 ,河南省农业科学院植物保护研究所/河南省农作物病虫害防治重点实验室/农业农村部华北南部作物有害生物综合治理重点实验室,郑州 450002
,河南省农业科学院植物保护研究所/河南省农作物病虫害防治重点实验室/农业农村部华北南部作物有害生物综合治理重点实验室,郑州 450002Complete Nucleotide Sequence Analysis and Genetic Characterization of the Sweet potato feathery mottle virus O and RC Strains Isolated from China
QIN YanHong, WANG YongJiang, WANG Shuang, QIAO Qi, TIAN YuTing, ZHANG DeSheng, ZHANG ZhenChen ,Institute of Plant Protection, Henan Academy of Agricultural Sciences/Henan Key Laboratory of Crop Pest Control/ IPM Key Laboratory in Southern Part of North China, Ministry of Agriculture and Rural Affairs, Zhengzhou 450002
,Institute of Plant Protection, Henan Academy of Agricultural Sciences/Henan Key Laboratory of Crop Pest Control/ IPM Key Laboratory in Southern Part of North China, Ministry of Agriculture and Rural Affairs, Zhengzhou 450002通讯作者:
责任编辑: 岳梅
收稿日期:2019-11-7接受日期:2019-11-28网络出版日期:2020-06-01
| 基金资助: |
Received:2019-11-7Accepted:2019-11-28Online:2020-06-01
作者简介 About authors
秦艳红,E-mail: qinyanhong6040@163.com。

摘要
关键词:
Abstract
Keywords:
PDF (901KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
秦艳红, 王永江, 王爽, 乔奇, 田雨婷, 张德胜, 张振臣. 甘薯羽状斑驳病毒O株系和RC株系中国分离物全基因组 序列分析及其遗传特征[J]. 中国农业科学, 2020, 53(11): 2207-2218 doi:10.3864/j.issn.0578-1752.2020.11.007
QIN YanHong, WANG YongJiang, WANG Shuang, QIAO Qi, TIAN YuTing, ZHANG DeSheng, ZHANG ZhenChen.
0 引言
【研究意义】甘薯羽状斑驳病毒(Sweet potato feathery mottle virus,SPFMV)属于马铃薯Y病毒科(Potyviridae)马铃薯Y病毒属(Potyvirus)病毒,是侵染甘薯的重要病毒之一,在全世界甘薯产区和主要甘薯品种上均有发生[1]。SPFMV的寄主范围较窄,仅侵染旋花科植物[2,3],其传播介体为蚜虫,靠蚜虫以非持久方式进行传播[4]。SPFMV单独侵染甘薯时一般产生轻微花叶症状或无症状表现,对甘薯产量影响较小[5],但其在田间常与甘薯褪绿矮化病毒(Sweet potato chlorotic stunt virus,SPCSV)混合侵染甘薯,产生甘薯复合病毒病(sweet potato virus disease,SPVD),表现为植株严重矮化,叶片畸形、花叶、褪绿、明脉等症状,SPFMV的含量比单独侵染时增加600倍,对甘薯造成严重危害[5,6]。根据核苷酸序列及在寄主上的症状类型,SPFMV可划分为SPFMV普通株系(Ordinary,SPFMV-O)、褐裂株系(Russet Crack,SPFMV-RC)和东非株系(East Africa,SPFMV-EA)3个株系[1,6-7]。获得SPFMV中国主要株系类型的基因组全序列,对该病毒的遗传进化及分子变异研究具有重要意义。【前人研究进展】1990年,李汝刚等[8]利用血清学方法首次从我国北京、江苏、四川和山东等地区的甘薯样品上检测到SPFMV。目前,SPFMV在我国发生比较普遍,在南方、北方和长江中下游三大甘薯产区的多个省(直辖市)均有分布[9,10]。QIN等[10,11]对SPFMV的CP基因进行了分子变异分析,明确了我国SPFMV的株系类型,目前我国甘薯上存在O、RC和EA株系,O株系是我国的优势株系,分布最为广泛,其次是RC株系。1997年,SAKAI等[12]利用RT-PCR方法从日本甘薯上克隆了SPFMV RC株系S分离物的基因组全序列,对该分离物的基因组结构和编码蛋白进行了分析,SPFMV与马铃薯Y病毒属其他成员的结构相似,包含一个开放阅读框(open reading frame,ORF),经切割加工产生P1、HC-Pro、P3、6K1、CI、6K2、VPg、NIa-Pro、NIb和CP 10个成熟蛋白,是基因组最大的马铃薯Y病毒属成员。2009年,KREUZE等[13]利用深度测序结合RT-PCR方法从秘鲁获得了EA株系Piu3分离物的基因组全序列。2010年,YAMASAKI等[14]从日本获得了O株系的两个分离物Ordinary和10-O的基因组全序列,并对Ordinary和10-O分离物及RC株系的代表性分离物S和EA株系的代表性分离物EA-Piu3等的基因组结构、序列相似性等进行了比较分析。随后,韩国、西班牙、澳大利亚、东帝汶、美国、日本、巴西、南非、阿根廷、秘鲁、肯尼亚和乌干达等多个国家和地区的不同分离物基因组全序列陆续被报道[15,16,17,18]。与其他马铃薯Y病毒属病毒一样,SPFMV在P3内部通过移码策略编码一个P3N-PIPO蛋白。近年来的研究表明,SPFMV等4种侵染甘薯的马铃薯Y病毒属病毒在P1蛋白内通过移码还编码一个具有基因沉默抑制子功能的P1N-PISPO蛋白[16,19-20]。【本研究切入点】目前,SPFMV在中国仅有部分序列报道,尚未见关于SPFMV-O株系和SPFMV-RC株系中国分离物基因组全序列的报道。【拟解决的关键问题】从我国甘薯样品中克隆获得SPFMV-O和SPFMV-RC株系中国分离物全基因组序列,将两个株系的中国分离物与GenBank中登录的其他国家分离物进行比较分析,了解SPFMV-O和SPFMV-RC株系中国分离物的基因组结构及遗传进化特征。1 材料与方法
试验于2015—2016年在河南省农业科学院植物保护研究所完成。1.1 材料
1.1.1 植物材料 2015年4月,在河南省南阳市唐河县采集植株矮化、叶脉发黄、叶片皱缩等症状的甘薯病毒病样品1份,将甘薯植株种植于25—28℃防虫温室中,16 h光照、8 h黑暗培养。同时采集甘薯茎蔓上部幼嫩叶片保存于-70℃冰箱中备用。1.1.2 主要试剂 Trizol总RNA提取试剂盒购自生工生物工程(上海)股份有限公司;Reverse Transcriptase XL反转录试剂盒、2×Premix LA Taq DNA聚合酶、dNTP、T4 DNA连接酶、pMD19-T载体和DL5000分子量标准购自宝生物工程(大连)有限公司;DNA胶回收试剂盒和柱式质粒小量抽提试剂盒均购自美国OMEGA公司;大肠杆菌TG1感受态细胞为笔者实验室制备并保存于-70℃冰箱;其他生化常用试剂为进口或国产分析纯。
1.1.3 引物设计与合成 根据GenBank中登录的SPFMV-O株系和SPFMV-RC株系不同分离物的基因组全序列(表1)设计了2对简并引物和3对特异性引物,引物由生工生物工程(上海)股份有限公司合成,引物序列见表2。
Table 1
表1
表1本研究所用分离物的名称、株系、来源和登录号信息
Table 1
| 序号Number | 分离物名称Isolate name | 株系Strain | 地理来源Geographic origin | 登录号Accession number |
|---|---|---|---|---|
| 1 | RC-Ch1 | RC | 中国China | KY296451 |
| 2 | O-Ch1 | O | 中国China | KY296450 |
| 3 | CW137 | RC | 韩国South Korea | KP115608 |
| 4 | IS90 | RC | 韩国South Korea | KP115610 |
| 5 | GJ122 | O | 韩国South Korea | KP115609 |
| 6 | S | RC | 日本Japan | D86371 |
| 7 | 10-O | O | 日本Japan | AB439206 |
| 8 | Ordinary | O | 日本Japan | AB465608 |
| 9 | 17-O | O | 日本Japan | AB509454 |
| 10 | 11-8 | RC | 美国USA | MH782223 |
| 11 | 95-204R | RC | 美国USA | MH782224 |
| 12 | 11-1 | RC | 美国USA | MH778648 |
| 13 | TFSW-1J | RC | 美国USA | MH782227 |
| 14 | 95-2T | O | 美国USA | MH782225 |
| 15 | Aus7D | RC | 澳大利亚Australia | MF572046 |
| 16 | Aus12D | RC | 澳大利亚Australia | MF572049 |
| 17 | Aus8-8CA | RC | 澳大利亚Australia | MG656422 |
| 18 | Aus11D | RC | 澳大利亚Australia | MF572048 |
| 19 | Aus15-21CA | RC | 澳大利亚Australia | MG656429 |
| 20 | Aus11-13CA | RC | 澳大利亚Australia | MG656425 |
| 21 | Aus6-3B | RC | 澳大利亚Australia | MG656420 |
| 22 | Aus9-9B-2 | RC | 澳大利亚Australia | MG656423 |
| 23 | Aus10-10D | RC | 澳大利亚Australia | MG656424 |
| 24 | Aus13-18B-1 | RC | 澳大利亚Australia | MG656427 |
| 25 | Aus4D | RC | 澳大利亚Australia | MF572047 |
| 26 | Aus3D | RC | 澳大利亚Australia | MF572052 |
| 27 | Aus7-7B | RC | 澳大利亚Australia | MG656421 |
| 28 | Aus2D | RC | 澳大利亚Australia | MF572051 |
| 29 | Aus4-3A | RC | 澳大利亚Australia | MG656418 |
| 30 | Aus9D | RC | 澳大利亚Australia | MF572054 |
| 31 | Aus14-20CA | RC | 澳大利亚Australia | MG656428 |
| 32 | Aus12-16B | RC | 澳大利亚Australia | MG656426 |
| 33 | Aus16-24A | RC | 澳大利亚Australia | MG656430 |
| 34 | Aus3-2BC | RC | 澳大利亚Australia | MG656417 |
| 35 | Aus17-11D | O | 澳大利亚Australia | MG656431 |
| 36 | Aus5-3B | O | 澳大利亚Australia | MG656419 |
| 37 | Aus13B | O | 澳大利亚Australia | MF572050 |
| 38 | Aus1-2B | O | 澳大利亚Australia | MG656415 |
| 39 | Aus2-2BC | O | 澳大利亚Australia | MG656416 |
| 40 | UNB-01 | RC | 巴西Brazil | MF185715 |
| 41 | TM33C | O | 东帝汶East Timor | MF572055 |
| 42 | TM64B | O | 东帝汶East Timor | MF572053 |
| 43 | TM66B | O | 东帝汶East Timor | MF572056 |
| 44 | Ruk73 | O | 乌干达Uganda | KP729265 |
| 45 | RC-Arg | RC | 阿根廷Argentina | KF386014 |
| 46 | O-Arg | O | 阿根廷Argentina | KF386013 |
| 47 | SRF109a | O | 肯尼亚Kenya | MH264535 |
| 48 | ZA-O | O | 南非South Africa | KT069222 |
| 49 | Piu3 | EA | 秘鲁Peru | FJ155666 |
| 50 | AM-MB2 | 西班牙Spain | KU511268 |
新窗口打开|下载CSV
Table 2
表2
表2引物序列及目的片段大小
Table 2
| 扩增片段 Amplified fragment | 引物名称 Primer name | 引物序列 Primer sequence (5′→3′) | 目的片段大小 Expected size (bp) |
|---|---|---|---|
| O-1/RC-1 | 1-F | AATAACACAACWCAAYACAACAYAASAAAACT | 4784/4711 |
| 4711-R | GTGATATCATCTAARCTYTGATG | ||
| O-2 | 4689-F | CATCARAGYTTAGATGATATCAC | 5692 |
| 10380-R | CATATCGCGCAAGACTCATATC | ||
| O-3 | SPFMV-O-4160-F | AAGCGACAGAGCAACAGACTTATGTACTA | 1124 |
| SPFMV-O-5284-R | GATATTGCGTTGTAAATTCAACCTCACGTC | ||
| RC-2 | SPFMV-RC-4260-F | AAGTTTGTGAATATGCTACTAGTCATCA | 3814 |
| SPFMV-RC-8220-R | TACCTTCTAGGTCTGTGATCAGTTTAGAC | ||
| RC-3 | SPFMV-RC-7580-F | CCGATAGCTAGCGTCATATGCCATCTCG | 3426 |
| SPFMV-11000-R | TTGGCTCGATCACGAACCAAAAAGGCT |
新窗口打开|下载CSV
1.2 方法
1.2.1 总RNA提取 称取约0.1 g甘薯病样叶片,在液氮中迅速研磨成粉末,按照Trizol总RNA提取试剂盒说明书提取甘薯病样叶片总RNA,保存于-70℃冰箱中备用。1.2.2 RT-PCR扩增基因组序列 第一链cDNA合成参照Reverse Transcriptase XL反转录试剂盒说明书进行,以OligodT为引物,以总RNA为模板合成cDNA。反转录反应体系(10 μL):5×AMV Buffer 2 μL、OligodT引物1 μL、dNTP mixture(10 mmol·L-1)1 μL、AMV反转录酶0.5 μL、RNA酶抑制剂0.5 μL、RNA模板5 μL;42℃保温60 min,16℃保温5 min。PCR反应体系(50 μL):cDNA 5 μL、2×Premix LA Taq DNA聚合酶25 μL、上游和下游引物(10 μmol·L-1)各1 μL、加ddH2O补足至50 μL。反应条件:94℃预变性3 min,94℃变性30 s,53℃退火30 s,根据目的条带大小不同,72℃延伸2—6 min,循环30次后,72℃延伸10 min。RT-PCR扩增产物经1%琼脂糖凝胶电泳检测。将电泳鉴定正确的目的条带利用OMEGA公司的琼脂糖凝胶回收试剂盒对目的片段进行切胶纯化。
1.2.3 PCR产物的克隆及测序 参照TaKaRa公司pMD19-T载体说明书,将纯化的目的片段分别连接至pMD19-T载体,转化至大肠杆菌TG1感受态细胞。经菌液PCR鉴定筛选到阳性克隆,每个样品选取3—5个克隆进行测序,测序由生工生物工程(上海)股份有限公司完成,由宝生物工程(大连)有限公司进行5′和3′端RACE试验。利用BLAST比对测序结果。
1.2.4 全基因组序列拼接及分析 利用DNAMAN软件对测序成功的基因片段进行序列拼接,利用软件MEGA 7.0将SPFMV-O-Ch1和SPFMV-RC-Ch1分离物与GenBank上登录的其他分离物(表1)进行多序列比对分析,利用邻接法(neighbor-joining,NJ)构建系统进化树,系统进化树中各分支置信度(Bootstrap)进行1 000次重复分析。基因重组分析采用Recombination Detection Program(RDP v5.3)软件,利用RDP5软件中的RDP、GENECONV、Chimaera、MaxChi、BootScan、SiScan和3Seq 7种方法对序列进行重组分析,以P≤0.05为标准,当3种以上方法同时检测到重组时,认为其为有意义的重组事件[21,22,23]。
2 结果
2.1 RT-PCR扩增
利用设计的1-F/4711-R、4689-F/10380-R、SPFMV- O-4160-F/SPFMV-O-5284-R、SPFMV-RC-4260-F/ SPFMV- RC-8220-R、SPFMV-RC-7580-F/SPFMV-11000-R等引物扩增SPFMV-O株系中国分离物和SPFMV-RC株系中国分离物的不同基因组片段,扩增产物经1%琼脂糖凝胶电泳分析,结果表明获得了预期大小的目的条带,条带大小分别约为4 790 bp(O-1)、1 270 bp(O-2)、5 700 bp(O-3)、4 710 bp(RC-1)、3 820 bp(RC-2)、3 420 bp(RC-3)(图1)。图1
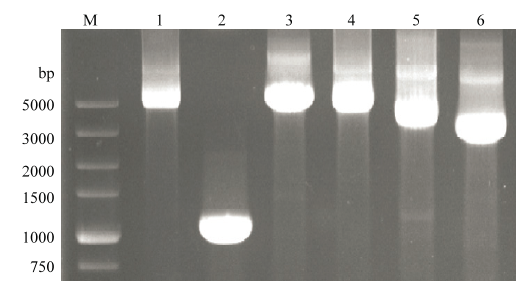 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1O-Ch1和RC-Ch1分离物基因组不同片段RT-PCR产物
M:DL5000分子量标准 DL5000 marker;1:O-1扩增片段Amplification of O-1;2:O-2扩增片段Amplification of O-2;3:O-3扩增片段Amplification of O-3;4:RC-1扩增片段Amplification of RC-1;5:RC-2扩增片段Amplification of RC-2;6:RC-3扩增片段Amplification of RC-3
Fig. 1RT-PCR products of different genomic segments of O-Ch1 and RC-Ch1 isolates
2.2 SPFMV-O和SPFMV-RC株系中国分离物基因组全序列克隆及结构分析
重组质粒经PCR鉴定为阳性,对阳性克隆进行序列测定,通过RACE试验获得了两个分离物的5′和3′末端序列,确保引物及重叠区域碱基无变异,拼接后获得了SPFMV-O和SPFMV-RC株系中国分离物的全基因组序列,分别命名为SPFMV-O-Ch1(GenBank登录号为KY296450)和SPFMV-RC-Ch1(GenBank登录号为KY296451)。SPFMV-O-Ch1和SPFMV-RC- Ch1与已报道的其他分离物基因组结构特征相一致,3′末端均包含一个poly (A)尾序,编码一个大的多聚蛋白,加工产生10个成熟蛋白,从N端到C端依次为P1、HC-Pro、P3、6K1、CI、6K2、VPg、NIa-Pro、NIb和CP。除多聚蛋白外,还通过移码策略分别编码一个P1N-PISPO和P3N-PIPO蛋白。SPFMV-O-Ch1基因组全长为10 922 nt,5′和3′非翻译区(UTR)分别含有114和251 nt,多聚蛋白基因位于基因组的115—10 671处,由10 557 nt组成,多聚蛋白大小为3 518 aa。SPFMV-RC-Ch1基因组全长为10 851 nt,5′和3′非翻译区(UTR)分别含有121和248 nt,多聚蛋白基因位于基因组的122—10 603处,由10 482 nt组成,多聚蛋白含3 493 aa。SPFMV-O-Ch1编码的P1蛋白含有689 aa,SPFMV-RC-Ch1编码的P1蛋白含有664 aa,两者大小不同。两个分离物编码的其他蛋白大小相同,依次为HC-Pro(458 aa)、P3(352 aa)、6K1(52 aa)、CI(643 aa)、6K2(53 aa)、VPg(192 aa)、NIa-Pro(243 aa)、NIb(521 aa)和CP(315 aa)。2.3 基因组序列一致性分析
利用DNAMAN软件对SPFMV-O-Ch1、SPFMV-RC-Ch1分离物以及GenBank中登录的其他48个分离物基因组序列进行比对分析(表3),结果表明SPFMV-O-Ch1与SPFMV-RC-Ch1之间的全序列核苷酸一致性较低,为87.3%,多聚蛋白的核苷酸和氨基酸一致性分别为88.2%和94.0%,属于不同的株系。SPFMV-O-Ch1与GenBank中登录的48个分离物基因组全序列核苷酸一致性为86.0%—95.8%,与乌干达的Ruk73分离物一致性最高(95.8%),与美国的11-1分离物一致性最低(86.0%)。多聚蛋白的比对结果表明,SPFMV-O-Ch1与48个分离物核苷酸一致性范围为87.0%—95.8%,与Ruk73分离物一致性最高,为95.8%,与11-1和澳大利亚Aus3-2BC分离物一致性最低,为87.0%;氨基酸一致性范围为92.1%—97.6%,与阿根廷O-Arg分离物一致性最高,为97.6%,与肯尼亚SRF109a分离物一致性最低,为92.1%。Table 3
表3
表3O-Ch1、RC-Ch1与其他分离物核苷酸和氨基酸序列一致性
Table 3
| 基因组区域 Genome region | O-Ch1蛋白大小及与48个分离物比对 Protein size of O-Ch1 and comparison with 48 isolates | RC-Ch1蛋白大小及与48个分离物比对 Protein size of RC-Ch1 and comparison with 48 isolates | O-Ch1与RC-Ch1比对 Comparison of O-Ch1 and RC-Ch1 | ||||
|---|---|---|---|---|---|---|---|
| 蛋白大小 (氨基酸) Protein size (aa) | 核苷酸 一致性 nt identity (%) | 氨基酸 一致性 aa identity (%) | 蛋白大小 (氨基酸) Protein size (aa) | 核苷酸 一致性 nt identity (%) | 氨基酸 一致性 aa identity (%) | 核苷酸(氨基酸) 一致性 nt (aa) identity (%) | |
| 全长Complete sequence | - | 86.0-95.8 | - | - | 85.9-98.7 | - | 87.3 (-) |
| 多聚蛋白Polyprotein | 3518 | 87.0-95.8 | 92.1-97.6 | 3493 | 87.0-98.8 | 92.8-99.3 | 88.2 (94.0) |
| P1 | 689 | 83.4-97.8 | 81.2-96.4 | 664 | 82.8-99.0 | 80.6-99.2 | 85.4 (83.9) |
| HC | 458 | 83.3-98.2 | 91.9-98.9 | 458 | 83.4-99.0 | 92.1-100.0 | 84.0 (93.2) |
| P3 | 352 | 93.2-98.5 | 95.5-98.6 | 352 | 92.8-99.3 | 94.3-99.4 | 93.8 (96.0) |
| 6K1 | 52 | 92.9-100.0 | 92.3-100.0 | 52 | 91.7-98.7 | 94.2-100.0 | 94.2 (98.1) |
| CI | 643 | 89.0-98.0 | 97.0-99.7 | 643 | 89.7-98.9 | 97.5-99.8 | 89.9 (98.0) |
| 6K2 | 53 | 91.2-98.7 | 94.3-100.0 | 53 | 89.3-98.7 | 96.2-100.0 | 91.8 (98.1) |
| NIa-VPg | 192 | 78.1-97.7 | 90.6-100.0 | 192 | 83.9-99.3 | 89.6-100.0 | 84.4 (97.4) |
| NIa-Pro | 243 | 83.4-96.3 | 93.4-98.8 | 243 | 79.0-98.6 | 92.6-99.6 | 85.9 (96.3) |
| NIb | 521 | 86.3-97.3 | 93.1-98.5 | 521 | 85.3-98.7 | 92.9-99.8 | 89.5 (96.2) |
| CP | 315 | 76.7-96.6 | 94.0-99.7 | 315 | 79.9-98.9 | 95.6-99.7 | 92.5 (96.8) |
新窗口打开|下载CSV
SPFMV-RC-Ch1与其他48个分离物全基因组核苷酸一致性为85.9%—98.7%,与韩国的IS90分离物一致性最高(98.7%),与澳大利亚的Aus1-2B分离物一致性最低(85.9%)。多聚蛋白的比对结果表明,SPFMV-RC-Ch1与韩国IS90分离物的核苷酸和氨基酸一致性最高,分别为98.8%和99.3%,与澳大利亚Aus1-2B分离物的核苷酸和氨基酸一致性最低,分别为87.0%和92.8%。
在SPFMV编码的10个蛋白中,P1蛋白变异最大,O-Ch1、RC-Ch1分离物与48个分离物氨基酸序列一致性分别为81.2%—96.4%和80.6%—99.2%,而且不同株系P1蛋白的大小不同,属于O株系的O-Ch1分离物P1蛋白包含689 aa,属于RC株系的RC-Ch1分离物P1蛋白包含664 aa,而属于EA株系的Piu3分离物P1蛋白包含724 aa。最保守的是CI蛋白,O-Ch1、RC-Ch1分离物与48个分离物氨基酸序列一致性分别为97.0%—99.7%和97.5%—99.8%。
2.4 系统进化关系分析
基于SPFMV多聚蛋白基因核苷酸序列构建的系统进化树分析表明,O-Ch1分离物与来自乌干达的O株系Ruk73分离物形成一个小分支,亲缘关系最近,与其他属于O株系的分离物在一个大分支上,而与来自秘鲁的EA株系Piu3分离物亲缘关系也较近。RC-Ch1分离物与来自韩国RC株系的IS90分离物聚类到同一个小分支上,亲缘关系最近,与属于RC株系其他31个分离物形成一个大分支。对变异最大的P1蛋白基因核苷酸序列构建进化树,结果表明O-Ch1分离物与阿根廷的O-Arg、日本的Ordinary及17-O等属于O株系的分离物形成一个小分支,亲缘关系较近。RC-Ch1分离物与RC株系的IS90分离物(韩国)聚类在同一个小分支上,亲缘关系最近(图2)。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2SPFMV分离物多聚蛋白和P1蛋白核苷酸序列遗传进化分析
Fig. 2Phylogenetic trees reconstructed using the nucleotide sequences of polyprotein and P1 protein of SPFMV isolates
比较保守的CI蛋白核苷酸序列进化树分析结果显示,O-Ch1分离物与来自东帝汶的O株系TM66B分离物亲缘关系最近,RC-Ch1分离物与IS90分离物亲缘关系最近。对CP蛋白核苷酸序列构建进化树,结果与P1蛋白系统进化树相似,O-Ch1与O-Arg、10-O、Ordinary及17-O等属于O株系的分离物形成一个小分支,亲缘关系较近,RC-Ch1与IS90分离物在同一个小分支上,亲缘关系最近(图3)。
图3
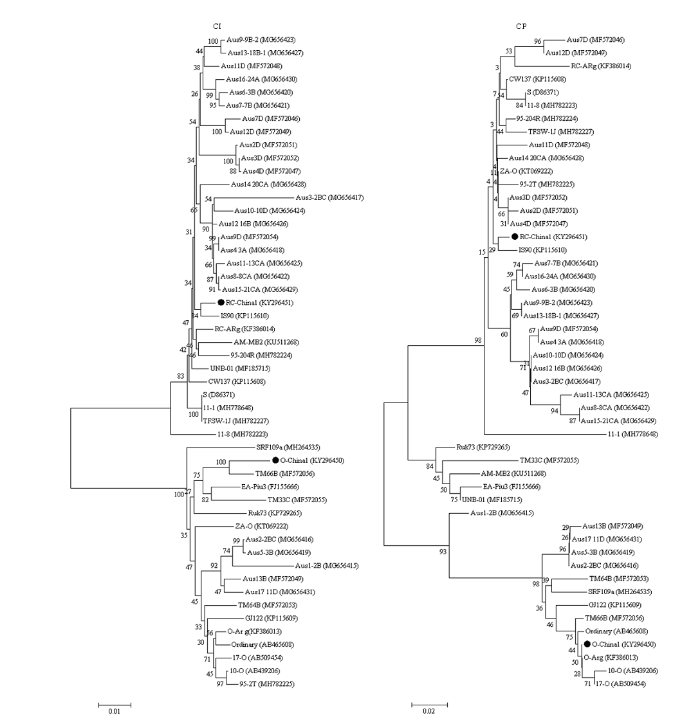 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3SPFMV分离物CI蛋白和CP蛋白核苷酸序列遗传进化分析
Fig. 3Phylogenetic trees reconstructed using the nucleotide sequences of CI and CP proteins of SPFMV isolates
综上所述,基于O-Ch1分离物基因组不同编码区域构建的系统进化树,亲缘关系最近的分离物不尽相同;而RC-Ch1分离物基因组不同编码区域构建的系统进化树分析结果相一致,均与来自韩国RC株系的IS90分离物亲缘关系最近。推测O-Ch1分离物可能发生了重组。
2.5 基因组全序列重组分析
为了明确我国的O-Ch1、RC-Ch1分离物是否存在潜在的重组事件,利用RDP5软件中的7种算法(P≤0.05为显著),将O-Ch1、RC-Ch1与GenBank登录的其他48个分离物基因组全序列进行重组分析。结果表明(表4),在O-Ch1分离物中检测到3个潜在的重组事件(其他分离物重组事件未列出)。重组事件1发生在7 731—9 710 nt,获得7种算法支持,最低P值为1.095×10-44(3Seq),最高P值为8.317× 10-17(GENECONV),主要亲本为日本的O株系Ordinary分离物,次要亲本为东帝汶的O株系TM33C分离物。重组事件2发生在135—10 012 nt,获得4种算法支持,最低P值为1.561×10-56(3Seq),最高P值为7.437× 10-3(Chimaera),主要亲本为秘鲁的EA株系Piu3分离物,次要亲本为澳大利亚的O株系Aus5-3B分离物。重组事件3发生在4 825—6 948 nt,获得3种算法支持,最低P值为1.104×10-3(MaxChi),最高P值为1.256×10-2(3Seq),主要亲本为澳大利亚的O株系Aus13B分离物,次要亲本为秘鲁的EA株系Piu3分离物。RC-Ch1分离物未检测到重组事件发生。Table 4
表4
表4O-Ch1分离物的重组分析
Table 4
| 序号 Number | 主要亲本 Major parent | 次要亲本 Minor parent | 位置 Position (nt) | P值P-Value | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RDP | GENECONV | BootScan | MaxChi | Chimaera | SiScan | 3Seq | ||||
| 1 | Ordinary- AB465608 | TM33C- MF572055 | 7731- 9710 | 5.583×10-33 | 8.317×10-17 | 7.407×10-31 | 3.758×10-20 | 3.466×10-22 | 2.430×10-25 | 1.095×10-44 |
| 2 | EA-Piu3- FJ155666 | Aus5-3B- MG656419 | 135- 10012 | - | - | - | 8.783×10-4 | 7.437×10-3 | 1.803×10-20 | 1.561×10-56 |
| 3 | Aus13B- MF572050 | EA-Piu3- FJ155666 | 4825- 6948 | - | - | - | 1.104×10-3 | 1.071×10-2 | - | 1.256×10-2 |
新窗口打开|下载CSV
3 讨论
SPFMV是甘薯上分布最广泛的病毒之一,其可以与SPCSV发生协生作用,形成毁灭性病害SPVD。自我国首次检测到SPFMV的发生以来,SPFMV在我国逐渐扩散蔓延,成为甘薯上最重要的病毒之一。国内对SPFMV开展了多项研究,主要集中在病毒的分离纯化[2,24-25]、生物学性状[26]、抗体制备[27]、检测技术[28,29,30,31,32,33,34,35,36,37,38]、分布及遗传多样性[9-11,39-40]等方面,而对SPFMV中国分离物的基因组序列测定还未开展相关研究。笔者研究室前期对SPFMV的发生、分布及分子变异等进行了研究,明确了我国SPFMV的主要株系类型为O和RC,在此基础上,继续对我国SPFMV O株系和RC株系的基因组开展了相关研究,利用RT-PCR方法结合RACE技术获得了这两个主要株系的基因组全序列,获得的O-Ch1和RC-Ch1分离物是中国SPFMV完整基因组的首次报道。对O-Ch1和RC-Ch1两个分离物的基因组结构及编码的主要蛋白分析发现,O株系和RC株系编码的P1蛋白大小不同,不同株系(分离物)之间P1蛋白的氨基酸一致性最低,变异最大;而其余蛋白大小均相同,氨基酸一致性高于P1蛋白。YAMASAKI等[14]对SPVC和SPFMV的O、RC和EA 3个株系代表分离物的基因组进行了比较分析,表明不同株系之间P1蛋白的核苷酸和氨基酸一致性低于其余9个蛋白;KWAK等[15]对韩国SPFMV等5种侵染甘薯的马铃薯Y病毒属病毒基因组分析结果表明,无论是种间还是株系间,P1蛋白的变异均最大。本研究结果与前人的报道[14,15]相一致。
早期的研究表明,依据CP基因序列分析,SPFMV可分为C、O、RC和EA 4个株系,C株系与其他3个株系遗传关系较远,已将其作为新种重新命名为甘薯病毒C(Sweet potato virus C,SPVC),EA株系仅在东非的一些地区存在,而O和RC株系不存在地理局限性[41]。近年来,随着种质资源的扩散和种薯的调运,EA株系现在已不仅局限于东非地区,我国已报道了EA株系的存在[10]。本研究获得的O-Ch1和RC-Ch1两个分离物分别与O株系和RC株系的分离物聚类成一个分支,O-Ch1变异较大,推测可能是由于O株系为我国的优势株系,地理分布上最广泛,更易随着频繁种质资源交换和调运,而发生变异重组等。
SPFMV经常发生种内重组,不同株系之间重组很常见,比如对10-O分离物的分析结果表明,该分离物是由O、RC和EA 3个株系重组形成的,P1蛋白是重组的热点区域[20]。韩国的GJ122分离物与10-O分离物类似,也发生了株系间重组,在P1蛋白区域内存在重组位点,是由O株系和RC株系重组产生的[15]。本研究获得的O-Ch1分离物可能是由O株系和EA株系重组产生的,重组位点发生在NIa-NIb基因、P1基因、CI基因和CP基因处。由于田间常常发生SPFMV不同株系混合侵染现象,不同株系基因组之间容易发生重组变异,从而增强病毒群体的适应能力,种内重组可能是造成SPFMV株系分化较复杂的主要原因,不同株系之间的重组还可能形成新的株系,甚至产生更多的新种等,分析病毒的重组并明确重组热点区域对了解病毒的进化至关重要。
4 结论
完成了甘薯羽状斑驳病毒O株系和RC株系中国分离物基因组全序列测定。基因组结构分析结果显示,我国两个分离物与其他国家分离物的基因组结构相一致。两个分离物之间序列相似性较低,变异较大,分别属于两个不同的株系。遗传进化树分析结果表明,SPFMV-O-Ch1与O株系的分离物形成一个分支,SPFMV-RC-Ch1与RC株系的分离物形成一个分支,基因组重组分析结果显示,O-Ch1分离物检测到3个重组事件,RC-Ch1未检测到重组事件。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1094/PDIS-93-9-0933URL [本文引用: 2]
[本文引用: 2]
[本文引用: 2]
DOI:10.1023/A:1025172402230URL [本文引用: 1]

The 3
 -proximal part (1.8
-proximal part (1.8 kb) of the Sweet potato feathery mottle virus (SPFMV) genome was studied in four SPFMV isolates collected from farmers' fields in western Uganda (SPFMV-Bny), eastern Uganda (SPFMV-Sor) and Bagamoyo district, Tanzania (SPFMV-TZ1 and SPFMV-TZ2). Unlike the other three SPFMV isolates, SPFMV-Sor was not detected with the polyclonal antisera to SPFMV. It showed moderately high coat protein (CP) nucleotide (93.3–96.7%) and amino acid (93.6–96.8%) sequence identity to the isolates of the SPFMV strain group C. In contrast, identities (78.1–80.1%, and 79.9–83.1%) to isolates of the SPFMV strain groups O, RC, and the East African (EA) strain group were low. Similar to some isolates (SPFMV-CH2 and SPFMV-6) of strain group C, but different from other SPFMV isolates, SPFMV-Sor contained a deletion of 6 nucleotides in the CP-encoding region (CP amino acid positions 62–63). Phylogenetic analysis of the CP sequences indicated that SPFMV-Sor belongs to the SPFMV strain group C that has not been reported from Africa. Sequence data were obtained for the first time from Tanzanian SPFMV isolates in this study, and phylogenetic analysis indicated that they belong to the strain group EA, which is unique to East Africa.
kb) of the Sweet potato feathery mottle virus (SPFMV) genome was studied in four SPFMV isolates collected from farmers' fields in western Uganda (SPFMV-Bny), eastern Uganda (SPFMV-Sor) and Bagamoyo district, Tanzania (SPFMV-TZ1 and SPFMV-TZ2). Unlike the other three SPFMV isolates, SPFMV-Sor was not detected with the polyclonal antisera to SPFMV. It showed moderately high coat protein (CP) nucleotide (93.3–96.7%) and amino acid (93.6–96.8%) sequence identity to the isolates of the SPFMV strain group C. In contrast, identities (78.1–80.1%, and 79.9–83.1%) to isolates of the SPFMV strain groups O, RC, and the East African (EA) strain group were low. Similar to some isolates (SPFMV-CH2 and SPFMV-6) of strain group C, but different from other SPFMV isolates, SPFMV-Sor contained a deletion of 6 nucleotides in the CP-encoding region (CP amino acid positions 62–63). Phylogenetic analysis of the CP sequences indicated that SPFMV-Sor belongs to the SPFMV strain group C that has not been reported from Africa. Sequence data were obtained for the first time from Tanzanian SPFMV isolates in this study, and phylogenetic analysis indicated that they belong to the strain group EA, which is unique to East Africa.[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.1007/s00705-012-1503-8URL [本文引用: 3]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
DOI:10.1016/j.virol.2009.03.024URL [本文引用: 1]
[本文引用: 3]
[本文引用: 4]
DOI:10.1128/JVI.02360-15URL [本文引用: 2]
DOI:10.1094/PDIS-08-17-1156-REURL [本文引用: 1]
DOI:10.1094/PDIS-12-17-1972-REURL [本文引用: 1]
DOI:10.1094/PDIS-07-11-0550URL [本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
