 ,, 莫双榕, 钱莹, 臧栋楠, 杨帆, 蒋红亮, 武媛, 丁海东
,, 莫双榕, 钱莹, 臧栋楠, 杨帆, 蒋红亮, 武媛, 丁海东 ,扬州大学生物科学与技术学院,江苏扬州 225009
,扬州大学生物科学与技术学院,江苏扬州 225009Screening of Interacting Protein of Tomato SIVQ6 by GST Pull-Down
YUAN GuiBo ,, MO ShuangRong, QIAN Ying, ZANG DongNan, YANG Fan, JIANG HongLiang, WU Yuan, DING HaiDong
,, MO ShuangRong, QIAN Ying, ZANG DongNan, YANG Fan, JIANG HongLiang, WU Yuan, DING HaiDong ,College of Bioscience and Biotechnology, Yangzhou University, Yangzhou 225009, Jiangsu
,College of Bioscience and Biotechnology, Yangzhou University, Yangzhou 225009, Jiangsu通讯作者:
责任编辑: 李莉
收稿日期:2019-10-17接受日期:2020-01-29网络出版日期:2020-08-01
| 基金资助: |
Received:2019-10-17Accepted:2020-01-29Online:2020-08-01
作者简介 About authors
原贵波,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (3057KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
原贵波, 莫双榕, 钱莹, 臧栋楠, 杨帆, 蒋红亮, 武媛, 丁海东. 应用GST pull-down技术筛选番茄SIVQ6互作蛋白[J]. 中国农业科学, 2020, 53(15): 3146-3157 doi:10.3864/j.issn.0578-1752.2020.15.014
YUAN GuiBo, MO ShuangRong, QIAN Ying, ZANG DongNan, YANG Fan, JIANG HongLiang, WU Yuan, DING HaiDong.
0 引言
【研究意义】在植物生长过程中,高温是常见的环境胁迫因子,对植物的生长发育、品质及产量有着极其重要的影响。随着全球气候变暖,温室效应加剧,严重影响了农业和农产品的安全。番茄作为世界上最受欢迎和广泛消费的蔬菜之一,限制其生产力的主要因素就是高温,高温影响各种核酸、蛋白质和细胞骨架结构的稳定性,并改变细胞内酶的催化效率,导致代谢失衡[1,2],它对番茄的营养和繁殖阶段产生不利影响,最终降低产量和果实品质[3,4]。根据DING等[5]在番茄中鉴定出26种SlVQ蛋白,发现SlVQ6在番茄不同组织器官中均为高效表达,在拟南芥中显示异位过表达SlVQ6能够降低高温耐受性。因此,SlVQ6可能在番茄生长发育过程中尤其在响应高温信号传导过程中起到重要作用。【前人研究进展】VQ蛋白家族是一类植物特有的蛋白,含有保守VQ基序(FxxxVQxxTG)的植物特异性转录调节子家族被命名为VQ蛋白,这些含有VQ基序的蛋白质,广泛分布于不同物种中[6,7,8]。VQ基序对VQ蛋白的功能或作用至关重要。例如,VQ基序影响蛋白质定位和蛋白质-蛋白质相互作用[9,10,11,12,13]。VQ蛋白调节植物的生长和发育、种子发育,响应不同胁迫和光形态建成[7]。在拟南芥中,HAIKU1(VQ14)调节胚乳生长和种子大小[10]。AtVQ8突变体表现出淡绿色和发育不良,表明AtVQ8调节叶绿体发育或光系统组装[7]。AtVQ18和AtVQ26通过与ABI5转录因子相互作用进而在拟南芥种子萌发和早期幼苗建立的过程中发挥作用[14]。过表达AtVQ29植株的下胚轴长度在远红光或弱光情况下显著长于野生型,而缺失突变株的下胚轴长度显著短于野生型,表明AtVQ29在植物光形态建成中发挥重要作用。VQ29可与光敏色素结合因子(PIF1)互作,进而影响下游基因的转录表达[13]。越来越多的证据表明,VQPs不仅参与植物生长发育过程如种子萌发、胚轴伸长、光形态建成等,还参与植物的环境胁迫应答反应如盐害、干旱、病原菌等。WANG等[15]通过对拟南芥VQ12和VQ29研究,发现过量表达这两种基因的植株对病原菌B. cinerea高度敏感,而突变株抗病性则显著提高。HU等[12]研究发现过表达AtVQ9对NaCl敏感,而其突变体在NaCl处理下表现出较高的萌发率和较好的生长力。目前,关于VQPs的作用机理主要集中在两方面:其一是与WRKY互作;其二是被MAPK磷酸化[16]。SIB1和SIB2可与WRKY33的保守区域WRKY结合,提高后者与W-box的结合能力,正调控植物的抗病能力[11],而AtVQ9与WRKY8互作则负调控植物耐盐能力[12]。LEI等[17]研究表明AtVQ20通过与AtWRKY2/AtWRKY34的相互作用调控花粉发育。AtVQ14同样是与WRKY家族来共同调节种子大小和胚乳生长[10]。AtVQ21/MKS1首先作为AtMPK4的底物被发现,AtVQ21可与AtMPK4和AtWRKY33形成相互作用复合体,当受到生物胁迫后,AtVQ21与AtWRKY33复合体从AtMPK4激酶中释放出来,迅速与抗毒素基因PAD3的启动子结合,调控基因表达,从而调节植物生长和抗病性[18,19]。近年来,越来越多的植物VQ蛋白被鉴定出来,在水稻[20]、拟南芥[7]、玉米[21]、大豆[22]、葡萄[23]、大白菜[24]等中分别鉴定出39、34、61、74、18和57个VQ基因。最近,在苹果中确认了49种MdVQ蛋白[25],且这些MdVQs在非生物胁迫(如干旱、高温等)中发挥重要作用。【本研究切入点】何洁[26]研究表明SlVQ6极有可能参与番茄中的高温胁迫,且通过酵母双杂交验证了SlVQ6能够与SlMPK1相互作用。但对于SlVQ6的分子机理还不清楚。为了进一步研究SlVQ6的生物学功能,构建pGEX-4T-1-PC-SlVQ6载体,并通过大肠杆菌表达纯化,从而获得带有GST标签的融合蛋白GST-SlVQ6,将其作为本研究中的诱饵蛋白,筛选出与之互作的其他蛋白。【拟解决的关键问题】本研究通过体外磷酸化确定SlVQ6是否是SlMPK1的底物,并通过GST pull-down技术从番茄中筛选出与SlVQ6相互作用的蛋白,通过质谱分析鉴定这些蛋白并结合生物信息学预测其功能,为进一步研究SlVQ6介导的高温应答信号通路奠定基础。1 材料与方法
试验于2018年在扬州大学生物科学与技术学院完成。1.1 试验材料
番茄种子由扬州大学生物科学与技术学院509实验室保存;pGEX-4T-1质粒为扬州大学实验室保存;TOP10菌株、DNA聚合酶(货号PC12)、Arctic-Express TM表达菌购自钟鼎生物;DNA胶回收试剂盒、纯化质粒提取试剂盒购自康为世纪生物科技有限公司;限制性内切酶购自TaKaRa公司;0.22 μm无菌滤器和透析袋购自Millipore公司;GST亲和层析胶(购自金斯瑞)。1.2 pGEX-4T-1-PC-SlVQ6载体构建
设计SlVQ6的正、反向引物,提取番茄叶片cDNA,并以其作为模板,扩增SlVQ6,琼脂糖凝胶电泳鉴定后切胶回收,与PMD?19-T Vector连接转化,测序验证后,将获得的目的基因SlVQ6经BamHⅠ和NotⅠ的酶切后,与载体pGEX-4T-1连接,从而获得重组质粒pGEX-4T-1-PC-SlVQ6。1.3 pGEX-4T-1-PC-SlVQ6原核蛋白的表达纯化
1.3.1 转化 将pGEX4T-1-PC-SlVQ6重组质粒转化至大肠杆菌中培养转化,涂板过夜培养,挑取转化平板上的单菌落接种于含50 μg·mL-1 AMP的3 mL LB培养液中,37℃ 220 r/min振摇至菌液浓度OD600为0.6—0.8。1.3.2 IPTG诱导表达 加入IPTG至终浓度为0.5 mmol·L-1,11℃ 220 r/min振摇诱导融合蛋白表达,获得带有GST标签的融合蛋白GST-SlVQ6,经12% SDS-PAGE分析目标蛋白主要存在的位置后,利用GST柱对融合蛋白进行亲和纯化,通过低压层析系统,将溶液上样至GST Binding-Buffer预平衡的GST亲和层析柱,并冲洗,GST Elution-Buffer(20 mmol·L-1 Tris-HCl、50 mmol·L-1 GSH和0.15 mol·L-1 NaCl,pH8.0)洗脱目的蛋白,将收集的流出液加入透析袋中,用20 mmol·L-1 Tris-HCl和0.10 mol·L-1 NaCl(pH 8.0)进行透析,再次通过GST柱亲和纯化目标蛋白GST-SIVQ6,并进行12% SDS-PAGE分析,以确定获得融合蛋白的纯度。
1.4 SlMPK1磷酸化SIVQ6的体外验证
将3 μg His-SlMPK1蛋白与2 μg GST-SlVQ6在激酶反应缓冲液(50 μmol·L-1 ATP、[γ-32P]ATP(每个反应2 μ Ci)、25 mmol·L-1 Tris-HCl pH 7.5、20 mmol·L-1 MgCl2和1 mmol·L-1 DTT)中混合。以GST蛋白作为阴性对照。30℃ 30 min,使用12%(w/v)SDS-PAGE分离蛋白质。电泳完成后切下所需凝胶,为了彻底除去游离的[γ-32P]ATP,配置50 mL 5%三氯乙酸(TCA)和1%焦磷酸钠(NaPPi)混合液,将凝胶转入清洗2 h(期间更换4次溶液),并对凝胶进行干燥处理,最后使用Typhoon9410 Phosphor-Imager(GE Healthcare)对凝胶成像。1.5 筛选与SIVQ6相互作用的蛋白
挑选生长力旺盛、均匀饱满籽粒的野生型M14番茄种子,经75%酒精、15% NaClO和无菌水消毒后,播种在灭菌的1/2MS培养基上生长。待苗长至2—3周龄时,取少许叶片迅速放入液氮中,(加提取缓冲液)放置数小时后,配置蛋白提取液,提取番茄总蛋白。将50 μg GST-SlVQ6(或GST,阴性对照)加入GST sepharose 4B beads中,并用结合缓冲液(50 mmol·L-1磷酸钠和300 mmol·L-1氯化钠)充分洗涤。取10 mg番茄叶粗蛋白提取物,与固定于GST sheperose 4B beads上的GST在4℃共孵育4 h后,离心取上清液,从而除去可能存在的非特异性吸附的影响。将5 mg上清液蛋白质提取物加入GST-SlVQ6包被的beads中,并在4℃孵育过夜。充分洗涤后,加入1×SDS loading buffer煮沸,通过12% SDS-PAGE凝胶电泳分离样品。将凝胶用CBB R-250染色,切下整个泳道,脱色,干燥并用胰蛋白酶消化。质谱分析:在AB SCIEX Triple TOF 5600+LC/ MS/MS上分析胰蛋白酶消化物。通过C18柱(100 μm×3 cm,C18,3 μm,150?)进行肽分离,然后脱盐10 min。70 min的洗脱梯度(5%—35%乙腈)用于分析型ChromXP C18色谱柱(75 μm×15 cm,C18,3 μm,120?)。通过5600系统(AB SCIEX)结合纳米升喷雾Ⅲ离子源(AB SCIEX,USA)分析数据,将原始MS/MS文件数据提交至ProteinPilot TM软件5.0(Applied Biosystems,Sciex)进行分析。使用的搜索参数设置如下:(1)样本类型:识别鉴定;(2)消化:胰蛋白酶;(3)仪器:Triple-TOF 5600;(4)Cys烷基化:碘乙酰胺;(5)数据库:Uniprot。
1.6 生物信息学分析
通过对GO富集与KEGG富集筛选出的与SlVQ6互作的37个蛋白进行分析。GO主要是对基因和蛋白质在分子功能、生物过程和细胞成分3个方面进行描述。KEGG PATHWAY[27]数据库是一个代谢通路的集合,包含多个方面的分子间相互作用和反应网络。STRING[28]是一个搜索已知蛋白与预测蛋白之间相互作用的系统。包括蛋白质之间直接的物理作用与蛋白质之间间接的功能相关性。通过STRING Version: 11.0在线软件对蛋白互作进行验证(2 结果
2.1 构建pGEX-4T-1-PC-SlVQ6载体
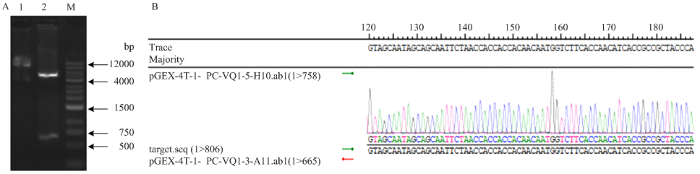
根据SlVQ6的基因序列,设计含有BamHⅠ和NotⅠ酶切位点的正反向引物,扩增SlVQ6的ORF全长编码序列,1%琼脂糖凝胶电泳检测,将测序正确的质粒双酶切,与pGEX-4T-1-PC载体连接。通并进行双酶切鉴定。结果显示,得到载体DNA片段和相应的目的片段2个片段(图1-A),与预知片段大小完全吻合,测序结果显示重组质粒中插入的序列完全正确(图1-B),表明pGEX-4T-1-PC-SlVQ6重组载体构建成功。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1融合表达载体的构建
A:重组质粒pGEX-4T-1-PC-SlVQ6的双酶切鉴定,M:Marker;1:酶切前质粒;2:酶切后质粒。B:pGEX-4T-1-PC-SlVQ6的部分序列比对
Fig. 1Construction of fusion expression vector
A: Double enzyme digestion of recombinant plasmid pGEX-4T-1-PC-SlVQ6, M: Marker; 1: Pre-digested plasmid; 2: Plasmid after digestion. B: The partial sequence alignment of pGEX-4T-1-PC-SlVQ6
2.2 带有GST标签的融合蛋白GST-SIVQ6的获得
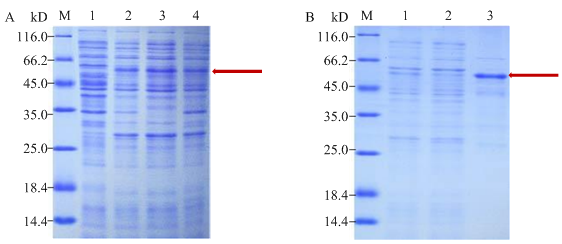
成功获得带有GST标签的融合表达载体pGEX- 4T-1-PC-SlVQ6(图1)。首先富集诱导表达的大肠杆菌,超声后取上清液,并进行重悬,重悬后的上清经过12%的SDS-PAGE检测,考马斯亮蓝显色后发现目标蛋白主要存在于上清液中(图2-A);因此,需将上清液进行亲和层析纯化,从而获取目的融合蛋白。用GST柱进行亲和层析纯化,将最终经过透析收集到的融合蛋白进行聚丙烯酰胺凝胶电泳检测(图2-B),用GST-Sepharose 4B bead亲合层析柱纯化后可得到大量大小为54 kD的融合蛋白GST-SlVQ6。经纯化获得的GST-SIVQ6融合蛋白将被用于体外磷酸化和pull-down试验。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2融合蛋白的表达纯化
A:SDS-PAGE分析Arctic Express中的表达形式,M:Marker;1:未被诱导;2:诱导;3:0.5 mmol·L-1 IPTG诱导上清液;4:0.5 mmol·L-1 IPTG诱导沉淀;箭头:GST-SlVQ6蛋白。B:融合蛋白纯化的SDS-PAGE,M:Marker;1:未被纯化;2:流出液;3:洗脱液;箭头:GST-SlVQ6蛋白
Fig. 2Expression and purification of fusion protein
A: SDS-PAGE of expression form analysis in Arctic Express, M: Marker; 1: Un-induced; 2: Induced; 3: Supernatant of induction with 0.5 mmol·L-1 IPTG; 4: Precipitate of induction with 0.5 mmol·L-1 IPTG; Arrow: GST-SlVQ6 protein. B: SDS-PAGE of fusion protein purification, M: Marker; 1: Un-purified; 2: Flow through; 3: Elution; Arrow: GST-SlVQ6 protein
2.3 体外验证SIMPK1磷酸化下游蛋白SIVQ6
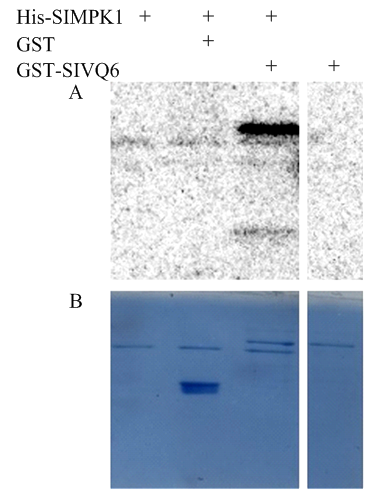
前期研究通过Y2H已证明SlVQ6能够与SlMPK1相互作用,为了进一步探索SlVQ6和SlMPK1之间的关系,将保存的His-SlMPK1和本研究获得的GST-SlVQ6重组蛋白进行体外磷酸化试验(图3),结果表明,His-SlMPK1可使GST-S1VQ6磷酸化,但不能够磷酸化GST蛋白,且GST-SlVQ6不存在自磷酸化的现象。表明SlVQ6是SlMPK1的磷酸化底物。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3SlMPK1磷酸化纯化的SlVQ6
A:His-SlMPK1和GST-SlVQ6用于带有32P标记的磷酸化反应,GST用作阴性对照,单独添加GST-SlVQ6证明了GST-SlVQ6的自磷酸化反应;B:内参参照考马斯亮蓝染色
Fig. 3In-gel analysis of phosphorylation of purified SlVQ6 by SlMPK1
A: His-tagged SlMPK1 and GST-tagged SlVQ6 were used in phosphorylation reactions with 32P labeling, GST was used as a negative control, and the autophosphorylation of GST-SlVQ6 was proved by adding GST-SlVQ6 alone; B: Coomassie blue staining in the below panel
2.4 GST pull-down及质谱法筛选与SIVQ6相互作用的蛋白
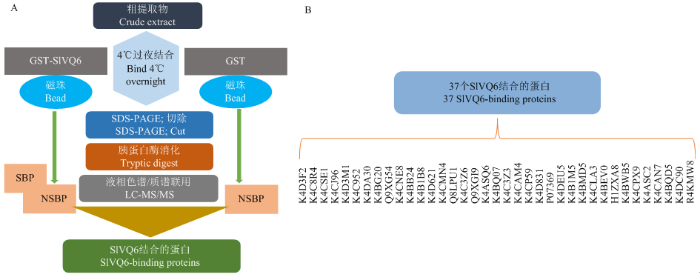
为研究SlVQ6可能的作用机制,根据CHANG等[29]研究方法对番茄细胞裂解液进行pull-down研究。提取番茄叶片总蛋白,并与带有GST标签的纯化过后的融合蛋白GST-SlVQ6孵育,以GST-SlVQ6为诱饵蛋白(空GST为阴性对照),利用GST-SlVQ6 pull-down试验筛选番茄叶片组织蛋白中与SlVQ6结合的蛋白(图4-A),并经SDS-PAGE分离、液相色谱-串联质谱(LC-MS/MS)鉴定以及Mascot与蛋白数据库检索,共鉴定出37个SlVQ6结合的蛋白(图4-B和表1),包括蛋白激酶(receptor for activated C kinase 1B,RACK1B)。37个SlVQ6结合蛋白的具体信息如(表1)。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4GST pull down分析SlVQ6互作蛋白
A:GST pull down分析与SlVQ6相互作用的蛋白质的流程图。将GST-SlVQ6(或GST,阴性对照)添加至GST sheperose 4B beads,将番茄叶片的粗蛋白提取液添加beads,并在4℃孵育过夜。将样品煮沸,用12% SDS-PAGE凝胶分离,回收条带,脱色,干燥,胰蛋白酶消化,并进行LC-MS/MS分析。SBP:特异性结合蛋白;NSBP:非特异性结合蛋白。B:鉴定出37个SlVQ6特异性结合蛋白
Fig. 4GST pull down analysis of SlVQ6-interacting proteins
A: The flow chart of GST pull down analysis of SlVQ6-interacting proteins. GST-SlVQ6 (or GST, a negative control) was added to GST sheperose 4B beads and crude protein extract of tomato leaves was added to the beads and incubated for overnight at 4℃. The samples were boiled and then separated by 12% SDS-PAGE gels. The bands were cut out, destained, dried, digested using trypsin, and the LC-MS/MS analysis was performed. SBP: Specific binding proteins; NSBP: Non-specific binding proteins. B: Thirty-seven of SlVQ6-specific binding proteins were identified
Table 1
表1
表1通过GST pull-down鉴定的37个SlVQ6特异性结合蛋白
Table 1
| 蛋白质ID UniProt ID | 拟南芥ID At ID | 基因IDa Gene ID | 描述b Description |
|---|---|---|---|
| R4KMW8 | ATCG00490 | 二磷酸核酮糖羧化酶大链Ribulose bisphosphate carboxylase large chain | |
| K4DC90 | AT2G24200 | Solyc12g010020 | 胞质氨肽酶Cytosol aminopeptidase |
| K4BQD5 | AT5G58420 | Solyc04g016350 | 40S核糖体蛋白S4 (RPS4D) 40S ribosomal protein S4 (RPS4D) |
| K4CAN7 | AT2G20450 | Solyc06g083820 | 60S核糖体蛋白l14(RPL14A) 60S ribosomal protein L14 (RPL14A) |
| K4ASC2 | AT1G65930 | Solyc01g005560 | 异柠檬酸脱氢酶/ NADP+异柠檬酸脱氢酶,推定 Isocitrate dehydrogenase / NADP+ isocitrate dehydrogenase, putative |
| K4CPX9 | AT1G32990 | Solyc08g083350 | PRPL11;核糖体的结构组成 PRPL11; Structural constituent of ribosome |
| K4BWB5 | AT5G14320 | Solyc05g005880 | 核糖体蛋白S13,叶绿体(CS13) 30S ribosomal protein S13, chloroplast (CS13) |
| H1ZXA8 | AT3G12580 | 热休克蛋白70亚型2 Heat shock protein 70 isoform 2 | |
| K4BEV0 | AT3G04770 | Solyc03g019780 | RPSAb;核糖体的结构组成 RPSAb; Structural constituent of ribosome |
| K4CLA3 | AT4G33010 | Solyc08g065220 | AtGLDP1(拟南芥黄曲霉毒素P-蛋白1)AtGLDP1 (Arabidopsis thaliana glycine decarboxylase P-protein 1) |
| K4BMD5 | AT1G74070 | Solyc03g119860 | 肽基脯氨酸顺反异构酶亲环素型家族蛋白 Eptidyl-prolyl cis-trans isomerase cyclophilin-type family protein |
| K4B1M5 | AT5G49840 | Solyc01g102990 | ATP结合/ATP酶/核苷三磷酸酶/核苷酸结合 ATP binding/ATPase/nucleoside-triphosphatase/ nucleotide binding |
| K4DEU5 | AT1G48630 | Solyc12g040510 | RACK1B_AT;核苷酸结合 RACK1B_AT; Nucleotide binding |
| P07369 | AT1G29930 | Solyc03g005780 | CAB1,AB140,CAB140,LHCB1.3 |
| K4D831 | AT2G37270 | Solyc11g042610 | ATRPS5B;核糖体的结构组成ATRPS5B; Structural constituent of ribosome |
| K4CP59 | AT4G11650 | Solyc08g080590 | ATOSM34|ATOSM34(渗调蛋白34)ATOSM34|ATOSM34 (osmotin 34) |
| K4CAM4 | AT4G28730 | Solyc06g083690 | 谷氨酸家族蛋白 Glutaredoxin family protein |
| K4C3Z3 | AT3G62410 | Solyc06g009630 | CP12-2,CP12|CP12-2;蛋白结合 CP12-2, CP12|CP12-2; protein binding |
| K4BQ07 | AT2G43560 | Solyc04g015040 | 免疫亲和蛋白Immunophilin |
| K4ASQ6 | AT5G06480 | Solyc01g006900 | MD-2相关脂质识别含域蛋白MD-2-related lipid recognition domain-containing protein |
| Q9XGI9 | AT2G27450 | Solyc11g068540 | NLP1,ATNLP1,CPA;N-氨基甲酰腐胺酰胺酶NLP1, ATNLP1, CPA; N-carbamoylputrescine amidase |
| K4C3Z6 | AT4G02080 | Solyc06g009660 | ASAR1,ATSARA1C,ATSAR2|ATSAR2;GTP结合 ASAR1, ATSARA1C, ATSAR2|ATSAR2; GTP binding |
| Q8LPU1 | AT4G11650 | Solyc08g080620 | ATOSM34|ATOSM34(渗调蛋白34)ATOSM34|ATOSM34 (osmotin 34) |
| K4CMN4 | AT2G35040 | Solyc08g075160 | AICARFT/IMPCHase双酶家族蛋白AICARFT/IMPCHase bienzyme family protein |
| K4D621 | AT1G12050 | Solyc11g012160 | 延胡索酰乙酰乙酸酶,推定 Fumarylacetoacetase, putative |
| K4B1B8 | AT2G47170 | Solyc01g100870 | ARF1A1C;GTP结合/磷脂酶激活剂/蛋白结合ARF1A1C; GTP binding / phospholipase activator/ protein binding |
| K4BB24 | AT5G66530 | Solyc02g085100 | Aldose 1-变旋酶家族蛋白 Aldose 1-epimerase family protein |
| K4CNE8 | AT4G17040 | Solyc08g077890 | ATP依赖的Clp蛋白酶水解亚基,推定 ATP-dependent Clp protease proteolytic subunit, putative |
| Q9XG54 | AT1G76690 | Solyc10g086220 | OPR2,ATOPR2|OPR2 |
| K4BG20 | AT3G49910 | Solyc03g044000 | 60S核糖体蛋白L26(RPL26A)60S ribosomal protein L26 (RPL26A) |
| K4DA30 | AT5G22300 | Solyc11g068730 | NIT4;3-氰基丙氨酸水合酶/氰基丙氨酸腈水解酶NIT4; 3-cyanoalanine hydratase/cyanoalanine nitrilase |
| K4C952 | AT3G11710 | Solyc06g073330 | ATP结合/氨酰基-tRNA连接酶/赖氨酸-tRNA连接酶ATP binding/aminoacyl-tRNA ligase/lysine-tRNA ligase |
| K4D3M1 | AT3G09630 | Solyc10g084350 | 60S核糖体蛋白L4/L1(RPL4A)60S ribosomal protein L4/L1 (RPL4A) |
| K4CJ96 | AT2G44050 | Solyc08g015660 | COS1;6,7-二甲基-8-核苷鲁嗪合酶COS1; 6,7-dimethyl-8-ribityl lumazine synthase |
| K4CSE1 | AT3G25530 | Solyc09g018790 | GHBDH,ATGHBDH;3-羟丁酸脱氢酶GHBDH, ATGHBDH; 3-hydroxybutyrate dehydrogenase |
| K4C8R4 | AT3G04120 | Solyc06g071920 | GAPC,GAPC-1,GAPC1|GAPC1 |
| K4D3F2 | AT3G52960 | Solyc10g083650 | 2类过氧化物酶,推定Peroxiredoxin type 2, putative |
新窗口打开|下载CSV
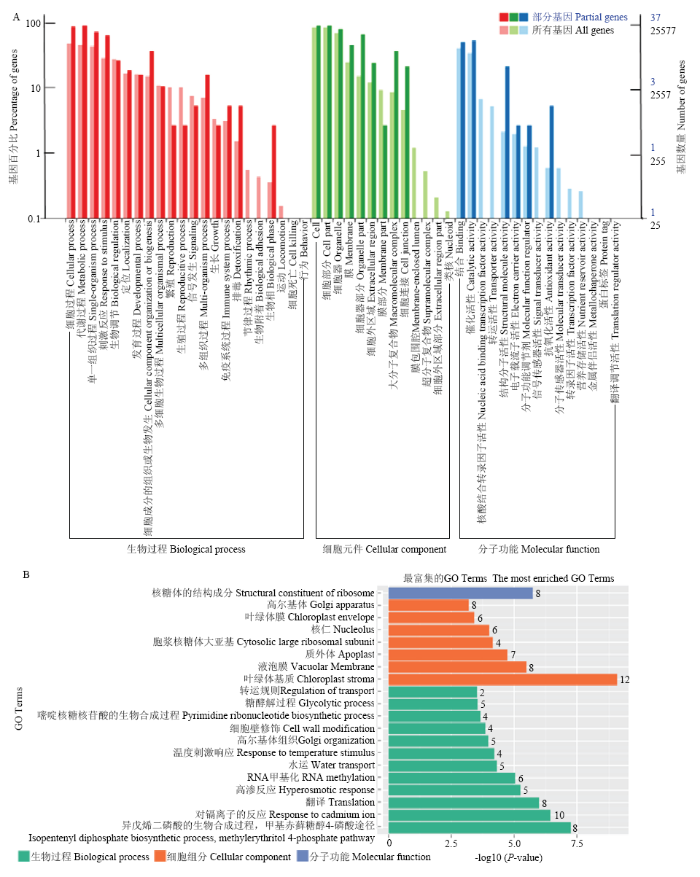
2.5 SlVQ6互作蛋白的GO富集分析
将筛选出的37个互作蛋白通过OmicsBean在线软件进行GO富集分析,得出这些蛋白在生物过程、细胞组成、分子功能均发挥作用。其中几乎所有的蛋白都参与细胞过程、代谢过程、细胞、细胞部分、细胞器组成以及核结合、催化活性等(图5-A)。从富集程度最高的功能来看,在发挥分子功能的区域全部都是与核糖体的结构成分有关(图5-B),说明与SlVQ6互作的蛋白可能介导高温等胁迫反应。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5SlVQ6互作蛋白的GO分析
A、B:SlVQ6互作蛋白GO富集功能分析(生物过程、细胞组成和分子功能)
Fig. 5GO analysis of SlVQ6 interacting protein
A, B: GO functional enrichment analysis of SlVQ6 interacting proteins (Biological process, Cellular component, Molecular function)
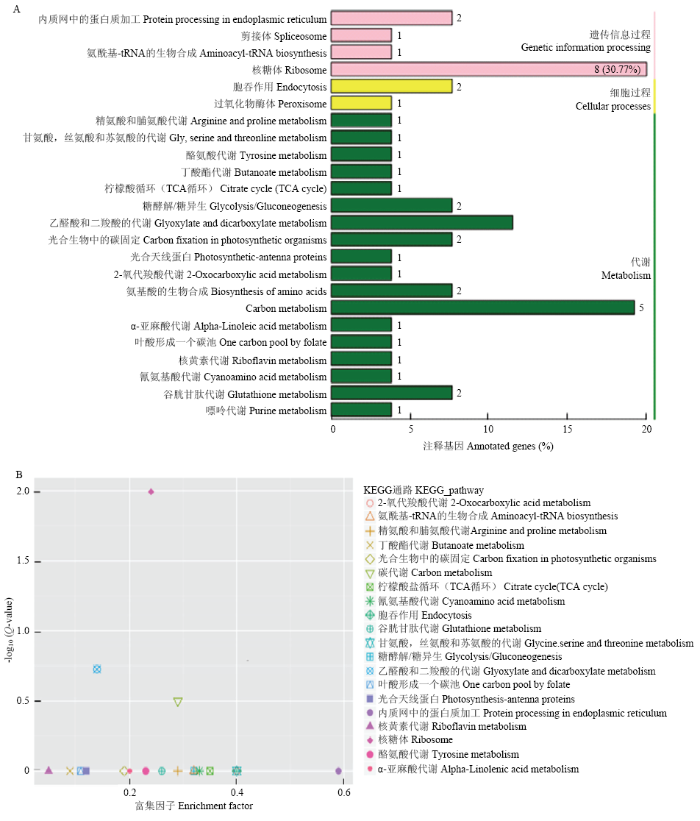
2.6 SIVQ6互作蛋白的KEGG富集分析
通过对SlVQ6互作蛋白进行KEGG分析,发现有8个蛋白参与了核糖体通路,占比达到30%以上,超过20个蛋白参与了代谢过程,少部分蛋白参与了细胞过程(图6-A),表明这些蛋白可能在基因遗传过程、细胞过程、代谢方面发挥发挥重要的作用。对差异表达基因KEGG分析后显示内质网中的蛋白过程基因富集和核糖体通路途径富集程度最高(图6-B)。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6SlVQ6互作蛋白的KEGG分析
A:SlVQ6互作蛋白的KEGG信号通路功能分类;B:KEEG对SlVQ6互作蛋白的富集途径统计,差异表达基因通过散点图展示,KEEG富集程度通过富集因子、q值和富集到此通路的基因数量来衡
Fig. 6KEGG analysis of SlVQ6 interacting proteins
A: KEGG signaling pathway function classification of SlVQ6 interacting proteins; B: Statistics of pathway enrichment of SlVQ6 interacting proteins by KEEG. Differentially expressed genes are displayed by scatter plots. The degree of KEEG enrichment is measured by the enrichment factor, q value, and the number of genes enriched in this pathway
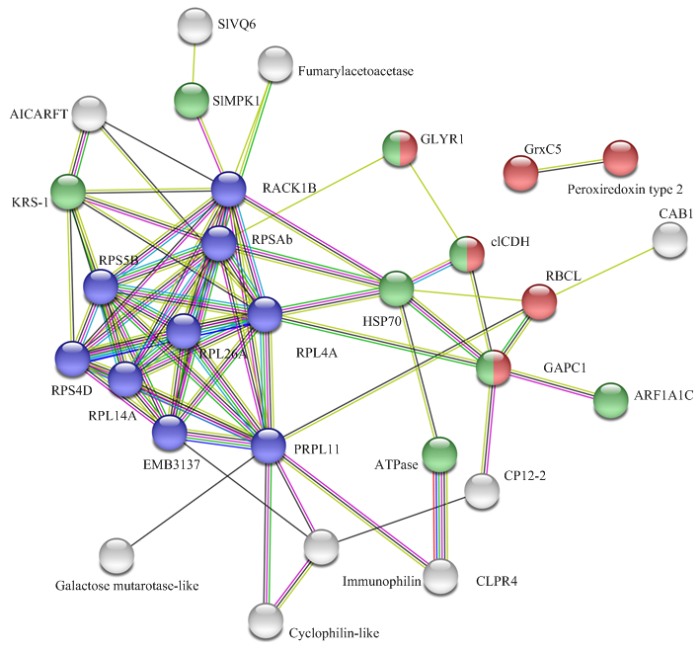
2.7 蛋白互作分析
通过STRING在线软件分析SlVQ6互作蛋白,互作网络中共有28个蛋白得到数据支持,表明试验可靠(图7)。图中蓝色节点为核糖体蛋白,处于整个互作网络中最重要的位置,能够与较多的蛋白相互作用。绿色节点和红色节点分别代表核苷酸结合蛋白与具有氧化还原酶活性蛋白,其中HSP70和KRS-1分别能与7个和6个其他蛋白相互作用,表明这些蛋白在响应高温等非生物胁迫过程中可能发挥着重要的作用。同时可以清晰看出SlVQ6与SlMPK1在本次分析中同样具有相互作用,进一步支持了MAPK级联系统参与SlVQ6介导的信号通路的结论。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7番茄中SlVQ6蛋白的推定相互作用网络
使用STRING工具集成了功能交互网络模型,并将可信度参数设置为0.40阈值。蓝色节点:核糖体蛋白;绿色节点:核苷酸结合;红色节点:氧化还原酶活性
Fig. 7Putative interaction network of SlVQ6 proteins in tomato
Functional interacting network models were integrated using the STRING tool, and the confidence parameters were set at a 0.40 threshold. Blue node: Ribosomal protein; Green node: Nucleotide binding; Red node: Oxidoreductase activity
3 讨论
3.1 VQ蛋白通过与WRKY与MAPK互作在植物体中发挥重要作用
含有VQ基序的蛋白质(称为VQ蛋白质)是一类具有保守和单个短FxxhVQxhTG氨基酸序列基序的植物特异性蛋白质,其中x代表任何氨基酸,h代表疏水性残基[30]。VQ蛋白的一级结构在VQ基序以外的区域中具有高度多样性,表明VQ蛋白具有不同的相互作用体。而植物VQ蛋白和其他生物中的蛋白序列之间具有很小的相似性,表明VQ家族对于植物是特异性的[7]。自发现第一种VQ蛋白质SIGMA FACTOR-BINDING PROTEIN1(SIB1,也称为VQ23)以来[31],已经对9种VQ蛋白质进行了功能鉴定,这些蛋白在植物防御反应,胁迫耐受性以及生长和发育中起着不同的作用[8]。例如,VQ23/SIB1功能丧失的植物对灰霉病菌和丁香假单胞菌的抗性均受损,而VQ23过表达株系在被病原体感染后可以减轻疾病症状;钙调节蛋白结合蛋白AtCAMBP25(AtVQ15),负调节植物渗透胁迫反应[32];AtVQ9在响应盐胁迫时与AtWRKY8拮抗相互作用[33]。HU等[11]先前的研究表明含有VQ基序的蛋白质(例如SIB1和SIB2)可以识别WRKY结构域的C末端并激活拟南芥中WRKY33的DNA结合活性。其次,VQ蛋白通过保守的V和Q残基与WRKY转录因子相互作用而参与WRKY介导的基因表达[34],这些结果表明植物VQ蛋白在WRKY介导的基因表达网络中起关键作用[23]。通过与WRKY转录因子相互作用,是研究VQ蛋白家族的结构和功能最为广泛的的方法。VQ蛋白的另一个重要特征是作为丝裂原活化蛋白激酶(MAPK)的下游底物而参与MAPK介导的信号通路,调控植物的胁迫反应及生长发育等。MAPK级联是真核生物中的通用信号传导模块,并且在多种途径中发挥作用[34]。VQ21作为MPK4的一个底物而参与MPK4介导的抗性通路。AtVQ21和WRKY33可以与AtMPK4桥接以影响植物生长和抗病性[18,19]。MPK3和/或MPK6可以磷酸化10种AtVQ蛋白,包括VQ4(MVQ1)、VQ13(MVQ2)、VQ33(MVQ3)、VQ19(MVQ4)、VQ11(MVQ5)、VQ31(MVQ6)、VQ32(MVQ7)、VQ6(MVQ8)、VQ14(MVQ9)和VQ9(MVQ10)[29],综上所述,VQ蛋白在多种植物体中发挥着重要的作用。3.2 SlVQ6与SlPMK1的互作研究
PECHER等[34]研究表明AtVQ4是AtMPK6的磷酸化底物。SlVQ6与AtVQ4同源,SlMPK1与AtMPK6同源[30]。在之前的研究中,通过Y2H验证了SlVQ6与SlMPK1的相互作用后,进一步通过体外磷酸化试验得出SlVQ6为SlMPK1的下游底物,表明SlVQ6可能在SlMPK1介导的信号通路中起到关键作用[26],为研究VQ6在番茄高温胁迫反应提供了强有力的支持。3.3 SlVQ6与37个互作蛋白的鉴定
SlVQ6因其在不同组织中的最高表达水平引起注意[5]。SlVQ6与AtVQ4(MVQ1)同源,MVQ1以VQ-基序依赖性方式抑制PAMP诱导的基因表达,充当PAMP诱导对病原体的抗性的反应的负调节因子[30]。在番茄中,SlVQ6在所有组织中均有表达,其中红色果实中的表达水平最高,而绿色果实中的表达水平最低[5]。已知SlVQ6可以响应盐,干旱和寒冷胁迫等。然而,SlVQ6的具体功能仍不清楚。为了研究SlVQ6的可能作用机制,本研究进行了pull down研究,通过LC-MS/MS鉴定了37个SIVQ6相互作用蛋白(图4-B)。由于SlVQ6和SlMPK1之间已知的相互作用,将SlMPK1添加到由STRING构建的蛋白质网络中。有趣的是,SlMPK1是SIVQ6与其他蛋白质之间的联系纽带,该网络表明SlVQ6在基因转录和翻译中的重要性(图7)。本研究鉴定出蛋白激酶RACK1B(Solyc12g040510),它能够与SlVQ6相互作用。拟南芥中的RACK1A、RACK1B和RACK1C编码的活化C激酶1(RACK1)蛋白的受体充当MAPK支架蛋白并将MPK3/MPK6级联连接至上游异源三聚体G蛋白信号传导,AtVQ12与RACK1C相互作用[35]。此外,还报道了AtVQ8与RACK1C相互作用[7]。因此,本研究推测RACK1蛋白与SlVQ6之间存在相互作用,MPK6可能参与这种相互作用。还不确定番茄中RACK1和SlMPK1之间是否存在直接相互作用。迄今为止,已经鉴定SlVQ6的37个不同的相互作用蛋白,它们互作机制和生理功能仍需要进一步研究。4 结论
SlVQ6是番茄VQ蛋白家族中一个重要的蛋白,通过蛋白表达纯化获得带有GST标签的GST-SlVQ6融合蛋白,进而由体外磷酸化得出SlVQ6作为SlMPK1的下游底物而参与番茄中的信号通路。GST pull-down及LC-MS/MS技术从番茄中筛选出37个与SlVQ6互作的蛋白,这些蛋白参与不同的代谢通路及分子功能,可能在响应高温等胁迫反应中发挥着重要的作用。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:22236506 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URLPMID:12466104 [本文引用: 1]
URLPMID:31479880 [本文引用: 3]
URLPMID:20040062 [本文引用: 1]
URLPMID:22535423 [本文引用: 6]
DOI:10.1104/pp.15.00788URLPMID:26220951 [本文引用: 2]

The VQ motif-containing proteins (designated as VQ proteins) are a class of plant-specific proteins with a conserved and single short FxxhVQxhTG amino acid sequence motif. VQ proteins regulate diverse developmental processes, including responses to biotic and abiotic stresses, seed development, and photomorphogenesis. In this Update, we summarize and discuss recent advances in our understanding of the regulation and function of VQ proteins and the role of the VQ motif in mediating transcriptional regulation and protein-protein interactions in signaling pathways. Based on the accumulated evidence, we propose a general mechanism of action for the VQ protein family, which likely defines a novel class of transcriptional regulators specific to plants.
URLPMID:21203436 [本文引用: 1]
DOI:10.1111/j.1365-313X.2010.04271.xURLPMID:20545893 [本文引用: 3]

Arabidopsis seed size is regulated by the IKU pathway that includes IKU2 (a leucine-rich repeat kinase) and MINI3 (a WRKY transcription factor). We report the cloning of the IKU1 (At2g35230) gene. iku1 mutants cause reduced endosperm growth and the production of small seeds. IKU1 encodes a protein containing a VQ motif, which is a motif specific to plants. IKU1 is expressed in the early endosperm and its progenitor, the central cell. Restoration of IKU1 function in the endosperm is sufficient to rescue seed size. A genomic construct carrying mutations in the VQ motif failed to complement the iku1 mutation, suggesting an essential role for the VQ motif. IKU1 interacts with MINI3 in the yeast two-hybrid system, consistent with an IKU1 function in the IKU-MINI pathway. Our data support the proposition that endosperm development is an important determinant of seed size.
DOI:10.1105/tpc.111.090571URLPMID:21990940 [本文引用: 3]

Necrotrophic pathogens are important plant pathogens that cause many devastating plant diseases. Despite their impact, our understanding of the plant defense response to necrotrophic pathogens is limited. The WRKY33 transcription factor is important for plant resistance to necrotrophic pathogens; therefore, elucidation of its functions will enhance our understanding of plant immunity to necrotrophic pathogens. Here, we report the identification of two WRKY33-interacting proteins, nuclear-encoded SIGMA FACTOR BINDING PROTEIN1 (SIB1) and SIB2, which also interact with plastid-encoded plastid RNA polymerase SIGMA FACTOR1. Both SIB1 and SIB2 contain an N-terminal chloroplast targeting signal and a putative nuclear localization signal, suggesting that they are dual targeted. Bimolecular fluorescence complementation indicates that WRKY33 interacts with SIBs in the nucleus of plant cells. Both SIB1 and SIB2 contain a short VQ motif that is important for interaction with WRKY33. The two VQ motif-containing proteins recognize the C-terminal WRKY domain and stimulate the DNA binding activity of WRKY33. Like WRKY33, both SIB1 and SIB2 are rapidly and strongly induced by the necrotrophic pathogen Botrytis cinerea. Resistance to B. cinerea is compromised in the sib1 and sib2 mutants but enhanced in SIB1-overexpressing transgenic plants. These results suggest that dual-targeted SIB1 and SIB2 function as activators of WRKY33 in plant defense against necrotrophic pathogens.
URLPMID:23706819 [本文引用: 3]
URLPMID:24569844 [本文引用: 2]
URLPMID:29771466 [本文引用: 1]
DOI:10.1038/srep14185URLPMID:26394921 [本文引用: 1]

Arabidopsis VQ motif-containing proteins have recently been demonstrated to interact with several WRKY transcription factors; however, their specific biological functions and the molecular mechanisms underlying their involvement in defense responses remain largely unclear. Here, we showed that two VQ genes, VQ12 and VQ29, were highly responsive to the necrotrophic fungal pathogen Botrytis cinerea. To characterize their roles in plant defense, we generated amiR-vq12 transgenic plants by using an artificial miRNA approach to suppress the expression of VQ12, and isolated a loss-of-function mutant of VQ29. Phenotypic analysis showed that decreasing the expression of VQ12 and VQ29 simultaneously rendered the amiR-vq12 vq29 double mutant plants resistant against B. cinerea. Consistently, the B. cinerea-induced expression of defense-related PLANT DEFENSIN1.2 (PDF1.2) was increased in amiR-vq12 vq29. In contrast, constitutively-expressing VQ12 or VQ29 confered transgenic plants susceptible to B. cinerea. Further investigation revealed that VQ12 and VQ29 physically interacted with themselves and each other to form homodimers and heterodimer. Moreover, expression analysis of VQ12 and VQ29 in defense-signaling mutants suggested that they were partially involved in jasmonate (JA)-signaling pathway. Taken together, our study indicates that VQ12 and VQ29 negatively regulate plant basal resistance against B. cinerea.
URLPMID:25763630 [本文引用: 1]
URLPMID:28635025 [本文引用: 1]
URLPMID:18650934 [本文引用: 2]
URLPMID:25082411 [本文引用: 2]
DOI:10.1016/j.gene.2013.08.023URLPMID:23958655 [本文引用: 1]

WRKY transcription factors are encoded by a large gene superfamily with a broad range of roles in plants. Proteins containing a short VQ (FxxxVQxLTG) motif have been recently shown to interact with WRKY transcription factors, implying that AtVQ proteins are important in the plant defense responses in Arabidopsis, either as positive or negative cofactors of WRKY transcription factors. Thirty-nine Oryza sativa genes containing the VQ motif (OsVQs) were identified and the genome structures of OsVQ proteins were characterized through genome-wide analysis in rice. Also, phylogenetic tree analysis was performed with the VQ domain of Arabidopsis and rice. The expression patterns of these OsVQ genes in plants under several stress treatments were assessed, specifically, following infection with the bacterial pathogen Xanthomonas oryzae pv. oryzae (Xoo), treatment with abscisic acid (ABA), or exposure to drought. The cellular localization of a few OsVQ proteins was examined using rice protoplast system. Based on our results, we suggest that OsVQ proteins function as important co-regulators during the plant defense response to biotic and abiotic stresses.
URLPMID:26779214 [本文引用: 1]
URLPMID:24727126 [本文引用: 1]
URLPMID:26124765 [本文引用: 2]
DOI:10.3390/ijms161226127URLPMID:26633387 [本文引用: 1]

Previous studies have showed that the VQ motif-containing proteins in Arabidopsis thaliana and Oryza sativa play an important role in plant growth, development, and stress responses. However, little is known about the functions of the VQ genes in Brassica rapa (Chinese cabbage). In this study, we performed genome-wide identification, characterization, and expression analysis of the VQ genes in Chinese cabbage, especially under adverse environment. We identified 57 VQ genes and classified them into seven subgroups (I-VII), which were dispersedly distributed on chromosomes 1 to 10. The expansion of these genes mainly contributed to segmental and tandem duplication. Fifty-four VQ genes contained no introns and 50 VQ proteins were less than 300 amino acids in length. Quantitative real-time PCR showed that the VQ genes were differentially expressed in various tissues and during different abiotic stresses and plant hormone treatments. This study provides a comprehensive overview of Chinese cabbage VQ genes and will benefit the molecular breeding for resistance to stresses and disease, as well as further studies on the biological functions of the VQ proteins.
URLPMID:29807593 [本文引用: 1]
[D].
[本文引用: 2]
[D].
[本文引用: 2]
[本文引用: 1]
URLPMID:27924014 [本文引用: 1]
DOI:10.1128/JVI.01835-08URLPMID:19211746 [本文引用: 2]

Tumor necrosis factor alpha (TNF-alpha) activates the nuclear factor kappaB (NF-kappaB) signaling pathway that regulates expression of many cellular factors playing important roles in innate immune responses and inflammation in infected hosts. Poxviruses employ many strategies to inhibit NF-kappaB activation in cells. In this report, we describe a poxvirus host range protein, CP77, which blocked NF-kappaB activation by TNF-alpha. Immunofluorescence analyses revealed that nuclear translocation of NF-kappaB subunit p65 protein in TNF-alpha-treated HeLa cells was blocked by CP77. CP77 did so without blocking IkappaBalpha phosphorylation, suggesting that upstream kinase activation was not affected by CP77. Using GST pull-down, we showed that CP77 bound to the NF-kappaB subunit p65 through the N-terminal six-ankyrin-repeat region in vitro. CP77 also bound to Cullin-1 and Skp1 of the SCF complex through a C-terminal 13-amino-acid F-box-like sequence. Both regions of CP77 are required to block NF-kappaB activation. We thus propose a model in which poxvirus CP77 suppresses NF-kappaB activation by two interactions: the C-terminal F-box of CP77 binding to the SCF complex and the N-terminal six ankyrins binding to the NF-kappaB subunit p65. In this way, CP77 attenuates innate immune response signaling in cells. Finally, we expressed CP77 or a CP77 F-box deletion protein from a vaccinia virus host range mutant (VV-hr-GFP) and showed that either protein was able to rescue the host range defect, illustrating that the F-box region, which is important for NF-kappaB modulation and binding to SCF complex, is not required for CP77's host range function. Consistently, knocking down the protein level of NF-kappaB did not relieve the growth restriction of VV-hr-GFP in HeLa cells.
URLPMID:29678861 [本文引用: 3]
DOI:10.1016/s0014-5793(02)02388-8URLPMID:11943170 [本文引用: 1]

Sigma factor binding proteins are involved in modifying the promoter preferences of the RNA polymerase in bacteria. We found the nuclear encoded protein (SibI) that is transported into chloroplasts and interacts specifically with the region 4 of Sig1 in Arabidopsis. SibI and its homologue, T3K9.5 are novel proteins, which are not homologous to any protein of known function. The expression of sibI was tissue specific, light dependent, and developmentally timed. We suggest the transcriptional regulation by sigma factor binding proteins to function in the plastids of higher plant.
DOI:10.1111/j.1365-313X.2004.02062.xURLPMID:15086802 [本文引用: 1]

A clone for a novel Arabidopsisthaliana calmodulin (CaM)-binding protein of 25 kDa (AtCaMBP25) has been isolated by using a radiolabelled CaM probe to screen a cDNA expression library derived from A. thaliana cell suspension cultures challenged with osmotic stress. The deduced amino acid sequence of AtCaMBP25 contains putative nuclear localization sequences and shares significant degree of similarity with hypothetical plant proteins only. Fusion of the AtCaMBP25 coding sequence to reporter genes targets the hybrid protein to the nucleus. Bacterially expressed AtCaMBP25 binds, in a calcium-dependent manner, to a canonical CaM but not to a less conserved isoform of the calcium sensor. AtCaMBP25 is encoded by a single-copy gene, whose expression is induced in Arabidopsis seedlings exposed to dehydration, low temperature or high salinity. Transgenic plants overexpressing AtCaMBP25 exhibits an increased sensitivity to both ionic (NaCl) and non-ionic (mannitol) osmotic stress during seed germination and seedling growth. By contrast, transgenic lines expressing antisense AtCaMBP25 are significantly more tolerant to mannitol and NaCl stresses than the wild type. Thus, the AtCaMBP25 gene functions as a negative effector of osmotic stress tolerance and likely participates in stress signal transduction pathways.
DOI:10.1111/tpj.12159URLPMID:23451802 [本文引用: 1]

The WRKY transcription factors have been demonstrated to play crucial roles in regulating stress responses; however, the exact mechanisms underlying their involvement in stress responses are not fully understood. Arabidopsis WRKY8 was predominantly expressed in roots and was highly upregulated by salt treatment. Disruption of WRKY8 rendered plants hypersensitive to salt, showing delayed germination, inhibited post-germination development and accelerated chlorosis. Further investigation revealed that WRKY8 interacted with VQ9, and their interaction decreased the DNA-binding activity of WRKY8. The VQ9 protein was exclusively localized in the nucleus, and VQ9 expression was strongly responsive to NaCl treatment. Mutation of VQ9 enhanced tolerance to salt stress, indicating that VQ9 acts antagonistically with WRKY8 to mediate responses to salt stress. The antagonist functions of WRKY8 and VQ9 were consistent with an increased or reduced Na(+)/K(+) concentration ratio, as well as contrasting expression patterns of downstream stress-responsive genes in salt-stressed wrky8 and vq9 mutants. Moreover, chromatin immunoprecipitation (ChIP) assays showed that WRKY8 directly bound the promoter of RD29A under salt conditions. These results provided strong evidence that the VQ9 protein acts as a repressor of the WRKY8 factor to maintain an appropriate balance of WRKY8-mediated signaling pathways to establish salinity stress tolerance.
DOI:10.1111/nph.12817URLPMID:24750137 [本文引用: 3]

Mitogen-activated protein kinase (MAPK) cascades play key roles in plant immune signalling, and elucidating their regulatory functions requires the identification of the pathway-specific substrates. We used yeast two-hybrid interaction screens, in vitro kinase assays and mass spectrometry-based phosphosite mapping to study a family of MAPK substrates. Site-directed mutagenesis and promoter-reporter fusion studies were performed to evaluate the impact of substrate phosphorylation on downstream signalling. A subset of the Arabidopsis thaliana VQ-motif-containing proteins (VQPs) were phosphorylated by the MAPKs MPK3 and MPK6, and renamed MPK3/6-targeted VQPs (MVQs). When plant protoplasts (expressing these MVQs) were treated with the flagellin-derived peptide flg22, several MVQs were destabilized in vivo. The MVQs interact with specific WRKY transcription factors. Detailed analysis of a representative member of the MVQ subset, MVQ1, indicated a negative role in WRKY-mediated defence gene expression - with mutation of the VQ-motif abrogating WRKY binding and causing mis-regulation of defence gene expression. We postulate the existence of a variety of WRKY-VQP-containing transcriptional regulatory protein complexes that depend on spatio-temporal VQP and WRKY expression patterns. Defence gene transcription can be modulated by changing the composition of these complexes - in part - through MAPK-mediated VQP degradation.
URLPMID:26697044 [本文引用: 1]
