 ,, 林世强, 倪冬昕, 伊恒杰, 刘江洪, 杨志坚, 郑金贵
,, 林世强, 倪冬昕, 伊恒杰, 刘江洪, 杨志坚, 郑金贵 ,福建农林大学农学院/作物生物技术福建省高校重点实验室/作物遗传育种与综合利用教育部重点实验室,福州 350002
,福建农林大学农学院/作物生物技术福建省高校重点实验室/作物遗传育种与综合利用教育部重点实验室,福州 350002Cloning and Function Characterization of Chalcone Synthase Gene AgCHS1 in Ampelopsis grossedentata
XU Ming ,, LIN ShiQiang, NI DongXin, YI HenJie, LIU JiangHong, YANG ZhiJian, ZHENG JinGui
,, LIN ShiQiang, NI DongXin, YI HenJie, LIU JiangHong, YANG ZhiJian, ZHENG JinGui ,College of Agriculture, Fujian Agriculture and Forestry University/Key Laboratory of Crop Biotechnology in Fujian Province University/Key Laboratory of Ministry of Education for Genetics, Breeding and Multiple Utilization of Crops, Fuzhou 350002
,College of Agriculture, Fujian Agriculture and Forestry University/Key Laboratory of Crop Biotechnology in Fujian Province University/Key Laboratory of Ministry of Education for Genetics, Breeding and Multiple Utilization of Crops, Fuzhou 350002通讯作者:
责任编辑: 赵伶俐
收稿日期:2020-05-14接受日期:2020-07-15网络出版日期:2020-12-16
| 基金资助: |
Received:2020-05-14Accepted:2020-07-15Online:2020-12-16
作者简介 About authors
许明,Tel:0591-83789231;E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2908KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
许明, 林世强, 倪冬昕, 伊恒杰, 刘江洪, 杨志坚, 郑金贵. 藤茶查尔酮合成酶基因AgCHS1的克隆及功能鉴定[J]. 中国农业科学, 2020, 53(24): 5091-5103 doi:10.3864/j.issn.0578-1752.2020.24.012
XU Ming, LIN ShiQiang, NI DongXin, YI HenJie, LIU JiangHong, YANG ZhiJian, ZHENG JinGui.
开放科学(资源服务)标识码(OSID):

0 引言
【研究意义】藤茶学名为显齿蛇葡萄(Ampelopsis grossedentata W.T.Wan),属于葡萄科蛇葡萄属藤本植物,主要分布在中国福建、云南、广东、广西、贵州和湖南等长江以南的山区,民间将其作为茶药两用植物应用已有数百年的历史[1]。藤茶的主要有效成分是以二氢杨梅素为主的黄酮类化合物,具有抗氧化、保肝护肝、降血压和抗肿瘤等多种功效[1,2,3]。研究表明,品种、季节和栽培条件等多种因素都会造成藤茶黄酮含量出现较大差异,从而影响藤茶产品的质量甚至疗效[3,4,5]。因此,开展藤茶黄酮类化合物合成关键基因的克隆、功能鉴定及其调控机制研究,对藤茶药用品质提升及其资源持续高效利用具有重要意义。【前人研究进展】黄酮类化合物(又称类黄酮)的生物合成途径在国内外已有了较为广泛地研究,其合成途径的主要步骤已基本探明[6]。查尔酮合成酶(chalcone synthase,CHS)属于III型聚酮合成酶(polyketide synthase,PKS)家族,是类黄酮合成途径中的第1个关键结构酶,负责催化3分子丙二酰CoA与1分子的对-香豆酰辅酶A缩合形成柚皮素查尔酮,并进一步生成黄酮醇、异黄酮和花青素等各类黄酮类化合物[7]。CHS广泛存在于高等植物中,目前CHS已经在烟草[8]、虎杖[9]、苹果[10]、桑树[11]、雪莲[12]、地钱[13]、香雪兰[14]等多种植物中被克隆,并明确了其功能。CHS主要以多基因家族形式存在[15],不同植物中数目不等,例如在水稻中可能有27个CHS [16],烟草中至少有5个成员[8],而金鱼草中只有1个[17]。近期LI等[18]利用高通量转录组测序技术,注释了多条与藤茶CHS相关的unigene,暗示CHS在藤茶中也是以多基因家族形式存在。对不同植物CHS的分子系统进化研究发现,CHS超基因家族的起源较早,但在长期进化过程中,CHS家族成员内部发生了多样的遗传改变,使其功能上可能各有分工[7]。典型的CHS催化生成查尔酮,进入花青素合成途径,而其他成员则催化生成一些与植物抗逆相关的产物[15]。增强或抑制不同植物CHS的表达,能改变植物花色或抗逆能力[14,19-20]。此外,CHS的表达受发育阶段和环境因子的影响,在不同组织和器官中的表达模式也存在差异[21,22]。【本研究切入点】前期研究已从藤茶中克隆和注释了一部分CHS,并对其序列进行了生物信息学分析[18,23],但对其功能的研究尚未见报道。【拟解决的关键问题】本研究基于转录组测序基础,从藤茶中克隆得到1条CHS的编码区序列,命名为AgCHS1,通过生物信息学分析、组织表达特异性分析、体外酶活性检测和烟草遗传转化试验,探讨该基因在藤茶类黄酮生物合成过程中的作用,为进一步研究藤茶类黄酮高效累积的分子机理奠定基础。1 材料与方法
试验于2017年9月至2020年5月在福建农林大学农学院作物生物技术福建省高校重点实验室进行。1.1 植物材料与试剂
藤茶引自福建尤溪县下村林场,经福建农林大学植物学教研室鉴定为葡萄科蛇葡萄属显齿蛇葡萄。在同一植株上分别采集幼叶、成熟叶、老叶、茎、花和根,液氮速冻后研磨成粉末,然后分成2份,一部分用于总RNA提取,另一部分冻干后贮存于-80℃冰箱,用于总黄酮的提取与含量测定。植物总RNA提取试剂盒、质粒提取DNA和胶回收试剂盒购自北京艾德莱生物科技有限公司。限制性内切酶、T4 DNA连接酶、克隆载体pMD-18T、原核表达载体pET28a-pGTf2、大肠杆菌感受态细胞(DH5α)、ExTaq DNA聚合酶购自宝生物工程(大连)有限公司。反转录试剂盒和Sybr Green qPCR Master Mix购自BBI生命科学有限公司。二氢杨梅素、槲皮素对照品购自上海同田生物技术股份有限公司,丙二酰辅酶A、柚皮素购自美国Sigma公司,对-香豆酰辅酶A购自德国MicroCombiChem公司。引物合成和测序均由上海生工有限公司合成。
1.2 基因组DNA、总RNA的提取和第一链cDNA合成
采用CTAB法提取藤茶叶片基因组DNA。藤茶不同组织RNA的提取参照笔者实验室前期建立的方法[24]进行,烟草总RNA的提取参考植物RNA提取试剂盒说明书进行。通过1%琼脂糖凝胶电泳检测DNA和总RNA的完整性,用Nanodrop 2000分光光度计测定样品浓度和纯度。利用PrimeScript? RT reagent试剂盒(Takara)将总RNA反转录为第一链cDNA。1.3 AgCHS1的克隆与序列分析
根据前期藤茶叶片转录组的测序结果(NCBI登录号:SRP241021),设计5′端分别带BamH I和Sac I酶切位点的AgCHS1特异性引物AgCHS1-F/R(表1),分别以藤茶叶片DNA和cDNA为模板,利用ExTaq DNA聚合酶进行PCR扩增,反应条件为:94℃预变性5 min;94℃ 30 s,54℃ 30 s,72℃ 60 S,30个循环;72℃延伸10 min。PCR产物经琼脂糖凝胶电泳后回收目的片段,将回收产物与pMD18-T载体连接并转化大肠杆菌感受态,挑选阳性克隆进行测序。Table 1
表1
表1本试验中所用的引物序列
Table 1
| 引物名称 Primer name | 序列(5'-3') Primer sequence (5'-3') | 用途 Purpose |
|---|---|---|
| AgCHS1-F | GGATCCATGGTGTCGGTGCAGGAAATCAGA | ORF序列扩增、载体构建 |
| AgCHS1-R | GAGCTCTCAGTGAGCCAGTGGTGCAGAC | Amplification of ORF sequence and vector construction |
| AgCHS1-P1 | ACTCCAGCCAACTGTGTCCA | 转基因烟草的PCR鉴定 |
| AgCHS1-P2 | TGAGACCCACTTCACGCAA | PCR identification of transgenic tobacco |
| AgCHS1-qF | GCCTCAAACCCTCCGTCA | 实时荧光定量PCR Quantitative real-time PCR |
| AgCHS1-qR | GACCCACGAGTGAATCCAAGT | Quantitative real-time PCR |
| NtEF1α-qF | AATTTTGACCAAGATCGACAGG | 烟草内参基因 Control |
| NtEF1α-qR | CAGCAACAGTTTGACGCATG | Reference gene in tobacco |
| AgGAPDH-qF | CATCTCAGCCCCAAGCAA | 藤茶内参基因 |
| AgGAPDH-qR | GTGGCAGTAATGGAGTGAACAG | Reference gene in A.grossedentata |
新窗口打开|下载CSV
将测序所得序列在NCBI上进行同源性比对(
1.4 AgCHS1重组蛋白原核表达及纯化
用BamHⅠ和SacⅠ双酶切含有AgCHS1的pMD-18T载体,回收AgCHS1片段,连接到用相同内切酶消化的pET28a-pGTf2载体上,将连接产物导入大肠杆菌BL21(DE3)感受态中,鉴定出阳性单菌落并测序。确定序列及载体组装无误后,使用0.4 mmol?L-1 IPTG诱导表达目的蛋白,诱导表达条件及步骤参照LIN等[25]的方法。采用Bradford法测定蛋白浓度。1.5 AgCHS1体外酶活性试验
AgCHS1酶活性测定参照YAHYAA等[10]的方法并略作修改,250 μL反应体系包括15 μmol·L-1的对-香豆酰辅酶A、56 μmol·L-1的丙二酰辅酶A、0.1 mol·L-1磷酸钾缓冲液(pH 7.0)以及2.0 μg纯化的AgCHS1。将上述反应体系置于35℃下反应30 min后,加入等体积乙酸乙酯抽提2次,于12 000 r/min离心10 min;取上清液真空干燥后,加入250 μL 50%(v/v)的甲醇水溶液溶解残留。通过UPLC(Waters,2695)对酶促反应产物进行初步分析,再利用HPLC-MS(Thermo Fisher,LCQ Fleet)对产物进行鉴定。液相条件:液相柱BEH C18,粒径1.7 μm, 2.1 mm×100 mm;柱温35℃;进样量2 μL。A相:1.0%乙酸;B相:100%乙腈;流速:0.3 mL?min-1。质谱条件:离子源为电喷雾(ESI-),毛细管温度300℃,鞘气(N2)15 arb,喷雾电压-4.47 kV,检测电压1.5 kV,时间100 ms,碰撞气体He,母离子选择宽度:3 u,采集范围100—600 m/z。1.6 基因表达分析
根据所获得AgCHS1的序列,采用Primer5.0软件设计荧光定量PCR引物(表1)。藤茶的内参基因为GAPDH[26],烟草的内参基因为EF1α。荧光定量反应体系为:SybrGreen qPCR Master Mix(2×)10 μL,10 μmol·L-1上、下游引物各0.4 μL,cDNA 2 μL,加ddH2O至终体积20 μL。反应在ABI StepOne型荧光定量PCR仪上进行,程序为95℃变性3 min,95℃变性5 s,60℃退火延伸30 s,45个循环,于60℃收集荧光,反应结束后分析荧光值变化曲线和熔解曲线,每个反应重复3次,采用2-ΔΔCt算法计算基因相对表达量。1.7 植物表达载体构建及转基因烟草的获得
利用BamH Ⅰ和Sac I双酶切pBI121载体,回收载体大片段,与1.3中回收的AgCHS1片段连接,将连接产物转化E.coli DH5α感受态细胞,通过酶切鉴定阳性克隆,得到植物表达载体pBI-35S:AgCHS1,采用冻融法将其导入根癌农杆菌EHA105中,按照NING等[27]的方法进行烟草遗传转化。提取T0代抗性植株叶片DNA,用引物AgCHS1-P1/P2进行PCR验证,阳性植株连续自交种植2代后获得T2代植株,用于后续的检测。1.8 黄酮类物质的提取与测定
藤茶不同器官总黄酮的提取参照LI等[18]介绍的提取方法,然后采用硝酸铝比色法[28],检测样品溶液在324 nm波长的吸收值,通过二氢杨梅素的标准曲线换算样品中的总黄酮含量。烟草花瓣花青素的提取与测定按照WANG等[29]的方法,并略作修改。取50 mg野生型和转基因烟草的花瓣(鲜重,M),加入1 mL酸化甲醇(含1% HCl(v/v)),黑暗条件下提取24 h,10 000 r/min离心10 min,取上清液,用紫外可见光分光光度计(Thermo Scientific,Evolution 201)测定530 nm与657 nm处的吸光值(A530和A657)。相对花青素含量(Q)用公式(Q=(A530-0.25×A 657)M)计算。烟草花瓣黄酮醇的提取与测定参照TIRUMALAI等[30]的方法进行,并略作修改。取100 mg新鲜的烟草花瓣在液氮中研磨成粉末,加入2.0 mL 75%乙醇提取3 h,滤液干燥后,溶解于1.0 mL 75%乙醇中。取0.1 mL提取液,加入0.5 mL 5% NaNO2,摇匀放置6 min,再加10% Al(NO3)3,摇匀放置6 min,最后加入2.5 mL 1.0 mol·L-1 NaOH溶液中止反应,放置15 min后,在波长510 nm处测定吸光值。按照槲皮素的标准曲线换算样品中总黄酮醇含量。2 结果
2.1 AgCHS1的克隆和生物信息学分析
根据转录组测序信息,设计特异性引物,以藤茶叶片cDNA为模板,扩增得到的AgCHS1 ORF序列(GenBank登录号为MT316507)长度为1 182 bp,编码393个氨基酸。以基因组DNA为模板,扩增得到的gDNA序列(GenBank登录号为MT316508)长度为1 315 bp(图1-A),与ORF序列比对后发现,AgCHS1具有一个内含子(133 bp),外显子1为178 bp,外显子2为1 004 bp,内含子遵循GT/AG规则(图1-B)。图1
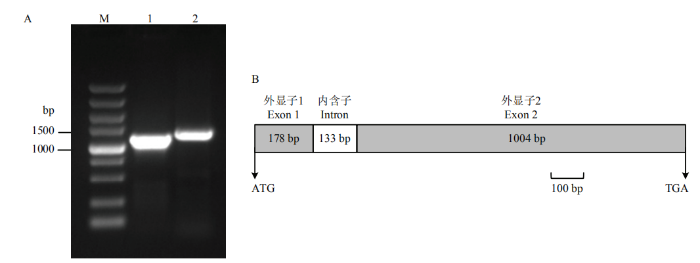 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1藤茶AgCHS1的克隆与结构分析
A:AgCHS1的PCR扩增产物,M:DL5000;1:ORF扩增产物;2:基因组DNA扩增产物。B:AgCHS1的结构
Fig. 1Cloning and structure analysis of the AgCHS1
A: PCR amplification product of AgCHS1, M: DL5000; 1: The amplification product of ORF; 2: The amplification product of genomic DNA; B: Structure of AgCHS1
通过ProtParam在线分析AgCHS1蛋白理化性质,其理论分子量为42.8 kD,理论等电点(pI)为6.15,分子式为C1908H3059N513O565S19,带负电荷的氨基酸(Asp+Glu)为48个,带正电荷的氨基酸(Arg+Lys)为43个,不稳定系数为34.71,属于稳定性蛋白。平均亲水性(GRAVY)为-0.045<0,表明AgCHS1属于亲水性蛋白。
利用DNAMAN软件将AgCHS1与葡萄、水稻、拟南芥、烟草等常见植物的CHS进行序列多重比对,发现该蛋白与葡萄CHS的氨基酸序列相似度最高,达到96.7%,与陆地棉、水稻、拟南芥、烟草的相似度分别为89.1%、82.2%、84.5%和86.5%,与付明等[23]报道的藤茶CHS氨基酸序列存在较大差异,二者相似度为88.1%。AgCHS1含有查尔酮合成酶家族典型的Cys-His-Asn三联活性中心位点和2个高度保守的苯丙氨酸残基Phe215和Phe265(图2),这些位点与查尔酮合成酶底物特异性相关[29]。此外,AgCHS1还具有丙二酰辅酶A结合位点和2个高度保守的CHS特征序列RLMMYQQGCFAGGTVLR和GVLFGFGPGL(图2)。进一步说明本研究所克隆的序列为藤茶CHS的序列。
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2AgCHS1与其他物种CHS氨基酸序列比对分析
AgCHS1:藤茶Ampelopsis grossedentata;VvCHS:葡萄 Vitis vinifera NP_001267879.1;GhCHS1:陆地棉 Gossypium hirsutum CV72638;OsCHS:水稻 Oryza sativa CAA61955.1;AtCHS:拟南芥Arabidopsis thaliana AAA32771.1;NtCHS:烟草Nicotiana tabacum ANA78328.1。▲:Cys-His-Asn三联活性中心;★:表示苯丙氨酸残基Phe215和Phe265;方框为CHS高度保守特征序列,下划线为丙二酰CoA结合位点 ▲: Cys-His-Asn triple active center; ★: phenylalanine residues Phe215 and Phe265; the highly conserved CHS signature sequence are boxed with black line; the malonyl-CoA binding site are underlined
Fig. 2Amino acid sequence alignment of AgCHS1 with other CHS enzymes
采用MEGA 6软件邻接法(Neighbor-Joining,NJ),对包括藤茶在内的14种植物的CHS蛋白序列进行聚类分析,构建系统进化树。结果显示,藤茶AgCHS1与同科的葡萄、山葡萄进化关系最近,与圆叶葡萄、茶树、陆地棉的进化关系也较近,而与白鲜、甜橙等进化关系较远(图3)。
图3
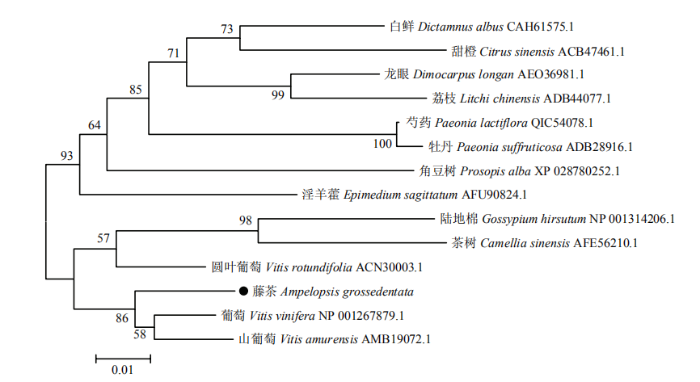 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3AgCHS1与其他植物CHS蛋白的系统进化树分析
Fig. 3Phylogenetic tree analysis of AgCHS2 protein with other plant CHS proteins
2.2 藤茶不同器官AgCHS1的表达及总黄酮含量
对AgCHS1在藤茶幼叶、成熟叶、老叶、茎、花和根中的表达进行分析。结果显示,AgCHS1在藤茶所有器官中均有表达。其中,成熟叶和花中的表达量最高,在老叶中的表达量最低。在叶片3个不同发育时期,AgCHS1的表达水平呈先升后降的趋势。总黄酮含量变化趋势与AgCHS1的表达变化基本一致(图4),二者在0.05水平上呈显著正相关(相关系数为0.9224),说明AgCHS1的表达在藤茶总黄酮积累过程中可能发挥着重要作用。图4
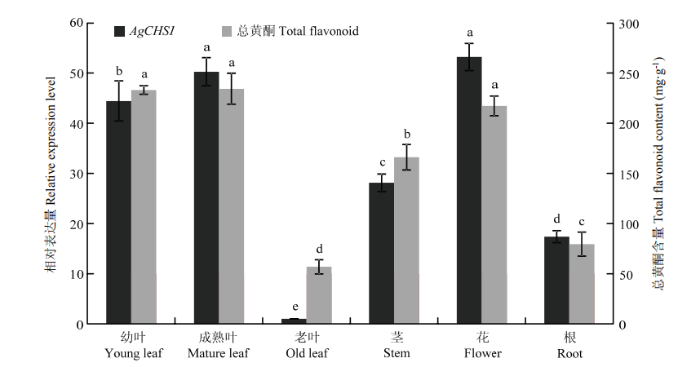 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4藤茶不同器官的AgCHS1表达与总黄酮含量变化
不同字母表示在P<0.05水平差异显著。下同
Fig. 4The change of relative expression of AgCHS1 and total flavonoid content in different organs of A. grossedentata
Different letters indicate statistical difference (P<0.05). The same as below
2.3 AgCHS1的原核表达、纯化与酶活性测定
将重组质粒pET28a-AgCHS1/pGTf2转化至大肠杆菌E.coli BL21,经IPTG诱导,超声裂解和Ni亲和层析柱纯化,得到纯化的AgCHS1重组蛋白。经SDS-PAGE确定其分子量约为43 kD(包括His标签),与预期分子量大小一致(图5)。通过Bradford法测定AgCHS1蛋白含量为0.75 mg?mL-1。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5大肠杆菌表达重组AgCHS1蛋白的Ni柱纯化
M:蛋白Marker;1:离心后的裂解液上清;2:裂解液上清经过Ni柱的流穿;3:Binding buffer洗脱;4:Binding buffer + 60 mmol?L-1咪唑洗脱;5:Binding buffer + 90 mmol?L-1咪唑洗脱;5:Binding buffer + 250 mmol?L-1咪唑洗脱
Fig. 5Ni column purification of recombinant AgCHS1 protein expressed in E. coli
M: Protein Marker; 1: Centrifuged supernatant; 2: Flow through; 3: binding buffer elution; 4: Binding buffer + 60 mmol?L-1 imidazole elution; 5: Binding buffer + 90 mmol?L-1 imidazole elution; 5: Binding buffer + 250 mmol?L-1 imidazole elution
在体外酶促反应中,当以对-香豆酰辅酶A与丙二酰辅酶A为底物时,AgCHS1能有效催化柚皮素查尔酮合成,然后柚皮素查尔酮自发异构化为柚皮素。HPLC-MS结果显示,AgCHS1酶促反应产物色谱峰的保留时间出现在6.8 min,与柚皮素标准品的保留时间一致(图6-A、B),而空载蛋白对照组未观察到目标峰(图6-C)。柚皮素标准品二级质谱中母离子峰的质荷比(m/z)为271,其子离子峰m/z为151和177(图6-D、E),通过文献检索和标准品比对,确定酶促反应产物二级质谱中具有相应的特征离子峰(图6-F、G)。说明AgCHS1编码的蛋白具有查尔酮合成酶活性,在体外能够催化对-香豆酰辅酶A生成柚皮素。
图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6重组蛋白AgCHS1体外酶促产物的HPLC-MS鉴定
A:重组蛋白AgCHS1酶促产物的色谱图;B:柚皮素标准品色谱图;C:空载体对照;D—E:柚皮素标准品的质谱图;F—G:重组蛋白AgCHS1酶促产物的质谱图
Fig. 6HPLC-MS identification of in vitro enzymatic reaction product of recombinant protein AgCHS1
HPLC chromatograms of the products in the reactions of AgCHS1 recombinant protein (A); naringen instandard (B) and the empty control (C); D-E: The masss pectrometric spectrum of reaction products; F-G: The mass spectrometric spectrum of the naringenin standard
2.4 AgCHS1在烟草中的过量表达
构建由组成型启动子CaMV35S调控AgCHS1表达的植物表达载体,采用农杆菌介导法将其转到早花烟草中,获得5株独立的卡那霉素抗性植株。经PCR检测5株全部呈阳性(图7-A),其中2株(OE3和OE4)的花瓣颜色比野生型对照明显加深(图7-B),且该表型在后代能稳定传递。花青素含量测定结果显示,过表达株系OE3、OE4花瓣中的花青素含量分别比野生型对照提高56.6%和25.3%(图7-C,P<0.05)。进一步通过荧光定量PCR检测发现,AgCHS1在株系OE3和OE4花瓣中的转录水平要远高于野生型株系(图7-D),且其表达量与花青素含量相一致,说明AgCHS1的过量表达促进了烟草花瓣的花青素生物合成。另外,CHS同时也是植物黄酮醇合成途径上游编码酶基因,其上调表达可能影响黄酮醇的累积,因此对转基因烟草花瓣的黄酮含量进行测定(图7-E)。结果显示,在上述两个过表达株系中,虽然OE3的花瓣颜色比对照加深更明显,但其黄酮醇含量与野生型对照没有显著差异,而株系OE4却比对照提高了39.1%(P<0.05)。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7过量表达AgCHS1对烟草花色、花青素和黄酮醇含量的影响
A:转基因烟草的PCR鉴定;B:花瓣表型;C:花瓣AgCHS1表达量;D:花瓣相对花青素含量;E:花瓣的相对黄酮醇含量
M:DL1000 DNA标准分子量;P:质粒对照,WT:野生型;OE1—OE5:转基因株系。不同字母表示在P<0.05水平差异显著
Fig. 7Efects of overexpression AgCHS1 on the color, anthocyanin and flavonol content of tobacco
A: PCR identification of transgenic tobacco; B: Petal phenotypes; C: AgCHS1 expression in petal; D: Relative anthocyanin content; E: Relative flavonol content;
M: DNA marker DL1000; P: Plasmid control; WT: Wild type; OE1-5: different transgenic lines. Different letters indicate statistical difference (P<0.05)
3 讨论
CHS是高等植物中分布最广泛的类型III PKS,具有相对保守的基因结构和较高的序列相似性[31,32]。本研究根据藤茶叶片转录组测序结果设计引物,克隆获得AgCHS1,与多数物种CHS的大小基本一致[8,9,10,11,12,13,14]。目前报道的多数植物CHS序列只含有1个内含子,但也有少数例外,比如金鱼草CHS含有2个内含子[17],紫丁香CHS没有内含子[29],而在小立碗藓(Physcomitrella patens)CHS基因家族的14个成员中,存在外显子-内含子的多种组合类型[33]。本研究克隆的藤茶AgCHS1基只含有1个内含子,这与多数报道[8,13-14,32]的植物CHS结构相类似。序列多重比对发现,AgCHS1包含查尔酮合成酶家族的特征序列以及高度保守的活性氨基酸残基,与葡萄、水稻、拟南芥、烟草等常见植物的CHS氨基酸序列有较高的相似性(82.2%—96.7%),证实AgCHS1属于结构保守型CHS基因。另外,AgCHS1与付明等[23]报道的藤茶CHS上存在一定的序列差异,但它们的特征性保守位点基本相同(结果未列),据此判断它们属于藤茶CHS家族的不同成员,可能在藤茶类黄酮生物合成过程中发挥相似的作用。以上这些结果说明AgCHS1在序列上与其他植物的CHS有诸多相似之处,属于典型的查尔酮合成酶家族基因。CHS的体外酶活性于1972年首次在欧芹悬浮细胞培养物中被检测到[34]。本研究通过原核表达重组蛋白的方法,对AgCHS1重组蛋白的体外酶活特性进行了分析,发现该重组蛋白可以催化对-香豆酰辅酶A与丙二酰辅酶A的缩合反应,形成柚皮素,说明AgCHS1具有查尔酮合成酶的活性。值得注意的是,酶促产物除了主产物柚皮素之外,还存在少量的副产物(图6-A),类似的情况也出现在小立碗藓[35]、地钱[36]等植物CHS的体外酶促反应中,推测该副产物可能是CHS酶促反应早期的脱轨产物Bisnoryangonin(BNY)或4-Coumaroyltriaceticacid lactone(CTAL)[37],有待于进一步鉴定。另据报道,CHS起始底物较为宽泛,但对脂肪族酰基辅酶A和芳香族酰基辅酶A具有明显偏好性[14,38]。例如,对小立碗藓PpCHS的体外活性分析表明,该酶的底物除了对-香豆酰辅酶A外,还可以是肉桂酰辅酶A或二氢香豆酰辅酶A,但在这些底物中,PpCHS更偏好对-香豆酰辅酶A[35]。同样,黄芩CHS的底物特异性分析也表明,CHS可以同时接受苯甲酰辅酶A、苯乙酰辅酶A、异戊基辅酶A和异丁酰辅酶A等芳香族和脂肪族酰基辅酶A,生成多种反应产物[38]。鉴于上述这些发现,需要进一步研究AgCHS1对不同底物的催化效率,以明确其体内的生理作用底物。
黄酮类物质与植物花色关系密切[39],CHS作为类黄酮合成途径的上游编码酶基因,其基因表达量的增加或减少都可能导致植物花色的变化。在矮牵牛中过量表达小苍兰CHS,可显著增加黄酮醇和花青苷的含量,并使花瓣由原来的白色变为粉红色[14]。在烟草中过量表达紫丁香CHS,可使花瓣颜色明显加深,花青素含量提高50%以上[29]。相反地,抑制烟草[19]、大丽花[40,41]和三环草[42]等植物中的CHS表达,可导致花青素含量降低,花色随之变淡或出现完全白色的花。本研究将AgCHS1转入野生型烟草中过表达,结果显示转基因植株花瓣颜色由淡紫色变成紫红色,花青素含量也相应的增加,此结果与前人报道的结果基本一致。值得注意的是,在本研究获得的2个烟草过表达转基因株系中,株系OE3花青素含量的增加幅度大于株系OE4,但其黄酮醇含量却没有增加,而株系OE4却提高了39.1%,这可能与黄酮醇和花青素合成调控方式较为复杂有关。黄酮醇和花青素处于类黄酮合成途径不同的代谢分支,它们的合成除了受上游CHS、F3H等酶的影响,还与下游DFR和FLS两种酶对分支点中间产物二氢黄酮醇的竞争有关[43]。
4 结论
从藤茶克隆到查尔酮合成酶基因AgCHS1,该基因编码蛋白具有典型的CHS家族蛋白保守结构域,在体外能够催化对-香豆酰辅酶A生成柚皮素,过量表达该基因可以提高烟草的花青素和黄酮醇的含量,说明AgCHS1可能在藤茶类黄酮的生物合成中发挥重要作用。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1021/acs.jafc.7b01155URLPMID:28534405 [本文引用: 2]

Dihydromyricetin (DMY), a flavanonol compound found as the most abundant and bioactive constituent in vine tea (Ampelopsis grossedentata), possesses numerous biological activities. In the present study, an HPLC-MS/MS method for the determination of DMY in tissues, urine, and feces was developed and applied to the tissue distribution and excretion study after oral administration in rats, and the metabolic profile of DMY was further investigated using UPLC-QTOF-MS. The results indicated that DMY could be distributed rapidly in various tissues and highly in the gastrointestinal tract. The elimination of DMY occurred rapidly as well, and most unconverted forms were excreted in feces. A total of eight metabolites were identified in urine and feces, while metabolites were barely found in plasma. The predicted metabolic pathways including reduction, dehydroxylation, methylation, glucuronidation, and sulfation were proposed. The present findings may provide the theoretical basis for evaluating the biological activities of DMY and will be helpful for its future development and application.
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/jxb/erx177URLPMID:28922752 [本文引用: 1]

Flavonoids are a signature class of secondary metabolites formed from a relatively simple collection of scaffolds. They are extensively decorated by chemical reactions including glycosylation, methylation, and acylation. They are present in a wide variety of fruits and vegetables and as such in Western populations it is estimated that 20-50 mg of flavonoids are consumed daily per person. In planta they have demonstrated to contribute to both flower color and UV protection. Their consumption has been suggested to presenta wide range of health benefits. Recent technical advances allowing affordable whole genome sequencing, as well as a better inventory of species-by-species chemical diversity, have greatly advanced our understanding as to how flavonoid biosynthesis pathways vary across species. In parallel, reverse genetics combined with detailed molecular phenotyping is currently allowing us to elucidate the functional importance of individual genes and metabolites and by this means to provide further mechanistic insight into their biological roles. Here we provide an inventory of current knowledge of pathways of flavonoid biosynthesis in both the model plant Arabidopsis thaliana and a range of crop species, including tomato, maize, rice, and bean.
[本文引用: 2]
[本文引用: 2]
DOI:10.1007/s00344-016-9646-6URL [本文引用: 4]
DOI:10.1007/s00425-008-0845-7URL [本文引用: 2]

A type III polyketide synthase cDNA and the corresponding gene (PcPKS2) were cloned from Polygonum cuspidatum Sieb. et Zucc. Sequencing results showed that the ORF of PcPKS2 was interrupted by three introns, which was an unexpected finding because all type III PKS genes studied so far contained only one intron at a conserved site in flowering plants, except for an Antirrhinum majus chalcone synthase gene. Besides the unusual gene structure, PcPKS2 showed some interesting characteristics: (1) the CHS “gatekeepers” Phe215 and Phe265 are uniquely replaced by Leu and Cys, respectively; (2) recombinant PcPKS2 overexpressed in Escherichia coli efficiently afforded 4-coumaroyltriacetic acid lactone (CTAL) as a major product along with bis-noryangonin (BNY) and p-hydroxybenzalacetone at low pH; however, it effectively yielded p-hydroxybenzalacetone as a dominant product along with CTAL and BNY at high pH. Beside p-hydroxybenzalacetone, CTAL and BNY, a trace amount of naringenin chalcone could be detected in assays at different pH. Furthermore, 4-coumaroyl-CoA and feruloyl-CoA were the only cinnamoyl-CoA derivatives accepted as starter substrates. PcPKS2 did not accept isobutyryl-CoA, isovaleryl-CoA or acetyl-CoA as substrate. DNA gel blot analysis indicated that there are two to four PcPKS2 copies in the P. cuspidatum genome. RNA gel blot analysis revealed that PcPKS2 is highly expressed in the rhizomes and in young leaves, but not in the roots of the plant. PcPKS2 transcripts in leaves were induced by pathogen infection, but not by wounding.
DOI:10.1016/j.phytochem.2017.04.022URLPMID:28482241 [本文引用: 3]

Apple (Malus x domestica Brokh.) is a widely cultivated deciduous tree species of significant economic importance. Apple leaves accumulate high levels of flavonoids and dihydrochalcones, and their formation is dependent on enzymes of the chalcone synthase family. Three CHS genes were cloned from apple leaves and expressed in Escherichia coli. The encoded recombinant enzymes were purified and functionally characterized. In-vitro activity assays indicated that MdCHS1, MdCHS2 and MdCHS3 code for proteins exhibiting polyketide synthase activity that accepted either p-dihydrocoumaroyl-CoA, p-coumaroyl-CoA, or cinnamoyl-CoA as starter CoA substrates in the presence of malonyl-CoA, leading to production of phloretin, naringenin chalcone, and pinocembrin chalcone. MdCHS3 coded a chalcone-dihydrochalcone synthase enzyme with narrower substrate specificity than the previous ones. The apparent Km values of MdCHS3 for p-dihydrocoumaryl-CoA and p-coumaryl-CoA were both 5.0 muM. Expression analyses of MdCHS genes varied according to tissue type. MdCHS1, MdCHS2 and MdCHS3 expression levels were associated with the levels of phloretin accumulate in the respective tissues.
DOI:10.1016/j.plaphy.2017.03.014URLPMID:28355585 [本文引用: 2]

Chalcone synthase (CHS) is the pivotal enzyme that catalyzes the first committed step of the phenylpropanoid pathway leading to flavonoids. Here, five CHS genes were determined in mulberry (Morus atropurpurea Roxb.). Interestingly, phylogenetic analysis tended to group three MaCHSs in the stilbene synthase (STS) family and initially annotated these as MaSTSs. A co-expression system that harbored a 4-coumarate:CoA ligase gene and one of the candidate genes was established to determine the functions of this novel gene family. The fermentation result demonstrated that MaSTS in fact encoded a CHS enzyme, and was consequently retermed MaCHS. Tissue-specific expression analysis indicated that MaCHS1/MaCHS2 was highly abundant in fruit, and MaCHS4 had significant expression in root bark, stem bark and old leaves, while MaCHS3 and MaCHS5 were more expressed in old leaves. Subcellular localization experiments showed that MaCHS was localized to the cytoplasm. Transcription levels suggested MaCHS genes were involved in a series of defense responses. Over-expression of MaCHS in transgenic tobacco modified the metabolite profile, and resulted in elevated tolerance to a series of environmental stresses. This study comprehensively evaluated the function of MaCHS genes and laid the foundation for future research on MaCHS in mulberry.
[本文引用: 2]
[本文引用: 2]
DOI:10.1007/s00299-014-1702-8URLPMID:25404490 [本文引用: 3]

KEY MESSAGE: A chalcone synthase gene ( PaCHS ) was isolated and functionally characterized from liverwort. The ectopic expression of PaCHS in Marchantia paleacea callus raised the flavonoids content. Chalcone synthase (CHS; EC 2.3.1.74) is pivotal for the biosynthesis of flavonoid and anthocyanin pigments in plants. It produces naringenin chalcone by condensing one p-coumaroyl- and three malonyl-coenzyme A thioesters through a polyketide intermediate that is cyclized by intramolecular Claisen condensation. Although CHSs of higher plants have been extensively studied, enzyme properties of the CHSs in liverworts have been scarcely characterized. In this study, we report the cloning and characterization of CHS (designated as PaCHS) from the liverwort Plagiochasma appendiculatum. The gene product was 60-70 % identical with chalcone synthases from other species, and contained the characteristic conserved Cys-His-Asn catalytic triad. The recombinant PaCHS was able to catalyze p-coumaroyl-CoA and malonyl-CoA to generate naringenin in vitro. Heterologously expressed PaCHS protein showed similar kinetic properties to those of higher plant CHS. The ectopic expression of PaCHS in Marchantia paleacea callus raised the content of the total flavonoids. These results suggested that PaCHS played a key role in the flavonoids biosynthesis in liverworts. Furthermore, when the thallus of P. appendiculatum was treated with abiotic stress inducers methyl jasmonate, salicylic acid and abscisic acid, PaCHS expression was enhanced. This is the first time that a CHS in liverworts has been functionally characterized.
DOI:10.1371/journal.pone.0119054URLPMID:25742495 [本文引用: 6]

Chalcone synthase (CHS) catalyzes the first committed step in the flavonoid biosynthetic pathway. In this study, the cDNA (FhCHS1) encoding CHS from Freesia hybrida was successfully isolated and analyzed. Multiple sequence alignments showed that both the conserved CHS active site residues and CHS signature sequence were found in the deduced amino acid sequence of FhCHS1. Meanwhile, crystallographic analysis revealed that protein structure of FhCHS1 is highly similar to that of alfalfa CHS2, and the biochemical analysis results indicated that it has an enzymatic role in naringenin biosynthesis. Moreover, quantitative real-time PCR was performed to detect the transcript levels of FhCHS1 in flowers and different tissues, and patterns of FhCHS1 expression in flowers showed significant correlation to the accumulation patterns of anthocyanin during flower development. To further characterize the functionality of FhCHS1, its ectopic expression in Arabidopsis thaliana tt4 mutants and Petunia hybrida was performed. The results showed that overexpression of FhCHS1 in tt4 mutants fully restored the pigmentation phenotype of the seed coats, cotyledons and hypocotyls, while transgenic petunia expressing FhCHS1 showed flower color alteration from white to pink. In summary, these results suggest that FhCHS1 plays an essential role in the biosynthesis of flavonoid in Freesia hybrida and may be used to modify the components of flavonoids in other plants.
[本文引用: 2]
[本文引用: 2]
DOI:10.1007/s10265-016-0871-7URLPMID:27878652 [本文引用: 1]

The enzymes of the chalcone synthase family are also known as type III polyketide synthases (PKS), and produce a series of secondary metabolites in bacteria, fungi and plants. In a number of plants, genes encoding PKS comprise a large multigene family. Currently, detailed reports on rice (Oryza sativa) PKS (OsPKS) family genes and tissue expression profiling are limited. Here, 27 candidate OsPKS genes were identified in the rice genome,and 23 gene structures were confirmed by EST and cDNA sequencing; phylogenetic analysis has indicated that these 23 OsPKS members could be clustered into three groups (I-III). Comparative analysis has shown OsPKS08 and OsPKS26 could be classified with the CHS genes of other species. Two members OsPKS10 and OsPKS21 were grouped into anther specific chalcone synthase-like (ASCL) clade. Intron/exon structure analysis revealed that nearly all of the OsPKS members contained one phase-1 intron at a conserved Cys. Analysis of chromosomal localization and genome distribution showed that some of the members were distributed on a chromosome as a cluster. Expression data exhibited widespread distribution of the rice OsPKS gene family within plant tissues, suggesting functional diversification of the OsPKS genes. Our results will contribute to future study of the complexity of the OsPKS gene family in rice.
DOI:10.1007/BF00333273URL [本文引用: 2]
DOI:10.1186/s12870-020-2324-7URLPMID:32228461 [本文引用: 3]

BACKGROUND: Leaves of the medicinal plant Ampelopsis grossedentata, which is commonly known as vine tea, are used widely in the traditional Chinese beverage in southwest China. The leaves contain a large amount of dihydromyricetin, a compound with various biological activities. However, the transcript profiles involved in its biosynthetic pathway in this plant are unknown. RESULTS: We conducted a transcriptome analysis of both young and old leaves of the vine tea plant using Illumina sequencing. Of the transcriptome datasets, a total of 52.47 million and 47.25 million clean reads were obtained from young and old leaves, respectively. Among 471,658 transcripts and 177,422 genes generated, 7768 differentially expressed genes were identified in leaves at these two stages of development. The phenylpropanoid biosynthetic pathway of vine tea was investigated according to the transcriptome profiling analysis. Most of the genes encoding phenylpropanoid biosynthesis enzymes were identified and found to be differentially expressed in different tissues and leaf stages of vine tea and also greatly contributed to the biosynthesis of dihydromyricetin in vine tea. CONCLUSIONS: To the best of our knowledge, this is the first formal study to explore the transcriptome of A. grossedentata. The study provides an insight into the expression patterns and differential distribution of genes related to dihydromyricetin biosynthesis in vine tea. The information may pave the way to metabolically engineering plants with higher flavonoid content.
[本文引用: 2]
DOI:10.1007/s00299-015-1751-7URLPMID:25632925 [本文引用: 1]

KEY MESSAGE: EaCHS1 functions in the tolerance of plantlets to salinity stress by maintaining ROS homeostasis. Chalcone synthase (CHS) is an essential enzyme in the biosynthesis of flavonoids. Expression of CHS is governed by a wide range of environmental stimuli, including UV light, pathogen attack, and circadian clocks. However, little research exists on the relationship between CHS and salinity stress. In this work, we constructed separate overexpression and RNA interference vectors of EaCHS1, and transferred them into tobacco. Overexpression of EaCHS1 increased the production of downstream flavonoids and the expressions of related genes in the phenylpropanoid pathway. It also improved resistance to salinity stress during seed germination and root development. In contrast, heterologous silencing of endogenous CHS in tobacco by a conserved EaCHS1 fragment had opposite effect. Together, our results indicated that changing the expression level of EaCHS1 in plants alters the accumulation of flavonoids and regulates plantlet tolerance to salinity stress by maintaining ROS homeostasis.
DOI:10.1021/acs.jafc.7b05617URLPMID:29494770 [本文引用: 1]

Eggplant ( Solanum melongena L.) fruits accumulate flavonoids in their cuticle and epidermal cells during ripening. Although many mutants available in model plant species, such as Arabidopsis thaliana and Medicago truncatula, are enabling the intricacies of flavonoid-related physiology to be deduced, the mechanisms whereby flavonoids influence eggplant fruit physiology are unknown. Virus-induced gene silencing (VIGS) is a reliable tool for the study of flavonoid function in fruit, and in this study, we successfully applied this technique to downregulate S. melongena chalcone synthase gene ( SmCHS) expression during eggplant fruit ripening. In addition to the expected change in fruit color attributable to a lack of anthocyanins, several other modifications, including differences in epidermal cell size and shape, were observed in the different sectors. We also found that silencing of CHS gene expression was associated with a negative gravitropic response in eggplant fruits. These observations indicate that epidermal cell expansion during ripening is dependent upon CHS expression and that there may be a relationship between CHS expression and gravitropism during eggplant fruit ripening.
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
[本文引用: 3]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s13238-011-1032-3URL [本文引用: 1]

Genome duplication in E. coli is carried out by DNA polymerase III, an enzyme complex consisting of ten subunits. Investigations of the biochemical and structural properties of DNA polymerase III require the expression and purification of subunits including alpha, epsilon, theta, gamma, delta', delta, and beta separately followed by in vitro reconstitution of the pol III core and clamp loader. Here we propose a new method for expressing and purifying DNA polymerase III components by utilizing a protein co-expression strategy. Our results show that the subunits of the pol III core and those of the clamp loader can be co-expressed and purified based on inherent interactions between the subunits. The resulting pol III core, clamp loader and sliding clamp can be reconstituted effectively to perform DNA polymerization. Our strategy considerably simplifies the expression and purification of DNA polymerase III and provides a feasible and convenient method for exploring other multi-subunit systems.
[本文引用: 1]
[本文引用: 1]
DOI:10.1111/j.1438-8677.2012.00571.xURL [本文引用: 1]

Definitive allocation of function requires the introduction of genetic mutations and analysis of their phenotypic consequences. Novel, rapid and convenient techniques or materials are very important and useful to accelerate gene identification in functional genomics research. Here, over-expression of PmFT (Prunus mume), a novel FT orthologue, and PtFT (Populus tremula) lead to shortening of the tobacco life cycle. A series of novel short life cycle stable tobacco lines (3050 days) were developed through repeated self-crossing selection breeding. Based on the second transformation via a gusA reporter gene, the promoter from BpFULL1 in silver birch (Betula pendula) and the gene (CPC) from Arabidopsis thaliana were effectively tested using short life cycle tobacco lines. Comparative analysis among wild type, short life cycle tobacco and Arabidopsis transformation system verified that it is optional to accelerate functional gene studies by shortening host plant material life cycle, at least in these short life cycle tobacco lines. The results verified that the novel short life cycle transgenic tobacco lines not only combine the advantages of economic nursery requirements and a simple transformation system, but also provide a robust, effective and stable host system to accelerate gene analysis. Thus, shortening tobacco life cycle strategy is feasible to accelerate heterologous or homologous functional gene identification in genomic research.
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.gene.2017.09.002URLPMID:28890377 [本文引用: 4]

The flower color of Syringa oblata Lindl., which is often modulated by the flavonoid content, varies and is an important ornamental feature. Chalcone synthase (CHS) catalyzes the first key step in the flavonoid biosynthetic pathway. However, little is known about the role of S. oblata CHS (SoCHS) in flavonoid biosynthesis in this species. Here, we isolate and analyze the cDNA (SoCHS1) that encodes CHS in S. oblata. We also sought to analyzed the molecular characteristics and function of flavonoid metabolism by SoCHS1. We successfully isolated the CHS-encoding genomic DNA (gDNA) in S. oblata (SoCHS1), and the gene structural analysis indicated it had no intron. The opening reading frame (ORF) sequence of SoCHS1 was 1170bp long and encoded a 389-amino acid polypeptide. Multiple sequence alignment revealed that both the conserved CHS active site residues and CHS signature sequence were in the deduced amino acid sequence of SoCHS1. Crystallographic analysis revealed that the protein structure of SoCHS1 is highly similar to that of FnCHS1 in Freesia hybrida. The quantitative real-time polymerase chain reaction (PCR) performed to detect the SoCHS1 transcript expression levels in flowers, and other tissues revealed the expression was significantly correlated with anthocyanin accumulation during flower development. The ectopic expression results of Nicotiana tabacum showed that SoCHS1 overexpression in transgenic tobacco changed the flower color from pale pink to pink. In conclusion, these results suggest that SoCHS1 plays an essential role in flavonoid biosynthesis in S. oblata, and could be used to modify flavonoid components in other plant species.
DOI:10.1093/jxb/erz264URLPMID:31145783 [本文引用: 1]

MicroRNAs are a class of non-coding small RNAs involved in the negative regulation of gene expression, which play critical roles in developmental and metabolic pathways. Studies in several plants have identified a few microRNAs and other small RNAs that target regulators of the phenylpropanoid metabolic pathway called the MYB transcription factors. However, it is not well understood how sRNA-mediated regulation of MYBs influences the accumulation of specific secondary metabolites. Using sRNA sequencing, degradome analysis, mRNA sequencing, and proteomic analysis, we establish that grape lines with high anthocyanin content express two MYB-targeting microRNAs abundantly, resulting in the differential expression of specific MYB proteins. miR828 and miR858 target coding sequences of specific helix motifs in the mRNA sequences of MYB proteins. Targeting by miR828 caused MYB RNA decay and the production of a cascade of secondary siRNAs that depend on RNA-dependent RNA polymerase 6. MYB suppression and cascade silencing was more robust in grape lines with high anthocyanin content than in a flavonol-rich grape line. We establish that microRNA-mediated silencing targeted the repressor class of MYBs to promote anthocyanin biosynthesis in grape lines with high anthocyanins. We propose that this process regulates the expression of appropriate MYBs in grape lines to produce specific secondary metabolites.
URL [本文引用: 1]

Plant-specific type III polyketide synthase (PKS) produces a variety of plant secondary metabolites with notable structural diversity and biological activity. So far 14 plant-specific type III PKS have been identified according to their enzymatic products, and the corresponding genes have been cloned and characterized. The differences among the various PKS are mainly in their substrate specificities, the number of their condensation reactions, and the type of ring closure of their products. However, numerous studies have revealed the common features among the plant-specific type III PKS, which include sequence homology, similar gene structure, conserved amino acid residues in the reaction center, enzymatic characteristics and reaction mechanism. We briefly reviewed 14 plant-specific type III PKS to better understand genetic and metabolic engineering of plant-specific type III PKS.
URL [本文引用: 1]

Plant-specific type III polyketide synthase (PKS) produces a variety of plant secondary metabolites with notable structural diversity and biological activity. So far 14 plant-specific type III PKS have been identified according to their enzymatic products, and the corresponding genes have been cloned and characterized. The differences among the various PKS are mainly in their substrate specificities, the number of their condensation reactions, and the type of ring closure of their products. However, numerous studies have revealed the common features among the plant-specific type III PKS, which include sequence homology, similar gene structure, conserved amino acid residues in the reaction center, enzymatic characteristics and reaction mechanism. We briefly reviewed 14 plant-specific type III PKS to better understand genetic and metabolic engineering of plant-specific type III PKS.
DOI:10.3864/j.issn.0578-1752.2014.01.014URL [本文引用: 2]

【Objective】 The objectives of this study were to determine the structures of CHS genes in Camellia sinensis (CsCHS) by obtaining gDNA sequences of these genes, and to analyze single nucleotide polymorphism. Besides, association analysis was also carried out in order to find potential SNP sites in CsCHS which would influence the polyphenol content in tea plant.【Method】 Based on CsCHS sequences uploaded to NCBI, specific primers were designed using primer 3.0 software. Genomic DNA and cDNA of tea leaves were used as templates in polymerase chain reaction (PCR) to obtain gDNA and cDNA sequences of CsCHS1, CsCHS2 and CsCHS3, respectively. Gene structures were determined through blasting gDNA and cDNA sequences. The putative amino acid sequences were analyzed by bioinformatics softwares, such as Compute pI/Mw, SOPMA, and so on. Single nucleotide polymorphism of CsCHS were analyzed in 57 cultivars with great variation polyphenol contents. In order to obtain the coding region of CsCHS genes, PCR reactions with specific primers were carried out by using cDNA of individual tea cultivars as template. TASSEL software was introduced in association analysis.【Result】 cDNA sequences of CsCHS1, CsCHS2 and CsCHS3 were 1 277 bp, 1 320 bp and 1 242 bp, respectively. And an open reading frame (ORF) of 1 170 bp was found in each CsCHS gene. gDNA sequences of CsCHS1, CsCHS2, and CsCHS3 were 1 600 bp, 1 330 bp and 1 607 bp, respectively. By comparing gDNA and cDNA sequences of each CsCHS gene, combined with GT-AG rule, it was determined that CsCHS1 and CsCHS2 have two exons and one intron, respectively. And the intron in CsCHS1 is 323 bp, compared with 356 bp in CsCHS3. While no interruption region was found in CsCHS2, this might prove there is no intron in CsCHS2. Deduced amino acid sequences analysis suggests that identity of amino acid sequences are 92.6%-95.4%. All of the conserved amino acids found in CHS protein subfamily were also found in these deduced sequences. Furthermore, bioinformatics analysis showed highly similarities among CsCHS1, CsCHS2 and CsCHS3 protein structures. There are 71 SNP sites in CsCHS1’s coding region, SNP frequency was 1SNP/16.48 bp, and no Indel was found. A total of 55 SNP sites were found in CsCHS2’s coding region, suggesting one SNP in every 21.27 bp. And the nucleotide diversity (π) in CsCHS1 (0.01088) was significantly higher than that of CsCHS2 (0.00530). By correlation analysis, two SNP sites were positioned thought to be related to polyphenol content in tea plant in CsCHS1 and 4 in CsCHS2. No further analysis referring to CsCHS3 was carried out due to low success rate of PCR reaction.【Conclusion】 Both CsCHS1 and CsCHS3 were determined to be conserved CHS genes, and bioinformatics analysis of protein structures results show that CsCHS1, CsCHS2 and CsCHS3 are similar. CsCHS1 and CsCHS3 are active, there should be hot spots of mutation in their coding regions.
DOI:10.3864/j.issn.0578-1752.2014.01.014URL [本文引用: 2]

【Objective】 The objectives of this study were to determine the structures of CHS genes in Camellia sinensis (CsCHS) by obtaining gDNA sequences of these genes, and to analyze single nucleotide polymorphism. Besides, association analysis was also carried out in order to find potential SNP sites in CsCHS which would influence the polyphenol content in tea plant.【Method】 Based on CsCHS sequences uploaded to NCBI, specific primers were designed using primer 3.0 software. Genomic DNA and cDNA of tea leaves were used as templates in polymerase chain reaction (PCR) to obtain gDNA and cDNA sequences of CsCHS1, CsCHS2 and CsCHS3, respectively. Gene structures were determined through blasting gDNA and cDNA sequences. The putative amino acid sequences were analyzed by bioinformatics softwares, such as Compute pI/Mw, SOPMA, and so on. Single nucleotide polymorphism of CsCHS were analyzed in 57 cultivars with great variation polyphenol contents. In order to obtain the coding region of CsCHS genes, PCR reactions with specific primers were carried out by using cDNA of individual tea cultivars as template. TASSEL software was introduced in association analysis.【Result】 cDNA sequences of CsCHS1, CsCHS2 and CsCHS3 were 1 277 bp, 1 320 bp and 1 242 bp, respectively. And an open reading frame (ORF) of 1 170 bp was found in each CsCHS gene. gDNA sequences of CsCHS1, CsCHS2, and CsCHS3 were 1 600 bp, 1 330 bp and 1 607 bp, respectively. By comparing gDNA and cDNA sequences of each CsCHS gene, combined with GT-AG rule, it was determined that CsCHS1 and CsCHS2 have two exons and one intron, respectively. And the intron in CsCHS1 is 323 bp, compared with 356 bp in CsCHS3. While no interruption region was found in CsCHS2, this might prove there is no intron in CsCHS2. Deduced amino acid sequences analysis suggests that identity of amino acid sequences are 92.6%-95.4%. All of the conserved amino acids found in CHS protein subfamily were also found in these deduced sequences. Furthermore, bioinformatics analysis showed highly similarities among CsCHS1, CsCHS2 and CsCHS3 protein structures. There are 71 SNP sites in CsCHS1’s coding region, SNP frequency was 1SNP/16.48 bp, and no Indel was found. A total of 55 SNP sites were found in CsCHS2’s coding region, suggesting one SNP in every 21.27 bp. And the nucleotide diversity (π) in CsCHS1 (0.01088) was significantly higher than that of CsCHS2 (0.00530). By correlation analysis, two SNP sites were positioned thought to be related to polyphenol content in tea plant in CsCHS1 and 4 in CsCHS2. No further analysis referring to CsCHS3 was carried out due to low success rate of PCR reaction.【Conclusion】 Both CsCHS1 and CsCHS3 were determined to be conserved CHS genes, and bioinformatics analysis of protein structures results show that CsCHS1, CsCHS2 and CsCHS3 are similar. CsCHS1 and CsCHS3 are active, there should be hot spots of mutation in their coding regions.
DOI:10.1007/s11103-009-9565-zURL [本文引用: 1]

Enzymes of the chalcone synthase (CHS) superfamily catalyze the production of a variety of secondary metabolites in bacteria, fungi and plants. Some of these metabolites have played important roles during the early evolution of land plants by providing protection from various environmental assaults including UV irradiation. The genome of the moss, Physcomitrella patens, contains at least 17 putative CHS superfamily genes. Three of these genes (PpCHS2b, PpCHS3 and PpCHS5) exist in multiple copies and all have corresponding ESTs. PpCHS11 and probably also PpCHS9 encode non-CHS enzymes, while PpCHS10 appears to be an ortholog of plant genes encoding anther-specific CHS-like enzymes. It was inferred from the genomic locations of genes comprising it that the moss CHS superfamily expanded through tandem and segmental duplication events. Inferred exon–intron architectures and results from phylogenetic analysis of representative CHS superfamily genes of P. patens and other plants showed that intron gain and loss occurred several times during evolution of this gene superfamily. A high proportion of P. patens CHS genes (7 of 14 genes for which the full sequence is known and probably 3 additional genes) are intronless, prompting speculation that CHS gene duplication via retrotransposition has occurred at least twice in the moss lineage. Analyses of sequence similarities, catalytic motifs and EST data indicated that a surprisingly large number (as many as 13) of the moss CHS superfamily genes probably encode active CHS. EST distribution data and different light responsiveness observed with selected genes provide evidence for their differential regulation. Observed diversity within the moss CHS superfamily and amenability to gene manipulation make Physcomitrella a highly suitable model system for studying expansion and functional diversification of the plant CHS superfamily of genes.
DOI:10.1016/0014-5793(72)80679-3URLPMID:4646877 [本文引用: 1]
DOI:10.1016/j.phytochem.2006.09.030URLPMID:17083952 [本文引用: 2]

Since the early evolution of land plants from primitive green algae, flavonoids have played an important role as UV protective pigments in plants. Flavonoids occur in liverworts and mosses, and the first committed step in the flavonoid biosynthesis is catalyzed by chalcone synthase (CHS). Although higher plant CHSs have been extensively studied, little information is available on the enzymes from bryophytes. Here we report the cloning and characterization of CHS from the moss, Physcomitrella patens. Taking advantage of the available P. patens EST sequences, a CHS (PpCHS) was cloned from the gametophores of P. patens, and heterologously expressed in Escherichia coli. PpCHS exhibited similar kinetic properties and substrate preference profile to those of higher plant CHS. p-Coumaroyl-CoA was the most preferred substrate, suggesting that PpCHS is a naringenin chalcone producing CHS. Consistent with the evolutionary position of the moss, phylogenetic analysis placed PpCHS at the base of the plant CHS clade, next to the microorganism CHS-like gene products. Therefore, PpCHS likely represents a modern day version of one of the oldest CHSs that appeared on earth. Further, sequence analysis of the P. patens EST and genome databases revealed the presence of a CHS multigene family in the moss as well as the 3'-end heterogeneity of a CHS gene. Of the 19 putative CHS genes, 10 genes are expressed and have corresponding ESTs in the databases. A possibility of the functional divergence of the multiple CHS genes in the moss is discussed.
DOI:10.1016/j.plaphy.2018.01.030URLPMID:29428820 [本文引用: 1]

Chalcone synthases (CHSs) of the type III polyketide synthases (PKSs), catalyze the formation of a tetraketide intermediate from a CoA-tethered starter and malonyl-CoA but use different cyclization mechanisms to produce distinct chemical scaffolds. Herein, we characterized CHS and CHS-like enzymes (designated MpCHS and MpCHSL1, 2 and 3) from Marchantia paleacea and determined the crystal structure of MpCHSL1. MpCHS catalyzed a Claisen condensation to form chalcone, while MpCHSLs catalyzed the formation of lactonized alpha-pyrones in vitro. Based on the structural, mutational and in vitro biochemical analyses, we established that MpCHSL1 is structurally and functionally closer to prototype CHS than stilbene synthase, and characterized the structural basis for the functional diversity of the type III PKSs. A chalcone-forming mutant of MpCHSL1 was build directed by the structural information. These findings pave the way for future studies to elucidate the functional diversity of type III PKSs in liverwort.
DOI:10.1038/s41467-020-14558-9URLPMID:32054839 [本文引用: 1]

Land plants produce diverse flavonoids for growth, survival, and reproduction. Chalcone synthase is the first committed enzyme of the flavonoid biosynthetic pathway and catalyzes the production of 2',4,4',6'-tetrahydroxychalcone (THC). However, it also produces other polyketides, including p-coumaroyltriacetic acid lactone (CTAL), because of the derailment of the chalcone-producing pathway. This promiscuity of CHS catalysis adversely affects the efficiency of flavonoid biosynthesis, although it is also believed to have led to the evolution of stilbene synthase and p-coumaroyltriacetic acid synthase. In this study, we establish that chalcone isomerase-like proteins (CHILs), which are encoded by genes that are ubiquitous in land plant genomes, bind to CHS to enhance THC production and decrease CTAL formation, thereby rectifying the promiscuous CHS catalysis. This CHIL function has been confirmed in diverse land plant species, and represents a conserved strategy facilitating the efficient influx of substrates from the phenylpropanoid pathway to the flavonoid pathway.
DOI:10.1006/bbrc.2000.3920URLPMID:11112437 [本文引用: 2]

Substrate specificity of recombinant chalcone synthase (CHS) from Scutellaria baicalensis (Labiatae) was investigated using chemically synthesized aromatic and aliphatic CoA esters. It was demonstrated for the first time that CHS converted benzoyl-CoA to phlorobenzophenone (2,4,6-trihydroxybenzophenone) along with pyrone by-products. On the other hand, phenylacetyl-CoA was enzymatically converted to an unnatural aromatic polyketide, phlorobenzylketone (2, 4,6-trihydroxyphenylbenzylketone), whose structure was finally confirmed by chemical synthesis. Furthermore, in agreement with earlier reports, S. baicalensis CHS also accepted aliphatic CoA esters, isovaleryl-CoA and isobutyryl-CoA, to produce phloroacylphenones. In contrast, hexanoyl-CoA only afforded pyrone derivatives without formation of a new aromatic ring. It was noteworthy that both aromatic and aliphatic CoA esters were accepted in the active site of the enzyme as a starter substrate for the complex condensation reaction. The low substrate specificity of CHS thus provided further insight into the structure and function of the enzyme.
DOI:10.3864/j.issn.0578-1752.2019.11.010URL [本文引用: 1]

【Objective】 The object of this study was to determine the relationship between anthocyanins and flower colors of bud mutation in Camellia japonica, so as to provide the scientific basis for the bud mutation breeding of flower colors in C. japonica. 【Method】 Flower colors in C. japonica cultivars and their bud mutation cultivars were measured by CIE L*a*b* scale, and anthocyanin components and contents were measured by high-performance liquid chromatography coupled with diode array detection (HPLC-DAD) and ultra-performance liquid chromatography quadrupole-time-of-flight mass spectrometry (UPLC-Q-T OF-MS). The relationship between flower colors and anthocyanins was explored by multiple liner regression analyses. 【Result】 Seven anthocyanins were detected in C. japonica cultivars and their bud mutation cultivars, which were cyanidin-3-O-β-galactoside (Cy3Ga),cyanidin-3-O-β-glucoside (Cy3G),cyanidin-3-O-(6-O-(E)-caffeoyl)-β-galactoside (Cy3GaECaf), cyanidin-3-O-(6-O-(E)- caffeoyl)-β-glucoside (Cy3GECaf), cyanidin-3-O-(6-O-(Z)-p-coumaroyl)-β-glucoside (Cy3GZpC), cyanidin-3-O-(6-O-(E)-p- coumaroyl)-β-galactoside (Cy3GaEpC) and cyanidin-3-O-(6-O-(E)-p-coumaroyl)-β-glucoside (Cy3GEpC). Among the bud mutation cultivars of C. japonica, anthocyanins were not detected in white petals, and anthocyanins in red petals were identical with that in pink petals, but the contents of anthocyanin components and total anthocyanin in red petals were far higher than that in pink petals. The main anthocyanin components were Cy3G and Cy3GEpC in red and pink petals. The proportion of Cy3G and Cy3Ga in red petals were larger than that in pink petals, while the proportion of else anthocyanins, such as Cy3GEpC, were smaller than that in pink petals.【Conclusion】 Among the bud mutation cultivars of C. japonica, the greater the contents of various anthocyanin components and total anthocyanin were, the deeper the red of petal were. Cy3G, Cy3Ga and Cy3GEpC were the main anthocyanins which determined the flower colors of bud mutation in C. japonica, and the accumulation of their contents enhanced the red color of petals.
DOI:10.3864/j.issn.0578-1752.2019.11.010URL [本文引用: 1]

【Objective】 The object of this study was to determine the relationship between anthocyanins and flower colors of bud mutation in Camellia japonica, so as to provide the scientific basis for the bud mutation breeding of flower colors in C. japonica. 【Method】 Flower colors in C. japonica cultivars and their bud mutation cultivars were measured by CIE L*a*b* scale, and anthocyanin components and contents were measured by high-performance liquid chromatography coupled with diode array detection (HPLC-DAD) and ultra-performance liquid chromatography quadrupole-time-of-flight mass spectrometry (UPLC-Q-T OF-MS). The relationship between flower colors and anthocyanins was explored by multiple liner regression analyses. 【Result】 Seven anthocyanins were detected in C. japonica cultivars and their bud mutation cultivars, which were cyanidin-3-O-β-galactoside (Cy3Ga),cyanidin-3-O-β-glucoside (Cy3G),cyanidin-3-O-(6-O-(E)-caffeoyl)-β-galactoside (Cy3GaECaf), cyanidin-3-O-(6-O-(E)- caffeoyl)-β-glucoside (Cy3GECaf), cyanidin-3-O-(6-O-(Z)-p-coumaroyl)-β-glucoside (Cy3GZpC), cyanidin-3-O-(6-O-(E)-p- coumaroyl)-β-galactoside (Cy3GaEpC) and cyanidin-3-O-(6-O-(E)-p-coumaroyl)-β-glucoside (Cy3GEpC). Among the bud mutation cultivars of C. japonica, anthocyanins were not detected in white petals, and anthocyanins in red petals were identical with that in pink petals, but the contents of anthocyanin components and total anthocyanin in red petals were far higher than that in pink petals. The main anthocyanin components were Cy3G and Cy3GEpC in red and pink petals. The proportion of Cy3G and Cy3Ga in red petals were larger than that in pink petals, while the proportion of else anthocyanins, such as Cy3GEpC, were smaller than that in pink petals.【Conclusion】 Among the bud mutation cultivars of C. japonica, the greater the contents of various anthocyanin components and total anthocyanin were, the deeper the red of petal were. Cy3G, Cy3Ga and Cy3GEpC were the main anthocyanins which determined the flower colors of bud mutation in C. japonica, and the accumulation of their contents enhanced the red color of petals.
DOI:10.1007/s00425-011-1473-1URLPMID:21735195 [本文引用: 1]

Proteomics facilitates our understanding of cellular processes and network functions in the plant defense response during abiotic and biotic stresses. Here, we demonstrate that the ectopic expression of the Capsicum annuum antimicrobial protein CaAMP1 gene in Arabidopsis thaliana confers enhanced tolerance to methyl viologen (MV)-induced oxidative stress, which is accompanied by lower levels of lipid peroxidation. Quantitative comparative proteome analyses using two-dimensional gel electrophoresis coupled with mass spectrometry identified some of the oxidative stress- and disease-related proteins that are differentially regulated by CaAMP1 overexpression in Arabidopsis leaves. Antioxidant- and defense-related proteins, such as 2-cys peroxiredoxin, L-ascorbate peroxidase, peroxiredoxin, glutathione S-transferase and copper homeostasis factor, were up-regulated in the CaAMP1 transgenic leaf tissues. In contrast, GSH-dependent dehydroascorbate reductase and WD-40 repeat family protein were down-regulated by CaAMP1 overexpression. In addition, CaAMP1 overexpression enhanced resistance to Pseudomonas syringae pv. tomato (Pst) DC3000 infection and also H(2)O(2) accumulation in Arabidopsis. The identified antioxidant- and defense-related genes were differentially expressed during MV-induced oxidative stress and Pst DC3000 infection. Taken together, we conclude that CaAMP1 overexpression can regulate the differential expression of defense-related proteins in response to environmental stresses to maintain reactive oxygen species (ROS) homeostasis.
DOI:10.1007/s00425-018-2862-5URLPMID:29455261 [本文引用: 1]

MAIN CONCLUSION: During maize somatic embryogenesis, suppression of phytoglobins (Pgbs) reduced ABA levels leading to ethylene-induced programmed cell death in the developing embryos. These effects modulate embryonic yield depending on the cellular localization of specific phytoglobin gene expression. Suppression of Zea mays phytoglobins (ZmPgb1.1 or ZmPgb1.2) during somatic embryogenesis induces programmed cell death (PCD) by elevating nitric oxide (NO). While ZmPgb1.1 is expressed in many embryonic domains and its suppression results in embryo abortion, ZmPgb1.2 is expressed in the basal cells anchoring the embryos to the embryogenic tissue. Down-regulation of ZmPgb1.2 is required to induce PCD in these anchor cells allowing the embryos to develop further. Exogenous applications of ABA could reverse the effects caused by the suppression of either of the two ZmPgbs. A depletion of ABA, ascribed to a down-regulation of biosynthetic genes, was observed in those embryonic domains where the respective ZmPgbs were repressed. These effects were mediated by NO. Depletion in ABA content increased the transcription of genes participating in the synthesis and response of ethylene, as well as the accumulation of ethylene, which influenced embryogenesis. Somatic embryo number was reduced by high ethylene levels and increased with pharmacological treatments suppressing ethylene synthesis. The ethylene inhibition of embryogenesis was linked to the production of reactive oxygen species (ROS) and the execution of PCD. Integration of ABA and ethylene in the ZmPgb regulation of embryogenesis is proposed in a model where NO accumulates in ZmPgb-suppressing cells, decreasing the level of ABA. Abscisic acid inhibits ethylene biosynthesis and the NO-mediated depletion of ABA relieves this inhibition causing ethylene to accumulate. Elevated ethylene levels trigger production of ROS and induce PCD. Ethylene-induced PCD in the ZmPgb1.1-suppressing line [ZmPgb1.1 (A)] leads to embryo abortion, while PCD in the ZmPgb1.2-suppressing line [ZmPgb1.2 (A)] results in the elimination of the anchor cells and the successful development of the embryos.
DOI:10.1007/s11032-011-9653-zURL [本文引用: 1]

Chalcone synthase (CHS) is the key enzyme in an early stage of the flavonoid biosynthetic pathway. In the present study, a full-length cDNA clone for CHS was isolated from flower tepals of the liliaceous ornamental Tricyrtis sp., in which tepals have many reddish-purple spots resulting from accumulation of cyanidin derivatives. The deduced amino acid sequence of the isolated cDNA clone, designated TrCHS1 (accession number AB478624 in the GenBank/EMBL/DDBJ databases), shows 79.4-91.4% identity with those of previously reported CHS genes. An RNA interference (RNAi) construct targeting TrCHS1 was introduced by Agrobacterium-mediated transformation in order to alter the flower color of Tricyrtis sp. Seven transgenic plants that produced flowers could be classified into three types according to flower color phenotype: one transgenic plant had tepals with as many reddish-purple spots as non-transgenic plants (Type I); one had tepals with reduced numbers of reddish-purple spots (Type II); and five had completely white tepals without any spots (Type III). High-performance liquid chromatography analysis showed that tepals of Type III transgenic plants did not accumulate detectable amounts of anthocyanidins. In addition, TrCHS1 mRNA levels in tepals of Type II and Type III transgenic plants decreased substantially compared with non-transgenic plants, as determined by quantitative real-time reverse transcription-polymerase chain reaction analysis. Our results indicate the validity of RNAi suppression of the flavonoid biosynthetic pathway genes for flower color alteration in Tricyrtis sp. To the best of our knowledge, this is the first report on flower color alteration by genetic transformation in monocotyledonous ornamentals.
DOI:10.1073/pnas.1515294113URL [本文引用: 1]
