 ,1,2, 程丽1, 郭月1, 龙卫华1, 高建芹1, 浦惠明
,1,2, 程丽1, 郭月1, 龙卫华1, 高建芹1, 浦惠明 ,1,*, 张洁夫1,2, 陈松1
,1,*, 张洁夫1,2, 陈松1Development and application of the marker for imidazolinone-resistant gene in Brassica napus
HU Mao-Long ,1,2, CHENG Li1, GUO Yue1, LONG Wei-Hua1, GAO Jian-Qin1, PU Hui-Ming
,1,2, CHENG Li1, GUO Yue1, LONG Wei-Hua1, GAO Jian-Qin1, PU Hui-Ming ,1,*, ZHANG Jie-Fu1,2, CHEN Song1
,1,*, ZHANG Jie-Fu1,2, CHEN Song1通讯作者:
收稿日期:2020-03-2接受日期:2020-06-2网络出版日期:2020-06-14
| 基金资助: |
Received:2020-03-2Accepted:2020-06-2Online:2020-06-14
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (1420KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
胡茂龙, 程丽, 郭月, 龙卫华, 高建芹, 浦惠明, 张洁夫, 陈松. 油菜抗咪唑啉酮类除草剂基因标记的开发与应用[J]. 作物学报, 2020, 46(10): 1639-1646. doi:10.3724/SP.J.1006.2020.04056
HU Mao-Long, CHENG Li, GUO Yue, LONG Wei-Hua, GAO Jian-Qin, PU Hui-Ming, ZHANG Jie-Fu, CHEN Song.
油菜是我国第一大油料作物, 产油量占国产油料作物总产的55%以上, 发展油菜生产对维护我国食用油供给安全具有重要的战略意义[1]。然而, 随着我国农村经济的快速发展, 劳动力成本大幅提高, 油菜比较效益下降, 农户种植油菜积极性不高, 生产管理粗放, 草害日益严重, 已成为制约我国油菜生产的重要因素。生产上使用的稳杀得、盖草能等除草剂能有效防除油菜田单子叶杂草, 对减轻杂草危害和提高油菜产量起到一定的作用。但对绝大多数阔叶杂草, 目前生产上尚未开发出安全高效的除草剂, 我国油菜田化学除草面积非常有限。开发抗性基因, 选育种植对特定除草剂具有选择抗性的油菜品种, 是解决油菜化除最经济有效的途径[2]。
咪唑啉酮(imidazolinone, IMI)类除草剂是应用较广泛的一类除草剂。该类除草剂具有选择性强、广谱、高活性等优点, 可进行播前混土、苗前土表处理以及茎叶喷雾, 除草剂经植物根与叶吸收后可有效防除一年生禾本科与阔叶杂草, 也能防除多年生杂草[3]。但该类除草剂对一般不具有抗(耐)除草剂特性的农作物本身也会产生药害, 极大限制了其使用时间和空间, 如需要在农作物播种前一段时间使用才能避免产生药害。培育抗(耐)除草剂品种可减少药害、拓宽现有咪唑啉酮类除草剂的使用范围。通过定向诱变IMIs的靶标酶乙酰乳酸合酶(acetolactate synthase, ALS)氨基酸变异, 可引起除草剂与ALS结合方式的变化产生抗性植物[4]。参照模式植物拟南芥ALS氨基酸序列, 已报道的抗性位点中主要涉及Ala122、Pro197、Ala205、Asp376、Arg377、Trp574、Ser653、Gly654等氨基酸, 其中对IMIs产生抗性的氨基酸突变位点主要有Pro122、Trp574、Ser653和Gly654 [5,6,7,8,9,10]。而已发现抗IMI的油菜突变位点主要是Trp574和Ser653 [11,12,13,14]。前期我们获得了自然突变的抗IMI油菜M9, 其抗性浓度是除草剂有效杀草浓度的2~3倍, 抗性稳定、具有利用价值[15,16,17]。胡茂龙等[18]从M9中克隆到抗性基因BnALS1R发现, 抗性基因BnALS1R与野生型基因在第1913位点存在1处SNP, 导致ALS1的第638位丝氨酸残基被天冬酰胺酸替代。抗性基因BnALS1R的发现为我国提供了具有自主知识产权的抗除草剂基因, 通过常规的杂交、回交等育种手段可将该基因转育到栽培品种中, 但需要喷施除草剂逐代筛选鉴定, 其过程经历多代的杂交和回交, 育种周期较长, 而且该基因为不完全显性基因, 表型选择难以分辨纯合和杂合抗性基因型, 更增加了育种的周期和难度。因此, 若能开发到检测抗性功能基因分子标记, 利用分子标记辅助选择(marker-assisted selection, MAS)技术鉴定基因型, 指导田间育种, 对于抗除草剂基因快速转育、加快育种进程具有重要意义。
竞争性等位基因PCR (kompetitive allele specific PCR, KASP)标记技术是基于引物末端碱基的特异匹配来对SNPs和InDels (Insertions and Deletions, 插入和缺失)进行高精度的双等位基因分型方法, 是由LGC (Laboratory of the Government Chemist)公司(http://www.lgcgroup.com/)设计和创制, 具有通量高、成本低、准确度高等优点, 已经成为新一代的SNP分型标记, 在水稻、小麦、玉米、大豆等作物的基因精细定位、MAS育种、种质资源鉴定等方面被广泛应用[19,20,21,22]。但迄今, KASP标记在油菜研究上鲜有报道, 特别是有关检测油菜抗除草剂基因在国内外未见报道。本研究根据M9中BnALS1R基因第1913位点的SNP变异, 设计开发了KASP标记, 能够高效、精确选择抗除草剂基因BnALS1R的基因型, 加快了油菜材料的抗性稳定进程, 以期为油菜抗除草剂MAS育种提供技术支撑。
1 材料与方法
1.1 试验材料
M9-2、M9-14为含有抗性基因BnALS1R的M9后代系选材料, M9是浦惠明等[17]在油菜和大豆多年轮作的试验田中发现的抗IMI自然突变体。3075R和N341为江苏省农业科学院经济作物研究所选育的MICMS细胞质雄性不育双低恢复系, 均经过多代自交, 各项性状稳定。F1-2是以N341为母本、抗性材料M9-2为父本杂交F1代, F1-3是以3075R为母本、抗性材料M9-14为父本杂交F1代。2个F1自交衍生获得F2群体, 用于BnALS1R基因型在分离群体中检测鉴定。10M169为带有抗性基因BnALS1R的MICMS恢复系, 对应不育系宁A7均由江苏省农业科学院经济作物研究所选育保存。1.2 分子试剂与除草剂
由英国LGC公司合成KASP反应试剂、KASP引物及特定的FAM和HEX荧光接头序列。由上海英俊(Invitrogen)生命技术有限公司合成其他常规引物, DNA测序委托南京钟鼎生物技术有限公司完成。DNA分子量标准、DNA回收试剂盒购自天根生化科技北京有限公司, 高保真性DNA聚合酶KOD-Plus及PCR试剂购自东洋纺(上海)生物科技有限公司。IMI类除草剂为山东先达化工有限公司生产的5.0%“豆施乐”水剂。1.3 田间试验与除草剂处理
油菜材料种植于江苏省农业科学院南京溧水植物科学基地油菜隔离繁殖区, 试验严格控制花粉外漂, 以常规方法栽培管理。油菜花期用M9-2、M9-14与MICMS双低恢复系3075R和N341配制F1组合, 次年春自交获得F2世代, 秋播鉴定分离世代的抗性表现。同时用抗性恢复系10M169与不育系宁A7以人工杂交、网罩隔离和天然授粉3种方法生产抗性杂交种, 秋播鉴定F1和亲本的抗性表现。油菜三至四叶期用浓度为90 g a.i. hm-2除草剂处理, 处理前取少许叶片保存于-80℃冰箱, 用于DNA提取。21 d后调查抗性表型。处理后油菜植株生长良好, 无药害表现, 为抗除草剂材料, 抗性表型用R表示; 处理后油菜植株生长受到抑制, 最终死亡, 为感除草剂材料, 抗性表型用S表示。1.4 分子标记设计与KASP分型
M9与常规油菜宁油16、宁油18编码ALS家族中BnALS1的核苷酸序列存在1处SNP[12], 根据该处SNP可开发KASP分子标记检测抗性基因BnALS1R。将己克隆的BnALS1R基因序列与野生型序列比对, 明确SNP在油菜BnALS1基因的第1913位。以候选SNP位点为中心提取两侧50 bp侧翼序列, 设计2组KASP分子标记引物, 每组标记由3条引物组成, 其中2条特异引物3'末端为等位变异碱基G/A, 且在5'端分别连接英国LGC公司KASP反应试剂特定的FAM和HEX荧光接头序列。通用引物的序列选择要保证扩增片段在60~120 bp (表1)。Table 1
表1
表1抗性基因BnALS1R的KASP分子标记引物序列
Table 1
| KASP名称KASP ID | 引物Primer (5°-3°) |
|---|---|
| KBA1R19681913A | X-allele: AACATGTGTTACCGATGATCCCAAG Y-allele: GAACATGTGTTACCGATGATCCCAAA Common: CTTAGTGCGACCATCCCCTTCT |
| KBA1R19681913B | X-allele: TATTACATCTTTGAAAGTGCCACCAC Y-allele: GTTATTACATCTTTGAAAGTGCCACCAT Common: CAGGACCATACCTGTTGGATGTGATA |
新窗口打开|下载CSV
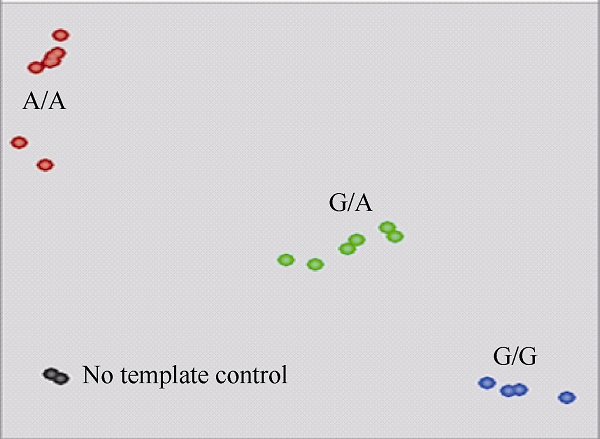
采用CTAB法并稍作修改提取DNA, 用1% (v/w)的琼脂糖电泳和分光光度法检测其纯度[2]。KASP反应体系包含模板DNA (20 ng μL-1) 2.5 μL、2×KASP Master mix 2.5 μL、KASP Assay mix 0.07 μL。PCR反应条件为94℃ 15 min; 94℃ 20 s, 61~55℃ 1 min, 每个循环退火温度降低0.6℃, 共10个循环; 94℃ 20 s, 55℃ 1 min, 共26个循环。若扩增效果不理想可加循环, 每次加3个循环, 最多3次。KASP反应在LGC Hydrocycler-16进行, 完成反应后利用扫描仪Pherastar对KASP产物读取荧光数据, 荧光扫描的结果会自动转化成图形。利用BMG PHERAstar仪器检测荧光信号并查看分型情况。若分型不充分, 则继续扩增, 每3个循环查看分型情况, 直至分型完全。根据分析结果确定待测样本中抗性基因BnALS1R的基因型, 在X轴附近显示蓝色的样本基因型为连接FAM荧光标签序列的等位基因型G/G, 为不含抗性基因BnALS1R的纯合体; 在Y轴附近显示红色样本的基因型为连接HEX荧光标签序列的等位基因型A/A, 为含抗性基因BnALS1R的纯合体; 中间显示绿色样本的基因型为杂合等位基因型G/A, 为含抗性基因BnALS1R的杂合体; 显示黑色的样本为空白对照。
1.5 基因克隆与测序
利用Primer Premier 5.0软件根据甘蓝型油菜BnALS1 (GenBank登录号为Z11524)基因序列设计正向引物ALS1-5F: 5'-TGGATATTGACGGTGATGG-3'和反向引物ALS1-5R: 5'-CGAGTACGTCTGGGAACAA-3', 通过PCR特异扩增一段含有突变位点(G/A)的BnALS1基因片段, 长度为537 bp。PCR反应体系包含DNA模板5 μL、10×酶反应缓冲液5 μL、25 mmol L-1 MgSO4 2 μL、2 mmol L-1 dNTPs 5 μL、10 μmol L-1引物各5 μL、1 U μL-1 KOD-Plus Taq酶1 μL, 加ddH2O水至50 μL。反应程序为94℃ 5 min; 94℃ 30 s, 60℃ 30 s, 72℃ 30 s, 35个循环。经1.2% (v/w)琼脂糖凝胶电泳分离, 用DNA凝胶回收试剂盒将目的片段回收纯化, 采用上下游引物双向测序。通过DNAMAN 6.0、Sequencher、DNASTAR等生物信息学软件分析测序结果, 确定样本中BnALS1R的基因型。1.6 应用标记检测F2群体基因型和F1杂种纯度
于苗期提取1.3中的2个F2分离群体和3种不同授粉条件下的F1杂交种叶片DNA, 利用标记KBA1R19681913B引物进行KASP分型。根据基因分型结果, 统计F1杂种的纯度和F2群体的基因型分离比例, 并对F2群体基因分型结果进行卡平方测验。2 结果与分析
2.1 BnALS1R基因KASP标记的开发与验证
油菜突变体M9中的抗性基因BnALS1R与常规油菜的BnALS1在第1913处存在SNP位点(G/A), 属于错义突变位点(丝氨酸/天冬酰胺), 携带A等位变异为含抗除草剂基因BnALS1R的抗性材料, 携带G等位变异的为不含抗除草剂基因BnALS1R的敏感材料[18]。由于油菜ALS基因家族核酸序列的高度同源性, 特别是BnALS1和BnALS3 (GenBank登录号为Z11524、Z11526)在核酸水平的同源性达98% [23,24], 因此本研究选择在第1913突变位点上下游2个基因序列有差异的位置设计2组特异性KASP标记引物KBA1R19681913A和KBA1R19681913B (表1)。选取含有抗性基因的M9后代株系M9-2、M9-14和不含抗性基因的MICMS双低恢复系3075R和N341, 以及它们杂交获得的F1代和F2群体单株共18个油菜材料, 利用设计的KASP 标记进行多态型筛选与小群体验证。多次验证结果发现, 标记KBA1R19681913A在亲本和小群体中没有多态性, 而标记KA1R19681913B具有多态性, 其中KBA1R19681913B-X-allele检测携带敏感基因的等位变异, KBA1R19681913B-Y-allele检测携带抗性基因的等位变异, 上游共同引物KBA1R19681913B-common与BnALS3存在3处碱基差异(图1)。从图2可以看出, 该标记可以清晰地把3种基因型分开, 其中靠近Y轴的红点为携带A等位变异的抗IMI类除草剂基因BnALS1R纯合型材料, 如M9后代株系M9-2、M9-14等; 靠近X轴的蓝点为携带G等位变异的不含抗IMI类除草剂基因BnALS1R纯合型材料, 如MICMS双低恢复系3075R和N341等; 中间的绿点为携带G/A等位变异的含抗IMI类除草剂基因BnALS1R杂合型材料, 如F1代和F2群体单株M318-5、M318-7和M318-9等, 黑点为空白对照(图2和表2)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1油菜BnALS1与BnALS3基因序列比对与多态性引物位置
倒三角形表示突变位点。箭头线表示引物位置和扩增方向。
Fig. 1Sequencing alignment of BnALS1 and BnALS3 genes and loci of polymorphic primers in rapeseed
The single-site mutation is denoted by inverted triangle. The arrow lines represent primer sites and amplification directions.
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2KASP标记KBA1R19681913B在小群体中分型图
Fig. 2Allelic discrimination plot of KASP KBA1R19681913B genotyping in the small population
Table 2
表2
表2KASP标记KALS1R19681913B在小群体中的分型结果与抗性表型
Table 2
| 材料 Material | 标记分型 Genotyping | 测序结果 Sequecing | 抗性特性 Resistance | 材料来源 Source |
|---|---|---|---|---|
| M9-2 | A | A | R | M9后代株系 M9 progeny line |
| M9-14 | A | A | R | M9后代株系 M9 progeny line |
| N341 | G | G | S | MICMS恢复系 MICMS restorer line |
| 3075R | G | G | S | MICMS恢复系 MICMS restorer line |
| F1-2 | G/A | G/A | R | F1(N341×M9-2) F1 hybrid |
| F1-3 | G/A | G/A | R | F1(3075R×M9-14) F1 hybrid |
| M318-1 | G | G | S | F2(N341×M9-2)单株 F2 plant |
| M318-2 | A | A | R | F2(N341×M9-2)单株 F2 plant |
| M318-3 | G | G | S | F2(N341×M9-2)单株 F2 plant |
| M318-4 | A | A | R | F2(N341×M9-2)单株 F2 plant |
| M318-5 | G/A | G/A | R | F2(N341×M9-2)单株 F2 plant |
| M318-6 | A | A | R | F2(N341×M9-2)单株 F2 plant |
| M318-7 | G/A | G/A | R | F2(N341×M9-2)单株 F2 plant |
| M318-8 | A | A | R | F2(N341×M9-2)单株 F2 plant |
| M318-9 | G/A | G/A | R | F2(N341×M9-2)单株 F2 plant |
| M318-10 | A | A | R | F2(N341×M9-2)单株 F2 plant |
| M318-11 | A | A | R | F2(N341×M9-2)单株 F2 plant |
| M318-12 | G/A | G/A | R | F2(N341×M9-2)单株 F2 plant |
新窗口打开|下载CSV
为了进一步验证KASP标记引物的特异性和准确性, 设计1对引物特异扩增含有突变位点(G/A)的BnALS1R基因片段进行双向测序。所有材料的PCR产物测序结果与KASP标记分型结果一致(表2)。油菜田间苗期抗除草剂表型鉴定的结果也发现, 基因型与抗除草剂鉴定表型结果吻合(表2)。因此, 本研究获得了检测油菜抗IMI类除草剂基因BnALS1R的KASP分子标记引物, 能够精确鉴定出抗性基因BnALS1R的3种基因型。
2.2 群体中标记KBA1R19681913B检测效果及抗性分析
利用标记KBA1R19681913B检测2个F2群体(3075R/M9-14和N341/M9-2)基因型。2个F2群体中标记检测效果良好, 呈现3种基因型(图3)。携带抗IMI类除草剂基因BnALS1R的纯合和杂合基因型的F2单株都表现抗除草剂, 而所有不携带抗IMI类除草剂基因BnALS1R纯合基因型的F2群体单株均表现感除草剂。其中F2(3075R/M9-14)群体共分析29个单株, 有7个单株为抗性基因BnALS1R的纯合体, 8个单株为不含抗性基因BnALS1R的纯合体, 14个单株为抗性基因BnALS1R的杂合型, 3种基因型符合1︰2︰1 (χ2 = 0.012, P = 0.994)。F2 (N341/M9-2)群体共分析50个单株, 有16个单株为抗性基因BnALS1R的纯合体, 11个单株为不含抗性基因BnALS1R的纯合体, 23个单株为抗性基因BnALS1R的杂合型, 3种基因型符合1︰2︰1 (χ2 = 0.085, P = 0.958)。2个群体均符合单基因遗传分离规律, 标记检测结果与苗期喷施除草剂鉴定表型完全一致, 即标记KBA1R19681913B与抗/感除草剂表型完全共分离, 同时能检测到抗性杂合基因型, 表明标记KBA1R19681913B可用于抗IMI类除草剂育种的MAS选择(表3)。图3
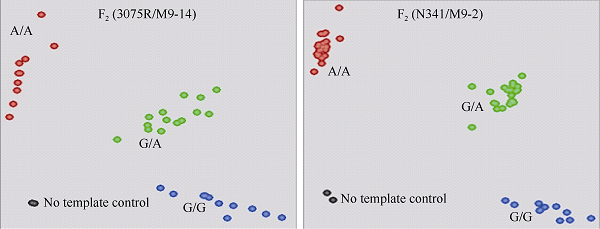 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3标记KBA1R19681913B在2个F2群体及亲本中的基因分型
Fig. 3Allelic discrimination plot of KASP KBA1R19681913B genotyping in two F2 populations and their parents
Table 3
表3
表3标记在F2群体中基因型分型
Table 3
| 杂交组合 Cross | 基因型Genotype | 总株数 Total | 卡平方值 χ2 -value | P值 P- value | ||
|---|---|---|---|---|---|---|
| G/G | G/A | A/A | ||||
| F2 (3075R/M9-14) | 8 | 14 | 7 | 29 | 0.012 | 0.994 |
| F2 (N341/M9-2) | 11 | 23 | 16 | 50 | 0.085 | 0.958 |
新窗口打开|下载CSV
2.3 利用标记KBA1R19681913B辅助选择抗性恢复系
本研究以M9-2和M9-14为抗性基因供体亲本, MICMS恢复系3075R和N341为轮回亲本, 分别对3075R/M9-14和N341/M9-2组合后代的抗性植株进行回交转育, 回交2代, 每一代均对杂交种检测保留抗性基因BnALS1R的杂合型单株, 与轮回亲本回交; 对BC2F2单株保留抗性基因BnALS1R纯合单株, 再自交一代, 结合农艺和品质性状选择, 获得除草剂抗性稳定BC2F3 株系。同时, 由于3075R和N341均为带有不育细胞质的恢复系, 因此在回交后代连续选择可育单株自交并进行抗性鉴定, 就育成抗咪唑啉酮的MICMS恢复系。目前已从2个回交后代群体选择到生长势强、成熟期适中的200多个抗性株系, 可为选育抗除草剂杂交种提供亲本。2.4 标记KBA1R19681913B在杂交种纯度鉴定中的利用
利用携带抗性基因BnALS1R的MICMS恢复系10M169与不育系宁A7在3种不同授粉条件下配置F1杂种, 通过利用分子标记KBA1R19681913B对未经IMI类除草剂咪唑乙烟酸处理和经过喷药处理的菜苗进行纯度鉴定发现, 在网罩隔离授粉和自然授粉条件下, 除草剂未处理的F1杂种纯度分别为84%和80%, 均达不到85%的国标要求[25], 而经过除草剂处理的F1杂种纯度分别为97%和96%, 且标记鉴定结果与杂交种田间鉴定纯度结果吻合, 表明标记KBA1R19681913B可用于抗性杂交油菜种子纯度提前鉴定(表4)。因此, 在生产上如果通过KBA1R19681913B标记鉴定的杂交种子纯度达不到国标要求, 可对F1杂种苗期进行除草剂处理, 以达到提高种子纯度的目的, 同时兼顾大田除草, 一举多得。Table 4
表4
表4标记KBA1R19681913B在鉴定杂交种子纯度的应用
Table 4
| 授粉方式 Different pollination conditions | 除草剂处理 IMI treatment | 鉴定株数 Numbers of plants | 标记鉴定纯度 Seed purity identification by the markers (%) | 田间鉴定纯度 Seed purity identification in the field (%) | 杂种纯度提高 Increased seed purity after IMI treatment (%) |
|---|---|---|---|---|---|
| 人工杂交授粉 Artificial hybridization | 喷药Treated | 117 | 100 | 100 | — |
| 不喷药Untreated | 90 | 100 | 100 | — | |
| 网罩隔离授粉 Hybridization in tents | 喷药Treated | 130 | 97 | 97 | 13 |
| 不喷药Untreated | 194 | 84 | 84 | — | |
| 天然授粉 Natural pollination | 喷药Treated | 165 | 96 | 96 | 16 |
| 不喷药Untreated | 155 | 80 | 80 | — |
新窗口打开|下载CSV
3 讨论
杂草是影响油菜生产的重要生物灾害之一, 喷施除草剂是防治草害的有效手段。加拿大、澳大利亚、法国、德国等油菜主产区早以通过种植抗IMI油菜实施杂草化学防控[4]。然而我国缺乏自主知识产权的油菜抗性基因, 至今无法应用IMI类除草剂进行化学除草[26,27]。前期我们从M9中克隆获得抗性基因BnALS1R, 该基因的突变位点与Swanson等[11]报道的PM1突变位点相同, 属于除草剂靶标酶ALS的Ser653位点突变, 但BnALS1R是我国自主研发具有知识产权的抗性基因, 不受国际知识产权保护的限制[2]。本研究基于该突变位点的SNP, 在国内外首次开发了检测该抗性基因的KASP标记KBA1R19 681913B, 将该标记在F2群体进行验证, 并应用到抗性恢复系选育和杂交种纯度鉴定中, 发现该KASP标记能够快速、精确区分出BnALS1R的3种基因型和真假杂种。显然, 以上研究可加快我国抗IMI油菜的选育和推广应用步伐。便宜、高效且准确的基因SNP分型方法一直是遗传学家和育种家追求的技术手段。目前SNP分型技术有RFLP、CAPS等传统的标记方法, 也有高通量测序和基因芯片等现代方法[28,29,30]。前者涉及到繁琐的分子杂交和酶切实验, 操作流程复杂、通量不高; 后者具有快速高效、结果可靠等优势, 但费用相对昂贵。同时芯片技术主要用于对百个以上的SNP检测, 对少量SNP或单个基因的SNP进行检测时, KASP技术具有经济高效的优势[31]。与基于芯片的Illumina GoldenGate相比, 使用KASP分型用于MAS回交选择比使用其他高通量平台节省7.9%~46.1%的费用[31]。KASP技术价格低廉、高效灵活特性[32], 已在水稻、小麦、玉米、大豆等作物中得到应用[19-22,33]。在油菜中, Chang等[34]开发了源于黑芥(Brassica nigra)的抗根肿病基因Rcr6的KASP标记, 并应用于基因的精细定位和同源性关系分析。Fu等[35]利用BSR-Seq (bulked segregant RNA Sequencing)技术开发了多个KASP标记, 对油菜抗黑胫病(blackleg)基因Rlm1进行了精细定位, 获得了候选基因BnA07G27460D和可用于MAS选择的KASP标记。但迄今在国内外未见有关油菜抗IMI类除草剂基因分型的报道。本研究获得了高效、精确检测抗除草剂基因BnALS1R基因型的KASP标记KBA1R19681913B, 与之前的检测该基因型的AS-PCR标记相比, 本研究获得的标记在实际应用中成本更低、通量更高[36]。另外, KASP标记仅需PCR和荧光检测2个步骤, 无需对产物进行凝胶电泳检测, 彻底杜绝了电泳中常用的染色剂溴化乙锭对人体的危害和环境的污染。综上, 本研究获得的BnALS1R基因分型方法具有简便、快速、通量高、环境友好的显著优点, 更加适用于育种家在遗传大群体中对BnALS1R不同基因型的鉴定以及MAS育种。
咪唑啉酮类除草剂的靶标ALS是植物支链氨基酸合成的关键酶。除草剂通过抑制ALS酶活性, 导致支链氨基酸合成受阻, 达到杀草目的。ALS基因碱基位点变异引起编码氨基酸的改变产生抗性基因, 可使敏感作物产生除草剂抗性。在获得抗性基因后, 开发合适的分子标记鉴定抗性基因型是进行MAS选择的前提, 此方面前人有些研究。Thompson等[37]开发了检测鹰嘴豆(Cicer arietinum L.)抗性位点Ala205的KASP标记; Kadaru等[38]开发了检测水稻抗性位点Ser653和Gly654的AS-PCR标记; Lee等[39]开发了检测大麦抗性位点Ser653的AS-PCR标记。然而, 与鹰嘴豆染色体组(2n=16)、水稻(2n=24)和大麦(2n=14)不同, 甘蓝型油菜为异源四倍体(2n=38), 基因组庞大, 包括A、C两套基因组。基因组进化的高度重排导致甘蓝型油菜相关基因多以基因家族的形式存在[40]。同一家族基因核酸序列高度同源, IMI的靶基因ALS也不例外。位于基因组A的BnALS3和基因组C的BnALS1核酸同源性达98%, 这给KASP标记检测BnALS1增加了难度, 因为在进行PCR扩增BnALS1过程中会受到同源序列BnALS3的干扰[23,24]。为此, 在本研究中为明确KASP标记KBA1R19681913B基因分型的准确性, 我们另外设计了特异性引物进行双向测序, 测序结果与KASP分型结果一致(表2)。同时, 用该标记对2个F2群体进行检测发现, 基因型分离比遵循单基因遗传模式, 证明标记KBA1R19681913B可以有效、准确检测BnALS1R的3种不同基因型。有意思的是, KBA1R19681913B的共用引物恰好锚定在BnALS1和BnALS3的3个SNP上(图1), 而无多态性的KBA1R19681913A共用引物仅锚定在BnALS1和BnALS3的1个SNP (数据未列出)。这给了我们启示, 即将来在油菜中开发KASP标记要特别重视共用引物序列的特异性。
参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
DOI:10.3864/j.issn.0578-1752.2012.20.022URL [本文引用: 3]

【目的】分离和鉴定自然突变的油菜乙酰乳酸合成酶抑制剂类除草剂抗性突变体M9抗性基因,阐明抗性产生的分子基础。【方法】通过配置杂交组合研究其遗传规律,应用同源序列法克隆油菜ALS家族基因,采用杂交转育和小孢子培养技术将抗性基因转育到陆奥-五十铃细胞质雄性不育(MICMS)恢复系中,进行PCR鉴定。【结果】该突变体抗性由1个显性核基因控制。克隆获得BnALS1—BnALS3,核酸序列比对发现,M9抗性是由BnALS1的点突变使蛋白序列的第638位丝氨酸(AGT)残基被天冬酰胺酸(AAT)替代所致,将此抗性基因命名为BnALS1R。抗性基因BnALS1R转育到MICMS恢复系中的PCR检测表明,所有抗除草剂的恢复系都携带有BnALS1R。【结论】突变体M9抗性由1个显性核基因控制,BnALS1第638位丝氨酸突变成天冬酰胺酸是抗性产生的分子基础。
[本文引用: 3]
DOI:10.3864/j.issn.0578-1752.2015.24.006URL [本文引用: 1]

33是抗咪唑乙烟酸谷子(Setaria italica)品种,在生产实践中通过喷施咪唑乙烟酸除草剂可达到间苗、除草的目的,适应规模化、集约化种植。论文旨在通过研究咪唑乙烟酸对冀谷33生长发育的影响及对后茬作物的安全性,为冀谷33的应用和推广提供依据。【方法】试验于2013年5月至2015年6月在河北省农林科学院谷子研究所郄马试验站进行,试验设置0、56.25、75、112.5、150、225 g a.i./hm2 6种咪唑乙烟酸剂量水平。对冀谷33生长发育的影响试验在田间进行,用药后7、10、13、16、20、25和30 d,分别调查统计苗高和鲜重;成熟期调查株高、穗长和穗径,并统计小区产量;并对小米、谷壳、植株和土壤中的残留进行检测。对后茬作物的影响试验在网室进行,研究其药后30、60 d和药后1年播种的8种后茬作物白菜、冀谷19、冀谷33、甜菜、高粱、玉米、小麦、棉花的安全性。【结果】56.25—225 g a.i./hm2剂量下,冀谷33正常生长,无明显药害。药后不同时期对冀谷33苗高、鲜重的影响结果表明,喷药谷子苗高和鲜重与对照相比差异不显著;药后13—16 d处于拔节期初期,为苗高敏感期,药后20—25 d处于拔节期末期、孕穗期初期,为苗重敏感期,之后抑制逐渐解除,收获时冀谷33生长发育正常,产量高于对照。只有喷施225 g a.i./hm2咪唑乙烟酸的植株中检测到(0.01±0.006) mg·kg-1少量残留,低于检测限LOQ,其余处理均未检出。随着咪唑乙烟酸施药时间的增长,对后茬作物的抑制作用有所减轻,在药后1年播种的后茬作物中,株高差异显著,仅对白菜、高粱产量有显著影响。在夏谷区喷施咪唑乙烟酸的田地,冀谷33可与玉米、小麦、棉花轮作种植。【结论】施用56.25—225 g a.i./hm2咪唑乙烟酸对冀谷33安全,对其生长发育和产量没有显著影响。随着咪唑乙烟酸施药时间的增长,对后茬作物的抑制作用有所减轻,冀谷33可与玉米、小麦、棉花轮作种植。在生产中,要注意控制药量,合理轮作。]]>
[本文引用: 1]
DOI:10.1002/ps.993URLPMID:15627242 [本文引用: 2]

Imidazolinone herbicides, which include imazapyr, imazapic, imazethapyr, imazamox, imazamethabenz and imazaquin, control weeds by inhibiting the enzyme acetohydroxyacid synthase (AHAS), also called acetolactate synthase (ALS). AHAS is a critical enzyme for the biosynthesis of branched-chain amino acids in plants. Several variant AHAS genes conferring imidazolinone tolerance were discovered in plants through mutagenesis and selection, and were used to create imidazolinone-tolerant maize (Zea mays L), wheat (Triticum aestivum L), rice (Oryza sativa L), oilseed rape (Brassica napus L) and sunflower (Helianthus annuus L). These crops were developed using conventional breeding methods and commercialized as Clearfield* crops from 1992 to the present. Imidazolinone herbicides control a broad spectrum of grass and broadleaf weeds in imidazolinone-tolerant crops, including weeds that are closely related to the crop itself and some key parasitic weeds. Imidazolinone-tolerant crops may also prevent rotational crop injury and injury caused by interaction between AHAS-inhibiting herbicides and insecticides. A single target-site mutation in the AHAS gene may confer tolerance to AHAS-inhibiting herbicides, so that it is technically possible to develop the imidazolinone-tolerance trait in many crops. Activities are currently directed toward the continued improvement of imidazolinone tolerance and development of new Clearfield* crops. Management of herbicide-resistant weeds and gene flow from crops to weeds are issues that must be considered with the development of any herbicide-resistant crop. Thus extensive stewardship programs have been developed to address these issues for Clearfield* crops.
DOI:10.3389/fpls.2018.01014URLPMID:30061911 [本文引用: 1]

Tribenuron-methyl (TBM), an acetohydroxyacid synthase (AHAS)-inhibiting herbicide, can be used as an efficient chemical hybridization agent to induce male sterility for practical utilization of heterosis in rapeseed (Brassica napus L.). Utilization of rapeseed mutants harboring herbicide-resistant AHAS alleles as the male parent can simplify the hybrid seed production protocol. Here we characterized a novel TBM-resistant mutant K5 derived from an elite rapeseed variety, Zhongshuang No. 9 (ZS9), by ethyl methyl sulfonate mutagenesis. Comparative analysis of three BnAHAS genes (BnAHAS1, BnAHAS2, and BnAHAS3) between the mutant K5 and ZS9 identified a C-to-T transition at 544 from the translation start site in BnAHAS1 in K5 (This resistant allele is referred to as BnAHAS1(544T) ), which resulted in a substitution of proline with serine at 182 in BnAHAS1. Both ZS9 and K5 plants could be induced complete male sterility under TBM treatment (with 0.10 and 20 mgL(-1) of TBM, respectively). The relationship between TBM-induced male sterility (Y) and the relative AHAS activity of inflorescences (X) could be described as a modified logistic function, Y = 100-A/(1+Be((-)(KX)())) for the both genotypes, although the obtained constants A, B, and K were different in the functions of ZS9 and K5. Transgenic Arabidopsis plants expressing BnAHAS1(544T) exhibited a higher TBM resistance of male reproductive organ than wild type, which confirmed that the Pro-182-Ser substitution in BnAHAS1 was responsible for higher TBM-resistance of male reproductive organs. Taken together, our findings provide a novel valuable rapeseed mutant for hybrid breeding by chemical hybridization agents and support the hypothesis that AHAS should be the target of the AHAS-inhibiting herbicide TBM when it is used as chemical hybridization agent in rapeseed.
DOI:10.1007/s00122-014-2415-7URLPMID:25504538 [本文引用: 1]

KEY MESSAGE: Identification and molecular analysis of four tribenuron-methyl resistant mutants in Brassica napus , which would be very useful in hybrid production using a Chemically induced male sterility system. Chemically induced male sterility (CIMS) systems dependent on chemical hybridization agents (CHAs) like tribenuron-methyl (TBM) represent an important approach for practical utilization of heterosis in rapeseed. However, when spraying the female parents with TBM to induce male sterility the male parents must be protected with a shield to avoid injury to the stamens, which would otherwise complicate the seed production protocol and increase the cost of hybrid seed production. Here we report the first proposed application of a herbicide-resistant cultivar in hybrid production, using a CIMS system based on identifying four TBM-resistant mutants in Brassica napus. Genetic analysis indicated that the TBM resistance was controlled by a single dominant nuclear gene. An in vitro enzyme activity assay for acetohydroxyacid synthase (AHAS) suggested that the herbicide resistance is caused by a gain-of-function mutation in a copy of AHAS genes. Comparative sequencing of the mutants and wild type BnaA.AHAS.a coding sequences identified a C-to-T transition at either position 535 or 536 from the translation start site, which resulted in a substitution of proline with serine or leucine at position 197 according to the Arabidopsis thaliana protein sequence. An allele-specific dCAPS marker developed from the C536T variation co-segregated with the herbicide resistance. Transgenic A. thaliana plants expressing BnaA.ahas3.a conferred herbicide resistance, which confirmed that the P197 substitution in BnaA.AHAS.a was responsible for the herbicide resistance. Moreover, the TBM-resistant lines maintain normal male fertility under TBM treatment and can be of practical value in hybrid seed production using CIMS.
DOI:10.1007/s00425-017-2817-2URLPMID:29170911 [本文引用: 1]

MAIN CONCLUSION: The acetohydroxy acid synthase S627N mutation confers herbicide tolerance in rice, and the rice variety containing this mutation produces good yields. This variety is commercially viable at Shanghai and Jiangsu regions in China. Weedy rice is a type of rice that produces lower yields and poorer quality grains than cultivated rice. It plagues commercial rice fields in many countries. One strategy to control its proliferation is to develop rice varieties that are tolerant to specific herbicides. Acetohydroxy acid synthase (AHAS) mutations have been found to confer herbicide tolerance to rice. Here, we identified a single mutation (S627N) in AHAS from an indica rice variety that conferred tolerance against imidazolinone herbicides, including imazethapyr and imazamox. A japonica rice variety (JD164) was developed to obtain herbicide tolerance by introducing the mutated indica ahas gene. Imidazolinone application was sufficient to efficiently control weedy rice in the JD164 field. Although the imazethapyr treatment caused dwarfing in the JD164 plants, it did not significantly reduce yields. To determine whether the decrease of the ahas mRNA expression caused the dwarfism of JD164 after imazethapyr application, we detected the ahas mRNA level in plants. The abundance of the ahas mRNA in JD164 increased after imidazolinone application, thus excluding the mRNA expression level as a possible cause of dwarfism. Activity assays showed that the mutated AHAS was tolerant to imidazolinone but the catalytic efficiency of the mutated AHAS decreased in its presence. Moreover, the activity of the mutated AHAS decreased more in the presence of imazethapyr than in the presence of imazamox. We observed no difference in the AHAS secondary structures, but homology modeling suggested that the S627N mutation enabled the substrate to access the active site channel in AHAS, resulting in imidazolinone tolerance. Our work combined herbicides with a rice variety to control weedy rice and showed the mechanism of herbicide tolerance in this rice variety.
DOI:10.1002/ps.3278URL [本文引用: 1]

BACKGROUND: Wild radish, a problem weed worldwide, is a severe dicotyledonous weed in crops. In Australia, sustained reliance on ALS-inhibiting herbicides to control this species has led to the evolution of many resistant populations endowed by any of several ALS mutations. The molecular basis of ALS-inhibiting herbicide resistance in a novel resistant population was studied. RESULTS: ALS gene sequencing revealed a previously unreported substitution of Tyr for Ala at amino acid position 122 in resistant individuals of a wild radish population (WARR30). A purified subpopulation individually homozygous for the Ala-122-Tyr mutation was generated and characterised in terms of its response to the different chemical classes of ALS-inhibiting herbicides. Whole-plant dose-response studies showed that the purified subpopulation was highly resistant to chlorsulfuron, metosulam and imazamox, with LD50 or GR50 R/S ratio of > 1024, > 512 and > 137 respectively. The resistance to imazypyr was found to be relatively moderate (but still substantial), with LD50 and GR50 R/S ratios of > 16 and > 7.8 respectively. In vitro ALS activity assays showed that Ala-122-Tyr ALS was highly resistant to all tested ALS-inhibiting herbicides. CONCLUSION: The molecular basis of ALS-inhibiting herbicide resistance in wild radish population WARR30 was identified to be due to an Ala-122-Tyr mutation in the ALS gene. This is the first report of an amino acid substitution at Ala-122 in the plant ALS that confers high-level and broad-spectrum resistance to ALS-inhibiting herbicides, a remarkable contrast to the known mutation Ala-122-Thr endowing resistance to imidazolinone herbicide. Copyright (c) 2012 Society of Chemical Industry
Online [
URL [本文引用: 1]
DOI:10.3390/plants8100382URL [本文引用: 1]
DOI:10.1007/BF00290837URLPMID:24225680 [本文引用: 2]

In vitro microspore mutagenesis and selection was used to produce five fertile double-haploid imidazolinone-tolerant canola plants. The S2 plants of three of the mutants were resistant to at least the field-recommended levels of Assert and Pursuit. One mutant was tolerant to between five and ten times the field-recommended rates of Pursuit and Scepter. Two semi-dominant mutants, representing two unlinked genes, were combined to produce an F1 hybrid which was superior in imidazolinone tolerance to either of the heterozygous mutants alone. Evaluation of the mutants under field conditions indicated that this hybrid and the original homozygous mutants could tolerate at least two times the field-recommended rates of Assert. The field results indicated the mutants were unaffected in seed yield, maturity, quality and disease tolerance. These genes represent a potentially valuable new herbicide resistance system for canola, which has little effect on yield, quality or maturity. The mutants could be used to provide tolerance to several imidazolinones including Scepter, Pursuit and Assert.
DOI:10.1007/BF00290445URLPMID:7891655 [本文引用: 2]

Acetohydroxy acid synthase (AHAS) is an essential enzyme for many organisms as it catalyzes the first step in the biosynthesis of the branched-chain amino acids valine, isoleucine, and leucine. The enzyme is under allosteric control by these amino acids. It is also inhibited by several classes of herbicides, such as the sulfonylureas, imidazolinones and triazolopyrimidines, that are believed to bind to a relic quinone-binding site. In this study, a mutant allele of AHAS3 responsible for sulfonylurea resistance in a Brassica napus cell line was isolated. Sequence analyses predicted a single amino acid change (557 Trp-->Leu) within a conserved region of AHAS. Expression in transgenic plants conferred strong resistance to the three classes of herbicides, revealing a single site essential for the binding of all the herbicide classes. The mutation did not appear to affect feedback inhibition by the branched-chain amino acids in plants.
[本文引用: 1]
DOI:10.1016/S2095-3119(17)61659-9URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 2]

]]>
[本文引用: 2]
DOI:10.1007/s11032-015-0227-3URL [本文引用: 2]
DOI:10.1007/s10142-016-0486-zURLPMID:26922174 [本文引用: 2]

The increases in the usage of nitrogen fertilizer result in deleterious impacts on the environment; thus, there is an urgent need to improve nitrogen use efficiency (NUE) in crops including rice (Oryza sativa L.). Attentions have focused on quantitative trait loci (QTL) mapping of NUE-related traits using single experimental population, but to date, very few studies have taken advantage of association mapping to examine hundreds of lines for identifying potentially novel QTLs in rice. Here, we conducted association analysis on NUE-related traits using a population containing 184 varieties, which were genotyped with 157 genome-wide simple sequence repeat (SSR) markers. We detected eight statistically significant marker loci associating with NUE-related traits, of which two QTLs at RM5639 and RM3628 harbored known NUE-related genes GS1;2 and AspAt3, respectively. At a novel NUE-related locus RM5748, we developed Kompetitive Allele Specific PCR (KASP) single nucleotide polymorphism (SNP) markers and searched for putative NUE-related genes which are close to the associated SNP marker. Based on a transcriptional map of N stress responses constructed by our lab, we evaluated expressions of the NUE-related genes in this region and validated their effect on NUE. Meanwhile, we analyzed NUE-related alleles of the eight loci that could be utilized in marker-assisted selection. Moreover, we estimated breeding values of all the varieties through genomic prediction approach that could be beneficial for rice NUE enhancement.
DOI:10.1007/s00122-017-2930-4URLPMID:28624908 [本文引用: 1]

KEY MESSAGE: Greenbug and Hessian fly are important pests that decrease wheat production worldwide. We developed and validated breeder-friendly KASP markers for marker-assisted breeding to increase selection efficiency. Greenbug (Schizaphis graminum Rondani) and Hessian fly [Mayetiola destructor (Say)] are two major destructive insect pests of wheat (Triticum aestivum L.) throughout wheat production regions in the USA and worldwide. Greenbug and Hessian fly infestation can significantly reduce grain yield and quality. Breeding for resistance to these two pests using marker-assisted selection (MAS) is the most economical strategy to minimize losses. In this study, doubled haploid lines from the Synthetic W7984 x Opata M85 wheat reference population were used to construct linkage maps for the greenbug resistance gene Gb7 and the Hessian fly resistance gene H32 with genotyping-by-sequencing (GBS) and 90K array-based single nucleotide polymorphism (SNP) marker data. Flanking markers were closely linked to Gb7 and H32 and were located on chromosome 7DL and 3DL, respectively. Gb7-linked markers (synopGBS773 and synopGBS1141) and H32-linked markers (synopGBS901 and IWB65911) were converted into Kompetitive Allele Specific PCR (KASP) assays for MAS in wheat breeding. In addition, comparative mapping identified syntenic regions in Brachypodium distachyon, rice (Oryza sativa), and sorghum (Sorghum bicolor) for Gb7 and H32 that can be used for fine mapping and map-based cloning of the genes. The KASP markers developed in this study are the first set of SNPs tightly linked to Gb7 and H32 and will be very useful for MAS in wheat breeding programs and future genetic studies of greenbug and Hessian fly resistance.
DOI:10.1007/s00122-015-2551-8URLPMID:26081946 [本文引用: 1]

DOI:10.1186/s12864-015-1531-3URLPMID:25903750 [本文引用: 2]

BACKGROUND: Soybean cyst nematode (SCN) is the most economically devastating pathogen of soybean. Two resistance loci, Rhg1 and Rhg4 primarily contribute resistance to SCN race 3 in soybean. Peking and PI 88788 are the two major sources of SCN resistance with Peking requiring both Rhg1 and Rhg4 alleles and PI 88788 only the Rhg1 allele. Although simple sequence repeat (SSR) markers have been reported for both loci, they are linked markers and limited to be applied in breeding programs due to accuracy, throughput and cost of detection methods. The objectives of this study were to develop robust functional marker assays for high-throughput selection of SCN resistance and to differentiate the sources of resistance. RESULTS: Based on the genomic DNA sequences of 27 soybean lines with known SCN phenotypes, we have developed Kompetitive Allele Specific PCR (KASP) assays for two Single nucleotide polymorphisms (SNPs) from Glyma08g11490 for the selection of the Rhg4 resistance allele. Moreover, the genomic DNA of Glyma18g02590 at the Rhg1 locus from 11 soybean lines and cDNA of Forrest, Essex, Williams 82 and PI 88788 were fully sequenced. Pairwise sequence alignment revealed seven SNPs/insertion/deletions (InDels), five in the 6th exon and two in the last exon. Using the same 27 soybean lines, we identified one SNP that can be used to select the Rhg1 resistance allele and another SNP that can be employed to differentiate Peking and PI 88788-type resistance. These SNP markers have been validated and a strong correlation was observed between the SNP genotypes and reactions to SCN race 3 using a panel of 153 soybean lines, as well as a bi-parental population, F5-derived recombinant inbred lines (RILs) from G00-3213xLG04-6000. CONCLUSIONS: Three functional SNP markers (two for Rhg1 locus and one for Rhg4 locus) were identified that could provide genotype information for the selection of SCN resistance and differentiate Peking from PI 88788 source for most germplasm lines. The robust KASP SNP marker assays were developed. In most contexts, use of one or two of these markers is sufficient for high-throughput marker-assisted selection of plants that will exhibit SCN resistance.
DOI:10.1007/BF00264210URLPMID:1896019 [本文引用: 2]

The Brassica napus rapeseed cultivar Topas contains an acetohydroxyacid synthase (AHAS) multigene family consisting of five members (AHAS 1-5). DNA sequence analysis indicate that AHAS1 and AHAS3 share extensive homology. They probably encode the AHAS enzymes essential for plant growth and development. AHAS2 has diverged significantly from AHAS1 and AHAS3 and has unique features in the coding region of the mature polypeptide, transit peptide and upstream non-coding DNA, which raises the possibility that it has a distinct function. AHAS4 and AHAS5 have interrupted coding regions and may be defective. The complexity of the AHAS multigene family in the allotetraploid species B. napus is much greater than reported for Arabidopsis thaliana and Nicotiana tabacum. Analysis of the presumptive progenitor diploid species B. campestris and B. oleracea indicated that AHAS2, AHAS3 and AHAS4 originate from the A genome, whereas AHAS1 and AHAS5 originate from the C genome. Further variation within each of the AHAS genes in these species was found.
URLPMID:1303798 [本文引用: 2]

Brassica species possess the most complex acetohydroxyacid synthase (AHAS) multigene family reported for plants. The AHAS genes code for an essential enzyme in branched-chain amino acid biosynthesis. In the allotetraploid species, B. napus, four (AHAS1-4) of the five AHAS genes have been cloned and sequenced. The transcripts were examined by RNase protection assays using gene-specific, antisense RNA probes. Only AHAS1, AHAS2 and AHAS3 were shown to be expressed in B. napus and one of the diploid progenitor species B. campestris or B. oleracea. AHAS1 and AHAS3 are highly conserved genes that presumably code for the essential AHAS housekeeping functions. They were expressed as low abundance mRNA in all somatic and reproductive tissues examined. AHAS2, which is structurally distinct from all other plant AHAS genes, was only expressed in mature ovules and extraembryonic tissues of immature seeds. This study provides direct evidence for multiple AHAS isoforms in plants and for an AHAS gene which is developmentally regulated in a tissue-specific manner. The discovery raises questions concerning the functional significance of AHAS in seed development.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]

以转基因抗除草剂油菜Q3为花粉供体材料,油菜远缘杂草为花粉受体材料,在自然传粉和人工辅助授粉条件下研究甘蓝型油菜与十字花科杂草间的基因漂移频率。结果表明,以转基因油菜为父本,十字花科杂草荠菜、碎米荠、播娘蒿、诸葛菜、风花菜、遏蓝菜和菜为母本,杂交高度不亲和,基因漂移率为0 % ,无生态风险,但对野芥菜的基因漂移率高达0 .885 %。野芥菜是我国大部分地区的常见杂草,种类繁多,分布范围广,大面积种植转基因抗除草剂油菜对野生芥菜的基因污染应引起高度重视。
[本文引用: 1]
DOI:10.1046/j.1365-313x.1993.04020403.xURLPMID:8106085 [本文引用: 1]

A set of mapping markers have been designed for Arabidopsis thaliana that correspond to DNA fragments amplified by the polymerase chain reaction (PCR). The ecotype of origin of these amplified fragments can be determined by cleavage with a restriction endonuclease. Specifically, 18 sets of PCR primers were synthesized, each of which amplifies a single mapped DNA sequence from the Columbia and Landsberg erecta ecotypes. Also identified was at least one restriction endonuclease for each of these PCR products that generates ecotype-specific digestion patterns. Using these co-dominant cleaved amplified polymorphic sequences (CAPS), an Arabidopsis gene can be unambiguously mapped to one of the 10 Arabidopsis chromosome arms in a single cross using a limited number of F2 progeny.
DOI:10.1046/j.1365-313x.1998.00123.xURLPMID:9628032 [本文引用: 1]

Numerous techniques in plant molecular genetic analysis, such as mapping and positional cloning techniques, rely on the availability of molecular markers that can differentiate between alleles at a particular locus. PCR-based cleaved amplified polymorphic sequences (CAPS) markers have been widely used as a means of rapidly and reliably detecting a single-base change that creates a unique restriction site in one of a pair of alleles. However, the majority of single-nucleotide changes do not create such sites and thus cannot be used to create CAPS markers. In this paper, a modification of the CAPS technique that allows detection of most single-nucleotide changes by utilizing mismatched PCR primers is described. The mismatches in the PCR primers, in combination with the single-nucleotide change, create a unique restriction site in one of the alleles.
DOI:10.1046/j.1365-313x.1998.00124.xURLPMID:9628033 [本文引用: 1]

PCR-based detection of single nucleotide polymorphisms is a powerful tool for the plant geneticist. Cleaved amplified polymorphic sequence analysis is the most widely used approach for the detection of single nucleotide polymorphisms. However, this technique is limited to mutations which create or disrupt a restriction enzyme recognition site. This paper presents a modification of this technique where mismatches in a PCR primer are used to create a polymorphism based on the target mutation. This technique is useful for following known mutations in segregating populations and genetic mapping of isolated DNAs used for positional based cloning of new genes. In addition, a computer program has been developed that facilitates the design of these PCR primers.
URL [本文引用: 2]
DOI:10.1007/s11032-013-9917-xURL [本文引用: 1]
DOI:10.3724/SP.J.1006.2018.01600URL [本文引用: 1]

Heterodera glycines Ichinohe)是严重危害世界范围大豆生产的害虫, 采用合理轮作和种植抗病品种可有效控制损失。为了开展分子标记辅助选择以加速抗病品种培育, 本研究针对前期发现的与大豆胞囊线虫3号小种(SCN3)抗性显著关联的非同义变异SNP位点Map-5149, 开发高通量、低成本的新型分子标记—竞争性等位基因特异PCR标记(kompetitive allele specific PCR, KASP), GmSNAP11-5149。利用GmSNAP11-5149鉴定了来自8个国家的202份代表性大豆抗感资源, 发现141份材料携带抗病基因型GmSNAP11-5149-AA, 平均雌虫指数为6.2%, 极显著低于58份携带感病基因型GmSNAP11-5149-GG材料的雌虫指数(61.1%), 方差分析表明, GmSNAP11-5149与胞囊线虫的抗性显著相关(F=44.18, P<0.0001), 对抗病材料的选择效率达到92%, GmSNAP11-5149可作为一个实用的分子标记应用于辅助抗大豆胞囊线虫品种选育和抗病种质资源鉴定。]]>
[本文引用: 1]
DOI:10.1186/s12870-019-1844-5URLPMID:31142280 [本文引用: 1]

BACKGROUND: Clubroot, caused by Plasmodiophora brassicae Woronin, is a very important disease of Brassica species. Management of clubroot relies heavily on genetic resistance. In a cross of Brassica nigra lines PI 219576 (highly resistant, R) x CR2748 (highly susceptible, S) to clubroot, all F1 plants were resistant to clubroot. There was a 1:1 ratio of R:S in the BC1 and 3R:1S in the F2, which indicated that a single dominant gene controlled clubroot resistance in PI 219576. This gene was designated Rcr6. Mapping of Rcr6 was performed using genome sequencing information from A-genome of B. rapa and B-genome of B. nigra though bulked segregant RNA sequencing (BSR-Seq) and further mapping with Kompetitive Allele Specific PCR (KASP) analysis. RESULTS: Reads of R and S bulks from BSR-Seq were initially aligned onto B. rapa (A-genome; B. nigra has the B-genome) where Rcr6 was associated with chromosome A08. KASP analysis showed that Rcr6 was flanked by SNP markers homologous to the region of 14.8-15.4 Mb of chromosome A08. There were 190 genes annotated in this region, with five genes (Bra010552, Bra010588, Bra010589, Bra010590 and Bra010663) identified as encoding the toll-interleukin-1 receptor / nucleotide-binding site / leucine-rich-repeat (TIR-NBS-LRR; TNL) class of proteins. The reads from BSR-Seq were then aligned into a draft B-genome of B. nigra, where Rcr6 was mapped on chromosome B3. KASP analysis indicated that Rcr6 was located on chromosome B3 in a 0.5 Mb region from 6.1-6.6 Mb. Only one TNL gene homologous to the B. rapa gene Bra010663 was identified in the target region. This gene is a likely candidate for Rcr6. Subsequent analysis of the Rcr6 equivalent region based on a published B. nigra genome was performed. This gene is located into chromosome B7 of the published B-genome, homologous to BniB015819. CONCLUSION: Rcr6 was the first gene identified and mapped in the B-genome of Brassica species. It resides in a genomic region homologous to chromosome A08 of A-genome. Based on this finding, it could possibly integrate into A08 of B. napus using marker assisted selection with SNP markers tightly linked to Rcr6 developed in this study.
DOI:10.1038/s41598-019-51191-zURLPMID:31601933 [本文引用: 1]

The fungal pathogen Leptosphaeria maculans causes blackleg disease on canola and rapeseed (Brassica napus) in many parts of the world. A B. napus cultivar, 'Quinta', has been widely used for the classification of L. maculans into pathogenicity groups. In this study, we confirmed the presence of Rlm1 in a DH line (DH24288) derived from B. napus cultivar 'Quinta'. Rlm1 was located on chromosome A07, between 13.07 to 22.11 Mb, using a BC1 population made from crosses of F1 plants of DH16516 (a susceptible line) x DH24288 with bulked segregant RNA Sequencing (BSR-Seq). Rlm1 was further fine mapped in a 100 kb region from 19.92 to 20.03 Mb in the BC1 population consisting of 1247 plants and a F2 population consisting of 3000 plants using SNP markers identified from BSR-Seq through Kompetitive Allele-Specific PCR (KASP). A potential resistance gene, BnA07G27460D, was identified in this Rlm1 region. BnA07G27460D encodes a serine/threonine dual specificity protein kinase, catalytic domain and is homologous to STN7 in predicted genes of B. rapa and B. oleracea, and A. thaliana. Robust SNP markers associated with Rlm1 were developed, which can assist in introgression of Rlm1 and confirm the presence of Rlm1 gene in canola breeding programs.
DOI:10.3724/SP.J.1006.2013.01711URL [本文引用: 1]

油菜抗咪唑啉酮类除草剂基因BnALS1R是从抗性突变体M9中克隆获得,抗性基因BnALS1R与野生型基因BnALS1存在1处SNP,即乙酰乳酸合酶第638位丝氨酸残基被天冬酰胺酸替代。为获得油菜抗除草剂基因BnALS1R的分子标记,根据该处点突变,结合获得的BnALS3与BnALS1序列,开发30条等位基因特异PCR (allele-specific PCR,AS-PCR)引物,采用筛选出的3条AS-PCR引物在F2、BC1和BC2群体中进行PCR扩增。结果表明,该标记有效检测出群体中存在的3种基因型,其分离比分别为1∶2∶1、1∶1、1∶1,均遵循单基因遗传规律。应用该标记对获得的抗性恢复系进行PCR扩增,结果发现所有抗性恢复系均能扩增出抗性基因BnALS1R目的条带,表明3条标记引物可应用于抗性基因的检测。AS-PCR标记的获得将促进以抗性基因进行油菜抗除草剂分子标记辅助选择育种。]]>
[本文引用: 1]
DOI:10.1007/s00122-014-2320-0URL [本文引用: 1]
DOI:10.1007/s10681-008-9662-0URL [本文引用: 1]

Outcrossing or cross hybridization is a potential concern in herbicide-resistant crop management strategies such as in the Clearfield™ rice system. Recent studies have shown that the mutated acetolactate synthase (ALS) gene that confers resistance to imazethapyr (Newpath) herbicide can be transferred from Clearfield rice cultivars via cross pollination under field conditions to weedy red rice. Resistance of commercial Clearfield rice cultivars to imazethapyr is due to the presence of two point mutations in the ALS gene that result in amino acid substitutions from serine to asparagine (S to D) and glycine to glutamic acid (G to E). We report here development of a DNA-based method that involves application of allele-specific PCR assays to distinguish herbicide-susceptible and resistant ALS alleles in either homozygous or heterozygous genotypes produced from natural outcrosses between Clearfield varieties CL121, CL141, CL161 and weedy red rice. PCR assays that can distinguish between the homozygous and heterozygous imazethapyr-resistant S653D and G654E SNP alleles of the rice ALS gene were developed and evaluated. A total of 483 individual red rice plants were successfully screened for the presence of S653D SNP and another 145 F2 individuals from natural red rice×CL121 hybridizations were screened for the presence of the G654E SNP. The PCR-based assays produced during this study are simple, rapid, inexpensive, reproducible and require only standard PCR and electrophoretic instruments that can be applied toward outcrossing evaluation and effective weed management strategies for the Clearfield crop system.]]>
DOI:10.1073/pnas.1105612108URLPMID:21551103 [本文引用: 1]

Induced mutagenesis can be an effective way to increase variability in self-pollinated crops for a wide variety of agronomically important traits. Crop resistance to a given herbicide can be of practical value to control weeds with efficient chemical use. In some crops (for example, wheat, maize, and canola), resistance to imidazolinone herbicides (IMIs) has been introduced through mutation breeding and is extensively used commercially. However, this production system imposes plant-back restrictions on rotational crops because of herbicide residuals in the soil. In the case of barley, a preferred rotational crop after wheat, a period of 9-18 mo is required. Thus, introduction of barley varieties showing resistance to IMIs will provide greater flexibility as a rotational crop. The objective of the research reported was to identify resistance in barley for IMIs through induced mutagenesis. To achieve this objective, a sodium azide-treated M(2)/M(3) population of barley cultivar Bob was screened for resistance against acetohydroxy acid synthase (AHAS)-inhibiting herbicides. The phenotypic screening allowed identification of a mutant line showing resistance against IMIs. Molecular analysis identified a single-point mutation leading to a serine 653 to asparagine amino acid substitution in the herbicide-binding site of the barley AHAS gene. The transcription pattern of the AHAS gene in the mutant (Ser653Asn) and WT has been analyzed, and greater than fourfold difference in transcript abundance was observed. Phenotypic characteristics of the mutant line are promising and provide the base for the release of IMI-resistant barley cultivar(s).
DOI:10.1104/pp.110.169482URL [本文引用: 1]

Brassica napus is an allotetraploid (AACC) formed from the fusion of two diploid progenitors, Brassica rapa (AA) and Brassica oleracea (CC). Polyploidy and genome-wide rearrangement during the evolution process have resulted in genes that are present as multiple homologs in the B. napus genome. In this study, three B. napus homologous genes encoding endoplasmic reticulum-bound sn-glycerol-3-phosphate acyltransferase 4 (GPAT4) were identified and characterized. Although the three GPAT4 homologs share a high sequence similarity, they exhibit different expression patterns and altered epigenetic features. Heterologous expression in yeast further revealed that the three BnGPAT4 homologs encoded functional GPAT enzymes but with different levels of polypeptide accumulation. Complementation of the Arabidopsis (Arabidopsis thaliana) gpat4 gpat8 double mutant line with individual BnGPAT4 homologs suggested their physiological roles in cuticle formation. Analysis of gpat4 RNA interference lines of B. napus revealed that the BnGPAT4 deficiency resulted in reduced cutin content and altered stomatal structures in leaves. Our results revealed that the BnGPAT4 homologs have evolved into functionally divergent forms and play important roles in cutin synthesis and stomatal development.
