 ,, 郭涛
,, 郭涛 ,华南农业大学国家植物航天育种工程技术研究中心,广州 510642
,华南农业大学国家植物航天育种工程技术研究中心,广州 510642Genome-Wide Association Analysis for Rice Submergence Seedling Rate
SUN Kai, LI DongXiu, YANG Jing, DONG JiChi, YAN XianCheng, LUO LiXin, LIU YongZhu, XIAO WuMing, WANG Hui, CHEN ZhiQiang ,, GUO Tao
,, GUO Tao ,National Plant Space Breeding Engineering Technology Research Center, South China Agricultural University, Guangzhou 510642
,National Plant Space Breeding Engineering Technology Research Center, South China Agricultural University, Guangzhou 510642通讯作者:
收稿日期:2018-09-18接受日期:2018-11-12网络出版日期:2019-02-13
| 基金资助: |
Received:2018-09-18Accepted:2018-11-12Online:2019-02-13
作者简介 About authors
孙凯,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2955KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
孙凯, 李冬秀, 杨靖, 董骥驰, 严贤诚, 罗立新, 刘永柱, 肖武名, 王慧, 陈志强, 郭涛. 水稻耐淹成苗率相关性状全基因组的关联分析[J]. 中国农业科学, 2019, 52(3): 385-398 doi:10.3864/j.issn.0578-1752.2019.03.001
SUN Kai, LI DongXiu, YANG Jing, DONG JiChi, YAN XianCheng, LUO LiXin, LIU YongZhu, XIAO WuMing, WANG Hui, CHEN ZhiQiang, GUO Tao.
0 引言
【研究意义】水稻是世界上最重要的粮食作物,全世界近1/2的人口以稻米为主食。水稻也是中国最主要的粮食作物,中国2/3的人口以稻米为主粮,水稻播种面积占国内粮食种植面积的27%左右[1]。在水稻各种生产方式中,直播技术由于具有轻型、高效、节水、省工的优点,正逐渐成为水稻轻简高效栽培的理想方式。直播技术分水直播、湿直播和旱直播3种形式,其中,湿直播与旱直播成苗率较高,但田间操作难度大,且容易受到草害、鸟害和虫害影响,而水直播是直接将种子撒播于浅水层覆盖的土壤中,田间操作难度低,杂草危害轻,易于为农民接受,在中国水稻直播生产中得到了广泛应用。然而,水直播的种子萌发时处于淹水条件下,受到低氧胁迫,对品种的耐淹成苗能力具有较高要求,不适合直播的品种通常耐淹成苗能力差,田间秧苗群体不均匀,进而影响直播产量。水稻的胚芽鞘是极少数可以在缺氧条件下生长的植物组织之一[2]。缺氧环境促进水稻胚芽鞘的伸长,抑制幼茎和幼根的生长。水稻胚芽鞘通过快速伸长到达有氧环境,为胚芽和胚根的生长提供氧气。在氧气供应下,水稻种子胚芽和胚根的生长很大程度上受到种子活力的影响,种子活力越高,幼苗生长越快,抗逆性越强,田间群体表现越整齐。因此,水稻耐淹成苗能力受到胚芽鞘性状和种子活力性状的共同控制。在解析水稻耐淹成苗能力与胚芽鞘性状、种子活力性状关系的基础上,进一步发掘与耐淹成苗性状有关的遗传位点并获取分子标记,对于培育适合直播的水稻品种具有重要的意义。【前人研究进展】水稻耐淹萌发的遗传机制比较复杂,已有的研究表明不同水稻品种的耐淹萌发能力存在显著差异,耐缺氧的品种比不耐缺氧的品种有着更长的芽鞘[3,4],表明这一性状受到了遗传位点的控制。陈孙禄等[5]通过对粳稻80380与籼稻RP2334的高代回交自交系连锁分析,发现了4个控制耐淹萌发的QTL。SEPTININGSIH等[6]以淹水发芽21 d的存活率为指标检测到6个QTL,其中位于第7染色体的位点得到了另一个群体的验证,说明其控制耐淹萌发的真实性。芽鞘的生长与中胚轴的发育密切相关,LEE等[7]采取5 cm的深播胁迫,用Kasalath/Nip构建98个回交重组自交系对水稻幼苗中胚轴长度进行QTL定位,发现在第1、3、6染色体有3个关于中胚轴伸长的QTL,依次为qMel-1、qMel-3和qMel-6,其中qMel-1和qMel-6的贡献率分别为12.89%和8.28%,qMel-3的贡献率在2个独立重复中分别为37.56%和26.97%。已有的报道大部分基于双亲分离群体(如RIL、NIL、BIL等)进行遗传位点挖掘,受限于双亲遗传差异或分子标记密度,所能鉴定的耐淹萌发位点比较有限。全基因组关联分析(genome-wide association study,GWAS)以自然群体长期重组交换保留下来的位点间连锁不平衡(linkage disequilibrium,LD)为基础,通过检测表型数据和基因型数据的有效关联来寻找遗传位点[8,9],对于复杂数量性状位点的挖掘具有突出优势。目前,在水稻中展开了大量的GWAS研究,对水稻的代谢性状、叶绿素含量、抽穗期、穗型、种子发芽率、芒长、耐冷、柱头外露等很多性状展开了GWAS分析,解释了各个性状的表型变异和遗传基础[10,11,12,13,14]。同样,水稻种子耐低氧、耐淹相关性状也运用GWAS技术进行了研究,王洋等[15]用91对SSR标记在94个太湖流域核心种质组成的自然群体中采用关联分析的方法挖掘遗传控制位点,共找到6个SSR标记与耐淹萌发关联的位点。HSU等[16]利用高密度SNP标记对153份水稻品种进行关联研究,共找到88个与低氧萌发显著关联的SNPs位点,其中部分位点与前人报道相符。LEE等[17]报道了一个控制水稻淹水萌发的基因CIPK15,该基因的存在能够使水稻种子在淹水时快速生长出胚芽鞘以应对淹水胁迫,而该基因的功能缺失突变体淹水萌发时不能长出胚芽鞘。随着基因组测序技术的不断完善,利用测序获得极高密度SNP将显著提升GWAS策略在遗传位点鉴定方面的效率。【本研究切入点】耐淹成苗率是影响直播稻产量的重要性状。耐淹成苗率的高低与种子活力以及耐低氧萌发密切相关。现有关于水稻耐淹成苗的研究多是以芽鞘性状作为指标,而耐淹成苗率与种子活力的关系报道较少。此外,前人进行全基因组关联分析定位的与耐淹成苗相关的位点较少。【拟解决的关键问题】对影响耐淹成苗率的关键表型性状进行分析,挖掘相关的遗传位点和候选基因,为直播稻品种选育和耐淹成苗机理研究提供一定的理论和材料基础。1 材料与方法
1.1 试验材料
材料包括200份种质资源,来自于中国(主要来自广东)、菲律宾国际水稻研究所(IRRI)、越南、印度、尼泊尔、韩国等国家和地区,材料具体名称见电子版附表1。Table 1
表1
表1表型性状
Table 1
| 性状 Trait | 极小值 Min | 极大值 Max | 均值 Average | 标准误 Standard error | 标准差 Standard deviation | 变异系数 Coefficient of variation |
|---|---|---|---|---|---|---|
| 发芽率 Germination rate (%) | 64.9515 | 100.0000 | 95.8427 | 0.3259 | 4.9420 | 5.1564 |
| 发芽指数 Germination index | 32.6747 | 90.3761 | 59.6827 | 0.6815 | 10.3348 | 17.3162 |
| 活力指数 Vitality index | 0.1376 | 0.9656 | 0.3345 | 0.0071 | 0.1079 | 32.2571 |
| 胚芽鞘长 Coleoptile length (cm) | 0.9640 | 3.4622 | 2.3019 | 0.0302 | 0.4577 | 19.8836 |
| 胚芽鞘直径 Coleoptile diameter (mm) | 0.4544 | 0.6576 | 0.5287 | 0.0026 | 0.0387 | 7.3198 |
| 成苗率 Seedling rate (%) | 11.1111 | 97.7788 | 61.5295 | 1.3074 | 19.8212 | 32.2141 |
新窗口打开|下载CSV
1.2 正常条件下种子萌发试验
200份种质材料,每个材料设3次生物学重复,每个生物学重复取200粒种子(经过挑选,去除空粒、瘪粒及发霉的种子),共取600粒种子,放置在30℃恒温光照培养箱中培养7 d。测量发芽率、发芽指数、活力指数。发芽率=(发芽种子数/种子总数)×100%
发芽指数=∑(Gt/Dt),Gt指第t天新萌发的正常幼苗数,Dt指发芽的天数。
活力指数=∑(Gt/Dt)×幼苗的干重
1.3 低氧条件下种子萌发试验
200份种质材料,每个材料设3次生物学重复,每个生物学重复取20粒种子(经过挑选,去除空粒、瘪粒及发霉的种子),共取60粒种子,放置在灌满水的50 mL透明塑料管中,以此来模拟淹水的低氧环境,放置在30℃恒温光照培养箱中培养6 d。6 d后将种子扫描成图像,利用根系扫描仪(Epson LA2000)测量芽鞘长度、芽鞘直径。1.4 耐淹成苗试验
200份种质材料,每个材料设3次生物学重复,每个生物学重复取30粒种子(经过挑选,去除空粒、瘪粒及发霉的种子),共取90粒种子。在12 cm直径塑料花盆加3 cm厚的砂土,放置种子,种子上覆0.5 cm的砂土,加水至水深10 cm,自然条件下培养20 d,20 d后测量成苗率。以种子的叶伸出水面并具备正常的根为存活个体,调查存活种子占全部种子的比例为成苗率。成苗率=(成苗数/种子总数)×100%
1.5 简化基因组测序与全基因组关联分析
利用CTAB法进行DNA提取,检测合格后利用Illumina HiSeq测序平台,进行双末端(Paired-End)150测序。采用SAMTOOLS等软件进行群体SNP的检测。利用贝叶斯模型检测群体中的多态性位点,经过条件为dp2、Miss0.9、Maf0.01的过滤后,将得到的高质量SNP,利用ANNOVAR软件进行群体SNP注释。运用TreeBest软件计算距离矩阵,以此为基础,通过邻接法(neighbor-joining method)构建系统进化树。通过GCTA软件计算特征向量以及特征值,并利用R软件绘制PCA分布图。使用GEMMA对性状进行关联分析,通过关联的显著度(P-value),筛选出候选SNPs。1.6 转录组测序试验处理
选用籼稻品种玉针香与粳稻品种02428,分别在30℃有氧条件下培养3 d,第4天转入装满水的离心管中进行低氧处理1 d;同时在30℃的装满水的离心管中,低氧条件下培养3 d,第4天转入有氧条件培养1 d。每个处理设3个重复,每个重复每天取样1 g,锡箔纸包裹置于液氮中速冻,-80℃保存备用。对样品中的转录组RNA进行高通量测序,将得到的测序片段(reads)比对到参考序列上,计算参考序列的每个基因上reads的覆盖深度,并进行归一化,所得的结果用以表示基因的表达量。利用基于R语言的软件包edgeR,处理RNA-seq数据成对样本组间差异显著性。利用FDR与log2FC来筛选差异基因,筛选条件为FDR<0.05且|log2FC|>1。委托广州基迪奥生物科技有限公司构建RNA测序文库并完成高通量测序。1.7 数据处理
使用Excel2010进行数据的录入和整理,使用SPSS19.1对所有表型数据进行处理分析,计算表型的平均值、标准差等,并进行方差分析、相关性分析等。2 结果
2.1 200份材料的性状表现
200份材料在种子活力和低氧萌发性状方面具有丰富变异(表1)。其中,活力指数和耐淹成苗率的变异系数均超过了30%,而发芽率的变异系数最小,仅为5.1564%。与种子活力相关的发芽率、发芽指数、活力指数变异系数具有较大差别,发芽指数和活力指数变异系数显著高于发芽率,表明发芽指数和活力指数可以更好地体现材料间的差异。胚芽鞘长变异系数显著高于胚芽鞘直径,表明不同品种的胚芽鞘长度差别明显,而胚芽鞘直径的差别较小。耐淹成苗率在不同品种间也具有较大差异,说明研究材料在此性状上具有广泛的遗传分离。由性状频率分布图(图1)可见,上述性状均呈连续分布,表明是由微效多基因控制的数量性状。图1
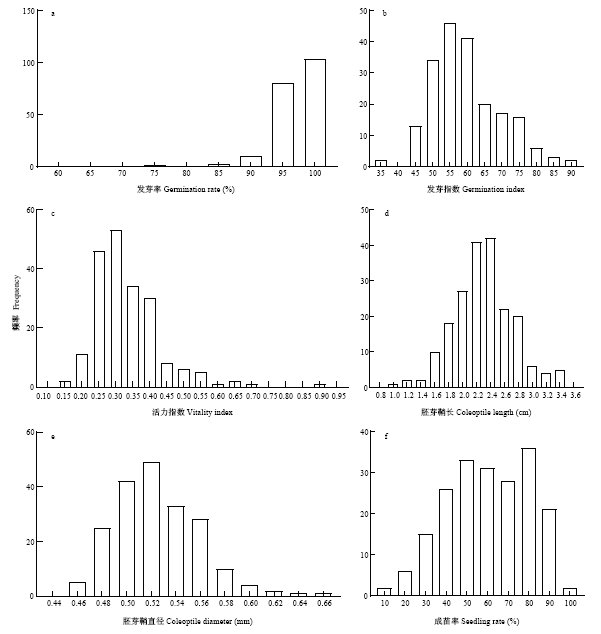 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1性状频率分布直方图
a:发芽率;b:发芽指数;c:活力指数;d:胚芽鞘长;e:胚芽鞘直径;f:耐淹成苗率
Fig. 1Histogram of trait frequency distribution
a: Germination rate; b: Germination index; c: Vitality index; d: Coleoptile length; e: Coleoptile diameter; f: Seedling rate
2.2 各性状间的相关性分析
相关分析表明,耐淹成苗率与发芽率、发芽指数、活力指数、胚芽鞘长度呈极显著正相关,其中,胚芽鞘长与耐淹成苗率的相关性最高,达0.271(表2)。此外,不同性状间表现出复杂的相关性,发芽率与发芽指数呈极显著正相关,与胚芽鞘长显著正相关,与胚芽鞘直径呈极显著负相关;发芽指数与发芽率、活力指数、胚芽鞘长均呈极显著正相关,与胚芽鞘直径呈极显著负相关;活力指数与发芽指数、胚芽鞘长呈极显著正相关;胚芽鞘长与发芽指数、活力指数呈极显著正相关,与发芽率、胚芽鞘直径显著正相关;胚芽鞘直径与胚芽鞘长显著正相关,与发芽率、发芽指数、活力指数呈极显著负相关。上述结果表明,胚芽鞘长和活力指数是影响耐淹成苗率的关键性状。Table 2
表2
表2表型相关性分析
Table 2
| 性状 Trait | 成苗率 Seedling rate | 发芽率 Germination rate | 发芽指数 Germination index | 活力指数 Vitality index | 胚芽鞘长 Coleoptile length | 胚芽鞘直径 Coleoptile diameter |
|---|---|---|---|---|---|---|
| 成苗率 Seedling rate | 1 | |||||
| 发芽率 Germination rate | 0.254** | 1 | ||||
| 发芽指数 Germination index | 0.229** | 0.422** | 1 | |||
| 活力指数Vitality index | 0.224** | 0.129 | 0.518** | 1 | ||
| 胚芽鞘长 Coleoptile length | 0.271** | 0.129* | 0.287** | 0.305** | 1 | |
| 胚芽鞘直径 Coleoptile diameter | -0.078 | -0.382** | -0.226** | 0.022 | 0.137* | 1 |
新窗口打开|下载CSV
2.3 胚芽鞘长和活力指数性状的全基因组关联位点检测
2.3.1 SNP获取 利用200份种质进行简化基因组测序,基因组的平均测序深度为14.16X,平均覆盖度为11.24%。利用SAMTOOLS软件检测共获得了652 457个SNP位点,经过条件为dp2、Miss0.9、Maf0.01的过滤后,最后共获得了161 657个高质量的SNP位点用于后续分析。
2.3.2 群体结构分析 利用SNP数据对200份种质的群体进行结构分析,可见此关联群体主要可分为2个亚群,第一个亚群包括6个材料;另外一个亚群包括其他194份材料(图2-a)。通过GCTA软件计算特征向量以及特征值,并利用R软件绘制PCA分布图(图2-b),PCA分析结果与系统进化树结果一致。
图2
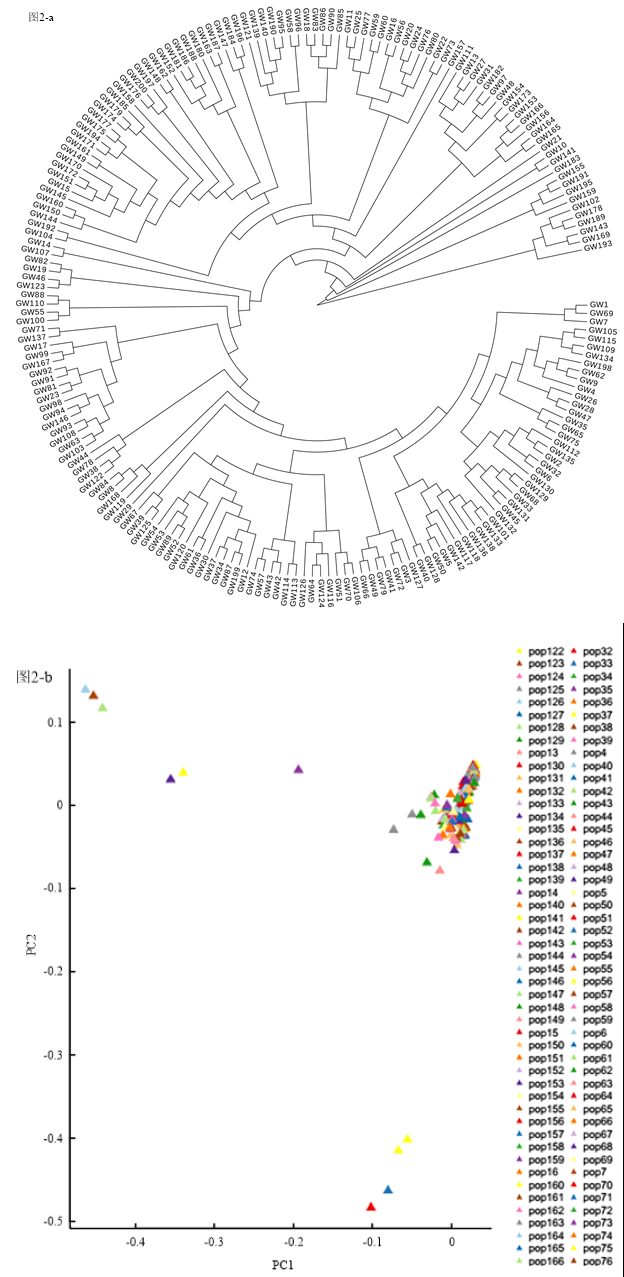 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2进化树及PCA分析结果
Fig. 2Phylogenetic tree and PCA analysis
2.3.3 活力指数关联位点鉴定 对200份材料的活力指数进行全基因组关联分析(图3)。左侧为曼哈顿图,图中水平的虚线表示显著性水平,当-lg10(P)值高于虚线时,认为该SNP与该性状显著关联;右侧为Q-Q图(Quantile-quantile plot),表示实际P值和无关联零假设期望P值的分布。Manhattan图与Q-Q图均表明在全基因组范围内存在与活力指数显著关联的位点。以-lg10(P)=6为阈值,共检测到了8个与活力指数显著关联的SNP位点(表3),分布在第1、2、5、6、10、11和12染色体上。依据LD衰减距离,对关联SNP位点上下游各100 kb区域进行了基因筛选,共筛选得到154个基因。依据基因组注释,筛选出6个与物质代谢、逆境胁迫相关的基因(表4)。
图3
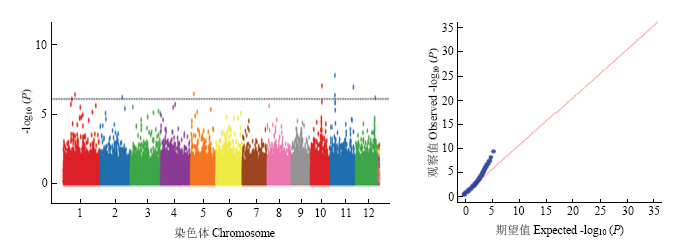 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3活力指数曼哈顿图及Q-Q图
Fig. 3Manhattan map and Q-Q map of vitality index
Table 3
表3
表3活力指数关联位点
Table 3
| 染色体 Chr. | SNP位置 Position (bp) | 等位基因 Allele | 峰值 Peak value | 已报道的位点 Known loci |
|---|---|---|---|---|
| 1 | 13783629 | C/T | 6.322302731 | |
| 2 | 26540479 | A/G | 6.142571185 | [18-19] |
| 5 | 3878312 | A/G | 6.384993065 | |
| 6 | 5542656 | C/T | 8.912458149 | [20] |
| 10 | 13589547 | C/T | 6.960402683 | [20-21] |
| 11 | 4608768 | A/G | 7.710957603 | |
| 11 | 26538371 | G/T | 6.863393569 | |
| 12 | 23449435 | T/C | 6.103889317 | [22-23] |
新窗口打开|下载CSV
Table 4
表4
表4活力指数关联位点候选基因
Table 4
| SNP位置 SNP position | 基因编号 Gene ID | 基因注释 Gene annotation |
|---|---|---|
| Chr.01_13783629 | Os01g0347500 | 木瓜蛋白酶样半胱氨酸蛋白酶 Papain-like cysteine proteinase |
| Chr.02_26540479 | Os02g0657000 | 含AP2结构域的蛋白质 AP2 domain-containing protein |
| Chr.02_26540479 | Os02g0658200 | 锌指,含有PHD型结构域的蛋白质 Zinc finger, PHD-type domain containing protein |
| Chr.02_26540479 | Os02g0658400 | Remorin,含有C末端结构域的蛋白质 Remorin, C-terminal domain containing protein |
| Chr.11_26538371 | Os11g0660300 | 热激蛋白DnaJ,含有N-末端结构域的蛋白质 Heat shock protein DnaJ, N-terminal domain containing protein |
| Chr.11_26538371 | Os11g0661200 | 糖苷水解酶 Glycoside hydrolasen |
新窗口打开|下载CSV
2.3.4 胚芽鞘长关联位点鉴定 图4为胚芽鞘长关联分析的曼哈顿图和Q-Q图(quantile-quantile plot)。以-lg10(P)=4为阈值,共检测到了15个与胚芽鞘长度显著关联的SNP位点,分布在第3、4、5、6、8和11染色体上(表5)。基于水稻基因组注释,依据LD衰退水平,本研究在关联 SNP位点上下游区域100 kb,共检测到296个基因。依据基因组注释,筛选出6个与种子生长发育以及逆境胁迫相关的基因(表6)。
图4
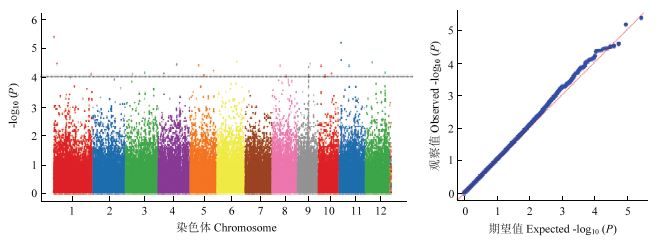 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4胚芽鞘长曼哈顿图和Q-Q图
Fig. 4Manhattan map and Q-Q map of coleoptile length
Table 5
表5
表5胚芽鞘长关联位点
Table 5
| 染色体 Chr. | SNP位置 Position (bp) | 等位基因 Allele | 峰值 Peak value | 已报道的位点 Known loci |
|---|---|---|---|---|
| 3 | 22007803 | C/T | 5.023909483 | |
| 4 | 6615471 | A/G | 4.342639196 | |
| 4 | 23562335 | T/C | 4.028691488 | |
| 5 | 15450007 | G/A | 4.777759954 | [24] |
| 6 | 757734 | A/G | 4.284055624 | |
| 6 | 9693092 | A/G | 4.340712509 | |
| 6 | 21932341 | T/C | 4.165186256 | [25-26] |
| 6 | 29822164 | T/A | 4.48136579 | [27-28] |
| 8 | 5689664 | C/T | 4.675971197 | [29] |
| 8 | 18019426 | T/C | 4.364278864 | |
| 10 | 3160014 | G/A | 4.652868342 | |
| 10 | 7504391 | T/C | 4.286580613 | |
| 10 | 17384859 | G/C | 4.84143398 | |
| 11 | 10896433 | T/C | 4.076205999 | |
| 11 | 18620615 | C/A | 4.169961251 |
新窗口打开|下载CSV
Table 6
表6
表6胚芽鞘长关联位点候选基因
Table 6
| SNP位置 SNP Position | 基因编号 Gene ID | 基因注释 Gene annotation |
|---|---|---|
| Chr.03_22007803 | Os03g0595600 | 谷胱甘肽转移酶 Glutathione transferase |
| Chr.03_22007803 | Os03g0592500 | 叶绿素a-b结合蛋白 Chlorophyll a-b binding protein |
| Chr.06_29822164 | Os06g0704300 | 锌指CCCH结构域的蛋白质 Zinc finger CCCH domain-containing protein |
| Chr.08_15689664 | Os08g0198200 | 天冬氨酸蛋白酶CDR1 Aspartic proteinase CDR1 |
| Chr.08_18019270 | Os08g0380100 | 含有BURP结构域的蛋白质 BURP domain-containing protein |
| Chr.10_17384859 | Os10g0466500 | 锌指蛋白CONSTANS-LIKE 4 Zinc finger protein CONSTANS-LIKE 4 |
| Chr.11_10896433 | Os11g0293900 | 植物含UBX结构域的蛋白质 Plant UBX domain-containing protein |
新窗口打开|下载CSV
2.3.5 候选基因表达模式分析 基于粳稻02428与籼稻玉针香转录组测序结果,获得上述13个基因在有氧、无氧及氧气转换下的基因表达热图(图5)。有3个基因表现出对氧气处理的敏感性,分别为Os02g0657000、Os03g0592500和Os08g0380100。进一步对这三个基因的表达进行分析(图6),可知Os02g0657000与Os08g0380100在02428和玉针香中表达模式相近,主要体现在:(1)有氧条件下的表达高于无氧;(2)从有氧转换到低氧条件,其表达显著降低;(3)从无氧转换到有氧,其表达上升。Os02g0657000与活力指数性状关联,编码AP2结构域蛋白的基因;Os08g0380100与胚芽鞘长性状关联,编码含BURP结构域蛋白的基因。Os03g0592500编码叶绿素a-b结合蛋白基因,与胚芽鞘长性状关联,该基因在2个材料中的表达模式有较大不同。在02428中,该基因在低氧转换到有氧时表达量显著上调,并且在有氧转换到低氧时表达量也有上调;在玉针香中,该基因同样在低氧转换到有氧时表达量明显上调,但是在有氧转换到低氧时表达量并无明显差异。
图5
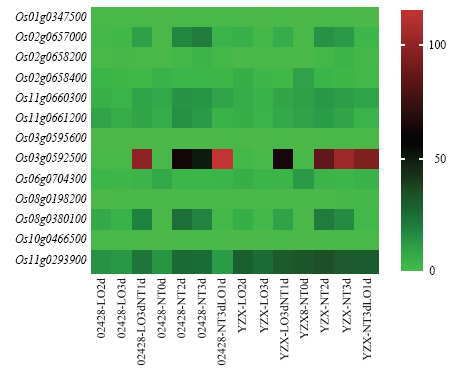 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5候选基因表达热图
Fig. 5Expression heatmap of candidate genes
02428-LO2d 02428低氧处理2 d;02428-LO3d 02428低氧处理3 d;02428-LO3dNT1d 02428低氧转有氧;02428-NT0d 02428处理前;02428-NT2d 02428有氧处理2 d;02428-NT3d 02428有氧处理3 d;02428-NT3dLO1d 02428有氧转低氧;YZX-LO2d 玉针香低氧处理2 d;YZX-LO3d 玉针香低氧处理3 d;YZX-LO3dNT1d玉针香低氧转有氧;YZX8-NT0d玉针香处理前;YZX-NT2d 玉针香有氧处理2 d;YZX-NT3d玉针香有氧处理3 d;YZX-NT3dLO1d玉针香有氧转低氧
02428-LO2d 02428 hypoxia treatment for 2 days; 02428-LO3d 02428 hypoxia treatment for 3 days; 02428-LO3dNT1d 02428 hypoxia to aerobic; 02428-NT0d 02428 before treatment; 02428-NT2d 02428 for aerobic treatment for 2 days; -NT3d 02428 aerobic treatment for 3 days; 02428-NT3dLO1d 02428 aerobic to hypoxia; YZX-LO2d jade needle fragrant for 2 days; YZX-LO3d jade needle fragrant for 3 days; YZX-LO3dNT1d Oxygen to aerobic; YZX8-NT0d before the treatment of jade needle; YZX-NT2d jade needle aerobic treatment for 2 days; YZX-NT3d jade needle aerobic treatment for 3 days; YZX-NT3dLO1d jade needle aerobic to hypoxia
图6
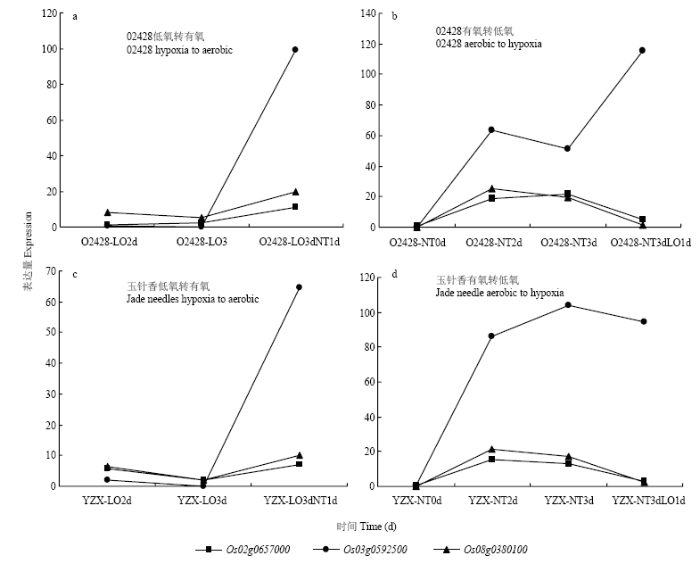 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6候选基因表达模式
Fig. 6Expression profile of candidate genes
3 讨论
3.1 活力指数和胚芽鞘性状是影响直播稻全苗率的关键性状
全苗率低一直是直播稻产量不高不稳的首要问题,它直接影响群体的起点苗数,进而影响群体质量。一般认为三叶期成苗数达到落田稻粒数70%以上,基本上达到全苗目标,但生产上往往达不到50%[30]。有****提出通过选用耐逆境发芽出苗较好的水稻品种可有效解决直播稻全苗问题。因此,种子活力和耐缺氧能力是直播稻品种选育的重要指标。直播稻在淹水萌发期间可以分为2个时期,第一个时期为胚芽鞘的快速生长,此时通过芽鞘的快速伸长突破水面以获取氧气,根和第一片叶的生长受到抑制[31,32];第二个时期为胚芽和胚根的形态建成,此时胚芽鞘停止生长,胚芽和胚根在胚芽鞘输送氧气的支持下快速发育,完成幼苗形态构成,种子活力在此过程起重要作用[33]。本研究结果表明,活力指数与芽鞘长的变异系数远大于其他表型,因此活力指数和芽鞘长可以更好地反映遗传差异。进一步分析后发现耐淹成苗率与活力指数以及胚芽鞘长度呈极显著正相关,与前人研究相符[33,34]。因此,高活力指数与发达的胚芽鞘是选育优良直播稻品种必须考虑的重要性状,对控制这些性状的遗传位点挖掘具有重要意义。
3.2 GWAS是鉴定活力指数与芽鞘长性状遗传位点的有效方法
本研究利用基于重测序的高密度SNP标记,对活力指数和胚芽鞘性状进行全基因组关联分析。与基于RIL、BIL、DH群体的QTL定位方法比较,全基因组关联省去构建家系的时间,所能鉴定的遗传位点更加精细,更适用于耐低氧萌发复杂性状的分析。本研究共检测到8个与活力指数显著关联的SNP位点,其中有4个位点与前人报道区间位于同一位置,第2染色体26540479位置与BI等[18]报道的CERK基因座位相同,同时也与SUGIYAMA等[19]报道的OsAGK2基因座位相同;第6染色体5542656位置与MOEDER等[20]报道的P0664C05.6基因座位相同;第10染色体13589547位置与MORI等[21]报道的ORC3基因座位相同,同时也与MIYOSHI等[22]报道的PLA1基因座位相同;第12染色体23449435位置与MIYOSHI等[22]报道的PLA1基因座位相同,同时也与ZENG等[23]报道的SPL11基因座位相同,这4个位点在前人研究中分别控制了水稻细胞的增殖发育、细胞程序性死亡以及水稻花粉管发育等性状。本研究同时检测到15个与胚芽鞘长度显著关联的SNP位点,有4个位点与前人报道区间位于同一位置,第5染色体15450007位置与LIANG等[24]报道的GA1基因座位相同;第6染色体21932341位置与SINGH等[25]报道的HSF17基因座位相同,同时也与ZHU等[26]报道的D35基因座位相同;第6染色体29822164位置与YONG等[27]报道的BWMK1基因座位相同,同时也与ZHONG等[28]报道的XYXT1基因座位相同;第8染色体5689664位置与CHEUNG等[29]报道的YchF1基因座位相同,这4个位点分别控制了水稻能量运转、抗病以及干旱胁迫、高温胁迫等性状。这些共定位的位点一方面验证了本研究结果的可靠性,另一方面也提供了更多遗传信息,前人并未对共定位位点与水稻种子活力和耐淹能力的性状进行描述,暗示这些位点可能具有多效性。此外,本研究也获得19个前人未报道的控制种子活力或耐低氧萌发位点,为候选基因的筛选提供了基础。3.3 水稻耐低氧萌发过程可能受到逆境胁迫、光合作用相关基因的调控
根据02428和YZX 2个材料的转录组测序结果,本研究对筛选出的13个关联位点区间候选基因在有氧条件、低氧条件及氧气条件转换下的表达模式进行了分析,发现有3个基因表现出对氧气处理的敏感性,分别为Os02g0657000、Os03g0592500和Os08g0380100。其中,Os02g0657000位于种子活力指数显著关联区间,该基因编码含AP2结构域蛋白的基因。在高等植物中,具有AP2结构域的转录因子在植物的开花发育以及抗逆过程中起重要作用[35],根据转录组结果可知,该基因在低氧条件下的表达量较低,而在有氧条件下表达量较高,因此,该基因的高表达对种子活力增强具有促进作用。Os03g0592500位于胚芽鞘长显著关联区间,为编码叶绿素a-b结合蛋白基因,此类蛋白复合体能捕获光能,并迅速把光能传到PSⅠ和光系统Ⅱ的反应中心,引起光化学反应,将光能转化为化学能[36],是植物进行光合作用的重要组成部分。该基因在两个材料中的表达模式有较大不同,在02428中,该基因在低氧转换到有氧时表达量显著上调,并且在有氧转换到低氧时表达量也有上调;在玉针香中,该基因同样在低氧转换到有氧时表达量明显上调,但是在有氧转换到低氧时表达量并无明显差异。在低氧转换到有氧的处理中,因种子在淹水萌发前期只进行芽鞘的快速生长,芽鞘并不含叶绿素,几乎不进行光合作用,转换到有氧条件后,茎秆以及叶片开始生长,叶绿素大量合成,光合作用增强,因此表达量大幅上升;而在有氧转换到低氧的处理时,种子在有氧条件下正常萌发生长,光合作用随着种子萌发即开始进行,转换到低氧后由于叶片已经长出,光合作用仍在进行,因此,表达量变化并不明显,推测该基因对水稻种子耐淹生长具有正调控作用。此外,由于低氧条件下02428的胚芽鞘生长快于玉针香,因此,该基因还可能是低氧条件下02428和玉针香胚芽鞘生长差异的影响因子。Os08g0380100位于胚芽鞘长显著关联区间,该基因编码含BURP结构域蛋白的基因,此类蛋白不仅与植物的生殖发育相关,而且在植物耐受非生物胁迫的过程中具有重要的作用[37]。根据转录组结果可知,该基因的表达模式与Os02g0657000相近,在低氧条件下的表达量较低,而在有氧条件下表达量较高,推测该基因的高表达抑制了胚芽鞘的生长,对种子耐低氧胁迫起负调控作用。4 结论
水稻耐淹成苗率与芽鞘长、活力指数呈极显著正相关,以种子活力、芽鞘长度为指示性状可以筛选出耐淹成苗能力较强的材料。全基因组关联分析、转录组分析与基因表达模式比较的联合应用可提高相关性状候选基因的筛选效率。水稻耐淹萌发过程可能受到与逆境胁迫、光合作用相关基因的调控。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
[本文引用: 1]
DOI:10.1023/A:1004250901931URL [本文引用: 1]

Seedling establishment of direct sown rice plants is less successful in flooded soil than in drained soil. This study was conducted to clarify the difference in morphogenesis of rice seeds sown in flooded and drained soils and to identify the morphological characteristics responsible for successful establishment of cultivars in flooded soil. Rice cultivars ASD1 and IR41996-50-2-1-3, superior in seedling establishment in flooded soil, and Mahsuri and IR72, non-superior (control), were sown at a depth of 25 mm in soil flooded with 25 mm of water or in drained soil. The coleoptile and 1st leaf emerged from the soil surface simultaneously in drained soil while in flooded soil the coleoptile emerged first. The coleoptile of superior cultivars, unlike the controls, elongated more in flooded soil than in drained soil. In flooded soil, the development of mesocotyl, 1st leaf, and roots were inhibited to a greater extent in the controls, than in the superior cultivars. In sealed flasks in which gas containing 0-21% O60 was exchanged daily, the superior cultivars developed longer coleoptiles than the controls at lower O60 concentrations. These findings suggest that the reason superior cultivars grow better in flooded soil than the controls is that the coleoptile elongates faster and longer in hypoxia and is able to reach the soil surface where O60 is available.
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00122-013-2057-1URLPMID:23417074Magsci [本文引用: 1]

AbstractWide adoption of direct-seeded rice practices has been hindered by poorly leveled fields, heavy rainfall and poor drainage, which cause accumulation of water in the fields shortly after sowing, leading to poor crop establishment. This is due to the inability of most rice varieties to germinate and reach the water surface under complete submergence. Hence, tolerance of anaerobic conditions during germination is an essential trait for direct-seeded rice cultivation in both rainfed and irrigated ecosystems. A QTL study was conducted to unravel the genetic basis of tolerance of anaerobic conditions during germination using a population derived from a cross between IR42, a susceptible variety, and Ma-Zhan Red, a tolerant landrace from China. Phenotypic data was collected based on the survival rates of the seedlings at 21 days after sowing of dry seeds under 10 cm of water. QTL analysis of the mapping population consisting of 175 F families genotyped with 118 SSR markers identified six significant QTLs on chromosomes 2, 5, 6, and 7, and in all cases the tolerant alleles were contributed by Ma-Zhan Red. The largest QTL on chromosome 7, having a LOD score of 14.5 and an of 31.7 %, was confirmed using a BCF population. The QTLs detected in this study provide promising targets for further genetic characterization and for use in marker-assisted selection to rapidly develop varieties with improved tolerance to anaerobic condition during germination. Ultimately, this trait can be combined with other abiotic stress tolerance QTLs to provide resilient varieties for direct-seeded systems.
DOI:10.1186/s12284-017-0173-2URLPMID:28710696 [本文引用: 1]

Abstract BACKGROUND: Direct-seeding cultivation by deep-seeding of seeds (drill seeding) is becoming popular due to the scarcity of land and labor. However, poor emergence and inadequate seedling establishment can lead to yield loss in direct-seeding cultivation by deep-sowing. In rice, mesocotyl and coleoptile are primarily responsible for seedling emergence from deeper levels of soil. RESULTS: Quantitative trait loci (QTLs) for mesocotyl and coleoptile length at 5-cm seeding depth were detected using 98 backcross inbred lines from a cross between Kasalath and Nipponbare. Three QTLs qMel-1, qMel-3, and qMel-6 for mesocotyl length were identified on chromosomes 1, 3, and 6, respectively, in two independent replicates. At two QTLs, qMel-1 and qMel-3, the Kasalath alleles increased mesocotyl length, whereas Nipponbare allele increased at qMel-6. The Nipponbare alleles at two QTLs (qCol-3 and qCol-5) increased the coleoptile length. Further, seeds of 54 chromosome segment substitution lines (CSSLs) from the cross between Kasalath and Nipponbare sown at 50002cm soil depth showed a significant positive correlation between seedling emergence and mesocotyl elongation (r0002>00020.6, P0002<00020.0001), but not with coleoptile elongation (r0002=00020.05, P0002=00020.7). Seedling emergence of Nipponbare, Kasalath, and the 3 of the 54 CSSLs rapidly decreased with increasing sowing depth. Seedling emergence at seeding depths of 7 and 100002cm was faster in Kasalath and CSSL-5 that harbored the Kasalath alleles across the qMel-1 and qMel-3 regions than in the other two CSSLs that contained a single QTL and Nipponbare alleles. CSSL-5 showed the longest mesocotyl among the 3 CSSLs, but no difference in coleoptile length was observed among the 3 CSSLs at seeding depths of 7 and 100002cm. CONCLUSION: Variation of mesocotyl elongation was found to be associated with seedling emergence at the seeding depth of 50002cm. To our knowledge, this is the first study performed using CSSLs to detect QTLs for mesocotyl or coleoptile elongation and to determine the effect of mesocotyl elongation on seedling emergence in rice. Our findings provides a foundation for developing rice cultivars that show higher seedling emergence after direct seeding by introgressing QTLs for mesocotyl elongation in rice breeding.
DOI:10.1016/j.tplants.2006.12.001URLPMID:17224302Magsci [本文引用: 1]

Linkage disequilibrium (LD) mapping in plants detects and locates quantitative trait loci (QTL) by the strength of the correlation between a trait and a marker. It offers greater precision in QTL location than family-based linkage analysis and should therefore lead to more efficient marker-assisted selection, facilitate gene discovery and help to meet the challenge of connecting sequence diversity with heritable phenotypic differences. Unlike family-based linkage analysis, LD mapping does not require family or pedigree information and can be applied to a range of experimental and non-experimental populations. However, care must be taken during analysis to control for the increased rate of false positive results arising from population structure and variety interrelationships. In this review, we discuss how suitable the recently developed alternative methods of LD mapping are for crops.
DOI:10.1038/nrg2612URLPMID:19584810 [本文引用: 1]

Abstract A major challenge in current biology is to understand the genetic basis of variation for quantitative traits. We review the principles of quantitative trait locus mapping and summarize insights about the genetic architecture of quantitative traits that have been obtained over the past decades. We are currently in the midst of a genomic revolution, which enables us to incorporate genetic variation in transcript abundance and other intermediate molecular phenotypes into a quantitative trait locus mapping framework. This systems genetics approach enables us to understand the biology inside the 'black box' that lies between genotype and phenotype in terms of causal networks of interacting genes.
DOI:10.3835/plantgenome2015.11.0115URL [本文引用: 1]

Abstract Panicle architecture determines the number of spikelets per panicle (SPP) and is highly associated with grain yield in rice (Oryza sativa L.). Understanding the genetic basis of panicle architecture is important for improving the yield of rice grain. In this study, we dissected panicle architecture traits into eight components, which were phenotyped from a germplasm collection of 529 cultivars. Multiple regression analysis revealed that the number of secondary branch (NSB) was the major factor that contributed to SPP. Genome-wide association analysis was performed independently for the eight particle architecture traits observed in the indica and japonica rice subpopulations compared with the whole rice population. In total, 30 loci were associated with these traits. Of these, 13 loci were closely linked to known panicle architecture genes, and 17 novel loci were repeatedly identified in different environments. An association signal cluster was identified for NSB and number of spikelets per secondary branch (NSSB) in the region of 31.6 to 31.7 Mb on chromosome 4. In addition to the common associations detected in both indica and japonica subpopulations, many associated loci were unique to one subpopulation. For example, Ghd7 and DST were specifically associated with panicle length (PL) in indica and japonica rice, respectively. Moreover, the Ghd7-mediated flowering genes Hd17 and Ehd1 were associated with the formation of panicle architecture in indica rice. These results suggest that different gene networks regulate panicle architecture in indica and japonica rice.
DOI:10.3389/fpls.2016.01270URLPMID:5002401 [本文引用: 1]

Rice is a short-day plant. Short-day length promotes heading, and long-day length suppresses heading. Many studies have evaluated rice heading in field conditions in which some individuals in the population were exposed to various day lengths, including short and long days, prior to a growth phase transition. In this study, we investigated heading date under natural short-day conditions (SD) and long-day conditions (LD) for 100s of accessions and separately conducted genome-wide association studies withinindicaandjaponicasubpopulations. Under LD, three and four quantitative trait loci (QTLs) were identified inindicaandjaponicasubpopulations, respectively, two of which were less than 80 kb from the known genesHd17andGhd7. But no common QTLs were detected in both subpopulations. Under SD, six QTLs were detected inindica, three of which were less than 80 kb from the known heading date genesGhd7,Ehd1, andRCN1. But no QTLs were detected injaponicasubpopulation.qHd3under SD andqHd4under LD were two novel major QTLs, which deserve isolation in the future. Eleven known heading date genes were used to test the power of association mapping at the haplotype level.Hd17,Ghd7,Ehd1, andRCN1were again detected at more significant level and three additional genes,Hd3a,OsMADS56, andGhd7.1, were detected. However, of the detected seven genes, only one gene,Hd17, was commonly detected in both subpopulations and two genes,Ghd7andGhd7.1, were commonly detected inindicasubpopulation under both conditions. Moreover, haplotype analysis identified favorable haplotypes ofGhd7andOsMADS56for breeding design. In conclusion, diverse heading date genes/QTLs betweenindicaandjaponicasubpopulations responded to SD and LD, and haplotype-level association mapping was more powerful than SNP-level association in rice.
DOI:10.1186/s12863-016-0340-2URLPMID:4727300 [本文引用: 1]

Seed dormancy is an adaptive trait employed by flowering plants to avoid harsh environmental conditions for the continuity of their next generations. In cereal crops, moderate seed dormancy could help prevent pre-harvest sprouting and improve grain yield and quality. We performed a genome wide association study (GWAS) for dormancy, based on seed germination percentage (GP) in freshly harvested seeds (FHS) and after-ripened seeds (ARS) in 350 worldwide accessions that were characterized with strong population structure of indica, japonica and Aus subpopulations. The germination tests revealed that Aus and indica rice had stronger seed dormancy than japonica rice in FHS. Association analysis revealed 16 loci significantly associated with GP in FHS and 38 in ARS. Three out of the 38 loci detected in ARS were also detected in FHS and 13 of the ARS loci were detected near previously mapped dormancy QTL. In FHS, three of the association loci were located within 100 kb around previously cloned GA/IAA inactivation genes such asGA2ox3,EUI1andGH3-2and one near dormancy gene,Sdr4. In ARS, an association signal was detected near ABA signaling geneABI5. No association peaks were commonly detected among the sub-populations in FHS and only one association peak was detected in both indica and japonica populations in ARS.Sdr4andGA2OX3haplotype analysis showed that Aus and indica II (IndII) varieties had stronger dormancy alleles whereas indica I (IndI) and japonica had weak or non-dormancy alleles. The association study and haplotype analysis together, indicate an involvement of independent genes and alleles contributing towards regulation and natural variation of seed dormancy among the rice sub-populations. The online version of this article (doi:10.1186/s12863-016-0340-2) contains supplementary material, which is available to authorized users.
.
DOI:10.1007/s12041-016-0679-1URLPMID:27659335 [本文引用: 1]

Awn is one of the most important domesticated traits in rice ( Oryza sativa ). Understanding the genetic basis of awn length is important for grain harvest and production, because long awn length is disadvantageous for both grain harvest and milling. We investigated the awn length of 529 rice cultivars and performed a Genomewide association studies (GWAS) in the indica and japonica subpopulations, and the whole population. In total, we found 17 loci associated with awn length. Of these loci, seven were linked to previously reported quantitative trait loci, and one was linked to the awn gene An-1 . Nine novel loci were repeatedly identified in different environments. One of the nine associations was identified in both the whole and japonica populations. Special interest was the detection of the most significant association SNP, sf0136352825, which was less than 95 kb from the seed shattering gene qSH1 . These results may provide potentially favourable haplotypes for molecular breeding in rice.
DOI:10.1016/j.molp.2017.01.001URLPMID:28110091 [本文引用: 1]
[D].
[本文引用: 1]
[D].
[本文引用: 1]
DOI:10.1186/s12284-015-0072-3URLPMID:4689725 [本文引用: 1]

Increasing numbers of rice farmers are adopting direct-seeding methods to save on costs associated with labor and transplanting. Successful seedling establishment in flooded conditions requires rapid coleoptile growth to ensure access to oxygen near the water surface. It is important that the natural variations in coleoptile growth of submerged rice plants are identified. Coleoptile responses of submerged plants at the germination stage were analyzed in diverse rice accessions and recombinant inbred lines. Several genomic regions identified from a genome-wide association analysis were significantly associated with anaerobic germination, with many that corresponded to published quantitative trait locus (QTL) intervals. In the recombinant inbred line population derived from a cross betweenjaponicaandindicavarieties, only one unique and strong signal explaining about 27 % of the phenotypic variation was detected. Distinct haplotypes associated with variations in coleoptile length were identified in diverse germplasm. We demonstrated the value of combining genome-wide association analysis and biparental QTL mapping approaches to identify chromosomal regions regulating coleoptile elongation in submerged rice plants. The significant genomic regions detected in this study are potential candidates for incorporation into elite cultivars to improve seedling survival during anaerobic germination. Future studies that map the QTLs and investigate the effects and functions of candidate genes may lead to new rice varieties that can be used in direct-seeding systems. The online version of this article (doi:10.1186/s12284-015-0072-3) contains supplementary material, which is available to authorized users.
DOI:10.1126/scisignal.2000333URLPMID:19809091 [本文引用: 1]

Flooding is a widespread natural disaster that leads to oxygen (O(2)) and energy deficiency in terrestrial plants, thereby reducing their productivity. Rice is unusually tolerant to flooding, but the underlying mechanism for this tolerance has remained elusive. Here, we show that protein kinase CIPK15 [calcineurin B-like (CBL)-interacting protein kinase] plays a key role in O(2)-deficiency tolerance in rice. CIPK15 regulates the plant global energy and stress sensor SnRK1A (Snf1-related protein kinase 1) and links O(2)-deficiency signals to the SnRK1-dependent sugar-sensing cascade to regulate sugar and energy production and to enable rice growth under floodwater. Our studies contribute to understanding how rice grows under the conditions of O(2) deficiency necessary for growing rice in irrigated lowlands.
[本文引用: 1]
DOI:10.1093/pcp/pch199URLPMID:15653795 [本文引用: 1]

PII protein in bacteria is a sensor for 2-oxoglutarate and a transmitter for glutamine signaling. We identified an OsGlnB gene that encoded a bacterial PII-like protein in rice. Yeast two-hybrid analysis showed that an OsGlnB gene product interacted with N-acetylglutamate kinase 1 (OsNAGK1) and PII-like protein (OsGlnB) itself in rice. In cyanobacteria, NAGK is a key enzyme in arginine biosynthesis. Transient expression of OsGlnB cDNA or OsNAGK1 cDNA fused with sGFP in rice leaf blades strongly suggested that the PII-like protein as well as OsNAGK1 protein is located in chloroplasts. Both OsGlnB and OsNAGK1 genes were expressed in roots, leaf blades, leaf sheaths and spikelets of rice, and these two genes were coordinately expressed in leaf blades during the life span. Thus, PII-like protein in rice plants is potentially able to interact with OsNAGK1 protein in vivo. This finding will provide a clue to the precise physiological function of PII-like protein in rice.
DOI:10.1371/journal.pgen.1007066URLPMID:5708612 [本文引用: 1]

Since the first plant cyclic nucleotide-gated ion channel (CNGC), HvCBT1, was identified as a calmodulin binding protein, more than a decade has passed and a substantial amount of work has been done to understand the molecular nature and function of these channel proteins. Based on electrophysiological and heterologous expression analyses, plant CNGCs function as non-selective cation channels... [Show full abstract]
DOI:10.1016/j.gene.2005.03.047URLPMID:15939553 [本文引用: 1]

The origin recognition complex (ORC) protein plays a critical role in DNA replication through binding to sites (origins) where replication commences. The protein is composed of six subunits (ORC1 to 6) in animals and yeasts. Our knowledge of the ORC protein in plants is, however, much less complete. We have performed cDNA cloning and characterization of ORC subunits in rice ( Oryza sativa L. cv. Nipponbare) in order to facilitate study of plant DNA replication mechanisms. Our previous report provided a description of a gene, ORC1 ( OsORC1), that encodes one of the protein subunits. The present report extends this initial analysis to include the genes that encode four other rice ORC subunits, OsORC2, 3, 4 and 5. Northern hybridization analyses demonstrated the presence of abundant transcripts for all OsORC subunits in shoot apical meristems (SAM) and cultured cells, but not in mature leaves. Interestingly, only OsORC5 showed high levels of expression in organs in which cell proliferation is not active, such as flag leaves, the ears and the non-tip roots. The pattern of expression of OsORC2 also differed from other OsORC subunits. When cell proliferation was temporarily halted for 6 10 days by removal of sucrose from the growth medium, expression of OsORC1, OsORC3, OsORC4 and OsORC5 was substantially reduced. However, the level of expression of OsORC2 remained constant. We suggest from these results that expression of OsORC1, 3, 4 and 5 are correlated with cell proliferation, but the expression of OsORC2 is not.
DOI:10.1073/pnas.2636936100URLPMID:321774 [本文引用: 2]

http://www.pnas.org/cgi/doi/10.1073/pnas.2636936100
DOI:10.1105/tpc.104.025171URLPMID:15377756 [本文引用: 1]

The rice (Oryza sativa) spotted leaf11 (spl11) mutant was identified from an ethyl methanesulfonate-mutagenized indica cultivar IR68 population and was previously shown to display a spontaneous cell death phenotype and enhanced resistance to rice fungal and bacterial pathogens. Here, we have isolated Spl11 via a map-based cloning strategy. The isolation of the Spl11 gene was facilitated by the identification of three additional spl11 alleles from an IR64 mutant collection. The predicted SPL11 protein contains both a U-box domain and an armadillo (ARM) repeat domain, which were demonstrated in yeast and mammalian systems to be involved in ubiquitination and protein-protein interactions, respectively. Amino acid sequence comparison indicated that the similarity between SPL11 and other plant U-box-ARM proteins is mostly restricted to the U-box and ARM repeat regions. A single base substitution was detected in spl11, which results in a premature stop codon in the SPL11 protein. Expression analysis indicated that Spl11 is induced in both incompatible and compatible rice-blast interactions. In vitro ubiquitination assay indicated that the SPL11 protein possesses E3 ubiquitin ligase activity that is dependent on an intact U-box domain, suggesting a role of the ubiquitination system in the control of plant cell death and defense.
DOI:10.1016/j.plantsci.2018.05.019URL [本文引用: 1]

Nitrogen is a key nutrient for plant growth and development. Plants regulate nitrogen availability and uptake efficiency through controlling root architecture. While the heterotrimeric G protein complex is an important element to regulate root morphology, it remains unknown whether the G protein regulates the root architecture in response to nitrogen supply. We used rice and Arabidopsis G protein mutants to study the root architecture in response to different nitrogen concentrations. We found that nitrogen inhibits root horizontal projection area (network area), root perimeter, total length, but not root diameter (average root width). Nitrogen influenced bushiness and root spatial distribution by inhibiting horizontal growth and promoting vertical expansion. The dynamic changes of the rice G protein mutant DK22 at different concentrations of nitrogen from day 7 to day 9 were different from the wild type with regard to bushiness and spatial distribution. The agb1 -2 mutant in Arabidopsis lacked the inhibitory effect of nitrate on root growth. The heterotrimeric G protein complex regulates the inhibitory effect on root growth caused by high nitrogen supply and root spatial distribution in response to different nitrogen concentrations.
URL [本文引用: 1]

Elevated temperatures affect the growth and reproduction of crop plants and thus have become concern worldwide. Hsp101/ClpB protein is a major molecular chaperone, performing dis-aggregation of protein aggregates formed during heat stress. In rice, OsHsfA6a binds to the promoter of OsHsp101/ClpB-C and regulates its expression. In this study, analysis of C-terminal domains of ClassA OsHsfs revealed the presence ofromatic,ydrophobic,cidic (AHA) anduclearxportignal (NES) motifs in all the members. Using deletion constructs, we show that the activation potential of OsHsfA6a is confined in the C-terminal activation domain comprising of AHA and NES sequences. The results obtained in yeast were complemented with transient expression of reporter in protoplast (TERP) based assay. Detailed analysis of OsHsfA6a splice variants shows the presence of one full version and a DBD truncated smaller version whose existence needs experimental evidences. Phylogeny analysis revealed that OsHsfA6a has diverged from A6a/A6b forms of Arabidopsis and tomato and has no expressologs. OsHsfA6anetwork was enriched in MAP kinases along with Hsp70 and Hsp90 proteins. Thus, it appears that regulation of OsClpB-C by HsfA6a is unique in rice and activation potential of OsHsfA6a resides in the single AHA motif located in the C-terminal domain.
DOI:10.1104/pp.105.072306URLPMID:16299167 [本文引用: 1]

The mechanisms of viral diseases are a major focus of biology. Despite intensive investigations, how a plant virus interacts with host factors to cause diseases remains poorly understood. The Rice dwarf virus (RDV), a member of the genus Phytoreovirus, causes dwarfed growth phenotypes in infected rice (Oryza sativa) plants. The outer capsid protein P2 is essential during RDV infection of insects and thus influences transmission of RDV by the insect vector. However, its role during RDV infection within the rice host is unknown. By yeast two-hybrid and coimmunoprecipitation assays, we report that P2 of RDV interacts with ent-kaurene oxidases, which play a key role in the biosynthesis of plant growth hormones gibberellins, in infected plants. Furthermore, the expression of ent-kaurene oxidases was reduced in the infected plants. The level of endogenous$\text{GA}_{1}$(a major active gibberellin in rice vegetative tissues) in the RDV-infected plants was lower than that in healthy plants. Exogenous application of GA3to RDV-infected rice plants restored the normal growth phenotypes. These results provide evidence that the P2 protein of RDV interferes with the function of a cellular factor, through direct physical interactions, that is important for the biosynthesis of a growth hormone leading to symptom expression. In addition, the interaction between P2 and rice ent-kaurene oxidase-like proteins may decrease phytoalexin biosynthesis and make plants more competent for virus replication. Moreover, P2 may provide a novel tool to investigate the regulation of GA metabolism for plant growth and development.
[本文引用: 1]
DOI:10.1093/pcp/pcy003URLPMID:29325159 [本文引用: 1]

Secondary walls are mainly composed of cellulose, hemicelluloses (xylan and glucomannan) and lignin, and are deposited in some specialized cells, such as tracheary elements, fibers and other sclerenchymatous cells. Secondary walls provide strength to these cells, which lend mechanical support and protection to the plant body and, in the case of tracheary elements, enable them to function as... [Show full abstract]
DOI:10.1074/jbc.M110.172080URLPMID:20876569 [本文引用: 1]

YchF is a subfamily of the Obg family in the TRAFAC class of P-loop GTPases. The wide distribution of YchF homologues in both eukarya and bacteria suggests that they are descendents of an ancient protein, yet their physiological roles remain unclear. Using the OsYchF1-OsGAP1 pair from rice as the prototype, we provide evidence for the regulation of GTPase/ATPase activities and RNA binding capacity of a plant YchF (OsYchF1) by its regulatory protein (OsGAP1). The effects of OsGAP1 on the subcellular localization/cycling and physiological functions of OsYchF1 are also discussed. The finding that OsYchF1 and OsGAP1 are involved in plant defense response might shed light on the functional roles of YchF homologues in plants. This work suggests that during evolution, an ancestral P-loop GTPase/ATPase may acquire new regulation and function(s) by the evolution of a lineage-specific regulatory protein.
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s10681-009-0014-5URL [本文引用: 1]

Direct seeding of rice is increasingly being practiced in both rainfed and irrigated areas because of labor shortage for transplanting and opportunities for crop intensification. However, poor crop establishment remains a major obstacle facing its large-scale adoption in areas prone to flooding. Screening of over 8,000 gene bank accessions and breeding lines identified a few tolerant genotypes. One of these, Khao Hlan On, was selected for mapping QTLs associated with tolerance using a backcross population with IR64 as a recurrent parent. Survival of BC 2 F 2 lines varied from 0 to 68%, and averaged about 28%. A linkage map of 1475.7 cM with an average interval of 11.9 cM was constructed using 135 polymorphic SSRs and 1 indel marker. Five putative QTLs were detected, on chromosomes 1 ( qAG-1-2 ), 3 ( qAG-3-1 ), 7 ( qAG-7-2 ), and 9 ( qAG-9-1 and qAG-9-2 ), explaining 17.9 to 33.5% of the phenotypic variation, and with LOD scores of 5.69 20.34. Khao Hlan On alleles increased tolerance of flooding during germination for all the QTLs. Graphical genotyping of the lines with highest and lowest survival verified the detected QTLs that control tolerance and some QTLs co-localize with previously identified QTLs for traits relevant to tolerance, which warrant further studies.
[D].
[本文引用: 2]
[D].
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
