 ,1,*, 王立志1, 杨贤莉1, 李波2, 母伟杰3, 董世晨3, 车韦才3, 李忠杰1, 迟力勇1, 李明贤1, 张喜娟1, 姜辉2, 李锐1, 赵茜1, 李文华
,1,*, 王立志1, 杨贤莉1, 李波2, 母伟杰3, 董世晨3, 车韦才3, 李忠杰1, 迟力勇1, 李明贤1, 张喜娟1, 姜辉2, 李锐1, 赵茜1, 李文华 ,2,*
,2,*Detection of QTLs controlling cold tolerance at bud bursting stage by using a high-density SNP linkage map in japonica rice
JIANG Shu-Kun ,1,*, WANG Li-Zhi1, YANG Xian-Li1, LI Bo2, MU Wei-Jie3, DONG Shi-Chen3, CHE Wei-Cai3, LI Zhong-Jie1, CHI Li-Yong1, LI Ming-Xian1, ZHANG Xi-Juan1, JIANG Hui2, LI Rui1, ZHAO Qian1, LI Wen-Hua
,1,*, WANG Li-Zhi1, YANG Xian-Li1, LI Bo2, MU Wei-Jie3, DONG Shi-Chen3, CHE Wei-Cai3, LI Zhong-Jie1, CHI Li-Yong1, LI Ming-Xian1, ZHANG Xi-Juan1, JIANG Hui2, LI Rui1, ZHAO Qian1, LI Wen-Hua ,2,*
,2,*通讯作者:
收稿日期:2019-12-08接受日期:2020-03-24网络出版日期:2020-08-12
| 基金资助: |
Received:2019-12-08Accepted:2020-03-24Online:2020-08-12
| Fund supported: |
作者简介 About authors
E-mail:sk_jiang@126.com, E-mail:sk_jiang@126.com

摘要
关键词:
Abstract
Keywords:
PDF (3422KB)元数据多维度评价相关文章导出EndNote|Ris|Bibtex收藏本文
本文引用格式
姜树坤, 王立志, 杨贤莉, 李波, 母伟杰, 董世晨, 车韦才, 李忠杰, 迟力勇, 李明贤, 张喜娟, 姜辉, 李锐, 赵茜, 李文华. 基于高密度SNP遗传图谱的粳稻芽期耐低温QTL鉴定[J]. 作物学报, 2020, 46(8): 1174-1184. doi:10.3724/SP.J.1006.2020.92066
JIANG Shu-Kun, WANG Li-Zhi, YANG Xian-Li, LI Bo, MU Wei-Jie, DONG Shi-Chen, CHE Wei-Cai, LI Zhong-Jie, CHI Li-Yong, LI Ming-Xian, ZHANG Xi-Juan, JIANG Hui, LI Rui, ZHAO Qian, LI Wen-Hua.
作为世界上最重要的粮食作物之一, 水稻起源于热带、亚热带, 相较小麦、大麦等作物, 对低温更加敏感[1]。冷害是全球性自然灾害, 在世界许多国家均有发生, 尤以澳大利亚南部、日本、巴西、韩国、朝鲜为甚, 大规模冷害的发生会造成数十上百亿公斤粮食的损失[2,3,4,5,6]。我国多数稻作区均有低温冷害发生, 在华南和长江中下游的双季稻区, 春季的“倒春寒”和秋季的“寒露风”常常引起低温冷害[7]。东北地区平均3~4年就会遭遇一次大范围的冷害, 造成水稻大量减产, 危害粮食安全[8]。水稻萌发期、芽期、苗期、孕穗期、开花期和灌浆期易受低温冷害影响[9,10], 与产量有直接或间接关系。20世纪90年代以来, 我国的水稻生产发生了较大的变化。一方面, 水稻生产目标从追求产量向降低成本、提高经济效益转变; 另一方面, 随着我国经济的快速发展, 农村主要劳动力向城镇转移, 农村劳动力紧缺已成为水稻生产发展的主要限制因素。在这样的背景下, 水稻直播由于省时、省工和节约成本, 深得农民的欢迎, 并迅速推广。据不完全统计, 水稻直播面积在安徽省已达56.7万公顷, 湖北省为49万公顷, 江西省为43.3万公顷, 江苏省为69.3万公顷, 浙江省为37.3万公顷, 广东省为10万公顷, 黑龙江省的直播稻面积为40万公顷[11,12]。华南双季稻区早稻直播和东北高寒稻区的单季稻直播往往会遇到春季低温天气, 造成缺苗断垄, 导致严重减产[9,10,11,12]。这就要求用于直播的水稻种子有较强的芽期耐低温能力。而上述稻作区一直以育秧移栽为主, 芽期耐低温能力长期不作为育种目标, 致使许多高产、优质的水稻品种由于芽期耐低温能力弱而不能用作直播稻, 限制了直播稻的生产。因此, 水稻芽期耐冷性是其生长发育过程中不可忽视的重要性状。
自从20世纪30年代, 科研人员开始研究水稻冷害以来, 已在冷害发生的气象学原因、水稻耐冷的生理机制、遗传解析和基因克隆等方面积累了大量的研究成果[1,13-19]。但与萌发期、苗期和孕穗期冷害相比, 人们在水稻芽期耐冷性方面的研究稍显不足, 目前主要开展的是芽期耐冷基因的QTL定位研究。严长杰等[20]利用南京11和巴利拉的DH群体, 在7号染色体上鉴定出1个芽期冷害QTL。Zhang等[21]利用Lemont和特青的重组自交系群体, 在3号、7号和11号染色体上检测到3个控制水稻芽期耐冷性的QTL。乔永利等[22]利用密阳23和吉冷1号的F2:3群体, 在2号、4号和7号染色体上检测到3个芽期耐冷QTL。陈玮等[23]利用Lemont和特青的重组自交系群体, 在1号、3号、7号和11号染色体上检测到4个控制水稻芽期耐冷性的QTL。张露霞等[24]利用Asominori和IR24的重组自交系群体和相应的染色体片段置换系, 在5号和12号染色体上检测到3个芽期耐冷QTL。巩迎军等[25]利用越光和Kasalath的回交重组自交系群体, 在4号、6号和11号染色体上检测到4个芽期耐冷QTL。林静等[26]利用以籼稻9311为受体、粳稻日本晴为供体的染色体片段置换系, 在5号和7号染色体上鉴定出4个芽期耐冷QTL。Baruah等[27]利用栽培稻A58和野生稻W107的重组自交系群体, 在1号、11号和12号染色体上检测到了3个芽期耐冷QTL。Ji等[28]利用TN1和春江06的DH群体, 在2号、4号和11号染色体上鉴定出3个芽期耐冷QTL。周勇等[29]利用籼稻9311为受体、粳稻日本晴为供体的染色体单片段置换系, 在除1号和11号染色体以外的10条染色体上检测出18个芽期耐冷QTL。杨洛淼等[30]利用空育131和东农422的重组自交系群体, 在4号染色体上检测到1个芽期耐冷QTL。朱金燕等[31]利用广陆矮4号为受体, 日本晴为供体的染色体片段置换系, 在1号、6号、8号、9号和10号染色体上检测到8个芽期耐冷QTL。Yang等[32]利用华粳籼74为受体, 南洋占为供体的染色体片段置换系, 在5号和6号染色体上鉴定了2个芽期耐冷QTL。Zhang等[33]利用249份籼稻种质资源材料结合5K水稻芯片进行芽期耐冷的GWAS分析, 在1号、3号、4号、5号、6号、10号和12号染色体上共检测到13个芽期耐冷相关区域。
虽然人们对芽期耐冷性进行了较多的QTL鉴定分析, 而且在水稻的12条染色体上均检测到了QTL的存在, 但鉴定到的QTL多为微效QTL, 进行精细定位和后续研究难度较大。同时, 由于研究所用的遗传群体和试验环境等方面的不同, 研究结果不尽相同, 因此有必要采用与以往不同的遗传群体和更高密度的遗传图谱来对已报道的芽期耐冷QTL进行验证, 并检测新的芽期耐冷位点。本研究利用丽江新团黑谷和沈农265构建的重组自交系群体, 结合重测序技术构建的高密度bin遗传图谱, 挖掘控制芽期耐冷性的QTL, 以期为直播稻的耐冷设计育种提供有用的“基因资源”, 同时为阐明水稻芽期耐冷的遗传和分子机制提供参考。
1 材料与方法
1.1 试验材料
2004年, 以芽期耐冷性强的云南地方粳稻品种丽江新团黑谷(LTH)为母本, 芽期耐冷性弱的北方超级稻品种沈农265 (SN265)为父本配制杂交组合, 2005—2013年分别在辽宁沈阳(2005—2006, 2008—2009)、海南三亚(2008)和黑龙江哈尔滨(2011—2015)三地, 采用单粒传法(single seed descent, SSD)套袋自交11代获得包含144个株系的稳定遗传重组自交系群体(recombinant inbred lines, RILs)。参照Huang等[34]报道的重测序手段, 采用“滑动窗口”法构建了丽江新团黑谷和沈农265的重组自交系群体bin遗传图谱。该bin连锁图谱共包含58,738个重组热点, 平均每个株系405个重组热点。最终产生了2818个重组bin标记(图1-A), 这些标记覆盖了大部分的重组热点(图1-B), 平均每条染色体上覆盖235个bin标记, 标记区间平均为128.8 kb[35]。
图1
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图1丽江新团黑谷-沈农265重组自交系群体的物理图谱(A)和基因型(B)
物理图谱位置基于日本晴参考基因组(MSU Rice Genome Annotation Project Release 7), 黄色表示沈农265; 蓝色表示丽江新团黑谷。
Fig. 1Physical map (A) and genotype (B) of Lijiangxintuanheigu-shennong 265 (LTH-SN265) RIL population
Physical position is based on MSU Rice Genome Annotation Project Release 7 sequence. Yellow: SN265 genotype; Blue: LTH genotype.
1.2 芽期耐冷性评价方法
亲本丽江新团黑谷(LTH)、沈农265 (SN265)和144份重组自交系群体材料存放于种子储藏柜中待用, 储藏条件为温度5°C, 相对湿度为10%。评价芽期耐冷性参照韩龙植等[36]的方法略作修改, 每个株系3次重复, 利用3次的平均值进行后续分析。首先将种子从储藏柜中取出, 在50°C恒温干燥箱内高温处理48 h, 使其充分干燥并打破种子休眠。每次重复挑选饱满粒100粒, 经0.1%的氯化汞溶液消毒10 min, 自来水冲洗3~4次, 去离子水洗涤3次, 于垫双层滤纸的直径9 cm培养皿中, 加入10 mL去离子水。盖上培养皿盖, 在30°C恒温培养箱(上海一恒MGC-250)内催芽2~3 d。然后从恒温箱中取出, 用去离子水冲洗1~2次。从发芽的种子中精心挑选芽长约1~2 cm的种子50粒, 种植于边长2 cm的营养钵盆中, 钵盆中事先放置配制好的营养育苗土。28°C光照培养箱(上海一恒MGC-450HP)内放置1 d后, 转入大型人工气候箱(日本三江平原项目援建霜冻害实验室)内处理48 h, 温度条件为2°C。处理后, 将试验材料转移到温度为20~30°C, 并有阳光的地方, 使其恢复生长, 每天适量浇水。恢复10 d后, 调查秧苗生长状态。按照图2的标准给处理后的秧苗恢复状态赋值, 共分为5级: 恢复后幼芽枯死的赋值为0; 恢复后幼芽停止发育, 出现畸形的赋值为1; 恢复后有一定生长能力, 叶色较淡, 但发育非常缓慢的赋值为2; 恢复后生长能力较强, 叶色淡, 发育较缓慢的赋值为3; 恢复后叶色较深, 能够快速生长的赋值为4。并按照不同级别进行加权计分。芽期耐冷级别=(0×0级的株数+1×1级的株数+2×2级的株数+3×3级的株数+4×4级的株数)/处理的材料总株数。图2
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图2水稻芽期冷害评价等级标准
Fig. 2Evaluation criteria for chilling injury at rice bud stage SN265: Shennong 265; LTH: Lijiangxintuanheigu.
1.3 芽期耐冷性的QTL鉴定和QTL效应分析
采用R/qtl的CIM方法进行QTL定位, 采用mqmpermutation命令进行排列组合1000次的LOD阈值(α=0.05)确定, 当实际求得的LOD值大于LOD阈值时, 就认为该区段存在1个QTL, 其置信区间为LOD峰值向下1个LOD值单位的区间[37]。为了进一步验证本试验定位结果的准确性, 同时评价筛选到的QTL累加效应, 利用“选择作图”策略进行QTL验证和累加效应分析[38]。首先将控制芽期耐冷的其他QTL位点根据分子标记数据进行非增效基因的固定, 以此来避免其他QTL对后续分析的影响, 然后在RILs群体中筛选目标QTL区间的重组交换单株, 进而对QTL进行验证和累加效应分析。
2 结果与分析
2.1 亲本的芽期耐冷性比较及群体的芽期耐冷性分布
课题组前期的研究表明, 丽江新团黑谷与沈农265在多个不同时期的耐冷相关性状上存在显著差异, 丽江新团黑谷具有更优秀的综合耐冷特性。本研究中, 2个亲本在2°C低温处理2 d并恢复10 d后, 沈农265的平均芽期耐冷等级仅为0.813, 而丽江新团黑谷高达3.188 (图3和图4), 差异达到极显著水平。重组自交系群体的芽期耐冷性等级分布范围为0.313~3.688, 平均值为1.699, 标准差为0.767。整体上呈现近似正态分布, 并表现出较强的超亲分离(图4)。图3
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图3亲本丽江新团黑谷和沈农265芽期不经冷处理(A)和冷处理后(B)的比较
Fig. 3Comparison of no cold treatment (A) and cold treatment (B) at bud bursting stage between LTH and SN265
图4
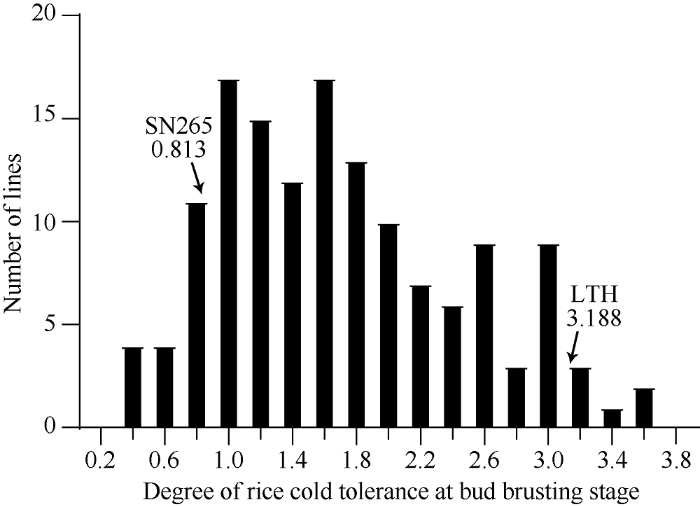 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图4重组自交系群体的芽期耐冷性分布
Fig. 4Distribution of cold tolerance at bud bursting stage in LTH-SN265 RILs
2.2 群体芽期耐冷性的QTL分布与效应
利用R/qtl软件共检测到5个控制水稻芽期耐冷性的QTL, 分布在水稻的1号、3号、9号和11号染色体上, 这些QTL的增效等位基因均来自于丽江新团黑谷(表1和图5)。表型贡献率最大的主效QTL是qCTB11b (LOD值为24.01), 位于11号染色体长臂端的21.24 Mb~22.03 Mb之间, 区间大小0.79 Mb。另外两个贡献率大于15%的主效QTL是qCTB9 (LOD值为5.36)和qCTB11a (LOD值为8.58), 分别位于水稻第9染色体上19.90 Mb~22.30 Mb之间和11号染色体上8.53 Mb~9.93 Mb之间, 区间大小分别为2.40 Mb和1.40 Mb。两个微效QTL是qCTB1 (LOD值为3.26)和qCTB3 (LOD值为3.05), 分别位于水稻1号染色体上0.43 Mb~0.93 Mb之间和3号染色体上29.40 Mb~32.87 Mb之间, 区间大小分别为0.50 Mb和0.61 Mb。没能从沈农265中检测到芽期耐冷性的增效等位QTL。Table 1
表1
表1利用丽江新团黑谷-沈农265的重组自交系群体检测的芽期耐冷QTL信息
Table 1
| 数量性状 基因座 QTL | 染色体 Chr. | 峰值位置a Peak pos.a | 峰值标记 Peak marker | QTL区间QTL interval | LOD值 LOD value | 表型贡献率 Var. (%) | 加性 效应 Add. | 增效等位 基因来源 Positive allele | |
|---|---|---|---|---|---|---|---|---|---|
| 物理图谱 Physical (Mb) | 定位区间 Location interval (Mb) | ||||||||
| qCTB1 | 1 | 0.73 | Bin01-002 | 0.43-0.93 | 0.50 | 3.26 | 8.20 | 0.22 | LTH |
| qCTB3 | 3 | 29.40 | Bin03-281 | 29.40-32.87 | 3.47 | 3.05 | 8.00 | 0.21 | LTH |
| qCTB9 | 9 | 21.45 | Bin09-185 | 19.90-22.30 | 2.40 | 5.36 | 16.60 | 0.34 | LTH |
| qCTB11a | 11 | 9.71 | Bin11-087 | 8.53-9.93 | 1.40 | 8.58 | 24.10 | 0.39 | LTH |
| qCTB11b | 11 | 21.80 | Bin11-141 | 21.24-22.03 | 0.79 | 24.01 | 53.50 | 0.64 | LTH |
新窗口打开|下载CSV
图5
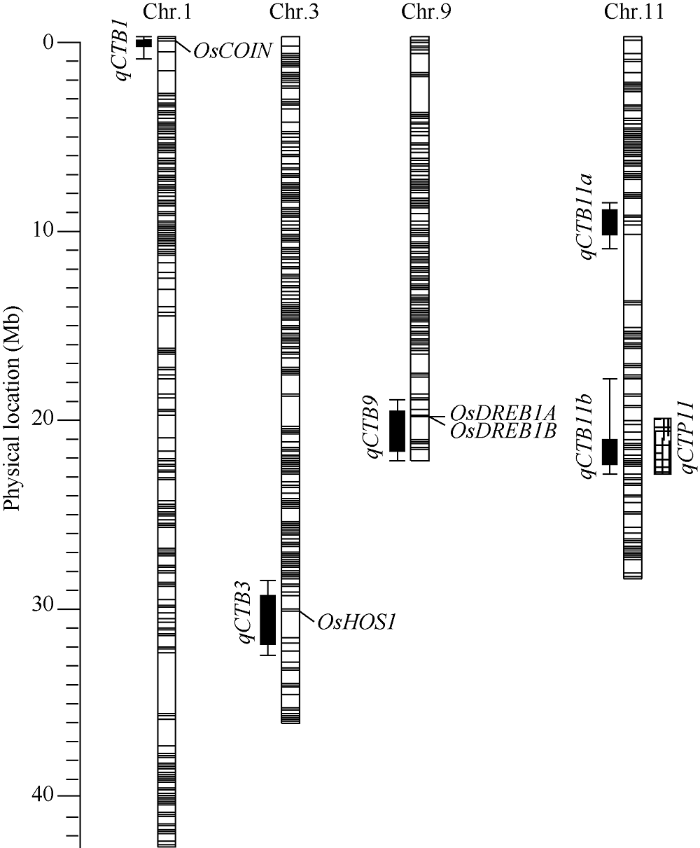 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图5控制芽期耐冷性的QTL在染色体上的位置分布
OsCOIN: 低温诱导的锌指蛋白[46]; OsHOS1: 参与冷胁迫的E3泛素连接酶[47]; OsDREB1A, OsDREB1B: 低温诱导的AP2/ EREBP转录因子基因[48]; qCTP11: 芽期耐冷QTL[27]。
Fig. 5Chromosome location of putative QTL for cold tolerance at bud bursting stage in LTH-SN265 RILs
OsCOIN: cold inducible zinc finger protein[46]; OsHOS1: a rice E3-Ubiquitin Ligase in the modulation of cold stress response[47]; OsDREB1A, OsDREB1B: cold inducible AP2/EREBP transcription factor gene[48]; qCTP11: a QTL for cold tolerance at bud stage[27].
2.3 芽期耐冷性QTL的验证和累加效应分析
我们对检测到的芽期耐冷QTL进行了验证并评价了这些QTL的互作和累加效应。首先, 将控制芽期耐冷性的其他QTL利用分子标记辅助进行非增效基因的固定, 然后在重组自交系群体中筛选目标区间(待评价QTL区间)为增效等位基因的单株, 进而对这5个QTL进行验证。随后, 利用类似的策略, 通过标记辅助选择出不同QTL组合的单株, 进行QTL的互作分析和累加效应评价。不导入任何芽期耐冷QTL株系的芽期低温抗性等级为0.80 (图6和图7-A, X), 含有1个QTL株系的芽期低温抗性等级从1.03到2.33 (图6和图7-C~F), 含有qCTB11b的株系具有最高的芽期低温抗性等级(图6和图7-G)。含有2个QTL株系的芽期低温抗性等级从1.67到2.82 (图6和图7-H~M), 含有qCTB11b株系的芽期低温抗性等级普遍较高(2.44~2.82), 2个QTL之间的作用总体上表现为简单的加性效应, 互作效应较弱。由于受群体规模的影响, 并没有覆盖到全部的3个QTL聚合类型, 筛选到的6个类型含有3个QTL株系的芽期低温抗性等级从1.33到2.98 (图6和图7-N~S), 与两个QTL聚合的规律类似, 含有qCTB11b株系的芽期低温抗性等级普遍较高(2.52~2.98), 3个QTL之间的作用总体上也表现为简单的加性效应。4个QTL聚合的株系检测到3个类型, 均含有qCTB11a和qCTB11b, 它们的芽期低温抗性等级从3.00到3.06 (图6和图7-T~V)。共检测到同时聚合5个QTL的株系8个, 它们的芽期低温抗性等级从2.94到3.50 (图6和图7-W)。通过上述分析, 基本确定了芽期耐低温QTL在重组自交系群体中聚合时主要表现为加性效应。图6
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图6芽期耐冷QTL的验证和聚合效应分析使用的重组单株基因型
“-”: 缺失。
Fig. 6Genotype of recombinant plant for validating cold tolerance QTL and analyzing pyramiding effect at bud busting stage
“-”: missing.
图7
 新窗口打开|下载原图ZIP|生成PPT
新窗口打开|下载原图ZIP|生成PPT图7控制芽期耐冷性的QTL效应及在LTH-SN265重组自交系群体中的聚合效果分析
A: 亲本沈农265 (SN265); B: 亲本丽江新团黑谷(LTH); C~G: qCTB1、qCTB3、qCTB9、qCTB11a和qCTB11b的芽期耐冷效应; H: qCTB1和qCTB3的芽期耐冷聚合效应; I: qCTB3和qCTB9的芽期耐冷聚合效应; J: qCTB9和qCTB11a的芽期耐冷聚合效应; K: qCTB11a和qCTB11b的芽期耐冷聚合效应; L: qCTB1和qCTB11b的芽期耐冷聚合效应; M: qCTB9和qCTB11b的芽期耐冷聚合效应; N: qCTB1、qCTB3和qCTB9的芽期耐冷聚合效应; O: qCTB3、qCTB9和qCTB11a的芽期耐冷聚合效应; P: qCTB1、qCTB3和qCTB11a的芽期耐冷聚合效应; Q: qCTB1、qCTB11a和qCTB11b的芽期耐冷聚合效应; R: qCTB3、qCTB11a和qCTB11b的芽期耐冷聚合效应; S: qCTB9、qCTB11a和qCTB11b的芽期耐冷聚合效应; T: qCTB1、qCTB3、qCTB11a和qCTB11b的芽期耐冷聚合效应; U: qCTB1、qCTB9、qCTB11a和qCTB11b的芽期耐冷聚合效应; V: qCTB3、qCTB9、qCTB11a和qCTB11b的芽期耐冷聚合效应; W: qCTB1、qCTB3、qCTB9、qCTB11a和qCTB11b的芽期耐冷聚合效应; X: 无QTL的株系表型。
Fig. 7Effect of cold tolerance QTL and pyramiding effect at bud stage in LTH-SN265 RILs
A: Shennong 265 (SN265); B: Lijiangxintuanheigu (LTH); C-G: Cold tolerance effect of qCTB1, qCTB3, qCTB9, qCTB11a, and qCTB11b at bud stage; H: Cold tolerance cluster effect of qCTB1 and qCTB3 at bud stage; I: Cold tolerance cluster effect of qCTB3 and qCTB9 at bud stage; J: Cold tolerance cluster effect of qCTB9 and qCTB11a at bud stage; K: Cold tolerance cluster effect of qCTB11a and qCTB11b at bud stage; L: Cold tolerance cluster effect of qCTB11a and qCTB11b at bud stage; M: Cold tolerance cluster effect of qCTB9 and qCTB11b at bud stage; N: Cold tolerance cluster effect of qCTB1, qCTB3, and qCTB9 at bud stage; O: Cold tolerance cluster effect of qCTB3, qCTB9, and qCTB11a at bud stage; P: Cold tolerance cluster effect of qCTB1, qCTB3, and qCTB11a at bud stage; Q: Cold tolerance cluster effect of qCTB1, qCTB11a, and qCTB11b at bud stage; R: Cold tolerance cluster effect of qCTB3, qCTB11a, and qCTB11b at bud stage; S: Cold tolerance cluster effect of qCTB9, qCTB11a, and qCTB11b at bud stage; T: Cold tolerance cluster effect of qCTB1, qCTB3, qCTB11a, and qCTB11b at bud stage; U: Cold tolerance cluster effect of qCTB1, qCTB9, qCTB11a and qCTB11b at bud stage; V: Cold tolerance cluster effect of qCTB3, qCTB9, qCTB11a, and qCTB11b at bud stage; W: Cold tolerance cluster effect of qCTB1, qCTB3, qCTB9, qCTB11a, and qCTB11b at bud stage; X: none QTL line.
3 讨论
3.1 水稻芽期耐冷性评价方法的比较
水稻芽期耐冷能力是决定水稻直播成苗率和整齐度的重要农艺性状, 尤其是在北方稻区和南方双季稻早稻进行直播栽培的重要抗逆性状。近年来, 随着直播技术的改良, 越来越多的地区在水稻直播时选择“芽种直播”或“盲谷播种”, 使得芽期耐冷特性成为决定品种是否适宜直播栽培的关键因素。因此, 准确评价试验材料的芽期耐冷特性就变得越来越重要。芽期耐冷鉴定方法中比较常用的是利用低温培养箱处理萌芽后的种子, 并利用秧苗成活率作为评价指标, 这种方法简单、直观、结果可靠[22,23,24,25,26,27,28,29,30,31,32,33]。本研究对这种方法进行了进一步的改进, 主要包括以下几个方面: 一是低温处理的温度和时间, 前人报道中, 多采用5°C 5 d [22,23,24,25,26,27,28,29,30,31,32,33], 本研究采用了2°C 2 d, 既满足了低温处理的低温条件, 又缩短了处理时间。二是低温处理后的秧苗培养方式, 前人多使用全程水培[22,23,24,25], 本研究选择土培, 这样更接近水稻直播生产实际。三是芽期冷害等级的评价方式, 前人多是简单地将处理缓苗后的秧苗成活率作为芽期耐冷性鉴定的指标[22,23,24,25], 本研究为了更全面地反映每一个株系的综合抗性, 采用了不同级别加权合计的方式评价某一株系整体的耐冷水平。通过上述改进, 不仅缩短了处理时间, 而且还提高了鉴定的精准度。3.2 重测序技术和高密度bin标记遗传图谱在水稻QTL分析中的应用
随着重测序成本的大幅下降和测序数据利用技术的日益成熟, 利用重测序技术结合“滑动窗口”法构建bin遗传图谱已成为水稻[39,40,41]、玉米[42,43]、高粱[44]等作物农艺性状遗传研究的重要技术手段。与传统的分子标记相比, 重测序获得的SNP标记具有高基因组覆盖度和高密度的优势, 它不仅显著提高了QTL鉴定和定位的准确性和精确性, 而且能够为后续的基因功能研究提供相对较小的染色体区间参考。本研究利用包含2818个bin标记的遗传图谱分别将控制水稻芽期耐冷性的QTL定位到了0.50 Mb~2.40 Mb的区间内, 最小的物理区间分别是500 kb (qCTB1)和790 kb (qCTB11b), 相对较大的物理区间也仅分别是1.40 Mb (qCTB11a)、2.40 Mb (qCTB9)和3.47 Mb (qCTB3), 这远远小于传统分子标记高达5~10 cM的定位区间(约4 Mb~10 Mb)。500 kb左右的QTL定位区间虽然离直接完成基因克隆尚有一定的差距[45], 但是也为后续的精细定位提供了足够小的物理区间。此外, 传统的QTL分析作图群体主要是籼-粳亚种间杂交得到的, 少数为籼-籼交群体, 很少使用两个粳稻作为亲本材料的遗传群体, 主要是因为粳稻亚种内的多态性远远小于籼稻亚种内, 与籼-粳亚种间的多态性相比, 更是差距极大[46]。重测序技术成为解决这一问题的有效手段, 使得利用粳-粳交群体也可以构建高密度的遗传图谱, 为研究粳稻食味品质、孕穗期障碍型冷害等要求双亲均为粳稻的实验提供了极大的便利。3.3 耐冷QTL全部来自LTH的可能原因及利用RIL群体进行QTL聚合评估的可行性
本研究中发现SN265和LTH构建的重组自交系群体存在芽期耐冷性的超亲分离, 但检测到的耐冷QTL全部来自LTH, 其原因主要是QTL检测时, 为了尽可能降低假阳性, 使用了相对较高的LOD阈值(3.0)。如果将LOD阈值降低到2.4, 能检测到一个可能的QTL-qCTB7, 其LOD值仅为2.45, 表型贡献率很小, 仅为5.8%, 加性效应值为-0.13。该QTL的耐冷增效等位基因来自沈农265, 但由于表型贡献率太小, 还需要构建该QTL的高世代回交群体进行真伪验证。此外, 对检测的QTL进行效应评价, 尤其是对QTL进行聚合效果分析是QTL用于水稻育种实践的重要前提。但是, 一般而言, 由于重组自交系中导入片段相对较多, 排除遗传背景的噪音影响难度很大, 很可能影响QTL累加效应评估。所以, 重组自交系较少用于QTL的累加效应评估。本研究采用了“选择作图”的方法进行QTL的累加效应分析, 该方法的关键就是充分发挥已构建的高密度物理图谱和详细的群体基因型数据将目标QTL以外的其他QTL进行基因型固定, 以尽可能排除遗传背景的影响。我们之前也成功利用这一方法分析了水稻根系的QTL聚合效果[38]。当然, 以选择作图分析QTL聚合效果的能力肯定不如近等基因系或染色体片段代换系, 但在没有更高级的遗传群体时, 仍不失为一种有效手段。3.4 本研究鉴定的水稻芽期耐低温QTL与其他研究结果的比较和育种应用潜力
随着农村劳动力的大规模转移和农业生产成本的不断攀升, 水稻直播生产越来越受到黑龙江省地广人稀的农业主产区农户的重视。但是, 由于独特的寒地生态环境, 黑龙江水稻直播不得不面对春季的低温环境。水稻芽期耐冷性是在低温下水稻幼芽细胞维持生活的能力, 从外观上表现为幼芽诱发和维持绿苗的能力, 是水稻直播必具备的重要农艺性状[22]。本文从芽期耐冷性强的云南地方粳稻品种丽江新团黑谷中, 鉴定出5个控制芽期耐低温能力的QTL, 同QTL数据库(Q-TARO: http://qtaro.abr.affrc. go.jp/)搜集整理的耐冷相关QTL比对发现, qCTB1附近有一个已知的受冷诱导的锌指蛋白基因OsCOIN, 该基因过量表达可以增强水稻耐冷性, 同时增加脯氨酸含量[47]。qCTB3区间内包含一个已知的耐冷相关基因OsHOS1, 它通过与ICE1互作来参与水稻的冷胁迫反应[48]。qCTB9区间内包含2个成簇存在的耐冷相关基因, 分别是OsDREB1A和OsDREB1B, 二者均受冷诱导, 参与水稻的冷胁迫反应[49]。qCTB11a附近没有发现已经鉴定的冷害相关QTL和基因, 是一个新发现的位点。主效QTL- qCTB11b与Baruah等[27]鉴定的芽期耐冷QTL- qCTP11位于相同的区间, qCTP11来源于北海道地方品种A58。综上可以看出, 控制水稻芽期耐冷性的QTL在不同遗传背景条件下具有一定的重演性, 而且第11染色体上鉴定的主效位点qCTB11b具有较大的后续研究价值。我们也利用“选择作图”的方法评价了这些QTL的育种潜力和聚合效果, 发现芽期耐低温QTL主要表现为加性效应, 非常适于水稻的分子辅助育种。而且主效QTL-qCTB11b不仅自身具有很大的遗传效应, 而且在与其他QTL聚合时, 常常表现出更加突出的改良效果, 极具育种应用价值。4 结论
构建了丽江新团黑谷和沈农265重组自交系群体包含2818个bin标记的遗传图谱。利用此群体和bin标记图谱, 共检测到5个芽期耐冷QTL, 分布在水稻的1号、3号、9号和11号染色体上, 表型贡献率分别为8.0%、8.2%、16.6%、24.4%和53.5%, 增效等位基因均来自耐冷亲本丽江新团黑谷。主效QTL-qCTB11b位于11号染色体长臂端的17.97 Mb~23.03 Mb之间。利用“选择作图”策略进行了QTL验证和累加效应分析, 明确了可以通过QTL的累加聚合实现芽期耐冷能力的遗传改良, 聚合的增效QTL越多, 耐冷能力等级越高。参考文献 原文顺序
文献年度倒序
文中引用次数倒序
被引期刊影响因子
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.5923/j.food.20120205.05URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 1]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 2]
[本文引用: 1]
[本文引用: 1]
DOI:10.1073/pnas.0805303105URLPMID:18719107

Tolerance to abiotic stress is an important agronomic trait in crops and is controlled by many genes, which are called quantitative trait loci (QTLs). Identification of these QTLs will contribute not only to the understanding of plant biology but also for plant breeding, to achieve stable crop production around the world. Previously, we mapped three QTLs controlling low-temperature tolerance at the germination stage (called low-temperature germinability). To understand the molecular basis of one of these QTLs, qLTG3-1 (quantitative trait locus for low-temperature germinability on chromosome 3), map-based cloning was performed, and this QTL was shown to be encoded by a protein of unknown function. The QTL qLTG3-1 is strongly expressed in the embryo during seed germination. Before and during seed germination, specific localization of beta-glucuronidase staining in the tissues covering the embryo, which involved the epiblast covering the coleoptile and the aleurone layer of the seed coat, was observed. Expression of qLTG3-1 was tightly associated with vacuolation of the tissues covering the embryo. This may cause tissue weakening, resulting in reduction of the mechanical resistance to the growth potential of the coleoptile. These phenomena are quite similar to the model system of seed germination presented by dicot plants, suggesting that this model may be conserved in both dicot and monocot plants.
URLPMID:30120236
DOI:10.1016/j.plantsci.2010.04.004URL
DOI:10.1038/ncomms14788URLPMID:28332574

Low temperature is a major factor limiting rice productivity and geographical distribution. Improved cold tolerance and expanded cultivation to high-altitude or high-latitude regions would help meet growing rice demand. Here we explored a QTL for cold tolerance and cloned the gene, CTB4a (cold tolerance at booting stage), encoding a conserved leucine-rich repeat receptor-like kinase. We show that different CTB4a alleles confer distinct levels of cold tolerance and selection for variation in the CTB4a promoter region has occurred on the basis of environmental temperature. The newly generated cold-tolerant haplotype Tej-Hap-KMXBG was retained by artificial selection during temperate japonica evolution in cold habitats for low-temperature acclimation. Moreover, CTB4a interacts with AtpB, a beta subunit of ATP synthase. Upregulation of CTB4a correlates with increased ATP synthase activity, ATP content, enhanced seed setting and improved yield under cold stress conditions. These findings suggest strategies to improve cold tolerance in crop plants.
DOI:10.1016/j.cell.2015.01.046URLPMID:25728666

Rice is sensitive to cold and can be grown only in certain climate zones. Human selection of japonica rice has extended its growth zone to regions with lower temperature, while the molecular basis of this adaptation remains unknown. Here, we identify the quantitative trait locus COLD1 that confers chilling tolerance in japonica rice. Overexpression of COLD1(jap) significantly enhances chilling tolerance, whereas rice lines with deficiency or downregulation of COLD1(jap) are sensitive to cold. COLD1 encodes a regulator of G-protein signaling that localizes on plasma membrane and endoplasmic reticulum (ER). It interacts with the G-protein alpha subunit to activate the Ca(2+) channel for sensing low temperature and to accelerate G-protein GTPase activity. We further identify that a SNP in COLD1, SNP2, originated from Chinese Oryza rufipogon, is responsible for the ability of COLD(jap/ind) to confer chilling tolerance, supporting the importance of COLD1 in plant adaptation.
DOI:10.1073/pnas.1819769116URLPMID:30808744 [本文引用: 1]

Rice (Oryza sativa L.) is a chilling-sensitive staple crop that originated in subtropical regions of Asia. Introduction of the chilling tolerance trait enables the expansion of rice cultivation to temperate regions. Here we report the cloning and characterization of HAN1, a quantitative trait locus (QTL) that confers chilling tolerance on temperate japonica rice. HAN1 encodes an oxidase that catalyzes the conversion of biologically active jasmonoyl-L-isoleucine (JA-Ile) to the inactive form 12-hydroxy-JA-Ile (12OH-JA-Ile) and fine-tunes the JA-mediated chilling response. Natural variants in HAN1 diverged between indica and japonica rice during domestication. A specific allele from temperate japonica rice, which gained a putative MYB cis-element in the promoter of HAN1 during the divergence of the two japonica ecotypes, enhances the chilling tolerance of temperate japonica rice and allows it to adapt to a temperate climate. The results of this study extend our understanding of the northward expansion of rice cultivation and provide a target gene for the improvement of chilling tolerance in rice.
URL [本文引用: 1]

利用籼稻品种南京11和粳稻品种巴利拉的F1花药进行组织培养,获得67个加倍单倍体植株(Doubled haploid, DH)。 以该群体构建了一张包含131个RFLP标记的水稻分子图谱。 以芽期的死苗率为指标, 评定亲本及各 DH系在低温(4~5℃)条件下的耐冷性。结果表明:死苗率在DH群体中呈双峰连续分布, 表明芽期耐冷性是由主基因控制的数量性状。将死苗率作为数量性状进行QTL的区间作图分析, 发现在第7染色体上G379b-RG4区间存在有与耐冷性有关的基因(Cts7) 。
URL [本文引用: 1]

利用籼稻品种南京11和粳稻品种巴利拉的F1花药进行组织培养,获得67个加倍单倍体植株(Doubled haploid, DH)。 以该群体构建了一张包含131个RFLP标记的水稻分子图谱。 以芽期的死苗率为指标, 评定亲本及各 DH系在低温(4~5℃)条件下的耐冷性。结果表明:死苗率在DH群体中呈双峰连续分布, 表明芽期耐冷性是由主基因控制的数量性状。将死苗率作为数量性状进行QTL的区间作图分析, 发现在第7染色体上G379b-RG4区间存在有与耐冷性有关的基因(Cts7) 。
DOI:10.1016/j.plantsci.2004.09.021URL [本文引用: 1]
URL [本文引用: 6]

以籼粳交密阳23号/吉冷1号的F2:3 代200个家系作为作图群体,构建了一张含有97个微卫星 (SSR)标记的分子连锁图谱。在5℃低温条件下,对F3家系进行芽期耐冷性鉴定,并利用SSR标记进行了芽期耐冷性数量性状位点(QTL)分析。研究结果表明,芽期耐冷性在F3家系群中呈单峰连续分布,表现为由多基因控制的数量性状;共检测到与芽期耐冷性有关的QTL 3个,分别位于第2、4 和7染色体上,对表型变异的贡献率范围为11.5%~20.5%。其中,位于第4染色体RM273~RM303的qCTBP4对表型变异的贡献率最大。
URL [本文引用: 6]

以籼粳交密阳23号/吉冷1号的F2:3 代200个家系作为作图群体,构建了一张含有97个微卫星 (SSR)标记的分子连锁图谱。在5℃低温条件下,对F3家系进行芽期耐冷性鉴定,并利用SSR标记进行了芽期耐冷性数量性状位点(QTL)分析。研究结果表明,芽期耐冷性在F3家系群中呈单峰连续分布,表现为由多基因控制的数量性状;共检测到与芽期耐冷性有关的QTL 3个,分别位于第2、4 和7染色体上,对表型变异的贡献率范围为11.5%~20.5%。其中,位于第4染色体RM273~RM303的qCTBP4对表型变异的贡献率最大。
[本文引用: 5]
[本文引用: 5]
[本文引用: 5]
[本文引用: 5]
[本文引用: 5]
[本文引用: 5]
DOI:10.3969/j.issn.1001-7216.2010.03.004URL [本文引用: 3]

以籼稻品种9311为受体、粳稻品种日本晴为供体构建的95个染色体片段置换系为材料, 在5℃低温条件下进行芽期耐冷性鉴定。结果表明,6个置换系低温处理后的成苗率与受体亲本9311有一定差异,其耐冷性略强于9311。利用代换作图法共鉴定出4个与芽期耐冷性相关的QTL,分别位于水稻第5和第7染色体上。其中qCTB51、qCTB52和qCTB53分别被定位在第5染色体RM267与RM1237、RM2422与RM6054及RM3321与RM1054之间遗传距离分别为21.3 cM、27.4 cM和12.7 cM的置换片段上;qCTB7被定位在第7染色体RM11-RM2752区间遗传距离为6.8 cM的置换片段上。
DOI:10.3969/j.issn.1001-7216.2010.03.004URL [本文引用: 3]

以籼稻品种9311为受体、粳稻品种日本晴为供体构建的95个染色体片段置换系为材料, 在5℃低温条件下进行芽期耐冷性鉴定。结果表明,6个置换系低温处理后的成苗率与受体亲本9311有一定差异,其耐冷性略强于9311。利用代换作图法共鉴定出4个与芽期耐冷性相关的QTL,分别位于水稻第5和第7染色体上。其中qCTB51、qCTB52和qCTB53分别被定位在第5染色体RM267与RM1237、RM2422与RM6054及RM3321与RM1054之间遗传距离分别为21.3 cM、27.4 cM和12.7 cM的置换片段上;qCTB7被定位在第7染色体RM11-RM2752区间遗传距离为6.8 cM的置换片段上。
DOI:10.1007/s10681-008-9753-yURL [本文引用: 6]

The present study was conducted to understand the pattern of variation and the genetic bases for cold tolerance at the early growth stage in Asian rice. The genetic variation was investigated at the germination, plumule and seedling stages among 57 strains including cultivated rice (Oryza sativa ssp. indica and ssp. japonica) and its wild progenitor (Oryza rufipogon). The significant differentiation of cold tolerance was observed among the taxonomically divided groups. At the germination stage, both indica and japonica subspecies tended to be more tolerant than O. rufipogon, whereas at the plumule and seedling stages, ssp. japonica tended to be more tolerant than ssp. indica and O. rufipogon. Furthermore, in cold tolerance at the plumule stage, the clinal variation across the latitude of origins was observed within O. rufipogon and ssp. japonica, suggesting that the current pattern of variation seems to have been shaped by both their phylogenetic histories and on-going adaptation to the local environments. QTL analysis between O. sativa ssp. japonica (tolerant) and O. rufipogon (susceptible) revealed five putative QTLs for cold tolerance at the plumule and seedling stages but not at the germination stage. Substitution mapping was also carried out to precisely locate the two major QTLs for cold tolerance at the plumule stage, which could be used for improvement of tolerance to cold stress in ssp. indica.
DOI:10.1016/S1672-6308(09)60028-7URL [本文引用: 3]

A doubled haploid (DH) population consisted of 120 lines, derived from a cross between an indica variety, TN1, and a japonica variety, Chunjiang 06, was used to identify QTLs controlling rice cold tolerance at the plumule and 3-leaf-seedling stages by using the QTLNetwork software. The percentages of normal plumules after 4°C treatments for 7 d, 9 d, 11 d, and 14 d, as well as the cold stress tolerance index (CSTI) and the withering index (WI) of rice seedling were investigated. A total of five single-effect QTLs, each for percentages of normal plumules after 4°C treatments for 9 d, 11 d and 14 d, and CSTI and WI, respectively were identified. The QTLs for the percentages of normal plumules after low temperature treatments for 9 d, 11 d and 14 d were on chromosomes 4, 2 and 11, accounting for 14.1%, 17.3% and 21.5% of the phenotypic variation, respectively. QTLs for CSTI and WI were on chromosomes 10 and 1, respectively. Two pairs of epistatic loci were identified, but none of the epistatic loci involved the single-effect QTLs. The RM528–RM340 interval on chromosome 6 interacted with the RM278–RM3919B interval on chromosome 9 for CSTI, and the epistatic interaction accounted for 17.7% of the phenotypic variation. A pair of epistatic loci was identified for WI, the RM246–RM5461 interval on chromosome 1 interacted with the ISA–RM447 interval on chromosome 8, which accounted for 22.6% of the phenotypic variation.
DOI:10.3969/j.issn.10017216.2013.04.007URL [本文引用: 3]

对籼型两系骨干亲本9311(受体)和粳稻日本晴(供体)构建的单片段代换系群体,在芽期和苗期分别进行低温处理,以芽期处理后的恢复成活率和苗期卷叶程度作为耐冷性的评价指标进行耐冷性评价。结果表明,日本晴在芽期和苗期耐冷性极显著强于9311,不同代换系芽期和苗期耐冷性也存在极显著差异,芽期和苗期耐冷性较强的代换系X724、X732、X733较好地保持了9311的农艺性状,其产量等性状的一般配合力优于9311;鉴定出18个芽期耐冷性QTL和5个苗期耐冷性QTL,分布于12条染色体上的20个代换片段中,第12染色体的RM1261-RM519和RM17区域同时检测出芽期和苗期耐冷性QTL;芽期耐冷性QTL qCTP9、qCTP11.2、qCTP12.1和苗期耐冷性QTL qCTS1.1、qCTS1.2具有较大加性效应,对应代换系耐冷性较强。
DOI:10.3969/j.issn.10017216.2013.04.007URL [本文引用: 3]

对籼型两系骨干亲本9311(受体)和粳稻日本晴(供体)构建的单片段代换系群体,在芽期和苗期分别进行低温处理,以芽期处理后的恢复成活率和苗期卷叶程度作为耐冷性的评价指标进行耐冷性评价。结果表明,日本晴在芽期和苗期耐冷性极显著强于9311,不同代换系芽期和苗期耐冷性也存在极显著差异,芽期和苗期耐冷性较强的代换系X724、X732、X733较好地保持了9311的农艺性状,其产量等性状的一般配合力优于9311;鉴定出18个芽期耐冷性QTL和5个苗期耐冷性QTL,分布于12条染色体上的20个代换片段中,第12染色体的RM1261-RM519和RM17区域同时检测出芽期和苗期耐冷性QTL;芽期耐冷性QTL qCTP9、qCTP11.2、qCTP12.1和苗期耐冷性QTL qCTS1.1、qCTS1.2具有较大加性效应,对应代换系耐冷性较强。
[本文引用: 3]
[本文引用: 3]
DOI:10.3724/SP.J.1259.2015.00338URL [本文引用: 3]

水稻(Oryza sativa)芽期耐冷性是其生长发育过程中不可忽视的重要数量性状, 易受遗传背景的干扰和环境因素的影响; 利用单片段代换系(SSSLs)能减少遗传背景的干扰。该研究以85个单片段代换系为材料, 其受体亲本为广陆矮4号, 供体亲本为日本晴。通过单因素方差分析和Dunnett’s多重比较, 分析单片段代换系与受体亲本之间芽期耐冷性的差异, 并对代换片段上的芽期耐冷QTL进行鉴定。以P≤0.001为阈值共检测到8个芽期耐冷QTL, 分别分布在第1、6、8、9和10号染色体上, 其中4个QTL通过代换作图被初步定位。这些QTL加性效应均表现为增效作用, 在2个年度间其加性效应值的变化范围分别为14%-44%和10%-45%, 加性效应百分率的变化范围分别为700% -2 200%和500%-2 250%, 其中qCTP9-2在2个年度间的加性效应均最高, 分别为44%和45%。研究结果对进一步发掘和利用新的水稻芽期耐冷QTL具有重要意义。
DOI:10.3724/SP.J.1259.2015.00338URL [本文引用: 3]

水稻(Oryza sativa)芽期耐冷性是其生长发育过程中不可忽视的重要数量性状, 易受遗传背景的干扰和环境因素的影响; 利用单片段代换系(SSSLs)能减少遗传背景的干扰。该研究以85个单片段代换系为材料, 其受体亲本为广陆矮4号, 供体亲本为日本晴。通过单因素方差分析和Dunnett’s多重比较, 分析单片段代换系与受体亲本之间芽期耐冷性的差异, 并对代换片段上的芽期耐冷QTL进行鉴定。以P≤0.001为阈值共检测到8个芽期耐冷QTL, 分别分布在第1、6、8、9和10号染色体上, 其中4个QTL通过代换作图被初步定位。这些QTL加性效应均表现为增效作用, 在2个年度间其加性效应值的变化范围分别为14%-44%和10%-45%, 加性效应百分率的变化范围分别为700% -2 200%和500%-2 250%, 其中qCTP9-2在2个年度间的加性效应均最高, 分别为44%和45%。研究结果对进一步发掘和利用新的水稻芽期耐冷QTL具有重要意义。
DOI:10.1007/s11032-016-0520-9URL [本文引用: 3]
DOI:10.1007/s00299-017-2247-4URLPMID:29322237 [本文引用: 3]

KEY MESSAGE: A region containing three genes on chromosome 1 of indica rice was associated with cold tolerance at the bud burst stage; these results may be useful for breeding cold-tolerant lines. Low temperature at the bud burst stage is one of the major abiotic stresses limiting rice growth, especially in regions where rice seeds are sown directly. In this study, we investigated cold tolerance of rice at the bud burst stage and conducted a genome-wide association study (GWAS) based on the 5K rice array of 249 indica rice varieties widely distributed in China. We improved the method to assess cold tolerance at the bud burst stage in indica rice, and used severity of damage (SD) and seed survival rate (SR) as the cold-tolerant indices. Population structure analysis demonstrated that the Chinese indica panel was divided into three subgroups. In total, 47 significant single-nucleotide polymorphism (SNP) loci associated with SD and SR, were detected by association mapping based on mixed linear model. Because some loci overlapped between SD and SR, the loci contained 13 genome intervals and most of them have been reported previously. A major QTL for cold tolerance on chromosome 1 at the position of 31.6 Mb, explaining 13.2% of phenotypic variation, was selected for further analysis. Through LD decay, GO enrichment, RNA-seq data, and gene expression pattern analyses, we identified three genes (LOC_Os01g55510, LOC_Os01g55350 and LOC_Os01g55560) that were differentially expressed between cold-tolerant and cold-sensitive varieties, suggesting they may be candidate genes for cold tolerance. Together, our results provide a new method to assess cold tolerance in indica rice, and establish the foundation for isolating genes related to cold tolerance that could be used in rice breeding.
DOI:10.1101/gr.089516.108URLPMID:19420380 [本文引用: 1]

The next-generation sequencing technology coupled with the growing number of genome sequences opens the opportunity to redesign genotyping strategies for more effective genetic mapping and genome analysis. We have developed a high-throughput method for genotyping recombinant populations utilizing whole-genome resequencing data generated by the Illumina Genome Analyzer. A sliding window approach is designed to collectively examine genome-wide single nucleotide polymorphisms for genotype calling and recombination breakpoint determination. Using this method, we constructed a genetic map for 150 rice recombinant inbred lines with an expected genotype calling accuracy of 99.94% and a resolution of recombination breakpoints within an average of 40 kb. In comparison to the genetic map constructed with 287 PCR-based markers for the rice population, the sequencing-based method was approximately 20x faster in data collection and 35x more precise in recombination breakpoint determination. Using the sequencing-based genetic map, we located a quantitative trait locus of large effect on plant height in a 100-kb region containing the rice
DOI:10.3390/ijms21041284URL [本文引用: 1]
[本文引用: 1]
[本文引用: 1]
DOI:10.1093/bioinformatics/btq565URLPMID:20966004 [本文引用: 1]

MOTIVATION: R/qtl is free and powerful software for mapping and exploring quantitative trait loci (QTL). R/qtl provides a fully comprehensive range of methods for a wide range of experimental cross types. We recently added multiple QTL mapping (MQM) to R/qtl. MQM adds higher statistical power to detect and disentangle the effects of multiple linked and unlinked QTL compared with many other methods. MQM for R/qtl adds many new features including improved handling of missing data, analysis of 10,000 s of molecular traits, permutation for determining significance thresholds for QTL and QTL hot spots, and visualizations for cis-trans and QTL interaction effects. MQM for R/qtl is the first free and open source implementation of MQM that is multi-platform, scalable and suitable for automated procedures and large genetical genomics datasets. AVAILABILITY: R/qtl is free and open source multi-platform software for the statistical language R, and is made available under the GPLv3 license. R/qtl can be installed from http://www.rqtl.org/. R/qtl queries should be directed at the mailing list, see http://www.rqtl.org/list/. CONTACT: kbroman@biostat.wisc.edu.
DOI:10.3969/j.issn.10017216.2014.06.005URL [本文引用: 2]

为揭示水稻移栽后新生根系相关特性的遗传基础,以笹锦、北陆129及其杂交衍生的回交重组自交系群体为材料,构建包含对应的由245个分子标记组成的遗传图谱对水稻移栽后的新生根相关性状进行分析。共检测到10个控制根平均直径、总根长、平均根表面积、平均根长和根数等性状的QTL。这些QTL分布在第1、2、5、9和11染色体上,并在第9和11染色体上成簇分布,表型贡献率为10.7%~28.5%。随后利用“选择作图”策略对主效QTL进行了验证分析。其中,位于第11染色体上标记C477和G320B之间(5 461 121-6 686 166 bp)的同时控制根长、根表面积和根数的QTL qRL11.1,qASA11和qRN11.1是新发现的位点,具有一定的研究与应用价值。
DOI:10.3969/j.issn.10017216.2014.06.005URL [本文引用: 2]

为揭示水稻移栽后新生根系相关特性的遗传基础,以笹锦、北陆129及其杂交衍生的回交重组自交系群体为材料,构建包含对应的由245个分子标记组成的遗传图谱对水稻移栽后的新生根相关性状进行分析。共检测到10个控制根平均直径、总根长、平均根表面积、平均根长和根数等性状的QTL。这些QTL分布在第1、2、5、9和11染色体上,并在第9和11染色体上成簇分布,表型贡献率为10.7%~28.5%。随后利用“选择作图”策略对主效QTL进行了验证分析。其中,位于第11染色体上标记C477和G320B之间(5 461 121-6 686 166 bp)的同时控制根长、根表面积和根数的QTL qRL11.1,qASA11和qRN11.1是新发现的位点,具有一定的研究与应用价值。
DOI:10.1371/journal.pone.0017595URLPMID:21390234 [本文引用: 1]

Huge efforts have been invested in the last two decades to dissect the genetic bases of complex traits including yields of many crop plants, through quantitative trait locus (QTL) analyses. However, almost all the studies were based on linkage maps constructed using low-throughput molecular markers, e.g. restriction fragment length polymorphisms (RFLPs) and simple sequence repeats (SSRs), thus are mostly of low density and not able to provide precise and complete information about the numbers and locations of the genes or QTLs controlling the traits. In this study, we constructed an ultra-high density genetic map based on high quality single nucleotide polymorphisms (SNPs) from low-coverage sequences of a recombinant inbred line (RIL) population of rice, generated using new sequencing technology. The quality of the map was assessed by validating the positions of several cloned genes including GS3 and GW5/qSW5, two major QTLs for grain length and grain width respectively, and OsC1, a qualitative trait locus for pigmentation. In all the cases the loci could be precisely resolved to the bins where the genes are located, indicating high quality and accuracy of the map. The SNP map was used to perform QTL analysis for yield and three yield-component traits, number of tillers per plant, number of grains per panicle and grain weight, using data from field trials conducted over years, in comparison to QTL mapping based on RFLPs/SSRs. The SNP map detected more QTLs especially for grain weight, with precise map locations, demonstrating advantages in detecting power and resolution relative to the RFLP/SSR map. Thus this study provided an example for ultra-high density map construction using sequencing technology. Moreover, the results obtained are helpful for understanding the genetic bases of the yield traits and for fine mapping and cloning of QTLs.
DOI:10.1073/pnas.1306579110URLPMID:23940322 [本文引用: 1]

The growing world population and shrinkage of arable land demand yield improvement of rice, one of the most important staple crops. To elucidate the genetic basis of yield and uncover its associated loci in rice, we resequenced the core recombinant inbred lines of Liang-You-Pei-Jiu, the widely cultivated super hybrid rice, and constructed a high-resolution linkage map. We detected 43 yield-associated quantitative trait loci, of which 20 are unique. Based on the high-density physical map, the genome sequences of paternal variety 93-11 and maternal cultivar PA64s of Liang-You-Pei-Jiu were significantly improved. The large recombinant inbred line population combined with plentiful high-quality single nucleotide polymorphisms and insertions/deletions between parental genomes allowed us to fine-map two quantitative trait loci, qSN8 and qSPB1, and to identify days to heading8 and lax panicle1 as candidate genes, respectively. The quantitative trait locus qSN8 was further confirmed to be days to heading8 by a complementation test. Our study provided an ideal platform for molecular breeding by targeting and dissecting yield-associated loci in rice.
DOI:10.3389/fpls.2017.01223URLPMID:28747923 [本文引用: 1]

Mapping major quantitative trait loci (QTL) responsible for rice seed germinability under low temperature (GULT) can provide valuable genetic source for improving cold tolerance in rice breeding. In this study, 124 rice backcross recombinant inbred lines (BRILs) derived from a cross indica cv. Changhui 891 and japonica cv. 02428 were genotyped through re-sequencing technology. A bin map was generated which includes 3057 bins covering distance of 1266.5 cM with an average of 0.41 cM between markers. On the basis of newly constructed high-density genetic map, six QTL were detected ranging from 40 to 140 kb on Nipponbare genome. Among these, two QTL qCGR8 and qGRR11 alleles shared by 02428 could increase GULT and seed germination recovery rate after cold stress, respectively. However, qNGR1 and qNGR4 may be two major QTL affecting indica Changhui 891germination under normal condition. QTL qGRR1 and qGRR8 affected the seed germination recovery rate after cold stress and the alleles with increasing effects were shared by the Changhui 891 could improve seed germination rate after cold stress dramatically. These QTL could be a highly valuable genetic factors for cold tolerance improvement in rice lines. Moreover, the BRILs developed in this study will serve as an appropriate choice for mapping and studying genetic basis of rice complex traits.
DOI:10.1186/s12864-015-2242-5URLPMID:26691201 [本文引用: 1]

BACKGROUND: To safeguard the food supply for the growing human population, it is important to understand and exploit the genetic basis of quantitative traits. Next-generation sequencing technology performs advantageously and effectively in genetic mapping and genome analysis of diverse genetic resources. Hence, we combined re-sequencing technology and a bin map strategy to construct an ultra-high-density bin map with thousands of bin markers to precisely map a quantitative trait locus. RESULTS: In this study, we generated a linkage map containing 1,151,856 high quality SNPs between Mo17 and B73, which were verified in the maize intermated B73 x Mo17 (IBM) Syn10 population. This resource is an excellent complement to existing maize genetic maps available in an online database (iPlant, http://data.maizecode.org/maize/qtl/syn10/ ). Moreover, in this population combined with the IBM Syn4 RIL population, we detected 135 QTLs for flowering time and plant height traits across the two populations. Eighteen known functional genes and twenty-five candidate genes for flowering time and plant height trait were fine-mapped into a 2.21-4.96 Mb interval. Map expansion and segregation distortion were also analyzed, and evidence for inadvertent selection of early flowering time in the process of mapping population development was observed. Furthermore, an updated integrated map with 1,151,856 high-quality SNPs, 2,916 traditional markers and 6,618 bin markers was constructed. The data were deposited into the iPlant Discovery Environment (DE), which provides a fundamental resource of genetic data for the maize genetic research community. CONCLUSIONS: Our findings provide basic essential genetic data for the maize genetic research community. An updated IBM Syn10 population and a reliable, verified high-quality SNP set between Mo17 and B73 will aid in future molecular breeding efforts.
URLPMID:26593310 [本文引用: 1]
DOI:10.1093/jxb/ers205URLPMID:22859680 [本文引用: 1]

The productivity of sorghum is mainly determined by agronomically important traits. The genetic bases of these traits have historically been dissected and analysed through quantitative trait locus (QTL) mapping based on linkage maps with low-throughput molecular markers, which is one of the factors that hinder precise and complete information about the numbers and locations of the genes or QTLs controlling the traits. In this study, an ultra-high-density linkage map based on high-quality single nucleotide polymorphisms (SNPs) generated from low-coverage sequences (~0.07 genome sequence) in a sorghum recombinant inbred line (RIL) population was constructed through new sequencing technology. This map consisted of 3418 bin markers and spanned 1591.4 cM of genome size with an average distance of 0.5 cM between adjacent bins. QTL analysis was performed and a total of 57 major QTLs were detected for eight agronomically important traits under two contrasting photoperiods. The phenotypic variation explained by individual QTLs varied from 3.40% to 33.82%. The high accuracy and quality of this map was evidenced by the finding that genes underlying two cloned QTLs, Dw3 for plant height (chromosome 7) and Ma1 for flowering time (chromosome 6), were localized to the correct genomic regions. The close associations between two genomic regions on chromosomes 6 and 7 with multiple traits suggested the existence of pleiotropy or tight linkage. Several major QTLs for heading date, plant height, numbers of nodes, stem diameter, panicle neck length, and flag leaf width were detected consistently under both photoperiods, providing useful information for understanding the genetic mechanisms of the agronomically important traits responsible for the change of photoperiod.
[本文引用: 1]
[本文引用: 1]
[本文引用: 3]
DOI:10.1007/s00425-007-0548-5URLPMID:17549515 [本文引用: 3]

Rice (Oryza sativa L.) plant is sensitive to chilling, particularly at early stages of seedling development. Here a novel cold-inducible gene, designated OsCOIN (Oryza sativa cold-inducible), was isolated and characterized. Results showed that OsCOIN protein, a RING finger protein, was localized in both nuclear and cytoplasm membrane. OsCOIN is expressed in all rice organs and strongly induced by low temperature, ABA, salt and drought. Over-expression of OsCOIN in transgenic rice lines significantly enhanced their tolerance to cold, salt and drought, accompanied by an up-regulation of OsP5CS expression and an increase of cellular proline level.
DOI:10.1007/s11103-013-0092-6URLPMID:23780733 [本文引用: 3]

Plants can cope with adverse environmental conditions through the activation of stress response signalling pathways, in which the proteasome seems to play an important role. However, the mechanisms underlying the proteasome-mediated stress response in rice are still not fully understood. To address this issue, we have identified a rice E3-ubiquitin ligase, OsHOS1, and characterized its role in the modulation of the cold stress response. Using a RNA interference (RNAi) transgenic approach we found that, under cold conditions, the RNAi::OsHOS1 plants showed a higher expression level of OsDREB1A. This was correlated with an increased amount of OsICE1, a master transcription factor of the cold stress signalling. However, the up-regulation of OsDREB1A was transient and the transgenic plants did not show increased cold tolerance. Nevertheless, we could confirm the interaction of OsHOS1 with OsICE1 by Yeast-Two hybrid and bi-molecular fluorescence complementation in Arabidopsis protoplasts. Moreover, we could also determine through an in vitro degradation assay that the higher amount of OsICE1 in the transgenic plants was correlated with a lower amount of OsHOS1. Hence, we could confirm the involvement of the proteasome in this response mechanism. Taken together our results confirm the importance of OsHOS1, and thus of the proteasome, in the modulation of the cold stress signalling in rice.
DOI:10.1371/journal.pone.0047275URLPMID:23077584 [本文引用: 1]

The clustered genes C-repeat (CRT) binding factor (CBF)1/dehydration-responsive element binding protein (DREB)1B, CBF2/DREB1C, and CBF3/DREB1A play a central role in cold acclimation and facilitate plant resistance to freezing in Arabidopsis thaliana. Rice (Oryza sativa L.) is very sensitive to low temperatures; enhancing the cold stress tolerance of rice is a key challenge to increasing its yield. In this study, we demonstrate chilling acclimation, a phenomenon similar to Arabidopsis cold acclimation, in rice. To determine whether rice CBF/DREB1 genes participate in this cold-responsive pathway, all putative homologs of Arabidopsis DREB1 genes were filtered from the complete rice genome through a BLASTP search, followed by phylogenetic, colinearity and expression analysis. We thereby identified 10 rice genes as putative DREB1 homologs: nine of these were located in rice genomic regions with some colinearity to the Arabidopsis CBF1-CBF4 region. Expression profiling revealed that six of these genes (Os01g73770, Os02g45450, Os04g48350, Os06g03670, Os09g35010, and Os09g35030) were similarly expressed in response to chilling acclimation and cold stress and were co-expressed with genes involved in cold signalling, suggesting that these DREB1 homologs may be involved in the cold response in rice. The results presented here serve as a prelude towards understanding the function of rice homologs of DREB1 genes in cold-sensitive crops.
