 ,, 叶佐东, 范双旗, 陈金顶, 张静远, 朱梦娇, 赵明秋
,, 叶佐东, 范双旗, 陈金顶, 张静远, 朱梦娇, 赵明秋 ,华南农业大学兽医学院,广州 510642
,华南农业大学兽医学院,广州 510642Screen of MicroRNAs in Classical Swine Fever Virus-Infected PK-15 Cells and the Regulation of Virus Replication by miR-214
DENG ShaoFeng ,, YE ZuoDong, FAN ShuangQi, CHEN JinDing, ZHANG JingYuan, ZHU MengJiao, ZHAO MingQiu
,, YE ZuoDong, FAN ShuangQi, CHEN JinDing, ZHANG JingYuan, ZHU MengJiao, ZHAO MingQiu ,College of Veterinary Medicine, South China Agricultural University, Guangzhou 510642
,College of Veterinary Medicine, South China Agricultural University, Guangzhou 510642通讯作者:
责任编辑: 林鉴非
收稿日期:2018-04-18接受日期:2018-09-7网络出版日期:2018-11-01
| 基金资助: |
Received:2018-04-18Accepted:2018-09-7Online:2018-11-01

摘要
关键词:
Abstract
Keywords:
PDF (785KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
邓少锋, 叶佐东, 范双旗, 陈金顶, 张静远, 朱梦娇, 赵明秋. PK-15细胞中与CSFV感染相关的microRNAs筛选及 miR-214的功能研究[J]. 中国农业科学, 2018, 51(21): 4157-4168 doi:10.3864/j.issn.0578-1752.2018.21.014
DENG ShaoFeng, YE ZuoDong, FAN ShuangQi, CHEN JinDing, ZHANG JingYuan, ZHU MengJiao, ZHAO MingQiu.
0 引言
【研究意义】猪瘟(classical swine fever, CSF)是由猪瘟病毒(classical swine fever virus, CSFV)引起的猪的一种重大烈性传染性疾病[1],被世界动物卫生组织(Office International Des Epizooties,OIE)列为法定报告动物传染病之一[2],严重危害着我国养猪业的健康发展,造成重大经济损失。CSFV的致病机制十分复杂,因此,从不同角度深入研究其致病过程有利于促进对CSFV致病机制的认识。microRNA(miRNA)是一类非编码单链小分子RNA,通过与其靶mRNA分子的3′端非编码区域互补匹配抑制该mRNA分子的翻译,从而对靶蛋白表达进行调控[3]。miRNA在病毒致病性以及病毒与宿主的相互作用方面发挥重要作用,但是对CSFV相关的miRNA研究较少。本研究通过生物信息学等试验方法筛选出可能与猪瘟病毒相关的miRNA,并对其进行功能研究,从宿主细胞的角度研究猪瘟病毒与细胞之间相互作用的关系,为猪瘟病毒的致病机理提供理论依据。【前人研究进展】研究证实miRNA参与许多重要生物学过程,包括调控信号通路[4,5,6]、促进机体发育[7]、诱导细胞凋亡[8,9,10]、参与免疫反应[11,12,13]以及在病毒感染中发挥作用。研究表明,有些miRNA能够直接以病毒基因组为靶位,调节病毒的复制[14,15]。有些miRNA能介导宿主的抗病毒活性[16,17]。目前对于CSFV相关的miRNA研究较少,如miRNA-let-7c可下调CSFV的RNA复制水平[18]。作为与CSFV同属黄病毒科的HCV的相关miRNA的研究已经十分深入[14, 19-20],对研究CSFV相关miRNA的作用有很大指导意义。【本研究切入点】miRNA在病毒感染中发挥重要作用,但目前对于CSFV相关的miRNA研究较少,而作为与CSFV同属黄病毒科的HCV的研究已十分成熟,对研究CSFV相关miRNA的作用有很大指导意义。【拟解决的关键问题】通过生物信息学和试验方法筛选出可能与CSFV感染相关的miRNA及其功能研究,从宿主细胞的角度来研究CSFV与细胞相互作用的关系,为研究其致病机理提供有价值的科学资料。1 材料与方法
试验于2015年9月至2016年5月在华南农业大学兽医学院完成。1.1 试验材料
猪源肾脏细胞(PK-15)、CSFV石门株和大肠杆菌DH5α感受态细胞由华南农业大学兽医学院微生物学与免疫学教研室保存。pmirGLO Dual-Luciferase miRNA Target Expression Vector由华南农业大学动物科学学院王翀教授惠赠。1.2 试验方法
1.2.1 病毒接种和滴度测定 PK-15细胞传代于培养瓶中生长到80%—90%时,用PBS洗涤细胞2次,加入CSFV病毒液,置于含5% CO2的37℃培养箱中孵育4—6 h后,弃去病毒液,用PBS洗涤细胞2次,加入细胞维持培养基继续培养,72 h后收获病毒液,于-80℃保存备用。用无血清无抗生素的DMEM将上述所获得的CSFV病毒液分别做10倍连续梯度稀释至10-10,再分别将每个梯度的病毒液按0.1 mL/孔接种4孔细胞,同时设空白细胞对照。接种CSFV后继续培养5 d,取出细胞培养板进行间接免疫荧光检测,并计算CSFV感染滴度。
1.2.2 细胞总RNA的提取与检测 弃去细胞培养液,用PBS洗涤细胞2次,加1 mL的胰酶消化并收集细胞至1.5 mL离心管中,参考Omega公司的总RNA提取试剂盒说明书提取细胞总RNA,再经过1%琼脂糖凝胶电泳鉴定提取的总RNA的完整性,并用超微量分光光度计测定RNA的浓度和纯度。
1.2.3 制作miRNA表达谱芯片 miRNA表达谱芯片试验是由广州锐博公司完成。根据miRBase version 19.0数据库中326条猪miRNA合成探针,采用原位合成技术制作表达谱芯片。miRNA表达谱芯片制作完成后进行扫描。
1.2.4 荧光定量PCR检测基因的表达 分别采用相应的反应体系和条件,根据PrimeScript miRNA cDNA Synthesis Kit说明书和PrimeScript? RT Master Mix说明书进行反转录反应合成miRNA cDNA和病毒基因或蛋白基因cDNA。然后根据SYBR? Premix Ex Taq? II说明书进行qRT-PCR检测基因的表达。
1.2.5 生物信息学预测miRNA的靶标蛋白 根据miRNA表达谱芯片筛选出来CSFV感染PK-15后表达变化的miRNA,用Target Scan、RNA22、Pictar等生物信息学软件预测miRNA的靶蛋白。
1.2.6 重组质粒的构建 提取PK-15细胞总RNA并反转录为cDNA作为模板,以上游引物:5′CCGCTCG AGGCGGCCGCCTGAGTACCAGAGAAAGAGG3′,下游引物:5′TGCTCTAGA TCAGAGTACCAT ACTG AGTG3′,扩增TRADD 3′UTR基因。胶回收PCR产物和表达载体,以Xho I和Xba I进行双酶切。酶切产物用1%琼脂糖凝胶电泳分离并回收目的片段。按T4 DNA连接酶说明书进行目的片段与载体质粒的连接,连接产物转化DH5α感受态细胞,挑取单菌落进行菌落PCR,同时送测序公司测序。选取菌落PCR和测序正确的菌落,按照Omega无内毒素质粒抽提试剂盒说明书进行质粒抽提。
1.2.7 双荧光素酶报告基因检测试验 将PK-15细胞接种至24孔培养板,待细胞生长密度至80%左右时,按照Lipofectamine 2000试剂说明书进行转染。将合成的miRNA 模拟物、模拟物对照、miRNA 抑制物、抑制物对照分别与pmirGLO-TRADD 3′UTR质粒共转染,以单独转染pmirGLO-TRADD 3′UTR质粒重组质粒为对照。转染24 h后,用GloMax生物化学发光仪检测荧光素酶活性。
1.2.8 免疫印迹试验(Western blot) 裂解细胞提取总蛋白,用BCA蛋白浓度测定试剂盒对蛋白样品进行定量。蛋白样品与含有5%的β-巯基乙醇上样缓冲液充分混合后,放入95℃水浴锅中加热5 min,进行蛋白变性。变性后的蛋白80 V电泳2 h,采用湿法转膜(200 mA、1 h)将蛋白转印到PVDF膜上,用含5%吐温的PBS(PBST)室温漂洗转印后的PVDF膜3次,每次10 min,加入含5%脱脂奶粉的PBST封闭液37℃摇床摇动封闭1 h。用PBST洗涤3次后4℃过夜孵育一抗,次日取出孵育后的PVDF膜用PBST漂洗3次,37℃条件下孵育HRP标记的荧光二抗1 h,漂洗3次后,扫描PVDF膜。
1.2.9 流式细胞术 按照Annexin V-FITC细胞凋亡检测试剂盒说明书方法,检测细胞凋亡率。用PBS洗涤细胞2次,加入适量胰酶消化后,弃去胰酶,用1 mL细胞培养液吹打混匀细胞,转移到离心管内,1 000 r/min离心5 min,弃上清,收集细胞,用PBS轻轻重悬细胞并计数。取5—10万重悬的细胞,1 000 r/min离心5 min,弃上清,加入200 μL Annexin V-FITC结合液和10 μL碘化丙锭(PI)染色液,轻轻混匀,室温避光孵育10—20 min,随后置于冰浴中。进行流式细胞仪检测,Annexin V-FITC为绿色荧光,PI为红色荧光。
1.2.10 数据统计与分析 所有实验都至少重复3次,获得的数据用平均值±标准差表示,利用GraphPad Prism软件进行统计学作图和差异显著性分析。统计学分析采用t检验(*P<0.05;**P<0. 01;***P<0.001),当P<0.05时认为有统计学差异。
2 结果
2.1 PK-15细胞中与CSFV感染相关的miRNA的初步筛选
为了检测CSFV体外感染PK-15细胞后miRNA表达的变化,用CSFV石门株感染PK-15,24 h后进行miRNA表达谱芯片试验。结果表明,CSFV感染PK-15细胞后,诱导细胞miRNA差异表达。与未感染CSFV的对照组相比,CSFV感染PK-15细胞共引起69条miRNAs表达变化,其中包括39条miRNAs表达量上调,30条miRNAs表达量下调(表1、表2)。这表明CSFV感染PK-15细胞后,宿主细胞miRNA表达发生变化。Table 1
表1
表1CSFV感染PK-15细胞后表达量上调的miRNA
Table 1
| 编号 ID | miRNA名称 Name | log2比率 log2 ratio | P值 P-value |
|---|---|---|---|
| MIMAT0006786 | ssc-miR-140-3p | 0.48443 | 0.00026 |
| MIMAT0002147 | ssc-miR-214 | 0.42896 | 0.00001 |
| MIMAT0013955 | ssc-miR-335 | 0.38587 | 0.00001 |
| MIMAT0002120 | ssc-miR-125b | 0.09519 | 0.00043 |
| MIMAT0025368 | ssc-miR-194a | 0.31888 | 0.01203 |
| MIMAT0013944 | ssc-miR-342 | 0.31845 | 0.03668 |
| MIMAT0010192 | ssc-miR-215 | 0.30804 | 0.04995 |
| MIMAT0022956 | ssc-miR-202-3p | 0.29007 | 0.00009 |
| MIMAT0007759 | ssc-miR-185 | 0.22220 | 0.00035 |
| MIMAT0013912 | ssc-miR-208b | 0.21017 | 0.00771 |
| MIMAT0013876 | ssc-miR-191 | 0.20254 | 0.03214 |
| MIMAT0025360 | ssc-miR-31 | 0.20020 | 0.01347 |
| MIMAT0002119 | ssc-miR-122 | 0.19622 | 0.01318 |
| MIMAT0007758 | ssc-miR-130a | 0.19402 | 0.02421 |
| MIMAT0013916 | ssc-miR-34c | 0.19369 | 0.01302 |
| MIMAT0017385 | ssc-miR-1839-3p | 0.19364 | 0.03377 |
| MIMAT0013906 | ssc-miR-664-5p | 0.16827 | 0.00425 |
| MIMAT0013880 | ssc-miR-423-5p | 0.16298 | 0.00001 |
| MIMAT0002134 | ssc-miR-24-3p | 0.16088 | 0.01132 |
| MIMAT0007757 | ssc-miR-34a | 0.16044 | 0.00131 |
| MIMAT0017956 | ssc-miR-382 | 0.15206 | 0.00146 |
| MIMAT0013954 | ssc-miR-1285 | 0.14589 | 0.04690 |
| MIMAT0025356 | ssc-let-7d-5p | 0.14434 | 0.01134 |
| MIMAT0017376 | ssc-miR-365-5p | 0.14349 | 0.03370 |
| MIMAT0028156 | ssc-miR-7140-3p | 0.13804 | 0.00007 |
| MIMAT0015300 | ssc-miR-30a-3p | 0.13721 | 0.02233 |
| MIMAT0028144 | ssc-miR-7134-3p | 0.13182 | 0.00483 |
| MIMAT0013956 | ssc-miR-500 | 0.13164 | 0.02126 |
| MIMAT0013910 | ssc-miR-192 | 0.12760 | 0.00862 |
| MIMAT0013922 | ssc-miR-130b | 0.12228 | 0.02580 |
| MIMAT0013903 | ssc-miR-885-3p | 0.11877 | 0.00246 |
| MIMAT0022921 | ssc-miR-139-3p | 0.11860 | 0.00302 |
| MIMAT0028152 | ssc-miR-7138-3p | 0.11640 | 0.03755 |
| MIMAT0006018 | ssc-miR-99b | 0.11097 | 0.01450 |
| MIMAT0028155 | ssc-miR-7140-5p | 0.09369 | 0.00232 |
| MIMAT0013920 | ssc-miR-424-5p | 0.09366 | 0.00090 |
| MIMAT0022922 | ssc-miR-30c-3p | 0.08348 | 0.01393 |
| MIMAT0013894 | ssc-miR-193a-5p | 0.08314 | 0.00302 |
| MIMAT0010188 | ssc-miR-450a | 0.03115 | 0.00119 |
新窗口打开|下载CSV
Table 2
表2
表2CSFV感染PK-15细胞后表达下调的miRNA
Table 2
| 编号 ID | miRNA名称 Name | log2比率 log2 ratio | P值 P-value |
|---|---|---|---|
| MIMAT0025385 | ssc-miR-1249 | -2.15272 | 0.00001 |
| MIMAT0013934 | ssc-miR-361-3p | -1.95208 | 0.00001 |
| MIMAT0017952 | ssc-miR-296-5p | -1.05927 | 0.00001 |
| MIMAT0013937 | ssc-miR-1306-5p | -0.95394 | 0.00001 |
| MIMAT0013941 | ssc-miR-532-3p | -0.68756 | 0.00001 |
| MIMAT0017980 | ssc-miR-4339 | -0.6224 | 0.00001 |
| MIMAT0013871 | ssc-miR-30d | -0.30603 | 0.00001 |
| MIMAT0002165 | ssc-miR-21 | -0.27854 | 0.00001 |
| MIMAT0025381 | ssc-miR-671-5p | -0.2569 | 0.00001 |
| MIMAT0017974 | ssc-miR-484 | -0.248 | 0.00001 |
| MIMAT0020586 | ssc-miR-129b | -0.22805 | 0.00008 |
| MIMAT0018379 | ssc-miR-149 | -0.21029 | 0.00004 |
| MIMAT0015708 | ssc-miR-744 | -0.20892 | 0.00386 |
| MIMAT0017965 | ssc-miR-1224 | -0.18271 | 0.00033 |
| MIMAT0013900 | ssc-miR-345-3p | -0.17812 | 0.00828 |
| MIMAT0025365 | ssc-miR-150 | -0.15152 | 0.00775 |
| MIMAT0020596 | ssc-miR-1343 | -0.14743 | 0.00461 |
| MIMAT0013935 | ssc-miR-1277 | -0.13761 | 0.00482 |
| MIMAT0010186 | ssc-miR-133a-3p | -0.13388 | 0.03942 |
| MIMAT0013959 | ssc-miR-129a | -0.12737 | 0.01689 |
| MIMAT0013939 | ssc-miR-339-5p | -0.11957 | 0.01352 |
| MIMAT0025384 | ssc-miR-874 | -0.11957 | 0.00046 |
| MIMAT0028147 | ssc-miR-7136-5p | -0.11865 | 0.00367 |
| MIMAT0013909 | ssc-miR-92b-3p | -0.10387 | 0.02389 |
| MIMAT0017962 | ssc-miR-4332 | -0.09669 | 0.03493 |
| MIMAT0013918 | ssc-miR-425-3p | -0.09066 | 0.00617 |
| MIMAT0013869 | ssc-miR-133b | -0.08309 | 0.00758 |
| MIMAT0013904 | ssc-miR-365-3p | -0.06645 | 0.03768 |
| MIMAT0022958 | ssc-miR-296-3p | -0.06567 | 0.04572 |
| MIMAT0013947 | ssc-miR-935 | -0.05012 | 0.01506 |
新窗口打开|下载CSV
2.2 CSFV上调miR-214的表达水平
miRNA表达谱芯片的结果显示,在上调的39条miRNAs中,miR-214表达差异最明显,故选取miR-214作为研究对象,研究其在CSFV体外感染中的功能与作用。为了验证基因芯片数据结果,采用qRT-PCR方法验证miR-214在CSFV感染的PK-15细胞中的表达情况。结果显示,CSFV感染PK-15细胞后,随着时间推移,miR-214表达水平逐渐升高(图1),与表达谱芯片结果一致,表明CSFV感染促进了miR-214的表达。图1
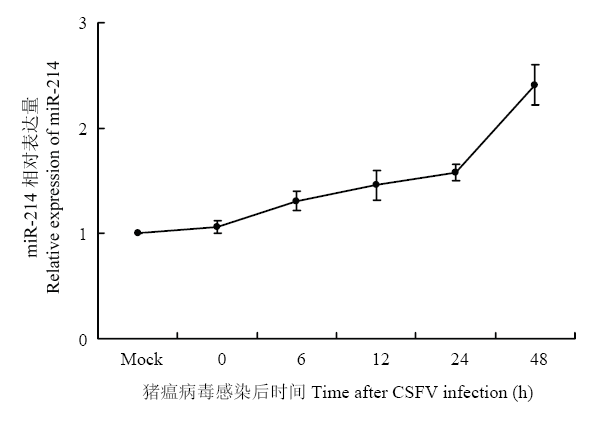 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1CSFV感染PK-15细胞后miR-214的表达水平
Fig. 1Relative expression of miR-214 in CSFV-infected PK-15 cells
2.3 miR-214模拟物与抑制剂效果检测
根据miRBase下载miR-214的序列并合成其模拟物及对应的抑制剂。将miR-214模拟物和抑制剂分别转染PK-15,24 h后提取细胞总RNA,qRT-PCR检测细胞内miR-214的表达量。结果显示,miR-214模拟物转染能够极显著提高细胞内miR-214的表达水平(图2-A),抑制剂则相应的显著抑制miR-214的表达水平(图2-B)。表明合成的miR-214模拟物及抑制剂可以进行后续试验。图2
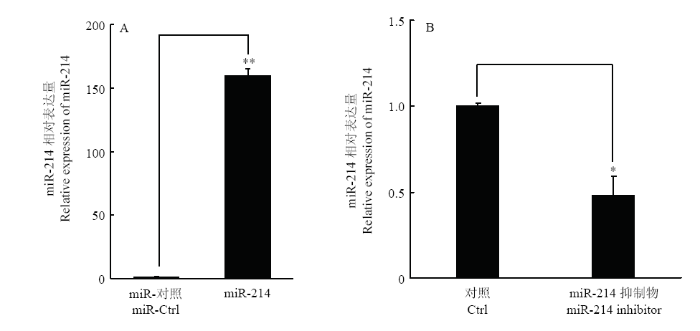 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2miR-214模拟物与抑制剂转染PK-15细胞后miR-214的表达
A. miR-214模拟物转染细胞后miR-214的表达量;B. miR-214抑制剂转染细胞后miR-214的表达量
Fig. 2The expression of miR-214 after transfection with miR-214 mimics or miR-214 inhibitor in PK-15 cells
A. The expression of miR-214 in cells after transfected with miR-214; B. The expression of miR-214 in cells after transfected with miR-214 inhibitor
2.4 miR-214促进CSFV复制
为了确定miR-214对CSFV复制的影响,将miR-214 模拟物和抑制剂分别转染PK-15细胞,转染后24 h,以MOI=0.1接种CSFV石门株,继续培养48 h后,提取细胞总RNA,用qRT-PCR方法检测CSFV NS5B mRNA表达水平。收集细胞培养液上清,间接免疫荧光(IFA)检测病毒滴度。结果表明,miR-214模拟物能够显著促进CSFV的复制及病毒滴度(图3-A),而miR-214抑制剂显著抑制了CSFV的复制及病毒滴度(图3-B),表明miR-214促进了CSFV RNA复制。图3
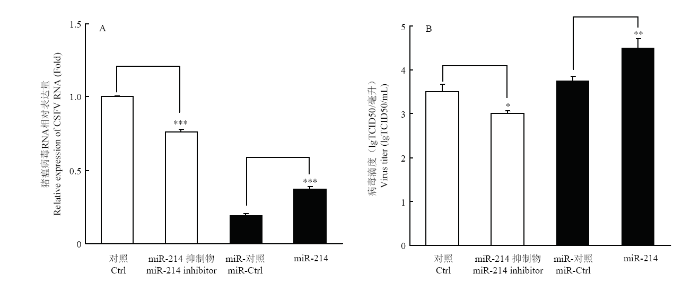 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3miR-214对CSFV复制的影响
A. miR-214对CSFV NS5B mRNA表达量的影响;B. miR-214对CSFV病毒滴度的影响
Fig. 3The effects of miR-214 on CSFV replication
A. The influence of miR-214 on the expression of CSFV NS5B mRNA; B. The effect of miR-214 on the virus titer of CSFV
2.5 荧光素酶报告基因系统验证miR-214的靶蛋白TRADD
为了进一步确认miR-214能够靶向作用于TRADD,将TRADD的3′UTR区域构建入荧光素酶报告基因表达载体。分别共转染重组质粒与miR-214模拟物和miR-214抑制剂,24 h后检测荧光素酶活性。结果显示,转染miR-214 模拟物能够显著下调荧光素酶报告基因的表达(图4-A),而转染miR-214抑制剂能够显著上调荧光素酶报告基因的表达(图4-B),表明miR-214能够靶向作用于TRADD的3′UTR,进一步证明了miR-214对TRADD的靶向作用。图4
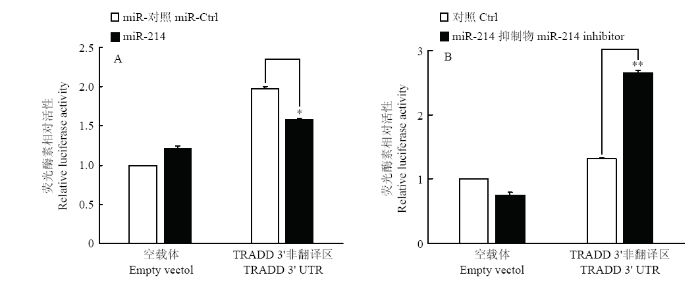 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4miR-214对pmirGLO-TRADD 3′UTR荧光素酶活性的影响
Fig. 4The effects of miR-214 on luciferase activity of pmirGLO-TRADD 3′UTR
2.6 生物信息学软件预测miR-214的靶蛋白
通过TargetScan、RNA22、Pictar等生物信息学软件预测miRNA的靶蛋白,发现,TRADD的3′UTR区域存在miR-214潜在作用靶点(图5)。图5
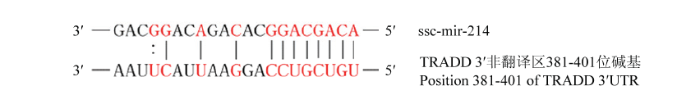 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5miR-214与TRADD潜在作用位点
图中红色标注部分为miR-214与TRADD的3′非翻译区互补配对的碱基对
Fig. 5The potential interaction site between miR-214 and TRADD
The red labels in the figure is the complementary base pairing between mir-214 and TRADD 3'-UTR
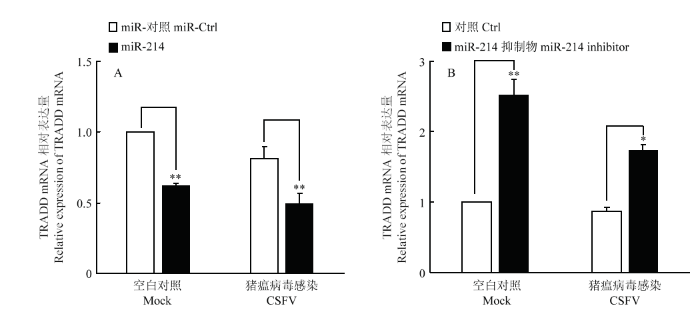
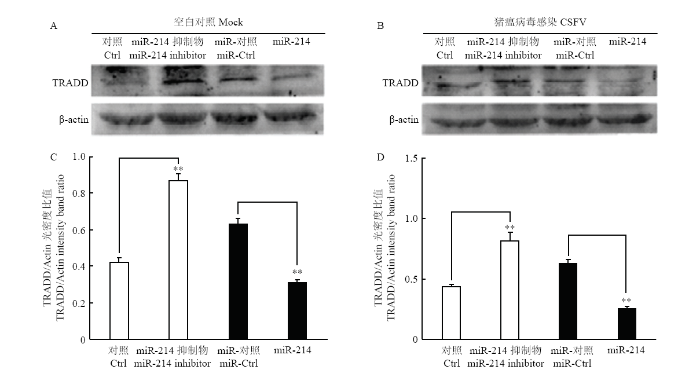
2.7 miR-214抑制TRADD的表达
通过上述试验,证明了miR-214对TRADD的靶向作用。为了进一步探究miR-214对TRADD表达的影响,将miR-214 模拟物及其抑制剂转染细胞,24 h后以MOI=0.1接种CSFV石门株,同步设立不感染组作为对照。 48 h后收集细胞提取总RNA,用qRT-PCR方法检测潜在靶蛋白TRADD的mRNA表达水平,验证miR-214对TRADD表达的影响。结果显示,miR-214模拟物能够显著下调TRADD mRNA表达(图6-A),而miR-214抑制剂则显著上调TRADD mRNA表达(图6-B)。同时裂解细胞提取总蛋白,用鼠源TRADD单抗和鼠源β-actin单抗进行Western blot,检测TRADD的蛋白表达水平。结果显示,miR-214模拟物抑制了TRADD的蛋白表达水平,而miR-214抑制剂则促进了TRADD的蛋白表达(图7)。综上试验结果表明,miR-214能抑制TRADD的表达。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6miR-214对TRADD表达的影响
A.转染miR-214 对TRADD mRNA表达的影响;B.转染miR-214抑制物对TRADD mRNA表达的影响
Fig. 6The Effects of miR-214 on the expression of TRADD
A. Effect of transfection of mir-214 on TRADD mRNA expression; B. Effect of transfection of mir-214 inhibitor on TRADD mRNA expression
图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7miR-214对TRADD蛋白表达的影响
A.转染miR-214模拟物对TRADD 蛋白表达的影响;B.转染miR-214抑制剂对TRADD 蛋白表达的影响;C和D分别为A和B光密度比值的统计学分析
Fig. 7The Effects of miR-214 on the expression of TRADD
A. Effect of transfection of mir-214 on TRADD protein expression; B. Effect of transfection of mir-214 inhibitor on TRADD protein expression; C and D are the statistical analysis of the intensity band ratio of A and B respectively
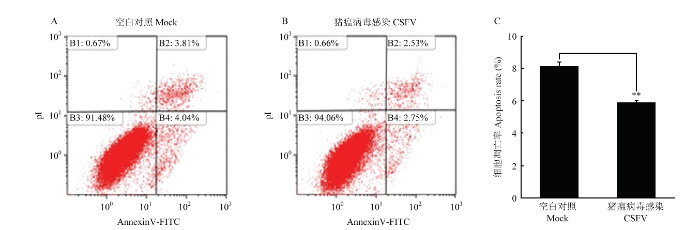
2.8 CSFV体外感染抑制PK-15细胞凋亡
有研究表明,CSFV在体外感染过程中通过抑制细胞凋亡实现持续感染[21]。在本研究中,首先验证CSFV对细胞凋亡的影响。PK-15细胞在6孔板中培养生长至80%,接种CSFV(MOI=1),同步设立未感染为对照组,感染24 h后,收集细胞,用流式细胞术检测细胞凋亡情况。结果显示,与对照组相比,CSFV感染PK-15细胞后抑制了细胞的凋亡(图8)。图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8CSFV感染PK-15细胞对细胞凋亡的影响
A. PK-15细胞未感染CSFV的凋亡率;B. PK-15细胞感染CSFV的凋亡率;C.CSFV感染组与对照组凋亡率的统计学分析
Fig. 8The effect of CSFV infection on apoptosis in PK-15 cells
A. Apoptosis rate of PK-15 cells without infected with CSFV; B. Apoptosis rate of PK -15 cells infected with CSFV; C. Statistical analysis of apoptosis rate of CSFV infection group and control group
2.9 miR-214抑制PK-15细胞凋亡
试验证明CSFV感染PK-15细胞后抑制细胞凋亡,有研究表明,TRADD与肿瘤坏死因子受体相互作用,能诱导细胞凋亡[22,23],而miR-214能抑制TRADD的表达。为了验证在CSFV体外感染过程中,miR-214是否参与调控细胞凋亡,PK-15细胞转染miR-214模拟物,用流式细胞术检测细胞凋亡情况。结果显示,miR-214模拟物能够抑制细胞凋亡(图9-A)。为了进一步验证在CSFV感染过程中,miR-214对凋亡的影响,将miR-214 模拟物和抑制剂及其相应的阴性对照转染PK-15细胞,转染24 h后,以MOI=0.1接种CSFV石门株,48 h后用流式细胞术检测细胞凋亡情况。结果显示,在未感染CSFV的PK-15细胞中,转染miR-214模拟物下调了细胞凋亡率,而转染miR-214抑制剂能够上调细胞凋亡率。同样,在CSFV感染组中,得到一致的结果,即转染miR-214模拟物能够下调细胞凋亡率,而转染miR-214抑制剂能够上调细胞凋亡率(图9-B、C)。由此推测,在CSFV感染过程中,miR-214可能参与抑制细胞凋亡。图9
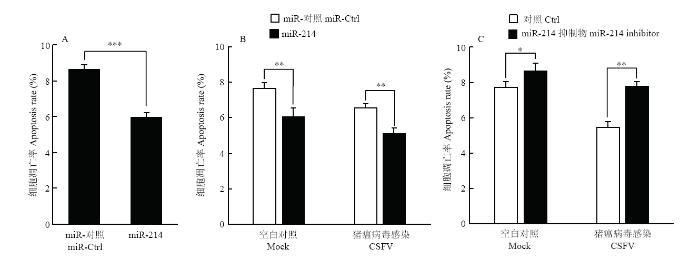 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9miR-214及其抑制剂对PK-15细胞凋亡的影响
A. miR-214对PK-15细胞凋亡的影响;B. miR-214对在CSFV感染条件下PK-15细胞凋亡的影响;C. miR-214抑制剂对在CSFV感染条件下PK-15细胞凋亡的影响
Fig. 9Effects of miR-214 and miR-214 inhibitors on apoptosis of PK-15 cells
A. The effect of miR-214 on apoptosis of PK-15 cells; B. The effect of mir-214 on apoptosis of PK-15 cells infected with CSFV; C. The effect of miR-214 inhibitors on apoptosis of PK-15 cells infected with CSFV
3 讨论
miRNA是一类调节性非编码RNA分子,广泛存在于植物、动物以及动物病毒的基因组,主要参与基因转录后调控。近年来大量研究表明,细胞内的miRNA在调控宿主细胞基因及病毒复制中发挥重要作用[20,24],但是大部分研究都是基于人和小鼠等模式动物上,对猪源miRNA的研究较少。此外,对于HCV的miRNA的研究较为深入,为HCV致病机理研究方面提供了很多理论支持。展开对CSFV相关miRNA的研究,为揭示其致病机理开辟了新的方向,也为防控CSFV及寻找药物靶位提供了新的策略。本研究中,以PK-15细胞为研究对象,通过表达谱筛选出与CSFV密切相关的差异表达的细胞miRNA,为研究miRNA介导调控CSFV感染致病机制打下前期基础。表达谱试验共筛选出69条差异表达的miRNA,但是与未感染CSFV的细胞组相比,大部分miRNA表达变化差异不大,差异水平在1.5倍左右,最多达到4倍,与其他病毒的miRNA表达谱结果相比,变化差异不是非常明显。推测其原因,可能跟病毒及种属有关,也可能细胞miRNA表达量相对较高,导致病毒感染后表达虽然有变化,但是变化比例较小。如果联合全基因组小RNA高通量测序,得到的差异结果可能更明显。另外,也可能与CSFV病毒自身感染PK-15细胞的特性有关,miRNA的调控水平可能较微弱。
本试验选择miR-214为研究对象,因为在筛选CSFV上调的39条miRNA当中,miR-214的表达差异最大。此外,miR-214的靶蛋白TRADD对凋亡具有调节作用。前期的研究表明猪瘟病毒能够抑制凋亡,达到持续感染的目的[21,25]。已经有大量的研究报道miR-214参与调控细胞凋亡,如miR-214能直接靶向于Bcl2l2,通过抑制Bcl2l2蛋白表达水平,从而诱导凋亡[26];在鼻咽癌组织或细胞系中高表达miR-214,而抑制miR-214表达,能够上调Bim蛋白的表达,从而促进细胞凋亡抑制癌细胞增殖[27]。因此推测,宿主miR-214在CSFV感染的PK-15细胞中参与调控细胞凋亡。
细胞凋亡是细胞Ⅰ型死亡程序,细胞凋亡启动细胞程序性死亡后不利于病毒在细胞内的存活和持续性感染[28]。有研究表明,CSFV感染细胞后,可以通过抑制细胞凋亡从而促进病毒的复制[21]。通过生物信息学手段预测miR-214的靶蛋白中,发现TRADD存在miR-214结合位点,推测可能是miR-214的靶蛋白之一。死亡受体介导的细胞凋亡包括FASL- FAS、TRAIL-DR4以及TNFα-TNFR1途径[29,30],TRADD作为TNFα-TNFR1途径中一个接头分子,对下游凋亡信号激活及分子招募至关重要。因此本试验对TNFα-TNFR1介导的凋亡进行探讨,结果证明在CSFV感染中高表达miR-214能够一定程度上抑制TRADD表达,抑制PK-15细胞凋亡。表明CSFV体外感染中抑制细胞凋亡的机制可能与CSFV上调的miR-214有关。
综上所述,本研究筛选出CSFV感染诱导差异表达的miRNA,为以后研究miRNA介导调控CSFV感染和从miRNA方面揭示CSFV致病机理奠定了基础。此外,细胞凋亡作为宿主抗病毒感染的第一道防线,是宿主细胞自身受外界刺激下启动的死亡程序,不利于病毒复制。但许多病毒却可以利用细胞某些调控机制延迟或者抑制细胞发生凋亡,从而有利于自身复制。本试验初步揭示了CSFV诱导的miR-214的功能和作用,表明在CSFV感染过程中miR-214可能参与调控细胞凋亡,从而有利于CSFV在细胞内的复制和持续性感染,为了解CSFV的致病机制提供新的方向,也为防控CSFV感染寻找新的作用靶位提供一个新的思路。
4 结论
利用miRNA表达谱芯片筛选CSFV感染PK-15细胞后差异表达的miRNAs,结果表明,CSFV感染PK-15细胞诱导69条差异表达的猪源miRNAs,其中包括上调表达的39条,下调表达的30条。CSFV感染PK-15细胞后上调miR-214表达,且miR-214抑制PK-15细胞凋亡,促进CSFV RNA的复制。miR-214靶向于TRADD蛋白,作用于TRADD 3′UTR。即CSFV感染宿主细胞后,通过促进miR-214的表达从而抑制TRADD的表达水平,达到抑制细胞凋亡的目的,使CSFV实现持续感染。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1016/S0378-1135(00)00137-1URLPMID:10785320 [本文引用: 1]

Classical swine fever virus is a spherical enveloped particle of about 40 60 nm in diameter with a single stranded RNA genome of about 12,300 bases with positive polarity, classified as a pestivirus within the family Flaviviridae. Natural hosts are domestic and wild pigs. The virus causes one of the most severe diseases in pigs world wide with grave economic consequences. The clinical picture of classical swine fever is variable, depending on the age of the affected animals and viral virulence. The virus is well characterised and reliable laboratory diagnostic procedures are available. In many parts of the world live attenuated vaccines are being used as a safe and efficient prophylactic tool. However, in EU Member States and several other countries vaccination is prohibited and CSF is controlled by a strict stamping out policy. In order to overcome the disadvantages of conventional vaccination inactivated marker vaccines have been developed that enable the distinction between vaccinated and infected animals. Whether these vaccines will be accepted as an additional tool in the framework of the stamping out policy is not yet decided.
DOI:10.1016/S0378-1135(00)00144-9URLPMID:10785327 [本文引用: 1]

In 1997, the pig husbandry in the Netherlands was struck by a severe epidemic of classical swine fever (CSF). During this epidemic 429 CSF-infected herds were depopulated and ≈1300 herds were slaughtered pre-emptively. In addition millions of pigs of herds not CSF-infected were killed for welfare reasons (over crowding or overweight). In this paper, we describe the course of the epidemic and the measures that were taken to control it. The first outbreak was detected on 4 February 1997 in the pig dense south-eastern part of the Netherlands. We estimate that CSF virus (CSFV) had already been present in the country by that time for 5–7 weeks and that the virus had been introduced into ≈39 herds before the eradication campaign started. This campaign consisted of stamping-out infected herds, movement restrictions and efforts to diagnose infected herds as soon as possible. However, despite these measures the rate at which new outbreaks were detected continued to rise. The epidemic faded out only upon the implementation of additional measures such as rapid pre-emptive slaughter of herds in contact with or located near infected herds, increased hygienic measures, biweekly screening of all herds by veterinary practitioners, and reduction of the transportation movements for welfare reasons. The last infected herd was depopulated on 6 March 1998.
DOI:10.1016/j.cell.2009.01.002URL [本文引用: 1]
DOI:10.1038/ng1953URL [本文引用: 1]
[本文引用: 1]
DOI:10.1128/JVI.02136-07URLPMID:2258704 [本文引用: 1]

Abstract The Epstein-Barr virus (EBV)-encoded latent membrane protein 1 (LMP1) is a functional homologue of the tumor necrosis factor receptor family and contributes substantially to the oncogenic potential of EBV through activation of nuclear factor kappaB (NF-kappaB). MicroRNAs (miRNAs) are a class of small RNA molecules that are involved in the regulation of cellular processes such as growth, development, and apoptosis and have recently been linked to cancer phenotypes. Through miRNA microarray analysis, we demonstrate that LMP1 dysregulates the expression of several cellular miRNAs, including the most highly regulated of these, miR-146a. Quantitative reverse transcription-PCR analysis confirmed induced expression of miR-146a by LMP1. Analysis of miR-146a expression in EBV latency type III and type I cell lines revealed substantial expression of miR-146a in type III (which express LMP1) but not in type I cell lines. Reporter studies demonstrated that LMP1 induces miR-146a predominantly through two NF-kappaB binding sites in the miR-146a promoter and identified a role for an Oct-1 site in conferring basal and induced expression. Array analysis of cellular mRNAs expressed in Akata cells transduced with an miR-146a-expressing retrovirus identified genes that are directly or indirectly regulated by miR-146a, including a group of interferon-responsive genes that are inhibited by miR-146a. Since miR-146a is known to be induced by agents that activate the interferon response pathway (including LMP1), these results suggest that miR-146a functions in a negative feedback loop to modulate the intensity and/or duration of the interferon response.
DOI:10.1016/j.atherosclerosis.2014.05.364URLPMID:25722434 [本文引用: 1]

OBJECTIVE: In this study, we attempted to uncover the functional impact of microRNA-22 (miR-22) and its target gene in smooth muscle cell (SMC) differentiation and delineate the molecular mechanism involved. APPROACH AND RESULTS: miR-22 was found to be significantly upregulated during SMC differentiation from embryonic stem cells and adventitia stem/progenitor cells. Enforced expression of miR-22 by its mimic, while knockdown of miR-22 by its antagomiR, promotes or inhibits SMC differentiation from embryonic stem cells and adventitia stem/progenitor cells, respectively. Expectedly, miR-22 overexpression in stem cells promoted SMC differentiation in vivo. Methyl CpG-binding protein 2 (MECP2) was predicted as one of the top targets of miR-22. Interestingly, the gene expression levels of MECP2 were significantly decreased during SMC differentiation, and MECP2 was dramatically decreased in miR-22 overexpressing cells but significantly increased when miR-22 was knockdown in the differentiating stem cells. Importantly, luciferase assay showed that miR-22 substantially inhibited wild-type, but not mutant MECP2-3' untranslated region-luciferase activity. In addition, modulation of MECP2 expression levels affects multiple SMC-specific gene expression in differentiated embryonic stem cells. Mechanistically, our data showed that MECP2 could transcriptionally repress SMC gene expression through modulating various SMC transcription factors, as well as several proven SMC differentiation regulators. Evidence also revealed that enrichment of H3K9 trimethylation around the promoter regions of the SMC differentiation regulators genes were significantly increased by MECP2 overexpression. Finally, miR-22 was upregulated by platelet-derived growth factor-BB and transforming growth factor-beta through a transcriptional mechanism during SMC differentiation. CONCLUSIONS: miR-22 plays an important role in SMC differentiation, and epigenetic regulation through MECP2 is required for miR-22 mediated SMC differentiation.
DOI:10.14348/molcells.2014.0147URLPMID:4275704 [本文引用: 1]

Abstract In our previous study, miRNA-183, a miRNA in the miR-96-182-183 cluster, was significantly over-expressed in esophageal squamous cell carcinoma (ESCC). In the present study, we explored the oncogenic roles of miR-183 in ESCC by gain and loss of function analysis in an esophageal cancer cell line (EC9706). Genome-wide mRNA microarray was applied to determine the genes that were regulated directly or indirectly by miR-183. 3'UTR luciferase reporter assay, RT-PCR, and Western blot were conducted to verify the target gene of miR-183. Cell culture results showed that miR-183 inhibited apoptosis (p < 0.05), enhanced cell proliferation (p < 0.05), and accelerated G1/S transition (p < 0.05). Moreover, the inhibitory effect of miR-183 on apoptosis was rescued when miR-183 was suppressed via miR-183 inhibitor (p < 0.05). Western blot analysis showed that the expression of programmed cell death 4 (PDCD4), which was predicted as the target gene of miR-183 by microarray profiling and bioinformatics predictions, decreased when miR-183 was over-expressed. The 3'UTR luciferase reporter assay confirmed that miR-183 directly regulated PDCD4 by binding to sequences in the 3'UTR of PDCD4. Pearson correlation analysis further confirmed the significant negative correlation between miR-183 and PDCD4 in both cell lines and in ESCC patients. Our data suggest that miR-183 might play an oncogenic role in ESCC by regulating PDCD4 expression.
DOI:10.1016/S0960-9822(03)00250-1URLPMID:12725740 [本文引用: 1]

MicroRNAs (miRNAs) are small regulatory RNAs that are between 21 and 25 nucleotides in length and repress gene function through interactions with target mRNAs . The genomes of metazoans encode on the order of several hundred miRNAs [3], but the processes they regulate have been defined for only two in [4]. We searched for new inhibitors of apoptotic cell death by testing existing collections of P element insertion lines for their ability to enhance a small-eye phenotype associated with eye-specific expression of the Drosophila cell death activator Reaper. Here we report the identification of the Drosophila miRNA mir-14 as a cell death suppressor. Loss of mir-14 enhances Reaper-dependent cell death, whereas ectopic expression suppresses cell death induced by multiple stimuli. Animals lacking mir-14 are viable. However, they are stress sensitive and have a reduced lifespan. Mir-14 mutants have elevated levels of the apoptotic effector caspase Drice, suggesting one potential site of action. Mir-14 also regulates fat metabolism. Deletion of mir-14 results in animals with increased levels of triacylglycerol and diacylglycerol, whereas increases in mir-14 copy number have the converse effect. We discuss possible relationships between these phenotypes.
DOI:10.1172/JCI114950URLPMID:12930946 [本文引用: 1]

Micro-RNAs are a class of small non-coding regulatory RNAs that impair translation by imperfect base pairing to mRNAs. For analysis of their cellular function we injected different miRNA-specific DNA antisense oligonucleotides in Drosophila embryos. In four cases we observed severe interference with normal development, one had a moderate impact and six oligonucleotides did not cause detectable phenotypes. We further used the miR-13a DNA antisense oligonucleotide as a PCR primer on a cDNA library template. In this experimental way we identified nine Drosophila genes, which are characterised by 3' untranslated region motifs that allow imperfect duplex formation with miR-13 or related miRNAs. These genes, which include Sos and Myd88, represent putative targets for miRNA regulation. Mutagenesis of the target motif of two genes followed by transfection in Drosophila Schneider 2 (S2) cells and subsequent reporter gene analysis confirmed the hypothesis that the binding potential of miR-13 is inversely correlated with gene expression.
DOI:10.1101/gad.1522907URL [本文引用: 1]
DOI:10.1016/j.cell.2007.03.008URLPMID:17382377 [本文引用: 1]

T cell sensitivity to antigen is intrinsically regulated during maturation to ensure proper development of immunity and tolerance, but how this is accomplished remains elusive. Here we show that increasing miR-181a expression in mature T cells augments the sensitivity to peptide antigens, while inhibiting miR-181a expression in the immature T cells reduces sensitivity and impairs both positive and negative selection. Moreover, quantitative regulation of T cell sensitivity by miR-181a enables mature T cells to recognize antagonists—the inhibitory peptide antigens—as agonists. These effects are in part achieved by the downregulation of multiple phosphatases, which leads to elevated steady-state levels of phosphorylated intermediates and a reduction of the T cell receptor signaling threshold. Importantly, higher miR-181a expression correlates with greater T cell sensitivity in immature T cells, suggesting that miR-181a acts as an intrinsic antigen sensitivity “rheostat” during T cell development.
DOI:10.1016/j.immuni.2007.10.009URL [本文引用: 1]
DOI:10.1186/1742-4690-5-17URLPMID:18241347 [本文引用: 2]

The RNA interference mechanism utilizes short RNA duplexes to either suppress or induce target gene expression. Post-transcriptional regulation mediated by microRNA is an integral component of innate antiviral defense. The magnitude and the efficiency of viral restriction guided by RNA-based defenses, as well as the full physiological implication of host-pathogen engagement, constitute exciting areas of investigation in the biology of non-coding RNAs.
DOI:10.1126/science.1108784URL [本文引用: 1]
DOI:10.4049/jimmunol.179.8.5082URL [本文引用: 1]
DOI:10.1073/pnas.0605298103URLPMID:16885212 [本文引用: 1]

Abstract Activation of mammalian innate and acquired immune responses must be tightly regulated by elaborate mechanisms to control their onset and termination. MicroRNAs have been implicated as negative regulators controlling diverse biological processes at the level of posttranscriptional repression. Expression profiling of 200 microRNAs in human monocytes revealed that several of them (miR-146a/b, miR-132, and miR-155) are endotoxin-responsive genes. Analysis of miR-146a and miR-146b gene expression unveiled a pattern of induction in response to a variety of microbial components and proinflammatory cytokines. By means of promoter analysis, miR-146a was found to be a NF-kappaB-dependent gene. Importantly, miR-146a/b were predicted to base-pair with sequences in the 3' UTRs of the TNF receptor-associated factor 6 and IL-1 receptor-associated kinase 1 genes, and we found that these UTRs inhibit expression of a linked reporter gene. These genes encode two key adapter molecules downstream of Toll-like and cytokine receptors. Thus, we propose a role for miR-146 in control of Toll-like receptor and cytokine signaling through a negative feedback regulation loop involving down-regulation of IL-1 receptor-associated kinase 1 and TNF receptor-associated factor 6 protein levels.
URL [本文引用: 1]

【Objective】The study was to construct a recombinant vector bearing the let-7c gene and to research on potential regulation of classical swine fever virus(CSFV)by let-7.【Method】let-7c was selected by Bioinformatics of RNA22.Two pairs of single DNA chain encoding pre-let-7c were synthesized,and were ligated to the expression vector pGenesil-1.1-dBm2.The recombinant plasmid pGene-pre-let-7c was constructed and transfected into the swine umbilicus vein endothelial cells(SUVEC)by liposome after extraction and purification.The positive clones were selected by G418 screening.We also analyzed the expression of CSFV through the approach of real1717time fluorescence quantitative RT1717PCR and MTT was performed to detect cell proliferatime.【Result】The pGene-pre-let-7c was successfully constructed and pre-let-7c was expressed in porcine vascular endothelial cell line.【Conclusion】let-7c can inhibit the duplication of CSFV.
.
URL [本文引用: 1]

【Objective】The study was to construct a recombinant vector bearing the let-7c gene and to research on potential regulation of classical swine fever virus(CSFV)by let-7.【Method】let-7c was selected by Bioinformatics of RNA22.Two pairs of single DNA chain encoding pre-let-7c were synthesized,and were ligated to the expression vector pGenesil-1.1-dBm2.The recombinant plasmid pGene-pre-let-7c was constructed and transfected into the swine umbilicus vein endothelial cells(SUVEC)by liposome after extraction and purification.The positive clones were selected by G418 screening.We also analyzed the expression of CSFV through the approach of real1717time fluorescence quantitative RT1717PCR and MTT was performed to detect cell proliferatime.【Result】The pGene-pre-let-7c was successfully constructed and pre-let-7c was expressed in porcine vascular endothelial cell line.【Conclusion】let-7c can inhibit the duplication of CSFV.
DOI:10.1371/journal.ppat.1003248URLPMID:3635988 [本文引用: 1]

Upon recognition of viral components by pattern recognition receptors, such as the toll-like receptors (TLRs) and retinoic acid-inducible gene I (RIG-I)-like helicases, cells are activated to produce type I interferon (IFN) and proinflammatory cytokines. These pathways are tightly regulated by the host to prevent an inappropriate cellular response, but viruses can modulate these pathways to proliferate and spread. In this study, we revealed a novel mechanism in which hepatitis C virus (HCV) evades the immune surveillance system to proliferate by activating microRNA-21 (miR-21). We demonstrated that HCV infection upregulates miR-21, which in turn suppresses HCV-triggered type I IFN production, thus promoting HCV replication. Furthermore, we demonstrated that miR-21 targets two important factors in the TLR signaling pathway, myeloid differentiation factor 88 (MyD88) and interleukin-1 receptor-associated kinase 1 (IRAK1), which are involved in HCV-induced type I IFN production. HCV-mediated activation of miR-21 expression requires viral proteins and several signaling components. Moreover, we identified a transcription factor, activating protein-1 (AP-1), which is partly responsible for miR-21 induction in response to HCV infection through PKC??/JNK/c-Jun and PKC??/ERK/c-Fos cascades. Taken together, our results indicate that miR-21 is upregulated during HCV infection and negatively regulates IFN-?? signaling through MyD88 and IRAK1 and may be a potential therapeutic target for antiviral intervention.
DOI:10.1101/sqb.2006.71.022URLPMID:17381319 [本文引用: 2]

Abstract microRNAs (miRNAs) are small RNAs that in general down-regulate the intracellular abundance and translation of target mRNAs. We noted that sequestration of liver-specific miR-122 by modified antisense oligonucleotides resulted in a dramatic loss of hepatitis C virus (HCV) RNA in cultured human liver cells. A binding site for miR-122 was predicted to reside close to the 5' end of the viral genome, and its functionality was tested by mutational analyses of the miRNA-binding site in viral RNA, resulting in reduced intracellular viral RNA abundance. Importantly, ectopic expression of miR-122 molecules that contained compensatory mutations restored viral RNA abundance, revealing a genetic interaction between miR-122 and the viral RNA genome. Studies with replication-defective viral RNAs demonstrated that miR-122 affected mRNA abundance by positively modulating RNA replication. In contrast, interaction of miR-122 with the 3'-noncoding region (3'NCR) of the cellular mRNA encoding the cationic amino acid transporter CAT-1 resulted in the down-regulation of CAT-1 protein abundance. These findings provide evidence that a specific miRNA can regulate distinct target mRNAs in both a positive and negative fashion. The positive role of miR-122 in viral replication suggests that this miRNA could be targeted for antiviral therapy.
DOI:10.1099/vir.0.19637-0URLPMID:15039545 [本文引用: 3]

Abstract Infection with virulent strains of classical swine fever virus (CSFV) results in an acute haemorrhagic disease of pigs, characterized by disseminated intravascular coagulation, thrombocytopenia and immunosuppression, whereas for less virulent isolates infection can become chronic. In view of the haemorrhagic pathology of the disease, the effects of the virus on vascular endothelial cells was studied by using relative quantitative PCR and ELISA. Following infection, there was an initial and short-lived increase in the transcript levels of the proinflammatory cytokines interleukins 1, 6 and 8 at 3 h followed by a second more sustained increase 24 h post-infection. Transcription levels for the coagulation factor, tissue factor and vascular endothelial cell growth factor involved in endothelial cell permeability were also increased. Increases in these factors correlated with activation of the transcription factor NF-kappaB. Interestingly, the virus produced a chronic infection of endothelial cells and infected cells were unable to produce type I interferon. Infected cells were also protected from apoptosis induced by synthetic ouble-stranded RNA. These results demonstrate that, in common with the related pestivirus bovine viral diarrhoea virus, CSFV can actively block anti-viral and apoptotic responses and this may contribute to virus persistence. They also point to a central role for infection of vascular endothelial cells during the pathogenesis of the disease, where a proinflammatory and procoagulant endothelium induced by the virus may disrupt the haemostatic balance and lead to the coagulation and thrombosis seen in acute disease.
[本文引用: 1]
DOI:10.1016/S0092-8674(03)00521-XURLPMID:1288792012887920 [本文引用: 1]

Apoptosis induced by TNF-receptor I (TNFR1) is thought to proceed via recruitment of the adaptor FADD and caspase-8 to the receptor complex. TNFR1 signaling is also known to activate the transcription factor NF-κB and promote survival. The mechanism by which this decision between cell death and survival is arbitrated is not clear. We report that TNFR1-induced apoptosis involves two sequential signaling complexes. The initial plasma membrane bound complex (complex I) consists of TNFR1, the adaptor TRADD, the kinase RIP1, and TRAF2 and rapidly signals activation of NF-κB. In a second step, TRADD and RIP1 associate with FADD and caspase-8, forming a cytoplasmic complex (complex II). When NF-κB is activated by complex I, complex II harbors the caspase-8 inhibitor FLIP L and the cell survives. Thus, TNFR1-mediated-signal transduction includes a checkpoint, resulting in cell death (via complex II) in instances where the initial signal (via complex I, NF-κB) fails to be activated.
DOI:10.1128/JVI.00456-10URLPMID:20554777 [本文引用: 1]

MicroRNAs (miRNAs) are a class of noncoding RNAs of lengths ranging from 18 to 23 nucleotides (nt) that play critical roles in a wide variety of biological processes. There is a growing amount of evidence that miRNAs play critical roles in intricate host-pathogen interaction networks, but the involvement of miRNAs during influenza viral infection is unknown. To determine whether the cellular miRNAs play an important role in H1N1 influenza A viral infections, 3' untranslated region (UTR) reporter analysis was used to identify putative miRNA targets in the influenza virus genome, and virus proliferation analysis was used to detect the effect of the screened miRNAs on the replication of H1N1 influenza A virus (A/WSN/33) in MDCK cells. The results showed that miRNA 323 (miR-323), miR-491, and miR-654 inhibit replication of the H1N1 influenza A virus through binding to the PB1 gene. Moreover mutational analysis of the predicted miRNA binding sites showed that the three miRNAs bind to the same conserved region of the PB1 gene. Intriguingly, despite the fact that the miRNAs and PB1 mRNA binding sequences are not a perfect match, the miRNAs downregulate PB1 expression through mRNA degradation instead of translation repression. This is the first demonstration that cellular miRNAs regulate influenza viral replication by degradation of the viral gene. Our findings support the notion that any miRNA has antiviral potential, independent of its cellular function, and that the cellular miRNAs play an important role in the host, defending against virus infection.
DOI:10.1099/vir.0.016576-0URLPMID:20007358 [本文引用: 1]

Classical swine fever virus (CSFV) causes severe disease in pigs associated with leukopenia, haemorrhage and fever. We show that CSFV infection protects endothelial cells from apoptosis induced by the dsRNA mimic, pIpC, but not from other apoptotic stimuli, FasL or staurosporine. CSFV infection inhibits pIpC-induced caspase activation, mitochondrial membrane potential loss and cytochrome c release as well as the pro-apoptotic effects of truncated Bid (tBid) overexpression. The CSFV proteins Npro and Erns both contribute to CSFV inhibition of apoptosis. We conclude that CSFV infection can inhibit apoptotic signalling at multiple levels, including at the caspase-8 and the mitochondrial checkpoints. By supporting viral replication, endothelial cells may promote CSFV pathogenesis.
DOI:10.1016/j.febslet.2013.01.016URLPMID:23337879 [本文引用: 1]

MiR-214 has been shown to inhibit cell growth, migration and invasion. Here we demonstrate that ectopic expression of miR-214 reduces cell survival, induces apoptosis and enhances sensitivity to cisplatin through directly inhibiting Bcl2l2 expression in cervical cancer HeLa and C-33A cells. Further analysis reveals that apoptosis induced by miR-214 is correlated with increased expression of Bax, caspase-9, caspase-8 and caspase-3. Moreover, we show that miR-214 is regulated by DNA methylation and histone deacetylation. Taken together, these data suggest that miR-214 might be a candidate target for the development of novel therapeutic strategies to treat cervical cancer. (C) 2013 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
DOI:10.1371/journal.pone.0086149URLPMID:24465927 [本文引用: 1]

MicroRNA-214 (MiR-214) is aberrantly expressed in several human tumors such as ovarian cancer and breast cancer. However, the role of miR-214 in nasopharyngeal carcinoma (NPC) is still unknown. In this study, we report that miR-214 was overexpressed in NPC cell lines and tissues. Silencing of miR-214 by LNA-antimiR-214 in NPC cells resulted in promoting apoptosis and suppressing cell proliferationin vitro, and suppressed tumor growth in nude micein vivo. Luciferase reporter assay was performed to identify Bim as a direct target of miR-214. Furthermore, this study showed that low Bim expression in NPC tissues correlated with poor survival of NPC patients. Taken together, our findings suggest that miR-214 plays an important role in NPC carcinogenesis.
DOI:10.1146/annurev.micro.51.1.565URLPMID:9343360 [本文引用: 1]

Annu Rev Microbiol. 1997;51:565-92. Review
DOI:10.1152/physrev.00013.2006URL [本文引用: 1]
DOI:10.1016/j.cell.2009.08.021URLPMID:19737514 [本文引用: 1]

The complex process of apoptosis is orchestrated by caspases, a family of cysteine proteases with unique substrate specificities. Accumulating evidence suggests that cell death pathways are finely tuned by multiple signaling events, including direct phosphorylation of caspases, whereas kinases are often substrates of active caspases. Importantly, caspase-mediated cleavage of kinases can terminate prosurvival signaling or generate proapoptotic peptide fragments that help to execute the death program and facilitate packaging of the dying cells. Here, we review caspases as kinase substrates and kinases as caspase substrates and discuss how the balance between cell survival and cell death can be shifted through crosstalk between these two enzyme families.
