 ,, 苏菁, 杨健源, 韦小燕, 陈凯玲, 陈珍, 陈深, 朱小源
,, 苏菁, 杨健源, 韦小燕, 陈凯玲, 陈珍, 陈深, 朱小源 ,广东省农业科学院植物保护研究所/广东省植物保护新技术重点实验室,广州 510640
,广东省农业科学院植物保护研究所/广东省植物保护新技术重点实验室,广州 510640Analysis of Magnaporthe oryzae Avirulent Genes in the Infected Hybrid Rice Combinations Derived from a Sterile Line of Guang 8 A
WANG WenJuan ,, SU Jing, YANG JianYuan, WEI XiaoYan, CHEN KaiLing, CHEN Zhen, CHEN Shen, ZHU XiaoYuan
,, SU Jing, YANG JianYuan, WEI XiaoYan, CHEN KaiLing, CHEN Zhen, CHEN Shen, ZHU XiaoYuan ,Plant Protection Research Institute, Guangdong Academy of Agricultural Sciences/Guangdong Provincial Key Laboratory of High Technology for Plant Protection, Guangzhou 510640
,Plant Protection Research Institute, Guangdong Academy of Agricultural Sciences/Guangdong Provincial Key Laboratory of High Technology for Plant Protection, Guangzhou 510640通讯作者:
第一联系人:
收稿日期:2018-06-22接受日期:2018-08-9网络出版日期:2018-12-26
| 基金资助: |
Received:2018-06-22Accepted:2018-08-9Online:2018-12-26

摘要
关键词:
Abstract
Keywords:
PDF (1317KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
汪文娟, 苏菁, 杨健源, 韦小燕, 陈凯玲, 陈珍, 陈深, 朱小源. 源于广8 A杂交稻组合的稻瘟病菌无毒基因型分析[J]. 中国农业科学, 2018, 51(24): 4633-4646 doi:10.3864/j.issn.0578-1752.2018.24.005
WANG WenJuan, SU Jing, YANG JianYuan, WEI XiaoYan, CHEN KaiLing, CHEN Zhen, CHEN Shen, ZHU XiaoYuan.
0 引言
【研究意义】水稻(Oryza sativa)是世界上最重要的粮食作物之一,全球一半以上的人口以稻米作为主食[1]。由稻瘟病菌(Magnaporthe oryzae;无性态:Pyricularia oryzae)引起的稻瘟病是全球水稻生产上最具毁灭性的病害之一[2],严重威胁着粮食产量,每年由该病害引起的水稻产量损失高达10%—30%,严重时甚至颗粒无收[3,4,5]。培育和种植抗病品种是控制该病害最经济与环保的措施[6,7]。以野败型优质籼稻不育系广8 A配组选育的广8优165、广8优169和广8优2168等杂交稻组合,是华南稻区近年来广泛推广种植的优质杂交稻组合[8]。随着该系列组合的连续大面积推广种植,其抗瘟性逐渐下降。探明侵染广8 A杂交稻组合的稻瘟病菌无毒基因类型及其变异情况,可为该稻区抗病品种的轮换及新品种的选育提供科学依据。【前人研究进展】水稻对稻瘟病菌的抗性符合“基因对基因”学说,即在水稻品种中存在一个抗性基因,在稻瘟病菌中也对应存在一个无毒基因[9];只有在稻瘟病菌的无毒基因与寄主抗性基因相互作用的前提下,水稻品种才表现出抗瘟性[10,11]。近年来,在水稻抗瘟基因和稻瘟病菌无毒基因的鉴定与分析方面,取得了重要的研究进展。目前,已有PWL1、PWL2、Avr1-CO39、AvrPita、ACE1、AvrPiz-t、AvrPia、AvrPii、AvrPik/km/kp、AvrPib、AvrPi9及AvrPi54等12个无毒基因完成了克隆[12,13,14,15]。并且,根据已克隆无毒基因的序列信息开发特异性分子标记,在鉴定稻瘟病菌群体中无毒基因的类型与变异方面已有大量的研究报道。李祥晓等[16]利用7个稻瘟病菌无毒基因的特异性引物,对177个来自黑龙江省的稻瘟病菌单孢菌株进行PCR检测,结果表明7个无毒基因以不同的出现频率在黑龙江省不同地区均有分布;王世维等[17]利用6个已克隆无毒基因分子标记,对辽宁省26个稻瘟病菌单孢菌株进行PCR检测及无毒基因PCR产物测序分析,结果表明在辽宁稻区稻瘟病菌中AvrPik、AvrPiz-t和AvrPita分布相对广泛;朱名海等[18]利用6个无毒基因分子标记对南繁区稻瘟病菌无毒基因的分布情况进行分析,结果表明除了无毒基因AvrPia在南繁非核心区的稻瘟病菌株中未检测到外,其他5个无毒基因在所检测的稻瘟病菌中均存在,其中无毒基因AvrPik的分布频率为100%。另一方面,随着国内外水稻抗稻瘟病近等基因系和单基因系的育成,国内****已开始利用这些单基因系鉴别系统开展稻瘟病菌致病小种变异的研究[19,20,21,22]。【本研究切入点】深入阐明分离自主栽品种稻瘟病菌小种的变异特征,需要从病原菌致病性变异和无毒基因型分析两方面开展研究。为准确探明侵染华南主推品种广8 A杂交稻组合的稻瘟病菌所携带的无毒基因类型及变异情况,本研究结合抗病单基因系品种致病性鉴定和已知无毒基因的扩增和测序,对采集自广8 A杂交稻组合的稻瘟病菌进行分析,研究不同菌株中无毒基因的类型及其变异情况。【拟解决的关键问题】通过25个稻瘟病抗性单基因系对采集自广8 A杂交稻组合的27个稻瘟病菌株进行致病型测定;根据8个已克隆无毒基因(Avr1-CO39、AvrPita、AvrPii、AvrPi9、AvrPiz-t、AvrPia、AvrPik/km/kp及AvrPib)功能性分子标记的检测,明确源自广8 A杂交稻组合的稻瘟病菌无毒基因或其等位基因的存在/缺失情况,并对扩增的无毒基因产物进行测序,比较分析测序菌株间无毒基因序列的变异特征;比较分离自广8 A杂交稻组合的稻瘟病菌菌株与其他来源菌株的无毒基因类型,为华南稻区抗瘟品种的合理布局与稻瘟病的有效控制提供依据。1 材料与方法
试验于2017年在广东省农业科学院植物保护研究所试验基地完成。1.1 病原菌材料、繁殖及产孢
病原菌来源:2012—2016年,从广东不同稻区的杂交稻主导品种广8 A杂交稻组合上采集穗颈瘟病样,从中分离纯化稻瘟病菌菌株。田间采集的穗颈瘟标样,用0.5%的强氯精消毒2 min,随后用无菌水清洗,置于灭菌的培养皿中保湿培养约3 d,利用孢子振落法进行病原菌株的单孢分离。共分离获得稻瘟病菌单孢菌株41个(表1),所有菌株保存于广东省农业科学院植物保护研究所。
Table 1
表1
表141个稻瘟病菌菌株及其来源
Table 1
| 采集地区 Sampling location | 采集品种 Sampling cultivar | 菌株编号 Strain number | 采集地区 Sampling location | 采集品种 Sampling cultivar | 菌株编号 Strain number | |
|---|---|---|---|---|---|---|
| 河源 Heyuan | 广8优165 Guang8you165 | GD12-535 | 河源 Heyuan | 广8优169 Guang8you169 | GD15-259 | |
| 阳江 Yangjiang | 广8优169 Guang8you169 | GD12-716 | 河源 Heyuan | 广8优169 Guang8you169 | GD15-270 | |
| 阳江 Yangjiang | 广8优169 Guang8you169 | GD12-721 | 河源 Heyuan | 广8优165 Guang8you165 | GD15-291 | |
| 阳江 Yangjiang | 广8优169 Guang8you169 | GD12-730 | 河源 Heyuan | 广8优2168 Guang8you2168 | GD15-304 | |
| 河源 Heyuan | 广8优169 Guang8you169 | GD13-493 | 河源 Heyuan | 广8优165 Guang8you165 | GD15-315 | |
| 阳江 Yangjiang | 广8优169 Guang8you169 | GD13-621 | 河源 Heyuan | 广8优188 Guang8you188 | GD15-316 | |
| 阳江 Yangjiang | 广8优169 Guang8you169 | GD13-626 | 云浮 Yunfu | 广8优169 Guang8you169 | GD15-436 | |
| 茂名 Maoming | 广8优169 Guang8you169 | GD13-659 | 阳江 Yangjiang | 广8优169 Guang8you169 | GD15-506 | |
| 阳江 Yangjiang | 广8优188 Guang8you188 | GD13-3009 | 阳江 Yangjiang | 广8优2168 Guang8you2168 | GD15-520 | |
| 阳江 Yangjiang | 广8优2168 Guang8you2168 | GD13-3024 | 阳江 Yangjiang | 广8优169 Guang8you169 | GD15-523 | |
| 河源 Heyuan | 广8优2168 Guang8you2168 | GD14-070 | 阳江 Yangjiang | 广8优169 Guang8you169 | GD15-530 | |
| 云浮 Yunfu | 广8优169 Guang8you169 | GD14-298 | 阳江 Yangjiang | 广8优169 Guang8you169 | GD15-542 | |
| 阳江 Yangjiang | 广8优169 Guang8you169 | GD14-349 | 阳江 Yangjiang | 广8优165 Guang8you165 | GD15-543 | |
| 阳江 Yangjiang | 广8优169 Guang8you169 | GD14-366 | 茂名 Maoming | 广8优169 Guang8you169 | GD15-586 | |
| 阳江 Yangjiang | 广8优169 Guang8you169 | GD14-368 | 河源 Heyuan | 广8优165 Guang8you165 | GD16-246 | |
| 阳江 Yangjiang | 广8优169 Guang8you169 | GD14-372 | 河源 Heyuan | 广8优188 Guang8you188 | GD16-272 | |
| 阳江 Yangjiang | 广8优165 Guang8you165 | GD14-376 | 河源 Heyuan | 广8优165 Guang8you165 | GD16-280 | |
| 阳江 Yangjiang | 广8优169 Guang8you169 | GD14-381 | 阳江 Yangjiang | 广8优169 Guang8you169 | GD16-327 | |
| 阳江 Yangjiang | 广8优165 Guang8you165 | GD14-401 | 阳江 Yangjiang | 广8优2168 Guang8you2168 | GD16-334 | |
| 河源 Heyuan | 广8优2168 Guang8you2168 | GD15-023 | 从化 Conghua | 广8优169 Guang8you169 | GD16-3071 | |
| 韶关 Shaoguan | 广8优金占 Guang8youjinzhan | GD15-236 |
新窗口打开|下载CSV
菌株活化及产孢:将保存的菌株转移至酵母固体培养基,25℃左右的生化培养箱中活化培养7 d以上;然后将菌丝体转接到玉米培养基上繁殖,25℃左右培养约13 d。用消毒过的无菌水洗去玉米粒表面的菌丝,将玉米粒置于消毒的搪瓷盘中(25 cm×19 cm×2 cm),上面覆盖一层湿纱布,然后在日光灯下光照培养3—4 d,进行产孢。
1.2 致病型测定
植物材料:25个抗稻瘟病单基因系分别为IRBLa-A(Pia)、IRBLi-F5(Pii)、IRBLks-F5(Piks)、IRBLk-Ka(Pik)、 IRBLkp-K60(Pikp)、IRBLkh-K3(Pikh)、IRBLz-Fu(Piz)、IRBLz5-CA(Pi2)、IRBLzt-T(Piz-t)、IRBLta-K1(Pita)、IRBLb-B(Pib)、IRBLt-K59(Pit)、IRBLsh-S(Pish)、IRBL1-CL(Pi1)、IRBL3-CP4(Pi3)、IRBL5-M(Pi5)、IRBL7-M(Pi7)、IRBL9-W(Pi9)、IRBL12-M(Pi12)、IRBL19-A(Pi19)、IRBLkm-Ts(Pikm)、IRBL20-IR24(Pi20)、IRBLta2-Re(Pita2)、IRBL11-Zh(Pi11)、NIL-e1(Pi50),其中NIL-e1是ZHU等[23]以丽江新团黑谷为背景,经过6次回交及6次自交发展而成;含Pi50的抗稻瘟病近等基因系,由广东省农业科学院植物保护研究所选育,其余单基因系由国际水稻研究所选育。感病对照:丽江新团黑谷(LTH)。植物材料种植:将水稻种子催芽后穴播于30 cm×20 cm×5 cm规格的搪瓷盆里,每盆播25个材料,每个材料播种量约为10粒种子。稻苗采取旱育栽培,长至1叶1心期用硫酸铵施肥,每盆施肥量为0.5 g,接种前共需施肥3次。待稻苗长至3.5—4叶龄时,进行人工喷雾接种。接种及调查:用无菌水洗下玉米粒上的孢子,用2层塑料细纱网隔去除玉米残渣,配成适量浓度的孢子液,一般将孢子液浓度调至1×105个/mL,进行人工喷雾接种。接种后置于遮光密闭的培养箱中暗培养,在25℃及相对湿度达95%以上的条件下保湿24 h,之后转至玻璃温室,在25—28℃下保湿培养至稻苗发病,一般在接种7 d左右观察稻苗的整体发病情况进行调查。病级调查方法按照国际水稻研究所稻瘟病圃苗瘟分级标准进行:0—3级为抗(R);4—9级为感(S)。1.3 稻瘟病菌DNA的提取
将分离纯化的稻瘟病菌单孢菌株在CM(完全培养基)固体培养基上活化培养,挑取适量菌丝块接到CM液体培养基中,于28℃摇床,160 r/min振荡培养3—5 d,收集菌丝体;用Omega公司的真菌DNA抽提试剂盒,提取菌丝体的基因组DNA。1.4 无毒基因分子标记检测
引物设计:8个已克隆稻瘟病菌的无毒基因(Avr1- CO39、AvrPita、AvrPii、AvrPi9、AvrPiz-t、AvrPia、AvrPik、AvrPib)中,AvrPib的特异引物根据GenBank中AvrPib基因序列,利用软件Primer Premier 5.0设计;其他7个无毒基因的特异引物参照SELISANA等[24]开发的序列。所有引物都在生工生物工程(上海)股份有限公司(广州生工)合成。引物序列详见表2。Table 2
表2
表2本研究中用于检测无毒基因的引物
Table 2
| 引物 Primer | 引物序列 Sequence (5′-3′) | 预期片段大小 Expected fragment size (bp) | 目的 Purpose |
|---|---|---|---|
| Avr1-CO39 F | TGCCGCATTTTGCTAACCG | 994 | 检测Avr1-CO39启动子及CDS To detect Avr1-CO39 promoter and CDS |
| Avr1-CO39 R | GCGAATCCATAGACAAGGAC | ||
| AvrPita F | CAGGCATACATTGGAGAGCC | 1549 | 检测AvrPita启动子及CDS To detect AvrPita promoter and CDS |
| AvrPita R | CCCTCCATTCCAACACTAAC | ||
| AvrPii F | GGTAGATATCCGCTGACTGG | 839 | 检测AvrPii启动子及CDS To detect AvrPii promoter and CDS |
| AvrPii R | ACTGTCCGCCGCTCGTTTGG | ||
| AvrPi9 F | ATGCAGTTCTCTCAGATCCTC | 342 | 检测AvrPi9 CDS To detect AvrPi9 CDS |
| AvrPi9 R | CTACCAGTGCGTCTTTTCGAC | ||
| AvrPiz-t F | GTTGCGATTATGATCCGTCG | 1144 | 检测AvrPiz-t启动子及CDS To detect AvrPiz-t promoter and CDS |
| AvrPiz-t R | GTACTCTAGCAAACGACCGG | ||
| AvrPia F | CAGAGAAACGGACTTGGAGG | 1220 | 检测AvrPia启动子及CDS To detect AvrPia promoter and CDS |
| AvrPia R | GGTATACACGTACGGTAGGG | ||
| AvrPik-CDE F | TCCTGCTGCTAACTCCATTC | ~1000 | 检测AvrPik-CDE型启动子及CDS To detect the type of AvrPik-CDE promoter and CDS |
| AvrPik-CDE R | GGGTACAGGAATACCAGG | ||
| AvrPik-D F | ACCCTAACTTTTTCGACC | 286 | 检测AvrPik-D型CDS To detect the type of AvrPik-D CDS |
| AvrPik-D R | TCAACCAAGCGTAAACCTCG | ||
| AvrPib F | ATGCCGACAATGCGAGGTAT | 1598 | 检测AvrPib启动子及CDS To detect AvrPib promoter and CDS |
| AvrPib R | GGACAAGGGAGGCAAATCTAAC |
新窗口打开|下载CSV
无毒基因分子标记检测:PCR反应总体积为20 μL,其中包括稻瘟病菌基因组DNA(20—30 ng·μL-1)1 μL;10×PCR Mixture10 μL,正、反向引物(10 μmol·L-1)各0.5 μL,ddH2O 8 μL。PCR扩增程序:94℃预变性3 min,94℃变性30 s,56—60℃(根据不同引物而定)退火30 s,72℃延伸1 min/kb,共设置35次循环,最后72℃延伸5 min。PCR产物于1%—1.2%的琼脂糖凝胶中电泳检测。
1.5 部分无毒基因序列测序分析
利用高保真酶KOD-Plus(东洋纺,日本)进行PCR扩增,并对产物进行目的片段的纯化,将纯化的PCR产物送到广州英潍捷基公司进行测序。采用Lasergene 7.0软件中的SeqMan(http://www.dnastar.com/)进行全部测序序列的比对与拼接;并采用DNAStar MegAlign ClustalV软件对有差异的核苷酸序列进行比较分析。2 结果
2.1 分离自广8优组合的菌株致病型
利用25个水稻抗稻瘟病单基因系和感病对照品种丽江新团黑谷,对其中采集自广8 A杂交稻组合的27个稻瘟病菌株进行致病型测定。结果表明,这些菌株对25个单基因系的无毒性频率具有丰富的多样性(表3)。所测定的菌株对其中的5个单基因系NIL-e1(Pi50)、IRBL9-W(Pi9)、IRBLz5-CA (Pi2)、IRBLkh-K3(Pikh)和IRBLb-B(Pib)表现高无毒性频率(>85%);其中,所有菌株对单基因系NIL-e1(Pi50)和IRBL9-W(Pi9)均表现无毒。然而,这些菌株对其中的7个单基因系IRBLt-K59(Pit)、IRBL12-M(Pi12)、IRBLks-F5(Piks)、IRBLa-A(Pia)、IRBL3-CP4(Pi3)、IRBL11-Zh(Pi11)、IRBL19-A(Pi19)表现相对较低的无毒性频率(<20%)。上述结果表明,分离自广8 A杂交稻组合的稻瘟病菌对不同的抗瘟单基因系表现出不同的致病型。Table 3
表3
表3不同菌株对25个抗稻瘟单基因系的致病型
Table 3
| 菌株 Strain | IRBLs单基因系对菌株的反应 Reactions of IRBLs to the strains | LTH | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | ||
| Pia | Pii | Piks | Pik | Pikp | Pikh | Piz | Piz5 | Pizt | Pita | Pib | Pit | Pish | Pi1 | Pi3 | Pi5 | Pi7 | Pi9 | Pi12 | Pi19 | Pikm | Pi20 | Pita2 | Pi11 | Pi50 | ||
| GD12-535 | S | S | S | S | S | R | S | S | R | R | R | S | S | S | S | S | S | R | S | S | S | S | R | S | R | S |
| GD13-3009 | S | R | S | R | R | R | R | R | S | S | R | R | R | S | R | R | R | R | S | S | R | S | S | S | R | S |
| GD13-3024 | S | S | S | R | R | S | R | R | R | S | R | S | S | S | S | S | R | R | S | S | S | S | S | S | R | S |
| GD13-493 | S | S | S | R | S | R | S | R | S | S | R | S | S | S | S | S | S | R | S | S | S | S | R | S | R | S |
| GD13-621 | S | R | S | R | R | R | S | R | R | S | R | S | R | R | S | R | R | R | S | S | R | S | R | S | R | S |
| GD13-659 | S | S | S | R | S | R | S | R | S | S | R | S | S | R | S | S | S | R | S | S | S | S | S | S | R | S |
| GD14-070 | S | S | S | R | R | R | R | R | S | S | R | S | S | S | S | S | R | R | S | S | R | S | S | S | R | S |
| GD14-349 | S | S | S | R | R | R | R | R | R | R | R | S | S | R | S | S | R | R | S | S | R | S | R | S | R | S |
| GD14-366 | S | R | S | R | R | R | R | R | R | R | R | S | R | S | S | S | R | R | S | R | S | R | S | S | R | S |
| GD14-368 | S | R | S | S | R | S | R | R | R | R | S | S | R | R | S | S | S | R | S | R | S | S | S | S | R | S |
| GD14-376 | S | S | S | S | S | S | S | R | R | S | R | S | S | S | S | S | S | R | S | S | S | S | S | S | R | S |
| GD14-401 | S | R | S | R | R | R | R | R | S | S | R | S | S | S | S | S | R | R | S | S | S | S | S | S | R | S |
| GD15-23 | S | S | S | S | S | R | R | R | S | S | R | S | S | S | S | S | S | R | S | S | S | S | S | S | R | S |
| GD15-259 | S | S | S | S | S | R | R | R | S | S | R | S | S | S | S | S | S | R | S | S | S | S | S | S | R | S |
| GD15-270 | S | S | S | S | S | R | S | R | R | S | R | S | S | S | S | R | S | R | S | S | S | S | S | S | R | S |
| GD15-291 | S | S | S | R | R | R | S | R | R | S | R | S | S | S | S | S | R | R | S | S | S | S | S | S | R | S |
| GD15-315 | R | R | R | R | S | R | R | R | R | R | R | S | R | R | R | R | R | R | R | R | R | R | S | R | R | S |
| GD15-436 | S | S | S | S | S | R | R | R | S | S | R | S | S | S | S | R | S | R | S | S | S | S | S | S | R | S |
| GD15-506 | S | S | S | S | S | S | S | R | R | S | S | S | R | S | S | S | S | R | S | S | S | R | R | R | R | S |
| GD15-523 | S | S | S | R | S | R | S | R | S | S | S | S | S | S | S | S | S | R | S | S | S | S | S | S | R | S |
| GD15-530 | S | R | S | R | R | R | R | S | S | R | R | S | S | R | S | S | S | R | S | S | S | S | S | S | R | S |
| GD15-586 | S | R | S | R | S | R | R | S | S | S | R | S | S | R | S | S | S | R | S | S | S | S | S | S | R | S |
| GD16-272 | R | R | S | R | S | R | R | R | R | R | R | S | R | R | S | R | R | R | S | R | R | R | R | S | R | S |
| GD16-280 | R | S | R | R | S | R | R | R | R | R | R | S | S | S | R | R | S | R | S | R | S | R | R | S | R | S |
| GD16-3071 | S | S | S | S | S | R | S | R | R | S | R | S | S | S | S | S | S | R | S | S | S | R | R | R | R | S |
| GD16-327 | S | S | S | R | S | R | S | R | S | S | S | S | S | R | S | R | S | R | S | S | S | S | S | S | R | S |
| GD16-334 | S | S | S | S | S | R | R | R | S | S | R | S | S | S | S | S | S | R | S | S | S | S | S | S | R | S |
| 无毒基因频率 The frequency of avirulent genes (%) | 11.1 | 33.3 | 7.4 | 63.0 | 37.0 | 85.2 | 59.3 | 88.9 | 51.9 | 29.6 | 85.2 | 3.7 | 25.9 | 33.3 | 11.1 | 29.6 | 37.0 | 100.0 | 3.7 | 18.5 | 22.2 | 22.2 | 29.6 | 11.1 | 100.0 | |
新窗口打开|下载CSV
2.2 稻瘟病菌8个无毒基因在广8优组合分离菌株中的单倍型
为了确定试验菌株中8个已克隆无毒基因的单倍型,利用SELISANA等[24]开发的7个无毒基因的分子标记及本文设计开发的AvrPib分子标记(表2),通过PCR扩增,对27个供试菌株进行无毒基因单倍型分析。部分菌株无毒基因的PCR扩增结果如图1所示。在27个供试菌株中均能检测到无毒基因AvrPi9的片段;有26个菌株能检测到AvrPik-CDE基因片段,其中经AvrPik-D型引物检测,有12个菌株能检测到AvrPik-D型。相反地,在任何菌株中都扩增不到Avr1-CO39、AvrPii和AvrPia。无毒基因AvrPiz-t和AvrPib在所检测的27个菌株中可扩增出3种带型:扩增预期大小的条带、无扩增条带和有转座子插入的条带,但只在其中的4个菌株中检测到预期大小的目的基因片段。无毒基因AvrPita在所检测的27个菌株中可扩增出2种带型:扩增预期大小的条带和无扩增条带,仅在其中的3个菌株中检测到AvrPita预期大小的片段。图1
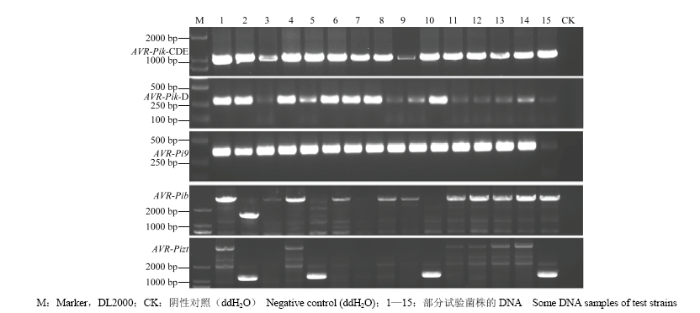 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1基于无毒基因分子标记的稻瘟病菌菌株无毒基因检测
Fig. 1Detection of Avr genes in M. oryzae strains based on molecular markers of avirulent genes
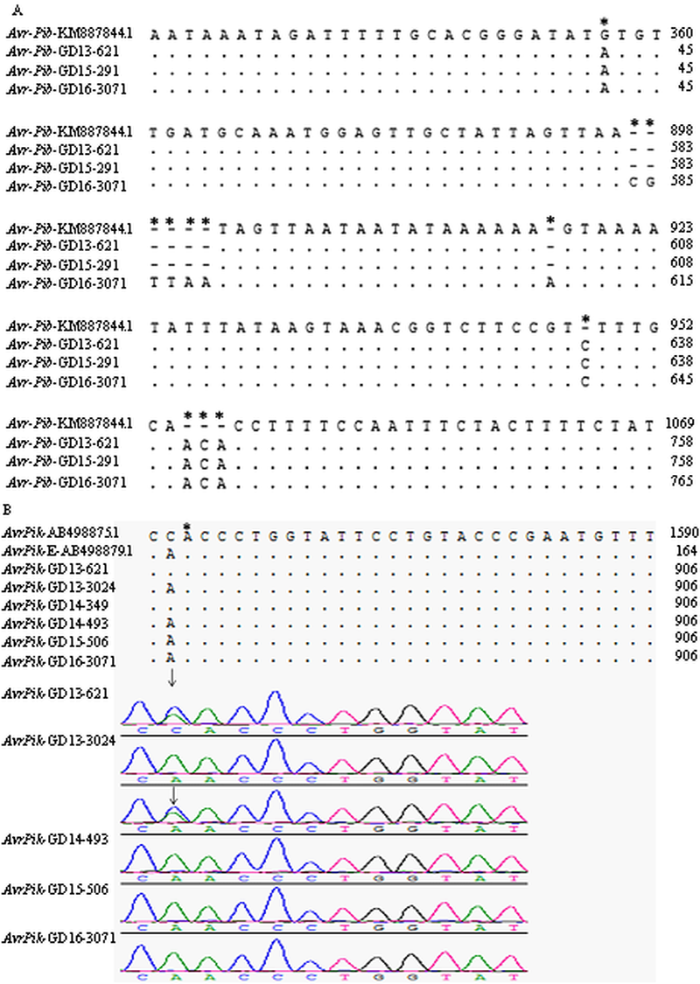
对GD13-621、GD13-3024、GD14-349、GD14-493、GD15-291、GD15-506与GD16-3071等7个菌株进行相应无毒基因全长或CDS区域的PCR产物测序,该7个菌株分别能检测到AvrPita、AvrPi9、AvrPiz-t、AvrPik或AvrPib等无毒基因。结果表明,所测序7个菌株的AvrPi9基因编码区序列和5个测序菌株包含启动子区的AvrPiz-t基因序列,与其各自无毒基因的序列完全一致(GenBank登录号分别为KM004023和EU837058);3个菌株包含启动子区的AvrPita基因编码区序列与参照AvrPita序列无差异(GenBank登录号为AF207841.1)。然而,3个菌株的AvrPib基因序列与参照AvrPib的序列(GenBank登录号KM887844.1)相比,在基因的上游区域存在2个SNP的差异及3个连续碱基的插入,其中菌株GD16-3071比较特殊,存在连续6个碱基的插入(图2-A);6个菌株AvrPik仅在编码区存在1个SNP的差异即136(C/A),导致氨基酸的错义突变46(H/N),表现为菌株携带AvrPik-D或AvrPik-E型;菌株GD13-621和GD14-349在该位点出现了双峰的现象,可能这2个菌株中同时含有AvrPik-D型和AvrPik-E型基因(图2-B);在同一个菌株中同时存在AvrPik-D型和AvrPik-E型的情况,在YOSHIDA等[12]的文中未见报道。
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2测序菌株的序列与参照AvrPib或AvrPik序列比对
Fig. 2Sequences alignment of AvrPib or AvrPik from tested strains to their standard sequences
2.3 稻瘟病菌中Avr基因的存在与各自R基因致病型间的相关性
稻瘟病菌菌株中无毒基因的存在与相应抗病基因致病型间的相关性分析,是基于无毒基因特异性分子标记确定稻瘟病菌致病谱的重要基础。将无毒基因分子标记检测的稻瘟病菌无毒基因型与抗稻瘟病单基因系分析的病原菌致病型进行比较,结果表明所检测的27个菌株都含有无毒基因AvrPi9,并且在致病型监测试验中,这27个菌株对含Pi9的抗瘟单基因系IRBL9-W都显示无毒性。含有无毒基因AvrPiz-t的菌株对其相应的抗瘟单基因系IRBLzt-T也都表现无毒性;然而,一些含AvrPita和AvrPib的菌株却能侵染其对应的抗病基因单基因系。另外,有些不含有相应无毒基因的菌株对IRBLi-F5和IRBLa-A单基因系显示无毒性(如GD13-3009、GD13-632和GD15-315等);有4个菌株(GD14-366、GD15-315、GD16-272和GD16-280)不含有对应的无毒基因,却对其中的4—5个抗瘟单基因系IRBLi-F5、IRBLzt-T、IRBLta-K1、IRBLb-B或IRBLa-A表现出一致的无毒性(表4)。Table 4
表4
表4稻瘟病菌菌株无毒基因单倍型与致病型的比较
Table 4
| 菌株 Strain | 对Pi9的反应 Reactions to Pi9 | 对Pik不同等位基因的反应 Reactions to different Pik alleles | 对Pii的反应 Reactions to Pii | 对Piz-t的反应 Reactions to Piz-t | 对Pita的反应 Reactions to Pita | 对Pib的反应 Reactions to Pib | 对Pia和Pi-CO39的反应 Reactions to Pia and Pi-CO39 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IRBL9 -W | AVR单倍型 AVR haplotype | IRBLk -Ka | IRBLkp -K60 | IRBLkh -K3 | IRBL1 -CL | IRBL7 -M | IRBLkm -Ts | AVR单倍型 AVR haplotype | IRBLi -F5 | AVR单倍型 AVR haplotype | IRBLzt -T | AVR单倍型 AVR haplotype | IRBLta -K1 | AVR单倍型 AVR haplotype | IRBLb -B | AVR单倍型 AVR haplotype | IRBLa -A | AVR单倍型 AVR haplotype | ||
| Avr-Pia | Avr1-CO39 | |||||||||||||||||||
| GD12-535 | R | + | S | S | R | S | S | S | D | S | - | R | - | R | - | R | - | S | - | - |
| GD13-3009 | R | + | R | R | R | S | R | R | D | R | - | S | - | S | - | R | - | S | - | - |
| GD13-3024 | R | + | R | R | S | S | R | S | - | S | - | R | + | S | + | R | - | S | - | - |
| GD13-493 | R | + | R | S | R | S | S | S | - | S | - | S | - | S | + | R | - | S | - | - |
| GD13-621 | R | + | R | R | R | S | R | R | D | R | - | R | + | S | + | R | + | S | - | - |
| GD13-659 | R | + | R | S | R | S | S | S | - | S | - | S | - | S | - | R | - | S | - | - |
| GD14-349 | R | + | R | R | R | S | R | R | D | S | - | R | - | R | + | R | - | S | - | - |
| GD14-366 | R | + | R | R | R | S | R | S | D | R | - | R | - | R | - | R | - | S | - | - |
| GD14-368 | R | + | S | R | S | S | S | S | - | R | - | R | - | R | - | S | - | S | - | - |
| GD14-376 | R | + | S | S | S | S | S | S | - | S | - | R | + | S | - | R | - | S | - | - |
| GD14-401 | R | + | R | R | R | S | R | S | D | R | - | S | - | S | - | R | - | S | - | - |
| GD14-70 | R | + | R | R | R | S | R | R | D | S | - | S | - | S | - | R | - | S | - | - |
| GD15-23 | R | + | S | S | R | S | S | S | - | S | - | S | - | S | - | R | - | S | - | - |
| GD15-259 | R | + | S | S | R | S | S | S | - | S | - | S | - | S | - | R | - | S | - | - |
| GD15-270 | R | + | S | S | R | S | S | S | - | S | - | R | + | S | - | R | - | S | - | - |
| GD15-291 | R | + | R | R | R | S | R | S | D | S | - | R | + | S | - | R | + | S | - | - |
| GD15-315 | R | + | R | S | R | S | R | R | D | R | - | R | - | R | - | R | - | R | - | - |
| GD15-436 | R | + | S | S | R | S | S | S | - | S | - | S | - | S | - | R | - | S | - | - |
| GD15-506 | R | + | S | S | S | S | S | S | - | S | - | R | + | S | - | S | + | S | - | - |
| GD15-523 | R | + | R | S | R | S | S | S | - | S | - | S | - | S | - | S | - | S | - | - |
| GD15-530 | R | + | R | R | R | S | S | S | D | R | - | S | - | R | - | R | - | S | - | - |
| GD15-586 | R | + | R | S | R | S | S | S | - | R | - | S | - | S | - | R | - | S | - | - |
| GD16-272 | R | + | R | S | R | S | R | R | D | R | - | R | - | R | - | R | - | R | - | - |
| GD16-280 | R | + | R | S | R | S | S | S | D | S | - | R | - | R | - | R | - | R | - | - |
| GD16-3071 | R | + | S | S | R | S | S | S | - | S | - | R | + | S | - | R | + | S | - | - |
| GD16-327 | R | + | R | S | R | S | S | S | - | S | - | S | - | S | - | S | - | S | - | - |
| GD16-334 | R | + | S | S | R | S | S | S | - | S | - | S | - | S | - | R | - | S | - | - |
新窗口打开|下载CSV
2.4 源自广8 A杂交稻组合以及其他主栽品种的稻瘟病菌无毒基因型比较
为了进一步比较源自广8 A杂交稻组合的稻瘟病菌菌株与其他品种来源菌株致病性的差异,选取25个抗性单基因系中的11个品种对源自广8优组合的41个菌株进行致病性测定。这11个品种所含的抗病基因常见于华南稻区水稻品种中。结果表明,所测试的41个菌株对11个抗稻瘟病单基因系的致病性具有丰富的多样性,对其中的4个单基因系IRBL9-W(Pi9)、NIL-e1(Pi50)、IRBLkh-K3(Pikh)及IRBL1-CL(Pi1)表现高的无毒性频率(>92%),相反地,对其中的2个单基因系IRBLzt-T(Piz-t)和IRBLta2-Re(Pita2)表现较低的无毒性频率(<40%)(表5)。Table 5
表5
表541个菌株对11个抗稻瘟病单基因系的致病性
Table 5
| 采集品种 Sampling cultivar | 菌株 Strain | 11个单基因系对菌株的反应 Reactions of 11 blast-resistant lines to the strains | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
| Pi9 | Pi2 | Piz | Pikh | Pi1 | Pikp | Pita2 | Pish | Pii | Piz-t | Pi50 | ||
| 广8优165 Guang8you165 | GD12-535 | R | R | S | R | R | S | R | S | S | R | R |
| 广8优169 Guang8you169 | GD12-716 | R | R | R | R | R | R | R | R | S | S | R |
| 广8优169 Guang8you169 | GD12-721 | R | R | R | R | R | S | S | S | S | S | R |
| 广8优169 Guang8you169 | GD12-730 | R | R | S | R | R | S | S | S | S | S | R |
| 广8优169 Guang8you169 | GD13-493 | R | R | S | R | R | S | R | S | S | S | R |
| 广8优169 Guang8you169 | GD13-621 | R | R | R | R | R | R | R | R | R | R | R |
| 广8优169 Guang8you169 | GD13-626 | R | R | R | R | R | R | S | R | S | R | R |
| 广8优169 Guang8you169 | GD13-659 | R | S | S | R | R | S | S | S | S | S | R |
| 广8优188 Guang8you188 | GD13-3009 | R | R | S | R | R | R | S | S | S | S | R |
| 广8优2168 Guang8you2168 | GD13-3024 | R | R | R | R | R | R | S | R | R | S | R |
| 广8优2168 Guang8you2168 | GD14-070 | R | R | R | R | R | S | S | S | S | S | R |
| 广8优169 Guang8you169 | GD14-298 | R | R | R | R | R | R | R | R | R | R | R |
| 广8优169 Guang8you169 | GD14-349 | R | R | S | R | R | R | R | S | S | S | R |
| 广8优169 Guang8you169 | GD14-366 | R | S | R | R | S | R | S | R | R | S | R |
| 广8优169 Guang8you169 | GD14-368 | R | S | R | R | R | R | S | R | R | R | R |
| 广8优169 Guang8you169 | GD14-372 | R | S | R | R | S | S | S | R | R | S | R |
| 广8优165 Guang8you165 | GD14-376 | R | R | S | R | R | S | S | R | S | R | R |
| 广8优169 Guang8you169 | GD14-381 | R | R | R | R | R | R | S | S | S | S | R |
| 广8优165 Guang8you165 | GD14-401 | R | S | R | R | R | R | S | S | S | S | R |
| 广8优2168 Guang8you2168 | GD15-023 | R | R | S | R | R | S | S | S | S | S | R |
| 广8优金占 Guang8youjinzhan | GD15-236 | R | S | R | R | R | S | S | R | R | S | R |
| 广8优169 Guang8you169 | GD15-259 | R | S | S | R | R | S | S | S | S | S | R |
| 广8优169 Guang8you169 | GD15-270 | R | R | S | S | R | S | S | S | R | R | R |
| 广8优165 Guang8you165 | GD15-291 | R | S | S | R | R | R | R | R | R | S | R |
| 广8优2168 Guang8you2168 | GD15-304 | R | R | R | R | R | R | S | S | S | R | R |
| 广8优165 Guang8you165 | GD15-315 | R | S | R | R | R | R | S | R | R | S | R |
| 广8优188 Guang8you188 | GD15-316 | R | S | R | R | R | R | R | R | R | S | R |
| 广8优169 Guang8you169 | GD15-436 | R | R | S | R | R | S | S | S | S | S | R |
| 广8优169 Guang8you169 | GD15-506 | R | R | R | R | R | S | S | R | R | S | R |
| 广8优2168 Guang8you2168 | GD15-520 | R | R | S | R | R | S | S | S | R | S | R |
| 广8优169 Guang8you169 | GD15-523 | R | R | R | R | R | S | S | S | S | S | R |
| 广8优169 Guang8you169 | GD15-530 | R | S | R | R | R | R | R | R | R | R | R |
| 广8优169 Guang8you169 | GD15-542 | R | R | R | R | R | R | S | S | R | S | R |
| 广8优165 Guang8you165 | GD15-543 | R | R | R | R | R | R | R | R | R | S | R |
| 广8优169 Guang8you169 | GD15-586 | R | S | R | R | R | S | S | S | S | S | R |
| 广8优165 Guang8you165 | GD16-246 | R | S | S | R | R | R | R | S | S | S | S |
| 广8优188 Guang8you188 | GD16-272 | R | R | R | R | R | R | R | R | S | S | R |
| 广8优165 Guang8you165 | GD16-280 | R | R | R | R | R | R | R | R | R | R | R |
| 广8优169 Guang8you169 | GD16-327 | R | R | S | R | R | S | S | S | S | S | R |
| 广8优2168 Guang8you2168 | GD16-334 | R | R | S | R | R | S | S | S | S | S | R |
| 广8优169 Guang8you169 | GD16-3071 | R | R | S | R | S | S | R | S | S | R | R |
| 无毒基因频率 The frequency of avirulent genes (%) | 100.00 | 68.29 | 58.54 | 97.56 | 92.68 | 51.22 | 34.15 | 43.90 | 41.46 | 26.83 | 97.56 | |
新窗口打开|下载CSV
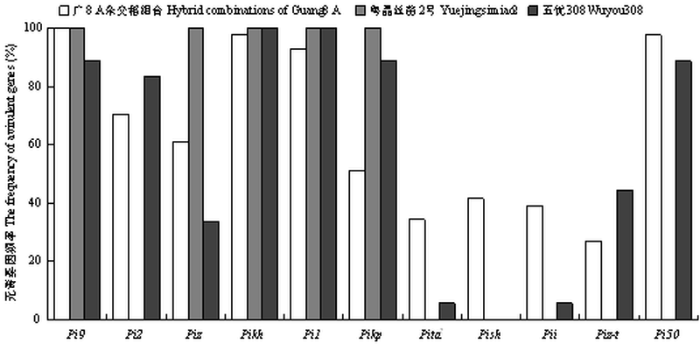
将11个抗稻瘟病单基因系鉴别品种对源自常规稻粤晶丝苗2号、杂交稻五优308和本研究的广8 A杂交稻组合的稻瘟病菌致病型分析进行比较,结果表明分离自广8 A杂交稻组合、粤晶丝苗2号[21]和五优308[22]的菌株对其中的3个单基因系IRBL9-W(Pi9)、IRBLkh-K3(Pikh)和IRBL1-CL(Pi1)表现出一致的较高无毒性频率(>88%);对其中的1个单基因系IRBLta2-Re(Pita2)表现出一致较低的无毒性频率(<40%)。另外,不同来源菌株间的致病性存在一定的差异,分离自粤晶丝苗2号的菌株对含抗病基因Pikp与Piz单基因系的无毒基因频率高达100%,但所测试的菌株可以侵染含抗病基因Pi50、Pish、Pi2及Piz-t的单基因系;分离自五优308的菌株对含抗病基因Pi2、Pikp和Pi50的无毒基因频率达83%以上,但所测试菌株都能侵染含Pish的单基因系;分离自广8 A杂交稻系列组合的菌株对含抗病基因Piz-t的无毒基因频率较低(26.83%)(图3)。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3源自不同水稻品种的稻瘟病菌无毒基因频率比对
Fig. 3Comparison the frequency of different Avr genes in M. oryzae from different rice cultivars
3 讨论
3.1 无毒基因监测与品种布局
近年来,水稻稻瘟病在华南稻区区域性或间歇性暴发成灾,多次出现一个或几个品种在部分稻区稻瘟病暴发,造成粮食的大面积减产甚至绝收[22,25]。监测本地区主栽水稻品种稻瘟病菌无毒基因的类型及发生情况,可为抗病品种在基因型水平上的合理布局与利用提供科学依据。侵染华南不同主栽品种的稻瘟病菌无毒基因型具有共性及差异性。分离自粤晶丝苗2号、五优308和广8优组合的稻瘟病菌对单基因IRBL9-W(Pi9)、IRBLkh-K3(Pikh)和IRBL1-CL(Pi1)表现出一致的高无毒性频率(>88%),对单基因系IRBLta2-Re(Pita2)和IRBLi-F5(Pii)表现出较低的无毒性频率(<40%);分离自广8优组合的稻瘟病菌对单基因系NIL-e1(Pi50)均表现无毒,分离自粤晶丝苗2号的稻瘟病菌不侵染含抗病基因Pikp及Piz单基因系,分离自五优308的稻瘟病菌对含抗病基因Pi2的品种呈现较高的无毒性频率。在上述3类品种均感病的稻区,可以布局含Pi9、Pikh或Pi1的抗病品种,而上述品种由于其无毒基因型存在一定的差异,可考虑在同一稻区进行轮换使用,以减轻病原菌无毒基因型单一化的选择压力。前人研究表明,抗瘟基因Pi50与Pi9在我国水稻育种上具有重要的应用价值,ZHU等[23]对抗瘟基因Pi50的研究表明,该基因对华南地区稻瘟病菌表现出广谱抗性;李进斌等[26]分析了22个抗病基因在云南3个稻区的抗性,表明Pi9和Pi2在该3个稻区具有良好的抗性;张国民等[27]分析了24个抗病基因在我国寒地稻区的抗性,表明Pi9对该稻区的稻瘟病菌具有广谱抗性。因此,含Pi50或Pi9的水稻品种抗瘟性好,可与广8 A杂交稻组合进行轮换使用。
3.2 无毒基因变异机制
“植物-病原菌”的相互作用是两者协同进化的主要推动力[28],目前存在两种寄主与病原菌的进化机制,即“军备竞赛”和“阵地战模式”机制[29]。无毒基因AvrPik与Pik等位基因间呈现竞争性的阶梯型共进化模式[30],该基因的突变主要是编码区单碱基的突变引起氨基酸的变异。在本研究中检测到了已报道的AvrPik-D型和AvrPik-E型等位基因,其中AvrPik-D型可被抗病基因Pik、Pik-p、Pik-m和Pik-h所识别;AvrPik-E型可被抗病基因Pik、Pikh和Pikm识别[30]。本研究在26个菌株中都能检测到AvrPik-CDE基因片段,利用AvrPik-D型引物在其中的12个菌株能检测到AvrPik-D型;说明含Pik、Pikp、Pikm、Pikh及其等位基因的水稻品种可抵御侵染广8 A杂交稻组合的稻瘟病菌。汪文娟等[31]在华南杂交稻裕优132、恒丰优华占、吉丰优华占等组合中分别检测到Pik或Pikp;这些品种可与广8 A杂交稻组合在生产上轮换使用。无毒基因AvrPib等位基因在华南稻区所受的自然选择压较强,致使华南稻区AvrPib等位基因具有丰富的遗传多样性;且从华南到东北AvrPib的出现频率逐渐增加[13]。在本研究中,AvrPib在所检测的27个菌株中可扩增出3种带型:扩增预期大小的条带、无扩增条带和有转座子插入的条带;但分别只在其中的4个菌株中检测到预期大小的目的基因片段;且其序列与已克隆的无毒基因在基因编码区的上游存在SNP或几个碱基的缺失,证明AvrPib具有较强的变异能力,遗传多样性丰富,与前人的研究结果相符[13]。
3.3 无毒基因型分析
基于抗稻瘟病单基因系的苗期致病性分析,结合已克隆无毒基因对病原菌的单倍型监测,可有效地用于稻瘟病菌群的致病谱分析及水稻品种有效抗病基因的推断[16,32]。据报道,利用不同遗传背景的品种创建的抗稻瘟病单基因系,其鉴定结果受不同品种的影响较大[33]。基于无毒基因型分子标记的诊断,可对抗稻瘟病单基因系鉴定结果进行科学的补充。本研究利用水稻抗稻瘟病单基因系和无毒基因分子标记对病原菌进行了无毒基因型分析,发现个别单基因系的检测结果与无毒基因标记检测的结果存在差异,如有4个菌株(GD14-366、GD15-315、GD16-272和GD16-280)不含有对应的无毒基因,却对其中的4—5个抗瘟单基因系表现出一致的无毒性;推断存在2种可能的原因:一种是物理原因,可能是PCR扩增效果、植株生长情况或接种效果不理想等;另一种是品种内在原因,这2个抗瘟单基因系可能含有其他未知功能的抗瘟基因。从抗稻瘟病单基因系鉴定及无毒基因型分子鉴定水平了解稻瘟病菌无毒基因的类型,从而推断水稻品种的抗瘟基因型,可为水稻品种的合理布局与抗瘟基因的有效利用提供重要的参考;并可对相应稻区稻瘟病的流行进行有效预警,为稻瘟病的及时防控提供科学的理论依据,从而避免稻瘟病的大暴发。4 结论
抗稻瘟病单基因系致病型监测显示,含抗瘟基因Pi9、Pikh和Pi50的单基因系对分离自广8 A杂交稻组合的稻瘟病菌菌株具有较高的抗病性,无毒基因AvrPi9与AvrPik在侵染广8 A杂交稻组合的稻瘟病菌中分布较为广泛;且在该组合种植区的稻瘟菌群体中遗传结构稳定,推广含Pi9或Pik的水稻品种可减轻稻瘟病的发生。分离自粤晶丝苗2号、五优308和广8 A杂交稻组合的稻瘟病菌间具有不同的无毒基因类型及频率,这3个品种或组合感病区域适合种植含Pi9或Pikh或Pi1的品种。其中,在广8优组合感病的区域,可轮换种植粤晶丝苗2号和五优308;在粤晶丝苗2号感病的种植区,可轮换种植与广8 A杂交稻组合及五优308抗瘟基因类型相近的品种。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1007/978-90-481-2465-7_1URL [本文引用: 1]

RICE is life, for most people living in Asia. Rice has shaped the cultures, diets and economies of thousands of millions of people. For more than half of humanity rice is life (Fig. 1.1). Considering its important position, the United Nations designated year 2004 as the International Year of Rice. Devoting a year to a commodity was unprecedented in United Nations history. However, the 57th session of the United Nations General Assembly noted that rice is the staple food of more than half the world’s population, affirmed the need to heighten the awareness of the role of rice in alleviating poverty and malnutrition and reaffirmed the need to focus world attention on the role rice can play in providing food security and eradicating poverty and declared the year 2004 as the International Year of Rice (adopted on December16, 2002; www.67fao.67org/67ag/67irc ).
[本文引用: 1]
DOI:10.1016/j.tibtech.2008.12.002URLPMID:19187990Magsci [本文引用: 1]

Rice is the staple diet of more than three billion people. Yields must double over the next 40 years if we are to sustain the nutritional needs of the ever-expanding global population. Between 10% and 30% of the annual rice harvest is lost due to infection by the rice blast fungus Magnaporthe oryzae. Evaluation of genetic and virulence diversity of blast populations with diagnostic markers will aid disease management. We review the M. oryzae species-specific and cultivar-specific avirulence determinants and evaluate efforts towards generating durable and broad-spectrum resistance in single resistant cultivars or mixtures. We consider modern usage of fungicides and plant defence activators, assess the usefulness of biological control and categorize current approaches towards blast-tolerant genetically modified rice.
DOI:10.1007/s11033-012-2318-0URLPMID:23184051Magsci [本文引用: 1]

Blast disease caused by the fungal pathogen Magnaporthe oryzae is the most severe diseases of rice. Using classical plant breeding techniques, breeders have developed a number of blast resistant cultivars adapted to different rice growing regions worldwide. However, the rice industry remains threatened by blast disease due to the instability of blast fungus. Recent advances in rice genomics provide additional tools for plant breeders to improve rice production systems that would be environmentally friendly. This article outlines the application of conventional breeding, tissue culture and DNA-based markers that are used for accelerating the development of blast resistant rice cultivars. The best way for controlling the disease is to incorporate both qualitative and quantitative genes in resistant variety. Through conventional and molecular breeding many blast-resistant varieties have been developed. Conventional breeding for disease resistance is tedious, time consuming and mostly dependent on environment as compare to molecular breeding particularly marker assisted selection, which is easier, highly efficient and precise. For effective management of blast disease, breeding work should be focused on utilizing the broad spectrum of resistance genes and pyramiding genes and quantitative trait loci. Marker assisted selection provides potential solution to some of the problems that conventional breeding cannot resolve. In recent years, blast resistant genes have introgressed into Luhui 17, G46B, Zhenshan 97B, Jin 23B, CO39, IR50, Pusa1602 and Pusa1603 lines through marker assisted selection. Introduction of exotic genes for resistance induced the occurrence of new races of blast fungus, therefore breeding work should be concentrated in local resistance genes. This review focuses on the conventional breeding to the latest molecular progress in blast disease resistance in rice. This update information will be helpful guidance for rice breeders to develop durable blast resistant rice variety through marker assisted selection.
DOI:10.1371/journal.pone.0057196URLPMID:3582606 [本文引用: 1]

Outbreaks of rice blast have been a threat to the global production of rice. Members of the Magnaporthe grisea species complex cause blast disease on a wide range of gramineous hosts, including cultivated rice and other grass species. Recently, based on phylogenetic analyses and mating tests, isolates from crabgrass were separated from the species complex and named M. grisea. Then other isolates from grasses including rice were named as M. oryzae. Here, we collected 103 isolates from 11 different species of grasses in Korea and analyzed their phylogenetic relationships and pathogenicity. Phylogenetic analyses of multilocus sequences and DNA fingerprinting revealed that the haplotypes of most isolates were associated with their hosts. However, six isolates had different haplotypes from the expectation, suggesting potential host shift in nature. Results of pathogenicity tests demonstrated that 42 isolates from crabgrass and 19 isolates from rice and other grasses showed cross-infectivity on rice and crabgrass, respectively. Interestingly, we also found that the isolates from rice had a distinct deletion in the calmodulin that can be used as a probe.
DOI:10.3389/fpls.2015.00886URLPMID:4644793 [本文引用: 1]

Rice is a staple and most important security food crop consumed by almost half of the world鈥檚 population. More rice production is needed due to the rapid population growth in the world. Rice blast caused by the fungus,Magnaporthe oryzaeis one of the most destructive diseases of this crop in different part of the world. Breakdown of blast resistance is the major cause of yield instability in several rice growing areas. There is a need to develop strategies providing long-lasting disease resistance against a broad spectrum of pathogens, giving protection for a long time over a broad geographic area, promising for sustainable rice production in the future. So far, molecular breeding approaches involving DNA markers, such as QTL mapping, marker-aided selection, gene pyramiding, allele mining and genetic transformation have been used to develop new resistant rice cultivars. Such techniques now are used as a low-cost, high-throughput alternative to conventional methods allowing rapid introgression of disease resistance genes into susceptible varieties as well as the incorporation of multiple genes into individual lines for more durable blast resistance. The paper briefly reviewed the progress of studies on this aspect to provide the interest information for rice disease resistance breeding. This review includes examples of how advanced molecular method have been used in breeding programs for improving blast resistance. New information and knowledge gained from previous research on the recent strategy and challenges towards improvement of blast disease such as pyramiding disease resistance gene for creating new rice varieties with high resistance against multiple diseases will undoubtedly provide new insights into the rice disease control.
DOI:10.1016/j.crvi.2015.03.001URLPMID:25843222 [本文引用: 1]

Rice blast caused by Magnaporthe oryzae is one of the most devastating diseases of rice around the world and crop losses due to blast are considerably high. Many blast resistant rice varieties have been developed by classical plant breeding and adopted by farmers in various rice-growing countries. However, the variability in the pathogenicity of the blast fungus according to environment made blast disease a major concern for farmers, which remains a threat to the rice industry. With the utilization of molecular techniques, plant breeders have improved rice production systems and minimized yield losses. In this article, we have summarized the current advanced molecular techniques used for controlling blast disease. With the advent of new technologies like marker-assisted selection, molecular mapping, map-based cloning, marker-assisted backcrossing and allele mining, breeders have identified more than 100 Pi loci and 350 QTL in rice genome responsible for blast disease. These Pi genes and QTLs can be introgressed into a blast-susceptible cultivar through marker-assisted backcross breeding. These molecular techniques provide timesaving, environment friendly and labour-cost-saving ways to control blast disease. The knowledge of host lant interactions in the frame of blast disease will lead to develop resistant varieties in the future.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1371/journal.ppat.1001261URLPMID:3024261 [本文引用: 1]

Abstract Surface recognition and penetration are among the most critical plant infection processes in foliar pathogens. In Magnaporthe oryzae, the Pmk1 MAP kinase regulates appressorium formation and penetration. Its orthologs also are known to be required for various plant infection processes in other phytopathogenic fungi. Although a number of upstream components of this important pathway have been characterized, the upstream sensors for surface signals have not been well characterized. Pmk1 is orthologous to Kss1 in yeast that functions downstream from Msb2 and Sho1 for filamentous growth. Because of the conserved nature of the Pmk1 and Kss1 pathways and reduced expression of MoMSB2 in the pmk1 mutant, in this study we functionally characterized the MoMSB2 and MoSHO1 genes. Whereas the Momsb2 mutant was significantly reduced in appressorium formation and virulence, the Mosho1 mutant was only slightly reduced. The Mosho1 Momsb2 double mutant rarely formed appressoria on artificial hydrophobic surfaces, had a reduced Pmk1 phosphorylation level, and was nonresponsive to cutin monomers. However, it still formed appressoria and caused rare, restricted lesions on rice leaves. On artificial hydrophilic surfaces, leaf surface waxes and primary alcohols-but not paraffin waxes and alkanes- stimulated appressorium formation in the Mosho1 Momsb2 mutant, but more efficiently in the Momsb2 mutant. Furthermore, expression of a dominant active MST7 allele partially suppressed the defects of the Momsb2 mutant. These results indicate that, besides surface hydrophobicity and cutin monomers, primary alcohols, a major component of epicuticular leaf waxes in grasses, are recognized by M. oryzae as signals for appressorium formation. Our data also suggest that MoMsb2 and MoSho1 may have overlapping functions in recognizing various surface signals for Pmk1 activation and appressorium formation. While MoMsb2 is critical for sensing surface hydrophobicity and cutin monomers, MoSho1 may play a more important role in recognizing rice leaf waxes.
DOI:10.1105/tpc.109.066324URLPMID:19454732 [本文引用: 2]

To subvert rice (Oryza sativa) host defenses, the devastating ascomycete fungus pathogen Magnaporthe oryzae produces a battery of effector molecules, including some with avirulence (AVR) activity, which are recognized by host resistance (R) proteins resulting in rapid and effective activation of innate immunity. To isolate novel avirulence genes from M. oryzae, we examined DNA polymorphisms of secreted protein genes predicted from the genome sequence of isolate 70-15 and looked for an association with AVR activity. This large-scale study found significantly more presence/absence polymorphisms than nucleotide polymorphisms among 1032 putative secreted protein genes. Nucleotide diversity of M. oryzae among 46 isolates of a worldwide collection was extremely low, suggestive of recent pathogen dispersal. However, no association between DNA polymorphism ano AVR was identified. Therefore, we used genome resequencing of Ina 168, an M. oryzae isolate that contains nine AVR genes. Remarkably, a total of 1.68 Mb regions, comprising 316 candidate effector genes, were present in Ina168 but absent in the assembled sequence of isolate 70-15. Association analyses of these 316 genes revealed three novel AVR genes, AVR-Pia, AVR-Pii, and AVR-Pik/km/kp, corresponding to five previously known AVR genes, whose products are recognized inside rice cells possessing the cognate R genes. AVR-Pia and AVR-Pii have evolved by gene gain/loss processes, whereas AVR-Pik/km/kp has evolved by nucleotide substitutions and gene gain/loss.
[本文引用: 3]
[本文引用: 1]
DOI:10.3389/fpls.2016.01140URLPMID:4976503 [本文引用: 1]

Rice blast caused byMagnaporthe oryzaeis one of the most important diseases of rice.Pi54, a rice gene that imparts resistance toM. oryzaeisolates prevalent in India, was already cloned but its avirulent counterpart in the pathogen was not known. After decoding the whole genome of anavirulentisolate ofM. oryzae, we predicted 11440 protein coding genes and then identified four candidate effector proteins which are exclusively expressed in the infectious structure, appresoria.In silicoprotein modeling followed by interaction analysis between Pi54 protein model and selected four candidate effector proteins models revealed that Mo-01947_9 protein model encoded by a gene located at chromosome 4 ofM. oryzae, interacted best at the Leucine Rich Repeat domain of Pi54 protein model. Yeast-two-hybrid analysis showed that Mo-01947_9 protein physically interacts with Pi54 protein.Nicotiana benthamianaleaf infiltration assay confirmed induction of hypersensitive response in the presence ofPi54gene in a heterologous system. Genetic complementation test also proved that Mo-01947_9 protein induces avirulence response in the pathogen in presence ofPi54gene. Here, we report identification and cloning of a new fungal effector gene which interacts with blast resistance genePi54in rice.
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 3]
[本文引用: 3]
DOI:10.1007/s00122-012-1787-9URLPMID:22270148 [本文引用: 2]

The deployment of broad-spectrum resistance genes is the most effective and economic means of controlling blast in rice. The cultivar Er-Ba-Zhan (EBZ) is a widely used donor of blast resistance in South China, with many cultivars derived from it displaying broad-spectrum resistance against blast. Mapping in a set of recombinant inbred lines bred from the cross between EBZ and the highly blast-susceptible cultivar Liangjiangxintuanheigu (LTH) identified in EBZ a blast resistance gene on each of chromosomes 1 ( Pish ), 6 ( Pi2 / Pi9 ) and 12 ( Pita / Pita - 2 ). The resistance spectrum and race specificity of the allele at Pi2 / Pi9 were both different from those present in other known Pi2 / Pi9 carriers. Fine-scale mapping based on a large number of susceptible EBZ02×02LTH F 2 and EBZ02×02LTH BC 1 F 2 segregants placed the gene within a 53-kb segment, which includes Pi2 / Pi9 . Sequence comparisons of the LRR motifs of the four functional NBS-LRR genes within Pi2 / Pi9 revealed that the EBZ allele is distinct from other known Pi2 / Pi9 alleles. As a result, the gene has been given the designation Pi50 (t).
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s10681-015-1465-5URL [本文引用: 1]

A set of 63 rice blast pathogen, Magnaporthe oryzae isolates from different cultivars in different rice growing regions of Eastern India was surveyed for the presence of nine known avirulence genes, ...
[本文引用: 1]
