 ,, 马宗桓, 张元霞, 张娟, 卢世雄, 张志强, 赵鑫, 吴玉霞, 毛娟
,, 马宗桓, 张元霞, 张娟, 卢世雄, 张志强, 赵鑫, 吴玉霞, 毛娟 ,甘肃农业大学园艺学院,兰州 730070
,甘肃农业大学园艺学院,兰州 730070Identification and Expression Analysis of LBD Gene Family in Grape
HE HongHong ,, MA ZongHuan, ZHANG YuanXia, ZHANG Juan, LU ShiXiong, ZHANG ZhiQiang, ZHAO Xin, WU YuXia, MAO Juan
,, MA ZongHuan, ZHANG YuanXia, ZHANG Juan, LU ShiXiong, ZHANG ZhiQiang, ZHAO Xin, WU YuXia, MAO Juan ,College of Horticulture, Gansu Agricultural University, Lanzhou, 730070
,College of Horticulture, Gansu Agricultural University, Lanzhou, 730070通讯作者:
责任编辑: 李莉
收稿日期:2018-04-28接受日期:2018-06-25网络出版日期:2018-11-01
| 基金资助: |
Received:2018-04-28Accepted:2018-06-25Online:2018-11-01

摘要
关键词:
Abstract
Keywords:
PDF (3245KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
何红红, 马宗桓, 张元霞, 张娟, 卢世雄, 张志强, 赵鑫, 吴玉霞, 毛娟. 葡萄LBD基因家族的鉴定与表达分析[J]. 中国农业科学, 2018, 51(21): 4102-4118 doi:10.3864/j.issn.0578-1752.2018.21.009
HE HongHong, MA ZongHuan, ZHANG YuanXia, ZHANG Juan, LU ShiXiong, ZHANG ZhiQiang, ZHAO Xin, WU YuXia, MAO Juan.
0 引言
【研究意义】葡萄(Vitis vinifera)是一个重要的经济作物,在全世界具有很高的经济价值,种植区域广泛。然而中国西北地区存在许多不利于葡萄生长和结果的因素(如寒冷、干旱、盐碱等)[1],有针对性地进行非生物胁迫基因的研究和改造,可以抵御不利于葡萄生长的自然环境,从而提高和改善葡萄的产量和品质。【前人研究进展】转录因子(transcription factors,TF)基因家族在植物的一些生物合成过程(如生长、发育、信号转导和对环境压力胁迫反应)中起着重要的作用。利用生物信息学研究葡萄基因家族的功能已在葡萄中大量报道,如MIKC、ARF、WRKY、SnRK2、CIPK、OVATE和bHLH等基因家族[2,3,4,5,6,7,8]。侧生器官边界域(lateral organ boundaries domain,LBD)基因也称作AS2/LOB基因,是一类植物中所特有的转录因子基因家族[9],该基因家族参与高等植物侧生器官形态建成,特别是在侧生器官与顶端分生组织之间边界的形态建成起到关键作用[10,11,12,13,14,15]。在调控植物生长发育进程中,尤其在响应逆境胁迫中具有积极作用[16,17,18]。LBD基因包含3个特定的保守结构域,分别是类似锌指的CX2CX6CX3C结构域、类似亮氨酸拉链(LX6LX3LX6L)的“螺旋卷曲”(coiled coil)二级结构和甘氨酸-丙氨酸-丝氨酸(GAS)区域[18,19]。目前,LBD基因已经在许多植物中被鉴定,如拟南芥43个[10]、玉米44个[20]、毛果杨57个[21]、大麦24个[22]、辣椒45个[18]、番茄46个[16]和蒺藜苜蓿56个[23]。在拟南芥中,AtLOB(AtASL4)可调控拟南芥叶片早期的发育,AtLBD6(AtAS2)可抑制近轴面区域的细胞增殖[24],使叶片发育成两面对称的平展叶,AtLBD受生长素和细胞分裂素诱导并调控拟南芥的激素响应[25,26,27,28,29,30],AtLBD37—AtLBD39通过响应硝酸盐的合成,影响花青苷形成和氮素代谢[31,32]。在柑桔中,LBD1同源物可能通过调控参与扩增、生物合成和降解的细胞壁代谢基因的表达进而调控对细菌溃疡病的耐药性[33,34]。多种植物关于LBD基因芯片的表达分析表明该基因家族会响应不同的生物或非生物胁迫。番茄中,LBD基因在调控非生物胁迫时发挥重要作用[16]。苎麻中,16个BnLBDs能够增加植株在高温环境下的耐受力[35]。苜蓿中,幼苗期进行干旱处理,有一部分LBD基因在根的形态建成过程中变化差异较大[23]。辣椒中,LBD基因中ClassⅠ在响应高温胁迫时明显低于ClassⅡ,且在不同时长的高温胁迫下其表达量存在差异[17]。大豆中,GmLBD12受干旱、低温、盐碱和各种激素的强烈诱导[36]。【本研究切入点】目前,葡萄LBD基因家族虽有报道[37,38],但其主要是对基因芯片表达谱数据的分析研究,定量分析研究较少[38],尤其是对逆境胁迫下表达的分析鲜见报道。【拟解决的关键问题】本研究利用生物信息学方法探究VvLBD基因家族的分布、基因结构与表达分析,通过芯片表达谱进行基因在非生物胁迫下的功能预测,分析VvLBD基因家族在试管苗葡萄叶片中的表达情况,为VvLBD基因家族响应逆境胁迫时的功能奠定理论基础。1 材料与方法
1.1 试验材料
试验于2016—2017年进行。荧光定量所用试验材料为‘红地球’葡萄(Vitis vinifera ‘Red Globe’)试管苗,保存于甘肃农业大学果树生理与生物技术实验室。将试管苗单芽茎段接于固体GS培养基上,分别置于LED白光下培养35 d后,分别用50 μmol·L-1 ABA、400 mmol·L-1 NaCl以及10%PEG处理24 h,每个处理设置3组重复,等体积蒸馏水处理为对照。基因芯片表达谱所用试验数据取自于GEO数据库中的GSE31594和GSE31662,其试验材料为‘赤霞珠’葡萄(Cabernet Sauvignon)。GSE31662数据是‘赤霞珠’软化的绿色浆果在培养皿中离体培养,在培养皿中加入0.3 mol·L-1蔗糖+脱落酸(ABA)作为处理,未处理的作为对照(CK1),每个处理设置3个重复,在处理后的3和10 d分别进行取样。GSE31594数据是‘赤霞珠’生长在水培滴灌系统中,用120 mmol·L-1(10﹕1 NaCl﹕CaCl)、聚乙二醇(PEG)和低温(5℃)分别进行处理,未处理的作为对照(CK2),每个处理设置3个重复。分别在1、4、8和24 h采集叶片,其中24 h不用低温处理。
1.2 葡萄LBD基因家族成员鉴定
根据文献获得拟南芥(Arabidopsis thaliana)的LBD基因ID[18]。并从NCBI网站(https://www.ncbi.nlm. nih.gov/nuccore/NM_113577.4)获得每一条基因对应的CDS与Full-length的序列。将获得的CDS序列提交到葡萄基因库中,同源搜索VvLBD基因,获取已知序列的基因号、CDS长度、氨基酸数目、cDNA和Full-length,保留长度大于1 000 bp的序列。并运用DANMAN进行多重比对,依据LBD基因家族所具有的特定结构域,手动去除不含有LBD特定结构域或者结构域不完整的序列。利用ExPASy(http://web.expasy.org/protparam/)在线软件对葡萄LBD蛋白的分子量和等电点进行预测。从葡萄基因组网站获取染色体基本信息,选取LBD基因的位置信息,利用MapInspect程序绘制其染色体物理位置图。1.3 葡萄LBD基因家族进化、基因结构、motif和亚细胞定位分析
利用Clustal X对葡萄和TAIR(http://www. arabidopsis.org/)中拟南芥LBD蛋白进行多序列比对,使用MEGA5.0构建LBD家族系统发育树[35],Bootstrap值设置为1 000。在线软件GSDS2.0(http://gsds.cbi.pku.edu.cn/)用于分析基因结构[39]。用DNAMAN进行结构域序列多重比对,采用(http://meme-suite.org/tools/meme)进行motif序列分析;采用WoLF PSORT进行亚细胞定位(http://www. genscript.com/wolf-psort.html)。1.4 葡萄LBD基因家族基因芯片表达模式分析
从NCBI的GEO数据库中下载葡萄RNASeq数据,登录号为GSE31662和GSE31594,用VvLBD核酸序列为探针检索出序列相同的Affymetrix GeneChip 16K基因ID,然后提取VvLBD在各种非生物胁迫下的RNASeq数据,通过Excel对数据进行log2转换,最后用HemI制作表达热图(heatmap)。1.5 实时荧光定量PCR
对VvLBD基因家族的CDS序列进行引物设计(表1)。cDNA合成用Prime Script RT Reagent Kit(Perfect Real Time)试剂盒(TaKaRa)。反转录产物-20℃保存备用。实时荧光定量PCR(real-time fluorescent quantitative polymerase chain reaction, qRT-PCR)应用Bio-Rad iCycler iQ实时定量PCR仪进行扩增,以葡萄UBQ为内参[40],扩增体系为2 μL cDNA、上下游引物各1 μL、SYBR 10 μL反应MIX和6 μL ddH2O。反应程序为95℃ 30 s;95℃ 5 s,60℃ 34 s,95℃ 15 s,60℃ 60 s,95℃ 15 s,共40个循环。3次重复,数据用Excel软件分析。Table 1
表1
表1VvLBD基因家族表达分析的实时荧光定量引物
Table 1
| 基因Gene | 上游引物Forward primer (5′-3′) | 下游引物Reverse primer (5′-3′) |
|---|---|---|
| VvLBD1 | CAGCAGGAGACTGACAGCATGTTC | GATAGAAGCCAAGAGTGAGTTCCAACC |
| VvLBD2 | GCCGTGGATGCAGTGCTCAG | GCTTGGACTTAGAAGAACGCTTGAATG |
| VvLBD3 | TGGCCTTCATCTCCACCGTACC | CCGCTCCGAACACAGGATTCAC |
| VvLBD4 | GAGCTAGTGAACATGCAATGCCAAC | GCTGTCATCGAGGAAGAAGGAGTTAC |
| VvLBD5 | AGAGGAGCCTGCCTTACCAACTC | AGCTGAGGTAGTGTTGAAGGTTGTTG |
| VvLBD6 | TCCTCCACCTCCTCTACCTCTACC | CCTGTTGTGCTTGAAGGCAGTATTG |
| VvLBD7 | GCTGCGGTTACCATTGCTTATGAAG | TGTAGATTAACAACCTGCTGCTGGAG |
| VvLBD8 | GCCTTGCAGCAGCAGGTAGC | TGCGGCGATACTGAGTTCATGTAAC |
| VvLBD9 | TTCGCTAATGTCCACAAGGTCTTCG | ACGGCATCTTCTCGATGTGCAAC |
| VvLBD10 | TTCGTTAATGTCCACAAGGTCTTCGG | AATCAGACCAACACAGCCATAGACG |
| VvLBD11 | GTCCTTCATCTCTGCTGTTCCTGAG | TCACGGTCCGGCCACACG |
| VvLBD12 | AGCAGCAGCAGCAGCATCAG | TCTCTTCCTCGTCTTCGTCCTCATC |
| VvLBD13 | AGTTCTCCATCGTTCACAAGGTCTTC | GCAGGTATGATATGGCTCCGACAC |
| VvLBD14 | CAGCACAGCCACCAGCACAG | GGAGGAAGAGGAGTTAGGAGGAAGAG |
| VvLBD15 | CTCAACCAACAGGCTCTGCTACTATC | AAGGAGGTGGCTGTTCATTCATTAGG |
| VvLBD16 | AGCAGCAGCAGCCACAATATCAG | TCCATGACAAGATTGCCATCCTCAAG |
| VvLBD17 | GATGCCTTCTCCTTCTCTTCCAACTG | GTTCCTGCAACGCCTTGACAATG |
| VvLBD18 | CTTGCAGCTCTGAGGATGGAATACG | AGCTAGAATACAACTCTCATCGCATCG |
| VvLBD19 | ATTCGTGATCCAGTCTATGGTTGTGC | ATCTGTCCAGGAAGAAGCTTGCATTAG |
| VvLBD20 | GCCAATGCCACCGTCTTCCTC | CACAAGCCTCGTAGAGAAGAGAACTG |
| VvLBD21 | GCCACCAATGCTGACCTGATCC | CTCCACCTCCACCTCCATGACTC |
| VvLBD22 | GATGGTAACGCCGCCTTCACTC | ACCTCTGGAGCCACCGAACTG |
| VvLBD23 | CCTGTCTATGGCTGTGTTGCTCATG | GCGGTGCCAGAGAAGGAATGC |
| VvLBD24 | GTCCGTCAGCGATGGCACATG | ATAAGGAATGACCGACGATGAAGAAGG |
| VvLBD25 | CCTTCTTCCAACTTCAACAACTCCTTG | GGTCGGATGGAGAGCGGTAGAG |
| VvLBD26 | TGCGGATGAGTTGTAATGGCTGTC | GCGAGGAAGACGGTGGCATTG |
| VvLBD27 | TCGTTACGCCTGCAATGAGATGTC | GCTTCCTCCTCCTTCATTACCAATCC |
| VvLBD28 | GCAGCGGTGGACACAGTTCTC | CAGCGAGTATGCGTCACGTAGC |
| VvLBD29 | TAGGAGAAGATGTGCTCAGGACTGTG | TGCTTCATACACCATGCTACTCACTG |
| VvLBD30 | CCAACTCCGCCAACTCCAGATG | CTGTGGATGGTGGTGGTGATTGTG |
| UBI | GCTCGCTGTTTTGCAGTTCTAC | AACATAGGTGAGGCCGCACTT |
新窗口打开|下载CSV
1.6 试验数据统计与分析
采用Excel进行数据统计与分析,采用单因素(One-way ANOVO)的Duncan’s法进行显著性差异分析,显著性水平为P<0.05,并作图。2 结果
2.1 葡萄LBD基因家族的蛋白质理化性质分析
经同源搜索获得30个VvLBD,分别命名为VvLBD1—VvLBD30。通过对VvLBD染色体进行定位分析(图1),发现该家族30个基因分布于葡萄11条染色体上(Chr.01、Chr.03、Chr.06、Chr.07、Chr.08、Chr.13、Chr.14、Chr.15、Chr.16、Chr.17和Chr.18),其中第13染色体上有7个基因(VvLBD13、VvLBD14、VvLBD15、VvLBD16、VvLBD17、VvLBD18和VvLBD19)。在第3、第8和第18染色体上分别含有1个基因,有2个基因分布在未知染色体上。VvLBD1蛋白序列最长,包含386个氨基酸残基,VvLBD13蛋白序列最短,含127个氨基酸残基。分子量为14.19636—43.30087 kD;等电点介于4.77—9.28。外显子数介于1—4,VvLBD12的外显子数是4,VvLBD30的外显子数是1,外显子数为2的有18个基因,外显子数为3的有10个基因(表2)。说明该转录因子基因家族是一个相对保守的基因家族。图1
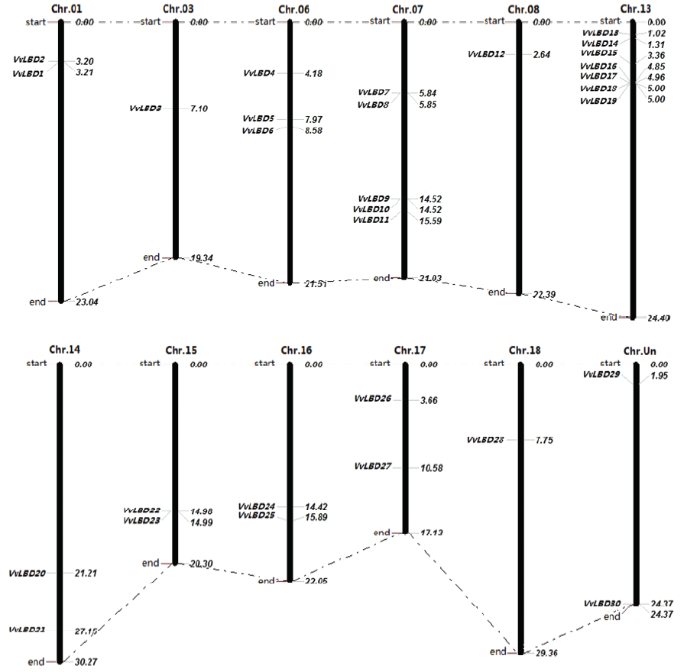 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1葡萄LBD基因家族在染色体上的位置
Fig. 1The chromosome location of the LBD gene family in Vitis vinifera
Table 2
表2
表2葡萄中LBD转录因子家族信息
Table 2
| 基因 Gene | 基因登录号 Gene accession No. | 染色体定位 Chromosome location | 长度 Length of CDS (bp) | 基因全长 Full length genomic (bp) | 分子量 Molecular weight (kD) | 等电点 pI | 氨基酸 Amino acid | 外显子 Exon | cDNA全长 FL-cDNA (bp) | 拟南芥同源基因Arabidopsis homologous gene | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 登录号 Accession No. | 名称 Gene name | E值 E-value | ||||||||||
| VvLBD1 | GSVIVT01011895001 | Chr.01:3210000..3211373 | 1158 | 1374 | 43.30087 | 8.96 | 386 | 3 | 1158 | AT1G68510 | LBD42 | 7e-05 |
| VvLBD2 | GSVIVT01011896001 | Chr.01:3202459..3205665 | 726 | 1076 | 26.97486 | 9.28 | 242 | 2 | 998 | AT1G68510 | LBD42 | 7e-05 |
| VvLBD3 | GSVIVT01037853001 | Chr.03:7098160..7099838 | 615 | 1679 | 22.87014 | 6.73 | 205 | 3 | 714 | AT4G37540 | LBD39 | 1e-09 |
| VvLBD4 | GSVIVT01025128001 | Chr.06:4180823..4182134 | 609 | 1312 | 22.02887 | 4.77 | 202 | 2 | 953 | AT2G28500 | LBD11 | 8e-14 |
| VvLBD5 | GSVIVT01024662001 | Chr.06:7971774..7972539 | 492 | 766 | 18.36397 | 6.40 | 164 | 2 | 644 | AT1G31320 | LBD4 | 0.003 |
| VvLBD6 | GSVIVT01024592001 | Chr.06:8584134..8585865 | 690 | 1735 | 24.92138 | 8.58 | 229 | 3 | 1529 | AT2G30340 | LBD13 | 6e-18 |
| VvLBD7 | GSVIVT01028294001 | Chr.07:5837981..5838834 | 699 | 854 | 25.72175 | 5.87 | 232 | 2 | 750 | AT2G42430 | LBD16 | 0.018 |
| VvLBD8 | GSVIVT01028295001 | Chr.07:5846731..5847540 | 588 | 810 | 21.46149 | 7.54 | 195 | 2 | 588 | AT2G42430 | LBD16 | 0.018 |
| VvLBD9 | GSVIVT01003547001 | Chr.07:14517830..14519654 | 426 | 1825 | 15.69194 | 8.60 | 142 | 2 | 513 | AT5G66870 | LBD36 | 2e-09 |
| VvLBD10 | GSVIVT01003548001 | Chr.07:14524923..14526703 | 663 | 1781 | 23.96578 | 9.16 | 220 | 2 | 765 | AT5G66870 | LBD36 | 2e-09 |
| VvLBD11 | GSVIVT01000141001 | Chr.07:15589316..15590604 | 687 | 1289 | 24.96159 | 8.30 | 229 | 2 | 1167 | AT4G37540 | LBD39 | 0.005 |
| VvLBD12 | GSVIVT01029979001 | Chr.08:2636746..2653598 | 777 | 16853 | 27.99185 | 5.85 | 259 | 4 | 777 | AT3G03760 | LBD20 | 7e-27 |
| VvLBD13 | GSVIVT01032752001 | Chr.13:1022036..1023024 | 381 | 989 | 14.19636 | 8.80 | 127 | 3 | 417 | AT2G30130 | LBD12 | 4e-09 |
| VvLBD14 | GSVIVT01032714001 | Chr.13:1310001..1311298 | 675 | 1298 | 24.53103 | 8.99 | 225 | 3 | 956 | AT2G30340 | LBD13 | 2e-11 |
| VvLBD15 | GSVIVT01016500001 | Chr.13:3355781..3356736 | 660 | 955 | 24.29509 | 5.08 | 219 | 2 | 849 | AT2G42430 | LBD16 | 8e-08 |
| VvLBD16 | GSVIVT01016335001 | Chr.13:4853462..4854800 | 594 | 1339 | 21.75785 | 5.67 | 197 | 2 | 677 | AT2G28500 | LBD11 | 3e-16 |
| VvLBD17 | GSVIVT01016332001 | Chr.13:4961089..4962007 | 498 | 919 | 18.35293 | 8.21 | 166 | 2 | 498 | AT2G28500 | LBD11 | 3e-16 |
| VvLBD18 | GSVIVT01016329001 | Chr.13:4999327..4999957 | 444 | 631 | 16.23288 | 8.72 | 148 | 2 | 505 | AT3G11090 | LBD21 | 8e-04 |
| VvLBD19 | GSVIVT01016328001 | Chr.13:5001203..5002266 | 420 | 1064 | 15.73712 | 8.50 | 140 | 2 | 462 | AT3G11090 | LBD21 | 8e-04 |
| VvLBD20 | GSVIVT01031035001 | Chr.14:21210990..21212344 | 564 | 1355 | 20.29311 | 8.98 | 187 | 3 | 1124 | AT1G68510 | LBD42 | 7e-05 |
| VvLBD21 | GSVIVT01032415001 | Chr.14:27152774..27157752 | 606 | 4979 | 22.07197 | 8.52 | 201 | 3 | 720 | AT3G27650 | LBD25 | 0.003 |
| VvLBD22 | GSVIVT01027621001 | Chr.15:14983162..14985661 | 795 | 2500 | 28.19431 | 8.12 | 264 | 3 | 924 | AT2G45420 | LBD18 | 2e-21 |
| VvLBD23 | GSVIVT01027620001 | Chr.15:14992324..14994786 | 585 | 2463 | 21.17114 | 6.50 | 195 | 2 | 585 | AT2G42430 | LBD16 | 1.1 |
| VvLBD24 | GSVIVT01018486001 | Chr.16:14419741..14420915 | 672 | 1175 | 24.58742 | 7.65 | 223 | 2 | 672 | AT5G66870 | LBD36 | 3e-11 |
| VvLBD25 | GSVIVT01010625001 | Chr.16:15894465..15895136 | 462 | 672 | 17.36371 | 9.08 | 154 | 2 | 462 | AT3G26620 | LBD23 | 7e-19 |
| VvLBD26 | GSVIVT01008284001 | Chr.17:3660239..3661508 | 684 | 1270 | 24.77241 | 8.05 | 228 | 3 | 1035 | AT3G02550 | LBD41 | 0.005 |
| VvLBD27 | GSVIVT01007677001 | Chr.17:10578569..10581053 | 468 | 2485 | 16.93792 | 6.81 | 156 | 3 | 670 | AT5G63090 | LOB | 3e-22 |
| VvLBD28 | GSVIVT01009360001 | Chr.18:7746214..7747271 | 669 | 1058 | 24.10543 | 8.54 | 222 | 2 | 939 | AT3G49940 | LBD38 | 3e-07 |
| VvLBD29 | GSVIVT01013631001 | Chr.Un:1949856..1952420 | 528 | 2565 | 19.30889 | 7.68 | 176 | 2 | 1646 | AT1G16530 | LBD3 | 2e-10 |
| VvLBD30 | GSVIVT01006269001 | Chr.Un:24370759..24373006 | 651 | 2248 | 23.99094 | 8.25 | 217 | 1 | 1243 | AT1G65620 | LBD6 | 3e-47 |
新窗口打开|下载CSV
2.2 葡萄LBD基因家族多序列比对分析
通过对30个VvLBD蛋白序列进行多序列比对,发现VvLBD蛋白序列的N端都含有1个由15个氨基酸组成的保守CX2CX6CX3C序列(图2),预测其可能直接参与下游基因顺式元件的结合,说明30个基因家族功能域完整。ClassⅠ类蛋白结构域C端含有赖氨酸组成的类似亮氨酸拉链LX6LX3LX6L螺旋卷曲结构(图2)。大部分VvLBD蛋白都具有GAS保守结构,VvLBD4—VvLBD19、VvLBD21—VvLBD25、VvLBD27、VvLBD29—VvLBD30的序列是GAS结构,GAS保守结构区域中,VvLBD1、VvLBD2、VvLBD3、VvLBD11、VvLBD20和VvLBD26的序列是GRA,VvLBD28的序列是GRS结构,进一步说明该基因家族序列高度保守,在葡萄体内可能行使相似的功能。图2
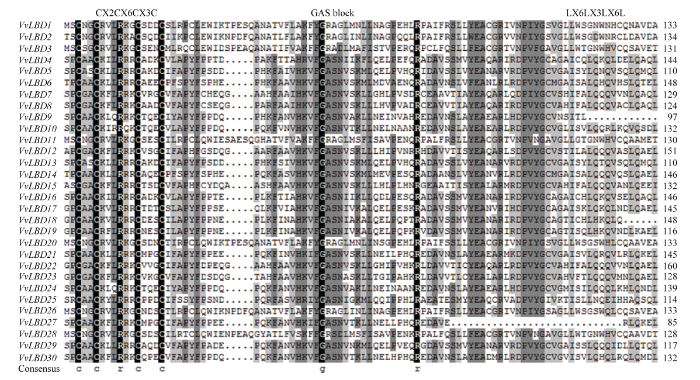 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2葡萄LBD家族保守结构域比对
Fig. 2Alignment of conserved domains of LBD family in Vitis vinifera
2.3 葡萄LBD基因家族基因结构和进化分析
通过对VvLBD基因家族基因进行结构分析,结果表明,30个VvLBD基因分为两类,分为为ClassⅠ和ClassⅡ,分别含有23个和7个LBD蛋白。在葡萄LBD基因家族中有10个旁系同源系基因,其中有4对旁系同源系基因VvLBD4/VvLBD16、VvLBD5/VvLBD13、VvLBD6/VvLBD14、VvLBD18/VvLBD19的自展值高于90。如图3所示,基因结构分析发现葡萄LBD基因结构内含子数均小于3个,外显子数约为4个,但VvLBD30不含内含子。有18个基因(VvLBD2、VvLBD4—VvLBD5、VvLBD7—VvLBD11、VvLBD15—VvLBD19、VvLBD23—VvLBD25、VvLBD28—VvLBD29)仅含1个内含子,有10个基因(VvLBD1、VvLBD3、VvLBD6、VvLBD13—VvLBD14、VvLBD20—VvLBD22、VvLBD26—VvLBD27)含有2个内含子,只有VvLBD12含有3个内含子。聚类关系较近的基因具有相似的基因结构,如7个ClassⅡ基因中的VvLBD2、VvLBD11和VvLBD28含有1个内含子,VvLBD1、VvLBD3、VvLBD20和VvLBD26含有2个内含子,且内含子结构高度相似。在10个旁系同源系中自展值大于90的4对基因结构也高度相似,分别为VvLBD4/VvLBD16、VvLBD5/VvLBD13、VvLBD6/VvLBD14和VvLBD18/VvLBD19。图3
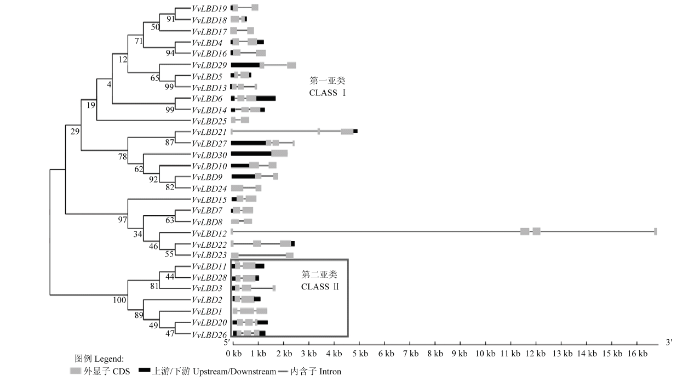 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3葡萄LBD家族进化树及基因结构
Fig. 3The phylogenetic tree and gene structure of LBD family in Vitis vinifera
为进一步了解VvLBD基因家族的进化特性,利用拟南芥LBD基因家族成员与葡萄LBD基因家族成员构建系统进化树(图4)。将葡萄ClassⅠ分为Ⅰa、Ⅰb、Ⅰc、Ⅰd和Ⅰe等5个亚类,分别包含6、2、6、4和5个VvLBD家族成员,ClassⅡ分为Ⅱa和Ⅱb 2个亚类,分别包含3和4个VvLBD家族成员,说明VvLBD基因家族成员之间即高度保守,又存在一定的差异,这与基因家族的功能有相似之处。
图4
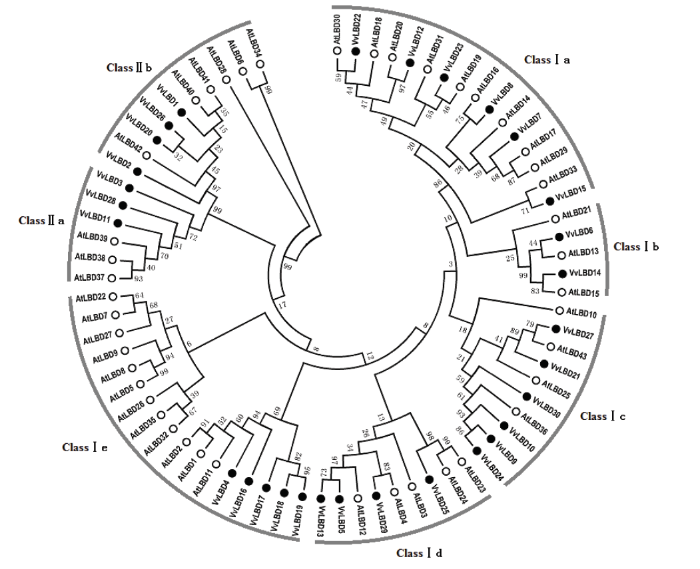 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4葡萄(●)与拟南芥(○)LBD转录因子的系统进化树分析
Fig. 4The unrooted phylogenetic tree of LBD transcription factors in Vitis vinifera(●) and Arabidopsis(○)
2.4 葡萄LBD基因家族motif序列分析
VvLBD基因家族中最主要的motif有3个(图5-A):C端均含有一个保守的半胱氨酸残基组成的锌指结构CX2CX6CX3C(motif 2)基序,推测其可能直接与基因顺式作用元件相接合;一个完全保守的GAS(Gly-Ala-Ser)保守结构(motif 1);N端为一个类似亮氨酸拉链的螺旋卷曲结构亮氨酸拉链式基序LX6LX3LX6L(motif 3),可能参与转录因子的二聚化过程。图5-B为MEME搜索得到的8个模体,motif 1和motif 2在所有成员中均有出现,motif 3只出现在大多数ClassⅠ成员当中,motif 4分布在ClassⅡ的所有成员中,motif 5出现在ClassⅡ的4个基因中,motif 6出现在ClassⅡ的3个基因中,motif 7主要出现在ClassⅠ中的部分成员中,motif 8只出现在ClassⅠ中的第四对旁系同源系的2个基因中。虽然不同亚族中包含motif种类和motif的排列顺序不同,但是同一亚族中,均包含相同的motif排列,这可能与该基因家族功能的表达方面有很大的关系。图5
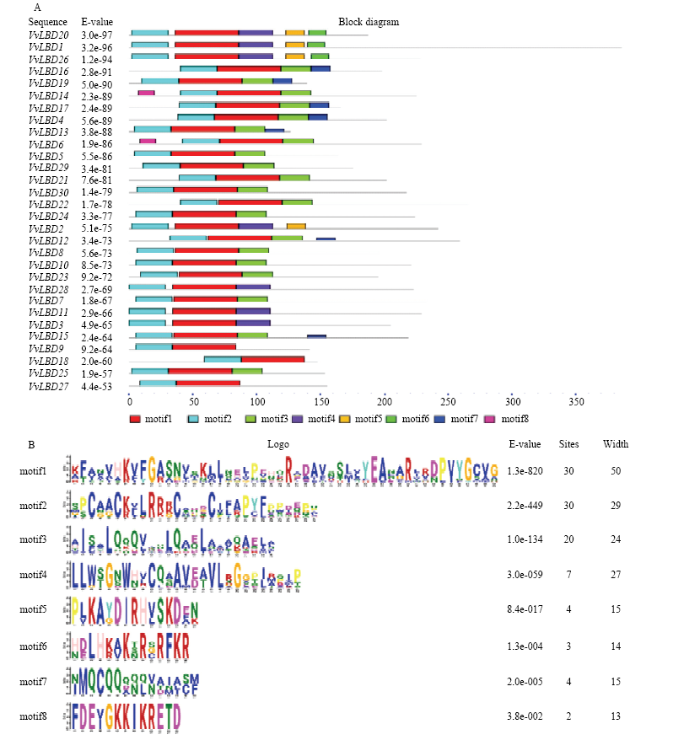 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5葡萄LBD基因家族的序列分析
A:葡萄LBD基因家族motif分析;B:MEME预测的8个保守位点LOGO图
Fig. 5LBD gene family sequence analysis in Vitis vinifera
A: LBD gene family motif analysis in Vitis vinifera; B: LOGO of 8 conserved motif of LOB domain
2.5 葡萄LBD基因家族亚细胞定位和蛋白质二级结构分析
VvLBD基因家族主要集中在叶绿体、线粒体和细胞核中进行表达(表3),说明该基因家族主要存在于光合、呼吸代谢较强的器官中。其中VvLBD基因家族在细胞质中表达的基因只有VvLBD1、VvLBD2、VvLBD5、VvLBD6、VvLBD12、VvLBD15、VvLBD16和VvLBD22;在叶绿体中除了VvLBD1和VvLBD21没有表达外,其余基因均有表达;在细胞核中除了VvLBD16、VvLBD19、VvLBD20、VvLBD26中没有表达,其余基因均有表达;在线粒体中除了VvLBD6、VvLBD7、VvLBD12、VvLBD13、VvLBD17、VvLBD19、VvLBD21、VvLBD22、VvLBD27和VvLBD29这10个基因没有表达外,其他基因均有表达;在过氧化物酶体、液泡、内质网、高尔基体和细胞基中都没有表达;在细胞骨架中表达的基因只有VvLBD12,说明其可能参与了细胞壁的形成。Table 3
表3
表3葡LBD基因亚细胞定位预测
Table 3
| 名称 Name | 细胞质 Cytoplasm | 叶绿体 Chloroplast | 细胞核 Nucleus | 线粒体 Mitochondria | 过氧化物 酶体 Peroxisome | 细胞骨架 Cytoskeleton | 液泡 Vacuole | 内质网 Endoplasmic network | 细胞膜 Plasma membrane | 高尔基体 Golgi apparatus | 细胞基 Extracellular matrix | 细胞核和细胞质 Nuclear and cytoplasmic | 线粒体和细胞质 Mitochondrial and cytoplasmic | 细胞核和细胞膜 Nuclear and plasma membrane |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| VvLBD1 | 1.5 | 0 | 6.5 | 4.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.5 | 3 | 0 |
| VvLBD2 | 2 | 3 | 3 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD3 | 0 | 2 | 8 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD4 | 0 | 5 | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD5 | 1 | 3 | 6 | 1.5 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1.5 | 0 |
| VvLBD6 | 6 | 1 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD7 | 0 | 5 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD8 | 0 | 8 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD9 | 0 | 6 | 4.5 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| VvLBD10 | 0 | 7 | 4 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| VvLBD11 | 0 | 3 | 9 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD12 | 3 | 5 | 2 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 |
| VvLBD13 | 0 | 12 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD14 | 0 | 3 | 6 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4.5 |
| VvLBD15 | 1 | 2 | 5 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD16 | 1 | 8 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD17 | 0 | 11 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD18 | 0 | 4 | 5.5 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.5 | 0 | 0 |
| VvLBD19 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD20 | 0 | 2 | 0 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD21 | 0 | 0 | 13 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD22 | 4 | 3 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD23 | 0 | 10 | 2 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD24 | 0 | 1 | 4.5 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 0 |
| VvLBD25 | 0 | 2 | 5.5 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3.5 | 0 | 0 |
| VvLBD26 | 0 | 11 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD27 | 0 | 4 | 8.5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 0 |
| VvLBD28 | 0 | 5 | 6.5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 |
| VvLBD29 | 0 | 11 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| VvLBD30 | 0 | 3 | 9 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
新窗口打开|下载CSV
VvLBD基因家族编码蛋白质的二级结构主要有α-螺旋、β-转角与不规则卷曲。VvLBD基因家族编码的30个蛋白主要是α-螺旋(20.66%—56.36%)和不规则卷曲(26.77%—53.72%),β-转角较少(0— 12.96%)(表4)。
Table 4
表4
表4葡萄LBD蛋白二级结构分析
Table 4
| 名称Name | α-螺旋Alpha helix (%) | β-转角Beta turn (%) | 不规则卷曲Random coil (%) |
|---|---|---|---|
| VvLBD1 | 39.38 | 6.74 | 36.01 |
| VvLBD2 | 20.66 | 6.20 | 53.72 |
| VvLBD3 | 37.56 | 8.78 | 39.02 |
| VvLBD4 | 45.05 | 2.97 | 41.58 |
| VvLBD5 | 41.46 | 1.83 | 39.63 |
| VvLBD6 | 49.34 | 3.49 | 37.12 |
| VvLBD7 | 48.28 | 6.90 | 38.79 |
| VvLBD8 | 39.49 | 7.69 | 38.97 |
| VvLBD9 | 33.80 | 8.45 | 30.28 |
| VvLBD10 | 56.36 | 4.55 | 27.37 |
| VvLBD11 | 32.89 | 11.84 | 40.35 |
| VvLBD12 | 33.98 | 3.47 | 44.02 |
| VvLBD13 | 52.76 | 0.00 | 26.77 |
| VvLBD14 | 43.11 | 4.44 | 44.89 |
| VvLBD15 | 47.95 | 6.85 | 33.33 |
| VvLBD16 | 29.95 | 5.58 | 41.62 |
| VvLBD17 | 39.76 | 2.41 | 42.77 |
| VvLBD18 | 37.84 | 6.76 | 41.22 |
| VvLBD19 | 45.71 | 6.43 | 37.14 |
| VvLBD20 | 27.81 | 11.23 | 38.50 |
| VvLBD21 | 31.34 | 9.95 | 42.79 |
| VvLBD22 | 33.71 | 6.82 | 49.24 |
| VvLBD23 | 51.79 | 7.69 | 33.33 |
| VvLBD24 | 39.91 | 8.97 | 32.29 |
| VvLBD25 | 51.30 | 6.49 | 28.57 |
| VvLBD26 | 32.46 | 10.96 | 38.60 |
| VvLBD27 | 37.82 | 7.69 | 45.51 |
| VvLBD28 | 41.11 | 12.96 | 33.33 |
| VvLBD29 | 42.61 | 4.55 | 42.05 |
| VvLBD30 | 49.77 | 5.99 | 36.41 |
新窗口打开|下载CSV
2.6 葡萄LBD基因家族表达模式分析
从葡萄基因组数据库中克隆出30个LBD基因,对其中29条VvLBD(VvLBD17未在芯片数据检索到)基因进行芯片表达模式的分析(图6),在ABA胁迫下,VvLBD9和VvLBD26的表达量明显上调,而VvLBD1、VvLBD4、VvLBD5、VvLBD7、VvLBD21、VvLBD25的表达量明显下调;在盐、PEG、低温胁迫下,VvLBD2、VvLBD3、VvLBD6、VvLBD15、VvLBD16、VvLBD18、VvLBD19、VvLBD22、VvLBD23、VvLBD28和VvLBD29的表达量明显上调,VvLBD4、VvLBD21和VvLBD25的表达量明显下调。此外,逆境胁迫时间的长短对VvLBD基因家族的表达也存在不同的影响,如VvLBD1在ABA胁迫4 h的表达和对照(CK1)无差异,而在ABA处理8 h的表达量明显比对照(CK1)下调趋势;VvLBD27在盐和PEG处理24 h后表达量明显高于对照(CK2)。VvLBD23在1 h盐胁迫下的表达量和对照(CK2)相同,而在4、5和6 h盐胁迫下的表达量明显比对照(CK2)有所下调,说明该基因家族成员响应不同逆境胁迫和不同胁迫时间的反应机制之间存在差异。图6
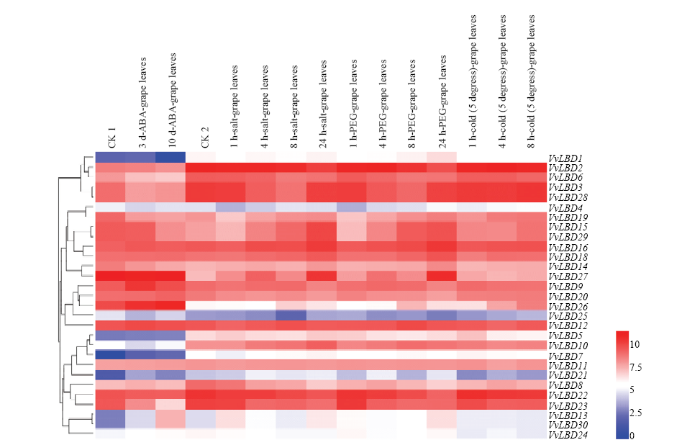 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6葡萄LBD基因表达谱
图中用深蓝、浅蓝、无色、浅红、深红五色代表基因表达水平。蓝色表示基因表达弱,红色表示基因表达强
Fig. 6Expression profile of LBD gene in Vitis vinifera
Dark blue, light blue, colorless, light red and dark red are used to represent gene expression levels. Blue indicates weak gene expression, red indicates strong gene expression
2.7 葡萄LBD基因的实时荧光定量分析
VvLBD基因家族受NaCl、ABA和PEG的诱导,各处理间表达量具有较显著的差异,在400 mmol·L-1 NaCl处理24 h条件下,VvLBD基因家族明显上调表达(图7)。VvLBD1—VvLBD3、VvLBD5—VvLBD6、VvLBD9—VvLBD10、VvLBD13—VvLBD14、VvLBD20—VvLBD22、VvLBD24、VvLBD 26—VvLBD30在50 μmol·L-1 ABA、400 mmol·L-1 NaCl和10% PEG处理24 h后,均呈下调表达。VvLBD12和VvLBD19在ABA、NaCl和PEG处理后,均呈上调表达。VvLBD19在PEG处理后,呈上调表达。VvLBD8、VvLBD11—VvLBD12、VvLBD15—VvLBD17在NaCl处理后,呈现出明显上调表达,而在ABA和PEG处理后,呈现出明显下调表达。VvLBD12在ABA、NaCl和PEG处理24 h后,均呈上调表达,且在NaCl处理后,表达量是对照的8倍;在ABA处理后,表达量是对照的5倍;在PEG处理后,表达量是对照的3倍。结果表明,在VvLBD基因家族成员中,部分基因受400 mmol·L-1 NaCl、50 μmol·L-1 ABA、10%PEG的强烈诱导;部分基因对400 mmol·L-1 NaCl、50 μmol·L-1 ABA和10%PEG的胁迫并不敏感。图7
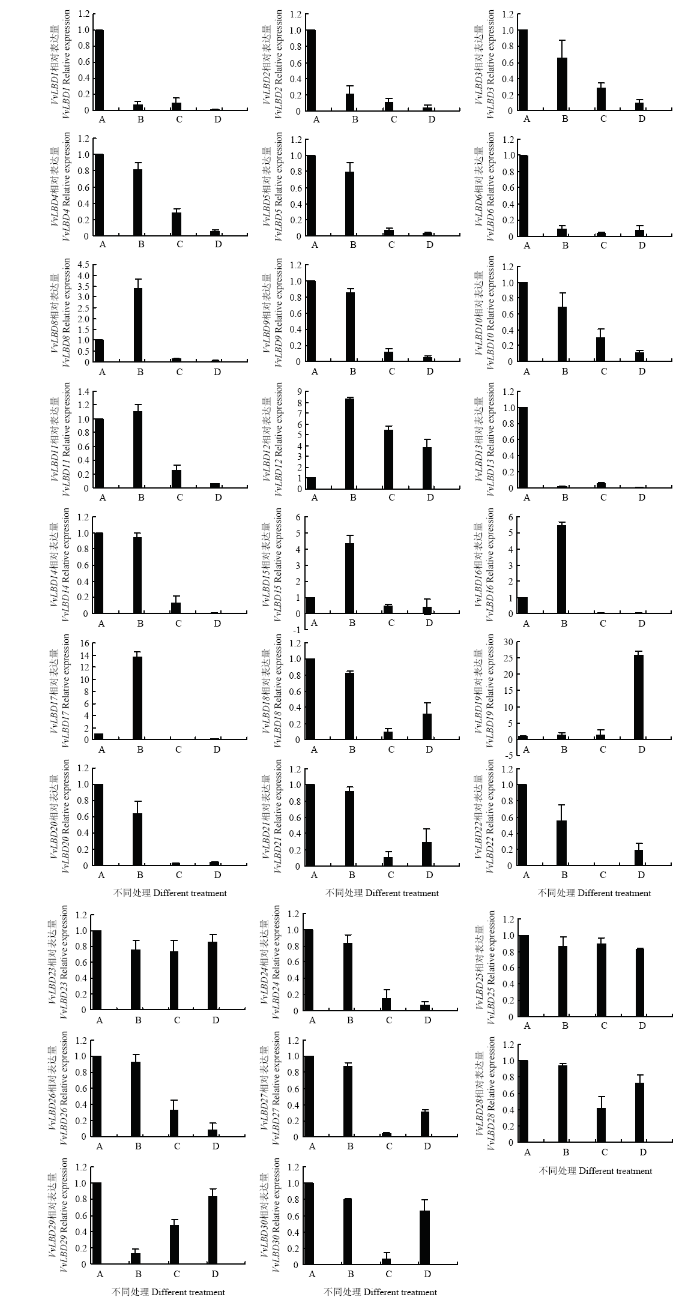 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7葡萄LBD基因在不同处理下的表达
A:对照;B:400 mmol·L-1 NaCl处理24 h;C:50 μmol·L-1 ABA处理24 h;D:10%PEG处理24 h
Fig.7VvLBD gene expression under different treatment
A: Control; B: 24 h treatment by 400 mmol·L-1 NaCl; C: 24 h treatment by 50 μmol·L-1 ABA; D: 24 h treatment by 10%PEG
3 讨论
本研究通过已知拟南芥LBD蛋白序列从葡萄基因组数据库中同源搜索到30个VvLBD基因,GRIMPLET等[38]研究结果表明,VvLBD基因家族成员有50个,并对VvLBD基因家族的生物信息学分析,利用基因芯片数据分析、组织表达和逆境胁迫等多方面功能的预测,但是通过比较分析发现,其中大部分基因结构是不完整的,因此未对基因的功能进行确切的验证。本研究通过去掉一些结构域不存在的序列后,用实时荧光定量法对葡萄试管苗进行了非生物胁迫的相关功能验证。结果表明,30个VvLBD基因在染色体上所处的位置均有所差异,理化性质也存在一定的差异。通过VvLBD基因家族多序列比对,发现它与拟南芥、玉米、番茄具有相似的结构域,推测VvLBD基因与它们可能具有相似的功能[16, 20-21]。在ClassⅠ的VvLBD基因家族成员中,LOB 结构域的C端大部分包含有一个类似亮氨酸拉链的LX6LX3LX6L螺旋卷曲二级结构,这可能与转录因子间的二聚化相关[16,17],而VvLBD基因家族中7个ClassⅡ成员均不含该结构域,这与普通烟草中研究结果有相似的特点[41]。本试验对VvLBD基因家族和拟南芥基因家族进行聚类分析,结果表明,在葡萄中ClassⅠ有对自展值在95以上的直系同源系基因VvLBD12/AtLBD20。THATCHER等[15]研究结果表明,AtLBD20在拟南芥中对尖孢镰刀杆菌的防御过程中发挥着负调控作用。ZHU等[43]认为 AtLBD16(At2G42430)和AtLBD18(At2G45420)的表达在拟南芥中受生长素诱导,同时聚类于ClassⅠ中,相对应的葡萄LBD同源基因VvLBD8和VvLBD22可能具有相似的功能。VvLBD基因结构分析表明聚类关系较近的基因具有相似的基因结构,这与番茄LBD基因家族的聚类分析结果相近[16]。对VvLBD基因家族编码蛋白质二级结构进行预测,发现该基因编码蛋白质以α-螺旋和不规则卷曲为主。motif序列分析表明,在VvLBD基因家族中主要存在3个保守序列,分别是GAS保守结构域、锌指结构域和亮氨酸拉链卷曲螺旋基序[10,41-42]。说明该基因家族与水稻、玉米和大麦的LBD基因家族高度相似。本研究对葡萄LBD基因芯片数据进行分析,结果表明,在非生物胁迫条件下,该基因家族在葡萄果皮和叶片中表达,在ABA胁迫下,‘赤霞珠’浆果果皮中VvLBD26的表达量最高。在盐、ABA、PEG和低温胁迫下,在‘赤霞珠’叶片中,VvLBD2、VvLBD3、VvLBD22和VvLBD28的表达量最高,其中VvLBD2、VvLBD3和VvLBD28都在ClassⅡ亚族中,研究结果与拟南芥、水稻等植物LBD家族的表达模式相似[17,43]。但是,不同时长的逆境胁迫可以使葡萄LBD基因有不一样的表达情况,如:VvLBD1在ABA胁迫4 h的表达和对照(CK1)无差异,而在ABA处理8 h的表达量明显比对照(CK1)下调;VvLBD23在1 h盐胁迫下的表达量和对照(CK2)一样,而在4、5和6 h盐胁迫下的表达量明显比对照(CK2)有所下调(图6)。
AtLBD蛋白参与调控植物生长发育的多种过程,在胚胎形成、侧生组织发育、花青苷合成以及氮代谢过程报道的较多[44,45],然而对于LBD基因应对非生物胁迫响应的研究较少。在ClassⅠa亚族中的VvLBD23和ClassⅠd亚族中的VvLBD25,在50 μmol·L-1 ABA、400 mmol·L-1 NaCl和10% PEG处理24 h后,各处理间的表达没有显著差异。葡萄LBD8、LBD12、LBD15—LBD17,在400 mmol·L-1 NaCl处理24 h后呈现明显上调趋势,葡萄LBD1—LBD6、LBD9—LBD10、LBD 13—LBD14、LBD18、LBD20—LBD22、LBD24、LBD26—LBD30,在50 μmol·L-1 ABA、400 mmol·L-1 NaCl和10%PEG处理24 h后呈现出下调趋势。只VvLBD19在10%PEG处理24 h后基因表达呈现上调趋势,且表达量是对照的26倍。GRIMPLET等[38]研究结果表明VviLBDId5在浆果果皮中表达,并且在外源ABA处理下表现出正调控作用,而在本文的定量分析中发现在外源ABA处理下VvLBD14的表达量并没有显著的变化(VviLBDId5对应着本文研究的VvLBD14)。以上研究结果与CAO等[37]有关VvLBD基因家族关于逆境胁迫的研究成果有相似之处。本研究表明,在ClassⅠ和ClassⅡ2个亚族中的VvLBD基因对400 mmol·L-1 NaCl胁迫24 h后上调表达较为强烈,而对于ABA、PEG的胁迫下的相对表达下调趋势较为明显。推测VvLBD基因家族的ClassⅠ和ClassⅡ 2个亚族都参与了逆境胁迫应答,且在不同植物中LBD基因存在不同的调控机制。
4 结论
获得30个VvLBD基因家族成员,且含有保守结构域,可分为2大类和7个亚类。除VvLBD12在细胞骨架中表达以外,其他基因主要在叶绿体、线粒体和细胞核中表达。在响应不同时长不同逆境胁迫时,不同的基因起到不同的表达调控作用。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
.
[本文引用: 1]
DOI:10.1104/pp.108.131052URL [本文引用: 1]
DOI:10.1007/s00299-014-1622-7URLPMID:24792421 [本文引用: 1]

Abstract KEY MESSAGE: Our study has identified and analyzed the VvARF gene family that may be associated with the development of grape berry and other tissues. Auxin response factors (ARFs) are transcription factors that regulate the expression of auxin responsive genes through specific binding to auxin response elements (AuxREs). The ARF genes are represented by a large multigene family in plants. Until now, many ARF families have been characterized based on genome resources. However, there is no specialized research about ARF genes in grapevine (Vitis vinifera). In this study, a comprehensive bioinformatics analysis of the grapevine ARF gene family is presented, including chromosomal locations, phylogenetic relationships, gene structures, conserved domains and expression profiles. Nineteen VvARF genes were identified and categorized into four groups (Classes 1, 2, 3 and 4). Most of VvARF proteins contain B3, AUX_RESP and AUX_IAA domains. The VvARF genes were widely expressed in a range of grape tissues, and fruit had higher transcript levels for most VvARFs detected in the EST sources. Furthermore, analysis of expression profiles indicated some VvARF genes may play important roles in the regulation of grape berry maturation processes. This study which provided basic genomic information for the grapevine ARF gene family will be useful in selecting candidate genes related to tissue development in grapevine and pave the way for further functional verification of these VvARF genes.
URL [本文引用: 1]
DOI:10.16420/j.issn.0513-353x.2015-0912URLMagsci [本文引用: 1]

<p>以‘宝石无核’(<a name="OLE_LINK18">Ruby Seedless</a>)葡萄试管苗为材料,采用RT-PCR技术,克隆得到8个葡萄SnRK2家族基因。序列分析发现,该家族基因蛋白结构在N端相对保守,而在C端极其特异;聚类结果表明葡萄SnRK2基因家族可以分为3个亚家族;对这8个基因所编码的蛋白质进行分析发现,其富含酸性氨基酸,且均为亲水蛋白;基因组结构分析发现<em>VvSnRK2.2</em>和<em>VvSnRK2.8</em>含有10个外显子,其他6个均含9个外显子;对蛋白二级结构分析发现8个基因编码的蛋白主要以α–螺旋、β–转角和不规则卷曲为主;亚细胞定位预测,8个基因主要定位于细胞质中。顺式作用元件分析表明,除<em>VvSnRK2.1、VvSnRK2.2、VvSnRK2.6</em>外,其他基因顺式作用元件包含ABRE、DRE/CRT、LRTE中的一个或多个。定量PCR分析表明,<em>VvSnRK2</em>的表达存在组织差异性,<em>VvSnRK2.7</em>在根中表达水平最高,是叶片的3.8倍,<em>VvSnRK2.8</em>在茎中表达水平最高,是叶片的5.0倍。0 ~–4 ℃处理后,表达水平下调幅度最小的为<em>VvSnRK2.2</em>,<em>VvSnRK2.7</em>下调幅度较大,<em>VvSnRK2.8</em>的表达水平为0;30 ℃处理后<em>VvSnRK2.2</em>和<em>VvSnRK2.5</em>上调表达,分别为对照的3.8倍和3.6倍;<em>VvSnRK2.1</em>和<em>VvSnRK2.2</em>与盐胁迫调节紧密相关,<em>VvSnRK2.5</em>与干旱胁迫调节密切相关。</p>
DOI:10.16420/j.issn.0513-353x.2015-0912URLMagsci [本文引用: 1]

<p>以‘宝石无核’(<a name="OLE_LINK18">Ruby Seedless</a>)葡萄试管苗为材料,采用RT-PCR技术,克隆得到8个葡萄SnRK2家族基因。序列分析发现,该家族基因蛋白结构在N端相对保守,而在C端极其特异;聚类结果表明葡萄SnRK2基因家族可以分为3个亚家族;对这8个基因所编码的蛋白质进行分析发现,其富含酸性氨基酸,且均为亲水蛋白;基因组结构分析发现<em>VvSnRK2.2</em>和<em>VvSnRK2.8</em>含有10个外显子,其他6个均含9个外显子;对蛋白二级结构分析发现8个基因编码的蛋白主要以α–螺旋、β–转角和不规则卷曲为主;亚细胞定位预测,8个基因主要定位于细胞质中。顺式作用元件分析表明,除<em>VvSnRK2.1、VvSnRK2.2、VvSnRK2.6</em>外,其他基因顺式作用元件包含ABRE、DRE/CRT、LRTE中的一个或多个。定量PCR分析表明,<em>VvSnRK2</em>的表达存在组织差异性,<em>VvSnRK2.7</em>在根中表达水平最高,是叶片的3.8倍,<em>VvSnRK2.8</em>在茎中表达水平最高,是叶片的5.0倍。0 ~–4 ℃处理后,表达水平下调幅度最小的为<em>VvSnRK2.2</em>,<em>VvSnRK2.7</em>下调幅度较大,<em>VvSnRK2.8</em>的表达水平为0;30 ℃处理后<em>VvSnRK2.2</em>和<em>VvSnRK2.5</em>上调表达,分别为对照的3.8倍和3.6倍;<em>VvSnRK2.1</em>和<em>VvSnRK2.2</em>与盐胁迫调节紧密相关,<em>VvSnRK2.5</em>与干旱胁迫调节密切相关。</p>
URL [本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 1]
URL [本文引用: 3]
[本文引用: 1]
DOI:10.1007/s00299-016-2057-0URLPMID:27686461 [本文引用: 1]

KEY MESSAGE: An unambiguous nomenclature is proposed for the twenty-eight-member LOB domain transcription factor family in Brachypodium . Expression analysis provides unique transcript patterns that are characteristic of a wide range of organs and plant parts. LOB (lateral organ boundaries)-domain proteins define a family of plant-specific transcription factors involved in developmental processes from embryogenesis to seed production. They play a crucial role in shaping the plant architecture through coordinating cell fate at meristem to organ boundaries. Despite their high potential importance, our knowledge of them is limited, especially in the case of monocots. In this study, we characterized LOB domain protein coding genes (LBDs) of Brachypodium distachyon, a model plant for grasses, and present their phylogenetic relationships and an overall spatial expression study. In the Brachypodium genome database, 28 LBDs were found and then classified based on the presence of highly conserved LOB domain motif. Their transcript amounts were measured via quantitative real-time RT-PCR in 37 different plant parts from root tip to generative organs. Comprehensive phylogenetic analysis suggests that there are neither Brachypodium- nor monocot-specific lineages among LBDs, but there are differences in terms of complexity of subclasses between monocots and dicots. Although LBDs in Brachypodium have wide variation of tissue-specific expression and relative transcript levels, overall expression patterns show similarity to their counterparts in other species. The varying transcript profiles we observed support the hypothesis that Brachypodium LBDs have diverse but conserved functions in plant organogenesis.
[本文引用: 1]
[本文引用: 1]
DOI:10.4161/psb.22097URLPMID:22786889 [本文引用: 2]

Over the last two decades, several transcription factor gene families have been identified with some of them characterized in detail for their roles on transcriptional regulation of plant defense responses against pest or pathogen attack. We have recently added another transcription factor gene family to this list through the characterization of the LATERAL ORGAN BOUNDARIES (LOB) DOMAIN (LBD)-CONTAINING PROTEIN20 (LBD20). We showed LBD20 acts as a repressor of a subset of jasmonate mediated defenses and in susceptibility to the root-infecting fungal pathogen Fusarium oxysporum. However, possible roles for other members of this gene family in plant defense are currently unknown. Here we searched publically available microarray expression data and provide an overview of the expression patterns of selected members of the LBD gene family for their response to other fungal pathogens and soil nematodes. Distinct expression patterns of the LBD genes suggest that certain members of this gene family have previously undescribed roles in plant defense.
DOI:10.3864/j.issn.0578-1752.2013.12.011Magsci [本文引用: 6]

【目的】从番茄全基因组中鉴定LBD基因,并进行基因进化、基因结构、染色体定位以及组织表达和诱导表达分析,为番茄LBD基因的功能研究与利用奠定基础。【方法】利用番茄基因组数据库,通过生物信息学手段,鉴定番茄LBD家族成员;采用MEGA5软件进行系统进化树分析;通过perl程序、MapDRAW及GSDS工具进行基因结构及染色体定位分析;利用已有的番茄芯片数据进行组织表达谱和基因表达响应分析。【结果】系统分析鉴定了 46 个番茄LBD 家族基因,根据基因结构及系统进化分析将其分成class I与class II两类,细分为5个亚家族(Ia、Ib、Ic、Id与II)。基因定位表明,12 条染色体中的 10 条均有LBD 基因,该基因家族的分布具有广泛性。各个发育阶段表达谱的分析结果表明,LBD 家族基因具有不同的组织表达模式,在各个发育时期均有LBD 基因的表达。基因响应分析发现,不同LBD基因响应不同的外界信号。【结论】通过全基因组分析,番茄LBD家族基因包括 46 个成员,在进化上分为两大类,5 个亚家族,分布于 10 条染色体上,组织表达模式及基因响应具有多样性。这些信息为番茄LBD基因家族的功能分析奠定了基础。
DOI:10.3864/j.issn.0578-1752.2013.12.011Magsci [本文引用: 6]

【目的】从番茄全基因组中鉴定LBD基因,并进行基因进化、基因结构、染色体定位以及组织表达和诱导表达分析,为番茄LBD基因的功能研究与利用奠定基础。【方法】利用番茄基因组数据库,通过生物信息学手段,鉴定番茄LBD家族成员;采用MEGA5软件进行系统进化树分析;通过perl程序、MapDRAW及GSDS工具进行基因结构及染色体定位分析;利用已有的番茄芯片数据进行组织表达谱和基因表达响应分析。【结果】系统分析鉴定了 46 个番茄LBD 家族基因,根据基因结构及系统进化分析将其分成class I与class II两类,细分为5个亚家族(Ia、Ib、Ic、Id与II)。基因定位表明,12 条染色体中的 10 条均有LBD 基因,该基因家族的分布具有广泛性。各个发育阶段表达谱的分析结果表明,LBD 家族基因具有不同的组织表达模式,在各个发育时期均有LBD 基因的表达。基因响应分析发现,不同LBD基因响应不同的外界信号。【结论】通过全基因组分析,番茄LBD家族基因包括 46 个成员,在进化上分为两大类,5 个亚家族,分布于 10 条染色体上,组织表达模式及基因响应具有多样性。这些信息为番茄LBD基因家族的功能分析奠定了基础。
DOI:10.16420/j.issn.0513-353x.2015-0971URLMagsci [本文引用: 4]

利用生物信息学的方法,从辣椒基因组中鉴定出45个LBD基因,这些基因分布于辣椒9条染色体上。该家族成员内含子数整体上不超过3个,结构相对简单。进化关系显示辣椒LBD基因可分为ClassⅠ和ClassⅡ两大类,细分为Ⅰa、Ⅰb、Ⅰc、Ⅰd、Ⅰe、Ⅱa和Ⅱb等7个亚类。不同组织和发育时期的表达模式研究发现,该基因家族具有一定的时空表达特异性。qRT-PCR结果表明,热激胁迫可以明显激活或抑制部分LBD基因的表达,其中ClassⅡ类基因较ClassⅠ类对高温具有更高的敏感性。
DOI:10.16420/j.issn.0513-353x.2015-0971URLMagsci [本文引用: 4]

利用生物信息学的方法,从辣椒基因组中鉴定出45个LBD基因,这些基因分布于辣椒9条染色体上。该家族成员内含子数整体上不超过3个,结构相对简单。进化关系显示辣椒LBD基因可分为ClassⅠ和ClassⅡ两大类,细分为Ⅰa、Ⅰb、Ⅰc、Ⅰd、Ⅰe、Ⅱa和Ⅱb等7个亚类。不同组织和发育时期的表达模式研究发现,该基因家族具有一定的时空表达特异性。qRT-PCR结果表明,热激胁迫可以明显激活或抑制部分LBD基因的表达,其中ClassⅡ类基因较ClassⅠ类对高温具有更高的敏感性。
DOI:10.1016/j.tplants.2010.09.009URLPMID:20961800 [本文引用: 4]

(Loundaries omain genes associated with mutant phenotypes related to almost all aspects of plant development, including embryo, root, leaf, and inflorescence development have been functionally characterized. These novel insights contribute to a better understanding of the molecular definition of boundaries between organs or boundaries between organs and meristems and the regulation of these processes by environmental cues and phytohormones.
DOI:10.1104/pp.010926URL [本文引用: 1]
[本文引用: 2]
[D].
[本文引用: 2]
[D].
[本文引用: 2]
[本文引用: 1]
DOI:10.7606/j.issn.1000-4025.2014.10.2176URL [本文引用: 2]

LBD是植物中所特有的转录因子基因家族,在调控植物侧生组织发育、营养代谢以及响应逆境胁迫等方面具有重要作用。该研究利用生物信息学手段,从全基因组水平筛选和鉴定了蒺藜苜蓿LBD基因家族,并对基因结构、系统进化、进化压力、保守域、染色体定位以及基因表达模式等进行了分析。研究结果共鉴定出2类5亚类共计56个蒺藜苜蓿LBD家族基因,在8条染色体上均有分布,但分布不均匀。该家族成员外显子数目都不超过2个,结构简单,基因间在进化时存在负向选择作用。基因表达模式分析发现,该家族成员的表达具有一定的时空特异性,并受干旱和氮素调控。该研究结果对蒺藜苜蓿LBD基因功能研究及进化分析具有重要的意义。
DOI:10.7606/j.issn.1000-4025.2014.10.2176URL [本文引用: 2]

LBD是植物中所特有的转录因子基因家族,在调控植物侧生组织发育、营养代谢以及响应逆境胁迫等方面具有重要作用。该研究利用生物信息学手段,从全基因组水平筛选和鉴定了蒺藜苜蓿LBD基因家族,并对基因结构、系统进化、进化压力、保守域、染色体定位以及基因表达模式等进行了分析。研究结果共鉴定出2类5亚类共计56个蒺藜苜蓿LBD家族基因,在8条染色体上均有分布,但分布不均匀。该家族成员外显子数目都不超过2个,结构简单,基因间在进化时存在负向选择作用。基因表达模式分析发现,该家族成员的表达具有一定的时空特异性,并受干旱和氮素调控。该研究结果对蒺藜苜蓿LBD基因功能研究及进化分析具有重要的意义。
DOI:10.1007/s12374-009-9048-4URL [本文引用: 1]

LATERALORGAN BOUNDARIES DOMAIN ( LBD ) genes, a novel plant-specific family, play specific roles in plant development. Although function of ASYMMETRIC LEAVES2 ( AS2 ), a LBD gene, was extensively studied in Arabidopsis of dicots, little is known on the role of its ortholog in rice of monocots. In this study, a LBD gene that shares higher homology with Arabidopsis AS2 gene was identified in rice and it was designated as OsAS2 . Its transcripts were detected throughout predicted leaf primordia in shoot apical meristem (SAM), leaf primordia, and young leaves. Overexpression of the OsAS2 gene inhibited shoot differentiation, promoted cell division, and delayed cell differentiation in rice calli. Transgenic plants with OsAS2 gene showed the aberrant twisted leaves, which lack auricle, and leaf structure was abnormal. Furthermore, a few genes involved in shoot meristem development were upregulated in transgenic plants. Our results suggest that proper expression of the OsAS2 gene is required for shoot differentiation and leaf development in rice.
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1271/bbb.60681URLPMID:17485849 [本文引用: 1]

In Arabidopsis thaliana, each member of a large family of AS2/LOB (ASYMMETRIC LEAVES 2/LATERAL ORGAN BOUNDARIES) genes encodes a plant specific protein. They are highly homologous to one other. A mutational lesion in the representative AS2 gene results in the development of anomalous asymmetric leaves, implying that these family members commonly play some roles in plant development. In this study, we found that ectopic overexpression of ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) in transgenic plants displayed a markedly anomalous architecture during the development of adult plants. Then we found that among AS2/LOB family members, ASL9 is distinct from the others in that it is exclusively regulated by the plant hormone cytokinin in a manner dependent on His-Asp phosphorelay signal transduction. We further found that when supplied externally in a medium, cytokinin specifically affected the growth properties of ASL9-ox seedlings. Taken together, the results of this study suggest that the cytokinin-induced ASL9 gene is implicated in regulation of the development of Arabidopsis thaliana.
URL [本文引用: 1]
DOI:10.1016/j.tplants.2015.10.010URLPMID:26616195 [本文引用: 1]

LBDgenes encode a family of plant-specific transcription factors that are essential for lateral organ development in higher plants. Functional diversity of the LBD family exceeds lateral organ boundaries formation by defining pollen development, plant regeneration, photomorphogenesis, pathogen response, and some specific developmental processes in grass, wood, and fruit species. The LBD-dependent developmental programs depend on the cooperative action of the LBD-mediated molecular pathways, which comprise a range of newly identified upstream regulators, protein partners, and downstream targets of LBD family members. The integration of developmental changes induced by phytohormones or environmental stimuli requires the functions of LBD proteins.
[本文引用: 1]
DOI:10.1093/mp/sst176URLPMID:24398629 [本文引用: 1]
DOI:10.1007/s10535-016-0588-4URL [本文引用: 2]

Auxin controls numerous processes in plant development and auxin/indoleacetic acid ( Aux / IAA ), an auxin response factor ( ARF ), and a lateral organ boundaries domain ( LBD ) were considered as early auxin response transcription factors (TFs). Till now, no Aux / IAA , ARF , and LBD TFs were identified in ramie ( Boehmeria nivea L. Gaud). In this study, we used Arabidopsis and mulberry sequences as query to search against the ramie transcriptome database and the searched sequences were analyzed for a full-length coding sequence. In total, we obtained 16 BnAux / IAA , 14 BnARF , and 16 BnLBD TFs on which evolutionary analysis and expression profiling were conducted. Analysis of sequence conservation revealed close evolution relationships between ramie and mulberry. Expression analysis shows these genes were actively expressed in major ramie tissues, and several were auxin responsive. The expressions of these genes were also investigated under drought and a high temperature, main abiotic stresses during ramie life cycle. We found that most genes of the three families were stress-responsive and showed distinct expression patterns under the drought and high temperature stresses.
[本文引用: 1]
DOI:10.1007/s12041-016-0660-zURLPMID:27659322 [本文引用: 2]

In plants, the transcription factor families have been implicated in many important biological processes. These processes include morphogenesis, signal transduction and environmental stress responses. Proteins containing the lateral organ boundaries domain (LBD), which encodes a zinc finger-like domain are only found in plants. This finding indicates that this unique gene family regulates only plant-specific biological processes. LBD genes play crucial roles in the growth and development of plants such as Arabidopsis , Oryza sativa , Zea mays , poplar, apple and tomato. However, relatively little is known about the LBD genes in grape ( Vitis vinifera ). In this study, we identified 40 LBD genes in the grape genome. A complete overview of the chromosomal locations, phylogenetic relationships, structures and expression profiles of this gene family during development in grape is presented here. Phylogenetic analysis showed that the LBD genes could be divided into classes I and II, together with LBDs from Arabidopsis . We mapped the 40 LBD genes on the grape chromosomes (chr1 hr19) and found that 37 of the predicted grape LBD genes were distributed in different densities across 12 chromosomes. Grape LBDs were found to share a similar intron/exon structure and gene length within the same class. The expression profiles of grape LBD genes at different developmental stages were analysed using microarray data. Results showed that 21 grape LBD genes may be involved in grape developmental processes, including preveraison, veraison and ripening. Finally, we analysed the expression patterns of six LBD genes through quantitative real-time polymerase chain reation analysis. The six LBD genes showed differential expression patterns among the three representative grape tissues, and five of these genes were found to be involved in responses to mannitol, sodium chloride, heat stress and low temperature treatments. To our knowledge, this is the first study to analyse the LBD gene family in grape and provides valuable information for classification and functional investigation of this gene family.
[本文引用: 4]
DOI:10.1007/s11033-012-1700-2URLPMID:22711305 [本文引用: 1]

Abstract (HAK) transporter gene family constitutes the largest family that functions as potassium transporter in plant and is important for various cellular processes of plant life. In spite of their physiological importance, systematic analyses of ZmHAK genes have not yet been investigated. In this paper, we indicated the isolation and characterization of ZmHAK genes in whole-genome wide by using bioinformatics methods. A total of 27 members (ZmHAK1 mHAK27) of this family were identified in maize genome. ZmHAK genes were distributed in all the maize 10 chromosomes. These genes expanded in the maize genome partly due to tandem and segmental duplication events. Multiple alignment and motif display results revealed major maize ZmHAK proteins share all the three conserved domains. Phylogenetic analysis indicated -elements involved in Ca response, abiotic stress adaption, light and circadian rhythms regulation and seed development were observed in the promoters of ZmHAK genes. Expression data mining suggested maize ZmHAK genes have temporal and spatial expression pattern. In all, these results will provide molecular insights into the potassium transporter research in maize.
DOI:10.7501/j.issn.0253-2670.2017.06.023URL [本文引用: 1]

目的筛选适用于显齿蛇葡萄Ampelopsis grossedentata实时荧光定量PCR(real-time fluorescence quantitative PCR,qRT-PCR)的内参基因。方法设计简并引物,采用RT-PCR从显齿蛇葡萄中克隆6个候选内参基因片段,包括肌动蛋白基因(Actin)、3-磷酸甘油醛脱氢酶基因(GAPDH)、18 S核糖体rRNA基因(18 S-rRNA)、α-微管蛋白基因(α-Tubulin)、β-微管蛋白基因(β-Tubulin)和多聚泛素酶基因(UBQ)。应用qRT-PCR技术检测这6个候选内参基因在显齿蛇葡萄不同组织器官中(茎尖、嫩叶、成熟叶、老叶、茎和根)的表达情况。借助Ge Norm、Norm Finder和Best Keeper 3种统计学软件综合评价6个内参基因的表达稳定性。通过苯丙氨酸解氨酶基因(Ag PAL)的表达分析对筛选出的内参基因进行稳定性验证。结果 18 S-rRNA、GAPDH和Actin的表达稳定性较好,适合作为显齿蛇葡萄不同组织基因表达研究的内参基因,以这3个基因为内参基因分析Ag PAL基因的相对表达量,结果发现它们在各组织器官中的表达变化趋势基本一致。结论确定显齿蛇葡萄qRT-PCR分析的合适内参基因,为后续相关基因表达研究奠定基础。
DOI:10.7501/j.issn.0253-2670.2017.06.023URL [本文引用: 1]

目的筛选适用于显齿蛇葡萄Ampelopsis grossedentata实时荧光定量PCR(real-time fluorescence quantitative PCR,qRT-PCR)的内参基因。方法设计简并引物,采用RT-PCR从显齿蛇葡萄中克隆6个候选内参基因片段,包括肌动蛋白基因(Actin)、3-磷酸甘油醛脱氢酶基因(GAPDH)、18 S核糖体rRNA基因(18 S-rRNA)、α-微管蛋白基因(α-Tubulin)、β-微管蛋白基因(β-Tubulin)和多聚泛素酶基因(UBQ)。应用qRT-PCR技术检测这6个候选内参基因在显齿蛇葡萄不同组织器官中(茎尖、嫩叶、成熟叶、老叶、茎和根)的表达情况。借助Ge Norm、Norm Finder和Best Keeper 3种统计学软件综合评价6个内参基因的表达稳定性。通过苯丙氨酸解氨酶基因(Ag PAL)的表达分析对筛选出的内参基因进行稳定性验证。结果 18 S-rRNA、GAPDH和Actin的表达稳定性较好,适合作为显齿蛇葡萄不同组织基因表达研究的内参基因,以这3个基因为内参基因分析Ag PAL基因的相对表达量,结果发现它们在各组织器官中的表达变化趋势基本一致。结论确定显齿蛇葡萄qRT-PCR分析的合适内参基因,为后续相关基因表达研究奠定基础。
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
DOI:10.1093/bioinformatics/btm113URLPMID:17392330 [本文引用: 2]

The database of poplar transcription factors (DPTF) is a plant transcription factor (TF) database containing 2576 putative poplar TFs distributed in 64 families. These TFs were identified from both computational prediction and manual curation. We have provided extensive annotations including sequence features, functional domains, GO assignment and expression evidence for all TFs. In addition, DPTF contains cross-links to the Arabidopsis and rice transcription factor databases making it a unique resource for genome-scale comparative studies of transcriptional regulation in model plants. Availiability: DPTF is available at http://dptf.cbi.pku.edu.cn.
DOI:10.1038/cr.2012.63URLPMID:3391013 [本文引用: 1]

The remarkable regeneration capability of plant tissues or organs under culture conditions has underlain an extensive practice for decades. The initial step in plant in vitro regeneration often involves the induction of a pluripotent cell mass termed callus, which is driven by the phytohormone auxin and occurs via a root development pathway. However, the key molecules governing callus formation remain unknown. Here we demonstrate that Arabidopsis LATERAL ORGAN BOUNDARIES DOMAIN (LBD)/ASYMMETRIC LEAVES2-LIKE (ASL) transcription factors are involved in the control of callus formation program. The four LBD genes downstream of AUXIN RESPONSE FACTORs (ARFs), LBD16, LBD17, LBD18 and LBD29, are rapidly and dramatically induced by callus-inducing medium (CIM) in multiple organs. Ectopic expression of each of the four LBD genes in Arabidopsis is sufficient to trigger spontaneous callus formation without exogenous phytohormones, whereas suppression of LBD function inhibits the callus formation induced by CIM. Moreover, the callus triggered by LBD resembles that induced by CIM by characteristics of ectopically activated root meristem genes and efficient regeneration capacity. These findings define LBD transcription factors as key regulators in the callus induction process, thereby establishing a molecular link between auxin signaling and the plant regeneration program.
[本文引用: 1]
