 ,, 曹坳程
,, 曹坳程 ,中国农业科学院植物保护研究所植物病虫害生物学国家重点实验室,北京 100193
,中国农业科学院植物保护研究所植物病虫害生物学国家重点实验室,北京 100193Function of Copper-Resistant Gene copA of Ralstonia solanacearum
WANG XiaoNing, LIANG Huan, WANG Shuai, FANG WenSheng, XU JingSheng, FENG Jie, XU Jin ,, CAO AoCheng
,, CAO AoCheng ,State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193
,State Key Laboratory for Biology of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193通讯作者:
收稿日期:2018-10-30接受日期:2018-11-29网络出版日期:2019-03-01
| 基金资助: |
Received:2018-10-30Accepted:2018-11-29Online:2019-03-01
作者简介 About authors
王晓宁,E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1138KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王晓宁, 梁欢, 王帅, 方文生, 许景升, 冯洁, 徐进, 曹坳程. 青枯菌铜抗性基因copA 的功能[J]. 中国农业科学, 2019, 52(5): 837-848 doi:10.3864/j.issn.0578-1752.2019.05.006
WANG XiaoNing, LIANG Huan, WANG Shuai, FANG WenSheng, XU JingSheng, FENG Jie, XU Jin, CAO AoCheng.
0 引言
【研究意义】由茄科雷尔氏菌(Ralstonia solanacearum )引起的植物细菌性青枯病(bacterial wilt of plants)是一种世界性重大病害[1,2]。由于广阔的地理分布和广泛的生态与寄主适应性,青枯菌可侵染54科450余种植物[3],造成产量损失15%—95%,是农业生产上的重要限制因子[4,5]。铜是生物有机体必备的微量元素,在电子传递链中可以作为电子供体和受体。同时在微生物体内,铜是许多酶和蛋白的重要组分,在许多重要的氧化和还原过程中发挥作用[6,7]。然而,铜的过量积累会促进羟基自由基的产生,从而造成严重的细胞损伤[8]。因此铜制剂作为抗菌剂在防治细菌性病害中起着重要作用。含铜杀菌剂广泛使用造成的持续选择压力,致使包括肠杆菌属(Enterobacter 、Hormaeche 和Edwards )、假单胞菌属(Pseudomonas )和黄单胞菌属(Xanthomonas )等多个种的细菌群体中出现了铜抗性或耐受性菌株[8,9,10]。因此,解析青枯菌铜抗性的分子机制有助于青枯菌铜抗性菌株的防治。【前人研究进展】在植物病原细菌中,第一个阐述较为清晰的铜抗性编码系统是丁香假单胞菌(P. syringae pv. tomato )中的copABCDRS 基因簇,copRS 是双组分调控系统,调控下游结构基因copABCD 的表达。丁香假单胞菌采用累积隔离的策略来应对铜胁迫,周质蛋白CopA可结合10.9±1.2铜原子,周质蛋白CopC可结合0.6±0.1铜原子,CopB为外膜蛋白,CopD为内膜转运蛋白[6,9,11]。大肠杆菌(Escherichia coli )采用外排的作用方式达到抗铜的目的。大肠杆菌存在两套铜抗性系统,其中位于染色体上的copA -cueO 和cusCFBA 基因赋予大肠杆菌在好氧条件下中等铜浓度的铜耐受性和厌氧条件下的高铜浓度下铜耐受性,位于质粒上的pcoABCDRS 基因系统负责提供高铜浓度的铜耐受性[12,13]。大肠杆菌CopA蛋白属于P-型ATP酶,负责将过量的铜从细胞质中输出到周质空间。而CusCFBA编码蛋白属于抗性结节化家族成员,负责将铜从细胞质周围排出体外[13]。其中CusA、CusB和CusC参与组装成质子动力外排泵,将过多的铜泵出细胞质外。pcoA 是pco 基因簇的核心蛋白,编码多铜氧化酶家族蛋白[14]。青枯菌中copA 与copB 的起始密码子重叠,采用CD-search对青枯菌Po82菌株copA 进行蛋白质序列分析,结果表明CopA蛋白属于CopA铜抗性蛋白家族,具有铜结合位点和典型的TaT信号序列(双精氨酸转运途径),表明青枯菌铜抗性的机制为铜转运机制。【本研究切入点】在前期研究中,发现青枯菌Po82菌株的大质粒上携带了与丁香假单胞菌铜抗性编码cop 系统、大肠杆菌抗性编码pco 系统同源的基因簇[15]。该菌株于含亚抑制铜浓度水平(0.8 mmol·L-1 CuSO4)的NA上生长48 h后,形成了典型的黑色菌落,推测该菌株可能采用了与丁香假单胞菌相同的应对铜胁迫策略,将铜“隔离/累积”于周质空间。【拟解决的关键问题】为了进一步深入解析copA 是否参与Po82菌株的铜抗性,采用反向遗传学的研究手段,探明copA 与青枯菌铜抗性、致病性等表型生物学特征之间的关系,为进一步研究青枯菌寄主及环境适应性进化的分子机理打下基础,为青枯病的防治提供理论依据。1 材料与方法
试验于2017年11月至2018年8月在中国农业科学院植物保护研究所植物病虫害生物学国家重点实验室完成。1.1 菌株、试剂与仪器
供试菌株:青枯菌2号小种Po82菌株,由笔者实验室收集保存;TIANamp Bacteria DNA Kit由天根生化科技有限公司提供,Ex Taq酶由宝日医生物技术有限公司提供,Assembly试剂盒由北京中美泰和生物技术有限公司提供。荧光定量PCR仪为美国AB公司产品,紫外分光光度计为德国AJ公司产品。1.2 菌株培养条件
供试菌株材料详见表1。青枯菌及其基因缺失菌株在NA(nutrient agar)培养基(葡萄糖10 g·L-1,酵母浸粉0.5 g·L-1,蛋白胨5 g·L-1,牛肉膏3 g·L-1,蔗糖10 g·L-1,琼脂18 g·L-1)28℃培养。大肠杆菌在LB培养基(氯化钠10 g·L-1,胰蛋白胨10 g·L-1,酵母提取物5 g·L-1,琼脂16 g·L-1)37℃培养。所用抗生素浓度:庆大霉素50 μg·mL-1,卡纳霉素50 μg·mL-1,氨苄青霉素50 μg·mL-1。Table 1
表1
表1本试验所用菌株及质粒
Table 1
| 菌株和质粒 Strains and plasmids | 特征 Characteristics | 来源 Source | |
|---|---|---|---|
| 菌株 Strain | R. solanacearum Po82 | Wild-type potato strain; Phylotype Ⅱ, sequevar. 4 (race 3 biovar. 2) | 本实验室The laboratory |
| E. coli DH5α | mcrA φ80 lacZΔM15, recA1, endA1 | 本研究This study | |
| ΔPo82copA | copA deletion strain, Gmr | 本研究This study | |
| PBBR-ΔPo82copA | copA complentmentary strain, Gmr, Ampr | 本研究This study | |
| 质粒 Plasmid | pKMS1-gm (+U) | Cloning vector, Gmr, Kanr | 本实验室The laboratory |
| pBBR1MCS-4 | Ampr, lacZ alpha | 本实验室The laboratory | |
| pKMS1-copA-gm | Gmr, for copA gene deletion | 本研究This study | |
| pBBR-copA | Ampr, for copA gene complementation | 本研究This study | |
新窗口打开|下载CSV
1.3 序列分析与系统进化树的构建
从GenBank数据库中获得23个与Po82菌株铜抗性copA 基因同源的基因序列,利用MAGA6构建系统进化树,并以Pseudomas syringas 的copA 为外组,采用邻接法(neighbor-joining,NJ)构建系统进化树,自检重复值设置为1 000[16]。1.4 基因缺失及互补载体构建
NA培养基上培养Po82菌株48—72 h,经PCR验证(759f/760r)后,挑取单菌落于NB培养基中,28℃,180 r/min培养至OD600=0.8—1.0,提取基因组DNA(TIANamp Bacteria DNA Kit)。根据NCBI数据库中发表的Po82全基因组序列(https://www.ncbi. nlm.nih.gov/nuccore/CP002820),分别选取copA 上下游各1 000—1 100 bp左右的序列为靶标,用Blast和Primer 5.0软件设计引物copAupf/r和copAdownf/r,利用高保真酶(TakaRa Ex Taq? )扩增,使用USER酶将扩增的copA 上下游片段与经Pac I酶切的pKMS1-gm质粒连接,连接顺序为Up-gm-Down[17]。敲除载体构建后经酶切和测序验证。Po82基因组为模板,设计引物copA Hf/r扩增copA 的整个开放阅读框。设计引物PBBRf/r,通过PCR扩增线性化质粒pBBR1mcs-4。根据同源重组双交换的原理,利用Assembly试剂盒将copA 扩增基因连接在载体质粒上。互补载体构建后,经引物copATestf/r扩增验证和测序验证。
1.5 基因缺失及互补菌株构建
采用同源重组双交换的原理构建copA 的缺失菌株[18]。通过自然转化法将构建好的敲除载体转化到青枯菌Po82野生型菌株中,筛选出庆大霉素与蔗糖抗性,卡那霉素敏感菌株,分别用759f/760r、copAf/copAr和copAupf/copAdownr进行PCR验证后,测序验证。采用电转的方法构建互补菌株[19]。互补载体转化到缺失菌株中,筛选出具有氨苄青霉素和庆大霉素抗性的菌株,经759f/760r和copAf/r引物PCR验证后,测序验证。1.6 铜抗性最小抑制浓度(minimal inhibition concentration,MIC)测定
Po82野生型、copA 基因缺失菌株及互补菌株置于NB培养基中,28℃培养至对数生长期(OD600=0.6—0.8),以比浊法稀释至105 cfu/mL,取5 μL菌液分别涂在含不同浓度CuSO4的NA培养平板上(0.4、0.6、0.8、1.0、1.2 mmol·L-1)。28℃培养48—72 h,以菌落能正常生长的最大CuSO4浓度作为MIC值,每个处理设置3次重复[15]。1.7 基因表达量测定
挑取NA固体培养基上的青枯菌单菌落于NB培养基中振荡培养(28℃,180 r/min),当菌生长到OD600为0.3—0.4时,分别吸取1 mL菌液至NB培养基和含不同浓度CuSO4的NB培养基中振荡培养,尽量使两份菌液的生长条件一致。摇菌到OD600为0.8时,离心收集菌体用于RNA的提取。RNA的提取采用总TRIzol法(Invitrogen,USA)[20]。cDNA的反转录采用PrimerScript RT reagent Kit with gDNA Eraser(TaKaRa)试剂盒。RT-qPCR采用SYBR Premix Ex TaxⅡ(TaKaRa)试剂盒。荧光定量的仪器为ABI 7500 Real-time Detection System(Applied Biosystems,USA)。PCR反应程序:95℃ 30 s;95℃ 5 s,60℃ 34 s,设置40个循环。用标准曲线评价引物的扩增效率,用熔解曲线评价引物的特异性。用内参基因gyrB 对RNA样品进行均一化(表2)。相对表达量的计算采用公式为2-ΔΔCt[21]。每个菌株设置3个重复,每个试验重复3次。Table 2
表2
表2本试验所用引物序列
Table 2
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 用途Purpose |
|---|---|---|
| 759f | GTCGCCGTCAACTCACTTTCC | 青枯菌特异性验证 Specificity verification of R. solanacearum |
| 760r | GTCGCCGTCAGCAATGCGGAATCG | |
| copA Upf | GGTCTTAAUCCCGTGAGGTTGGGAGGTGA | 扩增copA 上游片段 Amplification of upstream of copA |
| copA Upr | GGCATTAAUTGCTCGACTTTCACAGCGGA | |
| copA Downf | GGACTTAAUGATCGAGTTTTCCTTACGTAA | 扩增copA 下游片段 Amplification of downstream of copA |
| copA Downr | GGGTTTAAUTCACGCAGCGCCGTGTAGTCC | |
| copAf | ATATGGCGGCCTCATCATCG | 基因缺失菌株验证 Verification of gene deletion strain |
| copAr | CGAGAAAAGCCCTGTCCAGT | |
| pBRRf | CATTAGGCACCCCAGGCTTTAC | 质粒线性化 Plasmid linearization |
| pBRRr | CCTCTTCGCTATTACGCCAGC | |
| copA Hf | CCTCGAGGTCGACGGTATCGATATGCGCAGCAATCGTGCATCCCGCC | copA 扩增 Amplification of copA |
| copA Hr | GAACTAGTGGATCCCCCGGGCTTCAGGCCACCACCACTTCGCGGAAC | |
| copA Testf | GCCGTTTGTGATGGCTTCC | 基因互补菌株验证 Verification of gene complementary strain |
| copA Testr | CTTATTCAGGCGTAGCACCAGG | |
| gyrBf | GACCTTCCAGGGGTTGATCG | RT-qPCR |
| gyrBr | TCTCCCCCAGCCCCTTATAC | |
| copAf | ATATGGCGGCCTCATCATCG | |
| copAr | CGAGAAAAGCCCTGTCCAGT | |
| hrpBf | GAAGTGGCCGCCCATATC | |
| hrpBr | GCTTGCGGTAGCCCTTGA | |
| hrpGf | GGACACATTCCACGTTCTGCA | |
| hrpGr | CCATGAAATTCGCCGTATTGA | |
| popAf | TTCAGGAGCTTCACCAGGTCT | |
| popAr | CAACACCAATGGCAACTCCAA |
新窗口打开|下载CSV
1.8 生长曲线测定
将Po82野生型、copA 基因缺失菌株及其相应互补菌株在NB培养基中培养至OD600=0.6—0.8,稀释到OD600=0.3,按1﹕100的体积比接种到不同浓度的CuSO4培养基中(0、0.6 mmol·L-1)。培养于28℃,180 r/min,每3 h测定细胞OD600值,每组重复3次。1.9 致病性测定
采用伤根接种法测定copA 基因缺失对青枯菌致病力的影响[22]。用NA培养基培养野生型菌株及copA 缺失株48—72 h,灭菌水洗脱菌株,用比浊法稀释到107cfu/mL。番茄感病品种“中杂9号”生长至7—8叶龄期时,在离植株1 cm处的土壤中切下3 cm深的裂口,每棵植株浇注30 mL浓度为107 cfu/mL菌液。培养条件为光周期L﹕D=16 h﹕8 h,温度为32/30℃。每个菌株接种20株,每个试验重复3次。病情指数分为5级:0级=无症状;1级=1片叶萎蔫;2级=2—3片叶萎蔫;3级=4片及以上叶片萎蔫;4级=整株叶片萎蔫[23]。1.10 代谢活性测定
利用Biolog代谢芯片技术评价青枯菌的代谢能力。将纯培养的野生型Po82菌株、copA 基因缺失菌株和互补菌株接种至NA固体培养基上28℃培养48 h备用。将菌落释放到IFA接种液中,调节浊度85%—95%,采用GENⅢ 96孔微孔板28℃培养(图1),测定培养48、72、96、120、144 h后各个菌株的代谢情况[24]。每个处理设置3个重复,试验重复3次。图1
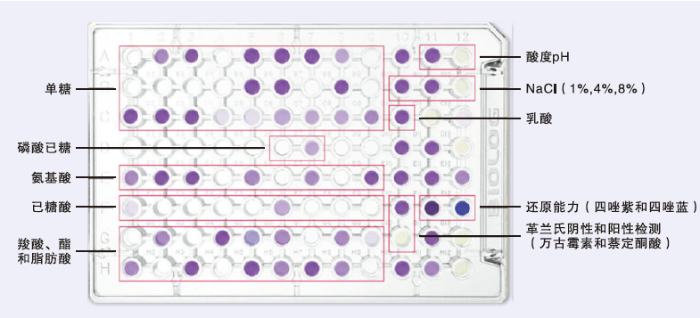 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1Gen III鉴定板组成
单糖:Monosaccharide;磷酸己糖:Phosphohexose;氨基酸:Amino acid;己糖酸:Hexonic acid;羧酸、酯和脂肪酸:Carboxylic acid, ester and fatty acid;酸度:Acidity;乳酸:Lactic acid;还原能力(四唑紫和四唑蓝):Reducing power (tetrazolium violet and tetrazolium blue);革兰氏阴性和阳性检测(万古霉素和萘啶酮酸):Gram-negative and positive detection (vancomycin and nalidixic acid)
Fig. 1Constituent components of GEN Ⅲ MicroPlant
1.11 运动性测定
在含0.5%琼脂(g·L-1)的SMM半固体培养基上测定野生型和基因缺失菌株的运动性[25]。青枯菌在NB培养基中培养,生长至对数期OD600=0.6—0.8时,离心收集细胞,用灭菌水洗涤两次,稀释至OD600=0.3。取5 μL菌液垂直加在SMM培养基,28℃培养3—5 d,不倒置。青枯菌形成白色晕圈,通过测定平板菌落直径分析菌株的运动性,每个菌株4个重复。2 结果
2.1 copA 系统进化分析
为深入探究青枯菌铜抗性编码基因簇中copA 的系统进化关系,以耐金属贪铜菌(Cupriavidus metallidurans )、稻黄单胞菌(Xanthomonas oryzae )、大肠杆菌和假单胞菌中携带的该基因为参照,构建了系统进化树(图2)。结果显示,copA 在青枯菌群体中广泛存在,copA 在亲缘关系上与耐金属贪铜菌最为紧密,与稻黄单胞菌、丁香假单胞菌和大肠杆菌的亲缘关系较远。图2
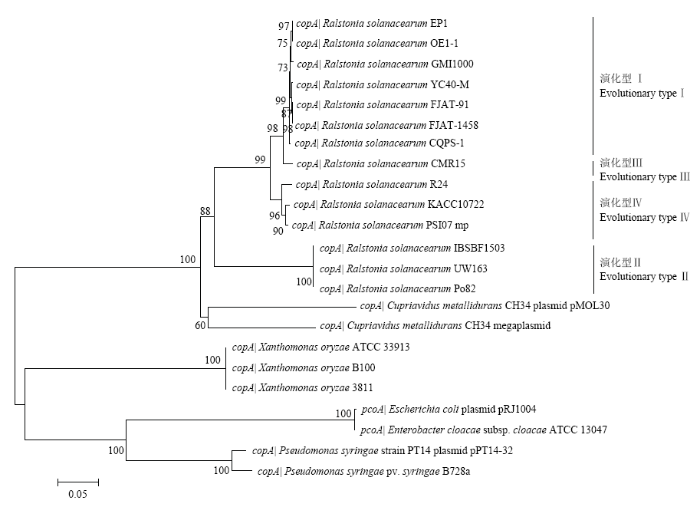 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2基于copA 构建的系统进化树
Fig. 2Phylogenetic tree based on copA sequence
2.2 copA 基因缺失菌株和互补菌株的构建
将构建好的敲除载体pKMS1-copA -gm扩增后提取质粒,使用限制性内切酶Sal I和Bam H I进行双酶切验证,copA 的左右臂长度分别约为1 000和1 010 bp,加上中间的gm 共2 986 bp,pKMS1-copA-gm载体片段总长度为9.2 kb,酶切后剩余部分片段约为6.4 kb,双酶切图谱如图3-A。酶切结果与测序比对得到copA 基因缺失载体。将copA 基因缺失载体转化到野生型菌株,筛选出庆大霉素抗性和蔗糖抗性的菌株,PCR验证(图3-C、3-D)和测序验证后,最终获得基因缺失菌株。图3
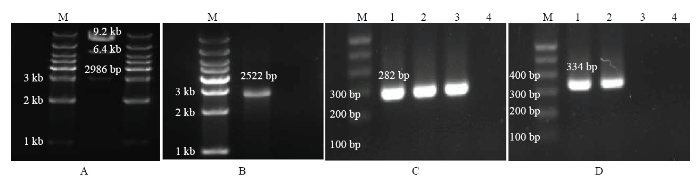 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3copA 基因缺失及互补菌株PCR检测
A:缺失载体酶切验证图The enzyme cutting verification diagram of deletion plasmid;B:互补载体验证图(copA Testf/r)The verification diagram of complementary carrier (copA Testf/r);C:敲除及互补菌株引物检测(759f/760r;1—4:Po82野生型菌株、copA 缺失菌株、copA互补菌株、空白对照)The detection of deletion and complementary strains (759f/760r; 1-4: Po82 wild-type strain, copA gene deletion strain, copA complementary strain and the control);D:敲除及互补菌株引物检测(copAf/r;1—4:Po82野生型菌株、copA 互补菌株、copA 缺失菌株、空白对照)The detection of deletion and complementary strains (copAf/r; 1-4: Po82 wild-type strain, copA complementary strain, copA gene deletion strain and the control)
Fig. 3PCR detection of copA gene deletion and complementary strains
copA 的扩增片段与互补载体连接后,PCR(图3-B)与测序验证。将重组互补载体pBBR-copA转化入基因缺失菌株后,构建copA 互补菌株,PCR(图3-C、3-D)和测序验证最终获得copA 互补菌株。
2.3 copA 缺失对青枯菌铜离子耐受能力的影响
为评价copA 在青枯菌铜抗性中的作用,构建了copA 基因缺失菌株及其相应互补菌株,采用MIC法测定青枯菌的铜抗性水平。Po82野生型菌株的MIC值为1.2 mmol·L-1,copA 基因缺失菌株的MIC值降至0.8 mmol·L-1,铜抗性水平下降了33.3%,copA 互补菌株恢复了与野生型菌株相当的抗铜能力。2.4 copA 的诱导表达分析
Po82菌株在常规(非铜胁迫)培养条件下,其携带的copA 在不同生长阶段均以本底水平表达;在不同CuSO4浓度的胁迫下,与对照组相比,copA 表达量随CuSO4诱导浓度的增加而增加,差异显著。其中基因表达量最高的CuSO4浓度为1.0 mmol·L-1,是对照组基因表达量的85.6倍(图4)。图4
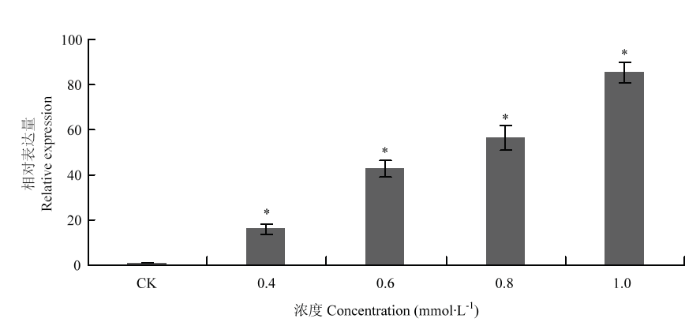 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4copA 诱导表达情况
柱上星号表示不同诱导浓度下,基因表达量差异显著(P <0.05)
Fig. 4Induced expression of copA
The asterisk on the column indicates significant difference in gene expression under different inductive concentrations (P <0.05)
2.5 copA 对青枯菌在铜逆境条件下生长的影响
在NA培养基中,与野生型菌株Po82相比,copA 基因缺失菌株ΔPo82copA在对数生长期的生长速率显著下降,基因缺失菌株与互补菌株之间无显著差异。随着培养时间的增加,基因缺失菌株及互补菌株的生长速率较野生型菌株之间无显著差异(图5-A)。在添加了0.6 mmol·L-1 CuSO4的NA培养基中,与野生型菌株相比,copA 基因缺失菌株在迟缓期已达显著差异,对数期生长速率显著下降(图5-B)。互补菌株的生长速率较低,互补菌株的构建是将copA 的开放阅读框连接在互补质粒pBBR1mcs-4载体的lacZa ,RT-qPCR结果显示copA 持续表达(结果未显示),因此推测可能是因为互补菌株中copA 的过量表达导致生长速率下降。图5
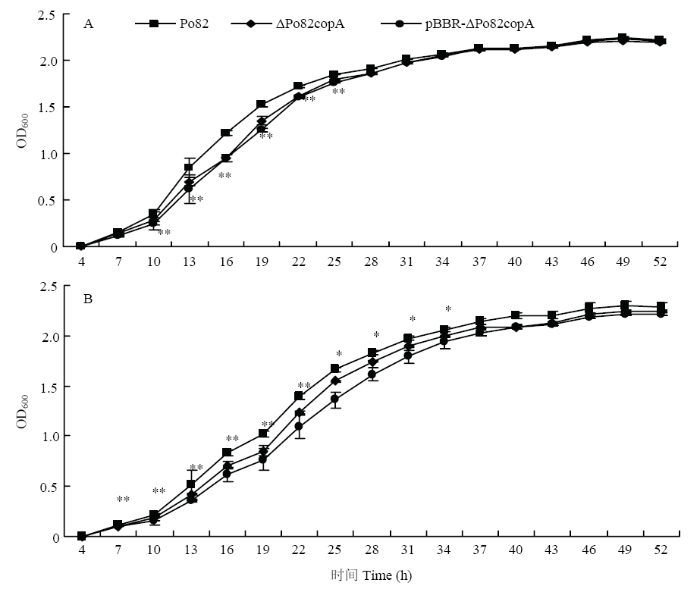 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5青枯菌野生型菌株、copA 基因缺失和互补菌株在不同培养基中生长曲线的测定
A:NA培养基NA medium;B:添加0.6 mmol·L-1 CuSO4的NA培养基 NA medium added 0.6 mmol·L-1 CuSO4
*表示相同处理下基因缺失菌株与野生型菌株差异显著(0.01<P ≤0.05);**表示相同处理下基因缺失菌株与野生型菌株差异极显著(P <0.01)。下同
Fig. 5Growth curve test of the wild-type Po82, ΔPo82copA and pBBR-ΔPo82copA strains
* indicates significant difference between the mutant strain and wild-type strain under the same treatment (0.01<P ≤0.05); ** indicates extremely significant difference between the mutant strain and wild-type strain under the same treatment (P <0.01). The same as below
2.6 copA 的缺失对青枯菌致病性的影响
采用伤根法接种番茄植株探究copA 对青枯菌致病性的影响。结果显示,接种第6天开始,野生型菌株与基因缺失菌株开始出现发病萎蔫现象,二者无显著差异。接种第7—10天,基因缺失菌株与野生型菌株之间差异显著。接种第10天,基因缺失菌株的病情指数为45.3,病情指数较野生型Po82菌株下降了11.7%。接种13 d后恢复与野生型菌株相当的致病力(图6)。互补菌株恢复了其部分的致病力。图6
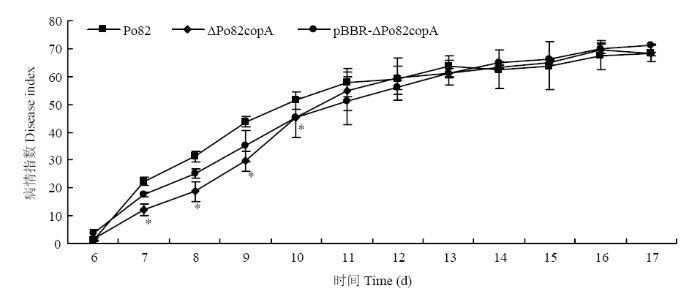 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6青枯菌野生型菌株、copA 基因缺失和互补菌株接种番茄后致病力测定
Fig. 6Pathogenicity test of the wild-type Po82, ΔPo82copA and pBBR-ΔPo82copA strains on tomato
采用RT-qPCR测定copA 基因缺失菌株中与致病相关基因的表达情况,结果显示与野生型菌株相比,hrpB 、hrpG、ripX 的表达量显著下调(图7)。
图7
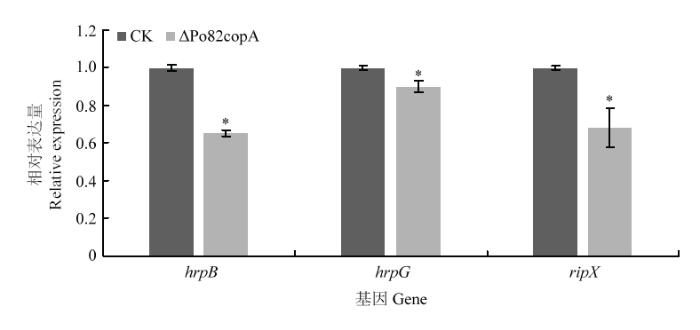 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7青枯菌野生型菌株、copA 基因缺失菌株致病相关基因表达分析
Fig. 7Expression analysis of pathogenic genes in the wild-type Po82 and ΔPo82copA strains
2.7 copA 基因缺失对青枯菌代谢活性的影响
为了探究copA 对青枯菌代谢通路的影响,采用Biolog代谢芯片技术测定了野生型菌株和copA 基因缺失、互补菌株对96种底物(包括糖类、氨基酸、脂肪酸、羧酸)的代谢利用情况。结果表明,copA 基因缺失菌株对α -D-葡萄糖(微孔板C1)、D-海藻糖(微孔板A4)等碳源和L-丙氨酸(微孔板E3)、葡萄糖醛酰胺(微孔板F6)等氮源在代谢上有明显差距(图8)。图 8
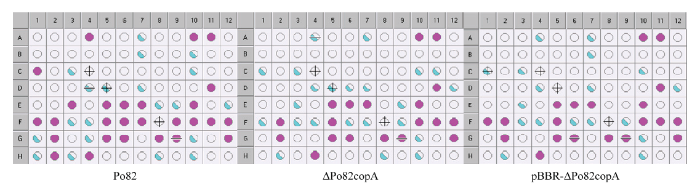 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图 8Biolog代谢芯片测定
紫色孔定义为阳性,浅于紫色的孔定义为边界值。该图分别表示野生型菌株、基因缺失菌株ΔPo82copA和互补菌株pBBR-ΔPo82copA在培养96 h后的代谢结果。α -D-葡萄糖(微孔板C1)在3种菌株的OD590分别为0.632、0.521、0.372;D-海藻糖(微孔板A4)在3种菌株的OD590分别为0.620、0.480、0.383;L-丙氨酸(微孔板E3)在3种菌株的OD590分别为0.786、0.370、0.436;葡萄糖醛酰胺(微孔板F6)在3种菌株的OD590分别为0.566、0.388、0.380
Fig. 8Biolog metabolic microarray detection
Purple pore is defined as positive and pores shallower than purple are defined as boundary values. The picture indicates the metabolic results of Po82, ΔPo82copA and pBBR-ΔPo82copA strains in 96 h, respectively. The OD590 of α -D-glucose (micropore plate C1) in the three strains is 0.632, 0.521 and 0.372, respectively. The OD590 of D-trehalose (micropore plate A4) in the three strains is 0.620, 0.480 and 0.383, respectively. The OD590 of L-alanine (micropore plate E3) in the three strains is 0.786, 0.370 and 0.436, respectively. The OD590 of glucuronamide (micropore plate F6) in the three strains is 0.566, 0.388 and 0.380, respectively
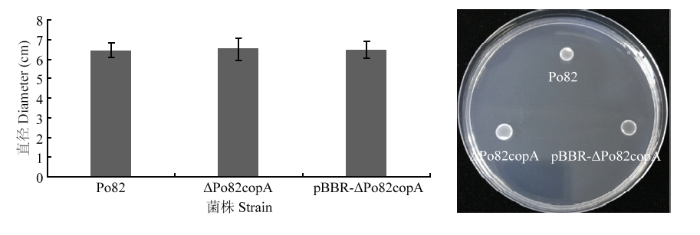
2.8 copA 基因缺失对青枯菌运动性的影响
运动性在细菌致病过程中发挥着重要作用。将野生型菌株、copA 基因缺失和互补菌株接种至5% SMM半固体培养基上,用十字交叉法测定运动性(图9),并进行统计学分析[26]。与野生型菌株相比,缺失和互补菌株运动能力变化差异不显著。图9
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9青枯菌野生型菌株、copA 基因缺失和互补菌株的运动性
Fig. 9The mobility of the wild-type Po82, ΔPo82copA and pBBR-ΔPo82copA strains
3 讨论
生境中铜胁迫水平的变化可诱导细菌中铜抗性编码系统的转录表达,进而通过外排、隔离/累积以及介导细胞膜结构改变等机制,抵御过量铜对自身正常生理过程的干扰。细菌携带的铜抗性编码系统大都存在于接合质粒等可移动遗传因子上(mobile genetic elements,MGEs),如copABCD 之于丁香假单胞菌的pP14-32质粒,pcoABCD 之于大肠杆菌的pRJ1004质粒。相关研究表明,铜抗性编码基因簇可通过质粒接合等水平基因转移事件赋予相同生态位中敏感菌株以铜抗性。ALTIMIRA等[27]基于土壤宏基因组的研究结果显示,3份不同地理来源的铜污染农田土中均可检测到copA ;反之,无污染农田土中则检测不到该基因。耐金属贪铜菌携带了两套copABCD 铜抗性编码系统,一套位于大质粒(等同于第二染色体),另一套位于小质粒pMOL30。青枯菌Po82菌株中单一拷贝的copABCD 基因簇同样位于大质粒上,基于copA 系统进化分析的结果显示,青枯菌在系统进化关系上与耐金属贪铜菌同源性最高。PRIOR等[28]基于egl 、hrpB 和mutS 等青枯菌持家基因的系统进化关系,将青枯菌进一步划分为与地理起源密切相关的4个演化型:演化型I对应亚洲分支菌株;演化型Ⅱ对应美洲分支菌株;演化型Ⅲ对应非洲分支菌株;演化型Ⅳ对应印尼分支菌株。本研究基于copA 构建的系统进化树与基于上述持家基因构建的进化树具有相同的分支结构[5],表明该基因可能是青枯菌核心基因组成员,随青枯菌祖先基因共同进化。RT-qPCR结果显示copA 的表达受胞外铜离子信号的诱导,其表达量在亚抑制浓度范围内(≤1.0 mmol·L-1)随铜离子浓度的增加而上调,证实copA 是青枯菌Po82菌株铜抗性所必需的。基因反向遗传学研究手段进一步探明了该基因缺失后,Po82菌株的铜耐受水平显著降低;pBBR-ΔPo82copA恢复部分铜抗性水平。本研究中copA 的编码产物为可能的周质蛋白,其缺失导致Po82菌株的铜耐受性下降,但MIC值仍高于不携带copABCD 基因簇的铜敏感青枯菌株Po40的0.4 mmol·L-1,推测原因可能为:(1)cop 基因簇中的copB 、copC 和copD 编码产物参与了宿主菌Po82应对铜胁迫的应答反应;(2)Po82菌株中携带的Cus(RND)和P-ATPase等铜代谢基因(簇)[29]与cop铜抗性基因簇相互协同以应对外界铜逆境的胁迫。但采用何种方式相互作用,尚需进一步深入研究。
ZHANG等[30]报道微生物铜抗性基因与致病性具有一定联系,荧光假单胞菌(Pseudomonas fluorescens )cueA 基因缺失菌株在甜菜和豌豆植株根部竞争性定殖的能力降低。在本研究中,copA 基因缺失菌株对番茄的致病力与野生型菌株相比显著下降;RT-qPCR结果同时显示基因缺失菌株Ⅲ型分泌系统相关调控基因与效应子编码基因hrpB 、hrpG 和ripX 的表达量显著下降,表明铜抗性编码基因copA 部分参与了青枯菌的致病过程。Ⅲ型分泌系统在青枯菌成功完成侵染过程中发挥着至关重要的作用,相关研究也已证实青枯菌通过外膜受体蛋白PrhA(plant regulator of hrp )感知寄主信号,并通过级联依次将信号传递并激活prhR 、prhJ 、hrpG 和hrpB 等转录调控相关基因的表达。hrpB 基因编码产物通过与Ⅲ型分泌系统装置蛋白及效应蛋白编码基因上游的顺式作用元件结合,启动Ⅲ型分泌系统的组装形成及Ⅲ型效应子的泌出[31,32,33]。铜抗性编码基因copA 如何与Ⅲ型分泌系统调控基因hrpG 与hrpB 相互作用,并参与青枯菌的致病过程,值得进一步深入研究。Biolog代谢分析表明,基因缺失菌株ΔPo82copA对α -D-葡萄糖、D-海藻糖等碳源的代谢利用效率显著降低,以上碳源存在于植株维管束和胞间液流中,青枯菌通过自身分泌的胞外酶分解代谢利用这些碳源,有助于在寄主维管束中侵染。因此,青枯菌对这些碳源的代谢利用程度关系到青枯菌对寄主的侵染过程。这一结果也佐证了基因缺失菌株ΔPo82copA的致病力较野生型降低。
4 结论
对青枯菌铜抗性编码基因copA 进行了功能分析,该基因在青枯菌铜胁迫应答、致病性等生物学功能上发挥着重要作用,研究结果有助于进一步深入解析青枯菌铜抗性的分子调控机制,并可为铜抗性菌株的有效治理提供理论依据。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.3864/j.issn.0578-1752.2013.14.006URLMagsci [本文引用: 1]

植物细菌性青枯病是由茄科雷尔氏菌(Ralstonia solanacearum Yabuuchi et al.)引起的一种世界性重大病害。作为复合种,青枯菌在与寄主长期协同进化过程中,表现出广泛的生态和寄主适应性。Fengan和Prior提出青枯菌演化型分类框架,用以描述青枯菌种以下的遗传多样性。青枯菌种内表型特征差异的本质是其基因组较其它植物病原细菌更为复杂和更具可塑性。笔者对近期青枯菌遗传多样性和致病基因组学方面的研究进展进行了归纳和讨论。
.
DOI:10.3864/j.issn.0578-1752.2013.14.006URLMagsci [本文引用: 1]

植物细菌性青枯病是由茄科雷尔氏菌(Ralstonia solanacearum Yabuuchi et al.)引起的一种世界性重大病害。作为复合种,青枯菌在与寄主长期协同进化过程中,表现出广泛的生态和寄主适应性。Fengan和Prior提出青枯菌演化型分类框架,用以描述青枯菌种以下的遗传多样性。青枯菌种内表型特征差异的本质是其基因组较其它植物病原细菌更为复杂和更具可塑性。笔者对近期青枯菌遗传多样性和致病基因组学方面的研究进展进行了归纳和讨论。
DOI:10.1128/mBio.00359-16URLPMID:27073091 [本文引用: 1]

The plant pathogenRalstonia solanacearumuses a large repertoire of type III effector proteins to succeed in infection. To clarify the function of effector proteins in host eukaryote cells, we expressed effectors in yeast cells and identified seven effector proteins that interfere with yeast growth. One of the effector proteins, RipAY, was found to share homology with the ChaC family proteins that function as -glutamyl cyclotransferases, which degrade glutathione (GSH), a tripeptide that plays important roles in the plant immune system. RipAY significantly inhibited yeast growth and simultaneously induced rapid GSH depletion when expressed in yeast cells. Thein vitroGSH degradation activity of RipAY is specifically activated by eukaryotic factors in the yeast and plant extracts. Biochemical purification of the yeast protein identified that RipAY is activated by thioredoxin TRX2. On the other hand, RipAY was not activated by bacterial thioredoxins. Interestingly, RipAY was activated by planth-type thioredoxins that exist in large amounts in the plant cytosol, but not by chloroplasticm-,f-,x-,y- andz-type thioredoxins, in a thiol-independent manner. The transient expression of RipAY decreased the GSH level in plant cells and affected the flg22-triggered production of reactive oxygen species (ROS) and expression of pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) marker genes inNicotiana benthamianaleaves. These results indicate that RipAY is activated by host cytosolic thioredoxins and degrades GSH specifically in plant cells to suppress plant immunity. Ralstonia solanacearumis the causal agent of bacterial wilt disease of plants. This pathogen injects virulence effector proteins into host cells to suppress disease resistance responses of plants. In this article, we report a biochemical activity ofR. solanacearumeffector protein RipAY. RipAY can degrade GSH, a tripeptide that plays important roles in the plant immune system, with its -glutamyl cyclotransferase activity. The high GSH degradation activity of RipAY is considered to be a good weapon for this bacterium to suppress plant immunity. However, GSH also plays important roles in bacterial tolerance to various stresses and growth. Interestingly, RipAY has an excellent safety mechanism to prevent unwanted firing of its enzyme activity in bacterial cells because RipAY is specifically activated by host eukaryotic thioredoxins. This study also reveals a novel host plant protein acting as a molecular switch for effector activation.
URL [本文引用: 1]

【目的】研究植物青枯菌 (Ralstonia solanacearum)aac基因编码的蛋白是否具有降解细菌群体感应信号分子的功能。【方法】PCR扩增获得青枯菌GMI1000菌株的aac基 因,将aac基因克隆到pET-5a原核表达载体上,在大肠杆菌中表达出AAC融合蛋白,将AAC融合蛋白与胡萝卜软腐欧氏杆菌(Erwinia carotovorasp.carotovora)混合后,共接种于马铃薯块茎,研究细菌致病力的变化。【结果】克隆了全长的aac基因,完成了aac基 因原核表达载体的构建,研究了AAC融合蛋白对细菌致病力的影响。【结论】青枯菌aac基因编码的蛋白具有减弱胡萝卜软腐欧氏致病力的功能,为开发新的控 害策略提供了依据。
URL [本文引用: 1]

【目的】研究植物青枯菌 (Ralstonia solanacearum)aac基因编码的蛋白是否具有降解细菌群体感应信号分子的功能。【方法】PCR扩增获得青枯菌GMI1000菌株的aac基 因,将aac基因克隆到pET-5a原核表达载体上,在大肠杆菌中表达出AAC融合蛋白,将AAC融合蛋白与胡萝卜软腐欧氏杆菌(Erwinia carotovorasp.carotovora)混合后,共接种于马铃薯块茎,研究细菌致病力的变化。【结果】克隆了全长的aac基因,完成了aac基 因原核表达载体的构建,研究了AAC融合蛋白对细菌致病力的影响。【结论】青枯菌aac基因编码的蛋白具有减弱胡萝卜软腐欧氏致病力的功能,为开发新的控 害策略提供了依据。
.
[本文引用: 1]
.
[本文引用: 2]
.
[本文引用: 2]
DOI:10.1099/mic.0.058487-0URLPMID:22918892 [本文引用: 1]

Copper is an essential cofactor of various enzymes, but free copper is highly toxic to living cells. To maintain cellular metabolism at different ambient copper concentrations, bacteria have evolved specific copper homeostasis systems that mostly act as defence mechanisms. As well as under free-living conditions, copper defence is critical for virulence in pathogenic bacteria. Most bacteria synthesize P-type copper export ATPases as principal defence determinants when copper concentrations exceed favourable levels. In addition, many bacteria utilize resistance-nodulation-cell division (RND)-type efflux systems and multicopper oxidases to cope with excess copper. This review summarizes our current knowledge on copper-sensing transcriptional regulators, which we assign to nine different classes. Widespread one-component regulators are CueR, CopY and CsoR, which were initially identified in Escherichia coli, Enterococcus hirae and Mycobacterium tuberculosis, respectively. CueR activates homeostasis gene expression at elevated copper concentrations, while CopY and CsoR repress their target genes under copper-limiting conditions. Besides these one-component systems, which sense the cytoplasmic copper status, many Gram-negative bacteria utilize two-component systems, which sense periplasmic copper concentrations. In addition to these well-studied transcriptional factors, copper control mechanisms acting at the post-transcriptional and the post-translational levels will be discussed.
DOI:10.3389/fmicb.2017.00912URLPMID:5447758 [本文引用: 2]

Copper homeostasis has been extensively studied in mammals, bacteria, and yeast, but it has not been well-documented in filamentous fungi. In this report, we investigated the basis of copper tolerance in the model fungusAspergillus nidulans. Three genes involved in copper homeostasis have been characterized. First,crpAtheA. nidulansortholog ofCandida albicans CaCRP1gene encoding a PI-type ATPase was identified. The phenotype ofcrpAdeletion led to a severe sensitivity to Cu+2toxicity and a characteristic morphological growth defect in the presence of high copper concentration. CrpA displayed some promiscuity regarding metal species response. The expression pattern ofcrpAshowed an initial strong elevation of mRNA and a low continuous gene expression in response to long term toxic copper levels. Coinciding with maximum protein expression level, CrpA was localized close to the cellular surface, however protein distribution across diverse organelles suggests a complex regulated trafficking process. Secondly,aceAgene, encoding a transcription factor was identified and deleted, resulting in an even more extreme copper sensitivity than the crpA mutant. Protein expression assays corroborated that AceA was necessary for metal inducible expression of CrpA, but not CrdA, a putative metallothionein the function of which has yet to be elucidated.
[本文引用: 2]
[本文引用: 1]
DOI:10.1007/s10565-013-9262-1URLPMID:3847284 [本文引用: 1]

Copper is a metallic element that is crucial for cell metabolism; however, in extended concentrations, it is toxic for all living organisms. The dual nature of copper has forced organisms, including bacteria, to keep a tight hold on cellular copper content. This challenge has led to the evolution of complex mechanisms that on one hand enable them to deliver the essential element and on the other to protect cells against its toxicity. Such mechanisms have been found in both eukaryotic and prokaryotic cells. In bacteria a number of different systems such as extra- and intracellular sequestration, enzymatic detoxification, and metal removal from the cell enabling them to survive in the presence of high concentration of copper have been identified. Gram-negative bacteria, due to their additional compartment, need to deal with both cytoplasmic and periplasmic copper. Therefore, these bacteria have evolved intricate and precisely regulated systems which interact with each other. In this review the active mechanisms of copper resistance at their molecular level are discussed.
[本文引用: 1]
DOI:10.1074/jbc.M104122200URLPMID:11399769 [本文引用: 2]

Abstract Copper is essential but can be toxic even at low concentrations. Coping with this duality requires multiple pathways to control intracellular copper availability. Three copper-inducible promoters, controlling expression of six copper tolerance genes, were recently identified in Escherichia coli. The cue system employs an inner membrane copper transporter, whereas the cus system includes a tripartite transporter spanning the entire cell envelope. Although cus is not essential for aerobic copper tolerance, we show here that a copper-sensitive phenotype can be observed when cus is inactivated in a cueR background. Furthermore, a clear copper-sensitive phenotype for the cus system is revealed in the absence of O(2). These results indicate that the cue pathway, which includes a copper exporter, CopA, and a periplasmic oxidase, CueO, is the primary aerobic system for copper tolerance. During anaerobic growth, however, copper toxicity increases, and the independent cus copper exporter is also necessary for full copper tolerance. We conclude that the cytosolic (CueR) and periplasmic (CusRS) sensor systems differentially regulate copper export systems in response to changes in copper and oxygen availability. These results underscore the increased toxicity of copper under anaerobic conditions and the complex adaptation of copper export in E. coli.
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
DOI:10.14088/j.cnki.issn0439-8114.2016.21.059URL [本文引用: 1]

从NCBI网站筛选到97条蔷薇科植物PGIP基因序列,用邻位相接法构建系统进化树,分析他们之间的亲缘关系。结果表明,蔷薇科植物PGIP基因可聚为17个类群,这与传统的分类结果基本一致,初步探讨了吐根树、耆叶梅、Cercocarpus ledifolius、Purshia tridentate、Horkelia cuueata和卡塔林硬木的亲缘关系,对进一步研究蔷薇科植物之间进化与亲缘关系提供参考。
DOI:10.14088/j.cnki.issn0439-8114.2016.21.059URL [本文引用: 1]

从NCBI网站筛选到97条蔷薇科植物PGIP基因序列,用邻位相接法构建系统进化树,分析他们之间的亲缘关系。结果表明,蔷薇科植物PGIP基因可聚为17个类群,这与传统的分类结果基本一致,初步探讨了吐根树、耆叶梅、Cercocarpus ledifolius、Purshia tridentate、Horkelia cuueata和卡塔林硬木的亲缘关系,对进一步研究蔷薇科植物之间进化与亲缘关系提供参考。
DOI:10.1186/1471-2199-9-70URLPMID:18673530 [本文引用: 1]

Background The rapid increase in whole genome fungal sequence information allows large scale functional analyses of target genes. Efficient transformation methods to obtain site-directed gene replacement, targeted over-expression by promoter replacement, in-frame epitope tagging or fusion of coding sequences with fluorescent markers such as GFP are essential for this process. Construction of vectors for these experiments depends on the directional cloning of two homologous recombination sequences on each side of a selection marker gene. Results Here, we present a USER Friendly cloning based technique that allows single step cloning of the two required homologous recombination sequences into different sites of a recipient vector. The advantages are: A simple experimental design, free choice of target sequence, few procedures and user convenience. The vectors are intented for Agrobacterium tumefaciens and protoplast based transformation technologies. The system has been tested by the construction of vectors for targeted replacement of 17 genes and overexpression of 12 genes in Fusarium graminearum. The results show that four fragment vectors can be constructed in a single cloning step with an average efficiency of 84% for gene replacement and 80% for targeted overexpression. Conclusion The new vectors designed for USER Friendly cloning provided a fast reliable method to construct vectors for targeted gene manipulations in fungi.
.
[本文引用: 1]
DOI:10.1094/MPMI.2002.15.10.1058URLPMID:12437304 [本文引用: 1]

A functional analysis of an 11-kb-long region of the genome of the plant-pathogenic bacterium Ralstonia solanacearum, previously identified as an alternative codon usage region (ACUR), reveals that it was probably acquired through horizontal gene transfer. This ACUR encodes an insertion sequence and eight potential proteins, one of which is partially homologous with a host-specificity factor from a plant-pathogenic Erwinia sp., and another, PopP1, which is homologous to members of the YopJ/AvrRxv family of type III-secreted bacterial effectors controlling interaction between bacteria and their hosts. The analysis of mutants affecting all except one of the genes identified in the ACUR showed that only the popP1-deficient strain had an altered phenotype in plant infection tests. This mutant strain became pathogenic to a Petunia line that is resistant to the wild-type strain. Therefore, popP1 behaves as a typical avirulence gene. We demonstrate that PopP1 protein is secreted and that secretion of this protein requires a functional type III-secretion pathway. In contrast to the structural genes for other type III-secreted proteins identified in R. solanacearum, transcription of the popP1 gene is not coregulated with transcription of hrp genes but is constitutive.
[本文引用: 1]
[本文引用: 1]
DOI:10.1006/meth.2001.1262URL [本文引用: 1]
.
[本文引用: 1]
.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1128/JB.188.10.3697-3708.2006URLPMID:1482862 [本文引用: 1]

Abstract Ralstonia solanacearum, a soilborne plant pathogen of considerable economic importance, invades host plant roots from the soil. Qualitative and quantitative chemotaxis assays revealed that this bacterium is specifically attracted to diverse amino acids and organic acids, and especially to root exudates from the host plant tomato. Exudates from rice, a nonhost plant, were less attractive. Eight different strains from this heterogeneous species complex varied significantly in their attraction to a panel of carbohydrate stimuli, raising the possibility that chemotactic responses may be differentially selected traits that confer adaptation to various hosts or ecological conditions. Previous studies found that an aflagellate mutant lacking swimming motility is significantly reduced in virulence, but the role of directed motility mediated by the chemotaxis system was not known. Two site-directed R. solanacearum mutants lacking either CheA or CheW, which are core chemotaxis signal transduction proteins, were completely nonchemotactic but retained normal swimming motility. In biologically realistic soil soak virulence assays on tomato plants, both nonchemotactic mutants had significantly reduced virulence indistinguishable from that of a nonmotile mutant, demonstrating that directed motility, not simply random motion, is required for full virulence. In contrast, nontactic strains were as virulent as the wild-type strain was when bacteria were introduced directly into the plant stem through a cut petiole, indicating that taxis makes its contribution to virulence in the early stages of host invasion and colonization. When inoculated individually by soaking the soil, both nontactic mutants reached the same population sizes as the wild type did in the stems of tomato plants just beginning to wilt. However, when tomato plants were coinoculated with a 1:1 mixture of a nontactic mutant and its wild-type parent, the wild-type strain outcompeted both nontactic mutants by 100-fold. Together, these results indicate that chemotaxis is an important trait for virulence and pathogenic fitness in this plant pathogen.
DOI:10.1186/1471-2180-12-193URLPMID:3496636 [本文引用: 1]

Background Copper mining has led to Cu pollution in agricultural soils. In this report, the effects of Cu pollution on bacterial communities of agricultural soils from Valparaiso region, central Chile, were studied. Denaturing gradient gel electrophoresis (DGGE) of the 16S rRNA genes was used for the characterization of bacterial communities from Cu-polluted and non-polluted soils. Cu-resistant bacterial strains were isolated from Cu-polluted soils and characterized. Results DGGE showed a similar high number of bands and banding pattern of the bacterial communities from Cu-polluted and non-polluted soils. The presence of copA genes encoding the multi-copper oxidase that confers Cu-resistance in bacteria was detected by PCR in metagenomic DNA from the three Cu-polluted soils, but not in the non-polluted soil. The number of Cu-tolerant heterotrophic cultivable bacteria was significantly higher in Cu-polluted soils than in the non-polluted soil. Ninety two Cu-resistant bacterial strains were isolated from three Cu-polluted agricultural soils. Five isolated strains showed high resistance to copper (MIC ranged from 3.1 to 4.7 mM) and also resistance to other heavy metals. 16S rRNA gene sequence analyses indicate that these isolates belong to the genera Sphingomonas, Stenotrophomonas and Arthrobacter. The Sphingomonas sp. strains O12, A32 and A55 and Stenotrophomonas sp. C21 possess plasmids containing the Cu-resistance copA genes. Arthrobacter sp. O4 possesses the copA gene, but plasmids were not detected in this strain. The amino acid sequences of CopA from Sphingomonas isolates (O12, A32 and A55), Stenotrophomonas strain (C21) and Arthrobacter strain (O4) are closely related to CopA from Sphingomonas, Stenotrophomonas and Arthrobacter strains, respectively. Conclusions This study suggests that bacterial communities of agricultural soils from central Chile exposed to long-term Cu-pollution have been adapted by acquiring Cu genetic determinants. Five bacterial isolates showed high copper resistance and additional resistance to other heavy metals. Detection of copA gene in plasmids of four Cu-resistant isolates indicates that mobile genetic elements are involved in the spreading of Cu genetic determinants in polluted environments.
[本文引用: 1]
DOI:10.1074/jbc.M104122200URLPMID:11399769 [本文引用: 1]

Abstract Copper is essential but can be toxic even at low concentrations. Coping with this duality requires multiple pathways to control intracellular copper availability. Three copper-inducible promoters, controlling expression of six copper tolerance genes, were recently identified in Escherichia coli. The cue system employs an inner membrane copper transporter, whereas the cus system includes a tripartite transporter spanning the entire cell envelope. Although cus is not essential for aerobic copper tolerance, we show here that a copper-sensitive phenotype can be observed when cus is inactivated in a cueR background. Furthermore, a clear copper-sensitive phenotype for the cus system is revealed in the absence of O(2). These results indicate that the cue pathway, which includes a copper exporter, CopA, and a periplasmic oxidase, CueO, is the primary aerobic system for copper tolerance. During anaerobic growth, however, copper toxicity increases, and the independent cus copper exporter is also necessary for full copper tolerance. We conclude that the cytosolic (CueR) and periplasmic (CusRS) sensor systems differentially regulate copper export systems in response to changes in copper and oxygen availability. These results underscore the increased toxicity of copper under anaerobic conditions and the complex adaptation of copper export in E. coli.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1046/j.1365-2958.2000.01851.xURLPMID:10792714 [本文引用: 1]

As in many other Gram-negative plant pathogenic bacteria, the Ralstonia solanacearum hrp genes are involved in the production of a type III secretion apparatus that allows the translocation of PopA protein to the external medium. Here, we show that hrp genes are also involved in the biogenesis of pili that are mainly composed of the HrpY protein. These pili are produced at one pole of the bacterium and are also released into the external medium where they can form very long straight bundles. An hrpY mutant is defective in pilus production, impaired in interactions with plants and in secretion of the PopA protein but not in attachment to plant cells.
