 ,2,*
,2,*Mechanism of BnaBZR1 and BnaPIF4 regulating photosynthetic efficiency in oilseed rape (Brassica napus L.) under poor light
FENG Tao1,2, TAN Hui1,2, GUAN Mei2, GUAN Chun-Yun ,2,*
,2,*通讯作者:
收稿日期:2019-12-16接受日期:2020-03-24网络出版日期:2020-08-12
| 基金资助: |
Received:2019-12-16Accepted:2020-03-24Online:2020-08-12
| Fund supported: |
作者简介 About authors
E-mail:812298771@qq.com。

摘要
关键词:
Abstract
Keywords:
PDF (10855KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
冯韬, 谭晖, 官梅, 官春云. BnaBZR1和BnaPIF4基因调控甘蓝型油菜弱光光效的机制[J]. 作物学报, 2020, 46(8): 1146-1156. doi:10.3724/SP.J.1006.2020.94198
FENG Tao, TAN Hui, GUAN Mei, GUAN Chun-Yun.
植物中转录因子PIF4和BZR1互作是植物油菜素内酯信号与光信号互作的核心节点[1], 同时二者互作也是介导植物对弱光照等非生物胁迫响应的节点[2]。油菜素内酯通过一系列信号转导调节BZR1和BZR2 (BES1)的表达和磷酸化[3], 并通过磷酸化等方式串联与之互作的其他因子调控下游基因表达, 调控植物生长, 最后形成完整的反馈回路平衡植物内源油菜素内酯的合成[4]。拟南芥和水稻中BZR1/2以单体或复合体形式与PIF4互作, 介导下游的基因转录调控[5]。植物中PIF4蛋白在光照条件下稳定性较弱, 极易发生泛素化降解, 因此植物PIF4在转录水平的调控也是相关信号互作中重要的调节方式。
甘蓝型油菜是世界范围内最重要的油料作物之一[6], 但其具有极为复杂的异源二倍体基因组[7], 不同甘蓝型油菜对低光等非生物胁迫的抗性有极大的多态性[8], 但油菜相关BnaPIF4和BnaBZR1的表达调控与功能均不清楚。前期研究发现, 从甘蓝型油菜湘油15中克隆BnaPIF4基因发现其A03染色体上的同源拷贝相较测序品种中双11号存在着内含子突变[9], 这表明在甘蓝型油菜中BnaPIF4基因可能存在着更多的突变。甘蓝型油菜品系XY881和XY883由相同亲本湘油15经辐照诱变后连续自交筛选而得, 其中XY881基本保持了湘油15的原有特征, XY883则表现为更高的光合效率, 更高的种子含油量和油酸含量, 但同时种子含油量的稳定性较低、对低光寡照胁迫更为敏感。在XY881和XY883中进行BnaPIF4和BnaBZR1基因克隆、序列特征、表达和功能分析, 对探明油菜中BnaPIF4和BnaBZR1的表达调控规律及其在低光照胁迫中扮演的角色具有重要意义, 同时也将为阐明甘蓝型油菜对低光照胁迫响应存在差异的分子机制提供一些新的理论支持。
本文克隆了两品系甘蓝型油菜中BnaPIF4和BnaBZR1基因并进行了完整的比较分析, 确定了甘蓝型油菜品系XY883中BnaPIF4和BnaBZR1基因存在的可变剪接和启动子插入突变, 并通过酵母杂交、表达分析和植物遗传转化等方式确认了XY883中BnaPIF4和BnaBZR1基因突变对基因表达和蛋白翻译的影响, 并初步确定了BnaPIF4和BnaBZR1在拟南芥和油菜中的功能。
1 材料与方法
1.1 植物材料与培养
甘蓝型油菜品种(系)湘油15、XY881、XY883以及野生型拟南芥由国家油料改良中心(湖南)提供。分别于湖南农业大学耘园试验基地和室内植物光照培养箱中(KBW240, BINDER, 美国) (16 h/8 h光周期)种植受试材料, 以常规水肥管理。在植物培养箱中设置光源100 μmol m-2 s-1模拟强光照, 20 μmol m-2 s-1模拟弱光照, 试验基地中以遮光网遮光模拟弱光胁迫。1.2 取样和测试
分别于甘蓝型油菜品系XY881和XY883种子萌发后45、120、200和220 d对根、茎、叶、花、角果及完成遗传转化后的甘蓝型油菜和T2代拟南芥叶片随机取样, 以液氮速冻后储存于-80℃冰箱, 用于RNA和蛋白提取。对经不同处理的甘蓝型油菜品系XY881和XY883完全成熟的种子随机取样, 以索氏抽提测试含油量, 以气相色谱法[10]测试脂肪酸组成。1.3 DNA提取、RNA提取与cDNA合成
使用CTAB-PVP提取液从甘蓝型油菜叶片中提取基因组DNA。使用TRIzol RNA Extraction Kit (TransGen Biotech Co., Ltd.)从甘蓝型油菜和拟南芥样品中提取总RNA。使用Easy Script First-Strand cDNA Synthesis SuperMix Kit (TransGen Biotech Co., Ltd.)进行第1链cDNA合成。1.4 基因克隆
从BRAD数据库(Table 1
表1
表1基因克隆引物
Table 1
| 引物 Primer | 克隆产物 Product of cloning | 序列 Sequence (5'-3') |
|---|---|---|
| PIF4_mF | mRNA of BnaPIF4 | ATAGATCTCATCCCTAAAGA |
| PIF4_mR | TATGTTCAAAAGATAGCCTTAG | |
| BZR1_mF | mRAN of BnaBZR1 | GGAGAAGGAAAGAGAGATTC |
| BZR1_mR | TTGAGAGAAACAAAATGGGC | |
| PIF4_cF | CDS of BnaPIF4 | ATGGAACACCAAGGTTGGAG |
| PIF4_cR | CTAACGGGGACCGTCGG | |
| BZR1_cF | CDS of BnaBZR1 | ATGACGTCAGATGGAGCTACG |
| BZR1_cR | TCAACCACGAGCTTTGCC | |
| PIF4_pF | Promoter of BnaPIF4 | ATTGAAACCGATTGTAAGGA |
| PIF4_pR | AGAAACAAAGGAGCATAAAG | |
| BZR1_pF | Promoter of BnaBZR1 | GGTTATTTTCAAATAATGGATG |
| BZR1_pR | CTTGAGCTCTTAGCCCTGTG | |
| PIF4_uF | 5'-UTR of BnaPIF4 | ATAGATCTCATCCCTAAAGA |
| PIF4_uR | GTCAGATCTCAGATTTGGAAAGC | |
| PIF4_dF | Full-length gene of BnaPIF4 | ATAGATCTCATCCCTAAAGAAG |
| PIF4_dR | TTTTGACAAACTAAACCAGG |
新窗口打开|下载CSV
1.5 RT-qPCR与WB
以RT-qPCR检测BnaPIF4和BnaBZR1基因表达, 以WB检测BnaPIF4、BnaBZR1及1,5-二磷酸核酮糖羧化酶(Ribulose-1,5-bisphosphate carboxylase, RuBPCase)等蛋白表达。以BnaAction为内参基因, 采用2-ΔΔCt法以RT-qPCR检测BnaPIF4和BnaBZR1各拷贝表达, 根据序列特征设计所需引物(表2), 各试验均3次技术重复。Table 2
表2
表2RT-qPCR引物
Table 2
| 引物 Primer | 基因 Gene | 序列 Sequence (5'-3') |
|---|---|---|
| ActF | BnaActin | GGTTGGGATGGACCAGAAGG |
| ActR | TCAGGAGCAATACGGAGC | |
| PIF4F | BnaPIF4_cds | CTGTGCTGTTGTGCTTAC |
| PIF4R | AGTCTCTACATAAACCCATAGG | |
| U1_F | BnaPIF4_UTR_U01 | CTGTGCTGTTGTGCTTAC |
| U1_R | AGTCTCTACATAAACCCATAGG | |
| U2_F | BnaPIF4_UTR_U02 | CTGTGCTGTTGTGCTTAC |
| U2_R | TTCCTCTCACTTGCTCTC | |
| U3_F | BnaPIF4_UTR_U03 | GAGAGCAAGTGAGAGGAA |
| U3_R | CAGAAGCTGAAGTAGTAGAAG | |
| BZR1F | BnaBZR1 | CTCTCATCTCCAACTTCCAA |
| BZR1R | GACTCATCACACTCAGGTATA |
新窗口打开|下载CSV
分别以HA-tag (ab18181, Abcam, Inc., 美国)(1:1000)和RbcL抗体(AS03037, Agrisera, Ins., 瑞典) (1:2000)为一抗进行WB, 检测连接HA标签的BnaPIF4蛋白和RuBPCase蛋白, 以ECL发光法进行印迹, 随后以GS800扫描仪(Bio-Rad Company, 美国)扫描蛋白光密度。
1.6 载体构建与遗传转化
用于拟南芥和甘蓝型油菜BnaPIF4和BnaBZR1遗传转化的pRI101-AN载体来源于国家油料改良中心(湖南)。分别以3种BnaPIF4基因的5°-UTR替换pRI101-AN载体中的AtADH的5'-UTR序列, 随后将携带HA标签的BnaPIF4 CDS序列插入pRI101-AN载体。以农杆菌GV3103侵染介导进行拟南芥和甘蓝型油菜的遗传转化。
1.7 酵母杂交实验
分别以pGAD-T7作为酵母单杂交和双杂交的AD质粒, 以pGBK-T7为酵母双杂交的BD质粒, 以pHIS2为酵母单杂交的BD质粒, 共转化酵母菌株HF7c用于鉴定BnaPIF4和BnaBZR1之间的互作关系。2 结果与分析
2.1 弱光和2,4-BL处理下XY881与XY883重要农艺性状
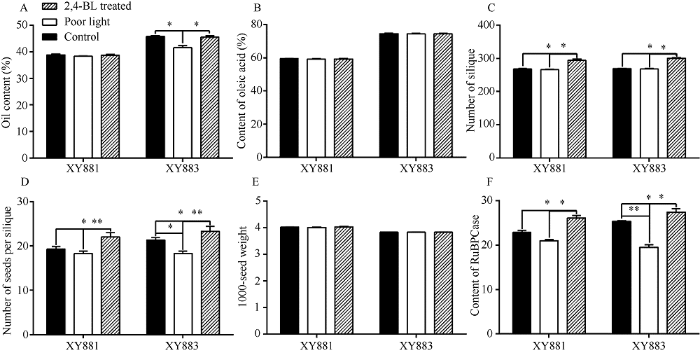
对大田栽培的XY881和XY883分别进行遮光和2,4-BL处理, 并统计其有效光合叶RuBPCase活性、全株总角果数、每角果种子粒数、成熟种子含油量、油酸含量及种子千粒重(图1)。图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1弱光和2,4-BL处理下XY881和XY883重要农艺性状
A: 种子含油量; B: 种子油酸含量; C: 单株总角果数; D: 每角果种子粒数; E: 成熟种子千粒重; F: 叶片RuBPCase含量。*P < 0.05, **P < 0.01。
Fig. 1Investigation of important agronomic traits of XY881 and XY883 under poor light and 2,4-BL treatment
A: oil content of seed; B: oleic acid content of seed; C: total number of silique per plant; D: number of seeds per silique; E: 1000-seed weight of mature seeds; F: RuBPCase content in leaves. *P < 0.05, **P < 0.01.
遮光处理下XY881和XY883表现出明显的敏感性差异, XY883叶片中RuBPCase含量下降较XY881更明显, 与之对应的XY883每角果中种子粒数和种子含油量也由明显下降, 而XY881每角果粒数和种子含油量均无明显下降。2,4-BL处理对XY881与XY883的影响规律相似, 油菜叶片中RuBPCase含量明显上升, 光合作用加强, 与之对应的全株总角果数和每角果种子粒数明显增多, 但整体而言XY883受影响更明显。无论弱光照胁迫或2,4-BL处理均未对XY881和XY883种子千粒重及种子油脂中油酸比例。由此推测弱光和2,4-BL处理下主要通过影响光合作用水平, 调控XY881和XY883种子油脂合成的底物, 该过程中XY881和XY883存在油菜素内酯信号响应因子表达水平差异。
2.2 两品系甘蓝型油菜(XY881和XY883)中BnaBZR1和BnaPIF4基因克隆和结构分析
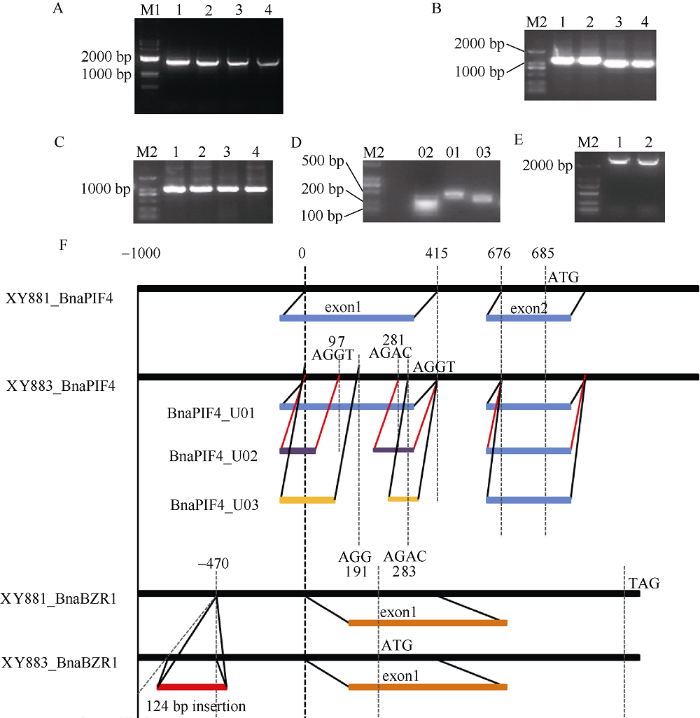
为进一步理解BnaPIF4和BnaBZR1在XY881和XY883对弱光和2,4-BL响应差异中的作用, 本文从XY881和XY883中分别克隆BnaPIF4和BnaBZR1的全长mRNA和全长基因, 并从中亚克隆启动子、CDS和5°-UTR等元件, 并对两品系中BnaPIF4和BnaBZR1基因结构进行分析(图2)。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2XY881和XY883中BnaPIF4和BnaBZR1基因结构
A: BnaPIF4和BnaBZR1全长mRNA; B: BnaPIF4和BnaBZR1全长CDS; C: BnaPIF4和BnaBZR1启动子; D: BnaPIF4三种5'-UTR; E: BnaPIF4全长基因; F: XY881和XY883中BnaPIF4和BnaBZR1突变结构示意。1: XY881中BnaPIF4; 2: XY883中BnaPIF4; 3: XY881中BnaBZR1; 4: XY883中BnaBZR1; 01~03: XY883中BnaPIF4三种5'-UTRs; M1: 2K plus DNA marker; M2: 2K DNA marker。
Fig. 2Different gene structures of BnaPIF4 and BnaBZR1 in XY881 and XY883
A: cloning of mRNA of BnaPIF4 and BnaBZR1; B: cloning of CDS of BnaPIF4 and BnaBZR1; C: cloning of promoter of BnaPIF4 and BnaBZR1; D: cloning of 5'-UTR of BnaPIF4; E: cloning of full-length BnaPIF4 gene; F: the mutation structure of BnaPIF4 and BnaBZR1 in XY881 and XY883. 1: BnaPIF4 of XY881; 2: BnaPIF4 of XY883; 3: BnaBZR1 of XY881; 4: BnaBZR1 of XY883; 01-03: three 5'-UTRs of BnaPIF4 from XY883; M1: 2K plus DNA marker; M2: 2K DNA marker.
基因结构分析显示, XY881中BnaPIF4和BnaBZR1基因与亲本湘油15一致, 而XY883中BnaPIF4和BnaBZR1基因分别存在可变剪接和启动子插入突变。XY883中BnaPIF4可变剪接共形成3种含有不同5°-UTR的转录产物, 分别为U01 (424 bp)、U02 (239 bp)和U03 (332 bp) (图2-D); 该可变剪接均发生于亲本BnaPIF4基因的第一外显子内, 3个5°-UTR拷贝具有相同的剪接边界序列(AGGT和AGAC), 第一剪接点分别位于+97 bp、+191 bp和+415 bp位置。XY883中BnaBZR1基因在启动子-470 bp位置内存在一个124 bp长度的富A/T碱基插入突变。
2.3 BnaBZR1启动子插入突变功能
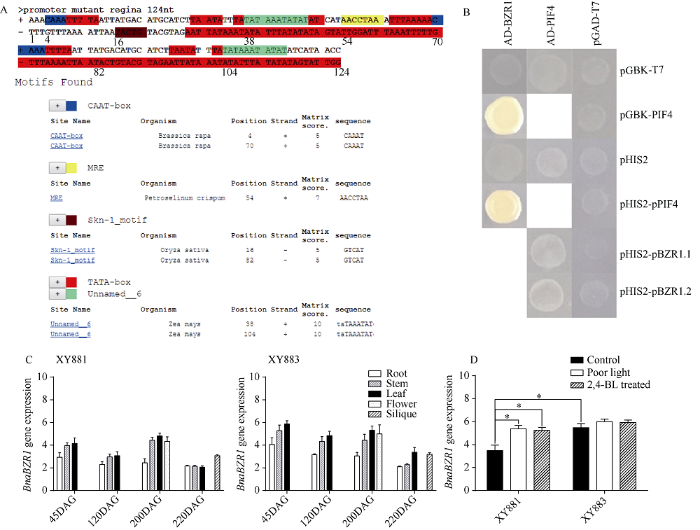
为进一步确认XY883中BnaBZR1基因启动子区域的插入突变有何功能, 分别以PlantCARE数据库(图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3BnaBZR1启动子插入突变、基因表达和BnaBZR1与BnaPIF4互作
A: BnaBZR1启动子插入突变序列分析; B: BnaBZR1和BnaPIF4酵母杂交分析; C: XY881和XY883中BnaBZR1时空表达; D: 弱光和2,4-BL处理下XY881和XY883中BnaBZR1表达。DAG表示种子萌发后天数。
Fig. 3Insertion mutation in the BnaBZR1 promoter changed the BnaBZR1 gene expression, did not change the interaction of BnaBZR1 with BnaPIF4
A: sequence analysis of insertion mutation regions in the promoter of BnaBZR1; B: analysis of the interaction between BnaBZR1 and BnaPIF4 by yeast hybridization; C: the temporal and spatial gene expression of BnaBZR1 in XY881 and XY883; D: the expression of BnaBZR1 in XY881 and XY883 under poor light stress and 2,4-BL treatment. DAG means days after seed germination.
分析发现, BnaBZR1基因启动子区124 bp的插入突变中含有大量的启动子元件, 包含多个TATA盒、2个CAAT盒、1个MRE 结构域及2个Skn-1结构域。酵母杂交结果表明, BnaBZR1与BnaPIF4蛋白可相互结合, BnaBZR1能结合BnaPIF4启动子, 而BnaPIF4不能结合BnaBZR1启动子, BnaBZR1基因启动子插入突变不影响BnaPIF4与BnaBZR1启动子的结合关系。XY883中BnaBZR1启动子插入突变会明显提高其表达量, 苗期和苔期XY883中BnaBZR1表达明显高于XY881, 叶片和茎等光合作用部位XY883中BnaBZR1表达明显高于XY881。XY883中BnaBZR1表达稳定, 不受低光和2,4-BL的明显诱导, 而XY881中BnaBZR1表达受到低光和2,4-BL明显诱导, 刺激后XY881中BnaBZR1表达明显提高, 这表明BnaBZR1启动子插入突变可能提高BnaBZR1本底表达水平, 同时造成BnaBZR1表达对环境的响应减弱, 插入突变序列影响BnaBZR1表达具体通过何种方式尚需进一步研究, 由插入突变中大量启动子元件来看, 该插入突变可能引入了上游转录因子的结合位点, 加强了启动, 但同时由于启动子元件的增加造成光响应和油菜素内酯响应的转录因子结合位点竞争抑制, 降低了BnaBZR1表达对低光和2,4-BL的响应。
2.4 可变剪接影响BnaPIF4基因表达和翻译
通过RT-qPCR检测XY881和XY883中BnaPIF4基因的时空表达规律以及在弱光胁迫和2,4-BL处理下BnaPIF4不同转录产物表达的响应(图4-A, B)。将BnaPIF4可变剪接形成的不同转录产物分别克隆到pRI101-AN载体上, 重组载体结构如图4-C, 随后将重组载体转化拟南芥, 观察表型并检测转基因拟南芥中BnaPIF4表达情况及光合酶RuBPCase含量(图4-D~F)。图4
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4BnaPIF4可变剪接对基因表达和翻译的影响
A: XY881和XY883中BnaPIF4时空表达; B: 弱光和2,4-BL处理下XY881和XY883中BnaPIF4表达; C: 含不同5'-UTRs重组pRI101-AN 载体结构; D: 转基因拟南芥中BnaPIF4 mRNA和蛋白表达; E: 转BnaPIF4拟南芥表型; F: 转BnaPIF4拟南芥RuBPCase含量。DAG表示种子萌发后天数。
Fig. 4Alternative splicing affects BnaPIF4 gene expression and protein translation
A: temporal and spatial gene expression of BnaPIF4 in XY881 and XY883; B: the expression of BnaPIF4 in XY881 and XY883 under poor light stress and 2,4-BL treatment; C: construction of pRI101-AN vector with different 5'-UTRs; D: the mRNA expression of BnaPIF4 and the protein expression of BnaPIF4; E: the phenotype of transgenic BnaPIF4 Arabidopsis thaliana; F: the RuBPCase protein content of transgenic BnaPIF4 Arabidopsis thaliana. DAG means days after seed germination.
时空表达检测发现XY881和XY883中BnaPIF4总表达量模式无明显差异, BnaPIF4表达均为随生育进程表达量逐渐降低, 有明显的组织特异性, 主要表达于地上部分, 叶中表达量最高。低光照和2,4-BL处理诱导BnaPIF4表达, 且XY883中BnaPIF4表达量增幅远大于XY881, 进一步分析发现BnaPIF4不同转录产物对弱光和2,4-BL的响应具有明显差别, 其中转录本U01 (具有U01结构5°- UTR, 即亲本无突变转录产物)响应最明显, U03转录产物次之(具有U03结构5°-UTR), U02转录产物(具有U02结构5°-UTR)无明显响应。拟南芥遗传转化发现不同的5°-UTR在35S启动子驱动下不影响BnaPIF4的CDS转录, 但不同的5°-UTR对BnaPIF4蛋白合成具有显著影响, 其中U01与对照AtADH5具有相近的高水平BnaPIF4蛋白合成, U02和U03具有明显偏低的BnaPIF4蛋白合成量。转BnaPIF4基因拟南芥表现出叶片狭长、茎伸长、早花及结实减少等表型, 转不同的BnaPIF4转录产物, 效应具有差异, U01转录产物影响最大, U03次之, U02影响最小, 叶片光合关键酶RuBPCase含量的下降也表现U01最明显, U03次之, U02相对野生型无明显下降。BnaPIF4本身是bHLH家族重要的转录因子, 其下游存在大量的响应基因, BnaPIF4不同转录产物的蛋白合成能力具有差异, 由此推测, 环境因素可能影响XY883中BnaPIF4的剪接过程, 调控不同转录产物的比例, 一方面在转录水平响应BnaPIF4上游因子的调控, 一方面在蛋白水平传导和改变对下游响应基因的调控水平, 通过自身在转录和翻译上平衡的调节来调控XY883对环境的综合响应, 这可能是XY883相对亲本湘油15在正常光照条件下具有更高的光合效能且对弱光等环境因素更敏感的原因之一。
2.5 甘蓝型油菜和拟南芥中BnaPIF4和BnaBZR1功能
为进一步验证BnaPIF4和BnaBZR1的功能, 分别对拟南芥和甘蓝型油菜亲本湘油15进行遗传转化, 观察过表达BnaPIF4和BnaBZR1后的表型(图5)。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5转基因拟南芥和转基因油菜表型
A: 转基因拟南芥; B: 转基因拟南芥RuBPCase含量; C: 转基因拟南芥种子含油量; D: 转基因油菜; E: 转基因油菜中BnaPIF4和BnaBZR1表达; F: 转基因油菜RuBPCase含量。
Fig. 5Phenotype of transgenic Arabidopsis thaliana and B. napus L.
A: the phenotype of transgenic BnaPIF4 and BnaBZR1 Arabidopsis thaliana; B: the RuBPCase protein content of transgenic BnaPIF4 and BnaBZR1 Arabidopsis thaliana; C: the oil content of transgenic Arabidopsis thaliana seed; D: the plant of transgenic B. napus L.; E: the gene expression of BnaPIF4 and BnaBZR1 in transgenic B. napus L.; F: the RuBPCase content of leaves of transgenic B. napus L.
转BnaPIF4基因拟南芥相较野生型表现出叶片狭长、茎伸长、早花且结实率降低, 叶片中RuBPCase含量明显下降的表型, 成熟种子的含油量也明显下降, BnaPIF4不同转录产物的效应存在差异, 转U01和U03拟南芥种子含油量下降明显, 转U02拟南芥种子含油量则无明显下降, 这与3种转录物对拟南芥叶片RuBPCase含量的影响一致。转BnaBZR1基因拟南芥相较野生型具有明显更大的生物量, 生育期适度缩短, 总结实量上升, 但叶片RuBPCase含量和种子含油量无明显变化。共转化BnaPIF4和BnaBZR1基因拟南芥仍保持转BnaPIF4基因拟南芥叶片狭长和早花的特征, 但植株生物量明显提升, 叶片RuBPCase含量和种子含油量相对野生型无明显变化。综合来看, 在拟南芥中BnaPIF4表现为光合作用的负调控因子; BnaBZR1表现生长促进作用, 提升拟南芥生物量; BnaBZR1对BnaPIF4的光合作用负效应具有拮抗作用。在亲本湘油15中过表达BnaPIF4会让其主茎相对更细长, 叶片也更狭长, 开花略有提前; 在湘油15中过表达BnaBZR1会提高其生物量, 提早花期, 增加开花量, 但均无统计学差异; 湘油15过表达BnaPIF4和BnaBZR1后叶片RuPBCase含量亦无明显变化。整体而言, BnaPIF4和BnaBZR1过表达对甘蓝型油菜的影响和拟南芥基本一致, 但对甘蓝型油菜的影响远小于拟南芥, 这可能与甘蓝型油菜复杂的双二倍体基因组中存在大量同源拷贝有关, 单一基因过表达影响远不如拟南芥明显。
3 讨论
甘蓝型油菜品系XY883和XY881均来源于亲本湘油15, XY881基本保持了亲本特性, XY883的RuBPCase含量、种子含油量、种子油酸含量和结实量相较亲本均明显提升, 弱光胁迫对XY881和湘油15影响较小, 而对XY883影响十分明显, 这表明XY883和XY881的表型差异可能源于光响应元件的差异。2,4-BL对XY881和XY883的调控规律基本一致, 均提升RuBPCase含量, 促进光合, 但影响幅度差异明显, 2,4-BL对BnaPIF4和BnaBZR1基因具有一致的诱导作用, 诱导效应存在品系差异[11], 这表明BnaPIF4和BnaBZR1的表达调控介导油菜素甾类信号和光信号通路的互作可能是XY881和XY883光合作用和弱光敏感性等差异的原因。植物中BZR1是油菜素内酯信号通路中的关键转录因子, 其通过核质穿梭调控下游靶基因表达, 并通过反馈回路影响植物油菜素甾醇合成来调节植物的生长[12], 可见油菜中BnaBZR1自身的表达调控同样可以影响油菜素内酯信号途径。XY883中BnaBZR1启动子中的插入突变引入了多个具有转录因子特征的元件, 本底表达水平明显高于XY881, 但弱光胁迫和2,4-BL处理对XY883中BnaBZR1的诱导效应较弱, 造成在胁迫和2,4-BL处理下两品系油菜中BnaBZR1表达水平接近, 这可能是由于启动子插入突变之后增强了启动, 同时对上游转录调控因子产生竞争性结合, 抑制了弱光和2,4-BL的响应元件工作, 但具体的分子机制尚需进一步探索, 另一方面, XY883中BnaBZR1对弱光和2,4-BL钝感, 导致内源油菜素内酯合成的BnaBZR1反馈调节失效, 造成XY883对弱光照等不利环境因素的调节能力下降。
PIF4是植物光信号通路中的核心转录因子之一, 它通过响应光质变化[13]和热胁迫[14]等调节植物植物开花和生长等生物学过程。PIF4是介导光信号与植物激素途径相互作用的关键转录因子, 如与赤霉素信号[15]、油菜素内酯信号[16]和生长素信号[17]等产生互作。在XY883中BnaPIF4选择性剪接形成3种具有不同5'-UTR的转录物, 弱光和2,4-BL对这3种转录物的诱导效应具有明显差别, 在转基因拟南芥中这3种转录物也表现明显不一致的翻译效率, 并导致转基因拟南芥产生不同的RuBPCase含量, 这表明XY883中BnaPIF4的剪接调控可能是其与XY881存在明显光合水平和弱光敏感性差异的原因。XY883中BnaPIF4通过可变剪接形成不同转录物, 造成转录水平和翻译水平的变化量存在明显差异, 在正常光照下XY883中BnaPIF4低翻译水平的U02和U03转录物相对比例较高, 弱光下高翻译水平的U01转录物比例明显上升, 而BnaPIF4在拟南芥和油菜中都表现出光效负调控因子的特征, 这可能是XY883在常光下表现明显高于亲本光合效能, 而弱光下光合效能明显降低的原因。
酵母杂交实验表明, BnaBZR1蛋白与BnaPIF4相互作用, BnaBZR1与BnaPIF4的启动子相互作用, 但BnaPIF4不与BnaBZR1的启动子相互作用, 这表明BnaBZR1可能调节BnaPIF4的表达, 而BnaPIF4可能不直接调控BnaBZR1的表达。PIF4和BZR的互作是植物油菜素内酯信号和光信号互作的关键节点, BnaBZR1与BnaPIF4的互作模式可能意味着油菜中油菜素内酯信号与光信号的互作存在方向性偏好。前期研究显示在甘蓝型油菜中BnaPIF4和BnaBZR1均存在大量同源拷贝[9,18], 因此在功能上可能存在大量的冗余, 二者在甘蓝型油菜光效调节中可能存在更复杂的互作网络, 需要进一步对二者同源拷贝及上下游调控因子进行发掘和探索。
总体来看, 甘蓝型油菜中涉及油菜素内酯和光信号互作因子存在着基因多态性, 由此介导的油菜素内酯信号途径和光信号途径互作等方面差异可能是不同品系甘蓝型油菜光合效能差异的原因, 两信号途径之间互作的平衡可能是筛选稳定的高产油油菜品种的重要指标。
4 结论
甘蓝型油菜品系XY883相对亲本湘油15, 分别存在BnaPIF4的可变剪接和BnaBZR1启动子的插入突变, 导致两基因在不同条件下的表达出现不同于亲本的变化规律。BnaPIF4在拟南芥和油菜中表现光合负效应因子的特点, 而BnaBZR1可对此负效应形成拮抗, 拟南芥受BnaPIF4和BnaBZR1遗传转化影响较甘蓝型油菜更大。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
DOI:10.1038/ncb2545URLPMID:22820378 [本文引用: 1]

Plant growth is coordinately regulated by environmental and hormonal signals. Brassinosteroid (BR) plays essential roles in growth regulation by light and temperature, but the interactions between BR and these environmental signals remain poorly understood at the molecular level. Here, we show that direct interaction between the dark- and heat-activated transcription factor phytochrome-interacting factor 4 (PIF4) and the BR-activated transcription factor BZR1 integrates the hormonal and environmental signals. BZR1 and PIF4 interact with each other in vitro and in vivo, bind to nearly 2,000 common target genes, and synergistically regulate many of these target genes, including the PRE family helix-loop-helix factors required for promoting cell elongation. Genetic analysis indicates that BZR1 and PIFs are interdependent in promoting cell elongation in response to BR, darkness or heat. These results show that the BZR1-PIF4 interaction controls a core transcription network, enabling plant growth co-regulation by the steroid and environmental signals.
DOI:10.1038/ncb2546URLPMID:22820377 [本文引用: 1]

Brassinosteroid and gibberellin promote many similar developmental responses in plants; however, their relationship remains unclear. Here we show that BR and GA act interdependently through a direct interaction between the BR-activated BZR1 and GA-inactivated DELLA transcription regulators. GA promotion of cell elongation required BR signalling, whereas BR or active BZR1 suppressed the GA-deficient dwarf phenotype. DELLAs directly interacted with BZR1 and inhibited BZR1-DNA binding both in vitro and in vivo. Genome-wide analysis defined a BZR1-dependent GA-regulated transcriptome, which is enriched with light-regulated genes and genes involved in cell wall synthesis and photosynthesis/chloroplast function. GA promotion of hypocotyl elongation requires both BZR1 and the phytochrome-interacting factors (PIFs), as well as their common downstream targets encoding the PRE-family helix-loop-helix factors. The results demonstrate that GA releases DELLA-mediated inhibition of BZR1, and that the DELLA-BZR1-PIF4 interaction defines a core transcription module that mediates coordinated growth regulation by GA, BR and light signals.
URLPMID:21802346 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
URL [本文引用: 1]
DOI:10.1126/science.1253435URLPMID:25146293 [本文引用: 1]

Oilseed rape (Brassica napus L.) was formed ~7500 years ago by hybridization between B. rapa and B. oleracea, followed by chromosome doubling, a process known as allopolyploidy. Together with more ancient polyploidizations, this conferred an aggregate 72x genome multiplication since the origin of angiosperms and high gene content. We examined the B. napus genome and the consequences of its recent duplication. The constituent An and Cn subgenomes are engaged in subtle structural, functional, and epigenetic cross-talk, with abundant homeologous exchanges. Incipient gene loss and expression divergence have begun. Selection in B. napus oilseed types has accelerated the loss of glucosinolate genes, while preserving expansion of oil biosynthesis genes. These processes provide insights into allopolyploid evolution and its relationship with crop domestication and improvement.
DOI:10.1002/jsfa.8562URLPMID:28732144 [本文引用: 1]

BACKGROUND: Vegetable growers in Arctic areas must increasingly rely on market strategies based on regional origin and product quality. Swede roots (rutabaga) were grown in a phytotron to investigate the effect of high latitude light conditions on sensory quality and some health and sensory-related compounds. Experimental treatments included modifications of 24 h natural day length (69 degrees 39' N) by moving plants at daily intervals to dark chambers with either no light, fluorescent growth light and/or low intensity photoperiod extension. RESULTS: Shortening the photosynthetic light period to 12 h produced smaller roots than 15.7 h and 18 h, with highest scores for bitter and sulfur taste, and lowest scores for sweetness, acidic taste and fibrousness. The photoperiod in combination with the photosynthetic light period also had an influence on glucosinolate (GLS) contents, with lowest concentrations in 24 h natural light and highest in 12 h natural light. Concentrations of vitamin C, glucose, fructose and sucrose were not significantly influenced by any of the treatments. CONCLUSION: High latitude light conditions, with long photosynthetic light periods and 24 h photoperiod, can enhance sweet/less bitter taste and reduce GLS contents in swede roots, compared to growth under short day conditions. This influence of light conditions on eating quality may benefit marketing of regional products from high latitudes. (c) 2017 Society of Chemical Industry.
[本文引用: 2]
[本文引用: 2]
URLPMID:18504157 [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/s1534-5807(02)00153-3URLPMID:11970900 [本文引用: 1]

Plant steroid hormones, brassinosteroids (BRs), are perceived by a cell surface receptor kinase, BRI1, but how BR binding leads to regulation of gene expression in the nucleus is unknown. Here we describe the identification of BZR1 as a nuclear component of the BR signal transduction pathway. A dominant mutation bzr1-1D suppresses BR-deficient and BR-insensitive (bri1) phenotypes and enhances feedback inhibition of BR biosynthesis. BZR1 protein accumulates in the nucleus of elongating cells of dark-grown hypocotyls and is stabilized by BR signaling and the bzr1-1D mutation. Our results demonstrate that BZR1 is a positive regulator of the BR signaling pathway that mediates both downstream BR responses and feedback regulation of BR biosynthesis.
DOI:10.1016/j.cub.2008.12.046URLPMID:19185498 [本文引用: 1]

Stomata are pores on the surfaces of leaves that regulate gas exchange between the plant interior and the atmosphere [1]. Plants adapt to changing environmental conditions in the short term by adjusting the aperture of the stomatal pores, whereas longer-term changes are accomplished by altering the proportion of stomata that develop on the leaf surface [2, 3]. Although recent work has identified genes involved in the control of stomatal development [4], we know very little about how stomatal development is modulated by environmental signals, such as light. Here, we show that mature leaves of Arabidopsis grown at higher photon irradiances show significant increases in stomatal index (S.I.) [5] compared to those grown at lower photon irradiances. Light quantity-mediated changes in S.I. occur in red light, suggesting that phytochrome photoreceptors [6] are involved. By using a genetic approach, we demonstrate that this response is dominated by phytochrome B and also identify a role for the transcription factor, PHYTOCHROME-INTERACTING FACTOR 4 (PIF4) [7]. In sum, we identify a photoreceptor and downstream signaling protein involved in light-mediated control of stomatal development, thereby establishing a tractable system for investigating how an environmental signal modulates stomatal development.
DOI:10.1038/nature10928URLPMID:22437497 [本文引用: 1]

Plant growth and development are strongly affected by small differences in temperature. Current climate change has already altered global plant phenology and distribution, and projected increases in temperature pose a significant challenge to agriculture. Despite the important role of temperature on plant development, the underlying pathways are unknown. It has previously been shown that thermal acceleration of flowering is dependent on the florigen, FLOWERING LOCUS T (FT). How this occurs is, however, not understood, because the major pathway known to upregulate FT, the photoperiod pathway, is not required for thermal acceleration of flowering. Here we demonstrate a direct mechanism by which increasing temperature causes the bHLH transcription factor PHYTOCHROME INTERACTING FACTOR4 (PIF4) to activate FT. Our findings provide a new understanding of how plants control their timing of reproduction in response to temperature. Flowering time is an important trait in crops as well as affecting the life cycles of pollinator species. A molecular understanding of how temperature affects flowering will be important for mitigating the effects of climate change.
DOI:10.1038/nature06520URLPMID:18216857 [本文引用: 1]

Cell elongation during seedling development is antagonistically regulated by light and gibberellins (GAs). Light induces photomorphogenesis, leading to inhibition of hypocotyl growth, whereas GAs promote etiolated growth, characterized by increased hypocotyl elongation. The mechanism underlying this antagonistic interaction remains unclear. Here we report on the central role of the Arabidopsis thaliana nuclear transcription factor PIF4 (encoded by PHYTOCHROME INTERACTING FACTOR 4) in the positive control of genes mediating cell elongation and show that this factor is negatively regulated by the light photoreceptor phyB (ref. 4) and by DELLA proteins that have a key repressor function in GA signalling. Our results demonstrate that PIF4 is destabilized by phyB in the light and that DELLAs block PIF4 transcriptional activity by binding the DNA-recognition domain of this factor. We show that GAs abrogate such repression by promoting DELLA destabilization, and therefore cause a concomitant accumulation of free PIF4 in the nucleus. Consistent with this model, intermediate hypocotyl lengths were observed in transgenic plants over-accumulating both DELLAs and PIF4. Destabilization of this factor by phyB, together with its inactivation by DELLAs, constitutes a protein interaction framework that explains how plants integrate both light and GA signals to optimize growth and development in response to changing environments.
DOI:10.1101/gad.243675.114URLPMID:25085420 [本文引用: 1]

Signaling by the hormones brassinosteroid (BR) and gibberellin (GA) is critical to normal plant growth and development and is required for hypocotyl elongation in response to dark and elevated temperatures. Active BR signaling is essential for GA promotion of hypocotyl growth and suppresses the dwarf phenotype of GA mutants. Cross-talk between these hormones occurs downstream from the DELLAs, as GA-induced destabilization of these GA signaling repressors is not affected by BRs. Here we show that the light-regulated PIF4 (phytochrome-interacting factor 4) factor is a phosphorylation target of the BR signaling kinase BRASSINOSTEROID-INSENSITIVE 2 (BIN2), which marks this transcriptional regulator for proteasome degradation. Expression of a mutated PIF41A protein lacking a conserved BIN2 phosphorylation consensus causes a severe elongated phenotype and strongly up-regulated expression of the gene targets. However, PIF41A is not able to suppress the dwarf phenotype of the bin2-1 mutant with constitutive activation of this kinase. PIFs were shown to be required for the constitutive BR response of bes1-D and bzr1-1D mutants, these factors acting in an interdependent manner to promote cell elongation. Here, we show that bes1-D seedlings are still repressed by the inhibitor BRZ in the light and that expression of the nonphosphorylatable PIF41A protein makes this mutant fully insensitive to brassinazole (BRZ). PIF41A is preferentially stabilized at dawn, coinciding with the diurnal time of maximal growth. These results uncover a main role of BRs in antagonizing light signaling by inhibiting BIN2-mediated destabilization of the PIF4 factor. This regulation plays a prevalent role in timing hypocotyl elongation to late night, before light activation of phytochrome B (PHYB) and accumulation of DELLAs restricts PIF4 transcriptional activity.
DOI:10.1073/pnas.1110682108URLPMID:22123947 [本文引用: 1]

At high ambient temperature, plants display dramatic stem elongation in an adaptive response to heat. This response is mediated by elevated levels of the phytohormone auxin and requires auxin biosynthesis, signaling, and transport pathways. The mechanisms by which higher temperature results in greater auxin accumulation are unknown, however. A basic helix-loop-helix transcription factor, PHYTOCHROME-INTERACTING FACTOR 4 (PIF4), is also required for hypocotyl elongation in response to high temperature. PIF4 also acts redundantly with its homolog, PIF5, to regulate diurnal growth rhythms and elongation responses to the threat of vegetative shade. PIF4 activity is reportedly limited in part by binding to both the basic helix-loop-helix protein LONG HYPOCOTYL IN FAR RED 1 and the DELLA family of growth-repressing proteins. Despite the importance of PIF4 in integrating multiple environmental signals, the mechanisms by which PIF4 controls growth are unknown. Here we demonstrate that PIF4 regulates levels of auxin and the expression of key auxin biosynthesis genes at high temperature. We also identify a family of SMALL AUXIN UP RNA (SAUR) genes that are expressed at high temperature in a PIF4-dependent manner and promote elongation growth. Taken together, our results demonstrate direct molecular links among PIF4, auxin, and elongation growth at high temperature.
[本文引用: 1]
[本文引用: 1]
