 ,1,2, 荐红举1, 邱晓2, 李加纳
,1,2, 荐红举1, 邱晓2, 李加纳 ,1,*
,1,*Regulatory mechanism of the seed coat color gene BrTT1 in Brassica rapa L.
WANG Yan-Hua ,1,2, JIAN Hong-Jiu1, QIU Xiao2, LI Jia-Na
,1,2, JIAN Hong-Jiu1, QIU Xiao2, LI Jia-Na ,1,*
,1,*通讯作者:
收稿日期:2020-02-17接受日期:2020-07-2网络出版日期:2020-11-12
| 基金资助: |
Received:2020-02-17Accepted:2020-07-2Online:2020-11-12
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (11248KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
王艳花, 荐红举, 邱晓, 李加纳. 白菜型油菜粒色主效基因BrTT1的调控机制分析[J]. 作物学报, 2020, 46(11): 1678-1689. doi:10.3724/SP.J.1006.2020.04036
WANG Yan-Hua, JIAN Hong-Jiu, QIU Xiao, LI Jia-Na.
油菜是世界上重要的油料作物, 菜籽油是重要的食用油和生物柴油来源, 菜籽饼粕是优良的动物蛋白饲料[1,2,3,4]。白菜型油菜作为在世界范围内广泛种植的芸薹属油料作物之一, 包含许多优异、稳定的天然黄籽资源。自20世纪70年代以来, 国内外许多研究者对油菜黄籽性状的大量研究表明, 在相同遗传背景下, 黄籽油菜的含油量、蛋白质含量均高于黒籽油菜, 且黄籽油菜具有皮壳率低、木质素和多酚含量低、抗营养物质(植酸、单宁、芥子碱、硫苷等)含量低等一系列优良品质性状[5,6,7,8,9,10,11,12,13,14,15,16,17,18,19]。通过对这些黄籽材料粒色性状的研究, 可以为白菜型黄籽油菜育种方案的制订提供指导, 促进白菜型油菜品种的改良。同时由于甘蓝型油菜中不存在天然的黄籽资源, 白菜作为甘蓝型油菜的供体亲本之一, 研究者常利用种间杂交或人工合成等方法将白菜型油菜中的黄籽性状导入甘蓝型油菜[20,21,22,23,24,25,26]。通过对白菜型油菜黄籽性状的研究, 以期为甘蓝型黄籽油菜遗传机理的分析奠定基础。
油菜种子的种皮色素、大小、成分影响油菜的产量和品质[24,25]。大黄油菜是白菜型油菜中一种天然的黄籽资源, 其粒色鲜黄, 黄籽性状能够稳定遗传, 且具有大粒、含油量高、自交亲和性好等优点, 是研究油菜粒色性状的良好材料[26]。目前对拟南芥透明种皮的形成机理较为清楚, 可为黄籽油菜种皮色素积累的研究提供借鉴, 但黄籽油菜种皮色素的形成及调控机理仍有待研究。Li等[27]对拟南芥和白菜型油菜种皮结构的解剖学观察证实了拟南芥与白菜型油菜种皮结构不同, 拟南芥与油菜种皮色素积累的方式应该存在潜在的差异。BrTT1作为油菜中重要的粒色调控基因, 研究其调控机制具有重要意义[28,29,30,31]。然而, 目前关于BrTT1及其他粒色主效基因在白菜型油菜粒色调控机制解析的报道较少。在前期研究中, 通过全基因组重测序结合遗传连锁图谱对白菜型黄籽油菜大黄的粒色基因进行了精细定位, 并对粒色候选基因进行克隆及功能分析, 初步鉴定了BrTT1为控制大黄粒色的主效基因[30]。本研究将搜集不同来源的白菜型油菜黄、黑籽材料, 分析BrTT1在其他白菜型油菜的序列差异, 并对BrTT1在大黄油菜中的功能进行验证, 初步明确了该转录因子在类黄酮代谢路径中可能调控的靶基因, 揭示BrTT1下游粒色调控网络。解析BrTT1对白菜型油菜粒色的分子调控机制, 以期为白菜型和甘蓝型油菜粒色研究奠定基础。
1 材料与方法
1.1 供试材料
不同遗传背景的白菜型油菜共22份, 主要包括粒色纯合的6份黄籽材料(Y1~Y6)、9份红褐籽材料(R1~R9)和7份黑籽材料(B1~B7), 均由西南大学油菜工程研究中心提供(附表1)。Supplementary Table 1
附表1
附表1不同来源的白菜型油菜黄、红、黑籽材料
Supplementary Table 1
| 编号 No. | 材料名称和来源 Material name and origin | 种子粒色表型 Phenotype of the seeds | 编号 No. | 材料名称和来源 Material name and origin | 种子粒色表型 Phenotype of the seeds |
|---|---|---|---|---|---|
| Y1 | 大黄(中国青海) Dahuang (Qinghai, China) | 纯黄籽 Yellow seeds | R6 | 18B167-168 (中国青海) 18B167-168 (Qinghai, China) | 红褐籽 Red or brown seeds |
| Y2 | 18B07-08 (中国青海) 18B07-08 (Qinghai, China) | 纯黄籽 Yellow seeds | R7 | 18B171-172 (中国青海) 18B171-172 (Qinghai, China) | 红褐籽 Red or brown seeds |
| Y3 | 彭波黄(中国青海) Pengbo yellow (Qinghai, China) | 纯黄籽 Yellow seeds | R8 | 18B141-142 (中国青海) 18B141-142 (Qinghai, China) | 红褐籽 Red or brown seeds |
| Y4 | 18B001-002 (中国青海) 18B001-002 (Qinghai, China) | 纯黄籽 Yellow seeds | R9 | 18B51-52 (中国青海) 18B51-52 (Qinghai, China) | 红褐籽 Red or brown seeds |
| Y5 | Sarson (印度) Sarson (India) | 纯黄籽 Yellow seeds | B1 | 浩油 11号(中国青海) Haoyou 11 (Qinghai, China) | 纯黑籽 Black seeds |
| Y6 | 18BCan01 (加拿大) 18BCan01 (Canada) | 纯黄籽 Yellow seeds | B2 | 芦花小油菜(中国青海) Luhua rapeseed (Qinghai, China) | 纯黑籽 Black seeds |
| R1 | 18B017-018 (中国青海) 18B017-018 (Qinghai, China) | 红褐籽 Red or brown seeds | B3 | 街子小油菜(中国青海) Jiezi rapeseed (Qinghai, China) | 纯黑籽 Black seeds |
| R2 | 18B021-022 (中国青海) 18B021-022 (Qinghai, China) | 红褐籽 Red or brown seeds | B4 | 18B077-078 (中国青海) 18B077-078 (Qinghai, China) | 纯黑籽 Black seeds |
| R3 | 18B105-106 (中国甘肃) 18B105-106 (Gansu, China) | 红褐籽 Red or brown seeds | B5 | 麻玉小油菜(中国青海) Mayu rapeseed (Qinghai, China) | 纯黑籽 Black seeds |
| R4 | 18B131-132 (中国青海) 18B131-132 (Qinghai, China) | 红褐籽 Red or brown seeds | B6 | 18B161-162 (中国青海) 18B161-162 (Qinghai, China) | 纯黑籽 Black seeds |
| R5 | 18B165-166 (中国青海) 18B165-166 (Qinghai, China) | 红褐籽 Red or brown seeds | B7 | 18BCan02 (加拿大) 18BCan02 (Canada) | 纯黑籽 Black seeds |
新窗口打开|下载CSV
1.2 不同遗传背景白菜型油菜中BrTT1的克隆及序列分析
选取Y1~Y6、R1~R9、B1~B7籽粒饱满的种子, 用纸盘发苗, 温度设置为22~25℃, 浇足水, 暗光培养2~3 d, 再移到光照培养室, 待长出4片叶子, 选取单株嫩叶, 采用CTAB (Cetyltrimethylammonium Ammonium Bromide)法[32]提取DNA, 利用紫外分光光度计检测每个单株的DNA浓度, 并稀释至50 ng μL-1, 冻于-20℃保存备用。采用TA克隆的方法[33]获得BrTT1序列信息。取油菜授粉后21、28、35、42、49、56 d的种子, 取样后放-80℃冰箱备用, 采用北京Biomed柱式小量植物总RNA抽提试剂盒提取RNA。采用Geneious Basic 4.8.5软件进行基因序列的比对; 利用MEGA6.0软件及NCBI数据库BLASTP分析BrTT1序列。1.3 BrTT1互作蛋白预测
在BRAD (1.4 酵母双杂分析BrTT1互作蛋白
采用Primer Premier 5.0软件设计引物Yb-F/-R (附表2), 克隆BrTT1的CDS序列, 以pGBKT7为载体构建融合表达诱饵载体pGBKT7-BrTT1; 采用Primer Premier 5.0软件设计基因特异引物(附表2)分别克隆白菜型油菜类黄酮代谢相关基因BrTT2、BrTT3、BrTT4、BrTT8、BrTT18、BrTTG1、BrTTG2、BrTT10、BrTT7、BrTT5的CDS序列, 以pGADT7为载体构建融合表达捕获载体pGADT7-BrTT2、pGADT7-BrTT3、pGADT7-BrTT4、pGADT7-BrTT8、pGADT7-BrTT18、pGADT7-BrTTG1、pGADT7-BrTTG2、pGADT7-BrTT10、pGADT7-BrTT7、pGADT7-BrTT5与pGBKT7-BrTT1质粒分别共转入酵母细胞Y2HGold, 并涂布在SD/-Leu/-Trp/-Ade/-His及SD/-Leu/-Trp/-Ade/-His/+X-α-gal的平板上, 在30℃恒温培养3 d, 同时设阳性对照组(pGADT7-T + pGBKT7-53)和阴性对照组(pGADT7-T + pGBKT7-laminC), 筛选BrTT1的互作蛋白。Supplementary Table 2
附表2
附表2用于载体构建的引物序列
Supplementary Table 2
| 引物名称 Primer name | 正向引物 Forward primer (5°-3°) | 反向引物 Reverse primer (5°-3°) |
|---|---|---|
| Yb-F/-R | CCGGAATTCATGTTTTCATCACTCTCAAACCACT | CGCGGATCCTTAAAAGTGCGTTTCAGAGACAGA |
| T2-F/-R | CCGGAATTCATGATGAGAAAGAGAGAAAGTAGTA | CGCGGATCCCTAACAATTAAAGTCCCAGAGACAA |
| T3-F/-R | CCGGAATTCATGATTCTGGAGGAGAAAGATGCAT | CCCATCGATTTAGCTATCTGAACGTTTTGGATCG |
| T7-F/-R | CCGGAATTCATGGTGATGGGTACACCGTCTTCGT | CGCGGATCCTCAAACAGGAACGCTGTGCAAGACC |
| T8-F/-R | CCGGAATTCATGGATGAATTAAGTATTATACCGT | CGCGGATCCCTAGAGTTTATTATTATATATGATT |
| T18-F/-R | CCGGAATTCATGGGTAGCAAGATGCTGTTCAGTT | CGCGGATCCTTAGCTTGCTTCTGGCAATGTGATC |
| TG1-F/-R | CCGGAATTCATGGACAACTCAGCTCCGGACTCCT | CGCGGATCCTCAAACTCTAAGGAGCTGCATTTTG |
| TG2-F/-R | CCGGAATTCATGGAGGTGAAAGAGAGTAAGAGAG | CGCGGATCCTCAAATGGCTTGATTAGAATGTTGT |
| T10-F/-R | CCGGAATTCATGACAAGCACTGATCAGACCACCG | CGCGGATCCTTAGGCCCATCGGTTTGTCTTAAAG |
| T12-F/-R | CCGGAATTCATGGGGAGGAAGACGTGGTTCGACG | CGCGGATCCTCAGAAGAGATAATCAAGTATCTGA |
| T5-F/-R | CCGGAATTCATGAGACATTCTTGCTGTTATAAAC | CGCGGATCCCTAAAGGGACTGACCAAAAGAGACA |
| F/R-TT1 | ATGTTTTCATCACTCTCAAACCACT | TTAAAAGTGCGTTTCAGAGACAGA |
| gCb-F/-R | CACCATGTTTTCATCACTCTCAAACCACT | AAAGTGCGTTTCAGAGACAGA |
| F35S3ND/CB-R | GGAAGTTCATTTCATTTGGAGAG | AAAGTGCGTTTCAGAGACAGA |
| FBTT1I | GGATCCGACGTCGAGATCTATAAACATTCCC | |
新窗口打开|下载CSV
1.5 超量和抑制表达BrTT1载体构建及遗传转化
采用Primer Premier 5.0软件设计引物gCb-F/-R (附表2), 以白菜型油菜褐籽材料cDNA为模板, 采用TA克隆方法获得BrTT1基因的CDS序列, Axygen胶回收试剂盒回收目的基因, 采用Gateway方法将胶回收片段连入骨干载体pEarlyGate101。连接产物转入大肠杆菌DH5α, 挑选阳性克隆送擎科生物测序。采用液氮冷激的方法将阳性克隆转化农杆菌菌株感受态细胞GV3101, 构建BrTT1超量表达载体p35S::BrTT1。以pFGC5941为骨干载体, 构建BrTT1干扰载体pBBrTT1I, 构建方法参照Ma等[34]。参考Cardoza和Stewart [35]的农杆菌介导的遗传转化体系, 将构建好的表达载体送武汉双螺旋生物公司完成遗传转化试验。1.6 RNA提取和荧光定量分析方法
根据BRAD数据库信息, 利用白菜型油菜中已知类黄酮途径参考基因的CDS序列设计定量引物(附表3), 采集田间或温室不同发育时期(开花后21、28、35、42、48、56 d)的黄、黑籽种子, 用液氮冷冻后, 保存于-80℃超低温冰箱备用。待所有不同发育时期的种子取样完成后, 使用EASYspin RNA Rapid Plant Kit (Biomed)提取RNA, 使用PrimeScript RT reagent Kit with gDNA Eraser (TaKaRa, http://www.takara.com.cn/)合成cDNA。qRT-PCR反应体系为20 μL, 包含10 μL 2× Promega GoTaq qPCR Master Mix、1 μL 20 μmol L-1正向引物、1 μL 20 μmol L-1反向引物、100 ng cDNA第一链, 添加ddH2O至20 μL。反应程序为94℃ 2 min; 94℃ 3 s, 60℃ 30 s, 40个循环; 60~95℃, 熔解曲线分析。使用2-ΔΔCt法计算基因的表达量[36], 28S rRNA基因作为内参基因。Supplementary Table 3
附表3
附表3用于差异表达分析的引物序列
Supplementary Table 3
| 引物名称 Primer name | 正向引物 Forward primer (5°-3°) | 反向引物 Reverse primer (5°-3°) |
|---|---|---|
| 28S | ATAACCGCATCAGGTCTCCAAG | CCTCAGAGCCAATCCTTTTCC |
| BrTT1 | TCGCTACAACAATCTTCAGATGCACA | TCCTGCACCCTTCAACGCAGC |
| BrTT2 | AGCTGGTCTCAAGAGGTGTGGCA | AGCCTCCCAGCTATCAACGACC |
| BrTT3 | CAGGATGGATGTATTTCATGTCG | TGTGCCTCGTTACGAGTGATAG |
| BrTT4 | GACTACTACTTCCGCATCACCAACAG | GCCTAGCTTAGGGACTTCAACAACC |
| BrTT5 | CTTCCTCGGTGGCGCAGGTG | ACACAGTTCTCCGTTACTTTCTCTGA |
| BrTT6 | CGAGAAAGAGGCACTCACCAATG | TGAACCTCCCGTTGCTCAGATA |
| BrTT7 | GTGGTTGCCGCCTCTAAATC | CTAGCTCGCGCATGAGTGTTC |
| BrTT8 | GGCTGAAGAGGCTGCGTCGG | GTGCTGTGCAAGCCCTCGCT |
| BrTT10 | GCGACTGTGCCAAGAAACGGT | CCCCACGTGAGATGTCTATCAAAGTG |
| BrTT12 | GCTCCACAGAGACATACGAGCCG | ACGGTGACGAAGCTGAGCATGTA |
| BrTT16 | TGCTCACATCGGTCTCATCGTCT | GCTCGTGTGGAGGAATGGAGGC |
| BrTT18 | AAGAAGAGCTCGAGAGCATCAAC | GCATACTTCTCCTTCTCTTCCAC |
| BrTTG1 | TCCTCCGGCGACTTCCTCCG | GCTGCGTCTCCACCACGGAC |
| BrTTG2 | AAACCTAAAGCAAAGCTTGTCTCCCA | ACTTCCTTTGACTTGCTTCTGTCCGT |
| BrBAN | TTAACTGGGCATACCCAATCTC | TGCATTTCTTTCCGGGTAATC |
新窗口打开|下载CSV
2 结果与分析
2.1 芸薹属作物中TT1同源基因家族成员的序列差异分析
在BRAD (http://brassicadb.org/brad/)数据库下载获取芸薹属作物TT1同源基因家族成员, 包括甘蓝型油菜BnTT1-1和BnTT1-2、白菜型油菜BrTT1、甘蓝BoTT1、芥菜型油菜BjTT1的氨基酸序列, 采用ClustalW (http://www.ebi.ac.uk/clustalw)及GEN EDOC软件进行氨基酸序列比对分析。其中BnTT1-1与BnTT1-2的氨基酸序列存在9处差异, 一致性达95%, 而BrTT1与BnTT1-1的氨基酸序列存在4处差异, BrTT1与BnTT1-2的氨基酸序列存在7处差异, 相似度均大于97.5% (图1)。白菜型油菜BrTT1与甘蓝型油菜BnTT1-1的相似度大于甘蓝型油菜BnTT1-1与BnTT1-2之间的相似度, 且芸薹属作物不同种之间的氨基酸序列相似度均大于95%, 说明BrTT1在进化过程中有一定的序列保守性。图1
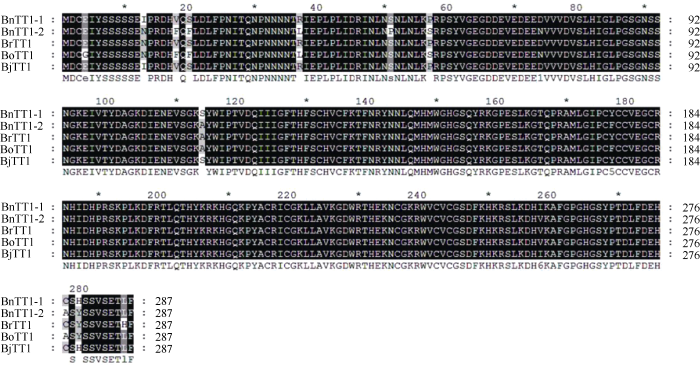 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1BrTT1同源基因氨基酸序列比对
Fig. 1BrTT1 homologous genes amino acid sequences alignment
2.2 不同遗传背景白菜型油菜中BrTT1的克隆及表达分析
从22份不同遗传背景的白菜型油菜(包括6个黄籽材料Y1~Y6、9个红褐籽材料R1~R9和7份黑籽材料B1~B7)中克隆了BrTT1基因序列(图2-A, B)。根据BRAD数据库信息可知, 该基因包含有2个外显子和1个内含子。Geneious软件对测序得到的BrTT1序列比对结果表明, 所有的黑籽材料(B1~B7) BrTT1-B序列一致; 9份不同来源的红褐籽材料(R1~R9)扩增获得的BrTT1-R序列表现一致, 且与黑籽材料BrTT1-B序列相比, BrTT1-R在第834 bp处(以起始密码子ATG为起点)多1个碱基A, 979 bp处存在1处碱基替换T→C; 黄籽材料Y2、Y3、Y4、Y5、Y6扩增得到的BrTT1-Y序列与前期对大黄油菜粒色主效基因BrTT1的研究结果一致, 黄籽序列相同; 与BrTT1-B序列相比, BrTT1-Y内含子区域存在17处SNP (single nucleotide polymorphisms)差异、2处片段缺失, 在外显子区域存在7处SNP差异, 分别是A→C、T→A、G→T、A→T、G→A、C→A、A→T, 发生有义突变的为A→C、C→A、A→T, 第2个外显子中的最后2个SNP (CNP, ANP)导致BrTT1-Y的DNA结合区域发生2个氨基酸变化(S氨基, H氨基), 这可能导致BrTT1-Y的功能丢失[31]。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2不同粒色的BrTT1基因的核苷酸序列比对
A: 不同粒色的白菜型油菜种子表型(黑色、红褐色和黄色); B: 来自不同粒色的 BrTT1的核苷酸序列。TT1-B: 从黑籽中扩增得到的BrTT1-B序列; TT1-R: 从红褐籽种子中扩增得到的BrTT1-R序列; TT1-Y: 从黄籽中扩增得到的BrTT1-Y序列。核苷酸多态性通过颜色突出显示, 共有序列下方的线代表内含子区域。标尺 = 5000 μm。
Fig. 2Nucleotide polymorphisms of BrTT1 genes from different seed color lines
A: phenotypes of B. rapa lines (black, reddish-brown, and yellow) with different seed colors; B: nucleotide sequences of B. rapa BrTT1 from the three lines with different seed color. TT1-B was BrTT1-B sequence amplified from the black seeds; TT1-R was BrTT1-R sequence amplified from the reddish-brown seeds; TT-Y was BrTT1-Y sequence amplified from the yellow seeds. Nucleotide polymorphisms were highlighted by color, the line below the consensus sequence represents the intron region. Bar = 5000 μm.
2.3 BrTT1在白菜型油菜不同粒色形成期种子中的表达分析
BrTT1在红褐、黑籽种子中的表达量均高于黄籽中, 且黑籽和红褐籽种子中BrTT1的表达没有显著差异(图3)。表明BrTT1-Y的内含子中2个片段的缺失以及外显子的SNP差异可能对BrTT1在种子形成期的表达具有重要影响。前期研究已知BrTT1全长序列为4586 bp, 包含启动子片段1796 bp、终止子下游序列986 bp、编码区序列1804 bp。等位基因序列分析发现, BrTT1在黄、褐籽的启动子序列相同[37]。因此推测BrTT1内含子中片段缺失以及外显子的SNP差异会影响到BrTT1-Y mRNA的剪接或稳定性。说明BrTT1在白菜型油菜油菜中粒色调控过程中均发挥重要的作用。图3
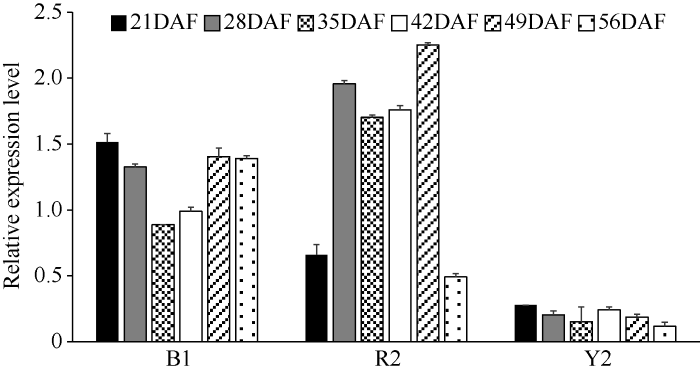 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3BrTT1基因在3种不同粒色种子形成期的表达分析
B1: 黑色种子系; R2: 红褐籽种子系; Y2: 黄籽种子系。DAF: 开花后天数。
Fig. 3Expression analysis of BrTT1 in the three types of B. rapa lines
B1: a black seed line; R2: a reddish-brown seed line; Y2: a yellow seed line. DAF: days after flowering.
2.4 BrTT1互作蛋白预测
将BrTT1 (Bra028067)信息提交至STRING (图4
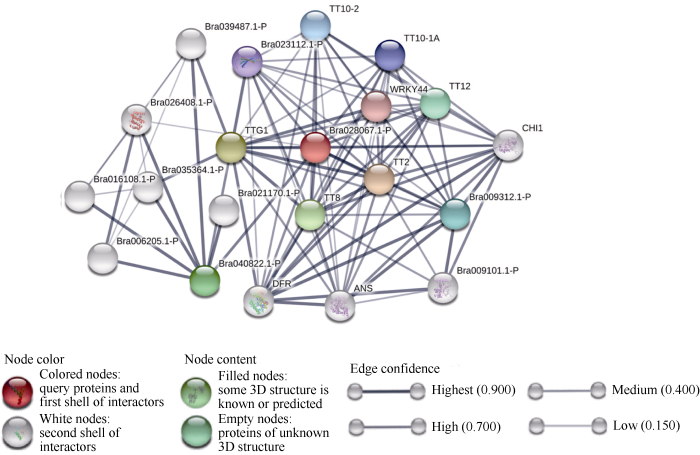 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4白菜型油菜BrTT1的互作蛋白预测分析
Fig. 4Protein-predictive analysis of BrTT1 in B. rapa
Table 1
表1
表1预测白菜型油菜BrTT1的互作蛋白的信息
Table 1
| 基因ID Gene ID | 蛋白类型 Protein type | 打分值 Score | 基因ID Gene ID | 蛋白类型 Protein type | 打分值Score |
|---|---|---|---|---|---|
| Bra027457 | BrTT3 | 0.996 | Bra005210 | BrTTG2-A, WRKY44 | 0.814 |
| Bra035532 | BrTT2 | 0.992 | Bra023112 | BrTTG2-B | 0.814 |
| Bra009770 | BrTTG1 | 0.909 | Bra026408 | Uncharacterized protein | 0.607 |
| Bra037887 | BrTT8 | 0.894 | Bra013652 | BrTT18 | 0.568 |
| Bra009312 | BrTT7 | 0.885 | Bra016108 | Uncharacterized protein | 0.568 |
| Bra040822 | Uncharacterized protein | 0.882 | Bra009101 | Uncharacterized protein | 0.568 |
| Bra003361 | BrTT12 | 0.881 | Bra039487 | Uncharacterized protein | 0.434 |
| Bra007142 | BrTT5 | 0.822 | Bra035364 | Uncharacterized protein | 0.434 |
| Bra037510 | BrTT10-2 | 0.821 | Bra006205 | Uncharacterized protein | 0.379 |
| Bra020720 | BrTT10-1A | 0.821 | Bra021170 | Uncharacterized protein | 0.365 |
新窗口打开|下载CSV
2.5 酵母双杂交筛选BrTT1互作蛋白
分别设计可能与BrTT1互作的类黄酮代谢基因(打分值在0.568~0.996)的特异引物(附表3), 克隆各基因CDS序列分别构建AD载体, 克隆BrTT1基因CDS序列构建pGBKT7-BrTT1载体。将各捕获载体分别与诱饵载体pGBKT7-BrTT1共转化到酵母菌株Y2H Gold中, 同时设阳性对照组(pGADT7-T + pGBKT7-53)和阴性对照组(pGADT7-T + pGBKT7-laminC)。由图5可知, BrTT1与BrTT2、BrTT3、BrTTg1均有较强的互作, 而与类黄酮途径其他蛋白则没有发生互作, 推测BrTT1在类黄酮途径中可能的互作蛋白有BrTT2、BrTT3、BrTTg1。图5
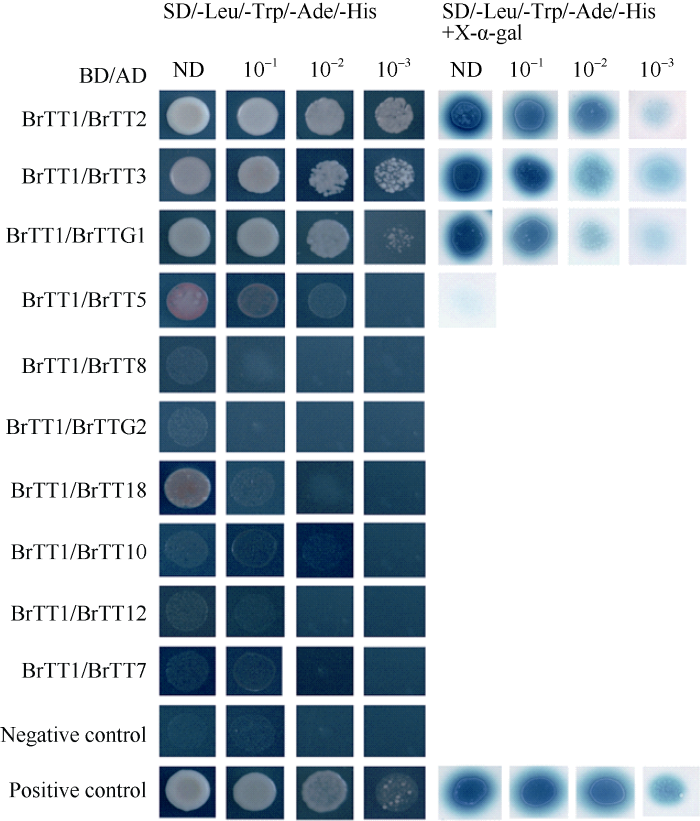 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5酵母双杂筛选BrTT1可能的互作蛋白
酵母细胞涂布于-Leu/-Trp/-Ade/-His的培养基, pGADT7- T/pGBKT7-53作为阳性对照, pGADT7-T/pGBKT7-laminC作为阴性对照。X-α-gal的染色深度表示互作强弱。
Fig. 5Screening for interactors of BrTT1 by a yeast two-hybrid system
Yeast cells were co-transformed and plated on -Leu/-Trp/-Ade/-His plates, with pGADT7-T/pGBKT7-53 as the positive control and pGADT7-T/pGBKT7-laminC as the negative control. X-α-gal activity was determined by colony-lift filter assay; the color intensity of the colonies depicts the qualitative binding strength of the interaction.
2.6 转基因株系中BrTT1的差异表达
为进一步探索BrTT1的功能, 本研究构建了BrTT1的超量表达载体p35S::BrTT1和RNAi干扰载体pBTT1转化白菜型油菜浩油11号。并通过PCR检测鉴定获得干扰表达BrTT1的转基因株系(附图1)和超量表达BrTT1的转基因株系(附图2)。在干扰株系中, BrTT1基因的表达显著低于对照株系B (图6-A), 从而导致干扰株系转基因种子种皮色素积累减少(图6-B), 表明干扰BrTT1可以减少转基因种子中黑色素的生物合成和沉积。与非转基因褐籽材料相比, 过量表达BrTT1所得转基因种子中, BrTT1的表达量及成熟种子的粒色并无显著差异(图6-A, B), 表明在BrTT1白菜型油菜粒色中具有重要的调节功能。附图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图1PCR扩增检测RNAi BrTT1得到的阳性转基因植株
检测引物为F35S3ND/FBTT1I (
Supplementary Fig. 1PCR amplification of a transgene fragment in transgenic plants with BrTT1 down-regulated by RNAi using genomic DNA isolated from the seedlings as templates
The primer of F35S3ND/FBTT1I (
附图2*
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT附图2*PCR扩增检测过表达 BrTT1得到的阳性转基因植株
检测引物为F35S3ND/FBTT1I (
Supplementary Fig. 2PCR amplification of a transgene fragment in transgenic plants over-expressing BrTT1 using genomic DNA isolated from the seedlings as templates
The primer of F35S3ND/CB-R (
图6
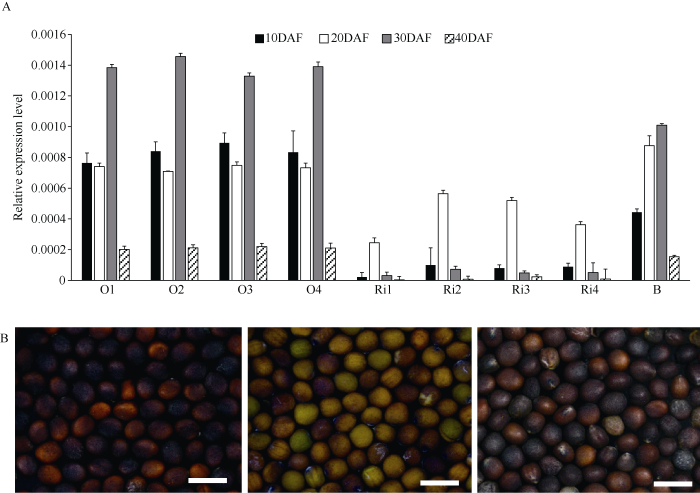 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6过量和干扰表达BrTT1基因
A: 转基因种子中BrTT1的差异表达; O1~O4: 过量表达转基因株系; Ri1~Ri4: 干扰表达转基因株系; B: 非转基因黑籽材料; DAF: 开花后天数。B: 左、中、右分别为过量表达、干扰表达BrTT1基因、非转基因褐籽对照成熟种子表型。标尺 = 5000 μm。
Fig. 6Expression of BrTT1 in RNAi and over-expression lines
A: differential expression analysis of BrTT1 in seeds of the transgenic lines; O1-O4: over-expression lines; Ri1-Ri4: RNAi expression lines; B: non-transgenic lines as the control; DAF: days after flowering. B: left, middle and right represent seed color phenotype in transgenic lines of over-expression, RNAi expression, and the non-transgenic black seed lines, respectively. Bar = 5000 μm.
2.7 转基因株系中类黄酮代谢相关基因的差异表达
为进一步了解BrTT1可能的调控活性, 取过表达和抑制表达BrTT1后, 转基因株系发育阶段的种子(开花后10、20、30、40 d)为材料, 通过qRT-PCR表达谱分析了类黄酮途径基因的表达水平。根据差异表达情况, 可将这些基因分为3类(图7), 第1类为正调控表达基因, 主要包括BrTTG1、BrTT3、BrTT4、BrBAN、BrTT18、BrTT6和BrTTG2, 即BrTT1的下调表达降低了这些基因的表达, 而BrTT1的上调表达则提高了它们在发育中种子的表达, 表明这组基因可能受到BrTT1的正向调控; 第2类包括BrTT2、BrTT7和BrTT10, 其表达模式与第1类相反, 即下调BrTT1表达可增加这些基因的表达, 而上调BrTT1则可降低这组基因表达, 表明这些基因受BrTT1负向调控; 第3组包括BrTT5、BrTT8、BrTT12和BrTT16, 其表达对BrTT1表达的变化无反应, 在2种不同类型的转基因品系中均未观察到明显的变化, 表明这些基因未受BrTT1调控。图7
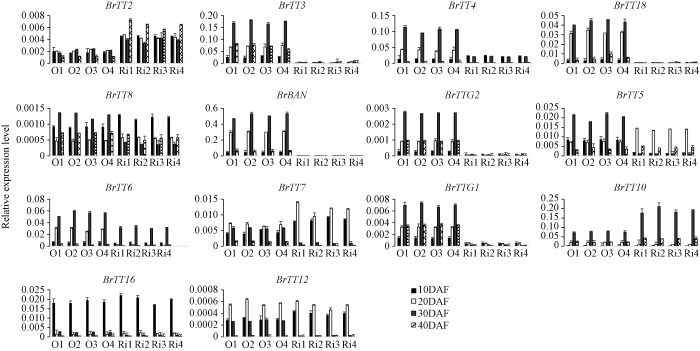 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7RNAi和超量表达BrTT1基因的转基因株系中类黄酮相关基因的表达分析
O1~O4: 过量表达转基因株系; Ri1~Ri4: 干扰表达转基因株系; DAF: 开花后天数。
Fig. 7Expression analysis of genes involved in the flavonoid biosynthesis in BrTT1-RNAi and over-expression lines
O1-O4: over-expression lines; Ri1-Ri4: RNAi expression lines; DAF: days after flowering.
3 讨论
原花色素是一种广泛存在于植物果实、树叶、种皮中的植物色素, 其生物合成及合成途径中主要基因功能的解析已在拟南芥、葡萄等植物中有较详细的研究, 主要由公共苯丙烷途径、类黄酮途径及原花色素途径组成的复合途径[38,39,40,41,42,43,44,45,46]。油菜的种皮颜色是由原花色素决定, 且黄籽油菜种皮中不含有原花色素[47,48], 在油菜中原花色素的合成路径及调控机制尚未完全清楚。模式植物拟南芥原花色素代谢途径主要涉及到12个结构基因、6个转录因子及3个转运蛋白的调控[49]。植物类黄酮代谢反应犹如一个链条式结构, 该结构中涉及的每一个酶的突变都可能导致整个代谢路径的中断, 原花色素积累受阻[50,51]。在白菜型油菜中, BrTT1被认为是调节种皮色素的生物合成和沉积的主要转录因子[30,31]。本研究对22份不同粒色(黑籽、红褐籽和黄籽)的白菜型油菜品系中BrTT1进行了克隆测序。序列比较表明, BrTT1序列在不同粒色材料中存在固定的差异位点。与黑籽和红褐籽中的BrTT1相比, 来自黄籽的BrTT1序列在2个外显子区域共有7个SNP位点, 而在内含子区域具有2个片段缺失, 这可能导致黄籽中BrTT1调节活性丧失。qRT-PCR分析表明, BrTT1在黄籽品系和RNAi转基因系中的表达非常低。这些结果进一步证明, BrTT1在白菜型油菜粒色性状中具有重要的调控作用, 白菜型种子粒色表型可以通过BrTT1序列多态性来确定, 与前研究结果在白菜和甘蓝型油菜中沉默TT1或下调表达TT1, 导致种皮中黄酮类化合物的减少, 从而形成黄籽的研究结果相一致[28,29,30,31]。在芥菜型油菜中, 从不同的粒色材料(黄籽和褐籽)克隆了BjTT1, 并鉴定到8个单核苷酸多态性(SNP)位点, 但未检测到BjTT1的差异表达[28]。拟南芥AtTT1作为调节因子能够与转录因子R2R3-MYB (AtTT2)和WD40转录因子(AtTTG1)相互作用, 从而调节结构基因如AtTT4 (CHS)、AtTT3 (DFR)和AtTT18 (LDOX)的表达[52,53,54]。酵母双杂分析表明, BrTT1可以与BrTT2、BrTT3和BrTTG1相互作用, 已有研究表明在拟南芥中TT1与TT2和TTG1的相互作用, 其中TT2的异位表达可以部分恢复tt1突变体的表型[49]。此外, 本研究检测到BrTT1与BrTT3的相互作用, 在白菜型油菜中, BrTT3 (BrDFR)编码二氢黄酮醇4-还原酶, BrTT3受2个转录因子BrMYB2-2和BrTT8调控[55,56]。尽管通过酵母双杂系统检测到白菜型油菜中BrTT1与BrTT3的相互作用, 且干扰和过量表达BrTT1的转基因品系中BrTT3的表达受到了明显的影响, 但仍需要更多试验来进一步证明BrTT1和BrTT3在白菜型油菜中是否存在相互作用。
在甘蓝型油菜中, TT4、TT5、TT6、TT3、TT18和BAN在黄籽种子中表达量较低, 黄籽与褐籽之间的表达差异显著。此外, BnTT1的沉默表达降低类黄酮合成相关基因的表达, 如TT3、TT4、TT5、TT6、TT7和TT18[31]。本研究表明, 白菜型RNAi转基因品系中下调BrTT1的表达导致BrTTG1、BrTTG2、BrTT3、BrTT4、BrTT6、BrBAN和BrTT18的表达降低。BrTTG1和BrTTG2编码2个转录因子(WD40和WRKY), BrTT4编码查尔酮合酶(CHS), 这是类黄酮途径中催化丙二酰辅酶A和4-香豆酰辅酶A缩合的第一种酶[39,40,41]。BrTT6、BrTT3、BrTT18和BrBAN是类黄酮生物合成早期和中期重要的结构基因。这些基因的表达均受到BrTT1的正向调控。此外, 与BrTT1的表达呈负向调控的结构基因主要包括BrTT2、BrTT7和BrTT10。BrTT7编码黄烷酮3'-羟化酶(F3'H), TT10编码漆酶15 (LAC15), 将原花青素(PA)聚合成更大的聚合物并进一步氧化原花色素, 从而使种皮呈现褐色[57], 这一类基因受BrTT1负调控。根据我们之前对大黄油菜中相应BrTT的功能表达谱的研究, BrTT4、BrTT5、BrTT6、BrTT3、BrTT18和BAN在黄籽种子中表达较低, 但在近等基因系褐籽种子中表达较高。该结果与本研究RNAi转基因系中类黄酮代谢BrTT的下调表达的结果相一致。进一步表明BrTT1是调节白菜型油菜粒色性状的关键基因之一。BrTT1的调控活性的分子解析为芸薹属作物种皮中类黄酮的生物合成研究提供借鉴。
4 结论
前期研究对白菜型黄籽油菜大黄的粒色性状进行了基因精细定位、克隆及拟南芥中遗传转化分析, 初步明确了大黄油菜粒色性状受1对隐性基因BrTT1控制。本研究在此基础上明确BrTT1在其他白菜型油菜黄籽、红褐籽、黑籽品种中的序列及表达特性, 同一粒色材料所得BrTT1序列完全一致, 且不同粒色材料存在固定的差异位点, 为更多不同遗传背景的白菜型油菜粒色研究奠定基础。通过酵母双杂及超量和干扰BrTT1的表达, 解析BrTT1控制粒色可能的顺式作用元件, BrTT1与R2R3-MYB转录因子(TT2)及TTG1转录因子互作影响类黄酮合成路径结构基因TT3的表达, 初步解析BrTT1调控油菜粒色的分子机制, 为白菜型油菜优质育种提供新思路, 同时也为甘蓝型油菜粒色调控网络的研究提供借鉴。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s10535-005-0053-2URL [本文引用: 1]
DOI:10.1023/A:1018646223643URL [本文引用: 1]
DOI:10.3923/jbs.2004.731.734URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1021/jf00056a020URL [本文引用: 1]
URLPMID:28855913 [本文引用: 1]
[本文引用: 1]
DOI:10.1139/g07-068URLPMID:17893725 [本文引用: 1]

Yellow seed is one of the most important traits of Brassica napus L. Efficient selection of the yellow-seed trait is one of the most important objectives in oilseed rape breeding. Two recombinant inbred line (RIL) populations (RIL-1 and RIL-2) were analyzed for 2 years at 2 locations. Four hundred and twenty SSR, RAPD, and SRAP marker loci covering 1744 cM were mapped in 26 linkage groups of RIL-1, while 265 loci covering 1135 cM were mapped in 20 linkage groups of RIL-2. A total of 19 QTLs were detected in the 2 populations. A major QTL was detected adjacent to the same marker (EM11ME20/200) in both maps in both years. This major QTL could explain 53.71%, 39.34%, 42.42%, 30.18%, 24.86%, and 15.08% of phenotypic variation in 6 combinations (location x year x population). BLASTn analysis of the sequences of the markers flanking the major QTL revealed that the homologous region corresponding to this major QTL was anchored between genes At5g44440 and At5g49640 of Arabidopsis thaliana chromosome 5 (At C5). Based on comparative genomic analysis, the bifunctional gene TT10 is nearest to the homologue of EM11ME20/200 on At C5 and can be considered an important candidate gene for the major QTL identified here. Besides providing an effective strategy for marker-assisted selection of the yellow-seed trait in B. napus, our results also provide important clues for cloning of the candidate gene corresponding to this major QTL.
DOI:10.1007/BF00293352URLPMID:24270566 [本文引用: 1]

The inheritance of seed colour was investigated in the progenies of crosses between seven yellow seeded forms and a brown seeded one of turnip rape (Brassica campestris L.). Seed colour differences were found to be determined in each case by one or two genes with epistatic effect. Moreover, independent inheritance of hilum colour was observed. Seed colour was predominantly but not completely determined by the maternal parent. In total, two genes for seed colour with epistatic (Br 1 br 1; Br 6 br 6) and four with hypostatic effect (Br 3 br 3; Br 4 br 4; Br 5 br 5; Br 7 br 7) as well as one gene for hilum colour (Br 8 br 8) were distinguished. Two alleles for yellow seed colour (br 1 (1) and br 1 (2) ) were present in the locus Br 1 br 1, whereas in the locus Br 6 br 6 one allele was found for yellow seed colour (br 6 (1) ) and another for light hilum colour (br 6 (2) ). The possibilities of multiple allelism are discussed.
DOI:10.4141/cjps79-100URL [本文引用: 1]
DOI:10.1371/journal.pone.0044145URLPMID:22984469 [本文引用: 1]

Yellow seed is a desirable quality trait of the Brassica oilseed species. Previously, several seed coat color genes have been mapped in the Brassica species, but the molecular mechanism is still unknown. In the present investigation, map-based cloning method was used to identify a seed coat color gene, located on A9 in B. rapa. Blast analysis with the Arabidopsis genome showed that there were 22 Arabidopsis genes in this region including at4g09820 to at4g10620. Functional complementation test exhibited a phenotype reversion in the Arabidopsis thaliana tt8-1 mutant and yellow-seeded plant. These results suggested that the candidate gene was a homolog of TRANSPARENT TESTA8 (TT8) locus. BrTT8 regulated the accumulation of proanthocyanidins (PAs) in the seed coat. Sequence analysis of two alleles revealed a large insertion of a new class of transposable elements, Helitron in yellow sarson. In addition, no mRNA expression of BrTT8 was detected in the yellow-seeded line. It indicated that the natural transposon might have caused the loss in function of BrTT8. BrTT8 encodes a basic/helix-loop-helix (bHLH) protein that shares a high degree of similarity with other bHLH proteins in the Brassica. Further expression analysis also revealed that BrTT8 was involved in controlling the late biosynthetic genes (LBGs) of the flavonoid pathway. Our present findings provided with further studies could assist in understanding the molecular mechanism involved in seed coat color formation in Brassica species, which is an important oil yielding quality trait.
DOI:10.1111/j.1469-8137.2004.01217.xURLPMID:15720617 [本文引用: 1]

Proanthocyanidins are oligomeric and polymeric end products of the flavonoid biosynthetic pathway. They are present in the fruits, bark, leaves and seeds of many plants, where they provide protection against predation. At the same time they give flavor and astringency to beverages such as wine, fruit juices and teas, and are increasingly recognized as having beneficial effects on human health. The presence of proanthocyanidins is also a major quality factor for forage crops. The past 2 years have seen important breakthroughs in our understanding of the biosynthesis of the building blocks of proanthocyanidins, the flavan-3-ols (+)-catechin and (-)-epicatechin. However, virtually nothing is known about the ways in which these units are assembled into the corresponding oligomers in vivo. Molecular genetic approaches are leading to an understanding of the regulatory genes that control proanthocyanidin biosynthesis, and this information, together with increased knowledge of the enzymes specific for the pathway, will facilitate the genetic engineering of plants for introduction of value-added nutraceutical and forage quality traits.
DOI:10.1007/s11240-014-0593-2URL [本文引用: 1]
URLPMID:18470187 [本文引用: 1]
DOI:10.1007/BF03191854URL [本文引用: 1]
URLPMID:17846742 [本文引用: 2]
[本文引用: 2]
DOI:10.2135/cropsci1973.0011183X001300010039xURL [本文引用: 2]
DOI:10.1007/s00122-011-1754-xURL [本文引用: 1]

The development of yellow-seeded cultivars in Brassica rapa (B. rapa) would improve the quality and quantity of available oil. The identification and mapping of the seed coat color gene may aid in the development of yellow-seeded cultivars and facilitate introgression of the yellow-seeded gene into desirable Brassica napus (B. napus) lines through marker-assisted selection. In the current study, we investigated the inheritance of a yellow-seeded landrace in B. rapa, "Dahuang'', originating from the Qinghai-Tibetan plateau. Genetic analysis revealed that the phenotype of the yellow-seeded trait in Dahuang is controlled by one recessive gene, termed Brsc1. Mapping of the Brsc1 gene was subsequently conducted in a BC1 population comprised 456 individuals, derived from (Dahuang x 09A-126) x Dahuang. From a survey of 256 amplified fragment length polymorphism (AFLP) primer combinations, 10 tightly linked AFLP markers were obtained. The closest AFLP markers flanking Brsc1, Y10 and Y06, were 0.2 and 0.4 cM away, respectively. Subsequently, using simple sequence repeat (SSR) markers in the reference map, the Brsc1 gene was mapped on A09 in B. rapa. Blast analysis revealed that seven AFLP markers showed sequence homology to A09 of B. rapa, wherein six AFLP markers in our map were in the same order as those in A09 of B. rapa. The two closest markers, Y10 and Y06, delimited the Brsc1 gene within a 2.8 Mb interval. Furthermore, Y05 and Y06, the two closest AFLP markers on one side linked to Brsc1, were located in scaffold 000059 on A09 of B. rapa, whereas the closet AFLP marker on the opposite side of Brsc1, Y10, was located in scaffold000081 on A09 of B. rapa. Molecular markers developed from these studies may facilitate marker-assisted selection (MAS) of yellow-seeded lines in B. rapa and B. napus and expedite the process of map-based cloning of Brsc1.
DOI:10.3724/SP.J.1006.2010.01634URL [本文引用: 3]

TT1 gene encodes a WIP domain protein with Zn-finger, which is essential for seed coat development and organ color in Arabidopsis, its mutation causes transparent testa. In order to study molecular mechanism of seed coat color in rapeseed, TT1 gene was cloned from Brassica juncea using homology-based cloning and RACE (rapid-amplification of cDNA ends) strategy. A modified allele-specific PCR procedure developed for assaying single nucleotide polymorphisms of TT1 gene in Brassica juncea. In this study, cloning and characterization of the orthologous TT1 gene from Brassica juncea were reported. The full length DNA sequence of TT1 gene was 2 197 bp with one intron. The TT1 cDNA sequence was 1 412 bp in length, 903 bp of which was open reading frames (ORF) encoding a deduced polypeptide of 300 amino acids with a predicted molecular weight of 33.97 kD and an isoelectric point of 6.99. The above-mentioned genomic sequences showed 99% identity with the TT1 gene from Brassica napus, and 85% identity with the TT1 gene from Arabidopsis. RT-PCR analysis showed that TT1 expressed in the seed coats of both the black- and the yellow-seeded B. juncea lines. Comparison of sequences of Sichuan Yellow (yellow seed) and purple-leaf mustard (black seed) revealed nucleotide variation at the eight sites which all are located within the coding region of the gene. However, the mutations of the TT1 gene at these sites had no effect on the seed coat color in B. juncea through comparing the sequences of NILB and purple-leaf mustard. The allele-specific PCR of the TT1 gene could distinguish purple-leaf mustard from Sichuan Yellow.]]>
DOI:10.1016/j.plantsci.2016.10.012URLPMID:27964783 [本文引用: 2]

TRANSPARENT TESTA1 (TT1) is a zinc finger protein that contains a WIP domain. It plays important roles in controlling differentiation and pigmentation of the seed coat endothelium, and can affect the expression of early biosynthetic genes and late biosynthetic genes of flavonoid biosynthesis in Arabidopsis thaliana. In Brassica napus (AACC, 2n=38), the functions of BnTT1 genes remain unknown and few studies have focused on their roles in fatty acid (FA) biosynthesis. In this study, BnTT1 family genes were silenced by RNA interference, which resulted in yellow rapeseed, abnormal testa development (a much thinner testa), decreased seed weight, and altered seed FA composition in B. napus. High-throughput sequencing of genes differentially expressed between developing transgenic B. napus and wild-type seeds revealed altered expression of numerous genes involved in flavonoid and FA biosynthesis. As a consequence of this altered expression, we detected a marked decrease of oleic acid (C18:1) and notable increases of linoleic acid (C18:2) and alpha-linolenic acid (C18:3) in mature transgenic B. napus seeds by gas chromatography and near-infrared reflectance spectroscopy. Meanwhile, liquid chromatography-mass spectrometry showed reduced accumulation of flavonoids in transgenic seeds. Therefore, we propose that BnTT1s are involved in the regulation of flavonoid biosynthesis, and may also play a role in FA biosynthesis in B. napus.
DOI:10.1371/journal.pone.0166464URLPMID:27829069 [本文引用: 4]

A yellow seed coat is a desirable agronomic trait in the seeds of oilseed-type Brassica crops. In this study, we identified a candidate gene for seed coat color in Dahuang, a landrace of Brassica rapa. A previous study of Dahuang mapped the seed coat color gene Brsc1 to a 2.8-Mb interval on chromosome A9 of B. rapa. In the present study, the density of the linkage map for Brsc1 was increased by adding simple sequence repeat (SSR) markers, and the candidate region for Brsc1 was narrowed to 1.04 Mb. In addition, whole-genome resequencing with bulked segregant analysis (BSA) was conducted to identify candidate intervals for Brsc1. A genome-wide comparison of SNP profiles was performed between yellow-seeded and brown-seeded bulk samples. SNP index analyses identified a major candidate interval on chromosome A9 (A09:18,255,838-18,934,000, 678 kb) containing a long overlap with the target region recovered from the fine mapping results. According to gene annotation, Bra028067 (BrTT1) is an important candidate gene for Brsc1 in the overlapping region. Quantitative reverse transcription (qRT)-PCR revealed that BrTT1 mainly functions in the seed. Point mutations and small deletions in BrTT1 were found between yellow- and brown-seeded Dahuang plants. Collectively, the expression and sequence analysis results provide preliminary evidence that BrTT1 is a candidate gene for the seed coat color trait in Dahuang.
[本文引用: 5]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1007/s00299-002-0560-yURLPMID:12789436 [本文引用: 1]

An efficient protocol for the production of transgenic Brassica napus cv. Westar plants was developed by optimizing two important parameters: preconditioning time and co-cultivation time. Agrobacterium tumefaciens-mediated transformation was performed using hypocotyls as explant tissue. Two variants of a green fluorescent protein (GFP)-encoding gene--mGFP5-ER and eGFP--both under the constitutive expression of the cauliflower mosaic virus 35S promoter, were used for the experiments. Optimizing the preconditioning time to 72 h and co-cultivation time with Agrobacterium to 48 h provided the increase in the transformation efficiency from a baseline of 4% to 25%. With mGFP5-ER, the transformation rate was 17% and with eGFP it was 25%. Transgenic shoots were selected on 200 mg/l kanamycin. Rooting efficiency was 100% on half-strength Murashige and Skoog medium with 10 g/l sucrose and 0.5 mg/l indole butyric acid in the presence of kanamycin.
DOI:10.1038/nprot.2008.73URL [本文引用: 1]
[本文引用: 1]
.
[本文引用: 1]
URLPMID:3386631 [本文引用: 1]
DOI:10.1105/tpc.111.083667URLPMID:21441431 [本文引用: 2]

The model organism Arabidopsis thaliana is readily used in basic research due to resource availability and relative speed of data acquisition. A major goal is to transfer acquired knowledge from Arabidopsis to crop species. However, the identification of functional equivalents of well-characterized Arabidopsis genes in other plants is a nontrivial task. It is well documented that transcriptionally coordinated genes tend to be functionally related and that such relationships may be conserved across different species and even kingdoms. To exploit such relationships, we constructed whole-genome coexpression networks for Arabidopsis and six important plant crop species. The interactive networks, clustered using the HCCA algorithm, are provided under the banner PlaNet (http://aranet.mpimp-golm.mpg.de). We implemented a comparative network algorithm that estimates similarities between network structures. Thus, the platform can be used to swiftly infer similar coexpressed network vicinities within and across species and can predict the identity of functional homologs. We exemplify this using the PSA-D and chalcone synthase-related gene networks. Finally, we assessed how ontology terms are transcriptionally connected in the seven species and provide the corresponding MapMan term coexpression networks. The data support the contention that this platform will considerably improve transfer of knowledge generated in Arabidopsis to valuable crop species.
DOI:10.1023/a:1005921914384URLPMID:9526507 [本文引用: 2]

To identify DNA sequences of the Arabidopsis thaliana chalcone synthase gene (CHS) concerned with induction by UV-B and UV-A/blue light, AtCHS promoter constructions were assayed by transient expression in protoplasts prepared from two different lines of cultured A. thaliana cells. The protoplasts responded similarly to A. thaliana leaf tissue in light-dependent CHS transcript accumulation. The reporter enzyme beta-glucuronidase (GUS) was used to monitor light-responsive promoter activity. A 1972 bp promoter conferred UV-B and UV-A/blue light induction of GUS activity. Deletion to 164 bp resulted in reduced promoter strength but retention of responsiveness to UV-B and UV-A/blue light. Further deletion abolished transcriptional activity. The 164 bp promoter contains sequences closely resembling LRUPcCHS, (light-responsive unit of the Petroselinum crispum CHS promoter). This A. thaliana CHS promoter region, designated LRUAtCHS, was sufficient to confer UV-B and UV-A/blue light responsiveness to a heterologous core promoter. Mutation of sequences in LRUAtCHS corresponding to the ACGT element and the MYB recognition element of LRUPcCHS resulted in inactivation of the 164 bp and 335 bp promoter deletions. However, the mutant 668 bp promoter retained residual UV-B and UV-A/blue light-induced expression, indicating the presence of additional functional sequences upstream of -335. Mutation of a single G-box-like sequence around -442 had no effect on light responsiveness, indicating that it does not function in light regulation of this promoter. Since no difference in responsiveness to UV-B and UV-A/blue light was observed with any promoter variant, we conclude that the two phototransduction pathways regulate transcription factors which interact with common promoter elements. The results from-our analysis of a A. thaliana light-responsive promoter will facilitate the study of light-dependent gene regulation by genetic means in Arabidopsis thaliana.
DOI:10.1105/tpc.110.074161URLPMID:20511296 [本文引用: 2]

Defects in phenylpropanoid biosynthesis arising from deficiency in hydroxycinnamoyl CoA:shikimate hydroxycinnamoyl transferase (HCT) or p-coumaroyl shikimate 3'-hydroxylase (C3'H) lead to reduced lignin, hyperaccumulation of flavonoids, and growth inhibition in Arabidopsis thaliana. It was previously reported that flavonoid-mediated inhibition of auxin transport is responsible for growth reduction in HCT-RNA interference (RNAi) plants. This conclusion was based on the observation that simultaneous RNAi silencing of HCT and chalcone synthase (CHS), an enzyme essential for flavonoid biosynthesis, resulted in less severe dwarfing than silencing of HCT alone. In an attempt to extend these results using a C3'H mutant (ref8) and a CHS null mutant (tt4-2), we found that the growth phenotype of the ref8 tt4-2 double mutant, which lacks flavonoids, is indistinguishable from that of ref8. Moreover, using RNAi, we found that the relationship between HCT silencing and growth inhibition is identical in both the wild type and tt4-2. We conclude from these results that the growth inhibition observed in HCT-RNAi plants and the ref8 mutant is independent of flavonoids. Finally, we show that expression of a newly characterized gene bypassing HCT and C3'H partially restores both lignin biosynthesis and growth in HCT-RNAi plants, demonstrating that a biochemical pathway downstream of coniferaldehyde, probably lignification, is essential for normal plant growth.
URLPMID:26347569 [本文引用: 1]
[本文引用: 1]
DOI:10.1104/pp.109.152801URLPMID:20357139 [本文引用: 1]

Three genes encoding flavonoid 3'-hydroxylase (F3'H) in apple (Malus x domestica), designated MdF3'HI, MdF3'HIIa, and MdF3'HIIb, have been identified. MdF3'HIIa and MdF3'HIIb are almost identical in amino acid sequences, and they are allelic, whereas MdF3'HI has 91% nucleotide sequence identity in the coding region to both MdF3'HIIa and MdF3'HIIb. MdF3'HI and MdF3'HII genes are mapped onto linkage groups 14 and 6, respectively, of the apple genome. Throughout the development of apple fruit, transcriptional levels of MdF3'H genes along with other anthocyanin biosynthesis genes are higher in the red-skinned cv Red Delicious than that in the yellow-skinned cv Golden Delicious. Moreover, patterns of MdF3'H gene expression correspond to accumulation patterns of flavonoids in apple fruit. These findings suggest that MdF3'H genes are coordinately expressed with other genes in the anthocyanin biosynthetic pathway in apple. The functionality of these apple F3'H genes has been demonstrated via their ectopic expression in both the Arabidopsis (Arabidopsis thaliana) transparent testa7-1 (tt7) mutant and tobacco (Nicotiana tabacum). When grown under nitrogen-deficient conditions, transgenic Arabidopsis tt7 seedlings expressing apple F3'H regained red color pigmentation and significantly accumulated both 4'-hydrylated pelargonidin and 3',4'-hydrylated cyanidin. When compared with wild-type plants, flowers of transgenic tobacco lines overexpressing apple F3'H genes exhibited enhanced red color pigmentation. This suggests that the F3'H enzyme may coordinately interact with other flavonoid enzymes in the anthocyanin biosynthesis pathway.
URLPMID:12376625 [本文引用: 1]
URLPMID:11193333 [本文引用: 1]
DOI:10.1111/j.1467-7652.2004.00094.xURLPMID:17147621 [本文引用: 1]

Flavonoids are secondary metabolites that are specific to higher plants. PAP1, a member of the family of MYB domain transcription factors in Arabidopsis, is a positive regulator of the biosynthesis of anthocyanin. We show here that a chimeric PAP1 repressor, in which the EAR-motif repression domain from SUPERMAN was fused to PAP1, suppressed the expression of four flavonoid biosynthetic genes, namely CHS, DFR, LDOX, and BAN, in siliques, and inhibited the accumulation of proanthocyanidin, even in the presence of an endogenous positive regulator, such as TT2. This suppression resulted in the formation of light yellow seeds in 35S::PAP1SRDX transgenic plants. Our results indicate that PAP1 has the potential ability to regulate the biosynthesis not only of anthocyanin but also of proanthocyanidin. Our gene silencing system, using chimeric repressors, appears to be a useful tool for the manipulation of the biosynthesis of secondary metabolites in plants.
DOI:10.1111/j.1365-313X.2007.03373.xURLPMID:18036197 [本文引用: 1]

In all higher plants studied to date, the anthocyanin pigment pathway is regulated by a suite of transcription factors that include Myb, bHLH and WD-repeat proteins. However, in Arabidopsis thaliana, the Myb regulators remain to be conclusively identified, and little is known about anthocyanin pathway regulation by TTG1-dependent transcriptional complexes. Previous overexpression of the PAP1 Myb suggested that genes from the entire phenylpropanoid pathway are targets of regulation by Myb/bHLH/WD-repeat complexes in Arabidopsis, in contrast to other plants. Here we demonstrate that overexpression of Myb113 or Myb114 results in substantial increases in pigment production similar to those previously seen as a result of over-expression of PAP1, and pigment production in these overexpressors remains TTG1- and bHLH-dependent. Also, plants harboring an RNAi construct targeting PAP1 and three Myb candidates (PAP2, Myb113 and Myb114) showed downregulated Myb gene expression and obvious anthocyanin deficiencies. Correlated with these anthocyanin deficiencies is downregulation of the same late anthocyanin structural genes that are downregulated in ttg1 and bHLH anthocyanin mutants. Expression studies using GL3:GR and TTG1:GR fusions revealed direct regulation of the late biosynthetic genes only. Functional diversification between GL3 and EGL3 with regard to activation of gene targets was revealed by GL3:GR studies in single and double bHLH mutant seedlings. Expression profiles for Myb and bHLH regulators are also presented in the context of pigment production in young seedlings.
DOI:10.1111/j.1365-313X.2011.04603.xURLPMID:21477081 [本文引用: 2]

Wild type seed coats of Arabidopsis thaliana are brown due to the accumulation of proanthocyanidin pigments (PAs). The pigmentation requires activation of phenylpropanoid biosynthesis genes and mutations in some of these genes cause a yellow appearance of seeds, termed transparent testa (tt) phenotype. The TT1 gene encodes a WIP-type zinc finger protein and is expressed in the seed coat endothelium where most of the PAs accumulate in wild type plants. In this study we show that TT1 is not only required for correct expression of PA-specific genes in the seed coat, but also affects CHS, encoding the first enzyme of flavonoid biosynthesis. Many steps of this pathway are controlled by complexes of MYB and BHLH proteins with the WD40 factor TTG1. We demonstrate that TT1 can interact with the R2R3 MYB protein TT2 and that ectopic expression of TT2 can partially restore the lack in PA production in tt1. Reduced seed coat pigmentation was obtained using a TT1 variant lacking nuclear localisation signals. Based on our results we propose that the TT2/TT8/TTG1 regulon may also comprise early genes like CHS and discuss steps to further unravel the regulatory network controlling flavonoid accumulation in endothelium cells during A. thaliana seed development.
[本文引用: 1]
[本文引用: 1]
URLPMID:15255866 [本文引用: 1]
DOI:10.1016/j.scienta.2012.01.013URL [本文引用: 1]
DOI:10.1016/s1360-1385(99)01405-3URLPMID:10322557 [本文引用: 1]

Modification of the plant primary cell wall is required for both cell expansion and for developmental events, such as fruit softening, where cell size remains static but where wall loosening is an important feature. Recent studies suggest that the cellulose-xyloglucan network is targeted by similar enzymatic activities in both expanding cells and ripening fruit but that unique isoforms are expressed in each process. Disassembly of this structural network probably involves the concerted and synergistic action of suites of these enzyme families, where one family of cell wall modifying proteins might mediate the activity of another, providing the basis for orchestrating ordered cell wall restructuring and turnover.
DOI:10.1371/journal.pone.0161394URLPMID:27548675 [本文引用: 1]

Rapid cycling Brassica rapa, also known as Wisconsin Fast Plants, are a widely used organism in both K-12 and college science education. They are an excellent system for genetics laboratory instruction because it is very easy to conduct genetic crosses with this organism, there are numerous seed stocks with variation in both Mendelian and quantitative traits, they have a short generation time, and there is a wealth of educational materials for instructors using them. Their main deficiency for genetics education is that none of the genetic variation in RCBr has yet been characterized at the molecular level. Here we present the first molecular characterization of a gene responsible for a trait in Fast Plants. The trait under study is purple/nonpurple variation due to the anthocyaninless locus, which is one of the Mendelian traits most frequently used for genetics education with this organism. We present evidence that the DFR gene, which encodes dihyroflavonol 4-reductase, is the candidate gene for the anthocyaninless (ANL) locus in RCBr. DFR shows complete linkage with ANL in genetic crosses with a total of 948 informative chromosomes, and strains with the recessive nonpurple phenotype have a transposon-related insertion in the DFR which is predicted to disrupt gene function.
DOI:10.1016/j.gene.2014.08.013URLPMID:25108127 [本文引用: 1]

Flavonoids including anthocyanins provide flower and leaf colors, as well as other derivatives that play diverse roles in plant development and interactions with the environment. Dihydroflavonol 4-reductase (DFR) is part of an important step in the flavonoid biosynthetic pathway of anthocyanins. This study characterized 12 DFR genes of Brassica rapa and investigated their association with anthocyanin coloration, as well as cold and freezing stress in several genotypes of B. rapa. Comparison of sequences of these genes with DFR gene sequences from other species revealed a high degree of homology. Constitutive expression of the genes in several pigmented and non-pigmented lines of B. rapa demonstrated correlation with anthocyanin accumulation for BrDFR8 and 9. Conversely, BrDFR2, 4, 8 and 9 only showed very high responses to cold stress in pigmented B. rapa samples. BrDFR1, 3, 5, 6 and 10 responded to cold and freezing stress treatments, regardless of pigmentation. BrDFRs were also shown to be regulated by two transcription factors, BrMYB2-2 and BrTT8, contrasting with anthocyanin accumulation and cold and freezing stress. Thus, the above results suggest that these genes are associated with anthocyanin biosynthesis and cold and freezing stress tolerance and might be useful resources for development of cold and/or freezing stress resistant Brassica crops with desirable colors as well. These findings may also facilitate exploration of the molecular mechanism that regulates anthocyanin biosynthesis and its response to abiotic stresses.
URLPMID:23613820 [本文引用: 1]
