 ,四川省农业科学院作物研究所/农业农村部西南地区小麦生物学与遗传育种重点实验室,成都 610066
,四川省农业科学院作物研究所/农业农村部西南地区小麦生物学与遗传育种重点实验室,成都 610066Genome-Wide Identification and Expression Analysis of PIN Genes Family in Wheat
LIU PeiXun, WAN HongShen, ZHENG JianMin, LUO JiangTao, PU ZongJun ,Crop Research Institute, Sichuan Academy of Agricultural Sciences/Key Laboratory of Wheat Biology and Genetic Improvement on Southwestern China, Ministry of Agriculture and Rural Areas, Chengdu 610066
,Crop Research Institute, Sichuan Academy of Agricultural Sciences/Key Laboratory of Wheat Biology and Genetic Improvement on Southwestern China, Ministry of Agriculture and Rural Areas, Chengdu 610066通讯作者:
责任编辑: 杨鑫浩
收稿日期:2019-09-4网络出版日期:2020-06-16
| 基金资助: |
Received:2019-09-4Online:2020-06-16
作者简介 About authors
刘培勋,Tel:028-84504231;E-mail:littlefarmer@163.com。

摘要
关键词:
Abstract
Keywords:
PDF (2968KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
刘培勋, 万洪深, 郑建敏, 罗江陶, 蒲宗君. 小麦PIN基因家族的鉴定及表达分析[J]. 中国农业科学, 2020, 53(12): 2321-2330 doi:10.3864/j.issn.0578-1752.2020.12.001
LIU PeiXun, WAN HongShen, ZHENG JianMin, LUO JiangTao, PU ZongJun.
0 引言
【研究意义】小麦是全球重要的粮食作物,中国作为世界上最大的小麦生产国和消费国,其生产对保障国家粮食安全具有重要意义。小麦籽粒硬度是国内外小麦分类和定价的重要因素之一,是当前专用小麦育种的重要指标。不同硬度小麦面粉用途不同,硬麦适用于制作面包和面条等,软麦适用于制作饼干、糕点和酿酒等。明确小麦籽粒硬度遗传基础,对于改良小麦品质,培育具有不同硬度的小麦品种具有重大意义。【前人研究进展】前人研究发现,小麦籽粒硬度受多个遗传因子控制并受环境影响。GREENWELL等[1]首次用SDS方法从软质小麦水洗淀粉中分离获得了与籽粒硬度相关的Friabilin蛋白,其分子量为15 kD,该蛋白位于小麦5DS染色体臂的Ha位点,在软粒小麦中大量表达,在硬质小麦中含量极少或者缺失[2]。小麦籽粒硬度受PIN(puroindoline)基因控制[3],包括Pina和Pinb,2个基因的氨基酸序列相似性达60%,共同作用影响籽粒柔软度。当二者同时存在时,小麦籽粒表现为软质,当Pina缺失或Pinb发生变异时,小麦籽粒表现为硬质。HEINZE等[4]将PIN基因转入硬粒小麦(Triticum turgidum L. var. durum),导致其籽粒品质发生改变并显著降低了磨粉所需能量。GASPARIS等[5]运用RNA介导的基因沉默技术使PIN基因沉默,发现PIN蛋白显著减少,小麦籽粒硬度增加。WILEY等[6]运用组织印记等技术,发现小麦PIN蛋白的合成和积累仅发生在籽粒胚乳细胞中。世界各地****对不同地区小麦资源PIN基因的变异类型开展了研究并发现了大量变异类型[7,8,9,10]。随着对PIN基因研究的深入,与硬度相关的grain softness protein-1(gsp-1)也被分离出来,该基因与PIN基因高度同源,且共同存在于5DS染色体上,3个基因的物理顺序为PinB-PinA-Gsp1[11]。此外,WILKINSON等[12]在普通小麦cDNA中发现一种与puroindoline b核苷酸序列相似性高达70%的类PIN基因,称为puroindoline b-2。陈锋等[13]发现该基因有4种变异类型,分别为Pinb-v1、Pinb-v2、Pinb-v3和Pinb-v4。利用缺体材料将其分别定位为7DL、7BL、7B和7AL上。研究表明,小麦籽粒硬度不仅由基因决定,还受环境及栽培措施等的影响。王月福等[14]研究了氮肥用量对不同小麦品种籽粒蛋白质组分含量变化及加工品质的影响,结果表明,增施氮肥能够显著提高籽粒蛋白质各组分的含量,提高湿面筋含量和沉降值,从而增加籽粒硬度。李友军等[15]研究表明,氮钾配合施用可降低小麦品种豫麦50和郑麦9023的籽粒硬度。然而,其改变小麦籽粒硬度的分子遗传基础尚不明确。【本研究切入点】目前,对于小麦籽粒硬度的研究主要集中于PIN基因(Pina和Pinb),对其所属基因家族的同源基因的研究尚不全面。【拟解决的关键问题】本研究以小麦基因组参考序列(iwgsc_refseqv1.0 2018)为基础,鉴定PIN基因家族成员,分析基因分布、基因结构、保守结构域、顺式作用元件和系统进化关系等,明确小麦PIN家族基因在不同组织不同发育阶段的表达特异性和不同硬度小麦样本中的表达模式,为解析该基因功能奠定基础,为小麦遗传改良提供理论参考。1 材料与方法
1.1 试验材料和处理方法
小麦品种蜀麦969和川麦601来自于四川省。蜀麦969是筋力最强、硬度最高的四川主推品种之一,川麦601是筋力最弱、硬度最低的品种之一。于2018年10月至2019年5月种植于四川省农业科学院德阳试验基地(31°14′N,104°24′E)。播种时各施75 kg·hm-2的磷肥(过磷酸钙)和钾肥(硫酸钾)作为基肥,常规组施150 kg·hm-2的尿素,低氮组施75 kg·hm-2的尿素,尿素的施用分为2次,一半作为基肥施用;另一半作为拔节肥施用。在开花30 d时,收取饱满的种子,液氮速冻后,-80℃保存备用,用于提取RNA。使用单粒谷物测定仪KSCS4100检测成熟小麦种子籽粒硬度指数,使用近红外谷物品质分析仪Infratec TM 1241检测粗蛋白含量。1.2 小麦PIN基因家族的鉴定和系统发育分析
从UniProt数据库[16]下载小麦已知PIN蛋白序列[17]和大麦hordoindoline序列[18],作为搜索小麦同源基因的探针序列(电子附表1)。小麦全基因组序列、蛋白序列和注释文件来源于URGI[19]。根据小麦PIN蛋白序列和大麦hordoindoline序列,使用HMMER3.0软件[20,21]建立HMM模型,在小麦全基因组蛋白序列中搜索同源序列。同时利用本地BLASTP程序[22],同源比对小麦蛋白序列,进行鉴定,同时满足2种鉴定方法的被认为是小麦PIN基因家族成员。对鉴定出的小麦PIN基因,利用Clustal X 2.0进行序列比对,利用MEGA 7.0软件[23]最大似然法构建进化树。1.3 小麦PIN基因的序列分析
使用PFAM和CDD和数据库[24,25]对鉴定出的小麦PIN基因进行保守结构域鉴定。利用ExPASy进行预测小麦PIN基因家族蛋白序列的分子量和等电点[26]。使用MEME在线程序[27]鉴定蛋白保守motif,通过GSDS在线软件[28]分析基因结构。此外,使用PlantCARE软件[29]对每个基因编码序列上游2 000 bp的DNA序列分析顺式元件,并运用TBtools工具[30]进行可视化作图。1.4 小麦PIN基因的表达分析
从Wheat Expression Browser(powered by expVIP)[31,32]下载小麦品种中国春在不同时期不同组织的转录组表达数据,用于分析小麦PIN基因的表达模式,采用实时荧光定量PCR对小麦PIN基因的表达进行鉴定。使用Omega biotek公司植物RNA提取试剂盒(R6827)提取开花后30 d的小麦籽粒RNA,并反转录成cDNA。根据小麦PIN基因序列,利用Oligo 7软件设计定量PCR引物(表1),由上海生工生物工程有限公司合成。实时荧光定量PCR反应体系(20 μL)包括10 μL SYBR?Green Realtime PCR Master Mix、1 μL primer #1 (10 μmol·L-1)、1 μL primer #2 (10 μmol·L-1)、1 μL Template DNA和4 μL PCR grade water。反应程序为95℃ 2 min;95℃ 15 s,54℃ 30 s(同时收集信号值),40个循环,溶解曲线分析为54℃—95℃。使用β-Actin作为相对定量的内参基因,采用2-ΔΔCt方法进行数据计算,基因的相对表达量使用(平均值±标准差)表示。Table 1
表1
表1TaPINs实时荧光定量PCR分析所用引物
Table 1
| 基因名称 Gene name | 正向引物序列 Forward primer sequence (5′-3′) | 反向引物序列 Revers primer sequence (5′-3′) |
|---|---|---|
| TaPIN6 | TATGCCGCTCTCTTGGGT | GATCGCCTTGGATTGATG |
| TaPIN7 | AGCTATGCAAGCTCCCAC | CACAACTTCTCTTCCCCC |
| TaPIN8 | TATGCCGCTCTCTTGGTT | GATCGCCTTGGATTGATG |
| TaPIN9 | AGCTCCTTGGGGAGTGTT | CAGGTTCTTGGCTTCTTG |
| TaPIN14 | GGCGGTGAAGGGTTTTTC | GCTATCGGGCGTAGTTGC |
| TaPIN15 | AAGGATTATGTGATGGAG | GCTGGTAACACTGGTCTA |
| TaPIN16 | AAAGAAGTGCCGATGTGAGG | GCTGAAAGCCAAAGACGC |
| TaPIN19 | TGTGAACAAGAAGCCCTA | TGCTGAAAACCAAAGATG |
| TaPIN10 | TGAGCATGAGGTTCGGGA | TTGCACTTTGAGGGGAGG |
| β-Actin | ATGTACCGTGGTGATGTT | CCTGGTGGCTGGTAGTTG |
新窗口打开|下载CSV
2 结果
2.1 小麦PIN基因的鉴定和序列分析
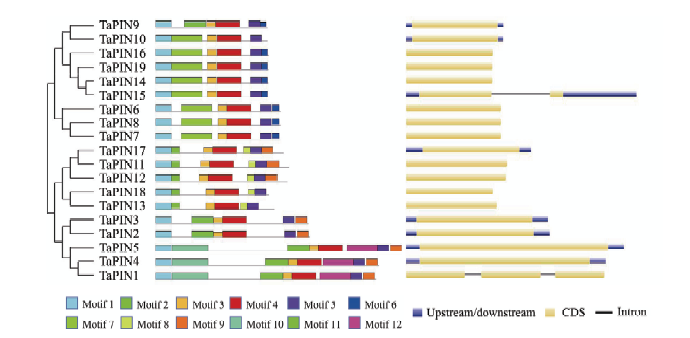
通过BLAST和Hidden Markov Model方法从小麦基因组数据库中共鉴定到19个小麦PIN家族基因,并随机命名为TaPIN1—TaPIN19,这些基因集中成簇分布于1B/1D、5A/5B/5D、7A/7B/7D染色体(电子附表2),编码148—327个氨基酸,相对分子量为16.39—37.19 kD,等电点为6.35—9.34(电子附表3)。Conserved Domains search分析结果表明,所有TaPINs基因都包含AAI_SS结构域(cl07890),该结构域是AAI_LTSS超级家族的特征,因此,PINs基因属于AAI_LTSS超级家族成员。AAI_LTSS超级家族为高等植物特有,包括谷物型阿尔法淀粉酶抑制剂(alpha amylase inhibitor)、脂质转移蛋白(lipid transfer protein)、种子储存蛋白(seed storage protein)和其他类似蛋白。Pfam数据库进一步分析表明,19个PIN蛋白结构域的Top Hit不同,其中,9个蛋白(TaPIN6—TaPIN10、TaPIN14—TaPIN16和TaPIN19)为Tryp_alpha_amyl(PF00234.22),剩下的10个(TaPIN1—TaPIN5、TaPIN11—TaPIN13、TaPIN17—TaPIN18)为Gliadin(PF13016.6)(电子附表4)。2.2 小麦PIN基因的系统发育分析、蛋白保守结构域及基因结构分析
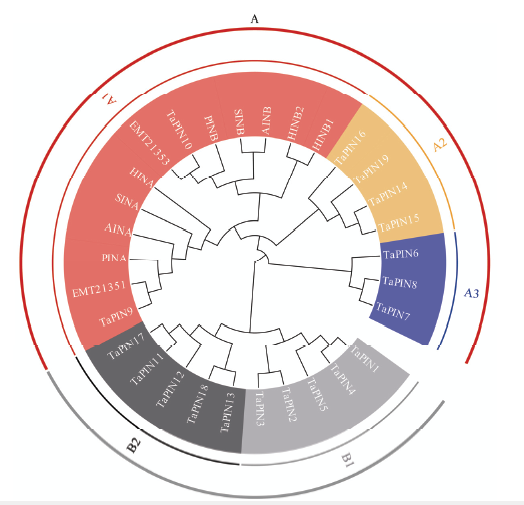
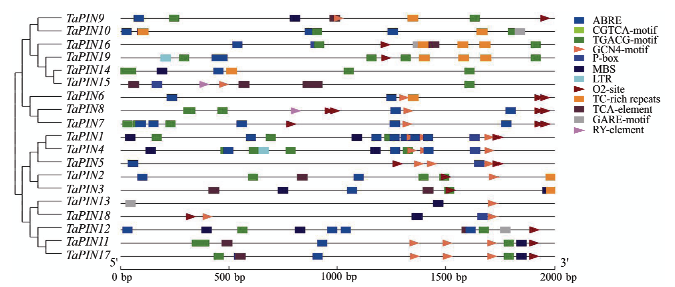
19个TaPINs蛋白序列的系统发育分析显示该基因家族可以分为A和B两大类,其中,作为参考的小麦PIN蛋白序列和大麦hordoindoline序列与A类聚为同一类。A类又进一步分为A1、A2和A3亚类,A1包括TaPIN9和TaPIN10,A2包括TaPIN14、TaPIN15、TaPIN16和TaPIN19,A3包括TaPIN6、TaPIN7和TaPIN8;B类可进一步分为B1和B2亚类,B1包括TaPIN11、TaPIN12、TaPIN13、TaPIN17和TaPIN18,B2包括TaPIN1、TaPIN2、TaPIN3、TaPIN4和TaPIN5(图1)。为了研究小麦PIN基因的结构特征,根据其进化关系进行保守motif分析,通过MEME数据库共鉴定12个保守motif(图2),所有序列均包含motif1、motif3、motif4和motif5,其中,属于PIN基因特征之一的富色氨酸区域位于motif3。A类中独有motif6和motif7,而B类中特有motif 8—motif12。如图2所示,除TaPIN15有1个长的内含子和TaPIN1有2个短内含子外,大部分TaPINs基因仅有1个外显子,没有内含子。19个TaPINs基因上游序列含多个顺式作用元件(图3),包括与抗逆相关的脱落酸作用元件(ABRE)、茉莉酸甲酯作用元件(CGTCA-motif和TGACG- motif)、干旱胁迫反应元件(MBS)、水杨酸作用元件(TCA-element)、抗逆元件(TC-rich repeats)、赤霉素反应元件(P-box和GARE-motif)和低温反应元件(LTR);与种子发育相关的胚乳发育元件(GCN4_motif)、蛋白代谢调控元件(O2-site)和种子特异调控元件(RY-element)。丰度最大的3种元件为ABRE、CGTCA和TGACG,均与抗逆性有关。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1TaPINs蛋白序列构建的系统发育树
PINA和PINB为已报道的小麦(Triticum aestivum)PIN蛋白,AINA和AINB为已报道的燕麦(Avena sativa)中同源蛋白,SINA和SINB为已报道的黑麦(Secale cereale)中同源蛋白,HINA和HINB为已报道的大麦(Hordeum vulgare)中同源蛋白,EMT21351和EMT21353为已报道的粗山羊草(Aegilops tauschii)中同源蛋白
Fig. 1Phylogenetic tree of TaPINs using the complete protein sequences
PINA and PINB were puroindoline proteins reported in Triticum aestivum, AINA and AINB were homologous proteins in Avena sativa, SINA and SINB in Secale cereale, HINA and HINB in Hordeum vulgare, EMT21351 and EMT21353 in Aegilops tauschii, respectively
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图219个TaPINs蛋白的保守序列和基因结构图
因TaPIN15太长(17 881 bp),在作图时手动删除了中间的部分内含子,最终保留了1 200 bp,以便于基因结构展示
Fig. 2The conserved motifs and gene structure analysis of 19 TaPINs
The sectional intron of TaPIN15 gene was deleted manually, as it was too long (17881 bp), and only 1200 bp was retained for showing the gene structure
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图319个TaPINs基因顺式作用元件分析
ABRE:脱落酸作用元件;CGTCA-motif:茉莉酸甲酯作用元件;TGACG-motif:茉莉酸甲酯作用元件;GCN4_motif:胚乳发育元件;P-box:赤霉素反应元件;MBS:干旱胁迫反应元件;LTR:低温反应元件;O2-site:蛋白代谢调控元件;TC-rich repeats:抗逆元件;TCA-element:水杨酸作用元件;GARE-motif:赤霉素反应元件;RY-element:种子特异调控元件
Fig. 3Cis-acting element analysis of 19 TaPINs
ABRE: cis-acting element involved in the abscisic acid responsiveness; CGTCA-motif: cis-acting regulatory element involved in the MeJA-responsiveness; TGACG-motif: cis-acting regulatory element involved in the MeJA-responsiveness; GCN4_motif: cis-regulatory element involved in endosperm expression; P-box: Gibberellin-responsive element; MBS: MYB binding site involved in drought-inducibility; LTR: cis-acting element involved in low-temperature responsiveness; O2-site: cis-acting regulatory element involved in zein metabolism regulation; TC-rich repeats: cis-acting element involved in defense and stress responsiveness; TCA-element: cis-acting element involved in salicylic acid responsiveness; GARE-motif: Gibberellin-responsive element; RY-element: cis-acting regulatory element involved in seed-specific regulation
2.3 小麦PIN基因在不同组织、不同时期的表达分析
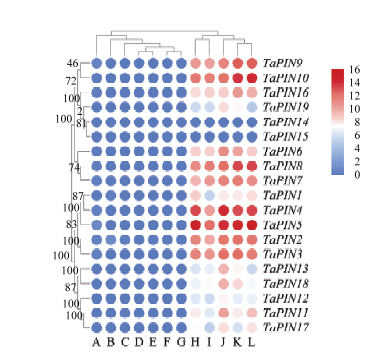
利用小麦转录组数据分析了小麦品种中国春在不同组织和发育时期的表达(图4和电子附表5),19个TaPINs基因在小麦籽粒中的相对表达量显著高于其他组织部位,而在根茎叶等组织中表达量低,几乎不表达。在小麦拔节期和开花期几乎不表达,而在籽粒形成期在籽粒中大量表达表明TaPINs主要在籽粒发育过程中起作用。不同基因成员间表达差异显著,TaPIN14、TaPIN15和TaPIN12等相对表达量较低。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4TaPINs基因在小麦品种中国春不同发育时期不同组织中的表达情况
A:三叶期的根;B:抽穗期的穗子;C:开花期的穗子;D:开花期的茎;E:开花后2 d的籽粒;F:五叶期的根;G:七叶期的叶子;H:开花后14 d的籽粒;I:开花后20 d的糊粉层;J:开花后10 d的胚乳;K:开花后20 d的胚乳;L:开花后30 d的胚乳
Fig. 4Expression profiles of TaPINs in different growth stages and different tissues of Chinese Spring
A: Root_3 leaf stage; B: Spike_emergence; C: Spike_anthesis; D: Stem_anthesis; E: Grain_2dpa; F: Shoot_5 leaf stage; G: Leaf_7 leaf stage; H: Grain_14dp; I: Aleurone layer_20dpa; J: Endosperm_10dpa; K: Endosperm_20dpa; L: Endosperm_30dpa
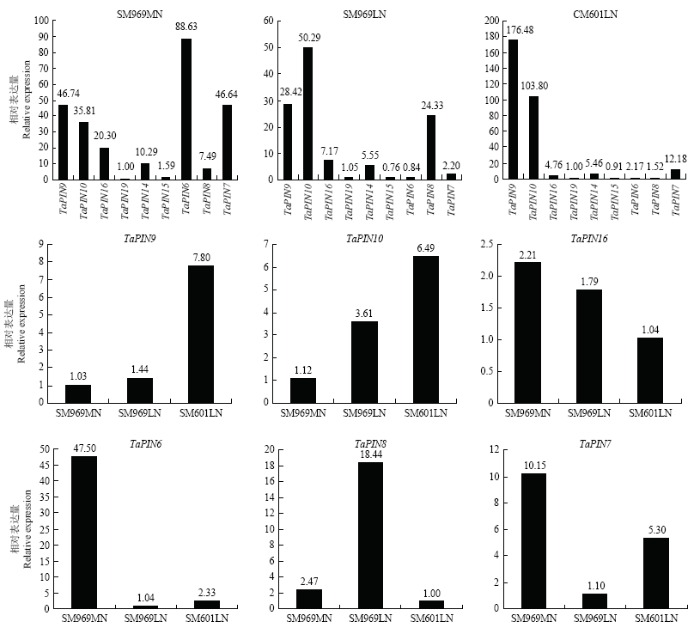
2.4 小麦PIN基因在不同硬度小麦中的相对表达分析
选取了3个不同籽粒硬度的小麦样品(SM969MN籽粒硬度指数为40.2,粗蛋白含量为13.4%;SM969LN硬度指数为35.3,粗蛋白含量为11.4%;CM601LN籽粒硬度指数为25.4,粗蛋白含量为10.7%),分析了TaPINs基因在不同硬度小麦中的表达模式(图6),在3个样品中,TaPIN9和TaPIN10均表达量较高,但随着施氮量降低或是在弱筋小麦品种中,其他几个基因表达量相对较低,而TaPIN9和TaPIN10表达比例非常高。比较几个表达量相对较低的基因的表达模式发现,仅有TaPIN9和TaPIN10的表达量随籽粒硬度的减低,相对表达量上调。TaPIN16和TaPIN6则呈现出相反的趋势,随籽粒硬度的降低,表达量下调。其他几个基因(TaPIN14、TaPIN15和TaPIN19)相对表达量很低。3 讨论
根据系统发育树,可将本研究鉴定出的19个小麦PIN基因分为A和B两大类,通过同源比对和查阅小麦基因组蛋白注释可知,TaPIN9为Pina,TaPIN10为Pinb,属于A类。Pfam数据库分析表明,19个PIN蛋白结构域的Top Hit不同,A类的9个基因为Tryp_alpha_amyl(protease inhibitor/seed storage/LTP family),B类的10个基因为Gliadin(Cys-rich Gliadin N-terminal),推测至少A类的9个基因在影响小麦籽粒硬度中发挥相似功能,而B类则与另一类与PIN基因高度同源的基因类别Avenin-like b(AVNLB)高度相关[33]。A类又分为3个亚类,A1亚类包含了Pina和Pinb。A2亚类包括TaPIN16、TaPIN19、TaPIN14和TaPIN15,该亚类与Pinb亲缘关系较近,且与之前报道的Pinb的同源基因puroindoline b-2的变异类型数量吻合,均为4个,推测其为已报道的puroindolineb-2。前人利用小麦中国春缺体材料对该类基因进行了定位,认为Pinb-2v1位于7DL,Pinb-2v2位于7BL,Pinb-2v3位于7B,Pinb-2v4位于7AL[34]。本研究利用生物信息学方法,在小麦中国春的基因组中进行鉴定,得出TaPIN14和TaPIN15均位于7A,TaPIN16位于7B,TaPIN19位于7D染色体,与前人研究略有差异。A3亚类包括TaPIN6、TaPIN7和TaPIN8,分别位于5A、5B和5D染色体,蛋白序列比对发现,TaPIN8为已报道的gsp-1,但位于5A和5B的同源序列TaPIN6 和TaPIN7之前未见报道。A2和A3亚类均在小麦的3套染色体中存在,而A1则只存在于5D染色体,这可能是由于小麦漫长的进化过程中发生了大量基因重组(转座因子插入、基因组缺失、重复和逆序)造成[35]。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5TaPINs基因家族中A类部分基因相对表达分析
图中数值为3次重复平均值。SM969MN:蜀麦969,常规氮处理;SM969LN:蜀麦969,低氮处理;CM601LN:川麦601,低氮处理
Fig. 5Relative expression of TaPINs belong to class A
The value in the figure is the average value of three replicates. SM969MN: SM969, Medium nitrogen application; SM969LN: SM969, Low nitrogen treatment; CM601LN: CM601, Low nitrogen treatment
鉴定出的19个小麦PIN基因与近缘种大麦、燕麦[36]、黑麦[37]中已报道的同源蛋白共同构建的进化树见图1中A1亚类。可见,4个麦类作物均含有该基因,其他麦类未发布完整基因组,因此,其含有PIN同源基因的数量未知。PINA与粗山羊草中的EMT21351聚在一起,PINB与EMT21353聚在一起,且蛋白序列几乎一致,再次验证了小麦D基因组来源于粗山羊草。从该基因的进化分析来看,粗山羊草与小麦亲缘关系最近,燕麦和黑麦次之,大麦与小麦亲缘最远。
通过对该基因家族编码区上游2 000 bp的DNA序列进行顺式作用元件分析,发现启动子区除了种子发育相关元件外,还存在大量抗逆相关元件,推测该类基因除影响籽粒硬度外,还参与小麦植株后期的抗性调控。DHATWALIA等[38]认为puroindoline在植物种子中具有抗菌活性,可能是一种membranotoxin膜毒素,在植物抵御微生物病原体的防御机制中发挥作用。KIM等[39]研究表明,puroindoline具有体外抗菌活性,通过转基因手段进行过表达,证明其是一种有效的抗真菌蛋白。HANEY等[40]对puroindoline抗菌活性进行了检测,表明puroindoline衍生的多肽均与DNA结合,阻断体内大分子的合成。虽然序列信息和前人研究均表明puroindoline参与抗菌活动,但基于该基因家族仅限于在种子内表达,其具体对哪些病害起作用,其调控方式等尚需进一步试验来明确。
目前,对Pina和Pinb的研究尚不能完全解释小麦籽粒硬度变异的机制。AYALA等[41]研究了西班牙南部安达鲁西亚小麦地方品种puroindoline基因的分子特征,发现13个硬质小麦样本,仅有7个可以用Pina-D1或Pinb-D1突变来解释。NIRMAL等[42]对硬籽粒小麦的基因型进行了鉴定,表明除了Pina和Pinb变异,还存在其他基因影响小麦硬度。CHEN等[43]利用电子显微镜分析的硬质和软质节节麦,不能用PIN等位基因的序列变异来解释。DARLINGTON等[44]对3个硬质大麦和3个软质大麦中的puroindoline的同源基因hordoindoline进行了序列比对,大麦中硬质与软质品种中该基因并没有差异。这些研究表明硬度是受多基因控制的数量性状。除Pina和Pinb表达量较高外,其他几个同源基因表达量非常低,对籽粒硬度的影响有限。除Pina和Pinb之外,该基因家族其他成员同样具有AAI_SS功能结构域,其在籽粒硬度调控中也可能具有共同作用,如果这些基因的表达量提升到相近水平后,能否改良籽粒硬度性状,尚需进一步试验来明确。
4 结论
在小麦全基因组范围中共鉴定出19个TaPINs基因家族成员,可分为两大类。与已报到的其他麦类作物同源基因构建的系统发育表明,从该基因来看,小麦和粗山羊草亲缘关系最近,其次是燕麦、黑麦和大麦。该基因家族均只在籽粒中表达,但部分基因表达量很低。小麦籽粒硬度的调节以Pina和Pinb为主,推测其他基因家族成员也具有相似功能,但受表达量低的限制,对籽粒硬度影响较小。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
DOI:10.1007/BF00838714URLPMID:24193708 [本文引用: 1]

The Mr 15000 protein associated with water-washed wheat starch granules from soft wheats was shown to be heterogeneous: it could be divided into a fraction containing one or morealpha-amylase inhibitor subunits and a fraction largely composed of a previously uncharacterised polypeptide(s) referred to as the
[本文引用: 1]
DOI:10.1016/j.jcs.2016.08.016URL [本文引用: 1]
DOI:10.1093/jxb/err103URL [本文引用: 1]

The RNAi-mediated silencing of Pina and Pinb, the two genes responsible for the grain texture of allohexaploid wheat, was induced and analysed in two wheat cultivars, Kontesa and Torka. A characterization of the two genes in non-transgenic plants revealed that Pinb carries a point mutation, designated Pinb-D1c in both cultivars. This mutation does not influence transcript abundance or protein content. Two silencing cassettes of the hpRNA type were constructed and used for stable transformation via Agrobacterium. In total, 43 transgenic lines representing the two cultivars were obtained, transformed with the silencing cassettes for Pina or for Pinb or co-transformed with both cassettes. The relative transcript levels of the two genes in the same progeny plant were found to be similar, independent of the silencing cassette used. The reduction in the Pina and Pinb transcript levels in the segregating T(1) progeny of Kontesa and Torka transformed with one of the silencing cassettes exceeded 80%. Co-transformation with the silencing cassettes for both genes resulted in a reduction of over 91% of Pina and Pinb transcripts in some segregating T(1) progeny of Kontesa. The silencing was transmitted to the T(4) kernel generation of the T(3) lines. A significant reduction or lack of both puroindoline proteins in the silenced lines correlated with an essential increase in grain hardness. The discussion covers some new insights into the function of the Pin genes, including the simultaneous silencing of both, independent of the siRNA signal.
DOI:10.1007/s11103-007-9139-xURL [本文引用: 1]
URL [本文引用: 1]

Grain texture is one of the characteristics of bread wheat (Triticum aestivum L.) quality and has an important influence on wheat milling and processing quality. Puroindoline gene, mainly controlling grain texture of bread wheat, locates on the short arm of dismal 5D chromosome and is divided into Pina and Pinb genes tightly linking together. Pina and Pinb with the coding sequence of 447 bp have no any intron and form the basis of molecular genetics of grain texture in bread wheat. Up to now, lots of haplotypes of Pina and Pinb genes have been identified in bread wheat and Aegilops tauschii. Of them, Pina-D1b allele with hardest grain texture among the known haplotypes has been found to possess a large 15 380 bp deletion beginning after nt 23 inside of Pina coding sequence in comparison with the wild type. Besides the obvious influence on grain texture, puroindoline gene plays a significant role on other aspects of wheat quality so that some Pina and Pinb alleles have significantly different quality characteristics, e.g. Pinb-D1b have slightly superior quality of steamed bread, pan bread and Chinese noodles to Pina-D1b. Gsp-1 gene tightly linking with Pina and Pinb possesses more haplotypes than puroindoline genes while has no significant effect on grain texture according to lots of reports so far. Pinb-2v genes with approximate 70% identity to Pinb gene at the DNA level have been found to possess four variants very recently that locate on 7A, 7B and 7D chromosome, respectively, which play a minor role in controlling grain texture. This study has further provided useful information on understanding the molecular basis of grain texture and Chinese wheat improvement.
URL [本文引用: 1]

Grain texture is one of the characteristics of bread wheat (Triticum aestivum L.) quality and has an important influence on wheat milling and processing quality. Puroindoline gene, mainly controlling grain texture of bread wheat, locates on the short arm of dismal 5D chromosome and is divided into Pina and Pinb genes tightly linking together. Pina and Pinb with the coding sequence of 447 bp have no any intron and form the basis of molecular genetics of grain texture in bread wheat. Up to now, lots of haplotypes of Pina and Pinb genes have been identified in bread wheat and Aegilops tauschii. Of them, Pina-D1b allele with hardest grain texture among the known haplotypes has been found to possess a large 15 380 bp deletion beginning after nt 23 inside of Pina coding sequence in comparison with the wild type. Besides the obvious influence on grain texture, puroindoline gene plays a significant role on other aspects of wheat quality so that some Pina and Pinb alleles have significantly different quality characteristics, e.g. Pinb-D1b have slightly superior quality of steamed bread, pan bread and Chinese noodles to Pina-D1b. Gsp-1 gene tightly linking with Pina and Pinb possesses more haplotypes than puroindoline genes while has no significant effect on grain texture according to lots of reports so far. Pinb-2v genes with approximate 70% identity to Pinb gene at the DNA level have been found to possess four variants very recently that locate on 7A, 7B and 7D chromosome, respectively, which play a minor role in controlling grain texture. This study has further provided useful information on understanding the molecular basis of grain texture and Chinese wheat improvement.
DOI:10.1007/s00122-008-0893-1URL [本文引用: 1]

The Hardness (Ha) locus controls grain texture and affects many end-use properties of wheat (Triticum aestivum L.). The Ha locus is functionally comprised of the Puroindoline a and b genes, Pina and Pinb, respectively. The lack of Pin allelic diversity is a major factor limiting Ha functional analyses and wheat quality improvement. In order to create new Ha alleles, a 630 member M2 population was produced in the soft white spring cultivar Alpowa using ethylmethane sulfonate mutagenesis. The M2 population was screened to identify new alleles of Pina and Pinb. Eighteen new Pin alleles, including eight missense alleles, were identified. F2 populations for four of the new Pin alleles were developed after crossing each back to non-mutant Alpowa. Grain hardness was then measured on F2:3 seeds and the impact of each allele on grain hardness was quantified. The tested mutations were responsible for between 28 and 94% of the grain hardness variation and seed weight and vigor of all mutation lines was restored among the F2 populations. Selection of new Pin alleles following direct phenotyping or direct sequencing is a successful approach to identify new Ha alleles useful in improving wheat product quality and understanding Ha locus function.
DOI:10.1016/j.jcs.2008.03.007URL [本文引用: 1]

Abstract
Transcripts encoding three novel variant forms of puroindoline b have been identified in developing seeds of wheat. These show 57–60% sequence identity with the wild type form of Pin b but all lack one of the three tryptophan residues present in the “tryptophan loop” region of the wild type protein. Counts of ESTs and array analysis indicate that the transcripts encoding variant forms of Pin b are about an order of magnitude less abundant than those encoding wild type Pin b while array analysis also shows that expression of the variant form 1 declines more rapidly than that of the wild type form during the later stages of grain development. The gene(s) encoding variant form 1, named Pinb-A2, were mapped to the long arm of chromosome 7A of bread wheat where they show linkage to novel QTLs for hardness which have been identified in two doubled haploid populations derived from crosses between hard parental cultivars (Shamrock × Shango, Malacca × Charger).DOI:10.1016/j.jtbi.2015.04.001URLPMID:25865523 [本文引用: 1]

Kernel hardness determined by two tightly linked Puroindoline genes, Pina-D1 and Pinb-D1, located on chromosome 5DS define commercially important characteristics, uses, major grades and export markets of wheat. This study was conducted to characterize Pina-D1 and Pinb-D1 alleles, in fifteen synthetic hexaploid wheats (SHWs) and its relation with grain hardness. Additionally, in silico functional analyses of puroindoline-a protein was conducted for better understanding of their putative importance in grain quality. Six different Pina-D1 alleles were identified in the SHWs, of which three i.e. Pina-D1a, Pina-D1c and Pina-D1d were already known whereas the other three had new sequence polymorphisms and were designated as Pina-D1w, Pina-D1x and Pina-D1y. Three different Pinb-D1 alleles were identified which have been reported earlier and no novel sequence polymorphism was detected. It was concluded that despite some primary, secondary and 3D structure variations, ligand binding sites and disulfide bonds discrepancies, the main features of PINA, i.e. the tryptophan-rich domain, the cysteine backbone, the signal peptide and basic identity of the proteins were all conserved. In silico analysis showed that puroindolines having binding capacity with small parts of prolamins causing celiac disease of human, however their potential role is not obvious. Conclusively, the new Pina-D1 alleles with modest effect on grain hardness, and insight into their functional and structural characteristics are important findings and their putative role in celiac disease require further studies to validate.
DOI:10.1016/j.jcs.2006.07.011URL [本文引用: 1]

Abstract
While the commercially important trait of grain hardness in wheat is largely determined by the puroindoline proteins, a number of studies suggest additional loci, and a role for the grain softness protein-1 (GSP-1), encoded by the gene Gsp-1 tightly linked to the puroindoline genes, remains ambiguous. To investigate their role individual Gsp-1 genes were cloned from several accessions of durum, diploid and hexaploid wheat, rather than sequencing of genomic DNA PCR products. Analysis of clones identified >1 restriction fragment length polymorphisms (RFLPs) in some 2n progenitors, >2 RFLP types in several durums and >3 in common wheat. DNA sequencing identified 30 different Gsp-1 haplotypes from various 2n, 4n and 6n wheats, exhibiting extensive single nucleotide polymorphisms (SNPs) and a 3-base deletion, all but three haplotypes being novel. Shared SNPs between various diploids, indicated an ancient origin of this gene family, but certain aspects were unique to durums, and existence of multigenes in at least some genomes was confirmed through single plant analysis of select accessions of Aegilops speltoides, durum and common wheat. Most interestingly, despite the great sequence diversity, the functionally important amino acids involved in lipid binding, i.e., the 10 cysteines and two tryptophans, were retained in all putative proteins. The results suggest that these genes may be functionally important, particularly in durums which lack puroindolines, and may have major roles in plant defence and only a minor influence on grain texture.DOI:10.1016/j.jcs.2008.03.007URL [本文引用: 1]

Abstract
Transcripts encoding three novel variant forms of puroindoline b have been identified in developing seeds of wheat. These show 57–60% sequence identity with the wild type form of Pin b but all lack one of the three tryptophan residues present in the “tryptophan loop” region of the wild type protein. Counts of ESTs and array analysis indicate that the transcripts encoding variant forms of Pin b are about an order of magnitude less abundant than those encoding wild type Pin b while array analysis also shows that expression of the variant form 1 declines more rapidly than that of the wild type form during the later stages of grain development. The gene(s) encoding variant form 1, named Pinb-A2, were mapped to the long arm of chromosome 7A of bread wheat where they show linkage to novel QTLs for hardness which have been identified in two doubled haploid populations derived from crosses between hard parental cultivars (Shamrock × Shango, Malacca × Charger).DOI:10.1007/s00122-009-1195-yURL [本文引用: 1]

Puroindoline a and b proteins soften the endosperm of wheat kernels. When the underlying puroindoline genes are altered by mutation or are deleted, kernels become harder. Thus, puroindoline a and b (Pina and Pinb) play an important role in wheat quality and utilization. Recently, additional Pinb genes have been reported. In the present report, we provide corroborating coding and additional 5′ and 3′ flanking sequence for three Pinb variants: Pinb-2v1, Pinb-2v2, and Pinb-2v3. Additionally, a new Pinb variant, Pinb-2v4, is reported. All four variants were physically mapped using Chinese Spring (CS) diteolosomics, nullisomic–tetrasomics, and CS-Cheyenne disomic substitution lines. Results place Pinb-2v1 on 7DL, Pinb-2v2 on 7BL, Pinb-2v3 on 7B, and Pinb-2v4 on 7AL. Pinb-2v1 and Pinb-2v4 were present in all cvs. examined: CS, Cheyenne, Recital, Wichita and Winsome. Pinb-2v2 was present in CS and Recital; Pinb-2v3 was present in Cheyenne, Wichita, and Winsome. These results are not wholly consistent with prior research and additional studies will be required to reconcile discrepancies. The discovery of Pinb-2v4 and the mapping of all four variants will contribute to a better understanding of gene duplication events in wheat and their bearing on wheat kernel texture and grain utilization.
URL [本文引用: 1]

研究了氮肥用量对不同小麦品种籽粒蛋白质组分含量变化及加工品质的影响 ,分析了蛋白质各组分含量和比例与加工品质的关系。结果表明 ,增施氮肥能够显著提高籽粒蛋白质各组分的含量 ,但对各组分含量提高的幅度存在差异 ,其中清蛋白、球蛋白和谷蛋白随着施氮量的增加 ,所占的比例升高 ,而醇溶蛋白和剩余蛋白则随着施氮量的增加 ,所占比例下降。增施氮肥能够显著提高湿面筋含量和沉降值 ,延长面团形成时间和稳定时间 ,提高吸水率。面团吸水率、形成时间和稳定时间与麦谷蛋白含量均呈极显著正相关。发现增施氮肥提高了小麦籽粒蛋白质含量并改变了蛋白质各组分所占的比例 ,增施氮肥是改善小麦加工品质的重要原因。
URL [本文引用: 1]

研究了氮肥用量对不同小麦品种籽粒蛋白质组分含量变化及加工品质的影响 ,分析了蛋白质各组分含量和比例与加工品质的关系。结果表明 ,增施氮肥能够显著提高籽粒蛋白质各组分的含量 ,但对各组分含量提高的幅度存在差异 ,其中清蛋白、球蛋白和谷蛋白随着施氮量的增加 ,所占的比例升高 ,而醇溶蛋白和剩余蛋白则随着施氮量的增加 ,所占比例下降。增施氮肥能够显著提高湿面筋含量和沉降值 ,延长面团形成时间和稳定时间 ,提高吸水率。面团吸水率、形成时间和稳定时间与麦谷蛋白含量均呈极显著正相关。发现增施氮肥提高了小麦籽粒蛋白质含量并改变了蛋白质各组分所占的比例 ,增施氮肥是改善小麦加工品质的重要原因。
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/nar/gkw1099URLPMID:27899622 [本文引用: 1]

The UniProt knowledgebase is a large resource of protein sequences and associated detailed annotation. The database contains over 60 million sequences, of which over half a million sequences have been curated by experts who critically review experimental and predicted data for each protein. The remainder are automatically annotated based on rule systems that rely on the expert curated knowledge. Since our last update in 2014, we have more than doubled the number of reference proteomes to 5631, giving a greater coverage of taxonomic diversity. We implemented a pipeline to remove redundant highly similar proteomes that were causing excessive redundancy in UniProt. The initial run of this pipeline reduced the number of sequences in UniProt by 47 million. For our users interested in the accessory proteomes, we have made available sets of pan proteome sequences that cover the diversity of sequences for each species that is found in its strains and sub-strains. To help interpretation of genomic variants, we provide tracks of detailed protein information for the major genome browsers. We provide a SPARQL endpoint that allows complex queries of the more than 22 billion triples of data in UniProt (http://sparql.uniprot.org/). UniProt resources can be accessed via the website at http://www.uniprot.org/.
DOI:10.1111/j.1742-4658.2006.05528.xURLPMID:17076702 [本文引用: 1]

Puroindoline proteins were purified from selected UK-grown hexaploid wheats. Their identities were confirmed on the basis of capillary electrophoresis mobilities, relative molecular mass and N-terminal amino acid sequencing. Only one form of puroindoline-a protein was found in those varieties, regardless of endosperm texture. Three allelic forms of puroindoline-b protein were identified. Nucleotide sequencing of cDNA produced by RT-PCR of isolated mRNA indicated that these were the 'wild-type', found in soft wheats, puroindoline-b containing a Gly-->Ser amino acid substitution (position 46) and puroindoline-b containing a Trp-->Arg substitution (position 44). The latter two were found in hard wheats. Microheterogeneity, due to short extensions and/or truncations at the N-terminus and C-terminus, was detected for both puroindoline-a and puroindoline-b. The type of microheterogeneity observed was more consistent for puroindoline-a than for puroindoline-b, and may arise through slightly different post-translational processing pathways. A puroindoline-b allele corresponding to a Leu-->Pro substitution (position 60) was identified from the cDNA sequence of the hard variety Chablis, but no mature puroindoline-b protein was found in this or two other European varieties known to possess this puroindoline-b allele. Wheats possessing the puroindoline-b proteins with point mutations appeared to contain lower amounts of puroindoline protein. Such wheats have a hard endosperm texture, as do wheats from which puroindoline-a or puroindoline-b are absent. Our results suggest that point mutations in puroindoline-b genes may confer hard endosperm texture through accumulation of allelic forms of puroindoline-b proteins with altered functional properties and/or through lower amounts of puroindoline proteins.
DOI:10.1139/g02-008URLPMID:12033628 [本文引用: 1]

Endosperm texture has a tremendous impact on the end-use quality of wheat (Triticum aestivum L.). Cultivars of barley (Hordeum vulgare L.), a close relative of wheat, also vary measurably in grain hardness. However, in contrast to wheat, little is known about the genetic control of barley grain hardness. Puroindolines are endosperm-specific proteins found in wheat and its relatives. In wheat, puroindoline sequence variation controls the majority of wheat grain texture variation. Hordoindolines, the puroindoline homologs of barley, have been identified and mapped. Recently, substantial allelic variation was found for hordoindolines among commercial barley cultivars. Our objective was to determine the influence of hordoindoline allelic variation upon grain hardness and dry matter digestibility in the 'Steptoe' x 'Morex' mapping population. This population is segregating for hordoindoline allele type, which was measured by a HinA/HinB/Gsp composite marker. One-hundred and fifty lines of the 'Steptoe' x 'Morex' population were grown in a replicated field trial. Grain hardness was estimated by near-infrared reflectance (NIR) and measured using the single kernel characterization system (SKCS). Variation attributable to the HinA/HinB/Gsp locus averaged 5.7 SKCS hardness units (SKCS U). QTL analysis revealed the presence of several areas of the genome associated with grain hardness. The largest QTL mapped to the HinA/HinB/Gsp region on the short arm of chomosome 7 (5H). This QTL explains 22% of the SKCS hardness difference observed in this study. The results indicate that the Hardness locus is present in barley and implicates the hordoindolines in endosperm texture control.
DOI:10.1186/s13059-018-1491-4URLPMID:30115101 [本文引用: 1]

The Wheat@URGI portal has been developed to provide the international community of researchers and breeders with access to the bread wheat reference genome sequence produced by the International Wheat Genome Sequencing Consortium. Genome browsers, BLAST, and InterMine tools have been established for in-depth exploration of the genome sequence together with additional linked datasets including physical maps, sequence variations, gene expression, and genetic and phenomic data from other international collaborative projects already stored in the GnpIS information system. The portal provides enhanced search and browser features that will facilitate the deployment of the latest genomics resources in wheat improvement.
DOI:10.1371/journal.pcbi.1002195URLPMID:22039361 [本文引用: 1]

Profile hidden Markov models (profile HMMs) and probabilistic inference methods have made important contributions to the theory of sequence database homology search. However, practical use of profile HMM methods has been hindered by the computational expense of existing software implementations. Here I describe an acceleration heuristic for profile HMMs, the
[本文引用: 1]
DOI:10.1093/nar/25.17.3389URLPMID:9254694 [本文引用: 1]

The BLAST programs are widely used tools for searching protein and DNA databases for sequence similarities. For protein comparisons, a variety of definitional, algorithmic and statistical refinements described here permits the execution time of the BLAST programs to be decreased substantially while enhancing their sensitivity to weak similarities. A new criterion for triggering the extension of word hits, combined with a new heuristic for generating gapped alignments, yields a gapped BLAST program that runs at approximately three times the speed of the original. In addition, a method is introduced for automatically combining statistically significant alignments produced by BLAST into a position-specific score matrix, and searching the database using this matrix. The resulting Position-Specific Iterated BLAST (PSI-BLAST) program runs at approximately the same speed per iteration as gapped BLAST, but in many cases is much more sensitive to weak but biologically relevant sequence similarities. PSI-BLAST is used to uncover several new and interesting members of the BRCT superfamily.
DOI:10.1093/molbev/msw054URLPMID:27004904 [本文引用: 1]

We present the latest version of the Molecular Evolutionary Genetics Analysis (Mega) software, which contains many sophisticated methods and tools for phylogenomics and phylomedicine. In this major upgrade, Mega has been optimized for use on 64-bit computing systems for analyzing larger datasets. Researchers can now explore and analyze tens of thousands of sequences in Mega The new version also provides an advanced wizard for building timetrees and includes a new functionality to automatically predict gene duplication events in gene family trees. The 64-bit Mega is made available in two interfaces: graphical and command line. The graphical user interface (GUI) is a native Microsoft Windows application that can also be used on Mac OS X. The command line Mega is available as native applications for Windows, Linux, and Mac OS X. They are intended for use in high-throughput and scripted analysis. Both versions are available from www.megasoftware.net free of charge.
DOI:10.1093/nar/gkv1344URLPMID:26673716 [本文引用: 1]

In the last two years the Pfam database (http://pfam.xfam.org) has undergone a substantial reorganisation to reduce the effort involved in making a release, thereby permitting more frequent releases. Arguably the most significant of these changes is that Pfam is now primarily based on the UniProtKB reference proteomes, with the counts of matched sequences and species reported on the website restricted to this smaller set. Building families on reference proteomes sequences brings greater stability, which decreases the amount of manual curation required to maintain them. It also reduces the number of sequences displayed on the website, whilst still providing access to many important model organisms. Matches to the full UniProtKB database are, however, still available and Pfam annotations for individual UniProtKB sequences can still be retrieved. Some Pfam entries (1.6%) which have no matches to reference proteomes remain; we are working with UniProt to see if sequences from them can be incorporated into reference proteomes. Pfam-B, the automatically-generated supplement to Pfam, has been removed. The current release (Pfam 29.0) includes 16 295 entries and 559 clans. The facility to view the relationship between families within a clan has been improved by the introduction of a new tool.
DOI:10.1093/nar/gkw1129URLPMID:27899674 [本文引用: 1]

NCBI's Conserved Domain Database (CDD) aims at annotating biomolecular sequences with the location of evolutionarily conserved protein domain footprints, and functional sites inferred from such footprints. An archive of pre-computed domain annotation is maintained for proteins tracked by NCBI's Entrez database, and live search services are offered as well. CDD curation staff supplements a comprehensive collection of protein domain and protein family models, which have been imported from external providers, with representations of selected domain families that are curated in-house and organized into hierarchical classifications of functionally distinct families and sub-families. CDD also supports comparative analyses of protein families via conserved domain architectures, and a recent curation effort focuses on providing functional characterizations of distinct subfamily architectures using SPARCLE: Subfamily Protein Architecture Labeling Engine. CDD can be accessed at https://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml.
DOI:10.1093/nar/gkg563URLPMID:12824418 [本文引用: 1]

The ExPASy (the Expert Protein Analysis System) World Wide Web server (http://www.expasy.org), is provided as a service to the life science community by a multidisciplinary team at the Swiss Institute of Bioinformatics (SIB). It provides access to a variety of databases and analytical tools dedicated to proteins and proteomics. ExPASy databases include SWISS-PROT and TrEMBL, SWISS-2DPAGE, PROSITE, ENZYME and the SWISS-MODEL repository. Analysis tools are available for specific tasks relevant to proteomics, similarity searches, pattern and profile searches, post-translational modification prediction, topology prediction, primary, secondary and tertiary structure analysis and sequence alignment. These databases and tools are tightly interlinked: a special emphasis is placed on integration of database entries with related resources developed at the SIB and elsewhere, and the proteomics tools have been designed to read the annotations in SWISS-PROT in order to enhance their predictions. ExPASy started to operate in 1993, as the first WWW server in the field of life sciences. In addition to the main site in Switzerland, seven mirror sites in different continents currently serve the user community.
DOI:10.1093/bioinformatics/btt248URL [本文引用: 1]

Genome-wide expression analysis can result in large numbers of clusters of co-expressed genes. Although there are tools for ab initio discovery of transcription factor-binding sites, most do not provide a quick and easy way to study large numbers of clusters. To address this, we introduce a web tool called MEME-LaB. The tool wraps MEME (an ab initio motif finder), providing an interface for users to input multiple gene clusters, retrieve promoter sequences, run motif finding and then easily browse and condense the results, facilitating better interpretation of the results from large-scale datasets.
DOI:10.1093/bioinformatics/btu817URLPMID:25504850 [本文引用: 1]

UNLABELLED: : Visualizing genes' structure and annotated features helps biologists to investigate their function and evolution intuitively. The Gene Structure Display Server (GSDS) has been widely used by more than 60 000 users since its first publication in 2007. Here, we reported the upgraded GSDS 2.0 with a newly designed interface, supports for more types of annotation features and formats, as well as an integrated visual editor for editing the generated figure. Moreover, a user-specified phylogenetic tree can be added to facilitate further evolutionary analysis. The full source code is also available for downloading. AVAILABILITY AND IMPLEMENTATION: Web server and source code are freely available at http://gsds.cbi.pku.edu.cn. CONTACT: gaog@mail.cbi.pku.edu.cn or gsds@mail.cbi.pku.edu.cn SUPPLEMENTARY INFORMATION: Supplementary data are available at Bioinformatics online.
DOI:10.1093/nar/30.1.325URLPMID:11752327 [本文引用: 1]

PlantCARE is a database of plant cis-acting regulatory elements, enhancers and repressors. Regulatory elements are represented by positional matrices, consensus sequences and individual sites on particular promoter sequences. Links to the EMBL, TRANSFAC and MEDLINE databases are provided when available. Data about the transcription sites are extracted mainly from the literature, supplemented with an increasing number of in silico predicted data. Apart from a general description for specific transcription factor sites, levels of confidence for the experimental evidence, functional information and the position on the promoter are given as well. New features have been implemented to search for plant cis-acting regulatory elements in a query sequence. Furthermore, links are now provided to a new clustering and motif search method to investigate clusters of co-expressed genes. New regulatory elements can be sent automatically and will be added to the database after curation. The PlantCARE relational database is available via the World Wide Web at http://sphinx.rug.ac.be:8080/PlantCARE/.
[本文引用: 1]
DOI:10.1126/science.aar6089URLPMID:30115782 [本文引用: 1]

The coordinated expression of highly related homoeologous genes in polyploid species underlies the phenotypes of many of the world's major crops. Here we combine extensive gene expression datasets to produce a comprehensive, genome-wide analysis of homoeolog expression patterns in hexaploid bread wheat. Bias in homoeolog expression varies between tissues, with ~30% of wheat homoeologs showing nonbalanced expression. We found expression asymmetries along wheat chromosomes, with homoeologs showing the largest inter-tissue, inter-cultivar, and coding sequence variation, most often located in high-recombination distal ends of chromosomes. These transcriptionally dynamic genes potentially represent the first steps toward neo- or subfunctionalization of wheat homoeologs. Coexpression networks reveal extensive coordination of homoeologs throughout development and, alongside a detailed expression atlas, provide a framework to target candidate genes underpinning agronomic traits in wheat.
DOI:10.1104/pp.15.01667URLPMID:26869702 [本文引用: 1]

The majority of transcriptome sequencing (RNA-seq) expression studies in plants remain underutilized and inaccessible due to the use of disparate transcriptome references and the lack of skills and resources to analyze and visualize these data. We have developed expVIP, an expression visualization and integration platform, which allows easy analysis of RNA-seq data combined with an intuitive and interactive interface. Users can analyze public and user-specified data sets with minimal bioinformatics knowledge using the expVIP virtual machine. This generates a custom Web browser to visualize, sort, and filter the RNA-seq data and provides outputs for differential gene expression analysis. We demonstrate expVIP's suitability for polyploid crops and evaluate its performance across a range of biologically relevant scenarios. To exemplify its use in crop research, we developed a flexible wheat (Triticum aestivum) expression browser (www.wheat-expression.com) that can be expanded with user-generated data in a local virtual machine environment. The open-access expVIP platform will facilitate the analysis of gene expression data from a wide variety of species by enabling the easy integration, visualization, and comparison of RNA-seq data across experiments.
DOI:10.1073/pnas.1812855115URLPMID:30530679 [本文引用: 1]

Fifteen full-length wheat grain avenin-like protein coding genes (TaALP) were identified on chromosome arms 7AS, 4AL, and 7DS of bread wheat with each containing five genes. Besides the a- and b-type ALPs, a c type was identified in the current paper. Both a and b types have two subunits, named x and y types. The five genes on each of the three chromosome arms consisted of two x-type genes, two y-type genes, and one c-type gene. The a-type genes were typically of 520 bp in length, whereas the b types were of 850 bp in length, and the c type was of 470 bp in length. The ALP gene transcript levels were significantly up-regulated in Blumeria graminis f. sp. tritici (Bgt)-infected wheat grain caryopsis at early grain filling. Wild emmer wheat [(WEW), Triticum dicoccoides] populations were focused on in our paper to identify allelic variations of ALP genes and to study the influence of natural selection on certain alleles. Consequently, 25 alleles were identified for TdALP-bx-7AS, 13 alleles were identified for TdALP-ax-7AS, 7 alleles were identified for TdALP-ay-7AS, and 4 alleles were identified for TdALP-ax-4AL Correlation studies on TdALP gene diversity and ecological stresses suggested that environmental factors contribute to the ALP polymorphism formation in WEW. Many allelic variants of ALPs in the endosperm of WEW are not present in bread wheat and therefore could be utilized in breeding bread wheat varieties for better quality and elite plant defense characteristics.
DOI:10.1007/s00122-009-1195-yURLPMID:19911160 [本文引用: 1]

Puroindoline a and b proteins soften the endosperm of wheat kernels. When the underlying puroindoline genes are altered by mutation or are deleted, kernels become harder. Thus, puroindoline a and b (Pina and Pinb) play an important role in wheat quality and utilization. Recently, additional Pinb genes have been reported. In the present report, we provide corroborating coding and additional 5' and 3' flanking sequence for three Pinb variants: Pinb-2v1, Pinb-2v2, and Pinb-2v3. Additionally, a new Pinb variant, Pinb-2v4, is reported. All four variants were physically mapped using Chinese Spring (CS) diteolosomics, nullisomic-tetrasomics, and CS-Cheyenne disomic substitution lines. Results place Pinb-2v1 on 7DL, Pinb-2v2 on 7BL, Pinb-2v3 on 7B, and Pinb-2v4 on 7AL. Pinb-2v1 and Pinb-2v4 were present in all cvs. examined: CS, Cheyenne, Recital, Wichita and Winsome. Pinb-2v2 was present in CS and Recital; Pinb-2v3 was present in Cheyenne, Wichita, and Winsome. These results are not wholly consistent with prior research and additional studies will be required to reconcile discrepancies. The discovery of Pinb-2v4 and the mapping of all four variants will contribute to a better understanding of gene duplication events in wheat and their bearing on wheat kernel texture and grain utilization.
DOI:10.1105/tpc.104.029181URLPMID:15749759 [本文引用: 1]

The Hardness (Ha) locus controls grain hardness in hexaploid wheat (Triticum aestivum) and its relatives (Triticum and Aegilops species) and represents a classical example of a trait whose variation arose from gene loss after polyploidization. In this study, we investigated the molecular basis of the evolutionary events observed at this locus by comparing corresponding sequences of diploid, tertraploid, and hexaploid wheat species (Triticum and Aegilops). Genomic rearrangements, such as transposable element insertions, genomic deletions, duplications, and inversions, were shown to constitute the major differences when the same genomes (i.e., the A, B, or D genomes) were compared between species of different ploidy levels. The comparative analysis allowed us to determine the extent and sequences of the rearranged regions as well as rearrangement breakpoints and sequence motifs at their boundaries, which suggest rearrangement by illegitimate recombination. Among these genomic rearrangements, the previously reported Pina and Pinb genes loss from the Ha locus of polyploid wheat species was caused by a large genomic deletion that probably occurred independently in the A and B genomes. Moreover, the Ha locus in the D genome of hexaploid wheat (T. aestivum) is 29 kb smaller than in the D genome of its diploid progenitor Ae. tauschii, principally because of transposable element insertions and two large deletions caused by illegitimate recombination. Our data suggest that illegitimate DNA recombination, leading to various genomic rearrangements, constitutes one of the major evolutionary mechanisms in wheat species.
DOI:10.1186/1471-2229-13-190URL [本文引用: 1]
DOI:10.1007/s00438-014-0894-5URLPMID:25120168 [本文引用: 1]

Among cereals, Avena sativa is characterized by an extremely soft endosperm texture, which leads to some negative agronomic and technological traits. On the basis of the well-known softening effect of puroindolines in wheat kernel texture, in this study, indolines and their encoding genes are investigated in Avena species at different ploidy levels. Three novel 14 kDa proteins, showing a central hydrophobic domain with four tryptophan residues and here named vromindoline (VIN)-1,2 and 3, were identified. Each VIN protein in diploid oat species was found to be synthesized by a single Vin gene whereas, in hexaploid A. sativa, three Vin-1, three Vin-2 and two Vin-3 genes coding for VIN-1, VIN-2 and VIN-3, respectively, were described and assigned to the A, C or D genomes based on similarity to their counterparts in diploid species. Expression of oat vromindoline transgenes in the extra-hard durum wheat led to accumulation of vromindolines in the endosperm and caused an approximate 50 % reduction of grain hardness, suggesting a central role for vromindolines in causing the extra-soft texture of oat grain. Further, hexaploid oats showed three orthologous genes coding for avenoindolines A and B, with five or three tryptophan residues, respectively, but very low amounts of avenoindolines were found in mature kernels. The present results identify a novel protein family affecting cereal kernel texture and would further elucidate the phylogenetic evolution of Avena genus.
[本文引用: 1]
DOI:10.1111/j.1439-0434.2012.01881.xURL [本文引用: 1]

Puroindolines (PINs) are the main components of the wheat grain hardness locus (Ha) and have in vitro antimicrobial activity against bacteria and fungi. Here, we examined the effect of variation in PINA and/or PINB content upon Penicillium sp. seed fungal growth inhibition. The Penicillium sp. assays were germination assays performed after incubating seeds in Penicillium sp. contaminated soil. The first set of wheat genotypes consisted of two sets of transgenic isolines created in the varieties Bobwhite and Hi-Line having over-expression of PINA and/or PINB. The second set of genotypes consisted of near-isogenic lines (NILs) varying for mutations in PINA or PINB created in the varieties Explorer and Hank. After incubation in Penicillium sp.-infected soil, transgenic wheat seeds over-expressing PINA in both Hi-Line and Bobwhite and both PINs in Hi-Line exhibited significantly reduced fungal infection and increased germination. No significant differences in Penicillium sp. infection or germination rates were observed in seeds of the NILs. The results indicate that puroindolines native role in seeds is to increase seed viability and that when over-expressed as transgenes, the puroindolines are effective antifungal proteins.
[本文引用: 1]
DOI:10.1016/j.jcs.2016.07.017URL [本文引用: 1]
DOI:10.1371/journal.pone.0164746URLPMID:27741295 [本文引用: 1]

Puroindoline (Pina and Pinb) genes control grain texture or hardness in wheat. Wild-type/soft alleles lead to softer grain while a mutation in one or both of these genes results in a hard grain. Variation in hardness in genotypes with identical Pin alleles (wild-type or mutant) is known but the molecular basis of this is not known. We now report the identification of wheat genotypes with hard grain texture and wild-type/soft Pin alleles indicating that hardness in wheat may be controlled by factors other than mutations in the coding region of the Pin genes. RNA-Seq analysis was used to determine the variation in the transcriptome of developing grains of thirty three diverse wheat genotypes including hard (mutant Pin) and soft (wild type) and those that were hard without having Pin mutations. This defined the role of pin gene expression and identified other candidate genes associated with hardness. Pina was not expressed in hard wheat with a mutation in the Pina gene. The ratio of Pina to Pinb expression was generally lower in the hard non mutant genotypes. Hardness may be associated with differences in Pin expression and other factors and is not simply associated with mutations in the PIN protein coding sequences.
DOI:10.1007/s00122-005-0047-7URL [本文引用: 1]

DNA from six hexaploid, tetraploid and diploid species of Aegilops with the C, D, S, M and U genomes was amplified with specific PCR primers to identify sequences encoding puroindolines (Pins) a and b and grain softness protein (GSP), all of which are encoded by genes at the Ha (hardness) locus, with Ae. tauschii (DD) and bread wheat (T. aestivum) (AABBDD) cv Hiline being studied as controls. Seven new allelic forms of Pin a and Pin b were identified, including forms with mutations within or close to the tryptophan motif. In addition, five new forms of GSP were detected. In all species both genomic DNA from leaves and cDNA from developing grain were analysed. This revealed the presence of both silent genes (with premature stop codons) and multiple genes, with the latter being confirmed by Southern blot analysis. Freeze fracture analysis demonstrated that all except one accession (Ae. sharonensis) were soft textured. However, this difference cannot be accounted for by the sequences of the Pin alleles present in this line.
DOI:10.1023/A:1013691530675URL [本文引用: 1]

Grain texture in barley is an important quality character as soft-textured cultivars have better malting quality. In wheat, texture is considered to be determined by the puroindolines, a group of basic hydrophobic proteins present on the surface of the starch granule. Hard wheats have been proposed to lack puroindoline a or to have mutant forms of puroindoline b which do not bind to the granule surface. Analysis of six barley cultivars (three soft-textured and three hard) showed that all contained proteins homologous to wheat puroindoline b, but PCR analysis failed to show any differences in amino acid sequences similar to those which have been proposed to determine textural differences in wheat. Southern blot analysis showed two hordoindoline b genes which were isolated and shown to encode proteins with 94% sequence identity. Expression of hordoindoline b mRNA occurred in the starchy endosperm and aleurone layer of the developing seed, but not in the embryo. Analysis of seven soft- and six hard-textured barley varieties showed that all contained hordoindoline a except two hard varieties (Sundance, Hart) which were subsequently shown to both lack hordoindoline a mRNA. It was therefore concluded that there is not a clear relationship between the presence of hordoindoline a and grain texture in barley.
