 ,1
,1Expression and Function Analysis of Endocuticle Structural Glycoprotein Gene LmAbd-2 in Locusta migratoria
JIA Pan1,2, ZHANG Jing1,2, YANG Yang1,2, LIU WeiMin1, ZHANG JianZhen1, ZHAO XiaoMing ,1
,1通讯作者:
收稿日期:2018-09-29接受日期:2018-11-15网络出版日期:2019-02-16
| 基金资助: |
Received:2018-09-29Accepted:2018-11-15Online:2019-02-16
作者简介 About authors
贾盼,Tel:0351-7018871;E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (3259KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
贾盼, 张晶, 杨洋, 刘卫敏, 张建珍, 赵小明. 飞蝗内表皮结构糖蛋白基因LmAbd-2的表达与功能分析[J]. 中国农业科学, 2019, 52(4): 651-660 doi:10.3864/j.issn.0578-1752.2019.04.007
JIA Pan, ZHANG Jing, YANG Yang, LIU WeiMin, ZHANG JianZhen, ZHAO XiaoMing.
0 引言
【研究意义】昆虫体壁主要由上表皮(epicuticle)、外表皮(exocuticle)、内表皮(endocuticle)和真皮细胞层(epidermal cell)组成[1],其在维持虫体外部形态、防止体内水分蒸发、阻止病菌入侵等方面具有重要作用[2]。昆虫在蜕皮过程中首先合成上表皮和外表皮(在蜕皮前合成),然后在蜕皮后逐渐合成内表皮[3]。表皮蛋白(cuticle protein,CP)是昆虫体壁的重要组成成分,其编码基因在昆虫基因组总基因中所占比例超过1%[4],在昆虫生长发育、表皮物理特性以及虫体形态构成中具有重要作用[1]。飞蝗(Locusta migratoria)是一种渐变态世界性农业害虫,对RNA干扰(RNA interference,RNAi)具有敏感性,是基于RNAi研究基因功能的理想模型。因此,以飞蝗蜕皮后表皮蛋白基因为研究对象,探讨其分子特性、表达模式及生物学功能,可为蜕皮后表皮蛋白基因在渐变态昆虫蜕皮过程中参与内表皮形成的作用机制研究提供依据。【前人研究进展】目前,随着黑腹果蝇(Drosophila melanogaster)、赤拟谷盗(Tribolium castaneum)、家蚕(Bombyx mori)、冈比亚按蚊(Anopheles gambiae)、褐飞虱(Nilaparvata lugens)、烟草天蛾(Manduca sexta)等昆虫基因组测序的完成,越来越多的CPs或CP-like蛋白被鉴定[5,6,7,8,9,10]。根据所含基序不同,这些CPs被分为不同家族,如数目最多的CPR家族(包括RR-1、RR-2和RR-3 3个亚类)、CPAPs家族、Tweedle家族、CPF/CPFL家族、CPG家族以及CPH家族等。在双翅目黑腹果蝇、鞘翅目赤拟谷盗、鳞翅目家蚕、半翅目褐飞虱等昆虫中CPR家族表皮蛋白在表皮分化、鞘翅片层结构形成、翅原基发育以及虫体生长发育等方面具有重要作用[10,11,12,13,14,15,16,17]。同样,CPAPs家族表皮蛋白在昆虫体形和生长发育中具有重要作用[18,19,20]。目前,对其他家族表皮蛋白基因的功能研究也取得了重要进展,如甜菜夜蛾(Spodoptera exigua)CPG家族蛋白基因(CPG316、CPG860和CPG4855)的缺失导致表皮黑化变硬和蜕皮致死[21];LU等干扰褐飞虱未分组表皮蛋白(不属于任何已分类家族)基因NlCP21.92(主要表达于蜕皮后时期)可导致虫体发育异常或致死,并且影响内表皮的形 成[17];家蚕蜕皮后高表达表皮蛋白基因BmorCPH24在维持虫体形态和内表皮的形成中具有重要作用[16]。在蝗虫中,20世纪90年代,丹麦科学家ANDERSEN[22]通过双向电泳结合质谱分析方法从沙漠蝗(Schistocerca gregaria)成虫腹部表皮中鉴定出了8个内表皮结构蛋白(endocuticle structural protein),分别命名为SgAbd-1、SgAbd-2、SgAbd-3、SgAbd-4、SgAbd-5、SgAbd-6、SgAbd-8、SgAbd-9。在飞蝗中,根据这8个内表皮结构蛋白氨基酸序列笔者课题组前期基于飞蝗转录组数据库获得了相应的同源蛋白序列[23],分别命名为LmAbd-1、LmAbd-2、LmAbd-3、LmAbd-4、LmAbd-5、LmAbd-6、LmAbd-8、LmAbd-9,它们均属于CPR家族RR-1亚类,并且研究发现LmAbd-5主要表达于外胚层形成的组织中,可能参与飞蝗内表皮的形成[24]。然而,这些表皮蛋白基因在飞蝗蜕皮过程中的具体功能尚不清楚。【本研究切入点】目前,尽管表皮蛋白基因在模式昆虫生长发育中的功能研究取得了较大进展,但对于渐变态昆虫飞蝗表皮蛋白的研究主要集中于表皮蛋白的鉴定和分类,对其编码基因的表达模式及功能尚缺乏系统研究,尤其是蜕皮后表皮蛋白基因在内表皮结构形成中具有怎样的作用机制有待进一步研究。【拟解决的关键问题】以重要农业害虫——飞蝗为研究对象,基于转录组数据获得的内表皮蛋白基因LmAbd-2,研究其分子特性、组织和时期表达特性,并基于RNAi方法结合H&E染色以及透射电镜技术分析其生物学功能,探讨其在飞蝗蜕皮过程中对内表皮结构形成的影响,为阐明蜕皮后表皮蛋白在内表皮形成中的作用机制提供依据。1 材料与方法
试验于2017—2018年在山西大学应用生物学研究所完成。1.1 供试昆虫与试剂
将飞蝗虫卵(购自河北沧州蝗虫养殖基地)包裹于已扎孔透明塑料袋中, 置于昆虫恒温培养室内进行孵化,孵化后转移至纱笼(30 cm×30 cm×30 cm)内,控制昆虫培养室内环境:温度为(30±2)℃,相对湿度为(40±10)%,光周期为14 h﹕10 h(L﹕D),以新鲜小麦苗和麦麸饲喂。试剂:动物组织RNA提取试剂(RNAisoTM Plus)、cDNA反转录试剂(Reverse Transcriptase M-MLV,5×Reverse Transcriptase M-MLV Buffer,Recombinant RNase Inhibitor,dNTP Mixture)、实时荧光定量试剂(TB GreenTM Premix Ex TaqTM Ⅱ)购自TaKaRa公司;dsRNA合成试剂盒(T7 RiboMAXTM Express RNAi System)购自Promega公司;RNase Free H2O等其他试剂购自生工生物(上海)有限公司。
1.2 LmAbd-2 cDNA序列分析
根据飞蝗转录组LmAbd-2的cDNA编码序列,克隆测序后,利用NCBI中的Open Reading Frame Finder(ORF Finder)(1.3 序列比对与系统发育分析
在NCBI网站下载不同昆虫物种Abd-2氨基酸序列,利用MEGA 6.0软件中neighbor-joining(NJ)方法将LmAbd-2氨基酸序列与沙漠蝗、德国小蠊(Blattella germanica)、赤拟谷盗、斜纹夜蛾(Spodoptera litura)、褐飞虱等不同目物种的同源氨基酸序列进行聚类分析,独立分析1 000次,数值代表bootstrap估算值。不同目物种Abd-2序列和飞蝗LmAbd-2序列的GenBank登录号见表1。Table 1
表1
表1本研究中用于构建系统进化树的物种和Abd-2的GenBank登录号
Table 1
| 目Order | 物种Species | 基因Gene | GenBank登录号 GenBank accession number |
|---|---|---|---|
| 鳞翅目Lepidoptera | 玉带凤蝶Papilio polytes | PpAbd-2 | XP_013141188.1 |
| 柑橘凤蝶Papilio xuthus | PxAbd-2 | NP_001299525.1 | |
| 棉铃虫Helicoverpa armigera | HaAbd-2 | XP_021201029.1 | |
| 斜纹夜蛾Spodoptera litura | SlAbd-2 | XP_022816544.1 | |
| 半翅目Hemiptera | 臭虫Cimex lectularius | ClAbd-2 | XP_014247089.1 |
| 褐飞虱Nilaparvata lugens | NlAbd-2 | XP_022195400.1 | |
| 直翅目Orthoptera | 飞蝗Locusta migratoria | LmAbd-2 | ASQ42722.1 |
| 沙漠蝗Schistocerca gregaria | SgAbd-2 | Q7M4F3.1 | |
| 蜚蠊目Blattaria | 堆砂白蚁Cryptotermes secundus | CsAbd-2 | XP_023704658.1 |
| 内华达古白蚁Zootermopsis nevadensis | ZnAbd-2 | XP_021932412.1 | |
| 德国小蠊Blattella germanica | BgAbd-2 | PSN37147.1 | |
| 鞘翅目Coleoptera | 山松甲虫Dendroctonus ponderosae | DpAbd-2 | XP_019764279.1 |
| 蜣螂Onthophagus taurus | OtAbd-2 | XP_022906571.1 | |
| 天牛Anoplophora glabripennis | AgAbd-2 | XP_018574122.1 | |
| 赤拟谷盗Tribolium castaneum | TcAbd-2 | XP_974125.1 |
新窗口打开|下载CSV
1.4 LmAbd-2组织特异性和时期表达分析
组织特异性表达分析:解剖发育整齐的5龄第2天(N5D2)飞蝗若虫的体壁(integument,IN)、前肠(foregut,FG)、中肠(midgut,MG)、后肠(hindgut,HG)、胃盲囊(gastric caeca,GC)、马氏管(Malpighian tubules,MT)、脂肪体(fat body,FB)和翅芽(wing pad,WP)8个不同组织,设置4个生物学重复,每个重复3头虫体。时期表达分析:选取并解剖4龄第1—6天飞蝗若虫(N4D1—N4D6)、5龄第1—8天飞蝗若虫(N5D1—N5D8)和成虫第1—4天(AD1—AD4)第2腹节处体壁组织,设4个生物学重复,每3头虫体组织为一个生物学重复,组织样品冻存于液氮中。利用动物组织RNA提取试剂RNAisoTM Plus(TaKaRa公司)提取上述不同组织和不同发育时期体壁组织的总RNA,使用1%的琼脂糖凝胶电泳检测RNA的质量,利用Nanodrop 2000对总RNA进行定量。以1 μg总RNA为模板,使用cDNA反转录试剂(TaKaRa公司)合成cDNA模板。采用荧光定量PCR方法(reverse-transcription quantitative PCR,RT-qPCR)检测LmAbd-2在5龄第2天飞蝗不同组织部位及不同发育时期体壁中的表达情况。RT-qPCR反应体系和程序参考文献[25]。β-actin(KX276642)作为内参基因,引物见表2(由上海生工生物公司合成)。Table 2
表2
表2本研究中所用引物
Table 2
| 基因 Gene | 引物序列Primer sequence (5′-3′) | 用途 Application | 产物长度 Product length (bp) |
|---|---|---|---|
| LmAbd-2 | F:taatacgactcactatagggATGCAGCTGTTGGTATGCCT | dsLmAbd-2合成 dsLmAbd-2 synthesis | 196 |
| R:taatacgactcactatagggGCTCCTGGGCGGCGATGCCG | |||
| GFP | F:taatacgactcactatagggTGGAGAGGGTGAAGG | dsGFP 合成 dsGFP synthesis | 571 |
| R:taatacgactcactatagggGGGCAGATTGTGTGGAC | |||
| LmAbd-2 | F:GTGTTTGACCCTGCTGGTG | RT-qPCR | 106 |
| R:CCGTTGCCGTTTCGTATT | |||
| β-actin | F:CGAAGCACAGTCAAAGAGAGGTA | 156 | |
| R:GCTTCAGTCAAGAGAACAGGATG |
新窗口打开|下载CSV
1.5 基于RNAi的LmAbd-2生物学功能分析
为了探讨LmAbd-2在飞蝗表皮形成中的作用,利用T7 RiboMAXTM Express试剂盒(Promega公司)体外合成该基因双链RNA(dsRNA),以绿色荧光蛋白基因(GFP)为对照。利用微量注射器向4龄第3天(N4D3)若虫第2—3腹节处注射10 μg dsLmAbd-2和dsGFP,正常饲养;若飞蝗正常蜕皮后,再向5龄第4天(N5D4)若虫注射10 μg的dsLmAbd-2和dsGFP,24 h后利用1.4中RT-qPCR方法检测LmAbd-2沉默效率,其余若虫正常饲养、观察表型。1.6 表皮显微和超微结构观察
为了观察干扰LmAbd-2后对飞蝗表皮结构的影响,取5龄蜕皮后72 h的对照组和处理组腹部表皮样品(第3腹节)于3%戊二醛溶液中4℃固定过夜。固定的样品进行石蜡包埋、切片,然后用苏木精和伊红(hematoxylin-eosin staining,H&E)染色[26,27]、封片后利用显微镜(Olympus BX51,Japan)观察表皮结构。为进一步从超微结构分析LmAbd-2对飞蝗内表皮形成的影响,用透射电镜技术观察表皮结构变化。用于透射电镜观察的腹部表皮样品固定、切片制备和染色等方法参考文献[25]。
1.7 数据分析
基因相对表达量采用2-ΔΔCt法[28]进行分析,采用Student t-test方法进行差异表达分析,*表示在P<0.05水平差异显著,**表示在P<0.01水平差异显著,***表示在P<0.001水平差异显著。2 结果
2.1 LmAbd-2的序列结构特征
根据飞蝗转录组数据库获得LmAbd-2 cDNA编码序列并进行了克隆和测序验证,其编码序列长为689 bp,其中ORF为459 bp, 编码152个氨基酸,预测其分子量为16.98 kD,等电点为5.28,为偏酸性表皮蛋白。序列分析发现其含有1个信号肽和1个几丁质结合域(chitin binding domain 4,ChtBD4)(图1-A),并且具有RR-1保守基序,属于CPR家族RR-1亚类(图1-B)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1LmAbd-2的序列分析
A:LmAbd-2的序列分析Sequence analysis of LmAbd-2;下划线为信号肽The underline: signal peptide,方框内为几丁质结合域The box: chitin binding domain 4, ChtBD4。B:Weblogo分析LmAbd-2保守基序Analysis of conserved motifs by Weblogo (
Fig. 1Sequence analysis of LmAbd-2
2.2 Abd-2序列比对与系统发育树分析
多序列比对分析发现LmAbd-2与沙漠蝗、褐飞虱、赤拟谷盗和棉铃虫(Helicoverpa armigera)等Abd-2序列具有较高相似性,其中与沙漠蝗SgAbd-2的氨基酸序列相似度高达92.5%(信号肽除外),几丁质结合域序列完全一致(图2-A)。同时发现Abd-2 N末端存在一个保守基序PTPPPIP,且第2位苏氨酸(T)是O-连接糖基化位点(O-GalNAc)(图2-A),与SgAbd-2类似,是一个糖基化蛋白[22]。将获得的飞蝗Abd-2氨基酸序列与其他昆虫的Abd-2氨基酸序列进行聚类分析,结果显示LmAbd-2与沙漠蝗SgAbd-2聚为一支,具有较近的亲缘关系(图2-B)。图2
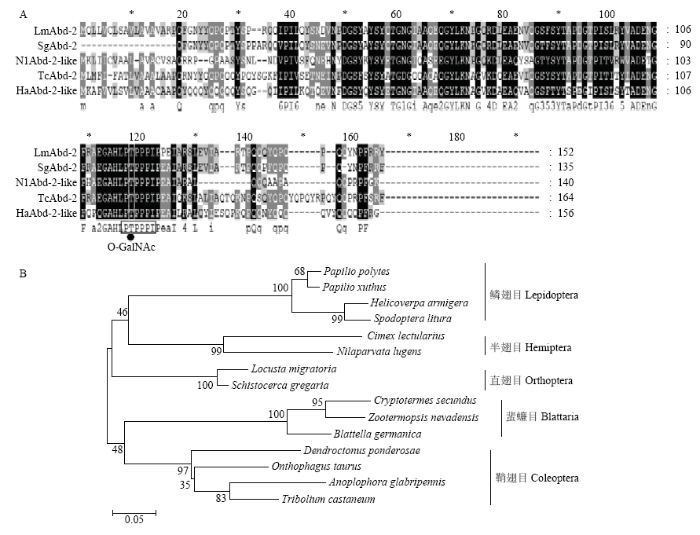 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2LmAbd-2与其他昆虫表皮蛋白Abd-2多序列比对与系统聚类分析
A:LmAbd-2与其他昆虫表皮蛋白Abd-2多序列比对 Multiple sequence alignments of LmAbd-2 and Abd-2 from different insect species;B:LmAbd-2与其他昆虫Abd-2的系统聚类分析 The phylogenetic analysis of LmAbd-2 and Abd-2 from different insect species
Fig. 2Multiple sequence alignments and phylogenetic analysis of LmAbd-2 and Abd-2 from different insect species
2.3 LmAbd-2组织特异性和发育时期表达
RT-qPCR分析结果显示LmAbd-2在体壁中高表达,在其他组织中低表达或不表达(图3-A)。发育时期结果显示,LmAbd-2在4龄(N4D1—N4D2)、5龄(N5D1—N5D3)和成虫早期(AD1—AD3)呈周期性高表达,其他时期表达较低(图3-B),这种表达模式与内表皮形成时间一致,推测其可能参与飞蝗内表皮结构的形成。图3
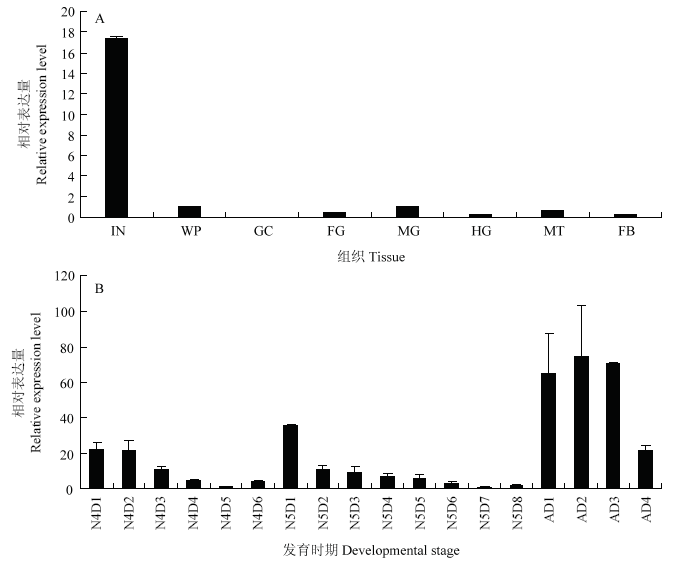 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3LmAbd-2在飞蝗不同组织和不同发育时期的表达
A:LmAbd-2在5龄飞蝗不同组织中的表达 The relative expression of LmAbd-2 in different tissues of 5th instar nymphs。IN:体壁 Integument;WP:翅芽 Wing pads;GC:胃盲囊 Gastric caeca;FG:前肠 Foregut;MG:中肠 Midgut;HG:后肠 Hindgut;MT:马氏管 Malpighian tubules;FB:脂肪体 Fat body;B:LmAbd-2在飞蝗4龄(第1—6天)、5龄(第1—8天)和成虫早期(第1—4天)不同发育天数的表达The relative expression of LmAbd-2 in different days of 4th instar nymphs (Day 1 to day 6), 5th instar nymphs (Day 1 to day 8), and early stages of adult (Day 1 to day 4)
Fig. 3The relative expression of LmAbd-2 in different tissues and different developmental stages
2.4 LmAbd-2的生物学功能
为了探讨LmAbd-2在飞蝗表皮形成中的作用,体外合成dsRNA并向4龄第3天飞蝗若虫注射10 μg的dsLmAbd-2和dsGFP,正常饲养至蜕皮。结果发现处理组和对照组均能正常蜕皮至5龄且无肉眼可见表型(LmAbd-2被显著沉默)。向该5龄期若虫再次分别注射10 μg的dsLmAbd-2和dsGFP,结果显示再次注射dsLmAbd-2的若虫和对照组仍均能正常蜕皮,尽管LmAbd-2在蜕至成虫72 h后表达量显著降低(处理组中LmAbd-2表达量显著降低了97.8%)(图4-A),但未出现肉眼可见的异常表型。H&E染色结果显示,与对照组相比,注射dsLmAbd-2的处理组表皮层结构变薄(减少约40%)(图4-B);透射电镜结果显示,与对照组相比,处理组内表皮片层数减少(约减少6层),使得整个表皮层结构变薄(图4-C),表明LmAbd-2参与飞蝗内表皮结构的形成。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4LmAbd-2 对飞蝗表皮结构的影响
A:若虫期连续注射dsLmAbd-2蜕皮至成虫72 h后LmAbd-2的相对表达量 The relative expression of LmAbd-2 determined by RT-qPCR after injected with dsLmAbd-2 72 h。B:注射dsRNA后,蜕皮至成虫72 h的表皮显微结构 After injection of dsRNA, microstructure and magnification of cuticle by H&E staining;红色为细胞质及细胞外基质,蓝色为细胞核 Cytoplasm and extracellular matrix are stained red with eosin, nuclei are stained dark blue with hematoxylin (Scale bars, 20 μm),Cut:表皮Cuticle;Ec:表皮细胞Epidermal cell。C:注射dsRNA后,蜕皮至成虫72 h的表皮超微结构观察 Ultrastructure observation of adult cuticle at 72 h after injected with dsRNA。Epi:上表皮Epicuticle;Exo:外表皮Exocuticle;Endo:内表皮Endocuticle
Fig. 4Effect of LmAbd-2 on the formation of cuticle by H&E staining and TEM after injection of dsRNA
3 讨论
体壁是覆盖在昆虫身体表面的保护性和支撑性的结构,其主要成分是表皮蛋白和几丁质,其成分的缺失或含量变化都可能影响体壁结构的形成,进而影响昆虫的正常生长发育。本文根据飞蝗转录组数据库获得一个蜕皮后表皮蛋白基因LmAbd-2,其编码蛋白序列特征分析发现含有一个RR-1基序(GxFxYxxPDGxxxxVxYxADENGYQPxGAHLP),与沙漠蝗SgAbd-2的氨基酸序列具有92.5%的相似度(信号肽除外),均属于CPR家族RR-1亚类[4]。随着基因组学和转录组学的发展,除直翅目飞蝗和沙漠蝗外,有很多昆虫物种的Abd-2或Abd-2 like被鉴定,如鳞翅目棉铃虫、半翅目褐飞虱、蜚蠊目德国小蠊、以及鞘翅目赤拟谷盗等。然而,这些基因仅仅是序列在NCBI中报道,尚未有功能研究报道。有研究报道,RR-1亚类的表皮蛋白主要存在于柔软表皮中,如双翅目和鳞翅目幼虫表皮、蝗虫节间膜和内表皮中[6,22]。昆虫在蜕皮过程中表皮依次形成片层结构,其中蜕皮前主要形成上表皮和外表皮,而内表皮主要在蜕皮后开始形成和沉积,蜕皮后约72 h完全形成[29]。组织和时期表达结果显示LmAbd-2主要在飞蝗体壁中高表达,而且发现其在所试飞蝗各个龄期内表皮形成时期呈周期性高表达(图3),暗示其可能参与飞蝗内表皮的合成和沉积。利用飞蝗对RNAi的敏感性,本文首先在4龄若虫期沉默LmAbd-2的表达,随后在5龄若虫期连续干扰LmAbd-2的表达,虽然均未出现肉眼可见的异常表型,但从显微和超微结构分析发现内表皮片层结构减少导致表皮层结构变薄(图4),表明LmAbd-2参与飞蝗内表皮结构的形成。该基因与家蚕假设表皮蛋白基因BmorCPH24、褐飞虱未分组表皮蛋白基因NlCP21.92以及飞蝗其他家族表皮蛋白基因LmNCP1功能类似[16-17,27],均在昆虫内表皮结构形成具有重要作用。
根据表皮片层结构形成时期的不同,ANDERSEN利用MALDI-MS分析方法从沙漠蝗蜕皮后表皮组织中鉴定出8种RR-1亚类蜕皮后表皮蛋白[22],课题组前期从飞蝗转录组数据库中鉴定到与这8种沙漠蝗蜕皮后表皮蛋白同源序列[23],其中研究发现LmAbd-5参与飞蝗内表皮的形成[24]。然而,沉默LmAbd-5的表达后,结果与LmAbd-2类似均未引起飞蝗肉眼可见的异常表型。PAN等[10]利用RNAi方法对褐飞虱表皮蛋白基因功能研究发现仅部分基因产生致死表型,推测表皮蛋白可能与其他表皮蛋白相互作用而存在互补效应。在飞蝗中,这8种蜕皮后表皮蛋白在蜕皮过程中可能存在协同或互补作用,但尚有待进一步的研究。
4 结论
飞蝗LmAbd-2是一个内表皮结构糖蛋白基因,其编码的蛋白含有一个信号肽和一个几丁质结合域ChtBD4,属于CPR家族RR-1亚类;LmAbd-2主要在飞蝗体壁组织中高表达,而且在所试飞蝗各龄期内表皮形成时期呈周期性高表达;连续在若虫期干扰LmAbd-2后未出现肉眼可见的表型,但内表皮片层结构减少,表明其参与飞蝗内表皮结构的形成。(责任编辑 岳梅)
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 2]
DOI:10.1016/j.asd.2004.05.006URLPMID:18089034 [本文引用: 1]

Since nearly all adult insects fly, the cuticle has to provide a very efficient and lightweight skeleton. Information is available about the mechanical properties of cuticle—Young's modulus of resilin is about 1 MPa, of soft cuticles about 102kPa to 50 MPa, of sclerotised cuticles 1–20 GPa; Vicker's Hardness of sclerotised cuticle ranges between 25 and 8002kgf02mm; density is 1–1.3 kg m—and one of its components, chitin nanofibres, the Young's modulus of which is more than 150 GPa. Experiments based on fracture mechanics have not been performed although the layered structure probably provides some toughening. The structural performance of wings and legs has been measured, but our understanding of the importance of buckling is lacking: it can stiffen the structure (by elastic postbuckling in wings, for example) or be a failure mode. We know nothing of fatigue properties (yet, for instance, the insect wing must undergo millions of cycles, flexing or buckling on each cycle). The remarkable mechanical performance and efficiency of cuticle can be analysed and compared with those of other materials using material property charts and material indices. Presented in this paper are four: Young's modulus—density (stiffness per unit weight), specific Young's modulus—specific strength (elastic hinges, elastic energy storage per unit weight), toughness—Young's modulus (fracture resistance under various loading conditions), and hardness (wear resistance). In conjunction with a structural analysis of cuticle these charts help to understand the relevance of microstructure (fibre orientation effects in tendons, joints and sense organs, for example) and shape (including surface structure) of this fibrous composite for a given function. With modern techniques for analysis of structure and material, and emphasis on nanocomposites and self-assembly, insect cuticle should be the archetype for composites at all levels of scale.
DOI:10.1016/j.ibmb.2017.11.003URLPMID:29117500 [本文引用: 1]

react-text: 218 Exploiting specific targets is of specific interest in developing eco-friendly pesticides. We isolated, purified and characterized a novel beta-N-acetyl-D-hexosaminidase (OfHex1) from the fifth instar larva integument of the Asian corn borer, Ostrinia furnacalis (Guen茅e). OfHex1 was purified 1468-fold to homogeneity with an activity yield of 20% by four column chromatography steps. Under... /react-text react-text: 219 /react-text [Show full abstract]
DOI:10.1016/j.ibmb.2010.02.001URLPMID:20171281 [本文引用: 2]

The availability of whole genome sequences of several arthropods has provided new insights into structural cuticular proteins (CPs), in particular the distribution of different families, the recognition that these proteins may comprise almost 2% of the protein coding genes of some species, and the identification of features that should aid in the annotation of new genomes and EST libraries as they become available. Twelve CP families are described: CPR (named after the Rebers and Riddiford Consensus); CPF (named because it has a highly conserved region consisting of about forty-four amino acids); CPFL (like the CPFs in a conserved C-terminal region); the TWDL family, named after a picturesque phenotype of one mutant member; four families in addition to TWDL with a preponderance of low complexity sequence that are not member of the families listed above. These were named after particular diagnostic features as CPLCA, CPLCG, CPLCW, CPLCP. There are also CPG, a lepidopteran family with an abundance of glycines, the apidermin family, named after three proteins in Also described are common motifs and features. Four unusual CPs are discussed in detail. Data that facilitated the analysis of sequence variation of single CP genes in natural populations are analyzed.
[本文引用: 1]
[本文引用: 2]
DOI:10.1016/j.ibmb.2008.05.007URLPMID:19280704 [本文引用: 1]

Many kinds of cuticular proteins are found in a single insect species and their numbers and features are diversified among insects. Because there are so many cuticular proteins and so much sequence variation among them, an overview of cuticular protein gene is needed. Recently, a complete silkworm genome sequence was obtained through the integration of data from two whole genome sequence projects performed independently in 2004. To identify cuticular protein genes in the silkworm Bombyx mori exhaustively, we searched both the Bombyx whole genome sequence as well as various EST libraries, and found 220 putative cuticular protein genes. We also revised the annotation of the gene model, and named each identified cuticular protein based on its motif. The phylogenetic tree of cuticular protein genes among B. mori, Drosophila melanogaster, and Apis mellifera revealed that duplicate cuticular protein clusters have evolved independently among insects. Comparison of EST libraries and northern blot analyses showed that the tissue- and stage-specific expression of each gene was intricately regulated, even between adjacent genes in the same gene cluster. This study reveals many novel cuticular protein genes as well as insights into cuticular protein gene regulation.
DOI:10.1016/j.ibmb.2014.06.004URLPMID:4143468 [本文引用: 1]

The arthropod cuticle is a composite, bipartite system, made of chitin filaments embedded in a proteinaceous matrix. The physical properties of cuticle are determined by the structure and the interactions of its two major components, cuticular proteins (CPs) and chitin. The proteinaceous matrix consists mainly of structural cuticular proteins. The majority of the structural proteins that have been described to date belong to the CPR family, and they are identified by the conserved R&R region (Rebers and Riddiford Consensus). Two major subfamilies of the CPR family RR-1 and RR-2, have also been identified from conservation at sequence level and some correlation with the cuticle type. Recently, several novel families, also containing characteristic conserved regions, have been described. The package HMMER v3.0 (http://hmmer.janelia.org/) was used to build characteristic profile Hidden Markov Models based on the characteristic regions for 8 of these families, (CPF, CPAP3, CPAP1, CPCFC, CPLCA, CPLCG, CPLCW, Tweedle). In brief, these families can be described as having: CPF (a conserved region with 44 amino acids); CPAP1 and CPAP-3 (analogous to peritrophins, with 1 and 3 chitin-binding domains, respectively); CPCFC (2 or 3 C-x(5)-C repeats); and four of five low complexity (LC) families, each with characteristic domains. Using these models, as well as the models previously created for the two major subfamilies of the CPR family, RR-1 and RR-2 (Karouzou et al., 2007), we developed CutProtFam-Pred, an on-line tool (http://bioinformatics.biol.uoa.gr/CutProtFam-Pred) that allows one to query sequences from proteomes or translated transcriptomes, for the accurate detection and classification of putative structural cuticular proteins. The tool has been applied successfully to diverse arthropod proteomes including a crustacean (Daphnia pulex) and a chelicerate (Tetranychus urticae), but at this taxonomic distance only CPRs and CPAPs were recovered. (C) 2014 Elsevier Ltd. All rights reserved.
DOI:10.1016/j.ibmb.2014.12.010URLPMID:25576653 [本文引用: 1]

61248 cuticular protein (CP) genes were annotated in Manduca sexta.61Orthology was established between M.02sexta and Bombyx mori CP genes.61Gene expression was analyzed via 52 RNAseq libraries.61Diverse expression was observed across various tissues and developmental stages.
DOI:10.1073/pnas.1716951115URL [本文引用: 3]

The cuticle, mainly composed of chitin and cuticular proteins (CPs), is a multifunctional structure of arthropods. CPs usually account for >1% of the total insect proteins encoded in the genome. Why does an insect need so many different CPs? In this study, we use comprehensive large-scale technologies to study the full complement of CPs and their functions in the brown planthopper (BPH). A total of 32 of the 140 BPH CP genes are found to be essential for nymph/adult development, egg production, or embryo development; in addition, redundant and complementary functions of CPs are revealed. Cuticle, mainly composed of chitin and cuticular proteins (CPs), is a multifunctional structure of arthropods. CPs usually account for >1% of the total insect proteins. Why does an insect encode so many different CP genes in the genome? In this study, we use comprehensive large-scale technologies to study the full complement of CPs (i.e., the CP-ome) of the brown planthopper (BPH),Nilaparvata lugens, a major rice plant pest. Eight CP families (CPR, CPF, TWDL, CPLCP, CPG, CPAP1, CPAP3, and CPAPn) including 140 proteins in BPH, in which CPAPn is a CP family that we discovered. The CPG family that was considered to be restricted to the Lepidoptera has also been identified in BPH. As reported here, CPLCP family members are characterized by three conserved sequence motifs. In addition, we identified a testis protein family with a peritrophin A domain that we named TPAP. We authenticated the real existence of 106 proteins among the 140 CPs. RNA interference (RNAi) experiments were conducted against 135 CP genes in early- and late-instar nymphs and newly emerged female adults, demonstrating that 32 CPs were essential for BPH normal development or egg production. Combined RNAi experiments suggested redundant and complementary functions of the large number of CPs. Transcriptomic data revealed that the CP genes were expressed in a tissue-specific manner, and there were four clusters of developmental expression patterns. This study gives a comprehensive understanding of the roles of CPs in an insect cuticle.
DOI:10.1073/pnas.0607616103PMID:17075064 [本文引用: 1]

Body shape determination represents a critical aspect of morphogenesis. In the course of investigating body shape regulation in Drosophila, we have identified a dominant mutation,$TweedleD^{1}$$(TwdlD^{1})$, that alters overall dimensions at the larval and pupal stages. Characterization of the affected locus led to the discovery of a gene family that has 27 members in Drosophila and is found only among insects. Analysis of gene expression at the RNA and protein levels revealed gene-specific temporal and spatial patterns in ectodermally derived tissues. In addition, light microscopic studies of fluorescently tagged proteins demonstrated that Tweedle proteins are incorporated into larval cuticular structures. This demonstration that a mutation in a Drosophila cuticular protein gene alters overall morphology confirms a role for the fly exoskeleton in determining body shape. Furthermore, parallels between these findings and studies of cuticle collagen genes in Caenorhabditis elegans suggest that the exoskeleton influences body shape in diverse organisms.
DOI:10.1371/journal.pgen.1002682URLPMID:22570623 [本文引用: 1]

Insect cuticle is composed primarily of chitin and structural proteins. To study the function of structural cuticular proteins, we focused on the proteins present in elytra (modified forewings that become highly sclerotized and pigmented covers for the hindwings) of the red flour beetle,Tribolium castaneum. We identified two highly abundant proteins, TcCPR27 (10 kDa) and TcCPR18 (20 kDa), which are also present in pronotum and ventral abdominal cuticles. Both are members of the Rebers and Riddiford family of cuticular proteins and contain RR2 motifs. Transcripts for both genes dramatically increase in abundance at the pharate adult stage and then decline quickly thereafter. Injection of specific double-stranded RNAs for each gene into penultimate or last instar larvae had no effect on larval arval, larval upal, or pupal dult molting. The elytra of the resulting adults, however, were shorter, wrinkled, warped, fenestrated, and less rigid than those from control insects. TcCPR27-deficient insects could not fold their hindwings properly and died prematurely approximately one week after eclosion, probably because of dehydration. TcCPR18-deficient insects exhibited a similar but less dramatic phenotype. Immunolocalization studies confirmed the presence of TcCPR27 in the elytral cuticle. These results demonstrate that TcCPR27 and TcCPR18 are major structural proteins in the rigid elytral, dorsal thoracic, and ventral abdominal cuticles of the red flour beetle, and that both proteins are required for morphogenesis of the beetle's elytra. Primitive insects have two pairs of membranous flight wings, but during the evolution of the beetle lineage the forewings lost their flight function and became modified as hard, rigid covers called elytra for protection of soft body parts of the abdomen and also the delicate flexible hindwings, which retained their flight function. This transformation is manifested by a greatly thickened and rigid (sclerotized) exoskeletal cuticle secreted by the forewing epidermis. We demonstrate that this evolutionary modification is accompanied by the incorporation of two highly abundant structural proteins into the elytral cuticle, namely TcCPR18 and TcCPR27. Depletion of these proteins by RNA interference results in malformation and weakening of the elytra, culminating in insect death. These proteins are also abundant in hard cuticle from other regions such as the pronotum and ventral abdomen, but are absent in soft cuticles, and therefore may function as key determinants of rigid cuticle. Expression of such proteins at high levels in the modified forewing appears to have been a fundamental evolutionary step in the transformation of the membranous wing into a thickened and rigid elytron in the Coleoptera.
DOI:10.1534/genetics.113.158766URLPMID:24514903 [本文引用: 1]

Cuticular proteins (CPs) are crucial components of the insect cuticle. Although numerous genes encoding cuticular proteins have been identified in known insect genomes to date, their functions in maintaining insect body shape and adaptability remain largely unknown. In the current study, positional cloning led to the identification of a gene encoding an RR1-type cuticular protein, BmorCPR2, highly expressed in larval chitin-rich tissues and at the mulberry leaf-eating stages, which is responsible for the silkworm stony mutant. In the Dazao-stony strain, the BmorCPR2 allele is a deletion mutation with significantly lower expression, compared to the wild-type Dazao strain. Dysfunctional BmorCPR2 in the stony mutant lost chitin binding ability, leading to reduced chitin content in larval cuticle, limitation of cuticle extension, abatement of cuticle tensile properties, and aberrant ratio between internodes and intersegmental folds. These variations induce a significant decrease in cuticle capacity to hold the growing internal organs in the larval development process, resulting in whole-body stiffness, tightness, and hardness, bulging intersegmental folds, and serious defects in larval adaptability. To our knowledge, this is the first study to report the corresponding phenotype of stony in insects caused by mutation of RR1-type cuticular protein. Our findings collectively shed light on the specific role of cuticular proteins in maintaining normal larval body shape and will aid in the development of pest control strategies for the management of Lepidoptera.
DOI:10.1038/srep10484URLPMID:25994234 [本文引用: 1]

In the insect cuticle, structural proteins (CPs) and the polysaccharide chitin are the major components. It has been hypothesized that CPs are cross-linked to other CPs and possibly to chitin by quinones or quinone methides produced by the laccase2-mediated oxidation of N-acylcatechols. In this study we investigated functions of TcCP30, the third most abundant CP in protein extracts of elytra (wing covers) from Tribolium castaneum adults. The mature TcCP30 protein has a low complexity and highly polar amino acid sequence. TcCP30 is localized with chitin in horizontal laminae and vertically oriented columnar structures in rigid cuticles, but not in soft and membranous cuticles. Immunoblot analysis revealed that TcCP30 undergoes laccase2-mediated cross-linking during cuticle maturation in vivo, a process confirmed in vitro using recombinant rTcCP30. We identified TcCPR27 and TcCPR18, the two most abundant proteins in the elytra, as putative cross-linking partners of TcCP30. RNAi for the TcCP30 gene had no effect on larval and pupal growth and development. However, during adult eclosion, ~70% of the adults were unable to shed their exuvium and died. These results support the hypothesis that TcCP30 plays an integral role as a cross-linked structural protein in the formation of lightweight rigid cuticle of the beetle.
DOI:10.1371/journal.pgen.1004963URLPMID:25664770 [本文引用: 1]

Abstract Insect cuticle is composed mainly of structural proteins and the polysaccharide chitin. The CPR family is the largest family of cuticle proteins (CPs), which can be further divided into three subgroups based on the presence of one of the three presumptive chitin-binding sequence motifs denoted as Rebers-Riddiford (R&R) consensus sequence motifs RR-1, RR-2 and RR-3. The TcCPR27 protein containing the RR-2 motif is one of the most abundant CPs present both in the horizontal laminae and in vertical pore canals in the procuticle of rigid cuticle found in the elytron of the red flour beetle, Tribolium castaneum. Depletion of TcCPR27 by RNA interference (RNAi) causes both unorganized laminae and pore canals, resulting in malformation and weakening of the elytron. In this study, we investigated the function(s) of another CP, TcCPR4, which contains the RR-1 motif and is easily extractable from elytra after RNAi to deplete the level of TcCPR27. Transcript levels of the TcCPR4 gene are dramatically increased in 3 d-old pupae when adult cuticle synthesis begins. Immunohistochemical studies revealed that TcCPR4 protein is present in the rigid cuticles of the dorsal elytron, ventral abdomen and leg but not in the flexible cuticles of the hindwing and dorsal abdomen of adult T. castaneum. Immunogold labeling and transmission electron microscopic analyses revealed that TcCPR4 is predominantly localized in pore canals and regions around the apical plasma membrane protrusions into the procuticle of rigid adult cuticles. RNAi for TcCPR4 resulted in an abnormal shape of the pore canals with amorphous pore canal fibers (PCFs) in their lumen. These results support the hypothesis that TcCPR4 is required for achieving proper morphology of the vertical pore canals and PCFs that contribute to the assembly of a cuticle that is both lightweight and rigid.
DOI:10.1534/genetics.117.300300URLPMID:28923848 [本文引用: 3]

The genetic basis of body shape and coloration patterns on caterpillars is often assumed to be regulated separately, but it is possible that common molecules affect both types of trait simultaneously. Here we examine the genetic basis of a spontaneous cuticle defect in silkworm, where larvae exhibit a bamboo-like body shape and decreased pigmentation. We performed linkage mapping and mutation screening to determine the gene product that affects body shape and coloration simultaneously. In these mutant larvae we identified a null mutation in BmorCPH24, a gene encoding a cuticular protein with low complexity sequence. Spatiotemporal expression analyses showed that BmorCPH24 is expressed in the larval epidermis postecdysis. RNAi-mediated knockdown and CRISPR/Cas9-mediated knockout of BmorCPH24 produced the abnormal body shape and the inhibited pigment typical of the mutant phenotype. In addition, our results showed that BmorCPH24 may be involved in the synthesis of endocuticle and its disruption-induced apoptosis of epidermal cells that accompanied the reduced expression of R&R-type larval cuticle protein genes and pigmentation gene Wnt1. Strikingly, BmorCPH24, a fast-evolving gene, has evolved a new function responsible for the assembly of silkworm larval cuticle and has evolved to be an indispensable factor maintaining the larval body shape and its coloration pattern. This is the first study to identify a molecule whose pleiotropic function affects the development of body shape and color patterns in insect larvae.
DOI:10.1016/j.ibmb.2018.06.001URL [本文引用: 3]

Using transcriptome analysis of tissues of the brown planthopper (BPH), Nilaparvata lugens , we identified a gene tentatively designated NlCP21. 92 that was expressed at high levels in the integument. Spatiotemporal expression profiling with quantitative PCR and Western blotting verified its integument-specific expression and showed periodic expression during molting. The open reading frame was GC-rich and encoded a hydrophobic polypeptide. The polypeptide contained AAPA/V repeat motifs and other sequence features similar to several reported cuticular proteins but lacked an R&R consensus and other chitin-binding domains. Double-stranded RNA ediated RNA interference of the NlCP21.92 gene resulted in abnormal and lethal morphological phenotypes, and transmission electron microscopy revealed the corresponding ultrastructural defects. Immunohistochemical staining demonstrated that the NlCP21.92 protein was primarily located in the procuticle. Our results suggest that NlCP21.92 is a novel ungrouped cuticular protein essential for normal endocuticle formation.
DOI:10.1074/jbc.m112.359984URLPMID:3375561 [本文引用: 1]

The epidermis and internal tubular organs, such as gut and lungs, are exposed to a hostile environment. They form an extracellular matrix to provide epithelial integrity and to prevent contact with pathogens and toxins. In arthropods, the cuticle protects, shapes, and enables the functioning of organs. During development, cuticle matrix is shielded from premature degradation; however, underlying molecular mechanisms are poorly understood. Previously, we identified the conserved obstructor multigene-family, which encodes chitin-binding proteins. Here we show that Obstructor-A is required for extracellular matrix dynamics in cuticle forming organs. Loss of obstructor-A causes severe defects during cuticle molting, wound protection, tube expansion and larval growth control. We found that Obstructor-A interacts and forms a core complex with the polysaccharide chitin, the cuticle modifier Knickkopf and the chitin deacetylase Serpentine. Knickkopf protects chitin from chitinase-dependent degradation and deacetylase enzymes ensure extracellular matrix maturation. We provide evidence that Obstructor-A is required to control the presence of Knickkopf and Serpentine in the extracellular matrix. We propose a model suggesting that Obstructor-A coordinates the core complex for extracellular matrix protection from premature degradation. This mechanism enables exoskeletal molting, tube expansion, and epithelial integrity. The evolutionary conservation suggests a common role of Obstructor-A and homologs in coordinating extracellular matrix protection in epithelial tissues of chitinous invertebrates.
DOI:10.3864/j.issn.0578-1752.2015.01.08URL [本文引用: 1]

【目的】获得飞蝗(Locusta migratoria)表皮蛋白 Obstructor(Obst)家族基因的 cDNA 序列,并研究其序列特征和 mRNA 表达特性,探讨其生物学功能,为害虫防治提供新的分子靶标。【方法】采用生物信息学方法搜索飞蝗转录组数据库获得 Obst 家族基因 cDNA 片段,并进行 BLAST 分析得到 Obst 家族基因的 cDNA 序列;采用 RACE 技术扩增该家族基因的3′cDNA 序列,拼接后获得全长;SignalP 在线软件分析蛋白的信号肽,SMART 网站预测其功能域,并使用 Mega 5.10软件中 Neighbor-Joining 方法,与黑腹果蝇(Drosophila melanogaster) Obst 家族基因和赤拟谷盗(Tribolium castaneum)CPAP3家族基因(Obst 家族基因的同源基因)氨基酸序列进行聚类分析;采用 real-time quantitative PCR(qPCR)方法检测 LmObst 家族基因在5龄若虫不同组织部位和不同龄期体壁的表达情况,绘制表达图谱;采用 RNA 干扰(RNAi)技术探讨 LmObsts 对飞蝗发育的影响。【结果】在飞蝗转录组数据库中搜索得到8个Obst 家族基因的cDNA片段,通过NCBI进行BLAST分析显示与赤拟谷盗CPAP3、黑腹果蝇 Obst 高度同源,属于 LmObst 家族基因片段,其中5个是全长序列,3个序列缺失3′端;采用 RACE 技术获得3′末端 cDNA 序列;将得到的8个 LmObst 家族基因全长序列进行功能域分析,发现具有 Obst 家族表皮蛋白的特点,即有1个信号肽与3个几丁质结合域 ChtBD2;并与黑腹果蝇、赤拟谷盗同源基因进行进化树的构建,根据进化树分析结果,分别命名为 LmObst-A1、LmObst-A2、LmObst-B、LmObst-C、LmObst-D1、LmObst-D2、LmObst-E1和 LmObst-E2。qPCR 结果显示 LmObst-E1和 LmObst-E2在前肠和后肠高特异性表达,LmObst-D1在体壁和前肠高表达,其他 LmObsts 在体壁或外胚层内陷形成的前肠和后肠高表达,在胃盲囊、中肠、马氏管和脂肪体中低表达;LmObst 家族基因在5龄若虫17
DOI:10.3864/j.issn.0578-1752.2015.01.08URL [本文引用: 1]

【目的】获得飞蝗(Locusta migratoria)表皮蛋白 Obstructor(Obst)家族基因的 cDNA 序列,并研究其序列特征和 mRNA 表达特性,探讨其生物学功能,为害虫防治提供新的分子靶标。【方法】采用生物信息学方法搜索飞蝗转录组数据库获得 Obst 家族基因 cDNA 片段,并进行 BLAST 分析得到 Obst 家族基因的 cDNA 序列;采用 RACE 技术扩增该家族基因的3′cDNA 序列,拼接后获得全长;SignalP 在线软件分析蛋白的信号肽,SMART 网站预测其功能域,并使用 Mega 5.10软件中 Neighbor-Joining 方法,与黑腹果蝇(Drosophila melanogaster) Obst 家族基因和赤拟谷盗(Tribolium castaneum)CPAP3家族基因(Obst 家族基因的同源基因)氨基酸序列进行聚类分析;采用 real-time quantitative PCR(qPCR)方法检测 LmObst 家族基因在5龄若虫不同组织部位和不同龄期体壁的表达情况,绘制表达图谱;采用 RNA 干扰(RNAi)技术探讨 LmObsts 对飞蝗发育的影响。【结果】在飞蝗转录组数据库中搜索得到8个Obst 家族基因的cDNA片段,通过NCBI进行BLAST分析显示与赤拟谷盗CPAP3、黑腹果蝇 Obst 高度同源,属于 LmObst 家族基因片段,其中5个是全长序列,3个序列缺失3′端;采用 RACE 技术获得3′末端 cDNA 序列;将得到的8个 LmObst 家族基因全长序列进行功能域分析,发现具有 Obst 家族表皮蛋白的特点,即有1个信号肽与3个几丁质结合域 ChtBD2;并与黑腹果蝇、赤拟谷盗同源基因进行进化树的构建,根据进化树分析结果,分别命名为 LmObst-A1、LmObst-A2、LmObst-B、LmObst-C、LmObst-D1、LmObst-D2、LmObst-E1和 LmObst-E2。qPCR 结果显示 LmObst-E1和 LmObst-E2在前肠和后肠高特异性表达,LmObst-D1在体壁和前肠高表达,其他 LmObsts 在体壁或外胚层内陷形成的前肠和后肠高表达,在胃盲囊、中肠、马氏管和脂肪体中低表达;LmObst 家族基因在5龄若虫17
[本文引用: 1]
DOI:10.1038/s41598-017-16435-wURLPMID:29167522 [本文引用: 1]

Abstract The beet armyworm, Spodoptera exigua (Hubner), is one of the major crop pests and is a target for current pest control approaches using insecticides. In this study three cuticular protein genes CPG316, CPG860 and CPG4855 have been cloned from 0 h pupal integument of S. exigua through race PCR Strategy. The deduced amino acid sequences were found to contain the RR-2 consensus region of other insect cuticular proteins and construct phylogenetic trees for each protein. Using quantitative RT-PCR, the developmental expression of the three genes through several larval and the early pupal stages was studied. All three genes contribute to the endocuticle although CPG316 may have a different role from the other two genes. All three newly isolated genes were analyzed and their functions were determined by using direct injection of the dsRNA into early 5 th instar larvae. All genes are expressed in the larvae and early pupae but in different patterns. Furthermore, phenotypic results show that these genes have differing effects on the development of cuticle, its flexibility and a big role in metamorphosis in both larval and pupal stages.
[本文引用: 4]
DOI:10.1038/srep45462URLPMID:5377371 [本文引用: 2]

Abstract Many types of cuticular proteins are found in a single insect species, and their number and features are very diversified among insects. The cuticle matrix consists of many different proteins that confer the physical properties of the exoskeleton. However, the number and properties of cuticle proteins in Locusta migratoria remain unclear. In the present study, Illumina sequencing and de novo assembly were combined to characterize the transcriptome of L. migratoria. Eighty-one cuticular protein genes were identified and divided into five groups: the CPR family (51), Tweedle (2), CPF/CPFLs (9), CPAP family (9), and other genes (10). Based on the expression patterns in different tissues and stages, most of the genes as a test were distributed in the integument, pronotum and wings, and expressed in selected stages with different patterns. The results showed no obvious correlation between the expression patterns and the conservative motifs. Additionally, each cluster displayed a different expression pattern that may possess a different function in the cuticle. Furthermore, the complexity of the large variety of genes displayed differential expression during the molting cycle may be associated with cuticle formation and may provide insights into the gene networks related to cuticle formation.
DOI:10.3864/j.issn.0578-1752.2017.10.007URL [本文引用: 2]

【目的】基于飞蝗(Locusta migratoria)转录组数据库获得内表皮蛋白(endocuticle structural glycoprotein)基因Lm Abd-5的c DNA序列,分析该基因的序列特征和m RNA表达特性,采用RNAi方法分析其生物学功能,探讨其在飞蝗表皮形成中的作用,为害虫防治提供新的分子靶标。【方法】采用生物信息学方法搜索飞蝗转录组数据库,获得Lm Abd-5 c DNA全长序列并克隆验证;采用Signal P在线软件分析蛋白的信号肽,利用SMART网站预测其功能域,使用MEGA 7.0软件中neighbor-joining(NJ)方法,与其他昆虫同源序列进行聚类分析;采用reverse-transcription quantitative PCR(RT-q PCR)方法检测Lm Abd-5在飞蝗5龄第2天若虫不同组织部位和5龄不同发育时期体壁组织中的表达情况,揭示其组织和时期表达模式;采用RNA干扰(RNAi)技术及透射电镜技术(TEM)观察沉默Lm Abd-5后对飞蝗生长发育和表皮结构的影响。【结果】通过搜索得到Lm Abd-5 c DNA全长序列并进行了克隆和测序验证,获得520 bp全长c DNA序列,其中ORF为303 bp;基因结构分析显示该基因含有3个外显子;功能域分析发现其含有1个信号肽和1个几丁质结合域(chitin binding domain 4,Cht BD4),与沙漠蝗、白蚁、赤拟谷盗等的Abd-5结构类似;BLAST分析结果表明Abd-5在昆虫中高度保守,飞蝗与沙漠蝗Abd-5序列一致度高达81%;对其保守基序进行Web Logo分析发现Lm Abd-5属于表皮蛋白CPR家族中的RR-1亚类;聚类分析结果显示,Lm Abd-5与沙漠蝗和白蚁的Abd-5显示出较近的亲缘关系;RT-q PCR结果显示Lm Abd-5在前肠、后肠、气管和体壁等由外胚层形成的组织中高表达,而在胃盲囊、中肠、马氏管、脂肪体和翅芽中低表达或不表达;不同时期表达分析发现,Lm Abd-5在4龄若虫蜕皮后0—72 h(5龄早期)具有高表达,其中蜕皮后72 h时达到最大表达量,随后表达量急剧降低(96—168 h),其表达时期与内表皮形成时
DOI:10.3864/j.issn.0578-1752.2017.10.007URL [本文引用: 2]

【目的】基于飞蝗(Locusta migratoria)转录组数据库获得内表皮蛋白(endocuticle structural glycoprotein)基因Lm Abd-5的c DNA序列,分析该基因的序列特征和m RNA表达特性,采用RNAi方法分析其生物学功能,探讨其在飞蝗表皮形成中的作用,为害虫防治提供新的分子靶标。【方法】采用生物信息学方法搜索飞蝗转录组数据库,获得Lm Abd-5 c DNA全长序列并克隆验证;采用Signal P在线软件分析蛋白的信号肽,利用SMART网站预测其功能域,使用MEGA 7.0软件中neighbor-joining(NJ)方法,与其他昆虫同源序列进行聚类分析;采用reverse-transcription quantitative PCR(RT-q PCR)方法检测Lm Abd-5在飞蝗5龄第2天若虫不同组织部位和5龄不同发育时期体壁组织中的表达情况,揭示其组织和时期表达模式;采用RNA干扰(RNAi)技术及透射电镜技术(TEM)观察沉默Lm Abd-5后对飞蝗生长发育和表皮结构的影响。【结果】通过搜索得到Lm Abd-5 c DNA全长序列并进行了克隆和测序验证,获得520 bp全长c DNA序列,其中ORF为303 bp;基因结构分析显示该基因含有3个外显子;功能域分析发现其含有1个信号肽和1个几丁质结合域(chitin binding domain 4,Cht BD4),与沙漠蝗、白蚁、赤拟谷盗等的Abd-5结构类似;BLAST分析结果表明Abd-5在昆虫中高度保守,飞蝗与沙漠蝗Abd-5序列一致度高达81%;对其保守基序进行Web Logo分析发现Lm Abd-5属于表皮蛋白CPR家族中的RR-1亚类;聚类分析结果显示,Lm Abd-5与沙漠蝗和白蚁的Abd-5显示出较近的亲缘关系;RT-q PCR结果显示Lm Abd-5在前肠、后肠、气管和体壁等由外胚层形成的组织中高表达,而在胃盲囊、中肠、马氏管、脂肪体和翅芽中低表达或不表达;不同时期表达分析发现,Lm Abd-5在4龄若虫蜕皮后0—72 h(5龄早期)具有高表达,其中蜕皮后72 h时达到最大表达量,随后表达量急剧降低(96—168 h),其表达时期与内表皮形成时
DOI:10.1016/j.ibmb.2017.11.001URLPMID:29113754 [本文引用: 2]

Abstract During growth and development of insects, the steroid hormone 20-Hydroxyecdysone (20E) regulates the molting process through activation of a series of genes including E74, E75 and HR3 by the 20E receptor EcR. Here, we analyzed the function of LmHR3 in the migratory locust Locusta migratoria. By sequence comparison, we first identified and characterized the putative nuclear receptor protein (LmHR3) based on L. migratoria transcriptome data. The full length cDNA is 2272 bp long encoding a protein of 455 amino acids that contains a DNA binding domain (zinc finger) and a ligand binding domain. Phylogenetic analyses showed that LmHR3 has a high homology with the ortholog from Blattaria. RT-qPCR results revealed that LmHR3 has a low level expression in the early days of 5th instar nymphs, and then increases and peaks at day 6, followed by a decrease to low levels before ecdysis. The LmHR3, hence, coincides with the profile of circulating 20E levels. Indeed, we show that transcription of LmHR3 is induced by 20E in vivo, and significantly suppressed by successfully knocking down expression of LmEcR. After injection of dsRNA for LmHR3 (dsLmHR3) at day 2 of earlier instar nymphs (3rd and 4th instar) and final instar nymphs (5th instar), none of the nymphs were able to molt normally, and eventually died. Chitin staining and ultra-structural analysis showed that both the synthesis of the new cuticle and the degradation of the old cuticle were blocked in the dsLmHR3 treated nymphs. Especially, chitin synthesis genes (LmUAP1 and LmCHS1) and chitinase genes (LmCHT5 and LmCHT10) were significantly down-regulated in the dsLmHR3 treatment group. Together, our results suggest that LmHR3 is involved in the control of chitin synthesis and degradation during L. migratoria molting.
DOI:10.1111/1744-7917.12342URLPMID:27430427 [本文引用: 1]

Abstract The cuticle, an essential structure for insects, is produced from cuticular proteins and chitin via a series of biochemical reactions. Tweedle genes are important members of the cuticular protein family and have four conserved motifs binding to chitin. Tweedle family genes have been found to play a profound effect on cuticle development. Here, we report that the cuticular protein gene LmTwdl1 of Locusta migratoria belongs to the Tweedle family. In situ hybridization showed that LmTwdl1 is localized to epidermal cells of the cuticle. The expression patterns of LmTwdl1 showed low expression in the cuticle during the early and middle stages of the fifth-instar nymphs; in contrast, its expression rapidly increased in the late stages of fifth-instar nymphs. We performed RNA interference to examine the function of LmTwdl1 in locusts. Silencing of LmTwdl1 resulted in high mortality during the molting process before the next stage. Also, the epicuticle of nymphs failed to molt, tended to be thinner and the arrangement of chitin in the procuticle appeared to be disordered compare to the control group. These results demonstrate that LmTwdl1 plays a critical role in molting, which contributes to a better understanding of the distinct functions of the Tweedle family in locusts.
DOI:10.3864/j.issn.0578-1752.2018.07.008URL [本文引用: 2]

【目的】基于飞蝗(Locusta migratoria)转录组数据库搜索并克隆获得飞蝗表皮蛋白基因Lm NCP1(nymph cuticle protein 1),分析其序列特征和表达特性,通过蜕皮激素(20-hydroxyecdysone,20E)诱导和干扰20E受体基因Lm Ec R表达研究其表达调控,并基于RNA干扰(RNAi)方法分析其生物学功能,为阐明该基因在飞蝗表皮发育过程中的作用提供理论依据。【方法】依据飞蝗转录组数据库获得表皮蛋白基因Lm NCP1,结合RT-PCR技术克隆获得其全长开放阅读框(ORF)序列,并结合飞蝗基因组序列分析其基因结构及序列特征;使用MEGA 6.0软件中邻接法(neighbor-joining,NJ),与其他昆虫同源序列聚类分析,并根据聚类结果进行命名;利用reverse-transcription quantitative PCR(RT-q PCR)分析其在5龄飞蝗不同组织和不同发育天数的基因表达特性;通过体内注射20E诱导以及干扰20E受体基因Lm Ec R的表达,RT-q PCR检测该基因的表达情况;基于RNAi结合HE染色方法分析其生物学功能。【结果】通过搜索获得表皮蛋白基因Lm NCP1,并进行了克隆和测序验证,获得457 bp的c DNA序列,其全长ORF序列为306 bp,编码101个氨基酸。氨基酸序列分析表明该基因编码的蛋白含有1个信号肽,不含几丁质结合域,但包含3个重复基序,Weblogo分析结果显示其在物种间具有保守性。系统进化树分析显示该蛋白与蜚蠊目蟑螂(Blaberus craniifer)Bc NCP1序列一致度最高,聚为一支。根据聚类结果将该蛋白命名为Lm NCP1(Gen Bank登录号:MF326211)。RT-q PCR结果显示Lm NCP1在5龄若虫体壁中高表达,在翅芽、前肠和脂肪体中表达次之,在中肠、后肠、胃盲囊和马氏管中表达量较低或不表达;Lm NCP1在5龄早期(N5D1和N5D2)高表达,随后表达量逐渐降低(N5D3—N5D6),到下一次蜕皮前表达有所升高(N5D7),其表达量动态与内表皮形成时间一致。20E诱导不同时间结果显示,与对照组相比,Lm NCP1在20E诱导1 h和3 h后表达量显著上调,分别上调了1.3倍和0.9倍;利用RNAi方法干扰Lm Ec R 48 h和72 h后,与对照组相比,Lm NCP1表达量均显著降低,分别降低了71%和87%。利用RNAi方法在4龄和5龄第2天分次注射ds Lm NCP1后,飞蝗仍能够正常蜕皮,无致死表型,但对其表皮进行HE染色发现,与对照组相比,其内表皮形成减少,导致表皮显著变薄,表皮层厚度约减少了33%。【结论】飞蝗表皮蛋白Lm NCP1含有3个物种间保守的重复基序,不含几丁质结合域,属于其他家族蛋白;Lm NCP1主要在体壁中高表达,而且响应Lm Ec R介导的20E信号通路调控;RNAi试验表明,Lm NCP1在飞蝗蜕皮过程中参与内表皮的沉积。
DOI:10.3864/j.issn.0578-1752.2018.07.008URL [本文引用: 2]

【目的】基于飞蝗(Locusta migratoria)转录组数据库搜索并克隆获得飞蝗表皮蛋白基因Lm NCP1(nymph cuticle protein 1),分析其序列特征和表达特性,通过蜕皮激素(20-hydroxyecdysone,20E)诱导和干扰20E受体基因Lm Ec R表达研究其表达调控,并基于RNA干扰(RNAi)方法分析其生物学功能,为阐明该基因在飞蝗表皮发育过程中的作用提供理论依据。【方法】依据飞蝗转录组数据库获得表皮蛋白基因Lm NCP1,结合RT-PCR技术克隆获得其全长开放阅读框(ORF)序列,并结合飞蝗基因组序列分析其基因结构及序列特征;使用MEGA 6.0软件中邻接法(neighbor-joining,NJ),与其他昆虫同源序列聚类分析,并根据聚类结果进行命名;利用reverse-transcription quantitative PCR(RT-q PCR)分析其在5龄飞蝗不同组织和不同发育天数的基因表达特性;通过体内注射20E诱导以及干扰20E受体基因Lm Ec R的表达,RT-q PCR检测该基因的表达情况;基于RNAi结合HE染色方法分析其生物学功能。【结果】通过搜索获得表皮蛋白基因Lm NCP1,并进行了克隆和测序验证,获得457 bp的c DNA序列,其全长ORF序列为306 bp,编码101个氨基酸。氨基酸序列分析表明该基因编码的蛋白含有1个信号肽,不含几丁质结合域,但包含3个重复基序,Weblogo分析结果显示其在物种间具有保守性。系统进化树分析显示该蛋白与蜚蠊目蟑螂(Blaberus craniifer)Bc NCP1序列一致度最高,聚为一支。根据聚类结果将该蛋白命名为Lm NCP1(Gen Bank登录号:MF326211)。RT-q PCR结果显示Lm NCP1在5龄若虫体壁中高表达,在翅芽、前肠和脂肪体中表达次之,在中肠、后肠、胃盲囊和马氏管中表达量较低或不表达;Lm NCP1在5龄早期(N5D1和N5D2)高表达,随后表达量逐渐降低(N5D3—N5D6),到下一次蜕皮前表达有所升高(N5D7),其表达量动态与内表皮形成时间一致。20E诱导不同时间结果显示,与对照组相比,Lm NCP1在20E诱导1 h和3 h后表达量显著上调,分别上调了1.3倍和0.9倍;利用RNAi方法干扰Lm Ec R 48 h和72 h后,与对照组相比,Lm NCP1表达量均显著降低,分别降低了71%和87%。利用RNAi方法在4龄和5龄第2天分次注射ds Lm NCP1后,飞蝗仍能够正常蜕皮,无致死表型,但对其表皮进行HE染色发现,与对照组相比,其内表皮形成减少,导致表皮显著变薄,表皮层厚度约减少了33%。【结论】飞蝗表皮蛋白Lm NCP1含有3个物种间保守的重复基序,不含几丁质结合域,属于其他家族蛋白;Lm NCP1主要在体壁中高表达,而且响应Lm Ec R介导的20E信号通路调控;RNAi试验表明,Lm NCP1在飞蝗蜕皮过程中参与内表皮的沉积。
DOI:10.1006/meth.2001.1262URL [本文引用: 1]
DOI:10.1016/j.ibmb.2009.12.005URLPMID:20060042 [本文引用: 1]

The exoskeleton of insects (cuticle) is an assembly of chitin and cuticle proteins. Its physical properties are determined largely by the proteins it contains, and vary widely with developmental stages and body regions. The genes encoding cuticle proteins are therefore good models to study the molecular mechanisms of signalling by ecdysteroids and juvenile hormones, which regulate molting and metamorphosis in insects. This review summarizes the studies of hormonal regulation of insect cuticle protein genes, and the recent progress in the analysis of the regulatory sequences and transcription factors important for their expression.
