 ,, 刘淑娴, 杨宗桃, 王彤, 程光远, 商贺阳, 徐景升
,, 刘淑娴, 杨宗桃, 王彤, 程光远, 商贺阳, 徐景升 ,*福建农林大学国家甘蔗工程技术研究中心 / 农业农村部福建甘蔗生物学与遗传育种重点实验室 / 教育部作物遗传育种与综合利用重点实验室, 福建福州 350002
,*福建农林大学国家甘蔗工程技术研究中心 / 农业农村部福建甘蔗生物学与遗传育种重点实验室 / 教育部作物遗传育种与综合利用重点实验室, 福建福州 350002Sugarcane PsbS subunit response to Sugarcane mosaic virus infection and its interaction with 6K2 protein
ZHANG Hai ,, LIU Shu-Xian, YANG Zong-Tao, WANG Tong, CHENG Guang-Yuan, SHANG He-Yang, XU Jing-Sheng
,, LIU Shu-Xian, YANG Zong-Tao, WANG Tong, CHENG Guang-Yuan, SHANG He-Yang, XU Jing-Sheng ,*Sugarcane Research & Development Center, China Agricultural Technology System, Fujian Agriculture and Forestry University / Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture and Rural Affairs / Key Laboratory of Ministry of Education for Genetics, Breeding and Multiple Utilization of Crops, Fuzhou 350002, Fujian, China
,*Sugarcane Research & Development Center, China Agricultural Technology System, Fujian Agriculture and Forestry University / Key Laboratory of Sugarcane Biology and Genetic Breeding, Ministry of Agriculture and Rural Affairs / Key Laboratory of Ministry of Education for Genetics, Breeding and Multiple Utilization of Crops, Fuzhou 350002, Fujian, China通讯作者:
收稿日期:2020-02-10接受日期:2020-04-15网络出版日期:2020-11-12
| 基金资助: |
Received:2020-02-10Accepted:2020-04-15Online:2020-11-12
| Fund supported: |
作者简介 About authors
E-mail:

摘要
关键词:
Abstract
Keywords:
PDF (10421KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
张海, 刘淑娴, 杨宗桃, 王彤, 程光远, 商贺阳, 徐景升. 甘蔗PsbS亚基应答甘蔗花叶病毒侵染及其与6K2蛋白的互作研究[J]. 作物学报, 2020, 46(11): 1722-1733. doi:10.3724/SP.J.1006.2020.04030
ZHANG Hai, LIU Shu-Xian, YANG Zong-Tao, WANG Tong, CHENG Guang-Yuan, SHANG He-Yang, XU Jing-Sheng.
甘蔗(Saccharum spp. hybrid)是我国重要的糖料作物, 我国食糖约90%来自甘蔗[1,2]。同时, 甘蔗还是重要生物质能源作物[3,4,5]。甘蔗花叶病是发生最普遍、危害最严重的一类病毒病性病害, 严重影响甘蔗产量和品质[6,7,8]。甘蔗花叶病的病原主要有甘蔗花叶病毒(Sugarcane mosaic virus, SCMV)、高粱花叶病毒(Sorghum mosaic virus, SrMV)和甘蔗条纹花叶病毒(Sugarcane streak mosaic virus, SCSMV) 3种[9,10], 均为马铃薯Y病毒科(Potyviridae)病毒[7,11-19]。SCMV和SrMV属于马铃薯Y病毒属(Potyvirus)[20,21], SCSMV是该科新属, 即禾病毒属(Poacevirus)[22,23,24]。近年来, 我国蔗区SrMV和SCSMV的检出率呈上升趋势[23,25], 但是SCMV依然是主要病原, 且在全世界普遍发生, 威胁甘蔗产业[14,26-28]。
SCMV、SrMV和SCSMV的基因组由约为10,000 nt的单链正义RNA组成, 编码2个多聚蛋白, 最终水解成11个成熟蛋白, 分别为P1、HC-Pro、P3、P3N-PIPO、6K1、CI、6K2、VPg、NIa-Pro、NIb和CP[29,30,31,32,33,34,35]。其中, 6K2蛋白在马铃薯Y病毒科病毒侵染中具有重要作用, 参与病毒的复制和胞间移动[32,35-38]。该蛋白以衣被蛋白I (coatomer protein complex I, COPI)和COPII依赖方式形成[39,40,41]诱导内质网(endoplasmic reticulum, ER)形成复制囊泡[41,42,43], 将病毒RNA、蛋白以及病毒复制所需的一些宿主蛋白包裹其中[44,45,46]。这些囊泡在细胞内沿着微丝移动[41], 并通过6K2与CI互作到达胞间连丝(plasmodesmata, PD), 然后通过PD进入未感染的相邻细胞[47], 或通过早期分泌途径沿着微丝达到叶绿体, 与叶绿体外膜融合, 进行病毒基因组高效复制[48]。病毒侵染造成叶绿素减少[49,50,51], 光合电子传递速率和同化效率降低[52,53], 甚至破坏叶绿体结构[49,54], 严重影响植物的光合作用[27,49,55]。病毒侵染也影响植物的非光化学猝灭(non-photochemical quenching, NPQ) [49,51,55], 但其潜在的分子机制尚不清楚。NPQ是植物在长期进化过程中发展的一种光保护机制, 植物藉此将过剩光能转化为热能耗散, 以减少过强光照的损伤[56,57,58,59,60,61,62,63]。光合系统II (Photosystem II, PSII)的PsbS亚基在NPQ起关键作用, 在缺失该基因的拟南芥突变体中, 光捕获和光合作用未受影响, 但是NPQ严重受抑[62,64]。进一步研究表明, 过量光照导致PsbS质子化, 进而诱导一系列生理生化反应如叶黄素循环、LHCII的磷酸化以及LHCII在PSI和PSII间的迁移, 调节激发能在2个光合系统上的合理分配, 以保护光合膜免遭受强光危害[63,65-69]; 同时, PsbS还可以通过与LHCII三聚体蛋白直接互作来增强过剩光能的猝灭从而保护光合膜[70]。
在前期研究工作中, 我们以SCMV编码的6K2蛋白为诱饵, 利用酵母双杂交(yeast two hybrid, Y2H)技术从甘蔗ROC22叶片的 cDNA 酵母文库中筛选并克隆了甘蔗的PsbS编码基因, 命名为ScPsbS (GenBank登录号为MN167889), 并利用Y2H验证了ScPsbS与SCMV-6K2的互作[71]。ScPsbS的开放读码框(open reading frame, ORF)长度为798 bp, 编码长度为 265 aa的蛋白。在本研究中, 我们对ScPsbS进行了深入的生物信息学分析, 利用定量PCR技术分析了ScPsbS组织特异性与应答SCMV侵染的表达模式, 利用双分子荧光互补(bimolecular fluorescent complimentary, BiFC)技术进一步验证了ScPsbS与SCMV-6K2 的互作关系, 并对其在SCMV侵染中的作用做了探讨。本研究为阐明病毒影响植物NPQ的分子机制及甘蔗抗花叶病育种提供了基础数据和实验依据。
1 材料与方法
1.1 材料及处理方法
SCMV-FZ1病毒株系[72]、甘蔗品种Badila和本氏烟(Nicotiana benthamiana)由福建农林大学农业农村部福建甘蔗生物学与遗传育种重点实验室提供。本氏烟用于亚细胞定位和BiFC实验, 在光周期为16 h光照/8 h暗, 光照强度为130 μmol m-2 s-1, 温度为22℃和空气湿度为60%的条件下培养。参照翟玉山等[73]的方法, 研究目的基因ScPsbS应答SCMV侵染的表达模式。采用腋芽快繁技术培养Badila组培苗, 培养条件为光强200 μmol m-2 s-1, 光周期16 h光照/8 h暗, 温度28℃, 空气湿度60%。待组培苗长至15~25 cm、出现4~5片完全展开的叶片时, 摩擦接种SCMV, 设置3个重复, 每个重复3株, 在光培养期间的第1小时后接种, 使用磷酸缓冲液(pH 7.0)摩擦接种对照植株, 取接种部位, 使用SCMV-CP基因特异引物(表1)检测接种是否成功。分别在接种后0 h、4 h、8 h、12 h、1 d、3 d、5 d、7 d取样。从福建农林大学隔离网室中选取长势一致的伸长期甘蔗品种Badila健康植株, 于早上9点取未成熟叶心叶、成熟叶片正一叶(甘蔗植株由上到下第一个有可见肥厚带的叶片)、初衰叶片正七叶、未成熟节间第三节间、完成形态建成的第八节间和根, 用于基因的组织特异性表达实验, 设3个生物学重复, 每个生物学重复设置3株。取样后用液氮速冻, 置-80℃冰箱保存备用。Table 1
表1
表1本研究使用的引物
Table 1
| 引物名称 Primer name | 引物序列 Primer sequence (5′-3′) | 策略 Strategy |
|---|---|---|
| 221-ScPsbS-F | GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCGCAGTCCATGCT | 双分子荧光互补载体构建 |
| 221-ScPsbS-R | GGGGACCACTTTGTACAAGAAAGCTGGGTCCTACTCTTCGTCCTCGC | Vector generation for BiFC |
| ScPsbS-qF | CTCCATCATCGGCGAGATCATCAC | 定量PCR |
| ScPsbS-qR | GGCGGCGACGAAGAAGAAGAG | Real-time-qPCR |
| GAPDH-F | CACGGCCACTGGAAGCA | 内参基因 |
| GAPDH-R eEF-1α-F eEF-1α-R | TCCTCAG GGTTCCTGATGCC TTTCACACTTGGAGTGAAGCAGAT GACTTCCTTCACAATCTCATCATAA | Reference gene 内参基因 Reference gene |
| SCMV-CP-F | TACAGAGAGACACACAGCTG | SCMV检测 |
| SCMV-CP-R | ACGCTACACCAGAAGACACT | Detection of SCMV |
新窗口打开|下载CSV
1.2 ScPsbS蛋白的生物信息学分析
利用ProtParam (1.3 RNA提取和cDNA合成
将采集的样品在液氮中研磨成粉末, 采用TRIzol (Invotrigen, USA)试剂按照其说明书提取样品总RNA, 使用1.0%琼脂糖凝胶电泳检测RNA质量。利用Evo-MLV反转录试剂盒(艾科瑞, 中国), 参照其说明书, 将RNA反转录成cDNA。1.4 载体构建
利用Gateway技术, 将ScPsbS基因构建到pEarleygate-202-YC和pEarleygate-201-YN载体中, 获得ScPsbS-YC和YN-ScPsbS载体。用ScPsbS目的片段, 以黄色荧光蛋白(yellow fluorescence protein, YFP)为标记, 构建ScPsbS蛋白的亚细胞定位载体ScPsbS-YFP。所有构建的载体都经过上海生工生物工程有限公司测序验证。亚细胞定位载体SCMV-6K2-CFP以及用于BiFC实验的SCMV-6K2-YN和SCMV-6K2-YC均来自本课题组前期研究工作[71]。1.5 ScPsbS的亚细胞定位
参照Cheng等[32]的方法, 将含有植物表达载体ScPsbS-YFP的农杆菌GV3101以及等比例混合的含有植物表达载体SCMV-6K2-CFP和ScPsbS-YFP的农杆菌GV3101分别注射到健康的本氏烟叶片, 48 h后在激光共聚焦显微镜(Leica TCS SP5II)下观察本氏烟叶片表皮细胞中SCMV-6K2蛋白的定位以及ScPsbS蛋白和SCMV-6K2蛋白的共定位。CFP的激发波长为442 nm, 采集波长为450~500 nm; GFP的激发波长为514 nm, 采集波长为530~590 nm; 叶绿素自荧光的激发光波长为552 nm, 采集波长为 650~680 nm。图像采用数字采集, 使用LSM 2.6.3软件处理。1.6 BiFC验证ScPsbS与SCMV-6K2的互作
参照朱海龙等[74]的方法将含有ScPsbS-YC和SCMV-6K2-YN, ScPsbS-YN和SCMV-6K2-YC的农杆菌GV3101等比例混合, 分别注射入健康的本氏烟叶片中。在正常条件下培养2~3 d后在激光共聚焦显微镜(Leica TCS SP5II)下观察照相。YFP的激发光波长为514 nm, 捕获波长为530~590 nm?图像采用数字采集, 并用LSM 2.6.3软件处理。1.7 ScPsbS的RT-qPCR表达分析
根据ScPsbS基因的ORF序列, 设计特异性实时荧光定量PCR引物, 以供试材料的cDNA为模板, 以GAPDH基因和eEF-1α基因(表1)为内参基因[75], 利用SYBR Green PCR Master Mix Kit (Roche, 日本)试剂盒, 参照其说明书配制反应体系。在7500型实时荧光定量PCR仪(ABI, USA)上完成PCR扩增, 反应程序为50℃ 2 min; 95℃ 10 min; 95℃ 15 s; 60℃ 1 min, 40个循环。每个样品设置3次重复, 以DEPC处理的无菌水作为空白对照, 采用2-ΔΔCt算法进行基因表达水平分析, 使用统计软件SPSS 12.0对基因表达差异进行显著性分析。2 结果与分析
2.1 ScPsbS蛋白的生物信息学分析
ProtParam分析表明, ScPsbS蛋白的分子量为27.81 kD, 等电点为9.07; 不稳定系数为28.47, 为稳定蛋白; 脂溶指数为100.53, 总平均亲水性是0.238, 可能是疏水性蛋白。GOR4预测表明, ScPsbS中无规则卷曲所占的比例最高, 为47.17%; 其次是α螺旋, 占42.64%; 延伸链所占比例是10.19%; 无β–折叠结构。SignalP 5.0分析表明, ScPsbS蛋白不含信号肽, 是非分泌蛋白。跨膜结构域预测结果表明, ScPsbS具有4个跨膜结构域。将ScPsbS蛋白序列提交SWISS-MODEL, 以蛋白质数据库中的菠菜PsbS (SMTL ID: 4ri2.1)A链晶体分子[76]为模板, 对ScPsbS、玉米(Zea mays)的ZmPsbS (NP_0011 05228.2)、水稻(Oryza sativa)的OsPsbS (XP_ 015621169.1)和拟南芥(Arabidopsis thaliana)的AtPsbS (NP_973971.1)进行同源建模结构预测。由于PsbS亚基是以二聚体执行其生物学功能的[76], 因此结构预测以二聚体的形式体现。预测结果表明, ScPsbS、玉米ZmPsbS、水稻OsPsbS、拟南芥AtPsbS与模板菠菜PsbS蛋白的氨基酸序列一致性分别为83.89%、84.36%、85.31%和88.21%,空间结构类似(图1)。图1
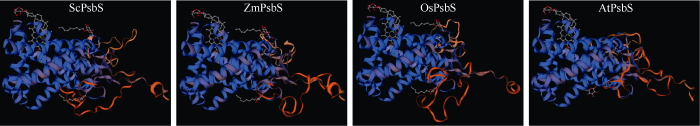 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1基于SWISS-MODEL的ScPsbS、ZmPsbS、OsPsbS和AtPsbS蛋白三维建模
ZmPsbS (NP_001105228.2): 玉米的PsbS亚基; OsPsbS (XP_ 015621169.1): 水稻的PsbS亚基; AtPsbS (NP_973971.1): 拟南芥的PsbS亚基。
Fig. 1Three-dimensional structure of ScPsbS, ZmPsbS, OsPsbS, and AtPsbS protein based on SWISS-MODEL
ZmPsbS (NP_001105228.2) is from Zea mays; OsPsbS (XP_ 015621169.1) is from Oryza sativa; AtPsbS (NP_973971.1) is from Arabidopsis thaliana.
保守结构域分析表明, ScPsbS编码的蛋白包含2个PsbS保守结构域, 分别在88~132、192~249位(图2), 确定ScPsbS属于PsbS基因家族。PsbS蛋白由核基因编码, 其N端具有转运肽, 靶向叶绿体内囊体腔, 进入叶绿体后在保守结构域AXA (X为任意氨基酸)后切割[77,78]。使用ChloroP对ScPsbS进行转运肽预测分析表明, ScPsbS蛋白N端的66 aa片段为转运肽, 其64~66位氨基酸为AAA (图2)。
图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2ScPsbS基因的核苷酸序列及其编码的氨基酸序列
*: 终止密码子。黑色下画线部分为转运肽; 红色下画线部分为PsbS结构域; 红色标记的氨基酸为靶向叶绿体前体蛋白切割位点的保守结构域。↓: 切割位点。
Fig. 2Nucleotide sequence of the ScPsbS gene and its deduced amino acid sequence
*: stop codon. The black underlined sequence indicates the transit peptide. The red underlined part indicates the PsbS domains. The red font marked amino acids indicate the conserved motif between transit peptide and mature protein in the precursor protein targeting to thylakoid lumen. ↓ indicates the digestion site.
2.2 ScPsbS蛋白的氨基酸序列同源性分析和系统进化树分析
通过NCBI 网站的Blastp和Phytozome网站的Proteome Blastp搜索不同物种的PsbS同源序列, 结果显示, ScPsbS蛋白与高粱(Sorghum bicolor, XP_ 002456704.1)、玉米、谷子(Setaria italic, XP_004970 722.1)、黍(Panicum hallii, XP_025818476.1)、二穗短柄草(Brachypodium distachyon, XP_003564 708.1)、小麦(Triticum aestivum, CDM85166.1)和水稻的PsbS相似性分别为98%、95%、84%、89%、81%、80%和79%。其在PsbS结构域的氨基酸序列高度保守, 而在N端的转运肽由于物种的不同而表现出差异, 且C3和C4植物有明显的差异(图3)。图3
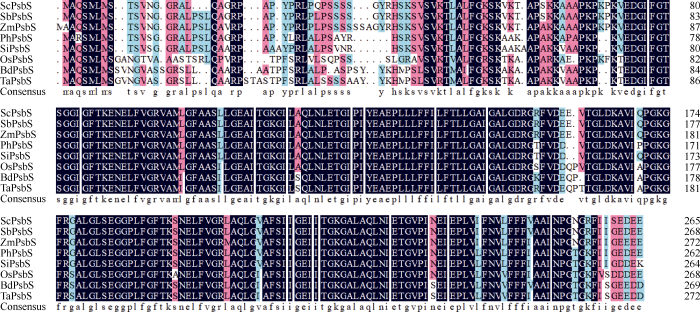 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3甘蔗ScPsbS与其他单子叶植物PsbS蛋白的氨基酸序列比对
高粱: SbPsbS (XP_002456704.1); 玉米: ZmPsbS (NP_ 001105228.2); 黍: PhPsbS (XP_025818476.1); 谷子: SiPsbS (XP_ 004970722.1); 水稻: OsPsbS (XP_015621169.1); 二穗短柄草: BdPsbS (XP_003564708.1); 小麦: TaPsbS (CDM85166.1)。
Fig. 3Amino acid sequence alignment of ScPsbS and PsbSs of other monocotyledon species
Sorghum bicolor: SbPsbS (XP_002456704.1); Zea mays: ZmPsbS (NP_001105228.2); Panicum hallii: PhPsbS (XP_025818476.1); Setaria italic: SiPsbS (XP_004970722.1); Oryza sativa: OsPsbS (XP_015621169.1); Brachypodium distachyon: BdPsbS (XP_ 003564708.1); Triticum aestivum: TaPsbS (CDM85166.1).
以小立碗藓(Physcomitrella patens)的PsbS蛋白序列作为外源序列, 使用ClustalX和MEGA 7.0的ML (maximum likelihood, LG+G)法构建系统进化树, 分析ScPsbS蛋白与其他物种PsbS蛋白的进化关系。结果表明, 双子叶植物番茄(Solanum lycopersicum)、烟草(Nicotiana tabacum)、拟南芥(Arabidopsis thaliana)、葡萄(Vitis vinifera)、三叶杨(Populus trichocarpa)、大豆(Glycine max)、苜蓿(Medicago sativa)形成群I; 单子叶植物甘蔗、高粱、玉米、谷子、狗尾草(Setaria viridis)、黍、柳枝稷(Panicum virgatum)、水稻、二穗短柄草、短柄草(Brachypodium sylvaticum)、小麦、大麦(Hordeum vulgare)形成群II, 同时单子叶中的C3植物和C4植物分别形成亚群II-1和II-2 (图4)。这表明, 在遗传进化上PsbS蛋白在单子叶植物和双子叶植物之间, 以及在单子叶中的C3植物和C4植物之间存在明显分化。
图4
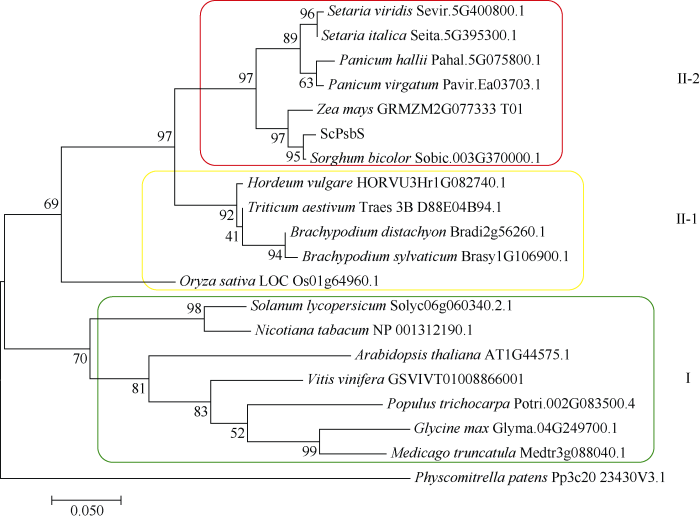 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4ScPsbS与其他物种PsbS蛋白的系统进化树分析
Fig. 4Phylogenetic tree analysis of ScPsbS protein and PsbS proteins from other plant species
2.3 ScPsbS的亚细胞定位
亚细胞定位试验表明, ScPsbS-YFP的黄色荧光信号与叶绿体的荧光信号重合(图5-A), 表明ScPsbS定位于叶绿体, 与拟南芥PsbS的叶绿体定位结果一致[64,79]。Y2H表明ScPsbS与SCMV-6K2互作[65], 因此本研究探讨了ScPsbS和SCMV-6K2的共同定位情况。结果表明, ScPsbS-YFP的黄色荧光信号与6K2-CFP的青色荧光蛋白信号重合, 部分共定位信号与叶绿体荧光信号重合, 且这些叶绿体均分布在细胞膜周围(图5-B)。图5
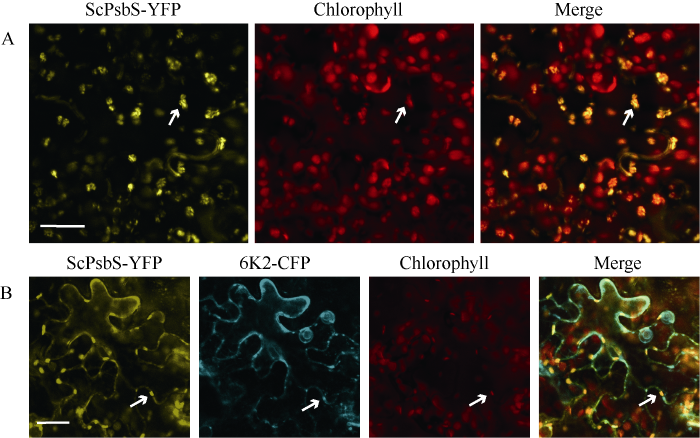 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5ScPsbS-YFP在本氏烟表皮细胞中的定位
A: ScPsbS-YFP亚细胞定位; B: ScPsbS-YFP和6K2-CFP亚细胞共定位。白色箭头表示叶绿体, 标尺= 25 μm。
Fig. 5Subcellular localization of ScPsbS fused with YFP in the epidermal cells of N. benthamiana
A: subcellular localization of ScPsbS-YFP; B: subcellular colocalization of ScPsbS-YFP with 6K2-CFP. White arrows indicate the chloroplasts; bar = 25 μm.
2.4 ScPsbS与SCMV-6K2的互作验证
BiFC试验结果表明, 共注射的YN-6K2和ScPsbS-YC, YN-ScPsbS和6K2-YC组合都分别产生黄色荧光信号(图6), 说明ScPsbS与SCMV-6K2互作, 与我们前期Y2H试验结果一致[71], 进一步证明了ScPsbS与SCMV-6K2互作。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6BiFC检测ScPsbS与SCMV-6K2的互作
A: YC融合于ScPsbS的C末端, YN融合于SCMV-6K2的N末端; B: YN融合于ScPsbS的N末端, YC融合于SCMV-6K2的C末端。将YN-6K2和ScPsbS-YC(A), YN-ScPsbS和6K2-YC(B)分别共注射到本氏烟叶片中进行瞬时表达, 48 h后激光共聚焦观察。标尺= 25 μm。
Fig. 6BiFC assays for protein-protein interaction between ScPsbS and SCMV-6K2
A: the C-terminal half of YFP was fused to the C-terminal of ScPsbS to generate ScPsbS-YC, while the N-terminal half of YFP was fused to the N-terminla of SCMV-6K2 to generate YN-6K2; B: the N-terminal half of YFP was fused to the N-terminal of ScPsbS to generate YN- ScPsbS, while the C-terminal half of YFP was fused to the C-terminal of SCMV-6K2 to generate 6K2-YC. Plasmids combination of YN-6K2 plus ScPsbS-YC (A), YN-ScPsbS plus SCMV-6K2-YC (B) were individually co-injected into N. benthamiana leaves for transient expression. The fluorescent signal was monitored by confocal microscopy at 48 h after infiltration; bar = 25 μm.
2.5 ScPsbS基因的组织特异性表达及应答SCMV侵染的表达模式
以健康Badila植株为材料, 研究ScPsbS基因在不同组织表达的特异性, 荧光定量PCR结果表明, ScPsbS基因在不同组织中的表达具有显著差异(图7): 在正一叶中相对表达量最高, 正七叶和心叶次之, 茎和根中表达量极低。正一叶是完全展开的光合作用最旺盛的叶片, 正七叶为初衰叶, 心叶基本是分生组织, 茎和根中则基本无光合作用, 因此ScPsbS的表达与其所在组织的光合能力呈正相关。图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7ScPsbS基因在甘蔗不同组织中的表达模式
误差线为每组处理的标准误差(n = 3)。LR: 心叶; +1 L: 正一叶; +7 L: 正七叶; +3 I: 第三节间; +8 I: 第八节间; R: 根。
Fig. 7Expression profile of ScPsbS in different sugarcane tissues
The error bars represent the standard error of each treatment group (n = 3). LR: leaf roll; +1 L: +1 leaf; +7 L: +7 leaf; +3 I: +3 internode; +8 I: +8 Internode; R: root.
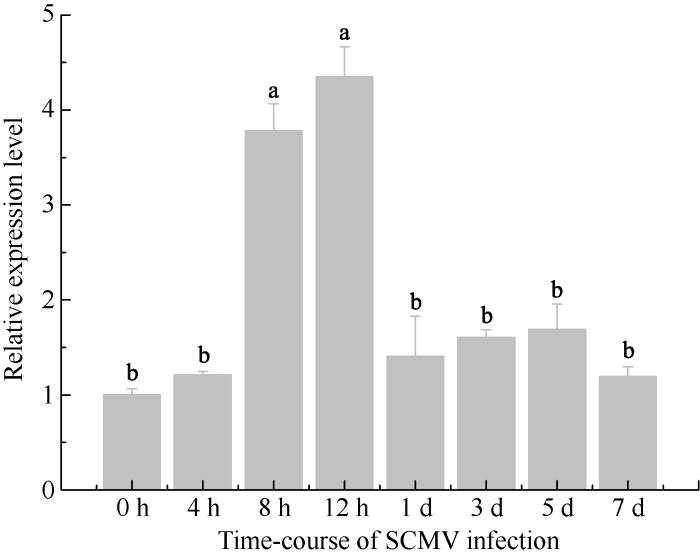
以SCMV-FZ1接种的Badila叶片为材料[73], 研究SCMV侵染对ScPsbS基因表达的影响。使用SCMV-CP基因特异引物(表1)检测SCMV接种甘蔗, 扩增出目的片段, 表明接种成功。荧光定量PCR结果表明, SCMV侵染对ScPsbS基因表达影响显著, 与对照相比, ScPsbS基因在侵染早期显著上调, 然后下调至略高于对照的水平, 但是差异不显著(图8)。
图8
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图8ScPsbS基因应答SCMV侵染的表达模式
误差线为每组处理的标准误差(n = 3)。
Fig. 8Expression profile of ScPsbS under the infection of SCMV
The error bars represent the standard error of each treatment group (n = 3).
3 讨论
甘蔗是生长于热带亚热带的C4植物, 具有很高的光合效能[80]。甘蔗在光照强度最高的中午时段, 存在光抑制现象[81], 但是对净光合效率影响不大[82], 主要是因为甘蔗具有较强的光呼吸和光抑制自我保护能力, 相应地, 其NPQ也在中午出现峰值[80]。在甘蔗伸长期, 正一叶是光合作用最旺盛的叶片[81]。本研究对ScPsbS的组织特异性表达分析(图7)也表明, ScPsbS的表达可能与其所在组织的光合能力相关。由于PsbS是植物NPQ的关键因子[62,64,67,70], 因此推测这种相关源自以NPQ为主的光保护。植物病毒基因组简单, 必须与寄主因子互作才能在寄主体内建立系统性侵染[82,83]。本课题组在前期研究中, 利用Y2H验证了SCMV-6K2与ScPsbS的互作[71], 本研究利用BiFC技术进一步证明了它们的互作(图 6)。SCMV-6K2定位于内质网和叶绿体[71], ScPsbS定位于叶绿体(图5-A), SCMV-6K2与ScPsbS的部分共定位信号出现在叶绿体上(图5-B)。叶绿体是马铃薯Y病毒属病毒高效复制的场所, 比如芜菁花叶病毒(Turnip mosaic virus, TuMV)利用6K2与Syp71互作[48], 烟草脉带花叶病毒(Tobacco vein banding mosaic virus, TVBMV)利用6K2与NbPsbO1互作[84], 促进病毒基因组复制。因此, 我们推测SCMV可能利用6K2与ScPsbS互作, 使SCMV的复制复合体定位于叶绿体, 有利于SCMV基因组的高效复制。供试材料Badila极易感染甘蔗花叶病, ScPsbS在SCMV侵染早期表达显著上调(图8), 一方面有助于SCMV在寄主体内建立系统性侵染, 另一方面则是寄主对SCMV侵染的抗性反应。甘蔗花叶病的主要症状是出现花叶、植株矮小、产量和品质下降等, 主要是因为SCMV侵染导致叶绿素减少[50]、叶绿体破坏[85], 进而光合作用受损。在SCMV侵染后期, ScPsbS表达量迅速下降(图8)至略高于对照, 但无显著性差异。Lei等[55]对黄瓜花叶病毒(Cucumber mosaic virus, CMV)侵染烟草的研究表明, 在侵染后的第8天和第12天, 感病叶片与健康叶片相比, NPQ略高但无显著差异, 推测是烟草应对CMV侵染的抗逆反应[49]。ScPsbS与SCMV-6K2互作于细胞膜上(图6), 改变了ScPsbS的叶绿体定位, 因此可能抑制NPQ, 使甘蔗的光合作用失去保护, 加剧叶绿体的损伤, 导致叶片黄化, 进而吸引蚜虫等昆虫载体传播病毒[14,86-90]。
4 结论
ScPsbS基因在成熟叶片中表达量最高, 在SCMV侵染早期上调表达。ScPsbS蛋白定位于叶绿体, 可与SCMV-6K2共定位于叶绿体。ScPsbS蛋白与SCMV-6K2互作于细胞膜。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
DOI:10.1007/s00705-008-0072-3URLPMID:18438601

A survey of cultivated hybrid sugarcane (Saccharum inter-specific hybrid) and noble sugarcane (Saccharum officinarum) in southern China for the presence of Sugarcane mosaic virus (SCMV), Sorghum mosaic virus (SrMV) and Sugarcane streak mosaic virus (SCSMV) was conducted by RT-PCR from the years 2003 to 2006. SCMV and SrMV, but not SCSMV, were found. A high incidence of SCMV and SrMV coinfection was revealed in both hybrid and noble sugarcanes. All coinfected plants showed mosaic symptom, whereas plants infected with a single virus were symptomatic or asymptomatic. It appears that virus mixtures are more virulent than single infections. The nucleotide sequences of the coat protein (CP) gene of 33 SCMV and 10 SrMV isolates from this study were compared to those of CP genes of SCMV and SrMV reported in GenBank. One hundred and seventy-three SCMV isolates, with the exception of MDB and Abaca strains, can be grouped into five groups, which include three previously known groups, the sugarcane (SCE), maize (MZ), and Thailand groups, and two newly identified groups, the noble sugarcane (NSCE) and Brazil groups. Twenty-two SrMV isolates were divided into two groups, HS (hybrid sugarcane) and NS (noble sugarcane) groups. Five out of eight SrMV hybrid isolates belonged to the HS group, and two SrMV noble isolates and three hybrid isolates were within the NS group. Interestingly, the three hybrid isolates within the NS group were isolated from hybrid sugarcane co-infected with SCMV. This indicates that SCMV helps the NS group SrMV to infect hybrid sugarcane.
DOI:10.1016/j.bbrc.2015.09.015URLPMID:26362180 [本文引用: 1]

Sugarcane (Saccharum sp. hybrid) provides the main source of sugar for humans. Sugarcane mosaic disease (SMD) is a major threat to sugarcane production. Currently, control of SMD is mainly dependent on breeding resistant cultivars through hybridization, which is time-consuming. Understanding the mechanism of viral infection may facilitate novel strategies to breed cultivars resistant to SMD and to control the disease. In this study, a wide interaction was detected between the viral VPg protein and host proteins. Several genes were screened from sugarcane cDNA library that could interact with Sugarcane streak mosaic virus VPg, including SceIF4E1 and ScELC. ScELC was predicted to be a cytoplasmic protein, but subcellular localization analysis showed it was distributed both in cytoplasmic and nuclear, and interactions were also detected between ScELC and VPg of SCMV or SrMV that reveal ScELC was widely used in the SMD pathogen infection process. ScELC and VPgs interacted in the nucleus, and may function to enhance the viral transcription rate. ScELC also interacted with SceIF4E2 both in the cytoplasm and nucleus, but not with SceIF4E1 and SceIF4E3. These results suggest that ScELC may be essential for the function of SceIF4E2, an isomer of eIF4E.
DOI:10.1006/mpev.1998.0535URLPMID:10051385 [本文引用: 1]

The nucleic acid of a serologically distinct potyvirus, originally isolated out of sugar cane from Pakistan, was reverse transcribed and the 3' terminal 2000 bp was PCR amplified, cloned, and sequenced. Phylogenetic comparisons of viruses representing each genus of the Potyviridae show that the Pakistani isolate is most closely related to the rymoviruses wheat streak mosaic virus (WSMV) and brome streak mosaic virus. We therefore propose that this new virus species be named sugar cane streak mosaic virus to reflect its similarity to WSMV. The phylogenetic data also show that the genus Rymovirus contains at least two unique evolutionary lineages. Thus the current taxonomy, based on transmission vector, is paraphyletic. We present an analysis of the taxonomic relationships among members of the family and propose a classification that both resolves the paraphyly and more accurately represents the evolutionary history of the Potyviridae.
URLPMID:1657820 [本文引用: 1]
DOI:10.1007/s12355-013-0279-9URL [本文引用: 1]
DOI:10.1007/s00705-011-1090-0URLPMID:21927898 [本文引用: 2]

The complete nucleotide sequences of three sugarcane streak mosaic virus (SCSMV) isolates found in sugarcane plants imported from Japan (JP1, JP2) and Indonesia (ID) into China were determined. Sequence analysis showed that, based on the whole genome sequence and the nucleotide sequences encoding the ten mature proteins, the three SCSMV isolates shared identities of 81.62%-81.67% and 76.39%-85.93%, respectively, with a Pakistan (PAK) isolate of SCSMV. Phylogenetic analysis indicated that SCSMV-JP1, -JP2 and -ID were clustered in a subgroup separate from the previously described SCSMV-PAK and -India isolates, and may thus represent a new strain of SCSMV.
DOI:10.1007/s11262-010-0457-8URL [本文引用: 1]

Sugarcane streak mosaic virus (SCSMV-PAK) is determined to be 9782 nucleotides in length, excluding the 3′ poly(A) tail, and it comprises a large open reading frame encoding a polyprotein of 3130 amino acid residues. The deduced polyprotein is likely to be cleaved at nine putative protease sites by three viral proteases to ten mature proteins. Conserved motifs of orthologous proteins of other potyviruses are identified in corresponding positions of SCSMV-PAK. The genomic organization is virtually identical to the genera Ipomovirus, Potyvirus, Rymovirus, and Tritimovirus in the family Potyviridae. Sequence analyses indicate that the SCSMV-PAK genomic sequence is different from those of Sugarcane mosaic virus and Sorghum mosaic virus, two viruses with very similar symptoms and host range to SCSMV-PAK. SCSMV-PAK shares 52.7% identity with Triticum mosaic virus (TriMV) and 26.4–31.5% identities with species of the existing genera and unassigned viruses in the Potyviridae at the polyprotein sequence level. Phylogenetic analyses of the polyprotein and deduced mature protein amino acid sequences reveal that SCSMV, together with TriMV, forms a distinct group in the family at the genus level. Therefore, SCSMV should represent a new genus, Susmovirus, in the Potyviridae.]]>
URL [本文引用: 1]

以饲料甜菜和糖甜菜杂种F1与3个亲本对照为试验材料,对各材料的主要性状进行比较研究.试验结果表明,杂种F1代在根产量、粗蛋白含量、粗脂肪含量3个性状上表现强杂种优势;粗纤维含量、含糖率与抗病性杂种优势弱;根产量与含糖率(r=-0.818*)、含糖率与粗纤维含量(r=0.907**)、粗蛋白含量与粗脂肪含量(r=0.954**)、根产量与粗纤维(r=0.736*)4对性状相关程度高,规律明显.同一性状正交、反交杂种优势不同,根产量、粗蛋白含量、粗脂肪含量在正交杂种中杂种优势明显;含糖率、粗纤维含量、干物质含量在反交杂种中优势明显.
[本文引用: 1]
DOI:10.1094/PDIS-04-18-0581-REURLPMID:30207899 [本文引用: 1]

A viral metagenomics study of the sugarcane virome in Florida was carried out in 2013 to 2014 to analyze occurrence of known and potentially new viruses. In total, 214 sugarcane leaf samples were collected from different commercial sugarcane (Saccharum interspecific hybrids) fields in Florida and from other Saccharum and related species taken from two local germplasm collections. Virion-associated nucleic acids (VANA) metagenomics was used for detection and identification of viruses present within the collected leaf samples. VANA sequence reads were obtained for 204 leaf samples and all four previously reported sugarcane viruses occurring in Florida were detected: Sugarcane yellow leaf virus (SCYLV, 150 infected samples out of 204), Sugarcane mosaic virus (1 of 204), Sugarcane mild mosaic virus (13 of 204), and Sugarcane bacilliform virus (54 of 204). High prevalence of SCYLV in Florida commercial fields and germplasm collections was confirmed by reverse-transcription polymerase chain reaction. Sequence analyses revealed the presence of SCYLV isolates belonging to two different phylogenetic clades (I and II), including a new genotype of this virus. This viral metagenomics approach also resulted in the detection of a new sugarcane-infecting mastrevirus (recently described and named Sugarcane striate virus), and two potential new viruses in the genera Chrysovirus and Umbravirus.
URLPMID:32529501 [本文引用: 1]
URLPMID:30806574 [本文引用: 1]
DOI:10.1016/s0168-1702(01)00220-9URLPMID:11226583 [本文引用: 1]
DOI:10.1093/nar/gkw441URLPMID:27185887 [本文引用: 1]

The Potyviridae comprise the largest and most important family of RNA plant viruses. An essential overlapping ORF, termed pipo, resides in an internal region of the main polyprotein ORF. Recently, expression of pipo was shown to depend on programmed transcriptional slippage at a conserved GAAAAAA sequence, resulting in the insertion of an extra A into a proportion of viral transcripts, fusing the pipo ORF in frame with the 5' third of the polyprotein ORF. However, the sequence features that mediate slippage have not been characterized. Using a duplicate copy of the pipo slip site region fused into a different genomic location where it can be freely mutated, we investigated the sequence requirements for transcriptional slippage. We find that the leading G is not strictly required, but increased flanking sequence GC content correlates with higher insertion rates. A homopolymeric hexamer is optimal for producing mainly single-nucleotide insertions. We also identify an overabundance of G to A substitutions immediately 3'-adjacent to GAAAAAA in insertion-free transcripts, which we infer to result from a 'to-fro' form of slippage during positive-strand synthesis. Analysis of wild-type and reverse complement sequences suggests that slippage occurs preferentially during synthesis of poly(A) and therefore occurs mainly during positive-strand synthesis.
[本文引用: 1]
URLPMID:28852157 [本文引用: 3]

The coding sequence of P3N-PIPO was cloned by fusion PCR from Sugarcane mosaic virus (SCMV), a main causal agent of sugarcane (Saccharum spp. hybrid) mosaic disease. SCMV P3N-PIPO preferentially localized to the plasma membrane (PM) compared with the plasmodesmata (PD), as demonstrated by transient expression and plasmolysis assays in the leaf epidermal cells of Nicotiana benthamiana. The subcellular localization of the P3N-PIPO mutants P3N-PIPOT1 and P3N-PIPOT2 with 29 and 63 amino acids deleted from the C-terminus of PIPO, respectively, revealed that the 19 amino acids at the N-terminus of PIPO contributed to the PD localization. Interaction assays showed that the 63 amino acids at the C-terminus of PIPO determined the P3N-PIPO interaction with PM-associated Ca(2+)-binding protein 1, ScPCaP1, which was isolated from the SCMV-susceptible sugarcane cultivar Badila. Like wild-type P3N-PIPO, P3N-PIPOT1 and P3N-PIPOT2 could translocate to neighbouring cells and recruit the SCMV cylindrical inclusion protein to the PM. Thus, interactions with ScPCaP1 may contribute to, but not determine, SCMV Pm3N-PIPO's localization to the PM or PD. These results also imply the existence of truncated P3N-PIPO in nature.
DOI:10.1099/0022-1317-73-1-1URLPMID:1730931 [本文引用: 1]
URLPMID:18408156 [本文引用: 1]
URLPMID:31657467 [本文引用: 2]
DOI:10.1105/tpc.18.00281URLPMID:30150314

Infection of plant cells by RNA viruses leads to the generation of organelle-like subcellular structures that contain the viral replication complex. During Turnip mosaic virus (TuMV) infection of Nicotiana benthamiana, the viral membrane protein 6K2 plays a key role in the release of motile replication vesicles from the host endoplasmic reticulum (ER). Here, we demonstrate that 6K2 contains a GxxxG motif within its predicted transmembrane domain that is vital for TuMV infection. Replacement of the Gly with Val within this motif inhibited virus production, and this was due to a relocation of the viral protein to the Golgi apparatus and the plasma membrane. This indicated that passage of 6K2 through the Golgi apparatus is a dead-end avenue for virus infection. Impairing the fusion of transport vesicles between the ER and the Golgi apparatus by overexpression of the SNARE Sec22 protein resulted in enhanced intercellular virus movement. Likewise, expression of nonfunctional, Golgi-located synaptotagmin during infection enhanced TuMV intercellular movement. 6K2 copurified with VTI11, a prevacuolar compartment SNARE protein. An Arabidopsis thaliana vti11 mutant was completely resistant to TuMV infection. We conclude that TuMV replication vesicles bypass the Golgi apparatus and take an unconventional pathway that may involve prevacuolar compartments/multivesicular bodies for virus infection.
DOI:10.1104/pp.17.01484URLPMID:29089395

Plant viruses move from the initially infected cell to adjacent cells through plasmodesmata (PDs). To do so, viruses encode dedicated protein(s) that facilitate this process. How viral proteins act together to support the intercellular movement of viruses is poorly defined. Here, by using an infection-free intercellular vesicle movement assay, we investigate the action of CI (cylindrical inclusion) and P3N-PIPO (amino-terminal half of P3 fused to Pretty Interesting Potyviridae open reading frame), the two PD-localized potyviral proteins encoded by Turnip mosaic virus (TuMV), in the intercellular movement of the viral replication vesicles. We provide evidence that CI and P3N-PIPO are sufficient to support the PD targeting and intercellular movement of TuMV replication vesicles induced by 6K2, a viral protein responsible for the generation of replication vesicles. 6K2 interacts with CI but not P3N-PIPO. When this interaction is impaired, the intercellular movement of TuMV replication vesicles is inhibited. Furthermore, in transmission electron microscopy, vesicular structures are observed in connection with the cylindrical inclusion bodies at structurally modified PDs in cells coexpressing 6K2, CI, and P3N-PIPO. CI is directed to PDs through its interaction with P3N-PIPO. We hypothesize that CI serves as a docking point for PD targeting and the intercellular movement of TuMV replication vesicles. This work contributes to a better understanding of the roles of different viral proteins in coordinating the intercellular movement of viral replication vesicles.
URLPMID:30538165 [本文引用: 1]
DOI:10.1128/JVI.01329-08URLPMID:18842721 [本文引用: 1]

Single-stranded positive-sense RNA viruses induce the biogenesis of cytoplasmic membranous vesicles, where viral replication takes place. However, the mechanism underlying this characteristic vesicular proliferation remains poorly understood. Previously, a 6-kDa potyvirus membrane protein (6K) was shown to interact with the endoplasmic reticulum (ER) and to induce the formation of the membranous vesicles. In this study, the involvement of the early secretory pathway in the formation of the 6K-induced vesicles was investigated in planta. By means of live-cell imaging, it was found that the 6K protein was predominantly colocalized with Sar1, Sec23, and Sec24, which are known markers of ER exit sites (ERES). The localization of 6K at ERES was prevented by the coexpression of a dominant-negative mutant of Sar1 that disables the COPII activity or by the coexpression of a mutant of Arf1 that disrupts the COPI complex. The secretion of a soluble secretory marker targeting the apoplast was arrested at the level of the ER in cells overexpressing 6K or infected by a potyvirus. This blockage of protein trafficking out of the ER by 6K and the distribution of 6K toward the ERES may account for the aggregation of the 6K-bound vesicles. Finally, virus infection was reduced when the accumulation of 6K at ERES was inhibited by impairing either the COPI or COPII complex. Taken together, these results imply that the cellular COPI and COPII coating machineries are involved in the biogenesis of the potyvirus 6K vesicles at the ERES for viral-genome replication.
DOI:10.1128/JVI.00503-15URLPMID:25878114 [本文引用: 1]

UNLABELLED: Positive-sense RNA viruses remodel host cell endomembranes to generate quasi-organelles known as
DOI:10.1128/JVI.00819-09URLPMID:19656892 [本文引用: 3]

Nicotiana benthamiana plants were agroinoculated with an infectious cDNA clone of Turnip mosaic virus (TuMV) that was engineered to express a fluorescent protein (green fluorescent protein [GFP] or mCherry) fused to the viral 6K2 protein known to induce vesicle formation. Cytoplasmic fluorescent discrete protein structures were observed in infected cells, corresponding to the vesicles containing the viral RNA replication complex. The vesicles were motile and aligned with microfilaments. Intracellular movement of the vesicles was inhibited when cells were infiltrated with latrunculin B, an inhibitor of microfilament polymerization. It was also observed that viral accumulation in the presence of this drug was reduced. These data indicate that microfilaments are used for vesicle movement and are necessary for virus production. Biogenesis of the vesicles was further investigated by infecting cells with two recombinant TuMV strains: one expressed 6K2GFP and the other expressed 6K2mCherry. Green- and red-only vesicles were observed within the same cell, suggesting that each vesicle originated from a single viral genome. There were also vesicles that exhibited sectors of green, red, or yellow fluorescence, an indication that fusion among individual vesicles is possible. Protoplasts derived from TuMV-infected N. benthamiana leaves were isolated. Using immunofluorescence staining and confocal microscopy, viral RNA synthesis sites were visualized as punctate structures distributed throughout the cytoplasm. The viral proteins VPg-Pro, RNA-dependent RNA polymerase, and cytoplasmic inclusion protein (helicase) and host translation factors were found to be associated with these structures. A single-genome origin and presence of protein synthetic machinery components suggest that translation of viral RNA is taking place within the vesicle.
DOI:10.1099/0022-1317-73-11-2785URLPMID:1431807 [本文引用: 1]

The complete RNA genome of turnip mosaic potyvirus (TuMV) was amplified by seven consecutive reverse transcriptase-polymerase chain reactions and cloned into pUC9. The viral RNA is 9830 nucleotides long and contains a single open reading frame (ORF) of 9489 bases encoding a large polyprotein of 3863 amino acids with a calculated M(r) of 358,000. The non-coding region (NCR) preceding the ORF is 129 nucleotides long and has a high AU content (70%). Its predicted secondary structure is characterized by a hairpin loop with a free energy loss of -69.9 kJ/mol. The termination codon is followed by an AU-rich NCR of 209 bases, excluding the poly(A) tail. Seven potential nuclear inclusion a proteinase (NIa-Pro) recognition heptapeptides are found in the polyprotein. Their sequences agree with consensus potyviral NIa-Pro cleavage sequences except for that at the 6K-VPg site, which is characterized by a glutamic acid residue preceding the hydrolysed peptide bond. The TuMV proteins are similar to their corresponding potyviral proteins.
URLPMID:17670821 [本文引用: 1]
URLPMID:17079311 [本文引用: 1]
DOI:10.1016/j.virol.2007.12.014URLPMID:18222516 [本文引用: 1]

Tandem affinity purification was used in Arabidopsis thaliana to identify cellular interactors of Turnip mosaic virus (TuMV) RNA-dependent RNA polymerase (RdRp). The heat shock cognate 70-3 (Hsc70-3) and poly(A)-binding (PABP) host proteins were recovered and shown to interact with the RdRp in vitro. As previously shown for PABP, Hsc70-3 was redistributed to nuclear and membranous fractions in infected plants and both RdRp interactors were co-immunoprecipitated from a membrane-enriched extract using RdRp-specific antibodies. Fluorescently tagged RdRp and Hsc70-3 localized to the cytoplasm and the nucleus when expressed alone or in combination in Nicotiana benthamiana. However, they were redistributed to large perinuclear ER-derived vesicles when co-expressed with the membrane binding 6K-VPg-Pro protein of TuMV. The association of Hsc70-3 with the RdRp could possibly take place in membrane-derived replication complexes. Thus, Hsc70-3 and PABP2 are potentially integral components of the replicase complex and could have important roles to play in the regulation of potyviral RdRp functions.
URLPMID:26958722 [本文引用: 1]
DOI:10.3389/fmicb.2013.00351URLPMID:24409170 [本文引用: 1]

To successfully infect plants, viruses replicate in an initially infected cell and then move to neighboring cells through plasmodesmata (PDs). However, the nature of the viral entity that crosses over the cell barrier into non-infected ones is not clear. The membrane-associated 6K2 protein of turnip mosaic virus (TuMV) induces the formation of vesicles involved in the replication and intracellular movement of viral RNA. This study shows that 6K2-induced vesicles trafficked toward the plasma membrane and were associated with plasmodesmata (PD). We demonstrated also that 6K2 moved cell-to-cell into adjoining cells when plants were infected with TuMV. 6K2 was then fused to photo-activable GFP (6K2:PAGFP) to visualize how 6K2 moved intercellularly during TuMV infection. After activation, 6K2:PAGFP-tagged vesicles moved to the cell periphery and across the cell wall into adjacent cells. These vesicles were shown to contain the viral RNA-dependent RNA polymerase and viral RNA. Symplasmic movement of TuMV may thus be achieved in the form of a membrane-associated viral RNA complex induced by 6K2.
DOI:10.1371/journal.ppat.1003378URLPMID:23696741 [本文引用: 2]

All positive-strand RNA viruses induce the biogenesis of cytoplasmic membrane-bound virus factories for viral genome multiplication. We have previously demonstrated that upon plant potyvirus infection, the potyviral 6K2 integral membrane protein induces the formation of ER-derived replication vesicles that subsequently target chloroplasts for robust genome replication. Here, we report that following the trafficking of the Turnip mosaic potyvirus (TuMV) 6K2 vesicles to chloroplasts, 6K2 vesicles accumulate at the chloroplasts to form chloroplast-bound elongated tubular structures followed by chloroplast aggregation. A functional actomyosin motility system is required for this process. As vesicle trafficking and fusion in planta are facilitated by a superfamily of proteins known as SNAREs (soluble N-ethylmaleimide-sensitive-factor attachment protein receptors), we screened ER-localized SNARES or SNARE-like proteins for their possible involvement in TuMV infection. We identified Syp71 and Vap27-1 that colocalize with the chloroplast-bound 6K2 complex. Knockdown of their expression using a Tobacco rattle virus (TRV)-based virus-induced gene silencing vector showed that Syp71 but not Vap27-1 is essential for TuMV infection. In Syp71-downregulated plant cells, the formation of 6K2-induced chloroplast-bound elongated tubular structures and chloroplast aggregates is inhibited and virus accumulation is significantly reduced, but the trafficking of the 6K2 vesicles from the ER to chloroplast is not affected. Taken together, these data suggest that Syp71 is a host factor essential for successful virus infection by mediating the fusion of the virus-induced vesicles with chloroplasts during TuMV infection.
DOI:10.1111/ppl.1997.100.issue-2URL [本文引用: 5]
[本文引用: 2]
[本文引用: 2]
DOI:10.5423/PPJ.OA.04.2019.0120URLPMID:31632226 [本文引用: 2]

Tomato yellow leaf curl China virus is a species of the widespread geminiviruses. The infection of Nicotiana benthamiana by Tomato yellow leaf curl China virus (TYLCCNV) causes a reduction in photosynthetic activity, which is part of the viral symptoms. betaC1 is a viral factor encoded by the betasatellite DNA (DNAbeta) accompanying TYLCCNV. It is a major viral pathogenicity factor of TYLCCNV. To elucidate the effect of betaC1 on plants' photosynthesis, we measured the relative chlorophyll (Chl) content and Chl fluorescence in TYLCCNV-infected and betaC1 transgenic N. benthamiana plants. The results showed that Chl content is reduced in TYLCCNV A-infected, TYLCCNV A plus DNAbeta (TYLCCNV A + beta)-infected and betaC1 transgenic plants. Further, changes in Chl fluorescence parameters, such as electron transport rate, F v /F m , NPQ, and qP, revealed that photosynthetic efficiency is compromised in the aforementioned N. benthamiana plants. The presense of betaC1 aggravated the decrease of Chl content and photosynthetic efficiency during viral infection. Additionally, the real-time quantitative PCR analysis of oxygen evolving complex genes in photosystem II, such as PsbO, PsbP, PsbQ, and PsbR, showed a significant reduction of the relative expression of these genes at the late stage of TYLCCNV A + beta infection and at the vegetative stage of betaC1 transgenic N. benthamiana plants. In summary, this study revealed the pathogenicity of TYLCCNV in photosynthesis and disclosed the effect of betaC1 in exacerbating the damage in photosynthesis efficiency by TYLCCNV infection.
URLPMID:21708634 [本文引用: 1]

We examined the effects of geminivirus infection on fitness components and on photosynthetic properties of the host plant, Eupatorium makinoi, grown at two irradiance levels in a natural-light greenhouse. Under the low-light condition (13% full sunlight), more than a half of the infected plants died during the 9-mo experiment, while most of uninfected plants survived. Growth rate was also lowered by infection. At high light (50% full sunlight), by contrast, virus infection did not cause mortality despite slight decrease in growth rate. Flowering occurred only at high light, and reproductive outputs of the plants were markedly reduced by the infection. Infected leaves had distinct yellow variegations and, when compared with uninfected leaves, they showed (1) comparable light-saturated photosynthetic rate per unit area, but (2) lower initial slope of light-response curve of photosynthesis on an incident irradiance basis. The lower initial slope was mainly due to reduction of light-harvesting chlorophyll-protein complexes in the variegated parts. Since the differences in plant performance, depending both on infection and on growth irradiance, were largely explained by the differences in growth rate and/or plant size, the reduced photosynthetic production in the infected plants would be a major factor explaining the inferior performance of the host plants.
DOI:10.1111/j.1399-3054.2008.01189.xURLPMID:19140890 [本文引用: 1]

We examined the responses of the photosynthetic and respiratory electron transport and antioxidant systems in cell organelles of cucumber (Cucumis sativus L.) and tomato (Lycopersicon esculentum Mill.) leaves to infection of cucumber mosaic virus (CMV) by comparing the gas exchange, Chl fluorescence, respiratory electron transport, superoxide dismutase (SOD, EC 1.15.1.1) and ascorbate-glutathione (AsA-GSH) cycle enzymes and the production of H(2)O(2) in chloroplasts, mitochondria and soluble fraction in virus-infected and non-infected leaves. Long-term CMV infection resulted in decreased photosynthesis and respiration rates. Photosynthetic electron flux to carbon reduction, respiratory electron transport via both complex I and complex II and also the Cyt respiration rate all significantly decreased, while photosynthetic alternative electron flux and alternative respiration significantly increased. These changes in electron transport were accompanied by a general increase in the activities of SOD/AsA-GSH cycle enzymes followed by an increased H(2)O(2) accumulation in chloroplasts and mitochondria. These results demonstrated that disturbance of photosynthetic and respiratory electron transport by CMV also affected the antioxidative systems, thereby leading to oxidative stress in various organelles.
DOI:10.1016/S2095-3119(19)62572-4URL [本文引用: 1]
DOI:10.1007/s00299-016-2083-yURLPMID:27904946 [本文引用: 3]

KEY MESSAGE: Leaf chlorosis induced by plant virus infection has a short fluorescence lifetime, which reflects damaged photosynthetic complexes and degraded chloroplasts. Plant viruses often induce chlorosis and necrosis, which are intimately related to photosynthetic functions. Chlorophyll fluorescence lifetime measurement is a valuable noninvasive tool for analyzing photosynthetic processes and is a sensitive indicator of the environment surrounding the fluorescent molecules. In this study, our central goal was to explore the effect of viral infection on photosynthesis by employing chlorophyll fluorescence lifetime imaging (FLIM), steady-state fluorescence, non-photochemical quenching (NPQ), transmission electron microscopy (TEM), and pigment analysis. The data indicated that the chlorophyll fluorescence lifetime of chlorotic leaves was significantly shorter than that of healthy control leaves, and the fitted short lifetime component of chlorophyll fluorescence of chlorotic leaves was dominant. This dominant short lifetime component may result from damage to the structure of thylakoid, which was confirmed by TEM. The NPQ value of chlorotic leaves was slightly higher than that of healthy green leaves, which can be explained by increased neoxanthin, lutein and violaxanthin content relative to chlorophyll a. The difference in NPQ is slight, but FLIM can provide simple and direct characterization of PSII structure and photosynthetic function. Therefore, this technique shows great potential as a simple and rapid method for studying mechanisms of plant virus infection.
DOI:10.1146/annurev.arplant.58.032806.103946URLPMID:17288534 [本文引用: 1]

Reactive oxygen species (ROS) and reactive nitrogen species (RNS) are produced in many places in living cells and at an increased rate during biotic or abiotic stress. ROS and RNS participate in signal transduction, but also modify cellular components and cause damage. We first look at the most common ROS and their properties. We then consider the ways in which the cell can regulate their production and removal. We critically assess current knowledge about modifications of polyunsaturated fatty acids (PUFAs), DNA, carbohydrates, and proteins and illustrate this knowledge with case stories wherever possible. Some oxidative breakdown products, e.g., from PUFA, can cause secondary damage. Other oxidation products are secondary signaling molecules. We consider the fate of the modified components, the energetic costs to the cell of replacing such components, as well as strategies to minimize transfer of oxidatively damaged components to the next generation.
DOI:10.1146/annurev.arplant.58.032806.103844URLPMID:19575582 [本文引用: 1]

Plants and algae often absorb too much light-more than they can actually use in photosynthesis. To prevent photo-oxidative damage and to acclimate to changes in their environment, photosynthetic organisms have evolved direct and indirect mechanisms for sensing and responding to excess light. Photoreceptors such as phototropin, neochrome, and cryptochrome can sense excess light directly and relay signals for chloroplast movement and gene expression responses. Indirect sensing of excess light through biochemical and metabolic signals can be transduced into local responses within chloroplasts, into changes in nuclear gene expression via retrograde signaling pathways, or even into systemic responses, all of which are associated with photoacclimation.
DOI:10.1021/bi036219iURLPMID:15157112 [本文引用: 1]

The photogeneration of hydroxyl radicals (OH(*)) in photosystem II (PSII) membranes was studied using EPR spin-trapping spectroscopy. Two kinetically distinguishable phases in the formation of the spin trap-hydroxyl (POBN-OH) adduct EPR signal were observed: the first phase (t(1/2) = 7.5 min) and the second phase (t(1/2) = 30 min). The generation of OH(*) was found to be suppressed in the absence of the Mn-complex, but it was restored after readdition of an artificial electron donor (DPC). Hydroxyl radical generation was also lost in the absence of oxygen, whereas it was stimulated when the oxygen concentration was increased. The production of OH(*) during the first kinetic phase was sensitive to the presence of SOD, whereas catalase and EDTA diminished the production of OH(*) during the second kinetic phase. The POBN-OH adduct EPR signal during the first phase exhibits a similar pH-dependence as the ability to oxidize the non-heme iron, as monitored by the Fe(3+) (g = 8) EPR signal: both EPR signals gradually decreased as the pH value was lowered below pH 6.5 and were absent at pH 5. Sodium formate decreases the production of OH(*) in intact and Mn-deleted PSII membranes. Upon illumination of PSII membranes, both superoxide, as measured by EPR signal from the spin trap-superoxide (EMPO-OOH) adduct, and H(2)O(2), measured colormetrically, were generated. These results indicated that OH(*) is produced on the electron acceptor side of PSII by two different routes, (1) O(2)(*)(-), which is generated by oxygen reduction on the acceptor side of PSII, interacts with a PSII metal center, probably the non-heme iron, to form an iron-peroxide species that is further reduced to OH(*) by an electron from PSII, presumably via Q(A)(-), and (2) O(2)(*)(-) dismutates to form free H(2)O(2) that is then reduced to OH(*) via the Fenton reaction in the presence of metal ions, the most likely being Mn(2+) and Fe(2+) released from photodamaged PSII. The two different routes of OH(*) generation are discussed in the context of photoinhibition.
URLPMID:23583332 [本文引用: 1]
DOI:10.1021/bi0494020URLPMID:15222740 [本文引用: 1]

Oxygenic photosynthesis in plants involves highly reactive intermediates and byproducts that can damage the photosynthetic apparatus and other chloroplast constituents. The potential for damage is exacerbated when the amount of absorbed light exceeds the capacity for light energy utilization in photosynthesis, a condition that can lead to decreases in photosynthetic efficiency. A feedback de-excitation mechanism (qE), measured as a component of nonphotochemical quenching of chlorophyll fluorescence, regulates photosynthetic light harvesting in excess light in response to a change in thylakoid lumen pH. qE involves de-excitation of the singlet excited state of chlorophyll in the light-harvesting antenna of photosystem II, thereby minimizing the deleterious effects of high light via thermal dissipation of excess excitation energy. While the physiological importance of qE has been recognized for many years, a description of its physical mechanism remains elusive. We summarize recent biochemical and spectroscopic results that have brought us closer to the goal of a mechanistic understanding of this fundamental photosynthetic regulatory process.
DOI:10.1016/j.tplants.2010.10.001URLPMID:21050798 [本文引用: 1]

Sunlight damages photosynthetic machinery, primarily photosystem II (PSII), and causes photoinhibition that can limit plant photosynthetic activity, growth and productivity. The extent of photoinhibition is associated with a balance between the rate of photodamage and its repair. Recent studies have shown that light absorption by the manganese cluster in the oxygen-evolving complex of PSII causes primary photodamage, whereas excess light absorbed by light-harvesting complexes acts to cause inhibition of the PSII repair process chiefly through the generation of reactive oxygen species. As we review here, PSII photodamage and the inhibition of repair are therefore alleviated by photoprotection mechanisms associated with avoiding light absorption by the manganese cluster and successfully consuming or dissipating the light energy absorbed by photosynthetic pigments, respectively.
DOI:10.1038/35000131URLPMID:10667783 [本文引用: 3]

Photosynthetic light harvesting in plants is regulated in response to changes in incident light intensity. Absorption of light that exceeds a plant's capacity for fixation of CO2 results in thermal dissipation of excitation energy in the pigment antenna of photosystem II by a poorly understood mechanism. This regulatory process, termed nonphotochemical quenching, maintains the balance between dissipation and utilization of light energy to minimize generation of oxidizing molecules, thereby protecting the plant against photo-oxidative damage. To identify specific proteins that are involved in nonphotochemical quenching, we have isolated mutants of Arabidopsis thaliana that cannot dissipate excess absorbed light energy. Here we show that the gene encoding PsbS, an intrinsic chlorophyll-binding protein of photosystem II, is necessary for nonphotochemical quenching but not for efficient light harvesting and photosynthesis. These results indicate that PsbS may be the site for nonphotochemical quenching, a finding that has implications for the functional evolution of pigment-binding proteins.
DOI:10.1074/jbc.M402461200URLPMID:15033974 [本文引用: 2]

The biochemical, biophysical, and physiological properties of the PsbS protein were studied in relation to mutations of two symmetry-related, lumen-exposed glutamate residues, Glu-122 and Glu-226. These two glutamates are targets for protonation during lumen acidification in excess light. Mutation of PsbS did not affect xanthophyll cycle pigment conversion or pool size. Plants containing PsbS mutations of both glutamates did not have any rapidly inducible nonphotochemical quenching (qE) and had similar chlorophyll fluorescence lifetime components as npq4-1, a psbS deletion mutant. The double mutant also lacked a characteristic leaf absorbance change at 535 nm (DeltaA535), and PsbS from these plants did not bind dicyclohexylcarbodiimide (DCCD), a known inhibitor of qE. Mutation of only one of the glutamates had intermediate effects on qE, chlorophyll fluorescence lifetime component amplitudes, DCCD binding, and DeltaA535. Little if any differences were observed comparing the two single mutants, suggesting that the glutamates are chemically and functionally equivalent. Based on these results a bifacial model for the functional interaction of PsbS with photosystem II is proposed. Furthermore, based on the extent of qE inhibition in the mutants, photochemical and nonphotochemical quenching processes of photosystem II were associated with distinct chlorophyll fluorescence life-time distribution components.
URLPMID:27249196 [本文引用: 3]
DOI:10.1093/jxb/eri056URLPMID:15611143 [本文引用: 2]

The PsbS protein of photosystem II functions in the regulation of photosynthetic light harvesting. Along with a low thylakoid lumen pH and the presence of de-epoxidized xanthophylls, PsbS is necessary for photoprotective thermal dissipation (qE) of excess absorbed light energy in plants, measured as non-photochemical quenching of chlorophyll fluorescence. What is known about PsbS in relation to the hypothesis that this protein is the site of qE is reviewed here.
DOI:10.1111/j.1365-313X.2009.04051.xURLPMID:19843315

It is commonly accepted that the photosystem II subunit S protein, PsbS, is required for the dissipation of excess light energy in a process termed 'non-photochemical quenching' (NPQ). This process prevents photo-oxidative damage of photosystem II (PSII) thus avoiding photoinhibition which can decrease plant fitness and productivity. In this study Arabidopsis plants lacking PsbS (the npq4 mutant) were found to possess a competent mechanism of excess energy dissipation that protects against photoinhibitory damage. The process works on a slower timescale, taking about 1 h to reach the same level of NPQ achieved in the wild type in just a few minutes. The NPQ in npq4 was found to display very similar characteristics to the fast NPQ in the wild type. Firstly, it prevented the irreversible light-induced closure of PSII reaction centres. Secondly, it was uncoupler-sensitive, and thus triggered by the DeltapH across the thylakoid membrane. Thirdly, it was accompanied by significant quenching of the fluorescence under conditions when all PSII reaction centres were open (F(o) state)(.) Fourthly, it was accompanied by NPQ-related absorption changes (DeltaA535). Finally, it was modulated by the presence of the xanthophyll cycle carotenoid zeaxanthin. The existence of a mechanism of photoprotective energy dissipation in plants lacking PsbS suggests that this protein plays the role of a kinetic modulator of the energy dissipation process in the PSII light-harvesting antenna, allowing plants to rapidly track fluctuations of light intensity in the environment, and is not the primary cause of NPQ or a direct carrier of the pigment acting as the non-photochemical quencher.
URLPMID:20615387 [本文引用: 1]
DOI:10.1126/science.1082833URLPMID:12624254
DOI:10.3724/SP.J.1260.2012.20107URL [本文引用: 1]

光合膜是地球上捕获、转换和利用太阳能的关键场所,光合膜的活动所提供的能源、粮食及氧气,是人类世界赖以生存的基础。经过35亿年的进化,光合膜已经进化成了一个高度精密的结构,色素分子高密度结合并合理排列,具有高精度的能级耦联网络和高效率的能量传递系统,这使得光合膜成为自然界中能够最高效地吸收和传递太阳能、并能在常温常压下高效地将太阳能转换成化学能和还原势的色素蛋白复合体体系。由于这一特性,光合膜被认为是最有潜力的固定太阳能的新材料,并为研究新型光电转换器件提供了新思路和新理论。因此,长期以来,光合膜的结构-功能关系研究及其功能模拟,特别是执行固定和转化太阳能第一步的光系 统Ⅱ,在新能源的利用中吸引了大量的研究力量,取得了突飞猛进的进展。本文总结了近年来关于光系统Ⅱ的结构与功能,以及光合膜对环境的感应和功能调节机制等方面的研究进展。
[本文引用: 1]
DOI:10.1104/pp.15.00361URLPMID:26069151 [本文引用: 2]

Light is the primary energy source for photosynthetic organisms, but in excess, it can generate reactive oxygen species and lead to cell damage. Plants evolved multiple mechanisms to modulate light use efficiency depending on illumination intensity to thrive in a highly dynamic natural environment. One of the main mechanisms for protection from intense illumination is the dissipation of excess excitation energy as heat, a process called nonphotochemical quenching. In plants, nonphotochemical quenching induction depends on the generation of a pH gradient across thylakoid membranes and on the presence of a protein called PHOTOSYSTEM II SUBUNIT S (PSBS). Here, we generated Physcomitrella patens lines expressing histidine-tagged PSBS that were exploited to purify the native protein by affinity chromatography. The mild conditions used in the purification allowed copurifying PSBS with its interactors, which were identified by mass spectrometry analysis to be mainly photosystem II antenna proteins, such as LIGHT-HARVESTING COMPLEX B (LHCB). PSBS interaction with other proteins appears to be promiscuous and not exclusive, although the major proteins copurified with PSBS were components of the LHCII trimers (LHCB3 and LHCBM). These results provide evidence of a physical interaction between specific photosystem II light-harvesting complexes and PSBS in the thylakoids, suggesting that these subunits are major players in heat dissipation of excess energy.
DOI:10.3390/ijms20163867URL [本文引用: 5]
[本文引用: 1]
[本文引用: 1]
DOI:10.3724/SP.J.1006.2019.94002URL [本文引用: 2]

Saccharum spp. hybrid)中NDH复合体应答及参与甘蔗花叶病毒(Sugarcane mosaic virus, SCMV)的侵染尚未见报道。本研究克隆了甘蔗NDH复合体的O亚基, 命名为ScNdhO, 其开放读码框(open reading frame, ORF)长度为471 bp, 编码长度为156 aa的蛋白。生物信息学分析表明, ScNdhO为稳定的亲水性蛋白, 不存在信号肽, 无跨膜结构域; 蛋白二级结构元件多为无规则卷曲, 具有典型的NDH复合体O亚基结构域; 系统进化树分析表明, 该蛋白属于NDH复合体O亚基蛋白家族。实时荧光定量 PCR分析发现, ScNdhO基因的表达具有明显的组织特异性, 在成熟甘蔗叶片中的表达量最高, 在茎中的表达量最低, 在根中几乎不表达; ScNdhO基因在SCMV侵染早期上调表达, 后期下调表达。亚细胞定位分析表明, ScNdhO定位于叶绿体。酵母双杂交(yeast two hybrid, Y2H)和双分子荧光互补(bimolecular fluorescence complementation, BiFC)实验表明, ScNdhO与SCMV-VPg互作。推测ScNdhO被SCMV选择性利用, 可能参与SCMV基因组复制及花叶病症状的产生。]]>
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1038/srep07042URLPMID:25391499 [本文引用: 1]

Sugarcane (Saccharum spp. hybrids) is a world-wide cash crop for sugar and biofuel in tropical and subtropical regions and suffers serious losses in cane yield and sugar content under salinity and drought stresses. Although real-time quantitative PCR has a numerous advantage in the expression quantification of stress-related genes for the elaboration of the corresponding molecular mechanism in sugarcane, the variation happened across the process of gene expression quantification should be normalized and monitored by introducing one or several reference genes. To validate suitable reference genes or gene sets for sugarcane gene expression normalization, 13 candidate reference genes have been tested across 12 NaCl- and PEG-treated sugarcane samples for four sugarcane genotypes using four commonly used systematic statistical algorithms termed geNorm, BestKeeper, NormFinder and the deltaCt method. The results demonstrated that glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and eukaryotic elongation factor 1-alpha (eEF-1a) were identified as suitable reference genes for gene expression normalization under salinity/drought-treatment in sugarcane. Moreover, the expression analyses of SuSK and 6PGDH further validated that a combination of clathrin adaptor complex (CAC) and cullin (CUL) as reference should be better for gene expression normalization. These results can facilitate the future research on gene expression in sugarcane under salinity and drought stresses.
URLPMID:26258636 [本文引用: 2]
DOI:10.1016/0014-5793(94)80513-xURLPMID:8150081 [本文引用: 1]

The intrinsic 22 kDa polypeptide associated with photosystem II (psbS protein) was found to be able to bind chlorophyll. Extraction of isolated photosystem II membranes with octyl-thioglucopyranoside, followed by repetitive electrophoresis under partially denaturing conditions gave only one green band. It contained both chlorophyll a and chlorophyll b, exhibited an absorption maximum at 674 nm and a 77 K fluorescence peak at 675 nm. The chlorophyll-protein band contained a single polypeptide of 22 kDa. Based on these results and on previous protein sequence comparisons, it is suggested that the psbS protein is a chlorophyll a/b binding polypeptide and should thus be denoted CP22.
URLPMID:1360412 [本文引用: 1]
DOI:10.1016/j.bpj.2017.09.034URLPMID:29211990 [本文引用: 1]

Nonphotochemical quenching is the protective mechanism against overexcitation of photosystem II, triggered by excess DeltapH in photosynthetic membranes. The light-harvesting complexes (LHCs), the de-epoxidation of violaxanthin to zeaxanthin, and the photosystem II subunit S (PsbS) work in synergy for an optimized multilevel response. Understanding the fine details of this synergy has proven challenging to scientific research. Here, we employ large-scale, all-atom molecular simulations and beyond experimental insight, we proceed a step further in identifying the PsbS dynamics that could possibly be associated with this synergy. For the first time, to our knowledge, we probe the distinct behavior of PsbS under DeltapH that probes the details of the potential dimer-to-monomer transition, and in a violaxanthin/zeaxanthin-rich membrane, at an all-atom resolution. We propose that the lumen-exposed residues, threonine 162 and glutamic acid 173, form stabilizing hydrogen bonds between the PsbS monomers only at high lumen pH, whereas at low pH (excess DeltapH) this interaction is lost, and leads to higher flexibility of the protein and potentially to the dimer-to-monomer transition. Lastly, we discuss how conformational changes under the presence of DeltapH/zeaxanthin are related to the PsbS role in the current nonphotochemical quenching model in the literature. For the latter, we probe a PsbS-monomeric LHCII association. The association is proposed to potentially alter the monomeric LHCII sensitivity to DeltapH by changing the pKa values of interacting LHCII residues. This serves as an example where protonation-ligation events enhance protein-protein interactions fundamental to many life processes.
DOI:10.1016/S0378-4290(99)00069-6URL [本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1146/annurev-phyto-080614-120001URLPMID:25938276 [本文引用: 1]

A successful infection by a plant virus results from the complex molecular interplay between the host plant and the invading virus. Thus, dissecting the molecular network of virus-host interactions advances the understanding of the viral infection process and may assist in the development of novel antiviral strategies. In the past decade, molecular identification and functional characterization of host factors in the virus life cycle, particularly single-stranded, positive-sense RNA viruses, have been a research focus in plant virology. As a result, a number of host factors have been identified. These host factors are implicated in all the major steps of the infection process. Some host factors are diverted for the viral genome translation, some are recruited to improvise the viral replicase complexes for genome multiplication, and others are components of transport complexes for cell-to-cell spread via plasmodesmata and systemic movement through the phloem. This review summarizes current knowledge about host factors and discusses future research directions.
DOI:10.1006/viro.1997.8634URLPMID:9234949 [本文引用: 1]

The yeast LexA interaction trap was used to screen a cDNA library from Arabidopsis thaliana in order to identify proteins that interact with the viral protein genome linked (VPg)-proteinase of turnip mosaic potyvirus. The screen allowed the isolation of four candidate cDNA clones. Clones pHC4, pHC21, and pHC40 were partially sequenced but no homologies to known proteins were found. However, the amino acid sequence deduced from the complete nucleotide sequence of pSW56 revealed that it was the eukaryotic initiation factor (iso) 4E [eIF(iso)4E]. Deletion analysis indicated that the VPg domain was involved in the interaction with the plant protein. Interaction between the viral protein and the cellular protein was confirmed by ELISA-based binding experiments. eIF(iso)4E plays an essential role in the initiation of the translation of capped mRNAs and its association with VPg would point to a role of the viral protein in the translation of the virus.
URLPMID:28230184 [本文引用: 1]
DOI:10.1016/S0065-3527(10)76001-2URLPMID:20965070

Virus infection may damage the plant, and plant defenses are effective against viruses; thus, it is currently assumed that plants and viruses coevolve. However, and despite huge advances in understanding the mechanisms of pathogenicity and virulence in viruses and the mechanisms of virus resistance in plants, evidence in support of this hypothesis is surprisingly scant, and refers almost only to the virus partner. Most evidence for coevolution derives from the study of highly virulent viruses in agricultural systems, in which humans manipulate host genetic structure, what determines genetic changes in the virus population. Studies have focused on virus responses to qualitative resistance, either dominant or recessive but, even within this restricted scenario, population genetic analyses of pathogenicity and resistance factors are still scarce. Analyses of quantitative resistance or tolerance, which could be relevant for plant-virus coevolution, lag far behind. A major limitation is the lack of information on systems in which the host might evolve in response to virus infection, that is, wild hosts in natural ecosystems. It is presently unknown if, or under which circumstances, viruses do exert a selection pressure on wild plants, if qualitative resistance is a major defense strategy to viruses in nature, or even if characterized genes determining qualitative resistance to viruses did indeed evolve in response to virus infection. Here, we review evidence supporting plant-virus coevolution and point to areas in need of attention to understand the role of viruses in plant ecosystem dynamics, and the factors that determine virus emergence in crops.
DOI:10.1111/j.1364-3703.2004.00240.xURLPMID:20565624

SUMMARY Aphids are the most common vector of plant viruses. Mechanisms of transmission are best understood by considering the routes of virus movement in the aphid (circulative versus non-circulative) and the sites of retention or target tissues (e.g. stylets, salivary glands). Capsid proteins are a primary, but not necessarily sole, viral determinant of transmission. A summary is presented of the taxonomic affiliations of the aphid transmitted viruses, including 8 families, 18 genera, and taxonomically unassigned viruses.
DOI:10.1093/aesa/90.5.521URL
URLPMID:23407835 [本文引用: 1]
