 ,1, 刘永伟
,1, 刘永伟 ,2, 杨军峰3, 张双喜4, 于太飞1, 陈隽1, 陈明1, 周永斌1, 马有志1, 徐兆师
,2, 杨军峰3, 张双喜4, 于太飞1, 陈隽1, 陈明1, 周永斌1, 马有志1, 徐兆师 ,1, 付金东
,1, 付金东 ,1
,1Identification and Analysis of Salt Tolerance of Wheat Transcription Factor TaWRKY33 Protein
ZHANG HuiYuan ,1, LIU YongWei
,1, LIU YongWei ,2, YANG JunFeng3, ZHANG ShuangXi4, YU TaiFei1, CHEN Jun1, CHEN Ming1, ZHOU YongBin1, MA YouZhi1, XU ZhaoShi
,2, YANG JunFeng3, ZHANG ShuangXi4, YU TaiFei1, CHEN Jun1, CHEN Ming1, ZHOU YongBin1, MA YouZhi1, XU ZhaoShi ,1, FU JinDong
,1, FU JinDong ,1
,1通讯作者:
第一联系人:
收稿日期:2018-06-27接受日期:2018-09-12网络出版日期:2018-12-26
| 基金资助: |
Received:2018-06-27Accepted:2018-09-12Online:2018-12-26

摘要
关键词:
Abstract
Keywords:
PDF (2880KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张惠媛, 刘永伟, 杨军峰, 张双喜, 于太飞, 陈隽, 陈明, 周永斌, 马有志, 徐兆师, 付金东. 小麦转录因子基因TaWRKY33的耐盐性分析[J]. 中国农业科学, 2018, 51(24): 4591-4602 doi:10.3864/j.issn.0578-1752.2018.24.001
ZHANG HuiYuan, LIU YongWei, YANG JunFeng, ZHANG ShuangXi, YU TaiFei, CHEN Jun, CHEN Ming, ZHOU YongBin, MA YouZhi, XU ZhaoShi, FU JinDong.
0 引言
【研究意义】作物在生命周期内会不可避免的遭受干旱和高盐等非生物逆境胁迫,影响作物正常的生长,由此造成减产,品质降低,严重影响作物的经济产量。植物在进化过程中形成了一个复杂的信号转导网络[1,2],因此,研究植物对逆境信号的感知、传递以及适应性响应的分子机制,对于阐明植物适应逆境机理,提高作物抗性具有重要意义。【前人研究进展】转录因子(transcription factors,TFs)是信号传递和基因表达调控过程的重要环节。根据基序保守性将植物体众多转录因子[3]分成包括WRKY在内的若干个家族[1,2]。WRKY转录因子在植物中分布广泛,通过信号网络参与多个植物生命过程的调节。WRKY转录因子的N端含有WRKY结构域,在少数WRKY蛋白中其被WRRY、WSKY、WKRY、WVKY或WKKY代替[4]。WRKY家族可以被进一步分为3个亚族GroupⅠ、GroupⅡ和GroupⅢ;依据系统发育数据分析,高等植物WRKY家族GroupⅡ被精确地分为Ⅱa+Ⅱb、Ⅱc和Ⅱd+Ⅱe[5,6]。据报道,WRKY转录因子家族广泛参与植物的生物胁迫响应[7]。近几年,WRKY转录因子通过ABA介导对植物逆境的应答,在受到非生物胁迫信号时,能够调控下游抗逆相关功能基因的表达,增强植株的抗逆性[8,9,10]。过表达OsWRKY45提高了水稻对盐和干旱的抗性[11]。拟南芥AtWRKY46调节渗透胁迫应答和气孔运动[12]。过表达和RNA干扰试验表明大豆GmWRKY27通过抑制其下游抗逆负调因子GmNAC29的表达,从而提高转基因大豆发状根对盐和干旱的抗性[13]。因此,深入研究WRKY转录因子基因的作用机制对提高作物抗性有着重要的现实意义。一般蛋白质行使功能时,需要与其他蛋白质或者其他分子相互作用才能完成。因而,在蛋白质互作水平上研究蛋白质作用机制对理解蛋白质功能具有重要的意义。研究发现,蛋白激酶级联反应OsMKK4-OsMPK3/OsMPK6调控了水稻WRKY转录因子OsWRKY53的激活活性,从而提高植物抗性[14]。目前,关于小麦WRKY转录因子与其他蛋白之间互作的研究还很少。因此,在小麦中寻找与WRKY转录因子互作的蛋白,对于进一步了解小麦WRKY转录因子的作用机制具有重要意义。【本研究切入点】近年来,对WRKY转录因子的研究除抗病之外还集中在发芽、衰老和响应非生物胁迫几方面。然而,大多数还是集中在模式植物,小麦耐盐WRKY蛋白还鲜有报道。【拟解决的关键问题】HE等[15]从小麦中克隆获得一个抗逆相关WRKY转录因子基因TaWRKY33,在ABA和干旱应答信号网络中发挥重要作用。本研究拟通过实时荧光定量PCR鉴定TaWRKY33在盐胁迫下的表达表达模式,通过转基因小麦和转基因拟南芥研究其对盐的响应;利用酵母双杂交技术以pGBKT7-TaWRKY33作为诱饵筛选小麦cDNA文库,得到可能与其互作的候选蛋白,为研究小麦WRKY转录因子的耐盐性提供线索。1 材料与方法
1.1 植物材料及生长环境
小白麦(Triticum aestivum L.)获赠于中国农业科学院作物科学研究所景蕊莲研究员。播种及生长条件参考文献[15],通过对温室中2周龄的小麦植株进行盐(150 mmol·L-1 NaCl)处理,并分别于0、0.5、1、2、6、12和24 h取样,液氮迅速冷冻,-80℃保存备用。1.2 小麦原生质体制备及双荧光素酶表达系统
提前配置纤维素酶解液,原生质体制备参考ABEL等[16]。选取1周龄小白麦幼嫩的叶片,切成0.5—1 mm,浸入纤维素酶解液中,黑暗条件下,真空泵抽30 min;然后黑暗条件下室温50 r/min酶解约3 h;用W5(154 mmol·L-1 NaC1、125 mmol·L-1 CaC12、5 mmol·L-1 KC1和5 mmol·L-1 glucose,pH5.6)溶液润湿的60—100目筛子过滤含有原生质体的酶解液,1 030 r/min离心1 min,收集原生质体;再用适量的预冷的W5温和重悬,冰上静置30 min后1 030 r/min离心1 min,用适量的MMG重悬原生质体,使之终浓度为2×105个/mL。双荧光素酶系统质粒由中国农业科学院生物所林浩研究员转赠。将报告因子质粒和效应因子质粒共转入110 μL的原生质体,质粒总量为15—20 μg,然后加入1.1×体积的PEG4000溶液,轻柔混合,室温诱导转化30 min。最后用400 μL的W5溶液混合终止反应,1 030 r/min离心1 min,取上清,加入1 mL W5溶液,离心1 min,取上清,加入200 μL W5溶液,23℃黑暗孵育16 h。遵照双荧光素酶检测试剂盒Dual- Luciferase? Reporter Assay System(Promega,E1910)分别检测萤火虫荧光素酶(firefly luciferase)和海肾荧光素酶(renilla luciferase)。瞬时表达试验重复3次。
1.3 载体的构建
将经EcoRⅠ酶切的酵母双杂诱饵载体pGBKT7(Clontech,美国)和TaWRKY33进行连接,并转化大肠杆菌TOP10,筛选正确的阳性克隆。拟南芥过表达载体的构建:将TaWRKY33与经NcoⅠ酶切的pCAMBIA1302载体连接,形成pCAMBIA1302-TaWRKY33重组子,并转染到GV3101根癌农杆菌中。
小麦双元表达载体的构建:将TaWRKY33插入到由泛素启动子驱动的pAHC25载体中,bar作为植物中的筛选标记基因。将构建好的pWMB110- TaWRKY33转染到EHA105农杆菌中。
1.4 酵母感受态细胞的制备及自激活的验证
按照试剂盒说明书(MATCHMAK2 ER pLexA two-Hybrid User Manual,yeast Protocols Handbook)制备酵母感受态。TaWRKY33诱饵载体的自激活验证参考文献[17]。1.5 酵母双杂系统筛选核库及阳性克隆的分析
挑取SD/-Trp单缺平板的单克隆于800 μL的-Trp单缺液体培养基中,30℃,230 r/min振荡培养8 h,用PCR技术进行菌液检测。PCR产物经1%琼脂糖凝胶电泳,将带有pGBKT7-TaWRKY33重组质粒的正确酵母菌液制备酵母感受态细胞见于太飞等[17]。核库筛选过程见参考文献[17]。用1 mL YPDA液体培养基培养阳性酵母单克隆,用于PCR检测。将插入片段大小在1 000 bp左右的酵母单克隆的质粒转化到Trans10(TransGen,北京)感受态,菌液测序结果通过DNAMAN软件进行多重序列比对后在NCBI网站进行BLAST同源性分析。
1.6 荧光定量qRT-PCR分析
用试剂盒(TransGen Biotech,北京)提取不同时间盐处理的小麦样品RNA,反转录合成第一链cDNA。设计TaWRKY33引物(TaWRKY33F:5′-GCGAGATGC TCAGGGAGGTG-3′,反向引物序列TaWRKY33R:5′-ACGGCTGGTTGTTGTAGTGGC-3′)进行表达模式分析,反应体系及扩增程序参考文献[15]。以小麦Actin 2为内参,每个反应3次重复。按照2-ΔΔCT法分析目的基因的相对表达量及其标准差。1.7 相对电导率
分别取自然条件下生长和0.6%盐处理条件下两叶期野生型冬小麦和过表达冬小麦叶片0.1 g,浸入20 mL蒸馏水中,抽真空30 min后室温静置2 h,测得起始电导率值S1。然后将试管沸水煮30 min,待冷却至室温测得最终电导率值S2。相对电导率计算公式为REC = S1×100/S2。2 结果
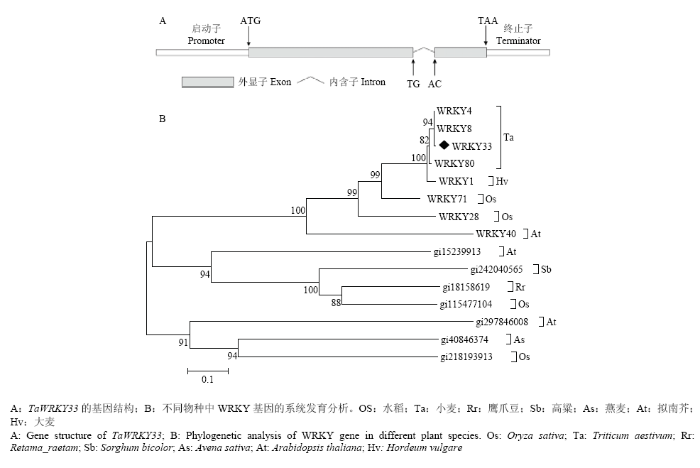
2.1 TaWRKY33结构及编码氨基酸的保守域分析
前期研究中,从小麦中获得一个干旱响应的WRKY转录因子基因TaWRKY33[15],通过进一步解析TaWRKY33的结构及编码氨基酸的特点。发现TaWRKY33含有2个外显子和1个内含子(图1-A),内含子含有典型的GT-AG序列。经Ensembl Plants基因组数据库BLAST分析,TaWRKY33位于小麦6B染色体上。WRKY转录因子家族序列十分保守,通过在线BLAST分析TaWRKY33蛋白序列,得到CDD中与其WRKY基序(WRKY motif)相似度较高的不同物种的WRKY基因。
通过构建系统发育树,发现小麦TaWRKY33转录因子与TaWRKY4、TaWRKY8相似度很高,与OsWRKY28和OsWRKY71均属于GroupⅡ-a亚组,拟南芥gi15239913、水稻gi115477104、鹰爪豆gi18158619和高粱gi242040565处于同一进化分支,为旁系同源基因,可能由同一祖先进化而来;而与拟南芥gi297846008和另外2个基因虽保守域相同但相似性较低。
2.2 TaWRKY33启动子序列的分析
TaWRKY33对干旱、ABA胁迫均有响应[15]。为进一步解析挖掘TaWRKY33在逆境响应方面的功能,首先分析了TaWRKY3启动子的顺式元件。通过对起始密码子ATG上游1 281 bp序列的分析显示,TaWRKY33的启动子区含有多种与逆境应答相关的元件,例如元件HSE(热响应元件)、ABRE(ABA应答元件)、BOXI与Sp1(光应答元件)、MBS(非生物胁迫诱导的MYB结合元件)和TC-rich repeats(与防御和逆境响应相关的元件)等(图2),说明TaWRKY3启动子可能对干旱、ABA以外的胁迫有响应。另外还包含与发育相关的顺式元件(图2)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1TaWRKY33的基因结构及编码蛋白的保守域和同源性分析
Fig. 1Gene structure, WRKY motif and phylogenetic analysis of TaWRKY33
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2TaWRKY33启动子顺式元件分析
Fig. 2Cis-element analysis of TaWRKY33 promoter
2.2 TaWRKY33受盐胁迫的诱导表达
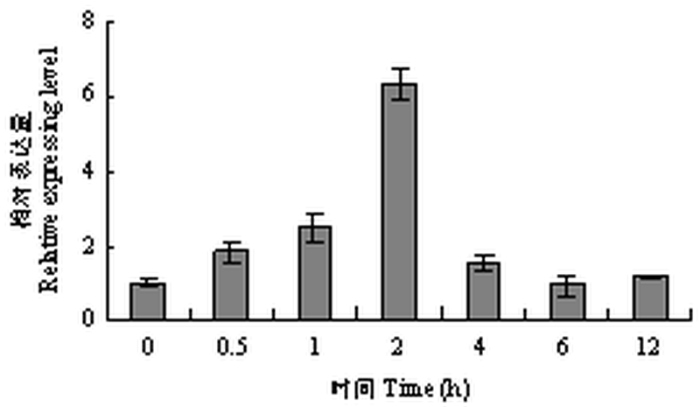
为进一步研究TaWRKY33对盐胁迫的响应,用qRT-PCR分析NaCl处理下的表达模式。结果表明,TaWRKY33在盐胁迫处理下被缓慢诱导表达(图3)。转录水平在150 mmol·L-1 NaCl盐处理后0.5 h时开始上升,在2 h时相对转录水平达到最大值。此后表达量迅速下降,处理后12 h相对表达水平逐渐恢复到本底水平。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3TaWRKY33在150 mmol·L-1 NaCl盐处理条件下的表达模式分析
Fig. 3The expression pattern of TaWRKY33 under 150 mmol·L-1 NaCl treatment
2.3 TaWRKY33提高了转基因拟南芥在盐胁迫下根的伸长
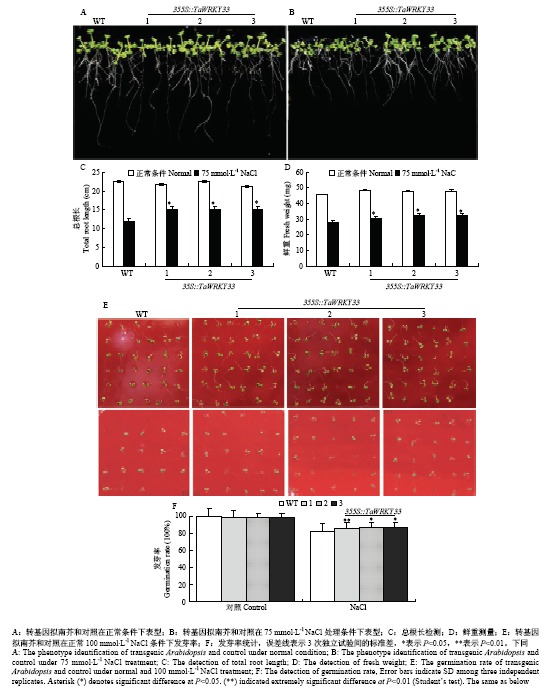
为进一步研究TaWRKY33的耐盐性,通过对过表达TaWRKY33拟南芥植株进行研究。发现过表达拟南芥和野生型拟南芥在正常条件下生长状态良好,表型无明显差异,表明TaWRKY33在正常生长条件下不会影响植物的生长和发育。通过进一步观察并统计在NaCl处理条件下过表达拟南芥和野生型拟南芥根伸长情况,在正常培养条件下,与野生型拟南芥相比,过表达拟南芥根长无明显差异(图4)。在含有75 mmol·L-1 NaCl培养基上,与野生型拟南芥相比,过表达拟南芥的根长较长,达到显著水平;而在含100 mmol·L-1 NaCl的培养基上,过表达拟南芥总根与野生型拟南芥的总根长均受到明显的抑制,过表达拟南芥的根长比野生型拟南芥的根长略有优势,说明TaWRKY33过表达影响了转基因拟南芥的发芽率(图4-E—图4-F)和根的生长,但在幼苗期对植株的耐盐性只在低盐浓度下有明显效果(图4-A—图4-D)。
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4盐胁迫条件下拟南芥的总根长和发芽率
Fig. 4Total root length of Arabidopsis under salt stress
2.4 WRKY33蛋白作为核转录激活因子
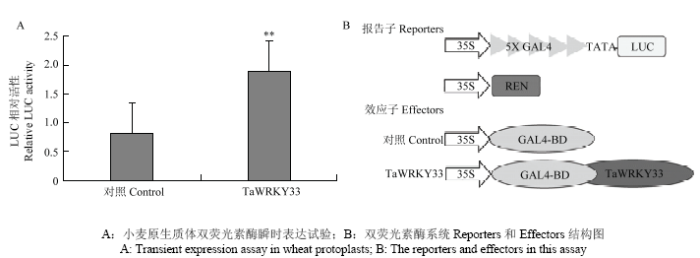
WRKY类转录因子分为3个亚组。TaWRKY33蛋白属于第二亚组,含有一个保守的WRKY结构域和一个C2H2锌指结构。小麦原生质体瞬时表达证明WRKY33定位在细胞核[15]。为进一步探究其在细胞内的转录活性,利用瞬时双荧光酶系统检测TaWRKY33转录因子在小麦原生质体中的转录活性。融合表达5×GAL4结合位点的Firefly luciferase(LUC)基因作为报告因子,由35S启动子驱动表达的Renilla luciferase(REN)基因作为内参。与此同时,TaWRKY33蛋白融合GAL4结合结构域组成效应子。与对照相比,TaWRKY33明显激活了相关荧光素酶的活性(图5),说明TaWRKY33在植物细胞中具有潜在的转录激活活性。2.5 诱饵载体的构建及自激活检测
为解析TaWRKY33的抗性机理,利用酵母双杂筛选其可能的候选互作蛋白。将TaWRKY33编码序列构建到pGBKT7诱饵载体,用PCR技术进行菌液检测,琼脂糖电泳得到与目的片段大小相符的单一条带约1 100 bp(图6),经测序和序列比对,提取正确的重组质粒pGBKT7-TaWRKY33。为检测TaWRKY33的自激活活性,将pGBKT7- TaWRKY33重组质粒转入酵母感受态中,然后涂布于SD/-Trp、SD/-Trp/-Ade和SD/-Trp/-His的固体培养基上,倒置培养2—4 d(图6),只在SD/-Trp培养基长出酵母菌落,而SD/-Trp/-Ade和SD/-Trp/-His平板上没有菌落长出,表明pGBKT7-TaWRKY33成功转入酵母感受态细胞,且TaWRKY33无自激活活性。
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5TaWRKY33激活荧光素酶活性
Fig. 5TaWRKY33 upregulates relative luciferase activity
图6
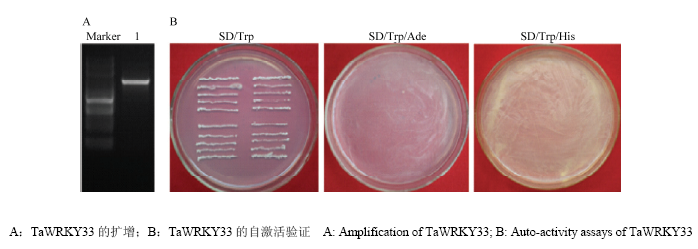 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6TaWRKY33的扩增及自激活验证
Fig. 6Amplification and Auto-activity assays of TaWRKY33
2.6 TaWRKY33互作蛋白的筛选
WRKY转录因子多与其他转录因子、激酶相互作用,共同参与植物的抗逆过程。为了进一步解析TaWRKY33的抗性机理,利用酵母双杂系统筛选其可能的候选蛋白。利用pGBKT7-TaWRKY33诱饵载体筛选小麦cDNA文库,将转化后的酵母细胞涂布于营养缺陷型SD/-Trp/-Leu/-His/-Ade固体平板上,在30℃培养箱中倒置培养4 d左右,挑取直径大于2 mm的单克隆,分别用YPDA培养基活化并点涂于SD/-Trp/- Leu/-His/-Ade/X-α-gal平板上,筛选蓝色阳性克隆(图7-B)。图7
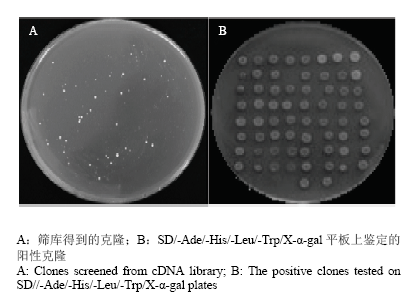 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7TaWRKY33互作蛋白的筛选
Fig. 7Screening of TaWRKY33-interacting proteins
2.7 互作蛋白的序列比对分析
将酵母阳性克隆进行菌液PCR检测,结果显示,筛出的互作基因的片段大部分集中在1 000 bp左右(图8)。提取扩增条带大小在750 bp以上的单克隆酵母质粒,进而转化TOP10大肠杆菌并测序,经BLAST比对(表1),发现筛选出的互作候选蛋白包括介导细胞内代谢物质交换的蛋白、 植物抗逆应答及蛋白修复功能相关蛋白和其他 一些编码功能未知蛋白,这些基因可能参与植物 能量代谢、胁迫响应、防御、生长发育等一些生理过程。图8
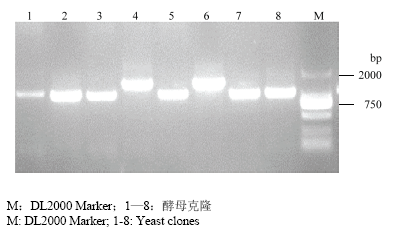 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8候选克隆的PCR检测
Fig. 8PCR identification of candidate clones
2.8 转TaWRKY33小麦的耐盐性鉴定
通过对1周龄的T3转TaWRKY33小麦和对照科农199(KN199)进行盐胁迫处理,结果显示,在正常生长条件下,转基因小麦的生长趋势与对照相比没有显著差异(图9-A),然而,在盐处理条件下,对照组小麦的萎蔫程度要显著高于转基因小麦(图9-B)。通过对T3转基因小麦和对照KN199进行qRT-PCR分析,结果显示,在转TaWRKY33小麦的2个株系中,TaWRKY33的表达模式显著高于对照KN199。表明TaWRKY33能够在转基因小麦中稳定表达(图9-C)。此时,对盐处理一周的小麦进行钠离子含量、鲜重以及相对电导率进行测定,在正常生长条件下,上述指标在转基因小麦与对照KN199之间无显著差异,而在盐处理条件下,转基因小麦拥有低的相对钠离子含量/电导率以及相对高的鲜重(图9-D—图9-F)。3 讨论
3.1 WRKY与多种蛋白互作行使功能
WRKY转录因子通过与含有W-box顺式元件以及MAPK、MAPKK、14-3-3蛋白、钙调素、组蛋白去乙酰化酶、抗性蛋白和其他WRKY转录因子相互作用发挥功能,在调节植物许多生命过程的信号网络中都很重要[17]。早期研究表明,WRKY转录因子主要在调节植物应答病原体侵染和特定发育过程中发挥作用[17]。但是,随着研究的不断深入,发现一些WRKY转录因子同样参与植物对非生物胁迫的应答[18,19,20,21]。目前,从干旱处理的小麦转录重测序中找到48个干旱响应的WRKY基因[15]。早先研究发现,拟南芥转录因子WRKY25和WRKY33增加了拟南芥的耐盐性[22,23,24];此外,近年研究表明小麦WRKY2和WRKY19通过提高下游相关基因的表达量提高了转基因拟南芥的盐和干旱抗性[34]。在水稻中,OsWRKY11的过表达提高了其对高温和干旱的抗性[25]。图9
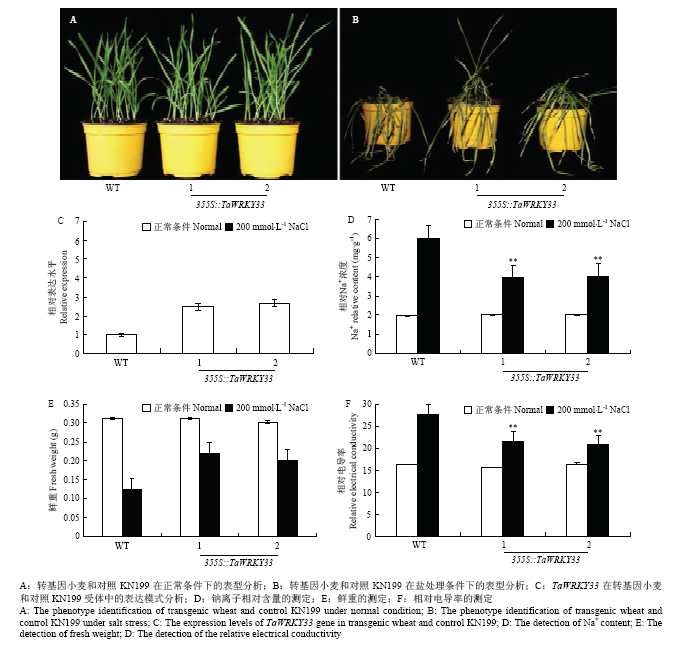 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9转TaWRKY33小麦耐盐性的鉴定与分析
Fig. 9Identification and analysis of salt tolerance of TaWRKY33 transgenic wheat
3.2 候选蛋白的生物功能
为了进一步解析TaWRKY33的抗逆机制,本研究筛选了其可能的互作蛋白(表1),包括电控阴离子通道蛋白(VDAC)。VDAC是一类广泛存在于真菌、植物和动物线粒体外膜的蛋白。在水稻、烟草、百脉根、马铃薯、拟南芥和小麦中都有发现[29]。植物VDAC除了在生长发育发面发挥作用,在生物和非生物应答方面也具有功能[29]。因此,TaWRKY33转录因子可能通过与VDAC互作,在植物抗逆调节中发挥着至关重要的作用。另外,有研究表明,乙醇酸氧化酶和ATP合成酶是植物光呼吸反应中重要的2种酶。植物可以通过光呼吸反应调节来增强植物对干旱的抵抗能力[27]。泛素结合酶E2能调控泛素化过程进而调节下游不同靶蛋白。在小麦中已有报导E2连接酶(TaU4)是抗斑枯病的负调因子[30]。另外,当植物受到干旱和高温等非生物胁迫时,E2连接酶的表达可以增强植物的抗逆能力,能对一些损伤或错误折叠的蛋白进行修复[30,31,32,33]。本研究利用酵母双杂筛选出的候选蛋白多数参与了植物生长、发育,防御生物胁迫及抵抗非生物胁迫等生物过程。这些也说明了TaWRKY33转录因子可能通过与不同蛋白相互作用,在植物抗逆调节中发挥功能。Table 1
表1
表1候选基因的BLAST分析结果及其推测的功能
Table 1
| TaWRKY33的互作蛋白 The proteins interacting with TaWRKY33 | GenBank登录号 GenBank accession No. | 功能 Functions |
|---|---|---|
| 烯醇化酶Enolase 乙醇酸氧化酶Glycolate oxidase | KC342469 AAB82143 | 糖酵解过程Glycolytic process 光呼吸反应中的关键酶,能氧化乙醇酸形成乙醛酸 One of the key enzymes in photorespiration where it oxidizes glycolate to glyoxylate |
| 1,5-二磷酸核酮糖羧化酶/加氧酶小亚基 Chloroplast ribulose-1,5-bisphosphate carboxylase/ oxygenase small subunit (rbcS) gene | KT288199 | 主要在固定CO2中对D-核酮糖二磷酸羧化以及对戊糖底物的氧化裂解 To catalyze the primary CO2 fixation step in the reductive pentose phosphate pathway |
| 叶绿素a/b结合蛋白 Chlorophyll a/b-binding protein | AAT81763 | 在光合作用中利用光能同化二氧化碳 To utilizie light energy to assimilate carbon dioxide in photosynthesis |
| ATP合成酶γ亚基ATP synthase gamma subunit | ADC33136 | 与其他亚基组成ATP合成酶 To form the ATP synthase with other subunits |
| 半胱氨酸蛋白酶抑制剂 Cysteine proteinase inhibitor | AY062608 | 抑制蛋白氨基酸肽链内切酶的水解作用,维持细胞蛋白的构象 Protect cells from inappropriate endogenous or external proteolysis and may regulate intra-or extracellular protein breakdown |
| 泛素结合酶2E2 | XM_003568254 | 参与蛋白的降解途径和DNA的修复途径 To be involved in the ubiquitin-mediated protein degradation pathway and the DNA repair pathway |
| 电压依赖性阴离子通道VDAC3 | X82148 | 电压门控离子通道活性Vo ltage-gated anion channel activity |
| 铝胁迫基因AIP | ACV44213 | 受到铝、干旱以及高盐胁迫上调表达 Up-regulated by the imposition of aluminum, high-salt, and drought stress |
| WD40重复蛋白WD40 尿酸氧化酶Urate oxidase 未知蛋白Unknown protein | NM_124800 DQ365938 | 控制植物表皮特征,并且能通过与其他蛋白互作来调控下游基因的表达 Controls plant epidermal traits, and the combination and interaction of these regulatory proteins determine the set of downstream genes to be expressed 参与尿酸合成第一步 This protein is involved in step 1 of the subpathway that synthesizes (S)-allantoin from urate 小麦染色体3B基因组片段 Triticum aestivum chromosome 3B, genomic scaffold |
新窗口打开|下载CSV
3.3 遗传学分析TaWRKY33的耐盐性
越来越多研究表明,过表达逆境应答转录因子对提高作物非生物胁迫抗性是一种可行性策略[26,27]。ABA在植物非生物胁迫应答中发挥重要的作用[15],一系列转录因子和它们的靶基因都参与介导ABA的信号转导,调节许多分子细胞过程。ABI5是ABI1/2和AtWRKY40下游的正向调节因子,作为植物ABA信号转导途径的重要组;RD29A受干燥、低温、高盐和外源性ABA诱导。然而,ABI5和RD29A在转基因拟南芥中的表达水平明显提高[15]。发现TaWRKY33可能在ABA和干旱抗逆应答的信号网络中发挥重要作用[15]。为进一步研究TaWRKY33是否参与了耐盐响应,分析了盐胁迫下的表达模式,数据显示盐胁迫激活了TaWRKY33的转录表达;而且在盐处理下,TaWRKY33的转基因过表达株系的总根长明显高于野生型拟南芥(图4)。据MAHAJAN等[28]报道,很多提高干旱抗性的转录因子对盐胁迫也有正调作用。近期研究显示,小麦TaWRKY93是一个早期盐诱导表达基因,同时具有抗旱特性。过表达TaWRKY33的小麦,可能激活了下游逆境应答基因的表达,表现出一定的耐盐性。因此,一些WRKY转录因子基因可能同时参与了植物对干旱和盐胁迫的调控。这些结果为TaWRKY33在抗逆研究中提供了新方向。4 结论
小麦TaWRKY33受盐胁迫的诱导表达,TaWRKY33的过表达提高了转基因拟南芥在种子萌发期和苗期对低盐的耐性。TaWRKY33转录因子可能通过与其他蛋白相互作用参与转基因拟南芥耐盐调节过程。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 2]
DOI:10.1111/j.1744-7909.2011.01062.xURLPMID:21676172Magsci [本文引用: 2]

You-Zhi Ma (Corresponding author)Plants have acquired sophisticated stress response systems to adapt to changing environments. It is important to understand plants鈥 stress response mechanisms in the effort to improve crop productivity under stressful conditions. The AP2/ERF transcription factors are known to regulate diverse processes of plant development and stress responses. In this study, the molecular characteristics and biological functions of AP2/ERFs in a variety of plant species were analyzed. AP2/ERFs, especially those in DREB and ERF subfamilies, are ideal candidates for crop improvement because their overexpression enhances tolerances to drought, salt, freezing, as well as resistances to multiple diseases in the transgenic plants. The comprehensive analysis of physiological functions is useful in elucidating the biological roles of AP2/ERF family genes in gene interaction, pathway regulation, and defense response under stress environments, which should provide new opportunities for the crop tolerance engineering.
[本文引用: 1]
[本文引用: 1]
DOI:10.1104/pp.107.114041URLPMID:18337489 [本文引用: 1]

Tobacco (Nicotiana tabacum) is a member of the Solanaceae, one of the agronomically most important groups of flowering plants. We have performed an in silico analysis of 1.15 million gene-space sequence reads from the tobacco nuclear genome and report the detailed analysis of more than 2,500 tobacco transcription factors (TFs). The tobacco genome contains at least one member of each of the 64 well-characterized TF families identified in sequenced vascular plant genomes, indicating that evolution of the Solanaceae was not associated with the gain or loss of TF families. However, we found notable differences between tobacco and non-Solanaceae species in TF family size and evidence for both tobacco- and Solanaceae-specih'c subfamily expansions. Compared with TF families from sequenced plant genomes, tobacco has a higher proportion of ERF/ AP2, C2H2 zinc finger, homeodomain, GRF, TCP, zinc finger homeodomain, BES, and STERILE APETALA (SAP) genes and novel subfamilies of BES, C2H2 zinc finger, SAP, and NAC genes. The novel NAC subfamily, termed TNACS, appears restricted to the Solanaceae, as they are absent from currently sequenced plant genomes but present in tomato (Solanum lycopersicum), pepper (Capsicum annuum), and potato (Solanum tuberosum). They constitute approximately 25% of NAC genes in tobacco. Based on our phylogenetic studies, we predict that many of the more than 50 tobacco group IX ERF genes are involved in jasmonate responses. Consistent with this, over two-thirds of group IX ERF genes tested showed increased mRNA levels following jasmonate treatment. Our data are a major resource for the Solanaceae and fill a void in studies of TF families across the plant kingdom.
DOI:10.1186/1471-2148-5-1URLPMID:544883Magsci [本文引用: 1]

pAbstract/p pBackground/p pWRKY proteins are newly identified transcription factors involved in many plant processes including plant responses to biotic and abiotic stresses. To date, genes encoding WRKY proteins have been identified only from plants. Comprehensive search for WRKY genes in non-plant organisms and phylogenetic analysis would provide invaluable information about the origin and expansion of the WRKY family./p pResults/p pWe searched all publicly available sequence data for WRKY genes. A single copy of the WRKY gene encoding two WRKY domains was identified from itGiardia lamblia/it, a primitive eukaryote, itDictyostelium discoideum/it, a slime mold closely related to the lineage of animals and fungi, and the green alga itChlamydomonas reinhardtii/it, an early branching of plants. This ancestral WRKY gene seems to have duplicated many times during the evolution of plants, resulting in a large family in evolutionarily advanced flowering plants. In rice, the WRKY gene family consists of over 100 members. Analyses suggest that the C-terminal domain of the two-WRKY-domain encoding gene appears to be the ancestor of the single-WRKY-domain encoding genes, and that the WRKY domains may be phylogenetically classified into five groups. We propose a model to explain the WRKY familys origin in eukaryotes and expansion in plants./p pConclusions/p pWRKY genes seem to have originated in early eukaryotes and greatly expanded in plants. The elucidation of the evolution and duplicative expansion of the WRKY genes should provide valuable information on their functions./p
DOI:10.1023/A:1020780022549URLPMID:12602888Magsci [本文引用: 1]

WRKY proteins are a recently identified class of DNA-binding proteins that recognize the TTGAC(C/T) W-box elements found in the promoters of a large number of plant defense-related genes. With oligo molecules containing the W-box sequences as probes, we detected a number of WRKY DNA-binding activities in Arabidopsis that were induced by salicylic acid (SA). Search of the Arabidopsis genome identifies 72 genes encoding proteins characteristic of WRKY DNA-binding transcription factors that can be divided into three groups based on the number and structures of their WRKY zinc-finger motifs. Northern blotting analysis revealed that 49 of the 72 AtWRKY genes were differentially regulated in the plants infected by an avirulent strain of the bacterial pathogen Pseudomonas syringae or treated by SA. These pathogen- and/or SA-regulated WRKY genes can be further categorized into groups based on their expression patterns in both wild-type plants and mutants defective in defense signaling pathways. Inspection of the 5 sequences upstream of the predicated translation start sites revealed a substantial enrichment of W boxes in the promoters of pathogen- and/or SA-regulated Arabidopsis WRKY genes. These results suggest that defense-regulated expression of WRKY genes involves extensive transcriptional activation and repression by its own members of the transcription factor superfamily.
DOI:10.1016/j.pbi.2007.04.014URLPMID:17468040Magsci [本文引用: 1]

Plants must adapt to drought stress to survive. The phytohormone abscisic acid (ABA) is produced under drought stress conditions and is essential for the response to drought stress. The ABA level plays an important role in the response, and several enzymes for ABA biosynthesis and catabolism have been identified. Physiological studies have shown that several metabolites accumulate and function as osmolytes under drought stress conditions. Many drought-inducible genes with various functions have been identified, and transgenic plants that harbor these genes have shown increased tolerance to drought.
DOI:10.1007/s00299-014-1634-3URLPMID:24913125 [本文引用: 1]

Key message The expression of LcWRKY5 was induced significantly by salinity, mannitol and cutting treatments. Arabidopsis- overexpressing LcWRKY5greatly increased dehydration tolerance by regulating...
DOI:10.1105/tpc.105.035568URLPMID:16214899 [本文引用: 1]

To understand the gene network controlling tolerance to cold stress, we performed an Arabidopsis thaliana genome transcript expression profile using Affymetrix GeneChips that contain 鈭24,000 genes. We statistically determined 939 cold-regulated genes with 655 upregulated and 284 downregulated. A large number of early cold-responsive genes encode transcription factors that likely control late-responsive genes, suggesting a multitude of transcriptional cascades. In addition, many genes involved in chromatin level and posttranscriptional regulation were also cold regulated, suggesting their involvement in cold-responsive gene regulation. A number of genes important for the biosynthesis or signaling of plant hormones, such as abscisic acid, gibberellic acid, and auxin, are regulated by cold stress, which is of potential importance in coordinating cold tolerance with growth and development. We compared the cold-responsive transcriptomes of the wild type and inducer of CBF expression 1 (ice1), a mutant defective in an upstream transcription factor required for chilling and freezing tolerance. The transcript levels of many cold-responsive genes were altered in the ice1 mutant not only during cold stress but also before cold treatments. Our study provides a global picture of the Arabidopsis cold-responsive transcriptome and its control by ICE1 and will be valuable for understanding gene regulation under cold stress and the molecular mechanisms of cold tolerance.
DOI:10.1016/j.envexpbot.2008.07.002URL [本文引用: 1]

The WRKY transcriptional factor superfamily regulates diverse functions, including processes such as plant development and stress response. In this study, we have shown that the rice WRKY45 ( OsWRKY45) expression is markedly induced in response to stress-related hormone abscisic acid (ABA) and various stress factors, e.g., application of NaCl, PEG, mannitol or dehydration, treatment with 0 C and 42 C as well as infection by Pyricularia oryzae Cav. and Xanthomonas oryzae pv. oryzae. Together, these results indicate that the OsWRKY45 may be involved in the signal pathways of both biotic and abiotic stress response. Further analyses of 35S: OsWRK45 Arabidopsis plants have shown that ectopic, constitutive over-expression of the OsWRKY45 transgene confers a number of properties to transgenic plants. These properties include significantly increased expression of PR genes, enhanced resistance to the bacterial pathogen Pseudomonas syringae tomato DC3000, enhanced tolerance to salt and drought stresses, decreased sensitivity toward ABA signalling during seed germination and post-germination processes, and modulation of ABA/stress-regulated genes during drought induction. In addition, higher levels of OsWRKY45 expression in transgenic plants correlate positively with the strength of the abiotic and biotic responses mentioned above. More specifically, the decreased ABA sensitivities, the enhanced disease resistance and drought tolerances may be attributed, in part, to stomatal closure and induction of stress-related genes during drought induction. The relationship between OsWRKY45 expression and ABA signalling is discussed.
DOI:10.1111/tpj.12538URLPMID:24773321 [本文引用: 1]

SummaryDrought and salt stress severely inhibit plant growth and development; however, the regulatory mechanisms of plants in response to these stresses are not fully understood. Here we report that the expression of a WRKY transcription factor WRKY46 is rapidly induced by drought, salt and oxidative stresses. T鈥揇NA insertion of WRKY46 leads to more sensitivity to drought and salt stress, whereas overexpression of WRKY46 (OV46) results in hypersensitivity in soil-grown plants, with a higher water loss rate, but with increased tolerance on the sealed agar plates. Stomatal closing in the OV46 line is insensitive to ABA because of a reduced accumulation of reactive oxygen species (ROS) in the guard cells. We further find that WRKY46 is expressed in guard cells, where its expression is not affected by dehydration, and is involved in light-dependent stomatal opening. Microarray analysis reveals that WRKY46 regulates a set of genes involved in cellular osmoprotection and redox homeostasis under dehydration stress, which is confirmed by ROS and malondialdehyde (MDA) levels in stressed seedlings. Moreover, WRKY46 modulates light-dependent starch metabolism in guard cells via regulating QUA-QUINE STARCH (QQS) gene expression. Taken together, we demonstrate that WRKY46 plays dual roles in regulating plant responses to drought and salt stress and light-dependent stomatal opening in guard cells.
DOI:10.1111/tpj.12879URLPMID:25990284 [本文引用: 1]

Summary Soybean ( Glycine max ) is an important crop for oil and protein resources worldwide. The molecular mechanism of the abiotic stress response in soybean is largely unclear. We previously identified multiple stress-responsive WRKY genes from soybean. Here, we further characterized the roles of one of these genes, GmWRKY27 , in abiotic stress tolerance using a transgenic hairy root assay. GmWRKY27 expression was increased by various abiotic stresses. Over-expression and RNAi analysis demonstrated that GmWRKY27 improves salt and drought tolerance in transgenic soybean hairy roots. Measurement of physiological parameters, including reactive oxygen species and proline contents, supported this conclusion. GmWRKY27 inhibits expression of a downstream gene GmNAC29 by binding to the W oxes in its promoter region. The GmNAC29 is a negative factor of stress tolerance as indicated by the performance of transgenic hairy roots under stress. GmWRKY27 interacts with GmMYB174, which also suppresses GmNAC29 expression and enhances drought stress tolerance. The GmWRKY27 and GmMYB174 may have evolved to bind to neighbouring cis elements in the GmNAC29 promoter to co-reduce promoter activity and gene expression. Our study discloses a valuable mechanism in soybean for regulation of the stress response by two associated transcription factors. Manipulation of these genes should facilitate improvements in stress tolerance in soybean and other crops.
DOI:10.1371/journal.pone.0098737URLPMID:24892523 [本文引用: 1]

WRKY transcription factors and mitogen-activated protein kinase (MAPK) cascades have been shown to play pivotal roles in the regulation of plant defense responses. We previously reported that OsWRKY53-overexpressing rice plants showed enhanced resistance to the rice blast fungus. In this study, we identified OsWRKY53 as a substrate of OsMPK3/OsMPK6, components of a fungal PAMP-responsive MAPK cascade in rice, and analyzed the effect of OsWRKY53 phosphorylation on the regulation of basal defense responses to a virulence race of rice blast fungus Magnaporthe oryzae strain Ina86-137. An in vitro phosphorylation assay revealed that the OsMPK3/OsMPK6 activated by OsMKK4 phosphorylated OsWRKY53 recombinant protein at its multiple clustered serine-proline residues (SP cluster). When OsWRKY53 was coexpressed with a constitutively active mutant of OsMKK4 in a transient reporter gene assay, the enhanced transactivation activity of OsWRKY53 was found to be dependent on phosphorylation of the SP cluster. Transgenic rice plants overexpressing a phospho-mimic mutant of OsWRKY53 (OsWRKY53SD) showed further-enhanced disease resistance to the blast fungus compared to native OsWRKY53-overexpressing rice plants, and a substantial number of defense-related genes, including pathogenesis-related protein genes, were more upregulated in the OsWRKY53SD-overexpressing plants compared to the OsWRKY53-overexpressing plants. These results strongly suggest that the OsMKK4-OsMPK3/OsMPK6 cascade regulates transactivation activity of OsWRKY53, and overexpression of the phospho-mimic mutant of OsWRKY53 results in a major change to the rice transcriptome at steady state that leads to activation of a defense response against the blast fungus in rice plants.
DOI:10.1186/s12870-016-0806-4URLPMID:27215938 [本文引用: 10]

Drought stress is one of the major causes of crop loss. WRKY transcription factors, as one of the largest transcription factor families, play important roles in regulation of many plant processes, including drought stress response. However, far less information is available on drought-responsive WRKY genes in wheat (Triticum aestivumL.), one of the three staple food crops. Forty eight putative drought-induced WRKY genes were identified from a comparison betweende novotranscriptome sequencing data of wheat without or with drought treatment.TaWRKY1andTaWRKY33from WRKY Groups III and II, respectively, were selected for further investigation. Subcellular localization assays revealed that TaWRKY1 and TaWRKY33 were localized in the nuclei in wheat mesophyll protoplasts. Various abiotic stress-relatedcis-acting elements were observed in the promoters ofTaWRKY1andTaWRKY33. Quantitative real-time PCR (qRT-PCR) analysis showed thatTaWRKY1was slightly up-regulated by high-temperature and abscisic acid (ABA), and down-regulated by low-temperature.TaWRKY33was involved in high responses to high-temperature, low-temperature, ABA and jasmonic acid methylester (MeJA). Overexpression ofTaWRKY1andTaWRKY33activated several stress-related downstream genes, increased germination rates, and promoted root growth inArabidopsisunder various stresses.TaWRKY33transgenicArabidopsislines showed lower rates of water loss thanTaWRKY1transgenicArabidopsislines and wild type plants during dehydration. Most importantly,TaWRKY33transgenic lines exhibited enhanced tolerance to heat stress. The functional roles highlight the importance of WRKYs in stress response. The online version of this article (doi:10.1186/s12870-016-0806-4) contains supplementary material, which is available to authorized users.
DOI:10.1111/j.1365-313X.1994.00421.xURLPMID:8180625 [本文引用: 1]

Summary An improved protocol is reported to isolate and transiently transform mesophyll protoplasts of Arabidopsis thaliana. Transfected leaf protoplasts support high levels of expression of the bacterial reporter gene coding for β‐glucuronidase (GUS), under the control of the cauliflower mosaic virus (CaMV) 35S promoter. Transient expression of GUS activity was monitored spectrophotometrically and reached a maximum between 18 and 48 h after polyethylene glycol (PEG)‐mediated DNA uptake. Histochemical staining for GUS activity revealed reproducible transformation frequencies between 40 and 60%, based on the number of protoplasts survived. To demonstrate the applicability of the transient expression system, the subcellular localization of GUS proteins tagged with different nuclear polypeptides was studied in transfected mesophyll protoplasts, revealing nuclear compartmentalization of the chimeric GUS enzymes. Furthermore, Arabidopsis mesophyll protoplasts support auxin‐mediated induction of chloramphenicol acetyl‐transferase (CAT) activity when transfected with a transcriptional fusion between the CAT reporter gene and the early auxin‐inducible PS‐IAA4/5 promoter. Hence, the method allows in vivo analysis of promoter activity and subcellular localization of fusion proteins in a homologous transformation system.
[本文引用: 5]
[本文引用: 5]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.tplants.2010.02.006Magsci [本文引用: 1]

<p id="">WRKY transcription factors are one of the largest families of transcriptional regulators in plants and form integral parts of signalling webs that modulate many plant processes. Here, we review recent significant progress in WRKY transcription factor research. New findings illustrate that WRKY proteins often act as repressors as well as activators, and that members of the family play roles in both the repression and de-repression of important plant processes. Furthermore, it is becoming clear that a single WRKY transcription factor might be involved in regulating several seemingly disparate processes. Mechanisms of signalling and transcriptional regulation are being dissected, uncovering WRKY protein functions via interactions with a diverse array of protein partners, including MAP kinases, MAP kinase kinases, 14-3-3 proteins, calmodulin, histone deacetylases, resistance proteins and other WRKY transcription factors. WRKY genes exhibit extensive autoregulation and cross-regulation that facilitates transcriptional reprogramming in a dynamic web with built-in redundancy.</p>
[本文引用: 1]
DOI:10.1007/s00425-010-1258-yURLPMID:20811906 [本文引用: 1]

In this study, two WRKY genes were isolated from Erysiphe necator-resistant Chinese wild Vitis pseudoreticulata W. T. Wang 'Baihe-35-1', and designated as VpWRKY1 (GenBank accession no. GQ884198) and VpWRKY2 (GenBank accession no. GU565706). Nuclear localization of the two proteins was demonstrated in onion epidermal cells, while trans-activation function was confirmed in the leaves of 'Baihe-35-1'. Expression of VpWRKY1 and VpWRKY2 was induced rapidly by salicylic acid treatment in 'Baihe-35-1'. Expression of VpWRKY1 and VpWRKY2 was also induced rapidly by E. necator infection in 11 grapevine genotypes; the maximum induction of VpWRKY1 was greater in E. necator-resistant grapevine genotypes than in susceptible ones post E. necator inoculation. Furthermore, ectopic expression of VpWRKY1 or VpWRKY2 in Arabidopsis enhanced resistance to powdery mildew Erysiphe cichoracearum, and enhanced salt tolerance of transgenic plants. VpWRKY2 also enhanced cold tolerance of transgenic plants. In addition, the two proteins were shown to regulate the expression of some defense marker genes in Arabidopsis and grapevine. The data suggest that VpWRKY1 and VpWRKY2 may underlie the resistance in transgenic grapevine to E. necator and tolerance to salt and cold stresses.
DOI:10.1111/j.1467-7652.2008.00336.xURL [本文引用: 1]

Abstract WRKY-type transcription factors have multiple roles in the plant defence response and developmental processes. Their roles in the abiotic stress response remain obscure. In this study, 64 GmWRKY genes from soybean were identified, and were found to be differentially expressed under abiotic stresses. Nine GmWRKY proteins were tested for their transcription activation in the yeast assay system, and five showed such ability. In a DNA-binding assay, three proteins (GmWRKY13, GmWRKY27 and GmWRKY54) with a conserved WRKYGQK sequence in their DNA-binding domain could bind to the W-box (TTGAC). However, GmWRKY6 and GmWRKY21, with an altered sequence WRKYGKK, lost the ability to bind to the W-box. The function of three stress-induced genes, GmWRKY13, GmWRKY21 and GmWRKY54, was further investigated using a transgenic approach. GmWRKY21-transgenic Arabidopsis plants were tolerant to cold stress, whereas GmWRKY54 conferred salt and drought tolerance, possibly through the regulation of DREB2A and STZ/Zat10. Transgenic plants over-expressing GmWRKY13 showed increased sensitivity to salt and mannitol stress, but decreased sensitivity to abscisic acid, when compared with wild-type plants. In addition, GmWRKY13-transgenic plants showed an increase in lateral roots. These results indicate that the three GmWRKY genes play differential roles in abiotic stress tolerance, and that GmWRKY13 may function in both lateral root development and the abiotic stress response.
[本文引用: 1]
DOI:10.1007/s00299-008-0666-yURLPMID:19125253 [本文引用: 1]

The WRKY family is one of the major groups of plant-specific transcriptional regulators. Arabidopsis thaliana WRKY25, which is induced by heat stress, is one of the group I WRKY proteins and responds to both abiotic and biotic stress. This study has examined the regulatory role of WRKY25 using wrky25 mutant and over-expressing WRKY25 transgenic A. thaliana . After 45C for different time periods, wrky25 null mutants showed a moderate increase in thermosensitivity with decreased germination, reduced hypocotyl and root growth, and enhanced conductivity compared to those of wide-type, while WRKY25 over-expressed transgenic seeds exhibited enhanced thermotolerance. Northern blot analysis of wrky25 mutants and WRKY25 over-expressing plants identified putative genes regulated by WRKY25. In consistence with the implication of WRKY25 in heat tolerance, the expression level of six heat-inducible genes and two oxidative stress-responsive genes was more or less down-regulated in wrky25 mutants during heat stress. Among them, heat shock protein Hsp101 , heat shock transcription factor HsfB2a , and cytosolic ascrobate peroxidase APX1 were reduced more obviously than other detected genes. Meanwhile, over-expression of WRKY25 increased the expression of HsfA2 , HsfB1 , HsfB2a , and Hsp101 slightly or moderately. Together, these findings reveal that WRKY25 plays a partial role in thermotolerance.
[本文引用: 1]
DOI:10.1007/s11103-008-9408-3URLPMID:18839316 [本文引用: 1]

Previous microarray analyses of Arabidopsis roots identified two closely related WRKY transcription factors ( WRKY25 and WRKY33 ) among the transcripts that increased in abundance following treatment with NaCl. Here, we report further characterization of these genes, which we found to be inducible by a variety of abiotic stresses in an SOS-pathway independent manner, although WRKY33 induction was dependent on ABA signaling. Transcripts of both genes were detected in roots and leaves, while specific patterns of enrichment were observed in stems and floral buds for WRKY25 and WRKY33 , respectively. We also identified upstream intergenic regions from each gene that were sufficient to confer stress-inducible expression on a reporter gene. However, the stress sensitivity of wrky25 null mutants did not differ from wild-type under any assay condition, while wrky33 null mutants and wrky25wrky33 double mutants showed only a moderate increase in NaCl-sensitivity, suggesting functional redundancy with other transcription factors. Nevertheless, overexpression of WRKY25 or WRKY33 was sufficient to increase Arabidopsis NaCl tolerance, while increasing sensitivity to ABA. Through microarray analyses of relevant genotypes, we identified 31 and 208 potential downstream targets of WRKY25 and WRKY33, respectively, most of which contained a W-box in their upstream regions.
DOI:10.1007/s00299-008-0614-xMagsci [本文引用: 2]

<a name="Abs1"></a>An <i>OsWRKY11</i> gene, which encodes a transcription factor with the WRKY domain, was identified as one of the genes that was induced by both heat shock and drought stresses in seedlings of rice (<i>Oryza sativa</i> L.). To determine if overexpression of <i>OsWRKY11</i> confers heat and drought tolerance, <i>OsWRKY11</i> cDNA was fused to the promoter of <i>HSP101</i> of rice and introduced into a rice cultivar Sasanishiki. Overexpression of <i>OsWRKY11</i> was induced by heat treatment. After heat pretreatment, the transgenic lines showed significant heat and drought tolerance, as indicated by the slower leaf-wilting and less-impaired survival rate of green parts of plants. They also showed significant desiccation tolerance, as indicated by the slower water loss in detached leaves. Our results indicate that the <i>OsWRKY11</i> gene plays a role in heat and drought stress response and tolerance, and might be useful for improvement of stress tolerance.
DOI:10.1270/jsbbs.61.121Magsci [本文引用: 1]

The WRKY transcription factors belong to a lame protein family characterized by the conserved WRKY domain. These factors have been identified to play biological functions in various plant developmental processes. WRKY proteins are also known to be involved in regulating plant responses to pathogen attack and stress-related hormones. In this study, we report the isolation and characterization of the gene (TaWRKY45) for the wheat WRKY45 transcription factor. Amino acid sequence alignment and phylogenetic analyses demonstrated that the TaWRKY45 protein is orthologous to rice OsWRKY45. Our analysis of its expression in wheat indicated that TaWRKY45 was constitutively expressed in various organs and throughout the lifetime of the plant. We observed that TaWRKY45 was upregulated in response to benzothiadiazole (BTH), a plant immune system strengthner, and Fusarium graminearum, which is a causal fungus for Fusarium head blight (FHB). The constitutive overexpression of the TaWRKY45 transgene conferred an enhanced resistance against F. graminearum to transenic wheat plants grown under greenhouse conditions. These results indicate that TaWRKY45 is involved M the defense systems for the biotic stressors in wheat and that it may be potentially utilized to improve the disease resistance of wheat.
URL [本文引用: 2]

Salinity stress limits crop yield affecting plant growth and restricting the use of land. As world population is increasing at alarming rate, agricultural land is shrinking due to industrialization and/or habitat use. Hence, there is a need to utilize salt affected land to meet the food requirement. Although some success has been achieved through conventional breeding but its use is limited due...
[本文引用: 2]
DOI:10.1007/s10495-013-0845-3URLPMID:23568336 [本文引用: 1]

Voltage-dependent anion channels (VDACs), known as outer mitochondrial membrane proteins, are present in all eukaryotic cells. In mammals, they are now recognized to play crucial roles in the regulation of metabolic and energetic functions of mitochondria as well as in mitochondria-mediated apoptosis, in association with various proteins and non-protein modulators. Although there is much less information available for plant than for animal VDACs, their similar electrophysiological and topological properties suggest that some common functions are conserved among eukaryotic VDACs. Recently, it has been revealed that plant VDACs also have various important physiological functions not only in developmental and reproductive processes, but also in biotic and abiotic stress responses, including programmed cell death. In this review, we summarize recent findings about the sequence motifs, localization, and function of plant VDACs and discuss these results in the light of recent advances in research on animal VDACs.
DOI:10.1038/srep35683URL [本文引用: 1]

Mycosphaerella graminicola(Zymoseptoria triticicommonly known as Septoria), the causal agent of Septoria Leaf Blotch (STB), is considered one of the major threats to European wheat production. Previous studies have shown the importance of ubiquitination in plant defence against a multitude of pathogens. However the ubiquitination machinery in wheat is under studied, particularly E2 enzymes that have the ability to control the ubiquitination and thereby the fate of many different target proteins. In this study we identify an E2 enzyme,Triticum aestivumUbiquitin conjugating enzyme 4 (TaU4) that functions in wheat defence against Septoria. We demonstrate TaU4 to be a bona fide E2 enzyme through an E2 charging assay. TaU4 localises in both the cytoplasm and nucleus, therefore potentially interacting with E3 ligases and substrate proteins in multiple compartments. Virus Induced Gene Silencing of TaU4 in wheat leaves resulted in delayed development of disease symptoms, reduced Septoria growth and reproduction. We conclude that TaU4 is a novel negative regulator of defence against Septoria.
[本文引用: 1]
DOI:10.1016/S0014-5793(97)00509-7URLPMID:9202147 [本文引用: 1]

Abstract A clone of an ubiquitin-conjugating enzyme (UBC) was isolated from a lambda-ZAP-cDNA library, generated from mRNA of tomato (Lycopersicon esculentum) cells grown in suspension for 3 days. The open reading frame called LeUBC1, encodes for a polypeptide with a predicted molecular mass of 21.37 kDa, which was confirmed by bacterial overexpression and SDS-PAGE. Database searches with LeUBC1 showed highest sequence similarities to UBC1 of bovine and yeast. By Southern blot analysis LeUBC1 was identified as a member of a small E2 subfamily of tomato, presumably consisting of at least two members. As revealed by Northern blot analysis LeUBC1 is constitutively expressed in an exponentially growing tomato cell culture. In response to heat shock an increase in LeUBC1-mRNA was detectable. A strong accumulation of the LeUBC1-transcript was observed by exposure to heavy metal stress which was performed by treatment with cadmium chloride (CdCl2). The cellular uptake of cadmium was controlled via ICP-MS measurements. The data suggest that like in yeast, in plants a certain subfamily of UBC is specifically involved in the proteolytic degradation of abnormal proteins as result of stress.
DOI:10.1023/A:1022289509702URL

The induction of general phenylpropanoid and flavonoid glycoside pathways and the consequential accumulation of UV protective flavonoids so far have been regarded as the only major metabolic response to UV light. Phenylalanine ammonia-lyase (PAL) is a key enzyme of phenylpropanoid metabolism. Inducible in vivo footprints in the pal -1 promoter defined the motif CTCCAACAACCCCTTC as a cis-element that transmits the UV signal. In order to clone its corresponding trans-acting element, three copies of the motif were synthesized and used as a bait of yeast one-hybrid system. A number of genes coding ubiquitin-conjugating enzyme-like proteins (E2) were isolated from a rice cDNA library by the system. The rice (Oryza sativa L) E2 (RE2) was high homology to other E2 of different species and can be induced by UV light. RE2 can react with nuclear extraction protein and binds the pal-1 promoter cis-element. These results suggest that RE2 takes part in plant response to UV stress through regulating the phenylpropanoid metabolism.
DOI:10.1111/j.1365-3040.2012.02480.xURLPMID:22220579

WRKY-type transcription factors are involved in multiple aspects of plant growth, development and stress response. WRKY genes have been found to be responsive to abiotic stresses; however, their roles in abiotic stress tolerance are largely unknown especially in crops. Here, we identified stress-responsive WRKY genes from wheat (Triticum aestivum L.) and studied their functions in stress tolerance. Forty-three putative TaWRKY genes were identified and two multiple stress-induced genes, TaWRKY2 and TaWRKY19, were further characterized. TaWRKY2 and TaWRKY19 are nuclear proteins, and displayed specific binding to typical cis-element W box. Transgenic Arabidopsis plants overexpressing TaWRKY2 exhibited salt and drought tolerance compared with controls. Overexpression of TaWRKY19 conferred tolerance to salt, drought and freezing stresses in transgenic plants. TaWRKY2 enhanced expressions of STZ and RD29B, and bound to their promoters. TaWRKY19 activated expressions of DREB2A, RD29A, RD29B and Cor6.6, and bound to DREB2A and Cor6.6 promoters. The two TaWRKY proteins may regulate the downstream genes through direct binding to the gene promoter or via indirect mechanism. Manipulation of TaWRKY2 and TaWRKY19 in wheat or other crops should improve their performance under various abiotic stress conditions.
