 ,山西农业大学农学院,山西太谷 030800
,山西农业大学农学院,山西太谷 030800Expression Analysis of the Chlorophyll Biosynthesis Structural Genes in Green and White Foxtail Millet [Setaria italica (L.) Beauv]
ZHANG Bin, LI Meng, LIU Jing, WANG JunJie, HOU SiYu, LI HongYing, HAN YuanHuai ,College of Agriculture, Shanxi Agricultural University, Taigu 030800, Shanxi
,College of Agriculture, Shanxi Agricultural University, Taigu 030800, Shanxi通讯作者:
责任编辑: 杨鑫浩
收稿日期:2019-11-3网络出版日期:2020-06-16
| 基金资助: |
Received:2019-11-3Online:2020-06-16
作者简介 About authors
张彬,E-mail:abingood@126.com。

摘要
关键词:
Abstract
Keywords:
PDF (2690KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张彬, 李萌, 刘晶, 王俊杰, 侯思宇, 李红英, 韩渊怀. 绿小米和白小米谷子籽粒叶绿素合成途径结构基因的表达分析[J]. 中国农业科学, 2020, 53(12): 2331-2339 doi:10.3864/j.issn.0578-1752.2020.12.002
ZHANG Bin, LI Meng, LIU Jing, WANG JunJie, HOU SiYu, LI HongYing, HAN YuanHuai.
0 引言
【研究意义】米色是消费者衡量小米品质的重要指标,小米米色包括黄色、绿色、白色等多种性状,其中,黄色小米所占比例最高。近年来,绿小米作为一类“营养-功能型”新米种,备受人们青睐,一度被认为是继沁州黄之后身价最高的小米品种[1]。然而对于绿小米米色形成的机制却鲜有报道,研究谷子籽粒中叶绿素合成代谢调控机制,可为今后通过基因工程手段提高谷物中叶绿素含量,方便人们通过膳食摄入更多的叶绿素奠定重要的理论基础。【前人研究进展】谷子(Setaria italica (L.) Beauv),在8 700年前被人们驯化[2],具有生育期短、耐旱、耐贫瘠、稳产性强等特点[3],是重要的杂粮作物。谷子去壳后称为小米,含有丰富的营养物质,如必需氨基酸、脂肪酸、蛋白质、维生素、矿物质等[4,5]。米色具有多样性,可直接反应小米品质的优劣。黄小米中黄色素的主要成分是类胡萝卜素,且以叶黄素和玉米黄质为主要组成成分。绿小米中的黄色素含量虽低,但其叶绿素含量较高[5]。叶绿素(Chlorophyll)包括叶绿素a(Chla)和叶绿素b(Chlb),是叶片和果实色素积累的重要色素之一,其代谢过程受多个基因控制,叶绿素生物合成途径结构基因的突变可能导致其编码的酶失活进而使得植物中叶绿素含量降低,表型上会表现出泛黄、变白、斑马叶和条纹叶等特征[6,7]。半个世纪的基础研究和临床应用表明叶绿素对人体具有消炎、造血、去臭、解毒的药理作用,并且对于癌症的治疗也有一定的辅助疗效[8]。对于叶绿素生物合成相关基因的研究始于孟德尔时期,整个合成过程可分为2个阶段:L-谷氨酰tRNA至原卟啉Ⅸ的生物合成和原卟啉Ⅸ至叶绿素的生物合成,包括18种酶参与的15步酶促反应[9,10]。高等植物的叶绿素生物合成途径已较为明确,主要集中在拟南芥[11]、水稻[12]等植物中,且在拟南芥中完成其全部基因的克隆。拟南芥叶绿素生物合成过程有15种酶参与反应,并分离出27个编码这些酶的基因[13,14]。【本研究切入点】随着人们生活水平的提高,对饮食健康价值的理念越来越强,小米作为药食同源性食品越来越受到重视,谷子品质改良必然成为今后谷子育种工作的重要任务,因此,明确优异米质米色形成的分子机制势在必行。然而多年来对于谷子的相关研究主要集中在营养品质和种质方面,而不同颜色小米米色的形成机制尚未明确。【拟解决的关键问题】本研究通过对比分析绿色和白色小米叶绿素含量,小米籽粒内部超微结构以及叶绿素生物合成途径相关基因的表达模式,对绿色小米米色形成的分子机制进行研究,为揭示谷物籽粒中叶绿素的合成提供理论依据。1 材料与方法
1.1 试验材料
以绿小米大青谷(DQG)、露米青谷(LMQG)和白小米牛毛白(NMB)为试验材料,其中大青谷和露米青谷为绿色小米谷子品种,对照材料牛毛白为白色小米谷子品种。试验材料于2018年5月种植于山西农业大学农作站试验田(112°55′E,37°42′N),土壤类型为褐壤土。采用随机区组设计方法,设置3个生物重复种植材料。待籽粒成熟时收获置于50 mL离心管,并立即放入液氮冷冻,-80℃保存备用。1.2 主要试剂与仪器
RNAiso、PrimeScriptTM RT reagent Kit with gDNA Eraser试剂盒、SYBR Premix Ex Taq II,SYBR Premix Ex Taq II 试剂、PrimeSTAR HS聚合酶均购自Takara公司;Epon LX112试剂盒(美国Williston公司);丙酮、乙醇、甲醇、戊二醛、OSO4、磷酸缓冲液、乙酸铀酰和柠檬酸铅均购自上海国药集团化学试剂有限公司。非接触式色差仪兼光泽仪VS450(美国Xrite公司);Hitachi TEM H-7500(日本Hitachi公司)。
1.3 米色测定
运用色差仪对米色(L*、a*、b*)进行测定,测定前剔除发育不良、黑粒及碎粒等。每个样品进行3次技术重复,取平均值。在这个测量系统中,测量参数L*代表亮度,a*代表红色/绿色度,数值为正值表示为红色,数值为负值表示为绿色;b*值代表黄色/蓝色度,正值代表黄色,负值代表蓝色[15]。1.4 叶绿素含量的测定
将成熟收获后的谷子进行脱壳处理,称取0.2 g,用液氮研磨成粉末,置于10 mL离心管中,分别加入6 mL丙酮和乙醇(2﹕1,v/v)混合溶液进行提取,室温避光条件下振荡24—48 h,直至组织变白,12 000 r/min离心10 min,以乙醇和丙酮混合液为参比,测定645和663 nm处吸光值,每个品种重复3次。参照ARNON等[16]方法计算。Chla(mg·g-1)=(12.72A663—2.59A645)×V/W
Chlb(mg·g-1)=(22.88A645—4.68A663)×V/W
Chl(mg·g-1)=Chla+Chlb = 20.29A645+8.02A663
式中,V:提取液体积;W:样品重量。
1.5 叶绿素合成基因表达模式分析
选取处于各品种灌浆中期(籽粒中乳状物质固化,胚处于粉状物质中)谷子,去壳后提取籽粒RNA,以1 μg RNA作为模板反转录为cDNA;利用Primer Premier 5.0软件设计定量引物(电子附表1),引物由北京六合华大基因科技股份有限公司合成。运用Bio-Rad CFX96TM Touch实时定量PCR系统,SYBR Premix Ex Taq Ⅱ试剂,进行荧光定量PCR。试验设置3次生物学重复,每个生物学重复设置3次技术重复,采用2-ΔΔCt法计算表达量[17]。1.6 SiCAO的克隆
从不同谷子幼苗叶片中提取各材料中的基因组DNA,使用PrimeSTAR HS聚合酶对不同米色品种中的SiCAO进行PCR扩增(引物为SiCAO-F:5′-CAA CAACCCCTGCTCATTCG-3′和SiCAO-R:5′-GTGAT CCGTCGCTAGGTA-3′),测序,通过Multalin进行氨基酸序列的多重比对(http://mult alin.Toulouse.inra. fr/mult alin/mult alin.html)。1.7 透射电子显微镜分析
同样选取处于灌浆中期的各品种籽粒,参照KECK等[18]研究方法对绿米和白米小米籽粒进行透射电镜分析。从2种小米上切出1 mm×1 mm的新鲜片段,4℃置于4%戊二醛中8 h,然后用0.1 mmol·L-1磷酸盐缓冲液冲洗,经1% OSO4固定2 h后,用乙醇和丙酮进行脱水,并将其包埋在Epon LX112试剂盒中。为了便于透射电镜观察,将超薄切片(60 nm)用甲醇乙酸铀酰和柠檬酸铅进行染色。所有透射电镜图像都是用Hitachi TEM H-7500设备拍摄。1.8 数据分析
使用Excel 2016进行数据处理,SPSS 20.0软件对数据进行方差分析,相关图表使用OriginPro 8.0软件绘制。2 结果
2.1 不同品种米色表型分析比较
通过对绿色和白色小米谷子品种米色表型进行对比分析(图1),3个小米品种米色表型差异显著。由表1色差仪测得数据显示,2种绿色小米(L*<50)的亮度值均低于白色小米(L*>60),可能是由于白色小米的反射性更强导致。比较红绿度指标a*值结果显示,大青谷和露米青谷的a*值分别为2.66和2.9,且趋向于负值,而牛毛白的a*值为4.99,显著高于2种绿色小米,表明大青谷和露米青谷比牛毛白更绿,这与观察到的表型差异结果一致。图1
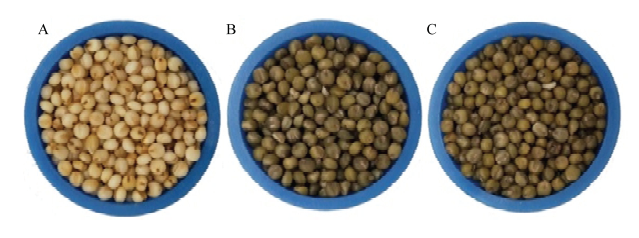 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1白小米和绿小米米色表型差异比较
A:牛毛白;B:大青谷;C:露米青谷
Fig. 1The phenotype comparison between white and green millet varieties
A: Niumaobai; B: Daqinggu; C: Lumiqinggu
Table 1
表1
表1不同谷子品种的米色指标
Table 1
| 小米品种Millet varieties | 亮度值L* | 黄/蓝度值a* | 红/绿度值b* |
|---|---|---|---|
| NMB | 66.91±0.12a | 4.99±0.10a | 28.95±0.26a |
| DQG | 43.92±0.50b | 2.66±0.14b | 22.97±0.29c |
| LMQG | 41.10±0.18c | 2.93±0.07b | 25.31±0.48b |
新窗口打开|下载CSV
2.2 不同米色品种小米叶绿素含量比较
通过对各品种籽粒中总叶绿素、叶绿素a及叶绿素b含量进行测定分析(图2),2个绿色小米中的Chla、Chlb和Chl含量均显著高于白色小米,其中,露米青谷总叶绿素含量最高为0.102 mg·g-1,大青色略低为0.097 mg·g-1,而牛毛白中总叶绿素含量仅为0.016 mg·g-1,且在白色小米中Chlb的含量仅为0.006 mg·g-1。图2
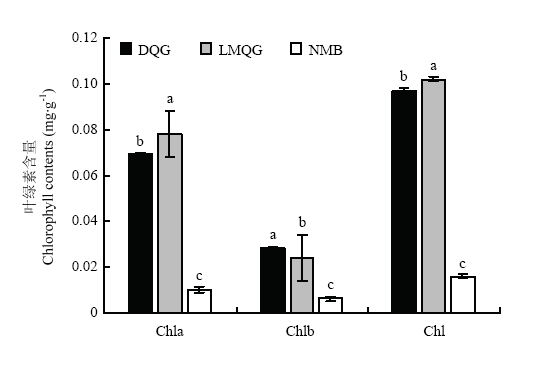 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2不同米色品种小米中叶绿素含量比较
NMB:牛毛白;DQG:大青谷;LMQG:露米青谷。不同字母表示差异显著(Dunnett检验,P<0.05)。下同
Fig. 2The comparison of chlorophyll contents in different millet color varieties
NMB: Niumaobai; DQG: Daqinggu; LMQG: Lumiqinggu. Different letters indicate a statistically significant difference (Dunnett’s test, P<0.05). The same as below
2.3 叶绿素生物合成途径结构基因在不同米色品种谷子籽粒中的表达模式分析
为了从分子水平上探索导致绿小米和白小米颜色表型差异的原因,利用qRT-PCR的方法检测了对照组谷子品种籽粒中18个叶绿素生物合成结构基因的表达模式(图3),位于叶绿素生物合成途径上游部分的结构基因在2个品种中均有较高的表达量,其中只有SiHEMA2在白米品种中的表达量高于绿色品种,SiPPXII、SiCHLM在2个绿米品种中的表达均显著高于白米品种,其他结构基因的在各品种间的表达都没有表现出明显差异。下游途径中,除SiPORA在3个品种中的表达水平基本一致外,其余结构基因都表现出明显的品种间差异,值得注意的是,SiMTC、SiDVR和SiCHIG在白米品种中的表达要显著高于绿米品种,与此相反,处于代谢途径最下游的SiCAO虽然在2个绿米品种中都有高丰度的表达,但在白米品种中基本检测不到该基因的表达。综上所述,推测SiCAO的差异表达可能是造成绿小米叶绿素含量显著高于白小米的主要原因。图3
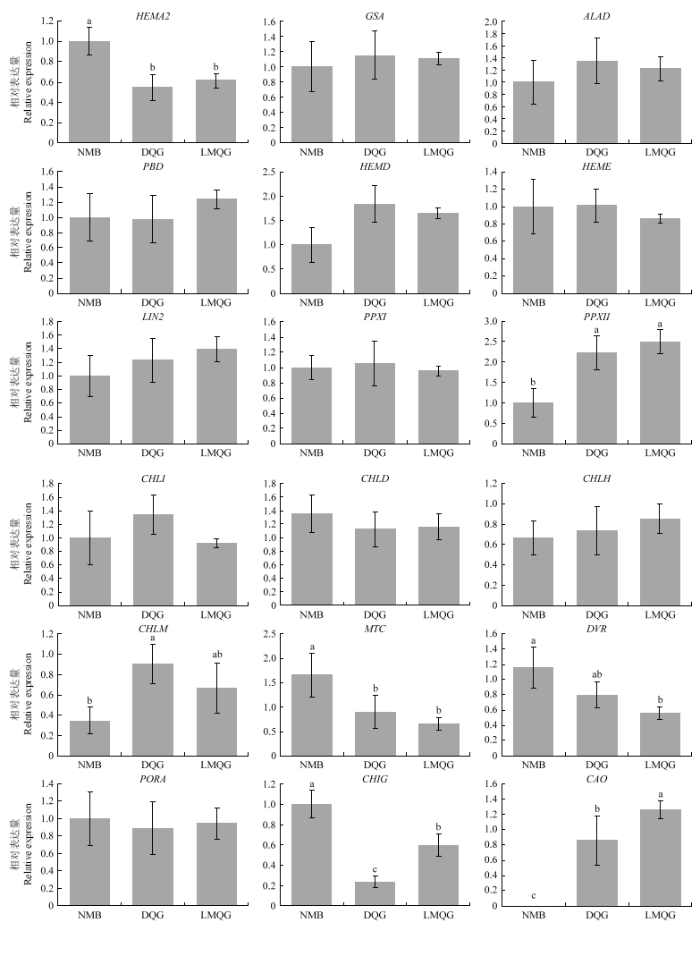 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3白色品种NMB与绿色品种DQG和LMQG叶绿素生物合成基因表达量分析
Fig. 3Quantitative real-time RT-PCR analysis of the expression profiling of the chlorophyll biosynthetic genes in white cultivate NMB and green cultivates DQG and LMQG
2.4 谷子中SiCAO的克隆及蛋白序列比对分析
从这3种基因型中分别克隆出开放阅读框为1 626 bp的SiCAO,编码541个氨基酸。对NMB、DQG和LMQG编码的SiCAO蛋白进行序列比对分析(图4),3个品种的SiCAO蛋白序列分别在第171、184、195、286和318个氨基酸处存在差异。图4
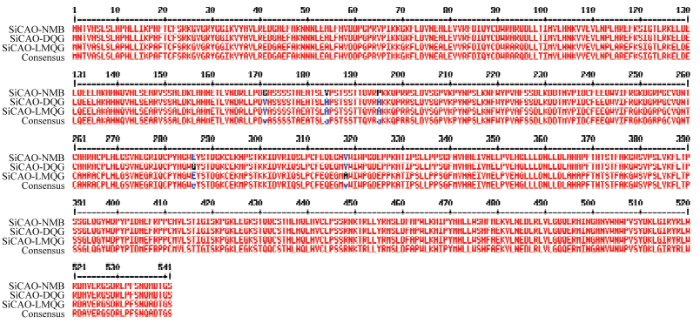 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4牛毛白(NMB)、大青谷(DQG)和露米青谷(LMQG)SiCAO蛋白序列比对
Fig. 4Protein sequence alignment of SiCAO in NMB, DQG and LMQG
2.5 不同米色品种成熟期谷子籽粒质体超微结构比较
通过对白色和绿色小米谷子品种灌浆中期籽粒内部透射电镜结果的观察比较(图5),发现灌浆中期谷子籽粒胚乳层主要由圆形或椭圆形的淀粉体构成,且为单粒淀粉体。与白小米相比绿小米中的淀粉体个体较大,着色很深。另外,绿小米淀粉体周围充满了质体小球及圆球体,而白小米淀粉体周围很少,推测绿色谷子可能通过灌浆后期淀粉体与质体小球的相对含量、淀粉体向有色体的转化对色素积累产生影响。图5
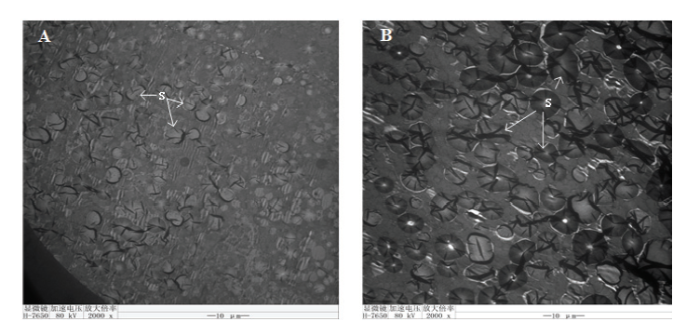 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5白小米和绿小米籽粒超微结构
A:白小米籽粒;B:绿小米籽粒。S:淀粉体
Fig. 5Ultrastructure of white and green millet grains
A: Grain of white millet; B: Grain of green millet. S: Starch
3 讨论
谷子是中国北方重要的杂粮和抗旱作物,谷子脱壳后的小米营养价值丰富,除含有蛋白质、脂肪、碳水化合物外,还富含多种维生素和铁、钙、磷等人体必需的矿质元素[19]。据谷子种质资源数据库记录,小米米色包括金黄色、黄色、淡黄色、白色、绿色、橙黄色、灰色等在内共16种颜色[20]。其中,绿小米因具有特殊的食疗价值且资源稀缺,其商品价值也越来越高[1]。高等植物中的绿色来源于叶绿素,本研究2个绿米品种籽粒中均有较高的叶绿素含量,而白米品种籽粒中的叶绿素含量仅为0.016 mg·g-1。显然叶绿素在大青谷和露米青谷2个品种籽粒中的过量积累是使得它们米色呈现绿色的主要原因。叶绿素生物合成整个途径中一个或部分结构基因的突变或表达量的变化都可以引起植物中叶绿素含量的变化[21,22]。为了进一步从分子水平上揭示绿小米米色形成的分子机制,本研究以白小米作为对照,运用qRT-PCR技术对对照谷子品种籽粒中叶绿素生物合成途径结构基因的表达模式进行了分析。植物中ALA的合成是四吡咯化合物合成途径的限速步骤,它决定叶绿素合成途径的总流量[23]。HEMA家族基因编码的谷氨酰tRNA还原酶在ALA的合成中起着重要作用。HEMA1受光诱导,主要在光合组织中表达,而HEMA2在非光合组织中表达[24],因此,本研究主要针对非光组织中表达的HEMA2进行研究。同时,HEMA2的表达表明植物中需要血红素、西罗卡因和光敏色素[25]。本研究SiHEMA2在白小米品种籽粒中的表达明显高于2个绿小米品种,这表明在白小米中四吡咯化合物的代谢更可能流向的是血红素而不是叶绿素。叶绿素与血红素的生物合成具有共同的前期步骤,ProtoⅨ由原卟啉原氧化酶(PPXI和PPXII)编码,是血红素和叶绿素生物合成的分支点。后期镁螯合酶催化Mg2+插入ProtoⅨ形成Mg-ProtoⅨ,然后再生成叶绿素,铁螯合酶催化Fe2+形成血红素[11]。其中SiPPXII在绿小米中表达量显著高于白小米,表明绿小米较白小米积累了较多的叶绿素合成前体物质。镁螯合酶由3个亚基组成(CHLI、CHLD和CHHL),它们各自的酶基因在3个品种谷子籽粒中的表达水平相似。
下游基因SiCHLM和SiCAO在绿米籽粒中的表达都显著高于白米,特别值得注意的是,SiCAO在白米品种中基本不表达。CAO(叶绿素a氧化酶)是催化Chla转化为Chlb的必要酶[26,27]。在水稻中,克隆出2个编码CAO的基因,其中OsCAO1由光诱导,但OsCAO2仅在黑暗中表达,两者独立发挥作用[28]。研究表明,当拟南芥中不存在AtCAO时,没有或极少检测到Chlb[29]。本研究中,SiCAO在白米品种籽粒中的不表达同样导致在白小米中只检测到极低水平的Chlb,由于白色小米中SiCAO的不表达水平抑制叶绿素的生物合成,最终产生白色表型。下游途径中SiMTC、SiDVRS、SiCHIG在绿米品种中的低表达可能是由于绿小米里SiCAO的过量表达,促进了Chlb的合成,从而拉动整个代谢途径向叶绿素合成的方向走,而较多的叶绿素的积累又会对前面代谢反应有反馈抑制作用。因此,推测SiCAO在2种米色品种中的极显著差异表达是导致绿小米和白小米米色差异的主要原因。
质体是植物细胞特有的细胞器,根据颜色、结构和代谢物积累的不同,分为叶绿体、有色体、白色体及淀粉体等[30]。对花卉、果实和蔬菜成熟过程中质体的形成和分化研究主要集中在叶绿体向其他类型质体转化方面,伴随着叶绿体的降解消失,逐步积累类胡萝卜素至成熟期形成有色体。除此之外,很多情况下有色体也由淀粉体转化而来,尤其是在非光合组织中[31]。之前的研究表明灌浆中期是谷子籽粒开始积累色素的转色期,因此,选用这个时期的谷子籽粒材料,并利用透射电镜对其内部结构进行观察分析[32],结果显示,谷子籽粒中的淀粉主要为单粒淀粉体,有别于水稻的复粒淀粉体,与小麦和高粱的单粒淀粉体类似[32,33]。与白小米相比,绿小米籽粒中的淀粉体颗粒大且颜色深,周围充满了质体小球和圆球体。处于灌浆期绿色小米籽粒中的这些大颗粒淀粉体在后期成熟过程逐步将转化为更多的有色体,结合之前的报道,由于绿小米籽粒中的滞绿基因SiSGR表达量受到抑制,进而影响绿小米籽粒成熟过程中叶绿素的降解[34]。因此,绿小米中很可能由于生物合成途径SiCAO的过量表达与降解途径SiSGR的低表达共同促进了其籽粒在后期成熟时,淀粉体向有色体转化过程中叶绿素的积累,而白小米中SiCAO的不表达导致其籽粒中叶绿素含量极低,使得观察到的淀粉体在绿小米中着色深,白小米着色浅。
4 结论
由于SiCAO在绿米品种籽粒中的过量表达和在白米品种籽粒中的不表达导致叶绿素含量的显著差异,进而造成绿小米和白小米颜色表型差异。SiCAO在绿色小米籽粒中积累叶绿素而呈现绿色表型中发挥作用。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 2]
[本文引用: 2]
DOI:10.1038/nbt.2195URL [本文引用: 1]

Foxtail millet (Setaria italica), a member of the Poaceae grass family, is an important food and fodder crop in arid regions and has potential for use as a C-4 biofuel. It is a model system for other biofuel grasses, including switchgrass and pearl millet. We produced a draft genome (similar to 423 Mb) anchored onto nine chromosomes and annotated 38,801 genes. Key chromosome reshuffling events were detected through collinearity identification between foxtail millet, rice and sorghum including two reshuffling events fusing rice chromosomes 7 and 9, 3 and 10 to foxtail millet chromosomes 2 and 9, respectively, that occurred after the divergence of foxtail millet and rice, and a single reshuffling event fusing rice chromosome 5 and 12 to foxtail millet chromosome 3 that occurred after the divergence of millet and sorghum. Rearrangements in the C-4 photosynthesis pathway were also identified.
DOI:10.1073/pnas.1115430109URL [本文引用: 1]

It is generally understood that foxtail millet and broomcorn millet were initially domesticated in Northern China where they eventually became the dominant plant food crops. The rarity of older archaeological sites and archaeobotanical work in the region, however, renders both the origins of these plants and their processes of domestication poorly understood. Here we present ancient starch grain assemblages recovered from cultural deposits, including carbonized residues adhering to an early pottery sherd as well as grinding stone tools excavated from the sites of Nanzhuangtou (11.5-11.0 cal kyBP) and Donghulin (11.0-9.5 cal kyBP) in the North China Plain. Our data extend the record of millet use in China by nearly 1,000 y, and the record of foxtail millet in the region by at least two millennia. The patterning of starch residues within the samples allow for the formulation of the hypothesis that foxtail millets were cultivated for an extended period of two millennia, during which this crop plant appears to have been undergoing domestication. Future research in the region will help clarify the processes in place.
[本文引用: 1]
DOI:10.15302/J-FASE-2015054URL [本文引用: 2]
DOI:10.1093/pcp/pcg064URLPMID:12773632 [本文引用: 1]

We have previously generated a large pool of T-DNA insertional lines in rice. In this study, we screened those T-DNA pools for rice mutants that had defective chlorophylls. Among the 1,995 lines examined in the T2 generation, 189 showed a chlorophyll-deficient phenotype that segregated as a single recessive locus. Among the mutants, 10 lines were beta-glucuronidase (GUS)-positive in the leaves. Line 9-07117 has a T-DNA insertion into the gene that is highly homologous to XANTHA-F in barley and CHLH in Arabidopsis: This OsCHLH gene encodes the largest subunit of the rice Mg-chelatase, a key enzyme in the chlorophyll branch of the tetrapyrrole biosynthetic pathway. In the T2 and T3 generations, the chlorina mutant phenotypes are co-segregated with the T-DNA. We have identified two additional chlorina mutants that have a Tos17 insertion in the OsCHLH gene. Those phenotypes were co-segregated with Tos17 in the progeny. GUS assays and RNA blot analysis showed that expression of the OsCHLH gene is light inducible, while TEM analysis revealed that the thylakoid membrane of the mutant chloroplasts is underdeveloped. The chlorophyll content was very low in the OschlH mutants. This is the first report that T-DNA insertional mutagenesis can be used for functional analysis of rice genes.
DOI:10.1105/tpc.104.027276URLPMID:15632054 [本文引用: 1]

Chlorophyll metabolism has been extensively studied with various organisms, and almost all of the chlorophyll biosynthetic genes have been identified in higher plants. However, only the gene for 3,8-divinyl protochlorophyllide a 8-vinyl reductase (DVR), which is indispensable for monovinyl chlorophyll synthesis, has not been identified yet. In this study, we isolated an Arabidopsis thaliana mutant that accumulated divinyl chlorophyll instead of monovinyl chlorophyll by ethyl methanesulfonate mutagenesis. Map-based cloning of this mutant resulted in the identification of a gene (AT5G18660) that shows sequence similarity with isoflavone reductase genes. The mutant phenotype was complemented by the transformation with the wild-type gene. A recombinant protein encoded by AT5G18660 was expressed in Escherichia coli and found to catalyze the conversion of divinyl chlorophyllide to monovinyl chlorophyllide, thereby demonstrating that the gene encodes a functional DVR. DVR is encoded by a single copy gene in the A. thaliana genome. With the identification of DVR, finally all genes required for chlorophyll biosynthesis have been identified in higher plants. Analysis of the complete genome of A. thaliana showed that it has 15 enzymes encoded by 27 genes for chlorophyll biosynthesis from glutamyl-tRNA(glu) to chlorophyll b. Furthermore, identification of the DVR gene helped understanding the evolution of Prochlorococcus marinus, a marine cyanobacterium that is dominant in the open ocean and is uncommon in using divinyl chlorophylls. A DVR homolog was not found in the genome of P. marinus but found in the Synechococcus sp WH8102 genome, which is consistent with the distribution of divinyl chlorophyll in marine cyanobacteria of the genera Prochlorococcus and Synechococcus.
[本文引用: 1]
[本文引用: 1]
DOI:10.1104/pp.122.1.49URLPMID:10631248 [本文引用: 1]

5-aminolevulinic acid (ALA) is a precursor in the biosynthesis of tetrapyrroles including chlorophylls and heme. The formation of ALA involves two enzymatic steps which take place in the chloroplast in plants. The first enzyme, glutamyl-tRNA reductase, and the second enzyme, glutamate-1-semialdehyde-2,1-aminomutase, are encoded by the nuclear HEMA and GSA genes, respectively. To assess the significance of the HEMA gene for chlorophyll and heme synthesis, transgenic Arabidopsis plants that expressed antisense HEMA1 mRNA from the constitutive cauliflower mosaic virus 35S promoter were generated. These plants exhibited varying degrees of chlorophyll deficiency, ranging from patchy yellow to total yellow. Analysis indicated that these plants had decreased levels of chlorophyll, non-covalently bound hemes, and ALA; their levels were proportional to the level of glutamyl-tRNA reductase expression and were inversely related to the levels of antisense HEMA transcripts. Plants that lacked chlorophyll failed to survive under normal growth conditions, indicating that HEMA gene expression is essential for growth.
DOI:10.1073/pnas.95.21.12719URLPMID:9770552 [本文引用: 1]

Chlorophyll b is an ubiquitous accessory pigment in land plants, green algae, and prochlorophytes. Its biosynthesis plays a key role in the adaptation to various light environments. We isolated six chlorophyll b-less mutants by insertional mutagenesis by using the nitrate reductase or argininosuccinate lyase genes as tags and examined the rearrangement of mutant genomes. We found that an overlapping region of a nuclear genome was deleted in all mutants and that this encodes a protein whose sequence is similar to those of methyl monooxygenases. This coding sequence also contains putative binding domains for a [2Fe-2S] Rieske center and for a mononuclear iron. The results demonstrate that a chlorophyll a oxygenase is involved in chlorophyll b formation. The reaction mechanism of chlorophyll b formation is discussed.
DOI:10.1007/s11103-004-2331-3URL [本文引用: 2]

Chlorophyll (Chl) has unique and essential roles in photosynthetic light-harvesting and energy transduction, but its biosynthesis, accumulation and degradation is also associated with chloroplast development, photomorphogenesis and chloroplast-nuclear signaling. Biochemical analyses of the enzymatic steps paved the way to the identification of their encoding genes. Thus, important progress has been made in the recent elucidation of almost all genes involved in Chl biosynthesis and breakdown. In addition, analysis of mutants mainly in Arabidopsis, genetically engineered plants and the application of photo-reactive herbicides contributed to the genetic and regulatory characterization of the formation and breakdown of Chl. This review highlights recent progress in Chl metabolism indicating highly regulated pathways from the synthesis of precursors to Chl and its degradation to intermediates, which are not longer photochemically active.
DOI:10.1007/s11103-005-2066-9URL [本文引用: 1]

Chlorophyll b is synthesized from chlorophyll a by chlorophyll a oxygenase. We have identified two genes (OsCAO1 and OsCAO2) from the rice genome that are highly homologous to previously studied chlorophyll a oxygenase (CAO) genes. They are positioned in tandem, probably resulting from recent gene duplications. The proteins they encode contain two conserved functional motifs – the Rieske Fe–sulfur coordinating center and a non-heme mononuclear Fe-binding site. OsCAO1 is induced by light and is preferentially expressed in photosynthetic tissues. Its mRNA level decreases when plants are grown in the dark. In contrast, OsCAO2 mRNA levels are higher under dark conditions, and its expression is down-regulated by exposure to light. To elucidate the physiological function of the CAO genes, we have isolated knockout mutant lines tagged by T-DNA or Tos17. Mutant plants containing a T-DNA insertion in the first intron of the OsCAO1 gene have pale green leaves, indicating chlorophyll b deficiency. We have also isolated a pale green mutant with a Tos17 insertion in that OsCAO1 gene. In contrast, OsCAO2 knockout mutant leaves do not differ significantly from the wild type. These results suggest that OsCAO1 plays a major role in chlorophyll b biosynthesis, and that OsCAO2 may function in the dark.
DOI:10.1016/j.tplants.2005.05.005URL [本文引用: 1]
DOI:10.1111/j.1432-1033.1996.00323.xURLPMID:8647070 [本文引用: 1]

All living organisms contain tetrapyrroles. In plants, chlorophyll (chlorophyll a plus chlorophyll b) is the most abundant and probably most important tetrapyrrole. It is involved in light absorption and energy transduction during photosynthesis. Chlorophyll is synthesized from the intact carbon skeleton of glutamate via the C5 pathway. This pathway takes place in the chloroplast. It is the aim of this review to summarize the current knowledge on the biochemistry and molecular biology of the C5-pathway enzymes, their regulated expression in response to light, and the impact of chlorophyll biosynthesis on chloroplast development. Particular emphasis will be placed on the key regulatory steps of chlorophyll biosynthesis in higher plants, such as 5-aminolevulinic acid formation, the production of Mg(2+)-protoporphyrin IX, and light-dependent protochlorophyllide reduction.
DOI:10.1016/j.jcs.2014.10.009URL [本文引用: 1]
DOI:10.1104/pp.24.1.1URLPMID:16654194 [本文引用: 1]
DOI:10.1006/meth.2001.1262URL [本文引用: 1]
DOI:10.1104/pp.46.5.692URLPMID:16657531 [本文引用: 1]

A nuclear mutation of Glycine max (soybean) segregates 1:2:1 in regard to chlorophyll content. The heterozygous (LG) leaf blade contains about one-half the pigment content of the wild type (DG) per gram fresh weight. A lethal yellow (LY) type contains about 1 to 2% of the DG leaf pigment values. The chlorophyll a/b ratio in the LG is about 5 compared to about 2 in the DG. Protein/leaf values are lower in the LG and LY types when compared to DG. The LG plastid lamellae contain more protein/chlorophyll, cytochromes/chlorophyll, and quinones/chlorophyll than the DG. P(700)/chlorophyll values are similar in the DG and LG types.The chlorophyll-depleted LG and LY types had less total acyl lipids per leaf weight when compared to the DG type. Similar amounts of sulfolipid and phosphatidyl glycerol per protein residue weight were found in the LG and DG plastids; however, the monogalactosyl and digalactosyl diglycerides were reduced in the LG paralleling the chlorophyll depletion.Thin sections of leaf tissue show similar-sized LG and DG plastids but reduced grana formation in the LG. The LY has very few grana and very small grana compared to either DG or LG. The two characteristic particles revealed in higher plant chloroplasts by freeze-etching are about 15% smaller in the LG compared to the DG plants.
[本文引用: 1]
[本文引用: 1]
DOI:10.3724/SP.J.1006.2016.00019URL [本文引用: 1]

Evaluation of genetic diversity could benefit the identification of optimal parental combinations for obtaining segregating offspring with maximum genetic variability, and facilitate the introgression of favorable genes from various germplasm into commercial cultivars. In this study, foxtail millet core collections including 878 world-wide accessions were evaluated through phenotypic analysis of 15 agronomic traits. Main conclusions are as follows: (1) Chinese accessions present higher level of phenotypic diversity, especially in grain weight per main stem, panicle length, panicle diameter, plant height, stem node number and growth period; phenotypic diversity is lower in elite cultivars than in traditional landraces;gains of breeding progress in foxtail millet are expressed in reduced plant height and panicle length, coupling with increased diameter of main stem, stem node number, panicle diameter, grain weight per main stem, panicle weight per main stem and growth period; (2)from the cluster analysis, foreign resources are divided into three categories according to the geographical origins, the first category mainly includes accessions from northeast European countries, the second cluster is mainly collected from North America and Africa, and the third group includes varieties mainly from East Asia, South Asia, African, and European countries; Chinese collections is able to be divided into three groups of spring-sowing, summer-sowing and southern ecotypes; (3) comprehensive assessment of phenotypic traits based on principal component analysis (PCA) and step regression analysis demonstrates that leaf sheath color, bristle length, hull color, grain color, plant height, spike length, stem diameter and grain weight per plant can be used as main identification indicators of foxtail millet phenotypic variations. This research will benefit the utilization of foxtail millet resources in variety breeding practices in the future.
DOI:10.3724/SP.J.1006.2016.00019URL [本文引用: 1]

Evaluation of genetic diversity could benefit the identification of optimal parental combinations for obtaining segregating offspring with maximum genetic variability, and facilitate the introgression of favorable genes from various germplasm into commercial cultivars. In this study, foxtail millet core collections including 878 world-wide accessions were evaluated through phenotypic analysis of 15 agronomic traits. Main conclusions are as follows: (1) Chinese accessions present higher level of phenotypic diversity, especially in grain weight per main stem, panicle length, panicle diameter, plant height, stem node number and growth period; phenotypic diversity is lower in elite cultivars than in traditional landraces;gains of breeding progress in foxtail millet are expressed in reduced plant height and panicle length, coupling with increased diameter of main stem, stem node number, panicle diameter, grain weight per main stem, panicle weight per main stem and growth period; (2)from the cluster analysis, foreign resources are divided into three categories according to the geographical origins, the first category mainly includes accessions from northeast European countries, the second cluster is mainly collected from North America and Africa, and the third group includes varieties mainly from East Asia, South Asia, African, and European countries; Chinese collections is able to be divided into three groups of spring-sowing, summer-sowing and southern ecotypes; (3) comprehensive assessment of phenotypic traits based on principal component analysis (PCA) and step regression analysis demonstrates that leaf sheath color, bristle length, hull color, grain color, plant height, spike length, stem diameter and grain weight per plant can be used as main identification indicators of foxtail millet phenotypic variations. This research will benefit the utilization of foxtail millet resources in variety breeding practices in the future.
DOI:10.1007/s10725-013-9829-0URL [本文引用: 1]

"White Dove" is a mutant in kale (Brassica oleracea var. acephala f. tricolor), which exhibits a mutant albino phenotype in the interior of the plant under low temperature conditions. Chlorophyll content in "White Dove" was dramatically reduced under low temperature conditions, while the content in "Green Dove" decreased slightly under the same conditions. The levels of five chlorophyll precursors suggested that chlorophyll biosynthesis in white kale was inhibited by low temperature stress at the step of Pchlide. However, Mg-Proto IX was not inhibited in white kale grown under low temperature conditions. The results of quantitative RT-PCR illustrated that the chlorophyll biosynthetic genes in the white cultivar were dramatically down-regulated by low temperature stress from the step of POR, while CISC and DBB1B in the white cultivar were dramatically induced under low temperature conditions. The results of transmission electron microscopy analysis showed that there were normal chloroplasts in the young leaves of white kale grown at 20 A degrees C, whereas proplastids were observed in white kale grown at 5 A degrees C. These results strongly suggested that low-temperature stress significantly inhibited plastid development in the young leaves of white kale, and repressed chlorophyll biosynthesis at the step of Pchlide by down-regulating the expression of downstream chlorophyll biosynthetic genes, resulting in undifferentiated proplastids and the albino phenotype observed in young leaves. Several genes associated with chlorophyll accumulation were also affected by low temperature conditions in white kale, especially CISC and DBB1B.
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S1360-1385(03)00064-5URLPMID:12758040 [本文引用: 1]

Regulation of tetrapyrrole biosynthesis is crucial to plant metabolism. The two pivotal control points are formation of the initial precursor, 5-aminolaevulinic acid (ALA), and the metal-ion insertion step: chelation of Fe(2+) into protoporphyrin IX leads to haem and phytochromobilin, whereas insertion of Mg(2+) is the first step to chlorophyll. Recent studies with mutants and transgenic plants have demonstrated that perturbation of the branch point affects ALA formation. Moreover, one of the signals that controls the expression of genes for nuclear-encoded chloroplast proteins has been shown to be Mg-protoporphyrin-IX. Here, we discuss the regulation of branch-point flux and the relative contributions of the haem and chlorophyll branches to the regulation of ALA synthesis and thus to flow through the tetrapyrrole pathway.
DOI:10.1046/j.1365-313x.1996.9060867.xURLPMID:8696365 [本文引用: 1]

In plants tetrapyrrole synthesis is initially light regulated on the level of 5-aminolevulinate (ALA) synthesis. ALA is formed from glutamate in three enzymatic steps. Glutamyl tRNA reductase (GluTR) catalyses the NADPH-dependent reduction of glutamyl tRNA to glutamate 1-semialdehyde. GluTR is encoded by a low-copy gene family consisting of three to four genes. Three different cDNA clones are presented. Full-length clones BHA1 and 87 differ in the length of the 3' untranslated region and code for a 58.5 kDa protein. The sequence of the partial clone, BHA13, contains at least 87 base mismatches in the coding region for the mature GluTR resulting in 11 amino acid substitutions. Synthesis of a recombinant mature and a truncated GluTR lacking the first 19 amino terminal amino acids in Escherichia coli lead only in the latter case to complementation of an E. coli hemA mutant. Steady-state level of BHA1- and BHA13-specific mRNA encoding GluTR were analysed by Northern blot hybridization using cDNA-specific oligo nucleotides and quantitative reverse transcriptase-polymerase chain reaction. Accumulation of the two RNA species is light induced in greening barley and controlled during cellular development. In contrast to BHA13, BHA1 transcripts are present in roots and are elevated after cytokinin treatment of dark-grown seedlings. Furthermore, BHA1 mRNA shows oscillation under circadian growth conditions. GluTR transcript levels correlate with the capacity for ALA synthesis indicating that the rate-limiting substrate flux through the ALA synthesizing pathway can be at least partially attributed to GluTR expression. Consequences of the initial control of the chlorophyll metabolic pathway on the level of ALA formation are discussed.
DOI:10.1146/annurev.arplant.57.032905.105448URL [本文引用: 1]
DOI:10.1007/s004250100593URLPMID:11678270 [本文引用: 1]

Plant tetrapyrrole metabolism is located in two different organelles and distributes end products into the whole cell. A complex regulatory network is involved to prevent metabolic imbalance and inefficient allocation of intermediates as well as to correlate the metabolic activities with organelle development. This review presents new findings about the control of tetrapyrrole biosynthesis and addresses the question of which regulatory principles are involved in controlling the expression of the participating enzymes and the metabolic flow in the entire pathway. It is suggested that functional organelles are required for nuclear gene expression and that metabolic signals participate in a signalling cascade transferring information from plastids to the nucleus. Recent reports about plastid-localised control mechanisms for plant tetrapyrrole metabolism are summarised and compared with results obtained in experiments on nucleus-plastid communication.
DOI:10.1046/j.1365-313x.2000.00672.xURLPMID:10758481 [本文引用: 1]

Chlorophyll (Chl) biosynthesis and degradation are the only biochemical processes on Earth that can be directly observed from satellites or other planets. The bulk of the Chls is found in the light-harvesting antenna complexes of photosynthetic organisms. Surprisingly little is known about the biosynthesis of Chl b, which is the second most abundant Chl pigment after Chl a. We describe here the expression and properties of the chlorophyllide a oxygenase gene (CAO) from Arabidopsis thaliana, which is apparently the key enzyme in Chl b biosynthesis. The recombinant enzyme produced in Escherichia coli catalyses an unusual two-step oxygenase reaction that is the 'missing link' in the chlorophyll cycle of higher plants.
DOI:10.1007/s11103-005-2066-9URL [本文引用: 1]

Chlorophyll b is synthesized from chlorophyll a by chlorophyll a oxygenase. We have identified two genes (OsCAO1 and OsCAO2) from the rice genome that are highly homologous to previously studied chlorophyll a oxygenase (CAO) genes. They are positioned in tandem, probably resulting from recent gene duplications. The proteins they encode contain two conserved functional motifs – the Rieske Fe–sulfur coordinating center and a non-heme mononuclear Fe-binding site. OsCAO1 is induced by light and is preferentially expressed in photosynthetic tissues. Its mRNA level decreases when plants are grown in the dark. In contrast, OsCAO2 mRNA levels are higher under dark conditions, and its expression is down-regulated by exposure to light. To elucidate the physiological function of the CAO genes, we have isolated knockout mutant lines tagged by T-DNA or Tos17. Mutant plants containing a T-DNA insertion in the first intron of the OsCAO1 gene have pale green leaves, indicating chlorophyll b deficiency. We have also isolated a pale green mutant with a Tos17 insertion in that OsCAO1 gene. In contrast, OsCAO2 knockout mutant leaves do not differ significantly from the wild type. These results suggest that OsCAO1 plays a major role in chlorophyll b biosynthesis, and that OsCAO2 may function in the dark.
DOI:10.1073/pnas.96.18.10507URLPMID:10468639 [本文引用: 1]

Chlorophyll b is synthesized from chlorophyll a and is found in the light-harvesting complexes of prochlorophytes, green algae, and both nonvascular and vascular plants. We have used conserved motifs from the chlorophyll a oxygenase (CAO) gene from Chlamydomonas reinhardtii to isolate a homologue from Arabidopsis thaliana. This gene, AtCAO, is mutated in both leaky and null chlorina1 alleles, and DNA sequence changes cosegregate with the mutant phenotype. AtCAO mRNA levels are higher in three different mutants that have reduced levels of chlorophyll b, suggesting that plants that do not have sufficient chlorophyll b up-regulate AtCAO gene expression. Additionally, AtCAO mRNA levels decrease in plants that are grown under dim-light conditions. We have also found that the six major Lhcb proteins do not accumulate in the null ch1-3 allele.
DOI:10.3724/SP.J.1006.2017.01290URL [本文引用: 1]
DOI:10.3724/SP.J.1006.2017.01290URL [本文引用: 1]
DOI:10.1016/j.jcs.2018.11.005URL [本文引用: 1]
[D].
[本文引用: 2]
[D].
[本文引用: 2]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1134/S1021443717040045URL [本文引用: 1]
