 ,, 杨官显, 王意程, 姜生辉, 王楠, 陈学森
,, 杨官显, 王意程, 姜生辉, 王楠, 陈学森 ,山东农业大学园艺科学与工程学院/作物生物学国家重点实验室,山东泰安 271018
,山东农业大学园艺科学与工程学院/作物生物学国家重点实验室,山东泰安 271018Apple MdMYB32 Inhibits the Anthocyanin Biosynthesis by Its Own EAR Inhibitory Sequence
XU HaiFeng ,, YANG GuanXian, WANG YiCheng, JIANG ShengHui, WANG Nan, CHEN XueSen
,, YANG GuanXian, WANG YiCheng, JIANG ShengHui, WANG Nan, CHEN XueSen ,College of Horticulture Science and Engineering, Shandong Agricultural University/State Key Laboratory of Crop Biology, Tai’an 271018, Shandong
,College of Horticulture Science and Engineering, Shandong Agricultural University/State Key Laboratory of Crop Biology, Tai’an 271018, Shandong通讯作者:
第一联系人:
收稿日期:2018-06-22接受日期:2018-07-16网络出版日期:2018-12-26
| 基金资助: |
Received:2018-06-22Accepted:2018-07-16Online:2018-12-26

摘要
关键词:
Abstract
Keywords:
PDF (2643KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
许海峰, 杨官显, 王意程, 姜生辉, 王楠, 陈学森. 苹果MdMYB32通过自身EAR抑制序列抑制花青苷的生物合成[J]. 中国农业科学, 2018, 51(24): 4690-4699 doi:10.3864/j.issn.0578-1752.2018.24.009
XU HaiFeng, YANG GuanXian, WANG YiCheng, JIANG ShengHui, WANG Nan, CHEN XueSen.
0 引言
【研究意义】水果和观赏作物器官的着色是一种重要的经济特性,而花青苷通常是器官着色的重要原因,如鲜红色、红色、蓝色和紫色[1]。花青苷作为类黄酮的一种,具有抗氧化,预防心血管疾病以及抑制肥胖等功能[2,3]。MYB蛋白是植物较大的转录因子家族,在花青苷合成中发挥重要的作用。因此,研究MYB家族中MdMYB32在花青苷生物合成中的功能,对完善花青苷合成代谢机理具有重要意义。【前人研究进展】花青苷合成受紫外照射、低温、干旱等环境因素诱导,且涉及多种结构基因和转录因子[4,5],前人研究表明MYB-bHLH-WD40转录复合体能够控制花青苷的合成,其中MYB家族和bHLH家族在调控花青苷合成中的作用在模式植物中得到了广泛的研究[6,7]。最近,关于水果方面花青苷合成取得了一些进展,在葡萄中,与拟南芥AtMYB75同源的VvMYBA1和VvMYBA2被发现能够调控葡萄花青苷的合成[8],类似的同源基因在草莓、梨和桃中也被发现,分别是FaMYB10、PyMYB10和PpMYB10[9,10]。在苹果中,MdMYB1和MdMYBA参与了光诱导下花青苷合成[11,12],ESPLEY等[13,14]研究发现MdMYB10能够调控红肉苹果中花青苷合成。笔者课题组于2006年率先构建了新疆红肉苹果与栽培苹果品种的杂种分离群体[15,16,17,18,19,20],以此群体为试验材料,许海峰等[21]分析了杂种一代4个红肉苹果株系类黄酮和花青苷含量及相关基因的表达;JI等[22,23]探讨了氮对愈伤组织花青苷生物合成的影响,并对红色和黄色愈伤组织进行了RNA-seq分析;而WANG等[24]则对F1分离群体中的绿色和红色单株各20株进行了RNA-seq分析,筛选到了与花青苷合成相关的差异表达基因;此外,XU等[25]和WANG等[26]还分别研究了MdMYB16和MdMYB12参与花青苷合成的分子机理。【本研究切入点】EAR抑制序列的发现,为花青苷合成调控提供了新的视角。拟南芥AtMYB4是第一个被发现含有EAR抑制序列的负调控因子,它通过直接结合靶基因的启动子来发挥抑制其表达的作用[27,28],草莓FaMYB1蛋白也由于存在一个类似的EAR序列,对花青苷合成具有抑制作用[29]。在苹果中,关于MYB中EAR抑制序列对花青苷负调控研究尚未明确。【拟解决的关键问题】本研究以新疆红肉苹果杂种一代优系‘红脆6号’为试材,克隆了含有EAR序列的MdMYB32,测定其在不同果实及在不同胁迫处理下的表达水平与花青素的关系,通过酵母单杂和Chip-PCR分析其互作关系,并通过转基因验证其在花青苷合成中的功能,旨在为进一步完善花青苷合成代谢机理提供参考。1 材料与方法
试验于2017年在山东农业大学园艺科学与工程学院进行。1.1 植物材料和处理
植物材料为从新疆红肉苹果(Malus sieversii f. neidzwetzkyana)中的‘塔尔阿尔玛’与‘烟富3号’(M. domestica cv. Fuji)杂种一代选育出的‘红脆6号’‘红脆8号’和‘红脆9号’优株及‘紫红3号’苹果诱导的红肉愈伤组织(MS+1 mg·L-16-BA+0.3 mg·L-1NAA培养基上培养)。红肉苹果愈伤组织诱导、培育方法参照JI等[22]。取继代生长16 d的红肉愈伤组织,分别用4℃和150 mmol?L-1 NaCl处理24 h,每隔6 h取样一次,-80℃保存备用。1.2 总RNA的提取与qRT-PCR
RNA提取及qRT-PCR参考XU等[25]方法。RNA提取试剂盒(DP432)、反转录试剂盒(KR106)、SYBR染料(FP205)均购自北京天根公司,荧光定量每个样品设3个重复,以苹果Actin为内参,每个基因扩增均伴有内参同时扩增,默认条件下读取Ct值,采用2-ΔΔCT方法进行数据分析[30]。MdActin荧光定量引物为5′-TGACCGAATGAGCAAGGAAATTACT-3′和5′-TA CTCAGCTTTGGCAATCCACATC-3′,MdMYB32荧光定量引物为5′-CTGGAACACTCACGTCAAGC-3′,MdMYB10荧光定量引物为5′-GGAAACAGGTGGTC ATTGATTG-3′和5′-GGCTGAGGTCTTATCACATTG GT-3′,MdDFR荧光定量引物为5′-GATAGGGTTTGA GTTCAAGTA-3′和5′-TCTCCTCAGCAGCCTCAGT TTTCT-3′,MdANS荧光定量引物为5′-GTGTCATGCA CCTTGTGAACC-3′和5′-GTAGTCCTCCCACTCAAG CTG-3′,MdUFGT荧光定量引物为5′-CCGCCCTTCC AAACACTCT和5′-GAGCTCTATGTCCTCCTGCG-3′。1.3 花青苷提取和吸光度测定
花青苷提取参照JI[22]等方法,并略作改动。称取0.5 g植物材料在液氮中研磨成粉,用20 mL 1%(v/v)HCl-甲醇在4℃黑暗条件下萃取24 h,12 000 r/min离心10 min后保留上清液,用紫外分光光度计测定上清液在530 nm处吸光度,最后花青苷的相对含量用Abs/g(吸光度/质量)来表示。1.4 overlap PCR技术敲除MdMYB32蛋白C端EAR抑制序列
参考XU等[25]方法,通过overlap PCR技术敲除MdMYB32蛋白C端EAR(GDLNLDLSIG)抑制序列。具体引物设计如下,F1:5′-ATGGGGAGAGCACCTTGTT-3′
R1:5′-ACGGCTCTAAACACTTGTATTTCTGCT GTT-3′
F2:5′-AATACAAGTGTTTAGAGCCGTTTCAG TCCAA-3′
R2:5′-TCAACCTGGCAGCACAAA-3′
以‘红脆6号’苹果cDNA为模板,用F1和R1引物通过PCR扩增获得MdMYB32-1序列,用F2和R2引物通过PCR扩增获得MdMYB32-2序列,将扩增获得的MdMYB32-1和MdMYB32-2产物1﹕1混合,以它们为模板用F1和R2引物通过PCR扩增获得的产物即为没有EAR抑制序列的MdMYB32,暂命名为LESMdMYB32。
1.5 MdMYB32和LESMdMYB32红肉苹果愈伤组织的转化
用引物5′-GTCGACATGGGGAGAGCACCTTGT T-3′和5′-GGATCCTCAACCTGGCAGCACAAA-3′扩增得到MdMYB32的编码框序列,用上述overlap-PCR方法扩增得到LESMdMYB32的编码框序列,琼脂糖凝胶电泳,回收目的条带,连接到PLB零背景克隆载体。用Sal I和BamH I对克隆载体和植物表达载体pRI101分别进行双酶切,构建重组表达载体pRI101-MdMYB32和pRI101-LESMdMYB32。将重组质粒导入农杆菌LBA4404,在28℃下,用30 mL含50 μg·mL-1卡那霉素和50 μg·mL-1利福平的YEP液体培养基培养重组农杆菌至OD600=0.6,12 000 r/min离心收集菌体,用30 mL ddH2O悬浮,加入乙酰丁香酮并使其浓度为100 μmol·L-1,得到侵染液。取生长16 d的红肉苹果愈伤组织浸到侵染液中,室温振荡25 min,取愈伤组织置于含1 mg·L-16-BA+0.3 mg·L-1NAA的MS固体培养基上,28℃暗培养2 d。随后转移到含1 mg·L-16-BA+ 0.3 mg·L-1NAA+50 μg·m L-1卡那霉素+250 μg·mL-1羧苄青霉素的MS固体培养基上暗培养5周左右。1.6 酵母单杂交
用引物5′-CATATGATGGGGAGAGCACCTTGT T-3′和5′-GGATCCTCAACCTGGCAGCACAAA-3′扩增MdMYB32的编码框序列,用引物5′-GAATTCGGC TAGTTCTGTATTATGGTTGATA-3′和5′-GAGCTCT GATAATGTAACGTTGATTTTATAGTAG-3′扩增MdANS的启动子序列,分别连接PLB零背景载体。用Nde I和BamH I对MdMYB32-PLB和pGADT7分别进行双酶切,用EcoR I和Sac I对MdANS pro-PLB和pHIS2分别进行双酶切,构建pGADT7-MdMYB32和pHIS2-MdANS pro重组载体。按照YeastmakerTM Yeast Transformation System 2试剂盒(Clontech)说明书方法,将pHIS2-MdANS pro重组载体转化到酵母Y187感受态细胞,细胞培养在-T-H+3-AT培养基中 (-Trp/His,Clontech),筛选抑制酵母生长的3-AT浓度,然后将这两种重组质粒共转化到Y187酵母感受态细胞,细胞培养在-T-H-L+3-AT培养基中(-Trp/ His/Leu,Clontech),观察酵母生长情况。1.7 Chip-PCR
Chip-PCR试验根据HE等[31]方法,并略作改动。按照Chip Kit(Upstate,Lake Placid,NY,USA)里说明书方法进行交联、解交联、免疫沉淀和洗脱,免疫抗体使用GFP抗体(Abmart,Shanghai,China),最后用PCR检测。PCR引物为:5′-CTGGTATGTACCATTCA CTTGTTG-3′和5′-CGATCATCCTCCGAGTGTC-3′。1.8 数据分析
荧光定量数据用Excel 2007进行作图和标准差分析,用DPS 7.05软件(http://www.chinadps.net)进行显著性检验,用MEGA 5软件构建系统发育进化树,分析方法采用Construct/Test Neighbor-Joining Tree,用Clustal X软件分析蛋白序列。2 结果
2.1 MdMYB32在不同苹果果实中的表达水平
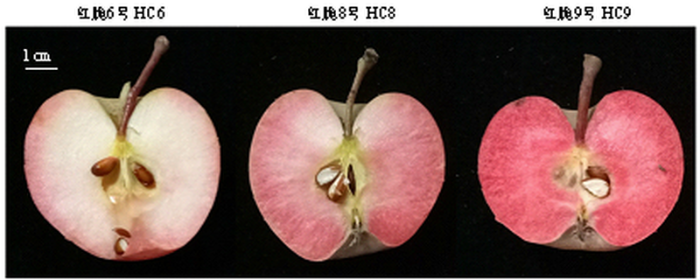
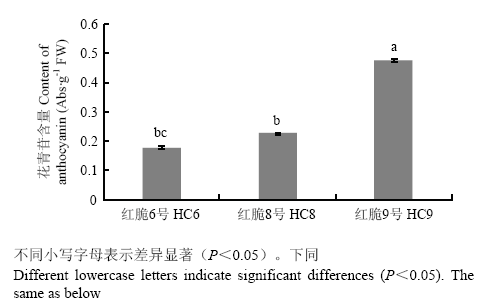
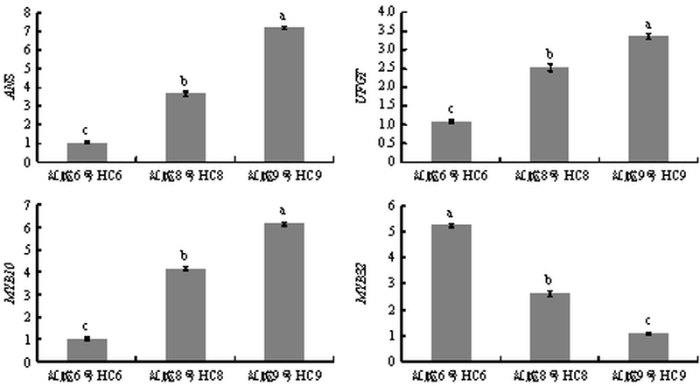
图1为‘红脆6号’(HC6)‘红脆8号’(HC8)和‘红脆9号’(HC9)优株果实成熟期横截面图。由图2和图3可得,果实成熟期花青苷相对含量和花青苷合成结构基因ANS及UFGT的相对表达水平高低顺序为‘红脆9号’>‘红脆8号’>‘红脆6号’,控制红肉苹果花青苷合成关键转录因子MYB10的相对表达水平也是‘红脆9号’>‘红脆8号’>‘红脆6号’,但MYB32的表达水平却与之相反,为‘红脆6号’>‘红脆8号’>‘红脆9号’。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1苹果果实成熟期横截面图
Fig. 1Sectional drawing of apples in the mature period
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图23个苹果株系中花青苷相对含量
Fig. 2Relative content of anthocyanin in 3 apple strains
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3三个苹果株系中花青苷合成结构基因及相关转录因子相对表达水平
Fig. 3Relative expression of structural genes associated with anthocyanin synthesis and related transcription factors in 3 apple strains
2.2 MdMYB32的进化树和蛋白序列分析
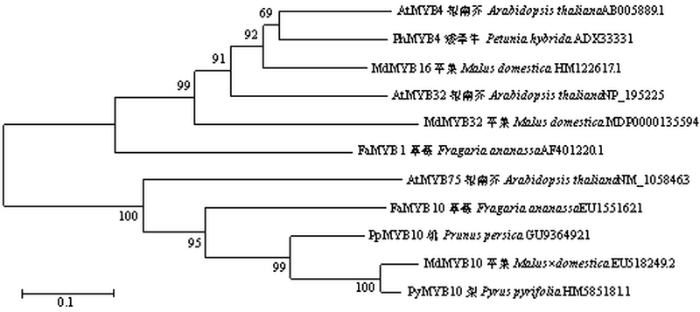
如图4所示,促进花青苷合成的AtMYB75、FaMYB10、PpMYB10、MdMYB10和PyMYB10都在同一个进化枝上,抑制花青苷合成的AtMYB4、PhMYB4、MdMYB16、AtMYB32和FaMYB1在同一个进化枝上,而MdMYB32与AtMYB32等在同一个进化枝,推测其可能具有抑制花青苷合成的功能(图4中比例尺0.1代表每10个氨基酸有1个不同)。由图5可看出在MdMYB32蛋白C端存在一个10个氨基酸长度的EAR抑制序列。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4MYB32和相关MYB蛋白系统进化树分析
Fig. 4Phylogenetic tree of MYB32 and related MYB protein
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5MYB32和相关蛋白序列分析
Fig. 5Sequence analysis of MYB32 and related protein
2.3 冷胁迫和盐胁迫处理红肉愈伤MdMYB32的表达水平分析
由图6可知,4℃和150 mmol?L-1 NaCl处理均能抑制MdMYB32的表达,其中4℃低温处理后MdMYB32的表达水平逐渐降低,而150 mmol?L-1 NaCl处理后MdMYB32的表达水平先降低后又轻微回升。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6冷胁迫(A)和盐胁迫(B)处理下MdMYB32的相对表达水平
Fig. 6Relative expression of MdMYB32 treated with cold stress or salt stress
2.4 MdMYB32和LESMdMYB32在红肉愈伤中过表达分析
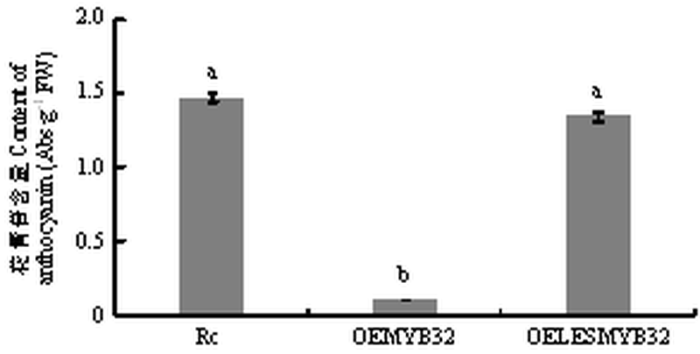
红肉愈伤组织(Rc)、过表达MdMYB32的红肉愈伤(OEMYB32)和过表达LESMdMYB32(敲除EAR抑制序列的MdMYB32)的红肉愈伤(OELESMYB32)如图7所示。由图8和图9可知,在红肉愈伤中过表达MdMYB32能够抑制花青苷的合成和ANS的表达,对DFR和UFGT表达水平没有影响,但敲除MdMYB32的EAR抑制序列后,在红肉愈伤中过表达LESMdMYB32不能够抑制花青苷的合成,且对DFR、ANS和UFGT的表达水平没有影响。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7红肉愈伤和过表达MYB32及LESMYB32愈伤
Fig. 7Red-fleshed callus and the red-fleshed callus that overexpressing MYB32 or LESMYB32
图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图83种愈伤花青相对苷含量
Fig. 8Relative content of anthocyanin in 3 kinds of callus
图9
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图93种愈伤花青苷生物合成相关基因相对表达水平
Fig. 9Relative expression level of related genes involved of anthocyanin biosynthesis in 3 callus
2.5 MdMYB32和LESMdMYB32与MdANS启动子的酵母单杂交和Chip-PCR分析
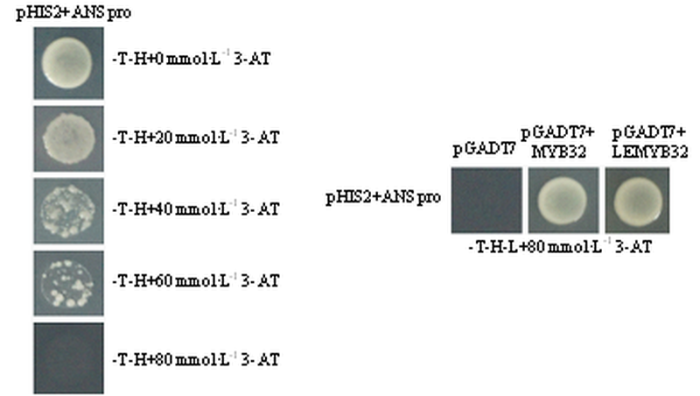
由图10酵母单杂交可知,pHIS2+ANS pro单转Y187后培养在-T-H培养基中,通过3-AT筛选抑制酵母生长的浓度,最后在80 mmol?L-1 3-AT中酵母不能生长,因此ANS启动子的自激活浓度为80 mmol?L-1 3-AT,将pHIS2+ANS pro分别和pGADT7空载体、pGADT7+MYB32、pGADT7+LESMYB32共转Y187后培养在-T-H-L+80 mmol?L-1 3-AT中,共转pHIS2+ ANS pro和pGADT7空载体的不能生长,而共转pHIS2+ANS pro和pGADT7+MYB32以及pHIS2+ ANS pro和pGADT7+LESMYB32的均能生长,因此,MdMYB32和LESMdMYB32均能结合MdANS的启动子。图10
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图10MdMYB32和LESMdMYB32与MdANS启动子的酵母单杂交分析
Fig. 10Yeast one-hybrid analysis in MdMYB32, LESMdMYB32 and MdANS promoter
图11
 新窗口打开|下载原图ZIP|生成PPT
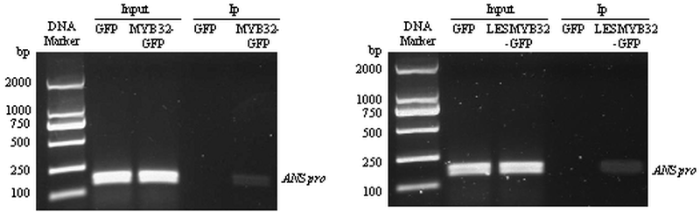
新窗口打开|下载原图ZIP|生成PPT图11MdMYB32和LESMdMYB32与MdANS启动子的Chip-PCR分析
Fig. 11Chip-PCR analysis in MdMYB32, LESMdMYB32 and MdANS promoter
HARTMANN等[32]研究表明花青苷合成结构基因启动子上的MRE(MYB recognition element)能够被MYB转录因子识别。为了进一步验证MdMYB32和LESMdMYB32与MdANS启动子的互作关系,在MdANS启动子上-1 365到-1 371处,发现了一个MRE元件,因此,选取包含MRE元件的一段序列(-1 302到-1 475)用作Chi-PCR分析。由图11可知,在Input中,GFP、MYB32-GFP和LESMYB32-GFP都有条带,而在Ip中,只有MYB32-GFP和LESMYB32-GFP有条带,因此,MYB32和LESMYB32均能结合MdANS的启动子。
3 讨论
3.1 MdMYB32能够结合MdANS启动子抑制花青苷的合成
在烟草中异源表达草莓FaMYB1时能抑制花青苷合成,是第一个被发现抑制花青苷合成的MYB转录因子[29]。在矮牵牛中,PhMYB27能够结合bHLH蛋白,作用于MBW复合体抑制花青苷的合成[33],类似的作用模式在FaMYB1、VvMYBC2和MtMYB2中也被发现[34,35,36],它们不能结合靶基因的启动子,但它们能够与bHLH蛋白结合,作用于MBW复合体从而发挥抑制作用。AtMYB4和PhMYB4与上述不同,它们能够直接结合靶基因的启动子从而发挥抑制作用[28,37],XU等[25]将MYB类抑制因子分为两类,一种作用于MBW复合体发挥功能,类似于FaMYB1等;另一种能直接结合靶基因启动子发挥功能,类似于AtMYB4。在本文中,发现MdMYB32在花青苷含量高的苹果中表达水平较低,而在花青苷含量较低的苹果中表达水平较高,这暗示着MdMYB32可能参与调控花青苷合成,且冷胁迫和盐胁迫能够抑制MdMYB32的表达,在红肉愈伤中过表达MdMYB32发现,它能够抑制MdANS的表达和花青苷的合成,进一步通过酵母单杂和Chip-PCR分析,MdMYB32能够结合MdANS的启动子,暗示着MdMYB32能直接结合花青苷合成结构基因MdANS的启动子从而抑制花青苷的合成,这与AtMYB4等类似。3.2 MdMYB32通过自身C端EAR抑制序列发挥功能
EAR基序是EAR型转录抑制子的共同特征,EAR基序内氨基酸残基的变化会导致抑制功能减少或丧失[38,39,40]。拟南芥AtERF3通过其C末端的EAR基序能抑制AtERF5与植物中GCC box的结合,然而,当EAR基序(LDLNLAP)中D突变成A时,这种抑制功能就丧失了[39];在拟南芥中过表达ZAT7能增加盐胁迫抗性,而ZAT7蛋白的EAR基序突变或缺失降低了转基因植物中这种增加的抗性[41];近年来,根据EAR型转录抑制子的结构和功能,开发了一种嵌合抑制基因沉默技术,将EAR基序添加到活化剂的C末端作为嵌合蛋白融合,通过转基因导入植物来实现定性功能缺失[42,43]。本研究通过overlap PCR技术敲除MdMYB32蛋白C端的EAR基序(GDLNLDLSIG),暂命名为LESMdMYB32,酵母单杂交和Chip-PCR实验表明MdMYB32和LESMdMYB32都能够结合MdANS的启动子,说明EAR序列的存在与否不影响MdMYB32与MdANS启动子的结合,然而在红肉愈伤中过表达LESMdMYB32后,发现它不能够抑制花青苷的合成,因此,MdMYB32通过自身的EAR抑制基序来发挥功能。4 结论
本研究发现MdMYB32蛋白C端存在一个EAR抑制序列,盐胁迫和冷胁迫均能够抑制其表达,酵母单杂和Chip-PCR分析表明MdMYB32能够结合MdANS的启动子。在红肉愈伤中过表达MdMYB32,能通过自身的EAR抑制基序抑制花青苷的合成。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00217-008-0824-zURL [本文引用: 1]

Total phenolics, flavonoids, anthocyanins, cyanidin-3- O -glucoside (Cy-3-glu) and antioxidant capacity of Chinese bayberry fruit ( Myrica rubra Sieb. and Zucc.) differed among the four cultivars “Baizhong” (white), “Fenhong” (pink), “Wuzhong” (red) and “Biqi” (dark red). Antioxidant capacity determined by both the ferric reducing antioxidant power (FRAP) assay and 2,2-diphenyl-2-picrylhydrazyl (DPPH . ) radical scavenging capacity was significantly correlated with the antioxidant components in the fruit, and directly related to fruit color. Cy-3-glu accounted for at least 82, 38, and 12% of the total antioxidant capacity in “Biqi”, “Wuzhong” and “Fenhong” fruits, respectively. No detectable Cy-3-glu was found in “Baizhong” fruit. Greater fruit maturity was associated with higher levels of all the bioactive components and antioxidant capacity. Significant increases were also found during postharvest storage of “Biqi” fruit held at either 20 °C for 2days or 0 °C for 5days. However, these levels decreased during a 2-day shelf-life at 20 °C after 5days at 0 °C. These results show that storage and shelf-life conditions are important if health-based bioactive components of bayberry fruit are to be maintained after harvest.
DOI:10.1007/s11130-013-0349-xURLPMID:23605674 [本文引用: 1]

Chinese bayberry (Myrica rubra Sieb. et Zucc.) is a subtropical fruit tree native to China and other Asian countries, and culture of this Myricaceae plant has been recorded in Chinese history for more than 2000 years. Bayberry fruit is delicious with attractive color, flavor, and high economic value. Compared with other berries, bayberry fruit is a rich source of cyanidin-3-glucoside (C3G, e.g., 64.8 mg/100 g fresh weight in 'Biqi' cultivar), which accounts for at least 85 % of the anthocyanins in the fruit. Bayberry is also a plant with high medicinal value since different organs have been used historically as folk medicines. Research efforts suggest bayberry extracts contain antioxidants that exhibit bioactivities counteracting inflammation, allergens, diabetes, cancer, bacterial infection, diarrhea and other health issues. Bayberry compounds have been isolated and characterized to provide a better understanding of the chemical mechanisms underlying the biological activities of bayberry extracts and to elaborate the structure-activity relationships. As the identification of compounds progresses, studies investigating the in vivo metabolism and bioavailability as well as potential toxicity of bayberry extracts in animal models are receiving more attention. In addition, breeding and genetic studies of bayberry with high accumulation of health-benefiting compounds may provide new insight for the bayberry research and industry. This review is focused on the main medicinal properties reported and the possible pharmaceutically active compounds identified in different bayberry extracts.
[本文引用: 1]
DOI:10.1007/s00425-014-2129-8URLPMID:25074586 [本文引用: 1]

Main conclusion Our studies showed that an apple B-box protein, MdCOL11, the homolog of AtBBX22, is involved in UV-B- and temperature-induced anthocyanin biosynthesis in apple peel. Anthocyanin is...
DOI:10.1104/pp.109.147322URLPMID:19906891 [本文引用: 1]

The color of tomato fruit is mainly determined by carotenoids and flavonoids. Phenotypic analysis of an introgression line (IL) population derived from a cross between Solanum lycopersicum 'Moneyberg' and the wild species Solanum chmielewskii revealed three ILs with a pink fruit color. These lines had a homozygous S. chmielewskii introgression on the short arm of chromosome 1, consistent with the position of the y (yellow) mutation known to result in colorless epidermis, and hence pink-colored fruit, when combined with a red flesh. Metabolic analysis showed that pink fruit lack the ripening-dependent accumulation of the yellow-colored flavonoid naringenin chalcone in the fruit peel, while carotenoid levels are not affected. The expression of all genes encoding biosynthetic enzymes involved in the production of the flavonol rutin from naringenin chalcone was down-regulated in pink fruit, suggesting that the candidate gene underlying the pink phenotype encodes a regulatory protein such as a transcription factor rather than a biosynthetic enzyme. Of 26 MYB and basic helix-loop-helix transcription factors putatively involved in regulating transcription of genes in the phenylpropanoid and/or flavonoid pathway, only the expression level of the MYB12 gene correlated well with the decrease in the expression of structural flavonoid genes in peel samples of pink- and red-fruited genotypes during ripening. Genetic mapping and segregation analysis showed that MYB12 is located on chromosome 1 and segregates perfectly with the characteristic pink fruit color. Virus-induced gene silencing of SlMYB12 resulted in a decrease in the accumulation of naringenin chalcone, a phenotype consistent with the pink-colored tomato fruit of IL1b. In conclusion, biochemical and molecular data, gene mapping, segregation analysis, and virus-induced gene silencing experiments demonstrate that the MYB12 transcription factor plays an important role in regulating the flavonoid pathway in tomato fruit and suggest strongly that SlMYB12 is a likely candidate for the y mutation.
DOI:10.1111/j.1365-313X.2010.04465.xURLPMID:21235651 [本文引用: 1]

Summary We present an investigation of anthocyanin regulation over the entire petunia plant, determining the mechanisms governing complex floral pigmentation patterning and environmentally induced vegetative anthocyanin synthesis. DEEP PURPLE ( DPL ) and PURPLE HAZE ( PHZ ) encode members of the R2R3-MYB transcription factor family that regulate anthocyanin synthesis in petunia, and control anthocyanin production in vegetative tissues and contribute to floral pigmentation. In addition to these two MYB factors, the basic helix oopelix (bHLH) factor ANTHOCYANIN1 (AN1) and WD-repeat protein AN11, are also essential for vegetative pigmentation. The induction of anthocyanins in vegetative tissues by high light was tightly correlated to the induction of transcripts for PHZ and AN1 . Interestingly, transcripts for PhMYB27, a putative R2R3-MYB active repressor, were highly expressed during non-inductive shade conditions and repressed during high light. The competitive inhibitor PhMYBx (R3-MYB) was expressed under high light, which may provide feedback repression. In floral tissues DPL regulates vein-associated anthocyanin pigmentation in the flower tube, while PHZ determines light-induced anthocyanin accumulation on exposed petal surfaces (bud-blush). A model is presented suggesting how complex floral and vegetative pigmentation patterns are derived in petunia in terms of MYB, bHLH and WDR co-regulators.
DOI:10.1007/s00122-008-0840-1URLPMID:18651125 [本文引用: 1]

As a result of natural hybridization and human selection over millennia, the skin colors of grapes have become greatly diversified. The color is determined by the quantity and composition of anthocyanins. Color-skinned cultivars accumulate anthocyanins in their skins, whereas white-skinned cultivars do not. Myb -related transcription-factor genes such as VvmybA1 regulate anthocyanin biosynthesis. VvMYBA2r , VlmybA1-1 , VlmybA1-2 , and VlmybA2 , which are homologs of VvmybA1 , also regulate anthocyanin biosynthesis. In this study, we isolated a novel Myb -related sequence, VlmybA1-3 , from cultivars of Vitis labruscana ( Vitis vinifera Vitis labrusca ) by means of inverse PCR, and confirmed by means of transient gene expression assay that the gene regulates anthocyanin biosynthesis in grape berry skin. Seedlings of V. labruscana with two functional haplotypes at a region of berry color loci accumulated more anthocyanins than seedlings with a single functional haplotype. In addition, we investigated the haplotypes at the region in 35 cultivars (both V. vinifera and V. labruscana ), and found certain typical characteristics. These findings will contribute to the selection of seedlings with high anthocyanin quantities in breeding programs for wine and table grapes, and will help elucidate the origin and evolution of Vitis species.
URL [本文引用: 1]
URL [本文引用: 1]
DOI:10.1104/pp.106.088104URLPMID:17012405 [本文引用: 1]

Anthocyanins are secondary metabolites found in higher plants that contribute to the colors of flowers and fruits. In apples (Malus domestica Borkh.), several steps of the anthocyanin pathway are coordinately regulated, suggesting control by common transcription factors. A gene encoding an R2R3 MYB transcription factor was isolated from apple (cv Cripps' Pink) and designated MdMYB1. Analysis of the deduced amino acid sequence suggests that this gene encodes an ortholog of anthocyanin regulators in other plants. The expression of MdMYB1 in both Arabidopsis (Arabidopsis thaliana) plants and cultured grape cells induced the ectopic synthesis of anthocyanin. In the grape (Vitis vinifera) cells MdMYB1 stimulated transcription from the promoters of two apple genes encoding anthocyanin biosynthetic enzymes. In ripening apple fruit the transcription of MdMYB1 was correlated with anthocyanin synthesis in red skin sectors of fruit. When dark-grown fruit were exposed to sunlight, MdMYB1 transcript levels increased over several days, correlating with anthocyanin synthesis in the skin. MdMYB1 gene transcripts were more abundant in red skin apple cultivars compared to non-red skin cultivars. Several polymorphisms were identified in the promoter of MdMYB1. A derived cleaved amplified polymorphic sequence marker designed to one of these polymorphisms segregated with the inheritance of skin color in progeny from a cross of an unnamed red skin selection (a sibling of Cripps' Pink) and the non-red skin cultivar Golden Delicious. We conclude that MdMYB1 coordinately regulates genes in the anthocyanin pathway and the expression level of this regulator is the genetic basis for apple skin color.
DOI:10.1093/pcp/pcm066URLPMID:17526919 [本文引用: 1]

Red coloration of apple (Malus x domestica) skin is an important determinant of consumer preference and marketability. Anthocyanins are responsible for this coloration, and their accumulation is positively correlated with the expression level of anthocyanin biosynthetic genes. Regulation of expression of these genes is believed to be controlled by MYB transcription factors, and the MYB transcription factors involved in the activation of anthocyanin biosynthetic genes have been isolated in various plants. In the present study, we isolated and characterized a MYB transcription factor gene (MdMYBA) from apple skin. Characterization of MdMYBA demonstrated that (i) MdMYBA expression was specifically regulated depending on the tissue and cultivar/species; (ii) its expression level was much higher in a deep-red cultivar ('Jonathan') than in a pale-red cultivar ('Tsugaru'); (iii) when cauliflower mosaic virus 35S::MdMYBA was introduced into the cotyledons of apple seedlings by means of a transient assay, reddish-purple spots were induced, and MdMYBA also induced anthocyanin accumulation in reproductive tissues of transgenic tobacco; (iv) the expression of MdMYBA was induced by UV-B irradiation and low-temperature treatment, both of which are known to be important in the promotion of anthocyanin accumulation in apple skin; (v) MdMYBA bound specifically to an anthocyanidin synthase (MdANS) promoter region in a gel-shift assay; and (vi) MdMYBA was mapped to the near region of the BC226-STS (a1) marker for the red skin color locus (R(f)). These results suggest that MdMYBA is a key regulatory gene in anthocyanin biosynthesis in apple skin.
DOI:10.1111/j.1365-313X.2006.02964.xURLPMID:1865000 [本文引用: 1]

Anthocyanin concentration is an important determinant of the colour of many fruits. In apple (Malus domestica), centuries of breeding have produced numerous varieties in which levels of anthocyanin pigment vary widely and change in response to environmental and developmental stimuli. The apple fruit cortex is usually colourless, although germplasm does exist where the cortex is highly pigmented due to the accumulation of either anthocyanins or carotenoids. From studies in a diverse array of plant species, it is apparent that anthocyanin biosynthesis is controlled at the level of transcription. Here we report the transcript levels of the anthocyanin biosynthetic genes in a red-fleshed apple compared with a white-fleshed cultivar. We also describe an apple MYB transcription factor,MdMYB10, that is similar in sequence to known anthocyanin regulators in other species. We further show that this transcription factor can induce anthocyanin accumulation in both heterologous and homologous systems, generating pigmented patches in transient assays in tobacco leaves and highly pigmented apple plants following stable transformation with constitutively expressed MdMYB10. Efficient induction of anthocyanin biosynthesis in transient assays by MdMYB10 was dependent on the co-expression of two distinct bHLH proteins from apple, MdbHLH3 and MdbHLH33. The strong correlation between the expression ofMdMYB10and apple anthocyanin levels during fruit development suggests that this transcription factor is responsible for controlling anthocyanin biosynthesis in apple fruit; in the red-fleshed cultivar and in the skin of other varieties, there is an induction ofMdMYB10expression concurrent with colour formation during development. Characterization ofMdMYB10has implications for the development of new varieties through classical breeding or a biotechnological approach.
DOI:10.1105/tpc.108.059329URL [本文引用: 1]

Mutations in the genes encoding for either the biosynthetic or transcriptional regulation of the anthocyanin pathway have been linked to color phenotypes. Generally, this is a loss of function resulting in a reduction or a change in the distribution of anthocyanin. Here, we describe a rearrangement in the upstream regulatory region of the gene encoding an apple (Malus domestica) anthocyanin-regulating transcription factor, MYB10. We show that this modification is responsible for increasing the level of anthocyanin throughout the plant to produce a striking phenotype that includes red foliage and red fruit flesh. This rearrangement is a series of multiple repeats, forming a minisatellite-like structure that comprises five direct tandem repeats of a 23-bp sequence. This MYB10 rearrangement is present in all the red foliage apple varieties and species tested but in none of the white fleshed varieties. Transient assays demonstrated that the 23-bp sequence motif is a target of the MYB10 protein itself, and the number of repeat units correlates with an increase in transactivation by MYB10 protein. We show that the repeat motif is capable of binding MYB10 protein in electrophoretic mobility shift assays. Taken together, these results indicate that an allelic rearrangement in the promoter of MYB10 has generated an autoregulatory locus, and this autoregulation is sufficient to account for the increase in MYB10 transcript levels and subsequent ectopic accumulation of anthocyanins throughout the plant.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s11240-014-0609-yURL [本文引用: 3]

We have induced callus tissues from one R6/R6 homozygous genotype red-fleshed apple individual which was the hybrid offspring of Malus sieversii f .niedzwetzkyana and ‘Fuji’, and investigated the effect of auxin alone and auxin combined with cytokinin or nitrogen deficiency on anthocyanin synthesis. In callus culture, auxin alone significantly inhibited anthocyanin biosynthesis with the increase of auxin concentration. The inhibitory effect of 2,4-dichlorophenoxyacetic acid (2,4-D) on anthocyanin accumulation was about tenfold stronger than naphthalene acetic acid. Anthocyanin regulatory genes ( MdMYB10 and MdbHLH3 ) and structural genes were dramatically suppressed by 0.602mg/L 2,4-D. The inhibitory effect of auxin on anthocyanin biosynthesis was influenced by cytokinins 6-benzylaminopurine (BAP) and thidiazuron (TDZ) as well as nitrogen deficiency. Auxin and cytokinin displayed the interaction in controlling anthocyanin biosynthesis. Co-treatment of auxin and cytokinin (BAP or TDZ) significantly enhanced the cytokinin-induced increase in anthocyanin levels but too high auxin concentration strongly inhibited anthocyanin synthesis even in the presence of cytokinin. Nitrogen deficiency could reverse the inhibition of anthocyanin accumulation by auxin.
[本文引用: 1]
[本文引用: 1]
[本文引用: 4]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S1360-1385(01)01915-XURLPMID:11286899 [本文引用: 2]

Mounting evidence indicates that members of the large family of plant MYB proteins are involved in the transcriptional regulation of an array of metabolic and developmental processes. Recently, the Arabidopsis thaliana MYB, AtMYB4, was shown to regulate the accumulation of the UV-protectant compound sinapoylmalate by repressing the transcription of the gene encoding the phenylpropanoid enzyme cinnamate 4-hydroxylase. AtMYB4 is thus a key regulator of phenylpropanoid pathway gene expression, and is the first example of a MYB protein that functions as a transcriptional repressor.
DOI:10.1046/j.1365-313X.2001.01154.xURLPMID:11722774 [本文引用: 2]

Summary Fruit ripening is characterized by dramatic changes in gene expression, enzymatic activities and metabolism. Although the process of ripening has been studied extensively, we still lack valuable information on how the numerous metabolic pathways are regulated and co-ordinated. In this paper we describe the characterization of FaMYB1 , a ripening regulated strawberry gene member of the MYB family of transcription factors. Flowers of transgenic tobacco lines overexpressing FaMYB1 showed a severe reduction in pigmentation. A reduction in the level of cyanidin 3-rutinoside (an anthocyanin) and of quercetin-glycosides (flavonols) was observed. Expression of late flavonoid biosynthesis genes and their enzyme activities were aversely affected by FaMYB1 overexpression. Two-hybrid assays in yeast showed that FaMYB1 could interact with other known anthocyanin regulators, but it does not act as a transcriptional activator. Interestingly, the C-terminus of FaMYB1 contains the motif pdLNL D / E Lxi G / S . This motif is contained in a region recently proposed to be involved in the repression of transcription by AtMYB4, an Arabidopsis MYB protein. Our results suggest that FaMYB1 may play a key role in regulating the biosynthesis of anthocyanins and flavonols in strawberry. It may act to repress transcription in order to balance the levels of anthocyanin pigments produced at the latter stages of strawberry fruit maturation, and/or to regulate metabolite levels in various branches of the flavonoid biosynthetic pathway.
[本文引用: 1]
DOI:10.1126/science.1107580URLPMID:2925132 [本文引用: 1]

Abstract Brassinosteroid (BR) homeostasis and signaling are crucial for normal growth and development of plants. BR signaling through cell-surface receptor kinases and intracellular components leads to dephosphorylation and accumulation of the nuclear protein BZR1. How BR signaling regulates gene expression, however, remains unknown. Here we show that BZR1 is a transcriptional repressor that has a previously unknown DNA binding domain and binds directly to the promoters of feedback-regulated BR biosynthetic genes. Microarray analyses identified additional potential targets of BZR1 and illustrated, together with physiological studies, that BZR1 coordinates BR homeostasis and signaling by playing dual roles in regulating BR biosynthesis and downstream growth responses.
DOI:10.1007/s11103-004-6910-0URLPMID:15821875Magsci [本文引用: 1]

Chalcone synthase (CHS), chalcone flavanone isomerase (CFI), flavanone 3-hydroxylase (F3H) and flavonol synthase (FLS) catalyze successive steps in the biosynthetic pathway leading to the production of flavonols. We show that in Arabidopsis thaliana all four corresponding genes are coordinately expressed in response to light, and are spatially coexpressed in siliques, flowers and leaves. Light regulatory units (LRUs) sufficient for light responsiveness were identified in all four promoters. Each unit consists of two necessary elements, namely a MYB-recognition element (MRE) and an ACGT-containing element (ACE). C1 and Sn, a R2R3-MYB and a BHLH factor, respectively, known to control tissue specific anthocyanin biosynthesis in Z. mays , were together able to activate the AtCHS promoter. This activation of the CHS promoter required an intact MRE and a newly identified sequence designated R response element (RRE AtCHS ) containing the BHLH factor consensus binding site CANNTG. The RRE was dispensable for light responsiveness, and the ACE was not necessary for activation by C1/Sn. These data suggest that a BHLH and a R2R3-MYB factor cooperate in directing tissue-specific production of flavonoids, while an ACE-binding factor, potentially a BZIP, and a R2R3-MYB factor work together in conferring light responsiveness.
DOI:10.4161/psb.29526URLPMID:24642943 [本文引用: 1]

The diversity of pigmentation patterns observed in plants occurs due to the spatial distribution and accumulation of colored compounds, which may also be associated with structural changes to the tissue. Anthocyanins are flavonoids that provide red/purple/blue coloration to plants, often forming complex patterns such as spots, stripes, and vein-associated pigmentation, particularly in flowers. These patterns are determined by the activity of MYB-bHLH-WDR (MBW) transcription factor complexes, which activate the anthocyanin biosynthesis genes, resulting in anthocyanin pigment accumulation. Recently, we established that the MBW complex controlling anthocyanin synthesis acts within a gene regulation network that is conserved within at least the Eudicots. This network involves hierarchy, reinforcement, and feedback mechanisms that allow for stringent and responsive regulation of the anthocyanin biosynthesis genes. The gene network and mobile nature of the WDR and R3-MYB proteins provide exciting new opportunities to explore the basis of pigmentation patterning, and to investigate the evolutionary history of the MBW components in land plants.
DOI:10.1093/jxb/erq344URLPMID:21041370Magsci [本文引用: 1]

Abstract Proanthocyanidins (PAs) are agronomically important biopolymers in higher plants composed primarily of catechin and epicatechin units. The biosynthesis of these natural products is regulated by transcription factors including proteins of the R2R3MYB class. To gain insight into the genetic control of the catechin and epicatechin branches of the PA pathway in forage legumes, here the effects of the expression of FaMYB1, a flavonoid R2R3MYB repressor from strawberry, in Lotus corniculatus (birdsfoot trefoil), were tested. It was found that in leaves of T(0) transgenic lines the degree of PA inhibition correlated with the level of FaMYB1 expression. These effects were heritable in the transgene-positive plant T(1) generation and were tissue specific as the suppression of proanthocyanidin biosynthesis was most pronounced in mesophyll cells within the leaf, whereas other flavonoid and phenolic compounds were substantially unaltered. The data suggest that FaMYB1 may counter-balance the activity of the endogenous transcriptional MYB-bHLH-WD40 (MBW) complex promoting proanthocyanidin biosynthesis via the catechin and epicatechin branches and that FaMYB1 does not interfere with the expression levels of a resident R2R3MYB activator of PAs. It is proposed that in forage legumes leaf cell commitment to synthesize proanthocyanidins relies on the balance between the activity of activator and repressor MYBs operating within the MBW complex.
DOI:10.1105/tpc.15.00476URLPMID:26410301 [本文引用: 1]

Abstract Accumulation of anthocyanins and proanthocyanidins (PAs) is limited to specific cell types and developmental stages, but little is known about how antagonistically acting transcriptional regulators work together to determine temporal and spatial patterning of pigmentation at the cellular level, especially for PAs. Here, we characterize MYB2, a transcriptional repressor regulating both anthocyanin and PA biosynthesis in the model legume Medicago truncatula. MYB2 was strongly upregulated by MYB5, a major regulator of PA biosynthesis in M. truncatula and a component of MYB-basic helix loop helix-WD40 (MBW) activator complexes. Overexpression of MYB2 abolished anthocyanin and PA accumulation in M. truncatula hairy roots and Arabidopsis thaliana seeds, respectively. Anthocyanin deposition was expanded in myb2 mutant seedlings and flowers accompanied by increased anthocyanin content. PA mainly accumulated in the epidermal layer derived from the outer integument in the M. truncatula seed coat, starting from the hilum area. The area of PA accumulation and ANTHOCYANIDIN REDUCTASE expression was expanded into the seed body at the early stage of seed development in the myb2 mutant. Genetic, biochemical, and cell biological evidence suggests that MYB2 functions as part of a multidimensional regulatory network to define the temporal and spatial pattern of anthocyanin and PA accumulation linked to developmental processes. 2015 American Society of Plant Biologists. All rights reserved.
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
DOI:10.2307/3871536URLPMID:11752389 [本文引用: 2]

Aux/IAA genes are early auxin response genes that encode short-lived nuclear proteins with four conserved domains, referred to as I, II, III, and IV. Arabidopsis Aux/IAA proteins repressed transcription on auxin-responsive reporter genes in protoplast transfection assays. Mutations in domain II resulted in increased repression, whereas mutations in domains I and III partially relieved repression. Aux/IAA proteins fused to a heterologous DNA binding domain were targeted to promoters of constitutively expressed reporter genes and actively repressed transcription in an auxin-responsive and dose-dependent manner. In comparison with an unfused luciferase protein, luciferase fused to Aux/IAA proteins displayed less luciferase activity, which further decreased in the presence of auxin in transfected protoplasts. Domain II mutations increased and domain I mutations decreased luciferase activity with the fusion proteins. These results suggested that Aux/IAA proteins function as active repressors by dimerizing with auxin response factors bound to auxin response elements and that early auxin response genes are regulated by auxin-modulated stabilities of Aux/IAA proteins.
DOI:10.1105/tpc.017384URLPMID:14742873 [本文引用: 1]

Aux/IAA proteins are short-lived nuclear proteins that repress expression of primary/early auxin response genes in protoplast transfection assays. Repression is thought to result from Aux/IAA proteins dimerizing with auxin response factor (ARF) transcriptional activators that reside on auxin-responsive promoter elements, referred to as AuxREs. Most Aux/IAA proteins contain four conserved domains, designated domains I, II, III, and IV. Domain II and domains III and IV play roles in protein stability and dimerization, respectively. A clear function for domain I had not been established. Results reported here indicate that domain I in Aux/IAA proteins is an active repression domain that is transferable and dominant over activation domains. An LxLxL motif within domain I is important for conferring repression. The dominance of Aux/IAA repression domains over activation domains in ARF transcriptional activators provides a plausible explanation for the repression of auxin response genes via ARF-Aux/IAA dimerization on auxin-responsive promoters.
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s11103-008-9319-3URLPMID:18347915 [本文引用: 1]

Q-type C 2 H 2 zinc finger proteins (ZFPs) form a subfamily of transcription factors that contain a plant-specific QALGGH amino acid motif. A total of 47 expressed Q-type C 2 H 2 zinc finger genes in bread wheat ( Triticum aestivum ) (designated TaZFP) were identified from the current databases. Protein sequence analysis for the presence of ERF-associated amphiphilic repressor (EAR) motif sequences from known transcriptional repressors revealed that 26% of the TaZFP subfamily members contain an EAR motif. Quantitative RT-PCR analysis of the mRNA distribution of 44 TaZFP genes in various organs revealed that 30 genes were predominantly expressed in the roots. The majority of the TaZFP genes showed significant changes in their mRNA levels during leaf development and aging. Expression of 37 TaZFP genes in the leaves and roots responded to drought stress at least in one organ with 74% of the drought-responsive TaZFP genes being down-regulated in the drought-stressed roots. In contrast, only 6 out of the 44 TaZFP genes showed expression changes in the leaves with sucrose treatment. Expression of 50% of the drought-responsive TaZFP genes in the leaves (16 genes analysed) did not respond to ABA treatment, indicating that some TaZFP genes are involved in ABA-independent signalling pathways. These results indicate that the Q-type TaZFP subfamily is likely to have an important role in wheat roots and is enriched with members that are potentially involved in regulating cellular activities during changes of the physiological status of plant cells, as it occurs during drought stress or leaf development/aging.
