 ,1,2
,1,2Development and Application of the Functional Marker for the Broad-Spectrum Blast Resistance Gene PigmR in Rice
WANG FangQuan1,2, CHEN ZhiHui1,2, XU Yang1,2, WANG Jun1,2, LI WenQi1,2, FAN FangJun1,2, CHEN LiQin1, TAO YaJun1,2, ZHONG WeiGong1, YANG Jie ,1,2
,1,2通讯作者:
责任编辑: 李莉
收稿日期:2018-11-18接受日期:2019-01-18网络出版日期:2019-03-16
| 基金资助: |
Received:2018-11-18Accepted:2019-01-18Online:2019-03-16
作者简介 About authors
王芳权,Tel:025-84390320;E-mail: wfqjaas@163.com。

摘要
关键词:
Abstract
Keywords:
PDF (3044KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王芳权, 陈智慧, 许扬, 王军, 李文奇, 范方军, 陈丽琴, 陶亚军, 仲维功, 杨杰. 水稻广谱抗稻瘟病基因PigmR功能标记的开发及应用[J]. 中国农业科学, 2019, 52(6): 955-967 doi:10.3864/j.issn.0578-1752.2019.06.001
WANG FangQuan, CHEN ZhiHui, XU Yang, WANG Jun, LI WenQi, FAN FangJun, CHEN LiQin, TAO YaJun, ZHONG WeiGong, YANG Jie.
0 引言
【研究意义】水稻是全球重要的粮食作物之一,保证水稻高产稳产具有重要的生产和社会意义。稻瘟病是危害水稻稳产、优质和安全生产的主要因素之一,严重时可导致水稻减产40%—50%,甚至颗粒无收[1]。近几年,江淮稻区稻瘟病频发,给国家造成大量经济损失[2]。培育与利用抗病品种是解决稻瘟病危害最经济有效的方式[3],而这离不开抗病基因的挖掘与利用。【前人研究进展】目前,已经定位得到了100多个水稻稻瘟病抗性相关基因,其中30多个基因被成功克隆,包括Pib[4]、Pi-ta[5]、Pi54[6]、pi21[7]、bsr-d1[8]、Piz-t[9]、Pi9[10]、Pi50[11,12]、Pigm[13,14]、Ptr[15]等。PigmR是一个广谱抗稻瘟病基因,来源于谷梅4号的Pigm/Pi2-Pi9基因簇[13,14]。Pigm/Pi2-Pi9基因簇在不同品种或资源材料中的R基因拷贝数不同,例如,日本晴含有7个R基因(R1、R2、R3、R9、R10、R12和R13),9311含有5个R基因(R1、R2、R3、R11和R12),谷梅4号含有13个R基因(R1—R13)[14]。在谷梅4号的Pigm/Pi2-Pi9基因簇中,R4、R6和R8能形成完整的转录产物。R6为抗病基因,是具有功能的Pigm,称为PigmR,在日本晴、9311等大多数品种或种质中不存在等位基因;R4与PigmR在蛋白质水平上存在4个氨基酸的差异,不具有功能,称为Pigm-R4;R8为感病基因,称为PigmS。研究发现,PigmR对来源于世界各地的50个稻瘟病供试菌株表现抗性[14,16],具有抗谱广、抗性强的特点,有很好的应用前景。传统的水稻抗稻瘟病遗传育种是通过对植株进行稻瘟病自然诱发或人工接种的方式进行表型鉴定,人力物力投入大,且受外界因素影响大,鉴定结果往往存在较大误差。分子标记辅助选择是提高抗稻瘟病水稻育种效率的有效途径[17,18]。【本研究切入点】目前,已报道的针对抗病基因PigmR的标记大多基于Pigm/Pi2-Pi9基因簇开发,如Indel587、Pigm-4、G1408、Pigm-SM等[19,20,21,22],与PigmR存在一定的分离概率,在分子标记辅助选择育种过程中,可能存在连锁累赘或丢失目标基因的问题[20]。功能标记是基于基因内突变位点开发的,与基因完全共分离,能有效解决连锁标记的问题。【拟解决的关键问题】本研究根据抗病基因PigmR的序列特点,设计并优化功能标记。利用功能标记进行分子标记辅助选择,以获得穗颈瘟抗性显著改善的水稻新材料,加快该基因在抗稻瘟病水稻育种中的应用。1 材料与方法
1.1 试验材料
水稻材料包括PigmR供体材料谷梅4号在内的水稻亲本材料229份,丽江新团黑谷单基因系材料29份,2017—2018年江苏省迟熟中粳预备试验材料216份和早熟晚粳区域试验(早熟组)材料24份。遗传群体为南粳53045/谷梅4号的BC1F2和BC1F3群体。29份丽江新团黑谷单基因系材料由国际水稻研究所提供,其他试验材料均由江苏省农业科学院收集和保存。水稻材料均种植于试验田,以常规方法栽培。1.2 PCR扩增与电泳检测
采用CTAB法提取水稻样品的基因组DNA。应用Oligo 7软件设计分子标记引物(由Invitrogen公司合成),进行PCR扩增。PCR反应的体系为基因组DNA 2 μL、10×PCR buffer 2 μL、MgCl2(5 mmol·L-1)2 μL、dNTP(2 mmol·L-1)2 μL、上游引物(2 μmol·L-1)2 μL、下游引物(2 μmol·L-1)2 μL、Taq酶0.2 μL和ddH2O 7.8 μL。扩增条件为94℃ 5 min;94℃ 30 s,55℃ 30 s,72℃ 30 s,35个循环;72℃ 10 min。扩增产物经琼脂糖凝胶电泳分离,用凝胶成像仪拍照,并记录结果。1.3 序列分析
谷梅4号Pigm/Pi2-Pi9基因簇参考序列的GenBank登录号为KU904633[14]。采用Snapgene 2.3.2软件进行核酸序列分析。1.4 表型鉴定
在江苏省农业科学院试验田进行稻瘟病表型鉴定。稻瘟病的供试菌株为江苏省农业科学院植物保护研究所分离得到的江苏省稻瘟病代表菌株(2018-4、2018-65、2018-102、2018-222和2018-241)的混合菌。将供试菌株移植RCA(玉米粉40 g、稻秆50 g和琼脂20 g)培养基上,25℃培养7 d,用黑光灯照射72 h,待稻瘟病菌产生孢子后,再用无菌水洗下,配成10×10倍显微镜下每视野30—40个孢子的悬浮液。采用人工注射接种法进行水稻穗颈瘟的抗性鉴定,于水稻抽穗前3—4 d注射混合菌株,每穗注射1 mL菌液,做好标记。在水稻灌浆成熟后进行抗性调查,参照国际水稻研究所分级标准调查穗颈瘟抗性级别[23]。2 结果
2.1 PigmR序列分析及功能标记的设计
Pigm基因簇中,PigmR与Pigm-R4、PigmS高度同源且紧密连锁,PigmR为有功能的Pigm,Pigm-R4不具有功能,PigmS为感病基因。通过序列比对发现,PigmR与PigmS的编码区存在82个碱基差异,PigmR与Pigm-R4的编码区存在6个碱基差异,其中第2 557、2 572、2 579、2 881位为PigmR与PigmS、Pigm-R4均存在差异的碱基(图1)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1PigmR与部分同源基因在变异位点的序列比对
Fig. 1Sequence alignment of the variation sites between PigmR and parts of its homologous genes
为了开发PigmR特异的功能标记,根据PigmR与PigmS、Pigm-R4均存在的差异位点,设计了8对基因特异检测引物(表1),其中GMR-1、GMR-2、GMR-3和GMR-4用于特异检测PigmR,GMS-1、GMS-2、GMS-3和GMS-4用于特异检测PigmS。部分引物中引入了碱基错配,GMR-2F、GMS-2F、GMR-4F和GMS-4F引物5′-3′的倒3位碱基由A错配为C,GMR-4R和GMS-4R引物5′-3′的倒3位碱基由G错配为C。PigmR在很多品种中不存在,因此,还设计了2对特异检测内参基因Actin1的分子标记(表1),以排除样品、试验操作等过程中引起的假阴性。
Table 1
表1
表1分子标记引物
Table 1
| 标记名称 Marker name | 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 理论产物长度 Expected size (bp) |
|---|---|---|---|
| GMR-1 | GMR-1F | TTCCTTTCTCTTTATCATAATT | 180 |
| GMR-1R | AGATTCCAACCTGCACTTGCCT | ||
| GMS-1 | GMS-1F | GTCCTTGATCTTTATCAGAAAG | 180 |
| GMS-1R | TGGTTCCAACCTGCAGAATCTG | ||
| GMR-2 | GMR-2F | TTCCTTTCTCTTTATCATAcTT | 180 |
| GMR-2R | AGATTCCAACCTGCACTTGCCT | ||
| GMS-2 | GMS-2F | GTCCTTGATCTTTATCAGAcAG | 180 |
| GMS-2R | TGGTTCCAACCTGCAGAATCTG | ||
| GMR-3 | GMR-3F | AGTTCTACTTACGGAGGAGC | 98 |
| GMR-3R | AGAATTATGATAAAGAGAAAGGAA | ||
| GMS-3 | GMS-3F | AGATCAACTTATTGAGCAGC | 98 |
| GMS-3R | AGCTTTCTGATAAAGATCAAGGAC | ||
| GMR-4 | GMR-4F | AGTTCTACTTACGGAGGcGC | 98 |
| GMR-4R | AGAATTATGATAAAGAGAAAGcAA | ||
| GMS-4 | GMS-4F | AGATCAACTTATTGAGCcGC | 98 |
| GMS-4R | AGCTTTCTGATAAAGATCAAGcAC | ||
| Actin1-1 | Actin1-1F | ACAAAGTTTTCAACCGGCCTA | 146 |
| Actin1-1R | CTGGTACCCTCATCAGGCATC | ||
| Actin1-2 | Actin1-2F | AGTCTGGCCCATCCATTGTG | 149 |
| Actin1-2R | CGGTTGAAAACTTTGTCCACGCTA |
新窗口打开|下载CSV
2.2 功能标记的检测及优化
以谷梅4号、日本晴和黄华占的基因组为模板,对设计的分子标记进行PCR筛选(图2)。结果发现,GMR-1、GMR-2和GMR-4 3个标记对谷梅4号、日本晴和黄华占3个品种均无条带;GMS-1、GMS-2和GMS-3 3个标记能在谷梅4号和日本晴中扩增出条带;GMR-3和GMS-4仅能在谷梅4号中扩增出条带。根据设计,GMR-3特异检测的是抗病基因PigmR,GMS-4特异检测的是感病基因PigmS,因此,选取GMR-3进行条件优化。图2
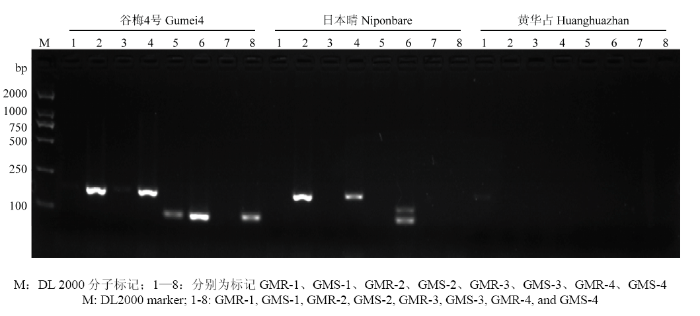 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2PigmR分子标记的扩增结果
M:DL 2000分子标记;1—8:分别为标记GMR-1、GMS-1、GMR-2、GMS-2、GMR-3、GMS-3、GMR-4、GMS-4
Fig. 2The PCR amplification products of molecular maker for PigmR
M: DL2000 marker; 1-8: GMR-1, GMS-1, GMR-2, GMS-2, GMR-3, GMS-3, GMR-4, and GMS-4
为进一步分析GMR-3对PigmR的特异性,将PigmR与Pi9、Pi2和Piz-t进行比较,发现GMR-3上下游引物能特异匹配PigmR(图1)。
PigmR在很多品种或材料中不存在,因此还设计了特异检测内参基因Actin1的分子标记(表1),以排除样品、试验操作等过程中引起的假阴性。检测结果发现,Actin1-1和Actin1-2均能对谷梅4号、日本晴和黄华占扩增出特异明亮条带(图3)。为优化引物的浓度配比,选取GMR-3和Actin1-1引物,设计12个不同浓度试验(图4)。结果发现,0.4 μmol·L-1 GMR-3与0.1 μmol·L-1 Actin1-1组合浓度扩增出PigmR和Actin1特征条带的效率相当,效果最优(图4)。将0.4 μmol·L-1 GMR-3和0.1 μmol·L-1 Actin1-1引物混合组成的分子标记命名为GMRA。携带PigmR的样品能被GMRA同时扩增出146和98 bp条带,不携带PigmR的样品仅能被GMRA扩增出146 bp条带。
图3
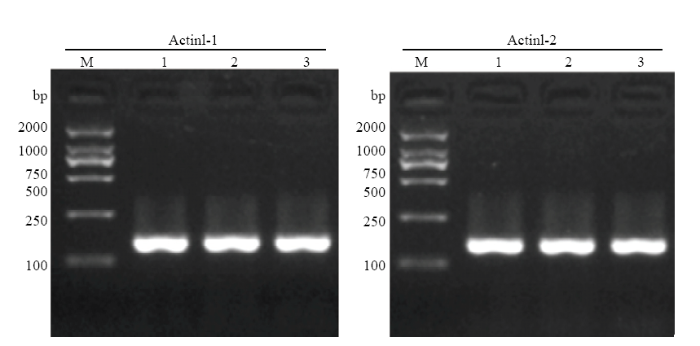 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3Actin1鉴定分子标记的扩增结果
M:DL 2000分子标记;1—3分别表示谷梅4号、日本晴和黄华占
Fig. 3PCR amplification products of molecular maker for Actin1
M: DL2000 marker; 1-3 represent Gumei 4, Niponbare, and Huanghuazhan
图4
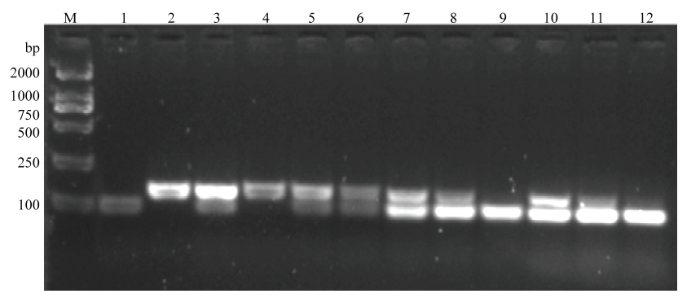 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4PigmR鉴定引物的优化
M:DL2000;1—12:12个不同引物浓度组合,分别为0.2 μmol·L-1 GMR-3+0 μmol·L-1Actin1-1、0 μmol·L-1 GMR-3+0.2 μmol·L-1 Actin1-1、0.2 μmol·L-1 GMR-3+0.2 μmol·L-1 Actin1-1、0.1 μmol·L-1 GMR-3+0.1 μmol·L-1 Actin1-1、0.2 μmol·L-1 GMR-3+0.1 μmol·L-1 Actin1-1、0.2 μmol·L-1 GMR-3+0.05 μmol·L-1 Actin1-1、0.4 μmol·L-1 GMR-3+0.1 μmol·L-1 Actin1-1、0.4 μmol·L-1 GMR-3+0.05 μmol·L-1 Actin1-1、0.4 μmol·L-1 GMR-3+0.025 μmol·L-1 Actin1-1、0.8 μmol·L-1 GMR-3+0.1 μmol·L-1 Actin1-1、0.8 μmol·L-1 GMR-3+0.05 μmol·L-1 Actin1-1和0.8 μmol·L-1 GMR-3+0.025 μmol·L-1 Actin1-1
Fig. 4Optimization of identification primers for PigmR
M: DL2000 marker; 1-12: Represent the combination with the different concentration of primers, including 0.2 μmol·L-1 GMR-3+0 μmol·L-1 Actin1-1, 0 μmol·L-1 GMR-3+0.2 μmol·L-1 Actin1-1, 0.2 μmol·L-1 GMR-3+0.2 μmol·L-1 Actin1-1, 0.1 μmol·L-1 GMR-3+0.1 μmol·L-1 Actin1-1, 0.2 μmol·L-1 GMR-3+0.1 μmol·L-1 Actin1-1, 0.2 μmol·L-1 GMR-3+0.05 μmol·L-1 Actin1-1, 0.4 μmol·L-1 GMR-3+0.1 μmol·L-1 Actin1-1, 0.4 μmol·L-1 GMR-3+0.05 μmol·L-1 Actin1-1, 0.4 μmol·L-1 GMR-3+0.025 μmol·L-1 Actin1-1, 0.8 μmol·L-1 GMR-3+0.1 μmol·L-1 Actin1-1, 0.8 μmol·L-1 GMR-3+0.05 μmol·L-1 Actin1-1, and 0.8 μmol·L-1 GMR-3+0.025 μmol·L-1 Actin1-1
2.3 GMRA标记检测水稻材料
利用GMRA标记检测229份水稻亲本材料,结果表明,只有谷梅4号能检测到PigmR条带,其他粳稻和籼稻品种均不能扩增出98 bp特征条带(图5和表2)。为进一步验证GMRA的特异性,还检测了29份丽江新团黑谷单基因系,发现这些材料均无PigmR特征条带,表明GMRA有很好的特异性,且能区分PigmR与Pi9、Piz、Piz-t等同源性高的基因(电子附图1和表3)。图5
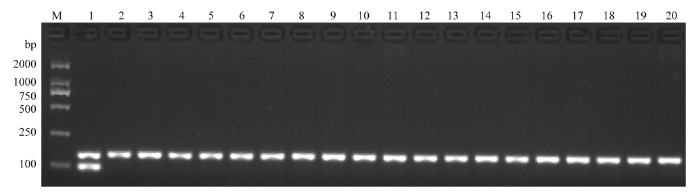 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5GMRA标记检测水稻品种/品系
M:DL2000分子标记;1—20:分别为谷梅4号、日本晴、黄华占、IR36、玉针香、南京16号、Kasalath、连粳7号、南粳9108、武运粳24号、华粳5号、扬育粳2号、南粳44、南粳5055、金粳818、苏秀867、武运粳21号、武运粳27号、淮稻5号、南粳53045
Fig. 5Molecular detection for partial varieties by GMRA
M: DL2000 marker; 1-20: Represent Gumei 4, Niponbare, Huanghuazhan, IR36, Yuzhenxiang, Nanjing 16, Kasalath, Liangeng 7, Nangeng 9108, Wuyungeng 24, Huageng 5, Yangyugeng 2, Nangeng 44, Nangeng 5055, Jingeng 818, Suxiu 867, Wuyungeng 21, Wuyungeng 27, Huaidao 5, and Nangeng53045
Table 2
表2
表2GMRA标记鉴定部分水稻材料
Table 2
| 编号 No. | 品种 Variety | 粳/籼稻 Geng/Xian | 带型 Type | 编号 No. | 品种 Variety | 粳/籼稻 Geng/Xian | 带型 Type | |
|---|---|---|---|---|---|---|---|---|
| 1 | 谷梅4号 Gumei 4 | X | 2 | 40 | 盐粳10号 Yandao 10 | G | 1 | |
| 2 | 日本晴 Niponbare | G | 1 | 41 | 盐粳11号 Yangeng 11 | G | 1 | |
| 3 | 黄华占 Huanghuazhan | X | 1 | 42 | 镇稻99 Zhendao 99 | G | 1 | |
| 4 | IR36 | X | 1 | 43 | 扬粳113 Yanggeng 113 | G | 1 | |
| 5 | 玉针香 Yuzhenxiang | X | 1 | 44 | WP16 | G | 1 | |
| 6 | 南京16号 Nanjing 16 | X | 1 | 45 | WP59 | G | 1 | |
| 7 | Kasalath | X | 1 | 46 | 泗稻15号 Sidao 15 | G | 1 | |
| 8 | 连粳7号 Liangeng 7 | G | 1 | 47 | 连糯1号 Liannuo 1 | G | 1 | |
| 9 | 南粳9108 Nangeng 9108 | G | 1 | 48 | 皖粳糯1号 Liangengnuo 1 | G | 1 | |
| 10 | 武运粳24号 Wuyungeng 24 | G | 1 | 49 | 圣稻19 Shengdao 19 | G | 1 | |
| 11 | 华粳5号 Huageng 5 | G | 1 | 50 | JX14-3 | G | 1 | |
| 12 | 扬育粳2号 Yangyugeng 2 | G | 1 | 51 | JX14-4 | G | 1 | |
| 13 | 南粳44 Nangeng 44 | G | 1 | 52 | K11 | G | 1 | |
| 14 | 南粳45 Nangeng 45 | G | 1 | 53 | N14-109 | G | 1 | |
| 15 | 南粳46 Nangeng 46 | G | 1 | 54 | 宁粳7号 Ninggeng 7 | G | 1 | |
| 16 | 南粳5055 Nangeng 5055 | G | 1 | 55 | YJN1 | G | 1 | |
| 17 | 金粳818 Jingeng 818 | G | 1 | 56 | 常粳144 Changgeng 144 | G | 1 | |
| 18 | 苏秀867 Suxiu 867 | G | 1 | 57 | 常农粳4号 Changnonggeng 4 | G | 1 | |
| 19 | 武运粳21号 Wuyungeng 21 | G | 1 | 58 | 常农粳6号 Changnonggeng 6 | G | 1 | |
| 20 | 武运粳27号 Wuyungeng 27 | G | 1 | 59 | 常农粳7号 Changnonggeng 7 | G | 1 | |
| 21 | 淮稻5号 Huaidao 5 | G | 1 | 60 | Feng1326 | G | 1 | |
| 22 | 南粳53045 Nangeng53045 | G | 1 | 61 | 华粳3号 Huageng 3 | G | 1 | |
| 23 | 武运粳29号 Wuyungeng 29 | G | 1 | 62 | 华粳4号 Huageng 4 | G | 1 | |
| 24 | 楚粳39 Chugeng 39 | G | 1 | 63 | 华粳6号 Huageng 6 | G | 1 | |
| 25 | 丰粳3227 Fenggeng 3227 | G | 1 | 64 | 华瑞稻1号 Huaruidao 1 | G | 1 | |
| 26 | 华粳7号 Huageng 7 | G | 1 | 65 | 淮1188 Huai 1188 | G | 1 | |
| 27 | 淮稻7号 Huaidao 7 | G | 1 | 66 | 淮208 Huai 208 | G | 1 | |
| 28 | 淮糯11号 Huainuo 11 | G | 1 | 67 | 淮稻10号 Huaidao 10 | G | 1 | |
| 29 | 淮优粳2号 Huaiyougeng 2 | G | 1 | 68 | 淮稻11号 Huaidao 11 | G | 1 | |
| 30 | 连粳11号 Liangeng 11 | G | 1 | 69 | 淮稻13号 Huaidao 13 | G | 1 | |
| 31 | 连粳12号 Liangeng 12 | G | 1 | 70 | 淮稻8号 Huaidao 8 | G | 1 | |
| 32 | 连粳4号 Liangeng 4 | G | 1 | 71 | 淮稻9号 Huaidao 9 | G | 1 | |
| 33 | 连粳6号 Liangeng 6 | G | 1 | 72 | 淮粳096 Huaigeng 096 | G | 1 | |
| 34 | 连粳147729 Liangeng 147729 | G | 1 | 73 | 淮香稻15号 Huaixiangdao 15 | G | 1 | |
| 35 | 农香软米 Nongxiangruanmi | G | 1 | 74 | 津稻263 Jindao 263 | G | 1 | |
| 36 | 泗稻785 Sidao 785 | G | 1 | 75 | 连粳9号 Liangeng 9 | G | 1 | |
| 37 | 苏秀326 Suxiu 326 | G | 1 | 76 | 连粳10号 Liangeng 10 | G | 1 | |
| 38 | 徐稻3号 Xudao 3 | G | 1 | 77 | 南粳40 Nangeng 40 | G | 1 | |
| 39 | 盐稻11号 Yandao 11 | G | 1 | 78 | 南粳41 Nangeng 41 | G | 1 | |
| 79 | 南粳42 Nangeng 42 | G | 1 | 119 | 镇稻7号 Zhendao 7 | G | 1 | |
| 80 | 南粳49 Nangeng 49 | G | 1 | 120 | 镇稻88 Zhendao 88 | G | 1 | |
| 81 | 宁2600 Ning 2600 | G | 1 | 121 | 镇稻9424 Zhendao 9424 | G | 1 | |
| 82 | 宁粳2号 Ninggeng 2 | G | 1 | 122 | 宁粳1号 Ninggeng 1 | G | 1 | |
| 83 | 宁粳4号 Ninggeng 4 | G | 1 | 123 | 宁粳3号 Ninggeng 3 | G | 1 | |
| 84 | 宁粳5号 Ninggeng 5 | G | 1 | 124 | 南粳47 Nangeng 47 | G | 1 | |
| 85 | 迁稻11-72 Qiandao 11-72 | G | 1 | 125 | 通粳981 Tonggeng 981 | G | 1 | |
| 86 | 泗稻12号 Sidao 12 | G | 1 | 126 | 武运粳7号 Wuyungeng 7 | G | 1 | |
| 87 | 苏沪香粳 Suhuxianggeng | G | 1 | 127 | 武运粳19号 Wuyungeng 19 | G | 1 | |
| 88 | 苏粳5号 Sugeng 5 | G | 1 | 128 | 武运粳23号 Wuyungeng 23 | G | 1 | |
| 89 | 苏秀10号 Suxiu 10 | G | 1 | 129 | 常农粳3号 Changnonggeng 3 | G | 1 | |
| 90 | 武粳13号 Wugeng 13 | G | 1 | 130 | 常农粳5号 Changnonggeng 5 | G | 1 | |
| 91 | 武粳15号 Wugeng 15 | G | 1 | 131 | 苏粳8号 Sugeng 8 | G | 1 | |
| 92 | 武粳4号 Wugeng 4 | G | 1 | 132 | 苏香粳2号 Suxianggeng 2 | G | 1 | |
| 93 | 武陵粳1号 Wulinggeng 1 | G | 1 | 133 | 嘉33 Jia 33 | G | 1 | |
| 94 | 武香粳14号 Wuxianggeng 14 | G | 1 | 134 | 嘉991 Jia 991 | G | 1 | |
| 95 | 武香粳9号 Wuxianggeng 9 | G | 1 | 135 | 镇稻10号 Zhendao 10 | G | 1 | |
| 96 | 武育粳18号 Wuyugeng 18 | G | 1 | 136 | 镇稻16号 Zhendao 16 | G | 1 | |
| 97 | 武育粳3号 Wuyugeng 3 | G | 1 | 137 | 镇稻17号 Zhendao 17 | G | 1 | |
| 98 | 武运粳11号 Wuyungeng 11 | G | 1 | 138 | 镇稻18号 Zhendao 18 | G | 1 | |
| 99 | 兴化紫稻 Xinghuazidao | G | 1 | 139 | 苏粳9号 Sugeng 9 | G | 1 | |
| 100 | 徐稻8号 Xudao 8 | G | 1 | 140 | 宁2602 Ning 2602 | G | 1 | |
| 101 | 徐稻9号 Xudao 9 | G | 1 | 141 | 宁2604 Ning 2604 | G | 1 | |
| 102 | 盐稻8号 Yandao 8 | G | 1 | 142 | 嘉58 Jia 58 | G | 1 | |
| 103 | 盐稻9号 Yandao 9 | G | 1 | 143 | 苏05-1176 Su 05-1176 | G | 1 | |
| 104 | 盐粳16号 Yangeng 16 | G | 1 | 144 | 苏12-130 Su 12-130 | G | 1 | |
| 105 | 盐粳9号 Yangeng 9 | G | 1 | 145 | 沪香软268 Huxiangruan 268 | G | 1 | |
| 106 | 扬9709 Yang9700 | G | 1 | 146 | 沪香软386 Huxiangruan 386 | G | 1 | |
| 107 | 扬辐粳1号 Yangfugeng 1 | G | 1 | 147 | 软大穗 Ruandaoli | G | 1 | |
| 108 | 扬辐粳8号 Yangfugeng 8 | G | 1 | 148 | 大粒糯 Dalinuo | G | 1 | |
| 109 | 扬粳4227 Yanggeng 4227 | G | 1 | 149 | 太湖香粳稻 Taihuxianggengdao | G | 1 | |
| 110 | 扬粳4308 Yanggeng 4308 | G | 1 | 150 | JX14-5 | G | 1 | |
| 111 | 扬粳805 Yanggeng 805 | G | 1 | 151 | 南粳43 Nangeng 43 | G | 1 | |
| 112 | 扬农210 Yangnong210 | G | 1 | 152 | 农垦57 Nongken 57 | G | 1 | |
| 113 | 扬中稻1号 Yangzhongdao 1 | G | 1 | 153 | 农垦58 Nongken 58 | G | 1 | |
| 114 | 镇糯19号 Zhennuo 19 | G | 1 | 154 | 秀水63 Xiushui 63 | G | 1 | |
| 115 | 镇稻12号 Zhendao 12 | G | 1 | 155 | 徐81698 Xu 81698 | G | 1 | |
| 116 | 镇稻13号 Zhendao 13 | G | 1 | 156 | 徐稻4号 Xudao 4 | G | 1 | |
| 117 | 镇稻15号 Zhendao 15 | G | 1 | 157 | 盐稻980 Yandao 980 | G | 1 | |
| 118 | 镇稻1号 Zhendao 1 | G | 1 | 158 | 盐丰2号 Yanfeng 2 | G | 1 | |
| 159 | 扬粳9224 Yanggeng 9224 | G | 1 | 195 | 14CZY43 | G | 1 | |
| 160 | 扬育粳116 Yangyugeng 116 | G | 1 | 196 | 14CZY47 | G | 1 | |
| 161 | 银玉2239 Yinyu 2239 | G | 1 | 197 | 14CZY49 | G | 1 | |
| 162 | 镇稻615 Zhendao 615 | G | 1 | 198 | 14CZY53 | G | 1 | |
| 163 | 隆粳968 Longgeng 968 | G | 1 | 199 | 14CZY60 | G | 1 | |
| 164 | 大华香糯 Dahuaxiangnuo | G | 1 | 200 | 14CZY65 | G | 1 | |
| 165 | 华粳8号 Huageng 8 | G | 1 | 201 | 14CZY71 | G | 1 | |
| 166 | 盐粳13号 Yangeng 13 | G | 1 | 202 | 14CZY73 | G | 1 | |
| 167 | 盐粳15号 Yangeng 15 | G | 1 | 203 | 14CZY83 | G | 1 | |
| 168 | 南粳505 Nangeng 505 | G | 1 | 204 | 14CZY84 | G | 1 | |
| 169 | 盐丰稻2号 Yanfengdao 2 | G | 1 | 205 | 14CZY85 | G | 1 | |
| 170 | 软香玉 Ruanxiangyu | G | 1 | 206 | 14CZY86 | G | 1 | |
| 171 | 苏香粳100 Suxianggeng 100 | G | 1 | 207 | 14CZY87 | G | 1 | |
| 172 | 苏2100 Su 2100 | G | 1 | 208 | 15ZZY2 | G | 1 | |
| 173 | 沪粳137 Hugeng 137 | G | 1 | 209 | 15ZZY7 | G | 1 | |
| 174 | 沪香粳165 Huxianggeng 165 | G | 1 | 210 | 15ZZY8 | G | 1 | |
| 175 | 松粳9号 Songgeng 9 | G | 1 | 211 | 15ZZY79 | G | 1 | |
| 176 | 14CZY1 | G | 1 | 212 | 15ZZY89 | G | 1 | |
| 177 | 14CZY8 | G | 1 | 213 | 15ZZY16 | G | 1 | |
| 178 | 14CZY11 | G | 1 | 214 | 15ZZY48 | G | 1 | |
| 179 | 14CZY13 | G | 1 | 215 | 15ZZY53 | G | 1 | |
| 180 | 14CZY15 | G | 1 | 216 | 15ZZY58 | G | 1 | |
| 181 | 14CZY16 | G | 1 | 217 | 15ZZY73 | G | 1 | |
| 182 | 14CZY17 | G | 1 | 218 | 15ZZY90 | G | 1 | |
| 183 | 14CZY21 | G | 1 | 219 | 15ZZQ6 | G | 1 | |
| 184 | 14CZY22 | G | 1 | 220 | 15ZZQ12 | G | 1 | |
| 185 | 14CZY23 | G | 1 | 221 | 15CZY3 | G | 1 | |
| 186 | 14CZY28 | G | 1 | 222 | 15CZY31 | G | 1 | |
| 187 | 14CZY30 | G | 1 | 223 | 15CZY65 | G | 1 | |
| 188 | 14CZY31 | G | 1 | 224 | 15CZY70 | G | 1 | |
| 189 | 14CZY36 | G | 1 | 225 | 15CZY71 | G | 1 | |
| 190 | 14CZY37 | G | 1 | 226 | 15CZY77 | G | 1 | |
| 191 | 14CZY38 | G | 1 | 227 | 15CZY84 | G | 1 | |
| 192 | 14CZY39 | G | 1 | 228 | 15CZY86 | G | 1 | |
| 193 | 14CZY40 | G | 1 | 229 | 15CZY91 | G | 1 | |
| 194 | 14CZY41 | G | 1 |
新窗口打开|下载CSV
Table 3
表3
表3GMRA标记鉴定部分丽江新团黑谷单基因系
Table 3
| 编号No. | 名称Designation | 目标基因Target gene | 带型Type |
|---|---|---|---|
| 1 | IRBLa-A | Pia | 1 |
| 2 | IRBLa-C | Pia | 1 |
| 3 | IRBLi-F5 | Pii | 1 |
| 4 | IRBLks-F5 | Pik-s | 1 |
| 5 | IRBLks-S | Pik-s | 1 |
| 6 | IRBLk-ka | Pik | 1 |
| 7 | IRBLkp-K60 | Pik-p | 1 |
| 8 | IRBLkh-K3 | Pik-h | 1 |
| 9 | IRBLz-Fu | Piz | 1 |
| 10 | IRBLzt-T | Piz-t | 1 |
| 11 | IRBLta-K1 | Pita | 1 |
| 12 | IRBLta-CT2 | Pita | 1 |
| 13 | IRBLb-B | Pib | 1 |
| 14 | IRBLt-K59 | Pit | 1 |
| 15 | IRBLsh-S | Pish | 1 |
| 16 | IRBLsh-B | Pish | 1 |
| 17 | IRBL1-CL | Pi1 | 1 |
| 18 | IRBL3-CP4 | Pi3 | 1 |
| 19 | IRBL5-M | Pi5(t) | 1 |
| 20 | IRBL7-M | Pi7(t) | 1 |
| 21 | IRBL9-W | Pi9 | 1 |
| 22 | IRBL12-M | Pi12(t) | 1 |
| 23 | IRBL19-A | Pi19 | 1 |
| 24 | IRBLkm-Ts | Pik-m | 1 |
| 25 | IRBL20-IR24 | Pi20 | 1 |
| 26 | IRBLta2-Pi | Pita2 | 1 |
| 27 | IRBLta2-Re | Pita2 | 1 |
| 28 | IRBLta-CP1 | Pita | 1 |
| 29 | IRBL11-Zh | Pi11(t) | 1 |
| 30 | 丽江新团黑谷 Lijiangxintuanheigu | - | 1 |
| 31 | 谷梅4号 Gumei 4 | Pigm | 2 |
新窗口打开|下载CSV
以上结果表明,针对抗病基因的特异序列设计的GMRA标记能特异检测谷梅4号携带的PigmR,可用于分子标记辅助选择育种。
为进一步明确PigmR在江苏粳稻育种中的利用情况,用GMRA标记检测了江苏省迟熟中粳预备试验中间材料216份、早熟晚粳区域试验(早熟组)中间材料24份,共鉴定到3份中间材料携带PigmR,表明PigmR在江苏粳稻育种中已经使用,但使用频率仍然很低。这些中间材料可以作为良好的供体材料,在粳稻育种中进一步利用。
2.4 GMRA标记在育种中的应用
利用GMRA标记对南粳53045/谷梅4号的BC1F2群体的320个单株进行检测,共检测到198个单株携带PigmR,部分单株检测结果见图6。保留携带PigmR的单株,待成熟后按单株收种。种植BC1F3代株系,在部分株系中各选取10个单株进行检测。若10个单株均表现为携带PigmR,根据分离比例,该株系为PigmR纯合系;若10个单株中有部分单株不携带PigmR,表明该株系的PigmR分离。本研究共检测了19个株系的190个单株,6个系的PigmR纯合,13个系的PigmR分离(图7)。随机选取38个单株进行穗颈瘟接种发现,携带PigmR的单株均对穗颈瘟表现为抗或中抗;不携带PigmR的单株均表现为高感(图8和电子附表1)。以上结果表明,GMRA功能标记可以准确鉴定PigmR,具有很好的特异性,能有效用于分子标记辅助选育抗稻瘟病水稻新品种。图6
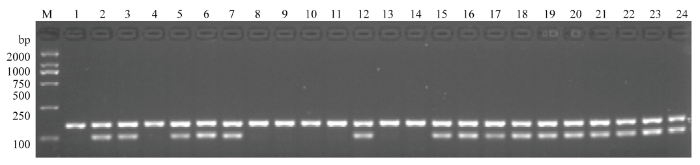 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6GMRA标记检测南粳53045/谷梅4号的BC1F2群体单株
M:DL2000分子标记;1—24:南粳53045/谷梅4号的BC1F2群体部分单株
Fig. 6The molecular detection of BC1F2 of Nangeng53045/Gumei 4 by GMRA
M: DL2000 marker; 1-24: Represent partial BC1F2 plants of Nangeng53045/Gumei 4
图7
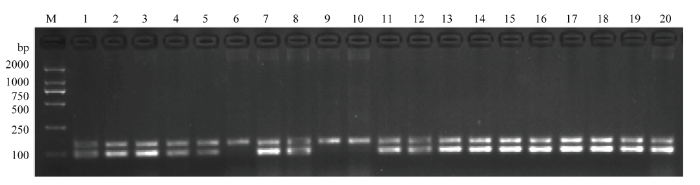 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7GMRA标记检测南粳53045/谷梅4号的BC1F3群体部分单株
M:DL2000分子标记;1—20:BC1F3群体部分单株
Fig. 7The molecular detection of BC1F3 of Nangeng53045/Gumei 4 by GMRA
M: DL2000 marker; 1-20: Represent partial BC1F3 plants of Nangeng53045/Gumei 4
图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8南粳53045/谷梅4号的BC1F3群体穗颈瘟鉴定结果
PigmR-:不携带PigmR;PigmR+:携带PigmR
Fig. 8Infection of rice panicle blast in BC1F3 plants of Nangeng53045/Gumei 4
PigmR-: The plants without PigmR; PigmR+: The plants with PigmR
3 讨论
稻瘟病是世界上最严重的水稻病害之一,被称为水稻的“癌症”。稻瘟病菌的生理小种组成复杂,群体大,具有高度的多样性和较强的变异性[24,25]。目前,推广品种仅携带一些抗谱较窄的抗病基因,容易因病菌种群结构的演化及新小种的产生而丧失抗性[26]。例如,近几年,在长江中下游地区广泛利用且具有较好抗性的Pi-ta等基因已经有抗性减弱的趋势。因此,利用具有广谱持久抗性的基因对于水稻抗稻瘟病育种具有重要的生产实践意义。Pigm是一个广谱持久抗稻瘟病基因(簇),对来源于全国不同地区稻瘟病的代表性小种抗性频率高达91.9%,在生产上具有广泛的应用前景[16, 26-28]。然而,Pigm基因簇与品质相关基因Wx和ALK、生育期相关基因Hd1和Hd3等农艺性状相关基因连锁[29,30,31]。而且,在谷梅4号的Pigm基因簇中,含有13个R基因,同时含有抗病基因PigmR和感病基因PigmS,以及与PigmR高度同源但无功能的Pigm-R4[12]。Pigm-R4、PigmS和PigmR紧密连锁,使得PigmR的利用显得困难重重。利用PigmR改良水稻稻瘟病抗性的同时保留优异农艺性状,需要经过多代的回交筛选,而在此过程中选用的分子标记非常关键。
目前,已经报道的针对抗病基因PigmR的标记大多基于其基因簇开发,如Indel587、Pigm-4、G1408、Pigm-SM等[14, 19-22],与PigmR存在一定的分离概率,在分子标记辅助选择育种过程中可能存在连锁累赘或丢失目标基因的问题[20]。利用Pigm基因簇内Pigm-R4、PigmS和PigmR的碱基差异,设计的分子标记GMR-3为功能标记,该标记能区分PigmR与Pi9、Piz、Piz-t等同源性较高的基因。GMR-3为显性标记,即能扩增出条带的样品携带PigmR,不能扩增出条带的样品不携带PigmR。在基因的分子检测过程中,往往会因为DNA样品质量、试验试剂的差异、试验操作人员的失误等原因而导致PCR的扩增失败,而显性标记不能像等位基因特异PCR、CAPS标记一样区分等位基因而避免这种假阴性[32]。因此,通过设计内参基因的检测标记,与显性标记同时对样品进行检测,有利于避免这种假阴性。由于GMR-3和Actin1-1扩增产物差异较大,引物GMR-3F、GMR-3R、Actin1-1F和Actin1-1R可以在一个PCR中进行,携带PigmR的样品能被GMRA同时扩增出146和98 bp条带,不携带PigmR的样品仅能被GMRA扩增出146 bp条带。该方法可以简单可靠的对样品进行鉴定。通过对229份育种材料检测发现,GMRA可以特异扩增来源于谷梅4号的PigmR,特异性显著。以优质粳稻品系南粳53045为背景,利用GMRA进行分子标记辅助选择,成功获得了穗颈瘟抗性显著提高的新株系,表明GMRA能用于分子标记辅助选择育种中。因此,本研究设计的功能标记GMRA可以有效改善对PigmR的鉴定效果及在育种的应用。
4 结论
根据PigmR与Pigm-R4、PigmS的碱基差异设计和优化的功能标记GMRA能有效区分PigmR与Pi9、Piz、Piz-t等同源性较高的基因,具有很强的特异性,且可以有效避免因PCR扩增失败引起的假阴性。PigmR在江苏粳稻育种中已经使用,但使用频率仍然很低,筛选获得的材料可以作为PigmR资源应用于粳稻抗稻瘟病育种中。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/j.crvi.2014.08.007URLPMID:25444707 [本文引用: 1]

A critical investigation was conducted to find out the effect of neck blast disease on yield-contributing characters, and seed quality traits of aromatic rice in Bangladesh. Both healthy and neck-blast-infected panicles of three aromatic rice cultivars (high-yielding and local) were collected and investigated at Plant Pathology Division, Bangladesh Rice Research Institute (BRRI), Gazipur, Bangladesh. All of the tested varieties were highly susceptible to neck blast disease under natural conditions, though no leaf blast symptoms appear on leaves. Neck blast disease increased grain sterility percentages, reduced grain size, yield and quality traits of seeds. The degrees of yield and seed quality reduction depended on disease severity and variety's genetic make-up. Unfilled grains were the main source of seed-borne pathogen, especially for blast in the seed lot. Transmission of blast pathogen from neck (panicle base) to seed was very poor. These findings are important, especially concerning the seed certification programme in which seed lots are certified on the basis of field inspection. Finally, controlled experiments are needed to draw more critical conclusions.
DOI:10.15889/j.issn.1002-1302.2016.08.042 [本文引用: 1]

2014年稻瘟病在江苏全省偏重发生,局部大流行,自然发生程度为近30年最重,严重影响水稻高产稳产。分析了稻瘟病重发原因:水稻品种抗性下降,气候条件极其有利,部分地区防治质量不高,栽培措施复杂。提出了稻瘟病防控对策的“五个改进”:改进工作思路,加强协作攻关;改进监测办法,建立预警体系;改进防控对策,实施综合治理;改进用药技术,提高防治水平;改进服务方式,推进措施到位。
DOI:10.15889/j.issn.1002-1302.2016.08.042 [本文引用: 1]

2014年稻瘟病在江苏全省偏重发生,局部大流行,自然发生程度为近30年最重,严重影响水稻高产稳产。分析了稻瘟病重发原因:水稻品种抗性下降,气候条件极其有利,部分地区防治质量不高,栽培措施复杂。提出了稻瘟病防控对策的“五个改进”:改进工作思路,加强协作攻关;改进监测办法,建立预警体系;改进防控对策,实施综合治理;改进用药技术,提高防治水平;改进服务方式,推进措施到位。
DOI:10.1007/s40003-011-0003-5URL [本文引用: 1]

Rice ( Oryza sativa ) plays a significant role in achieving global food security. However, it suffers from several biotic and abiotic stresses that seriously affect its production. Rice blast caused by hemibiotropic fungal pathogen Magnaporthe oryzae is one of the most widespread and devastating diseases of rice. The crop rice is vulnerable to this pathogen from seedlings to adult plant stages affecting leaves, nodes, collar, panicles and roots. This disease can be effectively managed through host resistance. Of the 100 blast resistance genes, identified and mapped in different genotypes of rice, 19 genes have been cloned and characterized at the molecular level. Most of these genes belong to nucleotide binding sites and leucine rich repeats. Besides more than 350 quantitative trait loci (QTLs) have also been identified in the rice genome. These blast resistance genes and QTLs have been successfully mobilized in the commercial cultivars by using standard plant breeding techniques and also by marker assisted backcross breeding. With the advent of latest molecular biology techniques and our understanding of the basic mechanisms of Magnaporthe -rice pathosystem, the strategies for broad-spectrum resistance to M. oryzae can be designed in future.
DOI:10.1080/00222935808697054URLPMID:22667447 [本文引用: 1]

Abstract The Pib gene in rice confers resistance to a wide range of races of the rice blast pathogen, Magnaporthe oryzae, including race IE1k that overcomes Pita, another broad-spectrum resistance gene. In this study, the presence of Pib was determined in 164 rice germplasm accessions from a core subset of the National Small Grains Collection utilizing DNA markers and pathogenicity assays. The presence of Pib was evaluated with two simple sequence repeat (SSR) markers and a dominant marker (Pib-dom) derived from the Pib gene sequence. Pathogenicity assays using two avirulent races (IE1k and IB1) and a virulent race (IB54) were performed to verify the resistance responses of accessions. Of the 164 accessions evaluated, 109 contained the Pib gene as determined using both SSR markers and pathogenicity assays, albeit different haplotypes were detected. The remaining 52 germplasm accessions were different in their responses to the blast races IB54, IE1k, and IB1, thus indicating the presence of R gene(s) other than Pib. The accessions characterized in this study could be used for marker-assisted breeding to improve blast resistance in indica and japonica cultivars worldwide.
DOI:10.2307/3871103URLPMID:11090207 [本文引用: 1]

The rice blast resistance (R) gene Pi-ta mediates gene-for-gene resistance against strains of the fungus Magnaporthe grisea that express avirulent alleles of AVR-Pita. Using a map-based cloning strategy, we cloned Pi-ta, which is linked to the centromere of chromosome 12. Pi-ta encodes a predicted 928-amino acid cytoplasmic receptor with a centrally localized nucleotide binding site. A single-copy gene, Pi-ta shows low constitutive expression in both resistant and susceptible rice. Susceptible rice varieties contain $pi\text{-}ta^{-}$ alleles encoding predicted proteins that share a single amino acid difference relative to the Pi-ta resistance protein: serine instead of alanine at position 918. Transient expression in rice cells of a $Pi\text{-}ta^{+}$ R gene together with $AVR\text{-}Pita^{+}$ induces a resistance response. No resistance response is induced in transient assays that use a naturally occurring $pi\text{-}ta^{-}$ allele differing only by the serine at position 918. Rice varieties reported to have the linked $Pi\text{-}ta^{2}$ gene contain Pi-ta plus at least one other R gene, potentially explaining the broadened resistance spectrum of $Pi\text{-}ta^{2}$ relative to Pi-ta. Molecular cloning of the AVR-Pita and Pi-ta genes will aid in deployment of R genes for effective genetic control of rice blast disease.
DOI:10.1007/BF03323441URL [本文引用: 1]

Blast disease caused by Magnaporthe oryzae is one of the important biotic stresses of rice. So far more than 85 blast resistance genes have been identified of these more than 14 have already been cloned. A broad spectrum rice blast resistance gene Pi - k h was cloned from the rice line Tetep. The gene was named Pi - k h based on the earlier reports on its genetic analysis in various rice lines. However, with the advances in molecular genetics and genomics of rice, the Pik locus has now been mapped more precisely. Since there are two reports on the mapping of Pi - k h gene from different rice lines, there is some confusion in the naming of this gene. In this report the name of Pi - k h gene cloned from the rice line Tetep has been designated as per the standard guidelines of Committee on Gene Symbolization, Nomenclature and Linkage (CGSNL) and its physical location on rice chromosome 11, which is 2.5 Mbp away from the Pik locus mapped recently. Hence Pi - k h gene cloned from Tetep is now designated as Pi54 .
DOI:10.1126/science.1175550URLPMID:19696351 [本文引用: 1]

Blast disease is a devastating fungal disease of rice, one of the world's staple foods. Race-specific resistance to blast disease has usually not been durable. Here, we report the cloning of a previously unknown type of gene that confers non--race-specific resistance and its successful use in breeding. Pi21 encodes a proline-rich protein that includes a putative heavy metal--binding domain and putative protein-protein interaction motifs. Wild-type Pi21 appears to slow the plant's defense responses, which may support optimization of defense mechanisms. Deletions in its proline-rich motif inhibit this slowing. Pi21 is separable from a closely linked gene conferring poor flavor. The resistant pi21 allele, which is found in some strains of japonica rice, could improve blast resistance of rice worldwide.
DOI:10.1016/j.cell.2017.06.008URLPMID:28666113 [本文引用: 1]

Abstract Rice feeds half the world's population, and rice blast is often a destructive disease that results in significant crop loss. Non-race-specific resistance has been more effective in controlling crop diseases than race-specific resistance because of its broad spectrum and durability. Through a genome-wide association study, we report the identification of a natural allele of a C 2 H 2 -type transcription factor in rice that confers non-race-specific resistance to blast. A survey of 3,000 sequenced rice genomes reveals that this allele exists in 10% of rice, suggesting that this favorable trait has been selected through breeding. This allele causes a single nucleotide change in the promoter of the bsr-d1 gene, which results in reduced expression of the gene through the binding of the repressive MYB transcription factor and, consequently, an inhibition of H 2 O 2 degradation and enhanced disease resistance. Our discovery highlights this novel allele as a strategy for breeding durable resistance in rice. Copyright 2017 Elsevier Inc. All rights reserved.
DOI:10.1094/MPMI-19-1216URLPMID:17073304 [本文引用: 1]

The rice blast resistance (R) genes Pi2 and Piz-t confer broad-spectrum resistance against different sets of Magnaporthe grisea isolates. We first identified the Pi2 gene using a map-based cloning strategy. The Pi2 gene is a member of a gene cluster comprising nine gene members (named Nbs1-Pi2 to Nbs9-Pi2) and encodes a protein with a nucleotide-binding site and leucine-rich repeat (LRR) domain. Fine genetic mapping, molecular characterization of the Pi2 susceptible mutants, and complementation tests indicated that Nbs4-Pi2 is the Pi2 gene. The Piz-t gene, a Pi2 allele in the rice cultivar Toride 1, was isolated based on the Pi2 sequence information. Complementation tests confirmed that the family member Nbs4-Piz-t is Piz-t. Sequence comparison revealed that only eight amino-acid changes, which are confined within three consecutive LRR, differentiate Piz-t from Pi2. Of the eight variants, only one locates within the xxLxLxx motif. A reciprocal exchange of the single amino acid between Pi2 and Piz-t did not convert the resistance specificity to each other but, rather, abolished the function of both resistance proteins. These results indicate that the single amino acid in the xxLxLxx motif may be critical for maintaining the recognition surface of Pi2 and Piz-t to their respective avirulence proteins.
DOI:10.1534/genetics.105.044891URLPMID:16387888 [本文引用: 1]

Abstract The broad-spectrum rice blast resistance gene Pi9 was cloned using a map-based cloning strategy. Sequencing of a 76-kb bacterial artificial chromosome (BAC) contig spanning the Pi9 locus led to identification of six tandemly arranged resistance-like genes with a nucleotide-binding site (NBS) and leucine-rich repeats (LRRs) (Nbs1-Pi9-Nbs6-Pi9). Analysis of selected Pi9 deletion mutants and transformation of a 45-kb fragment from the BAC contig into the susceptible rice cultivar TP309 narrowed down Pi9 to the candidate genes Nbs2-Pi9 and Nbs3-Pi9. Disease evaluation of the transgenic lines carrying the individual candidate genes confirmed that Nbs2-Pi9 is the Pi9 gene. Sequence comparison analysis revealed that the six paralogs at the Pi9 locus belong to four classes and gene duplication might be one of the major evolutionary forces contributing to the formation of the NBS-LRR gene cluster. Semiquantitative reverse transcriptase (RT)-PCR analysis showed that Pi9 was constitutively expressed in the Pi9-resistant plants and was not induced by blast infection. The cloned Pi9 gene provides a starting point to elucidate the molecular basis of the broad-spectrum disease resistance and the evolutionary mechanisms of blast resistance gene clusters in rice.
DOI:10.1007/s00122-012-1787-9URLPMID:22270148 [本文引用: 1]

The deployment of broad-spectrum resistance genes is the most effective and economic means of controlling blast in rice. The cultivar Er-Ba-Zhan (EBZ) is a widely used donor of blast resistance in South China, with many cultivars derived from it displaying broad-spectrum resistance against blast. Mapping in a set of recombinant inbred lines bred from the cross between EBZ and the highly blast-susceptible cultivar Liangjiangxintuanheigu (LTH) identified in EBZ a blast resistance gene on each of chromosomes 1 ( Pish), 6 ( Pi2/ Pi9) and 12 ( Pita/ Pita- 2). The resistance spectrum and race specificity of the allele at Pi2/ Pi9 were both different from those present in other known Pi2/ Pi9 carriers. Fine-scale mapping based on a large number of susceptible EBZ 01— LTH F and EBZ 01— LTH BCF segregants placed the gene within a 53-kb segment, which includes Pi2/ Pi9. Sequence comparisons of the LRR motifs of the four functional NBS-LRR genes within Pi2/ Pi9 revealed that the EBZ allele is distinct from other known Pi2/ Pi9 alleles. As a result, the gene has been given the designation Pi50(t).
DOI:10.1007/s00122-015-2579-9URLPMID:26183036 [本文引用: 2]

Key message We characterized a novel blast resistance gene Pi50 at the Pi2/9 locus; Pi50 is derived from functional divergence of duplicated genes. The unique features of Pi50 should facilitate its use in rice breeding and improve our understanding of the evolution of resistance specificities. Abstract Rice blast disease, caused by the fungal pathogen Magnaporthe oryzae , poses constant, major threats to stable rice production worldwide. The deployment of broad-spectrum resistance ( R ) genes provides the most effective and economical means for disease control. In this study, we characterize the broad-spectrum R gene Pi50 at the Pi2/9 locus, which is embedded within a tandem cluster of 12 genes encoding proteins with nucleotide-binding site and leucine-rich repeat (NBS RR) domains. In contrast with other Pi2/9 locus, the Pi50 cluster contains four duplicated genes ( Pi50_NBS4_1 to 4 ) with extremely high nucleotide sequence similarity. Moreover, these duplicated genes encode two kinds of proteins (Pi50_NBS4_1/2 and Pi50_NBS4_3/4) that differ by four amino acids. Complementation tests and resistance spectrum analyses revealed that Pi50_NBS4_1/2 , not Pi50_NBS4_3/4 , control the novel resistance specificity as observed in the Pi50 near isogenic line, NIL-e1. Pi50 shares greater than 96% amino acid sequence identity with each of three other R proteins, i.e., Pi9, Piz-t, and Pi2, and has amino acid changes predominantly within the LRR region. The identification of Pi50 with its novel resistance specificity will facilitate the dissection of mechanisms behind the divergence and evolution of different resistance specificities at the Pi2/9 locus.
DOI:10.1007/s00122-006-0338-7URLPMID:16832648 [本文引用: 2]

Abstract The identification and utilization of broad-spectrum resistance genes have been proven the most effective and economical approach to control rice blast disease. To understand the molecular mechanism of broad-spectrum resistance to rice blast, we conducted genetic and fine mapping analysis of the blast resistance gene in a Chinese rice variety: Gumei 4 (GM4) identified with broad-spectrum resistance and used in rice breeding for blast resistance for more than 20 years. Genetic and mapping analysis indicated that blast resistance to nine isolates of different Chinese races in GM4 was controlled by the same dominant locus designated as Pigm(t) that was finely mapped to an approximately 70-kb interval between markers C5483 and C0428 on chromosome 6, which contains five candidate NBS--LRR disease resistance genes. The allelism test showed that Pigm(t) was either tightly linked or allelic to Pi2 and Pi9, two known blast resistance genes. Mapping information also indicated that another blast resistance gene Pi26(t) might also be located at the same region. Candidate genes were identified by sequence analysis of the Nipponbare and Pi9 locus and the corresponding region in GM4. Sequence divergence of candidate genes was observed between GM4 and model varieties Nipponbare and 9311, and Pi9. Our current study provides essential information and new genetic resource for the cloning of functional resistance gene(s) and for marker-assisted selection in rice breeding for broad-spectrum blast resistance.
DOI:10.1126/science.aai8898URLPMID:28154240 [本文引用: 6]

Abstract Crop breeding aims to balance disease resistance with yield, however single resistance (R) genes can lead to resistance breakdown and R gene pyramiding may impact growth fitness. Here we report that the rice Pigm locus contains a cluster of genes encoding nucleotide-binding leucine-rich repeat (NLR) receptors that confer durable resistance to the fungus Magnaporthe oryzae without yield penalty. In the cluster, PigmR confers broad-spectrum resistance, whereas PigmS competitively attenuates PigmR homodimerization to suppress resistance. PigmS expression, and thus PigmR-mediated resistance, are subjected to tight epigenetic regulation. PigmS increases seed production to counteract the yield cost induced by PigmR Therefore, our study reveals a mechanism balancing high disease resistance and yield through epigenetic regulation of paired antagonistic NLRs, providing a tool to develop elite crop varieties. Copyright 2017, American Association for the Advancement of Science.
DOI:10.1038/s41467-018-04369-4URL [本文引用: 1]

Quantitative trait loci (QTL) play important roles in controlling rice blast disease. In the present study, 10 field isolates of the races IA1, IB1, IB17, and IC1 of US rice blast fungus Magnaporthe oryzae collected in 1996 and 2009 were used to identify blast resistance QTL with a recombinant inbred line (RIL) population consisting of 227 F7 individuals derived from the cross of rice (Oryza... [Show full abstract]
DOI:10.1371/journal.pone.0126130URLPMID:26030358 [本文引用: 2]

Rice blast caused by Magnaporthe oryzae is the most devastating disease of rice and poses a serious threat to world food security. In this study, the distribution and effectiveness of 18 R genes in 277 accessions were investigated based on pathogenicity assays and molecular markers. The results showed that most of the accessions exhibited some degree of resistance (resistance frequency, RF >50%). Accordingly, most of the accessions were observed to harbor two or more R genes, and the number of R genes harbored in accessions was significantly positively correlated with RF. Some R genes were demonstrated to be specifically distributed in the genomes of rice sub-species, such as Pigm, Pi9, Pi5 and Pi1, which were only detected in indica-type accessions, and Pik and Piz, which were just harbored in japonica-type accessions. By analyzing the relationship between R genes and RF using a multiple stepwise regression model, the R genes Pid3, Pi5, Pi9, Pi54, Pigm and Pit were found to show the main effects against M. oryzae in indica-type accessions, while Pita, Pb1, Pik, Pizt and Pia were indicated to exhibit the main effects against M. oryzae in japonica-type accessions. Principal component analysis (PCA) and cluster analysis revealed that combination patterns of major R genes were the main factors determining the resistance of rice varieties to M. oryzae, such as 'Pi9+Pi54', 'Pid3+Pigm', 'Pi5+Pid3+Pigm', 'Pi5+Pi54+Pid3+Pigm', 'Pi5+Pid3' and 'Pi5+Pit+Pid3' in indica-type accessions and 'Pik+Pib', 'Pik+Pita', 'Pik+Pb1', 'Pizt+Pia' and 'Pizt+Pita' in japonica-type accessions, which were able to confer effective resistance against M. oryzae. The above results provide good theoretical support for the rational utilization of combinations of major R genes in developing rice cultivars with broad-spectrum resistance.
DOI:10.7668/hbnxb.2011.03.001URLMagsci [本文引用: 1]

主效抗稻瘟病基因<em>Pita</em>和<em>Pib</em>在我国很多稻区表现高水平的稻瘟病抗性,被广泛应用于我国的水稻育种和生产。但这2个基因在国内品种资源中的分布及利用情况缺乏详细的资料,致使育种利用上存在着盲目性。本研究利用<em>Pita</em>和<em>Pib</em>基因的功能标记,检测和分析了我国115份水稻地方品种的<em>Pita</em>和<em>Pib</em>基因型。结果表明,<em>Pita</em>在我国浙江、福建、广东、广西、贵州、四川、安徽、江西、河南、河北、吉林均有分布,在江苏、上海、山东、湖南、湖北、辽宁没有发现;而只有来自四川的地方品种桂东籼和来自河南的德国稻携带<em>Pib</em>,其他省均未发现。在江苏的主要推广粳稻品种中,晚粳品种几乎都携带这两个基因,部分迟熟中粳携带这两个基因,中熟中粳多数不携带。
DOI:10.7668/hbnxb.2011.03.001URLMagsci [本文引用: 1]

主效抗稻瘟病基因<em>Pita</em>和<em>Pib</em>在我国很多稻区表现高水平的稻瘟病抗性,被广泛应用于我国的水稻育种和生产。但这2个基因在国内品种资源中的分布及利用情况缺乏详细的资料,致使育种利用上存在着盲目性。本研究利用<em>Pita</em>和<em>Pib</em>基因的功能标记,检测和分析了我国115份水稻地方品种的<em>Pita</em>和<em>Pib</em>基因型。结果表明,<em>Pita</em>在我国浙江、福建、广东、广西、贵州、四川、安徽、江西、河南、河北、吉林均有分布,在江苏、上海、山东、湖南、湖北、辽宁没有发现;而只有来自四川的地方品种桂东籼和来自河南的德国稻携带<em>Pib</em>,其他省均未发现。在江苏的主要推广粳稻品种中,晚粳品种几乎都携带这两个基因,部分迟熟中粳携带这两个基因,中熟中粳多数不携带。
URL [本文引用: 1]

稻瘟病是危害水稻最严重的病害之一,选育抗病品种是防治该病害最有效的方法。Bsr-d1是对稻瘟病菌具有广谱抗性的一个重要基因。为提高Bsr-d1基因在育种中的选择效率,根据Bsr-d1与其感病等位基因bsr-d1在功能区域存在的单核苷酸差异,设计和筛选出Bsr-d1基因不同类型的基因功能标记CAPs5-1和3Bsr-d1/3bsr-d1,结合测序分析验证,可准确鉴定出Bsr-d1的不同基因型。利用3Bsr-d1/3bsr-d1对34份籼稻品种、江苏历年来主要推广的110份粳稻品种、其他省份的13份粳稻品种、148份太湖流域地方粳稻资源和19份太湖流域地方籼稻资源进行了Bsr-d1基因型检测,筛选到携带Bsr-d1基因的籼型资源11份,271份粳型资源中均不携带Bsr-d1基因,这也说明Bsr-d1主要分布在籼型水稻资源中,在粳型资源中几乎不存在。本研究为Bsr-d1基因的育种利用和分子标记辅助选择奠定了基础。
URL [本文引用: 1]

稻瘟病是危害水稻最严重的病害之一,选育抗病品种是防治该病害最有效的方法。Bsr-d1是对稻瘟病菌具有广谱抗性的一个重要基因。为提高Bsr-d1基因在育种中的选择效率,根据Bsr-d1与其感病等位基因bsr-d1在功能区域存在的单核苷酸差异,设计和筛选出Bsr-d1基因不同类型的基因功能标记CAPs5-1和3Bsr-d1/3bsr-d1,结合测序分析验证,可准确鉴定出Bsr-d1的不同基因型。利用3Bsr-d1/3bsr-d1对34份籼稻品种、江苏历年来主要推广的110份粳稻品种、其他省份的13份粳稻品种、148份太湖流域地方粳稻资源和19份太湖流域地方籼稻资源进行了Bsr-d1基因型检测,筛选到携带Bsr-d1基因的籼型资源11份,271份粳型资源中均不携带Bsr-d1基因,这也说明Bsr-d1主要分布在籼型水稻资源中,在粳型资源中几乎不存在。本研究为Bsr-d1基因的育种利用和分子标记辅助选择奠定了基础。
DOI:10.16819/j.1001-7216.2018.7135URL [本文引用: 2]

【目的】Pigm是一个广谱的稻瘟病抗性基因,源自持久抗性品种谷梅4号,与Piz、Piz-t、Pi2、Pi9和Pi40等互为复等位基因但抗谱存在差异。为了更好地在分子辅助选择育种中利用Pigm基因,开发与Pigm特异性标记具有重要意义。【方法】在已有文献报道定位结果的基础上,通过随机测序获得了一段谷梅4号基因组的特异序列,并据此开发了一组用于筛选Pigm基因的分子标记,进一步选取江淮稻区3个不同生态型代表性粳稻品种作为受体,利用分子标记辅助选择结合对抗性基因的背景检测将Pigm基因导入受体品种。【结果】Pigm-4标记位于Pigm基因簇内部,与抗病功能元件Pigm R紧密连锁,对不同类型的品种检测发现该标记特异性强,且利用该标记可将Pigm与Piz、Piz-t、Pi2、Pi9以及Pi40区分开来。对受体亲本稻瘟病抗性基因的检测和接种结果分析发现江淮稻区粳稻品种虽然携带了Pib、Pi54、Pita、Pb1中的2~3个基因,但是对强毒力的稻瘟病小种抗性普遍不强,而3种代表性粳稻背景下导入Pigm基因均可显著提高其对穗颈瘟的抗性水平。【结论】Pigm可以作为抗稻瘟病粳稻育种的有利基因资源加以利用,而Pigm-4是分子标记辅助筛选Pigm的优异标记。
DOI:10.16819/j.1001-7216.2018.7135URL [本文引用: 2]

【目的】Pigm是一个广谱的稻瘟病抗性基因,源自持久抗性品种谷梅4号,与Piz、Piz-t、Pi2、Pi9和Pi40等互为复等位基因但抗谱存在差异。为了更好地在分子辅助选择育种中利用Pigm基因,开发与Pigm特异性标记具有重要意义。【方法】在已有文献报道定位结果的基础上,通过随机测序获得了一段谷梅4号基因组的特异序列,并据此开发了一组用于筛选Pigm基因的分子标记,进一步选取江淮稻区3个不同生态型代表性粳稻品种作为受体,利用分子标记辅助选择结合对抗性基因的背景检测将Pigm基因导入受体品种。【结果】Pigm-4标记位于Pigm基因簇内部,与抗病功能元件Pigm R紧密连锁,对不同类型的品种检测发现该标记特异性强,且利用该标记可将Pigm与Piz、Piz-t、Pi2、Pi9以及Pi40区分开来。对受体亲本稻瘟病抗性基因的检测和接种结果分析发现江淮稻区粳稻品种虽然携带了Pib、Pi54、Pita、Pb1中的2~3个基因,但是对强毒力的稻瘟病小种抗性普遍不强,而3种代表性粳稻背景下导入Pigm基因均可显著提高其对穗颈瘟的抗性水平。【结论】Pigm可以作为抗稻瘟病粳稻育种的有利基因资源加以利用,而Pigm-4是分子标记辅助筛选Pigm的优异标记。
URL [本文引用: 3]

本发明公开了一种用于检测谷梅4号抗稻瘟病基因Pigm(t)的分子标记InDel587,本发明提供的分子标记InDel587位于第6染色体10484756-10484985bp处,与抗稻瘟病基因Pigm(t)的定位区间10367733-1042226bp相距65.2kb,紧密连锁,而且标记InDel587多态率高,87.2%的受体亲本与供体亲本谷梅4号之间存在多态;InDel587的PCR产物差异大,能快速检测出材料的标记基因型。通过用分子标记检测待检材料,可以判断其是否携带抗病基因Pigm(t)。
URL [本文引用: 3]

本发明公开了一种用于检测谷梅4号抗稻瘟病基因Pigm(t)的分子标记InDel587,本发明提供的分子标记InDel587位于第6染色体10484756-10484985bp处,与抗稻瘟病基因Pigm(t)的定位区间10367733-1042226bp相距65.2kb,紧密连锁,而且标记InDel587多态率高,87.2%的受体亲本与供体亲本谷梅4号之间存在多态;InDel587的PCR产物差异大,能快速检测出材料的标记基因型。通过用分子标记检测待检材料,可以判断其是否携带抗病基因Pigm(t)。
.
[本文引用: 1]
[本文引用: 1]
.
URL [本文引用: 2]

本发明提供了一种稻瘟病抗性基因Pi1功能特异性分子标记及其应用,是通过引物对SEQ02ID02NO.1和SEQ02ID02NO.2从水稻基因组DNA中扩增出与稻瘟病抗性基因Pi1呈特异性带型的分子标记。本发明所提供的稻瘟病抗性基因Pi1基因特异性分子标记具有重要的应用价值,利用此标记可以提高该基因在种质资源筛选、分子标记辅助选择育种、基因聚合育种,以及转基因育种中利用的效率。
.
URL [本文引用: 2]

本发明提供了一种稻瘟病抗性基因Pi1功能特异性分子标记及其应用,是通过引物对SEQ02ID02NO.1和SEQ02ID02NO.2从水稻基因组DNA中扩增出与稻瘟病抗性基因Pi1呈特异性带型的分子标记。本发明所提供的稻瘟病抗性基因Pi1基因特异性分子标记具有重要的应用价值,利用此标记可以提高该基因在种质资源筛选、分子标记辅助选择育种、基因聚合育种,以及转基因育种中利用的效率。
[本文引用: 1]
DOI:10.3969/j.issn.1008-0384.2016.05.019URL [本文引用: 1]

种植抗性品种是预防水稻稻瘟病最有效、最经济和环保的方式之一。目前农业生产上推广种植的抗病品种在数年内抗性减弱,甚至丢失,这可能与寄主体内的抗性基因丧失或相对应的无毒基因发生变异有关。本文综述了稻瘟病菌的致病机理、稻瘟病菌发生变异的原因、稻瘟病菌无毒基因发现与克隆以及稻瘟病的相关防治策略,以期为稻瘟病防治提供理论基础。
DOI:10.3969/j.issn.1008-0384.2016.05.019URL [本文引用: 1]

种植抗性品种是预防水稻稻瘟病最有效、最经济和环保的方式之一。目前农业生产上推广种植的抗病品种在数年内抗性减弱,甚至丢失,这可能与寄主体内的抗性基因丧失或相对应的无毒基因发生变异有关。本文综述了稻瘟病菌的致病机理、稻瘟病菌发生变异的原因、稻瘟病菌无毒基因发现与克隆以及稻瘟病的相关防治策略,以期为稻瘟病防治提供理论基础。
URL [本文引用: 1]

【目的】鉴定不同地区谷瘟病菌()所含无毒基因的类型,确定无毒基因在菌株中的分布及变异情况,为深入研究谷瘟病菌无毒基因变异机制提供参考。【方法】从中国北方谷子主产区不同区域内采集并分离76个谷瘟病菌的单孢菌株,提取其基因组DNA,根据目前已成功克隆的稻瘟病菌的7个无毒基因的核苷酸序列设计特异性引物,进行PCR扩增及电泳检测,并对部分菌株的无毒基因进行测序分析。【结果】在76个谷瘟病菌中,无毒基因ACE1、Avr-pita、Avr1-CO39和AvrPiz-t的扩增率为100%,无毒基因Avr-pik、Avr-pia和Avr-pii的扩增率分别为63.2%、42.1%和21.1%。在谷瘟病菌菌株P11和P34中,Avr1-CO39的扩增条带较预期片段大490 bp,测序结果发现菌株P11和P34中的Avr1-CO39基因序列完全一致,均在启动子区插入了490 bp核苷酸,该插入序列与non-LTR retrotransposon: Mg-SINE的相似度达99.16%。Avr-pita的测序结果发现,谷瘟病菌菌株中的Avr-pita基因序列变异较为丰富,其变异形式主要为单核苷酸的变异,包括单碱基的插入、缺失及多位点的SNP。Avr-pia的变异类型主要为整个无毒基因的缺失,经测序验证等位基因序列分为4种类型。Avr-pia-A与参考序列(AB498873.1)一致,包含10个菌株;Avr-pia-B包含20个菌株,在-116、-109和-16 bp处分别存在C/T、G/T和C/A变异,但与参考序列的CDS区序列相同;Avr-pia-C仅包含菌株P10,在+150 bp处存在T/G变异,但为同义突变;Avr-pia-D仅包含菌株P18,在+212 bp位点处存在C/T变异,导致该变异位点由编码苏氨酸突变为编码异亮氨酸。谷瘟病菌Avr-pii包含3种等位基因类型。Avr-pii-A型与参考序列(AB498874.1)一致,共包含14个菌株;Avr-pii-B型和Avr-pii-C型分别在+139和+64 bp处存在A/G变异,核苷酸的变异导致该位点由编码苏氨酸改为丙氨酸。Avr-pii-B型和Avr-pii-C型变异均为首次报道。单元型分析表明,AG2包含23个菌株,占供试菌株的30.2%,为优势单元型。【结论】明确了不同地区的谷瘟病菌中无毒基因ACE1、Avr-pita、Avr1-CO39和AvrPiz-t不存在地理来源的差异;而无毒基因Avr-pik、Avr-pia和Avr-pii在各地分布有差异。谷瘟病菌AG2单元型为优势单元型,其次是单元型AG1和AG5。
URL [本文引用: 1]

【目的】鉴定不同地区谷瘟病菌()所含无毒基因的类型,确定无毒基因在菌株中的分布及变异情况,为深入研究谷瘟病菌无毒基因变异机制提供参考。【方法】从中国北方谷子主产区不同区域内采集并分离76个谷瘟病菌的单孢菌株,提取其基因组DNA,根据目前已成功克隆的稻瘟病菌的7个无毒基因的核苷酸序列设计特异性引物,进行PCR扩增及电泳检测,并对部分菌株的无毒基因进行测序分析。【结果】在76个谷瘟病菌中,无毒基因ACE1、Avr-pita、Avr1-CO39和AvrPiz-t的扩增率为100%,无毒基因Avr-pik、Avr-pia和Avr-pii的扩增率分别为63.2%、42.1%和21.1%。在谷瘟病菌菌株P11和P34中,Avr1-CO39的扩增条带较预期片段大490 bp,测序结果发现菌株P11和P34中的Avr1-CO39基因序列完全一致,均在启动子区插入了490 bp核苷酸,该插入序列与non-LTR retrotransposon: Mg-SINE的相似度达99.16%。Avr-pita的测序结果发现,谷瘟病菌菌株中的Avr-pita基因序列变异较为丰富,其变异形式主要为单核苷酸的变异,包括单碱基的插入、缺失及多位点的SNP。Avr-pia的变异类型主要为整个无毒基因的缺失,经测序验证等位基因序列分为4种类型。Avr-pia-A与参考序列(AB498873.1)一致,包含10个菌株;Avr-pia-B包含20个菌株,在-116、-109和-16 bp处分别存在C/T、G/T和C/A变异,但与参考序列的CDS区序列相同;Avr-pia-C仅包含菌株P10,在+150 bp处存在T/G变异,但为同义突变;Avr-pia-D仅包含菌株P18,在+212 bp位点处存在C/T变异,导致该变异位点由编码苏氨酸突变为编码异亮氨酸。谷瘟病菌Avr-pii包含3种等位基因类型。Avr-pii-A型与参考序列(AB498874.1)一致,共包含14个菌株;Avr-pii-B型和Avr-pii-C型分别在+139和+64 bp处存在A/G变异,核苷酸的变异导致该位点由编码苏氨酸改为丙氨酸。Avr-pii-B型和Avr-pii-C型变异均为首次报道。单元型分析表明,AG2包含23个菌株,占供试菌株的30.2%,为优势单元型。【结论】明确了不同地区的谷瘟病菌中无毒基因ACE1、Avr-pita、Avr1-CO39和AvrPiz-t不存在地理来源的差异;而无毒基因Avr-pik、Avr-pia和Avr-pii在各地分布有差异。谷瘟病菌AG2单元型为优势单元型,其次是单元型AG1和AG5。
[本文引用: 2]

稻瘟病是水稻上最严重的真菌病害之一。由于多数抗瘟基因具有较高的小种特异性,并且稻瘟菌小种变异频繁,水稻品种抗瘟性极易丧失,因此发掘和利用广谱抗性基因是控制稻瘟病的最有效途径之一。本文综述了水稻对稻瘟病广谱抗性基因特性、作用机制与育种应用及其最新研究进展,为今后水稻-稻瘟病互作的分子机制研究和广谱抗性基因的育种利用提供思路。
[本文引用: 2]

稻瘟病是水稻上最严重的真菌病害之一。由于多数抗瘟基因具有较高的小种特异性,并且稻瘟菌小种变异频繁,水稻品种抗瘟性极易丧失,因此发掘和利用广谱抗性基因是控制稻瘟病的最有效途径之一。本文综述了水稻对稻瘟病广谱抗性基因特性、作用机制与育种应用及其最新研究进展,为今后水稻-稻瘟病互作的分子机制研究和广谱抗性基因的育种利用提供思路。
DOI:10.3724/SP.J.1006.2013.01927URLMagsci

<p><span ><em>Pigm</em></span><em><span >是</span><span >Pi2</span></em><em><span >基因簇的等位或紧密连锁基因。本研究</span></em><span >构建了</span><span >4</span><span >个不同遗传背景下</span><em><span >Pigm</span></em><span >和</span><em><span >Pi2</span></em><span >的系列近等基因系,</span><span >204</span><span >个菌株苗期接种结果显示其抗性频率均超过</span><span >70%</span><span >,但</span><em><span >Pigm</span></em><span >和</span><em><span >Pi2</span></em><span >的抗谱重叠度仅为</span><span >54.4%~65.7%</span><span >,<a name="OLE_LINK19">聚合</a></span><span><em><span >Pi1/Pigm</span></em><span >和</span><em><span >Pi1</span></em><span >/<em>Pi2</em></span><span >杂种的抗性频率均超过</span><span >90%</span><span >。</span></span><span >穗瘟人工接种及病圃自然诱发鉴定表现与苗期接种一致的发病趋势。农艺性状调查结果显示获得的近等基因系与其轮回亲本基本相似,存在较少的累赘连锁。表明</span><em><span >Pigm</span></em><span >是一个与</span><em><span >Pi2</span></em><span >抗谱差异明显的广谱抗性基因,对稻瘟病抗性育种具有重要应用价值。</span></p>
DOI:10.3724/SP.J.1006.2013.01927URLMagsci

<p><span ><em>Pigm</em></span><em><span >是</span><span >Pi2</span></em><em><span >基因簇的等位或紧密连锁基因。本研究</span></em><span >构建了</span><span >4</span><span >个不同遗传背景下</span><em><span >Pigm</span></em><span >和</span><em><span >Pi2</span></em><span >的系列近等基因系,</span><span >204</span><span >个菌株苗期接种结果显示其抗性频率均超过</span><span >70%</span><span >,但</span><em><span >Pigm</span></em><span >和</span><em><span >Pi2</span></em><span >的抗谱重叠度仅为</span><span >54.4%~65.7%</span><span >,<a name="OLE_LINK19">聚合</a></span><span><em><span >Pi1/Pigm</span></em><span >和</span><em><span >Pi1</span></em><span >/<em>Pi2</em></span><span >杂种的抗性频率均超过</span><span >90%</span><span >。</span></span><span >穗瘟人工接种及病圃自然诱发鉴定表现与苗期接种一致的发病趋势。农艺性状调查结果显示获得的近等基因系与其轮回亲本基本相似,存在较少的累赘连锁。表明</span><em><span >Pigm</span></em><span >是一个与</span><em><span >Pi2</span></em><span >抗谱差异明显的广谱抗性基因,对稻瘟病抗性育种具有重要应用价值。</span></p>
URL [本文引用: 1]

根据广谱抗瘟基因Pigm精细定位结果,筛选获得共显性InDel标记DG-3在Pigm基因供体亲本谷梅4号与9个籼稻受体亲本之间存在明显且稳定的多态性。在T98B×谷梅4号和R640×谷梅4号2个组合的BC1F1群体中随机取单株进行稻瘟病抗性表型及基因型分析,结果表明,无论是田间病圃鉴定,还是室内接种鉴定,分子标记DG-3对这2个组合回交群体的抗稻瘟病表型选择效率达95%以上,说明DG-3与Pigm基因紧密连锁。在此基础上,采用分子标记辅助选择与回交育种相结合的方法,利用DG-3在9个组合的回交群体中选择含有目的基因的单株分别与相应轮回亲本逐代回交,分别获得了9个组合的BC3F1群体,为进一步连续回交定向改良轮回亲本的稻瘟病抗性奠定了基础。
URL [本文引用: 1]

根据广谱抗瘟基因Pigm精细定位结果,筛选获得共显性InDel标记DG-3在Pigm基因供体亲本谷梅4号与9个籼稻受体亲本之间存在明显且稳定的多态性。在T98B×谷梅4号和R640×谷梅4号2个组合的BC1F1群体中随机取单株进行稻瘟病抗性表型及基因型分析,结果表明,无论是田间病圃鉴定,还是室内接种鉴定,分子标记DG-3对这2个组合回交群体的抗稻瘟病表型选择效率达95%以上,说明DG-3与Pigm基因紧密连锁。在此基础上,采用分子标记辅助选择与回交育种相结合的方法,利用DG-3在9个组合的回交群体中选择含有目的基因的单株分别与相应轮回亲本逐代回交,分别获得了9个组合的BC3F1群体,为进一步连续回交定向改良轮回亲本的稻瘟病抗性奠定了基础。
DOI:10.1111/nph.13538URLPMID:26147403 [本文引用: 1]

Summary Rice cultivars have been adapted to favorable ecological regions and cropping seasons. Although several heading date genes have separately made contributions to this adaptation, the roles of gene combinations are still unclear. We employed a map-based cloning approach to isolate a heading date gene, which coordinated the interaction between Ghd7 and Ghd8 to greatly delay rice heading. We resequenced these three genes in a germplasm collection to analyze natural variation. Map-based cloning demonstrated that the gene largely affecting the interaction between Ghd7 and Ghd8 was Hd1 . Natural variation analysis showed that a combination of loss-of-function alleles of Ghd7 , Ghd8 and Hd1 contributes to the expansion of rice cultivars to higher latitudes; by contrast, a combination of pre-existing strong alleles of Ghd7 , Ghd8 and functional Hd1 (referred as SSF) is exclusively found where ancestral Asian cultivars originated. Other combinations have comparatively larger favorable ecological scopes and acceptable grain yield. Our results indicate that the combinations of Ghd7 , Ghd8 and Hd1 largely define the ecogeographical adaptation and yield potential in rice cultivars. Breeding varieties with the SSF combination are recommended for tropical regions to fully utilize available energy and light resources and thus produce greater yields.
DOI:10.1007/s12374-015-0425-xURL [本文引用: 1]

Rice flowers after a lengthy vegetative growth. During the vegetative growth period flowering is inhibited by several independent pathways. Whereas Grain number, plant height, and heading date 7 (Ghd7), Heading date 1 (Hd1), Heading date 5 (Hd5), Heading date 6 (Hd6), and Heading date 16 (Hd16) preferentially function to delay flowering under long day conditions, Oryza sativa Phytochrome B (OsPhyB), Oryza sativa CONSTANS-like 4 (OsCOL4), SUPERNUMERARY BRACT (SNB) and Oryza sativa INDETERMINATE SPIKELET 1 (OsIDS1) independently inhibit flowering regardless of day length. After sufficient vegetative growth, flowering signals are produced in the leaves due to reduced expression of the inhibitors. In addition, Hd1 becomes a flowering promoter when the day length becomes shorter. Long-day specific activators OsMADS50 and OsDof12, and a constitutive activators Oryza sativa INDETERMINATE 1 (OsId1), Early heading date 4 (Ehd4), and miR172, are accumulated in the leaves when plants are grown sufficiently. Several circadian clock genes are also involved in floral transition, including Oryza sativa GIGANTEA (OsGI), Heading date 2 (Hd2), and Heading date 17 (Hd17). Floral transition is also controlled by photoreceptors and chromatin remodeling factors. Most of the upstream signals are transferred to Early heading date 1 (Ehd1) that is a positive regulator of Heading data 3a (Hd3a) and Rice FT 1 (RFT1), which are transferred to the shoot apical meristem to induce the reproductive transition.
DOI:10.3389/fpls.2018.00035URL [本文引用: 1]

The heading date is a vital factor in achieving a full rice yield. Cultivars with particular flowering behaviors have been artificially selected to survive in the long-day and low-temperature conditions of Northeast China. To dissect the genetic mechanism responsible for heading date in rice populations from Northeast China, association mapping was performed to identify major controlling loci. A genome-wide association study (GWAS) identified three genetic loci,Hd1,Ghd7, andDTH7, using general and mixed linear models. The three genes were sequenced to analyze natural variations and identify their functions. Loss-of-function alleles of these genes contributed to early rice heading dates in the northern regions of Northeast China, while functional alleles promoted late rice heading dates in the southern regions of Northeast China. Selecting environmentally appropriate allele combinations in new varieties is recommended during breeding. Introducing the earlyindicarice鈥檚 genetic background into Northeastjaponicarice is a reasonable strategy for improving genetic diversity.
DOI:10.1111/pbr.12088URL [本文引用: 1]

Soft rice with low amylose content (AC) ranging by 5-15% is a unique type with special eating and appearance quality and has become popular in the rice market. We resequenced the Wx-mp, a key allele from Milky Princess, a Japanese low AC variety, and found that the +473 mutation in exon 4 is the key mutation in both Wx-mp and its ancestor allele, Wx-mq from Milky Queen. Based on this functional mutation, an allele-specific PCR (AS-PCR) marker was developed and proven in a breeding population derived from a cross between a Chinese late variety Nan Keng 46 (Wx-mp/Wx-mp) and an early line Ning 63121(Wx-b/Wx-b). Based on the marker-aided selection by our newly developed AS-PCR marker for Wx-mp and the known ST10 marker for Stvb-i, a total of 12 Wx-mp homozygotes were selected from 198 F-2 progenies, and four of them were immune to rice stripe virus (RSV) with averagely 11.3 days earlier heading than Nan Keng 46 without significant change in grain yield.
