | 6.6 |
| C | 13 | 2.5 | 6 | 1.1 | 2 | 0.4 | 18 | 3.4 | 6 | 1.1 | 21 | 4.0 |
| D | 2 | 0.4 | 9 | 1.7 | 3 | 0.6 | 38 | 7.2 | 51 | 9.6 | 19 | 3.6 |
| E | 8 | 1.5 | 12 | 2.3 | 2 | 0.4 | 24 | 4.5 | 35 | 6.6 | 16 | 3.0 |
| F | 1 | 0.2 | 26 | 4.9 | 5 | 0.9 | 12 | 2.3 | 6 | 1.1 | 20 | 3.8 |
| G | 16 | 3.0 | 10 | 1.9 | 19 | 3.6 | 20 | 3.8 | 23 | 4.3 | 23 | 4.3 |
| H | 1 | 0.2 | 29 | 5.5 | 0 | 0 | 20 | 3.8 | 3 | 0.6 | 12 | 2.3 |
| I | 10 | 1.9 | 10 | 1.9 | 0 | 0 | 12 | 2.3 | 7 | 1.3 | 38 | 7.2 |
| K | 0 | 0 | 3 | 0.6 | 8 | 1.5 | 19 | 3.6 | 3 | 0.6 | 8 | 1.5 |
| L | 22 | 4.2 | 87 | 16.4 | 9 | 1.7 | 21 | 4.0 | 15 | 1.3 | 49 | 9.2 |
| M | 0 | 0 | 10 | 1.9 | 1 | 0.2 | 4 | 0.8 | 2 | 0.4 | 4 | 0.8 |
| N | 1 | 0.2 | 22 | 4.2 | 5 | 0.9 | 14 | 2.6 | 21 | 4.0 | 22 | 4.2 |
| P | 8 | 1.5 | 7 | 1.3 | 22 | 4.2 | 5 | 0.9 | 148 | 27.9 | 44 | 8.3 |
| Q | 5 | 0.9 | 48 | 9.1 | 6 | 1.1 | 68 | 12.8 | 24 | 4.5 | 25 | 4.7 |
| R | 9 | 1.7 | 27 | 5.1 | 3 | 0.6 | 10 | 1.9 | 6 | 1.1 | 14 | 2.6 |
| S | 56 | 10.6 | 68 | 12.8 | 36 | 6.8 | 30 | 5.7 | 44 | 8.3 | 40 | 7.5 |
| T | 40 | 7.5 | 44 | 8.3 | 9 | 1.7 | 26 | 4.9 | 61 | 11.5 | 74 | 14.0 |
| V | 89 | 16.8 | 35 | 6.6 | 9 | 1.7 | 31 | 5.8 | 33 | 6.2 | 41 | 7.7 |
| W | 0 | 0 | 1 | 0.2 | 1 | 0.2 | 4 | 0.8 | 6 | 1.1 | 5 | 0.9 |
| Y | 0 | 0 | 16 | 3.0 | 3 | 0.6 | 9 | 1.7 | 10 | 1.9 | 20 | 3.8 |
新窗口打开|下载CSV
2.3 可可毛色二孢全基因组候选分泌蛋白的功能预测
将预测的可可毛色二孢候选分泌蛋白在NCBI数据库进行比对分析发现,216个分泌蛋白描述为功能未知的推定蛋白,其余336个则有明确的功能描述。在已有功能描述的这些分泌蛋白中,注释功能主要集中在细胞壁组分降解相关的酶类,如蛋白酶、糖基水解酶、纤维素酶、角质酶、果胶裂解酶等,占比41.7%;此外还有CFEM结构域蛋白、FAD结合结构域蛋白、LysM结构域蛋白等结构域蛋白,以及与致病侵染相关的坏死诱导相关蛋白以及几丁质结合蛋白等(表4)。并且上述候选分泌蛋白在序列长度、等电点、分子量和脂肪族氨基酸指数等方面均存在差异,推测其可能参与不同的生理活动。Table 4
表4
表4可可毛色二孢部分分泌蛋白生化特性与功能注释
Table 4
| 基因编号 Gene ID | 蛋白长度 Length (aa) | 分子量 Molecular weight (kD) | 等电点 pI | 脂肪族氨基酸指数 Aliphatic index | 功能注释 Function |
|---|---|---|---|---|---|
| evm.model.scaffold_1.1884 | 265 | 28.57 | 4.57 | 70.38 | 糖基水解酶Glycosyl hydrolases family |
| evm.model.scaffold_7.188 | 334 | 36.85 | 4.62 | 71.89 | 纤维素酶Cellulase |
| evm.model.scaffold_6.309 | 322 | 36.16 | 5.96 | 80.90 | 水解酶家族Alpha/beta hydrolase family |
| evm.model.scaffold_3.829 | 549 | 59.13 | 5.16 | 82.00 | 羧酸酯酶Carboxylesterase family |
| evm.model.scaffold_5.474 | 324 | 35.76 | 4.96 | 83.36 | 过氧化物酶Peroxidase |
| evm.model.scaffold_5.945 | 348 | 39.24 | 5.23 | 69.20 | 酪氨酸酶Tyrosinase |
| evm.model.scaffold_13.28 | 251 | 26.69 | 5.34 | 82.03 | 角质酶Cutinase |
| evm.model.scaffold_6.540 | 395 | 42.03 | 5.39 | 74.63 | 天冬氨酸蛋白酶Aspartyl protease |
| evm.model.scaffold_8.302 | 265 | 27.76 | 5.55 | 65.92 | 脂肪酶GDSL-like Lipase |
| evm.model.scaffold_4.1035 | 581 | 62.25 | 4.80 | 87.04 | 氧化还原酶GMC oxidoreductase |
| evm.model.scaffold_3.522 | 252 | 26.19 | 4.13 | 74.64 | 果胶酸裂解酶Pectate lyase |
| evm.model.scaffold_2.202 | 377 | 40.24 | 5.22 | 73.37 | 肽酶Peptidase family |
| evm.model.scaffold_2.1494 | 201 | 20.37 | 4.41 | 60.45 | WSC结构域蛋白WSC domain protein |
| evm.model.scaffold_11.25 | 430 | 45.29 | 4.01 | 63.14 | PAN结构域蛋白PAN domain protein |
| evm.model.scaffold_1.947 | 254 | 28.05 | 8.38 | 57.01 | 坏死诱导蛋白Necrosis inducing protein |
| evm.model.scaffold_4.1274 | 186 | 19.83 | 4.43 | 61.51 | LysM结构域蛋白LysM domain protein |
| evm.model.scaffold_10.213 | 122 | 12.15 | 4.33 | 111.15 | FAD结构域蛋白FAD domain protein |
| evm.model.scaffold_1.937 | 247 | 26.14 | 5.05 | 84.45 | Cupin结构域蛋白Cupin domain protein |
| evm.model.scaffold_14.112 | 246 | 23.24 | 3.90 | 71.14 | CFEM结构域蛋白CFEM domain protein |
| evm.model.scaffold_11.308 | 412 | 40.74 | 5.14 | 53.98 | 几丁质结合蛋白Chitin binding protein |
新窗口打开|下载CSV
2.4 分泌蛋白预测信号肽的生物学功能分析
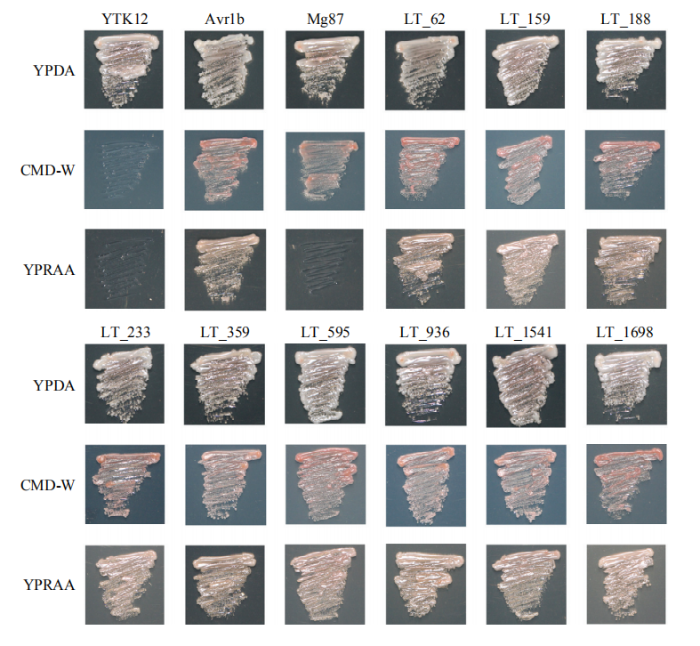
借助pSUC2系统[30,31]验证候选分泌蛋白的信号肽是否具有分泌功能。SUC2编码一个果糖苷酶,可将蔗糖、棉籽糖等多糖酶解生成葡萄糖、果糖等单糖。将9个候选可可毛色二孢分泌蛋白信号肽融合至SUC2蛋白的氨基端,转至酵母突变体YTK12中,若pSUC2融合载体成功转化到酵母中,则能在CMD-W培养基中正常生长。随后挑去生长的单斑,在棉籽糖培养基(YPRAA)上划线,若预测的分泌蛋白信号肽能够引导SUC2向胞外分泌,则可分泌到酵母细胞外将YPRAA培养基中的棉籽糖分解成单糖,酵母突变体就能正常生长,反之,则不会生长。结果如图6所示,阴性对照Mg87酵母菌不能在YPRAA培养基上存活,阳性对照Avr1b的酵母菌可以在YPRAA培养基生长,而预测的9个分泌蛋白融合载体转化的酵母可在YPRAA培养基上生长,表明上述分泌蛋白信号肽具有分泌活性,是典型的分泌蛋白。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6分泌蛋白信号肽功能验证
Fig. 6Functional validation of signal peptide of secreted proteins
2.5 可可毛色二孢候选分泌蛋白基因的表达分析
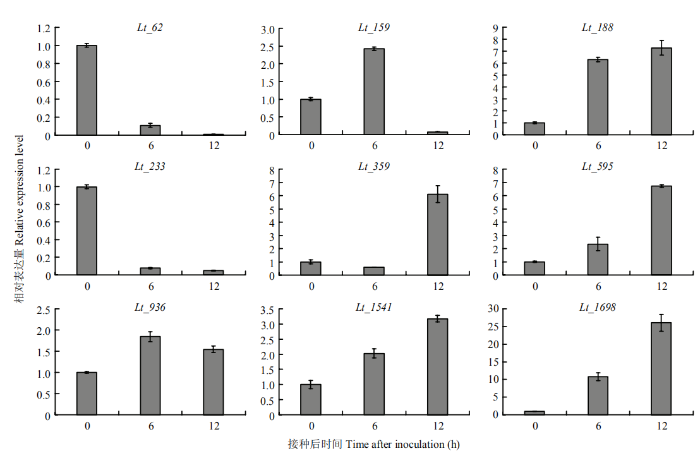
在可可毛色二孢侵染初期,Lt_62和Lt_233的表达是持续下降的趋势;Lt_159在接种6 h后的表达量是接种0 h的2.5倍,随后表达下调;Lt_188、Lt_595、Lt_936、Lt_1541和Lt_1698的表达持续上调;而Lt_359在接种12 h才表现出诱导表达(图7)。结果表明,上述分泌蛋白可能在可可毛色二孢的侵染初期发挥一定的作用。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图79个候选分泌蛋白基因在可可毛色二孢侵染过程中的相对表达量
Fig. 7Relative expression level of 9 putative secreted protein genes during L. theobromae infection
3 讨论
在与宿主植物的协同进化过程中,病原真菌衍化出多种攻击宿主的策略[11,32]。病原菌会通过分泌大量蛋白质来干扰宿主细胞功能并诱导其免疫反应,从而促进病原菌的侵染[14,33]。因此,利用全基因组测序信息,研究不同病原菌中分泌蛋白的数量、类型和特征以探究其致病机理尤为重要[34,35,36]。葡萄座腔病菌全基因组测序的完成和公布为该菌激发子、致病蛋白和效应子以及与宿主葡萄的互作研究提供了重要的数据支撑[22]。基于病原真菌分泌蛋白的典型结构特征,周晓罡等利用生物信息学分析软件已对病原真菌以及卵菌等的分泌蛋白进行了预测[17,18,19,20,21]。本文基于以上研究,运用5种生物学软件进行预测分析,保证了测试结果的准确性。LIU等研究表明,在侵染过程中,大豆疫霉(Phytophthora sojae)分泌蛋白PsIsc1和大丽轮枝菌(Verticillium dahliae)分泌蛋白VdIsc1可作为水解酶分解宿主植物的异分支酸合酶,破坏宿主的水杨酸代谢途径,终止水杨酸介导的免疫反应从而使宿主更为感病[35]。值得关注的是,上述2种分泌蛋白均缺乏分泌信号肽。因此,对真菌分泌蛋白的筛选鉴定,不仅需要通过生物信息学的手段并结合生物学试验进行验证,还需要结合双向电泳(2-DE)和质谱为基础的蛋白质组学等技术进行多维度的挖掘[36]。基于可可毛色二孢全基因组的12 902个蛋白序列,本研究预测得到552个经典分泌蛋白,占比4.3%。大多数候选分泌蛋白属于小型蛋白(图3),其信号肽长度多集中在18—20 aa(图4),能够被Sp I型信号肽识别并切割,这与前人所报道的有关植物病原真菌、卵菌等分泌蛋白无明显差异[16,17,18,19,20,21]。分泌蛋白信号肽切割位点-3—+3位使用频率最多是A、L、A、A、P、T(表3),切割位点上氨基酸使用种类较大丽轮枝菌[16]要多。切割位点-3和-1位的氨基酸使用种类最少,分别是16和18种(表3),这也与其他病原菌不同[17,18,19,20,21]。氨基酸使用频率变化最大的在-1位,范围从0(H,组氨酸;I,异亮氨酸)到73%(A,丙氨酸),与大丽轮枝菌、马铃薯晚疫病菌(Phytophthora infestans)情况类似[16,17,18,19,20,21],说明这一切割位点位置的保守性。
对于可可毛色二孢候选分泌蛋白功能注释分析发现,39%预测为功能未知的假定蛋白,说明该菌候选蛋白的特异性,其功能有待于进一步验证分析。植物病原真菌可能最初分泌细胞壁降解酶类来消化细胞壁的阻碍,以促进其侵入[37]。在有功能描述的候选分泌蛋白中(表4),其功能主要集中于降解细胞壁组分的酶类,如纤维素酶、果胶裂解酶和果胶酶等,这与其他病原真菌分泌蛋白的功能相似[17,18,19,20,21]。此外,预测有LysM结构域蛋白、几丁质结合蛋白、坏死诱导蛋白等蛋白(表4),这些蛋白可能参与了可可毛色二孢的致病过程[38,39]。随后,借助于pSUC2系统[30,31]对所选分泌蛋白信号肽进行了功能验证,随机挑选的9个候选蛋白信号肽均正常发挥作用,说明本研究预测方法的精准度较高(图6)。进一步通过qPCR表达分析(图7),验证了候选分泌蛋白基因在可可毛色二孢侵染初期不同的表达模式,推测其在病原菌致病过程中发挥着不同的作用。
4 结论
基于病原真菌分泌蛋白具有的典型特征,利用在线生物信息学程序从可可毛色二孢全基因组中共预测获得552个经典分泌蛋白,多属于小型蛋白。其信号肽氨基酸长度分布广泛,氨基酸组成中非极性、疏水的氨基酸使用频率最高。功能注释主要集中在细胞壁组分降解相关的酶类、与致病侵染相关的坏死诱导相关蛋白以及几丁质结合蛋白等。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1126/science.324_721bURLPMID:19423798 [本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
URLPMID:14616074 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:26512660 [本文引用: 2]
DOI:10.1186/1471-2164-11-105URL [本文引用: 1]
URLPMID:31171472 [本文引用: 2]
DOI:10.1038/nature05286URLPMID:17108957 [本文引用: 1]

Many plant-associated microbes are pathogens that impair plant growth and reproduction. Plants respond to infection using a two-branched innate immune system. The first branch recognizes and responds to molecules common to many classes of microbes, including non-pathogens. The second responds to pathogen virulence factors, either directly or through their effects on host targets. These plant immune systems, and the pathogen molecules to which they respond, provide extraordinary insights into molecular recognition, cell biology and evolution across biological kingdoms. A detailed understanding of plant immune function will underpin crop improvement for food, fibre and biofuels production.
URLPMID:26162502 [本文引用: 2]
URLPMID:26343014 [本文引用: 2]
URLPMID:28356329 [本文引用: 1]
DOI:10.3864/j.issn.0578-1752.2011.15.009URL [本文引用: 4]

【目的】预测并分析大丽轮枝菌基因组范围内的分泌蛋白,为大丽轮枝菌分泌蛋白致病机理的研究奠定基础。【方法】利用已公布的大丽轮枝菌全基因组序列,组合使用生物信息学软件SignalP、TargetP、TMHMM、Big-pi和PROSITE,预测大丽轮枝菌基因组范围内所有分泌蛋白,定义为分泌组。统计分析分泌组中蛋白N-端信号肽特点;应用碳水化合物活性酶类数据库和病原菌-寄主互作蛋白数据库对分泌组蛋白进行注释,预测分泌组中潜在果胶酶、纤维素酶和病原菌寄主互作蛋白;利用真菌激发子的保守结构域,预测分泌组中潜在的激发子蛋白集;应用BLASTP程序比较分析大丽轮枝菌和黑白轮枝菌分泌组,获得大丽轮枝菌相对于黑白轮枝菌特异的分泌蛋白。【结果】大丽轮枝菌分泌组共有922个蛋白。信号肽分析表明,以19个氨基酸为信号肽的蛋白数目最多,非极性氨基酸丙氨酸的出现频率最高,而有带电侧链的氨基酸天冬氨酸和谷氨酸的出现频率最低,信号肽的-3和-1位置上的氨基酸相对保守。大丽轮枝菌分泌组含有158个潜在的碳水化合物活性酶类,其中,包括10个果胶水解酶和14个果胶裂解酶;190个潜在的病原菌-寄主互作蛋白、97个含有RxLx[EDQ]模体的蛋白和52个富含半胱氨酸的小分子量分泌蛋白;58个相对于黑白轮枝菌分泌组特异的蛋白。【结论】本文建立了预测大丽轮枝菌分泌组蛋白的方法。分泌组蛋白信号肽长度具有高度的变异性,氨基酸组成多为脂肪族氨基酸,序列在C-端结构域较为保守。分泌组中包含大量潜在的果胶降解酶、病原菌-寄主互作蛋白、RxLx[EDQ]模体蛋白和富含半胱氨酸的小分子量蛋白等致病相关蛋白。
DOI:10.3864/j.issn.0578-1752.2011.15.009URL [本文引用: 4]

【目的】预测并分析大丽轮枝菌基因组范围内的分泌蛋白,为大丽轮枝菌分泌蛋白致病机理的研究奠定基础。【方法】利用已公布的大丽轮枝菌全基因组序列,组合使用生物信息学软件SignalP、TargetP、TMHMM、Big-pi和PROSITE,预测大丽轮枝菌基因组范围内所有分泌蛋白,定义为分泌组。统计分析分泌组中蛋白N-端信号肽特点;应用碳水化合物活性酶类数据库和病原菌-寄主互作蛋白数据库对分泌组蛋白进行注释,预测分泌组中潜在果胶酶、纤维素酶和病原菌寄主互作蛋白;利用真菌激发子的保守结构域,预测分泌组中潜在的激发子蛋白集;应用BLASTP程序比较分析大丽轮枝菌和黑白轮枝菌分泌组,获得大丽轮枝菌相对于黑白轮枝菌特异的分泌蛋白。【结果】大丽轮枝菌分泌组共有922个蛋白。信号肽分析表明,以19个氨基酸为信号肽的蛋白数目最多,非极性氨基酸丙氨酸的出现频率最高,而有带电侧链的氨基酸天冬氨酸和谷氨酸的出现频率最低,信号肽的-3和-1位置上的氨基酸相对保守。大丽轮枝菌分泌组含有158个潜在的碳水化合物活性酶类,其中,包括10个果胶水解酶和14个果胶裂解酶;190个潜在的病原菌-寄主互作蛋白、97个含有RxLx[EDQ]模体的蛋白和52个富含半胱氨酸的小分子量分泌蛋白;58个相对于黑白轮枝菌分泌组特异的蛋白。【结论】本文建立了预测大丽轮枝菌分泌组蛋白的方法。分泌组蛋白信号肽长度具有高度的变异性,氨基酸组成多为脂肪族氨基酸,序列在C-端结构域较为保守。分泌组中包含大量潜在的果胶降解酶、病原菌-寄主互作蛋白、RxLx[EDQ]模体蛋白和富含半胱氨酸的小分子量蛋白等致病相关蛋白。
[本文引用: 6]
[本文引用: 6]
[本文引用: 6]
[本文引用: 6]
[本文引用: 6]
[本文引用: 6]
DOI:10.1111/mpp.12405URLPMID:27010366 [本文引用: 6]

Asian soybean rust (ASR), caused by the obligate biotrophic fungus Phakopsora pachyrhizi, can cause losses greater than 80%. Despite its economic importance, there is no soybean cultivar with durable ASR resistance. In addition, the P. pachyrhizi genome is not yet available. However, the availability of other rust genomes, as well as the development of sample enrichment strategies and bioinformatics tools, has improved our knowledge of the ASR secretome and its potential effectors. In this context, we used a combination of laser capture microdissection (LCM), RNAseq and a bioinformatics pipeline to identify a total of 36 350 P. pachyrhizi contigs expressed in planta and a predicted secretome of 851 proteins. Some of the predicted secreted proteins had characteristics of candidate effectors: small size, cysteine rich, do not contain PFAM domains (except those associated with pathogenicity) and strongly expressed in planta. A comparative analysis of the predicted secreted proteins present in Pucciniales species identified new members of soybean rust and new Pucciniales- or P. pachyrhizi-specific families (tribes). Members of some families were strongly up-regulated during early infection, starting with initial infection through haustorium formation. Effector candidates selected from two of these families were able to suppress immunity in transient assays, and were localized in the plant cytoplasm and nuclei. These experiments support our bioinformatics predictions and show that these families contain members that have functions consistent with P. pachyrhizi effectors.
[本文引用: 6]
DOI:10.1093/dnares/dsx040URLPMID:29036669 [本文引用: 7]

Botryosphaeriaceae are an important fungal family that cause woody plant diseases worldwide. Recent studies have established a correlation between environmental factors and disease expression; however, less is known about factors that trigger these diseases. The current study reports on the 43.3 Mb de novo genome of Lasiodiplodia theobromae and five other genomes of Botryosphaeriaceae pathogens. Botryosphaeriaceous genomes showed an expansion of gene families associated with cell wall degradation, nutrient uptake, secondary metabolism and membrane transport, which contribute to adaptations for wood degradation. Transcriptome analysis revealed that genes involved in carbohydrate catabolism, pectin, starch and sucrose metabolism, and pentose and glucuronate interconversion pathways were induced during infection. Furthermore, genes in carbohydrate-binding modules, lysine motif domain and the glycosyl hydrolase gene families were induced by high temperature. Among these genes, overexpression of two selected putative lignocellulase genes led to increased virulence in the transformants. These results demonstrate the importance of high temperatures in opportunistic infections. This study also presents a set of Botryosphaeriaceae-specific effectors responsible for the identification of virulence-related pathogen-associated molecular patterns and demonstrates their active participation in suppressing hypersensitive responses. Together, these findings significantly expand our understanding of the determinants of pathogenicity or virulence in Botryosphaeriaceae and provide new insights for developing management strategies against them.
[本文引用: 2]
[本文引用: 2]
DOI:10.1006/jmbi.2000.4315URLPMID:11152613 [本文引用: 2]

We describe and validate a new membrane protein topology prediction method, TMHMM, based on a hidden Markov model. We present a detailed analysis of TMHMM's performance, and show that it correctly predicts 97-98 % of the transmembrane helices. Additionally, TMHMM can discriminate between soluble and membrane proteins with both specificity and sensitivity better than 99 %, although the accuracy drops when signal peptides are present. This high degree of accuracy allowed us to predict reliably integral membrane proteins in a large collection of genomes. Based on these predictions, we estimate that 20-30 % of all genes in most genomes encode membrane proteins, which is in agreement with previous estimates. We further discovered that proteins with N(in)-C(in) topologies are strongly preferred in all examined organisms, except Caenorhabditis elegans, where the large number of 7TM receptors increases the counts for N(out)-C(in) topologies. We discuss the possible relevance of this finding for our understanding of membrane protein assembly mechanisms. A TMHMM prediction service is available at http://www.cbs.dtu.dk/services/TMHMM/.
URLPMID:11287675 [本文引用: 2]
URLPMID:31515291 [本文引用: 1]
URLPMID:12876315 [本文引用: 2]
Humana Press,
[本文引用: 1]
[本文引用: 3]
DOI:10.1094/MPMI-09-15-0200-RURLPMID:26927000 [本文引用: 3]

Ustilaginoidea virens (Cooke) Takah (telemorph Villosiclava virens) is an ascomycetous fungus that causes rice false smut, one of the most important rice diseases. Fungal effectors often play essential roles in host-pathogen coevolutionary interactions. However, little is known about the functions of U. virens effectors. Here, we performed functional studies on putative effectors in U. virens and demonstrated that 13 of 119 putative effectors caused necrosis or necrosis-like phenotypes in Nicotiana benthamiana. Among them, 11 proteins were confirmed to be secreted, using a yeast secretion system, and the corresponding genes are all highly induced during infection, except UV_44 and UV_4753. Eight secreted proteins were proven to trigger cell death or defenses in rice protoplasts and the secretion signal of these proteins is essential for their cell death-inducing activity. The ability of UV_44 and UV_1423 to trigger cell death is dependent on the predicted serine peptidase and ribonuclease catalytic active sites, respectively. We demonstrated that UV_1423 and UV_6205 are N-glycosylated proteins, which glycosylation has different impacts on their abilities to induce cell death. Collectively, the study identified multiple secreted proteins in U. virens with specific structural motifs that induce cell death or defense machinery in nonhost and host plants.
[本文引用: 1]
DOI:10.1016/j.pbi.2019.02.001URLPMID:30878771 [本文引用: 1]

Plant plasma membrane pattern recognition receptors are key to microbe sensing and activation of immunity to microbial invasion. Plants employ several types of such receptors that differ mainly in the structure of their ectodomains and the presence or absence of a cytoplasmic protein kinase domain. Plant immune receptors do not function as single entities, but form larger complexes which undergo compositional changes in a ligand-dependent manner. Here, we highlight current knowledge of molecular mechanisms underlying receptor complex dynamics and regulation, and cover early signaling networks implicated in the activation of generic plant immune responses. We further discuss how an increasingly comprehensive set of immune receptors may be employed to engineer crop plants with enhanced, durable resistance to microbial infection.
[本文引用: 1]
URLPMID:25156390 [本文引用: 2]
[本文引用: 2]
[本文引用: 2]
DOI:10.1016/j.fgb.2014.08.011URLPMID:25192612 [本文引用: 1]

Carbohydrate-Active enZymes (CAZymes) form particularly interesting targets to study in plant pathogens. Despite the fact that many CAZymes are pathogenicity factors, oomycete CAZymes have received significantly less attention than effectors in the literature. Here we present an analysis of the CAZymes present in the Phytophthora infestans, Ph. ramorum, Ph. sojae and Pythium ultimum genomes compared to growth of these species on a range of different carbon sources. Growth on these carbon sources indicates that the size of enzyme families involved in degradation of cell-wall related substrates like cellulose, xylan and pectin is not always a good predictor of growth on these substrates. While a capacity to degrade xylan and cellulose exists the products are not fully saccharified and used as a carbon source. The Phytophthora genomes encode larger CAZyme sets when compared to Py. ultimum, and encode putative cutinases, GH12 xyloglucanases and GH10 xylanases that are missing in the Py. ultimum genome. Phytophthora spp. also encode a larger number of enzyme families and genes involved in pectin degradation. No loss or gain of complete enzyme families was found between the Phytophthora genomes, but there are some marked differences in the size of some enzyme families.
DOI:10.1093/femsre/fuu003URLPMID:25725011 [本文引用: 1]

Fungal cell walls play dynamic functions in interaction of fungi with their surroundings. In pathogenic fungi, the cell wall is the first structure to make physical contact with host cells. An important structural component of fungal cell walls is chitin, a well-known elicitor of immune responses in plants. Research into chitin perception has sparked since the chitin receptor from rice was cloned nearly a decade ago. Considering the widespread nature of chitin perception in plants, pathogens evidently evolved strategies to overcome detection, including alterations in the composition of cell walls, modification of their carbohydrate chains and secretion of effectors to provide cell wall protection or target host immune responses. Also non-pathogenic fungi contain chitin in their cell walls and are recipients of immune responses. Intriguingly, various mutualists employ chitin-derived signaling molecules to prepare their hosts for the mutualistic relationship. Research on the various types of interactions has revealed different molecular components that play crucial roles and, moreover, that various chitin-binding proteins contain dissimilar chitin-binding domains across species that differ in affinity and specificity. Considering the various strategies from microbes and hosts focused on chitin recognition, it is evident that this carbohydrate plays a central role in plant-fungus interactions.
URLPMID:25589417 [本文引用: 1]
