 , 王亚琴
, 王亚琴
Expression Pattern and Protein Localization of a Yellow-Green Leaf 6 (YGL6) Gene in Rice (Oryza sativa)
SHIJun-Qiong , WANGYa-Qin
, WANGYa-Qin
通讯作者:
收稿日期:2017-09-23
接受日期:2018-01-8
网络出版日期:2018-01-29
版权声明:2018作物学报编辑部作物学报编辑部
基金资助:
作者简介:
-->
展开
摘要
关键词:
Abstract
Keywords:
-->0
PDF (1882KB)元数据多维度评价相关文章
本文引用格式导出EndNoteRisBibtex收藏本文-->
水稻叶色突变是比较常见的, 突变基因往往直接或间接影响叶绿素的合成和降解, 从而影响水稻光合作用, 造成植株减产甚至死亡。近年来, 大量研究结果显示, 叶色突变体是植物光合作用机制、叶绿素生物合成途径、叶绿体的结构功能和遗传发育调控机理、作物标记性状等研究的理想材料[1,2,3]。
水稻叶绿素缺乏突变类型丰富, 目前已鉴定突变体90多个, 除第12染色体外, 其他染色体均发现了叶绿素缺乏突变基因(http://www.shigen.nig.ac. jp/rice/oryzabaseV4/)。虽然其大部分已被定位, 但仅少数被克隆, 它们分别参与叶绿素的生物合成、降解和叶绿体的发育[4,5-12]。叶色突变的机制复杂, 除叶绿素代谢和叶绿体发育引起外, 还有质-核信号转导途径受阻[13]、血红素的反馈调节紊乱[14]、光敏色素调控受阻[15]、叶绿体蛋白转运受阻[16]、ATP转运蛋白基因[17]等其他突变机制。随着新的水稻叶色突变体的发现, 叶色突变相关基因的分子机理研究将更加透彻。
叶绿体RNA结合蛋白是一类定位于叶绿体, 参与叶绿体RNA代谢的由核基因编码的蛋白。叶绿体RNA结合蛋白与细胞核或细胞质中的RNA结合蛋白具有类似的功能, 包括参与RNA的稳定和RNA转录后的加工过程等。此外, 叶绿体RNA结合蛋白对细胞核与叶绿体之间的信息传递和功能协调起重要作用。本研究利用EMS诱变恢复系缙恢10号, 从其后代中获得一份稳定遗传的黄绿叶突变体ygl6, 该突变体叶片在苗期呈黄绿色, 分蘖后期至成熟期转为淡绿色。图位克隆及测序结果表明Os12g23180为候选基因, 遗传互补实验证实黄绿叶基因YGL6为Os12g23180。根据gramene网站(http://www. gramene.org/)提供的信息, YGL6蛋白属于RNA结合蛋白类, 利用qRT-PCR分析表明YGL6基因仅在绿色组织如心叶、成熟叶、叶鞘和绿色颖壳中表达, 并受光照条件的诱导。构建YGL6与GFP的融合表达载体转化水稻原生质体发现YGL6与GFP的融合蛋白定位于叶绿体, 因此YGL6极有可能是叶绿体中的RNA结合蛋白, 并参与了水稻叶绿体的发育过程。本研究初步揭示了YGL6的基因功能, 为进一步研究奠定了基础, 并丰富了水稻突变体库, 深化了当前对叶绿体发育过程的研究。
1 材料与方法
1.1 实验材料
黄绿叶突变体ygl6由EMS诱变缙恢10号 (Jinhui 10)所得, 经多代培养已遗传稳定。缙恢10号是西南大学水稻研究所选育的优良籼稻恢复系, 以其作为野生型对照。1.2 基因组互补载体的设计、构建与转化
由于pCAMBIA1301上的单酶切位点在YGL6基因的基因组DNA序列均存在, 并且扩增片段较长, 则采用重组酶的方法进行。引物6HBF1和6HBR2分别与载体pCAMBIA1301 BamH I和Hind III酶切后的片段有15 bp的重叠; 引物6HBR1和6HBF2有15 bp的重叠用于连接前后2个扩增片段的重叠片段。引物6HBF1、6HBR2、6HBF2、6HBR1的序列分别为5°-cggtacccggggatcAACATGGTCAGGCACA GTGATGAGAT-3°、5°-AGATAGGAGATGGTAGCAG GAGGCTG-3°、5°-TACCATCTCCTATCTCTGACTTC AGTAGTGC-3°和5°-GGCCAGTGCCAAGCTGGTC AAGCTCCCTGGTGAGGCTAT-3°。载体参照Shi等[18]的方法构建与转化。1.3 YGL6蛋白的进化分析
参照Ren等[19]的方法分析YGL6蛋白的进化。1.4 基因表达分析
参照Shi等[18]的方法进行RNA的提取和纯化、cDNA的合成和qRT-PCR的表达分析。1.5 光合色素含量的测定
以野生型、突变体和互补植株为材料, 上午9:00在种植小区中间随机选择长势相对一致的5个单株, 测定叶片中部的光合色素含量。称取0.1 g叶片剪碎装入离心管, 加25 mL丙酮∶无水乙醇(1∶1, v/v) 混合液封口暗处理24~48 h, 其间经常摇动, 每个样品重复3次。用分光光度计测定470 nm、645 nm和663 nm的光吸收值, 参考Lichtenthaler[20]的方法计算。1.6 亚细胞定位
选择载体pTCK303为载体骨架, 其上的Ubiquitin为启动子, BamH I和Sac I为双酶切位点。设计引物扩增YGL6基因的整个CDS片段, 前引物ATG前加入来自pTCK303 BamH I酶切后Ubiquitin侧15 bp, 后引物不含终止密码子。再设计1对引物扩增GFP基因的整个CDS片段, 前引物与YGL6的后引物有15 bp的重叠, 后引物加入来自pTCK303 Sac I酶切后NOS侧15 bp碱基。引物6OEPF1、6OEPR1、6GFPF1和6GFPR1的序列分别为5°-tacttc tgcaggatcATGGCAGCAACAGCCTCCCT-3°、5°-tcacc atgacgctgaCGAGCTTCTTGCC-3°、5°-tcagcgtcatggtga GCAAGGGCGAGGA-3°和5°-CAAATGTTTGAAC GGTTACTTGTACAGCTCGTCCATGCCG-3°。参照Ren等[19]转水稻原生质体的亚细胞定位方法。2 结果与分析
2.1 YGL6基因组互补验证
为了验证通过图位克隆筛选出的候选基因Os12g23180就是YGL6基因[18], 构建了基因组互补载体, 将野生型中Os12g23180基因组片段包括上游3500 bp、编码框和下游1500 bp序列利用重组酶整合到pCAMBIA1301表达载体中, 然后通过农杆菌介导法遗传转化突变体ygl6。在T0获得的转基因植株中, 共观察到12株转基因植株的突变体表型得以恢复(图1-A, B)。同时通过提取DNA后测序比对发现, 12株转基因植株在突变位点均出现了双峰, 证实互补载体成功转入突变体, 使其表型得以恢复正常(图1-C)。分析表明恢复表型的转基因植株的叶绿素a、叶绿素b和类胡萝卜素含量均达到了野生型的色素含量水平(图1-D)。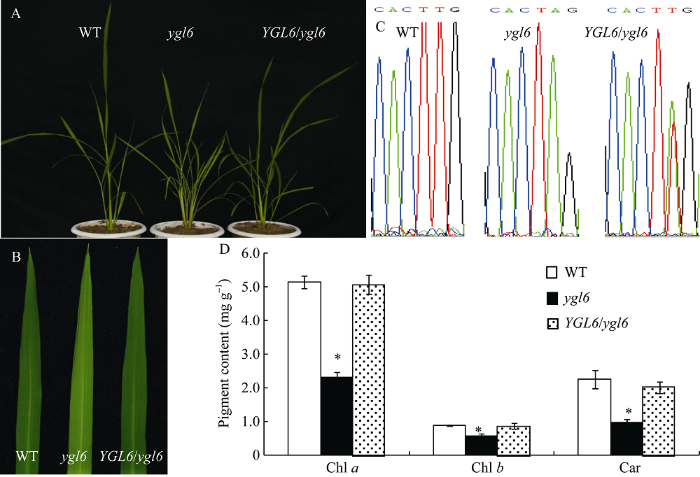 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图1YGL6基因的互补验证
A: WT、ygl6和转基因互补植株; B: WT、ygl6和转基因互补植株的叶片; C: WT、ygl6和转基因互补植株突变位点的测序结果; D: WT、ygl6和转基因互补植株的色素含量。*表示在0.05水平上差异显著。
-->Fig. 1Functional complementation analysis of YGL6
A: phenotypes of WT, ygl6 mutant, and transgenic plant (ygl6/YGL6); B: leaves of WT, ygl6 mutant, and transgenic plant; C: the YGL6 sequencing of WT, ygl6 mutant, and transgenic plant; D: pigment contents of WT, ygl6 mutant, and transgenic plant. *P﹤0.05.
-->
2.2 YGL6蛋白质功能注释与进化分析
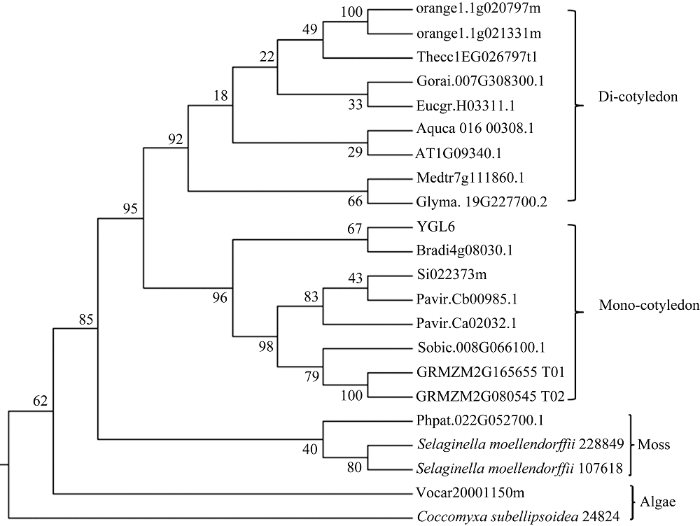
通过比对水稻基因组序列, 在其他位置未发现与YGL6基因高度相似的序列, 表明YGL6基因是单拷贝基因。利用NCBI网站对YGL6蛋白质BLASTp分析表明, YGL6基因编码NAD(P)-结合的Rossmann折叠超家族蛋白质, 属于短链脱氢酶/还原酶家族, 推断为异黄酮还原酶、糖脱水酶和mRNA结合蛋白等。YGL6蛋白质具有多个保守结构域, 如SDR-a1结构域、NAD(P)-结合位点、UDP-葡萄糖-4-异构酶C末端亚基、短链脱氢酶、RNA结合蛋白、NAD依赖的异构酶家族等。YGL6蛋白在网站gramene (http://www.gramene. org/)预测是一个RNA结合蛋白, 生物信息学分析该蛋白质定位于叶绿体, 系统进化树分析表明其与其他物种的叶绿体茎环结合蛋白聚类在一起。从系统进化中可以看出, 该基因属于高保守低拷贝, 从藻类、苔藓到高等植物均含有此蛋白, 并且藻类、苔藓、单子叶和双子叶植物各自组成一个分支(图2)。
 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图2YGL6基因的进化分析
-->Fig. 2Phylogenetic analysis of the YGL6
-->
2.3 YGL6基因的表达分析
通过qRT-PCR检测, YGL6基因在苗期除根外, 心叶、成熟叶和叶鞘中均有表达, 尤以心叶的表达量最高, 达到成熟叶的6倍多(图3-A); 在孕穗期, 仅在叶片和叶鞘中有表达, 在心叶的表达量最高; 在抽穗期的绿色颖壳中也有表达(图3-B)。以上结果表明YGL6基因在整个生育期的绿色部位表达, 尤以心叶的表达量最高。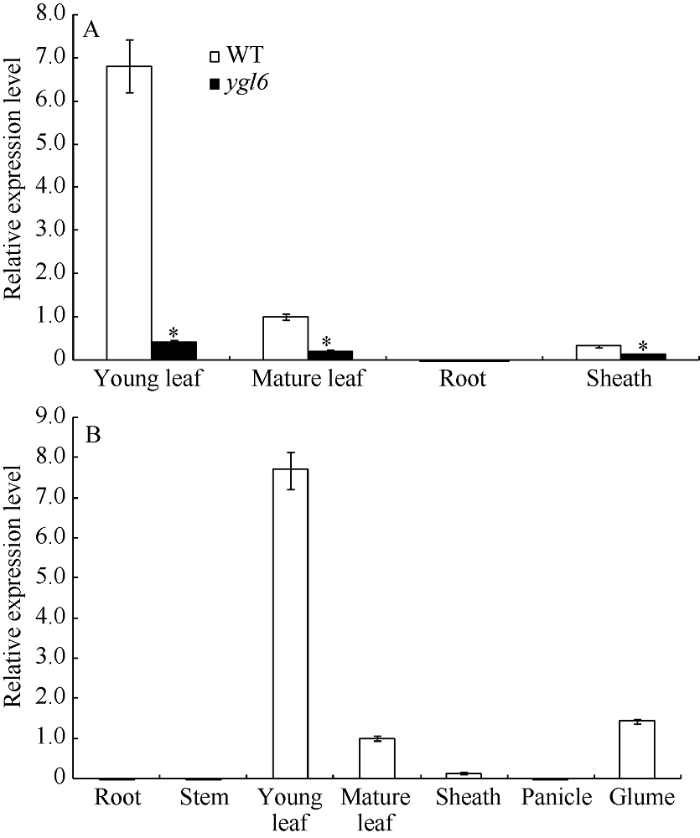 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图3YGL6基因表达分析
A: 苗期YGL6基因在WT和ygl6的表达分析; B: 孕穗期YGL6基因在WT的表达分析。*表示在0.05水平上差异显著。
-->Fig. 3Expression analysis of YGL6 by qRT-PCR
A: expression level of YGL6 at seedling stage in WT and the YGL6 mutant; B: expression level of YGL6 at booting stage in WT. *P﹤0.05.
-->
2.4 YGL6基因表达受光照的影响
对连续暗培养14 d的水稻幼苗光照处理表明, 随着光照时间的延长, YGL6基因的表达量不断上升(图4-A)。对连续光培养14 d的水稻幼苗进行黑暗处理表明暗培养0.5 h表达量下降, 但随后表达量逐渐上升, 在4 h时达到最大表达量, 随后逐渐下降, 在24 h时降到最低表达量(图4-B)。以上结果表明YGL6基因的表达受光照的诱导。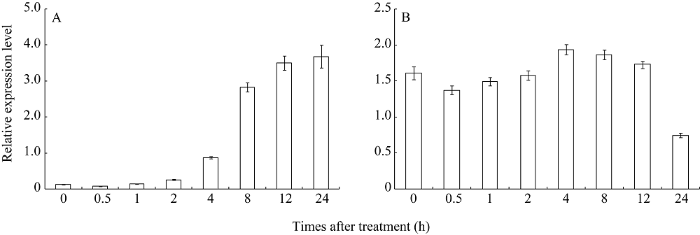 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图4光照对YGL6基因表达的分析
A: 连续黑暗培养后在不同光照时间YGL6基因的表达; B: 连续光照培养后在不同黑暗时间YGL6基因的表达。
-->Fig. 4qRT-PCR analysis of the expression level of YGL6
A: expression level of YGL6 in wild-type plants at different hours transferred to continuous light from continuous darkness; B: expression level of YGL6 in wild-type plants at different hours transferred to continuous darkness from continuous light.
-->
2.5 YGL6蛋白的亚细胞定位
亚细胞定位分析网站(http://www.cbs.dtu.dk/ services/SignalP/)预测YGL6蛋白定位于叶绿体, 为了验证YGL6蛋白是否定位于叶绿体, 构建了35S-YGL6ORF-GFP和35S-GFP融合瞬时表达载体。将瞬时表达载体转化水稻原生质体, 用绿色荧光蛋白GFP做指示。结果表明, 在转35S-GFP原生质体, 除液泡外绿色荧光信号在整个细胞中均能观察到(图5-A~D); 而在转35S-YGL6ORF-GFP的原生质体中, GFP的绿色荧光信号(图5-E)和叶绿体自发的红色荧光信号(图5-F)重叠为黄色(图5-H), 说明YGL6蛋白定位在叶绿体。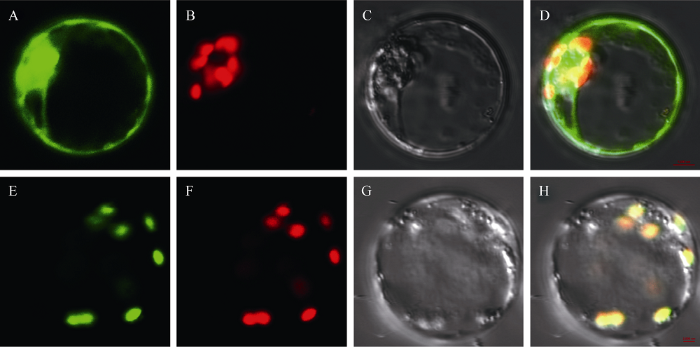 显示原图|下载原图ZIP|生成PPT
显示原图|下载原图ZIP|生成PPT图5YGL6蛋白的亚细胞定位
A~D: 阴性对照(35S-GFP); E~H: YGL6蛋白定位(35S-YGL6ORF-GFP)。
-->Fig. 5Subcellular location of YGL6 protein using rice protoplasts
A-D: negative control (35S-GFP); E-H: subcellular location of YGL6 protein (35S-YGL6ORF-GFP).
-->
3 讨论
在众多被鉴定的叶色突变体中, 仅有少数基因被克隆。叶绿素生物合成和叶绿体发育是一个相当复杂的过程, 目前已克隆的水稻叶色突变基因直接或间接影响叶绿素代谢或者叶绿体的发育。直接参与编码叶绿素合成与降解的酶基因有编码镁离子螯合酶H、D和I亚基的OsCHLH[7]、OsCHLD和OsCHLI[11]基因, 叶绿素酸酯氧化酶基因OsCAO1、OsCAO2[8], 叶绿素合成酶基因ygl1[12]。调控叶绿体发育的基因有鸟苷酸激酶基因virescent-2[9,10], 三角状五肽重复蛋白基因OsPPR1[21], 叶绿体RNA聚合酶基因OsRpoTp[22]等。此外已成功克隆出的叶色相关基因还有持绿突变体基因SGR (stay green rice)[23], 叶绿素b还原酶基因NYC1 (non-yellow coloring1)[24], 叶绿素b还原酶的同源基因NYC1-LIKE[25]等。通过图位克隆和基因互补实验表明YGL6基因是一个位于第12染色体的SDR超家族基因, 虽然目前已发现的叶色突变相关基因有90多个, 但在第12染色体鲜有叶色突变的报道, Shi等首次报道了ygl6突变体[18], 本研究证实了YGL6是一个调控叶色突变的基因。最近有研究表明, 叶绿体有不同的RNA池, 其中有些RNA被翻译, 而其他的并不被翻译[26]。细菌RNA转录后即被翻译, 剪切、RNA编辑、内含子剪接等转录后加工在细菌中较少存在[27]。然而, 叶绿体的初级RNA产物均需要不同形式的转录后加工[28]。细菌的多顺反子直接进行翻译, 而叶绿体的多顺反子需要进一步剪切产生更小的多顺反子或单顺反子RNA, 有些还需要进一步的剪切或编辑产生成熟的RNA[28,29]。多数质体RNA的成熟需要低特异性的核酸酶, 加工的程度通常由RNA结合蛋白和RNA的二级结构决定[28,29]。叶绿体RNA代谢需要数量众多的RNA结合蛋白的参与, YGL6具有mRNA结合结构域, 亚细胞定位结果显示YGL6蛋白定位于叶绿体。综合上述结果可以确定YGL6基因是一个新的调控水稻叶绿体RNA代谢的基因, 有助于更进一步理解和完善水稻叶色突变的机制。
利用NCBI网站(https://www.ncbi.nlm.nih.gov/)对YGL6蛋白质BLAST分析表明, YGL6基因编码NAD(P)-结合的Rossmann折叠超家族蛋白质, 属于一个短链脱氢酶/还原酶家族 (short-chain dehydrogenase/reductase, SDR), 多数被鉴定的SDR被推断为异黄酮还原酶、糖脱水酶和mRNA结合蛋白等。Baker等研究表明菠菜一个mRNA结合蛋白CSP41和核苷-糖异构酶及羟类固醇脱氢酶同源[30]。CSP41是一类分子量为41 kD的叶绿体茎环结合蛋白, 目前已对多个物种的CSP41进行了克隆和功能分析。最早报道的CSP41a在体外可以结合到petD mRNA的3°末端, 具有序列特异的茎环结构结合能力和核酸内切酶活性[31]。随后研究表明, CSP41蛋白也在质体编码的RNA PEP (plastid-encoded polymerase)或者质体核糖体中存在, 可能参与RNA的转录或翻译过程[32,33]。但最近的研究却不能证实CSP41是PEP复合物的一部分[34]。研究表明CSP41缺乏已知的核酸内切酶基序, 但属于SDR超家族, 实验表明SDR基序可能是新类型的核酸内切酶基序[35]。CSP41的蛋白功能具有多样性, 并且已报道的CSP41蛋白均来自其他物种, 在水稻中尚无报道。水稻究竟行使以上的哪种或哪几种功能, 还是其他的未知功能, 以及YGL6蛋白质是否为CSP蛋白, 需要进一步的相关实验探究。
4 结论
黄绿叶基因YGL6为Os12g23180, 与其他物种的叶绿体茎环结合蛋白聚类在一起。YGL6基因仅在绿色组织如心叶、成熟叶、叶鞘和绿色颖壳中表达, 尤其以心叶的表达量最高, 其表达还受光照的诱导。YGL6蛋白被定位于叶绿体。我们认为YGL6通过蛋白产物的RNA结合功能参与叶绿体的发育过程。本研究为进一步研究YGL6的基因功能奠定了基础, 并丰富了水稻突变体库, 深化了当前对叶绿体的发育过程的研究。The authors have declared that no competing interests exist.
作者已声明无竞争性利益关系。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
| [1] | . Photosystem I (PS I) is a large membrane protein complex that catalyzes the first step of solar conversion, the light-induced transmembrane electron transfer, and generates reductants for CO 2 assimilation. It consists of 12 different proteins and 127 cofactors that perform light capturing and electron transfer. The function of PS I includes inter-protein electron transfer between PS I and smaller soluble electron transfer proteins. The structure of PS I is discussed with respect to the potential docking sites for the soluble electron acceptors, ferredoxin/flavodoxin, at the stromal side and the soluble electron donors, cytochrome c 6/plastocyanin, at the luminal side of the PS I complex. Furthermore, the potential interaction sites with the peripheral antenna proteins are discussed. |
| [2] | . Nuclear genes control plastid differentiation in response to developmental signals, environmental signals, and retrograde signals from plastids themselves. In return, plastids emit signals that are essential for proper expression of many nuclear photosynthetic genes. Accumulation of magnesium-protoporphyrin IX (Mg-Proto), an intermediate in chlorophyll biosynthesis, is a plastid signal that represses nuclear transcription through a signaling pathway that, in Arabidopsis, requires the GUN4 gene. GUN4 binds the product and substrate of Mg-chelatase, an enzyme that produces Mg-Proto, and activates Mg-chelatase. Thus, GUN4 participates in plastid-to-nucleus signaling by regulating Mg-Proto synthesis or trafficking. |
| [3] | . |
| [4] | . |
| [5] | . Differentiation of proplastids into functionally active chloroplasts is one of the most significant changes in cellular organization associated with leaf development in higher plants. This process involves activation of a large number of nuclear and chloroplast genes. A central question, therefore, concerns the nature and origin of the signals that initiate and control this process. The rice nuclear mutant, virescent-1 ( v 1 ), is temperature-conditional and develops chlorotic leaves when grown at restrictive temperatures. We report here the effects of v 1 mutation on the expressions of plastid and nuclear genes during leaf development. In the wild-type rice seedlings, the transcripts of the plastid RNA polymerase gene ( rpoB ) and ribosomal protein genes ( rps7, rps15 ) accumulated during a strictly limited period of early leaf development, prior to the accumulation of the transcripts of photosynthetic genes ( rbcL, RbcS, psbA, Lhc ). This period coincides very closely with the leaf developmental stage (late P4) at which the V 1 gene gives the signal that determines the virescent phenotype. On the contrary, in the v 1 seedlings grown at a restrictive temperature (20 C), this stage-specific accumulation of the rpo and rps transcripts was missing. Instead, the accumulation of these transcripts occurred during a later stage of leaf maturation. In such mutant seedlings, the expression of other plastid genes ( psbA, rbcL , 16S rDNA ) was strongly suppressed and the normal chloroplast development was disturbed. These data indicate that the V 1 gene controls the timing of expression of the key plastid genes for the transcription/ translation apparatus that are essential for the subsequent activation of other plastid genes. |
| [6] | . Summary During early chloroplast differentiation, the regulation of the plastid genetic system including transcription and translation differs greatly from that in the mature chloroplast, suggesting the existence of a stage-dependent mechanism that regulates the chloroplast genetic system during this period. The virescent-1 ( v 1 ) mutant of rice ( Oryza sativa ) is temperature-conditional and develops chlorotic leaves under low-temperature conditions. We reported previously that leaf chlorosis in the v 1 mutant is caused by blockage of the activation of the chloroplast genetic system during early leaf development. Here we identify the V 1 gene, which encodes a chloroplast-localized protein NUS1. Accumulation of NUS1 specifically occurred in the pre-emerged immature leaves, and is enhanced by low-temperature treatment. The C-terminus of NUS1 shows structural similarity to the bacterial antitermination factor NusB, which is known to play roles in the regulation of ribosomal RNA transcription. The RNA-immunoprecipitation and gel mobility shift assays indicated that NUS1 binds to several regions of chloroplast RNA including the upstream leader region of the 16S rRNA precursor. In the leaves of the NUS1-deficient mutant, accumulation of chloroplast rRNA during early leaf development was impaired and chloroplast translation/transcription capacity was severely suppressed under low temperature. Our results suggest that NUS1 is involved in the regulation of chloroplast RNA metabolism and promotes the establishment of the plastid genetic system during early chloroplast development under cold stress conditions. |
| [7] | . Abstract We have previously generated a large pool of T-DNA insertional lines in rice. In this study, we screened those T-DNA pools for rice mutants that had defective chlorophylls. Among the 1,995 lines examined in the T2 generation, 189 showed a chlorophyll-deficient phenotype that segregated as a single recessive locus. Among the mutants, 10 lines were beta-glucuronidase (GUS)-positive in the leaves. Line 9-07117 has a T-DNA insertion into the gene that is highly homologous to XANTHA-F in barley and CHLH in Arabidopsis: This OsCHLH gene encodes the largest subunit of the rice Mg-chelatase, a key enzyme in the chlorophyll branch of the tetrapyrrole biosynthetic pathway. In the T2 and T3 generations, the chlorina mutant phenotypes are co-segregated with the T-DNA. We have identified two additional chlorina mutants that have a Tos17 insertion in the OsCHLH gene. Those phenotypes were co-segregated with Tos17 in the progeny. GUS assays and RNA blot analysis showed that expression of the OsCHLH gene is light inducible, while TEM analysis revealed that the thylakoid membrane of the mutant chloroplasts is underdeveloped. The chlorophyll content was very low in the OschlH mutants. This is the first report that T-DNA insertional mutagenesis can be used for functional analysis of rice genes. |
| [8] | . |
| [9] | . The rice virescent-2 mutant (ν |
| [10] | . Summary Guanylate kinase (GK) is a critical enzyme in guanine nucleotide metabolism pathways, catalyzing the phosphorylation of (d)GMP to (d)GDP. Here we show that a novel gene, VIRESCENT 2 ( V2 ), encodes a new type of GK (designated pt/mtGK) that is localized in plastids and mitochondria. We initially identified the V2 gene by positional cloning of the rice v2 mutant. The v2 mutant is temperature-sensitive and develops chlorotic leaves at restrictive temperatures. The v2 mutation causes inhibition of chloroplast differentiation; in particular, it disrupts the chloroplast translation machinery during early leaf development [ Sugimoto etal. (2004) Plant Cell Physiol. 45, 985]. In the bacterial and animal species studied to date, GK is localized in the cytoplasm and participates in maintenance of the guanine nucleotide pools required for many fundamental cellular processes. Phenotypic analysis of rice seedlings with RNAi knockdown of cytosolic GK (designated cGK) showed that cGK is indispensable for the growth and development of plants, but not for chloroplast development. Thus, rice has two types of GK, as does Arabidopsis, suggesting that higher plants have two types of GK. Our results suggest that, of the two types of GK, only pt/mtGK is essential for chloroplast differentiation. |
| [11] | . |
| [12] | . Chlorophyll (Chi) synthase catalyzes esterification of chlorophyllide to complete the last step of Chl biosynthesis. Although the Chl synthases and the corresponding genes from various organisms have been well characterized, Chl synthase mutants have not yet been reported in higher plants. In this study, a rice (Oryza Sativa) Chl-deficient mutant, yellow-green leaf1 (ygl1), was isolated, which showed yellow-green leaves in young plants with decreased Chl synthesis, increased level of tetrapyrrole intermediates, and delayed chloroplast development. Genetic analysis demonstrated that the phenotype of ygl1 was caused by a recessive mutation in a nuclear gene. The ygl1 locus was mapped to chromosome 5 and isolated by map-based cloning. Sequence analysis revealed that it encodes the Chl synthase and its identity was verified by transgenic complementation. A missense mutation was found in a highly conserved residue of YGL1 in the ygl1 mutant, resulting in reduction of the enzymatic activity. YGL1 is constitutively expressed in all tissues, and its expression is not significantly affected in the ygl1 mutant. Interestingly, the mRNA expression of the cab1R gene encoding the Chl a/b-binding protein was severely suppressed in the ygl1 mutant. Moreover, the expression of some nuclear genes associated with Chl biosynthesis or chloroplast development was also affected in ygl1 seedlings. These results indicate that the expression of nuclear genes encoding various chloroplast proteins might be feedback regulated by the level of Chl or Chl precursors. |
| [13] | . |
| [14] | . Abstract The aurea (au) and yellow-green-2 (yg-2) mutants of tomato (Solanum lycopersicum L.) are unable to synthesize the linear tetrapyrrole chromophore of phytochrome, resulting in plants with a yellow-green phenotype. To understand the basis of this phenotype, we investigated the consequences of the au and yg-2 mutations on tetrapyrrole metabolism. Dark-grown seedlings of both mutants have reduced levels of protochlorophyllide (Pchlide) due to an inhibition of Pchlide synthesis. Feeding experiments with the tetrapyrrole precursor 5-aminolevulinic acid (ALA) demonstrate that the pathway between ALA and Pchlide is intact in au and yg-2 and suggest that the reduction in Pchlide is a result of the inhibition of ALA synthesis. This inhibition was independent of any deficiency in seed phytochrome, and experiments using an iron chelator to block heme synthesis demonstrated that both mutations inhibited the degradation of the physiologically active heme pool, suggesting that the reduction in Pchlide synthesis is a consequence of feedback inhibition by heme. We discuss the significance of these results in understanding the chlorophyll-deficient phenotype of the au and yg-2 mutants. |
| [15] | . Abstract Photosynthetic organisms must achieve a delicate balance between the light energy absorbed by chlorophyll and their capacity to channel that energy into productive photochemical reactions. Release of excess absorbed energy in the cell can cause lethal photooxidative damage. We identified a basic helix-loop-helix (bHLH) transcription factor, designated PHYTOCHROME-INTERACTING FACTOR 1 (PIF1), that negatively regulates chlorophyll biosynthesis. pif1 mutant seedlings accumulate excess free protochlorophyllide when grown in the dark, with consequent lethal bleaching upon exposure to light. PIF1 interacts specifically with the photoactivated conformer of phytochromes A and B, suggesting a signaling pathway by which chlorophyll biosynthetic rates are tightly controlled during the critical initial emergence of seedlings from subterranean darkness into sunlight. |
| [16] | . NADPH:protochlorophyllide oxidoreductase (POR) A is a key enzyme of chlorophyll biosynthesis in angiosperms. It is nucleus-encoded, synthesized as a larger precursor in the cytosol and imported into the plastids in a substrate-dependent manner. Plastid envelope membrane proteins, called protochlorophyllide-dependent translocon proteins, Ptcs, have been identified that interact with pPORA during import. Among them are a 16-kDa ortholog of the previously characterized outer envelope protein Oep16 (named Ptc16) and a 33-kDa protein (Ptc33) related to the GTP-binding proteins Toc33 and Toc34 of Arabidopsis. In the present work, we studied the interactions and roles of Ptc16 and Ptc33 during pPORA import. Radiolabeled Ptc16/Oep16 was synthesized from a corresponding cDNA and imported into isolated Arabidopsis plastids. Crosslinking experiments revealed that import of 35 S-Oep16/Ptc16 is stimulated by GTP. 35 S-Oep16/Ptc16 forms larger complexes with Toc33 but not Toc34. Plastids of the ppi1 mutant of Arabidopsis lacking Toc33, were unable to import pPORA in darkness but imported the small subunit precursor of ribulose-1,5-bisphosphate carboxylase/oxygenase (pSSU), precursor ferredoxin (pFd) as well as pPORB which is a close relative of pPORA. In white light, partial suppressions of pSSU, pFd and pPORB import were observed. Our results unveil a hitherto unrecognized role of Toc33 in pPORA import and suggest photooxidative membrane damage, induced by excess Pchlide accumulating in ppi1 chloroplasts because of the lack of pPORA import, to be the cause of the general drop of protein import. |
| [17] | . A mutation in the Arabidopsis gene STARIK leads to dwarfism and chlorosis of plants with an altered morphology of leaf and cell nuclei. We show that the STARIK gene encodes the mitochondrial ABC transporter Sta1 that belongs to a subfamily of Arabidopsis half-ABC transporters. The severity of the starik phenotype is suppressed by the ectopic expression of the STA2 homolog; thus, Sta1 function is partially redundant. Sta1 supports the maturation of cytosolic Fe/S protein in atm1 yeast, substituting for the ABC transporter Atm1p. Similar to Atm1p-deficient yeast, mitochondria of the starik mutant accumulated more nonheme, nonprotein iron than did wild-type organelles. We further show that plant mitochondria contain a putative L-cysteine desulfurase. Taken together, our results suggest that plant mitochondria possess an evolutionarily conserved Fe/S cluster biosynthesis pathway, which is linked to the intracellular iron homeostasis by the function of Atm1p-like ABC transporters. |
| [18] | . Abstract A novel rice (Oryza sativa L.) mutant derived from the ethyl methane sulfonate-treated restorer line Jinhui10 was identified that exhibited stable inheritance and was tentatively named yellowgreen leaf 6 (ygl6). Leaves of ygl6 were yellowgreen at the seedling stage and pale green at the jointing stage. The contents of chlorophyll a, chlorophyll b, and carotenoids were significantly lower in the mutant than those in the wildtype (WT) at the seedling and heading stages. Leaf mesophyll cells of the ygl6 mutant and WT showed no obvious differences in ultrastructure. However, ygl6 chloroplasts developed abnormally with poorly developed thylakoids and fewer grana stacks. Genetic analysis demonstrated that the mutation was controlled by a single recessive gene. Genetic mapping of the mutant gene was conducted using 1997 recessive individuals from the F2 segregating population. YGL6 was mapped to the centromeric region of chromosome 12 between the insertion/deletion primers Ind23 and Ind37, with an interval of 143 kb. Sequence analysis of the candidate genes within the delimited region showed a single base-pair change in Os12 g23180 that corresponded to a codon transition from leucine to a stop codon. Results of RNA interference knockdown of YGL6 suggested that Os12 g23180 is the candidate gene. Further characterization of YGL6 will provide insights into the molecular mechanisms of chloroplast development. |
| [19] | . |
| [20] | . Abstract Publisher Summary This chapter presents detailed information on chlorophylls and carotenoids to give practical directions toward their quantitative isolation and determination in extracts from leaves, chloroplasts, thylakoid particles, and pigment proteins. The chapter focuses on the spectral characteristics and absorption coefficients of chlorophylls, pheophytins, and carotenoids, which are the basis for establishing equations to quantitatively determine them. Therefore, the specific absorption coefficients of the pigments are re-evaluated. This is achieved by using a two-beam spectrophotometer of the new generation, which allows programmed automatic recording and printing out of the proper wavelengths and absorbancy values. Several procedures have been developed for the separation of the photosynthetic pigments, including column (CC), paper (PC), and thin-layer chromatography (TLC) and high-pressure liquid chromatography (HPLC). All chloroplast carotenoids exhibit a typical absorption spectrum that is characterized by three absorption maxima (violaxanthin, neoxanthin) or two maxima with one shoulder (lutein and 尾-carotene) in the blue spectral region. |
| [21] | |
| [22] | . Abstract We isolated and characterized two rice genes, OsRpoTp and OsRpoTm, that encode putative phage-type RNA polymerases. Predicted amino acid sequences showed high homology of these genes to known RpoT genes. A transient expression assay using green fluorescent protein indicated that the encoded proteins were localized to plastids and mitochondria, respectively. We demonstrated by reverse transcription-PCR experiments and immunoblot analysis that OsRpoTp expression occurred at an early stage of leaf development, prior to the transcript accumulation of the genes that were transcribed by the nuclear-encoded plastid RNA polymerase (NEP). Expression analyses of the chloroplast-deficient rice mutant, virescent-1, showed a discrepancy between OsRpoTp protein accumulation and the level of transcripts of NEP-transcribed genes. Our results suggest that NEP activation is regulated by a process after transcription, and is affected by the developmental state of chloroplast biogenesis. |
| [23] | . Abstract Loss of green color in leaves results from chlorophyll (Chl) degradation in chloroplasts, but little is known about how Chl catabolism is regulated throughout leaf development. Using the staygreen (sgr) mutant in rice (Oryza sativa), which maintains greenness during leaf senescence, we identified Sgr, a senescence-associated gene encoding a novel chloroplast protein. Transgenic rice overexpressing Sgr produces yellowish-brown leaves, and Arabidopsis thaliana pheophorbide a oxygenase-impaired mutants exhibiting a stay-green phenotype during dark-induced senescence have reduced expression of Sgr homologs, indicating that Sgr regulates Chl degradation at the transcriptional level. We show that the leaf stay-greenness of the sgr mutant is associated with a failure in the destabilization of the light-harvesting chlorophyll binding protein (LHCP) complexes of the thylakoid membranes, which is a prerequisite event for the degradation of Chls and LHCPs during senescence. Transient overexpression of Sgr in Nicotiana benthamiana and an in vivo pull-down assay show that Sgr interacts with LHCPII, indicating that the Sgr-LHCPII complexes are formed in the thylakoid membranes. Thus, we propose that in senescing leaves, Sgr regulates Chl degradation by inducing LHCPII disassembly through direct interaction, leading to the degradation of Chls and Chl-free LHCPII by catabolic enzymes and proteases, respectively. |
| [24] | . |
| [25] | . |
| [26] | . After endosymbiosis, organelles lost most of their initial genome. Moreover, expression of the few remaining genes became tightly controlled by the nucleus through trans-acting protein factors that are required for post-transcriptional expression (maturation/stability or translation) of a single (or a few) specific organelle target mRNA(s). Here, we characterize the nucleus-encoded TDA1 factor, which is specifically required for translation of the chloroplast atpA transcript that encodes subunit of ATP synthase in Chlamydomonas reinhardtii. The sequence of TDA1 contains eight copies of a degenerate 38-residue motif, that we named octotrico peptide repeat (OPR), which has been previously described in a few other trans-acting factors targeted to the C. reinhardtii chloroplast. Interestingly, a proportion of the untranslated atpA transcripts are sequestered into high-density, non-polysomic, ribonucleoprotein complexes. Our results suggest that TDA1 has a dual function: (i) trapping a subset of untranslated atpA transcripts into non-polysomic complexes, and (ii) translational activation of these transcripts. We discuss these results in light of our previous observation that only a proportion of atpA transcripts are translated at any given time in the chloroplast of C. reinhardtii. |
| [27] | . Small RNA regulators (sRNAs) have been identified in a wide range of bacteria and found to play critical regulatory roles in many processes. The major families of sRNAs include true antisense RNAs, synthesized from the strand complementary to the mRNA they regulate, sRNAs that also act by pairing but have limited complementarity with their targets, and sRNAs that regulate proteins by binding to and affecting protein activity. The sRNAs with limited complementarity are akin to eukaryotic microRNAs in their ability to modulate the activity and stability of multiple mRNAs. In many bacterial species, the RNA chaperone Hfq is required to promote pairing between these sRNAs and their target mRNAs. Understanding the evolution of regulatory sRNAs remains a challenge; sRNA genes show evidence of duplication and horizontal transfer but also could be evolved from tRNAs, mRNAs or random transcription. |
| [28] | . |
| [29] | . Abstract No abstract. I believe abstracts are not used for "updates". |
| [30] | . Spinach CSP41 is part of a protein complex that binds to the 3' untranslated region (UTR) of petD precursor-mRNA, a chloroplast gene encoding subunit IV of the cytochrome b6/f complex. CSP41 cleaves the 3'-UTR of petD mRNA within the stem-loop structure, suggesting a key role in the control of chloroplast mRNA stability. We discovered that CSP41 is homologous to nucleotide-sugar epimerases and hydroxysteroid dehydrogenases while seeking distant homologs of these enzymes with a hidden Markov model-based search of Genpept. This analysis identified Synechocystis ORF, Accession 1652543 as a homolog. Subsequent analyses show that spinach CSP41 and Arabidopsis thaliana 2765081 are homologous to the Synechocystis ORF. Information from the solved 3D structures of epimerases and dehydrogenases and our motif analysis of these enzymes is used to predict domains on CSP41 that are important in binding and metabolism of mRNA. Cyanobacteria are among the earliest life forms, indicating that the divergence from a common ancestor of nucleotide-sugar epimerases and an mRNA binding protein with ribonuclease activity was ancient. |
| [31] | . |
| [32] | . |
| [33] | . |
| [34] | . Chloroplast mRNA populations are characterized by overlapping transcripts derived by processing from polycistronic precursors. The mechanisms and functional significance of these processing events are poorly understood. We describe a pentatricopeptide repeat (PPR) protein, PPR10, whose binding defines mRNA segments derived from two transcription units in maize chloroplasts. PPR10 interacts in vivo and in vitro with two intergenic RNA regions of similar sequence. The processed 5090005 and 3090005 RNA termini in these regions overlap by approximately 25 nucleotides. The PPR10-binding sites map precisely to these overlapping sequences, and PPR10 is required specifically for the accumulation of RNAs with these termini. These findings show that PPR10 serves as a barrier to RNA decay from either the 5090005 or 3090005 direction and that a bound protein provides an alternative to an RNA hairpin as a barrier to 3090005 exonucleases. The results imply that protein 090004caps090005 at both 5090005 and 3090005 ends can define the termini of chloroplast mRNA segments. These results, together with recent insights into bacterial RNA decay, suggest a unifying model for the biogenesis of chloroplast transcript populations and for the determinants of chloroplast mRNA stability. |
| [35] | . Abstract CSP41 (chloroplast stem-loop-binding protein of 41 kDa), a chloroplast endonuclease belonging to the SDR superfamily, preferentially cleaves stem-loop-containing RNAs in vitro. This potentially directs it to the 3'-ends of mature chloroplast mRNAs, which generally possess such structures. To understand the basis for this discrimination, the RNA elements directing CSP41 cleavage of petD RNA in vitro were dissected. Substrates containing fully base-paired stem-loops were optimal substrates, whereas deletion of part of the stem-loop decreased activity by 100-fold, and deletion of the distal arm of the stem-loop abolished cleavage, even in substrates containing the primary CSP41 cleavage site. Competition assays showed that the decrease in activity resulted from decreased affinity for the RNA by CSP41. Mutations of the residues at the scissile bond and mutations and deletions at the terminal loop of the stem had a moderate effect on activity but no effect on cleavage site specificity, suggesting that CSP41 has no sequence specificity. Titration of ethidium bromide into the assay decreased activity to a basal level of approximately 18%, and introduction of a single base bulge into either arm of the stem-loop decreased cleavage at the primary cleavage site by up to 70%. This suggests that changing the structure of the helical stem has a mild effect on activity. Deletion analysis of CSP41 suggests that the specificity domain lies in the first 73 amino acids of the protein, a domain that also contains a putative dehydrogenaselike mononucleotide binding motif. These results are consistent with a broad role for CSP41 in the degradation of stem-loop-containing mRNAs. |
