 ,1,**, 练旺民1,**, 占小登1, 徐海明3, 张迎信1, 程式华1, 楼向阳
,1,**, 练旺民1,**, 占小登1, 徐海明3, 张迎信1, 程式华1, 楼向阳 ,1,*, 曹立勇
,1,*, 曹立勇 ,1,2,*, 洪永波
,1,2,*, 洪永波 ,1,*
,1,*Genetic dissection of the bacterial blight disease resistance in super hybrid rice RILs using genome-wide association study
ZHAO Hai-Han ,1,**, LIAN Wang-Min1,**, ZHAN Xiao-Deng1, XU Hai-Ming3, ZHANG Ying-Xin1, CHENG Shi-Hua1, LOU Xiang-Yang
,1,**, LIAN Wang-Min1,**, ZHAN Xiao-Deng1, XU Hai-Ming3, ZHANG Ying-Xin1, CHENG Shi-Hua1, LOU Xiang-Yang ,1,*, CAO Li-Yong
,1,*, CAO Li-Yong ,1,2,*, HONG Yong-Bo
,1,2,*, HONG Yong-Bo ,1,*
,1,*通讯作者: *洪永波, E-mail:hongyongbo@caas.cn;曹立勇, E-mail:caoliyong@caas.cn;楼向阳, E-mail:louxiangyang01@caas.cn
第一联系人:
收稿日期:2020-12-24接受日期:2021-04-14网络出版日期:2021-07-06
| 基金资助: |
Corresponding authors: *E-mail:hongyongbo@caas.cn;E-mail:caoliyong@caas.cn;E-mail:louxiangyang01@caas.cn
First author contact:
Received:2020-12-24Accepted:2021-04-14Published online:2021-07-06
| Fund supported: |
作者简介 About authors
赵海涵,E-mail:haihan3354@163.com

摘要
白叶枯病是水稻生产最严重的细菌性病害, 挖掘新的白叶枯病抗性基因资源并培育抗病品种是控制该病害的重要方法。本研究利用父母本抗性差异较大的协优9308衍生的139个重组自交系群体作为遗传材料, 人工接种不同白叶枯菌后的病斑长度作为连续型表型, 结合经DNA深度测序获得的476,505个单核苷酸多态性(single-nucleotide polymorphism, SNP)标记进行全基因组关联分析(genome-wide associated study, GWAS)。结果表明在P < 1×10-4下, 4个菌株处理后共鉴定到109个与白叶枯抗性显著关联的SNPs位点, 解释表型变异率59.78%~63.29%。其中CR4接种发现了25个SNP位点其贡献率为61.00%, 在这些SNP位点附近共筛选到19个基因, 其中有2个为NBS-LRR抗病相关基因(LOC_Os11g43420和LOC_Os11g45930)。表达分析验证发现该2个基因在抗性亲本中恢9308的表达量分别为感病母本协青早B的4.42倍和8.86倍, 表明其可能在正调控白叶枯病抗性机制中发挥重要作用。进化树分析发现这2个候选基因与已克隆的抗性基因属于不同的亚组, 表明可能是新基因。本研究为进一步挖掘白叶枯抗性基因和培育抗病品种提供了理论基础和基因资源。
关键词:
Abstract
Bacterial blight caused by Xanthomonas oryzae pv. oryzae is the most destructive bacterial disease in rice production, and cultivating new disease-resistance variety by mining and utilizing the novel disease resistance genes is one of the most effective ways to control this disease. In our study, a population of 139 recombinant inbred lines (RILs) derived from super hybrid rice Xieyou 9308, between the parents of which there was genetic difference in resistance to bacterial blight, were inoculated with four Xoo strains. A genome-wide association study (GWAS) was carried out using 476,505 SNPs for the lesion length as a quantitative phenotype. The result revealed that a total of 109 significant SNPs including two genes encoding NBS-LRR containing proteins were detected to be significantly difference at P < 1×10-4 for four Xoo strains-treated lesions, accounting for phenotype variation of 59.78%-63.29%, respectively. Furthermore, 25 SNPs located in/nearby 19 candidate genes were identified by the CR4 inoculation, accounting for 61.00%. Relative expression analysis of two selected candidate genes, LOC_Os11g43420 and LOC_Os11g45930, demonstrated their higher expression levels in the resistant cultivar Zhonghui 9308 than in the susceptible variety Xieqingzao B, suggesting that these two genes might positively regulated bacterial blight resistance. Phylogenetic tree analysis indicated that these two genes were different from the previous cloned resistant genes in evolution, suggesting they were novel disease resistance genes. These results lays a theoretical basis and provides genetic resources for future resistant breeding in rice.
Keywords:
PDF (7462KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
赵海涵, 练旺民, 占小登, 徐海明, 张迎信, 程式华, 楼向阳, 曹立勇, 洪永波. 水稻协优9308重组自交系群体白叶枯病抗性的全基因组关联分析. 作物学报, 2022, 48(1): 121-137 DOI:10.3724/SP.J.1006.2022.02090
ZHAO Hai-Han, LIAN Wang-Min, ZHAN Xiao-Deng, XU Hai-Ming, ZHANG Ying-Xin, CHENG Shi-Hua, LOU Xiang-Yang, CAO Li-Yong, HONG Yong-Bo.
水稻是世界上重要的粮食作物, 保证其高产、稳产对国家的粮食安全至关重要。然而全球每年因病害引起的产量损失平均在20%以上[1]。由稻黄单孢水稻致病变种(Xanthomonas oryzae pv. oryzae, Xoo)引起的水稻白叶枯病是世界水稻生产中危害最严重的细菌性病害。Xoo通过在水稻叶片的维管组织中定植和传播而引起白叶枯病, 从而影响光合作用最终导致水稻严重减产[2,3,4]。育种实践表明挖掘抗性基因并培育抗病新品种是防治该病最经济有效的手段。
水稻白叶枯病抗性基因主要分为主效抗病基因介导的质量性状抗性和微效多基因控制的数量性状抗性。目前发现水稻白叶枯抗性基因位点有44个[5], 其中包含12个已经克隆的水稻白叶枯R基因, 分别是Xa1、Xa3/Xa26、Xa4、xa5、Xa7、Xa10、xa13、Xa21、Xa23、Xa25、Xa27和xa41(t)[6,7,8,9,10,11,12,13,14,15,16,17,18]。R基因介导的植物抗病防御机制, 主要是由基因自身抗病基因编码的抗病蛋白识别相应的由病原菌无毒基因(Avr)编码的无毒蛋白后, 进一步激活植物体内抗病信号途径引发的防卫反应, 在侵染部位产生局部的细胞和组织过敏性坏死(hypersensitive response, HR)反应, 最终抵御病原菌的进一步侵染及扩散[19,20]。植物抗病基因编码的NBS-LRR保守蛋白结构域(nucleotide binding site and leucine rich repeat)可特异性识别病原菌分泌的效应子(Effector)从而激发更高级别的抗性反应[21,22]。已报道的白叶枯病抗性基因大多包含编码NBS-LRR蛋白结构域[23], 例如已克隆的基因Xa1和Xa21[9]。对4号染色体长臂上Xa1基因附近的抗病基因分析揭示了该区域富含NBS-LRR序列[24]。Xa21是第一个克隆的白叶枯病广谱抗性基因, Ronald等[25]利用123个DNA标记和985个随机引物对含有Xa21的近等基因系进行RFLP和RAPD分析, 发现3个分子标记RG103、RAPD818和RAPD248与11号染色体上的抗性位点Xa21共分离。Song等[17]进一步利用分子标记RG103作为探针筛选到8个包含Xa21的阳性克隆, 其中有7个片段与Xa21共分离, 随后分别进行遗传转化得到转基因阳性植株, 抗性鉴定发现9.6 kp片段编码1025个氨基酸的LRR类受体激酶产生完全抗性。然而根据“基因对基因”假说, R基因和Avr基因的互作表现为品种对小种的专化性抗性[26], 随着Xa4、Xa21等抗性基因导入水稻栽培品种且大面积种植, 导致新的白叶枯菌致病生理小种不断出现和进化以及主栽水稻品种逐渐失去抗性; 或者多数白叶枯主抗性基因影响生长发育难以直接应用于抗病育种实践[27]。因此亟需应用新的技术手段发掘更多广谱、持久抗性基因并应用于水稻抗病育种中。
多数白叶枯抗性研究基于传统的定位手段如数量性状位点(QTL)定位[28,29]。Yang等[30]利用中抗白叶枯的粳稻品种春江06和感病品种TN1及其双单倍体(double haploid, DH)群体进行QTL分析, 共检测到3个QTL, 总效应为54.4%; Chen等[31]利用多亲本高代互交(multiparent advanced generation inter-cross, MAGIC)群体关联定位白叶枯病抗性QTL, 共检测到7个白叶枯病抗性QTL, 但定位到的QTL大多遗传背景效应明显, 位点与环境存在互作效应, 限制了育种应用的潜力; 且连锁作图方法的标记密度小, 定位精度不高, 难以找到单一位点的多个等位基因和深入挖掘一些受环境影响的微效基因和不同亚群间可能存在的微效位点。借助于高通量测序技术, 全基因组关联分析(genome-wide association study, GWAS)通过计算连锁不平衡(linkage disequilibrium, LD)用于解析复杂农艺性状的遗传背景[32]。Huang等[33]通过GWAS挖掘水稻产量性状相关位点, 开发连锁分子标记, 可直接用于水稻品种的产量性状改良。
为了深入挖掘水稻白叶枯病抗性位点, 本研究通过单粒传方法构建了感病母本协青早B和抗病父本中恢9308的极端重组自交系群体(F13), 对该群体进行不同年份、4个不同白叶枯菌株接种的表型调查。结合深度测序获得的单核苷酸多态性(single- nucleotide polymorphism, SNP)标记, 应用EMMAX软件进行全基因组关联分析, 该软件以混合线性模型(mix linear model, MLM)为基础并能最大程度减少群体结构对分析结果的影响, 期望找到与水稻白叶枯抗性显著关联的抗性基因, 为后续的水稻抗病育种提供基因资源。
1 材料与方法
1.1 试验群体
试验材料为来源于超级稻‘协优9308’的重组自交系群体(recombination inbred lines, RILs), 杂交组合原始亲本为‘协青早B’和‘中恢9308’, 通过单粒传法连续13代获得重组自交系。2019和2020年在中国水稻研究所富阳试验基地(30°32′N, 120°12′E)种植协优9308重组自交系群体, 包括139个重组自交系株系及2个原始亲本。所有的重组自交系材料种子经单株收种后均采用秧盘育苗, 30 d后移栽于大田, 每个编号种植5行, 每行8株, 同时种植水稻白叶枯病感病对照品种‘金刚30’。1.2 水稻白叶枯病抗性鉴定及分析
白叶枯病抗性鉴定采用剪叶法, 在水稻孕穗期接种4个不同的白叶枯生理菌株(PXO96、PXO61、PXO99、CR4), 其中PXO96、PXO61和PXO99三个生理菌株来自于菲律宾, CR4生理菌株来自于中国浙江省。白叶枯菌在NA培养基(酵母浸膏1 g, 牛肉膏3 g, 蛋白胨5 g, 蔗糖10 g, 琼脂粉15 g, pH 6.0~ 6.5)于28℃培养2 d, 用无菌水稀释至1×109个细菌 mL-1 (OD600=0.8)。每个生理菌株接种1行, 每个编号至少接种完全展开的叶子20片。接种后不打药剂处理, 其余按常规田间管理, 2019年和2020年田间接种后分别在第15天和第21天测量病斑长度和叶片总长。白叶枯病抗性分级标准参考方中达的第3版《植病研究方法》, 水稻剑叶发病程度分级标准: 其中0级为免疫, 无病斑; 1级为高抗, 病斑长度不超过3 cm; 2级为中抗, 病斑长度小于总叶片长度的25%; 3级为中感, 病斑长度小于总叶片长度的50%; 4级为高感, 病斑长度小于总叶片长度的75%; 5级为感病, 全叶发病。
Microsoft Excel软件统计每一个编号下的菌株表型数据并处理离群值。利用R语言对2个环境4个菌株下叶片病斑长度的平均值、标准差、频率分布等进行描述性统计分析。利用SPSS统计软件进行成对样本相关和t检验, 确定同一年份下2个生理菌株数据之间是否存在显著性相关和差异。
1.3 基因分型和群体结构分析
采用CTAB方法从水稻叶片组织中提取基因组DNA, 在诺和生物技术(天津)有限公司使用二代测序技术(next-generation sequencing)完成重测序工作。双亲和139个重组自交系分别采用100×和10×覆盖率。日本晴基因组(os-Nipponbare-reference- IRGSP-1.0)作为参考基因组序列, 使用BWA软件完成序列读数(sequence reads)和参考基因组的序列比对(sequence alignment); 应用PLINK软件过滤最小等位基因频率(minimum allele frequency, MAF) < 0.05, 基因型缺失率< 0.2的SNP标记, 并对该群体进行主成分分析(principal components analysis, PCA)。1.4 连锁不平衡衰减分析
通过连锁不平衡(linkage disequilibrium, LD)衰减曲线反映位点间由连锁不平衡到连锁平衡的演变过程, 可用于辅助关联分析。利用协优9308重组自交系群体的重测序数据得到约50万SNP, 通过PopLDdecay软件计算连锁不平衡衰减并绘制该群体在整个基因组上的LD衰减图, 同一个染色体上2个位点之间的距离以1000 bp为基本分类单元计算平均LD。1.5 全基因组关联分析
应用EMMAX软件, 选择可有效控制群体结构和亲缘关系的混合线性模型(mixed linear model, MLM)进行全基因组关联分析。其中全基因组测序结果用于评估重组自交系中个体的遗传相似性, 得到亲缘关系矩阵(kinship matrix); ADMIXTURE软件计算作为协变量的群体结构Q矩阵。结果通过R语言qqman包绘制曼哈顿图和Q-Q图以及由Bonferroni校正得到显著性水平。所有的显著标记解释表型变异值均通过SAS软件的逐步回归模型计算。1.6 基因组注释
P值小于阈值1×10-4的标记设置为显著位点, 参照日本晴序列注释图谱(the Nipponbare reference genome, IRGSP 1.0), 在每个位点内-log10(P值)峰值的SNP上、下游连锁不平衡衰减距离的区间范围内挖掘候选基因。并参照水稻基因组功能注释网站(1.7 候选基因定量PCR及进化分析
为了验证候选基因是否在调控白叶枯抗性发挥作用, 根据水稻基因组功能注释网站提供的编码区序列设计定量引物。协青早B和中恢9308在接菌前后不同时间点进行田间取样, 利用TRIzol试剂提取总RNA, 经反转录后的cDNA作为模板, 采用Roche的Fast Essentia DNA Green试剂在CFX96 (Bio-Rad)进行定量PCR。提取已克隆的11个白叶枯抗病基因和4个显著相关的抗病候选基因编码的蛋白序列, 用Clustal X软件进行一致性比较, 并利用MEGA软件将比对结果转化成MEGA格式, 利用邻间法模型(Bootstrap = 1000)构建进化树。
2 结果与分析
2.1 4种不同白叶枯菌的统计分析和正态分布
139个重组自交系和双亲经4个白叶枯生理菌株(分别是2019年PXO96和CR4与2020年PXO61和PXO99接种后的平均病斑长度的表型统计特征数见表1。双亲协青早B和中恢9308分别表现为感病和抗病(图1), 4个不同生理菌株处理139个RIL群体及双亲并对病斑长度数据进行正态检验, 发现PXO96、PXO99和CR4菌株处理后的病斑长度均服从正态分布(图2), 且该数据在群体中均表现为超亲分离, 说明该性状符合多基因控制的数量性状遗传[34,35,36,37], 适用于全基因组关联分析。然而接种PXO61后发现其群体均值较小(1.2 cm)不服从正态分布。进一步对白叶枯病抗性分级数据统计发现接种PXO61多表现为完全抗病(图3)。2个样本t测验对两亲本均值差异显著性检验的结果表明, PXO96和CR4生理菌株处理组(2019年)与PXO61和PXO99生理菌株处理组(2020年)均存在极显著差异(P < 0.01) (表1)。由此推断协青早B和中恢9308重组自交系可作为水稻白叶枯抗性接种群体并用于关联分析。Table 1
表1
表1139个重组自交系及双亲接种4个生理菌株后的病斑长度统计特征数
Table 1
| Xoo菌株 Xoo strain | 年份/区域 Year/location | 协青早B病斑长度(P1) Lesion length of XB (P1) (cm) | 中恢9308病斑长度(P2) Lesion length of R9308 (P2) (cm) | 亲本差异(P1-P2)的显著性P值 Significant P-value of parental difference (P1-P2) | 重组自交系 Recombinant inbred line | 变异系数 CV (%) | |
|---|---|---|---|---|---|---|---|
| 均值±标准差 Mean ± SD (cm) | 变化范围 Range (cm) | ||||||
| PXO96 | 2019/Hangzhou | 21.6 | 5.4 | <0.001** | 13.7±7.0 | 0.2-37.7 | 51.9 |
| CR4 | 2019/Hangzhou | 21.6 | 5.3 | <0.001** | 7.7±4.2 | 0.2-20.3 | 54.5 |
| PXO61 | 2020/Hangzhou | 19.4 | 0.2 | 0.002** | 1.2±2.4 | 0-19.4 | 203.0 |
| PXO99 | 2020/Hangzhou | 10.4 | 0.8 | <0.001** | 5.6±3.6 | 0.1-21.8 | 64.2 |
新窗口打开|下载CSV
图1
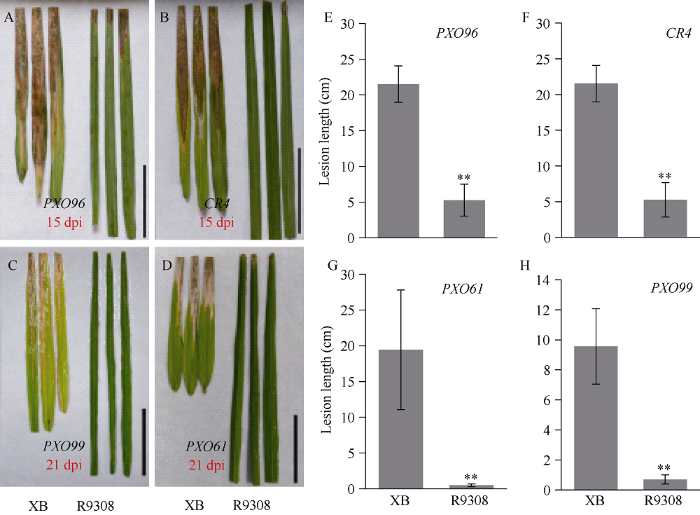 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1协B/中恢9308接种4个生理菌株的白叶枯表型和病斑长度
A~D为双亲协青早B (左)和中恢9308 (右)分别接种4种生理菌株(PXO96、CR4、PXO61和PXO99), 15 d和21 d后调查病斑长度, 标尺为10 cm。图E~H为双亲接种不同生理菌株的平均病斑长度(cm)。**表示P ≤ 0.01 (Student’s t-test)。
Fig. 1Phenotype and lesion length of XB/R9308 after inoculating of four Xoo strains
A-D indicate the phenotype of XB and R9308 after inoculating of four Xoo strains (PXO96, CR4, PXO61, and PXO99) and measure the lesion length after 15 dpi (days post inoculation) and 21 dpi. Bar: 10 cm. E-H indicate the average lesion length after inoculating four Xoo strains of parents, respectively. Data presents are the mean ± SD from the independent experiments. **: P ≤ 0.01 (Student’s t-test).
图2
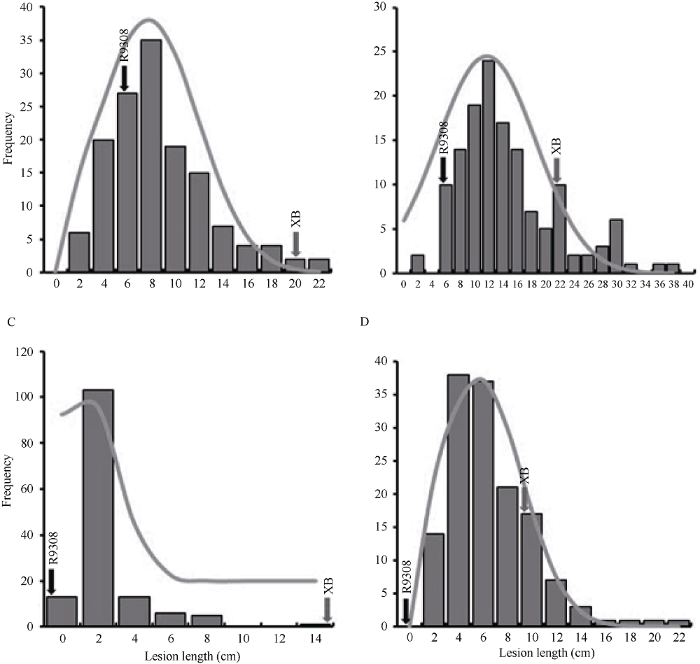 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2139个重组自交系及双亲接种4个生理菌株的病斑长度的频率分布图
A~D分别来自于4种生理菌株: A: PXO96; B: CR4; C: PXO61; D: PXO99。图中横坐标表示平均病斑长度(cm), 纵坐标表示分布频数。黑色和红色箭头分别表示中恢93008和协青早B病斑长度对应横坐标的位置, 线条表示为数据的正态分布曲线。
Fig. 2Histogram of phenotypic frequency distribution for four Xoo strains in 139 RILs and two parents
A-D come from four Xoo strains of PXO96, CR4, PXO61, and PXO99, respectively. The X-axis represents a mean lesion length, and the Y-axis represents the corresponding frequency. Black and red arrows mean the lesion length of R9308 and XB, respectively. Light grey line indicates the normal distribution curve.
图3
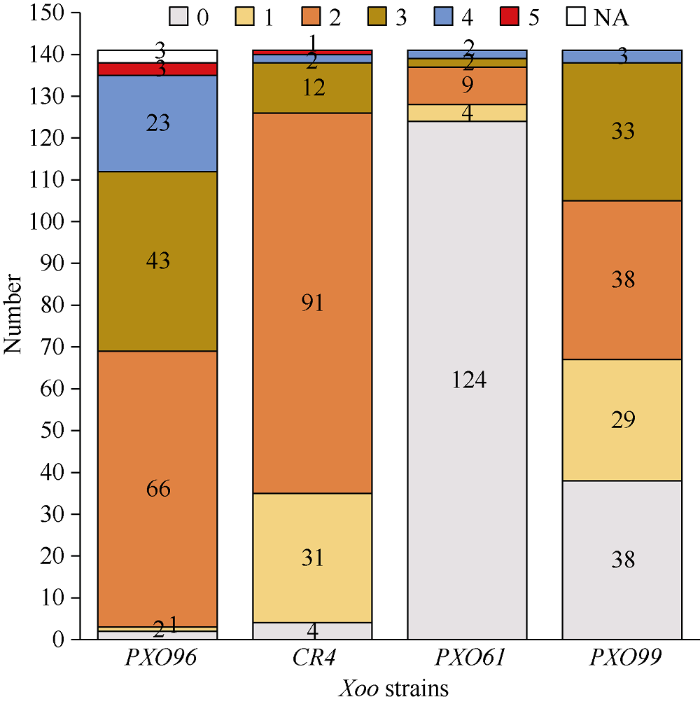 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3139个重组自交系及双亲接种4个生理菌株的白叶枯抗性等级分布图
利用139 RIL和双亲在4种Xoo生理菌株中抗性等级数据进行分组: 根据每个水稻编号的平均病斑长度(LL)划分等级: 其中LL = 0 cm, 0 cm ≤ LL < 3 cm, LL/总叶片长度< 25%, LL/总叶片长度< 50%, LL/总叶片长度< 75%和整叶发病分别代表抗病(0级)、高抗(1级)、中抗(2级)、中感(3级)、高感(4级)和易感(5级); NA代表缺失值。
Fig. 3Bar chart of bacterial blight resistance levels for four Xoo strains in 139 RILs and two parents
Number of accessions in the following reactions to four Xoo races: LL = 0 cm, 0 cm ≤ LL < 3 cm, LL/Total Leaf Length < 25%, LL/Total Leaf Length < 50%, LL/Total Leaf Length < 75%, and LL/Total leaf lesion represented resistant (level 0), highly resistant (level 1), moderately resistant (level 2), moderately susceptible (level 3), highly susceptible (level 4), and susceptible (level 5), respectively. LL indicates mean lesion length of each accession.
另外, 同一年份接种的发病表型间(即2019年的PXO96和CR4间与2020年的PXO61和PXO99间)存在显著相关性(P < 0.01), 且成对数据比较表明成对性状间差异显著(P < 0.001) (数据未列入正文表格)。
2.2 基因型和主成分分析
使用GATK软件完成SNPs测定(SNPs calling), 得到8,000,000个SNPs。PLINK对表型和测序后的基因型数据筛选后得到1,980,581个SNPs标记, 去除在双亲中不存在基因差异的位点最终得到476,505个SNPs标记。标记统计表明基因分型比率为98.8%且标记非均匀分布在12条染色体上: 1号染色体标记分布最低为3.7%, 10号染色体分布最高为6.6%; 最小等位基因频率在0.2~0.5之间; 等位基因缺失频率在0.00~0.10之间。为了判断关联分析是否受到群体结构的影响, PCA分析结果显示该群体的不存在明显分群, 可直接用于后续分析。
2.3 水稻全基因组的连锁不平衡分析
水稻全重组自交系群体的基因组连锁不平衡约在10,000 kb后衰减至接近0 (图4)。LD衰减距离是用于判断分析的检测效力和精度的一个常用指标, 定义为在同一条染色体2个位点的连锁不平衡系数(the average squared pairwise correlation coefficient, r2)降低到最大值一半时的物理距离。计算结果预测群体全基因组LD衰减距离大约在2000 kb。Zhou等[38]和Mather等[39]对协优9308群体的连锁不平衡衰减距离计算为1000 kb左右, 均大于已报道的在100 kb到200 kb之间的水稻自然群体预测区间。图4
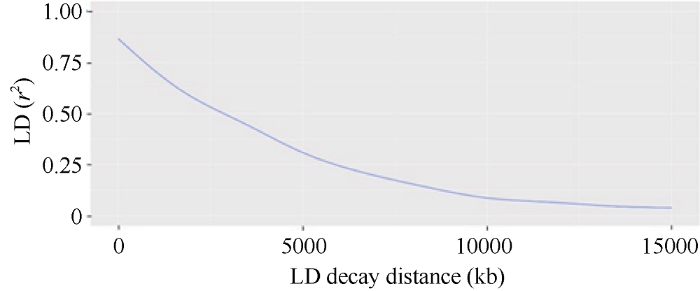 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4全基因组范围内的连锁不平衡衰减图
LD衰减图显示了476,506 SNPs在整个基因组中的连锁不平衡(r2)随距离(kb)的衰减。
Fig. 4Pattern of LD decay with average distance
Figure shows that decay of LD (r2) with distance (kb) between SNPs.
2.4 水稻白叶枯病抗性基因的关联定位
协优9308重组自交系群体接种4个白叶枯菌株(PXO96、CR4、PXO99和PXO61)后的病斑长度作为连续型表型结合最终的SNPs标记进行关联分析, 曼哈顿图(图5)发现Q+K模型可有效的控制假阳性, 在P < 1×10-4下共检测到109个显著关联的SNPs (表2), 参照日本晴基因组注释信息共得到87个基因。逐步回归算法计算显著位点解释的表型变异百分比, 以R2值由大到小排序输出, 并输出每个回归方程的回归系数。109个显著标记中有32个、25个和52个分别来自PXO96、CR4和PXO99生理菌株, 分别解释表型变异率为61.00%、59.78%和63.29% (因不服从正态假定接种PXO61的关联分析结果暂不作报告)。图5
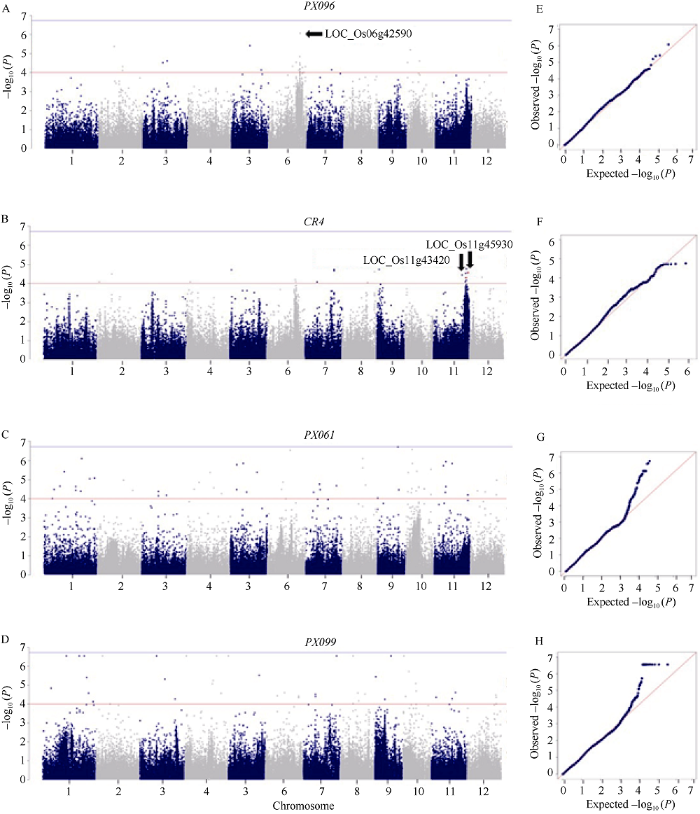 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5139个重组自交系及双亲接种4个生理菌株的GWAS分析
A~D分别来自于4种生理菌株: A: PXO96; B: CR4; C: PXO61; D: PXO99。横坐标对应水稻全基因组, 纵坐标表示-log10 (P值), 蓝色线条表示Bonferroni矫正的P值, 橙色线条表示-log10 (P值)大于4.0的位点, 可判断与抗性关联。E~H分别绘制了对应的Q-Q图: E: PXO96; F: CR4; G: PXO61; H: PXO99。横坐标表示为期望的-log10 (P值), 纵坐标表示为观测的-log10 (P值)。红线作为参考线。
Fig. 5Genome-wide association study of lesion length after inoculating four Xoo strains in 139 RILs and two parents
Manhattan plot (A-D) are plotted with the leaf lesion length after inoculating four Xoo strains of PXO96, CR4, PXO61, and PXO99, respectively. The X-axis represents the whole rice genome and the y-axis represents the -log10 (P-value). The blue line represents a Bonferroni corrected P-value. The orange line representing a stringent criterion of -log10 (P-value) > 4.0 under four environments was used for determining the association significance of four Xoo strains. Q-Q plot (E-H): E: PXO96; F: CR4; G: PXO61; H: PXO99. The horizontal axis of Q-Q plot (E-H) represents the expected -log10 (P-value) while the vertical axis is the observed -log10 (P-value). Red line is the cutoff line.
Table 2
表2
表2全基因组关联分析检测的显著SNP位点和抗性相关候选基因的注释信息
Table 2
| 菌株 Xoo | SNP标记 SNP | 染色体 Chr. | 物理位置 Position | P值 P-value | 基因名称(MSU) Gene ID (MSU) | 基因注释 Gene annotation | |
|---|---|---|---|---|---|---|---|
| CR4 | 11_26502942_C_A | 11 | 26502942 | 1.71260×10-5 | LOC_Os11g43890 | WD domain, G-beta repeat domain containing protein, expressed | |
| CR4 | 9_1843902_G_A | 9 | 1843902 | 1.83954×10-5 | LOC_Os09g03660 | Retrotransposon protein, putative, unclassified, expressed | |
| CR4 | 7_23436266_C_T | 7 | 23436266 | 1.88459×10-5 | LOC_Os07g39114 | Expressed protein | |
| CR4 | 7_23436267_C_G | 7 | 23436267 | 1.88459×10-5 | |||
| CR4 | 7_23436291_C_T | 7 | 23436291 | 1.88459×10-5 | |||
| CR4 | 5_1383235_C_A | 5 | 1383235 | 1.92989×10-5 | LOC_Os05g03360 | Retrotransposon protein, putative, unclassified, expressed | |
| CR4 | 7_23436858_C_T | 7 | 23436858 | 2.13921×10-5 | LOC_Os07g39114 | Expressed protein | |
| CR4 | 7_23436871_A_T | 7 | 23436871 | 2.13921×10-5 | |||
| CR4 | 8_26662359_G_A | 8 | 26662359 | 2.44334×10-5 | LOC_Os08g42210 | Expressed protein | |
| CR4 | 11_27792921_A_G | 11 | 27792921 | 2.70257×10-5 | LOC_Os11g45930 | NBS-LRR type disease resistance protein, putative, expressed | |
| CR4 | 11_26056269_A_G | 11 | 26056269 | 2.90443×10-5 | LOC_Os11g43200 | Tropinone reductase 2, putative, expressed | |
| CR4 | 2_11969901_C_T | 2 | 11969901 | 3.20777×10-5 | LOC_Os02g20330 | Expressed protein | |
| CR4 | 11_23030967_G_A | 11 | 23030967 | 3.55194×10-5 | LOC_Os11g38760 | Retrotransposon protein, putative, LINE subclass, expressed | |
| CR4 | 12_10252946_G_T | 12 | 10252946 | 4.65692×10-5 | LOC_Os12g17880 | Armadillo/beta-catenin repeat protein-related, putative, expressed | |
| CR4 | 11_26220673_C_T | 11 | 26220673 | 5.00378×10-5 | LOC_Os11g43420 | LZ-NBS-LRR class RGA, putative, expressed | |
| CR4 | 6_23077582_G_A | 6 | 23077582 | 6.20380×10-5 | LOC_Os06g38890 | Transposon protein, putative, unclassified, expressed | |
| CR4 | 11_26410429_C_A | 11 | 26410429 | 6.81889×10-5 | LOC_Os11g43740 | OsMADS68 - MADS-box family gene with MIKC* type-box | |
| CR4 | 11_25840166_C_A | 11 | 25840166 | 7.97330×10-5 | LOC_Os11g42900 | Expressed protein | |
| CR4 | 11_25849255_T_C | 11 | 25849255 | 8.02272×10-5 | |||
| CR4 | 6_23441121_G_C | 6 | 23441121 | 8.04677×10-5 | LOC_Os06g39470 | Transferase family protein, putative, expressed | |
| CR4 | 2_1831384_A_G | 2 | 1831384 | 8.31200×10-5 | LOC_Os02g04180 | Transposon protein putative CACTA En/Spm sub-class expressed | |
| CR4 | 4_3331408_G_A | 4 | 3331408 | 8.31200×10-5 | LOC_Os04g06374 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| CR4 | 7_9599243_T_C | 7 | 9599243 | 8.31200×10-5 | LOC_Os07g16400 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| CR4 | 8_20838490_G_A | 8 | 20838490 | 9.02380×10-5 | LOC_Os08g33410 | Hypothetical protein | |
| CR4 | 8_20838500_T_C | 8 | 20838500 | 9.02380×10-5 | LOC_Os02g21320 | Retrotransposon protein putative unclassified expressed | |
| PXO96 | 6_25611297_T_C | 6 | 25611297 | 8.66084×10-7 | LOC_Os10g05510 | ||
| PXO96 | 5_14854774_G_A | 5 | 14854774 | 3.83500×10-6 | LOC_Os06g42010 | ||
| PXO96 | 2_12653706_C_A | 2 | 12653706 | 4.27352×10-6 | LOC_Os03g35500 | Expressed protein | |
| PXO96 | 10_2723114_A_G | 10 | 2723114 | 6.44760×10-6 | LOC_Os10g20020 | Expressed protein | |
| PXO96 | 6_25220112_T_G | 6 | 25220112 | 1.45559×10-5 | LOC_Os06g37790 | Transposon protein putative unclassified expressed | |
| PXO96 | 3_19680871_T_C | 3 | 19680871 | 2.49545×10-5 | LOC_Os10g01510 | Retrotransposon protein putative Ty3-gypsy subclass | |
| PXO96 | 10_10043595_C_T | 10 | 10043595 | 2.53663×10-5 | LOC_Os03g28010 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| PXO96 | 6_22366894_G_A | 6 | 22366894 | 2.89696×10-5 | LOC_Os06g42010 | Expressed protein | |
| PXO96 | 10_330955_C_T | 10 | 330955 | 2.98270×10-5 | LOC_Os06g42250 | Transposon protein putative CACTA En/Spm sub-class expressed | |
| PXO96 | 3_16101988_G_T | 3 | 16101988 | 3.07716×10-5 | LOC_Os06g42370 | Retrotransposon protein putative unclassified expressed | |
| PXO96 | 6_25219787_G_A | 6 | 25219787 | 3.14674×10-5 | LOC_Os06g33630 | Transposon protein putative unclassified expressed | |
| PXO96 | 6_25373580_A_G | 6 | 25373580 | 3.85706×10-5 | LOC_Os02g32680 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| PXO96 | 6_25462404_C_T | 6 | 25462404 | 4.00401×10-5 | LOC_Os06g49185 | Retrotransposon protein putative unclassified expressed | |
| PXO96 | 6_19579269_C_T | 6 | 19579269 | 4.66700×10-5 | LOC_Os06g45410 | Retrotransposon protein putative Ty3-gypsy subclass | |
| PXO96 | 2_19389629_A_G | 2 | 19389629 | 4.76090×10-5 | LOC_Os06g42310 | Lectin receptor-type protein kinase putative expressed | |
| PXO96 | 6_29801791_T_C | 6 | 29801791 | 4.82490×10-5 | LOC_Os06g42010 | Expressed protein | |
| PXO96 | 6_27455797_A_T | 6 | 27455797 | 5.46665×10-5 | LOC_Os06g42370 | MYB family transcription factor putative expressedMYB | |
| PXO96 | 6_25409464_C_T | 6 | 25409464 | 6.23488×10-5 | LOC_Os07g33300 | Beta-galactosidase precursor putative expressed | |
| PXO96 | 6_25226687_G_C | 6 | 25226687 | 6.81127×10-5 | LOC_Os05g40940 | Transposon protein putative unclassified expressed | |
| PXO96 | 6_25465479_C_T | 6 | 25465479 | 6.91484×10-5 | LOC_Os06g41120 | Retrotransposon protein putative unclassified expressed | |
| PXO96 | 7_19906050_T_A | 7 | 19906050 | 7.23635×10-5 | LOC_Os02g32520 | Expressed protein | |
| PXO96 | 5_24009603_G_A | 5 | 24009603 | 7.38552×10-5 | LOC_Os06g42350 | Retrotransposon protein putative unclassified expressed | |
| PXO96 | 6_24594013_A_C | 6 | 24594013 | 7.54406×10-5 | LOC_Os06g42560 | Expressed protein | |
| PXO96 | 2_19249914_C_T | 2 | 19249914 | 7.84868×10-5 | LOC_Os06g42020 | ERD1 protein chloroplast precursor putative expressed | |
| PXO96 | 6_25451288_C_G | 6 | 25451288 | 9.02863×10-5 | LOC_Os06g42350 | Transposon protein putative CACTA En/Spm sub-class expressed | |
| PXO96 | 6_25592476_C_T | 6 | 25592476 | 9.10497×10-5 | LOC_Os06g42560 | Tryptophan synthase beta chain 2 putative expressed | |
| PXO96 | 6_25233994_G_A | 6 | 25233994 | 9.17863×10-5 | LOC_Os06g42020 | CSLA9 - cellulose synthase-like family A expressed | |
| PXO96 | 6_25446478_G_A | 6 | 25446478 | 9.81605×10-5 | LOC_Os06g42350 | Transposon protein putative CACTA En/Spm sub-class expressed | |
| PXO96 | 6_25226517_G_A | 6 | 25226517 | 9.81690×10-5 | LOC_Os06g42010 | Transposon protein putative unclassified expressed | |
| PXO96 | 6_25226850_C_T | 6 | 25226850 | 9.81690×10-5 | |||
| PXO96 | 6_28762618_T_C | 6 | 28762618 | 9.93356×10-5 | LOC_Os06g47500 | Retrotransposon protein putative Ty3-gypsy subclass | |
| PXO96 | 6_27492113_G_A | 6 | 27492113 | 9.99334×10-5 | LOC_Os06g45470 | Expressed protein | |
| PXO99 | 1_19343496_G_A | 1 | 19343496 | 2.88795×10-7 | LOC_Os01g34960 | Retrotransposon putative centromere-specific | |
| PXO99 | 1_29753419_G_A | 1 | 29753419 | 2.88795×10-7 | LOC_Os01g51740 | Expressed protein | |
| PXO99 | 1_33782923_T_C | 1 | 33782923 | 2.88795×10-7 | LOC_Os01g58444 | Expressed protein | |
| PXO99 | 2_5398259_G_A | 2 | 5398259 | 2.88795×10-7 | LOC_Os02g10280 | Transposon protein putative unclassified expressed | |
| PXO99 | 3_13482702_G_A | 3 | 13482702 | 2.88795×10-7 | LOC_Os03g23790 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| PXO99 | 4_1315077_T_C | 4 | 1315077 | 2.88795×10-7 | LOC_Os04g03150 | Retrotransposon protein putative unclassified expressed | |
| PXO99 | 4_25911404_C_T | 4 | 25911404 | 2.88795×10-7 | LOC_Os04g43770 | Expressed protein | |
| PXO99 | 4_35500148_C_A | 4 | 35500148 | 2.88795×10-7 | LOC_Os04g59630 | Prenylcysteine oxidase 1 precursor putative expressed | |
| PXO99 | 7_26936907_G_A | 7 | 26936907 | 2.88795×10-7 | LOC_Os07g45120 | Expressed protein | |
| PXO99 | 8_10707525_C_A | 8 | 10707525 | 2.88795×10-7 | LOC_Os08g17480 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| PXO99 | 9_13062864_A_G | 9 | 13062864 | 2.88795×10-7 | LOC_Os09g21580 | OsFBX324 - F-box domain containing protein expressed | |
| PXO99 | 10_770633_A_C | 10 | 770633 | 2.88795×10-7 | LOC_Os10g02220 | Peptide transporter PTR2 putative expressed | |
| PXO99 | 10_4768566_T_C | 10 | 4768566 | 1.88802×10-6 | LOC_Os10g08820 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| PXO99 | 5_25082791_T_C | 5 | 25082791 | 3.02499×10-6 | LOC_Os05g43190 | Expressed protein | |
| PXO99 | 9_682155_C_T | 9 | 682155 | 3.65396×10-6 | LOC_Os09g01980 | Retrotransposon protein putative unclassified expressed | |
| PXO99 | 1_35729850_C_T | 1 | 35729850 | 3.96584×10-6 | LOC_Os01g61780 | Vacuolar ATP synthase 98 kDa subunit putative expressed | |
| PXO99 | 3_20089598_G_A | 3 | 20089598 | 4.86058×10-6 | LOC_Os03g36220 | Retrotransposon protein putative unclassified expressed | |
| PXO99 | 4_23009161_T_C | 4 | 23009161 | 5.65336×10-6 | LOC_Os04g38730 | Hypothetical protein | |
| PXO99 | 10_10280800_G_A | 10 | 10280800 | 8.21681×10-6 | LOC_Os10g20430 | Transposon protein putative CACTA En/Spm sub-class expressed | |
| PXO99 | 4_22488159_C_T | 4 | 22488159 | 8.54439×10-6 | LOC_Os04g37830 | Retrotransposon protein putative unclassified expressed | |
| PXO99 | 1_6763731_G_A | 1 | 6763731 | 1.45237×10-5 | LOC_Os01g12381 | GDSL-like lipase/acylhydrolase putative expressed | |
| PXO99 | 10_10622194_C_T | 10 | 10622194 | 2.05682×10-5 | LOC_Os10g20980 | Transposon protein putative CACTA En/Spm sub-class expressed | |
| PXO99 | 11_19490121_G_C | 11 | 19490121 | 2.45252×10-5 | LOC_Os11g32980 | Transposon protein putative unclassified expressed | |
| PXO99 | 6_15683763_C_T | 6 | 15683763 | 2.50426×10-5 | LOC_Os06g27700 | Retrotransposon protein putative unclassified expressed | |
| PXO99 | 2_11530742_G_A | 2 | 115307424 | 2.50426×10-5 | LOC_Os02g19710 | Retrotransposon protein putative unclassified expressed | |
| PXO99 | 8_14551707_C_T | 8 | 14551707 | 2.58147×10-5 | LOC_Os08g24080 | Retrotransposon protein putative unclassified | |
| PXO99 | 1_37300757_C_T | 1 | 37300757 | 2.64067×10-5 | LOC_Os01g64230 | Long cell-linked locus protein putative expressed | |
| PXO99 | 4_29550982_A_G | 4 | 29550982 | 2.67698×10-5 | LOC_Os04g49550 | RING-H2 finger protein ATL2A putative expressed | |
| PXO99 | 4_29550988_A_T | 4 | 29550988 | 2.67698×10-5 | |||
| PXO99 | 4_29550990_T_C | 4 | 29550990 | 2.67698×10-5 | |||
| PXO99 | 8_20326896_C_T | 8 | 20326896 | 2.96673×10-5 | LOC_Os08g32790 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| PXO99 | 7_9714575_A_T | 7 | 9714575 | 3.05773×10-5 | LOC_Os07g16560 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| PXO99 | 12_23369262_A_G | 12 | 23369262 | 3.32972×10-5 | LOC_Os12g38030 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| PXO99 | 6_9880157_A_G | 6 | 9880157 | 3.68628×10-5 | LOC_Os06g17040 | Retrotransposon protein putative unclassified expressed | |
| PXO99 | 6_27238355_G_T | 6 | 27238355 | 3.79721×10-5 | LOC_Os06g45030 | Transposon protein putative unclassified expressed | |
| PXO99 | 10_2015605_G_A | 10 | 2015605 | 3.98789×10-5 | LOC_Os10g04290 | Transposon protein putative CACTA En/Spm sub-class expressed | |
| PXO99 | 10_2015606_T_C | 10 | 2015606 | 3.98789×10-5 | |||
| PXO99 | 10_2015616_T_C | 10 | 2015616 | 3.98789×10-5 | |||
| PXO99 | 7_9797928_A_T | 7 | 9797928 | 4.00610×10-5 | LOC_Os07g16710 | Retrotransposon protein putative Ty3-gypsy subclass | |
| PXO99 | 12_23610190_C_T | 12 | 23610190 | 4.37810×10-5 | LOC_Os12g38460 | RNA recognition motif family protein expressed | |
| PXO99 | 11_3704859_G_A | 11 | 3704859 | 4.37973×10-5 | LOC_Os11g07350 | Expressed protein | |
| PXO99 | 6_27238423_T_C | 6 | 27238423 | 4.48254×10-5 | LOC_Os06g45030 | Transposon protein putative unclassified expressed | |
| PXO99 | 6_27238426_A_G | 6 | 27238426 | 4.48254×10-5 | LOC_Os06g45030 | Transposon protein putative unclassified expressed | |
| PXO99 | 11_16720749_G_A | 11 | 16720749 | 5.24152×10-5 | LOC_Os11g28890 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| PXO99 | 3_28507186_C_T | 3 | 28507186 | 5.35983×10-5 | LOC_Os03g49980 | Retrotransposon protein putative unclassified expressed | |
| PXO99 | 9_8088480_G_C | 9 | 8088480 | 5.59333×10-5 | LOC_Os09g13800 | Retrotransposon protein putative Ty1-copia subclass expressed | |
| PXO99 | 8_15034247_T_G | 8 | 15034247 | 6.82446×10-5 | LOC_Os08g24850 | Retrotransposon putative centromere-specific expressed | |
| PXO99 | 1_40956241_G_A | 1 | 40956241 | 7.49332×10-5 | LOC_Os01g70760 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| PXO99 | 1_40956243_C_T | 1 | 40956243 | 7.49332×10-5 | |||
| PXO99 | 1_40956268_A_G | 1 | 40956268 | 7.49332×10-5 | |||
| PXO99 | 8_10707195_C_A | 8 | 10707195 | 9.39077×10-5 | LOC_Os08g17480 | Retrotransposon protein putative Ty3-gypsy subclass expressed | |
| PXO99 | 12_23610183_G_A | 12 | 23610183 | 9.70588×10-5 | LOC_Os12g38460 | RNA recognition motif family protein expressed | |
新窗口打开|下载CSV
进一步统计与CR4、PXO96和PXO99关联的SNPs标记: 接种CR4后的重组自交系群体在P < 1×10-4水平检测到25个SNPs, 贡献率为61.00%并与19个基因关联, 其中有8个坐落在基因编码区。SNP_11_26502942_C_A的P值最小为1.7126×10-5, 位于11号染色体上编码WD结构域的蛋白。结合功能预测发现2个标记SNP_11_27792921_A_G (P=2.70257×10-5)和SNP_11_26220673_C_T (P = 5.00378×10-5)均编码典型的NBS-LRR抗病结构域蛋白; 其余基因主要编码肌钙蛋白还原酶、MADS家族蛋白和表达蛋白等。
在P < 1×10-6和1×10-5水平下, 接种PXO96后分别检测到1个和4个SNPs标记, 其中SNP_5_ 14854774_G_A (P=3.835×10-6)位于基因LOC_ Os05g25560编码区, 为糖基水解酶10家族蛋白; 在P < 1×10-4水平下, 鉴定到32个SNPs标记计贡献率为59.78%, 共得到26个基因; 其中标记SNP_2_19389629_A_G (P=4.7609×10-5)附近基因LOC_Os02g32680编码凝集素类受体蛋白激酶基因; 其余基因编码反转座子蛋白、转座子蛋白和表达蛋白等。
接种PXO99后在上述3个不同P值水平下分别检测到12个、20个和52个SNPs, 共解释表型变异率为63.29%并得到42个基因。在P < 1×10-6水平找到12个显著SNPs标记, 贡献率为33.08%并与12个基因关联。在P=2.89×10-7水平下发现SNP_4_ 35500148_C_A、SNP_9_13062864_A_G和SNP_ 10_770633_A_C分别位于4号、9号和10号染色体上基因编码异戊二烯氧化酶1、RING-H2蛋白、F-box蛋白和多肽转移酶蛋白; 其余基因主要编码ATP酶、反转座子蛋白和表达蛋白等。
2.5 候选抗病相关基因的表达分析和进化分析
为了验证新功能位点附近的NBS-LRR编码蛋白基因在调控白叶枯病抗性中发挥作用, 选择了3个显著关联的SNPs: SNP_11_26220673_C_T (P= 5.00378×10-5)、SNP_11_27792921_A_G (P=2.70257× 10-5)和SNP_6_25611297_T_C (P=8.66×10-7)。由此将位点附近的3个候选基因LOC_Os11g43420、LOC_Os11g45930和LOC_Os06g42590以及白叶枯病抗性基因Xa21作为标志基因, 利用实时定量PCR方法测定接种白叶枯菌前、后的表达量变化(图6)。结果显示该3个候选基因和Xa21在抗性亲本中恢9308中表达量均高于感病亲本协青早B。其中LOC_ Os11g43420和LOC_Os06g42590在接种前双亲间表达量无显著差异, 接种后1 d和4 d该基因在抗性亲本中恢9308显著上调表达; LOC_Os11g45930和Xa21在接种前后抗病亲本中均显著高于感病母本协青早B。图6
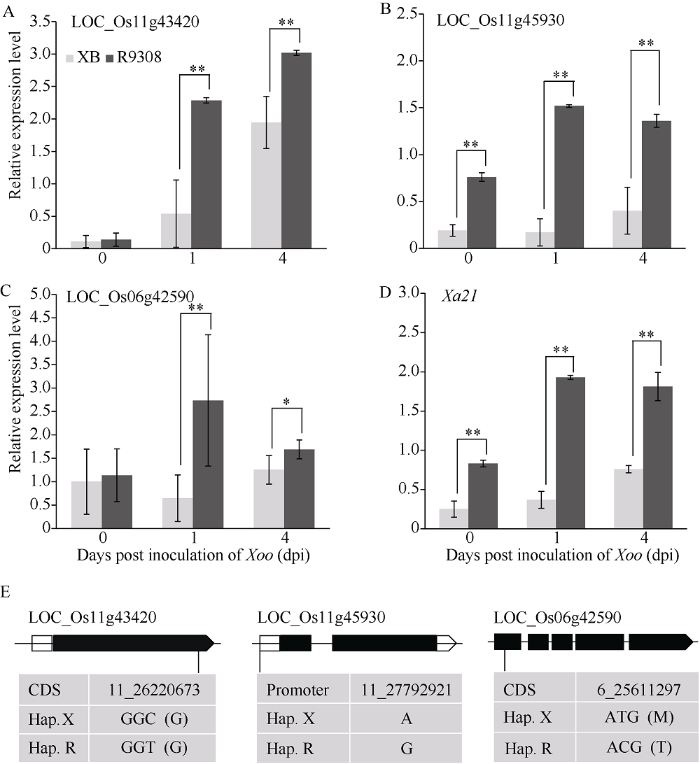 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6候选白叶枯抗性基因的相对表达量分析
A~D分别包括关联位点SNP_11_26220673_C_T (P = 5.00378× 10-5)、SNP_11_27792921_A_G (P = 2.70257×10-5)和SNP_6_ 25611297_T_C (P = 8.66×10-7)和Xa21基因的qRT-PCR分析。横坐标表示经过接菌时间: “0”表示未接生理菌株; “1”表示接种生理菌株当天; “4”表示接种生理菌株后4 d, 在相同的时间点对同一批次水稻植株的叶片进行取样。纵坐标表示以actin作为内参基因计算相对表达水平(folds)。E表示3个候选基因(LOC_Os11g43420、LOC_Os11g45930和LOC_Os06g42590) SNP突变分别在编码区和启动子区域。*表示P ≤ 0.05和**表示P ≤ 0.01 (Student’s t-test)。
Fig. 6Relative expression profile of candidate genes
A-D indicate the qRT-PCR results of four genes including SNP_11_ 26220673_C_T (P=5.00378×10-5), SNP_11_27792921_A_G (P=2.70257×10-5), SNP_ 6_25611297_T_C (P=8.66×10-7), and Xa21 respectively. The horizontal axis of each bar chart is the days post inoculation of Xoo strains “0” means the samples have not been inoculated; “1” means the samples have been inoculated for one day; “4” means the samples have been inoculated for four days. The inoculated leaves were collected from the same group at three different days with at least three biological repeats. The vertical axis is the relative expression level (folds) with actin as the internal reference gene. E indicates SNP polymorphism of three candidate genes (LOC_Os11g43420, LOC_Os11g45930, and LOC_Os06g42590) in coding sequence or promoter region. *: P ≤ 0.05 **: P ≤ 0.01 (Student’s t-test).
为了探明本研究得到的4个显著候选基因与已经克隆的Xa抗病基因的进化关系(表3), 进化树分析结果发现本研究找到的4个基因与Xa1、Xa14聚在同一组, 其中LOC_Os11g43420和LOC_ Os11g45930同源性最高且处于同亚组(图7)。
Table 3
表3
表3已克隆的水稻白叶枯病抗性基因
Table 3
| R基因 R gene | 位点 Loci (MSU format) | 染色体 Chr. | 物理位置 Position | 参考文献 Reference |
|---|---|---|---|---|
| Xa1 | LOC_Os04g53120 | 4 | 31638099-31644795 | Yoshimura et al. [9] |
| Xa3/Xa26 | LOC_Os11g47210 | 11 | 28399360-28402773 | Sun et al. [13] |
| Xa4 | LOC_Os11g46870 | 11 | 28142192-28148337 | Sun et al. [14] |
| xa5 | LOC_Os05g01710 | 5 | 437013-443270 | Blair et al. [11]; |
| Xa10 | LOC_Os11g37570 | 11 | 22181556-22181152 | Tian et al. [16] |
| xa13 | LOC_Os08g42350 | 8 | 26728795-26725898 | Chu et al. [10] |
| Xa21 | LOC_Os11g35500 | 11 | 20802924-20806518 | Song et al. [17 ]; |
| Xa23 | LOC_Os11g37620 | 11 | 22204676-22203734 | Wang et al. [15 ] |
| Xa25 | LOC_Os12g29220 | 12 | 17305326-17302073 | Triplettl et al [55]. |
| Xa27 | LOC_Os06g39810 | 6 | 23654303-23653851 | Gu et al. [12] |
| xa41(t) | LOC_Os11g31190 | 11 | 18171678-18174478 | Hutin et al. [6] |
新窗口打开|下载CSV
图7
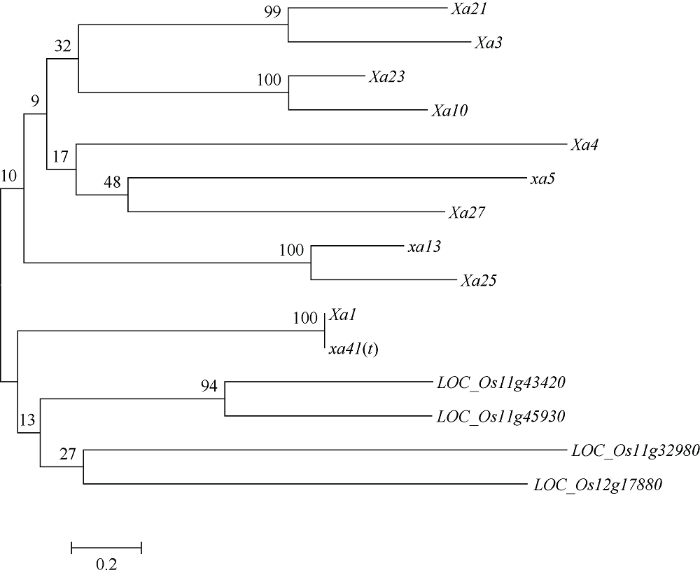 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7候选白叶枯抗性基因的进化分析
利用多重比对软件Clastal X将LOC_Os11g43420、LOC_Os11g45930、LOC_Os11g32980、LOC_Os12g17880、Xa1、Xa3/Xa26、Xa4、xa5、Xa10、xa13、Xa21、Xa23、Xa25、Xa27和xa41(t)比对后, 使用MEGA软件最大邻间法构建进化树。
Fig. 7Phylogenetic tree of bacterial blight candidate resistance genes in rice
Neighbor-joining tree are constructed using MEGA based on multiple sequence alignments made by Clastal X. Selected protein names and MSU loci are as follows: LOC_Os11g43420, LOC_Os11g45930, LOC_Os11g32980, LOC_Os12g17880, Xa1, Xa3/Xa26, Xa4, xa5, Xa10, xa13, Xa21, Xa23, Xa25, Xa27, and xa41(t).
3 讨论
在过去的20年中, 水稻抗白叶枯病主效抗性基因(R gene)的鉴定和定位研究以及抗性基因的大量利用极大地弥补了化学防治的缺点。然而传统的QTL定位获得的抗病基因大部分具有小种特异性[40,41], 难以在育种上应用, 少量白叶枯广谱抗性基因如Xa4和Xa21的大规模推广又导致病原菌进化从而抗性逐年丧失, 针对该问题本研究旨在利用高质量的SNP标记结合具有多年持久抗性的育种材料协优9308, 挖掘水稻白叶枯病的广谱、持久抗性基因来拓展抗病育种材料[42]。3.1 借助极端抗、感表型双亲构建作图群体
本研究选用的育种材料为协优9308衍生的重组自交系, 其父本9308系谱分析发现含有籼稻品种IR26血缘其包含白叶枯抗性基因的遗传背景[43], 在浙江省区试鉴定表现为中抗白叶枯病。原始亲本协青早B和中恢9308经同时接种生理菌株后分别表现为高感和高抗白叶枯病, 且病斑长度经t检验证实存在极显著差异(P < 0.01); 协优9308的139个重组自交系及双亲群体白叶枯病斑长度均呈现为连续表型且变异范围广, 出现抗、感双向超亲分离现象, 属于典型的数量性状遗传。Zhou等[38]和本研究计算全基因组的LD衰减距离约为2 MB范围。利用构建的重组自交系群体进行全基因关联分析可行。在文献中也已有报道, 例如, Kou等[44]在极端抗性和感性表型双亲的水稻定位群体中, 利用分子标记技术筛选到多个与已报道过抗病基因连锁的多态性标记。有趣的是, 在该研究中发现了LD衰减速度影响因素, 整合前人研究与本研究关于水稻不同群体的连锁不平衡分析, 发现由于不同的遗传背景, LD衰减速度存在较大的差异。我们的重组自交系群体比起野生稻有更大的LD衰减距离, 其在2000 kb范围内, 推断存在偏分离现象导致群体的遗传多样性下降, 位点间的相关性(连锁程度)加强[45]。3.2 白叶枯抗性全基因组关联分析结果分析和比较
相比于少量标记的QTL定位研究, 经高通量测序构建的高密度SNP标记可发现更多的关联位点。本研究协优9308衍生的重组自交系群体, 该群体遗传背景相对可控、主成分分析未发现明显的群体结构, 参考前人研究结果确定P < 1.0×10-4作为显著性筛选标准[46,47], 利用高效的混合线性模型该标准下共找到109个关联位点, 分别坐落于4号、9号、10号和11号染色体上。前人研究发现白叶枯主效抗性基因定位在1号、4号、5号、8号和11号染色体上, 其中在11号染色体上分布最多(前人研究发现大部分白叶枯抗性基因均编码NBS-LRR蛋白, 通常呈基因簇的形式出现, 且在调控水稻广谱抗病发挥重要作用, 2个含有NBS- LRR结构域的抗病基因显著关联且位于11号染色体上[15,17]。本研究中, CR4菌株处理出现最显著的位点SNP_11_26502942_C_A (P=1.7126×10-5), 并未在已发现的抗病候选基因中, 这可能与已知抗性基因及其附近的基因尚未注释有关[48], 有待后续的研究进一步功能验证。在PXO96和PXO99菌株处理组发现显著位点信号SNP_6_25611297_T_C和SNP_ 1_19343496_G_A, 其对应P值分别为8.66084×10-7和2.88795×10-7, 表明这些位点遗传效应较大可进一步加以功能验证。经GWAS分析找到2个NBS- LRR基因包括LOC_Os11g43420和LOC_ Os11g45930, 进化分析结果显示和已克隆的11个白叶枯抗性基因位于不同的亚组, 表明该2个基因可能为新型的白叶枯抗病候选基因, 有待进一步证实。本研究还找到了很多凝集素受体蛋白激酶(lectin receptor kinase)、纤维素合成酶(cellulose synthase)、MYB转录因子以及表达蛋白可能和持久抗性相关。
不同白叶枯生理菌株鉴定得到的SNPs进行基因注释, 发现所有SNPs位点均未发现与已克隆的白叶枯抗性基因重叠, 可能不同的生理小种以及遗传材料介导的抗病机制存在较大差异[41]。值得一提的是本研究4个生理菌株处理后均筛选到大量显著性标记编码转座子和反转录转座子蛋白, 过去研究发现该类蛋白可能通过顺式作用元件调控旁侧基因表达从而促进抗性基因的进化[49,50], 由此极大地丰富了白叶枯抗性基因的多样性。上述结果暗示白叶枯抗性遗传受主效位点的R基因以及微效多基因共同调控[51,52]。
3.3 编码典型抗病结构蛋白的初步基因功能验证
植物抗性基因中大多数包含NBS-LRR类抗病结构域[53], 本研究借助水稻基因组重测序构建的SNP图谱进行全基因组关联分析找到2个NBS-LRR基因, 对于进一步了解该基因的群体变异、揭示水稻抗病遗传机理、提高抗病育种的效率和水稻产量具有重要意义。本研究利用实时定量PCR对3个候选基因(显著性最高的SNP基因LOC_Os06g42590、包含NBS-LRR类抗病结构域基因LOC_Os11g43420和LOC_Os11g45930)以及广谱抗病基因Xa21作为标志基因进行了初步验证, 结果显示: 接种前LOC_ Os11g43420和LOC_Os06g42590表达量在双亲间无显著差异, 接种后在抗性亲本的表达量均高于感病亲本; LOC_Os11g45930和Xa21在接种前、后抗病和感病品种中的表达量存在显著差异(P < 0.01), 说明该3个候选基因可能正调控白叶枯抗性(图5)。进化分析结果显示2个NBS-LRR基因(LOC_ Os11g43420和LOC_Os11g45930)和已克隆的白叶枯抗性基因属于不同的亚组, 表明该2个基因为新型的白叶枯抗病候选基因, 为进一步功能验证奠定了基础。3.4 抗性关联分析结果中相关问题的探讨
本研究通过全基因组关联分析方法, 在3个生理菌株处理后, 检测到3个共同的SNP位点(2_ 11969901_C_T、2_12653706_C_A和2_11530742_ G_A), 但不属于已报道水稻白叶枯抗性基因关联的位点。其中CR4和PXO99菌株处理后, 分别在7号染色体7-9599243位点以及相邻位点7-9714575同时检测到P<1×10-5分子标记; PXO96和PXO99菌株处理后分别在10号染色体10-10043595位点以及相邻位点10-10280800同时检测到P<1×10-5分子标记(表2)。在3个生理菌株下也发现许多不同或不重叠的SNP位点, 这可能与不同的生理菌株被不同的抗性基因所识别, 且由抗性基因调控的一系列下游抗病信号通路相关基因也存在差异等有关。也许不同的生理菌株处理后得到了不同的SNP位点, 进一步验证了抗性基因存在小种专化性的假设[51]。同时, 本研究材料中的抗性可能是主效基因和微效多基因共同作用的结果, 由于重组自交系数目偏少, 本研究关联分析对小效应SNP解析效果可能不高, 需增大样本量以提高GWAS关联分析的功效。GWAS作为筛选工具, 可适当地放宽显著性标准, 结合基因和蛋白质在表型表达中调控分析, 为植物抗性研究和育种实践有效鉴定候选基因提供潜在的途径。然而, 目前对基因功能研究尚不够完善, 应用仍受限制, 若整合多个表达数据库创建基因共表达网络, 与GWAS紧密结合, 以辅助我们对植物基因组功能的认知, 更好地理解复杂的遗传问题[54]。
此外, PXO61菌株处理后表型未服从正态分布, 这可能该菌株毒性较弱且感病亲本协青早B发病程度较轻有关, 也可能由于白叶枯接种效果受田间环境影响较大, 该年度接种后出现的低温天气对发病进程存在较大影响, 其分析结果暂不列入本文结果讨论中。
4 结论
本研究通过全基因组关联分析获得109个与白叶枯抗性关联的显著性SNP标记, 进一步结合基因注释和实时定量PCR, 初步验证了2个NBS-LRR类抗病基因在抗病/感病亲本的表达情况; 研究结果为后续水稻白叶枯病抗性的遗传改良和分子机制研究提供了候选基因资源和遗传材料。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 3]
[本文引用: 3]
[本文引用: 2]
[本文引用: 4]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
