 ,1,6
,1,6Differential circRNA Analysis in the Spleen of Hu-sheep Lambs Infected with F17 Escherichia coli
ZOU ShuangXia1, JIN ChengYan1, BAO JianJun2, WANG Yue1, CHEN WeiHao1, WU TianYi1, WANG LiHong1, Lü XiaoYang1, GAO Wen1, WANG BuZhong3, ZHU GuoQiang4, DAI GuoJun1, SHI DongFang5, SUN Wei ,1,6
,1,6通讯作者:
责任编辑: 林鉴非
收稿日期:2018-09-18接受日期:2018-12-12网络出版日期:2019-03-16
| 基金资助: |
Received:2018-09-18Accepted:2018-12-12Online:2019-03-16
作者简介 About authors
邹双霞,E-mail: 1074432801@qq.com;Tel:18705271578。

摘要
关键词:
Abstract
Keywords:
PDF (1057KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
邹双霞, 金澄艳, 鲍建军, 王悦, 陈炜昊, 吴天弋, 王利宏, 吕晓阳, 高雯, 王步忠, 朱国强, 戴国俊, 师东方, 孙伟. 感染大肠杆菌F17湖羊羔羊脾脏中差异circRNA分析[J]. 中国农业科学, 2019, 52(6): 1090-1101 doi:10.3864/j.issn.0578-1752.2019.06.012
ZOU ShuangXia, JIN ChengYan, BAO JianJun, WANG Yue, CHEN WeiHao, WU TianYi, WANG LiHong, Lü XiaoYang, GAO Wen, WANG BuZhong, ZHU GuoQiang, DAI GuoJun, SHI DongFang, SUN Wei.
0 引言
【研究意义】羊大肠杆菌病是规模化羊场最为常见高发的细菌性疾病之一,传统沿用的抗生素治疗方案存在诸多缺陷。利用RNA-seq筛选与不腹泻绵羊大肠杆菌病有关的circRNA是分析绵羊抗病分子机制的基础,从而发现与抗病性状相关的候选基因。【前人研究进展】环状RNA(circular RNA,circRNA)是一种特殊的内源性非编码RNA(non-coding RNA,ncRNA),是继微小RNA(microRNA,miRNA)及长链非编码RNA(long noncoding RNA,lncRNA)后的RNA 家族又一研究新热点[1]。SANGER等[2]利用电镜首先在植物感染的类病毒中发现了这些以共价键形成的闭合环状单链RNA 分子,它们具有高度的热稳定性。1990年,研究人员在酿酒酵母中发现20S RNA 没有自由的5' 端和3' 端,通过电镜观察发现,这是呈环形的RNA分子[3,4]。随后人们陆续在丁型肝炎病毒中发现circRNA[5],也发现在小鼠的睾丸内含有性别决定基因(sex-determining region Y,Sry)转录而来的circRNA[6],还证实了circRNA 也存在于人体细胞[7]。虽然很早就发现circRNA 广泛存在于各种不同类型的细胞中,但在过去的几十年里,circRNA 的研究却始终进展缓慢,其生成机制和表达调节机制目前尚不完全清楚[8]。长久以来,circRNA 被认为是pre-mRNA 加工过程中选择性剪接产生的副产物。事实上仅很少的circRNA 被发现是选择性剪接过程中产生的外显子[9]。最近的研究[10]发现,circRNA 不是mRNA 成熟过程中的副产物,而与mRNA一样,是pre-mRNA 加工后的一个重要产物,并且circRNA 的加工机制能与mRNA 形成竞争。同时,经典的剪切信号和剪切机制对于反向剪切来说也是必需的[11]。【本研究切入点】目前,关于绵羊抗病方面的研究主要集中在疾病防治方面[12,13],而重要的抗病分子机制研究却鲜有报道。【拟解决的关键问题】本研究利用RNA-seq,首次筛选出对大肠杆菌F17菌株不腹泻与腹泻型个体中差异表达的circRNA,利用Miranda分析circRNA- miRNA-mRNA相互作用,寻找miRNA的靶基因,在此基础上利用q-PCR进行验证。本研究从circRNA层面上,加深了对绵羊拮抗大肠杆菌F17菌株的认识,同时有望确定绵羊拮抗大肠杆菌F17菌株的功能基因,解决中国地方羊品种对大肠杆菌病的抗病育种关键问题,为今后制订抗大肠杆菌病遗传选育策略奠定基础和提供理论依据。1 材料与方法
1.1 试验设计和样本采集
试验用羊于2016年12月购自江苏西来原生态农业有限公司。随机选择包括性别、生长发育良好、日龄体重相近的18只3日龄羔羊,并将羔羊全部隔离饲养。先用10%羔羊奶粉(不含有任何抗生素和微生态制剂)饲喂,以确保在试验前适应饮食需要。5日龄时开始饲喂12.5%羔羊奶粉和大肠杆菌F17菌液(4.6×108 CFU·mL-1),同时保证自由饮水。每天记录羔羊粪便形态[14],在部分羔羊持续腹泻2d后,将羔羊分为不腹泻组和腹泻组,并对羊进行安乐死。用RNAlater收集每只羔羊的肝脏、脾脏、十二指肠、空肠和回肠,并立即在液氮中冷冻带回实验室以进行RNA提取。因为主要研究羔羊的免疫状态,而脾脏是动物体最大的免疫器官,所以选用脾脏这个器官作为试验对象。1.2 文库建设和测序
分别从不腹泻组和腹泻组中选择了3个典型样本并从绵羊脾脏提取总RNA,使用NanoDrop 2000超微量分光光度计和Agilent 2100生物分析仪进行质控。用Ribo-Zero TM试剂盒(Epicenter, Madison, WI, USA)除去核糖体RNA。将RNA片段化(平均长度约为200bp),然后通过逆转录合成和纯化cDNA。使用Qubit?dsDNA HS测定试剂盒进行PCR扩增和纯化后,选择使用NEBNext?Ultra?RNA文库制备试剂盒进行文库构建。在上海欧易生物医学科技有限公司使用Illumina HiSeq 2500平台,对文库进行末端配对测序(测序读长为150 bp)。1.3 鉴定circRNA
circBase[15]数据库中只收录了人、小鼠、线虫、矛尾鱼和腔棘鱼5个物种的circRNA序列。由于绵羊不属于上述物种之一,使用CIRI[16]软件从头预测circRNA。根据circRNA在基因组上的位置,可以将circRNA分为以下5类:exonic circRNA、intronic circRNA、antisense circRNA、sense overlapping circRNA、intergenic circRNA。1.4 差异表达分析
差异表达分析旨在找出不同样本间存在差异表达的circRNA,得到差异表达circRNA之后,对其来源基因做Gene Ontology(GO)和KEGG Pathway显著性分析。DESeq[17]适用于有生物学重复的实验,可以进行样品组间的差异表达分析,获得两个生物学条件之间的差异表达circRNA列表;对于没有生物学重复的实验,则使用edgeR[18]进行差异表达分析,获得两个样品之间的差异表达circRNA列表。1.5 GO和KEGG通路分析
对差异表达转录本进行GO富集分析,对其功能进行描述(结合GO注释结果)。统计每个GO条目中所包括的差异转录本个数,并用Fisher's exact test计算每个GO条目中差异转录本富集的显著性。KEGG[19]是有关Pathway的主要公共数据库,利用KEGG数据库对差异转录本进行Pathway分析(结合KEGG注释结果),并用Fisher's exact test计算每个Pathway条目中差异转录本富集的显著性。1.6 circRNA-miRNA-mRNA互作研究
circRNA可以作为miRNA的靶标分子,受到miRNA的调节。由于circRNA包含多个miRNA结合位点,通过circRNA-miRNA相互作用分析,可以帮助解析作为海绵发挥作用的 circRNA的功能和作用机制。对于动物,使用Miranda [20,21]软件来预测与miRNA结合的circRNA以及miRNA的靶基因,根据miRNA靶基因的功能注释来阐明此部分circRNA的功能。1.7 验证差异转录本的表达水平
为验证筛选的DE circRNA在拮抗过程中发挥的作用,笔者用q-PCR检测了不腹泻和腹泻羔羊脾脏组织中DE circRNA和miRNA靶基因的表达水平,使用2-ΔΔCt方法将每个RNA的相对定量归一化为GAPDH,circRNA的引物见表1。Table 1
表1
表1GAPDH,DE circRNA和mRNA的引物
Table 1
| 基因符号Gene symbol | 引物序列Primer sequence | 产物长度Length of product (bp) | |
|---|---|---|---|
| 差异表达的环状RNA的引物 Primers of DE circRNA | circRNA_2125 | F:ATTGAATCACTTCTCTGTTGC | 129 |
| R:TAGGTGCTCAAAATAGGAC | |||
| circRNA_3832 | F:AGCCTCTCATCTGTACAC | 134 | |
| R:CAGTAACTGCCTAGAGCA | |||
| circRNA_7711 | F:ACAAAGATTCCATTGACAG | 101 | |
| R:ACCAAGAGGCTAGCAAGAC | |||
| circRNA_6710 | F:CAGATTACAGCTATGGCGA | 124 | |
| R:CCCTCATGATCTCATAGG | |||
| circRNA_6914 | F:TTGGCTGTTACTATCATGAG | 124 | |
| R:CTGAACTCTTAACTTGCA | |||
| circRNA_4030 | F:TGATGCAGATATTAAACCTC | 133 | |
| R:CCAATCTCGGATAACTTCAC | |||
| 差异表达的信使RNA的引物 Primers of DE mRNA | NEB | F:ATTACAGCTATCCACCCGAC | 149 |
| R:TGCCTTTTCCATTTCTAAG | |||
| UBE3B | F:TAAGATTGCCAGGAAACTGC | 133 | |
| R:AGCCAGGGACACGTACCAC | |||
| ADGRF2 | F:GGCGTTTACCTCTTTCTCG | 103 | |
| R:CAAGCTGCAAATAGAAAC | |||
| LAMA1 | F:AAATGATCGAAAAGGCTAC | 127 | |
| R:AACCGCCTTTTCCGTAGGAC | |||
| LTF | F:GAAAAGCGTATCCCAACCTG | 103 | |
| R:TTGAAGGCACCAGAATAAC | |||
| MGAT5 | F:CATCATCCACACCTACACG | 111 | |
| R:AACTGCAAGTCTCGTCCGC | |||
| TLN2 | F:ACGACGGTGGTTAAATAC | 125 | |
| R:AGTTGCCCATAGTCACTGGTC | |||
| ARHGAP30 | F:TCTTCAACCTGGGTCGCTC | 159 | |
| R:GCAGCCCCTCTGGTTCATC | |||
| SLC25A29 | F:GCGTCCTGGCTCTCCACCT | 125 | |
| R:CCCTGCCTCCCCGCGCTC | |||
| GAPDH引物 Primers of GAPDH | GAPDH-F | F:GTTCCACGGCACAGTCAAGG | 127 |
| R:ACTCAGCACCAGCATCACCC |
新窗口打开|下载CSV
1.8 统计分析
使用SPSS软件(版本20.0)分析所有数据,使用单因素方差分析(ANOVA)分析差异转录本的相对表达量,并使用Tukey检验进行多重比较。P<0.05被认为具有统计学意义。每组包含3个样品,每个试验重复3次。2 结果
2.1 绵羊脾脏中转录本的鉴定
在绘制参考序列后,鉴定出已知的7 730个circRNA。由图1可以看出,circRNA长度主要分布于200—900 bp,平均长度为1 943bp,GC含量大约在43.5%,统计circRNA的外显子数目主要为2—4个。统计circRNA序列中反向剪切位点的可变剪切信号(GT-AG)情况,并绘制图形,结果见下图2。统计circRNA类型见下图3,其中sense overlapping circRNA占92.11%,exonic circRNA占3.27%,intergenic circRNA占3.18%,intronic circRNA占0.88%,antisense circRNA占0.56%。将circRNA与基因元件进行比较,从而探索circRNA在基因组上的分布,统计各条染色体上或scaffold上预测得到的circRNA数目情况,并绘制图形,结果见下图4,其中主要分布在3条染色体上,分别是NC_019458.2(853)、NC_019459.2(772)、NC_019460.2(787)。图1
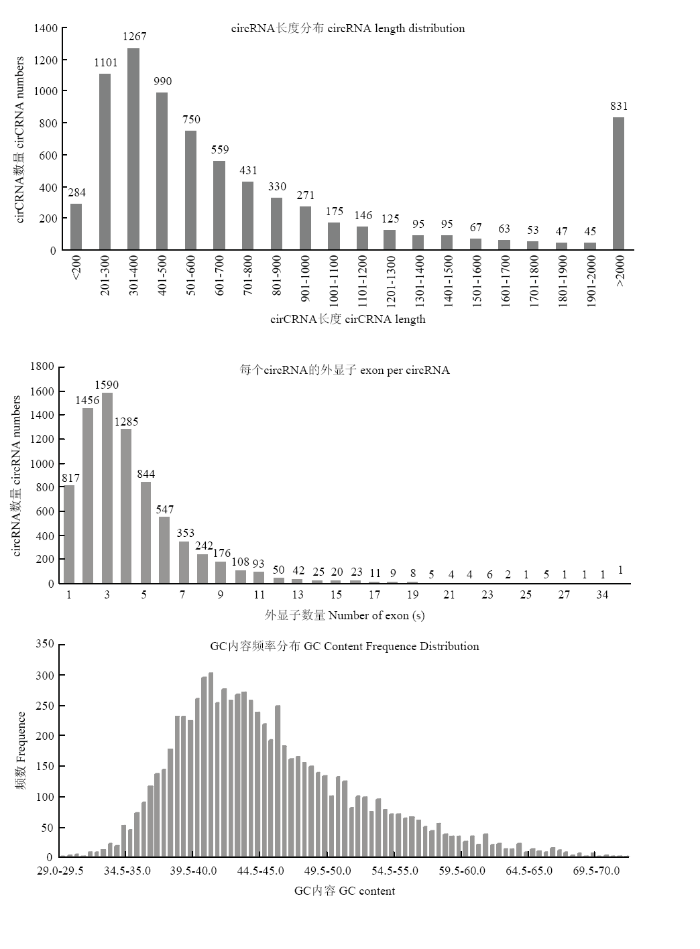 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1circRNA长度、GC含量、预测circRNA的外显子数目统计
Fig. 1circRNA length, GC content, predicting the number of exons in circRNA
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2circRNA剪切信号统计
Fig. 2circRNA shear signal statistics
图3
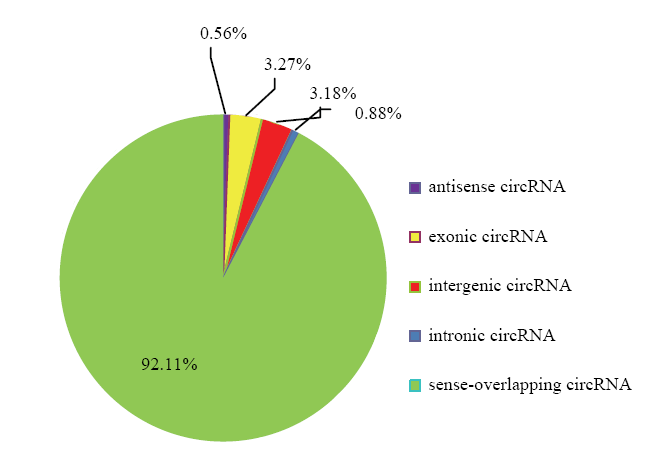 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3circRNA基因结构分布
Fig. 3circRNA gene structure map
图4
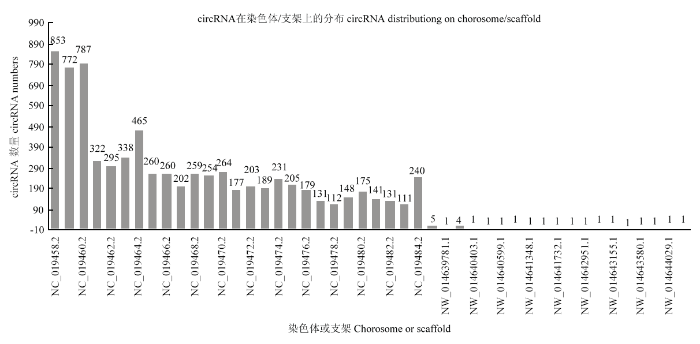 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4circRNA在各条染色体上或scaffold上数目分布
Fig. 4Distribution of circRNA on various chromosomes or on scaffold
2.2 分析和验证差异转录本
利用FPKM值估计circRNA转录物的表达水平,可以看出circRNA转录本表达水平相对很低(图5)。筛选出31个上调和29个下调的DE circRNA(图6)为了进一步验证RNA-seq的可靠性,随机选择6个DE circRNA,用q-PCR验证它们在不腹泻组和腹泻组羔羊体内的相对表达水平,发现与RNA-seq结果一致(图7),表明RNA-seq数据是可靠的。 这些分析还表明,高通量测序具有检测低表达水平(0 <FPKM <1)基因的优点。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5circRNA和mRNA转录本表达水平箱线
Fig. 5circRNA and mRNA transcript expression level box plot
图6
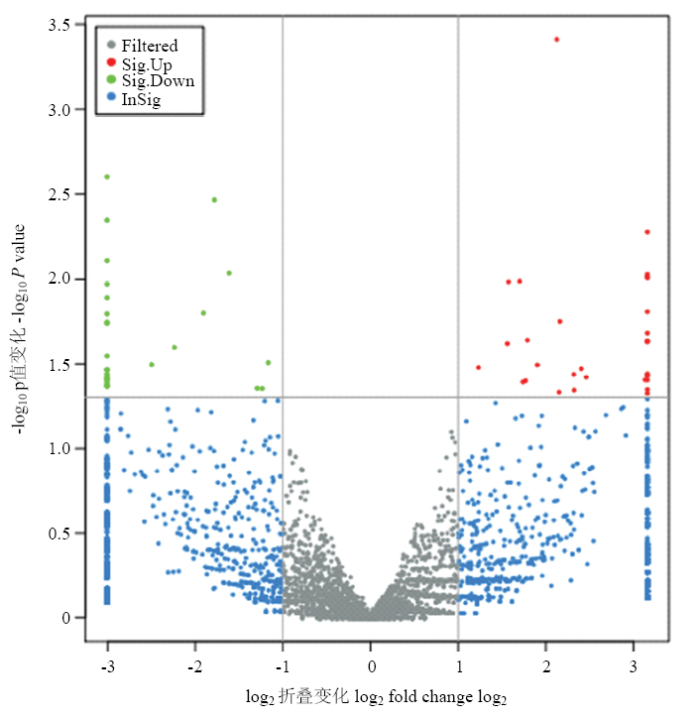 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6不腹泻和腹泻型羔羊之间差异表达的circRNAs
灰色为没有显著性差异的circRNA;红色为显著性上调的circRNA;绿色为显著性下调的circRNA;蓝色为差异倍数达到2倍以上,但在差异显著性检验中不显著的circRNA。横轴为log2 FoldChange的展示,纵轴方向为log10 P value的展示
Fig. 6circRNAs differentially expressed between lambs without diarrhea and diarrhea
Gray is a circRNA with no significant difference; red is a significantly up-regulated circRNA; green is a significantly down-regulated circRNA; blue is a doulog that is more than 2-fold different, but not significant in the significance test. The horizontal axis is the display of log2 FoldChange, and the vertical axis is the display of log10 P value
图7
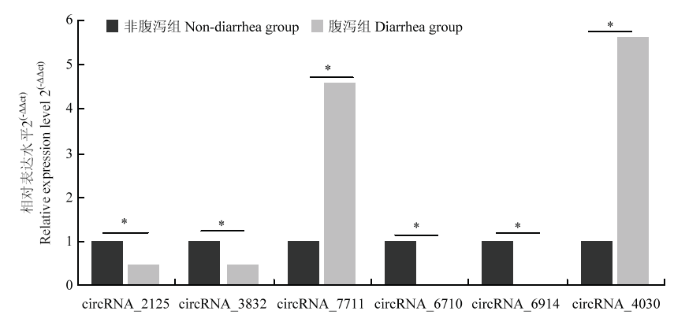 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7DE circRNA在不腹泻和腹泻羔羊体内的相对表达水平
Fig. 7Relative expression levels of DE circRNA in lambs without diarrhea and diarrhea
2.3 DE circRNA的GO功能注释和KEGG PATHWAY富集分析
DE circRNA与GO 数据库进行比对的结果表明,一共有60条circRNA被注释和分类到297个功能亚类中。图8显示了3种分类中DE circRNA数目大于2的GO条目。结果显示,绵羊oxidation-reduction process(GO:0055114)、transport(GO:0006810)、extracellular region(GO:0005576)、focal adhesion(GO:0005925)、extracellular exosome(GO:0070062)、extracellular space(GO:0005615)、zinc ion binding(GO:0008270)等7个功能亚类的circRNA较多,而其余的功能亚类的circRNA分布较少。DE circRNA与KEGG PATHWAY 数据库进行比对的结果表明,一共有60条circRNA被注释和归类到73个KEGG PATHWAY 中。图8显示了KEGG通路中DE circRNA数目大于2的KEGG条目。结果显示,绵羊Estrogen signaling pathway(path: ko04915)、Protein processing in endoplasmic reticulum(path:ko04141)、Regulation of actin cytoskeleton(path: ko04810)等3个KEGG Pathway的circRNA较多,而其余的KEGG Pathway的circRNA分布较少。图8
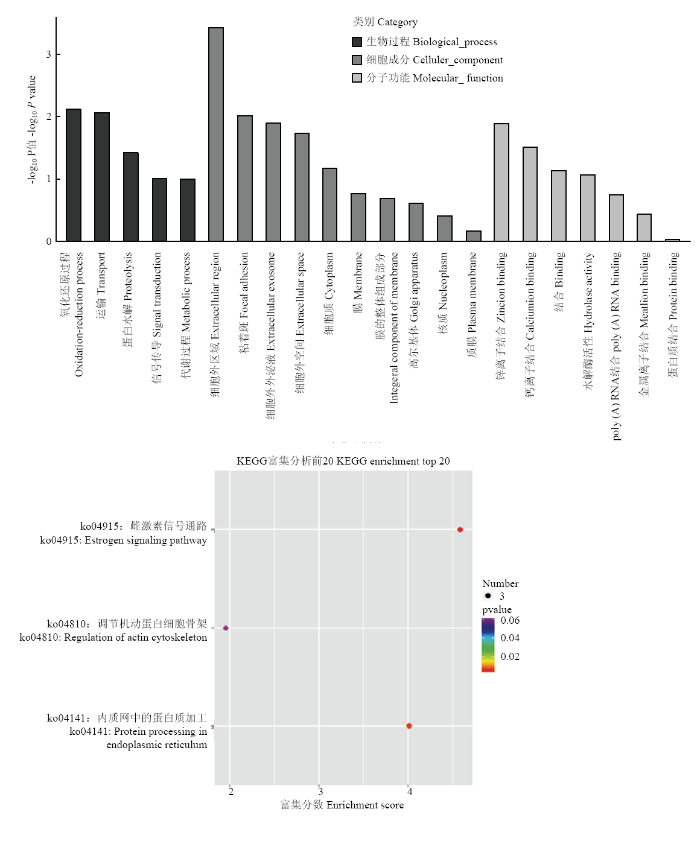 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8circRNA的GO功能注释(上)KEGG PATHWAY富集分析(下)
Fig. 8GORNA PATHWAY enrichment analysis (down) of GO function annotation (up) of circRNA
2.4 circRNA-miRNA-mRNA靶标关系预测
由于circRNA包含多个miRNA结合位点,因此可以使用miRNA靶基因预测的方法鉴定与miRNA结合的circRNA,笔者使用Miranda软件来预测与miRNA结合的circRNA以及miRNA的靶基因,根据miRNA靶基因的功能注释来阐明此部分circRNA的功能,预测详细结果见表2。Table 2
表2
表2circRNA-miRNA-mRNA靶标关系预测
Table 2
| 环状RNA circRNA | 最佳circRNA基因 Best gene of circRNA | P值 P vale | 微小RNA miRNA | 微小RNA的靶基因 Target gene of miRNA | 靶标基因转录ID Transcription ID of target gene |
|---|---|---|---|---|---|
| circRNA_6577 | LOC101111058 (Btnl 1) | 0.000190825 | oar-miR-381-5p | ||
| circRNA_7725 | 0.003873598 | oar-miR-1193-5p | NEB | XM_012137591.2 | |
| UBE3B | XM_004017436.3 | ||||
| circRNA_0309 | LOC101108092 (GSTM1) | 0.004205007 | oar-miR-370-3p | ADGRF2 | XM_004018870.3 |
| circRNA_2125 | LOC101115614 (NRAMP2) | 0.004205007 | oar-miR-370-3p | LAMA1 | XM_012103553.2 |
| circRNA_3832 | B2M | 0.004205007 | oar-miR-370-3p | LTF | NM_001024862.1 |
| circRNA_6577 | LOC101111058 (Btnl 1) | 0.004205007 | oar-miR-370-3p | MGAT5 | XM_012139230.2 |
| circRNA_7711 | 0.004205007 | oar-miR-370-3p | TLN2 | XM_012181407.2 | |
| circRNA_6577 | LOC101111058 (Btnl 1) | 0.006550584 | oar-miR-3956-3p | SLC25A29 | XM_015102051.1 |
| circRNA_6577 | LOC101111058 (Btnl 1) | 0.011765813 | oar-miR-370-5p |
新窗口打开|下载CSV
3 讨论
由于传统的分子生物学方法对circRNA数量和丰度的检测能效都非常有限,因此一直以来circRNA被认为是RNA异常剪接的产物[7]。近年来随着生物信息学的快速发展和高通量测序技术的不断革新,目前通过高通量测序和生物信息学分析已经在真核生物中鉴定了大量的circRNA[6,22-23],并发现circRNA可能在调控基因表达过程中发挥着重要作用[9,24-25]。研究发现大多数circRNA包含有miRNA结合位点,可以作为高效的竞争性内源RNA,有效吸附miRNA从而调控miRNA的靶基因[26,27],其具有以下4种生物学功能:miRNA海绵作用[25,26],作为蛋白质翻译模板[28,29],调控基因转录[30,31]以及通过竞争调控线性RNA的生成[32,33]。然而,迄今为止,关于羔羊腹泻的circRNA,特别是绵羊的研究报道很少。湖羊是一种具有高繁殖力和对湿热气候适应性强的中国特有品种,可以全年在室内饲养。在这项研究中,笔者不仅提供了绵羊腹泻过程中circRNA的第一个概况,还研究了circRNA在抗病过程中的可能作用。长期以来,羔羊腹泻对牧场造成了严重的经济损失。在研究中,发现circRNA的表达水平很低。由于circRNA包含多个miRNA结合位点,因此可以通过Miranda 软件使用miRNA靶基因预测的方法鉴定与6个circRNA结合的5个miRNA,并鉴定出其中4个circRNA的母基因,分别为Btnl 1、GSTM1、NRAMP2、B2M。
Btnl 1是T细胞活化和免疫疾病的关键抑制因子[34]。Btnl 1的作用机制不同于Btnl 2,BtnlA1,它是通过反受体接合直接抑制T细胞活化[34,35,36,37]。研究发现Btnl基因可能是肠道炎症的新型重要局部调节因子[38]。GSTM1编码谷胱甘肽-S-转移酶(glutathione-S- transferase,GST)M1酶,参与肺癌各种致癌物质的排毒[39],并且在保护细胞免受氧化应激方面发挥关键作用[40]。NRAMP2是一种金属转运蛋白,在锰缺乏的条件下,NRAMP2参与了高尔基体中Mn的再生,促进植物根系生长[41]。B2M编码主要组织相容性复合体(major histocompatibility complex, MHC)I类分子的β链,并在炎症和肿瘤细胞中上调[42]。
笔者也使用Miranda 软件预测了3个miRNA的靶基因,并且在不腹泻和腹泻组之间显著差异表达,分别为NEB、UBE3B、ADGRF2、LAMA1、LTF、MGAT5、TLN2、SLC25A29。
NEB基因编码伴肌动蛋白,是细胞骨架基质的巨大蛋白组分,与骨骼肌肌小节中的粗细肌丝共存,NEB基因的突变是杆状体肌病最常见的原因,约占50%[43]。泛素化需要3种酶的连续作用:活化酶(E1),结合酶(E2)和连接酶(E3)。UBE3B是泛素化连接酶(UBE3)成员之一,它和E2结合酶的独特组合提供了靶向特定蛋白质降解所需的高度底物特异性[44]。ADGRF2是粘附性G蛋白偶联受体家族成员之一,在细胞间与细胞-基质的粘附过程中扮演重要角色[45]。LAMA1突变可能与Poretti-Boltshauser综合征有关,有研究证明LAMA1缺陷可导致细胞骨架形态的改变[46]。LTF是转铁蛋白家族基因的成员,其蛋白质产物能够启动宿主防御,抵抗广泛的微生物感染,抗原活性[47]。MGAT5编码的蛋白质属于糖基转移酶家族,它是参与调控糖蛋白寡糖生物合成的最重要的酶之一,细胞表面糖蛋白上寡糖的改变引起细胞粘着或迁移行为的显著变化,这种酶的活性增加与侵袭性恶性肿瘤的发展有关[48]。Talins是将粘附分子的整合素家族连接到F-肌动蛋白的大的接头蛋白,Talin 1对于整联蛋白介导的细胞粘附是必需的,TLN2与Talin 1一样,被认为与独特的跨膜受体结合,在细胞外基质和肌动蛋白细胞骨架之间形成新的连接[49]。SLC25A29编码核编码的线粒体蛋白质,其是溶质运载体家族25(SLC25)线粒体运载体大家族的成员,SLC25A29的主要生理作用是将碱性氨基酸导入线粒体,进行线粒体蛋白质合成和氨基酸降解[50]。
GO是一种广泛用于研究基因功能关系的生物信息学工具。对60个DE circRNA进行GO分析,结果表明,绵羊oxidation-reduction process(GO:0055114)、transport(GO:0006810)、extracellular region(GO:0005576)、focal adhesion(GO:0005925)、extracellular exosome(GO: 0070062)、extracellular space(GO:0005615)、zinc ion binding(GO:0008270)等7个功能亚类富集到的circRNA较多。KEGG Pathway分析表明,信号通路如Estrogen signaling pathway(path:ko04915)、Protein processing in endoplasmic reticulum(path:ko04141)、Regulation of actin cytoskeleton(path:ko04810)可能是DE circRNA参与调控的重要途径,相关的circRNA可能潜在地参与菌毛黏附肠道黏膜的过程。然而,这些通路在抗病过程中的作用很大程度上仍然是未知的。
笔者发现总共60个已知的circRNA在不腹泻组和腹泻组之间显著差异表达,其中31个上调和29个下调。另外,我们确定了两组中共有1 942个新的circRNA。为了进一步验证RNA-Seq结果,采用q-PCR验证6个已知circRNA的表达水平,结果一致。
4 结论
通过对不腹泻组和腹泻组羔羊脾脏中circRNA表达谱的研究,了解其在绵羊抗病发生过程中具有调控作用。进一步发现circRNA的差异表达,预测了与circRNA竞争性结合的miRNA及其靶基因,初步构建circRNA-miRNA-mRNA网络互作模型。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.3969/j.issn.1008-8199.2014.03.021URL [本文引用: 1]

长链非编码RNAs(long non-coding RNAs,LncRNAs)是一类转录本长度超过200个核苷酸残基(nt)的RNA分子,位于细胞核或细胞质内,大多由RNA聚合酶Ⅱ转录,但不能编码蛋白质。越来越多的证据表明LncRNA基因也发生遗传变异,导致LncRNA基因存在着广泛的基因多态性,而大多数LrwRNA基因多态性与肿瘤的发生、转移和预后相关。文中结合国内外最新报道,对LncRNAs多态性与前列腺癌、肝癌、膀胱癌、急性淋巴细胞性白血病及甲状腺肿瘤的相关性研究进展作一综述,旨在为肿瘤的诊断和治疗提供新的思路。
DOI:10.3969/j.issn.1008-8199.2014.03.021URL [本文引用: 1]

长链非编码RNAs(long non-coding RNAs,LncRNAs)是一类转录本长度超过200个核苷酸残基(nt)的RNA分子,位于细胞核或细胞质内,大多由RNA聚合酶Ⅱ转录,但不能编码蛋白质。越来越多的证据表明LncRNA基因也发生遗传变异,导致LncRNA基因存在着广泛的基因多态性,而大多数LrwRNA基因多态性与肿瘤的发生、转移和预后相关。文中结合国内外最新报道,对LncRNAs多态性与前列腺癌、肝癌、膀胱癌、急性淋巴细胞性白血病及甲状腺肿瘤的相关性研究进展作一综述,旨在为肿瘤的诊断和治疗提供新的思路。
DOI:10.1073/pnas.73.11.3852URLPMID:1069269 [本文引用: 1]

Viroids are uncoated infectious RNA molecules pathogenic to certain higher plants. Four different highly purified viroids were studied. By ultracentrifugation, thermal denaturation, electron microscopy, and end group analysis the following features were established: (i) the molecular weight of cucumber pale fruit viroid from tomato is 110,000, of citrus exocortis viroid from Gynura 119,000, of citrus exocortis viroid from tomato 119,000, and of potato spindle tuber viroid from tomato 127,000. (ii) Viroids are single-stranded molecules. (iii) Viroids exhibit high thermal stability, cooperativity, and self-complementarity resulting in a rod-like native structure. (iv) Viroids are covalently closed circular RNA molecules.
DOI:10.1073/pnas.87.19.7628URLPMID:1699230 [本文引用: 1]

Circular RNA replicons have been reported in plants and, in one case, in animal cells. We describe such an element in yeast. In certain yeast strains, a 20S RNA species appears on transfer of cells to acetate medium. This phenotype shows cytoplasmic (non-Mendelian) inheritance and the 20S RNA is associated with 23-kDa protein subunits as a 32S particle. We demonstrate that yeast 20S RNA is an independent replicon with no homology to host genomic, mitochondrial, or 2- m plasmid DNA or to the L-A, L-BC, or M1double-stranded RNA viruses of yeast. The circularity of the 20S RNA is shown by the apparent absence of 3' and 5' ends, by two-dimensional gel electrophoresis, and by electron microscopy. Replication of yeast 20S RNA proceeds through an RNA-RNA pathway, and a 10,000-fold amplification occurs on shift to acetate medium. The copy number of 20S RNA is also reduced severalfold by the SKI gene products, a host antiviral system that also lowers the copy numbers of yeast double-stranded RNA viruses. Yeast 20S RNA and the hepatitis virus show some similarities.
DOI:10.1016/0092-8674(80)90505-XURLPMID:6986989 [本文引用: 1]

11S and 18S fractions of yeast mitochondrial RNAs, isolated by electrophoresis through agarose gels, have been found by electron microscopy to contain approximately 50% circular molecules. Circles in the 11S fraction have a contour length of 0.36 ± 0.02 μm, which is approximately equal to the length of the majority of linear molecules also present. Circles in the 18S fraction have an average length of 0.78 ± 0.11 μm. The size distribution is broader than for the 11S fraction, and we cannot exclude the possibility that more than one size class may be present. The 11S circular RNA forms circular R loops and RNA-DNA hybrids with DNA fragments of the oxi 3 region of mtDNA, which contains the structural gene for subunit I of cytochrome oxidase. As judged from the electron micrographs, the complete RNA participates in hybrid formation and the sequences coding for it appear to be continuous. Both 11S and 18S circles withstand treatment with DNAase and pronase. They are not eliminated by treatment with 1 M glyoxal in 50% formamide for 1 hr at 50°C. We conclude that they are covalently closed. The function of the circular RNAs is unknown. They may be active as mRNAs, storage forms, or arise in a cut-and-splice process which generates mRNAs from longer transcripts.
DOI:10.1038/323558a0 [本文引用: 1]
[本文引用: 2]
DOI:10.1006/excr.1993.1021URLPMID:7678559 [本文引用: 2]

Abstract We previously identified novel human ets-1 transcripts in which the normal order of exons is inverted, and demonstrated that although the order of exons is different than in the genomic DNA, splicing of these exons out of order occurs in pairs using genuine splice sites (1). Here we determine the structure of these novel transcripts, showing that they correspond to circular RNA molecules containing only exons in genomic order. These transcripts are stable molecules, localized in the cytoplasmic component of the cells. To our knowledge, this is the first case of circular transcripts being processed from nuclear pre-mRNA in eukaryotes. This new type of transcript might represent a novel aspect of gene expression and hold some interesting clues about the splicing mechanism.
DOI:10.1261/rna.047126.114URLPMID:25404635 [本文引用: 1]

It is now clear that there is a diversity of circular RNAs in biological systems. Circular RNAs can be produced by the direct ligation of 5' and 3' ends of linear RNAs, as intermediates in RNA processing reactions, or by "backsplicing," wherein a downstream 5' splice site (splice donor) is joined to an upstream 3' splice site (splice acceptor). Circular RNAs have unique properties including the potential for rolling circle amplification of RNA, the ability to rearrange the order of genomic information, protection from exonucleases, and constraints on RNA folding. Circular RNAs can function as templates for viroid and viral replication, as intermediates in RNA processing reactions, as regulators of transcription in cis, as snoRNAs, and as miRNA sponges. Herein, we review the breadth of circular RNAs, their biogenesis and metabolism, and their known and anticipated functions.
DOI:10.1371/journal.pone.0030733URLPMID:3270023 [本文引用: 2]

Most human pre-mRNAs are spliced into linear molecules that retain the exon order defined by the genomic sequence. By deep sequencing of RNA from a variety of normal and malignant human cells, we found RNA transcripts from many human genes in which the exons were arranged in a non-canonical order. Statistical estimates and biochemical assays provided strong evidence that a substantial fraction of the spliced transcripts from hundreds of genes are circular RNAs. Our results suggest that a non-canonical mode of RNA splicing, resulting in a circular RNA isoform, is a general feature of the gene expression program in human cells.
DOI:10.1016/j.molcel.2014.08.019URL [本文引用: 1]

Ashwal-Fluss et02al. provide evidence that circRNA production competes with linear mRNA splicing. They demonstrate that intronic sequences largely determine the production rate of circRNAs and identify a splice factor (muscleblind) that can promote circularization.
DOI:10.1016/j.celrep.2014.12.002URLPMID:25543144 [本文引用: 1]

Starke et al. conclusively demonstrate a circular configuration of two predicted circular RNAs and analyze the sequence requirements for exon circularization. Canonical splice signals are required for circularization, which together with splice inhibitor assays indicates that the canonical spliceosome machinery functions in the biogenesis of circular RNAs.
DOI:10.3969/j.issn.1673-4556.2017.04.109URL [本文引用: 1]

羊大肠杆菌病是规模养羊场最为常见高发的细菌性疾病之一,传统沿用的抗生素治疗方案存在诸多缺陷.基于对致病原生物学特性及流行病学特点深入研究的基础上,采取早期准确诊断及科学防治措施,在药物防治上达到理想效果,为业内防治羊大肠杆菌病提供借鉴.
DOI:10.3969/j.issn.1673-4556.2017.04.109URL [本文引用: 1]

羊大肠杆菌病是规模养羊场最为常见高发的细菌性疾病之一,传统沿用的抗生素治疗方案存在诸多缺陷.基于对致病原生物学特性及流行病学特点深入研究的基础上,采取早期准确诊断及科学防治措施,在药物防治上达到理想效果,为业内防治羊大肠杆菌病提供借鉴.
DOI:10.3969/J.ISSN.1671-6027.2017.06.071URL [本文引用: 1]

羊大肠杆菌病也称为新生羔羊腹泻病,是一种传染性极强的病症。大肠杆菌是羔羊体内常在的条件致病菌,当外界环境发生突变时极易引发大肠杆菌病,应加以重视。
DOI:10.3969/J.ISSN.1671-6027.2017.06.071URL [本文引用: 1]

羊大肠杆菌病也称为新生羔羊腹泻病,是一种传染性极强的病症。大肠杆菌是羔羊体内常在的条件致病菌,当外界环境发生突变时极易引发大肠杆菌病,应加以重视。
[本文引用: 1]
DOI:10.1261/rna.043687.113URL [本文引用: 1]
DOI:10.1186/s13059-014-0571-3URLPMID:25583365 [本文引用: 1]

Recent studies reveal that circular RNAs (circRNAs) are a novel class of abundant, stable and ubiquitous noncoding RNA molecules in animals. Comprehensive detection of circRNAs from high-throughput...
[本文引用: 1]
DOI:10.1093/bioinformatics/btp616URL [本文引用: 1]
DOI:10.1093/nar/gkm882URLPMID:2238879 [本文引用: 1]

KEGG (http://www.genome.jp/kegg/) is a database of biological systems that integrates genomic, chemical and systemic functional information. KEGG provides a reference knowledge base for linking genomes to life through the process of PATHWAY mapping, which is to map, for example, a genomic or transcriptomic content of genes to KEGG reference pathways to infer systemic behaviors of the cell or the organism. In addition, KEGG provides a reference knowledge base for linking genomes to the environment, such as for the analysis of drug-target relationships, through the process of BRITE mapping. KEGG BRITE is an ontology database representing functional hierarchies of various biological objects, including molecules, cells, organisms, diseases and drugs, as well as relationships among them. KEGG PATHWAY is now supplemented with a new global map of metabolic pathways, which is essentially a combined map of about 120 existing pathway maps. In addition, smaller pathway modules are defined and stored in KEGG MODULE that also contains other functional units and complexes. The KEGG resource is being expanded to suit the needs for practical applications. KEGG DRUG contains all approved drugs in the US and Japan, and KEGG DISEASE is a new database linking disease genes, pathways, drugs and diagnostic markers.
DOI:10.1371/journal.pbio.0020363URL [本文引用: 1]
DOI:10.1186/gb-2003-5-1-r1URLPMID:14709173 [本文引用: 1]

Abstract BACKGROUND: The recent discoveries of microRNA (miRNA) genes and characterization of the first few target genes regulated by miRNAs in Caenorhabditis elegans and Drosophila melanogaster have set the stage for elucidation of a novel network of regulatory control. We present a computational method for whole-genome prediction of miRNA target genes. The method is validated using known examples. For each miRNA, target genes are selected on the basis of three properties: sequence complementarity using a position-weighted local alignment algorithm, free energies of RNA-RNA duplexes, and conservation of target sites in related genomes. Application to the D. melanogaster, Drosophila pseudoobscura and Anopheles gambiae genomes identifies several hundred target genes potentially regulated by one or more known miRNAs. RESULTS: These potential targets are rich in genes that are expressed at specific developmental stages and that are involved in cell fate specification, morphogenesis and the coordination of developmental processes, as well as genes that are active in the mature nervous system. High-ranking target genes are enriched in transcription factors two-fold and include genes already known to be under translational regulation. Our results reaffirm the thesis that miRNAs have an important role in establishing the complex spatial and temporal patterns of gene activity necessary for the orderly progression of development and suggest additional roles in the function of the mature organism. In addition the results point the way to directed experiments to determine miRNA functions. CONCLUSIONS: The emerging combinatorics of miRNA target sites in the 3' untranslated regions of messenger RNAs are reminiscent of transcriptional regulation in promoter regions of DNA, with both one-to-many and many-to-one relationships between regulator and target. Typically, more than one miRNA regulates one message, indicative of cooperative translational control. Conversely, one miRNA may have several target genes, reflecting target multiplicity. As a guide to focused experiments, we provide detailed online information about likely target genes and binding sites in their untranslated regions, organized by miRNA or by gene and ranked by likelihood of match. The target prediction algorithm is freely available and can be applied to whole genome sequences using identified miRNA sequences.
DOI:10.18632/aging.100834PMID:26546448 [本文引用: 1]

Circular RNAs (circRNAs) have been identified in numerous species, including human, mouse, nematode, and coelacanth. They are believed to function as regulators of gene expression at least in part by sponging microRNAs. Here, we describe the identification of circRNAs in monkey (Rhesus macaque) skeletal muscle. RNA sequencing analysis was employed to identify and annotate 12,000 circRNAs, including numerous circular intronic RNAs (ciRNAs), from skeletal muscle of monkeys of a range of ages. Reverse transcription followed by real-time quantitative (q)PCR analysis verified the presence of these circRNAs, including the existence of several highly abundant circRNAs, and the differential abundance of a subset of circRNAs as a function of age. Taken together, our study has documented systematically circRNAs expressed in skeletal muscle and has identified circRNAs differentially abundant with advancing muscle age. We propose that some of these circRNAs might influence muscle function.
DOI:10.1371/journal.pgen.1001233URLPMID:2996334 [本文引用: 1]

Human genome-wide association studies have linked single nucleotide polymorphisms (SNPs) on chromosome 9p21.3 near the INK4/ARF (CDKN2a/b) locus with susceptibility to atherosclerotic vascular disease (ASVD). Although this locus encodes three well-characterized tumor suppressors, p16(INK4a), p15(INK4b), and ARF, the SNPs most strongly associated with ASVD are 120 kb from the nearest coding gene within a long non-coding RNA (ncRNA) known as ANRIL (CDKN2BAS). While individuals homozygous for the atherosclerotic risk allele show decreased expression of ANRIL and the coding INK4/ARF transcripts, the mechanism by which such distant genetic variants influence INK4/ARF expression is unknown. Here, using rapid amplification of cDNA ends (RACE) and analysis of next-generation RNA sequencing datasets, we determined the structure and abundance of multiple ANRIL species. Each of these species was present at very low copy numbers in primary and cultured cells; however, only the expression of ANRIL isoforms containing exons proximal to the INK4/ARF locus correlated with the ASVD risk alleles. Surprisingly, RACE also identified transcripts containing non-colinear ANRIL exonic sequences, whose expression also correlated with genotype and INK4/ARF expression. These non-polyadenylated RNAs resisted RNAse R digestion and could be PCR amplified using outward-facing primers, suggesting they represent circular RNA structures that could arise from by-products of mRNA splicing. Next-generation DNA sequencing and splice prediction algorithms identified polymorphisms within the ASVD risk interval that may regulate ANRIL splicing and circular ANRIL (cANRIL) production. These results identify novel circular RNA products emanating from the ANRIL locus and suggest causal variants at 9p21.3 regulate INK4/ARF expression and ASVD risk by modulating ANRIL expression and/or structure.
DOI:10.1371/journal.pgen.1003777URLPMID:24039610 [本文引用: 1]

Thousands of loci in the human and mouse genomes give rise to circular RNA transcripts; at many of these loci, the predominant RNA isoform is a circle. Using an improved computational approach for circular RNA identification, we found widespread circular RNA expression in Drosophila melanogaster and estimate that in humans, circular RNA may account for 1% as many molecules as poly(A) RNA. Analysis of data from the ENCODE consortium revealed that the repertoire of genes expressing circular RNA, the ratio of circular to linear transcripts for each gene, and even the pattern of splice isoforms of circular RNAs from each gene were cell-type specific. These results suggest that biogenesis of circular RNA is an integral, conserved, and regulated feature of the gene expression program. Last year, we reported that circular RNA isoforms, previously thought to be very rare, are actually a pervasive feature of eukaryotic gene expression programs; indeed, the major RNA isoform from hundreds of human genes is a circle. Previous novel RNA species that initially appeared to be special cases, of dubious biological significance, have subsequently proved to have critical, conserved biological roles. An almost universal characteristic of regulatory macromolecules is that they are themselves regulated during development and differentiation. Here, we show that the repertoire of genes expressing circular RNA, the relative levels of circular: linear transcripts from each gene, and even the pattern of splice isoforms of circular RNAs from each gene were cell-type specific, including examples of striking regulation. In humans, we estimate that circular RNA may account for about 1% as many molecules as poly(A) RNA. The ubiquity of circular RNA and its specific regulation could significantly alter our perspective on post-transcriptional regulation and the roles that RNA can play in the cell.
DOI:10.1038/nature11928URL [本文引用: 2]
DOI:10.1038/nature11993URLPMID:23446346 [本文引用: 2]

MicroRNAs (miRNAs) are important post-transcriptional regulators of gene expression that act by direct base pairing to target sites within untranslated regions of messenger RNAs1. Recently, miRNA activity has been shown to be affected by the presence of miRNA sponge transcripts, the so-called competing endogenous RNA in humans and target mimicry in plants(2-7). We previously identified a highly expressed circular RNA (circRNA) in human and mouse brain(8). Here we show that this circRNA acts as a miR-7 sponge; we term this circular transcript ciRS-7 (circular RNA sponge for miR-7). ciRS-7 contains more than 70 selectively conserved miRNA target sites, and it is highly and widely associated with Argonaute (AGO) proteins in a miR-7-dependent manner. Although the circRNA is completely resistant to miRNA-mediated target destabilization, it strongly suppresses miR-7 activity, resulting in increased levels of miR-7 targets. In the mouse brain, we observe overlapping co-expression of ciRS-7 and miR-7, particularly in neocortical and hippocampal neurons, suggesting a high degree of endogenous interaction. We further show that the testis-specific circRNA, sex-determining region Y (Sry)(9), serves as a miR-138 sponge, suggesting that miRNA sponge effects achieved by circRNA formation are a general phenomenon. This study serves as the first, to our knowledge, functional analysis of a naturally expressed circRNA.
DOI:10.1038/ncomms11215URLPMID:27050392 [本文引用: 1]

Circular RNAs (circRNAs) represent a class of widespread and diverse endogenous RNAs that may regulate gene expression in eukaryotes. However, the regulation and function of human circRNAs remain largely unknown. Here we generate ribosomal-depleted RNA sequencing data from six normal tissues and seven cancers, and detect at least 27,000 circRNA candidates. Many of these circRNAs are differently expressed between the normal and cancerous tissues. We further characterize one abundant circRNA derived from Exon2 of theHIPK3gene, termed circHIPK3. The silencing of circHIPK3 but notHIPK3mRNA significantly inhibits human cell growth. Via a luciferase screening assay, circHIPK3 is observed to sponge to 9 miRNAs with 18 potential binding sites. Specifically, we show that circHIPK3 directly binds to miR-124 and inhibits miR-124 activity. Our results provide evidence that circular RNA produced from precursor mRNA may have a regulatory role in human cells. Circular RNAs are formed from exon back-splicing, the significance of these endogenous RNAs is beginning to be unraveled. Here, the authors identify thousands of circular RNAs differentially expressed between normal and cancer tissues and show that an abundant circular RNA generated fromHIPK3regulates cell growth.
DOI:10.1126/science.7536344URLPMID:7536344 [本文引用: 1]

The ribosome scanning model predicts that eukaryotic ribosomal 40S subunits enter all messenger RNAs at their 5′ ends. Here, it is reported that eukaryotic ribosomes can initiate translation on circular RNAs, but only if the RNAs contain internal ribosome entry site elements. Long-repeating polypeptide chains were synthesized from RNA circles with continuous open reading frames. These results indicate that ribosomes can translate such RNA circles for multiple consecutive rounds and that the free 5′ end of a messenger RNA is not necessarily the entry point for 40S subunits.
DOI:10.1261/rna.048272.114URLPMID:25449546 [本文引用: 1]

Abstract While the human transcriptome contains a large number of circular RNAs (circRNAs), the functions of most circRNAs remain unclear. Sequence annotation suggests that most circRNAs are generated from splicing in reversed orders across exons. However, the mechanisms of this backsplicing are largely unknown. Here we constructed a single exon minigene containing split GFP, and found that the pre-mRNA indeed produces circRNA through efficient backsplicing in human and Drosophila cells. The backsplicing is enhanced by complementary introns that form double-stranded RNA structure to bring splice sites in proximity, but such structure is not required. Moreover, backsplicing is regulated by general splicing factors and cis-elements, but with regulatory rules distinct from canonical splicing. The resulting circRNA can be translated to generate functional proteins. Unlike linear mRNA, poly-adenosine or poly-thymidine in 3' UTR can inhibit circular mRNA translation. This study revealed that backsplicing can occur efficiently in diverse eukaryotes to generate circular mRNAs. 2015 Wang and Wang; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
DOI:10.1016/j.molcel.2013.08.017URLPMID:24035497 [本文引用: 1]

61Circular intronic RNAs accumulate in human cells owing to escape from debranching61Their processing depends on RNA motifs near 5′ splice site and branchpoint61ciRNAs regulate parent gene expression by modulating elongation Pol II activity
DOI:10.1038/nsmb.2959URL [本文引用: 1]
DOI:10.1261/rna.047126.114URLPMID:25404635 [本文引用: 1]

It is now clear that there is a diversity of circular RNAs in biological systems. Circular RNAs can be produced by the direct ligation of 5' and 3' ends of linear RNAs, as intermediates in RNA processing reactions, or by "backsplicing," wherein a downstream 5' splice site (splice donor) is joined to an upstream 3' splice site (splice acceptor). Circular RNAs have unique properties including the potential for rolling circle amplification of RNA, the ability to rearrange the order of genomic information, protection from exonucleases, and constraints on RNA folding. Circular RNAs can function as templates for viroid and viral replication, as intermediates in RNA processing reactions, as regulators of transcription in cis, as snoRNAs, and as miRNA sponges. Herein, we review the breadth of circular RNAs, their biogenesis and metabolism, and their known and anticipated functions.
DOI:10.1016/j.molcel.2014.08.019URL [本文引用: 1]

Ashwal-Fluss et02al. provide evidence that circRNA production competes with linear mRNA splicing. They demonstrate that intronic sequences largely determine the production rate of circRNAs and identify a splice factor (muscleblind) that can promote circularization.
DOI:10.4049/jimmunol.1000835URL [本文引用: 2]
DOI:10.4049/jimmunol.176.12.7354URLPMID:1626526 [本文引用: 1]

Abstract B7 family members regulate T cell activation and tolerance. Although butyrophilin proteins share sequence homology with the B7 molecules, it is unclear whether they have any function in immune responses. In the present study, we characterize an MHC class II gene-linked butyrophilin family member, butyrophilin-like 2 (BTNL2), the mutation of which has been recently associated with the inflammatory autoimmune diseases sarcoidosis and myositis. Mouse BTNL2 is a type I transmembrane protein with two pairs of Ig-like domains separated by a heptad peptide sequence. BTNL2 mRNA is highly expressed in lymphoid tissues as well as in intestine. To characterize the function of BTNL2, we produced a BTNL2-Ig fusion protein. It recognized a putative receptor whose expression on B and T cells was significantly enhanced after activation. BTNL2-Ig inhibited T cell proliferation and TCR activation of NFAT, NF-kappaB, and AP-1 signaling pathways. BTNL2 is thus the first member of the butyrophilin family that regulates T cell activation, which has implications in immune diseases and immunotherapy.
DOI:10.4049/jimmunol.178.3.1523URLPMID:17237401 [本文引用: 1]

Abstract Butyrophilin-like 2 (BTNL2) is a butyrophilin family member with homology to the B7 costimulatory molecules, polymorphisms of which have been recently associated through genetic analyses to sporadic inclusion body myositis and sarcoidosis. We have characterized the full structure, expression, and function of BTNL2. Structural analysis of BTNL2 shows a molecule with an extracellular region containing two sets of two Ig domains, a transmembrane region, and a previously unreported cytoplasmic tail. Unlike most other butyrophilin members, BTNL2 lacks the prototypical B30.2 ring domain. TaqMan and Northern blot analysis indicate BTNL2 is predominantly expressed in digestive tract tissues, in particular small intestine and Peyer's patches. Immunohistochemistry with BTNL2-specific Abs further localizes BTNL2 to epithelial and dendritic cells within these tissues. Despite its homology to the B7 family, BTNL2 does not bind any of the known B7 family receptors such as CD28, CTLA-4, PD-1, ICOS, or B and T lymphocyte attenuator. Because of its localization in the gut and potential role in the immune system, BTNL2 expression was analyzed in a mouse model of inflammatory bowel disease. BTNL2 is overexpressed during both the asymptomatic and symptomatic phase of the Mdr1a knockout model of spontaneous colitis. In functional assays, soluble BTNL2-Fc protein inhibits the proliferation of murine CD4(+) T cells from the spleen, mesenteric lymph node, and Peyer's patch. In addition, BTNL2-Fc reduces proliferation and cytokine production from T cells activated by anti-CD3 and B7-related protein 1. These data suggest a role for BTNL2 as a negative costimulatory molecule with implications for inflammatory disease.
DOI:10.4049/jimmunol.0900416URLPMID:20208008 [本文引用: 1]

Abstract Butyrophilin (BTN) genes encode a set of related proteins. Studies in mice have shown that one of these, BTN1A1, is required for milk lipid secretion in lactation, whereas butyrophilin-like 2 is a coinhibitor of T cell activation. To understand these disparate roles of BTNs, we first compared the expression and functions of mouse Btn1a1 and Btn2a2. Btn1a1 transcripts were not restricted to lactating mammary tissue but were also found in virgin mammary tissue and, interestingly, spleen and thymus. In confirmation of this, BTN1A1 protein was detected in thymic epithelial cells. By contrast, Btn2a2 transcripts and protein were broadly expressed. Cell surface BTN2A2 protein, such as the B7 family molecule programmed death ligand 1, was upregulated upon activation of T cells. We next examined the potential of both BTN1A1 and BTN2A2 to interact with T cells. Recombinant Fc fusion proteins of murine BTN2A2 and, surprisingly BTN1A1, bound to activated T cells, suggesting the presence of one or more receptors on these cells. Immobilized BTN-Fc fusion proteins, but not MOG-Fc protein, inhibited the proliferation of CD4 and CD8 T cells activated by anti-CD3. BTN1A1 and BTN2A2 also inhibited T cell metabolism, IL-2, and IFN-gamma secretion. Inhibition of proliferation was not abrogated by exogenous IL-2 but could be overcome following costimulation with high levels of anti-CD28 Ab. These data are consistent with a coinhibitory role for mouse BTNs, including BTN1A1, the BTN expressed in the lactating mammary gland and on milk lipid droplets.
DOI:10.1073/pnas.1010647108URL [本文引用: 1]

Although local regulation of T-cell responses by epithelial cells is increasingly viewed as important, few molecules mediating such regulation have been identified. Skint1, a recently identified member of the Ig-supergene family expressed by thymic epithelial cells and keratinocytes, specifies the murine epidermal intraepithelial lymphocyte (IEL) repertoire. Investigating whether Skint1-related molecules might regulate IEL in other compartments, this study focuses on buytrophilin-like 1 (Btnl1), which is conspicuously similar to Skint1 and primarily restricted to small intestinal epithelium. Btnl1 protein is mostly cytoplasmic, but surface expression can be induced, and in vivo Btnl1 can be detected adjacent to the IEL. In a newly developed culture system, enforced epithelial cell expression of Btnl1 attenuated the cells' response to activated IEL, as evidenced by suppression of IL-6 and other inflammatory mediators. These findings offer a unique perspective on emerging genetic data that Btnl genes may comprise novel and important local regulators of gut inflammation.
.
DOI:10.1080/10810730.2012.688245URLPMID:22888806 [本文引用: 1]

Genetic markers of lung cancer susceptibility, such as the common variant of the glutathione S-transferase Mu 1 gene (GSTM1-null), confer small probabilities of disease risk. The authors explored the influence of different approaches to communicating the small variations in risk associated with this biomarker. College smokers (N02=02128) imagined that they had the GSTM1 wild-type variant versus the GSTM1 null-type variant. The authors presented lung cancer risk in 6 ways that varied the risk format (absolute risk vs. incremental risk) and the presentation style of the information (no graphics vs. graphic display of foreground only vs. graphic display of foreground02+02background). Presentation style had minor effects. However, absolute risk information increased negative emotions more than did incremental risk information. Perceptions of risk and negative emotions were most profoundly affected by the difference between having the GSTM1 wild-type variant versus the GSTM1 null-type variant. The authors discuss implications for conveying small probabilities related to genetic risk.
DOI:10.1111/ctr.2009.23.issue-4URL [本文引用: 1]
DOI:10.3389/fpls.2014.00106URLPMID:3978347 [本文引用: 1]

Manganese (Mn), an essential trace element, is important for plant health. In plants, Mn serves as a cofactor in essential processes such as photosynthesis, lipid biosynthesis and oxidative stress. Mn deficient plants exhibit decreased growth and yield and are more susceptible to pathogens and damage at freezing temperatures. Mn deficiency is most prominent on alkaline soils with approximately one third of the world's soils being too alkaline for optimal crop production. Despite the importance of Mn in plant development, relatively little is known about how it traffics between plant tissues and into and out of organelles. Several gene transporter families have been implicated in Mn transport in plants. These transporter families include NRAMP (natural resistance associated macrophage protein), YSL (yellow stripe-like), ZIP (zinc regulated transporter/iron-regulated transporter [ZRT/IRT1]-related protein), CAX (cation exchanger), CCX (calcium cation exchangers), CDF/MTP (cation diffusion facilitator/metal tolerance protein), P-type ATPases and VIT (vacuolar iron transporter). A combination of techniques including mutant analysis and Synchrotron X-ray Fluorescence Spectroscopy can assist in identifying essential transporters of Mn. Such knowledge would vastly improve our understanding of plant Mn homeostasis.
DOI:10.1038/sj.ijir.3900333URLPMID:9727382 [本文引用: 1]

Accumulating evidence suggests that angiotensin-(1-7) is an important component of the renin-angiotensin system, having actions that are either identical to or opposite that of angiotensin II. Angiotensin I can be directly converted to angiotensin-(1-7), bypassing formation of angiotensin II. This pathway is under the control of three enzymes: neutral endopeptidases 24.11 (neprilysin) and 24.15 and prolyl-endopeptidase 24.26. Two of the three angiotensin-forming enzymes (neprilysin and endopeptidase 24.15) also contribute to the breakdown of bradykinin and the atrial natriuretic peptide. Furthermore, angiotensin-(1-7) is a major substrate for angiotensin-converting enzyme. These observations suggest that the process of biotransformation between the various Ang peptides of the renin-angiotensin system and other vasodepressor peptides are intertwined through this enzymatic pathway. Substantial evidence suggests that angiotensin-(1-7) stimulates the synthesis and release of vasodilator prostaglandins, and nitric oxide, while also augmenting the metabolic actions of bradykinin. In addition, angiotensin-(1-7) alters tubular sodium and bicarbonate reabsorption, decreases Na+-K+-ATPase activity, induces diuresis, and exerts a vasodilator effect. These physiologic effects of angiotensin-(1-7) favor a blood pressure-lowering effect. The majority of the data currently available suggest that an,angiotensin-(1-7) mediates its effects through a novel non-AT(1)/AT(2) receptor subtype.
DOI:10.1016/j.spen.2011.10.004URLPMID:22172418 [本文引用: 1]

Nemaline myopathy constitutes a continuous spectrum of primary skeletal muscle disorders named after the Greek word for thread, nema. The diagnosis is based on muscle weakness, combined with visualization of nemaline bodies on muscle biopsy. The patients' muscle weakness is usually generalized, but there may be a selective pattern of more pronounced weakness, and, most importantly, respiratory muscles may be especially weak. Histologically, additional features may coexist with the nemaline bodies. There are 7 known causative genes. The function of the most recently identified gene is unknown, but the other 6 encoded proteins are associated with the muscle thin filament. The 2 most common causes of nemaline myopathy are recessive mutations in nebulin and de novo dominant mutations in skeletal muscle -actin. At least 1 further gene remains to be identified. Patient care is based on managing the clinical symptoms. Animal models are helping to gain insight into pathogenesis, and a variety of therapeutic approaches are being investigated.
DOI:10.1074/jbc.272.21.13548URLPMID:9153201 [本文引用: 1]

The cellular protein E6AP functions as an E3 ubiquitin protein ligase in the E6-dependent ubiquitination of p53. E6AP is a member of a family of functionally related E3 proteins that share a conserved carboxyl-terminal region called the Hect domain. Although several different E2 ubiquitin-conjugating enzymes have been shown to function with E6AP in the E6-dependent ubiquitination of p53 , the E2s that cooperate with E6AP in the ubiquitination of its normal substrates are presently unknown. Moreover, the basis of functional cooperativity between specific E2 and Hect E3 proteins has not yet been determined.
DOI:10.1124/pr.114.009647URLPMID:25713288 [本文引用: 1]

Abstract The Adhesion family forms a large branch of the pharmacologically important superfamily of G protein-coupled receptors (GPCRs). As Adhesion GPCRs increasingly receive attention from a wide spectrum of biomedical fields, the Adhesion GPCR Consortium, together with the International Union of Basic and Clinical Pharmacology Committee on Receptor Nomenclature and Drug Classification, proposes a unified nomenclature for Adhesion GPCRs. The new names have ADGR as common dominator followed by a letter and a number to denote each subfamily and subtype, respectively. The new names, with old and alternative names within parentheses, are: ADGRA1 (GPR123), ADGRA2 (GPR124), ADGRA3 (GPR125), ADGRB1 (BAI1), ADGRB2 (BAI2), ADGRB3 (BAI3), ADGRC1 (CELSR1), ADGRC2 (CELSR2), ADGRC3 (CELSR3), ADGRD1 (GPR133), ADGRD2 (GPR144), ADGRE1 (EMR1, F4/80), ADGRE2 (EMR2), ADGRE3 (EMR3), ADGRE4 (EMR4), ADGRE5 (CD97), ADGRF1 (GPR110), ADGRF2 (GPR111), ADGRF3 (GPR113), ADGRF4 (GPR115), ADGRF5 (GPR116, Ig-Hepta), ADGRG1 (GPR56), ADGRG2 (GPR64, HE6), ADGRG3 (GPR97), ADGRG4 (GPR112), ADGRG5 (GPR114), ADGRG6 (GPR126), ADGRG7 (GPR128), ADGRL1 (latrophilin-1, CIRL-1, CL1), ADGRL2 (latrophilin-2, CIRL-2, CL2), ADGRL3 (latrophilin-3, CIRL-3, CL3), ADGRL4 (ELTD1, ETL), and ADGRV1 (VLGR1, GPR98). This review covers all major biologic aspects of Adhesion GPCRs, including evolutionary origins, interaction partners, signaling, expression, physiologic functions, and therapeutic potential. Copyright 2015 by The American Society for Pharmacology and Experimental Therapeutics.
[本文引用: 1]
DOI:10.1016/j.bjm.2017.05.008URLPMID:29132828 [本文引用: 1]

Abstract The evolution of microorganisms resistant to many medicines has become a major challenge for the scientific community around the world. Motivated by the gravity of such a situation, the World Health Organization released a report in 2014 with the aim of providing updated information on this critical scenario. Among the most worrying microorganisms, species from the genus Candida have exhibited a high rate of resistance to antifungal drugs. Therefore, the objective of this review is to show that the use of natural products (extracts or isolated biomolecules), along with conventional antifungal therapy, can be a very promising strategy to overcome microbial multiresistance. Some promising alternatives are essential oils of Melaleuca alternifolia (mainly composed of terpinen-4-ol, a type of monoterpene), lactoferrin (a peptide isolated from milk) and chitosan (a copolymer from chitin). Such products have great potential to increase antifungal therapy efficacy, mitigate side effects and provide a wide range of action in antifungal therapy. Copyright 2017 Sociedade Brasileira de Microbiologia. Published by Elsevier Editora Ltda. All rights reserved.
DOI:10.1158/1541-7786.MCR-17-0120URL [本文引用: 1]

Glioblastoma multiforme (GBM) is the most common primary malignant brain tumor and accounts for a significant proportion of all primary brain tumors. Median survival after treatment is around 15 months. Remodeling of N-glycans by the N-acetylglu- cosamine glycosyltransferase (MGAT5) regulates tumoral devel- opment. Here, perturbation of MGAT5 enzymatic activity by the small-molecule inhibitor 3-hydroxy-4,5-bis-benzyloxy-6-benzy- loxymethyl-2-phenyl2-oxo-2l5-[1,2]oxaphosphinane (PST3.1a) restrains GBM growth. In cell-based assays, it is demonstrated that PST3.1a alters the b1,6-GlcNAc N-glycans of GBM-initiating cells (GIC) by inhibiting MGAT5 enzymatic activity, resulting in the inhibition of TGFbR and FAK signaling associated with double- cortin (DCX) upregulation and increase oligodendrocyte lineage transcription factor 2 (OLIG2) expression. PST3.1a thus affects
DOI:10.1111/j.1742-4658.2009.06893.xURLPMID:19220457 [本文引用: 1]

http://doi.wiley.com/10.1111/j.1742-4658.2009.06893.x
DOI:10.1074/jbc.M114.547448URLPMID:24652292 [本文引用: 1]

Abstract The human genome encodes 53 members of the solute carrier family 25 (SLC25), also called the mitochondrial carrier family, many of which have been shown to transport carboxylates, amino acids, nucleotides, and cofactors across the inner mitochondrial membrane, thereby connecting cytosolic and matrix functions. In this work, a member of this family, SLC25A29, previously reported to be a mitochondrial carnitine/acylcarnitine- or ornithine-like carrier, has been thoroughly characterized biochemically. The SLC25A29 gene was overexpressed in Escherichia coli, and the gene product was purified and reconstituted in phospholipid vesicles. Its transport properties and kinetic parameters demonstrate that SLC25A29 transports arginine, lysine, homoarginine, methylarginine and, to a much lesser extent, ornithine and histidine. Carnitine and acylcarnitines were not transported by SLC25A29. This carrier catalyzed substantial uniport besides a counter-exchange transport, exhibited a high transport affinity for arginine and lysine, and was saturable and inhibited by mercurial compounds and other inhibitors of mitochondrial carriers to various degrees. The main physiological role of SLC25A29 is to import basic amino acids into mitochondria for mitochondrial protein synthesis and amino acid degradation.
