Chen Zhu


Basic Medical School, Guizhou University of Traditional Chinese Medicine, Guiyang 550025, Guizhou Province, China
Received: 27 October 2020; Revised: 14 December 2020; Published online: 21 May 2021
Foundation item: Supported by the Doctoral Fund of Guiyang University of Chinese Medicine (3043-043190047) and by the Science and Technology Innovation Talent Team in Guizhou Province (Qiankehepingtairencai [2020]5010)
Corresponding author: Chen Zhu, Tel/Fax: +86-851-85558309; E-mail: zhuchenln@163.com.
Abstract: [Objective] To study the regulatory effect of TetR family transcription factor Ms0606 on the drug resistance of Mycobacterium smegmatis.[Methods] Firstly, the growth curve was measured to detect the regulatory role of Ms0606 in mycobacterial resistance; electrophoretic mobility shift assay and DNase I footprinting assay were used to identify the conservative sequence recognized by the transcription factor Ms0606, and then explore its potential target genes. Secondly, reverse transcription-qPCR and β-galactosidase activity experiments were used to detect the regulatory effect of Ms0606 on the target gene Ms0608. The potential roles of Ms0606 in the regulation of drug resistance are further discussed.[Results] Overexpressing Ms0606 made mycobacteria more sensitive than the wild-type strain in response to isoniazid, whereas disrupting Ms0606 is resistant to isoniazid; Ms0606 could recognize a conserved 22 bp palindromic motif within the upstream region of its self-operon, using the palindrome sequence to search M. smegmatis genome, it is found that Ms0606 may regulate 5 potential target genes; Reverse transcription-qPCR and β-galactosidase activity experiments revealed Ms0606 acts as a repressor and negatively regulates Ms0608 expression, which may affect mycobacterial antibiotic resistance.[Conclusion] I have identified a new TetR family transcriptional regulator of Mycobacterium smegmatis encoded by Ms0606, which regulates mycobacterial sensitivity to isoniazid, and further characterized its target genes and its regulatory mechanism for mycobacterial resistance.
Keywords: transcription factormycobacteriaisoniazidantibiotic resistance
耻垢分枝杆菌中一个TetR家族转录因子负调控异烟肼的敏感性
朱晨


贵州中医药大学基础医学院, 贵州 贵阳 550025
收稿日期:2020-10-27;修改日期:2020-12-14;网络出版日期:2021-05-21
基金项目:贵州中医药大学博士基金(3043-043190047);贵州省科技创新人才团队(黔科合平台人才[2020]5010)
通信作者:朱晨, Tel/Fax: +86-851-85558309;E-mail: zhuchenln@163.com.
摘要:[目的] 研究TetR家族转录因子Ms0606对耻垢分枝杆菌耐药性的调控作用。[方法] 首先,通过测定生长曲线检测Ms0606在分枝杆菌耐药性中的调控作用;通过凝胶迁移阻滞实验和DNase I足迹法鉴定转录因子Ms0606识别的保守序列,进而探究其潜在的靶基因;其次,利用逆转录-qPCR和β-半乳糖苷酶活性实验检测Ms0606对靶基因Ms0608的调控作用,进一步探讨Ms0606调控分枝杆菌耐药性的分子机制。[结果] 与野生型菌株相比,Ms0606超表达菌株对异烟肼表现为敏感,Ms0606敲除菌株对异烟肼表现为抗性;Ms0606可以识别其自身操纵子上游区域内保守的22 bp回文序列,利用回文序列搜索耻垢分枝杆菌的基因组,发现Ms0606可能调控5个潜在靶基因;逆转录-qPCR和β-半乳糖苷酶活性实验显示Ms0606作为抑制子负调控Ms0608的表达,这可能会影响分枝杆菌对药物的耐药性。[结论] 鉴定了一种新的由Ms0606编码的耻垢分枝杆菌的TetR家族转录调控因子,该因子可调节分枝杆菌对异烟肼的敏感性,并进一步探讨其对靶基因的调控功能及对分枝杆菌耐药性的调控机制。
关键词:转录因子分枝杆菌异烟肼抗生素耐药性
Isoniazid (INH) is one of the major first-line drugs for treating a serious infectious disease-tuberculosis (TB), caused by Mycobacterium tuberculosis[1]. However, while anti-TB drugs have effectively inhibited the spread of TB, the problem of bacterial resistance is becoming more and more serious[2].
The mechanism of anti-TB drugs in Mycobacterium tuberculosis is very complex. Most of the mechanisms are the mutations in drug target genes, in addition to the barrier function of cell wall, the deactivation of antibiotics, changes in drug targets and the increase of drug efflux have been reported[3-4]. However, the regulatory mechanisms of drug resistance, particularly the regulators that directly mediate the drug resistance are not well understood in Mycobacterium tuberculosis.
Both Mycobacterium smegmatis and pathogenic Mycobacterium tuberculosis belong to the genus Mycobacterium, and have many similarities in physiology and genetics. However, M. smegmatis is a non-pathogenic strain with no harm to humans or animals and grows relatively quickly[5-6]. Therefore, these characteristics make M. smegmatis an important model strain for studying the drug resistance and transcriptional regulation mechanism of M. tuberculosis. Recently, the genome sequence of M. smegmatis has been completed, and nearly 500 regulators exist in the M. smegmatis genome, including a large group of TetR family regulators[6]. TetR/AcrR family transcription regulator contains the N-terminal DNA binding motif with a helix-turn-helix structure, and C-terminal has a ligand recognition domain, which often act as inhibitors in bacteria and paleontology[7]. It has been reported to mediate drug resistance by regulating some drug transport and resistance-related genes[8]. Therefore transcriptional regulation plays a vital role in the response of bacteria to environmental stress[9].
For example, the TetR in Escherichia coli negatively regulate the expression of genes related to tetracycline efflux pump and acts as a repressor[10]. The C-terminus of this family protein is a ligand-binding domain that can bind to tetracycline and transport the tetracycline to the outside of the cell, alleviating the inhibitory effect of TetR on the transport pump[11]. In M. smegmatis, LfrR is a transcriptional inhibitor of the TetR-like family, which controls the expression of the drug efflux pump LfrA negatively, thus reducing the efflux of drugs, and caused the strain to be sensitive to various drugs such as ciprofloxacin, norfloxacin and ethidium bromide[12]. Regulating the expression of drug-resistant genes may be a general characteristic of transcriptional repressors. In addition, TetR family transcriptional regulators usually regulate their own expression. Such as Ms6564 in Mycobacterium smegmatis regulates the expression of the regulator by binding with its own promoter, evidence show that Ms6564 might serve as a extensive regulator and may be involve in a global regulation of gene expression, including DNA damage repair and cell cycle[13]. However, what I know about transcription factors that respond to the antituberculosis drugs is still limited within the range of M. smegmatis and related species of mycobacterium.
In this research, I characterized a new TetR family regulator in M. smegmatis, which is encoded by Ms0606. The results showed that Ms0606 specifically bound to its self-promoter by identifying a 22 bp palindromic sequence motif separated by six nucleotides, and its overexpression significantly enhanced bacterial INH sensitivity, whereas its deletion resulted in INH resistance. Thus, I have identified a transcriptional repressor that regulates INH sensitivity in M. smegmatis.
1 Materials and methods 1.1 Strains, enzymes, reagents, plasmids The Escherichia coli BL21 strains and vector (pET-28a) needed for protein expression were purchased from Novagen (Germany). E. coli XR strains, pBT, pTRG vectors were obtained from Stratagene (USA). Restriction enzymes, T4 DNA ligase, and reagent required for PCR reaction were bought from TaKaRa Biotech (Dalian), all antibiotics chemical reagents were bought from Sigma company. PCR primers I used were obtained from Tsingke and Invitrogen (China).
1.2 Gene cloning, expression and purification of Ms0606 protein The related genes required in this study were amplified from the M. smegmatis mc2155 genome. Then the resulting products were treated with the corresponding restriction endonucleases, and purified with PCR DNA purification kit (BioFlux). The Ms0606 gene was cloned into the pET-28a (cm1) vector and transformed into E. coli BL21 strain, the correct sequencing strain was selected for expression, and the activated bacterial cells were grown in 1 L of Kan-containing Luria-Bertani (LB) culture until the OD600 was about 0.8–1.0 at 37 ℃. Then 0.5 mmol/L isopropy-β-D-thiogalactoside (IPTG) was added to induce protein expression at 30 ℃ for 4 h. Later the proteins were purified by Ni2+ column affinity chromatography. Firstly, the cells were centrifuged at 8000 r/min for 2 min and resuspended in binding buffer (10 mmol/L imidazole; 100 mmol/L NaCl; 20 mmol/L Tris-HCl, pH 8.0), then the supernatant after sonication was passed through Ni2+ (50 mmol/L NiSO4) treated His-gel beads. The non-target protein was washed by 10–15 mL buffer (100 mmol/L NaCl; 20 mmol/L Tris-HCl, pH 8.0; 40 mmol/L imidazole). Finally, the Ms0606 protein is eluted with 15–20 mL of elution buffer (100 mmol/L NaCl; 20 mmol/L Tris-HCl, pH 8.0; 250 mmol/L imidazole), then the protein is saved at 4 ℃ and SDS-PAGE gel was used to detect protein. The protein with higher purity and concentration was selected for dialysis and transferred to 100 μL aliquots per tube, the eluate was marked and stored at –80 ℃ for later use.
1.3 Bacterial one-hybrid experiment The coding sequence of Ms0606 was cloned into the pTRG vector, and the 500 bp upstream promoter region of Ms0606 (Ms0606p) and the control gene Ms0612 (Ms0612p) were cloned into the HIS3-aadA reporter gene upstream of the one-hybrid reporter vector pBXcmT[14]. Co-transformants were selected on a screening medium which contained 15 μg/mL tetracycline, 16 μg/mL streptomycin, 30 μg/mL kanamycin, 34 μg/mL chloramphenicol, and 20 mmol/L 3-amino-1, 2, 4-triazole. Bacterial one-hybrid experiment was conducted as previously described[14].
1.4 Electrophoretic mobility shift assays (EMSA) Long fragments over 500 bp were amplified by PCR from M. smegmatis mc2155 genomic DNA. Subsequently, the obtained products were purified by DNA purification kit and 5'-terminus labeled with the fluorescein isothiocyanate (FITC). The short segments below 50 bp were directly annealed in vitro. The EMSA assays were operated as reported procedure[13] with several changes. The 1 μL labeled DNA fragment were co-incubated with concentration gradient of the protein in a total volume of 20 μL EMSA buffer (50 mmol/L Tris-HCl pH 7.5, 50 mmol/L NaCl, 10 mmol/L MgCl2, 1 mmol/L DTT), then the mixtures were reacted at 4 ℃ for 30 min and conducted 5% native Polyacrylamide Gel Electrophoresis (5 mL 40% acrylamide, 31 mL ddH2O, 300 μL APS and 50 μL TEMED) at 150 V for 1–2 h. Finally it was put into the Typhoon Scanner (GE Healthcare) fluorescence to scan, then saved the results after analyzing with image processing software.
1.5 DNase I footprinting assay As mentioned above in reference [15]. The promoter region of the Ms0606 gene was amplified with primers labeled with FITC and purified with BioFlux DNA Purification kit. Under the same conditions as the above EMSAs, just add 1–2 μL of DNase I to the system and expand the reaction to 150 μL, then each mixture was co-incubated at 37 ℃ for 1–2 min. The final sample was performed by an Applied Biosystems 3730XL DNA instrument of Wuhan Qingke Company, and the electropherograms of sequencing analysis was analyzed and drawn with GENEMAPPER software (version 4.0).
1.6 Southern blotting The wild-type M. smegmatis mc2155 and Ms0606-deleted strains were collected in the mid-log phase, and the total DNA was extracted using the TIANGEN bacterial genomic DNA extraction kit (spin column type), then the qualified total DNA was digested by restriction enzyme Sph I, 0.8% agarose gel was used to concentrate and separate the total DNA. After the electrophoresis was completed, it was denatured with 0.4 mol/L NaOH. Finally, after the membrane was transferred, the reaction was carried out according to the instructions of the Roche digoxin labeling southern hybridization kit. The specific experimental method was referred to the published literature[16].
1.7 Quantitative PCRs analysis The real-time PCR analysis follow the procedure described above[17]. mRNA was extracted from M. smegmatis wild-type (Ms and Ms/pMV261) and Ms0606 recombinant strains (Ms/△Ms0606 and Ms/pMV261-Ms0606). Briefly, the strains were cultured to the OD600 is about 0.8, and the RNA is extracted by bacterial RNA kit (see the kit for details). 0.8% agarose gel is used to detect the purity and integrity of RNA samples. The cDNA obtained after reverse transcription is subjected to RT-qPCR reaction: 10 μL 2×SYBR Green Master Mix Rergent, 3 μL cDNA, 1 μL gene-specific primers, 6 μL deionized water. The reactions were reacted in a CFX96 instrument (Bio-Rad, USA) using the following protocol: 95 ℃ for 5 min; 35 cycles of 95 ℃ for 30 s, 60 ℃ for 30 s and 72 ℃ for 30 s. The Bio-Rad CFX Manager v.2.1 software was used to analyze the data. Expression levels of different genes were normalized by sigA and RT-qPCR datas were calculated using the 2-ΔΔCt method[18].
1.8 β-galactosidase activity analysis The β-galactosidase activity experiment was carried out in M. smegmatis, a series of pMV261-vectors were constructed with LacZ as the reporter gene, Ms0608p and Ms0612p were digested and ligated into pMV261-vector[19]. Recombinant plasmids pMV261-Ms0608p-lacZ and pMV261- Ms0612p-lacZ were transformed into the wild-type M. smegmatis and Ms0606-knockout strains. All report strains were grown in 7H9 liquid medium at 37 ℃ for two days until OD600 to 1.0, then cell suspension was collected and β-galactosidase activity was measured as before[20].
1.9 Determination of mycobacterial growth curves and the MIC of drugs When the Ms0606 recombinant strains grew to OD600 between 1.5 and 1.8, the fresh bacteria were transferred to 100 mL 7H9 medium adding the 5 μg/mL INH. Then the cultures (OD600=0.15) were shaken at 160 r/min for 3–4 d at 37 ℃ for further growth, and these samples were harvested and measured at the same time for OD600.
Minimal inhibitory concentration (MIC) determination using Kirby-Bauer test method[21] with several changes. The strain to be tested was coated on the 7H10 plate, and then 20 μL of 0.64, 1.28, 2.56, 5.12 μg/mL, 10.24 μg/mL INH were added to the filter paper with a diameter of 1 cm. After the filter paper was dried, the filter paper with different drug concentration was placed on the coated plate with bacteria, allowing the drug could spread on the plate. The plate was incubated at 37 ℃ for 48 h, and the inhibitory zone around the filter paper was observed. The size of the inhibition zone reflects the sensitivity of the tested bacteria to the INH, and is negatively correlated with the minimum inhibitory concentration (MIC) of the INH, that is, the inhibition zone is large and the MIC of INH for M. smegmatis strain is small.
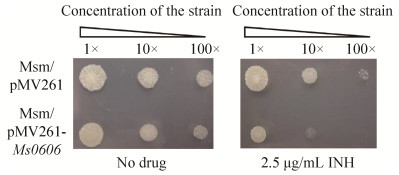
2 Results 2.1 Ms0606 potentially modulates the sensitivity of Mycobacterium smegmatis to INH In order to recognize the potential transcription factors regulating the drug resistance in Mycobacterium smegmatis, I selected the library of M. smegmatis mc2155 (Accession number CP000480) under the strong promoter hsp60. I cloned approximately-500 transcription factors of M. smegmatis into the overexpressing plasmid pMV261 and the recombinant strains were detected on 7H10 solid medium with 2.5 μg/mL INH. A hypothetical transcription factor encoded by Ms0606 was isolated, potentially leading to the INH sensitivity of M. smegmatis. As shown in Figure 1, when the recombinant strains were 10×fold diluted to two different concentrations and spotted on 7H10 plates without (left panel) or with 2.5 μg/mL INH (right panel), compared with the wild-type strain with blank vector pMV261(Msm/pMV261), the Ms0606-over expressed strain transformed with pMV261-Ms0606 (Msm/pMV261-Ms0606) was more sensitive to INH (Figure 1, right panel), the two recombinant mycobacterial strains grew equally on the plate without the drug (Figure 1, left panel).
 |
| Figure 1 The effect of Ms0606 on the INH sensitivity of M. smegmatis and its domain structure. |
| 图选项 |
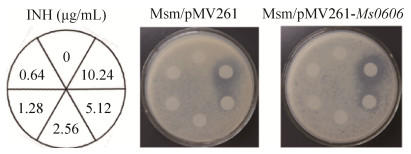
Furthermore, the minimal inhibitory concentrations (MICs) of M. smegmatis wild-type and Ms0606-overexpressed strains on INH were determined in the existence of INH with different dilution gradients. As shown in Figure 2 and Table 1, in the presence of INH in the culture medium, the inhibition zone diameter of INH to Ms0606- overexpressed strain was 25 mm, and the diameter of wild-type strain was 20 mm. This observation suggests that the Ms0606 could increase the INH sensitivity of M. smegmatis. A sequence analysis showed that the Ms0606 gene encodes a 209-residue protein, which contains a typical TetR N-terminal helix-turn-helix structure in the AcrR region (Figure 3). Therefore, Ms0606 belongs to the TetR/AcrR family of transcription factors[22].
 |
| Figure 2 The MIC determination in Msm/pMV261 and Msm/pMV261-Ms0606 strains. |
| 图选项 |
Table 1. Inhibition zone diameter of INH to M. smegmatis (Msm)
| Msm/pMV261 | Msm/pMV261-Ms0606 | |
| Inhibition zone diameter/mm | 20 | 25 |
| Inhibition zone of INH to Msm/pMV261 and Msm/pMV261- Ms0606 was measured by Kirby-Bauer test method. | ||
表选项
 |
| Figure 3 The domain structure of Ms0606 protein. |
| 图选项 |
To confirm the regulating effect of Ms0606 on the growth of M. smegmatis under the action of INH, I measured the growth curves of the wild-type (Msm/WT, Msm/pMV261), Ms0606-knockout strain (Msm/△Ms0606) and Ms0606-overexpressed strain (Msm/pMV261-Ms0606). As shown in Figure 4-A, Ms0606-overexpressed strain grew dramatically slowly than the wild-type strain in 7H9 liquid medium with 5 μg/mL INH (Figure 4-B), *P≤0.05. Besides, no significant difference was found in these two strains (Figure 4-A). But in contrast, the Ms0606-deleted M. smegmatis strain grew only a modest better than the wild-type strain under the same conditions (Figure 4-D), and these two strains were similar in the absence of INH (Figure 4-C).
 |
| Figure 4 Growth analysis of evaluating the effect of Ms0606 on INH sensitivity in M. smegmatis. Growth curves of wild-type (Msm/pMV261) and Ms0606-overexpressing (Msm/pMV261-Ms0606) strains in the absence (A) and presence of (B) 5 μg/mL INH; Growth curves of the wild-type (Msm/WT) and Ms0606-deleted (Msm/△Ms0606) strains in the absence (C) and presence of (D) 5 μg/mL INH. *: P≤0.05. |
| 图选项 |
Therefore, these results indicated that Ms0606 may be involved in the regulation of INH sensitivity in M. smegmatis.
2.2 Ms0606 binds its own promoter The binding of Ms0606 to its own promoter was also detected because most of the TetR family transcriptional factors presented a self-regulation mechanism in mycobacteria. To test this idea, firstly, a bacterial one-hybrid system was used[14] to detect DNA-protein interactions through transcriptional activation effects of the reporter genes HIS3 and aadA (Figure 5-A). When co-transformants were spotted on the plates with or without the selective drugs 3-AT and Strr (see Materials and Methods), if the regulatory factor cloned into the pTRG vector can activate the promoter cloned into the pBXcmT vector, the reporter genes would be induced and therefore the co- transformants grow well on the screening medium (+3-AT, +Strr). As shown in Figure 5-B, both the positive control (CK+) and pTRG-Ms0606/pBX-Ms0606p grew very well in the selective plate (Figure 5-B). On the contrary, no colony growth was observed for the negative control (CK–) and the self-activation control including either Ms0606 or the promoter alone, indicating that Ms0606 can bind with its own promoter, Ms0606p.
 |
| Figure 5 Ms0606 binds to its own promoter specifically. A: The putative promoter region of Ms0606 was cloned into the HIS3-aadA reporter genes upstream of the bacterial one-hybrid reporter vector pBXcmT; B: Bacterial one-hybrid assays; C: EMSA assays; D: EMSA assays for the specific binding of Ms0606 to its own promoter. |
| 图选项 |
Secondly, electrophoretic mobility shift assays (EMSAs) were used to further confirm the results in vitro. As shown in Figure 5-C, when 0.3 nmol/L promoter DNA (Ms0606p) and a control DNA substrate (Ms0612p) were incubated with Ms0606 protein, stable protein-DNA complex bands were observed as the protein concentration (1–8 μmol/L) in the reaction system increased (Figure 5-C, lanes 2–5), but not negative control Ms0612p (Figure 5-C, lanes 7–10). Additionally, a competition assay was performed to determine the specificity of Ms0606 binding with Ms0606p. As shown in Figure 5-D, when the concentration of unlabeled Ms0606p was gradually increased in the reaction system, the binding of FITC-labeled Ms0606p to Ms0606 protein could be competitively inhibited (Figure 5-D, lanes 7–9), but not unlabeled Ms0612p under the same conditions (Figure 5-D, lanes 10–12).
The above results indicated that Ms0606 can recognize its self-promoter specifically.
2.3 Ms0606 identifies a palindromic sequence motif To further map the DNA binding motif identified by Ms0606, several truncated DNA substrates covering the promoter region of Ms0606 were produced and designated as S1, S2, S3. As shown in Figure 6, an obvious DNA-protein complex was observed only for S3 (lanes 11–15) in EMSA assays, but not fragment S1 or S2 (lanes 1–10), indicating that the potential binding motif for Ms0606 should be in the region S3 fragment.
 |
| Figure 6 EMSA detected the DNA-binding fragments of Ms0606 in the upstream regions of Ms0606. |
| 图选项 |
Then DNase I footprinting assay was used to precisely recognize the binding motif of Ms0606. The FITC-labeled S3 fragment and Ms0606 protein were co-incubated and digested with DNase I. As shown in Figure 7-A, as the Ms0606 protein (0–4 μmol/L) concentration increased, the area around GAAACTGAATCTAACATC AGATTCAGGTTG protected by Ms0606 became more and more obvious (Figure 7-B). The region contained two inverted repeats (IRs) and separated from each other by six nucleotides.
 |
| Figure 7 Identification of the DNA motif bound for Ms0606 with Ms0606p. A: DNase I footprinting experiments; B: sequence structure of the protected DNA region; C: EMSA for the DNA-binding activity of Ms0606 to the Ms0606p1, Ms0606p2, Ms0606p3, Ms0606p4 and Ms0606p5. |
| 图选项 |
In order to further confirm the specificity of the motif identified by Ms0606. In the EMSA assays, I synthesized a series of about 40 bp fragments containing binding sites or motif mutations oligonucleotides (Figure 7-C). As shown in Figure 7-C, Ms0606 can't combine Ms0606p2 in which the two inverted repeats were mutated by random sequences (Figure 7-C, lanes 6–8). In contrast, neither a portion of the IRs nor a mutation in the interval sequence has eliminated their interaction (Figure 7-C, 10–12, 14–16, 18–20), and only one binding band was observed in mutants, suggesting that Ms0606 may be combined with the promoter in two steps.
In summary, my results indicate that Ms0606 can recognize a conserved 22 bp palindromic sequence motifs (CTGAATCTNNNNNNAGATTCAG).
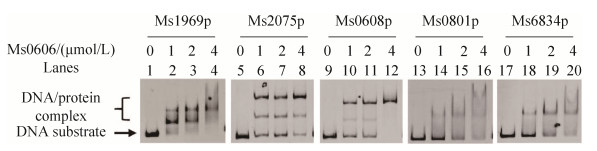
2.4 Several genes promoter region may contain the binding motif for Ms0606 Through the identification of the binding motif of Ms0606, I can find the potential target genes regulated by the protein Ms0606. The entire 500 bp upstream promoter region of the open reading frame was searched in the M. smegmatis genome for the binding motif of Ms0606 with partial base mismatch, five promoters (Ms1969, Ms2075, Ms0608, Ms0801 and Ms6834) was searched to contain the potential Ms0606 binding site, in addition to the self-promoter of Ms0606, indicating that these genes may be regulated by Ms0606. This leads us to assume that Ms0606 may be a regulator of these potential target genes. To test this assumption, the promoter DNA of Ms1969, Ms2075, Ms0608, Ms0801 and Ms6834 were synthesized and detected by EMSA assays. As shown in Figure 8, When putative promoters DNA and Ms0606 protein were co-incubated, with the increase of protein concentration (0–4 μmol/L), the promoter of Ms1969, Ms2075 and Ms0608 form a stable complex with Ms0606 (Figure 8, lanes 1–12). But in the promoter of Ms0801 and Ms6834, only faint shifted band was observed (Figure 8, lanes 13–20), suggesting that Ms0606 may have different ability to bind promoters.
 |
| Figure 8 EMSA detected the binding of Ms0606 with the promoters of Ms1969, Ms2075, Ms0608, Ms0801 and Ms6834. |
| 图选项 |
These findings suggest that the five genes may be the potential target gene of Ms0606 in M. smegmatis.
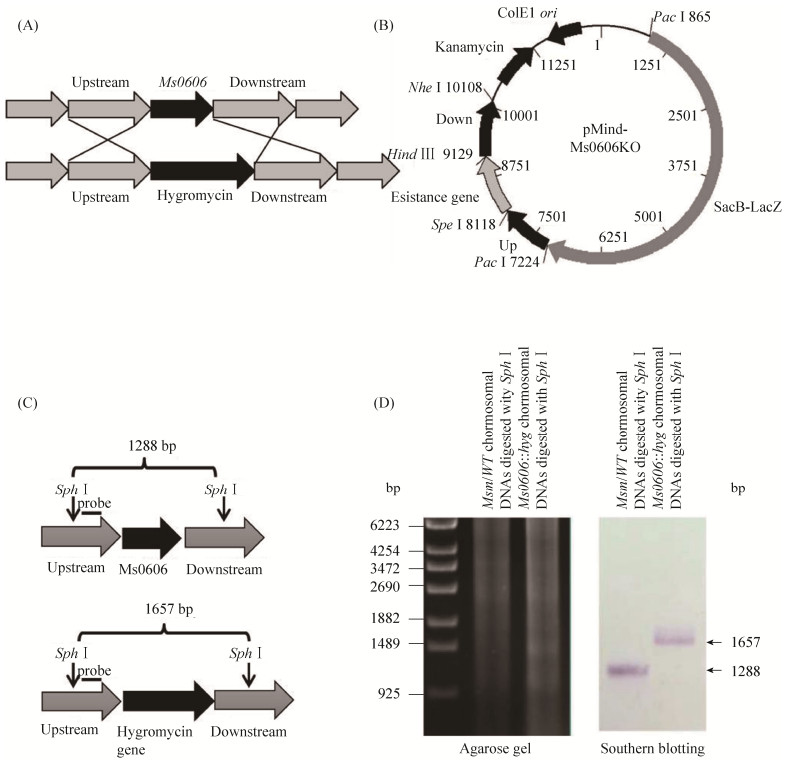
2.5 Ms0606 is a transcriptional repressor that negatively regulates the expression of the Ms0608 To further study the regulatory function of Ms0606, I used homologous recombination exchange to construct the Ms0606 knockout strain in the M. smegmatis strain (Figure 9-A). As shown in Figure 9-A, the 1 kb upstream and downstream of the Ms0606 gene was cloned into a pMind-derived[23] suicide plasmid by homologous recombination, then the constructed knockout vector was transferred into M.smegmatis wild-type strain to obtain Ms0606- deleted strain (Figure 9-B). Further Southern blotting[24] was used to verify whether Ms0606 was knocked out. As shown in Figure 9-C, a schematic diagram of genomic DNA of wild-type and Ms0606 knock-out strain after digestion with Sph I restriction enzyme, and then hybridized with digoxin-labeled probe. As a result, 1.3 kb were detected in wild-type strain, a band of 1.7 kb size was detected in the Ms0606-deleted strain, indicating that Ms0606 gene was successfully knocked out in the mutant strain (Figure 9-D).
 |
| Figure 9 Construction of the Ms0606 knockout strain in M. smegmatis and Southern blot analysis. A: schematic diagram of the recombination strategy; B: a map of the recombinant vector pMindMs0606KO; C: schematic diagram of the DNA fragments of the wild-type and Ms0606-deleted strain digested with the restriction enzyme Sph I; D: Southern blotting assays. |
| 图选项 |
Subsequently, I compared expression levels of the adjacent converse gene, Ms0608, in wild type strain and Ms0606 knock-out strain using RT-qPCR. As shown in Figure 10-A, compared with the wild-type strain, the expression level of Ms0608 was up-regulated more than twice (P < 0.05) in the Ms0606-knockout strain. Similarly, I detected the expression level of Ms0608 in Ms0606 overexpressing strain containing a pMV261-derived recombinant plasmid. Compared with the control strain, the expression of Ms0608 in the overexpressed strain was reduced to less than 0.5 times (*P < 0.05) (Figure 10-B), which coincided the previous result. Therefore, RT-qPCR experiments proved that Ms0606 acts as a transcriptional repressor, which negatively regulates the expression of Ms0608 gene.
 |
| Figure 10 The effect of the Ms0606 regulator on the expression of Ms0608. A: real-time quantitative PCR detection in the Ms0606-knockout strain; B: real-time quantitative PCR detection in the overexpressed strain; C: β-Galactosidase activity assay. *: P≤0.05. |
| 图选项 |
Further β-galactosidase activity assays confirm this conclusion, a series of promoter-lacZ reporter strains were constructed in M. smegmatis. As shown in Figure 10-B, compared with the CK-plasmid containing the non-promoter lacZ plasmid, the hsp60 promoter strongly up-regulated the expression of lacZ (CK+), indicating that the reporter system is working properly. As expected, compared with the wild type strain, the expression of lacZ under the Ms0608 promoter induced by the Ms0606 was increased 5-fold in the Ms0606-deleted M. smegmatis strain (*P < 0.05). This finding is consistent with the results of RT-qPCR described above.
Based on the above results, Ms0606 acts as a repressor and negatively regulates Ms0608 gene expression in M. smegmatis.
3 Discussion The molecular network of M. tuberculosis in response to anti-tuberculosis drugs and its inherent regulation mechanism of resistance to isoniazid are not clear[6]. In this study, I reported a TetR family regulator Ms0606 in M. smegmatis, overexpressing Ms0606 enhances the sensitivity of mycobacteria to INH, while destroying Ms0606 makes the mycobacteria slightly resistant to INH, which possible "Genetic compensation response (GCR)"[25], that is, after Ms0606 gene is mutated and completely loses its function, the bacteria will adopt a corresponding mechanism to increase the expression of other genes to replace the function of Ms0606. Therefore, I've uncovered a new transcriptional factor that affects the INH sensitivity of M. smegmatis.
The TetR/AcrR regulatory family is widely distributed in many bacteria and usually functions as a suppressor. It plays a regulatory role in bacterial resistance, pathogenicity, antibiotic synthesis and osmotic stress[7]. The acrR operon contains acrR, acrA and acrB, three genes in E. coli, the latter two are multidrug-resistant efflux pumps[26-27]. In contrast, Ms0606 contain a typical AcrR domain, but its target is unknown and will be elucidated. In this study, my EMSA data suggest that Ms0606 can bind to the promoter regions of some genes (Ms1969, Ms2075, Ms0608, Ms0801 and Ms6834) and act as a repressor, possibly regulating the expression of these genes.
To further explain the resistance of bacteria, I try to confirm the function of protein encoded by Ms0608 in M. smegmatis. Bioinformatic analysis indicates that Ms0608 belongs to vicinal-oxygen- chelate (VOC) superfamily, which contains motifs that provide a metal binding site to promote metal ions to participate in the catalysis of enzymes, including the glyoxalase I, type I extradiol dioxygenases and a group of antibiotic resistance proteins[28]. Although the proteins of this superfamily are functionally different, their structures are similar. Antimicrobial resistance proteins use multiple mechanisms to block the function of antibiotics. For example, bleomycin resistant protein (BLMA) inhibits its activity by directly binding to bleomycin[29], glyoxalase I and BLMA exhibit domain exchange between subunits. Whereas, three fosfomycin resistant proteins inactivated fosfomycin by modifying the fosfomycin molecule[30]. Moreover, I tried to identify orthologs of Ms0608 based on a BLAST search, as shown in Figure 11, Ms0608 protein was identified 79% amino acid sequence similarity with orthologs in M. tuberculosis H37Rv and M. bovis BCG (100% amino acids are identical throughout the length of the protein). It has been reported Rv0274 may encode a novel detoxification protein in M. tuberculosis[31], indicating Ms0608 protein may has a similar protective effect against toxins and antibiotics. I have proved that Ms0606 negatively regulates expression of Ms0608, although the details were not very clear, it is reasonable that Ms0606 may affect bacteria sensitivity to drugs by regulating the expression of Ms0608. In addition to Ms0608, while others (Ms1969, Ms2075, Ms0801 and Ms6834) may also affect drug resistance, which may play a role in multiple stress adaptations in mycobacteria. Therefore, the underlying mechanism of regulation of antibiotic resistance formation remains to be further studied in the future.
 |
| Figure 11 Ms0608 are conserved in mycobacterial species. A: the arrangement of Ms0608 and its adjacent genes in Mycobacterium genome; B: the Ms0608 protein orthologs in M. tuberculosis, M. bovis BCG (100% amino acid identity) and M. smegmatis (79% amino acid identity). |
| 图选项 |
4 Conclusion In summary, a TetR/AcrR family of transcription factors, Ms0606, was turned out to be a transcription repressor and negatively regulate INH resistance in M. smegmatis. Ms0606 specifically identified a 22 bp palindromic sequence motif separated by a 6 bp spacer. Notably, I found that Ms0606 regulates the expression of the Ms0608 gene. Apparently, my findings provide important clues for further studying a link of a transcriptional regulator Ms0606 and mechanism of antibiotic in mycobacteria.
References
| [1] | Vilchèze C, Jacobs WR. The mechanism of isoniazid killing: clarity through the scope of genetics. Annual Review of Microbiology, 2007, 61: 35-50. DOI:10.1146/annurev.micro.61.111606.122346 |
| [2] | Goldman RC, Plumley KV, Laughon BE. The evolution of extensively drug resistant tuberculosis (XDR-TB): history, status and issues for global control. Infectious Disorders Drug Targets, 2007, 7(2): 73-91. DOI:10.2174/187152607781001844 |
| [3] | Almeida da Silva PE, Palomino JC. Molecular basis and mechanisms of drug resistance in Mycobacterium tuberculosis: classical and new drugs. Journal of Antimicrobial Chemotherapy, 2011, 66(7): 1417-1430. DOI:10.1093/jac/dkr173 |
| [4] | Nikaido H. Multidrug resistance in bacteria. Annual Review of Biochemistry, 2009, 78(1): 119-146. DOI:10.1146/annurev.biochem.78.082907.145923 |
| [5] | Cole ST. Learning from the genome sequence of Mycobacterium tuberculosis H37Rv. FEBS Letters, 1999, 452(1/2): 7-10. |
| [6] | Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Krogh A, McLean J, Moule S, Murphy L, Oliver K, Osborne J, Quail MA, -A Rajandream M, Rogers J, Rutter S, Seeger K, Skelton J, Squares R, Squares S, Sulston JE, Taylor K, Whitehead S, Barrell BG. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature, 1998, 393(6685): 537-544. DOI:10.1038/31159 |
| [7] | Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang XD, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiology and Molecular Biology Reviews, 2005, 69(2): 326-356. |
| [8] | Nguyen L. Antibiotic resistance mechanisms in M. tuberculosis: an update. Archives of Toxicology, 2016, 90(7): 1585-1604. DOI:10.1007/s00204-016-1727-6 |
| [9] | Miravet-Verde S, Lloréns-Rico V, Serrano L. Alternative transcriptional regulation in genome-reduced bacteria. Current Opinion in Microbiology, 2017, 39: 89-95. DOI:10.1016/j.mib.2017.10.022 |
| [10] | Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nature Structural Biology, 2000, 7(3): 215-219. DOI:10.1038/73324 |
| [11] | Saenger W, Orth P, Kisker C, Hillen W, Hinrichs W. The tetracycline repressor-A paradigm for a biological switch. Angewandte Chemie International Edition, 2000, 39(12): 2042-2052. DOI:10.1002/1521-3773(20000616)39:12<2042::AID-ANIE2042>3.0.CO;2-C |
| [12] | Buroni S, Manina G, Guglierame P, Pasca MR, Riccardi G, De Rossi E. LfrR is a repressor that regulates expression of the efflux pump LfrA in Mycobacterium smegmatis. Antimicrobial Agents and Chemotherapy, 2006, 50(12): 4044-4052. DOI:10.1128/AAC.00656-06 |
| [13] | Yang M, Gao CH, Cui T, An JN, He ZG. A TetR-like regulator broadly affects the expressions of diverse genes in Mycobacterium smegmatis. Nucleic Acids Research, 2012, 40(3): 1009-1020. DOI:10.1093/nar/gkr830 |
| [14] | Guo MM, Feng H, Zhang J, Wang WQ, Wang Y, Li YQ, Gao CH, Chen HC, Feng Y, He ZG. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Research, 2009, 19(7): 1301-1308. DOI:10.1101/gr.086595.108 |
| [15] | Yang M, Gao CH, Hu JL, Zhao L, Huang QY, He ZG. InbR, a TetR family regulator, binds with isoniazid and influences multidrug resistance in Mycobacterium bovis BCG. Scientific Reports, 2015, 5: 13969. DOI:10.1038/srep13969 |
| [16] | Zhu C, Liu Y, Hu LH, Yang M, He ZG. Molecular mechanism of the synergistic activity of ethambutol and isoniazid against Mycobacterium tuberculosis. Journal of Biological Chemistry, 2018, 293(43): 16741-16750. DOI:10.1074/jbc.RA118.002693 |
| [17] | Li WH, He ZG. LtmA, a novel cyclic di-GMP-responsive activator, broadly regulates the expression of lipid transport and metabolism genes in Mycobacterium smegmatis. Nucleic Acids Research, 2012, 40(22): 11292-11307. DOI:10.1093/nar/gks923 |
| [18] | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods: San Diego, Calif, 2001, 25(4): 402-408. DOI:10.1006/meth.2001.1262 |
| [19] | Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF. New use of BCG for recombinant vaccines. Nature, 1991, 351(6326): 456-460. DOI:10.1038/351456a0 |
| [20] | Miller JH. Experiments in Molecular Genetics. New York: Cold Spring Harbor Laboratory Press, 1972: 352-355. |
| [21] | Esser VM, Elefson DE. Experiences with the kirby-bauer method of antibiotic susceptibility testing. American Journal of Clinical Pathology, 1970, 54(2): 193-198. DOI:10.1093/ajcp/54.2.193 |
| [22] | Deng WY, Li CM, Xie JP. The underling mechanism of bacterial TetR/AcrR family transcriptional repressors. Cellular Signalling, 2013, 25(7): 1608-1613. DOI:10.1016/j.cellsig.2013.04.003 |
| [23] | Blokpoel MCJ, Murphy HN, O'Toole R, Wiles S, Runn ESC, Stewart GR, Young DB, Robertson BD. Tetracycline-inducible gene regulation in Mycobacteria. Nucleic Acids Research, 2005, 33(2): e22. DOI:10.1093/nar/gni023 |
| [24] | Wang Y, Huang YX, Xue CL, He Y, He ZG. ClpR protein-like regulator specifically recognizes RecA protein-independent promoter motif and broadly regulates expression of DNA damage-inducible genes in Mycobacteria. Journal of Biological Chemistry, 2011, 286(36): 31159-31167. DOI:10.1074/jbc.M111.241802 |
| [25] | Rossi A, Kontarakis Z, Gerri C, Nolte H, H?lper S, Krüger M, Stainier DYR. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature, 2015, 524(7564): 230-233. DOI:10.1038/nature14580 |
| [26] | Ma D, Alberti M, Lynch C, Nikaido H, Hearst JE. The local repressor AcrR plays a modulating role in the regulation of acrAB genes of Escherichia coli by global stress signals. Molecular Microbiology, 1996, 19(1): 101-112. DOI:10.1046/j.1365-2958.1996.357881.x |
| [27] | Su CC, Rutherford DJ, Yu EW. Characterization of the multidrug efflux regulator AcrR from Escherichia coli. Biochemical and Biophysical Research Communications, 2007, 361(1): 85-90. DOI:10.1016/j.bbrc.2007.06.175 |
| [28] | He PQ, Moran GR. Structural and mechanistic comparisons of the metal-binding members of the vicinal oxygen chelate (VOC) superfamily. Journal of Inorganic Biochemistry, 2011, 105(10): 1259-1272. |
| [29] | Kumagai T, Sugiyama M. Protection of mammalian cells from the toxicity of bleomycin by expression of a bleomycin-binding protein gene from Streptomyces verticillus. The Journal of Biochemistry, 1998, 124(4): 835-841. DOI:10.1093/oxfordjournals.jbchem.a022187 |
| [30] | Ballestero-Téllez M, Docobo-Pérez F, Portillo-Calderón I, Rodríguez-Martínez JM, Racero L, Ramos-Guelfo MS, Blázquez J, Rodríguez-Ba?o J, Pascual A. Molecular insights into fosfomycin resistance in Escherichia coli. Journal of Antimicrobial Chemotherapy, 2017, 72(5): 1303-1309. |
| [31] | Rawat M, Heys J, Av-Gay Y. Identification and characterization of a diamide sensitive mutant of Mycobacterium smegmatis. FEMS Microbiology Letters, 2003, 220(2): 161-169. DOI:10.1016/S0378-1097(03)00127-7 |
