 ,, 王大江, 王昆
,, 王大江, 王昆 ,, 丛佩华
,, 丛佩华 ,, 李连文, 朴继成中国农业科学院果树研究所/农业农村部园艺作物种质资源利用重点实验室,辽宁兴城 125100
,, 李连文, 朴继成中国农业科学院果树研究所/农业农村部园艺作物种质资源利用重点实验室,辽宁兴城 125100Genetic Relationship and Structure Analysis of 15 Species of Malus Mill. Based on SNP Markers
GAO Yuan ,, WANG DaJiang, WANG Kun
,, WANG DaJiang, WANG Kun ,, CONG PeiHua
,, CONG PeiHua ,, LI LianWen, PIAO JiChengInstitute of Pomology, Chinese Academy of Agricultural Sciences/Key Laboratory of Horticultural Crops Germplasm Resources Utilization, Ministry of Agriculture and Rural Affairs of the People’s Republic of China, Xingcheng 125100, Liaoning
,, LI LianWen, PIAO JiChengInstitute of Pomology, Chinese Academy of Agricultural Sciences/Key Laboratory of Horticultural Crops Germplasm Resources Utilization, Ministry of Agriculture and Rural Affairs of the People’s Republic of China, Xingcheng 125100, Liaoning通讯作者:
责任编辑: 赵伶俐
收稿日期:2019-12-30接受日期:2020-06-10网络出版日期:2020-08-16
| 基金资助: |
Received:2019-12-30Accepted:2020-06-10Online:2020-08-16
作者简介 About authors
高源,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1608KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
高源, 王大江, 王昆, 丛佩华, 李连文, 朴继成. 基于高密度SNP标记的苹果属15种植物资源的亲缘关系与遗传结构分析[J]. 中国农业科学, 2020, 53(16): 3333-3343 doi:10.3864/j.issn.0578-1752.2020.16.011
GAO Yuan, WANG DaJiang, WANG Kun, CONG PeiHua, LI LianWen, PIAO JiCheng.
0 引言
【研究意义】苹果属(Malus Mill.)为蔷薇科(Rosaceae)植物,世界苹果属植物资源约35个种[1,2],主要分布于北温带,横跨欧亚大陆和北美洲;而原产中国的有27个种,其中野生近缘种21种、栽培种6种[3]。研究分析苹果属植物不同种的遗传结构,探讨其遗传多样性,是揭示物种的进化历史[4,5]、分析其进化潜力和未来命运[6]、探讨物种稀有或濒危原因[7,8]的重要方法,对制订种质资源保护策略、指导核心种质资源筛选和相关优异基因挖掘具有重要意义。【前人研究进展】分子标记是揭示不同品种间遗传多样性和亲缘关系的有效手段[9],RAPD[10]、AFLP[11]等分子标记曾被用于苹果的遗传多样性分析,SSR分子标记被认为是动植物中最重要的标记之一[12],在苹果遗传多样性研究中应用最为广泛[13]。SNP即单核苷酸多态性,是由单个核苷酸变异引起的DNA序列多态性,SNP标记是目前所有DNA分子标记中多样性最为丰富的标记。研究SNP最彻底、最精确的方法即为直接测定某特定区域的核苷酸序列,与参照基因组中对应区域的核苷酸序列进行比较,从而检测出具有多态性的单个核苷酸变异。SNP基因型鉴定已经被应用到许多作物的遗传结构分析、遗传多样性评价和遗传连锁图谱的构建[14,15]。CHAGNE等[16]利用EST-SNP标记的方法对苹果基因进行了研究。现代测序技术的发展,苹果全基因组测序的完成,对于SNP在苹果研究中的应用具有重要的推动作用。MICHELETTI等[17]对27份苹果属种质进行测序,并筛选出237个SNP标记研究260份苹果品种的遗传多样性并构建遗传连锁图谱。常源升等[18]、SUN等[19]和孙瑞[20]分别将SNP分子标记的方法用于苹果果形和果实品质相关基因的主基因分析与QTL定位分析,刘更森[21]利用SNP标记构建了苹果遗传图谱。随着苹果参考基因组的发表[22,23,24],结合现代测序技术,有利于在全基因组范围内寻找多态的SNP分子标记用于苹果属植物种质资源遗传多样性研究,对于其保存和利用具有重要意义[25]。SLAF-seq(Specific Locus Amplified Fragment Sequencing)是一种高通量的简化基因组深度测序技术,通过生物信息学方法设计最佳试验方案,在测序获得海量特异性长度的DNA片段(SLAF标签)基础上,在全基因组范围开发出大量特异性SNP标记。其已经被应用到多种作物,如水稻[26]、甘薯[27]、金花茶[28]和葡萄[29]等的SNP标记开发中,陶红霞[30]基于SLAF技术开发SNP构建苹果遗传连锁图谱。【本研究切入点】随着近些年国家苹果资源圃苹果属植物种质资源收集和保存工作的不断深入,国家苹果资源圃保存了大量待鉴定的苹果属植物种质资源,可在SLAF-seq基础上从全基因组开发SNP标记研究苹果属植物不同种的亲缘关系和遗传结构。【拟解决的关键问题】本研究在对国家苹果种质资源圃保存的15个种的427份苹果属植物种质资源进行高通量简化基因组测序基础上,开发SNP标记,解析15种苹果属植物种内和种间的亲缘关系和遗传结构,探讨不同种间的系统演化关系,为不同种苹果属植物的鉴定评价以及进一步收集和保存提供依据。1 材料与方法
1.1 材料
427份苹果种质材料中有25份取自国家果树种质公主岭寒地果树圃(吉林省公主岭),1份滇池海棠和1份沧江海棠取自国家果树种质云南特有果树及砧木圃(云南省昆明),其余400份材料均取自国家果树种质兴城梨、苹果圃(辽宁省兴城市)。427份种质分属于苹果属的15个种,供试各种名称及种质来源见表1。于2017年春季采集健康幼嫩叶片,叶片经硅胶干燥之后备用。Table 1
表1
表1用于SLAF测序分析的15个种苹果属植物种质资源
Table 1
| 序号 Code | 供试种 Species | 来源地 Origin | 数量 Number | 序号 Code | 供试种 Species | 来源地 Origin | 数量 Number | |
|---|---|---|---|---|---|---|---|---|
| 1 | 新疆野苹果 Malus sieversii | 新疆 Xinjiang | 161 | 5 | 花红 Malus asiatica | 黑龙江 Heilongjiang | 7 | |
| 2 | 中国苹果 Malus domestica subsp.chinensis | 新疆 Xinjiang | 2 | 甘肃 Gansu | 1 | |||
| 黑龙江 Heilongjiang | 2 | 河北 Hebei | 9 | |||||
| 甘肃 Gansu | 4 | 云南 Yunnan | 1 | |||||
| 河北 Hebei | 14 | 6 | 八棱海棠 Malus robusta | 河北 Hebei | 32 | |||
| 山西 Shanxi | 10 | 山西 Shanxi | 1 | |||||
| 山东 Shandong | 1 | 吉林 Jilin | 1 | |||||
| 3 | 山荆子 Malus baccata | 黑龙江 Heilongjiang | 47 | 7 | 陇东海棠 Malus kansuensis | 甘肃 Gansu | 4 | |
| 甘肃 Gansu | 3 | 8 | 垂丝海棠 Malus halliana | 甘肃 Gansu | 9 | |||
| 河北 Hebei | 10 | 9 | 山楂海棠 Malus komarovii | 吉林 Jilin | 1 | |||
| 山西 Shanxi | 14 | 10 | 变叶海棠 Malus toringoides | 四川 Sichuan | 2 | |||
| 内蒙古 Inner Mongolia | 41 | 云南 Yunnan | 1 | |||||
| 吉林 Jilin | 19 | 11 | 花叶海棠 Malus transitoria | 四川 Sichuan | 1 | |||
| 4 | 楸子 Malus prunifolia | 黑龙江 Heilongjiang | 5 | 12 | 丽江山荆子 Malus rockii | 云南 Yunnan | 1 | |
| 甘肃 Gansu | 4 | 13 | 滇池海棠 Malus yunnanensis | 云南 Yunnan | 1 | |||
| 河北 Hebei | 2 | 14 | 湖北海棠 Malus hupehensis | 云南 Yunnan | 1 | |||
| 山西 Shanxi | 8 | 15 | 沧江海棠 Malus ombrophila | 云南 Yunnan | 1 | |||
| 内蒙古 Inner Mongolia | 1 | |||||||
| 吉林 Jilin | 5 |
新窗口打开|下载CSV
1.2 基因组DNA的提取
采用德国QIAGEN的DNeasy Plant Mini Kit提取供试材料春季嫩叶的基因组DNA。分别用1%的琼脂糖凝胶电泳和超微量紫外分光光度计(美国DeNovix,DS-11型)检测其浓度和纯度,对照λDNA(40 ng·μL-1)将提取基因组的DNA浓度调整到100 ng·μL-1,-20℃保存备用。1.3 酶切预测
以2010年已经发表的苹果基因组作为参考基因组进行酶切预测。参考基因组信息:苹果(Malus pumila Mill.)基因组[22](http://www.ncbi.nlm.nih. gov/genome/?term=apple),组装出的基因组大小为1 874.77 Mb,GC含量为45.32%。利用SLAF-predict软件,通过苹果基因组进行方案预测,确定酶切组合并进行酶切。1.4 测序及数据开发
将获得酶切片段的3′端进行加A处理,在加有polyA的酶切片段上连接测序接头,经过PCR扩增和切胶回收目的片段,构建文库。在Illumina HiSeqTM2500(美国Illumina公司,HiSeq 2500型)上对检验合格的文库进行测序,测序结果经过去接头、低质量阅读框和污染处理而获得干净序列即SLAF标签。评估测序获得序列的GC含量和Q30指标,检验测序质量。在不同样品间有差异的SLAF标签即为多态性SLAF标签。通过BWA软件[31]将SLAF标签与苹果参考基因组[22]以及在不同样品间进行比对,将其定位到参考基因组上获得多态性的SLAF标签。利用GATK[32]和SAMtools[33]两种方法在多态性SLAF中开发SNP,筛选两种方法共同得到的SNP作为开发的SNP标记数据集。根据完整度>0.94,次要等位基因频率(MAF)>0.05过滤[34],筛选多态性的SNP,用于进一步的数据统计和分析。1.5 群体遗传结构和亲缘关系分析
基于筛选的多态性SNP,使用MEGA 7[35]的NJ(neighbor-joining)算法[36],构建苹果属不同种的系统进化树。不同种间的遗传距离大小用系统进化树的分支长度体现,长度越短即代表两份种质之间的亲缘关系越近。利用Admixture软件[37]进行群体遗传结构分析,假设样品的分群数(K)为1—15进行聚类,根据交叉验证错误率确定分群数。2 结果
2.1 建库和测序质量评估
利用SLAF-predict软件,参照2010年苹果基因组进行酶切预测,确定选择Rsa I+Hae III酶组合进行酶切,筛选SLAF标签长度范围为314—414 bp,共预测到151 808个SLAF标签,SLAF标签在基因组上基本分布均匀。以水稻的测序数据(http://rapdb.dna. affrc.go.jp/)为对照进一步评估酶切的有效性和酶切效率,将水稻的测序数据与苹果参考基因组进行比对,双端比对效率为95.19%,酶切效率为92.79%,酶切反应正常,SLAF建库正常。对所有供试苹果属植物种质测序共获得1 276.7 Mb的读长数据,各种质样品获得的读长数目在1 189 223—1 968 102 bp,测序平均Q30为91.58%,所有测序样品的Q30值均在80%以上;平均GC含量为40.04%,GC含量普遍较低。用于评估试验建库准确性的水稻测序获得0.45 Mb数据量。测序结果的碱基错误率低,测序数据达到要求。2.2 SLAF标签与多态性SNP标记筛选
对427份苹果属植物种质测序获得586 454个SLAF标签,平均测序深度为6.42 X。通过BWA软件比对获得463 612个多态性SLAF标签,开发5 896 021个群体SNP,根据完整度>0.94、MAF>0.05过滤,共得到46 460个多态性SNP位点,用于后续的群体结构分析。根据筛选的多态性SNP在染色体上的分布,绘制多态性的SNP在染色体上的分布图(图1)。图1
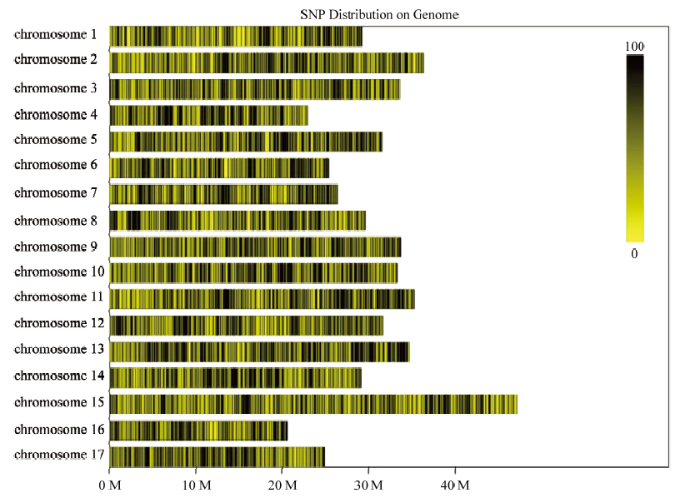 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1多态性的SNP在染色体上的分布
每一个黄色条带代表一条染色体,黑线代表多态性SNP定位的位置;横坐标为染色体长度,按照1 M的大小对基因组进行划分。黑色越深代表SNP标记数越多,颜色越深的区域即SNP标记集中分布的区域
Fig. 1The distribution of polymorphic SNP in 17 chromosomes
Every yellow band indicated one chromosome, and black line indicated the position of SNP. The abscissa is the length of the chromosome, and the genome was divided by every 1M. The darker position represented more SNPs, and the darker regions showed the centralized distribution area of SNPs
2.3 基于SNP标记的苹果属植物亲缘关系分析
以每个种作为一个种群,基于每个种群的SNP标记用MEGA7计算苹果属各种间的遗传距离(表2)。苹果属植物两两种间的遗传距离为0—1.000,其中花叶海棠与变叶海棠、丽江山荆子、滇池海棠、湖北海棠、沧江海棠、陇东海棠和山楂海棠间的遗传距离均为1.000;变叶海棠与湖北海棠、滇池海棠、沧江海棠、陇东海棠、丽江山荆子和山楂海棠间的遗传距离均为0.000;丽江山荆子与滇池海棠、湖北海棠、沧江海棠、陇东海棠和山楂海棠间的遗传距离均为0.000;滇池海棠与湖北海棠、沧江海棠,陇东海棠和山楂海棠间的遗传距离均为0.000,湖北海棠与沧江海棠、陇东海棠和山楂海棠间的遗传距离均为0.000;沧江海棠与陇东海棠和山楂海棠,陇东海棠和山楂海棠间的遗传距离均为0.000。以种间遗传距离做15个种的系统发育树(图2),15个种明显分为4个类群,类群Ⅰ为陇东海棠、山楂海棠、滇池海棠、沧江海棠、湖北海棠、丽江山荆子和变叶海棠,类群Ⅱ为山荆子,类群Ⅲ为垂丝海棠、楸子、花红和八棱海棠,类群Ⅳ为中国苹果、新疆野苹果和花叶海棠。Table 2
表2
表2基于SNP的苹果属植物15个种间的遗传距离
Table 2
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 0.000 | ||||||||||||||
| 2 | 0.030 | 0.000 | |||||||||||||
| 3 | 0.970 | 1.000 | 0.000 | ||||||||||||
| 4 | 0.600 | 0.606 | 0.394 | 0.000 | |||||||||||
| 5 | 0.030 | 0.000 | 1.000 | 0.606 | 0.000 | ||||||||||
| 6 | 0.030 | 0.000 | 1.000 | 0.606 | 0.000 | 0.000 | |||||||||
| 7 | 0.030 | 0.000 | 1.000 | 0.606 | 0.000 | 0.000 | 0.000 | ||||||||
| 8 | 0.187 | 0.167 | 0.833 | 0.571 | 0.167 | 0.167 | 0.167 | 0.000 | |||||||
| 9 | 0.030 | 0.000 | 1.000 | 0.606 | 0.000 | 0.000 | 0.000 | 0.167 | 0.000 | ||||||
| 10 | 0.953 | 0.981 | 0.019 | 0.398 | 0.981 | 0.981 | 0.981 | 0.821 | 0.981 | 0.000 | |||||
| 11 | 0.143 | 0.120 | 0.880 | 0.581 | 0.120 | 0.120 | 0.120 | 0.247 | 0.120 | 0.866 | 0.000 | ||||
| 12 | 0.030 | 0.000 | 1.000 | 0.606 | 0.000 | 0.000 | 0.000 | 0.167 | 0.000 | 0.981 | 0.120 | 0.000 | |||
| 13 | 0.134 | 0.111 | 0.889 | 0.582 | 0.111 | 0.111 | 0.111 | 0.241 | 0.111 | 0.874 | 0.204 | 0.111 | 0.000 | ||
| 14 | 0.279 | 0.265 | 0.735 | 0.550 | 0.265 | 0.265 | 0.265 | 0.343 | 0.265 | 0.727 | 0.321 | 0.265 | 0.317 | 0.000 | |
| 15 | 0.030 | 0.000 | 1.000 | 0.606 | 0.000 | 0.000 | 0.000 | 0.167 | 0.000 | 0.981 | 0.120 | 0.000 | 0.111 | 0.265 | 0.000 |
新窗口打开|下载CSV
图2
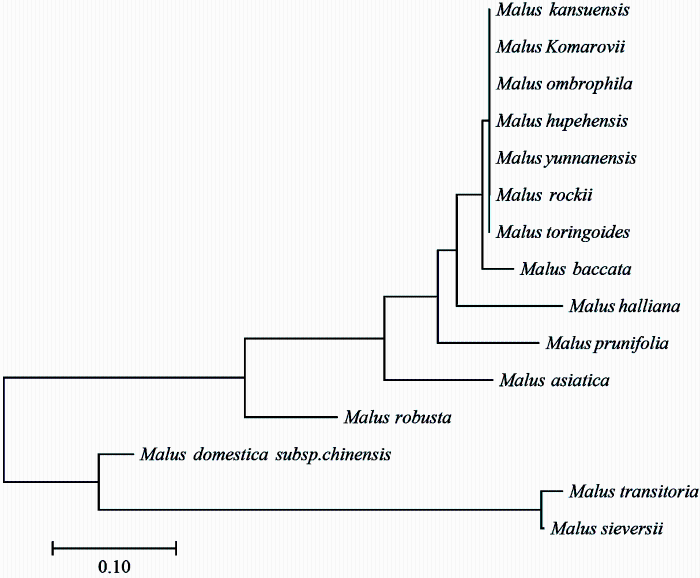 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2基于SNP位点的苹果属15个种的进化树
Fig. 2Polygenetic tree of 15 species of Malus Mill. based on SNP
2.4 基于SNP标记的苹果属植物的遗传结构分析
群体遗传结构分析能够获得个体的血统来源及其组成信息,是遗传关系分析的一种重要手段。运用Admixture软件,分析427份苹果属植物种质基于多态性SNP的遗传结构。分别假设427份种质的分群数(K值)为1—15(图3),进行聚类。根据交叉验证错误率,当K=5的时候,交叉验证错误率有明显降落,应该为确定分群数的一个关键点,首先确定所有供试苹果属植物的分群数为5,来自于5个可能的原始祖先。随着进一步的分化,当K=14时,交叉验证错误率最小,因此确定最佳分群数为14,反映了这些苹果属植物种质分化后可能来自于14个祖先。从K=1—14的群体遗传结构分析见图4。图3
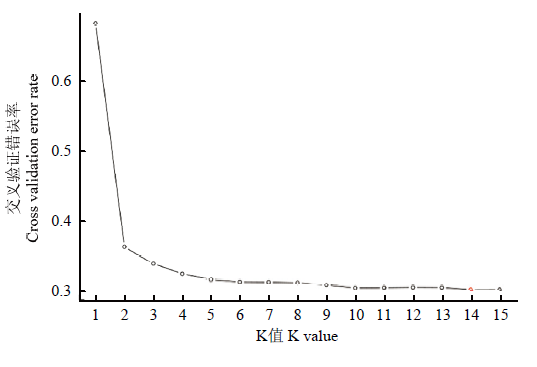 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3每个K值对应的交叉验证错误率
Fig. 3Cross validation error rates corresponding to every K values
图4
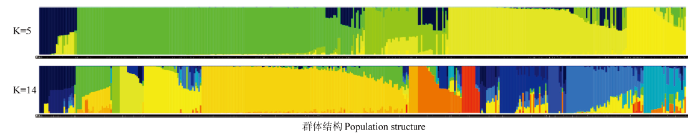 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4427份15种苹果属植物种质群体遗传结构(K=5和K=14)
每个条柱代
Fig. 4The genetic structure of 427 accessions of 15 species of Malus Mill. (K=5 and K=14)
Each bar represents one accession, and the abscissa is the code of germplasms corresponding to each bar. One color represents one group, and the ordinate is Q value 0.00-1.00
当确定分群数为5时,类群Ⅰ包含25份种质,有3份变叶海棠、3份垂丝海棠、1份丽江山荆子、1分新疆野苹果和17份山荆子,其中有7份山荆子的Q值为1.000,这7份山荆子代表该类群的基因库(蓝色基因库)。类群Ⅱ包含190份种质,有1份八棱海棠、1份垂丝海棠、4份花红、5份楸子、1份山荆子、20份中国苹果和158份新疆野苹果,其中有63份新疆野苹果和1份中国苹果(新疆)的Q值为1.000,这63份新疆野苹果和1份中国苹果代表该类群体的基因库(绿色基因库)。类群Ⅲ包含55份种质,有3份八棱海棠、1份沧江海棠、2份垂丝海棠、1份滇池海棠、12份花红、1份花叶海棠、4份陇东海棠、16份楸子、4份山定子、1份山楂海棠和10份中国苹果,其中有1份八棱海棠、1份花红、1份山荆子、1份中国苹果和1份楸子的Q值为1.000,这5份材料代表了该类群的基因库(淡绿色基因库)。类群Ⅳ包含119份种质,有2份八棱海棠、3份垂丝海棠、1份湖北海棠、2份楸子、2份新疆野苹果、2份中国苹果和107份山荆子,其中有26份山荆子的Q值为1.000,26份山荆子代表了该类群的基因库(黄色基因库)。类群Ⅴ包括39份种质,有28份八棱海棠、3份花红、2份楸子、4份山定子和1份中国苹果,其中16份八棱海棠和1份楸子的Q值为1.000,该17份种质代表了该类群的基因库(浅黄色基因库)。
当确定分群数为14时,类群1包含24份种质,有14份八棱海棠、3份花红、1份楸子、1份山荆子和5份中国苹果,其中有3份中国苹果(绵苹果)的Q值为0.9999,其代表了该类群的基因库(藏蓝色基因库)。类群2包含37份种质,有1份楸子、1份中国苹果和35份新疆野苹果,其中有2份新疆野苹果的Q值为0.9999,其代表了该类群的基因库(绿色基因库)。类群3包含5份种质,全部为山荆子,且全部为内蒙古收集的山荆子,其Q值全部为0.9999,其代表了该类群的基因库(浅绿色基因库)。类群4包含16份种质,有10份中国苹果、2份花红、2份楸子、1份八棱海棠和1份垂丝海棠,其中有5份中国苹果(2份绵苹果和3份槟子)的Q值为0.9999,其代表了该类群的基因库(浅黄色基因库)。类群5包含38份种质,有1份中国苹果,其余全部为山荆子,其中有6份山荆子的Q值为0.9999,此6份山荆子全部为内蒙古收集到的山荆子,其代表了该类群的基因库(黄色基因库)。类群6包含124份种质,除4份中国苹果、1份山荆子和2份楸子外,其余全部为新疆野苹果,且有11份新疆野苹果的Q值为0.9999,其代表了该类群的基因库(深黄色基因库)。类群7包含了6份种质,全部为新疆野苹果,且Q值全部为0.9999,该类群为由新疆野苹果全部代表的类群(土黄色基因库)。类群8包含28份种质,有2份花红、4份山荆子、8份中国苹果、3份垂丝海棠、8份楸子和3份八棱海棠,其中有1份中国苹果、1份山荆子、1份花红、1份楸子和2份八棱海棠的Q值为0.9999,其代表了该类群基因库(橘色基因库)。类群9包含12份种质,有4份陇东海棠、1份滇池海棠、1份沧江海棠、1份山楂海棠、1份花叶海棠、3份变叶海棠和1份楸子,其中除3份变叶海棠外的9份种质的Q值全部为0.9999,代表了该类群的基因库(红色基因库)。类群10包含13份种质,有4份八棱海棠和9份山荆子,其中占比最重的为深蓝色基因库,但无纯深蓝色基因库的种质,全部为多种基因库混杂的种质。类群11包含32份种质,有1份丽江山荆子、1份花红、1份中国苹果、4份垂丝海棠、2份新疆野苹果和23份山荆子,其中有9份山荆子和1份垂丝海棠的Q值为0.9999,9份山荆子来源于黑龙江和内蒙古,其代表了该类群的基因库(蓝色基因库)。类群12包含12份种质,由1份楸子和11份八棱海棠组成,其中有7份八棱海棠的Q值为0.9999,其代表了该类群的基因库(浅蓝色基因库)。类群13包含51份种质,除1份垂丝海棠、1份湖北海棠、1份八棱海棠、1份新疆野苹果和3份楸子外,其余全部为山荆子,其中有5份山荆子的Q值为0.9999,其代表了该类群的基因库(天蓝色基因库)。类群14包含28份种质,由10份花红、9份山荆子、3份中国苹果和6份楸子组成,其中2份楸子、2份花红和1份中国苹果的Q值≥0.9999,其代表了该类群的基因库(湖蓝色基因库)。只由一个种来代表一个基因库的类群有类群1、类群2、类群3、类群4、类群5、类群6、类群11、类群12、类群13,代表种分别为中国苹果(绵苹果)、新疆野苹果、山荆子(内蒙古)、中国苹果(绵苹果和槟子)、山荆子(内蒙古)、新疆野苹果、山荆子(黑龙江和内蒙古)、八棱海棠、山荆子(黑龙江)。
3 讨论
SNP变异通常由单个核苷酸碱基的替换、较小片段的插入或缺失引起[38],其类型有单个碱基的转换、颠换、掺入和缺失以及小片段的掺入缺失(InDel)。大多数的SNP并不直接体现在表型上,位于基因的非编码区,但却能够体现群体间的遗传和生物进化关系,可作为遗传标记而应用于动植物的群体遗传关系和进化研究[39,40]。在动植物研究的诸多方面均有应用,诸如果蝇和小鼠[41]等动物以及拟南芥[42]、水稻[43]和小麦[44]等植物。SLAF测序技术的发展促进了SNP标记在多种植物的种质资源鉴定、系统发育、遗传进化以及性状关联分析中的研究,诸如玉米[45]、小麦[46]、棉花[47]、茶树[48]、大豆[49]和葡萄[50]等。本研究在SLAF测序的基础之上开发了46 460个多态性的SNP标记,分析了15种苹果属植物的亲缘关系和遗传结构,探讨不同种间和种内的系统演化关系。基于种群间遗传距离的系统发育分析以及群体遗传结构分析均表明,15种苹果属植物分为4个基本的类群,基于种间遗传距离进行系统发育分析时,花叶海棠与新疆野苹果的遗传距离只有0.019,与群体遗传结构的分析结果有所偏差,有可能是供试的15个种在以种作为种群分析时,各种群内个体数量差异较大造成的分析结果有所偏差。
苹果属植物的遗传结构分析中,在K=5和K=14两种分群情况下,陇东海棠、变叶海棠、花叶海棠、滇池海棠、沧江海棠和山楂海棠均在同一类群中,基因来源和背景相似。6个野生种中,除山楂海棠主要分布在中国吉林省长白山地区,其余5个种在中国的西南地区均有野生分布。陇东海棠、变叶海棠、花叶海棠和山楂海棠叶片均有裂刻,只是裂刻的深浅不一。这6个苹果属植物的野生种可能具有相似的起源种,但在演化过程中逐渐出现分化。
当K=5时,类群2和类群3的中国苹果中均有来自于山荆子代表的纯基因库(蓝色基因库)基因,两个类群中各有1份中国苹果种质具有该类群的纯基因库,类群3的中国苹果含有极少量的新疆野苹果基因。此外,八棱海棠具有纯的基因库,其在遗传结构中的作用被突显出来。新疆野苹果和山荆子都有比较纯的基因库,中国苹果的基因并不全来自于新疆野苹果,而有山荆子基因的加入,并与栽培种的关系密切。
当K=14时的群体遗传结构分析表明,中国原产苹果属植物有9个比较纯的同源基因库,其中有2个为中国苹果的同源基因库,2个为新疆野苹果的同源基因库,3个为山荆子的同源基因库,中国苹果中的部分绵苹果和槟子、新疆野苹果、内蒙古和黑龙江的山荆子、八棱海棠代表了较为原始的基因来源。部分中国苹果的种质中有新疆野苹果和山荆子的基因背景,再次证明部分中国苹果在起源演化过程中有新疆野苹果和山荆子的参与,这与DUAN等[51]的结论一致。但中国苹果中还有一部分绵苹果(主要指彩苹)和槟子可以独立代表类群基因库,其基因库中并没有新疆野苹果的参与,其所在类群与山荆子以及苹果属植物的栽培种花红、楸子和八棱海棠密切相关。花红、楸子和八棱海棠与山荆子的亲缘关系更近,与苹果属植物其他野生种之间的亲缘关系也近于与新疆野苹果的亲缘关系。而根据李育农[3]的理论,中国苹果起源于新疆野苹果,在中国有近2 000年的栽培历史,其作为“西洋苹果”(Malus domestica Borkh.)引入中国之前的中国特有种,与“西洋苹果”有着不同的起源和演化过程。分析此种情况产生的原因,一是按照CAO等[52]的理论,人类活动可以造成SNP多态性的迅速降低。如果苹果属植物栽培种中国苹果起源于新疆野苹果,在从新疆野苹果向中国苹果的演化过程中,由于人为参与活动而使SNP多态性迅速降低,取而代之的是与其地理位置相近的苹果属其他种对其造成的影响;二是中国苹果中部分类型本身可能也是一个起源种,其起源演化过程与新疆野苹果可能并不相关。此次供试的中国苹果大多数来源于中国的华北地区,来源于废弃果园旁、靠近果园的山坡或者农家院,与新疆野苹果原始集中分布区相距甚远。即使是人类活动和鸟兽等对新疆野苹果向中国苹果的驯化过程有一定影响,也不足以使新疆野苹果基因在部分供试的中国苹果中几乎完全消失。因此,中国苹果与新疆野苹果的起源演化关系有待于进一步考究。随着测序技术的发展,全基因重测序成本的降低,全基因组范围零缺失的SNP标记挖掘将为相关研究提供更多的证据。
4 结论
利用SLAF技术快速挖掘覆盖全基因组的46 460个多态性SNP标记,对427份15种中国原产苹果属植物种内和种间的亲缘关系和遗传结构进行研究。15种苹果属植物分为4个基本的类群,一是山荆子类群,二是新疆野苹果和少数中国苹果类群,三是变叶海棠、花叶海棠、陇东海棠、山楂海棠、滇池海棠和沧江海棠类群,四是4个苹果属植物栽培种中国苹果、八棱海棠、花红和楸子类群。中国原产苹果属植物栽培种中国苹果中部分种质的起源演化过程有山荆子和新疆野苹果的参与,中国苹果与其他栽培种的亲缘关系密切,其与新疆野苹果的起源演化关系有待进一步考究。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]

Adiantum reniforme var. sinense is an economically important plant species endemic to China. It is only found in a few regions of the Chongqing Municipal Region. For decades, its distribution has been shrinking as a result of over-exploitation, which has caused endangerment of this species. To assist in efforts to effectively conserve this species, we investigated the genetic variation of six natural populations using isoelectric focusing in thin-layer polyacrylamide slab gels. Fourteen loci of five enzyme systems were detected, of which seven were polymorphic. The mean number of effective alleles per locus (Ae) = 1.778, the percentage of polymorphic loci (P) = 0.441, the mean expected heterozygosity (He)= 0.199, and the mean observed heterozygosity (Ho)= 0.235. The results revealed that low levels of genetic diversity existed within the natural populations of A. reniforme var. sinense in comparison with other species of ferns. Only 1.49% of the genetic variation occurred among populations whereas 98.51% existed within populations, suggesting a very low genetic divergence among the populations. The equilibrium state of the populations was measured using Hardy-Weinberg equilibrium and the Fixation Index (F). The results showed that a mixed mating system was possibly the main heterogamy of this species, and its endangerment was caused by overexploitation and habitat loss.
URL [本文引用: 1]

Adiantum reniforme var. sinense is an economically important plant species endemic to China. It is only found in a few regions of the Chongqing Municipal Region. For decades, its distribution has been shrinking as a result of over-exploitation, which has caused endangerment of this species. To assist in efforts to effectively conserve this species, we investigated the genetic variation of six natural populations using isoelectric focusing in thin-layer polyacrylamide slab gels. Fourteen loci of five enzyme systems were detected, of which seven were polymorphic. The mean number of effective alleles per locus (Ae) = 1.778, the percentage of polymorphic loci (P) = 0.441, the mean expected heterozygosity (He)= 0.199, and the mean observed heterozygosity (Ho)= 0.235. The results revealed that low levels of genetic diversity existed within the natural populations of A. reniforme var. sinense in comparison with other species of ferns. Only 1.49% of the genetic variation occurred among populations whereas 98.51% existed within populations, suggesting a very low genetic divergence among the populations. The equilibrium state of the populations was measured using Hardy-Weinberg equilibrium and the Fixation Index (F). The results showed that a mixed mating system was possibly the main heterogamy of this species, and its endangerment was caused by overexploitation and habitat loss.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/hdy.2008.35URLPMID:18461083 [本文引用: 1]

The last two decades have witnessed a remarkable activity in the development and use of molecular markers both in animal and plant systems. This activity started with low-throughput restriction fragment length polymorphisms and culminated in recent years with single nucleotide polymorphisms (SNPs), which are abundant and uniformly distributed. Although the latter became the markers of choice for many, their discovery needed previous sequence information. However, with the availability of microarrays, SNP platforms have been developed, which allow genotyping of thousands of markers in parallel. Besides SNPs, some other novel marker systems, including single feature polymorphisms, diversity array technology and restriction site-associated DNA markers, have also been developed, where array-based assays have been utilized to provide for the desired ultra-high throughput and low cost. These microarray-based markers are the markers of choice for the future and are already being used for construction of high-density maps, quantitative trait loci (QTL) mapping (including expression QTLs) and genetic diversity analysis with a limited expense in terms of time and money. In this study, we briefly describe the characteristics of these array-based marker systems and review the work that has already been done involving development and use of these markers, not only in simple eukaryotes like yeast, but also in a variety of seed plants with simple or complex genomes.
URLPMID:18721872 [本文引用: 1]
DOI:10.1007/s11295-011-0380-8URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:26437648 [本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[本文引用: 3]
URLPMID:28581499 [本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
URLPMID:19451168 [本文引用: 1]
URLPMID:20644199 [本文引用: 1]
DOI:10.1093/bioinformatics/btp352URLPMID:19505943 [本文引用: 1]

SUMMARY: The Sequence Alignment/Map (SAM) format is a generic alignment format for storing read alignments against reference sequences, supporting short and long reads (up to 128 Mbp) produced by different sequencing platforms. It is flexible in style, compact in size, efficient in random access and is the format in which alignments from the 1000 Genomes Project are released. SAMtools implements various utilities for post-processing alignments in the SAM format, such as indexing, variant caller and alignment viewer, and thus provides universal tools for processing read alignments. AVAILABILITY: http://samtools.sourceforge.net.
[本文引用: 1]
[本文引用: 1]
URLPMID:3447015 [本文引用: 1]
URLPMID:19648217 [本文引用: 1]
URL [本文引用: 1]

Single nucleotide polymorphisms (SNPs) are an abundant form of DNA variation which have a frequency of 1% or more throughout the genomes. SNPs consist of a single nucleotide base alteration including transition and transversion. They are stable and reliable mutation and are frequently referred to as bi-allelic makers. SNPs can be used conveniently for large-scale and high throughput genome analysis, in particular combining DNA chips and microarrays techniques. Therefore, SNPs provide a novel molecular marker system potentially useful for a wide range of biological disciplines. Here we briefly introduce the history and developments of SNP techniques, including its basic concept, its discovery and screening. We also discuss its applications in different research areas such as genetic mapping in mode animals and plants, DNA fingerprinting and its application in variety identification, species origin and relationship, linkage disequilibrium and associate analysis, and its application in population genetics. We anticipate that SNP markers will contribute greatly to the studies on population genetics, molecular breeding as well as evolutionary biology.
URL [本文引用: 1]

Single nucleotide polymorphisms (SNPs) are an abundant form of DNA variation which have a frequency of 1% or more throughout the genomes. SNPs consist of a single nucleotide base alteration including transition and transversion. They are stable and reliable mutation and are frequently referred to as bi-allelic makers. SNPs can be used conveniently for large-scale and high throughput genome analysis, in particular combining DNA chips and microarrays techniques. Therefore, SNPs provide a novel molecular marker system potentially useful for a wide range of biological disciplines. Here we briefly introduce the history and developments of SNP techniques, including its basic concept, its discovery and screening. We also discuss its applications in different research areas such as genetic mapping in mode animals and plants, DNA fingerprinting and its application in variety identification, species origin and relationship, linkage disequilibrium and associate analysis, and its application in population genetics. We anticipate that SNP markers will contribute greatly to the studies on population genetics, molecular breeding as well as evolutionary biology.
URLPMID:9582121 [本文引用: 1]
URLPMID:11733746 [本文引用: 1]
DOI:10.1073/pnas.0130101100URL [本文引用: 1]
URLPMID:12068090 [本文引用: 1]
DOI:10.1101/gr.2479404URLPMID:15342564 [本文引用: 1]

Dense coverage of the rice genome with polymorphic DNA markers is an invaluable tool for DNA marker-assisted breeding, positional cloning, and a wide range of evolutionary studies. We have aligned drafts of two rice subspecies, indica and japonica, and analyzed levels and patterns of genetic diversity. After filtering multiple copy and low quality sequence, 408,898 candidate DNA polymorphisms (SNPs/INDELs) were discerned between the two subspecies. These filters have the consequence that our data set includes only a subset of the available SNPs (in particular excluding large numbers of SNPs that may occur between repetitive DNA alleles) but increase the likelihood that this subset is useful: Direct sequencing suggests that 79.8% +/- 7.5% of the in silico SNPs are real. The SNP sample in our database is not randomly distributed across the genome. In fact, 566 rice genomic regions had unusually high (328 contigs/48.6 Mb/13.6% of genome) or low (237 contigs/64.7 Mb/18.1% of genome) polymorphism rates. Many SNP-poor regions were substantially longer than most SNP-rich regions, covering up to 4 Mb, and possibly reflecting introgression between the respective gene pools that may have occurred hundreds of years ago. Although 46.2% +/- 8.3% of the SNPs differentiate other pairs of japonica and indica genotypes, SNP rates in rice were not predictive of evolutionary rates for corresponding genes in another grass species, sorghum. The data set is freely available at http://www.plantgenome.uga.edu/snp.
DOI:10.1139/g03-027URLPMID:12834059 [本文引用: 1]

Single-nucleotide polymorphisms (SNPs) represent a new form of functional marker, particularly when they are derived from expressed sequence tags (ESTs). A bioinformatics strategy was developed to discover SNPs within a large wheat EST database and to demonstrate the utility of SNPs in genetic mapping and genetic diversity applications. A collection of > 90000 wheat ESTs was assembled into contiguous sequences (contigs), and 45 random contigs were then visually inspected to identify primer pairs capable of amplifying specific alleles. We estimate that homoeologue sequence variants occurred 1 in 24 bp and the frequency of SNPs between wheat genotypes was 1 SNP/540 bp (theta = 0.0069). Furthermore, we estimate that one diagnostic SNP test can be developed from every contig with 10-60 EST members. Thus, EST databases are an abundant source of SNP markers. Polymorphism information content for SNPs ranged from 0.04 to 0.50 and ESTs could be mapped into a framework of microsatellite markers using segregating populations. The results showed that SNPs in wheat can be discovered in ESTs, validated, and be applied to conventional genetic studies.
DOI:10.3864/j.issn.0578-1752.2018.04.003URL [本文引用: 1]

【Objective】 Understanding the genetic diversity and population structure of representative maize accessions are of importance in breeding practice for the guidance and reference. 【Method】A total of 344 maize inbred lines were selected, including American heterotic group, local germplasm, New germplasm used in maize breeding in China in recent years which were broadly representative. These lines were genotyped by 3 072 SNP markers which were developed by Maize Research Center, BAAFS to reveal the genetic diversity and population structure. 【Result】For 3 072 high-quality SNPs, the gene diversity averaged 0.442, ranging from 0.028 to 0.646, and the PIC averaged 0.344, ranging from 0.028 to 0.570. The result of population structure based on a model-based method indicated that these 344 lines could be divided into eight groups, including Lüda red cob, Huangzaosi improved lines, Iodent, Lancaster, P group, Improved Reid group, Reid and X group. The seven groups above were well-known, and the X group was selected from the populations constructed from X1132X. Among the eight groups, the Fst ranged from 0.319 to 0.512, and the genetic distance ranged from 0.229 to 0.514. AMOVA results indicated that 38.6% of the total genetic variation occurred among groups, 58.1% within groups and 3.3% within lines. PCA results showed that X group had higher genetic differentiation with Huangzaosi improved lines and Lancaster, but lower with Iodent. The genetic diversity of subpopulations indicated that with the increase of breeding years, the average of genetic diversity in each subpopulation was decreased, and among them, X group had the highest genetic diversity. Further analysis showed that the genetic diversity of core accessions in American heterotic group and local germplasm were higher decreased compared with that in P group and Improved Reid group. However, the genetic diversity of core accessions in X group was no decreased, which indicated that the core accessions of X group still maintained higher genetic diversity and had potential application in breeding.【Discussion】X group was different from the other seven known groups, which can be defined as an independent group. Furthermore, X group had further genetic relationship with Huangzaosi improved lines which indicated the strong heterosis pattern of "X group × Huangzaosi improved lines" had application potential.
DOI:10.3864/j.issn.0578-1752.2018.04.003URL [本文引用: 1]

【Objective】 Understanding the genetic diversity and population structure of representative maize accessions are of importance in breeding practice for the guidance and reference. 【Method】A total of 344 maize inbred lines were selected, including American heterotic group, local germplasm, New germplasm used in maize breeding in China in recent years which were broadly representative. These lines were genotyped by 3 072 SNP markers which were developed by Maize Research Center, BAAFS to reveal the genetic diversity and population structure. 【Result】For 3 072 high-quality SNPs, the gene diversity averaged 0.442, ranging from 0.028 to 0.646, and the PIC averaged 0.344, ranging from 0.028 to 0.570. The result of population structure based on a model-based method indicated that these 344 lines could be divided into eight groups, including Lüda red cob, Huangzaosi improved lines, Iodent, Lancaster, P group, Improved Reid group, Reid and X group. The seven groups above were well-known, and the X group was selected from the populations constructed from X1132X. Among the eight groups, the Fst ranged from 0.319 to 0.512, and the genetic distance ranged from 0.229 to 0.514. AMOVA results indicated that 38.6% of the total genetic variation occurred among groups, 58.1% within groups and 3.3% within lines. PCA results showed that X group had higher genetic differentiation with Huangzaosi improved lines and Lancaster, but lower with Iodent. The genetic diversity of subpopulations indicated that with the increase of breeding years, the average of genetic diversity in each subpopulation was decreased, and among them, X group had the highest genetic diversity. Further analysis showed that the genetic diversity of core accessions in American heterotic group and local germplasm were higher decreased compared with that in P group and Improved Reid group. However, the genetic diversity of core accessions in X group was no decreased, which indicated that the core accessions of X group still maintained higher genetic diversity and had potential application in breeding.【Discussion】X group was different from the other seven known groups, which can be defined as an independent group. Furthermore, X group had further genetic relationship with Huangzaosi improved lines which indicated the strong heterosis pattern of "X group × Huangzaosi improved lines" had application potential.
DOI:10.3724/SP.J.1006.2013.00727URL [本文引用: 1]

The 1E and 7E chromosomes of Thinopyrum elongatum carry important resistance genes to wheat Fusarium head blight. Mapping and utilization of these resistance genes require numerous molecular markers specific to the 1E or 7E chromosome. In this study, we developed 368 specific fragments of Thinopyrum elongatum 1E chromosome using SLAF-seq technique, and randomly selected 80 fragments to design specific primers. Finally, 20 1E-specific, 2 genome-specific, and 26 other specific molecular markers were obtained, with the efficiency up to 60%. All the newly developed markers could amplify the specific bands in different lines derived from wheat–Th. elongatum progenies. According to the cosegregation of the specific markers and elite traits, some markers detected could be closely linked to the genes corresponding to target traits.
DOI:10.3724/SP.J.1006.2013.00727URL [本文引用: 1]

The 1E and 7E chromosomes of Thinopyrum elongatum carry important resistance genes to wheat Fusarium head blight. Mapping and utilization of these resistance genes require numerous molecular markers specific to the 1E or 7E chromosome. In this study, we developed 368 specific fragments of Thinopyrum elongatum 1E chromosome using SLAF-seq technique, and randomly selected 80 fragments to design specific primers. Finally, 20 1E-specific, 2 genome-specific, and 26 other specific molecular markers were obtained, with the efficiency up to 60%. All the newly developed markers could amplify the specific bands in different lines derived from wheat–Th. elongatum progenies. According to the cosegregation of the specific markers and elite traits, some markers detected could be closely linked to the genes corresponding to target traits.
DOI:10.1111/pbr.12144URL [本文引用: 1]

Lacking of reference genome sequence for the development of stable molecular markers for specific chromosomes (intervals) remains to be a challenge in cotton, which was a necessary step in fine mapping of gene (QTL). In this study, the feasibility of development of single-nucleotide polymorphism (SNP) markers between CS-B14Sh (a substitution line for short arm of Chromosome 14) and TM-1 (the recurrent parent) was explored using next-generation sequencing (NGS) based on reduced representation libraries (RRLs). High-quality genome sequences, representing about 7.1%, 8.8% and 10.4% of the tetraploid cotton genome, were generated for TM-1, 3-79 (the donor parent) and CS-B14Sh, respectively. A total of 397 putative SNP markers were detected between CS-B14Sh and TM-1, and most (358) of them were also detected between TM-1 and 3-79. Allele-specific PCR method was used for validation of 40 SNP markers, and 27 of them showed polymorphism between TM-1 and CS-B14Sh, and a linkage group comprising of 25 SNP markers and five SSR markers was constructed. The order of SNP markers agreed well with the position of them on Chr05 of D genome, which further approved the truth of SNPs detected. The results suggested that the development of SNP markers in specific genome region using NGS was efficient in substitution or near-isogenic lines.
DOI:10.1186/1471-2164-16-1URL [本文引用: 1]
DOI:10.1111/nph.13626URLPMID:26479264 [本文引用: 1]

Present-day soybeans consist of elite cultivars and landraces (Glycine max, fully domesticated (FD)), annual wild type (Glycine soja, nondomesticated (ND)), and semi-wild type (semi-domesticated (SD)). FD soybean originated in China, although the details of its domestication history remain obscure. More than 500 diverse soybean accessions were sequenced using specific-locus amplified fragment sequencing (SLAF-seq) to address fundamental questions regarding soybean domestication. In total, 64,141 single nucleotide polymorphisms (SNPs) with minor allele frequencies (MAFs) > 0.05 were found among the 512 tested accessions. The results indicated that the SD group is not a hybrid between the FD and ND groups. The initial domestication region was pinpointed to central China (demarcated by the Great Wall to the north and the Qinling Mountains to the south). A total of 800 highly differentiated genetic regions and > 140 selective sweeps were identified, and these were three- and twofold more likely, respectively, to encompass a known quantitative trait locus (QTL) than the rest of the soybean genome. Forty-three potential quantitative trait nucleotides (QTNs; including 15 distinct traits) were identified by genome-wide association mapping. The results of the present study should be beneficial for soybean improvement and provide insight into the genetic architecture of traits of agronomic importance.
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/s41467-017-00336-7URL [本文引用: 1]
DOI:10.1186/s13059-014-0415-1URLPMID:25079967 [本文引用: 1]

BACKGROUND: Recently, many studies utilizing next generation sequencing have investigated plant evolution and domestication in annual crops. Peach, Prunus persica, is a typical perennial fruit crop that has ornamental and edible varieties. Unlike other fruit crops, cultivated peach includes a large number of phenotypes but few polymorphisms. In this study, we explore the genetic basis of domestication in peach and the influence of humans on its evolution. RESULTS: We perform large-scale resequencing of 10 wild and 74 cultivated peach varieties, including 9 ornamental, 23 breeding, and 42 landrace lines. We identify 4.6 million SNPs, a large number of which could explain the phenotypic variation in cultivated peach. Population analysis shows a single domestication event, the speciation of P. persica from wild peach. Ornamental and edible peach both belong to P. persica, along with another geographically separated subgroup, Prunus ferganensis. CONCLUSIONS: Our analyses enhance our knowledge of the domestication history of perennial fruit crops, and the dataset we generated could be useful for future research on comparative population genomics.
