 ,1, 王庆莲1, 夏瑾1, 王西成1, 宋志忠
,1, 王庆莲1, 夏瑾1, 王西成1, 宋志忠 ,1,2, 吴伟民1
,1,2, 吴伟民1Cloning, Characterization and Expression Analysis of K +/H + Antiporter Genes in Grape
WANG ZhuangWei ,1, WANG QingLian1, XIA Jin1, WANG XiCheng1, SONG ZhiZhong
,1, WANG QingLian1, XIA Jin1, WANG XiCheng1, SONG ZhiZhong ,1,2, WU WeiMin1
,1,2, WU WeiMin1通讯作者:
第一联系人:
责任编辑: 李莉
收稿日期:2018-05-14接受日期:2018-06-21网络出版日期:2018-12-01
| 基金资助: |
Received:2018-05-14Accepted:2018-06-21Online:2018-12-01

摘要
关键词:
Abstract
Keywords:
PDF (2543KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王壮伟, 王庆莲, 夏瑾, 王西成, 宋志忠, 吴伟民. 葡萄KEA家族基因的克隆、鉴定及表达分析[J]. 中国农业科学, 2018, 51(23): 4522-4534 doi:10.3864/j.issn.0578-1752.2018.23.011
WANG ZhuangWei, WANG QingLian, XIA Jin, WANG XiCheng, SONG ZhiZhong, WU WeiMin.
0 引言
【研究意义】钾素营养与果树生长、花开放及果实发育密切相关,但其分子基础研究报道鲜少。钾离子(K+)是植物细胞中含量最为丰富的阳离子之一,维持细胞渗透调节平衡,控制细胞的基础膜电位水平,在气孔运动、光合作用、蒸腾作用、信号传导和胁迫抗逆等生命过程中均起着至关重要的作用[1,2]。由于K+控制着细胞的基础膜电位水平,然而,功能活跃细胞的真实膜电位水平往往偏离K+的扩散平衡电位。另一方面,离子或分子跨膜运输过程的激活,则依赖于这一基本驱动力并大多同时受细胞内质子(H+)浓度的调控。细胞质膜质子泵和液泡膜质子泵控制着细胞内质子H+浓度的电化学势,是造成细胞膜电位偏离的重要原因[3,4,5,6]。由此可见,K+和H+浓度的动态平衡对于细胞和植物的生命过程具有至关重要的作用。【前人研究进展】阳离子-质子逆向转运体CPAs(cation proton antiporters)广泛存在于植物、动物、真菌和细菌中,主要定位于细胞质膜、液泡膜和植物中的细胞器膜,可将细胞中的Na+、K+、Li+等阳离子排出,并引起细胞内H+的内流和积累[7,8],维持K+和H+浓度的动态平衡。植物中,CPAs可介导细胞中离子和质子的动态平衡,在植物的离子平衡、生长发育和信号转导中起重要作用[7,8,9]。植物CPAs分为2个亚族:CPA1和CPA2,其中,CPA1包括NHX(Na+/H+ exchanger),CPA2包括KEA(K+ efflux antiporter,K+/H+逆向转运体)和CHX(Cation/H+ exchanger)[7,8,9]。特别地,植物CPAs中有关KEA亚族的研究最为稀少,主要体现在模式作物拟南芥中。2001年,MASER等[10]最早提出拟南芥中有6个AtKEA转运体基因,但其功能依然未知。最近研究表明,拟南芥AtKEA转运体定位于维管组织、保卫细胞和花萼等不同组织部位[11],在植物体内K+动态平衡和渗透调节中起主要作用[12,13];此外,有报道表明拟南芥AtKEA1-3转运体在叶绿体渗透调节、光合作用及pH调控等方面起关键作用[12,13,14,15]。然而,果树中有关K+/H+动态平衡问题的研究报道鲜少,仅见于蔷薇科梨基因组中KEA家族基因的鉴定及生物信息学分析[16],具体功能依然未知。【本研究切入点】近30年来,关于植物K+吸收转运以及质子泵的研究较为详尽,然而,对于同时转运K+和H+的K+/H+逆向转运体的研究鲜有报道。果树作物中,有关K+/H+动态平衡及KEA家族基因的功能依然未知。【拟解决的关键问题】本研究以‘白罗莎里奥’葡萄为材料,克隆并鉴定了4个葡萄KEA家族基因及其编码蛋白的特征,明确该家族基因在葡萄不同组织中的表达特征及对缺钾、ABA、NaCl和sorbitol等胁迫处理的响应,为研究果树K+/H+平衡与钾素动态营养机制提供了基因资源和理论基础。1 材料与方法
1.1 试验时间、地点
试验于2017年4—12月在江苏省农业科学院/江苏省高效园艺作物遗传改良实验室完成。1.2 试验材料与处理
供试材料为江苏省农业科学院溧水植物科学基地的8年生‘白罗莎里奥’葡萄(Rosario Bianco),树体健壮,南北行向,株行距为1 m×3 m,H型平棚架式,联栋大棚避雨设施栽培,常规田间管理。分别于特定日期采集8年生‘白罗莎里奥’的花蕾(4月30日,花序中部的花蕾)、花(5月3日,花序中部,盛开期)、新生叶片(5月24日,自顶端数起的第3片一年生新叶)、新生根(5月24日,近地表土层中侧根顶端的新生细根)、幼果(5月24日,幼果迅速膨大期)和熟果(9月7日,浆果成熟期)等组织材料,随后立即液氮冷冻并保存于-80℃冰箱中备用。胁迫试验供试材料为‘白罗莎里奥’试管幼苗,来源于江苏省农业科学院果树研究所浆果研究室。2016年8月,将生长情况一致的试管幼苗(长约12 cm)转接于GS液体培养基中,于人工气候箱(条件设置为25℃,200 μmol·m-2·s-1光照16 h;20℃,暗培养8 h)中预培养3 d,然后分别进行缺钾(用NaNO3和NaH2PO3分别代替GS配方中的KNO3和KH2PO4)、脱落酸(ABA,200 μmol·L-1)、氯化钠(NaCl,160 mmol·L-1)、山梨糖醇(sorbitol,320 mmol·L-1)等处理,每个处理6株幼苗。胁迫处理48 h后,分别采集不同处理后的幼苗地上部和根部材料,立即液氮冷冻并保存于-80℃冰箱中备用。1.3 葡萄KEA基因克隆
以拟南芥TAIR数据库(http://www.arabidopsis. org/browse/genefamily/index.jsp)的6个KEA家族基因的氨基酸序列为参考序列,在Phytozome grape genome database (http://www.phytozome.net)中检索葡萄基因组中可能的KEA家族基因。检索结果在Pfam(http://pfam.xfam.org/search)在线服务器预测功能结构域。根据Phytozome获得的葡萄VvKEA的CDS序列(coding sequence),分别设计引物,利用Prime STARTM HS DNA聚合酶(TaKaRa,大连)进行PCR扩增,测序验证后,提交NCBI GenBank获得登录号。1.4 葡萄KEA家族基因的生物信息学分析
在Phytozome葡萄基因组数据库中获得KEA家族各基因的CDS编码区序列及基因组DNA序列,然后通过Gene Structure Display(http://gsds.cbi.pku.edu. cn/index.php)在线服务器进行基因结构分析;利用在线软件TMpredict(http://ch.embnet.org/software/ TMPRED_form.html)分析葡萄KEA家族蛋白的跨膜结构域;使用在线服务器MEME(v4.8.1)(http://meme. nbcr.net/meme/cgi-bin/meme.cgi)预测葡萄KEA家族蛋白的保守结构域;使用在线工具ProtParam(http:// expasy.org/tools/protparam.html)评估KEA蛋白成员的理论等电点、分子量、稳定性等理化性质;利用在线软件SignalP4.0(http://www.cbs.dtu.dk/services/ SignalP-4.0/)预测KEA家族基因的信号肽情况;利用PSORT在线服务器(http://psort.hgc.jp/form.html)预测葡萄KEA家族基因的亚细胞定位;利用Phyre2在线服务器(http://www.sbg.bio.ic.ac.uk/phyre2/html/page. cgi?id=index)分析葡萄KEA家族蛋白的三级结构;利用ClustalX 2.0软件对葡萄KEA转运体与拟南芥(6个)、水稻(6个)、玉米(8个)、高粱(7个)、短柄草(5个)、杨树(4个)、梨(12个)、苹果(7个)等[16,17]已知物种的同源KEA转运体进行氨基酸比对分析,用分子进化遗传分析软件MEGA 7.0中的邻接法(Neighbor-joining)构建系统进化树;利用NCBI的dbEST在线数据库(http://www.ncbi nlm. nih.gov/dbEST/),通过BLASTN比对,检索葡萄KEA家族基因全长CDS序列和EST数据,选择匹配率大于95%、长度大于160 bp,并且E≤10-10的结果作为对应的EST序列,按照不同组织或器官来源进行分类,从而获得葡萄KEA家族基因的表达谱信息;为分析葡萄KEA家族基因的启动子区域,在Phytozome葡萄基因组数据库检索目的基因CDS区域起始密码子ATG的上游1.5 kb左右的片段为启动子区域,并通过PLACE(http://www.dna.affrc.go.jp/ PLACE/)在线软件分析启动子序列中含有的cis-顺式作用元件。1.5 总RNA提取与实时荧光定量PCR分析
分别采集实生幼苗和8年生葡萄树体不同部位的组织材料,液氮冷冻后-80℃冰箱保存,通过MiniBEST Plant RNA Extraction Kit(TaKaRa,大连)提取样品的总RNA,并利用PrimeScriptTM RT reagent Kit 反转录试剂盒(TaKaRa,大连)合成第一链cDNA作为模板,用于实时荧光定量PCR。利用NCBI/Primer- BLAST在线服务器,设计葡萄VvKEA的特异性表达引物(表1),以葡萄Ubiquitin(GenBank No. MH114011),通过ABI 7500实时荧光定量PCR仪检测VvKEA在葡萄树不同组织部位的表达特征。荧光染料使用SYBR Green(TaKaRa,大连),反应体系参照商品说明书的描述,反应程序为95℃ 30 s;95℃ 5 s,60℃ 34 s(40个循环);72℃ 10 s。每个样品进行3次重复,不同样品在实时荧光定量PCR仪获得相应的Ct值,经内参基因Ubiquitin均一化处理后,采用2-ΔΔCt法计算基因的相对表达量[18]。Table 1
表1
表1实时荧光定量PCR所用特异性引物
Table 1
| 基因 Gene | 引物 Primer (5’-3’) | 扩增产物大小Amplicon size (bp) |
|---|---|---|
| VvKEA1 | F: GTCAATGGCACTAACCCCCT R: TCATAAAAATGATGTACCTGAC | 171 |
| VvKEA2 | F: GGTACTGTTGCGGTGGTTCT R:TCATGAGAGCCTTGGACCATC | 114 |
| VvKEA3 | F: GGTGACGACTCCTGTCCTGT R: ACGGAAGCTCTCTCCTCATTC | 111 |
| VvKEA4 | F: GACTGATGAAGAAGCTTCCCT R: ACCAGCAGCCAAGTAACCAA | 121 |
| Ubiquitin | F: CCTCATCTTCGCTGGCAAAC R: GGTGTAGGTCTTCTTCTTGCG | 133 |
新窗口打开|下载CSV
1.6 数据显著性分析
所有数据通过SPSS 13.0(SPSS Chicago,美国)软件进行显著性分析,即葡萄幼苗在处理条件与对照条件下2个独立样品间进行t-检验(检验水平0.01<*P<0.05, **P<0.01)。2 结果
2.1 葡萄KEA家族基因的克隆
以6个拟南芥KEA家族基因的氨基酸序列为参考序列,在Phytozome grape genome database(http:// www.phytozome.net)葡萄基因组数据库中检索到4个KEA家族基因,检索结果在Pfam在线服务器预测到K/H交换结构域(K/H exchanger domain,PF00999)和TrkA-N domain(PF02254)功能结构域,均属于典型的KEA家族蛋白。检索获得各基因的电子序列后,以CDS序列始密码子ATG和终止密码子TAG所在位置22 bp左右的序列作为上下游引物,以‘白罗莎里奥’葡萄幼苗叶片RNA反转录获得的cDNA为模板,PCR扩增各个候选基因的CDS序列,分别连接到pGEM-T载体,转化大肠杆菌感受态菌株DH5α,筛选阳性转化子送上海生工公司测序。经测序验证后,获得‘白罗莎里奥’葡萄的KEA家族各基因的CDS序列及相应翻译得到的氨基酸序列,分别命名为VvKEA1—VvKEA4,提交NCBI GenBank获得登录号(表2)。Table 2
表2
表2葡萄KEA家族成员基本信息
Table 2
| 基因 Gene | GenBank 登录号 GenBank No. | 染色体定位 Chromosome location | 内含子数目 Intron No. | 基序数目 Motif No. | 亚族 Group | 等电点 pI | 跨膜区Transmemberane regions | 不稳定性指数 Instability index | 信号肽 Signal peptide |
|---|---|---|---|---|---|---|---|---|---|
| VvKEA1 | MH118299 | Chr.15:19993952..20009808 forward | 17 | 4 | Ⅰ | 6.60 | 13 | 33.27稳定Stable | 无No |
| VvKEA2 | MH118298 | Chr.Un:21024004..21038616 forward | 13 | 4 | Ⅰ | 6.37 | 13 | 32.04稳定Stable | 无No |
| VvKEA3 | MH118297 | Chr.16:14444939..14477574 reverse | 19 | 7 | Ⅱ | 5.83 | 12 | 28.87稳定Stable | 有,第1—26氨基酸 Yes, amino acids of 1-26 |
| VvKEA4 | MH118300 | Chr.11:1922070..1943368 forward | 19 | 7 | Ⅱ | 6.32 | 12 | 29.59稳定Stable | 无No |
新窗口打开|下载CSV
2.2 9种不同科属植物KEA家族成员的系统发育树
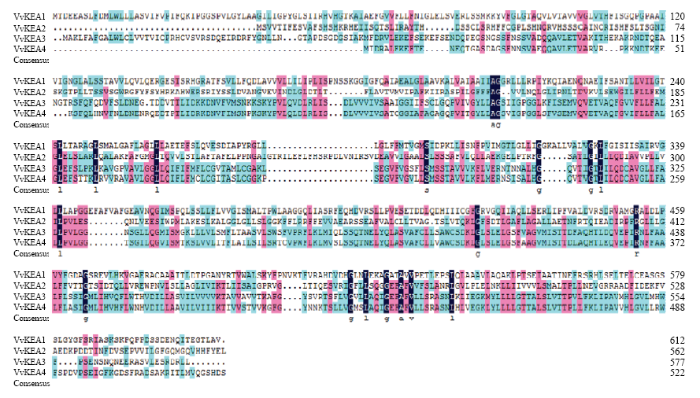
将葡萄(葡萄科)、拟南芥(十字花科)、水稻(禾本科)、玉米(禾本科)、高粱(禾本科)、短柄草(禾本科)、白杨(杨柳科)、梨(蔷薇科)和苹果(蔷薇科)9种不同科属的物种的KEA家族基因(表3),通过ClustalX 2.0进行氨基酸水平的多重序列比对。结果表明,供试物种的KEA家族基因之间具有较高的同源性,同源关系较近的两者之间的序列一致性均高于70%(数据未展示);9种植物56个KEA家族成员在氨基酸水平依然具有28.30%的一致性,在核苷酸水平具有33.10%的一致性(数据未展示);4个葡萄KEA家族成员在氨基酸水平具有42.20%的一致性(图1),在核苷酸水平具有57.03%的一致性(数据未展示)。Table 3
表3
表39种植物KEA家族蛋白信息
Table 3
| 物种 Species | 蛋白 Protein | GenBank登录号 GenBank No. | 编码区 CDS (bp) | 氨基酸数目 Amino acid No. | 分子量 Molecular weight (kD) |
|---|---|---|---|---|---|
| 葡萄(4) Vitis vinifera (4) | VvKEA1 | MH118299 | 1836 | 612 | 64.50 |
| VvKEA2 | MH118298 | 1689 | 563 | 61.41 | |
| VvKEA3 | MH118297 | 1731 | 577 | 62.48 | |
| VvKEA4 | MH118300 | 1569 | 522 | 56.21 | |
| 拟南芥(6) Arabidopsis thaliana (6) | AtKEA1 | At1g01790 | 3582 | 1194 | 128.03 |
| AtKEA2 | At4g00630 | 3525 | 1175 | 126.15 | |
| AtKEA3 | At4g04850 | 1914 | 638 | 69.04 | |
| AtKEA4 | At2g19600 | 1779 | 593 | 64.24 | |
| AtKEA5 | At5g51710 | 1707 | 569 | 61.59 | |
| AtKEA6 | At5g11800 | 1794 | 598 | 64.39 | |
| 水稻(6) Oryza sativa (6) | OsKEA1 | Os06g36590 | 1884 | 628 | 66.21 |
| OsKEA2 | Os05g31730 | 1362 | 453 | 46.15 | |
| OsKEA3 | Os08g43690 | 2454 | 817 | 87.82 | |
| OsKEA4 | Os09g37300 | 2484 | 827 | 88.97 | |
| OsKEA5 | Os04g58620 | 3465 | 1154 | 123.82 | |
| OsKEA6 | Os12g42300 | 2376 | 792 | 85.34 | |
| 玉米(8) Zea mays (8) | ZmKEA1 | GRMZM2G169114 | 1848 | 616 | 65.62 |
| ZmKEA2 | GRMZM2G171031 | 1872 | 624 | 66.03 | |
| ZmKEA3 | GRMZM2G020610 | 2232 | 744 | 80.54 | |
| ZmKEA4 | GRMZM2G430755 | 2460 | 820 | 88.35 | |
| ZmKEA5 | GRMZM2G105219 | 2424 | 808 | 86.86 | |
| ZmKEA6 | GRMZM2G058948 | 2529 | 843 | 90.14 | |
| ZmKEA7 | GRMZM2G093643 | 3099 | 1033 | 110.83 | |
| ZmKEA8 | GRMZM2G009715 | 1941 | 647 | 69.76 | |
| 高粱(7) Sorghum bicolor (7) | SbKEA1 | Sb01g048670 | 1734 | 578 | 61.85 |
| SbKEA2 | Sb10g022170 | 1860 | 620 | 65.76 | |
| SbKEA3 | Sb09g018910 | 1398 | 466 | 47.28 | |
| SbKEA4 | Sb07g024760 | 2454 | 818 | 88.35 | |
| SbKEA5 | Sb02g031690 | 2661 | 887 | 95.82 | |
| SbKEA6 | Sb08g021840 | 2340 | 780 | 83.76 | |
| SbKEA7 | Sb06g033310 | 3504 | 1168 | 124.64 | |
| 短柄草(5) Brachypodium distachyon (5) | BdKEA1 | Bradi1g37860 | 1830 | 610 | 65.10 |
| BdKEA2 | Bradi2g26740 | 1368 | 456 | 46.46 | |
| BdKEA3 | Bradi3g42260 | 2508 | 836 | 88.82 | |
| BdKEA4 | Bradi5g26820 | 3492 | 1164 | 123.70 | |
| BdKEA5 | Bradi4g01430 | 2337 | 779 | 83.60 | |
| 续表3 Continued table 3 | |||||
| 物种 Species | 蛋白 Protein | GenBank登录号 GenBank No. | 编码区 CDS (bp) | 氨基酸数目 Amino acid No. | 分子量 Molecular weight (kD) |
| 白杨(4) Populus trichocarpa (4) | PtKEA1 | Potri.002G157200.1 | 3645 | 1215 | 87.83 |
| PtKEA2 | Potri.014G080800.1 | 3648 | 1216 | 130.80 | |
| PtKEA3 | Potri.009G080800.1 | 2469 | 823 | 89.30 | |
| PtKEA4 | Potri.012G130500.1 | 1752 | 584 | 64.05 | |
| 梨(12) Pyrus bretschneideri (12) | PbKEA1 | Pbr001286.1 | 3678 | 1226 | 132.42 |
| PbKEA2 | Pbr004889.1 | 2229 | 743 | 80.16 | |
| PbKEA3 | Pbr007039.1 | 2298 | 766 | 82.83 | |
| PbKEA4 | Pbr009904.1 | 2301 | 767 | 82.91 | |
| PbKEA5 | Pbr015588.2 | 1674 | 558 | 60.26 | |
| PbKEA6 | Pbr019828.1 | 1767 | 589 | 62.99 | |
| PbKEA7 | Pbr031450.1 | 3183 | 1061 | 114.86 | |
| PbKEA8 | Pbr036088.1 | 2400 | 800 | 86.68 | |
| PbKEA9 | Pbr036420.2 | 1743 | 581 | 62.20 | |
| PbKEA10 | Pbr038070.1 | 1164 | 388 | 41.77 | |
| PbKEA11 | Pbr038477.1 | 2244 | 748 | 81.11 | |
| PbKEA12 | Pbr041608.1 | 2514 | 838 | 90.54 | |
| 苹果(7) Malus domestica (7) | MdKEA1 | MDP0000144878 | 2346 | 782 | 121.52 |
| MdKEA2 | MDP0000165222 | 3390 | 1130 | 134.60 | |
| MdKEA3 | MDP0000173207 | 2949 | 983 | 106.80 | |
| MdKEA4 | MDP0000206618 | 3009 | 1003 | 109.76 | |
| MdKEA5 | MDP0000244586 | 3726 | 1242 | 85.30 | |
| MdKEA6 | MDP0000818940 | 1743 | 581 | 41.59 | |
| MdKEA7 | MDP0000903895 | 1173 | 391 | 62.53 | |
新窗口打开|下载CSV
图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1葡萄KEA氨基酸序列一致性分析
Fig. 1Identify analysis of grape KEA family proteins
利用MEGA 7.0建立系统进化树,结果表明九种植物KEA家族基因在遗传进化关系上有差异,葡萄和拟南芥2种双子叶植物遗传距离上是较近的,葡萄VvKEA1、VvKEA2和VvKEA 4分别与拟南芥AtKEA2、AtKEA3和AtKEA 5紧密聚在一起,而葡萄VvKEA3则与梨PbrKEA5和苹果MdoKEA7紧密聚在一起(图2);水稻、玉米、高粱和短柄草4种禾本科植物KEA家族的相关成员,更倾向于聚在一起,而木本植物白杨、苹果、梨KEA家族更倾向于聚在一起,特别是同属蔷薇科的苹果和梨,遗传距离上更为相近(图2)。此外,9种植物KEA家族成员可分为2个亚族,GroupⅠ和Ⅱ,其中,葡萄VvKEA1和VvKEA2属于GroupⅠ,而VvKEA3和VvKEA4属于GroupⅡ(图2和表2)。
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图29种植物KEA家族蛋白的系统进化树
红色圆点标注为葡萄KEA家族蛋白,红色线条表明亚族GroupⅠ和Ⅱ的分界线
Fig. 2The phylogenetic tree of KEA family proteins from nine plants
Grape KEA family genes were highlight with red circle, and the red line indicated the boundary between GroupⅠandⅡ
2.3 葡萄KEA基因定位、编码蛋白特征与Motif分析
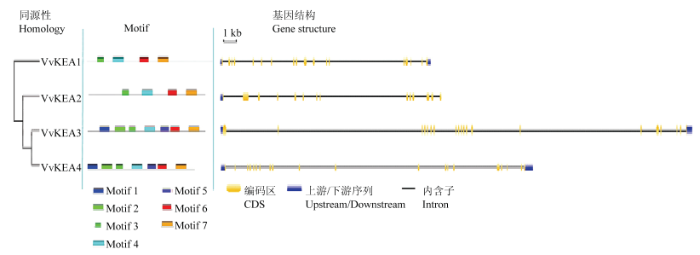
葡萄KEA家族基因定位于不同染色体上(VvKEA2具体染色体未知),含有13—19个长度不一的内含子(图3和表2);其中,VvKEA1基因CDS编码区最长,其次是VvKEA3和VvKEA2,VvKEA4最短,其编码氨基酸数目和分子量与CDS长度成正比(表2);葡萄KEA家族蛋白的等电点pI均<7.00,表明其含有的酸性氨基酸较多;葡萄KEA家族蛋白均含有12—13个跨膜区,均为稳定蛋白,且只有VvKEA3具有信号肽,位于第1—26氨基酸区域(表2)。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3葡萄KEA家族成员Motif和基因结构分析
Fig. 3Motif and gene structure analysis of grape KEA family members
保守基序分析结果表明葡萄VvKEA3和VvKEA4蛋白均含有7个Motif基序,即Motif1—Motif7,而VvKEA1和VvKEA2均只含有4个Motif基序,即Motif3、Motif4、Motif6和Motif7,缺少Motif1、Motif2和Motif 5(图3和表2);其中,Motif2、Motif3和Motif5最长,均含有50个特征氨基酸序列,而Motif6最短,含有31个氨基酸序列(图4)。蛋白三级结构预测分析表明所有葡萄KEA家族蛋白拥有相似的三级结构(图5),暗示该家族基因可能拥有相近的功能。
图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4不同Motif的特征序列及长度
Fig. 4Distinct characteristic sequence and length of Motif
图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5葡萄KEA家族蛋白三级结构预测
Fig. 5Tertiary structure prediction of grape KEA family members
2.4 葡萄KEA基因亚细胞定位预测及表达谱分析
亚细胞定位预测结果表明葡萄KEA家族基因定位趋势一致,均主要定位于细胞质膜,其次是内质网膜(VvKEA3除外)和液泡膜(VvKEA1除外)(表4)。EST表达谱分析结果表明,葡萄KEA家族基因可能在果实中的表达水平最高,其次是叶、种子、根和雌蕊,在花、果皮、花粉和木质部中亦有表达(图6)。Table 4
表4
表4葡萄KEA家族基因的亚细胞定位预测
Table 4
| 基因 Gene | 细胞质膜 Plasma membrane (%) | 内质网膜 Endoplasmic reticulum membrane (%) | 液泡膜 Vacuole membrane (%) |
|---|---|---|---|
| VvKEA1 | 85.72 | 14.28 | - |
| VvKEA2 | 85.72 | 7.14 | 7.14 |
| VvKEA3 | 92.86 | - | 7.14 |
| VvKEA4 | 78.58 | 7.14 | 14.28 |
新窗口打开|下载CSV
图6
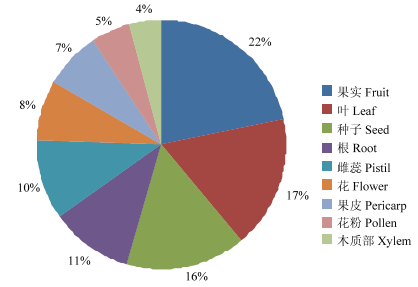 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6葡萄KEA家族基因的EST表达谱分析
Fig. 6Expression profiles analysis of EST sequence from grape KEA family genes
2.5 葡萄KEA基因启动子顺式作用元件预测
启动子区域cis-顺式作用元件预测结果表明,葡萄KEA家族基因启动子区域鉴定到至少15种的顺式作用元件,包括胁迫响应、营养和发育、激素响应、昼夜规律等不同生命活动相关的调控元件,且在不同VvKEA中数量差异显著(表5)。其中,5种(光感应、胚乳特异表达、水杨酸响应、热胁迫响应、防御和胁迫响应)转录因子在全部4个VvKEA的启动子中均能预测到,赤霉素响应作用元件在3个基因的启动子区域能检测到(除VvKEA3),厌氧感应、昼夜规律、茉莉酮酸甲酯、玉米蛋白代谢、乙烯响应和干旱响应相关的顺式作用元件均能在不同的2个VvKEA的启动子区域鉴定到,此外,脱落酸响应(VvKEA3)、低温感应(VvKEA3)和真菌激发子(VvKEA4)分别在一个VvKEA的启动子区域出现(表5)。Table 5
表5
表5葡萄KEA家族基因启动子顺式作用元件分析
Table 5
| 顺式作用元件 cis- regulatory elements | VvKEA1 | VvKEA2 | VvKEA3 | VvKEA4 |
|---|---|---|---|---|
| 光感应Light | 19 | 17 | 24 | 22 |
| 胚乳表达 Endosperm expression | 4 | 8 | 2 | 5 |
| 水杨酸响应Salicylic acid | 3 | 2 | 2 | 1 |
| 热胁迫Heat stress | 1 | 2 | 3 | 3 |
| 防御与胁迫 Defense and stress | 2 | 1 | 1 | 2 |
| 赤霉素响应Gibberellin | 2 | 3 | - | 2 |
| 厌氧感应 Anaerobic induction | - | 2 | - | 2 |
| 昼夜规律Circadian | 1 | - | - | 2 |
| 茉莉酮酸甲酯 Methyl jasmonate | - | - | 4 | 2 |
| 玉米蛋白代谢 Zein metabolism | 1 | 2 | - | - |
| 乙烯响应Ethylene | - | 1 | 1 | - |
| 干旱感应 Drought-inducibility | 1 | - | - | 2 |
| 脱落酸响应Abscisic acid | - | - | 4 | - |
| 低温感应Low temperature | - | - | 1 | - |
| 真菌激发子Fungal elicitor | - | - | - | 1 |
新窗口打开|下载CSV
2.6 葡萄KEA家族基因表达特征分析
通过实时荧光定量PCR分析,VvKEA1—VvKEA4在8年生‘白罗莎里奥’葡萄不同组织中的表达量有差异,VvKEA3在不同组织中的整体表达水平最高(图7),其他3个基因在葡萄不同组织中的表达水平较为接近,均显著低于VvKEA3;在不同组织中,VvKEA1在花器官中的表达量最高,VvKEA2和VvKEA3在幼果中的表达量最高,VvKEA4在果实和叶片中的表达量较高,其中,VvKEA3在幼果中的表达水平最高,约为其他3个基因的4—10倍(图7),暗示其可能在幼果发育过程中发挥作用。图7
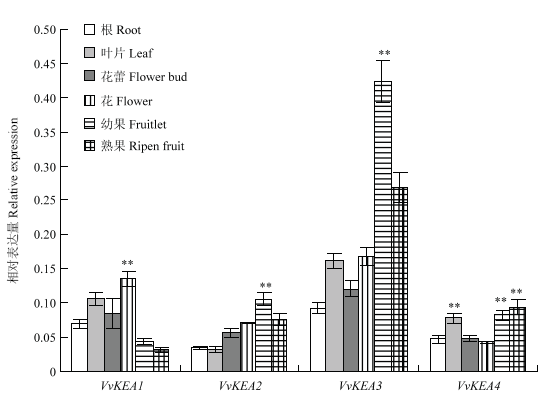 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7荧光定量RT-PCR检测VvKEA不同组织部位的表达特征
**表示在P<0.01水平差异极显著。下同
Fig. 7Quantitative RT-PCR expression analysis of VvKEA gene in different tissues
** indicates statistically extremely significant differences at P<0.01. The same as below
2.7 幼苗中葡萄KEA家族基因对胁迫处理的响应
幼苗中,VvKEA1—VvKEA4在转录水平对不同胁迫处理的响应有差异:VvKEA1—VvKEA4对ABA处理最为敏感,其表达水平在检测的地上部和根部均被显著诱导;VvKEA1在地上部及VvKEA3在地上部和根部的表达水平对缺钾处理较为敏感,其表达水平均显著被诱导;VvKEA3在地上部和根部及VvKEA4在根部对sorbitol渗透处理较为敏感,其表达水平显著被诱导;此外,VvKEA1—VvKEA4在转录水平对NaCl处理没有响应,其表达水平没有显著差异(图8)。图8
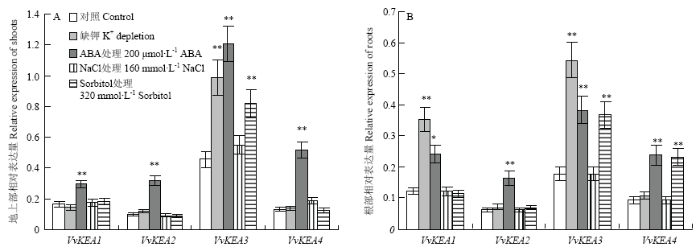 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8幼苗中VvKEA对缺钾、ABA、NaCl和sorbitol处理的响应
A:地上部相对表达量;B:根部相对表达量。*表示在P<0.05水平差异显著
Fig. 8Response of VvKEA gene under K+ depletion, ABA, NaCl and sorbitol stresses in strawberry seedlings
A: Relative expression in shoots; B: Relative expression in roots. * indicates statistically significant differences at P<0.05
3 讨论
果园中,适量的钾肥施用有助于果树发育、开花和果实品质形成[19,20,21,22],相关报道主要集中在生理生化层面,钾素营养分子基础的研究报道鲜少。植物中,KEA转运蛋白是一类维持体内K+动态平衡并参与叶绿体功能、光合作用和渗透调节作用的K+/H+逆向转运体,相关报道主要集中在模式作物拟南芥[8, 13-15]。果树中有关K+/H+动态平衡的研究依然是未知的。近年来,植物基因组测序技术迅速发展,为植物科学研究提供了丰富的基因资源。不同物种之间,同一家族基因成员之间数目差异显著。本研究在葡萄中克隆并鉴定了4个VvKEA转运体基因,与白杨(4)和草莓(5)中KEA成员数目接近,而远低于蔷薇科果树梨(12)和苹果(7)。此外,不同物种KEA家族成员在遗传进化关系上存在差异,2种双子叶植物(葡萄和拟南芥)KEA成员在遗传距离上是较近的,4种禾本科植物(水稻、玉米、高粱和短柄草)KEA成员更倾向于聚在一起,而3种木本植物(白杨、苹果和梨)KEA成员更倾向于聚在一起(图2),反映了不同物种在长期进化中依然是保守的,遗传距离也是较近的。在4个葡萄KEA转运体中,VvKEA1和VvKEA2同属于GroupⅠ,且具有6个相似的Motif,而VvKEA3和VvKEA4同属于GroupⅡ,具有7个相似的Motif(图2和图3),这些结果表明,进化关系上相近的(同一亚族)且具有类似蛋白Motif基序的两个转运体之间可能具有相似的功能。
前人研究表明拟南芥AtKEA1—AtKEA3主要定位于细胞质膜上,其中AtKEA1和AtKEA2定位于叶绿体膜上,AtKEA3定位于类囊体膜上[13,15],亚细胞定位预测结果表明葡萄VvKEA1—VvKEA4主要在细胞质膜上(表4),与KUNZ等[15]报道一致,需要进一步的试验验证。HAN等[11]报道指出拟南芥AtKEA1和AtKEA3主要在地上部表达,而其他4个成员基因在植物全身均有表达,与之情况一致的是,EST表达谱表明葡萄KEA家族基因在葡萄多种组织中均有表达,其中,在果实中的表达水平最高(图6);荧光定量PCR验证结果与表达谱分析是一致的,即在8年生‘白罗莎里奥’葡萄树不同组织中,3个基因(VvKEA2、VvKEA3和VvKEA4)的表达量均在果实中是最高的,特别是VvKEA3,其表达量在所有基因、不同检测组织中是最高的,暗示VvKEA3可能在果实发育中(特别是幼果时期)发挥重要作用的K+/H+逆向转运体。
植物KEA转运体在K+动态平衡、渗透调节、光合作用及pH调控中起主要作用[12,13,14,15]。荧光定量PCR结果表明葡萄幼苗中VvKEA1—VvKEA4在转录水平的表达量易受缺钾、ABA、NaCl或sorbitol等胁迫的诱导而上调,对不同胁迫处理的响应有差异(图8),其中作为葡萄KEA家族基因中整体表达量最为丰富的基因,VvKEA3对胁迫处理最为敏感,其表达水平极易受缺钾、ABA和sorbitol处理的影响而显著增强,与拟南芥[13]和大豆[23]中报道的现象类似;此外,CHEN等[23]报道表明大豆KEA基因易受NaCl处理诱导表达,然而,本研究中VvKEA1—VvKEA4对NaCl处理没有响应,说明禾本科植物与木本科植物同源家族基因的功能或调控机制方面可能存在差异;尽管VvKEA3与VvKEA4对NaCl处理没有响应,但对等渗作用的sorbitol处理较为敏感,直接表明葡萄VvKEA3与VvKEA4易受渗透作用的胁迫诱导,而不受盐离子作用(Na+),这些发现与拟南芥中AtKEA2和AtKEA5的报道是一致的[13]。
此外,cis-顺式作用元件可通过与启动子区域的关键元件结合来控制启动子效率,进而调控靶标基因的表达[24,25,26]。本文结果发现,葡萄KEA家族基因启动子区域鉴定到多种顺式作用元件(表5),特别地,VvKEA3启动子区域含有脱落酸(ABA)响应的作用元件,而该基因的表达水平在葡萄幼苗全身组织中是受ABA处理诱导的;VvKEA1—VvKEA4启动子区域均含有光感应、胚乳特异表达、水杨酸响应、热胁迫响应及防御与胁迫相关的作用元件,这些发现与同为钾离子转运体的KT/HAK/KU家族转运体的报道是类似的[23,24,25],也为进一步研究葡萄KEA转运体的功能及其调控机理提供了理论支持。
本文为研究果树K+/H+平衡及钾素动态平衡机制提供了分子基础,并为高效园艺作物的遗传改良与育种奠定了理论依据。
4 结论
从葡萄中克隆并鉴定了4个KEA家族基因,该家族基因主要在葡萄果实、叶片和种子中表达;葡萄KEA成员与拟南芥KEA成员同源性较高;VvKEA3在成年葡萄树中的整体表达水平最为丰富(幼果中最高)。幼苗中VvKEA3在转录水平受缺钾、ABA和sorbitol渗透胁迫的调控;VvKEA3是葡萄果实中重要的K+/H+逆向转运体。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1146/annurev.arplant.54.031902.134831URL [本文引用: 1]
DOI:10.1163/016942409X12598231567862URLPMID:3243584 [本文引用: 1]

61 Background and Aims Potassium transporters belonging to the KT/KUP/HAK family are important for various aspects of plant life including mineral nutrition and the regulation of development. Genes encoding these transporters are present in the genomes of all plants, but have not been found in the genomes of Protista or Animalia. The aim of this Botanical Briefing is to analyse the function of KT/KUP/HAK transporters from evolutionary, molecular and physiological perspectives. 61 Scope This Briefing covers the phylogeny and evolution of KT/KUP/HAK transporters, the role of transporters in plant mineral nutrition and potassium homeostasis, and the role of KT/KUP/HAK transporters in plant development.
DOI:10.1146/annurev.arplant.52.1.817URL [本文引用: 1]
DOI:10.1055/s-2001-19372URL [本文引用: 1]

Abstract: Since water spontaneously ionizes, protons cannot be removed from the medium: their free concentration in cells must be regulated through actively controlling H + -related transport across membranes, by active and passive buffering, and by setting a certain pH within the metabolic network. Whereas these are the basic tools that provide effective H + homeostasis, cellular compartmentation serves as an intermediate store into which protons can be shifted temporarily and from which protons can be regained when required. On the other hand, intracellular compartments can also serve as a final proton sink. pH regulation is not confined to intracellular spaces, but also comprises the apoplast. Whereas the pH of the cytosol is kept slightly alkaline at 7.2 to 7.5, with an average buffer capacity of 20 to 80 mM H + per pH unit, the apoplastic pH may vary among tissues but is always acidic, with values between pH 5 and 6 and with a buffer capacity in the lower millimolar range per pH unit. pH can be a signal and/or a messenger, a distinction not always clearly made. Here, "signal" should be understood as information about an ongoing or preceding process, whereas "messenger" would be the carrying of certain information that will lead to a change of state. As such, pH would signal light intensity changes, drought, lack of oxygen and the presence of symbiotic partners or microbial attackers. On the other hand, pH would be a messenger in situations where pH changes are preconditions for certain processes, e.g., the gravity response or for activation of certain transporters in stomatal movements, and possibly for growth. The function of pH as a cellular messenger raises the question of whether pH should be understood as a "second messenger" in the way this is done for Ca 2+ . In an effort to give a comprehensive answer to this problem, the different roles of Ca 2+ and H + in cellular signalling are discussed and a number of Ca 2+ /pH interactions are presented.
DOI:10.1016/S1360-1385(02)02240-9URL [本文引用: 1]
DOI:10.1093/aob/mci207URLPMID:4247022 [本文引用: 1]

Abstract BACKGROUND: pH regulation is the result of a complex interaction of ion transport, H+ buffering, H+-consuming and H+-producing reactions. Cells under anoxia experience an energy crisis; an early response thereof (in most tissues) is a rapid cytoplasmic acidification of roughly half a pH unit. Depending on the degree of anoxia tolerance, this pH remains relatively stable for some time, but then drops further due to an energy shortage, which, in concert with a general breakdown of transmembrane gradients, finally leads to cell death unless the plant finds access to an energy source. SCOPE: In this review the much-debated origin of the initial pH change and its regulation under anoxia is discussed, as well as the problem of how tissues deal with the energy crisis and to what extent pH regulation and membrane transport from and into the vacuole and the apoplast is a part thereof. CONCLUSIONS: It is postulated that, because a foremost goal of cells under anoxia must be energy production (having an anaerobic machinery that produces insufficient amounts of ATP), a new pH is set to ensure a proper functioning of the involved enzymes. Thus, the anoxic pH is not experienced as an error signal and is therefore not reversed to the aerobic level. Although acclimated and anoxia-tolerant tissues may display higher cytoplasmic pH than non-acclimated or anoxia-intolerant tissues, evidence for an impeded pH-regulation is missing even in the anoxia-intolerant tissues. For sufficient energy production, residual H+ pumping is vital to cope with anoxia by importing energy-rich compounds; however it is not vital for pH-regulation. Whereas the initial acidification is not due to energy shortage, subsequent uncontrolled acidosis occurring in concert with a general gradient breakdown damages the cell but may not be the primary event.
DOI:10.1074/jbc.M111.252650URL [本文引用: 3]
DOI:10.3389/fpls.2012.00025URLPMID:3355601 [本文引用: 4]

All organisms have evolved strategies to regulate ion and pH homeostasis in response to developmental and environmental cues. One strategy is mediated by monovalent cation–proton antiporters (CPA) that are classified in two superfamilies. Many CPA1 genes from bacteria, fungi, metazoa, and plants have been functionally characterized; though roles of plant CPA2 genes encoding K+-efflux antiporter (KEA) and cation/H+ exchanger (CHX) families are largely unknown. Phylogenetic analysis showed that three clades of the CPA1 Na+–H+ exchanger (NHX) family have been conserved from single-celled algae to Arabidopsis. These are (i) plasma membrane-bound SOS1/AtNHX7 that share ancestry with prokaryote NhaP, (ii) endosomal AtNHX5/6 that is part of the eukaryote Intracellular-NHE clade, and (iii) a vacuolar NHX clade (AtNHX1–4) specific to plants. Early diversification of KEA genes possibly from an ancestral cyanobacterium gene is suggested by three types seen in all plants. Intriguingly, CHX genes diversified from three to four members in one subclade of early land plants to 28 genes in eight subclades of Arabidopsis. Homologs from Spirogyra or Physcomitrella share high similarity with AtCHX20, suggesting that guard cell-specific AtCHX20 and its closest relatives are founders of the family, and pollen-expressed CHX genes appeared later in monocots and early eudicots. AtCHX proteins mediate K+ transport and pH homeostasis, and have been localized to intracellular and plasma membrane. Thus KEA genes are conserved from green algae to angiosperms, and their presence in red algae and secondary endosymbionts suggest a role in plastids. In contrast, AtNHX1–4 subtype evolved in plant cells to handle ion homeostasis of vacuoles. The great diversity of CHX genes in land plants compared to metazoa, fungi, or algae would imply a significant role of ion and pH homeostasis at dynamic endomembranes in the vegetative and reproductive success of flowering plants.
DOI:10.1016/j.pbi.2014.08.002URLPMID:25173972 [本文引用: 2]

The biochemical characterization of cation/H+ exchange has been known since 1985 [1], yet only recently have we begun to understand the contribution of individual exchangers to ion homeostasis in plants. One particularly important class of exchangers is the NHX-type that is associated with Na+ transport and therefore salinity tolerance. New evidence suggests that under normal growth conditions NHXs are critical regulators of K+ and pH homeostasis and have important roles, depending on their cellular localization, in the generation of turgor as well as in vesicular trafficking. Recent advances highlight novel and exciting functions of intracellular NHXs in growth and development, stress adaptation and osmotic adjustment. Here, we elaborate on new and emerging cellular and physiological functions of this group of H+-coupled cation exchangers.
DOI:10.1104/pp.126.4.1646URLPMID:11500563 [本文引用: 1]

Uptake and translocation of cationic nutrients play essential roles in physiological processes including plant growth, nutrition, signal transduction, and development. Approximately 5% of the Arabidopsis genome appears to encode membrane transport proteins. These proteins are classified in 46 unique families containing approximately 880 members. In addition, several hundred putative transporters have not yet been assigned to families. In this paper, we have analyzed the phylogenetic relationships of over 150 cation transport proteins. This analysis has focused on cation transporter gene families for which initial characterizations have been achieved for individual members, including potassium transporters and channels, sodium transporters, calcium antiporters, cyclic nucleotide-gated channels, cation diffusion facilitator proteins, natural resistance-associated macrophage proteins (NRAMP), and Zn-regulated transporter Fe-regulated transporter-like proteins. Phylogenetic trees of each family define the evolutionary relationships of the members to each other. These families contain numerous members, indicating diverse functions in vivo. Closely related isoforms and separate subfamilies exist within many of these gene families, indicating possible redundancies and specialized functions. To facilitate their further study, the PlantsT database (http://plantst.sdsc.edu) has been created that includes alignments of the analyzed cation transporters and their chromosomal locations.
DOI:10.1007/s11738-015-1845-4URL [本文引用: 2]

The functions of the Arabidopsis K + /H + antiporters (KEA) have only been partially determined, and further assessments of their biological roles are needed. In this report, we provide localization and expression analyses of six members of the KEA gene family in Arabidopsis . Promoter-driven -glucuronidase activity analyses in transgenic Arabidopsis revealed that all KEAs were detected in the vascular tissues of the plants. In the roots, AtKEA2 , 4 , 5 , and 6 were mainly localized to the steles; in addition, AtKEA4 was also found in the root tips. In the leaves, AtKEA1 , 2 , and 3 were localized to the leaf veins and petioles, whereas AtKEA4 , 5 , and 6 were preferentially localized to the trichomes. Furthermore, AtKEA1 , 2 , 3 , 4 , and 5 were expressed in the guard cells, but AtKEA6 was not. In the florescent organs, all six AtKEA genes were detected in the sepals; interestingly, AtKEA1 was the only member found in the pollen grains. In accordance with promoter localization, quantitative RT-PCR analyses indicated that AtKEA1 and AtKEA3 were mainly expressed in the shoots, whereas AtKEA2 , 4 , 5 , and 6 were expressed in the entire plant. The expression levels of AtKEA transcripts were affected by K + deficiency and NaCl or osmotic stress Additionally, AtKEAs were regulated by 2,4-dichlorophenoxyacetic acid, benzyladenine, and sucrose. These results provide important information that will support future research on the function and localization of the putative K + /H + antiporters in Arabidopsis .
DOI:10.1016/j.bbamem.2012.04.011URLPMID:22551943 [本文引用: 3]

78 Plant KEA proteins share high homology to bacterial KefC/B antiporters. 78 Full‐length AtKEA2 is inactive in yeast. 78 AtKEA2 without N-terminal domain complements yeast Nhx1p. 78 Purified and reconstituted short AtKEA2 protein has K+/H+ exchange activity. 78 Cation specificity of AtKEA2 is similar to EcKefC.
DOI:10.1371/journal.pone.0081463URLPMID:24278440 [本文引用: 7]

Abstract AtKEAs, homologs of bacterial KefB/KefC, are predicted to encode K(+)/H(+) antiporters in Arabidopsis. The AtKEA family contains six genes forming two subgroups in the cladogram: AtKEA1-3 and AtKEA4-6. AtKEA1 and AtKEA2 have a long N-terminal domain; the full-length AtKEA1 was inactive in yeast. The transport activity was analyzed by expressing the AtKEA genes in yeast mutants lacking multiple ion carriers. AtKEAs conferred resistance to high K(+) and hygromycin B but not to salt and Li(+) stress. AtKEAs expressed in both the shoot and root of Arabidopsis. The expression of AtKEA1, -3 and -4 was enhanced under low K(+) stress, whereas AtKEA2 and AtKEA5 were induced by sorbitol and ABA treatments. However, osmotic induction of AtKEA2 and AtKEA5 was not observed in aba2-3 mutants, suggesting an ABA regulated mechanism for their osmotic response. AtKEAs' expression may not be regulated by the SOS pathway since their expression was not affected in sos mutants. The GFP tagging analysis showed that AtKEAs distributed diversely in yeast. The Golgi localization of AtKEA3 was demonstrated by both the stably transformed seedlings and the transient expression in protoplasts. Overall, AtKEAs expressed and localized diversely, and may play roles in K(+) homeostasis and osmotic adjustment in Arabidopsis.
DOI:10.1038/ncomms6439URLPMID:25451040 [本文引用: 2]

Abstract Many photosynthetic organisms globally, including crops, forests and algae, must grow in environments where the availability of light energy fluctuates dramatically. How photosynthesis maintains high efficiency despite such fluctuations in its energy source remains poorly understood. Here we show that Arabidopsis thaliana K(+) efflux antiporter (KEA3) is critical for high photosynthetic efficiency under fluctuating light. On a shift from dark to low light, or high to low light, kea3 mutants show prolonged dissipation of absorbed light energy as heat. KEA3 localizes to the thylakoid membrane, and allows proton efflux from the thylakoid lumen by proton/potassium antiport. KEA3's activity accelerates the downregulation of pH-dependent energy dissipation after transitions to low light, leading to faster recovery of high photosystem II quantum efficiency and increased CO2 assimilation. Our results reveal a mechanism that increases the efficiency of photosynthesis under fluctuating light.
DOI:10.1073/pnas.1323899111URL [本文引用: 5]

Multiple K(+) transporters and channels and the corresponding mutants have been described and studied in the plasma membrane and organelle membranes of plant cells. However, knowledge about the molecular identity of chloroplast K(+) transporters is limited. Potassium transport and a well-balanced K(+) homeostasis were suggested to play important roles in chloroplast function. Because no loss-of-function mutants have been identified, the importance of K(+) transporters for chloroplast function and photosynthesis remains to be determined. Here, we report single and higher-order loss-of-function mutants in members of the cation/proton antiporters-2 antiporter superfamily KEA1, KEA2, and KEA3. KEA1 and KEA2 proteins are targeted to the inner envelope membrane of chloroplasts, whereas KEA3 is targeted to the thylakoid membrane. Higher-order but not single mutants showed increasingly impaired photosynthesis along with pale green leaves and severely stunted growth. The pH component of the proton motive force across the thylakoid membrane was significantly decreased in the kea1kea2 mutants, but increased in the kea3 mutant, indicating an altered chloroplast pH homeostasis. Electron microscopy of kea1kea2 leaf cells revealed dramatically swollen chloroplasts with disrupted envelope membranes and reduced thylakoid membrane density. Unexpectedly, exogenous NaCl application reversed the observed phenotypes. Furthermore, the kea1kea2 background enables genetic analyses of the functional significance of other chloroplast transporters as exemplified here in kea1kea2Na(+)/H(+) antiporter1 (nhd1) triple mutants. Taken together, the presented data demonstrate a fundamental role of inner envelope KEA1 and KEA2 and thylakoid KEA3 transporters in chloroplast osmoregulation, integrity, and ion and pH homeostasis.
DOI:10.1007/s00438-016-1215-yURLPMID:27193473 [本文引用: 2]

Abstract The monovalent cation proton antiporters (CPAs) play essential roles in plant nutrition, development, and signal transduction by regulating ion and pH homeostasis of the cell. The CPAs of plants include the Na(+)/H(+) exchanger, K(+) efflux antiporter, and cation/H(+) exchanger families. However, currently, little is known about the CPA genes in Rosaceae species. In this study, 220 CPA genes were identified from five Rosaceae species (Pyrus bretschneideri, Malus domestica, Prunus persica, Fragaria vesca, and Prunus mume), and 53 of which came from P. bretschneideri. Phylogenetic, structure, collinearity, and gene expression analyses were conducted on the entire CPA genes of pear. Gene expression data showed that 35 and 37 CPA genes were expressed in pear fruit and pollen tubes, respectively. The transcript analysis of some CPA genes under abiotic stress conditions revealed that CPAs may play an important role in pollen tubes growth. The results presented here will be useful in improving understanding of the complexity of the CPA gene family and will promote functional characterization in future studies.
[本文引用: 1]
[本文引用: 1]
DOI:10.1006/meth.2001.1262URL [本文引用: 1]
DOI:10.1081/PLN-200042179URL [本文引用: 1]

The effect of different potassium (K) rates on yield, fruit quality, and chemical composition of greenhouse-grown melon (Cucumis melo L., cv. Galia) was investigated. The experiment was carried out in a greenhouse similar to that used by farmers in the coastal Mediterranean region of Turkey. Control and three rates of potassium (0, 200, 400, 600 mg L611) were applied to cv. Galia in a randomized complete block design with 3 replications. Leaf and fruit quality analyses were carried out and yield was determined. Different rates of potassium had no effect on yield. However, fruit number and fruit firmness were higher than the control at the 400 and 600 mg L611 rates. Total soluble solids were higher than the control at the 600 mg L611 rate. Average fruit weight was lower than the control at the 600 mg L611 rate. Treatments did not affect nitrogen (N), phosphorus (P), calcium (Ca), magnesium (Mg), and zinc (Zn) contents of leaves, but leaf K content was higher than the control at all three rates. Iron (Fe) content was lower than the control at the 600 mg L611 rate and manganese (Mn) content was lower at the 400 and 600 mg L611rates. The data from our study indicate that 300 mg L611 K in the root zone was sufficient for optimum yield of greenhouse-grown Galia melons. In terms of total yield, it is not necessary to apply higher rates of K. However, results show that it is possible to improve fruit quality by applying as much as 600 mg L611 additional K to the plants without a reduction in yield.
DOI:10.1016/j.agwat.2005.04.018URL [本文引用: 1]

The effects of four irrigation water salinities (0.25, 2.5, 5 and 10 dS m 611) and three potassium fertilizer levels (0, 5 and 10 mmol l 611) on yield and some quality parameters of a native Central Anatolian tomato species ( Lycopersicon esculentum) were investigated under greenhouse conditions. A fully randomized factorial experiment was conducted between 13th of June and 11th of October 2002 at the Faculty Agricultural Experimental Station of Ankara University. Yield, fruit quality, drainage water salinity and evapotranspiration data were collected. It was found that both salinity and potassium fertilizer levels affected fresh fruit yield. The yield decreased with increasing salinity starting at salinity level of 2.5 dS m 611 and continued to 10 dS m 611 treatment. The interaction between salinity and K levels was significant, p < 0.05, but the changes in yield did not exhibit any clear result to state that K + levels had a direct effect on salinity–induced yield decrease. Plant biomass was affected only by the salinity levels of the irrigation water: the biomass decreased with increasing salinity. Increasing salinity levels resulted in smaller fruit size, higher soluble solid content (SSC) and decreased the pH of the fruit juice. Water consumption decreased with increasing salinity. Increasing salinity levels led to a decrease in water use efficiency (WUE).
DOI:10.1007/s10658-005-2332-3URL [本文引用: 1]

Abstract The effect of K fertigation through subsurface irrigation lines on processing tomato (Lycopersicon esculentum Mill.) fruit yield and quality was evaluated in four field trials in California from 2002–04. Fields had exchangeable soil K between 0.48 to 0.85 cmol·kg–1, with high exchangeable Mg (10.6 to 13.7 cmol·kg–1) and a history of yellow shoulder (YS, a fruit color disorder) occurrence. K treatments evaluated included seasonal amount applied (0 to 800 kg·ha–1), fertigation method (continuous versus weekly), and timing (early, mid or late season); foliar K treatments were also included in the 2002 trial. In two fields total and marketable fruit yield were significantly increased by K fertigation, and fruit color improvements were observed in all trials. Among color parameters improved by K fertigation were YS incidence, blended color, and L*, chroma, and hue of the shoulder region of fruit. K fertigation did not affect fruit soluble solids concentration. Yield increased only with fertigation treatments initiated during early fruit set. The effects of fertigation method and rate were inconsistent. Foliar K application was ineffective in increasing either fruit yield or quality.
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/S2095-3119(14)60918-7URL [本文引用: 3]

Sodium toxicity and potassium insufficient are important factors affecting the growth and development of soybean in saline soil. As the capacity of plants to maintain a high cytosolic, K+/Na+ ratio is the key determinant of tolerance under salt stress. The aims of the present study were to identify and analyse expression patterns of the soybean K+ efflux antiporter (KEA) gene and Na+/H+ antiporter (NHX) gene family, and to explore their roles under abiotic stress. As a result, 12 soybean GmKEAs genes and 10 soybean GmNHXs genes were identified and analyzed from soybean genome. Interestingly, the novel soybean KEA gene Glyma16g32821 which encodes 11 transmembrane domains were extremely up-regulated and remained high level until 48 h in root after the excessive potassium treatment and lack of potassium treatment, respectively. The novel soybean NHX gene Glyma09g02130 which encodes 10 transmembrane domains were extremely up-regulated and remained high level until 48 h in root with NaCl stress. Imaging of subcellular locations of the two new Glyma16g32821-GFP and Glyma09g02130-GFP fusion proteins indicated all plasma membrane localizations of the two novel soybean genes. The 3D structures indicated that the two soybean novel proteins Glyma09g02130 (NHX) and Glyma16g32821 (KEA) all belong to the cation/hydrogen antiporter family.
DOI:10.1007/s00438-008-0377-7URLPMID:18810495 [本文引用: 2]

KT/HAK/KUP potassium transporter protein-encoding genes constitute a large family in the plant kingdom. The KT/HAK/KUP family is important for various physiological processes of plant life. In this study, we identified 27 potential KT/HAK/KUP family genes in rice ( Oryza sativa ) by database searching. Analysis of these KT/HAK/KUP family members identified three conserved motifs with unknown functions, and 11 15 trans -membrane segments, most of which are conserved. A total of 144 putative cis -elements were found in the 2kb upstream region of these genes, of which a Ca 2+ -responsive cis -element, two light-responsive cis -elements, and a circadian-regulated cis -element were identified in the majority of the members, suggesting regulation of these genes by these signals. A comprehensive expression analysis of these genes was performed using data from microarrays hybridized with RNA samples of 27 tissues covering the entire life cycle from three rice genotypes, Minghui 63, Zhenshan 97, and Shanyou 63. We identified preferential expression of two OsHAK genes in stamen at 1day before flowering compared with all the other tissues. OsHAK genes were also found to be differentially upregulated or downregulated in rice seedlings subjected to treatments with three hormones. These results would be very useful for elucidating the roles of these genes in growth, development, and stress response of the rice plant.
DOI:10.4238/2015.January.30.21URLPMID:25730015 [本文引用: 2]

The KT/HAK/KUP family members encoding high-affinity potassium (K(+)) transporters mediate K(+) transport across the plasma membranes of plant cells to maintain plant normal growth and metabolic activities. In this paper, we identified 16 potassium transporter genes in the peach (Prunus persica) using the Hidden Markov model scanning strategy and searching the peach genome database. Utilizing the Arabidopsis KT/HAK/KUP family as a reference, phylogenetic analysis indicates that the KT/HAK/KUP family in the peach can be classified into 3 groups. Genomic localization indicated that 16 KT/HAK/KUP family genes were well distributed on 7 scaffolds. Gene structure analysis showed that the KT/HAK/KUP family genes have 6-9 introns. In addition, all of the KT/HAK/KUP family members were hydrophobic proteins; they exhibited similar secondary structure patterns and homologous tertiary structures. Putative cis-elements involved in abiotic stress adaption, Ca(2+) response, light and circadian rhythm regulation, and seed development were observed in the promoters of the KT/HAK/KUP family genes. Subcellular localization prediction indicated that the KT/HAK/KUP members were mainly located in the plasma membrane. Expression levels of the KT/HAK/ KUP family genes were much higher in the fruit and flower than those in the other 7 tissues examined, indicating that the KT/HAK/KUP family genes may have important roles in K(+) uptake and transport, which mainly contribute to flower formation and fruit development in the peach.
DOI:10.1007/s11033-012-1700-2URLPMID:22711305 [本文引用: 1]

Abstract (HAK) transporter gene family constitutes the largest family that functions as potassium transporter in plant and is important for various cellular processes of plant life. In spite of their physiological importance, systematic analyses of ZmHAK genes have not yet been investigated. In this paper, we indicated the isolation and characterization of ZmHAK genes in whole-genome wide by using bioinformatics methods. A total of 27 members (ZmHAK1 mHAK27) of this family were identified in maize genome. ZmHAK genes were distributed in all the maize 10 chromosomes. These genes expanded in the maize genome partly due to tandem and segmental duplication events. Multiple alignment and motif display results revealed major maize ZmHAK proteins share all the three conserved domains. Phylogenetic analysis indicated -elements involved in Ca response, abiotic stress adaption, light and circadian rhythms regulation and seed development were observed in the promoters of ZmHAK genes. Expression data mining suggested maize ZmHAK genes have temporal and spatial expression pattern. In all, these results will provide molecular insights into the potassium transporter research in maize.
