 ,1,2,**, 张春
,1,2,**, 张春 ,2,**, 王作平2, 邹华文
,2,**, 王作平2, 邹华文 ,1,*, 吴忠义
,1,*, 吴忠义 ,2,*
,2,*Cloning and functional analysis of ZmbHLH161 gene in maize
YANG Meng-Ting ,1,2,**, ZHANG Chun
,1,2,**, ZHANG Chun ,2,**, WANG Zuo-Ping2, ZOU Hua-Wen
,2,**, WANG Zuo-Ping2, ZOU Hua-Wen ,1,*, WU Zhong-Yi
,1,*, WU Zhong-Yi ,2,*
,2,*通讯作者:
收稿日期:2020-03-28接受日期:2020-06-2网络出版日期:2020-06-22
| 基金资助: |
Received:2020-03-28Accepted:2020-06-2Online:2020-06-22
| Fund supported: |

摘要
关键词:
Abstract
Keywords:
PDF (4689KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
杨梦婷, 张春, 王作平, 邹华文, 吴忠义. 玉米ZmbHLH161基因的克隆及功能研究[J]. 作物学报, 2020, 46(12): 2008-2016. doi:10.3724/SP.J.1006.2020.03022
YANG Meng-Ting, ZHANG Chun, WANG Zuo-Ping, ZOU Hua-Wen, WU Zhong-Yi.
玉米是我国第一大农作物, 种植面积3300多万公顷, 其总产量2亿多吨, 在我国粮食安全和国民经济发展中起着重要作用[1]。玉米起源于南美洲高温多湿的热带地区, 喜温、需水较多、耐旱性不强, 耐盐性较差[2]。如今, 随着人类活动范围的扩大, 生态环境也遭受着巨大的破环, 如土壤沙漠化、盐碱化、水流污染和地下水枯竭等, 而土壤干旱、盐碱化则严重影响玉米产量, 如何提高玉米对干旱、高盐等不利因素的抵抗能力越来越受到重视。
转录因子(transcription factors) 是RNA聚合酶在真核生物特异启动子上起始转录所需要的作用因子, 由DNA结合区、转录调控区、寡聚化位点和核定位信号区组成[3]。bHLH (碱性/螺旋-环-螺旋, basic/Helix-Loop-Helix)转录因子名称来自其结构中的bHLH基序, bHLH结构域由50~60个氨基酸组成, 其结构包括2个保守区域: 一是长度为10~15个氨基酸的碱性区域, 分布在多肽链的N端, 含有大量碱性氨基酸, 与DNA结合相关; 另一为长度40个氨基酸左右HLH区域, 分布在C端, 主要由疏水氨基酸残基构成, 利于HLH之间相互作用形成二聚体, 其中环的长度在不同bHLH蛋白中会有差异。1989年Murre等[4]从小鼠中首次鉴定出bHLH转录因子E12和E47, 之后越来越多bHLH转录因子在线虫、果蝇、小鼠和人类中得到了分离鉴定。Nuno等[5]发现, 在4.4亿年前的早期陆地植物中就已经存在bHLH转录因子家族。在研究较为深入的动物中, 依据其进化关系和DNA结合模式, 将bHLH蛋白分为6个组(A~F组)[6], 而植物中bHLH蛋白大多数都属于B组。与植物逆境抗性相关转录因子家族中, 研究较多的有bZIP、WRKY、AP2/EREBP和MYB四大类[7], bHLH转录因子家族在植物抗逆中的研究则相对滞后, 正因如此, 引起了科学家们广泛关注。bHLH转录因子是一个大家族, 在调节植物生长发育[8,9,10,11]、信号转导[12,13,14,15]和逆境胁迫响应[16,17,18,19]方面发挥着重要作用。其中bHLH转录因子ZmPTF1基因不仅能促进玉米侧根生长发育, 而且对促进ABA合成、信号转导和提高耐旱性等方面都起到正向调控作用[20]。目前, 在拟南芥和水稻中分别鉴定到了162个和167个bHLH家族成员[21,22]。Zhang等[23]利用全基因组分析在玉米中鉴定到208个bHLH蛋白, 并通过与拟南芥bHLH蛋白进行比较, 将这些蛋白划分为18个亚家族。而其他植物因为基因组测序工作开展得较晚, 因此对其基因组中的bHLH家族成员分析还未见报道。由于已知的植物bHLH基因序列主要来源于水稻和拟南芥这两种模式植物, 对其他植物bHLH蛋白的功能研究开展的还比较少。
前期工作中, 我们对玉米根系生长发育4个关键时期进行了转录组测序分析, 在相邻时期间均显著差异表达的转录因子中, 鉴定到唯一一个bHLH家族转录因子基因ZmbHLH161, 推测该基因可能在调控玉米根系生长发育过程中发挥重要的作用, 那么作为bHLH转录因子家族的成员之一, 它是否也参与逆境胁迫响应呢?为了证实这一猜测, 我们首先克隆了玉米ZmbHLH161基因, 对其表达模式、亚细胞定位以及在高盐、脱水、低温和干旱胁迫处理下的表达情况进行了研究; 然后将该基因在拟南芥中进行了异源表达, 利用转基因拟南芥株系进行相关耐逆性研究, 初步探索了玉米转录因子ZmbHLH161的功能, 旨在为玉米抗逆遗传改良提供参考依据。
1 材料与方法
1.1 植物材料及试剂
试验材料为玉米自交系B73、郑58、野生型拟南芥(Col-0)和农杆菌GV3101, 以上材料均为本实验室提供。试验所用试剂为大肠杆菌Trans1-T1、pEASY T5 Zero Clone载体、RNA提取试剂盒、逆转录试剂盒等均购于北京全式金公司; Ex Taq、LA Taq、SYBR Premix Ex Taq II (Tli RNaseH Plus)等购于TaKaRa公司; EDTA、蔗糖、NaCl、乙醇、Tris、磷酸氢二钠、磷酸二氢钠酚、氯仿、异戊醇、异丙醇、浓硫酸、PEG-4000、冰醋酸、浓盐酸、甘油、琼脂糖、蛋白胨、酵母粉和β-巯基乙醇等生化试剂均购自生工生物工程(上海)股份有限公司, 所有的测序工作都由生工生物工程(上海)股份有限公司完成。
试验所用设备为低温高速台式离心机(型号: MUITLFUGE X1R); 低温台式水平离心机(型号: SC-3612); PCR仪(型号: T100 Thermal Cycler); 激光共聚焦显微镜(型号: Leica TCS SP8)。
玉米材料处理: 挑选颗粒饱满的自交系B73种子种植于温室花盆中, 分别设置对照组、脱水处理组、盐(0.2 mol L-1 NaCl)处理组、冷(4℃)处理组、不浇水干旱处理组, 每组6盆, 每盆2株, 各设3个生物学重复。玉米幼苗正常生长至三叶一心时分别进行脱水、高盐、冷、干旱处理, 分别取脱水(将盆栽三叶期玉米幼苗整株取出, 用流水冲洗掉泥土, 用吸水纸吸收表面多余水分后, 室温放置, 自然脱水)、高盐、冷处理0、1、2、5、10和24 h后材料的叶片和根, -80℃冻存。不浇水干旱处理取0、3、7和12 d后的叶片和根(其中0 d为最后一次充分浇水时取样), -80℃冻存。组织差异表达材料取露地栽培自然生长至三叶期幼苗的根、叶以及抽雄期的根、茎、叶、雄穗、花丝、幼胚, -80℃冻存。
1.2 ZmbHLH161 cDNA的克隆及生物信息学分析
按照Transzol Up Plus RNA Kit试剂盒(北京全式金公司)说明提取玉米根组织的总RNA, 逆转录得到cDNA模板。依据ZmbHLH161基因信息利用Primer 5设计1号引物对(表1), 以逆转录的cDNA为模板进行PCR扩增, 反应程序为: 94℃预变性60 s; 98℃ 10 s, 68℃ 70 s, 30个循环; 72℃延伸10 min, 获得ZmbHLH161全长。PCR产物经胶回收后连接至T5 Zero载体, 获得单克隆后送生工生物工程(上海)股份有限公司进行菌落测序。各引物序列见表1。Table 1
表1
表1本试验中所用的引物
Table 1
| 引物对编号 Number of primer pairs | 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) |
|---|---|---|
| 1 | pZmbHLH161-F | AGCGTTAGTCACAGTTGTTGCC |
| pZmbHLH161-R | CGTAAGCTGGTCCAATCGGTAC | |
| 2 | pZmbHLH161SL-F | ATGAATTCATGGCGCTCGAAG (EcoR I) |
| pZmbHLH161SL-R | AATAAGCTTGTGGCCTGGCC (Hind III) | |
| 3 | pZmbHLH161RT-F | AGCATCAGCTCCTGTCCGTGTC |
| pZmbHLH161RT-R | CTCTCCATCTCGTCCGTGTTCC | |
| 4 | pZmbHLH161At-F | ATGAATTCAGCGTTAGTCACAGTTGTTGCC (EcoR I) |
| pZmbHLH161At-R | ATGGTACCTCAGTGGCCTGGCCCCCCGC (Kpn I) | |
| 5 | Actin-F | TACGAGATGCCTGATGGTCAGGTCA |
| Actin-R | TGGAGTTGTACGTGGCCTCATGGAC |
新窗口打开|下载CSV
用DNAMAN 8软件进行序列比对分析, 应用ProParam (
1.2.1 植物表达载体pYBA1132:ZmbHLH161:EGFP的构建 使用2号引物对进行扩增, 获得在5′端和3′端分别引入EcoR I、Hind III酶切位点的ZmbHLH161基因编码区, PCR产物经EcoR I、Hind III双酶切连接至PYBA1132表达载体上获得重组载体。将重组载体转化大肠杆菌(Trans1-T1)后, 挑取阳性克隆测序验证。
1.2.2 玉米原生质体的提取及转化 以玉米自交系郑58黄化苗胚芽鞘为材料, 依据Song-Dong[24]的方法进行改良后提取原生质体并转化。转化后的原生质体23℃黑暗孵育16 h后, 轻轻混匀后再加入终浓度为5 μmol L-1的DAPI (4,6-diamidino-2-phenylindole)染色5 min, PBS缓冲液洗涤后置于激光共聚焦显微镜下观看荧光情况。
1.3 玉米ZmbHLH161基因表达模式分析
按照试剂盒的说明提取地上部及根RNA, 逆转录后作为qPCR模板。设计3号特异引物对及内参基因Actin特异引物5号引物对。应用BIO-RAD CFX96 Real-Time System qPCR仪进行实时荧光扩增。反应体系如下: 10 μL 2×SYBR Premix Ex Taq、0.5 μL GSP (10 μmol L-1)、1 μL cDNA模板、8.5 μL ddH2O。PCR条件: 95℃ 30 s; 95℃ 10 s, 60℃ 20 s, 共40个循环; 融解曲线分析, Step 1: 95℃ 10 s; Step 2: 65℃ 1 min; Step 3: 95℃ 15 s。每组试验设3个生物学样本, 每个生物学样本设3个技术重复, 数据为3个生物学重复平均值±标准误。应用2-??CT方法分析ZmbHLH161在各组织中的相对表达情况[25]。逆境处理试验分别以0 h时, ZmbHLH161在幼苗根和叶片的表达量计为1, 组织差异表达试验中以抽雄期花丝的该基因表达量计为1。1.4 植物异源表达载体pYBA1132:ZmbHLH161的构建及拟南芥遗传转化
1.4.1 异源表达载体构建 设计4号引物对扩增获得含有酶切位点的ZmbHLH161编码区片段。PCR产物经EcoR I、Kpn I双酶切连接至pYBA1132表达载体上获得重组载体, 将重组载体转化大肠杆菌(Trans1-T1)后, 挑取阳性克隆进行菌体PCR检验及酶切验证, 并送生工生物工程(上海)股份有限公司测序。1.4.2 转化农杆菌GV3101及侵染拟南芥 pYBA1132: ZmbHLH161重组质粒转入农杆菌GV3101, 筛选鉴定后通过浸花法[26]侵染拟南芥幼苗。单株收取T0代种子, 经卡那霉素抗性筛选后获得T1代转基因拟南芥。提取T1代幼苗叶片基因组DNA为模板, 以4号引物对为引物进行PCR检测, 单株收获阳性植株种子并加代获得T3代纯合植株。
1.4.3 转基因拟南芥种子逆境处理 利用含NaCl、甘露醇的1/2 MS培养基对转基因拟南芥分别模拟盐和渗透胁迫。NaCl浓度梯度为0、0.10、0.15和0.18 mol L-1, 甘露醇浓度梯度为0、0.15、0.20和0.30 mol L-1, 调pH 5.8。逆境胁迫处理试验: 将拟南芥种子用消毒液消毒后, 将1/2 MS固体培养基上生长7 d的转基因和野生型拟南芥幼苗(根长相同), 分别移栽到含不同浓度NaCl、甘露醇的1/2 MS固体培养基上。培养基分为两部分, 野生型和转基因拟南芥各占一半, 各移6株, 设3个重复, 垂直放置于恒温培养箱中培养, 7 d后统计拟南芥根长。
2 结果与分析
2.1 玉米转录因子基因ZmbHLH161序列扩增与生物信息学分析
从玉米根中提取RNA, 经反转录获得cDNA, 以其为模,板用1号引物对扩增获得目的基因, 经测序表明, ZmbHLH161基因的cDNA序列全长1460 bp, 编码352个氨基酸。氨基酸序列比对表明, ZmbHLH161基因含有典型的HLH结构域(图1-A)。生物信息学分析表明, 其蛋白分子量为37.1 kD, 理论等电点pI为6.10; 蛋白二级结构无规则卷曲所占比例最大, 为42.05%, 其次是α螺旋(34.66%), 其空间结构预测结果如图1-B所示。该蛋白无信号肽, 为亲水性蛋白, 无跨膜结构区(图1-C)。亚细胞定位分析预测可能定位在细胞核。图1
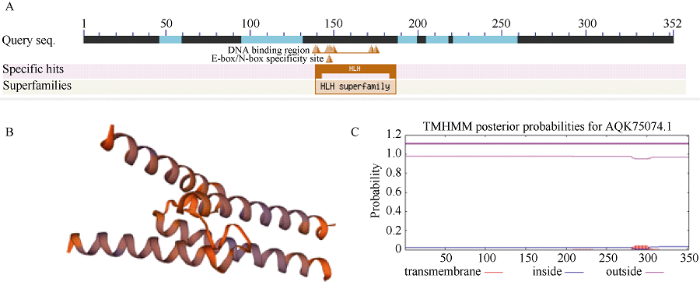 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1ZmbHLH161生物信息学分析
A: 氨基酸序列比对; B: 蛋白二级结构预测; C: 跨膜结构预测。
Fig. 1Bioinformatics analysis of ZmbHLH161
A: amino acid sequence alignment; B: protein secondary structure prediction; C: transmembrane structure prediction.
序列比对结果显示, ZmbHLH161 (Zm00001d018119)和ZmbHLH36 (Zm00001d052192)同水稻OsbHLH048 (Os02g0759000)同源性较高, 与拟南芥中的AT1G22490、AT1G72210 (AtbHLH96)和AT5G65320 (AtbHLH99)具有较高的相似性(图2)。
图2
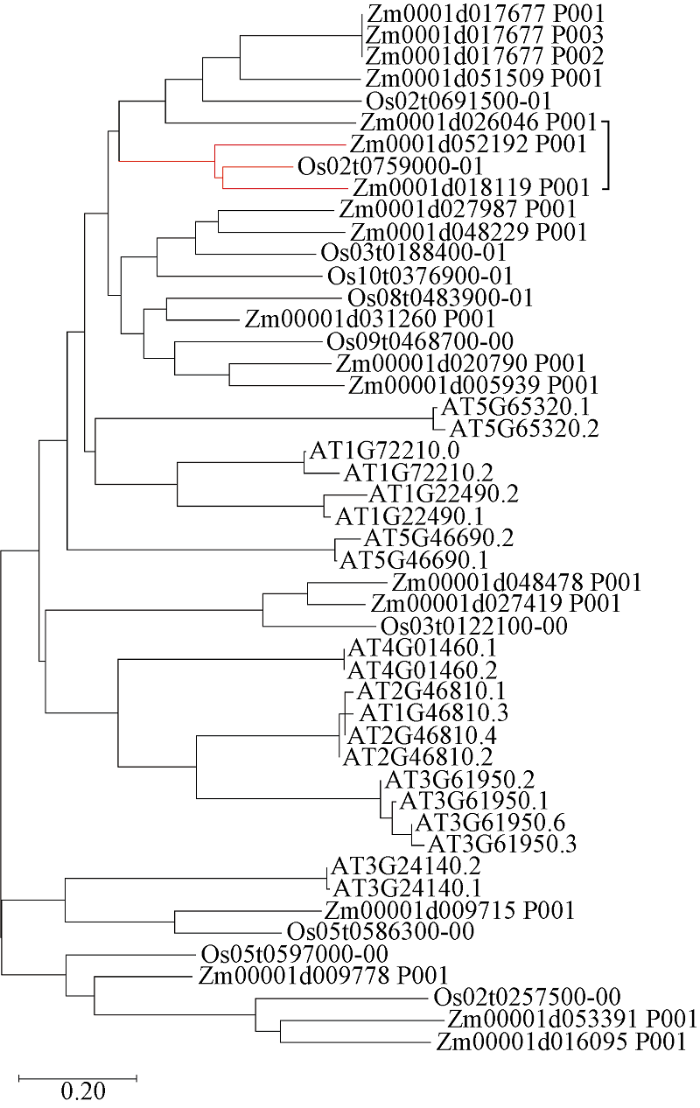 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2拟南芥、水稻和玉米bHLH转录因子氨基酸序列的系统发育树
Fig. 2Phylogenetic tree of the deduced amino acid sequences of the bHLH transcription factors in Arabidopsis, rice, and maize
启动子序列分析结果如图3, ZmbHLH161基因启动子含有多个与逆境胁迫相关的顺式作用元件, 如ABRE响应ABA胁迫、MYB响应干旱胁迫以及MYC响应ABA、低温胁迫。
图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3ZmbHLH161启动子序列分析
Fig. 3Promoter analysis of ZmbHLH161
2.2 亚细胞定位结果与分析
构建瞬时表达载体pYBA1132:ZmbHLH161:EGFP并转化玉米原生质体, 通过融合蛋白GFP荧光显示目的蛋白的亚细胞定位。原生质体经DAPI染色后置于激光共聚焦显微镜下观察发现(图4),转有绿色荧光蛋白空载体对照pYBA1132的玉米原生质体中, 整个细胞都有绿色荧光分布, 无明显细胞器定位特异性, 而转入目的基因的原生质体中, GFP绿色荧光只分布在细胞核, 并且与核特异染料DAPI的蓝色荧光完全重合, 表明ZmbHLH161定位于细胞核内。图4
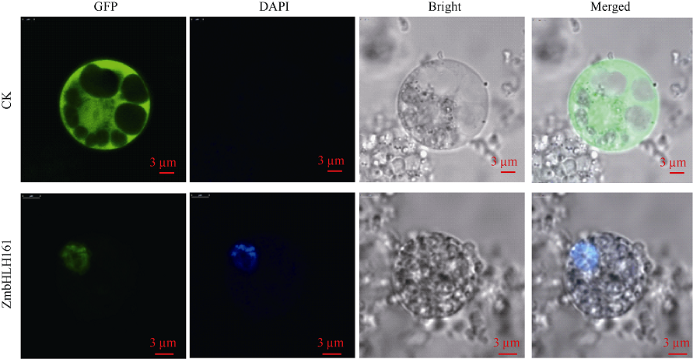 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4ZmbHLH161蛋白在玉米叶片原生质体中的亚细胞定位
GFP: GFP标记的ZmbHLH161在玉米原生质体的亚细胞定位; DAPI: DAPI标记的原生质体细胞核; Bright: 同一视野光镜下的原生质体; Merged: 光镜及荧光照片的叠加; CK: 转入空载体的原生质体; ZmbHLH161: 转入目的基因载体的原生质体; 标尺为3 μm。
Fig. 4Subcellular localization of ZmbHLH161 protein in maize leaf protoplasts
GFP: GFP-labeled ZmbHLH161 subcellular localization in maize protoplasts; DAPI: DAPI-labeled protoplast nuclei; Bright: protoplasts under the same field of view light microscope; Merged: superposition of light microscope and fluorescence photos; CK: transfer empty vector protoplasts; ZmbHLH161: transfer into protoplast of target vector; Bar = 3 μm.
2.3 玉米ZmbHLH161在不同组织中的表达情况
分别提取三叶期幼苗的根、叶, 抽雄期的根、茎、叶、雄穗、花丝和幼胚组织总RNA, 逆转录获得cDNA后进行qPCR分析, 结果表明, ZmbHLH161在不同组织中表达差异较大, 以抽雄期花丝中的表达量为标准进行比较, 生长旺盛期根中的表达量最高, 约为花丝中表达量的29.2倍; 其次是幼根和幼胚, 分别为8.3倍和5.4倍; 其他组织的表达量均低于花丝, 幼叶中的表达量最低(图5), 表明ZmbHLH161基因可能参与玉米根和幼胚的生长发育。图5
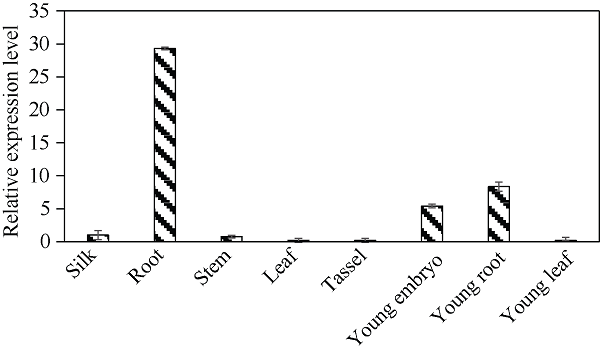 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5ZmbHLH161在玉米不同组织中的相对表达量
Fig. 5Relative expression level of ZmbHLH161 in different maize tissues
2.4 不同逆境处理对叶片和根中ZmbHLH161表达的影响
由图6可知, 不浇水干旱处理下, 叶片中ZmbHLH161的表达量在1~7 d一直呈下调趋势, 至12 d上调, 约为对照8.6倍, 而在根中则一直呈下调趋势。在脱水处理下, 叶片中ZmbHLH161的表达量在1 h内上调, 然后下调, 到5 h时约为对照的18倍, 随后上调并在10 h时达到峰值, 约为对照的82倍, 24 h时表达量约为对照的72倍; 而根中ZmbHLH161的表达量基本保持不变。低温处理下, 叶片中ZmbHLH161的表达量在1 h上调至对照1.9倍左右, 随后则是下调, 到24 h时为对照0.3倍; 根中先上调在2 h时达到峰值, 约为对照的1.4倍然后回落, 至24 h时约为对照的0.28倍。盐处理下, ZmbHLH161的表达量在叶片中急剧下调, 至24 h时约为对照的0.01倍; 根中先下调至5 h时约为对照的0.72倍, 后逐渐上调至24 h时, 约为对照1.5倍左右。以上结果表明, 玉米叶片中ZmbHLH161基因受到非生物胁迫干旱和脱水的诱导表达, 对冷和盐胁迫信号也有应答反应; 在玉米根中ZmbHLH161基因受到非生物胁迫干旱的诱导表达, 对冷和盐胁迫信号也有应答反应, 但不受脱水胁迫的诱导表达。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6实时荧光定量PCR分析不同处理条件下ZmbHLH161基因在叶片和根中的表达情况
A: 干旱处理; B: 脱水处理; C: 低温(4℃)处理; D: 高盐(200 mmol L-1 NaCl)处理。数据为3个生物学重复±标准差。
Fig. 6Real-time quantitative PCR analysis for expression level of ZmbHLH161 gene under different treatments in leaf and root
A: drought treatment; B: dehydration treatment; C: cold (4℃) treatment; D: high salt (200 mmol L-1 NaCl) treatment. The error bar represents ± SD of three biological replications.
2.5 转基因拟南芥阳性植株鉴定
提取卡那霉素抗性阳性植株叶片DNA进行PCR检测, 结果如图7所示, 阴性对照和野生型都无目的条带, 有3株转基因拟南芥含有目的基因。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7T1代拟南芥PCR检测
M: DL2000 marker; P: 阳性对照; W: 野生型拟南芥; N: 阴性对照; 1~3: T1代拟南芥。
Fig. 7PCR detection of T1 generation Arabidopsis
M: DL2000 marker; P: positive control; W: wild-type Arabidopsis; N: negative control; 1-3: T1Arabidopsis generation.
2.6 转基因拟南芥的耐盐性分析
0 mol L-1 NaCl浓度时野生型和转基因拟南芥均能正常生长, 当NaCl浓度为0.10 mol L-1和0.15 mol L-1时, 野生型和转基因拟南芥均能正常生长, 转基因拟南芥主根比野生型略长, 但差异不显著; 当NaCl浓度升为0.18 mol L-1时, 野生型和转基因拟南芥均不能正常生长, 叶片发黄枯死, 主根不生长, 无侧根(图8)。说明ZmbHLH161在转基因拟南芥响应盐胁迫过程中没有起到明显促进作用。图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8不同盐浓度下转基因拟南芥根长比较
A~D分别为0、0.10、0.15和0.18 mol L-1盐处理的生长情况; E: 不同盐浓度下拟南芥平均主根长度; WT: 野生型拟南芥; OE: 转ZmbHLH161拟南芥; 标尺为1.5 cm。
Fig. 8Root length of transgenic Arabidopsis thaliana on gradient NaCl concentrations
A-D indicates the seedling growth under the NaCl concentrations of 0, 0.10, 0.15, and 0.18 mol L-1; E: the average main root length; WT: wild type; OE: ZmbHLH161 transgenic Arabidopsis thaliana. Bar = 1.5 cm
2.7 转基因拟南芥的耐甘露醇分析
由图9可知, 0 mol L-1甘露醇浓度时, 野生型和转基因拟南芥均能正常生长, 根系发达; 当甘露醇浓度为0.15 mol L-1时, 野生型和转基因拟南芥均能正常生长, 但转基因拟南芥根长明显长于野生型, 且差异显著(P<0.05)。当甘露醇浓度为0.2 mol L-1时, 野生型和转基因拟南芥长势较弱, 叶片略微发黄, 转基因拟南芥主根比野生型略长, 差异显著(P<0.05)。当甘露醇浓度为0.3 mol L-1时, 2种拟南芥叶片明显变黄, 生长明显受到影响, 植株细小, 其中OE-1和OE-2拟南芥主根长度较野生型拟南芥呈差异极显著水平(P<0.01)。说明ZmbHLH161基因在拟南芥响应渗透胁迫过程中起到一定的促进作用。图9
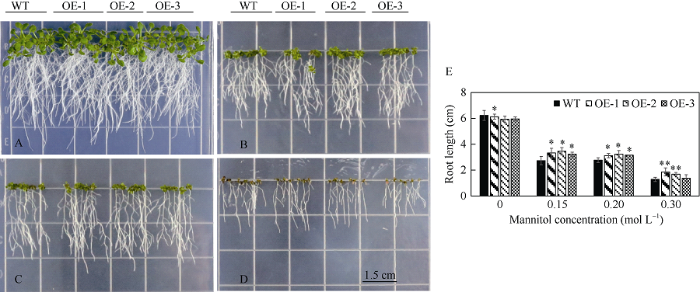 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图9不同甘露醇浓度下转基因拟南芥根长比较
A~D分别为0、0.15、0.2和0.3 mol L-1甘露醇处理的生长情况; E: 不同甘露醇浓度下拟南芥平均主根长度; WT: 野生型拟南芥; OE: 转ZmbHLH161拟南芥; 标尺为1.5 cm。*和**分别表示转基因拟南芥主根长度较野生型呈差异显著水平(P<0.05)和差异极显著水平(P<0.01)。
Fig. 9Root length of transgenic Arabidopsis thaliana on gradient mannitol concentrations
A-D indicates the seedling growth under the mannitol concentrations of 0, 0.15, 0.2, and 0.3 mol L-1; E: the average main root length of Arabidopsis at different mannitol concentrations; WT: wild type; OE: ZmbHLH161 of transgenic Arabidopsis thaliana; Bar = 1.5 cm. * and ** indicate that the root length of transgenic Arabidopsis thaliana is significantly different from that of wild type (P < 0.05) and the difference is extremely significant (P<0.01).
3 讨论
转录因子是能与真核基因顺式作用元件相互作用的DNA结合蛋白, 起激活或者抑制转录的作用。近年来, 越来越多的研究关注各种转录因子在响应植物胁迫中的作用, bHLH转录因子也是其中之一。在干旱研究方面, Chen等[27]通过农杆菌介导法将OsDREB1G和OsDREB2B在水稻中过表达, 其结果表明可以显著提高对缺水胁迫的耐受性。Rozenn等[28]报道了AtbHLH68缺失突变体对干旱胁迫的敏感性比野生型略高, 且AtbHLH68过表达株系显示出侧根形成缺陷和对干旱胁迫的耐受性显著提高。Yao等[29]将苦荞基因FtbHLH3在拟南芥中进行异源过表达, 结果显示其干旱和氧化耐受性增强, 这不仅归因于丙二醛(MDA)降低, 离子泄漏(IL)和活性氧(ROS)降低, 而且脯氨酸(Pro)含量高; 与野生型抗氧化剂活性相比较高, 转基因品系中的酶和光合作用效率更高。在盐胁迫研究方面, Li等[30]从东乡野生稻中分离并鉴定了bHLH蛋白基因OrbHLH001, 在拟南芥中过表达此基因后增强了转基因拟南芥植株对冰冻和盐胁迫的耐受性; Zhou等[31]从野生稻克隆并鉴定的基因OrbHLH2在拟南芥中的过表达赋予了其对盐和渗透胁迫的耐受性。Liu等[32]研究表明, 与野生型相比, 过表达bHLH122转基因拟南芥对干旱、NaCl和渗透胁迫表现出更大的抗性。相反, bHLH122突变体比野生型植物对NaCl和渗透胁迫更敏感。在低温胁迫研究方面, Xu等[33]研究发现, 中国野生葡萄VabHLH1或耐冷种“黑龙江苗”VvbHLH1转录因子过表达不影响转基因拟南芥植物生长和发育, 但增强了对冷胁迫的耐受性。烟草bHLH转录因子NtbHLH123, 响应冷胁迫(4℃), 过表达NtbHLH123烟草在冷胁迫下表现出电解质泄漏较低, 丙二醛含量降低, H2O2和ROS积累, 这有助于减轻冷胁迫处理后对细胞膜的氧化损伤, 从而增强了转基因烟草植株耐寒性[34]。本研究应用qPCR分析不同组织中ZmbHLH161表达情况, 结果表明该基因在不同时期多种组织中均有表达。其中以生长旺盛期根中表达量最高, 其次是幼根和幼胚, 因此推测ZmbHLH161可能参与根和幼胚生长发育调控。在ZmbHLH161转录起始密码子上游2 kb的DNA序列分析发现, 有多个与逆境胁迫、光反应等功能相关的顺式作用元件, 如ABRE、MYB、MYC等是响应盐、干旱、ABA、低温胁迫的响应元件。ZmbHLH161在脱水和干旱逆境处理中的叶片表达量上调程度最明显, 最高分别能达到对照的82.0倍和8.6倍, 推测ZmbHLH161参与植株体内干旱等逆境胁迫应答。利用不同浓度梯度NaCl和甘露醇处理ZmbHLH161过表达转基因拟南芥和野生型并统计根长, 结果表明, 甘露醇处理下转基因拟南芥主根比野生型长, 且差异显著(P<0.05); 而在盐胁迫处理试验中, 转基因株系根系生长与野生型无显著差异。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
Biol Teach
[本文引用: 1]
DOI:10.1016/0092-8674(89)90682-xURLPMID:2493990 [本文引用: 1]

Two cDNAs were isolated whose dimerized products bind specifically to a DNA sequence, kappa E2, located in the immunoglobulin kappa chain enhancer. Both cDNAs share a region of extensive identity to the Drosophila daughterless gene and obvious similarity to a segment in three myc proteins, MyoD, and members of the Drosophila achaete-scute and twist gene family. The homologous regions have the potential to form two amphipathic helices separated by an intervening loop. Remarkable is the stringent conservation of hydrophobic residues present in both helices. We demonstrate that this new motif plays a crucial role in both dimerization and DNA binding.
DOI:10.1093/molbev/msp288URLPMID:19942615 [本文引用: 1]

Basic helix-loop-helix (bHLH) proteins are a class of transcription factors found throughout eukaryotic organisms. Classification of the complete sets of bHLH proteins in the sequenced genomes of Arabidopsis thaliana and Oryza sativa (rice) has defined the diversity of these proteins among flowering plants. However, the evolutionary relationships of different plant bHLH groups and the diversity of bHLH proteins in more ancestral groups of plants are currently unknown. In this study, we use whole-genome sequences from nine species of land plants and algae to define the relationships between these proteins in plants. We show that few (less than 5) bHLH proteins are encoded in the genomes of chlorophytes and red algae. In contrast, many bHLH proteins (100-170) are encoded in the genomes of land plants (embryophytes). Phylogenetic analyses suggest that plant bHLH proteins are monophyletic and constitute 26 subfamilies. Twenty of these subfamilies existed in the common ancestors of extant mosses and vascular plants, whereas six further subfamilies evolved among the vascular plants. In addition to the conserved bHLH domains, most subfamilies are characterized by the presence of highly conserved short amino acid motifs. We conclude that much of the diversity of plant bHLH proteins was established in early land plants, over 440 million years ago.
DOI:10.1073/pnas.94.10.5172URLPMID:9144210 [本文引用: 1]

A natural (evolutionary) classification is provided for 242 basic helix-loop-helix (bHLH) motif-containing proteins. Phylogenetic analyses of amino acid sequences describe the patterns of evolutionary change within the motif and delimit evolutionary lineages. These evolutionary lineages represent well known functional groups of proteins and can be further arranged into five groups based on binding to DNA at the hexanucleotide E-box, the amino acid patterns in other components of the motif, and the presence/absence of a leucine zipper. The hypothesized ancestral amino acid sequence for the bHLH transcription factor family is given together with the ancestral sequences of the subgroups. It is suggested that bHLH proteins containing a leucine zipper are not a natural, monophyletic group.
[本文引用: 1]
[本文引用: 1]
DOI:10.1046/j.1365-313x.2003.01644.xURLPMID:12535353 [本文引用: 1]

Visual screening of a T-DNA mutagenised population of Arabidopsis thaliana for an absence of silique elongation lead to the isolation of the aborted microspores (ams) mutant that shows a sporophytic recessive male sterile phenotype. Homozygous mutant plants are completely devoid of mature pollen. Pollen degeneration occurs shortly after release of the microspores from the tetrad, prior to pollen mitosis I. Premature tapetum and microspore degeneration are the primary defects caused by this lesion, while a secondary effect is visualised in the stamen filaments, which are reduced in length and lie beneath the receptive stigma at flower opening. The disrupted gene was isolated and revealed a T-DNA element to be inserted into the eighth exon of a basic helix-loop-helix (bHLH) gene located on chromosome II. This protein sequence contains a basic DNA binding domain and two alpha helices separated by a loop, typical of a transcription factor belonging to the MYC sub family of bHLH genes. Therefore, AMS plays a crucial role in tapetal cell development and the post-meiotic transcriptional regulation of microspore development within the developing anther.
DOI:10.1371/journal.pone.0031325URLPMID:22363621 [本文引用: 1]

Grain size is a major yield component in rice, and partly controlled by the sizes of the lemma and palea. Molecular mechanisms controlling the sizes of these organs largely remain unknown. In this study, we show that an antagonistic pair of basic helix-loop-helix (bHLH) proteins is involved in determining rice grain length by controlling cell length in the lemma/palea. Overexpression of an atypical bHLH, named POSITIVE REGULATOR OF GRAIN LENGTH 1 (PGL1), in lemma/palea increased grain length and weight in transgenic rice. PGL1 is an atypical non-DNA-binding bHLH and assumed to function as an inhibitor of a typical DNA-binding bHLH through heterodimerization. We identified the interaction partner of PGL1 and named it ANTAGONIST OF PGL1 (APG). PGL1 and APG interacted in vivo and localized in the nucleus. As expected, silencing of APG produced the same phenotype as overexpression of PGL1, suggesting antagonistic roles for the two genes. Transcription of two known grain-length-related genes, GS3 and SRS3, was largely unaffected in the PGL1-overexpressing and APG-silenced plants. Observation of the inner epidermal cells of lemma revealed that are caused by increased cell length. PGL1-APG represents a new grain length and weight-controlling pathway in which APG is a negative regulator whose function is inhibited by PGL1.
DOI:10.4161/psb.23448URLPMID:23333962 [本文引用: 1]

We recently demonstrated that cell elongation in plants is regulated by a triantagonistic bHLH system, in which three bHLH proteins, Activator of Cell Elongation 1 (ACE1), Arabidopsis ILI1 binding BHLH 1 (AtIBH1) and Paclobutrazol Resistance 1 (PRE1), competitively regulate the expression of genes for cell elongation. Here we show that ATBS1 Interacting Factor 2 (AIF2), AIF3 and AIF4 interact with PRE1 and ACE1, similar to AtIBH1, and also negatively regulate cell elongation in the triantagonistic bHLH system. The expression of each AIF is constitutive or induced by light, but AtIBH1 expression is dependent on BR signaling and developmental phase. These results indicate that AIFs and AtIBH1 may play different roles in cell elongation in different signaling pathways.
DOI:10.1016/j.plantsci.2015.11.005URLPMID:26795149 [本文引用: 1]

To explore the transcription factors associated with carotenoid metabolism in citrus fruit, one transcription factor (CubHLH1) was selected through microarray screening in Satsuma mandarin (Citrus unshiu Marc.) fruit, which was treated with exogenous ethylene or gibberellin (GA), accelerating or retarding carotenoid accumulation in peel, respectively. The amino acid sequence of CubHLH1 has homology to Arabidopsis activation-tagged bri1 suppressor 1 (ATBS1) interacting factor (AIF), which is functionally characterized as a negative regulator of the brassinolide (BR) signalling pathway. Yeast two-hybrid analysis revealed that protein for CubHLH1 could interact with Arabidopsis and tomato ATBS1. Overexpression of CubHLH1 caused a dwarf phenotype in transgenic tomato (Solanum lycopersicum L.), suggesting that CubHLH1 has a similar function to Arabidopsis AIF. In the transgenic tomato fruit at ripening stage, the lycopene content was reduced along with the changes in carotenoid biosynthetic gene expression. The abscisic acid (ABA) content of all the transgenic tomato fruit was higher than that of the wild type. These results implied that CubHLH1 is considered to have a similar function to Arabidopsis AIFs and might be directly involved in carotenoid metabolism in mature citrus fruit.
URLPMID:12454087 [本文引用: 1]

Brassinosteroids (BRs) are a class of polyhydroxylated steroids that are important regulators of plant growth and development. We have identified three closely related basic helix-loop-helix (bHLH) transcription factors, BEE1, BEE2, and BEE3, as products of early response genes required for full BR response. Comparison of the phenotypes of plants that overexpress BEE1 with bee1 bee2 bee3 triple-knockout mutant plants suggests that BEE1, BEE2, and BEE3 are functionally redundant positive regulators of BR signaling. Expression of BEE1, BEE2, and BEE3 is also regulated by other hormones, notably abscisic acid (ABA), a known antagonist of BR signaling. Reduced ABA response in plants overexpressing BEE1 suggests that BEE proteins may function as signaling intermediates in multiple pathways.
DOI:10.1038/23500URLPMID:10466729 [本文引用: 1]

The phytochrome photoreceptor family directs plant gene expression by switching between biologically inactive and active conformers in response to the sequential absorption of red and farred photons. Several intermediates that act late in the phytochrome signalling pathway have been identified, but fewer have been identified that act early in the pathway. We have cloned a nuclear basic helix-loop-helix protein, PIF3, which can bind to non-photoactive carboxy-terminal fragments of phytochromes A and B and functions in phytochrome signalling in vivo. Here we show that full-length photoactive phytochrome B binds PIF3 in vitro only upon light-induced conversion to its active form, and that photoconversion back to its inactive form causes dissociation from PIF3. We conclude that photosensory signalling by phytochrome B involves light-induced, conformer-specific recognition of the putative transcriptional regulator PIF3, providing a potential mechanism for direct photoregulation of gene expression.
DOI:10.1093/emboj/21.10.2441URLPMID:12006496 [本文引用: 1]

Plants sense and respond to red and far-red light using the phytochrome (phy) family of photoreceptors. However, the mechanism of light signal transduction is not well defined. Here, we report the identification of a new mutant Arabidopsis locus, srl2 (short under red-light 2), which confers selective hypersensitivity to continuous red, but not far-red, light. This hypersensitivity is eliminated in srl2phyB, but not srl2phyA, double mutants, indicating that this locus functions selectively and negatively in phyB signaling. The SRL2 gene encodes a bHLH factor, designated PIF4 (phytochrome-interacting factor 4), which binds selectively to the biologically active Pfr form of phyB, but has little affinity for phyA. Despite its hypersensitive morphological phenotype, the srl2 mutant displays no perturbation of light-induced expression of marker genes for chloroplast development. These data suggest that PIF4 may function specifically in a branch of the phyB signaling network that regulates a subset of genes involved in cell expansion. Consistent with this proposal, PIF4 localizes to the nucleus and can bind to a G-box DNA sequence motif found in various light-regulated promoters.
DOI:10.1271/bbb.69.1042URLPMID:15914931 [本文引用: 1]

The jasmonic acid (JA)-responsive gene RERJ1 isolated from suspension-cultured rice cells encodes a transcription factor with a basic helix-loop-helix motif. In this study, we found that RERJ1 is also expressed in rice plants in response to JA, and that its expression in rice leaves is up-regulated by exposure to wounding and drought stress. It is also suggested that JA but not abscisic acid is involved in the up-regulation of RERJ1 expression caused by wounding and drought stress.
DOI:10.1007/s11103-007-9230-3URL [本文引用: 1]

Arabidopsis. The expression of AtAIB was transitorily induced by ABA and PEG, although its transcripts were accumulated in various organs. We provided evidence showing that AtAIB has transcriptional activation activity in yeast. Knockdown of AtAIB expression caused reduced sensitivity to ABA, whereas overexpression of this gene led to elevated sensitivity to ABA in cotyledon greening and seedling root growth. Furthermore, soil-grown plants overexpressing AtAIB showed increased drought tolerance. Taken together, these results suggested that AtAIB functions as a transcription activator involved in the regulation of ABA signaling in Arabidopsis.]]>
DOI:10.1007/s00438-009-0481-3URLPMID:19760256 [本文引用: 1]

In our previous microarray analysis of NaCl-treated Arabidopsis roots, we identified a basic-helix-loop-helix (bHLH) transcription factor, bHLH92 (At5g43650), as one of the transcripts showing the greatest fold-increase in abundance upon NaCl exposure. Here, we characterize the role of bHLH92 in the context of abiotic stress physiology and hormone responses. We observed that bHLH92 transcript abundance increases in response to NaCl, dehydration, mannitol, and cold treatments, and compared these responses to those of two closely related genes: bHLH41 and bHLH42. The NaCl-inducibility of bHLH92 was only partially dependent on abscisic acid (ABA) biosynthesis and SALT OVERLY SENSITIVE2 (SOS2) pathways. As compared to WT, root elongation of bhlh92 mutants was more sensitive to mannitol, and these mutants also showed increased electrolyte leakage following NaCl treatments. Overexpression of bHLH92 moderately increased the tolerance to NaCl and osmotic stresses. Finally, we identified at least 19 putative downstream target genes of bHLH92 under NaCl treatment using an oligonucleotide microarray. Together these data show that bHLH92 functions in plant responses to osmotic stresses, although the net contribution of bHLH92-regulated genes to stress tolerance appears relatively limited in proportion to what might be expected from its transcript expression pattern.
DOI:10.1101/gad.1077503URLPMID:12672693 [本文引用: 1]

Cold temperatures trigger the expression of the CBF family of transcription factors, which in turn activate many downstream genes that confer chilling and freezing tolerance to plants. We report here the identification of ICE1 (inducer of CBF expression 1), an upstream transcription factor that regulates the transcription of CBF genes in the cold. An Arabidopsis ice1 mutant was isolated in a screen for mutations that impair cold-induced transcription of a CBF3 promoter-luciferase reporter gene. The ice1 mutation blocks the expression of CBF3 and decreases the expression of many genes downstream of CBFs, which leads to a significant reduction in plant chilling and freezing tolerance. ICE1 encodes a MYC-like bHLH transcriptional activator. ICE1 binds specifically to the MYC recognition sequences in the CBF3 promoter. ICE1 is expressed constitutively, and its overexpression in wild-type plants enhances the expression of the CBF regulon in the cold and improves freezing tolerance of the transgenic plants.
DOI:10.1111/j.1365-313X.2007.03149.xURLPMID:17559517 [本文引用: 1]

Iron (Fe) deficiency is a major abiotic stress in crop production. Although responses to Fe deficiency in graminaceous plants, such as increased production and secretion of mugineic acid family phytosiderophores (MAs), have been described, the gene regulation mechanisms related to these responses are largely unknown. To elucidate the regulation mechanisms of the genes related to Fe acquisition in graminaceous plants, we characterized the Fe-deficiency-inducible basic helix-loop-helix transcription factor OsIRO2 in rice. In yeast cells, OsIRO2 functioned as a transcriptional activator. In rice, overexpression of OsIRO2 resulted in increased MAs secretion, whereas repression of OsIRO2 resulted in lower MAs secretion and hypersensitivity to Fe deficiency. Northern blots revealed that the expression of the genes involved in the Fe(III)-MAs transport system was dependent on OsIRO2. The expression of the genes for nicotianamine synthase, a key enzyme in MAs synthesis, was notably affected by the level of OsIRO2 expression. Microarray analysis demonstrated that OsIRO2 regulates 59 Fe-deficiency-induced genes in roots. Some of these genes, including two transcription factors upregulated by Fe deficiency, possessed the OsIRO2 binding sequence in their upstream regions. OsIRO2 possesses a homologous sequence of the Fe-deficiency-responsive cis-acting elements (IDEs) in its upstream region. We propose a novel gene regulation network for Fe-deficiency responses, including OsIRO2, IDEs and the two transcription factors.
DOI:10.1093/jxb/erz307URLPMID:31267122 [本文引用: 1]

Drought stress is the most important environmental stress limiting maize production. ZmPTF1, a phosphate starvation-induced basic helix-loop-helix (bHLH) transcription factor, contributes to root development and low-phosphate tolerance in maize. Here, ZmPTF1 expression, drought tolerance, and the underlying mechanisms were studied by using maize ZmPTF1 overexpression lines and mutants. ZmPTF1 was found to be a positive regulator of root development, ABA synthesis, signalling pathways, and drought tolerance. ZmPTF1 was also found to bind to the G-box element within the promoter of 9-cis-epoxycarotenoid dioxygenase (NCED), C-repeat-binding factor (CBF4), ATAF2/NAC081, NAC30, and other transcription factors, and to act as a positive regulator of the expression of those genes. The dramatically upregulated NCEDs led to increased abscisic acid (ABA) synthesis and activation of the ABA signalling pathway. The up-regulated transcription factors hierarchically regulate the expression of genes involved in root development, stress responses, and modifications of transcriptional regulation. The improved root system, increased ABA content, and activated ABA-, CBF4-, ATAF2-, and NAC30-mediated stress responses increased the drought tolerance of the ZmPTF1 overexpression lines, while the mutants showed opposite trends. This study describes a useful gene for transgenic breeding and helps us understand the role of a bHLH protein in plant root development and stress responses.
DOI:10.1105/tpc.151140URLPMID:14600211 [本文引用: 1]
DOI:10.1104/pp.106.080580URLPMID:16896230 [本文引用: 1]

The basic/helix-loop-helix (bHLH) transcription factors and their homologs form a large family in plant and animal genomes. They are known to play important roles in the specification of tissue types in animals. On the other hand, few plant bHLH proteins have been studied functionally. Recent completion of whole genome sequences of model plants Arabidopsis (Arabidopsis thaliana) and rice (Oryza sativa) allows genome-wide analysis and comparison of the bHLH family in flowering plants. We have identified 167 bHLH genes in the rice genome, and their phylogenetic analysis indicates that they form well-supported clades, which are defined as subfamilies. In addition, sequence analysis of potential DNA-binding activity, the sequence motifs outside the bHLH domain, and the conservation of intron/exon structural patterns further support the evolutionary relationships among these proteins. The genome distribution of rice bHLH genes strongly supports the hypothesis that genome-wide and tandem duplication contributed to the expansion of the bHLH gene family, consistent with the birth-and-death theory of gene family evolution. Bioinformatics analysis suggests that rice bHLH proteins can potentially participate in a variety of combinatorial interactions, endowing them with the capacity to regulate a multitude of transcriptional programs. In addition, similar expression patterns suggest functional conservation between some rice bHLH genes and their close Arabidopsis homologs.
DOI:10.1186/s12870-018-1441-zURLPMID:30326829 [本文引用: 1]

BACKGROUND: In plants, the basic helix-loop-helix (bHLH) transcription factors play key roles in diverse biological processes. Genome-wide comprehensive and systematic analyses of bHLH proteins have been well conducted in Arabidopsis, rice, tomato and other plant species. However, only few of bHLH family genes have been functional characterized in maize. RESULTS: In this study, our genome-wide analysis identified 208 putative bHLH family proteins (ZmbHLH proteins) in maize (Zea mays). We classified these proteins into 18 subfamilies by comparing the ZmbHLHs with Arabidopsis thaliana bHLH proteins. Phylogenetic analysis, conserved protein motifs, and exon-intron patterns further supported the evolutionary relationships among these bHLH proteins. Genome distribution analysis found that the 208 ZmbHLH loci were located non-randomly on the ten maize chromosomes. Further, analysis of conserved cis-elements in the promoter regions, protein interaction networks, and expression patterns in roots, leaves, and seeds across developmental stages, suggested that bHLH family proteins in maize are probably involved in multiple physiological processes in plant growth and development. CONCLUSION: We performed a genome-wide, systematic analysis of bHLH proteins in maize. This comprehensive analysis provides a useful resource that enables further investigation of the physiological roles and molecular functions of the ZmbHLH transcription factors.
DOI:10.1038/nprot.2007.199URLPMID:17585298 [本文引用: 1]

The transient gene expression system using Arabidopsis mesophyll protoplasts has proven an important and versatile tool for conducting cell-based experiments using molecular, cellular, biochemical, genetic, genomic and proteomic approaches to analyze the functions of diverse signaling pathways and cellular machineries. A well-established protocol that has been extensively tested and applied in numerous experiments is presented here. The method includes protoplast isolation, PEG-calcium transfection of plasmid DNA and protoplast culture. Physiological responses and high-throughput capability enable facile and cost-effective explorations as well as hypothesis-driven tests. The protoplast isolation and DNA transfection procedures take 6-8 h, and the results can be obtained in 2-24 h. The cell system offers reliable guidelines for further comprehensive analysis of complex regulatory mechanisms in whole-plant physiology, immunity, growth and development.
DOI:10.1006/meth.2001.1262URLPMID:11846609 [本文引用: 1]

The two most commonly used methods to analyze data from real-time, quantitative PCR experiments are absolute quantification and relative quantification. Absolute quantification determines the input copy number, usually by relating the PCR signal to a standard curve. Relative quantification relates the PCR signal of the target transcript in a treatment group to that of another sample such as an untreated control. The 2(-Delta Delta C(T)) method is a convenient way to analyze the relative changes in gene expression from real-time quantitative PCR experiments. The purpose of this report is to present the derivation, assumptions, and applications of the 2(-Delta Delta C(T)) method. In addition, we present the derivation and applications of two variations of the 2(-Delta Delta C(T)) method that may be useful in the analysis of real-time, quantitative PCR data.
DOI:10.1046/j.1365-313x.1998.00343.xURLPMID:10069079 [本文引用: 1]

The Agrobacterium vacuum infiltration method has made it possible to transform Arabidopsis thaliana without plant tissue culture or regeneration. In the present study, this method was evaluated and a substantially modified transformation method was developed. The labor-intensive vacuum infiltration process was eliminated in favor of simple dipping of developing floral tissues into a solution containing Agrobacterium tumefaciens, 5% sucrose and 500 microliters per litre of surfactant Silwet L-77. Sucrose and surfactant were critical to the success of the floral dip method. Plants inoculated when numerous immature floral buds and few siliques were present produced transformed progeny at the highest rate. Plant tissue culture media, the hormone benzylamino purine and pH adjustment were unnecessary, and Agrobacterium could be applied to plants at a range of cell densities. Repeated application of Agrobacterium improved transformation rates and overall yield of transformants approximately twofold. Covering plants for 1 day to retain humidity after inoculation also raised transformation rates twofold. Multiple ecotypes were transformable by this method. The modified method should facilitate high-throughput transformation of Arabidopsis for efforts such as T-DNA gene tagging, positional cloning, or attempts at targeted gene replacement.
DOI:10.1007/s10529-008-9811-5URLPMID:18779926 [本文引用: 1]

The DREB transcription factors, which specifically interact with C-repeat/DRE (A/GCCGAC), play an important role in plant abiotic stress tolerance by controlling the expression of many cold or/and drought-inducible genes in an ABA-independent pathway. We have isolated three novel rice DREB genes, OsDREB1E, OsDREB1G, and OsDREB2B, which are homologous to Arabidopsis DREB genes. The yeast one-hybrid assay indicated that OsDREB1E, OsDREB1G, and OsDREB2B can specifically bind to the C-repeat/DRE element. To elucidate the function of respective OsDREB genes, we have stably introduced these to rice by Agrobacterium-mediated transformation. Transgenic rice plants analysis revealed that over-expression of OsDREB1G and OsDREB2B in rice significantly improved their tolerance to water deficit stress, while over-expression of OsDREB1E could only slightly improved the tolerance to water deficit stress, suggesting that the OsDREBs might participate in the stress response pathway in different manners.
DOI:10.1111/ppl.12549URLPMID:28369972 [本文引用: 1]

Basic helix-loop-helix (bHLH) transcription factors are involved in a wide range of developmental processes and in response to biotic and abiotic stresses. They represent one of the biggest families of transcription factors but only few of them have been functionally characterized. Here we report the characterization of AtbHLH68 and show that, although the knock out mutant did not have an obvious development phenotype, it was slightly more sensitive to drought stress than the Col-0, and AtbHLH68 overexpressing lines displayed defects in lateral root (LR) formation and a significant increased tolerance to drought stress, likely related to an enhanced sensitivity to abscisic acid (ABA) and/or increased ABA content. AtbHLH68 was expressed in the vascular system of Arabidopsis and its expression was modulated by exogenously applied ABA in an organ-specific manner. We showed that the expression of genes involved in ABA metabolism [AtAAO3 (AtALDEHYDE OXIDASE 3) and AtCYP707A3 (AtABSCISIC ACID 8'HYDROXYLASE 3)], in ABA-related response to drought-stress (AtMYC2, AtbHLH122 and AtRD29A) or during LRs development (AtMYC2 and AtABI3) was de-regulated in the overexpressing lines. We propose that AtbHLH68 has a function in the regulation of LR elongation, and in the response to drought stress, likely through an ABA-dependent pathway by regulating directly or indirectly components of ABA signaling and/or metabolism.
DOI:10.3389/fpls.2017.00625URLPMID:28487715 [本文引用: 1]

bHLH (basic helix-loop-helix) transcription factors play important roles in the abiotic stress response in plants, but their characteristics and functions in tartary buckwheat (Fagopyrum tataricum), a flavonoid-rich cereal crop with a strong stress tolerance, have not been fully investigated. Here, a novel bHLH gene, designated FtbHLH3, was isolated and characterized. Expression analysis in tartary buckwheat revealed that FtbHLH3 was mainly induced by polyethylene glycol 6000 (PEG6000) and abscisic acid (ABA) treatments. Subcellular localization and a yeast one-hybrid assay indicated that FtbHLH3 has transcriptional activation activities. Overexpression of FtbHLH3 in Arabidopsis resulted in increased drought/oxidative tolerance, which was attributed to not only lower malondialdehyde (MDA), ion leakage (IL), and reactive oxygen species (ROS) but also higher proline (Pro) content, activities of antioxidant enzymes, and photosynthetic efficiency in transgenic lines compared to wild type (WT). Moreover, qRT-PCR analysis indicated that the expression of multiple stress-responsive genes in the transgenic lines was significantly higher than in WT under drought stress. In particular, the expression of AtNCED, a rate-limiting enzyme gene in ABA biosynthesis, was increased significantly under both normal and stress conditions. Additionally, an ABA-response-element (ABRE) was also found in the promoter regions. Furthermore, the transgenic Arabidopsis lines of the FtbHLH3 promoter had higher GUS activity after drought stress. In summary, our results indicated that FtbHLH3 may function as a positive regulator of drought/oxidative stress tolerance in transgenic Arabidopsis through an ABA-dependent pathway.
DOI:10.1007/s00299-010-0883-zURL [本文引用: 1]

Oryza rufipogon) is the northernmost wild rice in the world known to date and has extremely high cold tolerance and many other adversity-resistant properties. To identify the genes responsible for the high stress tolerance, we isolated and characterized a basic helix-loop-helix (bHLH) protein gene OrbHLH001 from Dongxiang Wild Rice. The gene encodes an ICE1-like protein containing multiple homopeptide repeats. Expression of OrbHLH001 is induced by salt stress and is predominant in the shoots of wild rice seedlings. Overexpression of OrbHLH001 enhanced the tolerance to freezing and salt stresses in transgenic Arabidopsis. Examination of the expression of cold-responsive genes in transgenic Arabidopsis showed that the function of OrbHLH001 differs from that of ICE1 and is independent of a CBF/DREB1 cold-response pathway.]]>
DOI:10.1016/j.jplph.2009.02.007URLPMID:19324458 [本文引用: 1]

Salt stress adversely affects plant growth and development. Some plants reduce the damage of high-salt stress by expressing a series of salt-responsive genes. Studies of the molecular mechanism of the salt-stress response have focused on the characterization of components involved in signal perception and transduction. In the present work, we cloned and characterized a basic helix-loop-helix (bHLH) encoding gene, OrbHLH2, from wild rice (Oryza rufipogon), which encodes a homologue protein of ICE1 in Arabidopsis. OrbHLH2 protein localized in the nucleus. Overexpression of OrbHLH2 in Arabidopsis conferred increased tolerance to salt and osmotic stress, and the stress-responsive genes DREB1A/CBF3, RD29A, COR15A and KIN1 were upregulated in transgenic plants. Abscisic acid (ABA) treatment showed a similar effect on the seed germination or transcriptional expression of stress-responsive genes in both wild type and OrbHLH2-overexpressed plants, which implies that OrbHLH2 does not depend on ABA in responding to salt stress. OrbHLH2 may function as a transcription factor and positively regulate salt-stress signals independent of ABA in Arabidopsis, which provides some useful data for improving salt tolerance in crops.
DOI:10.1111/nph.12607URL [本文引用: 1]

The Arabidopsis thaliana bHLH122 transcripts were strongly induced by drought, NaCl and osmotic stresses, but not by ABA treatment. Promoter::GUS analysis showed that bHLH122 was highly expressed in vascular tissues and guard cells. Compared with wild-type (WT) plants, transgenic plants overexpressing bHLH122 displayed greater resistance to drought, NaCl and osmotic stresses. In contrast, the bhlh122 loss-of-function mutant was more sensitive to NaCl and osmotic stresses than were WT plants.Microarray analysis indicated that bHLH122 was important for the expression of a number of abiotic stress-responsive genes. In electrophoretic mobility shift assay and chromatin immunoprecipitation assays, bHLH122 could bind directly to the G-box/E-box cis-elements in the CYP707A3 promoter, and repress its expression. Further, up-regulation of bHLH122 substantially increased cellular ABA levels.These results suggest that bHLH122 functions as a positive regulator of drought, NaCl and osmotic signaling.]]>
DOI:10.1007/s11033-014-3404-2URL [本文引用: 1]

Basic helix-loop-helix (bHLH)-type transcription factors play diverse roles in plant physiological response and stress-adaptive regulation network. Here, we identified one grapevine bHLH transcription factor from a cold-tolerant accession 'Heilongjiang seedling' of Chinese wild Vitis amurensis (VabHLH1) as a transcriptional activator involved in cold stress. We also compared with its counterpart from a cold-sensitive Vitis vinifera cv. Cabernet Sauvignon (VvbHLH1). These two putative proteins are characterized by the presence of the identically conserved regions of 54 amino acid residues of bHLH signature domain, and shared 99.1 % amino acid identity, whereas several stress-related cis-regulatory elements located in both promoter regions differed in types and positions. Expressions of two bHLHs in grapevine leaves were induced by cold stress, but evidently differ between two grapevine genotypes upon cold exposure. Two grapevine bHLH proteins were exclusively localized to the nucleus and exhibited strong transcriptional activation activities in yeast cells. Overexpression of either VabHLH1 or VvbHLH1 transcription factor did not affect the growth and development of transgenic Arabidopsis plants, but enhanced tolerance to cold stress. The improved tolerance in VabHLH1- or VvbHLH1-overexpressing Arabidopsis plants is associated with multiple physiological and biochemical changes that occurred during the time-course cold stress. These most common changes include the evaluated levels of proline, decreased amounts of malondialdehyde and reduced membrane injury as reflected by electrolyte leakage. VabHLH1 and VvbHLH1 displayed overlapping, but not identical, roles in activating the corresponding CBF cold signaling pathway, especially in regulating the expression of CBF3 and RD29A. Our findings demonstrated that two grapevine bHLHs act as positive regulators of the cold stress response, modulating the level of COR gene expression, which in turn confer tolerance to cold stress.
DOI:10.3389/fpls.2018.00381URLPMID:29643858 [本文引用: 1]

Cold stress is a major environmental factor that impairs plant growth and development, geographic distribution, and crop productivity. The C-repeat binding factor (CBF) regulatory pathway has an essential role in response to cold stress. Here, we characterized a bHLH transcription factor from Nicotiana tabacum, NtbHLH123, in response to cold stress (4 degrees C). Overexpression of NtbHLH123 enhanced cold tolerance in transgenic tobacco plants. Based on yeast one-hybrid, chromatin immunoprecipitation PCR, and transient expression analysis assays, NtbHLH123 binds directly to the G-box/E-box motifs in the promoter of the NtCBF genes and positively regulates their expression. Furthermore, NtbHLH123-overexpressing plants showed lower electrolyte leakage, reduced malondialdehyde contents, H2O2 and reactive oxygen species (ROS) accumulation under cold stress, which contributed to alleviating oxidative damage to the cell membrane after cold stress treatment. And NtbHLH123 increased stress tolerance by improving the expression of a number of abiotic stress-responsive genes to mediate the ROS scavenging ability and other stress tolerance pathways. Taken together, we present a model suggesting that NtbHLH123 is a transcriptional activator that functions as a positive regulator of cold tolerance by activating NtCBF, ROS scavenging-related, and stress-responsive genes.
