 ,, 李伟勋, ObaroakpoJOY, 逄晓阳
,, 李伟勋, ObaroakpoJOY, 逄晓阳 ,, 吕加平
,, 吕加平 ,中国农业科学院农产品加工研究所,北京 100193
,中国农业科学院农产品加工研究所,北京 100193CRISPR Locus Analysis of Lactobacillus casei
YANG Lan, YANG Yang ,, LI WeiXun, OBAROAKPO JOY, PANG XiaoYang
,, LI WeiXun, OBAROAKPO JOY, PANG XiaoYang ,, Lü JiaPing
,, Lü JiaPing ,Institute of Food Science and Technology,Chinese Academy of Agricultural Sciences, Beijing 100193
,Institute of Food Science and Technology,Chinese Academy of Agricultural Sciences, Beijing 100193通讯作者:
收稿日期:2018-08-3接受日期:2018-11-14网络出版日期:2019-02-13
| 基金资助: |
Received:2018-08-3Accepted:2018-11-14Online:2019-02-13
作者简介 About authors
杨兰,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1235KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
杨兰, 杨洋, 李伟勋, ObaroakpoJOY, 逄晓阳, 吕加平. 干酪乳杆菌CRISPR基因座分析[J]. 中国农业科学, 2019, 52(3): 521-529 doi:10.3864/j.issn.0578-1752.2019.03.012
YANG Lan, YANG Yang, LI WeiXun, OBAROAKPO JOY, PANG XiaoYang, Lü JiaPing.
0 引言
【研究意义】随着分子生物学和高通量测序技术的飞速发展,人类已经完成了数以千计乳酸菌的全基因组测序,但是目前对于乳酸菌全基因组的研究仍然面临两大艰巨任务,一是如何更精确解读基因组测序得到的海量数据;二是如何对乳酸菌基因组进行遗传修饰,更加突出乳酸菌的益生功能。近年来,研究火热的CRISPR/Cas9基因编辑技术为高效完成上述两大任务提供了强有力的工具。目前应用最多的CRISPR/Cas9系统是以酿脓链球菌(Streptococcus pyogenes)的spCas9为核心构建的,已经广泛应用于真核生物的基因组编辑[1,2,3]。然而它在乳酸菌中的应用仍然受到很大限制,主要原因是spCas9在多数乳酸菌内有较高的细胞毒性;另外,spCas9蛋白较大、异源蛋白密码子偏好性等也是限制该系统在乳酸菌成功应用的重要原因。对乳酸菌自身CRISPR系统进行深入研究,解析乳酸菌抵御噬菌体等外源遗传基因侵染的机制,对于后续开发适合于乳酸菌的CRISPR/Cas9基因编辑系统具有重要意义。另一方面,干酪乳杆菌作为具有益生功能的微生物发酵剂被广泛的用于食品发酵中,具有降血压[4]、调节肠道菌群[5]和提高机体免疫力[6]等促进健康的作用。深入研究该菌CRISPR系统行使免疫功能的机制,将为解决乳酸菌工业发酵过程中易受到噬菌体的侵染导致发酵失败提供重要的理论依据。【前人研究进展】CRISPR(clustered regularly interspaced short palindromic repeat)/Cas(CRISPR associated)系统是细菌的一种获得性免疫系统,用来抵御噬菌体、质粒等外源DNA的侵害[7]。CRISPR基因座包括CRISPR序列和Cas蛋白,CRISPR序列由重复序列(repeat)和间隔序列(spacer)交替组成[8]。在前导区的调控下,CRISPR序列转录为pre-crRNA[9,10],与一种反式编码的小RNA(tracrRNA)通过碱基配对形成双链RNA(dsRNA)区域,在Cas蛋白存在下被RNase III切割加工成熟,然后与细菌自身的Cas核酸酶形成核酸蛋白复合体[11]。在type-II型系统中,当入侵的外源DNA和crRNA序列配对结合,同时Cas9蛋白识别对应的特异性PAM序列,就能够对入侵的DNA进行靶向切割,从而得到破坏外源DNA、实现自我防御的目的[12]。CRISPR/Cas系统的机制和功能日益清晰,科学家逐渐意识到可以将其应用于基因编辑。目前,CRISPR/Cas编辑系统飞速发展,已经被迅速应用于小鼠[13,14,15]、大鼠[16]、斑马鱼[17,18]、秀丽隐杆线虫[19]、拟南芥[20,21]及大肠杆菌[22,23]等多个物种,成为基因组精准编辑的有力工具[24,25,26],但是目前在乳酸菌中利用此系统进行成功编辑的研究还较少。OH等[27]将CRISPR/Cas9系统引入罗伊氏乳杆菌中,结合RED重组技术实现了对乳酸菌基因组的成功编辑,这是CRISPR/Cas9系统在乳酸菌上应用的首次报道。但是该研究也存在一些问题,其Cas9蛋白载体的构建是一种组成型表达,而且构建敲除系统所用质粒较多,电转化效率不是很高,影响了基因编辑的效率。目前在乳酸菌领域使用的基因编辑方法仍然是传统的基于基因同源重组的策略,至少需要24 d才能获得一个基因敲除,这是相当繁琐且耗时的[28]。【本研究切入点】目前的CRISPR/Cas9系统应用于乳酸菌还存在很多限制因素,亟待对该系统进行改进优化。本研究以干酪乳杆菌(Lactobacillus casei)的CRISPR系统为切入点,深入研究该菌的CRISPR系统行使免疫功能的机制,解析lcCas9识别的PAM序列。【拟解决的关键问题】本研究对干酪乳杆菌不同菌株CRISPR系统进行深入分析,重点解析lcCas9蛋白识别的PAM序列,以期对现有CRISPR/Cas9系统进行改良,为开发适用于乳酸菌的CRISPR/Cas9基因编辑系统奠定理论基础。1 材料与方法
试验于2017年10月—2018年7月在中国农业科学院农产加工研究所乳品研究实验室进行。1.1 材料
NCBI Genbank(https://www.ncbi.nlm.nih.gov/)数据库中已公布全基因组序列的6株干酪乳杆菌。这些菌株包括:干酪乳杆菌BD-Ⅱ(NC_017474.1)、干酪乳杆菌BL23(NC_010999.1)、干酪乳杆菌LC2W (NC_017473.1)、干酪乳杆菌W56(NC_018641.1)、干酪乳杆菌ZHANG(NZ_CP001084.1/NC_014334.2)、干酪乳杆菌LOCK919(NC_021721.1)。1.2 方法
1.2.1 CRISPR系统结构分析 使用CRISPR-CAS++(1.2.2 TracrRNA位置预测 基于之前对大量tracrRNA同源物的研究[29],tracrRNA在CRISPR基因座内的位置并不是保守的,在4个典型的位置中发现了反重复序列,分别为:Cas9的上游,Cas9和Cas1之间以及CRISPR序列的上下游。为了预测新的tracrRNA,通过一系列序列比对筛选了CRISPR/Cas基因座非repeat-spacer序列,包括Cas9上游1 kb、Cas9-Cas1之间以及CRISPR序列上下游1 kb两条链的序列;验证CRISPR重复:反重复配对情况。将筛选出的反重复序列使用Promoter 2.0 prediction server(
1.2.3 DR序列的二级结构预测 在前导区的调控下,CRISPR基因座转录为单链RNA分子(pre- crRNA)[9,10]。由于CRISPR重复序列部分回文性质,其可能形成稳定的发夹二级结构[8,30]。使用RNAalifold web server(
1.2.4 间隔区多样性及同源性分析 通过祖先间隔区和保守间隔区的分布可以将CRISPR序列分型将间隔区序列进行,使用Clustal X进行序列相似性分析。将6株菌的spacer逐一进行blast,查找预期匹配的原间隔序列,最多允许3个不匹配,并对原间隔序列进行溯源。筛选去除来源于CRISPR序列的同源序列,选择来源于噬菌体或质粒的序列作为潜在的外源遗传元件[31,32,33]。
1.2.5 PAM序列预测及其可视化 使用CRISPR target(
2 结果
2.1 干酪乳杆菌CRISPR基因座结构
本研究的6株干酪乳杆菌中均只有一个已证实的CRISPR系统,同时在干酪乳杆菌ZHANG基因组上还存在5个疑似CRISPR区域,在干酪乳杆菌ZHANG质粒上存在1个疑似CRISPR区域,在干酪乳杆菌LOCK919质粒上存在一个疑似CRISPR区域,这些区域虽然存在简单的重复间隔区,但是均只有1—2个间隔区,且不存在Cas,本研究对此不进行深入分析(表1)。分析6株干酪乳杆菌确认的CRISPR系统,发现4个cas(cas9、cas1、cas2、csn2)位于CRISPR序列侧翼。6株菌的基因组上均含有标志性基因cas9,可以确定其CRISPR系统为type-ⅡA型。CRISPR序列最长的为1 424 bp,含有21个间隔序列,最短的为762 bp,含有11个间隔序列,重复序列均为36 bp,间隔序列为28—31 bp。对Cas蛋白序列保守型的比较分析表明,这些菌株的Cas蛋白之间具有高度的相似性。包括干酪乳杆菌BD-Ⅱ、干酪乳杆菌BL23、干酪乳杆菌LC2W、干酪乳杆菌W56、干酪乳杆菌ZHANG在内的菌株,他们的Cas蛋白序列相似性均达到了100%,仅LOCK919与他们有微小差别,但相似性仍然达到90%以上(图1)。图1
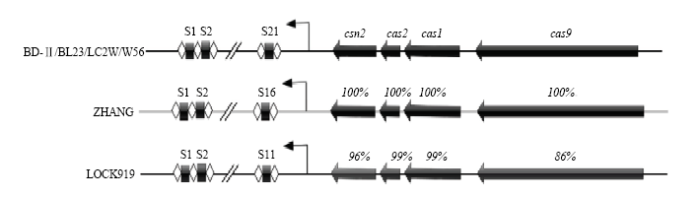 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1干酪乳杆菌CRISPR基因座结构
Fig. 1L. casei CRISPR locus structure
Table 1
表1
表16株干酪乳杆菌CRISPR序列情况
Table 1
| 菌株 Strain | CRISPR长度 Length of CRISPR | Spacer数量 Number of spacer | DR序列长度 Length of DR sequence | DR序列 DR sequence | Cas |
|---|---|---|---|---|---|
| L. casei ZHANG | 1092 | 16 | 36 | GTCTCAGGTAGATGTCGAATCAATCAGTTCAAGAGC | Cas9, Cas1, Cas2, Csn2 |
| L. casei ZHANG | 145 | 1 | 46 | GGGGTCCTTATGAGCAGGTTTCTGCGCCTGTTTGCGCGTTTCGAAA | 无 No |
| L. casei ZHANG | 109 | 1 | 27 | GGTCCTTACACGTAGGTTTCTGGTCTG | 无 No |
| L. casei ZHANG | 116 | 1 | 31 | CTTTGGTCGTTTAGGTTCGAGGTCCTTATGC | 无 No |
| L. casei ZHANG | 121 | 1 | 33 | CGGTTTCTAAACGCGTTCGCCACCCCAGAAACC | 无 No |
| L. casei ZHANG | 107 | 1 | 29 | GGTCCTTATGTGTAGGTTTCTGGGCCAGC | 无 No |
| L. casei ZHANG 质粒 plca36 | 78 | 1 | 24 | AAAGTCCGCATGACTTCGTTGAAA | 无 No |
| L. casei BD-Ⅱ | 1422 | 21 | 36 | GTCTCAGGTAGATGTCGAATCAATCAGTTCAAGAGC | Cas9, Cas1, Cas2, Csn2 |
| L. casei BL23 | 1422 | 21 | 36 | GTCTCAGGTAGATGTCGAATCAATCAGTTCAAGAGC | Cas9, Cas1, Cas2, Csn2 |
| L. casei LC2W | 1422 | 21 | 36 | GTCTCAGGTAGATGTCGAATCAATCAGTTCAAGAGC | Cas9, Cas1, Cas2, Csn2 |
| L. casei W56 | 1424 | 21 | 36 | GTCTCAGGTAGATGTCGAATCAATCAGTTCAAGAGC | Cas9, Cas1, Cas2, Csn2 |
| L. casei LOCK919 | 762 | 11 | 36 | GTCTCAGGTAGATGTCGAATCAATCAGTTCAAGAGC | Cas9, Cas1, Cas2, Csn2 |
| L. casei LOCK919 质粒 pLOCK919 | 146 | 2 | 26 | CGGGAAACCGAAAATCGGTCGCCCGC | 无 No |
新窗口打开|下载CSV
2.2 TracrRNA位置预测
对6株菌株tracrRNA的预测表明,其tracrRNA均位于Cas9与Cas1之间,与Cas转录方向相反(图2)。tracrRNA与repeat区有高达30 bp的配对,并成预测tracrRNA的启动子和终止子。启动子和转录终止子预测作为非必要的步骤,计算机模拟预测算法可以提供参考,若要获得准确的序列可以进行RNA深度测序验证。图2
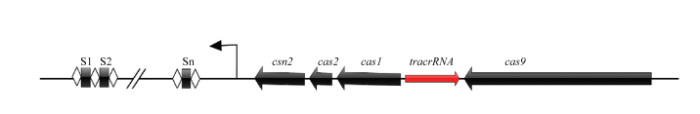 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2干酪乳杆菌tracrRNA在CRISPR基因座中的位置
Fig. 2Location of L. casei tracrRNA in the CRISPR locus
2.3 DR序列的二级结构分析
DR序列转录成非信使RNA,以特殊的结构发挥功能。随着转录的进行,连续的单链重复序列形成二级结构,茎可长达4—8个碱基,从而产生更稳定的RNA结构。为了评估干酪乳杆菌CRISPR重复序列形成稳定的RNA二级结构的可能性,使用RNAalifold来预测重复序列的分子内RNA结构。预测到的DR序列二级结构自由能为-2.50 kcal/mol,茎长度达7个碱基,结构稳定(图3)。图3
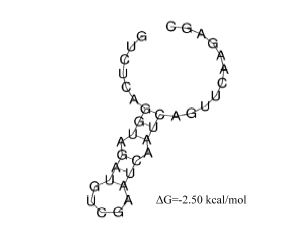 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3重复序列的RNA二级结构
Fig. 3Repeated RNA secondary structure
2.4 间隔区多样性分析
本研究发现40个独特的间隔区类型(图4),根据祖先间隔区及保守间隔区的位置,6株菌被分为3种CRISPR类型。其中BD-Ⅱ、BL23、LC2W分享完全相同的间隔区,W56与他们共享祖先间隔区,但是有3个间隔区不同,这3个不同的间隔区又与之前的序列高度相似。ZHANG与GENOTYPE 1型共享一些保守的间隔区,但是具有不完全相同的祖先间隔区及部分独特间隔区。LOCK919享有所有的独特间隔区。图4
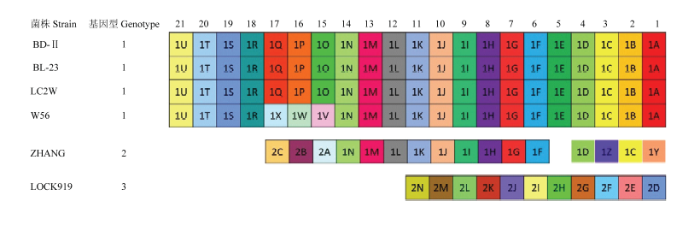 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图46株干酪乳杆菌CRISPR间隔区多样性分析
Fig. 4Analysis of the diversity of six isolates of L. casei
2.5 间隔序列同源性分析
为了确定CRISPR间隔区的可能起源,研究其与已知序列的同源性。得到与干酪乳杆菌的部分间隔区高度匹配的噬菌体或质粒序列,其中包括某些完全匹配(表2)。BD-Ⅱ、BL23、LC2W、W56的spacer21序列均与乳杆菌的质粒pREN 100%匹配。ZHANG的spacer14序列与乳酸乳球菌乳酸亚种和乳酸乳球菌乳脂亚种的质粒仅有一个碱基错配。Spacer15序列与巴克乳杆菌、副干酪乳杆菌、卡氏双球菌、植物乳杆菌、短乳酸杆菌、乳明串珠球菌、戊糖片球菌中的质粒有1—3个碱基错配。Table 2
表2
表2干酪乳杆菌spacer对应的原间隔区序列特点
Table 2
| 间隔 Spacer | 原间隔区 Original interval | 开放阅读框 Open reading box | 匹配性 Matching |
|---|---|---|---|
| BD-Ⅱ间隔区21 BD-Ⅱspacer21 | 乳杆菌质粒pREN Lactobacillus rennini plasmid pREN | 假定蛋白质 Assuming protein | 30/30 |
| BL23间隔区21 BL23 spacer21 | 乳杆菌质粒pREN Lactobacillus rennini plasmid pREN | 假定蛋白质 Assuming protein | 30/30 |
| LC2W 间隔区21 LC2W spacer21 | 乳杆菌质粒pREN Lactobacillus rennini plasmid pREN | 假定蛋白质 Assuming protein | 30/30 |
| W56 间隔区21 W56 spacer21 | 乳杆菌质粒pREN Lactobacillus rennini plasmid pREN | 假定蛋白质 Assuming protein | 30/30 |
| ZHANG间隔区14 ZHANG spacer14 | 乳酸乳球菌UL8质粒pUL8C Lactococcus lactis subsp. lactis strain UL8 plasmid pUL8C | ─ | 29/30 |
| 乳酸乳球菌乳脂亚种UC109 质粒pUC109F Lactococcus lactis subsp. cremoris strain UC109 plasmid pUC109F | ─ | 29/30 | |
| 乳酸乳球菌C10 质粒pC10A Lactococcus lactis subsp. lactis strain C10 plasmid pC10A | ─ | 29/30 | |
| 乳酸乳球菌KLDS 4.0325 质粒 unnamed2 Lactococcus lactis subsp. lactis KLDS 4.0325 plasmid unnamed2 | ─ | 29/30 | |
| 乳酸乳球菌质粒pCL2.1 Lactococcus lactis plasmid pCL2.1 | ─ | 29/30 | |
| ZHANG间隔区15 ZHANG spacer15 | 乳酸杆菌TMW 1.1992 质粒 pL11992-8 Lactobacillus backii strain TMW 1.1992 plasmid pL11992-8 | ─ | 29/30 |
| 副干酪乳杆菌质粒pLP5403 Lactobacillus paracasei plasmid pLP5403 | 假定蛋白质 Assuming protein | 29/30 | |
| 卡氏双球菌ATCC BAA-344 质粒 pPECL-1 Pediococcus claussenii ATCC BAA-344 plasmid pPECL-1 | ─ | 29/30 | |
| 植物乳杆菌质粒pXY3 Lactobacillus plantarum plasmid pXY3 | ORF4 | 29/30 | |
| 短乳酸杆菌质粒pLB925A01 Lactobacillus brevis plasmid pLB925A01 | ─ | 29/30 | |
| 乳明串珠球菌质粒pCI411 Leuconostoc lactis plasmid pCI411 | ─ | 29/30 | |
| 植物乳杆菌MF1298质粒19 Lactobacillus plantarum strain MF1298 plasmid unnamed19 | ─ | 28/30 | |
| 戊糖片球菌SRCM100892质粒pPC892-5 Pediococcus pentosaceus strain SRCM100892 plasmid pPC892-5 | ─ | 27/30 |
新窗口打开|下载CSV
Blast的结果中40个间隔区只有6个间隔区比对上了原间隔序列,这其中还包括一些非完全匹配序列。另外匹配到原间隔序列的spacer均为新获取的间隔区,祖先间隔区均未匹配到有效序列,这可能是由于数据库的不完整以及噬菌体质粒进化过程中发生突变所致。ZHANG的spacer14和spacer15匹配到了多株菌的质粒,这表明一个间隔序列可以抵抗多种外源元件的侵入。仅有几个间隔区能够匹配上编码蛋白的基因,ZHANG的spacer15匹配到了来源于植物乳杆菌pXY3质粒的ORF4,BD-Ⅱ、BL23、LC2W、W56的spacer21以及ZHANG的spacer来源于副干酪乳杆菌的质粒均匹配到的是假定的蛋白,其余间隔区均未匹配到编码蛋白的序列。结果表明,所有间隔序列的来源均为质粒,但是这并不能说明抗噬菌体的能力较弱,仅有较少的新间隔区在数据库中得到匹配,绝大多数的间隔区没有得到匹配,噬菌体数据库不完善成为了主要限制因素,blast结果只能成为细菌抗噬菌体能力的一种参考。
2.6 干酪乳杆菌Cas9蛋白识别PAM序列的预测
分析结果表明,干酪乳杆菌PAM序列的1、3位碱基偏好T/C、A/C,2、4位碱基对G、A的偏好性比较大,推测干酪乳杆菌CRISPR系统识别效率最高的PAM序列为TGMA(图5)。但是通过生物信息学分析的方法预测到的原间隔序列有限,所以这一方法可以对CRISPR系统识别的PAM进行初步预测,对于将这一系统应用于基因编辑,后续将通过分子试验进一步验证。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5干酪乳杆菌CRISPR系统识别PAM预测
Fig. 5Lactobacillus casei CRISPR system identification PAM prediction
3 讨论
CRISPR/Cas系统是细菌的获得性免疫系统,存在于46%细菌和84%古菌基因组中[31,34]。免疫功能由细菌基因组上的CRISPR序列和Cas蛋白执行,Cas1-Cas2复合物将新捕获的外源间隔序列整合入细菌的CRISPR区[35],间隔序列的插入代表着入侵者的特征遗传信息存储到宿主基因组。当入侵者再次侵染时,菌体自身的CRISPR系统会快速识别入侵者并引导相关Cas蛋白对入侵者DNA序列进行特异性切割破坏,从而发挥免疫作用。值得注意的是,间隔区在CRISPR基因座上的排列是按照进化时间顺序的,新获取的间隔区序列总是被整合到CRISPR位点的前导序列和第一个重复间隔单元之间[31]。因此,间隔区的位置可以代表他们被捕获时间的先后,这也为细菌提供了一种基于独特高变的基因座基因分型方法[36]。本研究通过分析Cas确定其均为type-ⅡA型系统,并且基于CRISPR序列,将6株菌分为3个基因型。通过对其Cas蛋白序列对比分析,发现6株菌的Cas蛋白相似性较高,ZHANG与BD-Ⅱ等菌株Cas蛋白相似度均为100%,但是其间隔区却不完全相同,仅共享一部分保守间隔区。LOCK919与其他菌株Cas蛋白相似度达到90%左右,但是他们的间隔区完全不同,保守的祖先间隔区意味着菌株之间具有较高的亲缘关系,而后来获得的间隔区由于相关菌株暴露于不同的外来侵入性DNA而不同。虽然间隔区的差异巨大,但是repeat以及tracrRNA和Cas蛋白比对结果相似,是高度保守的。通过对入侵者的靶标序列分析发现,原间隔序列侧翼位置上有一段2—7个碱基的前间隔序列邻近基序(Protospacer adjacent motif,PAM)[37]。现有研究表明,细菌CRISPR系统在获取新的间隔序列过程[38]和Cas9介导的靶序列特异性切割过程[10,39]中,PAM序列发挥着至关重要的作用。PAM存在于入侵者靶标序列侧翼但是在自身CRISPR序列中并不存在,这是宿主区分自我和非我的重要依据[40]。在不存在PAM的情况下,即使gRNA将Cas9蛋白引导到靶标基因的位置,Cas9蛋白也不会被激活,从而无切割活性。随着不同宿主来源的CRISPR系统被研究的逐渐深入,越来越多的PAM序列被解析出来。目前广泛使用的CRISPR/Cas9基因编辑系统是基于酿脓链球菌Cas9(SpCas9)为核心构建的,研究已证实该Cas9识别的经典PAM序列是5-NGG-3,进一步的研究表明5-NAG/NGA-3也是其识别的非经典PAM[40,41,42]。通过体外筛选试验,研究人员发现金黄色葡萄球菌Cas9(SaCas9)识别更长的PAM(5-NNGRR(T)-3),SaCas9体积小,有利于体内基因组编辑[43,44]。不同来源的Cas9基因序列相似性不同使他们识别不同的PAM序列,这使得可以不断探索不同物种Cas9蛋白所识别的PAM序列,丰富CRISPR工具库,提高基因编辑效率。本研究使用CRISPRtarget对干酪乳杆菌lcCas9所识别的PAM进行了初步预测。40个间隔序列在GenBank-Phage、RefSeq-Plasmid、RefSeq-Viral数据库中进行检索,仅有3个序列得到了有效匹配,但是其中2个匹配到了多达13个不同质粒。虽然间隔区是相同的,但是其匹配到的间隔区来源及错配数都是不同的,而且得到有效匹配的间隔序列均位于CRISPR的新获取间隔区,可靠性较强,表明TGMA非常有潜力成为高切割效率的PAM序列。
目前CRISPR/Cas9基因编辑方法在乳酸菌的应用并不广泛,受到了很多限制。对乳酸菌自身的CRISPR系统进行解析并基于此开发适合于乳酸菌的CRISPR/ cas9基因编辑系统,在乳酸菌遗传改良方面具有重要应用前景。本研究对干酪乳杆菌CRISPR基因座进行了全面的分析,预测了trcarRNA位置,并对PAM进行了初步预测,为解析干酪乳杆菌CRISPR系统提供了理论基础。同时也为解决乳酸菌工业发酵中噬菌体侵染这一产业难题提供了十分重要的理论依据,具有较高的应用价值和实际意义。
4 结论
6株干酪乳杆菌CRISPR系统均为type-ⅡA型,Cas基因序列和重复区序列保守,tracrRNA位于Cas9和Cas1蛋白之间。DR可以形成稳定的二级结构,根据间隔区序列可以将6株菌株分为3种基因型,TGMA为干酪乳杆菌CRISPR系统高效识别的PAM序列。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1021/sb500351fURLPMID:25458909 [本文引用: 1]

Abstract Actinobacteria, particularly those of genus Streptomyces, remain invaluable hosts for the discovery and engineering of natural products and their cognate biosynthetic pathways. However, genetic manipulation of these bacteria is often labor and time intensive. Here, we present an engineered CRISPR/Cas system for rapid multiplex genome editing of Streptomyces strains, demonstrating targeted chromosomal deletions in three different Streptomyces species and of various sizes (ranging from 20 bp to 30 kb) with efficiency ranging from 70 to 100%. The designed pCRISPomyces plasmids are amenable to assembly of spacers and editing templates via Golden Gate assembly and isothermal assembly (or traditional digestion/ligation), respectively, allowing rapid plasmid construction to target any genomic locus of interest. As such, the pCRISPomyces system represents a powerful new tool for genome editing in Streptomyces.
[本文引用: 1]
DOI:10.1128/AEM.01248-15URLPMID:26002895 [本文引用: 1]

Abstract To date, most genetic engineering approaches coupling the type II Streptococcus pyogenes clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system to lambda Red recombineering have involved minor single nucleotide mutations. Here we show that procedures for carrying out more complex chromosomal gene replacements in Escherichia coli can be substantially enhanced through implementation of CRISPR/Cas9 genome editing. We developed a three-plasmid approach that allows not only highly efficient recombination of short single-stranded oligonucleotides but also replacement of multigene chromosomal stretches of DNA with large PCR products. By systematically challenging the proposed system with respect to the magnitude of chromosomal deletion and size of DNA insertion, we demonstrated DNA deletions of up to 19.4 kb, encompassing 19 nonessential chromosomal genes, and insertion of up to 3 kb of heterologous DNA with recombination efficiencies permitting mutant detection by colony PCR screening. Since CRISPR/Cas9-coupled recombineering does not rely on the use of chromosome-encoded antibiotic resistance, or flippase recombination for antibiotic marker recycling, our approach is simpler, less labor-intensive, and allows efficient production of gene replacement mutants that are both markerless and "scar"-less. Copyright 2015, American Society for Microbiology. All Rights Reserved.
[本文引用: 1]
[本文引用: 1]
DOI:10.1155/2003/654907URLPMID:14631461 [本文引用: 1]

BACKGROUND: The aim of the present study was to investigate the effect of a probiotic beverage on gastrointestinal symptoms in patients with chronic constipation.METHODS: A double-blind, placebo-controlled, randomized study was conducted over a four-week period in patients with symptoms of chronic constipation (n=70). To all patients, 65 mL/day of a probiotic beverage containing Lactobacillus casei Shirota (LcS) or a sensorially identical placebo was administered. Patients completed a questionnaire on gastrointestinal symptoms, well-being and stool habits and underwent a medical examination weekly. Severity of constipation, flatulence and bloating was summarized into four categories (severe, moderately severe, mild and no symptoms).RESULTS: The consumption of LcS resulted in a significant improvement in self-reported severity of constipation and stool consistency, starting in the second week of the intervention phase (P<0.0001). Severe and moderately severe constipation was observed less in the LcS group. The occurrence and degree of flatulence or bloating sensation did not change. In the final examination, 89% of the LcS group and 56% of the placebo group showed a positive effect of their beverage on constipation (P=0.003). No adverse reactions were reported.CONCLUSIONS: The results indicate a beneficial effect on gastrointestinal symptoms of patients with chronic constipation. The administration of probiotic foodstuffs may be recommended as an adjunctive therapy of chronic constipation.
DOI:10.1016/j.jff.2015.05.021URL [本文引用: 1]

Lactobacillus casei Shirota (LcS) is a probiotic strain typically consumed with dairy products. In this study 142 hospitalized patients with symptoms of acute gastroenteritis were included. Patients in the LcS group received twice daily 6565ml of a commercial beverage containing L.65casei Shirota. The treatment with LcS resulted in a significantly decreased daily average (615.42 vs. 614.40) and cumulative rate of bowel movements (6132.49 vs. 6126.43) and improvement of the glomerular filtration rate (after 24 hours: 41.965±652.8 vs. 25.965±654.265ml/min, p65<650.01). Furthermore, treatment with LcS resulted in a significant decrease of CRP on days five, six and seven. Leukocyte counts decreased in all groups, while on day three the effect was significantly higher in the LcS group receiving antibiotics. In our study, L.65casei Shirota administration twice a day had positive effects on the reduction of bowel movements, improvement of kidney function and inflammation compared to the control group. Hence, we would suggest that additional LcS administration might be considered in those patients with acute gastroenteritis who present with high inflammation markers and/or acute impaired kidney function.
DOI:10.1093/nar/gkt157URLPMID:23470997Magsci [本文引用: 1]

Our knowledge of prokaryotic defense systems has vastly expanded as the result of comparative genomic analysis, followed by experimental validation. This expansion is both quantitative, including the discovery of diverse new examples of known types of defense systems, such as restriction-modification or toxin-antitoxin systems, and qualitative, including the discovery of fundamentally new defense mechanisms, such as the CRISPR-Cas immunity system. Large-scale statistical analysis reveals that the distribution of different defense systems in bacterial and archaeal taxa is non-uniform, with four groups of organisms distinguishable with respect to the overall abundance and the balance between specific types of defense systems. The genes encoding defense system components in bacterial and archaea typically cluster in defense islands. In addition to genes encoding known defense systems, these islands contain numerous uncharacterized genes, which are candidates for new types of defense systems. The tight association of the genes encoding immunity systems and dormancy- or cell death-inducing defense systems in prokaryotic genomes suggests that these two major types of defense are functionally coupled, providing for effective protection at the population level.
[本文引用: 2]
DOI:10.1128/JB.00187-07URL [本文引用: 2]

Expression of dev genes is important for triggering spore differentiation inside Myxococcus xanthus fruiting bodies. DNA sequence analysis suggested that dev and cas (CRISPR-associated) genes are cotranscribed at the dev locus, which is adjacent to CRISPR (clustered regularly interspaced short palindromic repeats). Analysis of RNA from developing M. xanthus confirmed that dev and cas genes are cotranscribed with a short upstream gene and at least two repeats of the downstream CRISPR, forming the dev operon. The operon is subject to strong, negative autoregulation during development by DevS. The dev promoter was identified. Its -35 and -10 regions resemble those recognized by M. xanthus sigma(A) RNA polymerase, the homolog of Escherichia coli sigma(70), but the spacer may be too long (20 bp); there is very little expression during growth. Induction during development relies on at least two positive regulatory elements located in the coding region of the next gene upstream. At least two positive regulatory elements and one negative element lie downstream of the dev promoter, such that the region controlling dev expression spans more than 1 kb. The results of testing different fragments for dev promoter activity in wild-type and devS mutant backgrounds strongly suggest that upstream and downstream regulatory elements interact functionally. Strikingly, the 37-bp sequence between the two CRISPR repeats that, minimally, are cotranscribed with dev and cas genes exactly matches a sequence in the bacteriophage Mx8 intP gene, which encodes a form of the integrase needed for lysogenization of M. xanthus.
[本文引用: 3]
DOI:10.1038/nature09886URLPMID:21455174 [本文引用: 1]

Abstract CRISPR/Cas systems constitute a widespread class of immunity systems that protect bacteria and archaea against phages and plasmids, and commonly use repeat/spacer-derived short crRNAs to silence foreign nucleic acids in a sequence-specific manner. Although the maturation of crRNAs represents a key event in CRISPR activation, the responsible endoribonucleases (CasE, Cas6, Csy4) are missing in many CRISPR/Cas subtypes. Here, differential RNA sequencing of the human pathogen Streptococcus pyogenes uncovered tracrRNA, a trans-encoded small RNA with 24-nucleotide complementarity to the repeat regions of crRNA precursor transcripts. We show that tracrRNA directs the maturation of crRNAs by the activities of the widely conserved endogenous RNase III and the CRISPR-associated Csn1 protein; all these components are essential to protect S. pyogenes against prophage-derived DNA. Our study reveals a novel pathway of small guide RNA maturation and the first example of a host factor (RNase III) required for bacterial RNA-mediated immunity against invaders.
DOI:10.1016/j.tibs.2009.05.002URLPMID:19646880Magsci [本文引用: 1]

genes. Three distinct stages are recognized in the CRISPR defense mechanism: (i) adaptation of the CRISPR via the integration of short sequences of the invaders as spacers; (ii) expression of CRISPRs and subsequent processing to small guide RNAs; and (iii) interference of target DNA by the crRNA guides. Recent analyses of key Cas proteins indicate that, despite some functional analogies, this fascinating prokaryotic system shares no phylogenetic relation with the eukaryotic RNA interference system.
DOI:10.1016/j.bbrc.2014.01.141URLPMID:24491566 [本文引用: 1]

The mammalian zygote-mediated CRISPR/Cas system can efficiently generate targeted genome-modified animals. However, this system is limited by the risk of off-target mutations. Here we show that offset-nicking by Cas9 nickase and paired gRNAs allows us to generate region deleted mice and targeted knock-in mice without off-target mutations.
DOI:10.1186/s13059-015-0653-xURLPMID:25924609 [本文引用: 1]

Although the CRISPR/Cas system has enabled one-step generation of knockout mice, low success rates of cassette knock-in limit its application range. Here we show that cloning-free, direct nuclear delivery of Cas9 protein complex with chemically synthesized dual RNAs enables highly efficient target digestion, leading to generation of knock-in mice carrying a functional cassette with up to 50% efficiency, compared with just 10% by a commonly used method consisting of Cas9 mRNA and single guide RNA. Our cloning-free CRISPR/Cas system facilitates rapid one-step generation of cassette knock-in mice, accelerating functional genomic research by providing various in vivo genetic tools.
DOI:10.1186/s12896-015-0144-xURLPMID:25997509 [本文引用: 1]

Background Clustered regulatory interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9)-mediated genome editing permits the rapid production of genetically engineered mice. To make the most of this innovative technology, a streamlined procedure is needed for the robust construction of CRISPR/Cas9 vectors, the efficient preparation of mouse oocytes, and refined genotyping strategies. Although we previously demonstrated the applicability of oocyte cryopreservation technologies and various genotyping methods in the production of transcription activator-like effector nuclease-mediated genome editing in mice, it has not yet been clarified whether these techniques can be applied to the CRISPR/Cas9-mediated generation of knockout mice. In this study, we investigated easy, efficient, and robust methods of creating knockout mice using several CRISPR/Cas9 systems. Results We constructed three types of CRISPR/Cas9 vectors expressing: 1) single guide RNA (gRNA) and Cas9 nuclease, 2) two gRNAs and Cas9 nickase, and 3) two gRNAs and FokI-dCas9, targeting the same genomic locus. These vectors were directly microinjected into the pronucleus of freeze-thawed fertilized oocytes, and surviving oocytes were transferred to pseudopregnant ICR mice. Cas9 nuclease resulted in the highest mutation rates with the lowest birth rates, while Cas9 nickase resulted in the highest birth rates with the lowest mutation rates. FokI-dCas9 presented well-balanced mutation and birth rates. Furthermore, we constructed a single all-in-one FokI-dCas9 vector targeting two different genomic loci, and validated its efficacy by blastocyst analysis, resulting in highly efficient simultaneous targeted mutagenesis. Conclusions Our report offers several choices of researcher-friendly consolidated procedures for making CRISPR/Cas9-mediated knockout mice, with sophisticated construction systems for various types of CRISPR vectors, convenient preparation of in vitro fertilized or mated freeze-thawed oocytes, and an efficient method of mutant screening.
DOI:10.1038/cr.2013.157URLPMID:24296780Magsci [本文引用: 1]

http://www.nature.com/doifinder/10.1038/cr.2013.157
DOI:10.1101/gr.186379.114URLPMID:26048245 [本文引用: 1]

Abstract The use of CRISPR/Cas9 as a genome-editing tool in various model organisms has radically changed targeted mutagenesis. Here, we present a high-throughput targeted mutagenesis pipeline using CRISPR/Cas9 technology in zebrafish that will make possible both saturation mutagenesis of the genome and large-scale phenotyping efforts. We describe a cloning-free single-guide RNA (sgRNA) synthesis, coupled with streamlined mutant identification methods utilizing fluorescent PCR and multiplexed, high-throughput sequencing. We report germline transmission data from 162 loci targeting 83 genes in the zebrafish genome, in which we obtained a 99% success rate for generating mutations and an average germline transmission rate of 28%. We verified 678 unique alleles from 58 genes by high-throughput sequencing. We demonstrate that our method can be used for efficient multiplexed gene targeting. We also demonstrate that phenotyping can be done in the F1 generation by inbreeding two injected founder fish, significantly reducing animal husbandry and time. This study compares germline transmission data from CRISPR/Cas9 with those of TALENs and ZFNs and shows that efficiency of CRISPR/Cas9 is sixfold more efficient than other techniques. We show that the majority of published "rules" for efficient sgRNA design do not effectively predict germline transmission rates in zebrafish, with the exception of a GG or GA dinucleotide genomic match at the 5' end of the sgRNA. Finally, we show that predicted off-target mutagenesis is of low concern for in vivo genetic studies. 2015 Varshney et al.; Published by Cold Spring Harbor Laboratory Press.
DOI:10.1101/gr.161638.113URLPMID:24179142Magsci [本文引用: 1]

Sequence-specific nucleases like TALENs and the CRISPR/Cas9 system have greatly expanded the genome editing possibilities in model organisms such as zebrafish. Both systems have recently been used to create knock-out alleles with great efficiency, and TALENs have also been successfully employed in knock-in of DNA cassettes at defined loci via homologous recombination (HR). Here we report CRISPR/Cas9-mediated knock-in of DNA cassettes into the zebrafish genome at a very high rate by homology-independent double-strand break (DSB) repair pathways. After co-injection of a donor plasmid with a short guide RNA (sgRNA) and Cas9 nuclease mRNA, concurrent cleavage of donor plasmid DNA and the selected chromosomal integration site resulted in efficient targeted integration of donor DNA. We successfully employed this approach to convert eGFP into Gal4 transgenic lines, and the same plasmids and sgRNAs can be applied in any species where eGFP lines were generated as part of enhancer and gene trap screens. In addition, we show the possibility of easily targeting DNA integration at endogenous loci, thus greatly facilitating the creation of reporter and loss-of-function alleles. Due to its simplicity, flexibility, and very high efficiency, our method greatly expands the repertoire for genome editing in zebrafish and can be readily adapted to many other organisms.
[本文引用: 1]
DOI:10.1038/cr.2013.123Magsci [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1021/sb500036qURLPMID:4277763Magsci [本文引用: 1]

In microbial communities, bacterial populations are commonly controlled using indiscriminate, broad range antibiotics. There are few ways to target specific strains effectively without disrupting the entire microbiome and local environment. Here, we use conjugation, a natural DNA horizontal transfer process among bacterial species, to deliver an engineered CRISPR interference (CRISPRi) system for targeting specific genes in recipient Escherichia coli cells. We show that delivery of the CRISPRi system is successful and can specifically repress a reporter gene in recipient cells, thereby establishing a new tool for gene regulation across bacterial cells and potentially for bacterial population control.
URLPMID:22745249 [本文引用: 1]

Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems provide bacteria and archaea with adaptive immunity against viruses and plasmids by using CRISPR RNAs (crRNAs) to guide the silencing of invading nucleic acids. We show here that in a subset of these systems, the mature crRNA that is base-paired to trans-activating crRNA (tracrRNA) forms a two-RNA structure that directs the CRISPR-associated protein Cas9 to introduce double-stranded (ds) breaks in target DNA. At sites complementary to the crRNA-guide sequence, the Cas9 HNH nuclease domain cleaves the complementary strand, whereas the Cas9 RuvC-like domain cleaves the noncomplementary strand. The dual-tracrRNA:crRNA, when engineered as a single RNA chimera, also directs sequence-specific Cas9 dsDNA cleavage. Our study reveals a family of endonucleases that use dual-RNAs for site-specific DNA cleavage and highlights the potential to exploit the system for RNA-programmable genome editing.
DOI:10.1038/nbt.2508URLPMID:23360965Magsci [本文引用: 1]

Here we use the clustered, regularly interspaced, short palindromic repeats (CRISPR)-associated Cas9 endonuclease complexed with dual-RNAs to introduce precise mutations in the genomes of Streptococcus pneumoniae and Escherichia coli. The approach relies on dual-RNA:Cas9-directed cleavage at the targeted genomic site to kill unmutated cells and circumvents the need for selectable markers or counter-selection systems. We reprogram dual-RNA:Cas9 specificity by changing the sequence of short CRISPR RNA (crRNA) to make single- and multinucleotide changes carried on editing templates. Simultaneous use of two crRNAs enables multiplex mutagenesis. In S. pneumoniae, nearly 100% of cells that were recovered using our approach contained the desired mutation, and in E. coli, 65% that were recovered contained the mutation, when the approach was used in combination with recombineering. We exhaustively analyze dual-RNA:Cas9 target requirements to define the range of targetable sequences and show strategies for editing sites that do not meet these requirements, suggesting the versatility of this technique for bacterial genome engineering.
DOI:10.1038/nrg3409URLPMID:23322222Magsci [本文引用: 1]

The article discusses the studies, "RNA-guided human genome engineering via Cas9," by P. Mali and colleagues, and "Multiplex genome engineering using CRISPR/Cas systems," by L. Cong and colleagues. Researchers of both studies attempt to determine the efficacy of CRISPR system. Both studies showed that the CRISPR system has comparable or superior genome-editing efficiency compared with systems based on transcription-activator-like effector nucleases (TALENs) or zinc finger nucleases (ZFNs).
[本文引用: 1]
DOI:10.1111/mmi.12678URLPMID:24942885 [本文引用: 1]

SummaryThe probiotic Lactobacillus casei catabolizes galacto-N-biose (GNB) and lacto-N-biose (LNB) by using a transport system and metabolic routes different from those of Bifidobacterium. L. casei contains a gene cluster, gnbREFGBCDA, involved in the metabolism of GNB, LNB and also N-acetylgalactosamine. Inactivation of gnbC (EIIC) or ptsI (Enzyme I) of the phosphoenolpyruvate : 塻ugar phosphotransferase system (PTS) prevented the growth on those three carbohydrates, indicating that they are transported and phosphorylated by the same PTSGnb. Enzyme activities and growth analysis with knockout mutants showed that GnbG (phospho- galactosidase) hydrolyses both disaccharides. However, GnbF (N-acetylgalactosamine-6P deacetylase) and GnbE (galactosamine-6P isomerase/deaminase) are involved in GNB but not in LNB fermentation. The utilization of LNB depends on nagA (N-acetylglucosamine-6P deacetylase), showing that the N-acetylhexosamine moieties of GNB and LNB follow different catabolic routes. A lacAB mutant (galactose-6P isomerase) was impaired in GNB and LNB utilization, indicating that their galactose moiety is channelled through the tagatose-6P pathway. Transcriptional analysis showed that the gnb operon is regulated by substrate-specific induction mediated by the transcriptional repressor GnbR, which binds to a 26 bp DNA region containing inverted repeats exhibiting a 2T/2A conserved core. The data represent the first characterization of novel metabolic pathways for human milk oligosaccharides and glycoconjugate structures in Firmicutes.
DOI:10.4161/rna.24321URLPMID:23563642Magsci [本文引用: 1]

CRISPR-Cas is a rapidly evolving RNA-mediated adaptive immune system that protects bacteria and archaea against mobile genetic elements. The system relies on the activity of short mature CRISPR RNAs (crRNAs) that guide Cas protein(s) to silence invading nucleic acids. A set of CRISPR-Cas, type II, requires a trans-activating small RNA, tracrRNA, for maturation of precursor crRNA (pre-crRNA) and interference with invading sequences. Following co-processing of tracrRNA and pre-crRNA by RNase III, dual-tracrRNA:crRNA guides the CRISPR-associated endonuclease Cas9 (Csn1) to cleave site-specifically cognate target DNA. Here, we screened available genomes for type II CRISPR-Cas loci by searching for Cas9 orthologs. We analyzed 75 representative loci, and for 56 of them we predicted novel tracrRNA orthologs. Our analysis demonstrates a high diversity in cas operon architecture and position of the tracrRNA gene within CRISPR-Cas loci. We observed a correlation between locus heterogeneity and Cas9 sequence diversity, resulting in the identification of various type II CRISPR-Cas subgroups. We validated the expression and co-processing of predicted tracrRNAs and pre-crRNAs by RNA sequencing in five bacterial species. This study reveals tracrRNA family as an atypical, small RNA family with no obvious conservation of structure, sequence or localization within type II CRISPR-Cas loci. The tracrRNA family is however characterized by the conserved feature to base-pair to cognate pre-crRNA repeats, an essential function for crRNA maturation and DNA silencing by dual-RNA:Cas9. The large panel of tracrRNA and Cas9 ortholog sequences should constitute a useful database to improve the design of RNA-programmable Cas9 as genome editing tool.
DOI:10.1126/science.1179555URLPMID:20056882 [本文引用: 1]

Microbes rely on diverse defense mechanisms that allow them to withstand viral predation and exposure to invading nucleic acid. In many Bacteria and most Archaea, clustered regularly interspaced short palindromic repeats (CRISPR) form peculiar genetic loci, which provide acquired immunity against viruses and plasmids by targeting nucleic acid in a sequence-specific manner. These hypervariable loci take up genetic material from invasive elements and build up inheritable DNA-encoded immunity over time. Conversely, viruses have devised mutational escape strategies that allow them to circumvent the CRISPR/Cas system, albeit at a cost. CRISPR features may be exploited for typing purposes, epidemiological studies, host-virus ecological surveys, building specific immunity against undesirable genetic elements, and enhancing viral resistance in domesticated microbes.
DOI:10.1126/science.1138140URLPMID:17379808 [本文引用: 3]

Clustered regularly interspaced short palindromic repeats (CRISPR) are a distinctive feature of the genomes of most Bacteria and Archaea and are thought to be involved in resistance to bacteriophages. We found that, after viral challenge, bacteria integrated new spacers derived from phage genomic sequences. Removal or addition of particular spacers modified the phage-resistance phenotype of the cell. Thus, CRISPR, together with associated cas genes, provided resistance against phages, and resistance specificity is determined by spacer-phage sequence similarity.
[本文引用: 1]
[本文引用: 1]
DOI:10.1126/science.1165771URLPMID:2695655 [本文引用: 1]

Abstract Horizontal gene transfer (HGT) in bacteria and archaea occurs through phage transduction, transformation, or conjugation, and the latter is particularly important for the spread of antibiotic resistance. Clustered, regularly interspaced, short palindromic repeat (CRISPR) loci confer sequence-directed immunity against phages. A clinical isolate of Staphylococcus epidermidis harbors a CRISPR spacer that matches the nickase gene present in nearly all staphylococcal conjugative plasmids. Here we show that CRISPR interference prevents conjugation and plasmid transformation in S. epidermidis. Insertion of a self-splicing intron into nickase blocks interference despite the reconstitution of the target sequence in the spliced mRNA, which indicates that the interference machinery targets DNA directly. We conclude that CRISPR loci counteract multiple routes of HGT and can limit the spread of antibiotic resistance in pathogenic bacteria.
DOI:10.1038/nmicrobiol.2017.92URLPMID:28581505 [本文引用: 1]

This year marks the tenth anniversary of the identification of the biological function of CRISPR-Cas as adaptive immune systems in bacteria. In just a decade, the characterization of CRISPR-Cas systems has established a novel means of adaptive immunity in bacteria and archaea and deepened our understanding of the interplay between prokaryotes and their environment, and CRISPR-based molecular machines have been repurposed to enable a genome editing revolution. Here, we look back on the historical milestones that have paved the way for the discovery of CRISPR and its function, and discuss the related technological applications that have emerged, with a focus on microbiology. Lastly, we provide a perspective on the impacts the field has had on science and beyond.
DOI:10.1371/journal.pone.0133661URLPMID:4521832 [本文引用: 1]

CRISPR-Cas systems constitute adaptive immune systems for antiviral defense in bacteria. We investigated the occurrence and diversity of CRISPR-Cas systems in 48 Bifidobacterium genomes to gain insights into the diversity and co-evolution of CRISPR-Cas systems within the genus and investigate CRISPR spacer content. We identified the elements necessary for the successful targeting and inference of foreign DNA in select Type II CRISPR-Cas systems, including the tracrRNA and target PAM sequence. Bifidobacterium species have a very high frequency of CRISPR-Cas occurrence (77%, 37 of 48). We found that many Bifidobacterium species have unusually large and diverse CRISPR-Cas systems that contain spacer sequences showing homology to foreign genetic elements like prophages. A large number of CRISPR spacers in bifidobacteria show perfect homology to prophage sequences harbored in the chromosomes of other species of Bifidobacterium, including some spacers that self-target the chromosome. A correlation was observed between strains that lacked CRISPR-Cas systems and the number of times prophages in that chromosome were targeted by other CRISPR spacers. The presence of prophage-targeting CRISPR spacers and prophage content may shed light on evolutionary processes and strain divergence. Finally, elements of Type II CRISPR-Cas systems, including the tracrRNA and crRNAs, set the stage for the development of genome editing and genetic engineering tools.
[本文引用: 1]
DOI:10.1038/ncomms2440URLPMID:23385575 [本文引用: 1]

Clustered regularly interspaced short palindromic repeats (CRISPR)-Cas systems provide adaptive immunity against phage via spacer-encoded CRISPR RNAs that are complementary to invasive nucleic acids. Here, we challenge Streptococcus thermophilus with a bacteriophage, and used PCR-based metagenomics to monitor phage-derived spacers daily for 15 days in two experiments. Spacers that target the host chromosome are infrequent and strongly selected against, suggesting autoimmunity is lethal. In experiments that recover over half a million spacers, we observe early dominance by a few spacer sub-populations and rapid oscillations in sub-population abundances. In two CRISPR systems and in replicate experiments, a few spacers account for the majority of spacer sequences. Nearly all phage locations targeted by the acquired spacers have a proto-spacer adjacent motif (PAM), indicating PAMs are involved in spacer acquisition. We detect a strong and reproducible bias in the phage genome locations from which spacers derive. This may reflect selection for specific spacers based on location and effectiveness.
DOI:10.1073/pnas.1208507109URLMagsci [本文引用: 1]

Clustered, regularly interspaced, short palindromic repeats (CRISPR)/CRISPR-associated (Cas) systems provide adaptive immunity against viruses and plasmids in bacteria and archaea. The silencing of invading nucleic acids is executed by ribonucleoprotein complexes preloaded with small, interfering CRISPR RNAs (crRNAs) that act as guides for targeting and degradation of foreign nucleic acid. Here, we demonstrate that the Cas9-crRNA complex of the Streptococcus thermophilus CRISPR3/Cas system introduces in vitro a double-strand break at a specific site in DNA containing a sequence complementary to crRNA. DNA cleavage is executed by Cas9, which uses two distinct active sites, RuvC and HNH, to generate site-specific nicks on opposite DNA strands. Results demonstrate that the Cas9-crRNA complex functions as an RNA-guided endonuclease with RNA-directed target sequence recognition and protein-mediated DNA cleavage. These findings pave the way for engineering of universal programmable RNA-guided DNA endonucleases.
DOI:10.1099/mic.0.023960-0URLPMID:19246744 [本文引用: 2]

Abstract Clustered regularly interspaced short palindromic repeats (CRISPR) and their associated CRISPR-associated sequence (CAS) proteins constitute a novel antiviral defence system that is widespread in prokaryotes. Repeats are separated by spacers, some of them homologous to sequences in mobile genetic elements. Although the whole process involved remains uncharacterized, it is known that new spacers are incorporated into CRISPR loci of the host during a phage challenge, conferring specific resistance against the virus. Moreover, it has been demonstrated that such interference is based on small RNAs carrying a spacer. These RNAs would guide the defence apparatus to foreign molecules carrying sequences that match the spacers. Despite this essential role, the spacer uptake mechanism has not been addressed. A first step forward came from the detection of motifs associated with spacer precursors (proto-spacers) of Streptococcus thermophilus, revealing a specific recognition of donor sequences in this species. Here we show that the conservation of proto-spacer adjacent motifs (PAMs) is a common theme for the most diverse CRISPR systems. The PAM sequence depends on the CRISPR-CAS variant, implying that there is a CRISPR-type-specific (motif-directed) choice of the spacers, which subsequently determines the interference target. PAMs also direct the orientation of spacers in the repeat arrays. Remarkably, observations based on such polarity argue against a recognition of the spacer precursors on transcript RNA molecules as a general rule.
DOI:10.1038/nbt.2647Magsci [本文引用: 1]

The Streptococcus pyogenes Cas9 (SpCas9) nuclease can be efficiently targeted to genomic loci by means of single-guide RNAs (sgRNAs) to enable genome editing(1-10). Here, we characterize SpCas9 targeting specificity in human cells to inform the selection of target sites and avoid off-target effects. Our study evaluates >700 guide RNA variants and SpCas9-induced indel mutation levels at >100 predicted genomic off-target loci in 293T and 293FT cells. We find that SpCas9 tolerates mismatches between guide RNA and target DNA at different positions in a sequence-dependent manner, sensitive to the number, position and distribution of mismatches. We also show that SpCas9-mediated cleavage is unaffected by DNA methylation and that the dosage of SpCas9 and sgRNA can be titrated to minimize off-target modification. To facilitate mammalian genome engineering applications, we provide a web-based software tool to guide the selection and validation of target sequences as well as off-target analyses.
DOI:10.1038/srep05405URLPMID:4066725 [本文引用: 1]

Abstract CRISPR/Cas9-mediated DNA cleavage (CCMDC) is becoming increasingly used for efficient genome engineering. Proto-spacer adjacent motif (PAM) adjacent to target sequence is one of the key components in the design of CCMDC strategies. It has been reported that NAG sequences are the predominant non-canonical PAM for CCMDC at the human EMX locus, but it is not clear whether it is universal at other loci. In the present study, we attempted to use a GFP-reporter system to comprehensively and quantitatively test the efficiency of CCMDC with non-canonical PAMs in human cells. The initial results indicated that the effectiveness of NGA PAM for CCMDC is much higher than that of other 14 PAMs including NAG. Then we further designed another three pairs of NGG, NGA and NAG PAMs at different locations in the GFP gene and investigated the corresponding DNA cleavage efficiency. We observed that one group of NGA PAMs have a relatively higher DNA cleavage efficiency, while the other groups have lower efficiency, compared with the corresponding NAG PAMs. Our study clearly demonstrates that NAG may not be the universally predominant non-canonical PAM for CCMDC in human cells. These findings raise more concerns over off-target effects in CRISPR/Cas9-mediated genome engineering.
[本文引用: 1]
[本文引用: 1]
