 ,1,2,3, 伍小方1,2,3, 黄伟1,3, 周定港1,3, 张大为1,3, 周美亮2, 张凯旋
,1,2,3, 伍小方1,2,3, 黄伟1,3, 周定港1,3, 张大为1,3, 周美亮2, 张凯旋 ,2,*, 严明理
,2,*, 严明理 ,1,3,*
,1,3,*Regulation of flavonoid pathway by BjuB.KAN4 gene in Brassica juncea
GAO Guo-Ying ,1,2,3, WU Xiao-Fang1,2,3, HUANG Wei1,3, ZHOU Ding-Gang1,3, ZHANG Da-Wei1,3, ZHOU Mei-Liang2, ZHANG Kai-Xuan
,1,2,3, WU Xiao-Fang1,2,3, HUANG Wei1,3, ZHOU Ding-Gang1,3, ZHANG Da-Wei1,3, ZHOU Mei-Liang2, ZHANG Kai-Xuan ,2,*, YAN Ming-Li
,2,*, YAN Ming-Li ,1,3,*
,1,3,*通讯作者:
收稿日期:2020-01-12接受日期:2020-04-15网络出版日期:2020-09-12
| 基金资助: |
Received:2020-01-12Accepted:2020-04-15Online:2020-09-12
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (2328KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
高国应, 伍小方, 黄伟, 周定港, 张大为, 周美亮, 张凯旋, 严明理. 芥菜型油菜BjuB.KAN4基因调控类黄酮的途径[J]. 作物学报, 2020, 46(9): 1322-1331. doi:10.3724/SP.J.1006.2020.04008
GAO Guo-Ying, WU Xiao-Fang, HUANG Wei, ZHOU Ding-Gang, ZHANG Da-Wei, ZHOU Mei-Liang, ZHANG Kai-Xuan, YAN Ming-Li.
原花青素(proanthocyanidins, PAs)是植物中广泛存在的一种类黄酮化合物, 具有决定植物器官颜色等作用。类黄酮生物合成途径由一系列酶催化完成, 包括苯丙氨酸脱氨酶(phenylalanin ammonialyase, PAL)、查尔酮合成酶(chalcone synthase, CHS)、查尔酮异构酶(chalcone isomerase, CHI)、黄烷酮3-羟化酶(flavanone 3-hydroxylase, F3H)、类黄酮3’-羟化酶(flavonoid 3’-hydroxylase, F3’H)和黄酮醇合酶(flavonol synthase, FLS)、4-二氢黄酮醇还原酶(dihydroflavonol 4-reductase, DFR)、花青素合成酶(anthocyanidin synthase, ANS)、花色素还原酶(anthocyanidin reductase, ANR)等[1]。由于类黄酮合成过程的结构基因已经被深入挖掘分析, 研究注意力逐渐转向调控基因, 对类黄酮生物合成网络的转录因子进行筛选鉴定和机理解析正成为研究的主要方向。
MYB转录因子广泛参与植物器官颜色的形成、次生代谢和环境胁迫反应等重要的生物学过程[2,3]。如Nesi等[4]发现, 拟南芥(Arabidopsis thaiiana L.) TT2基因编码一个R2R3-MYB蛋白, 是种子发育中的PAs积累的关键调控蛋白。Akhter等[5]发现, 水稻(Oryza sativa L.) MYB转录因子OsPL通过调控花青素代谢使种子耐热性和相关植物激素含量增加。Luo等[6]发现, 苦荞(Fagopyrum tataricum Gaertn.) R2R3-MYB转录因子FtMYB15在拟南芥中的过量表达使叶片和种皮中花青素和PAs含量增加。小麦(Triticum aestivum L.) TaPL1通过结合CHS、DFR、LDOX、UF3GT基因的启动子促进花青素合成[7]; 苦荞(Fagopyrum tataricum L.) FtMYB1和FtMYB2基因在烟草中的异源表达显著增加了PAs的积累[8]; 除MYB激活剂外, MYB阻遏物也参与调控类黄酮的生物合成, 它们可通过直接隔离特定的转录因子或间接抑制MBW复合物中其他基因的转录来破坏MBW复合物的活性, 从而阻止黄酮类化合物的合成, 如拟南芥R3-MYB转录因子AtCAPRICE (AtCPC)[9]和AtMYBL2[10]; 矮牵牛(Petunia hybrida Vilm) PtMYBx[11]等。此外, bHLH和WD40等转录因子也可以通过调控结构基因的表达来影响类黄酮的生物合成[12]。拟南芥中, KAN4基因编码的ABERRA NT TESTA SHAPE转录因子已被证明可以控制拟南芥种子的类黄酮途径, 该基因通常是拟南芥正在发育的种子中黄酮醇和PAs合成的正调节因子, 它可以与结构基因的启动子特异性结合, 正调节拟南芥种子中黄酮醇和PAs的含量, 而在成熟的拟南芥种子中, 该基因作为抑制子参与PAs的合成[13]。此外, Mcabee等[14]证明该基因参与调节植物胚珠体壁生长。然而, 油菜中KAN4参与类黄酮合成的研究未见报道。
芥菜型油菜(Brassica juncea)是芸薹属的重要油料作物。研究发现, 油菜种皮颜色差异是由于类黄酮生物合成途径中PAs在种皮中积累所致, 而类黄酮生物合成途径中涉及的基因表达主要受转录因子的控制[15,16,17]。因此探究芸薹属植物类黄酮生物合成途径转录因子的功能对阐明种皮颜色形成的分子机制和指导芸薹属育种具有重要意义。本研究从紫叶芥中克隆出BjuB.KAN4基因及其启动子, 结合实时定量荧光PCR分析该基因在油菜不同组织的表达模式, 利用毛状根体系和异源表达拟南芥对该基因进行一系列的功能验证, 为油菜PAs合成的分子机制研究和黄籽育种提供理论依据。
1 材料与方法
1.1 试验材料
芥菜型油菜紫叶芥(PM, 2n = 36, AABB)和四川黄籽(SY, 2n = 36, aabb)种子由湖南科技大学提供; 野生型拟南芥种子, 发根农杆菌(Agrobacterium rhizogenes) A4菌株, 根癌农杆菌(A. tumefaciens) GV3101菌株, 过表达载体pCAMBIA3301, 均由中国农业科学院作物科学研究所荞麦基因资源创新组提供。将PM、SY和拟南芥种子分别种于MS培养基和营养土中, 用于无菌苗和土培苗的获得。RNA提取试剂盒购自北京天根生化技术有限公司, cDNA反转录试剂盒和实时荧光定量PCR试剂盒购自南京诺唯赞生物科技有限公司。由北京博迈德生物技术有限公司合成引物和对菌液测序。1.2 实验方法
1.2.1 BjuB.KAN4基因及启动子克隆 以2周苗龄的PM幼苗叶片为材料, 用CTAB法提取总DNA,以RNA试剂盒提取总RNA, 参考试剂盒说明书反转录成cDNA。根据NCBI (NW008726050; NC027749; NC027761; NC027760; NC027768)和BRAD (
Table 1
表1
表1引物序列
Table 1
| 引物名称 Primer name | 引物序列 Primer sequences (5°-3°) | 引物用途 Purpose of primer |
|---|---|---|
| KAN4-B-F | AGTGAGATGATCATGTTCGAGTC | 基因克隆Gene cloning |
| KAN4-B-R | CAATTAGCACTTGAGAAGGGTTA | |
| KB-NcoIF | GGGGACTCTTGACCATGGTAATGATCATGTTCGAG | KAN4载体构建引物 KAN4 vector primers |
| KB-Eco91IR | GAAATTCGAGCTGGTCACCCAATTAGCACTTGAGAAGGG | |
| KB-pro-BamHIF | GGATCCATTGTCGTTGGTGACAGAAAC | KAN4启动子载体构建引物 KB-pro vector primer |
| KB-pro-NcoIR | CCATGGTTGGAGTTTTCAGAACTTTGGC | |
| Actin7-F | GCTGACCGTATGAGCAAAG | qRT-PCR检测引物 qRT-PCR detection primers |
| Actin7-R | AAGATGGATGGACCCGAC | |
| qKAN4-B-F | AGGACCCAAAGATCTCCTTGGT | |
| qKAN4-B-R | TTACACATAGTCAATCCCCCAACT | |
| qPAL-F | AGAGCTTTTGACCGGAGAGA | |
| qPAL-R | TTAATCACTCTTAACATATAGGAATGGGAG | |
| qCHS-F | TCTTCATATTGGACGAGATGAGGA | |
| qCHS-R | GCGTTTCTGTTCAAACAGGAA | |
| qCHI-F | CTTTGGAGCGACCATTAGAG | |
| qCHI-R | AGACAAAGCTTAACAAGAGAGGT | |
| qF3’H-F | TGATTGGGAATTAGCTGGAGGA | |
| qF3’H-R | AGTTAAATTTTAACCCGACCCGA | |
| qF3H-F | ATCTTGGAGGAGCCAATTACGT | |
| qF3H-R | ACACAAGGAGTCTAAGCGATGA | |
| qFLS-F | ACTAGGAATGTGATCGCACCA | |
| qFLS-R | TCAGAGGGATTAGGTTTACGG | |
| qDFR-F | TCTTTGGAACAGGTTTGAAGGA | |
| qDFR-R | TAAAGTGACAGGGAGAAAACCCT | |
| qANS-F | AAGCCGTTGCCTGAGA | |
| qANS-R | AGAGTTTCAGACTCAGACTTCA | |
| qBAN-F | GGTTTTTGTTGTTAGGGAAAGA | |
| qBAN-R | ATATGCTTACTCTGACAAAACAT |
新窗口打开|下载CSV
1.2.2 目的基因序列分析 通过DNAMAN软件将测序序列与下载序列比对, 使用NCBI-Blast在线分析获得不同物种该基因的同源蛋白序列, 用MEGA5.0软件绘制系统发育树, 树的每个节点bootstrap值为1000。
1.2.3 载体的构建和转化农杆菌感受态 采用同源重组法, 以Primer 5.0软件设计带有限制性核酸内切酶(Nco I和Eco91 I)识别位点的基因特异性引物用于重组载体pCAMBIA3301-BjuB.KAN4的构建(表1), 并将重组载体分别转化发根农杆菌和根癌农杆菌感受态细胞[18]。
1.2.4 油菜毛状根的诱导及鉴定 采用李隆等[19]的方法, 以2周苗龄的PM和SY幼苗为外植体进行毛状根的诱导。选取生长良好的毛状根提取DNA并进行PCR分子鉴定, 将阳性毛状根移入含100 mg mL-1 Cef的液体MS培养基培养, 设置3组生物学重复。
1.2.5 转基因油菜毛状根和拟南芥总黄酮含量检测
采用三氯化铝法[20]检测转基因油菜毛状根和拟南芥的总黄酮含量。以吸光度值Y为纵坐标, 芦丁浓度(mg L-1)为横坐标, 绘制标准曲线, Y = 0.0433X- 0.0038 (R2 = 0.9994)。用紫外分光光度计于420 nm测总黄酮的吸光度, 根据标准曲线方程计算总黄酮含量。
1.2.6 拟南芥遗传转化及纯合体筛选 采用蘸花法[21]侵染拟南芥植株。在MS固体培养基中加入25 mg mL-1草胺磷对T0代种子进行抗性筛选, 并提取叶片DNA进行阳性鉴定。
1.2.7 实时定量荧光PCR分析 提取PM和SY的根、茎、叶和转基因毛状根总RNA, 反转录获得cDNA, 利用实时定量荧光PCR (quantitative real-time PCR, qRT-PCR)检测油菜各组织、转基因毛状根和拟南芥中BjuB.KAN4的相对表达量, 以及对转基因毛状根中类黄酮合成相关酶基因的相对表达量进行分析。采用Microsoft Excel软件进行数据分析。以Actin7作为内参基因, 用相对表达量(relative expression) = 2-ΔΔCT的算法, 计算目标基因的相对表达量。qRT-PCR引物序列见表1。
1.2.8 转基因拟南芥原花青素含量分析 采用DMACA-HCl法[22]对T3代转基因拟南芥叶片进行原花青素含量检测。以吸光度值Y为纵坐标, 原花青素浓度(mg mL-1)为横坐标, 绘制标准曲线, Y = 0.9329X-0.0031 (R2 = 0.9996)。用紫外分光光度计于640 nm测原花青素的吸光度, 根据标准曲线方程计算样品中原花青素的含量。
1.2.9 转BjuB.KAN4pro::GUS拟南芥GUS染色
根据GUS染色法[23]利用GUS染色缓冲液对转BjuB.KAN4pro::GUS拟南芥染色。
2 结果与分析
2.1 BjuB.KAN4基因和启动子的克隆及序列分析
以PM的cDNA为模板克隆得到KAN4基因的CDS序列。经NCBI-Blast比对发现, 该基因序列与已经公布的拟南芥KAN4基因的2个mRNA序列(NM123627)和(NM001125891)相似性分别为85.94%和88.66%; 与甘蓝型油菜(B. napus, XM013870607)和白菜(B. rapa, XM009125437)的KAN4基因mRNA序列相似性均为96.39%。另外, 经BRAD-Blast比对显示, 该基因序列与B基因组相似度最高且分值为1098, 因此将该基因暂命名为BjuB.KAN4。与来源SY的BjuB.KAN4基因编码序列比对发现, 该基因在PM和SY中无序列差异。BjuB.KAN4基因的ORF长度为801 bp, 编码266个氨基酸, 预测蛋白分子量为29.94 kD, 等电点(pI)为9.14。通过NCBI-Blast比对分析, BjuB.KAN4蛋白与萝卜(Raphanus sativus,XP018449685) Rs.KAN4蛋白同源性最高, 达到93.58%。经NCBI-Conserved Domains分析, BjuB. KAN4蛋白包含一个MYB-like DNA结合结构域, 属于1R-MYB转录因子。将BjuB.KAN4蛋白与其他物种[如白菜、拟南芥、萝卜、亚麻荠(Camelina sativa)、野大豆(Glycine soja)、胡杨(Populus euphratica)、醉蝶花(Tarenaya hassleriana)、葡萄(Vitis vinifera)、玉米(Zea mays)等]的同源蛋白进行序列比对表明, 这些KAN4蛋白都含有一段高度保守的氨基酸序列(PRMRWTSTLHAHFVHAVQLLGGHERAT PKSVLELMNVKDLTLAHVKSHLQMYRT), 这段序列为MYB-like DNA结合结构域。以PM的DNA为模板克隆得到BjuB.KAN4基因上游1255 bp的启动子序列, 通过Plant-CARE分析启动子发现, 其含有激素响应元件、光响应元件和与分生组织表达有关的顺式作用元件, 为后续深入研究该转录因子在其他生物代谢途径的转录调控功能打下基础。2.2 BjuB.KAN4蛋白系统进化分析
使用MEGA5.0软件构建系统进化树, 以进一步探究BjuB.KAN4与其他物种中同源蛋白之间的进化关系(图1)。分析发现, BjuB.KAN4与白菜、萝卜、拟南芥等同类十字花科植物的KAN4蛋白亲缘关系较近, 与胡杨、大豆和玉米等植物亲缘关系较远。图1
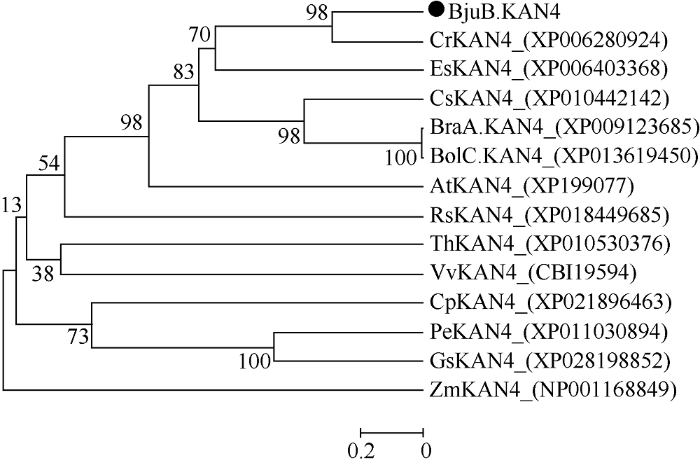 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1BjuB.KAN4与其他物种同源KAN4蛋白的系统发育树
AtKAN4: 拟南芥NP199077; BjuB.KAN4芥菜型油菜; BolC.KAN4: 甘蓝XP013619450; BraA.KAN4: 白菜XP009123685; CpKAN4: 番木瓜XP021896463; CrKAN4: 荠菜XP006280924; CsKAN4: 亚麻芥XP010442142; EsKAN4: 山嵛菜XP006403368; GsKAN4: 野大豆XP028198852; PeKAN4: 胡杨XP011030894; RsKAN4: 萝卜XP018449685; ThKAN4: 醉蝶花XP010530376; VvKAN4: 葡萄CBI19594; ZmKAN4: 玉米NP001168849。
Fig. 1Phylogenetic tree of BjuB.KAN4 and its homologous proteins from other species
AtKAN4: Arabidopsis thaliana NP199077; BjuB.KAN4: Brassica juncea; BolC.KAN4: Brassica oleracea XP013619450; BraA. KAN4: Brassica rapa XP009123685; CpKAN4: Carica papaya XP021896463; CrKAN4: Capsella rubella XP006280924; CsKAN4: Camelina sativa XP010442142; EsKAN4: Eutrema salsugineum XP006403368; GsKAN4: Glycine soja XP028198852; PeKAN4: Populus euphratica XP011030894; RsKAN4: Raphanus sativus XP018449685; ThKAN4: Tarenaya hassleriana XP010530376; VvKAN4: Vitis vinifera CBI19594; ZmKAN4: Zea mays NP001168849.
2.3 BjuB.KAN4基因的组织特异性表达分析
qRT-PCR分析发现, BjuB.KAN4在PM和SY的根茎叶组织中均有不同程度表达。BjuB.KAN4在PM和SY根中的表达量均显著高于茎和叶, 在PM中, 其在根中的表达量分别约为茎和叶的11倍和14倍(图2-A), 在SY中, BjuB.KAN4在根中的表达量分别约为茎和叶的5倍和10倍(图2-B)。说明BjuB.KAN4基因具有组织特异性, 在根中的表达量最高, 在叶中最低。图2
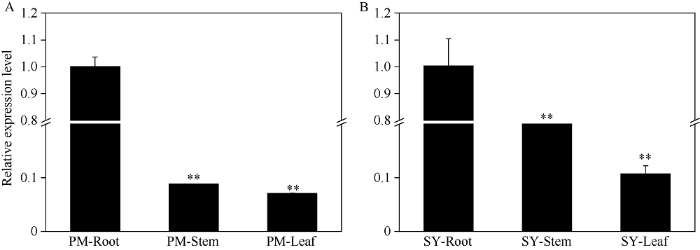 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2BjuB.KAN4基因的组织特异性表达分析
A: PM不同组织BjuB.KAN4的相对表达量; B: SY不同组织BjuB.KAN4的相对表达量。**表示在0.01水平差异显著。
Fig. 2Tissue-specific expression analysis of BjuB.KAN4 gene by qRT-PCR
A: relative expression of BjuB.KAN4 in different tissues of PM; B: relative expression of BjuB.KAN4 in different tissues of SY. ** Significant at P < 0.01.
2.4 毛状根的诱导及转基因根系鉴定
在试验成功获得转基因PM和SY的毛状根。qRT-PCR检测显示, BjuB.KAN4基因在PM (图3-C)和SY (图3-D)的阳性毛状根与野生型毛状根(A4)和转空载体(3301)的毛状根相比显著增加, 其中PM转基因毛状根中BjuB.KAN4基因的表达量是野生型的103倍, 而SY转基因毛状根中BjuB.KAN4基因的表达量是野生型的105倍, 说明目的基因能在毛状根中表达。图3
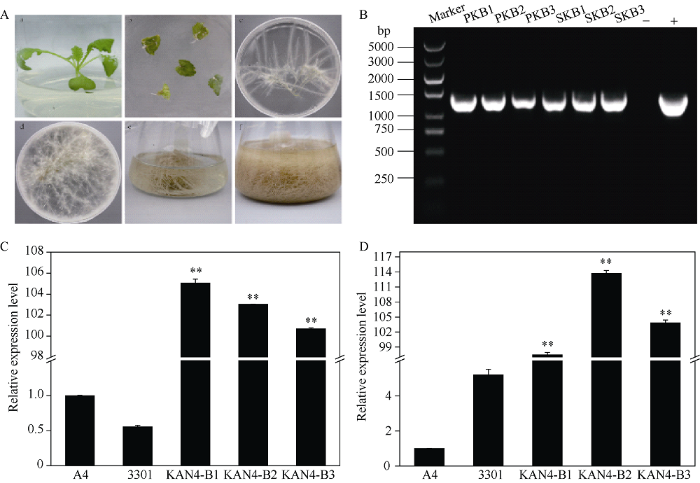 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3芥菜型油菜毛状根的诱导过程和转基因根系鉴定
A: 芥菜型油菜毛状根的诱导过程。B: PM和SY转基因毛状根DNA鉴定; PKB1-PKB3: PM阳性毛状根; SKB1-SKB3: SY阳性毛状根; -: H2O (阴性对照); +: 质粒(阳性对照)。C: PM毛状根中BjuB.KAN4的相对表达量。D: SY毛状根中BjuB.KAN4的相对表达量。A4: A4侵染的毛状根; 3301: 转pCAMBIA3301空载体毛状根; KAN4-B1~B3: 转基因毛状根。**表示在0.01水平差异显著。
Fig. 3Induction of hairy roots in Brassica juncea and identification of transgenic hairy roots
A: induction process of hairy roots in Brassica juncea. B: DNA identification of PM and SY transgenic hairy roots; PKB1-PKB3: positive hairy roots of PM; SKB1-SKB3: positive hairy roots of SY; -: H2O (negative control); +: plasmid (positive control). C: relative expression of BjuB.KAN4 in hairy roots of PM. D: relative expression of BjuB.KAN4 in hairy roots of SY. A4: hairy roots infected by A4; 3301: hairy roots transfected with pCAMBIA3301 empty vector; KAN4-B1-B3: transgenic hairy roots. ** Significant at P < 0.01.
2.5 转基因毛状根类黄酮合成途径关键酶基因相对表达量检测
以qRT-PCR检测显示, PM毛状根中Bju.CHS和Bju.ANS的表达量极显著增加, 分别是野生型的10.2倍和3.0倍, Bju.DFR的表达量显著增加1.67倍, 关键酶基因(Bju.CHI、Bju.F3’H和Bju.BAN)上升不显著, 另外一些关键酶基因(Bju.PAL、Bju.F3H和Bju.FLS)表达量下降不显著(均下降0.5倍左右)(图4-A)。SY毛状根中, Bju.CHS和Bju.FLS表达量极显著增加, 分别是野生型的10.0倍和5.3倍, Bju.DFR和Bju.BAN表达量显著增加, 分别是野生型的2.0倍和2.6倍, 其他关键酶基因(Bju.PAL、Bju.CHI、Bju.F3H、Bju.F3’H和Bju.ANS)的表达量下降不显著(分别下降0.15、0.20、0.36、0.37和0.80倍)(图4-B)。表明BjuB.KAN4基因有助于提高毛状根中类黄酮代谢途径部分关键酶基因的表达量, 从而促进类黄酮的生物合成, 但同时也下调了部分关键酶基因的表达。图4
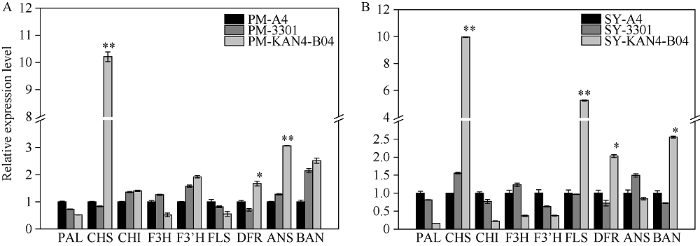 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4转基因毛状根中类黄酮代谢途径关键酶基因的相对表达量
A: PM转基因毛状根; B: SY转基因毛状根。A4: A4侵染的毛状根; 3301: 转pCAMBIA3301空载体毛状根; KAN4-B04: 转基因毛状根。内参基因: Actin7, 每组数据代
Fig. 4Relative expression of the key enzyme genes involved in flavonoids biosynthesis pathway in transgenic hairy roots
A: transgenic hairy roots of PM; B: transgenic hairy roots of SY. A4: hairy roots infected by A4; 3301: hairy roots transfected with pCAMBIA3301 empty vector; KAN4-B04: transgenic hairy roots. internal control: Actin7, error bars represent the standard deviation of triplicate runs for qRT-PCR. PAL: phenylalanine deaminase; CHS: chalcone synthase; CHI: chalcone isomerase; F3H: flavone 3-hydroxylase; F3’H: flavonoid 3’-hydroxylase; FLS: flavonol synthase; DFR: dihydroflavonol reductase; ANS: anthocyanin synthase; BAN: anthocyanin reductase. * Significant at P < 0.05; ** Significant at P < 0.01.
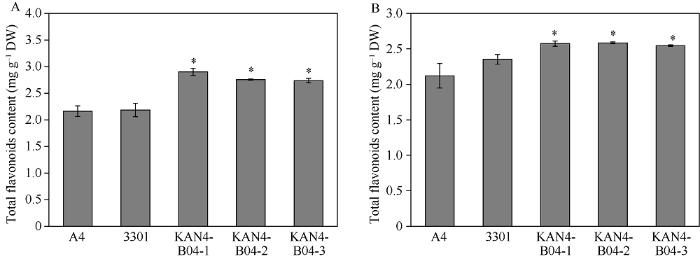
2.6 转基因毛状根总黄酮含量检测
PM转基因根系中总黄酮含量为2.798 mg g-1, 是野生型的1.3倍, SY中总黄酮含量为2.567 mg g-1, 是野生型的1.2倍, 转基因株系与野生型A4和转空载毛状根总黄酮含量相比均有显著增加(图5), 说明BjuB.KAN4基因参与调控芥菜型油菜毛状根中类黄酮的生物合成。图5
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5转基因毛状根中总黄酮含量检测
A: PM转基因毛状根; B: SY转基因毛状根。A4: A4侵染的毛状根; 3301: 转pCAMBIA3301空载体毛状根; KAN4-B04: 转基因毛状根。每组数据代
Fig. 5Measurement of the total flavonoids content in transgenic hairy roots
A: transgenic hairy roots of PM; B: transgenic hairy roots of SY. A4: hairy roots infected by A4; 3301: hairy roots transfected with pCAMBIA3301 empty vector; KAN4-B04: transgenic hairy roots. Each set of data represents the mean ± SD from three biological replicates. * Significant at P < 0.05.
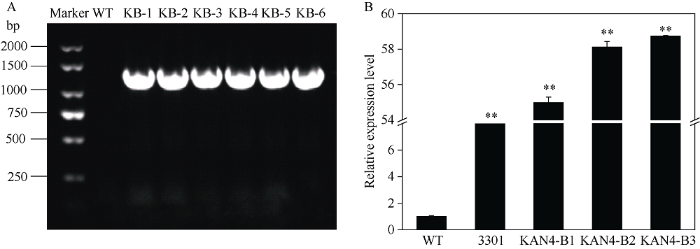
2.7 转基因拟南芥鉴定
提取拟南芥DNA进行阳性鉴定(图6-A), 并利用qRT-PCR检测BjuB.KAN4基因的相对表达量(图6-B)。结果说明, BjuB.KAN4基因成功转入拟南芥中, 且能够正常过量表达。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6过表达BjuB.KAN4基因拟南芥分子鉴定
A: 转基因拟南芥阳性PCR鉴定; B: 转基因拟南芥中BjuB.KAN4的相对表达量检测。WT: 野生型拟南芥; KB-1-6: 转基因阳性株系; 3301: 转空载体拟南芥; KAN4-B1-3: 转BjuB.KAN4基因拟南芥。**表示在0.01水平差异显著。
Fig. 6Identification of transgenic Arabidopsis plants overexpressing BjuB.KAN4
A: identification of transgenic Arabidopsis by PCR; B: relative expression of BjuB.KAN4 in transgenic Arabidopsis. WT: wild-type of Arabidopsis; KB-1-6: transgenic positive lines; 3301: Arabidopsis overexpressing empty vector; KAN4-B1-3: Arabidopsis lines overexpressing BjuB.KAN4 gene. ** Significant at P < 0.01.
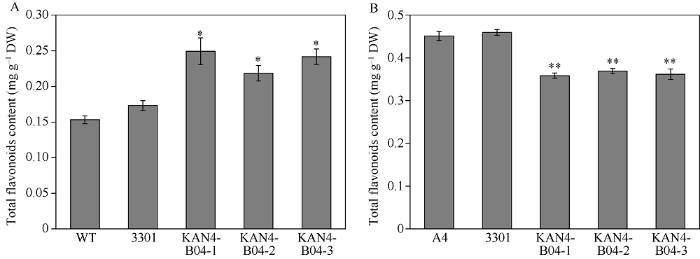
2.8 转基因拟南芥总黄酮和原花青素含量检测
转基因株系中总黄酮含量为0.237 mg g-1, 显著高于野生型和转空载体拟南芥, 为野生型的1.5倍, 转3301空载体的植株总黄酮含量略微增加但不显著(图7-A), 说明BjuB.KAN4基因参与调控拟南芥中类黄酮的生物合成。而拟南芥转基因植株原花青素含量为0.363 mg g-1, 较野生型含量下降了0.8倍且差异显著(图7-B), 推测该转录因子抑制了原花青素的生物合成途径, 同时可能对类黄酮代谢途径其他分支具有促进作用, 从而导致原花青素含量降低, 但总黄酮含量升高。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7转基因拟南芥总黄酮和原花青素含量检测
A: 转基因拟南芥总黄酮含量检测; B: 转基因拟南芥原花青素含量检测。WT: 野生型拟南芥; 3301: 转空载体拟南芥; KAN4-B04-1-3: 转BjuB.KAN4基因拟南芥; 每组数据代
Fig. 7Measurement of the total flavonoids and the proathocyanidins contents in transgenic Arabidopsis
A: measurement of the total flavonoids content in transgenic Arabidopsis; B: measurement of the proathocyanidins content in transgenic Arabidopsis. WT: wild-type Arabidopsis; 3301: Arabidopsis overexpressing empty vector; KAN4-B04-1-3: Arabidopsis lines overexpressing BjuB.KAN4 gene. Each set of data represents the mean ± SD of three biological replicates. * Significant at P < 0.05; ** Significant at P < 0.01.
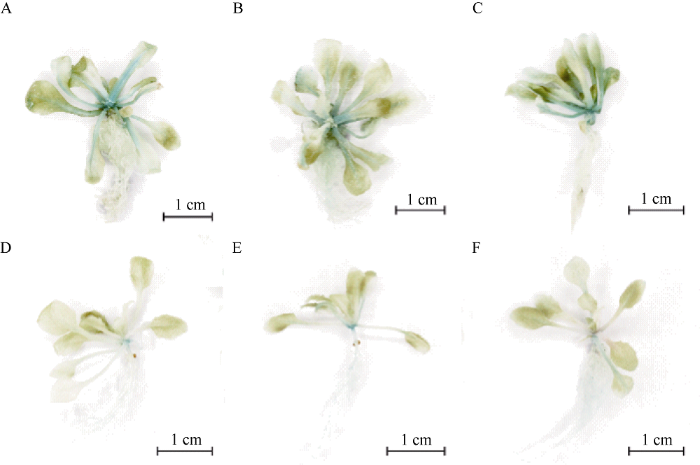
2.9 转基因拟南芥GUS组织化学检测
通过GUS染色发现, 转BjuB.KAN4pro::GUS的拟南芥主要在根、茎和叶的维管组织中呈现蓝色, 与转空载体3301的拟南芥相比染色较浅(图8), 推测BjuB.KAN4启动子主要在拟南芥的根茎叶的维管组织中表达。图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8转基因拟南芥组织化学检测
A, B, C: 转35Spro::GUS拟南芥的不同株系; D, E, F: 转BjuB.KAN4pro::GUS拟南芥的不同株系。
Fig. 8Histochemical staining of transgenic Arabidopsis plants
A, B, C: independent Arabidopsis lines overexpressing 35Spro::GUS; D, E, F: independent Arabidopsis lines overexpressing BjuB.KAN4pro::GUS.
3 讨论
基因在植物不同组织中具有特异性表达模式, 如草莓(Fragaria × ananassa Duch.) FaXTH1和FaXTH2基因具有果实特异性, 且能响应赤霉素、脱落酸和乙烯等激素诱导[24]; 拟南芥Athspr基因具有维管组织特异性, 其启动子包含对植物激素、光、生物和非生物胁迫以及对组织特异性表达响应的调控元件, 可能在维管发育和抵抗各种压力方面具有多种功能[25]。本研究发现, BjuB.KAN4基因在PM和SY根茎叶的表达量差异较大, 其在PM根中表达量分别是茎和叶的11倍和14倍, 在SY根中表达量分别是茎和叶的5倍和10倍(图2-A, B)。启动子作为调控基因表达的重要调控序列, 它能够通过调节特异性元件驱动基因表达。根据具有维管组织特异性的基因启动子一般包含激素、光和与分生组织表达有关的顺式作用元件等特点, 本研究通过GUS组织化学染色分析BjuB.KAN4在植株中的组织表达差异, 推测该基因可能在根茎叶的维管组织中表达(图8)。转录因子通过与结构基因启动子上的顺式元件结合来调控类黄酮生物合成途径, 其中MYB转录因子调控类黄酮生物合成途径已经在许多不同植物中得到阐明。Ma等[26]发现, 在杨树(P. tremula × tremuloides)中过表达MYB165和MYB194基因减少了花青素和PAs的积累。Zhai等[27]发现, 梨(Pyrus bretschneideri Rehd.)中PAP型MYB转录因子PbMYB10b可通过诱导PbDFR的表达来调节花色素和PAs合成途径; 另外, TT2型MYB转录因子PbMYB9不仅通过激活PbANR启动子充当PAs途径的特异性激活剂, 而且通过结合梨果实中的PbUFGT1启动子诱导花色素和黄酮醇的合成。Anwar等[28]发现, 中国水仙(Narcissus tazetta L. var. chinensis Roem) R2R3-MYB转录因子NtMYB2在烟草中异位表达显著减少了转基因烟草花中的色素沉着并改变了花的表型。在本研究中, BjuB.KAN4过表达的PM和SY毛状根株系总黄酮含量均显著高于对照组, 说明该基因对芥菜型油菜类黄酮合成具有促进作用(图3和图6)。将BjuB.KAN4基因在拟南芥中的异源表达发现, 转基因植株中的原花青素含量较野生型下降了0.8倍且差异显著, 说明BjuB.KAN4基因异源表达拟南芥使PAs含量降低(图7), 因此推测该基因的表达使类黄酮合成途径转向其他代谢物合成支路, 如生成黄酮醇类化合物等。
类黄酮合成途径由结构基因和调节基因协同调控完成。Huang等[29]研究发现, 葡萄(V. vinifera) VvMYBC2-L1基因通过调节关键酶基因VvDFR、VvLDOX和VvLAR1的表达下调了葡萄的PAs合成; Xu等[30]发现, 胡萝卜(Daucus carota L.) DcMYB6基因通过上调花青素相关的结构基因DcCHS和DcDFR来提高拟南芥中花青素的积累, 表明DcMYB6可以调节紫色胡萝卜中花青素的生物合成。Wang等[31]发现, 苹果(Malus pumila Mill.) MYB转录因子MdMYBPA1通过调节酶基因MdLAR、MdANR、MdUFGT和MdANS的表达, 将类黄酮合成途径从PAs转变为花青素的产生来响应低温。本研究通过对转基因毛状根进行关键酶基因表达量检测发现, 类黄酮代谢途径的关键酶基因Bju.CHS和Bju.DFR等的表达量在PM和SY的转基因根系中均显著增加, 说明BjuB.KAN4基因有助于提高毛状根中类黄酮代谢途径部分关键酶基因的表达量, 从而促进类黄酮的生物合成(图4和图5)。本研究通过毛状根体系和异源表达拟南芥初步探索了MYB转录因子BjuB.KAN4在植物类黄酮生物合成途径中的功能, 为植物原花青素形成的分子调控机理的研究提供借鉴。另外, BjuB.KAN4基因在油菜毛状根中通过结合Bju.CHS和Bju.DFR等关键酶基因启动子, 驱动结构基因表达参与调节毛状根中类黄酮的合成。国内外研究表明, PAs在油菜种皮中的积累决定
种皮的颜色, 本研究中异源表达BjuB.KAN4基因能够使植株PAs含量降低, 推测其对油菜种皮也具有降低PAs的功能, 因此减少了PAs合成的底物的消耗, 为油脂的合成提供了更多的底物, 帮助了种子含油量的提高, 为油菜高含油量育种提供了理论依据。
4 结论
BjuB.KAN4基因表达具有组织特异性, 在根中表达量显著高于叶和茎中。该基因可能在根茎叶的维管组织中表达。转BjuB.KAN4基因的毛状根和拟南芥中总黄酮含量均显著高于对照, 且转基因拟南芥原花青素含量下降, 推测该转录因子抑制原花青素的生物合成途径, 同时可能对类黄酮代谢途径其他分支具有促进作用。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
URLPMID:16669768 [本文引用: 1]
DOI:10.1104/pp.19.00523URLPMID:31213511 [本文引用: 1]

The original domesticated carrots (Daucus carota) are thought to have been purple, accumulating large quantities of anthocyanins in their roots. A quantitative trait locus associated with anthocyanin pigmentation in purple carrot roots has been identified on chromosome 3 and includes two candidate genes, DcMYB6 and DcMYB7 Here, we characterized the functions of DcMYB6 and DcMYB7 in carrots. Overexpression of DcMYB7, but not DcMYB6, in the orange carrot 'Kurodagosun' led to anthocyanin accumulation in roots. Knockout of DcMYB7 in the solid purple (purple periderm, phloem, and xylem) carrot 'Deep Purple' using the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 system resulted in carrots with yellow roots. DcMYB7 could activate the expression of its DcbHLH3 partner, a homolog of the anthocyanin-related apple (Malus x domestica) bHLH3, and structural genes in the anthocyanin biosynthetic pathway. We determined that the promoter sequence of DcMYB7 in nonpurple carrots was interrupted either by DcMYB8, a nonfunctional tandem duplication of DcMYB7, or by two transposons, leading to the transcriptional inactivation of DcMYB7 in nonpurple carrot roots. As a result, nonpurple carrots fail to accumulate anthocyanins in their roots. Our study supports the hypothesis that another genetic factor suppresses DcMYB7 expression in the phloem and xylem of purple peridermal carrot root tissues. DcMYB7 also regulated the glycosylation and acylation of anthocyanins by directly activating DcUCGXT1 and DcSAT1 We reveal the genetic factors conditioning anthocyanin pigmentation in purple versus nonpurple carrot roots. Our results also provide insights into the mechanisms underlying anthocyanin glycosylation and acylation.
DOI:10.1016/j.tplants.2010.06.005URLPMID:20674465 [本文引用: 1]

The MYB family of proteins is large, functionally diverse and represented in all eukaryotes. Most MYB proteins function as transcription factors with varying numbers of MYB domain repeats conferring their ability to bind DNA. In plants, the MYB family has selectively expanded, particularly through the large family of R2R3-MYB. Members of this family function in a variety of plant-specific processes, as evidenced by their extensive functional characterization in Arabidopsis (Arabidopsis thaliana). MYB proteins are key factors in regulatory networks controlling development, metabolism and responses to biotic and abiotic stresses. The elucidation of MYB protein function and regulation that is possible in Arabidopsis will provide the foundation for predicting the contributions of MYB proteins to the biology of plants in general.
URLPMID:11549766 [本文引用: 1]
DOI:10.1016/j.gene.2019.03.013URLPMID:30858135 [本文引用: 1]

Plants with purple leave attain interest because of their biological importance. A new rice mutant, purple leaf (pl) was isolated from an indicia cultivar Zhenong 34, which was induced by ethyl methane sulfonate (EMS) mutagenesis. The genetic analyses substantiated that pl was corroborated by one recessive allele and confirmed by map based cloning using Insertion-Deletion (InDel) markers located on the long arm of chromosome 5. DNAseq data of the candidate part showed one bp insertion ('C' insertion) at +901bp position in the 3rd exon of OsPL gene. The pl was characterized as purple leaves, sheaths and leaf senescence phenotype at late grain filling stage of growth cycle. It possessed abnormal cell with distorted chloroplasts, less chlorophyll, and increased anthocyanin content in leaves. The anthocyanin biosynthesis genes, OsPAL, OsCHS, OsANS, and OsMYB55 showed up-regulation in pl plants compared to wild type (WT). High super oxide dismutase enzyme (SOD), catalase enzyme activity (CAT), total soluble sugar (TSS) and malondialdehyde activity (MDA) were detected in the pl; contrastingly, photosynthesis linked genes were down-regulated. The germinated pl seeds showed comparatively higher temperature stress tolerance than WT. The phytohormones abscicic acid (ABA), jasmonic acid (JA) and indole acetic acid (IAA) content were increased significantly in the pl plants. This research work will be provided information on better understanding of the molecular mechanism toward the anthocyanin biosynthetic pathway in rice. Therefore, OsPL gene could be a good genetic tool in marker aided backcrossing or gene editing for improving the rice cultivation in future.
[本文引用: 1]
DOI:10.1016/j.bbrc.2015.12.001URLPMID:26692488 [本文引用: 1]

Transcriptional activation of anthocyanin biosynthesis genes in vegetative tissues of monocotyledonous plants is mediated by cooperative activity of one component from each of the following two transcription factor families: MYB encoded by PURPLE PLANT1/COLORED ALEURONE1 (PL1/C1), and basic helix-loop-helix (bHLH) encoded by RED/BOOSTER (R1/B1). In the present study, putative PL cDNA was cloned from the wheat (Triticum aestivum) cultivar Iksan370, which preferentially expresses anthocyanins in coleoptiles. Phylogenetic tree analysis of deduced amino acid sequences showed that a putative TaPL1 is highly homologous to barley (Hordeum vulgare) HvPL1, but is distinct from wheat TaC1. Transgenic Arabidopsis thaliana stably expressing putative TaPL1 accumulated anthocyanin pigments in leaves and up-regulated structural genes involved in both early and late anthocyanin biosynthesis steps. TaPL1 transcript levels in Iksan370 were more prominent in vegetative tissues such as young coleoptiles than in reproductive tissues such as spikelets. TaPL1 expression was significantly up-regulated by environmental stresses including cold, salt, and light, which are known to induce anthocyanin accumulation. These combined results suggest that TaPL1 is an active positive regulator of anthocyanin biosynthesis in wheat coleoptiles.
URLPMID:24730512 [本文引用: 1]
DOI:10.1093/mp/ssp030URLPMID:19825656 [本文引用: 1]

Single-repeat R3 MYB transcription factors like CPC (CAPRICE) are known to play roles in developmental processes such as root hair differentiation and trichome initiation. However, none of the six Arabidopsis single-repeat R3 MYB members has been reported to regulate flavonoid biosynthesis. We show here that CPC is a negative regulator of anthocyanin biosynthesis. In the process of using CPC to test GAL4-dependent driver lines, we observed a repression of anthocyanin synthesis upon GAL4-mediated CPC overexpression. We demonstrated that this is not due to an increase in nutrient uptake because of more root hairs. Rather, CPC expression level tightly controls anthocyanin accumulation. Microarray analysis on the whole genome showed that, of 37 000 features tested, 85 genes are repressed greater than three-fold by CPC overexpression. Of these 85, seven are late anthocyanin biosynthesis genes. Also, anthocyanin synthesis genes were shown to be down-regulated in 35S::CPC overexpression plants. Transient expression results suggest that CPC competes with the R2R3-MYB transcription factor PAP1/2, which is an activator of anthocyanin biosynthesis genes. This report adds anthocyanin biosynthesis to the set of programs that are under CPC control, indicating that this regulator is not only for developmental programs (e.g. root hairs, trichomes), but can influence anthocyanin pigment synthesis.
DOI:10.1111/j.1365-313X.2008.03565.xURLPMID:18532977 [本文引用: 1]

SUMMARY: In Arabidopsis, MYB transcription factors regulate flavonoid biosynthesis via the formation of protein complexes with a basic helix-loop-helix (bHLH) transcription factor and a WD40 repeat protein. Several R3-type single-MYB proteins (R3-MYB), such as CPC and TRY, act as negative regulators of the development of epidermal cells. However, such regulators of flavonoid biosynthesis have not yet been reported, to our knowledge. We show here that an R3-MYB protein, AtMYBL2, acts as a transcriptional repressor and negatively regulates the biosynthesis of anthocyanin in Arabidopsis. In an AtMYBL2 knockout line (mybl2), the expression of the DFR and TT8 genes was enhanced and resulted in the ectopic accumulation of anthocyanin, while ectopic expression of AtMYBL2 or of a chimeric repressor that is a dominant negative form of AtMYBL2 suppressed the expression of DFR and TT8, and the biosynthesis of anthocyanin. The expression of AtMYBL2 was detected in various tissues but not in those in which anthocyanin accumulated or TT8 was expressed. The minimal repression domain of AtMYBL2 was found to be the six amino acids (TLLLFR) at the carboxyl terminus, and TLLLFR appears to be a novel repression motif that is different from the ERF-associated amphiphilic repression (EAR) motif. The defective phenotype of mybl2 mutants was complemented by 35S:AtMYBL2 but enhanced by a truncated form of AtMYBL2 from which the repression domain had been deleted. AtMYBL2 bound directly to TT8 protein and this complex suppressed the expression of DFR and TT8. The repression activity of AtMYBL2 appears to play a critical role in the regulation of anthocyanin biosynthesis.
DOI:10.1105/tpc.113.122069URLPMID:24642943 [本文引用: 1]

Plants require sophisticated regulatory mechanisms to ensure the degree of anthocyanin pigmentation is appropriate to myriad developmental and environmental signals. Central to this process are the activity of MYB-bHLH-WD repeat (MBW) complexes that regulate the transcription of anthocyanin genes. In this study, the gene regulatory network that regulates anthocyanin synthesis in petunia (Petunia hybrida) has been characterized. Genetic and molecular evidence show that the R2R3-MYB, MYB27, is an anthocyanin repressor that functions as part of the MBW complex and represses transcription through its C-terminal EAR motif. MYB27 targets both the anthocyanin pathway genes and basic-helix-loop-helix (bHLH) ANTHOCYANIN1 (AN1), itself an essential component of the MBW activation complex for pigmentation. Other features of the regulatory network identified include inhibition of AN1 activity by the competitive R3-MYB repressor MYBx and the activation of AN1, MYB27, and MYBx by the MBW activation complex, providing for both reinforcement and feedback regulation. We also demonstrate the intercellular movement of the WDR protein (AN11) and R3-repressor (MYBx), which may facilitate anthocyanin pigment pattern formation. The fundamental features of this regulatory network in the Asterid model of petunia are similar to those in the Rosid model of Arabidopsis thaliana and are thus likely to be widespread in the Eudicots.
URLPMID:28575507 [本文引用: 1]
DOI:10.1111/pbi.2010.8.issue-9URL [本文引用: 1]
DOI:10.1111/j.1365-313X.2006.02717.xURLPMID:16623911 [本文引用: 1]

The Arabidopsis aberrant testa shape (ats) mutant produces a single integument instead of the two integuments seen in wild-type ovules. Cellular anatomy and patterns of marker gene expression indicate that the single integument results from congenital fusion of the two integuments of the wild type. Isolation of the ATS locus showed it to encode a member of the KANADI (KAN) family of putative transcription factors, previously referred to as KAN4. ATS was expressed at the border between the two integuments at the time of their initiation, with expression later confined to the abaxial layer of the inner integument. In an inner no outer (ino) mutant background, where an outer integument does not form, the ats mutation led to amorphous inner integument growth. The kan1kan2 double mutant exhibits a similar amorphous growth of the outer integument without affecting inner integument growth. We hypothesize that ATS and KAN1/KAN2 play similar roles in the specification of polarity in the inner and outer integuments, respectively, that parallel the known roles of KAN proteins in promoting abaxial identity during leaf development. INO and other members of the YABBY gene family have been hypothesized to have similar parallel roles in outer integument and leaf development. Together, these two hypotheses lead us to propose a model for normal integument growth that also explains the described mutant phenotypes.
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
四川农业大学硕士学位论文,
[本文引用: 1]
MS Thesis of Sichuan Agricultural University,
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1046/j.1365-313x.1998.00343.xURLPMID:10069079 [本文引用: 1]

The Agrobacterium vacuum infiltration method has made it possible to transform Arabidopsis thaliana without plant tissue culture or regeneration. In the present study, this method was evaluated and a substantially modified transformation method was developed. The labor-intensive vacuum infiltration process was eliminated in favor of simple dipping of developing floral tissues into a solution containing Agrobacterium tumefaciens, 5% sucrose and 500 microliters per litre of surfactant Silwet L-77. Sucrose and surfactant were critical to the success of the floral dip method. Plants inoculated when numerous immature floral buds and few siliques were present produced transformed progeny at the highest rate. Plant tissue culture media, the hormone benzylamino purine and pH adjustment were unnecessary, and Agrobacterium could be applied to plants at a range of cell densities. Repeated application of Agrobacterium improved transformation rates and overall yield of transformants approximately twofold. Covering plants for 1 day to retain humidity after inoculation also raised transformation rates twofold. Multiple ecotypes were transformable by this method. The modified method should facilitate high-throughput transformation of Arabidopsis for efforts such as T-DNA gene tagging, positional cloning, or attempts at targeted gene replacement.
[本文引用: 1]
[本文引用: 1]
湖南大学博士学位论文,
[本文引用: 1]
PhD Dissertation of Hunan University,
[本文引用: 1]
[本文引用: 1]
DOI:10.1016/j.plaphy.2014.02.019URLPMID:24675528 [本文引用: 1]

The vascular system--xylem, phloem and the cambium--is essential for water supply, nutrient transport, and physical support in higher plants. Although it is known that vascular-specific gene expression is regulated by cis-acting regulatory sequences in promoters, it is largely unknown how many regulatory elements exist and what their roles are in promoters. To understand the regulatory elements of vascular-specific promoters and their roles in vascular development, a T-DNA insertion mutant showing delayed growth and diminished resistance to environmental stress was isolated using promoter trap strategy. The novel gene, Arabidopsis thaliana heat shock protein-related (Athspr), was cloned from Arabidopsis ecotype C24. Strong GUS (beta-glucuronidase) staining in the original promoter trap line was found in the vascular tissues of all organs in the mutant. The Athspr promoter was cloned and fused with GUS and eGFP (enhanced green fluorescent protein) reporter genes to verify its vascular-specific expression in Arabidopsis. Further histochemical analysis in transgenic plants demonstrated a similar GUS expression pattern in the vascular tissues. In addition, ATHSPR-eGFP driven by Athspr promoter was observed in vascular bundles of the transgenic seedling roots. Finally, comparative analysis with promoter motifs from 37 genes involved in vascular development revealed that Athspr and all other promoters active in vascular tissues contained regulatory elements responding to phytohormones, light, biotic and abiotic stresses, as well as those regulating tissue-specific expression. These results demonstrated that the Athspr promoter has a vascular tissue-specific activity and Athspr may have multiple functions in vascular development and resistance against various stresses.
DOI:10.1111/tpj.14081URLPMID:30176084 [本文引用: 1]

The phenylpropanoid pathway leads to the production of many important plant secondary metabolites including lignin, chlorogenic acids, flavonoids, and phenolic glycosides. Early studies have demonstrated that flavonoid biosynthesis is transcriptionally regulated, often by a MYB, bHLH, and WDR transcription factor complex. In poplar, several R2R3 MYB transcription factors are known to be involved in flavonoid biosynthesis. Previous work determined that poplar MYB134 and MYB115 are major activators of the proanthocyanidin pathway, and also induce the expression of repressor-like MYB transcription factors. Here we characterize two new repressor MYBs, poplar MYB165 and MYB194, paralogs which comprise a subgroup of R2R3-MYBs distinct from previously reported poplar repressors. Both MYB165 and MYB194 repressed the activation of flavonoid promoters by MYB134 in transient activation assays, and both interacted with a co-expressed bHLH transcription factor, bHLH131, in yeast two-hybrid assays. Overexpression of MYB165 and MYB194 in hybrid poplar resulted in greatly reduced accumulation of several phenylpropanoids including anthocyanins, proanthocyanidins, phenolic glycosides, and hydroxycinnamic acid esters. Transcriptome analysis of MYB165- and MYB194-overexpressing poplars confirmed repression of many phenylpropanoid enzyme genes. In addition, other MYB genes as well as several shikimate pathway enzyme genes were downregulated by MYB165-overexpression. By contrast, leaf aromatic amino acid concentrations were greater in MYB165-overexpressing poplars. Our findings indicate that MYB165 is a major repressor of the flavonoid and phenylpropanoid pathway in poplar, and may also affect the shikimate pathway. The coordinated action of repressor and activator MYBs could be important for the fine tuning of proanthocyanidin biosynthesis during development or following stress.
DOI:10.1093/jxb/erv524URLPMID:26687179 [本文引用: 1]

Flavonoid compounds play important roles in the modern diet, and pear fruits are an excellent dietary source of these metabolites. However, information on the regulatory network of flavonoid biosynthesis in pear fruits is rare. In this work, 18 putative flavonoid-related MYB transcription factors (TFs) were screened by phylogenetic analysis and four of them were correlated with flavonoid biosynthesis patterns in pear fruits. Among these MYB-like genes, the specific functions of two novel MYB TFs, designated as PbMYB10b and PbMYB9, were further verified by both overexpression and RNAi transient assays. PbMYB10b, a PAP-type MYB TF with atypical motifs in its conserved region, regulated the anthocyanin and proanthocyanidin pathways by inducing the expression of PbDFR, but its function could be complemented by other MYB TFs. PbMYB9, a TT2-type MYB, not only acted as the specific activator of the proanthocyanidin pathway by activating the PbANR promoter, but also induced the synthesis of anthocyanins and flavonols by binding the PbUFGT1 promoter in pear fruits. The MYBCORE-like element has been identified in both the PbUFGT1 promoter and ANR promoters in most species, but it was not found in UFGT promoters isolated from other species. This finding was also supported by a yeast one-hybrid assay and thus enhanced the likelihood of the interaction between PbMYB9 and the PbUFGT1 promoter.
[本文引用: 1]
DOI:10.1111/nph.12557URLPMID:24147899 [本文引用: 1]

Flavonoids are secondary metabolites with multiple functions. In grape (Vitis vinifera), the most abundant flavonoids are proanthocyanidins (PAs), major quality determinants for fruit and wine. However, knowledge about the regulation of PA composition is sparse. Thus, we aimed to identify novel genomic regions involved in this mechanism. Expression quantitative trait locus (eQTL) mapping was performed on the transcript abundance of five downstream PA synthesis genes (dihydroflavonol reductase (VvDFR), leucoanthocyanidin dioxygenase (VvLDOX), leucoanthocyanidin reductase (VvLAR1), VvLAR2 and anthocyanidin reductase (VvANR)) measured by real-time quantitative PCR on a pseudo F1 population in two growing seasons. Twenty-one eQTLs were identified; 17 of them did not overlap with known candidate transcription factors or cis-regulatory sequences. These novel loci and the presence of digenic epistasis support the previous hypothesis of a polygenic regulatory mechanism for PA biosynthesis. In a genomic region co-locating eQTLs for VvDFR, VvLDOX and VvLAR1, gene annotation and a transcriptomic survey suggested that VvMYBC2-L1, a gene coding for an R2R3-MYB protein, is involved in regulating PA synthesis. Phylogenetic analysis showed its high similarity to characterized negative MYB factors. Its spatiotemporal expression profile in grape coincided with PA synthesis. Its functional characterization via overexpression in grapevine hairy roots demonstrated its ability to reduce the amount of PA and to down-regulate expression of PA genes.
DOI:10.1038/srep45324URLPMID:28345675 [本文引用: 1]

Carrots are widely grown and enjoyed around the world. Purple carrots accumulate rich anthocyanins in the taproots, while orange, yellow, and red carrots accumulate rich carotenoids in the taproots. Our previous studies indicated that variation in the activity of regulatory genes may be responsible for variations in anthocyanin production among various carrot cultivars. In this study, an R2R3-type MYB gene, designated as DcMYB6, was isolated from a purple carrot cultivar. In a phylogenetic analysis, DcMYB6 was grouped into an anthocyanin biosynthesis-related MYB clade. Sequence analyses revealed that DcMYB6 contained the conserved bHLH-interaction motif and two atypical motifs of anthocyanin regulators. The expression pattern of DcMYB6 was correlated with anthocyanin production. DcMYB6 transcripts were detected at high levels in three purple carrot cultivars but at much lower levels in six non-purple carrot cultivars. Overexpression of DcMYB6 in Arabidopsis led to enhanced anthocyanin accumulation in both vegetative and reproductive tissues and upregulated transcript levels of all seven tested anthocyanin-related structural genes. Together, these results show that DcMYB6 is involved in regulating anthocyanin biosynthesis in purple carrots. Our results provide new insights into the regulation of anthocyanin synthesis in purple carrot cultivars.
URLPMID:29978604 [本文引用: 1]
